β-Lactam-resistant Haemophilus influenzae is a clinical concern. A high prevalence (>40%) of β-lactamase-negative high-level ampicillin-resistant H. influenzae (high-BLNAR) isolates in Japan has been reported.
KEYWORDS: antimicrobial resistance, penicillin-binding proteins, Haemophilus influenzae
ABSTRACT
β-Lactam-resistant Haemophilus influenzae is a clinical concern. A high prevalence (>40%) of β-lactamase-negative high-level ampicillin-resistant H. influenzae (high-BLNAR) isolates in Japan has been reported. However, the reasons for the expansion are unknown. High-BLNAR strains possess an amino acid substitution, either Asn526Lys (group III) or Arg517His (group III-like) in addition to Ser385Thr, in penicillin-binding protein 3 (PBP3). To determine the current prevalence of high-BLNAR strains and the mechanisms behind their expansion in Japan, their prevalence, PBP3 types, multilocus sequence types, and susceptibilities to quinolones approved in Japan as alternatives were determined. Sixty percent of H. influenzae clinical isolates (62/104 isolates) were β-lactamase-negative ampicillin-resistant H. influenzae (BLNAR) strains. Among BLNAR isolates, 92% (57/62 isolates) were high-BLNAR strains. Most isolates were classified as belonging to group III, which contained many genotypes (11 PBP3 types and 25 sequence types). These results indicated that the expansion of high-BLNAR isolates was multiclonal and such strains are still predominant in Japanese clinical settings. One high-BLNAR isolate harbored the novel amino acid substitution Asn526Met in addition to Ser385Thr in PBP3, suggesting a new group (group IV). No quinolone-resistant H. influenzae isolates were identified. The MICs for the quinolones (moxifloxacin, garenoxacin, and tosufloxacin) were similar to that for levofloxacin, whereas sitafloxacin exhibited a lower MIC. However, we obtained 4 H. influenzae isolates with decreased quinolone susceptibility with the amino acid substitution Ser84Leu in GyrA, and 3 of those isolates were high-BLNAR isolates. In summary, this study shows that multiclonal high-BLNAR strains predominate in a Japanese university hospital. Isolates remain sensitive to quinolones, but vigilance is required to prevent the development of fluoroquinolone resistance in high-BLNAR strains.
INTRODUCTION
Haemophilus influenzae is a causal pathogen of community-acquired infections such as pneumonia, otitis media, sinusitis, and meningitis (1, 2). Recent 16S rRNA-targeted analysis of the microbiota of bronchoalveolar lavage fluid samples from patients with community-acquired pneumonia revealed that H. influenzae was the most prevalent bacterium detected, along with Streptococcus pneumoniae (3).
The increasing prevalence of β-lactam-resistant H. influenzae is a clinical concern worldwide. Two mechanisms are involved in the acquisition of β-lactam resistance in H. influenzae. One mechanism is the acquisition of β-lactamase genes such as blaTEM-1 and blaROB-1, and such strains are referred to as β-lactamase-positive ampicillin-resistant H. influenzae (BLPAR) strains (4, 5). BLPAR isolates with blaTEM-1 are predominant in Japan (6). The other mechanism is a mutation in ftsI, which encodes penicillin-binding protein 3 (PBP3), and strains are called β-lactamase-negative ampicillin-resistant H. influenzae (BLNAR) strains. Currently, BLNAR strains are more prevalent than BLPAR strains in many countries (6–11).
BLNAR strains are further classified into 4 groups, I, II, III, and III-like, as defined by the nature of amino acid substitutions in PBP3 (7, 12, 13). Isolates harboring the amino acid substitutions Arg517His and Asn526Lys near the conserved Lys-Thr-Gly (KTG) motif of PBP3 are classified as group I and group II, respectively. These are called β-lactamase-negative low-level ampicillin-resistant H. influenzae (low-BLNAR) isolates and exhibit decreased susceptibility to ampicillin, for which the MIC is typically 1 mg/liter, compared with β-lactamase-negative ampicillin-susceptible (BLNAS) isolates, with an ampicillin MIC of 0.25 mg/liter (12).
Isolates harboring amino acid substitutions near the Ser-Ser-Asn (SSN) motif, such as Ser385Thr, Ser357Asn, Met377Ile, and Leu389Phe, in addition to the amino substitution Asn526Lys, are classified as group III (12). Garcia-Cobos et al. reported a subgroup of group III, designated group III-like (7). Group III-like isolates possess the amino acid substitution Arg517His instead of the Asn526Lys substitution in group III (7). Group III and group III-like isolates are called β-lactamase-negative high-level ampicillin-resistant H. influenzae (high-BLNAR) strains and exhibit slightly higher ampicillin MICs (2 mg/liter) and >2-fold higher MICs for some cephems, such as cefuroxime, cefotaxime, and cefpodoxime, compared to low-BLNAR strains (7, 12).
A recent study indicated that BLNAR clinical isolates are largely classified as group III or group III-like through acquisition of Ar526His or Asn517His with Ser385Thr, respectively, because these substitutions are the main contributors of β-lactam resistance (14). In addition, BLNAR isolates with β-lactamase genes are referred to as β-lactamase-producing amoxicillin-clavulanate-resistant H. influenzae (BLPACR) strains (15–17).
In the past, low-BLNAR isolates were clinically predominant (10, 11, 15, 18, 19). However, high-BLNAR isolates have emerged since 1998 (19). Whereas low-BLNAR isolates constitute the majority of BLNAR strains in the United States and many European countries (7, 9–11, 14, 15), high-BLNAR isolates have significantly increased in prevalence in Japan and South Korea since 2005 (6, 8, 13, 20–23). According to the latest 2011 data in Japan, the prevalence of high-BLNAR isolates was 42.6% of H. influenzae isolates overall and 91.2% of all BLNAR isolates (22). These results reflect a serious issue in Japanese clinics with respect to the use of β-lactam antimicrobials for the treatment of H. influenzae infections. However, the situation with respect to the expansion of high-BLNAR strains in Japanese clinical settings has not been evaluated since 2011 (22), and the reasons for the high prevalence of high-BLNAR strains in Japan remain unknown.
Macrolides and quinolones (especially fluoroquinolones such as ciprofloxacin and levofloxacin) are used as alternative antimicrobials for the treatment of β-lactam-resistant H. influenzae infections. In addition to these agents, quinolones such as moxifloxacin and garenoxacin have been approved for treatment of respiratory infections and tosufloxacin for pediatric use in Japan. Furthermore, sitafloxacin is an approved quinolone in Japan for the treatment of severe bacterial infections, because it has greater antimicrobial activity against various bacteria than do other quinolones (24). It is important to understand the susceptibilities to the series of quinolones when making a secondary choice of antimicrobials for the treatment of infections caused by high-BLNAR isolates. In this study, we collected H. influenzae clinical isolates in a Japanese university hospital from 2016 to 2018 and investigated (i) the current prevalence of high-BLNAR strains, (ii) the clonality of high-BLNAR strains by genotypic analysis, and (iii) the susceptibility to various quinolones, to reveal the effectiveness of secondary-choice antimicrobials for treatment of high-BLNAR infections.
RESULTS
β-Lactam susceptibilities, PBP3 genogroups, and possession of β-lactamase genes.
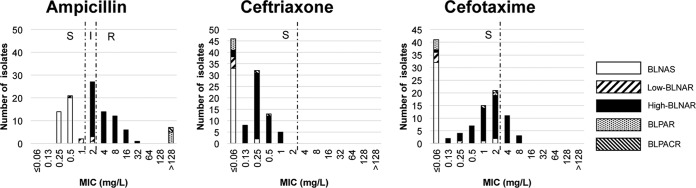
In total, 104 H. influenzae clinical isolates were obtained, all of which were nontypeable by serotyping. The ampicillin MIC50 and MIC90 values were 2 and 8 mg/liter, respectively, and the prevalence of ampicillin-resistant isolates was 37.5% (Fig. 1). The ceftriaxone MIC50 and MIC90 values were 0.13 and 0.5 mg/liter, respectively, and ceftriaxone-nonsusceptible isolates were not observed. The cefotaxime MIC50 and MIC90 values were 0.5 and 4 mg/liter, respectively, and the prevalence of cefotaxime-nonsusceptible isolates was 13.5%.
FIG 1.
Susceptibilities to β-lactams and the genetic types of β-lactam resistance in H. influenzae clinical isolates. S, susceptible; I, intermediate; R, resistant.
The 104 isolates were classified by BLNAS/BLNAR genotype based on amino acid substitutions in PBP3 (Table 1). Sixty-two H. influenzae isolates (59.6%) were identified as being BLNAR because they possessed either Arg517His or Asn526Lys in PBP3. There were no significant differences in data on patient gender, patient age, and specimen type between sources of BLNAS and BLNAR isolates (see Table S1 in the supplemental material).
TABLE 1.
Classification of BLNAS, low-BLNAR, and high-BLNAR isolates and their susceptibilities to β-lactamsa
| Genotype | Group | No. of strains | MIC (mg/liter) |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ampicillin |
Ceftriaxone |
Cefotaxime |
||||||||||||
| Geometric mean | MIC50 | MIC90 | Range | Geometric mean | MIC50 | MIC90 | Range | Geometric mean | MIC50 | MIC90 | Range | |||
| BLNAS | 35 | 0.4 | 0.5 | 0.5 | 0.35–0.5 | ≤0.06 | ≤0.06 | ≤0.06 | ≤0.06–0.25 | ≤0.06 | ≤0.06 | ≤0.06 | ≤0.06–2 | |
| Low-BLNAR | II | 5 | 1.15 | 1 | 2 | 0.5–2 | ≤0.06 | ≤0.06 | ≤0.06 | ≤0.06 | ≤0.06 | ≤0.06 | ≤0.06 | ≤0.06–0.25 |
| High-BLNAR | III | 47 | 3.77 | 4 | 8 | 2–16 | 0.28 | 0.25 | 1 | ≤0.06–1 | 1.34 | 2 | 4 | ≤0.06–8 |
| High-BLNAR | III-like | 9 | 5.44 | 8 | 16 | 2–16 | 0.16 | 0.25 | 0.25 | ≤0.06–0.5 | 2.59 | 4 | 8 | 0.25–8 |
Five BLPAR isolates and 2 BLPACR isolates possessed blaTEM-1 and were excluded from this table. The PBP3 type of the BLPACR isolates was group III. The high-BLNAR isolate SMHi90 was excluded because of its unique ftsI sequence (see Fig. S1 and S2 in the supplemental material).
The BLNAR isolates were classified into 4 groups (groups I, II, III, and III-like) on the basis of the presence of the amino acid substitutions Arg517His, Asn526Lys, and Ser385Thr in PBP3 (Table 2). Five isolates (4.8% of the total H. influenzae isolates) were low-BLNAR strains, and they belonged to group II, possessing Asn526Lys. There were no isolates belonging to group I, possessing Arg517His. In contrast, 57 isolates (54.8% of all H. influenzae isolates and 91.9% of BLNAR isolates) were high-BLNAR strains (Table 1). The high-BLNAR isolates were further divided into group III, possessing Ser385Thr and Asn526Lys, and group III-like, possessing Ser385Thr and Asn517His. Group III accounted for 45.2% of all H. influenzae isolates and 75.8% of BLNAR isolates, and group III-like accounted for 8.7% of all H. influenzae isolates and 14.5% of BLNAR isolates. One of the high-BLNAR isolates, SMHi90, was not classified into either group because it harbored Asn526Met, in addition to Ser385, in PBP3, resulting from a TGA insertion between nucleotide positions 1576 and 1578 of ftsI (Fig. S1); it exhibited a higher ampicillin MIC and similar cephalosporin MICs, compared to group III and group III-like isolates (Table 3).
TABLE 2.
Amino acid substitutions in PBP3 in BLNAR and BLPACR isolatesa
| Genotype | Group | No. of strains | MIC (mg/liter) |
Amino acid substitution(s) in PBP3 |
||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMP | CTR | CTX | D350 | S357 | M377 | S385 | L389 | V436 | A437 | A502 | R517 | T532 | N526 | A530 | V547 | A554 | G555 | Y557 | V562 | N569 | A586 | S594 | A595 | I601 | E603 | |||
| Low-BLNAR (n = 5) | II | 1 | 1 | ≤0.06 | ≤0.06 | K | ||||||||||||||||||||||
| 1 | 2 | ≤0.06 | 0.25 | K | S | I | L | G | ||||||||||||||||||||
| 1 | 2 | ≤0.06 | ≤0.06 | N | I | V | K | I | S | S | T | T | ||||||||||||||||
| 1 | 1 | ≤0.06 | ≤0.06 | N | I | V | K | I | L | S | S | T | T | |||||||||||||||
| 1 | 0.5 | ≤0.06 | ≤0.06 | N | S | V | K | I | L | S | S | T | T | D | ||||||||||||||
| High-BLNAR (n = 47) | III | 19 | 3.2b | 0.4b | 1.8b | N | N | I | T | F | K | I | L | S | ||||||||||||||
| 15 | 5b | 0.4b | 2.4b | N | N | I | T | F | K | I | L | S | S | T | T | D | ||||||||||||
| 3 | 3.2b | 0.1b | 0.5b | N | N | I | T | F | V | K | ||||||||||||||||||
| 3 | 4b | 0.2b | 0.6b | N | N | I | T | F | K | S | ||||||||||||||||||
| 1 | 2 | 0.25 | 0.5 | N | N | I | T | K | I | L | S | |||||||||||||||||
| 1 | 4 | 0.13 | 0.5 | N | N | I | T | F | T | K | I | S | ||||||||||||||||
| 1 | 2 | 0.25 | 2 | N | N | I | T | F | K | I | L | G | ||||||||||||||||
| 1 | 8 | 1 | 2 | N | N | I | T | F | A | K | I | L | S | |||||||||||||||
| 1 | 8 | 0.25 | 1 | N | N | I | T | F | K | I | L | S | D | |||||||||||||||
| 1 | 2 | ≤0.06 | 0.13 | N | N | I | T | F | V | K | I | S | D | |||||||||||||||
| 1 | 2 | 0.13 | 0.25 | N | N | T | V | K | I | S | S | T | T | D | ||||||||||||||
| High-BLNAR (n = 9) | III-like | 1 | 2 | ≤0.06 | 0.25 | N | N | I | T | H | S | I | L | N | ||||||||||||||
| 4 | 5.7b | 0.3b | 5.7b | N | N | I | T | F | H | S | I | H | S | |||||||||||||||
| 1 | 2 | 0.13 | 0.5 | N | N | T | H | I | E | H | S | S | T | T | ||||||||||||||
| 1 | 16 | 0.25 | 2 | N | N | I | T | F | H | I | T | H | L | D | ||||||||||||||
| 1 | 16 | 0.5 | 2 | N | N | I | T | F | H | I | E | H | S | S | T | T | ||||||||||||
| 1 | 4 | 0.25 | 1 | N | N | I | T | F | H | I | E | H | S | S | T | T | D | |||||||||||
| BLPACR (n = 2) | III | 2 | >128 | 0.25 or 0.5 | 1 or 2 | N | N | I | T | F | K | I | L | S | ||||||||||||||
Amino acid substitutions in the region from D350 to E603 in PBP3 were examined. Bold letters indicate amino acid substitutions classifying low- and high-BLNAR isolates and their groups. Amino acids at positions 379 to 381 and 512 to 514 were the Ser-Ser-Asn (SSN) motif and the Lys-Thr-Gly (KTG) motif, respectively. The high-BLNAR isolate SMHi90 was excluded because of its unique ftsI sequence (see Fig. S1 and S2). AMP, ampicillin; CTR, ceftriaxone; CTX, cefotaxime.
Geometric mean.
TABLE 3.
Characteristics of isolates with decreased susceptibility to quinolonesa
| Isolate | Genotype | Group | ST | Patient age (yr) | Patient sex | Specimen type | MIC (mg/liter) |
Mutation(s) in QRDR |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMP | CTR | CTX | LVX | MFX | GNX | TFX | SFX | GyrA | ParC | |||||||
| SMHi67 | BLNAS | ND | 66 | M | Sputum | 0.5 | 0.25 | 1 | 0.5 | 1 | 0.5 | 0.5 | 0.03 | Ser84Leu | Asn138Ser, Ser230Ala | |
| SMHi9 | High-BLNAR | III | 156 | 69 | M | Sputum | 2 | 0.25 | 1 | 0.13 | 0.25 | 0.13 | 0.13 | 0.03 | Ser84Leu, Glu142Lys | |
| SMHi18 | High-BLNAR | III | 1218 | 5 | M | Pharyngeal mucus | 8 | 0.5 | 1 | 0.13 | 0.25 | 0.25 | 0.13 | 0.03 | Ser84Leu | |
| SMHi90 | High-BLNAR | IV | 57 | 64 | F | Sputum | 32 | 0.25 | 2 | 0.13 | 0.25 | 0.25 | 0.13 | 0.015 | Ser84Leu | |
No amino acid substitutions in the QRDRs of GyrB and ParE were observed. M, male; F, female; AMP, ampicillin; CTR, ceftriaxone; CTX, cefotaxime; LVX, levofloxacin; MFX, moxifloxacin; GNX, garenoxacin; TFX, tosufloxacin; SFX, sitafloxacin; ND, not determined because of the failure of fucK amplification by PCR.
The associations between ampicillin, ceftriaxone, and cefotaxime susceptibilities and PBP3 genogroups were evaluated (Table 1 and Fig. 1). BLNAS isolates exhibited the lowest values for all parameters (MIC geometric mean, MIC50, MIC90, and MIC range) in the presence of the three antibiotics. The MIC parameters increased in the order of low-BLNAR isolates and then high-BLNAR isolates. The MICs of ampicillin and cefotaxime, but not ceftriaxone, for group III-like isolates were slightly higher than those for group III isolates. Isolate SMHi90, which harbored Asn526Met, exhibited MICs of 32, 0.25, and 2 mg/liter for ampicillin, ceftriaxone, and cefotaxime, respectively.
The gene blaTEM-1 was detected in 7 isolates (5 BLPAR isolates and 2 BLPACR isolates), and their ampicillin MICs were >128 mg/liter. BLPAR isolates exhibited MICs of ≤0.06 mg/liter and ≤0.06 or 2 mg/liter for ceftriaxone and cefotaxime, respectively, while BLPACR isolates exhibited corresponding values of 0.25 or 0.5 mg/liter and 2 or 1 mg/liter. None of the isolates possessed blaROB-1.
Amino acid sequence typing of PBP3, multilocus sequence typing, and phylogenetic analysis.
BLNAR isolates were classified in detail by PBP3 typing based on the partial amino acid sequences of the main region of PBP3, Asp350 to Glu611 (Table 2). Isolates of group II were divided into 5 PBP3 types. Isolates of groups III and III-like were divided into 11 and 6 PBP3 types, respectively. The high-BLNAR isolate SMHi90 was not classified into any genotype because it was found to possess multiple unique amino acid substitutions in PBP3, including Asn526Met, compared with other BLNAR isolates (Fig. S1A).
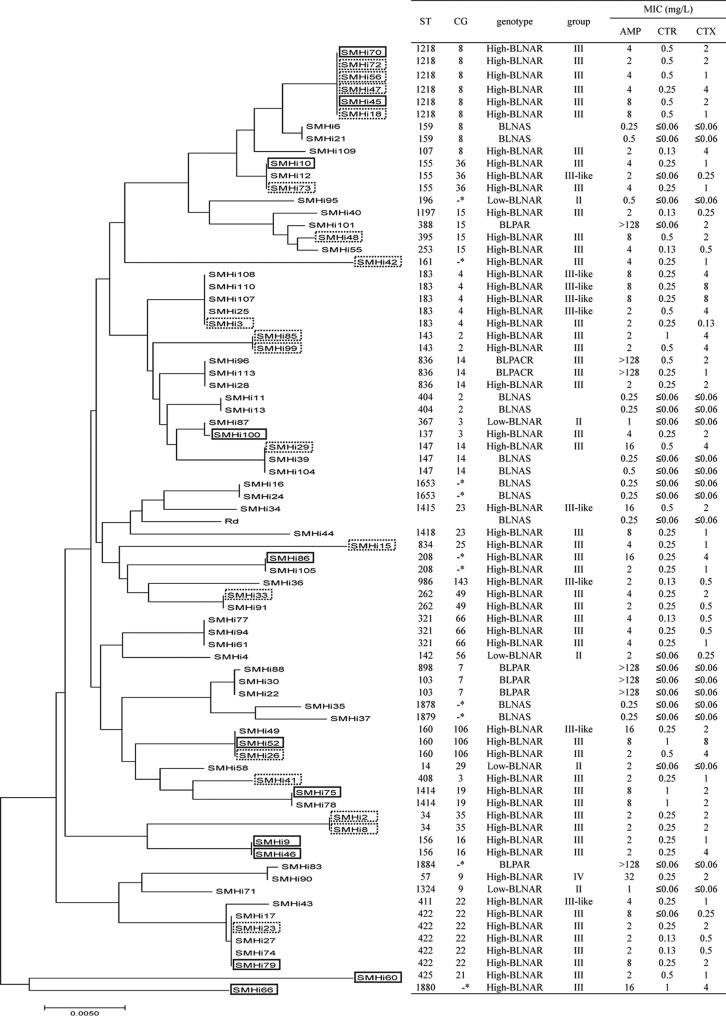
Multilocus sequence typing (MLST) analysis was performed for all BLNAR, BLPAR, and BLPACR isolates and 11 randomly selected BLNAS isolates. Four isolates could not be typed, because their fucK genes could not be amplified using the specific primer pair. The most prevalent sequence type (ST) was ST1218 (6 isolates), followed by ST422 and ST183 (5 isolates each). We observed 4 new STs, in 2 BLNAS isolates (ST1878 and ST1879), 1 high-BLNAR isolate (ST1880), and 1 BLPAR isolate (ST1881).
Isolates of the same ST mostly belonged to the same PBP3 genogroup. However, some STs (ST155, ST160, and ST183) contained both group III and group III-like isolates. ST147 contained a high-BLNAR isolate and 2 BLNAS isolates. Low- and high-BLNAR isolates were clustered into 34 STs and high-BLNAR isolates into 20 STs. We created a phylogenetic tree based on MLST data for 76 isolates, including BLNAR, BLPAR, and BLPACR isolates and 10 randomly selected BLNAS isolates, and the isolates grouped into many clusters (Fig. 2). A phylogenetic tree based on ftsI sequences (Asp350 to Asn611) of 80 isolates, including all BLNAR, BLPAR, and BLPACR isolates and 11 randomly selected BLNAS isolates, was also created. Most of the group III isolates belonged to 4 clusters (clusters A, B, C, and F), and group III-like isolates belonged to a single cluster (cluster D). Low-BLNAR, BLNAS, and BLPAR isolates constituted another cluster (cluster E) (Fig. S2).
FIG 2.
Phylogenetic tree based on MLST of H. influenzae clinical isolates. A neighbor-joining dendrogram of BLNAS (n = 10), low-BLNAR (n = 5), high-BLNAR (n = 54), BLPAR (n = 5), and BLPACR (n = 2) isolates investigated in this study was constructed. Four isolates (SMHi5, SMHi67, SMHi97, and SMHi102) were excluded because fucK was not amplified by PCR. Dotted rectangles indicate the most common type of amino acid substitution in group III, and solid rectangles indicate the second most common type. Asterisks indicate singletons.
Clarithromycin and quinolone susceptibilities and amino acid substitutions in quinolone-resistance-determining regions of GyrA, GyrB, ParC, and ParE.
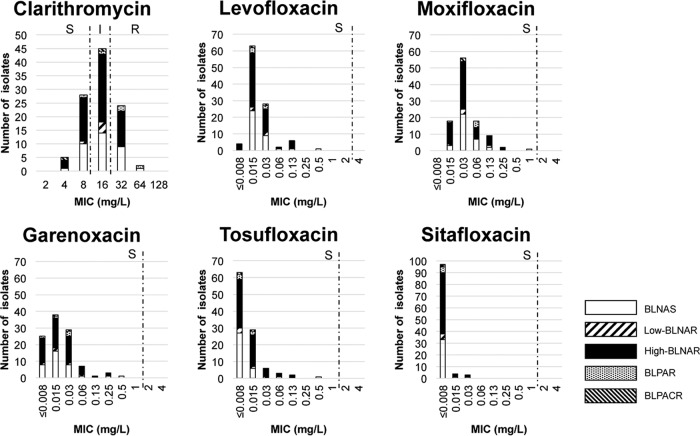
Susceptibility to clarithromycin and various quinolones was examined for the H. influenzae clinical isolates (Fig. 3). For clarithromycin, the MIC50 and MIC90 values were 16 and 32 mg/liter, respectively; 45 isolates (43.3%) were of intermediate resistance, and 26 isolates (25.0%) were resistant. For the quinolones, the MIC50 values were 0.015, 0.03, 0.015, ≤0.008, and ≤0.008 mg/liter and the MIC90 values were 0.03, 0.13, 0.06, 0.03, and ≤0.008 mg/liter for levofloxacin, moxifloxacin, garenoxacin, tosufloxacin, and sitafloxacin, respectively. No quinolone-nonsusceptible isolates were identified. Four isolates exhibited higher quinolone MICs than did the quinolone-susceptible isolates (Table 3). These 4 isolates exhibited 1 to 3 amino acid substitutions in the quinolone-resistance-determining regions (QRDRs) of GyrA and ParC but none in GyrB or ParE (Table 3).
FIG 3.
Susceptibilities to clarithromycin and various quinolones and the genetic patterns of β-lactam resistance in H. influenzae clinical isolates. S, susceptible; I, intermediate; R, resistant.
DISCUSSION
In this study, we revealed that the high prevalence of BLNAR isolates obtained in a Japanese university hospital between 2016 and 2018 could be attributed to an increase in high-BLNAR isolates. Although this study was performed in a single Japanese university hospital, the antimicrobial usage and the prevalence of β-lactam-resistant H. influenzae strains in this facility were similar to those determined in national surveillance in Japan (25, 26). Thus, this study should illustrate the status of expansion of β-lactam-resistant H. influenzae strains in Japanese clinical settings. The prevalence of BLNAR strains was similar to that in a previous study conducted at another site in Japan in 2011 (Table 4), although the clinical background was different from that in the present study (i.e., isolates used in the previous study were collected from pediatric acute otitis media cases) (22). This prevalence is distinct from that in other Asian and European countries, as well as the United States (Table 4). These results suggest that high-BLNAR isolates have been established in Japanese clinical settings since around 2005 (Table 4). Although a primary reason for the expansion of high-BLNAR isolates in Japan had not been elucidated, the relatively high levels of the use of β-lactams, including cephalosporins, in Japan could be a contributing factor (25).
TABLE 4.
Prevalence of high-BLNAR isolates in Japan and other countries
| Country | Year(s) of isolation | No. (%) of H. influenzae isolates |
High-BLNAR strains among BLNAR strains (%) | Reference | ||
|---|---|---|---|---|---|---|
| Total | BLNARa | High-BLNARb | ||||
| Japan | 2016–2018 | 104 | 62 (59.6) | 57 (54.8) | 91.9 | This study |
| 2011–2012 | 122 | 57 (46.7)c | 52 (42.6) | 91.2 | 22 | |
| 2004–2005 | 172 | 61 (35.5)c | 43 (25.0) | 70.5 | 23 | |
| 2003 | 264 | 172 (65.2)c | 74 (28.0) | 43.0 | 13 | |
| 2002–2004 | 457 | 211 (46.2)c | 121 (26.5) | 57.3 | 42 | |
| 2000–2009 | 508 | 247 (48.6)c | 159 (31.3) | 55.3 | 20 | |
| 1999 | 296 | 78 (26.3)c | 39 (13.1) | 50.0 | 15 | |
| 1995–2003 | 621 | 229 (36.9) | 77 (12.4) | 33.6 | 6 | |
| 1987–2000 | 162 | 22 (13.6)c | 3 (1.9) | 13.6 | 19 | |
| Korea | 2012 | 122 | 49 (40.2) | 18 (14.8) | 36.7 | 8 |
| Norway | 2007 | 808 | 116 (14.4) | 3 (0.4) | 2.6 | 14 |
| Korea | 2005–2006 | 540 | 33 (6.1) | 8 (1.5) | 33.3 | 21 |
| Portugal | 2001–2008 | 240 | 94 (39.1) | 2 (0.8) | 2.1 | 9 |
| Spain | 2001–2006 | 354 | 198 (55.9) | 12 (3.4) | 6.1 | 7 |
| France | 2000–2008 | 2,206 | 354 (16.0) | 3 (0.14) | 0.9 | 11 |
| Spain | 2000–2009 | 95 | 27 (28.4) | 0 (0) | 0 | 10 |
| Korea | 2000–2005 | 229 | 67 (29.3) | 0 (0) | 0 | 18 |
| United States | 1999 | 95 | 13 (13.7)c | 0 (0) | 0 | 15 |
Isolates with Arg517His or Asn526Lys substitutions in PBP3.
Isolates with Ser385Thr in addition to Arg517His or Asn526Lys substitutions in PBP3.
Defined by PCR.
To explore the mechanism underlying the increase in high-BLNAR isolates in Japan, we characterized the genotypes of the isolates. Most high-BLNAR isolates (82.5%) were group III or III-like, and the majority (72.3%) of the group III isolates fell into 2 PBP3 types. However, MLST analysis revealed that the 2 PBP3 types consisted of 20 STs with 15 clonal groups (CGs) and 3 singletons. Thus, we conclude that the high prevalence of high-BLNAR strains is due to expansion of multiple lineages and limited contribution by specific lineages.
In a 2014 study conducted in Norway, Skaare et al. classified 116 BLNAR isolates according to PBP3 type and MLST analyses (14). There were only 3 high-BLNAR isolates, 1 in group III (ST1197) and 2 in group III-like (ST160), reflecting the low prevalence of high-BLNAR isolates in that study. The authors showed that the majority (74.3%) of low-BLNAR isolates (most in group II) were classified into 3 PBP3 types (types A, B, and D, in that study) (14). In MLST analysis, approximately 71.6% of the low-BLNAR isolates were classified into 7 STs, and PBP3 types A, B, and D corresponded to ST14/ST367, ST369, and ST201, respectively (14). These results suggest that the expansion of low-BLNAR isolates was caused by a few specific lineages in Norway. This conclusion is in contrast to the multiclonal expansion of high-BLNAR strains in Japan shown in the present study.
In a previous study in South Korea, Park et al. performed pulsed-field gel electrophoresis (PFGE) and phylogenetic analyses of the entire ftsI gene in 29 low-BLNAR isolates and 20 high-BLNAR isolates obtained from pediatric nasopharyngeal specimens in three tertiary-care hospitals in 2012 (8). The isolates clustered into 26 groups, and there were 18 singletons in the ftsI phylogeny. There were no specific PFGE patterns among the BLNAS, low-BLNAR, or high-BLNAR isolates (8). The phylogenetic analysis based on MLST data in the present study revealed that several STs contained BLNAS and high-BLNAR isolates or group III and group III-like isolates in the same ST (Fig. 2). These results suggest that some lineages developed their β-lactam resistance by originating as BLNAS or low-BLNAR strains (groups I and II) and evolving to express a high-BLNAR phenotype. Interestingly, ST57 and ST397, which included group III high-BLNAR isolates in this study, were identified as including group II low-BLNAR or BLNAS isolates in a clinical setting in another country (Norway), suggesting the risk of subsequent evolution to become phenotypically high-BLNAR isolates (14).
Macrolides and quinolones are candidates for secondary choices to treat high-BLNAR infections. Clarithromycin lacks utility for such treatment because of the high prevalence (68.3%) of nonsusceptible isolates (Fig. 1). In contrast, quinolones are promising agents, because no quinolone-nonsusceptible isolates were observed. The prevalence of nonsusceptibility to quinolones is around 0.1 to 2% in Japan and other countries (27–31). These data indicate that the prevalence of quinolone-nonsusceptible H. influenzae strains has not increased. In addition, ceftriaxone should be considered a promising choice because no ceftriaxone-nonsusceptible isolates were detected.
We obtained 4 isolates (3.8%) of different STs that showed reduced susceptibility to quinolones (Table 3). The mechanism of the decrease in quinolone susceptibility involves amino acid substitutions in the QRDRs of GyrA (at positions Ser84 and Asp88) and ParC (at positions Gly82, Asp83, Ser84, and Glu88) in H. influenzae (28, 29, 32). All 4 isolates shared the amino acid substitution Ser84Leu in GyrA (Table 3). Another amino acid substitution, Glu142Lys, in GyrA was found in 1 isolate; however, this substitution is known not to contribute to quinolone nonsusceptibility (30). The Asn138Ser and Ser230Ala substitutions in ParC were also found in another isolate, and Asn138Ser is suggested not to contribute to quinolone resistance (28, 32). Although it is not known whether the Ser230Ala substitution in ParC contributes to quinolone resistance, higher quinolone MICs for the strain suggest that it is a candidate for contributing to resistance. Of note, 1 strain was isolated from a child, while H. influenzae isolates with decreased susceptibility to quinolones are mostly isolated from elderly people (28, 29).
Three of the isolates with decreased susceptibility to quinolones were high-BLNAR strains. This suggests that monitoring for the emergence of fluoroquinolone resistance in β-lactam-resistant H. influenzae is important for treatment in respiratory medicine as well as in pediatrics. It was reported that the prevalence of levofloxacin-resistant H. influenzae strains in Taiwan increased from 2% in 2004 to 24.3% in 2010 (33), and these data indicate that the inappropriate use of quinolones may result in the emergence of quinolone-resistant high-BLNAR isolates in the future.
One isolate, SMHi90, harbored multiple unique amino acid substitutions in PBP3 and exhibited a higher ampicillin MIC than did other high-BLNAR isolates. Thus, we propose that the ftsI sequence of SMHi90 represents a novel high-BLNAR group, group IV. The same ftsI sequence has been registered in the DNA Data Bank of Japan (DDBJ) genome database (accession number LC279277.1 [43]); it was derived from a H. influenzae clinical isolate from a region in Japan distinct from that in the present study. In addition, SMHi90 demonstrated decreased quinolone susceptibility. Thus, we need to be vigilant against the spread of novel emerging high-BLNAR strains.
MATERIALS AND METHODS
Ethics statement.
This study was approved by the institutional review board of Sapporo Medical University Hospital.
Bacterial isolates.
A total of 104 clinical H. influenzae isolates were obtained from Sapporo Medical University Hospital between May 2016 and January 2018. For each isolate, information was obtained regarding the patient's age and sex and the clinical source. The isolates were obtained from the following types of clinical specimens: sputum (n = 56), pharyngeal mucus (n = 16), nasal mucus (n = 9), nasal discharge (n = 7), otorrhea (n = 4), oral cavity (n = 3), tonsil (n = 3), bronchoalveolar lavage fluid (n = 2), and eye, colon, pleural effusion, and abdominal cavity drainage (n = 1 each). All isolates were grown for 24 to 48 h on chocolate agar II (Nippon Beckton-Dickinson, Tokyo, Japan) at 37°C, in a 5% CO2 atmosphere. These isolates were stored at −80°C using a Microbank system (Pro-Lab Diagnostics, Round Rock, TX). Identification of H. influenzae was achieved using a matrix-assisted laser desorption ionization (MALDI) Biotyper (Bruker Daltonics, Billerica, MA) and PCR amplification of the gene encoding outer membrane protein P6, ompP6, as described previously (15). Capsular serotypes were determined by PCR, as described previously (34).
Antimicrobial susceptibility.
MIC determination was carried out by the microdilution method, according to Clinical and Laboratory Standards Institute (CLSI) guidelines (35). Ampicillin (Wako Pure Chemical Industry, Tokyo, Japan), ceftriaxone (Tokyo Chemical Industry, Tokyo, Japan), cefotaxime (Tokyo Chemical Industry), clarithromycin (Taisho Pharmaceutical, Tokyo, Japan), levofloxacin (Daiichi-Sankyo, Tokyo, Japan), moxifloxacin (Bayer, Osaka, Japan), garenoxacin (Toyama Chemical, Tokyo, Japan), tosufloxacin (Toyama Chemical), and sitafloxacin (Daiichi-Sankyo) were used. Geometric means of ampicillin, ceftriaxone, and cefotaxime MICs were calculated as described previously (6). Strains with MICs exceeding 2 mg/liter for levofloxacin and/or 1 mg/liter for moxifloxacin were defined as quinolone nonsusceptible, according to CLSI guidelines (35). Because there are no CLSI definitions of breakpoints for tosufloxacin, garenoxacin, and sitafloxacin, strains with MICs exceeding 1 mg/liter were defined as being nonsusceptible, as stated by the Japanese Society of Chemotherapy (36).
Nucleotide sequences of ftsI genes and detection of β-lactamase genes.
Haemophilus influenzae clinical isolates were classified into groups I to III and III-like on the basis of amino acid substitutions in PBP3 revealed by DNA sequencing of the ftsI gene, as described in a previous report (14). Two pairs of primers for PCR amplification and DNA sequencing of ftsI (nucleotide positions 1048 to 1833) were designed in this study, using the sequence of the H. influenzae Rd strain genome (NCBI accession number NC000907) (37), i.e., ftsI-F2 (5′-GTGGTGGGTTATACGGATATTGATGG-3′) and ftsI-R2 (5′-CATAGGCACGAGCAATTTGTAAAGGT-3′), and ftsI-F3 (5′-TAAGCGGTAAAGAAATTGTGGA-3′) and ftsI-R3 (5′-ACGATGCTGCGCCAAACCGTGTGATGAAAC-3′). Genomic DNA was isolated from cells using the DNeasy kit (Qiagen, Hilden, Germany). PCR amplification was performed with a PCR system 9700 (Applied Biosystems, Waltham, MA) using Ex Taq polymerase (TaKaRa Shuzo, Kyoto, Japan), as follows: 2 min of denaturation at 94°C and 40 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, and extension at 68°C for 3 min. The amplified DNA fragments were sequenced using an Applied Biosystems 3730 DNA analyzer. BLNAS, low-BLNAR (groups I and II), and high-BLNAR (group III and III-like) isolates were defined as described previously (14). Each group was further divided by amino acid substitution typing of PBP3 (PBP3 types) as deduced from the amino acid sequences of PBP3 from Asp350 to Asn611, which are associated with β-lactam resistance, using methods described previously (6–11, 14, 18, 21). Detection of the β-lactamase genes blaTEM-1 and blaROB-1 was performed by PCR, as described previously (15).
Identification of mutations in the QRDRs of gyrA, gyrB, parC, and pare.
To clarify the mechanism of decreased susceptibility to quinolones, the nucleotide sequences of the QRDRs of the quinolone target genes, gyrA, gyrB, parC, and parE, were determined. PCR amplification was performed using Quick Taq HS DyeMix (Toyobo, Osaka, Japan). The specific primers used for PCR amplification and DNA sequencing of the QRDRs of gyrA, gyrB, and parE were described previously (38). The primers for amplifying the QRDR of parC, i.e., HiparC-F (5′-GTGCGTTGCCTTTTATCGGTGA-3′) and HiparC-R (5′-GAAGATTGATGTGGAAGCGCTGA-3′), were designed in this study using the sequence of the H. influenzae Rd strain genome (NCBI accession number NC000907) (37). The PCR products were analyzed as described above.
MLST.
STs were determined by MLST using 7 housekeeping genes (adk, atpG, frdB, fucK, mdh, pgi, and recA), as described previously (39). Allele numbers and STs were assigned using the MLST website (http://haemophilus.mlst.net). The CGs of these STs were defined by using the eBURSTv3 database (http://eburst.mlst.net). A phylogenetic tree based on MLST data for the H. influenzae BLNAR isolate was constructed based on the neighbor-joining method (40), using MEGA7 (41).
Accession number(s).
The nucleotide sequence of the high-BLNAR isolate SMHi90 was registered in the DDBJ database (accession number LC379877).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the Japan Initiative for Global Research Network on Infectious Diseases (J-GRID) from the Ministry of Education, Culture, Sport, Science, and Technology in Japan, the Japan Agency for Medical Research and Development (AMED), and MEXT for the Joint Research Program of the Research Center for Zoonosis Control, Hokkaido University. This work was also partly supported by JSPS KAKENHI (grant 17K15688) and the Yuasa Memorial Foundation. The funding sources did not play any role in the study design, in the collection, analysis, and interpretation of data, in the writing of the report, or in the decision to submit the article for publication.
We have no conflicts of interest to declare.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00851-18.
REFERENCES
- 1.Murphy TF, Apicella MA. 1987. Nontypable Haemophilus influenzae: a review of clinical aspects, surface antigens, and the human immune response to infection. Rev Infect Dis 9:1–15. doi: 10.1093/clinids/9.1.1. [DOI] [PubMed] [Google Scholar]
- 2.Saito M, Okada K, Takemori K, Yoshida SI. 2000. Clonal spread of an invasive strain of Haemophilus influenzae type b among nursery contacts accompanied by a high carriage rate of non-disease-associated strains. J Med Microbiol 49:845–847. doi: 10.1099/0022-1317-49-9-845. [DOI] [PubMed] [Google Scholar]
- 3.Yamasaki K, Kawanami T, Yatera K, Fukuda K, Noguchi S, Nagata S, Nishida C, Kido T, Ishimoto H, Taniguchi H, Mukae H. 2013. Significance of anaerobes and oral bacteria in community-acquired pneumonia. PLoS One 8:e63103. doi: 10.1371/journal.pone.0063103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vega R, Sadoff HL, Patterson MJ. 1976. Mechanisms of ampicillin resistance in Haemophilus influenzae type B. Antimicrob Agents Chemother 9:164–168. doi: 10.1128/AAC.9.1.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Medeiros AA, Levesque R, Jacoby GA. 1986. An animal source for the ROB-1β-lactamase of Haemophilus influenzae type b. Antimicrob Agents Chemother 29:212–215. doi: 10.1128/AAC.29.2.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanbongi Y, Suzuki T, Osaki Y, Senju N, Ida T, Ubukata K. 2006. Molecular evolution of β-lactam-resistant Haemophilus influenzae: 9-year surveillance of penicillin-binding protein 3 mutations in isolates from Japan. Antimicrob Agents Chemother 50:2487–2492. doi: 10.1128/AAC.01316-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia-Cobos S, Campos J, Lazaro E, Roman F, Cercenado E, Garcia-Rey C, Perez-Vazquez M, Oteo J, de Abajo F. 2007. Ampicillin-resistant non-β-lactamase-producing Haemophilus influenzae in Spain: recent emergence of clonal isolates with increased resistance to cefotaxime and cefixime. Antimicrob Agents Chemother 51:2564–2573. doi: 10.1128/AAC.00354-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park C, Kim KH, Shin NY, Byun JH, Kwon EY, Lee JW, Kwon HJ, Choi EY, Lee DG, Sohn WY, Kang JH. 2013. Genetic diversity of the ftsI gene in β-lactamase-nonproducing ampicillin-resistant and β-lactamase-producing amoxicillin-/clavulanic acid-resistant nasopharyngeal Haemophilus influenzae strains isolated from children in South Korea. Microb Drug Resist 19:224–230. doi: 10.1089/mdr.2012.0116. [DOI] [PubMed] [Google Scholar]
- 9.Barbosa AR, Giufre M, Cerquetti M, Bajanca-Lavado MP. 2011. Polymorphism in ftsI gene and β-lactam susceptibility in Portuguese Haemophilus influenzae strains: clonal dissemination of β-lactamase-positive isolates with decreased susceptibility to amoxicillin/clavulanic acid. J Antimicrob Chemother 66:788–796. doi: 10.1093/jac/dkq533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Puig C, Calatayud L, Marti S, Tubau F, Garcia-Vidal C, Carratala J, Linares J, Ardanuy C. 2013. Molecular epidemiology of nontypeable Haemophilus influenzae causing community-acquired pneumonia in adults. PLoS One 8:e82515. doi: 10.1371/journal.pone.0082515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dabernat H, Delmas C. 2012. Epidemiology and evolution of antibiotic resistance of Haemophilus influenzae in children 5 years of age or less in France, 2001–2008: a retrospective database analysis. Eur J Clin Microbiol Infect Dis 31:2745–2753. doi: 10.1007/s10096-012-1623-9. [DOI] [PubMed] [Google Scholar]
- 12.Ubukata K, Shibasaki Y, Yamamoto K, Chiba N, Hasegawa K, Takeuchi Y, Sunakawa K, Inoue M, Konno M. 2001. Association of amino acid substitutions in penicillin-binding protein 3 with β-lactam resistance in β-lactamase-negative ampicillin-resistant Haemophilus influenzae. Antimicrob Agents Chemother 45:1693–1699. doi: 10.1128/AAC.45.6.1693-1699.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hotomi M, Fujihara K, Billal DS, Suzuki K, Nishimura T, Baba S, Yamanaka N. 2007. Genetic characteristics and clonal dissemination of β-lactamase-negative ampicillin-resistant Haemophilus influenzae strains isolated from the upper respiratory tract of patients in Japan. Antimicrob Agents Chemother 51:3969–3976. doi: 10.1128/AAC.00422-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Skaare D, Anthonisen IL, Caugant DA, Jenkins A, Steinbakk M, Strand L, Sundsfjord A, Tveten Y, Kristiansen BE. 2014. Multilocus sequence typing and ftsl sequencing: a powerful tool for surveillance of penicillin-binding protein 3-mediated beta-lactam resistance in nontypeable Haemophilus influenzae. BMC Microbiol 14:131. doi: 10.1186/1471-2180-14-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hasegawa K, Yamamoto K, Chiba N, Kobayashi R, Nagai K, Jacobs MR, Appelbaum PC, Sunakawa K, Ubukata K. 2003. Diversity of ampicillin-resistance genes in Haemophilus influenzae in Japan and the United States. Microb Drug Resist 9:39–46. doi: 10.1089/107662903764736337. [DOI] [PubMed] [Google Scholar]
- 16.Ubukata K. 2003. Problems associated with high prevalence of multidrug-resistant bacteria in patients with community-acquired infections. J Infect Chemother 9:285–291. doi: 10.1007/s10156-003-0278-Y. [DOI] [PubMed] [Google Scholar]
- 17.Matic V, Bozdogan B, Jacobs MR, Ubukata K, Appelbaum PC. 2003. Contribution of β-lactamase and PBP amino acid substitutions to amoxicillin/clavulanate resistance in β-lactamase-positive, amoxicillin/clavulanate-resistant Haemophilus influenzae. J Antimicrob Chemother 52:1018–1021. doi: 10.1093/jac/dkg474. [DOI] [PubMed] [Google Scholar]
- 18.Kim IS, Ki CS, Kim S, Oh WS, Peck KR, Song JH, Lee K, Lee NY. 2007. Diversity of ampicillin resistance genes and antimicrobial susceptibility patterns in Haemophilus influenzae strains isolated in Korea. Antimicrob Agents Chemother 51:453–460. doi: 10.1128/AAC.00960-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qin L, Zhou ZY, Hu BJ, Yamamoto T, Watanabe H. 2012. Antimicrobial susceptibility and genetic characteristics of Haemophilus influenzae isolated from community-acquired respiratory tract infection patients in Shanghai City, China. J Infect Chemother 18:508–514. doi: 10.1007/s10156-012-0372-0. [DOI] [PubMed] [Google Scholar]
- 20.Hagiwara E, Baba T, Shinohara T, Nishihira R, Komatsu S, Ogura T. 2012. Antimicrobial resistance genotype trend and its association with host clinical characteristics in respiratory isolates of Haemophilus influenzae. Chemotherapy 58:352–357. doi: 10.1159/000343973. [DOI] [PubMed] [Google Scholar]
- 21.Bae S, Lee J, Kim E, Lee S, Yu J, Kang Y. 2010. Antimicrobial resistance in Haemophilus influenzae respiratory tract isolates in Korea: results of a nationwide acute respiratory infections surveillance. Antimicrob Agents Chemother 54:65–71. doi: 10.1128/AAC.00966-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Otsuka T, Kitami O, Kondo K, Ota H, Oshima S, Tsuchiya A, Shirai T, Fujii K, Nakamure M, Shoji Y, Nakamura H, Masuda Y, Komiyama K, Yoshida K, Ishikawa Y, Iwaya A, Takahashi S, Okazaki M, Hotomi M, Yamanaka N. 2013. Incidence survey of acute otitis media in children in Sado Island, Japan—Sado Otitis Media Study (SADOMS). PLoS One 8:e68711. doi: 10.1371/journal.pone.0068711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hashida K, Shiomori T, Hohchi N, Muratani T, Mori T, Udaka T, Suzuki H. 2008. Nasopharyngeal Haemophilus influenzae carriage in Japanese children attending day-care centers. J Clin Microbiol 46:876–881. doi: 10.1128/JCM.01726-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sato K, Hoshino K, Tanaka M, Hayakawa I, Osada Y. 1992. Antimicrobial activity of DU-6859, a new potent fluoroquinolone, against clinical isolates. Antimicrob Agents Chemother 36:1491–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muraki Y, Kitamura M, Maeda Y, Kitahara T, Mori T, Ikeue H, Tsugita M, Tadano K, Takada K, Akamatsu T, Yamada T, Yamada T, Shiraishi T, Okuda M. 2013. Nationwide surveillance of antimicrobial consumption and resistance to Pseudomonas aeruginosa isolates at 203 Japanese hospitals in 2010. Infection 41:415–423. doi: 10.1007/s15010-013-0440-0. [DOI] [PubMed] [Google Scholar]
- 26.Ministry of Health, Labour, and Welfare. 2016. Annual open report 2016 (all facilities): Japan Nosocomial Infections Surveillance (JANIS). Ministry of Health, Labour, and Welfare, Tokyo, Japan: https://janis.mhlw.go.jp/english/report/open_report/2016/3/1/ken_Open_Report_Eng_201600_clsi2012.pdf. [Google Scholar]
- 27.Yanagihara K, Kadota J, Aoki N, Matsumoto T, Yoshida M, Yagisawa M, Oguri T, Sato J, Ogasawara K, Wakamura T, Sunakawa K, Watanabe A, Iwata S, Kaku M, Hanaki H, Ohsaki Y, Watari T, Toyoshima E, Takeuchi K, Shiokoshi M, Takeda H, Miki M, Kumagai T, Nakanowatari S, Takahashi H, Utagawa M, Nishiya H, Kawakami S, Kobayashi N, Takasaki J, Mezaki K, Konosaki H, Aoki Y, Yamamoto Y, Shoji M, Goto H, Saraya T, Kurai D, Okazaki M, Niki Y, Yoshida K, Kawana A, Saionji K, Fujikura Y, Miyazawa N, Kudo M, Sato Y, Yamamoto M, Yoshida T, Nakamura M, Tsukada H, Imai Y, Tsukada A, Kawasaki S, Honma Y, Yamamoto T, Ban N, Mikamo H, Sawamura H, Miyara T, Toda H, Sato K, Nakamura T, Fujikawa Y, Mitsuno N, Mikasa K, Kasahara K, Sano R, Sugimoto K, Asari S, Nishi I, Toyokawa M, Miyashita N, Koguchi Y, Kusano N, Mihara E, Kuwabara M, Watanabe Y, Kawasaki Y, Takeda K, Tokuyasu H, Masui K, Negayama K, Hiramatsu K, Aoki Y, Fukuoka M, Magarifuchi H, Nagasawa Z, Suga M, Muranaka H, Morinaga Y, Honda J, Fujita M. 2015. Nationwide surveillance of bacterial respiratory pathogens conducted by the Surveillance Committee of Japanese Society of Chemotherapy, the Japanese Association for Infectious Diseases, and the Japanese Society for Clinical Microbiology in 2010: general view of the pathogens' antibacterial susceptibility. J Infect Chemother 21:410–420. doi: 10.1016/j.jiac.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 28.Yokota S, Ohkoshi Y, Sato K, Fujii N. 2008. Emergence of fluoroquinolone-resistant Haemophilus influenzae strains among elderly patients but not among children. J Clin Microbiol 46:361–365. doi: 10.1128/JCM.01561-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Puig C, Tirado-Velez JM, Calatayud L, Tubau F, Garmendia J, Ardanuy C, Marti S, de la Campa AG, Linares J. 2015. Molecular characterization of fluoroquinolone resistance in nontypeable Haemophilus influenzae clinical isolates. Antimicrob Agents Chemother 59:461–466. doi: 10.1128/AAC.04005-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shoji H, Shirakura T, Fukuchi K, Takuma T, Hanaki H, Tanaka K, Niki Y. 2014. A molecular analysis of quinolone-resistant Haemophilus influenzae: validation of the mutations in quinolone resistance-determining regions. J Infect Chemother 20:250–255. doi: 10.1016/j.jiac.2013.12.007. [DOI] [PubMed] [Google Scholar]
- 31.Critchley IA, Brown SD, Traczewski MM, Tillotson GS, Janjic N. 2007. National and regional assessment of antimicrobial resistance among community-acquired respiratory tract pathogens identified in a 2005–2006 U.S. faropenem surveillance study. Antimicrob Agents Chemother 51:4382–4389. doi: 10.1128/AAC.00971-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hirakata Y, Ohmori K, Mikuriya M, Saika T, Matsuzaki K, Hasegawa M, Hatta M, Yamamoto N, Kunishima H, Yano H, Kitagawa M, Arai K, Kawakami K, Kobayashi I, Jones RN, Kohno S, Yamaguchi K, Kaku M. 2009. Antimicrobial activities of piperacillin-tazobactam against Haemophilus influenzae isolates, including β-lactamase-negative ampicillin-resistant and β-lactamase-positive amoxicillin-clavulanate-resistant isolates, and mutations in their quinolone resistance-determining regions. Antimicrob Agents Chemother 53:4225–4230. doi: 10.1128/AAC.00192-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuo SC, Chen PC, Shiau YR, Wang HY, Lai JFs, Huang W, Lauderdale TL. 2014. Levofloxacin-resistant Haemophilus influenzae, Taiwan, 2004–2010. Emerg Infect Dis 20:1386–1390. doi: 10.3201/eid2008.140341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kilian M. 1976. A taxonomic study of the genus Haemophilus, with the proposal of a new species. J Gen Microbiol 93:9–62. doi: 10.1099/00221287-93-1-9. [DOI] [PubMed] [Google Scholar]
- 35.Clinical and Laboratory Standards Institute. 2017. Performance standards for antimicrobial susceptibility testing; 27th informational supplement. CLSI M100-S27. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 36.Kadota J, Ishii Y, Kusachi S, Kusano N, Niki Y, Higa F, Hiramatsu K, Hori S, Muratani T, Morita K. 2009. Clinical breakpoints in pulmonary infections, sepsis and urinary tract infections: addition of new antimicrobial agents. Jpn J Chemother 57:343–345. (In Japanese.) http://www.chemotherapy.or.jp/guideline/090728_point.pdf. [Google Scholar]
- 37.Fleischmann RD, Adams MD, White O, Clayton RA, Kirkness EF, Kerlavage AR, Bult CJ, Tomb JF, Dougherty BA, Merrick JM, McKenney K, Sutton G, FitzHugh W, Fields C, Gocayne JD, Scott J, Shirley R, Liu L, Glodek A, Kelley JM, Weidman JF, Phillips CA, Spriggs T, Hedblom E, Cotton MD, Utterback TR, Hanna MC, Nguyen DT, Saudek DM, Brandon RC, Fine LD, Fritchman JL, Fuhrmann JL, Geoghagen NSM, Gnehm CL, McDonald LA, Small KV, Fraser CM, Smith HO, Venter JC. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 38.Perez-Vazquez M, Roman F, Aracil B, Canton R, Campos J. 2004. Laboratory detection of Haemophilus influenzae with decreased susceptibility to nalidixic acid, ciprofloxacin, levofloxacin, and moxifloxacin due to gyrA and parC mutations. J Clin Microbiol 42:1185–1191. doi: 10.1128/JCM.42.3.1185-1191.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meats E, Feil EJ, Stringer S, Cody AJ, Goldstein R, Kroll JS, Popovic T, Spratt BG. 2003. Characterization of encapsulated and noncapsulated Haemophilus influenzae and determination of phylogenetic relationships by multilocus sequence typing. J Clin Microbiol 41:1623–1636. doi: 10.1128/JCM.41.4.1623-1636.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. [DOI] [PubMed] [Google Scholar]
- 41.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ohkoshi Y, Yokota S, Sato K, Hayashi T, Matsuda K, Kuwahara O, Akizawa H, Fujii N. 2008. Antibiotic susceptibility of Haemophilus influenzae strains isolated from various clinical sources in Hokkaido Prefecture, Japan. J Infect Chemother 14:93–98. doi: 10.1007/s10156-007-0583-Y. [DOI] [PubMed] [Google Scholar]
- 43.Kitaoka K, Kimura K, Kitanaka H, Banno H, Jin W, Wachino J, Arakawa Y. 2018. Carbapenem-nonsusceptible Haemophilus influenzae with penicillin-binding protein 3 containing an amino acid insertion. Antimicrob Agents Chemother 62:e00671-18. doi: 10.1128/AAC.00671-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.