Abstract
We provide an overview of groundbreaking studies that laid the foundation for our current understanding of exercise-induced mitochondrial biogenesis and its contribution to human skeletal muscle fitness. We highlight the mechanisms by which skeletal muscle responds to the acute perturbations in cellular energy homeostasis evoked by a single bout of endurance-based exercise and the adaptations resulting from the repeated demands of exercise training that ultimately promote mitochondrial biogenesis through hormetic feedback loops. Despite intense research efforts to elucidate the cellular mechanisms underpinning mitochondrial biogenesis in skeletal muscle, translating this basic knowledge into improved metabolic health at the population level remains a future challenge.
Exercise represents a major challenge to multiple whole-body homeostatic functions. In an effort to overcome this challenge, numerous responses take place at the cellular and systemic levels that operate to blunt the homeostatic threats generated by exercise-induced increases in muscle energy turnover and oxygen demand (Hawley et al. 2014). The capacity of skeletal muscle to adapt to repeated bouts of activity such that physical capacity is enhanced is termed exercise training. When considering endurance-based exercise (e.g., sustained activities that are >10 min duration and performed at 60%–90% of maximal oxygen uptake [VO2max] including sprint-interval training), the goals of such exercise are to induce an array of physiological and metabolic adaptations that enable an individual to increase the rate of energy production from both aerobic pathways, maintain tighter metabolic control (i.e., match adenosine triphosphate (ATP) production with ATP hydrolysis), minimize cellular perturbations, increase efficiency of motion, and improve the capacity of the trained musculature to resist fatigue (Hawley 2002). The mechanisms by which active muscle senses homeostatic perturbations and then translates them into improved function has been a topic of intense research for several decades. It is now recognized that a variety of cellular disruptions take place at the onset of exercise including (but not limited to) increased cytoplasmic free [Ca2+], increased free AMP (AMPf) and an increased adenosine diphosphate (ADP)/ATP ratio, reduced creatine phosphate (PCr) and glycogen levels, increased fatty acid and reactive oxygen/nitrogen species (ROS/RNS), acidosis, and altered redox state, including [NAD/NADH] (Hawley et al. 2014). Within the context of metabolic homeostasis, an array of regulatory networks are stimulated that sustain rates of ATP synthesis over time through the activation of rate-limiting enzymes controlling carbohydrate and fat catabolism. A long-standing question in exercise biology is how these acute disruptions in cellular signals that maintain energy supply also stimulate long-term adaptive processes that improve the ability of muscle to sustain a future contractile challenge. Indeed, this process of translating acute threats to essential homeostatic functions into positive adaptations is termed “hormesis” and underscores the principle of how exercise improves muscle health.
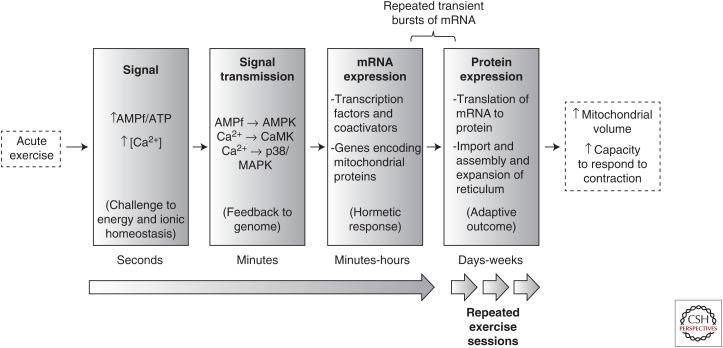
A key component of improved “muscle fitness” following exercise training is mitochondrial biogenesis, a highly coordinated process that requires the coordination of multiple cellular events, including transcription of two genomes (nuclear and mitochondrial), the synthesis of lipids and proteins, and the coordinated assembly of multisubunit protein complexes into a functional respiratory chain as reviewed by Hood (Hood 2001). This sequence of events ultimately results in an expansion of total muscle mitochondrial volume and this greater mitochondrial network improves the cell's ability to match ATP production with ATP hydrolysis, thereby minimizing the very disruptions to cellular homeostasis that occurred in the first place (Fig. 1). This review will present a brief historical overview of some of the seminal discoveries that have helped unravel the cellular events that underpin mitochondrial biogenesis as well as the more recent explosion of research into our current understanding of the signaling and genomic mechanisms regulating this process. We will focus on major breakthroughs in this area and for more detailed discussion of specific topics, the reader is referred to other reviews describing the various signaling networks involved in mitochondrial biogenesis following exercise (Wu et al. 1999; Lin et al. 2005; Hood et al. 2011).
Figure 1.
Schematic of the time course of events coupling contractile stimuli to mitochondrial biogenesis through hormetic feedback signals. Acute endurance-based exercise challenges the capacity of skeletal muscle to maintain both energy and calcium homeostasis (among other systems). Repeated increases in cytosolic (free AMP [AMPf]/adenosine triphosphate [ATP]) (because of ATP turnover) and calcium activate the AMP-activated protein kinase (AMPK) and Ca2+-sensitive signaling kinases, respectively, which activate specific proteins regulating gene transcription. Successive exercise sessions (i.e., endurance exercise training) results in repeated and transient increases in messenger RNA (mRNA) encoding mitochondrial proteins as well as transcriptional proteins, which likely enhance the genomic response to subsequent exercise challenges. The repeated drive for protein translation increases mitochondrial protein import and assembly, thereby expanding mitochondrial volume. The hormetic response is complete when the functional capacity of mitochondria has improved, which enables the muscle to better maintain energy and calcium homeostasis during future challenges. MAPK, Mitogen-activated protein kinase; CaMK, calmodulin-dependent protein kinase. (From Hood 2001; adapted, with permission.)
EXERCISE TRAINING–INDUCED INCREASES IN MITOCHONDRIA AND ENDURANCE CAPACITY
Early work in the 1950s on a variety of animal species revealed a strong relationship between the ability of a muscle to perform prolonged submaximal exercise and its content of respiratory enzymes (Paul and Sperling 1952; Lawrie 1953). Based on these observations, Holloszy (1967) hypothesized that such differences might be the result of an “adaptive process” that is linked to the level of habitual muscle contractile activity. To test this possibility, Holloszy subjected rats to a vigorous and intense program of treadmill running and showed exercise training induced an increase in skeletal muscle mitochondrial enzyme activities (Holloszy 1967). In that study, mitochondria from muscles of exercise-trained animals showed higher levels of respiratory control and tightly coupled oxidative phosphorylation compared with muscle from untrained animals. This increase in electron transport capacity was associated with a concomitant increase in the ability to produce ATP and was, in large part, responsible for the prolonged endurance running time observed after training. A key innovation of Holloszy’s exercise protocol was the use of intense interval run training, based on a recognition that any exercise signal needed to exceed a minimum “threshold stimulus” to induce metabolic and morphological adaptations in muscle. Indeed, the training regimen pioneered by Holloszy might be viewed as the original prototype for the current wave of sprint-intensity interval training protocols now widely accepted as a potent stimulus for physiological remodeling in humans (MacDougall et al. 1998; Parra et al. 2000; Rodas et al. 2000; Burgomaster et al. 2005; Perry et al. 2007, 2008; Yeo et al. 2008; MacInnis and Gibala 2016).
The work of Holloszy (1967) showed that skeletal muscle displays a remarkable plasticity, with the capacity to alter both the type and amount of protein in response to disruptions in cellular homeostasis induced by the habitual level of contractile activity. That study (Holloszy 1967) paved the way for subsequent work showing that increases in mitochondrial enzyme activity in rodent muscle following several weeks of exercise training were related to a greater mitochondrial number and size with more densely packed cristae (Gollnick and King 1969). A crucial finding was that the increased mitochondrial enzyme activities following training were not accompanied by increases in the cytosolic enzymes creatine kinase and adenylate kinase, giving credibility to the idea that improved ATP synthesis via oxygen-dependent pathways was a defining feature of exercise trained muscle (Oscai and Holloszy 1971). Subsequent studies in humans corroborated this hypothesis. Longitudinal exercise training (Varnauskas et al. 1970) and cross-sectional comparisons between trained and untrained men revealed higher maximal mitochondrial enzyme activities in samples taken from the vastus lateralis muscle (Booth and Narahara 1974) and greater mitochondrial volume based on electron microscopy techniques (Hoppeler et al. 1973) that were positively associated with training-induced improvements in whole-body maximal oxygen uptake (VO2max). Longitudinal studies also showed that endurance training increased mitochondrial content concurrent with mitochondrial enzyme activities and VO2max, but such an effect was attenuated in older individuals (Kiessling et al. 1974). These findings were the first to suggest that the muscle’s “plasticity capacity” may be compromised with aging. Nevertheless, these findings contributed to the belief that whole-body aerobic capacity was limited not only by the oxygen delivery/transport system (i.e., “central” cardiorespiratory factors) but potentially by “local” skeletal muscle mitochondrial content, consistent with the working muscle’s role as the end user of oxygen during ATP production.
In the 1970s in North America, a series of independent but related studies conducted largely in rodents contributed to a model that sought to explain how mitochondrial content was related to improved metabolic control in skeletal muscle during exercise. This model held that a greater mitochondrial content improved respiratory sensitivity to free ADP (ADPf), whereby a given rate of aerobic ATP production necessary to support muscle contraction would require a smaller increase in [ADPf] at the onset of exercise caused by the expanded mitochondrial volume (Oscai and Holloszy 1971; Gollnick et al. 1973; Holloszy and Booth 1976; Holloszy and Coyle 1984). Simultaneously, in Europe, this concept was further developed in studies of human skeletal muscle. Specifically, it was proposed that the accelerated induction of oxygen consumption at the onset of heavy exercise would be attenuated by exercise training, decreasing the rise in modulators of oxidative phosphorylation that would result in less stimulation of glycolysis and a greater proportion of energy being derived from fat oxidation (Gollnick and Saltin 1982). This premise was elegantly tested by a seminal investigation from Terjung’s group in 1987 in which they compared skeletal muscle from trained and untrained rats. This study showed that a greater training-induced increase in the maximal activity of cytochrome c oxidase (a marker of mitochondrial content) was associated with less of an increase in ADPf during acute muscle contraction in the face of less usage of PCr and flux of ATP to AMP and IMP as well as decreased glycolysis (Dudley et al. 1987). These associations were, at the time, viewed as compelling evidence for the major role that mitochondria played in overall metabolic control in skeletal muscle. Arising from these and other observations, renewed interest focused on the potential mechanisms by which exercise training stimulated mitochondrial biogenesis. Despite the complexity of exercise training–induced response, largely arising from training protocols of different intensity, volume, and duration, there was an association between the improvements in mitochondrial oxidative capacity and the intensity of training (Dudley et al. 1982), although differences in muscle fiber types sometimes confounded this observation (Hickson et al. 1976; Dudley et al. 1982). Nonetheless, work at that time stimulated great interest in the cellular signals generated during exercise and how they manifested in the greater training-induced mitochondrial content. However, it was not until the late 1980s that exercise was shown to stimulate the expression of nuclear genes that encoded mitochondrial proteins, albeit through unknown signaling cascades and genetic regulators.
THE DAWN OF MOLECULAR BIOLOGY
The Molecular Basis of Exercise Training–Induced Mitochondrial Biogenesis
One of the first studies to probe the molecular basis of how muscle contraction increases mitochondrial proteins was that of Williams et al. (1986). These researchers observed increases in cytochrome b messenger RNA (mRNA) as well as markers of mitochondrial content (citrate synthase [CS] activity and mitochondrial DNA) following a protocol of chronic electrical stimulation of rabbit tibialis anterior muscle. The increase in CS activity suggested contraction coordinated the expression of the nuclear genome with the mitochondrial genome (cytochrome b mRNA) (Williams et al. 1986). Such a premise was subsequently proven when Hood and coworkers (Hood et al. 1989) showed that chronic contraction of rat tibialis anterior coordinated the expression of a complex mitochondrial respiratory protein complex—cytochrome c oxidase—that is derived from both genomes. It was also verified that increases in both CS activity and mitochondrial DNA expression (mRNA) increased following short-term exercise training in rodents (Morrison et al. 1989). Such increases in CS activity following exercise training were later shown to be related to sustained increases in CS mRNA for at least 24 hours after a single bout of exercise but also after 1 week of training (Neufer and Dohm 1993), which further cemented the concept that protein translation stems from repeated but transient increases in mRNA content.
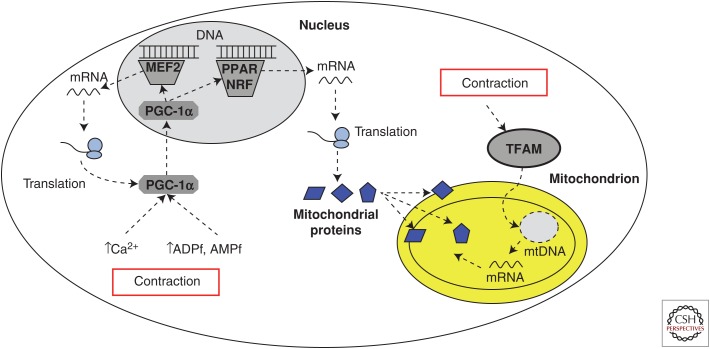
The next two decades marked the discovery of several transcription factors with important roles in the regulation and expression of the nuclear genes encoding mitochondrial proteins. It was recognized that specific families of mitochondrial proteins are regulated by specific transcription factors, including nuclear respiratory factors 1 and 2 (NRF-1 and NRF-2) that bind to the promoters and activate transcription of genes that specifically encode mitochondrial respiratory chain proteins (Chau et al. 1992; Kelly and Scarpulla 2004; Scarpulla 2006). Moreover, the belief that both nuclear and mitochondrial genomes may be coordinately regulated gained the ascendancy. Initially, it was found that NRF-1 activates expression of the nuclear gene that encodes mitochondrial transcription factor A (TFAM), which accumulates in the mitochondria and regulates transcription of the mitochondrial DNA (Fig. 2) (Gordon et al. 2001) (i.e., the mitochondrial genome). Given that not all promoters of genes transcribing mitochondrial proteins have NRF-1-binding sites, other transcription factors are obviously involved in contractile-modulated mitochondrial biogenesis, including the peroxisome proliferator-activated receptor coactivators (PPARs), which regulate expression of the mitochondrial fatty acid oxidative enzymes (Kelly and Scarpulla 2004). Another major breakthrough was the discovery of the peroxisome proliferator-activated receptor γ coactivator 1α (PGC-1α), an inducible coactivator that regulates the coordinated expression of mitochondrial proteins encoded in the nuclear and mitochondrial genomes (Puigserver et al. 1998; Wu et al. 1999; Hood 2001; Scarpulla 2002; Lin et al. 2005; Scarpulla 2006). An important functional feature of the PGC-1α coactivator is that it interacts with different transcription factors to activate distinct biological programs in different tissues rather than bind to DNA directly (Puigserver et al. 1998; Wu et al. 1999; Hood 2001; Scarpulla 2002; Lin et al. 2005; Martinez-Redondo et al. 2015). Indeed, the effect of PGC-1α on mitochondrial biogenesis is probably explained at least in part by the finding that in addition to being a coactivator of PPARγ, PGC-1α coactivates NRF-1 (Wu et al. 1999), and PPARα (Vega et al. 2000). PPARα plays a key role in the transcriptional control of the mitochondrial enzymes involved in the oxidation of fatty acids (Gulick et al. 1994), which may represent one mechanism by which exercise improves the capacity for fat oxidation. Similar roles in mitochondrial biogenesis were also discovered for PGC-1β (Lin et al. 2002a; Meirhaeghe et al. 2003; Lelliott et al. 2006).
Figure 2.
Transcriptional control of mitochondrial biogenesis. Stimuli generated during contraction are transmitted by specific signaling cascades to the transcriptional cofactor peroxisome proliferator-activated receptor γ coactivator (PGC)-1α, which translocates into the nucleus and coactivates several other transcription factors that regulate specific genetic programs encoding families of mitochondrial proteins. Both PGC-1α and a homologous isoform PGC-1β are involved but have been shown to be dispensable in this process (Ballmann et al. 2016), suggesting redundant signals exist that do not require these coactivators (not shown). Subsequent increases in steady-state messenger RNA (mRNA) drive protein translation that leads to expansion of the mitochondrial reticulum. An autoregulatory loop exists whereby PGC-1α induces its own expression leading to increases in PGC-1α protein following exercise. Contraction also induces the expression of mitochondrial DNA that encodes specific proteins within the electron transport chain. MEF, Myocyte enhancer factor; PPAR, peroxisome proliferator-activated receptor coactivator; NRF, nuclear respiratory factor; TFAM, mitochondrial transcription factor A; ADPf, free ADP; AMPf, free AMP; mtDNA, mitochondrial DNA.
With regard to exercise-induced skeletal muscle adaptation, PGC-1α has been proposed as a key regulator of mitochondrial biogenesis that responds to neuromuscular input and the prevailing contractile activity. A single bout of endurance exercise induces a rapid increase in PGC-1α mRNA and protein in both rodent (Baar et al. 2002; Terada et al. 2002; Meirhaeghe et al. 2003; Koves et al. 2005; Frier et al. 2011) and human skeletal muscle (Pilegaard et al. 2003; Norrbom et al. 2004; Watt et al. 2004; Cartoni et al. 2005; Russell et al. 2005; Hellsten et al. 2007; Mortensen et al. 2007; De Filippis et al. 2008; Mathai et al. 2008; Edgett et al. 2013; Cochran et al. 2014). Egan et al. (2010) reported a graded response whereby larger increases in PGC-1α mRNA in skeletal muscle of untrained humans were seen with energy-matched work bouts of short duration and high intensity (∼80% VO2peak) compared with prolonged duration, low-intensity (∼40% VO2peak) cycling. Similar findings were seen by Edgett et al. (2013), although the magnitude of PGC-1α mRNA increase was attenuated at exercise above VO2max. Moreover, the graded response of PGC-1α mRNA and exercise intensity observed in the study by Egan et al. (2010) was associated with a greater upstream phosphorylation of the AMP-activated protein kinase (AMPK), Ca2+/calmodulin-dependent protein kinase (CaMK)II, and the p38 mitogen-activated protein kinase (p38 MAPK), which are bona fide energy-exercise-sensitive kinases, and also regulate PGC-1α transcription and/or activity (discussed in a subsequent section) (Puigserver et al. 2001; Akimoto et al. 2005; Irrcher et al. 2008). Furthermore, a single bout of exercise has been associated with accumulation of PGC-1α in both nuclear (Wright et al. 2007; Little et al. 2010; Safdar et al. 2011) and mitochondrial (Safdar et al. 2011) compartments possibly following its translocation from the cytoplasm.
Endurance exercise training also increases PGC-1α mRNA expression as well as protein in both rodent and human skeletal muscle (Goto et al. 2000; Pilegaard et al. 2003; Russell et al. 2003; Short et al. 2003; Taylor et al. 2004; Garcia-Roves et al. 2006; Kuhl et al. 2006; Sriwijitkamol et al. 2006; Perry et al. 2010; Granata et al. 2016), further supporting its role in regulating mitochondrial biogenesis. The increase in PGC-1α protein with exercise training is likely related to the fact that PGC-1α autoregulates its own promoter through the transcription factor myocyte enhancer factor 2 (MEF2) (Fig. 2) (Handschin et al. 2003). Additional evidence for a key role for PGC-1α in exercise training comes from studies in which PGC-1α has been overexpressed in skeletal muscle resulting in a “classic” exercise-adaptation profile, including a large increase in functional mitochondria (Lin et al. 2002b), improvements in whole-body VO2max, a shift from carbohydrate to fat fuels during submaximal exercise, and improved endurance performance (Calvo et al. 2008). The notion that PGC-1α is essential for endurance training adaptation, however, has been challenged by findings that contraction-induced mitochondrial biogenesis is still observed in PGC-1α knockout mice (Leick et al. 2008) or double PGC-1α/β isoform knockouts (Ballmann et al. 2016). This biological redundancy indicates that perhaps the only obligatory response to exercise is the defense of homeostasis itself (Hawley et al. 2014) and serves to highlight the complex and extensive network of responses involved in exercise, involving many interdependent pathways with a high degree of cross talk, feedback regulation, and transient activation. Notwithstanding the results from “loss-of-function” models, it seems safe to place PGC-1α as a central player in orchestrating many of the oxidative adaptations to exercise. Of recent interest are the observations that commencing endurance-based exercise with low muscle glycogen availability results in greater activation of many signaling proteins with putative roles in mitochondrial biogenesis compared with when the same exercise is undertaken with normal glycogen concentration (Hawley and Morton 2014). These findings suggest that nutrient availability is a potent signal that can modulate the acute cellular responses to a single bout of exercise. When exercise sessions are repeated (i.e., training) in the face of low glycogen availability, the phenotypic adaptations resulting from such interventions are also augmented.
Recently, the tumor suppressor protein p53 has emerged as another transcription factor with a role in exercise-induced mitochondrial biogenesis in skeletal muscle. This protein is presumably activated by the AMPK and/or p38 MAPK (see subsequent discussion). Indeed, p53 knockout mice showed that p38 MAPK was not activated following exercise concomitant with an attenuated activation of AMPK as well as CaMKII (see below) (Saleem et al. 2014). Furthermore, p53 knockout mice display reduced endurance exercise capacity compared with wild-type mice, along with reduced subsarcolemmal and intermyofibrillar mitochondrial content and PGC-1α expression and an absence of mRNA transcripts related to mitochondrial biogenesis following exercise (Saleem et al. 2009). It has been suggested that p53 regulates exercise-induced mitochondrial biogenesis through its interaction with TFAM in the mitochondria, which coordinates expression of the mitochondrial genome (Bartlett et al. 2014).
Sending the Signal: From Kinase Activation to Increased Gene Expression
Although intense research efforts focused on several transcription factors involved in contraction-induced mitochondrial biogenesis, studies were also being undertaken to discover the “missing link” between upstream signaling cascades and the increased gene expression following exercise. In this regard, the AMPK was revealed to be an important regulator of PGC-1α. AMPK is a member of a metabolite-sensing protein kinase family that functions as a metabolic “fuel gauge” in skeletal muscle, consisting of an α catalytic and β and γ regulatory subunits that exist in multiple isoforms and are all required for full enzymatic activity. Seminal work discovered the AMPK was triggered in response to exercise-induced elevations in [AMPf] (Winder et al. 2000) and that this activation of AMPK was exercise intensity-dependent (Chen et al. 2003; Wadley et al. 2006). In this context, the insights of Holloszy (1967) made almost five decades ago are of relevance. In this study, Holloszy speculated that “the intracellular concentrations of numerous substances, including pyruvate, lactate, Pi, ADP, and AMP … may act as an inducer of the biosynthesis of the enzymes involved in mitochondrial electron transport.” Support for this contention is evidence from human work showing that the rapid and transient activation of AMPK after strenuous exercise is associated with peak elevations in [AMPf] (McConell et al. 2005). After a period of training, these rapid increases in [AMPf] and AMPK activation are attenuated during exercise undertaken at the same absolute (pretraining) work rate, which may, in part, explain why mitochondrial biogenesis slows and eventually reaches a plateau (McConell et al. 2005). This is consistent with the concept that muscle adaptation is a product of persistent disruptions to cellular homeostasis, which improves the ability of muscle to maintain cellular homeostasis during a subsequent exercise challenge (i.e., “hormesis”) and explains the attenuated activation of mRNA transcripts for mitochondrial biogenesis as training progresses (Perry et al. 2010), although this observation is equivocal (Pilegaard et al. 2003). Such observations provide, in part, mechanistic evidence for why training regimens must use a progressive overload stimulus to the contracting muscles once adaptation and/or performance has plateaued.
AMPK induces mitochondrial biogenesis partly by directly phosphorylating and activating PGC-1α (Jager et al. 2007) as well as the transcriptional repressor histone deacetylase 5 (HDAC5), which relieves inhibition of the transcription factor myocyte enhancer factor 2 (MEF2), a known regulator of the PGC-1α gene (McGee and Hargreaves 2010). Although whole-body deletion of AMP β2 reduces muscle AMPK activity and exercise capacity (Steinberg et al. 2010), deletion of the α subunit of AMPK does not impair exercise-induced mitochondrial biogenesis (Jorgensen et al. 2005; Fentz et al. 2015). These and the results of other studies clearly show that normal responses and adaptations to both acute exercise and chronic exercise training can be seen when one or more key pathways are absent or blocked, indicating that there is both biological redundancy and compensatory up-regulation of other exercise-induced pathways with putative roles in mitochondrial biogenesis.
A second critical signal that promotes mitochondrial biogenesis is the pulsatile increase in [Ca2+] observed at the onset of muscle contraction. Early work compared a variety of calcium signaling pathways, including calcineurin (CaN) and isoforms of calmodulin kinase (CaMK) that were known to be regulators of PGC-1α expression itself (CaN; Handschin et al. 2003) or mitochondrial biogenesis in nonexercise models (CaMK; Wu et al. 2002; Wright 2007). Studies inhibiting or deleting each protein in mouse models revealed that exercise-induced mitochondrial biogenesis was unaffected (Akimoto et al. 2004; Garcia-Roves et al. 2006), despite the fact that at least CaMK is activated in association with increased calcium spikes in human muscle during exercise (Rose and Hargreaves 2003; Rose et al. 2007). The consistent activation of these pathways during exercise suggests they have some role in the adaptive process to exercise, but that there exists physiological redundancy as with the PGC-1 isoforms (discussed previously).
The p38 MAPK is another calcium-signaling pathway that plays a role in contraction-induced mitochondrial biogenesis. The p38 MAPK phosphorylates and activates PGC-1α (Knutti et al. 2001; Puigserver et al. 2001; Fan et al. 2004) and translocates PGC-1α from the cytoplasm to the nucleus before increases in gene transcripts for mitochondrial proteins (Wright 2007). As discussed above, the translocation of PGC-1α first reported in rat muscle and subsequently verified in humans (Little et al. 2010) is thought to be a major mechanism by which PGC-1α activity was regulated by contraction. Indeed, the balance of evidence suggests that the AMPK and p38 MAPK are two important signaling cascades that converge on PGC-1α and consequently the regulation of mitochondrial biogenesis through contraction-mediated perturbations in muscle metabolites (i.e., [AMPf]/[ATP]) and ionic status (i.e., calcium handling). Overall, Ca2+ is a major contraction-induced stimulus for PGC-1α-mediated mitochondrial biogenesis (Chin 2005; Wright 2007), but the relative importance of each pathway following exercise remains to be determined.
Time Course of Events Leading to Exercise Training–Induced Mitochondrial Biogenesis
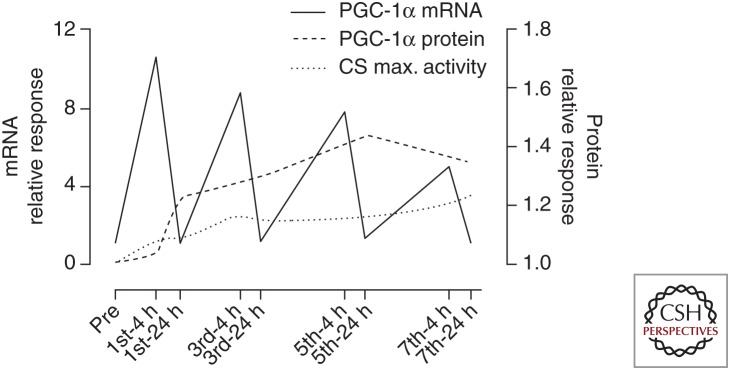
The time course of the exercise-induced increases in mRNA and protein are discordant, indicating that the extent to which a protein will increase in response to an adaptive stimulus cannot be predicted from the increase in mRNA alone. As previously noted (Baar et al. 2002), this makes the measurement of protein concentrations critical when studying the adaptive responses to exercise. It has been proposed that the exercise-induced increase in mitochondrial content is the result of the cumulative effects of repeated but transient increases in mRNA, encoding mitochondrial proteins that occurred in the recovery period from successive and additive exercise challenges (Williams and Neufer 1996; Pilegaard et al. 2000; Booth and Neufer 2006). To test this hypothesis, two short-term human training studies using serial muscle biopsy sampling throughout the training intervention have recently been reported (Perry et al. 2010; Egan et al. 2013). These investigations examined the early time course of several potential transcriptional, translational, and morphological processes that are involved in mitochondrial biogenesis during exercise training in human skeletal muscle. These studies used high-intensity interval training (HIIT) performed daily (14 sessions, 2 weeks) (Egan et al. 2013) or on every other day (nine sessions, 3 weeks) (Perry et al. 2010). Perry et al. (2010) showed an increase in exercise-induced mRNA before the initial increase in protein for specific mitochondrial enzymes and transcription proteins including both PGC-1 isoforms. However, both the timing and magnitude of mRNA and protein responses varied considerably for mRNA encoding transcriptional and mitochondrial proteins as well as the protein contents themselves, which was also shown by Egan et al. (2013). Furthermore, the mRNA response to exercise was attenuated as the muscle adapted to the exercise challenge, even as the training intensity increased (Perry et al. 2010), although this is not seen for all proteins (Egan et al. 2013) but may depend on the biopsy time-point. Taken collectively, these studies showed diverse time-dependent responses in genes encoding transcription factors and mitochondrial proteins, as well as protein accumulation of selected markers of mitochondrial biogenesis, and highlight how the induction and perpetuation of mitochondrial biogenesis involves time-dependent roles for different transcriptional targets. Furthermore, these studies showed, in humans, the important concept that early and rapid up-regulation of transcriptional proteins during training likely amplifies the “transcriptional sensitivity” (Perry et al. 2010) of the cell to a subsequent bout of exercise, thereby permitting sustained synthesis of mitochondrial proteins (Fig. 3). However, the precise role of increased mitochondrial content in mediating improved metabolic control early in training was challenged in the 1990s when reduced glycogenolysis and lactate accumulation were noted after only 5–7 days of training without concomitant increases in succinate dehydrogenase and CS (Green et al. 1992). The interplay between mitochondrial content and nonmitochondrial adaptations such as improved regulation of muscle blood flow, substrate transport, and posttranslational control of metabolic proteins remains an outstanding challenge to be resolved.
Figure 3.
Schematic representation of how repeated transient increases in the transcriptional cofactor peroxisome proliferator-activated receptor coactivator 1α (PGC-1α) messenger RNA (mRNA) after acute exercise sessions drives the continued accumulation of PGC-1α protein. Early increases in PGC-1α protein contribute to gradual increases in citrate synthase (CS) activity (a marker of mitochondrial biogenesis) by inducing expression of CS mRNA (not shown). Data was acquired throughout 2 weeks of high-intensity interval cycling training in human skeletal muscle. (From Perry et al. 2010; reproduced, with permission, from the authors.)
OTHER MECHANISMS PROMOTING MITOCHONDRIAL BIOGENESIS
Although considerable work has shown that additional regulation of mitochondrial biogenesis exists at the level of protein import into mitochondria following muscle contraction in rodents (Joseph et al. 2010; Hood et al. 2011), less attention has focused on posttranscriptional control at the level of protein translation or mRNA stability. Recent results suggest mRNA stability decreases with muscle contraction or exercise in rodents (Lai et al. 2010) along with increased mRNA-destabilizing proteins (Lai et al. 2010; Matravadia et al. 2013) and possibly with mRNA-stabilizing proteins (Lai et al. 2010). In this regard, it has been suggested that mRNA destabilization is actually beneficial for accelerating adaptive responses in a subsequent bout of contraction (Lai et al. 2010). Alternatively, automatic degradation following mRNA synthesis may ensure that tight control of mitochondrial biogenesis is maintained at all times to avoid excessive protein translation. Mitochondrial degradation through “mitophagy” may lead to a new era of research of how exercise may promote mitochondrial degradation as a “quality control” measure to facilitate regeneration as previously shown in rodents through a PGC1α-dependent manner (Vainshtein et al. 2015). Furthermore, mitochondrial biogenesis likely involves an extensive remodeling process whereby mitochondria are formed into a tubular reticulum particularly in intermyofibrillar mitochondria that shape them around the myofibrils (Ogata and Yamasaki 1997), which likely optimizes spatial distribution of energy. On this note, exercise training increases human muscle mitochondrial fission proteins but not fusion proteins (Perry et al. 2010; Egan et al. 2013), suggesting fission is an important component of the mitochondrial remodeling that likely takes place during reticulum elongation following training (Kirkwood et al. 1987). What is apparent is that the proteins that undergo cumulative responses to endurance-based exercise training are in a constant state of flux, the concentration of which reflects the balance between synthesis and degradation at any given time. In addition, those proteins with fast turnover rates that show rapid, but unsustained, changes in mRNA expression in response to exercise (e.g., transcription factors, activating cofactors, etc.) may be critical for amplifying the induction of genes encoding for proteins with longer half-lives that increase progressively with training. Finally, although the intense focus on AMP and calcium signal induction of mitochondrial biogenesis has dominated much of the literature, the role of ROS (Powers et al. 2011; Gomez-Cabrera et al. 2015), fatty acid ligands on PPARs (Gilde and Van Bilsen 2003), NAD/NADH via sirtuin-mediated deacetylation (White and Schenk 2012; Hood et al. 2016), and other signals have been implicated as additional regulators. However, the relative importance of each signal remains to be determined.
Since publication of the seminal investigation by Holloszy (1967), considerable effort from many independent laboratories around the world have advanced our understanding of how exercise activates cellular, molecular, and biochemical pathways with regulatory roles in the training-response adaptation. However, much work still remains before we fully understand how additional levels of control regulate the increased mitochondrial oxidative capacity that occurs with exercise training. The pioneering study of Holloszy (1967) laid the foundation on which much of our present knowledge of exercise training–induced mitochondrial biogenesis has been based, and paved the way for new and future discoveries in the field.
CONCLUDING REMARKS: FUTURE CHALLENGES—KNOWLEDGE TRANSLATION AND HEALTH PROMOTION
During the past two decades, the application of molecular techniques to exercise biology has led to the identification of multiple molecular pathways with key roles in promoting exercise training–induced mitochondrial biogenesis. However, linking many of these signaling cascades to defined metabolic responses and specific changes in gene expression in skeletal muscle that occur after exercise has proved more difficult. This is because many of these pathways are not linear but are instead part of a complex network, with a high degree of cross talk, feedback regulation, and transient activation (Hawley et al. 2014). The advances of various “omics” approaches along with the application of computational and systems biology approaches to problems in exercise biology should facilitate future progress in this domain, but this will require the integration of data from multiple platforms and require large sample sizes, big data sets, and expertise in computational biology to resolve the complex biology associated with diverse exercise responses (Zierath and Wallberg-Henriksson 2015). Although the health-promoting benefits of exercise have been recognized for centuries, direct evidence linking such effects to specific health outcomes and understanding how these effects exert their benefits in different populations remains elusive and a challenge for future research (Neufer et al. 2015). A major step toward this goal will be to identify the molecules that are altered in key organs/tissues in response to exercise training and how these responses differ between individuals (i.e., high versus low exercise responders). In this regard, gene variants may set the upper limit to an individual’s capacity to adapt to exercise training or even the desire/will to exercise. The interplay between genes and environment may alter our epigenome and impact our own health by modifying the transcriptional potential of a cell or organ to adapt to changes in any exercise training protocol (Zierath and Wallberg-Henriksson 2015). Future research in the field of exercise biology requires increasingly sophisticated approaches to understand the critical nodes of energy homeostasis and how these pathways are up-regulated in response to exercise training and disrupted in a number of inactivity-related disorders (Hawley et al. 2014). The results of such research will hopefully catalyze the advent of personalized exercise medicine by promoting health through improvements in individualized exercise prescription in combination with understanding the behavioral aspects of exercise adherence, which are the scope of this review. At present, the translation of our basic knowledge about exercise training–induced increases in mitochondrial biogenesis into improving the metabolic health of the general population has been mediocre, at best. Indeed, genetic and pharmacological manipulations of mitochondrial content or function have yielded conflicting results in muscle metabolic health, which remains a challenge to reconcile with the role of mitochondria in exercise-induced improvements in muscle fitness (see Hawley et al. 2014; Neufer et al. 2015). As such, this review extends previous challenges to the scientific community to bolster the translation of such knowledge to counter the epidemic of chronic inactivity–related diseases (Hawley et al. 2014; Neufer et al. 2015; Zierath and Wallberg-Henriksson 2015).
ACKNOWLEDGMENTS
The authors declare no conflict of interest. The writing of this review is supported, in part, by a Novo Nordisk Challenge Grant NNF14OC0011493 (to J.A.H.). Because of restrictions on the number of references, we have been unable to include important work by some of our peers, for which we apologize in advance.
Footnotes
Editors: Juleen R. Zierath, Michael J. Joyner, and John A. Hawley
Additional Perspectives on The Biology of Exercise available at www.perspectivesinmedicine.org
REFERENCES
- Akimoto T, Ribar TJ, Williams RS, Yan Z. 2004. Skeletal muscle adaptation in response to voluntary running in Ca2+/calmodulin-dependent protein kinase IV-deficient mice. Am J Physiol Cell Physiol 287: C1311–C1319. [DOI] [PubMed] [Google Scholar]
- Akimoto T, Pohnert SC, Li P, Zhang M, Gumbs C, Rosenberg PB, Williams RS, Yan Z. 2005. Exercise stimulates Pgc-1α transcription in skeletal muscle through activation of the p38 MAPK pathway. J Biol Chem 280: 19587–19593. [DOI] [PubMed] [Google Scholar]
- Baar K, Wende AR, Jones TE, Marison M, Nolte LA, Chen M, Kelly DP, Holloszy JO. 2002. Adaptations of skeletal muscle to exercise: Rapid increase in the transcriptional coactivator PGC-1. FASEB J 16: 1879–1886. [DOI] [PubMed] [Google Scholar]
- Ballmann C, Tang Y, Bush Z, Rowe GC. 2016. Adult expression of PGC-1α and -1β in skeletal muscle is not required for endurance exercise-induced enhancement of exercise capacity. Am J Physiol Endocrinol Metab 311: E928–E938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett JD, Close GL, Drust B, Morton JP. 2014. The emerging role of p53 in exercise metabolism. Sports Med 44: 303–309. [DOI] [PubMed] [Google Scholar]
- Booth FW, Narahara KA. 1974. Vastus lateralis cytochrome oxidase activity and its relationship to maximal oxygen consumption in man. Pflugers Arch 349: 319–324. [DOI] [PubMed] [Google Scholar]
- Booth FW, Neufer PD. 2006. Exercise genomics and proteomics. In ACSM’s advanced exercise physiology (ed. Tipton CM), pp. 623–651. Lippincott, Williams & Wilkins Baltimore. [Google Scholar]
- Burgomaster KA, Hughes SC, Heigenhauser GJ, Bradwell SN, Gibala MJ. 2005. Six sessions of sprint interval training increases muscle oxidative potential and cycle endurance capacity in humans. J Appl Physiol (1985) 98: 1985–1990. [DOI] [PubMed] [Google Scholar]
- Calvo JA, Daniels TG, Wang X, Paul A, Lin J, Spiegelman BM, Stevenson SC, Rangwala SM. 2008. Muscle-specific expression of PPARγ coactivator-1α improves exercise performance and increases peak oxygen uptake. J Appl Physiol (1985) 104: 1304–1312. [DOI] [PubMed] [Google Scholar]
- Cartoni R, Leger B, Hock MB, Praz M, Crettenand A, Pich S, Ziltener JL, Luthi F, Deriaz O, Zorzano A, et al. 2005. Mitofusins 1/2 and ERRα expression are increased in human skeletal muscle after physical exercise. J Physiol 567: 349–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau CM, Evans MJ, Scarpulla RC. 1992. Nuclear respiratory factor 1 activation sites in genes encoding the γ-subunit of ATP synthase, eukaryotic initiation factor 2α, and tyrosine aminotransferase. Specific interaction of purified NRF-1 with multiple target genes. J Biol Chem 267: 6999–7006. [PubMed] [Google Scholar]
- Chen ZP, Stephens TJ, Murthy S, Canny BJ, Hargreaves M, Witters LA, Kemp BE, McConell GK. 2003. Effect of exercise intensity on skeletal muscle AMPK signaling in humans. Diabetes 52: 2205–2212. [DOI] [PubMed] [Google Scholar]
- Chin ER. 2005. Role of Ca2+/calmodulin-dependent kinases in skeletal muscle plasticity. J Appl Physiol (1985) 99: 414–423. [DOI] [PubMed] [Google Scholar]
- Cochran AJ, Percival ME, Tricarico S, Little JP, Cermak N, Gillen JB, Tarnopolsky MA, Gibala MJ. 2014. Intermittent and continuous high-intensity exercise training induce similar acute but different chronic muscle adaptations. Exp Physiol 99: 782–791. [DOI] [PubMed] [Google Scholar]
- De Filippis E, Alvarez G, Berria R, Cusi K, Everman S, Meyer C, Mandarino LJ. 2008. Insulin-resistant muscle is exercise resistant: Evidence for reduced response of nuclear-encoded mitochondrial genes to exercise. Am J Physiol Endocrinol Metab 294: E607–E614. [DOI] [PubMed] [Google Scholar]
- Dudley GA, Abraham WM, Terjung RL. 1982. Influence of exercise intensity and duration on biochemical adaptations in skeletal muscle. J Appl Physiol 53: 844–850. [DOI] [PubMed] [Google Scholar]
- Dudley GA, Tullson PC, Terjung RL. 1987. Influence of mitochondrial content on the sensitivity of respiratory control. J Biol Chem 262: 9109–9114. [PubMed] [Google Scholar]
- Edgett BA, Foster WS, Hankinson PB, Simpson CA, Little JP, Graham RB, Gurd BJ. 2013. Dissociation of increases in PGC-1α and its regulators from exercise intensity and muscle activation following acute exercise. PLoS ONE 8: e71623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan B, Carson BP, Garcia-Roves PM, Chibalin AV, Sarsfield FM, Barron N, McCaffrey N, Moyna NM, Zierath JR, O’Gorman DJ. 2010. Exercise intensity-dependent regulation of peroxisome proliferator-activated receptor coactivator-1 mRNA abundance is associated with differential activation of upstream signalling kinases in human skeletal muscle. J Physiol 588: 1779–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan B, O’Connor PL, Zierath JR, O’Gorman DJ. 2013. Time course analysis reveals gene-specific transcript and protein kinetics of adaptation to short-term aerobic exercise training in human skeletal muscle. PLoS ONE 8: e74098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan M, Rhee J, St-Pierre J, Handschin C, Puigserver P, Lin J, Jaeger S, Erdjument-Bromage H, Tempst P, Spiegelman BM. 2004. Suppression of mitochondrial respiration through recruitment of p160 myb binding protein to PGC-1α: Modulation by p38 MAPK. Genes Dev 18: 278–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fentz J, Kjobsted R, Kristensen CM, Hingst JR, Birk JB, Gudiksen A, Foretz M, Schjerling P, Viollet B, Pilegaard H, et al. 2015. AMPKα is essential for acute exercise-induced gene responses but not for exercise training-induced adaptations in mouse skeletal muscle. Am J Physiol Endocrinol Metab 309: E900–E914. [DOI] [PubMed] [Google Scholar]
- Frier BC, Hancock CR, Little JP, Fillmore N, Bliss TA, Thomson DM, Wan Z, Wright DC. 2011. Reductions in RIP140 are not required for exercise- and AICAR-mediated increases in skeletal muscle mitochondrial content. J Appl Physiol (1985) 111: 688–695. [DOI] [PubMed] [Google Scholar]
- Garcia-Roves PM, Huss J, Holloszy JO. 2006. Role of calcineurin in exercise-induced mitochondrial biogenesis. Am J Physiol Endocrinol Metab 290: E1172–E1179. [DOI] [PubMed] [Google Scholar]
- Gilde AJ, Van Bilsen M. 2003. Peroxisome proliferator-activated receptors (PPARS): Regulators of gene expression in heart and skeletal muscle. Acta Physiol Scand 178: 425–434. [DOI] [PubMed] [Google Scholar]
- Gollnick PD, King DW. 1969. Effect of exercise and training on mitochondria of rat skeletal muscle. Am J Physiol 216: 1502–1509. [DOI] [PubMed] [Google Scholar]
- Gollnick PD, Saltin B. 1982. Significance of skeletal muscle oxidative enzyme enhancement with endurance training. Clin Physiol 2: 1–12. [DOI] [PubMed] [Google Scholar]
- Gollnick PD, Armstrong RB, Saltin B, Saubert CWt, Sembrowich WL, Shepherd RE. 1973. Effect of training on enzyme activity and fiber composition of human skeletal muscle. J Appl Physiol 34: 107–111. [DOI] [PubMed] [Google Scholar]
- Gomez-Cabrera MC, Salvador-Pascual A, Cabo H, Ferrando B, Vina J. 2015. Redox modulation of mitochondriogenesis in exercise. Does antioxidant supplementation blunt the benefits of exercise training? Free Radic Biol Med 86: 37–46. [DOI] [PubMed] [Google Scholar]
- Gordon JW, Rungi AA, Inagaki H, Hood DA. 2001. Effects of contractile activity on mitochondrial transcription factor A expression in skeletal muscle. J Appl Physiol (1985) 90: 389–396. [DOI] [PubMed] [Google Scholar]
- Goto M, Terada S, Kato M, Katoh M, Yokozeki T, Tabata I, Shimokawa T. 2000. cDNA cloning and mRNA analysis of PGC-1 in epitrochlearis muscle in swimming-exercised rats. Biochem Biophys Res Commun 274: 350–354. [DOI] [PubMed] [Google Scholar]
- Granata C, Oliveira RS, Little JP, Renner K, Bishop DJ. 2016. Training intensity modulates changes in PGC-1α and p53 protein content and mitochondrial respiration, but not markers of mitochondrial content in human skeletal muscle. FASEB J 30: 959–970. [DOI] [PubMed] [Google Scholar]
- Green HJ, Helyar R, Ball-Burnett M, Kowalchuk N, Symon S, Farrance B. 1992. Metabolic adaptations to training precede changes in muscle mitochondrial capacity. J Appl Physiol 72: 484–491. [DOI] [PubMed] [Google Scholar]
- Gulick T, Cresci S, Caira T, Moore DD, Kelly DP. 1994. The peroxisome proliferator-activated receptor regulates mitochondrial fatty acid oxidative enzyme gene expression. Proc Natl Acad Sci 91: 11012–11016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handschin C, Rhee J, Lin J, Tarr PT, Spiegelman BM. 2003. An autoregulatory loop controls peroxisome proliferator-activated receptor γ coactivator 1α expression in muscle. Proc Natl Acad Sci 100: 7111–7116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley JA. 2002. Adaptations of skeletal muscle to prolonged, intense endurance training. Clin Exp Pharmacol Physiol 29: 218–222. [DOI] [PubMed] [Google Scholar]
- Hawley JA, Morton JP. 2014. Ramping up the signal: Promoting endurance training adaptation in skeletal muscle by nutritional manipulation. Clin Exp Pharmacol Physiol 41: 608–613. [DOI] [PubMed] [Google Scholar]
- Hawley JA, Hargreaves M, Joyner MJ, Zierath JR. 2014. Integrative biology of exercise. Cell 159: 738–749. [DOI] [PubMed] [Google Scholar]
- Hellsten Y, Nielsen JJ, Lykkesfeldt J, Bruhn M, Silveira L, Pilegaard H, Bangsbo J. 2007. Antioxidant supplementation enhances the exercise-induced increase in mitochondrial uncoupling protein 3 and endothelial nitric oxide synthase mRNA content in human skeletal muscle. Free Radic Biol Med 43: 353–361. [DOI] [PubMed] [Google Scholar]
- Hickson RC, Heusner WW, Van Huss WD. 1976. Skeletal muscle enzyme alterations after sprint and endurance training. J Appl Physiol 40: 868–871. [DOI] [PubMed] [Google Scholar]
- Holloszy JO. 1967. Biochemical adaptations in muscle: Effects of exercise on mitochondrial oxygen uptake and respiratory enzyme activity in skeletal muscle. J Biol Chem 242: 2278–2282. [PubMed] [Google Scholar]
- Holloszy JO, Booth FW. 1976. Biochemical adaptations to endurance exercise in muscle. Annu Rev Physiol 38: 273–291. [DOI] [PubMed] [Google Scholar]
- Holloszy JO, Coyle EF. 1984. Adaptations of skeletal muscle to endurance exercise and their metabolic consequences. J Appl Physiol 56: 831–838. [DOI] [PubMed] [Google Scholar]
- Hood DA. 2001. Invited review: Contractile activity-induced mitochondrial biogenesis in skeletal muscle. J Appl Physiol (1985) 90: 1137–1157. [DOI] [PubMed] [Google Scholar]
- Hood DA, Zak R, Pette D. 1989. Chronic stimulation of rat skeletal muscle induces coordinate increases in mitochondrial and nuclear mRNAs of cytochrome-c-oxidase subunits. Eur J Biochem 179: 275–280. [DOI] [PubMed] [Google Scholar]
- Hood DA, Uguccioni G, Vainshtein A, D’Souza D. 2011. Mechanisms of exercise-induced mitochondrial biogenesis in skeletal muscle: Implications for health and disease. Compr Physiol 1: 1119–1134. [DOI] [PubMed] [Google Scholar]
- Hood DA, Tryon LD, Carter HN, Kim Y, Chen CC. 2016. Unravelling the mechanisms regulating muscle mitochondrial biogenesis. Biochem J 473: 2295–2314. [DOI] [PubMed] [Google Scholar]
- Hoppeler H, Luthi P, Claassen H, Weibel ER, Howald H. 1973. The ultrastructure of the normal human skeletal muscle. A morphometric analysis on untrained men, women and well-trained orienteers. Pflugers Arch 344: 217–232. [DOI] [PubMed] [Google Scholar]
- Irrcher I, Walkinshaw DR, Sheehan TE, Hood DA. 2008. Thyroid hormone (T3) rapidly activates p38 and AMPK in skeletal muscle in vivo. J Appl Physiol (1985) 104: 178–185. [DOI] [PubMed] [Google Scholar]
- Jager S, Handschin C, St-Pierre J, Spiegelman BM. 2007. AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1α. Proc Natl Acad Sci 104: 12017–12022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen SB, Wojtaszewski JF, Viollet B, Andreelli F, Birk JB, Hellsten Y, Schjerling P, Vaulont S, Neufer PD, Richter EA, et al. 2005. Effects of α-AMPK knockout on exercise-induced gene activation in mouse skeletal muscle. FASEB J 19: 1146–1148. [DOI] [PubMed] [Google Scholar]
- Joseph AM, Ljubicic V, Adhihetty PJ, Hood DA. 2010. Biogenesis of the mitochondrial Tom40 channel in skeletal muscle from aged animals and its adaptability to chronic contractile activity. Am J Physiol Cell Physiol 298: C1308–C1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly DP, Scarpulla RC. 2004. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes Dev 18: 357–368. [DOI] [PubMed] [Google Scholar]
- Kiessling KH, Pilstrom L, Bylund AC, Saltin B, Piehl K. 1974. Enzyme activities and morphometry in skeletal muscle of middle-aged men after training. Scand J Clin Lab Invest 33: 63–69. [DOI] [PubMed] [Google Scholar]
- Kirkwood SP, Packer L, Brooks GA. 1987. Effects of endurance training on a mitochondrial reticulum in limb skeletal muscle. Arch Biochem Biophys 255: 80–88. [DOI] [PubMed] [Google Scholar]
- Knutti D, Kressler D, Kralli A. 2001. Regulation of the transcriptional coactivator PGC-1 via MAPK-sensitive interaction with a repressor. Proc Natl Acad Sci 98: 9713–9718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koves TR, Li P, An J, Akimoto T, Slentz D, Ilkayeva O, Dohm GL, Yan Z, Newgard CB, Muoio DM. 2005. Peroxisome proliferator-activated receptor-γ co-activator 1α-mediated metabolic remodeling of skeletal myocytes mimics exercise training and reverses lipid-induced mitochondrial inefficiency. J Biol Chem 280: 33588–33598. [DOI] [PubMed] [Google Scholar]
- Kuhl JE, Ruderman NB, Musi N, Goodyear LJ, Patti ME, Crunkhorn S, Dronamraju D, Thorell A, Nygren J, Ljungkvist O, et al. 2006. Exercise training decreases the concentration of malonyl-CoA and increases the expression and activity of malonyl-CoA decarboxylase in human muscle. Am J Physiol Endocrinol Metab 290: E1296–E1303. [DOI] [PubMed] [Google Scholar]
- Lai RY, Ljubicic V, D’Souza D, Hood DA. 2010. Effect of chronic contractile activity on mRNA stability in skeletal muscle. Am J Physiol Cell Physiol 299: C155–C163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrie RA. 1953. The activity of the cytochrome system in muscle and its relation to myoglobin. Biochem J 55: 298–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leick L, Wojtaszewski JF, Johansen ST, Kiilerich K, Comes G, Hellsten Y, Hidalgo J, Pilegaard H. 2008. PGC-1α is not mandatory for exercise- and training-induced adaptive gene responses in mouse skeletal muscle. Am J Physiol Endocrinol Metab 294: E463–E474. [DOI] [PubMed] [Google Scholar]
- Lelliott CJ, Medina-Gomez G, Petrovic N, Kis A, Feldmann HM, Bjursell M, Parker N, Curtis K, Campbell M, Hu P, et al. 2006. Ablation of PGC-1β results in defective mitochondrial activity, thermogenesis, hepatic function, and cardiac performance. PLoS Biol 4: e369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Puigserver P, Donovan J, Tarr P, Spiegelman BM. 2002a. Peroxisome proliferator-activated receptor γ coactivator 1β (PGC-1β), a novel PGC-1-related transcription coactivator associated with host cell factor. J Biol Chem 277: 1645–1648. [DOI] [PubMed] [Google Scholar]
- Lin J, Wu H, Tarr PT, Zhang CY, Wu Z, Boss O, Michael LF, Puigserver P, Isotani E, Olson EN, et al. 2002b. Transcriptional co-activator PGC-1α drives the formation of slow-twitch muscle fibres. Nature 418: 797–801. [DOI] [PubMed] [Google Scholar]
- Lin J, Handschin C, Spiegelman BM. 2005. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab 1: 361–370. [DOI] [PubMed] [Google Scholar]
- Little JP, Safdar A, Cermak N, Tarnopolsky MA, Gibala MJ. 2010. Acute endurance exercise increases the nuclear abundance of PGC-1α in trained human skeletal muscle. Am J Physiol Regul Integr Comp Physiol 298: R912–R917. [DOI] [PubMed] [Google Scholar]
- MacDougall JD, Hicks AL, MacDonald JR, McKelvie RS, Green HJ, Smith KM. 1998. Muscle performance and enzymatic adaptations to sprint interval training. J Appl Physiol (1985) 84: 2138–2142. [DOI] [PubMed] [Google Scholar]
- MacInnis MJ, Gibala MJ. 2016. Physiological adaptations to interval training and the role of exercise intensity. J Physiol 10.113/JP273196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Redondo V, Pettersson AT, Ruas JL. 2015. The hitchhiker’s guide to PGC-1α isoform structure and biological functions. Diabetologia 58: 1969–1977. [DOI] [PubMed] [Google Scholar]
- Mathai AS, Bonen A, Benton CR, Robinson DL, Graham TE. 2008. Rapid exercise-induced changes in PGC-1α mRNA and protein in human skeletal muscle. J Appl Physiol (1985) 105: 1098–1105. [DOI] [PubMed] [Google Scholar]
- Matravadia S, Martino VB, Sinclair D, Mutch DM, Holloway GP. 2013. Exercise training increases the expression and nuclear localization of mRNA destabilizing proteins in skeletal muscle. Am J Physiol Regul Integr Comp Physiol 305: R822–R831. [DOI] [PubMed] [Google Scholar]
- McConell GK, Lee-Young RS, Chen ZP, Stepto NK, Huynh NN, Stephens TJ, Canny BJ, Kemp BE. 2005. Short-term exercise training in humans reduces AMPK signalling during prolonged exercise independent of muscle glycogen. J Physiol 568: 665–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee SL, Hargreaves M. 2010. Histone modifications and skeletal muscle metabolic gene expression. Clin Exp Pharmacol Physiol 37: 392–396. [DOI] [PubMed] [Google Scholar]
- Meirhaeghe A, Crowley V, Lenaghan C, Lelliott C, Green K, Stewart A, Hart K, Schinner S, Sethi JK, Yeo G, et al. 2003. Characterization of the human, mouse and rat PGC1β (peroxisome-proliferator-activated receptor-γ co-activator 1β) gene in vitro and in vivo. Biochem J 373: 155–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison PR, Biggs RB, Booth FW. 1989. Daily running for 2 wk and mRNAs for cytochrome c and α-actin in rat skeletal muscle. Am J Physiol 257: C936–C939. [DOI] [PubMed] [Google Scholar]
- Mortensen OH, Plomgaard P, Fischer CP, Hansen AK, Pilegaard H, Pedersen BK. 2007. PGC-1β is downregulated by training in human skeletal muscle: No effect of training twice every second day vs. once daily on expression of the PGC-1 family. J Appl Physiol (1985) 103: 1536–1542. [DOI] [PubMed] [Google Scholar]
- Neufer PD, Dohm GL. 1993. Exercise induces a transient increase in transcription of the GLUT-4 gene in skeletal muscle. Am J Physiol 265: C1597–C1603. [DOI] [PubMed] [Google Scholar]
- Neufer PD, Bamman MM, Muoio DM, Bouchard C, Cooper DM, Goodpaster BH, Booth FW, Kohrt WM, Gerszten RE, Mattson MP, et al. 2015. Understanding the cellular and molecular mechanisms of physical activity-induced health benefits. Cell Metab 22: 4–11. [DOI] [PubMed] [Google Scholar]
- Norrbom J, Sundberg CJ, Ameln H, Kraus WE, Jansson E, Gustafsson T. 2004. PGC-1α mRNA expression is influenced by metabolic perturbation in exercising human skeletal muscle. J Appl Physiol (1985) 96: 189–194. [DOI] [PubMed] [Google Scholar]
- Ogata T, Yamasaki Y. 1997. Ultra-high-resolution scanning electron microscopy of mitochondria and sarcoplasmic reticulum arrangement in human red, white, and intermediate muscle fibers. Anat Rec 248: 214–223. [DOI] [PubMed] [Google Scholar]
- Oscai LB, Holloszy JO. 1971. Biochemical adaptations in muscle. II: Response of mitochondrial adenosine triphosphatase, creatine phosphokinase, and adenylate kinase activities in skeletal muscle to exercise. J Biol Chem 246: 6968–6972. [PubMed] [Google Scholar]
- Parra J, Cadefau JA, Rodas G, Amigo N, Cusso R. 2000. The distribution of rest periods affects performance and adaptations of energy metabolism induced by high-intensity training in human muscle. Acta Physiol Scand 169: 157–165. [DOI] [PubMed] [Google Scholar]
- Paul MH, Sperling E. 1952. Cyclophorase system. XXIII: Correlation of cyclophorase activity and mitochondrial density in striated muscle. Proc Soc Exp Biol Med 79: 352–354. [DOI] [PubMed] [Google Scholar]
- Perry CG, Talanian JL, Heigenhauser GJ, Spriet LL. 2007. The effects of training in hyperoxia vs. normoxia on skeletal muscle enzyme activities and exercise performance. J Appl Physiol (1985) 102: 1022–1027. [DOI] [PubMed] [Google Scholar]
- Perry CGR, Heigenhauser GJF, Bonen A, Spriet LL. 2008. High-intensity aerobic interval training increases fat and carbohydrate metabolic capacities in human skeletal muscle. Appl Physiol Nutr Metab 33: 1112–1123. [DOI] [PubMed] [Google Scholar]
- Perry CG, Lally J, Holloway GP, Heigenhauser GJ, Bonen A, Spriet LL. 2010. Repeated transient mRNA bursts precede increases in transcriptional and mitochondrial proteins during training in human skeletal muscle. J Physiol 588: 4795–4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilegaard H, Ordway GA, Saltin B, Neufer PD. 2000. Transcriptional regulation of gene expression in human skeletal muscle during recovery from exercise. Am J Physiol Endocrinol Metab 279: E806–E814. [DOI] [PubMed] [Google Scholar]
- Pilegaard H, Saltin B, Neufer PD. 2003. Exercise induces transient transcriptional activation of the PGC-1α gene in human skeletal muscle. J Physiol 546: 851–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers SK, Nelson WB, Hudson MB. 2011. Exercise-induced oxidative stress in humans: Cause and consequences. Free Radic Biol Med 51: 942–950. [DOI] [PubMed] [Google Scholar]
- Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. 1998. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell 92: 829–839. [DOI] [PubMed] [Google Scholar]
- Puigserver P, Rhee J, Lin J, Wu Z, Yoon JC, Zhang CY, Krauss S, Mootha VK, Lowell BB, Spiegelman BM. 2001. Cytokine stimulation of energy expenditure through p38 MAP kinase activation of PPARγ coactivator-1. Mol Cell 8: 971–982. [DOI] [PubMed] [Google Scholar]
- Rodas G, Ventura JL, Cadefau JA, Cusso R, Parra J. 2000. A short training programme for the rapid improvement of both aerobic and anaerobic metabolism. Eur J Appl Physiol 82: 480–486. [DOI] [PubMed] [Google Scholar]
- Rose AJ, Hargreaves M. 2003. Exercise increases Ca2+-calmodulin-dependent protein kinase II activity in human skeletal muscle. J Physiol 553: 303–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose AJ, Frosig C, Kiens B, Wojtaszewski JF, Richter EA. 2007. Effect of endurance exercise training on Ca2+ calmodulin-dependent protein kinase II expression and signalling in skeletal muscle of humans. J Physiol 583: 785–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell AP, Feilchenfeldt J, Schreiber S, Praz M, Crettenand A, Gobelet C, Meier CA, Bell DR, Kralli A, Giacobino JP, et al. 2003. Endurance training in humans leads to fiber type-specific increases in levels of peroxisome proliferator-activated receptor-γ coactivator-1 and peroxisome proliferator-activated receptor-α in skeletal muscle. Diabetes 52: 2874–2881. [DOI] [PubMed] [Google Scholar]
- Russell AP, Hesselink MK, Lo SK, Schrauwen P. 2005. Regulation of metabolic transcriptional co-activators and transcription factors with acute exercise. FASEB J 19: 986–988. [DOI] [PubMed] [Google Scholar]
- Safdar A, Little JP, Stokl AJ, Hettinga BP, Akhtar M, Tarnopolsky MA. 2011. Exercise increases mitochondrial PGC-1α content and promotes nuclear-mitochondrial cross-talk to coordinate mitochondrial biogenesis. J Biol Chem 286: 10605–10617. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Saleem A, Adhihetty PJ, Hood DA. 2009. Role of p53 in mitochondrial biogenesis and apoptosis in skeletal muscle. Physiol Genomics 37: 58–66. [DOI] [PubMed] [Google Scholar]
- Saleem A, Carter HN, Hood DA. 2014. p53 is necessary for the adaptive changes in cellular milieu subsequent to an acute bout of endurance exercise. Am J Physiol Cell Physiol 306: C241–C249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpulla RC. 2002. Nuclear activators and coactivators in mammalian mitochondrial biogenesis. Biochim Biophys Acta 1576: 1–14. [DOI] [PubMed] [Google Scholar]
- Scarpulla RC. 2006. Nuclear control of respiratory gene expression in mammalian cells. J Cell Biochem 97: 673–683. [DOI] [PubMed] [Google Scholar]
- Short KR, Vittone JL, Bigelow ML, Proctor DN, Rizza RA, Coenen-Schimke JM, Nair KS. 2003. Impact of aerobic exercise training on age-related changes in insulin sensitivity and muscle oxidative capacity. Diabetes 52: 1888–1896. [DOI] [PubMed] [Google Scholar]
- Sriwijitkamol A, Ivy JL, Christ-Roberts C, DeFronzo RA, Mandarino LJ, Musi N. 2006. LKB1-AMPK signaling in muscle from obese insulin-resistant Zucker rats and effects of training. Am J Physiol Endocrinol Metab 290: E925–E932. [DOI] [PubMed] [Google Scholar]
- Steinberg GR, O’Neill HM, Dzamko NL, Galic S, Naim T, Koopman R, Jorgensen SB, Honeyman J, Hewitt K, Chen ZP, et al. 2010. Whole body deletion of AMP-activated protein kinase β2 reduces muscle AMPK activity and exercise capacity. J Biol Chem 285: 37198–37209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor EB, Hurst D, Greenwood LJ, Lamb JD, Cline TD, Sudweeks SN, Winder WW. 2004. Endurance training increases LKB1 and MO25 protein but not AMP-activated protein kinase activity in skeletal muscle. Am J Physiol Endocrinol Metab 287: E1082–E1089. [DOI] [PubMed] [Google Scholar]
- Terada S, Goto M, Kato M, Kawanaka K, Shimokawa T, Tabata I. 2002. Effects of low-intensity prolonged exercise on PGC-1 mRNA expression in rat epitrochlearis muscle. Biochem Biophys Res Commun 296: 350–354. [DOI] [PubMed] [Google Scholar]
- Vainshtein A, Tryon LD, Pauly M, Hood DA. 2015. Role of PGC-1α during acute exercise-induced autophagy and mitophagy in skeletal muscle. Am J Physiol Cell Physiol 308: C710–C719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varnauskas E, Bjorntorp P, Fahlen M, Prerovsky I, Stenberg J. 1970. Effects of physical training on exercise blood flow and enzymatic activity in skeletal muscle. Cardiovasc Res 4: 418–422. [DOI] [PubMed] [Google Scholar]
- Vega RB, Huss JM, Kelly DP. 2000. The coactivator PGC-1 cooperates with peroxisome proliferator-activated receptor α in transcriptional control of nuclear genes encoding mitochondrial fatty acid oxidation enzymes. Mol Cell Biol 20: 1868–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadley GD, Lee-Young RS, Canny BJ, Wasuntarawat C, Chen ZP, Hargreaves M, Kemp BE, McConell GK. 2006. Effect of exercise intensity and hypoxia on skeletal muscle AMPK signaling and substrate metabolism in humans. Am J Physiol Endocrinol Metab 290: E694–E702. [DOI] [PubMed] [Google Scholar]
- Watt MJ, Southgate RJ, Holmes AG, Febbraio MA. 2004. Suppression of plasma free fatty acids upregulates peroxisome proliferator-activated receptor (PPAR) α and δ and PPAR coactivator 1α in human skeletal muscle, but not lipid regulatory genes. J Mol Endocrinol 33: 533–544. [DOI] [PubMed] [Google Scholar]
- White AT, Schenk S. 2012. NAD+/NADH and skeletal muscle mitochondrial adaptations to exercise. Am J Physiol Endocrinol Metab 303: E308–E321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RS, Neufer PD. 1996. Regulation of gene expression in skeletal muscle by contractile activity. In The handbook of physiology, section 12, Exercise: Regulation and integration of multiple systems (ed. Rowell LB, Shepherd JT), pp. 1124–1150. Oxford University Press, New York. [Google Scholar]
- Williams RS, Salmons S, Newsholme EA, Kaufman RE, Mellor J. 1986. Regulation of nuclear and mitochondrial gene expression by contractile activity in skeletal muscle. J Biol Chem 261: 376–380. [PubMed] [Google Scholar]
- Winder WW, Holmes BF, Rubink DS, Jensen EB, Chen M, Holloszy JO. 2000. Activation of AMP-activated protein kinase increases mitochondrial enzymes in skeletal muscle. J Appl Physiol (1985) 88: 2219–2226. [DOI] [PubMed] [Google Scholar]
- Wright DC. 2007. Mechanisms of calcium-induced mitochondrial biogenesis and GLUT4 synthesis. Appl Physiol Nutr Metab 32: 840–845. [DOI] [PubMed] [Google Scholar]
- Wright DC, Han DH, Garcia-Roves PM, Geiger PC, Jones TE, Holloszy JO. 2007. Exercise-induced mitochondrial biogenesis begins before the increase in muscle PGC-1α expression. J Biol Chem 282: 194–199. [DOI] [PubMed] [Google Scholar]
- Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, Troy A, Cinti S, Lowell B, Scarpulla RC, et al. 1999. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell 98: 115–124. [DOI] [PubMed] [Google Scholar]
- Wu H, Kanatous SB, Thurmond FA, Gallardo T, Isotani E, Bassel-Duby R, Williams RS. 2002. Regulation of mitochondrial biogenesis in skeletal muscle by CaMK. Science 296: 349–352. [DOI] [PubMed] [Google Scholar]
- Yeo WK, Paton CD, Garnham AP, Burke LM, Carey AL, Hawley JA. 2008. Skeletal muscle adaptation and performance responses to once a day versus twice every second day endurance training regimens. J Appl Physiol (1985) 105: 1462–1470. [DOI] [PubMed] [Google Scholar]
- Zierath JR, Wallberg-Henriksson H. 2015. Looking ahead perspective: Where will the future of exercise biology take us? Cell Metab 22: 25–30. [DOI] [PubMed] [Google Scholar]