Abstract
Cockayne syndrome protein B (CSB) is a member of the SNF2/SWI2 ATPase family and is essential for transcription-coupled nucleotide excision DNA repair (TC-NER). CSB also plays critical roles in transcription regulation. CSB can hydrolyze ATP in a DNA-dependent manner, alter protein-DNA contacts and anneal DNA strands. How the different biochemical activities of CSB are utilized in these cellular processes have only begun to become clear in recent years. Mutations in the gene encoding CSB account for majority of the Cockayne syndrome cases, which result in extreme sun sensitivity, premature aging features and/or abnormalities in neurology and development. Here, we summarize and integrate recent biochemical, structural, single-molecule and somatic cell genetic studies that have advanced our understanding of CSB. First, we review studies on the mechanisms that regulate the different biochemical activities of CSB. Next, we summarize how CSB is targeted to regulate transcription under different growth conditions. We then discuss recent advances in our understanding of how CSB regulates transcription mechanistically. Lastly, we summarize the various roles that CSB plays in the different steps of TC-NER, integrating the results of different studies and proposing a model as to how CSB facilitates TC-NER.
INTRODUCTION
Cockayne syndrome protein B (CSB) was identified as an essential component of the transcription-coupled branch of nucleotide excision repair (TC-NER), a process that preferentially removes transcription-blocking DNA lesions (1–7). Without CSB, there is no preferential repair of lesions on the transcribed DNA strand. Mutations in the gene encoding the CSB protein account for the majority of Cockayne syndrome cases, a devastating premature aging disorder characterized by developmental and neurological defects as well as severe sun sensitivity (8–10). Previous work demonstrated that CSB is the first protein recruited to RNA polymerase II (RNA pol II) stalled at bulky DNA lesions, where it is required to initiate TC-NER and recruit downstream repair factors (11,12). How CSB mediates downstream repair factor recruitment and how CSB’s chromatin remodeling activity facilitates efficient repair and allows transcription to resume post-repair was unknown. Recent studies have provided mechanistic insights into how CSB and its biochemical activities may facilitate TC-NER (13,14).
CSB is found in a complex containing RNA pol II, and in vitro reconstitution assays as well as transcription profiling analyses suggest that CSB also plays a role in general transcription regulation (15–19). It was not until recently that direct evidence revealed CSB regulates transcription as an ATP-dependent chromatin remodeler (20). Additional studies suggest that CSB’s function in transcription regulation may underlie some of the neurological phenotypes of Cockayne syndrome (21–23).
CSB is also required for the relief of oxidative stress. Cells deficient in CSB are sensitive to oxidizing agents, accumulate more oxidative DNA lesions than CSB expressing cells, and display increased levels of intracellular reactive oxygen species (ROS) (24–27). Evidence for CSB in base excision repair (BER), the major repair pathway for oxidative DNA damage, has been provided by a number of groups, which report deficient repair of oxidative DNA lesions in vitro (26,28–30). Moreover, CSB has been shown to interact with several proteins involved in BER (27,31,32) and accumulates at sites of oxidative DNA damage (33). However, exactly how CSB participates in BER is unknown. It is important to note that oxidative DNA damage caused by exogenous or endogenous mechanism can also generate substrates that could potentially be repaired by TC-NER (34–36). Indeed, when considering the etiology of Cockayne syndrome, the repair of damaged DNA resulting from mechanisms other than UV irradiation may be more relevant, as CS patients exhibit numerous complications that cannot be attributed to sun exposure (8).
Here, we summarize recent findings on how CSB’s biochemical activities are regulated and discuss how CSB may use these different activities to accomplish its biological functions in transcription regulation and TC-NER. The importance of understanding how CSB functions within cells is highlighted by the severity of Cockayne syndrome phenotypes. Ultimately, the results of these studies may lead to therapeutic interventions for Cockayne syndrome patients.
REGULATION OF CSB’S BIOCHEMICAL ACTIVITY
CSB belongs to the SNF2/SWI2 family of ATP-dependent chromatin remodelers, and these proteins use ATP as energy to alter DNA-histone and/or DNA-protein contacts (37–39). CSB has demonstrated DNA- and nucleosome-stimulated ATP hydrolysis activities as well as DNA strand annealing and exchange activities (40–42). Importantly, CSB has been shown to alter nucleosome structure in an ATP-dependent manner (13,43). Here, we will review the recent advances in our understanding of the regulation of ATP-dependent chromatin remodeling by CSB.
The N-terminal region of CSB couples ATP hydrolysis to chromatin remodeling
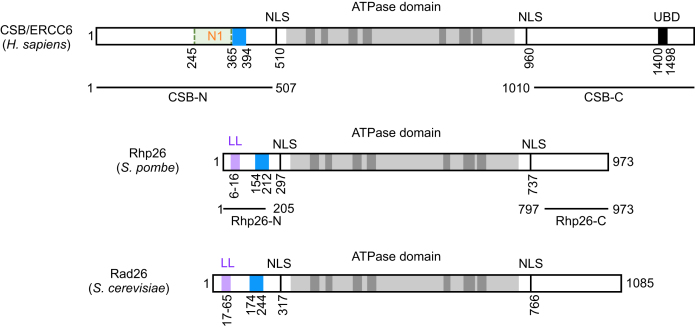
Using quantitative restriction enzyme accessibility assays, Cho et al. (2013) found that CSB exposes nucleosomal DNA in an ATP-dependent manner but does so with a maximal rate ten-times slower than that of the human remodeling complex ACF (13). Deletion of the first 454 amino acids abolishes CSB’s remodeling activity despite the fact that CSBΔ1–454 (CSBΔN) maintains its DNA- and nucleosome-stimulated ATP hydrolysis activity (Figure 1) (13). Further deletion analysis of the N-terminal region demonstrated that amino acids 245–365 are critical to couple ATP hydrolysis to chromatin remodeling, as CSBΔ245–365 (CSBΔN1) is devoid of any ATP-dependent chromatin remodeling activity, although it still is a robust DNA- and nucleosome-stimulated ATPase (Figure 1) (13). This region, termed the N1 region, is rich in basic amino acids without any recognizable motifs.
Figure 1.
Schematics of the human CSB/ERCC6 and its yeast homologs, Rhp26 and Rad26. ATPase domains are in gray. Leucine latch motifs (LL) are in purple and acidic rich regions are in blue. NLS, putative nuclear localization signal. UBD, ubiquitin binding domain.
NAP1-like histone chaperones interact with CSB and potentiate the ATP-dependent chromatin remodeling activity of CSB
To understand how CSB’s remodeling activity is regulated, Cho et al. (2013) identified the NAP1-like histone chaperones, NAP1L1 and NAP1L4, as new CSB binding proteins (13). These two proteins bind to CSB both in vitro and in cells, and the N1 region of CSB is critical for this interaction (13). Of great interest, NAP1L1 and NAP1L4 substantially increase CSB’s remodeling activity to a maximal site-exposure rate constant similar to that of ACF (13). Somatic cell genetics further demonstrated that chromatin remodeling by CSB and NAP1L4 is critical for the completion of TC-NER. It was shown that CSBΔN1 fails to completely rescue the UV sensitivity of CSB functional null cells; however, overexpressing NAP1L4 in the presence of CSBΔN1 fully complements the UV sensitivity (13). Interestingly, down-regulation of NAP1L2, the brain specific isoform of the NAP1-like proteins, is associated with neurodegenerative diseases, suggesting a biological significance to the CSB–NAP1L interactions in relation to Cockayne syndrome (44).
How do NAP1-like histone chaperones facilitate nucleosome remodeling by CSB? Lee et al. studied how CSB/NAP1L1 interact with DNA and remodel nucleosomes using single-molecule approaches, including protein induced fluorescence enhancement (PIFE) and fluorescence resonance energy transfer (FRET) (45). PIFE assays utilize a fluorophore attached to the DNA as a reporter of a protein binding on the DNA, and the intensity of a fluorophore is enhanced upon binding of a protein in the vicinity of the fluorophore (46). FRET is based on the excitation of a donor fluorophore and its concomitant energy transfer to a neighboring acceptor fluorophore, and the efficiency of this transfer is converted to an approximate distance between the two dyes.
Lee et al. found that CSB interacts with DNA in two principle ways: a rapid simple binding as revealed by PIFE and occasional gross DNA distortion detected by FRET (45). PIFE results indicated that CSB binds DNA at internal sites and ends without preference, in the presence or absence of ATP. In contrast to the rapid, simple DNA binding, ATP hydrolysis by CSB reduced the propensity of CSB to distort DNA. The latter result is similar to Beerens et al., in which scanning force microscopy (SFM) demonstrated that a shortening of contour length of a singly-nicked, circular DNA occurred upon CSB binding (47). The shortening of contour length presumably resulted from the wrapping of DNA on CSB. Similar to the gross changes observed by FRET, ATP hydrolysis by CSB reduced the frequency of this DNA wrapping event. However, the shortening of DNA contour length upon CSB binding observed by SFM was dependent on ATP binding, while the gross changes in DNA conformation detected by FRET is independent of ATP binding (45,47). Whether the difference between these two studies is due to the nature of the DNA used or if these are the same event remains to be determined.
Incubating CSB with NAP1L1 first, before mixing with DNA, induced rare and brief PIFE and no FRET events (45). Furthermore, PIFE events generated by premixing CSB with NAP1L1 contained well-defined borders in the fluorescence traces (with better defined DNA bound versus unbound states), in strong contrast to CSB alone, indicating NAP1L1 decreases CSB’s interaction with naked DNA. NAP1L1 on its own did not generate PIFE or FRET. Direct visualization of CSB-DNA interactions, by incubating Cy3-labeled CSB with immobilized DNA, revealed increasing Cy3 signals on DNA over time, indicating that multiple CSB molecules bind to a single DNA fragment. Similar to the PIFE results, inclusion of NAP1L1 reduced the number of CSB-DNA interactions, resulting in well-defined DNA bound vs. unbound states. Together, these results suggest that CSB may multimerize on DNA and that NAP1L1 decreases this tendency (45).
FRET was also used to monitor remodeling events in real time using mononucleosomes labeled on both DNA and histone H2A (45). CSB alone or CSB and NAP1L1 were incubated with immobilized mononucleosomes, and remodeling was initiated by adding ATP. Like the human ACF remodeling complex, nucleosome remodeling by CSB or by CSB plus NAP1L1 contains three distinct phases: activation, translocation, and pausing, with the activation step being rate limiting (45,48). Interestingly, the translocation steps induced by CSB and ACF both have a rate of about two base pairs per second. The major difference is that CSB has higher tendency to pause between two translocation events. Pre-incubating CSB with NAP1L1 increases the activation rate of CSB and decreases the number of pausing events during remodeling. Moreover, the distribution of FRET values created by CSB when NAP1L1 is present is narrower than with CSB alone, consistent with results from bulk experiments revealing that CSB creates more homogenous remodeled products when in complex with NAP1L1 (13,45).
Similarities and differences between CSB and the S. pombe homolog Rhp26
While the ATPase domains of CSB and its S. pombe homolog, Rhp26, are highly conserved, the flanking N- and C-terminal regions, which are suggested to regulate CSB’s enzymatic activity, are shorter and less conserved in Rhp26 (Figure 1) (49). In contrast to CSB, Rhp26 on its own has little remodeling activity (50). The N-terminal regions of each protein also function as auto-repressive modules for ATPase activity (50). However, the N-terminal region of CSB (amino acids 1–454) is also essential for the recognition of UV-induced DNA lesion-stalled transcription as well as ATP-dependent chromatin remodeling activity (13). Wang et al. identified a conserved ‘leucine latch’ motif in the N-terminus of Rhp26, and this short helix serves to lock Rhp26 in an inactive state (Figure 1) (50). It would be of interest to determine if ATP hydrolysis by Rhp26 unlocks this inactive state, similar to its human homolog CSB (51). Notably, while the N-terminal region of Rhp26 negatively regulates the remodeling activity of Rhp26, this region is dispensable for Rhp26 function in protecting cells from UV-irradiation, as Rhp26ΔN fully complements the UV sensitivity of ΔRhp26, in contrast to its human homolog CSB (13,50). These observations suggest that the highly regulated CSB-chromatin interaction mechanism that is used in mammals is dispensable in Schizosaccharomyces pombe.
The C-terminal regions of both CSB and Rhp26 function as positive regulatory regions, regardless of the detailed mechanism (Figure 1) (50,51). Like CSB, the C-terminal region positively regulates Rhp26’s ATPase activity, as Rhp26ΔC resulted in decreased ATPase activity (50). However, CSB contains a ubiquitin-binding domain (UBD) in its C-terminal region, which is absent in its yeast homologs (Figure 1). This domain is suggested to be critical for CSB function in TC-NER as well as transcription (52,53) and is likely specific to CSB’s role in multicellular organism development.
CSB IN TRANSCRIPTION REGULATION
Initial involvement of CSB in transcription regulation came from several lines of evidence. Assays using intact and permeabilized cells revealed that transcription elongation is reduced in CSB cells and that this defect can be complemented by extracts from normal cells (15). This study supported the hypothesis that Cockayne syndrome may in fact be a transcription syndrome in addition to a repair syndrome, and differences in the extent of gene expression defects might account for differences in the severity of Cockayne syndrome (54–57).
A role for CSB in transcription was further supported by in vitro reconstitution assays with purified components, which revealed that CSB can interact with RNA pol II in isolation and as part of an elongation complex, and that CSB can directly stimulate transcription elongation rates (16,17). Biochemical purification of CSB from whole cell extracts ultimately demonstrated that CSB is indeed part of a large complex (>700 kDa) that contains RNA pol II (18).
CSB in transcription regulation during replicative cell growth
Transcription profiling studies supported the notion that CSB participates in transcription regulation during normal cell growth and further revealed that this function of CSB is unlikely general but rather gene-specific (19,58). These expression studies implicated CSB in the regulation of genes involved in chromatin structure maintenance and remodeling as well as a variety of metabolic processes. Together, these findings supported the hypothesis that CSB’s role in regulating transcription may play a more significant role in the pathology of Cockayne syndrome than previously appreciated.
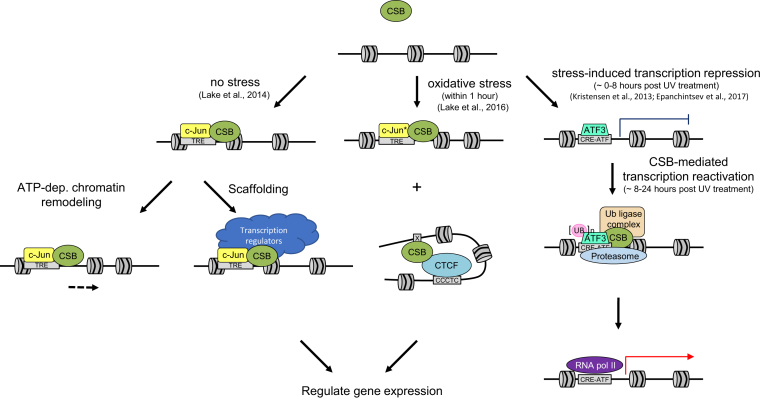
Deep sequencing of anti-CSB chromatin immunoprecipitated (ChIP-seq) DNA produced the first genome-wide map of CSB occupancy sites in cells during replicative growth (20). This study revealed that CSB is significantly enriched at promoters and enhancers, suggesting that CSB may function in transcription initiation, in addition to its previously demonstrated role in transcription elongation (17,18). Motif analysis of CSB-occupied sites revealed that CSB is enriched at sites containing the 12-O-tetradecanoylphorbol-13-acetate (TPA) response element (TRE), which contains the binding motif for activator protein 1 (AP-1). AP-1 is a family of bZIP transcription factors, which include Jun and Fos family members, and these proteins play critical roles in responding to environmental stimuli. CSB was shown to interact with c-Jun and this interaction was found to be critical for CSB recruitment to TRE-containing sites. Side-by side comparisons of wild-type CSB and a chromatin remodeling-deficient CSB derivative (CSBΔN1, Figure 1) revealed that CSB regulates nucleosome positioning around its binding sites and alters the expression of nearby genes (20). Therefore, this study provided the first direct evidence for a function of CSB in regulating transcription through its ATP-dependent chromatin remodeling activity (Figure 2). Nonetheless, not all genes near CSB-occupancy sites were dependent upon CSB’s remodeling activity, although gene expression was clearly impacted by CSB loss (Figure 2). Taken together, these observations demonstrate that CSB has both chromatin remodeling-dependent and –independent functions in transcription regulation (20). The remodeling independent function suggests that CSB may act as a scaffold to recruit transcriptional regulators (blue, Figure 2), similar to that of its function of recruiting repair factors in TC-NER (13). Combining tandem affinity purification (TAP) with mass spectrometry, Nicolai et al. identified 33 novel CSB interacting partners. These proteins include the SWI/SNF-related SMARCA family of proteins, the transcriptional activator MTA2, and the transcriptional repressors HDAC1 and GATA2A/B (59). Therefore, CSB may coordinate its own activity with other remodelers, histone modifying enzymes and transcription factors to regulate chromatin structure for transcriptional regulation.
Figure 2.
Models for transcription regulation by CSB under different growth conditions. (Left) Under non-stressed conditions, CSB is targeted to TREs by the transcription factor c-Jun, where CSB can regulate transcription by both remodeling-dependent and -independent mechanisms. (Middle) Targeting of CSB by c-Jun to TREs also occurs in cells following oxidative stress with an up-regulation of c-Jun after stress (c-Jun*). Additionally, following oxidative stress, CSB is also enriched at binding motifs for the architectural protein CTCF. CSB may regulate 3D chromatin structure by modulating CTCF-chromosome interactions to regulate gene expression. (Right) Following UV irradiation and DNA repair, ATF3 is ubiquitinated in a CSB- and CSA-dependent manner and degraded by the proteasome, allowing RNA pol II recruitment and transcription reactivation.
CSB in transcription regulation during oxidative stress
CSB has also been suggested to play a role in transcription regulation in response to oxidative stress (58). Kyng et al. conducted microarray analysis following H2O2 treatment in CSB-null or CSB ATPase mutant cell lines and, compared to wild type, found expression changes in 122 out of 6912 genes examined, including genes important for the stress response, transcription, translation, signal transduction, and the cell cycle (58). Lake et al. examined CSB occupancy at a genome-wide level after treatment with menadione, which induces oxidative stress (24). It was found that CSB occupancy was altered, with a significant increase at promoters from 2% in unstressed cells to 11% in cells experiencing oxidative stress, suggesting that CSB may regulate transcription initiation to mount a response to oxidative stress.
Motif analysis revealed that sites bound by CSB during oxidative stress are enriched for TREs, as in non-stressed cells, and that the percentage of CSB-bound TREs did not change (Figure 2) (24). However, binding motifs for the transcriptional regulator CCCTC-binding factor (CTCF) were substantially enriched upon oxidative stress, with an increase from 1% in non-stressed cells to 11% in oxidatively stressed cells. CTCF is involved in transcription regulation and is a key player in regulating long-range chromatin interactions (60,61). In vitro protein-interaction studies using purified proteins revealed that CSB and CTCF directly interact. Additionally, it was found that this interaction is enhanced in cells by oxidative stress (24). Using CTCF knockdown, it was found that the CTCF protein is needed to recruit CSB to sites containing the CTCF binding motif upon oxidative stress. Intriguingly, this was also found to be true for other stress-induced CSB occupied loci that do not contain the CTCF binding motif, suggesting that CTCF may also recruit CSB to sites that are near a CTCF binding motif, in addition to sites containing the CTCF motif. Reciprocally, CSB was found to increase CTCF-DNA interaction, both in vitro and in cells. Together, these results support the hypothesis that CSB may work with CTCF to organize 3-dimensional chromatin structure to efficiently regulate a transcriptional response to oxidative stress (Figure 2) (24). How CSB’s enzymatic and protein-recruitment activities contribute to this process remains to be determined.
Given that CSB is proposed to function in oxidative DNA damage repair, it is also possible that the interaction of CSB and CTCF may facilitate the formation of DNA repair hubs in the 3D chromatin space to efficiently remove oxidative DNA damage. It will be of great interest to determine the extent to which CSB is enriched at sites of oxidative DNA lesions by analyzing DNA mutation signatures associated with CSB-ChIPed DNA from cells exposed to oxidative stress.
CSB in transcription regulation in response to UV irradiation
Over thirty-five years ago, it was reported that RNA synthesis fails to recover after UV irradiation in cells from Cockayne syndrome patients (3). This was initially thought to be the consequence of a block in transcription elongation resulting from defective TC-NER. However, that model was challenged by experiments using in vitro transcription systems with nuclear extracts prepared from UV-irradiated or mock-irradiated normal human and Cockayne syndrome cells (62). From this study, it was found that there was a global defect in transcription initiation in Cockayne syndrome cells. This defect was associated with a loss of the hypophosphorylated, transcription-initiating form of RNA pol II and a concomitant increase in the hyperphosphorylated, transcription-elongating form of RNA pol II. Subsequent experiments using ChIP-qPCR confirmed the defect in transcription initiation and further revealed a decrease in the recruitment of RNA pol II to the promoters of certain genes (63).
Recent studies have suggested that the inability to reactivate transcription in Cockayne syndrome cells after UV irradiation may be due, in part, to an inability to relieve general transcriptional repression induced by UV irradiation (53,64). Kristensen et al. searched for common factor binding motifs near the promoters of a collection of UV-repressed genes and found that these genes contained binding sites for activating transcription factor 3 (ATF3), a transcriptional repressor that is activated in response to cellular stress (64–66). In CSB wild-type cells, ATF3 mRNA and protein levels increase and peak ∼8 h after UV irradiation, which corresponds with maximal repression of genes whose promoters are bound by ATF3 (64). Between approximately 12 and 24 h after UV irradiation, ATF3 levels decrease and ATF3 is removed from bound promoters, and this correlates with recruitment of RNA pol II as well as transcription resumption (Figure 2) (64). In CSB-deficient cells, however, the ATF3 protein and ATF3 occupancy at its target promoters remain high (64). This work suggested that CSB might be required to remove ATF3 from its target promoters to allow transcription resumption (Figure 2), and this hypothesis was subsequently tested (53). It was found that CSB collaborates with Cockayne syndrome protein A (CSA) to promote the ubiquitination and degradation of ATF3, thereby allowing transcription to resume (Figure 2). CSA is a WD-40 repeat-containing protein that is part of an E3-ubiquitin ligase complex along with DNA damage binding protein 1 (DDB1) and Cullin 4A (CUL4A) (64,67,68). Like CSB, mutations within CSA can also lead to Cockayne syndrome (69). Importantly, using an ATP-deficient CSB derivative (Q678E), these studies revealed that ATP hydrolysis by CSB is not necessary for recruitment of CSB to ATF3-occupied sites or ATF3 ubiquitination; however, ATP hydrolysis was necessary for recruitment of the proteasomal machinery and subsequent ATF3 turnover. It will be of great interest to determine if the chromatin remodeling deficient CSB derivative, CSBΔN1, can support ATF3 degradation, to examine the role that CSB may play in reorganizing the epigenetic landscape for the resumption of transcription initiation after UV-induced genotoxic stress.
It is, however, important to note that while CSB’s role in ATF3 release may contribute to transcription recovery after UV irradiation, it is unlikely to be the only mechanism. ATF3 is also induced by other genotoxins, such as ionizing radiation and alkylating agents, yet CSB null cells do not show the same level of sensitivity to these genotoxins as they do to UV irradiation (66,70–72).
By sequencing nascent transcripts after UV irradiation, Williamson et al., discovered that a short, non-coding RNA generated from the activating signal cointegrator 1 complex subunit 3 (ASCC3) is preferentially synthesized after UV irradiation (73). The ASCC3 protein produced from the full-length ASCC3 mRNA is a 3′-5′ DNA helicase that participates in the repair of alkylated DNA (74). This ASCC3 protein interacts with both CSB and RNA pol II (75). Intriguingly, the short, non-coding isoform, like CSB, is required for transcription resumption after UV irradiation (73). Moreover, the short and long RNA isoforms have antagonistic functions in the response to UV irradiation (73). Whether CSB-dependent and ASCC3-dependent transcription restart after UV irradiation are mechanistically related or represent two independent pathways remains to be determined.
REGULATION OF TC-NER BY CSB
CSB is critical for multiple steps of the TC-NER process. Lake et al. (2010) demonstrated that ATP hydrolysis by CSB is essential for CSB to become associated with UV-induced DNA lesion stalled RNA pol II, the critical first step of transcription-coupled DNA repair (51). However, chromatin remodeling by CSB is dispensable for this step, since the ATPase-proficient, yet remodeling-deficient CSBΔN1 mutant is capable of stably associating with chromatin at sites of lesion-stalled transcription after UV irradiation (13). Chromatin immunoprecipitation experiments followed by western blot analyses revealed that representative factors of the nucleotide excision repair machinery, the transcription elongation complex, and the CSA-ubiquitin ligase complex are recruited to UV-induced DNA lesion-stalled RNA pol II in the presence of the chromatin remodeling deficient CSBΔN1, arguing against the notion that the function of chromatin remodeling by CSB in TC-NER is to create access for factor binding (13). These observations, along with the work of Fousteri et al., indicate that CSB likely recruits factors for repair and transcription resumption through protein-protein interaction (11). Given that chromatin remodeling by CSB is required for steps after the recruitment of the TC-NER machinery, chromatin remodeling by CSB likely regulates the chromatin landscape for more efficient DNA repair and/or transcription resumption (13). NAP1-like proteins are also expected to contribute to these activities, as their overexpression in the presence of CSBΔN1 fully rescues the UV sensitivity resulting from the loss of CSB (13).
CSB without its last 273 amino acids (CSBdel) failed to complement the UV sensitivity of the CSB-deficient cell line CS1AN-sv (52). This mutant protein can interact with RNA polymerase II after UV irradiation and can recruit necessary NER factors; however, the DNA incision step of TC-NER is compromised in the CSBdel background (52). A ubiquitin binding domain (UBD) lies within the last 273 amino acids (Figure 1), and a CSB protein with point mutations that disrupt ubiquitin binding (LL to GG) also fails to fully complement the UV sensitivity of CS1AN-sv cells, although the effect on cell survival and RNA recovery is less severe than CSBdel. These observations reveal that ubiquitin binding by CSB is important for CSB function in TC-NER (52).
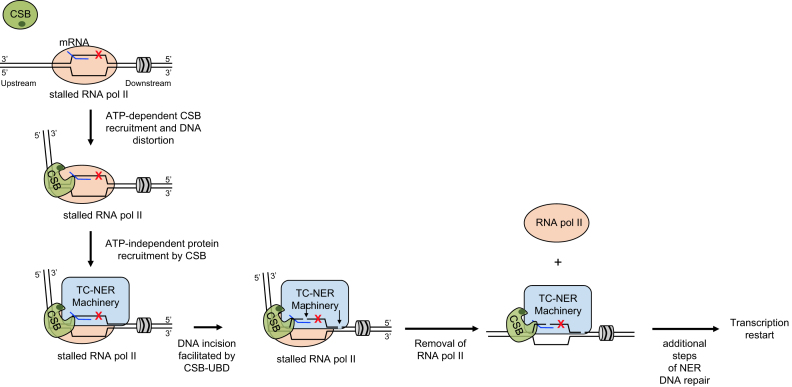
ATP hydrolysis by CSB is important for the initiation of TC-NER, however, the exact role of CSB in this process and how this protein recognizes and interacts with RNA pol II arrested at DNA lesions is unknown. The electron cryomicroscopy (cryo-EM) structure from Xu et al. revealed that the S. cerevisiae CSB ortholog, Rad26, binds DNA upstream of the RNA pol II elongation complex, making contacts with the upstream DNA duplex region and single-stranded DNA in the upstream fork of the transcription bubble (Figure 3) (14). Interestingly, Rad26 caused an 80° bend in the upstream duplex DNA, perhaps creating novel interaction surfaces to facilitate repair factor recruitment (Figure 3) (14). This DNA distortion observed by cryo-EM might be related to the DNA distortion previously observed by FRET and SFM (45,47).
Figure 3.
Model for how CSB integrates into different steps of TC-NER. The association of CSB with RNA pol II stalled at a bulky DNA lesion (red X) is stabilized. This step requires the DNA/nucleosome stimulated ATP hydrolysis activity of CSB. CSB interacts with both RNA pol II and duplex DNA upstream of RNA pol II as well as single-stranded DNA within the upstream fork of the transcription bubble. CSB generates an 80-degree bend in the upstream DNA and provides an interaction surface for recruiting additional TC-NER factors. CSB facilitates this step through protein-protein interactions, but not through its ATP-dependent chromatin remodeling activity. The C-terminal Ub-binding domain (dark green oval) of CSB is needed for efficient DNA incision. Dual incisions (arrows) happen in the presence of RNA polymerase II, which is subsequently removed to permit the remaining steps of the repair processes.
By modeling this Rad26 structure with the Snf2 remodeler bound to nucleosomes, Xu et al. proposed a model whereby Rad26 pulls the DNA template strand away from RNA pol II by translocating on the DNA duplex. This would lead to annealing the strands of the transcription bubble, consistent with the observed strand annealing activity of CSB (42), and promote the forward movement of RNA pol II (14). This work also supports a model in which CSB promotes transcription elongation by preventing backtracking and promoting forward movement of RNA pol II when it encounters a non-bulky transcription-stalling signal. However, CSB would fail to promote forward RNA pol II movement in the presence of bulky transcription-blocking lesions (14).
We would like to propose a model to account for CSB’s function in different steps of TC-NER, based on the collective work from the human and yeast homologs. CSB uses ATP hydrolysis to undergo a conformational change to probe chromatin for lesion-stalled RNA pol II (Figure 3) (14,51). Once discovered, the N-terminal, substrate-recognition domain would bind to the lesion-stalled RNA pol II complex. The association of CSB with lesion stalled transcription would lead to an 80° DNA bend. The CSB-chromatin association and resulting DNA conformation would be reinforced by interaction between chromatin and the C-terminal region of CSB, which is necessary for stable chromatin association (Figure 3) (14,51). Once stably associated, CSB would function as a scaffold, creating a platform to recruit factors needed to repair DNA and resume transcription (Figure 3) (11,14). This model is supported by the observations that the remodeling deficient CSBΔN1 protein is correctly recruited to DNA lesion-stalled RNA pol II and can initiate the recruitment of additional protein factors necessary for DNA repair (13). Subsequently, CSB would facilitate DNA incision by NER factors through its Ub-binding domain (UBD) (52). Removal of RNA pol II is not a prerequisite for the incision events, as strand excision can occur in a cell-free repair system with a stalled polymerase covering the DNA lesion (76).
Genome-wide studies have revealed that repair of transcription blocking lesions occurs as waves along gene bodies in the 5′-3′ direction (77,78). Importantly, by controlling these transcriptional waves with the reversible transcription elongation inhibitor 5,6-dicholor-1-β-d-ribofuranosybenzimidazole (DRB), Chiou et al. have provided strong evidence that a single transcription elongation complex does not progress along a template to engage multiple lesions, but rather dissociates from the template after the dual incisions. RNA pol II dissociation would expose the 3′ hydroxyl generated by the incision event to promote new DNA synthesis and ligation (78). RNA Pol II removal might be promoted by CSB translocation, by the helicase activity of XPB or XPD, or simply by instability created by fragment removal (Figure 3). Using in vitro reconstituted assays, Selby and Sancar did not observe CSB-mediated removal of lesion-stalled RNA pol II from DNA (40). However, given that NAP1L1 synergizes with CSB in chromatin remodeling (13), it would be of great interest to determine if the combined activities of CSB and NAP1L1 could perform this task. Additionally, whether the chromatin remodeling activity of CSB is used to create an epigenetic landscape that permits more efficient DNA repair or to facilitate transcription resumption after repair remains to be determined. Future studies examining the structure of the remodeling-deficient CSBΔN1-RNA pol II elongation complex (EC) will provide insights into the functions of ATP hydrolysis by CSB in TC-NER.
CONCLUSIONS
Over the recent years, we have learned much about how the biochemical activities of CSB are tightly regulated and are used to facilitate TC-NER and transcription regulation; however, there is still much to be learned. For example, given that the chromatin remodeling activity of CSB is crucial for efficient TC-NER, are there different requirements of CSB (or are additional proteins needed) for TC-NER at nucleosome dense as compared to nucleosome-free regions? Does the enzymatic activity of CSB in any way influence RNA pol II stalled at bulky DNA lesions to permit DNA repair? Does CSB play a role in resetting the epigenetic landscape after TC-NER for transcription resumption? Does CSB-dependent ubiquitinylation account for transcription regulation beyond ATF3? To what extent does CSB organize the three-dimensional chromatin structure to orchestrate DNA repair and transcription regulation during oxidative stress relief? Additional studies using structural, genomic and in vitro reconstituted systems will shed new light on these and other outstanding questions in the coming years.
FUNDING
National Institutes of Health [GM115888 to HYF, T32 GM-07229 to ELB]. HYF and RJL are partially supported by Cancer Center support Grant P30CA118100. Funding for open access charge: National Institutes of Health [GM115888].
Conflict of interest statement. None declared.
REFERENCES
- 1. Mellon I., Spivak G., Hanawalt P.C.. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell. 1987; 51:241–249. [DOI] [PubMed] [Google Scholar]
- 2. Bohr V.A., Smith C.A., Okumoto D.S., Hanawalt P.C.. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient that in the genome overall. Cell. 1985; 40:359–369. [DOI] [PubMed] [Google Scholar]
- 3. Mayne L.V., Lehmann A.R.. Failure of RNA synthesis to recover after UV irradiation: an early defect in cells from individuals with Cockayne's syndrome and xeroderma pigmentosum. Cancer Res. 1982; 42:1473–1478. [PubMed] [Google Scholar]
- 4. Venema J., van Hoffen A., Natarajan A.T., van Zeeland A.A., Mullenders L.H.F.. The residual repair capacity of xeroderma pigmentosum complementation group C fibroblasts is highly specific for transcriptionally active DNA. Proc. Natl. Acad. Sci. U.S.A. 1990; 18:443–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Troelstra C., Odijk H., de Wit J., Westerveld A., Thompson L.H., Bootsma D., Hoeijmakers J.H.. Molecular cloning of the human DNA excision repair gene ERCC-6. Mol. Cell. Biol. 1990; 10:5806–5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Troelstra C., van Gool A., de Wit J., Vermeulen W., Bootsma D., Hoeijmakers J.H.. ERCC6, a member of a subfamily of putative helicases, is involved in Cockayne's syndrome and preferential repair of active genes. Cell. 1992; 71:939–953. [DOI] [PubMed] [Google Scholar]
- 7. Hanawalt P.C., Spivak G.. Transcription-coupled DNA repair: two decades of progress and surprises. Nat. Rev. Mol. Cell. Biol. 2008; 9:958–970. [DOI] [PubMed] [Google Scholar]
- 8. Nance M.A., Berry S.A.. Cockayne syndrome: review of 140 cases. Am. J. Med. Genet. 1992; 42:68–84. [DOI] [PubMed] [Google Scholar]
- 9. Karikkineth A.C., Scheibye-Knudsen M., Fivenson E., Croteau D.L., Bohr V.A.. Cockayne syndrome: clinical features, model systems and pathways. Ageing Res. Rev. 2017; 33:3–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cockayne E.A. Dwarfism with retinal atrophy and deafness. Arch. Dis. Child. 1936; 11:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fousteri M., Vermeulen W., van Zeeland A.A., Mullenders L.H.. Cockayne syndrome A and B proteins differentially regulate recruitment of chromatin remodeling and repair factors to stalled RNA polymerase II in vivo. Mol. Cell. 2006; 23:471–482. [DOI] [PubMed] [Google Scholar]
- 12. van den Boom V., Citterio E., Hoogstraten D., Zotter A., Egly J.M., van Cappellen W.A., Hoeijmakers J.H., Houtsmuller A.B., Vermeulen W.. DNA damage stabilizes interaction of CSB with the transcription elongation machinery. J. Cell. Biol. 2004; 166:27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cho I., Tsai P.F., Lake R.J., Basheer A., Fan H.Y.. ATP-Dependent chromatin remodeling by cockayne syndrome protein B and NAP1-Like histone chaperones is required for efficient Transcription-Coupled DNA repair. PLos Genet. 2013; 9:e1003407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xu J., Lahiri I., Wang W., Wier A., Cianfrocco M.A., Chong J., Hare A.A., Dervan P.B., DiMaio F., Leschziner A.E. et al. . Structural basis for the initiation of eukaryotic transcription-coupled DNA repair. Nature. 2017; 551:653–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Balajee A.S., May A., Dianov G.L., Friedberg E.C., Bohr V.A.. Reduced RNA polymerase II transcription in intact and permeabilized Cockayne syndrome group B cells. Proc. Natl. Acad. Sci. U.S.A. 1997; 94:4306–4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Selby C.P., Sancar A.. Cockayne syndrome group B protein enhances elongation by RNA polymerase II. Proc. Natl. Acad. Sci. U.S.A. 1997; 94:11205–11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tantin D., Kansal A., Carey M.. Recruitment of the putative transcription-repair coupling factor CSB/ERCC6 to RNA polymerase II elongation complexes. Mol. Cell. Biol. 1997; 17:6803–6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. van Gool A.J., Citterio E., Rademakers S., van Os R., Vermeulen W., Constantinou A., Egly J.M., Bootsma D., Hoeijmakers J.H.. The Cockayne syndrome B protein, involved in transcription-coupled DNA repair, resides in an RNA polymerase II-containing complex. EMBO J. 1997; 16:5955–5965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Newman J.C., Bailey A.D., Weiner A.M.. Cockayne syndrome group B protein (CSB) plays a general role in chromatin maintenance and remodeling. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:9613–9618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lake R.J., Boetefuer E.L., Tsai P.F., Jeong J., Choi I., Won K.J., Fan H.Y.. The Sequence-Specific transcription factor c-Jun targets Cockayne syndrome protein B to regulate transcription and chromatin structure. PLos Genet. 2014; 10:e1004284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ciaffardini F., Nicolai S., Caputo M., Canu G., Paccosi E., Costantino M., Frontini M., Balajee A.S., Proietti-De-Santis L.. The Cockayne syndrome B protein is essential for neuronal differentiation and neuritogenesis. Cell Death Dis. 2014; 5:e1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang Y., Chakravarty P., Ranes M., Kelly G., Brooks P.J., Neilan E., Stewart A., Schiavo G., Svejstrup J.Q.. Dysregulation of gene expression as a cause of Cockayne syndrome neurological disease. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:14454–14459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang Y., Jones-Tabah J., Chakravarty P., Stewart A., Muotri A., Laposa R.R., Svejstrup J.Q.. Pharmacological bypass of Cockayne syndrome B function in neuronal differentiation. Cell Rep. 2016; 14:2554–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lake R.J., Boetefuer E.L., Won K.J., Fan H.Y.. The CSB chromatin remodeler and CTCF architectural protein cooperate in response to oxidative stress. Nucleic Acids Res. 2016; 44:2125–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pascucci B., Lemma T., Iorio E., Giovannini S., Vaz B., Iavarone I., Calcagnile A., Narciso L., Degan P., Podo F. et al. . An altered redox balance mediates the hypersensitivity of Cockayne syndrome primary fibroblasts to oxidative stress. Aging Cell. 2012; 11:520–529. [DOI] [PubMed] [Google Scholar]
- 26. Tuo J., Muftuoglu M., Chen C., Jaruga P., Selzer R.R., Brosh R.M. Jr., Rodriguez H., Dizdaroglu M., Bohr V.A.. The Cockayne Syndrome group B gene product is involved in general genome base excision repair of 8-hydroxyguanine in DNA. J. Biol. Chem. 2001; 276:45772–45779. [DOI] [PubMed] [Google Scholar]
- 27. Muftuoglu M., de Souza-Pinto N.C., Dogan A., Aamann M., Stevnsner T., Rybanska I., Kirkali G., Dizdaroglu M., Bohr V.A.. Cockayne syndrome group B protein stimulates repair of formamidopyrimidines by NEIL1 DNA glycosylase. J. Biol. Chem. 2009; 284:9270–9279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dianov G., Bischoff C., Sunesen M., Bohr V.A.. Repair of 8-oxoguanine in DNA is deficient in Cockayne syndrome group B cells. Nucleic Acids Res. 1999; 27:1365–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tuo J., Jaruga P., Rodriguez H., Dizdaroglu M., Bohr V.A.. The cockayne syndrome group B gene product is involved in cellular repair of 8-hydroxyadenine in DNA. J. Biol. Chem. 2002; 277:30832–30837. [DOI] [PubMed] [Google Scholar]
- 30. Tuo J., Jaruga P., Rodriguez H., Bohr V.A., Dizdaroglu M.. Primary fibroblasts of Cockayne syndrome patients are defective in cellular repair of 8-hydroxyguanine and 8-hydroxyadenine resulting from oxidative stress. FASEB J. 2003; 17:668–674. [DOI] [PubMed] [Google Scholar]
- 31. Tuo J., Chen C., Zeng X., Christiansen M., Bohr V.A.. Functional crosstalk between hOgg1 and the helicase domain of Cockayne syndrome group B protein. DNA Repair (Amst.). 2002; 1:913–927. [DOI] [PubMed] [Google Scholar]
- 32. Wong H.K., Muftuoglu M., Beck G., Imam S.Z., Bohr V.A., Wilson D.M. 3rd. Cockayne syndrome B protein stimulates apurinic endonuclease 1 activity and protects against agents that introduce base excision repair intermediates. Nucleic Acids Res. 2007; 35:4103–4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Menoni H., Hoeijmakers J.H., Vermeulen W.. Nucleotide excision repair-initiating proteins bind to oxidative DNA lesions in vivo. J. Cell Biol. 2012; 199:1037–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Iyama T., Lee S.Y., Berquist B.R., Gileadi O., Bohr V.A., Seidman M.M., McHugh P.J., Wilson D.M. 3rd. CSB interacts with SNM1A and promotes DNA interstrand crosslink processing. Nucleic Acids Res. 2015; 43:247–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Furuta T., Ueda T., Aune G., Sarasin A., Kraemer K.H., Pommier Y.. Transcription-coupled nucleotide excision repair as a determinant of cisplatin sensitivity of human cells. Cancer Res. 2002; 62:4899–4902. [PubMed] [Google Scholar]
- 36. Enoiu M., Jiricny J., Scharer O.D.. Repair of cisplatin-induced DNA interstrand crosslinks by a replication-independent pathway involving transcription-coupled repair and translesion synthesis. Nucleic Acids Res. 2012; 40:8953–8964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Eisen J.A., Sweder K.S., Hanawalt P.C.. Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res. 1995; 23:2715–2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Flaus A., Martin D.M., Barton G.J., Owen-Hughes T.. Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic. Acids. Res. 2006; 34:2887–2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lake R.J., Fan H.Y.. Structure, function and regulation of CSB: a multi-talented gymnast. Mech. Ageing Dev. 2013; 134:202–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Selby C.P., Sancar A.. Human transcription-repair coupling factor CSB/ERCC6 is a DNA-stimulated ATPase but is not a helicase and does not disrupt the ternary transcription complex of stalled RNA polymerase II. J. Biol. Chem. 1997; 272:1885–1890. [DOI] [PubMed] [Google Scholar]
- 41. Citterio E., Rademakers S., van der Horst G.T., van Gool A.J., Hoeijmakers J.H., Vermeulen W.. Biochemical and biological characterization of wild-type and ATPase-deficient Cockayne syndrome B repair protein. J. Biol. Chem. 1998; 273:11844–11851. [DOI] [PubMed] [Google Scholar]
- 42. Muftuoglu M., Sharma S., Thorslund T., Stevnsner T., Soerensen M.M., Brosh R.M. Jr., Bohr V.A.. Cockayne syndrome group B protein has novel strand annealing and exchange activities. Nucleic Acids Res. 2006; 34:295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Citterio E., Van Den Boom V., Schnitzler G., Kanaar R., Bonte E., Kingston R.E., Hoeijmakers J.H., Vermeulen W.. ATP-dependent chromatin remodeling by the Cockayne syndrome B DNA repair-transcription-coupling factor. Mol. Cell. Biol. 2000; 20:7643–7653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Li M.D., Burns T.C., Morgan A.A., Khatri P.. Integrated multi-cohort transcriptional meta-analysis of neurodegenerative diseases. Acta Neurol. Commun. 2014; 2:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lee J.Y., Lake R.J., Kirk J., Bohr V.A., Fan H.Y., Hohng S.. NAP1L1 accelerates activation and decreases pausing to enhance nucleosome remodeling by CSB. Nucleic Acids Res. 2017; 45:4696–4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hwang H., Myong S.. Protein induced fluorescence enhancement (PIFE) for probing protein-nucleic acid interactions. Chem. Soc. Rev. 2014; 43:1221–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Beerens N., Hoeijmakers J.H., Kanaar R., Vermeulen W., Wyman C.. The CSB protein actively wraps DNA. J. Biol. Chem. 2005; 280:4722–4729. [DOI] [PubMed] [Google Scholar]
- 48. Blosser T.R., Yang J.G., Stone M.D., Narlikar G.J., Zhuang X.. Dynamics of nucleosome remodelling by individual ACF complexes. Nature. 2009; 462:1022–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li S. Transcription coupled nucleotide excision repair in the yeast Saccharomyces cerevisiae: the ambiguous role of Rad26. DNA Repair (Amst.). 2015; 36:43–48. [DOI] [PubMed] [Google Scholar]
- 50. Wang L., Limbo O., Fei J., Chen L., Kim B., Luo J., Chong J., Conaway R.C., Conaway J.W., Ranish J.A. et al. . Regulation of the Rhp26 ERCC6/CSB chromatin remodeler by a novel conserved leucine latch motif. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:18566–18571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lake R.J., Geyko A., Hemashettar G., Zhao Y., Fan H.Y.. UV-induced association of the CSB remodeling protein with chromatin requires ATP-dependent relief of N-terminal autorepression. Mol. Cell. 2010; 37:235–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Anindya R., Mari P.O., Kristensen U., Kool H., Giglia-Mari G., Mullenders L.H., Fousteri M., Vermeulen W., Egly J.M., Svejstrup J.Q.. A ubiquitin-binding domain in Cockayne syndrome B required for transcription-coupled nucleotide excision repair. Mol. Cell. 2010; 38:637–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Epanchintsev A., Costanzo F., Rauschendorf M.A., Caputo M., Ye T., Donnio L.M., Proietti-de-Santis L., Coin F., Laugel V., Egly J.M.. Cockayne's syndrome A and B proteins regulate transcription arrest after genotoxic stress by promoting ATF3 degradation. Mol. Cell. 2017; 68:1054–1066. [DOI] [PubMed] [Google Scholar]
- 54. Chalut C., Moncollin V., Egly J.M.. Transcription by RNA polymerase II: a process linked to DNA repair. Bioessays. 1994; 16:651–655. [DOI] [PubMed] [Google Scholar]
- 55. Bootsma D., Hoeijmakers J.H.. The molecular basis of nucleotide excision repair syndromes. Mutat. Res. 1994; 307:15–23. [DOI] [PubMed] [Google Scholar]
- 56. Friedberg E.C., Bardwell A.J., Bardwell L., Wang Z., Dianov G.. Transcription and nucleotide excision repair–reflections, considerations and recent biochemical insights. Mutat. Res. 1994; 307:5–14. [DOI] [PubMed] [Google Scholar]
- 57. Brooks P.J. Blinded by the UV light: how the focus on transcription-coupled NER has distracted from understanding the mechanisms of Cockayne syndrome neurologic disease. DNA Repair (Amst.). 2013; 12:656–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kyng K.J., May A., Brosh R.M. Jr., Cheng W.H., Chen C., Becker K.G., Bohr V.A.. The transcriptional response after oxidative stress is defective in Cockayne syndrome group B cells. Oncogene. 2003; 22:1135–1149. [DOI] [PubMed] [Google Scholar]
- 59. Nicolai S., Filippi S., Caputo M., Cipak L., Gregan J., Ammerer G., Frontini M., Willems D., Prantera G., Balajee A.S. et al. . Identification of novel proteins co-purifying with Cockayne syndrome group B (CSB) reveals potential roles for CSB in RNA metabolism and chromatin dynamics. PLoS One. 2015; 10:e0128558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ong C.T., Corces V.G.. CTCF: an architectural protein bridging genome topology and function. Nat. Rev.Genet. 2014; 15:234–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lee B.K., Iyer V.R.. Genome-wide studies of CCCTC-binding factor (CTCF) and cohesin provide insight into chromatin structure and regulation. J. Biol. Chem. 2012; 287:30906–30913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Rockx D.A., Mason R., van Hoffen A., Barton M.C., Citterio E., Bregman D.B., van Zeeland A.A., Vrieling H., Mullenders L.H.. UV-induced inhibition of transcription involves repression of transcription initiation and phosphorylation of RNA polymerase II. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:10503–10508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Proietti-De-Santis L., Drane P., Egly J.M.. Cockayne syndrome B protein regulates the transcriptional program after UV irradiation. EMBO J. 2006; 25:1915–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kristensen U., Epanchintsev A., Rauschendorf M.A., Laugel V., Stevnsner T., Bohr V.A., Coin F., Egly J.M.. Regulatory interplay of Cockayne syndrome B ATPase and stress-response gene ATF3 following genotoxic stress. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:E2261–E2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hai T., Wolfgang C.D., Marsee D.K., Allen A.E., Sivaprasad U.. ATF3 and stress responses. Gene Expr. 1999; 7:321–335. [PMC free article] [PubMed] [Google Scholar]
- 66. Hai T., Hartman M.G.. The molecular biology and nomenclature of the activating transcription factor/cAMP responsive element binding family of transcription factors: activating transcription factor proteins and homeostasis. Gene. 2001; 273:1–11. [DOI] [PubMed] [Google Scholar]
- 67. Groisman R., Polanowska J., Kuraoka I., Sawada J., Saijo M., Drapkin R., Kisselev A.F., Tanaka K., Nakatani Y.. The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell. 2003; 113:357–367. [DOI] [PubMed] [Google Scholar]
- 68. Zhou H.X., Wang G.. Predicted structures of two proteins involved in human diseases. Cell Biochem. Biophys. 2001; 35:35–47. [DOI] [PubMed] [Google Scholar]
- 69. Lehmann A.R. Three complementation groups in Cockayne syndrome. Mut. Res. 1982; 106:347–356. [DOI] [PubMed] [Google Scholar]
- 70. Ranes M., Boeing S., Wang Y., Wienholz F., Menoni H., Walker J., Encheva V., Chakravarty P., Mari P.O., Stewart A. et al. . A ubiquitylation site in Cockayne syndrome B required for repair of oxidative DNA damage, but not for transcription-coupled nucleotide excision repair. Nucleic. Acids. Res. 2016; 44:5246–5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wei L., Nakajima S., Bohm S., Bernstein K.A., Shen Z., Tsang M., Levine A.S., Lan L.. DNA damage during the G0/G1 phase triggers RNA-templated, Cockayne syndrome B-dependent homologous recombination. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E3495–E3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wong H.K., Muftuoglu M., Beck G., Imam S.Z., Bohr V.A., Wilson D.M. 3rd. Cockayne syndrome B protein stimulates apurinic endonuclease 1 activity and protects against agents that introduce base excision repair intermediates. Nucleic Acids Res. 2007; 35:4103–4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Williamson L., Saponaro M., Boeing S., East P., Mitter R., Kantidakis T., Kelly G.P., Lobley A., Walker J., Spencer-Dene B. et al. . UV Irradiation Induces a Non-coding RNA that Functionally Opposes the Protein Encoded by the Same Gene. Cell. 2017; 168:843–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Dango S., Mosammaparast N., Sowa M.E., Xiong L.J., Wu F., Park K., Rubin M., Gygi S., Harper J.W., Shi Y.. DNA unwinding by ASCC3 helicase is coupled to ALKBH3-dependent DNA alkylation repair and cancer cell proliferation. Mol. Cell. 2011; 44:373–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Boeing S., Williamson L., Encheva V., Gori I., Saunders R.E., Instrell R., Aygun O., Rodriguez-Martinez M., Weems J.C., Kelly G.P. et al. . Multiomic analysis of the UV-induced DNA damage response. Cell Rep. 2016; 15:1597–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Selby C.P., Drapkin R., Reinberg D., Sancar A.. RNA polymerase II stalled at a thymine dimer: footprint and effect on excision repair. Nucleic Acids Res. 1997; 25:787–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Andrade-Lima L.C., Veloso A., Paulsen M.T., Menck C.F., Ljungman M.. DNA repair and recovery of RNA synthesis following exposure to ultraviolet light are delayed in long genes. Nucleic Acids Res. 2015; 43:2744–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Chiou Y.Y., Hu J., Sancar A., Selby C.P.. RNA polymerase II is released from the DNA template during transcription-coupled repair in mammalian cells. J. Biol. Chem. 2018; 293:2476–2486. [DOI] [PMC free article] [PubMed] [Google Scholar]