Abstract
A DNA-encoded chemical library (DECL) with 1.2 million compounds was synthesized by combinatorial reaction of 7 central scaffolds with two sets of 343 x 492 building blocks. Library screening by affinity capture revealed that for some target proteins the chemical nature of building blocks dominated the selection results, while for other proteins also the central scaffold crucially contributed to ligand affinity. Molecules based on a 3,5-bis(aminomethyl)-benzoic acid core structure bound to human serum albumin with a Kd of 6 nM, while compounds with the same substituents on an equidistant but flexible L-lysine scaffold showed a 140-fold lower affinity. A 18 nM tankyrase-1 binder featured l-lysine as linking moiety, while molecules based on d-Lysine or (2S,4S)-amino-l-proline had no detectable binding to the target. This work suggests that central scaffolds, predisposing the orientation of chemical building blocks towards the protein target, may enhance the screening productivity of encoded libraries.
Keywords: DNA-encoded chemical libraries, bifunctional ligands, tankyrase-1, human serum albumin
Introduction
The encoding of compounds with DNA tags, serving as amplifiable identification bar-codes, allows the facile construction and screening of large combinatorial chemical libraries[1]. The successful identification of binding molecules from DNA-encoded chemical libraries depends not only on the number and characteristics of the building blocks used for library construction, but also on library design[1]. We previously reported a DNA-encoded chemical library based on a conserved (S)-2,3-diaminopropanoic acid scaffold which had been combinatorially reacted with two sets of carboxylic acids that yielded nanomolar binders against various target proteins, including serum albumins and tankyrase-1[2]. These results motivated us to investigate how the geometry, stereochemistry and rigidity of a central scaffold influences the outcome of screening results and the binding affinity of selected compounds[3]. Here we describe the synthesis and characterization of a DNA-encoded chemical library[4] obtained by the combinatorial modification of seven central scaffolds, each bearing both an amine and an azide moiety. Using these orthogonal coupling sites for two diverse sets of building blocks (343 and 492 building blocks, respectively), eventually yielded a library with an overall size of 1.2 million compounds. Affinity measurements performed on selected hit compounds indicate that, for certain targets, the geometry and rigidity of the central scaffold can have a strong impact on the dissociation constants of the corresponding ligands[5].
Results
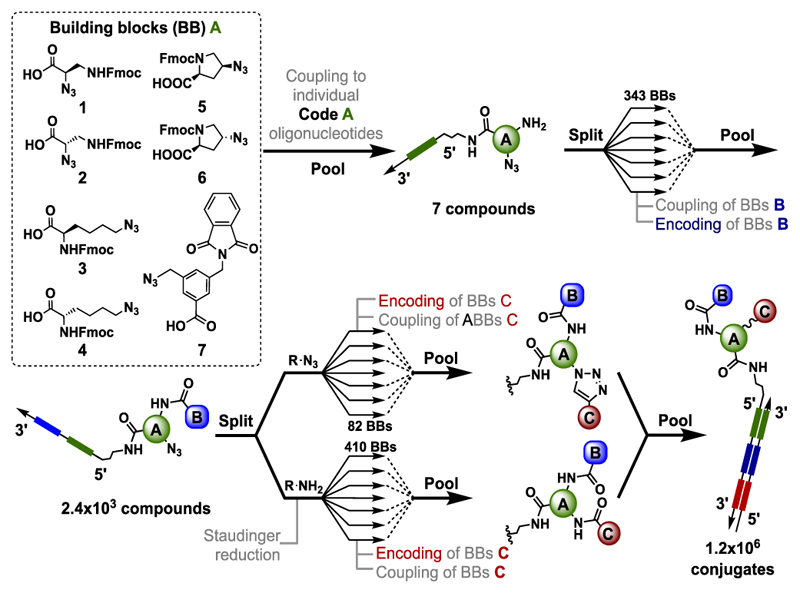
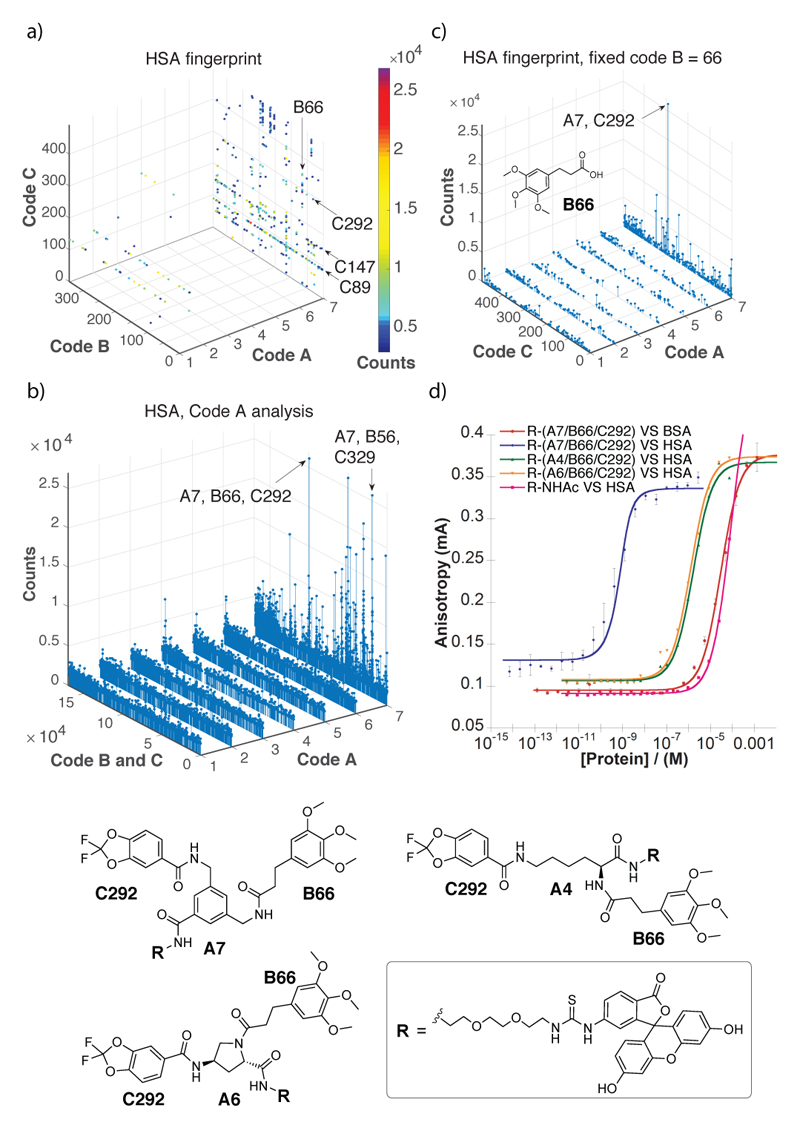
In this study, we explored whether subtle chemical variations of the central molecular “scaffold” impact the affinity and specificity of the discovered protein ligands. For this reason, we coupled seven trifunctional carboxylic acid derivatives to oligonucleotides, to enable the subsequent coupling and encoding of two sets of building blocks, resulting in a combinatorial library of 7 x 343 x 492 = 1,181,292 compounds [Figure 1]. The scaffolds included two stereo-defined protected derivatives of (R)-2-Azido-3-Aminopropionic acid (1) and (S)-2-Azido-3-Aminopropionic acid (2), the rigidified (2S, 4S)-azido- l-proline (5) and (2S, 4R)-azido- l-proline (6), ε-azido- d-lysine (3), ε-azido- l-lysine (4) and 3-(aminomethyl)-5-(azidomethyl) benzoic acid (7). The scaffolds each contained a protected primary amine group and an azide moiety. Using a split-and-pool protocol[7], featuring the reaction of amines with carboxylic acids[8] or the copper(I)-catalyzed alkyne-azide cycloaddition (CuAAC)[9] of azide derivatives with terminal alkynes, we constructed a library of structurally-related compounds. Screening experiments were performed with biotinylated proteins immobilized on streptavidin-coated magnetic beads[10]. Details of library synthesis and of library encoding procedures can be found in Supplementary Figure 1.1 and in the Supplementary Information. Figure 2a shows the results of a library selection performed with biotinylated human serum albumin (HSA), followed by high-throughput sequencing for the identification and relative quantification of the DNA barcodes[10]. These selection results are displayed in a pseudo-four dimensional space, using three dimensions for the definition of the identity of the three sets of building blocks (Code A, B and C), while spheres of different colors represent the fourth dimension (corresponding to the number of sequence counts for individual compounds) above a threshold of 3,000 counts. The plot indicates that the most highly enriched library members all contained the 3,5-bis(aminomethyl)benzoic acid scaffold (code A = 7), whereas there was no such preference in the pre-selected library and selections with a set of control proteins used in our laboratory [Supplementary Figures 6.2 and 6.3]. Figure 2b presents a graphical display of the selection counts for the library members, arranged with seven code A structures on the x-axis and the 343 x 492 = 168,756 combinations of code B and code C on the y-axis. Compound A7/B66/C292 is the most enriched compound [Figure 2]. The compound consisted of the 3,4-di(aminomethyl)benzoic acid scaffold, with 3-(3,4,5-trimethoxyphenyl)propanoic acid (B66), and 2,2-Difluoro-1,3-benzodioxole-5-carboxylic acid (C292). The same 3-(3,4,5-trimethoxyphenyl)propanoic acid found as building block B66 in the second reaction cycle position of compound A7/B66/C292, was also found in another combination. In particular the 3-(3,4,5-trimethoxyphenyl)propanoic acid moiety was found as building block C56 in the third reaction cycle position of a second highly enriched combination (A7/B329/C56). In a different representation of selection results, keeping code B constant (i.e., restricting the analysis to compounds based on the B66 building block), the preferential enrichment of library members based on the A7 scaffold (and of the A7/B66/C292 in particular) is visible [Figure 2c]. The dissociation constant (Kd) of a fluorescein derivative of A7/B66/C292 determined by concentration dependent fluorescence polarization experiments was Kd = 7 nM. In contrast, exchanging the A7 scaffold by two alternative diamines drastically decreased the affinity to HSA in agreement with the selection results. (Kd(A4/B66/C292) = 1.6 μM; Kd(A6/B66/C292) = 1.2 μM). The fluorophore-labeled and acetylated PEG2-diamino linker exhibited a residual dissociation constant higher than 100 μM [Figure 2d].
Figure 1.
Representation of the library synthesis scheme. Seven building blocks (1-7), carrying protected amine functions and azides, were coupled to amino-tagged oligonucleotides, each containing a central sequence (“code”), which unambiguously identifies the corresponding building block. After deprotection and using a split and pool procedure, carboxylic acids were coupled to the amines and the corresponding 343 building blocks “B” encoded by a ligation procedure. In a last synthesis step the azide moiety of the central scaffolds was either reacted with terminal alkynes or converted to a primary amine, allowing the formation of an amide bond with carboxylic acids. The final encoding step for building blocks “C” was performed using partially complementary oligonucleotides and Klenow polymerization, as described[6].
Figure 2.
Results of library selections against human serum albumin (HSA). (a) Selection fingerprint. The individual library members are unambiguously identified by their code A (ranging between 1 and 7), B (ranging between 1 and 343) and C (ranging between 1 and 492). The number of sequence counts for each compound is displayed as spheres of a different colour, with a cut-off threshold set at 3000 counts. In total, 59,596,519 sequence counts were read by high-throughput sequencing. (b) The Code A analysis shows how the different scaffolds (code A, ranging 1 and 7) display different enrichment factors for each building block A and B combination (Code B × Code, ranging between 1 and 1.7×105). (c) Selection fingerprint obtained fixing the code B = 66 (corresponding to 3-(3,4,5-trimethoxyphenyl)propanoic acid) with variable codes A (ranging between 1 and 7) and C (ranging between 1 and 492). This analysis shows the A7/B66/C292 as the most enriched combination of building blocks against HSA. (d) Fluorescence polarization (FP) measurement of FITC-conjugates of the most enriched combination of building blocks (B66/C292), featuring the preferred scaffold A7 (red and blue curves) or scaffolds A4 (green) and A6 (orange). A FITC conjugate of A7/B66/C292 was also tested against bovine serum albumin (BSA) a d revealed a double-digit micromolar dissociation constant (Kd) against that protein (pink curve).
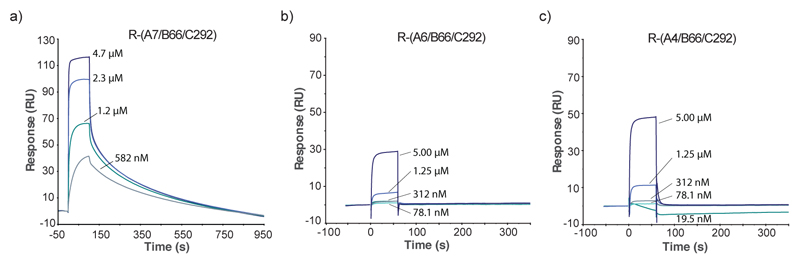
We confirmed the fluorescence polarization findings by studying the interaction of compounds A7/B66/C292, A6/B66/C292 and A4/B66/C292 with HSA using surface plasmon resonance on a BIAcore instrument. Figure 3 shows that compound A7/B66/C292 bound to its cognate target with a kinetic dissociation constant koff = 4.5x10-3 s-1. By contrast, the structural analogues featuring L-lysine and (2S,4R)-amino-l-proline as a central scaffold did not bind to HSA in the same experimental conditions [Figure 3], confirming the role played by the A7 3,5-bis(aminomethyl)-benzoic acid core structure.
Figure 3.
Binding properties of compound A7/B66/C292 analyzed by Surface Plasmon Resonance (SPR) against HSA immobilized on a BIAcore chip (CM5, 5770 RU). (a) SPR profile of compound R-(A7/B66/C292) at different concentration (5 μM, 1.25 μM, 312 nM, 78.1 nM, 19.5 nM). An overall fitting of the resulting curves (4.7μM, 2.3 μM, 1.2 μM and 582 nM) revealed a koff = 7.4x10-3 Ms-1 and a kon = 2.0x105 s-1M-1, resulting in a Kd = 36.6 nM (b) SPR profile of compound R-(A6/B66/C292) at different concentrations (5 μM, 1.25 μM, 312 nM, 78.1 nM, 19.5 nM). (c) SPR profile of compound R-(A4/B66/C292) at different concentrations (5 μM, 1.25 μM, 312 nM, 78.1 nM, 19.5 nM).
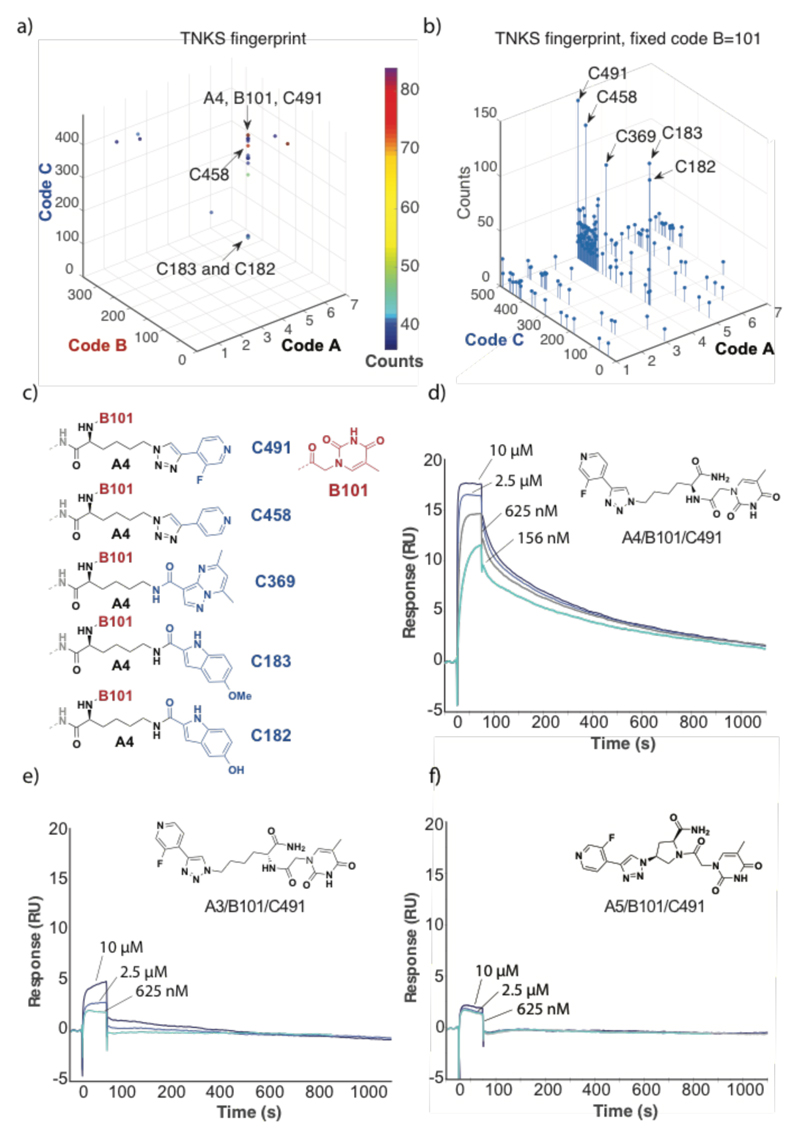
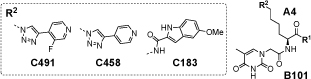
Library selections performed for human tankyrase-1 (TNKS-1) revealed a distinctive pattern of enriched compounds, featuring a preference for linker A4 (l-Lysine) and building block B101 (thymine-1-acetic acid) [Figure 4a]. The observed fingerprint was observed when selections were performed at various concentrations of Tween-20, providing confidence about the reproducibility of the screening procedure[1h] [Supplementary Figure 6.1]. A plot of selection results, emphasizing sequence counts for various library members featuring the B101 building block, highlighted the preferential enrichment of compounds with linker A4 and with certain conserved structural features of C building blocks [Figure 4b,c]. Synthesis of amide derivatives the most enriched compound A4/B101/C491 revealed a high-affinity binding to the cognate TNKS1 protein immobilized on a BIAcore chip (Kd = 15±8 nM), while the use of d-Lysine (A3/B101/C491) or of (2S,4S)-amino-l-proline (A5/B101/C491) as linker did not result in any detectable binding by SPR analysis [Figure 4d-f].
Figure 4.
Results of library selections against human tankyrase-1 (TNKS1). (a) Selection fingerprint, reavealing a preferential enrichment of compounds with A4 and B101. (b) Plot of sequence counts for library members, featuring B101 as preferred building block. The plot reveals a preferential enrichment of library members with linker A4 and building blocks C491, C453, C369, C183 and C182. The structures of the four most enriched compounds is shown in panel (c). The BIAcore profiles at various concentrations of ligand for the amide derivatives of A4/B101/C491, A3/B101/C491 and A5/B101/C491 are shown in panels (d), (e) and (f). Fitting of the sensograms for A4/B101/C491 in panel (d) yielded a koff = (2.1±0.8) x 10-3 s-1 and a kon = (1.7±0.2) x 105 s-1M-1, corresponding to a Kd = 15±8 nM.
We also explored the impact on affinity constants to tankyrase 1 for chemical modifications at the site, originally occupied by the linkage to DNA [Table 1 and Supplementary Figure 5.3]. While a simple amide derivative exhibited the best dissociation constant [Kd = 15 nM], the corresponding carboxylic acid or amides derived from 3-aminopropan-1-ol showed to a substantial decrease in binding affinity.
Table 1. Impact of chemical modification at the site of DNA coupling on dissociation constants.
| Compond ID[a] | R1 | R2 | Kd [b] |
|---|---|---|---|
| 45 (A4/B101/C491) | OH | C491 | 259 nM |
| 44 (A4/B101/C458) | OH | C458 | 307 nM |
| 54 (A4/B101/183) | OH | C183 | 258 nM |
| 46 (A4/B101/C491) | NH2 | C491 | 15 nM |
| 47 (A4/B101/C491) | NH(CH2)3OH | C491 | 6.0 μM |
| 55 (A4/B101/183) | NH(CH2)3OH | C183 | 2.4 μM |
The complete structures are reported in the Supplementary Information.
The SPR profiles are reported in the supplementary figure 5.3.
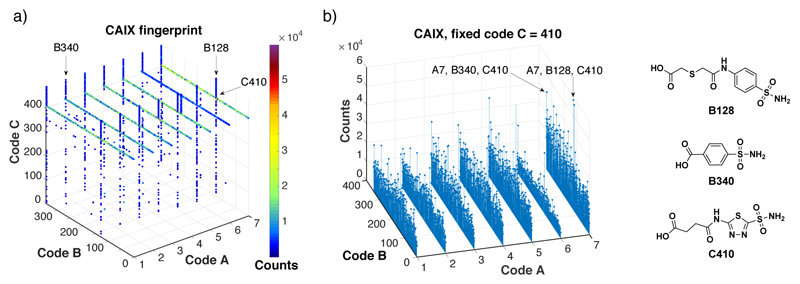
When selections were performed with targets, for which a specific building block dominates the ligand enrichment procedure, different fingerprint patterns were observed. Figure 5 shows the results of selections performed against carbonic anhydrase IX, a tumor-associated cell-surface marker[11]. This enzyme can efficiently be targeted by ligands containing aromatic or heteroaromatic sulfonamides[12]. Indeed, an efficient enrichment of building blocks B128, 340 and C410 was visible in the selection fingerprints. In this case, the role of the central scaffold was less important, as sulfonamide derivatives for all seven building blocks “A” could be efficiently enriched.
Figure 5.
Results of library selections against carbonic anhydrase IX (CAIX). (a) Selection fingerprint. The individual library members are unambiguously identified by their code A (ranging between 1 and 7), B (ranging between 1 and 343) and C (ranging between 1 and 492). The number of sequence counts for each compound is displayed as spheres of different colors, with a cut-off threshold set at 3000 counts. In total, 59,596,519 sequences were read by high-throughput DNA sequencing. (b) Selection fingerprint obtained fixing the code C = 410 (corresponding to acetazolamide) with variable codes A (ranging between 1 and 7) and B (ranging between 1 and 343). This picture shows how the scaffold does not affect the affinity for CAIX.
Conclusions
Collectively, the results of this study suggest that the chemical nature of central scaffolds, defining the orientation and flexibility of substituents blocks pointing towards the protein target of interest, may represent an important determinant of binding affinity. The examples of HSA and TNKS1 binders revealed that subtle differences in the chemical nature of central scaffolds can lead to substantial variations (i.e., > 100-fold differences) in binding affinity to the cognate target protein of interest. The nanomolar binders to HSA and to TNKS1 described in this paper may be useful for serum half-life prolongation purposes[13] and the TNKS1 binders as chemical probes for the biological characterization of TNKS1 function[14] although the selectivity to other poly-(ADP-ribose)polymerases remains to be established[15] We anticipate that new DNA-encoded chemical libraries will be designed in the future, exploiting novel designs for the spatial arrangement of building blocks.
Supplementary Material
Acknowledgements
Financial support from the ETH Zürich, the Swiss National Science Foundation (Grants 310030B_163479), Sinergia (CRS112_160699) and from the ERC Advanced Grant “ZAUBERKUGEL” (Grant agreement 670603) is gratefully acknowledged. The Structural Genomics Consortium is a registered charity (no. 1097737) that receives funds from AbbVie; Bayer Pharma AG; Boehringer Ingelheim; Canada Foundation for Innovation; Eshelman Institute for Innovation; Genome Canada; Innovative Medicines Initiative (EU/EFPIA) (ULTRA-DD grant no. 115766); Janssen; Merck & Co.; Novartis Pharma AG; Ontario Ministry of Economic Development and Innovation; Pfizer; São Paulo Research Foundation-FAPESP; Takeda; and the Wellcome Trust.
References
- [1].a) Brenner S, Lerner RA. Proc Natl Acad Sci USA. 1992;89:5381–5383. doi: 10.1073/pnas.89.12.5381. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Arico-Muendel C, Zhu Z, Dickson H, Parks D, Keicher J, Deng J, Aquilani L, Coppo F, Graybill T, Lind K, Peat A, et al. Antimicrob Agents Chemother. 2015;59:3450–3459. doi: 10.1128/AAC.00070-15. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Winssinger N. Chimia (Aarau) 2013;67:340–348. doi: 10.2533/chimia.2013.340. [DOI] [PubMed] [Google Scholar]; d) Blakskjaer P, Heitner T, Hansen NJ. Curr Opin Chem Biol. 2015;26:62–71. doi: 10.1016/j.cbpa.2015.02.003. [DOI] [PubMed] [Google Scholar]; e) Cuozzo JW, Centrella PA, Gikunju D, Habeshian S, Hupp CD, Keefe AD, Sigel EA, Soutter HH, Thomson HA, Zhang Y, Clark MA. Chembiochem. 2017;18:864–871. doi: 10.1002/cbic.201600573. [DOI] [PubMed] [Google Scholar]; f) Wichert M, Krall N, Decurtins W, Franzini RM, Pretto F, Schneider P, Neri D, Scheuermann J. Nat Chem. 2015;7:241–249. doi: 10.1038/nchem.2158. [DOI] [PubMed] [Google Scholar]; g) Samain F, Ekblad T, Mikutis G, Zhong N, Zimmermann M, Nauer A, Bajic D, Decurtins W, Scheuermann J, Brown PJ, Hall J, et al. J Med Chem. 2015;58:5143–5149. doi: 10.1021/acs.jmedchem.5b00432. [DOI] [PubMed] [Google Scholar]; h) Leimbacher M, Zhang Y, Mannocci L, Stravs M, Geppert T, Scheuermann J, Schneider G, Neri D. Chemistry. 2012;18:7729–7737. doi: 10.1002/chem.201200952. [DOI] [PubMed] [Google Scholar]; i) Franzini RM, Neri D, Scheuermann J. Acc Chem Res. 2014;47:1247–1255. doi: 10.1021/ar400284t. [DOI] [PubMed] [Google Scholar]; j) Salamon H, Klika Skopic M, Jung K, Bugain O, Brunschweiger A. ACS Chem Biol. 2016;11:296–307. doi: 10.1021/acschembio.5b00981. [DOI] [PubMed] [Google Scholar]; k) Neri D, Lerner R. Annu Rev Biochem. 2018;87:24. doi: 10.1146/annurev-biochem-062917-012550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Franzini RM, Ekblad T, Zhong N, Wichert M, Decurtins W, Nauer A, Zimmermann M, Samain F, Scheuermann J, Brown PJ, Hall J, et al. Angew Chem Int Ed Engl. 2015;54:3927–3931. doi: 10.1002/anie.201410736. [DOI] [PubMed] [Google Scholar]
- [3].Satz AL. ACS Comb Sci. 2016;18:415–424. doi: 10.1021/acscombsci.6b00001. [DOI] [PubMed] [Google Scholar]
- [4].a) Satz AL, Cai J, Chen Y, Goodnow R, Gruber F, Kowalczyk A, Petersen A, Naderi-Oboodi G, Orzechowski L, Strebel Q. Bioconjug Chem. 2015;26:1623–1632. doi: 10.1021/acs.bioconjchem.5b00239. [DOI] [PubMed] [Google Scholar]; b) Clark MA, Acharya RA, Arico-Muendel CC, Belyanskaya SL, Benjamin DR, Carlson NR, Centrella PA, Chiu CH, Creaser SP, Cuozzo JW, Davie CP, et al. Nat Chem Biol. 2009;5:647–654. doi: 10.1038/nchembio.211. [DOI] [PubMed] [Google Scholar]
- [5].a) Daguer JP, Zambaldo C, Ciobanu M, Morieux P, Barluenga S, Winssinger N. Chem Sci. 2015;6:739–744. doi: 10.1039/c4sc01654h. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Deng H, Zhou J, Sundersingh FS, Summerfield J, Somers D, Messer JA, Satz AL, Ancellin N, Arico-Muendel CC, Sargent Bedard KL, Beljean A, et al. ACS Med Chem Lett. 2015;6:919–924. doi: 10.1021/acsmedchemlett.5b00179. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Encinas L, O'Keefe H, Neu M, Remuinan MJ, Patel AM, Guardia A, Davie CP, Perez-Macias N, Yang H, Convery MA, Messer JA, et al. J Med Chem. 2014;57:1276–1288. doi: 10.1021/jm401326j. [DOI] [PubMed] [Google Scholar]
- [6].Mannocci L, Zhang Y, Scheuermann J, Leimbacher M, De Bellis G, Rizzi E, Dumelin C, Melkko S, Neri D. Proc Natl Acad Sci USA. 2008;105:17670–17675. doi: 10.1073/pnas.0805130105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Goodnow RA, Jr, Dumelin CE, Keefe AD. Nat Rev Drug Discov. 2017;16:131–147. doi: 10.1038/nrd.2016.213. [DOI] [PubMed] [Google Scholar]
- [8].Li Y, Gabriele E, Samain F, Favalli N, Sladojevich F, Scheuermann J, Neri D. ACS Comb Sci. 2016;18:438–443. doi: 10.1021/acscombsci.6b00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Haque MM, Peng X. Sci China Chem. 2013;57:215–231. [Google Scholar]
- [10].Decurtins W, Wichert M, Franzini RM, Buller F, Stravs MA, Zhang Y, Neri D, Scheuermann J. Nat Protoc. 2016;11:764–780. doi: 10.1038/nprot.2016.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].a) Muselaers S, Mulders P, Oosterwijk E, Oyen W, Boerman O. Immunotherapy. 2013;5:489–495. doi: 10.2217/imt.13.36. [DOI] [PubMed] [Google Scholar]; b Supuran CT. Nat Rev Drug Discov. 2008;7:168–181. doi: 10.1038/nrd2467. [DOI] [PubMed] [Google Scholar]
- [12].Alterio V, Esposito D, Monti SM, Supuran CT, De Simone G. J Enzyme Inhib Med Chem. 2018;33:151–157. doi: 10.1080/14756366.2017.1405263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Dumelin CE, Trussel S, Buller F, Trachsel E, Bootz F, Zhang Y, Mannocci L, Beck SC, Drumea-Mirancea M, Seeliger MW, Baltes C, et al. Angew Chem Int Ed Engl. 2008;47:3196–3201. doi: 10.1002/anie.200704936. [DOI] [PubMed] [Google Scholar]
- [14].a) Li X, Han H, Zhou MT, Yang B, Ta AP, Li N, Chen J, Wang W. Cell Rep. 2017;20:737–749. doi: 10.1016/j.celrep.2017.06.077. [DOI] [PubMed] [Google Scholar]; b) Wu X, Luo F, Li J, Zhong X, Liu K. Int J Oncol. 2016;48:1333–1340. doi: 10.3892/ijo.2016.3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Wahlberg E, Karlberg T, Kouznetsova E, Markova N, Macchiarulo A, Thorsell AG, Pol E, Frostell A, Ekblad T, Oncu D, Kull B, et al. Nat Biotechnol. 2012;30:283–288. doi: 10.1038/nbt.2121. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.