Abstract
Mycolactone is a bacteria-derived macrolide that blocks the biogenesis of a large array of secretory and integral transmembrane proteins (TMP) through potent inhibition of the Sec61 translocon. Here, we used quantitative proteomics to delineate the direct and indirect effects of mycolactone-mediated Sec61 blockade in living cells. In T lymphocytes, dendritic cells and sensory neurons, Sec61 substrates downregulated by mycolactone were in order of incidence: secretory proteins (with a signal peptide but no transmembrane domain), TMPs with a signal peptide (Type I) and TMPs without signal peptide and a cytosolic N terminus (Type II). TMPs without a signal peptide and the opposite N terminus topology (Type III) were refractory to mycolactone inhibition. This rule applied comparably to single- and multi-pass TMPs, and extended to exogenous viral proteins. Parallel to its broad-spectrum inhibition of Sec61-mediated protein translocation, mycolactone rapidly induced cytosolic chaperones Hsp70/Hsp90. Moreover, it activated an atypical endoplasmic reticulum stress response, differing from conventional unfolded protein response by the down-regulation of Bip. In addition to refining our mechanistic understanding of Sec61 inhibition by mycolactone, our findings thus reveal that Sec61 blockade induces proteostatic stress in the cytosol and the endoplasmic reticulum.
Keywords: Protein Translocation, Label-free quantification, Cell biology, Stress response, Protein Folding, Endoplasmic reticulum, Mycolactone, Protein translocation, Sec61, Stress
Mycolactone is a polyketide synthase-derived macrolide produced by Mycobacterium ulcerans, the skin pathogen causing Buruli ulcer disease (1). In addition to inducing local skin tissue destruction and analgesia, mycolactone diffuses in infected hosts to dampen immune responses at the systemic level (2). Recent findings demonstrate that mycolactone targets the central subunit of the Sec61 translocon, preventing import of newly synthesized Sec61 substrates into the endoplasmic reticulum (ER)1, and resulting in their cytosolic degradation by the ubiquitin-proteasome system (3, 4). Contrary to the Sec61 inhibitor cotransin, mycolactone is broadly active toward Sec61 substrates. In vitro assays of protein translocation (IVT) nevertheless identified several single- and multi-pass transmembrane proteins (TMPs) resisting its inhibitory action (3, 5, 6). The factors governing Sec61 substrate susceptibility or resistance to mycolactone are only partially understood.
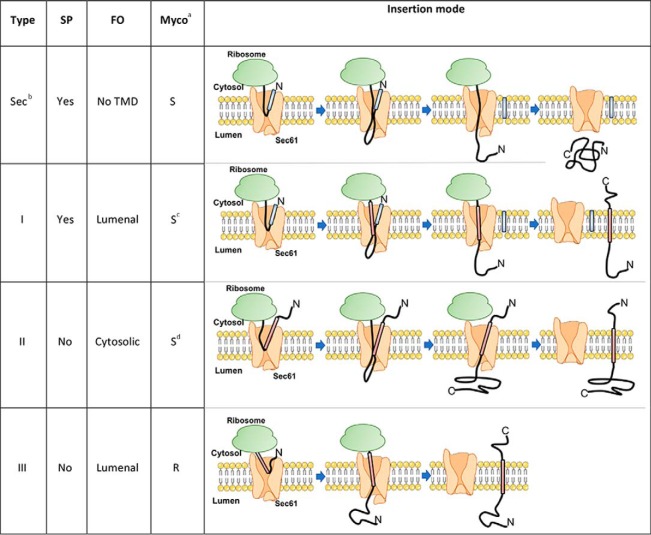
Sec61 substrates include secretory and integral transmembrane proteins (TMPs), which can be divided into Type I, II or III, according to the presence of a signal peptide (SP) and the orientation of the protein N terminus at the ER membrane (7) (Table I and Fig. 6). We classified as secretory all Sec61 substrates with a SP and without transmembrane domain (TMD). This encompasses secreted proteins and most ER-, Golgi-, endosome- and lysosome-resident proteins, as well as proteins containing a glycosylphosphatidylinisotol-anchoring motif. Type I TMPs contain a SP and at least one downstream TMD for initial insertion in the ER membrane, whereas Type II and Type III TMPs do not contain a SP. In Type I and Type III TMPs, the first N-terminal TMD is in a N-lumenal/C-cytosolic orientation at the ER membrane. In Type II TMPs, the N terminus of the initial TMD is on the lumenal side of the ER membrane. By testing the effects of mycolactone on the translocation of TMPs representing each category of Sec61 substrates in cell-free systems, McKenna et al. found that secretory proteins, Type I and Type II TMPs are generally susceptible to mycolactone-mediated Sec61 blockade (5, 6). Partial resistance to mycolactone was observed for some Type I TMPs, depending on their TMD hydrophobicity and lumenal domain size. In contrast, mycolactone had no effect on Type III TMP integration. Because Type III proteins differ from other Sec61 substrates in the way they insert into Sec61 (Table I), and were the only examples of mycolactone-resistant Sec61 substrates in IVT, the authors suggested that protein resistance to mycolactone essentially depends on how the protein initially engage the translocon. Whether this model applies to multi-pass TMPs, and explains all biological effects of mycolactone remained to be addressed.
Table I. Sec61 substrate classification used in this study.
The four types of Sec61 substrates are shown, with characteristic topogenic determinants and mode of insertion in Sec61: Signal peptide (SP, blue segment), final orientation (FO) of the first N-terminal transmembrane domain (TMD, pink). The differential susceptibility of each type of Sec61 sustrates to mycolactone-mediated Sec61 blockade, as proposed by McKenna et al. (6), is indicated (Myco).
aS: Susceptible; R: Resistant.
bSecretory.
cPartial resistance possible, depending on TMD hydrophobicity and lumenal domain size.
dType II TMPs with a short N-terminal domain may be trapped by mycolactone in an inverted orientation.
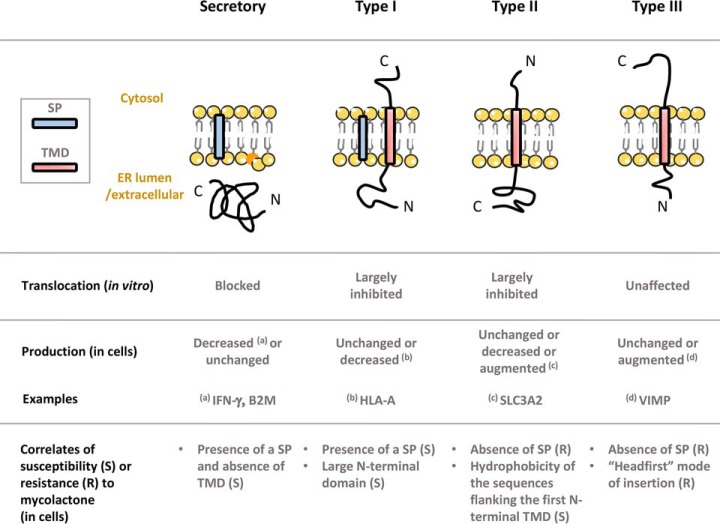
Fig. 6.
Diagram illustrating the differential effects of mycolactone on Sec61 client translocation (in vitro) and production in living cells.
In addition to inducing skin necrosis at the site of infection, bacterial production of mycolactone during infection has been associated with defective induction of pain and immune responses in infected hosts (3, 8–14). By profiling mycolactone-downregulated proteins in dendritic cells and Jurkat T cells exposed to mycolactone in vitro (3, 15), we were able to connect Sec61 blockade with alterations in immune cell functions such as cytokine production, cytokine signaling, antigen presentation and cell migration. With regard to analgesia, mycolactone was shown to activate type 2 angiotensin II receptors (AGTR2) expressed by sensory neurons, leading to cell hyperpolarization and defective pain transmission (12). Our observations that mycolactone prevents the release of inflammatory mediators by nervous cells in vitro, and development of inflammatory pain in vivo, indicated that mycolactone-mediated Sec61 blockade likely contributes to the analgesic properties of mycolactone (10, 16). Further, these data suggested that Sec61 blockade may alter the functional biology of sensory neurons beyond inflammation, and interfere with AGTR2 expression and signaling.
In the present study, we have analyzed the structure and content of mycolactone-susceptible proteomes in dendritic cells, T cells and sensory neurons. Our objectives were to (1) examine the relevance of the McKenna model in a biological setting, (2) delineate the impact of mycolactone on the proteome of sensory neurons and (3) characterize the secondary effects of Sec61 blockade in living cells.
EXPERIMENTAL PROCEDURES
Experimental Design
Table II outlines the conditions used to generate each proteomic dataset, with sample size (number of biological replicates), number and type of controls, and number of proteins that were reliably quantified and modulated by mycolactone. The proteomics data corresponding to Jurkat T cells (3) and dendritic cells (15) were generated previously. Detailed information on the materials and methods used to profile mycolactone-modulated proteins in these cells can be found in the cited references. In the present study, we performed an additional proteomic analysis to characterize the effects of mycolactone on sensory neurons. We used the mouse dorsal root ganglion cell line MED17.11 as it provides a convenient model for nociceptor cell biology (17), and exposed MED17.11 neurons to mycolactone in resting or LPS-activated conditions (supplemental Table S1). In T cells, dendritic cells and MED17.11 neurons, the mycolactone treatment conditions (dose, duration) were optimized prior to proteomic analyses to achieve maximal Sec61 inhibition without inducing cytotoxicity. Statistical methods for analysis are detailed in the Data Processing and Analysis paragraph.
Table II. Experimental conditions and result of proteomic studies.
aPhorbol myristate acetate/ionomycin.
bBiological replicates.
cTotal number (mycolactone-downregulated/mycolactone-upregulated).
Mycolactone
Natural mycolactone A/B was purified from M. ulcerans bacteria (strain 1615) (18), then quantified by spectrophotometry (λmax = 362 nm; log ε = 4.29) as previously described (19). Stock solutions were prepared in DMSO, and diluted 1000 x in culture medium immediately before use in cellular assays.
DNA Constructs
The DNA transcription template for IVT analysis of human SLC3A2 was PCR amplified from a plasmid using 5′-primers containing a T7 promoter, a Kozak sequence and a region complementary to the 5′-end of the gene (SLC3A2 in pENTR221, Genome Biology Unit cloning service, Biocenter Finland, University of Helsinki). The 3′-primers contained a stop-codon and a region complementary to the 3′-end of the gene. The PCR products were purified with a PCR Clean-up kit (Macherey-Nagel) before in vitro transcription. The pBABE-puro vector was kindly donated by Jay Morgenstern and Hartmut Land and distributed through Addgene (Cambridge, MA) (plasmid n°1764, (20)). Sec61 wt or mutant sequences were cloned into the pBABE-puro retroviral vector, for simultaneous translation of Sec61 and puromycin resistance gene in mouse lymphoma B cells.
Cell Cultures, Flow Cytometric Studies, and Viral Infection
MutuDCs (provided by Hans-Acha Orbea, University of Lausanne) were cultured in IMDM (Gibco), supplemented with 8% (v/v) FCS (Biowest-Biosera), 10 mm HEPES, 100 U/ml penicillin, 100 μg/ml streptomycin and 50 μm β-mercaptoethanol (all from Life Technologies). Flow cytometric studies of MutuDCs used anti-mouse MHC I (H2-Kb)-PE (eBioscience 12–5958-80), biotin-conjugated anti-mouse MHC II (I-A/I-E) (BD 553622) with APC-streptavidin (BD 554067), and anti-CD98 (Biolegend 128207). Flow cytometric acquisitions were conducted on an Accuri C6 (BD Biosciences) and analyzed by FlowJo software (TreeStar, Ashland, OR). To induce the UPR response, we used Tunicamycin 1 μm (Sigma T7765), Thapsigargin 1 μm (Sigma T9033) or 1 μm MG132 (Selleckchem S2619). MED17.11 (kindly provided by Mohammed Nassar, University of Sheffield) were cultured in DMEM/F12 Glutamax (Gibco), supplemented with 10% FCS (Biowest-Biosera), 10 ng/ml bFGF (Peprotech), 0.5 mm di-butryl cAMP (Sigma), 25 μm Forskolin (ApexBio Technology), 5 μg/ml Y-27632 (Focus Biomolecules), 100 ng/ml NGF (R&D Systems), 10 ng/ml GDNF (Peprotech) and 100 U/ml penicillin, 100 μg/ml streptomycin (Life Technologies). For Influenza A virus (IAV) infection assays, we used HEK293-Kb cells maintained in DMEM with 7.5% FBS in a 9% CO2 incubator. Recombinant IAV PR8 (A/Puerto Rico/8/34 H1N1) was grown in 10-day embryonated chicken eggs, and used as infectious allantoic fluid. HEK293 cells were resuspended in FCS-free acidified RPMI 1640 medium, infected with IAV at a multiplicity of infection of 10 at 37 °C for 1h, and then subcultured in the presence or absence of 125 nm mycolactone. At the indicated time points, an aliquot of 106 cells was removed, stained with antibodies and analyzed by flow cytometry using the following monoclonal antibodies: NA2–1C1 (anti-NA), H36–26 (anti-HA), O19 (anti-M2), BBM.1 (anti-beta 2 microglobulin), and HB54 (anti-HLA-A2). These antibodies were labeled with Pacific Orange, Alexa Fluor 647 or Alexa Fluor 488, using protein labeling kits from Life Technologies following the manufacturer's recommended protocols. Flow cytometric acquisitions were conducted on an LSR Fortessa X-20 flow cytometer (BD Biosciences) and analyzed by FlowJo software (TreeStar, Ashland, OR). Mouse v-Abl lymphoma B cells were kindly donated by Ludovic Deriano (Institut Pasteur, Paris). They were transduced with retroviruses prepared with the pBABE-Sec61-puro wt or R66G vectors as described in (3), and selected with 2 μg/ml puromycin over 1 week.
Quantitative Real-Time PCR (qPCR)
Total RNA was extracted from MutuDC using Qiazol lysis reagent (Qiagen), then purified using Qiagen RNeasy Mini Kit and digested with RNase-Free DNase set (Qiagen 79254) for 15 min at room temperature. First-strand cDNA was synthesized from 1 μg of total RNA with the high capacity cDNA reverse transcription kit (Applied Biosystems 4368814). Expression was quantified using Power SybR Green PCR Master Mix (Applied Biosystems 4367659) and gene-specific primers (supplemental Table S2). Amplification was performed in duplicate, from 5 ng of cDNA template in a final volume of 20 μl in a 96-well PCR plate. Amplification conditions were 2 min at 50 °C, 10 min at 95 °C, followed by 40 cycles of 15 s at 95 °C and 1 min at 60 °C on a StepOnePlus Real-time PCR System (Applied Biosystems). Results were normalized with the 2−ΔΔCt method by using Rpl-19 as an endogenous control.
IVT Assay
Protein translocation assays were performed as described in (21). DNA templates were transcribed with T7 Polymerase (New England Biolabs) for 1–2 h at 37 °C and used used without purification in subsequent translation/translocation reactions. The reactions were assembled at 0 °C in the presence of mycolactone or an equivalent volume of DMSO. Reactions included 35S-Methionine (Perkin Elmer, 2 μCi per 10 μl translation), RNasin (NEB, #M0314S) 10 U per 10 μl, and Sheep Rough Microsomes (SRM) (22). The amount of SRM was optimized to be 0.25 μl per 10 μl reaction. Translation was initiated by transferring the reactions to 32 °C for 60 min after which they were returned on ice. Reactions were then mixed with an equal volume of 2x High-Salt Buffer (50 mm Hepes, pH 7.8, 1 m KAc, 10 mm MgAc2). The samples were centrifuged at 49,000 rpm for 10 min at 4 °C in a S100-AT3 rotor (Thermo Scientific) through a sucrose cushion in 1 × High Salt Buffer (50 mm Hepes, pH 7.8, 0.5 m KAc, 5 mm MgAc2, 0.5 m sucrose) and the pelleted SRMs with associated translated nascent polypeptides were retrieved. The control reaction without SRMs was analyzed without pelleting. Endoglycosidase H treatment (500 U/reaction, 37 °C, overnight, NEB #P0702S) was performed with the manufacturer's buffer system, to demonstrate that differences in CD98 gel migration are based on glycosylation. After trichloroacetic acid precipitation, the synthesized polypeptides were analyzed by SDS-PAGE and autoradiography. SDS-PAGE analysis was performed either with 12% Tris/Tricine polyacrylamide gels containing 0.5% trichloroethanol (25) for stain-free total protein detection or TGX stain-free gradient gels (Bio-Rad). The dried gels were exposed on a storage phosphorus screen (GE Healthcare) and imaged on a Typhoon Trio phosphorimager (GE Healthcare).
Proteomic Analysis
MED17.11 cells (107 cells, n = 3) were treated with 25 nm mycolactone or DMSO vehicle for 16h, with or without activation with 10 nm LPS after 30 min of exposure to mycolactone. Cells in each condition were harvested and washed twice with PBS. The resulting cell pellets were re-suspended in 4 ml lysis buffer (1 mg/ml amphipol A8–35 (Anatrace) in 50 mm ammonium bicarbonate pH 8.0) and further processed as described in (23). Briefly, lysates were sonicated (three bursts of 15 s at an amplitude of 20%) and centrifuged for 15 min at 20,000 × g at 4 °C to remove insoluble material. The protein concentration in the supernatants was measured using a Bradford assay (Bio-Rad) and 1 ml of each sample containing ∼1 mg of total protein was used to continue the protocol. Proteins in each sample were reduced by addition of 20 mm DTT and incubation for 30 min at 55 °C, and then alkylated by addition of 40 mm iodoacetamide and incubation for 15 min at room temperature in the dark. Samples were acidified with 5% formic acid to pH 3.0 and precipitated proteins and amphipol were pelleted by centrifugation for 10 min at 20,000 × g at room temperature. The resulting protein pellet was washed once with 500 μl water and redissolved in 1 ml 50 mm ammonium bicarbonate pH 8.0. Proteins were digested with 4 μg LysC (Wako) (1/250, w/w) for 4h at 37 °C and then digested with 4 μg trypsin (Promega) (1/250, w/w) overnight at 37 °C. The resulting peptide mixture was acidified by addition of 10% trifluoroacetic acid (TFA) to pH 3.0 and samples were centrifuged for 10 min at 20,000 × g at room temperature to remove amphipol. Purified peptides were dried completely by vacuum drying, re-dissolved in loading solvent A (0.1% TFA in water/acetonitrile (98:2, v/v)) and 3 μg was injected for LC-MS/MS analysis on an Ultimate 3000 RSLCnano System (Thermo) in-line connected to a Q Exactive HF mass spectrometer equipped with a Nanospray Flex Ion source (Thermo). Trapping was performed at 10 μl/min for 4 min in solvent A (on a reverse-phase column produced in-house, 100 μm I.D. x 20 mm, 5 μm beads C18 Reprosil-Pur, Dr. Maisch) followed by loading the sample on a 40 cm column packed in the needle (produced in-house, 75 μm I.D. × 400 mm, 1.9 μm beads C18 Reprosil-HD, Dr. Maisch). Peptides were eluted by an increase in solvent B (0.1% formic acid in water/acetonitrile (2:8, v/v)) in linear gradients from 2% to 30% in 100 min, then from 30% to 56% in 40 min and finally from 56% to 99% in 5 min, all at a constant flow rate of 250 nl/min. The column temperature was kept constant at 50 °C (CoControl 3.3.05, Sonation). The mass spectrometer was operated in data-dependent mode, automatically switching between MS and MS/MS acquisition for the 16 most abundant ion peaks per MS spectrum. Full-scan MS spectra (375–1500 m/z) were acquired at a resolution of 60,000 after accumulation to a target value of 3,000,000 with a maximum fill time of 60 ms. The 16 most intense ions above a threshold value of 13,000 were isolated (window of 1.5 Thomson) for fragmentation at a normalized collision energy of 28% after filling the trap at a target value of 100,000 for maximum 80 ms. The S-lens RF level was set at 55 and we excluded precursor ions with single and unassigned charge states.
Data Processing and Analysis
Data analysis was performed with MaxQuant (version 1.6.0.16) (24) using the Andromeda search engine with default search settings including a false discovery rate set at 1% on both the peptide and protein level. Spectra were searched against the mouse proteins in the Uniprot/Swiss-Prot database (September 2017 version, www.uniprot.org, containing 16,840 entries) with a mass tolerance for precursor and fragment ions of 4.5 and 20 ppm, respectively, during the main search. Enzyme specificity was set as C-terminal to arginine and lysine, also allowing cleavage at proline bonds and a maximum of two missed cleavages. Variable modifications were set to oxidation of methionine residues and acetylation of protein N termini. Carbamidomethyl formation of cysteine residues was set as a fixed modification. Proteins with at least one unique or razor peptide were retained, then quantified by the MaxLFQ algorithm integrated in the MaxQuant software (25). A minimum ratio count of two unique or razor peptides was required for quantification. Further data analysis was performed with the Perseus software (version 1.5.4.1) after loading the protein groups file from MaxQuant. Proteins only identified by site, reverse database hits and potential contaminants were removed and replicate samples were grouped. Proteins with less than three valid values in at least one group were removed and missing values were imputed from a normal distribution around the detection limit. The statistical analysis to determine differentially expressed proteins was performed in R software (version 3.3.2) using the limma package. p values were corrected for multiple testing using the Benjamini-Hochberg method to obtain a False Discovery Rate (FDR). Proteins with a FDR ≤ 0,1 and a log2 mycolactone/control LFQ intensity fold change (log2 FC) > 0.5 were considered upregulated by mycolactone, whereas proteins with a FDR ≤ 0.1 and a log2 FC < -0.5 were considered downregulated.
Protein Annotation and Gene Ontology Analysis
Annotations of SP and TMD positions were downloaded from the Uniprot/Swiss-Prot database. Proteins with a single TMD located less than 20 amino acid residues from the C terminus were labeled as C-tail anchored proteins, and all proteins annotated with a mitochondrial localization were labeled as mitochondrial proteins. Other proteins with a SP and/or at least one TMD were considered Sec61 substrates and classified as secretory protein, Type I, II, or III TMP according to the criteria described in Table I. Proteins missing information needed for classification were excluded from the analysis. Gene ontology analysis was performed using the GoStats package on R software (version 3.3.2). A hypergeometric test was used to rank gene ontology terms (GOT) pertaining to biological processes, then redundant terms were removed using the REVIGO online software (26) and the four most significant terms were retained (Table III).
Table III. Biological process analysis of MutuDC proteins upregulated by mycolactone.
The biological processes that were most significantly enriched in “mycolactone-upregulated” proteins, compared to “all quantified” proteins, are listed. Proteins significantly upregulated by mycolactone in each category are shown with Uniprot accession number, FDR, variation extent of mycolactone/control, gene name and an indication of whether the protein is a Sec61 substrate inferred from www.uniprot.org. Upregulated (FDR ≤ 0.1; log2(Variation) > 0.5).
| Uniprot ID | FDRa | Variationb | Protein namec | Gene namec | Sec61 substrate |
|---|---|---|---|---|---|
| Response to unfolded protein******d | |||||
| P17879 | 9.42E-03 | 16.8 | Heat shock 70 kDa protein 1A;1B | Hspa1b;Hspa1a | |
| Q9JJK5 | 7.45E-05 | 9.17 | Homocysteine-responsive endoplasmic reticulum-resident ubiquitin-like domain member 1 protein | Herpud1 | Yes |
| Q9R099 | 2.05E-02 | 6.72 | Transducin beta-like protein 2 | Tbl2 | |
| Q9BCZ4 | 7.86E-03 | 3.95 | Selenoprotein S | Vimp | Yes |
| Q61699 | 7.70E-03 | 1.97 | Heat shock protein 105 kDa | Hsph1 | |
| Q3TDN2 | 2.03E-02 | 1.7 | FAS-associated factor 2 | Faf2 | |
| P07901 | 4.20E-03 | 1.65 | Heat shock protein HSP 90-alpha | Hsp90aa1 | |
| Q9QY76 | 3.70E-02 | 1.61 | Vesicle-associated membrane protein-associated protein B | Vapb | |
| P35821 | 5.61E-02 | 1.55 | Tyrosine-protein phosphatase non-receptor type 1 | Ptpn1 | |
| Q8BGQ7 | 2.79E-02 | 1.45 | Alanine–tRNA ligase, cytoplasmic | Aars | |
| Protein exit from endoplasmic reticulum*****d | |||||
| Q9BCZ4 | 7.86E-03 | 3.95 | Selenoprotein S | Vimp | Yes |
| O35166 | 1.75E-02 | 2.86 | Golgi SNAP receptor complex member 2 | Gosr2 | |
| P70295 | 1.52E-03 | 1.93 | Ancient ubiquitous protein 1 | Aup1 | Yes |
| P61620 | 1.35E-02 | 1.89 | Protein transport protein Sec61 subunit alpha isoform 1 | Sec61a1 | Yes |
| Q8CI04 | 3.28E-02 | 1.77 | Conserved oligomeric Golgi complex subunit 3 | Cog3 | |
| Q3TDN2 | 2.03E-02 | 1.7 | FAS-associated factor 2 | Faf2 | |
| P07901 | 4.20E-03 | 1.65 | Heat shock protein HSP 90-alpha | Hsp90aa1 | |
| Q9CQU3 | 5.68E-02 | 1.48 | Protein RER1 | Rer1 | Yes |
| tRNA aminoacylation for protein translation****d | |||||
| Q8BP47 | 2.57E-03 | 1.91 | Asparagine–tRNA ligase, cytoplasmic | Nars | |
| P26638 | 3.02E-03 | 1.78 | Serine–tRNA ligase, cytoplasmic | Sars | |
| Q9ER72 | 1.25E-02 | 1.84 | Cysteine–tRNA ligase, cytoplasmic | Cars | |
| Q9D0R2 | 1.32E-02 | 1.63 | Threonine–tRNA ligase, cytoplasmic | Tars | |
| Q8BMJ2 | 2.57E-02 | 1.63 | Leucine–tRNA ligase, cytoplasmic | Lars | |
| Q8BGQ7 | 2.79E-02 | 1.45 | Alanine–tRNA ligase, cytoplasmic | Aars | |
| Q8CGC7 | 3.73E-02 | 1.46 | Bifunctional glutamate/proline–tRNA ligase | Eprs | |
| Positive regulation of tyrosine kinase activity****d | |||||
| Q60823 | 8.73E-03 | 3.39 | RAC-beta serine/threonine-protein kinase | Akt2 | |
| P09535 | 7.12E-04 | 2.81 | Insulin-like growth factor II;Preptin | Igf2 | Yes |
| Q61699 | 7.70E-03 | 1.97 | Heat shock protein 105 kDa | Hsph1 | |
| Q8R0X7 | 8.57E-02 | 1.84 | Sphingosine-1-phosphate lyase 1 | Sgpl1 | Yes |
| Q9JM90 | 1.20E-02 | 1.73 | Signal-transducing adaptor protein 1 | Stap1 | |
| P08103 | 8.74E-02 | 1.63 | Tyrosine-protein kinase HCK | Hck | |
| Q91YI4 | 5.71E-03 | 1.59 | Beta-arrestin-2 | Arrb2 | |
| P35821 | 6.75E-02 | 1.55 | Tyrosine-protein phosphatase non-receptor type 1 | Ptpn1 | |
| P28867 | 5.61E-02 | 1.55 | Protein kinase C delta type | Prkcd | |
| O55143 | 3.28E-02 | 1.55 | Sarcoplasmic/ER calcium ATPase 2 | Atp2a2 | Yes |
aFalse Discovery Rate.
b(mycolactone/control) ratio of relative LFQ intensities.
cAccording to www.uniprot.org.
dHypergeometric test comparing the incidence of GOT between “upregulated” and “all quantified” proteins in MutuDCs
******, p < 10−6;
*****, p < 10−5;
****, p < 10−4.
Statistics
The Graphpad Prism software (6.0; La Jolla, CA) was used for statistical comparisons and graphical representations. Values of p ≤ 0.05 were considered significant.
RESULTS
Conserved and Variable Features of Mycolactone-induced Proteomic Alterations
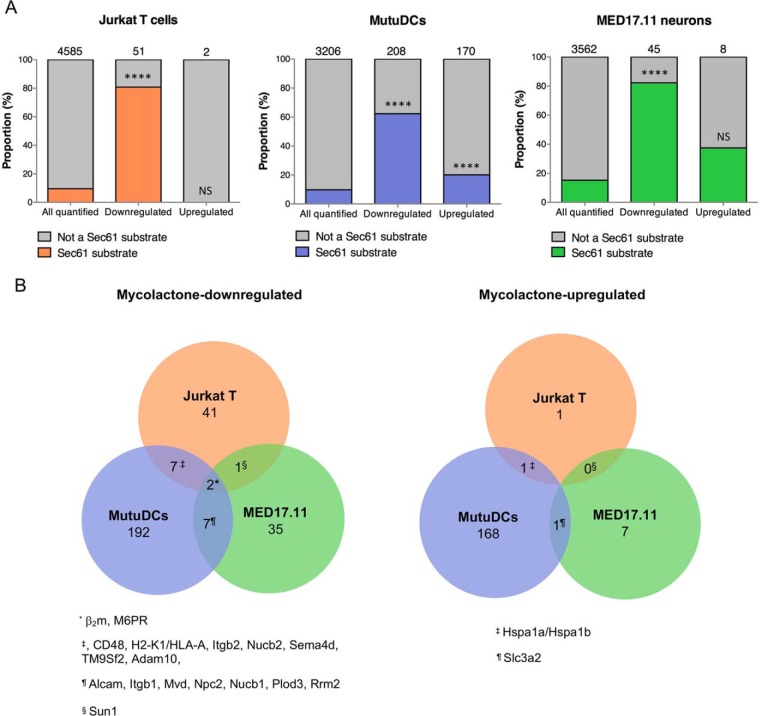
In previous studies using activated Jurkat T cells and MutuDCs, mycolactone-mediated Sec61 blockade impacted the cell proteome by downregulating a subset of proteins primarily composed of Sec61 substrates (3, 15). As shown in Fig. 1A, Sec61 substrates constituted 81 and 62% of mycolactone-downregulated proteins in Jurkat T cells and MutuDCs respectively, whereas their incidence in “all quantified” proteins was close to 10%. This distinctive alteration of the proteome was conserved in mycolactone-exposed MED17.11 neurons, in both resting and LPS-stimulated conditions (Figs. 1A and supplemental Fig. S1, supplemental Table S1). At the same time, we detected proteins that were significantly upregulated by mycolactone in each proteome. Although limited to 2 and 8 proteins, respectively, in Jurkat T cells and MED17.11 neurons exposed to mycolactone, 170 proteins were upregulated by mycolactone in MutuDCs, with a clear preference for Sec61 substrates (Fig. 1A). We next sought to compare mycolactone-upregulated and -downregulated proteins across cell types, by matching the genes of all proteins detected in Jurkat T cells with their mouse orthologs, which was possible for 2032 of 4585. We observed little overlap between mycolactone-altered proteins across Jurkat T cells, MutuDCs and MED17.11 neurons (Fig. 1B). Among proteins that were detected and conserved across species, those modulated by mycolactone in only one cell type were not modulated in others (supplemental Table S3). Together, our data in Figs. 1A and 1B thus support the view that mycolactone-mediated Sec61 blockade affect total proteomes in a cell-type specific manner, because of differences in Sec61 client turnover rates. Differences in the duration of mycolactone treatments may also account for variations in the magnitude of its inhibitory effects across experiments. Of note, beta-2 microglobulin (β2m, a component of the class I major histocompatibility complex) and cation-dependent mannose-6-phosphate receptor (M6PR) were downregulated by mycolactone in all cell types, thus representing potential markers of its activity.
Fig. 1.
Conserved and variable features of mycolactone-induced proteomic alterations. A, The proportion of Sec61 substrates in “all quantified” proteins is compared with that in “mycolactone downregulated” or “mycolactone-upregulated” proteins, in each cell type studied. Jurkat T cells (left), MutuDCs (middle) and MED17.11 cells (right) were treated with mycolactone or vehicle as control in the conditions outlined in Table II. Number of identified proteins in each subset are indicated on the top of each bar, **** p value < 0.0001, ns: not significant, Fisher exact test. B, Venn diagrams representing the overlap between mycolactone downregulated (left) or mycolactone-upregulated (right) proteins across cell types. Human proteins (Jurkat T cells) were matched to their mouse orthologues (MutuDCs and MED17.11 neurons). Proteins that were found downregulated or upregulated by mycolactone in 2 cell types or more are listed.
Primary Determinants of Sec61 Substrate Susceptibility or Resistance to Mycolactone
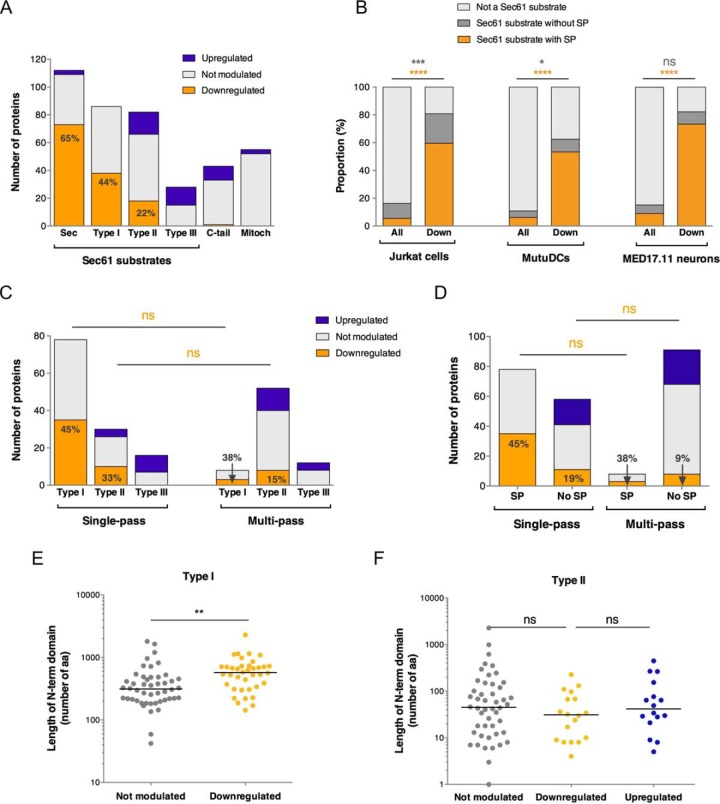
We initially analyzed the relative incidence of each category of Sec61 substrates (as defined in Table I) among mycolactone-downregulated proteins. Proteomic analyses were performed on cell extracts, and consequently secreted proteins could not be analyzed. Yet, >100 secretory, organelle-resident proteins were reliably quantified in MutuDCs, allowing statistical comparisons in the protein datasets from this cell type. The highest proportion of mycolactone-downregulated proteins was found in secretory proteins (65%), followed by Type I (44%), Type II (22%) and Type III (0%) TMPs (Fig. 2A). In all three studied cell types, presence of a SP in a Sec61 substrate was highly predictive of its down-regulation by mycolactone (Fig. 2B). The near complete absence of mycolactone-upregulated proteins in secretory proteins and Type I TMPs was consistent with SP-bearing Sec61 substrates being globally susceptible to mycolactone inhibition. At the opposite end of the spectrum, no Type III TMPs were downregulated by mycolactone. In this regard, Type III TMPs resembled C-terminal tail-anchored proteins and mitochondrial membrane proteins (Fig. 2A), which are not Sec61 substrates. This result supported McKenna's prediction that Type III TMPs resist mycolactone inhibition (6). Type II TMPs displayed an intermediate phenotype, with a relatively low incidence of mycolactone-downregulated proteins compared with secretory proteins and Type I TMPs, and the presence of mycolactone-upregulated proteins (Fig. 2A), leaving open the question of whether they contain mycolactone-resistant elements.
Fig. 2.
Primary determinants of Sec61 substrate susceptibility or resistance to mycolactone. A, Bar plot depicting the number of proteins that were upregulated, downregulated or not significantly modulated by mycolactone in MutuDCs. Sec: secretory proteins; C-tail: C-terminal tail-anchored proteins, depending on the Guided Entry of Tail-anchor (GET) pathway for insertion into the ER membrane; Mitoch: mitochondrial membrane proteins, depending on the TIM/TOM complexes for mitochondrial membrane insertion. B, The proportion of Sec61 substrates with a SP (secretory + Type I TMP) or without a SP (Type II/III TMPs) in ≪ all detected ≫ proteins (All) or downregulated proteins (Down) is shown for each cell type studied. Fisher exact tests comparing the proportions of Sec61 substrates with SP (orange) or without SP (gray) to all other proteins. *p < 0.05; *** p < 0.001; **** p < 0.0001. C, The proportions of mycolactone-downregulated proteins in each class of single-pass TMPs is shown compared with those in multi-pass TMPs. D, The proportion of mycolactone-downregulated proteins in single-pass TMPs with or without an SP is shown compared with those in multi-pass TMPs. Fisher exact test comparing the proportion of mycolactone-downregulated proteins in each subset. ns: not significant. E, F, Scatter dot plot representing the length (in amino acid residues) of the N-terminal domain before the first TMD in Type I (E) and Type II (F) TMPs of MutuDCs. A Mann-Whitney test was used to compare mean lengths in mycolactone-downregulated proteins and proteins not modulated by mycolactone. **p < 0.01; ns: no significant difference.
The relative incidence of mycolactone-downregulated proteins in Type I/II/III TMPs was comparable in single-pass and multi-pass TMPs (Fig. 2C). Similarly, to single-pass TMPs, multi-pass TMPs with a SP included a higher proportion of mycolactone-downregulated proteins than multi-pass TMPs without SP and did not contain any mycolactone-upregulated proteins (Fig. 2D). This observation suggested that Sec61 substrate susceptibility to mycolactone is primarily determined by the initial interaction between SP or first TMD of nascent polypeptides with the translocon, irrespective of the number of TMDs.
Interestingly, Type I TMPs were differentially downregulated by mycolactone. We found that the length of their N-terminal domain discriminated significantly mycolactone-downregulated proteins from the non-regulated ones (Fig. 2E). In line with McKenna et al. 's findings using genetically modified Type I TMPs (6) (Table I), this result indicated that Type I TMPs with a long N-terminal lumenal domain in the ER are relatively more sensitive to mycolactone. In contrast, N-terminal domain length did not affect Type II TMP's modulation by mycolactone (Fig. 2F).
Mycolactone-mediated Sec61 Blockade Prevents the Production of Viral Type I/II but Not Type III TMPs
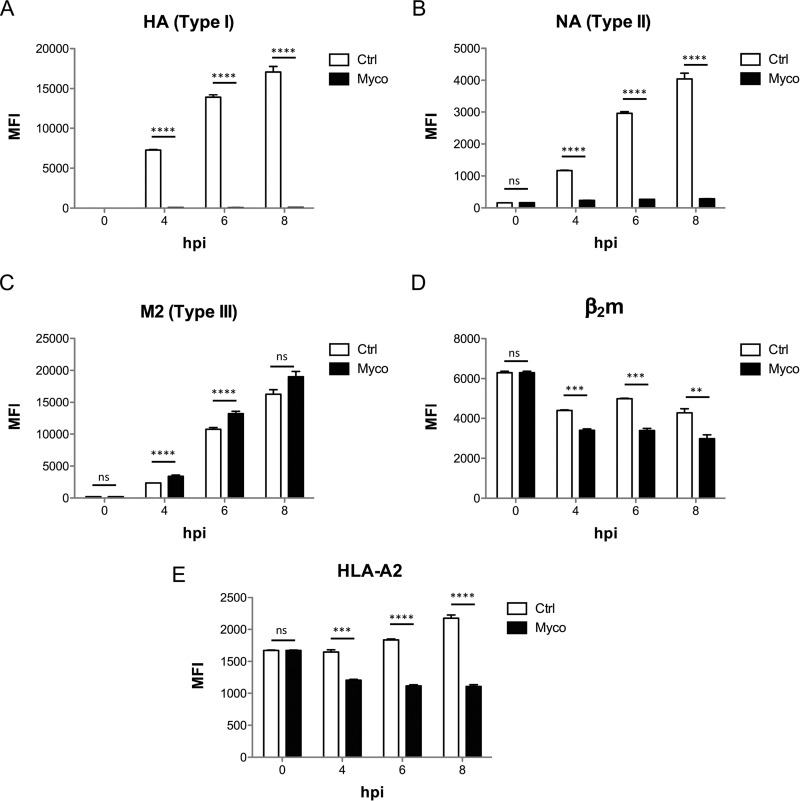
Having described the effects of mycolactone on the biogenesis of endogenous Sec61 substrates, we used influenza A virus (IAV) as a convenient model to study mycolactone's impact on production of virus-derived Sec61 substrates in infected cells. The IAV envelope contains three viral proteins, with Type I (HA), Type II (NA), and Type III (M2) topology. Mycolactone added to HEK293-Kb cells 1h post-infection with IAV efficiently prevented the cell surface expression of HA and NA, but not M2 protein (Figs. 3A–3C). In fact, the cell production of M2 was slightly elevated in mycolactone-treated cells, compared with controls (Fig. 3C), suggesting that inhibition of Type I/II TMP translocation may promote that of mycolactone-resistant Type III TMPs. Mycolactone treatment decreased concomitantly the cell surface expression of MHC Class I molecules, measured with antibodies specific for β2m or HLA-A2, which are constitutively synthesized by HEK293-Kb cells (Fig. 3D–3E). These data are consistent with previous findings, showing that MHC class I heavy chains and β2m are Sec61 substrates that are among the most susceptible to mycolactone inhibition (3, 15) (Fig. 1B).
Fig. 3.
Mycolactone-mediated Sec61 blockade prevents the production of viral Type I/II but not Type III TMPs. Surface expression of viral envelope proteins HA (A), NA (B) and M2 (C) as well as HLA components B2M (D) and HLA-A2b (E), in HEK293 cells infected with IAV for 1h prior to incubation with Mycolactone, or vehicle as control. MFI: mean fluorescence intensity; hpi: hours post infection. Data shown are MFI ± S.E. (n = 3) from one of two independent experiments, which gave similar results. MFIs of mycolactone-treated cells were compared with vehicle controls using a t test with Holm-Sidak correction for multiple testing. *p < 0.05; **p < 0.01; ***p < 0.001; ***p < 0.001; ns: no significant difference.
Low Doses of Mycolactone Upregulate the Transcription of Selected Sec61 Substrates
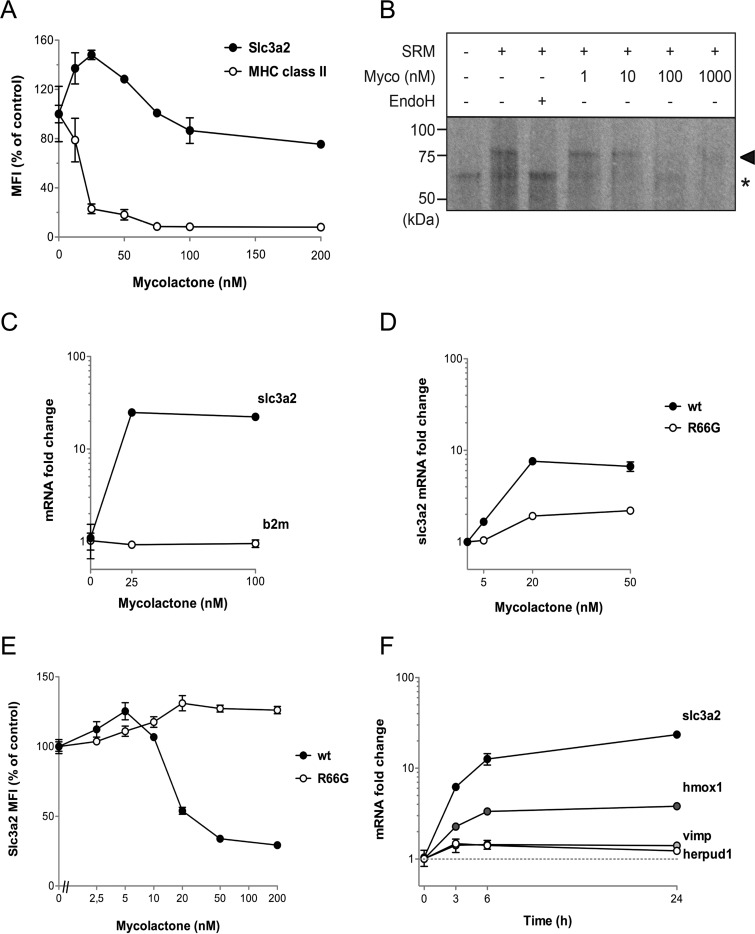
We were surprised to see in MutuDCs, but also in MED17.11 cells to a limited extent, the presence of mycolactone-upregulated Sec61 substrates (Fig. 1). In MutuDCs, most mycolactone-upregulated Sec61 substrates belonged to Type II or Type III subtypes of TMPs (Fig. 2A). CD98 (Slc3a2/Slc7a5) is a heterodimeric receptor contributing to amino acid transport and integrin signaling (27), of which both chains were significantly upregulated by mycolactone in MutuDCs (15). Notably, Slc3a2 was also upregulated in mycolactone-exposed MED17.11 neurons, in both resting and LPS-activated conditions (Fig. 1B, supplemental Table S1). Flow cytometric analysis of mycolactone-exposed MutuDCs revealed that the Slc3a2 dose-response curve displayed an unusual “bell” shape, with mycolactone doses <100 nm leading to increased surface expression of the receptor after 24 h (Fig. 4A). In comparison, (Type I) MHC class II expression was consistently suppressed by mycolactone, and fully abrogated by 100 nm mycolactone (Fig. 4A).
Fig. 4.
Low doses of mycolactone-upregulate the transcription of selected Sec61 substrates. A, Surface expression of Slc3a2 and MHC class II (I-A/I-E) in MutuDCs treated with the indicated doses of mycolactone for 24 h. B, Assay of Slc3a2 insertion in SRM, in the presence of increasing amounts of mycolactone (Myco). Membrane integration was assessed by analyzing the change in SDS-PAGE mobility and autoradiography. Correctly integrated, glycosylated SLC3A2 species are indicated with arrowheads and non-translocated, unglycosylated protein species with an asterisk. Endoglycosidase H (EndoH) treatment demonstrates that the change in SDS-PAGE migration is because of glycosylation. C, qRT-PCR comparing the expression slc3a2 and b2m in MutuDCs treated with 25 or 100 nm mycolactone for 24 h. D, qRT-PCR comparing the expression of slc3a2 in wt or R66G Sec61-expressing lymphoma B cells following a 24h treatment with 25 or 100 nm mycolactone. E, Effect of mycolactone on CD98 surface expression by lymphoma B cells overexpressing wild-type (wt) Sec61 or the mycolactone-resistant R66G Sec61 mutant. Cells were treated with the indicated doses of mycolactone for 24h, prior to flow cytometric analysis. F, Kinetic effects of mycolactone on transcript levels of slc3a2, hmox1, vimp and herpud1, as measured by qPCR in MutuDCs treated with 25 nm mycolactone for the indicated times. Data are mean ± S.E. (n = 3) from one of two independent experiments, which gave similar results.
In biochemical assays, 100 nm mycolactone fully blocked Slc3a2 membrane integration (Fig. 4B), demonstrating Slc3a2 susceptibility to mycolactone-mediated Sec61 blockade. Intriguingly, the increased expression of Slc3a2 by MutuDCs exposed to 25 nm mycolactone correlated with an acute upregulation of slc3a2 gene expression (Fig. 4C). This was a selective effect, as the same mycolactone treatment did not modify the levels of β2m transcripts (Fig. 4C). Low doses of mycolactone (<10 nm) triggered a comparable increase in slc3a2 gene and protein expression in a B-lymphoma cell line over-expressing wild-type (wt) Sec61 (Fig. 4D–4E). These effects were largely attenuated in B cells transduced with the mycolactone-resistant R66G mutant of Sec61 (Fig. 4D–4E), demonstrating the essential participation of Sec61 in Slc3a2 upregulation. Notably, transcription of Slc3a2 (Type II TMP), Herpud1 (Type II TMP), Hmox1 (not a Sec61 substrate) and Vimp (Type III TMP), all proteins upregulated by mycolactone in our proteomic analysis of MutuDCs, was increased in MutuDCs exposed to 25 nm mycolactone for only 3 h (Fig. 4F). From these data, we propose that mycolactone triggers a transcriptional stress response to ER translocation blockade encompassing the above described genes. Partial Sec61 inhibition by low mycolactone doses may explain the increased production of stress-induced Sec61 clients, despite their biochemical susceptibility to mycolactone.
Mycolactone-upregulated Proteins Outline Distinctive Stress Responses
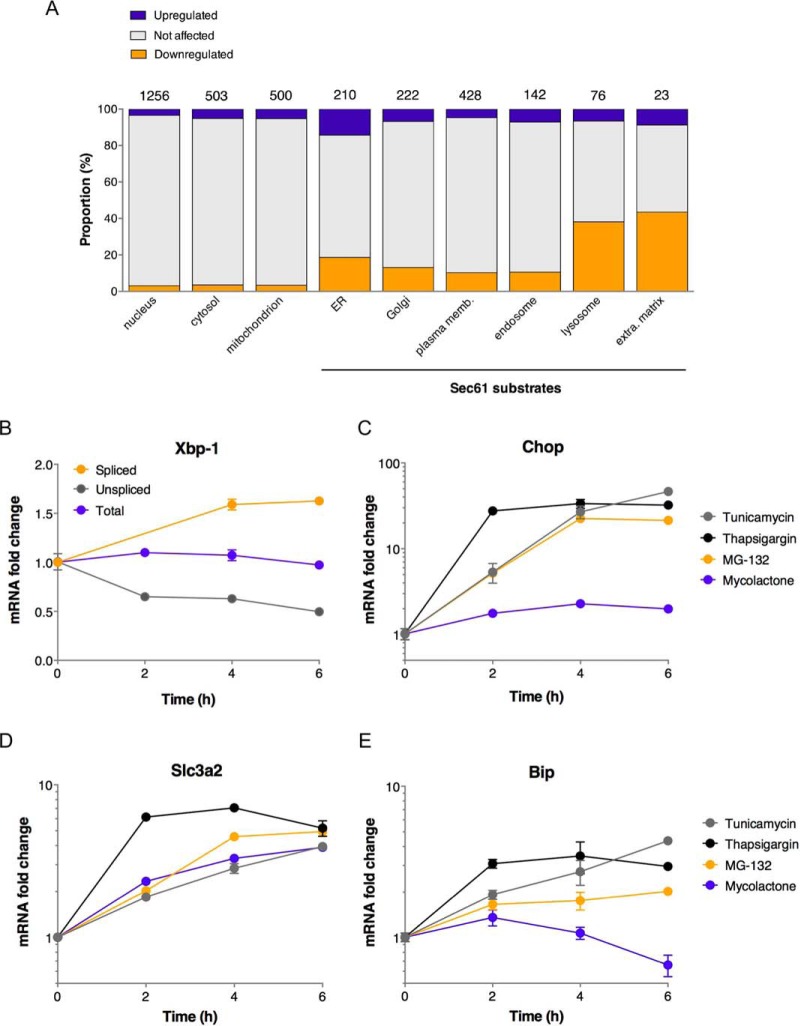
The data presented in Figs. 1 and 4 show that mycolactone triggered proteome-wide alterations, beyond Sec61 substrates. Hsp70 (Hspa1a/b), the stress-induced form of the Hsc70 molecular chaperone critical for nascent protein folding, was upregulated by mycolactone in both Jurkat T cells and MutuDCs (Fig. 1). Hsp90 (Hsp90aa/b1), which forms with Hsp70 a multichaperone machinery regulating proteostasis, was also upregulated by mycolactone in MutuDCs. In order to identify additional stress markers, we next analyzed in further detail all proteins upregulated by mycolactone in MutuDCs. Fig. 5A compares the distribution of mycolactone-upgulated, mycolactone-downregulated and non-modulated proteins in the different subcellular compartments of MutuDCs. Notably, mycolactone-upregulated proteins were most prevalent in the ER, but some were present in every cell compartment. A GOT analysis revealed that mycolactone-upregulated proteins were selectively enriched in markers of the ≪ unfolded protein response ≫ (UPR) and ≪ protein exit from the ER ≫, and to a lower extent with proteins involved in ≪ tRNA aminoacylation for protein translation≫ and ≪ positive regulation of tyrosine kinase activity ≫ (Table III). Together, these data suggested that Sec61 blockade triggers ER stress propagating to diverse physiological processes through the UPR (reviewed in (28)).
Fig. 5.
Mycolactone-upregulated proteins outline an atypical stress response. A, Proportion of mycolactone-downregulated or -upregulated Sec61 substrates across cell compartments in MutuDCs. Compartements primarily composed of Sec61 substrates are indicated. The numbers above the bars indicate the number of proteins in each category. B, Kinetics of mycolactone effects (25 nm) on MutuDC expression of total, spliced and unspliced Xbp-1. C–E, Differential effects of Tunicamycin (1 μm), Thapsigargin (1 μm), MG132 (1 μm) and mycolactone (25 nm) on expression of Chop, Slc3a2 and Bip in MutuDCs. Data are mean ± S.E. from biological triplicates.
ER-resident stress sensors IRE1α, PERK and ATF6 were not detected in our proteomic analyses, however as Type I (IRE1α and PERK) or Type II (ATF6) TMPs, they are predicted to be susceptible to mycolactone inhibition. When activated by ER stress, IRE1α splices Xbp1 mRNA, which can be monitored by quantitative real time PCR (29). We detected enhanced splicing of Xbp1 mRNA in MutuDCs exposed to mycolactone for 4h, indicating that the IRE1-α pathway was activated (Fig. 5B). Thapsigargin, tunicamycin, and MG132 are potent inducers of ER stress, which operate through inhibition of sarco-endoplasmic reticulum Ca2+ ATPases, protein glycosylation, and proteasome, respectively. Although less potent than thapsigargin (1 μm), mycolactone used at 25 nm was comparable to tunicamycin (1 μm) and superior to MG132 (1 μm) in capacity to induce Xbp-1 mRNA splicing (supplemental Fig. S3). ER stress-activated PERK phosphorylates eIF2α, which stimulates the translation of the ATF4 transcription factor. Together with activated ATF6, activated ATF4 induces the transcriptional upregulation of the C/EBP Homologous protein (Chop). Although less potent than canonical ER stressors in the conditions tested, mycolactone induced significant expression of Chop in MutuDCs after 2 h (Fig. 5C). Moreover, we identified 27 targets of the ATF4 and/or Chop transcription factors (30) within the 170 proteins upregulated by mycolactone in MutuDCs (Table IV), a significant enrichment compared with controls (p value<0.0001, Fisher exact test). ATF4/Chop targets upregulated by mycolactone included both chains of CD98 (Slc3a2/Slc7a5), Herpud1 and Hmox1. Fig. 5D shows that mycolactone was comparable to tunicamycin and MG132 for stimulation of Slc3a2 expression. In MED17.11 neurons, 2 of the 8 proteins upregulated by mycolactone were ATF4 targets (Table IV). Altogether, these data indicated that mycolactone robustly activates the ATF4/Chop branch of UPR. Importantly, although thapsigargin, tunicamycin, and MG132 all upregulated the expression of Bip, a master regulator of the UPR, mycolactone showed the opposite effect (Fig. 5D). Significant reduction in Bip transcript levels was reproducibly observed in MutuDCs exposed to mycolactone for longer than 4 h. We conclude that mycolactone triggers an atypical ER stress response, differing from conventional UPR by the down-regulation of Bip.
Table IV. Mycolactone upregulated proteins are enriched in targets of the ATF4 and CHOP transcription factor targets.
Mycolactone upregulated proteins (FDR ≤ 0.1; log2(Variation) > 0.5) of MutuDCs and MED17.11 neurons that are known targets of the ATF4 and CHOP transcription factors (37) are listed with Uniprot accession number, FDR, variation extent of mycolactone/control, gene name, presence of an ATF4 and/or CHOP binding site on their promoter region and type of Sec61 substrate type.
| MutuDCs | ||||||
|---|---|---|---|---|---|---|
| Uniprot ID | FDRa | Variationb | Protein namec | Gene name | CHOP/ATF4 | Sec61 substrate |
| Q3UM18 | 7.50E-02 | 43.87 | Large subunit GTPase 1 homolog | Lsg1 | ATF4 Only | No |
| Q9JJK5 | 7.45E-05 | 9.17 | Homocysteine-responsive endoplasmic reticulum-resident ubiquitin-like domain member 1 protein | Herpud1 | ATF4 Only | Type II TMP |
| P14901 | 2.11E-04 | 5.29 | Heme oxygenase 1 | Hmox1 | ATF4 Only | No |
| P10852 | 5.85E-04 | 3.50 | 4F2 cell-surface antigen heavy chain | Slc3a2 | ATF4 Only | Type II TMP |
| Q8BH04 | 1.36E-02 | 3.03 | Phosphoenolpyruvate carboxykinase [GTP], mitochondrial | Pck2 | ATF4 Only | No |
| Q61024 | 1.66E-03 | 2.85 | Asparagine synthetase [glutamine-hydrolyzing] | Asns | ATF4 Only | No |
| Q9Z127 | 7.67E-03 | 2.76 | Large neutral amino acids transporter small subunit 1 | Slc7a5 | Both | Type II TMP |
| Q64337 | 1.41E-04 | 2.67 | Sequestosome-1 | Sqstm1 | Both | No |
| P53995 | 7.62E-02 | 2.42 | Anaphase-promoting complex subunit 1 | Anapc1 | ATF4 Only | No |
| Q8CH25 | 1.56E-03 | 2.09 | SAFB-like transcription modulator | Sltm | ATF4 Only | No |
| Q8BP47 | 2.57E-03 | 1.91 | Asparagine–tRNA ligase, cytoplasmic | Nars | Both | No |
| Q6WKZ8 | 1.58E-02 | 1.89 | E3 ubiquitin-protein ligase UBR2 | Ubr2 | Both | No |
| Q9ER72 | 1.25E-02 | 1.84 | Cysteine–tRNA ligase, cytoplasmic | Cars | ATF4 Only | No |
| P26638 | 3.02E-03 | 1.78 | Serine–tRNA ligase, cytoplasmic | Sars | Both | No |
| Q9Z110 | 1.41E-02 | 1.77 | Delta-1-pyrroline-5-carboxylate synthase;Glutamate 5-kinase;Gamma-glutamyl phosphate reductase | Aldh18a1 | Both | No |
| Q9D0R2 | 1.32E-02 | 1.63 | Threonine–tRNA ligase, cytoplasmic | Tars | ATF4 Only | No |
| P59325 | 7.67E-03 | 1.63 | Eukaryotic translation initiation factor 5 | Eif5 | Both | No |
| Q8BMJ2 | 2.57E-02 | 1.63 | Leucine–tRNA ligase, cytoplasmic | Lars | Both | No |
| Q99K85 | 2.16E-03 | 1.60 | Phosphoserine aminotransferase | Psat1 | ATF4 Only | No |
| P18155 | 5.51E-03 | 1.59 | Bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase | Mthfd2 | Both | No |
| Q64131 | 7.39E-02 | 1.57 | Runt-related transcription factor 3 | Runx3 | ATF4 Only | No |
| Q61753 | 1.96E-02 | 1.54 | d-3-phosphoglycerate dehydrogenase | Phgdh | ATF4 Only | No |
| A2AN08 | 1.26E-02 | 1.51 | E3 ubiquitin-protein ligase UBR4 | Ubr4 | ATF4 Only | Type II or III TMP |
| Q3UPF5 | 2.33E-02 | 1.48 | Zinc finger CCCH-type antiviral protein 1 | Zc3hav1 | Both | No |
| Q8CGC7 | 3.73E-02 | 1.46 | Bifunctional glutamate/proline–tRNA ligase;Glutamate–tRNA ligase;Proline–tRNA ligase | Eprs | Both | No |
| Q9D898 | 3.66E-02 | 1.45 | Actin-related protein 2/3 complex subunit 5-like protein | Arpc5l | ATF4 Only | No |
| Q8BGQ7 | 2.79E-02 | 1.45 | Alanine–tRNA ligase, cytoplasmic | Aars | Both | No |
| MED17.11 sensory neurons | ||||||
| P10852 | 1.43E-02 | 1.709 | 4F2 cell-surface antigen heavy chain | Slc3a2 | ATF4 Only | Type II SP |
| Q9DB73 | 3.56E-02 | 1.556 | NADH-cytochrome b5 reductase 1 | Cyb5r1 | Both | Type II or III SP |
aFalse Discovery Rate.
b(mycolactone/control) ratio of relative LFQ intensities.
cAccording to www.uniprot.org.
DISCUSSION
The present work outlines distinctive proteomic alterations induced by mycolactone, resulting from primary and secondary effects on protein translocation blockade (summarized in Fig. 6). Regarding the direct consequences of Sec61 blockade, our integrated analysis of mycolactone's signature across cell types confirmed the predictions of McKenna's model e.g. (1) susceptibility of secretory proteins, (2) susceptibility of Type I TMPs, modulated by the size of their N-terminal TMD, and (3) resistance of Type III TMPs (Fig. 6). It remains to be determined whether a fraction of Type II TMPs may resist mycolactone inhibition. We detected mycolactone-upregulated Sec61 substrates within Type II TMPs, which supports this possibility. CD98 characterization nevertheless revealed that Type II TMPs upregulated by mycolactone are not necessarily resistant to mycolactone-mediated Sec61 blockade. Interestingly, a positive correlation was observed between the hydrophobicity of the sequences flanking the first N-terminal TMD in Type II TMPs and their susceptibility to mycolactone (supplemental Fig. S2). How hydrophobic signals in nascent polypeptides trigger Sec61 opening is not fully understood. In an inactive translocon, the Sec61 channel is occluded by a plug helix that must be displaced for protein translocation. The recent structure of an active, signal-engaged Sec61 suggests that ribosome binding triggers dynamic conformational changes in Sec61 that allow the insertion of hydrophobic signals in the central pore, while destabilizing the plug (30). Notably, amino acid substitutions conferring resistance to mycolactone all localize to the plug or lateral gate junction (3). The data in supplemental Fig. S2 thus suggest that mycolactone may operate by strengthening the molecular contacts between plug and lateral gate, thus increasing the hydrophobic threshold that is required for channel opening.
In accordance with the tissue-specific effects of mycolactone in patients with Buruli ulcer disease, the host proteins that were downregulated by mycolactone varied across cell types. Notably, β2m and M6PR were downregulated by mycolactone in T cells, dendritic cells and neurons, highlighting their potential interest as indicators of mycolactone activity. Virus envelope proteins were not different from endogenous TMPs in regard of their susceptibility to mycolactone-mediated Sec61 inhibition. Blocking Sec61 with mycolactone in Zika virus-infected cells efficiently prevented the cytopathic formation of ER-derived vacuoles (31). In the present work, mycolactone treatment of IAV-infected cells prevented production of Type I/II virus envelope glycoproteins, further illustrating the interest of mycolactone as a research tool to investigate Sec61 contribution to viral life cycles.
AGTR2 was not detected in our proteomic analysis of MED17.11 neurons (supplemental Table S1). However, as a Type III multi-pass TMP, AGTR2 is predicted to resist mycolactone-mediated Sec61 blockade. Only 8 proteins were upregulated by mycolactone in the conditions tested, suggesting that mycolactone induction of neuronal stress was minimal (Table II). Yet, we identified 45 proteins that were significantly downregulated by mycolactone (supplemental Table S1). Interestingly, a GOT analysis revealed that mycolactone-downregulated proteins were enriched in Sec61 clients mediating interactions between neurons and the extracellular matrix (ECM): Col1a1, Col3a1, Col5a1, Fn1, Itga6, Itgb1, Lamb1, Lamc1, and Sdc4 (p < 10−6; hypergeometric test comparing the incidence of GOT between “downregulated” and “all quantified” proteins). Given the importance of the ECM in neuronal structure and functions, alterations in ECM receptor activation may represent additional mechanisms by which Sec61 inhibition impairs pain signal integration and transmission by neurons (32).
A major new finding in this work was the description of a stress response to mycolactone-mediated Sec61 blockade manifesting through the transcriptional induction of several proteins, including Sec61 substrates. Despite the low number of mycolactone-upregulated proteins in Jurkat T cells and MED17.11 neurons under the conditions employed, Slc3a2 was upregulated in both MutuDCs and MED17.11 neurons, and Hsp90 in both MutuDCs and Jurkat T cells, suggesting that these proteins represent conserved markers of Sec61 blockade-driven stress response. Upregulation of Hsp70/Hsp90 in mycolactone-treated MutuDCs likely results from the cytosolic accumulation of mycolactone-susceptible Sec61 substrates blocked in translocation, which are unable to fold properly outside the oxidizing environment of the ER and without membrane insertion. Notably, a significant proportion of mycolactone-upregulated proteins were targets of the ATF4 and/or Chop transcription factors. These findings are consistent with those of Ogbechi et al., who reported recently that mycolactone-mediated Sec61 blockade drives ATF4 expression (33). However, in contrast to this study, we detected mycolactone-induced Xbp-1 splicing, indicative of ER stress. Our observation that mycolactone decreases Bip expression nevertheless highlighted a major difference between mycolactone-driven ER stress response and conventional UPR (Fig. 5E). Although the underlying mechanism is unclear, the 6h time to onset of Bip decrease suggests that it may result from secondary effects. Bip representing a major survival arm of the UPR, mycolactone-driven decline in Bip levels is likely to increase the cell susceptibility to ER stress-induced apoptosis. Mycolactone was recently proposed to promote BIM-dependent cell apoptosis through the mTORC2-Akt-FoxO3 axis (34). By transducing cells with a mycolactone-resistant Sec61 mutant, we were able to show that mycolactone cytotoxicity strictly depends on Sec61 inhibition (3). With mycolactone promoting a decrease in anti-apoptotic Bip and an increase in pro-apoptotic Chop, the data presented here thus support an additional scenario for mycolactone-induced toxicity, where mycolactone-mediated Sec61 blockade causes UPR-mediated apoptosis.
DATA AVAILABILITY
Proteomics data were deposited to the ProteomeXchange Consortium via the PRIDE partner repository (35, 36). The data sets corresponding to Jurkat T cells, MutuDCs and MED17.11 neurons are available under the identifiers PXD002971, PXD006103 and PXD007770. Annotated spectra were deposited in MS viewer with the following identifier keys: Jurkat: nnkr7jkmbl and xyczcpn2y0, MutuDCs: dmbpnhmpqb, MED17.11 neurons: lijxc0lue5. The proteomic analysis of mycolactone's effect on MutuDCs was performed with two time points (6 h and 24 h). Because only one protein was modulated after 6 h of mycolactone treatment, we only analyzed the 24 h time point (15).
Supplementary Material
Footnotes
* This work was supported by Institut Pasteur, Institut National de la Santé et de la Recherche Médicale (INSERM) and Fondation Raoul Follereau (C.D.). J.D.M. is the recipient of a doctoral fellowship from the Ecole Normale Supérieure de Lyon. F.I. is supported by Odysseus grant G0F8616N from the Research Foundation Flanders (FWO). J.W. and J.W.Y. are supported by the Division of Intramural Research, NIAID. V.O.P. is supported by the Academy of Finland (grant 289737), and the Sigrid Juselius Foundation.
 This article contains supplemental Figures and Tables. The authors declare that they have no competing interests.
This article contains supplemental Figures and Tables. The authors declare that they have no competing interests.
1 The abbreviations used are:
- ER
- endoplasmic reticulum
- AGTR2
- type 2 angiotensin II receptors
- bFGF
- basic fibroblast growth factor
- β2M
- beta 2 microglobulin
- cAMP
- cyclic adenosine monophosphate
- DMSO
- dimethyl sulfoxide
- DMEM
- Dulbecco's Modified Eagle's medium
- DTT
- 1,4-dithiothreitol
- ECM
- extracellular matrix
- FCS
- fetal calf serum
- FDR
- false discovery rate
- GOT
- Gene Ontology terms
- Hepes
- 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- Hsp
- heat shock protein
- IMDM
- Iscove's Modified Dulbecco's medium
- IAV
- influenza A virus
- LC-MS
- liquid chromatography-mass spectrometry
- ISR
- integrated stress response
- IVT
- in vitro protein translocation assay
- MHC
- major histocompatibility complex
- mTOR
- mammalian target of rapamycin
- PBS
- phosphate-buffered saline
- PCR
- polymerase chain reaction
- RPMI
- Roswell Park Memorial Institute medium
- SDS-PAGE
- sodium dodecyl sulfate polyacrylamide gel electrophoresis
- SEM
- standard error of the mean
- SP
- signal peptide
- SRM
- sheep rough microsomes
- TMD
- transmembrane domain
- TMP
- transmembrane protein
- UPR
- unfolded protein response
- wt
- wild type.
REFERENCES
- 1. Demangel C., Stinear T. P., and Cole S. T. (2009) Buruli ulcer: reductive evolution enhances pathogenicity of Mycobacterium ulcerans. Nat. Rev. Microbiol. 7, 50–60 [DOI] [PubMed] [Google Scholar]
- 2. Sarfo F. S., Phillips R., Wansbrough-Jones M., and Simmonds R. E. (2016) Recent advances: role of mycolactone in the pathogenesis and monitoring of Mycobacterium ulcerans infection/Buruli ulcer disease. Cell Microbiol. 18, 17–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Baron L., Paatero A. O., Morel J. D., Impens F., Guenin-Mace L., Saint-Auret S., Blanchard N., Dillmann R., Niang F., Pellegrini S., Taunton J., Paavilainen V. O., and Demangel C. (2016) Mycolactone subverts immunity by selectively blocking the Sec61 translocon. J. Exp. Med. 213, 2885–2896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hall B. S., Hill K., McKenna M., Ogbechi J., High S., Willis A. E., and Simmonds R. E. (2014) The pathogenic mechanism of the Mycobacterium ulcerans virulence factor, mycolactone, depends on blockade of protein translocation into the ER. PLoS Pathog. 10, e1004061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. McKenna M., Simmonds R. E., and High S. (2016) Mechanistic insights into the inhibition of Sec61-dependent co- and post-translational translocation by mycolactone. J. Cell Sci. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. McKenna M., Simmonds R. E., and High S. (2017) Mycolactone reveals the substrate-driven complexity of Sec61-dependent transmembrane protein biogenesis. J. Cell Sci. 130, 1307–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Goder V., and Spiess M. (2001) Topogenesis of membrane proteins: determinants and dynamics. FEBS letters 504, 87–93 [DOI] [PubMed] [Google Scholar]
- 8. Boulkroun S., Guenin-Mace L., Thoulouze M. I., Monot M., Merckx A., Langsley G., Bismuth G., Di Bartolo V., and Demangel C. (2010) Mycolactone suppresses T cell responsiveness by altering both early signaling and posttranslational events. J. Immunol. 184, 1436–1444 [DOI] [PubMed] [Google Scholar]
- 9. Coutanceau E., Decalf J., Martino A., Babon A., Winter N., Cole S. T., Albert M. L., and Demangel C. (2007) Selective suppression of dendritic cell functions by Mycobacterium ulcerans toxin mycolactone. J. Exp. Med. 204, 1395–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Guenin-Mace L., Baron L., Chany A. C., Tresse C., Saint-Auret S., Jonsson F., Le Chevalier F., Bruhns P., Bismuth G., Hidalgo-Lucas S., Bisson J. F., Blanchard N., and Demangel C. (2015) Shaping mycolactone for therapeutic use against inflammatory disorders. Sci. Transl. Med. 7, 289ra285. [DOI] [PubMed] [Google Scholar]
- 11. Guenin-Mace L., Carrette F., Asperti-Boursin F., Le Bon A., Caleechurn L., Di Bartolo V., Fontanet A., Bismuth G., and Demangel C. (2011) Mycolactone impairs T cell homing by suppressing microRNA control of L-selectin expression. Proc. Natl. Acad. Sci. U.S.A. 108, 12833–12838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Marion E., Song O. R., Christophe T., Babonneau J., Fenistein D., Eyer J., Letournel F., Henrion D., Clere N., Paille V., Guerineau N. C., Saint Andre J. P., Gersbach P., Altmann K. H., Stinear T. P., Comoglio Y., Sandoz G., Preisser L., Delneste Y., Yeramian E., Marsollier L., and Brodin P. (2014) Mycobacterial toxin induces analgesia in buruli ulcer by targeting the Angiotensin pathways. Cell 157, 1565–1576 [DOI] [PubMed] [Google Scholar]
- 13. Pahlevan A. A., Wright D. J., Andrews C., George K. M., Small P. L., and Foxwell B. M. (1999) The inhibitory action of Mycobacterium ulcerans soluble factor on monocyte/T cell cytokine production and NF-kappa B function. J. Immunol. 163, 3928–3935 [PubMed] [Google Scholar]
- 14. Phillips R., Sarfo F. S., Guenin-Mace L., Decalf J., Wansbrough-Jones M., Albert M. L., and Demangel C. (2009) Immunosuppressive signature of cutaneous Mycobacterium ulcerans infection in the peripheral blood of patients with Buruli Ulcer Disease. J. Infect Dis. [DOI] [PubMed] [Google Scholar]
- 15. Grotzke J. E., Kozik P., Morel J. D., Impens F., Pietrosemoli N., Cresswell P., Amigorena S., and Demangel C. (2017) Sec61 blockade by mycolactone inhibits antigen cross-presentation independently of endosome-to-cytosol export. Proc. Natl. Acad. Sci. U.S.A. 114, E5910–E5919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Isaac C., Mauborgne A., Grimaldi A., Ade K., Pohl M., Limatola C., Boucher Y., Demangel C., and Guenin-Mace L. (2017) Mycolactone displays anti-inflammatory effects on the nervous system. PLoS Negl. Trop. Dis. 11, e0006058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Doran C., Chetrit J., Holley M. C., Grundy D., and Nassar M. A. (2015) Mouse DRG Cell Line with Properties of Nociceptors. Plos One 10, e0128670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. George K. M., Chatterjee D., Gunawardana G., Welty D., Hayman J., Lee R., and Small P. L. (1999) Mycolactone: a polyketide toxin from Mycobacterium ulcerans required for virulence. Science 283, 854–857 [DOI] [PubMed] [Google Scholar]
- 19. Spangenberg T., and Kishi Y. (2010) Highly sensitive, operationally simple, cost/time effective detection of the mycolactones from the human pathogen Mycobacterium ulcerans. Chem. Commun. 46, 1410–1412 [DOI] [PubMed] [Google Scholar]
- 20. Morgenstern J. P., and Land H. (1990) Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 18, 3587–3596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sharma A., Mariappan M., Appathurai S., and Hegde R. S. (2010) In vitro dissection of protein translocation into the mammalian endoplasmic reticulum. Methods Mol. Biol. 619, 339–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vermeire K., Allan S., Provinciael B., Hartmann E., and Kalies K. U. (2015) Ribonuclease-neutralized pancreatic microsomal membranes from livestock for in vitro co-translational protein translocation. Anal. Biochem. 484, 102–104 [DOI] [PubMed] [Google Scholar]
- 23. Ning Z., Seebun D., Hawley B., Chiang C. K., and Figeys D. (2013) From cells to peptides: “one-stop” integrated proteomic processing using amphipols. J. Proteome Res. 12, 1512–1519 [DOI] [PubMed] [Google Scholar]
- 24. Cox J., and Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 25. Cox J., Hein M. Y., Luber C. A., Paron I., Nagaraj N., and Mann M. (2014) Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics 13, 2513–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Supek F., Bosnjak M., Skunca N., and Smuc T. (2011) REVIGO Summarizes and Visualizes Long Lists of Gene Ontology Terms. Plos One 6, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nguyen H. T., and Merlin D. (2012) Homeostatic and innate immune responses: role of the transmembrane glycoprotein CD98. Cell Mol. Life Sci. 69, 3015–3026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Arensdorf A. M., Diedrichs D., and Rutkowski D. T. (2013) Regulation of the transcriptome by ER stress: non-canonical mechanisms and physiological consequences. Front Genet. 4, 256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Oslowski C. M., and Urano F. (2011) Measuring ER stress and the unfolded protein response using mammalian tissue culture system. Methods Enzymol. 490, 71–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Voorhees R. M., and Hegde R. S. (2016) Structure of the Sec61 channel opened by a signal sequence. Science 351, 88–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Monel B., Compton A. A., Bruel T., Amraoui S., Burlaud-Gaillard J., Roy N., Guivel-Benhassine F., Porrot F., Genin P., Meertens L., Sinigaglia L., Jouvenet N., Weil R., Casartelli N., Demangel C., Simon-Loriere E., Moris A., Roingeard P., Amara A., and Schwartz O. (2017) Zika virus induces massive cytoplasmic vacuolization and paraptosis-like death in infected cells. EMBO J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kerrisk M. E., Cingolani L. A., and Koleske A. J. (2014) ECM receptors in neuronal structure, synaptic plasticity, and behavior. Progress Brain Res. 214, 101–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ogbechi J., Hall B. S., Sbarrato T., Taunton J., Willis A. E., Wek R. C., and Simmonds R. E. (2018) Inhibition of Sec61-dependent translocation by mycolactone uncouples the integrated stress response from ER stress, driving cytotoxicity via translational activation of ATF4. Cell Death Dis. 9, 397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bieri R., Scherr N., Ruf M. T., Dangy J. P., Gersbach P., Gehringer M., Altmann K. H., and Pluschke G. (2017) The Macrolide Toxin Mycolactone Promotes Bim-Dependent Apoptosis in Buruli Ulcer through Inhibition of mTOR. ACS Chem. Biol. 12, 1297–1307 [DOI] [PubMed] [Google Scholar]
- 35. Vizcaino J. A., Csordas A., del-Toro N., Dianes J. A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T., Xu Q. W., Wang R., and Hermjakob H. (2016) 2016 update of the PRIDE database and its related tools. Nucleic acids research 44, D447–D456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Vizcaino J. A., Deutsch E. W., Wang R., Csordas A., Reisinger F., Rios D., Dianes J. A., Sun Z., Farrah T., Bandeira N., Binz P. A., Xenarios I., Eisenacher M., Mayer G., Gatto L., Campos A., Chalkley R. J., Kraus H. J., Albar J. P., Martinez-Bartolome S., Apweiler R., Omenn G. S., Martens L., Jones A. R., and Hermjakob H. (2014) ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 32, 223–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Han J., Back S. H., Hur J., Lin Y. H., Gildersleeve R., Shan J., Yuan C. L., Krokowski D., Wang S., Hatzoglou M., Kilberg M. S., Sartor M. A., and Kaufman R. J. (2013) ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat. Cell Biol. 15, 481–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Proteomics data were deposited to the ProteomeXchange Consortium via the PRIDE partner repository (35, 36). The data sets corresponding to Jurkat T cells, MutuDCs and MED17.11 neurons are available under the identifiers PXD002971, PXD006103 and PXD007770. Annotated spectra were deposited in MS viewer with the following identifier keys: Jurkat: nnkr7jkmbl and xyczcpn2y0, MutuDCs: dmbpnhmpqb, MED17.11 neurons: lijxc0lue5. The proteomic analysis of mycolactone's effect on MutuDCs was performed with two time points (6 h and 24 h). Because only one protein was modulated after 6 h of mycolactone treatment, we only analyzed the 24 h time point (15).