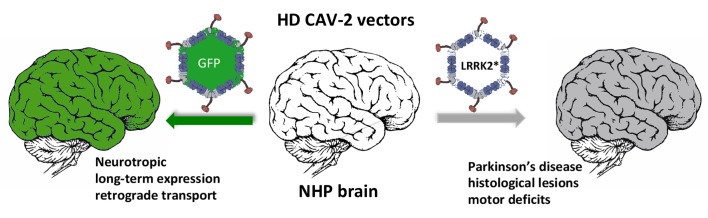
Abstract
Parkinson’s disease (PD) is the second most prevalent neurodegenerative disease among the elderly. To understand its pathogenesis and to test therapies, animal models that faithfully reproduce key pathological PD hallmarks are needed. As a prelude to developing a model of PD, we tested the tropism, efficacy, biodistribution, and transcriptional effect of canine adenovirus type 2 (CAV-2) vectors in the brain of Microcebus murinus, a nonhuman primate that naturally develops neurodegenerative lesions. We show that introducing helper-dependent (HD) CAV-2 vectors results in long-term, neuron-specific expression at the injection site and in afferent nuclei. Although HD CAV-2 vector injection induced a modest transcriptional response, no significant adaptive immune response was generated. We then generated and tested HD CAV-2 vectors expressing leucine-rich repeat kinase 2 (LRRK2) and LRRK2 carrying a G2019S mutation (LRRK2G2019S), which is linked to sporadic and familial autosomal dominant forms of PD. We show that HD-LRRK2G2019S expression induced parkinsonian-like motor symptoms and histological features in less than 4 months.
Keywords: Neuroscience
Keywords: Neurodegeneration, Parkinson’s disease
A viral vector expressing a Parkinson’s disease-associated variant of LRRK2 induces loss of dopaminergic cells and motor symptoms in primates in fewer than 4 months.
Introduction
Parkinson’s disease (PD) is a progressive disorder of the CNS that is primarily associated with impaired movement. PD likely develops over decades and is linked to the gradual loss of dopamine delivery to the striatum, predominantly via the loss of dopaminergic (DA) neurons in the substantia nigra pars compacta (SNpc) (1). In spite of intense research, PD etiology largely remains a mystery. In a minority of cases though, the characterization of familial forms has opened avenues into understanding PD pathophysiology. Among the handful of mutations linked to familial PD forms is a glycine-to-serine change at position 2019 (G2019S) in leucine-rich repeat kinase 2 (LRRK2 [also known as PARK8]). LRRK2G2019S is linked to hyperkinase activity (2) and phosphorylation of Rab proteins (3). LRRK2G2019S is found in approximately 30% of PD cases in North African Berbers and is the most common cause of late-onset, autosomal dominant, familial, and sporadic PD (4). LRRK2 is a multifunctional 285-kDa protein that contains a serine/threonine kinase domain, a GTPase domain, and protein-protein interaction domains (2, 5–8). LRRK2 is expressed in brain areas receiving DA innervation, such as the striatum, hippocampus, cortex, cerebellum, substantia nigra (SN), and ventral tegmental area (9–11).
Understanding the effect of LRRK2G2019S has been challenging because most LRRK2G2019S-expressing animals do not recapitulate many of the histological and behavioral hallmarks of PD (12, 13). An alternative to transgenesis is to deliver expression cassettes containing disease-associated cDNAs into the brain using viral vectors. In some cases, the vector should preferentially transduce neurons, allow long-term expression, distribute vector genomes to distal regions via axonal transport, be produced at high titers, and have a large cloning capacity. The last issue is essential in the case of LRRK2, whose cDNA is approximately 8 kbp. Helper-dependent (HD) canine adenovirus type 2 (CAV-2) vectors satisfy these and other criteria (14). For example, CAV-2 vectors preferentially transduce neurons from numerous species, including mice, rats, and dogs; additionally, in humans, CAV-2 vectors preferentially transduce neurons grown in 2D cultures, 3D neurospheres, and organotypic slices of cortical tissue (14–17). Following injection in the rodent and dog striatum, CAV-2 axonal transport into structures/neurons that project into the injection site is remarkably efficient (14–16). Notably, DA neurons in the rodent SNpc also express CAV-2–encoded foreign protein and normal cellular proteins for >1 year after injection (16–18).
As a prelude to developing a model of PD, we assayed the tropism, efficacy, transcriptional effect, and biodistribution of HD CAV-2 vectors in the striatum of Microcebus murinus. M. murinus is a nonhuman primate (NHP) that is readily bred in captivity, can live >10 years, and naturally shows some age-related signs of neurodegenerative diseases (19–23). We show that CAV-2 vectors lead to long-term, neuron-specific expression at the site of injection as well as in the ipsilateral and contralateral frontal cortex, SNpc, thalamus, and other regions. CAV-2 tropism and retrograde transport was consistent with the engagement of coxsackievirus and adenovirus receptor (CAR, ref. 24) at the presynapse (24–31). To test the potential of CAV-2 vectors to model PD, we generated vectors harboring 10-kbp expression cassettes containing LRRK2 or LRRK2G2019S. We show that LRRK2G2019S expression in the M. murinus brain can induce parkinsonian-like motor symptoms and histological lesions in less than 4 months, thus allowing evaluation of PD pathobiology, compounding factors, progression, and/or therapies.
Results
Preferential and widespread neuron transduction in the NHP brain.
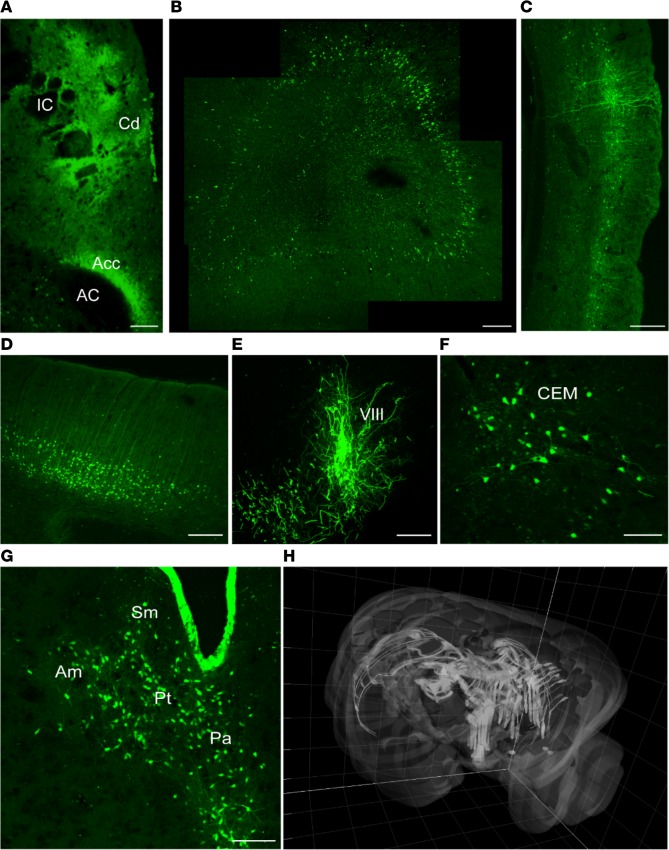
To test CAV-2 vector efficacy in the NHP brain, we unilaterally injected the M. murinus caudate nucleus with a HD vector expressing GFP (HD-GFP) (see Table 1 for details). At the indicated times, the animals were sacrificed and the brains were processed for GFP expression, immunohistochemistry, transcriptome analyses, and/or used to extract DNA. Fifty-micron-thick sections were screened by confocal microscopy to identify and quantify transduced cells throughout the CNS. At the injection site we found a dense concentration of GFP+ soma and processes (Figure 1A). GFP+ cell bodies and neurites were also present throughout the frontal (Figure 1, B and C) and occipital cortices (Figure 1D) in both hemispheres, in the SNpc of the injected hemisphere (Figure 1E), in the ipsilateral basal nuclei of Meynert (Figure 1F), and in other regions (Figure 1G). The dense GFP signal surrounding the injection site is consistent with transduction of striatal neurons and GFP-filled axons of neurons projecting therein. In a minority of animals, the GFP pattern suggested that some vector leaked into the ventricles/cerebral spinal fluid and transduced cells lining the ventricles (Figure 1G). To provide a global view of the regions transduced by a single injection of HD-GFP, we created a 3D image of the M. murinus brain and highlight areas that contained transduced cells (Figure 1H).
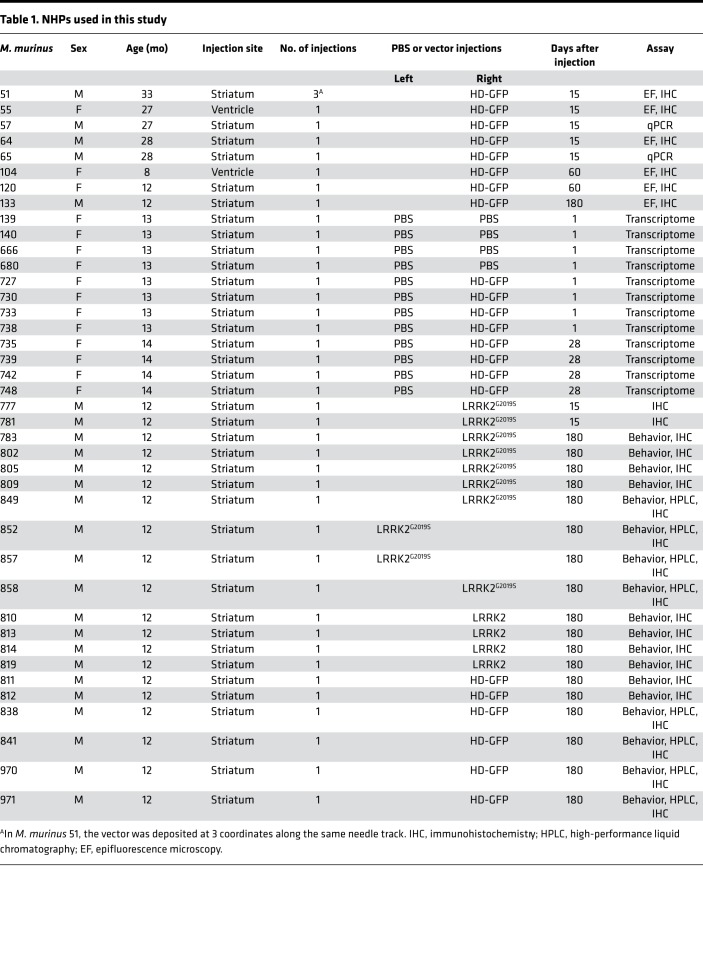
Table 1. NHPs used in this study.
Figure 1. Expression of GFP following injection of HD-GFP in the M.
murinusstriatum. The brains from 6 M. murinus, injected at 8–33 months of age (see Table 1), were used to determine the efficacy of GFP expression from a helper-dependent (HD) CAV-2 vector (HD-GFP). (A) Injection site. Juxtaposed images showing the caudate nucleus (Cd), internal capsule (IC), nucleus accumbens (Acc), and anterior commissural (AC). (B) Ipsilateral prefrontal cortex (4 images juxtaposed). (C) Ipsilateral pyriform cortex. (D) Contralateral frontal cortex (cell bodies are located in the second and fourth cortical layers). (E) Substantia nigra (VIII). (F) Basal nucleus of Meynert (central-medial nucleus [CEM]). (G) Thalamic nuclei: periventricular nucleus (Pa), nucleus parataenialis (Pt), stria medullaris (Sm), anterior-medial nucleus (Am). (H) 3D reconstruction of the brain showing distribution of the GFP signal in both hemispheres following injection into the caudate nucleus. Images in A, C, F, and G are from M. murinus 133, which was sacrificed 6 months after injection. Scale bar: 200 μm.
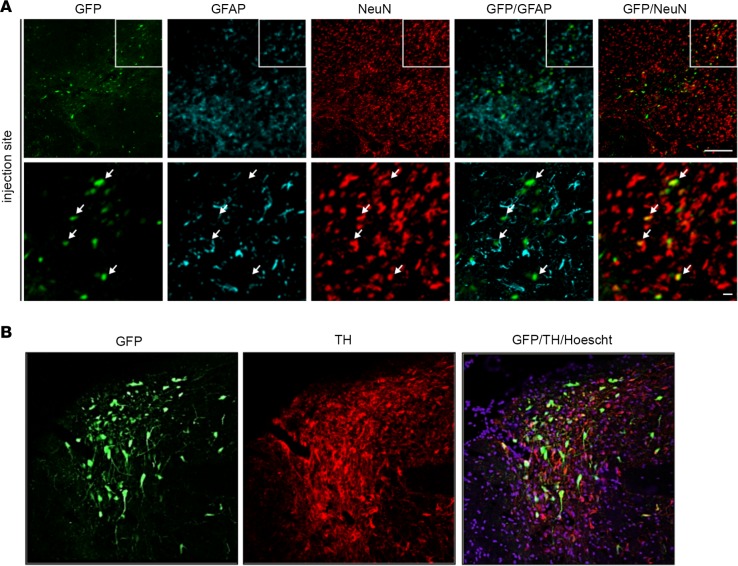
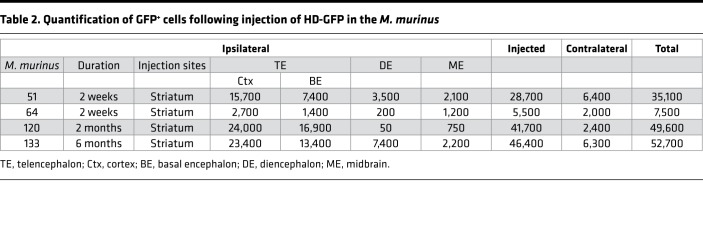
To identify the GFP+ cells, we stained sections with antibodies that preferentially identify neurons (anti-NeuN) or astrocytes (anti–glial fibrillary acidic protein [anti-GFAP]). We found that CAV-2 vectors preferentially transduced neurons (>90%) at the injection site (Figure 2A). Colabeling with tyrosine hydroxylase (TH) demonstrated that some GFP+ cells located in the SN were DA neurons (Figure 2B). To semiquantify HD-GFP transduction efficacy following striatal injection, we manually counted GFP+ cells throughout the parenchyma in animals killed at 2 weeks (n = 2), 2 months (n = 1), or 6 months (n = 1) (Table 2). While the number of animals/point was too small for statistical analyses, the data suggest that the total number of GFP+ neurons was at least as robust at 2 weeks as at 6 months. This is particularly relevant in the context of CAV-2 vector immunogenicity and demonstrates that the vectors are amendable to long-term expression of a potentially immunogenic protein (GFP) in the NHP brain. Anecdotally, the two animals that had the greatest number of GFP+ cells were also among the youngest when injected (12 months old). Because we were interested in transduction of neurons in the SNpc, we quantified TH+ and GFP+ neurons in this structure. Initially, we found that the M. murinus SNpc has approximately 3,000 TH+ neurons/hemisphere (n = 4 hemispheres). In M. murinus 51 and M. murinus 133, we detected approximately 2,100 GFP+ cells in the ipsilateral SNpc (Table 2). These data indicate a transduction efficacy of approximately 70% TH+ neurons/hemisphere following a CAV-2 vector deposit at a single coordinate in the caudate nucleus. Together, these data demonstrate that CAV-2 vectors preferentially transduce neurons, are transported to afferent structures in both hemispheres, and lead to stable expression of a foreign protein in the NHP brain.
Figure 2. Identification of HD-GFP–transduced cells in the M.
murinusbrain. Sections from M. murinus brains (n = 6 animals) were stained for neuronal and astrocytes specific markers to identify transduced cells. (A) Representative image from the injection site, caudate nucleus, showing GFP, glial fibrillary acidic protein (GFAP), and NeuN (neuronal nuclei antigen) expression followed by merged images. (B) Representative image from the substantia nigra (SN) showing GFP and tyrosine hydroxylase (TH) expression, followed by merged images. Scale bars: 100 μm (A, top) and 25 μm (A, bottom); and 100 μm (B).
Table 2. Quantification of GFP+ cells following injection of HD-GFP in the M. murinus.
GFP expression parallels vector biodistribution and CAR expression.
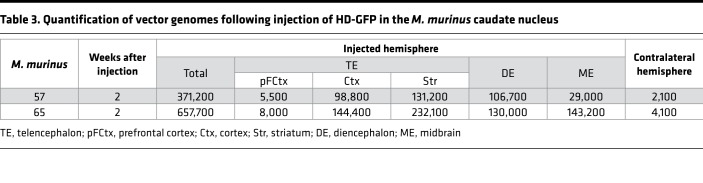
The quantification of GFP+ cells and use of 3D reconstruction generate a global view of transgene expression. However, these data do not provide a quantitative assessment of vector biodistribution because each neuron subtype could take up a different quantity of vector and, therefore, a single GFP+ cell could have multiple vector genomes. We therefore quantified vector genomes in distinct regions of two animals. Using qPCR on DNA isolated from various brain regions, we found that the highest number of vector genomes was in the basal forebrain, which corresponds to the injection site (Table 3). Notable levels of vectors were also found in the adjacent structures, such as the temporal and parietal cortices, the thalamus, and the midbrain. Fewer HD-GFP genomes were found in the prefrontal and occipital cortices, the cerebellum of the injected hemisphere, and in the olfactory bulb. Globally, vector genome biodistribution in M. murinus 65 mirrors GFP expression pattern in M. murinus 64.
Table 3. Quantification of vector genomes following injection of HD-GFP in the M. murinus caudate nucleus.
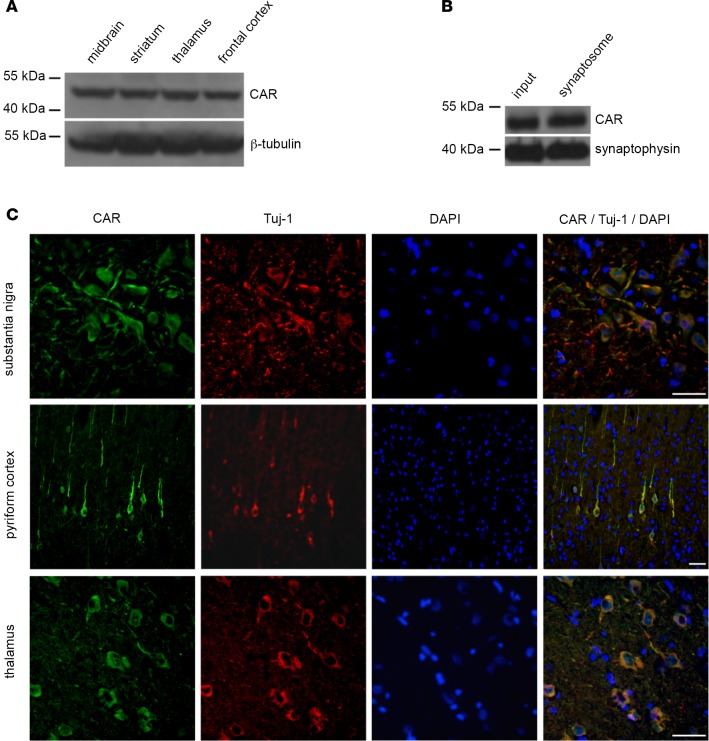
Ex vivo and in the rodent brain, CAV-2 engagement of CAR leads to neuron binding, internalization, and retrograde transport (24–31). To determine if the CAR expression pattern was consistent with CAV-2 tropism in the primate brain, we screened brains for CAR expression by immunoblotting and immunofluorescence. We found that CAR was readily detected in several regions of the M. murinus brain by immunoblotting (Figure 3A). However, for CAV-2 to be transported from the injection site to the soma of cells in other regions, CAR would need to be expressed on axonal projections. Consistent with our results in rodents (28), CAR was present in the synaptosome fraction in extracts of the M. murinus brain (Figure 3B). Using immunofluorescent staining of adult M. murinus brain sections, we found that CAR was expressed by neurons in the SNpc, cortex, and thalamus — regions that project into the caudate nucleus (Figure 3C). Together, these data suggest a physiological explanation for CAV-2 tropism and biodistribution via CAR engagement and axonal transport in a NHP brain.
Figure 3. Coxsackievirus and adenovirus receptor expression in the M.
murinusbrain. (A) Immunoblot analyses of coxsackievirus and adenovirus receptor (CAR) from the brains of M. murinus (n = 2 animals). β-Tubulin levels were used to normalize loading. (B) CAR levels in synaptosomes prepared from the cortex (n = 2 animals) and assayed for the presence of CAR. As a control, we used synaptophysin, a prototypic synapse protein. (C) CAR expression pattern, as assayed by immunofluorescence staining: SN, pyriform cortex, and thalamus (n = 3 animals). The 16-micron-thick floating sections were coincubated an anti-Tuj1 (β-III tubulin) antibody to identify neurons and with DAPI to label nuclei. Scale bar: 50 μm.
Effect of CAV-2 vectors on the brain transcriptome.
We previously showed that the transcriptional profile of human neurons grown in 2D and 3D cultures is differentially modified following interaction with HD human adenovirus type 5, lentivirus, and HD CAV-2 vectors (32). To address the transcriptional response in the more complex in vivo environment, we quantified changes in mRNA levels following HD-GFP versus PBS injection in the M. murinus caudate nucleus at 1 and 28 days after injection. We quantified transcriptional changes in the dorsal region of the frontal lobe, the striatum, and the midbrain (Figure 4 and Supplemental Tables 1 and 2; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.98202DS1). Twenty samples were analyzed for each brain to take into account the variation in time after injection and the regions from each hemisphere.
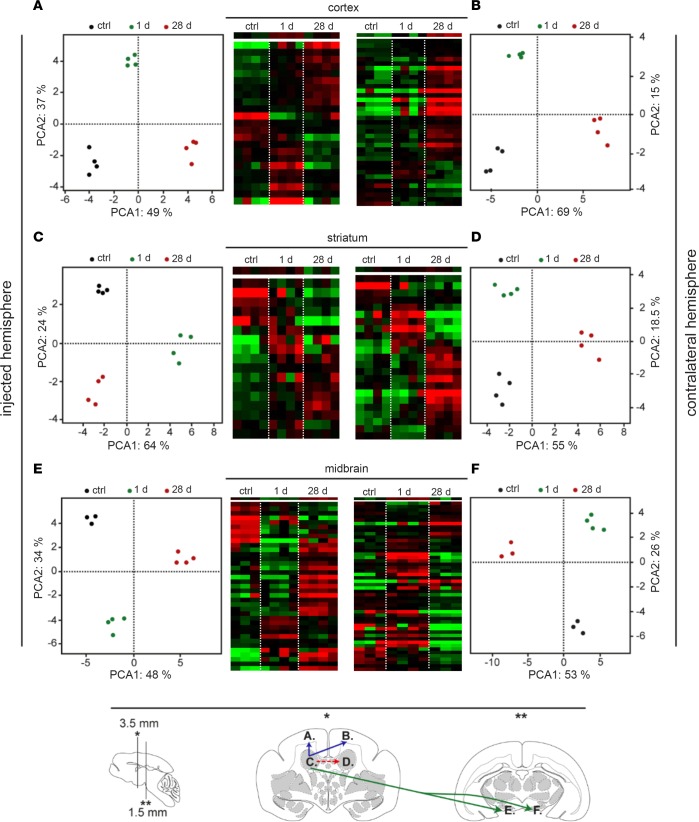
Figure 4. Transcriptional profiles of the most discriminant genes in the striatum, in the frontal lobe, and in the midbrain after HD-GFP injection.
Hierarchical clustering was obtained for each brain region: (A) right frontal cortex; (B) left frontal cortex; (C) injected (right) striatum; (D) contralateral striatum; (E) right midbrain; and (F) left midbrain. Each column corresponds to one animal, whereas each row corresponds to one gene (red: overexpressed genes; green: downregulated genes; black: genes without expression changes). Principal component analysis (PCA) of the 19–45 genes sorted by SAM or by ANOVA for each brain region analyzed. The x axis corresponds to the principal component 1 and the y axis to the principal component 2 (expressed in percentages). Projection of the individuals shows that each group of animal can be distinguished from the other irrespective of the brain structure. At the bottom of the figure, there is a schematic representation of the M. Murinus brain, indicating vector transport from the injected site, the right striatum, to the contralateral side (red hatched arrow), to the frontal cortex (blue arrow), and to the midbrain (green arrow). Brains were collected at 1 or 24 days after injection.
At 1 day after injection, transcriptional changes were detected in the 3 structures. By comparing sham-operated controls, injected caudate nuclei, and the contralateral caudate nuclei, we identified the differential expression of 17 genes in the striatum, 19 in the dorsal frontal lobe, and 40 in the midbrain (Supplemental Figure 1 and Supplemental Table 1). Overall, our analyses showed that vector injection induced transcriptional changes in genes involved in innate immunity, intracellular trafficking, and transcriptional regulations in the striatum. The transcriptional changes in the noninjected structures are consistent with the fast (~2 μm/s) retrograde transport of CAV-2 (31) to these regions. At 28 days after injection, approximately 100 genes were differentially expressed in each structure (Supplemental Table 2). In the injected striatum, 13 of the 19 modified transcripts suggested an initiation of an adaptive immune response. In contrast to those in the striatum, the transcripts in the frontal lobe of the injected hemisphere were similar to those found at day 1. In the midbrain of the injected hemisphere, 45 genes involved in a variety of functions, including trafficking and signaling, were differentially expressed. These data suggest that the transcriptional changes were linked to GFP expression.
Hierarchical clustering was used to identify gene families that changed in the frontal lobe, striatum, and midbrain as a function of the time after injection. Importantly, each group of genes displayed a distinct profile depending on the structure and the time after injection (Figure 4). Again, this pattern parallels the retrograde transport of HD-GFP from the caudate nucleus and the expression of GFP. We then applied a principal component analysis (PCA) to find patterns in the 6 groups (2 times × 3 structures) (Supplemental Tables 1 and 2). We found that two principal components were able to explain approximately 85% of the total information in the injected and left frontal cortex, 88% in the injected striatum, 73% in the contralateral striatum, and approximately 80% in the right and left midbrain. The variance inside each group did not result in an overlap among the groups, consistent with the different response following vector injection. This transcriptional profile suggests that HD-GFP injections may have induced an aborted innate immune response immediately after injection. Importantly, neurons are very poor antigen-presenting cells and cannot, to the best of current knowledge, initiate a T cell response (33). Because we see GFP expression for at least 6 months, this is consistent with the lack of an efficient vector- or transgene-specific immune response.
LRRK2G2019S expression leads to parkinsonian-like motor symptoms in M.
murinus. The above data provided an impetus for modeling PD in M. murinus. We therefore generated HD CAV-2 vectors expressing LRRK2 (HD-LRRK2) or LRRK2G2019S (HD-LRRK2G2019S) (see Supplemental Figure 2A for schema of vector). Initially, two animals were injected in the caudate nucleus with HD-LRRK2G2019S and killed 15 days after injection, and brain sections were incubated with anti-LRRK2 antibodies. We detected LRRK2 immunoreactivity in cells with neuron-like morphology in the injected hemisphere (Supplemental Figure 2B). By contrast, the rare LRRK2-immunoreactive cells in the contralateral hemisphere had astrocyte-like morphology. Interestingly, there was also a global and homogenous Iba1 and BRCA1 immunoreactivity throughout the injected caudate nucleus and putamen at 15 days after injection (Supplemental Figure 3) that was no longer detectable 6 months after injection. This widespread microglia immunoreactivity is inconsistent with a response against the vector injected into the caudate nucleus and suggests a LRRK2G2019S-induced microglia response.
The cardinal characteristics of PD are motor deficits, including bradykinesia, akinesia, rigidity, and resting tremor. We therefore tailored behavior assays for M. murinus to determine whether LRRK2 or LRRK2G2019S expression leads to parkinsonian-like motor symptoms. Prior to vector injections, the cohorts were subjected to hourglass, tower, and 6-tube tasks to obtain baseline readings (each animal served as its own control). Following the training, vectors were injected in the hemisphere controlling their dominant hand. The animals were assayed for motor control every week for 6 months after injection.
The hourglass assay consists of placing the animal in a transparent cylinder, inverting the cylinder, and measuring the time the animal needs to right itself. We found that GFP expression did not affect motor coordination compared with baseline performance. However, LRRK2 expression caused significant (P < 0.05) impairment, with a return to baseline scores after month 3 (see Figure 5A for cumulative data and Supplemental Figure 4A for group data at each week for 6 months). LRRK2G2019S expression induced significant (P < 0.05) delays in the ability of the animals to right themselves throughout the trial, and these delays were greater (P < 0.05) than those induced by LRRK2 expression.
Figure 5. Behavior analyses following GFP, LRRK2, or LRRK2G2019S expression.
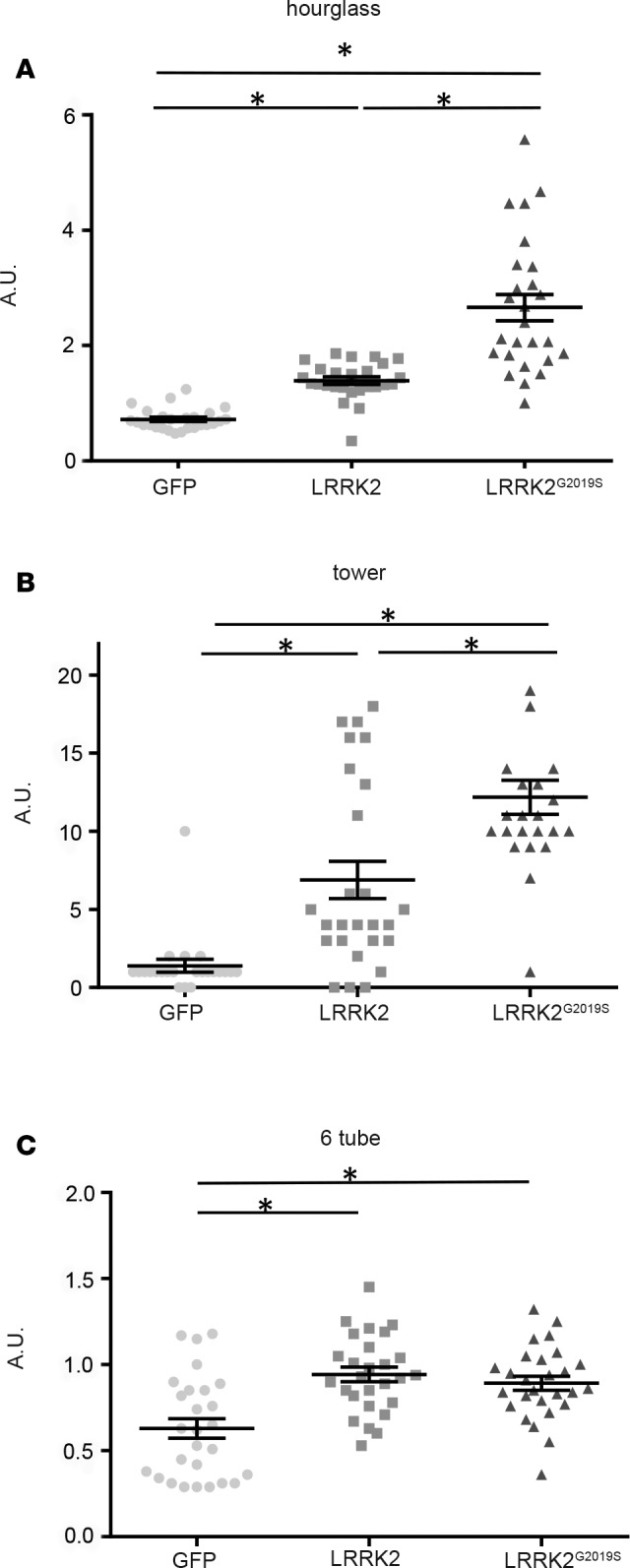
(A) Hourglass test (4 GFP-, 4 LRRK2-, and 8 LRRK2G2019S-expressing animals). (B) Tower task (6 GFP-, 4 LRRK2-, and 8 LRRK2G2019S-expressing animals). (C) Six-tube task (2 GFP-, 4 LRRK2-, and 8 LRRK2G2019S-expressing animals). Data are expressed as mean ± SEM. Nonparametric Kruskal-Wallis test: *P < 0.05. AU, arbitrary units; LRRK2, leucine-rich repeat kinase 2.
The tower task consists of placing the animal at the bottom of a tower with spiral rungs that are progressively further apart. The incentive to climb the rungs is fruit at the top. The performance of GFP-expressing animals was equal to that in their preinjection trials. However, LRRK2 and LRRK2G2019S expression significantly (P < 0.05) increased the time needed to reach the top compared with GFP expression (Figure 5B for cumulative data and Supplemental Figure 4B for group data at each week for 6 months). Moreover, while LRRK2 expression only caused increased climbing times, LRRK2G2019S expression also caused akinetic and bradykinetic responses, which resulted in significantly longer climbing times (P < 0.05) compared with those after LRRK2 expression.
The 6-tube task measures unconstrained manual dexterity. During the trial, the animal retrieved a raisin from inside 1 of 6 horizontal tubes. All animals showed a decreased response (longer times) immediately after injection (Supplemental Figure 4C). Thereafter, HD-GFP–injected animals improved their performance during the trial. By contrast, LRRK2 and LRRK2G2019S expression led to significantly (P < 0.05) slower recovery (Figure 5C). It is worth noting that M. murinus can have a dominant hand and that injections were unilateral. In most cases, LRRK2 and LRRK2G2019S expression caused animals to use their nondominant hand to retrieve raisins. However, we were unable to accurately quantify these events.
LRRK2G2019S expression leads parkinsonian-like histological anomalies.
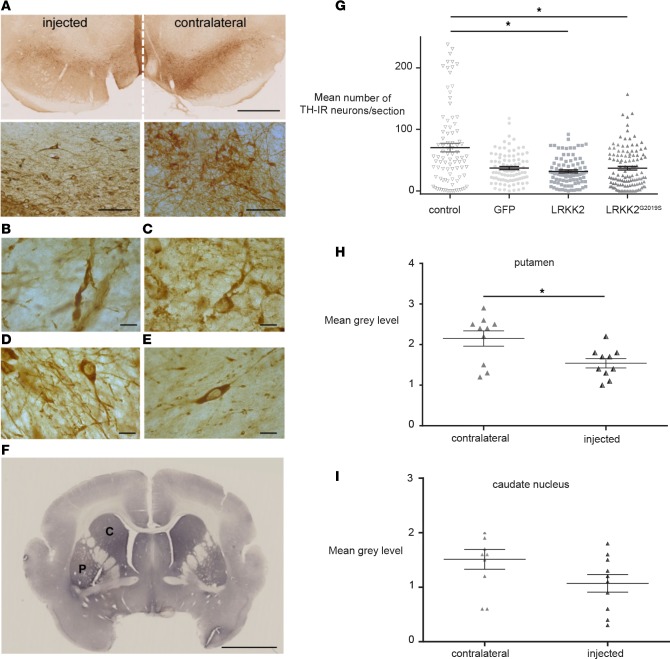
A key histological hallmark of PD is the progressive loss of DA neurons. To determine whether LRRK2 or LRRK2G2019S expression induced the degeneration of nigrostriatal DA neurons, we assayed TH immunoreactivity in brain sections from animals that were killed 6 months after injection. Compared with the contralateral hemisphere, LRRK2G2019S expression caused a striking loss of TH immunoreactivity, dystrophic neurites, and swollen axons in the SNpc of the injected hemisphere (Figure 6, A–F). By comparing the different cohorts, we found that LRRK2 and LRRK2G22019S expression caused a significant (P < 0.05) decrease in the number of TH+ nigral neurons (Figure 6G).
Figure 6. Changes in TH cell characteristics following LRRK2G2019S expression.
Representative images of (A) tyrosine hydroxylase (TH) immunoreactivity (dark brown staining) in a coronal section of the midbrain of LRRK2G2019S-expressing M. murinus (n = 8 animals). High-magnification images of TH+ cell bodies and projections in the hemisphere expressing LRRK2G2019S (left) versus the contralateral hemisphere (right) are shown below. Scale bars: 1 mm (top); 50 μm (bottom). (B) Swollen neurites and (C) dystrophic neurons due to LRRK2G2019S expression. (D) Neurons in control animals (n = 2). (E) Loss of neurites and (F) TH immunoreactivity due to LRRK2G2019S expression. Scale bars: 2.5 mm. (G) Quantification of TH+ cells in the substantia nigra of the LRRK2G2019S-expressing hemisphere versus controls (n = 3 mock-, 5 HD-GFP–, 7 HD-LRRK2G2019S–, and 4 HD-LRRK2-injected animals). TH+ neurons were quantified by counting cells in 1 of every 6 sections. The y axis represents the average number of TH+ neurons/section in all groups. Mann-Whitney test: *P < 0.05. (H) optical density of TH immunoreactivity in the putamen (n = 4 animals). (I) Optical density of TH immunoreactivity in the caudate nucleus (n = 3 animals). Data in H and I are expressed as mean ± SEM. Nonparametric Kruskal-Wallis test: *P < 0.05. C, caudate nucleus; P, putamen.
We then reasoned that, if the number of DA neurons were reduced in the injected hemisphere, this should correspond to a reduction in the density of TH+ nerve terminals. We therefore quantified TH immunoreactivity in sections containing the caudate nucleus and putamen from LRRK2G2019S-expressing animals. Consistent with the reduction in DA neurons, we found a significant (P < 0.05) reduction in TH immunoreactivity in the injected putamen (Figure 6H), but not in the caudate nucleus (Figure 6I), compared with the contralateral hemisphere. To further examine the effect of LRRK2G2019S expression on the nigrostriatal DA pathway, the levels of striatal dopamine and its metabolites 3,4-dihydroxyphenylacetic acid, homovanillic acid, 5-hydroxytryptamine, 5-hydroxyindoleacetic acid were quantified. However, LRRK2G2019S expression did not lead to lower levels of dopamine or its metabolites at 6 months after injection (Supplemental Figure 5).
Finally, we asked if LRRK2G2019S expression (Supplemental Figure 6A) was associated with tau or α-synuclein phosphorylation. Compared with contralateral hemispheres, we found increases in α-synucleinpS129 and phospho-tauSer396 immunoreactivity following LRRK2G2019S expression at 6 months after injection (Supplemental Figure 6, B and C, respectively).
Discussion
Because gene transfer to the human brain will be irreversible, this therapeutic approach creates unique challenges — in particular the need to address vector tropism, feasibility, efficacy, safety, and duration of expression. In this study, we evaluated the efficacy of CAV-2 vectors in the NHP brain as a prelude to expressing PD-causing proteins therein. We demonstrated robust transgene expression, duration, and biodistribution coupled with minimal transcriptional effect and immunogenicity in the M. murinus brain. Our results are also consistent with the use CAR, either on the soma or at the synapse, by CAV-2 to preferential infect neurons and be transported to efferent structures (14). We demonstrate that LRRK2G2019S expression leads to parkinsonian-like motor deficits and histological lesions, including the loss of TH+ neurons in the SN, a decrease of the density of TH+ fibers, the generation of swollen axons and dystrophic neurons, and the phosphorylation of tau and α-synuclein. We conclude that LRRK2G2019S expression in the M. murinus induced PD-associated motor symptoms and histological characteristics.
The M.
murinus as a PD model. The lack of relevant animal models that faithfully recapitulate human brain diseases has slowed the development of therapies. An ideal PD animal model should (a) have a normal complement of DA neurons at birth; (b) have more than 50% of these neurons be selectively and gradually lost in adulthood; (c) have motor deficits, preferably bradykinesia, rigidity, and resting tremor; (d) be based on a single mutation to allow robust propagation and facilitate crossing with enhancer or suppressor strains; and (e) have a disease course of a few months to allow rapid and inexpensive screening of therapeutic agents (12). Transgenic mice have often disappointed, possibly due to functional compensation during development, species-specific differences in disease mechanisms, immune responses, and/or in the anatomical organization (34, 35). Differences in general behavior and responses to common cognitive tests also make assays in rodents challenging for clinical evaluations. Therefore, studies using NHPs represent a “step to human” where discoveries could be readily translated into therapies (22, 36). However, the number of NHPs available for biomedical research, such as baboons, macaques, or marmosets, is limited, under increased regulatory regulations, and expensive (37). Conversely, self-sustaining breeding M. murinus colonies could provide a sufficient number of animals for trials that require large numbers to reach statistical significance. The M. murinus is small nocturnal primate, that is biologically, behaviorally, and socially much closer to humans than rodents. The M. murinus brain also has a structure and organization readily comparable to that of human brain. M. murinus have been maintained and bred successfully in captivity since the 1960s, with life span reaching >10 years; and this has contributed to the protection and the biodiversity of the wild populations. M. murinus reach maturity within the first year of life and have a relative high fecundity for a NHP (gestation, 2 months; 1–3 offspring/litter; weaning at 2 months; pubertal age, 6–8 months) (37). M. murinus are increasingly used to understand aging and Alzheimer’s disease (including amyloid-β vaccination) (38), MRI evaluation of cerebral atrophy (39), and identification of cognitive deficits (39, 40). In addition, because of captive breeding programs, M. murinus are one of the few NHPs approved by current European regulation. Finally, while drug-induced PD models (e.g., 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine or 6-hydroxydopamine) (41, 42) have significantly advanced our understanding of PD, they poorly recapitulate chronic and slow disease progression.
Our primary goals in delivering HD-LRRK2 and HD-LRRK2G2019S were to determine if overexpression leads to parkinsonian-like motor and histological lesions in NHPs. It is clear that the LRRK2G2019S M. murinus model fulfills many of the criteria needed to help advance our understanding of, and develop and validate therapies for, PD. Moreover, striatal injection of CAV-2 vectors could also allow one to better understand the complex circuitry in PD: for example, what is the role of pruning and regrowth of striatal projecting neurons into the basal ganglia? Does pruning and regrowth effect synaptic transmission? We found that unilateral injections in the caudate nucleus unexpectedly exhibited some cyclic motor deficits (Supplemental Figure 4). While unilateral injections allowed for internal comparison between hemispheres, they likely reduced the inception and maintenance of parkinsonian-like symptoms. It is possible that the cyclic motor deficits were due to compensation via the crosstalk between the SN and striata. Given the presence of cross-hemispheric DA neuron projections (43), M. murinus injected with HD-LRRK2G2019S could help address their functional significance. In addition, unilateral injections of HD-LRRK2G2019S may help us understand how the brain adapts during PD. For example, can cross-hemispheric DA neurons rescue unilateral striatal dysfunction, as suggested by our data? Are cross-hemispheric DA neuron projections more pronounced in primates than in rodents? In rats the current estimate is that approximately 5% of the neurons project to the contralateral hemisphere (44). It is reasonable to assume that the percentage is higher in the primate brain.
LRRK2G2019S expression induced parkinsonian-like histological changes in the DA neurons in the SN. It is encouraging that LRRK2G2019S expression induced a markedly decreased number of DA cells within 6 months, given that the LRRK2G2019S familial form of PD typically does not manifest itself until the fourth or fifth decade. The lack of significant difference in dopamine metabolite concentrations at 6 months after injection could be due to compensatory resprouting of the DA neuronal processes from the SNpc or from neurons from other nuclei/regions that project to the striatum. Our results are consistent with reports describing the association of fragmented axons with axonal spheroids and dystrophic neurites in LRRK2R1441G-transgenic mice (45) and other pathological changes in cell body and axons of transgenic mice overexpressing LRRK2G2019S (46). Axon perturbation could be due to LRRK2’s role in the regulation of Rab phosphorylation and neurite outgrowth (47).
Although LRRK2G2019S expression appears to be critical for the development of α-synuclein–induced neuropathology in mice, no LRRK2 transgenic model has developed α-synuclein inclusions. These observations suggest that α-synuclein produced at physiological levels in rodents is not affected by LRRK2 overexpression (48). Notably though, as primates age they produce greater amounts of endogenous α-synuclein in SN neurons, making them more susceptible to protein aggregates and degeneration. We are unaware of studies that compare endogenous levels of α-synuclein in rodents and primates. Here, we found that LRRK2G2019S expression led to increased α-synucleinpS129 as well as phospho-tauSer396 immunoreactivity. Tau binds tubulin to stabilize microtubules and promotes tubulin assembly into microtubules. The maintenance of axon morphology and transport of molecules and organelles depends on microtubule stabilization by tau, and altered tau function could block transport of organelles, neurofilaments, and vesicles (49). The presence of phospho-tau is a common feature in LRRK2 model systems (45, 47, 50) and is consistent with observations that LRRK2 mutations can be associated with tauopathies lacking α-synuclein Lewy bodies (51, 52).
CAV-2 vector characteristics.
Neurons in distal structures can readily take up CAV-2 vectors. It is likely that more vector is taken up by some structures due to the density of their projections near the injection site. This is likely one of the reasons that when CAV-2 vectors are injected into the striatum they efficiently infect DA neurons in the SN. Because the approximately 100-nm neutral-charged CAV-2 capsid (53) poorly diffuses in the brain parenchyma, it is likely that neurons are capable of continually taking up vector particles for an extended period, possibly for several days. In addition, axonal and synaptic CAR levels may also influence vector uptake from many regions and neuron subtypes. The transcriptional changes after HD-GFP injection were unique for each structure and at each time point, suggesting a dynamic process influenced by vector transport. The initial innate immune response is consistent with in vitro data obtained using human midbrain neuroprogenitors following HD-GFP infection (32). However, long-term transgene expression demonstrates that this transcriptional response does not lead to a notable adaptive immune response. Indeed, the transcriptional response at 28 days after injection suggested an evolution from an innate immune response toward tolerance. An adaptive response would upregulate IgMs, which have important roles in primary defense mechanisms; immunoglobulin λ-like polypeptide (IGLL3); HLA-DQA1 and HLA-DRB1, belonging to MHC II complex; and IGHA1, the major Ig class playing a role in the defense against local infection. By contrast, tolerance involves an upregulation of HLA-G, which is involved in antigen presentation and mediates protection against NK cells, cytotoxic T lymphocytes, and macrophages. Together, these data are consistent with the long-term CAV-2 vector–mediated transgene expression observed in the NHP brain. These data also set the stage for transcription-based analyses of the cell autonomous and nonautonomous effect of LRRK2G2019S expression by neurons in vivo. For example, characterizing the transcriptional networks induced by LRRK2G2019S in DA neurons of the SN could help identify therapeutic targets, as it has recently done with the PBX1 transcriptional network impaired in a mouse model of PD (54).
In conclusion, our results have notable fundamental and clinical implications in the context of modeling, preventing, or treating PD. While this study was focused on PD, numerous diseases may be amendable to modeling using CAV-2 vectors, regardless of whether they affect specific nuclei or the entire brain.
Methods
Animals
The M. murinus colony is housed at the primate facility at the University of Montpellier (license approval 34-05-026-FS). All procedures were in compliance with the rules of the European Community Council Directive (2010/63/EU) for the care and use of laboratory animals. Forty-seven animals were used in this study.
Vectors
Construction, purification, and storage of HD-GFP, a HD CAV-2 vector expressing GFP, have been previously described (16). Codon-optimized LRRK2G2019S cDNA, tagged at the N-terminus with a 3×FLAG epitope tag (initially provided by C. Ross, Johns Hopkins University, Baltimore, Maryland, USA) (55) is followed by an internal ribosome entry site sequence (IRES from pIRES, Clontech) and GFP, was cloned into a pGut-3 and then inserted into a pEJK25 containing the HD genomes as previously described (16). HD vectors used during this study were approximately 5 × 1011 physical particle (pp)/ml with an infectious to pp ratio of approximately 1:20.
Stereotaxic injections
Twenty-seven M. murinus (7 naive animals without injection, 6 control HD-GFP, 4 HD-LRRK2, and 10 HD-LRRK2G2019S) were used for behavioral and/or biogenic amine measurement and neuropathological analysis. Animals were anesthetized with 80 mg/kg ketamine/10 mg/kg xylazine by intramuscular injections. The animals were positioned in the stereotactic frame, an incision was made in the scalp, and a small hole was drilled unilaterally in the skull according to coordinates in M. murinus brain atlas (56). Needles were connected to a 10-μl Hamilton syringe and inserted into the putamen using the following coordinates: anterior, 6.5 mm from intra-aural line; lateral, 3 mm and –7 mm dorsoventral relative to the skull surface (56). HD-GFP, HD-LRRK2, or HD-LRRK2G2019S (109 pp of each vector, approximately 2 μl) was injected at a rate of 0.5 μl/min. The needle was left in place for additional 5 minutes before being slowly withdrawn. The skin was sterile sutured, and the M. murinus were monitored until they recovered from anesthesia. After surgery, primates were isolated in cage with food and water ad libitum.
Biodistribution study
Approximately 1 × 109 pp of HD-GFP were injected into the right caudate nucleus (anterior to inter-aural zero, +4.5 mm; lateral, +1.7 mm; ventral, –4.5 mm). Mock-treated animals were injected with carrier (PBS without Ca++/Mg++ and 10% glycerol) using mirror coordinates in the left hemisphere to control for surgery and injections. After analyses, we found that using these coordinates in a couple of female M. murinus that some vector was deposited in the lateral ventricle. After injections, the NHPs were isolated in ventilated cages with food and water ad libitum, checked twice a day for 3 days, and checked then daily for the duration of the protocol. The vast majority of animals appeared to recover completely from the surgery within 24 hours, and all recovered by 48 hours. All animals retained their pre-op weight and ate normally by 24 hours. All animals were sacrificed on the planned dates (Table 1).
Transcriptome analysis
Twelve 1-year-old female M. murinus were used for transcriptome experiments: 4 animals were used as sham-operated controls, 4 animals were injected with 109 pp of HD-GFP in the right and left caudate nuclei (mirror coordinates) and killed 1 day after injection, and 4 animals were injected in the right and left caudate nuclei and sacrificed 28 days after injection. After injections, primates were isolated in ventilated cages with food and water ad libitum. After euthanasia, each brain was cut in half along the midline. The right and the left hemispheres were dissected in 25- to 30-mg fragments corresponding to the principal anatomical regions and then were flash-frozen and temporarily stored at –80°C. All animals were sacrificed on the planned dates (Table 1).
RNA extraction and microarrays
Six regions were assayed: the right and left frontal cortices, the right and left striata, and the right and left midbrain. Total RNA was extracted from each region (25–30 mg) using the RNeasy Mini Kit (Qiagen) according to the manufacturer’s protocol. The quality of each purified RNA sample was checked using a NanoDrop Agilent BioAnalyzer (Agilent Technologies). Gene expression was detected by hybridization with human genome gene chips using an Affymetrix HG133 Plus 2. The processing and the data analysis of the brain samples were previously described (22). Briefly, total RNA was extracted from each brain sample and prepared for hybridization with the gene chips according to the manufacturer’s protocol and using Microarray Affymetrix Suite 5.0 to normalize and analyze the intensity value of each probe set. The primary expression files (*.cel) and (*.xls) are available at the GEO/NCBI repository (accession GSE102640, GSE102708, and GSE102709). Affymetrix microarray data were processed at the Microarray Core Facility of the Institute of Research of Biotherapy of Montpellier. Data were analyzed with the significance analysis of microarrays (SAM) method (57), which calculated a q value, i.e., the lowest false discovery rate, according to the Δ cutoffs. The differential expression of genes was also assessed by using ANOVA, Student’s 1-tailed t test, and PCA with the “R” software. The gene transcripts were assumed present in the brain sample when they were detected at least 3 times in all the brain samples (58).
Gene profiling and biological functions and pathways
The gene expression profile of each brain region (frontal cortex, striatum, and midbrain) and at the 2 time courses (1 and 28 days) were established using centered Pearson’s correlation (https://www.r-project.org/) and the Cluster and Treeview software programs (59). The involvement of the differentially expressed genes in biological functions, cell activities, and signaling pathways was assessed using the following databases: Ensembl (http://www.ensembl.org/index.html), Uniprot (https://www.uniprot.org/), Gene Ontology (http://www.geneontology.org/), KEGG (https://www.genome.jp/kegg/), and NCBI (https://www.ncbi.nlm.nih.gov/). The biological functions and pathways were also analyzed with the QIAGEN Ingenuity Pathway Analysis software (IPA; https://www.qiagenbioinformatics.com/).
Biodistribution of HD-GFP
Tissue collection, preparation, and analysis.
Primates were anesthetized with ketamine (80 mg/kg) and perfused transcardially with heparinized saline (0.9%) followed by 4% PFA in PBS, pH 7.4. Brains were post-fixed in 4% PFA and soaked in 30% sucrose in PBS, pH 7.4, at 4°C for at least 24 hours. Brains were embedded in OCT and cut rostrocaudally. One 50-μm-thick section and then three 16-μm-thick sections were cut. The 50-μm-thick sections were mounted with Vectashield H-1000 (AbCys) to assay for GFP expression. The 16-μm-thick sections were used for colorations (hematoxylin/eosin, Luxol fast blue/cresyl violet, Kluver, and Barrera) and immunohistology. Primary antibodies included anti-NeuN (Chemicon, clone A60), anti-GFAP (Dako, Cytomation), anti-TH (MilliporeSigma, Ab152), anti-Tuj-1 (R&D Systems, NL1195G), anti-CAR (R&D anti-human CXADR), and anti–α-synuclein antibody (T. Baron, Unité Maladies Neurodégénératives, Lyon, France). Synaptosomes were prepared as previously described (60). Floating 16-μm-thick sections were incubated with rabbit polyclonal anti-CAR (CAR-72) 1:300 in PBS containing 3% BSA and 0.3% TX-100. Secondary antibodies, anti-mouse Alexa Fluor 647 and anti-rabbit Alexa Fluor 555 (Molecular Probes), were diluted 1:300 in PBS containing 3% BSA and incubated for 1 hour at room temperature. After washing, floating sections were mounted in fluorescent mounting medium (DAKO).
Image acquisition and analysis.
Images were acquired by using an upright Zeiss laser scanning confocal microscope (Axiovert 100M) with LSM 510 (version 2.5) software. GFP and Alexa Fluor 488 signals were excited at 488 nm, and fluorescent emission was collected using a BP 505- to 550-nm filter; rhodamine, Cy3, and Alexa Fluor 555 signals were excited at 543 nm and were collected using a BP 565- to 615-nm filter. Cy5 and Alexa Fluor 647 signals were excited at 600 nm and collected using a BP 640- to 660-nm filter. A spectral analysis was used to identify GFP-specific signals to avoid false positive signal.
Quantification of neurons in the SNpc.
Brains from noninjected animals were fixed in 4% PFA, embedded in OCT, and cut into 16- or 50-μm-thick serial sections. The 16-μm-thick sections were stained with Luxol fast blue/cresyl violet, and neurons in the SN were quantified using stereology software (Mercator, Explora Nova). The 50-μm-thick sections were incubated with an anti-TH antibody and revealed using a fluorescent secondary antibody. Unbiased stereological method Mercator Pro software was used every 120 μm to quantify DA neurons in the SNpc.
Quantification of vector genomes
Brains were initially divided into left and right hemispheres. Each hemisphere was then dissected into frontal, parietal, temporal, and occipital cortex; olfactory bulb; basal nuclei (including the striatum); thalamus and hypothalamus; midbrain; and cerebellum. The tissue was dissociated mechanically, and total DNA was purified using a NucleoBond kit (Macherey-Nagel). qPCR standards were generated using dilutions of vector DNA and mixed with genomic DNA extracted from a noninjected M. murinus brain. Two primer sets were used, GFP 5′-CAGAAGAACGGCATCAAGGT-3′ (base pairs 472–491) and 5′-CTGGGTGCTCAGGTAGTGG-3′ (base pairs 597–615), for vector amplification; additionally, GAPDH, 5′-ACAGTCCATGCCATCACTGCC-3′ and 5′-GCCTGCTTCACCACCTTCTTG-3′, and β-actin, 5′-CTGCAGAATTCCAAAGGAG-3′ and 5′-CCAACCGTGAGAAGATGAC-3′, were used to normalize for genomic DNA copy number. qPCR was performed using the Light Cycler (Roche) and SYBR Green PCR kit (Qiagen) according to the manufacturers’ instructions. Data are expressed as a ratio to the genomic copies of GAPDH or β-actin.
Behavior analyses
Prior to injections, the animals were trained to perform the 3 motor tasks. We considered the performance before injection as the basal level (100%). Behavior analyses were performed every week for 6 months after injection.
Hourglass test.
This task was used to evaluate quantitatively the animal’s righting reflex as a measure for rigidity analogous to axial motor behavior (61). The animal was placed in a narrow Plexiglas cylinder (6 × 18 cm). One trial consisted of one natural 180-degree turn of the cylinder. After 30 seconds, a new trial started by turning the cylinder again. A test consisted of 5 subsequent trials. Animals were always turned upright after 30 seconds (e.g., when they had not turned upright). We measured the time taken by the M. murinus to return to the upright position. Maximum time noted was 30 seconds (if animals did not turn upright at all). The mean of the 3 fastest turns was used for the analysis. Because of the individual variability, a recovery index was calculated for each animal.
Tower test.
This task consisted of a tower with 6 levels of rungs, where the M. murinus have to jump from the ground to the seventh level with increasing difficulty. This test quantifies the animal jumping behavior as a measure of akinesia (61). The tower (35 × 35 × 180 cm) has a transparent Plexiglas front. It contains 6 levels of horizontal crossbars with an increasing distance varying from 10 to 50 cm to access the nest box. A camera was placed in front of the tower for video analysis. The M. murinus was placed in front of a sliding door at the bottom of the tower. During each test the mouse lemur could freely move around the 6 levels for 5 minutes. To motivate the animal, a reward was placed into the nest box. All animals were habituated to the tower before testing. A masked observer recorded the time needed to get into the nest box.
Six-tube task.
This task measures the unconstrained exploration of its peripersonal space (62). For each trial, the M. murinus was required to search for and retrieve a food reward (a raisin) that was placed inside 1 of the 6 tubes that were presented to the animal in a horizontal array. The animals had to jump on a small platform to reach the reward. The latency to retrieve the reward was recorded. Once the reward had been retrieved, or 30 seconds had passed, the array of tubes was removed and another tube was baited. The task consisted of a total of 30 trials, with the location of the reward changing in a fixed random order, such that each tube was baited a total of 5 times. The M. murinus had been pretrained on this task and was free to use either hand. The pretreatment score consisted of the time to remove all the rewards. Hand preference was also determined before surgery.
Histology
After the 6-month behavioral follow-up, the M. murinus were deeply anesthetized with ketamine (80 mg/kg) and perfused with saline followed by 4% PFA in 0.1 M phosphate buffer (pH 7.4). After post-fixation for 12 hours in 4% PFA and cryoprotection in 30% sucrose solution, brains were cut into 40-μm-thick coronal sections.
Immunohistochemistry and quantitative analysis
Coronal sections (40 μm thick) from brains were prepared and processed for immunohistochemistry with rabbit anti-TH antibody (Institute Jacques Boy) and anti-LRRK2 antibody (Abcam, ab133474), anti-tau-pSer396 (Novus Biologicals, NB100-92652), anti–α-synuclein (pS129) (Abcam, ab59264), anti-BRCA1 (Santa Cruz Biotechnology, SC642), and anti-Iba1 (Wako, NCNP24) (63). Briefly sections were quenched of endogenous peroxidase activity, permeabilized, and incubated with primary antibodies. Sections were processed with biotinylated anti-rabbit IgG or anti-mouse IgG antibodies and avidin-biotin complex coupled to horseradish peroxidase (Vectastain ABC, Vector Laboratories) and visualized with 3,3′-diaminobenzidine reagent (Vector Laboratories). Sections were mounted on Superfrost slides, dehydrated with increasing ethanol concentrations and xylene, and then covered using Permount (Thermo Fisher Scientific). Mercator pro software was used on every sixth section, a sampling adapted to the studied structure to count TH+ neurons in the left and right SNpc. Analyses was performed by a masked observer.
Reagent availability
CAV-2 vectors are available via the nonprofit plateforme de vectorologie de Montpellier.
Statistics
Data distribution and within-group variation were preliminarily assessed to guide our methodological choices. Nonparametric tests Kruskal-Wallis and Dunn’s multiple comparison tests were used to compared the 3 unmatched groups HD-GFP, HD-LRRK2, and HD-LRRK2G2019S, and comparison of contralateral and injected sides was added for each group for dopamine data. Changes were considered significant when P < 0.05. Data were plotted as mean ± SEM. During behavioral tests, animals that refused to perform the tests were not included for analyses. For quantitative TH analysis, 2 animals were excluded because entire the brain sections were not available. All statistical tests were verified by the bioinformatics service at CNRS BioCampus (SERANAD, http://http://www.igmm.cnrs.fr/en/service/service-danalyse-de-donnees-biologiques-complexes/).
Study approval
The studies in animals were reviewed and approved by the ethical committee of the Languedoc-Roussillon, France (CEEA-LR-1013, -1014, and -1015).
Author contributions
NMF, JMV, and EJK developed the study concept and design, supervised the study, and critically revised the manuscript. NS, AG, JD, BLS, and EJK designed and produced viral vectors. NMF performed primate surgery. NMF, SC, SGT, PF, and JMV performed the behavior test. NMF, NS, SC, SGT, PF, JMV, and EJK performed data analyses. CZ, VK, NMF, PF, and EJK acquired, analyzed, and interpreted histological data. PDD, PF, and NMF acquired, analyzed, and interpreted metabolite data. NMF, FJDM, NS, and EJK acquired, analyzed, and interpreted immunological data. GD, IS, and JMV acquired, analyzed, and interpreted transcriptome data. NS performed DNA extraction and PCR data analysis. PF and NMF performed statistical analyses. CZ and SS performed immunoblotting for CAR in synaptosomes. MRLP provided reagents and expertise in clinical PD physiology. NMF, NS, GD, JMV, and EJK wrote the manuscript.
Supplementary Material
Acknowledgments
We thank the members of our laboratories, in particular M. Lavigne and P. Aebischer, for constructive comments and help with figures throughout this study. We thank S. Rouland, J. Cuoq, and E. Huetter for their invaluable help with the M. murinus colony, T. Gostan (SERANAD) for help with statistical analyses, MGX, and the RHEM-Institute of Neurosciences of Montpellier platforms. We thank T. Baron for anti–α-synuclein antibody. We acknowledge the MRI imaging facility, a member of the national infrastructure France-BioImaging. This work was supported in part by France Parkinson (to EJK); BrainCAV (EC FP7 contract 292222) (to EJK, JMV, NMF, and IS); BrainVector (EC FP7 contract 286071) (to EJK and IS); the Université de Montpellier (to EJK and JMV); the Region Languedoc Roussillon (to EJK); Institut de Génétique Moléculaire de Montpellier (to EJK); LabEx EpiGenMed, an Investissements d’avenir program (ANR-10-LABX-12-01) (to EJK); and Mécanismes Moléculaires dans les Démences Neurodégénératives (to JMV).
Version 1. 07/25/2018
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: JCI Insight. 2018;3(14):e98202. https://doi.org/10.1172/jci.insight.98202.
Contributor Information
Nicolas Serratrice, Email: nico.serratrice@orange.fr.
Aurélie Gennetier, Email: aurelie.gennetier@hotmail.fr.
Gina Devau, Email: gina.devau@umontpellier.fr.
Sandra Cobo, Email: sandra_cobo@bio-rad.com.
Stéphanie G. Trouche, Email: stephanie.trouche@gmail.com.
Pascaline Fontès, Email: fontesp@univ-montp2.fr.
Charleine Zussy, Email: charleine.zussy@yahoo.fr.
Philippe De Deurwaerdere, Email: deurwaer@u-bordeaux.fr.
Sara Salinas, Email: sara.salinas@inserm.fr.
Bernard L. Schneider, Email: bernard.schneider@epfl.ch.
Isabella Saggio, Email: Isabella.Saggio@uniroma1.it.
Vasiliki Kalatzis, Email: vasiliki.kalatzis@inserm.fr.
Jean-Michel Verdier, Email: jean-michel.verdier@umontpellier.fr.
References
- 1.Surmeier DJ, Obeso JA, Halliday GM. Selective neuronal vulnerability in Parkinson disease. Nat Rev Neurosci. 2017;18(2):101–113. doi: 10.1038/nrn.2016.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gloeckner CJ, et al. The Parkinson disease causing LRRK2 mutation I2020T is associated with increased kinase activity. Hum Mol Genet. 2006;15(2):223–232. doi: 10.1093/hmg/ddi439. [DOI] [PubMed] [Google Scholar]
- 3.Steger M, et al. Phosphoproteomics reveals that Parkinson’s disease kinase LRRK2 regulates a subset of Rab GTPases. Elife. 2016;5:e12813. doi: 10.7554/eLife.12813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Healy DG, et al. Phenotype, genotype, and worldwide genetic penetrance of LRRK2-associated Parkinson’s disease: a case-control study. Lancet Neurol. 2008;7(7):583–590. doi: 10.1016/S1474-4422(08)70117-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo L, Gandhi PN, Wang W, Petersen RB, Wilson-Delfosse AL, Chen SG. The Parkinson’s disease-associated protein, leucine-rich repeat kinase 2 (LRRK2), is an authentic GTPase that stimulates kinase activity. Exp Cell Res. 2007;313(16):3658–3670. doi: 10.1016/j.yexcr.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ito G, et al. GTP binding is essential to the protein kinase activity of LRRK2, a causative gene product for familial Parkinson’s disease. Biochemistry. 2007;46(5):1380–1388. doi: 10.1021/bi061960m. [DOI] [PubMed] [Google Scholar]
- 7.Li X, Tan YC, Poulose S, Olanow CW, Huang XY, Yue Z. Leucine-rich repeat kinase 2 (LRRK2)/PARK8 possesses GTPase activity that is altered in familial Parkinson’s disease R1441C/G mutants. J Neurochem. 2007;103(1):238–247. doi: 10.1111/j.1471-4159.2007.04743.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith WW, Pei Z, Jiang H, Dawson VL, Dawson TM, Ross CA. Kinase activity of mutant LRRK2 mediates neuronal toxicity. Nat Neurosci. 2006;9(10):1231–1233. doi: 10.1038/nn1776. [DOI] [PubMed] [Google Scholar]
- 9.Taymans JM, Van den Haute C, Baekelandt V. Distribution of PINK1 and LRRK2 in rat and mouse brain. J Neurochem. 2006;98(3):951–961. doi: 10.1111/j.1471-4159.2006.03919.x. [DOI] [PubMed] [Google Scholar]
- 10.Simón-Sánchez J, Herranz-Pérez V, Olucha-Bordonau F, Pérez-Tur J. LRRK2 is expressed in areas affected by Parkinson’s disease in the adult mouse brain. Eur J Neurosci. 2006;23(3):659–666. doi: 10.1111/j.1460-9568.2006.04616.x. [DOI] [PubMed] [Google Scholar]
- 11.Melrose HL, et al. A comparative analysis of leucine-rich repeat kinase 2 (Lrrk2) expression in mouse brain and Lewy body disease. Neuroscience. 2007;147(4):1047–1058. doi: 10.1016/j.neuroscience.2007.05.027. [DOI] [PubMed] [Google Scholar]
- 12.Beal MF. Parkinson’s disease: a model dilemma. Nature. 2010;466(7310):S8–10. doi: 10.1038/466S8a. [DOI] [PubMed] [Google Scholar]
- 13.Dawson TM, Ko HS, Dawson VL. Genetic animal models of Parkinson’s disease. Neuron. 2010;66(5):646–661. doi: 10.1016/j.neuron.2010.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Junyent F, Kremer EJ. CAV-2--why a canine virus is a neurobiologist’s best friend. Curr Opin Pharmacol. 2015;24:86–93. doi: 10.1016/j.coph.2015.08.004. [DOI] [PubMed] [Google Scholar]
- 15.Soudais C, Laplace-Builhe C, Kissa K, Kremer EJ. Preferential transduction of neurons by canine adenovirus vectors and their efficient retrograde transport in vivo. FASEB J. 2001;15(12):2283–2285. doi: 10.1096/fj.01-0321fje. [DOI] [PubMed] [Google Scholar]
- 16.Soudais C, Skander N, Kremer EJ. Long-term in vivo transduction of neurons throughout the rat CNS using novel helper-dependent CAV-2 vectors. FASEB J. 2004;18(2):391–393. doi: 10.1096/fj.03-0438fje. [DOI] [PubMed] [Google Scholar]
- 17.Ord EN, et al. Combined antiapoptotic and antioxidant approach to acute neuroprotection for stroke in hypertensive rats. J Cereb Blood Flow Metab. 2013;33(8):1215–1224. doi: 10.1038/jcbfm.2013.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hnasko TS, et al. Cre recombinase-mediated restoration of nigrostriatal dopamine in dopamine-deficient mice reverses hypophagia and bradykinesia. Proc Natl Acad Sci USA. 2006;103(23):8858–8863. doi: 10.1073/pnas.0603081103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giannakopoulos P, et al. Quantitative analysis of tau protein-immunoreactive accumulations and beta amyloid protein deposits in the cerebral cortex of the mouse lemur, Microcebus murinus. Acta Neuropathol. 1997;94(2):131–139. doi: 10.1007/s004010050684. [DOI] [PubMed] [Google Scholar]
- 20.Bons N, Rieger F, Prudhomme D, Fisher A, Krause KH. Microcebus murinus: a useful primate model for human cerebral aging and Alzheimer’s disease? Genes Brain Behav. 2006;5(2):120–130. doi: 10.1111/j.1601-183X.2005.00149.x. [DOI] [PubMed] [Google Scholar]
- 21.Mestre-Francés N, Keller E, Calenda A, Barelli H, Checler F, Bons N. Immunohistochemical analysis of cerebral cortical and vascular lesions in the primate Microcebus murinus reveal distinct amyloid beta1-42 and beta1-40 immunoreactivity profiles. Neurobiol Dis. 2000;7(1):1–8. doi: 10.1006/nbdi.1999.0270. [DOI] [PubMed] [Google Scholar]
- 22.Abdel Rassoul R, et al. Distinct transcriptome expression of the temporal cortex of the primate Microcebus murinus during brain aging versus Alzheimer’s disease-like pathology. PLoS One. 2010;5(9):e12770. doi: 10.1371/journal.pone.0012770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Verdier JM, et al. Lessons from the analysis of nonhuman primates for understanding human aging and neurodegenerative diseases. Front Neurosci. 2015;9:64. doi: 10.3389/fnins.2015.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Freimuth P, Philipson L, Carson SD. The coxsackievirus and adenovirus receptor. Curr Top Microbiol Immunol. 2008;323:67–87. doi: 10.1007/978-3-540-75546-3_4. [DOI] [PubMed] [Google Scholar]
- 25.Loustalot F, Kremer EJ, Salinas S. The intracellular domain of the coxsackievirus and adenovirus receptor differentially influences adenovirus entry. J Virol. 2015;89(18):9417–9426. doi: 10.1128/JVI.01488-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zussy C, Salinas S. Study of adenovirus and CAR axonal transport in primary neurons. Methods Mol Biol. 2014;1089:71–78. doi: 10.1007/978-1-62703-679-5_5. [DOI] [PubMed] [Google Scholar]
- 27.Salinas S, et al. Disruption of the coxsackievirus and adenovirus receptor-homodimeric interaction triggers lipid microdomain- and dynamin-dependent endocytosis and lysosomal targeting. J Biol Chem. 2014;289(2):680–695. doi: 10.1074/jbc.M113.518365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zussy C, et al. Coxsackievirus adenovirus receptor loss impairs adult neurogenesis, synapse content, and hippocampus plasticity. J Neurosci. 2016;36(37):9558–9571. doi: 10.1523/JNEUROSCI.0132-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salinas S, et al. CAR-associated vesicular transport of an adenovirus in motor neuron axons. PLoS Pathog. 2009;5(5):e1000442. doi: 10.1371/journal.ppat.1000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Henaff D, Salinas S, Kremer EJ. An adenovirus traffic update: from receptor engagement to the nuclear pore. Future Microbiol. 2011;6(2):179–192. doi: 10.2217/fmb.10.162. [DOI] [PubMed] [Google Scholar]
- 31.Salinas S, Schiavo G, Kremer EJ. A hitchhiker’s guide to the nervous system: the complex journey of viruses and toxins. Nat Rev Microbiol. 2010;8(9):645–655. doi: 10.1038/nrmicro2395. [DOI] [PubMed] [Google Scholar]
- 32.Piersanti S, et al. Differentiated neuroprogenitor cells incubated with human or canine adenovirus, or lentiviral vectors have distinct transcriptome profiles. PLoS One. 2013;8(7):e69808. doi: 10.1371/journal.pone.0069808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liblau RS, Gonzalez-Dunia D, Wiendl H, Zipp F. Neurons as targets for T cells in the nervous system. Trends Neurosci. 2013;36(6):315–324. doi: 10.1016/j.tins.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 34.Hérodin F, Thullier P, Garin D, Drouet M. Nonhuman primates are relevant models for research in hematology, immunology and virology. Eur Cytokine Netw. 2005;16(2):104–116. [PubMed] [Google Scholar]
- 35.Tuszynski MH, Grill R, Jones LL, McKay HM, Blesch A. Spontaneous and augmented growth of axons in the primate spinal cord: effects of local injury and nerve growth factor-secreting cell grafts. J Comp Neurol. 2002;449(1):88–101. doi: 10.1002/cne.10266. [DOI] [PubMed] [Google Scholar]
- 36.Déglon N, Hantraye P. Viral vectors as tools to model and treat neurodegenerative disorders. J Gene Med. 2005;7(5):530–539. doi: 10.1002/jgm.707. [DOI] [PubMed] [Google Scholar]
- 37.Languille S, et al. The grey mouse lemur: a non-human primate model for ageing studies. Ageing Res Rev. 2012;11(1):150–162. doi: 10.1016/j.arr.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 38.Trouche SG, et al. Antibody response and plasma Abeta1-40 levels in young Microcebus murinus primates immunized with Abeta1-42 and its derivatives. Vaccine. 2009;27(7):957–964. doi: 10.1016/j.vaccine.2008.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kraska A, et al. Age-associated cerebral atrophy in mouse lemur primates. Neurobiol Aging. 2011;32(5):894–906. doi: 10.1016/j.neurobiolaging.2009.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trouche SG, Maurice T, Rouland S, Verdier JM, Mestre-Francés N. The three-panel runway maze adapted to Microcebus murinus reveals age-related differences in memory and perseverance performances. Neurobiol Learn Mem. 2010;94(1):100–106. doi: 10.1016/j.nlm.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carmichael O, Lockhart S. The role of diffusion tensor imaging in the study of cognitive aging. Curr Top Behav Neurosci. 2012;11:289–320. doi: 10.1007/7854_2011_176. [DOI] [PubMed] [Google Scholar]
- 42.Obeso JA, et al. The basal ganglia in Parkinson’s disease: current concepts and unexplained observations. Ann Neurol. 2008;64 Suppl 2:S30–S46. doi: 10.1002/ana.21481. [DOI] [PubMed] [Google Scholar]
- 43.Fox ME, et al. Cross-hemispheric dopamine projections have functional significance. Proc Natl Acad Sci USA. 2016;113(25):6985–6990. doi: 10.1073/pnas.1603629113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Geisler S, Zahm DS. Afferents of the ventral tegmental area in the rat-anatomical substratum for integrative functions. J Comp Neurol. 2005;490(3):270–294. doi: 10.1002/cne.20668. [DOI] [PubMed] [Google Scholar]
- 45.Li Y, et al. Mutant LRRK2(R1441G) BAC transgenic mice recapitulate cardinal features of Parkinson’s disease. Nat Neurosci. 2009;12(7):826–828. doi: 10.1038/nn.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ramonet D, et al. Dopaminergic neuronal loss, reduced neurite complexity and autophagic abnormalities in transgenic mice expressing G2019S mutant LRRK2. PLoS One. 2011;6(4):e18568. doi: 10.1371/journal.pone.0018568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.MacLeod D, Dowman J, Hammond R, Leete T, Inoue K, Abeliovich A. The familial Parkinsonism gene LRRK2 regulates neurite process morphology. Neuron. 2006;52(4):587–593. doi: 10.1016/j.neuron.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 48.Yue Z, Lachenmayer ML. Genetic LRRK2 models of Parkinson’s disease: Dissecting the pathogenic pathway and exploring clinical applications. Mov Disord. 2011;26(8):1386–1397. doi: 10.1002/mds.23737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spires-Jones TL, Stoothoff WH, de Calignon A, Jones PB, Hyman BT. Tau pathophysiology in neurodegeneration: a tangled issue. Trends Neurosci. 2009;32(3):150–159. doi: 10.1016/j.tins.2008.11.007. [DOI] [PubMed] [Google Scholar]
- 50.Melrose HL, et al. Impaired dopaminergic neurotransmission and microtubule-associated protein tau alterations in human LRRK2 transgenic mice. Neurobiol Dis. 2010;40(3):503–517. doi: 10.1016/j.nbd.2010.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rajput A, et al. Parkinsonism, Lrrk2 G2019S, and tau neuropathology. Neurology. 2006;67(8):1506–1508. doi: 10.1212/01.wnl.0000240220.33950.0c. [DOI] [PubMed] [Google Scholar]
- 52.Wszolek ZK, et al. Autosomal dominant parkinsonism associated with variable synuclein and tau pathology. Neurology. 2004;62(9):1619–1622. doi: 10.1212/01.WNL.0000125015.06989.DB. [DOI] [PubMed] [Google Scholar]
- 53.Schoehn G, et al. Three-dimensional structure of canine adenovirus serotype 2 capsid. J Virol. 2008;82(7):3192–3203. doi: 10.1128/JVI.02393-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Villaescusa JC, et al. A PBX1 transcriptional network controls dopaminergic neuron development and is impaired in Parkinson’s disease. EMBO J. 2016;35(18):1963–1978. doi: 10.15252/embj.201593725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smith WW, et al. Leucine-rich repeat kinase 2 (LRRK2) interacts with parkin, and mutant LRRK2 induces neuronal degeneration. Proc Natl Acad Sci USA. 2005;102(51):18676–18681. doi: 10.1073/pnas.0508052102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bons N, Silhol S, Barbié V, Mestre-Francés N, Albe-Fessard D. A stereotaxic atlas of the grey lesser mouse lemur brain (Microcebus murinus) Brain Res Bull. 1998;46(1-2):1–173. doi: 10.1016/S0361-9230(97)00458-9. [DOI] [PubMed] [Google Scholar]
- 57.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98(9):5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Duce JA, Podvin S, Hollander W, Kipling D, Rosene DL, Abraham CR. Gene profile analysis implicates Klotho as an important contributor to aging changes in brain white matter of the rhesus monkey. Glia. 2008;56(1):106–117. doi: 10.1002/glia.20593. [DOI] [PubMed] [Google Scholar]
- 59.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95(25):14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Phillips GR, et al. The presynaptic particle web: ultrastructure, composition, dissolution, and reconstitution. Neuron. 2001;32(1):63–77. doi: 10.1016/S0896-6273(01)00450-0. [DOI] [PubMed] [Google Scholar]
- 61.Verhave PS, Vanwersch RA, van Helden HP, Smit AB, Philippens IH. Two new test methods to quantify motor deficits in a marmoset model for Parkinson’s disease. Behav Brain Res. 2009;200(1):214–219. doi: 10.1016/j.bbr.2009.01.022. [DOI] [PubMed] [Google Scholar]
- 62.Marshall JW, Baker HF, Ridley RM. Contralesional neglect in monkeys with small unilateral parietal cortical ablations. Behav Brain Res. 2002;136(1):257–265. doi: 10.1016/S0166-4328(02)00138-9. [DOI] [PubMed] [Google Scholar]
- 63.Noristani HN, et al. RNA-seq analysis of microglia reveals time-dependent activation of specific genetic programs following spinal cord injury. Front Mol Neurosci. 2017;10:90. doi: 10.3389/fnmol.2017.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.