Abstract
Prenatal genetics has evolved over the last decade to include application of new ‘omics technologies to improve perinatal care. The clinical utility of these technologies when applied to direct fetal specimens from amniocentesis or chorionic villus sampling is being explored. In this review, we provide an overview of use of prenatal exome sequencing and role in evaluation of the structurally abnormal fetus, potential applications of genome sequencing, and finally, use of transcriptomics to assess placental and fetal well-being.
Keywords: Prenatal, Genomics, Exome, Transcriptome, Fetus
INTRODUCTION
The recent explosion of genomics into perinatal medicine has revolutionized prenatal diagnosis and fetal medicine. All women who carry a fetus with a structural birth defect are offered diagnostic testing with either amniocentesis or chorionic villus sampling. Previously, women undergoing diagnostic testing were limited to receiving results of G-banded karyotype only. In 2012, Wapner et al. showed increased diagnostic rate of 6% with use of chromosomal microarray (CMA) over standard karyotype in the setting of fetal abnormalities.1 This study changed clinical practice for prenatal diagnosis as all women are now offered CMA when a fetal abnormality is diagnosed. Attention has now turned to prenatal exome sequencing (ES; sequencing of the protein-coding regions of the genome) to determine utility in cases where CMA does not provide a diagnosis. Alterations in the exome cause the majority of Mendelian or single-gene disorders. Prenatal ES is now being offered in select cases on a research or, less commonly, on a clinical basis. genome sequencing, while on the horizon, has not yet been applied clinically because of the current difficulty in interpreting intronic regions of the genome. Finally, use of RNA-sequencing to interrogate the whole transcriptome promises to provide new insights into human development across gestation and new opportunities to monitor placental function noninvasively through maternal plasma cell-free RNA (cfRNA).
EXOME SEQUENCING: APPLICATION TO PRENATAL CARE OF ANOMALOUS FETUSES
Congenital abnormalities affect 2–4% of all infants and are responsible for 20.4% of perinatal deaths.2 Currently, prenatal diagnosis is limited to ultrasound followed by standard karyotype and microarray performed on amniocytes or chorionic villi.3 While microarray analysis increases diagnostic ability above standard karyotype, 70–80% (refs. 4,5) of anomalous fetuses with a normal karyotype also have a normal microarray and thus remain without a definitive diagnosis. (ES) is now being performed in routine clinical care of adults and pediatric patients with dysmorphic features and developmental delay showing a diagnostic yield of approximately 30%. Prenatally, ES has only recently been applied to direct fetal specimens from amniocentesis and chorionic villus sampling. Prenatal ES has the ability to identify a molecular diagnosis in select cases where standard genetic testing (karyotype, microarray, and targeted molecular panels) fail to reveal a diagnosis. However, despite the technical feasibility of genome and exome sequencing on prenatal samples, its performance still requires an invasive procedure with attendant risk of miscarriage. Moreover, there remain huge interpretive challenges. In fact, when analyzing the protein-coding regions of the genome, only 28% of genes intolerant to loss of function have a known function related to a human disease phenotype with even fewer having a prenatal phenotype.6 Many genes critical to human development have yet to be elucidated and the cause of birth defects and recurrent losses is largely unknown, leaving many families with uncertainty regarding recurrence risk in future pregnancies.7
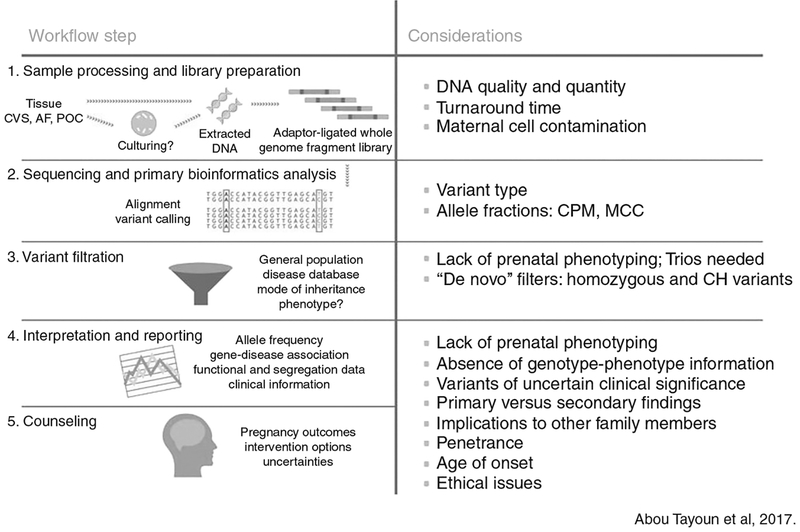
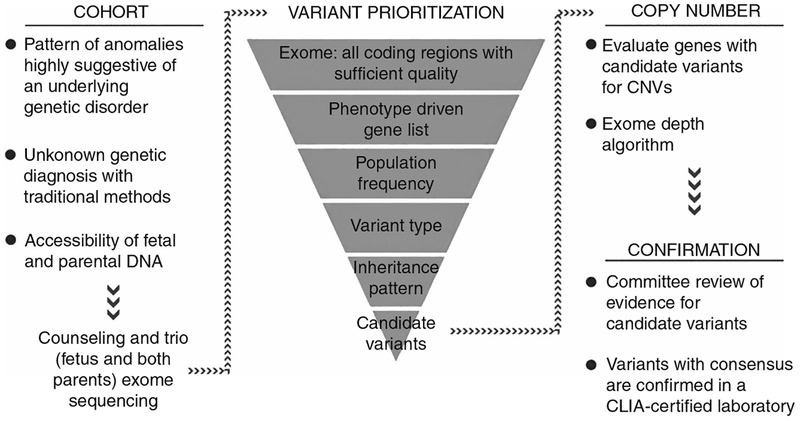
Several case series have been published showing the utility of ES in making a molecular diagnosis with a wide range reported (6.2–57.1%) dependent on a priori risk) (Table 1) (refs. 8–15). Because of selection bias, prospective studies are required to determine whether such a high yield will be observed in a larger, consecutive cohort. Prospective data has recently been presented in abstract form on 406 fetuses with various fetal structural anomalies (with normal karyotype and CMA results) in a UK cohort where trio ES was performed to identify diagnostic variants deemed to be causative of the prenatal phenotype.7,16 These data demonstrate an overall diagnostic yield of 6.2%. Similarly, in a prospective US cohort with various structural abnormalities, 7.7% (13/168) received a diagnosis with ES. Pathogenic/likely pathogenic variants were more common in fetuses with multisystem anomalies in both cohorts (16% in the UK cohort and 15.9% in the US cohort). Detection of diagnostic variants was less common in fetuses with all other types of isolated anomalies, including brain malformations. As more information becomes available on diagnostic capability of prenatal ES, strategies to incorporate this technology into clinical practice will be needed. The biggest challenges relate to variant interpretation and pre- and posttest counseling. Figs. 1 and 2 provides an overview of the process of ES, interpretation, and considerations specific to prenatal sequencing (Fig. 1, adapted from Abou Tayoun et al.17).
Table 1.
Summary of fetal exome sequencing publications with >5 fetuses included
| First author | Number of cases | Cohort description | Proband vs. trio | Pathogenic variant | Likely pathogenic variant |
|---|---|---|---|---|---|
| Yang et al., 2014 | 11 | Terminated anomalous fetus | Trio | 6 of 11 (54%) | — |
| Carss et al., 2014 | 30 | Prenatal sonographic anomalies | Trio | 3 of 30 (10%) | 5 of 30 (16.7%) |
| Drury et al., 2015 | 24 | Prenatal sonographic anomalies including NT ≥ 3.5mm | 14 Proband10 Trio | 5 of 24 (20.8%) | 1 of 24 (4.2%) |
| Alamillo et al., 2015 | 7 | Multiples sonographic anomalies termination or demise | Trio | 3 of 7 (42.9%) | 1 of 7 (14.3%) |
| Pangalos et al., 2017 | 14 | Prenatal sonographic anomalies | Proband only | 6 of 14 (42.9%) | — |
| Yates et al., 2017 | 84 | Demise or termination | 33 Proband/duo51 Trio/quad | 17 of 84 (20%) | 38 of 84 (45%) |
| Vora et al., 2017 | 15 | Multiple sonographic anomalies | Trio | 7 of 15 (46.7%) | 1 of 15 (6.7%) |
| Fu et al., 2018 | 196 | Prenatal sonographic anomalies | 34 Proband13 Trio | 47 of 196 (24%) | 25 of 196 (12.8%) |
Fig. 1.
Considerations specific to prenatal sequencing. CVS chorionic villus sampling, AF Amniotic fluid, POC Products of coneption, CPM Confined placental mosaicism, MCC Maternal cell contamination, CH Compound heterozygous
Fig. 2.
Approach to selection of prenatal exome sequencing candidates and variant interpretation. CNV, copy-number variant
In select prenatal cases in which other approaches to diagnosis have been uninformative, it may be appropriate to offer ES. Examples of such cases include recurrent or multiple congenital anomalies where standard genetic testing with karyotype and microarray have been normal. Prenatal ES also has a role in cases in which a fetus has structural abnormalities with reported consanguinity or homozygosity indicating relatedness on microarray. If the presenting disorder is highly genetically heterogeneous, ES is also potentially more cost-effective than sequencing individual genes using a targeted molecular panel.18 The American College of Medical Genetics and Genomics (ACMG), Society for Maternal–Fetal Medicine (SMFM), and International Society for Prenatal Diagnosis (ISPD) do not recommend ES for routine use for prenatal diagnosis.19 If prenatal ES is performed, pre- and posttest counseling by a provider with expertise in genetics is recommended.20
Targeted next-generation sequencing panels for different disorders
An alternate approach to ES is to send a targeted molecular panel specifically if the structural abnormality has a well-characterized phenotype. There are many reports showing the success of this approach.21,22 A targeted panel in comparison with ES may also be more cost-effective, have a faster turnaround time, and be more accessible in routine clinical care. The disadvantage is that many prenatal phenotypes are not well characterized because identification of phenotype is limited in many cases by ultrasound phenotype alone.
USING ES FOR DISCOVERY OF NOVEL CANDIDATE GENES
The use of ES prenatally has generated interest in identifying genes critical to human development. Some authors have identified novel candidate genes via use of prenatal ES and point to the utility of using extreme lethal prenatal phenotypes for gene discovery.23 These authors argue that it is likely that genes with orthologous embryonic lethal models in animal models will be found when sequencing extreme prenatal phenotypes. To increase confidence in the causality of the genes, cross-species phenotyping using animal models and functional studies using in vivo and in vitro animal models are recommended to validate the role of the gene in embryonic development.
Examples of novel candidate gene discovery using prenatal sequencing include discovery of KIF as a cause of fetal hydrolethalus and acrocallosal syndromes, two multiple malformation disorders with overlapping features that include polydactyly, brain abnormalities, and cleft palate.24 Putoux et al. used a combination of homozygosity mapping and targeted sequencing to identify truncating variants in four affected fetuses of consanguineous parents combined with finding a truncating variant of the same gene in eight unrelated fetuses. The gene was then modeled in zebrafish and results in zebrafish were consistent with the author’s hypothesis that the gene is involved in ciliary function because they showed KIF affected the sonic hedgehog pathway. Thomas et al. used the same approach to identify TCTN3 as the cause of orofaciodigital (OFD) syndrome IV (Mohr–Majewski syndrome).25
Prenatal ES identified other novel candidate genes in extreme prenatal phenotypes, including the following: (1) short-rib polydactyly syndrome (WDR60) (ref. 26); (2) a syndrome presenting with intrauterine growth restriction, severe microcephaly, renal cystic dysplasia/agenesis, and complex brain and genitourinary malformations (KIF14) (ref. 27); and (3) extreme microcephaly (MKL2) (ref. 28), among others. As prenatal ES is performed in abnormal fetuses, it is likely that genes critical to human development comprising the “developmentalome” will be identified.
A major challenge is that there is currently no shared prenatal database with genotype and phenotype information. A shared database would enable researchers to search for similar phenotypes and increase the confidence in pursuing novel gene discovery functional studies if there are multiple families with genotype/phenotype correlations. Similar to postnatal databases such as ClinGen and Matchmaker, researchers performing prenatal sequencing are encouraged to deposit all data in a shared de-identified database. Currently, a database where prenatal genotype/phenotype can be deposited and shared publicly does not exist but funding agencies should consider the importance of such a resource to the scientific community and prioritize its creation.
EXPANSION OF PRENATAL PHENOTYPES
Prenatal ES will inevitably expand phenotypes not previously described in the prenatal period. For example, Vora et al.14 identified a fetal presentation of scalp ear nipple syndrome (KCTDN1) and more recently identified a prenatal presentation of cerebellar ataxia, mental retardation, and disequilibrium syndrome 2 caused by pathogenic variants in the WDR81 gene (unpublished data). The prenatal presentation included hypoplastic cerebellum, abnormal cisterna magna, Arnold–Chiari malformation, omphalocele, broad thumbs, cystic hygroma, and ascites. This specific syndrome, characterized by quadrupedal locomotion and severe intellectual disability, was first described in seven members of a Turkish family,29 and related syndromes were described as early as 1917 (refs. 30,31). Homozygosity mapping and targeted genomic sequencing originally identified the gene in a consanguineous kindred.32 Identification of these variants with prenatal ES represents a phenotype expansion of this disorder into the prenatal period. Of note, another prenatal case of this syndrome was recently described in which a fetus with hydrocephalus and holoprosencephaly was found to have a homozygous Gly282Gly WDR81 pathogenic variant by exome sequencing of a terminated pregnancy.33 These cases illustrate the power of prenatal ES to elucidate prenatal presentations of previously postnatally diagnosed syndromes. This knowledge will ultimately improve our understanding of the pathophysiology of the disorder and potentially improve treatments and outcomes.
CHALLENGES OF PRENATAL EXOME SEQUENCING
Multiple challenges related to prenatal ES include (1) limited prenatal phenotypic information, (2) difficulties with variant interpretation, (3) counseling dilemmas, (4) current high cost, and (5) lack of providers with genetics expertise to provide adequate counseling and informed consent. In this article, we will focus on the specific challenge of limited prenatal phenotypic information with regard to use of sequencing. Other challenges have been thoroughly discussed in a recent review.34
Phenotypic classification is usually solely based on the prenatal ultrasound and/or fetal magnetic resonance image (MRI) findings. Accurate genotype–phenotype correlation is fundamental to improved prenatal diagnosis, but fetal pheno-typing using ultrasonography alone is a limitation to the use of prenatal ES because the phenotype is often incompletely developed. Thus, dysmorphological and pathologic evaluation of the fetus using fetal autopsy is essential. Dysmorphology examination, when combined with fetal autopsy and prenatal imaging using various modalities, can be hugely beneficial to accurate phenotypic classification, which can then improve our ability to identify causative variants.
Prenatal genome sequencing
Genome sequencing (GS) has been applied in select prenatal cases to show proof of principle but has not been applied clinically because of difficulty interpreting intronic/regulatory regions of the genome.35 However, GS provides complete coverage of all the exons17 because ES can suffer from insufficient coverage of certain coding exons (e.g., GC-rich exons), a limitation overcome by polymerase chain reaction (PCR)-free GS.36,37 GS offers the additional ability to detect copy-number variants (CNVs),38,39 other structural variants (SVs), and expansions of short tandem repeats40 at a much higher sensitivity than ES. GS is now gaining traction as a diagnostic and discovery strategy for adults and children with a suspected genetic disorder that remains undiagnosed after ES analysis, or as a first-line approach in lieu of ES.includes an exon capture step, which adds time and reagent costs, and biases against coverage in GC-rich regions. GS does not include this selection step, providing more uniform coverage which allows a lower mean read depth; offers ability to detect copy-number variants (CNVs) with higher resolution than CMA and more complex balanced rearrangements. Recent studies report that GS can detect up to 3% of protein-coding variants missed by ES. For example, GS studies have found more causative variants in coding and noncoding regions in autism.41 A study was recently published showing that GS on DNA obtained from cell pellet from 31 amniocenteses was comparable in quality with GS performed on cfDNA.42 It will not be long before noninvasive interrogation of the fetal genome will be clinically available. There are multiple challenges that we foresee, including how to apply this technology to improve prenatal care and how to interpret large datasets quickly.
ETHICAL, LEGAL, SOCIAL ISSUES OF ES/GS PRENATALLY
Because of the use of trio ES/GS where both parents and fetus are sequenced simultaneously, there is a chance of parental medically actionable incidental findings (i.e., BRCA), variants of uncertain significance in the fetus, and potential for identification of consanguinity and incest. In addition, other concerns have been raised including risks of genetic determinism regarding child rearing and undermining children’s future autonomy by removing the option of not knowing their genetic information.43 Because of the above issues, highly tailored genetic counseling by a provider with expertise in genetics is critical when using ES/GS. Although beyond the scope of this review, these issues are critical and other authors in recent reviews have discussed them in detail.34,44
TRANSCRIPTOMICS IN FETAL MEDICINE: NEW INSIGHTS INTO EARLY HUMAN DEVELOPMENT, FETAL ANOMALIES, AND PLACENTA-MEDIATED COMPLICATIONS
RNA-sequencing (RNA-seq) followed closely behind the expansion of DNA sequencing into the prenatal setting. The transcriptome is the sum of gene transcripts or RNAs in a cell during a specific physiological state or developmental stage. This simultaneous measurement of gene expression for many thousands of transcripts provides a snapshot of the functional elements of the genome that can be mined for biological information. Therefore, unlike the genome, it is dynamic and varies according to tissue type, developmental stage, and pathophysiological state.
Prior to NGS, transcriptomic studies were largely performed using hybridization-based techniques, such as gene expression microarrays. However, microarrays had the drawbacks of relying upon existing knowledge of the genome for selecting oligonucleotide sequences, limited dynamic range due to signal saturation, and cross-hybridization creating background signal.45 Early sequence-based approaches to the transcriptome utilized complementary DNA (cDNA), such as Sanger sequencing of cDNA of expressed sequence tag libraries, and chemical-tagged based methods such as serial analysis of gene expression (SAGE).
In 2008, the first applications of high-throughput NGS to study the transcriptome were published. There was early recognition of the advantages of RNA-seq with its high-throughput, single-base resolution, and low background noise.46,47 RNA-seq simultaneously maps transcribed regions and quantifies expression with large dynamic range, able to distinguish new isoforms and allelic expression, providing information on gene expression, splice variants, and allelic expression. RNA-sequencing can also analyze all species of RNA, including micro RNAs (miRNAs), transfer RNA (tRNA), and long noncoding RNA (lncRNAs). Furthermore, unlike Sanger sequencing, the cost is relatively low for large genomes.45
Since 2008, there has been an explosion in computational tools and workflows for RNA-seq, with continuously evolving practices in study design and bioinformatics.48–50 Most transcriptomics studies require many thousands of cells to obtain the required starting amount of RNA. However, with embryonic and fetal samples, large amounts of RNA may not be obtainable. Furthermore, many systems such as the placenta, are composed of a heterogeneous population of cells in which different cell types play distinct roles. The field is now responding to the challenges of these samples by pushing the boundaries of transcript detection and cell-level information with single-cell transcriptomics.51
NGS RNA-SEQ APPLICATIONS TO HUMAN DEVELOPMENT
The large scale of biological information obtainable from RNA-seq has stimulated researchers to characterize the transcriptome across a wide range of human tissues, generating publicly-accessible gene expression atlases such as Illumima’s Human BodyMap 2.0 (ref. 52), the RNA-seq Atlas,53 and the NIH Roadmap Epigenomics Consortium.54 However, the vast majority of tissues included in these atlases are of adult origin, with a notable paucity of perinatal tissues.
However, with the advent of single-cell RNA-Seq, even preimplantation embryos can be studied using NGS, providing novel human data on the first 7 days of human development. Petropoulos and colleagues used single-cell embryonic development, using data from 1529 individual cells from 88 human embryos.55 They were able to delineate the establishment of the trophectoderm, epiblast, and primitive endoderm lineages, and discovered unique features of X chromosome dosage compensation in humans.
Other researchers have investigated the postimplantation embryo during the crucial period of organogenesis with RNA-seq.56 Most birth defects have their origin during this critical period in first trimester. Until recently, very little was known about gene expression during human organogenesis due to restricted availability and small size. Existing studies of the postimplantation embryo were either performed on whole embryos and therefore lacked tissue-specific expression,57 or focused on a single site, such as the limb bud.58
In 2016, the first integrative transcriptomic atlas of organogenesis was created using human embryos.56 Gerrad and colleagues profiled the tissue expression in 15 tissues and organs from two human embryos, including brain, upper limb, heart, palate/tongue, liver, and lung. The investigators developed a computational method called lineage-guided principal component analysis (lgPCA), which incorporated known developmental lineages into the analysis of gene expression profiles from each organ. Their results revealed more than 6000 novel transcripts, over 90% of which were long noncoding RNAs (lncRNAs). They were able to infer their roles as fate programming transcription factors that guided gene expression across multiple tissues—so-called master regulators of organogenesis. This lineage-guided analysis allowed them to successfully predict the transcription factors known to be responsible for specific syndromes (e.g., SOX9 and campomelic dysplasia), suggesting new approaches to determining the etiology of many unresolved congenital disorders.
RNA-seq has also been applied to tracking brain development during fetal life. Zhong and colleagues performed single-cell RNA-seq of more than 2300 cells in the fetal prefrontal cortext from 8 to 26 weeks gestation.59 They identified 35 subtypes of cells within six main classes and traced their developmental trajectory through gestation. This transcriptomic approach to understanding the development of the human prefrontal cortex may contribute to our understanding of congenital neurological diseases and discovery of potential therapies.
Until now, knowledge of fetal development has largely relied on animal studies, but the power of RNA-seq to maximize gene expression information from picogram quantities of RNA will no doubt see the field grow exponentially. RNA-sequencing has the potential to provide additional information about the etiology of fetal abnormalities when ES and GS are uninformative. Further study on use of RNA-sequencing in this setting is needed.
TRANSCRIPTOMICS USING FETAL BIOFLUIDS
RNA-seq studies of human embryonic and fetal tissue are still relatively rare due to the restricted availability of these tissues. Hence, novel surrogate measures of fetal gene expression have been explored, such as amniotic fluid cell-free RNA.60 The amniotic fluid transcriptome was first described using micro-array technology in 2012 (ref. 61) when it was established that amniotic fluid supernatant contains gene transcripts from multiple organs including fetal brain, lung, and gastrointestinal tract. Gene expression in amniotic fluid varies with gestation in concordance with known maturation processes61 and dysre-gulated cardiovascular and neurodevelopment pathways can be found in different disease states.62,63 RNA-seq has been subsequently applied to amniotic fluid and shown to provide novel data on splice variants and upregulated physiological pathways.64 Further application of RNA-seq and systems biology approaches have established that the AF transcriptome exhibits unique cell/organ-selective expression patterns at different time points in pregnancy.65 Studying amniotic fluid cell-free RNA has technical challenges,64 but has greater translational potential than fetal tissue, as it represents a feasible clinical source of biomarkers of organ maturation and predictors of neonatal morbidity.
Other fetal fluids that have been studied with a transcriptomic approach include umbilical cord blood obtained immediately after birth. Microarray-based transcriptomics suggests that fetal development is significantly altered in the presence of maternal obesity66 and environmental exposures.67 To date, RNA-seq has been applied to study cord blood monocyte transcriptomes in preterm and term newborns,68 and no doubt will be expanded to study many more perinatal conditions.
PLACENTA TRANSCRIPTOMICS AND NONINVASIVE APPROACHES USING MATERNAL PLASMA CELL-FREE RNA
The placenta is the key organ unpinning embryo implantation, fetal nutrition, oxygenation, and development. It is also a complex endocrine organ that regulates maternal physiology. Disorders of placental implantation and function lead to some of the major causes of maternal and perinatal morbidity and mortality such as preeclampsia. Until systems biology approaches became available, the myriad functions of the placenta could only be studied one pathway at a time. Transcriptomics has allowed the complete gene expression profile of the placenta to be simultaneously captured. Saben and colleagues used RNA-seq to characterise the placental transcriptome on 20 healthy women who gave birth following uncomplicated pregnancies and identified several genes novel to placental biology.69 Other investigators have applied networks-based analysis, which accounts for coordinated gene expression within the transcriptome. In a study of 200 term placentas collected after birth, 17 coexpression networks models were identified that were dominated by growth, organ development, gas exchange, and immune response.70 A fetal growth-related placental gene signature was generated, high-lighting the potential of NGS approaches to provide new molecular insights into placental function.
However, postnatal assessment of the placenta has limited utility for addressing complications that have their origin in early pregnancy. Sampling of the placenta during pregnancy (chorionic villus sampling) is an invasive procedure that carries a small risk of pregnancy loss. However, it is possible to measure circulating placenta-derived nucleic acids in maternal plasma. Circulating RNA of fetoplacental origin was first isolated in 2000 (ref. 71) and is central to the translational potential of placental transcriptomics. Its uniquely accessible nature makes it theoretically possible to noninvasively monitor placental health and predict placenta-mediated complications such as preeclampsia and fetal growth restriction.
Many other conditions have been studied using microarray and PCR-based methods, including fetal hypoxia, preterm birth, placenta accreta, and early pregnancy complications (extensively reviewed by Whitehead et al.).72 However, these studies have not yet yielded any validated circulating RNA biomarkers to be adopted into clinical practice. RNA-sequencing, however, is producing rapid leaps in knowledge and may accelerate discovery and translational aspects of placental transcriptomics.
One of the major challenges with working with cell-free RNA in maternal plasma is the multitude of cell types that contribute to the circulating RNA pool. Koh et al. analyzed tissue-specific gene expression in the plasma of pregnant women using a combination of microarray, RNA-seq, and quantitative PCR. By focusing on tissue-specific genes, they could identify the relative contributions of different tissues to maternal plasma. Not surprisingly, they found that whole blood was the largest contributor to the cell-free RNA transcriptome (contributing 40% of all RNAs), and that the fetal contribution increased during gestation, contributing 0.4, 3.4, and 15.4% in first, second, and third trimester respectively. They established the important principles that specific longitudinal phenotypic changes can be tracked in both the mother and the fetus, and that it is possible to directly measure transcripts from a variety of fetal tissues in the maternal blood sample.
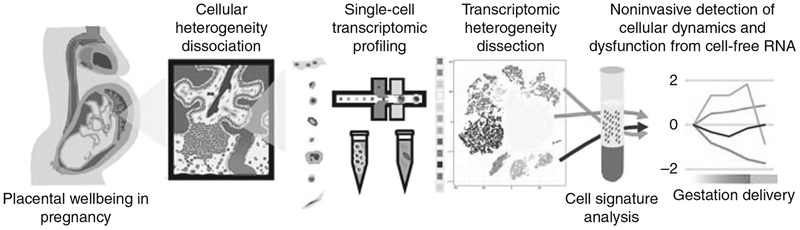
Single-cell RNA-seq has now been applied to the placenta to advance our understanding of placental biology and bring the field closer to the goal of noninvasive monitoring using maternal plasma. Tsang et al. used single-cell RNA-seq to characterize placental cell types and define cell type–specific gene signatures,73 before integrating these results with prior data from maternal plasma RNA (Fig. 3). Single-cell suspensions were created from placental tissue and 20,518 placental cells were subjected to large-scale droplet-based single-cell digital transcriptomic profiling. Cell-specific gene signatures were identified, revealing 12 major placental cell types that could be categorized as maternal or fetal in origin based on single-nucleotide polymorphism analysis. The cell types of maternal origin were decidual cells, uterine dendritic cells, and T-lymphocytes, while the fetal groups were vascular, stromal, macrophage-like, and trophoblastic. By integrating this information with datasets derived from cell-free RNA in maternal plasma, noninvasive monitoring of the cellular dynamics of the placenta could be performed.73 This work is an example of the power of NGS technologies and bioinformatics to improve our ability to care for women and their unborn babies.
Fig. 3. Single-cell transcriptomic profiling and the dissection of the cellular heterogeneity of human placenta (Tsang et al.73).
Cellular heterogeneity of the human placenta is dissected by droplet-based single-cell transcriptomic profiling. Cell type–specific signatures of different types of placental cells are identified and used to obtain information of cellular dynamics from maternal plasma RNA profiles in pregnancy and preeclampsia
FUNCTIONAL ANNOTATION FOR FETAL STUDIES
Gene expression profile data generated by NGS experiments on perinatal samples does require special consideration when it comes to choosing gene annotation sources and functional analysis tools. It is clear that the published literature on humans has a general skew toward adult biology, rather than fetal/developmental biology. For example, when using fetal gene expression data, physiologic cellular proliferation in the fetus produces many “false positive” cancer annotations within a propriety knowledgebase.74 This highlights the need for ongoing development of gene annotation resources with a developmental focus for ‘omics studies in perinatal medicine.75
FUTURE DIRECTIONS FOR TRANSCRIPTOMICS AND RNA-SEQ
There is enormous promise for NGS of RNA in a wide variety of applications in perinatal medicine. The field is not yet to reach clinical translation in the same way as DNA, but has wider potential because it provides us with information about tissue-specific function and is amenable to noninvasive methods. To date it has been used to study pre- and postimplantation embryology, stem cell biology, organogenesis, fetal maturation, and placental physiology. In the future, it may provide answers to longstanding questions regarding the origin of congenital anomalies, biomarkers of maturation and development, and allow noninvasive monitoring and targeted therapy for placenta-mediated complications of pregnancy.
CONCLUSIONS
Given the rapid advancement in use of ‘omic technologies in obstetrics, it is inevitable that the future for prenatal genomics will include the ability to interrogate the fetal genome and transcriptome noninvasively. Careful consideration is needed with introduction of these tools into prenatal care. Potential future uses may include targeted panels to identify conditions that necessitate immediate newborn treatment or use of cfRNA to screen pregnancies early that may be at risk of placentally mediated disorders. Given the feasibility of applying prenatal ‘omics on direct fetal specimens and noninvasively via cell-free DNA (covered extensively in a separate review), it is critical that genotype–phenotype information be deposited into shared databases so that interpretation and counseling regarding prenatal ‘omics data rapidly improves.
ACKNOWLEDGEMENTS
NICHD K23 HD088742 (PI: Vora). Figure 2 courtesy of Natasha Strande, PhD; Department of Pathology and Laboratory Medicine, University of North Carolina at Chapel Hill, Chapel Hill, NC
Footnotes
DISCLOSURE
The authors declare no conflict of interest.
REFERENCES
- 1.Wapner RJ, Martin CL, Levy B, et al. Chromosomal microarray versus karyotyping for prenatal diagnosis. N Engl J Med. 2012;367:2175–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murphy SL, Mathews TJ, Martin JA, et al. Annual summary of vital statistics: 2013–4. Pediatrics. 2017;139(6). [DOI] [PubMed] [Google Scholar]
- 3.Wellesley D, Dolk H, Boyd PA, et al. Rare chromosome abnormalities, prevalence and prenatal diagnosis rates from population-based congenital anomaly registers in Europe. Eur J Hum Genet. 2012;20:521–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hillman SC, McMullan DJ, Hall G, et al. Use of prenatal chromosomal microarray: prospective cohort study and systematic review and meta-analysis. Ultrasound Obstet Gynecol. 2013;41:610–20. [DOI] [PubMed] [Google Scholar]
- 5.Shaffer LG, Rosenfeld JA, Dabell MP, et al. Detection rates of clinically significant genomic alterations by microarray analysis for specific anomalies detected by ultrasound. Prenat Diagn. 2012;32:986–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McMullan DJ, Lord J, Eberhardt R, et al. Exome sequencing of 406 parental/fetal trios with structural abnormalities revealed by ultrasound in the UK Prenatal Assessment of Genomes and Exomes (PAGE) project. Paper presented at: European Society of Human Genetics; May 2017; Copenhagen, Denmark. [Google Scholar]
- 8.Jelin AC, Vora N. Whole exome sequencing: applications in prenatal genetics. Obstet Gynecol Clin North Am. 2018;45:69–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang Y, Muzny DM, Xia F, et al. Molecular findings among patients referred for clinical whole-exome sequencing. JAMA. 2014;312:1870–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carss KJ, Hillman SC, Parthiban V, et al. Exome sequencing improves genetic diagnosis of structural fetal abnormalities revealed by ultrasound. Hum Mol Genet. 2014;23:3269–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drury S, Williams H, Trump N, et al. Exome sequencing for prenatal diagnosis of fetuses with sonographic abnormalities. Prenat Diagn. 2015;35:1010–7. [DOI] [PubMed] [Google Scholar]
- 12.Alamillo CL, Powis Z, Farwell K, et al. Exome sequencing positively identified relevant alterations in more than half of cases with an indication of prenatal ultrasound anomalies. Prenat Diagn. 2015;35:1073–8. [DOI] [PubMed] [Google Scholar]
- 13.Yates CL, Monaghan KG, Copenheaver D, et al. Whole-exome sequencing on deceased fetuses with ultrasound anomalies: expanding our knowledge of genetic disease during fetal development. Genet Med. 2017;19:1171–8. [DOI] [PubMed] [Google Scholar]
- 14.Vora NL, Powell B, Brandt A, et al. Prenatal exome sequencing in anomalous fetuses: new opportunities and challenges. Genet Med. 2017;19:1207–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fu F, Li R, Li Y, et al. Whole exome sequencing as a diagnostic adjunct to clinical testing in fetuses with structural abnormalities. Ultrasound Obstet Gynecol. 2018;51:493–502. [DOI] [PubMed] [Google Scholar]
- 16.Wapner R, Petrovski S, Brennan K, et al. 8: Whole exome sequencing in the evaluation of fetal structural anomalies: a prospective study of sequential patients. Am J Obstet Gynecol. 2017;216:S5–S6. [Google Scholar]
- 17.Abou Tayoun AN, Spinner NB, Rehm HL, et al. Prenatal DNA sequencing: clinical, counseling, and diagnostic laboratory considerations. Prenat Diagn. 2018;38:26–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stark Z, Schofield D, Alam K, et al. Prospective comparison of the cost-effectiveness of clinical whole-exome sequencing with that of usual care overwhelmingly supports early use and reimbursement. Genet Med. 2017;19:867–74. [DOI] [PubMed] [Google Scholar]
- 19.Conesa A, Madrigal P, Tarazona S, et al. Erratum to: a survey of best practices for RNA-seq data analysis. Genome Biol. 2016;17:181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Committee on Genetics and the Society for Maternal-Fetal Medicine. Committee opinion no. 682: microarrays and next-generation sequencing technology: the use of advanced genetic diagnostic tools in obstetrics and gynecology. Obstet Gynecol. 2016;128:e262–e268. [DOI] [PubMed] [Google Scholar]
- 21.Hu P, Qiao F, Wang Y, et al. Clinical application of targeted next-generation sequencing on fetuses with congenital heart defects. Ultrasound Obstet Gynecol. 2018. March 13 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 22.Rasmussen M, Sunde L, Nielsen ML, et al. Targeted gene sequencing and whole-exome sequencing in autopsied fetuses with prenatally diagnosed kidney anomalies. Clin Genet. 2018;93:860–9. [DOI] [PubMed] [Google Scholar]
- 23.Filges I, Friedman JM. Exome sequencing for gene discovery in lethal fetal disorders–harnessing the value of extreme phenotypes. Prenat Diagn. 2015;35:1005–9. [DOI] [PubMed] [Google Scholar]
- 24.Putoux A, Thomas S, Coene KL, et al. KIF7 mutations cause fetal hydrolethalus and acrocallosal syndromes. Nat Genet. 2011;43:601–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thomas S, Legendre M, Saunier S, et al. TCTN3 mutations cause Mohr-Majewski syndrome. Am J Hum Genet. 2012;91:372–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McInerney-Leo AM, Schmidts M, Cortes CR, et al. Short-rib polydactyly and Jeune syndromes are caused by mutations in WDR60. Am J Hum Genet. 2013;93:515–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Filges I, Nosova E, Bruder E, et al. Exome sequencing identifies mutations in KIF14 as a novel cause of an autosomal recessive lethal fetal ciliopathy phenotype. Clin Genet. 2014;86:220–8. [DOI] [PubMed] [Google Scholar]
- 28.Ramos EI, Bien-Willner GA, Li J, et al. Genetic variation in MKL2 and decreased downstream PCTAIRE1 expression in extreme, fatal primary human microcephaly. Clin Genet. 2014;85:423–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Turkmen S, Demirhan O, Hoffmann K, et al. Cerebellar hypoplasia and quadrupedal locomotion in humans as a recessive trait mapping to chromosome 17p. J Med Genet. 2006;43:461–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Childs WJ. Across Asia Minor on Foot. Edinburgh: William Blackwood and Sons; 1917. [Google Scholar]
- 31.Hrdlicka A Children Who Run on all Fours and the Other Animal-like Behaviors in the Human Child. New York, NY: Whittlesey House; 1931. [Google Scholar]
- 32.Gulsuner S, Tekinay AB, Doerschner K, et al. Homozygosity mapping and targeted genomic sequencing reveal the gene responsible for cerebellar hypoplasia and quadrupedal locomotion in a consanguineous kindred. Genome Res. 2011;21:1995–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shamseldin HE, Kurdi W, Almusafri F, et al. Molecular autopsy in maternal-fetal medicine. Genet Med. 2018;20(4):420–427 [DOI] [PubMed] [Google Scholar]
- 34.Best S, Wou K, Vora N, et al. Promises, pitfalls and practicalities of prenatal whole exome sequencing. Prenat Diagn. 2018;38:10–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Talkowski ME, Ordulu Z, Pillalamarri V, et al. Clinical diagnosis by whole-genome sequencing of a prenatal sample. N Engl J Med. 2012;367:2226–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meienberg J, Bruggmann R, Oexle K, Matyas G. Clinical sequencing: is GS the better ES? Hum Genet. 2016;135:359–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Belkadi A, Bolze A, Itan Y, et al. Whole-genome sequencing is more powerful than whole-exome sequencing for detecting exome variants. Proc Natl Acad Sci USA. 2015;112:5473–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gilissen C, Hehir-Kwa JY, Thung DT, et al. Genome sequencing identifies major causes of severe intellectual disability. Nature. 2014;511:344–7. [DOI] [PubMed] [Google Scholar]
- 39.Handsaker RE, Van Doren V, Berman JR, et al. Large multiallelic copy number variations in humans. Nat Genet. 2015;47:296–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dolzhenko E, van Vugt J, Shaw RJ, et al. Detection of long repeat expansions from PCR-free whole-genome sequence data. Genome Res. 2017;27:1895–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Turner TN, Coe BP, Dickel DE, et al. Genomic patterns of de novo mutation in simplex autism. Cell. 2017;171:710–722e712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mao Q, Chin R, Xie W, et al. Advanced whole-genome sequencing and analysis of fetal genomes from amniotic fluid. Clin Chem. 2018;64:715–25. [DOI] [PubMed] [Google Scholar]
- 43.Donley G, Hull SC, Berkman BE. Prenatal whole genome sequencing: just because we can, should we? Hastings Cent Rep. 2012;42:28–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Botkin JR, Francis LP, Rose NC. Concerns about justification for fetal genome sequencing. Am J Bioeth. 2017;17:23–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mortazavi A, Williams BA, McCue K, et al. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–8. [DOI] [PubMed] [Google Scholar]
- 47.Sultan M, Schulz MH, Richard H, et al. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science. 2008;321:956–60. [DOI] [PubMed] [Google Scholar]
- 48.Schurch NJ, Schofield P, Gierlinski M, et al. How many biological replicates are needed in an RNA-seq experiment and which differential expression tool should you use? RNA. 2016;22:839–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Conesa A, Madrigal P, Tarazona S, et al. A survey of best practices for RNA-seq data analysis. Genome Biol. 2016;17:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sahraeian SME, Mohiyuddin M, Sebra R, et al. Gaining comprehensive biological insight into the transcriptome by performing a broad-spectrum RNA-seq analysis. Nat Commun. 2017;8:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kalisky T, Oriel S, Bar-Lev TH, et al. A brief review of single-cell transcriptomic technologies. Brief Funct Genomics. 2018;17:64–76. [DOI] [PubMed] [Google Scholar]
- 52.Vivante A, Mann N, Yonath H, et al. A dominant mutation in nuclear receptor interacting protein 1 causes urinary tract malformations via dysregulation of retinoic acid signaling. J Am Soc Nephrol. 2017;28:2364–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Krupp M, Marquardt JU, Sahin U, et al. RNA-Seq Atlas—a reference database for gene expression profiling in normal tissue by next-generation sequencing. Bioinformatics. 2012;28:1184–5. [DOI] [PubMed] [Google Scholar]
- 54.Roadmap Epigenomics C, Kundaje A, Meuleman W, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518: 317–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Petropoulos S, Edsgard D, Reinius B, et al. Single-cell RNA-seq reveals lineage and X chromosome dynamics in human preimplantation embryos. Cell. 2016;167:285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gerrard DT, Berry AA, Jennings RE, et al. An integrative transcriptomic atlas of organogenesis in human embryos. Elife. 2016;5:e15657 10.7554/eLife.15657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fang H, Yang Y, Li C, et al. Transcriptome analysis of early organogenesis in human embryos. Dev Cell. 2010;19:174–84. [DOI] [PubMed] [Google Scholar]
- 58.Cotney J, Leng J, Yin J, et al. The evolution of lineage-specific regulatory activities in the human embryonic limb. Cell. 2013;154:185–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhong S, Zhang S, Fan X, et al. A single-cell RNA-seq survey of the developmental landscape of the human prefrontal cortex. Nature. 2018;555:524–8. [DOI] [PubMed] [Google Scholar]
- 60.Zwemer LM, Bianchi DW The amniotic fluid transcriptome as a guide to understanding fetal disease. Cold Spring Harb Perspect Med. 2015;13:5(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hui L, Slonim DK, Wick HC, et al. The amniotic fluid transcriptome: a source of novel information about human fetal development. Obstet Gynecol. 2012;119:111–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hui L, Wick HC, Edlow AG, et al. Global gene expression analysis of term amniotic fluid cell-free fetal RNA. Obstet Gynecol. 2013;121:1248–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tarui T, Kim A, Flake A, et al. Amniotic fluid transcriptomics reflects novel disease mechanisms in fetuses with myelomeningocele. Am J Obstet Gynecol. 2017;217:587 e581–587 e510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zwemer LM, Hui L, Wick HC, Bianchi DW. RNA-Seq and expression microarray highlight different aspects of the fetal amniotic fluid transcriptome. Prenat Diagn. 2014;34:1006–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kamath-Rayne BD, Du Y, Hughes M, et al. Systems biology evaluation of cell-free amniotic fluid transcriptome of term and preterm infants to detect fetal maturity. BMC Med Genomics. 2015;8:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Edlow AG, Hui L, Wick HC, et al. Assessing the fetal effects of maternal obesity via transcriptomic analysis of cord blood: a prospective case-control study. BJOG. 2016;123:180–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Winckelmans E, Vrijens K, Tsamou M, et al. Newborn sex-specific transcriptome signatures and gestational exposure to fine particles: findings from the ENVIRONAGE birth cohort. Environ Health. 2017;16:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.de Jong E, Hancock DG, Hibbert J, et al. Identification of generic and pathogen-specific cord blood monocyte transcriptomes reveals a largely conserved response in preterm and term newborn infants. J Mol Med (Berl). 2018;96:147–57. [DOI] [PubMed] [Google Scholar]
- 69.Saben J, Zhong Y, McKelvey S, et al. A comprehensive analysis of the human placenta transcriptome. Placenta. 2014;35: 125–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Deyssenroth MA, Peng S, Hao K, et al. Whole-transcriptome analysis delineates the human placenta gene network and its associations with fetal growth. BMC Genomics. 2017;18:520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Poon LL, Leung TN, Lau TK, Lo YM. Presence of fetal RNA in maternal plasma. Clin Chem. 2000;46:1832–4. [PubMed] [Google Scholar]
- 72.Whitehead CL, Walker SP, Tong S. Measuring circulating placental RNAs to non-invasively assess the placental transcriptome and to predict pregnancy complications. Prenat Diagn. 2016;36: 997–1008. [DOI] [PubMed] [Google Scholar]
- 73.Tsang JCH, Vong JSL, Ji L, et al. Integrative single-cell and cell-free plasma RNA transcriptomics elucidates placental cellular dynamics. Proc Natl Acad Sci USA. 2017;114:E7786–E7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Edlow AG, Slonim DK, Wick HC, et al. The pathway not taken: understanding ‘omics data in the perinatal context. Am J Obstet Gynecol. 2015;213:59 e51–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wick HC, Drabkin H, Ngu H, et al. DFLAT: functional annotation for human development. BMC Bioinformatics. 2014;15:45. [DOI] [PMC free article] [PubMed] [Google Scholar]