Multidrug-resistant Acinetobacter baumannii infection has recently emerged as a worldwide clinical problem, and colistin is increasingly being used as a last-resort therapy. Despite its favorable bacterial killing, resistance and heteroresistance (HR) to colistin have been described.
KEYWORDS: Acinetobacter, PmrAB, colistin, heteroresistance
ABSTRACT
Multidrug-resistant Acinetobacter baumannii infection has recently emerged as a worldwide clinical problem, and colistin is increasingly being used as a last-resort therapy. Despite its favorable bacterial killing, resistance and heteroresistance (HR) to colistin have been described. The purpose of the present study was to investigate the role of the PmrAB regulatory pathway in laboratory-selected mutants representative of global epidemic strains. From three unrelated A. baumannii clinical strains (sequence types 2, 3, and 20), eight colistin-resistant mutants were selected. Half of the mutants showed HR to colistin according to the reference method (population analysis profiling), whereas the other half exhibited stable resistance. M12I mutation within pmrA and M308R, S144KLAGS, and P170L mutations for pmrB were associated with HR to colistin, while T235I, A226T, and P233S mutations within pmrB were associated with stable resistance. The transcript levels of the pmrCAB operon were upregulated in all the mutants. Compensatory mutations were explored for some mutants. A single mutant (T235I mutant) displayed a compensatory mutation through ISAba1 mobilization within the pmrB gene that was associated with the loss of colistin resistance. The mutant resistance phenotype associated with T235I was partially restored in a trans-complementation assay turning to HR. The level of colistin resistance was correlated with the level of expression of pmrC in the trans-complemented strains. This report shows the role of different mutations in the PmrAB regulatory pathway and warns of the development of colistin HR that could be present but not easily detected through routine testing.
INTRODUCTION
Acinetobacter baumannii has emerged as an opportunistic and medically important pathogen causing a broad range of nosocomial infections, particularly among the most vulnerable patients in the intensive care units. The most frequent infections are ventilator-associated pneumonia and bacteremia, both of which lead to high morbidity and mortality (1). A. baumannii is intrinsically resistant to several classes of antimicrobial agents and readily acquires resistance to other classes, leading to multidrug-resistant (MDR) A. baumannii isolates. Antimicrobial resistance provides them a selective ecological advantage that drives the clonal expansion of certain A. baumannii lineages, such as the three international clonal complexes I, II, and III (2).
Divalent cations bridging lipopolysaccharide (LPS) molecules stabilize the membrane through balancing the electrostatic net charges (3). This unique composition of the outer membrane makes the cell susceptible to cationic antimicrobial peptides (CAMPs). Polymyxin B and polymyxin E (colistin), biosynthesized by some strains of Paenibacillus polymyxa, are two FDA-approved antibiotics of the CAMP class that are increasingly used as the ultimate therapeutic agent in MDR Gram-negative bacteria such as A. baumannii. The mode of action of polymyxins is due to the amphipathic nature of these compounds. The positively charged cyclic group is thought to compete with divalent cations for binding to LPS, while the acyl chain is thought to disrupt both the outer and the inner membranes (4, 5). To survive exposure to these compounds, A. baumannii has developed two main antimicrobial resistance mechanisms: the loss of lipid A or its modification through enzymes (6–8). The former mechanism, which overthrows the dogma that LPS is essential in Gram-negative bacteria, is due to the inactivation of downstream lipid A biosynthetic genes. This mechanism, which is rare in clinical medicine, confers a high level of colistin resistance. The loss of lipid A leads, in turn, to a growth defect, higher fitness costs, virulence attenuation, and hypersensitivity to other classes of antibiotics (6, 9, 10). The second mechanism is mediated by the operon pmrCAB. Genetic changes in the PmrAB regulatory system lead to the activation of pmrA, pmrB, and pmrC, a phosphotransferase, which leads to the addition of phosphoethanolamine to lipid A (7, 8). Interestingly, A. baumannii may harbor a pmrC-like gene named eptA whose expression is increased in colistin-resistant strains (11). This mechanism reduces the net LPS negative charge, leading to a low to medium level of colistin resistance. The impact on virulence and fitness depends on the underlying mutations and could range from low to apparently unaffected (9, 10).
Colistin heteroresistance (HR) has been described previously (12). Colistin HR has been associated with prior colistin exposure (13). This phenomenon is underappreciated due to the use of different diagnostic methods and standards. A standard definition of HR has been recently determined (14). HR bacteria are defined as a single-species bacterial population displaying a heterogeneous response to an antibiotic, i.e., when the difference between the lowest concentration exhibiting maximum inhibition and the highest noninhibitory concentration is above 8-fold. The reference method to define microbial response to an antibiotic is population analysis profiling (PAP). Clinically, the situation may be critical when the majority of the heterogeneous population is susceptible to an antibiotic and a subpopulation is resistant. This may lead to therapeutic failures due to susceptibility categorization errors. To date, no mechanism has been described for colistin HR in A. baumannii. The purpose of the present study was to investigate the role of the pmrCAB operon in laboratory-selected mutants representative of clinical strains.
RESULTS
Mutations associated with colistin resistance.
Following exposure to colistin for 24 h, resistant mutants were obtained in the three clinical strains. Single nucleotide polymorphisms (SNPs) and indels could appear in the PmrAB regulatory pathway. Mutants derived from clinical isolates (Table 1) and obtained at extreme concentrations (and not those obtained at intermediate concentrations) were further sequenced in the pmrCAB operon to reveal putative changes. Based on targeted sequencing, two mutants were found for Ab1, four mutants were found for Ab2, and only one mutant was found for Ab3 (Table 2). A unique mutant of strain Ab2 contained a mutation in the response regulator domain of the pmrA gene, while the three others were mutated in the pmrB gene. No change was observed in the pmrC gene. Mutants of strain Ab3 displayed a unique mutation whatever the concentration used for the selection, whereas mutants of strain Ab2 harbored a mixed and more diverse population of mutants for each selection process. Mutants of strain Ab1 displayed an intermediate situation with a major mutant for each colistin concentration used for selection. Mutants in the histidine kinase domain of the pmrB gene had the highest MICs and became colistin resistant. In contrast, other mutants were still susceptible to colistin despite the screening performed at 10 mg/liter. Amino acid substitution in the PmrAB regulatory pathway is the prime suspect for colistin resistance, but other genetic changes could be responsible. To address this hypothesis, two pairs (Ab1/Ab1-8-R1 and Ab3/Ab3-2-R1) and one trio (Ab2/Ab2-1-R4/Ab2-1-R4-S1) of strains were sequenced. Genome-wide analysis of this subset confirmed the mutations observed using Sanger sequencing of the pmrCAB operon as well as the absence of nonsynonymous mutations outside the pmrCAB operon (data not shown). Only a synonymous mutation was observed in a putative siderophore for Ab1-8-R1. Two different mutations were observed in a noncoding region for Ab2-1-R4-S1 and Ab3-2-R1 (data not shown).
TABLE 1.
Clinical isolates used in this studya
| Isolate | Clinical source | STUO | STIP | CCUO/CCIP |
|---|---|---|---|---|
| Ab1 | Blood culture | 449 | 20 | 109/1 |
| Ab2 | Rectal swab | 208 | 2 | 92/2 |
| Ab3 | Skin swab | 106 | 3 | 187/3 |
UO and IP, University of Oxford and Institut Pasteur multilocus sequence typing schemes; ST, sequence type; CC, clonal complex.
TABLE 2.
Mutational analysis of the PmrAB regulatory pathwaya
| Strain | CST concn used for selection (mg/liter) | PmrA (224 aa) response regulator (aa 4–113) | PmrB (444 aa) |
CST MIC (mg/liter) |
Population analysis profile | ||||
|---|---|---|---|---|---|---|---|---|---|
| aa 1–217 | HisK (aa 218–279) | aa 278–325 | HATpaseC (aa 326–436) | BMD | Etest | ||||
| Ab1 | 0 | 2 | 0.125 | S | |||||
| Ab1-2-R1 | 2 | M308R | 2 | 0.25 | HR | ||||
| Ab1-8-R1 | 8 | S144KLAGS | 2 | 0.38 | HR | ||||
| Ab2 | 0 | 2 | 0.19 | S | |||||
| Ab2-1-R1 | 1 | M12I | 2 | 0.25 | HR | ||||
| Ab2-1-R4 | 1 | T235I | 8 | 6 | R | ||||
| Ab2-32-R1 | 32 | A226T | 8 | 4 | R | ||||
| Ab2-32-R3 | 32 | P170L | 2 | 0.38 | HR | ||||
| Ab3 | 0 | 2 | 0.25 | S | |||||
| Ab3-2-R1 | 2 | P233S | 16 | 8 | R | ||||
| Ab3-16-R1 | 16 | P233S | 8 | 4 | R | ||||
aa, amino acids; CST, colistin; BMD, broth microdilution; S, susceptible; R, resistant; HR, heteroresistant.
Mutations associated with the loss of colistin resistance.
The second aim was to assess whether compensatory mutations would occur using in vitro laboratory conditions and a one-step selection procedure. A single mutant, the one with the highest MIC, was screened for each resistant mutant derived from Ab1, Ab2, and Ab3. The loss of colistin resistance was observed through a compensatory mutation in Ab2-1-R4. An ISAba1 insertion sequence (1,188 bp in length) was inserted after the proline 170 codon within the pmrB gene and generated a 9-bp duplication at the extremities of the insertion site. Despite many attempts to isolate Ab3-2-R1 (48, 54, 161, 297, and 576 individual colonies tested per screen), the screened sampling failed to highlight the loss of colistin resistance. The screening procedure was unsuitable for isolates Ab1-2-R1 and Ab1-8-R1 that were HR mutants.
Mutants confirmed as resistant and HR to colistin.
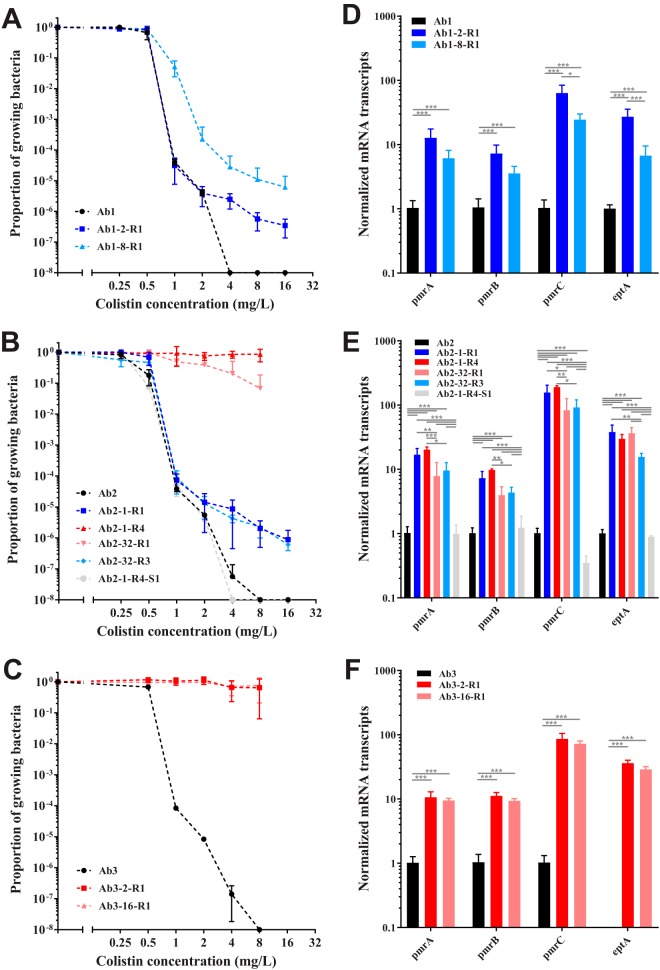
To confirm the character of colistin resistance of the mutants, PAP was carried out. First, the three clinical strains showed a homogeneous response to colistin in the susceptible range (Fig. 1A to C). Second, for many of the mutants, HR was suspected since MICs were similar to those of the parental strain. Mutants with an MIC of 2 or below exhibited heterogeneous responses (Fig. 1A and B) to colistin, while mutants with an MIC of 8 or above exhibited a clearly resistant population (Fig. 1B and C). Mutants harboring mutations outside the histidine kinase domain of the pmrB gene were confirmed as HR mutants. As additional changes could be suspected for the resistant subpopulation, PAP was performed on the resistant subpopulations for isolate Ab1-8-R1 and isolate Ab2-1-R1 grown in agar plates containing 16 mg/liter of colistin (data not shown). Both isolates remained HR, although the proportion of resistance for isolate Ab2-1-R1 was larger (10−2) than the original one (10−6). With regard to the loss of colistin resistance, isolate Ab2-1-R4-S1 displayed a phenotypic reversion. It had the same profile as the susceptible parental strain (Fig. 1B). The insertion sequence ISAba1 may have inactivated the pmrB gene. If the sensor protein is inactivated, the regulator protein cannot be upregulated, as well as the enzyme responsible for lipid A modification, the product of the pmrC gene.
FIG 1.
Response to colistin of resistant and heteroresistant mutants (A to C) and transcript levels of putative PmrA-regulated genes in sequentially derived isolates (D to F). Population analysis profiling (PAP) of isolates derived from Ab1 through Ab3 was conducted (A to C). Profiles were obtained by the spotting method (n = 3). Black curves and shapes indicate the parental isolate, blue curves and shapes indicate heteroresistant mutants, red curves and shapes indicate resistant mutants, and light gray curves and shapes indicate the mutant losing colistin resistance. For the transcriptional experiment (D to F), data represent the means from 3 independent experiments; error bars represent 1 SD. One-way analysis of variance with the Tukey multiple-comparison test was used to determine whether transcript levels from a gene differed among the strains. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Mutants associated with colistin resistance and HR.
To verify whether there was an association between mutants and the PmrAB regulatory pathway, it was decided to quantify the level of transcripts of previously described genes in longitudinally derived isolates. The aim was to study if one of these genes had a distinct expression according to the susceptibility to colistin (susceptible, HR, or resistant). As expected, the expression of the transcripts of the pmrCAB operon increased in both colistin-resistant and colistin-HR mutants (Fig. 1D to F). Unexpectedly, the levels of transcripts of pmrC were highly similar in colistin-resistant and colistin-HR strains derived from Ab2 (Fig. 1E). The role of colistin as an inducer of the expression of the transcripts of the pmrCAB operon was checked. Two stress concentrations of colistin (0.25 mg/liter and 2 mg/liter) were tested for Ab2 mutants, and no induction was observed (data not shown). In isolate Ab2-1-R4-S1, the transcript level of the pmrCAB operon came back to the level observed in the parental clinical isolate, confirming the role played by the insertion sequence within the pmrB gene. Furthermore, the transcript level of the pmrC gene remained even lower than the transcript level observed in the parental strain. The expression of the transcripts of eptA showed the same trend as that of the genes present in the pmrCAB operon, except that the transcripts of eptA were not constitutively expressed in Ab3 (Fig. 1F).
PmrAB mutants contributed to colistin resistance and colistin HR.
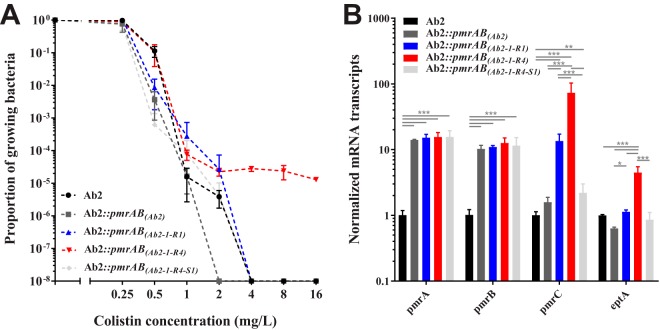
To check if amino acid substitutions identified by Sanger sequencing contributed to colistin resistance or HR, isolate Ab2 was complemented in trans with the wild-type and mutated pmrAB genes. Complementation of isolate Ab2 with the wild-type pmrAB genes did not alter the susceptibility to colistin (Fig. 2A). Complementation with an HR mutant or a disrupted mutant did not alter the susceptibility to colistin (Fig. 2A). Colistin resistance was only partially restored when isolate Ab2 was complemented with pmrAB(Ab2-1-R4). Complementation with a resistant mutant resulted in colistin HR (Fig. 2A). The levels of transcripts of the pmrA and pmrB genes in transformants complemented in trans with the wild-type and mutated pmrAB genes were equally upregulated (Fig. 2B). These results indicate that wild-type and mutated pmrAB genes were actually cloned with their own promoter. The level of transcripts of the pmrC gene was upregulated for the complemented resistant and HR mutants. Surprisingly, a slight and significant overexpression of pmrC was observed when the ISAba1-complemented strain was compared to its parental clinical strain. However, there was no difference when the ISAba1 complementation was compared to the wild-type complementation. This observation was likely due to the expression of genes on the vector or a normalization issue and not due to the ISAba1 insertion per se in the pmrB gene (Fig. 2B). The highest levels were observed for the resistant-mutant complementation. The level of colistin resistance was correlated to the level of expression of pmrC. This result was consistent with a direct regulation with PmrA when amino acid substitution occurred in the PmrAB regulatory pathway. The same trend was observed for eptA, except that the upregulation was less marked and held for the resistant mutant exclusively.
FIG 2.
Response to colistin of trans-complemented transformants (A) and transcript levels of putative PmrA-regulated genes in trans-complemented transformants (B). (A) PAP of transformants derived from trans-complemented Ab2 mutants. Profiles were obtained by the spotting method (n = 3). The black curve and circles indicate the parental isolate, the dark gray curve and shapes indicate a transformant complemented by the parental pmrAB genes from isolate Ab2, the blue curve and shapes indicate a transformant complemented by the heteroresistant pmrAB genes from isolate Ab2-1-R1, the red curve and shapes indicate a transformant complemented by the resistant pmrAB genes from isolate Ab2-1-R4, and the light gray curve and shapes indicate a transformant complemented by the pmrAB genes from isolate Ab2-1-R4-S1. For the transcriptional experiment (B), data represent the means from 3 independent experiments; error bars represent 1 SD. One-way analysis of variance with the Tukey multiple-comparison test was used to determine whether transcript levels from a gene differed among the transformants. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
DISCUSSION
HR is a phenomenon in which subpopulations of seemingly isogenic bacteria exhibit a range of susceptibilities to a particular antibiotic. HR can be intrinsic or acquired. In this study, colistin HR and colistin resistance were acquired through mutations in the PmrAB regulatory pathway. The trans-complementation assay suggested the role played by this two-component system and showed a direct correlation between expression of pmrC and colistin (hetero)resistance. However, this assay did not reveal the mechanism of HR in the four HR mutants, nor did it support the fact that the overexpression of pmrC was the explanation for HR. The assumed cis-regulation was likely different from this trans-assay. The possibility of manipulating these clinical strains was somewhat limited by their elevated antimicrobial resistance. This explains, in part, the need for a trans-complementation assay.
The one-step selection strategy generated both resistant and HR isolates. All mutations described in this study have been previously observed in clinical or laboratory strains except the two HR mutants derived from isolate Ab1, in which a 12-bp insertion or an M308R amino acid substitution was observed in pmrB. The fact that these two mutants had not been observed yet could be due to the process of selection for spontaneous mutation production chosen by the other authors (7, 8, 15, 16). The authors favored serial gradient strategies to promote stable mutants. As an example, Arroyo et al. were the first to identify the M12I mutation in the pmrA gene in the organism exhibiting a polymyxin B MIC of 4 mg/liter, compared to the parental MIC of 0.5 mg/liter. Their mutant was correctly classified as resistant, and the authors did not perform PAP as a result (8). Their process for selection may increase the proportion of resistant subpopulations for this specific mutant as observed with our isolate making an HR mutant fully resistant to colistin. This characteristic may explain why they did not succeed in obtaining a knockout for this mutant. Regarding the HR mutant derived from isolate Ab2 and presenting the P170L mutation, Pournaras et al. have related this mutation to colistin resistance before (MIC = 32 mg/liter) (17). Interestingly, Arroyo et al. have identified two clinical strains with mutations at the same position in the pmrB gene (8). One exhibited a different mutation, P170Q, leading to colistin resistance (MIC = 16 mg/liter). The other exhibited the same mutation but also the A80V mutation (MIC = 128 mg/liter). The detection of mutations appears to be insufficient for inferring phenotypes. To support this observation, Park et al. have studied colistin resistance without mutations in known colistin resistance genes (16). Although the P233S mutation observed in the Ab3 mutant seemed to be exclusively related to colistin resistance as demonstrated before (17–20), the discovery of a clinical compensation in a patient with compensatory mutations further restricts phenotype inference based on the genotype (18).
The level of colistin resistance is still low (1 to 4%) in Acinetobacter spp. (21, 22). The hypothesis of reduced virulence of resistant A. baumannii strains was previously raised in the literature, among other possibilities (23). The possibility of compensatory mutations restoring the virulence in colistin-resistant bacteria was noticed (24) and actually observed in a case study (25). The hypothesis of HR to explain the low prevalence of colistin resistance could not be ruled out. Two animal-based studies proved that Klebsiella pneumoniae and Enterobacter cloacae displaying undetected colistin HR could lead to treatment failure in murine infection (26, 27). HR might be an alternative to the costly colistin resistance. Transcriptome remodeling during infection and treatment has been performed, and a convergent change was observed for clinical isolates with pmrAB mutations (28). Most isolates originated from patients treated with colistin. All of the isolates were susceptible to colistin at an MIC of 0.5 mg/ml and had a differential expression of pmrC that is consistent with HR. In addition, this study allowed more strict definition of the Pmr regulon, in agreement with a preceding transcriptomic study (29). The PmrAB two-component regulatory system facilitates adaptation to diverse environments in many Gram-negative bacteria (30). The use of CAMPs may confer a cross-resistance to host CAMPs, and the A. baumannii PmrAB two-component regulatory system could be the culprit (31). HR due to pmrAB mutations may also have implications for immunogenicity. In Francisella tularensis, flmK leads to addition of galactosamine to lipid A, and its inactivation provided protective immunity (32). The galactosamine modification of lipid A has been described for A. baumannii as well (33).
Therapeutic concentrations of colistin range from 2 to 4 mg/liter in patients (34). Concentrations to prevent the selection of resistant mutants are considerably higher than therapeutic concentrations (35). It is not surprising that the rate of HR can vary from 18.7% to 100% (36). Disk diffusion or MIC gradient strips assays are typically used as screening methods (14). They might miss the HR phenotype because of the low bacterial load used in these methods, a load that is inappropriate to detect the (low-frequency) resistant subpopulation. In addition, colistin activity might be overestimated if tested in cation-adjusted Mueller-Hinton (MH) broths (as recommended) compared to interstitial space fluid-like media possibly causing a shift from susceptible to resistant in A. baumannii (37). On the other hand, adsorption of polymyxins to plastics might overestimate colistin MICs, which can be prevented by the use of low-protein-binding polypropylene (38). However, a study proved that reproducibility and robustness were superior for agar dilution compared to broth dilution methods (39). Both Clinical and Laboratory Standards Institute (CLSI) and EUCAST recommend the use of broth microdilution (BMD) as the only valid method for MIC testing (51). A standardized protocol globally accepted for colistin susceptibility testing would be more than welcome. The clinical relevance of HR was not proven at that time because of the difficulties in measuring the MIC (40) and in detecting HR a fortiori (14).
MATERIALS AND METHODS
Bacterial strains, growth conditions, and susceptibility testing.
A collection of 27 MDR A. baumannii isolates was collected at Geneva University Hospital (41). Multilocus sequence typing was performed to identify and select strains of different genetic backgrounds (42). Three clinical isolates (Ab1, Ab2, and Ab3), belonging to the international clones I, II, and III, were randomly selected (Table 1). All strains were stored in glycerol stocks at −80°C. Subcultures were performed on Mueller-Hinton agar medium (Becton Dickinson AG, Allschwil, Switzerland) at 37°C for 18 h under aerobic conditions unless described otherwise. Colistin susceptibility testing was done by BMD using cation-adjusted Mueller-Hinton broth medium (Becton Dickinson AG) according to the joint CLSI-EUCAST working group (51). The three parental isolates had colistin BMD MICs of 2 mg/liter. The CLSI and EUCAST breakpoints are the same for A. baumannii/colistin, but the need to continue with the revision of the methodology for testing and breakpoints is still reaffirmed (40). To date, an isolate is considered susceptible to colistin when the MIC is ≤2 mg/liter and resistant when above. MICs were also determined using Etest strips (bioMérieux SA, Marcy l'Etoile, France) on Mueller-Hinton agar plates (Becton Dickinson AG) inoculated with a 0.5 McFarland suspension.
Selection of colistin-resistant and colistin compensation mutants.
Isolates Ab1, Ab2, and Ab3 were subcultured in 5 ml of lysogeny broth (LB) (Becton Dickinson AG) containing colistin (Sigma-Aldrich, St. Louis, MO) at concentrations ranging from 0 to 32 mg/liter for 24 h at 37°C with shaking in 50-ml Falcon tubes (Corning Science Mexico, Tamaulipas, Mexico). In a one-step liquid-based culture, resistant mutants were isolated from LB plates containing 10 mg/liter of colistin selected at extreme concentrations (n = 4 per condition). Resistant mutants were stored as glycerol stocks at −80°C. Overnight cultures of resistant mutants were performed in 5 ml of LB medium at 37°C for 18 h, and a substantial number of clones (e.g., >100) were isolated. The loss of colistin resistance was screened by comparing growth on LB in the presence and absence of colistin. Mutants were isolated and saved (n = 4 per condition).
Bacterial lysis and Sanger sequencing of the pmrCAB operon.
Single colonies picked from LB agar plates were suspended in 100 μl of Tris-EDTA buffer (10 mM Tris and 1 mM EDTA, pH 8) containing acid-washed glass beads (Sigma-Aldrich). The cells were disrupted using an orbital shaker (Vortex Genius 3; IKA, Staufen, Germany) for 1 min, and the lysate was centrifuged at 3,500 × g for 3 min. Five microliters of 100-fold-diluted supernatant were directly used for PCR of the entire pmrCAB operon using previously described primers (see Table S1 in the supplemental material). The PCR products were purified using a QIAquick PCR purification kit (Qiagen, Hilden, Germany) according to the supplier's instructions and eluted in 30 μl of pure water. Both strands were completely sequenced with BigDye v3.1 on a 3730XL DNA analyzer (Life Technologies, Carlsbad, CA) using the same primers as were used for amplification (Table S1).
PAP.
Response to colistin in selected mutants was determined by population analysis profiling (PAP) using a modified spotting method (43). Briefly, a suspension of each strain was adjusted to 2.0 McFarland and serially 10-fold diluted to 10−7. Both isolates Ab1 and Ab2 and their mutants were streaked on MH agar alone and on a 2-fold series of plates containing colistin at concentrations of 0.25 to 16 mg/liter. Isolate Ab3 and mutants were streaked on MH agar alone and on a 2-fold series of plates containing colistin at concentrations of 0.5 to 8 mg/liter. For each condition, four 10-μl droplets of the four highest concentrations were spotted onto one plate and four 10-μl droplets of the lowest concentrations were spotted onto a second plate. Colonies were counted after 24 h of incubation at 37°C. Three biological replicates were performed. HR was considered when the antibiotic exhibiting the highest inhibitory effect was >8-fold higher than the highest noninhibitory concentration (14).
RT-qPCR experiments.
The selected clinical isolates and mutants were grown to the mid-logarithmic phase in cation-adjusted Mueller-Hinton broth. Suspensions were adjusted to an optical density of 0.01 at 595 nm and grown for 5 h with shaking at 37°C before harvesting. Isolation of total RNA and synthesis of cDNA were performed as recently described (44) except that Superscript II reverse transcriptase (RT) was used (Invitrogen, Carlsbad, CA). The primers for the PCR amplification of cDNA were retrieved from literature or designed using the primer3 program (45) (Table S1) and validated using MIQE guidelines (46). An Mx3005P quantitative PCR (qPCR) system (Agilent, Santa Clara, CA) was used for the quantification of cDNA. Triplicate PCRs were performed using Brilliant III Ultrafast SYBR green qPCR master mix (Agilent). The thermal profile was determined according to supplier instructions. To correct for differences in the amount of starting material, the rpoB gene was selected as a reference gene. Results were presented as ratios of gene expression between the target gene and the reference gene, which were obtained according to the method of Pfaffl et al. (47). Differences in the PCR efficiencies were corrected according to the LinRegPCR program (48), and assessment of PCR efficiencies was based on the median of individual values calculated for each amplicon. Biological triplicates were performed. One-way analysis of variance with the Tukey multiple-comparison test was used to determine whether transcript levels from a gene differed among the isolates (P ≥ 0.05).
Genome sequencing.
Whole-genome sequencing of the 7 A. baumannii isolates was performed using Illumina high-throughput sequencing technology: the Ab1 pair (Ab1/Ab1-8-R1), the Ab2 trio (Ab2/Ab2-1-R4/Ab2-1-R4-S1), and the Ab3 pair (Ab3/Ab3-2-R1). Extraction and purification of the genomic DNAs of the 7 isolates were achieved using an automated system (Nuclisens easyMag; bioMérieux SA, Marcy l'Etoile, France). DNA quantity and purity in extracts were controlled using a fluorometer (Qubit Molecular Probes, Eugene, OR) and a spectrophotometer (ND-2000; NanoDrop Technologies, Wilmington, DE). Library preparation was performed with the Nextera XT DNA sample preparation kit (Illumina, San Diego, CA) and Nextera XT index kit (Illumina) by following the manufacturer's recommendations. Library validation was performed on a 2100 bioanalyzer (Agilent) to control the distribution of fragmented DNA. Sequencing runs of paired-end 2 × 250 cycles with the MiSeq reagent kit v2 (500 cycles) (Illumina) on a MiSeq instrument (Illumina) was performed.
Genome assembly and mutation detection.
The genome was assembled using a version of the A5 pipeline called A5-miseq (49). Briefly, A5-miseq consists of three main stages: (i) read quality filtering and error correction, (ii) contig assembly and draft scaffolding with insert size, depth of coverage evaluation, and error correction by read pair mapping on contigs, and (iii) misassembly detection, conservative scaffolding, and error correction by read pair stringent mapping on contigs to generate the final consensus. The parental isolates were assembled in 50 to 77 scaffolds resulting in reference draft genomes, which were used to detect the next mutations of resistant and compensatory mutants. Punctual mutations supported by at least three reads after read pairs mapping were kept for following interpretation.
trans-Complementation assay.
The TelR cassette from the pMo130-TelR plasmid was PCR amplified and cloned into the SphI site of pABBR_MCS to create the plasmid pABBR-TelR. The pmrAB genes from Ab2, Ab2-1-R1, Ab2-1-R4, and Ab2-1-R4-S1 were PCR amplified and cloned into the multiple-cloning site (MCS) of pABBR-TelR. Cloning was carried out in Escherichia coli DH5α thermocompetent cells. The transformation protocol based on the heat shock method was performed as recommended by the supplier (New England BioLabs, Ipswich, MA). The antibiotics kanamycin (7.5 mg/liter) and ampicillin (100 mg/liter) were added as needed for the selection. The complementation plasmids were transformed by electroporation (25 μF, 1.8 kV, and 100 Ω in 0.2-mm cuvettes) as previously described (50). Transformants were selected in LB agar plates supplemented with tellurite at 60 mg/liter.
Accession number(s).
The genome sequences were deposited in the GenBank, ENA, and DDBJ databases under accession no. LNUX00000000 (Ab3), LNUY00000000 (Ab1), LNUZ00000000 (Ab2), LNVA00000000 (Ab3-2-R1), LNVB00000000 (Ab1-8-R1), LNVD00000000 (Ab2-1-R4-S1), and LNVE00000000 (Ab2-1-R4).
Supplementary Material
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00788-18.
REFERENCES
- 1.Dijkshoorn L, Nemec A, Seifert H. 2007. An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat Rev Microbiol 5:939–951. doi: 10.1038/nrmicro1789. [DOI] [PubMed] [Google Scholar]
- 2.Diancourt L, Passet V, Nemec A, Dijkshoorn L, Brisse S. 2010. The population structure of Acinetobacter baumannii: expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS One 5:e10034. doi: 10.1371/journal.pone.0010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nikaido H, Vaara M. 1985. Molecular basis of bacterial outer membrane permeability. Microbiol Rev 49:1–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vaara M, Vaara T. 1983. Polycations as outer membrane-disorganizing agents. Antimicrob Agents Chemother 24:114–122. doi: 10.1128/AAC.24.1.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hancock RE, Chapple DS. 1999. Peptide antibiotics. Antimicrob Agents Chemother 43:1317–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moffatt JH, Harper M, Harrison P, Hale JD, Vinogradov E, Seemann T, Henry R, Crane B, St Michael F, Cox AD, Adler B, Nation RL, Li J, Boyce JD. 2010. Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide production. Antimicrob Agents Chemother 54:4971–4977. doi: 10.1128/AAC.00834-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beceiro A, Llobet E, Aranda J, Bengoechea JA, Doumith M, Hornsey M, Dhanji H, Chart H, Bou G, Livermore DM, Woodford N. 2011. Phosphoethanolamine modification of lipid A in colistin-resistant variants of Acinetobacter baumannii mediated by the pmrAB two-component regulatory system. Antimicrob Agents Chemother 55:3370–3379. doi: 10.1128/AAC.00079-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arroyo LA, Herrera CM, Fernandez L, Hankins JV, Trent MS, Hancock RE. 2011. The pmrCAB operon mediates polymyxin resistance in Acinetobacter baumannii ATCC 17978 and clinical isolates through phosphoethanolamine modification of lipid A Antimicrob Agents Chemother 55:3743–3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beceiro A, Moreno A, Fernandez N, Vallejo JA, Aranda J, Adler B, Harper M, Boyce JD, Bou G. 2014. Biological cost of different mechanisms of colistin resistance and their impact on virulence in Acinetobacter baumannii. Antimicrob Agents Chemother 58:518–526. doi: 10.1128/AAC.01597-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wand ME, Bock LJ, Bonney LC, Sutton JM. 2015. Retention of virulence following adaptation to colistin in Acinetobacter baumannii reflects the mechanism of resistance. J Antimicrob Chemother 70:2209–2216. doi: 10.1093/jac/dkv097. [DOI] [PubMed] [Google Scholar]
- 11.Lesho E, Yoon EJ, McGann P, Snesrud E, Kwak Y, Milillo M, Onmus-Leone F, Preston L, St Clair K, Nikolich M, Viscount H, Wortmann G, Zapor M, Grillot-Courvalin C, Courvalin P, Clifford R, Waterman PE. 2013. Emergence of colistin-resistance in extremely drug-resistant Acinetobacter baumannii containing a novel pmrCAB operon during colistin therapy of wound infections. J Infect Dis 208:1142–1151. doi: 10.1093/infdis/jit293. [DOI] [PubMed] [Google Scholar]
- 12.Li J, Rayner CR, Nation RL, Owen RJ, Spelman D, Tan KE, Liolios L. 2006. Heteroresistance to colistin in multidrug-resistant Acinetobacter baumannii. Antimicrob Agents Chemother 50:2946–2950. doi: 10.1128/AAC.00103-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hawley JS, Murray CK, Jorgensen JH. 2008. Colistin heteroresistance in acinetobacter and its association with previous colistin therapy. Antimicrob Agents Chemother 52:351–352. doi: 10.1128/AAC.00766-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.El-Halfawy OM, Valvano MA. 2015. Antimicrobial heteroresistance: an emerging field in need of clarity. Clin Microbiol Rev 28:191–207. doi: 10.1128/CMR.00058-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adams MD, Nickel GC, Bajaksouzian S, Lavender H, Murthy AR, Jacobs MR, Bonomo RA. 2009. Resistance to colistin in Acinetobacter baumannii associated with mutations in the PmrAB two-component system. Antimicrob Agents Chemother 53:3628–3634. doi: 10.1128/AAC.00284-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park YK, Choi JY, Shin D, Ko KS. 2011. Correlation between overexpression and amino acid substitution of the PmrAB locus and colistin resistance in Acinetobacter baumannii. Int J Antimicrob Agents 37:525–530. doi: 10.1016/j.ijantimicag.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 17.Pournaras S, Poulou A, Dafopoulou K, Chabane YN, Kristo I, Makris D, Hardouin J, Cosette P, Tsakris A, De E. 2014. Growth retardation, reduced invasiveness, and impaired colistin-mediated cell death associated with colistin resistance development in Acinetobacter baumannii. Antimicrob Agents Chemother 58:828–832. doi: 10.1128/AAC.01439-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Snitkin ES, Zelazny AM, Gupta J, Palmore TN, Murray PR, Segre JA. 2013. Genomic insights into the fate of colistin resistance and Acinetobacter baumannii during patient treatment. Genome Res 23:1155–1162. doi: 10.1101/gr.154328.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim Y, Bae IK, Lee H, Jeong SH, Yong D, Lee K. 2014. In vivo emergence of colistin resistance in Acinetobacter baumannii clinical isolates of sequence type 357 during colistin treatment. Diagn Microbiol Infect Dis 79:362–366. doi: 10.1016/j.diagmicrobio.2014.03.027. [DOI] [PubMed] [Google Scholar]
- 20.Durante-Mangoni E, Del Franco M, Andini R, Bernardo M, Giannouli M, Zarrilli R. 2015. Emergence of colistin resistance without loss of fitness and virulence after prolonged colistin administration in a patient with extensively drug-resistant Acinetobacter baumannii. Diagn Microbiol Infect Dis 82:222–226. doi: 10.1016/j.diagmicrobio.2015.03.013. [DOI] [PubMed] [Google Scholar]
- 21.Gales AC, Jones RN, Sader HS. 2011. Contemporary activity of colistin and polymyxin B against a worldwide collection of Gram-negative pathogens: results from the SENTRY Antimicrobial Surveillance Program (2006–09). J Antimicrob Chemother 66:2070–2074. doi: 10.1093/jac/dkr239. [DOI] [PubMed] [Google Scholar]
- 22.Sader HS, Farrell DJ, Flamm RK, Jones RN. 2014. Antimicrobial susceptibility of Gram-negative organisms isolated from patients hospitalized in intensive care units in United States and European hospitals (2009–2011). Diagn Microbiol Infect Dis 78:443–448. doi: 10.1016/j.diagmicrobio.2013.11.025. [DOI] [PubMed] [Google Scholar]
- 23.López-Rojas R, Dominguez-Herrera J, McConnell MJ, Docobo-Perez F, Smani Y, Fernandez-Reyes M, Rivas L, Pachon J. 2011. Impaired virulence and in vivo fitness of colistin-resistant Acinetobacter baumannii. J Infect Dis 203:545–548. doi: 10.1093/infdis/jiq086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rolain JM, Roch A, Castanier M, Papazian L, Raoult D. 2011. Acinetobacter baumannii resistant to colistin with impaired virulence: a case report from France. J Infect Dis 204:1146–1147. doi: 10.1093/infdis/jir475. [DOI] [PubMed] [Google Scholar]
- 25.López-Rojas R, Jimenez-Mejias ME, Lepe JA, Pachon J. 2011. Acinetobacter baumannii resistant to colistin alters its antibiotic resistance profile: a case report from Spain. J Infect Dis 204:1147–1148. doi: 10.1093/infdis/jir476. [DOI] [PubMed] [Google Scholar]
- 26.Band VI, Crispell EK, Napier BA, Herrera CM, Tharp GK, Vavikolanu K, Pohl J, Read TD, Bosinger SE, Trent MS, Burd EM, Weiss DS. 2016. Antibiotic failure mediated by a resistant subpopulation in Enterobacter cloacae. Nat Microbiol 1:16053. doi: 10.1038/nmicrobiol.2016.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Band VI, Satola SW, Burd EM, Farley MM, Jacob JT, Weiss DS. 2018. Carbapenem-resistant Klebsiella pneumoniae exhibiting clinically undetected colistin heteroresistance leads to treatment failure in a murine model of infection. mBio 9:e02448-. doi: 10.1128/mBio.02448-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wright MS, Jacobs MR, Bonomo RA, Adams MD. 2017. Transcriptome remodeling of Acinetobacter baumannii during infection and treatment. mBio 8:e02193-. doi: 10.1128/mBio.02193-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheah SE, Johnson MD, Zhu Y, Tsuji BT, Forrest A, Bulitta JB, Boyce JD, Nation RL, Li J. 2016. Polymyxin resistance in Acinetobacter baumannii: genetic mutations and transcriptomic changes in response to clinically relevant dosage regimens. Sci Rep 6:26233. doi: 10.1038/srep26233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen HD, Groisman EA. 2013. The biology of the PmrA/PmrB two-component system: the major regulator of lipopolysaccharide modifications. Annu Rev Microbiol 67:83–112. doi: 10.1146/annurev-micro-092412-155751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Napier BA, Burd EM, Satola SW, Cagle SM, Ray SM, McGann P, Pohl J, Lesho EP, Weiss DS. 2013. Clinical use of colistin induces cross-resistance to host antimicrobials in Acinetobacter baumannii. mBio 4:e00021-. doi: 10.1128/mBio.00021-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kanistanon D, Hajjar AM, Pelletier MR, Gallagher LA, Kalhorn T, Shaffer SA, Goodlett DR, Rohmer L, Brittnacher MJ, Skerrett SJ, Ernst RK. 2008. A Francisella mutant in lipid A carbohydrate modification elicits protective immunity. PLoS Pathog 4:e24. doi: 10.1371/journal.ppat.0040024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pelletier MR, Casella LG, Jones JW, Adams MD, Zurawski DV, Hazlett KR, Doi Y, Ernst RK. 2013. Unique structural modifications are present in the lipopolysaccharide from colistin-resistant strains of Acinetobacter baumannii. Antimicrob Agents Chemother 57:4831–4840. doi: 10.1128/AAC.00865-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grégoire N, Mimoz O, Megarbane B, Comets E, Chatelier D, Lasocki S, Gauzit R, Balayn D, Gobin P, Marchand S, Couet W. 2014. New colistin population pharmacokinetic data in critically ill patients suggesting an alternative loading dose rationale. Antimicrob Agents Chemother 58:7324–7330. doi: 10.1128/AAC.03508-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cai Y, Li R, Liang B, Bai N, Liu Y, Wang R. 2010. In vitro antimicrobial activity and mutant prevention concentration of colistin against Acinetobacter baumannii. Antimicrob Agents Chemother 54:3998–3999. doi: 10.1128/AAC.00264-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cai Y, Chai D, Wang R, Liang B, Bai N. 2012. Colistin resistance of Acinetobacter baumannii: clinical reports, mechanisms and antimicrobial strategies. J Antimicrob Chemother 67:1607–1615. doi: 10.1093/jac/dks084. [DOI] [PubMed] [Google Scholar]
- 37.Matzneller P, Strommer S, Osterreicher Z, Mitteregger D, Zeitlinger M. 2015. Target site antimicrobial activity of colistin might be misestimated if tested in conventional growth media. Eur J Clin Microbiol Infect Dis 34:1989–1994. doi: 10.1007/s10096-015-2441-7. [DOI] [PubMed] [Google Scholar]
- 38.Karvanen M, Malmberg C, Lagerback P, Friberg LE, Cars O. 2017. Colistin is extensively lost during standard in vitro experimental conditions. Antimicrob Agents Chemother 61:e00857-. doi: 10.1128/AAC.00857-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Turlej-Rogacka A, Xavier BB, Janssens L, Lammens C, Zarkotou O, Pournaras S, Goossens H, Malhotra-Kumar S. 2018. Evaluation of colistin stability in agar and comparison of four methods for MIC testing of colistin. Eur J Clin Microbiol Infect Dis 37:345–353. doi: 10.1007/s10096-017-3140-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prim N, Rivera A, Coll P, Mirelis B. 2018. Is colistin susceptibility testing finally on the right track? Antimicrob Agents Chemother 62:e02067-17 (Letter.) doi: 10.1128/AAC.02067-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cherkaoui A, Emonet S, Renzi G, Schrenzel J. 2015. Characteristics of multidrug-resistant Acinetobacter baumannii strains isolated in Geneva during colonization or infection. Ann Clin Microbiol Antimicrob 14:42. doi: 10.1186/s12941-015-0103-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bartual SG, Seifert H, Hippler C, Luzon MA, Wisplinghoff H, Rodriguez-Valera F. 2005. Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J Clin Microbiol 43:4382–4390. doi: 10.1128/JCM.43.9.4382-4390.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pfeltz RF, Schmidt JL, Wilkinson BJ. 2001. A microdilution plating method for population analysis of antibiotic-resistant staphylococci. Microb Drug Resist 7:289–295. doi: 10.1089/10766290152652846. [DOI] [PubMed] [Google Scholar]
- 44.Charretier Y, Kohler T, Cecchini T, Bardet C, Cherkaoui A, Llanes C, Bogaerts P, Chatellier S, Charrier JP, Schrenzel J. 2015. Label-free SRM-based relative quantification of antibiotic resistance mechanisms in Pseudomonas aeruginosa clinical isolates. Front Microbiol 6:81. doi: 10.3389/fmicb.2015.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG. 2012. Primer3—new capabilities and interfaces. Nucleic Acids Res 40:e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 47.Pfaffl MW, Horgan GW, Dempfle L. 2002. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res 30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ruijter JM, Ramakers C, Hoogaars WM, Karlen Y, Bakker O, van den Hoff MJ, Moorman AF. 2009. Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res 37:e45. doi: 10.1093/nar/gkp045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Coil D, Jospin G, Darling AE. 2015. A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics 31:587–589. doi: 10.1093/bioinformatics/btu661. [DOI] [PubMed] [Google Scholar]
- 50.Jacobs AC, Thompson MG, Gebhardt M, Corey BW, Yildirim S, Shuman HA, Zurawski DV. 2014. Genetic manipulation of Acinetobacter baumannii. Curr Protoc Microbiol 35:6G.2.1–6G.211. [DOI] [PubMed] [Google Scholar]
- 51.European Committee on Antimicrobial Susceptibility Testing. 2016. Recommendations for MIC determination of colistin (polymyxin E) as recommended by the joint CLSI-EUCAST Polymyxin Breakpoints Working Group. European Committee on Antimicrobial Susceptibility Testing. http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/General_documents/Recommendations_for_MIC_determination_of_colistin_March_2016.pdf Accessed 22 April 2018.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.