The worldwide proliferation of life-threatening metallo-β-lactamase (MBL)-producing Gram-negative bacteria is a serious concern to public health. MBLs are compromising the therapeutic efficacies of β-lactams, particularly carbapenems, which are last-resort antibiotics indicated for various multidrug-resistant bacterial infections.
KEYWORDS: β-lactamase, metallo-β-lactamase, metallo-β-lactamase inhibitors, Gram-negative bacteria, antibiotic resistance, β-lactam antibiotics
ABSTRACT
The worldwide proliferation of life-threatening metallo-β-lactamase (MBL)-producing Gram-negative bacteria is a serious concern to public health. MBLs are compromising the therapeutic efficacies of β-lactams, particularly carbapenems, which are last-resort antibiotics indicated for various multidrug-resistant bacterial infections. Inhibition of enzymes mediating antibiotic resistance in bacteria is one of the major promising means for overcoming bacterial resistance. Compounds having potential MBL-inhibitory activity have been reported, but none are currently under clinical trials. The need for developing safe and efficient MBL inhibitors (MBLIs) is obvious, particularly with the continuous spread of MBLs worldwide. In this review, the emergence and escalation of MBLs in Gram-negative bacteria are discussed. The relationships between different class B β-lactamases identified up to 2017 are represented by a phylogenetic tree and summarized. In addition, approved and/or clinical-phase serine β-lactamase inhibitors are recapitulated to reflect the successful advances made in developing class A β-lactamase inhibitors. Reported MBLIs, their inhibitory properties, and their purported modes of inhibition are delineated. Insights into structural variations of MBLs and the challenges involved in developing potent MBLIs are also elucidated and discussed. Currently, natural products and MBL-resistant β-lactam analogues are the most promising agents that can become clinically efficient MBLIs. A deeper comprehension of the mechanisms of action and activity spectra of the various MBLs and their inhibitors will serve as a bedrock for further investigations that can result in clinically useful MBLIs to curb this global menace.
INTRODUCTION
The alarming spread of antimicrobial resistance (AMR) presents a major challenge to public health worldwide (1, 2). The dissemination of AMR is spearheaded by increasing world trade, rising human and animal populations, economic factors, changing climatic conditions, and air travel, which are breaking down geographical borders between countries and continents, exposing humans and animals to diverse kinds of infections (3). The immense clinical benefits obtained from antimicrobials in the management of infectious diseases were eroded by the emergence of AMR in pathogens a few years after their introduction and adoption into clinical medicine (4, 5). As new antibiotics have been discovered and introduced into clinical use, a similar resistance cycle has ensued: resistance to cephalosporins due to the expression of extended-spectrum β-lactamases (ESBLs), carbapenem resistance due to carbapenemases, resistance to colistin due to the plasmid-mediated mcr-1 gene and chromosomal mutations, and tigecycline resistance due to chromosomal mutations (6, 7).
Gram-negative bacteria develop resistance to β-lactams through different mechanisms (8), including the production of enzymes called β-lactamases that hydrolyze the β-lactam ring. Resistance can also be developed through modification(s) of the normal penicillin binding proteins (PBPs), reduced porin expression leading to impermeability of the outer membrane, and active antibiotic expulsion from the bacteria through efflux pump systems. The presence of one or more of these mechanisms in microorganisms can lead to β-lactam resistance (8–11).
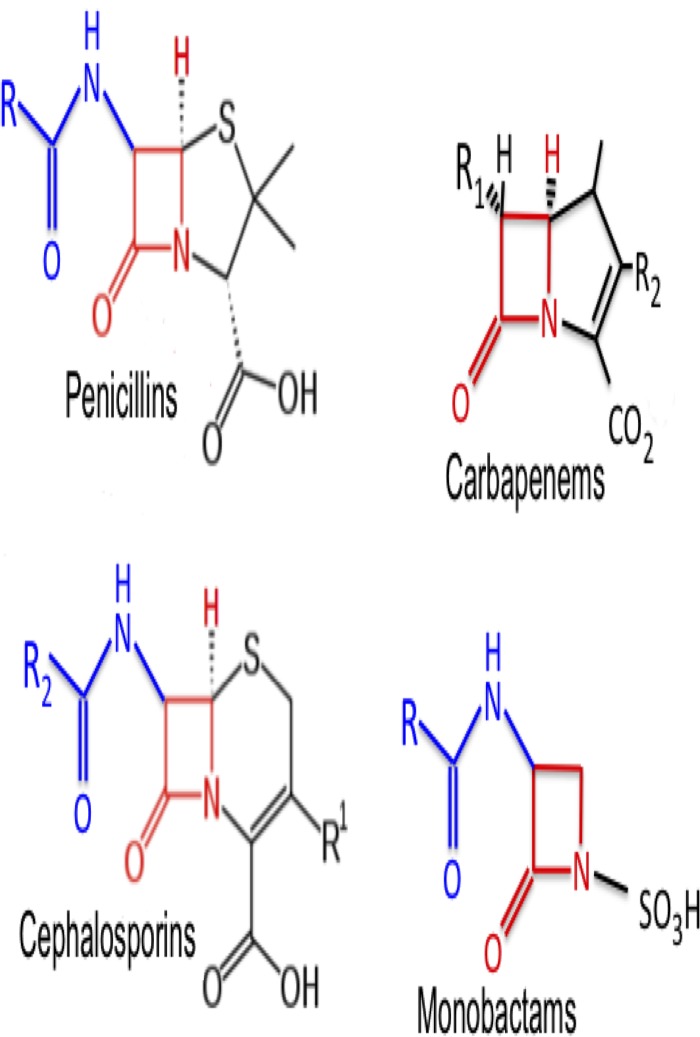
Bacterial β-lactamases are members of an enzyme family capable of impairing the efficacy of β-lactam antibiotics such as penicillins, cephalosporins, monobactams, and carbapenems (Fig. 1) (12, 13) by hydrolyzing their β-lactam rings (14). There are two globally accepted classification schemes for β-lactamases. The first scheme is based on the amino acid sequence of the enzyme and comprises four classes, i.e., classes A, B, C, and D. The second scheme is based on the enzyme's functionality or substrate and includes three major groups: group 1, cephalosporinases (class C); group 2, serine β-lactamases (SBLs) (classes A and D); and group 3, metallo-β-lactamases (class B). Each of these groups is further subdivided into several different subgroups (15, 16). Class A, C, and D β-lactamases possess the amino acid serine at their active site and are thus known as serine β-lactamases (SBLs), while Ambler class B β-lactamases contain one or two zinc ions at their active site and are thus called metallo-β-lactamases (MBLs).
FIG 1.
Structures of selected β-lactam antibiotics (penicillins, cephalosporins, monobactams, and carbapenems). Images were adapted from references 12 and 13.
The production of class B β-lactamases in bacteria, together with other resistance mechanisms, has narrowed down treatment options for infections caused by these pathogens (17). One approach for treating β-lactamase-producing bacterial infections is by combining existing β-lactam antibiotics with β-lactamase inhibitors (18). β-lactam/β-lactamase inhibitor combinations resulted in the restoration of the activity of β-lactam antibiotics against β-lactamase-producing bacteria. Such combination drugs that are currently approved by the FDA and available for clinical treatment (18) do not include Ambler class B (metallo-β-lactamase) inhibitors (MBLIs). There is thus far no MBLI in clinical use, which indicates that the presence of this class of β-lactamases in resistant bacteria needs much attention. Class B β-lactamases (MBLs) include plasmid-encoded enzymes such as imipenemase enzyme (IMP), Verona integron-encoded metallo-β-lactamase (VIM), Sao Paulo metallo-β-lactamase (SPM), German imipenemase (GIM), Seoul imipenemase (SIM), New Delhi metallo-β-lactamase (NDM), and Dutch imipenemase (DIM), which are being detected in Gram-negative bacteria increasingly and with considerable clinical impact (19).
Invariably, the increasing worldwide prevalence of MBLs is a major threat to global health care, affecting both community and hospital settings (20–23). Mortality rates of up to 50% have been reported for carbapenem-resistant Enterobacteriaceae (CRE), including MBL-positive infections (24, 25). Over the last decade, the prevalence of these drug-resistant bacterial infections has been increasing, affecting over a million patients worldwide. This has led to prolonged hospitalization, longer terms of disability, increased health care-associated costs, higher mortalities, etc. (23, 26, 27). The World Health Organization (WHO) recently listed CRE pathogens as “critical priority pathogens” for which novel and efficient antibiotics are required urgently (28).
The absence of clinical MBLIs for MBL-mediated drug-resistant infections, particularly those involving CREs, makes this class of β-lactamases especially important in infectious diseases. Hence, this review aims to map out advances made so far in finding MBLIs that can fight MBL producers and to point out challenges involved in designing clinically useful MBLIs.
METALLO-β-LACTAMASES
Metallo-β-lactamases (MBLs) confer resistance to all β-lactams except monobactams by using their active-site zinc ions to activate a nucleophilic water molecule, which opens the β-lactam ring (through hydrolysis) and renders it ineffective (29). For many decades, MBLs were considered to be clinically irrelevant enzymes that were chromosomally encoded in nonpathogenic organisms (30–33). This situation has changed with the increasing spread of NDM-, VIM-, and IMP-mediated AMR in Gram-negative pathogens, including Enterobacteriaceae. The above-mentioned enzymes are carried on chromosomes or plasmids and located on mobile genetic cassettes inserted into integrons (IMP and VIM) and/or bracketed by composite transposons (NDM) (34, 35). Transposons are DNA segments capable of moving to different positions in the genome of a single cell, while integrons are key elements that can shuttle genes between integrons on plasmids, therefore allowing the plasmids to transfer genetic material to different bacteria (36). These mechanisms mediate the spread of MBL-mediated resistance worldwide. A typical example is the dissemination of the NDM-1 MBL, first isolated from a Swedish patient transferred from India (37) in 2008, worldwide in diverse Enterobacteriaceae species. The incidence rate continues to grow (37).
MBLs are divided into three subclasses, namely, B1, B2, and B3 (Table 1), based on differences in their primary zinc coordination shell and their amino acid sequence. Sequence identity between subclass B1 and B2 enzymes ranges from 14 to 24%, while sequence identity between subclass B3 and both subclass B1 and B2 enzymes ranges from 2% to 14% (38). Subclass B1 possesses a binuclear active site, within which either one or two zinc ions can exist. B1 binds one zinc ion (Zn1) with three histidine residues (H116, H118, and H196) and a second zinc ion (Zn2) with three different residues, including a cysteine (D120, C221, and H263) in particular. Subclass B1 is a clinically relevant and notorious MBL subclass, comprising the largest number of MBLs located on plasmids. Thus, it is more likely to spread to other organisms to cause β-lactam-resistant clinical infections (39).
TABLE 1.
Historical timeline and characteristics of metallo-β-lactamases discovered up to December 2017
| MBL subclass and enzyme | Yr detected | Species in which first detected | No. of variants | Country(ies) of origin | β-Lactam antibiotic hydrolysis profile | Genetic location(s) | Accession no. | Reference(s) |
|---|---|---|---|---|---|---|---|---|
| B1 | ||||||||
| BcII | 1966 | Bacillus cereus | 7 | Uruguay | Broad spectrum β-lactams | Chromosome | M11189 | 32, 33, 89, 90 |
| CcrA or CfiA | 1990 | Bacteroides fragilis | 25 | England | Broad-spectrum β-lactams | Chromosome | AB087225 | 91, 92 |
| IMP | 1994 | Serratia marcescens | 67 | Japan | Broad-spectrum β-lactams | Plasmid, chromosome | HM036079 | 93, 94 |
| BlaB | 1998 | Elizabethkingia meningoseptica | 15 | —a | Broad-spectrum β-lactams | Chromosome | AF189298 | 95, 96, 97 |
| VIM | 1999 | Pseudomonas aeruginosa | 54 | Italy | Broad-spectrum β-lactams | Plasmid, chromosome | GU724868 | 94, 98 |
| IND | 1999 | Chryseobacterium indologenes | 16 | France | Broad-spectrum β-lactams | Plasmid, chromosome | EF394436 | 99 |
| EBR | 2002 | Empedobacter brevis | 1 | France | Narrow-spectrum cephalosporins | Chromosome | AF416700 | 100 |
| SPM | 2002 | P. aeruginosa | 1 | Brazil | Broad-spectrum β-lactams | Plasmid, chromosome | GU831565 | 101, 102 |
| Bla | 2003 | Bacillus anthracis | 2 | — | Broad-spectrum β-lactams | Chromosome | X62244 | 71 |
| GIM | 2004 | P. aeruginosa | 2 | Germany | Broad-spectrum β-lactams | Plasmid | JF414726 | 103 |
| SIM | 2005 | Acinetobacter baumannii | 2 | Korea | Broad-array β-lactams, narrow carbapenem | Plasmid, chromosome | GQ288397 | 104 |
| SLB | 2005 | Shewanella livingstonensis | 1 | Islands | Broad spectrum | Chromosome | AY590118 | 105 |
| SFB | 2005 | Shewanella frigidimarina | 1 | Islands | Narrow spectrum | Chromosome | AY590119 | 105 |
| PEDO-3 | 2015 | Pedobacter kyungheensis | 1 | United Kingdom | Broad-spectrum β-lactams | Chromosome | NG_049959 | 106 |
| JOHN | 2003 | Flavobacterium johnsoniae | 1 | — | Broad-spectrum β-lactams | Chromosome | AY02846 | 107 |
| CGB | 2002 | Chryseobacterium gleum | 1 | — | Broad-spectrum β-lactams | Chromosome | EF672680 | 108 |
| MUS | 2002 | Myroides odoratimimus | 2 | — | Broad spectrum, exception of aztreonam | Chromosome | AF441286 | 109, 110 |
| TUS | 2002 | Myroides odoratus | 1 | — | Large spectrum, exception of aztreonam | Chromosome | AF441287 | 109 |
| KHM | 2008 | Citrobacter freundii | 1 | Japan | Broad-spectrum β-lactam | Plasmid | AB364006 | 111 |
| DIM | 2010 | Pseudomonas stutzeri | 1 | Netherlands | Broad spectrum, exception of aztreonam | Plasmid | GU323019 | 112 |
| NDM | 2008 | Klebsiella pneumonia, Escherichia coli | 18 | India | Broad-spectrum β-lactams | Plasmid, chromosome | JQ080305 | 37, 113, 114, 115 |
| HMB | 2017 | P. aeruginosa | 1 | Germany | Broad-spectrum β-lactams | Chromosome | NG_052225 | 116 |
| FIM | 2012 | P. aeruginosa | 1 | Italy | Broad-spectrum β-lactams | Chromosome | JX570731 | 117 |
| MOC | — | M. odoratus | 1 | — | Broad-spectrum β-lactams | Chromosome | KX371616 | Unpublished |
| TMB | 2012 | Achromobacter sp. | 2 | Libya | Broad-spectrum β-lactams | Chromosome | FR771847 | 118, 119 |
| GRD23 | 2016 | Proteobacteria | 1 | Denmark | Broad-spectrum β-lactams | Plasmid | KU167043 | 120 |
| SPN79 | 2016 | Proteobacteria | 1 | Spain | Broad-spectrum β-lactams | Unknown | KU167036 | 120 |
| VarG | 2017 | Vibrio cholerae | 1 | — | Large spectrum β-lactams | Chromosome | AAF94716 | 121 |
| B2 | ||||||||
| CphA | 1990 | Aeromonas hydrophila | 17 | Italy | Narrow spectrum, more strictly carbapenems | Chromosome | AY261378 | 63, 122, 123 |
| ImiS | 1996 | Aeromonas veronii | 1 | United Kingdom | Narrow spectrum, more strictly carbapenems | Chromosome | Y10415 | 124, 125 |
| ImiH | 2003 | A. hydrophila | 1 | Australia | Narrow spectrum, more strictly carbapenems | Chromosome | AJ548797 | 126 |
| CEPH-A3 | — | A. veronii | 1 | — | Narrow spectrum, more strictly carbapenems | — | AY112998 | Unpublished |
| Sfh | 2003 | Serratia fonticola | 1 | Portugal | Narrow spectrum, more strictly carbapenems | Chromosome | AF197943 | 127 |
| B3 | ||||||||
| L-1 | 1982 | Stenotrophomonas maltophilia | 19 | — | Broad-spectrum β-lactams | Chromosome | AJ272109 | 128, 129, 130 |
| FEZ | 2000 | Legionella gormanii | 1 | — | Broad-spectrum β-lactams | Chromosome | Y17896 | 131 |
| GOB | 2000 | E. meningoseptica | 19 | France | Broad-spectrum β-lactams | Chromosome | AF090141 | 97, 132 |
| THIN-B | 2001 | Janthinobacterium lividum | 1 | Italy | Broad-spectrum β-lactams | Chromosome | AJ250876 | 133 |
| MBL1b | 2001 | Caulobacter crescentus | 1 | — | Broad-spectrum β-lactams | Chromosome | AJ315850 | 134 |
| CAU | 2002 | Caulobacter vibrioides | 2 | — | Broad-spectrum β-lactams | Chromosome | AJ308331 | 135 |
| BJP | 2006 | Bradyrhizobium japonicum | 1 | — | Broad-spectrum β-lactams | Chromosome | NP_772870 | 136 |
| AIM | 2012 | P. aeruginosa | 1 | Australia | Large spectrum, including aztreonam | Plasmid | AM998375 | 137, 138 |
| CAR | 2008 | Erwinia carotovora | 1 | — | Broad-spectrum β-lactams | Chromosome | NC_004547 | 139 |
| POM | 2011 | Pseudomonas otitidis | 1 | — | Broad-spectrum β-lactams | Chromosome | EU315252 | 140, 141 |
| PAM | 2013 | Pseudomonas alcaligenes | 1 | Japan | Broad-spectrum β-lactams | Chromosome | AB858498 | 142 |
| PEDO | 2015 | Pedobacter roseu, Pedobacter borealis | 2 | Denmark, Norway | Broad-spectrum β-lactams | Chromosome | NG_049957 | 106 |
| CPS | 2015 | Chryseobacterium piscium | 1 | United Kingdom | Broad-spectrum β-lactams | Chromosome | NG_048587 | 106, 143 |
| ESP | 2015 | Eristalis tenax | 2 | United Kingdom | Broad-spectrum β-lactams | Chromosome | NG_049088 | 106 |
| MSI | 2015 | Massilia oculi | 1 | Spain | Broad-spectrum β-lactams | Chromosome | NG_049323 | 106 |
| SPG | 2015 | Sphingomonas sp. | 1 | Norway | Broad-spectrum β-lactams | Chromosome | NG_050139 | 106 |
| Rm3 | 2016 | From soil sample | 1 | United Kingdom | Broad-spectrum β-lactams | — | KF485393 | 144 |
| LRA | 2009 | J. lividum | 8 | United States (Alaska) | Broad-spectrum β-lactams | — | EU408347 | 145 |
| SMB | 2011 | S. marcescens | 1 | Japan | Broad-spectrum β-lactams | Chromosome | AB636283 | 146 |
| GRD33 | 2016 | Gemmatimona | 1 | Denmark | Broad-spectrum β-lactams | Chromosome | KU167042 | 120 |
| CRD3 | 2016 | Erythrobacter | 1 | Denmark | Broad-spectrum β-lactams | Chromosome | KU167037 | 120 |
| DHT2 | 2016 | S. maltophilia | 1 | Germany | Broad-spectrum β-lactams | Chromosome | KU167035 | 120 |
| ALG6 | 2016 | J. lividum | 1 | Algeria | Broad-spectrum β-lactams | Chromosome | KU167038 | 120 |
| ALG11 | 2016 | Burkholderia | 1 | Algeria | Broad-spectrum β-lactams | — | KU167039 | 120 |
| EAM | 2012 | Erythrobacter aquimaris | 1 | France | Broad-spectrum β-lactams, specific for carbapenem | Chromosome | JN558591 | 147 |
| ECM | 2012 | Erythrobacter citreus | 1 | France | Broad spectrum, specific for carbapenem | Chromosome | JN558590 | 147 |
| EFM | 2012 | Erythrobacter flavus | 1 | France | Broad spectrum, specific for carbapenem | Chromosome | JN558587 | 147 |
| ELM | 2012 | Erythrobacter longus | 1 | France | Broad spectrum, specific for carbapenem | Chromosome | JN558589 | 147 |
| EVM | 2012 | Erythrobacter vulgaris | 1 | France | Broad spectrum, specific for carbapenem | Chromosome | JN558588 | 147 |
| SPR | 2014 | Serratia proteamaculans | 1 | United States | Broad spectrum, specific for carbapenem | — | CP000826 | Unpublished |
—, unpublished.
Subclass B2 MBLs have a Zn1 binding site with one altered residue (N116, H118, or H196), but retain a similar Zn2 site (D120, C221, H263); it selectively hydrolyzes carbapenems and possess the fewest members compared with the others. Subclass B3 MBLs have a varied Zn1 binding site (H/Q116, H118, or H196) and a distinctive Zn2 binding site that lacks a cysteine residue (D120, H121, or H263) (40). These structural variations in known and emerging MBLs offer a greater challenge to discovering potent MBLIs that can inhibit all MBLs. The historical timeline and characteristics of MBLs discovered up to 2017, retraced from different databases such as the Comprehensive Antibiotic Resistance Database (CARD) (41) and β-Lactamase Database (BLDB), are summarized in Table 1 (42).
MBLs were detected from diverse species of organisms, as shown in Table 1. However, subclass B2 was detected mostly from Aeromonas spp. Interestingly, most MBLs were initially identified in Europe (France, Germany, Denmark, the United Kingdom, Italy, etc.). Africa had the least discovered number of these MBLs, not because of their absence in the continent but rather because of a lack of proper awareness, skilled personnel, resistance diagnosis, and surveillance systems. Class B3 also contained the enzyme Adelaide imipenemase (AIM), which is plasmid borne (Table 1).
PHYLOGENETIC ANALYSIS AND METADATA OF MBLs
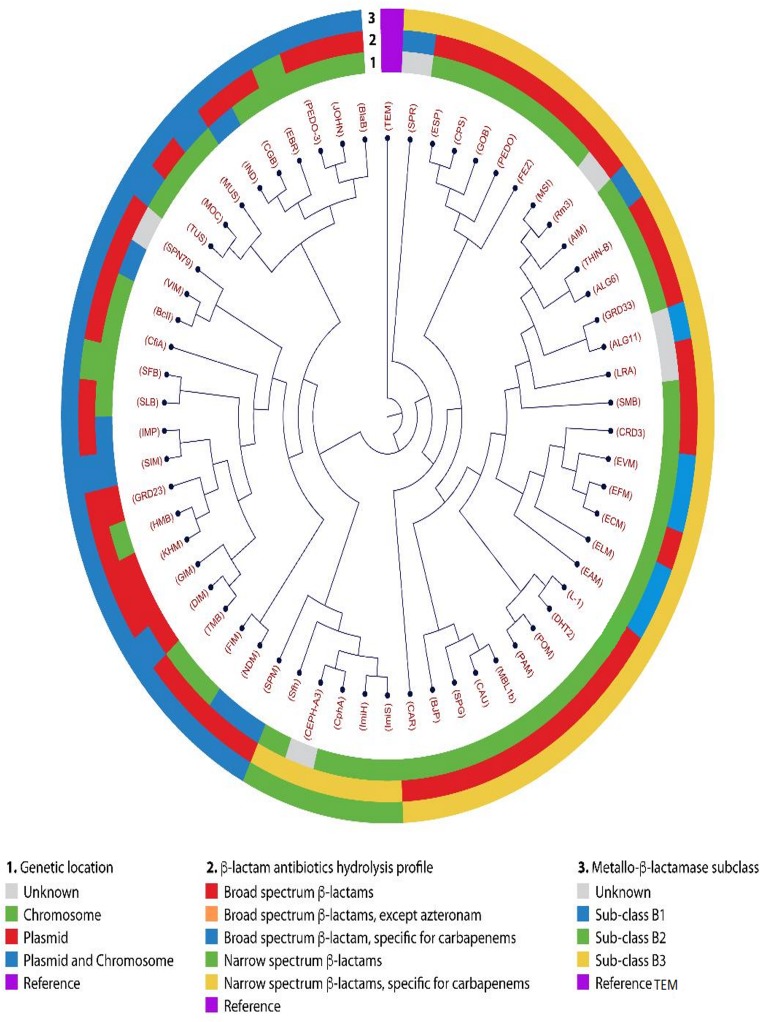
All the reported MBLs together with their available metadata are illustrated on a phylogenetic tree to demonstrate the associated characteristics of the different subclasses (B1, B2, and B3). Protein sequences of all reported MBLs (Table 1) were downloaded from GenBank using their accession numbers. They were then aligned using the default parameters/settings of classical sequence analysis with the CLC Genomics Workbench (version 10.1.1) software to generate an aligned file. The created aligned file was used to draw the maximum-likelihood phylogenetic tree to infer the evolutionary relationship using optimized parameters as follows: construction method, unweighted pair group method using average linkages (UPGMA); nucleotide substitution model, Jukes-Cantor; protein substitution model, WAG (198); transition/transversion ratio, 2; estimate substitution rate, yes; number of substitution rate, 4; perform bootstrap analysis, yes; replicates, 1,000. Metadata associated with the aligned sequences (Table 1) were imported to provide a comprehensive analysis of the generated phylogenetic tree. TEM-1 (Ambler class A β-lactamase) was used as the outgroup to root the tree to enable easy configuration of the phylogenetic distance between the MBL enzymes on the branches. The circular version of the tree is shown in Fig. 2.
FIG 2.
Graphical view of the phylogenetic and metadata of metallo-β-lactamases. The TEM-1 of Ambler class A β-lactamases was rooted and used as the outgroup in the tree. The patterns on the circular phylogram are as follows: 1, genetic location (inner layer); 2, β-lactam antibiotic hydrolysis profile (middle layer); and 3, metallo-β-lactamase (outer layer).
The phylogenetic analysis depicted a clear distinction between subclasses B1 and B3, as reported in the literature (43, 44), where they were more phylogenetically related to each other than to B2 (Fig. 2). Metadata analysis provided a deeper insight into the associated characteristics and distinctions between all the subclasses. Specifically, subclass B1 and B3 MBLs act on a broad spectrum of β-lactam antibiotics, while class B2 MBLs have a narrower β-lactam spectrum, affecting only carbapenems. This diversity further complicates the management of CRE infections in the clinical setting, making it a huge challenge for a single inhibitor to act efficiently on all MBL subgroups. The metadata also indicated that B2 and B3 were mostly chromosomal, while B1 was both plasmid and chromosomally mediated. Even though SPM was phylogenetically clustered with subclass B2, the metadata differentiated it into a distinct subgroup, as illustrated in Fig. 2, reiterating the importance of visualizing phylogenetic structures in relation to their metadata (45).
NOVEL MBLs RECENTLY DISCOVERED THROUGH GENOMICS AND METAGENOMICS
The advent of modern techniques such as next-generation and whole-genome sequencing as diagnostic tools in clinical microbiology has led to the discovery of novel genes and diverse mechanisms of drug resistance that could otherwise have been impossible to find using conventional diagnostic methods. Berglund and collaborators in 2017 (46) assumed that there is a large number of unexplored reservoirs of uncharacterized MBLs. Hence, with the aid of new computational methods based on hidden Markov models, they identified novel MBL genes of subclass B1 from bacterial genomes, plasmids, and metagenomic data. The findings predicted the existence of 76 novel genes of subclass B1. Based on the evolutionary origin of the genes, a phylogenetic analysis revealed that the subclass B1 could be differentiated into five groups (subclasses B1-1 to B1-5). However, these recently discovered novel MBLs are not listed in Table 1 due to the lack of proper nomenclature and very scant information or metadata (i.e., no information on country, bacteria from which the MBL was isolated, and genetic location).
These current findings by Berglund et al. (46) thus echo the diversity and proliferation of MBLs as well as their possible threat to clinically available antibiotic options, cautioning scientists to be prepared for the challenges that may occur in the future regarding the control and treatment of these life-threating pathogens. Therefore, there is an urgent need to find efficient and effective MBLIs to overcome this global scourge.
METALLO-β-LACTAMASE INHIBITORS
β-Lactam/β-lactamase inhibitor/adjuvant combinations have been developed and are in clinical use for the treatment of infections caused by SBLs, while others are in clinical trials (18) (Tables 2 and 3). These molecules fail to inhibit MBLs, thus urgently necessitating the development of inhibitors that target bacterial metalloenzymes.
TABLE 2.
Approved β-lactam/β-lactamase inhibitor combinations
| β-Lactam/β-lactamase inhibitor combination | Yr introduced into clinical use | Route of administration | Dosage regimen | Reference |
|---|---|---|---|---|
| Amoxicillin-clavulanate | 1981 | Oral | 20–40 mg/kg/day | 148 |
| Ticarcillin-clavulanate | 1985 | Intravenous | 80 mg/kg every 6–8 h | 149 |
| Ampicillin-sulbactam | 1997 | Intravenous | 100–200 mg/kg/day | 149 |
| Cefoperazone-sulbactam | Intravenous | 50–100 mg/kg/day | 150 | |
| Piperacillin-tazobactam | 1993 | Intravenous | 150–400 mg/kg/day | 151 |
| Ceftolozane-tazobactam | 2014 | Intravenous | 1 g/0.5 g every 8 h | 152 |
| Ceftazidime-avibactam | 2015 | Intravenous | 2.5-g infusion in 2 h every 8 h | 153 |
TABLE 3.
Clinical phases for β-lactam/β-lactamase inhibitor combinations
| β-Lactam/β-lactamase inhibitor combination | Clinical phase | Expected activity | Potential indicationsa | Reference(s) |
|---|---|---|---|---|
| Ceftaroline-avibactam | 2 | Active against Gram-negative ESKAPE and CDC pathogens | Bacterial infections | 154 |
| Aztreonam-avibactam | 2 | Active against Gram-negative ESKAPE and CDC pathogens | Complicated intra-abdominal infections | 155 |
| Imipenem-cilastatin-relebactam (MK-7655) | 3 | Active against Gram-negative ESKAPE and CDC pathogens | cUTIs, cIAIs, hospital-acquired and ventilator-associated bacterial pneumonia | 156 |
| Meropenem-vaborbactam (RPX-7009) (also known as carbavance) | New drug application submitted | Active against Gram-negative ESKAPE and CDC pathogens | cUTIs, hospital-acquired and ventilator-associated bacterial pneumonia, febrile neutropenia, bacteremia, infections caused by carbapenem-resistant Enterobacteriaceae | 157, 158 |
| Biapenem-vaborbactam (RPX-7009) | 1 | Active against Gram-negative ESKAPE and CDC pathogens | Anaerobic bacterial infection | 159 |
| ETX2514SUL | Active against Gram-negative ESKAPE pathogens | Infection caused by A. baumannii | 160 | |
| Cefepime-zidebactam | 1 | Active against Gram-negative ESKAPE pathogens, possible activity against CDC pathogens | cUTIs, hospital-acquired and ventilator-associated bacterial pneumonia | 161 |
cUTIs, complicated urinary tract infections; cIAIs, complicated intra-abdominal infections.
In areas with increasing proliferation of MBL-positive CREs, clavulanic acid, sulbactam, tazobactam, and avibactam, which are all serine-based β-lactamase inhibitors in clinical use (Table 2), do not provide much benefit (18). One approach to overcome the effects of MBLs is by designing specific inhibitors that can be coadministered with β-lactam antibiotics. Studies have been executed in this direction where a number of potent MBLIs have been identified, although none of these inhibitors has thus far reached clinical trials (47). The general challenge in identifying and developing broad-spectrum MBLIs stems from the structural and mechanistic differences within the three subclasses of MBLs. Reported MBLI groups include thiol and thioester derivatives (48), tetrazoles and hydroxamates (49, 50) sulfonic acid derivative s (51), β-lactam analogues derivatives (52), pyrroles-based inhibitors (53), pyridine dicarboxylates (54), peptides (55) natural products (56), nucleic acid (57), and metal chelators (58) (Table 4).
TABLE 4.
Types and characteristics of known metallo-β-lactamase inhibitors
| Metallo-β-lactamase inhibitor(s) | Source |
Ki (μM) |
IC50 (μM) |
Type of inhibition | Reference(s) | ||||
|---|---|---|---|---|---|---|---|---|---|
| B1 | B2 | B3 | B1 | B2 | B3 | ||||
| Carboxylates, tetrazoles, and hydroxamates | |||||||||
| Maleic acids | Synthetic | 0.4–120 | —a | — | 2.5–13 | — | — | Competitive | 67, 68 |
| Succinic acid | Synthetic | 3.3 | — | — | 0.0027–490 | — | 15–300 | Irreversible | 162, 163, 164 |
| Phthalic acid | Synthetic | — | — | — | 0.2–243 | — | — | — | 69, 70, 165 |
| N-Acylhydrazones | Synthetic | — | — | — | 1.02–24 | — | — | — | 166 |
| 2-Substituted 4,5-dihydrothiazole-4-carboxylic acid | Synthetic | 3.3–5.1 | — | — | 5–77 | — | — | — | 73 |
| 3-Mercapto-1,2,4-triazoles and N-acylated thiosemicarbazides | Synthetic | 11–75 | — | — | — | — | — | Mixed | 167 |
| N-Heterocyclic dicarboxylic acid derivatives | Synthetic | 0.6–6 | 3.5–7.1 | 0.7–2 | — | — | — | Mixed | 168 |
| Biphenyl tetrazoles | Synthetic | 0.6–1.6 | — | — | 0.3–860 | — | — | Competitive | 49, 169 |
| Hydroxamates (amino acid derived) | Synthetic | — | — | 6.1–18 | — | — | — | Competitive | 50, 170 |
| Phenazines | |||||||||
| SB 212021 | Streptomyces | — | — | — | 37 | — | 19 | Reversible | 171 |
| SB212305 | Streptomyces | — | — | — | 75 | — | 1 | Reversible | 171 |
| Trifluoromethyl ketones and alcohols | Synthetic | 30–1,000 | 6–217 | 1.5–5,000 | — | — | — | Irreversible for subclass B2 | 172 |
| Trifluoromethyl ketone 5b (3S) | Synthetic | 500 | 6 | 15 | — | — | — | Irreversible for subclass B2 | 172 |
| Trifluoromethyl ketone 5′a (3R) | Synthetic | 700 | 11 | 3 | — | — | — | Irreversible for subclass B2 | 172 |
| Trifluoromethyl ketone 5a (3S) | Synthetic | 300 | 44 | 1.5 | — | — | — | Irreversible for subclass B2 | 172 |
| Trifluoromethyl alcohol 4′b (2R, 3R), (2S, 3R) | Synthetic | 30 | 20 | >5,000 | — | — | — | Irreversible for subclass B2 | 172 |
| Trifluoromethyl alcohol 4b (2R, 3S), (2S, 3S) | Synthetic | 1,000 | 19 | >5,000 | — | — | — | Irreversible for subclass B2 | 172 |
| Trifluoromethyl alcohol 4′a (2R, 2R), (2S, 3R) | Synthetic | 700 | 217 | 35 | — | — | — | Irreversible for subclass B2 | 172 |
| β-Lactam analogues | |||||||||
| 1β-Methylcarbapenem | Synthetic | 0.0037–0.8 | — | 1 | <0.1–>10 | — | — | Reversible | 79 |
| Penicillin-derived inhibitors | Synthetic | — | — | — | 1.4–>200 | — | 0.1–>200 | — | 52 |
| Thioxo-cephalosporin derivatives | Synthetic | 29–720 | — | — | — | — | — | Competitive | 81, 173 |
| Cyclobutanone analogues of β-lactams | Synthetic | — | — | — | 122–1,000 | — | — | Reversible | 83 |
| β-Lactam substrates | Synthetic | 2,300 | — | — | — | — | — | Irreversible | 174, 175 |
| Peptides | |||||||||
| Cys-Val-His-Ser-Pro-Asn-Arg-Glu-Cys | Synthetic | — | — | 16/9 | — | — | — | Mixed | 176 |
| Homocysteinyl peptide | Synthetic | — | — | 0.0021–1 | — | — | — | Reversible competitive | 177 |
| Cysteinyl peptide | Synthetic | 3.0–1,000 | — | 0.9–3.7 | — | — | — | Reversible | 55 |
| Pyridine dicarboxylates | |||||||||
| 2-Picolinic acid | Synthetic | 54–95 | 5.7 | 29–62 | — | — | — | Competitive | 54 |
| Pyridine-2,4-dicarboxylic acid | Synthetic | 78–98 | 4.5 | 65–78 | — | — | — | Competitive | 54 |
| Pyridine monothiocarboxylic acid analogues | Natural products | — | — | — | 0.14–250 | — | 0.6–340 | Reversible | 86 |
| Natural products | |||||||||
| Flavonoids: galangin, quercetin | Plants | — | — | 18.5–185 | — | — | — | Irreversible | 76 |
| Tricyclic natural products: SB238569, SB236050, SB 236049 | Chaetomium fanicola | 17–88 | 3.4–15 | — | 0.7–256 | 2–29 | >1,000 | Competitive | 56 |
| Polyketides | Penicillium sp. | — | — | — | 88–95 | — | — | — | 77 |
| Triazoles and N-acylated thiosemicarbazides | |||||||||
| Sulfonyltriazole | Synthetic | 0.41–1.4 | — | — | 3.3–>56 | — | — | Competitive | 178 |
| NH-1,2,3-triazole inhibitors | Synthetic | 0.01–0.2 | — | — | 0.07–21 | — | — | Competitive and mixed | 179 |
| 4-Methyl-5-(trifluoromethyl)-4H-1,2,4-triazole-3-thiol | Synthetic | 970 | — | — | — | — | — | Competitive | 180 |
| 3-Mercapto-1,2,4-triazoles and N-acylated thiosemicarbazides | Synthetic | 11–75 | — | — | — | — | — | Mixed | 167 |
| Pyrrole derivatives | |||||||||
| Pyrrole-based inhibitors | Synthetic | 12–33 | — | — | — | — | — | Competitive | 53 |
| Tetrahydropyrimidine-2-thione and pyrrole derivatives | Synthetic | 19–235 | — | — | — | — | — | Competitive | 75 |
| Nucleic acids | |||||||||
| Single-stranded DNAs | DNA | 0.00031–0.0092 | — | — | 0.15–0.8 | — | — | Reversible and noncompetitive | 57, 181 |
| Double-stranded DNAs | DNA | 0.1–0.103 | — | — | 0.01–0.02 | — | — | Reversible | 57 |
| DNA nanoribbon | DNA | — | — | — | 3.3–40 | 11.7 | 5.7 | Nonintercalative binding | 182 |
| Sulfonic acid derivatives | |||||||||
| Bulcegin A | Microorganisms | 230 | — | 2.5 | — | — | — | Competitive | 183 |
| 4-Morpholinoethanesulfonic acid | Synthesis | 23,000 | — | — | — | — | — | Competitive | 51 |
| N-Arylsulfonyl hydrazones | Synthesis | 0.7–6.6 | — | — | 1.6–150 | — | — | Reversible competitive | 184 |
| Metal chelators | |||||||||
| Ca-EDTA | Synthesis | — | — | — | 28 | — | — | Noncompetitive | 73 |
| Aspergilomarasamine A | Aspergillus versicolor | — | — | — | 4–11 | — | — | Irreversible | 78, 185 |
| NOTA, DOTA, TPEN, DPA, NODAGA | Synthetic | — | — | — | — | — | — | — | |
| Dipicolinic acid | Synthetic | — | 5.7 | — | 0.14–250 | — | 0.6–340 | Competitive | 54, 86 |
| o-Phenanthroline | Synthetic | — | — | — | — | 55 | — | Reversible | |
| Thioester and thiol derivatives | |||||||||
| Thiodepsipeptides | Synthetic | — | — | — | 0.25–240 | — | — | 59 | |
| Thioesters and thiols | Synthetic | 0.01–4 | — | — | 0.0004–740 | — | — | Reversible competitive | 59, 186, 187 |
| Mercaptoacetic acid derivatives | Synthetic | 4 | — | — | 28–645 | 0.55–30 | 2–186 | Irreversible | 61, 188 |
| Mercaptophenylacetic acid derivatives | Synthetic | 0.02–1,500 | 144 | 0.3–0.6 | 22–479 | — | 1.95–8 | Reversible competitive | 189 |
| Mercaptocarboxylate | Synthetic | — | — | — | 1.1–16.4 | — | — | — | 190 |
| Mercaptophosphonate derivatives | Synthetic | 1–16 | 0.3–24 | 0.4–40 | — | — | — | Competitive | 191 |
| 1β-Methylcarbapenamens | Synthetic | 0.004–0.8 | — | 1 | >0.1–9 | — | — | 79 | |
| d-Captropil | Synthetic | 0.5–700 | 2.7–72 | — | 0.072–261.8 | — | — | Competitive | 64, 74, 192 |
| l-Captopril | Synthetic | 1.5–65 | 19–950 | — | 4.4–157.4 | — | — | Competitive | 64, 74, 193 |
| (R)-Thiomandelic acid | Synthetic | 0.03–0.8 | 144 | 0.3–0.6 | — | — | — | Reversible | 62, 194 |
| Various charged and neutral thiols | Synthetic | — | — | — | 1.3–200 | — | — | Slow-binding reversible | 195 |
| Dansyl-derived thiols | Synthetic | 0.14–1.3 | — | — | 0.7–6.3 | — | — | — | 196 |
| Penicillin-derived thiols | Synthetic | — | — | — | 1.4–106 | 0.1–72 | — | 52, 80 | |
| Bisthiazolidines derivatives | Synthetic | 3–84 | 0.3–29 | 1,041 | — | — | — | Competitive | 197 |
—, no data found.
THIOESTER, THIOL, AND CAPTOPRIL DERIVATIVES
Thiols and thiolcarboxylates are widely reported to be MBLIs (48, 59). In 1997, Goto et al. reported that the thiol esters 2-mercaptopropionic acid and mercaptoacetic acid are among the numerous thiol molecules that strongly and competitively inhibited IMP-1, a subclass B1 MBL (60). Payne et al. also demonstrated that a series of mercaptoacetic acid thiol ester derivatives inactivate the catalytic activity of class B MBLs such as BcII, CFiA, L-1, and CphA (61). Mass spectrometry analysis revealed that thiol esters inhibit MBLs by forming a disulfide bond with the active site of the enzymes (61).
Thiomandelic acid and derivatives were also found to be broad-spectrum MBLIs (62). During the establishment of a library of thiomandelic acid analogues, Mollard and coworkers realized that thiols with a carboxylic moiety improved the efficacy of the compounds, while the thiol group remained essential for the inhibitory effect. This was revealed by the substitution of the thiol group with a bromo, hydroxyl, or amidoxime function, and it was found that the thiomandelic acid analogues failed to inhibit the activity of the BcII MBL (62). The racemic thiomandelic acid exhibited greater inhibitory activity, with a Ki of 0.09 M, than the isomer thiomandelic acid, with Ki of 1.28 M, against the BcII enzyme. MBL members of subclass B1 were the most largely inhibited by thiomandelic acid and thiol-containing compounds; however, they were also effective against subclass B2 and B3 (48, 62, 63).
The captopril derivatives l- and d-captopril were investigated for their inhibitory activity against MBL by Heinz et al. (64). Both enantiomeric inhibitors demonstrated competitive inhibition against subclasses B1 and B2. Potential inhibitory activity of this group of MBLIs was observed against most class B β-lactamases; however, l-captopril exhibited weaker inhibition toward NDM-1, with a 50% inhibitory concentration (IC50) of 202 M, than d-captopril, which had an IC50 of 8 M (65). These captopril derivatives act by binding to the two zinc ions at the active site of the MBL enzymes, thus displacing the hydroxyl group that normally binds the two metal ions (65). Furthermore, Bai et al. synthesized analogues of d- and l-cysteine and reported that some cysteine or homocysteine derivatives could efficiently inactivate the NDM-1 enzyme. The design of these analogues was inspired by the potent inhibition of MBLs by captopril and derivatives (66). The most potent analogue exhibited an IC50 of 1 M, which was far more potent than the d-diastereoisomer captopril (IC50 = 8 M).
DICARBOXYLATE, CARBOXYLIC ACID, AND HYDROXAMATE DERIVATIVES
ME1071, a maleic acid derivative discovered by Meiji Seika Kaisha Ltd. (Tokyo, Japan) as a novel and specific MBLI, was evaluated in vitro in 2010 by Ishii et al. to determine its ability to potentiate the activity of biapenem and ceftazidime against IMP-1 and VIM-2 MBL-producing Pseudomonas aeruginosa strains (67). The in vivo efficacy of ME1071 was also determined in a murine model mimicking ventilator-associated pneumonia involving an MBL-producing P. aeruginosa strain. A dose of 100 mg/kg of biapenem alone or in combination with ME1071 was administered to infected mice, and the survival rates and bacterial burdens in the lungs were evaluated. Treatment of the infected mice with biapenem and ME1071 significantly resulted in longer survival and relatively lower bacterial burdens in the lungs than treatment with biapenem alone. These findings showed ME1071 as a potent and effective MBLI for treating ventilator-associated pneumonia infections (68).
Dicarboxylic acid group inhibitors, such as 3-amino and 3-alkoxy derivatives, 3-(4-hydroxypiperidin-1-yl) phthalic acid, and 3,6-bis(4-hydroxypiperidin-1-yl), showed inhibitory potency against diverse MBL-producing bacteria, particularly those in subclass B1 (69, 70). Studies have also shown the potential ability of hydroxamic acid to inactivate the catalytically essential zinc ions of the metalloprotease matrix linking with the peptide backbone, making them potential MBLIs (71). In 2016, Kim and collaborators (72) demonstrated that compounds with hydroxamic acid moieties exhibit reversible, competitive inhibitory activity against the Bla2 metallo-β-lactamase. This study showed that both sides of the dihydroxamic acids play an important role in the binding affinity, which explains the failure of the monohydroxamic acid-containing molecule to inhibit Bla2. Therefore, compounds containing dihydroxamic acid moieties may be promising MBLIs (72). The strong binding affinity of the complexes IMP-1 and 2-benzylthiazole-4-carboxylic acid was demonstrated by Chen et al. in 2012, where the IC50 was found to be 38 M, thus making this molecule a potential MBLI compound (73). Therefore, analogues were synthesized based on the structure of that molecule to improve the inhibitory activity against MBLs. The aromatic thiazole ring containing 2-benzyl exhibited greater inhibition (IC50 = 35 M) than the one with a phenyl group (IC50 > 200 M); on the other hand, the partially saturated 4,5-dihydrothiazole ring with a 2-benzyl group was inactive. The analogue (R)-2-phenyl-4,5-dihydrothiazole-4 carboxylic acid was the best compound of the group, inhibiting IMP-1 with an IC50 of 5.5 M, and was even more active than 2-benzylthiazole-4-carboxylic acid (73).
Cyclic boronates are broad-spectrum β-lactamase inhibitors which act against both serine- and zinc-based β-lactamases and target penicillin binding proteins (PBP) (74). This group of inhibitors was found to be active against subclass B1 MBLs, providing an avenue for the development of dual-action inhibitors targeting both serine- and zinc-based β-lactamases, in addition to possessing antimicrobial activity by inhibiting PBPs.
PYRROLE-BASED INHIBITORS
Synthesized pyrrole derivatives were investigated for their inhibitory properties against acquired IMP-1 MBL in P. aeruginosa and Klebsiella pneumoniae. Among the assayed compounds, six exhibited a good inhibitory effect, with Ki values ranging from 10 to 30 M (53). Hussein et al. also synthesized two sets of tetrahydropyrimidine-2-thione and pyrrole derivatives, and their ability to inhibit MBLs were examined against IMP-1; the recorded inhibition constant varied from 20 to 80 M (75). The interactions of this group of inhibitors with the MBLs are not yet well established, due in part to the absence of crystal structures of the enzyme-pyrrole derivatives. Modeling and docking studies of the most potent compound in the pyrrole derivative series indicated that they bind to the two zinc ions through the thiol anion, with sulfur-metal distances of 2.2 Å for zinc(I) and 2 Å for zinc(II) (53, 75). More data are required to elucidate the benefit of this group of inhibitors as potential MBLIs for clinical use.
NATURAL PRODUCTS
Natural products or metabolites play an important role in antimicrobial discovery. Diverse compounds from natural products exhibited good inhibitory activity against MBLs. A series of tricyclic products, SB238569, SB236050, and SB236049 from Chaetomium funicola, showed an inhibitory effect against CfiA, IMP-1, and BcII enzymes, with SB236049 as the lead compound exhibiting an IC50 of 2 M (56). These tricyclic molecules showed minimal or no inhibitory activity toward the angiotensin-converting enzyme (ACE), which is a mammalian metalloenzyme, thus predicting their specificity for class B MBLs. The flavonoids galangin and quercetin from Stenotrophomonas maltophilia irreversibly inhibited the L-1 MBL from S. maltophilia (76). Polyketides and derivatives from Penicillium spp. also showed activity against the clinically relevant subclass B1 enzyme NDM-1 (77). Aspergillomarasmine A (AMA), a natural product and metal-chelating agent produced by a fungus, recently identified by King et al., has shown an ability to inhibit the activities of the class B1 enzymes VIM-2 and NDM-1 (78). The ability of these natural products to avoid sequestration of human metalloenzymes would make them safer adjuvants. Nevertheless, systemic application of natural products as antimicrobial agents has been limited by toxicity.
β-LACTAMS STABLE AGAINST HYDROLYSIS BY MBLs
β-Lactam analogues that are stable against MBL hydrolysis have been identified. Nagano and coworkers in 1999 tested a variety of carbapenem analogues (1-methylcarbapenem conjugates) against MBLs (79) and showed that 1-methylcarbapenems containing dithiocarbamate, benzothienythio, or pyrrolidinylthio moieties at the C-2 position exhibited resistance to hydrolysis by series of MBL enzymes. The most promising compound among this group was J-110,441, a molecule possessing a benzothienylthio moiety at the C-2 position of 1-beta-methylcarbapenem, which inhibited the IPM-1, Ccra, L-1, and BcII MBLs with Ki values of 0.004, 0.2, 1, and 0.8 M, respectively. Interestingly J-110,441 also inhibited other classes of beta-lactamases (classes A and C) and thus possessed a broad-spectrum anti-MBL activity (79). Similarly, penicillin derivatives have also been reported as potent β-lactamase inhibitors targeting both serine- and zinc-based β-lactamases (52, 80), while thioxo derivatives of cephalosporins were found to be inhibitors of BcII MBLs (81). In order to develop simultaneous inactivation of serine- and zinc-based β-lactamases, series of cephalosporin analogues, reverse hydroxamates, and oximes were prepared and tested against MBLs to evaluate their inhibitory activities. The reverse hydroxamates were found to inhibit the GIM-1 MBL (82), whereas 6-alkylidene-2-substituted penam sulfones and cyclobutanone derivatives of β-lactams were also reported as inhibitors of Bla2 and IMP-1, respectively (80, 83). β-Lactam analogue inhibitors are likely to be broad spectrum, extending their activity to both classes of β-lactamases (serine- and zinc-based β-lactamases).
METAL CHELATORS
The use of zinc-chelating agents is an approach for inhibiting MBLs, as MBLs require one or two zinc ions in their active site to hydrolyze β-lactams (84). The utilization of zinc-chelating agents will interfere with the functionality of the zinc ions by sequestering them from the active site of MBLs, thus reducing the activity of the MBLs (58).
Several metal chelators with a strong affinity for zinc ions have shown inhibitory effects against different classes of MBLs. Docquier et al. demonstrated that VIM-2 was susceptible to inactivation by metal chelators, indicating that the zinc ions of this enzyme were probably loosely bound and thus subject to easy sequestration by zinc-chelating agents (84). Among the numerous metal-chelating agents, EDTA has been widely reported as a potential MBLI. It exhibited an IC50 of 27.9 M for the acquired IMP-1 enzymes (73). In addition, this molecule possesses essential antimicrobial properties, such as potentiation of the activity of other antibiotics through the disruption of the outer membrane of Gram-negative bacteria as well as the neutralization of enzymes and toxins produced by organisms. Due to its toxicity, EDTA was not considered suitable for therapeutic use (85). In 2013, Yoshizumi et al. evaluated the efficacy of calcium-EDTA (Ca-EDTA) as an MBLI, which was found to be less toxic than EDTA in in vivo assays using murine models. They discovered that imipenem's activity was restored in the presence of Ca-EDTA in all the tested P. aeruginosa isolates producing IMP and VIM enzymes but that activity failed to restored in all the non-MBL-producing strains (85). An in vivo study demonstrated significant reductions in bacterial CFU in the lungs of infected mice when treated with a combination of imipenem and Ca-EDTA. These findings suggest that Ca-EDTA could be used clinically due to having a lesser toxic effect than EDTA (85).
Dipicolinic acid is another metal-chelating agent that demonstrated good inhibitory effects against members of the three MBL classes, namely, CcrA, CphA, and L1 (86). Pyridine 2,4-dicarboxylic acid is also one of the numerous zinc-chelating agents that inhibited CphA (54). A natural fungal product, AMA, which is structurally similar to EDTA, was identified as a potent inhibitor of NDM-1 and VIM-2 in 2014 (78). Irreversible inhibition was observed with AMA after its removal by gel filtration; nevertheless, the activity of the subclass B1 enzyme was restored by adding an excess of ZnSO4, suggesting that AMA acts by metal sequestration. The observed survival rates of mice infected with lethal doses of K. pneumoniae strains producing NDM-1 were 95%, demonstrating that AMA prevented the hydrolysis of the β-lactam antibiotic in vivo (78). This potent MBLI also showed less toxicity than EDTA. In 2015, Somboro et al. evaluated two cyclic chelators, 1,4,7-triazacyclononane-triacetic acid (NOTA) and 1,4,7,10-tetra-azacyclododecane-tetra-acetic acid (DOTA), as potent MBLIs, with NOTA being the most promising agent (87). In 2016, Azumah et al. also investigated two acyclic zinc chelators, N,N,N′,N′-tetrakis(2-pyridylmethyl)ethylenediamine (TPEN) and di-(2-picolyl)amine (DPA), as well as peptide derivatives of 1,4,7-triazacyclononane-1-glutaric acid-4,7-diacetic acid (NODAGA), as MBLIs (88). These cyclic and acyclic metal chelators restored the activities of imipenem and meropenem against carbapenem-resistant bacteria producing NDM, IMP, and VIM enzymes, presumably by binding to the zinc ions of the enzymes (87, 88).
The greatest challenge to the clinical use of metal chelators is that they also inhibit human metalloenzymes such as matrix metalloproteinase, carbonic anhydrase, and carboxypeptidases, necessitating further investigations into their systemic effect on living tissues.
CONCLUSION AND FUTURE PERSPECTIVES
This review elucidates the diversity and alarming dissemination of MBLs causing carbapenem resistance. It is known that the discovery of efficient MBLIs is difficult, in part, due to the flexible active sites of the multiple enzymes and the challenges associated with targeting metalloenzymes. Therefore, the hunt for MBLIs should ideally be specific to bacterial metalloenzymes and circumvent other zinc-containing enzymes in humans. Inhibitors should either mimic the structures of the enzymes' substrates or allosterically inhibit the activity of the enzyme. The other challenge is that the performance of these inhibitors varied from one subclass to another. Worse still, structural variations are known to occur within the same subclass, rendering the discovery and development of MBLIs more challenging. Particularly, it will be a great feat to identify compounds that inhibit plasmid-encoded subclass B1 enzymes, which include the most widespread and clinically important MBLs.
Emphasis needs to be placed on designing and developing MBLIs to extend their inhibitory spectrum to the three classes of MBLs, restore the efficacy of available β-lactam antibiotics, and improve their pharmacological properties. Moreover, genomic data generated with current methods such as next-generation and whole-genome sequencing, coupled with phylogenetics and metadata, are currently one of the tools that give valuable insight into the identification and characterization of MBLs. They also help elucidate the global spread or epidemiology of these MBLs. These modern methods also have a quick turnaround time compared to traditional ones and should be explored to find lasting solutions to this menace. In-depth analysis and optimization of MBLIs, employing a multidisciplinary research approach involving advanced computational simulations and biochemical, microbiological, bioinformatic, and animal studies to develop novel metallo-β-lactamase inhibitors, are needed to precipitate the development of efficient clinical MBLI adjuvants.
Biographies

Anou M. Somboro, Pharm.D., M.Sc., Ph.D., received a Pharm.D. degree from the University of Bamako, Mali, and M.Sc. and Ph.D. degrees from the University of KwaZulu-Natal (UKZN), South Africa. He supported laboratory-based activities together with a team of HIV/tuberculosis researchers at the HIV/TB Research and Training Center (SEREFO) in Mali and at the National Institute of Allergic and Infectious Diseases, USA. Dr. Somboro currently works at the College of Health Sciences, UKZN, South Africa. His current projects include hunting for adjuvants that can modulate or synergize the efficacy of approved antibiotics to combat multidrug-resistant bacteria, particularly the design and development of novel metallo-β-lactamase inhibitors; he also participates in building strategies for the prevention and containment of antibiotic resistance based on the surveillance of antibiotic use and resistance in humans, the food chain, and associated environments (a One Health perspective).

John Osei Sekyere, B.Pharm., M.Phil., Ph.D., is a licensed pharmacist (B.Pharm.) trained at the Kwame Nkrumah University of Science and Technology, Ghana (2010). He studied veterinary and pharmaceutical microbiology at the same university for his M.Phil. degree (2014). He obtained his Ph.D. in clinical and molecular microbiology at the University of KwaZulu-Natal (UKZN), South Africa (2016), and worked at UKZN for a year as a postdoctoral fellow in antibiotic resistance genomics and One Health. Dr. Osei Sekyere is currently with the Department of Medical Microbiology, University of Pretoria, South Africa, where he supervises and undertakes research into antibiotic resistance mechanisms, the microbiome (gut-lung axis), and genomic epidemiology of Gram-negative bacteria, Gram-positive bacteria, and mycobacteria. His current projects include hunting for adjuvants that can modulate or synergize the efficacy of approved antibiotics to combat multidrug-resistant bacteria, probiotics and vaccine research, microbiome-immunity-drug interactions in the host, One Health antibiotic resistance surveillance and genomic epidemiology.

Daniel G. Amoako, B.Sc. (Hons.), M.Res., is a doctoral research student and the Lead Project Coordinator (Animal Health) at the Antimicrobial Research Unit (ARU), Discipline of Pharmaceutical Science, University of KwaZulu-Natal, Durban, South Africa. Mr. Amoako has a longstanding interest in drug development and microbial pathogenesis; his research applies pathogenomics and genome sequencing to better understand emerging pathogens and their mechanisms of resistance. He is currently involved in several large-scale bacterial whole-genome sequencing projects and actively translates microbial genomic data into genomic epidemiology for the benefit of South Africa's Public, Private, Food, Animal, and Environmental Health sector. He also applies metagenomics, transcriptomics, and microbiomics to investigate human health and disease. He received his B.Sc. (Hons.) in biochemistry from the University of Cape Coast in Ghana and an M.Res. degree in pharmaceutical science from the University of KwaZulu-Natal, South Africa.

Sabiha Y. Essack, B.Pharm., M.Pharm., Ph.D., received B.Pharm., M.Pharm., and Ph.D. degrees, awarded in 1989, 1994, and 2000, respectively, from the University of Durban-Westville. She is currently a Professor in Pharmaceutical Sciences and the South African Research Chair in Antibiotic Resistance and One Health at the University of KwaZulu-Natal (UKZN). Professor Essack established the Antimicrobial Research Unit (ARU) at UKZN. The ARU has as its overarching objective the optimization of antibiotic therapy in the management of infections in the context of escalating drug-resistant infections, both nationally within a dichotomous public and private health system and regionally in Africa, where communicable diseases remain a major cause of mortality, increasingly because of drug-resistant infections. Research in the ARU has adopted the One Health approach, investigating the molecular epidemiology of antibiotic resistance in human, (food) animal, and environmental health and delineating resistance phenotypes and genotypes by methods ranging from sensitivity testing to whole-genome sequencing.

Linda A. Bester, M.Tech.Vet., Ph.D., holds an M.Tech. degree in veterinary technology from the Tswane University of Technology and a Ph.D. from the University of KwaZulu-Natal (awarded in 2013). She specializes in both veterinary microbiology and laboratory animal sciences. She is currently a staff member of the Biomedical Resource Unit, School of Laboratory Medicine and Medical Sciences, at the University of KwaZulu-Natal, Durban, South Africa. Dr. Bester graduated under the mentorship of Professor Sabiha Essack, where she investigated antibiotic resistance via the food chain. Since then she has been a member of the Antimicrobial Research Unit (ARU) at UKZN under the directorship of Professor Essack. She has also established her own research theme looking at the dissemination of bacteria and antibiotic resistance in hospitals.
REFERENCES
- 1.Prestinaci F, Pezzotti P, Pantosti A. 2015. Antimicrobial resistance: a global multifaceted phenomenon. Pathog Glob Health 109:309–318. doi: 10.1179/2047773215Y.0000000030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Osei Sekyere J. 2016. Current state of resistance to antibiotics of last-resort in South Africa: a review from a public health perspective. Front Public Healt 4:209. doi: 10.3389/fpubh.2016.00209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Knobler S, Mahmoud A, Lemon S. 2006. A world in motion: the global movement of people, products, pathogens, and power. Inst Med 1:1–33. [Google Scholar]
- 4.Power E. 2006. Impact of antibiotic restrictions: the pharmaceutical perspective. Clin Microbiol Infect 12:25–34. [DOI] [PubMed] [Google Scholar]
- 5.Carlet J, Jarlier V, Harbarth S, Voss A, Goossens H, Pittet D. 2012. Ready for a world without antibiotics? The Pensières antibiotic resistance call To Action. Antimicrob Resist Infect Control 1:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Osei Sekyere J, Govinden U, Bester LA, Essack SY. 2016. Colistin and tigecycline resistance in carbapenemase-producing Gram negative bacteria: emerging resistance mechanisms and detection methods. J Appl Microbiol 121:601–617. doi: 10.1111/jam.13169. [DOI] [PubMed] [Google Scholar]
- 7.Osei Sekyere J, Asante J. 2018. Emerging mechanisms of antimicrobial resistance in bacteria and fungi: advances in the era of genomics. Future Microbiol 13:1–22. doi: 10.2217/fmb-2017-0172. [DOI] [PubMed] [Google Scholar]
- 8.Munita JM, Arias CA. 2016. Mechanisms of antibiotic resistance. Microbiol Spectr 4:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Livermore DM. 2003. Bacterial resistance: origins, epidemiology, and impact. Clin Infect Dis 36:S11–S23. doi: 10.1086/344654. [DOI] [PubMed] [Google Scholar]
- 10.Livermore DM. 1998. Beta-lactamase-mediated resistance and opportunities for its control. J Antimicrob Chemother 41:25–41. [DOI] [PubMed] [Google Scholar]
- 11.Osei Sekyere J, Amoako DG. 2017. Carbonyl cyanide m-chlorophenylhydrazine (CCCP) reverses resistance to colistin, but not to carbapenems and tigecycline in multidrug-resistant Enterobacteriaceae. Front Microbiol 8:228. doi: 10.3389/fmicb.2017.00228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bodey GP. 1990. Penicillins, monobactams, and carbapenems. Texas Hear Inst J 17:315. [PMC free article] [PubMed] [Google Scholar]
- 13.Hamed RB, Gomez-Castellanos JR, Henry L, Ducho C, McDonough MA, Schofield CJ. 2013. The enzymes of β-lactam biosynthesis. Nat Prod Rep 30:21–107. doi: 10.1039/C2NP20065A. [DOI] [PubMed] [Google Scholar]
- 14.Queenan AM, Bush K. 2007. Carbapenemases: the versatile β-lactamases. Clin Microbiol Rev 20:440–458. doi: 10.1128/CMR.00001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ambler RP. 1980. The structure of beta-lactamases. Philos Trans R Soc Lond B 289:321. doi: 10.1098/rstb.1980.0049. [DOI] [PubMed] [Google Scholar]
- 16.Bush K, Jacoby GA. 2010. Updated functional classification of beta-lactamases. Antimicrob Agents Chemother 54:969–976. doi: 10.1128/AAC.01009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bush K. 2013. Carbapenemases: partners in crime. J Glob Antimicrob Resist 1:7–16. doi: 10.1016/j.jgar.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 18.Bush K, Bradford PA. 2016. Beta-lactams and beta-lactamase inhibitors: an overview. Cold Spring Harb Perspect Med 6:a025247. doi: 10.1101/cshperspect.a025247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bebrone C. 2007. Metallo-β-lactamases (classification, activity, genetic organization, structure, zinc coordination) and their superfamily. Biochem Pharmacol 74:1686–1701. doi: 10.1016/j.bcp.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 20.Kelly AM, Mathema B, Larson EL. 2017. Carbapenem-resistant Enterobacteriaceae in the community: a scoping review. Int J Antimicrob Agents 50:127–134. doi: 10.1016/j.ijantimicag.2017.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Potter RF, D'Souza AW, Dantas G. 2016. The rapid spread of carbapenem-resistant Enterobacteriaceae. Drug Resist Updat 29:30–46. doi: 10.1016/j.drup.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gupta N, Limbago BM, Patel JB, Kallen AJ. 2011. Carbapenem-resistant enterobacteriaceae: epidemiology and prevention. Clin Infect Dis 53:60–67. doi: 10.1093/cid/cir202. [DOI] [PubMed] [Google Scholar]
- 23.Falagas ME, Tansarli GS, Karageorgopoulos DE, Vardakas KZ. 2014. Deaths attributable to carbapenem-resistant Enterobacteriaceae infections. Emerg Infect Dis 20:1170–1175. doi: 10.3201/eid2007.121004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van Duin D, Kaye KS, Neuner EA, Bonomo RA. 2013. Carbapenem-resistant Enterobacteriaceae: a review of treatment and outcomes. Diagn Microbiol Infect Dis 75:115–120. doi: 10.1016/j.diagmicrobio.2012.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim UJ, Kim HK, An JH, Cho SK, Park K-H, Jang H-C. 2014. Update on the epidemiology, treatment, and outcomes of carbapenem-resistant Acinetobacter infections. Chonnam Med J 50:37–44. doi: 10.4068/cmj.2014.50.2.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang X, Chen G, Wu X, Wang L, Cai J, Chan EW, Chen S, Zhang R. 2015. Increased prevalence of carbapenem resistant Enterobacteriaceae in hospital setting due to cross-species transmission of the blaNDM-1 element and clonal spread of progenitor resistant strains. Front Microbiol 6:595. doi: 10.3389/fmicb.2015.00595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pittet D, Allegranzi B, Storr J, Nejad SB, Dziekan G, Leotsakos A, Donaldson L. 2008. Infection control as a major World Health Organization priority for developing countries. J Hosp Infect 68:285–292. doi: 10.1016/j.jhin.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 28.World Health Organization. 2017. Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. World Health Organization, Geneva Switzerland. [Google Scholar]
- 29.Walsh TR, Toleman MA, Poirel L, Nordmann P. 2005. metallo-β-lactamases: the quiet before the storm? Clin Microbiol Rev 18:306–325. doi: 10.1128/CMR.18.2.306-325.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lim HM, Pene JJ, Shaw RW. 1988. Cloning, nucleotide sequence, and expression of the Bacillus cereus 5/B/6 β-lactamase II structural gene. J Bacteriol 170:2873–2878. doi: 10.1128/jb.170.6.2873-2878.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walsh TR, Hall L, Assinder SJ, Nichols WW, Cartwright SJ, MacGowan AP, Bennett PM. 1994. Sequence analysis of the L1 metallo-β-lactamase from Xanthomonas maltophilia. Biochim Biophys Acta 1218:199–201. doi: 10.1016/0167-4781(94)90011-6. [DOI] [PubMed] [Google Scholar]
- 32.Sabath LD, Abraham EP. 1966. Zinc as a cofactor for cephalosporinase from Bacillus cereus 569. Biochem J 98:11C. doi: 10.1042/bj0980011C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuwabara S, Abraham EP. 1967. Some properties of two extracellular beta-lactamases from Bacillus cereus 569/H. Biochem J 103:27C. doi: 10.1042/bj1030027C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cornaglia G, Giamarellou H, Rossolini GM. 2011. Metallo-beta-lactamases: a last frontier for beta-lactams? Lancet Infect Dis 11:381–393. doi: 10.1016/S1473-3099(11)70056-1. [DOI] [PubMed] [Google Scholar]
- 35.Laraki N, Galleni M, Thamm I, Riccio ML, Amicosante G, Frère JM, Rossolini GM. 1999. Structure of In31, a bla(IMP)-containing Pseudomonas aeruginosa integron phyletically related to In5, which carries an unusual array of gene cassettes. Antimicrob Agents Chemother 43:890–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bennett PM. 2008. Plasmid encoded antibiotic resistance: acquisition and transfer of antibiotic resistance genes in bacteria. Br J Pharmacol 153:S347–S357. doi: 10.1038/sj.bjp.0707607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, Walsh TR. 2009. Characterization of a new metallo-beta-lactamase gene, bla NDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother 53:5046–5054. doi: 10.1128/AAC.00774-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karsisiotis AI, Damblon CF, Roberts GCK. 2014. A variety of roles for versatile zinc in metallo-β-lactamases. Metallomics 6:1181–1197. doi: 10.1039/C4MT00066H. [DOI] [PubMed] [Google Scholar]
- 39.Galleni M, Lamotte-Brasseur J, Rossolini GM, Spencer J, Dideberg O, Frère JM, Amicosante G, Franceschini N, Bush K, Concha NO, Herzberg O, Livermore DM, Rasmussen BA, Rodrigues J, Saavedra MJ, Sutton B, Fabiane SM, Toney JH. 2001. Standard numbering scheme for class B β-lactamases. Antimicrob Agents Chemother 45:660–663. doi: 10.1128/AAC.45.3.660-663.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mojica MF, Bonomo RA, Fast W. 2016. B1-metallo-β-lactamases: where do we stand? Curr Drug Targets 17:1029–1050. doi: 10.2174/1389450116666151001105622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jia B, Raphenya AR, Alcock B, Waglechner N, Guo P, Tsang KK, Lago BA, Dave BM, Pereira S, Sharma AN. 2017. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res 45:D566–D573. doi: 10.1093/nar/gkw1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Naas T, Oueslati S, Bonnin RA, Dabos ML, Zavala A, Dortet L, Retailleau P, Iorga BI. 2017. Beta-lactamase database (BLDB)—structure and function. J Enzyme Inhib Med Chem 32:917–919. doi: 10.1080/14756366.2017.1344235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hall BG, Salipante SJ, Barlow M. 2004. Independent origins of subgroup Bl+B2 and subgroup B3 metallo-β-lactamases. J Mol Evol 59:133–141. doi: 10.1007/s00239-003-2572-9. [DOI] [PubMed] [Google Scholar]
- 44.Hall BG, Salipante SJ, Barlow M. 2003. The metallo-β-lactamases fall into two distinct phylogenetic groups. J Mol Evol 57:249–254. doi: 10.1007/s00239-003-2471-0. [DOI] [PubMed] [Google Scholar]
- 45.Asnicar F, Weingart G, Tickle TL, Huttenhower C, Segata N. 2015. Compact graphical representation of phylogenetic data and metadata with GraPhlAn. Peer J 3:e1029. doi: 10.7717/peerj.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Berglund F, Marathe NP, Österlund T, Bengtsson-Palme J, Kotsakis S, Flach C-F, Larsson DGJ, Kristiansson E. 2017. Identification of 76 novel B1 metallo-β-lactamases through large-scale screening of genomic and metagenomic data. Microbiome 5:134. doi: 10.1186/s40168-017-0353-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McGeary RP, Tan DT, Schenk G. 2017. Progress toward inhibitors of metallo-β-lactamases. Future Med Chem 9:673–691. doi: 10.4155/fmc-2017-0007. [DOI] [PubMed] [Google Scholar]
- 48.Tehrani KHME, Martin NI. 2017. Thiol-containing metallo-β-lactamase inhibitors resensitize resistant Gram-negative bacteria to meropenem. ACS Infect Dis 3:711–717. doi: 10.1021/acsinfecdis.7b00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Toney JH, Fitzgerald PM, Grover-Sharma N, Olson SH, May WJ, Sundelof JG, Vanderwall DE, Cleary K a, Grant SK, Wu JK, Kozarich JW, Pompliano DL, Hammond GG. 1998. Antibiotic sensitization using biphenyl tetrazoles as potent inhibitors of Bacteroides fragilis metallo-beta-lactamase. Chem Biol 5:185–196. doi: 10.1016/S1074-5521(98)90632-9. [DOI] [PubMed] [Google Scholar]
- 50.Walter MW, Valladares MH, Adlington RM, Amicosante G, Baldwin JE, Frère J-M, Galleni M, Rossolini GM, Schofield CJ. 1999. Hydroxamate inhibitors of Aeromonas hydrophila AE036 metallo-B-lactamase. Bioorg Chem 27:35–40. doi: 10.1006/bioo.1998.1111. [DOI] [Google Scholar]
- 51.Fitzgerald PMD, Wu JK, Toney JH. 1998. Unanticipated inhibition of the metallo-β-lactamase from Bacteroides fragilis by 4-morpholineethanesulfonic acid (MES): a crystallographic study at 1.85-Å resolution. Biochemistry 37:6791–6800. doi: 10.1021/bi9730339. [DOI] [PubMed] [Google Scholar]
- 52.Buynak JD, Chen H, Vogeti L, Gadhachanda VR, Buchanan CA, Palzkill T, Shaw RW, Spencer J, Walsh TR. 2004. Penicillin-derived inhibitors that simultaneously target both metallo- and serine-β-lactamases. Bioorg Med Chem Lett 14:1299–1304. doi: 10.1016/j.bmcl.2003.12.037. [DOI] [PubMed] [Google Scholar]
- 53.Mohamed MS, Hussein WM, McGeary RP, Vella P, Schenk G, Abd El-hameed RH. 2011. Synthesis and kinetic testing of new inhibitors for a metallo-β-lactamase from Klebsiella pneumonia and Pseudomonas aeruginosa. Eur J Med Chem 46:6075–6082. doi: 10.1016/j.ejmech.2011.10.030. [DOI] [PubMed] [Google Scholar]
- 54.Horsfall LE, Garau G, Liénard BMR, Dideberg O, Schofield CJ, Frère JM, Galleni M. 2007. Competitive inhibitors of the CphA metallo-β-lactamase from Aeromonas hydrophila. Antimicrob Agents Chemother 51:2136–2142. doi: 10.1128/AAC.00866-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bounaga S, Galleni M, Laws AP, Page MI. 2001. Cysteinyl peptide inhibitors of Bacillus cereus zinc β-lactamase. Bioorg Med Chem 9:503–510. doi: 10.1016/S0968-0896(00)00257-1. [DOI] [PubMed] [Google Scholar]
- 56.Payne DJ, Hueso-Rodríguez JA, Boyd H, Concha NO, Janson CA, Gilpin M, Bateson JH, Cheever C, Niconovich NL, Pearson S, Rittenhouse S, Tew D, Díez E, Pérez P, De la Fuente J, Rees M, Rivera-Sagredo A. 2002. Identification of a series of tricyclic natural products as potent broad-spectrum inhibitors of metallo-β-lactamases. Antimicrob Agents Chemother 46:1880–1886. doi: 10.1128/AAC.46.6.1880-1886.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shaw RW, Cottenoir M. March 2010. Inhibiton of metallo-beta-lactamase by double-stranded DNA. US patent 8,143,389.
- 58.Siemann S, Brewer D, Clarke AJ, Dmitrienko GI, Lajoie G, Viswanatha T. 2002. IMP-1 metallo-β-lactamase: effect of chelators and assessment of metal requirement by electrospray mass spectrometry. Biochim Biophys Acta 1571:190–200. doi: 10.1016/S0304-4165(02)00258-1. [DOI] [PubMed] [Google Scholar]
- 59.Greenlee ML, Laub JB, Balkovec JM, Hammond ML, Hammond GG, Pompliano DL, Epstein-Toney JH. 1999. Synthesis and SAR of thioester and thiol inhibitors of IMP-1 metallo-β-lactamase. Bioorg Med Chem Lett 9:2549–2554. doi: 10.1016/S0960-894X(99)00425-4. [DOI] [PubMed] [Google Scholar]
- 60.Goto M, Takahashi T, Yamashita F, Koreeda A, Mori H, Ohta M, Arakawa Y. 1997. Inhibition of the metallo-beta-lactamase produced from Serratia marcescens by thiol compounds. Biol Pharm Bull 20:1136–1140. doi: 10.1248/bpb.20.1136. [DOI] [PubMed] [Google Scholar]
- 61.Payne DJ, Bateson JH, Gasson BC, Proctor D, Khushi T, Farmer TH, Tolson DA, Bell D, Skett PW, Marshall AC, Reid R, Ghosez L, Combret Y, Marchand-Brynaert J. 1997. Inhibition of metallo-beta-lactamases by a series of mercaptoacetic acid thiol ester derivatives. Antimicrob Agents Chemother 41:135–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mollard C, Moali C, Papamicael C, Damblon C, Vessilier S, Amicosante G, Schofield CJ, Galleni M, Frère JM, Roberts GCK. 2001. Thiomandelic acid, a broad spectrum inhibitor of zinc β-lactam. Kinetic and spectroscopic studiesases. J Biol Chem 276:45015–45023. [DOI] [PubMed] [Google Scholar]
- 63.Liénard BMR, Garau G, Horsfall L, Karsisiotis AI, Damblon C, Lassaux P, Papamicael C, Roberts GCK, Galleni M, Dideberg O, Frere J-M, Schofield CJ. 2008. Structural basis for the broad-spectrum inhibition of metallo-beta-lactamases by thiols. Org Biomol Chem 6:2282–2294. doi: 10.1039/b802311e. [DOI] [PubMed] [Google Scholar]
- 64.Heinz U, Bauer R, Wommer S, Meyer-Klaucke W, Papamichaels C, Bateson J, Adolph HW. 2003. Coordination geometries of metal ions in d- or l-captopril-inhibited metallo-β-lactamases. J Biol Chem 278:20659–20666. doi: 10.1074/jbc.M212581200. [DOI] [PubMed] [Google Scholar]
- 65.Brem J, van Berkel SS, Zollman D, Lee SY, Gileadi O, McHugh PJ, Walsh TR, McDonough MA, Schofield CJ. 2016. Structural basis of metallo-β-lactamase inhibition by captopril stereoisomers. Antimicrob Agents Chemother 60:142–150. doi: 10.1128/AAC.01335-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cui-Gai Bai, Yin-Tong Xu, Ning-Ning Li, Jing-Han Wang, Cheng Yang, Yue Chen, Hong-Gang Zhou. 2015. Cysteine and its derivatives as New Delhi metallo-beta-lactamase-1 inhibitors. Curr Enzym Inhib 11:46–57. doi: 10.2174/1573408011666150408223245. [DOI] [Google Scholar]
- 67.Ishii Y, Eto M, Mano Y, Tateda K, Yamaguchi K. 2010. In vitro potentiation of carbapenems with ME1071, a novel metallo-beta-lactamase inhibitor, against metallo-beta-lactamase-producing Pseudomonas aeruginosa clinical isolates. Antimicrob Agents Chemother 54:3625–3629. doi: 10.1128/AAC.01397-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yamada K, Yanagihara K, Kaku N, Harada Y, Migiyama Y, Nagaoka K, Morinaga Y, Nakamura S, Imamura Y, Miyazaki T, Izumikawa K, Kakeya H, Hasegawa H, Yasuoka A, Kohno S. 2013. In vivo efficacy of biapenem with ME1071, a novel metallo-beta-lactamase (MBL) inhibitor, in a murine model mimicking ventilator-associated pneumonia caused by MBL-producing Pseudomonas aeruginosa. Int J Antimicrob Agents 42:238–243. doi: 10.1016/j.ijantimicag.2013.05.016. [DOI] [PubMed] [Google Scholar]
- 69.Hiraiwa Y, Saito J, Watanabe T, Yamada M, Morinaka A, Fukushima T, Kudo T. 2014. X-ray crystallographic analysis of IMP-1 metallo-β-lactamase complexed with a 3-aminophthalic acid derivative, structure-based drug design, and synthesis of 3,6-disubstituted phthalic acid derivative inhibitors. Bioorg Med Chem Lett 24:4891–4894. doi: 10.1016/j.bmcl.2014.08.039. [DOI] [PubMed] [Google Scholar]
- 70.Hiraiwa Y, Morinaka A, Fukushima T, Kudo T. 2013. Metallo-β-lactamase inhibitory activity of 3-alkyloxy and 3-amino phthalic acid derivatives and their combination effect with carbapenem. Bioorgc Med Chem 21:5841–5850. doi: 10.1016/j.bmc.2013.07.006. [DOI] [PubMed] [Google Scholar]
- 71.Materon IC, Queenan AM, Koehler TM, Bush K, Palzkill T. 2003. Biochemical characterization of beta-lactamases Bla1 and Bla2 from Bacillus anthracis. Antimicrob Agents Chemother 47:2040–2042. doi: 10.1128/AAC.47.6.2040-2042.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim S-K, Demuth M, Schlesinger SR, Kim SJ, Urbanczyk J, Shaw RW, Shin H. 2016. Inhibition of Bacillus anthracis metallo-β-lactamase by compounds with hydroxamic acid functionality. J Enzyme Inhib Med Chem 31:132–137. [DOI] [PubMed] [Google Scholar]
- 73.Chen P, Horton L, Mikulski R. 2012. 2-Substituted 4, 5-dihydrothiazole-4-carboxylic acids are novel inhibitors of metallo-β-lactamases. Bioorg Med Chem Lett 22:6229–6232. doi: 10.1016/j.bmcl.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Brem J, Cain R, Cahill S, McDonough MA, Clifton IJ, Jiménez-Castellanos J-C, Avison MB, Spencer J, Fishwick CWG, Schofield CJ. 2016. Structural basis of metallo-β-lactamase, serine-β-lactamase and penicillin-binding protein inhibition by cyclic boronates. Nat Commun 7:12406. doi: 10.1038/ncomms12406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hussein WM, Fatahala SS, Mohamed ZM, Mcgeary RP, Schenk G, Ollis DL, Mohamed MS. 2012. Synthesis and kinetic testing of tetrahydropyrimidine-2-thione and pyrrole derivatives as inhibitors of the metallo-beta-lactamase from Klebsiella pneumonia and Pseudomonas aeruginosa. Chem Biol Drug Des 80:500–515. doi: 10.1111/j.1747-0285.2012.01440.x. [DOI] [PubMed] [Google Scholar]
- 76.Denny BJ, Lambert PA, West PWJ. 2002. The flavonoid galangin inhibits the L1 metallo-beta-lactamase from Stenotrophomonas maltophilia. FEMS Microbiol Lett 208:21–24. doi: 10.1016/S0378-1097(01)00580-8,10.1111/j.1574-6968.2002.tb11054.x. [DOI] [PubMed] [Google Scholar]
- 77.Gan M, Liu Y, Bai Y, Guan Y, Li L, Gao R, He W, You X, Li Y, Yu L, Xiao C. 2013. Polyketides with New Delhi metallo-β-lactamase 1 inhibitory activity from Penicillium sp. J Nat Prod 76:1535–1540. doi: 10.1021/np4000944. [DOI] [PubMed] [Google Scholar]
- 78.King AM, Reid-Yu SA, Wang W, King DT, De Pascale G, Strynadka NC, Walsh TR, Coombes BK, Wright GD. 2014. Aspergillomarasmine A overcomes metallo-β-lactamase antibiotic resistance. Nature 510:503–506. doi: 10.1038/nature13445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nagano R, Adachi Y, Imamura H, Yamada K, Hashizume T, Morishima H. 1999. Carbapenem derivatives as potential inhibitors of various β-lactamases, including class B metallo-β-lactamases. Antimicrob Agents Chemother 43:2497–2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Beharry Z, Chen H, Gadhachanda VR, Buynak JD, Palzkill T. 2004. Evaluation of penicillin-based inhibitors of the class A and B β-lactamases from Bacillus anthracis. Biochem Biophys Res Commun 313:541–545. doi: 10.1016/j.bbrc.2003.11.158. [DOI] [PubMed] [Google Scholar]
- 81.Tsang WY, Dhanda A, Schofield CJ, Frère JM, Galleni M, Page MI. 2004. The inhibition of metallo-β-lactamase by thioxo-cephalosporin derivatives. Bioorg Med Chem Lett 14:1737–1739. doi: 10.1016/j.bmcl.2004.01.047. [DOI] [PubMed] [Google Scholar]
- 82.Ganta SR, Perumal S, Pagadala SRR, Samuelsen Ø, Spencer J, Pratt RF, Buynak JD. 2009. Approaches to the simultaneous inactivation of metallo- and serine-β-lactamases. Bioorg Med Chem Lett 19:1618–1622. doi: 10.1016/j.bmcl.2009.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Johnson JW, Gretes M, Goodfellow VJ, Marrone L, Heynen ML, Strynadka NCJ, Dmitrienko GI. 2010. Cyclobutanone analogues of β-lactams revisited: Insights into conformational requirements for inhibition of serine- and metallo-β-lactamases. J Am Chem Soc 132:2558–2560. doi: 10.1021/ja9086374. [DOI] [PubMed] [Google Scholar]
- 84.Docquier J-D, Lamotte-Brasseur J, Galleni M, Amicosante G, Frère J-M, Rossolini GM. 2003. On functional and structural heterogeneity of VIM-type metallo-beta-lactamases. J Antimicrob Chemother 51:257–266. doi: 10.1093/jac/dkg067. [DOI] [PubMed] [Google Scholar]
- 85.Yoshizumi A, Ishii Y, Livermore DM, Woodford N, Kimura S, Saga T, Harada S, Yamaguchi K, Tateda K. 2013. Efficacies of calcium-EDTA in combination with imipenem in a murine model of sepsis caused by Escherichia coli with NDM-1 Beta-lactamase. J Infect Chemother 19:992–995. doi: 10.1007/s10156-012-0528-y. [DOI] [PubMed] [Google Scholar]
- 86.Roll DM, Yang Y, Wildey MJ, Bush K, Lee MD. 2010. Inhibition of metallo-beta-lactamases by pyridine monothiocarboxylic acid analogs. J Antibiot (Tokyo) 63:255–257. doi: 10.1038/ja.2010.20. [DOI] [PubMed] [Google Scholar]
- 87.Somboro AM, Tiwari D, Bester LA, Parboosing R, Chonco L, Kruger HG, Arvidsson PI, Govender T, Naicker T, Essack SY. 2015. NOTA: a potent metallo-β-lactamase inhibitor. J Antimicrob Chemother 70:1594–1596. doi: 10.1093/jac/dku538. [DOI] [PubMed] [Google Scholar]
- 88.Azumah R, Dutta J, Somboro AM, Ramtahal M, Chonco L, Parboosing R, Bester LA, Kruger HG, Naicker T, Essack SY, Govender T. 2016. In vitro evaluation of metal chelators as potential metallo-β-lactamase inhibitors. J Appl Microbiol 120:860–867. doi: 10.1111/jam.13085. [DOI] [PubMed] [Google Scholar]
- 89.Carfi A, Pares S, Duee E, Galleni M, Duez C, Frère J-M, Dideberg O. 1995. The 3-D structure of a zinc metallo-beta-lactamase from Bacillus cereus reveals a new type of protein fold. EMBO J 14:4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Matagne A, Dubus A, Galleni M, Frère J-M. 1999. The β-lactamase cycle: a tale of selective pressure and bacterial ingenuity. Nat Prod Rep 16:1–19. doi: 10.1039/a705983c. [DOI] [PubMed] [Google Scholar]
- 91.Yang Y, Rasmussen BA, Bush K. 1992. Biochemical characterization of the metallo-beta-lactamase CcrA from Bacteroides fragilis TAL3636. Antimicrob Agents Chemother 36:1155–1157. doi: 10.1128/AAC.36.5.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rasmussen BA, Gluzman Y, Tally FP. 1991. Escherichia coli chromosomal mutations that permit direct cloning of the Bacteroides fragilis metallo-β-lactamase gene, ccrA. Mol Microbiol 5:1211–1219. doi: 10.1111/j.1365-2958.1991.tb01895.x. [DOI] [PubMed] [Google Scholar]
- 93.Osano E, Arakawa Y, Wacharotayankun R, Ohta M, Horii T, Ito H, Yoshimura F, Kato N. 1994. Molecular characterization of an enterobacterial metallo beta-lactamase found in a clinical isolate of Serratia marcescens that shows imipenem resistance. Antimicrob Agents Chemother 38:71–78. doi: 10.1128/AAC.38.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Oelschlaeger P, Ai N, DuPrez KT, Welsh WJ, Toney JH. 2010. Evolving carbapenemases: can medicinal chemists advance one step ahead of the coming storm? J Med Chem 53:3013–3027. doi: 10.1021/jm9012938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Vessillier S, Docquier J-D, Rival S, Frere J-M, Galleni M, Amicosante G, Rossolini GM, Franceschini N. 2002. Overproduction and biochemical characterization of the Chryseobacterium meningosepticum BlaB metallo-β-lactamase. Antimicrob Agents Chemother 46:1921–1927. doi: 10.1128/AAC.46.6.1921-1927.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rossolini G, Franceschini N, Riccio M, Mercuri P, Perilli M, Galleni M, Frere J, Amicosante G. 1998. Characterization and sequence of the Chryseobacterium (Flavobacterium) meningosepticum carbapenemase: a new molecular class B β-lactamase showing a broad substrate profile. Biochem J 332:145–152. doi: 10.1042/bj3320145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yum JH, Lee EY, Hur S-H, Jeong SH, Lee H, Yong D, Chong Y, Lee E-W, Nordmann P, Lee K. 2010. Genetic diversity of chromosomal metallo-β-lactamase genes in clinical isolates of Elizabethkingia meningoseptica from Korea. J Microbiol 48:358–364. doi: 10.1007/s12275-010-9308-5. [DOI] [PubMed] [Google Scholar]
- 98.Lauretti L, Riccio ML, Mazzariol A, Cornaglia G, Amicosante G, Fontana R, Rossolini GM. 1999. Cloning and characterization of bla VIM, a new integron-borne metallo-β-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob Agents Chemother 43:1584–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bellais S, Léotard S, Poirel L, Naas T, Nordmann P. 1999. Molecular characterization of a carbapenem-hydrolyzing β-lactamase from Chryseobacterium (Flavobacterium) indologenes. FEMS Microbiol Lett 171:127–132. [DOI] [PubMed] [Google Scholar]
- 100.Bellais S, Girlich D, Karim A, Nordmann P. 2002. EBR-1, a novel Ambler subclass B1 β-lactamase from Empedobacter brevis. Antimicrob Agents Chemother 46:3223–3227. doi: 10.1128/AAC.46.10.3223-3227.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Toleman MA, Simm AM, Murphy TA, Gales AC, Biedenbach DJ, Jones RN, Walsh TR. 2002. Molecular characterization of SPM-1, a novel metallo-β-lactamase isolated in Latin America: report from the SENTRY antimicrobial surveillance programme. J Antimicrob Chemother 50:673–679. doi: 10.1093/jac/dkf210. [DOI] [PubMed] [Google Scholar]
- 102.Gales AC, Menezes LC, Silbert S, Sader HS. 2003. Dissemination in distinct Brazilian regions of an epidemic carbapenem-resistant Pseudomonas aeruginosa producing SPM metallo-β-lactamase. J Antimicrob Chemother 52:699–702. doi: 10.1093/jac/dkg416. [DOI] [PubMed] [Google Scholar]
- 103.Castanheira M, Toleman MA, Jones RN, Schmidt FJ, Walsh TR. 2004. Molecular characterization of a β-lactamase gene, blaGIM-1, encoding a new subclass of metallo-β-lactamase. Antimicrob Agents Chemother 48:4654–4661. doi: 10.1128/AAC.48.12.4654-4661.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lee K, Yum JH, Yong D, Lee HM, Kim HD, Docquier J-D, Rossolini GM, Chong Y. 2005. Novel acquired metallo-β-lactamase gene, blaSIM-1, in a class 1 integron from Acinetobacter baumannii clinical isolates from Korea. Antimicrob Agents Chemother 49:4485–4491. doi: 10.1128/AAC.49.11.4485-4491.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Poirel L, Héritier C, Nordmann P. 2005. Genetic and biochemical characterization of the chromosome-encoded class B β-lactamases from Shewanella livingstonensis (SLB-1) and Shewanella frigidimarina (SFB-1). J Antimicrob Chemother 55:680–685. doi: 10.1093/jac/dki065. [DOI] [PubMed] [Google Scholar]
- 106.Gudeta DD, Bortolaia V, Amos G, Wellington EMH, Brandt KK, Poirel L, Nielsen JB, Westh H, Guardabassi L. 2016. The soil microbiota harbors a diversity of carbapenem-hydrolyzing β-lactamases of potential clinical relevance. Antimicrob Agents Chemother 60:151–160. doi: 10.1128/AAC.01424-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Naas T, Bellais S, Nordmann P. 2003. Molecular and biochemical characterization of a carbapenem-hydrolysing β-lactamase from Flavobacterium johnsoniae. J Antimicrob Chemother 51:267–273. doi: 10.1093/jac/dkg069. [DOI] [PubMed] [Google Scholar]
- 108.Bellais S, Naas T, Nordmann P. 2002. Genetic and biochemical characterization of CGB-1, an Ambler class B carbapenem-hydrolyzing β-lactamase from Chryseobacterium gleum. Antimicrob Agents Chemother 46:2791–2796. doi: 10.1128/AAC.46.9.2791-2796.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mammeri H, Bellais S, Nordmann P. 2002. Chromosome-encoded β-lactamases TUS-1 and MUS-1 from Myroides odoratus and Myroides odoratimimus (formerly Flavobacterium odoratum), new members of the lineage of molecular subclass B1 metalloenzymes. Antimicrob Agents Chemother 46:3561–3567. doi: 10.1128/AAC.46.11.3561-3567.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Al-Bayssari C, Gupta SK, Dabboussi F, Hamze M, Rolain J-M. 2015. MUS-2, a novel variant of the chromosome-encoded β-lactamase MUS-1, from Myroides odoratimimus. New Microbes New Infect 7:67–71. doi: 10.1016/j.nmni.2015.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sekiguchi J, Morita K, Kitao T, Watanabe N, Okazaki M, Miyoshi-Akiyama T, Kanamori M, Kirikae T. 2008. KHM-1, a novel plasmid-mediated metallo-β-lactamase from a Citrobacter freundii clinical isolate. Antimicrob Agents Chemother 52:4194–4197. doi: 10.1128/AAC.01337-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Poirel L, Rodríguez-Martínez J-M, Al Naiemi N, Debets-Ossenkopp YJ, Nordmann P. 2010. Characterization of DIM-1, an integron-encoded metallo-β-lactamase from a Pseudomonas stutzeri clinical isolate in the Netherlands. Antimicrob Agents Chemother 54:2420–2424. doi: 10.1128/AAC.01456-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Nordmann P, Poirel L, Toleman MA, Walsh TR. 2011. Does broad-spectrum β-lactam resistance due to NDM-1 herald the end of the antibiotic era for treatment of infections caused by Gram-negative bacteria? J Antimicrob Chemother 66:689–692. doi: 10.1093/jac/dkq520. [DOI] [PubMed] [Google Scholar]
- 114.Hornsey M, Phee L, Wareham DW. 2011. A novel variant, NDM-5, of the New Delhi metallo-β-lactamase in a multidrug-resistant Escherichia coli ST648 isolate recovered from a patient in the United Kingdom. Antimicrob Agents Chemother 55:5952–5954. doi: 10.1128/AAC.05108-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Nordmann P, Poirel L, Walsh TR, Livermore DM. 2011. The emerging NDM carbapenemases. Trends Microbiol 19:588–595. doi: 10.1016/j.tim.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 116.Pfennigwerth N, Lange F, Belmar Campos C, Hentschke M, Gatermann SG, Kaase M. 2017. Genetic and biochemical characterization of HMB-1, a novel subclass B1 metallo-β-lactamase found in a Pseudomonas aeruginosa clinical isolate. J Antimicrob Chemother 72:1068–1073. doi: 10.1093/jac/dkw554. [DOI] [PubMed] [Google Scholar]
- 117.Pollini S, Maradei S, Pecile P, Olivo G, Luzzaro F, Docquier J-D, Rossolini GM. 2013. FIM-1, a new acquired metallo-β-lactamase from a Pseudomonas aeruginosa clinical isolate from Italy. Antimicrob Agents Chemother 57:410–416. doi: 10.1128/AAC.01953-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.El Salabi A, Borra PS, Toleman MA, Samuelsen Ø, Walsh TR. 2012. Genetic and biochemical characterization of a novel metallo-β-lactamase, TMB-1, from an Achromobacter xylosoxidans strain isolated in Tripoli, Libya. Antimicrob Agents Chemother 56:2241–2245. doi: 10.1128/AAC.05640-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Suzuki S, Matsui M, Suzuki M, Sugita A, Kosuge Y, Kodama N, Ichise Y, Shibayama K. 2013. Detection of Tripoli metallo-β-lactamase 2 (TMB-2), a variant of bla TMB-1, in clinical isolates of Acinetobacter spp. in Japan. J Antimicrob Chemother 68:1441–1442. doi: 10.1093/jac/dkt031. [DOI] [PubMed] [Google Scholar]
- 120.Gudeta DD, Bortolaia V, Pollini S, Docquier J-D, Rossolini GM, Amos GCA, Wellington EMH, Guardabassi L. 2016. Expanding the repertoire of carbapenem-hydrolyzing metallo-β-lactamases by functional metagenomic analysis of soil microbiota. Front Microbiol 7:1985. doi: 10.3389/fmicb.2016.01985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lin H-TV, Massam-Wu T, Lin C-P, Wang Y-JA, Shen Y-C, Lu W-J, Hsu P-H, Chen Y-H, Borges-Walmsley MI, Walmsley AR. 2017. The Vibrio cholerae var regulon encodes a metallo-β-lactamase and an antibiotic efflux pump, which are regulated by VarR, a LysR-type transcription factor. PLoS One 12:e0184255. doi: 10.1371/journal.pone.0184255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Massidda O, Rossolini GM, Satta G. 1991. The Aeromonas hydrophila cphA gene: molecular heterogeneity among class B metallo-beta-lactamases. J Bacteriol 173:4611–4617. doi: 10.1128/jb.173.15.4611-4617.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Segatore B, Massidda O, Satta G, Setacci D, Amicosante G. 1993. High specificity of cphA-encoded metallo-beta-lactamase from Aeromonas hydrophila AE036 for carbapenems and its contribution to beta-lactam resistance. Antimicrob Agents Chemother 37:1324–1328. doi: 10.1128/AAC.37.6.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Walsh TR, Gamblin S, Emery DC, MacGowan AP, Bennett PM. 1996. Enzyme kinetics and biochemical analysis of ImiS, the metallo-β-lactamase from Aeromonas sobria 163a. J Antimicrob Chemother 37:423–431. doi: 10.1093/jac/37.3.423. [DOI] [PubMed] [Google Scholar]
- 125.Walsh TR, Neville WA, Haran MH, Tolson D, Payne DJ, Bateson JH, MacGowan AP, Bennett PM. 1998. Nucleotide and amino acid sequences of the metallo-β-lactamase, ImiS, from Aeromonas veronii bv. sobria. Antimicrob Agents Chemother 42:436–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Niumsup P, Simm AM, Nurmahomed K, Walsh TR, Bennett PM, Avison MB. 2003. Genetic linkage of the penicillinase gene, amp, and blrAB, encoding the regulator of β-lactamase expression in Aeromonas spp. J Antimicrob Chemother 51:1351–1358. doi: 10.1093/jac/dkg247. [DOI] [PubMed] [Google Scholar]
- 127.Saavedra MJ, Peixe L, Sousa JC, Henriques I, Alves A, Correia A. 2003. Sfh-I, a subclass B2 metallo-β-lactamase from a Serratia fonticola environmental isolate. Antimicrob Agents Chemother 47:2330–2333. doi: 10.1128/AAC.47.7.2330-2333.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ullah JH, Walsh TR, Taylor IA, Emery DC, Verma CS, Gamblin SJ, Spencer J. 1998. The crystal structure of the L1 metallo-β-lactamase from Stenotrophomonas maltophilia at 1.7 Å resolution. J Mol Biol 284:125–136. doi: 10.1006/jmbi.1998.2148. [DOI] [PubMed] [Google Scholar]
- 129.Yang Z, Liu W, Cui Q, Niu WK, Li H, Zhao XN, Wei X, Wang XS, Huang SM, Dong DR. 2014. Prevalence and detection of Stenotrophomonas maltophilia carrying metallo-β-lactamase blaL1 in Beijing, China. Front Microbiol 5:692. doi: 10.3389/fmicb.2014.00692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Saino Y, Kobayashi F, Inoue M, Mitsuhashi S. 1982. Purification and properties of inducible penicillin beta-lactamase isolated from Pseudomonas maltophilia. Antimicrob Agents Chemother 22:564–570. doi: 10.1128/AAC.22.4.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Boschi L, Mercuri PS, Riccio ML, Amicosante G, Galleni M, Frère J-M, Rossolini GM. 2000. The Legionella (Fluoribacter) gormanii metallo-β-lactamase: a new member of the highly divergent lineage of molecular-subclass B3 β-lactamases. Antimicrob Agents Chemother 44:1538–1543. doi: 10.1128/AAC.44.6.1538-1543.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bellais S, Aubert D, Naas T, Nordmann P. 2000. Molecular and biochemical heterogeneity of class B carbapenem-hydrolyzing β-lactamases in Chryseobacterium meningosepticum. Antimicrob Agents Chemother 44:1878–1886. doi: 10.1128/AAC.44.7.1878-1886.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Rossolini GM, Condemi MA, Pantanella F, Docquier J-D, Amicosante G, Thaller MC. 2001. Metallo-β-lactamase producers in environmental microbiota: new molecular class B enzyme in Janthinobacterium lividum. Antimicrob Agents Chemother 45:837–844. doi: 10.1128/AAC.45.3.837-844.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Simm AM, Higgins CS, Pullan ST, Avison MB, Niumsup P, Erdozain O, Bennett PM, Walsh TR. 2001. A novel metallo-β-lactamase, Mbl1b, produced by the environmental bacterium Caulobacter crescentus 1. FEBS Lett 509:350–354. doi: 10.1016/S0014-5793(01)03152-0. [DOI] [PubMed] [Google Scholar]
- 135.Docquier J-D, Pantanella F, Giuliani F, Thaller MC, Amicosante G, Galleni M, Frère J-M, Bush K, Rossolini GM. 2002. CAU-1, a subclass B3 metallo-β-lactamase of low substrate affinity encoded by an ortholog present in the Caulobacter crescentus chromosome. Antimicrob Agents Chemother 46:1823–1830. doi: 10.1128/AAC.46.6.1823-1830.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Stoczko M, Frère J-M, Rossolini GM, Docquier J-D. 2006. Postgenomic scan of metallo-β-lactamase homologues in rhizobacteria: identification and characterization of BJP-1, a subclass B3 ortholog from Bradyrhizobium japonicum. Antimicrob Agents Chemother 50:1973–1981. doi: 10.1128/AAC.01551-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Leiros H-KS, Borra PS, Brandsdal BO, Edvardsen KSW, Spencer J, Walsh TR, Samuelsen Ø. 2012. Crystal structure of the mobile metallo-β-lactamase AIM-1 from Pseudomonas aeruginosa: insights into antibiotic binding and the role of Gln157. Antimicrob Agents Chemother 56:4341–4353. doi: 10.1128/AAC.00448-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Yong D, Toleman MA, Bell J, Ritchie B, Pratt R, Ryley H, Walsh TR. 2012. Genetic and biochemical characterization of an acquired subgroup B3 metallo-β-lactamase gene, blaAIM-1, and its unique genetic context in Pseudomonas aeruginosa from Australia. Antimicrob Agents Chemother 56:6154–6159. doi: 10.1128/AAC.05654-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Stoczko M, Frère J-M, Rossolini GM, Docquier J-D. 2008. Functional diversity among metallo-β-lactamases: characterization of the CAR-1 enzyme of Erwinia carotovora. Antimicrob Agents Chemother 52:2473–2479. doi: 10.1128/AAC.01062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Thaller MC, Borgianni L, Di Lallo G, Chong Y, Lee K, Dajcs J, Stroman D, Rossolini GM. 2011. Metallo-β-lactamase production by Pseudomonas otitidis: a species-related trait. Antimicrob Agents Chemother 55:118–123. doi: 10.1128/AAC.01062-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Borgianni L, De Luca F, Thaller MC, Chong Y, Rossolini GM, Docquier J-D. 2015. Biochemical characterization of the POM-1 metallo-β-lactamase from Pseudomonas otitidis. Antimicrob Agents Chemother 59:1755–1758. doi: 10.1128/AAC.03843-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Suzuki M, Suzuki S, Matsui M, Hiraki Y, Kawano F, Shibayama K. 2014. A subclass B3 metallo-β-lactamase found in Pseudomonas alcaligenes. J Antimicrob Chemother 69:1430–1432. doi: 10.1093/jac/dkt498. [DOI] [PubMed] [Google Scholar]
- 143.Gudeta DD, Pollini S, Docquier J-D, Bortolaia V, Rossolini GM, Guardabassi L. 2016. Biochemical characterization of CPS-1, a subclass B3 metallo-β-lactamase from a Chryseobacterium piscium soil isolate. Antimicrob Agents Chemother 60:1869–1873. doi: 10.1128/AAC.01924-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Salimraj R, Zhang L, Hinchliffe P, Wellington EMH, Brem J, Schofield CJ, Gaze WH, Spencer J. 2016. Structural and biochemical characterization of Rm3, a subclass B3 metallo-β-lactamase identified from a functional metagenomic study. Antimicrob Agents Chemother 60:5828–5840. doi: 10.1128/AAC.00750-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Allen HK, Moe LA, Rodbumrer J, Gaarder A, Handelsman J. 2009. Functional metagenomics reveals diverse β-lactamases in a remote Alaskan soil. ISME J 3:243–251. doi: 10.1038/ismej.2008.86. [DOI] [PubMed] [Google Scholar]
- 146.Wachino J, Yoshida H, Yamane K, Suzuki S, Matsui M, Yamagishi T, Tsutsui A, Konda T, Shibayama K, Arakawa Y. 2011. SMB-1, a novel subclass B3 metallo-β-lactamase, associated with ISCR1 and a class 1 integron, from a carbapenem-resistant Serratia marcescens clinical isolate. Antimicrob Agents Chemother 55:5143–5149. doi: 10.1128/AAC.05045-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Girlich D, Poirel L, Nordmann P. 2012. Diversity of naturally occurring Ambler class B metallo-β-lactamases in Erythrobacter spp. J Antimicrob Chemother 67:2661–2664. doi: 10.1093/jac/dks289. [DOI] [PubMed] [Google Scholar]
- 148.Calver AD, Walsh NS, Quinn PF, Baran C, Lonergan V, Singh KP, Orzolek WS. 1997. Dosing of amoxicillin/clavulanate given every 12 hours is as effective as dosing every 8 hours for treatment of lower respiratory tract infection. Lower Respiratory Tract Infection Collaborative Study Group. Clin Infect Dis 024:570–574. [DOI] [PubMed] [Google Scholar]
- 149.McKinnon PS, Neuhauser MM. 1999. Efficacy and cost of ampicillin-sulbactam and ticarcillin-clavulanate in the treatment of hospitalized patients with bacterial infections. Pharmacother J Hum Pharmacol Drug Ther 19:724–733. doi: 10.1592/phco.19.9.724.31537. [DOI] [PubMed] [Google Scholar]
- 150.Li J, Lu Y, Hou J, Chen Y, Miao J, Jia Y, Hou J, Zhang X, Chen D, Hu W. 1997. Sulbactam/cefoperazone versus cefotaxime for the treatment of moderate-to-severe bacterial infections: results of a randomized, controlled clinical trial. Clin Infect Dis 24:498–505. doi: 10.1093/clinids/24.3.498. [DOI] [PubMed] [Google Scholar]
- 151.Sanders WE Jr, Sanders CC. 1996. Piperacillin/tazobactam: a critical review of the evolving clinical literature. Clin Infect Dis 22:107–123. doi: 10.1093/clinids/22.1.107. [DOI] [PubMed] [Google Scholar]
- 152.Sorbera M, Chung E, Ho CW, Marzella N. 2014. Ceftolozane/tazobactam: a new option in the treatment of complicated Gram-negative infections. Pharm Ther 39:825. [PMC free article] [PubMed] [Google Scholar]
- 153.Mazuski JE, Gasink LB, Armstrong J, Broadhurst H, Stone GG, Rank D, Llorens L, Newell P, Pachl J. 2016. Efficacy and safety of ceftazidime-avibactam plus metronidazole versus meropenem in the treatment of complicated intra-abdominal infection: results from a randomized, controlled, double-blind, phase 3 program. Clin Infect Dis 62:1380–1389. doi: 10.1093/cid/ciw133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wiskirchen DE, Crandon JL, Furtado GH, Williams G, Nicolau DP. 2011. In vivo efficacy of a human-simulated regimen of ceftaroline combined with NXL104 against extended-spectrum-β-lactamase (ESBL)-producing and non-ESBL-producing Enterobacteriaceae. Antimicrob Agents Chemother 55:3220–3225. doi: 10.1128/AAC.00024-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Biedenbach DJ, Kazmierczak K, Bouchillon SK, Sahm DF, Bradford PA. 2015. In vitro activity of aztreonam-avibactam against a global collection of Gram-negative pathogens from 2012-2013. Antimicrob Agents Chemother 59:4239–4248. doi: 10.1128/AAC.00206-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Hirsch EB, Ledesma KR, Chang K-T, Schwartz MS, Motyl MR, Tam VH. 2012. In vitro activity of MK-7655, a novel β-lactamase inhibitor, in combination with imipenem against carbapenem-resistant Gram-negative bacteria. Antimicrob Agents Chemother 56:3753–3757. doi: 10.1128/AAC.05927-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Lapuebla A, Abdallah M, Olafisoye O, Cortes C, Urban C, Quale J, Landman D. 2015. Activity of meropenem combined with RPX7009, a novel β-lactamase inhibitor, against Gram-negative clinical isolates in New York City. Antimicrob Agents Chemother 59:4856–4860. doi: 10.1128/AAC.00843-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Wenzler E, Gotfried MH, Loutit JS, Durso S, Griffith DC, Dudley MN, Rodvold KA. 2015. Meropenem-RPX7009 concentrations in plasma, epithelial lining fluid, and alveolar macrophages of healthy adult subjects. Antimicrob Agents Chemother 59:7232–7239. doi: 10.1128/AAC.01713-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Goldstein EJC, Citron DM, Tyrrell KL, Merriam CV. 2013. In vitro activity of biapenem plus RPX7009, a carbapenem combined with a serine β-lactamase inhibitor against anaerobic bacteria. Antimicrob Agents Chemother 57:2620–2630. doi: 10.1128/AAC.02418-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Durand-Réville TF, Guler S, Comita-Prevoir J, Chen B, Bifulco N, Huynh H, Lahiri S, Shapiro AB, McLeod SM, Carter NM. 2017. ETX2514 is a broad-spectrum β-lactamase inhibitor for the treatment of drug-resistant Gram-negative bacteria including Acinetobacter baumannii. Nat Microbiol 2:17104. doi: 10.1038/nmicrobiol.2017.104. [DOI] [PubMed] [Google Scholar]
- 161.Sader HS, Castanheira M, Huband M, Jones RN, Flamm RK. 2017. WCK 5222 (cefepime-zidebactam) antimicrobial activity tested against clinical isolates of Gram-negative bacteria collected worldwide (2015). Antimicrob Agents Chemother 61:1373–1385. doi: 10.1128/AAC.00072-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Toney JH, Hammond GG, Fitzgerald PMD, Sharma N, Balkovec JM, Rouen GP, Olson SH, Hammond ML, Greenlee ML, Gao Y-D. 2001. Succinic acids as potent inhibitors of plasmid-borne IMP-1 metallo-β-lactamase. J Biol Chem 276:31913–31918. doi: 10.1074/jbc.M104742200. [DOI] [PubMed] [Google Scholar]
- 163.Moloughney JG, Thomas DJ, Toney JH. 2005. Novel IMP-1 metallo-β-lactamase inhibitors can reverse meropenem resistance in Escherichia coli expressing IMP-1. FEMS Microbiol Lett 243:65–71. doi: 10.1016/j.femsle.2004.11.042. [DOI] [PubMed] [Google Scholar]
- 164.Olsen L, Jost S, Adolph H-W, Pettersson I, Hemmingsen L, Jørgensen FS. 2006. New leads of metallo-β-lactamase inhibitors from structure-based pharmacophore design. Bioorg Med Chem 14:2627–2635. doi: 10.1016/j.bmc.2005.11.046. [DOI] [PubMed] [Google Scholar]
- 165.Hiraiwa Y, Morinaka A, Fukushima T, Kudo T. 2009. metallo-β-lactamase inhibitory activity of phthalic acid derivatives. Bioorg Med Chem Lett 19:5162–5165. doi: 10.1016/j.bmcl.2009.07.018. [DOI] [PubMed] [Google Scholar]
- 166.Dmitrienko GI, Viswanatha T, Johnson JW, Ramadhar TR. September 2009. Inhibitors of class B and class D β-lactamases. Patent WO 2009/114921 A1.
- 167.Faridoon, Hussein WM, Vella P, Islam NU, Ollis DL, Schenk G, McGeary RP. 2012. 3-Mercapto-1,2,4-triazoles and N-acylated thiosemicarbazides as metallo-β-lactamase inhibitors. Bioorg Med Chem Lett 22:380–386. doi: 10.1016/j.bmcl.2011.10.116. [DOI] [PubMed] [Google Scholar]
- 168.Feng L, Yang K-W, Zhou L-S, Xiao J-M, Yang X, Zhai L, Zhang Y-L, Crowder MW. 2012. N-heterocyclic dicarboxylic acids: broad-spectrum inhibitors of metallo-β-lactamases with co-antibacterial effect against antibiotic-resistant bacteria. Bioorg Med Chem Lett 22:5185–5189. doi: 10.1016/j.bmcl.2012.06.074. [DOI] [PubMed] [Google Scholar]
- 169.Toney JH, Cleary KA, Hammond GG, Yuan X, May WJ, Hutchins SM, Ashton WT, Vanderwall DE. 1999. Structure-activity relationships of biphenyl tetrazoles as metallo-β-lactamase inhibitors. Bioorg Med Chem Lett 9:2741–2746. doi: 10.1016/S0960-894X(99)00458-8. [DOI] [PubMed] [Google Scholar]
- 170.Lienard BMR, Horsfall LE, Galleni M, Frère J-M, Schofield CJ. 2007. Inhibitors of the FEZ-1 metallo-β-lactamase. Bioorg Med Chem Lett 17:964–968. doi: 10.1016/j.bmcl.2006.11.053. [DOI] [PubMed] [Google Scholar]
- 171.Gilpin ML, Fulston M, Payne D, Cramp R, Hood I. 1995. Isolation and structure determination of two novel phenazines from a Streptomyces with inhibitory activity against metallo-enzymes, including metallo-β-lactamase. J Antibiot (Tokyo) 48:1081–1085. doi: 10.7164/antibiotics.48.1081. [DOI] [PubMed] [Google Scholar]
- 172.Walter MW, Felici A, Galleni M, Soto RP, Adlington RM, Baldwin JE, Frère J-M, Gololobov M, Schofield CJ. 1996. Trifluoromethyl alcohol and ketone inhibitors of metallo-β-lactamases. Bioorg Med Chem Lett 6:2455–2458. doi: 10.1016/0960-894X(96)00453-2. [DOI] [Google Scholar]
- 173.Tsang WY, Dhanda A, Schofield CJ, Page MI. 2004. Kinetics and mechanisms of hydrolysis and aminolysis of thioxocephalosporins. J Org Chem 69:339–344. doi: 10.1021/jo035471t. [DOI] [PubMed] [Google Scholar]
- 174.Thomas PW, Cammarata M, Brodbelt JS, Fast W. 2014. Covalent inhibition of New Delhi metallo-β-lactamase-1 (NDM-1) by cefaclor. Chembiochem 15:2541–2548. doi: 10.1002/cbic.201402268. [DOI] [PubMed] [Google Scholar]
- 175.Zervosen A, Valladares MH, Devreese B, Prosperi-Meys C, Adolph H, Mercuri PS, Vanhove M, Amicosante G, Van Beeumen J, Frère J. 2001. Inactivation of Aeromonas hydrophila metallo-β-lactamase by cephamycins and moxalactam. FEBS J 268:3840–3850. [DOI] [PubMed] [Google Scholar]
- 176.Sanschagrin F, Levesque RC. 2005. A specific peptide inhibitor of the class B metallo-beta-lactamase L-1 from Stenotrophomonas maltophilia identified using phage display. J Antimicrob Chemother 55:252–255. doi: 10.1093/jac/dkh550. [DOI] [PubMed] [Google Scholar]
- 177.Sun Q, Law A, Crowder MW, Geysen HM. 2006. Homo-cysteinyl peptide inhibitors of the L1 metallo-beta-lactamase, and SAR as determined by combinatorial library synthesis. Bioorg Med Chem Lett 16:5169–5175. doi: 10.1016/j.bmcl.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 178.Minond D, Saldanha SA, Subramaniam P, Spaargaren M, Spicer T, Fotsing JR, Weide T, Fokin VV, Sharpless KB, Galleni M, Bebrone C, Lassaux P, Hodder P. 2009. Inhibitors of VIM-2 by screening pharmacologically active and click-chemistry compound libraries. Bioorg Med Chem 17:5027–5037. doi: 10.1016/j.bmc.2009.05.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Weide T, Saldanha SA, Minond D, Spicer TP, Fotsing JR, Spaargaren M, Frère JM, Bebrone C, Sharpless KB, Hodder PS, Fokin VV. 2010. NH-1,2,3-triazole inhibitors of the VIM-2 metallo-beta-lactamase. ACS Med Chem Lett 1:150–154. doi: 10.1021/ml900022q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Vella P, Hussein WM, Leung EWW, Clayton D, Ollis DL, Mitić N, Schenk G, McGeary RP. 2011. The identification of new metallo-beta-lactamase inhibitor leads from fragment-based screening. Bioorg Med Chem Lett 21:3282–3285. doi: 10.1016/j.bmcl.2011.04.027. [DOI] [PubMed] [Google Scholar]
- 181.Kim SK, Sims CL, Wozniak SE, Drude SH, Whitson D, Shaw RW. 2009. Antibiotic resistance in bacteria: novel metalloenzyme inhibitors. Chem Biol Drug Des 74:343–348. doi: 10.1111/j.1747-0285.2009.00879.x. [DOI] [PubMed] [Google Scholar]
- 182.Ouyang X, Chang Y-N, Yang K-W, Wang W-M, Bai J-J, Wang J-W, Zhang Y, Wang S-Y, Xie B-B, Wang L. 2017. DNA nanoribbon as a potent inhibitor of metallo-β-lactamases. Chem Commun (Camb) 53:8878–8881. doi: 10.1039/C7CC04483F. [DOI] [PubMed] [Google Scholar]
- 183.Simm AM, Loveridge EJ, Crosby J, Avison MB, Walsh TR, Bennett PM. 2005. Bulgecin A: a novel inhibitor of binuclear metallo-beta-lactamases. Biochem J 387:585–590. doi: 10.1042/BJ20041542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Siemann S, Evanoff DP, Marrone L, Clarke AJ, Viswanatha T, Dmitrienko GI. 2002. N-arylsulfonyl hydrazones as inhibitors of IMP-1 metallo-β-lactamase. Antimicrob Agents Chemother 46:2450–2457. doi: 10.1128/AAC.46.8.2450-2457.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Koteva K, King AM, Capretta A, Wright GD. 2016. Total synthesis and activity of the metallo-β-lactamase inhibitor aspergillomarasmine A. Angew Chem 128:2250–2252. doi: 10.1002/ange.201510057. [DOI] [PubMed] [Google Scholar]
- 186.Hammond GG, Huber JL, Greenlee ML, Laub JB, Young K, Silver LL, Balkovec JM, Pryor KD, Wu JK, Leiting B, Pompliano DL, Toney JH. 1999. Inhibition of IMP-1 metallo-β-lactamase and sensitization of IMP-1-producing bacteria by thioester derivatives. FEMS Microbiol Lett 179:289–296. [DOI] [PubMed] [Google Scholar]
- 187.Klingler F-M, Wichelhaus TA, Frank D, Cuesta-Bernal J, El-Delik J, Müller HF, Sjuts H, Göttig S, Koenigs A, Pos KM. 2015. Approved drugs containing thiols as inhibitors of metallo-β-lactamases: strategy to combat multidrug-resistant bacteria. J Med Chem 58:3626–3630. doi: 10.1021/jm501844d. [DOI] [PubMed] [Google Scholar]
- 188.Kurosaki H, Yamaguchi Y, Higashi T, Soga K, Matsueda S, Yumoto H, Misumi S, Yamagata Y, Arakawa Y, Goto M. 2005. Irreversible Inhibition of metallo-β-lactamase (IMP-1) by 3-(3-mercaptopropionylsulfanyl) propionic acid pentafluorophenyl ester. Angew Chem Int Ed Engl 44:3861–3864. doi: 10.1002/anie.200500835. [DOI] [PubMed] [Google Scholar]
- 189.Payne DJ, Bateson JH, Gasson BC, Khushi T, Proctor D, Pearson SC, Reid R. 1997. Inhibition of metallo-β-lactamases by a series of thiol ester derivatives of mercaptophenylacetic acid. FEMS Microbiol Lett 157:171–175. doi: 10.1111/j.1574-6968.1997.tb12769.x. [DOI] [PubMed] [Google Scholar]
- 190.Concha NO, Janson CA, Rowling P, Pearson S, Cheever CA, Clarke BP, Lewis C, Galleni M, Frere J-M, Payne DJ. 2000. Crystal structure of the IMP-1 metallo β-lactamase from Pseudomonas aeruginosa and its complex with a mercaptocarboxylate inhibitor: binding determinants of a potent, broad-spectrum inhibitor. Biochemistry 39:4288–4298. doi: 10.1021/bi992569m. [DOI] [PubMed] [Google Scholar]
- 191.Lassaux P, Hamel M, Gulea M, Delbrück H, Mercuri PS, Horsfall L, Dehareng D, Kupper M, Frere J-M, Hoffmann K. 2010. Mercaptophosphonate compounds as broad-spectrum inhibitors of the metallo-β-lactamases. J Med Chem 53:4862–4876. doi: 10.1021/jm100213c. [DOI] [PubMed] [Google Scholar]
- 192.García-Sáez I, Hopkins J, Papamicael C, Franceschini N, Amicosante G, Rossolinill GM, Galleni M, Frère JM, Dideberg O. 2003. The 1.5-Å structure of Chryseobacterium meningosepticum zinc β-lactamase in complex with the inhibitor, d-captopril. J Biol Chem 278:23868–23873. doi: 10.1074/jbc.M301062200. [DOI] [PubMed] [Google Scholar]
- 193.King DT, Worrall LJ, Gruninger R, Strynadka NCJ. 2012. New Delhi metallo-β-lactamase: structural insights into β-lactam recognition and inhibition. J Am Chem Soc 134:11362–11365. doi: 10.1021/ja303579d. [DOI] [PubMed] [Google Scholar]
- 194.Damblon C, Jensen M, Ababou A, Barsukov I, Papamicael C, Schofield CJ, Olsen L, Bauer R, Roberts GCK. 2003. The inhibitor thiomandelic acid binds to both metal ions in metallo-β-lactamase and induces positive cooperativity in metal binding. J Biol Chem 278:29240–29251. doi: 10.1074/jbc.M301562200. [DOI] [PubMed] [Google Scholar]
- 195.Siemann S, Clarke AJ, Viswanatha T, Dmitrienko GI. 2003. Thiols as classical and slow-binding inhibitors of IMP-1 and other binuclear metallo-β-lactamases. Biochemistry 42:1673–1683. doi: 10.1021/bi027072i. [DOI] [PubMed] [Google Scholar]
- 196.Kurosaki H, Yamaguchi Y, Yasuzawa H, Jin W, Yamagata Y, Arakawa Y. 2006. Probing, inhibition, and crystallographic characterization of metallo-beta-lactamase (IMP-1) with fluorescent agents containing dansyl and thiol groups. ChemMedChem 1:969–972. doi: 10.1002/cmdc.200600115. [DOI] [PubMed] [Google Scholar]
- 197.Hinchliffe P, González MM, Mojica MF, González JM, Castillo V, Saiz C, Kosmopoulou M, Tooke CL, Llarrull LI, Mahler G. 2016. Cross-class metallo-β-lactamase inhibition by bisthiazolidines reveals multiple binding modes. Proc Natl Acad Sci U S A 113:E3745–E3754. doi: 10.1073/pnas.1601368113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Whelan S, Goldman N. 2001. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol 18:691–699. [DOI] [PubMed] [Google Scholar]