SUMMARY
Amongst the several arthropod-borne human flaviviral diseases, the recent outbreak of Zika virus (ZIKV) has caused devastating birth defects and neurological disorders, challenging the world with another major public health concern. We report here the refined structure of the mature ZIKV at a resolution of 3.1 Å as determined by cryo-electron microscopic single particle reconstruction. The improvement in the resolution, compared to previous enveloped virus structures, was because of optimized virus preparation methods and data processing techniques. The glycoprotein interactions and surface properties of ZIKV were compared with other mosquito-borne flavivirus structures. The largest structural differences and sequence variations occur at the glycosylation loop associated with receptor binding. Probable drug binding pockets were identified on the viral surface. These results also provide a structural basis for the design of vaccines against ZIKV.
Keywords: Zika virus, flavivirus comparisons, structure refinement, antiviral binding sites, receptor binding sites
eTOC blurb
The atomic structure of mature Zika virus at a resolution of 3.1 Å and its comparison with other mosquito-borne flavivirus structures revealed important structural components of the infection mechanism. These results also identified probable drug binding pockets and provides a structural basis for vaccine design.
INTRODUCTION
Zika virus (ZIKV) is a member of the Flaviviridae family, which consists of arthropod-borne, lipid-enveloped, positive, single-stranded RNA viruses. There are 52 recorded species in the flavivirus genus, many of which cause widespread morbidity and mortality of humans (Lindenbach and Rice, 2003). Amongst the major symptoms and illnesses caused by these viruses are fevers, headaches, diarrhea, nausea, encephalitis and severe neurological disorders. The most important disease-causing human viruses of this family are yellow fever virus (YFV) (Butler, 2016), dengue virus (DENV) (Guzman and Harris, 2015), West Nile virus (WNV) (Sejvar, 2016), Japanese encephalitis virus (JEV) (Wang and Liang, 2015), tick-borne encephalitis virus (TBEV) (Suss, 2008) and Zika virus (ZIKV) (Weaver et al., 2016). The recent ZIKV outbreak has been linked to microcephaly, a birth defect in newborn babies causing an abnormally small head (Brasil et al., 2016; Honein et al., 2017), and, in adults, Guillain-Barré syndrome (Cao-Lormeau et al., 2016; Passi et al., 2017). Based on structure-based phylogeny, ZIKV has greater similarity to the neurovirulent JEV and WNV than to DENV (Wang et al., 2017). Three-dimensional (3D) structure based functional analyses at atomic resolution should enhance identification of targets for rational design of vaccines and antiviral therapeutics against these viruses.
Flaviviruses have icosahedral symmetry with a diameter of about 500 Å and a genome of about 11,000 bases. The genome is packaged into a host-derived lipid membrane and encodes three structural proteins: (1) envelope (E), (2) pre-membrane (prM) and (3) capsid (C) as well as seven nonstructural proteins (Hasan et al., 2018; Sirohi and Kuhn, 2017). The glycoprotein shell consists of membrane anchored E (~500 amino acids) and M/prM proteins (~75/165 amino acids). Flavivirus structural proteins, E and prM/M play a role in early stages of viral infection, viral entry and receptor mediated endocytosis (Kaufmann and Rossmann, 2011; Perera-Lecoin et al., 2013; Smit et al., 2011). Both E and M/prM undergo large, pH-induced conformational changes during infection in which the spiky, immature particles consisting of 60 trimeric E:prM heterodimers change to smooth, mature particles consisting of 90 dimeric E:M heterodimers (Kuhn et al., 2002). Mature virus particles are formed after the cleavage of prM by the host protease furin to remove the pr peptide and expose the fusion loop. The mature particles subsequently become fusogenic and infectious upon removal of the pr peptide in the low pH environment of endosomes (Mukhopadhyay et al., 2005; Zhang et al., 2015; Zhang et al., 2004).
Cryo-electron microscopy (cryo-EM) structures of several flaviviruses have been determined starting with mature DENV structures at resolutions of 24 Å, 9.5 Å and 3.6 Å (Kuhn et al., 2002; Zhang et al., 2003a; Zhang et al., 2013b); immature DENV structures at resolutions of 16 Å, 12.5 Å and 6 Å (Kostyuchenko et al., 2013; Li et al., 2008; Zhang et al., 2003b; Zhang et al., 2004); fusogenic DENV structure at a resolution of 26 Å (Zhang et al., 2015); mature JEV structure at a resolution of 4.3 Å (Wang et al., 2017), mature TBEV structure at a resolution of 3.9 Å (Fuzik et al., 2018) and structures of immature and mature WNV at resolutions of 24 and 10.3 Å, respectively (Zhang et al., 2013a; Zhang et al., 2007). In addition, several flavivirus structures have been determined in complex with neutralizing antibodies (Barba-Spaeth et al., 2016; Cherrier et al., 2009; Cockburn et al., 2012a; Cockburn et al., 2012b; Dejnirattisai et al., 2016; Hasan et al., 2017; Kaufmann et al., 2009; Kaufmann et al., 2006; Kaufmann et al., 2010; Lok et al., 2008; Modis et al., 2005; Pokidysheva et al., 2006; Wang et al., 2013; Yu et al., 2009). The structure of mature ZIKV has been determined at a resolution of 3.8 Å (Sirohi et al., 2016) at room temperature and 3.7 Å at 40 °C (Kostyuchenko et al., 2016). Immature ZIKV structure has been determined to a resolution of 9.1 Å (Mangala Prasad et al., 2017). In the past two decades, the determination of these structures were milestones in understanding the function of E/prM proteins during infection.
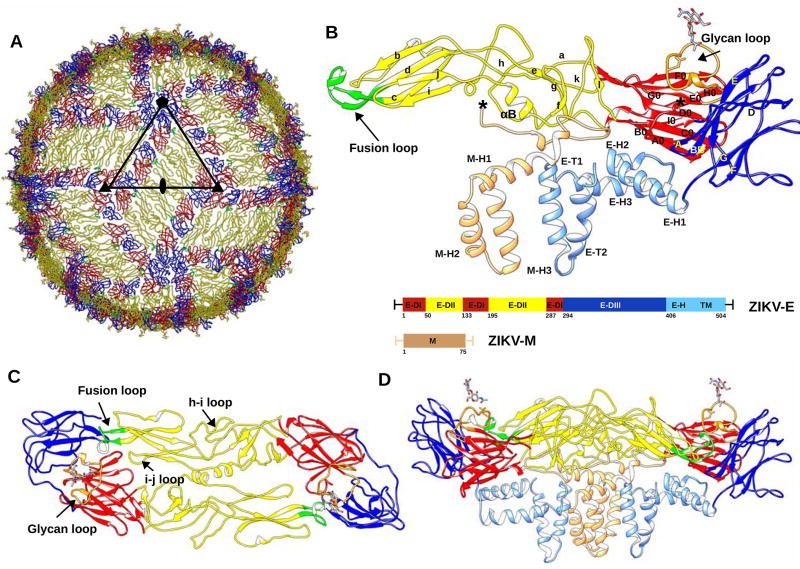
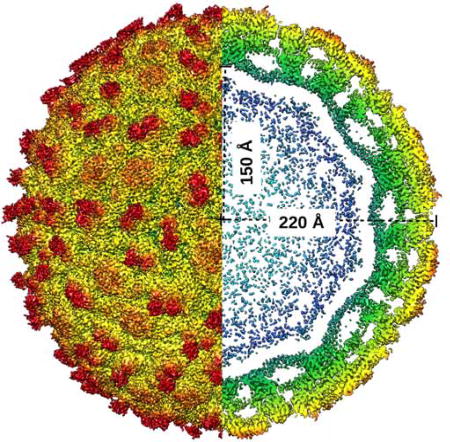
Mature ZIKV has an icosahedral, smooth surface covered by 180 copies each of the E and M protein (Kostyuchenko et al., 2016; Sirohi et al., 2016) (Fig. 1). The icosahedral asymmetric unit consists of three E–M heterodimers. Adjacent icosahedral asymmetric units form a herringbone pattern on the surface of the virus. The envelope protein, E, consists of three β-barrel domains: E-DI, E-DII, and E-DIII, which cover the virus capsid and are anchored on the internal membrane by two transmembrane helices: E-T1 and E-T2 via the three stem helices: E-H1, E-H2 and E-H3 (Rey et al., 1995). The N-terminal E-DI connects the dimerization domain E-DII and the C-terminal immunoglobulin-like domain E-DIII by two hinge regions (Table S1). The M protein has three helices: M-H1, M-H2 and M-H3. The helices M-H2 and M-H3 are transmembrane helices. The N-terminal residues of the M protein form a long loop making widespread interactions with the membrane side of E-DII (Figure 1). ZIKV E-DI is formed by nine β-strands, A0-I0, and three small 310 helices. Asn154 in the loop between β-strands E0 and F0 (146–166) is glycosylated and is implicated in neurovirulence (Annamalai et al., 2017). ZIKV E-DII has nine β-strands (a–i) and two α-helices, αA and αB. The hydrophobic loop between strands c and d contains the highly conserved fusion loop (98–109), important for membrane fusion during flavivirus infection (Mukhopadhyay et al., 2005). ZIKV E-DIII, implicated in receptor binding, consists of strands A–G. The hinge region between E-DI-DIII and E-DII plays a crucial role in domain movement and dimer to trimer transformation from immature to the mature and to the fusogenic form (Zhang et al., 2004).
Figure 1. Overall Structure of ZIKV.
(A) ZIKV structure highlighting the herringbone pattern formed by 6 E–M heterodimers. The icosahedral asymmetric unit, is outlined by a black triangle. The structures of the E proteins are shown as Cα backbone traces. Domains E-DI, E-DII and E-DIII of each E protein are shown in red, yellow and blue, respectively. The fusion loop of each E protein molecule is shown in green.
(B) Secondary structural elements of one E–M heterodimer are labelled. Secondary structure elements on E-DI, E-DII and E-DIII are labelled A0–I0, a–l and A–G, respectively. The stem and transmembrane helices (E-H1, E-H2, E-H3, E-T1, E-T2) of the E protein and the M protein (M-H1, M-H2, M-H3) are colored in light blue and light brown, respectively. Residue numbering and domain definitions are shown as a linear peptide. Domains E-DI, E-DII and E-DIII are colored as in Figure 1A.
Panels (C) and (D) show the top and side views of the dimer, respectively. The glycan loop, fusion loop, h–I loop and i–j loop are labelled. (See also Table S1.)
Here we report the 3.1 Å resolution structure and refinement of the mature ZIKV based on newly collected, single-particle, cryo-EM data. The refined atomic coordinates of ZIKV were then used to compare with other refined mosquito-borne flavivirus structures (DENV at 3.5 Å and JEV at 4.3 Å). Structure-based sequence alignments and differences in the landscape of surface exposed residues and accessible pockets shed light on structural adaptation to cellular tropism and escape from neutralizing antibodies.
RESULTS AND DISCUSSION
Cryo-EM Reconstruction and Refinement of the ZIKV Atomic Model
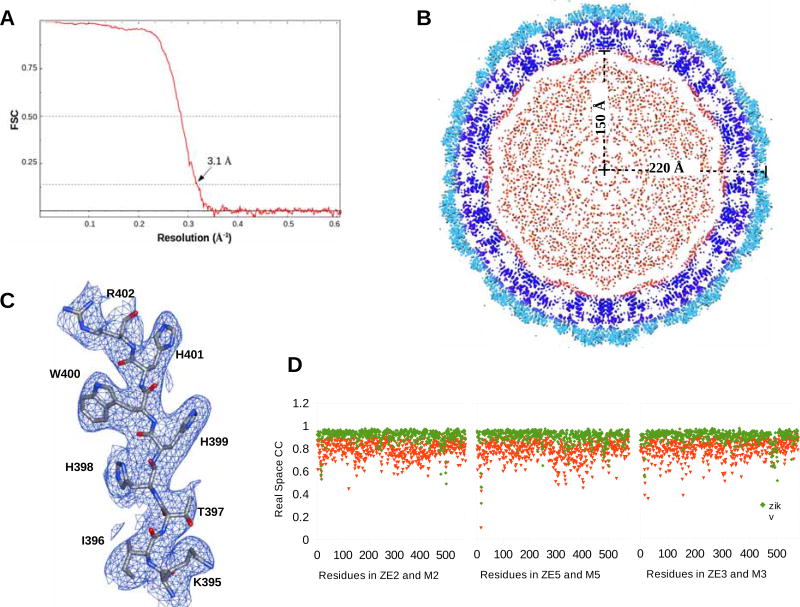
About 36,000 ZIKV particles (strain H/PF/2013) were selected from 2,085 cryo-EM micrographs based on two-dimensional (2D) classification. High resolution icosahedral refinement of orientation, center, astigmatism and magnification of the 2D projections were performed using the JSPR (Guo and Jiang, 2014) program. This generated a cryo-EM map at an average resolution of 3.1 Å, according to the 0.143 Fourier shell correlation (FSC) criterion (Figure 2) (Rosenthal and Henderson, 2003).
Figure 2. The Cryo-EM Reconstruction of ZIKV.
(A) Fourier Shell Coefficient plot versus resolution.
(B) A central section looking down a 5-fold axis of the cryo-EM electron potential map. The E-ectodomain (1–400), the E and M transmembrane regions and the core are colored light blue, dark blue and red, respectively.
(C) The cryo-EM map contoured at 1.5 σ around residues Lys395-Arg402.
(D) Plot showing real space correlation coefficient for the residues in the current structure (green) and the earlier structure (PDB ID:5IRE) (red).
Density-guided real-space refinement of the ZIKV model (PDB ID: 5IRE) into the 3.1 Å cryo-EM map was performed using the programs COOT (Emsley and Cowtan, 2004) and PHENIX.REFINE (Afonine et al., 2012). Nearly all the side chains were well-resolved in contrast to previous ZIKV structure determinations (Kostyuchenko et al., 2016; Sirohi et al., 2016). The model was refined against Fourier coefficients from the experimentally determined cryo-EM map placed into a cubic shaped box with cell dimensions of 830 Å. The cryo-EM map was Fourier inverted to give “observed” amplitudes and phases. These were compared with the calculated structure factors of the icosahedral model. Contrary to normal crystallographic refinement, where only amplitudes are observed, the cryo-EM map provided both amplitudes and phases. Thus the refinement minimized the phased maximum likelihood target function as implemented in PHENIX.REFINE (Pannu et al., 1998).
Reciprocal space refinement using electron scattering factors and no restraints to equalize the structure of independent monomers was alternated with real space refinement in PHENIX.REFINE (Afonine et al., 2012) and visual model improvements using COOT (Emsley and Cowtan, 2004). The refinement converged to an Rwork of 25.2 % and a real-space correlation coefficient between the observed and calculated electron potential maps in the vicinity of the fitted model of about 0.921. The data statistics for the whole virus map and the refinement statistics for the icosahedral asymmetric unit are summarized in Table 1.
Table 1.
Data and Refinement Statistics
| Statistics | |
|---|---|
|
| |
| Cell dimension a, b, c (Å) | 830.0, 830.0, 830.0 |
| Cell angles α, β, γ (°) | 90.0, 90.0, 90.0 |
| Resolution (Å) | 3.1 |
| No. of Fourier Coefficients | 33,257,143 |
| Rworkb | 25.2% |
| Number of non-hydrogen protein atoms (iasua) | 13256 |
| Average B factor (Å2) | 21 |
| r.m.s.d bond lengths (Å) from idealized values | 0.014 |
| r.m.s.d bond angles (°) from idealized values | 1.5 |
| Ramachandran Plot | |
| No. residues in favored + allowed regions | 1545 + 165 (99.65%) |
| No. of amino acid outliers | 6 (0.45%) |
iasu: icosahedral asymmetric unit,
r.m.s.d: root-mean-square-deviation, numbers were generated in Phenix (Afonine et al., 2012).
3D Structural Comparisons and Sequence Alignment
The Cα atoms of the refined ZIKV structure differ by a root mean square deviation (r.m.s.d) of 1.2 Å with respect to the previously determined structure (PDB ID: 5IRE). Differences as large as 1.9–3.0 Å were at several loop and transmembrane regions. The current model had an improved Rwork, and an improved real-space correlation coefficient, geometry and validation statistics relative to the earlier structure (Sirohi et al., 2016). Most of the loops, side chains and disulfide bonds unlike the earlier ZIKV structures are well resolved and easily recognizable (Figure 2). This provides information of the H-bond networks that control capsid assembly during different stages in the life cycle of the virus.
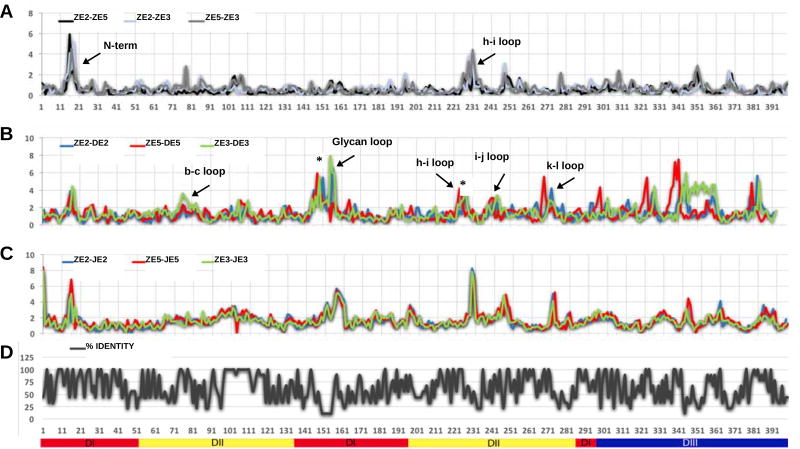
Although the overall fold of the three E monomers in the icosahedral asymmetric unit is similar, the immediate environment of each E monomer is different resulting in significant structural differences between the three independent E monomers. For the present discussion the three independent E monomers have been named E2, E3 and E5 according to whether the monomer is closest to a 2-fold, 3-fold or 5-fold icosahedral axis. Thus the E monomers in ZIKV, DENV2 (PDB ID: 3J27) and JEV (PDB ID: 5WSN) were identified as ZE2, ZE3, ZE5; DE2, DE3, DE5; JE2, JE3, JE5, respectively. The r.m.s.d between ZE2–ZE3, ZE2–ZE5 and ZE3–ZE5 were 0.74 Å, 0.61 Å, 0.77 Å, respectively (at a resolution of 3.1 Å) whereas in DENV, DE2–DE3, DE2–DE5, DE3–DE5, the r.m.s.d’s were 1.14 Å, 1.0 Å, 1.0 Å, respectively at a resolution of 3.5 Å (Figure 3). The smaller differences between independent ZIKV E monomers probably reflects the better resolution and optimized refinement.
Figure 3. Comparison of ZIKV, DENV and JEV Structures.
(A) R.m.s.d between equivalent Cα atoms of the three independent monomers in ZIKV (ZE2, ZE3, ZE5).
(B) R.m.s.d between equivalent Cα atoms of DENV monomers (DE2, DE3, DE5) and ZIKV E monomers.
(C) R.m.s.d between equivalent Cα atoms of JEV (JE2, JE3, JE5) monomers and ZIKV monomers.
(D) Plot showing percent sequence identity from multiple sequence alignment as in Figure S1.
The secondary structure elements with larger r.m.s.d’s are labeled in panels (A) and (B). Sequence insertions on ZIKV are marked with an asterisk on panel (B). In (A), (B) and (C) the x-axis shows amino acid residue numbering and the y-axis shows the r.m.s.d between Cα atoms. The secondary structure elements of the E-ectodomain are labelled below panel (D). (See also Figures S1 and S2.)
The largest differences (>6 Å for eight successive residues) between equivalent Cα atoms of the independent monomers in ZIKV occur in three specific regions. In contrast, no obvious large conformational differences (>6 Å for any one residue) were observed when comparing independent DENV monomers with each other or when comparing independent JEV monomers with each other. In retrospect this was not surprising because the refinement of the DENV structure was heavily dependent on averaging density between the independent monomers (Zhang et al., 2013b). Similarly, the deposited coordinates of the JEV structure (Wang et al., 2017) at a resolution of 4.3 Å show that there is no difference between the 3D structures of the three independent monomers, demonstrating that the refinement assumed the monomers had identical structures. Therefore, the three short regions, the N-terminal 20 residues and the loops between the strands h–i and i–j in E-DII that showed larger differences in ZIKV might be functionally significant. The E protein N-terminal region of ZIKV is surrounded by the glycan loop (145–165), the fusion loop (98–109) of a neighboring monomer and the N-terminal residues of the M protein. Thus, the structural differences between the ZIKV monomers can be attributed to this region being susceptible to large conformational changes during transformation from immature to mature forms.
The global root-mean-square deviation (r.m.s.d.) between Cα atoms of the ectodomain (E-DI, E-DII and E-DIII) of ZE2-DE2, ZE3-DE3, ZE5-DE5 are 1.78, 1.65 and 1.87 Å, respectively and the r.m.s.d between Cα atoms of the ectodomain (E-DI, E-DII and E-DIII) of ZE2-JE2, ZE3-JE3, ZE5-JE5 are 2.03, 2.02 and 1.98 Å, respectively (Figure 3). Structure-based sequence alignments of flavivirus E proteins show that strands A0–D0, strand I0, the fusion loop, the dimerization helix αB and the E-DI–DIII hinge contain large stretches of conserved residues (Figure S1 and Table S1). In contrast, the surface exposed loops (h–i loop, k–l loop, D–E loop and the glycan loop) are more variable (Figure S2). However, the neurovirulent flaviviruses (ZIKV, WNV, JEV) are mostly more conserved than in DENV and YFV (Figures S1 and S2).
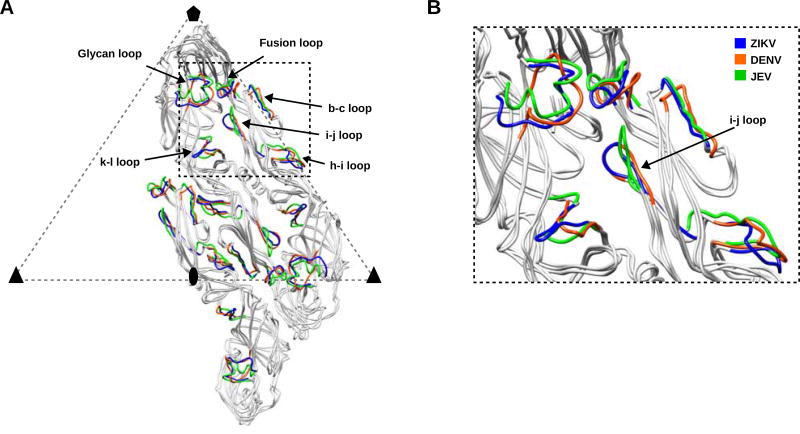
The surface-exposed E-DIII loops of ZIKV, DENV and JEV E proteins show a few large structural variations (~ 8 Å). These loops of E-DIII are implicated in receptor binding and form a unique signature of flavivirus cellular tropism and infectivity. The largest differences between ZIKV, DENV and JEV structures correspond to large sequence variations. These differences are in the glycan loop region and its immediate environment formed by the loops between strands b–c, h–i, i–j and k–l. These loops, located within the dimer-dimer interface, are surface-exposed and form an accessible pocket. The i–j loop has a similar closed conformation in ZIKV and DENV, whereas in JEV this loop has an open conformation (Wang et al., 2017) (Figure 4). The differences in these loop conformations might account for differences in binding of neutralizing antibodies. For example, neutralizing antibodies binding to these variable loop regions near the dimer-dimer interface are highly potent, ZIKV specific and show greater neutralization activity (Hasan et al., 2017).
Figure 4. Superposition of Cα Atoms of the E Monomers in One Asymmetric Unit of DENV and JEV on ZIKV.
(A) The E2, E3 and E5 monomers of ZIKV, DENV and JEV are colored in grey ribbons. Structural elements with maximum r.m.s.d’s in ZIKV, DENV and JEV are colored in blue, orange and green, respectively.
(B) The inset shows a region around the glycan loop and the differences in closed and open conformations of i–j loop in DENV and JEV respectively.
In ZIKV, the Asn154 glycosylation site (on the glycan loop) is relevant to neurovirulence (Annamalai et al., 2017). The conserved residues in all three neurovirulent viruses ZIKV, WNV and JEV (Asp67, Lys84, Asn238, Glu244, Glu276) are within these structurally variable regions (Figure S1). Binding of virus to host cell and subsequent entry occurs by sequential interactions with attachment factors and primary receptors (Perera-Lecoin et al., 2013). For example, in DENV negatively-charged glycosaminoglycan’s (GAG) such as heparan sulfate (Chen et al., 1997; Germi et al., 2002) and dendritic cell-specific intercellular adhesion molecule-3-grabbing non-integrin (DC-SIGN) (Tassaneetrithep et al., 2003) form the primary attachment factors. In DENV, the Asn67 glycosylation site, adjacent to a larger positively charged patch, is recognized by the carbohydrate recognition domain of DC-SIGN (Pokidysheva et al., 2006). The region described above with large structural differences in ZIKV is also adjacent to a positively charged patch near the conserved glycosylation site (Asn154) and might be one of the unique receptor binding sites. The positively charged patch in ZIKV is formed by residues Arg73, Arg99, Arg138, Lys166, Lys251, Arg252, Lys281 and Arg283 (Figure S3).
E-E and E–M Oligomer Interactions, Surface Properties and Pockets
The icosahedral mature virion envelope is formed on a lipid membrane by polar and hydrophobic interactions between various secondary structure elements in the E and M proteins (Tables S2, S3 and S4). The E-E interactions at the 2-fold in ZIKV mainly occur around helix αB (Table S4). This stretch has many conserved residues among flaviviruses (Figure S1). A second set of interactions between E monomers occurs between the fusion loop and the i–j loop in E-DII, the glycan loop in ED-I, and residues of strands A and B in E-DIII. The glycan loop between strands E0 and F0 of E-DI is about eight residues longer and proximal to the fusion loop in all three neurovirulent viruses (ZIKV, WNV and JEV) when compared to DENV. The close proximity of the glycan loop to the fusion loop hints that any interaction of the glycan loop with a putative receptor molecule on the host cell surface might promote the exposure of the fusion loop facilitating the formation of fusogenic trimers and thereby leading to membrane fusion.
The E–M heterodimers are held together by interactions between E-E, E–M and M-M proteins (Table S4). In JEV and DENV, these interactions are fewer and confined to the N-terminus of M protein in comparison to ZIKV, where the N-terminal loop, the helices M-H1 and M-H3, and the C-terminal transmembrane residues of the M-proteins are involved. This might account for the conformationally similar virus structure of mature ZIKV at higher temperatures (Kostyuchenko et al., 2016) in contrast to DENV (DENV2), which transforms to a bumpy structure at 37 °C (Fibriansah et al., 2013; Zhang et al., 2013c).
The E proteins in ZIKV pack tightly around the 5-fold vertex (Kostyuchenko et al., 2016) compared to DENV and JEV, stabilized by many interactions between side chains of E-DIII loops: C–D, F–G, the strands: A–C, F and E-DI-DIII hinge (Table S2). The 3-fold in ZIKV, DENV and JEV is lined by loops of E-DIII, E-DI-DII hinge, E-DI-DIII hinge and the strands G0 and H0 (Table S3). There are no interactions observed between transmembrane and stem helices of E and M proteins at either the 3-fold or 5-fold axes. Although, it seems reasonable to attribute the conformationally similar structures (no transformation to a bumpy structure at higher temperatures similar to DENV) of ZIKV at higher temperatures to its compact 5-fold interactions, recent mutational studies have shown that ZIKV infectivity decreases similar to other flaviviruses when incubated at 40 °C (Goo et al., 2016). Moreover, comparison of the interactions at the dimer interface to the interactions at the 3-fold and 5-fold axes clearly show that the ectodomain, stem helices and transmembrane interactions between E-E and E–M proteins contribute to a stable, mature structure (Table S4).
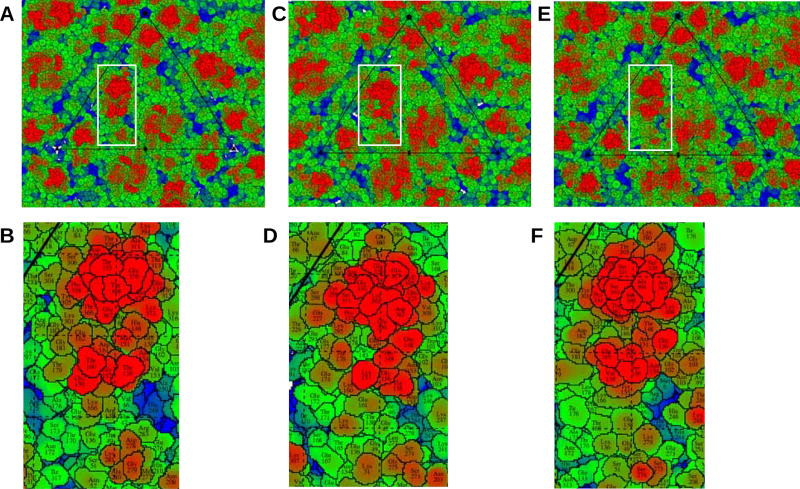
“Roadmaps” of surface exposed residues projected on the equatorial plane were calculated using the program RIVEM (Xiao and Rossmann, 2007) (Figure 5). Major differences can be observed in the landscape of these viruses depending on the size, shape, position and properties of surface exposed residues in ZIKV, DENV and JEV (Figure 5). For example, both in ZIKV and JEV, the region formed by E-DIII loops and the glycan loop adjacent to it (shown in red in Figure 5) is rich in serine and threonine residues, whereas DENV has a different residue distribution in the same region. The differences in these surface exposed motifs might direct these viruses to attach and infect a wide variety of different host cells. Identification and mutational studies of the residues forming these motifs could be a strategy towards generation of attenuation for successful flavivirus vaccine design.
Figure 5. Surface-Exposed Residues in ZIKV, DENV and JEV.
Roadmaps of surface-exposed residues in ZIKV (panels A and B), DENV (panels C and D) and JEV (panels E and F). Panels B, D, and F depict regions near the glycan loop as outlined by the white rectangles in panels A, C, and E, respectively. In (B) ZIKV and (F) JEV, the region formed by the E-DIII surface residues and the adjacent glycan loop (shown in red) is rich in serine and threonine residues, whereas (D) DENV has a different residue distribution in the same region. The shape of the motif formed by the surface exposed residues is similar in (B) ZIKV and (F) JEV. (See also Figures S3, S4, S5 and Tables S2, S3 and S4.)
The receptor binding domain E-DIII (Alhoot et al., 2013; Liao and Kielian, 2005), the stem helices (Hrobowski et al., 2005; Lok et al., 2012) and several regions enclosed by E-E and E–M dimers can be targets for the design of anti-viral agents (De La Guardia and Lleonart, 2014). Accessible pockets on the surface of the virus were calculated using the program 3V (Voss and Gerstein, 2010), which uses the rolling probe method for the calculation of volumes. The volume of accessible pockets in the space enclosed between E-E homodimer of ZIKV, DENV and JEV is 1840 Å3, 1538 Å3 and 3643 Å3 respectively (Figure S4). Several potential inhibitors, like tetracycline derivatives (Yang et al., 2007) and A5 (Kampmann et al., 2009), were identified to bind these pockets in DENV (De La Guardia and Lleonart, 2014). The differences in the residue environment and the size of accessible pockets in ZIKV, DENV and JEV can be utilized in developing strain-specific small molecule and peptide inhibitors.
Concluding Remarks
Flavivirus envelope protein, E, is the principal component of the virion surface and is required for host cell recognition, receptor binding and infection (Kaufmann and Rossmann, 2011; Perera-Lecoin et al., 2013; Smit et al., 2011). The three E monomers in the asymmetric unit of ZIKV have significant (>3σ, where σ is ~0.7Å) structural differences. Further comparisons with DENV and JEV identified regions of greater structural variability to coincide with sequence variability. The largest structural differences and highest sequence variations are observed in the glycan loop, the h–i loop, the hinge connecting E-DI and E-DII and the surface exposed loops of E-DIII (Figures 3 & 4 and Figures S1 & S2). Both structural and sequence variability, results in different surface characteristics. Detailed studies of interactions between the E and M proteins at the 2-fold, 3-fold and 5-fold axes identified stabilizing interactions in ZIKV that are missing in DENV and JEV (Figure S5 and Tables S2, S3 and S4) and, therefore, provide a starting point for mutational analyses.
STAR★METHODS
Detailed methods are provided in the online version of this paper and include the following:
CONTACT FOR REAGENT AND RESOURCE SHARING
- EXPERIMENTAL MODEL AND SUBJECT DETAILS
-
◦Species/strain of experimental models
-
◦Cultural conditions for microorganisms
-
◦
- METHOD DETAILS
-
◦Purification of mature ZIKV particles
-
◦Cryo-EM reconstruction and structure refinement of ZIKV
-
◦Multiple sequence alignments
-
◦3D superposition, residue contacts and calculation of surface properties
-
◦
QUANTIFICATION AND STATISTICAL ANALYSIS
DATA AND SOFTWARE AVAILABILITY
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| Zika Virus, strain H/PF/2013 | European Virus Archive | Ref SKU: 001v-EVA1545 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Blasticidin | Invivo-Gen | Cat. Code ant-bl-1 |
| DMEM Medium | ThermoFisher/Gibco | Ref #12800-082 |
| FBS Medium | Sigma | Cat # F2442 |
| 100xNEAA | ThermoFisher/Gibco | Ref # 11140-050 |
| Penicillin-Streptomycin | ThermoFisher | Cat # 15070063 |
| PEG8000 | SIGMA-ALDRICH | Product # P5413 |
| Tris | SIGMA-ALDRICH | Product # 11814273001 |
| NaCl | SIGMA-ALDRICH | Product # S7653 |
| EDTA | SIGMA-ALDRICH | Product # E6758 |
| K-tartrate | SIGMA-ALDRICH | Product # S2377 |
| Deposited Data | ||
| Coordinates | this manuscript | PDB ID 6CO8 |
| (Zhang et al., 2013b) | PDB ID 3J27 | |
| (Wang et al., 2017) | PDB ID 5WSN | |
| Map | this manuscript | EMDB 7543 |
| Experimental Models: Cell Lines, microbe strains | ||
| Vero-Furin (Vero cells that stably express furin) | (Mukherjee, S. et. al. 2016) | N/A |
| Zika Virus, strain H/PF/2013 | European Virus Archive | Ref SKU: 001v-EVA1545 |
| Software and Algorithms | ||
| APPION | (Lander et al., 2009) | http://emg.nysbc.org/redmine/projects/appion/wiki/Appion_Home |
| MOTIONCOR2 | (Zheng et al., 2017) | http://msg.ucsf.edu/em/software/motioncor2.html |
| CTFFIND4 | (Rohou and Grigorieff, 2015) | http://grigoriefflab.janelia.org/ctffind4 |
| RELION | (Scheres, 2012) | https://www2.mrc-lmb.cam.ac.uk/relion/index.php?title=Main_Page |
| JSPR | (Guo and Jiang, 2014; Yu et al., 2016) | http://jiang.bio.purdue.edu/jspr.php |
| COOT | (Emsley and Cowtan, 2004), | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| PHENIX | (Afonine et al., 2012) | https://www.phenix-online.org |
| HOMOLOGY | (Rossmann and Argos, 1976), | https://bilbo.bio.purdue.edu/~viruswww/Rossmann_home/softwares/kvz/strucomp.php |
| CONTACT | (Potterton et al., 2004) | http://www.ccp4.ac.uk/html/contact.html |
| 3V | (Voss and Gerstein, 2010) | http://3vee.molmovdb.org |
| RIVEM | (Xiao and Rossmann, 2007) | https://bilbo.bio.purdue.edu/~viruswww/Rossmann_home/softwares/river_programs/rivem.php |
| CHIMERA | (Pettersen et al., 2004) | https://www.cgl.ucsf.edu/chimera/ |
| Other | ||
| VIPR Sequence alignment | 2016 | https://www.viprbrc.org/brc/msa.spg?method=ShowCleanInputPage&decorator=rhabdo |
STAR★METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact Michael Rossmann (mr@purdue.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Species/strain of experimental models
Virus strain: Zika Virus, strain H/PF/2013
Cell lines: Vero-Furin cells (Vero cells that stably express furin), Sex of cell line-Female.
Cultural conditions for microorganisms
Cell culture
Vero-Furin cells (Mukherjee et al., 2016) below passage 20 were maintained in Dulbecco’s modified Eagle medium (DMEM) supplied with 10% fetal bovine serum (FBS), 1× non-essential amino acids (NEAA), 50 U/ml penicillin and streptomycin (PEN-STREP) and blasticidin (50 µg/ml media) at 37°C in a humidified incubator.
Virus growth
Approximately 1 × 109 Vero-Furin cells maintained in the culture conditions as stated above were infected after 80% confluency at a multiplicity of infection (MOI) of 0.1 with Zika virus (strain H/PF/2013) at 37°C (Sirohi et al., 2016) until 90% of cell cytopathic effect was observed (3–5 days). The media was collected every 12 hours and replaced with new growth media. The virus was further purified from samples collected 60 and 72 hours post-infection as described in the methods section.
METHOD DETAILS
Purification of Mature ZIKV Particles
ZIKV Strain H/PF/2013 was grown and purified according to the following protocol. Approximately 1 × 109 Vero-Furin cells (Mukherjee et al., 2016), grown in Dulbecco’s modified Eagle medium (DMEM) supplied with 10% fetal bovine serum (FBS), 1× non-essential amino acids (NEAA), 50 U/ml PEN-STREP and blasticidin (50 µg/ml media) were infected at a multiplicity of infection (MOI) of 0.1 with Zika virus (strain H/PF/2013) at 37°C (Sirohi et al., 2016). Virus particles were purified from media collected at 60 and 72 hours post-infection (Kuhn et al., 2002). Briefly, virus particles were precipitated from the media with 8% polyethylene glycol (PEG) 8000 overnight at 4°C, pelleted at 8891×g for 50 minutes at 4°C. Re-suspended particles were pelleted through a 24% sucrose cushion, re-suspended in 0.5mL NTE buffer (20mM Tris pH 8.0, 120mM NaCl, 1mM EDTA) and purified with a discontinuous gradient in 5% intervals from 35% to 10% K-tartrate, 20mM Tris pH 8.0, 1mM EDTA. Mature virus was extracted from the gradient, concentrated and buffer exchanged into NTE buffer.
Cryo-EM Reconstruction and Structure Refinement of ZIKV
Purified ZIKV preparation was frozen on lacey carbon EM grids (Ted Pella, Inc., Prod No. 01824). Micrographs were collected using a dose of 33.3 e−/Å2 on an FEI Titan Krios electron microscope equipped with a Gatan K2 Summit detector using a nominal magnification of 18,000 in the “super-resolution” mode, resulting in a pixel size of 0.81 Å. A total of 2085 images were collected, corrected for beam induced sample motion using MotionCor2 (Zheng et al., 2017) and CTF parameters estimated using CTFFIND4 (Rohou and Grigorieff, 2015). 108,963 particles were boxed out using the template-based picking module of the automated Appion method (Lander et al., 2009). Non-reference 2D classification was performed with Relion to select 35,842 particles (Scheres, 2012). The program jspr was used for initial model generation, orientation and center refinement of selected particles (Guo and Jiang, 2014; Yu et al., 2016) from 8-fold binned data. Further high-resolution icosahedral refinement for astigmatism, defocus, elliptical distortion and magnification with unbinned data using soft masks generated a cryo-EM map with an average resolution of 3.1 Å using the 0.143 Fourier shell correlation criterion (Rosenthal and Henderson, 2003).
Multiple Sequence Alignments
Multiple sequences of ZIKV, DENV, YFV, WNV and JEV were searched and analyzed using Virus Pathogen Database and Analysis Resource (ViPR) (https://www.viprbrc.org). The flavivirus sequences were selected based on the availability of complete genomes of flaviviruses that infect humans. Multiple sequence alignments were performed after removal of all duplicate sequences. The number of sequences that were aligned are: (a) ZIKV: 33; (b) DENV1: 385; (c) DENV2: 279; (d) DENV3: 213; (e) DENV4: 280; (f) JEV: 11; (g) YFV: 12; and (h) WNV: 50. From each subset a consensus sequence was calculated and all the consensus sequences were aligned.
3D Superposition, Residue Contacts and Calculation of Surface Properties
All structure superposition’s were performed using the program HOMOLOGY (Rossmann and Argos, 1976). The structural comparisons were made by superimposing individual E protein molecules. Interactions between the residues of E and M oligomers were calculated using the CCP4 program CONTACT (Potterton et al., 2004). Accessible surface pockets were calculated using the program 3V (Voss and Gerstein, 2010). Surface residues were plotted using the program RIVEM (Xiao and Rossmann, 2007). All the figures were prepared using the program CHIMERA (Pettersen et al., 2004).
QUANTIFICATION AND STATISTICAL ANALYSIS
Where appropriate, statistical details are given in the methods section and figure legends. Table 1 contains quantitative parameters related to data and refinement statistics.
DATA AND SOFTWARE AVAILABILITY
Cryo-EM reconstructed maps of ZIKV at 3.1Å were deposited with the EMDataBank (EMD-7543). The refined coordinates of ZIKV at 3.1Å have been deposited with the Protein Data Bank (accession code PDB ID 6CO8).
Supplementary Material
Highlights.
Structure and refinement of the mature Zika virus (ZIKV) at 3.1 Å resolution
Comparison of ZIKV with other mosquito-borne flavivirus structures
Largest structural differences at the glycan loop, important for receptor binding
Acknowledgments
The work described here was supported by NIH grant AI076331 to M.G.R. and R.J.K. and NIH grant AI073755 awarded to Michael S. Diamond (PI, Washington University in St. Louis) with a subaward to M.G.R. and R.J.K. We thank the Purdue Cryo-EM facility for their help and support and Yingyuan Sun (Nick) and Qianglin Fang for help with cryo-EM data collection. We are also grateful to the Purdue Rosen Center for Advanced Computing for research computing support. We thank S. Saif Hasan for discussions about the manuscript. We thank S. Kelly for help in preparing this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
A.S.M., G.B., and M.S. were involved in preparation of cell cultures and the optimization and purification of virus samples; M.S., F.L., and T.K. prepared samples for cryo-EM and collected and processed EM data; M.S. refined the ZIKV structure and performed the data analyses; M.S. and M.G.R. wrote the paper; R.J.K, F.L. and L.S. reviewed the text.
DECLARATION OF INTERESTS
The authors declare no competing financial interests.
References
- Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alhoot MA, Rathinam AK, Wang SM, Manikam R, Sekaran SD. Inhibition of dengue virus entry into target cells using synthetic antiviral peptides. Int. J. Med. Sci. 2013;10:719–729. doi: 10.7150/ijms.5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annamalai AS, Pattnaik A, Sahoo BR, Muthukrishnan E, Natarajan SK, Steffen D, Vu HLX, Delhon G, Osorio FA, Petro TM, et al. Zika virus encoding non-glycosylated envelope protein is attenuated and defective in neuroinvasion. J. Virol. 2017;91:e01348–01317. doi: 10.1128/JVI.01348-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barba-Spaeth G, Dejnirattisai W, Rouvinski A, Vaney MC, Medits I, Sharma A, Simon-Loriere E, Sakuntabhai A, Cao-Lormeau VM, Haouz A, et al. Structural basis of potent Zika-dengue virus antibody cross-neutralization. Nature. 2016;536:48–53. doi: 10.1038/nature18938. [DOI] [PubMed] [Google Scholar]
- Brasil P, Pereira JP, Moreira ME, Nogueira RMR, Damasceno L, Wakimoto M, Rabello RS, Valderramos SG, Halai UA, Salles TS, et al. Zika virus infection in pregnant women in Rio de Janeiro. N. Engl. J. Med. 2016;375:2321–2334. doi: 10.1056/NEJMoa1602412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler D. Fears rise over yellow fever's next move. Nature. 2016;532:155–156. doi: 10.1038/532155a. [DOI] [PubMed] [Google Scholar]
- Cao-Lormeau VM, Blake A, Mons S, Lastere S, Roche C, Vanhomwegen J, Dub T, Baudouin L, Teissier A, Larre P, et al. Guillain-Barré syndrome outbreak associated with Zika virus infection in French Polynesia: a case-control study. Lancet. 2016;387:1531–1539. doi: 10.1016/S0140-6736(16)00562-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Maguire T, Hileman RE, Fromm JR, Esko JD, Linhardt RJ, Marks RM. Dengue virus infectivity depends on envelope protein binding to target cell heparan sulfate. Nat. Med. 1997;3:866–871. doi: 10.1038/nm0897-866. [DOI] [PubMed] [Google Scholar]
- Cherrier MV, Kaufmann B, Nybakken GE, Lok SM, Warren JT, Chen BR, Nelson CA, Kostyuchenko VA, Holdaway HA, Chipman PR, et al. Structural basis for the preferential recognition of immature flaviviruses by a fusion-loop antibody. EMBO J. 2009;28:3269–3276. doi: 10.1038/emboj.2009.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockburn JJ, Navarro Sanchez ME, Fretes N, Urvoas A, Staropoli I, Kikuti CM, Coffey LL, Arenzana Seisdedos F, Bedouelle H, Rey FA. Mechanism of dengue virus broad cross-neutralization by a monoclonal antibody. Structure. 2012a;20:303–314. doi: 10.1016/j.str.2012.01.001. [DOI] [PubMed] [Google Scholar]
- Cockburn JJ, Navarro Sanchez ME, Goncalvez AP, Zaitseva E, Stura EA, Kikuti CM, Duquerroy S, Dussart P, Chernomordik LV, Lai CJ, et al. Structural insights into the neutralization mechanism of a higher primate antibody against dengue virus. EMBO J. 2012b;31:767–779. doi: 10.1038/emboj.2011.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Guardia C, Lleonart R. Progress in the identification of dengue virus entry/fusion inhibitors. Biomed. Res. Int. 2014;2014:825039. doi: 10.1155/2014/825039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejnirattisai W, Supasa P, Wongwiwat W, Rouvinski A, Barba-Spaeth G, Duangchinda T, Sakuntabhai A, Cao-Lormeau VM, Malasit P, Rey FA, et al. Dengue virus sero-cross-reactivity drives antibody-dependent enhancement of infection with Zika virus. Nature immunology. 2016;17:1102–1108. doi: 10.1038/ni.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Fibriansah G, Ng TS, Kostyuchenko VA, Lee J, Lee S, Wang JQ, Lok SM. Structural changes in dengue virus when exposed to a temperature of 37°C. J. Virol. 2013;87:7585–7592. doi: 10.1128/JVI.00757-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuzik T, Formanova P, Ruzek D, Yoshii K, Niedrig M, Plevka P. Structure of tick-borne encephalitis virus and its neutralization by a monoclonal antibody. Nat. Commun. 2018;9:436. doi: 10.1038/s41467-018-02882-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germi R, Crance JM, Garin D, Guimet J, Lortat-Jacob H, Ruigrok RW, Zarski JP, Drouet E. Heparan sulfate-mediated binding of infectious dengue virus type 2 and yellow fever virus. Virology. 2002;292:162–168. doi: 10.1006/viro.2001.1232. [DOI] [PubMed] [Google Scholar]
- Goo L, Dowd KA, Smith ARY, Pelc RS, DeMaso CR, Pierson TC. Zika virus is not uniquely stable at physiological temperatures compared to other flaviviruses. Mbio. 2016;7:e01396–01316. doi: 10.1128/mBio.01396-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Jiang W. Single particle cryo-electron microscopy and 3-D reconstruction of viruses. Methods Mol. Biol. 2014;1117:401–443. doi: 10.1007/978-1-62703-776-1_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman MG, Harris E. Dengue. Lancet. 2015;385:453–465. doi: 10.1016/S0140-6736(14)60572-9. [DOI] [PubMed] [Google Scholar]
- Hasan SS, Miller A, Sapparapu G, Fernandez E, Klose T, Long F, Fokine A, Porta JC, Jiang W, Diamond MS, et al. A human antibody against Zika virus crosslinks the E protein to prevent infection. Nat. Commun. 2017;8:14722. doi: 10.1038/ncomms14722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan SS, Sevvana M, Kuhn RJ, Rossmann MG. Structural biology of Zika virus and other flaviviruses. Nat. Struct. Mol. Biol. 2018;25:13–20. doi: 10.1038/s41594-017-0010-8. [DOI] [PubMed] [Google Scholar]
- Honein MA, Dawson AL, Petersen EE, Jones AM, Lee EH, Yazdy MM, Ahmad N, Macdonald J, Evert N, Bingham A, et al. Birth defects among fetuses and infants of US women with evidence of possible Zika virus infection during pregnancy. JAMA. 2017;317:59–68. doi: 10.1001/jama.2016.19006. [DOI] [PubMed] [Google Scholar]
- Hrobowski YM, Garry RF, Michael SF. Peptide inhibitors of dengue virus and West Nile virus infectivity. Virol. J. 2005;2:49. doi: 10.1186/1743-422X-2-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampmann T, Yennamalli R, Campbell P, Stoermer MJ, Fairlie DP, Kobe B, Young PR. In silico screening of small molecule libraries using the dengue virus envelope E protein has identified compounds with antiviral activity against multiple flaviviruses. Antiviral Res. 2009;84:234–241. doi: 10.1016/j.antiviral.2009.09.007. [DOI] [PubMed] [Google Scholar]
- Kaufmann B, Chipman PR, Holdaway HA, Johnson S, Fremont DH, Kuhn RJ, Diamond MS, Rossmann MG. Capturing a flavivirus pre-fusion intermediate. PLoS Pathog. 2009;5:e1000672. doi: 10.1371/journal.ppat.1000672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann B, Nybakken GE, Chipman PR, Zhang W, Diamond MS, Fremont DH, Kuhn RJ, Rossmann MG. West Nile virus in complex with the Fab fragment of a neutralizing monoclonal antibody. Proc. Natl. Acad. Sci. U.S.A. 2006;103:12400–12404. doi: 10.1073/pnas.0603488103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann B, Rossmann MG. Molecular mechanisms involved in the early steps of flavivirus cell entry. Microbes Infect. 2011;13:1–9. doi: 10.1016/j.micinf.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann B, Vogt MR, Goudsmit J, Holdaway HA, Aksyuk AA, Chipman PR, Kuhn RJ, Diamond MS, Rossmann MG. Neutralization of West Nile virus by cross-linking of its surface proteins with Fab fragments of the human monoclonal antibody CR4354. Proc. Natl. Acad. Sci. U.S.A. 2010;107:18950–18955. doi: 10.1073/pnas.1011036107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostyuchenko VA, Lim EXY, Zhang SJ, Fibriansah G, Ng TS, Ooi JSG, Shi J, Lok SM. Structure of the thermally stable Zika virus. Nature. 2016;533:425–428. doi: 10.1038/nature17994. [DOI] [PubMed] [Google Scholar]
- Kostyuchenko VA, Zhang Q, Tan JL, Ng TS, Lok SM. Immature and mature dengue serotype 1 virus structures provide insight into the maturation process. J. Virol. 2013;87:7700–7707. doi: 10.1128/JVI.00197-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn RJ, Zhang W, Rossmann MG, Pletnev SV, Corver J, Lenches E, Jones CT, Mukhopadhyay S, Chipman PR, Strauss EG, et al. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell. 2002;108:717–725. doi: 10.1016/s0092-8674(02)00660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander GC, Stagg SM, Voss NR, Cheng A, Fellmann D, Pulokas J, Yoshioka C, Irving C, Mulder A, Lau PW, et al. Appion: an integrated, database-driven pipeline to facilitate EM image processing. J. Struct. Biol. 2009;166:95–102. doi: 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Lok SM, Yu IM, Zhang Y, Kuhn RJ, Chen J, Rossmann MG. The flavivirus precursor membrane-envelope protein complex: structure and maturation. Science. 2008;319:1830–1834. doi: 10.1126/science.1153263. [DOI] [PubMed] [Google Scholar]
- Liao M, Kielian M. Domain III from class II fusion proteins functions as a dominant-negative inhibitor of virus membrane fusion. J. Cell Biol. 2005;171:111–120. doi: 10.1083/jcb.200507075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbach BD, Rice CM. Molecular biology of flaviviruses. Adv. Virus Res. 2003;59:23–61. doi: 10.1016/s0065-3527(03)59002-9. [DOI] [PubMed] [Google Scholar]
- Lok SM, Costin JM, Hrobowski YM, Hoffmann AR, Rowe DK, Kukkaro P, Holdaway H, Chipman P, Fontaine KA, Holbrook MR, et al. Release of dengue virus genome induced by a peptide inhibitor. PLoS One. 2012;7:e50995. doi: 10.1371/journal.pone.0050995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lok SM, Kostyuchenko V, Nybakken GE, Holdaway HA, Battisti AJ, Sukupolvi-Petty S, Sedlak D, Fremont DH, Chipman PR, Roehrig JT, et al. Binding of a neutralizing antibody to dengue virus alters the arrangement of surface glycoproteins. Nat. Struct. Mol. Biol. 2008;15:312–317. doi: 10.1038/nsmb.1382. [DOI] [PubMed] [Google Scholar]
- Mangala Prasad V, Miller AS, Klose T, Sirohi D, Buda G, Jiang W, Kuhn RJ, Rossmann MG. Structure of the immature Zika virus at 9 Å resolution. Nat. Struct. Mol. Biol. 2017;24:184–186. doi: 10.1038/nsmb.3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modis Y, Ogata S, Clements D, Harrison SC. Variable surface epitopes in the crystal structure of dengue virus type 3 envelope glycoprotein. J. Virol. 2005;79:1223–1231. doi: 10.1128/JVI.79.2.1223-1231.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S, Sirohi D, Dowd KA, Chen Z, Diamond MS, Kuhn RJ, Pierson TC. Enhancing dengue virus maturation using a stable furin over-expressing cell line. Virology. 2016;497:33–40. doi: 10.1016/j.virol.2016.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay S, Kuhn RJ, Rossmann MG. A structural perspective of the flavivirus life cycle. Nat. Rev. Microbiol. 2005;3:13–22. doi: 10.1038/nrmicro1067. [DOI] [PubMed] [Google Scholar]
- Pannu NS, Murshudov GN, Dodson EJ, Read RJ. Incorporation of prior phase information strengthens maximum-likelihood structure refinement. Acta Crystallogr. D Biol. Crystallogr. 1998;54:1285–1294. doi: 10.1107/s0907444998004119. [DOI] [PubMed] [Google Scholar]
- Passi D, Sharma S, Dutta SR, Ahmed M. Zika virus diseases - The new face of an ancient enemy as global public health emergency (2016): Brief review and recent updates. Int. J. Prev. Med. 2017;8:6. doi: 10.4103/2008-7802.199641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera-Lecoin M, Meertens L, Carnec X, Amara A. Flavivirus entry receptors: an update. Viruses. 2013;6:69–88. doi: 10.3390/v6010069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pokidysheva E, Zhang Y, Battisti AJ, Bator-Kelly CM, Chipman PR, Xiao CA, Gregorio GG, Hendrickson WA, Kuhn RJ, Rossmann MG. Cryo-EM reconstruction of dengue virus in complex with the carbohydrate recognition domain of DC-SIGN. Cell. 2006;124:485–493. doi: 10.1016/j.cell.2005.11.042. [DOI] [PubMed] [Google Scholar]
- Potterton L, McNicholas S, Krissinel E, Gruber J, Cowtan K, Emsley P, Murshudov GN, Cohen S, Perrakis A, Noble M. Developments in the CCP4 molecular-graphics project. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2288–2294. doi: 10.1107/S0907444904023716. [DOI] [PubMed] [Google Scholar]
- Rey FA, Heinz FX, Mandl C, Kunz C, Harrison SC. The envelope glycoprotein from tick-borne encephalitis virus at 2 Å resolution. Nature. 1995;375:291–298. doi: 10.1038/375291a0. [DOI] [PubMed] [Google Scholar]
- Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- Rossmann MG, Argos P. Exploring structural homology of proteins. J. Mol. Biol. 1976;105:75–95. doi: 10.1016/0022-2836(76)90195-9. [DOI] [PubMed] [Google Scholar]
- Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sejvar JJ. West Nile virus infection. Microbiol Spectr. 2016;4 doi: 10.1128/microbiolspec.EI10-0021-2016. EI10-0021-2016. [DOI] [PubMed] [Google Scholar]
- Sirohi D, Chen Z, Sun L, Klose T, Pierson TC, Rossmann MG, Kuhn RJ. The 3.8 Å resolution cryo-EM structure of Zika virus. Science. 2016;352:467–470. doi: 10.1126/science.aaf5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirohi D, Kuhn RJ. Zika virus structure, maturation, and receptors. J. Infect. Dis. 2017;216:S935–S944. doi: 10.1093/infdis/jix515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit JM, Moesker B, Rodenhuis-Zybert I, Wilschut J. Flavivirus cell entry and membrane fusion. Viruses. 2011;3:160–171. doi: 10.3390/v3020160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suss J. Tick-borne encephalitis in Europe and beyond--the epidemiological situation as of 2007. Euro. Surveill. 2008;13 [PubMed] [Google Scholar]
- Tassaneetrithep B, Burgess TH, Granelli-Piperno A, Trumpfheller C, Finke J, Sun W, Eller MA, Pattanapanyasat K, Sarasombath S, Birx DL, et al. DC-SIGN (CD209) mediates dengue virus infection of human dendritic cells. J. Exp. Med. 2003;197:823–829. doi: 10.1084/jem.20021840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss NR, Gerstein M. 3V: cavity, channel and cleft volume calculator and extractor. Nucleic Acids Res. 2010;38:W555–W562. doi: 10.1093/nar/gkq395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Liang G. Epidemiology of Japanese encephalitis: past, present, and future prospects. Ther. Clin. Risk Manag. 2015;11:435–448. doi: 10.2147/TCRM.S51168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Li SH, Zhu L, Nian QG, Yuan S, Gao Q, Hu Z, Ye Q, Li XF, Xie DY, et al. Near-atomic structure of Japanese encephalitis virus reveals critical determinants of virulence and stability. Nat. Commun. 2017;8:14. doi: 10.1038/s41467-017-00024-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Li L, Pennington JG, Sheng J, Yap ML, Plevka P, Meng G, Sun L, Jiang W, Rossmann MG. Obstruction of dengue virus maturation by Fab fragments of the 2H2 antibody. J. Virol. 2013;87:8909–8915. doi: 10.1128/JVI.00472-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver SC, Costa F, Garcia-Blanco MA, Ko AI, Ribeiro GS, Saade G, Shi PY, Vasilakis N. Zika virus: History, emergence, biology, and prospects for control. Antiviral Res. 2016;130:69–80. doi: 10.1016/j.antiviral.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C, Rossmann MG. Interpretation of electron density with stereographic roadmap projections. J. Struct. Biol. 2007;158:182–187. doi: 10.1016/j.jsb.2006.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JM, Chen YF, Tu YY, Yen KR, Yang YL. Combinatorial computational approaches to identify tetracycline derivatives as flavivirus inhibitors. PLoS One. 2007;2:e428. doi: 10.1371/journal.pone.0000428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Li K, Liu Y, Chen Z, Wang Z, Yan R, Klose T, Tang L, Jiang W. An algorithm for estimation and correction of anisotropic magnification distortion of cryo-EM images without need of pre-calibration. J. Struct. Biol. 2016;195:207–215. doi: 10.1016/j.jsb.2016.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu IM, Holdaway HA, Chipman PR, Kuhn RJ, Rossmann MG, Chen J. Association of the pr peptides with dengue virus at acidic pH blocks membrane fusion. J. Virol. 2009;83:12101–12107. doi: 10.1128/JVI.01637-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Chipman PR, Corver J, Johnson PR, Zhang Y, Mukhopadhyay S, Baker TS, Strauss JH, Rossmann MG, Kuhn RJ. Visualization of membrane protein domains by cryo-electron microscopy of dengue virus. Nat. Struct. Biol. 2003a;10:907–912. doi: 10.1038/nsb990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Kaufmann B, Chipman PR, Kuhn RJ, Rossmann MG. Membrane curvature in flaviviruses. J. Struct. Biol. 2013a;183:86–94. doi: 10.1016/j.jsb.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Ge P, Yu X, Brannan JM, Bi G, Zhang Q, Schein S, Zhou ZH. Cryo-EM structure of the mature dengue virus at 3.5-Å resolution. Nat. Struct. Mol. Biol. 2013b;20:105–110. doi: 10.1038/nsmb.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Sheng J, Austin SK, Hoornweg TE, Smit JM, Kuhn RJ, Diamond MS, Rossmann MG. Structure of acidic pH dengue virus showing the fusogenic glycoprotein trimers. J. Virol. 2015;89:743–750. doi: 10.1128/JVI.02411-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Sheng J, Plevka P, Kuhn RJ, Diamond MS, Rossmann MG. Dengue structure differs at the temperatures of its human and mosquito hosts. Proc. Natl. Acad. Sci. U.S.A. 2013c;110:6795–6799. doi: 10.1073/pnas.1304300110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Corver J, Chipman PR, Zhang W, Pletnev SV, Sedlak D, Baker TS, Strauss JH, Kuhn RJ, Rossmann MG. Structures of immature flavivirus particles. EMBO J. 2003b;22:2604–2613. doi: 10.1093/emboj/cdg270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Kaufmann B, Chipman PR, Kuhn RJ, Rossmann MG. Structure of immature West Nile virus. J. Virol. 2007;81:6141–6145. doi: 10.1128/JVI.00037-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhang W, Ogata S, Clements D, Strauss JH, Baker TS, Kuhn RJ, Rossmann MG. Conformational changes of the flavivirus E glycoprotein. Structure. 2004;12:1607–1618. doi: 10.1016/j.str.2004.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, Agard DA. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.