Group B streptococcus (GBS) is a bacterium associated with various diseases in humans and animals. This study describes the development of a strategy to label and visualize GBS using a fluorescence biomarker, termed GFPmut3. We show that this biomarker can be successfully applied to track the growth of bacteria in liquid medium, and it enables the detailed visualization of GBS in the context of live human cells in real-time microscopic analysis. The system for GFPmut3 expression in GBS described in this study provides a new tool for the visualization of this organism in diverse research applications.
KEYWORDS: group B streptococcus, Streptococcus agalactiae, green fluorescent protein, fluorescence detection, bacterial adhesion, CovS/CovR, uroepithelium
ABSTRACT
Group B streptococcus (GBS) is a Gram-positive bacterium associated with various diseases in humans and animals. Many studies have examined GBS physiology, virulence, and microbe-host interactions using diverse imaging approaches, including fluorescence microscopy. Strategies to label and visualize GBS using fluorescence biomarkers have been limited to antibody-based methods or nonspecific stains that bind DNA or protein; an effective plasmid-based system to label GBS with a fluorescence biomarker would represent a useful visualization tool. In this study, we developed and validated a green fluorescent protein (GFP)-variant-expressing plasmid, pGU2664, which can be applied as a marker to visualize GBS in experimental studies. The synthetic constitutively active CP25 promoter drives strong and stable expression of the GFPmut3 biomarker in GBS strains carrying pGU2664. GBS maintains GFPmut3 activity at different phases of growth. The application of fluorescence polarization enables easy discrimination of GBS GFPmut3 activity from the autofluorescence of culture media commonly used to grow GBS. Differential interference contrast microscopy, in combination with epifluorescence microscopy to detect GFPmut3 in GBS, enabled visualization of bacterial attachment to live human epithelial cells in real time. Plasmid pGU2664 was also used to visualize phenotypic differences in the adherence of wild-type GBS and an isogenic gene-deficient mutant strain lacking CovR (the control of virulence regulator) in adhesion assays. The system for GFPmut3 expression in GBS described in this study provides a new tool for the visualization of this organism in diverse research applications. We discuss the advantages and consider the limitations of this fluorescent biomarker system developed for GBS.
IMPORTANCE Group B streptococcus (GBS) is a bacterium associated with various diseases in humans and animals. This study describes the development of a strategy to label and visualize GBS using a fluorescence biomarker, termed GFPmut3. We show that this biomarker can be successfully applied to track the growth of bacteria in liquid medium, and it enables the detailed visualization of GBS in the context of live human cells in real-time microscopic analysis. The system for GFPmut3 expression in GBS described in this study provides a new tool for the visualization of this organism in diverse research applications.
INTRODUCTION
Streptococcus agalactiae (also known as group B streptococcus [GBS]) is a Gram-positive bacterium associated with various infections and diseases in humans and animals (1). Historically, GBS was a prominent veterinary pathogen of bovine mastitis (2); however, beginning in 1948, extensive use of tetracycline facilitated the transmission of distinct clones of GBS into humans (3), and now, GBS is a frequent colonizer of the gastrointestinal and urogenital tracts in healthy adults, with carriage rates of up to 30% (4–6). GBS is a major cause of neonatal disease related to vertical transmission of the bacteria from colonized mothers to infants. GBS-associated morbidities in pregnancy include urinary tract infections (UTIs), endometritis, chorioamnionitis, and sepsis (7). In neonates, GBS is commonly associated with sepsis and meningitis (1). Epidemiologically, GBS is also associated with significant morbidity and mortality in nonpregnant adults and elderly individuals, in whom GBS causes various skin and soft tissue infections, UTIs, pneumonia, arthritis, and sepsis (8). Research to develop a vaccine to prevent GBS infection is ongoing (9).
The development of various experimental models of GBS disease has enabled studies of the mechanisms of host colonization by GBS and virulence. Visualization of the bacteria by the application of microscopy approaches represents a key tool for studying GBS. Diverse imaging approaches, including confocal and epifluorescence (10–13), scanning electron, transmission electron, and immunoelectron (14–17), atomic force (18–21), and bright-field (22, 23) microscopy have enabled detailed investigations of host-microbe interactions, bacterial virulence strategies, and basic mechanisms of GBS physiology, including cell segregation processes (24). For example, numerous studies have used microscopic visualization techniques to study the effects of the GBS global virulence regulatory system termed CovS/CovR (23, 25, 26). Despite significant progress in understanding the mechanisms of host colonization by GBS, research advances in applying strategies to label and visualize GBS using fluorescent biomarkers have been principally focused on the development and application of DNA stains (which are often used to assess bacterial viability), nonspecific protein stains, and antibody-based immunostaining methods.
A common method used to label bacteria without the need for immunostaining exploits the expression of fluorescent proteins, such as green fluorescent protein (GFP). GFP in particular has been used extensively to successfully label a variety of low-GC-content Gram-positive bacteria (27–31) and has increased our understanding of bacterial cell biology and physiology (32–34). GFP exhibits intrinsic fluorescence; however, several studies have developed variants of GFP that offer several advantages, including increased fluorescence intensity (35–38). The so-called GFPmut3 variant exhibits 20-fold higher fluorescence intensity than wild-type (WT) GFP (39), and it is considered to be optimal for the expression of GFP in bacteria. The benefits of using fluorescently labeled bacteria include easy and rapid identification of bacteria in complex samples, the potential for tracking bacteria in vivo in live animals, the ability to explore spatial arrangements and subcellular locations of bacteria within eukaryotic host cells, and the potential to isolate and analyze host cells that carry fluorescent bacteria (40). Individual colonies of nonimmunostained fluorescent bacteria can easily be discerned from nonfluorescent bacteria on agar (30), and GFP is particularly useful as a fluorescent marker in fusion constructs to enable the monitoring of bacterial cellular processes (41). Thus, GFP is a versatile and widely used fluorescent marker for the study of bacterial physiology. Stable and high-level expression of a vector-borne fluorescent protein, such as GFP, in GBS would offer a useful tool with which to visualize this organism in various experimental systems.
Here, we developed a plasmid vector designed to express the fluorescent protein reporter gene gfpmut3 under the control of the synthetic, constitutively active CP25 promoter (42). We established the expression of GFPmut3 in GBS using plasmid pGU2664, which encodes spectinomycin resistance, and show its application as a reporter in bacterial growth assays. Further, we demonstrate the utility of fluorescence polarization methodology as a tool to discriminate between fluorescent signals from GBS (pGU2664) and autofluorescence that is encountered in growth media commonly used to culture GBS. Finally, we show that GFPmut3 fluorescence afforded by plasmid pGU2664 in GBS offers a useful approach for visualizing the bacteria in conjunction with host cells using a combination of differential interference and epifluorescence microscopy.
RESULTS
Plasmid stability under nonselective conditions.
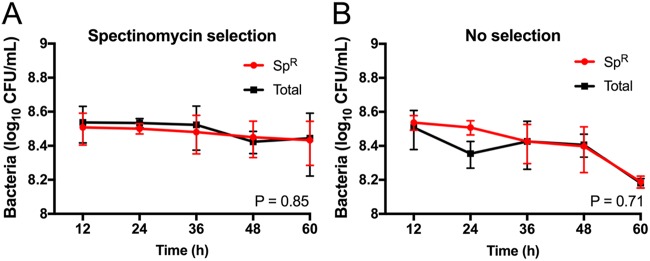
Using electroporation, we introduced GFPmut3 plasmid pGU2664 into GBS 874391 to create GBS strain GU2666 (GFP+), and we used the control non-GFPmut3 vector pDL278 to create GBS strain GU2672 (GFP−). To assess the relative stability of plasmid pGU2664 in GBS grown under nonselective conditions, we performed successive growth assays using liquid culture media and analyzed serial subcultures of GBS GU2666 grown with or without antibiotics for 100 generations (equivalent to 5 serial culture passages). We enumerated the bacteria at each point of subculture by quantifying colonies on selective and nonselective agar and compared the numbers of spectinomycin-resistant (Spr) cells to the total number of cells present. There was no significant difference (P = 0.71) between the total CFU and Spr CFU in serial cultures that were grown in the absence of antibiotic selective pressure (i.e., that did not select for the plasmid). Indeed, we observed that the majority of GBS GU2666 cells were Spr after at least 100 generations of growth, regardless of the presence of selection pressure (Fig. 1). These data show that in the absence of selection pressure, plasmid pGU2664 is stable in GBS under the growth conditions tested.
FIG 1.
Stability of plasmid pGU2664 in the absence of selection. (A and B) Numbers of Spr GBS (red) and the total numbers of GBS (black) recovered from growth conditions with (A) and without (B) antibiotic selection, respectively. Cells were diluted 1:1,000 into fresh THB medium (±Sp) and grown for 12 h in 5 successive culture passages, representing approximately 100 generations. The results presented are averages of the results from three independent experiments compared using AUC Student's t test, with bars showing the standard error.
GFP expression in GBS at the single-cell level.
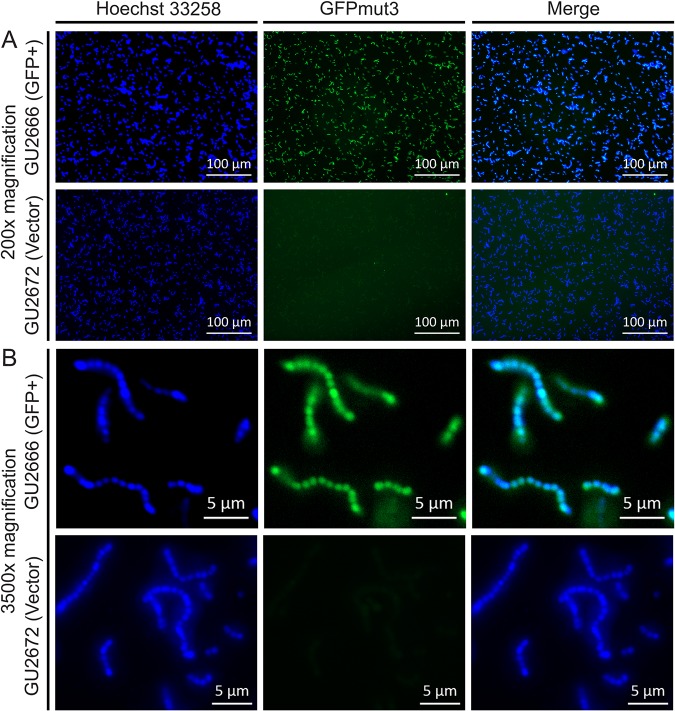
Overnight cultures of GBS GU2666 containing pGU2664 were analyzed using fluorescence microscopy to visualize GFPmut3 expression in GBS. We observed bright fluorescence in all GBS GU2666 cells examined under low-power (×200) and high-power (×3,500) magnification (Fig. 2A and B). GBS GU2672 containing the control vector pDL278 exhibited minimal fluorescence above background (Fig. 2A and B). Together, these data show that plasmid pGU2664 provides bright fluorescence to GBS that is equivalent among individual cells.
FIG 2.
Single-cell GFPmut3 fluorescence in GBS. Bright GFPmut3-borne fluorescence in GU2666 (GFP+) containing plasmid pGU2664 is shown above background fluorescence of GU2672 (GFP−) carrying control vector pDL278. The DNA stain Hoechst 33258, with excitation/emission maxima of 352/461 nm, compared to 501/511 nm of GFPmut3, shows the presence of cells. (A and B) Low-power (×200) (A) and high-power (×3,500) (B) magnifications, respectively.
GFP expression in GBS at the population level.
To evaluate populations of GBS grown as colonies on agar, we performed microscopy using an Olympus SZX16 stereomicroscope (Olympus Australia Pty Ltd.). Visualization of colonies showed that plasmid pGU2664 afforded stable and high-level fluorescence in GBS GU2666 compared to that in GBS GU2672 containing the empty vector control (Fig. 3). Interestingly, Escherichia coli DH5α containing pGU2664 exhibited sparse and irregular patterns of GFP expression (data not shown), consistent with low activity (at the population level) of the CP25 promoter in E. coli, as previously reported (42).
FIG 3.
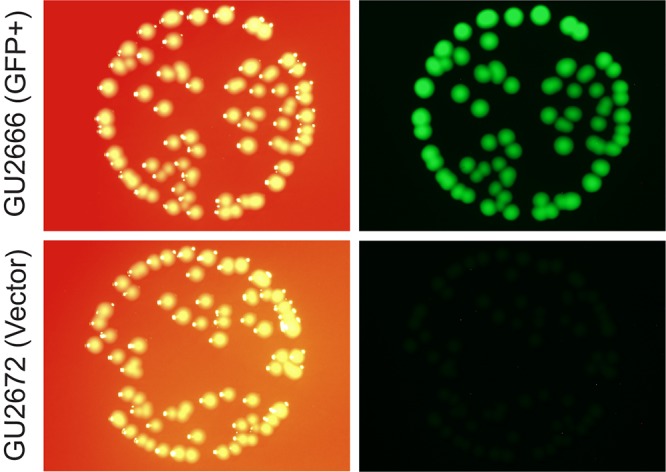
Population-level GFPmut3 fluorescence in GBS colonies. Bright GFPmut3-borne fluorescence in colonies of GBS strain GU2666 (GFP+) containing plasmid pGU2664 is shown, compared to background fluorescence of strain GU2672 (GFP−), carrying control empty vector pDL278. GBS colonies were grown on TH agar and imaged as described in Materials and Methods.
To assess the burden of pGU2664 expressing GFPmut3 on the physiology of GBS in more detail, we measured the growth of GBS 874391 by monitoring cell densities (using optical density at 600 nm [OD600]) in liquid culture in vitro while simultaneously monitoring GFPmut3 fluorescence intensity in a chemically defined medium (CDM) (43). The growth rates of GBS with plasmid pGU2664 were identical to those of GBS containing the empty vector pDL278 (Fig. 4A and B), suggesting that there is not a significant metabolic burden to GBS carrying plasmid pGU2664. Furthermore, we observed no differences in a comparison of either of the GBS strains carrying plasmid pGU2664, or pDL278, with parental GBS 874391 containing no plasmid at all (data not shown). In monitoring the fluorescence intensity of GFPmut3, we noted strong GFPmut3 emission that correlated with cell density, demonstrating the expression and stability of GFPmut3 across the exponential- and stationary-growth phases.
FIG 4.
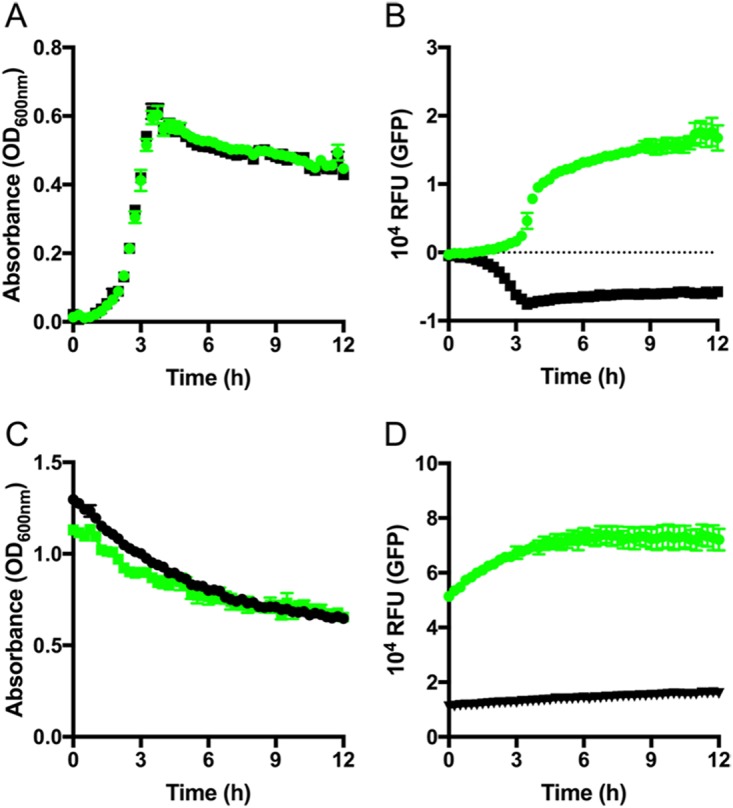
Measuring GBS growth in CDM using plasmid pGU2664. (A) Culture turbidity of GBS GU2666 (GFPmut3+ plasmid pGU2664; green) and GU2672 (empty vector pDL278; black) grown in CDM medium. (B) The fluorescence intensity of GFPmut3 was monitored simultaneously. (C and D) Turbidity (C) and GFPmut3 fluorescence (D) of stationary-phase bacteria incubated in PBS. Mean and SEM data are representative of at least three independent replicates. RFU, relative fluorescence units.
To further examine the fluorescence of GFPmut3 under stationary-phase conditions, GBS cells were pregrown overnight, washed in phosphate-buffered saline (PBS) to remove nutrients, and resuspended in PBS. These cells were then monitored for turbidity and fluorescence as described before. Consistent with the observations of single cells (described above), we observed stable and high-level fluorescence over a period of 12 h (Fig. 4C and D) from stationary-phase GBS populations during a period in which turbidity remained stable. Taken together, these results show that the activity of the GFPmut3-expressing vector is independent of active bacterial replication in a nutrient-rich environment, and that pGU2664 is a useful tool for marking GBS even where the bacteria are subjected to extended periods without fresh nutrients.
GFPmut3 expression in GBS in complex media.
Todd-Hewitt broth (THB) is a standard complete medium widely employed for the preparation of GBS for infection experiments using animal or cell culture-based methods. Therefore, we examined the expression of GFPmut3 fluorescence in GBS during growth in THB. Interestingly, we noted that THB exhibits a higher degree of background autofluorescence emission at 515 nm than either PBS or CDM used as described above (see Fig. S1A in the supplemental material). This may be due to a component of the medium, since the observed autofluorescence diminishes following incubation with non-GFPmut3 bacteria in a manner that is inversely proportional to increments in optical density (Fig. S1B and 5). To further elucidate this phenomenon, we monitored the growth of GBS in THB for 12 h using OD600 readings (Fig. 5A) while also simultaneously monitoring the fluorescence intensity for GFPmut3 and total polarized intensity (Fig. 5B and C) to eliminate the background interference caused by the medium. Strikingly, we observed that the use of total polarized intensity for fluorescence detection eliminated the interference due to the use of THB medium. This interference could also be excluded, in the absence of detection of polarized fluorescence emissions, by using negative-control correction of the fluorescence intensity of strain GU2672 containing the empty vector, which yielded a similar GFPmut3 emission profile (Fig. S1C).
FIG 5.
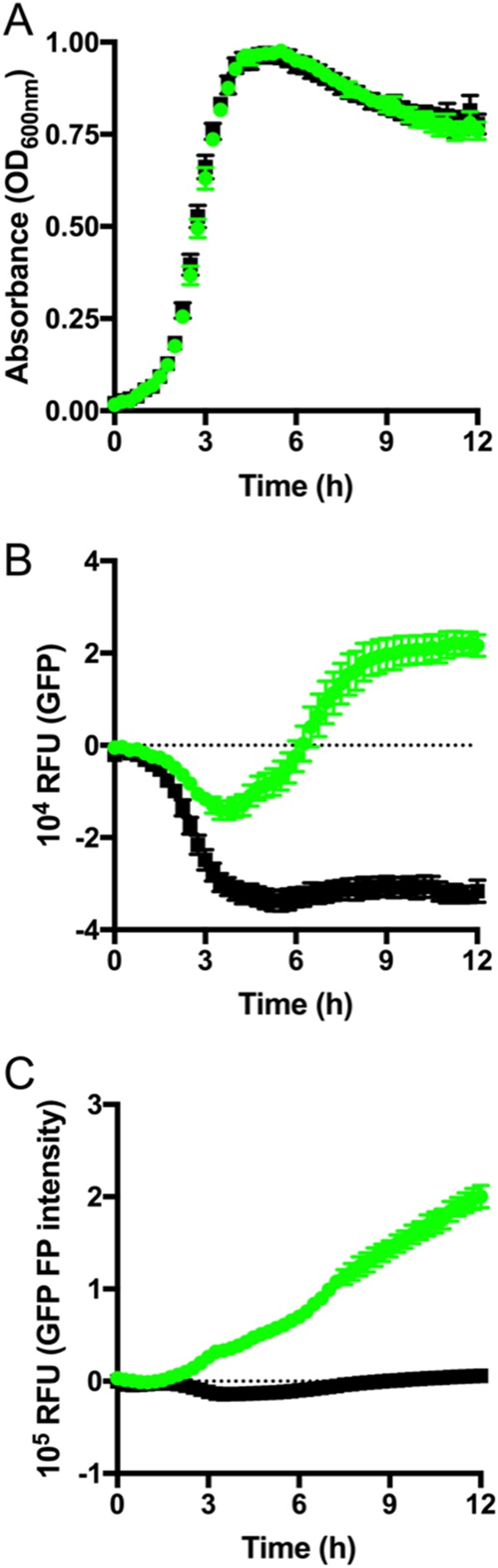
Measuring GBS growth in THB using plasmid pGU2664. (A) Culture turbidity of GBS GU2666 (GFPmut3+ plasmid pGU2664; green) and GU2672 (empty vector pDL278; black) grown in THB medium. (B) The fluorescence intensity of GFPmut3 was monitored simultaneously. (C) Fluorescence polarization (FP) intensity measurements were used to distinguish GFPmut3 intensity from background autofluorescence of THB medium. Mean and SEM data are representative of at least three independent replicates. RFU, relative fluorescence units.
Effect of antimicrobial compounds on GFPmut3 expression in GBS.
Quantification of fluorescence in GBS following exposure to antimicrobial compounds revealed that 0.02% sodium hypochlorite rendered the bacteria nonviable, according to colony count assays, but the cells continued to emit high levels of fluorescence (Fig. 6A and B). Sodium hypochlorite at 0.1% abolished the fluorescence in GBS cells that nonetheless remained intact. Sodium hypochlorite at >0.2% destroyed the bacteria; the cell morphology of GBS in these assays was ascertained using high-magnification bright-field and fluorescence microscopy (Fig. S2 for sodium hypochlorite assays; and data not shown). The exposure of GBS to chloramphenicol or gentamicin at a 1 mg · ml−1 concentration revealed no statistically significant perturbation of the GFPmut3 fluorescence signal in the bacteria despite a loss of viability (Fig. 7A and B). The exposure of GBS to dimethyl sulfoxide (DMSO; vehicle control for S-nitroso-N-acetyl-penicillamine [SNAP]) showed no effect on bacterial viability or GFPmut3 fluorescence under conditions containing up to 5.5% (vol/vol) DMSO (data not shown). In contrast, SNAP exhibited a dose-dependent effect on both GBS viability and GFPmut3 fluorescence in these assays (Fig. 7A and B). Together, these findings establish that high concentrations of sodium hypochlorite and SNAP kill GBS, which abolishes GFPmut3 fluorescence in the bacteria.
FIG 6.
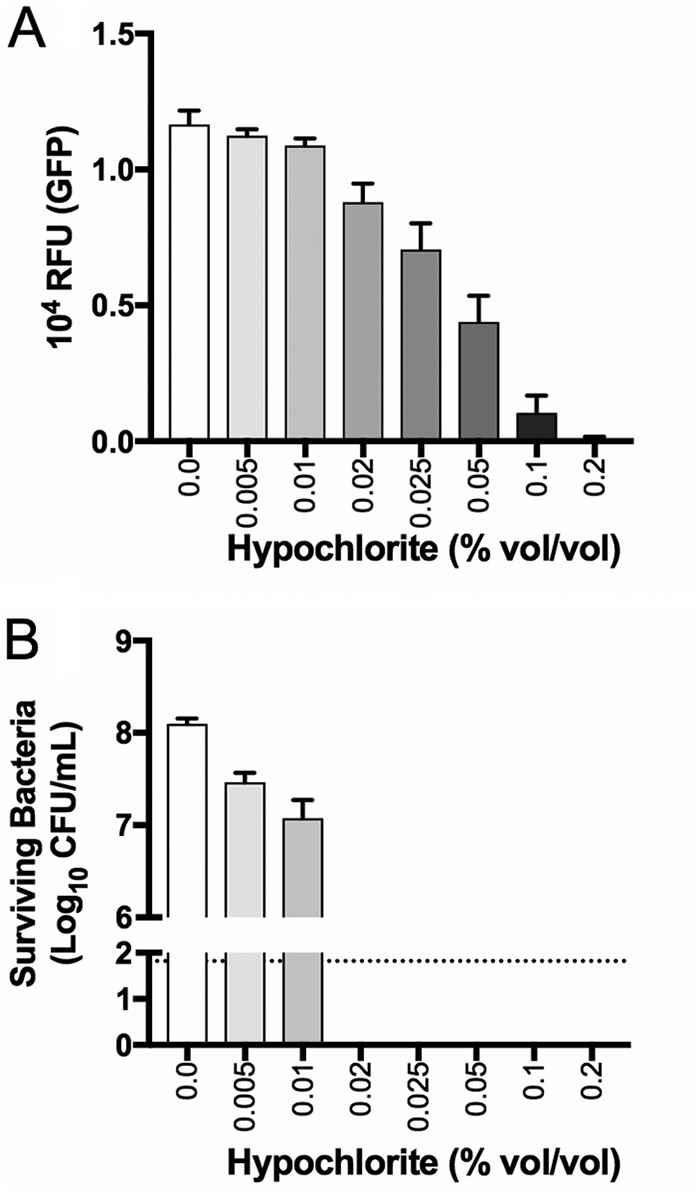
Effect of sodium hypochlorite on GBS viability and GFPmut3 fluorescence. GFPmut3+ GU2666 cells expressing GFP were exposed to a range of sodium hypochlorite concentrations for a period of 1 h. Subsequently, the bacteria were monitored for GFPmut3 fluorescence (A) and viability (B). The dotted line shows the detection limit for these assays. Bars show the SEM of the results from four independent experiments.
FIG 7.
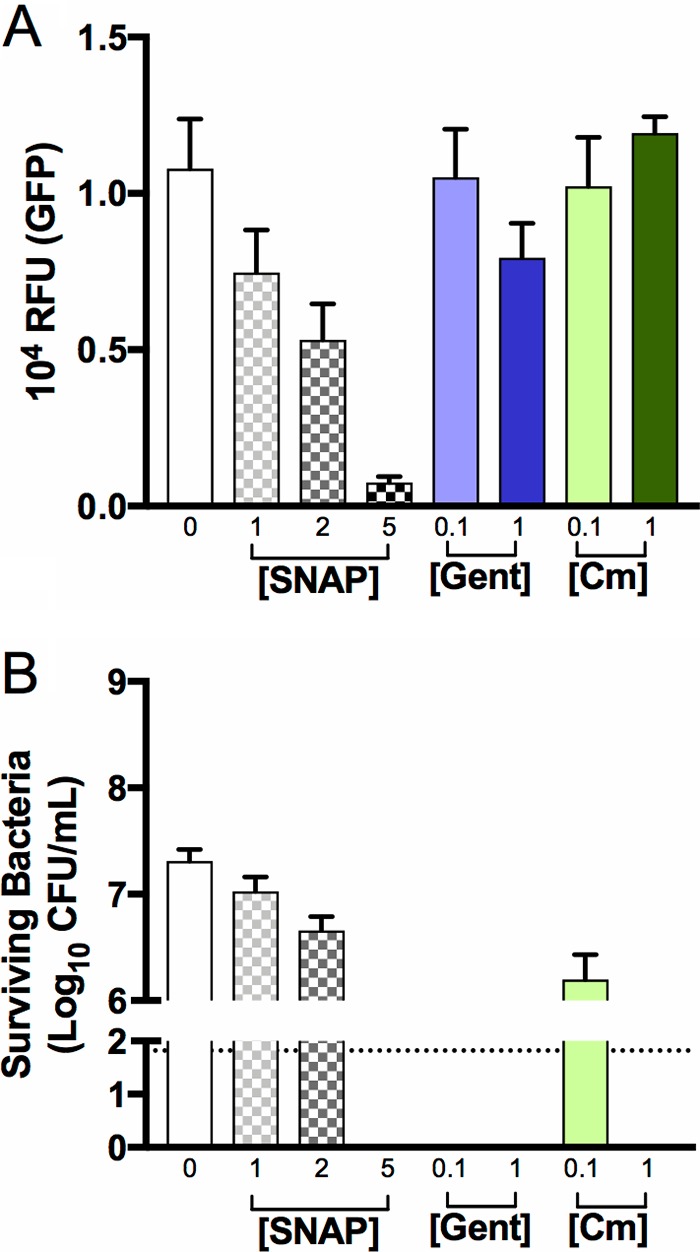
Effect of nitric oxide and distinct antibiotics on GBS viability and GFPmut3 fluorescence. GFPmut3+ GU2666 cells expressing GFP were exposed to SNAP (an NO donor) at various concentrations (in millimolar) or chloramphenicol or gentamicin (Gent) (in milligrams per milliliter), two antibiotics with distinct antibacterial effects (bacteriostatic and bactericidal, respectively), for a period of 24 h. Subsequently, the bacteria were monitored for GFPmut3 fluorescence (A) and viability (B). Bars show the SEM of three independent replicates.
Adhesion of GFPmut3-expressing GBS to human epithelial cells.
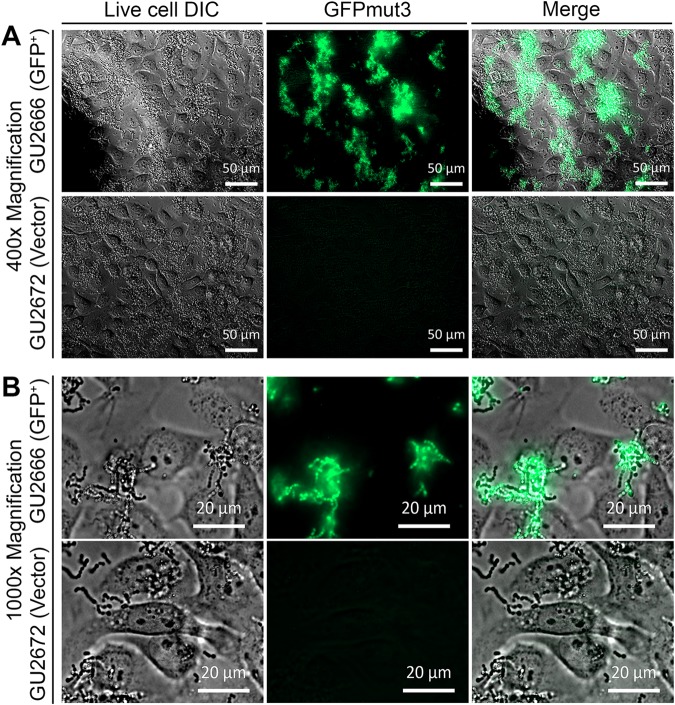
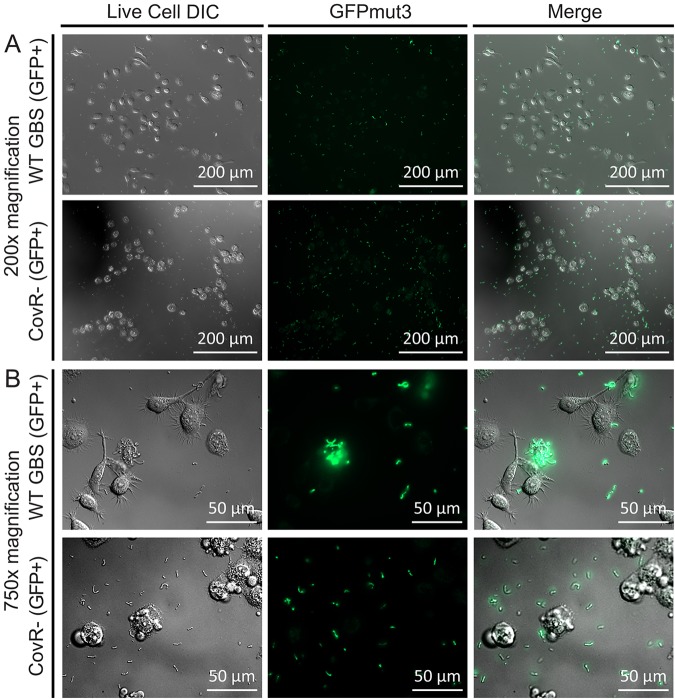
To evaluate GFPmut3-GBS in the context of host colonization, we used in vitro adhesion assays with human uroepithelial cells as a model of UTI and visualized the interaction in real time using live cells. Assays performed to validate plasmid pGU2664 as a fluorescent marker in GBS in the context of host cells showed uniform, stable, and high-level GFPmut3 expression in GBS GU2666 in association with human cells, compared to the control vector pDL278 in GU2672 that exhibited minimal fluorescence above background (Fig. 8). Irregular adhesive patterns of GBS to the cultured human bladder uroepithelial cells were observed following the 2-h incubation period. Video capture demonstrated many adhered GBS cells on the bladder uroepithelial cells amid a background of media and bacterial flowthrough (observed as free-floating chains of nonadhered GBS) within the culture chamber (see Movie S1 in the supplemental material). A comparison of the relative adhesion levels of WT GBS and a CovR− mutant that is known to be attenuated for adhesion to human epithelial cells (25) confirmed the attenuated phenotype of the mutant (Fig. 9). Relative quantification of the level of fluorescence in these cultures using pixel analysis confirmed a significant attenuation in the level of adhesion of the CovR− mutant compared to the WT strain (Fig. S3). Thus, plasmid pGU2664 is a useful tool for visualizing the interactions between GBS and live host cells.
FIG 8.
Plasmid pGU2664 enables visualization of GBS in association with human cells. Inoculation of 5637 uroepithelial cells with GU2666 (GFP+) containing plasmid pGU2664 compared to GU2672 (GFP−) carrying control vector pDL278. Incubations were for a period of 2 h based on an MOI of 100. (A and B) Low-power (×400) (A) and high-power (×1,000) (B) magnifications, which were captured preflow.
FIG 9.
Comparison of adhesion of WT and CovR− mutant GBS to human cells. Inoculation of 5637 uroepithelial cells with WT and CovR− mutant GBS (both carrying plasmid pGU2664) shows the attenuated adherence phenotype of the CovR− mutant based on the fluorescence detection of GFPmut3. Quantitation of the fluorescence signals in areas colocated with the human cells (cell-associated fluorescence) was achieved using the ImageJ software and compared to acellular areas (slide-associated fluorescence). The quantitated data reveal significantly more fluorescence signal colocated with epithelial cells inoculated with WT GBS versus the CovR− mutant (Fig. S3).
DISCUSSION
With the present study, we aimed to extend the range of available visualization tools for GBS by constructing and evaluating a plasmid-based system that can enable constitutive, high-level, growth phase-independent expression of GFPmut3 in GBS. We took advantage of the promoter probe vector pCP25 to merge CP25 and gfpmut3-derived GFPmut3 into pDL278, creating plasmid pGU2664. We have established the utility of plasmid pGU2664 in affording GFPmut3 expression in GBS using three systems: population-level analysis of GBS cultures, single-cell analysis of GBS, and cellular interaction analysis at the host-microbe interface with human cells. Plasmid pGU2664 affords visible expression of GFPmut3 in individual GBS cells and is stable over many generations of GBS replication in vitro in nutrient-rich media and in the absence of selection pressure. Spectrophotometric and temporal analyses of the expression of GFPmut3 in GBS at distinct phases of bacterial growth, achieved by the use of fluorescence polarization, validate plasmid pGU2664 as a fluorescence marker for GBS in cells maintained at stationary phase. The successful GFP tagging of GBS using a plasmid system in trans in this study, to afford GBS stable, high-level fluorescence, will be useful for various experimental systems.
Studies on GBS that have used fluorescence approaches to label the bacteria have principally applied labeling methods that incorporate DNA stains, such as LIVE/DEAD viability stains (23, 44–49), nonspecific protein stains, such as fluorescein isothiocyanate (FITC) (17, 50–53), or antibody-based immunostaining methods (12, 54, 55). Other previous studies have reported the expression of GFP in GBS, as follows: Périchon et al. examined transcriptional fusions with gfp as a reporter gene linked to the PI-2b pilus locus in GBS BM110 and A909 strains (54), Aymanns et al. used a transcriptional fusion with cfb to show that oxygen conditions can affect GBS fluorescence (56), and Cutting et al. investigated the expression of plasmid-borne GFP in GBS strain COH1 in trans (11). The development of plasmid pGU2664 in the current study provides a useful advancement for visualizing GBS, in that the expression of GFPmut3 in GBS(pGU2664) is independent of growth state or regulation by intrinsic transcriptional regulators of GBS. Appreciable high-level GFPmut3 expression in GBS strains carrying pGU2664 is easily visualized in single bacterial cells in comparison to the relatively low-level expression of GFP previously reported in GBS COH1 (11).
The reporter method developed in this study offers several advantages for imaging of GBS in different experimental contexts. In contrast to other approaches that have been used to visualize GBS, such as electron microscopy, which often require preparation techniques that kill or alter the structure of cells, plasmid pGU2664 enables the visualization of live GBS without the addition of exogenous substrates. Additional benefits include (i) potential application to real-time assays, such as flow cytometry for bacterial enumeration, (ii) independence of culture for bacterial enumeration, (iii) potential to detect viable nonculturable GBS, (iv) potential application to high-throughput methods, (v) high accuracy for some applications (relative quantitative fluorescence measures of GBS chain length, for example), and (vi) the capacity for visualization of subcellular location of GBS in host cells.
The results of adhesion assays with WT and CovR− mutant GBS strains in the present study show the utility of plasmid pGU2664 in host colonization studies. We used human uroepithelial cells to model GBS UTI, which is a similar approach to several previous studies (57–59). The present work, based on a fluorescence imaging approach, enabled live cell analysis to capture the interactions of GBS with living human cells in real time. Further work to characterize the interactions of GBS with live host cells under flow conditions similar to those reported here will be of interest. The imaging of GBS(pGU2664) in situ in tissue sections in animal model studies may also be possible. Theoretically, imaging GBS(pGU2664) in vivo in live animals could be possible, but more challenging, than using bioluminescent reporters that offer better tissue penetration than fluorescent proteins (60).
The limitations of the current study include the analysis of only a single GBS strain, 874391, which, given that different bacteria express fluorescent proteins differently (61), highlights the need to test this system in different GBS strains. The level of expression of GFP in bacteria may be affected by differential codon usage (62–64), mRNA stability (65), temperature, oxygen availability, pH, and other variables (37), none of which were tested in this study but would be of interest to investigate in GBS. GFP variants can express at different levels among different bacterial genera (30), and benchmarking of different GFP variants in GBS could be used to confirm optimal fluorescence. We accomplished GFPmut3 expression in GBS through the introduction of a plasmid, a common approach in many studies (31, 32, 66). An alternative approach could be to integrate the CP25-gfpmut3 construct into the GBS chromosome, akin to other studies (38, 66, 67), which would provide the advantage of not requiring continuous selection pressure to maintain the plasmid. Of note, however, are the data shown here of good stability of pGU2664 in GBS in vitro in the absence of selection pressure. Furthermore, chromosomal gfp integrants may be subject to unknown promoters in bacteria (68), integrate in multiple locations (69), and/or afford lower levels of fluorescence than bacteria carrying multicopy-number plasmids in trans. Additionally, designing chromosomal insertions without polar effects can be challenging. Finally, given the need for posttranslational oxidation for the maturation of GFP (30), it would also be of interest to examine the expression levels of GFPmut3 in GBS cultures grown aerobically versus anaerobically.
We showed that antimicrobial conditions, including exposure to hypochlorite, toxic radicals, or antibiotics, can cause a loss of viability of GBS and also abolish the GFPmut3 fluorescence signal in the bacteria. The implications of these findings relate to different (potentially stressful) environmental conditions. Overall, these observations suggest that where GBS cells are viable, they express GFPmut3. However, there are limitations to this general observation; we did observe strong fluorescence from dead intact GBS cells that had been treated with antibiotics. Exposure to oxidative products and free radicals, such as hypochlorite and nitric oxide, is likely to destroy the GFPmut3 protein more rapidly than antimicrobial conditions that render GBS nonviable through other (non-protein-targeting) mechanisms. This limitation may influence the utility of the GFPmut3 tag for detecting GBS in some experiments. Finally, we used monocultures of uroepithelial cells to model host-microbe interactions, and this has limitations compared to epithelial cell cocultures to model infection, as reviewed elsewhere (70). Interestingly, the irregular adhesive patterns of GBS to the cultured human bladder uroepithelial cells that we observed in this study are reminiscent of the mosaic pattern of E. coli adhesion known to occur with bladder uroepithelial cells (71). The molecular basis of adhesion of GBS to bladder uroepithelial cells is not known.
In summary, plasmid pGU2664 will enable new studies of GBS biology based on the detection of fluorescence. Further analysis of GFPmut3 as a biomarker will be vital to show its utility in diverse applications, including those aimed at better understanding GBS virulence.
MATERIALS AND METHODS
Bacterial strains, plasmids and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. The whole-genome-sequenced hypervirulent sequence type 17 (ST-17) type strain GBS 874391 (72) or its isogenic CovR-deficient derivative strain (25) was used. Plasmid pJRS9567 was kindly provided by Timothy Barnett and June Scott, Emory University. The E. coli-Gram-positive bacterial shuttle vector pDL278 was a gift from Gary Dunny (73). E. coli strains were routinely cultured in lysogeny broth (LB), and GBS strains were routinely cultured in THB (Thermo Fisher Scientific), on tryptone soya agar (TSA; Thermo Fisher Scientific) containing 5% horse blood (Thermo Fisher Scientific) or in CDM (43). Starter cultures were aerated at 200 rpm for 16 to 18 h prior to use. For strains carrying plasmids, the respective media were supplemented with antibiotics at the following concentrations: spectinomycin (Sp), 100 μg · ml−1; and chloramphenicol (Cm), 10 μg · ml−1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Characteristicsa | Reference(s) or source |
|---|---|---|
| Strains | ||
| E. coli DH5α | Cloning host; ϕlacZΔM15 Δ(lacZYA-argF) U169 recA1 endA1 hsdR17(rK− mK+) supE44 thi-1 gyrA96 relA1 | Bethesda Research Laboratories |
| GBS 874391 | WT serotype III sequence type 17 strain, vaginal isolate | 72, 81, 82 |
| GBS GU2400 | covR derivative of GBS 874391; Cmr | 25 |
| GBS GU2666 | GFP+ plasmid pGU2664 transformed into WT GBS strain 874391; Spr | This work |
| GBS GU2672 | pDL278 plasmid (empty vector) transformed into WT GBS strain 874391; Spr | This work |
| GBS GU2673 | pDL278 plasmid (empty vector) transformed into covR GBS strain GU2400; Spr Cmr | This work |
| GBS GU2676 | GFP+ plasmid pGU2664 transformed into covR GBS strain GU2400; Spr Cmr | This work |
| Plasmids | ||
| pLZ12 | E. coli-streptococcal shuttle vector; Cmr | 75 |
| pDL278 | E. coli-streptococcal shuttle vector; Spr | 73 |
| pJRS9567 | gfpmut3 plasmid; Cmr | June Scott |
| pGU2664 | gfpmut3 cloned into EcoRI site of pDL278; Spr | This work |
Cmr, chloramphenicol resistance; Spr, spectinomycin resistance.
DNA isolation procedures.
Plasmid DNA from E. coli DH5α derivatives was routinely isolated using the QIAprep spin plasmid miniprep kit (Qiagen), according to the manufacturer's instructions. GBS plasmid DNA was also isolated using this kit but with modification of the P1 buffer, which was supplemented with 30 mg · ml−1 lysozyme (catalog no. L6876; Sigma-Aldrich) and 100 U of mutanolysin (catalog no. M9901; Sigma-Aldrich) per 250 μl, and incubated for 1 h at 37°C prior to the addition of the P2 buffer.
DNA manipulations and electroporation of GBS.
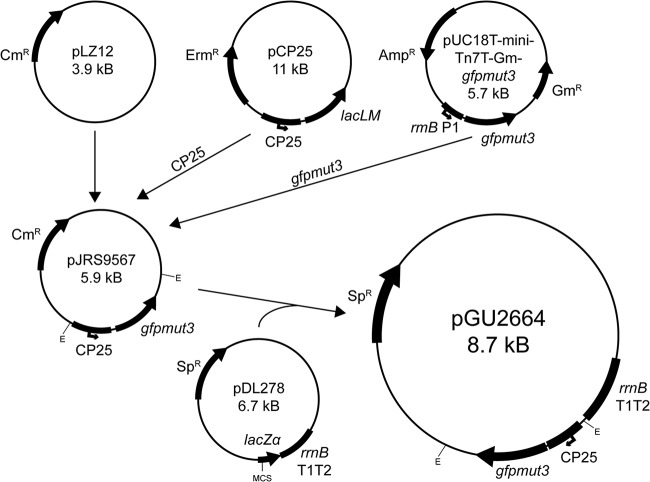
The scheme used to construct the GFPmut3-expressing plasmid pGU2664, which was designed to supply fluorescence to GBS in trans, is summarized in Fig. 10. The upper portion of Fig. 10 shows the GFPmut3-expressing plasmid pJRS9567 that contains a 108-bp XhoI+XbaI fragment from pCP25 (42) and a 748-bp XbaI+HindIII fragment from pUC18T-mini-Tn7T-Gm-gfpmut3 (74), both cloned into the Gram-positive shuttle vector pLZ12 (75). pJRS9567 was used to derive gfpmut3 for this study. Plasmid DNA was manipulated using restriction endonuclease EcoRI, alkaline phosphatase, and T4 DNA ligase, according to the manufacturer's specifications (Roche). Ligation reaction mixtures were transformed into E. coli DH5α, and the resultant recombinant plasmid was selected using a blue-white screen on LB agar containing Sp, isopropyl-β-d-thiogalactopyranoside (IPTG; 1 mM), and 5-bromo-4-chloro-3-indolyl-β-d-thiogalactopyranoside (X-Gal; 40 μg · ml−1). We constructed plasmid pGU2664 by excising a 1.9-kb EcoRI fragment containing the cp25-gfpmut3 fusion from pJRS9567 and cloning it into the EcoRI site of the multicloning site of pDL278, as shown in the lower portion of Fig. 10. We verified plasmid pGU2664 by restriction analysis and sequenced the insert in its entirety using the oligonucleotides listed in Table 2. Sequence reads across the 1.9-kb insert were mapped and assembled using Sequencher software. Electroporation of GBS with either plasmid pDL278 or pGU2664 was performed as described previously (57), and transformants were selected on THB agar containing Sp.
FIG 10.
Construction of plasmid pGU2664. Plasmid pJRS9567 comprises the pLZ12 (75) backbone and gfpmut3 under the control of the synthetic promoter CP25, and was kindly provided by Timothy Barnett and June Scott, Emory University. The gfpmut3 and cp25 fragments that were cloned into pJRS9567 originated from pUC18T-mini-Tn7T-Gm-gfpmut3 (74) and pCP25 (42), respectively. In the present study, a 1.9-kb EcoRI fragment from pJRS9567 was cloned into the EcoRI site of the multicloning site (MCS) of pDL278 to make plasmid pGU2664. The E. coli-Gram-positive bacterial shuttle vector pDL278 was a gift from Gary Dunny (73). E, EcoRI; Ermr, erythromycin resistance; Ampr, ampicillin resistance; Gmr, gentamicin resistance; Cmr, chloramphenicol resistance; Spr, spectinomycin resistance.
TABLE 2.
Oligonucleotides used in this study for sequencing of pGU2664
| Primer name | Sequence (5′–3′) |
|---|---|
| GFP_N-term_R1 | CCTTCACCCTCTCCACTGAC |
| GFP_C-term_F1 | AAAGGTATTGATTTTAAAGAAGATGGA |
| GFP_C-term_R2 | CCCAGCAGCTGTTACAAACTC |
| M13F | GGTGTAAAACGACGGCCAG |
| M13R | AGGAAACAGCTATGACCATG |
Plasmid stability assay.
The stability of plasmid pGU2664 in GBS strain GU2666 grown in the absence of antibiotics was determined essentially as previously described (28, 76). Briefly, plasmid-carrying GBS was cultured in THB, with or without selection (Sp), and was diluted 1:1,000 every 12 h in fresh selective or nonselective medium for a total of ∼100 generations; this equated to a total of 5 subcultures over a period of 2.5 days. Plasmid stability was determined every ∼20 generations (12 h) by enumerating the number of bacteria on TSA blood agar without antibiotics (e.g., the total CFU) compared to that on THB agar with antibiotics (e.g., Spr CFU). The experiments were repeated three times.
Fluorescence microscopy.
The expression of GFPmut3 in cells of GBS was analyzed using a Zeiss AxioImager.M2 microscope (Carl Zeiss MicroImaging) fitted with Plan-Apochromat ×20/0.8 and ×100/1.46 objective lenses, and an AxioCam MRm Rev.3 camera. We used the Zen 2012 SP2 imaging software for optical sectioning (77) and image acquisition. For cell imaging, GBS were grown overnight in THB with Sp; 0.5 ml (approximately 2 × 108 CFU) was washed three times in PBS (pH 7.4) (10,000 × g, 2 min, room temperature [RT]), stained for DNA using Hoechst 33258 (20 μg · ml−1 PBS; Sigma-Aldrich) for 5 min (at RT) and washed three times in PBS prior to resuspension in 10 μl of 0.2% n-propyl gallate (n-pg; in a 1:9 solution of PBS [pH 7.4] and glycerol). The cell suspensions were diluted 1:20 in fresh n-pg, and 7 μl was mounted under Zeiss high-performance coverslips (no. 11/2; 18 mm by 18 mm) using Valap (a mixture of Vaseline, lanolin, and paraffin wax) (78) and stored at 4°C until viewing. We imaged host-microbe interactions using human 5637 uroepithelial cells that were inoculated with GBS, as described below. To visualize the expression of GFPmut3 in colonies of GBS GU2666 on agar, we used a SZX16 stereomicroscope in conjunction with CellF software (Olympus Australia Pty Ltd.). Quantitation of fluorescence signals in some experiments was achieved using ImageJ software 1.51 (79).
Growth experiments using multimode plate reader.
Overnight THB cultures of GBS were harvested by centrifugation and washed three times in PBS (pH 7.4) subsequent to being diluted 1:100 into fresh CDM or THB (approximately 1 × 106 CFU per well). Then, 200-μl volumes of the cell suspensions were aliquoted in triplicate into the wells of a (flat-bottom) 96-well plate (catalog no. 655180; Greiner). Plates were sealed using gas-permeable Breathe-Easy membranes (catalog no. Z380059; Sigma-Aldrich) and incubated for 12 h in a CLARIOstar multimode plate reader (BMG Labtech) with an atmospheric control unit set at 37°C in an atmosphere containing 5% CO2. The optical density at 600 nm (OD600) was monitored every 15 min with agitation at 300 rpm between cycles with single orbital shaking, path length correction for 200-μl volumes, and orbital averaging from a 3-mm diameter with 10 flashes per well. The experiments were repeated independently at least three times.
Fluorescence measurements using multimode plate reader.
The GFPmut3 reporter has redshifted excitation maxima, exhibiting excitation and emission maxima of 501 nm and 511 nm, respectively (39). However, GFPmut3 can be monitored using standard FITC filter sets because of efficient excitation at 488 nm. For reference, the excitation and emission maxima for WT and other GFP variants reported in prior studies range from 395 nm to 501 nm and 508 nm to 535 nm, respectively (28, 30, 39); enhanced GFP (eGFP) has an excitation maximum of 488 nm (80). For plate reader assays, fluorescence intensity was monitored in parallel with OD600 growth measurements every 15 min. We used the linear variable filter monochromator of the CLARIOstar multimode plate reader to monitor GFPmut3 fluorescence using excitation at 470 nm with a 15-nm width and emission at 515 nm with a 20-nm width, and the gain was set at 1,700 for the detection of GFPmut3 in these assays. In addition to fluorescence intensity measurements, we also used fluorescence polarization to monitor GFPmut3 fluorescence and distinguish it from the background autofluorescence of THB medium. For polarization intensity measurements, parallel (Ipara) and perpendicular (Iperp) intensities were monitored using the excitation filter at 482 nm (16-nm width), dichroic filter low-pass (LP) at 504 nm, emission filter at 515 nm (30-nm width), gain A set at 951, gain B set at 1,113, focal height at 7.8 mm, and target millipolarization (mP) value of 35; the total fluorescence polarization intensity was calculated by the sum Ipara + 2Iperp.
Effect of cell death on GFPmut3 fluorescence in GBS.
To evaluate GFPmut3 fluorescence in GBS exposed to a variety of antimicrobial compounds, approximately 5 × 107 CFU of stationary-phase GU2666 cells were exposed to a range of concentrations of sodium hypochlorite, the endogenous nitric oxide donor S-nitroso-N-acetyl-penicillamine (SNAP; dissolved in DMSO), chloramphenicol, or gentamicin in 200 μl PBS. Vehicle control assays with various concentrations of DMSO were also performed in parallel to compare to SNAP-exposed GBS. The assays were performed using 96-well plates (Greiner) that were sealed using Breathe-Easy membranes (Sigma) for the entirety of incubation and fluorescence detection. The assays that contained sodium hypochlorite were incubated for a total period of 1 h, whereas those with SNAP, chloramphenicol, gentamicin, or DMSO were incubated for 24 h. GFPmut3 fluorescence was quantified as described above using a CLARIOstar multimode plate reader, and surviving bacteria were enumerated by colony counts on agar. The experiments were repeated independently at least three times.
Adhesion assays.
To evaluate GU2666 in the context of host cells, we used adhesion assays with uroepithelial cells and imaged the interactions in real time. Briefly, approximately 2 × 105 human 5637 uroepithelial cells were cultured in 200 μl of medium in μ-Slide I0.8 Luer chamber ibiTreat tissue culture slides (ibidi GmbH). The cells were grown for 24 h at 37°C in 5% CO2 in RPMI 1640 medium (Life Technologies) containing 10% fetal bovine serum, 25 mmol · liter−1 HEPES, 2 mmol · liter−1 l-glutamine, 100 mmol · liter−1 nonessential amino acids, and 1 mmol · liter−1 sodium pyruvate. Inoculations were performed with GBS at a multiplicity of infection (MOI) of approximately 100 bacteria per cell for 2 h. After 2 h of static incubation, adherent bacteria were imaged prior to, during, and after the flowing of 10 chamber volumes of RPMI medium through the slides. Flow conditions were generated by a perfusion set (50-cm length, 0.8-mm internal diameter 10-ml reservoirs; white) and were analyzed using low, medium, and high flow rates to visualize the removal of nonadherent bacteria from the chamber. The conditions of low flow (0.8 ml · min−1; shear stress, 0.2 dyne [dyn] · cm2; 500 Pa pressure), medium flow (1.6 ml · min−1; shear stress, 0.43 dyn · cm2; 900 Pa pressure), and high-flow (5.0 ml · min−1; shear stress, 1.25 dyn · cm2; 2.93 kPa pressure) were managed using a pump system and fluidic unit (ibidi GmbH). Images of the host cells and bacteria were captured live in real time using a Zeiss AxioObserver.Z1 microscope (Carl Zeiss MicroImaging) fitted with Plan-Apochromat ×20/0.8 and ×63/1.40 objective lenses and an AxioCam 506 camera. During imaging, the cells were maintained at 37°C using a Heating Insert P Lab-Tek S in combination with a TempModule S (PeCon GmbH). Images of live cells were captured using differential interference contrast (DIC) settings in combination with an eGFP filter (to detect GFPmut3) and Zen Pro (version 2) software.
Statistics.
The numbers of Spr colonies in GBS liquid cultures grown in the presence and absence of antibiotic selection were compared over time using area under the curve (AUC) analysis with Student's t tests. Group-wise data are displayed as the mean ± standard error of the mean (SEM). The experiments were repeated at least three times in independent assays. The statistical analyses were performed using GraphPad Prism version 6 and SPSS version 21.0.
Supplementary Material
ACKNOWLEDGMENTS
We thank Timothy Barnett for providing plasmid pJRS9567 and for helpful discussions.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01262-18.
REFERENCES
- 1.Le Doare K, Heath PT. 2013. An overview of global GBS epidemiology. Vaccine 31(Suppl 4):D7–D12. doi: 10.1016/j.vaccine.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 2.Keefe G. 2012. Update on control of Staphylococcus aureus and Streptococcus agalactiae for management of mastitis. Vet Clin North Am Food Anim Pract 28:203–216. doi: 10.1016/j.cvfa.2012.03.010. [DOI] [PubMed] [Google Scholar]
- 3.Da Cunha V, Davies MR, Douarre PE, Rosinski-Chupin I, Margarit I, Spinali S, Perkins T, Lechat P, Dmytruk N, Sauvage E, Ma L, Romi B, Tichit M, Lopez-Sanchez MJ, Descorps-Declere S, Souche E, Buchrieser C, Trieu-Cuot P, Moszer I, Clermont D, Maione D, Bouchier C, McMillan DJ, Parkhill J, Telford JL, Dougan G, Walker MJ, Consortium D, Holden MT, Poyart C, Glaser P. 2014. Streptococcus agalactiae clones infecting humans were selected and fixed through the extensive use of tetracycline. Nat Commun 5:4544. doi: 10.1038/ncomms5544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cools P, Jespers V, Hardy L, Crucitti T, Delany-Moretlwe S, Mwaura M, Ndayisaba GF, van de Wijgert JH, Vaneechoutte M. 2016. A multi-country cross-sectional study of vaginal carriage of group B streptococci (GBS) and Escherichia coli in resource-poor settings: prevalences and risk factors. PLoS One 11:e0148052. doi: 10.1371/journal.pone.0148052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schuchat A, Wenger JD. 1994. Epidemiology of group B streptococcal disease. Risk factors, prevention strategies, and vaccine development. Epidemiol Rev 16:374–402. [DOI] [PubMed] [Google Scholar]
- 6.Valkenburg-van den Berg AW, Sprij AJ, Oostvogel PM, Mutsaers JA, Renes WB, Rosendaal FR, Joep Dorr P. 2006. Prevalence of colonisation with group B streptococci in pregnant women of a multi-ethnic population in The Netherlands. Eur J Obstet Gynecol Reprod Biol 124:178–183. doi: 10.1016/j.ejogrb.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 7.Muller AE, Oostvogel PM, Steegers EA, Dorr PJ. 2006. Morbidity related to maternal group B streptococcal infections. Acta Obstet Gynecol Scand 85:1027–1037. doi: 10.1080/00016340600780508. [DOI] [PubMed] [Google Scholar]
- 8.Edwards MS, Baker CJ. 2005. Group B streptococcal infections in elderly adults. Clin Infect Dis 41:839–847. doi: 10.1086/432804. [DOI] [PubMed] [Google Scholar]
- 9.Kobayashi M, Vekemans J, Baker CJ, Ratner AJ, Le Doare K, Schrag SJ. 2016. Group B Streptococcus vaccine development: present status and future considerations, with emphasis on perspectives for low and middle income countries. F1000Res 5:2355. doi: 10.12688/f1000research.9363.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bolduc GR, Baron MJ, Gravekamp C, Lachenauer CS, Madoff LC. 2002. The alpha C protein mediates internalization of group B Streptococcus within human cervical epithelial cells. Cell Microbiol 4:751–758. doi: 10.1046/j.1462-5822.2002.00227.x. [DOI] [PubMed] [Google Scholar]
- 11.Cutting AS, Del Rosario Y, Mu R, Rodriguez A, Till A, Subramani S, Gottlieb RA, Doran KS. 2014. The role of autophagy during group B Streptococcus infection of blood-brain barrier endothelium. J Biol Chem 289:35711–35723. doi: 10.1074/jbc.M114.588657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lemire P, Houde M, Lecours MP, Fittipaldi N, Segura M. 2012. Role of capsular polysaccharide in group B Streptococcus interactions with dendritic cells. Microbes Infect 14:1064–1076. doi: 10.1016/j.micinf.2012.05.015. [DOI] [PubMed] [Google Scholar]
- 13.Pezzicoli A, Santi I, Lauer P, Rosini R, Rinaudo D, Grandi G, Telford JL, Soriani M. 2008. Pilus backbone contributes to group B Streptococcus paracellular translocation through epithelial cells. J Infect Dis 198:890–898. doi: 10.1086/591182. [DOI] [PubMed] [Google Scholar]
- 14.Erdogan S, Fagan PK, Talay SR, Rohde M, Ferrieri P, Flores AE, Guzman CA, Walker MJ, Chhatwal GS. 2002. Molecular analysis of group B protective surface protein, a new cell surface protective antigen of group B streptococci. Infect Immun 70:803–811. doi: 10.1128/IAI.70.2.803-811.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nizet V, Kim KS, Stins M, Jonas M, Chi EY, Nguyen D, Rubens CE. 1997. Invasion of brain microvascular endothelial cells by group B streptococci. Infect Immun 65:5074–5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tyrrell GJ, Kennedy A, Shokoples SE, Sherburne RK. 2002. Binding and invasion of HeLa and MRC-5 cells by Streptococcus agalactiae. Microbiology 148:3921–3931. doi: 10.1099/00221287-148-12-3921. [DOI] [PubMed] [Google Scholar]
- 17.Ulett GC, Webb RI, Ulett KB, Cui X, Benjamin WH, Crowley M, Schembri MA. 2010. Group B Streptococcus (GBS) urinary tract infection involves binding of GBS to bladder uroepithelium and potent but GBS-specific induction of interleukin 1alpha. J Infect Dis 201:866–870. doi: 10.1086/650696. [DOI] [PubMed] [Google Scholar]
- 18.Dover RS, Bitler A, Shimoni E, Trieu-Cuot P, Shai Y. 2015. Multiparametric AFM reveals turgor-responsive net-like peptidoglycan architecture in live streptococci. Nat Commun 6:7193. doi: 10.1038/ncomms8193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ho YR, Li CM, Yu CH, Lin YJ, Wu CM, Harn IC, Tang MJ, Chen YT, Shen FC, Lu CY, Tsai TC, Wu JJ. 2013. The enhancement of biofilm formation in group B streptococcal isolates at vaginal pH. Med Microbiol Immunol (Berl) 202:105–115. doi: 10.1007/s00430-012-0255-0. [DOI] [PubMed] [Google Scholar]
- 20.Hull JR, Tamura GS, Castner DG. 2008. Interactions of the streptococcal C5a peptidase with human fibronectin. Acta Biomaterials 4:504–513. doi: 10.1016/j.actbio.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saar-Dover R, Bitler A, Nezer R, Shmuel-Galia L, Firon A, Shimoni E, Trieu-Cuot P, Shai Y. 2012. d-Alanylation of lipoteichoic acids confers resistance to cationic peptides in group B streptococcus by increasing the cell wall density. PLoS Pathog 8:e1002891. doi: 10.1371/journal.ppat.1002891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carey AJ, Tan CK, Mirza S, Irving-Rodgers H, Webb RI, Lam A, Ulett GC. 2014. Infection and cellular defense dynamics in a novel 17β-estradiol murine model of chronic human group B streptococcus genital tract colonization reveal a role for hemolysin in persistence and neutrophil accumulation. J Immunol 192:1718–1731. doi: 10.4049/jimmunol.1202811. [DOI] [PubMed] [Google Scholar]
- 23.Patras KA, Wang NY, Fletcher EM, Cavaco CK, Jimenez A, Garg M, Fierer J, Sheen TR, Rajagopal L, Doran KS. 2013. Group B Streptococcus CovR regulation modulates host immune signalling pathways to promote vaginal colonization. Cell Microbiol 15:1154–1167. doi: 10.1111/cmi.12105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rajagopal L, Clancy A, Rubens CE. 2003. A eukaryotic type serine/threonine kinase and phosphatase in Streptococcus agalactiae reversibly phosphorylate an inorganic pyrophosphatase and affect growth, cell segregation, and virulence. J Biol Chem 278:14429–14441. doi: 10.1074/jbc.M212747200. [DOI] [PubMed] [Google Scholar]
- 25.Sullivan MJ, Leclercq SY, Ipe DS, Carey AJ, Smith JP, Voller N, Cripps AW, Ulett GC. 2017. Effect of the Streptococcus agalactiae virulence regulator CovR on the pathogenesis of urinary tract infection. J Infect Dis 215:475–483. doi: 10.1093/infdis/jiw589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whidbey C, Harrell MI, Burnside K, Ngo L, Becraft AK, Iyer LM, Aravind L, Hitti J, Waldorf KM, Rajagopal L. 2013. A hemolytic pigment of group B Streptococcus allows bacterial penetration of human placenta. J Exp Med 210:1265–1281. doi: 10.1084/jem.20122753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Freitag NE, Jacobs KE. 1999. Examination of Listeria monocytogenes intracellular gene expression by using the green fluorescent protein of Aequorea victoria. Infect Immun 67:1844–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grimm V, Gleinser M, Neu C, Zhurina D, Riedel CU. 2014. Expression of fluorescent proteins in bifidobacteria for analysis of host-microbe interactions. Appl Environ Microbiol 80:2842–2850. doi: 10.1128/AEM.04261-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martinez-Jaramillo E, Garza-Morales R, Loera-Arias MJ, Saucedo-Cardenas O, Montes-de-Oca-Luna R, McNally LR, Gomez-Gutierrez JG. 2017. Development of Lactococcus lactis encoding fluorescent proteins, GFP, mCherry and iRFP regulated by the nisin-controlled gene expression system. Biotech Histochem 92:167–174. doi: 10.1080/10520295.2017.1289554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Overkamp W, Beilharz K, Detert Oude Weme R, Solopova A, Karsens H, Kovacs A, Kok J, Kuipers OP, Veening JW. 2013. Benchmarking various green fluorescent protein variants in Bacillus subtilis, Streptococcus pneumoniae, and Lactococcus lactis for live cell imaging. Appl Environ Microbiol 79:6481–6490. doi: 10.1128/AEM.02033-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scott KP, Mercer DK, Glover LA, Flint HJ. 1998. The green fluorescent protein as a visible marker for lactic acid bacteria in complex ecosystems. FEMS Microbiol Ecol 26:219–230. doi: 10.1111/j.1574-6941.1998.tb00507.x. [DOI] [Google Scholar]
- 32.Bloemberg GV, O'Toole GA, Lugtenberg BJ, Kolter R. 1997. Green fluorescent protein as a marker for Pseudomonas spp. Appl Environ Microbiol 63:4543–4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Southward CM, Surette MG. 2002. The dynamic microbe: green fluorescent protein brings bacteria to light. Mol Microbiol 45:1191–1196. doi: 10.1046/j.1365-2958.2002.03089.x. [DOI] [PubMed] [Google Scholar]
- 34.Valdivia RH, Cormack BP, Falkow S. 2006. The uses of green fluorescent protein in prokaryotes. Methods Biochem Anal 47:163–178. [PubMed] [Google Scholar]
- 35.Pédelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS. 2006. Engineering and characterization of a superfolder green fluorescent protein. Nat Biotechnol 24:79–88. doi: 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- 36.Scholz O, Thiel A, Hillen W, Niederweis M. 2000. Quantitative analysis of gene expression with an improved green fluorescent protein. Eur J Biochem 267:1565–1570. doi: 10.1046/j.1432-1327.2000.01170.x. [DOI] [PubMed] [Google Scholar]
- 37.Shaner NC, Steinbach PA, Tsien RY. 2005. A guide to choosing fluorescent proteins. Nat Methods 2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 38.Suarez A, Guttler A, Stratz M, Staendner LH, Timmis KN, Guzman CA. 1997. Green fluorescent protein-based reporter systems for genetic analysis of bacteria including monocopy applications. Gene 196:69–74. doi: 10.1016/S0378-1119(97)00197-2. [DOI] [PubMed] [Google Scholar]
- 39.Cormack BP, Valdivia RH, Falkow S. 1996. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 40.Errampalli D, Leung K, Cassidy MB, Kostrzynska M, Blears M, Lee H, Trevors JT. 1999. Applications of the green fluorescent protein as a molecular marker in environmental microorganisms. J Microbiol Methods 35:187–199. doi: 10.1016/S0167-7012(99)00024-X. [DOI] [PubMed] [Google Scholar]
- 41.Phillips GJ. 2001. Green fluorescent protein–a bright idea for the study of bacterial protein localization. FEMS Microbiol Lett 204:9–18. doi: 10.1111/j.1574-6968.2001.tb10854.x. [DOI] [PubMed] [Google Scholar]
- 42.Jensen PR, Hammer K. 1998. The sequence of spacers between the consensus sequences modulates the strength of prokaryotic promoters. Appl Environ Microbiol 64:82–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moulin P, Patron K, Cano C, Zorgani MA, Camiade E, Borezee-Durant E, Rosenau A, Mereghetti L, Hiron A. 2016. The Adc/Lmb system mediates zinc acquisition in Streptococcus agalactiae and contributes to bacterial growth and survival. J Bacteriol 198:3265–3277. doi: 10.1128/JB.00614-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ackerman DL, Doster RS, Weitkamp JH, Aronoff DM, Gaddy JA, Townsend SD. 2017. Human milk oligosaccharides exhibit antimicrobial and antibiofilm properties against group B Streptococcus. ACS Infect Dis 3:595–605. doi: 10.1021/acsinfecdis.7b00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alkuwaity K, Taylor A, Heckels JE, Doran KS, Christodoulides M. 2012. Group B Streptococcus interactions with human meningeal cells and astrocytes in vitro. PLoS One 7:e42660. doi: 10.1371/journal.pone.0042660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cavaco CK, Patras KA, Zlamal JE, Thoman ML, Morgan EL, Sanderson SD, Doran KS. 2013. A novel C5a-derived immunobiotic peptide reduces Streptococcus agalactiae colonization through targeted bacterial killing. Antimicrob Agents Chemother 57:5492–5499. doi: 10.1128/AAC.01590-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.D'Urzo N, Martinelli M, Pezzicoli A, De Cesare V, Pinto V, Margarit I, Telford JL, Maione D, Members of the DEVANI Study Group. 2014. Acidic pH strongly enhances in vitro biofilm formation by a subset of hypervirulent ST-17 Streptococcus agalactiae strains. Appl Environ Microbiol 80:2176–2185. doi: 10.1128/AEM.03627-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fabbrini M, Sammicheli C, Margarit I, Maione D, Grandi G, Giuliani MM, Mori E, Nuti S. 2012. A new flow-cytometry-based opsonophagocytosis assay for the rapid measurement of functional antibody levels against group B Streptococcus. J Immunol Methods 378:11–19. doi: 10.1016/j.jim.2012.01.011. [DOI] [PubMed] [Google Scholar]
- 49.Oliveira L, Madureira P, Andrade EB, Bouaboud A, Morello E, Ferreira P, Poyart C, Trieu-Cuot P, Dramsi S. 2012. Group B streptococcus GAPDH is released upon cell lysis, associates with bacterial surface, and induces apoptosis in murine macrophages. PLoS One 7:e29963. doi: 10.1371/journal.pone.0029963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carlin AF, Lewis AL, Varki A, Nizet V. 2007. Group B streptococcal capsular sialic acids interact with siglecs (immunoglobulin-like lectins) on human leukocytes. J Bacteriol 189:1231–1237. doi: 10.1128/JB.01155-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chang YC, Olson J, Beasley FC, Tung C, Zhang J, Crocker PR, Varki A, Nizet V. 2014. Group B Streptococcus engages an inhibitory Siglec through sialic acid mimicry to blunt innate immune and inflammatory responses in vivo. PLoS Pathog 10:e1003846. doi: 10.1371/journal.ppat.1003846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cumley NJ, Smith LM, Anthony M, May RC. 2012. The CovS/CovR acid response regulator is required for intracellular survival of group B Streptococcus in macrophages. Infect Immun 80:1650–1661. doi: 10.1128/IAI.05443-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smith JM, Respess RH, Chaffin DG, Larsen B, Jackman SH. 2001. Differences in innate immunologic response to group B streptococcus between colonized and noncolonized women. Infect Dis Obstet Gynecol 9:125–132. doi: 10.1155/S1064744901000230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Périchon B, Szili N, du Merle L, Rosinski-Chupin I, Gominet M, Bellais S, Poyart C, Trieu-Cuot P, Dramsi S. 2017. Regulation of PI-2b pilus expression in hypervirulent Streptococcus agalactiae ST-17 BM110. PLoS One 12:e0169840. doi: 10.1371/journal.pone.0169840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Samen U, Eikmanns BJ, Reinscheid DJ, Borges F. 2007. The surface protein Srr-1 of Streptococcus agalactiae binds human keratin 4 and promotes adherence to epithelial HEp-2 cells. Infect Immun 75:5405–5414. doi: 10.1128/IAI.00717-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aymanns S, Mauerer S, van Zandbergen G, Wolz C, Spellerberg B. 2011. High-level fluorescence labeling of Gram-positive pathogens. PLoS One 6:e19822. doi: 10.1371/journal.pone.0019822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ipe DS, Ben Zakour NL, Sullivan MJ, Beatson SA, Ulett KB, Benjamin WHJ, Davies MR, Dando SJ, King NP, Cripps AW, Schembri MA, Dougan G, Ulett GC. 2015. Discovery and characterization of human-urine utilization by asymptomatic-bacteriuria-causing Streptococcus agalactiae. Infect Immun 84:307–319. doi: 10.1128/IAI.00938-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Leclercq SY, Sullivan MJ, Ipe DS, Smith JP, Cripps AW, Ulett GC. 2016. Pathogenesis of Streptococcus urinary tract infection depends on bacterial strain and beta-hemolysin/cytolysin that mediates cytotoxicity, cytokine synthesis, inflammation and virulence. Sci Rep 6:29000. doi: 10.1038/srep29000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tan CK, Carey AJ, Cui X, Webb RI, Ipe D, Crowley M, Cripps AW, Benjamin WH Jr, Ulett KB, Schembri MA, Ulett GC. 2012. Genome-wide mapping of cystitis due to Streptococcus agalactiae and Escherichia coli in mice identifies a unique bladder transcriptome that signifies pathogen-specific antimicrobial defense against urinary tract infection. Infect Immun 80:3145–3160. doi: 10.1128/IAI.00023-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Andreu N, Zelmer A, Wiles S. 2011. Noninvasive biophotonic imaging for studies of infectious disease. FEMS Microbiol Rev 35:360–394. doi: 10.1111/j.1574-6976.2010.00252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Baldridge GD, Burkhardt N, Herron MJ, Kurtti TJ, Munderloh UG. 2005. Analysis of fluorescent protein expression in transformants of Rickettsia monacensis, an obligate intracellular tick symbiont. Appl Environ Microbiol 71:2095–2105. doi: 10.1128/AEM.71.4.2095-2105.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gustafsson C, Govindarajan S, Minshull J. 2004. Codon bias and heterologous protein expression. Trends Biotechnol 22:346–353. doi: 10.1016/j.tibtech.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 63.Lithwick G, Margalit H. 2003. Hierarchy of sequence-dependent features associated with prokaryotic translation. Genome Res 13:2665–2673. doi: 10.1101/gr.1485203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Moszer I, Rocha EP, Danchin A. 1999. Codon usage and lateral gene transfer in Bacillus subtilis. Curr Opin Microbiol 2:524–528. doi: 10.1016/S1369-5274(99)00011-9. [DOI] [PubMed] [Google Scholar]
- 65.Catalão MJ, Figueiredo J, Henriques MX, Gomes JP, Filipe SR. 2014. Optimization of fluorescent tools for cell biology studies in Gram-positive bacteria. PLoS One 9:e113796. doi: 10.1371/journal.pone.0113796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Unge A, Tombolini R, Molbak L, Jansson JK. 1999. Simultaneous monitoring of cell number and metabolic activity of specific bacterial populations with a dual gfp-luxAB marker system. Appl Environ Microbiol 65:813–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cho JC, Kim SJ. 1999. Viable, but non-culturable, state of a green fluorescence protein-tagged environmental isolate of Salmonella Typhi in groundwater and pond water. FEMS Microbiol Lett 170:257–264. doi: 10.1111/j.1574-6968.1999.tb13382.x. [DOI] [PubMed] [Google Scholar]
- 68.Noah CW, Shaw CI, Ikeda JS, Kreuzer KS, Sofos JN. 2005. Development of green fluorescent protein-expressing bacterial strains and evaluation for potential use as positive controls in sample analyses. J Food Prot 68:680–686. doi: 10.4315/0362-028X-68.4.680. [DOI] [PubMed] [Google Scholar]
- 69.Newman KL, Almeida RP, Purcell AH, Lindow SE. 2003. Use of a green fluorescent strain for analysis of Xylella fastidiosa colonization of Vitis vinifera. Appl Environ Microbiol 69:7319–7327. doi: 10.1128/AEM.69.12.7319-7327.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Duell BL, Cripps AW, Schembri MA, Ulett GC. 2011. Epithelial cell coculture models for studying infectious diseases: benefits and limitations. J Biomed Biotechnol 2011:852419. doi: 10.1155/2011/852419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ofek I, Hasty DL, Doyle RJ, Ofek I. 2003. Bacterial adhesion to animal cells and tissues. ASM Press, Washington, DC. [Google Scholar]
- 72.Sullivan MJ, Forde BM, Prince DW, Ipe DS, Ben Zakour NL, Davies MR, Dougan G, Beatson SA, Ulett GC. 2017. Complete genome sequence of serotype III Streptococcus agalactiae sequence type 17 strain 874391. Genome Announc 5:e01107-17. doi: 10.1128/genomeA.01107-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.LeBlanc DJ, Lee LN, Abu-Al-Jaibat A. 1992. Molecular, genetic, and functional analysis of the basic replicon of pVA380-1, a plasmid of oral streptococcal origin. Plasmid 28:130–145. doi: 10.1016/0147-619X(92)90044-B. [DOI] [PubMed] [Google Scholar]
- 74.Choi KH, Schweizer HP. 2006. mini-Tn7 insertion in bacteria with single attTn7 sites: example Pseudomonas aeruginosa. Nat Protoc 1:153–161. doi: 10.1038/nprot.2006.24. [DOI] [PubMed] [Google Scholar]
- 75.Perez-Casal J, Caparon MG, Scott JR. 1991. Mry, a trans-acting positive regulator of the M protein gene of Streptococcus pyogenes with similarity to the receptor proteins of two-component regulatory systems. J Bacteriol 173:2617–2624. doi: 10.1128/jb.173.8.2617-2624.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cronin M, Knobel M, O'Connell-Motherway M, Fitzgerald GF, van Sinderen D. 2007. Molecular dissection of a bifidobacterial replicon. Appl Environ Microbiol 73:7858–7866. doi: 10.1128/AEM.01630-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Conchello JA, Lichtman JW. 2005. Optical sectioning microscopy. Nat Methods 2:920–931. doi: 10.1038/nmeth815. [DOI] [PubMed] [Google Scholar]
- 78.Cold Spring Harbor Laboratory. 2015. Recipe: Valap sealant. Cold Spring Harb Protoc doi: 10.1101/pdb.rec082917. [DOI] [Google Scholar]
- 79.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lichtman JW, Conchello JA. 2005. Fluorescence microscopy. Nat Methods 2:910–919. doi: 10.1038/nmeth817. [DOI] [PubMed] [Google Scholar]
- 81.Takahashi S, Nagano Y, Nagano N, Fujita K, Taguchi F, Okuwaki Y. 1993. Opsonisation of group B streptococci and restriction endonuclease digestion patterns of their chromosomal DNA. J Med Microbiol 38:191–196. doi: 10.1099/00222615-38-3-191. [DOI] [PubMed] [Google Scholar]
- 82.Takahashi S, Nagano Y, Nagano N, Hayashi O, Taguchi F, Okuwaki Y. 1995. Role of C5a-ase in group B streptococcal resistance to opsonophagocytic killing. Infect Immun 63:4764–4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.