Abstract
The wave of high-throughput technologies in genomics and phenomics are enabling data to be generated on an unprecedented scale and at a reasonable cost. Exploring the large-scale data sets generated by these technologies to derive biological insights requires efficient bioinformatic tools. Here we introduce an interactive, open-source web application (HTPmod) for high-throughput biological data modeling and visualization. HTPmod is implemented with the Shiny framework by integrating the computational power and professional visualization of R and including various machine-learning approaches. We demonstrate that HTPmod can be used for modeling and visualizing large-scale, high-dimensional data sets (such as multiple omics data) under a broad context. By reinvestigating example data sets from recent studies, we find not only that HTPmod can reproduce results from the original studies in a straightforward fashion and within a reasonable time, but also that novel insights may be gained from fast reinvestigation of existing data by HTPmod.
Dijun Chen et al. present HTPmod, a Shiny web application for modeling and visualization of large-scale genomic and phenomic datasets. The authors show that HTPmod can quickly reproduce analyses of high-throughput biological datasets and produce publication-quality figures.
Introduction
Over the last decade, technological advances in genomics (e.g., high-throughput sequencing, HTS) and phenomics (high-throughput plant phenotyping, HTP) have resulted in a tremendous increase of molecular and phenotypic data from large number of samples with a high-dimensional list of measurements. As a result, we can acquire an extensive range of phenotypes at organism-wide scale1,2, quantify the expression of tens of thousands of genes3–5, and measure the entire epigenome6,7 or regulatome8–10 simultaneously for hundreds to thousands of samples at a reasonable cost. The immense volume, variety, velocity, and veracity of high-throughput biological data generated by these technologies make it a big data problem11–13. In this regard, data handling and processing remain a major technical bottleneck when translating big biological data into knowledge.
Extracting hidden patterns and making accurate predictions from these massive data sets largely rely on machine-learning approaches14,15. From a computational point of view, machine learning methods are attractive in terms of their ability to derive predictive models without a need for strong assumptions about underlying mechanisms; hence they are especially useful to deal with certain biological questions of which our a priori knowledge is frequently unknown or insufficiently defined14. As a proof of concept, gene expression levels can be accurately predicted from a broad set of epigenetic features16–20 or binding profiles of diverse transcription factors (TFs)21–24 using various machine-learning-based approaches, although our knowledge about how the selected features determine the expression output is largely unknown. Modeling is, therefore, a key ingredient to derive novel biological insights by integrating large-scale data sets. Generally, a canonical machine learning workflow consists of the model fitting and evaluation. Although conceptually simple, applying adequate machine-learning algorithms to the large corpus of data remains an important challenge since it requires substantial computational expertise and effort. To our knowledge, an integrative web-based application for interactive exploration and interpretation of large-scale, high-dimensional data sets is not available to date. Here we present an interactive web application, HTPmod (http://www.epiplant.hu-berlin.de/shiny/app/HTPmod/), for high-throughput biological data modeling and visualization. By reinvestigating example data sets from recent studies, we demonstrate that HTPmod can be used for modeling and visualizing multiple types of omics data (such as phenomics, transcriptomics, metabolomics, and epigenomics data) under a broad context in a straightforward and an efficient fashion.
Results
Overview of the HTPmod application
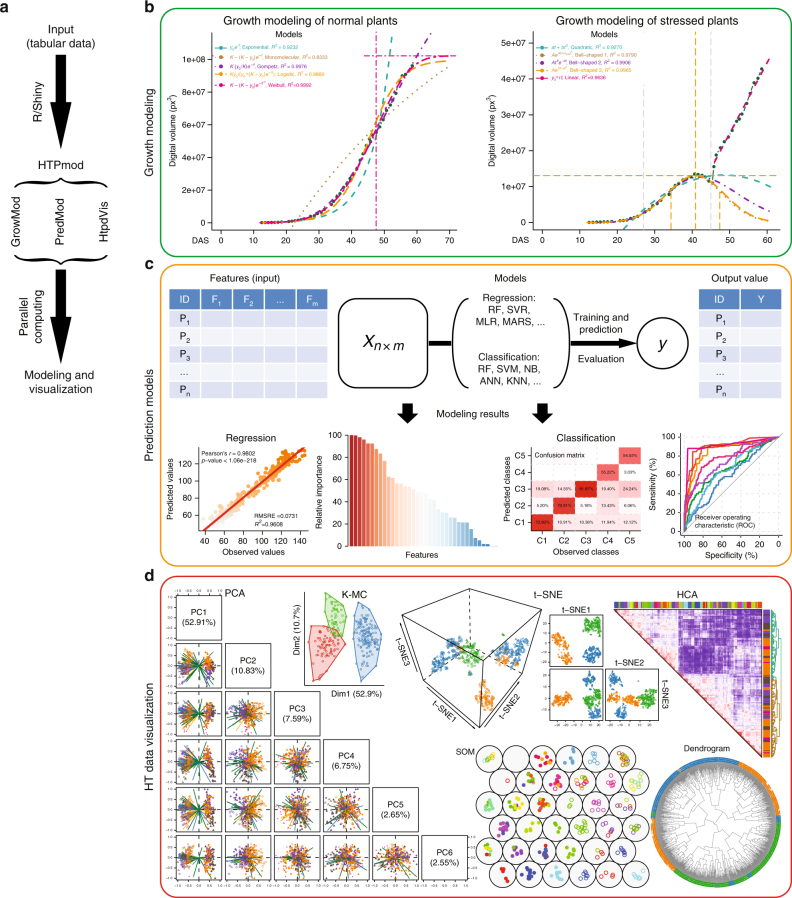
By integrating existing machine-learning approaches applied in high-throughput experiments1,25,26, HTPmod was implemented with the Shiny framework (http://shiny.rstudio.com/), which combines the computational power of R with friendly and interactive web interfaces. HTPmod provides three function modules for modeling (growMod and predMod) and visualizing (htpdVis) data especially from high-throughput experiments, such as HTP and HTS (Fig. 1 and Supplementary Fig. 1). Besides, HTPmod accepts the simplest table files as the only input (Fig. 1a and Supplementary Fig. 2) and supports the generation of various types of publication-quality graphics (Fig. 1b–d) and tables with possible customizations. Whenever possible, HTPmod adopts parallel computing to speed up analysis.
Fig. 1.
The HTPmod Shiny application for high-throughput data modeling and visualization. a The overall design and workflow of HTPmod. b The growMod module for plant growth modeling. Example results shown here are based on data from ref. 1. c The predMod application for predicting traits of interest from high-dimensional data using various prediction models. The upper panel shows the general workflow of predMod. The lower panel shows example output of regression (left) or classification (right) from predMod. d High-throughput data visualization with the htpdVis application. Example graphs are generated by htpdVis using data from refs. 1,25
The growMod module for plant growth modeling
The first module in HTPmod, growMod, was developed for plant growth modeling based on time-series data, e.g., from plant HTP experiments1,27. HTP is an ideal tool to study plant growth in a noninvasive way. We previously showed that the growth of barley (Hordeum vulgare) plants under normal and drought stress growth conditions follows a logistic curve and a bell-shaped curve, respectively1. In this study, we provided a graphical user interface (GUI) to perform growth modeling in an easy and efficient way (Fig. 1b). Generally, input data for growMod can be extracted from images by existing HTP image analysis software, such as IAP28 or PlantCV27,29. Image-derived features, such as plant height, project area and digital volume are some examples of traits that can be used to model plant growth. The growMod tool supports growth modeling for normal and stressed plants, which can be done either at single plant level or at group level (i.e., replicates in a group or a genotype). Moreover, we included several mechanistic growth models (including linear, bell-shaped, quadratic, exponential, monomolecular, logistic, Weibull and Gompertz curves; Supplementary Table 1) so that the performance of each model can be compared and evaluated (see Methods). Users can choose proper growth models to predict plant growth in their studies. Finally, biologically interpretable parameters can be derived from these models and can be further used for association mapping in a large population, allowing a deeper understanding of the performance and genetic basis of plant growth1.
The predMod module for prediction
The second module predMod was implemented with several supervised machine-learning models to relate input features (e.g., image data from HTP, and TF binding and histone modification data from HTS) to output quantities of interest (e.g., plant biomass, yield, stress status, or gene expression levels). The predMod tool is typically useful in situations where large amounts of data are available, with the aim to understand how a combination of factors (inputs) influence the output trait. In particular, the prediction models can be used for either regression (where output consists of numeric values) or classification (where output is a categorical class label). For instance, such prediction models have been widely used to predict the contribution of chromatin features to the change of gene expression18,21,30, to predict plant biomass from image-derived features25,27,31, to classify plants in different stress status1 or disease status32 based on image data, or to discriminate organ-specific target genes based on SELEX-seq data26. We integrated more than 30 widely used machine-learning approaches (Supplementary Table 2) into the predMod module, for regression or classification analyses (Fig. 1c). The prediction performance can be evaluated when multiple prediction models are selected18,25,30 (see Methods). Furthermore, feature importance and their prediction power can be extracted from the models18,21,25,30, which may aid for feature selection (e.g., to find potentially interesting features).
The htpdVis module for visualization
However, when there is no prior knowledge of the data investigated, unsupervised machine-learning approaches can be used to discover patterns from large data sets. To this end, we developed a third module, htpdVis, to explore and visualize large-scale, high-dimensional data using various unsupervised machine-learning approaches, such as principal component analysis (PCA), t-distributed stochastic neighbor embedding (t-SNE)33, self-organizing map, multidimensional scaling, K-means clustering or hierarchical cluster analysis with heatmaps (Fig. 1d). This module is particularly useful for exploration of hidden patterns and exploratory data mining from omics data sets such as phenome1, transcriptome34–36, or epigenome data37. For example, in PCA, the results of top principal components (PCs) are usually shown in a scatterplot where both the component scores (the transformed variable values of data points) and the factor loadings (the correlation coefficients between the observations [rows] and factors or features [columns]) are plotted in the same graphs (Fig. 1d). In addition, we also implemented the PCA with self-organizing map clustering approach, which is a useful way to visualize and explore multidimensional data sets, such as gene expression data across tissues in multiple species38–40. Notably, in the htpdVis module, different parameter settings can be used to generate diverse types of graphs with color and shape schema highlighting important data features (Fig. 1d).
Applications of HTPmod
To demonstrate the universal applications of HTPmod in data exploration and visualization, we provided various example data sets from recent studies (Supplementary Table 3) spanning phenomics1,25,27, metabolomics41, epigenomics37, regulatomics21,26 and transcriptomics42. We explored these data using the various functionalities implemented in our HTPmod system (see also online application for demonstrations). We showed that not only can HTPmod reproduce the corresponding findings of the original studies but also can gain novel insights from existing published data in a straightforward fashion and within a reasonable time (Supplementary Figs. 3-13).
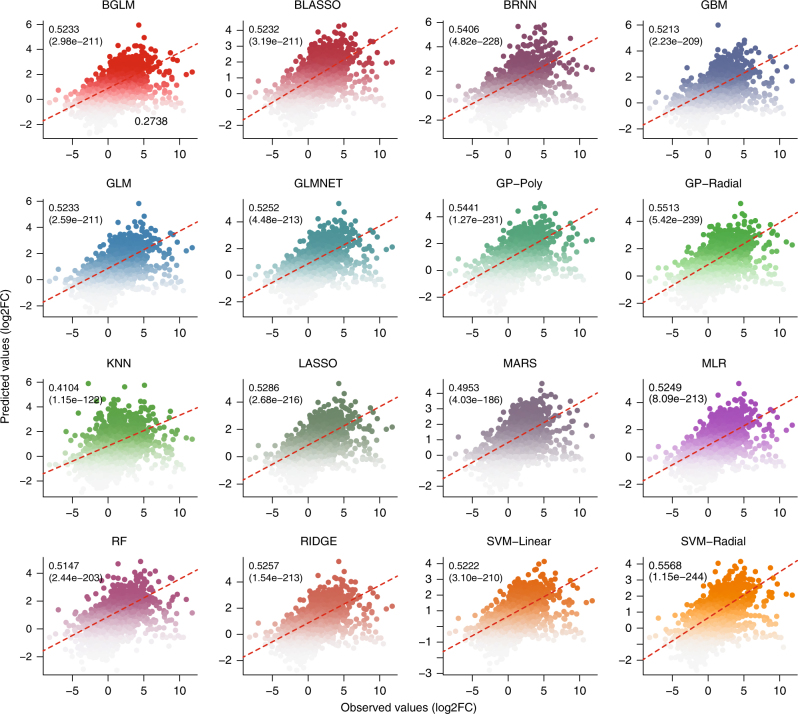
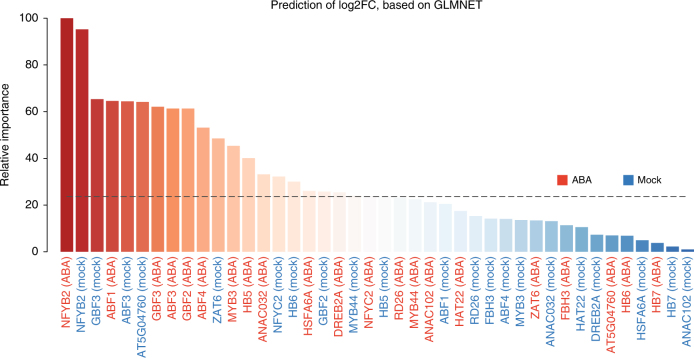
Here, we briefly described two case studies to show the power of HTPmod in data modeling and visualization. The first case study is to predict gene expression patterns using TF binding data in Arabidopsis thaliana, as shown in a recent study21. Briefly, we collected gene expression data from the supplemental data of ref. 21. and TF binding profiles from the Gene Expression Omnibus (GEO) database with an accession number GSE80568. The input data (consisting a matrix of TF binding score and expression changes for the differentially expressed genes) for HTPmod were prepared in a similar way as Song et al.21. We ran the predMod module with 16 regression models to relate TF binding strength to gene expression changes (log-transformed fold change [FC]) under ABA (phytohormone abscisic acid) treatment compared to mock. Strikingly, all the tested models show relatively comparable performance (Fig. 2 and Supplementary Fig. 7), implying that these models capture the intrinsic determinant of TF binding to the gene expression outcome. In addition, the relative feature importance determined by a glmnet regression model (Fig. 3) is consistent to the results presented in the original study21.
Fig. 2.
Prediction of gene expression changes using transcription factor binding data in Arabidopsis. Data obtained from ref. 21 and the full names of models referred to Supplementary Table 2. All prediction models with default parameter settings in predMod were used in the analysis. Pearson’s correlations and corresponding p-values (in parentheses) are shown
Fig. 3.
Relative importance of features in prediction of gene expression changes. GLMNET (lasso and elastic-net regularized generalized linear model) regression model (in predMod) was used to predict gene expression changes, using binding strength in both ABA- and mock-treated conditions. Barplot shows the relative importance of the binding features in the prediction. The result is consistent with that from the original study21
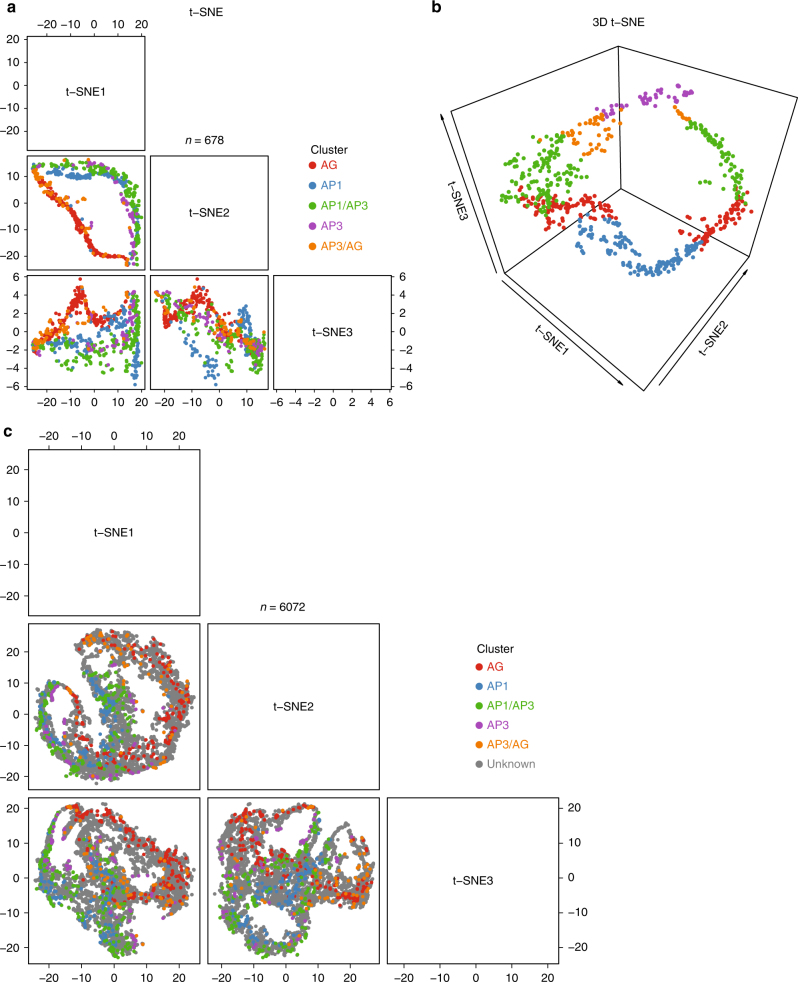
The second case study is to visualize floral organ-specific gene expression patterns42 by the htpdVis module. Domain-specific translatome data were obtained from the supplemental file of ref. 42. Based on analysis of variance (ANOVA), we identified 6072 genes that show significant spatiotemporal domain effects (p-value <0.05 based on ANOVA) with at least two-fold change (FC > 2) between different domains. We then filtered 678 domain-specific genes (see online document for more details) that were highly expressed in AP1-specific (specifying the sepal organ), AG-specific (carpel), AP1/AP3-common (petal), or AP3/AG-common (stamen) domains. We projected the data onto three dimensions via t-SNE plots based on htpdVis (Fig. 4a, b), which confirms that these organ-specific genes show well defined, distinct expression pattern. When adding more genes with unknown organ signature into visualization, we observed spatiotemporal gene expression trajectories during floral organ development (Fig. 4c). These observations provide an important starting point to investigate the mechanisms regulating organ differentiation in plants. In summary, the above results strongly support that HTPmod can make fast reproducible analysis without any programming demand.
Fig. 4.
Visualization of floral organ-specific transcriptome data in Arabidopsis42 via t-SNE plots33 using htpdVis. The pattern of organ-specific expression for genes with known organ signature is shown in the three-dimensional t-SNE plots in 2D (a) or 3D (b) views. c t-SNE plot in 2D view showing organ-specific expression pattern by adding more genes with unknown organ signature. Default parameter settings were used in all of these analyses
Discussion
In this work, we developed and characterized a web application for modeling and visualizing large-scale biological data sets. As implemented with the Shiny framework, the HTPmod application inherits the computational power as well as professional visualization of R. To avoid excessively long run-times, HTPmod also allows parallel computing to speed-up analysis whenever possible, facilitated by the BiocParallel package (http://bioconductor.org/packages/release/bioc/html/BiocParallel.html). The BiocParallel allows parallelization either on local web machine or on a cluster of computers using specific job schedulers. In short, HPTmod offers three modules (growMod, predMod, and htpdVis) for exploratory or interactive data mining with various omics data sets. An obviously distinctive feature of HTPmod is that it integrates widely used mathematical models (Supplementary Table 1) and machine-learning approaches (Supplementary Table 2) and runs them in a uniform way on a single data set, therefore allowing direct comparison and evaluation of the performance of different methods. However, different models may show distinct performance for a specific data set. In this respect, we may choose a model of interest or a model with the best performance in the analysis. Furthermore, model-derived knowledge, such as parameters to describe plant growth and performance1, and feature importance scores18,20,25, may allow important biological interpretation and be promising for providing novel insights.
In order to demonstrate that HTPmod is powerful for modeling and visualization of large-scale biological data in different contexts, we provided several case studies ranging from genomics to phenomics1,21,25–27,37,41,42 (Supplementary Table 3) and have shown that HTPmod is an easy-to-use tool that generates reproducible results in a very efficient way. Compared to existing analysis protocols38,43,44, HTPmod offers several advantages. First of all, HTPmod provides user friendly web interfaces to run a diverse set of models for data modeling and visualization based on a single input file, thus without the need of programming experience. Second, HTPmod can generate a variety of plots for publication purposes based on a single data set. Finally, HTPmod is open source and highly extendable. New prediction models can be easily integrated into HTPmod (see the online document). We will continue to integrate more prediction models or visualization/analysis components in the future. For example, deep learning is an emerging approach in the field of machine learning that can be used for image-based analytical tasks in plant phenotyping45–47. We believe that the data organization and visualization features offered by HTPmod are valuable for data scientists trying to apply deep learning to their HTP images.
As more and more big genomic and phenomic data sets are being or are going to be generated by large-scale, high-throughput experiments, the methodological framework for data modeling and visualization proposed in this work will have broadly potential applications. We anticipate that the plentiful output generated by HTPmod on a single data set will be useful to advance our views of a specific biological question under investigation. In summary, HTPmod is an open-source, interactive, and powerful web platform for large-scale biological data modeling and visualization.
Methods
Growth modeling (growMod)
With HTP data, image-derived features like plant height, projected area27 and digital volume1 can be considered as growth-related traits for growth modeling. In the growMod module, plant growth in control conditions can be modeled with six different mechanistic models: linear, exponential, monomolecular, logistic, Gompertz, and Weibull models (Supplementary Table 1). In order to fit these models using the linear regression function “lm” in R, the non-linear relationship of the models were first transformed into linearized forms (Supplementary Table 1). The growth traits are then fitted with the linearized models. Finally, the performance of models is assessed and compared based on their R2 and p-values. Some useful parameters can be derived from these models. For example, for the logistic model, the following three parameters are important to describe plant growth performance:1 (1) the intrinsic growth rate (R) that measures the speed of growth; (2) the inflection point (IP) that represents the time point when plant reaches the maximal speed of growth; and (3) the maximum final vegetative biomass (Kmax), which was estimated for each plant on the basis that the model could fit the data with the largest R2.
We also implemented several models to predict plant growth in in drought stress conditions1 (Supplementary Table 1). The modeling steps are divided into two parts: (1) growth before and during the stress phase and (2) re-growth during recovery phase. In the first phase, three different bell-shaped curves and a quadratic curve are fitted to the data, while in the recovery phase a simple linear model is used to characterize re-growth with the speed of re-growth (Rrec).
Prediction models (predMod) for regression or classification analysis
We included 32 widely used machine-learning approaches (Supplementary Table 2) into the predMod module, for regression or classification analysis purposes. Based on the powerful functionality of the caret R package and the uniform criteria for model performance evaluation (see below), predMod enables to run these models in a similar manner with comparable output.
Model performance
To evaluate the performance of the predictive models, we adopted a k-fold cross-validation strategy to check the prediction power of each model. Specifically, each data set will be randomly divided into a training set ((k − 1)/k of individuals) and a testing set (1/k of individuals). A specific model is first trained on the training data and then applied to make prediction for the testing data. The final performance of models is evaluated and compared based on the average prediction accuracies obtained from N resampling of the data set (N-times randomization), where both k and N are defined by users.
For regression models, their predictive performance can be measured by the Pearson correlation coefficient (PCC; r) between the predicted values and the observed values; and the coefficient of determination (R2) which equals to the fraction of variance explained by the model, defined as
where SSres and SStot are the sum of squares for residuals and the total sum of squares, respectively, the predicted and yi the observed value of the ith plant, is the mean value of the observed values; and the root mean squared relative error of cross-validation, defined as
where s denotes the sample size of the testing data set.
We repeated the cross-validation procedure ten times. The mean and standard deviation of the resulting R2 and RMSRE values were calculated across runs.
The predictive bias μ between the predicted and observed values, defined as
where n denotes the sample size of the data set. This bias indicates overestimation (μ > 0) or underestimation (μ > 0) of the target feature.
For classification models, their predictive performance can be measured by: (1) a confusion matrix, which is the contingency table of actual versus predicted class labels for each class, and is particularly helpful in the case of multiclass classification; (2) scalar characteristics as the accuracy, and average area under the ROC curve (see below); (3) a receiver operating characteristic (ROC) curve by plotting the true positive rate (TPR) against the false-positive rate (FPR) at various threshold settings, which is particularly helpful in two class problems; (4) a precision-recall curve (PRC)48 showing the tradeoff between precision and recall at different thresholds, which is particularly useful when the classes are very imbalanced.
Influence of features on prediction performance
We also developed several criteria to evaluate the relative importance of features for the prediction. For the models (including random forest, stochastic gradient boosting, classification and regression trees and multivariate adaptive regression spline) with built-in strategies to estimate the contribution of each variable to the prediction, the estimated measures of relative importance are scaled to the range between 0 (least important) and 100 (most important). Otherwise, the importance of each predictor is calculated individually using a filter approach as implemented in the caret R package.
Furthermore, the following criteria are also used to evaluate the importance of individual features and their redundancy in prediction. For regression, the ability of individual features to predict the response variable is calculated as the correlation coefficients (R2) between the predicted values and the actual values, which is termed as predictive power of the corresponding features. For classification problems, a greedy feature selection algorithm49 is conducted. Specifically, starting with the original set of n features, each feature is independently removed to produce n subsets of data with n − 1 features. Then the classification performance is computed with k-fold cross-validation and N-times randomizations, in the same way as described above, for each of these n subsets. The feature with least decreased the classification accuracy will be removed at this step. The above process is iterated until no feature can be removed. The classification performance driven by a specific combination of features can be visualized in a boxplot, with x-axis as the number of features and y-axis as cross-validation of classification accuracy.
Code availability
The HTPmod web-based application is freely available at http://www.epiplant.hu-berlin.de/shiny/app/HTPmod/. Users are encouraged deploy the HTPmod application at their own web server. The corresponding source code is available at https://github.com/htpmod/HTPmod-shinyApp and online document is available at https://github.com/htpmod/HTPmod-shinyApp/wiki.
Data availability
The processed example data sets used for demonstration purposes are provided alongside the HTPmod source code (https://github.com/htpmod/HTPmod-shinyApp).
Electronic supplementary material
Acknowledgements
This work was partially supported by the Leibniz Institute of Plant Genetics and Crop Plant Research (IPK), the National Natural Sciences Foundation of China (No. 31571366, 31771477), and the National Key Research and Development Program of China (2016YFA0501704). D.H. and M.C. are grateful to the support by the 111 Project and the Fundamental Research Funds for the Central Universities. K.K. wishes to thank the Alexander-von-Humboldt foundation and the Federal Ministry of Education and Research for support.
Author contributions
D.C. conceived and designed the study. M.C., C.K., and K.K. supervised the study. D.C. and L.F. implemented the Shiny application and conducted bioinformatics analysis. L.F. and D.H. assisted data collection and contributed to software testing. D.C. drafted the manuscript. All authors read and approved the final version of the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Dijun Chen, Email: chendijun2012@gmail.com.
Ming Chen, Email: mchen@zju.edu.cn.
Kerstin Kaufmann, Email: kerstin.kaufmann@hu-berlin.de.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s42003-018-0091-x.
References
- 1.Chen D, et al. Dissecting the phenotypic components of crop plant growth and drought responses based on high-throughput image analysis. Plant Cell. 2014;26:4636–4655. doi: 10.1105/tpc.114.129601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arend D, et al. Quantitative monitoring of Arabidopsis thaliana growth and development using high-throughput plant phenotyping. Sci. Data. 2016;3:160055. doi: 10.1038/sdata.2016.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsankov AM, et al. Transcription factor binding dynamics during human ES cell differentiation. Nature. 2015;518:344–349. doi: 10.1038/nature14233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gerstein MB, et al. Comparative analysis of the transcriptome across distant species. Nature. 2014;512:445–448. doi: 10.1038/nature13424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown JB, et al. Diversity and dynamics of the Drosophila transcriptome. Nature. 2014;512:393–399. doi: 10.1038/nature12962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kawakatsu T, et al. Epigenomic diversity in a global collection of Arabidopsis thaliana accessions. Cell. 2016;166:492–506. doi: 10.1016/j.cell.2016.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roadmap Epigenomics Consortium. et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–329. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neph S, et al. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature. 2012;489:83–90. doi: 10.1038/nature11212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malley RCO, et al. Cistrome and epicistrome features shape the regulatory DNA landscape. Cell. 2016;166:1598. doi: 10.1016/j.cell.2016.08.063. [DOI] [PubMed] [Google Scholar]
- 10.Sullivan AM, et al. Mapping and dynamics of regulatory DNA and transcription factor networks in A. thaliana. Cell Rep. 2014;8:2015–2030. doi: 10.1016/j.celrep.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 11.Schadt EE, Linderman MD, Sorenson J, Lee L, Nolan GP. Computational solutions to large-scale data management and analysis. Nat. Rev. Genet. 2010;11:647–657. doi: 10.1038/nrg2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tardieu F, Cabrera-Bosquet L, Pridmore T, Bennett M. Plant phenomics, from sensors to knowledge. Curr. Biol. 2017;27:R770–R783. doi: 10.1016/j.cub.2017.05.055. [DOI] [PubMed] [Google Scholar]
- 13.Houle D, Govindaraju DR, Omholt S. Phenomics: the next challenge. Nat. Rev. Genet. 2010;11:855–866. doi: 10.1038/nrg2897. [DOI] [PubMed] [Google Scholar]
- 14.Angermueller C, Pärnamaa T, Parts L, Oliver S. Deep learning for computational biology. Mol. Syst. Biol. 2016;12:1–16. doi: 10.15252/msb.20156651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singh A, Ganapathysubramanian B, Singh AK, Sarkar S. Machine learning for high-throughput stress phenotyping in plants. Trends Plant. Sci. 2016;21:110–124. doi: 10.1016/j.tplants.2015.10.015. [DOI] [PubMed] [Google Scholar]
- 16.Karlic R, Chung HR, Lasserre J, Vlahovicek K, Vingron M. Histone modification levels are predictive for gene expression. Proc. Natl Acad. Sci. USA. 2010;107:2926–2931. doi: 10.1073/pnas.0909344107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng C, et al. A statistical framework for modeling gene expression using chromatin features and application to modENCODE datasets. Genome Biol. 2011;12:R15. doi: 10.1186/gb-2011-12-2-r15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dong X, et al. Modeling gene expression using chromatin features in various cellular contexts. Genome Biol. 2012;13:R53. doi: 10.1186/gb-2012-13-9-r53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Costa IG, Roider HG, do Rego TG, de Carvalho FdeA. Predicting gene expression in T cell differentiation from histone modifications and transcription factor binding affinities by linear mixture models. BMC Bioinforma. 2011;12:S29. doi: 10.1186/1471-2105-12-S1-S29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Consortium EP, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song L, et al. A transcription factor hierarchy defines an environmental stress response network. Science (80-.). 2016;354:aag1550–aag1550. doi: 10.1126/science.aag1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmidt F, et al. Combining transcription factor binding affinities with open-chromatin data for accurate gene expression prediction. Nucleic Acids Res. 2017;45:54–66. doi: 10.1093/nar/gkw1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ouyang Z, Zhou Q, Wong WH. ChIP-Seq of transcription factors predicts absolute and differential gene expression in embryonic stem cells. Proc. Natl Acad. Sci. USA. 2009;106:21521–21526. doi: 10.1073/pnas.0904863106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang LQ, Li QZ, Su WX, Jin W. Predicting gene expression level by the transcription factor binding signals in human embryonic stem cells. Biosystems. 2016;150:92–98. doi: 10.1016/j.biosystems.2016.08.011. [DOI] [PubMed] [Google Scholar]
- 25.Chen, D. et al. Predicting plant biomass accumulation from image-derived parameters. Gigascience7 (2018). 10.1093/gigascience/giy001 [DOI] [PMC free article] [PubMed]
- 26.Smaczniak C, Muiño JM, Chen D, Angenent GC, Kaufmann K. Differences in DNA-binding specificity of floral homeotic protein complexes predict organ-specific target genes. Plant Cell. 2017;29:1822–1835. doi: 10.1105/tpc.17.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fahlgren N, et al. A versatile phenotyping system and analytics platform reveals diverse temporal responses to water availability in Setaria. Mol. Plant. 2015;8:1520–1535. doi: 10.1016/j.molp.2015.06.005. [DOI] [PubMed] [Google Scholar]
- 28.Klukas C, Chen D, Pape JM. Integrated analysis platform: an open-source information system for high-throughput plant phenotyping. Plant Physiol. 2014;165:506–518. doi: 10.1104/pp.113.233932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gehan MA, et al. PlantCVv2: Image analysis software for high-throughput plant phenotyping. PeerJ. 2017;5:e4088. doi: 10.7717/peerj.4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng C, et al. Understanding transcriptional regulation by integrative analysis of transcription factor binding data. Genome Res. 2012;22:1658–1667. doi: 10.1101/gr.136838.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang W, et al. Combining high-throughput phenotyping and genome-wide association studies to reveal natural genetic variation in rice. Nat. Commun. 2014;5:5087. doi: 10.1038/ncomms6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baranowski P, et al. Hyperspectral and thermal imaging of oilseed rape (Brassica napus) response to fungal species of the genus Alternaria. PLoS ONE. 2015;10:e0122913. doi: 10.1371/journal.pone.0122913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maaten LVanDer, Hinton G. Visualizing data using t-SNE. J. Mach. Learn. Res. 2008;1:267–284. [Google Scholar]
- 34.Chen J, et al. Dynamic transcriptome landscape of maize embryo and endosperm development. Plant Physiol. 2014;166:252–264. doi: 10.1104/pp.114.240689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Terol J, Tadeo F, Ventimilla D, Talon M. An RNA-Seq-based reference transcriptome for Citrus. Plant. Biotechnol. J. 2016;14:938–950. doi: 10.1111/pbi.12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhan J, et al. RNA sequencing of laser-capture microdissected compartments of the maize kernel identifies regulatory modules associated with endosperm cell differentiation. Plant Cell. 2015;27:513–531. doi: 10.1105/tpc.114.135657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang C, et al. Genome-wide analysis of local chromatin packing in Arabidopsis thaliana. Genome Res. 2015;25:246–256. doi: 10.1101/gr.170332.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chitwood DH, Maloof JN, Sinha NR. Dynamic transcriptomic profiles between tomato and a wild relative reflect distinct developmental architectures. Plant Physiol. 2013;162:537–552. doi: 10.1104/pp.112.213546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ranjan A, Townsley BT, Ichihashi Y, Sinha NR, Chitwood DH. An intracellular transcriptomic atlas of the giant coenocyte Caulerpa taxifolia. PLoS Genet. 2015;11:e1004900. doi: 10.1371/journal.pgen.1004900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ranjan A, et al. De novo assembly and characterization of the transcriptome of the parasitic weed dodder identifies genes associated with plant parasitism. Plant Physiol. 2014;166:1186–1199. doi: 10.1104/pp.113.234864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu G, et al. Rewiring of the fruit metabolome in tomato breeding. Cell. 2018;172:249–261. doi: 10.1016/j.cell.2017.12.019. [DOI] [PubMed] [Google Scholar]
- 42.Jiao Y, Meyerowitz EM. Cell-type specific analysis of translating RNAs in developing flowers reveals new levels of control. Mol. Syst. Biol. 2010;6:419. doi: 10.1038/msb.2010.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gómez J, et al. BioJS: an open source JavaScript framework for biological data visualization. Bioinformatics. 2013;29:1103–1104. doi: 10.1093/bioinformatics/btt100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tarca AL, Carey VJ, Chen X, Romero R, Drăghici S. Machine learning and its applications to biology. PLoS Comput. Biol. 2007;3:e116. doi: 10.1371/journal.pcbi.0030116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ubbens JR, Stavness I. Deep plant phenomics: a deep learning platform for complex plant phenotyping tasks. Front. Plant Sci. 2017;8:1190. doi: 10.3389/fpls.2017.01190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pound MP, et al. Deep machine learning provides state-of-the-art performance in image-based plant phenotyping. Gigascience. 2017;6:1–10. doi: 10.1093/gigascience/gix083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pound, M. P., Atkinson, J. A., Wells, D. M., Pridmore, T. P. & French, A. P. Deep learning for multi-task plant phenotyping. bioRxiv 204552 (2017). 10.1101/204552
- 48.Saito T, Rehmsmeier M. The precision-recall plot is more informative than the ROC plot when evaluating binary classifiers on imbalanced datasets. PLoS ONE. 2015;10:e0118432. doi: 10.1371/journal.pone.0118432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fuchs F, et al. Clustering phenotype populations by genome-wide RNAi and multiparametric imaging. Mol. Syst. Biol. 2010;6:370. doi: 10.1038/msb.2010.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The processed example data sets used for demonstration purposes are provided alongside the HTPmod source code (https://github.com/htpmod/HTPmod-shinyApp).