The growing need for cassava, a food and fuel crop, has led to a new plant breeding technique designed to accelerate breeding of cassava with modified starch.
Abstract
Crop diversification required to meet demands for food security and industrial use is often challenged by breeding time and amenability of varieties to genome modification. Cassava is one such crop. Grown for its large starch-rich storage roots, it serves as a staple food and a commodity in the multibillion-dollar starch industry. Starch is composed of the glucose polymers amylopectin and amylose, with the latter strongly influencing the physicochemical properties of starch during cooking and processing. We demonstrate that CRISPR-Cas9 (clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9)–mediated targeted mutagenesis of two genes involved in amylose biosynthesis, PROTEIN TARGETING TO STARCH (PTST1) or GRANULE BOUND STARCH SYNTHASE (GBSS), can reduce or eliminate amylose content in root starch. Integration of the Arabidopsis FLOWERING LOCUS T gene in the genome-editing cassette allowed us to accelerate flowering—an event seldom seen under glasshouse conditions. Germinated seeds yielded S1, a transgene-free progeny that inherited edited genes. This attractive new plant breeding technique for modified cassava could be extended to other crops to provide a suite of novel varieties with useful traits for food and industrial applications.
INTRODUCTION
Cassava (Manihot esculenta Crantz) is a multipurpose crop cultivated for food and as an industrial feedstock. Successful domestication of the species that originated from South America has promoted cassava to among the top five most important sources of carbohydrate globally. The starch-rich storage roots are a staple food in tropical and subtropical countries where the plant is relatively drought-tolerant and suitable for growth in marginal environments (1). The roots are also an important commodity and processed by a multibillion-dollar industry for use in manufacturing paper, beverages, biodegradable materials, animal feed, pharmaceuticals, and biofuels (2, 3). In western countries, the gluten-free property of cassava is further popularizing the crop as an alternative food source to help alleviate symptoms of celiac disease and gluten intolerance (4). This broadening interest in cassava led to a 60% increase in global harvest between 2000 and 2012 (2). However, with climate change and a world population projected to exceed 9 billion by 2050, there is a continual need to produce and distribute elite crop varieties, including cassava, to improve yields and adaptation (5, 6).
Cassava is an exceptional candidate for genetic engineering (7, 8), but implementation of new plant breeding technologies (NPBT), including the genome editing CRISPR (clustered regularly interspaced short palindromic repeats)–Cas9 (CRISPR-associated protein 9) system (9, 10), remains a challenge. Similar to many other crops, cassava is recalcitrant toward genetic transformation and in vitro regeneration (11, 12) and exhibits poor fertility in some breeding lines and farmer-preferred varieties (1, 13). Asynchronous maturation of the monoecious flowers means progeny are highly heterozygous and make conventional breeding programs for the introgression of traits very time-consuming (14–16). Moreover, flowering in a glasshouse environment seldom occurs, and sexual reproduction yields few seeds. Consequently, there is a necessity for accelerated flowering and improved breeding cycles in cassava (1, 13, 17) to fully capitalize on genome editing technology for agriculture (18–21). A NPBT combining genome editing to efficiently introduce homozygous mutations with accelerated flowering (17) will allow the production of a segregated progeny for rapid crop improvement even in glasshouse environments. Here, we describe such a NPBT for cassava and demonstrate its use for engineering starch biosynthesis, modifying the quality of starch in storage roots.
The importance of starch in food and nutrition, as well as for industrial processing, has motivated the diversification of crops with desired starch traits (22–24). Starch is composed of the glucose polymers (glucans) amylopectin and amylose and is stored as semicrystalline granules in chloroplasts of leaves and amyloplasts of storage organs. Amylopectin has a highly branched structure that gives rise to the crystallinity of the granule and typically constitutes most of the granule matrix. Amylose is a predominantly linear glucan polymer, and despite comprising only ~20% of the granule, it strongly determines the physicochemical properties of starch (24), including that in cassava (25). Amylose is synthesized by the glucosyltransferase GRANULE BOUND STARCH SYNTHASE (GBSS; Fig. 1A), which is tightly associated with starch granules (26) and is encoded by a highly conserved gene that is present as a single copy in cassava (27, 28). Amylose is not required to form the semicrystalline structure of the granule, and natural varieties of potato, maize, and wheat with amylose-free starch are cultivated for their desirable cooking and processing qualities (23, 29). This valuable amylose-free trait has also been created in cassava, through either transgenic RNA-based approaches (30–32) or the screening of self-pollinated lines for natural recessive mutants (33, 34). In Arabidopsis chloroplasts, GBSS localization on starch granules is mediated by PROTEIN TARGETING TO STARCH (PTST/PTST1) (Fig. 1A) (35). PTST1 is a 26-kDa plastidial protein containing an N-terminal coiled-coil domain and a C-terminal carbohydrate-binding module (CBM48) (36). Arabidopsis ptst1 knockout mutants produce amylose-free starch, similar to the gbss mutant. Although homologs of AtPTST1 have recently been identified, only AtPTST1 is involved in amylose biosynthesis (37). The effect of a PTST1 mutation in any crop species remains unknown. Here, we demonstrate the application of CRISPR-Cas9 to generate a catalog of plants with modified starch by targeted mutagenesis of GBSS or PTST1 and segregation under ex situ (glasshouse) conditions to yield transgene-free progeny with novel properties.
Fig. 1. Genome editing of MeGBSS and MePTST1.
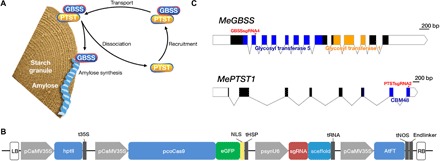
(A) Model for association between GBSS, PTST1, and starch granules as proposed by Seung et al. (35). Image recreated with permission. (B) Binary expression construct containing hptII (hygromycin B resistance) for plant selection; Cas9 codon optimized for cassava usage (pcoCas9) and fused to the eGFP reporter gene and directed to the nucleus via a nucleoplasmin nuclear localization signal (NLS); transcription terminated by sequence from a heat shock protein (tHSP) (56). A synthetic pU6 promoter [(psynU6; 10)] used to drive expression of the protospacer sequence [single guide RNA (sgRNA)] and synthetic scaffold (9) to generate the desired sgRNA. AtFT for early flowering is constitutively expressed by the CaMV35S promoter and terminated by nopaline synthase sequence (tNOS). Binary expression constructs named pCas9-sgGBSS-FT and pCas9-sgPTST-FT. Left and right borders (LB and RB) are shown. Diagram not to scale. (C) Gene maps of MeGBSS and MePTST1. Exons are depicted as blocks, and regions encoding functional domains are shaded. The target sites for the sgRNAs are indicated. Scale bars are shown.
RESULTS
Targeted mutagenesis of GBSS and PTST1 in transgenic cassava plants
Cassava plants harboring the transgene cassettes (pCas9-sgGBSS-FT or pCas9-sgPTST-FT; Fig. 1B) required for genome editing of GBSS or PTST1 (Fig. 1C) and accelerated flowering were successfully produced using stable transformation of embryogenic calli (fig. S1, A to D) (38). We confirmed in vivo expression of the transgenes (fig. S2A), as well as localization of Cas9 fused to enhanced green fluorescent protein (eGFP) in the nuclei of protoplasts (in a transient assay; fig. S2B) and stably transformed, regenerating somatic embryos (fig. S2C). A cleavage assay provided evidence that the designed ribonucleoprotein complexes encoded for in the cassettes were capable of DNA cleavage in vitro (fig. S2D). Nucleotide insertions/deletions (indels) resulting from Cas9-mediated cleavage and nonhomologous end joining (NHEJ) were also successfully identified following sequencing of plant DNA (Fig. 2, A and B).
Fig. 2. Sequence analysis of indels and predicted translation of reads in gbss and ptst lines.
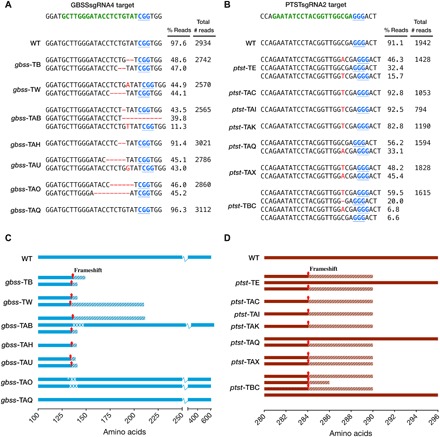
(A and B) PacBio SMRT sequencing of amplicons derived from gbss and ptst lines, respectively. The WT sequence is shown at the top of each alignment, with the protospacer adjacent motif (PAM; blue font and underlined) and sgRNA target site (green font). Nucleotide deletions are depicted as red dashes, and nucleotide insertions are in uppercase, red font. The number of reads per line is shown, together with the percentage of reads clustered according to indel sequence in the target site region. The most abundant sequence clusters for each line are shown. Low-abundant clusters (<5% of total reads) are not shown here since they are likely to be artifacts. Sequence data for WT are also shown as a control to determine conservation of sequence, following tissue culture and regeneration from callus. (C and D) Predicted polypeptides from gbss and ptst lines, respectively. The positions of frameshift mutations are indicated by red arrows, and hatched areas represent potentially translated regions until the next predicted termination codon. Predicted amino acid substitutions (white tick) and deletions (white cross) are indicated in gbss-TAB and gbss-TAO.
In most gbss lines, the majority of sequence reads had only two types of edited sequence, which presumably resulted from independent cleavage and repair of the GBSS alleles in the diploid genome (Fig. 2A). Line gbss-TAH appeared to have a homozygous gbss allele, whereas line gbss-TAB seemed to be chimeric based on the percentage of reads for each indel sequence (Fig. 2A). Translation predictions of the edited sequences revealed frameshift mutations that introduced premature termination codons (Fig. 2C). While deletions in most lines were between 1 and 9 base pairs (bp) in length, 40% of the reads in line gbss-TAB had a deletion of 56 bp (Fig. 2A). Much of this indel was in the downstream intron (Fig. 1C), and predicted translation of this read resulted in amino acid deletions (Val136, Ser137, Val138, and Glu139) at the cleavage site but followed by the native amino acid sequence. Thus, it is assumed that approximately 40% of the sequenced population may generate a shorter but possibly functional GBSS. The two mutated gbss-TAO alleles gave rise to predicted in-frame modifications (allele 1, an Asp133-to-Glu133 substitution and then deletions of Ser135 and Val136; allele 2, deletions of Thr134, Ser135, and Val136). Thus, despite successful targeted mutagenesis, a potentially functional GBSS protein may also be synthesized in the gbss-TAO line (Fig. 2C). No indels were detected in gbss-TAQ (Fig. 2A), and thus, an intact GBSS polypeptide was predicted (Fig. 2C).
Indels among the ptst lines were less variable than in the gbss lines, with the most prevalent indel sequence in multiple lines being a 1-bp insertion (Fig. 2B). Lines ptst-TAC, ptst-TAI, and ptst-TAK appeared to be homozygous, while ptst-TE and ptst-TBC were chimeric based on their more varied sequence population. Fifty-six percent, 32%, and 6.6% of the reads from ptst-TAQ, ptst-TE, and ptst-TBC, respectively, were wild-type (WT) sequence (Fig. 2B), predicting the presence of full-length PTST1 protein in these lines (Fig. 2D). The reason why the indel population in the ptst lines was less diverse than that in the gbss lines is unclear but possibly reflects the nature of NHEJ and/or sequence at these different genomic loci.
Unintended (“off-target”) cleavage and mutagenesis of genomic DNA by a Cas9-sgRNA complex could have a deleterious impact on plant morphology, physiology, and agronomic trait(s). We therefore investigated whether the sgRNAs designed to target GBSS and PTST1 may have triggered off-target cleavage in the genomes of the transgenic lines. Surveyor assays and Sanger sequencing of several predicted off-target sites were performed, but based on the results, no cleavage products or indels could be identified. Allelic differences were observed in some of the sequence data, which is typical for the heterozygous cassava genome, but no indels were identified at the predicted cleavage site of haplotypes (fig. S3).
gbss and ptst transgenic lines were cultivated in the glasshouse to maturity and appeared to have a phenotype similar to WT plants. Some variation in growth was expected between individuals within a line and between independent lines, seen here as an increase in plant height (fig. S4A) and reduction in fresh root mass (fig. S4B). Overall, these results show successful, targeted mutagenesis of GBSS and PTST1, and these plants formed storage roots in the glasshouse.
Genome editing of GBSS and PTST1 provides a catalog of starch mutants
Starch granules purified from harvested storage roots of glasshouse-cultivated gbss lines revealed that gbss-TAH starch stained with iodine gave the distinct brown color that is characteristic of amylose-free starch, in contrast to the blue-black stain of WT starch. gbss-TAB had an intermediate-staining phenotype (suggesting that amylose content was reduced but not absent), while gbss-TAO was indistinguishable from the WT (Fig. 3A). The absence of amylose in gbss-TAH starch was also evident after staining the cut storage root (Fig. 3B). Quantitative analysis of amylose content in starch from all gbss lines revealed that several gbss lines (gbss-TB, gbss-TW, gbss-TAH, and gbss-TAU) produced no detectable amylose (Fig. 3C). The starch from the gbss-TAQ line contained an average of 17% amylose, which was similar to the 18.5% measured in WT starch. This result is consistent with the indel profiling analysis (Fig. 2, A and C) that showed no detectable editing of GBSS in this line. The two allelic mutants in line gbss-TAO (Fig. 2A), as predicted, presumably gave rise to a mutant GBSS with reduced function, as an average of only 9.4% amylose was produced in this line. Despite the significantly reduced amylose content, it was not possible to visually distinguish this iodine-stained starch from the WT by light microscopy (Fig. 3A). The predicted amino acids modified in this line (Fig. 2C) could be useful for fundamental analysis of GBSS structure and function and generation of starch mutants in other crops. Despite the large, 56-bp deletion in gbss-TAB and predicted amino acid deletions (Fig. 2, A and C), it appears that this allelic form was capable of producing a functional GBSS, enabling amylose biosynthesis but to reduced levels (1.8% amylose; Fig. 3C) in the storage roots of this line.
Fig. 3. Amylose content and immunodetection of GBSS in gbss and ptst storage roots.
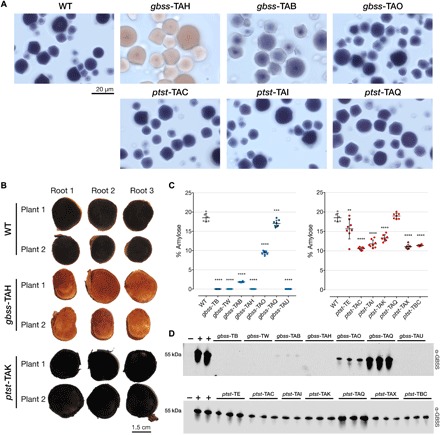
(A) Light microscopy images of iodine-stained purified starch granules of selected gbss and ptst lines. Blue-black staining is indicative of amylose-containing starch, while amylose-free starch stains red-brown. A visible intermediate staining was seen in line gbss-TAB (approximately 1.8% amylose), while a higher-percentage amylose (approximately 9.4%) in line gbss-TAO gave a WT-like phenotype. The 20-μm scale bar is representative for all panels. (B) Storage roots from two plants of WT, gbss-TAH, and ptst-TAK were harvested from 6-month-old glasshouse-cultivated material. Root sections were stained with iodine solution and immediately photographed. Coloration described above is evident in amylose-free and amylose-containing lines. (C) Percentage amylose content for gbss (left) and ptst (right) lines using iodine colorimetry as described for potato starch (66). n = 6 to 8 plants; means ± SD are shown. Statistical analysis using Tukey’s multiple comparisons test (****P < 0.0001, ***P < 0.001, and **P < 0.01) is shown. (D) Immunodetection of GBSS in granule-bound proteins extracted from selected plants of gbss (top) and ptst (bottom) lines. Equivalent loading of protein extracted from 0.5 mg of starch and detected using a polyclonal antibody raised against Arabidopsis GBSS. Granule-bound proteins from the Arabidopsis gbss mutant (negative control, lanes −) and from cassava WT (positive control, lanes +).
We conducted similar analyses with storage root starch extracted from the ptst lines. Most of these lines produced starch with reduced amylose content, with an average of 13.3% amylose compared to 18.5% in WT (Fig. 3C). Unlike some of the gbss lines, none of the ptst lines produced amylose-free starch. Moreover, no significant difference in amylose content was observed in the ptst-TAQ line, which, according to the sequence data, had only 33% of PTST1 alleles edited. However, a significant difference in amylose content was observed in the chimeric ptst-TE line (average 15.6% amylose), which had 62% of PTST1 edited and had a comparatively broad variation in amylose content (10.5 to 19.1%) between plants compared to other lines. The variable nature of NHEJ in some of the lines highlights the potential complications with associating genomic profiling and a phenotypic trait in Cas9-edited plants. These results also suggest that inactivation of both PTST1 alleles is required to significantly reduce the amount of amylose in cassava root starch. The more modest reduction in amylose in starches from the ptst lines compared to the gbss lines could not be visually distinguished from WT starch following iodine staining (Fig. 3, A and B).
Immunoblotting of starch granule–bound proteins extracted from purified starch confirmed that the four amylose-free gbss lines also lacked detectable GBSS protein (Fig. 3D). The amount of GBSS in starch from gbss-TAO was greatly reduced compared to the WT, suggesting that GBSS stability may be compromised by the predicted amino acid alterations in this line (Fig. 2B). This also appears to correlate with the reduced, but not completely absent, amylose content in this line. An even greater reduction of GBSS was observed in gbss-TAB, supporting the finding that amylose was also greatly reduced in this line (Fig. 3C) and that there was apparently only one functional but mutated allelic form (Fig. 2B). As expected, no change in the amount of GBSS was observed in gbss-TAQ (Fig. 3D). The amylose content of starch in these lines therefore correlated strongly with the amount of GBSS present in the starch granules. The ptst lines that had reduced amylose content also had a decreased amount of GBSS from purified starch (Fig. 3D). Mutating the CBM48 domain of PTST1 therefore affects the amount of GBSS, and thus amylose in cassava storage roots, but not to the same extent as previously observed in the Arabidopsis ptst1 mutant (35). These data suggest an association between the mutated alleles resulting from Cas9-mediated cleavage, GBSS levels, and amylose content in cassava storage roots. Multiple attempts were made to detect PTST1 with an antiserum raised against the recombinant cassava PTST1 protein, but the low specificity of the antiserum hampered result interpretation.
To determine whether targeted mutagenesis of GBSS or PTST1 had also altered amylopectin structure, we analyzed the chain length distributions (CLDs) of purified starch. CLDs were nearly identical between gbss and ptst lines and WT (fig. S5), suggesting that mutation of GBSS and PTST1 affecting amylose accumulation in transgenic cassava lines did not alter the amylopectin CLD. In addition, we also investigated whether starch granule size had been affected. Compared to WT starch, the two amylose-free lines gbss-TB and gbss-TAU had a significant increase in the number of large (>15 μm) granules (fig. S6). gbss-TAQ, which has a similar amylose content to WT, and gbss-TAO (9.4% amylose) had no significant change in any size category. There was no significant difference in the granule size of gbss-TW, despite having amylose-free starch. No significant increases in granule size were observed among the ptst mutants (fig. S6), which had greater content of amylose compared to the gbss mutants (Fig. 3C). These data require a more comprehensive analysis to determine the extent to which amylose content or other physiological factors contribute to granule size in cassava storage roots. Morphology analysis of purified starch from glasshouse-cultivated storage roots using scanning electron microscopy revealed no discernible difference in starch granules between gbss lines, ptst lines, and WT starch (fig. S7).
Physicochemical properties of modified cassava desired by some industries
The physicochemical properties of purified starch, which are factors critical in commercial processing, were analyzed for gbss and ptst lines (Fig. 4 and fig. S8). The amylose-free gbss lines (gbss-TB, gbss-TW, gbss-TAH, and gbss-TAU) had a different pasting profile compared to WT, with a lower peak temperature (the temperature at which maximum viscosity is achieved), higher peak viscosity (the maximum viscosity of the paste during heating), and higher final viscosity (the viscosity measured after cooling). The gbss lines with reduced amylose content had viscosity profiles that were intermediate between WT and amylose-free starches, although these lines (gbss-TAO and gbss-TAQ) also had greater biological variation (Fig. 4 and fig. S8). All starches had similar pasting onset temperatures (~65°C). Starch from the ptst lines had a pasting profile that was more similar to WT starch. However, a slightly higher peak viscosity was observed in lines ptst-TAC, ptst-TAI, ptst-TAK, and ptst-TAX (10.5 to 13.3% amylose content), compared to WT starch. An outlier in the ptst lines was the starch from ptst-TAQ, which had a similar amylose content (18.9%) compared to WT (18.5%) but had a much lower peak and final viscosity than all other samples tested (Fig. 4 and fig. S8). It is unclear why samples from this line behaved differently, but it could be a consequence of the lower harvested root mass (fig. S4B). This rheological profiling demonstrates that we have achieved desirable changes in the gelatinization behavior of starch through genome editing that is of interest for industrial applications.
Fig. 4. Physicochemical properties of gbss and ptst starch.
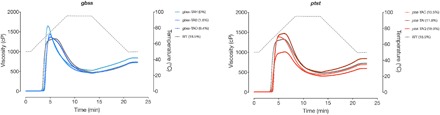
Viscosity measurements of purified starch from selected gbss and ptst lines. Amylose content for each line is provided in the graph legend. Temperature gradient is depicted as the gray dotted line against the secondary (right) y axis. Mean values (n = 3 to 4 plants) are plotted.
Accelerated ex situ breeding for transgene-free progeny with modified starch genes
Generating cassava with heritable allelic edits but lacking foreign DNA (that is, the T-DNA encoding the genome editing tools) requires the generation of a segregating population in which the T-DNA can be crossed out. Cassava seldom flowers in glasshouse conditions, but expression of AtFT in the glasshouse-grown transgenic cassava (Fig. 1B and fig. S2A) (17) permitted rapid flowering and heritability of edited lines. Plants from multiple gbss and ptst lines flowered within 12 weeks of glasshouse cultivation and continued to flower as the plants grew over the following months (Fig. 5A). In contrast, no WT plants flowered. Despite asynchronous maturation of pistillate and staminate flowers, we successfully crossed two pairs of gbss-TAH plants, yielding five seeds (Fig. 5B). Two independent crosses between gbss-TAO plants also yielded five seeds. Harvested seeds were sown in soil, and emerging seedlings (Fig. 5C) were screened for the presence of the T-DNA, as well as the inheritance of the mutated GBSS alleles. Polymerase chain reaction (PCR) amplification analysis confirmed that two progenies (named gbss-TAH S1-2 and gbss-TAO S1-2) were transgene-free (Fig. 5D). Furthermore, Sanger sequencing analysis revealed a 2-bp deletion in all sequenced alleles in the two gbss-TAH progenies (Fig. 5E), indicating the conservation of the homozygous gbss mutations in the S1 progeny. The two S1 progeny plants from the gbss-TAO crossing were heterozygous for 6- and 9-bp deletions (Fig. 5E). All the S1 progeny plants have indels identical to those identified in their respective parent populations (Fig. 2A). These data demonstrate successful and rapid inheritance of Cas9-edited genes in segregated transgene-free cassava.
Fig. 5. Early flowering in glasshouse-cultivated gbss lines and molecular characterization of S1 progeny.
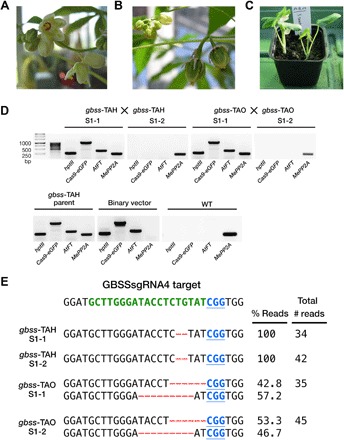
(A) Inflorescence on a gbss-TAB plant. (B) Developing fruit following manual pollination (selfing) on a gbss-TAH plant. (C) S1 progeny plantlets following manual pollination (selfing) and seed germination between gbss plants. (D) PCR amplification products of hptII, Cas9-eGFP, AtFT, and endogenous MePP2A. Reactions contained genomic DNA template from S1 progeny samples, a parent control (gbss-TAH), a vector control (binary vector), and WT. (E) Sequence of cloned amplicons derived from the GBSSsgRNA4 genomic target sequence in S1 lines. The WT sequence is shown at the top of the alignment and comprises the PAM (blue font and underlined) and the sgRNA target site (green font). Nucleotide deletions are depicted as red dashes. Number of reads and percentage distribution between indels from the sequenced population are provided.
DISCUSSION
Our data show that combined expression of Cas9-sgRNA and AtFT is a NPBT that can rapidly generate transgene-free progeny with inherited, mutated alleles in the economically important crop, cassava (Fig. 6). Through targeted mutagenesis of GBSS or PTST1, we generated homozygous, heterozygous, and possibly chimeric lines containing a spectrum of different amylose contents and provided an insight into the role of PTST1 in a crop species. The profiles that led to predicted deletions and substitutions of amino acids in GBSS (for example, gbss-TAO) have provided lines with interesting traits and segregating progeny to advance our understanding of amylose biosynthesis and GBSS-starch interactions. Amylose content strongly influences the physicochemical properties of starch, principally the pasting and gelatinization behavior, which are important factors particularly in commercial processing. Amylose-free starches generally have lower pasting temperatures and higher viscosity than normal starches (39, 40), which are characteristics also observed in our amylose-free gbss cassava starches and consistent with previous studies of reduced amylose cassava starch (30–34). We also observed an increase in peak viscosity in ptst starch lines with reduced amylose content (~13% versus 18.5% in WT). These alterations in rheological properties were less pronounced than those observed in amylose-free gbss starch and thus are intermediates with commercial potential; low-amylose starches of wheat, for example, are more desirable than both normal and amylose-free starch for producing pasta and noodles (41, 42). Modification of amylose content did not appear to have a negative impact on amylopectin but may be associated with increased granule size (43). Our capacity to modify the cassava genome for producing starches containing a range of different amylose contents provides a useful tool for engineering suitable starch properties for different food products and commercial processing.
Fig. 6. Schematic representation of conventional breeding and the designed NPBT for trait improvement.
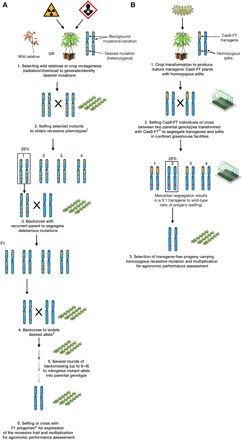
(A) Pathway for breeding of recessive traits using a wild relative or mutagenized plant as parent material. Multiple crosses are required to introgress a homozygous mutation into a farmer-preferred genotype. Several years are required to generate an improved variety. †Information about mutation and availability of genetic marker(s) could allow bypassing of the selfing step. ‡In the absence of marker(s), self-crosses are performed after every backcross to select offspring with the recessive trait phenotype. ΔAvoiding selfing should limit potential inbreeding depression. (B) NPBT described here for accelerated flowering and segregation of genome-edited lines. Agrobacterium-mediated stable transformation provides transgenic lines with different mutant populations. Several lines are screened (phenotype and genotype), and a single segregation from selfing or cross between two Cas9-FT–transformed genotypes can occur ex situ in a glasshouse environment, yielding T-DNA–free progeny with the homozygous recessive mutation.
Until now, the role of PTST1 had not been characterized in a plant species other than Arabidopsis. PTST1 is a CBM48-containing protein that was shown in Arabidopsis leaves to mediate GBSS localization to starch granules (35). We reveal that cassava with mutated PTST1 has both reduced GBSS protein and lower amylose content than WT, demonstrating that the protein also participates in amylose synthesis in cassava storage roots (Fig. 1A). Our ptst lines did not completely abolish amylose content in cassava roots, as it did in Arabidopsis ptst1 leaves. This difference is likely due to the fact that roots and leaves synthesize starch over different timescales. In Arabidopsis leaves, starch produced during the day is fully degraded during the subsequent night, as is the associated GBSS protein (44). Since leaf starch accumulation is confined to a single photoperiod during which new GBSS is also synthesized, amylose content is relatively low. In cassava storage roots, starch is synthesized throughout root bulking over a period of several weeks to months. The absence of PTST1 may therefore reduce the efficiency of amylose synthesis in cassava roots, but this is partially compensated by the longer time for amylose to accumulate compared with an Arabidopsis leaf. When starch accumulation in Arabidopsis occurs over longer timeframes [for example, in continuous light or in mutants like STARCH EXCESS 4 (sex4), in which starch degradation is impaired], amylose content increases to much higher levels (45). In such situations [that is, in the ptst sex4 double mutant (35)], the loss of PTST1 causes a low-amylose phenotype similar to what we observed in cassava roots.
The ability to efficiently modify genes in highly heterozygous crops, combined with accelerated time to flowering (46) to generate transgene-free progeny with a homozygous trait, should save several years over conventional breeding approaches and phenotypic recurrent selection (Fig. 6). It remains to be seen how genome-edited plants will be regulated in the European Union (47), but this NPBT should help with public perception of crop modification and be useful in applying transgene-free targeted mutagenesis in this recalcitrant species (48). Moreover, the use of wild relatives or parents from mutation-induced treatment [TILLING (Targeting Induced Local Lesions IN Genomes)] populations currently requires the screening of thousands of lines to identify suitable traits (Fig. 6A) (49, 50). The NPBT described here should allow widespread uptake of cost-effective, transgene-free crops with useful new traits to complement other breeding schemes (Fig. 6B). We are currently upscaling seed production to produce a catalog of gbss and ptst mutants for field-based characterization and breeding. The development of robust genetic transformation methods also complements this NPBT, offering the possibility of generating selected allelic forms directly in varieties that can be readily used by farmers and industries in countries where cassava biotechnology capacities have been enabled, for example, in the commercially important southern African landrace T200 (51, 52). Such programs should contribute to expanding crop diversity with precisely modified genomes for food security, commodity diversity, and sustainability necessary for global demands.
MATERIALS AND METHODS
Cassava protoplast culture and polyethylene glycol–mediated transformation
The protocol was modified from (53). In brief, 1-mm sections of cassava leaf tissue were incubated in enzyme solution [1.5% cellulose, 0.4% macerozyme, 0.4 M mannitol, 20 mM KCl, 20 mM MES (pH 5.7), and 10 mM CaCl2] and vacuum-infiltrated for 15 min. The samples were then incubated for 4 hours at 20°C, shaking gently at 50 rpm. The protoplasts were filtered through 100- and 50-μm sterile nylon mesh before centrifugation at 900 rpm for 2 min. The protoplast pellet was resuspended in wash solution [0.5 M mannitol, 4 mM MES (pH 5.7), and 20 mM KCl] and centrifuged as previously described. Protoplasts were resuspended in W5 solution [154 mM NaCl, 125 mM CaCl2, 5 mM KCl, and 2 mM MES (pH 5.7)] before performing viability assays using fluorescein diacetate and a microscope fitted with a GFP filter. Protoplast culture (100 μl of approximately 2 × 104 cells) was incubated with 10 μg of plasmid DNA and 40% (v/v) polyethylene glycol solution for 25 min at 20°C. Protoplasts were washed in W5 and incubated for 16 hours in media [1× Murashige and Skoog (MS) salts and vitamins, 1 parts per million (ppm) 1-naphthaleneacetic acid (NAA), 0.5 ppm 6-benzylaminopurine (BAP), 50 ppm digested casein, 0.2 M sucrose, and 0.1 M mannitol (pH 5.6)] before confocal microscopy analysis to observe Hoechst 33342–stained nuclei and transient expression of eGFP.
Construction of pCas9-sgGBSS-FT and pCas9-sgPTST-FT binary expression cassettes
The multigene binary cassettes were assembled using the Golden Gate cloning system (54, 55). Level 0 (L0) modules were assembled for four transcription units [termed level 1 (L1) modules]. Following digestion of L0 modules with Bsa I and ligation with T4 ligase, the hptII gene (hygromycin B resistance gene) was inserted between the CaMV35S promoter and terminator sequences to create the L1-F1-p35S-hptII-t35 vector required for plant selection. Similarly, a L1 module termed L1-F2-p35S-Cas9-eGFP-NLS-tHSP transcription unit was assembled from synthesized L0 modules. These L0 modules comprised the pCaMV35S promoter, a Cas9 derived from Streptococcus pyogenes codon optimized for expression in cassava, the eGFP reporter gene fused in-frame to a nucleoplasmin NLS, and a terminator sequence of heat shock protein (tHSP) (56). This L1 module was used for protoplast transfection. The sgRNA transcript unit (psynU6-sgRNA-Scaffold-Terminator) was synthesized in its entirety and cloned into a L1-F3 backbone vector to generate L1-F3-psynU6-sgRNA-Scaffold-Terminator. psynU6 is a synthetic promoter using the consensus sequence of the three U6 variants in Arabidopsis (10) and fused to a chimeric scaffold (9). The incorporated GBSSsgRNA4 sequence was 5′-GCTTGGGATACCTCTGTAT-3′, whereas for PTSTsgRNA2, it was 5′-GAATATCCTACGGTTGGCGA-3′. The fourth and final transcriptional unit used L0 modules of the pCaMV35S promoter, the FT coding sequence from Arabidopsis (17), and the Agrobacterium nopaline synthase (nos) terminator to create L1-F4-pCaMV35S-AtFT-tNOS. One-step Bpi I restriction enzyme digestion and ligation of the four L1 transcriptional units, a L2 backbone vector, and an endlinker (ELE-4) resulted in assembly of pCas9-sgGBSS-FT and pCas9-sgPTST-FT. Both expression plasmids were sequenced to verify the integrity of the cloned units. Multiple PAMs comprising the NGG nucleotide triplet were identified in the GBSS sequence of cv. 60444, and each was assessed by Basic Local Alignment Search Tool (BLAST) analysis and in silico profiling to determine suitability for gene disruption with minimal off-target effects. GBSSsgRNA4 targeting sequence in exon 2 and approximately 470 bp downstream of the transcription start site was selected for in vitro analysis. For PTST1, PTSTsgRNA2 was selected to target an exon at the 3′-end of the coding sequence. This region encodes a CBM48 domain required for interaction of PTST1 with starch (36); site-directed mutagenesis of this domain in Arabidopsis PTST1 abolishes starch binding (35).
Purification of Cas9-eGFP-NLS recombinant protein and in vitro cleavage assays
The Cas9-eGFP-NLS sequence was PCR-amplified from the L1 cassette (see above) using forward and reverse oligonucleotides with flanking Nde I and Xho I restriction enzyme recognition sites, respectively, using Q5 Hot Start High-Fidelity DNA Polymerase (New England Biolabs). Corresponding restriction enzyme digestion of the amplicons was followed by ligation (T4 Ligase, New England Biolabs) into the pET-21a+ vector, yielding a construct encoding Cas9-eGFP-NLS with a C-terminal His6 tag. Chemical transformation of ArcticExpress (DE3) RIL competent cells (Agilent Technologies) and selection on Luria agar plates containing gentamicin (20 mg/liter) and ampicillin (100 mg/liter) was used to select for plasmid-containing colonies. Plasmid integrity was confirmed following purification (GeneJET Plasmid Miniprep Kit, Thermo Fisher Scientific) and Sanger sequencing using vector- and fragment-specific oligonucleotides. Protein expression was induced by adding 1 mM isopropyl-β-d-thiogalactopyranoside to the cultures, which were then incubated at 18°C for 16 hours. Bacterial cells were lysed using a microfluidizer (Microfluidics), and affinity purification was performed using the ÄKTA pure 25 L System with a HisTrap HP nickel-Sepharose column (GE Healthcare Life Sciences). The in vitro assay that determines cleavage efficiency was performed as previously described (57). This protocol uses the purified Cas9-eGFP-NLS protein, a double-stranded DNA (dsDNA) PCR fragment containing the sgRNA target sites, and in vitro–transcribed sgRNA. Reaction products were analyzed by agarose gel electrophoresis.
Agrobacterium-mediated cassava transformation
Transformation was performed as described previously (38). In brief, friable, embryogenic calli (cv. 60444) were transformed with Agrobacterium (strain LBA4404) harboring the expression constructs described above. Following cocultivation, somatic embryogenic tissue was regenerated on MS-based media (58) containing hygromycin B for plant selection and the synthetic auxin NAA. Differentiating somatic embryos were transferred to MS-based media containing the synthetic cytokinin BAP to induce shoot development. Emerging juvenile shoots were propagated on standard media (lacking auxins or cytokinins) to establish in vitro plantlets. Following normal growth, the apical shoot tip was transferred to MS-based media containing hygromycin B to select further for transgenic material. Only plantlets positive for the plant selection marker (hptII) would form roots and were used for molecular analyses and propagated for glasshouse cultivation.
Cultivation of glasshouse-grown cassava plants
Transgenic and WT plantlets were propagated in vitro and transferred to soil as described previously (38). Cassava was cultivated under glasshouse conditions (60% humidity, 26°C, and 16-hour supplemental lighting) and fertilized fortnightly (Wuxal Universal Fertilizer). Plants were harvested after 6 months growth. Harvesting was performed toward the end of the photoperiod to maximize starch yields. Plant height (base of stem to apical shoot tip) was measured, and leaf numbers 1, 2, and 4 (counted from apical leaf) were isolated and frozen in liquid nitrogen before they were ground to a fine powder using pestle and mortar. The root mass of each plant was removed and weighed (in grams). The two largest storage roots from each plant were isolated, peeled, chopped, and briefly rinsed in water before they were frozen in liquid nitrogen and ground to a fine powder (IKA Analytical Mill). Tissue samples were stored in sterile vessels at −80°C.
Southern blot analysis to determine T-DNA integration
Genomic DNA was extracted from leaf tissue of putative transgenic lines using a modified protocol (59). Genomic DNA (10 μg) was digested with restriction enzymes, Hind III and Xba I, and prepared for Southern blotting as described previously (17). Digoxigenin (DIG)–labeled probe targeting hptII was used to confirm stable integration of T-DNA in the plant genome.
PCR amplification of T-DNA in putative transgenic lines
Three pairs of oligonucleotide primers specific to hptII, Cas9-eGFP, and AtFT were used in PCR to determine the presence of T-DNA integration in genomic DNA. Reactions contained 100 ng of genomic DNA, 0.5 μM forward primer, 0.5 μM reverse primer, 200 μM deoxynucleotide triphosphates (dNTPs), sterile distilled water (GibcoBRL), 1× DreamTaq buffer, and 1.5 U of DreamTaq (Thermo Fisher Scientific). Products were resolved on a 1% tris-acetate-EDTA agarose electrophoresis gel containing ethidium bromide and visualized using an ultraviolet transilluminator.
RNA extraction and semiquantitative reverse transcription PCR of transgenes
Total RNA from leaf tissue of glasshouse-cultivated plants was extracted as described previously (60). Total nucleic acid (2 μg), determined using NanoDrop, was treated with deoxyribonuclease, and a complementary DNA (cDNA) library was prepared using the RevertAid First Strand cDNA Synthesis Kit following the manufacturer’s guidelines (Thermo Fisher Scientific). Oligonucleotide primers specific to AtFT (qPCR-FT-F and qPCR-FT-R), GBSSsgRNA4 (GBSSsg4-F and sgRNA-R), PTSTsgRNA2 (PTSTsg2-F and sgRNA-R), and Cas9 (Cas9-F1 and Cas9-R) were used with 1.5 μl of cDNA and Fast SYBR Green Master Mix according to the manufacturer’s guidelines (Applied Biosystems). Data were analyzed using the 2−ΔCt equation (61), and transgene expression was determined relative to the endogenous MePP2A reference gene amplified using oligonucleotide primers PP2A-LP2 and PP2A-RP2.
PacBio SMRT sequencing to characterize indels
Genomic DNA was used as the template in PCR amplification of PTST1 and GBSS spanning the protospacer target site. Oligonucleotide primers specific to PTST1 and GBSS and incorporating PacBio barcodes were synthesized. Reaction conditions were 1× Q5 reaction buffer, 200 μM dNTPs, 0.5 μM PTST-F-BCn or GBSS-F-BCn, 0.5 μM PTST-R-BCn or GBSS-R-BCn, 200 ng of genomic DNA, 1 U of Q5 High-Fidelity DNA Polymerase (New England Biolabs), and sterile nuclease-free water. Conditions were as follows: initial step at 98°C (1 min 30 s); then 25 cycles of 98°C (10 s), 62°C (10 s), and 72°C (20 s); and a final step of 72°C for 2 min. Reactions were cleaned using the Wizard SV Gel and PCR Clean-Up System protocol (Promega) before quantification using the Quant-iT dsDNA Assay Kit (Invitrogen). Reactions were analyzed using PacBio SMRT sequencing [Functional Genomics Centre Zurich (FGCZ)]. To identify indels, the software tool Cas-Analyzer (62) was used in conjunction with PTST1 and GBSS sequences defined via BLASTn (63) on in-house genome data of cassava cv. 60444.
Screening for off-target cleavage by PTST and GBSS sgRNAs
Off-target sites for PTSTsgRNA2 and GBSSsgRNA4 were determined using the CRISPOR software (64) combined with next-generation sequencing (NGS) genome data of cv. 60444. Off-target sites with 3- to 5-nucleotide (nt) mismatches for GBSSsgRNA4 and with 4- to 5-nt mismatches for PTSTsgRNA2 were selected for further analysis. Specific oligonucleotide primers were designed to amplify a region of approximately 400 to 600 bp spanning each of the predicted off-target sites in each of the lines. PCR amplification consisted of 1× Q5 reaction buffer, 200 μM dNTPs, 0.5 μM forward primer, 0.5 μM reverse primer, 200 ng of genomic DNA, 1 U of Q5 High-Fidelity DNA Polymerase (New England Biolabs), and sterile nuclease-free water. Conditions were as follows: initial step at 98°C (1 min 30 s); then 25 cycles of 98°C (10 s), 60 to 67°C (depending on oligonucleotides Tm) (10 s), and 72°C (20 s); and a final step of 72°C for 2 min. Reactions were cleaned using the Wizard SV Gel and PCR Clean-Up System Protocol (Promega) before quantification using the Quant-iT dsDNA Assay Kit (Invitrogen). Amplicons for each off-target site were pooled and assessed using the Surveyor Mutation Detection Kit (Integrated DNA Technologies) following the manufacturer’s guidelines. To further screen predicted off-target sites, 50 ng of selected off-target PCR amplification products was cloned using the CloneJET PCR Cloning Kit (Thermo Fisher Scientific) and used to transform DH5α Escherichia coli following the manufacturer’s guidelines. Plasmids from 12 independent bacterial colonies for each of the cloned, selected, predicted off-target regions were Sanger-sequenced and data-mapped to similarly amplified and cloned WT control sequence.
Starch purification from cassava storage roots
The protocol was modified from (65). Powdered, frozen cassava root (5 cm3) was mixed with 30 ml of starch extraction medium [50 mM tris-HCl (pH 8), 0.2 mM EDTA (pH 8), and 0.5% (v/v) Triton X-100] before filtering through a 100-μm nylon mesh. The filtrate was spun at 3000g for 5 min at 20°C. The supernatant was discarded, and the starch pellet was resuspended in 5 ml of extraction medium. The suspension was filtered through a 70-μm nylon mesh, and the filtrate was overlaid on a 4-ml Percoll cushion [95% (v/v) Percoll and 5% (v/v) 0.5 M tris-HCl (pH 8)] contained in a 13-ml disposable sterile tube. Samples were spun for 5 min at 2500g. The supernatant and the Percoll cushion were vacuum-aspirated, and the remaining starch pellet was resuspended in 5 ml of 0.5% (w/v) SDS dissolved in sterile water. The samples were spun for 3 min at 4500g, and the supernatant was removed. The pellet was washed twice with sterile distilled water and then washed in 80% (v/v) ethanol before desiccation under vacuum for 36 hours and dry storage at 4°C.
Iodine staining of root tissue
The harvested storage roots were isolated, sliced, and sprayed with 10% Lugol solution (Sigma) and immediately photographed.
Scanning electron microscopy and light microscopy of starch granules
An aliquot (3 μl) of a suspension (20 mg/ml) of purified starch granules in sterile distilled water was placed on cleaned, glow-discharged coverslips and sputter-coated in gold. Images were collected using a Quanta 250 FEG microscope (FEI) at the ScopeM facility (FGCZ). For light microscopy samples, 90 μl of the granule suspension was mixed with Lugol solution. Samples were centrifuged for 30 s at 3000g, and the supernatant was discarded. The starch pellet was resuspended in 20 μl of 50% (v/v) sterile glycerol and observed using a 100× objective and AXIO Imager Z2 microscope (Zeiss).
Determination of amylose content in cassava storage roots
Purified root starch (5 mg) was transferred to a screw-top, sterile microcentrifuge tube, and 1 ml of sterile distilled water was added. Samples were vortexed to mix and then incubated in a thermoshaker at 95°C at 1000 rpm for 10 min. Gelatinized starch (5 μl) was immediately mixed with 200 μl of 10% (v/v) Lugol solution (Sigma) in a flat-bottom 96-well plate. Measurements at 550 and 618 nm were recorded on a microplate reader (Tecan). The percentage amylose content was calculated using the equation described for potato starch (% amylose = (3.5 − (5.1 × (OD618/OD550)))/((10.4 × (OD618/OD550)) − 19.9) × 100) (66).
Protein extraction and western immunodetection
Purified starch granules were resuspended at a concentration of 33 mg/ml in SDS–polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer [50 mM tris-HCl (pH 6.8), 2% (w/v) SDS, 100 mM dithiothreitol, 3% (v/v) glycerol, and 0.005% (w/v) bromophenol blue] and heated at 95°C for 10 min. Following a 60-min centrifugation, samples were loaded onto a 10% SDS-PAGE gel (Criterion TGX Precast Gel, Bio-Rad) and electrophoresed in tris-glycine-SDS running buffer. Proteins were transferred to low-fluorescence polyvinylidene difluoride membranes using the Trans-Blot Turbo Transfer System (Bio-Rad) and then incubated for 1 hour in blocking solution [5% (w/v) milk powder in 1× tris-buffered saline (TBS)]. The membrane was then incubated for 18 hours at 4°C in primary antibody diluted in blocking solution with 0.1% (v/v) Tween 20. The GBSS antibody was raised in rabbit (35) and used at a 1:1000 dilution. The membrane was washed four times for 5 min, each in 1× TBS and 0.1% (v/v) Tween 20 (TBST) at 20°C, before incubation in secondary antibody [donkey anti-rabbit conjugated to IRDye 800CW (LI-COR) used at 1:10,000 dilution] for 1 hour at 20°C. The membrane was washed four times for 5 min each in 1× TBST and then twice for 5 min in 1× TBS. The protein was visualized by infrared fluorescence using an Odyssey CLx Imaging System (LI-COR).
Chain length distribution
High-performance anion exchange chromatography with pulsed amperometric detection was used to measure CLD of gbss, ptst, and WT as described previously (65).
Starch granule size distribution
The protocol was adapted from (67). Purified root starch (5 mg) was suspended in 165 μl of sterile water, and 10 μl was transferred to a 96-well, flat-bottom plate containing 190 μl of water. Starch granules were detected using a flow cytometer. Moist starch granules have a refractive index of 1.52 (68). Here, we use a CytoFlex [settings: forward scatter (FSC) gain, 20; threshold FSC, 6000; max, 3000 events s−1; λ = 488 nm] to measure 50,000 granules per sample. The diameter was derived from the FSC value based on polymethyl methacrylate sizing microbeads (refractive index: 1.49; ø: 1, 4, 8, 20, and 30 μm) to determine the correlation of diameter to FSC value. Size gates were selected on the basis of standardization and WT biological data.
Rheometer analysis of starch pasting
Purified storage root starch (1.5 g) was resuspended in 18 ml of sterile distilled water and processed using a Modular Compact Rheometer (MCR92, Anton Paar GmbH) and associated software (RheoCompass). Starch suspensions were presheared at 960 rpm at 50°C for 10 s, held at 50°C for 50 s, heated to 95°C at 6°C/min, held at 95°C for 5 min, and then cooled to 50°C at 6°C/min before a final holding step at 50°C for 2 min. Suspension was constantly stirred at 160 rpm, and measurements for pasting temperature, peak viscosity, peak temperature, and final viscosity were recorded.
Pollination experiments and screening of S1 progeny for edits and T-DNA
Flowering, glasshouse-cultivated plants were maintained and manually pollinated as described previously (17). Seeds were germinated in soil, and genomic DNA was extracted from emerging leaf material [modified from (59)]. PCR amplification of regions of the T-DNA was performed (described in the “PCR amplification of T-DNA in putative transgenic lines” section). PCR amplification of GBSS alleles consisted of 1× Q5 reaction buffer, 200 μM dNTPs, 0.5 μM forward primer, 0.5 μM reverse primer, 200 ng of genomic DNA, 1 U of Q5 High-Fidelity DNA Polymerase (New England Biolabs), and sterile nuclease-free water. Conditions were as follows: initial step at 98°C (1 min 30 s); then 25 cycles of 98°C (10 s), 60 to 67°C (depending on oligonucleotides Tm) (10 s), and 72°C (20 s); and a final step of 72°C for 2 min. Reaction products were cloned using the CloneJET PCR Cloning Kit (Thermo Fisher Scientific) and used to transform DH5α E. coli following the manufacturer’s guidelines. Plasmids from independent bacterial colonies for each of the cloned GBSS amplicons were Sanger-sequenced (Microsynth AG), and the data were mapped to similarly amplified and cloned WT control sequence.
Statistical analysis
All statistical analyses were performed using GraphPad Prism version 7.0. Tukey’s multiple comparison test was performed, and n and P values were provided for each analysis.
Supplementary Material
Acknowledgments
S.E.B. thanks C. Rogers (Sainsbury Laboratory, University of Cambridge, UK) for access to Golden Gate cloning modules and M. A. Santana (Universidad Simón Bolívar, Venezuela) for training in protoplast cultivation. Scanning electron microscopy was performed by S. Rodighiero (ScopeM, ETH Zurich); PacBio SMRT sequencing was performed by A. Bratus, A. Patrignani, and W. Qi (FGCZ, Zurich). S.E.B. is grateful to Johannes Fütterer (ETH Zurich) for helpful discussions and advice throughout the project. E. J. de Oliveira (Embrapa, Brazil) is acknowledged for helpful discussions about the cassava breeding approaches. Funding: The project was supported by a PLANT FELLOWS post-doctoral fellowship (Marie Skłodowska Curie, EU FP7-COFUND) awarded to S.E.B. and ETH Zurich funds provided to W.G. Author contributions: S.E.B. and H.V. designed the fellowship project. S.E.B., D.S., and C.C. performed the experiments. D.M. contributed to data analysis and edited the figures. J.-E.K. processed NGS data. E.T. performed light microscopy. A.H. analyzed the flow cytometry data. I.Z. managed plant propagation and pollination experiments. D.S. and S.C.Z. provided advice regarding starch analysis. S.C.Z., W.G., and H.V. provided guidance. S.E.B. wrote the manuscript with contributions from all authors. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/9/eaat6086/DC1
Fig. S1. Molecular screening of transgenic plants.
Fig. S2. Efficacy of the genome editing constructs in vitro and in vivo.
Fig. S3. Screening of predicted off-target cleavage sites for GBSSsgRNA4 and PTSTsgRNA2.
Fig. S4. Morphology of glasshouse-cultivated gbss and ptst lines.
Fig. S5. CLDs in storage root starch of gbss and ptst lines.
Fig. S6. Flow cytometry analysis of starch granule size in gbss and ptst lines.
Fig. S7. Scanning electron microscopy of purified gbss and ptst starch granules.
Fig. S8. Physicochemical properties of gbss and ptst starch.
REFERENCES AND NOTES
- 1.H. Ceballos, C. H. Hershey, Cassava (Manihot esculenta Crantz), in Genetic Improvement of Tropical Crops, H. Campos, P. D. S. Caligari, Eds. (Springer International Publishing AG, 2017), pp. 129–180. [Google Scholar]
- 2.R. Howeler, N. Lutaladio, G. Thomas, Save and Grow: Cassava. A Guide to Sustainable Production Intensification (FAO, 2013). [Google Scholar]
- 3.Jansson C., Westerbergh A., Zhang J., Hu X., Sun C., Cassava, a potential biofuel crop in (the) People’s Republic of China. Appl. Energy 86, S95–S99 (2009). [Google Scholar]
- 4.Horstmann S. W., Lynch K. M., Arendt E. K., Starch characteristics linked to gluten-free products. Foods 6, E29 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Østerberg J. T., Xiang W., Olsen L. I., Edenbrandt A. K., Vedel S. E., Christiansen A., Landes X., Andersen M. M., Pagh P., Sandøe P., Nielsen J., Christensen S. B., Thorsen B. J., Kappel K., Gamborg C., Palmgren M., Accelerating the domestication of new crops: Feasibility and approaches. Trends Plant Sci. 22, 373–384 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Atlin G. N., Cairns J. E., Das B., Rapid breeding and varietal replacement are critical to adaptation of cropping systems in the developing world to climate change. Glob. Food Sec. 12, 31–37 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chavarriaga-Aguirre P., Brand A., Medina A., Prías M., Escobar R., Martinez J., Díaz P., López C., Roca W. M., Tohme J., The potential of using biotechnology to improve cassava: A review. In Vitro Cell. Dev. Biol. Plant 52, 461–478 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li K.-T., Moulin M., Mangel N., Albersen M., Verhoeven-Duif N. M., Ma Q., Zhang P., Fitzpatrick T. B., Gruissem W., Vanderschuren H., Increased bioavailable vitamin B6 in field-grown transgenic cassava for dietary sufficiency. Nat. Biotechnol. 33, 1029–1032 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J. A., Charpentier E., A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nekrasov V., Staskawicz B., Weigel D., Jones J. D. G., Kamoun S., Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat. Biotechnol. 31, 691–693 (2013). [DOI] [PubMed] [Google Scholar]
- 11.N. J. Baltes, J. Gil-Humanes, D. F. Voytas, Genome engineering and agriculture: Opportunities and challenges, in Progress in Molecular Biology and Translational Science, D. P. Weeks, B. Yang, Eds. (Elsevier Inc., 2017), pp. 1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Altpeter F., Springer N. M., Bartley L. E., Blechl A. E., Brutnell T. P., Citovsky V., Conrad L. J., Gelvin S. B., Jackson D. P., Kausch A. P., Lemaux P. G., Medford J. I., Orozco-Cárdenas M. L., Tricoli D. M., Van Eck J., Voytas D. F., Walbot V., Wang K., Zhang Z. J., Stewart C. N. Jr., Advancing crop transformation in the era of genome editing. Plant Cell 28, 1510–1520 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ceballos H., Jaramillo J. J., Salazar S., Pineda L. M., Calle F., Setter T., Induction of flowering in cassava through grafting. J. Plant Breed. Crop Sci. 9, 19–29 (2017). [Google Scholar]
- 14.Perera P. I. P., Quintero M., Dedicova B., Kularatne J. D. J. S., Ceballos H., Comparative morphology, biology and histology of reproductive development in three lines of Manihot esculenta Crantz (Euphorbiaceae: Crotonoideae). AoB Plants 5, pls046 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rudi N., Norton G. W., Alwang J. R., Asumugha G. N., Economic impact analysis of marker-assisted breeding for resistance to pests and post-harvest deterioration in cassava. Afr. J. Agric. Resour. Econ. 4, 110–122 (2010). [Google Scholar]
- 16.Bredeson J. V., Lyons J. B., Prochnik S. E., Wu G. A., Ha C. M., Edsinger-Gonzales E., Grimwood J., Schmutz J., Rabbi I. Y., Egesi C., Nauluvula P., Lebot V., Ndunguru J., Mkamilo G., Bart R. S., Setter T. L., Gleadow R. M., Kulakow P., Ferguson M. E., Rounsley S., Rokhsar D. S., Sequencing wild and cultivated cassava and related species reveals extensive interspecific hybridization and genetic diversity. Nat. Biotechnol. 34, 562–570 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Bull S. E., Alder A., Barsan C., Kohler M., Hennig L., Gruissem W., Vanderschuren H., FLOWERING LOCUS T triggers early and fertile flowering in glasshouse cassava (Manihot esculenta Crantz). Plants 6, E22 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hilscher J., Bürstmayr H., Stoger E., Targeted modification of plant genomes for precision crop breeding. Biotechnol. J. 12, 1600173 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Ma X., Mau M., Sharbel T. F., Genome editing for global food security. Trends Biotechnol. 36, 123–127 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Puchta H., Applying CRISPR/Cas for genome engineering in plants: The best is yet to come. Curr. Opin. Plant Biol. 36, 1–8 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Scheben A., Wolter F., Batley J., Puchta H., Edwards D., Towards CRISPR/Cas crops—Bringing together genomics and genome editing. New Phytol. 216, 682–698 (2017). [DOI] [PubMed] [Google Scholar]
- 22.M. N. Emmambux, J. R. N. Taylor, Starch: Nutritional and health aspects, in Carbohydrates in Food, A.-C. Eliasson, Ed. (CRC Press Taylor & Francis Group, 2016), pp. 579–626. [Google Scholar]
- 23.Jobling S., Improving starch for food and industrial applications. Curr. Opin. Plant Biol. 7, 210–218 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Sonnewald U., Kossmann J., Starches—From current models to genetic engineering. Plant Biotechnol. J. 11, 223–232 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Moorthy S. N., Ramanujan T., Variations in properties of starch in cassava varieties in relation to age of the crop. Starch 38, 58–61 (1986). [Google Scholar]
- 26.Shure M., Wessler S., Fedoroff N., Molecular identification and isolation of the Waxy locus in maize. Cell 35, 225–233 (1983). [DOI] [PubMed] [Google Scholar]
- 27.Salehuzzaman S. N. I. M., Jacobsen E., Visser R. G. F., Isolation and characterization of a cDNA encoding granule-bound starch synthase in cassava (Manihot esculenta Crantz) and its antisense expression in potato. Plant Mol. Biol. 23, 947–962 (1993). [DOI] [PubMed] [Google Scholar]
- 28.Aiemnaka P., Wongkaew A., Chanthaworn J., Nagashima S. K., Boonma S., Authapun J., Jenweerawat S., Kongsila P., Kittipadakul P., Nakasathien S., Sreewongchai T., Wannarat W., Vichukit V., López-Lavalle L. A. B., Ceballos H., Rojanaridpiched C., Phumichai C., Molecular characterization of a spontaneous waxy starch mutation in cassava. Crop Sci. 52, 2121–2130 (2012). [Google Scholar]
- 29.Santelia D., Zeeman S. C., Progress in Arabidopsis starch research and potential biotechnological applications. Curr. Opin. Biotechnol. 22, 271–280 (2011). [DOI] [PubMed] [Google Scholar]
- 30.Koehorst-van Putten H. J. J., Sudarmonowati E., Herman M., Pereira-Bertram I. J., Wolters A. M. A., Meima H., de Vetten N., Raemakers C. J. J. M., Visser R. G. F., Field testing and exploitation of genetically modified cassava with low-amylose or amylose-free starch in Indonesia. Transgenic Res. 21, 39–50 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raemakers K., Schreuder M., Suurs L., Furrer-Verhorst H., Vincken J.-P., de Vetten N., Jacobsen E., Visser R. G. F., Improved cassava starch by antisense inhibition of granule-bound starch synthase I. Mol. Breed. 16, 163–172 (2005). [Google Scholar]
- 32.Zhao S.-S., Dufour D., Sánchez T., Ceballos H., Zhang P., Development of waxy cassava with different biological and physico-chemical characteristics of starches for industrial applications. Biotechnol. Bioeng. 108, 1925–1935 (2011). [DOI] [PubMed] [Google Scholar]
- 33.Ceballos H., Sánchez T., Morante N., Fregene M., Dufour D., Smith A. M., Denyer K., Pérez J. C., Calle F., Mestres C., Discovery of an amylose-free starch mutant in cassava (Manihot esculenta Crantz). J. Agric. Food Chem. 55, 7469–7476 (2007). [DOI] [PubMed] [Google Scholar]
- 34.Morante N., Ceballos H., Sánchez T., Rolland-Sabaté A., Calle F., Hershey C., Gibert O., Dufour D., Discovery of new spontaneous sources of amylose-free cassava starch and analysis of their structure and techno-functional properties. Food Hydrocoll. 56, 383–395 (2016). [Google Scholar]
- 35.Seung D., Soyk S., Coiro M., Maier B. A., Eicke S., Zeeman S. C., PROTEIN TARGETING TO STARCH is required for localising GRANULE-BOUND STARCH SYNTHASE to starch granules and for normal amylose synthesis in Arabidopsis. PLOS Biol. 13, e1002080 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lohmeier-Vogel E. M., Kerk D., Nimick M., Wrobel S., Vickerman L., Muench D. G., Moorhead G. B. G., Arabidopsis At5g39790 encodes a chloroplast-localized, carbohydrate-binding, coiled-coil domain-containing putative scaffold protein. BMC Plant Biol. 8, 120 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seung D., Boudet J., Monroe J., Schreier T. B., David L. C., Abt M., Lu K.-J., Zanella M., Zeeman S. C., Homologs of PROTEIN TARGETING TO STARCH control starch granule initiation in Arabidopsis leaves. Plant Cell 29, 1657–1677 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bull S. E., Owiti J. A., Niklaus M., Beeching J. R., Gruissem W., Vanderschuren H., Agrobacterium-mediated transformation of friable embryogenic calli and regeneration of transgenic cassava. Nat. Protoc. 4, 1845–1854 (2009). [DOI] [PubMed] [Google Scholar]
- 39.Jane J., Chen Y. Y., Lee L. F., McPherson A. E., Wong K. S., Radosavljevic M., Kasemsuwan T., Effects of amylopectin branch chain length and amylose content on the gelatinization and pasting properties of starch. Cereal Chem. J. 76, 629–637 (1999). [Google Scholar]
- 40.Park I.-M., Ibáñez A. M., Zhong F., Shoemaker C. F., Gelatinization and pasting properties of waxy and non-waxy rice starches. Starch 59, 388–396 (2007). [Google Scholar]
- 41.Tanaka Y., Miura H., Fukushima M., Ito M., Nishio Z., Kim S.-J., Hashimoto N., Noda T., Takigawa S., Matsuura-Endo C., Yamauchi H., Physical properties of yellow alkaline noodles from near-isogenic wheat lines with different Wx protein deficiency. Starch 58, 186–195 (2006). [Google Scholar]
- 42.Sharma R., Sissons M. J., Rathjen A. J., Jenner C. F., The Null-4A allele at the waxy locus in durum wheat affects pasta cooking quality. J. Cereal Sci. 35, 287–297 (2002). [Google Scholar]
- 43.Rolland-Sabaté A., Sánchez T., Buléon A., Colonna P., Jaillais B., Ceballos H., Dufour D., Structural characterization of novel cassava starches with low and high-amylose contents in comparison with other commercial sources. Food Hydrocoll. 27, 161–174 (2012). [Google Scholar]
- 44.Smith S. M., Fulton D. C., Chia T., Thorneycroft D., Chapple A., Dunstan H., Hylton C., Zeeman S. C., Smith A. M., Diurnal changes in the transcriptome encoding enzymes of starch metabolism provide evidence for both transcriptional and posttranscriptional regulation of starch metabolism in Arabidopsis leaves. Plant Physiol. 136, 2687–2699 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zeeman S. C., Tiessen A., Pilling E., Kato K. L., Donald A. M., Smith A. M., Starch synthesis in Arabidopsis. Granule synthesis, composition, and structure. Plant Physiol. 129, 516–529 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Watson A., Ghosh S., Williams M. J., Cuddy W. S., Simmonds J., Rey M.-D., Hatta M. A. M., Hinchliffe A., Steed A., Reynolds D., Adamski N. M., Breakspear A., Korolev A., Rayner T., Dixon L. E., Riaz A., Martin W., Ryan M., Edwards D., Batley J., Raman H., Carter J., Rogers C., Domoney C., Moore G., Harwood W., Nicholson P., Dieters M. J., DeLacy I. H., Zhou J., Uauy C., Boden S. A., Park R. F., Wulff B. B. H., Hickey L. T., Speed breeding is a powerful tool to accelerate crop research and breeding. Nat. Plants 4, 23–29 (2018). [DOI] [PubMed] [Google Scholar]
- 47.M. Bobek, Case C‑528/16 (European Court of Justice, 2018). [Google Scholar]
- 48.Odipio J., Alicai T., Ingelbrecht I., Nusinow D. A., Bart R., Taylor N. J., Efficient CRISPR/Cas9 genome editing of Phytoene desaturase in Cassava. Front. Plant Sci. 8, 1780 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.M. W. Blair, M. A. Fregene, S. E. Beebe, H. Ceballos, Marker-assisted selection in common beans and cassava, in Marker-Assisted Selection: Current Status and Future Perspectives in Crops, Livestock, Forestry and Fish, E. P. Guimarães, J. Ruane, B. D. Scherf, A. Sonnino, J. D. Dargie, Eds. (FAO, 2007), pp. 81–115. [Google Scholar]
- 50.H. Ceballos, C. Hershey, L. A. Becerra-López-Lavalle, New approaches to cassava breeding, in Plant Breeding Reviews, J. Janick, Ed. (John Wiley & Sons Inc., 2012), pp. 427–504. [Google Scholar]
- 51.Chetty C. C., Rossin C. B., Gruissem W., Vanderschuren H., Rey M. E. C., Empowering biotechnology in southern Africa: Establishment of a robust transformation platform for the production of transgenic industry-preferred cassava. Nat. Biotechnol. 30, 136–143 (2013). [DOI] [PubMed] [Google Scholar]
- 52.Vanderschuren H., Strengthening African R&D through effective transfer of tropical crop biotech to African institutions. Nat. Biotechnol. 30, 1170–1172 (2012). [DOI] [PubMed] [Google Scholar]
- 53.Yoo S.-D., Cho Y.-H., Sheen J., Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572 (2007). [DOI] [PubMed] [Google Scholar]
- 54.Engler C., Gruetzner R., Kandzia R., Marillonnet S., Golden gate shuffling: A one-pot DNA shuffling method based on type IIs restriction enzymes. PLOS ONE 4, e5553 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Engler C., Youles M., Gruetzner R., Ehnert T.-M., Werner S., Jones J. D. G., Patron N. J., Marillonnet S., A golden gate modular cloning toolbox for plants. ACS Synth. Biol. 3, 839–843 (2014). [DOI] [PubMed] [Google Scholar]
- 56.Nagaya S., Kawamura K., Shinmyo A., Kato K., The HSP terminator of Arabidopsis thaliana increases gene expression in plant cells. Plant Cell Physiol. 51, 328–332 (2010). [DOI] [PubMed] [Google Scholar]
- 57.Anders C., Jinek M., In vitro enzymology of Cas9. Methods Enzymol. 546, 1–20 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Murashige T., Skoog F., A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 15, 473–497 (1962). [Google Scholar]
- 59.Soni R., Murray J. A. H., Isolation of intact DNA and RNA from plant tissues. Anal. Biochem. 218, 474–476 (1994). [DOI] [PubMed] [Google Scholar]
- 60.Chang S., Puryear J., Cairney J., A simple and efficient method for isolating RNA from pine trees. Plant Mol. Biol. Report. 11, 113–116 (1993). [Google Scholar]
- 61.Livak K. J., Schmittgen T. D., Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- 62.Park J., Lim K., Kim J.-S., Bae S., Cas-analyzer: An online tool for assessing genome editing results using NGS data. Bioinformatics 33, 286–288 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- 64.Haeussler M., Schönig K., Eckert H., Eschstruth A., Mianné J., Renaud J.-B., Schneider-Maunoury S., Shkumatava A., Teboul L., Kent J., Joly J.-S., Concordet J.-P., Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 17, 148 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.C. Hostettler, K. Kölling, D. Santelia, S. Streb, O. Kötting, S. C. Zeeman, Analysis of starch metabolism in chloroplasts, in Methods in Molecular Biology, R. P. Jarvis, Ed. (Springer, 2011), pp. 387–410. [DOI] [PubMed] [Google Scholar]
- 66.Hovenkamp-Hermelink J. H. M., De Vries J. N., Adamse P., Jacobsen E., Witholt B., Feenstra W. J., Rapid estimation of the amylose/amylopectin ratio in small amounts of tuber and leaf tissue of the potato. Potato Res. 31, 241–246 (1988). [Google Scholar]
- 67.Clédat D., Battu S., Mokrini R., Cardot P. J. P., Rice starch granule characterization by flow cytometry scattering techniques hyphenated with sedimentation field–flow fractionation. J. Chromatogr. A 1049, 131–138 (2004). [PubMed] [Google Scholar]
- 68.Wolf M. J., Ruggles V. J., MacMasters M. M., Refractive indices of wheat starch granules at various moisture levels determined with an interference microscope. Biochim. Biophys. Acta 57, 135–142 (1962). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/9/eaat6086/DC1
Fig. S1. Molecular screening of transgenic plants.
Fig. S2. Efficacy of the genome editing constructs in vitro and in vivo.
Fig. S3. Screening of predicted off-target cleavage sites for GBSSsgRNA4 and PTSTsgRNA2.
Fig. S4. Morphology of glasshouse-cultivated gbss and ptst lines.
Fig. S5. CLDs in storage root starch of gbss and ptst lines.
Fig. S6. Flow cytometry analysis of starch granule size in gbss and ptst lines.
Fig. S7. Scanning electron microscopy of purified gbss and ptst starch granules.
Fig. S8. Physicochemical properties of gbss and ptst starch.


