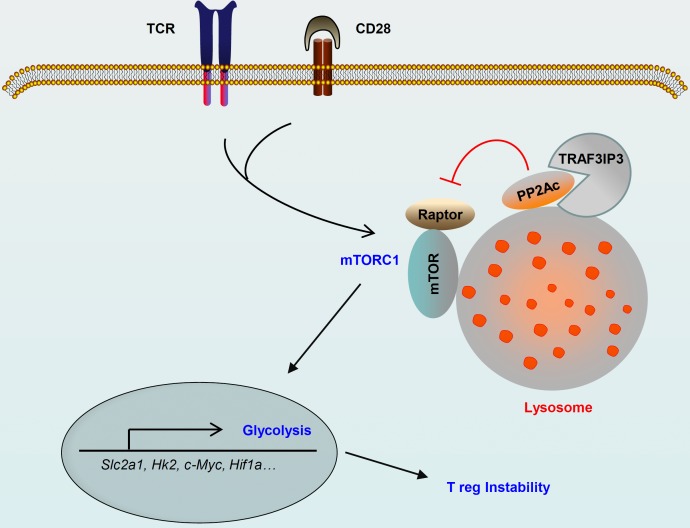
Metabolic programs are crucial for T reg cell stability and function. Yu et al. report that TRAF3IP3 acts as a pivotal regulator in controlling metabolic fitness to maintain T reg cell stability and function at the lysosome.
Abstract
Metabolic programs are crucial for regulatory T (T reg) cell stability and function, but the underlying mechanisms that regulate T reg cell metabolism are elusive. Here, we report that lysosomal TRAF3IP3 acts as a pivotal regulator in the maintenance of T reg cell metabolic fitness. T reg–specific deletion of Traf3ip3 impairs T reg cell function, causing the development of inflammatory disorders and stronger antitumor T cell responses in mice. Excessive mechanistic target of rapamycin complex 1 (mTORC1)–mediated hyper-glycolytic metabolism is responsible for the instability of TRAF3IP3-deficient T reg cells. Mechanistically, TRAF3IP3 restricts mTORC1 signaling by recruiting the serine-threonine phosphatase catalytic subunit (PP2Ac) to the lysosome, thereby facilitating the interaction of PP2Ac with the mTORC1 component Raptor. Our results define TRAF3IP3 as a metabolic regulator in T reg cell stability and function and suggest a lysosome-specific mTORC1 signaling mechanism that regulates T reg cell metabolism.
Graphical Abstract
Introduction
Regulatory T (T reg) cells are indispensable in preventing inflammatory disorders and establishing immunological homeostasis (Sakaguchi et al., 2008; Josefowicz et al., 2012). These cells also play critical roles in controlling antitumor immune responses and influence tumor immune surveillance (Bauer et al., 2014; Arce Vargas et al., 2017). Unlike CD4+ effector T cells, T reg cells rely on oxidative phosphorylation (OXPHOS) rather than glycolysis for the energy needed to support their expansion, survival, and function (Michalek et al., 2011; Coe et al., 2014; Newton et al., 2016). Elevated glycolysis leads to the expansion of highly proliferative T reg cells and the loss of Foxp3 in vivo, whereas blocking glycolytic metabolism promotes T reg cell generation (Shi et al., 2011; Zeng and Chi, 2017). Interestingly, the migration of activated T reg cells to inflamed tissue is dependent on the glycolytic pathway (Alon, 2017; Kishore et al., 2017). Thus, the dynamic regulation of cellular metabolic programs is central to T reg cell stability and functions.
Mechanistic target of rapamycin (mTOR) is a critical regulator of T reg cell identity (Chi, 2012; Zeng et al., 2013; Shrestha et al., 2015; Essig et al., 2017; Xu et al., 2017). mTOR functions in two different complexes, mTOR complex 1 (mTORC1) and mTORC2, which are distinguished by the scaffold proteins Raptor and Rictor, respectively (Zeng and Chi, 2017). Increased mTORC1 activity promotes T reg cell proliferation and instability, whereas loss of mTORC1 activity reduces T reg cell suppressive functions (Apostolidis et al., 2016; Zeng and Chi, 2017). Although the over-activation of mTORC2 destabilizes T reg cells and impairs the T reg–mediated suppression of Th1 and Tfh cell responses, mTORC2 is dispensable for T reg cell lineage stability and function (Shrestha et al., 2015; Zeng and Chi, 2017). Emerging studies reveal a central role for mTORC1 and mTORC2 in the glycolytic metabolism of T reg cells. For example, inflammatory signals emanating from TLR1 and TLR2 promote T reg cell glycolysis and proliferation in an mTORC1-dependent manner, but reduce the suppressive functions of T reg cells (Gerriets et al., 2016). The T reg–specific deletion of Atg7, a critical gene in autophagy, causes increased mTORC1 activity, which promotes hyper-glycolytic metabolism through the metabolic regulator c-Myc transcription factor and, in turn, destabilizes T reg cells (Wei et al., 2016). Loss of PTEN skews T reg cell metabolism toward glycolysis in an mTORC2-dependent manner, leading to disturbed T reg cell stability and function (Huynh et al., 2015). However, the underlying mechanisms that orchestrate the activation of mTOR signaling and the interplay between mTORC1 and mTORC2 in maintaining T reg cell metabolic fitness remain unclear.
The subcellular localization of distinct mTOR complexes is necessary for the precise spatial and temporal control of mTORC1 and mTORC2 activity (Betz and Hall, 2013). Prior studies have reported the localization of mTORC1 and mTORC2 to several distinct subcellular compartments in various organisms (Betz and Hall, 2013; Malik et al., 2013). Lysosomal translocation of mTORC1, which is dependent on amino acid influx, has been identified in various cell types (Kim et al., 2008; Sancak et al., 2010; Jewell et al., 2013). Notably, TCR-induced mTORC1 localization to the lysosome is essential for increased mTORC1 activity and glycolytic activity and contributes to the asymmetric division of CD8+ T cells (Pollizzi et al., 2016). mTORC2 localization in proximity to the mitochondria, ER, and mitochondria-associated ER membrane (MAM) has consistently been identified (Miyamoto et al., 2008; Giorgi et al., 2010; Betz and Hall, 2013). However, the physiological relevance of mTORC1 or mTORC2 subcellular localization in T reg cells has not been clearly established.
Here, we found that TRAF3IP3 actively restrained mTORC1 at the lysosome to restrict glycolytic metabolism and maintain T reg cell stability and function. This TRAF3IP3-mediated suppression of mTORC1 signaling and T reg cell glycolysis was dependent on the activity of the serine/threonine phosphatase PP2A. Mechanistically, TRAF3IP3 recruited the PP2A catalytic subunit (PP2Ac) to the lysosome and facilitated the interaction of PP2Ac with the mTORC1 component Raptor following TCR and CD28 stimulation. These findings identify TRAF3IP3 as a metabolic regulator in the maintenance of T reg cell identity and suggest a lysosome-specific mTORC1 signaling mechanism that regulates T reg cell metabolism.
Results
TRAF3IP3 is required for T reg cell maintenance and function
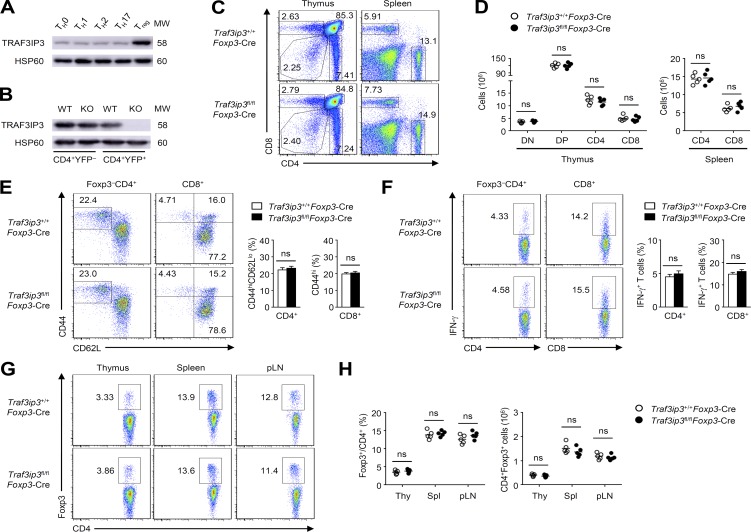
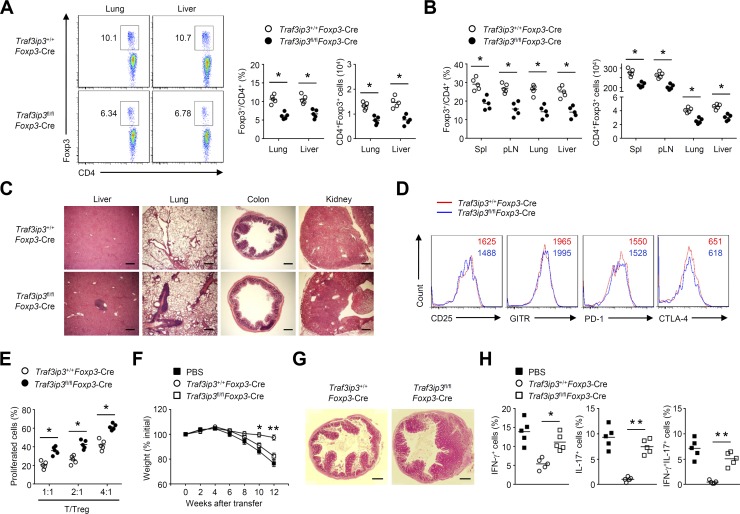
TRAF3IP3 is required for thymocyte development (Zou et al., 2015), but the potential role of TRAF3IP3 in different T cell subsets is still unknown. Interestingly, a comparison of different CD4+ T cell subsets generated in vitro revealed higher TRAF3IP3 expression in T reg cells (Fig. 1 A), which prompted us to investigate the role of TRAF3IP3 in T reg cell function. To this end, we crossed Traf3ip3-flox mice with Foxp3-Cre mice to generate Traf3ip3 T reg cell conditional knockout mice (Traf3ip3fl/flFoxp3-Cre), in which TRAF3IP3 was specifically deleted in T reg cells (Fig. 1 B). The 6-wk-old Traf3ip3fl/flFoxp3-Cre mice did not show obvious abnormalities in thymocyte development or peripheral T cell frequency (Fig. 1, C and D). The 8-wk-old wild-type (Traf3ip3+/+Foxp3-Cre) mice and Traf3ip3fl/flFoxp3-Cre mice had similar proportions of activated or memory-like CD4+ and CD8+ T cells in the spleen, as well as similar proportions of IFN-γ–producing CD4+ and CD8+ effector T (T eff) cells (Fig. 1, E and F). In addition, the percentage and number of T reg cells in the thymus, spleen, and peripheral lymph nodes of 6-wk-old Traf3ip3+/+Foxp3-Cre mice and Traf3ip3fl/flFoxp3-Cre mice were also comparable (Fig. 1, G and H). However, the percentage and number of T reg cells in the lung and liver from 6-wk-old Traf3ip3fl/flFoxp3-Cre mice were significantly lower than those from Traf3ip3+/+Foxp3-Cre mice (Fig. 2 A). Moreover, 8-mo-old Traf3ip3fl/flFoxp3-Cre mice showed remarkably lower frequency and number of T reg cells in the spleen, peripheral lymph nodes, lung, and liver (Fig. 2 B). Furthermore, 8-mo-old Traf3ip3fl/flFoxp3-Cre mice displayed autoimmune symptoms with lymphocytic infiltration into many nonlymphoid organs (Fig. 2 C), indicating that TRAF3IP3 in T reg cells prevents autoimmune responses.
Figure 1.
T cell development and homeostasis in Traf3ip3fl/flFoxp3-Cre mice. (A and B) IB analysis of TRAF3IP3 in various T cell subsets differentiated in vitro (A) or sorted CD4+YFP– and CD4+YFP+ splenocytes of Traf3ip3+/+Foxp3-Cre (wild-type [WT]) and Traf3ip3fl/flFoxp3-Cre mice (KO; B). (C) Flow cytometric analysis of the percentage of CD4+ and CD8+ T cells from the thymus and spleen of 6-wk-old mice. (D) Cell number of CD4–CD8– (DN), CD4+CD8+ (DP), CD4+CD8– (CD4), and CD4–CD8+ (CD8) in the thymus and CD4+ and CD8+ T cells in the spleen of 6-wk-old mice. (E) Flow cytometric analysis of the frequency of naive (CD44loCD62Lhi) and memory-like (CD44hiCD62Llo for CD4+ and CD44hi for CD8+ T cells) CD4+ and CD8+ T cells in total splenocytes from 8-wk-old Traf3ip3+/+Foxp3-Cre and Traf3ip3fl/flFoxp3-Cre mice. (F) Flow cytometric analysis of the percentage of IFN-γ–producing CD4+ and CD8+ T cells in the spleen from 8-wk-old Traf3ip3+/+Foxp3-Cre and Traf3ip3fl/flFoxp3-Cre mice. (G) Flow cytometric analysis of the percentage of CD4+Foxp3+ T cells in the thymus, spleen, and peripheral lymph node (pLN) from 6-wk-old Traf3ip3+/+Foxp3-Cre and Traf3ip3fl/flFoxp3-Cre mice. (H) Summary graphs of the percentage and number of CD4+Foxp3+ T cells in the thymus (Thy), spleen (Spl), and peripheral lymph node (pLN) from 6-wk-old Traf3ip3+/+Foxp3-Cre and Traf3ip3fl/flFoxp3-Cre mice. Data are representative of three independent experiments and are presented as mean ± SEM. ns, not statistically significant. Student’s t test. n = 5 in each group.
Figure 2.
TRAF3IP3 is required for T reg cell maintenance and function. (A) Flow cytometric analysis of CD4+Foxp3+ T cells in the lung and liver from 6-wk-old Traf3ip3+/+Foxp3-Cre and Traf3ip3fl/flFoxp3-Cre mice. (B) Flow cytometric analysis of CD4+Foxp3+ T cells in the spleen (Spl), peripheral lymph node (pLN), lung, and liver from 8-mo-old Traf3ip3+/+Foxp3-Cre and Traf3ip3fl/flFoxp3-Cre mice. (C) Hematoxylin and eosin staining of the indicated tissue sections of age- and sex-matched 8-mo-old Traf3ip3+/+Foxp3-Cre mice and Traf3ip3fl/flFoxp3-Cre mice. (D) Expression of surface markers on CD4+Foxp3+ T reg cells from the spleens of 8-wk-old Traf3ip3+/+Foxp3-Cre mice and Traf3ip3fl/flFoxp3-Cre mice. (E) In vitro suppressive activity of T reg cells. (F–H) Disease phenotype of RAG-1–deficient mice given adoptive transfer of wild-type naive CD45RBhi CD4+ T cells together with PBS or sorted T reg cells derived from 8-wk-old Traf3ip3+/+Foxp3-Cre or Traf3ip3fl/flFoxp3-Cre mice. (F) Body weight is presented relative to initial weight. (G) Hematoxylin and eosin staining of colon sections from recipient mice at 12 wk after adoptive transfer. (H) Frequency of cytokine-producing CD4+ T cells in the mesenteric lymph nodes from recipient mice at 12 wk after adoptive transfer. Bars, 500 µm. Data are representative of three or more independent experiments and are presented as mean ± SEM. ns, not statistically significant; *, P < 0.05; **, P < 0.01. n = 5 in each group.
We next examined whether TRAF3IP3 is required for T reg cell suppressive function. Although TRAF3IP3 deficiency did not alter the surface/transcriptional profiles in splenic T reg cells from 8-wk-old mice (Fig. 2 D and Fig. S1), T reg cell suppressive activity in vitro was impaired after TRAF3IP3 deletion (Fig. 2 E). Using a well-characterized adoptive transfer approach to measure T reg cell function in vivo (Chang et al., 2012), we observed that the transfer of TRAF3IP3-deficient T reg cells along with naive CD45RBhi CD4+ T cells resulted in gradual weight loss (Fig. 2 F), hyperplasia of the colonic mucosa (Fig. 2 G), and a greater frequency of memory and effector-like T cells (Fig. 2 H), whereas transferring of Traf3ip3+/+Foxp3-Cre T reg cells along with naive CD45RBhi CD4+ T cells did not (Fig. 2, F–H). Collectively, TRAF3IP3 is required for T reg cell maintenance and function.
T reg cell deletion of TRAF3IP3 boosts antitumor responses
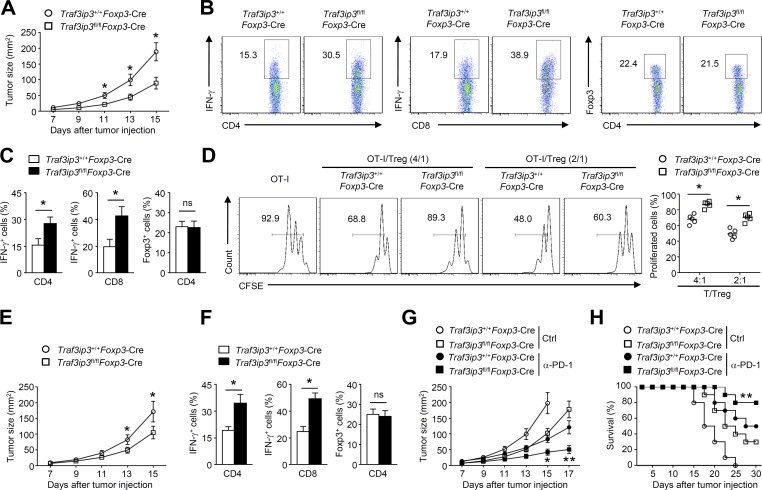
The findings that TRAF3IP3 deficiency suppresses T reg cell function indicated that targeting TRAF3IP3 might boost antitumor immune responses. We next examined the role of the T reg–specific deletion of TRAF3IP3 in regulating antitumor immunity in a B16-F10 melanoma model. Compared with Traf3ip3+/+Foxp3-Cre mice, Traf3ip3fl/flFoxp3-Cre mice exhibited a profound reduction in tumor size (Fig. 3 A) and an increased frequency of IFN-γ–producing CD4+ and CD8+ T eff cells infiltrating the tumors (Fig. 3, B and C). Although Traf3ip3+/+Foxp3-Cre and Traf3ip3fl/flFoxp3-Cre mice exhibited similar frequencies of tumor-infiltrating T reg cells (Fig. 3, B and C), the tumor-infiltrating Traf3ip3fl/flFoxp3-Cre T reg cells inhibited the proliferation of native T cells in vitro less potently than the tumor-infiltrating Traf3ip3+/+Foxp3-Cre T reg cells (Fig. 3 D). In the MC38 colon carcinoma model, TRAF3IP3 ablation in T reg cells also significantly suppressed tumor growth and enhanced antitumor immunity (Fig. 3, E and F). Remarkably, the combination of anti–PD-1 treatment and TRAF3IP3 deletion in T reg cells had a significant synergistic effect, leading to a profound reduction in the size of B16 tumors and tumor-induced lethality (Fig. 3, G and H). These data suggest that targeting TRAF3IP3 in T reg cells may be an approach for promoting antitumor T cell responses and improving tumor immunotherapy.
Figure 3.
TRAF3IP3 deletion in T reg cells boosts antitumor responses. (A) Tumor growth in 8-wk-old Traf3ip3+/+Foxp3-Cre mice and Traf3ip3fl/flFoxp3-Cre mice injected s.c. with B16-F10 melanoma cells (n = 8 mice per group). (B and C) Flow cytometric analysis of the frequency of IFN-γ–producing CD4+ or CD8+ T cells and Foxp3+CD4+ T cells in tumors from Traf3ip3+/+Foxp3-Cre mice and Traf3ip3fl/flFoxp3-Cre mice injected s.c. with B16-F10 melanoma cells (day 14 after injection). Data are presented as representative plots (B) and summary graphs (C). (D) CFSE-labeled OT-I T cells were co-cultured with OVA peptide (257–264) and splenocytes from wild-type mice for 24 h in the presence of the indicated ratios of T reg cells sorted from tumors of Traf3ip3+/+Foxp3-Cre mice or Traf3ip3fl/flFoxp3-Cre mice. (E) Tumor growth of 8-wk-old Traf3ip3+/+Foxp3-Cre mice and Traf3ip3fl/flFoxp3-Cre mice injected s.c. with MC38 colon cancer cells (n = 8 mice per group). (F) Flow cytometric analysis of the frequency of IFN-γ–producing CD4+ or CD8+ T cells and Foxp3+CD4+ T cells in tumors of Traf3ip3+/+Foxp3-Cre mice and Traf3ip3fl/flFoxp3-Cre mice injected s.c. with MC38 colon cancer cells (day 14 after injection). (G and H) Tumor growth (G) and survival curves (H) of 8-wk-old Traf3ip3+/+Foxp3-Cre mice and Traf3ip3fl/flFoxp3-Cre mice injected s.c. with B16-F10 melanoma cells (n = 10) followed by i.p. injection with PD-1 antibody on days 7, 10, and 13. Ctrl, control antibodies. Data are representative of at least three independent experiments and are presented as mean ± SEM. ns, not statistically significant; *, P < 0.05; **, P < 0.01. n = 5, 8, or 10 in each group.
TRAF3IP3 ablation affects T reg cell transcriptional programs and stability
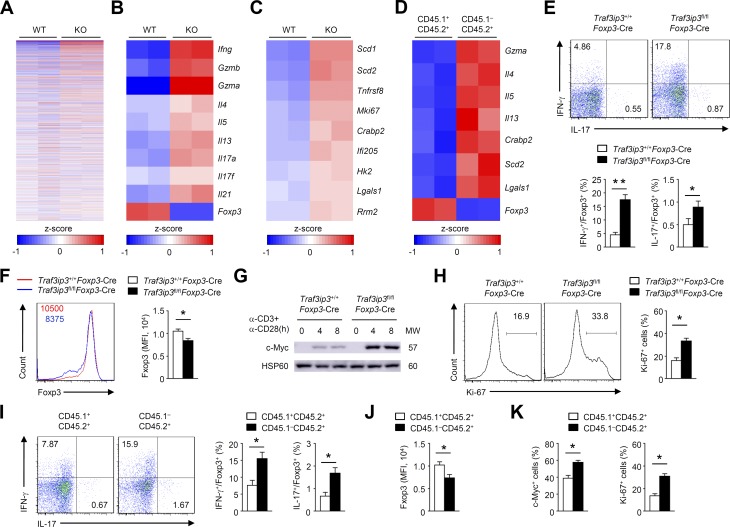
To explore TRAF3IP3-dependent transcriptional programs, we performed RNA sequencing using T reg cells activated in vitro (Fig. 4 A). Interestingly, TRAF3IP3-deficient T reg cells simultaneously acquired the expression of genes associated with effector cell differentiation, such as Ifng, Il4, Il13, Il17a, Il17f, and Il21 (Fig. 4 B). In contrast, the loss of TRAF3IP3 in T reg cells resulted in impaired transcription of the T reg cell signature gene Foxp3 (Fig. 4 B). Notably, the expression of c-Myc–associated genes, including Mki67 and Hk2, was up-regulated in TRAF3IP3-deficient T reg cells (Fig. 4 C), which was associated with T reg cell instability (Wei et al., 2016). We next investigated whether the dysregulated transcriptional programs in TRAF3IP3-deficient T reg cells were a cell-autonomous defect. We generated chimeric mice by reconstituting Rag1−/− mice with a mixture of bone marrow (BM) cells from Traf3ip3fl/flFoxp3-Cre (CD45.1−CD45.2+) mice and Traf3ip3+/+Foxp3-Cre (CD45.1+CD45.2+) mice and performed RNA sequencing using CD45.1−CD45.2+ and CD45.1+CD45.2+ T reg cells activated in vitro. TRAF3IP3-deficient T reg cells (CD45.1−CD45.2+) from the mixed chimeric mice showed higher levels of T eff– and c-Myc–associated genes, but lower level of Foxp3 (Fig. 4 D). These RNA sequencing analyses indicate a crucial role for TRAF3IP3 in regulating T reg cell transcriptional programs.
Figure 4.
TRAF3IP3 regulates T reg cell effector programs and stability. (A–C) Splenic T reg cells obtained from Traf3ip3+/+Foxp3-Cre (wild-type [WT]) mice and Traf3ip3fl/flFoxp3-Cre (KO) were stimulated with anti-CD3 and anti-CD28 antibodies for 24 h and subjected to RNA sequencing. (A) A heat map of highly variable genes (top 1,000). (B and C) Heat maps of up-regulated or down-regulated genes identified in TRAF3IP3-deficient T reg cells relative to that in wild-type T reg cells. (D) A heat map of indicated genes in TRAF3IP3-deficient (CD45.1−CD45.2+) and wild-type (CD45.1+CD45.2+) T reg cells (CD4+CD25+YFP+) from mixed chimeric mice stimulated with anti-CD3 and anti-CD28 antibodies for 24 h. Differentially expressed genes are normalized by z-score. (E and F) IFN-γ and IL-17 expression (E) or Foxp3 expression (F) in Traf3ip3+/+Foxp3-Cre and Traf3ip3fl/flFoxp3-Cre T reg cells stimulated with anti-CD3 and anti-CD28 antibodies for 24 h. (G) IB analysis of c-Myc in Traf3ip3+/+Foxp3-Cre and Traf3ip3fl/flFoxp3-Cre T reg cells stimulated as indicated. (H) Ki-67 expression in Traf3ip3+/+Foxp3-Cre and Traf3ip3fl/flFoxp3-Cre T reg cells stimulated with anti-CD3 and anti-CD28 antibodies for 24 h. (I and J) IFN-γ and IL-17 expression (I) or Foxp3 expression (J) in TRAF3IP3-deficient (CD45.1−CD45.2+) and wild-type (CD45.1+CD45.2+) T reg cells from mixed chimeric mice stimulated with anti-CD3 and anti-CD28 antibodies for 24 h. (K) Flow cytometric analysis of c-Myc or Ki-67 in TRAF3IP3-deficient (CD45.1−CD45.2+) and wild-type (CD45.1+CD45.2+) T reg cells from mixed chimeric mice stimulated for 8 or 24 h. Data are representative of at least three independent experiments and are presented as mean ± SEM. *, P < 0.05; **, P < 0.01. n = 5 in each group (E–K).
We next used an in vitro system to measure the cytokine profiles of T reg cells stimulated for 24 h with anti-CD3 and anti-CD28 antibodies. Compared with Traf3ip3+/+Foxp3-Cre T reg cells, Traf3ip3fl/flFoxp3-Cre T reg cells exhibited elevated IFN-γ and IL-17 protein levels after in vitro culture (Fig. 4 E and Fig. S2). In an in vitro system to examine T reg cell stability (Feng et al., 2014; Li et al., 2014), TRAF3IP3-deficient T reg cells exhibited lower levels of Foxp3 (Fig. 4 F). Notably, activated T reg cells from Traf3ip3fl/flFoxp3-Cre mice exhibited higher levels of c-Myc and the c-Myc–associated proliferation marker Ki-67 than T reg cells from Traf3ip3+/+Foxp3-Cre mice (Fig. 4, G and H). Consistently, TRAF3IP3-deficient T reg cells (CD45.1−CD45.2+) from the mixed chimeric mice showed much higher effector cytokine, but lower Foxp3 expression (Fig. 4, I and J), and displayed a substantial increase in c-Myc and Ki-67 expression (Fig. 4 K). Thus, TRAF3IP3 ablation affects T reg cell transcriptional programs and stability.
TRAF3IP3 restricts glycolytic metabolism in T reg cells
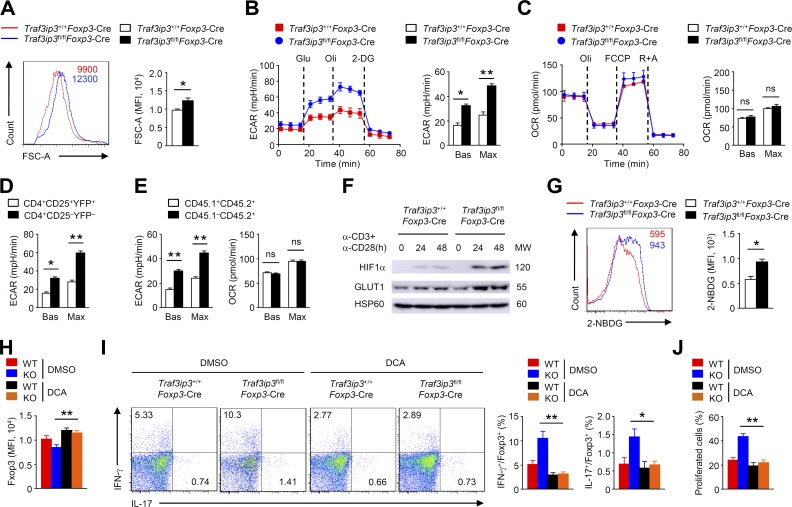
c-Myc is a crucial regulator of T cell glycolysis, which is critical for T cell growth (Wang et al., 2011). Indeed, in vitro–cultured Traf3ip3fl/flFoxp3-Cre T reg cells displayed increases in cell size (Fig. 5 A), which prompted us to examine the involvement of metabolic programs. Interestingly, TRAF3IP3-deficient T reg cells had significantly higher baseline and maximum glycolytic rates than the Traf3ip3+/+Foxp3-Cre T reg cells stimulated with TCR and CD28 (Fig. 5 B), indicating that TRAF3IP3 regulates T reg cell glycolysis. In contrast, activated TRAF3IP3-deficient and TRAF3IP3-sufficient T reg cells displayed similar OXPHOS rates, both at baseline and at maximum capacity (Fig. 5 C). We further confirmed that T reg cells were less glycolytic than conventional CD4+ T cells (Fig. 5 D), and TRAF3IP3-deficient T reg cells (CD45.1−CD45.2+) from the mixed chimeric mice had increased glycolytic rates and normal OXPHOS rates (Fig. 5 E). Notably, TCR and CD28 signals stimulated the expressions of metabolic master regulator HIF1-α, glucose transporter GLUT1 and glycolytic enzyme hexokinase 2, which were remarkably augmented in TRAF3IP3-deficient T reg cells (Fig. 5 F and Fig. S3). Consistently, direct measurements of glucose uptake in TCR and CD28-stimulated T reg cells showed about twofold increases in the uptake of the fluorescent glucose analogue 2-NBDG after TRAF3IP3 deletion (Fig. 5 G). These results suggest that TRAF3IP3 restricts T reg cell glycolytic metabolism.
Figure 5.
TRAF3IP3 restricts T reg cell glycolytic metabolism. (A) Flow cytometric analysis of FSC-A of Traf3ip3+/+Foxp3-Cre and Traf3ip3fl/flFoxp3-Cre T reg cells stimulated with anti-CD3 and anti-CD28 antibodies for 24 h. The numbers above the graphs indicate mean fluorescence intensity. (B) ECAR of Traf3ip3+/+Foxp3-Cre and Traf3ip3fl/flFoxp3-Cre T reg cells stimulated with anti-CD3 and anti-CD28 antibodies for 24 h under basal conditions (Bas) or at maximum (Max) with the addition of glucose (Glu), oligomycin (Oli), and 2-deoxy-d-glucose (2-DG). (C) Extracellular flux analysis of the OCRs of Traf3ip3+/+Foxp3-Cre and Traf3ip3fl/flFoxp3-Cre T reg cells stimulated with anti-CD3 and anti-CD28 antibodies for 24 h under basal conditions (Bas) or at maximum (Max) with the addition of oligomycin (Oli), the mitochondrial uncoupler FCCP, and rotenone plus antimycin A (R+A). (D) ECAR of T reg cells and conventional CD4+ T cells stimulated with anti-CD3 and anti-CD28 for 24 h. (E) ECAR of TRAF3IP3-deficient (CD45.1−CD45.2+) and wild-type (CD45.1+CD45.2+) T reg cells from mixed chimeric mice stimulated with anti-CD3 and anti-CD28 for 24 h. (F) IB analysis of the indicated proteins in Traf3ip3+/+Foxp3-Cre and Traf3ip3fl/flFoxp3-Cre T reg cells stimulated as indicated. (G) Traf3ip3+/+Foxp3-Cre and Traf3ip3fl/flFoxp3-Cre T reg cells stimulated with anti-CD3 and anti-CD28 antibodies for 36 h and analyzed by flow cytometry for glucose uptake (as measured by 2-NBDG uptake). (H and I) Flow cytometric analysis of Foxp3 expression (H) or IFN-γ and IL-17 expression (I) in Traf3ip3+/+Foxp3-Cre and Traf3ip3fl/flFoxp3-Cre T reg cells stimulated for 24 h with antibodies against CD3 and CD28 in the presence of DMSO or DCA. (J) In vitro suppression of T eff cells by Traf3ip3+/+Foxp3-Cre and Traf3ip3fl/flFoxp3-Cre T reg cells after incubation together at a T eff/T reg ratio of 2:1 in the presence of DMSO or DCA. The data shown are representative of three or more independent experiments and are presented as mean ± SEM. ns, not statistically significant; *, P < 0.05; **, P < 0.01; n = 4 in each group.
To examine the functional importance of glycolysis, we treated T reg cells with dichloroacetate (DCA) in the T reg cell stability assay in vitro; DCA targets pyruvate dehydrogenase kinase to shift glycolysis toward OXPHOS (Gerriets et al., 2015). Of note, DCA greatly restored Foxp3 expression but diminished IFN-γ and IL-17 production in TRAF3IP3-deficient T reg cells (Fig. 5, H and I). Moreover, DCA treatment increased the suppressive function of TRAF3IP3-deficient T reg cells (Fig. 5 J). Therefore, hyper-glycolytic metabolism of TRAF3IP3-deficient T reg cells results in the impaired T reg cell stability and function.
Aberrant mTORC1 activity leads to TRAF3IP3-deficient T reg cell instability
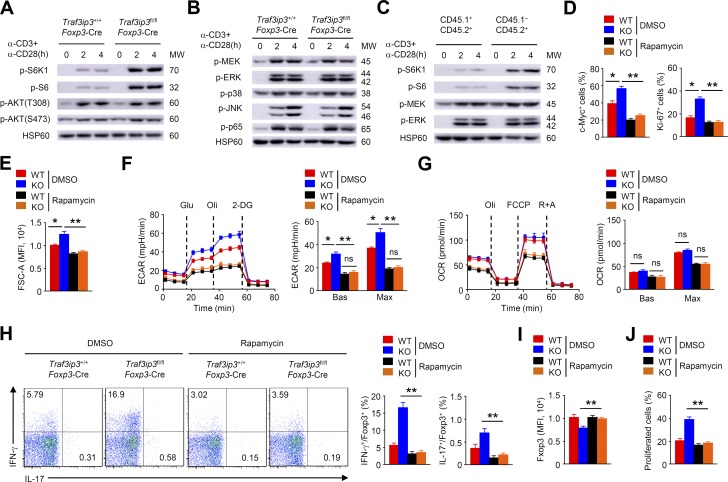
Given that mTOR signaling can destabilize T reg cells by disturbing their transcriptional programs and promoting glycolytic metabolism (Wei et al., 2016; Zeng and Chi, 2017), we sought to determine whether TRAF3IP3 regulates the activity of the mTOR signaling pathway. Indeed, immunoblot (IB) analysis revealed increased phosphorylation of ribosomal protein S6 kinase β-1 (S6K1) and ribosomal protein S6, indicative of mTORC1 activation, in Traf3ip3fl/flFoxp3-Cre T reg cells stimulated with anti-CD3 and anti-CD28 (Fig. 6 A). In contrast, AKT phosphorylation at Thr 308 or Ser473 in Traf3ip3fl/flFoxp3-Cre T reg cells was unaltered (Fig. 6 A). Furthermore, upon stimulation with anti-CD3 plus anti-CD28, Traf3ip3+/+Foxp3-Cre and Traf3ip3fl/flFoxp3-Cre T reg cells had similar level of MEK, ERK, JNK, p38, and p65 phosphorylation (Fig. 6 B). Consistent with our finding of elevated mTORC1 activity in TRAF3IP3-deficient T reg cells, Traf3ip3fl/flFoxp3-Cre T reg cells (CD45.1−CD45.2+) from the mixed chimeric mice displayed increases in S6 and S6K1 phosphorylation (Fig. 6 C). Therefore, TRAF3IP3 inhibits mTORC1 activation in T reg cells.
Figure 6.
Aberrant mTORC1 signaling leads to TRAF3IP3-deficient T reg cell instability. (A and B) IB analysis of the indicated proteins in Traf3ip3+/+Foxp3-Cre and Traf3ip3fl/flFoxp3-Cre T reg cells stimulated as indicated. (C) IB analysis of the indicated proteins in TRAF3IP3-deficient (CD45.1−CD45.2+) and wild-type (CD45.1+CD45.2+) T reg cells from mixed chimeric mice stimulated as indicated. (D) Flow cytometric analysis of c-Myc or Ki-67 expression in Traf3ip3+/+Foxp3-Cre (wild-type [WT]) and Traf3ip3fl/flFoxp3-Cre (KO) T reg cells stimulated for 8 or 24 h with antibodies against CD3 and CD28 in the presence of DMSO or rapamycin. (E–G) FSC-A (E), ECAR (F), and OCR (G) of Traf3ip3+/+Foxp3-Cre (WT) and Traf3ip3fl/flFoxp3-Cre (KO) T reg cells stimulated for 24 h with antibodies against CD3 and CD28 in the presence of DMSO or rapamycin. (H and I) Flow cytometric analysis of IFN-γ and IL-17 expression (H) or Foxp3 expression (I) in Traf3ip3+/+Foxp3-Cre and Traf3ip3fl/flFoxp3-Cre T reg cells stimulated for 24 h with antibodies against CD3 and CD28 in the presence of DMSO or rapamycin. (J) In vitro suppression of T eff cells by Traf3ip3+/+Foxp3-Cre and Traf3ip3fl/flFoxp3-Cre T reg cells after incubation together at a T eff/T reg ratio of 2:1 in the presence of DMSO or rapamycin. Data are representative of at least three independent experiments and are presented as mean ± SEM. ns, not statistically significant; *, P < 0.05; **, P < 0.01. n = 5 in each group.
To determine the contribution of elevated mTORC1 activity to TRAF3IP3-deficient T reg cell instability, we stimulated T reg cells from Traf3ip3+/+Foxp3-Cre and Traf3ip3fl/flFoxp3-Cre mice with anti-CD3 and anti-CD28 in the presence or absence of the mTORC1 inhibitor rapamycin. Rapamycin treatment considerably reduced c-Myc and Ki-67 expression in TRAF3IP3-deficient T reg cells (Fig. 6 D). In addition, rapamycin-treated TRAF3IP3-deficient T reg cells showed a substantial decrease in cell size (Fig. 6 E). Rapamycin treatment also lowered the extracellular acidification rate and erased the differences between TRAF3IP3-deficient and TRAF3IP3-sufficient T reg cells (Fig. 6 F), indicating a crucial role for mTORC1 in TRAF3IP3-dependent glycolytic homeostasis. TRAF3IP3-deficient and TRAF3IP3-sufficient T reg cells displayed similar OXPHOS rates, both in the absence and presence of rapamycin (Fig. 6 G), suggesting a dispensable role of TRAF3IP3 in regulating T reg cell OXPHOS. Importantly, rapamycin greatly diminished IFN-γ and IL-17 production, but restored Foxp3 expression in TRAF3IP3-deficient T reg cells (Fig. 6, H and I). Moreover, rapamycin treatment increased the suppressive function of TRAF3IP3-deficient T reg cells (Fig. 6 J). Together, these results suggest that aberrant mTORC1 activity contributes to TRAF3IP3-deficient T reg cell hyper-glycolysis and instability.
TRAF3IP3 is involved in the regulation of PP2A activity in T reg cells
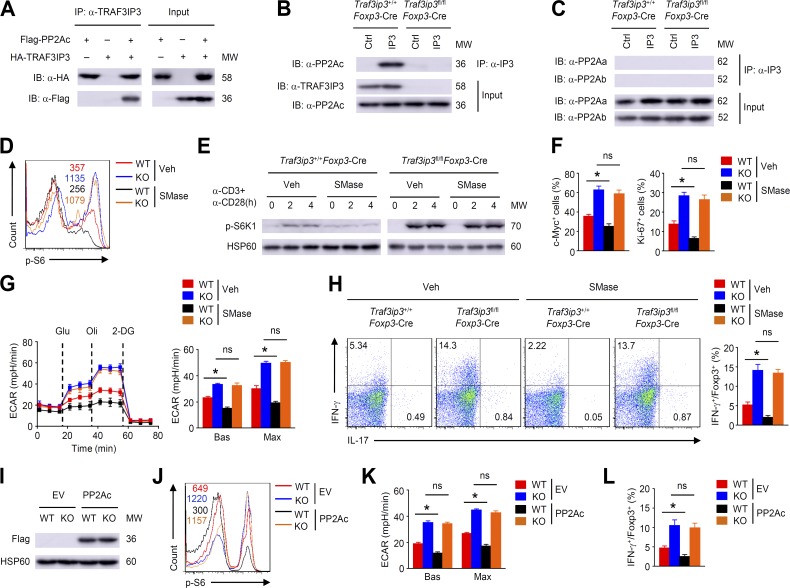
PP2A can maintain T reg cell metabolic programs and suppressive function by restraining mTORC1 activity (Apostolidis et al., 2016). Mass spectrometry analysis has indicated that TRAF3IP3 is associated with components of PP2A (Goudreault et al., 2009; Huttlin et al., 2017). Interestingly, transient transfection of HEK293T cells with the respective constructs demonstrated an interaction between TRAF3IP3 and PP2Ac (Fig. 7 A). Additionally, TRAF3IP3 was found to physically interact with PP2Ac, but not the scaffold A subunit (PP2Aa) or the regulatory B subunit (PP2Ab) in T reg cells (Fig. 7, B and C). Therefore, we asked whether TRAF3IP3 deficiency in T reg cells affects PP2A activity. Indeed, treating wild-type T reg cells with sphingomyelinase (SMase), which stimulates PP2A activation by inducing the production of ceramide (Dobrowsky et al., 1993), decreased S6 and S6K1 phosphorylation (Fig. 7, D and E). However, SMase treatment had a negligible effect on mTORC1 activation in TRAF3IP3-deficient T reg cells (Fig. 7, D and E). Moreover, although SMase-treated wild-type T reg cells displayed a substantial decrease in c-Myc and Ki-67 expression and glycolytic rates, this effect was not observed in TRAF3IP3-deficient T reg cells after SMase treatment (Fig. 7, F and G). Consistently, SMase did not alter IFN-γ and IL-17 production in TRAF3IP3-deficient T reg cells (Fig. 7 H). Therefore, TRAF3IP3 is involved in the regulation of PP2A activity in T reg cells.
Figure 7.
TRAF3IP3 is involved in the regulation of PP2A activity in T reg cells. (A) TRAF3IP3-PP2Ac coimmunoprecipitation (co-IP) assays using HEK293T cells transfected with the indicated expression vectors. (B and C) Lysates of Traf3ip3+/+Foxp3-Cre and Traf3ip3fl/flFoxp3-Cre T reg cells stimulated for 4 h were subjected to IP using an antibody against TRAF3IP3 (IP3) or control Ig (Ctrl). (D) Flow cytometric analysis of p-S6 expression in Traf3ip3+/+Foxp3-Cre (wild-type [WT]) and Traf3ip3fl/flFoxp3-Cre (KO) T reg cells stimulated with anti-CD3 and anti-CD28 antibodies for 4 h in the presence of SMase (0.5 U/ml) or vehicle (50% glycerol in PBS). The numbers above the graphs indicate mean fluorescence activity. (E) IB analysis of p-S6K1 expression in Traf3ip3+/+Foxp3-Cre or Traf3ip3fl/flFoxp3-Cre T reg cells stimulated as indicated. (F) Flow cytometric analysis of c-Myc and Ki-67 expression in Traf3ip3+/+Foxp3-Cre (wild-type [WT]) and Traf3ip3fl/flFoxp3-Cre (KO) T reg cells stimulated with anti-CD3 and anti-CD28 antibodies for 8 or 24 h in the presence of SMase or vehicle. (G) ECAR of Traf3ip3+/+Foxp3-Cre (wild-type [WT]) and Traf3ip3fl/flFoxp3-Cre (KO) T reg cells stimulated for 24 h with antibodies against CD3 and CD28 in the presence of SMase or vehicle. (H) Flow cytometric analysis of IFN-γ and IL-17 expression in Traf3ip3+/+Foxp3-Cre (wild-type [WT]) and Traf3ip3fl/flFoxp3-Cre (KO) T reg cells stimulated for 24 h with antibodies against CD3 and CD28 in the presence of SMase or vehicle. (I) IB analysis of Flag-PP2Ac in naive Traf3ip3+/+Cd4-Cre (wild-type [WT]) and Traf3ip3fl/flCd4-Cre (KO) CD4+ T cells transduced with either an empty vector (EV) or PP2Ac expression vector under T reg cell polarizing conditions. (J–L) p-S6 expression (J), ECAR (K), and cytokine expression (L) of transduced T reg cells from (I) stimulated for 4 (J) or 24 h (K and L) with antibodies against CD3 and CD28. Data are representative of three independent experiments and are presented as mean ± SEM. ns, not statistically significant; *, P < 0.05. n = 5 in each group.
To confirm the functional importance of PP2A, we transduced in vitro activated naive TRAF3IP3-deficient CD4+ T cells with PP2Ac and subsequently cultured these cells under T reg cell polarizing conditions (Fig. 7 I). PP2Ac-overexpressed wild-type T reg cells exhibited decreased S6 phosphorylation, glycolytic rates, and IFN-γ production (Fig. 7, J–L). However, PP2Ac overexpression in TRAF3IP3-deficient T reg cells did not significantly alter S6 phosphorylation, glycolytic rates and IFN-γ production (Fig. 7, J–L). These results suggest that TRAF3IP3 is required for PP2A function to inhibit mTORC1 signaling and glycolysis in T reg cells.
TRAF3IP3 mediates PP2A–mTORC1 axis at the lysosome
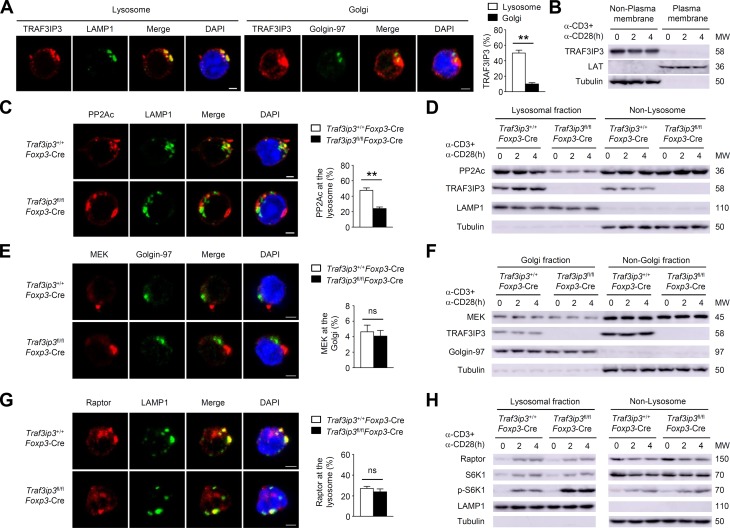
Given that mTORC1 localization to the lysosome is essential for its activation during T cell activation (Pollizzi et al., 2016), we asked whether the transmembrane protein TRAF3IP3 localizes to the lysosome and mediates the lysosomal recruitment of PP2A to inhibit mTORC1. Indeed, confocal microscopy assays revealed that ∼50% of TRAF3IP3 localized to the lysosome and only ∼10% of TRAF3IP3 localized to the Golgi in T reg cells (Fig. 8 A). However, TRAF3IP3 did not localize to the plasma membrane in T reg cells (Fig. 8 B). Notably, the lysosomal PP2Ac levels were lower in TRAF3IP3-deficient T reg cells (Fig. 8, C and D), indicating that TRAF3IP3 recruits PP2Ac to the lysosome in T reg cells. Only ∼5% of MEK localized to the Golgi and the constitutive Golgi localization of MEK was unaffected after TRAF3IP3 depletion in T reg cells (Fig. 8, E and F), suggesting a dispensable role of TRAF3IP3 in Golgi localization of MEK in T reg cells. Interestingly, Raptor and S6K1 were transferred to the lysosome following TCR and CD28 stimulation in a TRAF3IP3-independent manner (Fig. 8, G and H). Consistent with our finding of elevated mTORC1 activity in TRAF3IP3-deficient T reg cells, in response to TCR/CD28 stimulation, the inducible phosphorylation of S6K1 was enhanced at the lysosomes of TRAF3IP3-deficient T reg cells (Fig. 8 H).
Figure 8.
TRAF3IP3 facilitates lysosomal localization of PP2Ac in T reg cells. (A) Confocal microscopy analysis of TRAF3IP3, LAMP1 (a lysosomal marker), Golgin-97 (a Golgi marker), and DAPI (nuclear staining) in wild-type T reg cells stimulated for 4 h with antibodies against CD3 and CD28. Percent of fluorescent signal overlap of the indicated molecules is shown. (B) IB analysis of the indicated proteins in the plasma membrane and nonplasma membrane fractions of wild-type T reg cells stimulated as indicated. LAT, a plasma membrane marker. (C) Confocal microscopy analysis of PP2Ac, LAMP1, and DAPI in Traf3ip3+/+Foxp3-Cre or Traf3ip3fl/flFoxp3-Cre T reg cells stimulated for 4 h with antibodies against CD3 and CD28. (D) IB analysis of the indicated proteins in lysosomal or nonlysosomal extracts of Traf3ip3+/+Foxp3-Cre and Traf3ip3fl/flFoxp3-Cre T reg cells stimulated as indicated. (E) Confocal microscopy analysis of MEK, Golgin-97, and DAPI in Traf3ip3+/+Foxp3-Cre or Traf3ip3fl/flFoxp3-Cre T reg cells stimulated for 4 h with antibodies against CD3 and CD28. (F) IB analysis of the indicated proteins in Golgi or non-Golgi extracts of Traf3ip3+/+Foxp3-Cre and Traf3ip3fl/flFoxp3-Cre T reg cells stimulated as indicated. (G) Confocal microscopy analysis of Raptor, LAMP1, and DAPI in Traf3ip3+/+Foxp3-Cre or Traf3ip3fl/flFoxp3-Cre T reg cells stimulated for 4 h with antibodies against CD3 and CD28. (H) IB analysis of the indicated proteins in lysosomal or nonlysosomal extracts of Traf3ip3+/+Foxp3-Cre and Traf3ip3fl/flFoxp3-Cre T reg cells stimulated as indicated. Data are representative of three independent experiments and are presented as mean ± SEM. Bars, 5 µm. ns, not statistically significant; **, P < 0.01.
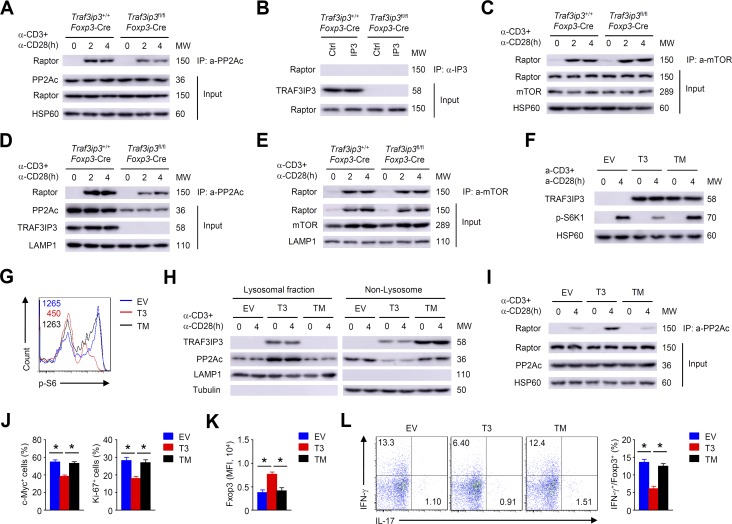
PP2A has been implicated in the regulation of mTORC1 activity through its association with Raptor in T reg cells (Apostolidis et al., 2016). Consistent with this finding, TCR- and CD28-stimulated binding of PP2Ac to Raptor was observed in wild-type T reg cells (Fig. 9 A). Indeed, the inducible association of PP2Ac with Raptor was reduced after TRAF3IP3 depletion (Fig. 9 A). However, the binding between TRAF3IP3 and Raptor in T reg cells was not detectable (Fig. 9 B). In addition, TCR- and CD28-stimulated Raptor–mTOR interaction was not altered after TRAF3IP3 depletion (Fig. 9 C). Importantly, the inducible binding of PP2Ac with Raptor in the lysosomal fraction was remarkably diminished in TRAF3IP3-deficient T reg cells (Fig. 9 D), whereas the inducible binding of mTOR with Raptor in the lysosomal fraction was not affected in TRAF3IP3-deficient T reg cells (Fig. 9 E). These data suggest that TRAF3IP3 facilitates the PP2Ac–Raptor interaction by recruiting PP2Ac to the lysosome in T reg cells.
Figure 9.
TRAF3IP3 mediates PP2A/mTORC1 axis at the lysosome of T reg cells. (A–C) IB and co-IP assays using whole cell lysates from Traf3ip3+/+Foxp3-Cre and Traf3ip3fl/flFoxp3-Cre T reg cells stimulated as indicated. (B) T reg cells stimulated for 4 h were subjected to IP using an antibody against TRAF3IP3 (IP3) or control Ig (Ctrl). (D and E) IB and co-IP assays using lysosomal membrane fractions from Traf3ip3+/+Foxp3-Cre and Traf3ip3fl/flFoxp3-Cre T reg cells stimulated as indicated. (F–L) In vitro activated naive TRAF3IP3-deficient CD4+ T cells were transduced with indicated retroviruses under T reg cell polarizing conditions. (F) CFP-positive cells were subsequently stimulated for IB analysis. EV, pCLXSN-CFP; T3, pCLXSN-TRAF3IP3-CFP; TM, pCLXSN mutant TRAF3IP3 lacking the TM-CFP. (G) Flow cytometric analysis of p-S6 expression in CFP-positive cells stimulated for 4 h. (H) IB analysis of the indicated proteins of CFP-positive cells stimulated as indicated. (I) IB and co-IP assays using whole cell lysates from CFP-positive cells stimulated as indicated. (J) Flow cytometric analysis of c-Myc or Ki-67 in CFP-positive cells stimulated for 8 or 24 h. (K and L) Flow cytometric analysis of Foxp3 (K), IFN-γ and IL-17 expression (L) in CFP-positive cells stimulated for 24 h. Data are representative of at least three independent experiments and are presented as mean ± SEM; *, P < 0.05.
To examine the role of the transmembrane domain (TM) of TRAF3IP3 in mTORC1 activity and T reg cell stability, we transduced naive TRAF3IP3-deficient CD4+ T cells with wild-type TRAF3IP3 (T3) or a mutant lacking the TM and subsequently cultured these cells under T reg cell polarizing conditions. Although TRAF3IP3 and its mutant were comparably expressed in the differentiated T reg cells, expression of wild-type TRAF3IP3, but not the mutant, suppressed mTORC1 activity (Fig. 9, F and G). Importantly, wild-type TRAF3IP3, but not the mutant, localized to the lysosome and recruited PP2Ac (Fig. 9 H). As expected, expression of wild-type TRAF3IP3, but not the mutant, increased the PP2Ac–Raptor interaction in TRAF3IP3-deficient T reg cells (Fig. 9 I). Furthermore, wild-type TRAF3IP3–expressing T cells, but not mutant-expressing T cells, displayed a substantial decrease in c-Myc and Ki-67 expression (Fig. 9 J) and showed much higher Foxp3 (Fig. 9 K), but lower IFN-γ expression (Fig. 9 L). Thus, the TM of TRAF3IP3 is required for its lysosomal localization and its function in mediating the lysosomal recruitment of PP2A to inhibit mTORC1 signaling in T reg cells.
Discussion
We identified TRAF3IP3 as a metabolic regulator of T reg cell stability and function. The genetic ablation of TRAF3IP3 in T reg cells greatly impaired their suppressive functions, resulting in the development of inflammatory disorders and stronger antitumor T cell responses. TRAF3IP3 deletion up-regulated the metabolic regulator c-Myc and promoted glycolytic metabolism in an mTORC1-dependent manner, leading to T reg cell instability. Mechanistically, TRAF3IP3 mediated the lysosomal recruitment of PP2Ac, and the resulting PP2Ac–Raptor interaction in turn restricted mTORC1 activity to maintain T reg cell stability and function.
In our study, TRAF3IP3 was observed to localize to the lysosome and Golgi, but not the plasma membrane, indicating that TRAF3IP3 is a transmembrane protein mainly localizing to the intracellular organelles in T reg cells. These findings suggest that TRAF3IP3 is unlikely to be involved in the regulation of T reg cell immunological synapses, explaining why the TRAF3IP3 deficiency did not have a global effect on T reg cell signaling. A compartmentalized BRAF–MEK–ERK signaling mechanism is regulated by TRAF3IP3 in thymocytes (Zou et al., 2015); however, TRAF3IP3 deletion did not affect the Golgi localization of MEK or the activation of MEK–ERK signaling in T reg cells. We found that TRAF3IP3 facilitated the lysosomal localization of PP2Ac to regulate mTORC1 signaling in T reg cells. These findings suggested that the function of TRAF3IP3 as a scaffold protein to regulate TCR downstream signaling may be cell type dependent.
Our results demonstrated an important role for TRAF3IP3 in mediating the metabolic control of T reg cells. In the absence of TRAF3IP3, T reg cells exhibited altered metabolic and cytokine profiles and lost the expression of Foxp3 and their suppressive functions. In resting T reg cells, mTORC1 was inactivated even in the absence of TRAF3IP3; however, the TCR and CD28 signals stimulated the activation of mTORC1 to drive T reg cell glycolysis. Therefore, a balance of mTORC1 signaling in resting and activating T reg cells might exist, but how it is regulated remains to be studied. In fact, TRAF3IP3 deficiency resulted in excessive mTORC1 activity, which contributed to the enhanced glycolytic metabolism in activating T reg cells. Pharmacological blockade of mTORC1 greatly diminished the glycolytic rates and restored the impaired stability of TRAF3IP3-deficient T reg cells. Overexpression of TRAF3IP3 rescued the defect in T reg cell stability by restraining mTORC1 activity. Our current findings establish TRAF3IP3 as a metabolic regulator in the control of mTORC1-dependent glycolysis to maintain T reg cell stability and function.
TCR-induced mTORC1 localization to the lysosome is important for mTORC1-dependent glycolytic metabolism during CD8+ T cell activation (Pollizzi et al., 2016), but whether the lysosome translocation of mTORC1 is involved in T reg cell identity remains unclear. We found that Raptor and S6K1 rapidly localized to the lysosome in response to TCR and CD28 stimulation, although how they are recruited to the lysosome in T reg cells remains to be determined. Importantly, the inducible phosphorylation of S6K1 was observed at the lysosome upon TCR and CD28 stimulation, suggesting the lysosome-specific activation of mTORC1 in T reg cells. Notably, TRAF3IP3-deficient T reg cells exhibited elevated mTORC1 activity at the lysosome, which was responsible for the hyper-glycolytic metabolism and T reg cell instability. Our findings provide an example for how mTORC1 signaling regulates glycolytic metabolism to maintain T reg cell stability and function in a specific subcellular compartment.
PP2A has been implicated in regulating mTOR pathway activation (Trotman et al., 2006; Liu et al., 2011). The potassium-mediated suppression of both mTORC1 and mTORC2 in effector T cells is dependent on the activity of PP2A (Eil et al., 2016). PP2A is also required for T reg cell function via the control of mTORC1 activity (Apostolidis et al., 2016). However, how PP2A specifically regulates the activation of mTORC1 but not mTORC2 in T reg cells is unclear. We obtained strong evidence that TRAF3IP3 localizes to the lysosome and mediates the lysosomal recruitment of PP2Ac. Although Raptor was transported to the lysosome upon TCR and CD28 stimulation in a TRAF3IP3-independent manner, decreased PP2Ac binding with Raptor at the lysosome, as well as increased mTORC1 activity, were observed in TRAF3IP3-deficient T reg cells. Importantly, the ability of TRAF3IP3 to mediate the lysosomal recruitment of PP2Ac and the PP2Ac–Raptor interaction relied on its TM. Thus, TRAF3IP3 facilitates the interaction of PP2Ac with Raptor at the lysosome, thereby allowing PP2A to inhibit mTORC1. On the other hand, TRAF3IP3 ablation in T reg cells had no effect on the mTORC2-dependent phosphorylation of AKT at Ser473. Lysosomal TRAF3IP3 is likely to be dispensable for mTORC2 activation, because mTORC2 principally localizes to the mitochondria, ER, and MAM (Betz and Hall, 2013). We concluded that PP2A activation in T reg cells primarily targeted mTORC1 signaling in a TRAF3IP3-dependent manner.
Collectively, these data indicate that TRAF3IP3 acts as a negative regulator of mTORC1 and glycolytic metabolism to maintain metabolic fitness in T reg cells. Our results highlight a lysosome-specific function of TRAF3IP3 in the regulation of mTORC1-mediated glycolysis in T reg cells. These findings also suggest that combining tumor immunotherapy with strategies that inhibit TRAF3IP3 could improve clinical efficacy. The identification of TRAF3IP3 as a metabolic regulator in T reg cells may also provide opportunities for therapeutic interventions in autoimmune diseases.
Materials and methods
Mice
Traf3ip3-floxed mice (in C57BL/6 background) were generated at Knockout Mouse Project (KOMP) using a LoxP targeting system (Zou et al., 2015). The Traf3ip3-floxed mice were crossed with Foxp3-YFP-Cre transgenic mice (The Jackson Laboratory) in C57BL/6 background to produce age-matched Traf3ip3+/+Foxp3-Cre and Traf3ip3fl/flFoxp3-Cre mice for experiments. The Traf3ip3-floxed mice were also crossed with Cd4-Cre transgenic mice (The Jackson Laboratory) in C57BL/6 background to produce age-matched Traf3ip3+/+Cd4-Cre and Traf3ip3fl/flCd4-Cre mice to prepare the naive TRAF3IP3-deficient CD4+ T cells. B6.SJL mice, Rag1-KO mice and OT-I TCR-transgenic mice in C57BL/6 background were from The Jackson Laboratory. Mice were maintained in a specific pathogen–free facility, and all animal experiments were in accordance with protocols approved by the Institutional Animal Care and Use Committee of Shanghai Jiao Tong University, School of Medicine.
Plasmids, antibodies, and reagents
Mouse wild-type TRAF3IP3 (T3) or mutant TRAF3IP3 lacking the TM were cloned into the retroviral vector pCLXSN-CFP. Flag-tagged mouse PP2Ac and HA-tagged mouse TRAF3IP3 were cloned into the pcDNA3-HA vector. Antibodies for HSP60 (H1), Tubulin (TU-02), and TRAF3IP3 (B-7) were from Santa Cruz Biotechnology. Antibodies for p-S6K, p-S6, p-AKT(T308), p-AKT(S473), MEK1/2, p-MEK1/2, p-p38, p-JNK, p-p65, p-ERK, HIF1-α, Golgin-97, LAT, mTOR, c-Myc, PP2Aa, PP2Ab, PP2Ac, and Raptor were purchased from Cell Signaling Technology. Antibodies for LAMP1 and GLUT1 were purchased from Abcam. Anti-PP2Ac (1D6) antibody for confocal analysis was from EMD Millipore. HRP-conjugated anti-HA antibody (3F10) was from Roche. Anti-FLAG (M2) antibody was from Sigma-Aldrich. Anti-mouse-PD-1 (clone J43) and isotype control IgG (hamster IgG) antibodies were from Bio X Cell. The fluorochrome-conjugated antibodies for CD4 (GK1.5), CD8 (53–6.7), CD44 (IM7), CD62L (MEL-14), Ki-67 (SolA15), Foxp3 (FJK-16s), IFN-γ (XMG1.2), IL-17 (eBio17B7), CD25 (CD25-4E3), GITR (eBioAITR), PD-1 (MIH4), CTLA-4 (14D3), and 2-(N-(7-Nitrobenz-2-oxa-1,3-diazol-4-yl)Amino)-2-Deoxyglucose (2-NBDG) were purchased from Thermo Fisher Scientific. Phospho-p44/42 MAPK (Erk1/2; Thr202/Tyr204; E10) Mouse mAb (Alexa Fluor 647 Conjugate), c-Myc (D84C12) Rabbit mAb (PE Conjugate), and phospho-S6 Ribosomal Protein (Ser235/236; D57.2.2E) XP Rabbit mAb (APC Conjugate) were purchased from Cell Signaling Technology. Rapamycin (R0395), okadaic acid (O9381; used at 10 nM), dichloroacetic acid (D54702), and sphingomyelinase (SMase; used at 0.5 units/ml) from Staphylococcus aureus (S8633) were purchased from Sigma-Aldrich.
Histology
Organs were removed from age- and sex-matched 8-mo-old Traf3ip3+/+Foxp3-Cre and Traf3ip3fl/flFoxp3-Cre mice or RAG-1–deficient mice given adoptive transfer of wild-type naive CD45RBhi CD4+ T cells together with sorted T reg cells, fixed in 10% neutral buffered formalin, embedded in paraffin, and sectioned for staining with hematoxylin and eosin.
Flow cytometry
Cells were stained in PBS containing 2% FBS with indicated fluorochrome-conjugated antibodies for surface marker analysis. To determine intracellular cytokine expression, cells were stimulated with phorbol 12-myristate 13-acetate, ionomycin, and monensin for 5 h. At the end of stimulation, cells were stained with the indicated fluorochrome-conjugated antibodies according to the manufacturer’s instructions (Thermo Fisher Scientific). Foxp3 staining was performed according to the manufacturer’s instructions (Thermo Fisher Scientific). For intracellular staining of p-S6 and p-ERK, stimulated T reg cells were fixed in Fix Buffer I (BD) for 10 min, followed by incubation in cold Perm Buffer III (BD) for 20 min, and then subjected to antibody staining and flow cytometric analysis.
T reg cell isolation and stimulation
T reg cells (CD4+CD25+YFP+) were isolated from the spleen of female age-matched Traf3ip3+/+Foxp3-Cre and Traf3ip3fl/flFoxp3-Cre mice (6–8 wk old). The purified T reg cells were stimulated with plate-bound anti-CD3 (1 µg/ml) and anti-CD28 (1 µg/ml) in replicate wells in 6-well plates (5 × 106 cells per well) for flow cytometric, IB, or RNA-sequencing analysis. In vitro suppressive activity of T reg cells, assessed as the proliferation of naive CD4+ T cells activated by anti-CD3 and anti-CD28 antibodies for 3 d in the presence of various ratios of T reg cells sorted from 8-wk-old Traf3ip3+/+Foxp3-Cre mice and Traf3ip3fl/flFoxp3-Cre mice, measured as dilution of the cytosolic dye CFSE. Naive CD4+ T cells were purified by flow cytometric cell sorting based on CD4+CD44loCD62Lhi markers. Naive CD4+ T cells were stimulated with anti-CD3 (5 µg/ml) plus anti-CD28 (1 µg/ml) under TH1 (5 µg/ml anti-IL4, 10 ng/ml IL-12), TH2 (5 µg/ml anti-IFN-γ, 20 ng/ml IL-4), TH17 (5 µg/ml anti-IL4, 5 µg/ml anti-IFN-γ, 15 ng/ml IL-6, 2.5 ng/ml TGF-β), and T reg (5 µg/ml anti-IL4, 5 µg/ml anti-IFN-γ, 5 ng/ml TGF-β) conditions (Zou et al., 2014).
Adoptive transfer of T cells
A total of 4 × 105 T eff cells (CD4+CD45RBhiCD25–) from B6.SJL (CD45.1+) congenic mice were mixed with 2 × 105 wild-type or TRAF3IP3-deficient T reg cells (8-wk-old Traf3ip3+/+Foxp3-Cre or Traf3ip3fl/flFoxp3-Cre mice) and were transferred i.p. into RAG-1–deficient mice. Mice were assessed for clinical signs of colitis weekly and were analyzed 12 wk after transfer.
Tumor models
B16-F10 melanoma cells and MC38 colon cancer cells were cultured in RPMI 1640 supplemented with 10% FBS. These tumor cells were injected s.c. into 8-wk-old Traf3ip3+/+Foxp3-Cre and Traf3ip3fl/flFoxp3-Cre mice (2×105 cells per mouse). The challenged mice were monitored for tumor growth, and the tumor size was expressed as tumor area. To minimize individual variations, 8 or 10 age- and sex-matched mice each group were used.
RNA-sequencing analysis
Fresh splenic T reg cells (CD4+CD25+YFP+) were isolated from 8-wk-old Traf3ip3+/+Foxp3-Cre and Traf3ip3fl/flFoxp3-Cre mice and stimulated with anti-CD3 and anti-CD28 for 24 h. Activated T reg cells were used for total RNA isolation with TRIzol (Invitrogen) and subjected to RNA-sequencing using Illumina Nextseq500 (75-bp paired end reads). The raw reads were aligned to the mouse reference genome (version mm10), using Tophat2 RNASeq alignment software. The mapping rate was 96% overall across all the samples in the dataset. The raw reads were processed using Hisat and HTSeq to generate read counts for every gene. The read counts were then normalized using R package DESeq2. The normalized read counts were then centered and scaled for each gene, generating z-scores. The z-scores basically represent how many standard deviations one particular count is from the average count for that gene. P values obtained from multiple tests were adjusted using Benjamini-Hochberg correction. Significant differentially expressed genes are defined by a Benjamini-Hochberg corrected P value cutoff of 0.05 and fold-change of ≥1. The RNA sequencing data reported in this paper are available under accession no. SRP131823 (NCBI Trace and Short-Read Archive).
BM chimera
We adoptively transferred lethally irradiated (950 rad) Rag1−/− mice (6–8 wk old) with the mixed BMs from Traf3ip3fl/flFoxp3-Cre (CD45.1−CD45.2+) mice and Traf3ip3+/+Foxp3-Cre (CD45.1+CD45.2+) mice. Splenic CD45.1−CD45.2+ and CD45.1+CD45.2+ T reg cells (CD4+CD25+YFP+) from Rag1−/− mice (8 wk after reconstitution) were isolated for other experiments.
Glycolytic and mitochondrial respiration rate measurement
Seahorse XFe96 Extracellular Flux Analyzer (Agilent Technologies) was used for metabolic experiments. Isolated T reg cells were stimulated for 24 h with anti-CD3 and anti-CD28. Stimulated T reg cells were seeded at a density of 3×105 per well. The extracellular acidification rate (ECAR) and the oxygen consumption rate (OCR) for each well were calculated, while the cells were subjected to the XF Cell Mito or the XF Glycolytic stress test using the following concentrations of injected compounds: 10 mM glucose, 2 µM oligomycin, 50 mM 2-DG, 1 µM FCCP, 0.5 µM rotenone/antimycin A. The XF Cell Mito and the XF Glycolytic stress test kits were purchased from Agilent Technologies.
IB and immunoprecipitation (IP)
Cells were washed with ice-cold PBS and lysed on ice for 30 min in RIPA buffer (50 mM Tris-HCl, pH 7.5; 135 mM NaCl; 1% NP-40; 0.5% sodium DOC; 1 mM EDTA; 10% glycerol) containing protease inhibitor (1:100, P8340; Sigma-Aldrich), 1 mM NaF, and 1 mM PMSF. Cell lysates were cleared by centrifugation, and supernatants were immunoprecipitated with the appropriate antibodies using protein A/G-agarose beads. Samples were then used for IB analysis with indicated antibodies. Lysosomal fractions were prepared using the Lysosome Isolation kit (Sigma-Aldrich) for IB analysis. Golgi fraction was isolated using a Golgi isolation kit (Sigma-Aldrich). Plasma membrane fraction was prepared using a plasma membrane protein extraction kit (ab65400; Abcam).
Retroviral transduction
Packaged pCLXSN retrovirus was produced and used to infect activated T cells. Purified naive TRAF3IP3-deficient CD4+ T cells were activated with plate-bound anti-CD3 (1 µg/ml) plus anti-CD28 (1 µg/ml) in 48-well plates for 24 h and then infected with the retrovirus in the presence of 10 µg/ml polybrene by spinning at 900 g for 90 min. Transduced CD4+ T cells were cultured under T reg cell polarizing conditions for 3 d. CFP-positive T cells were used for IB analysis and flow cytometric analysis.
Confocal microscopy
T reg cells were isolated from Traf3ip3+/+Foxp3-Cre and Traf3ip3fl/flFoxp3-Cre mice and stimulated with anti-CD3 and anti-CD28 for 24 h. Activated T reg cells were fixed and permeabilized for 20 min with Cytofix/Cytoperm solution (BD), blocked in 0.5% BSA in PBS, and then stained with primary antibody for 60 min, followed by staining with secondary antibodies for 30 min. Slides were mounted in antifade reagent with DAPI. Images were taken with a Leica SP8 confocal microscope and analyzed by Imaris (Bitplane).
Statistical analysis
Statistical analysis was performed using Prism software (GraphPad Prism version 6.01). Two-tailed unpaired Student’s t tests were performed, and data are presented as means ± SEM. One-way ANOVA, where applicable, was performed to determine whether an overall statistically significant change existed before the Student’s t test to analyze the difference between any two groups. P value <0.05 is considered statistically significant, and the level of significance was indicated as *, P < 0.05 and **, P < 0.01.
Online supplemental material
Fig. S1 shows the expressions of T reg cell–related genes in Traf3ip3+/+Foxp3-Cre and Traf3ip3fl/flFoxp3-Cre T reg cells. Fig. S2 shows the cytokine expression of gut T reg cells. Fig. S3 shows the expressions of metabolism related genes in Traf3ip3+/+Foxp3-Cre and Traf3ip3fl/flFoxp3-Cre T reg cells.
Supplementary Material
Acknowledgments
This study was supported by grants from the Recruitment Program of Global Experts of China, the National Natural Science Foundation of China (grant nos. 31670896, 31370904, 81671579), the National Key Research and Development Program (grant no. 2017YFA0104500), Shanghai Rising-Star Program (grant no. 16QA1403300), Shanghai Municipal Commission of Health and Family Planning (grant nos. 2018YQ08, 20174Y0049, and 20174Y0191), Shanghai Jiao Tong University “Program for young teachers” (grant no. KJ30214170006), Medical and Engineering Cross Research Foundation (grant no. YG2016QN77), and a grant from the U.S. National Institutes of Health (grant no. AI64639 to S.-C. Sun).
The authors declare no competing financial interests.
Author contributions: X. Yu, X. Teng, F. Wang, and Y. Zheng designed and performed the experiments, prepared the figures, and wrote part of the manuscript; G. Qu, Y. Zhou, Z. Hu, Z. Wu, Y. Chang, and L. Chen contributed to part of the experiments; H.-B. Li and B. Su contributed critical comments; L. Lu, Z. Liu, S.-C. Sun, and Q. Zou supervised the work and wrote the manuscript.
References
- Alon R. 2017. A Sweet Solution: Glycolysis-Dependent Treg Cell Migration. Immunity. 47:805–807. 10.1016/j.immuni.2017.11.006 [DOI] [PubMed] [Google Scholar]
- Apostolidis S.A., Rodríguez-Rodríguez N., Suárez-Fueyo A., Dioufa N., Ozcan E., Crispín J.C., Tsokos M.G., and Tsokos G.C.. 2016. Phosphatase PP2A is requisite for the function of regulatory T cells. Nat. Immunol. 17:556–564. 10.1038/ni.3390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arce Vargas F., Furness A.J.S., Solomon I., Joshi K., Mekkaoui L., Lesko M.H., Miranda Rota E., Dahan R., Georgiou A., Sledzinska A., et al. Lung TRACERx Consortium . 2017. Fc-Optimized Anti-CD25 Depletes Tumor-Infiltrating Regulatory T Cells and Synergizes with PD-1 Blockade to Eradicate Established Tumors. Immunity. 46:577–586. 10.1016/j.immuni.2017.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer C.A., Kim E.Y., Marangoni F., Carrizosa E., Claudio N.M., and Mempel T.R.. 2014. Dynamic Treg interactions with intratumoral APCs promote local CTL dysfunction. J. Clin. Invest. 124:2425–2440. 10.1172/JCI66375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betz C., and Hall M.N.. 2013. Where is mTOR and what is it doing there? J. Cell Biol. 203:563–574. 10.1083/jcb.201306041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J.H., Xiao Y., Hu H., Jin J., Yu J., Zhou X., Wu X., Johnson H.M., Akira S., Pasparakis M., et al. 2012. Ubc13 maintains the suppressive function of regulatory T cells and prevents their conversion into effector-like T cells. Nat. Immunol. 13:481–490. 10.1038/ni.2267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi H. 2012. Regulation and function of mTOR signalling in T cell fate decisions. Nat. Rev. Immunol. 12:325–338. 10.1038/nri3198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coe D.J., Kishore M., and Marelli-Berg F.. 2014. Metabolic regulation of regulatory T cell development and function. Front. Immunol. 5:590 10.3389/fimmu.2014.00590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrowsky R.T., Kamibayashi C., Mumby M.C., and Hannun Y.A.. 1993. Ceramide activates heterotrimeric protein phosphatase 2A. J. Biol. Chem. 268:15523–15530. [PubMed] [Google Scholar]
- Eil R., Vodnala S.K., Clever D., Klebanoff C.A., Sukumar M., Pan J.H., Palmer D.C., Gros A., Yamamoto T.N., Patel S.J., et al. 2016. Ionic immune suppression within the tumour microenvironment limits T cell effector function. Nature. 537:539–543. 10.1038/nature19364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essig K., Hu D., Guimaraes J.C., Alterauge D., Edelmann S., Raj T., Kranich J., Behrens G., Heiseke A., Floess S., et al. 2017. Roquin Suppresses the PI3K-mTOR Signaling Pathway to Inhibit T Helper Cell Differentiation and Conversion of Treg to Tfr Cells. Immunity. 47:1067–1082.e12. 10.1016/j.immuni.2017.11.008 [DOI] [PubMed] [Google Scholar]
- Feng Y., Arvey A., Chinen T., van der Veeken J., Gasteiger G., and Rudensky A.Y.. 2014. Control of the inheritance of regulatory T cell identity by a cis element in the Foxp3 locus. Cell. 158:749–763. 10.1016/j.cell.2014.07.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerriets V.A., Kishton R.J., Nichols A.G., Macintyre A.N., Inoue M., Ilkayeva O., Winter P.S., Liu X., Priyadharshini B., Slawinska M.E., et al. 2015. Metabolic programming and PDHK1 control CD4+ T cell subsets and inflammation. J. Clin. Invest. 125:194–207. 10.1172/JCI76012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerriets V.A., Kishton R.J., Johnson M.O., Cohen S., Siska P.J., Nichols A.G., Warmoes M.O., de Cubas A.A., MacIver N.J., Locasale J.W., et al. 2016. Foxp3 and Toll-like receptor signaling balance Treg cell anabolic metabolism for suppression. Nat. Immunol. 17:1459–1466. 10.1038/ni.3577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgi C., Ito K., Lin H.K., Santangelo C., Wieckowski M.R., Lebiedzinska M., Bononi A., Bonora M., Duszynski J., Bernardi R., et al. 2010. PML regulates apoptosis at endoplasmic reticulum by modulating calcium release. Science. 330:1247–1251. 10.1126/science.1189157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudreault M., D’Ambrosio L.M., Kean M.J., Mullin M.J., Larsen B.G., Sanchez A., Chaudhry S., Chen G.I., Sicheri F., Nesvizhskii A.I., et al. 2009. A PP2A phosphatase high density interaction network identifies a novel striatin-interacting phosphatase and kinase complex linked to the cerebral cavernous malformation 3 (CCM3) protein. Mol. Cell. Proteomics. 8:157–171. 10.1074/mcp.M800266-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttlin E.L., Bruckner R.J., Paulo J.A., Cannon J.R., Ting L., Baltier K., Colby G., Gebreab F., Gygi M.P., Parzen H., et al. 2017. Architecture of the human interactome defines protein communities and disease networks. Nature. 545:505–509. 10.1038/nature22366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh A., DuPage M., Priyadharshini B., Sage P.T., Quiros J., Borges C.M., Townamchai N., Gerriets V.A., Rathmell J.C., Sharpe A.H., et al. 2015. Control of PI(3) kinase in Treg cells maintains homeostasis and lineage stability. Nat. Immunol. 16:188–196. 10.1038/ni.3077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewell J.L., Russell R.C., and Guan K.L.. 2013. Amino acid signalling upstream of mTOR. Nat. Rev. Mol. Cell Biol. 14:133–139. 10.1038/nrm3522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josefowicz S.Z., Lu L.F., and Rudensky A.Y.. 2012. Regulatory T cells: mechanisms of differentiation and function. Annu. Rev. Immunol. 30:531–564. 10.1146/annurev.immunol.25.022106.141623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E., Goraksha-Hicks P., Li L., Neufeld T.P., and Guan K.L.. 2008. Regulation of TORC1 by Rag GTPases in nutrient response. Nat. Cell Biol. 10:935–945. 10.1038/ncb1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishore M., Cheung K.C.P., Fu H., Bonacina F., Wang G., Coe D., Ward E.J., Colamatteo A., Jangani M., Baragetti A., et al. 2017. Regulatory T Cell Migration Is Dependent on Glucokinase-Mediated Glycolysis. Immunity. 47:875–889.e10. 10.1016/j.immuni.2017.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Liang Y., LeBlanc M., Benner C., and Zheng Y.. 2014. Function of a Foxp3 cis-element in protecting regulatory T cell identity. Cell. 158:734–748. 10.1016/j.cell.2014.07.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu E., Knutzen C.A., Krauss S., Schweiger S., and Chiang G.G.. 2011. Control of mTORC1 signaling by the Opitz syndrome protein MID1. Proc. Natl. Acad. Sci. USA. 108:8680–8685. 10.1073/pnas.1100131108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik A.R., Urbanska M., Macias M., Skalecka A., and Jaworski J.. 2013. Beyond control of protein translation: what we have learned about the non-canonical regulation and function of mammalian target of rapamycin (mTOR). Biochim. Biophys. Acta. 1834:1434–1448. 10.1016/j.bbapap.2012.12.010 [DOI] [PubMed] [Google Scholar]
- Michalek R.D., Gerriets V.A., Jacobs S.R., Macintyre A.N., MacIver N.J., Mason E.F., Sullivan S.A., Nichols A.G., and Rathmell J.C.. 2011. Cutting edge: distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. J. Immunol. 186:3299–3303. 10.4049/jimmunol.1003613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto S., Murphy A.N., and Brown J.H.. 2008. Akt mediates mitochondrial protection in cardiomyocytes through phosphorylation of mitochondrial hexokinase-II. Cell Death Differ. 15:521–529. 10.1038/sj.cdd.4402285 [DOI] [PubMed] [Google Scholar]
- Newton R., Priyadharshini B., and Turka L.A.. 2016. Immunometabolism of regulatory T cells. Nat. Immunol. 17:618–625. 10.1038/ni.3466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollizzi K.N., Sun I.H., Patel C.H., Lo Y.C., Oh M.H., Waickman A.T., Tam A.J., Blosser R.L., Wen J., Delgoffe G.M., and Powell J.D.. 2016. Asymmetric inheritance of mTORC1 kinase activity during division dictates CD8(+) T cell differentiation. Nat. Immunol. 17:704–711. 10.1038/ni.3438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi S., Yamaguchi T., Nomura T., and Ono M.. 2008. Regulatory T cells and immune tolerance. Cell. 133:775–787. 10.1016/j.cell.2008.05.009 [DOI] [PubMed] [Google Scholar]
- Sancak Y., Bar-Peled L., Zoncu R., Markhard A.L., Nada S., and Sabatini D.M.. 2010. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 141:290–303. 10.1016/j.cell.2010.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L.Z., Wang R., Huang G., Vogel P., Neale G., Green D.R., and Chi H.. 2011. HIF1alpha-dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells. J. Exp. Med. 208:1367–1376. 10.1084/jem.20110278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha S., Yang K., Guy C., Vogel P., Neale G., and Chi H.. 2015. Treg cells require the phosphatase PTEN to restrain TH1 and TFH cell responses. Nat. Immunol. 16:178–187. 10.1038/ni.3076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotman L.C., Alimonti A., Scaglioni P.P., Koutcher J.A., Cordon-Cardo C., and Pandolfi P.P.. 2006. Identification of a tumour suppressor network opposing nuclear Akt function. Nature. 441:523–527. 10.1038/nature04809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Dillon C.P., Shi L.Z., Milasta S., Carter R., Finkelstein D., McCormick L.L., Fitzgerald P., Chi H., Munger J., and Green D.R.. 2011. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity. 35:871–882. 10.1016/j.immuni.2011.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J., Long L., Yang K., Guy C., Shrestha S., Chen Z., Wu C., Vogel P., Neale G., Green D.R., and Chi H.. 2016. Autophagy enforces functional integrity of regulatory T cells by coupling environmental cues and metabolic homeostasis. Nat. Immunol. 17:277–285. 10.1038/ni.3365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Huang Q., Wang H., Hao Y., Bai Q., Hu J., Li Y., Wang P., Chen X., He R., et al. 2017. The Kinase mTORC1 Promotes the Generation and Suppressive Function of Follicular Regulatory T Cells. Immunity. 47:538–551.e5. 10.1016/j.immuni.2017.08.011 [DOI] [PubMed] [Google Scholar]
- Zeng H., and Chi H.. 2017. mTOR signaling in the differentiation and function of regulatory and effector T cells. Curr. Opin. Immunol. 46:103–111. 10.1016/j.coi.2017.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H., Yang K., Cloer C., Neale G., Vogel P., and Chi H.. 2013. mTORC1 couples immune signals and metabolic programming to establish T(reg)-cell function. Nature. 499:485–490. 10.1038/nature12297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Q., Jin J., Hu H., Li H.S., Romano S., Xiao Y., Nakaya M., Zhou X., Cheng X., Yang P., et al. 2014. USP15 stabilizes MDM2 to mediate cancer-cell survival and inhibit antitumor T cell responses. Nat. Immunol. 15:562–570. 10.1038/ni.2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Q., Jin J., Xiao Y., Hu H., Zhou X., Jie Z., Xie X., Li J.Y., Cheng X., and Sun S.C.. 2015. T cell development involves TRAF3IP3-mediated ERK signaling in the Golgi. J. Exp. Med. 212:1323–1336. 10.1084/jem.20150110 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.