In their preceding paper, Wozniak et al. show that fertilization increases intracellular Ca2+ in Xenopus laevis eggs by activating an IP3 signaling cascade. Here, they reveal that Ca2+ subsequently opens the Cl− channel TMEM16A to allow Cl− efflux, cell depolarization, and fast block to polyspermy.
Abstract
In externally fertilizing animals, such as sea urchins and frogs, prolonged depolarization of the egg immediately after fertilization inhibits the entry of additional sperm—a phenomenon known as the fast block to polyspermy. In the African clawed frog Xenopus laevis, this depolarization is driven by Ca2+-activated Cl− efflux. Although the prominent Ca2+-activated Cl− currents generated in immature X. laevis oocytes are mediated by X. laevis transmembrane protein 16a (xTMEM16A) channels, little is known about the channels that contribute to the fast block in mature eggs. Moreover, the gamete undergoes a gross transformation as it develops from an immature oocyte into a fertilization-competent egg. Here, we report the results of our approach to identify the Ca2+-activated Cl− channel that triggers the fast block. By querying published proteomic and RNA-sequencing data, we identify two Ca2+-activated Cl− channels expressed in fertilization-competent X. laevis eggs: xTMEM16A and X. laevis bestrophin 2A (xBEST2A). By exogenously expressing xTMEM16A and xBEST2A in axolotl cells lacking endogenous Ca2+-activated currents, we characterize the effect of inhibitors on currents mediated by these channels. None of the inhibitors tested block xBEST2A currents specifically. However, 2-(4-chloro-2-methylphenoxy)-N-[(2-methoxyphenyl)methylideneamino]-acetamide (Ani9) and N-((4-methoxy)-2-naphthyl)-5-nitroanthranilic acid (MONNA) each reduce xTMEM16A currents by more than 70% while only nominally inhibiting those generated by xBEST2A. Using whole-cell recordings during fertilization, we find that Ani9 and MONNA effectively diminish fertilization-evoked depolarizations. Additionally, these inhibitors lead to increased polyspermy in X. laevis embryos. These results indicate that fertilization activates TMEM16A channels in X. laevis eggs and induces the earliest known event triggered by fertilization: the fast block to polyspermy.
Introduction
Fertilization of an egg by more than one sperm, a condition known as polyspermy, presents one of the earliest and most prevalent barriers to successful reproduction. In most sexually reproducing species, polyspermy causes chromosomal abnormalities that are embryonically lethal (Wong and Wessel, 2006). Eggs have evolved multiple strategies to combat the entry of sperm into an already-fertilized egg and thereby avoid such catastrophic consequences (Stricker, 1999; Wong and Wessel, 2006); however, the underlying molecular mechanisms are still poorly understood.
Two commonly used mechanisms to prevent polyspermy are referred to as the slow block and the fast block (Wong and Wessel, 2006). The slow block occurs minutes after fertilization and involves the exocytosis of cortical granules to create an impermeable extracellular matrix that surrounds the nascent zygote (Stricker, 1999; Wong and Wessel, 2006). This physical polyspermy block is used by all sexually reproducing species. In addition to the slow block, external fertilizers use a more immediate blockade: the fast block to polyspermy. Within seconds of fertilization, the fast block induces a depolarization of the egg membrane (Jaffe, 1976). Sperm can bind to, but not enter, a depolarized egg (Jaffe, 1976). Although the mechanisms underlying the slow block are largely understood (Wong and Wessel, 2006), we are just beginning to uncover the molecular pathways that prevent sperm entry into a depolarized egg.
As in all frogs, the fast block in the African clawed frog Xenopus laevis requires an increase of cytosolic Ca2+ and a depolarizing efflux of Cl− (Cross and Elinson, 1980; Grey et al., 1982; Webb and Nuccitelli, 1985; Kline, 1988; Runft et al., 1999; Glahn and Nuccitelli, 2003)—an event hypothesized to be mediated by a Ca2+-activated Cl− channel (CaCC; Hartzell et al., 2009; Cruz-Rangel et al., 2015). We have recently demonstrated that this cytosolic Ca2+ increase is mediated by inositol 1,4,5-triphosphate (IP3)–induced Ca2+ release from the ER (see Wozniak et al. in this issue). A requirement for a Cl− efflux was demonstrated in experiments recording the fast block in the presence of different external Cl− concentrations. Larger fertilization-evoked depolarizations were recorded from eggs inseminated in low extracellular Cl−, and smaller depolarizations were recorded from eggs inseminated in high extracellular Cl− (Grey et al., 1982; Webb and Nuccitelli, 1985). Furthermore, fertilization of eggs in solutions where the dominant extracellular halide (typically Cl−) was replaced with Br− or I− led to no changes in membrane polarization or hyperpolarizations with fertilization, respectively (Grey et al., 1982). Under these conditions, the magnitude and direction of the fertilization-evoked depolarization was directly linked to polyspermy. For example, multiple sperm penetrated all eggs inseminated in I−, which hyperpolarized at fertilization, compared with mostly monospermic inseminations in Cl− (Grey et al., 1982). Together, these experiments revealed a prominent role for a Cl− current in the fast block and underscored the importance of a fertilization-evoked depolarization for ensuring monospermic fertilization. Here, we sought to identify the CaCC that mediates the fast block in X. laevis.
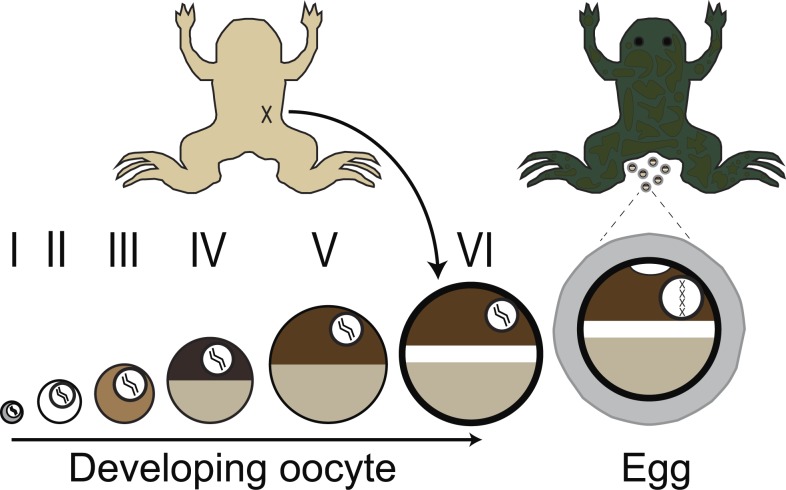
The channels expressed in the fertilization-competent X. laevis egg are not well studied, which is in stark contrast to the well-characterized channels found in the immature oocyte (Schroeder et al., 2008). Indeed, the oocytes and eggs of X. laevis are vastly different cells (Fig. 1; Rasar and Hammes, 2006). Immature oocytes are located in the ovary, are arrested in prophase I, and cannot be fertilized. In contrast, eggs are located outside the X. laevis female (after ovulation and laying), are arrested in metaphase II, and are fertilization competent. As an oocyte matures into an egg, many plasma membrane–localized ion channels and transporters are internalized, including the plasma membrane Ca2+-ATPase (El-Jouni et al., 2005); and Na+/K+ ATPase (Mohanty and Gupta, 2012). In addition, oocyte maturation induces intracellular proteins that closely interact with the plasma membrane, including components of the cytoskeleton, to undergo transformations in their structural contacts (Wylie et al., 1985). Therefore, experimental findings regarding prominent CaCCs, namely TMEM16A (Schroeder et al., 2008), in X. laevis oocytes cannot be directly applied to eggs in the absence of further testing, and thus it was necessary to study the CaCCs in eggs directly.
Figure 1.
Schematic depiction of gamete development in female X. laevis. Immature oocytes, ranging from the youngest (stage I) to the most developed (stage VI), are located within the ovaries. These oocytes can be surgically removed from the abdomen of the frog (shown in ventral view at top left) and are commonly used by electrophysiologists. Upon hormonal induction, stage VI oocytes mature into fertilization-competent eggs, which are laid by the frog (shown in dorsal view at top right). Oocytes and eggs differ with respect to membrane-localized proteins as well as the structure of the cytoskeleton.
We sought to identify the Cl− channel that mediates the fertilization-evoked depolarization of the fast block in X. laevis eggs. Using existing proteomic and transcriptomic data from X. laevis oocytes and eggs (Wühr et al., 2014; Session et al., 2016), we identified two candidate CaCCs: transmembrane protein 16 a (TMEM16A; Caputo et al., 2008; Schroeder et al., 2008; Yang et al., 2008) and bestrophin 2a (BEST2A; Qu and Hartzell, 2004; Kane Dickson et al., 2014). To distinguish between the currents produced by the X. laevis orthologues of these channels (X. laevis transmembrane protein 16a [xTMEM16A] and X. laevis BEST2A [xBEST2A]), we exogenously expressed each channel in axolotl oocytes, which lack endogenous Ca2+-activated currents. Using this isolated system, we independently characterized each CaCC using pharmacological inhibitors. By applying the most potent inhibitors to whole-cell recordings of X. laevis eggs during fertilization, we demonstrate that it is xTMEM16A, and not xBEST2A, that produces the depolarizing current of the fast block. Furthermore, these inhibitors lead to an increased incidence of polyspermy in X. laevis eggs. Thus, we describe the first known ion channel that mediates the fast block to polyspermy.
Materials and methods
Reagents
N-((4-methoxy)-2-naphthyl)-5-nitroanthranilic acid (MONNA) was purchased from Tocris and Sigma-Aldrich, 2-[(5-ethyl-1,6-dihydro-4-methyl-6-oxo-2-pyrimidinyl)thio]-N-[4-(4-methoxyphenyl)-2-thiazolyl]-acetamide (T16Ainh-A01) and 6-(1,1-dimethylethyl)-2-[(2-furanylcarbonyl)amino]-4,5,6,7-tetrahydrobenzo[b]thiophene-3-carboxylic acid (CaCCinh-A01) were purchased from Sigma-Aldrich, and 2-(4-chloro-2-methylphenoxy)-N-[(2-methoxyphenyl)methylideneamino]-acetamide (Ani9) from ChemDiv. Human chorionic gonadotropin was purchased from Henry Schien. All other materials, unless noted, were purchased from Thermo Fisher Scientific.
Proteomic and RNA-sequencing (RNA-seq) analysis
Paired-end raw RNA-seq reads from (Session et al., 2016) were downloaded from the NCBI Sequence Read Archive (www.ncbi.nlm.nih.gov/sra; accession nos. SRX1287719, SRX1287720, SRX1287721, and SRX1287707). Reads were aligned using HISAT2 (Kim et al., 2015) in paired-end mode with default parameters to the X. laevis v9.1 genome, obtained from Xenbase (www.xenbase.org), then assigned to genes using featureCounts (Liao et al., 2014) on Xenbase-annotated gene models in paired-end mode allowing multimappers (-p -M).
To identify channel genes, we assembled 25 relevant gene ontology (GO) terms that distinguished the following classes of channels: Cl−, Ca2+, Na+, and K+ (Dataset S1). To account for possible gaps in GO term annotation, all family members of any gene annotated into the calcium activated chloride channel category were also included in further analysis; for example, all TMEM16 family members, regardless of their GO annotation, were included in this analysis.
To estimate the number of channels in the egg, we combined the protein concentrations with the stoichiometry of the functional channel: two subunits for TMEM16A channels (Brunner et al., 2014) and five for BEST2A (Kane Dickson et al., 2014; Yang et al., 2014). We then assumed that X. laevis eggs are spherical, and we calculated their volume based on their measured diameter of 1.4 mm (Wozniak et al., 2017).
Solutions
Fertilization solutions
Modified Ringer’s (MR) had the following composition (in mM): 100 NaCl, 1.8 KCl, 2.0 CaCl2, 1.0 MgCl2, and 5.0 HEPES, pH 7.8; MR was filtered using a sterile, 0.2-µm polystyrene filter (Heasman et al., 1991). Development assays were performed in 33% MR (MR/3) with or without inhibitors. Fertilization recordings were made in our standard solution of 20% MR (also known as MR/5) with or without inhibitors, as indicated. After electrical recordings were made for fertilization experiments, embryos developed for 2 h in MR/3. Various recordings were made in the presence of inhibitors, made from concentrated stock solutions themselves made in water or DMSO. Final DMSO content was always <2% of the total volume.
Oocyte solutions
Oocyte Ringer’s 2 had the following composition (in mM): 82.5 NaCl, 2.5 KCl, 1 MgCl2, and 5 mM HEPES, pH 7.6 (Wallace et al., 1973).
Two-electrode voltage clamping
The two-electrode voltage clamp (TEVC) solution, ND96, had the following composition (in mM): 96 NaCl, 2 KCl, 1 MgCl2, 10 HEPES, pH 7.6. It was filtered with a sterile, 0.2-µm polystyrene filter (Schroeder et al., 2008).
Animals
All animal procedures were conducted using accepted standards of humane animal care and approved by the Animal Care and Use Committee at the University of Pittsburgh. X. laevis adults were obtained commercially (Research Resource Identifier NXR_0.0031; NASCO), as were axolotls, Ambystoma mexicanum (Research Resource Identifier AGSC_100A, Ambystoma Genetic Stock Center). X. laevis and axolotls were housed separately at 18°C with a 12-h/12-h light/dark cycle.
Collection of gametes, fertilization, and developmental assays
X. laevis and axolotl oocytes were collected by procedures similar to those described previously (Schroeder et al., 2008; Wozniak et al., 2017). In brief, ovarian sacs were obtained from X. laevis females anesthetized with a 30-min immersion in 1.0 g/liter tricaine-S (MS-222), pH 7.4, and axolotls euthanized via immersion in 3.6 g/liter tricaine-S, pH 7.4. For both sets of oocytes, ovarian sacs were manually pulled apart and incubated for 90 min in 1 mg/ml collagenase in ND96 supplemented with 5 mM sodium pyruvate and 10 mg/liter gentamycin. Collagenase was removed by repeated washes with oocyte Ringer’s 2, and healthy oocytes were sorted and stored at 14°C in ND96 with sodium pyruvate and gentamycin.
Eggs were collected from sexually mature X. laevis females as previously described (Wozniak et al., 2017). In brief, females were injected 1,000 IU human chorionic gonadotropin into their dorsal lymph sac and housed overnight for 12–16 h at 14–16°C. Typically, females began laying eggs within 2 h of moving to room temperature. Eggs were collected on dry Petri dishes and used within 10 min of being laid.
Sperm were harvested from testes of sexually mature X. laevis males, as previously described (Wozniak et al., 2017). After euthanasia by a 30-min immersion in 3.6 g/L tricaine-S, pH 7.4, testes were dissected. Cleaned testes were stored at 4°C in MR for usage on the day of dissection or in L-15 medium for use up to 1 wk later.
To create a sperm suspension for whole-cell recordings, ∼1/10 of a testis was minced in MR/5; if not used immediately, this solution was stored on ice and used for up to 1 h. No more than three sperm additions were added to a given egg during whole-cell recordings, and the total volume of sperm suspension added never exceeded 7.5% of the total fertilization solution. Eggs inseminated during whole-cell recordings were transferred to MR/3 for up to 16 h after insemination to monitor development. Development was assessed based on the appearance of cleavage furrows (Wozniak et al., 2017).
Polyspermy assays were performed as previously described (Grey et al., 1982). Approximately 20–35 eggs per treatment were incubated in MR/3 with or without indicated inhibitors for 10 min. One third of a testis was then minced in MR and added to eggs in each experimental condition. 30 min after insemination, eggs were washed three times in MR/3 and maintained in MR/3 for an additional 60–90 min. Polyspermy was assessed 90–120 min after sperm addition in developed embryos based on cleavage furrow symmetry (Elinson, 1975; Grey et al., 1982); embryos with a symmetrical pattern were denoted monospermic, and those with asymmetric furrows were deemed polyspermic.
Electrophysiology
Electrophysiology recordings were made using TEV-200A amplifiers (Dagan Co.) and digitized by Axon Digidata 1550A (Molecular Devices). Data were acquired with pClamp Software (Molecular Devices) at a rate of 5 kHz.
IP3-evoked currents were recorded in the TEVC configuration at −80 mV from X. laevis or axolotl oocytes. cDNA encoding the X. laevis xTMEM16A channel in the GEMHE vector was provided by Lily Jan (University of California, San Francisco, CA; Schroeder et al., 2008). cDNA encoding the xBEST2A channel was purchased from DNASU (Seiler et al., 2014) and engineered into the GEMHE vector with a carboxy-terminal Ruby tag (Kredel et al., 2009) using overlapping PCR and Gibson assembly methods. The sequences for all constructs were verified by automated Sanger sequencing (Gene Wiz). The xTMEM16A and xBEST2A cRNAs were transcribed using the T7 mMessage mMachine Ultra kit (Ambion), and membrane-anchored enhanced GFP (MemE) with the SP6 mMessage mMachine kit (Ambion). Defolliculated axolotl oocytes were injected with 5 ng of cRNA for xTMEM16A or xBest2A, as described previously (Schroeder et al., 2008). Both axolotl and X. laevis oocytes were injected with the photolabile IP3 analogue myo-inositol 1,4,5-trisphosphate, P4(5)-1-(2-nitrophenyl) ethyl ester (caged IP3). Each oocyte was injected with a 200 µM caged IP3 stock made in DDH2O to reach a final concentration of 5 µM within the oocyte (Schroeder et al., 2008), and incubated in the dark at 18°C for 1–5 h before recording. Pipettes of 1–8 MΩ resistance were pulled from borosilicate glass and filled with 1 M KCl. The nitrophenyl cage on IP3 was released by flash photolysis with a 250-ms exposure to light derived from the Ultra High Power White LED Illuminator (380–603 nm; Prizmatix) and guided by a liquid light source to the top of oocytes in our recording chambers (RC-26G; Warner Instruments). The bath solution was changed with the gravity-fed, pinch valve VC-8 solution changer (Warner Instruments). Background-subtracted peak currents were quantified from two consecutive recordings (one before and one with application of the tested inhibitors). The proportional difference between peak currents before and with inhibitor application for each oocyte was used to quantify the percent inhibition for each treatment. It is not possible to compare current amplitudes generated in different oocytes directly because of the innate variability of the experimental setup (e.g., positioning of the UV light or the exact amount of caged IP3 in each oocyte).
Fertilization-evoked depolarizations were recorded in the whole-cell configuration. Pipettes made from borosilicate glass were 5–20 MΩ resistance and filled with 1 M KCl. Resting and fertilization potentials were quantified ∼10 s before and after the depolarization, respectively. Depolarization rates for each recording were quantified by determining the maximum velocity of the quickest 1-mV shift in the membrane potential.
Imaging
Axolotl oocytes were imaged using a TCS SP5 confocal microscope (Leica Microsystems) equipped with a Leica 506224 5× objective. As a membrane control, oocytes were injected with cRNA for a MemE (Moriyoshi et al., 1996). Enhanced GFP was excited with a 488-nm visible laser, whereas Ruby was excited with a 561-nm laser. Using a galvo scanner with unidirectional (600 Hz) scanning, sequential frames were captured with 2× line averaging. Images were analyzed using LAS AF (version 3.0.0 build 834) software and ImageJ (National Institutes of Health; Schneider et al., 2012).
X. laevis eggs and embryos were imaged using a stereoscope (Leica Microsystems) equipped with a Leica 10447157 1× objective and DFC310 FX camera. Images were analyzed using LAS (version 3.6.0 build 488) software and Photoshop (Adobe).
Quantification and statistical analyses
All electrophysiology recordings were analyzed with Igor (WaveMetrics) and Excel (Microsoft). Data for each experimental condition are displayed in Tukey box plot distributions, where the box contains the data between 25% and 75% and the whiskers span 10–90%. Additionally, mean ± SEM values are reported for each experimental condition (Tables 1 and S1). All conditions contain trials that were conducted on multiple days with gametes from multiple females. T tests (one tailed for depolarization rates and two tailed for resting and fertilization potentials) were used to determine differences between inhibitor treatments. Depolarization rates were log10 transformed before statistical analysis. ANOVAs followed by post-hoc Honestly Significant Difference (HSD) tests were used to compare different currents recorded with the same inhibitors for uncaging experiments. Tukey tests were used to compare different currents recorded with the same inhibitors for uncaging experiments. ANOVAs followed by post-hoc Holm–Bonferroni tests were used to compare each inhibitor treatment to the control for development and polyspermy assays.
Table 1. Whole-cell recordings of fertilization using TMEM16A inhibitors.
| Resting potential (mV) | Fertilization potential (mV) | Sperm to depolarization (min) | Depolarization rate (mV/ms) | n | |
|---|---|---|---|---|---|
| Control (MR/5) | −19.2 ± 1.0 | 3.7 ± 2.3 | 4.9 ± 0.7 | 9.0 ± 3.4 | 30 |
| 10 µM MONNA | −12.8 ± 0.8 | n/a | n/a | n/a | 7 |
| 1 µM Ani9 | −14.7 ± 2.5 | 9.3 ± 5.2 | 5.4 ± 0.8 | 1.21 ± 1.15 | 5 |
Mean ± SEM values for each fertilization condition are shown. n reports the number of recordings from individual eggs. In conditions where fertilization failed to evoke any depolarization, n/a reflects that were no postdepolarization data to quantify postfertilization metrics.
Online supplemental material
Fig. S1 shows changes in RNA expression levels and protein concentrations of ion channels during X. laevis gamete development. Fig. S2 shows representative current traces evoked by IP3 uncaging experiments in various inhibitors. Table S1 shows inhibition of Ca2+-activated current using Cl− channel inhibitors. Dataset S1 lists GO terms used to identify channels. RNA-seq data from Session et al. (2016) of channels in X. laevis oocytes during developmental stages 1–2, 3–4, and 5–6 and in fertilization-competent eggs. Proteomics data from Wühr et al. (2014) from fertilization-competent X. laevis eggs.
Results
Two candidate CaCCs accumulate in the egg and are candidates for the trigger of the fast block
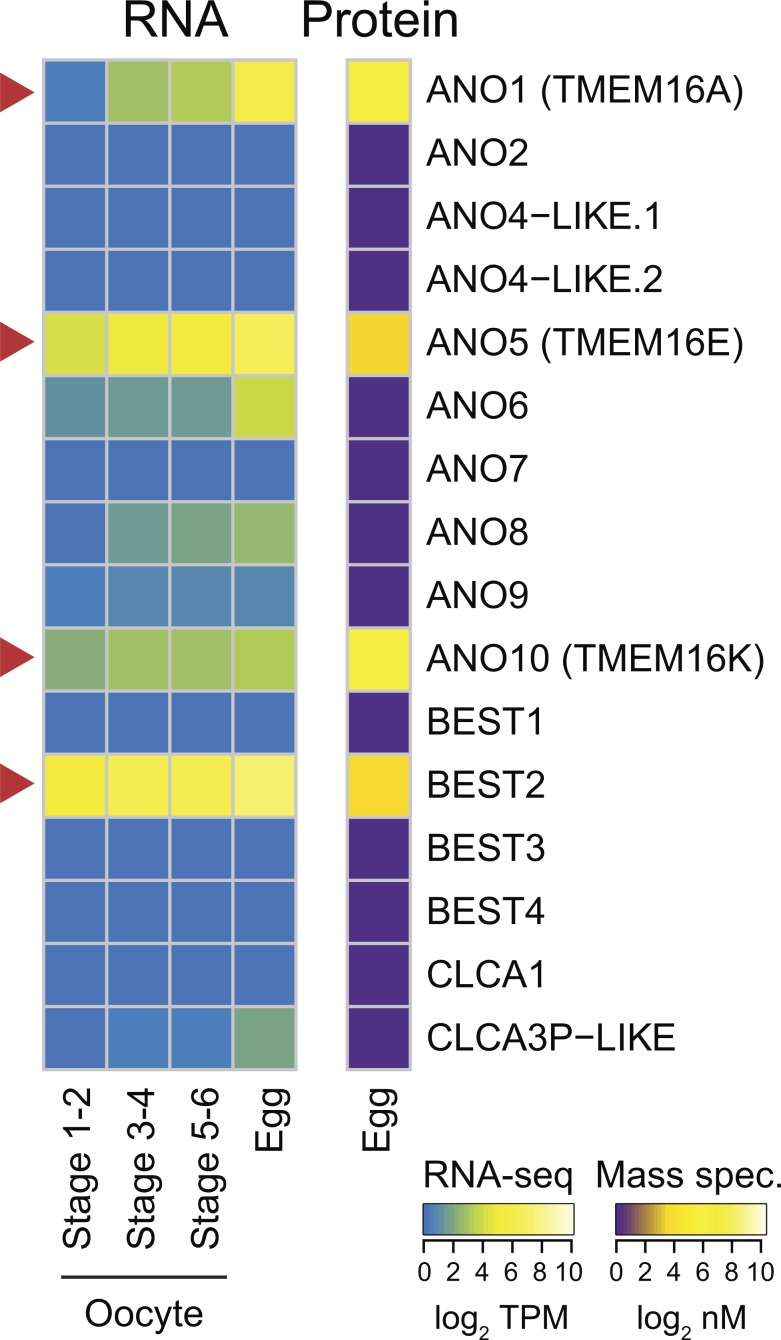
To identify candidate CaCCs that may trigger the fast block in X. laevis, we interrogated two previously published high-throughput gene expression datasets. First, we examined the proteome of fertilization-competent eggs (Wühr et al., 2014) and queried for all known ion channels (Fig. S1 and Dataset S1). Three protein families containing CaCCs have been characterized to date: the CLCAs, the bestrophins, and the transmembrane protein 16s (TMEM16/ANO; Huang et al., 2012). We discovered that only one member of the bestrophin family (xBEST2A) and three members of the TMEM16 family (xTMEM16A, xTMEM16E, and xTMEM16K) are represented in the egg proteome (Fig. 2). Second, we examined an RNA-seq time course in X. laevis oocytes and unfertilized eggs (Session et al., 2016). All four types of CaCC-encoding mRNA show increasing levels through gamete development, culminating in the egg (Fig. 2). Although ANO6 and CLCA3P-like mRNA are present, it is likely that they are expressed after fertilization to guide the developing embryo through the maternal-to-zygotic transition, because their proteins are not detected in the unfertilized egg (Tadros and Lipshitz, 2009; Lee et al., 2014).
Figure 2.
Expression of CaCCs in X. laevis oocytes and eggs. Heatmaps of expression levels of CaCCs at the developmental stages indicated. Right: Protein concentrations (from Wühr et al., 2014) as determined by mass spectrometry–based proteomics study (in log2 nanomolar). Left: Transcript levels (shown as transcripts per million [TPM]; from Session et al., 2016), as determined by RNA-seq–based transcriptome study. Arrowheads highlight CaCCs with proteins found in eggs.
Both xBEST2A and xTMEM16A were originally cloned from fertilization-incompetent X. laevis oocytes (Qu et al., 2003; Schroeder et al., 2008), and each has been characterized as plasma membrane localized (Qu et al., 2003; Schroeder et al., 2008; Jin et al., 2013; Gao da et al., 2016). Moreover, xTMEM16A is the prominent CaCC in X. laevis oocytes (Schroeder et al., 2008). In contrast, TMEM16E localizes to the ER, where it functions as a Ca2+-activated scramblase (Tran et al., 2014; Gyobu et al., 2015). TMEM16K similarly localizes to the ER (Hammer et al., 2015; Wanitchakool et al., 2017). Because both TMEM16E and TMEM16K proteins localize to the ER, they are not capable of passing the depolarizing Cl− current of the fast block and were therefore both excluded from further consideration. Together, these analyses suggest that the fast block to polyspermy in X. laevis eggs is mediated by either xBEST2A or xTMEM16A.
Uncaging IP3 activates xTMEM16A and xBEST2A
Having discovered two CaCCs as candidates for the channel that mediates the fast block in X. laevis eggs, we sought to distinguish between their currents in the context of fertilization. Studying the activities of xTMEM16A and xBEST2A independently necessitated their exogenous expression. For this purpose, we chose a highly tractable system that lacks endogenous Ca2+-activated currents: Ambystoma mexicanum (axolotl) oocytes.
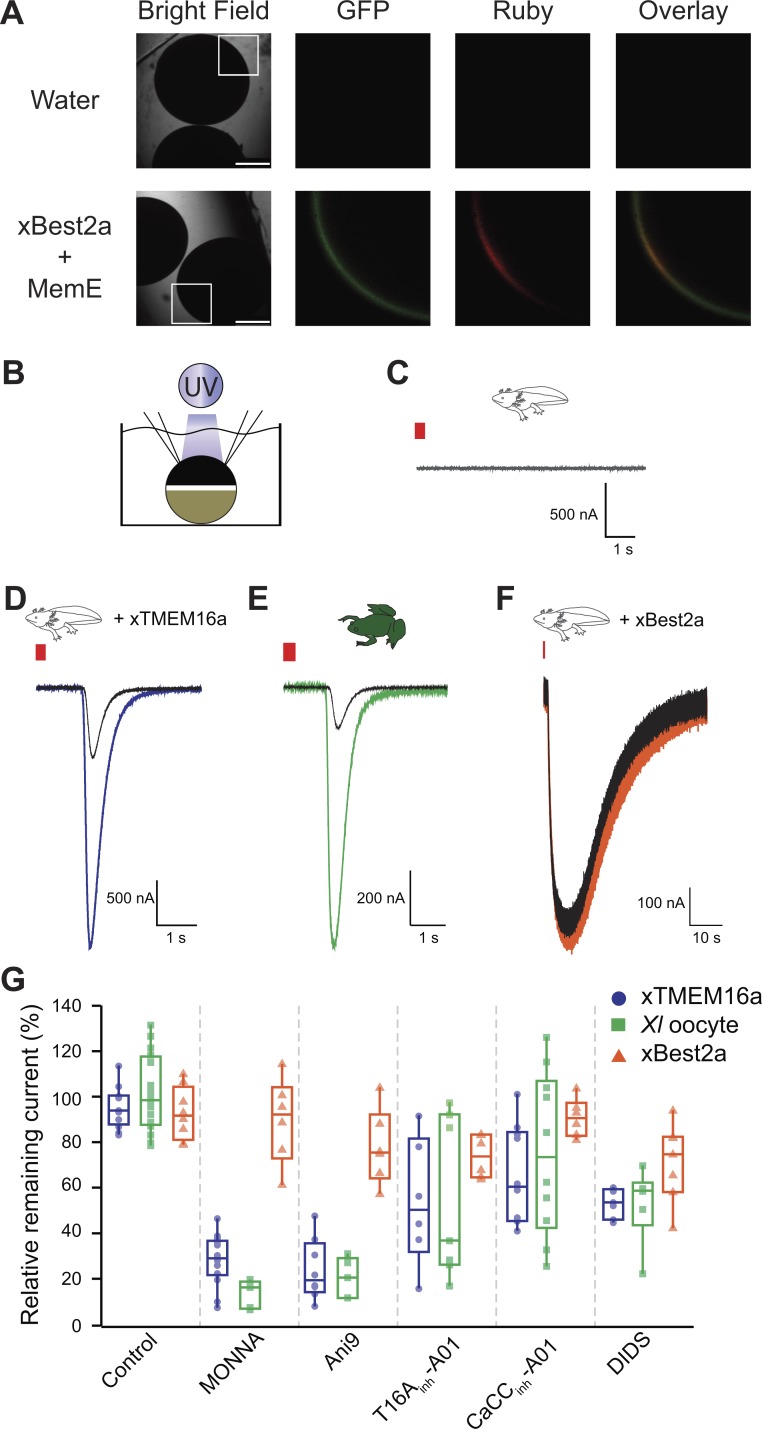
Although xTMEM16A was previously expressed in axolotl oocytes and currents generated in this context have been recorded (Schroeder et al., 2008), this is not the case for xBEST2A. We first confirmed xBEST2A is expressed in these oocytes. Confocal imaging of axolotl oocytes expressing both Ruby-tagged xBEST2A and the enhanced GFP–tagged membrane marker MemE (Moriyoshi et al., 1996) revealed that xBEST2A was indeed expressed in these cells and that it was transported proximal to the plasma membrane (Fig. 3 A). As expected, no fluorescence was detected in water-injected control oocytes (Fig. 3 A).
Figure 3.
MONNA and Ani9 inhibit TMEM16A-conducted Cl− currents. (A) Representative bright-field and fluorescence images of axolotl oocytes expressing Ruby-tagged xBEST2a and enhanced GFP–tagged MemE (reporter of plasma membrane). Boxes denote portions included in fluorescence images. Bars, 750 µm. Overlay is of GFP and Ruby images. (B) Schematic of experimental design: UV photolysis to uncage IP3 while conducting TEVC. (C–F) Current recordings from oocytes of axolotls (C, D, and F) or X. laevis (E), after injection with a photolabile caged-IP3 analogue, with clamping at −80 mV. Axolotl oocytes expressed no transgene (C), xTMEM16A (D), or xBEST2A (F). Wild-type X. laevis oocytes expressing endogenous channels (E). Typical current traces before and after uncaging, during (colored) control treatment, and (black) in the presence of 10 µM MONNA. Red bar denotes the 250-ms duration of UV exposure. (G) Tukey box plot distributions of the averaged proportion of current remaining after application of the indicated inhibitors, in axolotl oocytes expressing xTMEM16A (n = 6–14) or xBEST2A (n = 6–8), and in X. laevis oocytes expressing endogenous channels (n = 5–16). The central line represents the median value and the box denotes the data spread from 25–75%, and the whiskers reflect 10–90%.
To study the currents conducted by xTMEM16A and xBEST2A, we exploited their shared regulation by intracellular Ca2+ (Qu and Hartzell, 2004; Schroeder et al., 2008; Kane Dickson et al., 2014). Specifically, we used UV light application to photoactivate caged-IP3 in oocytes. This uncaging of IP3 induces Ca2+ release from the ER, thereby increasing the intracellular Ca2+ concentration and activating the channels (Fig. 3 B). Using the TEVC technique, we recorded Ca2+-activated Cl− currents ranging from 0.2 to 17 µA, with a mean of 5.6 ± 1.0 µA (n = 12) for xTMEM16A in axolotl oocytes and 0.5 ± 0.7 µA for xBEST2A in axolotl oocytes, as well as 6.9 ± 1.5 µA in X. laevis oocytes (n = 16). Importantly, we used the same splice variants of xTMEM16A and xBEST2A channels that are present in X. laevis eggs (Qu et al., 2003; Schroeder et al., 2008; Wühr et al., 2014). As shown previously (Schroeder et al., 2008), uncaging IP3 in wild-type axolotl oocytes does not elicit any Ca2+-induced currents (Fig. 3 C).
Using the uncaging system in conjunction with TEVC, we recorded whole-cell currents in the presence or absence of known channel inhibitor molecules. Our initial assessment of the effects of sequential uncaging events in the absence of inhibitors revealed no differences in current between axolotl oocytes expressing either of the channels or X. laevis oocytes expressing the endogenous channels (Table S1). This finding indicated that differences in Ca2+-evoked currents measured in the presence or absence of an inhibitor in this experimental system would reflect the efficacy of that inhibitor and enable us to characterize the efficacy of inhibitors in reducing xTMEM16A- or xBEST2A-mediated currents.
With this experimental design, we quantified the effects of five inhibitors on xTMEM16A- and xBEST2A-mediated currents (Table S1). Three of these (MONNA, Ani9, and T16ainh-A01) were previously reported to target human and/or mouse TMEM16A (Namkung et al., 2011; Oh et al., 2013; Seo et al., 2016), whereas CaCCinh-A01 is a general inhibitor of CaCCs (De La Fuente et al., 2008; Namkung et al., 2011). Although no BEST-specific inhibitor has been characterized to date, we included the broad-spectrum Cl− channel inhibitor DIDS, because it reportedly inhibits human bestrophin 1 channels with an affinity 160-fold higher than that for mouse TMEM16A (mTMEM16A; Liu et al., 2015).
MONNA and Ani9 inhibit xTMEM16A currents
To characterize the effects of each inhibitor, we applied them to axolotl oocytes expressing xTMEM16A or xBEST2A. In the case of xTMEM16A, both MONNA and Ani9 effectively reduced currents in the axolotl oocytes by over 70% (Figs. 3 D and S2; and Table S1), whereas T16Ainh-A01 and CaCCinh-A01 were much less effective (Figs. 3 G and S2; and Table S1). Unexpectedly, we found that 7.5 µM DIDS, a concentration well below the reported half maximal inhibitory concentration (IC50) for the drug on mTMEM16A (Liu et al., 2015), reduced xTMEM16A by almost 50% (Fig. S2 and Table S1).
In X. laevis oocytes, the prominent Ca2+-activated Cl− current is known to be generated by xTMEM16A channels (Schroeder et al., 2008). Comparison of the effects on xTMEM16A-mediated current in the axolotl oocytes to the endogenous TMEM16A-passed currents generated in X. laevis oocytes revealed that in nearly all cases, the efficacy of the inhibitors was very similar in the two test groups (Figs. 3 E and S2; and Table S1). The exception is that MONNA blocked significantly more xTMEM16A current in the X. laevis oocyte (87 ± 2%) than in the axolotl oocytes (72 ± 3%; P < 0.05, ANOVA with post-hoc HSD Tukey; Table S1). Collectively, these data demonstrate that only MONNA and Ani9 effectively inhibit xTMEM16A.
MONNA and Ani9 discriminate between currents generated by xTMEM16A and xBEST2A
Comparison of the effects of the five inhibitors on xBEST2A currents revealed that none had a significant effect (P > 0.05, ANOVA with post-hoc HSD Tukey; Figs. 3 G and S2; and Table S1). Most notably, currents generated in the presence of MONNA or Ani9 were no different than those produced in the control, confirming that these two compounds are specific for xTMEM16A. Furthermore, the lack of xBEST2A inhibition by MONNA and Ani9 demonstrates that these inhibitors do not interfere with the IP3-induced Ca2+ release pathway. Together, these results demonstrate that MONNA and Ani9 effectively target xTMEM16A channels but have only minimal effects on xBEST2A and the IP3 receptor, thereby providing a mechanism for discerning between xTMEM16A and xBEST2A currents during the fast block.
The TMEM16A mediated-current produces the fast block in X. laevis
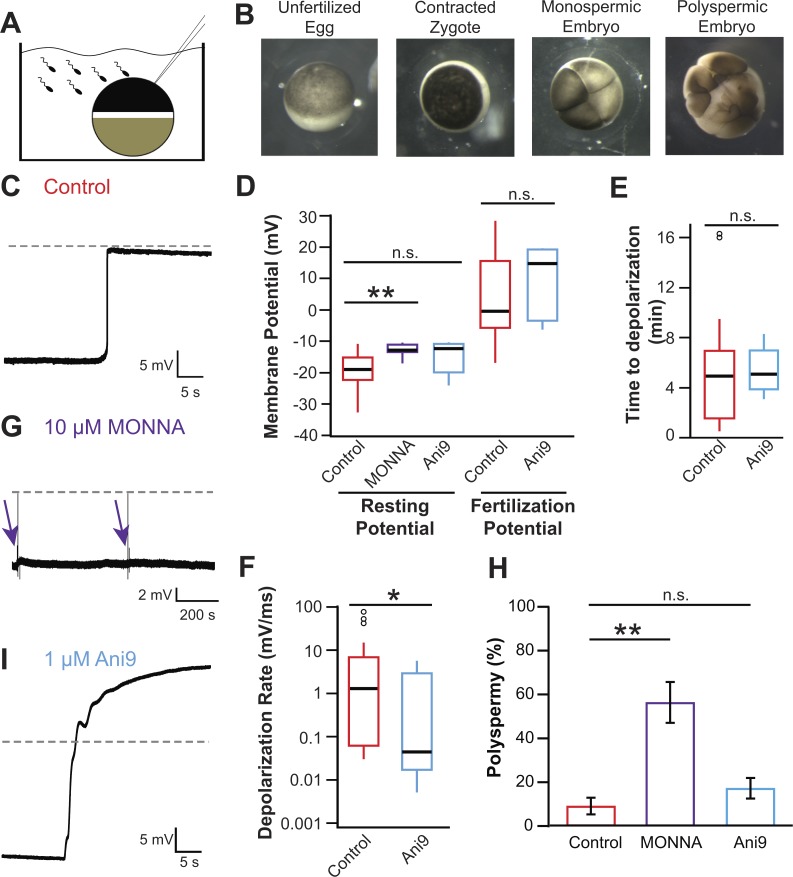
To study the fast block to polyspermy in X. laevis, we conducted whole-cell recording of eggs during fertilization (Fig. 4 A) and recorded currents up to 40 min after fertilization or until the cortex contracted (Fig. 4 B). Fig. 4 C depicts a typical fertilization-evoked depolarization that occurred after sperm addition. For eggs inseminated under control conditions, we found that: the resting potential was −19.2 ± 1.0 mV; the fertilization potential was 3.7 ± 2.3 mV (n = 30; Fig. 4 D and Table 1); the time between the addition of sperm and the onset of membrane depolarization was ∼4.9 ± 0.7 min (n = 30, Fig. 4 E, Table 1); and the mean rate of depolarization was 9.0 ± 3.4 mV/ms (n = 30; Fig. 4 F and Table 1).
Figure 4.
Fertilization activates TMEM16A to depolarize the egg. (A) Schematic depiction of experimental design showing whole-cell recordings made on X. laevis eggs during fertilization. (B) Images of an X. laevis egg before sperm addition (left), an egg ∼15 min after fertilization with animal pole contracted (center left), a monospermic embryo (center right), and a polyspermic embryo (right). (C, G, and I) Representative whole-cell recordings made during fertilization in control conditions (C), the presence of 10 µM MONNA (G), or the presence of 1 µM Ani9 (I). Dashed lines denote 0 mV, and arrows denote times at which sperm was applied to eggs in the presence of 10 µM MONNA. (D–F) Tukey box plot distributions of the resting and fertilization potentials in control conditions and with MONNA or Ani9 (D), the time between sperm application and depolarization in the absence and presence of Ani9 (E), and the depolarization rate in the absence and presence of Ani9 (F; n = 5–30, recorded over 2–16 experimental days per treatment). The central line represents the median value, and the box denotes the data spread from 25–75%, and the whiskers reflect 10–90%. (H) Proportion of polyspermic embryos out of total developed embryos in control, MONNA, and Ani9 (n = 3, recorded over three experiment days per treatment). n.s., P > 0.05; *, P < 0.05; **, P < 0.001.
To determine whether it is xTMEM16A or xBEST2A that conducts the depolarizing current responsible for the fast block, we inseminated eggs in the presence of MONNA or Ani9, each of which was expected to inhibit xTMEM16A but to have minimal effect on xBEST2a or IP3 receptors (Fig. 3 G and Table S1). In X. laevis eggs, inhibition of xTMEM16A using either inhibitor effectively diminished the fast block. In the presence of 10 µM MONNA, fertilization failed to evoke depolarization in seven independent experiments (Fig. 4 G); thus, this inhibitor completely abolished the fast block. Eggs incubated in MONNA had a significantly more positive resting potential than that of control eggs (−12.8 ± 0.8 mV vs. −19.2 ± 1.0 mV, t test, P < 0.001; Fig. 4 D and Table 1). However, this elevated resting potential did not interfere with fertilization; visual assessment revealed contraction of the animal pole followed by the appearance of cleavage furrows (Fig. 4 B), thus demonstrating that all eggs inseminated in the presence of MONNA-initiated embryonic development. Additionally, it has been shown that the majority of eggs fertilize at −11 mV in X. laevis (Nuccitelli et al., 1993). To determine whether inhibiting TMEM16A altered the polyspermy block, we compared the incidence of polyspermy in embryos inseminated under control conditions (MR/3 solution) or with 10 µM MONNA (Fig. 4 H and Table 2). Polyspermic fertilization was assessed based on the appearance of asymmetric cleavage furrows whereas symmetric furrows revealed monospermic fertilization (Fig. 4 B; Elinson, 1975; Grey et al., 1982). Significantly more embryos inseminated in MONNA were polyspermic compared with control (56 ± 9% [n = 3] vs. 9 ± 4% [n = 5], ANOVA, P < 0.01).
Table 2. Developmental assays using TMEM16A inhibitors.
| Condition | Undeveloped (%) | Developed (%) | Polyspermic (%) | n |
|---|---|---|---|---|
| Control (MR/3) | 0 ± 0 | 100 ± 0 | 9 ± 4 | 5 |
| 10 µM MONNA | 7 ± 2 | 93 ± 2 | 56 ± 9 | 3 |
| 1 µM Ani9 | 2 ± 2 | 98 ± 2 | 17 ± 5 | 3 |
Mean ± SEM values for each fertilization condition are shown. Of the embryos that developed cleavage furrows, embryos were categorized as monospermic or polyspermic based on cleavage furrow symmetry.
In the presence of 1 µM Ani9, the rate of the fertilization-evoked depolarization was significantly slower, thereby attenuating the fast block (1.2 ± 1.1 mV/ms with Ani9 [n = 5] vs. 9.0 ± 3.4 mV/ms in control [n = 30], t test, P < 0.05; Fig. 4, F and I). Because the rate of depolarization is proportional to the number of channels that are open, a slower rate reflects fewer channels being activated by fertilization. Based on the rates measured, we estimate that in the presence of 1 µM Ani9, 7.5-fold fewer channels were triggered to open by fertilization; i.e., only 13% of the channels that would be activated under normal conditions opened in Ani9. This 87% reduction in the number of open channels is consistent with the 80% inhibition of xTMEM1Aa channels measured when IP3 was uncaged in X. laevis and axolotl oocytes (Fig. 3 G and Table S1). No other metrics of the fast block differed significantly in recordings made in the presence versus absence of Ani9 (Fig. 4, D–F). More embryos inseminated in Ani9 were polyspermic compared with control (17 ± 5% [n = 3] vs. 9 ± 4% [n = 5]); however, this difference was not statistically significant (Fig. 4 H; P > 0.05, ANOVA).
Collectively, the ability of MONNA and Ani9 to effectively diminish fertilization-evoked depolarizations and increase the incidence of polyspermy demonstrates that TMEM16A channels produce the depolarizing current that mediates the fast block in X. laevis eggs.
Discussion
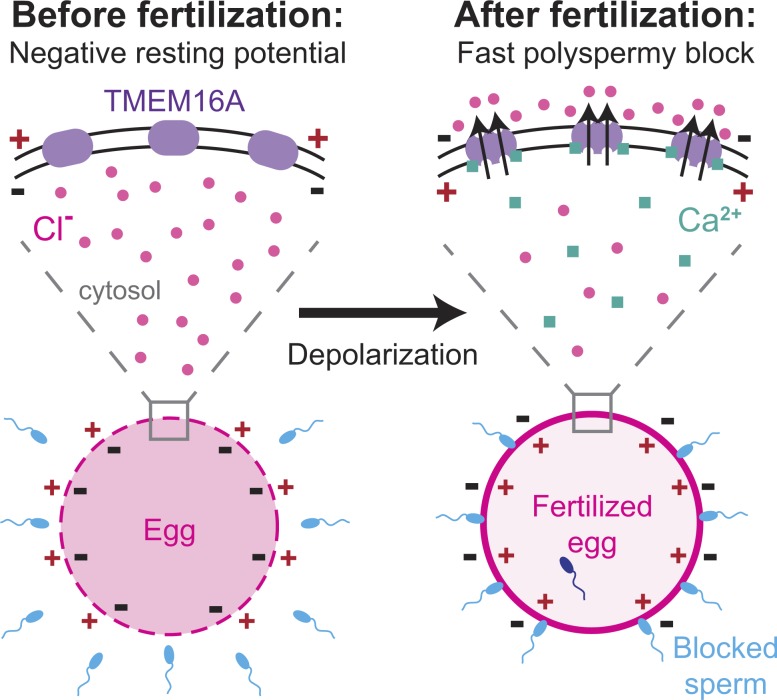
The fast block to polyspermy is one of the earliest and most prevalent events across species that undergo external fertilization. We have recently demonstrated that fertilization-signaled depolarization in X. laevis eggs requires activation of a phospholipase C that induces Ca2+ release from the ER in an IP3-mediated pathway (Wozniak et al., 2018). Here, we identify the CaCC that mediates the fast block in the African clawed frog X. laevis: xTMEM16A (Fig. 5). Given that an increase in the intracellular Ca2+ concentration and an efflux of Cl− are required for the fast block in all frogs and toads studied thus far (Cross and Elinson, 1980; Cross, 1981), we propose that the current produced by TMEM16A channels triggers the fast block to polyspermy in all anurans.
Figure 5.
Proposed model for fertilization signaled activation of TMEM16A. Before fertilization, X. laevis eggs have a negative resting potential, thereby electrically signaling to sperm that they can receive a male gamete. After fertilization, cytosolic Ca2+ increases to activate TMEM16Aa. An efflux of Cl− then depolarizes the egg, and this change in membrane potential blocks supernumerary sperm from entering the fertilized egg.
Our identification of xTMEM16A and xBEST2A as candidate CaCCs that may mediate the fast block is based on proteomics and transcriptomics. Indeed, both proteins are translated in high concentrations (∼22 × 109 xTMEM16A channels and 2 × 109 xBEST2A channels; see Materials and methods) in mature eggs. Given that both channel proteins are present in the egg, it was feasible that either or both could mediate the Ca2+-activated Cl− efflux that drives the fertilization-evoked depolarization of the fast block in X. laevis.
Our finding that 10 µM MONNA and 1 µM Ani9—concentrations higher than their published IC50 (Oh et al., 2013; Seo et al., 2016)—inhibit >70% of xTMEM16A channels in both axolotl and X. laevis oocytes yet are largely ineffective at reducing currents conducted by xBEST2A demonstrate that these inhibitors discriminate between our two candidate CaCCs. Both of these inhibitors are known to be highly specific for TMEM16A, with Ani9 failing to block even the closest relative of TMEM16A, TMEM16B (Seo et al., 2016). In contrast, T16inh-A01 and CaCCinh-A01 were much less effective at inhibiting either xTMEM16A or xBEST2A. The similarity between the pharmacological profiles of xTMEM16A currents recorded in axolotl oocytes and endogenous Ca2+-activated currents in X. laevis oocytes supports the hypothesis that the native Ca2+-activated Cl− currents in X. laevis oocytes are generated by xTMEM16A channels (Schroeder et al., 2008).
Although MONNA and Ani9 inhibited exogenously expressed xTMEM16A in axolotl oocytes to similar extents, MONNA was significantly more effective in reducing the endogenous Ca2+-activated currents of X. laevis oocytes (Fig. 4 F; P < 0.05, ANOVA with post-hoc HSD Tukey). The increased efficacy of MONNA with respect to endogenous xTMEM16A in the egg is consistent with the observed difference in its fertilization-induced electrical profile over that of Ani9 (i.e., with MONNA completely blocking depolarization and Ani9 merely slowing it). Although the mechanisms underlying TMEM16A inhibition by these chemically distinct agents have not yet been elucidated, we hypothesize that the differing effects of these inhibitors on X. laevis oocytes and eggs may reflect the strikingly different membrane environments of these two cell types. Furthermore, we speculate that the elevated resting potential recorded from eggs inseminated in the presence of MONNA reflects the altered resting state of these cells, consistent with a recent demonstration that TMEM16A activity plays a prominent role in Cl− homeostasis (He et al., 2017).
We find that fertilization in the presence of MONNA prevents fertilization-evoked depolarizations. Consistent with the role of a depolarization in ensuring monospermic fertilization, we report an increased incidence of polyspermy in the presence of MONNA. Furthermore, it is possible that the increased incidence of undeveloped eggs in MONNA (relative to the control) could also be caused by polyspermy, as the presence of multiple sperm could skew many different developmental programs. Together, these data highlight the importance of xTMEM16A mediating the fast block, thereby ensuring proper X. laevis embryonic development.
Despite the gross changes that the plasma membrane of a X. laevis oocyte undergoes as it matures into a fertilization-competent egg, it is evident that the xTMEM16A channels are retained in the plasma membrane. Although proteomics data reveal that X. laevis eggs express the xBEST2A channel, our data demonstrate that this channel does not contribute to the fast block. Based on its presence in the mature egg (Wühr et al., 2014) and its lack of contribution to the fast block, we speculate that xBEST2A is either desensitized or absent from the plasma membrane, as is the case for the plasma membrane Ca2+-ATPase (El-Jouni et al., 2005) and the Na+/K+ ATPase (Mohanty and Gupta, 2012).
In conclusion, the fertilization-induced activation of TMEM16A channels is the earliest known signaling event evoked by the sperm–egg interaction in X. laevis (Fig. 5). The discovery of a critical role for TMEM16A channels in fertilization lays a foundation for understanding how the membrane potential regulates fertilization. More broadly, TMEM16A channels regulate diverse processes ranging from epithelial secretions (Yang et al., 2008) to smooth muscle contraction (Huang et al., 2009; Bulley et al., 2012). These CaCCs are indispensable for human health (Rock et al., 2008). Because of their large size, ease and reproducibility for electrophysiology recordings, and years of study by developmental biologists and biophysicists alike, we propose that X. laevis fertilization may serve as a straightforward model system to study the physiological regulation of this critically important channel.
Supplementary Material
Acknowledgments
We thank B.L. Mayfield, E.R. Rochon, Z. Crowell, and R.E. Bainbridge for excellent technical assistance. We thank L.A. Jaffe and K.I. Kiselyov for helpful discussions and advice.
This work was supported by an Andrew Mellon Predoctoral Fellowship to K.L. Wozniak, the March of Dimes Foundation Basil O’Connor grant 5-FY16-307 to M.T. Lee, and National Institutes of Health grant R00HD69410 to A.E. Carlson.
The authors declare no competing financial interests.
Author contributions: K.L. Wozniak, M.T. Lee, and A.E. Carlson conceived of the research. K.L. Wozniak, W.A. Phelps, M. Tembo, M.T. Lee, and A.E. Carlson created the experiments, designed their implementation, planned analyses, and wrote the manuscript.
Merritt C. Maduke served as editor.
References
- Brunner J.D., Lim N.K., Schenck S., Duerst A., and Dutzler R.. 2014. X-ray structure of a calcium-activated TMEM16 lipid scramblase. Nature. 516:207–212. 10.1038/nature13984 [DOI] [PubMed] [Google Scholar]
- Bulley S., Neeb Z.P., Burris S.K., Bannister J.P., Thomas-Gatewood C.M., Jangsangthong W., and Jaggar J.H.. 2012. TMEM16A/ANO1 channels contribute to the myogenic response in cerebral arteries. Circ. Res. 111:1027–1036. 10.1161/CIRCRESAHA.112.277145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caputo A., Caci E., Ferrera L., Pedemonte N., Barsanti C., Sondo E., Pfeffer U., Ravazzolo R., Zegarra-Moran O., and Galietta L.J.. 2008. TMEM16A, a membrane protein associated with calcium-dependent chloride channel activity. Science. 322:590–594. 10.1126/science.1163518 [DOI] [PubMed] [Google Scholar]
- Cross N.L. 1981. Initiation of the activation potential by an increase in intracellular calcium in eggs of the frog, Rana pipiens. Dev. Biol. 85:380–384. 10.1016/0012-1606(81)90269-4 [DOI] [PubMed] [Google Scholar]
- Cross N.L., and Elinson R.P.. 1980. A fast block to polyspermy in frogs mediated by changes in the membrane potential. Dev. Biol. 75:187–198. 10.1016/0012-1606(80)90154-2 [DOI] [PubMed] [Google Scholar]
- Cruz-Rangel S., De Jesús-Pérez J.J., Contreras-Vite J.A., Pérez-Cornejo P., Hartzell H.C., and Arreola J.. 2015. Gating modes of calcium-activated chloride channels TMEM16A and TMEM16B. J. Physiol. 593:5283–5298. 10.1113/JP271256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Fuente R., Namkung W., Mills A., and Verkman A.S.. 2008. Small-molecule screen identifies inhibitors of a human intestinal calcium-activated chloride channel. Mol. Pharmacol. 73:758–768. 10.1124/mol.107.043208 [DOI] [PubMed] [Google Scholar]
- Elinson R.P. 1975. Site of sperm entry and a cortical contraction associated with egg activation in the frog Rana pipiens. Dev. Biol. 47:257–268. 10.1016/0012-1606(75)90281-X [DOI] [PubMed] [Google Scholar]
- El-Jouni W., Jang B., Haun S., and Machaca K.. 2005. Calcium signaling differentiation during Xenopus oocyte maturation. Dev. Biol. 288:514–525. 10.1016/j.ydbio.2005.10.034 [DOI] [PubMed] [Google Scholar]
- Gao da Y., Zhang B.L., Leung M.C., Au S.C., Wong P.Y., and Shum W.W.. 2016. Coupling of TRPV6 and TMEM16A in epithelial principal cells of the rat epididymis. J. Gen. Physiol. 148:161–182. 10.1085/jgp.201611626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glahn D., and Nuccitelli R.. 2003. Voltage-clamp study of the activation currents and fast block to polyspermy in the egg of Xenopus laevis. Dev. Growth Differ. 45:187–197. 10.1034/j.1600-0854.2004.00684.x [DOI] [PubMed] [Google Scholar]
- Grey R.D., Bastiani M.J., Webb D.J., and Schertel E.R.. 1982. An electrical block is required to prevent polyspermy in eggs fertilized by natural mating of Xenopus laevis. Dev. Biol. 89:475–484. 10.1016/0012-1606(82)90335-9 [DOI] [PubMed] [Google Scholar]
- Gyobu S., Miyata H., Ikawa M., Yamazaki D., Takeshima H., Suzuki J., and Nagata S.. 2015. A Role of TMEM16E Carrying a Scrambling Domain in Sperm Motility. Mol. Cell. Biol. 36:645–659. 10.1128/MCB.00919-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer C., Wanitchakool P., Sirianant L., Papiol S., Monnheimer M., Faria D., Ousingsawat J., Schramek N., Schmitt C., Margos G., et al. . 2015. A Coding Variant of ANO10, Affecting Volume Regulation of Macrophages, Is Associated with Borrelia Seropositivity. Mol. Med. 21:26–37. 10.2119/molmed.2014.00219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartzell H.C., Yu K., Xiao Q., Chien L.T., and Qu Z.. 2009. Anoctamin/TMEM16 family members are Ca2+-activated Cl- channels. J. Physiol. 587:2127–2139. 10.1113/jphysiol.2008.163709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M., Ye W., Wang W.J., Sison E.S., Jan Y.N., and Jan L.Y.. 2017. Cytoplasmic Cl- couples membrane remodeling to epithelial morphogenesis. Proc. Natl. Acad. Sci. USA. 114:E11161–E11169. 10.1073/pnas.1714448115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heasman J., Holwill S., and Wylie C.C.. 1991. Fertilization of cultured Xenopus oocytes and use in studies of maternally inherited molecules. Methods Cell Biol. 36:213–230. 10.1016/S0091-679X(08)60279-4 [DOI] [PubMed] [Google Scholar]
- Huang F., Rock J.R., Harfe B.D., Cheng T., Huang X., Jan Y.N., and Jan L.Y.. 2009. Studies on expression and function of the TMEM16A calcium-activated chloride channel. Proc. Natl. Acad. Sci. USA. 106:21413–21418. 10.1073/pnas.0911935106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F., Wong X., and Jan L.Y.. 2012. International Union of Basic and Clinical Pharmacology. LXXXV: calcium-activated chloride channels. Pharmacol. Rev. 64:1–15. 10.1124/pr.111.005009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe L.A. 1976. Fast block to polyspermy in sea urchin eggs is electrically mediated. Nature. 261:68–71. 10.1038/261068a0 [DOI] [PubMed] [Google Scholar]
- Jin X., Shah S., Liu Y., Zhang H., Lees M., Fu Z., Lippiat J.D., Beech D.J., Sivaprasadarao A., Baldwin S.A., et al. . 2013. Activation of the Cl- channel ANO1 by localized calcium signals in nociceptive sensory neurons requires coupling with the IP3 receptor. Sci. Signal. 6:ra73 10.1126/scisignal.2004184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane Dickson V., Pedi L., and Long S.B.. 2014. Structure and insights into the function of a Ca(2+)-activated Cl(-) channel. Nature. 516:213–218. 10.1038/nature13913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Langmead B., and Salzberg S.L.. 2015. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 12:357–360. 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline D. 1988. Calcium-dependent events at fertilization of the frog egg: injection of a calcium buffer blocks ion channel opening, exocytosis, and formation of pronuclei. Dev. Biol. 126:346–361. 10.1016/0012-1606(88)90145-5 [DOI] [PubMed] [Google Scholar]
- Kredel S., Oswald F., Nienhaus K., Deuschle K., Röcker C., Wolff M., Heilker R., Nienhaus G.U., and Wiedenmann J.. 2009. mRuby, a bright monomeric red fluorescent protein for labeling of subcellular structures. PLoS One. 4:e4391 10.1371/journal.pone.0004391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.T., Bonneau A.R., and Giraldez A.J.. 2014. Zygotic genome activation during the maternal-to-zygotic transition. Annu. Rev. Cell Dev. Biol. 30:581–613. 10.1146/annurev-cellbio-100913-013027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Smyth G.K., and Shi W.. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 30:923–930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Liu Y., Zhang H., Huang D., Qi J., Xu J., Gao H., Du X., Gamper N., and Zhang H.. 2015. Characterization of the effects of Cl− channel modulators on TMEM16A and bestrophin-1 Ca2+ activated Cl− channels. Pflugers Arch. 467:1417–1430. 10.1007/s00424-014-1572-5 [DOI] [PubMed] [Google Scholar]
- Mohanty B.K., and Gupta B.L.. 2012. A marked animal-vegetal polarity in the localization of Na(+),K(+) -ATPase activity and its down-regulation following progesterone-induced maturation. Mol. Reprod. Dev. 79:138–160. 10.1002/mrd.22012 [DOI] [PubMed] [Google Scholar]
- Moriyoshi K., Richards L.J., Akazawa C., O’Leary D.D., and Nakanishi S.. 1996. Labeling neural cells using adenoviral gene transfer of membrane-targeted GFP. Neuron. 16:255–260. 10.1016/S0896-6273(00)80044-6 [DOI] [PubMed] [Google Scholar]
- Namkung W., Phuan P.W., and Verkman A.S.. 2011. TMEM16A inhibitors reveal TMEM16A as a minor component of calcium-activated chloride channel conductance in airway and intestinal epithelial cells. J. Biol. Chem. 286:2365–2374. 10.1074/jbc.M110.175109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuccitelli R., Yim D.L., and Smart T.. 1993. The sperm-induced Ca2+ wave following fertilization of the Xenopus egg requires the production of Ins(1, 4, 5)P3. Dev. Biol. 158:200–212. 10.1006/dbio.1993.1179 [DOI] [PubMed] [Google Scholar]
- Oh S.J., Hwang S.J., Jung J., Yu K., Kim J., Choi J.Y., Hartzell H.C., Roh E.J., and Lee C.J.. 2013. MONNA, a potent and selective blocker for transmembrane protein with unknown function 16/anoctamin-1. Mol. Pharmacol. 84:726–735. 10.1124/mol.113.087502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Z., and Hartzell C.. 2004. Determinants of anion permeation in the second transmembrane domain of the mouse bestrophin-2 chloride channel. J. Gen. Physiol. 124:371–382. 10.1085/jgp.200409108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Z., Wei R.W., Mann W., and Hartzell H.C.. 2003. Two bestrophins cloned from Xenopus laevis oocytes express Ca(2+)-activated Cl(-) currents. J. Biol. Chem. 278:49563–49572. 10.1074/jbc.M308414200 [DOI] [PubMed] [Google Scholar]
- Rasar M.A., and Hammes S.R.. 2006. The physiology of the Xenopus laevis ovary. Methods Mol. Biol. 322:17–30. 10.1007/978-1-59745-000-3_2 [DOI] [PubMed] [Google Scholar]
- Rock J.R., Futtner C.R., and Harfe B.D.. 2008. The transmembrane protein TMEM16A is required for normal development of the murine trachea. Dev. Biol. 321:141–149. 10.1016/j.ydbio.2008.06.009 [DOI] [PubMed] [Google Scholar]
- Runft L.L., Watras J., and Jaffe L.A.. 1999. Calcium release at fertilization of Xenopus eggs requires type I IP3 receptors, but not SH2 domain-mediated activation of PLCgamma or G(q)-mediated activation of PLCbeta. Dev. Biol. 214:399–411. 10.1006/dbio.1999.9415 [DOI] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., and Eliceiri K.W.. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 9:671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder B.C., Cheng T., Jan Y.N., and Jan L.Y.. 2008. Expression cloning of TMEM16A as a calcium-activated chloride channel subunit. Cell. 134:1019–1029. 10.1016/j.cell.2008.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler C.Y., Park J.G., Sharma A., Hunter P., Surapaneni P., Sedillo C., Field J., Algar R., Price A., Steel J., et al. . 2014. DNASU plasmid and PSI:Biology-Materials repositories: resources to accelerate biological research. Nucleic Acids Res. 42(D1):D1253–D1260. 10.1093/nar/gkt1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo Y., Lee H.K., Park J., Jeon D.K., Jo S., Jo M., and Namkung W.. 2016. Ani9, A Novel Potent Small-Molecule ANO1 Inhibitor with Negligible Effect on ANO2. PLoS One. 11:e0155771 10.1371/journal.pone.0155771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Session A.M., Uno Y., Kwon T., Chapman J.A., Toyoda A., Takahashi S., Fukui A., Hikosaka A., Suzuki A., Kondo M., et al. . 2016. Genome evolution in the allotetraploid frog Xenopus laevis. Nature. 538:336–343. 10.1038/nature19840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stricker S.A. 1999. Comparative biology of calcium signaling during fertilization and egg activation in animals. Dev. Biol. 211:157–176. 10.1006/dbio.1999.9340 [DOI] [PubMed] [Google Scholar]
- Tadros W., and Lipshitz H.D.. 2009. The maternal-to-zygotic transition: a play in two acts. Development. 136:3033–3042. 10.1242/dev.033183 [DOI] [PubMed] [Google Scholar]
- Tran T.T., Tobiume K., Hirono C., Fujimoto S., Mizuta K., Kubozono K., Inoue H., Itakura M., Sugita M., and Kamata N.. 2014. TMEM16E (GDD1) exhibits protein instability and distinct characteristics in chloride channel/pore forming ability. J. Cell. Physiol. 229:181–190. 10.1002/jcp.24431 [DOI] [PubMed] [Google Scholar]
- Wallace R.A., Jared D.W., Dumont J.N., and Sega M.W.. 1973. Protein incorporation by isolated amphibian oocytes. 3. Optimum incubation conditions. J. Exp. Zool. 184:321–333. 10.1002/jez.1401840305 [DOI] [PubMed] [Google Scholar]
- Wanitchakool P., Ousingsawat J., Sirianant L., Cabrita I., Faria D., Schreiber R., and Kunzelmann K.. 2017. Cellular defects by deletion of ANO10 are due to deregulated local calcium signaling. Cell. Signal. 30:41–49. 10.1016/j.cellsig.2016.11.006 [DOI] [PubMed] [Google Scholar]
- Webb D.J., and Nuccitelli R.. 1985. Fertilization potential and electrical properties of the Xenopus laevis egg. Dev. Biol. 107:395–406. 10.1016/0012-1606(85)90321-5 [DOI] [PubMed] [Google Scholar]
- Wong J.L., and Wessel G.M.. 2006. Defending the zygote: search for the ancestral animal block to polyspermy. Curr. Top. Dev. Biol. 72:1–151. [DOI] [PubMed] [Google Scholar]
- Wozniak K.L., Mayfield B.L., Duray A.M., Tembo M., Beleny D.O., Napolitano M.A., Sauer M.L., Wisner B.W., and Carlson A.E.. 2017. Extracellular Ca2+ Is Required for Fertilization in the African Clawed Frog, Xenopus laevis. PLoS One. 12:e0170405 10.1371/journal.pone.0170405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak K.L., Tembo M., Phelps W.A., Lee M.T., and Carlson A.E.. 2018. PLC and IP3-evoked Ca2+ release initiate the fast block to polyspermy in Xenopus laevis eggs. J. Gen. Physiol. 10.1085/jgp.201812069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wühr M., Freeman R.M. Jr., Presler M., Horb M.E., Peshkin L., Gygi S., and Kirschner M.W.. 2014. Deep proteomics of the Xenopus laevis egg using an mRNA-derived reference database. Curr. Biol. 24:1467–1475. 10.1016/j.cub.2014.05.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wylie C.C., Brown D., Godsave S.F., Quarmby J., and Heasman J.. 1985. The cytoskeleton of Xenopus oocytes and its role in development. J. Embryol. Exp. Morphol. 89(Suppl):1–15. [PubMed] [Google Scholar]
- Yang T., Liu Q., Kloss B., Bruni R., Kalathur R.C., Guo Y., Kloppmann E., Rost B., Colecraft H.M., and Hendrickson W.A.. 2014. Structure and selectivity in bestrophin ion channels. Science. 346:355–359. 10.1126/science.1259723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.D., Cho H., Koo J.Y., Tak M.H., Cho Y., Shim W.S., Park S.P., Lee J., Lee B., Kim B.M., et al. . 2008. TMEM16A confers receptor-activated calcium-dependent chloride conductance. Nature. 455:1210–1215. 10.1038/nature07313 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.