Here, Wang et al. review available prostate cancer model systems, the prostate cancer genome atlas, cellular and functional heterogeneity in the tumor microenvironment, tumor-intrinsic and tumor-extrinsic mechanisms underlying therapeutic resistance, and technological advances focused on disease detection and management
Keywords: prostate cancer, therapy resistance, tumor microenvironment
Abstract
Despite the high long-term survival in localized prostate cancer, metastatic prostate cancer remains largely incurable even after intensive multimodal therapy. The lethality of advanced disease is driven by the lack of therapeutic regimens capable of generating durable responses in the setting of extreme tumor heterogeneity on the genetic and cell biological levels. Here, we review available prostate cancer model systems, the prostate cancer genome atlas, cellular and functional heterogeneity in the tumor microenvironment, tumor-intrinsic and tumor-extrinsic mechanisms underlying therapeutic resistance, and technological advances focused on disease detection and management. These advances, along with an improved understanding of the adaptive responses to conventional cancer therapies, anti-androgen therapy, and immunotherapy, are catalyzing development of more effective therapeutic strategies for advanced disease. In particular, knowledge of the heterotypic interactions between and coevolution of cancer and host cells in the tumor microenvironment has illuminated novel therapeutic combinations with a strong potential for more durable therapeutic responses and eventual cures for advanced disease. Improved disease management will also benefit from artificial intelligence-based expert decision support systems for proper standard of care, prognostic determinant biomarkers to minimize overtreatment of localized disease, and new standards of care accelerated by next-generation adaptive clinical trials.
The normal and neoplastic prostate
Prostate cancer is the most common noncutaneous cancer in men worldwide, with an estimated 1,600,000 cases and 366,000 deaths annually (Torre et al. 2015). Despite recent progress, prostate cancer remains a significant medical problem for the men affected, with overtreatment of inherently benign disease and inadequate therapies for metastatic prostate cancer. This review focuses on the current state of knowledge and summarizes opportunities to curb the morbidity and mortality of prostate cancer.
Prostate anatomy
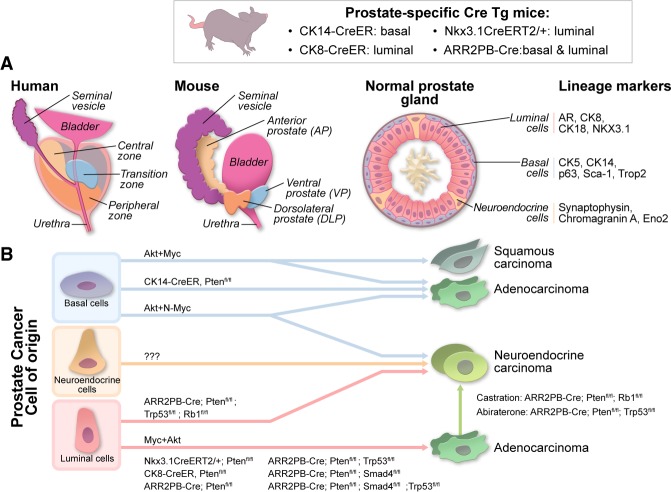
The human and mouse prostates exhibit anatomic differences as well as cellular similarities (Fig. 1A). On the basis of transcriptome profiles, the dorsolateral prostate in mice equates to the peripheral zone of the human prostate (Berquin et al. 2005), where ∼60%–75% of human prostate cancers arise (McNeal et al. 1988; Haffner et al. 2009). On the cellular level, both human and mouse prostates contain a pseudostratified epithelium with three types of terminally differentiated epithelial cells: luminal, basal, and neuroendocrine (van Leenders and Schalken 2003; Shen and Abate-Shen 2010). Although the cell of origin for prostate cancer remains an area of active investigation (Lee and Shen 2015; Strand and Goldstein 2015), luminal (Wang et al. 2009, 2013; Choi et al. 2012; Yoo et al. 2016) or basal (Lawson et al. 2007, 2010; Goldstein et al. 2010; Choi et al. 2012; Wang et al. 2013, 2014) phenotypes are observed in prostate cancer (Fig. 1B). Various model systems and techniques (e.g., flow cytometry sorting, ex vivo three-dimensional [3D] culture of prostate spheres, genetic lineage tracing, etc.) have documented the tumorigenic potential of both stem/progenitor and differentiated cells. The biological and clinical relevance of the cell of origin is not clear: One study concluded that luminal cell-derived prostate tumors are more aggressive and that a luminal cell signature carries a worse prognosis than basal cell-derived prostate cancer (Wang et al. 2013), whereas another study proposed that prostate cancers with a basal stem cell signature correlate with a more aggressive prostate cancer subtype (Smith et al. 2015). Larger prospective studies of these signatures are needed to determine their significance as prognostic biomarkers. The prostate epithelium's other cell types, such as fibroblasts, smooth muscle cells, endothelial cells, immune cells, autonomic nerve fibers, and associated ganglia, can influence the biology and clinical behavior of the prostate (see below; Barron and Rowley 2012).
Figure 1.
The normal and neoplastic prostate. (A) Comparison of human and mousse normal prostates. Anatomically, the human prostate contains three zones: (1) the peripheral zone, where ∼60%–75% of prostate cancers arise (McNeal et al. 1988; Haffner et al. 2009); the central zone; and (3) the transition zone (McNeal 1969, 1981, 1988). In contrast, the mouse prostate consists of the following distinct lobes: the anterior prostate (AP), the ventral prostate (VP), and the dorsolateral prostate (DLP) (Cunha et al. 1987). The luminal cells produce secretory proteins and are defined by expression of cytokeratin 8 (CK8) and CK18 and androgen receptor (AR). The basal cells are nestled between the basal lamina and luminal cells and express high levels of CK5 and p63 and very low levels of AR. Neuroendocrine cells, a small population of endocrine–paracrine cells located on the basal cell layer, express neuroendocrine markers such as synaptophysin and chromogranin A and do not express AR. (B) Prostate cancer cell of origin. Studies have demonstrated that both luminal cells and basal cells can serve as the cell of origin for prostate cancer; however, it remains unknown whether neuroendocrine cells can be transformed to generate prostate cancer. Overexpression of oncogenes such as constitutively active myristoylated AKT1 (myrAKT1) transforms normal human prostate epithelial cells into prostate cancer cells, which display prostate adenocarcinoma and squamous cell carcinoma phenotypes. In addition, N-Myc and myrAKT1 in normal prostate epithelial cells resulted in the formation of prostate adenocarcinoma and NEPC (neuroendocrine prostate cancer). Conditional inactivation of tumor suppressor genes Pten, Smad4, and Trp53 in both basal cells and luminal cells (ARR2PB-Cre), in basal cells (CK14-CreER), and in luminal cells (CK8-CreER) resulted the formation of prostate adenocarcinoma. Interestingly, inactivation of Pten, Rb1, and Trp53 resulted in the formation of NEPC. Castration in mice bearing Pten/Rb1-deficient prostate adenocarcinoma or abiraterone treatment of Pten/Trp53-deficient prostate adenocarcinoma resulted in the formation of NEPC.
Prostate neoplasia
Malignant transformation of the prostate follows a multistep process, initiating as prostatic intraepithelial neoplasia (PIN) followed by localized prostate cancer and then advanced prostate adenocarcinoma with local invasion, culminating in metastatic prostate cancer (Fig. 2; Shen and Abate-Shen 2010). The Gleason grading system, which was originally defined by Donald Gleason (Gleason and Mellinger 1974) based on histological patterns of prostate adenocarcinoma, has been refined over the years and is the most widely used grading system defining prostate cancer aggressiveness (Epstein et al. 2005, 2016). A central feature of prostate cancer is its hormone responsiveness, first recognized by Huggins and Hodges (1941), who reported that castration led to tumor regression in prostate cancer patients. Androgen deprivation therapy (ADT) using agents that block the androgen pathway is now the standard of care for prostate cancer. Resistance to ADT can develop, resulting in primary castration-resistant prostate cancer (CRPC) or metastatic CRPC (mCRPC). In recent years, androgen receptor (AR)-low or AR− aggressive variant prostate cancer with neuroendocrine features (NEPC) or small cell features (small cell prostate carcinoma) has increased in the clinic, which may relate to the use of potent AR antagonists. In addition, a subset of AR-independent tumors does not express markers of neuroendocrine differentiation (Bluemn et al. 2017). These variant cancers, which are completely unresponsive to ADT treatment, may emerge from clonal selection of rare pre-existing AR-low or AR− clones or the transdifferentiation of AR+ adenocarcinoma into AR-low and AR− tumors (Fig. 1B; Hu et al. 2015; Zou et al. 2017).
Figure 2.
Progression of prostate cancer and the development of mCRPC. The diagnosis of PIN is defined by luminal cell proliferation with dysplasia along the ducts. PIN in turn progresses to localized prostate adenocarcinoma, which then becomes locally invasive carcinoma as the basal cell layer is degraded and cancer cells invade through the basal lamina. Locally advanced prostate cancer metastasizes first to draining lymph nodes and then to distant organs, including the bones, liver, and lungs, with bone as the most common site of metastasis. In bone metastasis, there is a dynamic interaction between the cancer cells, osteoblasts, and osteoclasts, which results in a “vicious cycle” of bone formation and destruction—a process that supports cancer cell survival and tumor growth. AR-dependent localized advanced prostate adenocarcinoma can initially respond to ADT and then progress to CRPC. Localized advanced prostate adenocarcinoma can also display de novo resistance to ADT. Similarly, AR-dependent hormone-naïve metastatic tumors initially respond to ADT and then progress to mCRPC. AR-indifferent hormone-naïve metastatic tumors display de novo resistance. The treatment options for prostate cancer depend on tumor stage and previous treatments.
Metastatic prostate cancer
Metastatic disease is the leading cause of prostate cancer-associated deaths. Lymph nodes adjacent to the primary tumors are often the first site of metastases (Datta et al. 2010), followed by metastases to the liver, lungs, and bones (Fig. 2). Human prostate cancer bone metastases most often present as osteoblastic lesions with mixed osteolytic features, which cause severe pain, hypercalcemia, and frequent fractures.
Extensive effort has focused on understanding the biology of bone metastasis, with the goal of illuminating more effective treatment options for this lethal disease. Epithelial–mesenchymal transition (EMT) has been proposed to play a critical role in metastasis of various cancers, including prostate cancer, which has been reviewed extensively elsewhere, although its role in vivo is hotly debated (Kalluri and Weinberg 2009; Lamouille et al. 2014; Brabletz et al. 2018; Mittal 2018). Prostate cancer cells undergo EMT, disseminate into the circulation as circulating tumor cells (CTCs), and overcome several physical barriers in establishing bone metastasis, traversing sinusoid walls and bone marrow stroma and then migrating to the endosteal bone surface (Body et al. 2015) via sinusoids within the bone marrow cavity. Molecular and phenotypic characterization of CTCs, an extremely rare cell population with vast heterogeneity that may play a critical role in metastasis, has been a focus of mechanistic studies designed to understand cancer cell dissemination to distant organs (Aceto et al. 2015) and identify novel prognostic biomarkers (see “Outlook for Next-Generation Prostate Cancer Management”). Stromal cell-derived factor-1 (SDF-1 or CXCL12) and its receptor (CXCR4) have been implicated in the homing and invasion of metastatic tumor cells to the bone (Taichman et al. 2002). Correspondingly, Annexin A2 (or ANXA2), an anchor for SDF-1 that enables hematopoietic stem cells to locate and bind to the niche (Shiozawa et al. 2008), shows increased expression in prostate cancer cells, promotes recruitment into the bone marrow, and enhances proliferation and apoptosis resistance during chemotherapy (Jung et al. 2015). Moreover, αvβ3, an adhesion molecule integrin expressed in prostate cancer cells, binds the RGD peptide on extracellular matrix proteins to promote invasion into the bone endosteum (Barthel et al. 2013). Provocative recent work has shown that integrins in tumor-derived exosomes may determine organotropic metastasis (Hoshino et al. 2015). Activated RANK–RANKL signaling in prostate cancer cells is also implicated in the colonization of cancer cells in the bone (Jones et al. 2006).
Once prostate cancer cells colonize the bone marrow, interaction between cancer cells and the bone microenvironment results in a “vicious cycle” of bone formation and destruction—a process that supports cancer cell survival and tumor growth (Fig. 2). Growth factors secreted by prostate cancer cells, including endothelin 1 (ET-1), adrenomedullin, fibroblast growth factors (FGFs), platelet-derived growth factor (PDGF), and bone morphogenetic proteins (BMPs), can stimulate osteoblast activation to form new bone via paracrine signaling (Logothetis and Lin 2005; Guise et al. 2006; Body et al. 2015). In addition, tumor-secreted proteases, such as matrix metalloproteinases, prostate-specific antigen (PSA), and urokinase-type plasminogen activator, promote the release of osteoblast-inducing growth factors, including transforming growth factor β (TGF-β), insulin-like growth factors, and PDGF, to further promote osteoblast differentiation from mesenchymal stem cells. Subsequently, activated osteoblasts lead to increased RANKL concentrations and hypocalcemia as well as the release of parathyroid hormone in response to hypocalcemia, both of which induce osteoclast activation and subsequent release of factors such as TGF-β through osteoclast-mediated bone reabsorption. These host factors promote prostate cancer cell growth and survival, which in turn produce proteins such as parathyroid hormone-related protein, which drives osteoblast and stromal production of RANKL and down-regulation of osteoprotegerin, resulting in further activation of osteoclasts. The activated Wnt signaling pathway in prostate cancer cells also plays a role in promoting osteoblast differentiation (Hall et al. 2005). Prostate transmembrane protein androgen-induced-1 (Pmepa1), a gene induced by TGFβ1, was found to suppress prostate cancer metastasis to the bone by blocking TGF-β signaling via interaction with Smad2/3 and HECT E3 ubiquitin ligases (Fournier et al. 2015). Monoamine oxidase A (MAOA), a mitochondrial membrane-bound enzyme that catalyzes the degradation of biogenic and dietary monoamines by oxidative deamination, was demonstrated to play a role in the EMT process (Wu et al. 2014a) and promote bone metastasis through activation of paracrine Shh signaling in osteoblasts to induce the expression of RANKL and interleukin 6 (IL-6) (Wu et al. 2017). In summary, the growth of metastatic prostate cancer cells in the bone involves a dynamic bone remodeling process as a result of interactions between cancer cells, osteoblasts, and osteoclasts.
Model systems
Many model systems have been developed to study the genetics and biology of prostate cancer. Here we focus on novel models developed in recent years; details for established models are covered elsewhere (Shen and Abate-Shen 2010; Hensley and Kyprianou 2012; Ittmann et al. 2013; Grabowska et al. 2014). Tissue reconstitution models, originally developed to study epithelial–mesenchymal interaction in prostate organogenesis, use human or mouse prostate epithelial cells with rodent embryonic urogenital mesenchyme (UGM) or cancer-associated fibroblasts (CAFs) transplanted into immune-deficient mice (Shen and Abate-Shen 2010). Given the relative ease of genetic manipulation, this approach has been used to transform basal epithelium or immortalized human prostate epithelial cells by the overexpression of oncogenes (e.g., myristoylated AKT + ERG, myristoylated AKT + Myc, and myristoylated AKT + N-Myc), resulting in the formation of PIN, adenocarcinoma, NEPC, and squamous carcinoma (Fig. 1B). Since these tissue reconstitution models use subcutaneous or renal capsule implantation, further characterization of the tumor microenvironment (TME) in the derivative prostate tumors will be needed to determine how well they mirror the TME of human and genetically engineered mouse model (GEMM) prostate cancers (see also “Prostate Cancer Heterogeneity” and “Therapeutic Targeting of Cancer Cell-Intrinsic and TME Mechanisms”). Syngeneic mouse prostate epithelial cells and mouse embryonic UGM or CAFs in immune-competent hosts (e.g., C57BL/6 or FVB/NJ) may be one approach to better model TME biology, including the tumor-infiltrating immune cells. In classic prostate GEMMs, prostate epithelium has been engineered to express many oncogenic elements (e.g., Large T antigen, Myc, and ERG) and sustain deletion of various tumor suppressors (see “Genetic Predisposition, Genomics, and Epigenomes in Prostate Cancer” below; Ittmann et al. 2013). Some tumor suppressor genes can initiate (e.g., Nkx3.1 and Pten) and others promote (e.g., Smad4, Trp53, and Zbtb7a) progression of prostate cancer in combination with overexpression of oncogenes (e.g., Myc) or inactivation of other tumor suppressor genes (e.g., Pten) (Fig. 1B). Many of these prostate cancer GEMMs use the ARR2PB promoter to drive prostate-specific expression of Cre recombinase and transgenes encoding oncogenes (Wu et al. 2001). Other transcriptional regulatory elements from PSA, Nkx3.1, Hoxb13, and TMPRSS2 have been used to generate transgenic mice with constitutive (Hubbard et al. 2016) or ligand-dependent activation of Cre-ER recombinase—consisting of Cre fused to the estrogen receptor (ER) with mutated hormone-binding domains (PSA-Cre-ERT2, ARR2PB-Cre-ER, Probasin-MerCreMer, Nkx3.1-Cre-ERT2, and TMPRSS2-Cre-ERT2)—in the prostate by using synthetic ER ligand 4-hydroxytamoxifen (OHT) (Luchman et al. 2008; Ratnacaram et al. 2008; Birbach et al. 2009; Wang et al. 2009; Gao et al. 2016a). While these compound allelic GEMMs exhibit a full spectrum of disease evolution from PIN to invasive carcinoma with occasional metastasis (Ittmann et al. 2013), there are several limitations, including their costly and time-consuming nature and failure to recapitulate the metastatic features of human disease; that is, several models exhibit visceral metastasis to the lungs and liver, including Pten/Trp53 (Cho et al. 2014), Pten/Myc (Hubbard et al. 2016), and Pten/Trp53/Rb1 (Ku et al. 2017), and some show modest macroscopic bone metastases, including LADY/hepsin transgenic (Klezovitch et al. 2004), Pten/Trp53 telomerase-deficient (Ding et al. 2012), Hi-Myc (Magnon et al. 2013), and Pten/Trp53/Rb1 (Ku et al. 2017). Of note, metastatic tumors from LADY/hepsin-transgenic and Pten/Trp53/Rb1 models display neuroendocrine features, and those from the Pten/Trp53 telomerase-deficient model cannot be excluded from direct invasion of the spine by the primary tumors as suggested (Ittmann et al. 2013). The overall lack of highly penetrant bone metastasis GEMMs remains a major area for continued model refinement (Heyer et al. 2010) that will require a more thorough understanding of bone metastasis driver genes.
Another limitation of current modeling relates to the use of constitutively expressed prostate-specific Cre recombinase of oncogenic alleles in all Cre-expressing cells, which does not recapitulate the genesis and progression of human prostate cancer, where a few cells sustain initiating genetic aberrations followed by sequential genetic events during disease progression. The genesis issues may be addressed in part with minimal dosing of OHT to activate Cre-ER recombinase in fewer cells, as shown elsewhere (Boutin et al. 2017), or prostate injection of lentiviral-Cre with defined low multiplicity of infection (MOI) in mice harboring conditional alleles (Cho et al. 2014). Moreover, refinement of disease progression can be achieved with the combined use of Cre-LoxP and FLP-FRT systems to enable sequential activation of oncogenic alleles (Schonhuber et al. 2014). The generation of mice expressing prostate-specific codon-optimized Flippase recombinase (Flpo) and harboring FRT-flanked alleles is a key need for the development of the next generation of GEMMs. Recently, a mosaic cancer model system was developed to allow time-restricted perturbation of cell fate by combining GEMMs with LoxP alleles and FRT alleles, lentiviral expression of Flpo or Cre, and OHT-inducible Cre or Flpo recombinase (Genovese et al. 2017).
Additional technological advances are enabling the efficient generation of nongermline GEMMs. A highly efficient GEMM blastocyst injection system uses embryonic stem (ES) cells containing Probasin-Cre; conditional alleles of Pten, Trp53, and Smad4; and reporter alleles encoding mTmG and LSL-Luc (Lu et al. 2017a). The use of these ES cells provides opportunities for gene editing of additional prostate cancer-relevant alleles. Genome editing using CRISPR/Cas9 technology has allowed not only the rapid generation of germline modifications (e.g., gene deletions, point mutations, and translocations) or somatic modification of oncogenes and tumor suppressor genes in mice (Kersten et al. 2017) but also high-throughput functional screening with the CRISPR library (Dow 2015). Moreover, the mTmG allele and LST-Luc reporter allele allow for Cre-dependent green fluorescent protein (GFP) and luciferase expression in prostate epithelial cells as well as ubiquitous tdTomato expression in all other cells, which facilitates the visualization of cancer cells, stroma, and metastasis by fluorescence imaging and bioluminescence imaging. In this model, GFP+ cancer cells emerge at 3 mo of age and show dissemination to draining lymph nodes and the lungs. In addition, the use of blastocyst injection enables the simultaneous generation of many prostate cancer-prone mice, which can be enlisted into multiarm therapeutic testing (Lu et al. 2017a). Also, in vivo RNAi technology, particularly inducible shRNA expression in transgenic mice, enables time- and tissue-specific control of silencing of gene expression and affords an alternative gene inactivation approach to identify novel genes involved in tumor suppression or therapy resistance (Kersten et al. 2017).
Patient-derived xenograft (PDX) models also provide a complementary system for investigating the molecular mechanisms underlying tumor progression and therapeutic resistance, predicting clinical outcomes and informing treatment plans, and guiding drug development across many cancer types (Tentler et al. 2012; Aparicio et al. 2015), including prostate cancer (Lin et al. 2014). Unlike cancer cell lines, PDXs tend to maintain the histopathology, tumor heterogeneity, genomic aberrations, and transcriptome profiles of the original tumor. However, a recent report emphasizes that low-passage PDXs better recapitulate the original tumor features, since copy number alterations have been shown to accumulate rapidly during PDX passaging (Ben-David et al. 2017). Another limitation of PDXs is the lack of an intact immune system in the immune-deficient host into which they are typically grafted, which limits our ability to study how immune cells interact with cancer cells during tumor progression, investigate the development of therapy resistance, and test immunotherapies. The recent development of humanized mouse models, in which the mouse hematopoietic system is reconstituted with transplanted human CD34+ stem/progenitor cells, affords a significant opportunity to study the immunology of prostate cancer with these PDX models (Zitvogel et al. 2016). As PDX models require significant resources for establishment and characterization, the National Cancer Institute repository of patient-derived models (PDMs) comprised of PDXs and in vitro patient-derived cell cultures should provide researchers increased access to a diversity of human models.
Additional opportunities for disease modeling come from 3D in vitro organoid models of normal prostate epithelia or prostate cancer derived from human metastasis and CTCs (Gao et al. 2014), normal mouse and human prostate epithelia (Karthaus et al. 2014), and self-organizing stem cells from mouse CARNs (castration-resistant Nkx3.1-expressing cells) (Chua et al. 2014); these models can recapitulate in vivo the structural, functional, and genetic features of the prostate gland and the original disease (Dutta et al. 2017). Organoids, however, are limited by the lack of TME components (Clevers 2016), which may be addressed through coculture with other cell types in order to better model cancer cell–TME cross-talk in vitro. Additional methodological refinement is needed to address the facts that prostate organoids have been generated primarily from human metastatic tumors and CTCs and that the efficiency of generating organoids from luminal cells is extremely low compared with that from basal cells (Karthaus et al. 2014).
Overall, continued model refinement with new alleles and model characterization must remain a focus in the field, with the goal of recapitulating key features of the disease, particularly bone metastasis, as well as dissecting the role of TME components in tumor progression and therapy resistance (see “Cellular Heterogeneity in the TME” and “TME-Driven Mechanisms of Resistance to Conventional and Novel Cancer Therapies” below).
Genetic predisposition, genomics, and epigenomes in prostate cancer
Multiple studies, particularly epidemiological studies, twin studies, and large-scale genome-wide association studies (GWASs), have demonstrated a genetic component to the etiology of prostate cancer, which has been reviewed elsewhere (Eeles et al. 2014; Wallis and Nam 2015; Benafif and Eeles 2016; Cooney 2017; Benafif et al. 2018). Specifically, epidemiological studies have established that a family history of prostate cancer significantly increases risk (Goldgar et al. 1994; Lange 2010); twin studies have indicated that prostate cancer is among the most heritable cancers (Lichtenstein et al. 2000); GWASs have identified many prostate cancer susceptibility loci (Yeager et al. 2007; Eeles et al. 2008, 2009, 2013; Thomas et al. 2008; Gudmundsson et al. 2009; Yeager et al. 2009; Takata et al. 2010; Xu et al. 2012a; Schumacher et al. 2018), such as the risk-associated single-nucleotide polymorphism (SNP) rs339331 that increases expression of the cancer-promoting RFX6 gene through a functional interaction with the prostate cancer susceptibility gene HOXB13 (Huang et al. 2014); and genomic studies have identified familial mutations in HOXB13 (Breyer et al. 2012; Pritchard et al. 2016) and DNA repair genes such as BRCA2, ATM, CHEK2, BRCA1, RAD51D, and PALB2 (Pritchard et al. 2016). Moreover, differences in prostate cancer incidences and outcomes have been observed in men from different racial/ethnic groups, with men of African descent having the highest rates of incidence and mortality (Shenoy et al. 2016), which may partially be attributed to genetic factors (Huang et al. 2017).
Cataloging the genetic drivers of prostate cancer has been foundational to defining disease subtypes and associated therapeutic strategies. Several large-scale genomic studies in both primary prostate tumors and mCRPC have identified recurrent DNA copy number changes, mutations, rearrangements, and gene fusions (Table 1; Taylor et al. 2010; Barbieri et al. 2012; Grasso et al. 2012; Weischenfeldt et al. 2013; The Cancer Genome Atlas Research Network 2015; Beltran et al. 2016b; Fraser et al. 2017). Primary prostate tumors and mCRPC exhibit markedly increased genome-wide copy number alterations yet show only modestly increased mutations (Taylor et al. 2010; Grasso et al. 2012; Hieronymus and Sawyers 2012; The Cancer Genome Atlas Research Network 2015). Signature genetic alterations target the pathways of AR, PI3K–PTEN, WNT, and DNA repair and components of the cell cycle in nearly all metastatic prostate cancers and a high fraction of primary prostate cancers (Taylor et al. 2010; The Cancer Genome Atlas Research Network 2015; Robinson et al. 2015).
Table 1.
Common genetic aberrations in prostate cancers and their biological functions
E26 transformation-specific (ETS) fusions
The most common prostate cancer genomic alterations are translocations involving androgen-regulated promoters and the ETS family of transcription factors, such as ERG and the ETV genes (Sizemore et al. 2017). A recurrent gene fusion of the 5′ untranslated region of TMPRSS2 to ERG (TMPRSS2:ERG) was the first translocation discovered by Chinnaiyan and colleagues (Tomlins et al. 2005). TMPRSS2:ERG fusion is present in ∼50% of localized prostate cancers (Tomlins et al. 2009), and recurrent gene fusions are also found between TMPRSS2 and ETV1, ETV4, and ETV5. ETS2 deletion was found in approximately one-third of lethal mCRPCs, commonly through TMPRSS2:ERG fusions (Grasso et al. 2012). Notably, prostate-specific transgene expression of the truncated human ERG yields only minimal or weak PIN in GEMMs (Tomlins et al. 2007; Klezovitch et al. 2008), but another recent report illustrates that ERG overexpression alone can generate prostate cancer when mice are as old as 26 mo of age (Nguyen et al. 2015), which parallels the observation that ERG-driven human prostate cancers often take many years to develop. Furthermore, ERG overexpression combined with PTEN inactivation exhibits PIN with progression to prostate adenocarcinoma (Carver et al. 2009; King et al. 2009; Linn et al. 2015). Last, ERF, a member of the ETS transcription factor family found to be deleted or mutated in 1.5% of prostate cancer, acts as a transcriptional repressor that competes with ERG for binding to the ETS2 promoter (Bose et al. 2017; Huang et al. 2017), whose loss in part contributes to the aberration of ERG activation in prostate cancer.
NKX3.1
NKX3.1, a PSA-regulated homeobox gene, is frequently deleted in prostate cancer (He et al. 1997; Barbieri et al. 2012; Baca et al. 2013), and NKX3.1 haploinsufficiency is an initiating event in prostate carcinogenesis, as evidenced by multiple Nkx3.1 knockout GEMMs (Bhatia-Gaur et al. 1999; Abdulkadir et al. 2002).
MYC
Numerous studies have demonstrated an increase in MYC gene copy number in up to 50% of prostate cancer tumors (Jenkins et al. 1997; Beltran et al. 2016b; Kumar et al. 2016a) even at the PIN stage. The oncogenic role of MYC in prostate cancer has been substantiated in mice engineered to overexpress MYC in the prostate, resulting in PIN with progression to invasive adenocarcinoma (Ellwood-Yen et al. 2003). In addition, Myc functions as a driver in the metastatic Pten/Trp53-deficient RapidCaP GEMM (Nowak et al. 2015), and Myc activation in combination with Pten loss drives genomic instability and metastatic prostate cancer (Hubbard et al. 2016) in GEMMs.
Androgen pathway
AR signaling plays a central role in the development and function of the prostate. Studies using conventional approaches and next-generation sequencing have revealed that a majority of primary and metastatic prostate cancers harbors genomic alterations in the androgen signaling pathway, including AR amplification/mutations, gain of AR coactivator NCOA1/2, and loss of AR corepressor NCOR1/2 (Taplin et al. 1995; Visakorpi et al. 1995; Hodgson et al. 2005; Taylor et al. 2010), which contribute to castration resistance (discussed further below). In addition, AR genomic structural rearrangements were present in one-third of mCRPC tumors, resulting in aberrant expression of diverse AR variant species lacking the ligand-binding domain and resulting in persistent activation of AR signaling, such as AR variant 7 (AR-V7), which appears to drive disease progression (Antonarakis et al. 2014; Henzler et al. 2016). Notably, recurrent mutations in the AR collaborating factor FOXA1 have been documented in 3%–4% of both untreated localized prostate cancer and mCRPC; FOXA1 represses androgen signaling and promotes tumor growth (Zhang et al. 2011a; Barbieri et al. 2012; Grasso et al. 2012).
PI3K pathway
PTEN suppresses the PI3K–AKT–mammalian target of rapamycin (mTOR) pathway to regulate cell survival, proliferation, and energy metabolism. Loss of PTEN through deletion and mutation has an estimated frequency of 40% in prostate cancer and correlates with a greater Gleason score, poorer prognosis, and higher rate of metastasis (Pourmand et al. 2007; Taylor et al. 2010), consistent with the phenotype of Pten deletion in GEMMs (Wang et al. 2003). Deregulation of metabolic programs has been shown to impact tumor progression of Pten loss-induced prostate tumorigenesis. The metabolic transcriptional coactivator peroxisome proliferator-activated receptor γ coactivator 1α (PGC1α) was shown to induce a catabolic state and suppress prostate cancer metastasis through activation of an estrogen-related receptor α (ERRα)-dependent transcriptional program, as genetic inactivation of Pgc1a in Pten-deficient prostate tumors results in an increase in metastasis (Torrano et al. 2016). In addition, inactivation of pyruvate dehydrogenase E1α1 (Pdha1), a subunit of the pyruvate dehydrogenase complex that converts pyruvate to acetyl-CoA in the tricarboxylic acid cycle in mitochondria, was shown to significantly suppress Pten loss-driven prostate tumorigenesis through suppression of lipid biosynthesis (Bezzi et al. 2018). Finally, dietary factors have been implicated in driving metastasis—a high-fat diet activates SREBP, induces lipid accumulation, and provokes metastases in the indolent PTEN-null prostate cancer model (Chen et al. 2018). Notably, classical PI3K oncogenic aberrations found in diverse cancer types (e.g., PIK3CA mutation and AKT1/3 amplification) are altered in only a few percent of prostate cancers, limiting the application of targeted therapies in prostate cancer patients.
The TGF-β/SMAD4 pathway
Recurrent genetic alterations of key components in the TGF-β/SMAD4 pathway have been found in CRPC genomics (Grasso et al. 2012), consistent with our previous finding in GEMMs that codeletion of Pten and Smad4 generates rapidly progressive prostate cancer with metastasis to the lymph nodes and lungs (Ding et al. 2011, 2012). SMAD4 serves as a common downstream node of the TGF-β and BMP pathways and controls cell proliferation as well as TME remodeling (Ding et al. 2011; Wang et al. 2016a). Recently, in Pten-null GEMMs, loss of Tgfbr2 was found to accelerate, whereas loss of Bmpr2 impeded, tumor progress, consistent with a tumor suppressor role of Tgfbr2 (Lu et al. 2017b), indicating the antagonistic roles of the TGF-β and BMP pathways in Pten-deficient prostate cancer progression. Also, notably, telomerase reactivation in a genome-unstable mouse prostate cancer model was found to drive metastatic progression, partially by enrichment of genomic alterations of the TGF-β/SMAD4 network (Ding et al. 2012).
DNA repair pathways
Mutations in BRCA1 and BRCA2 predispose individuals to breast, ovarian, and prostate cancers (Farmer et al. 2005). Germline mutations in BRCA genes are associated with increased risk for prostate cancer or a more aggressive phenotype and worse outcomes (Pritchard et al. 2016; Barbieri et al. 2017; Sumanasuriya and De Bono 2018). Several independent genomic studies have revealed that 15%–35% of mCRPC contain DNA repair defects, including in BRCA1/2, ATM, ATR, and RAD51 (The Cancer Genome Atlas Research Network 2015; Robinson et al. 2015). Olaparib, a Food and Drug Administration (FDA)-approved oral PARP inhibitor for BRCA-deficient cancers (Bryant et al. 2005; Farmer et al. 2005), also shows promising clinical activity in cancers possessing mutations in other DNA repair genes (Lord and Ashworth 2016). In a phase II trial, olaparib treatment in mCRPC harboring defects in DNA repair genes showed high response rates (Mateo et al. 2015).
Genetic signatures of NEPC
Recent genetic studies revealed that mCRPC with neuroendocrine features commonly harbors RB1 and TP53 deficiencies and displays attenuated AR signaling compared with CRPC (Tan et al. 2014; Beltran et al. 2016b). Functional studies revealed that loss of RB1 and TP53 drives lineage plasticity, manifesting as a phenotypic shift from AR-dependent luminal epithelial cells to AR-independent neuroendocrine-like cells—a process driven by activation of the epigenetic reprogramming factors EZH2 and SOX2 (Ku et al. 2017; Mu et al. 2017). N-MYC, which is overexpressed or amplified in ∼40% of NEPCs, was identified as another driver of NEPC initiation (Beltran et al. 2011; Dardenne et al. 2016; Lee et al. 2016b).
Emerging genetic signatures
Recent studies identified new recurrent mutations of SPOP (11%–13%) in ETS fusion tumors (Barbieri et al. 2012; The Cancer Genome Atlas Research Network 2015), which defined a new prostate cancer subtype with the notable molecular features of increased DNA methylation and homogeneous gene expression patterns (The Cancer Genome Atlas Research Network 2015). SPOP encodes an E3 ubiquitin ligase component, and the mutated protein causes stabilization of oncogenic substrates such as MAPK8 (JNK), NCOA3, and DEK (Geng et al. 2013; Theurillat et al. 2014; Blattner et al. 2017). Additionally, three groups (Dai et al. 2017; Janouskova et al. 2017; Zhang et al. 2017) reported that wild-type SPOP promotes the ubiquitylation and proteasomal degradation of BET family proteins BRD2/3/4, and two of them found that SPOP mutated prostate tumors were resistant to BET inhibitors. A SPOP mutant GEMM confirmed the function of SPOP as a driver of prostate tumorigenesis through activation of both PI3K/mTOR and AR signaling and effective uncoupling of the normal negative feedback between these two pathways (Blattner et al. 2017). In 2015, ERG was identified as a SPOP degradation target in multiple prostate cancer cell lines (An et al. 2015; Gan et al. 2015), but, most recently, this finding was refuted by Shoag et al. (2018) in a SPOP-F133V GEMM. The SPOP molecular class displays loss of the chromatin remodeling factor CHD1 (Barbieri et al. 2012; Burkhardt et al. 2013), but these observations are in contrast to recent work demonstrating that CHD1 represents an essential effector of PTEN deficiency in prostate cancer (Zhao et al. 2017). Further study is warranted to evaluate CHD1 function in the SPOP mutant subtype. Another new genetically distinct subtype of prostate cancer was defined by hot spot mutations in IDH1 along with strongly elevated levels of genome-wide DNA hypermethylation; while of low incidence (1%), these IDH1 R132 mutant tumors define a distinct subgroup of early-onset prostate cancer that possesses fewer DNA copy number alterations or other canonical genomic lesions commonly found in most other prostate cancers (The Cancer Genome Atlas Research Network 2015). IDH1 and IDH2 mutations have been associated with a DNA methylation phenotype in other cancer types (Figueroa et al. 2010; Noushmehr et al. 2010), suggesting that IDH1 mutant prostate cancers might have oncogenic mechanisms similar to those in glioblastoma multiforme and acute myelogenous leukemia and may be sensitive to newly developed IDH1 targeted therapeutics.
Epigenetic deregulation
Deregulation of genes controlling epigenetic processes involved in DNA modification (e.g., methylation and hydroxymethylation), histone modification, or nucleosome remodeling can drive tumorigenesis in many cancer types (Dawson and Kouzarides 2012; Feinberg et al. 2016; Flavahan et al. 2017; Genovese et al. 2017), including prostate cancer (Albany et al. 2011; Jeronimo et al. 2011; Yegnasubramanian 2016).
DNA can be methylated by canonical DNA methyltransferase (DNMT) consisting of DNMT1, DNMT3A, and DNMT3B at the five position of the cytosine within CpG dinucleotides, which are often found in large clusters called CpG islands (Kulis and Esteller 2010; Lyko 2018). Methylated cytosine can be converted into 5-hydroxymethylcytosine (5hmC) by TET protein family members (i.e., TET1, TET2, and TET3), and 5hmC can be further oxidized to 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) (Branco et al. 2011). DNA methylation in normal cells ensures that gene expression and gene silencing are properly regulated. Aberrant DNA methylation—hypermethylation within promoter regions of tumor suppressor genes or global hypomethylation—contributes to transformation through silencing of tumor suppressor genes and genome instability, respectively. Recent studies uncovered a surprising function for DNMT in transcriptional activation through its interaction with TET proteins (Lyko 2018). DNMT1 has been shown to act as a tumor suppressor gene in early stage prostate cancer and an oncogene in late stage prostate cancer (Kinney et al. 2010), particularly in the metastasis process through regulation of EMT and cancer stem cell programs (Kinney et al. 2010; Lee et al. 2016a). Interestingly, TGF-β was shown to regulate the expression of DNMTs in prostate cancer, with their expression correlating with aggressiveness and recurrence (Zhang et al. 2011b). Both TET1 and TET2 were shown to play a tumor-suppressive role in prostate cancer through regulation of cell proliferation, migration, and invasion (Hsu et al. 2012; Nickerson et al. 2017).
Histone modification (e.g., acetylation, methylation, and phosphorylation) also plays a prominent role in normal and neoplastic processes through the regulation of gene expression (Jenuwein and Allis 2001; Allis and Jenuwein 2016; Audia and Campbell 2016). Genomic profiling has identified mutations in many epigenetic regulators and chromatin remodelers in up to 20% of primary prostate cancer and mCRPC. Mutant epigenetic regulators include ASXL1, KMT2C (MLL3), KMT2D (MLL2), KMT2A (MLL), KDM6A (UTX), SETD2, and SETDB1, and mutant chromatin remodelers include ARID1A, ARID4A, ARID2, SMARCA1, and other members of the SWI/SNF nucleosome remodeling complex. These mutations are significantly enriched in prostate tumors without ETS fusions or a driver mutation such as IDH1, SPOP, CUL3, or FOXA1. In primary tumors, these mutations are associated with higher Gleason scores (Grasso et al. 2012; Armenia et al. 2018). On the functional level, the MLL complex that interacts with AR via the menin–MLL subunit plays an important role in the development of CRPC and NEPC (Grasso et al. 2012; Malik et al. 2015). Therapeutic targeting of the interaction between menin and the MLL complex suppresses AR signaling and the growth of castration-naïve and castration-resistant tumors in the VCaP model (Malik et al. 2015). While the functional significance of ARID1A, ARID4A, ARID2, and SMARCA1 mutations are not known, the SWI/SNF complex has been shown to drive prostate tumorigenesis, thus implying a therapeutic strategy that targets interaction of the SWI/SNF complex with its interacting proteins. For example, BAF57, a subunit of the BAF57 SWI/SNF complex, directly interacts with AR and regulates the AR transcriptional program (Link et al. 2008); expressing the BAF57 inhibitory peptide (BIPep) in AR-positive cancer cell lines suppresses androgen-dependent cell proliferation. In addition, the function of the SWI/SNF complex was antagonized by the long noncoding RNA SChLAP1, which contributes to the oncogenic function of SChLAP1 (Prensner et al. 2013).
Members of the Polycomb group (PcG) protein complexes, which epigenetically repress transcriptional programs, can also contribute to prostate cancer. EZH2, a methyltransferase of Polycomb-repressive complex 2 (PRC2), which maintains the repressive histone mark H3K27me3, is often overexpressed in cancers and has been demonstrated to promote prostate cancer progression (Varambally et al. 2002) and castration resistance (Xu et al. 2012b). Loss of micoRNA-101, a negative regulator of EZH2 expression and functions, has been found in prostate cancer, resulting in overexpression of EZH2. BMI1, a component of PRC1, plays a role in basal prostate stem cell maintenance, marks a distinct population of castration-resistant luminal progenitor cells, and plays a documented role in prostate cancer initiation and progression (Lukacs et al. 2010; Yoo et al. 2016). Histone methyltransferase WHSC1 has been shown to be stabilized by AKT, leading to promotion of prostate cancer metastasis (Li et al. 2017b). Lysine-specific demethylase 1 (LSD1) functions as a transcriptional repressor of AR-regulated enhancers through H3K4 demethylation and as an AR-linked coactivator through interaction with CoREST and histone H3 Thr6 phosphorylation (H3T6ph) (Cai et al. 2011, 2014). LSD1 also promotes prostate cancer cell survival through activation of a gene network associated with a lethal prostate cancer independent of its demethylase function (Sehrawat et al. 2018) and promotes CRPC through epigenetic programming to induce CENPE expression (Liang et al. 2017).
Histone demethylases have also been implicated in prostate cancer. For example, JMJD1A recruits heterogeneous nuclear ribonucleoprotein F to promote alternative splicing of AR-V7 in prostate cancer cells (Fan et al. 2018); JMJD2A cooperates with ETV1 to drive prostate cancer initiation (Kim et al. 2016). Bromodomain-containing proteins, which recruit transcriptional regulatory complexes to acetylated chromatin, were shown to interact with AR (Asangani et al. 2014) and mediate the chromatin accessibility of BRD4 (Urbanucci et al. 2017).
This large number of genetic alterations uncovered by recent large-scale genomic studies has amplified the need to validate and functionally define their roles in primary prostate cancer, CRPC, and metastatic disease. In addition, these validations must occur in the context of the appropriate molecular subtype. Along these lines, there is critical need for GEMMs representing newly identified molecular subtypes, including the SPOP mutant, the IDH1 mutant, and AR−NE− subtypes. Another pressing need is the development of GEMMs with a high propensity to metastasize to bone, as currently only up to 17% of models display bone metastases and exhibit a less typical NEPC or sarcomatoid pathology (Grabowska et al. 2014). Moreover, androgen deprivation and relapse should be performed routinely in characterizing newly established prostate cancer GEMMs, as androgen independence may yield a better model for metastatic CRPC. The development of such refined multiallelic models should be guided by comparative genomics of primary versus bone metastatic tumors. Such investments will illuminate the key genetic events and effective therapeutic combinations for the molecular subsets encountered in the clinic.
Prostate cancer heterogeneity
Therapeutic advances in oncology have been shaped by a detailed catalog of genotypic variations between patients that informs responses to targeted treatments (Bedard et al. 2013). Similarly, intratumoral heterogeneity within a given patient is now recognized as an equally important factor in dictating drug response and disease relapse (Boutros et al. 2015; Kumar et al. 2016a). This intratumoral heterogeneity manifests on many levels and includes genomic and developmental cell variability within the cancer cell compartment as well as the diversity of numerous TME cell types and their complex heterotypic interactions.
Pathologic and genomic heterogeneity
Newly diagnosed prostate cancer commonly presents as multifocal disease with histopathologically distinct foci. Thus, a thorough pathologic review of the available specimen with all grades is critical for accurately describing the grading of biopsy samples and prostatectomy specimens in the clinical report (Beltran and Demichelis 2015). Inadequate sampling may lead to inaccurate clinical staging. Separate cancer foci in primary prostate cancers can also exhibit distinct genomic profiles; for instance, the coexistence of multiple cancer lineages harboring distinct ERG fusions within a single primary prostate cancer nodule (Cooper et al. 2015). To evaluate the molecular heterogeneity of primary prostate cancer, Boutros et al. (2015) performed genomic sequencing of multiple lesions in individual patients and identified novel alterations, including the recurrent focal amplification of MYCL and MYC genes, as well as known recurrent alterations, including loss of NKX3.1 and TP53. Strikingly, whole-genome sequencing of multifocal tumors revealed that very few copy number alterations were shared between pathologically identical tumor foci, consistent with the independent origins of these distinct foci (Boutros et al. 2015).
In light of this pathological and genomic heterogeneity, profiling studies can be limited in aiding accurate clinical decision-making, which often relies on a single biopsy for determining the molecular status of a specific prostate cancer case. Longitudinal sampling and comprehensive genomic and pathologic analyses of a patient with prostate cancer revealed that the lethal metastatic clone arose from a small low-grade primary tumor focus harboring PTEN and TP53 alterations rather than the bulk higher-grade primary cancer or a lymph node metastatic focus (Haffner et al. 2013). Another whole-genome study in primary and metastatic tumors longitudinally collected from four patients whose prostate cancers were lethal also tracked and identified the TP53 mutant subclone as an origin of metastatic expansion (Hong et al. 2015). To characterize the subclonal architecture of mCRPC, Gundem et al. (2015) performed whole-genome sequencing of 51 multifocal primary and metastatic tumors from 10 patients and discovered that metastasis derived from multiple clones that transfer between different metastatic sites or a single daughter clone that was seeded from another metastatic site. This study also uncovered that tumor suppressor gene alterations usually occurred as single events, whereas AR pathway gene mutations commonly involved simultaneous events that occur in multiple metastatic sites (Gundem et al. 2015). Overall, these studies show that, beyond a single biopsy, additional multifocal and longitudinal analyses of matched primary and metastatic tumors—coupled with liquid biopsies (of cell-free tumor DNA)—may be needed to better inform management of CRPC patients (Lohr et al. 2014).
Functional heterogeneity in prostate cancer cells
Prostate cancer heterogeneity also manifests on the functional level within the cancer cell population, particularly with respect to differentiation status and lineage plasticity. While cancer cells can exhibit different tumor-initiating capacities and self-renewal potential, the role of cancer stem cells in treatment responses remains an area of active study (Meacham and Morrison 2013). In the normal prostate, multipotent stem and progenitor cells have been identified in the basal epithelial compartment, which can give rise to basal, luminal, and neuroendocrine cells in mouse and human prostates (Goldstein et al. 2008, 2010). Lineage tracing studies in the mouse prostate revealed that both basal and luminal cells can serve as the cell of origin for prostate cancer and that deregulation of epithelial differentiation is a critical step for the initiation of prostate cancers of basal cell origin (Wang et al. 2009; Choi et al. 2012). Particularly, BMI1 has been identified as a key player in the regulation of the self-renewal of prostate stem cell and prostate cancer initiation, progression, and castration resistance (Lukacs et al. 2010; Zhu et al. 2018). In addition, PSA−/lo prostate cancer cells have been shown to possess self-renewal capability and initiate prostate tumorigenesis that is resistant to castration (Qin et al. 2012). In aggressive NEPC, increasing evidence suggests that neuroendocrine transdifferentiation represents an adaptive mechanism that enables resistance to ADT (Lin et al. 2014); various genetic and epigenetic alterations contribute to this process of lineage plasticity (Lee et al. 2016b; Ku et al. 2017; Mu et al. 2017; Zou et al. 2017). To add further complexity, some NEPC tumor regions can often be mixed in with typical adenocarcinoma cells (Epstein et al. 2014). Multiple studies using fluorescence in situ hybridization revexal the presence of the AR-regulated TMPRSS2–ERG genomic translocation in AR− NEPC (Lotan et al. 2011; Williamson et al. 2011), supporting the hypothesis that AR− prostate cancer arises directly from typical AR+ adenocarcinomas by transdifferentiation.
Cellular heterogeneity in the TME
Significant intratumoral heterogeneity is also reflected in the diversity of cell types and the composition of the extracellular matrix comprising the TME. TME cell types include CAFs, mesenchymal stem cells (MSCs), immune cells, and blood and lymphatic vascular cells (Fig. 3). TME composition plays essential roles in regulating cancer cell proliferation, angiogenesis, invasion, metastasis, immune evasion, and resistance to therapeutics (Hanahan and Weinberg 2011; Hanahan and Coussens 2012) and is mediated by signaling cross-talk between cancer cells and distinct stromal populations through direct cell contact and/or secreted factors such as cytokines, chemokines, and growth factors. In prostate cancer, various signaling molecules (e.g., androgen, FGFs, SRC, and TGF-β) are involved in these heterotypic and homotypic interaction networks across cancer cells and stromal cells (Egeblad et al. 2010; Karlou et al. 2010; Hanahan and Coussens 2012; Junttila and de Sauvage 2013). Intertumoral and intratumoral TME heterogeneity manifests in both cell type composition and differences in the phenotype and functional status of any individual cell type. Below, we catalog the many TME cell types and their functional roles in prostate cancer (Table 2).
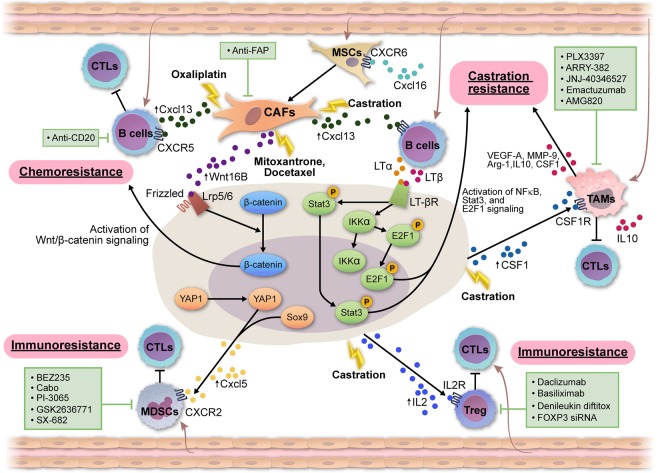
Figure 3.
The TME contributes to therapy resistance. (1) Chemoresistance. Cytotoxic chemotherapy (mitoxantrone and the docetaxel) induces WNT16B expression CAFs, which in turn activates WNT signaling in prostate cancer cells through binding to Lrp5/6 and Frizzle in a paracrine manner and subsequently promotes chemoresistance and tumor progression. Oxaliplatin induces Cxcl13 expression in CAFs, which promotes the recruitment of B cells to suppress immunogenic cell death induced by oxaliplatin; plasmocytes expressing immunoglobulin A, IL-10, and PD-L1 were identified as the immunosuppressive B cells that are directly involved in this process (Ammirante et al. 2014; Shalapour et al. 2015). (2) Castration resistance. Castration also induces Cxcl13 expression in CAFs, which promotes the recruitment of B cells. B-cell-derived lymphotoxin activates E2F/BMI1/Stat3 signaling to promote the development of CRPC. Castration also induced the expression of colony-stimulating factor 1 (Csf1) in prostate cancer cells to attract macrophages to promote the survival of prostate cancer cells. (3) Immunoresistance. Yap1 and Sox9 activation in prostate cancer cells leads to an increase in the expression of chemokine Cxcl5 and the subsequent recruitment of myeloid-derived suppressor cells (MDSCs) to promote prostate tumor progression and immunoresistance through multiple mechanisms, including the direct suppression of cytotoxic T cells. Castration induced the increased expression of IL-2 to recruit regulatory T cells (Tregs), which will limit the efficacy of the cytotoxic T cells. Various therapeutic agents have been used to target CAFs (Kakarla et al. 2012), B cells (Yuen et al. 2016), tumor-associated macrophages (TAMs) (Cannarile et al. 2017), MDSCs (Lu et al. 2017a), and Tregs (Liu et al. 2016).
Table 2.
Biological functions and clinical significance of cell types that are present in the prostate TME
MSCs are heterogeneous progenitor cells with pluripotent activities that contribute to the homeostasis of connective tissues such as bone, adipose, cartilage, and muscle (Pittenger et al. 1999; Uccelli et al. 2008). MSCs are recruited to the TME to become tumor-associated MSCs and CAFs (Kalluri 2016; Shi et al. 2017). MSCs can promote progression in multiple cancer types. For example, MSCs can promote metastasis of breast, gastric, and prostate cancers (Karnoub et al. 2007; Quante et al. 2011; Jung et al. 2013). CAFs are among the most abundant of the TME cell types (Quail and Joyce 2013; Augsten 2014; Kalluri 2016) and also promote oncogenic transformation, tumor proliferation, angiogenesis, invasion/metastasis, and drug resistance (Ayala et al. 2003; Yang et al. 2005; Giannoni et al. 2010; Liao et al. 2010; Barron and Rowley 2012; Hanahan and Coussens 2012; Quail and Joyce 2013; Kalluri 2016). Interestingly, a recent study demonstrated that colony-stimulating factor 1 receptor (CSF1R) blockade induced the expression of granulocytic chemokines such as Cxcl1 in CAFs to promote polymorphonuclear myeloid-derived suppressor cell (PMN-MDSC) recruitment into tumors. Correspondingly, the combination of a CSF1R inhibitor and a Cxcr2 inhibitor resulted in significantly reduced tumor growth (Kumar et al. 2017). Together, these findings suggest that knowledge of MSC and CAF biology and signaling could inform novel therapeutic strategies for many cancer types, including prostate cancer.
Lymphocytes are key cellular components in the mammalian adaptive immune system that protect the host from infectious pathogens, with various lymphocyte subtypes playing central roles in cancer biology and treatment (Gajewski et al. 2013). Several studies have been conducted to assess the association between lymphocytic infiltration and clinical parameters such as tumor stage and recurrence-free survival (Strasner and Karin 2015). A recent report analyzed the correlation of CD4+ helper T cells, CD8+ cytotoxic T cells, CD4+FOXP3+ regulatory T cells (Tregs), and CD8+FOXP3+ Tregs in tumor tissue with inflammation, types of atrophy, and indolent or lethal prostate cancer (Davidsson et al. 2013). These studies revealed that CD4+ Tregs, but not CD4+ T helper or CD8+ cytotoxic T cells, were associated with increased risk of lethality. Moreover, increased intratumoral CD20+ B cells were observed in high-risk tumors and are associated with disease recurrence or progression (Woo et al. 2014). That said, these immune profiles should be interpreted with caution, as the immune cell subtype, heterogeneity within immune cell subtypes, and functional state of immune cells should be audited to strengthen the predictive power of such profiles with respect to clinical outcomes. Moreover, all of these studies to date have been conducted in primary prostate tumors, underscoring the need for similar investigation of the metastatic TME.
Myeloid cells, the most abundant nucleated hematopoietic cells in the human body, are essential for the normal function of both the innate and adaptive immune systems. MDSCs and tumor-associated macrophages (TAMs) have emerged as important regulators of cancer progression, metastasis, and therapy resistance. MDSCs comprise a heterogeneous population of immature myeloid cells that accumulate in pathologic conditions such as cancer, owing to a partial block of its differentiation program in the myeloid lineage (Condamine et al. 2015; Kumar et al. 2016b). MDSCs were initially defined in murine models by the coexpression of CD11b and Gr-1 markers (Bronte et al. 1998; Talmadge and Gabrilovich 2013) and can be further separated into granulocytic MDSCs (CD11b+Ly6G+) and monocytic MDSCs (M-MDSCs; CD11b+Ly6C+). Human MDSCs express markers such as CD11b and CD33 but are mostly negative for human leukocyte antigen–antigen D-related and lineage-specific antigens, including CD3, CD19, and CD57 (Gabrilovich et al. 2012; Bronte et al. 2016), and can be separated into PMN-MDSCs and M-MDSCs (Table 2). These MDSCs possess potent immunosuppressive activity, play a major role in the suppression of immune responses in cancer through a variety of mechanisms (Gabrilovich and Nagaraj 2009), and have been implicated in the promotion of angiogenesis, tumor cell invasion, and metastases (Yang et al. 2004, 2008; Condamine et al. 2015; Kumar et al. 2016b). Furthermore, clinical findings have shown that the presence of MDSCs correlates with reduced survival in human cancers, including breast and colorectal cancers (Solito et al. 2011). MDSC abundance in the blood was found to correlate with circulating PSA levels in patients with prostate cancer (Vuk-Pavlović et al. 2010; Brusa et al. 2013). In addition, the level of blood M-MDSCs was found to correlate with negative prognostic markers such as elevated levels of lactate dehydrogenase and PSA in patients with mCRPC (Idorn et al. 2014).
Experimentally, GEMMs have highlighted the important role of MDSCs in prostate tumorigenesis and immune therapy resistance. Gr1+ myeloid cells, which may include CD11b+Gr1+ MDSCs, have been shown to play a role in tumor progression and the evasion of PTEN loss-induced cellular senescence and chemoresistance in cancer cells in a mouse model of indolent Pten-null prostate cancer (Di Mitri et al. 2014; Garcia et al. 2014). IL-6 has been implicated in the development of hormone-resistant prostate cancer using hormone-sensitive murine prostate cancer cell lines through the induction of MDSCs (Wu et al. 2012). In addition, MDSCs were shown to promote tumor initiation and progression in the Pten-null model (Garcia et al. 2014). In a metastatic Pten/Smad4-deficient GEMM, MDSCs were shown to play a critical role in tumor progression, with their recruitment to the TME driven in part through Yap1 signaling in the cancer cells (Wang et al. 2016a). These murine studies may be clinically relevant, as human primary prostate cancers with active Yap1 signaling also exhibit transcriptional signatures consistent with abundant MDSCs. Moreover, various therapies depleting MDSCs in this mouse prostate cancer model show significant anti-tumor activity (Wang et al. 2016a). Also, specific genotypes in prostate cancer cells may shape distinct immunocyte profiles in the TME to promote tumor progression through various mechanisms, as demonstrated in mouse models of prostate cancer engineered with loss of Pten alone or in combination with loss of Trp53, Zbtb7a, or Pml (Bezzi et al. 2018). Specifically, Ptenpc−/−Zbtb7apc−/− and Ptenpc−/− Trp53pc−/− tumors exhibit an immunologically “hot” TME with abundant immunocytes, whereas Ptenpc−/− Pmlpc−/− tumors display a “cold” TME with less intratumoral immune infiltration relative to the other genotypes. Moreover, while both Ptenpc−/−Zbtb7apc−/− and Ptenpc−/− Trp53pc−/− TMEs recruit MDSCs, there are differences in the types of cytokines and specific MDSC subtypes: Granulocytic MDSCs are recruited via Cxcl5 in Ptenpc−/− Zbtb7apc−/− tumors, and M-MDSCs are recruited via Cxcl17 in Ptenpc−/−Trp53pc−/− tumors.
TAMs, identified as Mac-1+(CD11b/CD18) and/or F4/80+ myeloid cells, also play important roles in the TME. TAMs can be classified into tumor-suppressive M1 macrophages or tumor-promoting M2 macrophages (Biswas and Mantovani 2010; Noy and Pollard 2014), which can be distinguished by the differential expression of transcription factors and surface molecules as well as differences in their cytokine profiles and metabolism (Murray 2017). Increased levels of TAMs are associated with poor prognosis in human cancers (Bingle et al. 2002). TAMs promote tumor progression, migration, and metastasis (Qian and Pollard 2010; Kitamura et al. 2015; Maolake et al. 2017; Linde et al. 2018), and depletion of TAMs has been shown to suppress tumor growth in multiple murine tumor models (Luo et al. 2006; Ries et al. 2014; Wu et al. 2014b). Mechanistically, the increased expression of CSF1 in cancer cells promotes TAM differentiation and survival, and TAMs promote tumor progression and metastasis through HIF1α-mediated VEGF and PDGF production and promote immunosuppression through IL-10. The clinical relevance of TAMs in prostate cancer progression has been evaluated in several studies (Craig et al. 2008; Gannon et al. 2009; Nonomura et al. 2011; Fujii et al. 2013; Gollapudi et al. 2013; Lanciotti et al. 2014) and is consistent with the protumorigenic effects of TAMs observed in other cancer types; some studies suggest a link between TAMs and disease recurrence (Gannon et al. 2009; Nonomura et al. 2011).
Therapeutic targeting of cancer cell-intrinsic and TME mechanisms
Current standard of care and emerging targeted therapies for prostate cancer
The treatment of prostate cancer depends on grade, stage, and age and ranges from active surveillance to a mix of surgery, chemotherapy, radiation, and/or ADT (Fig. 2; Litwin and Tan 2017). Localized cancers are stratified into three groups of low, intermediate, and high risk based on Gleason score (Rodrigues et al. 2012). Low-risk cancers (Gleason 3 + 3) are typically managed by active surveillance, as large randomized clinical trials show no mortality differences between active surveillance and radical prostatectomy or radiotherapy (Iversen et al. 1995; Wilt et al. 2012; Bill-Axelson et al. 2014; Hamdy et al. 2016; Sanyal et al. 2016; Wilt et al. 2017). At the other end of the spectrum are high-risk cancers (Gleason ≥8), which receive more aggressive treatment, including surgery and radiation-based therapies. A major treatment decision challenge in prostate cancer lies with intermediate-risk disease (e.g., Gleason 3 + 4), as these patients exhibit considerable differences in outcomes (see also “Outlook for Next-Generation Prostate Cancer Management”). Several proposed new classification systems have been developed to further classify these intermediate-risk cases into favorable and unfavorable subgroups (Serrano and Anscher 2016) based on clinical stage (Reese et al. 2012) or clinical characteristics such as the number of intermediate-risk factors (one vs. more than one), Gleason pattern (GS of 3 + 4 ≥ 7 vs. GS of 4 + 3 = 7), and percentage of positive biopsy cores (<50% vs. >50%) (Zumsteg and Zelefsky 2012). In addition, considerable efforts are focused on the development of biomarkers (e.g., transcriptome-based gene signatures) to more accurately predict disease aggressiveness and outcome. For patients who do receive treatment for localized prostate cancer and experience disease recurrence (defined by rising PSA), ADT is commonly used in combination with surgery or radiation. In the setting of metastatic disease, the initial treatment plan includes ADT, often with chemotherapy. ADT can involve two approaches: surgical castration (i.e., orchiectomy) or, more commonly, chemical castration with drugs targeting AR signaling regulated by the hypothalamic–pituitary–testicular axis (e.g., gonadotropin-releasing hormone agonists, AR antagonists, and CYP17A1 inhibitors).
Although most patients initially respond well to ADT, recurrence occurs in virtually all cases, leading to mCRPC. Until 2010, the gold standard treatment for CRPC was docetaxel chemotherapy (Quinn et al. 2017; Sumanasuriya and De Bono 2018). Another chemotherapy agent, cabazitaxel, was approved in 2010 for mCRPC patients previously treated with docetaxel and in 2017 for use at a lower dosage based on the results of two phase 3 randomized trials (de Bono et al. 2010; Sartor et al. 2016). In addition to chemotherapy using taxanes, treatment options for mCRPC have expanded significantly in the last decade. Potent second-generation anti-androgen FDA-approved therapies now include enzalutamide, abiraterone, and apalutamide as well as novel agents in clinical trials (e.g., EPI-506) (Vaishampayan et al. 2017) and in preclinical development (e.g., ASC-J9) (Wang et al. 2016b). The potent AR antagonists enzalutamide and apalutamide can increase the survival of patients with mCRPC (Scher et al. 2012; Beer et al. 2014) and localized CRPC (Smith et al. 2018), respectively. Abiraterone, a CYP17A1 inhibitor that blocks androgen production, also improves survival of patients with advanced prostate cancer with or without prior chemotherapy (de Bono et al. 2011; Ryan et al. 2013; Fizazi et al. 2017; James et al. 2017). Interestingly, Δ4-abiraterone (D4A), an abiraterone metabolite, inhibits multiple enzymes involved in DHT synthesis such as CYP17A1, 3βHSD, and SRD5A and displays a more potent anti-tumor activity than abiraterone, suggesting treatment with D4A as a more clinically effective therapeutic approach than treatment with abiraterone (Li et al. 2015). In addition, numerous FDA-approved and experimental therapies are available for the management of bone metastasis from prostate cancer; these therapies can delay or reduce skeletal-related events such as bone fractures and spinal cord compression. These agents target differentiation pathways of bone cells and include zoledronic acid (a bisphosphonate that binds to hydroxyapatite and impedes osteoclast-mediated resorption), antibodies for osteoprotegerin and parathyroid hormone-related protein, denosumab (a monoclonal antibody that targets RANKL), atrasentan (endothelin receptor antagonist), BMP antagonists such as Noggin and anti-BMP6, and radioactive drugs such as radium-223 (Body et al. 2015; Krzeszinski and Wan 2015).
Cancer immunotherapy
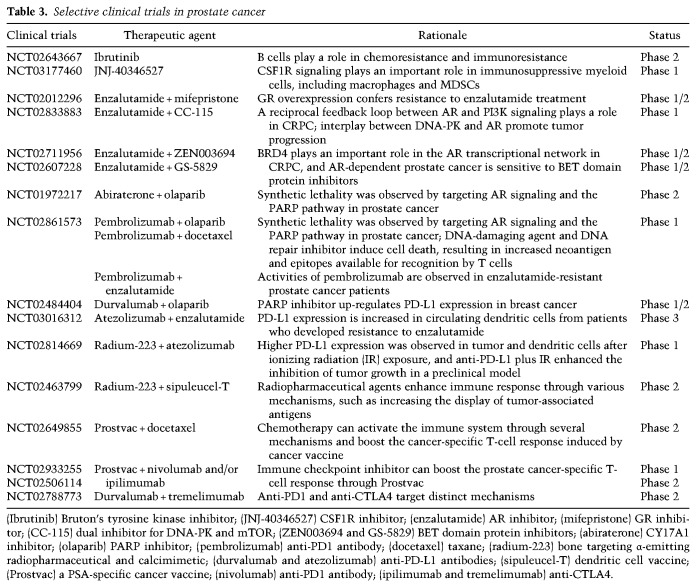
Intensive effort is focused on agents that modulate the immune response through the use of antibodies, small-molecule inhibitors, engineered immune cells, vaccines, and viruses to stimulate the patient's immune system to attack and destroy cancer cells. While durable therapeutic responses can be achieved in many types of advanced cancers, the majority of cases does not respond because of either “primary resistance,” in which cancers do not respond to initial therapy owing to a lack of active immune response, or “adaptive resistance,” in which a cancer is recognized by the immune system but induces immunosuppressive pathways in the tumor following an active immune attack on the tumor (Sharma et al. 2017). In addition, a small subset of initially responsive cancers may develop “acquired resistance,” resulting in tumor relapse (Ribas 2015; Restifo et al. 2016; McGray and Bramson 2017; Sharma et al. 2017). In mCRPC, robust immunotherapy regimens are not yet available (Maia and Hansen 2017). To date, the FDA-approved dendritic cell-based cancer vaccine sipuleucel-T has shown only modest survival benefit (Kantoff et al. 2010), and clinical trials with immune checkpoint inhibitors (e.g., anti-CTLA-4 and anti-PD-1) as single agents display minimal or no activity, consistent with primary or adaptive resistance mechanisms (Kwon et al. 2014; Graff et al. 2016; Beer et al. 2017). The prevailing view is that immunoresistance may be overcome by combined anti-CTLA-4 and anti–PD-1 regimens and/or synergistic therapies targeting immunosuppressive signals from myeloid cells (see “TME-Driven Mechanisms of Resistance to Conventional and Novel Cancer Therapies” below) and/or driver oncogenic signaling pathways.
Cancer cell-intrinsic mechanisms conferring therapeutic resistance
Various cancer cell-intrinsic mechanisms involving genetics, epigenetics, and metabolomics can dictate therapeutic responses and shape the composition of the TME. Several prostate cancer cell-intrinsic chemoresistance mechanisms include activation of ABCG2 (Robey et al. 2001; Imai et al. 2004; Patrawala et al. 2005), activation of PI3K signaling (Lee et al. 2004), loss of RAS-GTPase-activating protein DAB2IP (Wu et al. 2013), up-regulation of cancer stem cell-associated Notch and Hedgehog pathways (Domingo-Domenech et al. 2012), up-regulation of the NRF2 stress response pathway caused by KEAP1 loss (Zhang et al. 2010), and overexpression of ERG (Galletti et al. 2014). This section focuses on cancer cell-intrinsic mechanisms underlying resistance to ADT and immunotherapy.
AR-dependent castration resistance
Despite low circulating androgen levels under ADT, CRPC can sustain androgen signaling via increased intratumoral hormone synthesis, AR amplification, mutations, and/or dysregulated expression of AR coactivators and corepressors (Shen and Abate-Shen 2010; Watson et al. 2015). Targeting these mechanisms via the AR inhibitor enzalutamide or the CYP17A1 inhibitor abiraterone can improve overall survival in both localized and mCRPC patients, as described above. However, the expression of constitutively active AR splice variant AR-V7 in CTCs is predictive of resistance to abiraterone acetate or enzalutamide in men with mCRPC (Antonarakis et al. 2014), which has been further validated by a larger cohort (n = 202) of clinical study recently (Antonarakis et al. 2017). Of note, this hormone independence is associated with genetic alterations of the PTEN/PI3K pathway (The Cancer Genome Atlas Research Network 2015; Robinson et al. 2015), which cross-regulates with AR signaling and coordinately supports cancer cell survival (Petrylak et al. 2004; Carver et al. 2011; Mulholland et al. 2011). Indeed, combined inhibition of PI3K/AKT and AR signaling can provoke robust regressions in Pten-deficient GEMMs and human PDX models (Carver et al. 2011; Mulholland et al. 2011). Recently, however, Bluemn et al. (2017) revealed that inhibition of AR signaling can suppress PI3K/AKT signaling in metastatic disease. Specifically, they established an androgen-resistant/AR-negative cell line, LNCaPAPIPC (LNCaP-AR program-independent prostate cancer), derived from androgen-sensitive/PTEN-deficient prostate cancer cell line LNCaP cultured in androgen deprivation medium followed by long-term AR depletion (Bluemn et al. 2017). Notably, compared with the parental cell line, the LNCaPAPIPC line activated FGFR and MAPK signaling pathways but strongly suppressed PI3K/AKT signaling (Bluemn et al. 2017)—a finding that may dampen enthusiasm for PI3K targeting in mCRPC and instead enhance the usage of newly available androgen targeting drugs. Recent studies also uncovered additional factors boosting AR transcriptional activity, including RNF6 (Xu et al. 2009), SIAH2 (Qi et al. 2013), DNA-dependent protein kinases (DNA-PKcs) (Goodwin et al. 2013, 2015), bromodomain protein BRD4 (Asangani et al. 2014, 2016), TRIM24 (Groner et al. 2016), and insulin and keratinocyte growth factor (Culig 2004; Zhang et al. 2009). AR expression and transcriptional output are increased in the RB1-deficient cells through the activation of E2F1 to up-regulate AR mRNA and increase recruitment of AR to the promoters of its target genes (Sharma et al. 2010). AR protein stability is also stabilized by interaction with BMI1, which abrogates MDM2-mediated AR protein degradation, resulting in sustained AR signaling in prostate cancer cells (Yoo et al. 2016). In addition, AR plays a critical role in the regulation of anabolic pathways and biosynthesis through calcium/calmodulin-dependent protein kinase kinase 2 (CAMKK2) (Massie et al. 2011). Moreover, a gain-of-function mutation (N367T) in 3β-hydroxysteroid dehydrogenase type 1 (3βHSD1), an enzyme for the rate-limiting step in the conversion of adrenal-derived steroid dehydroepiandrosterone to DHT, resulted in an increase in DHT synthesis and the development of castration resistance in prostate cancer (Chang et al. 2013). Germline SNP at position 1245 of HSD3B1 (A → C conversion, SNP; rs1047303), which resulted in the gain-of-function mutant N367T, is associated with resistance to ADT (Hearn et al. 2016). Together, these mechanistic insights provide avenues for novel therapeutic strategies in combination with ADT.
Glucocorticoid receptor (GR)-dependent castration resistance
Up-regulation of the GR can cross-regulate AR target genes to confer resistance to enzalutamide or ARN-509 (Arora et al. 2013). Therefore, an early phase clinical trial of enzalutamide in combination with the GR antagonist mifepristone is currently being explored (ClinicalTrials.gov identifier: NCT02012296). A note of caution is warranted, since mifepristone binds with high affinity to AR and caused activation of its downstream signaling in an earlier single-agent phase II study (Taplin et al. 2008). An alternative approach may come from the observations that the tissue-specific enhancer regulating GR expression mediates adaptive and reversible AR bypass and that BET bromodomain inhibition can selectively perturb this enhancer and restore sensitivity to enzalutamide (Shah et al. 2017). AR bypass in CRPC may also involve the progesterone receptor and the mineralocorticoid receptor, which are steroid hormone nuclear receptors structurally related to AR and share substantial homology of the DNA-binding domain with AR (Lu et al. 2006; Watson et al. 2015).
AR-independent castration resistance
As described above, AR-independent NEPC, an aggressive subtype of CRPC, harbors deficiencies of TP53 and RB1 as well as amplification of N-myc (MYCN) and Aurora kinase A (AURKA). Recent findings in human and mouse prostate cancer models demonstrated that these genetic and consequently epigenetic alterations contribute to lineage plasticity, metastasis, and castration resistance (Lee et al. 2016b; Ku et al. 2017; Mu et al. 2017). In preclinical models, targeting N-MYC, AURKA, and EZH2 in NEPC has been an effective therapeutic approach. A recent mCRPC study identified emergence of an AR-null neuroendocrine-null phenotype with elevated FGF and MAPK pathway activity and demonstrated that pharmacologic inhibitors of MAPK or FGFR can repress the growth of prostate cancer that does not express AR and neuroendocrine markers in vitro and in vivo (Bluemn et al. 2017).
Cell-intrinsic mechanisms of immunoresistance
Several cell-intrinsic mechanisms of immunoresistance have been identified in preclinical models and patients receiving immunotherapy, although most of these observations were in cancer types other than prostate cancer (Pitt et al. 2016; Sharma et al. 2017). Cancer cell-intrinsic immunoresistance can result from a lack of tumor-specific antigen expression (Gubin 2014) or through decreased expression of or mutations in tumor-specific antigens (van Rooij et al. 2013; Schumacher and Schreiber 2015; Ruella et al. 2016). Cancer cell-intrinsic immunoresistance can also stem from defects in the antigen presentation machinery, including proteasome subunits, antigen processing-related transporter, β-2 microglobulin that is involved in human leukocyte antigen class I folding and transport, or the major histocompatibility complex itself (Marincola et al. 2000; Sucker et al. 2014); these defects contribute to the lack of T-cell responses observed in patients with primary resistance (Ribas 2015; McGray and Bramson 2017; Sharma et al. 2017) or acquired resistance (D'Urso et al. 1991; Restifo et al. 1996; Tran et al. 2016; Zaretsky et al. 2016). In addition, activation of the MYC, WNT, and MAPK pathways (Spranger et al. 2015; Casey et al. 2016) and loss of PTEN (Peng et al. 2016) have been implicated in primary and adaptive resistance in melanoma and T-lineage acute lymphoblastic leukemia. As the deregulation of these pathways occurs in a majority of advanced prostate cancers (Robinson et al. 2015), continued investigation of these alterations in immunoresistance of mCRPC is warranted. Similarly, multiple mutations in the interferon γ pathway (IFNGR1, IFNGR2, JAK1/2, and IRF1) have emerged as important regulators of primary, adaptive, and acquired immunoresistance in melanoma (Gao et al. 2016b; Zaretsky et al. 2016; Shin et al. 2017), justifying parallel investigations focused on the basis of the low response rates of mCRPC to immunotherapy.
TME-driven mechanisms of resistance to conventional and novel cancer therapies
Stroma–epithelium interactions play critical roles in the development of the prostate gland (Cunha et al. 1992) and can promote resistance to conventional and targeted cancer therapies and immunotherapy (Fig. 3; Table 2). Knowledge of these heterotypic interactions could lead to novel therapeutic approaches to improve clinical outcomes.
TME-mediated chemoresistance
With respect to chemoresistance mechanisms, WNT16B expression is induced in the TME after cytotoxic chemotherapy, which in turn activates WNT signaling in prostate cancer cells in a paracrine manner, promoting chemoresistance and tumor progression (Sun et al. 2012). Resistance to oxaliplatin, an immunogenic chemotherapeutic agent that is ineffective in aggressive prostate cancer, is mediated by B cells; accordingly, genetic or pharmacologic depletion of B cells restores therapeutic responsiveness in several mouse models of oxaliplatin-refractory prostate cancer (Shalapour et al. 2015). In addition, plasmocytes expressing immunoglobulin A, IL-10, and PD-L1 have been identified as the immunosuppressive B cells directly involved in this process.
Lymphocyte contributions to castration resistance and immunoresistance
ADT can induce B-cell and T-cell infiltration in the TME (Mercader et al. 2001b; Sorrentino et al. 2011). In a prostate cancer transplant model following castration, B-cell recruitment by cancer cell-secreted Cxcl13 promoted CRPC through lymphotoxin secretion and activation of IKKα/STAT3–BMI1 signaling (Ammirante et al. 2010, 2013). A phase 2 clinical trial (NCT02643667) of ibrutinib is currently being conducted as neoadjuvant therapy in localized prostate cancer to evaluate its toxicity and its effect on B-cell and T-cell infiltration (Table 3). In many human tumor types, immunosuppressive FoxP3+ Tregs are present in the TME (Woo et al. 2002; Ormandy et al. 2005; Chaudhary and Elkord 2016) and suppress effector T-cell responses (Josefowicz et al. 2012). In preclinical models of various cancer types, depletion of Tregs restores anti-tumor immunity (Linehan and Goedegebuure 2005; Viehl et al. 2006; Teng et al. 2010) and potentiates the efficacy of anti-PD-1 therapy (Sutmuller et al. 2001; Arce Vargas et al. 2017; Grinberg-Bleyer et al. 2017). In a Pten−/− mouse model of CRPC, castration increased the frequency and activity of antigen-specific CD8+ T cells following immunization; however, the concomitant rapid expansion of Tregs limited CD8+ effector cell function (Tang et al. 2012). This pattern is notable because a higher regulatory:effector T-cell ratio correlates with poor response to anti-CTLA-4 therapy in murine models and patients (Hamid et al. 2011). Ongoing clinical studies are assessing the impact of tumor-infiltrating Tregs on clinical outcomes for patients receiving immunotherapy agents such as anti-CD25 antibodies (daclizumab and basiliximab) and an anti-CD4 antibody (tregalizumab).
Table 3.
Selective clinical trials in prostate cancer
Myeloid cell contributions to castration resistance and immunoresistance
MDSCs and TAMs are powerfully immunosuppressive (Fig. 3). MDSC levels in peripheral blood correlate with response to immunotherapy and survival in cancer patients (Meyer et al. 2014; Santegoets et al. 2014). TAM-derived IL-6 was required for a phenotype of increased AR expression and castration resistance induced by BMP6 overexpression in cancer cells (Lee et al. 2013). Importantly, CSF1R inhibitors (PLX3397 or GW2580) in combination with ADT can reduce TAMs and myeloid cells, suppress CRPC growth (Escamilla et al. 2015), and enhance radiosensitivity of prostate cancer (Xu et al. 2013). In CRPC patients treated with combined prostate GVAX/ipilimumab immunotherapy, high numbers of M-MDSCs before treatment correlated with worse overall survival (Santegoets et al. 2014). However, a phase I trial in mCRPC combining ipilimumab with Prostvac, a vaccine containing PSA and a triad of costimulatory molecules, failed to show a similar correlation between MDSC levels and overall survival (Jochems et al. 2014). While a larger cohort will be needed to define the impact of MDSCs in mCRPC response to immunotherapy, emerging data from many cancer models, including prostate cancer, indicate that MDSC targeting agents such as CSF1R and p110γ inhibitors can potentiate the efficacy of various immunotherapies, including immune checkpoint inhibitors (Highfill et al. 2014; Motoshima et al. 2015; De Henau et al. 2016; Clavijo et al. 2017; Foubert et al. 2017; Lu et al. 2017a; Orillion et al. 2017), adoptive T-cell therapy (Kodumudi et al. 2012; Mok et al. 2014), and dendritic cell vaccination (Laborde et al. 2014). In addition, CpG-STAT3 siRNA conjugates targeting TLR9+ granulocytic MDSCs efficiently abrogated the immunosuppressive activity of MDSCs isolated from prostate cancer patients (Hossain et al. 2015). Given that CSF1R inhibitors and p110 inhibitors target both MDSCs and macrophages, the efficacy of these inhibitors in combination with immunotherapy may be due in part to the elimination of TAMs as well. A presurgical phase 1 clinical trial (NCT03177460) of JNJ-40346527, a CSF1R inhibitor, is currently being evaluated in men with high-risk localized prostate cancer followed by radical prostatectomy for its toxicity and its effect on immune modulation (Table 3). Several early phase “all-comers” clinical trials in advanced solid tumors (NCT02452424, NCT02777710, and NCT02880371) are testing the combination of CSF1R inhibition with checkpoint inhibitors. These studies are supported by recent mCRPC GEMM studies demonstrating dramatic responses when dual checkpoint inhibitors (anti–CTLA-4 and anti–PD-1) were combined with anti-MDSC targeting agents, including cabozantinib and BEZ235; p110 inhibitors (p110δ inhibitor PI-3065 and p110β inhibitor GSK2636771); and a Cxcr1/2 inhibitor (SX-682) (Lu et al. 2017a). Recently, the increased expression of VSIR (VISTA), an inhibitory immune checkpoint molecule, in TAMs after anti-CTLA-4 (ipilimumab) therapy in patients with prostate cancer pointed to a potential compensatory inhibitory pathway in prostate tumors after ipilimumab therapy; thus, VISTA may serve as a potential target for overcoming resistance to anti-CTLA-4 (Gao et al. 2017).
Outlook for next-generation prostate cancer management
Prognostic determination in newly diagnosed prostate cancer
An enduring unmet need is the accurate management of newly diagnosed prostate cancer. Despite the widespread use of PSA screening, four out of five recent randomized clinical trials showed little or no improvement in mortality associated with aggressive treatment of inherently benign disease (Andriole et al. 2009; Schroder et al. 2009, 2012; Ilic et al. 2013). The ongoing CAP and ProtecT trials of 450,000 men (ISRCTN92187251 and ISRCTN20141217), once completed, should provide more conclusive guidance regarding the value of PSA screening (Lane et al. 2010). The inability of clinical or pathologic parameters (PSA levels, TNM stage, and Gleason score) to accurately distinguish the few aggressive cancers from the many indolent cancers remains at the center of the overtreatment problem involving radical prostatectomy and radiation therapy. As noted above, while the management of cancers with a Gleason score of 6 versus those scored ≥8 is relatively straightforward (watchful waiting vs. surgery and/or radiotherapy, respectively), the management of disease with a Gleason score of 7 (3 + 4 or 4 + 3) remains a challenge, fueling efforts to identify molecular correlates of disease outcome. To date, the development of reliable markers has been hampered by the significant intratumor heterogeneity of disease in each patient. Prognostic signatures using transcriptome or copy number alteration data have been developed by comparing profiles in indolent (Gleason score ≤6) and aggressive (Gleason score ≥8) tumors to better predict outcomes (e.g., cancer death, recurrence, and metastasis) of intermediate-risk disease (Gleason score 7) (Cuzick et al. 2011, 2012; Penney et al. 2011; Erho et al. 2013; Irshad et al. 2013; Hieronymus et al. 2014; Sinnott et al. 2017). It is notable that these various signatures show little overlap of specific genes, emphasizing the need for independent validation studies. Additionally, novel biomarkers have been identified to predict aggressive disease in African American men with prostate cancer (Yamoah et al. 2015): Six genes (ERG, AMACR, SPINK1, NKX3-1, GOLM1, and AR) were found to differentially express in African American as compared with European American men; dysregulation of AMACR, ERG, FOXP1, and GSTP1 and mutations in NKX3-1 and RB1 were associated with a decreased risk of pT3 disease in African American men.
Several strategies have been developed to overcome the limitations of tissue-based analyses resulting from sampling bias of highly heterogeneous disease and address the practical challenge of repeat tissue collection in the same patient over long periods. The first strategy uses GEMMs with fully penetrant metastatic and nonmetastatic phenotypes to identify genes that drive metastasis, providing a cross-species filter to refine a human signature capable of predicting lethal outcomes and disease recurrence better than Gleason score and clinical parameters (Ding et al. 2011, 2012). The second strategy takes advantage of liquid biopsy technology to identify biomarkers involving CTCs, cell-free tumor DNA, microRNAs, and microvesicles isolated from the blood, urine, saliva, pleural effusions, and cerebrospinal fluid (Alix-Panabieres et al. 2012; Alix-Panabieres and Pantel 2014; Haber and Velculescu 2014; Yap et al. 2014; Siravegna et al. 2017; Wan et al. 2017). Baseline CTC count (Danila et al. 2007, 2011; de Bono et al. 2008; Scher et al. 2009; Goldkorn et al. 2014; Scher et al. 2015) and changes in post-treatment CTC count (Olmos et al. 2009; Scher et al. 2009; Goldkorn et al. 2014) were found to be prognostic factors for overall survival in prostate cancer patients with metastasis. Analysis of cell-free tumor DNA and tumor biopsy with next-generation sequencing in patients with prostate cancer who received second-generation anti-androgens have identified genomic aberrations in AR, RB1 loss, alterations in DNA damage repair genes and PI3K pathway genes, and activating mutations in the CTNNB1 gene, suggesting that cell-free tumor DNA can be used to monitor therapy response, identify emerging mechanisms of resistance (Joseph et al. 2013; Antonarakis et al. 2014; Carreira et al. 2014; Azad et al. 2015; Lallous et al. 2016; Wyatt et al. 2016; De Laere et al. 2017), predict progression-free survival (De Laere et al. 2017), and stratify patients for agents targeting DNA repair pathways (e.g., PARP inhibitors) (Annala et al. 2017). AR splicing variants (e.g., AR-V7) have drawn intense interest as a liquid biopsy prognostic biomarker for predicting therapy resistance. The presence of AR-V7 in CTCs, bone marrow biopsy, or plasma-derived exosomal RNA from mCRPC patients can predict response to enzalutamide or abiraterone treatment (Antonarakis et al. 2014; Efstathiou et al. 2015; Del Re et al. 2017), although a recent report failed to show the predictive potential of the presence of AR-V7 and AR-V9 in whole blood (To et al. 2018). The basis for these discrepancies may relate to the need for larger sample size to firmly establish whether these variants are useful prognostic biomarkers.
Advances in computational science have enabled the accumulation and integration of clinical information together with massive data sets, including genomic, transcriptomic, epigenomic, proteomic, and metabolomic profiles from biopsies, prostatectomies, and/or single cells (Wang and Navin 2015). Collectively, the integration of these approaches should better define disease variability and tumor evolution and lead to identification of biomarkers for managing newly diagnosed cases via robust prognostic determinant biomarkers, monitoring the emergence of therapeutic resistance, and guiding optimal therapy regimens for specific disease subsets. At the same time, a challenge remains in how to efficiently analyze and integrate these massive multidimensional data sets. Artificial intelligence approaches, including deep learning, enable computers to learn and improve continuously in performing a particular task with the accumulation of new data and associated outcomes (Silver et al. 2017). These approaches are yielding decision support systems showing promise in the diagnosis of eye diseases and pneumonia (Kermany et al. 2018) and the accurate discrimination of various cancer tissues, cancer subtypes, biomarkers, and immunohistochemical scores (Khosravi et al. 2018). Further development in artificial intelligence-driven algorithms is expected to accelerate the development of accurate biomarkers and management algorithms for predicting patient survival, responses to treatment, drug resistance, and minimal residual disease; that is, by adopting a biomarker-driven precision therapy approach and using predictive treatment biomarkers, physicians could more accurately assign patients to the best available standard of care that offers the maximal benefit for each patient. In addition, patients whose disease does not respond to the frontline standard of care can be matched into the best clinical trials that are most likely to benefit them—preferably in an adaptive clinical trial framework with longitudinal profiling.
Science-driven therapeutic development
The rapid development in computational approaches has identified and will continue to identify novel driver genes in prostate cancer. For example, the TMPRSS2-ERG translocation was identified by outlier gene expression analysis by Tomlins et al. (2005). In addition, cross-species genome-wide regulatory network (interactome) analyses for human and mouse prostate cancer not only identified FOXM1 and CENPF as synergistic master regulators of prostate cancer malignancy (Aytes et al. 2014) but also predicted drug efficacy in human cancer and identified drugs and drug combinations that inhibited the activity of FOXM1 and CENPF (Mitrofanova et al. 2015).
However, unlike the success of monotherapy or combination therapy in other cancer types, effective strategies have yet to emerge in the treatment of prostate cancer despite the development of checkpoint blockade immunotherapy, as discussed above (Kwon et al. 2014; Beer et al. 2017). Preclinical mechanistic studies have revealed novel combination strategies for the treatment of prostate cancer, leading to numerous clinical trials (Table 3). First, targeting androgen signaling in combination with novel targeted therapies is being explored in androgen-responsive tumors. Specifically, enzalutamide, which down-regulates BRCA1 expression in prostate cancer cells that have wild-type BRCA1, was found to potentiate response to the PARP inhibitor olaparib in preclinical models (Li et al. 2017a). A phase 2 randomized trial of olaparib combined with abiraterone (NCT01972217) provided clinical efficacy benefits in mCRPC patients (Clarke et al. 2018). AR inhibitors in combination with PI3K inhibitors targeting reciprocal negative regulation between AR and AKT signaling show synergy in preclinical models. Further clinical trials are needed to evaluate the efficacy of these treatment regimens in human prostate cancer patients. Phase 1/2 trials of enzalutamide in combination with BET domain protein inhibitors (ZEN003694 and GS-5829) are currently under way to target the BRD4 and AR cross-talk in mCRPC (NCT02711956 and NCT02607228). Second, ADT, which modulates the priming of tumor-specific adaptive immune responses (Mercader et al. 2001a; Drake et al. 2005; Sutherland et al. 2005; Morse and McNeel 2010), has led to a clinical trial (KEYNOTE-365) testing the potential synergy of anti-PD-1 (pembrolizumab) plus enzalutamide (NCT02861573) (Yu et al. 2017) and anti-PD-L1 (atezolizumab) plus enzalutamide (NCT03016312). Third, preclinical models have demonstrated that AR+ adenocarcinoma can transdifferentiate into AR-independent NEPC or small cell carcinoma; moreover, genes such as MYCN, BRN2 (also called POU3F2), SOX2, AURKA, and EZH2 have been shown to play a critical role in these androgen-insensitive tumors, and monotherapy targeting these genes (e.g., AURKA inhibitor) or combination therapy with an EZH2 inhibitor (GSK126 or EPZ-6438) and enzalutamide has shown therapeutic benefits in preclinical models (Beltran et al. 2011, 2016b; Dardenne et al. 2016; Lee et al. 2016b; Ku et al. 2017; Mu et al. 2017). A phase II study (NCT01799278) demonstrated that a subset of NEPC patients with clinical and pathologically defined features may benefit from single-agent AURKA inhibitor (alisertib) treatment (Beltran et al. 2016a). A longitudinal study of adenocarcinoma to NEPC or small cell progression would allow us to identify key driver genes in these processes and provide novel therapeutic targets for combination therapy.
Another promising avenue for new therapeutic strategies for prostate cancer is targeting DNA damage repair pathways. PARP inhibitors have yielded a high response rate in a subset of mCRPC patients with DNA repair defects (Mateo et al. 2015) and has been reported to induce PD-L1 expression in breast cancer (Jiao et al. 2017). Synthetic lethality was observed by targeting AR signaling and the PARP pathway in prostate cancer cells (Asim et al. 2017). In addition, enzalutamide in combination with CC-115, a dual inhibitor for DNA-PK and mammalian target of rapamycin (mTOR), is currently being tested in a phase 1 trial (NCT02833883) to target the cross-talk between AR signaling and DNA-PK. Defects in the MMR pathway, which are associated with microsatellite instability and high mutational load, were shown to correlate with clinical response to the anti-PD-1 agent pembrolizumab across 12 solid cancer types, including prostate cancer, resulting in FDA approval for pembrolizumab in MMR-defective cancers (Le et al. 2017). Ongoing clinical trials (for olaparib combined with PD-1 inhibitor pembrolizumab, PD-L1 inhibitor durvalumab, and abiraterone; NCT02861573 and NCT02484404) (Karzai et al. 2017; Yu et al. 2017) and future clinical trials will allow us to test the efficacy of agents targeting the DNA damage repair pathways in combinations with other therapies. Multiple resistance mechanisms to PARP inhibitors have been identified in ovarian and breast cancers, including secondary mutations in BRCA1/2 to restore the wild-type allele or the ORF that forms new non-wild-type isoforms and loss of 53BP1 (Lord and Ashworth 2013), and are likely to operate in prostate cancer.
Immunotherapy has transformed the standard of care for several malignancies, and a deeper understanding of the effects of conventional and targeted therapies on anti-tumor immunity has informed the design of combinations showing increased rates of complete and durable clinical responses (Gotwals et al. 2017). Ongoing clinical trials of immunotherapy in combination with other therapies are being conducted in prostate cancer, including the combination of the vaccine Prostvac with docetaxel (NCT02649855) or with the PD-1 inhibitor nivolumab and/or the CTLA-4 inhibitor ipilimumab (NCT02933255 and NCT02506114) and the combination of radium-223 with atezolizumab (NCT02814669) or sipuleucel-T (NCT02463799). In various preclinical cancer models, potent synergistic effects have been observed for agents targeting the immunosuppressive TME (MDSCs, TAMs, and Tregs) in combination with checkpoint inhibitors, prompting the launch of new clinical trials. Future studies should design combination trials based on a strong scientific rationales that include longitudinal biopsies (blood and tumor samples) from the treatment-naïve, pretreatment, on-treatment, and post-treatment (resistant) stages of disease to better understand the resistance mechanisms in prostate cancer, correlate response to genotypes, and identify prognostic biomarkers.
There are challenges associated with the development of effective combinations of conventional therapies, targeted therapies, and immunotherapies: Comprehensive understanding of the effects of these therapies on the patient's immune system is lacking; the efficacy, toxicity, and tolerability associated with combination therapies need to be determined through optimization of dosing regimens and sequencing, and approaches for prioritizing various combination therapies need to be developed (Gotwals et al. 2017). Given that the cancer genome and the TME coevolve during disease progression and treatment, it is important to model these interactions in refined genetic model systems as well as perform longitudinal omics analyses of patients under treatment and subsequently link all of these profiling data to clinical information to elucidate how genomic information and the TME landscape can inform and improve patient care (Chin et al. 2015). A deep understanding of prostate cancer biology and genomics, the advent of sophisticated profiling technology and artificial intelligence-based decision systems, and the capacity for multiple-armed adaptive clinical trials with longitudinal profiling all place the field in a position to save and improve the lives of many men with this disease.
Acknowledgments
We thank Christopher J. Logothetis, Filippo G. Giancotti, and Prasenjit Dey for critical reading of the manuscript and insightful comments. G.W. is supported by the Prostate Cancer Moon Shot and Institutional Research Grant (IRG) Program at the University of Texas MD Anderson Cancer Center. G.W. is also supported by the University of Texas Star Award and National Institutes of Health grant R00CA194289. D.Z. is supported by Prostate Cancer Foundation Young Investigator Award 17YOUN18. R.A.D. is supported by MD Anderson's Prostate Cancer Moon Shot.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.315739.118.
References
- Abdulkadir SA, Magee JA, Peters TJ, Kaleem Z, Naughton CK, Humphrey PA, Milbrandt J. 2002. Conditional loss of Nkx3.1 in adult mice induces prostatic intraepithelial neoplasia. Mol Cell Biol 22: 1495–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aceto N, Toner M, Maheswaran S, Haber DA. 2015. En route to metastasis: circulating tumor cell clusters and epithelial-to-mesenchymal transition. Trends Cancer 1: 44–52. [DOI] [PubMed] [Google Scholar]
- Albany C, Alva AS, Aparicio AM, Singal R, Yellapragada S, Sonpavde G, Hahn NM. 2011. Epigenetics in prostate cancer. Prostate Cancer 2011: 580318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alix-Panabieres C, Pantel K. 2014. Challenges in circulating tumour cell research. Nat Rev Cancer 14: 623–631. [DOI] [PubMed] [Google Scholar]
- Alix-Panabieres C, Schwarzenbach H, Pantel K. 2012. Circulating tumor cells and circulating tumor DNA. Annu Rev Med 63: 199–215. [DOI] [PubMed] [Google Scholar]
- Allis CD, Jenuwein T. 2016. The molecular hallmarks of epigenetic control. Nat Rev Genet 17: 487–500. [DOI] [PubMed] [Google Scholar]
- Ammirante M, Luo JL, Grivennikov S, Nedospasov S, Karin M. 2010. B-cell-derived lymphotoxin promotes castration-resistant prostate cancer. Nature 464: 302–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammirante M, Kuraishy AI, Shalapour S, Strasner A, Ramirez-Sanchez C, Zhang W, Shabaik A, Karin M. 2013. An IKKα–E2F1–BMI1 cascade activated by infiltrating B cells controls prostate regeneration and tumor recurrence. Genes Dev 27: 1435–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammirante M, Shalapour S, Kang Y, Jamieson CA, Karin M. 2014. Tissue injury and hypoxia promote malignant progression of prostate cancer by inducing CXCL13 expression in tumor myofibroblasts. Proc Natl Acad Sci 111: 14776–14781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An J, Ren S, Murphy SJ, Dalangood S, Chang C, Pang X, Cui Y, Wang L, Pan Y, Zhang X, et al. 2015. Truncated ERG oncoproteins from TMPRSS2-ERG fusions are resistant to SPOP-mediated proteasome degradation. Mol Cell 59: 904–916. [DOI] [PubMed] [Google Scholar]
- Andriole GL, Crawford ED, Grubb RL III, Buys SS, Chia D, Church TR, Fouad MN, Gelmann EP, Kvale PA, Reding DJ, et al. 2009. Mortality results from a randomized prostate-cancer screening trial. N Engl J Med 360: 1310–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annala M, Struss WJ, Warner EW, Beja K, Vandekerkhove G, Wong A, Khalaf D, Seppala IL, So A, Lo G, et al. 2017. Treatment outcomes and tumor loss of heterozygosity in germline DNA repair-deficient prostate cancer. Eur Urol 72: 34–42. [DOI] [PubMed] [Google Scholar]
- Antonarakis ES, Lu C, Wang H, Luber B, Nakazawa M, Roeser JC, Chen Y, Mohammad TA, Chen Y, Fedor HL, et al. 2014. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med 371: 1028–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonarakis ES, Lu C, Luber B, Wang H, Chen Y, Zhu Y, Silberstein JL, Taylor MN, Maughan BL, Denmeade SR, et al. 2017. Clinical significance of androgen receptor splice variant-7 mRNA detection in circulating tumor cells of men with metastatic castration-resistant prostate cancer treated with first- and second-line abiraterone and enzalutamide. J Clin Oncol 35: 2149–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio S, Hidalgo M, Kung AL. 2015. Examining the utility of patient-derived xenograft mouse models. Nat Rev Cancer 15: 311–316. [DOI] [PubMed] [Google Scholar]
- Arce Vargas F, Furness AJS, Solomon I, Joshi K, Mekkaoui L, Lesko MH, Miranda Rota E, Dahan R, Georgiou A, Sledzinska A, et al. 2017. Fc-Optimized anti-CD25 depletes tumor-infiltrating regulatory T cells and synergizes with PD-1 blockade to eradicate established tumors. Immunity 46: 577–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armenia J, Wankowicz SAM, Liu D, Gao J, Kundra R, Reznik E, Chatila WK, Chakravarty D, Han GC, Coleman I, et al. 2018. The long tail of oncogenic drivers in prostate cancer. Nat Genet 50: 645–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora VK, Schenkein E, Murali R, Subudhi SK, Wongvipat J, Balbas MD, Shah N, Cai L, Efstathiou E, Logothetis C, et al. 2013. Glucocorticoid receptor confers resistance to antiandrogens by bypassing androgen receptor blockade. Cell 155: 1309–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asangani IA, Dommeti VL, Wang X, Malik R, Cieslik M, Yang R, Escara-Wilke J, Wilder-Romans K, Dhanireddy S, Engelke C, et al. 2014. Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature 510: 278–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asangani IA, Wilder-Romans K, Dommeti VL, Krishnamurthy PM, Apel IJ, Escara-Wilke J, Plymate SR, Navone NM, Wang S, Feng FY, et al. 2016. BET bromodomain inhibitors enhance efficacy and disrupt resistance to AR antagonists in the treatment of prostate cancer. Mol Cancer Res 14: 324–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asim M, Tarish F, Zecchini HI, Sanjiv K, Gelali E, Massie CE, Baridi A, Warren AY, Zhao W, Ogris C, et al. 2017. Synthetic lethality between androgen receptor signalling and the PARP pathway in prostate cancer. Nat Commun 8: 374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audia JE, Campbell RM. 2016. Histone modifications and cancer. Cold Spring Harb Perspect Biol 8: a019521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augsten M. 2014. Cancer-associated fibroblasts as another polarized cell type of the tumor microenvironment. Front Oncol 4: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala G, Tuxhorn JA, Wheeler TM, Frolov A, Scardino PT, Ohori M, Wheeler M, Spitler J, Rowley DR. 2003. Reactive stroma as a predictor of biochemical-free recurrence in prostate cancer. Clin Cancer Res 9: 4792–4801. [PubMed] [Google Scholar]
- Aytes A, Mitrofanova A, Lefebvre C, Alvarez MJ, Castillo-Martin M, Zheng T, Eastham JA, Gopalan A, Pienta KJ, Shen MM, et al. 2014. Cross-species regulatory network analysis identifies a synergistic interaction between FOXM1 and CENPF that drives prostate cancer malignancy. Cancer Cell 25: 638–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azad AA, Volik SV, Wyatt AW, Haegert A, Le Bihan S, Bell RH, Anderson SA, McConeghy B, Shukin R, Bazov J, et al. 2015. Androgen receptor gene aberrations in circulating cell-free DNA: biomarkers of therapeutic resistance in castration-resistant prostate cancer. Clin Cancer Res 21: 2315–2324. [DOI] [PubMed] [Google Scholar]
- Baca SC, Prandi D, Lawrence MS, Mosquera JM, Romanel A, Drier Y, Park K, Kitabayashi N, MacDonald TY, Ghandi M, et al. 2013. Punctuated evolution of prostate cancer genomes. Cell 153: 666–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, Theurillat JP, White TA, Stojanov P, Van Allen E, Stransky N, et al. 2012. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet 44: 685–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri CE, Chinnaiyan AM, Lerner SP, Swanton C, Rubin MA. 2017. The emergence of precision urologic oncology: a collaborative review on biomarker-driven therapeutics. Eur Urol 71: 237–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barron DA, Rowley DR. 2012. The reactive stroma microenvironment and prostate cancer progression. Endocr Relat Cancer 19: R187–R204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthel SR, Hays DL, Yazawa EM, Opperman M, Walley KC, Nimrichter L, Burdick MM, Gillard BM, Moser MT, Pantel K, et al. 2013. Definition of molecular determinants of prostate cancer cell bone extravasation. Cancer Res 73: 942–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedard PL, Hansen AR, Ratain MJ, Siu LL. 2013. Tumour heterogeneity in the clinic. Nature 501: 355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beer TM, Armstrong AJ, Rathkopf DE, Loriot Y, Sternberg CN, Higano CS, Iversen P, Bhattacharya S, Carles J, Chowdhury S, et al. 2014. Enzalutamide in metastatic prostate cancer before chemotherapy. N Engl J Med 371: 424–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beer TM, Kwon ED, Drake CG, Fizazi K, Logothetis C, Gravis G, Ganju V, Polikoff J, Saad F, Humanski P, et al. 2017. Randomized, double-blind, phase III trial of ipilimumab versus placebo in asymptomatic or minimally symptomatic patients with metastatic chemotherapy-naive castration-resistant prostate cancer. J Clin Oncol 35: 40–47. [DOI] [PubMed] [Google Scholar]
- Beltran H, Demichelis F. 2015. Prostate cancer: intrapatient heterogeneity in prostate cancer. Nat Rev Urol 12: 430–431. [DOI] [PubMed] [Google Scholar]
- Beltran H, Rickman DS, Park K, Chae SS, Sboner A, MacDonald TY, Wang Y, Sheikh KL, Terry S, Tagawa ST, et al. 2011. Molecular characterization of neuroendocrine prostate cancer and identification of new drug targets. Cancer Discov 1: 487–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltran H, Danila D, Montgomery B, Szmulewitz R, Vaishampayan U, Armstrong A, Stein M, Hoimes C, Pinski J, Scher H, et al. 2016a. A phase 2 study of the aurora kinase A inhibitor alisertib for patients with neuroendocrine prostate cancer (NEPC). Ann Oncol 27: LBA29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltran H, Prandi D, Mosquera JM, Benelli M, Puca L, Cyrta J, Marotz C, Giannopoulou E, Chakravarthi BV, Varambally S, et al. 2016b. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat Med 22: 298–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benafif S, Eeles R. 2016. Genetic predisposition to prostate cancer. Br Med Bull 120: 75–89. [DOI] [PubMed] [Google Scholar]
- Benafif S, Kote-Jarai Z, Eeles RA. 2018. A review of prostate cancer genome wide association studies (GWAS). Cancer Epidemiol Biomarkers Prev 27: 845–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-David U, Ha G, Tseng YY, Greenwald NF, Oh C, Shih J, McFarland JM, Wong B, Boehm JS, Beroukhim R, et al. 2017. Patient-derived xenografts undergo mouse-specific tumor evolution. Nat Genet 49: 1567–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berquin IM, Min Y, Wu R, Wu H, Chen YQ. 2005. Expression signature of the mouse prostate. J Biol Chem 280: 36442–36451. [DOI] [PubMed] [Google Scholar]
- Bezzi M, Seitzer N, Ishikawa T, Reschke M, Chen M, Wang G, Mitchell C, Ng C, Katon J, Lunardi A, et al. 2018. Diverse genetic-driven immune landscapes dictate tumor progression through distinct mechanisms. Nat Med 24: 165–175. [DOI] [PubMed] [Google Scholar]
- Bhatia-Gaur R, Donjacour AA, Sciavolino PJ, Kim M, Desai N, Young P, Norton CR, Gridley T, Cardiff RD, Cunha GR, et al. 1999. Roles for Nkx3.1 in prostate development and cancer. Genes Dev 13: 966–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bill-Axelson A, Holmberg L, Garmo H, Rider JR, Taari K, Busch C, Nordling S, Haggman M, Andersson SO, Spangberg A, et al. 2014. Radical prostatectomy or watchful waiting in early prostate cancer. N Engl J Med 370: 932–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingle L, Brown NJ, Lewis CE. 2002. The role of tumour-associated macrophages in tumour progression: implications for new anticancer therapies. J Pathol 196: 254–265. [DOI] [PubMed] [Google Scholar]
- Birbach A, Casanova E, Schmid JA. 2009. A Probasin–MerCreMer BAC allows inducible recombination in the mouse prostate. Genesis 47: 757–764. [DOI] [PubMed] [Google Scholar]
- Biswas SK, Mantovani A. 2010. Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat Immunol 11: 889–896. [DOI] [PubMed] [Google Scholar]
- Blattner M, Liu D, Robinson BD, Huang D, Poliakov A, Gao D, Nataraj S, Deonarine LD, Augello MA, Sailer V, et al. 2017. SPOP mutation drives prostate tumorigenesis in vivo through coordinate regulation of PI3K/mTOR and AR signaling. Cancer Cell 31: 436–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bluemn EG, Coleman IM, Lucas JM, Coleman RT, Hernandez-Lopez S, Tharakan R, Bianchi-Frias D, Dumpit RF, Kaipainen A, Corella AN, et al. 2017. Androgen receptor pathway-independent prostate cancer is sustained through FGF signaling. Cancer Cell 32: 474–489 e476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Body JJ, Casimiro S, Costa L. 2015. Targeting bone metastases in prostate cancer: improving clinical outcome. Nat Rev Urol 12: 340–356. [DOI] [PubMed] [Google Scholar]
- Bose R, Karthaus WR, Armenia J, Abida W, Iaquinta PJ, Zhang Z, Wongvipat J, Wasmuth EV, Shah N, Sullivan PS, et al. 2017. ERF mutations reveal a balance of ETS factors controlling prostate oncogenesis. Nature 546: 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutin AT, Liao WT, Wang M, Hwang SS, Karpinets TV, Cheung H, Chu GC, Jiang S, Hu J, Chang K, et al. 2017. Oncogenic Kras drives invasion and maintains metastases in colorectal cancer. Genes Dev 31: 370–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutros PC, Fraser M, Harding NJ, de Borja R, Trudel D, Lalonde E, Meng A, Hennings-Yeomans PH, McPherson A, Sabelnykova VY, et al. 2015. Spatial genomic heterogeneity within localized, multifocal prostate cancer. Nat Genet 47: 736–745. [DOI] [PubMed] [Google Scholar]
- Brabletz T, Kalluri R, Nieto MA, Weinberg RA. 2018. EMT in cancer. Nat Rev Cancer 18: 128–134. [DOI] [PubMed] [Google Scholar]
- Branco MR, Ficz G, Reik W. 2011. Uncovering the role of 5-hydroxymethylcytosine in the epigenome. Nat Rev Genet 13: 7–13. [DOI] [PubMed] [Google Scholar]
- Breyer JP, Avritt TG, McReynolds KM, Dupont WD, Smith JR. 2012. Confirmation of the HOXB13 G84E germline mutation in familial prostate cancer. Cancer Epidemiol Biomarkers Prev 21: 1348–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronte V, Wang M, Overwijk WW, Surman DR, Pericle F, Rosenberg SA, Restifo NP. 1998. Apoptotic death of CD8+ T lymphocytes after immunization: induction of a suppressive population of Mac-1+/Gr-1+ cells. J Immunol 161: 5313–5320. [PMC free article] [PubMed] [Google Scholar]
- Bronte V, Brandau S, Chen SH, Colombo MP, Frey AB, Greten TF, Mandruzzato S, Murray PJ, Ochoa A, Ostrand-Rosenberg S, et al. 2016. Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards. Nat Commun 7: 12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brusa D, Simone M, Gontero P, Spadi R, Racca P, Micari J, Degiuli M, Carletto S, Tizzani A, Matera L. 2013. Circulating immunosuppressive cells of prostate cancer patients before and after radical prostatectomy: profile comparison. Int J Urol 20: 971–978. [DOI] [PubMed] [Google Scholar]
- Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T. 2005. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 434: 913–917. [DOI] [PubMed] [Google Scholar]
- Burkhardt L, Fuchs S, Krohn A, Masser S, Mader M, Kluth M, Bachmann F, Huland H, Steuber T, Graefen M, et al. 2013. CHD1 is a 5q21 tumor suppressor required for ERG rearrangement in prostate cancer. Cancer Res 73: 2795–2805. [DOI] [PubMed] [Google Scholar]
- Cai C, He HH, Chen S, Coleman I, Wang H, Fang Z, Chen S, Nelson PS, Liu XS, Brown M, et al. 2011. Androgen receptor gene expression in prostate cancer is directly suppressed by the androgen receptor through recruitment of lysine-specific demethylase 1. Cancer Cell 20: 457–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai C, He HH, Gao S, Chen S, Yu Z, Gao Y, Chen S, Chen MW, Zhang J, Ahmed M, et al. 2014. Lysine-specific demethylase 1 has dual functions as a major regulator of androgen receptor transcriptional activity. Cell Rep 9: 1618–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network. 2015. The molecular taxonomy of primary prostate cancer. Cell 163: 1011–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannarile MA, Weisser M, Jacob W, Jegg AM, Ries CH, Ruttinger D. 2017. Colony-stimulating factor 1 receptor (CSF1R) inhibitors in cancer therapy. J Immunother Cancer 5: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreira S, Romanel A, Goodall J, Grist E, Ferraldeschi R, Miranda S, Prandi D, Lorente D, Frenel JS, Pezaro C, et al. 2014. Tumor clone dynamics in lethal prostate cancer. Sci Transl Med 6: 254ra125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver BS, Tran J, Gopalan A, Chen Z, Shaikh S, Carracedo A, Alimonti A, Nardella C, Varmeh S, Scardino PT, et al. 2009. Aberrant ERG expression cooperates with loss of PTEN to promote cancer progression in the prostate. Nat Genet 41: 619–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver BS, Chapinski C, Wongvipat J, Hieronymus H, Chen Y, Chandarlapaty S, Arora VK, Le C, Koutcher J, Scher H, et al. 2011. Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell 19: 575–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey SC, Tong L, Li Y, Do R, Walz S, Fitzgerald KN, Gouw AM, Baylot V, Gutgemann I, Eilers M, et al. 2016. MYC regulates the antitumor immune response through CD47 and PD-L1. Science 352: 227–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang KH, Li R, Kuri B, Lotan Y, Roehrborn CG, Liu J, Vessella R, Nelson PS, Kapur P, Guo X, et al. 2013. A gain-of-function mutation in DHT synthesis in castration-resistant prostate cancer. Cell 154: 1074–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary B, Elkord E. 2016. Regulatory T cells in the tumor microenvironment and cancer progression: role and therapeutic targeting. Vaccines (Basel) 4: E28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Zhang J, Sampieri K, Clohessy JG, Mendez L, Gonzalez-Billalabeitia E, Liu XS, Lee YR, Fung J, Katon JM, et al. 2018. An aberrant SREBP-dependent lipogenic program promotes metastatic prostate cancer. Nat Genet 50: 206–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin L, Wargo JA, Spring DJ, Kantarjian H, Futreal PA. 2015. Cancer genomics in clinical context. Trends Cancer 1: 36–43. [DOI] [PubMed] [Google Scholar]
- Cho H, Herzka T, Zheng W, Qi J, Wilkinson JE, Bradner JE, Robinson BD, Castillo-Martin M, Cordon-Cardo C, Trotman LC. 2014. RapidCaP, a novel GEM model for metastatic prostate cancer analysis and therapy, reveals myc as a driver of Pten-mutant metastasis. Cancer Discov 4: 318–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi N, Zhang B, Zhang L, Ittmann M, Xin L. 2012. Adult murine prostate basal and luminal cells are self-sustained lineages that can both serve as targets for prostate cancer initiation. Cancer Cell 21: 253–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua CW, Shibata M, Lei M, Toivanen R, Barlow LJ, Bergren SK, Badani KK, McKiernan JM, Benson MC, Hibshoosh H, et al. 2014. Single luminal epithelial progenitors can generate prostate organoids in culture. Nat Cell Biol 16: 951–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke N, Wiechno P, Alekseev B, Sala N, Jones R, Kocak I, Chiuri VE, Jassem J, Flechon A, Redfern C, et al. 2018. Olaparib combined with abiraterone in patients with metastatic castration-resistant prostate cancer: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet Oncol 19: 975–986. [DOI] [PubMed] [Google Scholar]
- Clavijo PE, Moore EC, Chen J, Davis RJ, Friedman J, Kim Y, Van Waes C, Chen Z, Allen CT. 2017. Resistance to CTLA-4 checkpoint inhibition reversed through selective elimination of granulocytic myeloid cells. Oncotarget 8: 55804–55820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H. 2016. Modeling development and disease with organoids. Cell 165: 1586–1597. [DOI] [PubMed] [Google Scholar]
- Condamine T, Ramachandran I, Youn JI, Gabrilovich DI. 2015. Regulation of tumor metastasis by myeloid-derived suppressor cells. Annu Rev Med 66: 97–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooney KA. 2017. Inherited predisposition to prostate cancer: from gene discovery to clinical impact. Trans Am Clin Climatol Assoc 128: 14–23. [PMC free article] [PubMed] [Google Scholar]
- Cooper CS, Eeles R, Wedge DC, Van Loo P, Gundem G, Alexandrov LB, Kremeyer B, Butler A, Lynch AG, Camacho N, et al. 2015. Analysis of the genetic phylogeny of multifocal prostate cancer identifies multiple independent clonal expansions in neoplastic and morphologically normal prostate tissue. Nat Genet 47: 367–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig M, Ying C, Loberg RD. 2008. Co-inoculation of prostate cancer cells with U937 enhances tumor growth and angiogenesis in vivo. J Cell Biochem 103: 1–8. [DOI] [PubMed] [Google Scholar]
- Culig Z. 2004. Androgen receptor cross-talk with cell signalling pathways. Growth Factors 22: 179–184. [DOI] [PubMed] [Google Scholar]
- Cunha GR, Donjacour AA, Cooke PS, Mee S, Bigsby RM, Higgins SJ, Sugimura Y. 1987. The endocrinology and developmental biology of the prostate. Endocr Rev 8: 338–362. [DOI] [PubMed] [Google Scholar]
- Cunha GR, Alarid ET, Turner T, Donjacour AA, Boutin EL, Foster BA. 1992. Normal and abnormal development of the male urogenital tract. Role of androgens, mesenchymal-epithelial interactions, and growth factors. J Androl 13: 465–475. [PubMed] [Google Scholar]
- Cuzick J, Swanson GP, Fisher G, Brothman AR, Berney DM, Reid JE, Mesher D, Speights VO, Stankiewicz E, Foster CS, et al. 2011. Prognostic value of an RNA expression signature derived from cell cycle proliferation genes in patients with prostate cancer: a retrospective study. Lancet Oncol 12: 245–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuzick J, Berney DM, Fisher G, Mesher D, Moller H, Reid JE, Perry M, Park J, Younus A, Gutin A, et al. 2012. Prognostic value of a cell cycle progression signature for prostate cancer death in a conservatively managed needle biopsy cohort. Br J Cancer 106: 1095–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai XP, Gan WJ, Li XN, Wang SQ, Zhang W, Huang L, Liu SW, Zhong Q, Guo JP, Zhang JF, et al. 2017. Prostate cancer-associated SPOP mutations confer resistance to BET inhibitors through stabilization of BRD4. Nat Med 23: 1063–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danila DC, Heller G, Gignac GA, Gonzalez-Espinoza R, Anand A, Tanaka E, Lilja H, Schwartz L, Larson S, Fleisher M, et al. 2007. Circulating tumor cell number and prognosis in progressive castration-resistant prostate cancer. Clin Cancer Res 13: 7053–7058. [DOI] [PubMed] [Google Scholar]
- Danila DC, Anand A, Sung CC, Heller G, Leversha MA, Cao L, Lilja H, Molina A, Sawyers CL, Fleisher M, et al. 2011. TMPRSS2-ERG status in circulating tumor cells as a predictive biomarker of sensitivity in castration-resistant prostate cancer patients treated with abiraterone acetate. Eur Urol 60: 897–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dardenne E, Beltran H, Benelli M, Gayvert K, Berger A, Puca L, Cyrta J, Sboner A, Noorzad Z, MacDonald T, et al. 2016. N-Myc induces an EZH2-mediated transcriptional program driving neuroendocrine prostate cancer. Cancer Cell 30: 563–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta K, Muders M, Zhang H, Tindall DJ. 2010. Mechanism of lymph node metastasis in prostate cancer. Future Oncol 6: 823–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidsson S, Ohlson AL, Andersson SO, Fall K, Meisner A, Fiorentino M, Andren O, Rider JR. 2013. CD4 helper T cells, CD8 cytotoxic T cells, and FOXP3+ regulatory T cells with respect to lethal prostate cancer. Mod Pathol 26: 448–455. [DOI] [PubMed] [Google Scholar]
- Dawson MA, Kouzarides T. 2012. Cancer epigenetics: from mechanism to therapy. Cell 150: 12–27. [DOI] [PubMed] [Google Scholar]
- de Bono JS, Scher HI, Montgomery RB, Parker C, Miller MC, Tissing H, Doyle GV, Terstappen LW, Pienta KJ, Raghavan D. 2008. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res 14: 6302–6309. [DOI] [PubMed] [Google Scholar]
- de Bono JS, Oudard S, Ozguroglu M, Hansen S, Machiels JP, Kocak I, Gravis G, Bodrogi I, Mackenzie MJ, Shen L, et al. 2010. Prednisone plus cabazitaxel or mitoxantrone for metastatic castration-resistant prostate cancer progressing after docetaxel treatment: a randomised open-label trial. Lancet 376: 1147–1154. [DOI] [PubMed] [Google Scholar]
- de Bono JS, Logothetis CJ, Molina A, Fizazi K, North S, Chu L, Chi KN, Jones RJ, Goodman OB Jr., Saad F, et al. 2011. Abiraterone and increased survival in metastatic prostate cancer. N Engl J Med 364: 1995–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Henau O, Rausch M, Winkler D, Campesato LF, Liu C, Cymerman DH, Budhu S, Ghosh A, Pink M, Tchaicha J, et al. 2016. Overcoming resistance to checkpoint blockade therapy by targeting PI3Kγ in myeloid cells. Nature 539: 443–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Laere B, van Dam PJ, Whitington T, Mayrhofer M, Diaz EH, Van den Eynden G, Vandebroek J, Del-Favero J, Van Laere S, Dirix L, et al. 2017. Comprehensive profiling of the androgen receptor in liquid biopsies from castration-resistant prostate cancer reveals novel intra-AR structural variation and splice variant expression patterns. Eur Urol 72: 192–200. [DOI] [PubMed] [Google Scholar]
- Del Re M, Biasco E, Crucitta S, Derosa L, Rofi E, Orlandini C, Miccoli M, Galli L, Falcone A, Jenster GW, et al. 2017. The detection of androgen receptor splice variant 7 in plasma-derived exosomal RNA strongly predicts resistance to hormonal therapy in metastatic prostate cancer patients. Eur Urol 71: 680–687. [DOI] [PubMed] [Google Scholar]
- Di Mitri D, Toso A, Chen JJ, Sarti M, Pinton S, Jost TR, D'Antuono R, Montani E, Garcia-Escudero R, Guccini I, et al. 2014. Tumour-infiltrating Gr-1+ myeloid cells antagonize senescence in cancer. Nature 515: 134–137. [DOI] [PubMed] [Google Scholar]
- Ding Z, Wu CJ, Chu GC, Xiao Y, Ho D, Zhang J, Perry SR, Labrot ES, Wu X, Lis R, et al. 2011. SMAD4-dependent barrier constrains prostate cancer growth and metastatic progression. Nature 470: 269–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z, Wu CJ, Jaskelioff M, Ivanova E, Kost-Alimova M, Protopopov A, Chu GC, Wang G, Lu X, Labrot ES, et al. 2012. Telomerase reactivation following telomere dysfunction yields murine prostate tumors with bone metastases. Cell 148: 896–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingo-Domenech J, Vidal SJ, Rodriguez-Bravo V, Castillo-Martin M, Quinn SA, Rodriguez-Barrueco R, Bonal DM, Charytonowicz E, Gladoun N, de la Iglesia-Vicente J, et al. 2012. Suppression of acquired docetaxel resistance in prostate cancer through depletion of notch- and hedgehog-dependent tumor-initiating cells. Cancer Cell 22: 373–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dow LE. 2015. Modeling disease in vivo with CRISPR/Cas9. Trends Mol Med 21: 609–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake CG, Doody AD, Mihalyo MA, Huang CT, Kelleher E, Ravi S, Hipkiss EL, Flies DB, Kennedy EP, Long M, et al. 2005. Androgen ablation mitigates tolerance to a prostate/prostate cancer-restricted antigen. Cancer Cell 7: 239–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Urso CM, Wang ZG, Cao Y, Tatake R, Zeff RA, Ferrone S. 1991. Lack of HLA class I antigen expression by cultured melanoma cells FO-1 due to a defect in B2m gene expression. J Clin Invest 87: 284–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta D, Heo I, Clevers H. 2017. Disease modeling in stem cell-derived 3D organoid systems. Trends Mol Med 23: 393–410. [DOI] [PubMed] [Google Scholar]
- Eeles RA, Kote-Jarai Z, Giles GG, Olama AA, Guy M, Jugurnauth SK, Mulholland S, Leongamornlert DA, Edwards SM, Morrison J, et al. 2008. Multiple newly identified loci associated with prostate cancer susceptibility. Nat Genet 40: 316–321. [DOI] [PubMed] [Google Scholar]
- Eeles RA, Kote-Jarai Z, Al Olama AA, Giles GG, Guy M, Severi G, Muir K, Hopper JL, Henderson BE, Haiman CA, et al. 2009. Identification of seven new prostate cancer susceptibility loci through a genome-wide association study. Nat Genet 41: 1116–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eeles RA, Olama AA, Benlloch S, Saunders EJ, Leongamornlert DA, Tymrakiewicz M, Ghoussaini M, Luccarini C, Dennis J, Jugurnauth-Little S, et al. 2013. Identification of 23 new prostate cancer susceptibility loci using the iCOGS custom genotyping array. Nat Genet 45: 385–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eeles R, Goh C, Castro E, Bancroft E, Guy M, Al Olama AA, Easton D, Kote-Jarai Z. 2014. The genetic epidemiology of prostate cancer and its clinical implications. Nat Rev Urol 11: 18–31. [DOI] [PubMed] [Google Scholar]
- Efstathiou E, Titus M, Wen S, Hoang A, Karlou M, Ashe R, Tu SM, Aparicio A, Troncoso P, Mohler J, et al. 2015. Molecular characterization of enzalutamide-treated bone metastatic castration-resistant prostate cancer. Eur Urol 67: 53–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egeblad M, Nakasone ES, Werb Z. 2010. Tumors as organs: complex tissues that interface with the entire organism. Dev Cell 18: 884–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellwood-Yen K, Graeber TG, Wongvipat J, Iruela-Arispe ML, Zhang J, Matusik R, Thomas GV, Sawyers CL. 2003. Myc-driven murine prostate cancer shares molecular features with human prostate tumors. Cancer Cell 4: 223–238. [DOI] [PubMed] [Google Scholar]
- Epstein JI, Allsbrook WC Jr., Amin MB, Egevad LL, Committee IG. 2005. The 2005 International Society of Urological Pathology (ISUP) Consensus Conference on Gleason Grading of Prostatic Carcinoma. Am J Surg Pathol 29: 1228–1242. [DOI] [PubMed] [Google Scholar]
- Epstein JI, Amin MB, Beltran H, Lotan TL, Mosquera JM, Reuter VE, Robinson BD, Troncoso P, Rubin MA. 2014. Proposed morphologic classification of prostate cancer with neuroendocrine differentiation. Am J Surg Pathol 38: 756–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein JI, Egevad L, Amin MB, Delahunt B, Srigley JR, Humphrey PA, Grading C. 2016. The 2014 International Society of Urological Pathology (ISUP) Consensus Conference on Gleason Grading of Prostatic Carcinoma: definition of grading patterns and proposal for a new grading system. Am J Surg Pathol 40: 244–252. [DOI] [PubMed] [Google Scholar]
- Erho N, Crisan A, Vergara IA, Mitra AP, Ghadessi M, Buerki C, Bergstralh EJ, Kollmeyer T, Fink S, Haddad Z, et al. 2013. Discovery and validation of a prostate cancer genomic classifier that predicts early metastasis following radical prostatectomy. PLoS One 8: e66855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escamilla J, Schokrpur S, Liu C, Priceman SJ, Moughon D, Jiang Z, Pouliot F, Magyar C, Sung JL, Xu J, et al. 2015. CSF1 receptor targeting in prostate cancer reverses macrophage-mediated resistance to androgen blockade therapy. Cancer Res 75: 950–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan L, Zhang F, Xu S, Cui X, Hussain A, Fazli L, Gleave M, Dong X, Qi J. 2018. Histone demethylase JMJD1A promotes alternative splicing of AR variant 7 (AR-V7) in prostate cancer cells. Proc Natl Acad Sci 115: E4584–E4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, et al. 2005. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434: 917–921. [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Koldobskiy MA, Gondor A. 2016. Epigenetic modulators, modifiers and mediators in cancer aetiology and progression. Nat Rev Genet 17: 284–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, Li YS, Bhagwat N, Vasanthakumar A, Fernandez HF, et al. 2010. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 18: 553–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fizazi K, Tran N, Fein L, Matsubara N, Rodriguez-Antolin A, Alekseev BY, Ozguroglu M, Ye D, Feyerabend S, Protheroe A, et al. 2017. Abiraterone plus prednisone in metastatic, castration-sensitive prostate cancer. N Engl J Med 377: 352–360. [DOI] [PubMed] [Google Scholar]
- Flavahan WA, Gaskell E, Bernstein BE. 2017. Epigenetic plasticity and the hallmarks of cancer. Science 357: eaal2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foubert P, Kaneda MM, Varner JA. 2017. PI3Kγ activates integrin α4 and promotes immune suppressive myeloid cell polarization during tumor progression. Cancer Immunol Res 5: 957–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier PG, Juarez P, Jiang G, Clines GA, Niewolna M, Kim HS, Walton HW, Peng XH, Liu Y, Mohammad KS, et al. 2015. The TGF-β signaling regulator PMEPA1 suppresses prostate cancer metastases to bone. Cancer Cell 27: 809–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser M, Sabelnykova VY, Yamaguchi TN, Heisler LE, Livingstone J, Huang V, Shiah YJ, Yousif F, Lin X, Masella AP, et al. 2017. Genomic hallmarks of localized, non-indolent prostate cancer. Nature 541: 359–364. [DOI] [PubMed] [Google Scholar]
- Fujii T, Shimada K, Asai O, Tanaka N, Fujimoto K, Hirao K, Konishi N. 2013. Immunohistochemical analysis of inflammatory cells in benign and precancerous lesions and carcinoma of the prostate. Pathobiology 80: 119–126. [DOI] [PubMed] [Google Scholar]
- Gabrilovich DI, Nagaraj S. 2009. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol 9: 162–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabrilovich DI, Ostrand-Rosenberg S, Bronte V. 2012. Coordinated regulation of myeloid cells by tumours. Nat Rev Immunol 12: 253–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajewski TF, Schreiber H, Fu YX. 2013. Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol 14: 1014–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galletti G, Matov A, Beltran H, Fontugne J, Miguel Mosquera J, Cheung C, MacDonald TY, Sung M, O'Toole S, Kench JG, et al. 2014. ERG induces taxane resistance in castration-resistant prostate cancer. Nat Commun 5: 5548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan W, Dai X, Lunardi A, Li Z, Inuzuka H, Liu P, Varmeh S, Zhang J, Cheng L, Sun Y, et al. 2015. SPOP promotes ubiquitination and degradation of the ERG oncoprotein to suppress prostate cancer progression. Mol Cell 59: 917–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gannon PO, Poisson AO, Delvoye N, Lapointe R, Mes-Masson AM, Saad F. 2009. Characterization of the intra-prostatic immune cell infiltration in androgen-deprived prostate cancer patients. J Immunol Methods 348: 9–17. [DOI] [PubMed] [Google Scholar]
- Gao D, Vela I, Sboner A, Iaquinta PJ, Karthaus WR, Gopalan A, Dowling C, Wanjala JN, Undvall EA, Arora VK, et al. 2014. Organoid cultures derived from patients with advanced prostate cancer. Cell 159: 176–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao D, Zhan Y, Di W, Moore AR, Sher JJ, Guan Y, Wang S, Zhang Z, Murphy DA, Sawyers CL, et al. 2016a. A Tmprss2-CreERT2 knock-in mouse model for cancer genetic studies on prostate and colon. PLoS One 11: e0161084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Shi LZ, Zhao H, Chen J, Xiong L, He Q, Chen T, Roszik J, Bernatchez C, Woodman SE, et al. 2016b. Loss of IFN-γ pathway genes in tumor cells as a mechanism of resistance to anti-CTLA-4 therapy. Cell 167: 397–404.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Ward JF, Pettaway CA, Shi LZ, Subudhi SK, Vence LM, Zhao H, Chen J, Chen H, Efstathiou E, et al. 2017. VISTA is an inhibitory immune checkpoint that is increased after ipilimumab therapy in patients with prostate cancer. Nat Med 23: 551–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia AJ, Ruscetti M, Arenzana TL, Tran LM, Bianci-Frias D, Sybert E, Priceman SJ, Wu L, Nelson PS, Smale ST, et al. 2014. Pten null prostate epithelium promotes localized myeloid-derived suppressor cell expansion and immune suppression during tumor initiation and progression. Mol Cell Biol 34: 2017–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng C, He B, Xu L, Barbieri CE, Eedunuri VK, Chew SA, Zimmermann M, Bond R, Shou J, Li C, et al. 2013. Prostate cancer-associated mutations in speckle-type POZ protein (SPOP) regulate steroid receptor coactivator 3 protein turnover. Proc Natl Acad Sci 110: 6997–7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genovese G, Carugo A, Tepper J, Robinson FS, Li L, Svelto M, Nezi L, Corti D, Minelli R, Pettazzoni P, et al. 2017. Synthetic vulnerabilities of mesenchymal subpopulations in pancreatic cancer. Nature 542: 362–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannoni E, Bianchini F, Masieri L, Serni S, Torre E, Calorini L, Chiarugi P. 2010. Reciprocal activation of prostate cancer cells and cancer-associated fibroblasts stimulates epithelial–mesenchymal transition and cancer stemness. Cancer Res 70: 6945–6956. [DOI] [PubMed] [Google Scholar]
- Gleason DF, Mellinger GT. 1974. Prediction of prognosis for prostatic adenocarcinoma by combined histological grading and clinical staging. J Urol 111: 58–64. [DOI] [PubMed] [Google Scholar]
- Goldgar DE, Easton DF, Cannon-Albright LA, Skolnick MH. 1994. Systematic population-based assessment of cancer risk in first-degree relatives of cancer probands. J Natl Cancer Inst 86: 1600–1608. [DOI] [PubMed] [Google Scholar]
- Goldkorn A, Ely B, Quinn DI, Tangen CM, Fink LM, Xu T, Twardowski P, Van Veldhuizen PJ, Agarwal N, Carducci MA, et al. 2014. Circulating tumor cell counts are prognostic of overall survival in SWOG S0421: a phase III trial of docetaxel with or without atrasentan for metastatic castration-resistant prostate cancer. J Clin Oncol 32: 1136–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein AS, Lawson DA, Cheng D, Sun W, Garraway IP, Witte ON. 2008. Trop2 identifies a subpopulation of murine and human prostate basal cells with stem cell characteristics. Proc Natl Acad Sci 105: 20882–20887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein AS, Huang J, Guo C, Garraway IP, Witte ON. 2010. Identification of a cell of origin for human prostate cancer. Science 329: 568–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gollapudi K, Galet C, Grogan T, Zhang H, Said JW, Huang J, Elashoff D, Freedland SJ, Rettig M, Aronson WJ. 2013. Association between tumor-associated macrophage infiltration, high grade prostate cancer, and biochemical recurrence after radical prostatectomy. Am J Cancer Res 3: 523–529. [PMC free article] [PubMed] [Google Scholar]
- Goodwin JF, Schiewer MJ, Dean JL, Schrecengost RS, de Leeuw R, Han S, Ma T, Den RB, Dicker AP, Feng FY, et al. 2013. A hormone-DNA repair circuit governs the response to genotoxic insult. Cancer Discov 3: 1254–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin JF, Kothari V, Drake JM, Zhao S, Dylgjeri E, Dean JL, Schiewer MJ, McNair C, Jones JK, Aytes A, et al. 2015. DNA-PKcs-mediated transcriptional regulation drives prostate cancer progression and metastasis. Cancer Cell 28: 97–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotwals P, Cameron S, Cipolletta D, Cremasco V, Crystal A, Hewes B, Mueller B, Quaratino S, Sabatos-Peyton C, Petruzzelli L, et al. 2017. Prospects for combining targeted and conventional cancer therapy with immunotherapy. Nat Rev Cancer 17: 286–301. [DOI] [PubMed] [Google Scholar]
- Grabowska MM, DeGraff DJ, Yu X, Jin RJ, Chen Z, Borowsky AD, Matusik RJ. 2014. Mouse models of prostate cancer: picking the best model for the question. Cancer Metastasis Rev 33: 377–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graff JN, Alumkal JJ, Drake CG, Thomas GV, Redmond WL, Farhad M, Cetnar JP, Ey FS, Bergan RC, Slottke R, et al. 2016. Early evidence of anti-PD-1 activity in enzalutamide-resistant prostate cancer. Oncotarget 7: 52810–52817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, Quist MJ, Jing X, Lonigro RJ, Brenner JC, et al. 2012. The mutational landscape of lethal castration-resistant prostate cancer. Nature 487: 239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinberg-Bleyer Y, Oh H, Desrichard A, Bhatt DM, Caron R, Chan TA, Schmid RM, Klein U, Hayden MS, Ghosh S. 2017. NF-κB c-Rel is crucial for the regulatory T cell immune checkpoint in cancer. Cell 170: 1096–1108.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groner AC, Cato L, de Tribolet-Hardy J, Bernasocchi T, Janouskova H, Melchers D, Houtman R, Cato ACB, Tschopp P, Gu L, et al. 2016. TRIM24 is an oncogenic transcriptional activator in prostate cancer. Cancer Cell 29: 846–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubin MM. 2014. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature 515: 577–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudmundsson J, Sulem P, Gudbjartsson DF, Blondal T, Gylfason A, Agnarsson BA, Benediktsdottir KR, Magnusdottir DN, Orlygsdottir G, Jakobsdottir M, et al. 2009. Genome-wide association and replication studies identify four variants associated with prostate cancer susceptibility. Nat Genet 41: 1122–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guise TA, Mohammad KS, Clines G, Stebbins EG, Wong DH, Higgins LS, Vessella R, Corey E, Padalecki S, Suva L, et al. 2006. Basic mechanisms responsible for osteolytic and osteoblastic bone metastases. Clin Cancer Res 12: 6213s–6216s. [DOI] [PubMed] [Google Scholar]
- Gundem G, Van Loo P, Kremeyer B, Alexandrov LB, Tubio JMC, Papaemmanuil E, Brewer DS, Kallio HML, Hognas G, Annala M, et al. 2015. The evolutionary history of lethal metastatic prostate cancer. Nature 520: 353–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber DA, Velculescu VE. 2014. Blood-based analyses of cancer: circulating tumor cells and circulating tumor DNA. Cancer Discov 4: 650–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffner J, Potiron E, Bouye S, Puech P, Leroy X, Lemaitre L, Villers A. 2009. Peripheral zone prostate cancers: location and intraprostatic patterns of spread at histopathology. Prostate 69: 276–282. [DOI] [PubMed] [Google Scholar]
- Haffner MC, Mosbruger T, Esopi DM, Fedor H, Heaphy CM, Walker DA, Adejola N, Gurel M, Hicks J, Meeker AK, et al. 2013. Tracking the clonal origin of lethal prostate cancer. J Clin Invest 123: 4918–4922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall CL, Bafico A, Dai J, Aaronson SA, Keller ET. 2005. Prostate cancer cells promote osteoblastic bone metastases through Wnts. Cancer Res 65: 7554–7560. [DOI] [PubMed] [Google Scholar]
- Hamdy FC, Donovan JL, Lane JA, Mason M, Metcalfe C, Holding P, Davis M, Peters TJ, Turner EL, Martin RM, et al. 2016. 10-year outcomes after monitoring, surgery, or radiotherapy for localized prostate cancer. N Engl J Med 375: 1415–1424. [DOI] [PubMed] [Google Scholar]
- Hamid O, Schmidt H, Nissan A, Ridolfi L, Aamdal S, Hansson J, Guida M, Hyams DM, Gomez H, Bastholt L, et al. 2011. A prospective phase II trial exploring the association between tumor microenvironment biomarkers and clinical activity of ipilimumab in advanced melanoma. J Transl Med 9: 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Coussens LM. 2012. Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell 21: 309–322. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. 2011. Hallmarks of cancer: the next generation. Cell 144: 646–674. [DOI] [PubMed] [Google Scholar]
- He WW, Sciavolino PJ, Wing J, Augustus M, Hudson P, Meissner PS, Curtis RT, Shell BK, Bostwick DG, Tindall DJ, et al. 1997. A novel human prostate-specific, androgen-regulated homeobox gene (NKX3.1) that maps to 8p21, a region frequently deleted in prostate cancer. Genomics 43: 69–77. [DOI] [PubMed] [Google Scholar]
- Hearn JWD, AbuAli G, Reichard CA, Reddy CA, Magi-Galluzzi C, Chang KH, Carlson R, Rangel L, Reagan K, Davis BJ, et al. 2016. HSD3B1 and resistance to androgen-deprivation therapy in prostate cancer: a retrospective, multicohort study. Lancet Oncol 17: 1435–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensley PJ, Kyprianou N. 2012. Modeling prostate cancer in mice: limitations and opportunities. J Androl 33: 133–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henzler C, Li Y, Yang R, McBride T, Ho Y, Sprenger C, Liu G, Coleman I, Lakely B, Li R, et al. 2016. Truncation and constitutive activation of the androgen receptor by diverse genomic rearrangements in prostate cancer. Nat Commun 7: 13668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer J, Kwong LN, Lowe SW, Chin L. 2010. Non-germline genetically engineered mouse models for translational cancer research. Nat Rev Cancer 10: 470–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieronymus H, Sawyers CL. 2012. Traversing the genomic landscape of prostate cancer from diagnosis to death. Nat Genet 44: 613–614. [DOI] [PubMed] [Google Scholar]
- Hieronymus H, Schultz N, Gopalan A, Carver BS, Chang MT, Xiao Y, Heguy A, Huberman K, Bernstein M, Assel M, et al. 2014. Copy number alteration burden predicts prostate cancer relapse. Proc Natl Acad Sci 111: 11139–11144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Highfill SL, Cui Y, Giles AJ, Smith JP, Zhang H, Morse E, Kaplan RN, Mackall CL. 2014. Disruption of CXCR2-mediated MDSC tumor trafficking enhances anti-PD1 efficacy. Sci Transl Med 6: 237ra267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson MC, Astapova I, Cheng S, Lee LJ, Verhoeven MC, Choi E, Balk SP, Hollenberg AN. 2005. The androgen receptor recruits nuclear receptor corepressor (N-CoR) in the presence of mifepristone via its N and C termini revealing a novel molecular mechanism for androgen receptor antagonists. J Biol Chem 280: 6511–6519. [DOI] [PubMed] [Google Scholar]
- Hong MK, Macintyre G, Wedge DC, Van Loo P, Patel K, Lunke S, Alexandrov LB, Sloggett C, Cmero M, Marass F, et al. 2015. Tracking the origins and drivers of subclonal metastatic expansion in prostate cancer. Nat Commun 6: 6605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino A, Costa-Silva B, Shen TL, Rodrigues G, Hashimoto A, Tesic Mark M, Molina H, Kohsaka S, Di Giannatale A, Ceder S, et al. 2015. Tumour exosome integrins determine organotropic metastasis. Nature 527: 329–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain DM, Pal SK, Moreira D, Duttagupta P, Zhang Q, Won H, Jones J, D'Apuzzo M, Forman S, Kortylewski M. 2015. TLR9-targeted STAT3 silencing abrogates immunosuppressive activity of myeloid-derived suppressor cells from prostate cancer patients. Clin Cancer Res 21: 3771–3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu CH, Peng KL, Kang ML, Chen YR, Yang YC, Tsai CH, Chu CS, Jeng YM, Chen YT, Lin FM, et al. 2012. TET1 suppresses cancer invasion by activating the tissue inhibitors of metalloproteinases. Cell Rep 2: 568–579. [DOI] [PubMed] [Google Scholar]
- Hu CD, Choo R, Huang J. 2015. Neuroendocrine differentiation in prostate cancer: a mechanism of radioresistance and treatment failure. Front Oncol 5: 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q, Whitington T, Gao P, Lindberg JF, Yang Y, Sun J, Vaisanen MR, Szulkin R, Annala M, Yan J, et al. 2014. A prostate cancer susceptibility allele at 6q22 increases RFX6 expression by modulating HOXB13 chromatin binding. Nat Genet 46: 126–135. [DOI] [PubMed] [Google Scholar]
- Huang FW, Mosquera JM, Garofalo A, Oh C, Baco M, Amin-Mansour A, Rabasha B, Bahl S, Mullane SA, Robinson BD, et al. 2017. Exome sequencing of African-American prostate cancer reveals loss-of-function ERF mutations. Cancer Discov 7: 973–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard GK, Mutton LN, Khalili M, McMullin RP, Hicks JL, Bianchi-Frias D, Horn LA, Kulac I, Moubarek MS, Nelson PS, et al. 2016. Combined MYC activation and Pten loss are sufficient to create genomic instability and lethal metastatic prostate cancer. Cancer Res 76: 283–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huggins C, Hodges CV. 1941. Studies on prostatic cancer. I. The effect of castration, of estrogen and of androgen injection on serum phosphatases in metastatic carcinoma of the prostate. Cancer Res 1: 293–297. [DOI] [PubMed] [Google Scholar]
- Idorn M, Kollgaard T, Kongsted P, Sengelov L, Thor Straten P. 2014. Correlation between frequencies of blood monocytic myeloid-derived suppressor cells, regulatory T cells and negative prognostic markers in patients with castration-resistant metastatic prostate cancer. Cancer Immunol Immunother 63: 1177–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilic D, Neuberger MM, Djulbegovic M, Dahm P. 2013. Screening for prostate cancer. Cochrane Database Syst Rev 10.1002/14651858.CD004720.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai Y, Tsukahara S, Asada S, Sugimoto Y. 2004. Phytoestrogens/flavonoids reverse breast cancer resistance protein/ABCG2-mediated multidrug resistance. Cancer Res 64: 4346–4352. [DOI] [PubMed] [Google Scholar]
- Irshad S, Bansal M, Castillo-Martin M, Zheng T, Aytes A, Wenske S, Le Magnen C, Guarnieri P, Sumazin P, Benson MC, et al. 2013. A molecular signature predictive of indolent prostate cancer. Sci Transl Med 5: 202ra122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ittmann M, Huang J, Radaelli E, Martin P, Signoretti S, Sullivan R, Simons BW, Ward JM, Robinson BD, Chu GC, et al. 2013. Animal models of human prostate cancer: the consensus report of the New York meeting of the Mouse Models of Human Cancers Consortium Prostate Pathology Committee. Cancer Res 73: 2718–2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iversen P, Madsen PO, Corle DK. 1995. Radical prostatectomy versus expectant treatment for early carcinoma of the prostate. Twenty-three year follow-up of a prospective randomized study. Scand J Urol Nephrol Suppl 172: 65–72. [PubMed] [Google Scholar]
- James ND, de Bono JS, Spears MR, Clarke NW, Mason MD, Dearnaley DP, Ritchie AWS, Amos CL, Gilson C, Jones RJ, et al. 2017. Abiraterone for prostate cancer not previously treated with hormone therapy. N Engl J Med 377: 338–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janouskova H, El Tekle G, Bellini E, Udeshi ND, Rinaldi A, Ulbricht A, Bernasocchi T, Civenni G, Losa M, Svinkina T, et al. 2017. Opposing effects of cancer-type-specific SPOP mutants on BET protein degradation and sensitivity to BET inhibitors. Nat Med 23: 1046–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins RB, Qian J, Lieber MM, Bostwick DG. 1997. Detection of c-myc oncogene amplification and chromosomal anomalies in metastatic prostatic carcinoma by fluorescence in situ hybridization. Cancer Res 57: 524–531. [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. 2001. Translating the histone code. Science 293: 1074–1080. [DOI] [PubMed] [Google Scholar]
- Jeronimo C, Bastian PJ, Bjartell A, Carbone GM, Catto JW, Clark SJ, Henrique R, Nelson WG, Shariat SF. 2011. Epigenetics in prostate cancer: biologic and clinical relevance. Eur Urol 60: 753–766. [DOI] [PubMed] [Google Scholar]
- Jiao S, Xia W, Yamaguchi H, Wei Y, Chen MK, Hsu JM, Hsu JL, Yu WH, Du Y, Lee HH, et al. 2017. PARP inhibitor upregulates PD-L1 expression and enhances cancer-associated immunosuppression. Clin Cancer Res 23: 3711–3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jochems C, Tucker JA, Tsang KY, Madan RA, Dahut WL, Liewehr DJ, Steinberg SM, Gulley JL, Schlom J. 2014. A combination trial of vaccine plus ipilimumab in metastatic castration-resistant prostate cancer patients: immune correlates. Cancer Immunol Immunother 63: 407–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DH, Nakashima T, Sanchez OH, Kozieradzki I, Komarova SV, Sarosi I, Morony S, Rubin E, Sarao R, Hojilla CV, et al. 2006. Regulation of cancer cell migration and bone metastasis by RANKL. Nature 440: 692–696. [DOI] [PubMed] [Google Scholar]
- Josefowicz SZ, Lu LF, Rudensky AY. 2012. Regulatory T cells: mechanisms of differentiation and function. Annu Rev Immunol 30: 531–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph JD, Lu N, Qian J, Sensintaffar J, Shao G, Brigham D, Moon M, Maneval EC, Chen I, Darimont B, et al. 2013. A clinically relevant androgen receptor mutation confers resistance to second-generation antiandrogens enzalutamide and ARN-509. Cancer Discov 3: 1020–1029. [DOI] [PubMed] [Google Scholar]
- Jung Y, Kim JK, Shiozawa Y, Wang J, Mishra A, Joseph J, Berry JE, McGee S, Lee E, Sun H, et al. 2013. Recruitment of mesenchymal stem cells into prostate tumours promotes metastasis. Nat Commun 4: 1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung Y, Wang J, Lee E, McGee S, Berry JE, Yumoto K, Dai J, Keller ET, Shiozawa Y, Taichman RS. 2015. Annexin 2–CXCL12 interactions regulate metastatic cell targeting and growth in the bone marrow. Mol Cancer Res 13: 197–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junttila MR, de Sauvage FJ. 2013. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature 501: 346–354. [DOI] [PubMed] [Google Scholar]
- Kakarla S, Song XT, Gottschalk S. 2012. Cancer-associated fibroblasts as targets for immunotherapy. Immunotherapy 4: 1129–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalluri R. 2016. The biology and function of fibroblasts in cancer. Nat Rev Cancer 16: 582–598. [DOI] [PubMed] [Google Scholar]
- Kalluri R, Weinberg RA. 2009. The basics of epithelial–mesenchymal transition. J Clin Invest 119: 1420–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantoff PW, Higano CS, Shore ND, Berger ER, Small EJ, Penson DF, Redfern CH, Ferrari AC, Dreicer R, Sims RB, et al. 2010. Sipuleucel-T immunotherapy for castration-resistant prostate cancer. N Engl J Med 363: 411–422. [DOI] [PubMed] [Google Scholar]
- Karlou M, Tzelepi V, Efstathiou E. 2010. Therapeutic targeting of the prostate cancer microenvironment. Nat Rev Urol 7: 494–509. [DOI] [PubMed] [Google Scholar]
- Karnoub AE, Dash AB, Vo AP, Sullivan A, Brooks MW, Bell GW, Richardson AL, Polyak K, Tubo R, Weinberg RA. 2007. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature 449: 557–563. [DOI] [PubMed] [Google Scholar]
- Karthaus WR, Iaquinta PJ, Drost J, Gracanin A, van Boxtel R, Wongvipat J, Dowling CM, Gao D, Begthel H, Sachs N, et al. 2014. Identification of multipotent luminal progenitor cells in human prostate organoid cultures. Cell 159: 163–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karzai F, Madan RA, Owens H, Hankin A, Couvillon A, Houston ND, Fakhrejahani F, Bilusic M, Theoret MR, Cordes LM, et al. 2017. A phase II study of the anti-programmed death ligand-1 antibody durvalumab (D; MEDI4736) in combination with PARP inhibitor, olaparib (O), in metastatic castration-resistant prostate cancer (mCRPC). J Clin Oncol 35: 162–162. [Google Scholar]
- Kermany DS, Goldbaum M, Cai W, Valentim CCS, Liang H, Baxter SL, McKeown A, Yang G, Wu X, Yan F, et al. 2018. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell 172: 1122–1131.e9. [DOI] [PubMed] [Google Scholar]
- Kersten K, de Visser KE, van Miltenburg MH, Jonkers J. 2017. Genetically engineered mouse models in oncology research and cancer medicine. EMBO Mol Med 9: 137–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khosravi P, Kazemi E, Imielinski M, Elemento O, Hajirasouliha I. 2018. Deep convolutional neural networks enable discrimination of heterogeneous digital pathology images. EBioMedicine 27: 317–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TD, Jin F, Shin S, Oh S, Lightfoot SA, Grande JP, Johnson AJ, van Deursen JM, Wren JD, Janknecht R. 2016. Histone demethylase JMJD2A drives prostate tumorigenesis through transcription factor ETV1. J Clin Invest 126: 706–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King JC, Xu J, Wongvipat J, Hieronymus H, Carver BS, Leung DH, Taylor BS, Sander C, Cardiff RD, Couto SS, et al. 2009. Cooperativity of TMPRSS2-ERG with PI3-kinase pathway activation in prostate oncogenesis. Nat Genet 41: 524–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinney SR, Moser MT, Pascual M, Greally JM, Foster BA, Karpf AR. 2010. Opposing roles of Dnmt1 in early- and late-stage murine prostate cancer. Mol Cell Biol 30: 4159–4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura T, Qian BZ, Soong D, Cassetta L, Noy R, Sugano G, Kato Y, Li J, Pollard JW. 2015. CCL2-induced chemokine cascade promotes breast cancer metastasis by enhancing retention of metastasis-associated macrophages. J Exp Med 212: 1043–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klezovitch O, Chevillet J, Mirosevich J, Roberts RL, Matusik RJ, Vasioukhin V. 2004. Hepsin promotes prostate cancer progression and metastasis. Cancer Cell 6: 185–195. [DOI] [PubMed] [Google Scholar]
- Klezovitch O, Risk M, Coleman I, Lucas JM, Null M, True LD, Nelson PS, Vasioukhin V. 2008. A causal role for ERG in neoplastic transformation of prostate epithelium. Proc Natl Acad Sci 105: 2105–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodumudi KN, Weber A, Sarnaik AA, Pilon-Thomas S. 2012. Blockade of myeloid-derived suppressor cells after induction of lymphopenia improves adoptive T cell therapy in a murine model of melanoma. J Immunol 189: 5147–5154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzeszinski JY, Wan Y. 2015. New therapeutic targets for cancer bone metastasis. Trends Pharmacol Sci 36: 360–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku SY, Rosario S, Wang Y, Mu P, Seshadri M, Goodrich ZW, Goodrich MM, Labbe DP, Gomez EC, Wang J, et al. 2017. Rb1 and Trp53 cooperate to suppress prostate cancer lineage plasticity, metastasis, and antiandrogen resistance. Science 355: 78–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulis M, Esteller M. 2010. DNA methylation and cancer. Adv Genet 70: 27–56. [DOI] [PubMed] [Google Scholar]
- Kumar A, Coleman I, Morrissey C, Zhang X, True LD, Gulati R, Etzioni R, Bolouri H, Montgomery B, White T, et al. 2016a. Substantial interindividual and limited intraindividual genomic diversity among tumors from men with metastatic prostate cancer. Nat Med 22: 369–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V, Patel S, Tcyganov E, Gabrilovich DI. 2016b. The nature of myeloid-derived suppressor cells in the tumor microenvironment. Trends Immunol 37: 208–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V, Donthireddy L, Marvel D, Condamine T, Wang F, Lavilla-Alonso S, Hashimoto A, Vonteddu P, Behera R, Goins MA, et al. 2017. Cancer-associated fibroblasts neutralize the anti-tumor effect of CSF1 receptor blockade by inducing PMN-MDSC infiltration of tumors. Cancer Cell 32: 654–668.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon ED, Drake CG, Scher HI, Fizazi K, Bossi A, van den Eertwegh AJ, Krainer M, Houede N, Santos R, Mahammedi H, et al. 2014. Ipilimumab versus placebo after radiotherapy in patients with metastatic castration-resistant prostate cancer that had progressed after docetaxel chemotherapy (CA184-043): a multicentre, randomised, double-blind, phase 3 trial. Lancet Oncol 15: 700–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laborde RR, Lin Y, Gustafson MP, Bulur PA, Dietz AB. 2014. Cancer vaccines in the world of immune suppressive monocytes (CD14+HLA-DRlo/neg cells): the gateway to improved responses. Front Immunol 5: 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallous N, Volik SV, Awrey S, Leblanc E, Tse R, Murillo J, Singh K, Azad AA, Wyatt AW, LeBihan S, et al. 2016. Functional analysis of androgen receptor mutations that confer anti-androgen resistance identified in circulating cell-free DNA from prostate cancer patients. Genome Biol 17: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamouille S, Xu J, Derynck R. 2014. Molecular mechanisms of epithelial–mesenchymal transition. Nat Rev Mol Cell Biol 15: 178–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanciotti M, Masieri L, Raspollini MR, Minervini A, Mari A, Comito G, Giannoni E, Carini M, Chiarugi P, Serni S. 2014. The role of M1 and M2 macrophages in prostate cancer in relation to extracapsular tumor extension and biochemical recurrence after radical prostatectomy. Biomed Res Int 2014: 486798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane JA, Hamdy FC, Martin RM, Turner EL, Neal DE, Donovan JL. 2010. Latest results from the UK trials evaluating prostate cancer screening and treatment: the CAP and ProtecT studies. Eur J Cancer 46: 3095–3101. [DOI] [PubMed] [Google Scholar]
- Lange EM. 2010. Identification of genetic risk factors for prostate cancer: analytic approaches using hereditary prostate cancer families. In Male reproductive cancers: epidemiology, pathology and genetics (ed. Foulkes WD, Cooney KA), pp. 203–228. Springer, New York. [Google Scholar]
- Lawson DA, Xin L, Lukacs RU, Cheng D, Witte ON. 2007. Isolation and functional characterization of murine prostate stem cells. Proc Natl Acad Sci 104: 181–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson DA, Zong Y, Memarzadeh S, Xin L, Huang J, Witte ON. 2010. Basal epithelial stem cells are efficient targets for prostate cancer initiation. Proc Natl Acad Sci 107: 2610–2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, Lu S, Kemberling H, Wilt C, Luber BS, et al. 2017. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 357: 409–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Shen MM. 2015. Cell types of origin for prostate cancer. Curr Opin Cell Biol 37: 35–41. [DOI] [PubMed] [Google Scholar]
- Lee JT Jr., Steelman LS, McCubrey JA. 2004. Phosphatidylinositol 3′-kinase activation leads to multidrug resistance protein-1 expression and subsequent chemoresistance in advanced prostate cancer cells. Cancer Res 64: 8397–8404. [DOI] [PubMed] [Google Scholar]
- Lee GT, Jung YS, Ha YS, Kim JH, Kim WJ, Kim IY. 2013. Bone morphogenetic protein-6 induces castration resistance in prostate cancer cells through tumor infiltrating macrophages. Cancer Sci 104: 1027–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E, Wang J, Yumoto K, Jung Y, Cackowski FC, Decker AM, Li Y, Franceschi RT, Pienta KJ, Taichman RS. 2016a. DNMT1 regulates epithelial–mesenchymal transition and cancer stem cells, which promotes prostate cancer metastasis. Neoplasia 18: 553–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JK, Phillips JW, Smith BA, Park JW, Stoyanova T, McCaffrey EF, Baertsch R, Sokolov A, Meyerowitz JG, Mathis C, et al. 2016b. N-Myc drives neuroendocrine prostate cancer initiated from human prostate epithelial cells. Cancer Cell 29: 536–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Bishop AC, Alyamani M, Garcia JA, Dreicer R, Bunch D, Liu J, Upadhyay SK, Auchus RJ, Sharifi N. 2015. Conversion of abiraterone to D4A drives anti-tumour activity in prostate cancer. Nature 523: 347–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Karanika S, Yang G, Wang J, Park S, Broom BM, Manyam GC, Wu W, Luo Y, Basourakos S, et al. 2017a. Androgen receptor inhibitor-induced ‘BRCAness’ and PARP inhibition are synthetically lethal for castration-resistant prostate cancer. Sci Signal 10: eaam7479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Xue W, Yuan H, Dong B, Ding Y, Liu Y, Jiang M, Kan S, Sun T, Ren J, et al. 2017b. AKT-mediated stabilization of histone methyltransferase WHSC1 promotes prostate cancer metastasis. J Clin Invest 127: 1284–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Ahmed M, Guo H, Soares F, Hua JT, Gao S, Lu C, Poon C, Han W, Langstein J, et al. 2017. LSD1-mediated epigenetic reprogramming drives CENPE expression and prostate cancer progression. Cancer Res 77: 5479–5490. [DOI] [PubMed] [Google Scholar]
- Liao CP, Adisetiyo H, Liang M, Roy-Burman P. 2010. Cancer-associated fibroblasts enhance the gland-forming capability of prostate cancer stem cells. Cancer Res 70: 7294–7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, Koskenvuo M, Pukkala E, Skytthe A, Hemminki K. 2000. Environmental and heritable factors in the causation of cancer—analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med 343: 78–85. [DOI] [PubMed] [Google Scholar]
- Lin D, Wyatt AW, Xue H, Wang Y, Dong X, Haegert A, Wu R, Brahmbhatt S, Mo F, Jong L, et al. 2014. High fidelity patient-derived xenografts for accelerating prostate cancer discovery and drug development. Cancer Res 74: 1272–1283. [DOI] [PubMed] [Google Scholar]
- Linde N, Casanova-Acebes M, Sosa MS, Mortha A, Rahman A, Farias E, Harper K, Tardio E, Reyes Torres I, Jones J, et al. 2018. Macrophages orchestrate breast cancer early dissemination and metastasis. Nat Commun 9: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linehan DC, Goedegebuure PS. 2005. CD25+ CD4+ regulatory T-cells in cancer. Immunol Res 32: 155–168. [DOI] [PubMed] [Google Scholar]
- Link KA, Balasubramaniam S, Sharma A, Comstock CE, Godoy-Tundidor S, Powers N, Cao KH, Haelens A, Claessens F, Revelo MP, et al. 2008. Targeting the BAF57 SWI/SNF subunit in prostate cancer: a novel platform to control androgen receptor activity. Cancer Res 68: 4551–4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linn DE, Bronson RT, Li Z. 2015. Genetic interaction between Tmprss2-ERG gene fusion and Nkx3.1-loss does not enhance prostate tumorigenesis in mouse models. PLoS One 10: e0120628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litwin MS, Tan HJ. 2017. The diagnosis and treatment of prostate cancer: a review. JAMA 317: 2532–2542. [DOI] [PubMed] [Google Scholar]
- Liu C, Workman CJ, Vignali DA. 2016. Targeting regulatory T cells in tumors. FEBS J 283: 2731–2748. [DOI] [PubMed] [Google Scholar]
- Logothetis CJ, Lin SH. 2005. Osteoblasts in prostate cancer metastasis to bone. Nat Rev Cancer 5: 21–28. [DOI] [PubMed] [Google Scholar]
- Lohr JG, Adalsteinsson VA, Cibulskis K, Choudhury AD, Rosenberg M, Cruz-Gordillo P, Francis JM, Zhang CZ, Shalek AK, Satija R, et al. 2014. Whole-exome sequencing of circulating tumor cells provides a window into metastatic prostate cancer. Nat Biotechnol 32: 479–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord CJ, Ashworth A. 2013. Mechanisms of resistance to therapies targeting BRCA-mutant cancers. Nat Med 19: 1381–1388. [DOI] [PubMed] [Google Scholar]
- Lord CJ, Ashworth A. 2016. BRCAness revisited. Nat Rev Cancer 16: 110–120. [DOI] [PubMed] [Google Scholar]
- Lotan TL, Gupta NS, Wang W, Toubaji A, Haffner MC, Chaux A, Hicks JL, Meeker AK, Bieberich CJ, De Marzo AM, et al. 2011. ERG gene rearrangements are common in prostatic small cell carcinomas. Mod Pathol 24: 820–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu NZ, Wardell SE, Burnstein KL, Defranco D, Fuller PJ, Giguere V, Hochberg RB, McKay L, Renoir JM, Weigel NL, et al. 2006. International Union of Pharmacology. LXV. The pharmacology and classification of the nuclear receptor superfamily: glucocorticoid, mineralocorticoid, progesterone, and androgen receptors. Pharmacol Rev 58: 782–797. [DOI] [PubMed] [Google Scholar]
- Lu X, Horner JW, Paul E, Shang X, Troncoso P, Deng P, Jiang S, Chang Q, Spring DJ, Sharma P, et al. 2017a. Effective combinatorial immunotherapy for castration-resistant prostate cancer. Nature 543: 728–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Jin EJ, Cheng X, Feng S, Shang X, Deng P, Jiang S, Chang Q, Rahmy S, Chaudhary S, et al. 2017b. Opposing roles of TGFβ and BMP signaling in prostate cancer development. Genes Dev 31: 2337–2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luchman HA, Friedman HC, Villemaire ML, Peterson AC, Jirik FR. 2008. Temporally controlled prostate epithelium-specific gene alterations. Genesis 46: 229–234. [DOI] [PubMed] [Google Scholar]
- Lukacs RU, Memarzadeh S, Wu H, Witte ON. 2010. Bmi-1 is a crucial regulator of prostate stem cell self-renewal and malignant transformation. Cell Stem Cell 7: 682–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Zhou H, Krueger J, Kaplan C, Lee SH, Dolman C, Markowitz D, Wu W, Liu C, Reisfeld RA, et al. 2006. Targeting tumor-associated macrophages as a novel strategy against breast cancer. J Clin Invest 116: 2132–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyko F. 2018. The DNA methyltransferase family: a versatile toolkit for epigenetic regulation. Nat Rev Genet 19: 81–92. [DOI] [PubMed] [Google Scholar]
- Magnon C, Hall SJ, Lin J, Xue X, Gerber L, Freedland SJ, Frenette PS. 2013. Autonomic nerve development contributes to prostate cancer progression. Science 341: 1236361. [DOI] [PubMed] [Google Scholar]
- Maia MC, Hansen AR. 2017. A comprehensive review of immunotherapies in prostate cancer. Crit Rev Oncol Hematol 113: 292–303. [DOI] [PubMed] [Google Scholar]
- Malik R, Khan AP, Asangani IA, Cieslik M, Prensner JR, Wang X, Iyer MK, Jiang X, Borkin D, Escara-Wilke J, et al. 2015. Targeting the MLL complex in castration-resistant prostate cancer. Nat Med 21: 344–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maolake A, Izumi K, Shigehara K, Natsagdorj A, Iwamoto H, Kadomoto S, Takezawa Y, Machioka K, Narimoto K, Namiki M, et al. 2017. Tumor-associated macrophages promote prostate cancer migration through activation of the CCL22–CCR4 axis. Oncotarget 8: 9739–9751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marincola FM, Jaffee EM, Hicklin DJ, Ferrone S. 2000. Escape of human solid tumors from T-cell recognition: molecular mechanisms and functional significance. Adv Immunol 74: 181–273. [DOI] [PubMed] [Google Scholar]
- Massie CE, Lynch A, Ramos-Montoya A, Boren J, Stark R, Fazli L, Warren A, Scott H, Madhu B, Sharma N, et al. 2011. The androgen receptor fuels prostate cancer by regulating central metabolism and biosynthesis. EMBO J 30: 2719–2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateo J, Carreira S, Sandhu S, Miranda S, Mossop H, Perez-Lopez R, Nava Rodrigues D, Robinson D, Omlin A, Tunariu N, et al. 2015. DNA-repair defects and olaparib in metastatic prostate cancer. N Engl J Med 373: 1697–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGray AJR, Bramson J. 2017. Adaptive resistance to cancer immunotherapy. Adv Exp Med Biol 1036: 213–227. [DOI] [PubMed] [Google Scholar]
- McNeal JE. 1969. Origin and development of carcinoma in the prostate. Cancer 23: 24–34. [DOI] [PubMed] [Google Scholar]
- McNeal JE. 1981. The zonal anatomy of the prostate. Prostate 2: 35–49. [DOI] [PubMed] [Google Scholar]
- McNeal JE. 1988. Normal histology of the prostate. Am J Surg Pathol 12: 619–633. [DOI] [PubMed] [Google Scholar]
- McNeal JE, Redwine EA, Freiha FS, Stamey TA. 1988. Zonal distribution of prostatic adenocarcinoma. Correlation with histologic pattern and direction of spread. Am J Surg Pathol 12: 897–906. [DOI] [PubMed] [Google Scholar]
- Meacham CE, Morrison SJ. 2013. Tumour heterogeneity and cancer cell plasticity. Nature 501: 328–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercader M, Bodner BK, Moser MT, Kwon PS, Park ES, Manecke RG, Ellis TM, Wojcik EM, Yang D, Flanigan RC, et al. 2001a. T cell infiltration of the prostate induced by androgen withdrawal in patients with prostate cancer. Proc Natl Acad Sci 98: 14565–14570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercader M, Bodner BK, Moser MT, Kwon PS, Park ESY, Manecke RG, Ellis TM, Wojcik EM, Yang D, Flanigan RC, et al. 2001b. T cell infiltration of the prostate induced by androgen withdrawal in patients with prostate cancer. Proc Natl Acad Sci 98: 14565–14570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer C, Cagnon L, Costa-Nunes CM, Baumgaertner P, Montandon N, Leyvraz L, Michielin O, Romano E, Speiser DE. 2014. Frequencies of circulating MDSC correlate with clinical outcome of melanoma patients treated with ipilimumab. Cancer Immunol Immunother 63: 247–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrofanova A, Aytes A, Zou M, Shen MM, Abate-Shen C, Califano A. 2015. Predicting drug response in human prostate cancer from preclinical analysis of in vivo mouse models. Cell Rep 12: 2060–2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal V. 2018. Epithelial–mesenchymal transition in tumor metastasis. Annu Rev Pathol 13: 395–412. [DOI] [PubMed] [Google Scholar]
- Mok S, Koya RC, Tsui C, Xu J, Robert L, Wu L, Graeber T, West BL, Bollag G, Ribas A. 2014. Inhibition of CSF-1 receptor improves the antitumor efficacy of adoptive cell transfer immunotherapy. Cancer Res 74: 153–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse MD, McNeel DG. 2010. Prostate cancer patients on androgen deprivation therapy develop persistent changes in adaptive immune responses. Hum Immunol 71: 496–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motoshima T, Komohara Y, Horlad H, Takeuchi A, Maeda Y, Tanoue K, Kawano Y, Harada M, Takeya M, Eto M. 2015. Sorafenib enhances the antitumor effects of anti-CTLA-4 antibody in a murine cancer model by inhibiting myeloid-derived suppressor cells. Oncol Rep 33: 2947–2953. [DOI] [PubMed] [Google Scholar]
- Mu P, Zhang Z, Benelli M, Karthaus WR, Hoover E, Chen CC, Wongvipat J, Ku SY, Gao D, Cao Z, et al. 2017. SOX2 promotes lineage plasticity and antiandrogen resistance in TP53- and RB1-deficient prostate cancer. Science 355: 84–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulholland DJ, Tran LM, Li Y, Cai H, Morim A, Wang S, Plaisier S, Garraway IP, Huang J, Graeber TG, et al. 2011. Cell autonomous role of PTEN in regulating castration-resistant prostate cancer growth. Cancer Cell 19: 792–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray PJ. 2017. Macrophage polarization. Annu Rev Physiol 79: 541–566. [DOI] [PubMed] [Google Scholar]
- Nguyen LT, Tretiakova MS, Silvis MR, Lucas J, Klezovitch O, Coleman I, Bolouri H, Kutyavin VI, Morrissey C, True LD, et al. 2015. ERG activates the YAP1 transcriptional program and induces the development of age-related prostate tumors. Cancer Cell 27: 797–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson ML, Das S, Im KM, Turan S, Berndt SI, Li H, Lou H, Brodie SA, Billaud JN, Zhang T, et al. 2017. TET2 binds the androgen receptor and loss is associated with prostate cancer. Oncogene 36: 2172–2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonomura N, Takayama H, Nakayama M, Nakai Y, Kawashima A, Mukai M, Nagahara A, Aozasa K, Tsujimura A. 2011. Infiltration of tumour-associated macrophages in prostate biopsy specimens is predictive of disease progression after hormonal therapy for prostate cancer. BJU Int 107: 1918–1922. [DOI] [PubMed] [Google Scholar]
- Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, Pan F, Pelloski CE, Sulman EP, Bhat KP, et al. 2010. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell 17: 510–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak DG, Cho H, Herzka T, Watrud K, DeMarco DV, Wang VM, Senturk S, Fellmann C, Ding D, Beinortas T, et al. 2015. MYC drives Pten/Trp53-deficient proliferation and metastasis due to IL6 secretion and AKT suppression via PHLPP2. Cancer Discov 5: 636–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noy R, Pollard JW. 2014. Tumor-associated macrophages: from mechanisms to therapy. Immunity 41: 49–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmos D, Arkenau HT, Ang JE, Ledaki I, Attard G, Carden CP, Reid AH, A'Hern R, Fong PC, Oomen NB, et al. 2009. Circulating tumour cell (CTC) counts as intermediate end points in castration-resistant prostate cancer (CRPC): a single-centre experience. Ann Oncol 20: 27–33. [DOI] [PubMed] [Google Scholar]
- Orillion A, Hashimoto A, Damayanti N, Shen L, Adelaiye-Ogala R, Arisa S, Chintala S, Ordentlich P, Kao C, Elzey B, et al. 2017. Entinostat neutralizes myeloid-derived suppressor cells and enhances the antitumor effect of PD-1 inhibition in murine models of lung and renal cell carcinoma. Clin Cancer Res 23: 5187–5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ormandy LA, Hillemann T, Wedemeyer H, Manns MP, Greten TF, Korangy F. 2005. Increased populations of regulatory T cells in peripheral blood of patients with hepatocellular carcinoma. Cancer Res 65: 2457–2464. [DOI] [PubMed] [Google Scholar]
- Patrawala L, Calhoun T, Schneider-Broussard R, Zhou J, Claypool K, Tang DG. 2005. Side population is enriched in tumorigenic, stem-like cancer cells, whereas ABCG2+ and ABCG2- cancer cells are similarly tumorigenic. Cancer Res 65: 6207–6219. [DOI] [PubMed] [Google Scholar]
- Peng W, Chen JQ, Liu C, Malu S, Creasy C, Tetzlaff MT, Xu C, McKenzie JA, Zhang C, Liang X, et al. 2016. Loss of PTEN promotes resistance to T cell-mediated immunotherapy. Cancer Discov 6: 202–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penney KL, Sinnott JA, Fall K, Pawitan Y, Hoshida Y, Kraft P, Stark JR, Fiorentino M, Perner S, Finn S, et al. 2011. mRNA expression signature of Gleason grade predicts lethal prostate cancer. J Clin Oncol 29: 2391–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrylak DP, Tangen CM, Hussain MH, Lara PN Jr., Jones JA, Taplin ME, Burch PA, Berry D, Moinpour C, Kohli M, et al. 2004. Docetaxel and estramustine compared with mitoxantrone and prednisone for advanced refractory prostate cancer. N Engl J Med 351: 1513–1520. [DOI] [PubMed] [Google Scholar]
- Pitt JM, Vetizou M, Daillere R, Roberti MP, Yamazaki T, Routy B, Lepage P, Boneca IG, Chamaillard M, Kroemer G, et al. 2016. Resistance mechanisms to immune-checkpoint blockade in cancer: tumor-intrinsic and -extrinsic factors. Immunity 44: 1255–1269. [DOI] [PubMed] [Google Scholar]
- Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S, Marshak DR. 1999. Multilineage potential of adult human mesenchymal stem cells. Science 284: 143–147. [DOI] [PubMed] [Google Scholar]
- Pourmand G, Ziaee AA, Abedi AR, Mehrsai A, Alavi HA, Ahmadi A, Saadati HR. 2007. Role of PTEN gene in progression of prostate cancer. Urol J 4: 95–100. [PubMed] [Google Scholar]
- Prensner JR, Iyer MK, Sahu A, Asangani IA, Cao Q, Patel L, Vergara IA, Davicioni E, Erho N, Ghadessi M, et al. 2013. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat Genet 45: 1392–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard CC, Mateo J, Walsh MF, De Sarkar N, Abida W, Beltran H, Garofalo A, Gulati R, Carreira S, Eeles R, et al. 2016. Inherited DNA-repair gene mutations in men with metastatic prostate cancer. N Engl J Med 375: 443–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi J, Tripathi M, Mishra R, Sahgal N, Fazli L, Ettinger S, Placzek WJ, Claps G, Chung LW, Bowtell D, et al. 2013. The E3 ubiquitin ligase Siah2 contributes to castration-resistant prostate cancer by regulation of androgen receptor transcriptional activity. Cancer Cell 23: 332–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian BZ, Pollard JW. 2010. Macrophage diversity enhances tumor progression and metastasis. Cell 141: 39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Liu X, Laffin B, Chen X, Choy G, Jeter CR, Calhoun-Davis T, Li H, Palapattu GS, Pang S, et al. 2012. The PSA(−/lo) prostate cancer cell population harbors self-renewing long-term tumor-propagating cells that resist castration. Cell Stem Cell 10: 556–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail DF, Joyce JA. 2013. Microenvironmental regulation of tumor progression and metastasis. Nat Med 19: 1423–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quante M, Tu SP, Tomita H, Gonda T, Wang SS, Takashi S, Baik GH, Shibata W, Diprete B, Betz KS, et al. 2011. Bone marrow-derived myofibroblasts contribute to the mesenchymal stem cell niche and promote tumor growth. Cancer Cell 19: 257–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn DI, Sandler HM, Horvath LG, Goldkorn A, Eastham JA. 2017. The evolution of chemotherapy for the treatment of prostate cancer. Ann Oncol 28: 2658–2669. [DOI] [PubMed] [Google Scholar]
- Ratnacaram CK, Teletin M, Jiang M, Meng X, Chambon P, Metzger D. 2008. Temporally controlled ablation of PTEN in adult mouse prostate epithelium generates a model of invasive prostatic adenocarcinoma. Proc Natl Acad Sci 105: 2521–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese AC, Pierorazio PM, Han M, Partin AW. 2012. Contemporary evaluation of the National Comprehensive Cancer Network prostate cancer risk classification system. Urology 80: 1075–1079. [DOI] [PubMed] [Google Scholar]
- Restifo NP, Marincola FM, Kawakami Y, Taubenberger J, Yannelli JR, Rosenberg SA. 1996. Loss of functional β 2-microglobulin in metastatic melanomas from five patients receiving immunotherapy. J Natl Cancer Inst 88: 100–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restifo NP, Smyth MJ, Snyder A. 2016. Acquired resistance to immunotherapy and future challenges. Nat Rev Cancer 16: 121–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribas A. 2015. Adaptive immune resistance: how cancer protects from immune attack. Cancer Discov 5: 915–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ries CH, Cannarile MA, Hoves S, Benz J, Wartha K, Runza V, Rey-Giraud F, Pradel LP, Feuerhake F, Klaman I, et al. 2014. Targeting tumor-associated macrophages with anti-CSF-1R antibody reveals a strategy for cancer therapy. Cancer Cell 25: 846–859. [DOI] [PubMed] [Google Scholar]
- Robey RW, Medina-Perez WY, Nishiyama K, Lahusen T, Miyake K, Litman T, Senderowicz AM, Ross DD, Bates SE. 2001. Overexpression of the ATP-binding cassette half-transporter, ABCG2 (Mxr/BCrp/ABCP1), in flavopiridol-resistant human breast cancer cells. Clin Cancer Res 7: 145–152. [PubMed] [Google Scholar]
- Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, Montgomery B, Taplin ME, Pritchard CC, Attard G, et al. 2015. Integrative clinical genomics of advanced prostate cancer. Cell 162: 454. [DOI] [PubMed] [Google Scholar]
- Rodrigues G, Warde P, Pickles T, Crook J, Brundage M, Souhami L, Lukka H. 2012. Pre-treatment risk stratification of prostate cancer patients: a critical review. Can Urol Assoc J 6: 121–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruella M, Barrett DM, Kenderian SS, Shestova O, Hofmann TJ, Perazzelli J, Klichinsky M, Aikawa V, Nazimuddin F, Kozlowski M, et al. 2016. Dual CD19 and CD123 targeting prevents antigen-loss relapses after CD19-directed immunotherapies. J Clin Invest 126: 3814–3826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan CJ, Smith MR, de Bono JS, Molina A, Logothetis CJ, de Souza P, Fizazi K, Mainwaring P, Piulats JM, Ng S, et al. 2013. Abiraterone in metastatic prostate cancer without previous chemotherapy. N Engl J Med 368: 138–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santegoets SJ, Stam AG, Lougheed SM, Gall H, Jooss K, Sacks N, Hege K, Lowy I, Scheper RJ, Gerritsen WR, et al. 2014. Myeloid derived suppressor and dendritic cell subsets are related to clinical outcome in prostate cancer patients treated with prostate GVAX and ipilimumab. J Immunother Cancer 2: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal C, Aprikian AG, Cury FL, Chevalier S, Dragomir A. 2016. Management of localized and advanced prostate cancer in Canada: a lifetime cost and quality-adjusted life-year analysis. Cancer 122: 1085–1096. [DOI] [PubMed] [Google Scholar]
- Sartor AO, Oudard S, Sengelov L, Daugaard G, Saad F, Hansen S, Hjelm-Eriksson M, Jassem J, Thiery-Vuillemin A, Caffo O, et al. 2016. Cabazitaxel vs docetaxel in chemotherapy-naive (CN) patients with metastatic castration-resistant prostate cancer (mCRPC): a three-arm phase III study (FIRSTANA). J Clin Oncol 34: 5006. [DOI] [PubMed] [Google Scholar]
- Scher HI, Jia X, de Bono JS, Fleisher M, Pienta KJ, Raghavan D, Heller G. 2009. Circulating tumour cells as prognostic markers in progressive, castration-resistant prostate cancer: a reanalysis of IMMC38 trial data. Lancet Oncol 10: 233–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scher HI, Fizazi K, Saad F, Taplin ME, Sternberg CN, Miller K, de Wit R, Mulders P, Chi KN, Shore ND, et al. 2012. Increased survival with enzalutamide in prostate cancer after chemotherapy. N Engl J Med 367: 1187–1197. [DOI] [PubMed] [Google Scholar]
- Scher HI, Heller G, Molina A, Attard G, Danila DC, Jia X, Peng W, Sandhu SK, Olmos D, Riisnaes R, et al. 2015. Circulating tumor cell biomarker panel as an individual-level surrogate for survival in metastatic castration-resistant prostate cancer. J Clin Oncol 33: 1348–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonhuber N, Seidler B, Schuck K, Veltkamp C, Schachtler C, Zukowska M, Eser S, Feyerabend TB, Paul MC, Eser P, et al. 2014. A next-generation dual-recombinase system for time- and host-specific targeting of pancreatic cancer. Nat Med 20: 1340–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder FH, Hugosson J, Roobol MJ, Tammela TL, Ciatto S, Nelen V, Kwiatkowski M, Lujan M, Lilja H, Zappa M, et al. 2009. Screening and prostate-cancer mortality in a randomized European study. N Engl J Med 360: 1320–1328. [DOI] [PubMed] [Google Scholar]
- Schroder FH, Hugosson J, Roobol MJ, Tammela TL, Ciatto S, Nelen V, Kwiatkowski M, Lujan M, Lilja H, Zappa M, et al. 2012. Prostate-cancer mortality at 11 years of follow-up. N Engl J Med 366: 981–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher TN, Schreiber RD. 2015. Neoantigens in cancer immunotherapy. Science 348: 69–74. [DOI] [PubMed] [Google Scholar]
- Schumacher FR, Al Olama AA, Berndt SI, Benlloch S, Ahmed M, Saunders EJ, Dadaev T, Leongamornlert D, Anokian E, Cieza-Borrella C, et al. 2018. Association analyses of more than 140,000 men identify 63 new prostate cancer susceptibility loci. Nat Genet 50: 928–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehrawat A, Gao L, Wang Y, Bankhead A III, McWeeney SK, King CJ, Schwartzman J, Urrutia J, Bisson WH, Coleman DJ, et al. 2018. LSD1 activates a lethal prostate cancer gene network independently of its demethylase function. Proc Natl Acad Sci 115: E4179–E4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano NA, Anscher MS. 2016. Favorable vs unfavorable intermediate-risk prostate cancer: a review of the new classification system and its impact on treatment recommendations. Oncology (Williston Park) 30: 229–236. [PubMed] [Google Scholar]
- Shah N, Wang P, Wongvipat J, Karthaus WR, Abida W, Armenia J, Rockowitz S, Drier Y, Bernstein BE, Long HW, et al. 2017. Regulation of the glucocorticoid receptor via a BET-dependent enhancer drives antiandrogen resistance in prostate cancer. Elife 6: e27861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalapour S, Font-Burgada J, Di Caro G, Zhong Z, Sanchez-Lopez E, Dhar D, Willimsky G, Ammirante M, Strasner A, Hansel DE, et al. 2015. Immunosuppressive plasma cells impede T-cell-dependent immunogenic chemotherapy. Nature 521: 94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Yeow WS, Ertel A, Coleman I, Clegg N, Thangavel C, Morrissey C, Zhang X, Comstock CE, Witkiewicz AK, et al. 2010. The retinoblastoma tumor suppressor controls androgen signaling and human prostate cancer progression. J Clin Invest 120: 4478–4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P, Hu-Lieskovan S, Wargo JA, Ribas A. 2017. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell 168: 707–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen MM, Abate-Shen C. 2010. Molecular genetics of prostate cancer: new prospects for old challenges. Genes Dev 24: 1967–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenoy D, Packianathan S, Chen AM, Vijayakumar S. 2016. Do African-American men need separate prostate cancer screening guidelines? BMC Urol 16: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Du L, Lin L, Wang Y. 2017. Tumour-associated mesenchymal stem/stromal cells: emerging therapeutic targets. Nat Rev Drug Discov 16: 35–52. [DOI] [PubMed] [Google Scholar]
- Shin DS, Zaretsky JM, Escuin-Ordinas H, Garcia-Diaz A, Hu-Lieskovan S, Kalbasi A, Grasso CS, Hugo W, Sandoval S, Torrejon DY, et al. 2017. Primary resistance to PD-1 blockade mediated by JAK1/2 mutations. Cancer Discov 7: 188–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiozawa Y, Havens AM, Jung Y, Ziegler AM, Pedersen EA, Wang J, Wang J, Lu G, Roodman GD, Loberg RD, et al. 2008. Annexin II/annexin II receptor axis regulates adhesion, migration, homing, and growth of prostate cancer. J Cell Biochem 105: 370–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoag J, Liu DL, Blattner M, Sboner A, Park K, Deonarine L, Robinson BD, Mosquera JM, Chen Y, Rubin MA, et al. 2018. SPOP mutation drives prostate neoplasia without stabilizing oncogenic transcription factor ERG. J Clin Invest 128: 381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver D, Schrittwieser J, Simonyan K, Antonoglou I, Huang A, Guez A, Hubert T, Baker L, Lai M, Bolton A, et al. 2017. Mastering the game of Go without human knowledge. Nature 550: 354–359. [DOI] [PubMed] [Google Scholar]
- Sinnott JA, Peisch SF, Tyekucheva S, Gerke T, Lis R, Rider JR, Fiorentino M, Stampfer MJ, Mucci LA, Loda M, et al. 2017. Prognostic utility of a new mRNA expression signature of gleason score. Clin Cancer Res 23: 81–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siravegna G, Marsoni S, Siena S, Bardelli A. 2017. Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol 14: 531–548. [DOI] [PubMed] [Google Scholar]
- Sizemore GM, Pitarresi JR, Balakrishnan S, Ostrowski MC. 2017. The ETS family of oncogenic transcription factors in solid tumours. Nat Rev Cancer 17: 337–351. [DOI] [PubMed] [Google Scholar]
- Smith BA, Sokolov A, Uzunangelov V, Baertsch R, Newton Y, Graim K, Mathis C, Cheng D, Stuart JM, Witte ON. 2015. A basal stem cell signature identifies aggressive prostate cancer phenotypes. Proc Natl Acad Sci 112: E6544–E6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MR, Saad F, Chowdhury S, Oudard S, Hadaschik BA, Graff JN, Olmos D, Mainwaring PN, Lee JY, Uemura H, et al. 2018. Apalutamide treatment and metastasis-free survival in prostate cancer. N Engl J Med 378: 1408–1418. [DOI] [PubMed] [Google Scholar]
- Solito S, Bronte V, Mandruzzato S. 2011. Antigen specificity of immune suppression by myeloid-derived suppressor cells. J Leukoc Biol 90: 31–36. [DOI] [PubMed] [Google Scholar]
- Sorrentino C, Musiani P, Pompa P, Cipollone G, Di Carlo E. 2011. Androgen deprivation boosts prostatic infiltration of cytotoxic and regulatory T lymphocytes and has no effect on disease-free survival in prostate cancer patients. Clin Cancer Res 17: 1571–1581. [DOI] [PubMed] [Google Scholar]
- Spranger S, Bao R, Gajewski TF. 2015. Melanoma-intrinsic β-catenin signalling prevents anti-tumour immunity. Nature 523: 231–235. [DOI] [PubMed] [Google Scholar]
- Strand DW, Goldstein AS. 2015. The many ways to make a luminal cell and a prostate cancer cell. Endocr Relat Cancer 22: T187–T197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasner A, Karin M. 2015. Immune infiltration and prostate cancer. Front Oncol 5: 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sucker A, Zhao F, Real B, Heeke C, Bielefeld N, Mabetaen S, Horn S, Moll I, Maltaner R, Horn PA, et al. 2014. Genetic evolution of T-cell resistance in the course of melanoma progression. Clin Cancer Res 20: 6593–6604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumanasuriya S, De Bono J. 2018. Treatment of advanced prostate cancer-a review of current therapies and future promise. Cold Spring Harb Perspect Med 8: a030635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Campisi J, Higano C, Beer TM, Porter P, Coleman I, True L, Nelson PS. 2012. Treatment-induced damage to the tumor microenvironment promotes prostate cancer therapy resistance through WNT16B. Nat Med 18: 1359–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland JS, Goldberg GL, Hammett MV, Uldrich AP, Berzins SP, Heng TS, Blazar BR, Millar JL, Malin MA, Chidgey AP, et al. 2005. Activation of thymic regeneration in mice and humans following androgen blockade. J Immunol 175: 2741–2753. [DOI] [PubMed] [Google Scholar]
- Sutmuller RP, van Duivenvoorde LM, van Elsas A, Schumacher TN, Wildenberg ME, Allison JP, Toes RE, Offringa R, Melief CJ. 2001. Synergism of cytotoxic T lymphocyte-associated antigen 4 blockade and depletion of CD25+ regulatory T cells in antitumor therapy reveals alternative pathways for suppression of autoreactive cytotoxic T lymphocyte responses. J Exp Med 194: 823–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taichman RS, Cooper C, Keller ET, Pienta KJ, Taichman NS, McCauley LK. 2002. Use of the stromal cell-derived factor-1/CXCR4 pathway in prostate cancer metastasis to bone. Cancer Res 62: 1832–1837. [PubMed] [Google Scholar]
- Takata R, Akamatsu S, Kubo M, Takahashi A, Hosono N, Kawaguchi T, Tsunoda T, Inazawa J, Kamatani N, Ogawa O, et al. 2010. Genome-wide association study identifies five new susceptibility loci for prostate cancer in the Japanese population. Nat Genet 42: 751–754. [DOI] [PubMed] [Google Scholar]
- Talmadge JE, Gabrilovich DI. 2013. History of myeloid-derived suppressor cells. Nat Rev Cancer 13: 739–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan HL, Sood A, Rahimi HA, Wang W, Gupta N, Hicks J, Mosier S, Gocke CD, Epstein JI, Netto GJ, et al. 2014. Rb loss is characteristic of prostatic small cell neuroendocrine carcinoma. Clin Cancer Res 20: 890–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S, Moore ML, Grayson JM, Dubey P. 2012. Increased CD8+ T-cell function following castration and immunization is countered by parallel expansion of regulatory T cells. Cancer Res 72: 1975–1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taplin SH, Barlow W, Urban N, Mandelson MT, Timlin DJ, Ichikawa L, Nefcy P. 1995. Stage, age, comorbidity, and direct costs of colon, prostate, and breast cancer care. J Natl Cancer Inst 87: 417–426. [DOI] [PubMed] [Google Scholar]
- Taplin ME, Manola J, Oh WK, Kantoff PW, Bubley GJ, Smith M, Barb D, Mantzoros C, Gelmann EP, Balk SP. 2008. A phase II study of mifepristone (RU-486) in castration-resistant prostate cancer, with a correlative assessment of androgen-related hormones. BJU Int 101: 1084–1089. [DOI] [PubMed] [Google Scholar]
- Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, Arora VK, Kaushik P, Cerami E, Reva B, et al. 2010. Integrative genomic profiling of human prostate cancer. Cancer Cell 18: 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng MW, Ngiow SF, von Scheidt B, McLaughlin N, Sparwasser T, Smyth MJ. 2010. Conditional regulatory T-cell depletion releases adaptive immunity preventing carcinogenesis and suppressing established tumor growth. Cancer Res 70: 7800–7809. [DOI] [PubMed] [Google Scholar]
- Tentler JJ, Tan AC, Weekes CD, Jimeno A, Leong S, Pitts TM, Arcaroli JJ, Messersmith WA, Eckhardt SG. 2012. Patient-derived tumour xenografts as models for oncology drug development. Nat Rev Clin Oncol 9: 338–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theurillat JP, Udeshi ND, Errington WJ, Svinkina T, Baca SC, Pop M, Wild PJ, Blattner M, Groner AC, Rubin MA, et al. 2014. Prostate cancer. Ubiquitylome analysis identifies dysregulation of effector substrates in SPOP-mutant prostate cancer. Science 346: 85–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas G, Jacobs KB, Yeager M, Kraft P, Wacholder S, Orr N, Yu K, Chatterjee N, Welch R, Hutchinson A, et al. 2008. Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet 40: 310–315. [DOI] [PubMed] [Google Scholar]
- To SQ, Kwan EM, Fettke HC, Mant A, Docanto MM, Martelotto L, Bukczynska P, Ng N, Graham LK, Parente P, et al. 2018. Expression of androgen receptor splice variant 7 or 9 in whole blood does not predict response to androgen-axis-targeting agents in metastatic castration-resistant prostate cancer. Eur Urol 73: 818–821. [DOI] [PubMed] [Google Scholar]
- Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, et al. 2005. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 310: 644–648. [DOI] [PubMed] [Google Scholar]
- Tomlins SA, Laxman B, Dhanasekaran SM, Helgeson BE, Cao X, Morris DS, Menon A, Jing X, Cao Q, Han B, et al. 2007. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature 448: 595–599. [DOI] [PubMed] [Google Scholar]
- Tomlins SA, Bjartell A, Chinnaiyan AM, Jenster G, Nam RK, Rubin MA, Schalken JA. 2009. ETS gene fusions in prostate cancer: from discovery to daily clinical practice. Eur Urol 56: 275–286. [DOI] [PubMed] [Google Scholar]
- Torrano V, Valcarcel-Jimenez L, Cortazar AR, Liu X, Urosevic J, Castillo-Martin M, Fernandez-Ruiz S, Morciano G, Caro-Maldonado A, Guiu M, et al. 2016. The metabolic co-regulator PGC1α suppresses prostate cancer metastasis. Nat Cell Biol 18: 645–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. 2015. Global cancer statistics, 2012. CA Cancer J Clin 65: 87–108. [DOI] [PubMed] [Google Scholar]
- Tran E, Robbins PF, Lu YC, Prickett TD, Gartner JJ, Jia L, Pasetto A, Zheng Z, Ray S, Groh EM, et al. 2016. T-cell transfer therapy targeting mutant KRAS in cancer. N Engl J Med 375: 2255–2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uccelli A, Moretta L, Pistoia V. 2008. Mesenchymal stem cells in health and disease. Nat Rev Immunol 8: 726–736. [DOI] [PubMed] [Google Scholar]
- Urbanucci A, Barfeld SJ, Kytola V, Itkonen HM, Coleman IM, Vodak D, Sjoblom L, Sheng X, Tolonen T, Minner S, et al. 2017. Androgen receptor deregulation drives bromodomain-mediated chromatin alterations in prostate cancer. Cell Rep 19: 2045–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaishampayan U, Montgomery RB, Gordon MS, Smith DC, Barber K, de Haas-Amatsaleh A, Thapar N, Chandhasin C, Perabo F, Chi KN. 2017. 794PEPI-506 (ralaniten acetate), a novel androgen receptor (AR) N-terminal domain (NTD) inhibitor, in men with metastatic castration-resistant prostate cancer (mCRPC): phase 1 update on safety, tolerability, pharmacokinetics and efficacy. Ann Oncol 28: mdx370.011. [Google Scholar]
- van Leenders GJ, Schalken JA. 2003. Epithelial cell differentiation in the human prostate epithelium: implications for the pathogenesis and therapy of prostate cancer. Crit Rev Oncol Hematol 46 Suppl: S3–S10. [DOI] [PubMed] [Google Scholar]
- van Rooij N, van Buuren MM, Philips D, Velds A, Toebes M, Heemskerk B, van Dijk LJ, Behjati S, Hilkmann H, El Atmioui D, et al. 2013. Tumor exome analysis reveals neoantigen-specific T-cell reactivity in an ipilimumab-responsive melanoma. J Clin Oncol 31: e439–e442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, et al. 2002. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 419: 624–629. [DOI] [PubMed] [Google Scholar]
- Viehl CT, Moore TT, Liyanage UK, Frey DM, Ehlers JP, Eberlein TJ, Goedegebuure PS, Linehan DC. 2006. Depletion of CD4+CD25+ regulatory T cells promotes a tumor-specific immune response in pancreas cancer-bearing mice. Ann Surg Oncol 13: 1252–1258. [DOI] [PubMed] [Google Scholar]
- Visakorpi T, Hyytinen E, Koivisto P, Tanner M, Keinanen R, Palmberg C, Palotie A, Tammela T, Isola J, Kallioniemi OP. 1995. In vivo amplification of the androgen receptor gene and progression of human prostate cancer. Nat Genet 9: 401–406. [DOI] [PubMed] [Google Scholar]
- Vuk-Pavlović S, Bulur PA, Lin Y, Qin R, Szumlanski CL, Zhao X, Dietz AB. 2010. Immunosuppressive CD14+HLA-DRlow/- monocytes in prostate cancer. Prostate 70: 443–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallis CJ, Nam RK. 2015. Prostate cancer genetics: a review. EJIFCC 26: 79–91. [PMC free article] [PubMed] [Google Scholar]
- Wan JCM, Massie C, Garcia-Corbacho J, Mouliere F, Brenton JD, Caldas C, Pacey S, Baird R, Rosenfeld N. 2017. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer 17: 223–238. [DOI] [PubMed] [Google Scholar]
- Wang Y, Navin NE. 2015. Advances and applications of single-cell sequencing technologies. Mol Cell 58: 598–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Gao J, Lei Q, Rozengurt N, Pritchard C, Jiao J, Thomas GV, Li G, Roy-Burman P, Nelson PS, et al. 2003. Prostate-specific deletion of the murine Pten tumor suppressor gene leads to metastatic prostate cancer. Cancer Cell 4: 209–221. [DOI] [PubMed] [Google Scholar]
- Wang X, Kruithof-de Julio M, Economides KD, Walker D, Yu H, Halili MV, Hu YP, Price SM, Abate-Shen C, Shen MM. 2009. A luminal epithelial stem cell that is a cell of origin for prostate cancer. Nature 461: 495–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZA, Mitrofanova A, Bergren SK, Abate-Shen C, Cardiff RD, Califano A, Shen MM. 2013. Lineage analysis of basal epithelial cells reveals their unexpected plasticity and supports a cell-of-origin model for prostate cancer heterogeneity. Nat Cell Biol 15: 274–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZA, Toivanen R, Bergren SK, Chambon P, Shen MM. 2014. Luminal cells are favored as the cell of origin for prostate cancer. Cell Rep 8: 1339–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Lu X, Dey P, Deng P, Wu CC, Jiang S, Fang Z, Zhao K, Konaparthi R, Hua S, et al. 2016a. Targeting YAP-dependent MDSC infiltration impairs tumor progression. Cancer Discov 6: 80–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Lin W, Lin C, Li L, Sun Y, Chang C. 2016b. ASC-J9((R)) suppresses castration resistant prostate cancer progression via degrading the enzalutamide-induced androgen receptor mutant AR-F876L. Cancer Lett 379: 154–160. [DOI] [PubMed] [Google Scholar]
- Watson PA, Arora VK, Sawyers CL. 2015. Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nat Rev Cancer 15: 701–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weischenfeldt J, Simon R, Feuerbach L, Schlangen K, Weichenhan D, Minner S, Wuttig D, Warnatz HJ, Stehr H, Rausch T, et al. 2013. Integrative genomic analyses reveal an androgen-driven somatic alteration landscape in early-onset prostate cancer. Cancer Cell 23: 159–170. [DOI] [PubMed] [Google Scholar]
- Williamson SR, Zhang S, Yao JL, Huang J, Lopez-Beltran A, Shen S, Osunkoya AO, MacLennan GT, Montironi R, Cheng L. 2011. ERG-TMPRSS2 rearrangement is shared by concurrent prostatic adenocarcinoma and prostatic small cell carcinoma and absent in small cell carcinoma of the urinary bladder: evidence supporting monoclonal origin. Mod Pathol 24: 1120–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilt TJ, Brawer MK, Jones KM, Barry MJ, Aronson WJ, Fox S, Gingrich JR, Wei JT, Gilhooly P, Grob BM, et al. 2012. Radical prostatectomy versus observation for localized prostate cancer. N Engl J Med 367: 203–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilt TJ, Jones KM, Barry MJ, Andriole GL, Culkin D, Wheeler T, Aronson WJ, Brawer MK. 2017. Follow-up of prostatectomy versus observation for early prostate cancer. N Engl J Med 377: 132–142. [DOI] [PubMed] [Google Scholar]
- Woo EY, Yeh H, Chu CS, Schlienger K, Carroll RG, Riley JL, Kaiser LR, June CH. 2002. Cutting edge: regulatory T cells from lung cancer patients directly inhibit autologous T cell proliferation. J Immunol 168: 4272–4276. [DOI] [PubMed] [Google Scholar]
- Woo JR, Liss MA, Muldong MT, Palazzi K, Strasner A, Ammirante M, Varki N, Shabaik A, Howell S, Kane CJ, et al. 2014. Tumor infiltrating B-cells are increased in prostate cancer tissue. J Transl Med 12: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Wu J, Huang J, Powell WC, Zhang J, Matusik RJ, Sangiorgi FO, Maxson RE, Sucov HM, Roy-Burman P. 2001. Generation of a prostate epithelial cell-specific Cre transgenic mouse model for tissue-specific gene ablation. Mech Dev 101: 61–69. [DOI] [PubMed] [Google Scholar]
- Wu CT, Hsieh CC, Lin CC, Chen WC, Hong JH, Chen MF. 2012. Significance of IL-6 in the transition of hormone-resistant prostate cancer and the induction of myeloid-derived suppressor cells. J Mol Med 90: 1343–1355. [DOI] [PubMed] [Google Scholar]
- Wu K, Xie D, Zou Y, Zhang T, Pong RC, Xiao G, Fazli L, Gleave M, He D, Boothman DA, et al. 2013. The mechanism of DAB2IP in chemoresistance of prostate cancer cells. Clin Cancer Res 19: 4740–4749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JB, Shao C, Li X, Li Q, Hu P, Shi C, Li Y, Chen YT, Yin F, Liao CP, et al. 2014a. Monoamine oxidase A mediates prostate tumorigenesis and cancer metastasis. J Clin Invest 124: 2891–2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Schulte BC, Zhou Y, Haribhai D, Mackinnon AC, Plaza JA, Williams CB, Hwang ST. 2014b. Depletion of M2-like tumor-associated macrophages delays cutaneous T-cell lymphoma development in vivo. J Invest Dermatol 134: 2814–2822. [DOI] [PubMed] [Google Scholar]
- Wu JB, Yin L, Shi C, Li Q, Duan P, Huang JM, Liu C, Wang F, Lewis M, Wang Y, et al. 2017. MAOA-dependent activation of Shh-IL6-RANKL signaling network promotes prostate cancer metastasis by engaging tumor-stromal cell interactions. Cancer Cell 31: 368–382. [DOI] [PubMed] [Google Scholar]
- Wyatt AW, Azad AA, Volik SV, Annala M, Beja K, McConeghy B, Haegert A, Warner EW, Mo F, Brahmbhatt S, et al. 2016. Genomic alterations in cell-free DNA and enzalutamide resistance in castration-resistant prostate cancer. JAMA Oncol 2: 1598–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Shimelis H, Linn DE, Jiang R, Yang X, Sun F, Guo Z, Chen H, Li W, Chen H, et al. 2009. Regulation of androgen receptor transcriptional activity and specificity by RNF6-induced ubiquitination. Cancer Cell 15: 270–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Mo Z, Ye D, Wang M, Liu F, Jin G, Xu C, Wang X, Shao Q, Chen Z, et al. 2012a. Genome-wide association study in Chinese men identifies two new prostate cancer risk loci at 9q31.2 and 19q13.4. Nat Genet 44: 1231–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Wu ZJ, Groner AC, He HH, Cai C, Lis RT, Wu X, Stack EC, Loda M, Liu T, et al. 2012b. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science 338: 1465–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Escamilla J, Mok S, David J, Priceman S, West B, Bollag G, McBride W, Wu L. 2013. CSF1R signaling blockade stanches tumor-infiltrating myeloid cells and improves the efficacy of radiotherapy in prostate cancer. Cancer Res 73: 2782–2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamoah K, Johnson MH, Choeurng V, Faisal FA, Yousefi K, Haddad Z, Ross AE, Alshalafa M, Den R, Lal P, et al. 2015. Novel biomarker signature that may predict aggressive disease in African American men with prostate cancer. J Clin Oncol 33: 2789–2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, DeBusk LM, Fukuda K, Fingleton B, Green-Jarvis B, Shyr Y, Matrisian LM, Carbone DP, Lin PC. 2004. Expansion of myeloid immune suppressor Gr+CD11b+ cells in tumor-bearing host directly promotes tumor angiogenesis. Cancer Cell 6: 409–421. [DOI] [PubMed] [Google Scholar]
- Yang F, Tuxhorn JA, Ressler SJ, McAlhany SJ, Dang TD, Rowley DR. 2005. Stromal expression of connective tissue growth factor promotes angiogenesis and prostate cancer tumorigenesis. Cancer Res 65: 8887–8895. [DOI] [PubMed] [Google Scholar]
- Yang L, Huang J, Ren X, Gorska AE, Chytil A, Aakre M, Carbone DP, Matrisian LM, Richmond A, Lin PC, et al. 2008. Abrogation of TGFβ signaling in mammary carcinomas recruits Gr-1+CD11b+ myeloid cells that promote metastasis. Cancer Cell 13: 23–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap TA, Lorente D, Omlin A, Olmos D, de Bono JS. 2014. Circulating tumor cells: a multifunctional biomarker. Clin Cancer Res 20: 2553–2568. [DOI] [PubMed] [Google Scholar]
- Yeager M, Orr N, Hayes RB, Jacobs KB, Kraft P, Wacholder S, Minichiello MJ, Fearnhead P, Yu K, Chatterjee N, et al. 2007. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet 39: 645–649. [DOI] [PubMed] [Google Scholar]
- Yeager M, Chatterjee N, Ciampa J, Jacobs KB, Gonzalez-Bosquet J, Hayes RB, Kraft P, Wacholder S, Orr N, Berndt S, et al. 2009. Identification of a new prostate cancer susceptibility locus on chromosome 8q24. Nat Genet 41: 1055–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yegnasubramanian S. 2016. Prostate cancer epigenetics and its clinical implications. Asian J Androl 18: 549–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo YA, Roh M, Naseem AF, Lysy B, Desouki MM, Unno K, Abdulkadir SA. 2016. Bmi1 marks distinct castration-resistant luminal progenitor cells competent for prostate regeneration and tumour initiation. Nat Commun 7: 12943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu EY, Wu H, Schloss C. 2017. Phase 1b/2 keynote-365 trial: pembrolizumab (pembro) combination therapy in metastatic castration-resistant prostate cancer (mCRPC). J Clin Oncol 35: TPS5089. [Google Scholar]
- Yuen GJ, Demissie E, Pillai S. 2016. B lymphocytes and cancer: a love-hate relationship. Trends Cancer 2: 747–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaretsky JM, Garcia-Diaz A, Shin DS, Escuin-Ordinas H, Hugo W, Hu-Lieskovan S, Torrejon DY, Abril-Rodriguez G, Sandoval S, Barthly L, et al. 2016. Mutations associated with acquired resistance to PD-1 blockade in melanoma. N Engl J Med 375: 819–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Altuwaijri S, Deng F, Chen L, Lal P, Bhanot UK, Korets R, Wenske S, Lilja HG, Chang C, et al. 2009. NF-κB regulates androgen receptor expression and prostate cancer growth. Am J Pathol 175: 489–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Singh A, Yegnasubramanian S, Esopi D, Kombairaju P, Bodas M, Wu H, Bova SG, Biswal S. 2010. Loss of Kelch-like ECH-associated protein 1 function in prostate cancer cells causes chemoresistance and radioresistance and promotes tumor growth. Mol Cancer Ther 9: 336–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Wang L, Wu D, Chen H, Chen Z, Thomas-Ahner JM, Zynger DL, Eeckhoute J, Yu J, Luo J, et al. 2011a. Definition of a FoxA1 Cistrome that is crucial for G1 to S-phase cell-cycle transit in castration-resistant prostate cancer. Cancer Res 71: 6738–6748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Chen L, Helfand BT, Jang TL, Sharma V, Kozlowski J, Kuzel TM, Zhu LJ, Yang XJ, Javonovic B, et al. 2011b. TGF-β regulates DNA methyltransferase expression in prostate cancer, correlates with aggressive capabilities, and predicts disease recurrence. PLoS One 6: e25168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang PZ, Wang DJ, Zhao Y, Ren SC, Gao K, Ye ZQ, Wang SQ, Pan CW, Zhu YS, Yan YQ, et al. 2017. Intrinsic BET inhibitor resistance in SPOP-mutated prostate cancer is mediated by BET protein stabilization and AKT–mTORC1 activation. Nat Med 23: 1055–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D, Lu X, Wang G, Lan Z, Liao W, Li J, Liang X, Chen JR, Shah S, Shang X, et al. 2017. Synthetic essentiality of chromatin remodelling factor CHD1 in PTEN-deficient cancer. Nature 542: 484–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Zhao D, Yan L, Jiang W, Kim JS, Gu B, Liu Q, Wang R, Xia B, Zhao JC, et al. 2018. BMI1 regulates androgen receptor in prostate cancer independently of the polycomb repressive complex 1. Nat Commun 9: 500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zitvogel L, Pitt JM, Daillere R, Smyth MJ, Kroemer G. 2016. Mouse models in oncoimmunology. Nat Rev Cancer 16: 759–773. [DOI] [PubMed] [Google Scholar]
- Zou M, Toivanen R, Mitrofanova A, Floch N, Hayati S, Sun Y, Le Magnen C, Chester D, Mostaghel EA, Califano A, et al. 2017. Transdifferentiation as a mechanism of treatment resistance in a mouse model of castration-resistant prostate cancer. Cancer Discov 7: 736–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumsteg ZS, Zelefsky MJ. 2012. Short-term androgen deprivation therapy for patients with intermediate-risk prostate cancer undergoing dose-escalated radiotherapy: the standard of care? Lancet Oncol 13: e259–e269. [DOI] [PubMed] [Google Scholar]