Abstract
Infection with the apicomplexan protozoan parasite Toxoplasma gondii is an ongoing public health problem. The parasite's ability to invade and replicate within the host cell is dependent on many effectors, such as dense granule proteins (GRAs) released from the specialized organelle dense granules, into host cells. GRAs have emerged as important determinants of T. gondii pathogenesis. However, the functions of some GRAs remain undefined. In this study, we used CRISPR-Cas9 technique to disrupt 17 GRA genes (GRA11, GRA12 bis, GRA13, GRA14, GRA20, GRA21, GRA28-31, GRA33-38, and GRA40) in the virulent T. gondii RH strain. The CRISPR-Cas9 constructs abolished the expression of the 17 GRA genes. Functional characterization of single ΔGRA mutants was achieved in vitro using cell-based plaque assay and egress assay, and in vivo in BALB/c mice. Targeted deletion of these 17 GRA genes had no significant effect neither on the in vitro growth and egress of the mutant strains from the host cells nor on the parasite virulence in the mouse model of infection. Comparative analysis of the transcriptomics data of the 17 GRA genes suggest that GRAs may serve different functions in different genotypes and life cycle stages of the parasite. In sum, although these 17 GRAs might not be essential for RH strain growth in vitro or virulence in mice, they may have roles in other strains or parasite stages, which warrants further investigations.
Keywords: Toxoplasma gondii, CRISPR-Cas9, dense granule proteins (GRAs), host-pathogen interaction, virulence
Introduction
Toxoplasma gondii is an obligate intracellular apicomplexan protozoan parasite that can infect all warm-blooded animals, mainly through oral and congenital infections (Tenter et al., 2000; Elmore et al., 2010; Zhou et al., 2011). This parasite has a significant medical and socioeconomic importance because it infects over two billion people worldwide. It is the causative agent of toxoplasmosis, an important zoonotic disease that can cause serious, even fatal health consequences in immunocompromised individuals, such as AIDS patients and organ transplant recipients (Liu et al., 2008; Xiao et al., 2010; Chemoh et al., 2015). Infection in immunocompetent individuals are generally asymptomatic, however the parasite persists as bradyzoites-containing cysts in brain and muscle tissues for many years. Primary infection in pregnant women can lead to fetus death, deformity, abortion, and long-term damage of the eye and central nervous system (Hill and Dubey, 2002).
The remarkable ability of T. gondii to invade and colonize virtually all nucleated cells (Morisaki et al., 1995), and to adopt a successful intracellular lifestyle depends partly on the sequential secretion of effectors from the specialized secretory organelles micronemes, rhoptries and dense granules (Håkansson et al., 2001; Boothroyd and Dubremetz, 2008; Nadipuram et al., 2016). T. gondii tachyzoites enter the host cells through an active invasion mechanism and replicate within a membrane-bound parasitophorous vacuole (PV) inside the surrogate host cell cytoplasm (Coppens et al., 2006; Nam, 2009). After release from the host cell, newly formed parasites invade new mammalian host cells, and the replication cycle starts again. Most of the dense granule proteins (GRAs) are destined to the PV and parasitophorous vacuole membrane (PVM), and contribute to the biogenesis and maturation of the PV, and nutrient acquisition (Mercier and Cesbron-Delauw, 2015).
Some GRA proteins have the ability to traffic to the host cytoplasm or nucleus and interfere with host cell signaling pathways. For example, GRA6 is a polymorphic dense granule protein and activates the host transcription nuclear factor of activated T cells 4 (NFAT4) in order to manipulate host immune responses to maximize the parasite virulence in a strain-specific manner (Ma et al., 2014). GRA15 is another strain-specific effector that activates NF-κB pathway and induces IL-12 secretion in type II, but not type I or III genotypes. GRA15-deficient type II strain cannot activate NF-κB pathway or induces IL-12 secretion, hence GRA15-deficient type II strains grow faster compared with wild-type strains (Rosowski et al., 2011). GRA16 and GRA24 also manipulate host gene expression and signaling pathways (Bougdour et al., 2013; Braun et al., 2013). GRA17 mediates the transfer of small molecules between the host cell and PV, and maintains the stability of the PV (Gold et al., 2015). GRA7, GRA25, and GRA39 are also virulence factor and can interfere with host cell signaling pathways (Alaganan et al., 2014; Shastri et al., 2014; Nadipuram et al., 2016). GRA22 and GRA41 are involved in the parasite egress (Okada et al., 2013; LaFavers et al., 2017).
Despite the wealth of information regarding T. gondii's 40+ GRAs including sequence variation and expression of GRA coding genes, and their roles in the infection process, the contribution of many GRAs to the parasite growth and virulence are still unclear. The CRISPR-Cas9 system provides a novel and promising tool for editing T. gondii genes (Shen et al., 2014) and the genome (Sidik et al., 2014; Wang et al., 2016a; Shen et al., 2017). Using CRISPR-Cas9 to target T. gondii's GRA genes may ultimately offer a new approach to achieving functional cure to toxoplasmosis. In this study, we used the CRISPR-Cas9 technique to edit 17 GRA genes, namely GRA11, GRA12 bis, GRA13, GRA14, GRA20, GRA21, GRA28-31, GRA33-38, and GRA40 in the virulent T. gondii RH strain and examined the effects of gene loss on the parasite's ability to grow and exit from host cells in vitro and to cause death in mice.
Materials and methods
Parasite and cell cultures
Tachyzoites of T. gondii RH strain (type I) were maintained in vitro by passages in human foreskin fibroblast (HFF, ATCC, Manassas, VA, USA). HFF cells were grown in 75-cm2 tissue culture flasks containing Dulbecco's Modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 10 μg/ml gentamycin, at 37°C under a 5% CO2 atmosphere. To purify tachyzoites, infected HFF cells were lysed through a 27-gauge needle and the tachyzoites were filtered using a 5 μm pore size Millipore filter. The number of purified parasites was counted using a hemocytometer under a phase-contrast microscopy.
Construction of GRA knockout T. gondii strains using CRISPR-Cas9
GRA knockout strains were constructed by using CRISPR-Cas9 as previously described (Wang et al., 2016b). All plasmids, primers, and gRNAs used in this study are listed in Table S1. Briefly, sgRNA of each GRA was engineered into pSAG1::CAS9-U6::sgUPRT by PCR using the Q5 Mutagenesis Kit (NEB). Positive plasmid was extracted with Endo-Free Plasmid DNA Mini Kit Protocols (OMEGA). The resistance cassettes (DHFR*-Ts) were amplified from the plasmid pUPRT-DHFR-D and purified by agarose gel electrophoresis. About 40 μg positive plasmids and 15 μg purified DHFR*-Ts amplicons were co-transfected into freshly harvested T. gondii RH tachyzoites by electroporation. GRA-deficient tachyzoites were selected with pyrimethamine and examined by PCR analysis. Stable clones were confirmed by reverse transcription PCR (RT-PCR) by comparison to WT strains. Total RNA was extracted from tachyzoites of wild type (WT) or ΔGRA mutant T. gondii strains using TRIzol (Invitrogen). Reverse transcription was performed using a PrimeScriptTM 1st Strand cDNA Synthesis Kit (TaKaRa). The central region of each target gene cDNA was amplified by RT-PCR using specific primers (see Table S1 in the Supplemental Material).
Assessment of parasite growth using plaque assay
The growth rates of individual GRA-deficient and WT RH strains were determined in HFF cells. Cells were grown to confluence overnight in 6-well cell culture plates. The cells were then infected with tachyzoites of ΔGRA mutant and WT RH strains (~200 tachyzoites/well). Cells were incubated for 3 h to allow the parasites to enter the host cells and were then washed twice with sterile 1 × phosphate-buffered saline (PBS) to remove unbound parasites, and fresh medium was added. The plates were incubated for 7 days at 37°C in 5% CO2 environment. Then, the culture medium was removed and infected HFF cells were fixed with 4% paraformaldehyde in PBS (pH 7.4) for 15 min at ambient temperature. Then, fixed cells were incubated with crystal violet staining solution (2% [wt/vol] crystal violet, pH 7.4) for 10 min at ambient temperature. The size of plaques (i.e., areas in the cell culture devoid of cells caused by the proliferating parasites) was determined using inverted microscope as previously described (Wang et al., 2017). This experiment was performed in triplicate and the experiments were repeated three independent times.
Egress assay
Asexual reproduction of T. gondii culminates with the egress of the newly formed tachyzoites from the surrogate host cell and subsequent parasite invasion into new host cells. Therefore, parasite egress represents a very important event which is indispensable for the parasite dissemination in the host body. In order to determine the role of the GRAs in this important process, we examined the effect of ionophore A23187 modulating Ca2+ homeostasis on the egress of the parasite from the host cells. Briefly, HFF cells were incubated with 105 freshly harvested T. gondii tachyzoites in 2 ml culture medium for 3 h, followed by washing twice with DMEM medium to remove the unbound parasites. After 30–36 h of incubation, the wells were washed twice with sterile PBS and 3 μM calcium ionophore A23187 (Sigma) diluted in DMSO were added to the HFF cells. Live cell microscopy was used to monitor the timing of parasite egress from HFF cells infected with the WT strain compared with HFF cells infected with the mutant strains after addition of 3 μM calcium ionophore A23187.
Mouse infection with GRA-deficient strains
Specific-pathogen-free (SPF) inbred female BALB/c mice (8 weeks-old) were purchased from Center of Laboratory Animals, Lanzhou Veterinary Research Institute, Lanzhou, China. Mice (5 mice/cage) were housed in a SPF environment within the animal care facility during the experiment. Each mouse was injected intraperitoneally with 200 freshly harvested tachyzoites of GRA-deficient or WT RH strain (10 mice per strain). The negative control mice were injected with an equal amount PBS only. All mice were monitored daily for the signs of illness and time of death.
Bioinformatics analysis of T. gondii dense granule proteins
Information on the genomic features (signal peptide, the number of exons and transmembrane domains) and time-series expression data of the GRA genes by the parasite cell cycle phases, parasite life cycle stages (the oocyst, tachyzoite, and bradyzoites), and the parasite genotypes were downloaded from ToxoDB (http://ToxoDB.org; Gajria et al., 2007). GRA gene expression data were processed using Robust Multiarray Average (RMA) algorithm of the Partek Genomics Suite package (Partek, Inc, St Louis, MO, USA).
Statistical analysis
Statistical analysis for the in vitro and in vivo experiments was carried out using Graphpad Software package (GraphPad Software, La Jolla, CA). All experiments in the present study were conducted with at least 3 replicate and data were shown as means ± standard deviations (SD). Significant differences between means were determined by Student's t-test. P-values below 0.05 were considered significant.
Results and discussion
In this study, we assessed the impact of targeted disruption of the individual 17 GRA genes (GRA11, GRA12 bis, GRA13, GRA14, GRA20, GRA21, GRA28-31, GRA33-38, and GRA40) on the ability of the virulent T. gondii RH tachyzoites to grow and exit from the cultured HFF cells. We also examined the impact of the disruption of GRA genes on the virulence of T. gondii in BALB/c mice.
Generation of single ΔGRA knockout RH strains
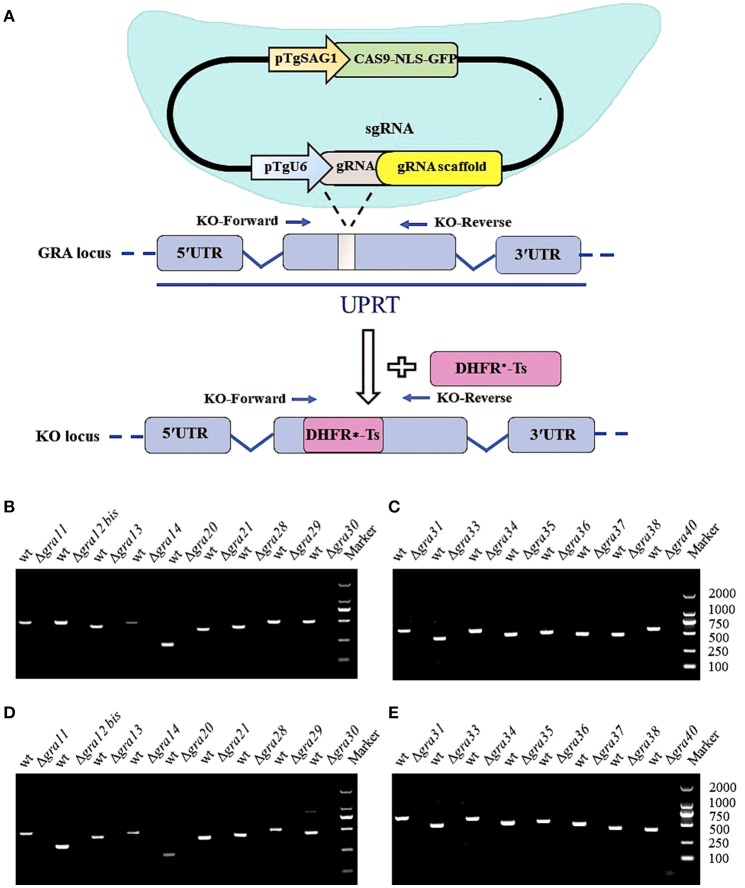
CRISPR-Cas9 technique was used to disrupt the GRA genes in type I RH strain. We designed RNA-guided CRISPR-Cas9 targeting T. gondii 17 GRA genes individually. The larger pyrimethamine-resist fragment (DHFR*-Ts) was designed for inserting into the sgRNA-targeted coding region causing frameshift mutation for GRA proteins (Figure 1A). Then, single clones were identified by PCR, and small fragment (~500 bp) was not amplified in GRA-deficient strains due to the insertion of the large DHFR*-Ts fragment with short extension time, however it was detected in the WT strain (Figures 1B,C). RT-PCR was used to amplify cDNA products, reverse transcribed from mRNA, of each GRA gene. Our result showed that CRISPR-Cas9 abolished the expression of GRA genes in the transfected strains compared with the WT strains (Figures 1D,E), indicating that the target GRA genes were mutated at the Cas9 cleavage sites and that 17 ΔGRA mutant strains were successfully generated.
Figure 1.
Overview of the CRISPR/Cas9 system and mutation analysis of Toxoplasma gondii dense granule proteins (GRA) genes. (A) Schematic representation of CRISPR-Cas9 system used for disrupting the 17 GRAs genes by insertion of DHFR*-Ts cassette. (B,C) Diagnostic PCR shows GRA gene disruption in the mutants compared to the wild-type strain. The KO-forward and KO-reverse primers were used to amplify the small fragment with 30 s extension time. (D,E) RT-PCR of mRNA from parental RH (WT) and GRA-deficient strains, showing that the 17 GRA's coding sequences were successfully disrupted. Marker denotes the DNA ladder.
Deletion of GRA genes did not affect the parasite growth and egress
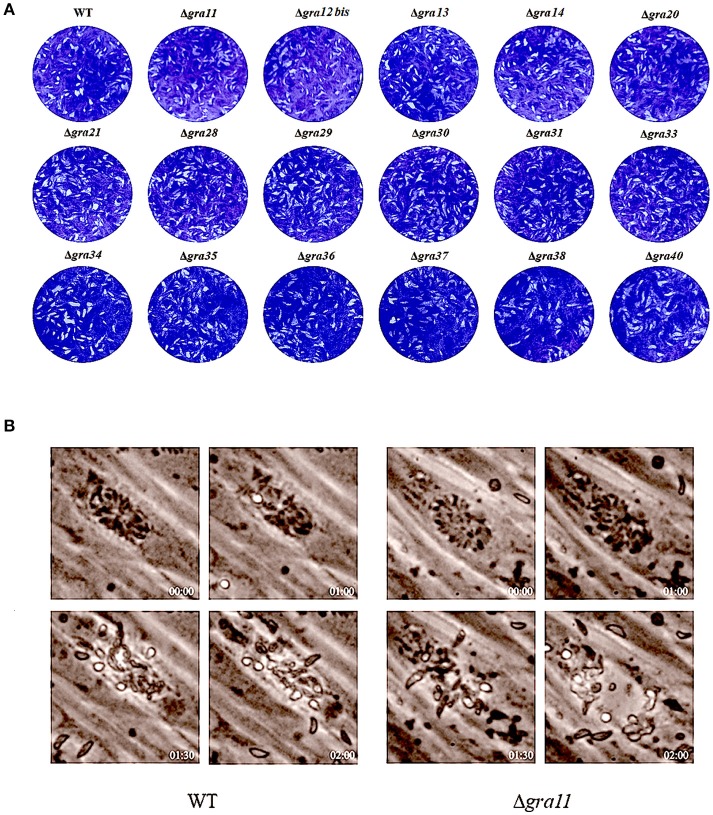
After confirming knockout by RT-PCR, we compared the plaque formation in HFF monolayers grown in 6-well culture plates 7 days after infection with 200 tachyzoites of wild-type and ΔGRA mutant strains by conventional crystal violet staining and microscopic examination. As shown in Figure 2A, no significant differences in the size of plaques were observed in cells infected with wild-type compared to cells infected with ΔGRA mutant strains (p = 0.1582). Next, we evaluated the role of GRAs in the parasite egress. The infected HFF cells were treated with 3 μM calcium ionophore A23187 and the timing of the parasite exit from the cells was monitored over 5 min (Figure 2B). No significant difference in the parasites egress was observed between ΔGRA mutants and WT strains. The 17 GRA KO strains and the WT strains remained within the PV after stimulation with DMSO. Calcium is a critical mediator of T. gondii invasion and egress processes. The calcium ionophore A23187 can stimulate the parasite to exit from the PV to the extracellular space (Arrizabalaga et al., 2004; Caldas et al., 2010). GRA41 can regulate the timing of egress and the sensitivity to calcium (LaFavers et al., 2017). However, none of the 17 GRA mutant strains tested in the present study seem to be responsive to the action of the A23187. In agreement with our results, a previous study has shown that deletion of 15 rhoptry organelle proteins ROPs (ROP10, ROP11, ROP15, ROP20, ROP23, ROP31, ROP32, ROP33, ROP34, ROP35, ROP36, ROP40, ROP41, ROP46, and ROP47) did not suppress the parasite's ability to grow in HFF cells or alter its pathogenicity for BALB/c mice (Wang et al., 2017). This lack of effect of GRA deletion on the phenotype of single parasite mutants argues for possible redundancy of function for GRAs.
Figure 2.
Phenotypic characterization of GRA Knockouts in vitro. (A) Two hundred freshly harvested T. gondii tachyzoites of WT RH strains and GRA-deficient RH strains per well were added to monolayers of HFF cells in 6-well culture plates. After 7 days, the number of plaques caused by the parasite's proliferation was counted using a microscope. No differences were detected in the number of plaques produced by wild-type (WT) RH strain vs. any of the GRA knockout strains. (B) Representative images show the parasite egress of the parental WT RH strain and one of the GRA mutant RH strains (Δgra11). Live cell imaging showed a similar egress pattern between WT and all mutant parasites after addition of 3 μM calcium ionophore A23187 at the indicated time points.
GRA deletion did not alter the parasite virulence in mice
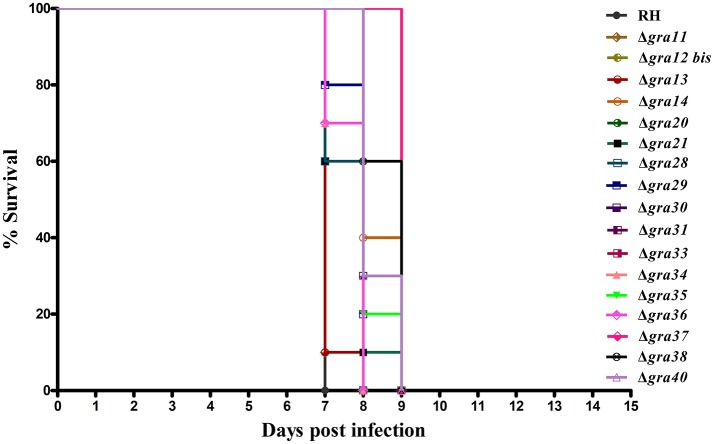
We tested whether deletion of GRA genes in T. gondii RH strains would cause any changes in the parasite virulence. We inoculated female BALB/c mice intraperitoneally with 200 T. gondii tachyzoites (wild-type or mutants) and monitored the mortality and signs of illness on a daily basis. All mice died within 7–9 days (Figure 3), suggesting that the 17 GRA genes tested in the present study might not contribute to the parasite virulence during infection. Previous studies have shown that some of dense granule proteins such as GRA25 may regulate the production of CCL2 and CCL1 in macrophages and thus affect virulence, which was different between Type II and Type III strains (Shastri et al., 2014). GRA39 is also an important virulence factor in Type II strains (Nadipuram et al., 2016). However, our results did not show any significant differences between WT and ΔGRA mutant strains. The fact that ΔGRA strains did not display any reduction or loss of pathogenicity in mice suggests that deletion of a single gene might not be enough to influence the highly virulent RH strain in which the knockout was performed. More research is required to elucidate the impact of individual or multiple GRA gene deletion on the phenotype and virulence of T. gondii strains. It is possible that these GRA genes have roles in the pathogenesis of T. gondii, but in T. gondii strains of other genotypes or in other hosts. Disruption of these GRAs in avirulent and more physiologically relevant cystogenic strains may allow the assessment of more subtle roles in the virulence, alteration in cyst formation or tissue tropism.
Figure 3.
Survival of BALB/c mice infected with Toxoplasma gondii wild-type or GRA-deficient RH strains. The mice were injected i.p., with 200 freshly harvested tachyzoites of the indicated strains. Ten mice were used per parasite strain. The survival time was recorded daily until all the mice have died within 7–9 days post challenge.
Sequence and expression analyses of GRAs in T. gondii
More than one-third of T. gondii mRNAs exhibit a tightly regulated expression pattern, probably driven by the diverse cell-cycle-dependent processes mediated by the parasite in the course of infection (Behnke et al., 2010). We mapped the transcriptomic data of the 17 GRAs available in ToxoDB and found that the expression profile of the majority of GRAs do not follow a particular cell cycle pattern, and that GRA11 and GRA12 bis expression level was low (Figure 4A). Bioinformatics features of GRAs, such as the number of exons, signal peptide and transmembrane domains are summarized in Table 1. The majority of the known GRAs include a signal peptide at the N terminus and a single transmembrane domain in the C-terminal part of the protein. We found that some GRAs encode multi-exons (even eight) and a small amount of GRAs do not have a signal peptide or a transmembrane domain. The signal peptide plays an important role in protein targeting and protein translocation in and eukaryotic cells and is considered as a feature of the GRA proteins that enter the secretory pathway (Hakimi and Bougdour, 2015). Most of the previously identified GRA proteins have been predicted to contain classical or non-classical signal peptide, such as GRA1, GRA2, GRA6, GRA9, GRA16, and GRA21, whereas other GRAs (GRA10, GRA15, GRA20, and GRA22) did not (Mercier and Cesbron-Delauw, 2015). However, a few GRAs do not seem to depend on signal peptide to enter the secretory pathway, such as GRA5, which is secreted into the PV as a unique soluble protein and then binds to the PVM (Gendrin et al., 2008). Some GRA proteins lack the transmembrane domain, typically GRA19, GRA20, and GRA21, and may interact with the PVM through protein-protein interactions (Hsiao et al., 2013). Other GRAs (38, 39, and 40) lack the transmembrane domains or other identifiable sequences for membrane association, suggesting they are soluble protein of the PV (Nadipuram et al., 2016). It is possible that GRAs that lack signal peptide or transmembrane domain perform their roles by as yet unknown mechanisms.
Figure 4.
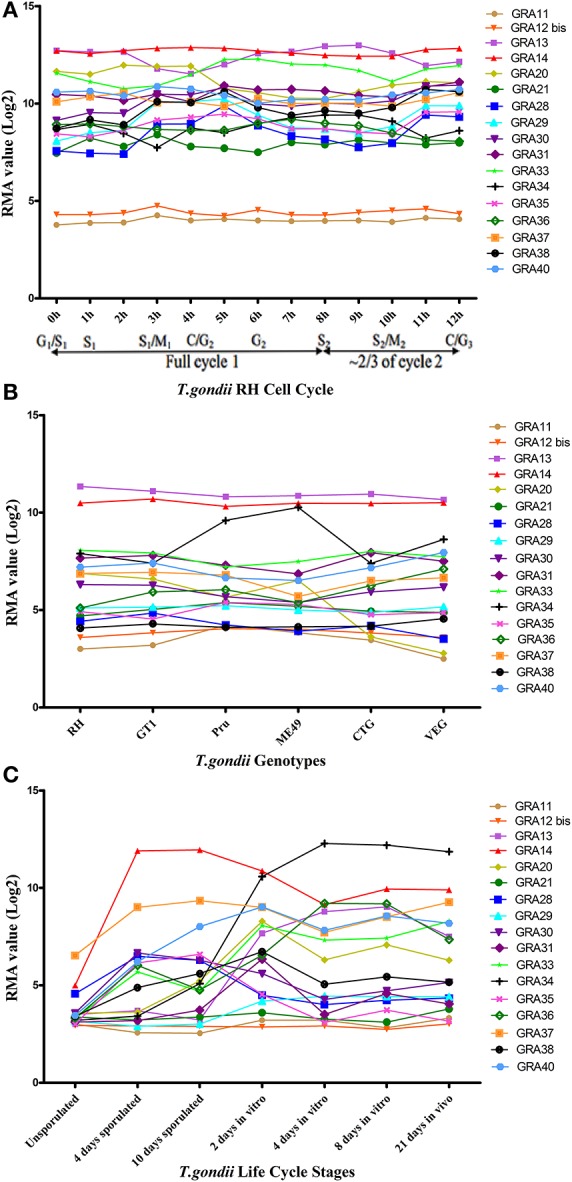
The trend charts of the distinct expression profiles of Toxoplasma gondii GRAs. (A) Time-series expression profile of 17 GRA genes of T. gondii RH strain by cell cycle phases of the parasite as described by Behnke et al. (2010). (B) Transcriptomic expression profiles of 17 GRA genes in Type I (RH and GT1), Type II (Pru and ME49), and Type III (CTG and VEG) strains. (C) Transcriptomic profiles of 17 GRA genes related to the parasite life cycle stages (oocyst, tachyzoite and bradyzoite). Expression profile of 17 GRA genes of the oocysts recovered from cat feces, at 0 day (unsporulated), 4 days (4 day sporulated), and 10 days (10 day sporulated), tachyzoites grown for 2 days in HFF cells (2 day in vitro), bradyzoites grown in HFF cells for 4 days and 8 days (4 day in vitro and 8 day in vitro), and 21 days tissue cyst-containing bradyzoites harvested from infected mouse brains (21 day in vivo). Each line represents the expression value of the corresponding gene. The data were obtained from ToxoDB (36 release) and the graph was generated using GraphPad Prism version 5.0.
Table 1.
Bioinformatics features of GRA proteins of Toxoplasma gondii.
| Name | Gene ID | Product description | Exons | Phenotype value | TMHMMa | Predicted signal peptide | Acute infection b | Chronic infection b |
|---|---|---|---|---|---|---|---|---|
| GRA11 | TGGT1_212410 | Dense granule protein GRA11 | 1 | ND | Yes | Yes | 5.97 | 5.21 |
| GRA12 bis | TGGT1_275850 | Dense granule protein GRA12 | 3 | 1.31 | No | Yes | 3.00 | 2.63 |
| GRA13 | TGGT1_237880 | Hypothetical protein | 1 | 0.76 | Yes | Yes | 120.50 | 34.28 |
| GRA14 | TGGT1_239740 | Dense granule protein GRA14 | 1 | 2.00 | No | Yes | 116.36 | 53.11 |
| GRA20 | TGGT1_200010 | Hypothetical protein | 2 | 2.54 | No | No | 37.97 | 30.74 |
| GRA21 | TGGT1_241610 | Hypothetical protein | 2 | ND | No | Yes | 18.40 | 13.56 |
| GRA28 | TGGT1_231960 | Putative omega secalin | 3 | 1.48 | No | Yes | 33.28 | 3.58 |
| GRA29 | TGGT1_269690 | Hypothetical protein | 1 | 1.54 | Yes | Yes | 47.08 | 77.60 |
| GRA30 | TGGT1_232000 | Hypothetical protein | 2 | 2.29 | Yes | Yes | 33.46 | 44.61 |
| GRA31 | TGGT1_220240 | Hypothetical protein | 1 | 0.90 | Yes | Yes | 33.92 | 22.57 |
| GRA33 | TGGT1_247440 | Hypothetical protein | 1 | 1.72 | Yes | Yes | 48.06 | 45.42 |
| GRA34 | TGGT1_203290 | Hypothetical protein | 1 | 2.26 | No | Yes | 64.29 | 248.97 |
| GRA35 | TGGT1_226380 | Hypothetical protein | 1 | 1.98 | Yes | Yes | 48.74 | 32.29 |
| GRA36 | TGGT1_213067 | Hypothetical protein | 1 | −0.21 | Yes | Yes | 83.85 | 24.24 |
| GRA37 | TGGT1_236890 | Hypothetical protein | 2 | 2.10 | Yes | No | 60.01 | 34.21 |
| GRA38 | TGGT1_312420 | Hypothetical protein | 4 | −1.15 | No | Yes | 27.17 | 17.82 |
| GRA40 | TGGT1_219810 | Hypothetical protein | 7 | 0.69 | Yes | No | 114.88 | 97.16 |
ND, Not determined.
Prediction of transmembrane helices was performed using the TMHMM program version 2.0.
Gene expression levels of fragments per kilobase of exon model per million mapped reads (FPKM) at acute (10 days post infection) and chronic infection (28 days post infection). Source: http://www.toxodb.org/toxo/ [accessed 26 July 2018].
We next analyzed the transcriptomic levels of 17 GRAs in different T. gondii genotypes (Type I, II, and III), and found that GRA20 and GRA34 were significantly different among the three genotypes (Figure 4B). This difference may be because these two GRAs play different roles in different strains. Levels of the transcriptomic expression of 17 GRA genes across T. gondii developmental stages are presented (Figure 4C). Our analysis showed that most GRAs are differentially expressed at different life cycle stages, but GRA11 and GRA21. The expression of some GRA genes can be specifically up- or down-regulated during parasite development (Fritz et al., 2012). For instance, the expression of GRA7 was significantly reduced in in vitro 4 day bradyzoites compared to that in the in vitro 2 day tachyzoites and then restored to near tachyzoite's levels in the in vitro 21 day bradyzoites (Buchholz et al., 2011). The expression of GRA4, GRA6, and GRA8 has been reported to be reduced or even non-detectable in the bradyzoites (Ferguson, 2004). The different expression levels of some GRAs in different life cycle stages indicate that the roles played by GRAs may play parasite stage-dependent.
Conclusion
GRAs play key roles in modulating host-parasite interactions, such as parasite vacuole remodeling, nutrient uptake and manipulation of host signaling pathways (Nadipuram et al., 2016). In this study, 17 ΔGRA mutant T. gondii strains were successfully generated using CRISPR-Cas9 technique. The role of these 17 GRA genes in the pathogenicity of T. gondii RH strain was investigated in vitro and in vivo. We report here that no significant difference was detected between ΔGRA knockouts and wild-type RH strains. These findings indicate that GRA genes examined in this study are not absolutely essential for T. gondii RH virulence, suggesting that other virulence factors may be involved in these processes or that virulence of T. gondii is the result of a multigene effect. We also investigated the patterns of gene expression and bioinformatics features of the 17 GRAs, by parasite cell cycle phases, life cycle stages and genotypes. Results indicated that the expression of GRAs can vary across life cycle stages or genotypes of T. gondii. Functional analysis of these GRAs in other parasite strains or life cycle forms is therefore of high importance and may further elucidate the pathogenic role of GRAs in T. gondii infection.
Ethics statement
The study was approved by the Animal Administration and Ethics Committee of Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Science (Permit No. LVRIAEC-2017-006). All mice were handled humanely in strict accordance with the Guidelines and Animal Ethics Procedures of the People's Republic of China.
Author contributions
X-QZ, J-LW, and HE designed the study and critically revised the manuscript. M-JB performed the experiments, analyzed data, and drafted the manuscript. Q-LL, KC, and L-BN participated in the implementation of the study. All the authors read and approved the final manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Professor Bang Shen (Huazhong Agricultural University) for providing the pSAG1::CAS9-U6::sgUPRT and pUPRT-DHFR-D vectors.
Footnotes
Funding. Project financial support was kindly provided by the International Science and Technology Cooperation Project of Gansu Province (Grant No. 17JR7WA031), by the Elite Program of Chinese Academy of Agricultural Sciences, and by the Agricultural Science and Technology Innovation Program (ASTIP) (Grant No. CAAS-ASTIP-2016-LVRI-03).
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2018.00300/full#supplementary-material
References
- Alaganan A., Fentress S. J., Tang K., Wang Q., Sibley L. D. (2014). Toxoplasma GRA7 effector increases turnover of immunity-related GTPases and contributes to acute virulence in the mouse. Proc. Natl. Acad. Sci. U.S.A. 111, 1126–1131. 10.1073/pnas.1313501111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrizabalaga G., Ruiz F., Moreno S., Boothroyd J. C. (2004). Ionophore-resistant mutant of Toxoplasma gondii reveals involvement of a sodium/hydrogen exchanger in calcium regulation. J. Cell. Biol. 165, 653–662. 10.1083/jcb.200309097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnke M. S., Wootton J. C., Lehmann M. M., Radke J. B., Lucas O., Nawas J., et al. (2010). Coordinated progression through two subtranscriptomes underlies the tachyzoite cycle of Toxoplasma gondii. PloS. ONE 5:e12354. 10.1371/journal.pone.0012354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boothroyd J. C., Dubremetz J. F. (2008). Kiss and spit: the dual roles of Toxoplasma rhoptries. Nat. Rev. Microbiol. 6, 79–88. 10.1038/nrmicro1800 [DOI] [PubMed] [Google Scholar]
- Bougdour A., Durandau E., Brenier-Pinchart M. P., Ortet P., Barakat M., Kieffer S., et al. (2013). Host cell subversion by Toxoplasma GRA16, an exported dense granule protein that targets the host cell nucleus and alters gene expression. Cell. Host. Microbe 13, 489–500. 10.1016/j.chom.2013.03.002 [DOI] [PubMed] [Google Scholar]
- Braun L., Brenier-Pinchart M. P., Yogavel M., Curt-Varesano A., Curt-Bertini R. L., Hussain T., et al. (2013). A Toxoplasma dense granule protein, GRA24, modulates the early immune response to infection by promoting a direct and sustained host p38 MAPK activation. J. Exp. Med. 210, 2071–2086. 10.1084/jem.20130103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchholz K. R., Fritz H. M., Chen X., Durbin-Johnson B., Rocke D. M., Ferguson D. J., et al. (2011). Identification of tissue cyst wall components by transcriptome analysis of in vivo and in vitro Toxoplasma gondii bradyzoites. Eukaryot. Cell. 10, 1637–1647. 10.1128/EC.05182-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldas L. A., de Souza W., Attias M. (2010). Microscopic analysis of calcium ionophore activated egress of Toxoplasma gondii from the host cell. Vet. Parasitol. 167, 8–18. 10.1016/j.vetpar.2009.09.051 [DOI] [PubMed] [Google Scholar]
- Chemoh W., Sawangjaroen N., Siripaitoon P., Andiappan H., Hortiwakul T., Sermwittayawong N., et al. (2015). Toxoplasma gondii – prevalence and risk factors in HIV-infected patients from Songklanagarind hospital, Southern Thailand. Front. Microbiol. 6:1304. 10.3389/fmicb.2015.01304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppens I., Dunn J. D., Romano J. D., Pypaert M., Zhang H., Boothroyd J. C., et al. (2006). Toxoplasma gondii sequesters lysosomes from mammalian hosts in the vacuolar space. Cell 125, 261–274. 10.1016/j.cell.2006.01.056 [DOI] [PubMed] [Google Scholar]
- Elmore S. A., Jones J. L., Conrad P. A., Patton S., Lindsay D. S., Dubey J. P. (2010). Toxoplasma gondii: epidemiology, feline clinical aspects, and prevention. Trends. Parasitol. 26, 190–196. 10.1016/j.pt.2010.01.009 [DOI] [PubMed] [Google Scholar]
- Ferguson D. J. (2004). Use of molecular and ultrastructural markers to evaluate stage conversion of Toxoplasma gondii in both the intermediate and definitive host. Int. J. Parasitol. 34:347–360. 10.1016/j.ijpara.2003.11.024 [DOI] [PubMed] [Google Scholar]
- Fritz H. M., Buchholz K. R., Chen X., Durbin-Johnson B., Rocke D. M., Conrad P. A., et al. (2012). Transcriptomic analysis of Toxoplasma development reveals many novel functions and structures specific to sporozoites and oocysts. PloS. ONE 7:e29998. 10.1371/journal.pone.0029998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajria B., Bahl A., Brestelli J., Dommer J., Fischer S., Gao X., et al. (2007). ToxoDB: an integrated Toxoplasma gondii database resource. Nucl. Acids. Res. 36, D553–D556. 10.1093/nar/gkm981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrin C., Mercier C., Braun L., Musset K., Dubremetz J. F., Cesbron-Delauw M. F. (2008). Toxoplasma gondii uses unusual sorting mechanisms to deliver transmembrane proteins into the host-cell vacuole. Traffic 9, 1665–1680. 10.1111/j.1600-0854.2008.00793.x [DOI] [PubMed] [Google Scholar]
- Gold D. A., Kaplan A. D., Lis A., Bett G. C., Rosowski E. E., Cirelli K. M., et al. (2015). The Toxoplasma dense granule proteins GRA17 and GRA23 mediate the movement of small molecules between the host and the parasitophorous vacuole. Cell. Host. Microbe 17, 642–652. 10.1016/j.chom.2015.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Håkansson S., Charron A. J., Sibley L. D. (2001). Toxoplasma evacuoles: a two-step process of secretion and fusion forms the parasitophorous vacuole. EMBO J. 20, 3132–3144. 10.1093/emboj/20.12.3132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakimi M. A., Bougdour A. (2015). Toxoplasma's ways of manipulating the host transcriptome via secreted effectors. Curr. Opin. Microbiol. 26, 24–31. 10.1016/j.mib.2015.04.003 [DOI] [PubMed] [Google Scholar]
- Hill D., Dubey J. P. (2002). Toxoplasma gondii: transmission, diagnosis and prevention. Clin. Microbiol. Infect. 8, 634–640. 10.1046/j.1469-0691.2002.00485.x [DOI] [PubMed] [Google Scholar]
- Hsiao C. H. C., Luisa Hiller N., Haldar K., Knoll L. J. (2013). A HT/PEXEL motif in Toxoplasma dense granule proteins is a signal for protein cleavage but not export into the host cell. Traffic 14, 519–531. 10.1111/tra.12049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFavers K. A., Márquez-Nogueras K. M., Coppens I., Moreno S. N. J., Arrizabalaga G. (2017). A novel dense granule protein, GRA41, regulates timing of egress and calcium sensitivity in Toxoplasma gondii. Cell. Microbiol. 19:e12749. 10.1111/cmi.12749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Yuan Z., Gao S., Liu B., Liu X. (2008). Detection of antibody IgG against Toxoplasma gondii from pregnant/parturient women and cancer patients in Changchun region. J. Pathog. Biol. 3, 122–123. 10.13350/j.cjpb.2008.02.018 [DOI] [Google Scholar]
- Ma J. S., Sasai M., Ohshima J., Lee Y., Bando H., Takeda K., et al. (2014). Selective and strain-specific NFAT4 activation by the Toxoplasma gondii polymorphic dense granule protein GRA6. J. Exp. Med. 211, 2013–2032. 10.1084/jem.20131272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier C., Cesbron-Delauw M. F. (2015). Toxoplasma secretory granules: one population or more? Trends. Parasitol. 31, 60–71. 10.1016/j.pt.2014.12.002 [DOI] [PubMed] [Google Scholar]
- Morisaki J. H., Heuser J. E., Sibley L. D. (1995). Invasion of Toxoplasma gondii occurs by active penetration of the host cell. J. Cell. Sci. 108, 2457–2464. [DOI] [PubMed] [Google Scholar]
- Nadipuram S. M., Kim E. W., Vashisht A. A., Lin A. H., Bell H. N., Coppens I., et al. (2016). In vivo biotinylation of the Toxoplasma parasitophorous vacuole reveals novel dense granule proteins important for parasite growth and pathogenesis. MBio 7:e00808-16. 10.1128/mBio.00808-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam H. W. (2009). GRA proteins of Toxoplasma gondii: maintenance of host-parasite interactions across the parasitophorous vacuolar membrane. Korean. J. Parasitol. 47, S29–S37. 10.3347/kjp.2009.47.S.S29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada T., Marmansari D., Li Z. M., Adilbish A., Canko S., Ueno A., et al. (2013). A novel dense granule protein, GRA22, is involved in regulating parasite egress in Toxoplasma gondii. Mol. Biochem. Parasitol. 189, 5–13. 10.1016/j.molbiopara.2013.04.005 [DOI] [PubMed] [Google Scholar]
- Rosowski E. E., Lu D., Julien L., Rodda L., Gaiser R. A., Jensen K. D., et al. (2011). Strain-specific activation of the NF-κB pathway by GRA15, a novel Toxoplasma gondii dense granule protein. J. Exp. Med. 208, 195–212. 10.1084/jem.20100717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shastri A. J., Marino N. D., Franco M., Lodoen M. B., Boothroyd J. C. (2014). GRA25 is a novel virulence factor of Toxoplasma gondii and influences the host immune response. Infect. Immun. 82, 2595–2605. 10.1128/IAI.01339-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B., Brown K., Long S., Sibley L. D. (2017). Development of CRISPR/Cas9 for efficient genome editing in Toxoplasma gondii. Methods. Mol. Biol. 1498, 79–103. 10.1007/978-1-4939-6472-7_6 [DOI] [PubMed] [Google Scholar]
- Shen B., Brown K. M., Lee T. D., Sibley L. D. (2014). Efficient gene disruption in diverse strains of Toxoplasma gondii using CRISPR/Cas9. MBio. 5:e01114–14. 10.1128/mBio.01114-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidik S. M., Hackett C. G., Tran F., Westwood N. J., Lourido S. (2014). Efficient genome engineering of Toxoplasma gondii using CRISPR/Cas9. PLoS ONE 9:e100450. 10.1371/journal.pone.0100450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenter A. M., Heckeroth A. R., Weiss L. M. (2000). Toxoplasma gondii: from animals to humans. Int. J. Parasitol. 30, 1217–1258. 10.1016/S0020-7519(00)00124-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. L., Huang S. Y., Behnke M. S., Chen K., Shen B., Zhu X. Q. (2016a). The past, present, and future of genetic manipulation in Toxoplasma gondii. Trends. Parasitol. 32, 542–553. 10.1016/j.pt.2016.04.013 [DOI] [PubMed] [Google Scholar]
- Wang J. L., Huang S. Y., Li T. T., Chen K., Ning H. R., Zhu X. Q. (2016b). Evaluation of the basic functions of six calcium-dependent protein kinases in Toxoplasma gondii using CRISPR-Cas9 system. Parasitol. Res. 115, 697–702. 10.1007/s00436-015-4791-6 [DOI] [PubMed] [Google Scholar]
- Wang J. L., Li T. T., Elsheikha H. M., Chen K., Zhu W. N., Yue D. M., et al. (2017). Functional characterization of rhoptry kinome in the virulent Toxoplasma gondii RH strain. Front. Microbiol. 8:84. 10.3389/fmicb.2017.00084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y., Yin J., Jiang N., Xiang M., Hao L., Lu H., et al. (2010). Seroepidemiology of human Toxoplasma gondii infection in China. BMC. Infect. Dis. 10:4. 10.1186/1471-2334-10-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Chen Z., Li H. L., Zheng H., He S., Lin R. Q., et al. (2011). Toxoplasma gondii infection in humans in China. Parasit. Vectors 4:165. 10.1186/1756-3305-4-165 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.