The antimicrobial armamentarium of a bacterium is a major asset for colonizing competitive environments. Bacteriocins comprise a subset of these compounds. Pyocins are an example of such antibacterial proteins produced by Pseudomonas aeruginosa, killing other P. aeruginosa strains. A large group of these molecules show a modular protein architecture that includes a receptor-binding domain for initial target cell attachment and a killer domain. In this study, we have shown that a novel modular pyocin (PaeM4) that kills target bacteria via interference with peptidoglycan assembly takes advantage of the HxuC heme receptor. Cells can protect themselves from killing by the presence of a dedicated immunity partner, an integral inner membrane protein that adopts a transmembrane topology distinct from that of proteins currently known to provide immunity against such toxin activity. Understanding the receptors with which pyocins interact and how immunity to pyocins is achieved is a pivotal step toward the rational design of bacteriocin cocktails for the treatment of P. aeruginosa infections.
KEYWORDS: protein antibiotic, pyocin, polymorphic toxin, toxin-immunity module, bacterial antagonism, TonB-dependent receptor, lipid II
ABSTRACT
Pyocins are bacteriocins secreted by Pseudomonas aeruginosa, and they assist in the colonization of different niches. A major subset of these antibacterial proteins adopt a modular organization characteristic of polymorphic toxins. They include a receptor-binding domain, a segment enabling membrane passage, and a toxin module at the carboxy terminus, which eventually kills the target cells. To protect themselves from their own products, bacteriocin-producing strains express an immunity gene concomitantly with the bacteriocin. We show here that a pyocin equipped with a phylogenetically distinct ColM toxin domain, PaeM4, mediates antagonism against a large set of P. aeruginosa isolates. Immunity to PaeM4 is provided by the inner membrane protein PmiC, which is equipped with a transmembrane topology not previously described for the ColM family. Given that strains lacking a pmiC gene are killed by PaeM4, the presence of such an immunity partner likely is a key criterion for escaping cellular death mediated by PaeM4. The presence of a TonB box in PaeM4 and enhanced bacteriocin activity under iron-poor conditions strongly suggested the targeting of a TonB-dependent receptor. Evaluation of PaeM4 activities against TonB-dependent receptor knockout mutants in P. aeruginosa PAO1 revealed that the heme receptor HxuC (PA1302) serves as a PaeM4 target at the cellular surface. Because other ColM-type pyocins may target the ferrichrome receptor FiuA, our results illustrate the versatility in target recognition conferred by the polymorphic nature of ColM-type bacteriocins.
IMPORTANCE The antimicrobial armamentarium of a bacterium is a major asset for colonizing competitive environments. Bacteriocins comprise a subset of these compounds. Pyocins are an example of such antibacterial proteins produced by Pseudomonas aeruginosa, killing other P. aeruginosa strains. A large group of these molecules show a modular protein architecture that includes a receptor-binding domain for initial target cell attachment and a killer domain. In this study, we have shown that a novel modular pyocin (PaeM4) that kills target bacteria via interference with peptidoglycan assembly takes advantage of the HxuC heme receptor. Cells can protect themselves from killing by the presence of a dedicated immunity partner, an integral inner membrane protein that adopts a transmembrane topology distinct from that of proteins currently known to provide immunity against such toxin activity. Understanding the receptors with which pyocins interact and how immunity to pyocins is achieved is a pivotal step toward the rational design of bacteriocin cocktails for the treatment of P. aeruginosa infections.
INTRODUCTION
Bacteria secrete a variety of molecules to gain hold in competitive environments, and the active killing of neighboring cells is a key strategy facilitating the occupation of a certain niche (1). Antagonistic interactions between microorganisms occur in contact-dependent and contact-independent ways, and a tremendous variety of inhibitory mediators, including bacteriocins and antibiotics, have been described to date (2). Whereas the latter compounds usually exhibit a broad target spectrum, bacteriocin killing is confined to related bacteria, often belonging to the same genus or species as the producer strain (2–4). These ribosomally synthesized peptides and proteins have been well studied in Gram-negative bacteria in particular, with colicins (Escherichia coli) and pyocins (Pseudomonas aeruginosa) serving as model systems (5, 6). Given that bacteriocins allow treatment of bacterial infections in animals (7, 8) and that several bacteriocins, including colicins and certain pyocins, are amenable to large-scale production in plants (9–11), these compounds constitute promising leads for the development of new therapeutic agents (8, 12).
Pyocins differ greatly in size and mechanism of action, and to date several such antibacterial proteins have been identified in P. aeruginosa and other Pseudomonas species. Well-studied groups of Pseudomonas bacteriocins include R-type and F-type tailocins and lectin-like (L-type) bacteriocins; a third large group of P. aeruginosa bacteriocins consists of the S-type pyocins (6, 13). The latter proteins are equipped with a receptor-binding domain, a segment or domain enabling membrane passage, and a toxin domain at the carboxy terminus. To circumvent suicidal expression in producer strains, immunity genes are expressed concomitantly with the bacteriocin. The encoded immunity partners temporarily form a complex with the bacteriocin's toxin module (14) or are inserted in the inner membrane to enable bacteriocin secretion without causing harm to the producer cell (15, 16). Cognate immunity genes are usually located downstream of the bacteriocin genes, allowing efficient coexpression. In general, the encoded proteins selectively provide immunity to “matching” toxin partners only. To date, a large number of toxin functionalities have been described for these modular pyocins, carrying killer modules acting in the periplasm (pore formation or lipid II hydrolysis) or in the cytoplasm (DNase or RNase activities) (5, 6). S-type pyocins typically take advantage of TonB-dependent receptors to gain access to target cells. The latter outer membrane proteins (OMPs) facilitate the binding and transport of iron-bound siderophores to the periplasm (17, 18). This receptor hijacking by pyocins was characterized in detail for pyocin S2; part of the bacteriocin's receptor-binding domain displays structural similarity to the ferripyoverdine and induces the conformational changes needed for import of this siderophore by its receptor (the type I ferripyoverdine transporter FpvAI), tricking the transporter to initiate pyocin translocation (19). Other examples of such pyocin targets are the ferripyochelin transporter FptA (pyocin S5) and the type II ferripyoverdine transporter FpvAII (pyocin S3) (20, 21). Interestingly, different pyocin receptor-binding domains may be coupled to different toxin/immunity modules, validating these modular bacteriocins as a class of polymorphic toxins (6, 22, 23).
A subset of these modular bacteriocins in pseudomonads are those equipped with a ColM domain (24), a toxin module that was first identified in colicin M of E. coli (25, 26) but actually occurs in a wide variety of proteobacterial genera (27), including other gammaproteobacteria (Pectobacterium) (28), as well as betaproteobacteria (Burkholderia) (29). The ColM domain acts in the periplasm and provokes cellular killing through degradation of lipid II (30, 31). Based on phylogeny, 2 large subgroups of Pseudomonas ColM bacteriocins were previously discerned (27). For a number of Pseudomonas ColM bacteriocins (subtype α), piracy of the ferrichrome receptor FiuA was demonstrated (32), indicating an OMP that is also targeted by colicin M in E. coli (FhuA) (25), although the possibility that Pseudomonas bacteriocins from this subtype may also target other receptors cannot be excluded. Pectocins of Pectobacterium also belong to the ColMα subtype but target FusA, a TonB-dependent receptor mediating the uptake of iron-bound plant ferredoxin (33). Immunity to Pseudomonas ColMα bacteriocins is provided by integral membrane proteins named PmiA (for Pseudomonas ColM-type immunity, type A), consisting of 4 transmembrane helices (TMHs) (15). The actual mechanism by which these proteins provide immunity remains unclear. The cell surface target of Pseudomonas ColMβ bacteriocins remains unknown at this point, and such bacteriocin genes are lacking in P. aeruginosa genomes. Immunity to ColMβ-type bacteriocins was demonstrated in Burkholderia but not yet in Pseudomonas and is provided by an integral membrane protein with 3 TMHs (Burkholderia) or a periplasmic module anchored in the inner membrane (Burkholderia and Pseudomonas) (PmiB) (29). More recently, a phylogenetically distinct P. aeruginosa ColM bacteriocin, PaeM4, was functionally identified (10). Through inspection of the genomic context of paeM4, a downstream open reading frame (pmiC) in the opposite direction was retained as a candidate immunity partner (27).
In this study, we explore the occurrence of the paeM4 locus in P. aeruginosa genomes, and we demonstrate the functionality of PmiC as a PaeM4 immunity partner. We show that PaeM4 does not use FiuA as a target receptor, in agreement with earlier suggestions (10), but takes advantage of another TonB-dependent OMP.
RESULTS AND DISCUSSION
Distribution of the pyocin M4 locus in P. aeruginosa genomes.
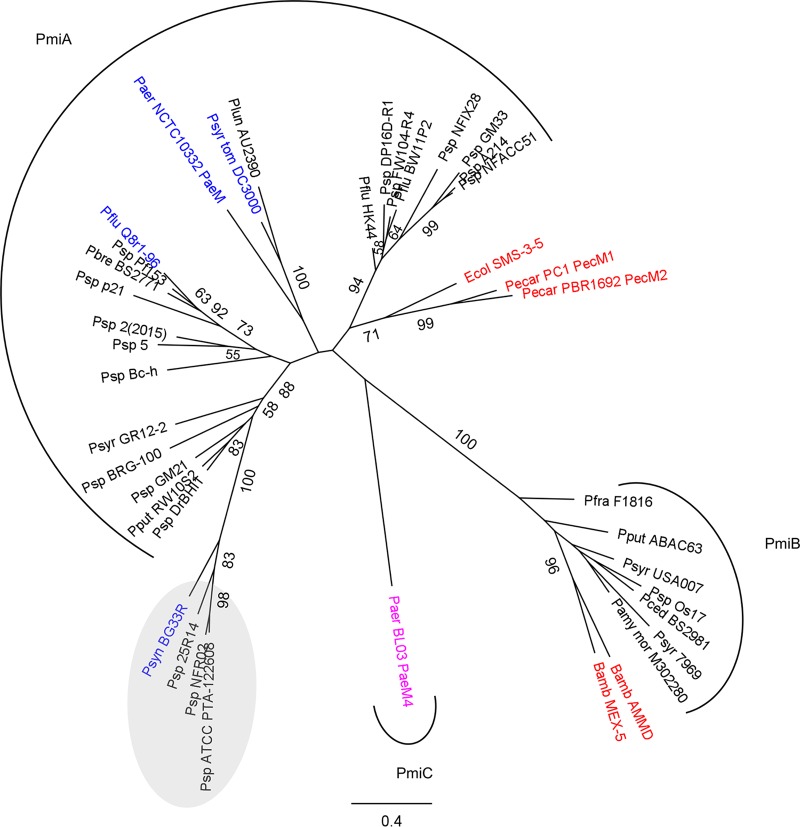
Based on homology searches using the ColM domain of pyocin M4 (named PaeM4) from P. aeruginosa BL03 (10) and previously characterized ColM bacteriocins as search queries, pyocin genes with high sequence similarity (>99% pairwise amino acid [AA] identity of the encoded proteins) were retrieved from ∼3% of the P. aeruginosa genomes (based on 2,665 assembled genomes) (Table 1). PaeM4 (342 AAs) is distinct from the less abundant PaeM (289 AAs, present in <1% of P. aeruginosa genomes) (Fig. 1), and thus far no P. aeruginosa isolate carrying both ColM-type pyocin genes could be identified. The ColM domains of PaeM (P. aeruginosa NCTC 10332) and PaeM4 (P. aeruginosa BL03) share 29% AA sequence identity, whereas their amino-terminal receptor-binding regions cannot be meaningfully aligned. The catalytic motif in the ColM domain of PaeM4 can be recognized as DxYD(x5)QR, slightly deviating from the equivalent motif from colicin M and PaeM, DxYD(x5)HR (27, 31, 34). However, it was found previously that limited variation in this sequence motif does not necessarily affect the bacteriocin function of ColM-type toxins (29). Furthermore, the presence of a proline-rich TonB box at the amino terminus of PaeM4 suggests that this pyocin likely targets a TonB-dependent receptor (27, 35), as was demonstrated previously for PaeM (32) and several other modular S-type pyocins (6).
TABLE 1.
Occurrence of the paeM4 locus in P. aeruginosa genomes and susceptibility to PaeM4
| paeM4 locus | Presence of paeM4 locus in P. aeruginosa genomes (%) | No. (%) of strains with paeM4 locus/total no. of strains in strain panel | No. of strains susceptible to PaeM4/no. of strains with paeM4 locus in strain panel |
|---|---|---|---|
| paeM4-pmiC | 3.04 | 2/75 (2.67) | 0/2 |
| pmiC (orphan) | 27.88 | 33/75 (44.00) | 2/33 |
| Absent | 69.08 | 40/75 (53.33) | 40/40 |
FIG 1.
Phylogeny of ColM domains derived from characterized (blue) and putative (black) Pseudomonas bacteriocins and characterized bacteriocins in other genera (red). The distinct ColM-type bacteriocin from P. aeruginosa BL03 is shown in pink. In the maximum likelihood phylogenetic tree (PhyML, with the JTT substitution model) of ColM domains, highly homologous Pseudomonas sequences (>75% pairwise AA identity for full-length proteins) are represented by one sequence only. Bootstrap values (percentages of 1,000 replicates) higher than 50 are shown at the branches. The scale bar represents 0.4 substitutions per site. Pseudomonas representatives are grouped by arcs specifying the immunity partner present. Proteins retrieved from strains in which the ColM domain is part of a bacteriocin integrating 2 toxin domains are grouped in a gray oval. Genes encoding such hybrid bacteriocins are not accompanied by a (known) putative ColM-type immunity partner. Bamb, Burkholderia ambifaria; Ecol, Escherichia coli; Paer, Pseudomonas aeruginosa; Pamy mor, Pseudomonas amygdali pv. morsprunorum; Pbre, Pseudomonas brenneri; Pced, Pseudomonas cedrina; Pecar, Pectobacterium carotovorum; Pflu, Pseudomonas fluorescens; Pfra, Pseudomonas fragi; Plun, Pseudomonas lundensis; Pput, Pseudomonas putida; Psp, Pseudomonas sp.; Psyn, Pseudomonas synxantha; Psyr, Pseudomonas syringae; Psyr tom, Pseudomonas syringae pv. tomato.
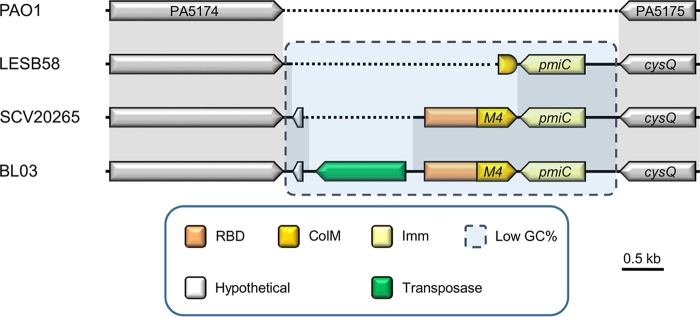
The paeM4 gene is part of a short region (∼2.5 kb) with a GC content markedly lower than average (∼46% versus ∼67%); it consistently arises at the same locus but differs from the genomic position of paeM, which occurs in exoU-containing genomic island A (24). In contrast, this ColM-type pyocin is sandwiched between a gene encoding a β-ketoacyl synthase and the 3′,5′-bisphosphate nucleotidase gene cysQ (orthologues PA5174 and PA5175, respectively, in Pseudomonas aeruginosa PAO1) (Fig. 2). Downstream and in the opposite direction of paeM4 is located a candidate immunity gene, previously termed pmiC (27). The encoded protein (232 AAs) is distinct from the PmiA ColM-type immunity partners; whereas the latter host 4 TMHs (15), PmiC adopts 6 predicted TMHs (Fig. 3A) (36). By genome analysis, a putative ColM immunity partner, PmiB, with a periplasmic domain was previously identified in a small set of pseudomonads; however, no pmiB immunity genes can be retrieved from P. aeruginosa genomes (27, 29) (Fig. 1). The paeM4 locus may also be loaded with a transposase, such as in P. aeruginosa BL03, although this organization appears to be quite rare. In ∼28% of P. aeruginosa genomes, such as in P. aeruginosa LESB58, only pmiC is present, accompanied by a short remnant of paeM4 (Fig. 2). Immunity genes not accompanied by an adjacent full-length bacteriocin are generally referred to as orphans. Such orphan immunity genes were studied previously in the context of different polymorphic toxin systems mediating antagonism via cell-to-cell contact (37–39).
FIG 2.
Representative genetic organizations of the paeM4 locus in Pseudomonas aeruginosa strains. Synteny is represented by sequence conservation (gray shading). Genes are represented by arrows, and a paeM4 fragment is shown as a shorter rounded shape. Functions of genes and gene parts (if known) are specified by color in the legend. The receptor-binding domain (RBD) of PaeM4 includes the TonB box that is part of a short region at the amino terminus of the encoded protein. The candidate immunity partner (Imm) PmiC is encoded downstream of paeM4, in the opposite direction. Dotted lines indicate the lack of an equivalent nucleotide sequence.
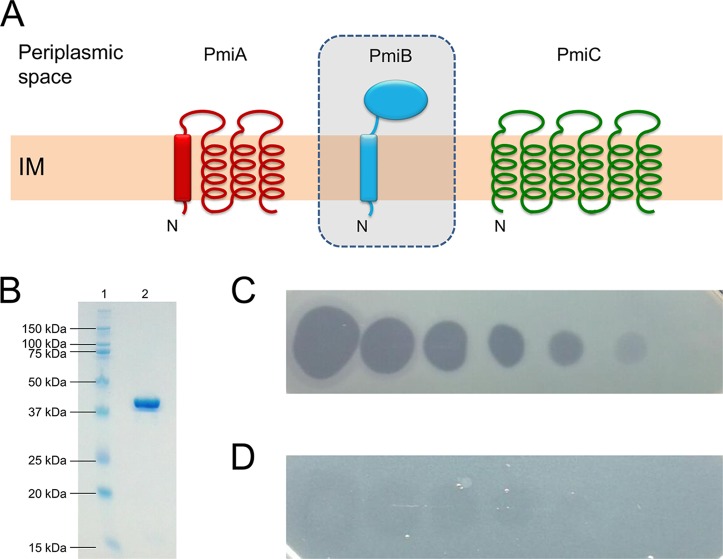
FIG 3.
(A) Membrane topology model of ColM-type immunity proteins in pseudomonads. PmiA carries 4 predicted TMHs, the first of which may act as a Sec- or Tat-dependent signal sequence (rectangle). The putative ColM immunity partner PmiB (in shaded box) has a periplasmic domain and is anchored in the inner membrane (IM), but PmiB-encoding genes are absent from P. aeruginosa genomes. PmiC has 6 TMHs and occurs exclusively in P. aeruginosa. N indicates the amino terminus. Pseudomonas ColM-type immunity proteins were drawn as in reference 27. (B) SDS-PAGE gel of purified recombinant PaeM4. Lane 1, Kaleidoscope ladder, with sizes indicated in kilodaltons; lane 2, purified His-tagged PaeM4 (∼41 kDa; predicted size, 39.99 kDa). (C and D) Spot assay of 10-fold serial dilutions of PaeM4 (initial concentration, 1 mg/ml) against P. aeruginosa Br993 on CAA medium, in the absence (C) or presence (D) of 50 μM FeCl3.
PaeM4 represents a broadly active bacteriocin.
The paeM4 gene (locus tag Q057_04089, from P. aeruginosa BL03) was cloned in expression vector pET28a(+). Previously, it was found that a coexpressed immunity gene protecting cells from colicin M toxin function is not needed when such bacteriocins are expressed in the cytoplasm of E. coli (24, 28, 29); therefore, the candidate immunity gene pmiC was not included in the construct. The bacteriocin gene was cloned to encode a carboxy-terminal His-tagged protein, since an amino-terminal fusion product would interfere with the import-related function of the amino-terminal translocation segment of the pyocin (15, 34, 35). After transformation into E. coli BL21(DE3), pyocin gene expression was induced, cells were harvested, soluble proteins were isolated, and the recombinant protein was purified (Fig. 3B). His-tagged pyocin displayed (weak) antagonistic activity against P. aeruginosa PAO1, as demonstrated earlier (10). A total of 56% of the strains (42/75 strains) in our P. aeruginosa test panel were killed by PaeM4 (Table 1; also see Table S1 in the supplemental material), a PaeM4 killing frequency very similar to that noted by Paškevičius and colleagues (53%) (10). Bactericidal activity was evident under iron-poor conditions (Casamino Acids [CAA] medium), whereas PaeM4 function was almost completely abrogated in the presence of iron (50 μM FeCl3) (Fig. 3C and D). This finding suggests that PaeM4 takes advantage of an outer membrane receptor that is controlled by the ferric uptake regulator (Fur) (40). Similar to several other modular pyocins (20, 21, 32, 41, 42), this correlates with the targeting of a TonB-dependent receptor involved in iron acquisition (see above).
PmiC confers immunity to PaeM4 killing.
The putative PaeM4 immunity partner, pmiC (locus tag Q057_04090, from P. aeruginosa BL03), was cloned in shuttle vector pJB3Tc20 and introduced into the PaeM4-susceptible strain P. aeruginosa PA7 (43) via electroporation. Via spot-on-lawn assays, transformants were tested for altered PaeM4 sensitivity. When equipped with pmiC, transformants were no longer susceptible to PaeM4, confirming the immunity function of PmiC (Fig. 4A and B). In contrast, when pmiA, providing immunity to PaeM from P. aeruginosa NCTC 10332 (15), was introduced into PA7, transformants remained as sensitive to PaeM4 as before (Fig. 4C). Conversely, pmiC could not immunize cells against the PaeM function in P. aeruginosa PA7 (15) (Fig. 4D to F). These observations are in line with the generally high specificity and selectivity of immunity partners in silencing toxin functions (14, 44–46).
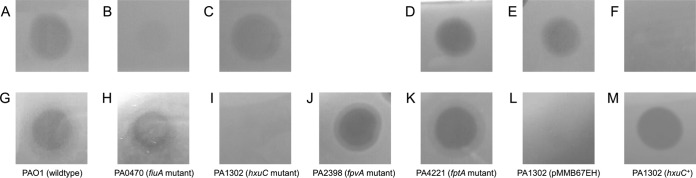
FIG 4.
(A to C) Spot assays of PaeM4 (10 μl; 0.08 mg/ml) against P. aeruginosa PA7 equipped with pJB3Tc20 (empty plasmid) (control) (A), expressing pmiCBL03 (B), or expressing pmiANCTC10332 (C). (D to F) Spot assays of PaeM (10 μl; 0.4 mg/ml) against P. aeruginosa PA7 equipped with pJB3Tc20 (empty plasmid) (D), expressing pmiCBL03 (E), or expressing pmiANCTC10332 (F). (G to M) Spot assays of PaeM4 (10 μl; 1 mg/ml) against P. aeruginosa PAO1 and selected TonB-dependent transporter mutants, i.e., wild-type PAO1 (G), PA0470 (fiuA) (H), PA1302 (hxuC) (I), PA2398 (fpvA) (J), PA4221 (fptA) (K), PA1302 equipped with pMMB67EH (empty vector) (control) (L), and PA1302 expressing hxuC (pCMPG6293) (M). Three biological repeats and 3 technical repeats were performed for every test.
In a next step, all isolates of the strain panel tested for PaeM4 susceptibility were evaluated for the presence of the paeM4 locus via PCR, using a primer couple situated on the β-ketoacyl synthase gene (forward) and cysQ (reverse) (Table 1 and Fig. 2). Amplicon sizes of ∼620 bp (based on the genome sequence of PAO1), ∼2,020 bp (LESB58), and ∼3,100 bp (SCV20265) corresponded to the absence and the presence of a partial and complete paeM4 locus, respectively (gel electrophoresis data not shown). Partial paeM4 loci (33/75 strains [44.00%]) were somewhat more abundant than expected from P. aeruginosa genome sequence data in GenBank (27.9%) (Table 1). Interestingly, all strains from the panel lacking pmiC were killed by PaeM4 (Table S1). Conversely, PaeM4-insensitive isolates were equipped with a partial or complete paeM4 region, suggesting that the pmiC-encoded immunity is also functional in vivo. Two strains of our P. aeruginosa panel (CF_PA41 [47] and CPHL2000 [48]) were sensitive to PaeM4, despite carrying a pmiC orphan immunity gene (based on PCR screening). In the corresponding amplicons, no single-nucleotide polymorphisms (such as frameshifts) that might account for a loss of immunity function could be detected (data not shown). Overall, pmiC genes coupled to paeM4 display a high degree of sequence conservation (>97% pairwise AA identity of the encoded proteins), whereas conservation with and among pmiC orphans is lower (∼76% AA identity with PmiCBL03 and >93% AA identity among PmiC orphans). Previously, it was noted that homologous orphan pyocin immunity genes with significant sequence similarity may offer immunity to noncognate toxins (15, 49), suggesting the functionality of orphan pmiC genes. It should be emphasized that significant DNA sequence variation can be noted in the regions preceding the orphan pmiC genes, which may in turn exert transcriptional effects. The expression, functionality, and role of orphan (pmiC) immunity genes in strains escaping pyocin killing will be discussed in a separate publication.
PaeM4 targets an uptake system for heme.
The presence of a TonB box at the amino-terminal end of PaeM4 and the observation that PaeM4 bacteriocin activity is significantly enhanced under iron-poor conditions strongly suggest the exploitation of a TonB-dependent receptor for target cell attachment and subsequent transfer to the periplasm. Strain PAO1, which is susceptible to PaeM4 and PaeM, contains a total of 36 TonB-dependent transporters (50), and defined mutants (51) of each of them were tested for altered pyocin susceptibility. As demonstrated previously (10), PAO1 mutant PA0470 (defective in the PaeM receptor, fiuA) was still sensitive to PaeM4 (Fig. 4G and H). This can be expected, given the quite different receptor-binding domains of PaeM and PaeM4. Other mutants also displayed PaeM4 susceptibility patterns similar to that of the wild-type strain, except for PA1302 (Fig. 4I to K; also see Table S1 in the supplemental material). The latter mutant is altered in the TonB-dependent receptor gene hxuC, which is involved in the uptake of heme, in line with the hypothesis regarding the targeting of an iron-dependent receptor by PaeM4. This OMP is part of the 3-gene cluster hxuCBA, which was previously studied in more detail in Haemophilus influenzae for the import of heme-hemopexin (52) and was shown to act as a virulence factor (53). The role of PA1302 was validated via complementation; when hxuC was cloned in shuttle vector pMMB67EH and transformed to PA1302, the PaeM4 susceptibility phenotype (spot assay) was restored (Fig. 4L and M).
The hxuC gene can be readily retrieved from all sequenced P. aeruginosa strains and appears to be part of the P. aeruginosa core genome (54). Furthermore, HxuC displays a high degree of conservation (>95% pairwise AA identity among orthologues in P. aeruginosa). Therefore, the presence of a pmiC immunity gene likely is the primary determinant accounting for PaeM4 resistance, a hypothesis that is supported by the results of the PaeM4 locus screening in this study. This observation may also be of future interest for designing bacteriocin cocktails (12). Preference should be given to the design of (chimeric) pyocins uniting (i) toxin modules for which immunity genes rarely occur and (ii) receptor-binding domains targeting TonB-dependent OMPs, which are part of the P. aeruginosa core genome and are highly conserved. In this perspective, the results obtained here suggest that the receptor-binding domain of PaeM4 constitutes an interesting candidate to include in the design of chimeric pyocins, in contrast to its toxin module (given the frequent occurrence of pmiC). Recently, it was shown that polymorphism in BamA, an insertase accounting for the assembly of proteins in the outer membrane, determines which Pseudomonas strains are killed by lectin-like bacteriocins (55). Also, polymorphism of the FiuA receptor was postulated to account for differences in susceptibility to the highly similar PaeM bacteriocins (from P. aeruginosa strains JJ692 and NCTC10332) (10, 12).
MATERIALS AND METHODS
Genome searches and phylogenetic analysis.
Genes encoding putative ColM bacteriocins in P. aeruginosa (2,665 assembled P. aeruginosa genomes; data collection on 10 December 2017) were identified by BLAST homology searches using the National Center for Biotechnology (NCBI) nonredundant database. ColM domains of previously characterized ColM-type bacteriocins were used as a search query (Pfam PF14859) (27). Similarly, P. aeruginosa genomes were analyzed for the presence of genes encoding PmiC immunity proteins (no Pfam domain). Regions upstream and downstream of paeM4 genes were analyzed further. progressiveMauve was used to align paeM4 loci (56), multiple sequence alignments were generated with MUSCLE (proteins) (57) and MAFFT (DNA sequences) (58), and phylogenetic analysis was executed with PhyML (1,000 bootstrap replicates) using the JTT substitution model (59). Transmembrane regions and topology were predicted using TOPCONS (http://topcons.cbr.su.se) (60) and TMHMM (http://www.cbs.dtu.dk/services/TMHMM) (36).
Bacterial strains and media.
P. aeruginosa isolates were grown in LB medium (MP Biomedicals) or CAA medium (BD Bacto), and E. coli was grown in LB medium. P. aeruginosa PAO1 and transposon mutants altered in genes encoding (putative) TonB-dependent receptors were obtained from the Manoil laboratory (University of Washington) (51); other strains originated from an in-house collection and J. P. Pirnay (Queen Astrid Military Hospital, Belgium) (48). The presence of the transposon insert in the correct gene in each of the PAO1 mutants was tested and confirmed via PCR (data not shown). All strains used in this study are summarized in Table S1. Strains were routinely grown at 37°C, with shaking at 200 rpm. Plasmids were propagated in E. coli DH5α (BIOKÉ), and E. coli BL21(DE3) (VWR International) was used for the production of recombinant bacteriocins. Bacterial growth media were solidified with agar (1.5%; Invitrogen). Filter-sterilized (0.22-μm filters; Sarstedt) additives, i.e., kanamycin (50 μg/ml; Sigma-Aldrich), gentamicin (15 μg/ml; TCI Europe NV), tetracycline (15 to 150 μg/ml; Sigma-Aldrich), isopropyl β-d-thiogalactopyranoside (IPTG) (0.1 to 1 mM; Formedium), and FeCl3 (50 μM; Sigma-Aldrich), were added when needed. Bacterial strains were stored on plates at 4°C or in glycerol (50% [vol/vol]; VWR International) at −80°C.
DNA methods and plasmid construction.
Genomic DNA was extracted from overnight cell cultures using the Puregene Yeast/Bact. kit B (Qiagen), and synthetic genes and primers were obtained from IDT DNA. The gene encoding bacteriocin PaeM4 from P. aeruginosa BL03 (locus tag Q057_04089) and the candidate immunity gene pmiC from P. aeruginosa BL03 (locus tag Q057_04050) were PCR amplified with Q5 polymerase (New England BioLabs) using a T100 thermal cycler (Bio-Rad). Primers are listed in Table 2. Subsequently, PCR amplicons were purified with the GenElute PCR cleanup kit (Sigma-Aldrich) and digested with NcoI and XhoI (for paeM4) or PstI and EcoRI (for pmiCBL03) (restriction enzymes were from New England BioLabs). After purification with the PCR cleanup kit, paeM4 and pmiC were ligated in pET28a(+) and the shuttle vector pJB3Tc20 (61), respectively, using T4 DNA ligase (Invitrogen). Ligation reactions were performed overnight at 16°C. The following day, products were transformed into E. coli DH5α via heat shock. After selection on LB plates containing the appropriate antibiotic, transformants were validated for the presence of the insert via colony PCR using Taq polymerase (New England BioLabs) with the primer pairs PGPRB-10249/PGPRB-10250 (pET28a) and PGPRB-10255/PGPRB-10256 (pJB3Tc20). Plasmids with the insert were isolated with the Nucleospin Plasmid EasyPure kit (Macherey-Nagel) and sequence verified by Sanger sequencing (GATC Biotech, Constance, Germany). A similar procedure was followed for the cloning of PA1302 from P. aeruginosa PAO1 in pMMB67EH (62), using restriction enzymes EcoRI and HindIII. The pyocin-encoding plasmids pCMPG6283 (containing paeM4) and pCMPG6250 (containing paeM from P. aeruginosa NCTC10332), which was constructed previously (15), were transformed into E. coli BL21(DE3), with selection on kanamycin. Plasmids containing the immunity genes pmiCBL03 (pCMPG6287) and pmiANCTC10332 (pCMPG6252), which was constructed previously (15), were electroporated to sucrose-competent (63) P. aeruginosa cells using a GenePulser Xcell (Bio-Rad), with selection on tetracycline. pCMPG6293 (hxuCPAO1 in pMMB67EH) was electroporated into P. aeruginosa PAO1 mutant PA1302, with selection on gentamicin. All plasmids used in this study are summarized in Table S2. Plasmids without inserts were used as controls in subsequent assays. All enzymes (DNA polymerases, restriction enzymes, and T4 DNA ligase) were used according to the supplier's specifications. The preparation of competent E. coli and heat shock transformation of E. coli were performed following standard procedures (64).
TABLE 2.
Primers used in this study
| Primer name | Sequencea | Purpose |
|---|---|---|
| PGPRB-10249 | TGGCAGCAGCCAACTCAGCTT | Insert validation in pET28a |
| PGPRB-10250 | TATAGGCGCCAGCAACCGCA | Insert validation in pET28a |
| PGPRB-10255 | GCTCACTCATTAGGCACCC | Insert validation in pJB3Tc20 |
| PGPRB-10256 | GGTAACGCCAGGGTTTTC | Insert validation in pJB3Tc20 |
| PGPRB-10409 | AGCGGATAACAATTTCACACAGG | Insert validation in pMMB67EH |
| PGPRB-10410 | TCTGTATCAGGCTGAAAATCTTCTCTC | Insert validation in pMMB67EH |
| PGPRB-10434 | TGGCTACCATGGGTCCAATGGAATTCCCTCCAACTAC | Cloning of paeM4BL03 in pET28a |
| PGPRB-10436 | TGGCTACTCGAGCAACTTATCTCCAGTGCCTGAT | Cloning of paeM4BL03 in pET28a |
| PGPRB-10440 | TGGCTACTGCAGCTTGCAGGACGTAGAATGCATGAG | Cloning of pmiCBL03 in pJB3Tc20 |
| PGPRB-10441 | TGGCTAGAATTCCTATCTCTTGATAGGTGCTTTCTCATCAC | Cloning of pmiCBL03 in pJB3Tc20 |
| PGPRB-10448 | GCCGCTGGTATACAAGAAGGA | Presence of paeM4 locus in P. aeruginosa genomes |
| PGPRB-10449 | GAGGAGTTCACCGTCAACGT | Presence of paeM4 locus in P. aeruginosa genomes |
| PGPRB-10473 | TGGCTAGAATTCAGTCACTTGGGGAGAGTTCCAGCATGAC | Cloning of hxuC in pMMB67EH |
| PGPRB-10474 | TGGCTAAAGCTTCCAATACTTCATGCTCGGCGACAACCG | Cloning of hxuC in pMMB67EH |
Restriction sites incorporated in primers are shown in bold, as follows: AAGCTT, HindIII; CCATGG, NcoI; CTCGAG, XhoI; CTGCAG, PstI; GAATTC, EcoRI.
The presence of a partial or complete paeM4 locus in strains from our P. aeruginosa strain panel was evaluated by PCR screening (in case no genomic information was available). Primers PGPRB-10463 and PGPRB-10464 were selected based on DNA sequence conservation in the paeM4-flanking genes PA5174 and PA5175 (P. aeruginosa PAO1). Q5 polymerase was used for PCR amplification. Fragments to be sequenced were purified with the PCR cleanup kit.
Expression and purification of His-tagged bacteriocin.
Plasmids containing pyocin-encoding genes (PaeM [pCMPG6250] and PaeM4 [pCMPG6283]) were transformed into E. coli BL21(DE3) via heat shock. Recombinant His-tagged proteins were generated as follows. First, 5-ml volumes of overnight E. coli cultures were transferred to 500-ml Erlenmeyer flasks and incubated at 37°C until the optical density at 600 nm (OD600) reached ∼0.7. Next, cell cultures were cooled, supplemented with IPTG (final concentration of 1 mM), and incubated overnight at 20°C, with shaking. The following day, cells were collected via centrifugation at 5,000 × g (20 min at 4°C) using a Beckman Coulter Avanti J-E ultracentrifuge, supernatants were discarded, and pellets were frozen overnight at −20°C. The next day, cell pellets were thawed, resuspended in lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole [pH 7.8]; VWR International and Sigma-Aldrich) (5 ml of buffer per 1 g of pellet), and sonicated using a Branson digital sonifier (amplitude of 20%; 10 cycles of 30 s on and 30 s off). Cell lysates were treated with Pierce universal nuclease (0.01 U/μl; Invitrogen) for 1 h at 37°C and then were centrifuged at 10,000 × g for 30 min at 4°C, to remove residual debris. Supernatants containing soluble proteins were filtered (0.22-μm filters) and validated for the presence of recombinant proteins by SDS-PAGE (Invitrogen), with subsequent Coomassie blue staining (InstantBlue; Expedeon). Samples containing His-tagged proteins were applied to 5-ml HisTrap HP columns (GE Healthcare) and purified by affinity chromatography using an Äkta Purifier (GE Healthcare). Bacteriocins were eluted with a linear gradient of imidazole (10 to 500 mM), using lysis buffer as the running buffer. Fractions that eluted at high imidazole concentrations were validated for the presence of recombinant pyocin by SDS-PAGE, using the Precision Plus Protein Kaleidoscope ladder (Bio-Rad) as a size standard, and were dialyzed with Tris buffer (50 mM, 200 mM NaCl [pH 7.5]; Sigma-Aldrich) overnight. Protein samples were subsequently concentrated using Vivaspin filters (Sartorius) and were purified via gel filtration on a HiLoad Superdex 200 preparative grade 16/600 column (GE Healthcare), using Tris buffer as the running buffer. Concentrations of purified bacteriocins were calculated by absorbance measurements at 280 nm, using a ND-1000 spectrophotometer (Thermo Scientific). Calculated molar extinction coefficients for the His-tagged pyocins PaeM and PaeM4 were 73,340 M−1 cm−1 and 77,810 M−1 cm−1, respectively.
Bacteriocin spot assay.
The bacteriocin activity of PaeM4 and PaeM was evaluated using the soft agar overlay assay (65). Purified and filter-sterilized recombinant proteins (10-μl volumes; 1 mg/ml) were spotted onto lawns of test bacteria. After air drying of the bacteriocin spots, petri dishes were incubated overnight. The following day, plates were evaluated for the presence of halos, i.e., zones lacking bacterial growth, which were indicative of antagonistic activity. Dialysis buffer was used as a negative control. Spot assays were performed with bacteriocin purified from 3 independent batches. All tests were routinely performed in triplicate (independent repeats) on CAA plates, which were supplemented with IPTG, tetracycline, or FeCl3 if needed. For evaluation of the contributions of immunity genes and FeCl3 to bacteriocin susceptibility, serial dilution series were used for comparison.
Supplementary Material
ACKNOWLEDGMENTS
M.G.K.G. is a postdoctoral fellow (grant 12M4618N) supported by the Fonds voor Wetenschappelijk Onderzoek (FWO) Vlaanderen. This work was supported by Krediet aan Navorser 1529718N from FWO Vlaanderen. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
We thank René De Mot for critical reading and suggestions.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00716-18.
REFERENCES
- 1.Hibbing ME, Fuqua C, Parsek MR, Peterson SB. 2010. Bacterial competition: surviving and thriving in the microbial jungle. Nat Rev Microbiol 8:15–25. doi: 10.1038/nrmicro2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chassaing B, Cascales E. 2018. Antibacterial weapons: targeted destruction in the microbiota. Trends Microbiol 26:329–338. doi: 10.1016/j.tim.2018.01.006. [DOI] [PubMed] [Google Scholar]
- 3.Hegarty JW, Guinane CM, Ross RP, Hill C, Cotter PD. 2016. Bacteriocin production: a relatively unharnessed probiotic trait? F1000Res 5:2587. doi: 10.12688/f1000research.9615.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Riley MA, Robinson SM, Roy CM, Dorit RL. 2013. Rethinking the composition of a rational antibiotic arsenal for the 21st century. Future Med Chem 5:1231–1242. doi: 10.4155/fmc.13.79. [DOI] [PubMed] [Google Scholar]
- 5.Cascales E, Buchanan SK, Duché D, Kleanthous C, Lloubès R, Postle K, Riley M, Slatin S, Cavard D. 2007. Colicin biology. Microbiol Mol Biol Rev 71:158–229. doi: 10.1128/MMBR.00036-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghequire MG, De Mot R. 2014. Ribosomally encoded antibacterial proteins and peptides from Pseudomonas. FEMS Microbiol Rev 38:523–568. doi: 10.1111/1574-6976.12079. [DOI] [PubMed] [Google Scholar]
- 7.Scholl D. 2017. Phage tail-like bacteriocins. Annu Rev Virol 4:453–467. doi: 10.1146/annurev-virology-101416-041632. [DOI] [PubMed] [Google Scholar]
- 8.Behrens HM, Six A, Walker D, Kleanthous C. 2017. The therapeutic potential of bacteriocins as protein antibiotics. Emerg Top Life Sci 1:65–74. doi: 10.1042/ETLS20160016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schulz S, Stephan A, Hahn S, Bortesi L, Jarczowski F, Bettmann U, Paschke AK, Tusé D, Stahl CH, Giritch A, Gleba Y. 2015. Broad and efficient control of major foodborne pathogenic strains of Escherichia coli by mixtures of plant-produced colicins. Proc Natl Acad Sci U S A 112:E5454–E5460. doi: 10.1073/pnas.1513311112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paškevičius Š, Starkevič U, Misiūnas A, Vitkauskienė A, Gleba Y, Ražanskienė A. 2017. Plant-expressed pyocins for control of Pseudomonas aeruginosa. PLoS One 12:e0185782. doi: 10.1371/journal.pone.0185782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schneider T, Hahn-Löbmann S, Stephan A, Schulz S, Giritch A, Naumann M, Kleinschmidt M, Tusé D, Gleba Y. 2018. Plant-made Salmonella bacteriocins salmocins for control of Salmonella pathovars. Sci Rep 8:4078. doi: 10.1038/s41598-018-22465-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghequire MGK, De Mot R. 2018. Turning over a new leaf: bacteriocins going green. Trends Microbiol 26:1–2. doi: 10.1016/j.tim.2017.11.001. [DOI] [PubMed] [Google Scholar]
- 13.Ghequire MG, De Mot R. 2015. The tailocin tale: peeling off phage tails. Trends Microbiol 23:587–590. doi: 10.1016/j.tim.2015.07.011. [DOI] [PubMed] [Google Scholar]
- 14.Joshi A, Grinter R, Josts I, Chen S, Wojdyla JA, Lowe ED, Kaminska R, Sharp C, McCaughey L, Roszak AW, Cogdell RJ, Byron O, Walker D, Kleanthous C. 2015. Structures of the ultra-high-affinity protein-protein complexes of pyocins S2 and AP41 and their cognate immunity proteins from Pseudomonas aeruginosa. J Mol Biol 427:2852–2866. doi: 10.1016/j.jmb.2015.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghequire MG, Kemland L, De Mot R. 2017. Novel immunity proteins associated with colicin M-like bacteriocins exhibit promiscuous protection in Pseudomonas. Front Microbiol 8:93. doi: 10.3389/fmicb.2017.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rasouliha BH, Ling H, Ho CL, Chang MW. 2013. A predicted immunity protein confers resistance to pyocin S5 in a sensitive strain of Pseudomonas aeruginosa. Chembiochem 14:2444–2446. doi: 10.1002/cbic.201300410. [DOI] [PubMed] [Google Scholar]
- 17.Cornelis P. 2010. Iron uptake and metabolism in pseudomonads. Appl Microbiol Biotechnol 86:1637–1645. doi: 10.1007/s00253-010-2550-2. [DOI] [PubMed] [Google Scholar]
- 18.Cornelis P, Dingemans J. 2013. Pseudomonas aeruginosa adapts its iron uptake strategies in function of the type of infections. Front Cell Infect Microbiol 3:75. doi: 10.3389/fcimb.2013.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.White P, Joshi A, Rassam P, Housden NG, Kaminska R, Goult JD, Redfield C, McCaughey LC, Walker D, Mohammed S, Kleanthous C. 2017. Exploitation of an iron transporter for bacterial protein antibiotic import. Proc Natl Acad Sci U S A 114:12051–12056. doi: 10.1073/pnas.1713741114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elfarash A, Dingemans J, Ye L, Hassan AA, Craggs M, Reimmann C, Thomas MS, Cornelis P. 2014. Pore-forming pyocin S5 utilizes the FptA ferripyochelin receptor to kill Pseudomonas aeruginosa. Microbiology 160:261–269. doi: 10.1099/mic.0.070672-0. [DOI] [PubMed] [Google Scholar]
- 21.Baysse C, Meyer JM, Plesiat P, Geoffroy V, Michel-Briand Y, Cornelis P. 1999. Uptake of pyocin S3 occurs through the outer membrane ferripyoverdine type II receptor of Pseudomonas aeruginosa. J Bacteriol 181:3849–3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jamet A, Nassif X. 2015. New players in the toxin field: polymorphic toxin systems in bacteria. mBio 6:e00285-15. doi: 10.1128/mBio.00285-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sharp C, Bray J, Housden NG, Maiden MCJ, Kleanthous C. 2017. Diversity and distribution of nuclease bacteriocins in bacterial genomes revealed using hidden Markov models. PLoS Comput Biol 13:e1005652. doi: 10.1371/journal.pcbi.1005652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barreteau H, Bouhss A, Fourgeaud M, Mainardi JL, Touzé T, Gérard F, Blanot D, Arthur M, Mengin-Lecreulx D. 2009. Human- and plant-pathogenic Pseudomonas species produce bacteriocins exhibiting colicin M-like hydrolase activity towards peptidoglycan precursors. J Bacteriol 191:3657–3664. doi: 10.1128/JB.01824-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Braun V, Helbig S, Patzer SI, Pramanik A, Römer C. 2015. Import and export of bacterial protein toxins. Int J Med Microbiol 305:238–242. doi: 10.1016/j.ijmm.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 26.Braun V, Helbig S, Patzer SI. 2012. Import of periplasmic bacteriocins targeting the murein. Biochem Soc Trans 40:1449–1455. doi: 10.1042/BST20120175. [DOI] [PubMed] [Google Scholar]
- 27.Ghequire MGK, Buchanan SK, De Mot R. 2018. The ColM family, polymorphic toxins breaching the bacterial cell wall. mBio 9:e02267-17. doi: 10.1128/mBio.02267-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grinter R, Milner J, Walker D. 2012. Ferredoxin containing bacteriocins suggest a novel mechanism of iron uptake in Pectobacterium spp. PLoS One 7:e33033. doi: 10.1371/journal.pone.0033033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ghequire MG, De Mot R. 2015. Distinct colicin M-like bacteriocin-immunity pairs in Burkholderia. Sci Rep 5:17368. doi: 10.1038/srep17368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.El Ghachi M, Bouhss A, Barreteau H, Touzé T, Auger G, Blanot D, Mengin-Lecreulx D. 2006. Colicin M exerts its bacteriolytic effect via enzymatic degradation of undecaprenyl phosphate-linked peptidoglycan precursors. J Biol Chem 281:22761–22772. doi: 10.1074/jbc.M602834200. [DOI] [PubMed] [Google Scholar]
- 31.Barreteau H, Bouhss A, Gérard F, Duché D, Boussaid B, Blanot D, Lloubès R, Mengin-Lecreulx D, Touzé T. 2010. Deciphering the catalytic domain of colicin M, a peptidoglycan lipid II-degrading enzyme. J Biol Chem 285:12378–12389. doi: 10.1074/jbc.M109.093583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ghequire MGK, Kemland L, Anoz-Carbonell E, Buchanan SK, De Mot R. 2017. A natural chimeric Pseudomonas bacteriocin with novel pore-forming activity parasitizes the ferrichrome transporter. mBio 8:e01961-16. doi: 10.1128/mBio.01961-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grinter R, Josts I, Mosbahi K, Roszak AW, Cogdell RJ, Bonvin AM, Milner JJ, Kelly SM, Byron O, Smith BO, Walker D. 2016. Structure of the bacterial plant-ferredoxin receptor FusA. Nat Commun 7:13308. doi: 10.1038/ncomms13308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barreteau H, Tiouajni M, Graille M, Josseaume N, Bouhss A, Patin D, Blanot D, Fourgeaud M, Mainardi JL, Arthur M, van Tilbeurgh H, Mengin-Lecreulx D, Touzé T. 2012. Functional and structural characterization of PaeM, a colicin M-like bacteriocin produced by Pseudomonas aeruginosa. J Biol Chem 287:37395–37405. doi: 10.1074/jbc.M112.406439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grinter R, Roszak AW, Cogdell RJ, Milner JJ, Walker D. 2012. The crystal structure of the lipid II-degrading bacteriocin syringacin M suggests unexpected evolutionary relationships between colicin M-like bacteriocins. J Biol Chem 287:38876–38888. doi: 10.1074/jbc.M112.400150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sonnhammer EL, von Heijne G, Krogh A. 1998. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol 6:175–182. [PubMed] [Google Scholar]
- 37.Poole SJ, Diner EJ, Aoki SK, Braaten BA, t' Kint de Roodenbeke C, Low DA, Hayes CS. 2011. Identification of functional toxin/immunity genes linked to contact-dependent growth inhibition (CDI) and rearrangement hotspot (Rhs) systems. PLoS Genet 7:e1002217. doi: 10.1371/journal.pgen.1002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koskiniemi S, Garza-Sánchez F, Sandegren L, Webb JS, Braaten BA, Poole SJ, Andersson DI, Hayes CS, Low DA. 2014. Selection of orphan Rhs toxin expression in evolved Salmonella enterica serovar Typhimurium. PLoS Genet 10:e1004255. doi: 10.1371/journal.pgen.1004255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kirchberger PC, Unterweger D, Provenzano D, Pukatzki S, Boucher Y. 2017. Sequential displacement of type VI secretion system effector genes leads to evolution of diverse immunity gene arrays in Vibrio cholerae. Sci Rep 7:45133. doi: 10.1038/srep45133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cornelis P, Matthijs S, Van Oeffelen L. 2009. Iron uptake regulation in Pseudomonas aeruginosa. Biometals 22:15–22. doi: 10.1007/s10534-008-9193-0. [DOI] [PubMed] [Google Scholar]
- 41.Denayer S, Matthijs S, Cornelis P. 2007. Pyocin S2 (Sa) kills Pseudomonas aeruginosa strains via the FpvA type I ferripyoverdine receptor. J Bacteriol 189:7663–7668. doi: 10.1128/JB.00992-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Elfarash A, Wei Q, Cornelis P. 2012. The soluble pyocins S2 and S4 from Pseudomonas aeruginosa bind to the same FpvAI receptor. Microbiologyopen 1:268–275. doi: 10.1002/mbo3.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roy PH, Tetu SG, Larouche A, Elbourne L, Tremblay S, Ren Q, Dodson R, Harkins D, Shay R, Watkins K, Mahamoud Y, Paulsen IT. 2010. Complete genome sequence of the multiresistant taxonomic outlier Pseudomonas aeruginosa PA7. PLoS One 5:e8842. doi: 10.1371/journal.pone.0008842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Papadakos G, Wojdyla JA, Kleanthous C. 2012. Nuclease colicins and their immunity proteins. Q Rev Biophys 45:57–103. doi: 10.1017/S0033583511000114. [DOI] [PubMed] [Google Scholar]
- 45.Klein A, Wojdyla JA, Joshi A, Josts I, McCaughey LC, Housden NG, Kaminska R, Byron O, Walker D, Kleanthous C. 2016. Structural and biophysical analysis of nuclease protein antibiotics. Biochem J 473:2799–2812. doi: 10.1042/BCJ20160544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wojdyla JA, Fleishman SJ, Baker D, Kleanthous C. 2012. Structure of the ultra-high-affinity colicin E2 DNase-Im2 complex. J Mol Biol 417:79–94. doi: 10.1016/j.jmb.2012.01.019. [DOI] [PubMed] [Google Scholar]
- 47.Dingemans J, Ye L, Hildebrand F, Tontodonati F, Craggs M, Bilocq F, De Vos D, Crabbé A, Van Houdt R, Malfroot A, Cornelis P. 2014. The deletion of TonB-dependent receptor genes is part of the genome reduction process that occurs during adaptation of Pseudomonas aeruginosa to the cystic fibrosis lung. Pathog Dis 71:26–38. doi: 10.1111/2049-632X.12170. [DOI] [PubMed] [Google Scholar]
- 48.Pirnay JP, Bilocq F, Pot B, Cornelis P, Zizi M, Van Eldere J, Deschaght P, Vaneechoutte M, Jennes S, Pitt T, De Vos D. 2009. Pseudomonas aeruginosa population structure revisited. PLoS One 4:e7740. doi: 10.1371/journal.pone.0007740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dingemans J, Ghequire MG, Craggs M, De Mot R, Cornelis P. 2016. Identification and functional analysis of a bacteriocin, pyocin S6, with ribonuclease activity from a Pseudomonas aeruginosa cystic fibrosis clinical isolate. Microbiologyopen 5:413–423. doi: 10.1002/mbo3.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ghysels B, Ochsner U, Möllman U, Heinisch L, Vasil M, Cornelis P, Matthijs S. 2005. The Pseudomonas aeruginosa pirA gene encodes a second receptor for ferrienterobactin and synthetic catecholate analogues. FEMS Microbiol Lett 246:167–174. doi: 10.1016/j.femsle.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 51.Held K, Ramage E, Jacobs M, Gallagher L, Manoil C. 2012. Sequence-verified two-allele transposon mutant library for Pseudomonas aeruginosa PAO1. J Bacteriol 194:6387–6389. doi: 10.1128/JB.01479-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zambolin S, Clantin B, Chami M, Hoos S, Haouz A, Villeret V, Delepelaire P. 2016. Structural basis for haem piracy from host haemopexin by Haemophilus influenzae. Nat Commun 7:11590. doi: 10.1038/ncomms11590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morton DJ, Seale TW, Madore LL, VanWagoner TM, Whitby PW, Stull TL. 2007. The haem-haemopexin utilization gene cluster (hxuCBA) as a virulence factor of Haemophilus influenzae. Microbiology 153:215–224. doi: 10.1099/mic.0.2006/000190-0. [DOI] [PubMed] [Google Scholar]
- 54.Cornelis P, Bodilis J. 2009. A survey of TonB-dependent receptors in fluorescent pseudomonads. Environ Microbiol Rep 1:256–262. doi: 10.1111/j.1758-2229.2009.00041.x. [DOI] [PubMed] [Google Scholar]
- 55.Ghequire MGK, Swings T, Michiels J, Buchanan SK, De Mot R. 2018. Hitting with a BAM: selective killing by lectin-like bacteriocins. mBio 9:e02138-17. doi: 10.1128/mBio.02138-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jones DT, Taylor WR, Thornton JM. 1992. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8:275–282. [DOI] [PubMed] [Google Scholar]
- 60.Tsirigos KD, Peters C, Shu N, Käll L, Elofsson A. 2015. The TOPCONS web server for consensus prediction of membrane protein topology and signal peptides. Nucleic Acids Res 43:W401–W407. doi: 10.1093/nar/gkv485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Blatny JM, Brautaset T, Winther-Larsen HC, Haugan K, Valla S. 1997. Construction and use of a versatile set of broad-host-range cloning and expression vectors based on the RK2 replicon. Appl Environ Microbiol 63:370–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Christen M, Kulasekara HD, Christen B, Kulasekara BR, Hoffman LR, Miller SI. 2010. Asymmetrical distribution of the second messenger c-di-GMP upon bacterial cell division. Science 328:1295–1297. doi: 10.1126/science.1188658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Choi KH, Kumar A, Schweizer HP. 2006. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J Microbiol Methods 64:391–397. doi: 10.1016/j.mimet.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 64.Green MR, Sambrook J. 2012. Molecular cloning: a laboratory manual, 4th ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 65.Hockett KL, Baltrus DA. 2017. Use of the soft-agar overlay technique to screen for bacterially produced inhibitory compounds. J Vis Exp (119):55064. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.