Abstract
This review summarizes our current understanding of translation in prokaryotes, focusing on the mechanistic and structural aspects of each phase of translation: initiation, elongation, termination, and ribosome recycling. The assembly of the initiation complex provides multiple checkpoints for messenger RNA (mRNA) and start-site selection. Correct codon–anticodon interaction during the decoding phase of elongation results in major conformational changes of the small ribosomal subunit and shapes the reaction pathway of guanosine triphosphate (GTP) hydrolysis. The ribosome orchestrates proton transfer during peptide bond formation, but requires the help of elongation factor P (EF-P) when two or more consecutive Pro residues are to be incorporated. Understanding the choreography of transfer RNA (tRNA) and mRNA movements during translocation helps to place the available structures of translocation intermediates onto the time axis of the reaction pathway. The nascent protein begins to fold cotranslationally, in the constrained space of the polypeptide exit tunnel of the ribosome. When a stop codon is reached at the end of the coding sequence, the ribosome, assisted by termination factors, hydrolyzes the ester bond of the peptidyl-tRNA, thereby releasing the nascent protein. Following termination, the ribosome is dissociated into subunits and recycled into another round of initiation. At each step of translation, the ribosome undergoes dynamic fluctuations between different conformation states. The aim of this article is to show the link between ribosome structure, dynamics, and function.
Translation is the last step in gene expression, during which the coding sequence of mRNA is translated into the amino-acid sequence of a protein. Translation is a highly dynamic process that entails four major phases: initiation, elongation, termination, and ribosome recycling. During each phase, ribosomes form transient complexes with auxiliary translation factors that facilitate protein synthesis. In addition to the compositional dynamics of translating ribosome complexes, conformational fluctuations of the ribosome and the translation factors play an important role in promoting the directionality of the process. A major challenge is to understand how the loosely coupled motions of the translational components lead to rapid and accurate protein production. Here, we summarize recent results of biochemical, biophysical, and structural work that dissected the order of events at each step of translation in bacteria, identified dynamic components, and captured structures of individual intermediates. Understanding the dynamics of ribosome complexes during translation could ultimately reveal how macromolecular machines navigate through the available conformational space and how their dynamics translates into function.
INITIATION
During translation initiation, the ribosome recruits an mRNA and selects the start codon of the open reading frame (ORF) (for recent reviews, see Milon and Rodnina 2012; Duval et al. 2015; Gualerzi and Pon 2015; see also Merrick and Pavitt 2018). In bacteria, translation initiation occurs cotranscriptionally, with the RNA polymerase (RNAP) and the ribosome physically interacting with each other (Kohler et al. 2017). The ribosome binds to the ribosome binding site (RBS) of the mRNA as soon as it emerges from the RNAP. Inhibition of translation leads to increased RNAP pausing, suggesting that transcription and translation are kinetically coupled (Landick et al. 1985; Proshkin et al. 2010). So far, almost nothing is known about the mechanism of initiation in the transcription–translation complex, a molecular machine denoted as the expressome (Kohler et al. 2017). Similarly, very little is known about initiation on mRNAs that are engaged in polysomes (Mitarai et al. 2008; Espah Borujeni and Salis 2016), as most of the mechanistic knowledge comes from studies that used free mRNAs not attached to the RNAP or to a preceding ribosome. Further studies are needed to determine whether initiation in expressomes or polysomes follows the same mechanism as initiation by the pioneering ribosome on free mRNA.
Among the different types of mRNAs found in prokaryotes, mRNAs containing the Shine–Dalgarno (SD) sequence are particularly well studied. They usually have an extended 5′ untranslated region (5′UTR) and an SD sequence located 8–10 nt upstream of the start codon (usually AUG). During SD-led initiation, the small subunit ([SSU], 30S in bacteria) is recruited to the RBS through interactions between the SD sequence and the complementary anti-SD (aSD) sequence in 16S ribosomal RNA (rRNA). Initiation on SD-led mRNAs is promoted by initiation factors IF1, IF2, and IF3. These bacterial factors display activities that resemble those of eIF1A, eIF2, and eIF1 in eukaryotes, respectively, but there is very little sequence homology between these prokaryotic and eukaryotic initiation factors. IF2 is homologous with eukaryotic initiation factor eIF5B. IF1 enhances the activities of IF2 and IF3. IF2 is a GTPase that recruits the initiator fMet-tRNAfMet. IF3 interferes with subunit association, ensures the fidelity of fMet-tRNAfMet selection over the elongator aminoacyl-tRNAs (aa-tRNAs), and helps to discriminate against mRNAs with unfavorable translation initiation regions (TIRs) (Milon and Rodnina 2012; Duval et al. 2015; Gualerzi and Pon 2015, and references therein).
However, not all mRNAs have an SD sequence. mRNAs lacking the SD sequence exist in most bacteria and archaea (Tolstrup et al. 2000; Weiner et al. 2000; Ma et al. 2002; Chang et al. 2006). The number of SD-led genes among 162 completed prokaryotic genomes varies from ∼12% to 90%, suggesting a significant number of non-SD-led or leaderless mRNAs (Chang et al. 2006). Very little is known about initiation on non-SD-led mRNAs except that the 5′UTR is usually unfolded and the AUG start codon resides in a single-stranded mRNA region (Scharff et al. 2011). In archaea and some bacteria, internal ORFs of multicistronic mRNAs are more likely to have an SD sequence than the leading ORF; genes with an AUG start codon are more likely to have an SD sequence than those with GUG or UUG start codons (Ma et al. 2002; Chang et al. 2006).
Another group of mRNAs comprises leaderless mRNAs that lack a 5′UTR. Such mRNAs are widespread in a variety of bacteria (Zheng et al. 2011) and may play an important role in regulating the stress response (Grill et al. 2000; Vesper et al. 2011). A major determinant for leaderless initiation is the presence of an AUG start codon close to the 5′ end of the mRNA (Krishnan et al. 2010). Leaderless mRNAs bind to 70S ribosomes directly; recruitment of fMet-tRNAfMet is facilitated by IF2 and IF3 (Grill et al. 2000; Yamamoto et al. 2016). Whereas IF2 can bind in a similar way to either the 30S subunit or 70S ribosome (Goyal et al. 2015), IF3 must move from its binding site on the 30S subunit on 50S subunit joining. Binding of IF3 to 70S ribosomes promotes their dissociation into subunits. This raises the question how IF3 can promote initiation on 70S ribosomes without splitting them into subunits. Recent results suggest that after dissociating from its 30S site on 50S subunit joining, IF3 may remain bound at the noncanonical binding site on the 50S subunit, which would allow the factor to act in leaderless initiation without promoting the dissociation of the 70S ribosome into subunits (Goyal et al. 2017). After translating the first ORF of a polycistronic mRNA, the ribosome can also reinitiate downstream at a second ORF using a 70S-scanning mechanism that requires fMet-tRNAfMet and IF3 (Yamamoto et al. 2016).
Translation initiation on SD-led mRNAs in Escherichia coli proceeds through three main assembly intermediates (Fig. 1) (Milon and Rodnina 2012; Duval et al. 2015; Gualerzi and Pon 2015). The SSU, IF1, IF2, IF3, and fMet-tRNAfMet form a labile 30S preinitiation complex (30S PIC). As soon as mRNA is recruited, start codon recognition converts the 30S PIC into the stable 30S initiation complex (30S IC). Joining of the large subunit ([LSU], 50S in bacteria) triggers the dissociation of the initiation factors, the accommodation of fMet-tRNAfMet in the P site, and the formation of the mature 70S IC, which is ready for translation elongation. The assembly pathway of the 30S PIC does not follow a strict order of factor addition. The factors can bind to the SSU independently of each other. However, there is a kinetically preferred sequence of factor association in the order IF3 and IF2, then IF1, followed by the recruitment of fMet-tRNAfMet through IF2 (Fig. 1) (Milon et al. 2012). Occasionally, fMet-tRNAfMet can form an IF2•GTP/fMet-tRNAfMet complex (Tsai et al. 2012), but this complex does not constitute an obligatory delivery pathway for fMet-tRNAfMet (Milon et al. 2010). The mRNA can bind to the SSU at any time, independent of the presence of the initiation factors (Studer and Joseph 2006; Milon et al. 2012). The association rate depends on the properties of the mRNA, such as the presence of secondary structures in the RBS, as well as the mRNA concentration (Studer and Joseph 2006). Codon recognition changes the conformation of the complex (Milon et al. 2008, 2012; Simonetti et al. 2008; Julian et al. 2011), stabilizes tRNA binding and destabilizes IF3 binding (Milon et al. 2012; Qin et al. 2012; Elvekrog and Gonzalez 2013; Hussain et al. 2016). IF3 changes its position on the ribosome in response to codon recognition (Hussain et al. 2016).
Figure 1.
Kinetic model of translation initiation. (Top) Assembly of the 30S preinitiation complex (PIC) and 30S initiation complex (IC). Arrival times are calculated using experimentally measured bimolecular association rate constants and the in vivo concentrations of initiation factors in E. coli. Residence times are calculated from the measured dissociation rate constants of the individual components; mRNA binding is shown as a last step, but can occur at any step of the assembly pathway, independent of the presence of initiation factors or fMet-tRNAfMet. Recognition of the start codon signifies the transition to the 30S IC (based on data in Milon et al. 2012). (Middle) Formation and maturation of the 70S IC. After subunit joining, IF3 may remain loosely bound to a site on the large subunit (LSU) (based on data in Goyal et al. 2017). (Bottom) Checkpoints of mRNA selection. From an mRNA-centric point of view, structured mRNAs can be recruited to the platform of the small subunit (SSU), unfold, and then accommodate in the mRNA-binding channel of the SSU (based on data in Milon et al. 2008 and Milon and Rodnina 2012).
The next major step entails the LSU docking onto the 30S IC (Fig. 1). Rapid docking depends on the presence of IF1, IF3, IF2•GTP, and fMet-tRNAfMet (Antoun et al. 2006; Milon et al. 2008; Goyal et al. 2015). In addition, the rate of subunit joining is attenuated by the mRNA depending on the sequence of the RBS, for example on the strength of the SD–aSD interactions and the length of the spacer between the SD and the start codon (Milon et al. 2008). After GTP hydrolysis by IF2, fMet-tRNAfMet accommodates in the P site (Grigoriadou et al. 2007; Milon et al. 2008; Goyal et al. 2015). Displacement of IF3 from its 30S binding site and dissociation of IF1 and IF2 from the complex allows the ribosome to make intersubunit bridges and leads to formation of the mature 70S IC (Fig. 1) (Fabbretti et al. 2007; Chen et al. 2015; Goyal et al. 2015, 2017; Liu and Fredrick 2015; MacDougall and Gonzalez 2015). The irreversible steps of start-codon recognition and GTP hydrolysis promote conformational changes of the 30S subunit and induce rotation of the two subunits relative to each other (Allen et al. 2005; Myasnikov et al. 2005; Marshall et al. 2009; Julian et al. 2011; Coureux et al. 2016; Sprink et al. 2016).
A key question is which features of the mRNA determine its translational efficiency. In bacteria, the RBS spans nucleotides –20 to +15 around the translation start codon. Translational efficiency is modulated by the nature of the codon used for initiation (AUG, GUG, or UUG), the SD sequence and the spacer between the SD sequence and the start codon, the mRNA secondary structure near the start site, and A/U-rich elements in the mRNA that are recognized by the SSU protein bS1. bS1, which is the largest and most acidic ribosomal protein, is required for the binding and unfolding of structured mRNAs (Duval et al. 2013; Byrgazov et al. 2015). The relative contribution of each specific element is not clear. The available on-line tools used to estimate translational efficiency from the thermodynamic properties of the RBS yield predictions that are quite good for engineered mRNAs (Salis et al. 2009; Kosuri et al. 2013; Reeve et al. 2014; Bonde et al. 2016). However, in natural mRNAs, each element of the RBS alone appears to have limited effect and can modulate the efficiency of initiation only within a certain context.
A more holistic approach conceptualizes the initiation pathway as comprising a sequence of kinetic checkpoints (Fig. 1) (Milon and Rodnina 2012; Duval et al. 2015; Gualerzi and Pon 2015). In this view, the initiation efficiency is determined by kinetic partitioning between the forward steps on the pathway toward the mature 70S IC, and the backward or rejection steps. The structure and thermodynamic stability of the RBS affect the association (step 1) and unfolding (step 2) of the mRNA. The identity of the start codon determines the stability of the codon–anticodon complex (step 3). Finally, the overall conformation of the 30S IC, which is modulated by the sequence context of the RBS, defines the rate of LSU joining (step 4). The kinetic model can explain any variations in the translational efficiency of different mRNAs. If the rate constants of the elemental steps are known, the translational efficiency can be predicted. In the few cases where such measurements were possible, the calculated value matched well with the directly measured translational efficiency (Milon et al. 2008, 2012). However, for most mRNAs the elemental rate constants are unknown, which hinders the use of the kinetic parameters as descriptors in global bioinformatics analysis. Although the mechanism of translation initiation is generally quite different in pro- and eukaryotes, the principles of kinetic partitioning most likely play a major role in start-site selection in eukaryotes as well (see Sokabe and Fraser 2018).
ELONGATION
Elongation entails repetitive cycles of decoding, peptide bond formation, and translocation. Elongation begins as soon as the second codon of the ORF becomes accessible for reading by elongator aa-tRNAs and ends when the ribosome arrives at the stop codon. The basic mechanism of elongation is very similar in prokaryotes and eukaryotes (see Dever et al. 2018) and is facilitated by homologous translation factors (EF-Tu/eEF1A, EF-G/eEF2, EF-P/eIF5A, SelB/EFsec), with some notable additions, such as eEF3, which is found in fungi.
Decoding
During decoding, the ribosome translates the sequence of codons in an mRNA into the amino acid sequence of a protein. A codon exposed in the A site is recognized by aa-tRNAs, which in bacteria are delivered to the ribosome in a ternary complex with EF-Tu and GTP. The initial recruitment of the EF-Tu•GTP/aa-tRNA complex occurs through the interactions with the bL12 stalk of the ribosome (Kothe et al. 2004; Diaconu et al. 2005). Interaction of the aa-tRNA anticodon with the mRNA codon in the decoding site of the SSU triggers GTP hydrolysis by EF-Tu. After Pi release, EF-Tu rearranges into the guanosine diphosphate (GDP)-bound form and releases the aa-tRNA. The aa-tRNA accommodates in the A site of the LSU, while EF-Tu•GDP dissociates from the ribosome (Rodnina et al. 2017).
The structures of several key intermediates of decoding, which were initially identified by biochemical and biophysical studies, have been determined by cryoelectron microscopy (cryo-EM). Currently, a sequence of snapshots of cognate decoding is available for EF-Tu•GTP/Phe-tRNAPhe (Loveland et al. 2017) and SelB•GTP/Sec-tRNASec (Fischer et al. 2016). In contrast to EF-Tu, which is a general translation factor that directs every elongator aa-tRNA to the A site, SelB is a specialized elongation factor that is responsible for the delivery of the 21st natural proteinogenic amino acid, selenocysteine (Forchhammer et al. 1989). Whereas the two cryo-EM structures capture somewhat different intermediates on the decoding pathway, and some details appear specific for EF-Tu or SelB, the overall sequence of rearrangements is remarkably similar. Both reports (Fischer et al. 2016; Loveland et al. 2017) identify an early decoding intermediate where the ternary complex is bound to the SSU, but the anticodon does not yet base-pair with the codon (Fig. 2). These structures have an open SSU-domain conformation similar to or even more open than in ribosomes with a vacant A site (Fischer et al. 2016; Loveland et al. 2017). The 16S rRNA residues forming the local core in the SSU decoding center point away from the codon–anticodon complex, although there seem to be subtle differences in the orientation of different nucleotides between the EF-Tu and SelB structures. In both structures, the key residue A1492 is in the “flipped-out” (Ogle et al. 2001) open form oriented away from the codon–anticodon complex. A1493 is in a semi-open orientation in the ribosome–EF-Tu complex, but in an open orientation in the SelB complex; also the mobility of G530 of 26S rRNA appears to differ between the two complexes (Fischer et al. 2016; Loveland et al. 2017).
Figure 2.
Structural mechanism of decoding as visualized by cryoelectron microscopy (cryo-EM). (Top) Intermediates of cognate decoding by elongation factor (EF)-Tu•GDP–Phe-tRNAPhe. (Left) Schematic of the cognate codon–anticodon interaction between the UUC mRNA codon and the AAG anticodon of Phe-tRNAPhe. Other panels show decoding intermediates from the T state prior to codon reading, A*/T, where the codon has been recognized but EF-Tu did not move onto the sarcin-ricin loop (SRL) of the SSU and A/T state with the correct codon–anticodon interaction and EF-Tu docked on the SRL. Insets on top show the orientation of the G-domain of EF-Tu relative to the SRL. GCP, nonhydrolyzable GTP analog GDPCP. Insets at the bottom show the codon–anticodon complex and the key residues of 16S ribosomal RNA (rRNA) interacting with it. (Middle) Same as above for a near-cognate pair with a single G–U mismatch in the second position of the codon–anticodon complex. (Bottom) Intermediates of cognate decoding by SelB•GDPNP/Sec-tRNASec. GNP, nonhydrolyzable GTP analog GDPNP; IB, initial binding prior to codon reading; CR, codon reading complex in which the anticodon of the tRNA comes into the proximity of the codon, but prior to base pairing; GA, GTPase-activated complex analogous to the A/T state. (Figure was prepared using structure coordinates from Fischer et al. 2016 and Loveland et al. 2017, PDB 5UYK, 5UYL, 5UYM, 5UYN, 5UYP, 5UYQ, 5LZB, 5LZC and 5LZD.)
The second intermediate of the SelB complex captures an early decoding state in which only a single potential base pair is formed between codon and anticodon, whereas in the EF-Tu-bound complex the anticodon is fully base-paired with the codon; in both cases, the ribosome remains in an open conformation. In the EF-Tu complex, the GTP-binding pocket remains distant from the LSU (Loveland et al. 2017), and thus the GTPase activity of EF-Tu, which requires an interaction of EF-Tu with the sarcin-ricin loop (SRL) of the 23S rRNA as a GTPase-activating element, remains low (Maracci et al. 2014). In contrast to EF-Tu, SelB interacts with the LSU in both intermediates prior to codon recognition, but the contact with the SRL is blocked by Sec-tRNASec (Fischer et al. 2016). Interestingly, a similar protective interaction of aa-tRNA with the SRL was identified among decoding intermediates of eukaryotic translation, where it blocks the access of eEF1A to the GTPase-activating center (Budkevich et al. 2014).
After the codon–anticodon interaction has been established, the 16S rRNA residues at the decoding center change their orientation to the “flipped-in” (Ogle et al. 2001) conformation and close on the codon–anticodon complex (Fischer et al. 2016; Loveland et al. 2017), consistent with the local rearrangements inferred from comparisons of SSU structures with or without A-site tRNA anticodon stem-loops (ASLs) (Ogle et al. 2001). In the EF-Tu complex, G530 appears to act as a latch that fastens the codon–anticodon helix into the decoding center. The local closure of the decoding center coincides with the SSU domain closure, which drags the tRNA and EF-Tu toward the SRL. The magnitude of the conformational changes that take place on domain closure (Fischer et al. 2016) appears to be even larger than those previously reported for the SSU–ASL complex (Ogle et al. 2001). The aa-tRNA becomes distorted on codon recognition (Valle et al. 2003; Schmeing et al. 2009; Schmeing et al. 2011; Fischer et al. 2016; Loveland et al. 2017). In solution, tRNA can adopt such distorted tRNA conformations spontaneously within less than a microsecond (Fischer et al. 2016). The ribosome seems to stabilize specific subsets of conformations in a given state, depending on the interactions at the decoding center. Docking of the G-domain of EF-Tu activates GTP hydrolysis, the irreversible step that separates initial selection from the subsequent proofreading step.
GTP hydrolysis in EF-Tu and other translational GTPases relies on the universally conserved His residue (His84 in E. coli EF-Tu) in the Switch II region and on the conserved Asp residue (Asp21 in EF-Tu) in the P loop (Maracci et al. 2014). The SRL of the LSU acts as GTPase activator (Wool et al. 1992; Schmeing et al. 2009). The interaction of the SRL with His84 shifts the pKa value of His84 in such a way that the side chain becomes positively charged at neutral pH and positions the nucleophilic water molecule for the attack on the γ-phosphate of GTP (Adamczyk and Warshel 2011; Wallin et al. 2013; Aqvist and Kamerlin 2015). Computer simulations favor the reaction mechanism with an early proton transfer from water to the γ-phosphate, followed by nucleophilic attack by a hydroxide ion, a scenario that appears to be consistent with the lack of pH-dependence of GTP hydrolysis at near-neutral pH and a negligible kinetic solvent isotope effect (Maracci et al. 2014). The conserved Asp21 complexed to Mg2+ may contribute to the acceleration of GTP hydrolysis by “pushing” the negative charge toward His84 (Aqvist and Kamerlin 2015). Thus, GTP hydrolysis is primarily governed by the electrostatics of the reaction center (Adamczyk and Warshel 2011; Prasad et al. 2013; Maracci et al. 2014; Aqvist and Kamerlin 2015). Also, ribosomal protein bL12 contributes to the GTPase activation through an as-yet-undetermined mechanism (Mohr et al. 2002; Diaconu et al. 2005). The mechanism of GTP hydrolysis is likely to be conserved in all translational GTPases, such as EF-Tu, EF-G, SelB, IF2, and RF3 and their eukaryotic homologs.
During decoding, the ribosome has to select an aa-tRNA cognate to the given codon from the pool of different aa-tRNAs. The fidelity of aa-tRNA selection is high, with error frequencies of 10−3 or less (Drummond and Wilke 2009). One important unresolved question is how the ribosome responds to codon–anticodon mismatches. The early model based on the comparison of the 30S–mRNA complexes with cognate or near-cognate ASLs suggested that mismatches disturb SSU domain closure (Ogle et al. 2002). However, high-resolution crystal structures of non- and near-cognate ribosome–mRNA–tRNA complexes all showed an identical local and global arrangement of the SSU (Demeshkina et al. 2012; Rozov et al. 2015, 2016a,b). Similarly, in the cryo-EM structure, near-cognate EF-Tu•GTP/Lys-tRNA (anticodon UUU, which makes a second position mismatch to the AGA codon) induced the same local and global conformational changes as EF-Tu•GTP/Phe-tRNAPhe on the cognate UUC codon (Loveland et al. 2017). However, only a minor portion of the near-cognate ternary complexes adopts the GTPase-activated state, whereas the majority is rejected at the initial selection stage prior to GTP hydrolysis (Rodnina et al. 2017). Thus, it is possible that the available structures of the near-cognate complexes provide snapshots of translational errors, rather than of regular decoding intermediates. The visualized complexes represent those that passed the selection screens of the ribosome and will result in the incorporation of an incorrect amino acid into the protein. In contrast, the structures of the rejected majority of near-cognate complexes may be similar to that of the complexes prior to codon–anticodon pairing. Notably, such complexes must be even less stable than the pre-codon-recognition intermediates of the cognate complexes, as they appear too transient to be captured by crystallography or cryo-EM. It should be emphasized that the structures of near-cognate complexes are extremely valuable, as they show how tautomerization or the presence of tRNA modifications help the mismatched complexes to adopt the geometry that the ribosome recognizes as correct.
After GTP hydrolysis and a slightly delayed Pi release (Kothe and Rodnina 2006), EF-Tu rearranges into the GDP-bound conformation. Molecular-dynamic simulations suggested that this may propel the 3′ end of aa-tRNA toward the A site on the LSU into a position where the tRNA elbow interacts with H89 of the LSU (Noel and Whitford 2016). Aa-tRNA may fluctuate between the two conformational states until EF-Tu is released. The accommodation of the tRNA 3′ end in the A site constitutes a separate step (Geggier et al. 2010). This notion is supported by recent biochemical data (Ieong et al. 2016; Ranjan and Rodnina 2017), although the two steps are not kinetically resolved for every tRNA. The accommodation is slower for near-cognate, compared to cognate aa-tRNA, which provides a second chance to reject a tRNA with mismatches in the codon–anticodon complex in the proofreading stage (Rodnina et al. 2017).
Peptide Bond Formation
In the peptidyl transferase center of the ribosome, peptidyl-tRNA in the P site and aa-tRNA in the A site react to form a peptide bond. In comparison with the reaction between model substrates in solution, the reaction on the ribosome is accelerated about 107-fold (Sievers et al. 2004). The ribosome's active site is comprised of rRNA (Ban et al. 2000; Polikanov et al. 2014), and thus the ribosome is the largest known RNA-catalyst and the only known natural ribozyme that has polymerase activity. Unlike protein enzymes, the ribosome does not provide catalytic groups with pKa values at neutral pH (Youngman et al. 2004; Bieling et al. 2006). The catalysis is mainly entropic (Sievers et al. 2004). The ribosome facilitates the reaction by ordering water molecules, positioning of rRNA and tRNA residues, and electrostatic shielding (Sharma et al. 2005; Wallin and Aqvist 2010).
Peptide bond formation proceeds by nucleophilic attack of the amino group of aa-tRNA on the carbonyl carbon of the ester bond in peptidyl-tRNA. In solution, the reaction is expected to have two intermediates, a zwitterionic tetrahedral intermediate (T±), that is deprotonated and forms a second intermediate (T−), and then decomposes to form the reaction products (Satterthwait and Jencks 1974). A comprehensive analysis of heavy-atom kinetic isotope effects indicated that the ribosome alters the reaction pathway in such a way that the T± intermediate no longer accumulates. Proton transfer from the attacking nitrogen and formation of the tetrahedral intermediate take place during the rate-limiting step, while tetrahedral intermediate breakdown happens in a separate rapid step (Hiller et al. 2011). Analysis of kinetic solvent isotope effects showed that in the rate-limiting transition state three protons move in a concerted manner (Kuhlenkoetter et al. 2011). Notably, there are several water molecules within the peptidyl transferase center that can exchange protons (Polikanov et al. 2014). Also the 2′OH group of A76 of the P-site tRNA (Zaher et al. 2011) and the 2′OH of A2451 of 23S rRNA (Erlacher et al. 2006) contribute to proton transfer and/or stabilize the charges developing as the reaction proceeds.
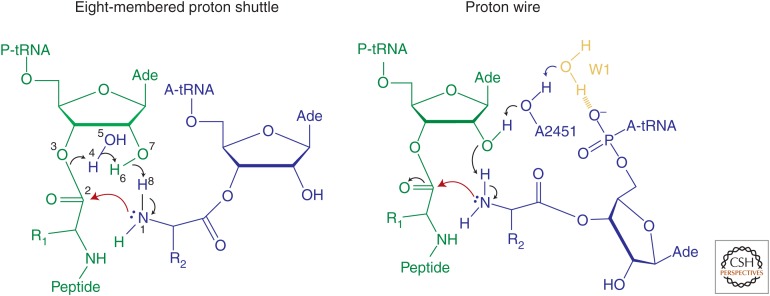
Currently there are two models that account for the movement of protons in the active site during peptide bond formation (Fig. 3). One model, referred to as the eight-membered proton shuttle (Wallin and Aqvist 2010), suggests that the attack of the α-amino group on the ester carbonyl carbon results in an eight-membered transition state in which a proton from the α-amino group is received by the 2′OH group of A76, which at the same time donates its proton to the carbonyl oxygen by way of an adjacent water molecule (Kuhlenkoetter et al. 2011). Protonation of the 3′OH is an independent rapid step. The alternative model, referred to as the “proton wire” model, suggests that one of the water molecules residing in the peptidyl transferase center is partially negatively charged by the vicinity of the deprotonated amino-terminal α-amino group of ribosomal protein bL27 and of the negatively charged 5′-phosphate oxygen of the A-site A76 (Polikanov et al. 2014). Upon formation of a tetrahedral intermediate, the emerging positive and negative charges may become separated in space and delocalized over the pockets containing water molecules. The two models account for the three protons moving in a concerted manner in the rate-limiting transition state, but disagree on the exact proton transfer pathway. One argument against the proton wire model (Polikanov et al. 2014) is that deletion of bL27 has no effect on peptide bond formation (Maracci et al. 2015). Furthermore, the role of the 2′OH of A2451 of the 23S rRNA, which plays an essential role in the proton wire model, has not been tested at conditions of rapid peptide bond formation (Erlacher et al. 2006). On the other hand, the proton shuttle model has a less optimal stereochemistry than the proton wire model (Polikanov et al. 2014).
Figure 3.
Models for proton transfer during peptide bond formation. Reaction schemes are shown for the eight-membered proton shuttle and proton wire mechanisms. P-site tRNA is shown in green, A-site tRNA in blue, and 2′OH of A2451 in black. The nucleophilic attack is depicted by red arrows. In the eight-membered proton shuttle, the attack of the α-amino group on the ester carbonyl carbon results in an eight-membered rate-limiting transition state in which a proton from the α-amino group is received by the 2′OH group of A76, which at the same time donates its proton to the carbonyl oxygen via an adjacent water molecule (Kuhlenkoetter et al. 2011). In the proton wire model, the proton from the α-amino group is received by the 2′OH group of A76, which in turn donates a proton to the 2′OH of A2451, and then to a water molecule (W1), which is partially negatively charged (Polikanov et al. 2014). Both models account for the concerted movement of three protons in the rate-limiting transition state (Kuhlenkoetter et al. 2011). (From Polikanov et al. 2014; adapted, with permission, from Nature Publishing Group under a Creative Commons license.)
The reactivities of natural amino acids in the peptidyl transferase reaction differ substantially (Wohlgemuth et al. 2008). Nevertheless, the ribosome can make peptides with most amino acid combinations without the help of any additional auxiliary factors. One notable exception is the synthesis of poly-Pro stretches with three or more consecutive prolines or of distinct XPPX sequences with two prolines flanked by specific amino acids (Hersch et al. 2013; Peil et al. 2013; Woolstenhulme et al. 2013). During synthesis of such peptides the ribosome stalls because of a low rate of peptide bond formation (e.g., for the PPP motif the ribosome is stalled after incorporation of the second Pro) (Ude et al. 2013). The stalling is alleviated by EF-P (Doerfel et al. 2013; Ude et al. 2013), a specialized translation factor that enters the E site of the ribosome and acts by entropic steering of the P- and A-site substrates toward their catalytically productive orientation in the peptidyl transferase center (Fig. 4) (Doerfel et al. 2015; Huter et al. 2017). The eukaryotic homolog of EF-P, eIF5A, also accelerates the formation of poly-Pro chains (Gutierrez et al. 2013). eIF5A appears to have additional functions in elongation and termination (Schuller et al. 2017; also see Dever et al. 2018). EF-P does not have such a broad functionality, because EF-P recognizes the tRNA in the P site and favors interactions with tRNAPro and tRNAfMet, but is considerably less active with other tRNAs (Katoh et al. 2016).
Figure 4.
Action of elongation factor P (EF-P) on ribosomes stalled at polyproline stretches. (A) Ribosomes stall during translation of proteins containing three consecutive prolines (red stars). Binding of the peptidyl-tRNA (green) to the P site is destabilized, which (B) can lead to peptidyl-tRNA drop-off. (C) The all-trans or all-cis conformations of polyprolines of the nascent chain are not possible because of a steric clash with G2061 (gray) within the tunnel wall. Peptidyl-tRNA is destabilized and prevents accommodation of the A-site tRNA (orange) and peptide bond formation. (D) Ribosomes stalled on polyproline stretches are recognized by EF-P (pink), which binds within the E-site region and stabilizes the peptidyl-tRNA. EF-P binding is facilitated via contacts with the L1 stalk and the P-site tRNA as well as E-site codon. (E) Interaction of the ε(R)-β-lysyl-hydroxylysine with the CCA-end of P-site tRNAPro stabilizes the P-site tRNA as well as the nascent chain, by forcing the prolines to adopt an alternative conformation that passes into the ribosomal exit tunnel. (F) Thus, an optimal geometry between the nascent chain and the aminoacyl-tRNA in the A site is achieved and peptide bond formation can occur. (From Huter et al. 2017; adapted, with permission, from Elsevier © 2017.)
EF-P and eIF5A are posttranslationally modified (for review see Lassak et al. 2016). The modification is essential for function (Doerfel et al. 2013; Gutierrez et al. 2013; Ude et al. 2013), but varies between different groups of bacteria and eukaryotes. In E. coli, Lys34 of EF-P is posttranslationally modified to a lysyl-lysine (Yanagisawa et al. 2010). In Pseudomonas aeruginosa and Shewanella oneidensis Arg32 (which occupies the position of E. coli Lys34) is rhamnosylated (Lassak et al. 2015), whereas EF-P from Bacillus subtilis bears a 5-aminopentanol moiety attached to Lys32 (Rajkovic et al. 2016). In eukaryotes, the conserved lysine residue of eIF5A is modified to hypusine (Cooper et al. 1983).
Translocation
After peptide bond formation, the ribosomal subunits move relative to each other, from the nonrotated (N) state with the two tRNAs bound to the P and A sites both on the SSU and LSU, to the rotated (R) state with the tRNAs bound in hybrid P/E and A/P states. At the same time, the uL1 stalk moves from an open to a closed conformation toward the P-site tRNA (for review, see Frank and Gonzalez 2010). Ribosomes in the pretranslocation state (PRE) fluctuate between N and R states. Molecular dynamics simulations suggest that ribosome rotation and the concomitant internal movement of the SSU head and body domains (referred to as head swiveling) are intrinsically very rapid and can occur in the microsecond time scale (Bock et al. 2013). However, tRNA translocation does not occur spontaneously because the interactions of the two tRNAs with the ribosome restrict the movement. In the posttranslocation (POST) state the ribosomes are predominantly in the N state with tRNAs in their classical positions.
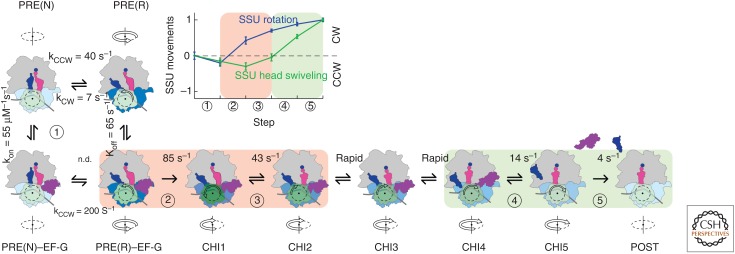
Translocation is promoted by EF-G at the cost of GTP hydrolysis. Movement of the tRNAs and the mRNA during translocation is a multistep process (Fig. 5). Current models distinguish up to eight discrete steps based on structural information as well as ensemble and single-molecule kinetic studies using a large variety of fluorescence reporters and fluorescence resonance energy transfer (FRET) pairs (Guo and Noller 2012; Adio et al. 2015; Belardinelli et al. 2016a; Sharma et al. 2016b; Wasserman et al. 2016). In addition to PRE and POST states, the ribosome can adopt several intermediate conformations that differ in the positions of the tRNAs with respect to the SSU head and body domains and the A- and P-site loops on the LSU. The tRNA positions correlate with the degree of subunit rotation and the SSU head domain swiveling. These states are referred to as chimeric (CHI) states (e.g., ap/P and pe/E [Ramrath et al. 2013]; ap/ap [Zhou et al. 2014]; or noncanonical states identified by smFRET [Chen et al. 2011; Adio et al. 2015; Wasserman et al. 2016]).
Figure 5.
Kinetic model of translocation. The rotation states of the small subunit (SSU) relative to the large subunit (LSU) (gray) are indicated by color intensity of the SSU body (light blue for nonrotated, N; dark blue for rotated, R). The swiveling motions of the SSU head are depicted by a color gradient from light green (classical nonswiveled SSU head position) to forest green (maximum degree of SSU head domain swiveling relative to the SSU body domain) (Belardinelli et al. 2016a). Peptidyl- and deacylated tRNA in the pretranslocation state (PRE) complex are shown in magenta and blue, respectively. EF-G (purple) is shown in two conformations, a compact (Lin et al. 2015) and an extended one after engagement with the ribosome (Ramrath et al. 2013; Zhou et al. 2014). The rates of transitions between PRE(N) and PRE(R) and PRE(N)–EF-G and PRE(R)–EF-G (kCW and kCCW at 37°C) are modified from data in Sharma et al. (2016b). The rate constants for the kinetically defined steps 1, 2, 3, 4, and 5 are from ensemble kinetics studies at 37°C (Belardinelli et al. 2016a). Step 1, EF-G binding. Step 2, GTP hydrolysis (Rodnina et al. 1997; Savelsbergh et al. 2003) and opening of the SSU because of the opposite movements of the SSU head and body domains (see inset) (Belardinelli et al. 2016a). Step 3, unlocking of the tRNA–mRNA complex on the SSU, which is rate-limiting for tRNA movement and Pi release from EF-G (Savelsbergh et al. 2003). The existence of rapid transitions between steps 3 and 4 was shown using stalled translocation intermediates (Savelsbergh et al. 2003; Holtkamp et al. 2014). Translocation intermediates (CHI1 to CHI4) are adopted from smFRET data (Adio et al. 2015). The posttranslocation (POST) state may entail further conformational substates (Wasserman et al. 2016). The light red background indicates complexes undergoing unlocking; the light green background shows complexes that move toward relocking. (Inset) Distinct timing of counterclockwise (CCW) and clockwise (CW) movements of the SSU body relative to LSU (blue symbols) and of the SSU head (green symbols) (Belardinelli et al. 2016a). (From Belardinelli et al. 2016b; with permission from Taylor & Francis and Creative Commons Public Domain licensing.)
After EF-G binding to the ribosome, the SSU head and body domains move in the same counterclockwise (CCW) direction relative to the LSU, which is referred to as forward, because it corresponds with the direction of translocation (Fig. 5) (Guo and Noller 2012; Belardinelli et al. 2016b; Wasserman et al. 2016). EF-G hydrolyzes GTP, but retains the Pi (Savelsbergh et al. 2003, 2005). Then, the SSU body begins moving backward in the clockwise (CW) direction, whereas the SSU head remains in the forward-swiveled state (Guo and Noller 2012; Belardinelli et al. 2016b; Wasserman et al. 2016). This may open the decoding region sufficiently to uncouple the tRNAs from the interactions with the ribosome elements that hold the mRNA and the tRNA anticodons in the A and P site, respectively. This explains how CHI states are formed: while the tRNA positions on the SSU head domain are retained, the SSU body moves, leading to the displacement of the tRNAs to the CHI states.
After the unlocking of the codon–anticodon complexes from the SSU, the SSU head domain starts to move backward (Guo and Noller 2012; Belardinelli et al. 2016b; Wasserman et al. 2016). The tRNAs adopt their canonical POST positions in P and E sites and EF-G releases Pi (Savelsbergh et al. 2003). Next, the E-site tRNA moves further away from the P-site tRNA, which is accompanied by the loss of the E-site codon–anticodon interaction, while the SSU head moves further backward (Adio et al. 2015; Belardinelli et al. 2016a). Finally, the dissociation of E-site tRNA and EF-G restores the N state with a classical P/P position of the peptidyl-tRNA. The role of EF-G is to accelerate and stabilize the R state of the ribosome, to unlock the tRNA–mRNA complex from the SSU, and to ensure the forward movement of the tRNAs by inserting EF-G domain 4 into the A site on the SSU. In other words, EF-G acts as a ratchet to rectify the Brownian motions of the ribosome into directed movement and promotes a key conformational rearrangement of the SSU that disrupts its interactions with the tRNAs, thereby allowing for rapid tRNA translocation (for review see Belardinelli et al. 2016b).
Translocation and Recoding
While moving along the mRNA the ribosome may encounter structures such as stem-loops or pseudoknots. In most cases, the ribosome helicase composed of ribosomal proteins uS3, uS4, and uS5 ensures mRNA unwinding by a combination of active and passive helicase mechanisms (Takyar et al. 2005; Qu et al. 2011). However, mRNA secondary structure elements in combination with slippery sequences can lead to –1 frameshifting. Slippery sites are mRNA sequences where codons in the 0- and –1-frame code for the same tRNAs. Ensemble kinetics and single-molecule studies indicate that –1 frameshifting on the mRNA coding for the infectious bronchitis virus (IBV) protein 1a/1b (which works in vivo in both eukaryotic and bacterial systems [Napthine et al. 2003]) or E. coli dnaX occurs during translocation of two tRNAs attached to the slippery site codons (Caliskan et al. 2014; Chen et al. 2014; Kim et al. 2014; Yan et al. 2015). Recoding in eukaryotes is described in Dever et al. (2018). The secondary structure element disturbs the ribosome dynamics during translocation. While early steps of translocation proceed with unperturbed efficiency, the backward swiveling of the SSU head domain is impeded by the structure obstacle in the mRNA (Caliskan et al. 2014; Kim et al. 2014). Ribosomes that are stalled in an unlocked state are apparently less stringent in maintaining the reading frame and can slip in the –1 direction.
Cotranslational Protein Folding
During elongation, the nascent peptide moves through the polypeptide exit tunnel of the ribosome, which spans from the peptidyl transferase center to the cytoplasmic surface of the LSU (Fig. 6). The exit tunnel is about 100 Å long and can accommodate 30 or more amino acids, depending on the structure of the peptide, before the nascent chain emerges from the ribosome. Emerging nascent proteins can start folding within the tunnel, which can accommodate secondary structure elements or even small domains (for reviews see Balchin et al. 2016; Rodnina 2016; Thommen et al. 2017). While traveling through the exit tunnel, the peptide can adopt a nonnative compact structure, which is rearranged to a native form when the whole domain emerges from the exit port (Holtkamp et al. 2015). The ribosome affects protein folding by imposing a vectorial folding pathway and by coupling folding to the pace of translation. Initial folding events are restricted by the confined space within the exit tunnel and are further modulated by interactions between the peptide emerging from the exit tunnel and the surface of the LSU. The relationship between the rate of translation and the direction of protein folding is quite complex. The presence of rare codons in the mRNA slows down translation, alters the kinetics of cotranslational folding, and changes the distribution of protein conformations in the resulting mature protein pool (Clarke and Clark 2008; Tsai et al. 2008; Zhang et al. 2009; Siller et al. 2010; Spencer et al. 2012; Yu et al. 2015; Buhr et al. 2016; Sharma et al. 2016a). Computer modeling suggests that changes of the translation rate can coordinate local folding rates and induce or prevent misfolding (O’Brien et al. 2014; Trovato and O’Brien 2017). Cotranslational protein folding is also affected by the proteins that bind in the vicinity of the exit port, such as the chaperone trigger factor (Gloge et al. 2014; Balchin et al. 2016).
Figure 6.
Cotranslational protein folding. Callouts summarize the potential effects at each step (Rodnina 2016). U, unfolded state; C, compact transient state or folding intermediate; N, native fold.
TERMINATION
Termination occurs when the ribosome encounters a stop codon in the mRNA. In bacteria, stop codons are recognized by the termination (or release) factors RF1 and RF2, which read the codons UAG/UAA and UGA/UAA, respectively. Another termination factor, RF3, facilitates turnover of RF1 and RF2 but is not required for peptidyl-tRNA hydrolysis. Crystal and cryo-EM structures as well as smFRET studies show that RF1 and RF2 stabilize the ribosome in the N state (Rawat et al. 2003, 2006; Korostelev et al. 2008, 2010; Weixlbaumer et al. 2008), whereas RF3 alone stabilizes the R state (Gao et al. 2007; Jin et al. 2011). When RF3 binds to the RF1–ribosome complex, RF1 changes its position and RF3 adopts a different conformation than in complexes with just one factor bound (Pallesen et al. 2013).
The mechanism of termination entails three steps: recognition of the stop codon, hydrolysis of the ester bond of the peptidyl-tRNA (these two steps are accomplished by RF1 or RF2) and dissociation of RF1/RF2 with the help of RF3. RF1 and RF2 select the respective stop codons by conserved recognition motifs, PVT in RF1 or SPF in RF2. Crystal structures identified the principal elements involved in the recognition (Korostelev et al. 2008, 2010; Laurberg et al. 2008; Weixlbaumer et al. 2008; Santos et al. 2013). The uracil in the first position of all three stop codons is recognized by the amino terminus of helix α5 of either RF1 or RF2. In RF1, interactions of the Thr residue in the PVT motif restrict reading to an A at the second position of the codon. In contrast, in RF2 the Ser residue in the SPV motif can bind to both A and G. The ability of RF1 to read both A and G in the third codon position is explained by a rotation of the side-chain amide of a Gln residue that is conserved in RF1, but not in RF2. RF2 has a hydrophobic Val residue at the homologous position and is restricted to an A in the third codon position.
Peptidyl-tRNA hydrolysis is catalyzed by the peptidyl transferase center of the ribosome with the help of the GGQ motif that is conserved in RF1 and RF2. The Gln residue of the GGQ motif is methylated; the modification increases the rate of peptide release, in particular for RF2. The reaction is expected to move through a tetrahedral intermediate, which breaks down and forms free peptide and deacylated tRNA (Jin et al. 2010). Proton inventories and computer simulations indicate that the transition state of the RF2-dependent hydrolysis reaction involves only one proton in flight (Trobro and Aqvist 2009; Kuhlenkoetter et al. 2011). Mutational analysis, pH/rate dependencies, and the lack of a D2O effect are consistent with two possible reaction mechanisms (Kuhlenkoetter et al. 2011; Indrisiunaite et al. 2015). The attacking water molecule could donate a proton to a hydroxide ion that facilitates both proton transfer and nucleophilic attack. Alternatively, a hydroxide ion could act as an attacking group in which the proton in flight could be transferred to the 2′OH directly or through another water molecule. The 2′OH of the P-site substrate is vital for orienting the nucleophile in a hydrogen-bonding network productive for catalysis (for review, see Rodnina 2013).
RF3 is required to release RF1/RF2 from the ribosome, but the mechanism of RF3 action is controversial (Zavialov et al. 2001, 2002; Pallesen et al. 2013; Koutmou et al. 2014; Peske et al. 2014; Shi and Joseph 2016). RF3 has a higher binding affinity for GDP than for GTP and can be activated by the ribosome through accelerating the nucleotide exchange. The original model for the RF3 mechanism relied on measurements of the affinity of RF3 for GTP and GDP, which suggested that at cellular concentrations of GTP/GDP most RF3 should be in the GDP-bound form (Zavialov et al. 2001). This model suggested that RF3•GDP can bind to the ribosome in a complex with RF1 or RF2 only after the peptide is released, thereby forming an unstable encounter complex. Subsequent dissociation of GDP would lead to a stable high-affinity complex with RF3 in the nucleotide-free state. The subsequent binding of GTP to RF3 would promote RF1/RF2 dissociation. In the final step of the model, RF3 hydrolyzes GTP and dissociates from the ribosome (Zavialov et al. 2001, 2002). However, more direct measurements of the RF3 affinity for GTP and GDP did not confirm a large preference of RF3 for GDP (Koutmou et al. 2014; Peske et al. 2014). Whereas GDP binding is favored, the affinity difference to GTP is only 10-fold, which suggests that at cellular conditions, where GTP is present in large excess over GDP, RF3 adopts the GTP-bound form. Furthermore, RF3•GTP binding is independent of peptide release, because a catalytically inactive RF2 mutant activates nucleotide exchange in RF3 (Peske et al. 2014). Because GTP binding to RF3 in the ribosome–RF2–RF3 complex is rapid, the lifetime of the putative apo-RF3 state (5 ms at cellular concentrations of RF3 [Peske et al. 2014]) is too short to have physiological significance. Peptide release results in the stabilization of the RF3•GTP–ribosome complex, thereby promoting the dissociation of RF1/2, followed by GTP hydrolysis and dissociation of RF3•GDP from the ribosome (Peske et al. 2014). Further work will be required to clarify the differences and come to a unifying model of translation termination.
Interestingly, the mechanism of translation termination appears different in prokaryotes and eukaryotes, where only two factors, eRF1 and eRF3, are responsible for termination on all three codons. In eukaryotes, eRF1 and eRF3 form a stable complex, which is recruited to the stop codon. Peptide release requires GTP hydrolysis by eRF3. Further steps of termination and ribosome recycling in eukaryotes require factors that do not exist in prokaryotes, such as ABCE1 (Rli1) (for reviews, see Dever and Green 2012; Jackson et al. 2012; see also Hellen 2018). The functional significance of these differences between prokaryotes and eukaryotes remains unclear.
RIBOSOME RECYCLING
After termination, the ribosomes still contain mRNA and tRNA, which have to be released to allow for the reuse of ribosomal subunits in the next round of initiation. In bacteria, subunit splitting is catalyzed by the ribosome recycling factor (RRF) and EF-G. RRF binds to the A site of the ribosome (Gao et al. 2005) and stabilizes a fully rotated state of the ribosome, with the P-site tRNA in the hybrid P/E binding state (Dunkle et al. 2011). GTP hydrolysis by EF-G promotes the push of RRF against the key intersubunit bridge, thereby promoting subunit splitting (Gao et al. 2005). An earlier suggestion that EF-G facilitates a translocation-like movement of RRF that acts as a tRNA mimic has been refuted by biochemical (Peske et al. 2005) and structural studies (see Fu et al. 2016 for references).
Efficient ribosome recycling occurs when RRF binds to the post-termination complex before EF-G is recruited (Borg et al. 2016). In principle, RRF and EF-G can bind independently of each other, but binding of EF-G•GTP in the absence of RRF leads to futile GTP hydrolysis without ribosome splitting (Seo et al. 2004; Savelsbergh et al. 2009; Borg et al. 2016). The sequence of subsequent events is currently a matter of debate. One model suggests that GTP hydrolysis by EF-G and the subsequent delayed Pi release result in the ribosome splitting into subunits. The resulting SSU still carries the mRNA and tRNA. Dissociation of tRNA is promoted by IF3, while mRNA is exchanged spontaneously. This sequence of events is supported by extensive kinetic experiments from different groups (Seo et al. 2004; Peske et al. 2005; Savelsbergh et al. 2009; Borg et al. 2016), smFRET study (Prabhakar et al. 2017), and time-resolved cryo-EM (Fu et al. 2016). However, in all these experiments, model mRNAs were used that contained an SD sequence that could stabilize the mRNA binding. An alternative pathway was recently suggested for mRNAs that do not contain an SD sequence in the vicinity of the stop codon (which is a normal case after termination) (Chen et al. 2017). In the latter case, GTP hydrolysis by EF-G was suggested to promote mRNA release, which would be a novel, unexpected function for EF-G. This is followed by tRNA dissociation and finally subunit splitting (Chen et al. 2017). Future experiments will show which order of events is correct or whether multiple different dissociation pathways are possible.
CONCLUSIONS AND PERSPECTIVES
The most important result of the last decade of studies on prokaryotic translation mechanisms is the view of the ribosome as a dynamic molecular machine, which changes its conformations at each step of translation. The movements of ribosome elements, including subunit rotations, flexing of SSU domains, or large movements of the S1 and L12 stalks, are inherently rapid and are gated by its ligands, such as tRNAs and translation factors. We can now describe the choreography of many steps of translation using a combination of ensemble kinetics and single-molecule techniques. Once the order of events is established, crystallography and cryo-EM can provide structures of intermediates. In the future, it would be exciting to obtain structural studies in a time-resolved fashion without the use of antibiotics or nonhydrolyzable GTP analogs that are now used to block intermediates (e.g., by performing time-resolved cryo-EM or XFEL [X-ray free-electron laser] crystallography). An improvement of the time resolution of single-molecule techniques may yield additional, yet uncharacterized transient intermediates. Tracking translation in living cells is another emerging direction. Thus, it is likely that soon we will have a near-complete 3D picture of prokaryotic translation resolved in time and space and possibly in living cells. The major challenge for years to come is to understand how eukaryotic ribosomes work. Whereas, in principle, the same experimental approaches can be used to study translation in bacteria and eukaryotes, the technical hurdles of reconstructing functional pathways for each step of translation are extremely high and amplified by a larger number of accessory proteins, extensive heterogeneity of components and complexes, and probably a larger degree of conformational mobility of translational components in eukaryotes. Solving these technical challenges and dissecting the detailed structural and functional mechanisms of the eukaryotic ribosome will constitute a milestone toward understanding translation and its regulation in health and disease.
ACKNOWLEDGMENTS
I apologize to those authors whose work was not cited because of space limitations. I am indebted to Cristina Maracci for the help with figure preparation. I thank Akanksha Goyal, Cristina Maracci, and Wolfgang Wintermeyer for critical reading of the manuscript. The work in my laboratory is supported by the Max Planck Society and Grants of the Deutsche Forschungsgemeinschaft (SFB 860 and FOR 1805).
Footnotes
Editors: Michael B. Mathews, Nahum Sonenberg, and John W.B. Hershey
Additional Perspectives on Translation Mechanisms and Control available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Adamczyk AJ, Warshel A. 2011. Converting structural information into an allosteric-energy-based picture for elongation factor Tu activation by the ribosome. Proc Natl Acad Sci 108: 9827–9832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adio S, Senyushkina T, Peske F, Fischer N, Wintermeyer W, Rodnina MV. 2015. Fluctuations between multiple EF-G-induced chimeric tRNA states during translocation on the ribosome. Nat Commun 6: 7442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen GS, Zavialov A, Gursky R, Ehrenberg M, Frank J. 2005. The cryo-EM structure of a translation initiation complex from Escherichia coli. Cell 121: 703–712. [DOI] [PubMed] [Google Scholar]
- Antoun A, Pavlov MY, Lovmar M, Ehrenberg M. 2006. How initiation factors tune the rate of initiation of protein synthesis in bacteria. EMBO J 25: 2539–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aqvist J, Kamerlin SC. 2015. The conformation of a catalytic loop is central to GTPase activity on the ribosome. Biochemistry 54: 546–556. [DOI] [PubMed] [Google Scholar]
- Balchin D, Hayer-Hartl M, Hartl FU. 2016. In vivo aspects of protein folding and quality control. Science 353: aac4354. [DOI] [PubMed] [Google Scholar]
- Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289: 905–920. [DOI] [PubMed] [Google Scholar]
- Belardinelli R, Sharma H, Caliskan N, Cunha CE, Peske F, Wintermeyer W, Rodnina MV. 2016a. Choreography of molecular movements during ribosome progression along mRNA. Nat Struct Mol Biol 23: 342–348. [DOI] [PubMed] [Google Scholar]
- Belardinelli R, Sharma H, Peske F, Wintermeyer W, Rodnina MV. 2016b. Translocation as continuous movement through the ribosome. RNA Biol 13: 1197–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieling P, Beringer M, Adio S, Rodnina MV. 2006. Peptide bond formation does not involve acid-base catalysis by ribosomal residues. Nat Struct Mol Biol 13: 423–428. [DOI] [PubMed] [Google Scholar]
- Bock LV, Blau C, Schroder GF, Davydov II, Fischer N, Stark H, Rodnina MV, Vaiana AC, Grubmuller H. 2013. Energy barriers and driving forces in tRNA translocation through the ribosome. Nat Struct Mol Biol 20: 1390–1396. [DOI] [PubMed] [Google Scholar]
- Bonde MT, Pedersen M, Klausen MS, Jensen SI, Wulff T, Harrison S, Nielsen AT, Herrgard MJ, Sommer MO. 2016. Predictable tuning of protein expression in bacteria. Nat Methods 13: 233–236. [DOI] [PubMed] [Google Scholar]
- Borg A, Pavlov M, Ehrenberg M. 2016. Complete kinetic mechanism for recycling of the bacterial ribosome. RNA 22: 10–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budkevich TV, Giesebrecht J, Behrmann E, Loerke J, Ramrath DJ, Mielke T, Ismer J, Hildebrand PW, Tung CS, Nierhaus KH, et al. 2014. Regulation of the mammalian elongation cycle by subunit rolling: A eukaryotic-specific ribosome rearrangement. Cell 158: 121–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhr F, Jha S, Thommen M, Mittelstaet J, Kutz F, Schwalbe H, Rodnina MV, Komar AA. 2016. Synonymous codons direct cotranslational folding toward different protein conformations. Mol Cell 61: 341–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrgazov K, Grishkovskaya I, Arenz S, Coudevylle N, Temmel H, Wilson DN, Djinovic-Carugo K, Moll I. 2015. Structural basis for the interaction of protein S1 with the Escherichia coli ribosome. Nucleic Acids Res 43: 661–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caliskan N, Katunin VI, Belardinelli R, Peske F, Rodnina MV. 2014. Programmed -1 frameshifting by kinetic partitioning during impeded translocation. Cell 157: 1619–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang B, Halgamuge S, Tang SL. 2006. Analysis of SD sequences in completed microbial genomes: Non-SD-led genes are as common as SD-led genes. Gene 373: 90–99. [DOI] [PubMed] [Google Scholar]
- Chen C, Stevens B, Kaur J, Cabral D, Liu H, Wang Y, Zhang H, Rosenblum G, Smilansky Z, Goldman YE, et al. 2011. Single-molecule fluorescence measurements of ribosomal translocation dynamics. Mol Cell 42: 367–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Petrov A, Johansson M, Tsai A, O’Leary SE, Puglisi JD. 2014. Dynamic pathways of −1 translational frameshifting. Nature 512: 328–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Kaledhonkar S, Sun M, Shen B, Lu Z, Barnard D, Lu TM, Gonzalez RL Jr, Frank J. 2015. Structural dynamics of ribosome subunit association studied by mixing-spraying time-resolved cryogenic electron microscopy. Structure 23: 1097–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Kaji A, Kaji H, Cooperman BS. 2017. The kinetic mechanism of bacterial ribosome recycling. Nucleic Acids Res 45: 10168–10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke TFt, Clark PL. 2008. Rare codons cluster. PLoS ONE 3: e3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper HL, Park MH, Folk JE, Safer B, Braverman R. 1983. Identification of the hypusine-containing protein hy+ as translation initiation factor eIF-4D. Proc Natl Acad Sci 80: 1854–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coureux PD, Lazennec-Schurdevin C, Monestier A, Larquet E, Cladiere L, Klaholz BP, Schmitt E, Mechulam Y. 2016. Cryo-EM study of start codon selection during archaeal translation initiation. Nat Commun 7: 13366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demeshkina N, Jenner L, Westhof E, Yusupov M, Yusupova G. 2012. A new understanding of the decoding principle on the ribosome. Nature 484: 256–259. [DOI] [PubMed] [Google Scholar]
- Dever TE, Green R. 2012. The elongation, termination, and recycling phases of translation in eukaryotes. Cold Spring Harb Perspect Biol 4: a013706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Dever TE, Dinman JD, Green R. 2018. Translation elongation and recoding in eukaryotes. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaconu M, Kothe U, Schlunzen F, Fischer N, Harms JM, Tonevitsky AG, Stark H, Rodnina MV, Wahl MC. 2005. Structural basis for the function of the ribosomal L7/12 stalk in factor binding and GTPase activation. Cell 121: 991–1004. [DOI] [PubMed] [Google Scholar]
- Doerfel LK, Wohlgemuth I, Kothe C, Peske F, Urlaub H, Rodnina MV. 2013. EF-P is essential for rapid synthesis of proteins containing consecutive proline residues. Science 339: 85–88. [DOI] [PubMed] [Google Scholar]
- Doerfel LK, Wohlgemuth I, Kubyshkin V, Starosta AL, Wilson DN, Budisa N, Rodnina MV. 2015. Entropic contribution of elongation factor P to proline positioning at the catalytic center of the ribosome. J Am Chem Soc 137: 12997–13006. [DOI] [PubMed] [Google Scholar]
- Drummond DA, Wilke CO. 2009. The evolutionary consequences of erroneous protein synthesis. Nat Rev Genet 10: 715–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunkle JA, Wang L, Feldman MB, Pulk A, Chen VB, Kapral GJ, Noeske J, Richardson JS, Blanchard SC, Cate JH. 2011. Structures of the bacterial ribosome in classical and hybrid states of tRNA binding. Science 332: 981–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duval M, Korepanov A, Fuchsbauer O, Fechter P, Haller A, Fabbretti A, Choulier L, Micura R, Klaholz BP, Romby P, et al. 2013. Escherichia coli ribosomal protein S1 unfolds structured mRNAs onto the ribosome for active translation initiation. PLoS Biol 11: e1001731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duval M, Simonetti A, Caldelari I, Marzi S. 2015. Multiple ways to regulate translation initiation in bacteria: Mechanisms, regulatory circuits, dynamics. Biochimie 114: 18–29. [DOI] [PubMed] [Google Scholar]
- Elvekrog MM, Gonzalez RL Jr. 2013. Conformational selection of translation initiation factor 3 signals proper substrate selection. Nat Struct Mol Biol 20: 628–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlacher MD, Lang K, Wotzel B, Rieder R, Micura R, Polacek N. 2006. Efficient ribosomal peptidyl transfer critically relies on the presence of the ribose 2′-OH at A2451 of 23S rRNA. J Am Chem Soc 128: 4453–4459. [DOI] [PubMed] [Google Scholar]
- Espah Borujeni A, Salis HM. 2016. Translation initiation is controlled by RNA folding kinetics via a ribosome drafting mechanism. J Am Chem Soc 138: 7016–7023. [DOI] [PubMed] [Google Scholar]
- Fabbretti A, Pon CL, Hennelly SP, Hill WE, Lodmell JS, Gualerzi CO. 2007. The real-time path of translation factor IF3 onto and off the ribosome. Mol Cell 25: 285–296. [DOI] [PubMed] [Google Scholar]
- Fischer N, Neumann P, Bock LV, Maracci C, Wang Z, Paleskava A, Konevega AL, Schroder GF, Grubmuller H, Ficner R, et al. 2016. The pathway to GTPase activation of elongation factor SelB on the ribosome. Nature 540: 80–85. [DOI] [PubMed] [Google Scholar]
- Forchhammer K, Leinfelder W, Bock A. 1989. Identification of a novel translation factor necessary for the incorporation of selenocysteine into protein. Nature 342: 453–456. [DOI] [PubMed] [Google Scholar]
- Frank J, Gonzalez RL Jr. 2010. Structure and dynamics of a processive Brownian motor: The translating ribosome. Annu Rev Biochem 79: 381–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z, Kaledhonkar S, Borg A, Sun M, Chen B, Grassucci RA, Ehrenberg M, Frank J. 2016. Key intermediates in ribosome recycling visualized by time-resolved cryoelectron microscopy. Structure 24: 2092–2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao N, Zavialov AV, Li W, Sengupta J, Valle M, Gursky RP, Ehrenberg M, Frank J. 2005. Mechanism for the disassembly of the posttermination complex inferred from cryo-EM studies. Mol Cell 18: 663–674. [DOI] [PubMed] [Google Scholar]
- Gao H, Zhou Z, Rawat U, Huang C, Bouakaz L, Wang C, Cheng Z, Liu Y, Zavialov A, Gursky R, et al. 2007. RF3 induces ribosomal conformational changes responsible for dissociation of class I release factors. Cell 129: 929–941. [DOI] [PubMed] [Google Scholar]
- Geggier P, Dave R, Feldman MB, Terry DS, Altman RB, Munro JB, Blanchard SC. 2010. Conformational sampling of aminoacyl-tRNA during selection on the bacterial ribosome. J Mol Biol 399: 576–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloge F, Becker AH, Kramer G, Bukau B. 2014. Co-translational mechanisms of protein maturation. Curr Opin Struct Biol 24: 24–33. [DOI] [PubMed] [Google Scholar]
- Goyal A, Belardinelli R, Maracci C, Milon P, Rodnina MV. 2015. Directional transition from initiation to elongation in bacterial translation. Nucleic Acids Res 43: 10700–10712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal A, Belardinelli R, Rodnina MV. 2017. Non-canonical binding site for bacterial initiation factor 3 on the large ribosomal subunit. Cell Rep 20: 3113–3122. [DOI] [PubMed] [Google Scholar]
- Grigoriadou C, Marzi S, Kirillov S, Gualerzi CO, Cooperman BS. 2007. A quantitative kinetic scheme for 70 S translation initiation complex formation. J Mol Biol 373: 562–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grill S, Gualerzi CO, Londei P, Blasi U. 2000. Selective stimulation of translation of leaderless mRNA by initiation factor 2: Evolutionary implications for translation. EMBO J 19: 4101–4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualerzi CO, Pon CL. 2015. Initiation of mRNA translation in bacteria: Structural and dynamic aspects. Cell Mol Life Sci 72: 4341–4367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Noller HF. 2012. Rotation of the head of the 30S ribosomal subunit during mRNA translocation. Proc Natl Acad Sci 109: 20391–20394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez E, Shin BS, Woolstenhulme CJ, Kim JR, Saini P, Buskirk AR, Dever TE. 2013. eIF5A promotes translation of polyproline motifs. Mol Cell 51: 35–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Hellen CUT. 2018. Translation termination and ribosome recycling in eukaryotes. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hersch SJ, Wang M, Zou SB, Moon KM, Foster LJ, Ibba M, Navarre WW. 2013. Divergent protein motifs direct elongation factor P-mediated translational regulation in Salmonella enterica and Escherichia coli. MBio 4: e00180-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller DA, Singh V, Zhong M, Strobel SA. 2011. A two-step chemical mechanism for ribosome-catalysed peptide bond formation. Nature 476: 236–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtkamp W, Cunha CE, Peske F, Konevega AL, Wintermeyer W, Rodnina MV. 2014. GTP hydrolysis by EF-G synchronizes tRNA movement on small and large ribosomal subunits. EMBO J 33: 1073–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtkamp W, Kokic G, Jager M, Mittelstaet J, Komar AA, Rodnina MV. 2015. Cotranslational protein folding on the ribosome monitored in real time. Science 350: 1104–1107. [DOI] [PubMed] [Google Scholar]
- Hussain T, Llacer JL, Wimberly BT, Kieft JS, Ramakrishnan V. 2016. Large-scale movements of IF3 and tRNA during bacterial translation initiation. Cell 167: 133–144.e113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huter P, Arenz S, Bock LV, Graf M, Frister JO, Heuer A, Peil L, Starosta AL, Wohlgemuth I, Peske F, et al. 2017. Structural basis for polyproline-mediated ribosome stalling and rescue by the translation elongation factor EF-P. Mol Cell 68: 515–527.e516. [DOI] [PubMed] [Google Scholar]
- Ieong KW, Uzun U, Selmer M, Ehrenberg M. 2016. Two proofreading steps amplify the accuracy of genetic code translation. Proc Natl Acad Sci 113: 13744–13749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indrisiunaite G, Pavlov MY, Heurgue-Hamard V, Ehrenberg M. 2015. On the pH dependence of class-1 RF-dependent termination of mRNA translation. J Mol Biol 427: 1848–1860. [DOI] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CU, Pestova TV. 2012. Termination and post-termination events in eukaryotic translation. Adv Protein Chem Struct Biol 86: 45–93. [DOI] [PubMed] [Google Scholar]
- Jin H, Kelley AC, Loakes D, Ramakrishnan V. 2010. Structure of the 70S ribosome bound to release factor 2 and a substrate analog provides insights into catalysis of peptide release. Proc Natl Acad Sci 107: 8593–8598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Kelley AC, Ramakrishnan V. 2011. Crystal structure of the hybrid state of ribosome in complex with the guanosine triphosphatase release factor 3. Proc Natl Acad Sci 108: 15798–15803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julian P, Milon P, Agirrezabala X, Lasso G, Gil D, Rodnina MV, Valle M. 2011. The cryo-EM structure of a complete 30S translation initiation complex from Escherichia coli. PLoS Biol 9: e1001095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh T, Wohlgemuth I, Nagano M, Rodnina MV, Suga H. 2016. Essential structural elements in tRNA(Pro) for EF-P-mediated alleviation of translation stalling. Nat Commun 7: 11657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HK, Liu F, Fei J, Bustamante C, Gonzalez RL Jr, Tinoco I Jr. 2014. A frameshifting stimulatory stem loop destabilizes the hybrid state and impedes ribosomal translocation. Proc Natl Acad Sci 111: 5538–5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler R, Mooney RA, Mills DJ, Landick R, Cramer P. 2017. Architecture of a transcribing-translating expressome. Science 356: 194–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korostelev A, Asahara H, Lancaster L, Laurberg M, Hirschi A, Zhu J, Trakhanov S, Scott WG, Noller HF. 2008. Crystal structure of a translation termination complex formed with release factor RF2. Proc Natl Acad Sci 105: 19684–19689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korostelev A, Zhu J, Asahara H, Noller HF. 2010. Recognition of the amber UAG stop codon by release factor RF1. EMBO J 29: 2577–2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosuri S, Goodman DB, Cambray G, Mutalik VK, Gao Y, Arkin AP, Endy D, Church GM. 2013. Composability of regulatory sequences controlling transcription and translation in Escherichia coli. Proc Natl Acad Sci 110: 14024–14029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kothe U, Rodnina MV. 2006. Delayed release of inorganic phosphate from elongation factor Tu following GTP hydrolysis on the ribosome. Biochemistry 45: 12767–12774. [DOI] [PubMed] [Google Scholar]
- Kothe U, Wieden HJ, Mohr D, Rodnina MV. 2004. Interaction of helix D of elongation factor Tu with helices 4 and 5 of protein L7/12 on the ribosome. J Mol Biol 336: 1011–1021. [DOI] [PubMed] [Google Scholar]
- Koutmou KS, McDonald ME, Brunelle JL, Green R. 2014. RF3:GTP promotes rapid dissociation of the class 1 termination factor. RNA 20: 609–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan KM, Van Etten WJ III, Janssen GR. 2010. Proximity of the start codon to a leaderless mRNA’s 5′ terminus is a strong positive determinant of ribosome binding and expression in Escherichia coli. J Bacteriol 192: 6482–6485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlenkoetter S, Wintermeyer W, Rodnina MV. 2011. Different substrate-dependent transition states in the active site of the ribosome. Nature 476: 351–354. [DOI] [PubMed] [Google Scholar]
- Landick R, Carey J, Yanofsky C. 1985. Translation activates the paused transcription complex and restores transcription of the trp operon leader region. Proc Natl Acad Sci 82: 4663–4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassak J, Keilhauer EC, Furst M, Wuichet K, Godeke J, Starosta AL, Chen JM, Sogaard-Andersen L, Rohr J, Wilson DN, et al. 2015. Arginine-rhamnosylation as new strategy to activate translation elongation factor P. Nat Chem Biol 11: 266–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassak J, Wilson DN, Jung K. 2016. Stall no more at polyproline stretches with the translation elongation factors EF-P and IF-5A. Mol Microbiol 99: 219–235. [DOI] [PubMed] [Google Scholar]
- Laurberg M, Asahara H, Korostelev A, Zhu J, Trakhanov S, Noller HF. 2008. Structural basis for translation termination on the 70S ribosome. Nature 454: 852–857. [DOI] [PubMed] [Google Scholar]
- Lin J, Gagnon MG, Bulkley D, Steitz TA. 2015. Conformational changes of elongation factor G on the ribosome during tRNA translocation. Cell 160: 219–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Fredrick K. 2015. Roles of helix H69 of 23S rRNA in translation initiation. Proc Natl Acad Sci 112: 11559–11564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loveland AB, Demo G, Grigorieff N, Korostelev AA. 2017. Ensemble cryo-EM elucidates the mechanism of translation fidelity. Nature 546: 113–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Campbell A, Karlin S. 2002. Correlations between Shine–Dalgarno sequences and gene features such as predicted expression levels and operon structures. J Bacteriol 184: 5733–5745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDougall DD, Gonzalez RL Jr. 2015. Translation initiation factor 3 regulates switching between different modes of ribosomal subunit joining. J Mol Biol 427: 1801–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maracci C, Peske F, Dannies E, Pohl C, Rodnina MV. 2014. Ribosome-induced tuning of GTP hydrolysis by a translational GTPase. Proc Natl Acad Sci 111: 14418–14423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maracci C, Wohlgemuth I, Rodnina MV. 2015. Activities of the peptidyl transferase center of ribosomes lacking protein L27. RNA 21: 2047–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall RA, Aitken CE, Puglisi JD. 2009. GTP hydrolysis by IF2 guides progression of the ribosome into elongation. Mol Cell 35: 37–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Merrick WC, Pavitt GD. 2018. Protein synthesis initiation in eukaryotic cells. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a033092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milon P, Rodnina MV. 2012. Kinetic control of translation initiation in bacteria. Crit Rev Biochem Mol Biol 47: 334–348. [DOI] [PubMed] [Google Scholar]
- Milon P, Konevega AL, Gualerzi CO, Rodnina MV. 2008. Kinetic checkpoint at a late step in translation initiation. Mol Cell 30: 712–720. [DOI] [PubMed] [Google Scholar]
- Milon P, Carotti M, Konevega AL, Wintermeyer W, Rodnina MV, Gualerzi CO. 2010. The ribosome-bound initiation factor 2 recruits initiator tRNA to the 30S initiation complex. EMBO Rep 11: 312–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milon P, Maracci C, Filonava L, Gualerzi CO, Rodnina MV. 2012. Real-time assembly landscape of bacterial 30S translation initiation complex. Nat Struct Mol Biol 19: 609–615. [DOI] [PubMed] [Google Scholar]
- Mitarai N, Sneppen K, Pedersen S. 2008. Ribosome collisions and translation efficiency: Optimization by codon usage and mRNA destabilization. J Mol Biol 382: 236–245. [DOI] [PubMed] [Google Scholar]
- Mohr D, Wintermeyer W, Rodnina MV. 2002. GTPase activation of elongation factors Tu and G on the ribosome. Biochemistry 41: 12520–12528. [DOI] [PubMed] [Google Scholar]
- Myasnikov AG, Marzi S, Simonetti A, Giuliodori AM, Gualerzi CO, Yusupova G, Yusupov M, Klaholz BP. 2005. Conformational transition of initiation factor 2 from the GTP- to GDP-bound state visualized on the ribosome. Nat Struct Mol Biol 12: 1145–1149. [DOI] [PubMed] [Google Scholar]
- Napthine S, Vidakovic M, Girnary R, Namy O, Brierley I. 2003. Prokaryotic-style frameshifting in a plant translation system: Conservation of an unusual single-tRNA slippage event. EMBO J 22: 3941–3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noel JK, Whitford PC. 2016. How EF-Tu can contribute to efficient proofreading of aa-tRNA by the ribosome. Nat Commun 7: 13314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien EP, Ciryam P, Vendruscolo M, Dobson CM. 2014. Understanding the influence of codon translation rates on cotranslational protein folding. Acc Chem Res 47: 1536–1544. [DOI] [PubMed] [Google Scholar]
- Ogle JM, Brodersen DE, Clemons WM Jr, Tarry MJ, Carter AP, Ramakrishnan V. 2001. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 292: 897–902. [DOI] [PubMed] [Google Scholar]
- Ogle JM, Murphy FV, Tarry MJ, Ramakrishnan V. 2002. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell 111: 721–732. [DOI] [PubMed] [Google Scholar]
- Pallesen J, Hashem Y, Korkmaz G, Koripella RK, Huang C, Ehrenberg M, Sanyal S, Frank J. 2013. Cryo-EM visualization of the ribosome in termination complex with apo-RF3 and RF1. eLife 2: e00411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peil L, Starosta AL, Lassak J, Atkinson GC, Virumae K, Spitzer M, Tenson T, Jung K, Remme J, Wilson DN. 2013. Distinct XPPX sequence motifs induce ribosome stalling, which is rescued by the translation elongation factor EF-P. Proc Natl Acad Sci 110: 15265–15270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peske F, Rodnina MV, Wintermeyer W. 2005. Sequence of steps in ribosome recycling as defined by kinetic analysis. Mol Cell 18: 403–412. [DOI] [PubMed] [Google Scholar]
- Peske F, Kuhlenkoetter S, Rodnina MV, Wintermeyer W. 2014. Timing of GTP binding and hydrolysis by translation termination factor RF3. Nucleic Acids Res 42: 1812–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polikanov YS, Steitz TA, Innis CA. 2014. A proton wire to couple aminoacyl-tRNA accommodation and peptide-bond formation on the ribosome. Nat Struct Mol Biol 21: 787–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabhakar A, Capece MC, Petrov A, Choi J, Puglisi JD. 2017. Post-termination ribosome intermediate acts as the gateway to ribosome recycling. Cell Rep 20: 161–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad RB, Plotnikov NV, Lameira J, Warshel A. 2013. Quantitative exploration of the molecular origin of the activation of GTPase. Proc Natl Acad Sci 110: 20509–20514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proshkin S, Rahmouni AR, Mironov A, Nudler E. 2010. Cooperation between translating ribosomes and RNA polymerase in transcription elongation. Science 328: 504–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin D, Liu Q, Devaraj A, Fredrick K. 2012. Role of helix 44 of 16S rRNA in the fidelity of translation initiation. RNA 18: 485–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X, Wen JD, Lancaster L, Noller HF, Bustamante C, Tinoco I Jr. 2011. The ribosome uses two active mechanisms to unwind messenger RNA during translation. Nature 475: 118–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajkovic A, Hummels KR, Witzky A, Erickson S, Gafken PR, Whitelegge JP, Faull KF, Kearns DB, Ibba M. 2016. Translation control of swarming proficiency in Bacillus subtilis by 5-amino-pentanolylated elongation factor P. J Biol Chem 291: 10976–10985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramrath DJ, Lancaster L, Sprink T, Mielke T, Loerke J, Noller HF, Spahn CM. 2013. Visualization of two transfer RNAs trapped in transit during elongation factor G-mediated translocation. Proc Natl Acad Sci 110: 20964–20969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjan N, Rodnina MV. 2017. Thio-modification of tRNA at the wobble position as regulator of the kinetics of decoding and translocation on the ribosome. J Am Chem Soc 139: 5857–5864. [DOI] [PubMed] [Google Scholar]
- Rawat UB, Zavialov AV, Sengupta J, Valle M, Grassucci RA, Linde J, Vestergaard B, Ehrenberg M, Frank J. 2003. A cryo-electron microscopic study of ribosome-bound termination factor RF2. Nature 421: 87–90. [DOI] [PubMed] [Google Scholar]
- Rawat U, Gao H, Zavialov A, Gursky R, Ehrenberg M, Frank J. 2006. Interactions of the release factor RF1 with the ribosome as revealed by cryo-EM. J Mol Biol 357: 1144–1153. [DOI] [PubMed] [Google Scholar]
- Reeve B, Hargest T, Gilbert C, Ellis T. 2014. Predicting translation initiation rates for designing synthetic biology. Front Bioeng Biotechnol 2: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodnina MV. 2013. The ribosome as a versatile catalyst: Reactions at the peptidyl transferase center. Curr Opin Struct Biol 23: 595–602. [DOI] [PubMed] [Google Scholar]
- Rodnina MV. 2016. The ribosome in action: Tuning of translational efficiency and protein folding. Protein Sci 25: 1390–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodnina MV, Savelsbergh A, Katunin VI, Wintermeyer W. 1997. Hydrolysis of GTP by elongation factor G drives tRNA movement on the ribosome. Nature 385: 37–41. [DOI] [PubMed] [Google Scholar]
- Rodnina MV, Fischer N, Maracci C, Stark H. 2017. Ribosome dynamics during decoding. Philos Trans R Soc Lond B Biol Sci 372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozov A, Demeshkina N, Westhof E, Yusupov M, Yusupova G. 2015. Structural insights into the translational infidelity mechanism. Nat Commun 6: 7251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozov A, Demeshkina N, Khusainov I, Westhof E, Yusupov M, Yusupova G. 2016a. Novel base-pairing interactions at the tRNA wobble position crucial for accurate reading of the genetic code. Nat Commun 7: 10457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozov A, Westhof E, Yusupov M, Yusupova G. 2016b. The ribosome prohibits the G*U wobble geometry at the first position of the codon-anticodon helix. Nucleic Acids Res 44: 6434–6441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salis HM, Mirsky EA, Voigt CA. 2009. Automated design of synthetic ribosome binding sites to control protein expression. Nat Biotechnol 27: 946–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos N, Zhu J, Donohue JP, Korostelev AA, Noller HF. 2013. Crystal structure of the 70S ribosome bound with the Q253P mutant form of release factor RF2. Structure 21: 1258–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterthwait AC, Jencks WP. 1974. The mechanism of the aminolysis of acetate esters. J Am Chem Soc 96: 7018–7031. [DOI] [PubMed] [Google Scholar]
- Savelsbergh A, Katunin VI, Mohr D, Peske F, Rodnina MV, Wintermeyer W. 2003. An elongation factor G-induced ribosome rearrangement precedes tRNA-mRNA translocation. Mol Cell 11: 1517–1523. [DOI] [PubMed] [Google Scholar]
- Savelsbergh A, Mohr D, Kothe U, Wintermeyer W, Rodnina MV. 2005. Control of phosphate release from elongation factor G by ribosomal protein L7/12. EMBO J 24: 4316–4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savelsbergh A, Rodnina MV, Wintermeyer W. 2009. Distinct functions of elongation factor G in ribosome recycling and translocation. RNA 15: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharff LB, Childs L, Walther D, Bock R. 2011. Local absence of secondary structure permits translation of mRNAs that lack ribosome-binding sites. PLoS Genet 7: e1002155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeing TM, Voorhees RM, Kelley AC, Gao YG, Murphy FVt, Weir JR, Ramakrishnan V. 2009. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science 326: 688–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeing TM, Voorhees RM, Kelley AC, Ramakrishnan V. 2011. How mutations in tRNA distant from the anticodon affect the fidelity of decoding. Nat Struct Mol Biol 18: 432–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuller AP, Wu CC, Dever TE, Buskirk AR, Green R. 2017. eIF5A functions globally in translation elongation and termination. Mol Cell 66: 194–205 e195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo HS, Kiel M, Pan D, Raj VS, Kaji A, Cooperman BS. 2004. Kinetics and thermodynamics of RRF, EF-G, and thiostrepton interaction on the Escherichia coli ribosome. Biochemistry 43: 12728–12740. [DOI] [PubMed] [Google Scholar]
- Sharma PK, Xiang Y, Kato M, Warshel A. 2005. What are the roles of substrate-assisted catalysis and proximity effects in peptide bond formation by the ribosome? Biochemistry 44: 11307–11314. [DOI] [PubMed] [Google Scholar]
- Sharma AK, Bukau B, O’Brien EP. 2016a. Physical origins of codon positions that strongly influence cotranslational folding: A framework for controlling nascent-protein folding. J Am Chem Soc 138: 1180–1195. [DOI] [PubMed] [Google Scholar]
- Sharma H, Adio S, Senyushkina T, Belardinelli R, Peske F, Rodnina MV. 2016b. Kinetics of spontaneous and EF-G-accelerated rotation of ribosomal subunits. Cell Rep 16: 2187–2196. [DOI] [PubMed] [Google Scholar]
- Shi X, Joseph S. 2016. Mechanism of translation termination: RF1 dissociation follows RF3 dissociation from the ribosome. Biochemistry 55: 6344–6354. [DOI] [PubMed] [Google Scholar]
- Sievers A, Beringer M, Rodnina MV, Wolfenden R. 2004. The ribosome as an entropy trap. Proc Natl Acad Sci 101: 7897–7901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siller E, DeZwaan DC, Anderson JF, Freeman BC, Barral JM. 2010. Slowing bacterial translation speed enhances eukaryotic protein folding efficiency. J Mol Biol 396: 1310–1318. [DOI] [PubMed] [Google Scholar]
- Simonetti A, Marzi S, Myasnikov AG, Fabbretti A, Yusupov M, Gualerzi CO, Klaholz BP. 2008. Structure of the 30S translation initiation complex. Nature 455: 416–420. [DOI] [PubMed] [Google Scholar]
- *.Sokabe M, Fraser CS. 2018. Toward a kinetic understanding of eukaryotic translation. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer PS, Siller E, Anderson JF, Barral JM. 2012. Silent substitutions predictably alter translation elongation rates and protein folding efficiencies. J Mol Biol 422: 328–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprink T, Ramrath DJ, Yamamoto H, Yamamoto K, Loerke J, Ismer J, Hildebrand PW, Scheerer P, Burger J, Mielke T, et al. 2016. Structures of ribosome-bound initiation factor 2 reveal the mechanism of subunit association. Sci Adv 2: e1501502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer SM, Joseph S. 2006. Unfolding of mRNA secondary structure by the bacterial translation initiation complex. Mol Cell 22: 105–115. [DOI] [PubMed] [Google Scholar]
- Takyar S, Hickerson RP, Noller HF. 2005. mRNA helicase activity of the ribosome. Cell 120: 49–58. [DOI] [PubMed] [Google Scholar]
- Thommen M, Holtkamp W, Rodnina MV. 2017. Co-translational protein folding: Progress and methods. Curr Opin Struct Biol 42: 83–89. [DOI] [PubMed] [Google Scholar]
- Tolstrup N, Sensen CW, Garrett RA, Clausen IG. 2000. Two different and highly organized mechanisms of translation initiation in the archaeon Sulfolobus solfataricus. Extremophiles 4: 175–179. [DOI] [PubMed] [Google Scholar]
- Trobro S, Aqvist J. 2009. Mechanism of the translation termination reaction on the ribosome. Biochemistry 48: 11296–11303. [DOI] [PubMed] [Google Scholar]
- Trovato F, O’Brien EP. 2017. Fast protein translation can promote co- and posttranslational folding of misfolding-prone proteins. Biophys J 112: 1807–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CJ, Sauna ZE, Kimchi-Sarfaty C, Ambudkar SV, Gottesman MM, Nussinov R. 2008. Synonymous mutations and ribosome stalling can lead to altered folding pathways and distinct minima. J Mol Biol 383: 281–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai A, Petrov A, Marshall RA, Korlach J, Uemura S, Puglisi JD. 2012. Heterogeneous pathways and timing of factor departure during translation initiation. Nature 487: 390–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ude S, Lassak J, Starosta AL, Kraxenberger T, Wilson DN, Jung K. 2013. Translation elongation factor EF-P alleviates ribosome stalling at polyproline stretches. Science 339: 82–85. [DOI] [PubMed] [Google Scholar]
- Valle M, Zavialov A, Li W, Stagg SM, Sengupta J, Nielsen RC, Nissen P, Harvey SC, Ehrenberg M, Frank J. 2003. Incorporation of aminoacyl-tRNA into the ribosome as seen by cryo-electron microscopy. Nat Struct Biol 10: 899–906. [DOI] [PubMed] [Google Scholar]
- Vesper O, Amitai S, Belitsky M, Byrgazov K, Kaberdina AC, Engelberg-Kulka H, Moll I. 2011. Selective translation of leaderless mRNAs by specialized ribosomes generated by MazF in Escherichia coli. Cell 147: 147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallin G, Aqvist J. 2010. The transition state for peptide bond formation reveals the ribosome as a water trap. Proc Natl Acad Sci 107: 1888–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallin G, Kamerlin SC, Aqvist J. 2013. Energetics of activation of GTP hydrolysis on the ribosome. Nat Commun 4: 1733. [DOI] [PubMed] [Google Scholar]
- Wasserman MR, Alejo JL, Altman RB, Blanchard SC. 2016. Multiperspective smFRET reveals rate-determining late intermediates of ribosomal translocation. Nat Struct Mol Biol 23: 333–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner J III, Herrmann R, Browning GF. 2000. Transcription in Mycoplasma pneumoniae. Nucl Acids Res 28: 4488–4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weixlbaumer A, Jin H, Neubauer C, Voorhees RM, Petry S, Kelley AC, Ramakrishnan V. 2008. Insights into translational termination from the structure of RF2 bound to the ribosome. Science 322: 953–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlgemuth I, Brenner S, Beringer M, Rodnina MV. 2008. Modulation of the rate of peptidyl transfer on the ribosome by the nature of substrates. J Biol Chem 283: 32229–32235. [DOI] [PubMed] [Google Scholar]
- Wool IG, Gluck A, Endo Y. 1992. Ribotoxin recognition of ribosomal RNA and a proposal for the mechanism of translocation. Trends Biochem Sci 17: 266–269. [DOI] [PubMed] [Google Scholar]
- Woolstenhulme CJ, Parajuli S, Healey DW, Valverde DP, Petersen EN, Starosta AL, Guydosh NR, Johnson WE, Wilson DN, Buskirk AR. 2013. Nascent peptides that block protein synthesis in bacteria. Proc Natl Acad Sci 110: E878–E887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H, Wittek D, Gupta R, Qin B, Ueda T, Krause R, Yamamoto K, Albrecht R, Pech M, Nierhaus KH. 2016. 70S-scanning initiation is a novel and frequent initiation mode of ribosomal translation in bacteria. Proc Natl Acad Sci 113: E1180–E1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan S, Wen JD, Bustamante C, Tinoco I Jr. 2015. Ribosome excursions during mRNA translocation mediate broad branching of frameshift pathways. Cell 160: 870–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa T, Sumida T, Ishii R, Takemoto C, Yokoyama S. 2010. A paralog of lysyl-tRNA synthetase aminoacylates a conserved lysine residue in translation elongation factor P. Nat Struct Mol Biol 17: 1136–1143. [DOI] [PubMed] [Google Scholar]
- Youngman EM, Brunelle JL, Kochaniak AB, Green R. 2004. The active site of the ribosome is composed of two layers of conserved nucleotides with distinct roles in peptide bond formation and peptide release. Cell 117: 589–599. [DOI] [PubMed] [Google Scholar]
- Yu CH, Dang Y, Zhou Z, Wu C, Zhao F, Sachs MS, Liu Y. 2015. Codon usage influences the local rate of translation elongation to regulate co-translational protein folding. Mol Cell 59: 744–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaher HS, Shaw JJ, Strobel SA, Green R. 2011. The 2′-OH group of the peptidyl-tRNA stabilizes an active conformation of the ribosomal PTC. EMBO J 30: 2445–2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavialov AV, Buckingham RH, Ehrenberg M. 2001. A posttermination ribosomal complex is the guanine nucleotide exchange factor for peptide release factor RF3. Cell 107: 115–124. [DOI] [PubMed] [Google Scholar]
- Zavialov AV, Mora L, Buckingham RH, Ehrenberg M. 2002. Release of peptide promoted by the GGQ motif of class 1 release factors regulates the GTPase activity of RF3. Mol Cell 10: 789–798. [DOI] [PubMed] [Google Scholar]
- Zhang G, Hubalewska M, Ignatova Z. 2009. Transient ribosomal attenuation coordinates protein synthesis and co-translational folding. Nat Struct Mol Biol 16: 274–280. [DOI] [PubMed] [Google Scholar]
- Zheng X, Hu GQ, She ZS, Zhu H. 2011. Leaderless genes in bacteria: Clue to the evolution of translation initiation mechanisms in prokaryotes. BMC Genomics 12: 361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Lancaster L, Donohue JP, Noller HF. 2014. How the ribosome hands the A-site tRNA to the P site during EF-G-catalyzed translocation. Science 345: 1188–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]