Rapid, efficient genetic engineering of Streptomyces strains is critical for genome mining of novel natural products (NPs) as well as strain improvement. Here, a novel and high-efficiency Streptomyces genome editing tool is established based on the FnCRISPR-Cpf1 system, which is an attractive and powerful alternative to the S. pyogenes CRISPR-Cas9 system due to its unique features. When combined with HDR or NHEJ, FnCpf1 enables the creation of gene(s) deletion with high efficiency. Furthermore, a ddCpf1-based integrative CRISPRi platform is established for simple, multiplex transcriptional repression. Of importance, FnCpf1-based genome editing proves to be a highly efficient tool for genetic modification of some important industrial Streptomyces strains (e.g., S. hygroscopicus SIPI-KF) that cannot utilize the SpCRISPR-Cas9 system. We expect the CRISPR-Cpf1-assisted genome editing tool to accelerate discovery and development of pharmaceutically active NPs in Streptomyces as well as other actinomycetes.
KEYWORDS: Streptomyces, CRISPR, FnCpf1, genome editing, multiplex repression
ABSTRACT
Streptomyces has a strong capability for producing a large number of bioactive natural products and remains invaluable as a source for the discovery of novel drug leads. Although the Streptococcus pyogenes CRISPR-Cas9-assisted genome editing tool has been developed for rapid genetic engineering in Streptomyces, it has a number of limitations, including the toxicity of SpCas9 expression in some important industrial Streptomyces strains and the need for complex expression constructs when targeting multiple genomic loci. To address these problems, in this study, we developed a high-efficiency CRISPR-Cpf1 system (from Francisella novicida) for multiplex genome editing and transcriptional repression in Streptomyces. Using an all-in-one editing plasmid with homology-directed repair (HDR), our CRISPR-Cpf1 system precisely deletes single or double genes at efficiencies of 75 to 95% in Streptomyces coelicolor. When no templates for HDR are present, random-sized DNA deletions are achieved by FnCpf1-induced double-strand break (DSB) repair by a reconstituted nonhomologous end joining (NHEJ) pathway. Furthermore, a DNase-deactivated Cpf1 (ddCpf1)-based integrative CRISPRi system is developed for robust, multiplex gene repression using a single customized crRNA array. Finally, we demonstrate that FnCpf1 and SpCas9 exhibit different suitability in tested industrial Streptomyces species and show that FnCpf1 can efficiently promote HDR-mediated gene deletion in the 5-oxomilbemycin-producing strain Streptomyces hygroscopicus SIPI-KF, in which SpCas9 does not work well. Collectively, FnCpf1 is a powerful and indispensable addition to the Streptomyces CRISPR toolbox.
IMPORTANCE Rapid, efficient genetic engineering of Streptomyces strains is critical for genome mining of novel natural products (NPs) as well as strain improvement. Here, a novel and high-efficiency Streptomyces genome editing tool is established based on the FnCRISPR-Cpf1 system, which is an attractive and powerful alternative to the S. pyogenes CRISPR-Cas9 system due to its unique features. When combined with HDR or NHEJ, FnCpf1 enables the creation of gene(s) deletion with high efficiency. Furthermore, a ddCpf1-based integrative CRISPRi platform is established for simple, multiplex transcriptional repression. Of importance, FnCpf1-based genome editing proves to be a highly efficient tool for genetic modification of some important industrial Streptomyces strains (e.g., S. hygroscopicus SIPI-KF) that cannot utilize the SpCRISPR-Cas9 system. We expect the CRISPR-Cpf1-assisted genome editing tool to accelerate discovery and development of pharmaceutically active NPs in Streptomyces as well as other actinomycetes.
INTRODUCTION
Streptomyces species, as Gram-positive and high-G+C% soil actinomycetes, have evolved to produce a large number of important bioactive natural products, such as insecticides, immunosuppressors, and antitumor agents (1–3). Recent large-scale bacterial genome sequencing analysis has revealed that ∼90% of secondary metabolite biosynthetic gene clusters (BGCs) in streptomycetes are silent or cryptic under laboratory conditions and are thus regarded as a treasure for the discovery of novel drug leads (4–6). Convenient and efficient Streptomyces genetic engineering will significantly advance functional genomic research and genome mining of bioactive compounds as well as strain improvement (7–10).
The CRISPR-Cas system is a widely distributed adaptive immunity system against invading mobile genetic elements in archaea and bacteria (11–14). The class 2 CRISPR-Cas system, consisting of a single multidomain effector paired with guide RNA, has been developed as a powerful genome editing tool for a broad spectrum of organisms due to its simplicity and versatility (15–17). As a type of fast and high-throughput genome editing tools, CRISPR-based systems have been widely employed for fundamental research and development of advanced cell factories in the microbial systems, such as Escherichia coli, Saccharomyces cerevisiae, and Streptomyces (18–21). Previous studies have demonstrated that the Streptococcus pyogenes CRISPR-Cas9 system (SpCas9) can be utilized for high-efficiency multiplex genome editing by homologous recombination (HR)- or nonhomologous end joining (NHEJ)-mediated double-strand-break (DSB) repair and reversible transcriptional gene expression in multiple model and industrial Streptomyces species (22–27). However, SpCas9-based DNA editing has some shortcomings to be addressed. For instance, a G-rich protospacer-adjacent motif (PAM) sequence (5′-NGG-3′) is required for SpCas9 to recognize a target site (28, 29). Although this PAM is rather frequently distributed across the Streptomyces genomes (GC content, >70%; ca. 260 unique genomic targets per 1,000 bp, which is the average size of Streptomyces coelicolor genes; see Table S1 in the supplemental material), it is not always available in the desired loci, particularly in AT-rich regions. Furthermore, complex plasmid construction for independent expression of multiple single guide RNAs (sgRNA) in the SpCas9-mediated editing system is not convenient for multiplex genome editing and transcriptional regulation. Notably, our earlier study indicated that the SpCas9-based editing plasmid did not work in several industrial Streptomyces strains, such as the 5-oxomilbemycin-producing strain Streptomyces hygroscopicus SIPI-KF. A similar phenomenon has also been observed in Corynebacterium glutamicum, in which the native SpCas9 cannot be constitutively expressed due to its toxicity in the recipient cells (30, 31). These limitations of SpCas9-based editing tool propel us to explore a novel, alternative CRISPR-Cas system for high-efficiency genetic engineering of Streptomyces species.
Cpf1 (also known as Cas12a), a new class 2 type V CRISPR-Cas RNA-guided endonuclease, was recently identified to possess efficient genomic DNA cleavage activity in bacterial, plant, and mammalian cells (32–35). Three Cpf1 family proteins, including FnCpf1 from Francisella novicida, AsCpf1 from Acidaminococcus species strain BV3L6, and LbCpf1 from a Lachnospiraceae bacterium, have been studied most intensively (34–36). Over the past 2 years, Cpf1 has been rapidly developed as a powerful genome editing tool in many organisms due to its unique features distinct from SpCas9.
(i) Cpf1 recognizes T-rich PAM sequences, i.e., 5′-TTTV-3′ for AsCpf1/LbCpf1 and 5′-TTV-3′ for FnCpf1 (V = A/G/C) (34). This feature can expand the genomic DNA scope that can be targeted. In addition, studies have demonstrated that Cpf1 could provide higher levels of target specificity than SpCas9 in mammalian cells (37, 38). These results suggest that Cpf1 may be an attractive alternative tool for genetic modification of actinomycetal genomes (particularly AT-rich regions).
(ii) Cpf1 is guided by a single CRISPR RNA (crRNA) without trans-activating crRNA (tracrRNA) and harbors a distinct RNase activity for pre-crRNA processing (35, 39, 40). Additionally, the Cpf1 crRNA (19-nucleotide [nt] direct repeat and a 23- to 25-nt spacer) is shorter than that of SpCas9 chimeric sgRNA by ∼60 nt (40). These advantages of Cpf1 make it easy to perform multiplex genome editing and transcriptional regulation by using only a single, customized crRNA array. Recently, the CRISPR-Cpf1 system was used to simultaneously edit up to four genes by NHEJ repair in rice and mammalian cells (35, 41). Furthermore, a DNase-deactivated Cpf1 (ddCpf1) paired with a single crRNA array was also employed for inducible transcriptional activation of up to three genes in human cells and multiplex gene repression of up to four genes in E. coli (42, 43).
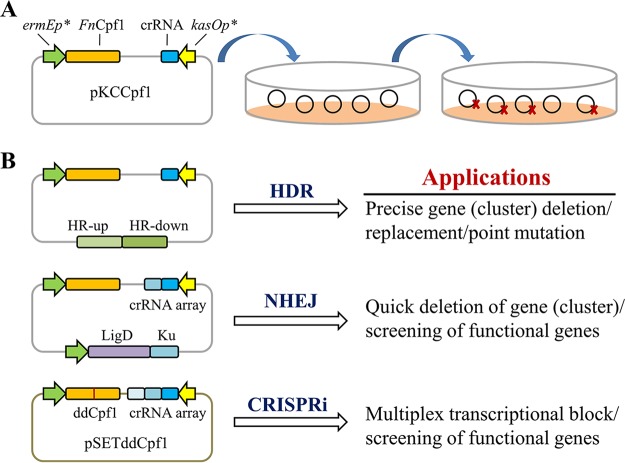
Thus far, Cpf1-mediated genome editing has been demonstrated in a few bacteria, including E. coli, C. glutamicum, Cyanobacteria, Yersinia pestis, and Mycobacterium smegmatis (31, 33, 44). The goal of the present study was to develop a high-efficiency CRISPR-Cpf1 system for genome editing as well as multiplex gene repression in the industrial important microbe, Streptomyces. First, we established an all-in-one FnCpf1-based plasmid for targeted genomic DNA cleavage (Fig. 1A). It should be noted that AsCpf1 and LbCpf1 were not chosen for genome editing of Streptomyces due to their excessively low PAM occurrence frequencies compared to FnCpf1 (Table S1). Second, the FnCpf1-assisted editing system was developed for precise single or double gene deletions by homology-directed repair (HDR) and random-sized deletions of target genes by the reconstituting NHEJ system (Fig. 1B). Third, a ddCpf1-based integrative CRISPR interference (CRISPRi) system was also designed for robust and multiplex gene expression control (Fig. 1B). Finally, we demonstrated that the FnCpf1-mediated editing system is an alternative and powerful tool for industrial Streptomyces strains in which the S. pyogenes CRISPR-Cas9 system does not work well.
FIG 1.
Overview of FnCpf1-assisted multiplex genome editing and transcriptional repression in Streptomyces. (A) Design of FnCpf1-mediated genome editing plasmid. Two strong promoters, ermEp* and kasOp*, were used to drive the expression of FnCpf1 and crRNA, respectively. (B) Broad applications of FnCpf1-assisted genome editing and ddCpf1-based gene repression in Streptomyces. HR, homology arm; HDR, homology-directed repair; NHEJ, nonhomologous end joining; CRISPRi, CRISPR interference.
RESULTS
Design and functionality of an all-in-one FnCpf1-editing system in Streptomyces.
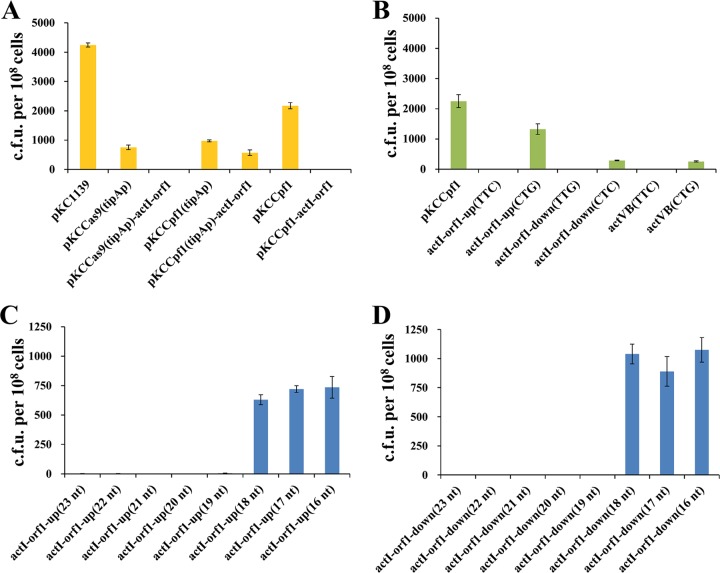
To develop an FnCpf1-mediated genome editing tool in Streptomyces, an all-in-one CRISPR-Cpf1 editing plasmid, pKCCpf1(tipAp), in which the expressions of the codon-optimized sco-cpf1 gene and target-specific crRNA were driven by the inducible promoter tipAp and the strong promoter kasOp* (45), respectively, was constructed. The editing plasmid pKCCpf1(tipAp), harboring the temperature-sensitive replicon pSG5, could be quickly eliminated when the edited strains were cultivated at 37°C (46). This feature would significantly facilitate FnCpf1-assisted iterative genome editing. To confirm that FnCpf1 was functional, we constructed the editing plasmid pKCCpf1(tipAp)-actI-orf1, which specifically targeted the actI-orf1 gene from the biosynthetic pathway of the blue-colored antibiotic actinorhobin (ACT) in the model strain S. coelicolor. It should be noted that a 5′-TTC-3′ PAM sequence in actI-orf1 was chosen to test FnCpf1-mediated genome cleavage activity. As a positive control, SpCas9-derived editing plasmid pKCCas9(tipAp)-actI-orf1 was also constructed. Then, five plasmids, namely, the control plasmid pKC1139, pKCCas9(tipAp) (without target-specific sgRNA expression cassette), pKCCas9(tipAp)-actI-orf1, pKCCpf1(tipAp) (without target-specific crRNA expression cassette), and pKCCpf1(tipAp)-actI-orf1 were introduced individually into S. coelicolor by conjugal transfer. It was observed that introduction of pKCCas9(tipAp)-actI-orf1 into strain M145 caused a 99.6% decrease in exoconjugant numbers compared to pKCCas9(tipAp). However, only a 41.5% decrease in exoconjugant numbers was caused when pKCCpf1(tipAp)-actI-orf1 was introduced into M145, indicating that the activity of the CRISPR-Cpf1 system was not sufficient for targeted genome cleavage (Fig. 2A). To address this problem, the strong constitutive promoter ermEp* was used to replace tipAp for FnCpf1 expression, thus generating plasmids pKCCpf1 and pKCCpf1-actI-orf1. Compared with pKCCpf1, introduction of pKCCpf1-actI-orf1 into M145 generated only very few exoconjugants (12 ± 3 per 108 recipient cells), with a nearly 100% killing frequency (Fig. 2A). The results clearly indicate that FnCpf1 expression in S. coelicolor is less toxic and the all-in-one CRISPR-Cpf1 system can efficiently mediate targeted DNA cleavage, which is a prerequisite for site-directed genetic modification by HDR or NHEJ.
FIG 2.
Optimization of the FnCpf1-mediated genome editing system for targeted DNA cleavage in S. coelicolor. (A) Growth of S. coelicolor expressing SpCas9 or FnCpf1, with or without the combined expression of crRNA targeting actI-orf1. The coexpression of FnCpf1 under the control of ermEp* but not tipAp and target-specific crRNA (pKCCpf1-actI-orf1) can efficiently induce DSBs and thus leads to significantly reduced exoconjugant survival. The plasmids pKC1139 and pKCCas9(tipAp) were used as the negative controls, and pKCCas9(tipAp)-actI-orf1 was used as the positive control. (B) Effects of different PAM sequences on FnCpf1 cleavage activity. The plasmid pKCCpf1 was used as the control. The bases in parentheses represent the PAM sequences. Six PAMs located in two genes, actI-orf1 and actVB, were tested. (C, D) Effects of spacer lengths on FnCpf1 cleavage activity. The numbers in parentheses represent the spacer lengths.
It has been previously reported that FnCpf1 could cleave target DNA specifically at the YTV (Y = T/C and V = A/G/C) PAM located on the 5′-end of the protospacer (40, 47). Here, PAM sequence preference of FnCpf1 was further checked in S. coelicolor. We designed six crRNAs targeting 5′-TTV-3′ or 5′-CTV-3′ PAM-preceded sequences located in two genes, actI-orf1 and actVB. It was observed that all 5′-TTV-3′ PAM-targeted crRNAs could result in significantly reduced exoconjugant formation, by several orders of magnitude, while 5′-CTV-3′ PAM-targeted crRNAs caused only 41.1 to 88.6% decreases in exoconjugant numbers compared with the control plasmid (Fig. 2B). Furthermore, we found that 86.7 to 100% of the obtained exoconjugants produced blue-pigmented ACT (see Fig. S1 in the supplemental material), indicating that most of the colonies had not undergone targeted Cpf1-mediated DNA cleavage. These results revealed that the 5′-CTV-3′ PAM is capable of mediating DNA interference but not suitable for high-efficiency genome editing, and 5′-TTV-3′ PAM would be the preferable choice for genetic engineering in Streptomyces.
The mature crRNA of the CRISPR-Cpf1 system is composed of a 19-nt direct repeat and 23 to 25 nt of spacer sequence (34). It has been demonstrated that to achieve efficient DNA cleavage in vitro, a minimum of 17 nt of guide sequence is required (34, 48). To determine the minimum spacer length of crRNA for efficient genomic DNA cleavage in S. coelicolor, we designed a series of crRNAs with different spacer lengths (16 to 23 nt), which targeted two different sequences located in actI-orf1. The results showed that FnCpf1 required at least 19 nt of spacer sequence to maintain efficient RNA-guided DNA cleavage activity in S. coelicolor (Fig. 2C and D).
FnCpf1-assisted single-gene and double-gene deletions by HDR.
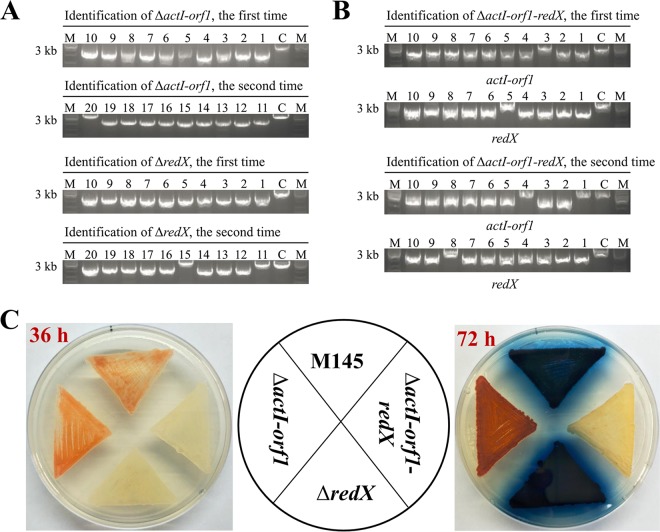
We first validated the CRISPR-Cpf1 system in S. coelicolor by in-frame deleting single or double genes based on homology-directed DSB repair (HDR). Two genes, actI-orf1 and redX, from the biosynthetic pathways of the blue-colored antibiotic ACT and the red-pigmented antibiotic prodiginine (RED), respectively, were chosen for deletion trials. Three editing plasmids, including pKCCpf1-actI-orf1-HR (targeting actI-orf1 only), pKCCpf1-redX-HR (targeting redX only), and pKCCpf1-actI-orf1-redX-HR (targeting both actI-orf1 and redX), were constructed and introduced individually into M145 by conjugal transfer. The lengths of homolog arms (HA) used for actI-orf1 and redX deletion were 1,017/1,033 bp and 1,019/1,033 bp (up/downstream), respectively. The experiments were repeated twice. The numbers of exoconjugants for actI-orf1, redX, and actI-orf1 and redX deletions were 54 ± 5, 41 ± 6, and 23 ± 4, respectively. Then, 10 colonies were randomly picked to determine the editing efficiency by PCR analysis for each trial, and the experiments were repeated twice. As shown in Fig. 3A and B, the deletion rates of actI-orf1 and redX were 95% (10/10 + 9/10) and 90% (10/10 + 8/10), respectively. The efficiency of simultaneous deletion of both genes also reached 75% (8/10 + 7/10). As expected, visual analysis on agar plates showed that disruption of actI-orf1 resulted in complete loss of blue pigment (ACT) formation while deletion of redX led to loss of red color (RED) production. The mutant with simultaneous deletion of both genes does not produce ACT and RED any more (Fig. 3C). It could be therefore concluded that similar to the S. pyogenes CRISPR-Cas9 system, the F. novicida CRISPR-Cpf1 system is capable of HDR-dependent multiplex genome engineering in S. coelicolor.
FIG 3.
FnCpf1-assisted in-frame deletions of single genes and simultaneous deletion of double genes. (A) Identification of actI-orf1 and redX deletion mutants (ΔactI-orf1 and ΔredX). Ten exoconjugants were selected and verified by PCR. The experiments were repeated twice. C and M represent the wild-type S. coelicolor M145 and 1-kb DNA ladder, respectively. (B) Identification of actI-orf1 and redX double deletion mutants (ΔactI-orf1-redX). (C) Effects of single or double deletion of actI-orf1 and redX on ACT and RED biosynthesis. The mutants were grown on R2YE plates and photographed from the back at 36 and 72 h.
FnCpf1-assisted random-sized deletion by the reconstituted NHEJ system.
The repair of endonuclease-induced DSBs by the NHEJ pathway can give rise to small insertions and deletions (indels) in genomes, which thus lead to inactivation of target genes (49–51). Compared to HDR-dependent genome editing that requires two homologous arms as the template for DSB repair, template-free NHEJ repair is imprecise but more convenient. Unfortunately, most streptomycetes lack a complete NHEJ pathway, which is composed of a DNA ligase D (LigD) and its cognate Ku protein. Here, three different NHEJ systems from Mycobacterium smegmatis, Streptomyces daghestanicus, and Pseudomonas putida, named Msm-LK, Sda-LK, and Ppu-LK, respectively, were selected to promote FnCpf1-induced DSB repair in S. coelicolor. The reasons why we selected the three NHEJ pathways were as follows. First, Msm-LK exhibited higher DSB repair activity than the other two NHEJ systems from Mycobacterium tuberculosis and Bacillus subtilis in E. coli (52). Second, Sda-LK is the only known NHEJ system of Streptomyces origin (NCBI GenPept ID of ligD and ku are WP_086839147.1 and WP_086839677.1, respectively). Finally, Ppu-LK possesses an NHEJ repair mechanism different from that of Msm-LK and Sda-LK, and the genome of P. putida has a high G+C content (61.9%), similar to Streptomyces species (50). These features promote Ppu-LK as a considerable alternative for reconstituting complete NHEJ system in S. coelicolor.
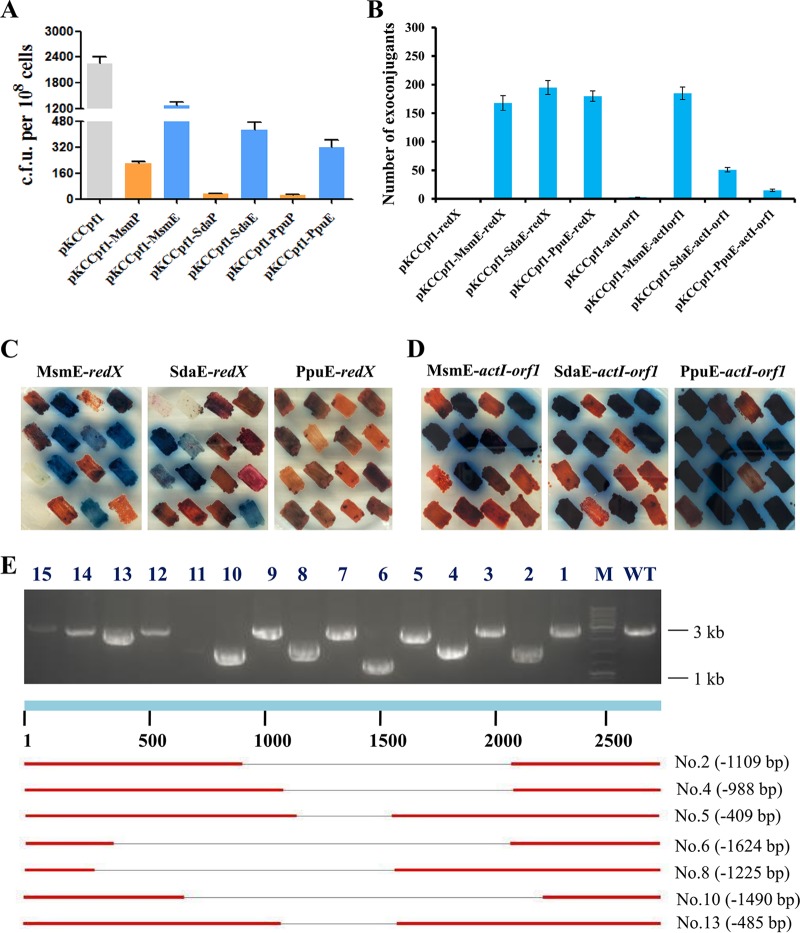
To develop an all-in-one CRISPR-Cpf1 editing plasmid containing the reconstituted NHEJ system, ligD and the cognate ku gene were directly ligated to pKCCpf1. Two different strong promoters, ermEp* and gadphp (53), were used, respectively, to drive coexpression of these two genes. As such, six plasmids, including pKCCpf1-MsmP, pKCCpf1-MsmE, pKCCpf1-SdaP, pKCCpf1-SdaE, pKCCpf1-PpuP, and pKCCpf1-PpuE, were constructed. As the first step, the toxicity of foreign NHEJ pathways was tested. It was observed that overexpression of these three different NHEJ systems under the control of gadphp all resulted in dramatically decreased formation of exoconjugants. The exoconjugant numbers were reduced by 90.2% to 98.9%, compared with the control (pKCCpf1). In contrast, expression of the NHEJ systems driven by ermEp* led to a decrease in exoconjugant numbers by 43.8% to 85.9%, which was acceptable for S. coelicolor genome editing. Additionally, we noticed that Msm-LK showed the lowest cell toxicity among three tested NHEJ systems.
Then, redX and actI-orf1 were chosen for template-free deletion trials. In total, six editing plasmids harboring both the NHEJ systems driven by ermEp* and the crRNA expression cassettes targeting redX or actI-orf1, including pKCCpf1-MsmE-redX, pKCCpf1-SdaE-redX, pKCCpf1-PpuE-redX, pKCCpf1-MsmE-actI-orf1, pKCCpf1-SdaE-actI-orf1, and pKCCpf1-PpuE-actI-orf1, were constructed. As controls, pKCCpf1-redX and pKCCpf1-actI-orf1 with the redX- and actI-orf1-targeted crRNA expression cassettes cloned in pKCCpf1, were also constructed, respectively. We observed that compared with the controls (pKCCpf1-redX and pKCCpf1-actI-orf1) no exoconjugants grew out, and introduction of these editing plasmids harboring different NHEJ systems significantly increased exoconjugant numbers (Fig. 4B). More precisely, the numbers of exoconjugants were increased from 0 to 168 ± 13 (MsmE-LK), 195 ± 12 (SdaE-LK), and 180 ± 9 (PpuE-LK) for editing redX. Meanwhile, the exoconjugant numbers for editing actI-orf1 were increased from 0 to 185 ± 11 (MsmE-LK), 51 ± 4 (SdaE-LK), and 16 ± 2 (PpuE-LK). These results indicated that the three foreign NHEJ systems were functional in S. coelicolor. Fifteen colonies were randomly picked to determine the editing efficiency by visual analysis on the R2YE plate for each experiment, and the tests were repeated twice. The results showed that the editing efficiencies for disruption of redX (the blue clones, producing no red-pigmented RED) were 0% (0/15 + 0/15, PpuE-redX), 33.3% (5/15 + 5/15, SdaE-redX), and 50% (7/15 + 8/15, MsmE-redX), respectively. In contrast, the efficiencies for inactivation of actI-orf1 (the red clones, producing no blue-pigmented ACT) were 20% (3/15 + 3/15, PpuE-actI-orf1), 43.3% (7/15 + 6/15, SdaE-actI-orf1), and 56.7% (9/15 + 8/15, MsmE-actI-orf1) (Fig. 5C and D). To further validate these results, PCR analysis combined with DNA sequencing was performed and primers were designed to amplify the ∼2,800-bp DNA fragments covering the cleavage site of the MsmE-redX-crRNA. In total, 15 exoconjugants were selected and tested. As shown in Fig. 4E, the deletions were precisely located at the target sites but varied in size (from 409 to 1,624 bp). These results clearly demonstrated that Msm-LK, the NHEJ pathway from M. smegmatis, exhibited the lowest cell toxicity and the highest NHEJ activity (∼50% DSB repair efficiency) in S. coelicolor.
FIG 4.
Rapid gene inactivation by the CRISPR/Cpf1 system combined with reconstituted NHEJ pathways in S. coelicolor. (A) Growth of S. coelicolor expressing FnCpf1 with three different reconstituted NHEJ pathways. Based on pKCCpf1, six editing plasmids containing the reconstituted NHEJ pathways, namely, pKCCpf1-MsmP, pKCCpf1-MsmE, pKCCpf1-SdaP, pKCCpf1-SdaE, pKCCpf1-PpuP, and pKCCpf1-PpuE, were constructed, in which the coexpression of ligD and ku genes from M. smegmatis, S. daghestanicus, or P. putida was driven by the strong promoter gadphp or ermEp*. (B) Effects of FnCpf1-induced DSB repair by three different NHEJ pathways on two target loci, redX and actI-orf1. (C, D) Phenotypic analysis of redX or actI-orf1 inactivation by FnCpf1 combined with the NHEJ systems. The strains with redX inactivation produced only blue-pigmented ACT on R2YE plates (imaged at 48 h). In contrast, these strains with actI-orf1 inactivation produced only red-pigmented RED on R2YE plates (imaged at 72 h). Fifteen random exoconjugants were randomly picked for phenotypic observations; the strain in the top right corner represents the wild-type in each group. Due to the good data repeatability, we show the results of the first phenotypic analysis only. (E) Large random-sized deletions in the redX gene caused by FnCpf1 combined with the Msm-NHEJ system.
FIG 5.
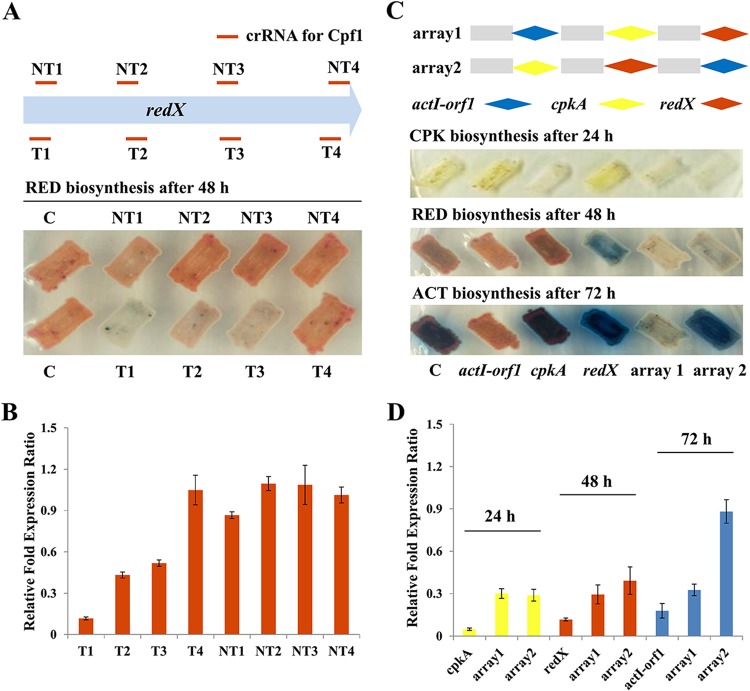
ddCpf1-mediated multiplex gene repression in S. coelicolor. (A, B) ddCpf1-mediated efficient block of redX transcriptional elongation. crRNAs targeting the T or NT strand of the redX coding region are illustrated. The phenotypic (A) and transcriptional (B) analyses were carried out after growth on R2YE agar for 48 h. Error bars indicate the standard deviations from three independent biological replicates. (C, D) Multiplex gene repression with ddCpf1 and a single customized crRNA array. Three target genes (cpkA, actI-orf1, and redX) were selected for simultaneous repression. The order of crRNAs targeting three genes is designated array 1 or array 2. The transcriptional levels of each gene were analyzed in the engineered strains expressing ddCpf1 with individual crRNA or crRNA arrays, and the strain expressing only ddCpf1 was used as the control. The phenotypic and transcriptional analysis was carried out for cpkA, redX, and actI-orf1 at 24, 48, and 72 h, respectively.
Next, to check whether the combination of the CRISPR-Cpf1 system and the reconstituted NHEJ pathway (Msm-LK) could be used to delete large DNA fragments (e.g., the antibiotic biosynthetic pathway) guided by a pair of crRNAs, the RED biosynthetic gene cluster (BGC) was selected and the editing plasmid pKCCpf1-MsmE-RED-BGC harboring two crRNA expression cassettes targeting redX and redG simultaneously were constructed (see Fig. S2A in the supplemental material). The plasmid was introduced into the wild-type strain M145, and then the exoconjugants were selected on MS agar plates (see Materials and Methods for descriptions of media). Visual analysis on R2YE plates showed that 30% (4/15 + 5/15) of tested colonies failed to produce the red-pigmented RED (see Fig. S2B in the supplemental material), indicating the existence of mutations within the RED BGC. However, further analysis by PCR amplification and DNA sequencing revealed that only 10% (1/15 + 2/15) colonies showed different sizes of deletions in the RED BGC (Fig. S2C). Collectively, the data presented here demonstrated that FnCpf1-assisted NHEJ system from M. smegmatis can efficiently mediate random-sized deletions of single genes in S. coelicolor; meanwhile, this system could also be employed for large-size deletions of secondary metabolite BGCs, but with a relatively low editing efficiency.
ddCpf1-mediated multiplex gene repression using a single crRNA array.
A reversible dCas9-based CRISPRi system was previously developed for efficient gene expression regulation in S. coelicolor (25). However, this system has two limitations that need to be overcome. First, the replicative plasmid pKC1139, which was used to express the CRISPR-dCas9 system, may lead to fluctuation of transcriptional repression of target genes due to the plasmid structural and segregational instability (46, 54, 55). Second, simultaneous repression of multiple targets with dCas9 is limited by the time-consuming procedure of the construction of multiple independently expressed, large single guide RNA (sgRNA) expression cassettes. In contrast, a DNase-deactivated Cpf1 (ddCpf1) harboring the E1006A mutation in the RuvC domain of FnCpf1 retains precursor crRNA processing but not DNA cleavage activity and can process a single customized CRISPR array for simplified, multiplex gene expression control (42, 43, 56). Additionally, whole-transcriptomic analysis revealed that ddCpf1-based gene repression showed no significant off-target effects in E. coli (42). In the present study, we developed a robust, multiplex CRISPRi system with chromosomal integration expression of ddCpf1 and crRNA(s) using the integrative plasmid pSET152. Again, the redX gene was chosen as a test case. Four crRNAs targeting the template DNA strand (T strand) and four targeting the nontemplate DNA strand (NT strand) in the open reading frame (ORF) of redX were designed. Phenotypic and transcriptional analysis showed that three of four crRNAs targeting the T strand enabled effective gene repression from 48.2% to 88.3%, and crRNA targeting the site nearest to the start codon exhibited the highest repression activity (Fig. 5A and B). The crRNAs targeting the end of the T strand or targeting the NT strand were less effective or not effective at all for transcriptional repression (Fig. 5A and B). Overall, these results indicate that the ddCpf1-based CRISPRi system can efficiently mediate repression of transcription elongation and the repression is more effective when the crRNAs specifically target the T strand of the target gene, which is just the reverse of the data obtained from the dCas9-based CRISPRi system in S. coelicolor (25).
Subsequently, three genes, including redX, actI-orf1, and cpkA (from the BGC of the yellow-pigmented cryptic polyketone [CPK]), were chosen to test ddCpf1-mediated multiplex gene repression. Prior to multiplex repression, single gene repression for these three genes was first investigated. Three crRNAs were designed for targeting the T strands of the ORFs of redX, actI-orf1, and cpkA, separately, and three corresponding plasmids, pSETddCpf1-redX, pSETddCpf1-actI-orf1, and pSETddCpf1-cpkA, were constructed. Phenotypic analysis showed that individual introduction of three CRISPRi editing plasmids could significantly reduce the production of the corresponding antibiotics (Fig. 5C). Furthermore, the transcriptional levels of three target genes in strains with the corresponding CRISPRi plasmid were repressed, with repression efficiencies of 82.1 to 95.2% (Fig. 5D). Then, two crRNA arrays expressing pre-crRNAs in the orders of actI-orf1-cpkA-redX (array 1) and cpkA-redX-actI-orf1 (array 2) were constructed using 19-nt direct repeats and 23-nt spacers for multiplex gene repression. It was observed that production of three pigmented antibiotics (CPK, RED, and ACT) and mRNA levels of their respective biosynthetic genes were significantly decreased in the engineered strain with the coexpression of array 1 and ddCpf1 compared to the wild-type strain expressing only ddCpf1, with repression efficiencies of 69.9% for cpkA, 70.6% for redX, and 67.4% for actI-orf1 (Fig. 5C and D). Interestingly, when array 2 was used to mediate multiplex gene regulation, the last crRNA unit targeting actI-orf1 resulted in only a slight reduction of ACT biosynthesis and it gave a repression efficiency of just 11.8% at the transcriptional level, indicating an obvious order effect of actI-orf1-targeted crRNA on repression efficiency.
In total, there are six combinations of arrays for three crRNA in different orders. To systematically explore the effects of the crRNA orders on repression efficiency, the other four arrays (arrays 3 to 6) were also constructed and tested (see Fig. S3A in the supplemental material). The results revealed that, as shown in Fig. S3A and B, similar to the results obtained from arrays 1 and 2, the effects of different crRNA orders on repression efficiency were observed only for the crRNA targeting actI-orf1 but not for the other two tested crRNAs, targeting cpkA and redX. Transcriptional analysis showed that the highest actI-orf1 repression efficiency (76.6%) was observed when its crRNA unit was put immediately downstream of kasOp* (array 5). However, when the actI-orf1-targeted crRNA unit was put at the middle and last orders in the arrays, they resulted in much lower repression efficiencies, 37.7% for array 4 (middle), 51.6% for array 6 (middle), and 17.9% for array 3 (last) (Fig. S3B). Overall, the results presented here clearly indicated that the crRNA order in arrays might influence only the repression efficiency of specific genes. Additionally, we noticed that the repression strength of gene transcription using a crRNA array was lower than those using individual crRNAs for all of the three tested genes (Fig. 5D and S3B), which might be ascribed to the insufficient expression quantity of ddCpf1. In our multiplex repression test, ddCpf1 was used not only to process pre-crRNA to the mature crRNA but also to simultaneously target three loci, which was significantly different from the role of ddCpf1 in the single repression test. This reduced repression activity might be addressed by replacing ermEp* with the stronger promoter kasOp* or its derivatives (45, 57). Together, these results establish that the ddCpf1-based CRISPRi system is a convenient and powerful tool for efficient repression of multiple genes in S. coelicolor.
Applicability of the F. novicida CRISPR-Cpf1 system in other Streptomyces species.
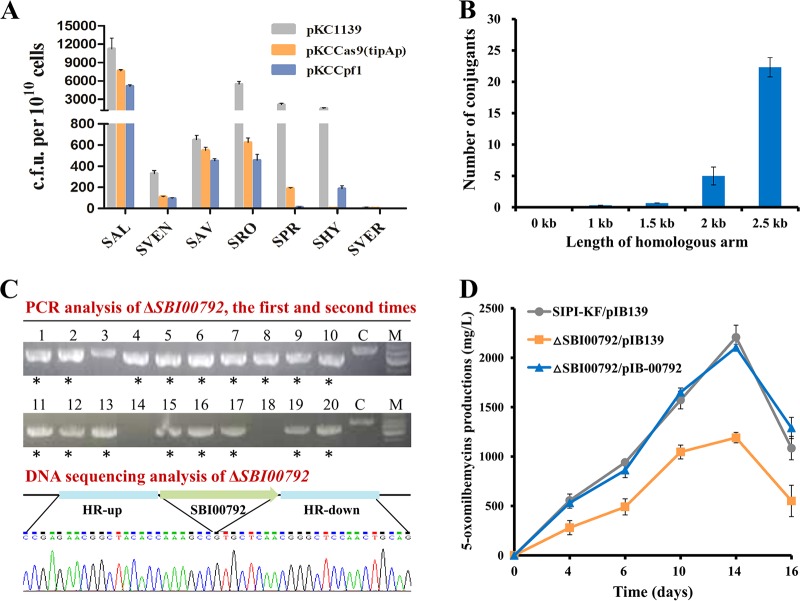
To determine whether the FnCpf1-assisted genome editing tool is applicable to Streptomyces species other than S. coelicolor, as the first step, pKCCpf1 was introduced into seven model or industrial Streptomyces strains, including S. albus J1074, S. venezuelae ATCC 10712, S. avermitilis NRRL8165, S. roseosporus SIPI-DT51, S. pristinaespiralis HCCB10218, S. hygroscopicus SIPI-KF, and S. verticillus SIPI-BL, for toxicity tests of FnCpf1 expression. Plasmids pKC1139 and pKCCas9(tipAp) were used as the controls. The results showed that among four Streptomyces strains tested, including S. albus J1074, S. venezuelae ATCC 10712, S. avermitilis NRRL8165, and S. roseosporus SIPI-DT51, introduction of both FnCpf1- and SpCas9-expressing plasmids reduced the numbers of exoconjugants in various degrees from 14.4% to 91.7% compared to pKC1139, but there was no obvious difference in transformation efficiencies between the two different CRISPR-Cas systems (Fig. 6A). However, introduction of pKCCas9(tipAp) generated 196 ± 5 exoconjugants, but very few colonies (5 ± 1) harboring the plasmid pKCCpf1 were obtained in S. pristinaespiralis HCCB10218. In contrast, introduction of pKCCpf1 into S. hygroscopicus SIPI-KF generated 204 ± 9 exoconjugants with >100-fold transformation efficiency compared to that of pKCCas9(tipAp) (Fig. 6A). Finally, no colonies were obtained when introducing pKC1139, pKCCas9(tipAp), or pKCCpf1 into S. verticillus SIPI-BL, which should be ascribed to the incompatibility of the temperature-sensitive replicon pSG5 in the recipient cells (Fig. 6A). Notably, similar results were previously observed in S. verticillus ATCC 15003, in which introduction of pKC1139 failed to yield any true exoconjugants (58). Together, these results clearly indicated that the two well-known Cas proteins, SpCas9 and FnCpf1, exhibited different suitability in the tested Streptomyces species.
FIG 6.
Applicability of the CRISPR-Cpf1 system in multiple model and industrial Streptomyces strains. (A) Growth of seven different Streptomyces species expressing only SpCas9 or FnCpf1. pKC1139 was used as the control. SAL, S. albus J1074; SVEN, S. venezuelae ATCC 10712; SAV, S. avermitilis NRRL8165; SRO, S. roseosporus SIPI-DT51; SPR, S. pristinaespiralis HCCB10218; SHY, S. hygroscopicus SIPI-KF; SVER, S. verticillus SIPI-BL. (B) Effects of homologous arm lengths on the exoconjugant numbers when deleting SBI00792 in S. hygroscopicus SIPI-KF. (C) PCR and DNA sequencing analysis of the SBI00792 deletion mutant (ΔSBI00792). C and M represent the parental strain KF and 1-kb DNA ladder, respectively. (D) 5-Oxomilbemycin A3/A4 production by the ΔSBI00792 mutant. Fermentation samples from the three S. hygroscopicus strains, including SIPI-KF/pIB139, the ΔSBJ00792 mutant with pIB139 (ΔSBJ00792/pIB139), and the complemented strain ΔSBJ00792/pIB-00792, were collected at 4, 6, 10, 14, and 16 days.
Subsequently, as an example, the Fn CRISPR-Cpf1 system was employed for HDR-mediated precise genome editing in the 5-oxomilbemycin-producing strain S. hygroscopicus SIPI-KF, in which the S. pyogenes CRISPR-Cas9 system did not work well, as described above. Here, we attempted to knock out a TetR-family regulatory gene, SBI00792, which is adjacent to the 5-oxomilbemycin BGC and may therefore be involved in 5-oxomilbemycin biosynthesis. Homologous arms (HAs) of 1 kb were first designed for SBI00792 deletion, but no exoconjugants could be obtained (Fig. 6B). So, the lengths of HAs were extended to 1.5, 2, and 2.5 kb to optimize DSB repair efficiency. As shown in Fig. 6B, along with the extension of HA lengths, the exoconjugant numbers were gradually increased. Notably, the repeated transformation trials demonstrated that introduction of the plasmid pKCCpf1-SBI00792-HA2.5 with two 2.5-kb HAs could stably generate more than 20 exoconjugants, which were enough for screening of positive mutants (Fig. 6B). These results indicated that HA lengths should be carefully considered for HDR-mediated gene deletion in some specific Streptomyces species.
Then, 10 colonies were randomly picked for PCR and DNA sequencing analysis, and this experiment was repeated twice. It was observed that the positive rate for in-frame deletion of SBI00792 was 85% (8/10 + 9/10), which was comparable to the data obtained for S. coelicolor (Fig. 6C). Phenotypic analysis showed that there was no growth difference between the parental strain SIPI-KF and the SBI00792 deletion mutant (see Fig. S4 in the supplemental material). Fermentation analysis showed that the inactivation of SBI00792 resulted in decreased production of 5-oxomilbemycin A3/A4 (the precursor of milbemycin oxime) by 50% compared with the parental strain (from 2,226 mg/liter to 1,120 mg/liter) and the complemented strain ΔSBI00792/pIB-00792 could successfully restore 5-oxomilbemycin production (Fig. 6D). These results suggested that SBI00792 was a positive regulator for 5-oxomilbemycin biosynthesis, whose detailed regulatory mechanism required to be further explored. Collectively, we demonstrated that FnCpf1 is capable of HDR-mediated targeted gene disruption in S. hygroscopicus SIPI-KF and can serve as a useful alternative for genome editing in other Streptomyces strains that cannot utilize the S. pyogenes CRISPR-Cas9 system.
DISCUSSION
In this study, the F. novicida CRISPR-Cpf1-based genome editing tool (cloned in an all-in-one plasmid) was successfully established to efficiently cleave Streptomyces genome, thereby promoting high-efficiency HDR- and NHEJ-dependent genetic engineering. Interestingly, we found that without the addition of inducer, the expression of SpCas9 driven by the inducible promoter tipAp, but not that of FnCpf1, was enough for efficient genomic DNA cleavage in S. coelicolor, suggesting that high expression levels of FnCpf1 are required for genome editing (26). Guided by previous studies in mammalian cell, the crRNA used in this work was designed to contain a 19-nt direct repeat and a 23-nt spacer preceded by a 5′-TTV-3′ PAM (34, 35). Here, we demonstrated that the PAM sequence could be relaxed from 5′-TTV-3′ to 5′-YTV-3′ (Y = T/C and V = A/G/C) for DNA cleavage in S. coelicolor. However, 5′-CTV-3′ PAM cannot mediate efficient genome editing due to its halfway DNA cleavage activity compared to 5′-TTV-3′ PAM. On the other hand, differing from a previous report that FnCpf1 enables complete DSB activity in vitro when paired with 17- to 25-nt spacer crRNA (48), our in vivo study revealed that the shortest length of spacers must be 19 nt for efficient genome cleavage. Therefore, the PAM sequence and crRNA spacer lengths require to be precisely determined in a specific organism before large-scale genome editing is conducted. Additionally, compared to the original crRNA spacer length (23 to 25 nt), the truncated spacer could reduce the price of primer synthesis for expression of a long crRNA array used in multiplex gene editing or expression regulation.
FnCpf1 can induce a site-specific DSB in the genome, which relies on repair by homologous recombination or NHEJ. In the present study, high efficiencies (75 to 95%) of the FnCpf1-based system for precise single and double genomic locus editing in the presence of HDR templates were achieved, which is comparable to the SpCas9-assisted editing system in Streptomyces (26, 27). Therefore, it is reasonable to believe that FnCpf1-mediated HDR could also be expanded to mediate deletion of large-size secondary metabolites BGCs (10 to 100 kb) or precise point mutations, as previously reported for SpCas9 (26). On the other hand, NHEJ-mediated DSBs repair results in stochastic indels at the target loci, but this strategy is very simple and convenient due to the lack of requirement for homology templates. Here, we demonstrated that in S. coelicolor, reconstituted NHEJ systems from actinomycetes, including Msm-LK and Sda-LK, exhibited higher DSB repair activities than did Ppu-LK from the Gram-negative bacterium P. putida. FnCpf1 combined with Msm-LK generated large, random-sized deletions in target genes from 0.4 to 1.6 kb. Interestingly, previous studies have showed that only very short indels (1 to 3 bp) were introduced by S. pyogenes CRISPR-Cas9 combined with Nocardia carnea NHEJ system in S. coelicolor (25). This difference might be due to the distinct cleavage mechanisms of these two different class 2 CRISPR-Cas systems, i.e., SpCas9 generates a PAM-proximal DSB with a blunt end, whereas FnCpf1 creates a PAM-distal DSB with a 5′ overhang (15, 59). Additionally, the efficiency for the deletion of large-size RED BGC (∼28 kb) by the FnCpf1-based Msm-LK system was low (only 10%) in S. coelicolor. However, in E. coli, the efficiencies for deleting 46-kb and 123-kb DNA fragments reached 46% and 36%, respectively, by combining CRISPR-Cas9 with Msm-LK system (52). Therefore, the developed FnCpf1-based Msm-LK system in Streptomyces still required to be further optimized, such as by fine-tuning the expression of the Msm-LK pathway and introducing other, more-efficient NHEJ systems.
In addition to the FnCpf1-mediated Streptomyces genome editing tool, in this study, a ddCpf1-based CRISPRi system was also developed for multiplex gene repression. Using this novel CRISPRi system, here we achieved simultaneous repression of three antibiotic biosynthetic genes at efficiencies of ∼70%. Here, the integrative plasmid pSET152 instead of the replicative plasmid pKC1139 was employed as the backbone for the expression of the CRISPRi system, which would theoretically enhance the stability of ddCpf1-based gene expression control. Additionally, the ΦC31 integration system in pSET152 has a broad host range for Streptomyces species (60), ensuring that our CRISPRi system can be widely used in the genus Streptomyces. Similar to the data obtained with E. coli, crRNA recognizing the T strand but not the NT strand of target genes could achieve efficient ddCpf1-based gene repression in Streptomyces (42, 61). However, we found that the crRNA order in a single array obviously influenced repression efficiency for actI-orf1 involved in ACT biosynthesis at 72 h after growth, which was not identical to that previously reported in E. coli (42). We speculated that the effect of different crRNA orders on repression efficiency of actI-orf1 transcription might be ascribed to the following three reasons. First, microarray analysis showed that actI-orf1 is strongly transcribed at 72 h after growth, but the other two tested genes, redX and cpkA, are basically not transcribed at that time (62). Second, ddCpf1 and crRNA array were under the control of the promoters ermEp* and kasOp*, respectively. Previous studies showed that the mRNA levels of these two promoters significantly decreased in the later growth stages (60 to 96 h) of S. coelicolor (45). Therefore, the expression levels of both ddCpf1 and crRNA array would be very low at 72 h, which might be the reason for the strong leakiness in actI-orf1 expression. Finally, the low expression level of ddCpf1 might also be insufficient for completely processing crRNA array, which thus led to the lower level of the crRNA for targeting actI-orf1 in the middle or last unit of array compared to that in the first unit of array. Nevertheless, we believe that the ddCpf1-based CRPISRi system can be broadly applicable to the study of essential genes and complex regulatory networks, as well as for quick screening of functional genes in Streptomyces (63).
Previously, it has been repeatedly reported that no transformant could be obtained due to the constitutive or leaky expression of a native SpCas9 in C. glutamicum (30, 31, 64). The toxic effect of SpCas9 expression could be overcome by three different strategies. First, the plasmid harboring a codon-optimized cas9 gene based on the bias of Streptomyces could be effectively transformed into C. glutamicum, for some unknown reasons (30). Second, two inducible promoters, Ptac and PprpD2, were utilized to strictly control the expression of the native cas9 gene (64). The strategy was also employed to overcome the toxicity of constitutive expression of the nuclease-deactivated SpCas9 (dCas9) in C. glutamicum (65). Finally, Jiang and coworkers successfully utilized the F. novicida CRISPR-Cpf1 system to achieve highly efficient genome editing in C. glutamicum, which skillfully bypassed the fine-tuning of SpCas9 and sgRNA expression (31). In the present study, we found that no exoconjugant could be obtained when pKCCas9(tipAp) was introduced into S. hygroscopicus SIPI-KF possibly due to the leaky expression of the codon-optimized cas9 under the control of the inducible promoter tipAp. Similarly, the toxic effect was effectively addressed by replacing SpCas9 with FnCpf1. Alternatively, other inducible promoters, including tetracycline-, glycerol-, and cumate-inducible systems, may be used to strictly control SpCas9 expression in S. hygroscopicus SIPI-KF (66). In contrast, the constitutive expression of FnCpf1 without a customized crRNA cassette resulted in serious cell toxicity in another industrial strain, S. pristinaespiralis HCCB10218, compared to SpCas9. These results suggested that the two well-known Cas proteins exhibited distinct toxicity effects in several specific Streptomyces strains. Notably, bioinformatical analysis showed that there was no typical SpCas9 or FnCpf1 crRNA sequence in the above two industrial Streptomyces strains. Therefore, SpCas9 or FnCpf1 toxicity is unlikely to be explained by the Cas endonuclease-mediated target or off-target DSBs. On the other hand, introduction of both dCas9- and ddCpf1-expressing plasmids reduced the numbers of exoconjugants compared to pKC1139 in both S. pristinaespiralis HCCB10218 and S. hygroscopicus SIPI-KF (see Fig. S5 in the supplemental material). Therefore, we speculated that SpCas9 or FnCpf1 may bind tightly to PAMs in the absence of crRNA, which thus results in different global transcriptional expression and influences bacterial growth to various degrees. The general toxic effects of Cas proteins should be further explored in the future. Nevertheless, from a practical perspective, FnCpf1 provides a useful complement to SpCas9 for high-efficiency genome editing and multiplex transcriptional regulation in Streptomyces. A diverse CRISPR toolbox will significantly advance metabolic engineering and natural product discovery in different kinds of industrial Streptomyces strains as well as other actinomycetes (9, 22, 67).
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
The strains and plasmids used in this study are listed in Table S2 in the supplemental material. S. coelicolor M145 and its derivatives were grown on MS medium (in grams/liter, soybean flour, 20; mannitol, 20; and agar, 20) for spore preparation and intergeneric conjugal transfer (46). Medium MS was also used for spore preparations of Streptomyces albus J1074, Streptomyces venezuelae ATCC 10712, and Streptomyces avermitilis NRRL8165. Medium RP (in grams/liter, tryptone, 5; yeast extract, 5; valine, 0.5; NaCl, 2; KH2PO4, 0.5; MgSO4·7H2O, 1; and agar, 20; pH 6.4), YMD medium (in grams/liter, malt extract, 10; yeast extract, 4; glucose, 4; and agar, 20), MB medium (in grams/liter, sucrose, 4; skim milk, 1; yeast extract, 2; malt extract, 5; and agar, 20; pH 7.0 to 7.2), and Isp4 medium [in grams/liter, starch, 10; tryptone, 1; yeast extract, 0.5; NaCl, 1; (NH4)2SO4, 2; K2HPO4, 1; CaCO3, 2; agar, 20; with 1 ml trace element solution; pH 7.0] were used for spore preparations of Streptomyces pristinaespiralis HCCB10218, Streptomyces roseosporus SIPI-DT51, Streptomyces hygroscopicus SIPI-KF, and Streptomyces verticillus SIPI-BL, respectively. When the CRISPR-Cas9 or CRISPR-Cpf1 genome editing tool was employed, M-Isp4 medium [in grams/liter, soybean flour, 5; mannitol, 5; starch, 5; tryptone, 2; yeast extract, 1; NaCl, 1; (NH4)2SO4, 2; K2HPO4, 1; CaCO3, 2; agar, 20; with 1 ml trace element solution; pH 7.0 to 7.2] with 10 mM or 60 mM MgCl2 was used for conjugal transfer between E. coli and Streptomyces. S. coelicolor and S. hygroscopicus cultures grown in liquid MB and YEME medium (in grams/liter, yeast extract, 3; peptone, 5; malt extract, 3; glucose, 10; sucrose, 340; and MgSO4·7H2O, 1.24), respectively, were harvested for total DNA isolation (46). All Streptomyces strains were cultivated at 30°C.
Escherichia coli DH5α was used for DNA cloning. E. coli ET12567/pUZ8002 or S17-1 was used for conjugal transfer from E. coli to Streptomyces. The detailed conditions of conjugal transfer are listed in Table S3 in the supplemental material. E. coli strains were grown at 37°C in Luria-Bertani (LB) medium or on LB agar plates. Antibiotics (50 μg/ml of ampicillin, apramycin, and kanamycin) were added when necessary.
Construction of Cpf1- and ddCpf1-based editing plasmids.
The cpf1 gene originating from Francisella novicida U112 was codon optimized based on the bias of S. coelicolor M145 (the optimized sequence, named “scocpf1,” is available in Data S1 in the supplemental material) and was synthesized. Then, the scocpf1 gene was cloned into the plasmid pKCCas9(tipAp) between the EcoRI and NdeI sites to replace the scocas9 gene (26), which is under the control of the inducible promoter tipAp, generating the plasmid pKCCpf1(tipAp)-E. Using the plasmid pAH91kasOp*-cmlR as the template (68), the crRNA scaffold, containing the strong promoter kasOp* and 19-nt direct repeat, was obtained by PCR amplification with the primers kasOp*-crRNA-fw/rev. The resulting DNA fragment was doubly digested by SpeI and XbaI and ligated into pKCCpf1(tipAp)-E to generate pKCCpf1(tipAp).
Based on pKCCpf1(tipAp), the plasmid pKCCpf1 containing the scocpf1 gene driven by the constitutive promoter ermEp* was constructed as follows. The ermEp* promoter was amplified from pIB139 using the primers ermEp*-fw/rev. The resulting DNA fragment was recombined into pKCCpf1(tipAp) that was digested by BamHI and NdeI, generating the plasmid pKCCpf1.
The plasmid pSETddCpf1, containing the DNase-dead Cpf1 (ddCpf1)-encoding gene, was constructed on the basis of the integrative plasmid pSET152 as follows. Using pKCCpf1 as the template, the upstream and downstream regions (884 and 3356 bp) of the CRISRP/Cpf1 expression cassette were obtained by PCR amplification using the primer pairs ddcpf1-up-fw/ddcpf1-up-E1006A-rev and ddcpf1-down-fw/rev (the primer ddcpf1-up-E1006A-rev contains the mutational DNA sequence for generating the point mutation E1006A in FnCpf1) and then ligated together by overlapping PCR with the primers ddcpf1-up-fw and ddcpf1-down-rev. The resulting DNA fragment was cloned into the EcoRI/XbaI-digested pSET152 by recombination, generating the plasmid pSETddCpf1. The E1006A mutation in pSETddCpf1 was confirmed by DNA sequencing.
On the basis of pKCCpf1, the plasmid pKCddCpf1, containing the ddCpf1-encoding gene, was constructed as follows. The ddcpf1 gene was amplified from pSETddCpf1 by PCR using the primers ddcpf1-fw/rev, followed by double digestion with EcoRI and NdeI. Then, the resulting DNA fragment was cloned into pKCCpf1 to yield pKCddCpf1. Similarly, based on pKCCas9(tipAp), the plasmid pKCdCas9(tipAp), containing the DNase-deficient cas9 (dcas9)-encoding gene was constructed. The dcas9 gene originating from S. pyogenes was codon optimized based on the bias of S. coelicolor M145 and synthesized. Then, the dcas9 gene was cloned into pKCCas9(tipAp) between the EcoRI and NdeI sites to replace the scocas9 gene (26), thus generating pKCdCas9(tipAp).
Cpf1-based gene editing plasmids and ddCpf1-based CRISPRi plasmids were constructed by introducing a 23-nt gene-specific spacer sequence into the crRNA scaffold of pKCCpf1 and pSETddCpf1. Briefly, using pKCCpf1 as the template, the crRNA expression cassettes were amplified with the forward primer 5′-CACTAGTN23ATCTACAACAGTAGAAATTTGG-3′ (N23 represents the 23-nt gene-specific spacer sequence) and the reverse primer (crRNA-rev). The PCR product was digested with NdeI and SpeI and ligated to NdeI/SpeI-digested pKCCpf1 and pSETddCpf1. Different Cpf1 or ddCpf1-based editing plasmids could be generated simply by just changing the N23 region of the forward primer. The primers used are listed in Table S4 in the supplemental material.
Construction of pKCCpf1-based NHEJ plasmids.
Based on pKCCpf1, a series of recombinant plasmids containing different NHEJ pathways driven by the strong promoter gadphp or ermEp* were constructed as follows. The Sda-ligD and Sda-ku genes from Streptomyces daghestanicus under the control of gadphp were synthesized and cloned into pKCCpf1 between HindIII and SpeI, thus generating pKCCpf1-SdaP. The promoter ermEp* was amplified from pIB139 using the primers ermEp*-fw (SpeI) and ermEp*-rev (EcoRV) and then ligated into pKCCpf1-SdaP to yield the plasmid pKCCpf1-SdaE. The Msm-ligD and Msm-ku genes from Mycobacterium smegmatis were amplified from the plasmid pZX09 using the primer pairs Msm-ligD-fw/rev and Msm-ku-fw/rev, respectively (52). The Msm-ligD and Msm-ku genes were doubly digested by EcoRV/XbaI and XbaI/HindIII, respectively, and ligated simultaneously into pKCCpf1-SdaP or pKCCpf1-SdaE between EcoRV and HindIII to generate pKCCpf1-MsmP or pKCCpf1-MsmE. Using the Pseudomonas putida genome as the template, the Ppu-ligD and Ppu-ku genes were amplified using the primer pairs Ppu-ligD-fw/rev and Ppu-ku-fw/rev. The Ppu-ligD and Ppu-ku genes were doubly digested by SpeI/XbaI and XbaI/HindIII, respectively, and cloned simultaneously into pKCCpf1 between SpeI and HindIII to yield pKCCpf1-Ppu. The promoter gadphp was amplified from pKCCpf1-SdaP using the primers gadphp-fw/rev and then ligated into pKCCpf1-Ppu between by SpeI and PmeI to yield pKCCpf1-PpuP. Similarly, the promoter ermEp* was amplified from pIB139 using the primers ermEp*-fw (SpeI) and ermEp*-rev (PmeI) and ligated into pKCCpf1-Ppu between SpeI and PmeI, generating pKCCpf1-PpuE.
To utilize FnCpf1-assisted NHEJ systems for random deletions of redX, actI-orf1, or RED biosynthetic gene cluster (BGC), the editing plasmids pKCCpf1-redX, pKCCpf1-actI-orf1, and pKCCpf1-RED BGC were first constructed as described above by introducing the gene-specific spacer sequence into the crRNA scaffold of pKCCpf1. This design was to check whether crRNAs were capable of efficiently mediating DSBs at the target loci in Streptomyces. Then, NHEJ-mediated editing plasmids were constructed as follows, taking pKCCpf1-MsmE-redX as an example to describe the process. In brief, the MsmE expression cassette containing the Msm-ligD and Msm-ku genes under the control of ermEp* was obtained by double digestion of pKCCpf1-MsmE with HindIII and SpeI and then ligated into pKCCpf1-redX to yield pKCCpf1-MsmE-redX. The other five NHEJ-mediated editing plasmids, including pKCCpf1-SdaE-redX, pKCCpf1-PpuE-redX, pKCCpf1-MsmE-actI-orf1, pKCCpf1-SdaE-actI-orf1, and pKCCpf1-PpuE-actI-orf1, were constructed similarly. The primers used are listed in Table S4.
Construction of the S. coelicolor and S. hygroscopicus mutants.
The ΔactI-orf1, ΔredX, and ΔactI-orf1-redX mutants with in-frame deletion of actI-orf1, redX, and both actI-orf1 and redX, respectively, were constructed using the CRISPR-Cpf1-assisted HDR strategy. Here, we took ΔactI-orf1 as an example to introduce the mutant construction process. Briefly, the upstream and downstream regions (1,019 and 1,033 bp) of actI-orf1 were obtained by PCR using the primer pairs Del-actI-orf1-up-fw/rev and Del-actI-orf1-down-fw/rev, respectively. Using the plasmid pKCCpf1 as the template, the crRNA expression cassette containing the strong promoter kasOp*, 19-nt repeat sequence, and 23-nt protospacer sequence targeting actI-orf1 was obtained by PCR amplification with primers actI-orf1-crRNA-fw/rev. Then, three DNA fragments were overlapped by PCR using the primers Del-actI-orf1-up-fw and actI-orf1-crRNA-rev, followed by double digestion with NdeI and SpeI. The resulting DNA fragment was ligated into pKCCpf1 to yield pKCCpf1-actI-orf1-HR, which was introduced into the wild-type S. coelicolor M145 by conjugal transfer. The resulting exoconjugants were subsequently grown on solid MS medium without apramycin at 37°C for two rounds to remove the plasmid pKCCpf1-actI-orf1-HR. The correct double crossovers were verified by colony PCR with the primers ID-actI-orf1-fw/rev, yielding the ΔactI-orf1 mutant. The ΔredX and ΔactI-orf1-redX mutants were constructed similarly.
The ΔRED-BGC mutant with deletion of the ∼28-kb RED BGC region between redX and redG was constructed by using the CRISPR-Cpf1-assisted NHEJ strategy. Briefly, using the plasmid pKCCpf1 as the template, the crRNA array expression cassette containing the strong promoter kasOp* and two crRNAs targeting redX and redG, respectively, was obtained by PCR amplification with primers crRNA-RED-BGC-fw and crRNA-rev, followed by double digestion with NdeI and SpeI. The resulting DNA fragment was cloned into pKCCpf1 to generate pKCCpf1-RED-BGC. Then, the ligD and ku expression cassette under the control of ermEp* from pKCCpf1-MsmE by double digestion with HindIII and SpeI was ligated into pKCCpf1-RED-BGC to generate pKCCpf1-MsmE-RED-BGC, which was introduced into the S. coelicolor wild-type strain M145. The resulting strains were subsequently grown on solid R2YE medium for phenotypic analysis. These strains, which produced only blue-pigmented actinorhodin (ACT), were cultivated on MS medium without apramycin at 37°C for two rounds to remove the plasmid pKCCpf1-MsmE-RED-BGC. The correct strains were verified by colony PCR with the primers ID-NHEJ-RED-BGC-fw/rev and DNA sequencing, yielding the ΔRED-BGC mutant. The primers used are listed in Table S4.
The ΔSBI00792 mutant with an in-frame deletion of SBI00792 was constructed on the basis of the 5-oxomilbemycin-producing strain S. hygroscopicus SIPI-KF, using the CRISPR-Cpf1-assisted HDR strategy. Briefly, the crRNA expression cassette was amplified from pKCCpf1 using the primers SBI00792-crRNA-fw/rev and recombined into pKCCpf1 to generate pKCCpf1-SBI00792. Then, the upstream and downstream regions (2,409 and 2,470 bp) of SBI00792 were obtained by PCR amplification using the primer pairs Del-SBI00792-up-2.5-fw/Del-SBI00792-up-rev and Del-SBI00792-down-fw/Del-SBI00792-down-2.5-rev, respectively. Two DNA fragments were ligated together by overlapping PCR using the primers Del-SBI00792-up-2.5-fw and Del-SBI00792-down-2.5-rev. The resulting DNA fragment was ligated into pKCCpf1-SBI00792 to yield pKCCpf1-SBI00792-HR2.5 by Gibson assembly (69), which was transferred into S. hygroscopicus SIPI-KF. The resulting strains were subsequently grown on solid MB medium without apramycin at 37°C for two rounds to remove the plasmid pKCCpf1-SBI00792-HR2.5. The correct double crossovers were verified by colony PCR with the primers ID-SBI00792-fw/rev, yielding the ΔSBI00792 mutant.
To confirm the role of SBI00792, the complemented strain ΔSBI00792/pIB-00792 was constructed. Briefly, using the S. hygroscopicus SIPI-KF genome as the DNA template, the gene SBI00792 was obtained by PCR amplification using the primers Com-SBI00792-fw/rev and doubly digested with EcoRI and NdeI. The resulting DNA fragment was cloned into the EcoRI/NdeI-digested pIB139, thus generating the plasmid pIB-00792. Then, the plasmid pIB-00792 was introduced into the ΔSBI00792 mutant, yielding the complemented strain ΔSBI00792/pIB-00792. As controls, two strains, SIPI-KF/pIB139 and ΔSBI00792/pIB139, were constructed by introducing the empty vector pIB139 into S. hygroscopicus SIPI-KF and the ΔSBI00792 mutant, respectively. The primers used are listed in Table S4.
Fermentation of S. hygroscopicus and HPLC analysis of 5-oxomilbemycin A3/A4 production.
For 5-oxomilbemycin A3/A4 production, S. hygroscopicus strains were first grown on solid MB medium at 30°C for 6 to 7 days and then inoculated into 25 ml seed medium (in grams/liter, sucrose, 20; meat peptone, 2.5; yeast extract, 5; and K2HPO4, 0.1; pH 7.2) in 250-ml Erlenmeyer flasks on an orbital shaker (240 rpm). After incubation at 28°C for 24 to 28 h, 1 ml of preculture was inoculated into 25 ml of fermentation medium (in gra,s/liter, sucrose, 120; cotton seed meal, 5; soybean flour, 5; K2HPO4, 0.5; FeSO4·7H2O, 0.1; ZnSO4·7H2O, 0.01; CuSO4·7H2O, 0.05; and CaCO3, 3) in 250-ml Erlenmeyer flasks. The pH of fermentation medium was adjusted to 7.2 prior to CaCO3 addition.
One-milliliter fermentation samples were collected at five time points (4, 6, 10, 14, and 16 days) and extracted with the same volume of ethanol for 1 to 2 h. Subsequently, the mixtures were centrifuged at 12,000 rpm for 10 min, and the supernatants (ethanol extracts) were directly analyzed by high-performance liquid chromatography (HPLC; 1100 series; Agilent) using a 4.6- by 150-mm Hypersil C18 column (Agilent). For HPLC detection, a mixture of acetonitrile and H2O (73:27, vol/vol) was used as the mobile phase, with a flow rate of 1.2 ml/min and retention time of 14 min of 5-oximilbemycin A3 and 20 min of 5-oximilbemycin A4. The eluates were monitored at 240 nm, and the column temperature was 30°C.
RNA preparation and RT-qPCR.
RNA preparation and quantitative real-time reverse transcription-PCR (RT-qPCR) analysis were performed as previously described (70). The primers used are listed in Table S4. S. coelicolor strains were grown on R2YE covered with sterilized plastic cellophane. Cultures were harvested for RNA preparation at different time points (24, 48, and 72 h). RT-qPCR analysis was performed in the MyiQ2 two-color real-time PCR detection system (Bio-Rad, USA) by using iQ SYBR green Supermix (Bio-Rad, USA). The reactions were conducted in triplicate for each transcript and repeated with three independent samples. The hrdB gene (SCO5820, encoding the principal sigma factor) was used as an internal control. The relative expression levels of tested genes were normalized to those of hrdB. The relative fold changes in the transcription of each gene were determined using the 2−ΔΔCT method (71). Error bars indicate the standard deviations from three independent biological replicates.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (31630003, 31570072, and 31770088), the National Postdoctoral Program for Innovative Talents (BX201700265), the Science and Technology Commission of Shanghai Municipality (18ZR1446700), and Project of Chinese Academy of Sciences (ZSYS-016). We also acknowledge support from the SA-SIBS Scholarship Program.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00827-18.
REFERENCES
- 1.Butler MS, Robertson AAB, Cooper MA. 2014. Natural product and natural product derived drugs in clinical trials. Nat Prod Rep 31:1612–1661. doi: 10.1039/C4NP00064A. [DOI] [PubMed] [Google Scholar]
- 2.Barka EA, Vatsa P, Sanchez L, Gaveau-Vaillant N, Jacquard C, Klenk HP, Clement C, Ouhdouch Y, van Wezel GP. 2016. Taxonomy, physiology, and natural products of actinobacteria. Microbiol Mol Biol Rev 80:1–43. doi: 10.1128/MMBR.00019-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hwang KS, Kim HU, Charusanti P, Palsson BO, Lee SY. 2014. Systems biology and biotechnology of Streptomyces species for the production of secondary metabolites. Biotechnol Adv 32:255–268. doi: 10.1016/j.biotechadv.2013.10.008. [DOI] [PubMed] [Google Scholar]
- 4.Baltz RH. 2017. Gifted microbes for genome mining and natural product discovery. J Ind Microbiol Biotechnol 44:573–588. doi: 10.1007/s10295-016-1815-x. [DOI] [PubMed] [Google Scholar]
- 5.Cimermancic P, Medema MH, Claesen J, Kurita K, Brown LCW, Mavrommatis K, Pati A, Godfrey PA, Koehrsen M, Clardy J, Birren BW, Takano E, Sali A, Linington RG, Fischbach MA. 2014. Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell 158:412–421. doi: 10.1016/j.cell.2014.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doroghazi JR, Albright JC, Goering AW, Ju KS, Haines RR, Tchalukov KA, Labeda DP, Kelleher NL, Metcalf WW. 2014. A roadmap for natural product discovery based on large-scale genomics and metabolomics. Nat Chem Biol 10:963–968. doi: 10.1038/nchembio.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li L, Jiang WH, Lu YH. 2017. New strategies and approaches for engineering biosynthetic gene clusters of microbial natural products. Biotechnol Adv 35:936–949. doi: 10.1016/j.biotechadv.2017.03.007. [DOI] [PubMed] [Google Scholar]
- 8.Luo YZ, Enghiad B, Zhao HM. 2016. New tools for reconstruction and heterologous expression of natural product biosynthetic gene clusters. Nat Prod Rep 33:174–182. doi: 10.1039/C5NP00085H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baltz RH. 2016. Genetic manipulation of secondary metabolite biosynthesis for improved production in Streptomyces and other actinomycetes. J Ind Microbiol Biotechnol 43:343–370. doi: 10.1007/s10295-015-1682-x. [DOI] [PubMed] [Google Scholar]
- 10.Weber T, Charusanti P, Musiol-Kroll EM, Jiang XL, Tong YJ, Kim HU, Lee SY. 2015. Metabolic engineering of antibiotic factories: new tools for antibiotic production in actinomycetes. Trends Biotechnol 33:15–26. doi: 10.1016/j.tibtech.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 11.Jackson SA, McKenzie RE, Fagerlund RD, Kieper SN, Fineran PC, Brouns SJJ. 2017. CRISPR-Cas: adapting to change. Science 356:eaal5056. doi: 10.1126/science.aal5056. [DOI] [PubMed] [Google Scholar]
- 12.Barrangou R, Horvath P. 2017. A decade of discovery: CRISPR functions and applications. Nat Microbiol 2:17092. doi: 10.1038/nmicrobiol.2017.92. [DOI] [PubMed] [Google Scholar]
- 13.Horvath P, Barrangou R. 2010. CRISPR/Cas, the immune system of bacteria and archaea. Science 327:167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- 14.Hille F, Richter H, Wong SP, Bratovic M, Ressel S, Charpentier E. 2018. The biology of CRISPR-Cas: backward and forward. Cell 172:1239–1259. doi: 10.1016/j.cell.2017.11.032. [DOI] [PubMed] [Google Scholar]
- 15.Shmakov S, Smargon A, Scott D, Cox D, Pyzocha N, Yan W, Abudayyeh OO, Gootenberg JS, Makarova KS, Wolf YI, Severinov K, Zhang F, Koonin EV. 2017. Diversity and evolution of class 2 CRISPR-Cas systems. Nat Rev Microbiol 15:169–182. doi: 10.1038/nrmicro.2016.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsu PD, Lander ES, Zhang F. 2014. Development and applications of CRISPR-Cas9 for genome engineering. Cell 157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barrangou R, Doudna JA. 2016. Applications of CRISPR technologies in research and beyond. Nat Biotechnol 34:933–941. doi: 10.1038/nbt.3659. [DOI] [PubMed] [Google Scholar]
- 18.Luo ML, Leenay RT, Beisel CL. 2016. Current and future prospects for CRISPR-based tools in bacteria. Biotechnol Bioeng 113:930–943. doi: 10.1002/bit.25851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choi KR, Lee SY. 2016. CRISPR technologies for bacterial systems: current achievements and future directions. Biotechnol Adv 34:1180–1209. doi: 10.1016/j.biotechadv.2016.08.002. [DOI] [PubMed] [Google Scholar]
- 20.Jiang WY, Bikard D, Cox D, Zhang F, Marraffini LA. 2013. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol 31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jakociunas T, Jensen MK, Keasling JD. 2016. CRISPR/Cas9 advances engineering of microbial cell factories. Metab Eng 34:44–59. doi: 10.1016/j.ymben.2015.12.003. [DOI] [PubMed] [Google Scholar]
- 22.Zhang MZM, Wong FT, Wang YJ, Luo SW, Lim YH, Heng E, Yeo WL, Cobb RE, Enghiad B, Ang EL, Zhao HM. 2017. CRISPR-Cas9 strategy for activation of silent Streptomyces biosynthetic gene clusters. Nat Chem Biol 13:607–609. doi: 10.1038/nchembio.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jia H, Zhang L, Wang T, Han J, Tang H. 2017. Development of a CRISPR/Cas9-mediated gene-editing tool in Streptomyces rimosus. Microbiology 163:1148–1155. doi: 10.1099/mic.0.000501. [DOI] [PubMed] [Google Scholar]
- 24.Zeng H, Wen SS, Xu W, He ZR, Zhai GF, Liu YK, Deng ZX, Sun YH. 2015. Highly efficient editing of the actinorhodin polyketide chain length factor gene in Streptomyces coelicolor M145 using CRISPR/Cas9-CodA(sm) combined system. Appl Microbiol Biotechnol 99:10575–10585. doi: 10.1007/s00253-015-6931-4. [DOI] [PubMed] [Google Scholar]
- 25.Tong YJ, Charusanti P, Zhang LX, Weber T, Lee SY. 2015. CRISPR-Cas9 based engineering of actinomycetal genomes. ACS Synth Biol 4:1020–1029. doi: 10.1021/acssynbio.5b00038. [DOI] [PubMed] [Google Scholar]
- 26.Huang H, Zheng GS, Jiang WH, Hu HF, Lu YH. 2015. One-step high-efficiency CRISPR/Cas9-mediated genome editing in Streptomyces. Acta Biochim Biophys Sin (Shanghai) 47:231–243. doi: 10.1093/abbs/gmv007. [DOI] [PubMed] [Google Scholar]
- 27.Cobb RE, Wang YJ, Zhao HM. 2015. High-efficiency multiplex genome editing of Streptomyces species using an engineered CRISPR/Cas system. ACS Synth Biol 4:723–728. doi: 10.1021/sb500351f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cong L, Ran FA, Cox D, Lin SL, Barretto R, Habib N, Hsu PD, Wu XB, Jiang WY, Marraffini LA, Zhang F. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cho JS, Choi KR, Prabowo CPS, Shin JH, Yang D, Jang J, Lee SY. 2017. CRISPR/Cas9-coupled recombineering for metabolic engineering of Corynebacterium glutamicum. Metab Eng 42:157–167. doi: 10.1016/j.ymben.2017.06.010. [DOI] [PubMed] [Google Scholar]
- 31.Jiang Y, Qian FH, Yang JJ, Liu YM, Dong F, Xu CM, Sun BB, Chen B, Xu XS, Li Y, Wang RX, Yang S. 2017. CRISPR-Cpf1 assisted genome editing of Corynebacterium glutamicum. Nat Commun 8:15179. doi: 10.1038/ncomms15179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zaidi SSEA, Mahfouz MM, Mansoor S. 2017. CRISPR-Cpf1: A new tool for plant genome editing. Trends Plant Sci 22:550–553. doi: 10.1016/j.tplants.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 33.Yan MY, Yan HQ, Ren GX, Zhao JP, Guo XP, Sun YC. 2017. CRISPR-Cas12a-assisted recombineering in bacteria. Appl Environ Microbiol 83:e00947-. doi: 10.1128/AEM.00947-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A, Koonin EV, Zhang F. 2015. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zetsche B, Heidenreich M, Mohanraju P, Fedorova I, Kneppers J, DeGennaro EM, Winblad N, Choudhury SR, Abudayyeh OO, Gootenberg JS, Wu WY, Scott DA, Severinov K, van der Oost J, Zhang F. 2017. Multiplex gene editing by CRISPR-Cpf1 using a single crRNA array. Nat Biotechnol 35:31–34. doi: 10.1038/nbt.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Swiat MA, Dashko S, den Ridder M, Wijsman M, van der Oost J, Daran JM, Daran-Lapujade P. 2017. FnCpf1: a novel and efficient genome editing tool for Saccharomyces cerevisiae. Nucleic Acids Res 45:12585–12598. doi: 10.1093/nar/gkx1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yan WX, Mirzazadeh R, Garnerone S, Scott D, Schneider MW, Kallas T, Custodio J, Wernersson E, Li YQ, Gao LY, Federova Y, Zetsche B, Zhang F, Bienko M, Crosetto N. 2017. BLISS is a versatile and quantitative method for genome-wide profiling of DNA double-strand breaks. Nat Commun 8:15058. doi: 10.1038/ncomms15058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kleinstiver BP, Tsai SQ, Prew MS, Nguyen NT, Welch MM, Lopez JM, McCaw ZR, Aryee MJ, Joung JK. 2016. Genome-wide specificities of CRISPR-Cas Cpf1 nucleases in human cells. Nat Biotechnol 34:869–874. doi: 10.1038/nbt.3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao P, Yang H, Rajashankar KR, Huang Z, Patel DJ. 2016. Type V CRISPR-Cas Cpf1 endonuclease employs a unique mechanism for crRNA-mediated target DNA recognition. Cell Res 26:901–913. doi: 10.1038/cr.2016.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fonfara I, Richter H, Bratovic M, Le Rhun A, Charpentier E. 2016. The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature 532:517–521. doi: 10.1038/nature17945. [DOI] [PubMed] [Google Scholar]
- 41.Wang MG, Mao YF, Lu YM, Tao XP, Zhu JK. 2017. Multiplex gene editing in rice using the CRISPR-Cpf1 system. Mol Plant 10:1011–1013. doi: 10.1016/j.molp.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 42.Zhang X, Wang J, Cheng Q, Zheng X, Zhao G. 2017. Multiplex gene regulation by CRISPR-ddCpf1. Cell Discov 3:17018. doi: 10.1038/celldisc.2017.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tak YE, Kleinstiver BP, Nunez JK, Hsu JY, Horng JE, Gong JY, Weissman JS, Joung JK. 2017. Inducible and multiplex gene regulation using CRISPR-Cpf1-based transcription factors. Nat Methods 14:1163–1166. doi: 10.1038/nmeth.4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ungerer J, Pakrasi HB. 2016. Cpf1 is a versatile tool for CRISPR genome editing across diverse species of Cyanobacteria. Sci Rep 6:39681. doi: 10.1038/srep39681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang WS, Li X, Wang J, Xiang SH, Feng XZ, Yang KQ. 2013. An engineered strong promoter for Streptomycetes. Appl Environ Microbiol 79:4484–4492. doi: 10.1128/AEM.00985-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kieser T, Bibb MJ, Butter MJ, Chater KF, Hopwood DA. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom. [Google Scholar]
- 47.Leenay RT, Maksimchuk KR, Slotkowski RA, Agrawal RN, Gomaa AA, Briner AE, Barrangou R, Beisel CL. 2016. Identifying and visualizing functional PAM diversity across CRISPR-Cas systems. Mol Cell 62:137–147. doi: 10.1016/j.molcel.2016.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lei C, Li SY, Liu JK, Zheng X, Zhao GP, Wang J. 2017. The CCTL (Cpf1-assisted Cutting and Taq DNA ligase-assisted Ligation) method for efficient editing of large DNA constructs in vitro. Nucleic Acids Res 45:e74. doi: 10.1093/nar/gkw1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Komor AC, Badran AH, Liu DR. 2017. CRISPR-based technologies for the manipulation of eukaryotic genomes. Cell 168:20–36. doi: 10.1016/j.cell.2016.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shuman S, Glickman MS. 2007. Bacterial DNA repair by non-homologous end joining. Nat Rev Microbiol 5:852–861. doi: 10.1038/nrmicro1768. [DOI] [PubMed] [Google Scholar]
- 51.Bowater R, Doherty AJ. 2006. Making ends meet: repairing breaks in bacterial DNA by non-homologous end-joining. PLoS Genet 2:93–99. doi: 10.1371/journal.pgen.0020008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zheng X, Li SY, Zhao GP, Wang J. 2017. An efficient system for deletion of large DNA fragments in Escherichia coli via introduction of both Cas9 and the non-homologous end joining system from Mycobacterium smegmatis. Biochem Biophys Res Commun 485:768–774. doi: 10.1016/j.bbrc.2017.02.129. [DOI] [PubMed] [Google Scholar]
- 53.Shao ZY, Rao GD, Li C, Abil Z, Luo YZ, Zhao HM. 2013. Refactoring the silent spectinabilin gene cluster using a plug-and-play scaffold. ACS Synth Biol 2:662–669. doi: 10.1021/sb400058n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Da Silva NA, Srikrishnan S. 2012. Introduction and expression of genes for metabolic engineering applications in Saccharomyces cerevisiae. FEMS Yeast Res 12:197–214. doi: 10.1111/j.1567-1364.2011.00769.x. [DOI] [PubMed] [Google Scholar]
- 55.Tyo KEJ, Ajikumar PK, Stephanopoulos G. 2009. Stabilized gene duplication enables long-term selection-free heterologous pathway expression. Nat Biotechnol 27:760-U115. doi: 10.1038/nbt.1555. [DOI] [PubMed] [Google Scholar]
- 56.Tang X, Lowder LG, Zhang T, Malzahn AA, Zheng X, Voytas DF, Zhong Z, Chen Y, Ren Q, Li Q, Kirkland ER, Zhang Y, Qi Y. 2017. A CRISPR-Cpf1 system for efficient genome editing and transcriptional repression in plants. Nat Plants 3:17103. doi: 10.1038/nplants.2017.103. [DOI] [PubMed] [Google Scholar]
- 57.Bai CX, Zhang Y, Zhao XJ, Hu YL, Xiang SH, Miao J, Lou CB, Zhang LX. 2015. Exploiting a precise design of universal synthetic modular regulatory elements to unlock the microbial natural products in Streptomyces. Proc Natl Acad Sci U S A 112:12181–12186. doi: 10.1073/pnas.1511027112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Galm U, Wang LY, Wendt-Pienkowski E, Yang RY, Liu W, Tao MF, Coughlin JM, Shen B. 2008. In vivo manipulation of the bleomycin biosynthetic gene cluster in Streptomyces verticillus ATCC15003 revealing new insights into its biosynthetic pathway. J Biol Chem 283:28236–28245. doi: 10.1074/jbc.M804971200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lewis KM, Ke AL. 2017. Building the class 2 CRISPR-Cas arsenal. Mol Cell 65:377–379. doi: 10.1016/j.molcel.2017.01.024. [DOI] [PubMed] [Google Scholar]
- 60.Baltz RH. 2012. Streptomyces temperate bacteriophage integration systems for stable genetic engineering of actinomycetes (and other organisms). J Ind Microbiol Biotechnol 39:661–672. doi: 10.1007/s10295-011-1069-6. [DOI] [PubMed] [Google Scholar]
- 61.Kim SK, Kim H, Ahn WC, Park KH, Woo EJ, Lee DH, Lee SG. 2017. Efficient transcriptional gene repression by type V-A CRISPR-Cpf1 from Eubacterium eligens. ACS Synth Biol 6:1273–1282. doi: 10.1021/acssynbio.6b00368. [DOI] [PubMed] [Google Scholar]
- 62.Huang J, Shi J, Molle V, Sohlberg B, Weaver D, Bibb MJ, Karoonuthaisiri N, Lih CJ, Kao CM, Buttner MJ, Cohen SN. 2005. Cross-regulation among disparate antibiotic biosynthetic pathways of Streptomyces coelicolor. Mol Microbiol 58:1276–1287. doi: 10.1111/j.1365-2958.2005.04879.x. [DOI] [PubMed] [Google Scholar]
- 63.Liu G, Chater KF, Chandra G, Niu GQ, Tan HR. 2013. Molecular regulation of antibiotic biosynthesis in Streptomyces. Microbiol Mol Biol Rev 77:112–143. doi: 10.1128/MMBR.00054-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu J, Wang Y, Lu YJ, Zheng P, Sun JB, Ma YH. 2017. Development of a CRISPR/Cas9 genome editing toolbox for Corynebacterium glutamicum. Microb Cell Fact 16:205. doi: 10.1186/s12934-017-0815-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cleto S, Jensen JVK, Wendisch VF, Lu TK. 2016. Corynebacterium glutamicum metabolic engineering with CRISPR interference (CRISPRi). ACS Synth Biol 5:375–385. doi: 10.1021/acssynbio.5b00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Myronovskyi M, Luzhetskyy A. 2016. Native and engineered promoters in natural product discovery. Nat Prod Rep 33:1006–1019. doi: 10.1039/C6NP00002A. [DOI] [PubMed] [Google Scholar]
- 67.Baltz RH. 2010. Streptomyces and Saccharopolyspora hosts for heterologous expression of secondary metabolite gene clusters. J Ind Microbiol Biotechnol 37:759–772. doi: 10.1007/s10295-010-0730-9. [DOI] [PubMed] [Google Scholar]
- 68.Li L, Zheng GS, Chen J, Ge M, Jiang WH, Lu YH. 2017. Multiplexed site-specific genome engineering for overproducing bioactive secondary metabolites in actinomycetes. Metab Eng 40:80–92. doi: 10.1016/j.ymben.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 69.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 70.Wang R, Mast Y, Wang J, Zhang WW, Zhao GP, Wohlleben W, Lu YH, Jiang WH. 2013. Identification of two-component system AfsQ1/Q2 regulon and its cross-regulation with GlnR in Streptomyces coelicolor. Mol Microbiol 87:30–48. doi: 10.1111/mmi.12080. [DOI] [PubMed] [Google Scholar]
- 71.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.