Abstract
Nuclear receptors are a superfamily of transcription factors restricted to animals. These transcription factors regulate a wide variety of genes with diverse roles in cellular homeostasis, development, and physiology. The origin and specificity of ligand binding within lineages of nuclear receptors (e.g., subfamilies) continues to be a focus of investigation geared toward understanding how the functions of these proteins were shaped over evolutionary history. Among early-diverging animal lineages, the retinoid × receptor (RXR) is first detected in the placozoan, Trichoplax adhaerens. To gain insight into RXR evolution, we characterized ligand- and DNA-binding activity of the RXR from T. adhaerens (TaRXR). Like bilaterian RXRs, TaRXR specifically bound 9-cis-retinoic acid, which is consistent with a recently published result and supports a conclusion that the ancestral RXR bound ligand. DNA binding site specificity of TaRXR was determined through protein binding microarrays (PBMs) and compared with human RXRα. The binding sites for these two RXR proteins were broadly conserved (~85% shared high-affinity sequences within a targeted array), suggesting evolutionary constraint for the regulation of downstream genes. We searched for predicted binding motifs of the T. adhaerens genome within 1000 bases of annotated genes to identify potential regulatory targets. We identified 648 unique protein coding regions with predicted TaRXR binding sites that had diverse predicted functions, with enriched processes related to intracellular signal transduction and protein transport. Together, our data support hypotheses that the original RXR protein in animals bound a ligand with structural similarity to 9-cis-retinoic acid; the DNA motif recognized by RXR has changed little in more than 1 billion years of evolution; and the suite of processes regulated by this transcription factor diversified early in animal evolution.
Keywords: DNA binding motif, nuclear receptor, protein binding microarray
Graphical abstract
INTRODUCTION
Nuclear receptors (NRs) are a diverse superfamily of metazoan transcription factors that regulate processes ranging from embryonic development to cellular differentiation and energetic metabolism (reviewed in Mangelsdorf, et al. 1995; Robinson-Rechavi, et al. 2003). Many NRs are activated by binding of specific ligands, such as steroids, thyroid hormones, retinoids, and fatty acids (Laudet, et al. 1992; Beato, et al. 1995; Mangelsdorf, et al. 1995; Escriva, et al. 1997; Blumberg and Evans 1998). The remaining “orphan” NRs may be activated by unknown endogenous or environmental ligands, be activated through alternative mechanisms (e.g., via phosphorylation), or possess constitutive activity (Giguere, et al. 1988; Power, et al. 1991; Marcus, et al. 1996; Escriva, et al. 1997). Activated NRs bind specific DNA response elements as monomers, homodimers, or heterodimers (reviewed by Khorasanizadeh and Rastinjad 2001). Upon binding to DNA response elements, NRs recruit coactivators or corepressors to modulate transcription of target genes. Progress toward elucidating the evolution of ligand diversity and specificity for NRs has accelerated in recent years, particularly through comparative studies involving more species from phylogenetically informative lineages. For example, integrated analyses of phylogeny, protein structure, and ligand affinity have resulted in novel and conflicting hypotheses for the evolution of ligands for the steroid hormone receptors (e.g., the estrogen receptor (ER), and "steroid receptor" (SR)) in bilaterian animals (e.g., Thornton 2003; Holzer, et al. 2017). Despite this progress, important theoretical and applied questions remain unanswered. Empirical identification of ligand specificity and ligand-mediated activation of NRs in aquatic animals remains a pressing concern due to the spectrum of pollutants that may function as potent mimics that disrupt endogenous NR signaling pathways.
Genes in the NR superfamily have been traditionally classified into six families, NR0 through NR6, by the Nuclear Receptors Nomenclature Committee (1999). Phylogenomic analyses of NR diversity over the past decade has shown that both protostomes and deuterostomes have at least one gene in each of these families, supporting the hypothesis that NRs had undergone extensive radiation prior to the divergence of these lineages. For example, Bertrand and colleagues (2004) analyzed the evolutionary relationships among NRs from nine bilaterian genomes and inferred that 25 NRs likely existed in the Urbilaterian (ancestral bilaterian) with at least one gene from each of the six families. Additional PCR-based surveys of NRs and analyses of sequenced genomes have identified patterns of lineage-specific diversification and have also revealed lineage-specific losses of several NRs since the Urbilaterian ancestor, particularly in insects and nematodes (Taubert, et al. 2011; Fahrbach, et al. 2012).
Because the NR superfamily had already diversified prior to the divergence of the protostomes and deuterostomes, several analyses have aimed to characterize NRs within phyla that diverged early from the animal stem (Reitzel and Tarrant 2009; Bridgham, et al. 2010; Reitzel, et al. 2011). Such studies sought to understand not only when the NR families originated, but also how the function of specific NRs evolved and diversified. Together, these studies have revealed that the diversification of the NR superfamily was gradual, with the original NR likely to have been similar to HNF4 (NR2A), and diversification of the NR2 family and the origins of the NR3 and NR6 families pre-dating the appearance of the ancestral Urbilaterian. Studies conducted within early animal lineages to assess ligand specificity and modulation of NR activity by ligands support a hypothesis that ligands may have been functionally important in early NR evolution. Recent results from species in the phylum Ctenophora, the lineage likely to have first diverged from the animal stem, have shown that the only NRs present have only a conserved ligand binding domain (LBD) but not a DNA binding domain (DBD) (Reitzel, et al. 2011). Studies of two NRs (an HNF4 homolog and a second NR2 family member) from the sponge Amphimedon queenslandica revealed that these NRs both bound a fatty acid ligand, and one of these NRs was ligand-activated in reporter assays (Bridgham, et al. 2010). A number of other studies have shown that sponge extracts can function as ligands for vertebrate NRs (FXR and PXR) (Fiorucci, et al. 2012). Within the other non-bilaterian animal phyla, the Cnidaria and Placozoa, NR ligand binding has only been demonstrated for the retinoid × receptor, as further discussed below (Kostrouch, et al. 1998; Fuchs, et al. 2014; Novotný, et al. 2017).
The retinoid × receptor (RXR, NR2B; USP in some insects) originated early in animal evolution with clear orthologs throughout the bilaterians as well as some cnidarians (medusozoans) and placozoans. RXR has not been reported from sponges or ctenophores but NR diversity has only been characterized within a few species in each of these phyla. RXR is generally classified as a "type II" NR: it forms heterodimers, resides within the nucleus bound to DNA response elements, and becomes transcriptionally active upon ligand binding and associated release of corepressors (reviewed by Sever and Glass 2013). In bilaterians, RXR exhibits promiscuous heterodimerization with a number NR1 family proteins, including PPAR, RAR, FXR, PXR, VDR, and TR (reviewed by Lefebvre, et al. 2010; Evans and Mangelsdorf 2014). The specific RXR heterodimerization partner influences the DNA binding behavior of the protein complex (Mader, et al. 1993; Laffitte, et al. 2000), thus influencing regulation of downstream genes. In addition, RXR has also been shown to bind as a tetramer and a monomer in vitro, although the physiological significance of this behavior has not been demonstrated (Kersten, et al. 1997; Kostrouch, et al. 1998). The combination of different partners and the corresponding differences in regulation of DNA correlate with the central roles for RXR in a diversity of cell functions from development to physiology (Evans and Mangelsdorf 2014). The ligands for RXR may be structurally diverse and influence the dimerization properties for this NR with other proteins. While it may not function as a physiological ligand, 9-cis-retinoic acid (9-cis-RA) binds RXR with high affinity and is the most commonly studied ligand (Heyman, et al. 1992). Studies in two cnidarian species, the box jellyfish Tripedalia cystophora (Kostrouch, et al. 1998) and the moon jelly Aurelia aurita (Fuchs, et al. 2014) have shown that the cnidarian RXR ortholog binds 9-cis-RA with high affinity and that ligand-based activation of RXR may be a central factor in mediating life history transitions in these species.
Trichoplax adhaerens, the originally described member of the phylum Placozoa, is a small, morphologically simple marine organism inhabiting pelagic marine environments around the world. Placozoa is an early-diverging phylum with a consensus position branching off the animal stem after sponges and ctenophores but prior to cnidarians, although its phylogenetic positions remains a subject of ongoing research (Nosenko, et al. 2013; Laumer, et al. 2017). Sequencing of the T. adhaerens genome in 2008 provided a critical data set to discern the evolutionary history of particular gene families (Srivastava, et al. 2008), including NRs (Baker 2008). Trichoplax adhaerens has four NRs and is the first animal lineage to have a well-supported RXR ortholog. Thus, studies of RXR from this species can provide a context for testing hypotheses regarding the antiquity of ligand and DNA binding within the NR2B subfamily. During development of this manuscript, it was reported that the RXR ortholog from T. adhaerens (TaRXR) can bind 9-cis-RA with high affinity (Novotny et al. 2017). Here, we further characterize the RXR ortholog from T. adhaerens including complementary studies of ligand binding, as well as new characterization of DNA binding and potential regulation of downstream genes. Because T. adhaerens lacks canonical RXR heterodimerization partners, these analyses of TaRXR binding also provide insights into the evolution of DNA binding by RXR before NR radiation in the bilaterians.
METHODS
Annotation and cloning of T. adhaerens RXR (TaRXR)
The NR complement from T. adhaerens, including an ortholog of RXR (JGI gene model 49897, TaRXR), was originally described by Baker (2008). The original annotation of the TaRXR was only a predicted gene model with no confirmation. We constructed a cDNA library (SMARTer RACE cDNA Amplification Kit, Clontech) with RNA isolated from a lab-reared colony of T. adhaerens in order to empirically determine the complete coding sequence for this gene. We used a nested primer design with two gene-specific "forward" primers (CGCGCCAAGTTGTGATGG and CCTGATGCCAAAGGATTGAATG) paired with the manufacturer-provided universal reverse primer to amplify the 3' end of TaRXR. Similarly, we used two gene-specific "reverse" primers (GGGATGTGCGATAGTAACCAA and TGCCGCAATTACTGAGAAAC) paired with the universal forward primer from the kit to amplify the 5' end. PCR amplicons were excised from 1% agarose gels, purified (QIAquick gel extraction, Qiagen), cloned (pGEM-T, Promega), and sequenced in both directions. Finally, we assembled the sequences in silico to produce a full-length transcript. We deposited the transcript into GenBank (Accession Number MG602679).
Phylogeny of nuclear receptors in Family 2
To more thoroughly assess the apparent absence of RXR from sponges (Phylum Porifera), we conducted BLAST-based searches of newly-available transcriptomes from 8 additional species that represented 4 classes: Class Hexactinellida: Aphrocallistes vastus; Class Demospongiae: Spongilla lacustris, Petrosia ficiformis, Pseudspongosorites suberitoides, Ircinia fasciculata Condrilla nucula; Class Homoscleromorpha: Corticium candelabrum; Class Calcarea: Sycon coactum (Riesgo, et al. 2014). Transcriptomic databases were queried using NR sequences from Trichoplax adhaerens (NR2-4), Tripedalia cystophora (RXR) and Amphimedon queenslandica (NR1-2) as a search set. Searches were constrained to return the top 10 sequences using the tBLASTn algorithm with a threshold E-value of e−10. These sequences were aligned with the query set using ClustalW, as implemented within BioEdit (Hall 1999). The alignment was manually edited in the case of clear errors, and trimmed to contain only the well-conserved DBD and most of the LBD beginning in helix 3 (as in Reitzel, et al. 2011). Sequences shorter than 40 (DBD) or 100 residues (LBD) were deleted. Maximum likelihood analysis were conducted with RAxML (v7.2.7) using the CIPRES Science Gateway v 3.3 (Miller, et al. 2011). Support for nodes is indicated as a percentage of 1000 bootstraps. Trees were visualized using FigTree v1.1.2 and edited using Adobe Illustrator (colors, fonts, and additional labels).
Expression constructs for protein expression
Using the TaRXR transcript sequence we determined from our RACE products, we amplified the full-length open reading frame (Forward primer: GCTTGCATGGAGGACAGATC, where underlined ATG is the start site; Reverse primer: CTGACCCACAATACAAGACAGC) and developed a protein expression construct using the Gateway cloning system (Thermo Fisher). We used pcDNA3.2/nV5-DEST (Invitrogen) generate a protein tagged with the v5 epitope at the amino terminus. We also obtained the human RXRα (HsRXRα) in the pDONR221 vector from the Harvard Medical School (Clone HsCD00079702). We subcloned this into pcDNA3.2/V5-DEST (Invitrogen) but included the endogenous stop codon to generate an untagged protein.
Ligand binding assays
[11,12-3H] 9-cis retinoic acid (“3H 9-cis-RA” hereafter), 52.9 Ci/mmol was obtained from PerkinElmer. Assay conditions were similar to those previously described (Tarrant, et al. 2011). Briefly, RXR proteins from human and T. adhaerens were expressed using the TnT T7 Quick Coupled Reticulocyte Lysate System (Promega) using 2 µg of plasmid per 50 µl reaction. Expression was confirmed by synthesizing the protein in the presence of 35S-labeled methionine. The labeled proteins were separated using SDS-PAGE, followed by fluorography (Amplify reagent, Amersham) and autoradiography. For binding studies, the proteins were expressed using unlabeled methionine and diluted 1:10 with TEG buffer (10 mM Tris, 1.5 mM EDTA, 10% v/v sterile glycerol, 1 mM DTT). Unprogrammed lysate, (“UPL” an in vitro expression reaction conducted with an empty expression plasmid rather than a receptor) was used to assess non-specific binding. Where specified in the results, the human RXR was mixed with UPL to adjust for the difference in expression between RXR from the human and T. adhaerens plasmids. Diluted proteins were incubated in triplicate overnight in glass tubes at 4°C with 3H 9-cis RA that was diluted in dimethylsulfoxide (2.5% solvent in assay) to give assay concentrations up to 25 nM. For competitive binding studies, triplicate glass tubes were incubated with 10 or 20 nM 3H 9-cis RA with or without a 100-fold excess of unlabeled 9-cis RA. The next morning, aliquots from each tube were incubated on ice (15 min, periodic vortexing) in a 1:1 mixture with 50 mg/ml dextran-coated charcoal. This mixture was then centrifuged at 14,000g for 1 min. A 40-µl aliquot of the supernatant was added to −4 ml of ScintiVerse II cocktail (Fisher Scientific) and counted on a Beckman 5000 liquid scintillation counter.
Protein binding microarray data generation and identification of binding motifs
DNA binding specificity of TaRXR was assessed using a custom protein binding microarray (PBM) ordered from Agilent Technologies (PBM2), which contains 15,000 spots (five replicates of ~3000 unique sequences). Microarray design, composition and exposure conditions followed those described by Boltin et al. (2010) and Fang et al. (2012). The v5-tagged TaRXR was expressed in Cos-7 cells. Eight hundred ng of crude nuclear extract were used for hybridization, followed by an overnight room temperature incubation in 1:100 dilution of anti-V5 antibody (mouse monoclonal, Invitrogen), and a 1 hr incubation in the secondary antibody (Dylight, Jackson ImmunoResearch). The slide was scanned at 543 (Cy3) and 633 (Cy5) nm (Supplemental Figure 1). Data were extracted for analysis of signal intensity for each sequence motif. Position weight matrices (PWMs) were generated using Weblogo (Crooks, et al. 2004). We compared the binding results for TaRXR with results from HsRXRα hybridized under the same conditions an identical microarray. Motif overlapping between T. adhaerens and human were compared using the program Venny (Oliveros 2007–2015).
Prediction of associated genes regulated by RXR and annotation based on function/GO
We used a custom bioinformatic pipeline to identify genes potentially regulated by RXR binding sites in the T. adhaerens genome (version 1.0, release date 8/18/2014). Briefly, the genome was screened using grep commands for a strict top binder (AGGTCAAAGGTCA), a consensus sequence from the top 10% of bound sequences ("strict", C A/C A/G A/G GGTCA), and the most common extended half-site (CAAAGGTCA) from the PBM hybridization data. Seventy base-pair regions of the genome with associated RXR binding sites were extracted and used in a subsequent BLASTn search against the T. adhaerens genome. The position information from the BLASTn search results was then used in combination with a custom Python script (Supplemental File 1) to isolate 1000 bases upstream and downstream of each site on each genome scaffold. These 2 kb nucleotide sequences were then used in BLASTx searches to identify annotated predicted proteins within these regions. This strategy was used in place of searching around the currently annotated start sites in the reference genome because these gene models are largely unvalidated and the start sites are unknown. Proteins with e-values below 1E-10 and an identity score greater than 95% were retained to identify possible function based on gene ontology (GO) groups. GO terms were clustered in the program REVIGO (Supek, et al. 2011) with the whole UniProt database used to reference the size of each term, and semantic similarities were determined using SimRel (Schlicker, et al. 2006). We also searched the T. adhaerens genome and the 2 kb windows for potential dimerization-specific RXR binding sites by using combinations of the two likely half-sites for this NR, AGGTCA and GGGTCA. We restricted these searches to DR1 sites only because these are common for the NR2 family. Lastly, we compared results from these searches with parallel queries using the human genome.
RESULTS
T. adhaerens RXR
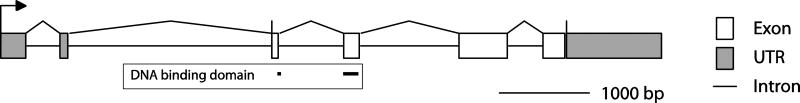
In silico assembly of sequenced PCR and RACE clones resulted in a transcript of 2,514 bp encoding RXR from T. adhaerens (TaRXR), with the open reading frame coding for a protein of 345 amino acids from position 406-1443. Mapping this transcript to the reference genome showed that TaRXR is composed of six exons spanning 7,236 bp of genomic DNA (scaffold2: 5291710-5298946; Figure 1). The first two exons exclusively encode 5' UTR, and the predicted translational start site is located on exon 3. The amino acids corresponding to the DNA binding domain (DBD) are located on exons 3 and 4, and the ligand binding domain (LBD) is located on exons 5 and 6. The stop site for TaRXR is located on exon 6, which is followed by 1,188 bp of 3' UTR with a poly(A) site at position 2,464. The DBD and LBD of TaRXR are both well conserved when compared with RXR sequences from other animals (~80% and ~65–75%, respectively).
Figure 1.
Gene structure for Trichoplax adhaerens RXR (TaRXR).
Phylogeny of nuclear receptors for Family 2
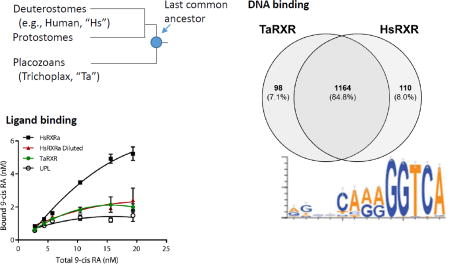
Queries of sponge transcriptomes with a set of six NRs resulted in largely overlapping results that differed primarily in the ordering of sequences returned (i.e., their relative e-values). Most sequences appeared to be incomplete, frequently missing either the DBD or LBD. For this reason, separate likelihood-based analyses were conducted for the DBD alone, LBD alone and DBD plus LBD. Results were qualitatively similar between these three analyses, so only the DBD plus LBD analyses are shown and used in all future analysis and discussion. Overall, analyses of sponge NRs resulted in a strongly supported clade that included the previously described HNF4 from A. queenslandica and was most similar to HNF4 (Supplemental Figure 2). The cnidarian and placozoan RXR sequences (from T. cystophora and T. adhaerens, respectively) were grouped together with strong support. Poor support was obtained for most other nodes, and no sponge genes emerged as clear RXR homologs.
Ligand binding assays
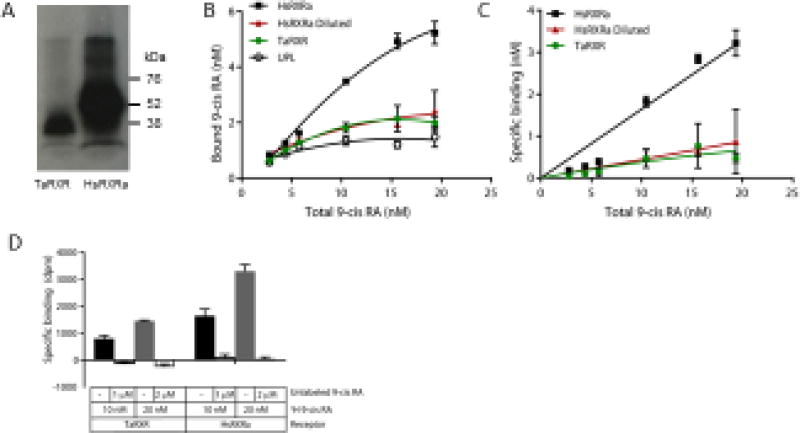
When expressed using in vitro transcription and translation and 35S-labeled methionine, the HsRXRα and TaRXR plasmids each produced a dominant band of the appropriate size, but the human plasmid yielded 4.2-fold more protein (Figure 2A). Both the human and T. adhaerens RXRs consistently bound tritiated 9-cis RA, in excess of binding to the “unprogrammed lysate” (UPL) control (Figure 2B, paired t-test on means p = 0.007). However, in this assay system, after subtracting this nonspecific binding, saturation was not observed, even at ligand concentrations up to 20 nM (Figure 2C). This may be due to high binding of 9-cis-RA to other proteins in the lysate mixture, resulting in ligand depletion. Thus, these assay conditions are not appropriate for measuring binding affinity and capacity, and can only give qualitative insight into ligand binding. In competitive binding assays, co-incubation with a 100-fold excess of unlabeled 9 cis-RA effectively decreased the bound ligand to background levels (Figure 2D).
Figure 2.
Characterization of 3H 9-cis retinoic acid (3H 9-cis RA) by the Trichoplax adhaerens RXR (TaRXR) in comparison with human RXRα (HsRXRa). (A) Proteins expressed in the presence of 35S methionine produced dominant bands of the expected size (TaRXR 39.4 kDa, HsRXRa: 56.7 kDa). (B) Total binding of HsRXR, TaRXR and unprogrammed lysate (UPL, non-specific) to varying concentrations of 3H 9-cis RA. Quadratic fitted curves shown. Mean and standard deviation are shown. Red symbols indicate the human RXRa diluted with UPL to produce an equivalent level of expressed protein. (C) Specific binding of HsRXR and TaRXR to 3H 9-cis RA, calculated from the data shown in B by subtracting the binding to the UPL. Non-linear curve fit one-site specific binding, as implemented within GraphPad Prism. (D) Competitive binding of 3H 9-cis RA to TaRXR and HsRXR after subtracting non-specific binding to UPL.
RXR binding motifs
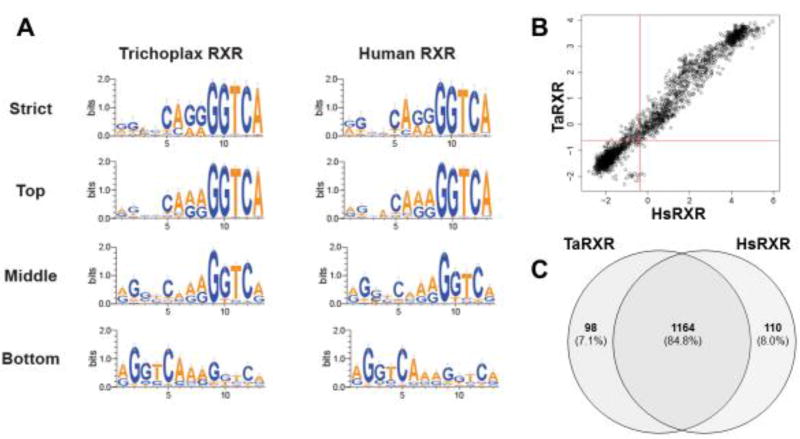
Custom PBMs were used to identify DNA binding motifs for TaRXR and to then compare these motifs with those bound by human RXRα. Bound sequences were ranked according to the signal intensity for each species. TaRXR bound a total of 1266 sequences (42%). The strongest binders contained a strongly conserved sequence (GGTCA) in the last five positions of the 3' half site and relatively well-conserved sequences in the four positions before these (Figure 3A). Weaker binding was measured for both halves of the DR1 sequence (middle and bottom binders), suggesting TaRXR may bind as a dimer despite a preference for the 3' half site. Human RXRα has been previously shown to bind a similar number of unique sequences (n = 1285) with the PWM showing predominant binding as a homodimer to two half sites as well as to the 3' half site in this PBM design (Fang, et al. 2012). In contrast, our results here using the same protein suggest strongest binding to the 3' half site; the difference is an artefactual one dependent on the precise number of sequences used to generate the PWM. The single top binding sequence for both human and TaRXR is an exact, full DR1 (AGGTCAAAGGTCA), suggesting that indeed both RXRs could be binding as homodimers. Binding motifs for human and T. adhaerens RXRs closely resemble one another for each grouping of binding strength. Overall specificity for binding is broadly correlated between the human and placozoan RXRs (Figure 3B). The similar specificity for binding by each RXR protein was mirrored by a large overlap in shared sequences bound in the PBM comparisons (Figure 3C). Approximately 84% of bound sequences are shared despite the vast evolutionary distance between these species.
Figure 3.
Binding site specificity of TaRXR and comparisons with HsRXRa assessed by protein binding microarray (PBM). (A) Weblogo summarizing the top 10% of sequences bound (strict), top 33% (top), middle 33% (middle), and bottom 33% (bottom) for TaRXR and HsRXRα bound sequences. (B) Overall specificity of TaRXR and HsRXRα over all sites bound is highly correlated. (C) Binding sites for these two RXR proteins are largely shared, with ~91% overlap of bound sequences.
Predicted RXR binding sites and Gene Ontology annotation
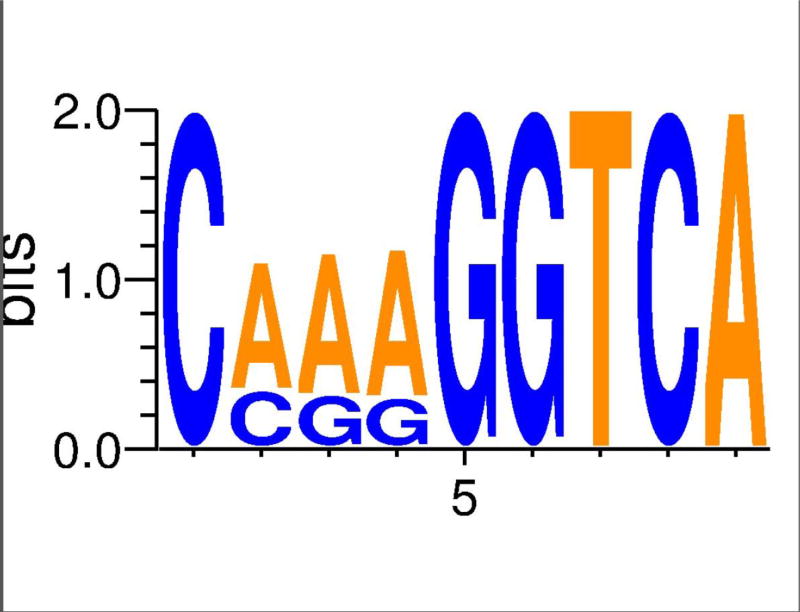
In total, our bioinformatic pipeline identified 1,567 potential "strict" RXR binding sites (i.e., match consensus motif from top 10% of PBM probes), 557 of which contain the most common extended half-site (i.e, CAAAGGTCA), within the T. adhaerens genome (Figure 4, Table 1). Of these, 685 RXR sites were within 1000 bases of a protein coding region, with approximately 10% of these assigned a potential function (Supplemental Table 2). The largest scaffold (Scaffold_1) in the T. adhaerens genome assembly had the highest number of these RXR binding sites (N = 109), although, 18 other scaffolds (39% of those recovered) had a higher RXR binding site occurrence per nucleotide. A relatively small scaffold (Scaffold_2888, length = 1338 bp) had the most RXR sites at ~4.5 per 1kb, while the average across scaffolds is ~1.4 per 10 kb (Supplemental Table 1). We further searched the placozoan genome for half-sites across the whole genome, in addition to regions adjacent to the RXR sites identified with the PBM data. Interestingly, these searches revealed that the first position in the half-site had a bias of 3:1 for A over G (Figure 4). Comparisons with the human genome for these individual binding sites suggest no strong bias in representation when genome size is accounted for (i.e., human genome is 60 times larger). Since potential dimer activity was detected in the PBM we searched for DR1 sites using the two common half-sites. Surprisingly, we only identified nine total, two of which were within 1 kb of an annotated gene (Table 1).
Figure 4.
Weblogo of each nucleotide across the 1,567 potential RXR binding sites identified in the the T. adhaerens genome.
Table 1.
Distribution of predicted RXR binding sites in the T. adhaerens genome and within 1 kb of an annotated gene. Identification of sites for possible RXR dimer binding was restricted to DR1-type binding sites composed of two half-sites that match the dominant sequences from PBM data (Figure 3A).
| Sequence motif for query (5' − 3') |
Name | Unique matches within 1kb of gene* |
Unique matches associated with RXR sites* |
Matches in T. adhaerens genome overall |
Unique matches in H. sapiens genome |
|---|---|---|---|---|---|
| CAAAGGTCA | Common top binder | 219 | 557 | 557 | 36,079 |
| C A/C A/G A/G GGTCA | Top 10% | 685 | 1,567 | 1,567 | 96,323 |
| AGGTCA | Half-site 1 (HS1) | 834 | 2,005 | 23,389 | 2,060,590 |
| GGGTCA | Half-site 2 (HS2) | 323 | 721 | 8,256 | 1,064,378 |
| AGGTCAnAGGTCA | DR1 (HS1-HS1) | 0 | 1 | 3 | 1,059 |
| GGGTCAnGGGTCA | DR1 (HS2-HS2) | 0 | 0 | 0 | 3,187 |
| AGGTCAnGGGTCA | DR1 (HS1-HS2) | 2 | 2 | 3 | 715 |
| GGGTCAnAGGTCA | DR1 (HS2-HS1) | 0 | 2 | 3 | 715 |
Values for all Half-site and DR1-type binding sites were estimated by screening 1KB regions flanking predicted RXR binding sites.
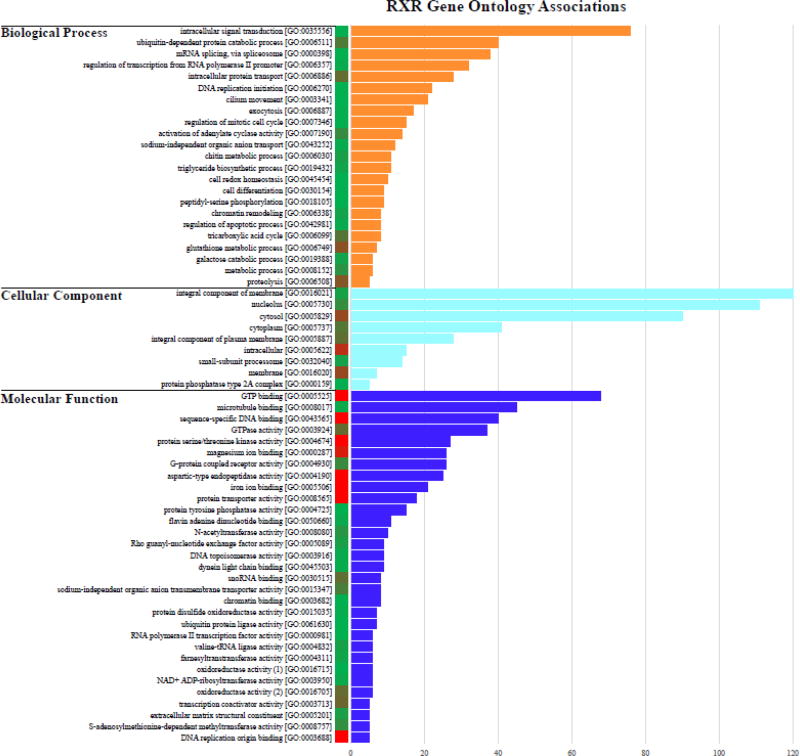
Many of the proteins near a predicted RXR binding site (Table 1) within the T. adhaerens genome could not be annotated using BLAST-based similarity searches against other taxa (574 of 648 proteins annotated only as “uncharacterized” or “predicted proteins”, Supplemental Table 2). Despite this, many of these proteins still contained GO information on UNIPROT, recovering 1,584 GO terms associated with the full set of 597 unique predicted proteins (Supplemental Table 3). The most abundant GO terms within the Biological Process domain were associated with intracellular signal transduction [GO:0035556], ubiquitin-dependent protein catabolic process [GO:0006511], and mRNA splicing visa spliceosome [GO:0000398]. The Cellular Component domain recovered the smallest number of GO terms, but included three of the most frequent terms: integral component of membrane [GO:0016021], nucleolus [GO:0005730], and cytosol [GO:005829]. The Molecular Function domain had the largest number of individual GO terms associated with predicted RXR target proteins, including GTP binding [GO:0005525], microtubule binding [GO:0008017], sequence-specific DNA binding [GO:0043565], and GTPase activity [GO:0003924] (Figure 5).
Figure 5.
Gene Ontology analysis of proteins identified through REVIGO with RXR binding sites within 1 kb of the start site. Values represent the abundance of selected GO terms; the full list of GO terms can be found in Supplemental Table 3. Colored column adjacent to sequence IDs correspond to REVIGO frequencies scores, with red representing GO terms that are more abundant/generic and green representing less abundant/rare GO terms.
DISCUSSION
Studies of nuclear receptor diversity and function from early-diverging animal phyla provide essential comparative data to determine historical patterns of gene evolution with particular molecular and physiological activities. NR-specific functions, including ligand-specific binding and cell-specific expression, are now well characterized in many bilaterian groups. Current understanding of the evolutionary origin of these genes and their ancestral functions has been heavily influenced by studies of individual NRs using a few species from early-diverging animal phyla. NRs in sponges and ctenophores and a majority of NRs from the cnidarians and placozoans are confidently placed in the NR2 family, suggesting that the ancestral NR resembled these proteins, particularly HNF4. RXR, as a member of NR2, also evolved quite early in animal evolution, likely at the ancestor of the Parahoxozoa, a group that includes Plazozoa, Cnidaria and Bilateria (terminology after Ryan, et al. 2010). Our analyses of NR complements from additional sponge species did not reveal any RXR homologs within sponges and consequently does not change the inferred timing for the evolution of RXR (Supplemental Figure 2). Wiens et al. (2003) previously reported that exposure to micromolar concentrations of all-trans retinoic acid induces tissue regression and up-regulates NR expression in the sponge Suberites domoncula. Given the absence of a clear retinoid receptor from the sponge lineage, the mechanism for these effects is still unclear.
Ligand binding assays within both placozoans (Novotný, et al. 2017 and this study) and cnidarians (Kostrouch, et al. 1998; Fuchs, et al. 2014) clearly support the hypothesis that the ancestral RXR bound ligand(s). This contrasts with vertebrate-type steroid receptors, in which the origins of ligand binding and activation are less clear (Thornton 2003; Holzer, et al. 2017). Within both protostome and deuterostome lineages, RXR homologs from many species are able to bind 9-cis RA. However the capacity for 9-cis RA binding has been lost from several lineages, most notably in the case of the insect ultraspiracle proteins (Iwema, et al. 2007), but also from the urochordate Halocynthis roretzi (Maeng, et al. 2012) and some RXR subtypes from the zebrafish Danio rerio (Jones, et al. 1995). As in other species, 9-cis RA is a useful pharmacological reagent to study the function of TaRXR, but it is not necessarily a physiological ligand. Identification of physiological ligands for RXR is a subject of active study (e.g., Rühl, et al. 2015 and references therein), and additional insight could be gained from assessing the activity of candidate ligands with RXRs from early-diverging animal lineages.
Evolutionary changes in the DNA binding domains of transcription factors are one molecular mechanism that can create shifts in the regulation of gene networks. Nonsynonymous mutations in these domains and other parts of the protein may result in amino acid substitutions that influence the particular sequence motifs bound (e.g., Balczarek, et al. 1997; Fang, et al. 2012; Sen, et al. 2013; Barrera, et al. 2016). Identification of NR binding sites has revealed subfamily-specific patterns of preferential binding to half sites with different spacing and orientation (e.g., Fang, et al. 2012). DNA binding motifs for TaRXR were highly similar to human RXRα with more than 84% of binding sites conserved. TaRXR bound DNA sequences that contained a majority of the conserved half-site AGGTCA, with only the most 5' position showing variation (A or G). The most strongly bound motif contained the canonical half-site. These results suggest extreme conservation of motifs for RXR orthologs over vast evolutionary distance. Indeed, binding sites for NR2 family members are generally conserved due to conservation of the contact amino acids in the DNA binding domain. However, subtle variations in the DNA binding motif for transcription factors can result in shifts in gene expression in particular cells types dependent on co-factors (Nakagawa, et al. 2013; Cheatle Jarvela, et al. 2014; Cary, et al. 2017).
Our queries of the T. adhaerens genome for potential RXR binding sites within 1 kb of the annotated genes suggested more than 600 potential downstream genes (of the total 11,514 predicted genes in the genome) could be regulated by this transcription factor. Of these, 192 predicted RXR binding sites were found adjacent to a predicted protein’s start codon; however, most of those proteins are uncharacterized. Of those with predicted functions, there were four locations within the T. adhaerens genome where RXR binding sites were found adjacent to start sites to the Eukaryotic translation initiation factor 3 (eIF-3) complex (B3RIH4) and two sites which may contain start sites for Acyl-coenzyme A oxidase (B3S9Y8). Recent research by Novotný, et al. (2017) showed that 9-cis-RA causes shifts in NR transcription, results in growth arrest in T. adhaerens, and induces expression of L-malate-NADP+ oxidoreductase. The annotations in the T. adhaerens genome remain largely unverified, thus our interpretations from these binding sites are certainly preliminary. Furthermore, these binding motifs may be shared with the other NR2 family genes in T. adhaerens (HNF4 and COUP-TF-like), as well as the ERR-like gene within the NR3 family. Future research to characterize the specific binding sites for these other three NRs and compare them to TaRXR would identify the potential degree of overlap for NR regulatory targets in this placozoan species.
It is of interest to determine whether TaRXR binds as a dimer or a monomer. The preference for a single half site from the PBM data suggest that Ta RXR may bind as a monomer. Particular bilaterian NRs have been shown to preferentially or exclusively bind to DNA as a monomer; e.g., NGFI-B (Meinke and Sigler 1999), TR (Quack and Carlberg 2001), and RORs (Schräder, et al. 1996). However, dimeric binding cannot necessarily be ascertained by PBMs. We also found a single half site preference for human RXR in our analysis, and mammalian RXRs are well-established to bind DNA as dimers, especially as heterodimers. Another perhaps more important question is, if TaRXR does bind DNA as a dimer, does it bind as a homo- or a heterodimer? There are just 4 NR genes in T. adhaerens. In addition to RXR, there is HNF4, which has been well established as an obligate homodimer that cannot heterodimerize with RXR in mammalian systems (Jiang, et al. 1995; Bogan, et al. 2000) and an ERR, which classically binds as a monomer (Johnston, et al. 1997). The last NR is COUP-TF-like, which is known to heterodimerize with RXR in bilaterians (e.g., Kliewer, et al. 1992). Co-immunoprecipitation studies to determine the potential dimerization activity of T. adhaerens NRs would be essential to determine how RXR may form protein-protein interactions. Identification of the first instance of heterodimerization among NRs is an important issue and requires additional studies.
Our study of ligand and DNA binding by TaRXR supports hypotheses that (1) ligand binding was present at the origin of RXR in animals, and (2) the DNA binding motif has changed little over more than 1 billion years of independent evolution. Although studies of steroid receptors have suggested that ligand binding may or may not have evolved after the evolution of the NR3 family (Eick and Thornton 2011; Markov and Laudet 2011), comparative studies of HNF4 in sponges suggest that specific ligand binding also was present early in animal evolution (Bridgham, et al. 2010). These data with NRs correspond with other transcription factors studied in early diverging phyla that have shown ligand specificity was present early animal evolution, e.g., canonical and non-canonical Wnt signaling (Lee, et al. 2006; Rigo-Watermeier, et al. 2012). Continued comparisons of transcription factor diversification and binding specificity will illuminate how ligand-regulated gene networks evolved in early animal evolution.
Supplementary Material
Acknowledgments
We would like to thank Andreas Heyland (University of Guelph) for providing T. adhaerens and for assisting in production of the cDNA library. We would also like to thank Joe Ryan (Whitney Laboratory) for access to transcriptomes to search for nuclear receptors in sponges and cnidarians. Support for AMT was provided by the Tropical Research Initiative and an Internal Research and Development Award from the Woods Hole Oceanographic Institution. AMR was supported by NIH award R15GM114740. JM was supported by NSF award 1536530 to AMR. DM-P, BF and FMS were supported by NIH award R01DK094707 to FMS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baker ME. Trichoplax, the simplest known animal, contains an estrogen-related receptor but no estrogen receptor: Implications for estrogen receptor evolution. Biochemical and Biophysical Research Communications. 2008;375 doi: 10.1016/j.bbrc.2008.08.047. [DOI] [PubMed] [Google Scholar]
- Balczarek KA, Lai ZC, Kumar S. Evolution of functional diversification of the paired box (Pax) DNA-binding domains. Molecular Biology and Evolution. 1997;14:829–842. doi: 10.1093/oxfordjournals.molbev.a025824. [DOI] [PubMed] [Google Scholar]
- Barrera LA, Vedenko A, Kurland JV, Rogers JM, Gisselbrecht SS, Rossin EJ, Woodard J, Mariani L, Kock KH, Inukai S, et al. Survey of variation in human transcription factors reveals prevalent DNA binding changes. Science. 2016;351:1450. doi: 10.1126/science.aad2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beato M, Herrlich P, Schutz G. Steroid hormone receptors: many actors in search of a plot. Cell. 1995;83:851–857. doi: 10.1016/0092-8674(95)90201-5. [DOI] [PubMed] [Google Scholar]
- Bertrand S, Brunet F, Escriva H, Parmentier G, Laudet V, Robinson-Rechavi M. Evolutionary genomics of nuclear receptors: from twenty-five ancestral genes to derived endocrine systems. Molecular Biology and Evolution. 2004;21:1923–1937. doi: 10.1093/molbev/msh200. [DOI] [PubMed] [Google Scholar]
- Blumberg B, Evans R. Orphan nuclear receptors--new ligands and new possibilities. Genes & Development. 1998;12:3149–3155. doi: 10.1101/gad.12.20.3149. [DOI] [PubMed] [Google Scholar]
- Bogan AA, Dallas-Yang Q, Ruse MD, Maeda Y, Jiang G, Nepomuceno L, Scanlan TS, Cohen FE, Sladek FM. Analysis of protein dimerization and ligand binding of orphan receptor HNF4α. Journal of Molecular Biology. 2000;302:831–851. doi: 10.1006/jmbi.2000.4099. [DOI] [PubMed] [Google Scholar]
- Bolotin E, Liao H, Ta TC, Yang C, Hwang-Verslues W, Evans JR, Jiang T, Sladek FM. Integrated approach for the identification of human hepatocyte nuclear factor 4α target genes using protein binding microarrays. Hepatology. 2010;51:642–653. doi: 10.1002/hep.23357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridgham JT, Eick GN, Larroux C, Deshpande K, Harms MJ, Gauthier MEA, Ortlund EA, Degnan BM, Thornton JW. Protein evolution by molecular tinkering: diversification of the nuclear receptor superfamily from a ligand-dependent ancestor. PLOS Biology. 2010;8:e1000497. doi: 10.1371/journal.pbio.1000497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cary GA, Cheatle Jarvela AM, Francolini RD, Hinman VF. Genome-wide use of high- and low-affinity Tbrain transcription factor binding sites during echinoderm development. Proceedings of the National Academy of Sciences. 2017;114:5854–5861. doi: 10.1073/pnas.1610611114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheatle Jarvela AM, Brubaker L, Vedenko A, Gupta A, Armitage BA, Bulyk ML, Hinman VF. Modular evolution of DNA-binding preference of a tbrain transcription factor provides a mechanism for modifying gene regulatory networks. Molecular Biology and Evolution. 2014;31:2672–2688. doi: 10.1093/molbev/msu213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia J-M, Brenner SE. WebLogo: A sequence logo generator. Genome Research. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eick GN, Thornton JW. Evolution of steroid receptors from an estrogen-sensitive ancestral receptor. Molecular and Cellular Endocrinology. 2011;334:31–38. doi: 10.1016/j.mce.2010.09.003. [DOI] [PubMed] [Google Scholar]
- Escriva H, Safi R, Hanni C, Langlois M-C, Saumitou-Laprade P, Stehelin D, Capron A, Pierce R, Laudet V. Ligand binding was acquired during the evolution of nuclear receptors. Proceedings of the National Academy of Science, USA. 1997;94:6803–6808. doi: 10.1073/pnas.94.13.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans RM, Mangelsdorf DJ. Nuclear receptors, RXR, and the big bang. Cell. 2014;157:255–266. doi: 10.1016/j.cell.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrbach SE, Smagghe G, Velarde RA. Insect nuclear receptors. Annual Review of Entomology. 2012;57:83–106. doi: 10.1146/annurev-ento-120710-100607. [DOI] [PubMed] [Google Scholar]
- Fang B, Mane-Padros D, Bolotin E, Jiang T, Sladek FM. Identification of a binding motif specific to HNF4 by comparative analysis of multiple nuclear receptors. Nucleic Acids Research. 2012;40:5343–5356. doi: 10.1093/nar/gks190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorucci S, Distrutti E, Bifulco G, D’Auria MV, Zampella A. Marine sponge steroids as nuclear receptor ligands. Trends in Pharmacological Sciences. 2012;33:591–601. doi: 10.1016/j.tips.2012.08.004. [DOI] [PubMed] [Google Scholar]
- Fuchs B, Wang W, Graspeuntner S, Li Y, Insua S, Herbst E-M, Dirksen P, Böhm A-M, Hemmrich G, Sommer F, et al. Regulation of polyp-to-jellyfish transition in Aurelia aurita. Current Biology. 2014;24:263–273. doi: 10.1016/j.cub.2013.12.003. [DOI] [PubMed] [Google Scholar]
- Giguere V, Yang N, Segui P, Evans R. Identification of a new class of steroid hormone receptors. Nature. 1988;331:91–94. doi: 10.1038/331091a0. [DOI] [PubMed] [Google Scholar]
- Hall TA. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nuclear Acids Symposium Series. 1999;41:95–98. [Google Scholar]
- Heyman RA, Mangelsdorf DJ, Dyck JA, Stein RB, Eichele G, Evans RM, Thaller C. 9-cis retinoic acid is a high affinity ligand for the retinoid × receptor. Cell. 1992;68:397–406. doi: 10.1016/0092-8674(92)90479-v. [DOI] [PubMed] [Google Scholar]
- Holzer G, Markov GV, Laudet V. Evolution of nuclear receptors and ligand signaling: Toward a soft key–lock model? In: Douglas F, Sophia T, editors. Current Topics in Developmental Biology. Academic Press; 2017. pp. 1–38. [DOI] [PubMed] [Google Scholar]
- Iwema T, Billas IM, Beck Y, Bonneton F, Nierengarten H, Chaumot A, Richards G, Laudet V, Moras D. Structural and functional characterization of a novel type of ligand-independent RXR-USP receptor. The EMBO Journal. 2007;26:3770–3782. doi: 10.1038/sj.emboj.7601810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang G, Nepomuceno L, Hopkins K, Sladek FM. Exclusive homodimerization of the orphan receptor hepatocyte nuclear factor 4 defines a new subclass of nuclear receptors. Molecular and Cellular Biology. 1995;15:5131–5143. doi: 10.1128/mcb.15.9.5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston SD, Liu X, Zuo F, Eisenbraun TL, Wiley SR, Kraus RJ, Mertz JE. Estrogen-related receptor α1 functionally binds as a monomer to extended half-site sequences including ones contained within estrogen-response elements. Molecular Endocrinology. 1997;11:342–352. doi: 10.1210/mend.11.3.9897. [DOI] [PubMed] [Google Scholar]
- Jones BB, Ohno CK, Allenby G, Boffa MB, Levin AA, Grippo JF, Petkovich M. New retinoid × receptor subtypes in zebra fish (Danio rerio) differentially modulate transcription and do not bind 9-cis retinoic acid. Molecular and Cellular Biology. 1995;15:5226–5234. doi: 10.1128/mcb.15.10.5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersten S, Gronemeyer H, Noy N. The DNA binding pattern of the retinoid × receptor Is regulated by ligand-dependent modulation of Its oligomeric state. Journal of Biological Chemistry. 1997;272:12771–12777. doi: 10.1074/jbc.272.19.12771. [DOI] [PubMed] [Google Scholar]
- Khorasanizadeh S, Rastinjad F. Nuclear-receptor interactions on DNA-response elements. Trends in Biochemical Sciences. 2001;26:384–390. doi: 10.1016/s0968-0004(01)01800-x. [DOI] [PubMed] [Google Scholar]
- Kliewer SA, Umesono K, Heyman RA, Mangelsdorf DJ, Dyck JA, Evans RM. Retinoid × receptor-COUP-TF interactions modulate retinoic acid signaling. Proceedings of the National Academy of Sciences. 1992;89:1448–1452. doi: 10.1073/pnas.89.4.1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostrouch Z, Kostrouchova M, Love W, Jannini E, Piatigorsky J, Rall J. Retinoic acid × receptor in the diploblast, Tripedalia cystophora. Proceedings of the National Academy of Science, USA. 1998;95 doi: 10.1073/pnas.95.23.13442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laffitte BA, Kast HR, Nguyen CM, Zavacki AM, Moore DD, Edwards PA. Identification of the DNA binding specificity and potential target genes for the farnesoid X-activated receptor. Journal of Biological Chemistry. 2000;275:10638–10647. doi: 10.1074/jbc.275.14.10638. [DOI] [PubMed] [Google Scholar]
- Laudet V, Hanni V, Coll J, Catzeflis F, Stehelin D. Evolution of the nuclear receptor gene superfamily. The EMBO Journal. 1992;11:1003–1013. doi: 10.1002/j.1460-2075.1992.tb05139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laumer CE, Gruber-Vodicka H, Hadfield MG, Pearse VB, Riesgo A, Marioni JC, Giribet G. Placozoans are eumetazoans related to Cnidaria. bioRxiv 2017 [Google Scholar]
- Lee PN, Pang K, Matus DQ, Martindale MQ. A WNT of things to come: Evolution of Wnt signaling and polarity in cnidarians. Seminars in Cell & Developmental Biology. 2006;17:157–167. doi: 10.1016/j.semcdb.2006.05.002. [DOI] [PubMed] [Google Scholar]
- Lefebvre P, Benomar Y, Staels B. Retinoid × receptors: common heterodimerization partners with distinct functions. Trends in Endocrinology & Metabolism. 2010;21:676–683. doi: 10.1016/j.tem.2010.06.009. [DOI] [PubMed] [Google Scholar]
- Mader S, Chen JY, Chen Z, White J, Chambon P, Gronemeyer H. The patterns of binding of RAR, RXR and TR homo- and heterodimers to direct repeats are dictated by the binding specificites of the DNA binding domains. The EMBO Journal. 1993;12:5029–5041. doi: 10.1002/j.1460-2075.1993.tb06196.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeng S, Lee JH, Choi S-C, Kim MA, Shin YK, Sohn YC. The retinoid × receptor in a marine invertebrate chordate: Evolutionary insights from urochordates. General and Comparative Endocrinology. 2012;178:380–390. doi: 10.1016/j.ygcen.2012.06.019. [DOI] [PubMed] [Google Scholar]
- Mangelsdorf D, Thummel C, Beato M, Herrlich P, Schatz G, Umesono K, Blumberg B, Chambon P, Evans R. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus S, Winrow C, Capone J, Rachubinski R. A p56lck ligand serves as a coactivator of an orphan nuclear hormone receptor. Journal of Biological Chemistry. 1996;271:27197–27200. doi: 10.1074/jbc.271.44.27197. [DOI] [PubMed] [Google Scholar]
- Markov GV, Laudet V. Origin and evolution of the ligand-binding ability of nuclear receptors. Molecular and Cellular Endocrinology. 2011;334:21–30. doi: 10.1016/j.mce.2010.10.017. [DOI] [PubMed] [Google Scholar]
- Meinke G, Sigler PB. DNA-binding mechanism of the monomeric orphan nuclear receptor NGFI-B. Nature Structural & Molecular Biology. 1999;6:471–477. doi: 10.1038/8276. [DOI] [PubMed] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. The CIPRES science gateway: a community resource for phylogenetic analyses. Proceedings of the 2011 TeraGrid Conference: Extreme Digital Discovery; Salt Lake City, Utah. 2016785: ACM; 2011. pp. 1–8. [Google Scholar]
- Nakagawa S, Gisselbrecht SS, Rogers JM, Hartl DL, Bulyk ML. DNA-binding specificity changes in the evolution of forkhead transcription factors. Proceedings of the National Academy of Sciences. 2013;110:12349–12354. doi: 10.1073/pnas.1310430110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosenko T, Schreiber F, Adamska M, Adamski M, Eitel M, Hammel J, Maldonado M, Müller WEG, Nickel M, Schierwater B, et al. Deep metazoan phylogeny: When different genes tell different stories. Molecular Phylogenetics and Evolution. 2013;67:223–233. doi: 10.1016/j.ympev.2013.01.010. [DOI] [PubMed] [Google Scholar]
- Novotný JP, Chughtai AA, Kostrouchová M, Kostrouchová V, Kostrouch D, Kaššák F, Kaňa R, Schierwater B, Kostrouchová M, Kostrouch Z. Trichoplax adhaerens reveals a network of nuclear receptors sensitive to 9-cis-retinoic acid at the base of metazoan evolution. Peer J. 2017;5:e3789. doi: 10.7717/peerj.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuclear Receptors Nomenclature Committee. A unified nomenclature system for the nuclear receptor superfamily. Cell. 1999;97:161–163. doi: 10.1016/s0092-8674(00)80726-6. [DOI] [PubMed] [Google Scholar]
- Oliveros JC. Venny. An interactive tool for comparing lists with Venn's diagrams. 2007–2015 http://bioinfogp.cnb.csic.es/tools/venny/index.html.
- Power R, Lydon J, Conneely O, O'Malley B. Dopamine activation of an orphan of the steroid receptor superfamily. Science. 1991;252:1546–1548. doi: 10.1126/science.2047861. [DOI] [PubMed] [Google Scholar]
- Quack M, Carlberg C. Single thyroid hormone receptor monomers are competent for co-activator-mediated transactivation. Biochemical Journal. 2001;360:387–393. doi: 10.1042/0264-6021:3600387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitzel AM, Pang K, Ryan JF, Mullikin JC, Martindale MQ, Baxevanis AD, Tarrant AM. Nuclear receptors from the ctenophore Mnemiopsis leidyi lack a zinc-finger DNA-binding domain: lineage-specific loss or ancestral condition in the emergence of the nuclear receptor superfamily? EvoDevo. 2011;2:3. doi: 10.1186/2041-9139-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitzel AM, Tarrant AM. Nuclear receptor complement of the cnidarian Nematostella vectensis: phylogenetic relationships and developmental expression patterns. BMC Evolutionary Biology. 2009;9:230. doi: 10.1186/1471-2148-9-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riesgo A, Farrar N, Windsor PJ, Giribet G, Leys SP. The analysis of eight transcriptomes from all poriferan classes reveals surprising genetic complexity in sponges. Molecular Biology and Evolution. 2014;31:1102–1120. doi: 10.1093/molbev/msu057. [DOI] [PubMed] [Google Scholar]
- Rigo-Watermeier T, Kraft B, Ritthaler M, Wallkamm V, Holstein T, Wedlich D. Functional conservation of Nematostella Wnts in canonical and noncanonical Wnt-signaling. Biology Open. 2012;1:43–51. doi: 10.1242/bio.2011021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson-Rechavi M, Escriva Garcia H, Laudet V. The nuclear receptor superfamily. Journal of Cell Science. 2003;116:585–586. doi: 10.1242/jcs.00247. [DOI] [PubMed] [Google Scholar]
- Rühl R, Krzyżosiak A, Niewiadomska-Cimicka A, Rochel N, Szeles L, Vaz B, Wietrzych-Schindler M, Álvarez S, Szklenar M, Nagy L, et al. 9-cis-13,14-dihydroretinoic acid is an endogenous retinoid acting as RXR ligand in mice. PLOS Genetics. 2015;11:e1005213. doi: 10.1371/journal.pgen.1005213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlicker A, Domingues FS, Rahnenführer J, Lengauer T. A new measure for functional similarity of gene products based on Gene Ontology. BMC Bioinformatics. 2006;7:302. doi: 10.1186/1471-2105-7-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schräder M, Danielsson C, Wiesenberg I, Carlberg C. Identification of natural monomeric response elements of the nuclear receptor RZR/ROR. They also bind COUP-TF homodimers. The Journal of Biological Chemsitry. 1996;271:19732–19736. doi: 10.1074/jbc.271.33.19732. [DOI] [PubMed] [Google Scholar]
- Sen P, Yang Y, Navarro C, Silva I, Szafranski P, Kolodziejska KE, Dharmadhikari AV, Mostafa H, Kozakewich H, Kearney D, et al. Novel FOXF1 mutations in sporadic and familial cases of alveolar capillary dysplasia with misaligned pulmonary veins imply a role for its DNA binding domain. Human Mutation. 2013;34:801–811. doi: 10.1002/humu.22313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sever R, Glass CK. Signaling by nuclear receptors. Cold Spring Harbor Perspectives in Biology. 2013;5 doi: 10.1101/cshperspect.a016709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava M, Begovic E, Chapman J, Putnam NH, Hellsten U, Kawashima T, Kuo A, Mitros T, Salamov A, Carpenter ML, et al. The Trichoplax genome and the nature of placozoans. Nature. 2008;454 doi: 10.1038/nature07191. [DOI] [PubMed] [Google Scholar]
- Supek F, Bošnjak M, Škunca N, Šmuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One. 2011;6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarrant AM, Behrendt L, Stegeman JJ, Verslycke T. Ecdysteroid receptor from the American lobster Homarus americanus: EcR/RXR isoform cloning and ligand-binding properties. General and Comparative Endocrinology. 2011;173:346–355. doi: 10.1016/j.ygcen.2011.06.010. [DOI] [PubMed] [Google Scholar]
- Taubert S, Ward JD, Yamamoto KR. Nuclear hormone receptors in nematodes: Evolution and function. Molecular and Cellular Endocrinology. 2011;334:49–55. doi: 10.1016/j.mce.2010.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton JW. Nuclear receptor diversity: phylogeny, evolution and endocrine disruption. Pure and Applied Chemistry. 2003;75 [Google Scholar]
- Wiens M, Batel R, Korzhev M, Müller W. Retinoid × receptor and retinoic acid response in the marine sponge Suberites domuncula. Journal of Experimental Biology. 2003;206:3261–3271. doi: 10.1242/jeb.00541. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.