Surfing motility is a novel form of surface adaptation exhibited by the nosocomial pathogen Pseudomonas aeruginosa in the presence of the glycoprotein mucin, which is found in high abundance at mucosal surfaces, especially those of the lungs of cystic fibrosis and bronchiectasis patients. Here, we investigated the adaptive antibiotic resistance of P. aeruginosa under conditions in which surfing occurs compared that in to cells undergoing swimming.
KEYWORDS: adaptive resistance, antibiotic resistance, cystic fibrosis, mucin, resistome, surfing
ABSTRACT
Surfing motility is a novel form of surface adaptation exhibited by the nosocomial pathogen Pseudomonas aeruginosa in the presence of the glycoprotein mucin, which is found in high abundance at mucosal surfaces, especially those of the lungs of cystic fibrosis and bronchiectasis patients. Here, we investigated the adaptive antibiotic resistance of P. aeruginosa under conditions in which surfing occurs compared that in to cells undergoing swimming. P. aeruginosa surfing cells were significantly more resistant to several classes of antibiotics, including aminoglycosides, carbapenems, polymyxins, and fluoroquinolones. This was confirmed by incorporation of antibiotics into growth medium, which revealed a concentration-dependent inhibition of surfing motility that occurred at concentrations much higher than those needed to inhibit swimming. To investigate the basis of resistance, transcriptome sequencing (RNA-Seq) was performed and revealed that surfing influenced the expression of numerous genes. Included among genes dysregulated under surfing conditions were multiple genes from the Pseudomonas resistome; these genes are known to affect antibiotic resistance when mutated. Screening transposon mutants in these surfing-dysregulated resistome genes revealed that several of these mutants exhibited changes in susceptibility to one or more antibiotics under surfing conditions, consistent with a contribution to the observed adaptive resistance. In particular, several mutants in resistome genes, including armR, recG, atpB, clpS, nuoB, and certain hypothetical genes, such as PA5130, PA3576, and PA4292, showed contributions to broad-spectrum resistance under surfing conditions and could be complemented by their respective cloned genes. Therefore, we propose that surfing adaption led to extensive multidrug adaptive resistance as a result of the collective dysregulation of diverse genes.
INTRODUCTION
The rise of antibiotic resistance is a global concern. As the number of new antibiotics being discovered declines and the extensive and sometimes inappropriate use of antibiotics continues, more patients suffer and die from infections caused by antibiotic-resistant bacteria (1, 2). Pseudomonas aeruginosa is categorized as a serious threat by the CDC (https://www.cdc.gov/drugresistance/biggest_threats.html) and a priority 1 critical pathogen by the World Health Organization (WHO) (3, 4). P. aeruginosa is a Gram-negative opportunistic pathogen that causes approximately 10% of all hospital-acquired infections, as well as chronic infections in the lungs of individuals with cystic fibrosis (CF) (5, 6). Therefore, there is a growing need to understand the mechanisms leading to antibiotic resistance in P. aeruginosa.
P. aeruginosa can deploy intrinsic, acquired, and adaptive resistance mechanisms (7, 8). Intrinsic resistance refers to the bacterium's natural qualities that allow it to evade the effects of antibiotics, and it is not induced by stress or by the presence of antibiotics (7). Intrinsic resistance includes low outer membrane permeability, which works in synergy with intrinsic levels of efflux pumps, β-lactamases, periplasmic enzymes, and other resistance mechanisms. Acquired resistance can be selected for by antibiotic exposure and occurs due to mutations or the acquisition of genetic elements such as plasmids, transposons, and integrons (7, 8). Adaptive resistance refers to resistance that occurs due to environmental circumstances, is thought to be due largely to transcriptional changes in genes that determine resistance/susceptibility, and is reversible when environmental circumstances (e.g., complex adaptive growth states such as swarming or biofilm formation or exposure to stresses, including antibiotics) are reversed (7). P. aeruginosa in the cystic fibrosis lung, especially during late stages, is thought to grow as biofilms (9). Biofilms represent a growth state in which bacteria grow as structured communities on surfaces and exhibit multidrug adaptive resistance (10).
Bacterial motility is a critical aspect of P. aeruginosa pathogenesis. Motility is needed for colonization of the host and the establishment of biofilms (11). It is also often coupled with the expression of virulence factors. P. aeruginosa has two known forms of polar locomotory appendages, a tail-like flagellum and a hair-like type IV pilus; these contribute to a diverse set of motile phenotypes or lifestyles. The three most highly studied forms of motility are swimming, twitching, and swarming (11, 12). Swimming motility involves the use of flagellar rotation to move within aqueous environments. Twitching depends on the type IV pilus to enable movement on solid surfaces through the extension and retraction of polar pili (11, 13). These types of movements are not accompanied by major changes in gene expression. In contrast, swarming motility is a complex adaptation that involves multicellular coordination to enable movement on semisolid surfaces in the presence of a poor nitrogen source (11, 13). This surface motility form is dependent on both pili and flagella, and it results in dendritic (P. aeruginosa strain PA14) or solar flare colonies (P. aeruginosa strain PAO1) on 0.4 to 0.6% (wt/vol) agar (11). A less-studied form of motility termed “sliding” occurs when Pseudomonas cells glide on solid surfaces independent of any appendages but dependent on the production of rhamnolipid surfactants to reduce surface tension (14). The conditions under which swarming motility occur have been proposed to reflect CF lung conditions due to the high similarities in composition (semiviscous surface, amino acids as a nitrogen source, and glucose as a carbon source); however, swarming models lack a major glycoprotein known as mucin found in the CF lungs and involved in regulating mucosal viscosity (11). By incorporating mucin into an artificial CF model, a novel form of motility termed “surfing” was discovered (11).
Mucin is secreted from mucosal and submucosal glands in the lungs and other mucosal surfaces (11). It contains a polypeptide core with branched oligosaccharide chains. Molecular crosslinking of its structure contributes to the viscoelastic properties of mucus. When mucin is added to medium that normally supports swimming or swarming, accelerated surface motility termed surfing occurs. Surfing depends on intact flagella but not type IV pili (11). Surfing colonies appear relatively circular, with thick white outer edges containing mostly nonflagellated cells piled on top of each other and a blue-green center with flagellated cells (11). Unlike swarming, surfing motility does not require such strict growth conditions and can occur in nutrient-rich or minimal medium, in the presence of ammonium as a nitrogen source, and at a range of viscosities/agar concentrations (ranging from 0.3% to 1.0% [wt/vol]). Mucin is proposed to act as a wetting agent or lubricant, and, unlike swarming or sliding, surfing does not depend on rhamnolipid production (11). It was suggested that surfing is a complex adaptive form of motility (11).
Here, we demonstrate that surfing cells exhibited multidrug adaptive resistance, dependent on the complex adaptive changes that accompanied this motility phenotype. Compared to swimming, surfing adaptive cells were significantly more resistant to several classes of antibiotics, including aminoglycosides, polymyxins, fluoroquinolones, and carbapenems. Screening mutants of resistome genes that were found to be dysregulated under surfing conditions revealed changes in susceptibility that may account for their contribution to the observed resistance.
RESULTS
Surfing cells exhibited broad-spectrum antibiotic resistance.
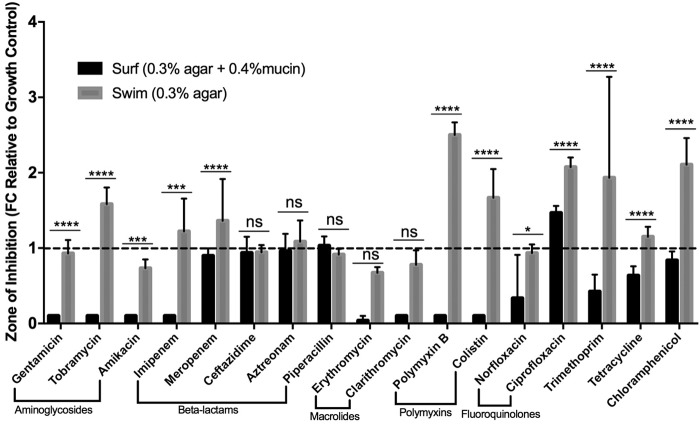
Disk diffusion assay results (Fig. 1) assessing how close surfing and swimming cells approached an antibiotic disk revealed a significant decrease in the zone of inhibition under surfing conditions (synthetic cystic fibrosis medium [SCFM] 0.3% agar, 0.4% mucin) compared to swimming (SCFM 0.3% agar). This was observed for 12 of the 17 antibiotics tested, with the exceptions of 3 β-lactams and 2 macrolides. Compared to swimming bacteria, surfing cells exhibited significant adaptive resistance to the tested aminoglycosides, carbapenems, polymyxins, fluoroquinolones, trimethoprim, tetracycline, and chloramphenicol, with complete resistance to 3 different aminoglycosides and to imipenem, clarithromycin, and the polymyxins.
FIG 1.
Multidrug adaptive resistance of surfing colonies. Zones of inhibition determined under swimming (0.3% agar) and surfing (0.3% agar 0.4% mucin) conditions in SCFM using the motility disk diffusion method with 17 different antibiotics. Statistical significance between swimming and surfing was determined using two-way analysis of variance (ANOVA). (n = 3). *, P < 0.5; **, P < 0.01; ***, P < 10−3; ****, P < 10−4.
Antibiotic incorporation assays to confirm adaptive resistance.
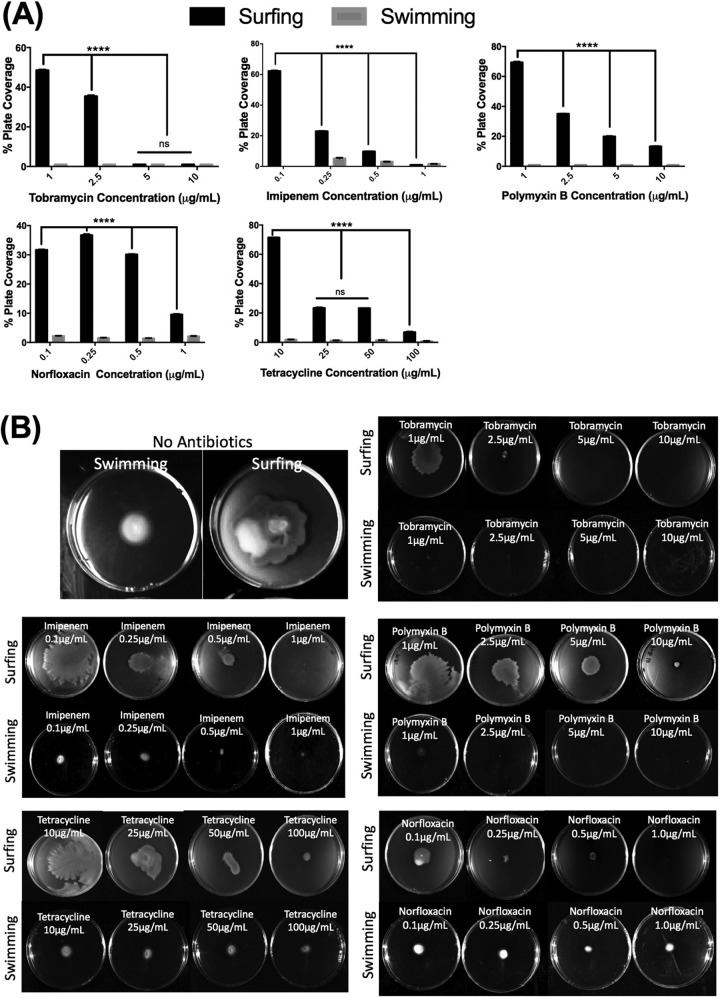
To further investigate the adaptive resistance of surfing colonies, 5 selected antibiotics were incorporated into growth plates to determine how they affect the initiation and propagation of motility colonies. Four of the selected antibiotics, polymyxin B, imipenem, tobramycin, and norfloxacin, were chosen as the representatives of their antibiotic class that showed the greatest (or complete) resistance under surfing conditions compared to swimming conditions and that showed no change in MIC in the presence of mucin in liquid media. Tetracycline was chosen since disk diffusion results were more consistent compared to trimethoprim and chloramphenicol. Antibiotic incorporation assays (Fig. 2), involving the 5 selected antibiotics, revealed a concentration-dependent inhibition of surfing motility and showed that surfing motility proceeded at antibiotic concentrations that completely inhibited swimming. For example, surfing occurred with 0.1 μM imipenem whereas swimming was completely abolished at this concentration. As the imipenem concentration increased, there was a clear reduction in the size of the surfing colony, and at a concentration of 1 μM imipenem, both surfing and swimming were completely inhibited. Indeed, for all five antibiotics tested, inhibition of surfing occurred with increasing concentrations but still occurred to some extent at concentrations much higher than those inhibiting swimming.
FIG 2.
Concentration-dependent inhibition of surfing motility. Surfing motility colonies of wild-type PA14 cells when the antibiotic at various concentrations was incorporated into 25 ml of SCFM agar containing 0.4% mucin (surfing) or no mucin (swimming). Incorporation assay results are described as the % plate coverage relative to that of the control with no antibiotics, measured using ImageJ. Significance analysis between surfing and swimming colonies was performed using two-way ANOVA. *, P < 0.5; **, P < 0.01; ***, P < 10−3; ****, P < 10−4.
Adaptive antibiotic resistance was not due to the presence of mucin alone.
To show that the observed resistances were attributable to the surfing adaptation rather than to the presence of mucin, we examined the effect of mucin on antibiotic activity. Mucin itself might have influenced antibiotic diffusion. Therefore, as a control, we assessed the effect of mucin on antibiotic diffusion by testing its effects in a disk diffusion format using 1.5% agar, under which conditions surfing did not occur. We observed that for 9 out of the 12 antibiotics for which surfing cells demonstrated resistance (and 3 of 5 for which they did not demonstrate resistance), there were no significant differences in the diffusion zones between agar plates with and without mucin, indicating that the influence of mucin on medium viscosity and antibiotic diffusion had no significant effect on diffusion (Table 1) or susceptibility. For those that did show a difference, tobramycin revealed increased susceptibility in the presence of mucin, the opposite of the effect of surfing conditions, while amikacin showed a partial but much lesser effect compared to that of surfing conditions, and ciprofloxacin showed a significant reduction in susceptibility. From this, we concluded that, with the possible exception of ciprofloxacin, the surfing adaptation, rather than altered antibiotic diffusion, was responsible for the observed multidrug adaptive resistance phenotype.
TABLE 1.
Impact of mucin addition on diffusion and antibiotic susceptibility
| Antibiotic | Zone of clearing (mm) on SCFM platesa |
||
|---|---|---|---|
| 1.5% agar | 1.5% agar + 0.4% mucin | P value | |
| Gentamicin | 8.7 | 8.0 | 1.0 |
| Tobramycin | 5.3 | 10.7 | <10−4 |
| Amikacin | 7.3 | 3.7 | <10−4 |
| Imipenem | 4.3 | 4.7 | >1.0 |
| Meropenem | 10.7 | 9.0 | 0.6 |
| Ceftazidime | 10.7 | 9.3 | 0.7 |
| Erythromycin | 4.0 | 9.3 | <10−4 |
| Clarithromycin | 2.7 | 6.3 | <10−4 |
| Aztreonam | 6.3 | 6.7 | >1.0 |
| Piperacillin | 8.0 | 6.7 | 0.7 |
| Polymyxin B | 3.0 | 3.0 | >1.0 |
| Colistin | 4.0 | 2.3 | 0.6 |
| Norfloxacin | 5.0 | 3.7 | 0.7 |
| Ciprofloxacin | 9.7 | 6.3 | 0.0003 |
| Trimethoprim | 5.7 | 5.7 | >1.0 |
| Tetracycline | 9.3 | 8.3 | 1.0 |
| Chloramphenicol | 9.7 | 10.3 | 1.0 |
Disk diffusion assays were performed on SCFM with 1.5% agar in the absence and presence of 0.4% mucin. Bacterial cultures were spread as a lawn, and the antibiotic disk was applied on top. The zones of inhibition were measured after overnight incubation at 37°C. P values were calculated using 2-way analysis of variance (ANOVA).
We also assessed the broth dilution MIC of P. aeruginosa PA14 (see Table S1 in the supplemental material) in the presence and absence of mucin, and we observed increased MIC values accompanying mucin addition for gentamicin, amikacin, and colistin, but no differences for other antibiotics in the same classes (tobramycin and polymyxin B). Conversely, for tetracycline, mucin actually increased susceptibility by 4-fold. Overall, these data suggested that the observed resistances for most antibiotics (Fig. 1 and 2) was likely due to the adaptation accompanying surfing motility rather than to the presence per se of mucin. For this reason, we investigated these adaptive changes in greater detail.
Surfing-mediated antibiotic resistance is associated with multiple resistome genes.
Transcriptome sequencing (RNA-Seq data) (NCBI GEO accession number GSE110044) revealed that surfing is an adaptation that strongly affected gene expression. RNA-Seq was performed on two different regions of surfing colonies, namely, the thick white edge and the blue-green center, and results were compared to those from swimming cells grown in SCFM medium without mucin. In total, there were 1,467 genes dysregulated at the edge and 2,078 genes dysregulated in the center, with 816 genes commonly dysregulated between the two, consistent with the strong phenotypic differences between the blue-green center and the thick white edge of surfing colonies (11). In particular, these global gene expression data confirmed that the surfing adaptation was considerably different from swarming in that out of the 1,467 genes dysregulated at the edge of the surfing colony, only 215 (14.6%) matched those previously found to be dysregulated during swarming (15), whereas out of the 2,078 dysregulated in the center, 217 (10.5%) overlapped with those from swarming cells. Overall, transcriptome analysis revealed that cells in the center appeared less motile (reduced flagellar gene expression) and generally exhibited lower expression of metabolic genes compared to those at the edge. Furthermore, in the center, quorum sensing appeared to be largely downregulated, especially the Pseudomonas quinolone signal (PQS) system, while the same PQS system was significantly upregulated at the edge of surfing colonies. These factors might have explained the substantial differences in gene expression profiles between edge and center cells. Intrinsic resistance genes, such as those encoding certain efflux proteins (namely, mexX, oprE, and oprG), were upregulated at the edge of surfing colonies.
To examine the possibility that adaptive resistance during surfing motility was due to the dysregulation of genes that influence resistance, literature searches were conducted. This revealed 119 genes that, when mutated, led to increased susceptibility (intrinsic resistance genes) and 252 genes that, when mutated, mediated antibiotic resistance; collectively, these form the resistomes for various antibiotics (16–22). Among the resistome genes, 65 were identified through RNA-Seq gene expression data from surfing cells, that matched the direction of dysregulation of expression levels expected if they were to have a potential role in surfing-mediated resistance. Available transposon mutants of these 65 resistome genes were tested for changes in susceptibility to certain antibiotics.
Table 2 shows the resistome genes dysregulated in the edge and/or center for which transposon mutants showed a change in susceptibility to at least one of the 5 tested antibiotics, based on an initial disk diffusion assay. Several of these genes showed a change in susceptibility to more than one antibiotic, possibly illustrating a contribution to broad-spectrum resistance. The mean zone of inhibition measurements are presented in Table S2 in the supplemental material. Five of the tested mutants, ΔrecG, ΔddaH, ΔarmR, ΔnalC, and ΔPA3667, were similarly dysregulated in the center and edge of a surfing colony, with recG and ddaH both upregulated and armR, nalC, and PA3667 downregulated. Complements of selected resistome mutants showed that this broad-spectrum effect could be significantly reversed either partially, completely or excessively (Table 3), and that overexpression of some of these genes also revealed a change in susceptibility to other antibiotics, as shown in Table 3. Reverse transcription-quantitative PCR (RT-qPCR) data (see Table S3 in the supplemental material) verified the direction of dysregulation shown in the RNA-Seq data for selected resistome genes.
TABLE 2.
Mutated resistome genes and their corresponding changes in antibiotic susceptibility relative to that of the wild type
| Geneb | Gene function(s)c | RNA-Seq fold change |
Antibiotic susceptibility of mutanta | |
|---|---|---|---|---|
| Center | Edge | |||
| armR | Antirepressor for MexR | −3.2 | −5.1 | TOBr, IMIr, PXBr, NFXr, TETr |
| atpB | ATP synthase A chain | −2.1 | NC | TOBr, NFXr |
| braB | Branched-chain amino acid transporter | −4.2 | NC | NFXr |
| ccmF | Cytochrome c-type biogenesis protein | −2.2 | NC | TOBr, PXBr, NFXr |
| ccoO1 | Cytochrome c oxidase, cbb3-type, CcoO subunit | −3.5 | NC | TOBr, NFXr |
| clpS | ATP-dependent Clp protease adaptor protein | NC | −2.3 | TOBr, IMIr, PXBr, NFXr |
| cycH | Cytochrome c-type biogenesis protein | NC | 2.2 | TOBs, PXBs |
| ddaH | Dimethylarginine dimethylaminohydrolase | 4.9 | 3.4 | IMIs, TETs |
| etfA | Electron transfer flavoprotein alpha subunit | −6.2 | NC | TOBr, PXBr, NFXr |
| gidA | Glucose-inhibited division protein A | −2.2 | NC | NFXr |
| htpX | Heat shock protein | −2.0 | NC | NFXr |
| mutS | DNA mismatch repair protein | −2.5 | NC | TOBr, PXBr |
| nalC | Transcriptional regulator | −5.3 | −2.7 | TOBr, PXBr, NFXr |
| nuoB | NADH dehydrogenase I chain B | −2.8 | NC | TOBr, IMIr, PXBr, NFXr |
| nuoF | NADH dehydrogenase I chain F | −2.4 | NC | TOBr |
| nuoG | NADH dehydrogenase I chain G | −2.1 | NC | TOBr, NFXr |
| PA1348 | Hypothetical protein | NC | −3.4 | IMIr, NFXr |
| PA1428 | Conserved hypothetical protein | −3.4 | NC | TOBr, NFXr |
| PA1513 | Hypothetical protein | NC | −3.0 | TETr |
| PA2047 | Probable transcriptional regulator | NC | −2.0 | TOBr, NFXr |
| PA2566 | Conserved hypothetical protein | NC | −5.0 | NFXr |
| PA2571 | Probable two-component sensor | NC | −2.7 | TOBr |
| PA3233 | Hypothetical protein | 2.5 | NC | NFXs |
| PA3576 | Hypothetical protein | NC | −2.9 | TOBr, NFXr, TETr |
| PA3667 | Probable pyridoxal-phosphate dependent enzyme | −1.7 | −2.5 | TETr |
| PA4292 | Probable phosphate transporter | −6.7 | NC | TOBr, IMIr, PXBr, NFXr |
| PA4429 | Probable cytochrome c1 precursor | −2.3 | NC | TOBr, PXBr, NFXr |
| PA4766 | Conserved hypothetical protein | NC | −2.3 | TOBr |
| PA4781 | Cyclic di-GMP phosphodiesterase | NC | −2.9 | TOBr, NFXr |
| PA5130 | Conserved hypothetical protein | NC | 2.4 | TOBs, IMIs, PXBs, NFXs, TETs |
| pchF | Pyochelin synthetase | −2.2 | NC | TOBr |
| pckA | Phosphoenolpyruvate carboxykinase | −2.6 | NC | TOBr |
| recG | ATP-dependent DNA helicase | 1.9 | 2.1 | TOBs, PXBs, NFXs, TETs |
| rph | RNase PH | −2.3 | NC | TOBr |
| serA | d-3-phosphoglycerate dehydrogenase | −12.4 | NC | TOBr, PXBr, NFXr |
| thiG | Thiamine biosynthesis protein, thiazole moiety | −2.9 | NC | IMIr, NFXr |
Relative to that of the wild type (WT). IMI, imipenem; TET, tetracycline; PXB, polymyxin B; TOB, tobramycin; NFX, norfloxacin; r, resistant; s, supersusceptible.
This list included 8 resistome genes that were similarly regulated in both the center and edge. A further 10 resistome genes that were dysregulated only at the edge of a surfing colony were affected in such a way as to influence resistance or susceptibility. Twenty resistome genes that were dysregulated only in the center of a surfing colony were affected in such a way as to influence antibiotic resistance or susceptibility.
See reference 23.
TABLE 3.
Impact of complementation of selected resistome mutants on antibiotic susceptibilitya
| Antibiotic concn (μg/disk) | Strain | Zone of inhibition (mm)b |
||||
|---|---|---|---|---|---|---|
| Imipenem | Tetracycline | Polymyxin B | Tobramycin | Norfloxacin | ||
| 10 | Wild-type | 5.7 | 5.0 | 5.6 | 3.3 | 1.0 |
| ΔrecG | 7.3 | 8.7c | 9.7d | 12.5f | 7.3f | |
| recG+ | 6.0 | 5.7 | 3.7 | 6.7c | 3.0 | |
| ΔddaH | 9.0d | 0e | 5.3 | 3.0 | 2.3 | |
| ddaH+ | 6.3 | 3.3 | 5.7 | 6.3 | 2.0 | |
| 100 | Wild-type | 12.3 | 6.7 | 10.3 | 12.0 | 14.7 |
| ΔPA1428 | 12.7 | 7.7 | 8.0 | 7.0e | 0.0f | |
| PA1428+ | 9.0c | 7.3 | 9.0 | 11.6 | 13.3 | |
| ΔPA2047 | 12.3 | 7.0 | 5.7 | 7.3d | 9.7e | |
| PA2047+ | 9.7 | 7.0 | 7.3 | 11.3 | 12.0 | |
| ΔthiG | 6.3f | 6.7 | 7.0 | 8.7 | 10.3d | |
| thiG+ | 9.0c | 8.7 | 8.3 | 11.0 | 15.0 | |
| ΔatpB | 9.7 | 4.0 | 8.0 | 8.3c | 9.7e | |
| atpB+ | 10.3 | 7.3 | 8.3 | 12.0 | 14.0 | |
| ΔPA3667 | 15.7 | 0.0f | 7.7 | 10.0 | 12.0 | |
| PA3667+ | 12.0 | 9.3 | 11.3c | 11.0 | 9.7e | |
| ΔPA3576 | 12.0 | 3.0c | 6.0 | 8.3c | 10.7c | |
| PA3576+ | 10.7 | 6.3 | 7.3 | 9.0c | 12.3 | |
| ΔPA3721 | 10 | 2d | 14.5f | 0f | 10d | |
| PA3721+ | 11.7 | 8.3 | 9.3 | 11.3 | 13.0 | |
| ΔclpS | 8.3c | 6.3 | 15f | 6.7e | 8.3f | |
| clpS+ | 11.3 | 13.0f | 11.5 | 11.7 | 12.3 | |
| ΔarmR | 0f | 0f | 1.0f | 6.3f | 0f | |
| armR+ | 12.3 | 12.3f | 10.3 | 12.7 | 15.0 | |
Results show the zone of inhibition (n = 3) of each mutant and that of its complemented equivalent against five antibiotics compared with that of the wild type (n = 6). Mutants of upregulated resistome genes were tested against 10 μg/disk of antibiotic and downregulated against 100 μg/disk of antibiotic. Standard deviations ranged between 0 and 2.5 mm.
Statistical significance relative to the wild type was determined using two-way ANOVA.
P < 0.5.
P < 0.01.
P < 10−3.
P < 10−4.
DISCUSSION
P. aeruginosa is a highly adaptable organism that exhibits diverse lifestyles, from coordinated forms of motility, like swarming, to community-based sessile structures, like biofilms. Previously, we described a new form of P. aeruginosa motility, known as surfing, under artificial cystic fibrosis-like conditions where the mucin content is high (11). Here we demonstrated that this novel form of motility is associated with multidrug adaptive resistance and is a complex adaptation influencing the expression of hundreds of genes. Both disk diffusion and antibiotic incorporation assays revealed that cells undergoing surfing were significantly more resistant to multiple antibiotics compared to those undergoing swimming, and the same concentrations of antibiotics that completely abolished swimming were found to be much less effective against surfing.
Antibiotic susceptibility was generally unaffected by the presence of mucin in the presence of high agar concentrations at which swimming and surfing do not occur, indicating that mucin had a minimal effect on the diffusion of most antibiotics (Table 1). For tobramycin, mucin significantly increased susceptibility, consistent with an enhancement of antibiotic diffusion, the opposite effect of mucin-mediated adaptive resistance under surfing conditions. For amikacin, the effect was opposite, with mucin mediating increased resistance but not to the extent of surfing motility conditions, which led to complete resistance. For ciprofloxacin, the result was more equivocal, since mucin under both high agar and surfing conditions led to partial resistance. Interestingly, norfloxacin, in the same antibiotic fluoroquinolone class, showed significant resistance only under surfing conditions. MIC assays also revealed that the observed adaptive resistance was dependent on surface growth associated with surfing adaption and not due merely to the presence of mucin. Indeed, experiments measuring the resistance of surfing and swimming colonies to antibiotics incorporated into agar plates not only confirmed that surfing cells were considerably more resistant to the 5 tested antibiotics (Fig. 2), but also that surfing cells could grow at concentrations above the liquid MICs in the presence of mucin (Table S1 in the supplemental material), which is again consistent with the concept of adaptive resistance.
RNA-Seq data on cells collected from the center of a surfing colony also revealed 10 genes—ccoO1, atpB, nuoB, PA4429, eftA, serA, ccmF, thiG, nuoF, and pckA—involved in metabolism and energy production that were downregulated, and for which mutants exhibited increased resistance to certain antibiotics. Three of these genes, ccoO1, atpB, and PA4429, have also been shown to be dysregulated under swarming conditions, as discussed previously (15, 24). Mutants for these 10 metabolic genes that were downregulated at the center of surfing colonies showed an increased resistance to norfloxacin and/or tobramycin. Aminoglycosides are taken up by energy-dependent mechanisms (25), and reduced metabolic activities have previously been shown to contribute to resistance to tobramycin in P. aeruginosa biofilms (26). Although norfloxacin has been shown to affect animal metabolism through interactions with cytochrome P450 (27), it has not been shown to affect metabolism in P. aeruginosa. Here, we have demonstrated that reduced expression levels of certain metabolic resistome genes in the center of the surfing colony may contribute to adaptive resistance against tobramycin and/or norfloxacin.
To explain the mechanisms behind surfing-mediated resistance, we explored the contribution of resistome genes found to be dysregulated in surfing through RNA-Seq and transposon mutant screens. In total, 36 resistome genes were identified as dysregulated under surfing conditions and that exhibited a change in susceptibility to certain antibiotics when mutated. Among the 36 resistome genes, there were 5 that showed the same direction of dysregulation (i.e., both down or upregulated) in both the center and edge of a surfing colony. RecG and ddaH were both upregulated in the surfing center and edge, and their mutants exhibited similarly reduced resistance to tetracycline. The mutant in recG (encoding an ATP-dependent DNA helicase) also exhibited increased susceptibility to polymyxin B, tobramycin, and norfloxacin. Tetracycline and tobramycin target protein synthesis through the 30S ribosomal submit, while polymyxin B targets the cell membrane, and norfloxacin targets DNA replication. The broad-spectrum activity observed by recG as a resistome gene against such diverse antibiotics may arise from its regulatory nature, in that recG transcriptionally regulates OxyR-controlled genes in Pseudomonas putida (28). Genes identified in the RecG regulon of P. putida included those encoding porins (oprE, oprD, and PP0883) and thioredoxin reductase (trxB), which is involved in stress coping mechanisms (28).
There were 3 genes, armR, nalC, and PA3667, that were downregulated in both regions of the surfing colony. NalC is known to negatively regulate the expression of armR, and ArmR inhibits the DNA-binding activity of MexR (29, 30). MexR negatively regulates expression of the mexAB-oprM operon, which encodes a major efflux pump in P. aeruginosa that is intrinsically involved in broad-spectrum antibiotic resistance (29). ArmR allosterically binds to MexR to alleviate its repression on the mexAB-oprM operon (29). Interestingly, Starr et al. revealed that a knockout mutant of armR still exhibited increased expression levels of the mexAB-oprM operon under certain conditions (30). Here, we showed that mutants in armR and nalC, which are both downregulated in surfing conditions, exhibited similar increases in resistance to tobramycin, norfloxacin, and polymyxin B. The observed increases in resistance to these antibiotics might be attributed in part to increased expression levels of the mexAB-oprM operon.
There were 11 genes dysregulated at the edge and 20 genes dysregulated at the center of a surfing colony that exhibited a change in susceptibility to at least one of the tested antibiotics when mutated compared to the wild type. PA5130 was a conserved hypothetical protein that was found to be upregulated in the surfing edge and that exhibited an increased susceptibility to all 5 of the tested antibiotics when mutated. The ATP-dependent protease adapter clpS, which was downregulated at the edge, exhibited significant increases in resistance to imipenem, polymyxin B, tobramycin, and norfloxacin. ClpS has been previously shown by our lab to contribute to antibiotic resistance, biofilm formation, and swarming motility (31). More specifically, a transposon mutant variant of clpS was observed to have increased resistance to β-lactams through the increased expression of β-lactamase (31). Here, it was shown that clpS also had an effect on resistance to imipenem, polymyxin B, tobramycin, and norfloxacin under surfing conditions.
Swarming is another complex form of motility exhibited by P. aeruginosa found to be involved with major transcriptional changes (15, 32), which are substantially distinct from the transcriptional profile of surfing cells. Swarming cells have also previously been shown to be resistant to multiple antibiotics, including polymyxin B, ciprofloxacin, and gentamicin, and pvdQ mutants influenced swarming-specific resistance (15, 32). Here, surfing was also found to be associated with resistance against these same antibiotics and several others. Among the resistome genes identified in this study to be dysregulated under surfing conditions that showed contributions to adaptive antibiotic resistance (Table 2), pchF (15, 24), atpB, ccoO1, and PA4429 (24) were also shown to be dysregulated under swarming conditions (15, 24). However, our preliminary studies of the swarming resistome (S.R. Coleman and R.E.W. Hancock, unpublished data) have indicated major differences compared to the surfing-associated resistome described here, and less than 15% of dysregulated genes were common in the two motility adaptations. Thus, the mechanistic overlap in swarming- and surfing-mediated adaptive resistance would appear to be minimal.
The biofilm growth state also leads to adaptive resistance (33, 34). Biofilms represent a very different adaptation being sessile rather than motile communities. Our preliminary analyses of gene expression in biofilm bacteria have suggested that there are considerable differences compared to surfing bacteria (D. Pletzer, E. Sun and R.E.W. Hancock, unpublished data) with only 22 to 34% common dysregulated genes, and thus we would again anticipate that different adaptations were involved in resistance. One possible overlap would be nalC, identified here as being dysregulated in surfing and as mediating surfing-associated polymyxin B, tobramycin and norfloxacin resistance. Mutants in nalC were found in biofilms formed by clinical strains of P. aeruginosa isolated from prosthetic valves, and the associated isolates were resistant to fluoroquinolones and carbapenems (34). Other surfing resistome genes identified in our study included nuoB, nuoF, and nuoG. The nuo operon, nuoA–N, which is involved in nitrate sensing, has previously been shown to be activated during biofilm formation and is important for regulating biofilm formation, as well as motility (35, 36), but its role in biofilm-mediated adaptive resistance was not studied. The other genes identified in this study as important for surfing-mediated resistance have not yet been shown to be involved in biofilm formation or related resistance.
In conclusion, surfing motility is a novel form of motility that results in a mucin-triggered lifestyle adaptation. Here, we demonstrated how surfing cells exhibit increased resistance, which can be attributed to a variety of transcriptomic changes resulting from that adaptation.
MATERIALS AND METHODS
Bacterial strains and complements.
All screens and assays were done using the Pseudomonas aeruginosa UCBPP-PA14 (37) wild-type strain. All mutants used were derivatives of this strain and were obtained from the PA14 Transposon Insertion Mutant Library (38). Complemented mutants were generated as follows. PCR primers listed in Table S3 were used to amplify the desired genes from strain PA14 genomic DNA. The amplified products were cloned into a TOPO vector using the Zero Blunt TOPO PCR cloning kit (Invitrogen). TOPO vectors containing amplified product were digested using two different enzymes, which differed depending on the gene of interest, and ligated into a pUCP18 vector containing the lac promoter. Vectors containing the desired genes were then transformed into their respective mutants.
Disk diffusion assay.
Disk diffusion assays were performed on synthetic cystic fibrosis media (SCFM) (39), which was prepared as described by Palmer et al. (39) without ammonia and with 0.3% agar and 0.4% (wt/vol) mucin (surfing conditions), or with 0.3% agar without mucin (swimming conditions). Control disk diffusion assays were performed on SCFM 1.5% agar with and without 0.4% mucin. Bacterial strains were grown in Luria broth (LB; Difco) liquid medium overnight and then subcultured to mid-log phase (optical density at 600 nm [OD600], 0.4 to 0.5). To assay motility, mid-log-phase cultures were spotted on agar surfaces at four points around an antibiotic disk (Fig. S1) impregnated with 10 μl of antibiotic at concentrations indicated in Table S5 in the supplemental material. Agar plates were air dried at 37°C for 30 min before inoculation and application of antibiotic disks. Once inoculated, plates were incubated at 37°C for 15 to 18 h. The zone of inhibition surrounding the antibiotic disk was measured in millimeters using a ruler. In the case of asymmetric zones of inhibition, the average of the four sides was taken. Disk diffusion controls or growth controls were spread as lawns on plates, and antibiotic disks were applied to the center. Two-way analysis of variance (ANOVA) was used to determine if any significant difference existed between surfing and swimming conditions. All statistical analysis was done using GraphPad Prism 7.
Antibiotic incorporation assay.
Incorporation assays were done on SCFM (39) using 0.3% agar with 0.4% mucin (surfing conditions) and 0.3% agar without mucin (swimming conditions). Antibiotics were added to the agar before solidification. Once hardened, plates were air dried for 30 min at 37°C before being inoculated with 1 μl of a subculture at an OD600 of 0.4 to 0.5. Plates were incubated at 37°C for 15 to 18 h. Spot inoculation involved stabbing bacteria midway into the agar. The percentage of area growth on the plates was measured using ImageJ. Two-way ANOVA was used to determine if significant differences occurred between the two conditions (surfing and swimming) and between concentrations for surfing.
Liquid MIC.
Liquid MIC assays were conducted as described by Wiegand et al. (20). This assay was performed in liquid SCFM (39) with and without 0.4% mucin. An inoculum of 2 to 7 ×105 cells was used. Significant differences between MICs were taken as a 3-fold or greater change.
RNA-Seq.
PA14 was grown in liquid LB medium overnight and subcultured to an OD600 of 0.4 to 0.5. Mid-log-phase cultures were used to inoculate SCFM (39) surfing and swimming plates, prepared as described above. Surfing plates were air dried for 30 min before inoculation with 1 μl of culture and incubation at 37°C for 15 to 18 h. Using sterile swabs, cells from the center and edge of a surfing colony and center of a swimming colony were collected into RNA Protect bacteria reagent (Qiagen). RNA extraction was conducted using an RNeasy minikit (Qiagen) according to the manufacturer's protocol. DNase treatment was performed using a Turbo DNA-free kit (Thermo Fisher), and rRNA depletion was performed using a RiboZero bacteria kit (Illumina). Single-end cDNA libraries were constructed using a Kapa stranded total RNA kit (Kapa Biosystems), and libraries were sequenced on an Illumina HiSeq 2500 instrument in rapid run mode with 100-bp reads that were base called and demultiplexed using built-in software on the sequencer. Fastq file quality control was performed using FastQC v0.11.5 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and MultiQC v0.8.dev0 (40). Fastq files were aligned to the UCBPP-PA14 genome (GenBank gene annotations) using Bowtie 2 (41). Bam-sam file conversion and sorting were performed with samtools (42). Read count tables were generated with htseq-count v2.5 (43). Differential expression analysis was performed using DESeq2 (44). Fold changes in surfing were calculated relative to swimming. Gene annotations were taken from the Pseudomonas Genome Database (23).
RT-qPCR.
RNA was collected as described for RNA-Seq. Reaction samples were prepared using the qScript one-step SYBR green RT-qPCR kit (QuantaBio) with 5 ng of RNA per 25 μl reaction mixture and amplified in a Roche LightCycler 96. Quantification analysis was done using the comparative CT method (45), using rpoD as the normalizing gene. All primers used for RT-qPCR are listed in Table S4 in the supplemental material.
Accession number(s).
Transcriptome sequences of gene expression in Pseudomonas aeruginosa UCBPP-PA14 grown in three conditions have been deposited in NCBI GEO under the accession number GSE110044.
Supplementary Material
ACKNOWLEDGMENTS
Research reported in this publication was supported by a Foundation grant from the Canadian Institutes for Health Research FDN-154287 and by a grant from Cystic Fibrosis (CF) Canada (award 2585). The content is solely the responsibility of the authors and does not necessarily represent the official views of the Canadian Institutes for Health Research. R.E.W.H. holds a Canada Research Chair in Health and Genomics and a UBC Killam Professorship.
E.S. performed antibiotic screens, resistome screens, generation of complements, and manuscript writing. E.E.G. analyzed and processed RNA-Seq data. R.F. prepared RNA samples for RNA-Seq. E.E.G. and R.F. also contributed to manuscript writing. N.L. contributed to the generation of complemented strains and performed liquid MIC assays. Conceptualization, acquisition of funding, discussion of results and extensive editing and review of the manuscript was performed by R.E.W.H.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00848-18.
REFERENCES
- 1.Bassetti M, Merelli M, Temperoni C, Astilean A. 2013. New antibiotics for bad bugs: where are we? Ann Clin Microbiol Antimicrob 12:22. doi: 10.1186/1476-0711-12-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ventola CL. 2015. The antibiotic resistance crisis: part 1: causes and threats. Pharm Ther 40:277–283. [PMC free article] [PubMed] [Google Scholar]
- 3.WHO. 2014. Antimicrobial resistance: global report on surveillance. 2014. WHO, Geneva, Switzerland: http://www.who.int/drugresistance/documents/surveillancereport/en/. [Google Scholar]
- 4.Garau J, Nicolau DP, Wullt B, Bassetti M. 2014. Antibiotic stewardship challenges in the management of community-acquired infections for prevention of escalating antibiotic resistance. J Glob Antimicrob Resist 2:245–253. doi: 10.1016/j.jgar.2014.08.002. [DOI] [PubMed] [Google Scholar]
- 5.Bennett JV. 1974. Nosocomial infections due to Pseudomonas. J Infect Dis 130:S4–S7. doi: 10.1093/infdis/130.Supplement.S4. [DOI] [PubMed] [Google Scholar]
- 6.van Ewijk BE, Wolfs TFW, Fleer A, Kimpen JLL, van der Ent CK. 2006. High Pseudomonas aeruginosa acquisition rate in CF. Thorax 61:641–642. doi: 10.1136/thx.2006.062372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taylor PK, Yeung ATY, Hancock REW. 2014. Antibiotic resistance in Pseudomonas aeruginosa biofilms: Towards the development of novel anti-biofilm therapies. J Biotechnol 191:121–130. doi: 10.1016/j.jbiotec.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 8.Breidenstein EBM, de la Fuente-Núñez C, Hancock REW. 2011. Pseudomonas aeruginosa: all roads lead to resistance. Trends Microbiol 19:419–426. doi: 10.1016/j.tim.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 9.Sousa AM, Pereira MO. 2014. Pseudomonas aeruginosa diversification during infection development in cystic fibrosis lungs—a review. Pathogens 3:680–703. doi: 10.3390/pathogens3030680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lewis K. 2001. Riddle of Biofilm Resistance. Antimicrob Agents Chemother 45:999–1007. doi: 10.1128/AAC.45.4.999-1007.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yeung ATY, Parayno A, Hancock REW. 2012. Mucin promotes rapid surface motility in Pseudomonas aeruginosa. mBio 3:e00073-12. doi: 10.1128/mBio.00073-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jain R, Kazmierczak BI. 2014. A conservative amino acid mutation in the master regulator FleQ renders Pseudomonas aeruginosa aflagellate. PLoS One 9:e97439. doi: 10.1371/journal.pone.0097439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harshey RM. 2003. Bacterial motility on a surface: many ways to a common goal. Annu Rev Microbiol 57:249–273. doi: 10.1146/annurev.micro.57.030502.091014. [DOI] [PubMed] [Google Scholar]
- 14.Murray TS, Kazmierczak BI. 2008. Pseudomonas aeruginosa exhibits sliding motility in the absence of type IV pili and flagella. J Bacteriol 190:2700–2708. doi: 10.1128/JB.01620-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Overhage J, Bains M, Brazas MD, Hancock REW. 2008. Swarming of Pseudomonas aeruginosa is a complex adaptation leading to increased production of virulence factors and antibiotic resistance. J Bacteriol 190:2671–2679. doi: 10.1128/JB.01659-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Breidenstein EBM, Khaira BK, Wiegand I, Overhage J, Hancock REW. 2008. Complex ciprofloxacin resistome revealed by screening a Pseudomonas aeruginosa mutant library for altered susceptibility. Antimicrob Agents Chemother 52:4486–4491. doi: 10.1128/AAC.00222-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fernández L, Gooderham WJ, Bains M, McPhee JB, Wiegand I, Hancock REW. 2010. Adaptive resistance to the “last hope” antibiotics polymyxin B and colistin in Pseudomonas aeruginosa is mediated by the novel two-component regulatory system ParR-ParS. Antimicrob Agents Chemother 54:3372–3382. doi: 10.1128/AAC.00242-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alvarez-Ortega C, Wiegand I, Olivares J, Hancock REW, Martínez JL. 2010. Genetic determinants involved in the susceptibility of Pseudomonas aeruginosa to beta-lactam antibiotics. Antimicrob Agents Chemother 54:4159–4167. doi: 10.1128/AAC.00257-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schurek KN, Marr AK, Taylor PK, Wiegand I, Semenec L, Khaira BK, Hancock REW. 2008. Novel genetic determinants of low-level aminoglycoside resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother 52:4213–4219. doi: 10.1128/AAC.00507-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wiegand I, Marr AK, Breidenstein EBM, Schurek KN, Taylor P, Hancock REW. 2008. Mutator genes giving rise to decreased antibiotic susceptibility in Pseudomonas aeruginosa. Antimicrob Agents Chemother 52:3810–3813. doi: 10.1128/AAC.00233-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dötsch A, Becker T, Pommerenke C, Magnowska Z, Jänsch L, Häussler S. 2009. Genomewide identification of genetic determinants of antimicrobial drug resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother 53:2522–2531. doi: 10.1128/AAC.00035-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gallagher LA, Shendure J, Manoil C. 2011. Genome-scale identification of resistance functions in Pseudomonas aeruginosa using Tn-seq. mBio 2:e00315-10. doi: 10.1128/mBio.00315-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Winsor GL, Griffiths EJ, Lo R, Dhillon BK, Shay JA, Brinkman FSL. 2016. Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas Genome Database. Nucleic Acids Res 44:D646–D653. doi: 10.1093/nar/gkv1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tremblay J, Déziel E. 2010. Gene expression in Pseudomonas aeruginosa swarming motility. BMC Genomics 11:587. doi: 10.1186/1471-2164-11-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bryan LE, Kwan S. 1983. Roles of ribosomal binding, membrane potential, and electron transport in bacterial uptake of streptomycin and gentamicin. Antimicrob Agents Chemother 23:835–845. doi: 10.1128/AAC.23.6.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walters SM, Dubey VS, Jeffrey NR, Dixon DR. 2010. Antibiotic-induced Porphyromonas gingivalis LPS release and inhibition of LPS-stimulated cytokines by antimicrobial peptides. Peptides 31:1649–1653. doi: 10.1016/j.peptides.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 27.McLellan RA, Drobitch RK, Monshouwer M, Renton KW. 1996. Fluoroquinolone antibiotics inhibit cytochrome P450-mediated microsomal drug metabolism in rat and human. Drug Metab Dispos 24:1134–1138. [PubMed] [Google Scholar]
- 28.Yeom J, Lee Y, Park W. 2012. ATP-dependent RecG helicase is required for the transcriptional regulator OxyR function in Pseudomonas species. J Biol Chem 287:24492–24504. doi: 10.1074/jbc.M112.356964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilke MS, Heller M, Creagh AL, Haynes CA, McIntosh LP, Poole K, Strynadka NCJ. 2008. The crystal structure of MexR from Pseudomonas aeruginosa in complex with its antirepressor ArmR. Proc Natl Acad Sci U S A 105:14832–14837. doi: 10.1073/pnas.0805489105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Starr LM, Fruci M, Poole K. 2012. Pentachlorophenol induction of the Pseudomonas aeruginosa mexAB-oprM efflux operon: involvement of repressors NalC and MexR and the antirepressor ArmR. PLoS One 7:e32684. doi: 10.1371/journal.pone.0032684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fernández L, Breidenstein EBM, Song D, Hancock REW. 2012. Role of intracellular proteases in the antibiotic resistance, motility, and biofilm formation of Pseudomonas aeruginosa. Antimicrob Agents Chemother 56:1128–1132. doi: 10.1128/AAC.05336-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang L, Zhang C, Gong F, Li H, Xie X, Xia C, Chen J, Song Y, Shen A, Song J. 2013. Influence of Pseudomonas aeruginosa pvdQ gene on altering antibiotic susceptibility under swarming conditions. Curr Microbiol 66:152–161. doi: 10.1007/s00284-012-0217-1. [DOI] [PubMed] [Google Scholar]
- 33.Høiby N, Bjarnsholt T, Givskov M, Molin S, Ciofu O. 2010. Antibiotic resistance of bacterial biofilms. Int J Antimicrob Agents 35:322–332. doi: 10.1016/j.ijantimicag.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 34.Domitrovic TN, Hujer AM, Perez F, Marshall SH, Hujer KM, Woc-Colburn LE, Parta M, Bonomo RA. 2016. Multidrug resistant Pseudomonas aeruginosa causing prosthetic valve endocarditis: a genetic-based chronicle of evolving antibiotic resistance. Open Forum Infect Dis 3:ofw188. doi: 10.1093/ofid/ofw188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Alst NE, Picardo KF, Iglewski BH, Haidaris CG. 2007. Nitrate sensing and metabolism modulate motility, biofilm formation, and virulence in Pseudomonas aeruginosa. Infect Immun 75:3780–3790. doi: 10.1128/IAI.00201-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Southey-Pillig CJ, Davies DG, Sauer K. 2005. Characterization of temporal protein production in Pseudomonas aeruginosa biofilms. J Bacteriol 187:8114–8126. doi: 10.1128/JB.187.23.8114-8126.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rahme LG, Stevens EJ, Wolfort SF, Shao J, Tompkins RG, Ausubel FM. 1995. Common virulence factors for bacterial pathogenicity in plants and animals. Science 268:1899–1902. doi: 10.1126/science.7604262. [DOI] [PubMed] [Google Scholar]
- 38.Liberati NT, Urbach JM, Miyata S, Lee DG, Drenkard E, Wu G, Villanueva J, Wei T, Ausubel FM. 2006. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc Natl Acad Sci U S A 103:2833–2838. doi: 10.1073/pnas.0511100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Palmer KL, Aye LM, Whiteley M. 2007. Nutritional cues control Pseudomonas aeruginosa multicellular behavior in cystic fibrosis sputum. J Bacteriol 189:8079–8087. doi: 10.1128/JB.01138-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ewels P, Magnusson M, Lundin S, Käller M. 2016. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32:3047–3048. doi: 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anders S, Pyl PT, Huber W. 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmittgen TD, Livak KJ. 2008. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.