Abstract
Plants have evolved strategies to cope with drought stress by maximizing physiological capacity and adjusting developmental processes such as flowering time. The WOX13 orthologous group is the most conserved among the clade of WOX homeodomain-containing proteins and is found to function in both drought stress and flower development. In this study, we isolated and characterized OsWOX13 from rice. OsWOX13 was regulated spatially in vegetative organs but temporally in flowers and seeds. Overexpression of OsWOX13 (OsWOX13-ov) in rice under the rab21 promoter resulted in drought resistance and early flowering by 7–10 days. Screening of gene expression profiles in mature leaf and panicles of OsWOX13-ov showed a broad spectrum of effects on biological processes, such as abiotic and biotic stresses, exerting a cross-talk between responses. Protein binding microarray and electrophoretic mobility shift assay analyses supported ATTGATTG as the putative cis-element binding of OsWOX13. OsDREB1A and OsDREB1F, drought stress response transcription factors, contain ATTGATTG motif(s) in their promoters and are preferentially expressed in OsWOX13-ov. In addition, Heading date 3a and OsMADS14, regulators in the flowering pathway and development, were enhanced in OsWOX13-ov. These results suggest that OsWOX13 mediates the stress response and early flowering and, thus, may be a regulator of genes involved in drought escape.
Keywords: drought, early flowering, escape, Hd3a, OsWOX13, vascular tissue
INTRODUCTION
Plants experience various types of drought stress during their life cycle. This stress is sometimes acute and short, and other times it is long and life-threatening. Plants respond to water deficiency by exhibiting many physiological and developmental changes (Shinozaki and Yamaguchi-Shinozaki, 2000; Yu et al., 2013). The immediate response to drought stress might be turgor pressure changes in guard cells via ion- and water-transport systems across membranes, which induces stomatal closure (Osakabe et al., 2014) to prevent water evaporation. Plants cope with the stress by inducing genes that directly protect against environmental stresses, as well as those that regulate gene expression (transcription factors) and signal transduction (phytohormones) in the stress response (Campo et al., 2014; Hu et al., 2008; Nakashima et al., 2007; Oh et al., 2005; Xiao et al., 2013). Plants produce oxidant species detoxifying enzymes such as superoxide dismutase, catalase, peroxidase, and glutathione reductase (Ouyang et al., 2010). To induce these stress-inducible genes, plants have at least two major pathways, an abscisic acid (ABA)-dependent and an ABA-independent pathway (TIAN et al., 2005; Todaka et al., 2015). A rice orthologue of the ABA receptor, OsPYL/RCAR5, has been identified as a positive regulator of the ABA signal transduction pathway together with other components such as OsPP2C30, SAPK2, and OREB1, and it might play a pivotal role in the ABA-related pathway (Kim et al., 2011).
Flowering is one of the key factors for plants to survive, through which plants hand down genetic components to the next generation. Along with the developmental stages, environmental factors such as day length (photoperiod) and temperature greatly influence flowering (Major, 1980). In terms of day length, plants are usually classified as short-day (SD) or long-day (LD) types, and day length-dependent florigens have been postulated to initiate flowering. In rice, Heading date 3a (Hd3a) and RICE FLOWERING LOCUS T1 (RFT1) are main florigenes under SD and LD, respectively (Hayama et al., 2003; Ishikawa et al., 2011). Heading date 1 (Hd1, counterpart of CO in Arabidopsis) acts as a transcription activator for Hd3a in SDs but as a repressor in the presence of functional O. sativa Phytochrome B in LDs (Hayama et al., 2003; Ishikawa et al., 2011). Early heading date 1 (Ehd1) has been identified as a rice-specific B-type response factor that acts in both LDs and SDs to promote Hd3a and RFT1 mRNA expression. The expression of Ehd1 is regulated by various repressors or activators, Ghd7, Ehd2, Ehd3, Ehd4, OsMADS51, OsLFL1, and OsPPR37/DTH7, in day length-dependent or -independent manners (Doi et al., 2004; Gao et al., 2014; Kim et al., 2007; Koo et al., 2013; Kwon et al., 2015; Matsubara et al., 2011; Peng et al., 2007; Tsuji et al., 2013; Wei et al., 2010; Xue et al., 2008; Yun et al., 2013). Florigenes are expressed in the phloem of rice leaf blades under short-day conditions and move to the SAM to initiate the floral transition. In rice, Hd3a associated with a 14-3-3 protein in the cytoplasm translocates to the nucleus and forms the florigen activation complex (Tsuji et al., 2013). The complex regulates a MADS box-containing transcription factor, Os07g0108900 (OsMADS15), which is a homolog of the Arabidopsis floral identity gene APETALA1.
Plants growing in many regions of the world demonstrate a plasticity of plant form in an altered environment (Aguirrezabal et al., 2006; Blázquez et al., 2003; Gray and Brady, 2016; Heschel and Riginos, 2005). Genetic and ecologic studies have revealed that plants have exploited these physiological aspects and evolved several strategies to maximize the ability to survive in harsh nature based on their genetic components (Ludlow, 1989; Mckay et al., 2003). In extreme condition, plants survive in dry environments with internal water deficits through drought tolerance (DT). Plants also use a drought avoidance (DA) strategy to maintain internal water in a dry environment by minimizing water loss and/or maximizing water uptake. The drought escape (DE) strategy is attained through a short life cycle or growing season. The DE response is characterized by early flowering, and plants adaptively shorten their life cycle to make seeds before severe stress leads to death (Riboni et al., 2013). Even within species, these strategies are differentially used according to their habitats, which involve distinct genetic components. In Arabidopsis, FRIGIDA (FRI), which was originally studied as a component of vernalization, encodes a transcription factor that activates flowering locus C (FLC) and regulates the flowering time response to drought stress in plants (Lovell et al., 2013; Searle et al., 2006). Weakly expressed FRI alleles confer a DE strategy owing to the fast growth, low water use efficiency and early flowering. By contrast, a DA strategy is conferred by functional genes that lead to late flowering, slow growth and efficient water use during photosynthesis (Lovell et al., 2013). In Arabidopsis, Riboni et al. (2013) have shown that the flower-promoting gene GIGANTEA (GI) and the florigen genes FLOWERING LOCUS T (FT) and TWIN SISTER OF FT (TSF) are key factors for the DE response mediating ABA-dependent responses, positively regulating flowering under long-day conditions. In rice, a short day plant, DE and DT are also observed in the genetic dissection of adaptive strategies to water stress (Xu et al., 2005). QTL data suggest that the genetic systems underlying the two adaptation strategies are largely non-overlapping.
The homeobox (HMB) is a 180-bp consensus DNA sequence that encodes a 60-amino-acid protein domain, the homeodomain (HD) consisting of three α-helices separated by a loop and a turn. Due to the DNA-binding structure, the HD-containing protein can bind to specific sequence of other genes and act as a transcription factor. Plant HMB genes have been implicated in various developmental processes and hormone response pathways throughout the plant life cycle (Chan et al., 1998; Hamant and Pautot, 2010; Jain et al., 2008). They shape the plant architecture and control the establishment and maintenance of the shoot and root apical meristem. They also regulate the transition to flowering and control the development of the ovule and fruit. In various plant species, the roles of several homeobox genes have also been implicated in abiotic stress responses (Bhattacharjee and Jain, 2013). Genome-wide scale and phylogenetic analyses of the rice HMB family in this family revealed well-supported subfamilies (Agalou et al., 2008; Jain et al., 2008). The number of rice HD TFs are further subdivided into 79 HB-PHD, 61 HD-ZIP and 17 HB-Other (Jin et al., 2016) according to their other secondary structural components. In addition to HD, each subfamily might contain one or more specialized domain(s). WUSCHEL (WUS), the first identified member of the subfamily, was first isolated from Arabidopsis, followed by rice (OsWUS) and maize (ZmWUS1, ZmWUS2). Among them, WUSCHEL-related homeobox (WOX) genes are plant-specific, and their products, WOX proteins, form a large subfamily (Deveaux et al., 2008; Nardmann et al., 2009; Zhang et al., 2010b). These proteins function in various physiological and developmental processes, especially embryonic patterning, stem cell maintenance and organ formation. Phylogenetic analysis has provided good support for the division of WOX subfamily into three separate orthologous groups (OGs), WOX1, WOX8 and WOX13, among which WOX13 OG is regarded as the most conserved clade (Deveaux et al., 2008; Lin et al., 2013; van der Graaff et al., 2009). The two Arabidopsis WOX13 genes, AtWOX13 and AtWOX14, function in organ initiation and development, most likely by preventing premature differentiation (Deveaux et al., 2008). Rice WOX genes are differentially expressed during the life cycle of rice based on microarray-based gene expression profiling in the rice Indica cultivar MH63 (Cheng et al., 2014).
In this study, we isolated OsWOX13 (OsWOX9B), which is induced during the acute dehydration response (Minh-Thu et al., 2013; Zhang et al., 2010b). OsWOX13 is differentially regulated in vegetative and reproductive organs, demonstrating spatially regulated expression in vascular tissue of leaf/stem/root and temporally regulated expression in flowers and developing seeds. Overexpression of OsWOX13 in rice under a water deficit specific promoter (Rab21) results in drought tolerance and early flowering in transgenic plants.
MATERIALS AND METHODS
Multiple sequence alignment and phylogenetic analysis
WOX genes from Arabidopsis, rice and maize were collected from Zhang et al. (2010) and edited due to the authors’ intention (Supplementary Table S1). Because OsNS1 and OsNS2 are duplications, only OsNS1/OsNS was used in the multiple sequence alignment and phylogenetic analysis. The conserved HD was detected using the blast program provided by NCBI at http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi (Marchler-Bauer et al., 2010) with default options.
Multiple sequence alignments of 48 HDs of WOX proteins or 8 full-length protein sequences of WOX13 OG were performed using ClustalW function in MEGA4.0.2 (Tamura et al., 2007) with default parameters and visualized with GeneDoc2.7.000 (Nicholas, 1997).
The evolutionary history of the WOX sub-family was inferred using the neighbor-joining method (Saitou and Nei, 1987) with the phylogeny function in the same MEGA4 software with bootstrap values (Felsenstein, 1985) inferred from 1000 replicates. The evolutionary distances were computed using the Poisson correction method (Zuckerkandl and Pauling, 1965) and are presented as the units of the number of amino acid substitutions per site. All positions containing gaps and missing data were eliminated from the dataset (complete deletion option).
Vector construction and transformation in rice
For construction of the overexpression and promoter analysis vector, the coding sequence of OsWOX13 and the 2-kb gDNA fragment upstream of the start codon were isolated (primer sequences are shown in Supplementary Table S2) and inserted into binary vectors by the Gateway technique (Invitrogen). OsWOX13 is controlled by rab21, a water deficit specific promoter isolated from rice (Mundy and Chua, 1988; Reddy et al., 2002). The constructs of the expression cassettes are shown in Supplementary Fig. S6B and S6C and were then introduced into Agrobacterium tumefaciens LBA 4404 by triparental mating and transfected into embryogenic calli from mature seeds (Oryza sativa L. cv. Ilmi) as previously described (Jang et al., 1999).
Plant growth, abiotic stress treatment and PCR
Rice seeds (Oryza sativa L. japonica) were geminated on MS0 (Murashige and Skoog) medium, incubated in a growth chamber (28°C, 2 days in dark followed by 2 days in the light), and then transferred into soil and grown in a greenhouse (16-h-light/8-h-dark cycle). Fourteen-day seedlings were then removed from the soil and air-dried in a growth chamber at 28°C with a humidity of 40% and continuous fluorescent light. The non-stressed control leaves were collected directly from in-soil plants. For other stresses, seedlings were washed to remove soil and maintained in fresh water. The control, heat and cold treatment were 28°C, 42°C and 4°C chamber, respectively. Sodium chloride was added to the water to a final concentration of 400 mM to induce salt stress. All leaves were collected after 3 h of each treatment. For plants growing in the field, mature leaves of the transgenic line 8-4-1 were collected at day 40, 33, and 25 prior to the heading date.
Total RNAs were extracted using TriReagent (Molecular Research Center, Inc.) according to the manufacturer’s manual. For first-strand cDNA synthesis, 5 μg of total RNA was reverse-transcribed using the RevertAid H Minus First Strand cDNA Synthesis kit (Fermentas) according to manufacturer’s manual (Chae et al., 2017). The cDNA mixture of rice was then diluted twice. Gene-specific primers were designed using the Primer Designer 4 software (Sci-ed. Software, NC) or Primer-BLAST tool (http://ncbi.nlm.nih.gov). PCR was performed in a 20-μl volume containing 1 μl rice cDNA aliquot, 0.25 pM gene-specific primers, and 10 μl 2x PCR master mix (GeneAll Biotechnology Co., ltd). The reaction included an initial 2-min denaturation at 94°C, followed by 25 to 35 cycles of PCR (95°C for 30 s, 55°C for 30 s, 72°C for 30 s). Subsequently, 9.5 μl of the reaction mixture was separated on a 1.5% agarose gel. The relative ratio to ubiquitin was determined by quantifying the band intensity using Multi-Gauge v2.3 (Fujifilm, Japan) and then dividing it by the corresponding ubi band. To determine the RT-PCR reproducibility, the experiment was repeated 3 times with 2 independent biological replicates.
Promoter analysis by GUS staining
Rice seeds (Oryza sativa L. japonica) were geminated and grown in a greenhouse as mentioned above. Samples were collected at different developmental stages due to experimental requirements, and the histochemical localization of β-glucuronidase (GUS) was examined using 5-bromo-4-chloro-3-indolyl β-D-glucuronidase (X-gluc) based on a standard protocol (Bomblies, 2000). For cytological analysis, plant tissues were embedded in Technovit 8100 resin (Heraeus Kulzer, http://www.kulzer-technik.de/) based on the manufacturer’s manual. Microscale sections were visualized using an Olympus Fluoview FV1000 confocal microscope (http://www.olympus-global.com/) with/without staining with 0.05% toluidine blue.
Cis-binding element identification
To determine the putative cis-binding element of OsWOX13, a protein binding microarray was carried out using E. coli-expressed protein. The coding sequence of the OsWOX13 gene was fused to DsRed and His tagged using pET32a (Novagen, Germany) as a backbone. The protein was induced by IPTG according to the manufacturer’s manual. The induced protein was then purified via the His tag using Ni-NTA resin from Qiagen. Then, the purified OsWOX13 -DsRed-His fusion protein was used in the 9-mer Protein Binding Microarray, Q9-PBM (Kim et al., 2009). The experiment was repeated twice with 2 independently prepared proteins.
Electrophoretic Mobility Shift Assay (EMSA)
EMSA was performed with 5′-biotinylated probes and unlabeled competitors according to the manufacturer’s manual (Thermo Scientific, USA) to confirm the binding ability of protein with putative cis-binding elements isolated from the protein binding microarray. Gel images were then quantified by Multi Gauge 2.3 (Fujifilm, Japan).
To acquire further detail about the strength of the binding affinity based on the Kd value, another EMSA experiment was conducted using Sybr Gold (S11494, Invitrogen) as an indicator. The reactions were carried out with various DNA substrate concentrations (0, 0.1, 0.2, 0.4, 0.8, 1.2, 2, 4, 6 μM) with 5 μg of OsWOX13. Binding was performed in binding buffer (10 mM Tris, 50 mM KCl, 1 mM DTT, pH 7.5) supplemented with 50 ng/L polyIC, 2.5% glycerol, 0.05% NP40, and 5 mM MgCl2 at room temperature for 1 h. After incubation, the reaction mixtures were added, which consisted of 2 μl 10X EMSA loading dye, and loaded on an 8% polyacrylamide gel. The gel was stained with Sybr Gold solution for 30 min and observed under UV light. The intensity of the DNA band was quantified using the GelPro3.0 analyzer program (MediaCybernetics Inc., USA) and transferred into SigmaPlot10.0 (Systat Software Inc., USA) to analyze the Kd value. The data were fit to the following equation: Y = Bmax*X/(Kd+X), where Bmax is the maximal binding and Kd is the concentration of ligand required to reach half-maximal binding.
Rice 3′-Tiling 135 k microarray analysis
For expression microarray analysis, non-transgenic and OsWOX13 overexpression rice seeds were germinated in soil, transferred into pots and grown in natural conditions outside the greenhouse from May to October, which is the main harvest period in Korea. Panicles were collected based on size, i.e., 1–3, 8–15, and 20.5–22 cm as P1, P8 and P21 cm; respectively. Moreover, panicles at 5-days after pollination (5DAP) were also collected to analyze the developing seed. Leaves (L) at the stage of 4 weeks before heading, which was around the date of panicle initiation, were used as a reference. Each tissue from non-transgenic plant was used as a control for the corresponding tissue from the overexpression line.
Total RNAs were extracted and used in the rice 3′-Tiling 135 k microarray experiment (designed by NimbleGen, http://www.nimblegen.com). The rice 3′-Tiling 135k microarray was designed using 31,439 genes (http://rapdb.lab.nig.ac.jp). Multiple analyses were performed with the limma package in the R computing environment (Smyth, 2004) as described previously (Chae et al., 2016). The package adopts the linear modeling approach implemented by lmFit and the empirical Bayesian statistics implemented by eBayes. Genes with an adjusted-p-value less than 0.05 were collected and further assessed for gene expression levels higher than 1 or lower than −1 for at least at one stage compared with the control.
To organize the genes in the context of pre-existing biological knowledge, the entire list of selected genes with the expression level was loaded into the Parametric Analysis of Gene Set Enrichment (PAGE) on agriGO (Du et al., 2010) with Oryza sativa as the analyzed species and other options as the default. Redundant GOs were then removed using REViGO (Supek et al., 2011) with an allowed similarity of 0.7.
To determine putative target genes of OsWOX13, the 2-kb long promoter regions of all rice genes in the microarray experiment were retrieved from RAP-DB (http://rapdb.dna.affrc.go.jp/). The genes with a promoter containing ATTGATTG were selected using an in-house Perl script. Overlap with preferential expressed genes was determined using Venn Diagrams provided by Oliveros (Oliveros, 2007).
Motif extraction from 2-kb promoters of the genes
Promoter region (2 kb) was extracted from the IRGSP1.0 database and searched for the ATTGATTG element.
RESULTS
OsWOX13 is a homeodomain-containing transcription factor
To classify WOX genes according to their structure, 126 WOX genes were collected from 17 model plants, including seven angiosperms (five dicots and two monocots), five gymnosperms, one lycopodiophyta, one bryophyte and two green algae (Supplementary Table S1). Before building a phylogenic tree, 65–69-amino-acid HDs of the translated proteins were aligned, and highly conserved amino acids were observed (Supplementary Fig. S1). To examine the evolutionary history of the WOX genes, we performed a phylogenetic analysis with the above HD sequences using the neighbor-joining (NJ) method. Figure 1 shows the NJ tree with three clearly distinguishable clades: WOX1, WOX8 and WOX13. Os01g0818400 was the only rice gene that clustered in WOX13 clade and was named OsWOX13. Among the genes in the WOX13 clade, OsWOX13 had the closest relationship with four genes named ZmWOX13A, 13B, 14A and 14B in maize (Supplementary Table S1; Zhang et al., 2010b).
Fig. 1. Domain analyses of WOX13 OG proteins. The evolutionary history was inferred from homeodomain sequences (Supplementary Fig. S1) using the neighbor-joining method.
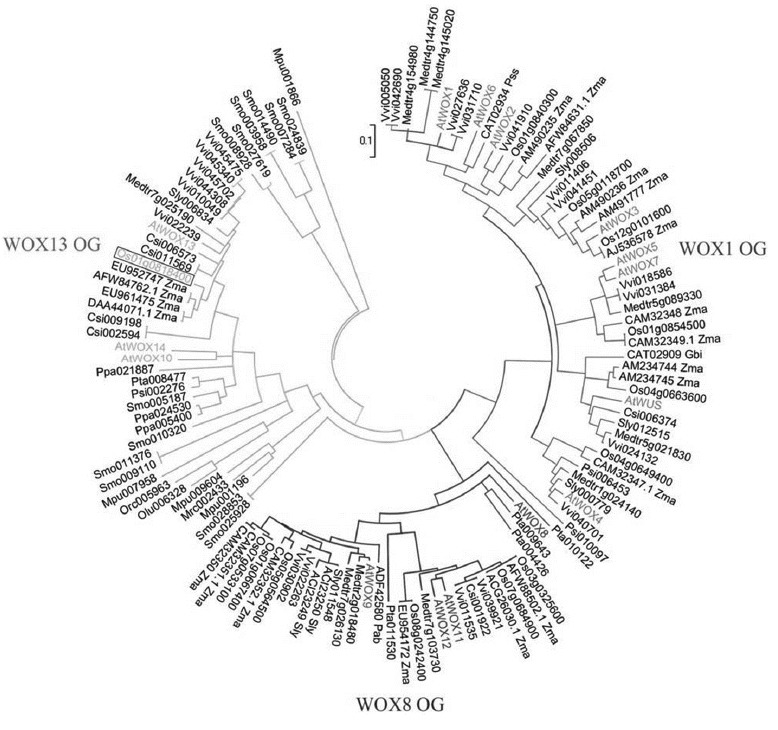
The evolutionary distances were computed using the Poisson correction method and are expressed as the number of amino acid substitutions per site. Members of WOX1, WOX8 and WOX13 orthologous groups are connected by blue, red and green lines; respectively. Arabidopsis genes in each OG are represented by OG-specific colored characters. Os01g0818400/OsWOX13 is colored and boxed.
To further investigate the structure of OsWOX13, the full-length sequence of eight WOX13s from three model plants were aligned: a dicot, Arabidopsis and two monocots, rice and maize (Supplementary Fig. S2). As reported by Deveaux et al. (2008), in addition to HD, WOX13 clade has a WOX13, a YxDpl and an ESExE motifs. OsWOX13 contains all four of the above domains and shares a monocot-WOX13-specific motif with maize proteins. Among three proteins from Arabidopsis, excluding the possible pseudogene AtWOX10 (Deveaux et al., 2008), OsWOX13 showed a closer relationship/more similarity to AtWOX13 than AtWOX14. The homeodomain of OsWOX13 shared 90% amino acid identity to that of AtWOX13 and only 74% with AtWOX14.
OsWOX13 is expressed in most vascular tissues related to development
To investigate the expression pattern of OsWOX13 during growth and development, we first collected information from our microarray data and observed the induction of OsWOX13 in root, different stages of panicle development, germinating seed and regeneration callus (Chae et al., 2016). Then, we examined expression by RT-PCR using tissues at different developmental stages (Supplementary Fig. S3). Although the degree of expression varied among the organs, OsWOX13 was detected in most examined tissue. It was especially highly expressed in young root, leaf sheath, around the shoot apex and in the panicle. This result agreed with the microarray data and suggested that OsWOX13 is expressed in developing organ/tissue.
To gather more details about the tissue-specific expression of OsWOX13, we created transgenic plants with the GUS reporter gene under the control of 2 kb of the OsWOX13 upstream regulatory sequence. No GUS staining was observed in dormant seed (Fig. 2A), but the scutellum showed a blue color after 2 days of imbibition (Fig. 2B), and the intensity increased in later stages of seed germination (Fig. 2C). OsWOX13 was expressed in the vascular tissue of 5-day leaf (Fig. 2D) and in the tiller with developed vascular tissues of 6-week-old seeding (Figs. 2J–2L). Primordia of crown roots from the main stem and developing tiller showed no GUS expression due to the absence of vascular bundles, VBs (Figs. 2K and 2L). In the node of an unelongated stem, GUS was strongly expressed in large VBs (Fig. 2N) and nodal vascular anatomoses (NVA, Fig. 2O). Large VBs from the lower node are surrounded by the NVA, which are a cyclic vascular system in the basal region of the nodes (Fig. 2N) and another large VB from the upper node became diffuse bundles (DVs) and enclosed the enlarged large VBs (Fig. 2O). The expression of OsWOX13 in all types of vascular tissue led to its presence at a high intensity, especially in unelongated stem.
Fig. 2. Histochemical analysis of OsWOX13p::GUS reporter gene expression in plant development.
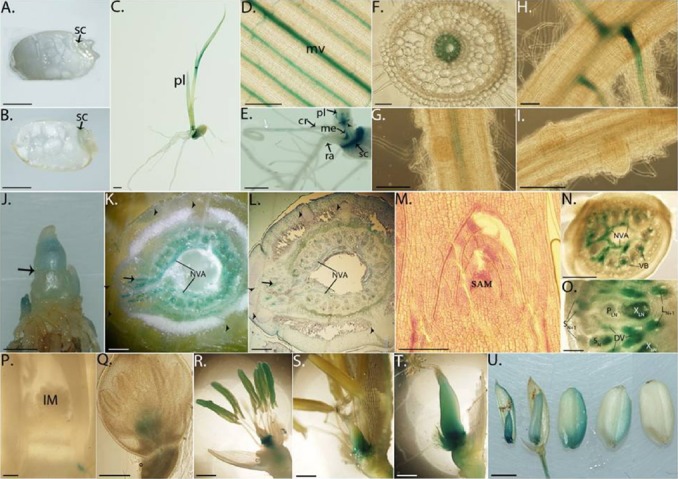
(A) Mature and (B) 2-day imbibed seeds. (C) A 5-day seedling. (D, E) Magnificent of a leaf blade (D) and longitudinal cut through a seed and stem (E) of a 5-day seedling. The white arrow in (E) indicates the position where GUS expression stopped on the crown root. (F) Cross-section of a crown root from a 7-day seedling. (G) Lateral root initiation in a 7-day seedling crown root. (H, I) Junctions with developed (H) and developing (I) lateral root in a 7-day seedling radicle. (J) Six-week-old stem with all developed leaves removed. The arrow shows the position of an auxiliary tiller where it is cross cut (K) and 12-μm-sectioned (L). The section in (L) was stained with toluidine blue. (M) The 12-μm cross-section of the SAM sample was stained with toluidine blue. (N) Hand section of a node from an unelongated stem. (O) Hand section of node Nth counted upward along an elongated stem. In node Nth, DVs only surround and connect to enlarged large VBs of leaf Nth at NVA. VBs of leaf (N+1)th are not enlarged and only pass through the node. (P) Panicles at the very early stage of inflorescent formation. (Q to T) Floret organs at stage (Q) 1 cm, (R) 12 cm, and (S) before and (T) after pollination. (U, left to right) Seeds at 2, 4, 6, 8, and 10DAP. Scale bar = 0.2 cm (a, b, c, e, j, t); 50 μm (F, H) or 200 μm (others). sc: scutellum, pl: primary leaf, ra: radicle, cr: crown root, me: mesocotyl, mv: main vien, NVA: nodal vascular anatomoses, VBs: vascular bundles, PLN and XLN: phloem and xylem of enlarged large VBs of leaf Nth, SN: enlarged small VBs of leaf Nth, L/SN+1: large/small VBs of leaf (N+1)th, DVs: diffuse vascular bundle, SAM: shoot apical meristem, IM: inflorescence meristem, DAP: day-after-pollination.
During root development, blue color was observed along the crown root and stopped at a distance far from the root tip (Fig. 2E). In the radicle, the GUS signal was not distributed evenly but accumulated in junctions with mature lateral root (LR, Fig. 2H). This expression pattern was also found in roots of mature plants, in which the GUS signal was distributed along the crown and young roots (Supplementary Fig. S4C) and accumulated in mature LR junctions in old roots. The highly specific expression of OsWOX13 led to its absence in apical meristem of root and shoot (RAM and SAM, Figs. 2E and 2M), where there was no vascular tissue. However, Supplementary Fig. S3 shows that this gene was strongly activated in the region around SAM, a special structure of the vascular system in the nodal portion of rice (Hoshikawa 1989; Yamaji and Ma, 2009; Yamaji et al. 2013).
Unlike the constitutive expression in vascular tissue of vegetative organs, OsWOX13 was expressed in a development-dependent manner in flowers and seeds. After the panicle developed, the GUS signal was consistently detected in the vascular tissue up to the receptacle (Fig. 2Q), but not in the early development stage (Fig. 2P). When the panicle reached 8–15 cm, it strongly accumulated in the anther and then disappeared in this organ at the stage before pollination, together with its increased expression in the pistil (Figs. 2R and 2S). After pollination, the whole pistil was stained blue and maintained up to 2 days after pollination (DAP). Then, the signal became weaker by the time of seed development and ceased at the stage of 10DAP (Figs. 2T and 2U). Thus, OsWOX13 is expressed during the initiation stage of male reproductive organ development but in the mature stage of the female organ. Additional expression information is provided in Supplementary Fig. S4.
OsWOX13 is induced by acute dehydration and its overexpression lines performed better against dehydration and water withdrawal
In an effort to understand how rice responds to drought stress, we observed the plant response to dehydration by the up- or down-regulation of many processes involved in inhibiting growth and development in leaf and root (Minh-Thu et al., 2013). In the data, 36 HD containing transcription factors were 1.6-fold up or down-regulated (Supplementary Fig. S5), including seven WUSCHEL-related homeobox (WOX) genes. The intensities of OsWOX13 leaf and root were enhanced 2 times higher than those at the initial stage and ranged from 6000 to 7000. In contrast, rab21 (Os11g0454300), a drought-inducible marker, was enhanced 4–5 times higher than OsWOX13, and its maximal intensities reached 23000 to 54000. RT-PCR analysis confirmed that OsWOX13 was up-regulated under drought stress in leaf and root (Fig. 3A). Markers for the dehydration response, rab21 and dip1, were also up-regulated after drought stress treatment. Although OsWOX13 was also induced by various stressors such as heat, cold and high salt, its induction was clearly induced by drought stress (Fig. 3B).
Fig. 3. Induction of OsWOX13 by abiotic stressors.
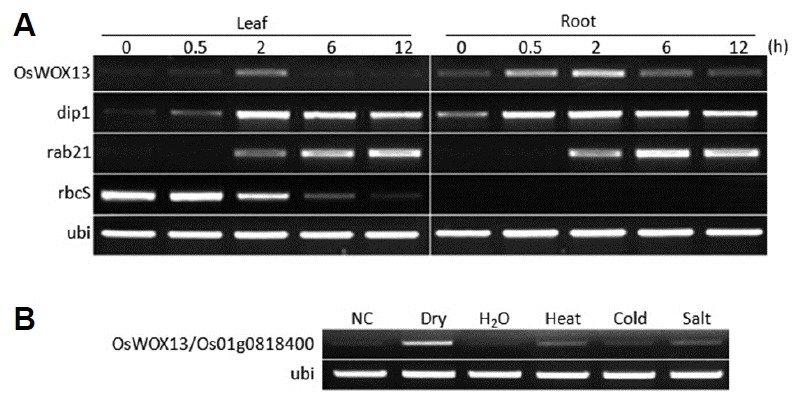
(A) RT-PCR analysis confirmed that Os01g0818400/OsWOX13 were moderately up-regulated under drought stress in leaf and root. (B) Leaves of 14-d greenhouse-grown seedlings were collected directly (NC) or subsequently the whole plant was air-dried (dry) in a growth chamber. Another set was removed from soil to water and kept in a chamber at 28°C (H2O), 42°C (heat) or 4°C (cold). Then, 400 mM NaCl was also added to the water to induce salt stress (salt). All leaves were collected after 3 h.
WOX genes are specifically expressed in plants, which is involved in the early phases of embryogenesis and lateral organ development in plants (Haecker et al., 2004). OsWOX13 is expressed in most vascular system with a high degree of variation according to the organ and induction during drought response. Since homeodomain TFs have been implicated in many aspects of development, we thought OsWOX13 might be involved in the modulation of development and the drought response. To test this hypothesis, we attempted to enhance the expression of the gene with the rab21 promoter, which is enhanced by drought (OsWOX13-ov, Supplementary Fig. S6A). The vector OsWOX13-ov contains bar as a selectable marker. It also contains GFP under the Wsi18 promoter to facilitate the selection of homozygotes at the seed stage. The vector was then introduced into embryogenic calli collected from mature seeds (Oryza sativa L. cv. Ilmi) as described in the methods. Initially, 37 stable transformants (T0) were collected, and the integration sites were determined by FSTVAL, a transgene flanking sequence validator (Kim et al., 2012). To avoid by genic localization of the transgene, transformants with a single copy in intergenic regions were selected, and 5 transformants were selected and passed through generation iterations (Supplementary Table S2).
To investigate whether the expression of OsWOX13 was correlated with abiotic stress tolerance, 4-week-old seedlings of the non-transgenic control (WT) and its 3 independent transgenic rice were subjected to drought-induced stress (Fig. 4). First, 4-week-old seedlings were kept in soil and then subjected to 1 day of drought. Under these conditions, all the leaves of the wild type plant had completely rolled and bent down, while transgenic plants still retained some opened and upright leaves (Fig. 4A, middle). In contrast to the wild type leaves, which were permanently damaged, approximately half of the OsWOX13-ov leaves were able to recover after 2 days of re-watering (Fig. 4A, right). Thus, transgenic plants experienced a faster recovery compared with the control plants. The overall survival rate was also higher in plants that overexpressed OsWOX13; thus, 1-1-1, 8-4-1, 30-5-1 lines showed survival rates of 83.3, 91.7, and 83.3%, respectively, after 7 days of re-watering, while wild type showed 54.2%.
Fig. 4. Seedlings of transgenic rice under dehydration and OsWOX13 expression pattern.
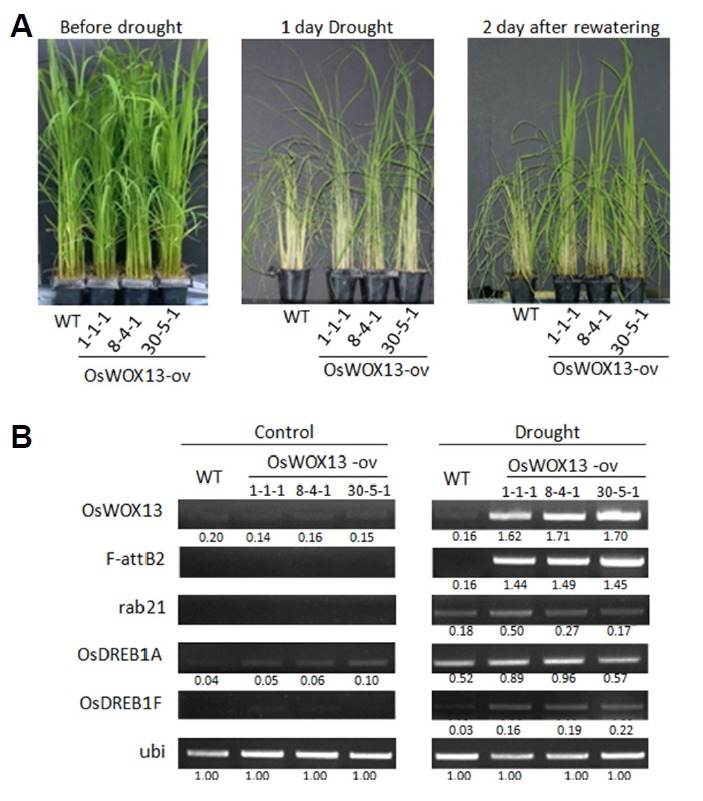
Seeds were germinated in soil (12 seeds/5x5x6 cm pot). Drought stress was conducted by water-withdrawal for 1 day in 2-week-old seedling followed by rewatering for 2 days (A). Leaves were collected, and the expression of OsWOX13 was examined (B). Bands were quantified and divided to assess the corresponding ubi to obtain the relative ratio to ubiquitin given as the number under each band. Given that ubi is the constitutive primer, all bands of ubi were calibrated to 1.00.
We tested the expression of OsWOX13 by RT-PCR using gene specific primers. OsWOX13 was weakly expressed under non-stressed conditions (Fig. 4B), while it was much more enhanced under acute dehydration. The band intensities of all the transgenic plants were strongly enhanced compared with the non-transgenic plants as expected. We tested the expression with transgene specific primers as indicated for F-attB2 in Supplementary Fig. S6, and the expression patterns were the same with those with gene-specific ones, suggesting the occurrence of gene expression under a drought-inducible promoter as intended.
Overexpression of OsWOX13 showed early flowering
The phenotypic appearance of flowers of transgenic plants were not different from those of wild type Ilmi. However, overexpression lines of OsWOX13 showed early flowering. Wild type Ilmi showed heading around Aug. 17th in the irrigated field in Korea, and the transgenic plants showed 7–10 days earlier flowering compared with the Ilmi wild type (Figs. 5A and 5B). We investigated the expression of OsWOX13 around the panicle-forming stages of mature leaf as early as 40 (Δ40), 33 (Δ33), and 25 (Δ25) days prior to the heading stage using 8-4-1 as a representative (OsWOX13-ov). The stages of 1 cm (P1cm, Δ19) and 8 cm (P8cm, Δ12)-long panicles were also included. As shown in Fig. 5A, OsWOX13 (F/R) in wild type was induced as early as 33 days prior to heading and maintained its expression up to P8cm. The expression of the gene in all transgenic lines, 1, 8, and 30, was induced 1 week earlier than wild type such that it was expressed as early as 40 days before of pollination. The expression of transgenes distinguished with the OsWOX13 F/attB2 primer set clearly showed that it was expressed around these stages. The promoter of rab21 was used for the vector and was also expressed even in irrigated fields (Fig. 5C).
Fig. 5. Overexpression of OsWOX13 (rab21 promoter) showed early flowering.
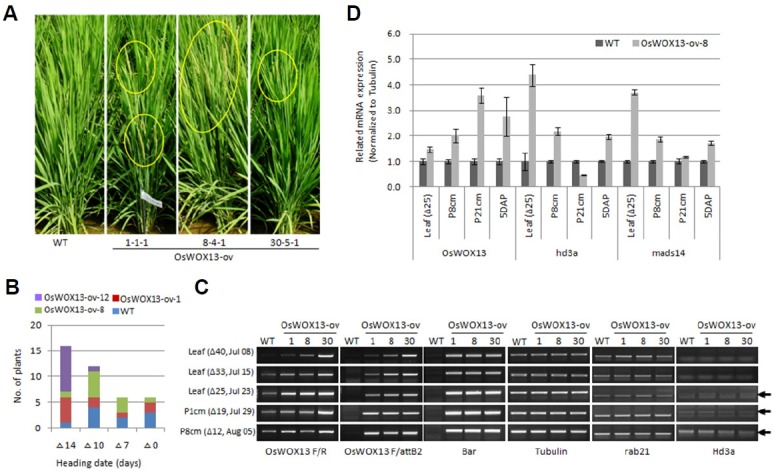
(A) Three T3lines showed the early flowering phenotype in the field. Wild type Ilmi showed heading around Aug. 17th in the irrigated field, and the transgenic plants showed 7–10-d earlier flowering compared with Ilmi. (B) Normal distribution of heading dates of transgenic and wild type plants. (C) Mature leaves were collected at day 40, 33, 25 (Δ40, Δ33, Δ25; respectively) prior to the heading date. The 1 cm (P1cm, Δ19) and 8 cm (P8cm, Δ12)-long panicles were also included. The transgenic line 8-4-1 was used as a representative (OsWOX13-ov). (D) The expression level of flowering-related genes by qPCR with tubulin as the expression standard.
It has been reported that Hd1 (Os06g0275000) acts either as an activator under short day conditions or a repressor under long day conditions during expression of the florigen, Hd3a (Os06g0157700). hd3a is expressed and translated in the phloem of the leaf, and its protein is transferred to the SAM to initiate the floral transition. Hd3a is associated with a 14-3-3 protein in the cytoplasm and translocated to the nucleus where it regulates OsMADS15 (Os07g0108900), a homolog of the Arabidopsis floral identity gene APETALA1 (Ishikawa et al., 2011; Tamaki et al., 2007; Tsuji et al., 2013). The expression level of florigen, Hd3a, was not observed as early as 40 to 33 days prior to heading (Fig. 5C). However, it was marginally induced in overexpression lines at 25 days prior to heading. Hd3a functions downstream of Os-MADS14 (Kim et al., 2007). We also quantified Hd3a and OsMADS14 by quantitative real-time RT-PCR using Tubulin (Tub) as a standard (Fig. 5D). Total RNA were prepared from the leaf (Δ25), P8cm (Δ12) and P21cm (Δ5) before the heading stage and 5 days after pollination (5DAP). The analysis showed an increase in Hd3a and OsMADS14 by 4 -16-fold in the leaf (Δ25) and P8cm of the OsWOX13-ov line compared with wild type, confirming the early flowering.
Genome-wide comparison of gene expression during flower development between wild type Ilmi and the OsWOX13 over line
To study how the genes were modulated during the leaf to panicle stages, genome-wide gene expression of wild type and OsWOX13-ov (8–4 lines) was performed using the Rice 3′-Tiling 135 k microarray designed from 31,439 genes (http://rapdb.lab.nig.ac.jp). To profile the gene expression, total RNA was extracted from leaf (Δ25), P1cm (Δ19), P8cm (Δ12), P21cm (Δ5) and 5DAP in wild type and OsWOX13-ov lines and hybridized as described in Methods. Two biological data replicates are deposited at NCBI (GSE55984). OsWOX13 was designated HMB4 in the database. When the expression of the leaf (Δ25) of wild type was compared, 4852, 4615, 4268, and 3822 genes were up-regulated at the stages of P1cm, P8cm, P21cm and 5DAP in wild type, respectively, by 2-fold with p-value less than 0.05. The down-regulated genes showed similar ranges of 5047, 4094, 3002, and 2492 at P1cm, P8cm, P21cm, and 5DAP, respectively. When the biological GO terms were searched, the 25 highest clade terms were enriched (Supplementary Fig. S7A). When the expression of leaf (Δ25) of OsWOX13-ov was compared, 4635, 4951, 4682 and 3990 genes were up-regulated and 5570, 4174, 3477 and 3202 gens were down-regulated at P1cm, P8cm, P21cm, and 5DAP of OsWOX13-ov, respectively, by 2-fold with p-value less than 0.05. The 22 highest clade terms or biological GO terms showed a similar pattern to those of wild type (Supplementary Fig. S7B). To represent the transition to the panicle from the leaf, development and reproduction, components such as GO:0022414 and GO:0032502 were highly enriched. The biological processes of the OsWOX13-ov line was very similar to those of wild type other than the term GO:0010143_cutin biosynthetic process, which was enriched at the P8cm stage. These data suggested that the OsWOX13-ov line had undergone a similar process as wild type.
Genome-wide gene expression showed the developmental transition from mature leaf to young seed in both Ilmi and OsWOX13-ov. We also measured the gene expression by stage-wise comparison between Ilmi and OsWOX13-ov. The expression was first compared to Ilmi leaf, and then the log 2-based data for OsWOX13-ov were subtracted from the corresponding data of the same stage of wild type (Supplementary Table S3). When the gene expression of OsWOX13-ov was compared to that of wild type, the 589, 315, 965, 472 and 369 genes were up-regulated and 163, 1091, 440, 485 and 277 genes were down-regulated by 2-fold at the leaf, P1cm, P8cm, P21cm and 5DAP stages, respectively (Supplementary Fig. S8).
When these genes were subjected to Gominer analysis as described in methods, the profiling of enriched GO terms in OsWOX13-ov was quite different from that of wild type (Table 1 and Supplementary Table S4). The up- or down- regulated genes in the stage of leaf (Δ25) for OsWOX13-ov compared with wild type revealed an enrichment, excluding the term GO:0009911_positive regulation of flower development, which was enriched. The genes in this category were Os01g0748800 (PEBP family protein), Os09g0420900 (BTB domain containing protein), Os02g0705500 (bHLH domain containing protein), Os02g0153500 (Protein kinase-like domain containing protein), and Os11g0547000 (Similar to FKF1), and they were increased 2–5-fold in the leaf of OsWOX13-ov compared with wild type, suggesting early flower initiation. In contrast, P8cm and 5DAP showed the most active modification in response to the overexpression of OsWOX13.
Table 1.
Biological processes enriched by 2-fold up-regulated genes in transgenic young panicles.
| Up regulated genes | REFLIST | Stagesa | |||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| P1cm | P8cm | P21cm | 5DAP | ||||||
|
| |||||||||
| No. of genes | No. of genes | P-value | No. of genes | P-value | No. of genes | P-value | No. of genes | P-value | |
| Regulation of metabolic process (GO:0019222) | 2909 | 46 | 4.18E-08 | 26 | 1.04E-03 | ||||
| Carbohydrate metabolic process (GO:0005975) | 1400 | 21 | 3.01E-02 | ||||||
| Negative regulation of catalytic activity (GO:0043086) | 241 | 9 | 1.02E-02 | ||||||
| Catabolic process (GO:0009056) | 2060 | 27 | 2.16E-02 | ||||||
| Negative regulation of cellular metabolic process (GO:0031324) | 303 | 10 | 9.14E-03 | ||||||
| Anchored component of plasma membrane (GO:0046658) | 208 | 8 | 8.51E-03 | ||||||
| Extracellular region (GO:0005576) | 794 | 29 | 2.81E-13 | ||||||
| Enzyme inhibitor activity (GO:0004857) | 243 | 9 | 8.55E-03 | ||||||
| Chitinase activity (GO:0004568) | 72 | 8 | 8.07E-06 | ||||||
| Regulation of jasmonic acid mediated signaling pathway (GO:2000022) | 18 | 4 | 4.77E-03 | 3 | 4.01E-02 | ||||
| Regulation of defense response (GO:0031347) | 42 | 5 | 5.48E-03 | 4 | 1.48E-02 | ||||
| Regulation of transcription, DNA-templated (GO:0006355) | 2273 | 36 | 7.12E-06 | 26 | 8.54E-06 | ||||
| Response to stimulus (GO:0050896) | 3509 | 26 | 2.95E-02 | ||||||
| Response to abscisic acid (GO:0009737) | 113 | 5 | 4.27E-02 | ||||||
| Rhythmic process (GO:0048511) | 14 | 3 | 1.90E-02 | ||||||
| Single-organism process (GO:0044699) | 8977 | 77 | 7.83E-03 | 84 | 1.53E-02 | ||||
| Chloroplast (GO:0009507) | 845 | 18 | 2.08E-03 | ||||||
| Response to heat (GO:0009408) | 43 | 5 | 1.13E-02 | ||||||
| Response to external stimulus (GO:0009605) | 151 | 8 | 6.84E-03 | ||||||
| Transmembrane transport (GO:0055085) | 1455 | 26 | 1.33E-03 | ||||||
| Membrane (GO:0016020) | 10519 | 91 | 3.79E-02 | ||||||
| Nutrient reservoir activity (GO:0045735) | 89 | 12 | 6.04E-10 | ||||||
| Substrate-specific transmembrane transporter activity (GO:0022891) | 955 | 19 | 7.40E-03 | ||||||
Stages are indicated as in Fig. 5C
At the P8cm stage, many related GO terms such as “regulation of metabolic process” (GO:0019222), “carbohydrate metabolic process” (GO:0005975), “catabolic process” (GO:0009056), and “regulation of transcription, DNA-templated” (GO:0006355), were enriched with up-regulated genes in OsWOX13-ov, suggesting that they provide the plant with energy for active growth. These results showed that gene expression and metabolism were positively regulated together at P8cm and P21cm of OsWOX13-ov. Interestingly biotic stress-related terms such as “regulation of defense response” (GO:0031347) and “regulation of jasmonic acid mediated signaling pathway” (GO:2000022) were significantly up-regulated at P8cm and P21cm in OsWOX13-ov. “Response to stimulus” (GO:0050896), “response to abscisic acid” (GO:0009737), and “rhythmic process” (GO:0048511) were up-regulated at P21cm prior to the pollination, but “response to stimulus” (GO:0050896) and “response to abscisic acid” (GO:0009737) were down-regulated at 5DAP of the OsWOX13-ov after suggesting many biological processes change after the stage of pollination. Similarly, chloroplast related genes such as “chloroplast” (GO:0009507), “photosystem II” (GO:0009523), and “photosynthetic membrane” (GO:0034357) were down-regulated from P8cm to P21 cm, but “chloroplast” (GO:0009507) was up-regulated at 5DAP in OsWOX13-ov.
Analysis of the protein binding microarray suggested that ATTG repeats might be cis-binding elements of OsWOX13 protein
To identify putative downstream genes of OsWOX13, we analyzed the cis-binding elements using a protein binding microarray (Kim et al., 2009). We constructed a fusion protein, OsWOX13-DsRed-6His, under the control of the T7 promoter in the pET-32a expression system and hybridized it to a quadruple 9-mer based protein binding microarray (Q9-PBM). The rank and signal intensity plot showed strong binding of OsWOX13 on Q9-PBM (Supplementary Fig. S9A), and the position weight matrix from strong binding ninemers was constructed (Supplementary Fig. S9B). These data revealed ATTGATTG as the DNA binding motif of OsWOX13 protein (Fig. 6A). To confirm the absolute presence of the two ATTG repeats, we designed an experiment using a 5′biotin probe that contains five repeats of ATTG and gradually replaced G with C in 5 competitors that made them remain 4-0 repeats of ATTG (Fig. 6B). An electromobility shift assay (EMSA) was performed with these probes. The probe ATTG_5x was strongly shifted by addition of OsWOX13 (Fig. 6C). When the ATTG_5x probe itself was used as a competitor, the band shift was almost inhibited, and the degree of inhibition was gradually disappeared as the number of replaced G was increased. Indeed, the competition activity could only occur when there were at least two repeats of ATTG in the sequence, suggesting that the ATTG repeats might be the cognate binding site.
Fig. 6. Analysis of the putative DNA binding sequence of OsWOX13.
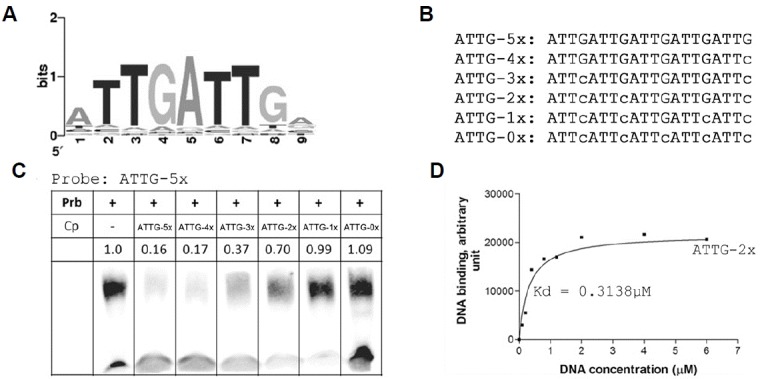
(A) LOGO of cis-acting element from the Q9-PBM results. (B) Sequences of non-biotinylated competitors (Cp) used in (c). (C) EMSA-based competition analysis. (D) DNA binding affinity graph of OsWOX13 protein with ATTG-2x non-biotinylated probe in an EMSA-based test.
To obtain more detail on the strength of the binding affinity represented by a number, we carried out an EMSA experiment followed by Sybr Gold staining to determine static kinetic constants. The intensity of the DNA binding band was quantified as described in Methods. The data were fit to the following equation: Y=Bmax*X/(Kd+X), where Bmax is the maximal binding and Kd is the concentration of ligand required to reach half-maximal binding. The calculated Kd value of ATTG_2x was approximately 0.3 μM (Fig. 6D), which is low enough to act as a DNA binding protein.
A survey of the RAP3.0 database (http://rapdb.dna.affrc.go.jp/) showed 5828 among 39102 genes carried the ATTGATTG motif in their 2-kb upstream regions. The number of genes that contained the motif and were up-regulated in OsWOX13-ov (H8) were 11, 11, 52, 11, and 35 at the leaf, P1cm, P8cm, P21cm, and 5DAP stages, ranging from 3–7% among the up-regulated genes (Supplementary Table S5). In contrast, among the down-regulated genes in the H8 line 5, 32, 7, 35 and 36 genes contained the ATTGATTG motif at the leaf, P1cm, P8cm, P21cm, and 5DAP stages, respectively.
Of 120 genes, 70 could be functionally assigned into 7 groups (Supplementary Fig. S10). The major group contained genes involved in cell defense and rescue (20%), notably, Os03g0639400, Os02g0786900, Os07g0240200, and Os01g0947000. Os02g0121700, Os10g0101100, and Os10g0465700 comprised the second largest group and are catalytic enzymes (19%). The third and fourth groups corresponded to signaling (11%) and regulation of transcription (10%). There were also 4 genes related to reproductive development (4%). Interestingly, Hd1-3 (Os08g0536300) was induced in P21cm of OsWOX13-ov. The protein is similar to Hd1, which is known to be a main regulator of the florigen Hd3a. Further results will be presented in the next section. The other two groups were transport (5%) and miscellaneous (3%). As these genes were induced through stages from leaf to 5DAP, the gene expression mechanisms involving OsWOX13 might be complex during the rice panicle forming processes.
To confirm the microarray experiments, RT-PCR were performed for the 9 genes, including Os03g0639400, Os03g0626700, Os04g0685400, Os12g0630500, and Os04g0437000 (Supplementary Fig. S11). These genes were up-regulated during at least one of the stages, as predicted by microarray, and they were especially highly induced more than 20 times at the P8cm stage in OsWOX13-ov. These findings were also confirmed in the following microarray experiments, suggesting that genes related to another development are also induced earlier than those in wild type.
Drought-responsive genes and genes involved in flowering might be targets of OsWOX13
Promoter scanning suggested that OsDREB1A and OsDREB1F have ATTGATTG motifs at -1557, and -1552, respectively from the transcript initiation site (Supplementary Table S5). We assessed whether they could be activated during abiotic stress-induced overexpression of OsWOX13. Compared to its expression in leaves of wild type, OsDREB1A was induced in line number 1 and 8 during drought-related stress. OsDREB1F was activated in all transgenic lines by drought stress (Fig. 4B). OsDREB1A and OsDREB1F have previously been reported to enhance stress tolerance in transgenic rice (Ito et al., 2006; Wang et al., 2008). Thus, the ability of the transgenic rice to tolerate drought in these lines might be a result of the activation of OsDREB1A and OsDREB1F by OsWOX13.
We also scanned the ATTGATTG motif among the 2-kb promoters of the genes related to rice flower development. As described above, Hd3a and OsMADS14 increased 4 -16-fold in the leaf (Δ25) of OsWOX13-ov compared with Ilmi, confirming the early flowering. The promoters of Hd3a and OsMADS14 contained the ATTGATTG motif in the promoters at -1902 and -1016, respectively, from the transcription starts. Taken together, these results suggested OsWOX13 might be involved in the process of drought response and flowering.
DISCUSSION
Plant flowering is critically influenced by photoperiodic pathways in the developmental stage from vegetation to reproduction. The various environmental stresses, such as drought, cold, and UV, among others, affect the process by modulating gene expression (Kumar et al., 2012; Peltzer et al., 2002; Riboni et al., 2013; Shinozaki and Yamaguchi-Shinozaki, 2000; Yu et al., 2013). Each stress, including drought, is mediated by mechanisms through positive and negative regulators (Bernier and Périlleux, 2005). Plants respond to stress through either DE by showing an early flowering phenotype or DT by delayed flowering. Their genetic components seem be maximally adapted according to the local habitat. In an effort to understand how rice responds to acute dehydration, we found that the leaf and root tissues of rice responded to dehydration by up- or down-regulating many processes involved in inhibiting growth and development, while some osmoprotectant and antioxidant pathways, which might help the plant endure harsh conditions, were immediately up-regulated (Minh-Thu et al., 2013). In addition, various transcription factors, including Myb and the homeodomain family, were induced in 2 h. Since homeodomain TFs have been implicated in many aspects of development, we thought the enhanced expression of OsWOX13 under the control of rab21 promoter (5–10 folds high) might provide some clues to elucidate the function of OsWOX13 related to water deficit stress.
OsWOX13 was regulated spatially in vegetative organs but temporally in flowers and seeds. Overexpression of OsWOX13 (OsWOX13-ov) resulted in drought resistance, and several plants showed early flowering. Our data raise the possibility that OsWOX13 might be a regulator of the genes involved in drought escape and enhance drought resistance.
OsWOX13 is similar in structure but shares few similarities with AtWOX13 and AtWOX14 in expression pattern
WUSCHEL (WUS) was first isolated from Arabidopsis and then rice and maize, but its functions in development might be divergent (Deveaux et al., 2008; Nardmann et al., 2009). WUS is restrictedly expressed in the organizing center (OC) located below the stem cells of the shoot apical meristem and regulates the size of the shoot meristem by maintaining the appropriate number of pluripotent stem cells in each shoot meristem in Arabidopsis (Ikeda et al., 2009). Subsequently, WUSCHEL-related homeobox (WOX) genes form a large subfamily and are plant-specific (Deveaux et al., 2008; Nardmann et al., 2009; Zhang et al., 2010b). Phylogenetic analysis provided good support for the division of the WOX subfamily into three separate orthologous groups (OGs), WOX1, WOX8 and WOX13, in which WOX13 OG is regarded as the most conserved clade (Deveaux et al., 2008; Lin et al., 2013; van der Graaff et al., 2009). These proteins function in various physiological and developmental processes, especially embryonic patterning, stem cell maintenance and organ formation. Members of the WOX genes in Arabidopsis, such as AtWOX2, 3, 6, 9, and 11, are expressed in restricted region (Zhang et al., 2010b). AtWOX5 expressed in the cells of the quiescent center in the root maintains stem cells in the root apical meristem (RAM) and stem cell maintenance capability (Zhang et al., 2010b).
Rice WOX genes, in contrast, are less understood. In contrast to the case of WUS homologs, QHB/OsWOX5 shares the same expression region and similar stem cell maintenance capability as its Arabidopsis homolog AtWOX5 (Zhang et al., 2010b). OsWOX3 is also excluded from the shoot apical meristem (Dai et al., 2007) as PRS1/AtWOX3 (Shimizu et al., 2009) and OsWOX11 has been shown to be able to activate the emergence and growth of crown root by recruiting a histone acetyltransferase complex (Zhang et al., 2010b; Zhou et al., 2017).
Cheng et al. (2014) reported that four genes (OsWUS, OsNS1/OsNS2, OsWOX3 and OsWOX9A) are specifically expressed during panicle and endosperm development, and six genes (OsWOX5, OsWOX9B, OsWOX9D, OsWOX11, OsWOX12A and OsWOX12B) are preferentially expressed in seeds (72 h after imbibition) during root emergence or growth. OsWOX13 was named because of its similarity in protein structure to AtWOX13 (Supplementary Fig. S2 and Fig. 1). WOX13 OG has also been reported as the most conserved clade in plants. However, the expression pattern shows minimal overlap between rice and Arabidopsis members. OsWOX13 is differentially regulated in vegetative and reproductive organs, demonstrating spatially regulated expression in vascular tissue of leaf/stem/root and temporally regulated expression in flowers and developing seeds. At-WOX13 is restricted to differentiating organs, and At-WOX14 is linked to the early stages of organ and tissue development (Deveaux et al., 2008). Both of them are no longer active in the mature organs. In contrast, OsWOX13 expression could only be detected in active vascular tissue, and no signal was observed in the root/shoot apical meristem or lateral root/shoot at the primordia and emergence stages (Fig. 2). OsWOX13 was strongly expressed in young organs and still maintained in mature ones (Supplementary Fig. S4). The only commonality in expression of these 3 genes is their presence in vascular tissue. This difference might be the result of other components, such as the monocot-specific WOX13 motif.
Enhancement of OsWOX13 not only endows drought resistance but also triggers floral development resulting in early flowering
Previous studies have demonstrated that overexpression of OsDREB1A under the CaMV 35S promoter causes both Arabidopsis and rice to grow slowly because of the constitutively high expression of the transgene (Dubouzet et al., 2003; Ito et al., 2006). Overexpression of OsNAC6 under the maize ubiquitin promoter has also been shown to reduce the plant growth rate; however, this trend can be reversed when OsNAC6 or the cold-inducible promoter LIP9 is used instead (Nakashima et al., 2007). OsWOX13 is actively expressed in vascular tissues. Interestingly, it is induced by several stressors, such as drought, heat, cold and salt, suggesting it is involved in responses to abiotic stresses (Fig. 3). Indeed, the intensities of OsWOX13 in the microarray data using total RNA from leaf and root treated for 2 h with acute dehydration were 6225 and 7299, respectively, and enhanced approximately 2-fold. In contrast, those of rab21 were enhanced to 22796 and 40185 in these organs and 3–5-fold in the same period, confirming that rab21 is induced by water deficit and ABA (Minh-Thu et al., 2013; Mundy and Chua, 1988; Oh et al., 2005). When OsWOX13 was under the control of the rab21 promoter, the overexpression lines showed strong enhancement of the gene in response to acute dehydration (Fig. 4). In addition, these plants showed less severe symptoms in observations of the degree of leaf folding and in water withdrawal and re-watering experiments. The induction of DREB1A and DREB1F also suggested that overexpression of the gene conferred drought resistance.
The HD protein family in rice is large, and its members are primarily involved in developmental processes. However, the expression levels of these genes are also modulated during abiotic stress. When Jain et al. (2008) performed a microarray analysis of 7-d-old seedlings that had been subjected to desiccation, salt or cold, a total of 37 homeobox genes were found to be differentially expressed under at least one stress condition. The expression of OsWOX13 was induced both by desiccation (Jain et al., 2008) and by acute dehydration (Minh-Thu et al., 2013). Previously, only one homeobox transcription factor (BIDH1) had been reported to function during the abiotic stress response in rice (Luo et al., 2005). However, BIDH1 overexpression has been shown to enhance the plant’s sensitivity to salt and oxidative stress, resulting in increased cellular damage in response to these stressors. Thus, OsWOX13 is the only homeobox gene in rice that has been reported to enhance tolerance to stress. Expression profiling of panicle development showed an enhancement of oxidative stress in young transgenic panicles (Supplementary Fig. S8 and Supplementary Table S4). As oxidative stress is one of the results of drought responses and OsWOX13 was up-regulated during drought (in this report) and salt stresses (unpublished data), these data support its involvement in the stress pathway.
Interestingly, OsWOX13 was induced in mature leaves as early as 40 days prior to heading and inflorescence tissue of transgenic plants under normal conditions, resulting in 7–10-day-earlier flowering and suggesting that OsWOX13 might be involved in not only the drought response but also developmental processes such as flowering (Fig. 5). Quantitative PCR clearly showed that the expression of Hd3a and Os-MADS14 increased 4 -16-fold in the leaf of the OsWOX13-ov line as early as 25 days before heading. Rice is a facultative SD type plant, and hd3a and RFT1 are the main florigenes under SD and LD, respectively (Hayama et al., 2003; Ishikawa et al., 2011). These data strongly suggest that flowering is initiated earlier in OsWOX13-ov than wt plant. This finding has also been confirmed in genome-wide comparisons with samples from the stage of leaf (Δ25), which revealed an enrichment of the term GO:0009911_positive regulation of flower development (Supplementary Table S4). These findings strongly suggest the involvement of OsWOX13 in responses not only to abiotic stress but also developmental processes.
OsWOX13 might be involved in multiple signaling pathways
The expression profiling of panicle development showed an enhancement of terms related to water deprivation, salt stress, osmotic stress, and abscisic acid in young transgenic panicles in OsWOX13-ov, suggesting that the over-expressing line responded to abiotic stress (Supplementary Table S4). In addition, terms related to biotic stress such as biotic stimulus defense, including response to fungus and hormones such as jasmonic acid and ethylene, were highly enriched. These observations are in agreement with previous results demonstrating a cross-talk between abiotic and biotic stresses in many overexpression lines. The signaling is exerted through complicated and dynamic networks with many members such as calcium, redox homeostasis, membranes, G-proteins, MAPKs, plant hormones and transcription factors (Knight and Knight, 2001; Sharma et al., 2013). The members of these components could be activated by overlapping receptors/sensors such that cross-talk occurs among stresses or stimuli but produces distinct final outputs that are specific to each stimulus. It is not uncommon that transcription factors are main factors in the crosstalk between signaling pathways. For example, the ethylene-responsive transcription factor SUB1A is induced by flooding by repressing gibberellic acid (GA) signaling (Fukao et al., 2008; Fukao et al., 2011; Xu et al., 2006). SUB1A is also implicated in drought tolerance by induction of the expression of ET-responsive and ABA-inducible transcripts, which aid in recovery from cellular dehydration. Interestingly, SUB1A prevents oxidative damage by restricting the accumulation of ROS (Voesenek and Bailey-Serres, 2013). Some other transcription factors, such as WRKY13 and OsbHLH068, are involved in abiotic and biotic stress signaling and are antagonistic (Chen et al., 2017; Xiao et al., 2013).
Cross-talk between signaling pathways can also be mediated by ROS and possibly influence developmental processes (Kotchoni and Gachomo, 2006; Lamb and Dixon, 1997; Mittler et al., 2011; Noctor et al., 2014; Orozco-Cardenas and Ryan, 1999; Petrov and Van Breusegem, 2012) because ROS seems to be associated with many different signaling events. In our data, the GO:0045730_respiratory burst and GO:0002679_respiratory burst involved in the defense response are also highly enriched terms, which suggests that the level of ROS had increased and might influence other signaling pathways (Supplementary Table S4). These data suggest that the level of ROS seem to be enhanced and might be interpreted as a pathogen infection. The sources of ROS are photosynthetic and respiratory electron transport chains, NADPH oxidase, photorespiration, amine oxidase, and cell wall-bound peroxidases. NADPH oxidases play a crucial role in ROS production during pathogen infection (Doke, 1983; Kadota et al., 2015; Torres and Dangl, 2005) The exact differentiation of the signals has not been determined. Plant cells might recognize the increased oxidative stress and interpret it as biotic stress and increase gene induction in the category of defense responses producing prominent cross-talk between abiotic and biotic stresses. It is noteworthy that SA involvement in endogenous signaling in plant defense against pathogens induces flowering in many species belonging to the Lemnaceae. This phenomenon has been implicated in the stress-induced flowering of A. thaliana (Wada et al., 2010). As plant disease resistance occurs as a hypersensitive response (HR) at the site of attempted pathogen invasion and oxidative burst, it is strongly dependent on the resistance of the cultivar. Yeast two-hybrid screening using the C-terminal region of tobacco NADPH-oxidase D, NtRBOHD, identified a 14-3-3 protein as an interactor (Elmayan et al., 2007).
OsWOX13 might contribute to cross-talk by modulating gene regulation
As indicated by the overexpression of transcription factors such as SUB1A and WRKY13, the pathway to drought tolerance and early flowering might be more gene-specific. Our analysis to identify the motif of OsWOX13 using Q9-PBM suggested that many genes might be directly activated by OsWOX13 through the ATTGATTG motif (Supplementary Table S5 and Fig. 4). A motif search in UniPROBE also provided various GA or AT-rich motifs of homeodomain containing transcription factors such as UP00615B_1 and UP00158A_1 in human and mouse, respectively (the_brain.bwh.harvard.edu; (Hume et al., 2014)). Especially, UP00158A_1 contains AATTAATTA and ATTA repeats and showed a base (A to G) difference with ATTG repeats in the ATTGATTG motif in our analysis.
The genes that respond to biotic stress processes and contain an ATTGATTG motif in their promoters are Os01g0946700 (glucan endo-1,3-beta-glucosidase), Os02g0638650 (pathogenesis-related transcriptional factor and ERF domain containing protein), Os07g0240200 (Beta-1,3 glucanase precursor), and Os12g0481700 (disease resistance protein family protein), which were enhanced in OsWOX13-ov. The genes OsDREB1A and OsDREB1F have previously been reported to enhance stress tolerance in transgenic plants (Ito et al., 2006; Wang et al., 2008), and the promoters of these genes contain the ATTGATTG motif. Compared to its expression in Ilmi leaves, OsDREB1A was highly expressed in line number 1 and 8 during drought-related stress. Its expression was also enhanced in all three OsWOX13-ov transgenic lines during salt-related stress (data not shown). OsDREB1F was activated in all transgenic lines by drought stress (Fig. 4). These data suggest that OsWOX13 might be involved in the regulation of gene expression in a pathway-specific manner.
The observation that the promoters of Hd3a and Os-MADS14 contain this motif raises the possibility that the expression of these genes might be under the control of OsWOX13. Indeed, and the expression of these genes was enhanced in the OsWOX13-ov lines (Figs. 5C and 5D). Taken together, these data suggest that the drought response and early flowering might be regulated through OsWOX13 control of the genes. OsWOX13 might also be involved in later reproductive development through genes containing the motif in their promoters. They were Os11g0684000, similar to AtMYB21; Os10g0552600, similar to Tfm5; and Os03g0830500, similar to petunia germination pollen-specific proteins (PGPS/D12). AtMYB21 acts together with AtMYB24 and AtMYB57 to regulate the development of stamen filaments in Arabidopsis in a GA-dependent manner (Cheng et al., 2009; Mandaokar et al., 2006; Song et al., 2011). Tfm5 has been used to enhance the early fruiting in tomato (Jeong et al., 1999; Lytovchenko et al., 2011; Santino et al., 1997). Slightly higher induction of Osc6 and significant up-regulation of OsCP1 has been seen in P8cm, which corresponds to the formation of uninucleated gametophytes (Itoh et al., 2005). This result is in line with reports showing that Osc6 and OsCP1 are specific for the microspore stage (Lee et al., 2004; Tsuchiya et al., 1992; Zhang et al., 2010a) and impose a possible effect of OsWOX13 during the development of anther, suggesting that OsWOX13 is involved in multiple pathways, which awaits further analysis.
OsWOX13 might modulate drought resistance and induction of flowering
Plants seem to harbor genetic systems conferring both dehydration avoidance and drought escape strategies. Rice is generally sensitive to drought, and the mechanisms leading to drought tolerance are complex both genetically and physiologically (Blum, 2002). Many rice cultivars tend to delay flowering in response to drought treatment (Wopereis et al., 1996). However, DE and DT are also observed in the genetic dissection of adaptive strategies to water stress (Xu et al., 2005) in rice. As observed in Arabidopsis, during reproductive development, flowering seems to be severely affected by drought stress, and DE might be activated to secure the production of offspring for the survival of species (Ma et al., 2014; Riboni et al., 2013). Flowering of Arabidopsis, a model dicot plant, is regulated by several environmental factors such as day length, vernalization, and hormones. Under a long day (LD), the flowering time is regulated by CONSTANS (CO) in terms of expression, protein stability and activity (Valverde et al., 2004). CO protein, a zinc finger transcription factor, acts during long-day conditions to facilitate the transcription of the FT gene, an equivalent of Hd3a in rice. The FT protein interacts with a bZIP transcription factor, FD, which is expressed in the shoot apex and induces SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1), MADS box and AP transcription factors, which are crucial for floral morphology (Abe et al., 2005; Coen and Meyerowitz, 1991; Pelaz et al., 2000; Searle et al., 2006; Yun et al., 2013). In Arabidopsis, abscisic acid is required for the DE response, positively regulating flowering under long-day conditions and showing the central role of the flower-promoting gene GI and the florigen genes FT and TSF in the DE response. The role of ABA in regulating the floral transition was initially proposed as an inhibitor in flowering based on the early-flowering phenotype of an ABA-deficient mutant (Koornneef, 1994). However, early flowering was also observed in constitutively activated ABA signaling mutants, and later flowering was observed in the mutants of ABA biosynthesis under LD conditions (Riboni et al., 2013; Rubio et al., 2009), suggesting a positive role of ABA in flowering.
The mode of transcription factors in floral transition regarding photoperiod could also be complex in rice. Recently, a bZIP transcriptional factor, O. sativa ABA responsive element binding factor 1 (OsABF1), was shown as a suppressor of floral transition in a photoperiod-independent manner (Zhang et al., 2016) when gene expression was controlled by the maize ubiquitin promoter (Pubi). However, knockdown of both OsABF1 and its closest homologous gene, OsbZIP40, in rice (Oryza sativa) by RNA interference resulted in a significantly earlier flowering phenotype. As the OsWOX13 was constructed under rab21 in our study, the mode of expression of OsWOX13 might be influenced by the internal ABA quantity under physiological conditions (Mundy and Chua, 1988). However, the promoter effect might be minimal, as our 5 other WOX vector constructs under the same promoter did not lead to early flowering (data not shown). Although the early flowering phenotype was observed without drought conditions, our data suggest that OsWOX13 might be involved both in drought resistance and in the induction of flowering. OsWOX13 might be the mediator of these pleiotropic effects.
Supplementary data
ACKNOWLEDGMENTS
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (YKK, grant no. NRF-2018R1D1A1B07049348, JSK, grant no. NRF-2018 R1D1A1B07049288 and NRF-2017R1A6A3A01076563).
Footnotes
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- Abe M., Kobayashi Y., Yamamoto S., Daimon Y., Yamaguchi A., Ikeda Y., Ichinoki H., Notaguchi M., Goto K., Araki T. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science. 2005;309:1052–1056. doi: 10.1126/science.1115983. [DOI] [PubMed] [Google Scholar]
- Agalou A., Purwantomo S.Övernäs EJohannesson H., Zhu X., Estiati A., de Kam R.J., Engström P., Slamet-Loedin I.H., Zhu Z. A genome-wide survey of HD-Zip genes in rice and analysis of drought-responsive family members. Plant Mol Biol. 2008;66:87–103. doi: 10.1007/s11103-007-9255-7. [DOI] [PubMed] [Google Scholar]
- Aguirrezabal L., BOUCHIER-COMBAUD S., Radziejwoski A., Dauzat M., Cookson S.J., Granier C. Plasticity to soil water deficit in Arabidopsis thaliana: dissection of leaf development into underlying growth dynamic and cellular variables reveals invisible phenotypes. Plant Cell Environ. 2006;29:2216–2227. doi: 10.1111/j.1365-3040.2006.01595.x. [DOI] [PubMed] [Google Scholar]
- Bernier G., Périlleux C. A physiological overview of the genetics of flowering time control. Plant Biotechnol J. 2005;3:3–16. doi: 10.1111/j.1467-7652.2004.00114.x. [DOI] [PubMed] [Google Scholar]
- Bhattacharjee A., Jain M. Plant Acclimation to Environmental Stress. Springer; 2013. Homeobox genes as potential candidates for crop improvement under abiotic stress; pp. 163–176. [Google Scholar]
- Blázquez M.A., Ahn J.H., Weigel D. A thermosensory pathway controlling flowering time in Arabidopsis thaliana. Nat Genet. 2003;33:168. doi: 10.1038/ng1085. [DOI] [PubMed] [Google Scholar]
- Blum A. Drought tolerance-is it a complex trait? Field screening for drought tolerance in crop plants with emphasis on rice. 2002:17–22. [Google Scholar]
- Bomblies K. Whole mount GUS staining. Arabidopsis: A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2000. pp. 243–245. [Google Scholar]
- Campo S., Baldrich P., Messeguer J., Lalanne E., Coca M., San Segundo B. Overexpression of a calcium-dependent protein kinase confers salt and drought tolerance in rice by preventing membrane lipid peroxidation. Plant Physiol. 2014;165:688–704. doi: 10.1104/pp.113.230268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae S., Kim J.S., Jun K.M., Lee S.-B., Kim M.S., Nahm B.H., Kim Y.-K. Analysis of genes with alternatively spliced transcripts in the leaf, root, panicle and seed of rice using a long oligomer microarray and RNA-Seq. Mol Cells. 2017;40:714–730. doi: 10.14348/molcells.2017.2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae S., Kim J.S., Jun K.M., Pahk Y.-M., Kim M.-J., Lee S.-B., Park H.-M., Lee T.-H., Nahm B.H., Kim Y.-K. Analysis of representative organ-specific genes and promoters of rice using a 3′ORF-oriented long oligomer microarray. J Plant Biol. 2016;59:579–593. [Google Scholar]
- Chan R.L., Gago G.M., Palena C.M., Gonzalez D.H. Homeoboxes in plant development. Biochimica et Biophysica Acta. 1998;1442:1–19. doi: 10.1016/s0167-4781(98)00119-5. [DOI] [PubMed] [Google Scholar]
- Chen H.-C., Hsieh-Feng V., Liao P.-C., Cheng W.-H., Liu L.-Y., Yang Y.-W., Lai M.-H., Chang M.-C. The function of OsbHLH068 is partially redundant with its homolog, AtbHLH112, in the regulation of the salt stress response but has opposite functions to control flowering in Arabidopsis. Plant Mol Biol. 2017;94:531–548. doi: 10.1007/s11103-017-0624-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H., Song S., Xiao L., Soo H.M., Cheng Z., Xie D., Peng J. Gibberellin acts through jasmonate to control the expression of MYB21, MYB24, and MYB57 to promote stamen filament growth in Arabidopsis. PLoS Genet. 2009;5:e1000440. doi: 10.1371/journal.pgen.1000440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S., Huang Y., Zhu N., Zhao Y. The rice WUSCHEL-related homeobox genes are involved in reproductive organ development, hormone signaling and abiotic stress response. Gene. 2014;549:266–274. doi: 10.1016/j.gene.2014.08.003. [DOI] [PubMed] [Google Scholar]
- Coen E.S., Meyerowitz E.M. The war of the whorls: genetic interactions controlling flower development. Nature. 1991;353:31. doi: 10.1038/353031a0. [DOI] [PubMed] [Google Scholar]
- Dai M., Hu Y., Zhao Y., Liu H., Zhou D.-X. A WUSCHEL-LIKE HOMEOBOX gene represses a YABBY gene expression required for rice leaf development. Plant Physiol. 2007;144:380–390. doi: 10.1104/pp.107.095737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveaux Y., Toffano-Nioche C., Claisse G., Thareau V., Morin H., Laufs P., Moreau H., Kreis M., Lecharny A. Genes of the most conserved WOX clade in plants affect root and flower development in Arabidopsis. BMC Evol Biol. 2008;8:291. doi: 10.1186/1471-2148-8-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi K., Izawa T., Fuse T., Yamanouchi U., Kubo T., Shimatani Z., Yano M., Yoshimura A. Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev. 2004;18:926–936. doi: 10.1101/gad.1189604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doke N. Generation of superoxide anion by potato tuber protoplasts during the hypersensitive response to hyphal wall components of Phytophthora infestans and specific inhibition of the reaction by suppressors of hypersensitivity. Physiol Plant Pathol. 1983;23:359–367. [Google Scholar]
- Dubouzet J.G., Sakuma Y., Ito Y., Kasuga M., Dubouzet E.G., Miura S., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt-and cold-responsive gene expression. Plant J. 2003;33:751–763. doi: 10.1046/j.1365-313x.2003.01661.x. [DOI] [PubMed] [Google Scholar]
- Elmayan T., Fromentin J., Riondet C., Alcaraz G., BLEIN J.P., SIMON-PLAS F. Regulation of reactive oxygen species production by a 14-3-3 protein in elicited tobacco cells. Plant, Cell Environ. 2007;30:722–732. doi: 10.1111/j.1365-3040.2007.01660.x. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Fukao T., Harris T., Bailey-Serres J. Evolutionary analysis of the Sub1 gene cluster that confers submergence tolerance to domesticated rice. Ann Bot. 2008;103:143–150. doi: 10.1093/aob/mcn172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukao T., Yeung E., Bailey-Serres J. The submergence tolerance regulator SUB1A mediates crosstalk between submergence and drought tolerance in rice. Plant Cell. 2011;23:412–427. doi: 10.1105/tpc.110.080325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H., Jin M., Zheng X.-M., Chen J., Yuan D., Xin Y., Wang M., Huang D., Zhang Z., Zhou K. Days to heading 7, a major quantitative locus determining photoperiod sensitivity and regional adaptation in rice. Proc Natl Acad Sci USA. 2014;111:16337–16342. doi: 10.1073/pnas.1418204111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray S.B., Brady S.M. Plant developmental responses to climate change. Dev Biol. 2016;419:64–77. doi: 10.1016/j.ydbio.2016.07.023. [DOI] [PubMed] [Google Scholar]
- Haecker A., Groß-Hardt R., Geiges B., Sarkar A., Breuninger H., Herrmann M., Laux T. Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development. 2004;131:657–668. doi: 10.1242/dev.00963. [DOI] [PubMed] [Google Scholar]
- Hamant O., Pautot V. Plant development: a TALE story. C R Biol. 2010;333:371–381. doi: 10.1016/j.crvi.2010.01.015. [DOI] [PubMed] [Google Scholar]
- Hayama R., Yokoi S., Tamaki S., Yano M., Shimamoto K. Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature. 2003;422:719. doi: 10.1038/nature01549. [DOI] [PubMed] [Google Scholar]
- Heschel M.S., Riginos C. Mechanisms of selection for drought stress tolerance and avoidance in Impatiens capensis (Balsaminaceae) Am J Bot. 2005;92:37–44. doi: 10.3732/ajb.92.1.37. [DOI] [PubMed] [Google Scholar]
- Hu H., You J., Fang Y., Zhu X., Qi Z., Xiong L. Characterization of transcription factor gene SNAC2 conferring cold and salt tolerance in rice. Plant Mol Biol. 2008;67:169–181. doi: 10.1007/s11103-008-9309-5. [DOI] [PubMed] [Google Scholar]
- Hume M.A., Barrera L.A., Gisselbrecht S.S., Bulyk M.L. UniPROBE, update 2015: new tools and content for the online database of protein-binding microarray data on protein-DNA interactions. Nucleic Acids Res. 2014;43:D117–D122. doi: 10.1093/nar/gku1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M., Mitsuda N., Ohme-Takagi M. Arabidopsis WUSCHEL is a bifunctional transcription factor that acts as a repressor in stem cell regulation and as an activator in floral patterning. Plant Cell. 2009;21:3493–3505. doi: 10.1105/tpc.109.069997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa R., Aoki M., Kurotani K.-i., Yokoi S., Shinomura T., Takano M., Shimamoto K. Phytochrome B regulates Heading date 1 (Hd1)-mediated expression of rice florigen Hd3a and critical day length in rice. Mol Genet Genomics. 2011;285:461–470. doi: 10.1007/s00438-011-0621-4. [DOI] [PubMed] [Google Scholar]
- Ito Y., Katsura K., Maruyama K., Taji T., Kobayashi M., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. Functional analysis of rice DREB1/CBF-type transcription factors involved in cold-responsive gene expression in transgenic rice. Plant Cell Physiol. 2006;47:141–153. doi: 10.1093/pcp/pci230. [DOI] [PubMed] [Google Scholar]
- Itoh J.I., Nonomura K.I., Ikeda K., Yamaki S., Inukai Y., Yamagishi H., Kitano H., Nagato Y. Rice plant development: from zygote to spikelet. Plant Cell Physiol. 2005;46:23–47. doi: 10.1093/pcp/pci501. [DOI] [PubMed] [Google Scholar]
- Jain M., Tyagi A.K., Khurana J.P. Genome-wide identification, classification, evolutionary expansion and expression analyses of homeobox genes in rice. FEBS J. 2008;275:2845–2861. doi: 10.1111/j.1742-4658.2008.06424.x. [DOI] [PubMed] [Google Scholar]
- Jang I.-C., Nahm B.H., Kim J.-K. Subcellular targeting of green fluorescent protein to plastids in transgenic rice plants provides a high-level expression system. Mol Breed. 1999;5:453–461. [Google Scholar]
- Jeong D.-H., Sung S.-K., An G. Molecular cloning and characterization of CONSTANS-like cDNA clones of the Fuji apple. J Plant Biol. 1999;42:23–31. [Google Scholar]
- Jin J., Tian F., Yang D.-C., Meng Y.-Q., Kong L., Luo J., Gao G. PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2016;45:D1040–D1045. doi: 10.1093/nar/gkw982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadota Y., Shirasu K., Zipfel C. Regulation of the NADPH oxidase RBOHD during plant immunity. Plant Cell Physiol. 2015;56:1472–1480. doi: 10.1093/pcp/pcv063. [DOI] [PubMed] [Google Scholar]
- Kim S.L., Lee S., Kim H.J., Nam H.G., An G. OsMADS51 is a short-day flowering promoter that functions upstream of Ehd1, OsMADS14, and Hd3a. Plant Physiol. 2007;145:1484–1494. doi: 10.1104/pp.107.103291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.-J., Lee T.-H., Pahk Y.-M., Kim Y.-H., Park H.-M., Do Choi Y., Nahm B.H., Kim Y.-K. Quadruple 9-mer-based protein binding microarray with DsRed fusion protein. BMC Mol Biol. 2009;10:91. doi: 10.1186/1471-2199-10-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Hwang H., Hong J.-W., Lee Y.-N., Ahn I.P., Yoon I.S., Yoo S.-D., Lee S., Lee S.C., Kim B.-G. A rice orthologue of the ABA receptor, OsPYL/RCAR5, is a positive regulator of the ABA signal transduction pathway in seed germination and early seedling growth. J Exp Bot. 2011;63:1013–1024. doi: 10.1093/jxb/err338. [DOI] [PubMed] [Google Scholar]
- Kim J.S., Kim J., Lee T.-H., Jun K.M., Kim T.H., Kim Y.-H., Park H.-M., Jeon J.-S., An G., Yoon U.-H. FSTVAL: a new web tool to validate bulk flanking sequence tags. Plant Methods. 2012;8:19. doi: 10.1186/1746-4811-8-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight H., Knight M.R. Abiotic stress signalling pathways: specificity and cross-talk. Trends Plant Sci. 2001;6:262–267. doi: 10.1016/s1360-1385(01)01946-x. [DOI] [PubMed] [Google Scholar]
- Koo B.-H., Yoo S.-C., Park J.-W., Kwon C.-T., Lee B.-D., An G., Zhang Z., Li J., Li Z., Paek N.-C. Natural variation in OsPRR37 regulates heading date and contributes to rice cultivation at a wide range of latitudes. Mol Plant. 2013;6:1877–1888. doi: 10.1093/mp/sst088. [DOI] [PubMed] [Google Scholar]
- Koornneef M. Arabidopsis. Cold Spring Harbor Laboratory Press; 1994. Arabidopsis genetics; pp. 89–120. [Google Scholar]
- Kotchoni S.O., Gachomo E.W. The reactive oxygen species network pathways: an essential prerequisite for perception of pathogen attack and the acquired disease resistance in plants. J Biosci. 2006;31:389–404. doi: 10.1007/BF02704112. [DOI] [PubMed] [Google Scholar]
- Kumar A., Mishra P., Kumari K., Panigrahi K. Environmental stress influencing plant development and flowering. Front Biosci. 2012;4:1315–1324. doi: 10.2741/s333. [DOI] [PubMed] [Google Scholar]
- Kwon C.-T., Koo B.-H., Kim D., Yoo S.-C., Paek N.-C. Casein kinases I and 2α phosphorylate Oryza sativa pseudo-response regulator 37 (OsPRR37) in photoperiodic flowering in rice. Mol Cells. 2015;38:81–88. doi: 10.14348/molcells.2015.2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb C., Dixon R.A. The oxidative burst in plant disease resistance. Ann Rev Plant Biol. 1997;48:251–275. doi: 10.1146/annurev.arplant.48.1.251. [DOI] [PubMed] [Google Scholar]
- Lee S., Jung K.-H., An G., Chung Y.-Y. Isolation and characterization of a rice cysteine protease gene, OsCP1, using T-DNA gene-trap system. Plant Mol Biol. 2004;54:755–765. doi: 10.1023/B:PLAN.0000040904.15329.29. [DOI] [PubMed] [Google Scholar]
- Lin H., Niu L., McHale N.A., Ohme-Takagi M., Mysore K.S., Tadege M. Evolutionarily conserved repressive activity of WOX proteins mediates leaf blade outgrowth and floral organ development in plants. Proc Natl Acad Sci USA. 2013;110:366–371. doi: 10.1073/pnas.1215376110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell J.T., Juenger T.E., Michaels S.D., Lasky J.R., Platt A., Richards J.H., Yu X., Easlon H.M., Sen S., McKay J.K. Pleiotropy of FRIGIDA enhances the potential for multivariate adaptation. Proc Biol Sci. 2013;280:20131043. doi: 10.1098/rspb.2013.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludlow M.M. Strategies of response to water stress. SPB Academic Publishers; 1989. pp. 269–281. [Google Scholar]
- Luo H., Song F., Zheng Z. Overexpression in transgenic tobacco reveals different roles for the rice homeodomain gene OsBIHD1 in biotic and abiotic stress responses. J Exp Bot. 2005;56:2673–2682. doi: 10.1093/jxb/eri260. [DOI] [PubMed] [Google Scholar]
- Lytovchenko A., Eickmeier I., Pons C., Osorio S., Szecowka M., Lehmberg K., Arrivault S., Tohge T., Pineda B., Anton M.T. Tomato fruit photosynthesis is seemingly unimportant in primary metabolism and ripening but plays a considerable role in seed development. Plant Physiol. 2011;157:1650–1663. doi: 10.1104/pp.111.186874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., Sukiran N.L., Ma H., Su Z. Moderate drought causes dramatic floral transcriptomic reprogramming to ensure successful reproductive development in Arabidopsis. BMC Plant Biol. 2014;14:164. doi: 10.1186/1471-2229-14-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Major D. Environmental effects on flowering. Hybridization of crop plants. 1980:1–15. [Google Scholar]
- Mandaokar A., Thines B., Shin B., Markus Lange B., Choi G., Koo Y.J., Yoo Y.J., Choi Y.D., Choi G., Browse J. Transcriptional regulators of stamen development in Arabidopsis identified by transcriptional profiling. Plant J. 2006;46:984–1008. doi: 10.1111/j.1365-313X.2006.02756.x. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A., Lu S., Anderson J.B., Chitsaz F., Derbyshire M.K., DeWeese-Scott C., Fong J.H., Geer L.Y., Geer R.C., Gonzales N.R. CDD: a conserved domain database for the functional annotation of proteins. Nucleic Acids Res. 2010;39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara K., Yamanouchi U., Nonoue Y., Sugimoto K., Wang Z.X., Minobe Y., Yano M. Ehd3, encoding a plant homeodomain finger-containing protein, is a critical promoter of rice flowering. Plant J. 2011;66:603–612. doi: 10.1111/j.1365-313X.2011.04517.x. [DOI] [PubMed] [Google Scholar]
- Mckay J.K., Richards J.H., Mitchell-Olds T. Genetics of drought adaptation in Arabidopsis thaliana: I. Pleiotropy contributes to genetic correlations among ecological traits. Mol Ecol. 2003;12:1137–1151. doi: 10.1046/j.1365-294x.2003.01833.x. [DOI] [PubMed] [Google Scholar]
- Minh-Thu P.-T., Hwang D.-J., Jeon J.-S., Nahm B.H., Kim Y.-K. Transcriptome analysis of leaf and root of rice seedling to acute dehydration. Rice. 2013;6:38. doi: 10.1186/1939-8433-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R., Vanderauwera S., Suzuki N., Miller G., Tognetti V.B., Vandepoele K., Gollery M., Shulaev V., Van Breusegem F. ROS signaling: the new wave? Trend Plant Sci. 2011;16:300–309. doi: 10.1016/j.tplants.2011.03.007. [DOI] [PubMed] [Google Scholar]
- Mundy J., Chua N.-H. Abscisic acid and water-stress induce the expression of a novel rice gene. EMBO J. 1988;7:2279–2286. doi: 10.1002/j.1460-2075.1988.tb03070.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K., Tran L.S.P., Van Nguyen D., Fujita M., Maruyama K., Todaka D., Ito Y., Hayashi N., Shinozaki K., Yamaguchi-Shinozaki K. Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J. 2007;51:617–630. doi: 10.1111/j.1365-313X.2007.03168.x. [DOI] [PubMed] [Google Scholar]
- Nardmann J., Reisewitz P., Werr W. Discrete shoot and root stem cell-promoting WUS/WOX5 functions are an evolutionary innovation of angiosperms. Mol Biol Evol. 2009;26:1745–1755. doi: 10.1093/molbev/msp084. [DOI] [PubMed] [Google Scholar]
- Nicholas K.B. GeneDoc: analysis and visualization of genetic variation. Embnew news. 1997;4:14. [Google Scholar]
- Noctor G., Mhamdi A., Foyer C.H. The roles of reactive oxygen metabolism in drought: not so cut and dried. Plant Physiol. 2014;164:1636–1648. doi: 10.1104/pp.113.233478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S.-J., Song S.I., Kim Y.S., Jang H.-J., Kim S.Y., Kim M., Kim Y.-K., Nahm B.H., Kim J.-K. Arabidopsis CBF3/DREB1A and ABF3 in transgenic rice increased tolerance to abiotic stress without stunting growth. Plant Physiol. 2005;138:341–351. doi: 10.1104/pp.104.059147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveros J.C. VENNY. An interactive tool for comparing lists with Venn Diagrams. 2007 http://bioinfogp.cnb.csic.es/tools/venny/index.html.
- Orozco-Cardenas M., Ryan C.A. Hydrogen peroxide is generated systemically in plant leaves by wounding and systemin via the octadecanoid pathway. Proc Natl Acad Sci USA. 1999;96:6553–6557. doi: 10.1073/pnas.96.11.6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osakabe Y., Osakabe K., Shinozaki K., Tran L.-S.P. Response of plants to water stress. Front Plant Scie. 2014;5:86. doi: 10.3389/fpls.2014.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang S.Q., Liu Y.F., Liu P., Lei G., He S.J., Ma B., Zhang W.K., Zhang J.S., Chen S.Y. Receptor-like kinase OsSIK1 improves drought and salt stress tolerance in rice (Oryza sativa) plants. Plant J. 2010;62:316–329. doi: 10.1111/j.1365-313X.2010.04146.x. [DOI] [PubMed] [Google Scholar]
- Pelaz S., Ditta G.S., Baumann E., Wisman E., Yanofsky M.F. B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature. 2000;405:200. doi: 10.1038/35012103. [DOI] [PubMed] [Google Scholar]
- Peltzer D., Dreyer E., Polle A. Differential temperature dependencies of antioxidative enzymes in two contrasting species: Fagus sylvatica and Coleus blumei. Plant Physiol Biochem. 2002;40:141–150. [Google Scholar]
- Peng L.-T., Shi Z.-Y., Li L., Shen G.-Z., Zhang J.-L. Ectopic expression of OsLFL1 in rice represses Ehd1 by binding on its promoter. Biochem Biophys Res Commun. 2007;360:251–256. doi: 10.1016/j.bbrc.2007.06.041. [DOI] [PubMed] [Google Scholar]
- Petrov V.D., Van Breusegem F. Hydrogen peroxide—a central hub for information flow in plant cells. AoB plants. 2012;2012 doi: 10.1093/aobpla/pls014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy A.R., Ramakrishna W., Sekhar A.C., Ithal N., Babu P.R., Bonaldo M., Soares M., Bennetzen J.L. Novel genes are enriched in normalized cDNA libraries from drought-stressed seedlings of rice (Oryza sativa L. subsp indica cv Nagina 22) Genome. 2002;45:204–211. doi: 10.1139/g01-114. [DOI] [PubMed] [Google Scholar]
- Riboni M., Galbiati M., Tonelli C., Conti L. GIGANTEA enables drought escape response via abscisic acid-dependent activation of the florigens and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1. Plant Physiol. 2013;162:1706–1719. doi: 10.1104/pp.113.217729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio S., Rodrigues A., Saez A., Dizon M.B., Galle A., Kim T.-H., Santiago J., Flexas J., Schroeder J.I., Rodriguez P.L. Triple loss of function of protein phosphatases type 2C leads to partial constitutive response to endogenous abscisic acid. Plant Physiol. 2009;150:1345–1355. doi: 10.1104/pp.109.137174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Santino C.G., Stanford G.L., Conner T.W. Developmental and transgenic analysis of two tomato fruit enhanced genes. Plant Mol Biol. 1997;33:405–416. doi: 10.1023/a:1005738910743. [DOI] [PubMed] [Google Scholar]
- Searle I., He Y., Turck F., Vincent C., Fornara F., Kröber S., Amasino R.A., Coupland G. The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev. 2006;20:898–912. doi: 10.1101/gad.373506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma R., De Vleesschauwer D., Sharma M.K., Ronald P.C. Recent advances in dissecting stress-regulatory crosstalk in rice. Mol Plant. 2013;6:250–260. doi: 10.1093/mp/sss147. [DOI] [PubMed] [Google Scholar]
- Shimizu R., Ji J., Kelsey E., Ohtsu K., Schnable P.S., Scanlon M.J. Tissue specificity and evolution of meristematic WOX3 function. Plant Physiol. 2009;149:841–850. doi: 10.1104/pp.108.130765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K., Yamaguchi-Shinozaki K. Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signaling pathways. Curr Opin Plant Biol. 2000;3:217–223. [PubMed] [Google Scholar]
- Smyth G.K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:1–25. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- Song S., Qi T., Huang H., Ren Q., Wu D., Chang C., Peng W., Liu Y., Peng J., Xie D. The jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect jasmonate-regulated stamen development in Arabidopsis. Plant Cell. 2011;23:1000–1013. doi: 10.1105/tpc.111.083089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek F., Bošnjak M., Škunca N., Šmuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PloS one. 2011;6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaki S., Matsuo S., Wong H.L., Yokoi S., Shimamoto K. Hd3a protein is a mobile flowering signal in rice. Science. 2007;316:1033–1036. doi: 10.1126/science.1141753. [DOI] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tian X.H., Li X.P., Zhou H.L., Zhang J.S., Gong Z.Z., Chen S.Y. OsDREB4 genes in rice encode AP2-containing proteins that bind specifically to the dehydration-responsive element. J Int Plant Biol. 2005;47:467–476. [Google Scholar]
- Todaka D., Shinozaki K., Yamaguchi-Shinozaki K. Recent advances in the dissection of drought-stress regulatory networks and strategies for development of drought-tolerant transgenic rice plants. Front Plant Sci. 2015;6:84. doi: 10.3389/fpls.2015.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres M.A., Dangl J.L. Functions of the respiratory burst oxidase in biotic interactions, abiotic stress and development. Curr Opin Plant Biol. 2005;8:397–403. doi: 10.1016/j.pbi.2005.05.014. [DOI] [PubMed] [Google Scholar]
- Tsuchiya T., Toriyama K., Nasrallah M.E., Ejiri S.-i. Isolation of genes abundantly expressed in rice anthers at the microspore stage. Plant Mol Biol. 1992;20:1189–1193. doi: 10.1007/BF00028907. [DOI] [PubMed] [Google Scholar]
- Tsuji H., Taoka K.-i., Shimamoto K. Florigen in rice: complex gene network for florigen transcription, florigen activation complex, and multiple functions. Curr Opin Plant Biol. 2013;16:228–235. doi: 10.1016/j.pbi.2013.01.005. [DOI] [PubMed] [Google Scholar]
- Valverde F., Mouradov A., Soppe W., Ravenscroft D., Samach A., Coupland G. Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science. 2004;303:1003–1006. doi: 10.1126/science.1091761. [DOI] [PubMed] [Google Scholar]
- van der Graaff E., Laux T., Rensing S.A. The WUS homeobox-containing (WOX) protein family. Genome Biol. 2009;10:248. doi: 10.1186/gb-2009-10-12-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voesenek L., Bailey-Serres J. Flooding tolerance: O2 sensing and survival strategies. Curr Opin Plant Biol. 2013;16:647–653. doi: 10.1016/j.pbi.2013.06.008. [DOI] [PubMed] [Google Scholar]
- Wada K.C., Kondo H., Takeno K. Obligatory short-day plant, Perilla frutescens var. crispa can flower in response to low-intensity light stress under long-day conditions. Physiol Plant. 2010;138:339–345. doi: 10.1111/j.1399-3054.2009.01337.x. [DOI] [PubMed] [Google Scholar]
- Wang Q., Guan Y., Wu Y., Chen H., Chen F., Chu C. Overexpression of a rice OsDREB1F gene increases salt, drought, and low temperature tolerance in both Arabidopsis and rice. Plant Mol Biol. 2008;67:589–602. doi: 10.1007/s11103-008-9340-6. [DOI] [PubMed] [Google Scholar]
- Wei X., Xu J., Guo H., Jiang L., Chen S., Yu C., Zhou Z., Hu P., Zhai H., Wan J. DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously. Plant Physiol. 2010;153:1747–1758. doi: 10.1104/pp.110.156943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wopereis M., Kropff M., Maligaya A., Tuong T. Drought-stress responses of two lowland rice cultivars to soil water status. Field Crops Res. 1996;46:21–39. [Google Scholar]
- Xiao J., Cheng H., Li X., Xiao J., Xu C., Wang S. Rice WRKY13 regulates cross talk between abiotic and biotic stress signaling pathways by selective binding to different cis-elements. Plant Physiol. 2013;163:1868–1882. doi: 10.1104/pp.113.226019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Lafitte H., Gao Y., Fu B., Torres R., Li Z. QTLs for drought escape and tolerance identified in a set of random introgression lines of rice. Theoretical Appl Genet. 2005;111:1642–1650. doi: 10.1007/s00122-005-0099-8. [DOI] [PubMed] [Google Scholar]
- Xu K., Xu X., Fukao T., Canlas P., Maghirang-Rodriguez R., Heuer S., Ismail A.M., Bailey-Serres J., Ronald P.C., Mackill D.J. Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature. 2006;442:705. doi: 10.1038/nature04920. [DOI] [PubMed] [Google Scholar]
- Xue W., Xing Y., Weng X., Zhao Y., Tang W., Wang L., Zhou H., Yu S., Xu C., Li X. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet. 2008;40:761. doi: 10.1038/ng.143. [DOI] [PubMed] [Google Scholar]
- Yu L., Chen X., Wang Z., Wang S., Wang Y., Zhu Q., Li S., Xiang C. Arabidopsis enhanced drought tolerance1/HOMEODOMAIN GLABROUS11 confers drought tolerance in transgenic rice without yield penalty. Plant Physiol. 2013;162:1378–1391. doi: 10.1104/pp.113.217596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun D., Liang W., Dreni L., Yin C., Zhou Z., Kater M.M., Zhang D. OsMADS16 genetically interacts with OsMADS3 and OsMADS58 in specifying floral patterning in rice. Mol Plant. 2013;6:743–756. doi: 10.1093/mp/sst003. [DOI] [PubMed] [Google Scholar]
- Zhang D., Liang W., Yin C., Zong J., Gu F., Zhang D. OsC6, encoding a lipid transfer protein, is required for postmeiotic anther development in rice. Plant Physiol. 2010a;154:149–162. doi: 10.1104/pp.110.158865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Zong J., Liu J., Yin J., Zhang D. Genome-wide analysis of WOX gene family in rice, sorghum, maize, Arabidopsis and poplar. J Int Plant Biol. 2010b;52:1016–1026. doi: 10.1111/j.1744-7909.2010.00982.x. [DOI] [PubMed] [Google Scholar]
- Zhang C., Liu J., Zhao T., Gomez A., Li C., Yu C., Li H., Lin J., Yang Y., Liu B. A drought-inducible bZIP transcription factor OsABF1 delays reproductive timing in rice. Plant Physiol. 2016;171:334–343. doi: 10.1104/pp.16.01691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S., Jiang W., Long F., Cheng S., Yang W., Zhao Y., Zhou D.-X. Rice homeodomain protein WOX11 recruits a histone acetyltransferase complex to establish programs of cell proliferation of crown root meristem. Plant Cell. 2017;29:1088–1104. doi: 10.1105/tpc.16.00908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuckerkandl E., Pauling L. Evolving genes and proteins. Elsevier; 1965. Evolutionary divergence and convergence in proteins; pp. 97–166. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


