Antimalarial drug resistance developed in Plasmodium falciparum has become a problem for malaria control. Evaluation of drug resistance is the first step for effective malaria control.
KEYWORDS: Plasmodium falciparum, antimalarial drug, artemisinin, resistance, polymorphism, haplotype
ABSTRACT
Antimalarial drug resistance developed in Plasmodium falciparum has become a problem for malaria control. Evaluation of drug resistance is the first step for effective malaria control. In this study, we investigated the gene mutations of P. falciparum using blood samples from returned Chinese migrant workers in order to identify drug resistance-associated molecular markers. These workers returned from Africa and Southeast Asia (SEA) during 2011 to 2016. Polymorphisms in pfcrt, pfmdr1, and k13-propeller genes and the haplotype patterns of Pfcrt and Pfmdr1 were analyzed. The results showed the presence of four haplotypes of Pfcrt codons 72 to 76, including CVMNK (wild type), SVMNT and CVIET (mutation types), and CV M/I N/E K/T (mixed type), with 50.57%, 1.14%, 25.00%, and 23.30% prevalence, respectively. For Pfmdr1, N86Y (22.28%) and Y184F (60.01%) were the main prevalent mutations (mutations are underlined). The prevalence of mutation at position 550, 561, 575, and 589 of K13-propeller were 1.09%, 0.54%, 0.54%, and 0.54%, respectively. These data suggested that Pfcrt, Pfmdr1, and K13-propeller polymorphisms are potential markers to assess drug resistance of P. falciparum in China, Africa, and SEA.
INTRODUCTION
Malaria is a life-threatening infectious disease caused by Plasmodium parasites. It is prevalent in the tropics and subtropics, especially in sub-Saharan Africa and Southeast Asia (SEA). It is estimated that approximately 216 million cases and 445,000 deaths due to malaria occurred worldwide in 2016, the majority of which were found in Africa and SEA (1). Although the global incidence and mortality of malaria were decreased in recent years (1), potential threats of pathogenic Plasmodium infections are persistent due to increasing population mobility (2–4). In China, imported malaria has been increased in recent years, mainly due to returning overseas workers from the regions of Africa and SEA (5). This is a challenge for the 2020 goal of eliminating malaria in China (2, 3). Emerging drug resistance/tolerance in Plasmodium falciparum has posed an additional threat. Surveillance of multidrug-resistant falciparum malaria would be a critical step to control malaria (6, 7).
It has been documented that P. falciparum has developed drug resistance/tolerance to nearly all currently used antimalarial drugs, including chloroquine (CQ) and artemisinin (ART) (8–10). Isolates with P. falciparum CQ-resistance (CQR) were originally detected in Thailand and Columbia in the early 1960s (8) and then in Africa (11). Currently, artemisinin-based combination therapies (ACTs) are considered the first-line antimalarial drugs for malaria treatment. Although ACTs are commonly used in Africa, SEA, South America, and China, various antimalarial resistances are not to be ignored. Recently, several studies detected ART resistance in SEA (12, 13). In comparison with P. falciparum isolates from SEA, the isolates from the China-Myanmar border are probably much more resistant (14). As a potential molecular marker for ART resistance in SEA, the k13-propeller gene (PlasmoDB PF3D7_1343700) was also reported in Africa (13, 15). Surveillance of imported malaria, particularly multidrug-resistant falciparum malaria, would be a primary mission in the process of controlling and eliminating malaria (6, 7).
Genetic alterations, such as those in pfcrt and pfmdr1 genes, have been used as drug resistance molecular markers (3, 16–18). Several Pfcrt mutations at codons 72 to 76 are associated with CQR in P. falciparum isolates from Africa, SEA, and South America (18–20). Some Pfmdr1 mutations are associated with the resistance to CQ (17), mefloquine, quinine, and halofantrine (21). Polymorphisms of K13-propeller, among which four drug resistance-associated mutations (C580Y, R539T, I543T, and Y493H) have been verified in Asia (12), are associated with drug resistance (12, 13). K13-propeller has been identified as a key causal determinant of ART resistance in SEA.
In this study, we investigated the mutations/polymorphisms in pfcrt, pfmdr1, and k13-propeller genes of P. falciparum imported from Africa and SEA to Wuhan, Central China. Our findings may provide a clue to prevent the spread of drug-resistant P. falciparum in Africa, SEA, and China.
RESULTS
General information.
A total of 230 migrant workers returned from Africa and Southeast Asia were diagnosed as malaria patients during 2011 to 2016, 211 with uncomplicated P. falciparum infections, 8 with Plasmodium vivax infections, 7 with Plasmodium ovale infections, 3 with Plasmodium malariae infections, and 1 P. falciparum and Toxoplasma gondii mixed infection from Uganda. Blood samples from 211 uncomplicated P. falciparum infections were collected from 85, 49, 47, 20, 4, and 6 patients returning from West Africa, South Africa, Central Africa, East Africa, North Africa and SEA, respectively. The samples from the area where malaria is endemic in West Africa, South Africa, Central Africa and East Africa accounted for 95.26% (201/211) of the samples, and a combination of Angola (13.74%, 29/211), Nigeria (13.74%, 29/211), Congo (10.90%, 23/211), and Liberia (9.95%, 21/211) was responsible for 48.34% (102/211) of the samples. The parasitemia of P. falciparum isolates ranged from 100 to 501,300 asexual parasites/μl, with a geometric mean of 76,181.25 parasites/μl.
Mutation prevalence of Pfcrt and Pfmdr1.
We obtained 209 PCR products for the pfcrt gene in genomic DNA (gDNA) and 176 sequencing results (84.21%, 176/209) from 211 malaria patients with uncomplicated P. falciparum malaria infections (for a list of primers used, see Table 1). The results showed the presence of polymorphisms in Pfcrt at codon 72 to 76 (Fig. 1). Collectively, 73.86% (130/176) of isolates carry the Pfcrt K76 allele in Africa (Table 2 and 3). There were four haplotypes of Pfcrt coding amino acids 72 to 76, including CVMNK (wild type), SVMNT and CVIET (mutation types), and CV M/I N/E K/T (mixed type), with 50.57%, 1.14%, 25.00%, and 23.30% prevalence, respectively (mutations are underlined). For patients with cerebral malaria, the haplotypes of Pfcrt were 62.5% (5/8) CVMNK, 25% (2/8) CVIET, and the haplotypes of 12.5% (1/8) were undetected. The Pfcrt haplotype CVIET was identified in three out of four samples from patients with recrudescence; the haplotype of the remaining one sample was undetected. For the only death case, the haplotype of Pfcrt was wild type. A considerably decreasing trend in prevalence of the Pfcrt CVIET haplotype (Z = 2.724, P = 0.006) was observed over the survey schedule (Table 3). Prevalence of the CVIET haplotype decreased from 57.14% in 2011 to 28% in 2012 but later increased to 52.17% in 2013 and then finally reduced to 12.24% in 2016 (Table 3).
TABLE 1.
Primers for genotyping pfcrt, pfmdr1, and k13-propeller genes
| Gene (ID) | PCR round | Primer | Sequence (5′–3′) | Primer binding region |
Size (bp) | |
|---|---|---|---|---|---|---|
| Start | End | |||||
| pfcrt (PF3D7_0709000) | Primary | Pfcrt_Outer P1 | CCGTTAATAATAAATACACGCAG | −86 | −64 | 547 |
| Pfcrt_Outer P2 | CGGATGTTACAAAACTATAGTTACC | 436 | 460 | |||
| Secondary | Pfcrt_Inner P1 | TGTGCTCATGTGTTTAAACTT | 307 | 327 | 145 | |
| Pfcrt_Inner P2 | CAAAACTATAGTTACCAATTTTG | 429 | 451 | |||
| pfmdr1 (PF3D7_0523000) | Primary | Pfmdr1(1)-N1F | TTAAATGTTTACCTGCACAACATAGAAAATT | 137 | 167 | 612 |
| Pfmdr1(1)-N1R | CTCCACAATAACTTGCAACAGTTCTTA | 722 | 748 | |||
| Secondary | Pfmdr1(1)-N2F | TGTATGTGCTGTATTATCAGGA | 183 | 204 | 526 | |
| Pfmdr1(1)-N2R | CTCTTCTATAATGGACATGGTA | 687 | 708 | |||
| Primary | Pfmdr1(2)-N1F | AATTTGATAGAAAAAGCTATTGATTATAA | 3019 | 3047 | 880 | |
| Pfmdr1(2)-N1R | TATTTGGTAATGATTCGATAAATTCATC | 3871 | 3898 | |||
| Secondary | Pfmdr1(2)-N2F | GAATTATTGTAAATGCAGCTTTA | 3068 | 3090 | 799 | |
| Pfmdr1(2)-N2R | GCAGCAAACTTACTAACACG | 3847 | 3866 | |||
| k13-propeller (PF3D7_1343700) | Primary | PfK13_outF | GGGAATCTGGTGGTAACAGC | 65 | 84 | 2,097 |
| PfK13_outR | CGGAGTGACCAAATCTGGGA | 2142 | 2161 | |||
| Secondary | PfK13_inF2 | TCAACAATGCTGGCGTATGTG | 1398 | 1418 | 501 | |
| PfK13_inR2 | TGATTAAGGTAATTAAAAGCTGCTCC | 1873 | 1898 | |||
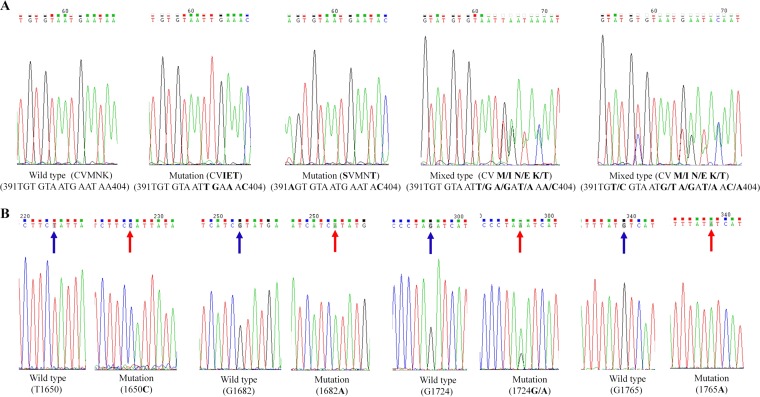
FIG 1.
Sequence profile of PCR products from pfcrt (A) and k13-propeller (B) genes. Shading with blue and red arrows represents the nucleotide wild type and mutation, respectively.
TABLE 2.
Haplotypes of Pfcrt and Pfmdr1 in different countriesa
| Areab | Countryf | Pfcrtc |
Pfmdr1c |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total no. of isolates | WTd (CVMNK) (no. [%]) | Mutation type (no. [%]) |
Mixed type (CV M/I N/E K/T) (%) | Total no. of isolates | WT (NY) (no. [%]) | Mutation type (no. [%]) |
Mixed type (no. [%]) |
||||||
| CVIET | SVMNT | YY | NF | YF | N Y/F | Y Y/F | |||||||
| WA | Nigeria | 25 | 13 (52.00) | 4 (16.00) | 0 (0.00) | 8 (32.00) | 26 | 11 (42.31) | 0 (0.00) | 8 (30.77) | 4 (15.38) | 3 (11.54) | 0 (0.00) |
| Liberia | 15 | 1 (6.67) | 9 (60.00) | 0 (0.00) | 5 (33.33) | 20 | 4 (20.00) | 2 (10.00) | 7 (35.00) | 7 (35.00) | 0 (0.00) | 0 (0.00) | |
| Guinea | 8 | 0 (0.00) | 5 (62.50) | 0 (0.00) | 3 (37.50) | 9 | 2 (22.22) | 0 (0.00) | 6 (66.67) | 0 (0.00) | 0 (0.00) | 1 (11.11) | |
| Sierra Leone | 2 | 1 (50.00) | 1 (50.00) | 0 (0.00) | 0 (0.00) | 6 | 0 (0.00) | 1 (16.67) | 3 (50.00) | 1 (16.67) | 0 (0.00) | 1 (16.67) | |
| Ghana | 4 | 3 (75.00) | 0 (0.00) | 0 (0.00) | 1 (25.00) | 6 | 2 (33.33) | 0 (0.00) | 3 (50.00) | 1 (16.67) | 0 (0.00) | 0 (0.00) | |
| Ivory Coast | 3 | 1 (100.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) | 4 | 0 (0.00) | 0 (0.00) | 4 (100.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) | |
| Benin | 3 | 2 (66.67) | 0 (0.00) | 0 (0.00) | 1 (33.33) | 3 | 1 (33.33) | 0 (0.00) | 1 (33.33) | 1 (33.33) | 0 (0.00) | 0 (0.00) | |
| Niger | 3 | 3 (100.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) | 3 | 2 (66.67) | 0 (0.00) | 0 (0.00) | 0 (0.00) | 1 (33.33) | 0 (0.00) | |
| Burkina Faso | 1 | 1 (100.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) | 1 | 0 (0.00) | 0 (0.00) | 1 (100.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) | |
| Mali | 1 | 0 (0.00) | 0 (0.00) | 0 (0.00) | 1 (100.00) | 1 | 0 (0.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) | 1 (100.00) | 0 (0.00) | |
| Subtotal | 65 | 27 (41.54) | 19 (29.23) | 0 (0.00) | 19 (29.23) | 79 | 22 (27.85) | 3 (3.80) | 33 (41.77) | 14 (17.72) | 5 (6.33) | 2 (2.53) | |
| SA | Angola | 24 | 10 (41.67) | 10 (41.67) | 0 (0.00) | 4 (16.67) | 29 | 13 (44.83) | 2 (6.90) | 9 (31.03) | 0 (0.00) | 3 (10.34) | 2 (6.90) |
| Zambia | 10 | 9 (90.00) | 0 (0.00) | 0 (0.00) | 1 (10.00) | 10 | 6 (60.00) | 0 (0.00) | 4 (40.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) | |
| Mozambique | 9 | 8 (88.89) | 1 (11.11) | 0 (0.00) | 0 (0.00) | 9 | 3 (33.33) | 0 (0.00) | 5 (55.56) | 1 (11.11) | 0 (0.00) | 0 (0.00) | |
| Subtotal | 43 | 27 (62.79) | 11 (25.58) | 0 (0.00) | 5 (11.63) | 48 | 22 (45.83) | 2 (4.17) | 18 (37.50) | 1 (2.08) | 3 (6.25) | 2 (4.17) | |
| CA | Congo | 22 | 12 (54.55) | 5 (22.73) | 0 (0.00) | 5 (22.73) | 20 | 6 (30.00) | 4 (20.00) | 5 (25.00) | 2 (10.00) | 3 (15.00) | 0 (0.00) |
| EG | 11 | 7 (63.64) | 3 (27.27) | 0 (0.00) | 1 (9.09) | 10 | 2 (20.00) | 0 (0.00) | 3 (30.00) | 5 (50.00) | 0 (0.00) | 0 (0.00) | |
| Cameroon | 7 | 6 (85.71) | 0 (0.00) | 0 (0.00) | 1 (14.29) | 6 | 3 (50.00) | 0 (0.00) | 2 (33.33) | 1 (16.67) | 0 (0.00) | 0 (0.00) | |
| Gabon | 3 | 1 (33.33) | 0 (0.00) | 0 (0.00) | 2 (66.67) | 3 | 2 (66.67) | 0 (0.00) | 0 (0.00) | 1 (33.33) | 0 (0.00) | 0 (0.00) | |
| CAR | 1 | 1 (100.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) | 1 | 0 (0.00) | 0 (0.00) | 1 (100.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) | |
| Subtotal | 44 | 27 (61.36) | 8 (18.18) | 0 (0.00) | 9 (20.45) | 40 | 13 (32.50) | 4 (10.00) | 11 (27.50) | 9 (22.50) | 3 (7.50) | 0 (0.00) | |
| EA | Uganda | 5 | 1 (20.00) | 1 (20.00) | 0 (0.00) | 3 (60.00) | 6 | 0 (0.00) | 1 (16.67) | 4 (66.67) | 0 (0.00) | 1 (16.67) | 0 (0.00) |
| South Sudan | 3 | 1 (33.33) | 0 (0.00) | 0 (0.00) | 2 (66.67) | 3 | 2 (66.67) | 0 (0.00) | 1 (33.33) | 0 (0.00) | 0 (0.00) | 0 (0.00) | |
| Tanzania | 4 | 3 (75.00) | 0 (0.00) | 0 (0.00) | 1 (25.00) | 4 | 0 (0.00) | 2 (50.00) | 1 (25.00) | 0 (0.00) | 1 (25.00) | 0 (0.00) | |
| Ethiopia | 2 | 0 (0.00) | 1 (50.00) | 0 (0.00) | 1 (50.00) | 2 | 0 (0.00) | 0 (0.00) | 1 (50.00) | 1 (50.00) | 0 (0.00) | 0 (0.00) | |
| Rwanda | 1 | 1 (100.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) | 1 | 1 (100.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) | |
| Subtotal | 15 | 6 (40.00) | 2 (13.33) | 0 (0.00) | 7 (46.67) | 16 | 3 (18.75) | 3 (18.75) | 7 (43.75) | 1 (6.25) | 2 (12.50) | 0 (0.00) | |
| NA | Sudan | 3 | 1 (33.33) | 1 (33.33) | 0 (0.00) | 1 (33.33) | 3 | 0 (0.00) | 0 (0.00) | 1 (33.33) | 1 (33.33) | 1 (33.33) | 0 (0.00) |
| Libya | 1 | 0 (0.00) | 1 (100.00) | 0 (0.00) | 0 (0.00) | 1 | 0 (0.00) | 0 (0.00) | 0 (0.00) | 1 (100.00) | 0 (0.00) | 0 (0.00) | |
| Subtotal | 4 | 1 (25.00) | 2 (50.00) | 0 (0.00) | 1 (25.00) | 4 | 0 (0.00) | 0 (0.00) | 1 (25.00) | 2 (50.00) | 1 (25.00) | 0 (0.00) | |
| SEA | Indonesia | 3 | 1 (33.33) | 0 (0.00) | 2 (66.67) | 0 (0.00) | 3 | 2 (66.67) | 0 (0.00) | 1 (33.33) | 0 (0.00) | 0 (0.00) | 0 (0.00) |
| Burma | 2 | 0 (0.00) | 2 (100.00) | 0 (0.00) | 0 (0.00) | 2 | 2 (100.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) | |
| Laos | 0 | 0 (—e) | 0 (—) | 0 (—) | 0 (—) | 1 | 1 (100.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) | |
| Subtotal | 5 | 1 (20.00) | 2 (40.00) | 2 (40.00) | 0 (0.00) | 6 | 5 (83.33) | 0 (0.00) | 1 (16.67) | 0 (0.00) | 0 (0.00) | 0 (0.00) | |
| All areas | Total | 176 | 89 (50.57) | 44 (25.00) | 2 (1.14) | 41 (23.30) | 193 | 65 (33.68) | 12 (6.22) | 71 (36.79) | 27 (13.99) | 14 (7.25) | 4 (2.07) |
The haplotypes were constructed considering codon positions 72 to 76 of Pfcrt and codon positions 86 and 184 of Pfmdr1.
WA, SA, CA, EA, NA, and SEA represent West Africa, South Africa, Central Africa, East Africa, North Africa, and Southeast Asia, respectively.
Amino acid mutations are underlined.
WT, wild type.
—, no data.
EG, Equatorial Guinea; CAR, Central African Republic.
TABLE 3.
Haplotypes of Pfcrt and Pfmdr1 during 2011 to 2016a
| Year | Samples | Pfcrtc |
Pfmdr1c |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of isolates |
WTb (CVMNK) (no. [%]) | Mutation (no. [%]) |
Mixed type (CV M/I N/E K/T) (no. [%]) | No. of isolates |
WT (NY) (no. [%]) | Mutation (no. [%]) |
Mixed type (no. [%]) |
||||||||
| PCR positive | Sequenced | CVIET | SVMNT | PCR positive | Sequenced | YY | NF | YF | N Y/F | Y Y/F | |||||
| Total | 211 | 209 | 176 | 89 (50.57) | 44 (25.00) | 2 (1.14) | 41 (23.30) | 211 | 193 | 65 (33.68) | 12 (6.22) | 71 (36.79) | 27 (13.99) | 14 (7.25) | 4 (2.07) |
| 2011 | 7 | 7 | 7 | 3 (42.86) | 4 (57.14) | 0 (0.00) | 0 (0.00) | 7 | 6 | 2 (33.33) | 0 (0.00) | 1 (16.67) | 2 (33.33) | 1 (16.67) | 0 (0.00) |
| 2012 | 33 | 33 | 25 | 13(52.00) | 7 (28.00) | 1 (4.00) | 4 (16.00) | 33 | 30 | 8 (26.67) | 4 (13.33) | 11 (36.67) | 4 (13.33) | 1 (3.33) | 2 (6.67) |
| 2013 | 45 | 43 | 23 | 7 (30.43) | 12 (52.17) | 1 (4.35) | 3 (13.04) | 45 | 42 | 10 (23.81) | 2 (4.76) | 20 (47.62) | 8 (19.05) | 1 (2.38) | 1 (2.38) |
| 2014 | 37 | 37 | 33 | 18 (54.55) | 7 (21.21) | 0 (0.00) | 8 (24.24) | 37 | 37 | 14 (37.84) | 3 (8.11) | 12 (32.43) | 7 (18.92) | 1 (2.70) | 0 (0.00) |
| 2015 | 39 | 39 | 39 | 19 (48.72) | 8 (20.51) | 0 (0.00) | 12 (30.77) | 39 | 35 | 9 (25.71) | 0 (0.00) | 15 (42.86) | 6 (17.14) | 4 (11.43) | 1 (2.86) |
| 2016 | 50 | 50 | 49 | 29 (59.18) | 6 (12.24) | 0 (0.00) | 14 (28.57) | 50 | 43 | 22 (51.16) | 3 (6.98) | 12 (27.91) | 0 (0.00) | 6 (13.95) | 0 (0.00) |
| Z (observed value) | 1.252 | 2.724 | 1.902 | 1.721 | 2.274 | 1.11 | 1.091 | 1.923 | 2.397 | 1.59 | |||||
| P value | 0.211 | 0.006d | 0.057 | 0.085 | 0.023d | 0.267 | 0.275 | 0.054 | 0.017d | 0.112 | |||||
The haplotypes were constructed considering codon positions 72 to 76 of Pfcrt and codon positions 86 and 184 of Pfmdr1.
WT, wild type.
Amino acid mutations are underlined.
The difference is statistically significant.
We successfully obtained sequences of 91.47% (193/211) of pfmdr1-N1 and 98.58% (208/211) of pfmdr1-N2 nested PCR products, which were generated from 211 isolates. For Pfmdr1, 77.72% (150/193) of isolates carried the N86 wild-type allele. N86Y and Y184F in Pfmdr1-N1 were the main prevalent mutations detected at 22.28% and 60.01%, respectively. No mutations in Pfmdr1-N2 at positions 1034, 1042, 1109, or 1246 were detected (Tables 2 and 3). Six haplotypes coding amino acids 84 and 184 of Pfmdr1, including NY (wild type), YY, NF, and YF (mutation type), N Y/F and Y Y/F (mixed type), were found (Tables 2 and 3). The haplotypes of Pfmdr1 from the patients with recrudescence were NY (25%, 1/4), YY (25%, 1/4), NF (25%, 1/4), and YF (25%, 1/4). For individuals with cerebral malaria, the haplotypes of Pfmdr1 were 50% (4/8) NY, 37.5% (3/8) YF, and haplotypes of 12.5% (1/8) were undetected. For the death case, the Pfmdr1 haplotype was wild type. Considerably increasing trends in the prevalence of Pfmdr1 NY (Z = 2.274, P = 0.023) and N Y/F (Z = 2.397, P = 0.017) were observed during 2011 to 2016 (Table 3). Pfmdr1 NY was maintained at 33.33% in 2011 and decreased to 23.81% in 2013 but later increased to 51.16% in 2016. Pfmdr1 N Y/F decreased from 16.67% in 2011 to 2.38% in 2013 but later increased to 11.43% in 2015 and finally increased to 13.95% in 2016 (Table 3). Novel mutations, including nonsynonymous and synonymous mutation of Pfmdr1 in P. falciparum isolates, were identified (Table 4).
TABLE 4.
Polymorphisms of Pfmdr1 and K13-propeller in Plasmodium falciparum isolates
| Gene (ID) | Referencea |
Mutationb |
No. of isolates |
Prevalence (% [95% CI])d | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Codon position | AAc | Codon | AA | Codon | Base position | PCR positive | Sequenced | With mutation | ||
| pfmdr1 (PF3D7_0523000) | 102 | G | ggt | G | ggC | 306 | 4 | 2.07 (0.06–4.08) | ||
| 102 | G | ggt | G | ggC/T | 306 | 4 | 2.07 (0.06–4.08) | |||
| 130 | E | gaa | K/E | A/Gaa | 388 | 211 | 193 | 1 | 0.52 (−0.49–1.53) | |
| 156 | D | gat | N | Aat | 466 | 1 | 0.52 (−0.49–1.53) | |||
| 182 | G | ggt | G | ggG | 546 | 1 | 0.52 (−0.49–1.53) | |||
| 182 | G | ggt | G | ggG/T | 546 | 1 | 0.52 (−0.49–1.53) | |||
| 1069 | T | act | T | acG | 3207 | 11 | 5.29 (2.25–8.33) | |||
| 1069 | T | act | T | acG/T | 3207 | 2 | 0.96 (−0.37–2.29) | |||
| 1113 | G | ggt | A | gCt | 3338 | 211 | 208 | 1 | 0.48 (−0.46–1.42) | |
| 1142 | P | cct | P | ccA | 3426 | 1 | 0.48 (−0.46–1.42) | |||
| 1157 | T | aca | T | acG | 3471 | 1 | 0.48 (−0.46–1.42) | |||
| 1196 | D | gat | N | Aat | 3586 | 2 | 0.96 (−0.37–2.29) | |||
| 1226 | F | ttt | Y | tAt | 3677 | 1 | 0.48 (−0.46–1.42) | |||
| 1230 | G | gga | G | ggC | 3690 | 1 | 0.48 (−0.46–1.42) | |||
| k13-propeller (PF3D7_1343700) | 550 | S | tct | S | tcC | 1650 | 199 | 184 | 2 | 1.09 (−0.41–2.59) |
| 561 | R | cgt | H | cAt | 1682 | 1 | 0.54 (−0.52–1.6) | |||
| 575 | R | aga | R/K | aA/Ga | 1724 | 1 | 0.54 (−0.52–1.6) | |||
| 589 | V | gtc | I | Atc | 1765 | 1 | 0.54 (−0.52–1.6) | |||
Reference sites are underlined.
Mutation sites are underlined.
AA, amino acid residue.
CI, confidence interval.
Analysis of mutation in the k13-propeller gene.
We obtained 199 (94.31%, 199/211) PCR products from the k13-propeller gene in gDNA and 184 sequencing results (92.46%, 184/199) from 211 malaria patients with uncomplicated P. falciparum infections. The results showed that there were single-nucleotide polymorphisms (SNPs) in k13-propeller (Table 4), including SNPs 550, 561, 575, and 589 (Fig. 1). Synonymous mutations at position 550 were found in samples from Liberia (0.54%, 1/184), West Africa, and Mozambique (0.54%, 1/184), South Africa. The nonsynonymous mutations R561H and V589I were found in samples from Rwanda, East Africa (0.54%, 1/184), and Ivory Coast, West Africa (0.54%, 1/184). For mixed types, the R575R/K mutation was found in samples from Gabon, Central Africa (0.54%, 1/184). No mutations were detected at positions 474, 476, 493, 508, 527, 533, 537, 539, 543, 553, 568, 574, 578, and 580 of the K13-propeller gene.
DISCUSSION
In the current study, we investigated the drug resistance-associated mutations of P. falciparum from Chinese migrant workers returned from Africa and SEA to Wuhan, Central China during 2011 to 2016, using genomic DNA from their blood samples. We found the presence of four haplotypes coding amino acids 72 to 76 of Pfcrt, including CVMNK (wild type), SVMNT and CVIET (mutation types), CV M/I N/E K/T (mixed type), with 50.57%, 1.14%, 25.00%, and 23.30% prevalence, respectively. NY (33.68%) and NF (36.79%) were the main prevalent haplotypes in the Pfmdr1 gene. The prevalence of mutations at position 550, 561, 575, and 589 of K13-propeller was 1.09%, 0.54%, 0.54% and 0.54%, respectively. These findings provide information on Pfcrt, Pfmdr1, and K13-propeller polymorphisms from imported P. falciparum isolates in Wuhan to assess drug resistance-associated molecular markers in China, Africa, and SEA, leading to control of imported P. falciparum malaria in Wuhan, Central China.
In the study, we found four haplotypes coding amino acids 72 to 76 of Pfcrt, CVMNK, SVMNT, CVIET and CV M/I N/E K/T, with a moderately high (51.46%) prevalence of Pfcrt mutations SVMNT, CVIET, and CV M/I N/E K/T, suggesting high levels of in vivo resistance to CQ in Africa. Thus, CQ is no longer a priority to treat falciparum malaria. For CQR P. falciparum, two principal haplotypes, with the amino acid sequences CVIET and CV M/I N/E K/T, are widely distributed. The SVMNT haplotype is particularly resistant to amodiaquine (AQ), while CVIET is less resistant to AQ. According to the variation in SVMNT prevalence and the decreasing trend of CVIET prevalence during 2011 to 2016 in our study, AQ remains an effective antimalarial drug. Furthermore, AQ is extensively used as a portion of artesunate-amodiaquine (AS-AQ) in Africa (22). It seems that the AS-AQ will be highly efficacious against malarial infections.
Five mutations of Pfmdr1 prevalent worldwide, N86Y, Y184F, S1034C, N1042D, and D1246Y, have been identified. The first two mutations are most prevalent in Asia and Africa, whereas the last three alleles are detected frequently in South American (23). In the present study, we found a predominance of Pfmdr1 N86Y (22.28%) and Y184F (60.01%) mutations, which is consistent with existing data on those of Africa. Furthermore, we found novel nonsynonymous mutations at position 130, 156, 1113, 1196 and 1226, and several synonymous mutations, including 102, 182, 1069, 1142, 1157, and 1230. The observed predominance of the NF and YF haplotypes in Africa, especially in West Africa, South Africa, and Central Africa, could be a result of selective pressure by treatment of severe malaria with CQ. N86Y might be more important because it is associated with resistance to AQ (23). A total of 77.72% (150/193) of isolates carry the N86 allele in Africa, indicating that these isolates are sensitive to AQ. Based on the alteration of NF and increasing prevalence of NY and N Y/F during 2011 to 2016, AS-AQ can be a recommended drug combination for malaria treatment. Artemether-lumefantrine (AL) has the best efficacy against isolates carrying the Pfcrt K76T mutation and the Pfmdr1 N86Y mutation. Both wild-type alleles (Pfcrt K76 and Pfmdr1 N86) are selected for reinfections after AL treatment (24, 25). In our study, these samples retained a high level of wild-type alleles in Pfcrt K76 and Pfmdr1 N86. In Africa, dihydroartemisinin-piperaquine (DHA-PIP), AS-AQ, and AL are the commonly used ACTs (24, 25). Such a strategy should be considered for treatment of imported falciparum malaria patients in China.
Based on use of a whole-genome high-throughput sequencing platform, the relationship between the mutations in K13-propeller and ART resistance has been established in vivo and in vitro (12). The polymorphisms in K13-propeller associated with ART resistance were surveyed in SEA, including the China-Myanmar border (26), Cambodia (27), Myanmar (28), Vietnam (29), Thailand (30, 31), and Bangladesh (32), and in Africa, including Equatorial Guinea (15), Senegal (33), Uganda (34), Western Kenya (35), Sub-Saharan Africa (36, 37), and Mayotte (38). These data indicate that the mutation profiles of K13-propeller are inconsistent between SEA and Africa (13). The major mutation in SEA is C580Y (12) and in Africa is A578S (13, 15). Although K13-propeller has been considered a marker of ART resistance in SEA, no ART resistance was found in Africa (13). In the present study, the hotspot mutations found at positions 493, 539, 543, and 580 in isolates from SEA (12) were not detected. Only two nonsynonymous mutations (R561H and V589I), one synonymous mutation (S550S), and one mixed mutation (R575R/K) in K13-propeller were found. The common African nonsynonymous A578S mutation (13, 15) was not detected either. Although only limited polymorphisms in P. falciparum K13-propeller from African countries were reported in the study, more data from continuous molecular surveillance is beneficial to prevent the spread of ART/ACT resistance and improve clinical malaria treatment in these countries and in China.
There are several limitations in this study. First, the data have limited information in terms of predicting drug response due to no data on posttreatment genotyping and in vitro susceptibility testing. Second, the majority of the samples were collected in Nigeria and Liberia in West Africa, followed by Angola in South Africa and Congo in Central Africa. The haplotype profiles of Pfcrt and Pfmdr1 are partially altered compared to those in the previous studies of Nigeria, Angola, and Congo (39–42). There were also several novel mutations and haplotypes found in these countries.
Conclusions.
The present study shows that the moderately prevalent polymorphism mutations of Pfcrt and Pfmdr1 linked to resistance to CQ and AQ and limited mutations of K13-propeller, which are potentially associated with ART resistance, are obviously observed from migrant workers in Wuhan, Central China. DHA-PIP and AS-AQ are recommended drugs for malaria treatment. Continuous surveillance with molecular markers from pfcrt, pfmdr1, and k13-propeller genes for CQ, AQ, and ART resistance is highly recommended.
MATERIALS AND METHODS
Collection of samples.
Blood samples (2 to 5 ml) were collected from patients with malaria in Wuhan Medical Treatment Center, Center for Disease Prevention and Control (Wuhan, China), and 14 hospitals in Wuhan from August 2011 to December 2016. Approximately 400 μl of blood was spotted on Whatman 3MM filter paper, air dried, and stored in an individually sealed polyethylene bag containing silica desiccant beads. The bags were stored at −20°C. These samples were subjected to One Step Malaria HRP2/pLDH (P.f/Pan) (Wondfo, Guangzhou, China) and Giemsa-stained thick and thin peripheral blood smear examination. Parasitemia (parasites/μl) was determined by counting the parasites during the erythrocytic stage against 200 leukocytes in the thick smears and multiplying by 8,000 as an estimated average total number of peripheral leukocytes for the individuals. The identities of Plasmodium spp. were confirmed by real-time fluorescent quantitative PCR. All positive cases with imported malaria were treated according to the malaria control manual compiled by the Ministry of Health Disease Control Bureau in China. This study was approved by the ethics committees of Hubei University of Medicine and Wuhan City Center for Disease Prevention and Control. Informed consent was obtained from all participating individuals.
Determination of P. falciparum gene mutations.
Genomic DNA (gDNA) from blood sample spots in filter papers was extracted using a TIANamp blood DNA kit (Tiangen Biotech Co., Ltd., Beijing, China) following the manufacturer's instruction with minor modification. Briefly, two pieces of 6 mm × 6 mm blood spot (approximately 130.6 μl) were used for gDNA extraction. The gDNA was eluted with 40 μl of elution buffer. Nested PCR was performed using the genomic DNA as the templates to amplify a 145-bp fragment of the pfcrt gene (PlasmoDB PF3D7_0709000) (3), two fragments (N1, 526 bp, and N2, 799 bp) of the pfmdr1 gene (PlasmoDB PF3D7_0523000) (43), and one fragment (501 bp) from the k13-propeller gene (PlasmoDB PF3D7_1343700) (15) in order to examine the mutations of C72S, M74I, N75E, and K76T in PfCRT, the mutations of N86Y, E130K, Y184F, S1034C, V1109I, N1042D, and D1246Y in Pfmdr1, and K13-propeller mutations at codons T474I, M476I, A481V, Y493H, T508N, P527T, G533S, N537I, R539T, I543T, P553L, R561H, V568G, P574L, A578S, and C580Y. The PCR primers for pfcrt, pfmdr1, and k13-propeller genes are listed in Table 1. The PCRs were set up following the published procedures (15, 43) with minor modifications. Briefly, for the first round of PCR, 1 μl gDNA template, 10 μl 2× Phusion PCR master mix (40 units/ml Phusion DNA polymerase, 400 μM deoxynucleoside triphosphate [dNTP] mixture, 2× Phusion high-fidelity [HF] buffer, and 3 mM Mg2+), 1 μl forward primer (10 μM), 1 μl reverse primer (10 μM), and 7 μl sterile ultrapure water were mixed and subjected to the following program: initial denaturation at 94°C for 3 min, followed by 30 cycles of 94°C for 30 s, 56°C for 30 s, and 60°C for 1 min, and then a final extension at 60°C for 5 min. For the second round of PCR, 2.0 μl products from the first round of PCR, 25 μl 2× Phusion PCR master mix, 2.0 μl forward primer (10 μM), 2.0 μl reverse primer (10 μM), and H2O (up to 50 μl) were mixed and subjected to the following program: initial denaturation at 95°C for 3 min, followed by 30 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s, and then a final extension at 72°C for 5 min. Then, 5-μl products from the second round of PCR were analyzed using 1.0% agarose gel electrophoresis. The major bands were isolated and purified for DNA sequencing by Genewiz (Soochow, China). The data were analyzed using DNAstar (DNAStar Inc., Madison, WI). The nucleotide and amino-acid sequences of Pfcrt, Pfmdr1, and K13-propeller from P. falciparum strain 3D7 were used as the references for alignment. Each novel mutation was confirmed by two additional independent PCRs and by bidirectional DNA sequencing.
Data analysis.
Data were analyzed using SPSS 18 (SPSS Inc., Chicago, IL). The number of samples with wild-type and mutant alleles was used to calculate allele frequency. The percentages were calculated using a 95% confidence interval calculator for proportions, as described previously (43). The variation tendency for haplotypes of Pfcrt and Pfmdr1 over the study period was evaluated by a Cochran-Armitage trend test using the XLSTAT software (Addinsoft, New York, NY). A P value of <0.05 was considered significant.
ACKNOWLEDGMENTS
We thank the Department of Schistosomiasis and Endemic Diseases, Wuhan Center for Disease Prevention and Control, and all participants who contributed their blood samples. Jiangtao Chen is a member of The Chinese Medical Aid Team to the Republic of Equatorial Guinea.
This study was supported by the Foundation for Innovative Research Team of Hubei University of Medicine (2014CXZ02 and FDFR201603), the Natural Science Foundation of Guangdong Province of China (2016A030313116), and the Education Agency Major Project of Hubei Province of China (D20162101).
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.WHO. 2017. World malaria report 2017. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 2.Liu Y, Hsiang MS, Zhou H, Wang W, Cao Y, Gosling RD, Cao J, Gao Q. 2014. Malaria in overseas labourers returning to China: an analysis of imported malaria in Jiangsu Province, 2001–2011. Malar J 13:29. doi: 10.1186/1475-2875-13-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou RM, Zhang HW, Yang CY, Liu Y, Zhao YL, Li SH, Qian D, Xu BL. 2016. Molecular mutation profile of pfcrt in Plasmodium falciparum isolates imported from Africa in Henan province. Malar J 15:265. doi: 10.1186/s12936-016-1306-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kimura M, Koga M, Hasegawa C, Mutoh Y, Kato Y, Maruyama H. 2017. Imported malaria in pregnant women experienced in Japan. J Infect Chemother 23:545–549. doi: 10.1016/j.jiac.2017.05.004. [DOI] [PubMed] [Google Scholar]
- 5.Feng J, Xiao H, Zhang L, Yan H, Feng X, Fang W, Xia Z. 2015. The Plasmodium vivax in China: decreased in local cases but increased imported cases from Southeast Asia and Africa. Sci Rep 5:8847. doi: 10.1038/srep08847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li K, Huang G, Zhang H, Lin W, Dong X, Pi Q, Pei S, Hu L. 2013. Epidemic situation and control strategy of imported malaria in Hubei Province from 2006 to 2011. Chin J Schisto Control 25:259–262. (In Chinese.) [PubMed] [Google Scholar]
- 7.Li K, Cai S, Lin W, Xia J, Pei S, Zhang H. 2016. Analysis of malaria epidemic situation and control in Hubei Province from 1974 to 2015. Chin J Schisto Control 28:393–396. (In Chinese.) doi: 10.16250/j.32.1374.2016017. [DOI] [PubMed] [Google Scholar]
- 8.Young MD, Contacos PG, Stitcher JE, Millar JW. 1963. Drug resistance in Plasmodium falciparum from Thailand. Am J Trop Med Hyg 12:305–314. doi: 10.4269/ajtmh.1963.12.305. [DOI] [PubMed] [Google Scholar]
- 9.Harinasuta T, Suntharasamai P, Viravan C. 1965. Chloroquine-resistant falciparum malaria in Thailand. Lancet ii:657–660. [DOI] [PubMed] [Google Scholar]
- 10.Amaratunga C, Witkowski B, Khim N, Menard D, Fairhurst RM. 2014. Artemisinin resistance in Plasmodium falciparum. Lancet Infect Dis 14:449–450. doi: 10.1016/S1473-3099(14)70777-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yung AP, Bennett NM. 1976. Chloroquine-resistant falciparum malaria in Papua New Guinea. Med J Aust 2:320–321. [PubMed] [Google Scholar]
- 12.Ariey F, Witkowski B, Amaratunga C, Beghain J, Langlois AC, Khim N, Kim S, Duru V, Bouchier C, Ma L, Lim P, Leang R, Duong S, Sreng S, Suon S, Chuor CM, Bout DM, Menard S, Rogers WO, Genton B, Fandeur T, Miotto O, Ringwald P, Le Bras J, Berry A, Barale JC, Fairhurst RM, Benoit-Vical F, Mercereau-Puijalon O, Menard D. 2014. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature 505:50–55. doi: 10.1038/nature12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Menard D, Khim N, Beghain J, Adegnika AA, Shafiul-Alam M, Amodu O, Rahim-Awab G, Barnadas C, Berry A, Boum Y, Bustos MD, Cao J, Chen JH, Collet L, Cui L, Thakur GD, Dieye A, Djalle D, Dorkenoo MA, Eboumbou-Moukoko CE, Espino FE, Fandeur T, Ferreira-da-Cruz MF, Fola AA, Fuehrer HP, Hassan AM, Herrera S, Hongvanthong B, Houze S, Ibrahim ML, Jahirul-Karim M, Jiang L, Kano S, Ali-Khan W, Khanthavong M, Kremsner PG, Lacerda M, Leang R, Leelawong M, Li M, Lin K, Mazarati JB, Menard S, Morlais I, Muhindo-Mavoko H, Musset L, Na-Bangchang K, Nambozi M, Niare K, Noedl H, et al. . 2016. A worldwide map of Plasmodium falciparum K13-propeller polymorphisms. N Engl J Med 374:2453–2464. doi: 10.1056/NEJMoa1513137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Z, Shrestha S, Li X, Miao J, Yuan L, Cabrera M, Grube C, Yang Z, Cui L. 2015. Prevalence of K13-propeller polymorphisms in Plasmodium falciparum from China-Myanmar border in 2007–2012. Malar J 14:168. doi: 10.1186/s12936-015-0672-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li J, Chen J, Xie D, Eyi UM, Matesa RA, Ondo Obono MM, Ehapo CS, Yang L, Yang H, Lin M. 2016. Limited artemisinin resistance-associated polymorphisms in Plasmodium falciparum K13-propeller and PfATPase6 gene isolated from Bioko Island, Equatorial Guinea. Int J Parasitol Drugs Drug Resist 6:54–59. doi: 10.1016/j.ijpddr.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Djimde A, Doumbo OK, Cortese JF, Kayentao K, Doumbo S, Diourte Y, Coulibaly D, Dicko A, Su XZ, Nomura T, Fidock DA, Wellems TE, Plowe CV. 2001. A molecular marker for chloroquine-resistant falciparum malaria. N Engl J Med 344:257–263. doi: 10.1056/NEJM200101253440403. [DOI] [PubMed] [Google Scholar]
- 17.Foote SJ, Kyle DE, Martin RK, Oduola AM, Forsyth K, Kemp DJ, Cowman AF. 1990. Several alleles of the multidrug-resistance gene are closely linked to chloroquine resistance in Plasmodium falciparum. Nature 345:255–258. doi: 10.1038/345255a0. [DOI] [PubMed] [Google Scholar]
- 18.Fidock DA, Nomura T, Talley AK, Cooper RA, Dzekunov SM, Ferdig MT, Ursos LM, Sidhu AB, Naude B, Deitsch KW, Su XZ, Wootton JC, Roepe PD, Wellems TE. 2000. Mutations in the P. falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol Cell 6:861–871. doi: 10.1016/S1097-2765(05)00077-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mehlotra RK, Fujioka H, Roepe PD, Janneh O, Ursos LM, Jacobs-Lorena V, McNamara DT, Bockarie MJ, Kazura JW, Kyle DE, Fidock DA, Zimmerman PA. 2001. Evolution of a unique Plasmodium falciparum chloroquine-resistance phenotype in association with pfcrt polymorphism in Papua New Guinea and South America. Proc Natl Acad Sci U S A 98:12689–12694. doi: 10.1073/pnas.221440898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sidhu AB, Verdier-Pinard D, Fidock DA. 2002. Chloroquine resistance in Plasmodium falciparum malaria parasites conferred by pfcrt mutations. Science 298:210–213. doi: 10.1126/science.1074045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reed MB, Saliba KJ, Caruana SR, Kirk K, Cowman AF. 2000. Pgh1 modulates sensitivity and resistance to multiple antimalarials in Plasmodium falciparum. Nature 403:906–909. doi: 10.1038/35002615. [DOI] [PubMed] [Google Scholar]
- 22.Lu F, Zhang M, Culleton RL, Xu S, Tang J, Zhou H, Zhu G, Gu Y, Zhang C, Liu Y, Wang W, Cao Y, Li J, He X, Cao J, Gao Q. 2017. Return of chloroquine sensitivity to Africa? Surveillance of African Plasmodium falciparum chloroquine resistance through malaria imported to China. Parasit Vectors 10:355. doi: 10.1186/s13071-017-2298-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Veiga MI, Dhingra SK, Henrich PP, Straimer J, Gnadig N, Uhlemann AC, Martin RE, Lehane AM, Fidock DA. 2016. Globally prevalent PfMDR1 mutations modulate Plasmodium falciparum susceptibility to artemisinin-based combination therapies. Nat Commun 7:11553. doi: 10.1038/ncomms11553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sisowath C, Stromberg J, Martensson A, Msellem M, Obondo C, Bjorkman A, Gil JP. 2005. In vivo selection of Plasmodium falciparum pfmdr1 86N coding alleles by artemether-lumefantrine (Coartem). J Infect Dis 191:1014–1017. doi: 10.1086/427997. [DOI] [PubMed] [Google Scholar]
- 25.Sisowath C, Petersen I, Veiga MI, Martensson A, Premji Z, Bjorkman A, Fidock DA, Gil JP. 2009. In vivo selection of Plasmodium falciparum parasites carrying the chloroquine-susceptible pfcrt K76 allele after treatment with artemether-lumefantrine in Africa. J Infect Dis 199:750–757. doi: 10.1086/596738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Z, Wang Y, Cabrera M, Zhang Y, Gupta B, Wu Y, Kemirembe K, Hu Y, Liang X, Brashear A, Shrestha S, Li X, Miao J, Sun X, Yang Z, Cui L. 2015. Artemisinin resistance at the China-Myanmar border and association with mutations in the K13 propeller gene. Antimicrob Agents Chemother 59:6952–6959. doi: 10.1128/AAC.01255-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Straimer J, Gnadig NF, Witkowski B, Amaratunga C, Duru V, Ramadani AP, Dacheux M, Khim N, Zhang L, Lam S, Gregory PD, Urnov FD, Mercereau-Puijalon O, Benoit-Vical F, Fairhurst RM, Menard D, Fidock DA. 2015. K13-propeller mutations confer artemisinin resistance in Plasmodium falciparum clinical isolates. Science 347:428–431. doi: 10.1126/science.1260867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tun KM, Imwong M, Lwin KM, Win AA, Hlaing TM, Hlaing T, Lin K, Kyaw MP, Plewes K, Faiz MA, Dhorda M, Cheah PY, Pukrittayakamee S, Ashley EA, Anderson TJ, Nair S, McDew-White M, Flegg JA, Grist EP, Guerin P, Maude RJ, Smithuis F, Dondorp AM, Day NP, Nosten F, White NJ, Woodrow CJ. 2015. Spread of artemisinin-resistant Plasmodium falciparum in Myanmar: a cross-sectional survey of the K13 molecular marker. Lancet Infect Dis 15:415–421. doi: 10.1016/S1473-3099(15)70032-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thuy-Nhien N, Tuyen NK, Tong NT, Vy NT, Thanh NV, Van HT, Huong-Thu P, Quang HH, Boni MF, Dolecek C, Farrar J, Thwaites GE, Miotto O, White NJ, Hien TT. 2017. K13 propeller mutations in Plasmodium falciparum populations in regions of malaria endemicity in Vietnam from 2009 to 2016. Antimicrob Agents Chemother 61:e01578-16. doi: 10.1128/AAC.01578-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Talundzic E, Okoth SA, Congpuong K, Plucinski MM, Morton L, Goldman IF, Kachur PS, Wongsrichanalai C, Satimai W, Barnwell JW, Udhayakumar V. 2015. Selection and spread of artemisinin-resistant alleles in Thailand prior to the global artemisinin resistance containment campaign. PLoS Pathog 11:e1004789. doi: 10.1371/journal.ppat.1004789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Putaporntip C, Kuamsab N, Kosuwin R, Tantiwattanasub W, Vejakama P, Sueblinvong T, Seethamchai S, Jongwutiwes S, Hughes AL. 2016. Natural selection of K13 mutants of Plasmodium falciparum in response to artemisinin combination therapies in Thailand. Clin Microbiol Infect 22:285.e1–285.e8. doi: 10.1016/j.cmi.2015.10.027. [DOI] [PubMed] [Google Scholar]
- 32.Mohon AN, Alam MS, Bayih AG, Folefoc A, Shahinas D, Haque R, Pillai DR. 2014. Mutations in Plasmodium falciparum K13 propeller gene from Bangladesh (2009–2013). Malar J 13:431. doi: 10.1186/1475-2875-13-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Torrentino-Madamet M, Fall B, Benoit N, Camara C, Amalvict R, Fall M, Dionne P, Ba Fall K, Nakoulima A, Diatta B, Dieme Y, Menard D, Wade B, Pradines B. 2014. Limited polymorphisms in k13 gene in Plasmodium falciparum isolates from Dakar, Senegal in 2012–2013. Malar J 13:472. doi: 10.1186/1475-2875-13-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cooper RA, Conrad MD, Watson QD, Huezo SJ, Ninsiima H, Tumwebaze P, Nsobya SL, Rosenthal PJ. 2015. Lack of artemisinin resistance in Plasmodium falciparum in Uganda based on parasitological and molecular assays. Antimicrob Agents Chemother 59:5061–5064. doi: 10.1128/AAC.00921-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lucchi NW, Komino F, Okoth SA, Goldman I, Onyona P, Wiegand RE, Juma E, Shi YP, Barnwell JW, Udhayakumar V, Kariuki S. 2015. In vitro and molecular surveillance for antimalarial drug resistance in Plasmodium falciparum parasites in wKenya reveals sustained artemisinin sensitivity and increased chloroquine sensitivity. Antimicrob Agents Chemother 59:7540–7547. doi: 10.1128/AAC.01894-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taylor SM, Parobek CM, DeConti DK, Kayentao K, Coulibaly SO, Greenwood BM, Tagbor H, Williams J, Bojang K, Njie F, Desai M, Kariuki S, Gutman J, Mathanga DP, Martensson A, Ngasala B, Conrad MD, Rosenthal PJ, Tshefu AK, Moormann AM, Vulule JM, Doumbo OK, Ter Kuile FO, Meshnick SR, Bailey JA, Juliano JJ. 2015. Absence of putative artemisinin resistance mutations among Plasmodium falciparum in Sub-Saharan Africa: a molecular epidemiologic study. J Infect Dis 211:680–688. doi: 10.1093/infdis/jiu467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kamau E, Campino S, Amenga-Etego L, Drury E, Ishengoma D, Johnson K, Mumba D, Kekre M, Yavo W, Mead D, Bouyou-Akotet M, Apinjoh T, Golassa L, Randrianarivelojosia M, Andagalu B, Maiga-Ascofare O, Amambua-Ngwa A, Tindana P, Ghansah A, MacInnis B, Kwiatkowski D, Djimde AA. 2015. K13-propeller polymorphisms in Plasmodium falciparum parasites from sub-Saharan Africa. J Infect Dis 211:1352–1355. doi: 10.1093/infdis/jiu608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Torrentino-Madamet M, Collet L, Lepere JF, Benoit N, Amalvict R, Menard D, Pradines B. 2015. K13-propeller polymorphisms in Plasmodium falciparum isolates from patients in Mayotte in 2013 and 2014. Antimicrob Agents Chemother 59:7878–7881. doi: 10.1128/AAC.01251-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Agomo CO, Oyibo WA, Sutherland C, Hallet R, Oguike M. 2016. Assessment of markers of antimalarial drug resistance in Plasmodium falciparum isolates from pregnant women in Lagos, Nigeria. PLoS One 11:e0146908. doi: 10.1371/journal.pone.0146908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Foumane Ngane V, Allico Djaman J, Culeux C, Piette N, Carnevale P, Besnard P, Fortes F, Basco LK, Tahar R. 2015. Molecular epidemiology of drug-resistant Plasmodium falciparum in Benguela province, Angola. Malar J 14:113. doi: 10.1186/s12936-015-0634-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mvumbi DM, Kayembe JM, Situakibanza H, Bobanga TL, Nsibu CN, Mvumbi GL, Melin P, De Mol P, Hayette MP. 2015. Falciparum malaria molecular drug resistance in the Democratic Republic of Congo: a systematic review. Malar J 14:354. doi: 10.1186/s12936-015-0892-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koukouikila-Koussounda F, Jeyaraj S, Nguetse CN, Nkonganyi CN, Kokou KC, Etoka-Beka MK, Ntoumi F, Velavan TP. 2017. Molecular surveillance of Plasmodium falciparum drug resistance in the Republic of Congo: four and nine years after the introduction of artemisinin-based combination therapy. Malar J 16:155. doi: 10.1186/s12936-017-1816-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li J, Chen J, Xie D, Eyi UM, Matesa RA, Obono MM, Ehapo CS, Yang L, Yang H, Lin M, Wu W, Wu K, Li S, Chen Z. 2015. Molecular mutation profile of Pfcrt and Pfmdr1 in Plasmodium falciparum isolates from Bioko Island, Equatorial Guinea. Infect Genet Evol 36:552–556. doi: 10.1016/j.meegid.2015.08.039. [DOI] [PubMed] [Google Scholar]