Significance
High-resolution data covering marine microbes and microeukaryotes are sparse, even though these organisms control global biogeochemical cycles. Here we present a dataset describing the microbial pro- and eukaryotic diversity along a 7,000-km transect from the Antarctic ice edge to the equator in the South Pacific Ocean. We show that (i) temperature is not a primary driver of richness gradients, (ii) prokaryotic richness increases with productivity, and (iii) oceanographic features can structure the diversity of pro- and eukaryotes. Our data have given us a better understanding of how diversity relates to dissolved inorganic nitrogen and productivity as well as insights into the potential shifts in the geographical range of marine microbe communities in light of the rapidly changing climate.
Keywords: prokaryotes, eukaryotes, Rapoport's rule, richness, latitude
Abstract
Marine microbes along with microeukaryotes are key regulators of oceanic biogeochemical pathways. Here we present a high-resolution (every 0.5° of latitude) dataset describing microbial pro- and eukaryotic richness in the surface and just below the thermocline along a 7,000-km transect from 66°S at the Antarctic ice edge to the equator in the South Pacific Ocean. The transect, conducted in austral winter, covered key oceanographic features including crossing of the polar front (PF), the subtropical front (STF), and the equatorial upwelling region. Our data indicate that temperature does not determine patterns of marine microbial richness, complementing the global model data from Ladau et al. [Ladau J, et al. (2013) ISME J 7:1669–1677]. Rather, NH4+, nanophytoplankton, and primary productivity were the main drivers for archaeal and bacterial richness. Eukaryote richness was highest in the least-productive ocean region, the tropical oligotrophic province. We also observed a unique diversity pattern in the South Pacific Ocean: a regional increase in archaeal and bacterial diversity between 10°S and the equator. Rapoport’s rule describes the tendency for the latitudinal ranges of species to increase with latitude. Our data showed that the mean latitudinal ranges of archaea and bacteria decreased with latitude. We show that permanent oceanographic features, such as the STF and the equatorial upwelling, can have a significant influence on both alpha-diversity and beta-diversity of pro- and eukaryotes.
Global climate change is impacting a wide variety of marine ecosystems (1, 2), and developing an adequate understanding of the base of the marine food web, including the richness of bacteria, archaea, and hetero-, mixo-, and phototrophic eukaryotes (3, 4), is an urgent priority if we are to predict its consequences (5). Ocean salinity and temperature have been shown to covary with species distributions, suggesting that oceanographic changes can shift geographic boundaries and alter phenology and community structures in organisms ranging from bacteria to planktonic eukaryotes and to higher trophic levels (6–9). Alterations in the biogeographic patterns of prokaryotic and eukaryotic microorganisms in the upper ocean could impact the efficiency of carbon (C) fixation, nutrient cycling, and ultimately the export of organic C to the deep ocean, as microorganisms are key biogeochemical regulators (4, 10, 11).
Much literature agrees on the consistent increase in richness from the poles toward the equator for all major groups of terrestrial, aquatic, and marine taxa [see Hillebrand (12); we note that there are exceptions, such as pinnipeds which peak at higher latitudes (13)]. However, diversity patterns for prokaryotic and unicellular eukaryotic organisms were underrepresented in the 600 latitudinal gradients assembled from the literature by Hillebrand (12). The latter noted that diversity gradients weaken when eukaryotic size decreases and that no gradients at all are expected when data are extrapolated to unicellular organisms. The large population sizes and high transportability, along with rapid generation times for prokaryotes and unicellular eukaryotes, increase the probability for repeated short-term reestablishment of populations in new habitats (14).
A growing body of evidence, however, suggests that pelagic marine microorganisms can exhibit large-scale biogeographical patterns that resemble those of multicellular macroorganisms (15–18). The findings of Pommier et al. (15) and Fuhrman et al. (18) showed that marine planktonic bacterial richness is correlated with temperature on a global scale. Ladau et al.’s (16) model outputs suggest that marine bacterial richness has a seasonal peak during the boreal and austral winters at temperate latitude. The work by Gilbert et al. (19) complemented the changes in diversity with season reported by Ladau et al. (16). The former concluded, from a study spanning 6 y, that bacterial richness peaked during winter in the English Channel. Their results highlighted that seasonal changes in environmental parameters (with day length explaining 65% of the variance) are in some cases more important than trophic interactions.
The mechanisms supporting biodiversity gradients [see Mittelbach et al. (20)] are better understood for larger-size taxa, and the controls for marine unicellular pro- and eukaryotic diversity trends remain more elusive due to the paucity of the spatial resolution (17, 21). Here we generated a high-resolution dataset in austral winter describing microbial diversity (on the surface and just below the thermocline) along with environmental parameters every half-degree along a 7,000-km transect spanning latitudes ∼66°S to 0°S and temperatures from −2 °C to 30 °C. We use this dataset to test the hypotheses concerning the ecological mechanisms controlling pro- and eukaryotic richness in the South Pacific Ocean that (H1) richness increases with increasing temperature (higher temperatures increase the rates of biochemical and metabolic pathways; e.g., Q10 coefficient) and (H2) richness increases with increasing productivity [more resources support higher numbers of species; Mittelbach et al. (22)]. In addition, we hypothesized that (H3) stable fronts and hydrographic features can act as control mechanisms on microbial assemblages (structuring of unique microbial oceanic provinces via ecological boundaries). Testing these hypotheses has given us a better understanding of how pro- and eukaryotic richness relates to ecosystem processes such as primary productivity as well as insights into the potential shifts in the geographical range of marine microbe communities in light of the rapidly changing global climate (5, 23).
Results
Physical and Biochemical Data from the Ice Edge to the Equator.
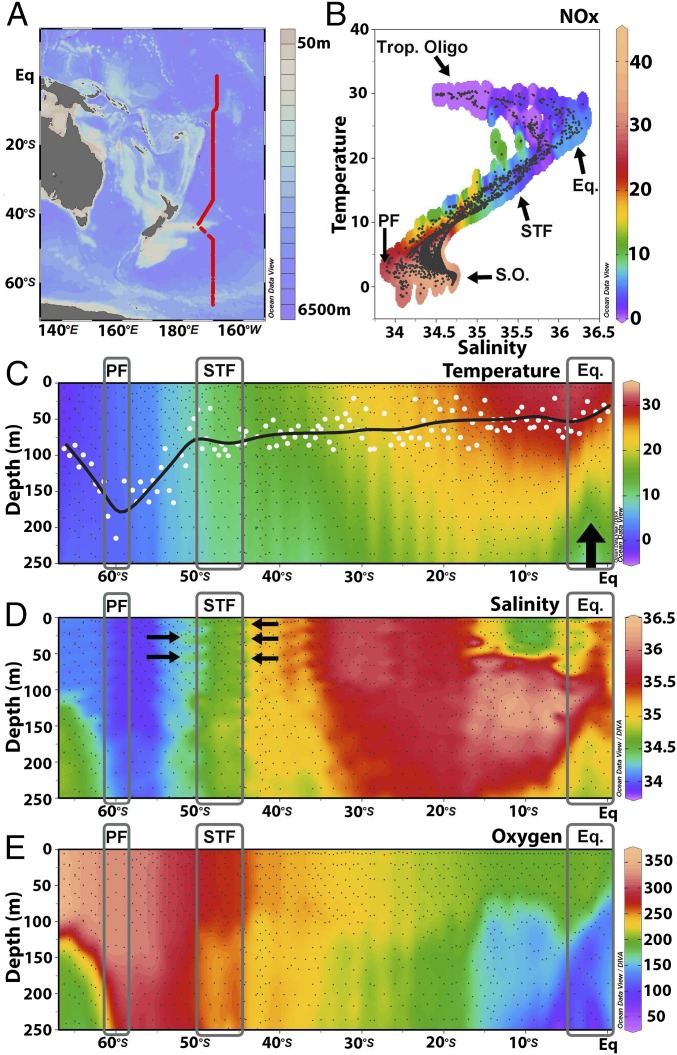
Sea surface temperatures increased gradually from −2 °C to 30 °C between 66°S and the equator (Fig. 1 A–C). Colder waters at a depth of ∼150 m near the equator showed a clear signal indicating equatorial upwelling (Fig. 1C). Lowest salinities were measured in the southern latitudes, with values around 34 practical salinity units (PSU), while they reached their maximum around 30°S at about 36.5 PSU. Prominent features along the P15S GO-SHIP transect are the polar front (PF) (∼60°S) and the subtropical front (STF) (∼43°S) (gray rectangles in Fig. 1 C and D). The PF separates the sub-Antarctic waters from the Antarctic circumpolar waters. The STF separates the colder, less saline, and nutrient-rich sub-Antarctic waters of the Southern Ocean from the warmer and saltier subtropical waters (Fig. 1 C and D). Clear temperature increases were measured passing the PF (from 0.8 to 1.42 °C). Both temperature and salinity increased when passing the STF (from 11.80 to 15.62 °C and from 34.6 to 35.2 PSU, respectively). Oxygen concentrations decreased from ∼350 µmol⋅kg−1 in the surface waters at higher latitudes to ∼200 µmol⋅kg−1 near the equator. Lower oxygen concentrations, between 50–100 µmol⋅kg−1, were measured near the equator at a depth of ∼150-m (Fig. 1E).
Fig. 1.
(A) The P15S GO-SHIP transect with bathymetry in color. The red line denotes 140 CTD (conductivity, temperature, depth) stations. (B) A plot of temperature (in degrees Centigrade) and salinity (in practical salinity units) using all depth data with NOx (NO3− + NO2−) concentrations (in micromoles per liter) overlaid in color. Black dots denote sampling depths where NOx concentrations were measured. The latitudinal transect could be divided in four oceanic provinces defined by Longhurst (25) and are denoted equatorial upwelling (Eq.), PF, Southern Ocean (S.O.), STF, and tropical oligotrophic province (Trop. Oligo). (C) Temperature profile to a 250-m depth along the transect. Filled white circles highlight MLDs (calculated as Δ0.2 °C; see Materials and Methods). The black line is a running average through the MLDs. The thick black arrow highlights the colder water and nutrient injection (seen in B) from equatorial upwelling. (D) Salinity profile (from bottle data; highlighted by black dots) to a 250-m depth. Black arrows denote salt fingers where warm, salty subtropical water intrudes between the cooler and fresher Southern Ocean waters. (E) Profile of oxygen concentrations in micromoles per kilogram (from bottle data; highlighted by black dots) to a 250-m depth. The PF, STF, and equatorial upwelling provinces are highlighted by gray rectangles.
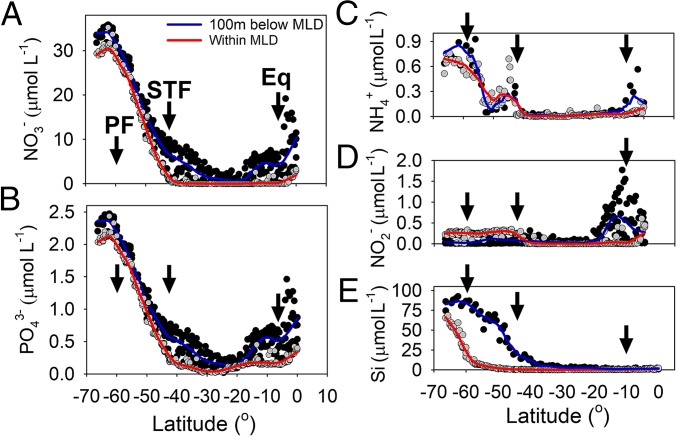
All nutrient concentrations within the mixed layer depth showed a similar latitudinal trend. Concentrations increased from south to north toward the PF and declined after the STF. Oligotrophic surface waters dominated the transect thereafter. Equatorial upwelling resulted in a local supply of new nutrients into the mixed-layer depth (MLD) (Fig. 2A). Nitrate (NO3−) concentrations within the mixed layer showed a steady decline after the PF, from 30 µmol⋅L−1 to <1 µmol⋅L−1 between 60°S and 40°S. NO3− concentrations increased up to 2 µmol⋅L−1 near the equator (Figs. 1B and 2A). Phosphate (PO43−) concentrations within the MLD decreased from 2.2 µmol⋅L−1 north of the PF to 0.15 µmol⋅L−1 at the STF. Thereafter PO43− decreased further, reaching the detection limit of the autoanalyzer. PO43− increased again from ∼25°S toward the equator, reaching 0.41 µmol⋅L−1 (Fig. 2B). Nitrite (NO2−) and ammonium (NH4+) concentrations were relatively elevated (up to 0.3 µmol⋅L−1) in the colder waters south of the STF. However, NO2− concentrations from the MLD to 100 m below the MLD were lower than the concentrations within the MLD. Both NO2− and NH4+ concentrations increased again toward the equator (Fig. 2 C and D). Silicate (Si) decreased rapidly north of the PF toward the STF, from 65.5 to 2 µmol⋅L−1, and the concentration stayed low (<0.5 µmol⋅L−1) in the tropical waters with a slight rise (∼1 µmol⋅L−1) toward the equator (Fig. 2E).
Fig. 2.
Nutrient data: NO3− (A), PO43− (B), NH4+(C), NO2− (D), and SiO4 (E) concentrations expressed in micromoles per liter along the P15S GO-SHIP transect. Gray circles are data from the surface to the base of the MLD (including all data within the MLD); thick red lines are negative exponential smoothing curves with a first polynomial degree. Black circles include the data from the base of the MLD to 100 m below the base of the MLD. Thick blue lines are negative exponential smoothing curves with a first polynomial degree. The black arrows on all panels highlight the oceanic PF, STF, and equatorial upwelling and divide the latitudinal transect into oceanic provinces defined by Longhurst (25).
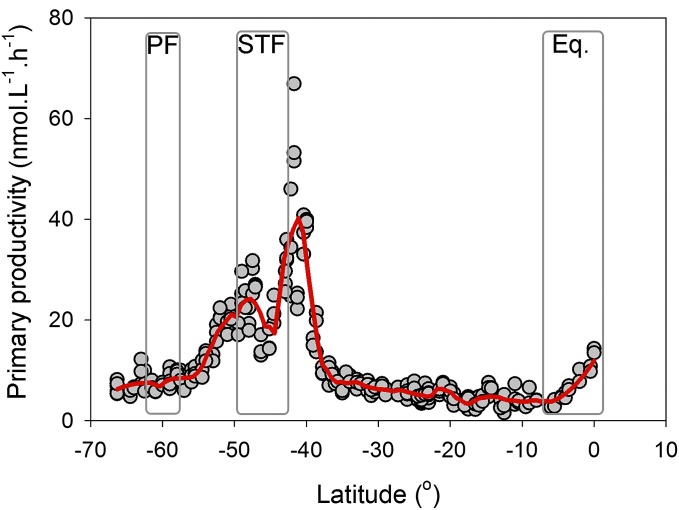
All measured biochemical parameters showed a similar latitudinal trend with a sharp decline north of the PF and north of the STF and a more modest increase near the equator. Chlorophyll a concentrations were elevated south of the STF and in the equatorial upwelling region. Transmission data [a proxy for particle concentration; see Karageorgis et al. (24)] showed elevated concentrations along the STF and the equatorial upwelling region (SI Appendix, Fig. S1 A and B). Primary productivity in the surface waters increased steadily from 5 to 30 nmol C⋅L−1⋅h−1 between ∼66°S and ∼50°S. We did not see an increase in productivity near the PF. South of the STF (around 43°S), productivity increased to a maximum of 77 nmol C⋅L−1⋅h−1; thereafter NO3−, NO2−, and NH4+ concentrations dropped to <0.02 µmol⋅L−1, and productivity rates decreased and remained low (∼5 nmol C⋅L−1⋅h−1) between 40°S and ∼10°S. Productivity rates increased north of 10°S (Fig. 3).
Fig. 3.
Surface primary productivity data (expressed in nanomoles of C per liter per hour) along the P15S GO-SHIP transect. The red line is a negative exponential smoothing curve. Gray rectangles highlight the PF, the STF, and the equatorial upwelling (Eq.).
Richness Data from the Ice Edge to the Equator.
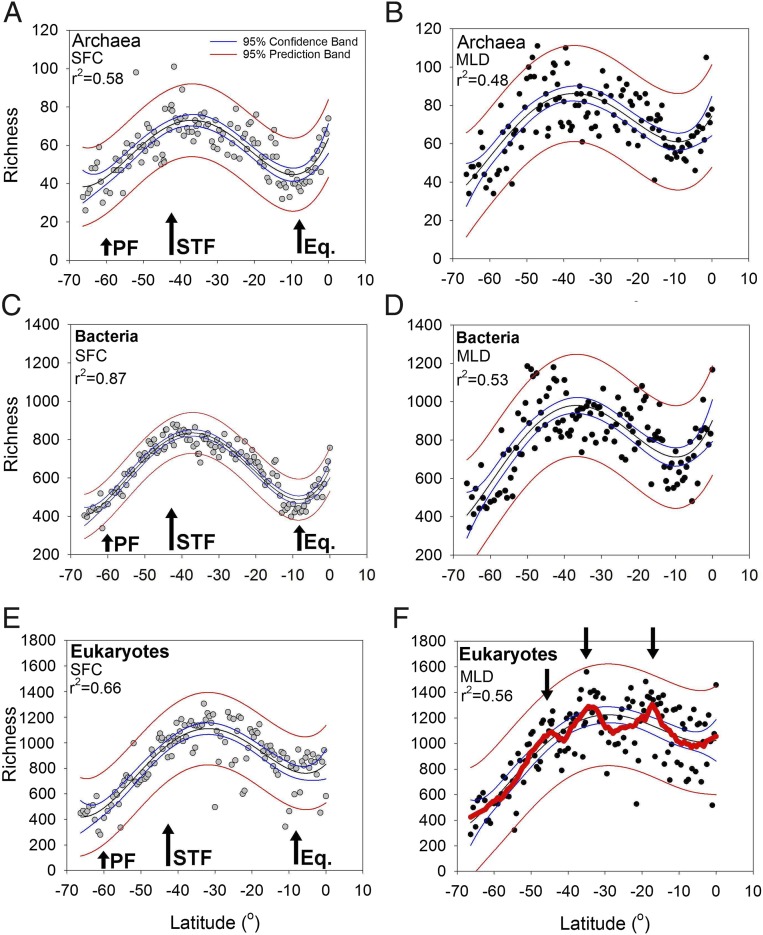
Our high-resolution data revealed that archaeal richness peaked at 100 operational taxonomic units (OTUs) at 41.7°S and at 75 OTUs at the equator in the surface waters (fourth polynomial fit r2 = 0.58; P < 0.0001) (Fig. 4A). OTU richness in the surface waters showed an approximate doubling from the ice edge (66°S; 30 OTUs) to the STF (43°S; 68 OTUs). We did not measure a distinct change in richness when we crossed the PF. However, the STF denoted a distinct physical barrier and separated the colder, higher latitudinal waters from the warmer, saltier subtropical waters (Fig. 1 C and D). North of the STF, the archaeal richness decreased to ∼30–40 OTUs around 10°S. Archaeal richness then showed a 2.5-fold increase from 10°S to the equator (linear regression P < 0.0001). The archaeal richness was significantly greater just below the MLD than in the surface waters (Dunn’s test P < 0.001, n = 131) but revealed a distribution similar to that in the surface samples (fourth polynomial fit r2 = 0.48; P < 0.0001) (Fig. 4B).
Fig. 4.
Richness gradients for archaea in the surface waters (gray circles) (A) and just below the MLD (filled black circles) (B), for bacteria in the surface waters (C) and just below the MLD (D), and for eukaryotes in the surface waters (E) and just below the MLD (F) along the P15S GO-SHIP transect. Fourth-order polynomial fits for all plots are shown by thin black lines. Blue lines represent 95% confidence bands, and thin red lines represent 95% predictions bands. The thick red line in F is a negative exponential smoothing curve with a first-polynomial degree to highlight the bimodal trend (also denoted by three black arrows in F). The PF, STF, and equatorial upwelling (Eq.) are shown by black arrows in A, C, and E.
Bacterial richness in the surface waters showed peaks near the STF and the equator (fourth polynomial fit r2 = 0.87; P < 0.0001) (Fig. 4C). We did not measure a distinct change in bacterial richness when we crossed the PF. However, bacterial richness approximately doubled from the sub-Antarctic waters to the STF. From the STF to 10°S, bacterial richness declined again from about 800 OTUs to 400 OTUs. Richness showed a steep and significant increase from 10°S to the equator (linear regression P < 0.0001). The bacterial richness was significantly greater just below the MLD than in the surface waters (Dunn’s test P < 0.001, n = 131). Richness below the MLD showed a trend similar to that of the surface samples but with a lower fit (fourth polynomial fit r2 = 0.53; P < 0.0001) (Fig. 4D). North of the STF the bacterial richness decreased to ∼400 OTUs at 10°S (linear regression P < 0.0001). Richness increased again up to 1,200 OTUs at the equator.
The eukaryotic richness in the surface waters peaked at 30°S with a modest increase near the equator (fourth polynomial fit r2 = 0.66; P < 0.0001) (Fig. 4E). We did not measure a distinct change in eukaryotic richness when we crossed the PF northwards. In contrast to observations in the surface waters, the eukaryotic richness just below the thermocline showed three richness peaks and an increase near the equator. A fourth-order polynomial fit (r2 = 0.56; P < 0.0001) (Fig. 5E) did not emphasize these eukaryotic trends, which are clearer when denoted by a negative exponential smoothing curve (thick red line in Fig. 4F). Richness peaked at 1,560 OTUs at 35°S and at 1,484 OTUs at 19°S. The richness was significantly greater just below the MLD than at the surface (Dunn’s test P < 0.001, n = 131).
Fig. 5.
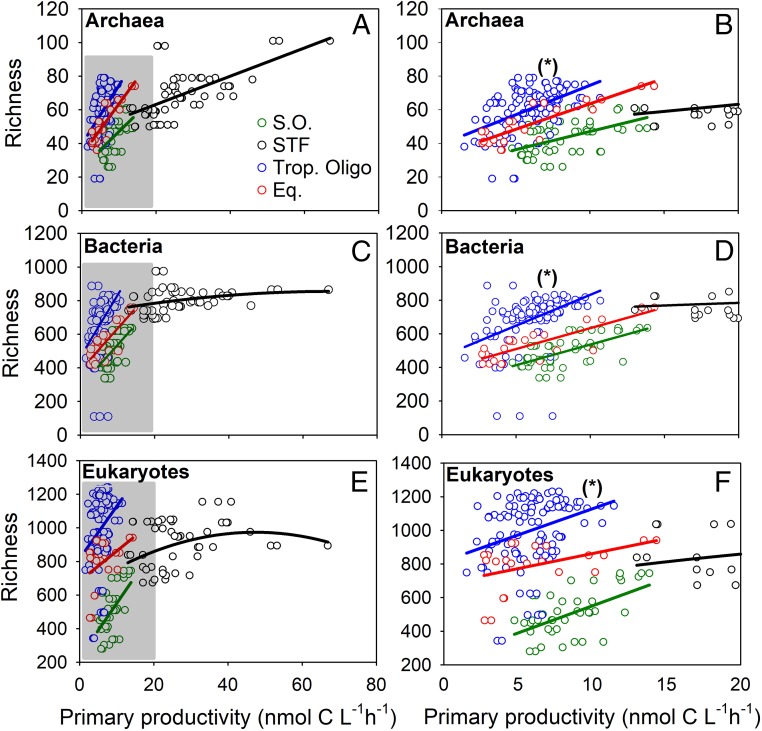
(A, C, and E) Surface primary productivity rates vs. richness data (total OTUs) in the distinct oceanic provinces in the South Pacific Ocean for archaea (A), bacteria (C), and eukaryotes (E). Gray rectangles in the archaeal (A), bacterial (B), and eukaryote (C) plots highlight the primary productivity data <20 nmol⋅L−1⋅h−1. (B, D, and F) Correlations with richness for these three domains. Data for the Southern Ocean are denoted by green circles (linear regression fits in green lines); data for the STF are denoted by open black circles (linear, quadratic, and cubic regression fits are denoted by black lines); data for the tropical oligotrophic province are denoted by blue circles (blue lines are linear regressions); and data for the equatorial upwelling province are denoted by red circles (red lines are the linear regression). All fits are significant with a P < 0.05. The asterisks in plots B, D, and F denote the finding that richness normalized to primary productivity is highest in the tropical oligotrophic region for archaea, bacteria, and eukaryotes for the productivity range <20 nmol⋅L−1⋅h−1. Note all replicate C-fixation data are plotted.
Fifteen predictor parameters were investigated to explain variation in pro- and eukaryotic richness using boosted regression trees (BRT) models and Spearman rank correlations. BRT model outputs using only the surface samples showed that oxygen concentrations, along with NH4+, salinity, the nanophytoplankton fraction, and primary productivity, were the main predictors for archaeal richness (Table 1). Salinity, the nanophytoplankton fraction, NH4+, and primary productivity were the main predictors describing patterns in bacterial richness (Table 1). Oxygen did not show a positive or negative relationship to richness (SI Appendix, Fig. S2A). Salinity, the nanophytoplankton component, and primary productivity all revealed positive relationships to archaeal and bacterial richness, whereas NH4+ showed a negative relationship (SI Appendix, Fig. S2A). Nanophytoplankton and primary productivity showed a strong positive relationship. Patterns in eukaryotic richness in surface waters were primarily explained by variability in Si, salinity, oxygen, the microphytoplankton, NH4+, and the nanophytoplankton (Table 1 and SI Appendix, Fig. S2A). Spearman correlations and BRTs showed that latitude and temperature were not significant drivers for archaeal and bacterial richness in the surface.
Table 1.
Relative influence of 16 predictor variables in explaining variation in pro- and eukaryotic richness in surface samples (n = 89) along the p15S GO-SHIP transect determined by BRT [see also Elith et al. (73)]
| Archaeal richness–SFC | Bacterial richness–SFC | Eukaryote richness–SFC | |||
| Predictor variable | % relative influence | Predictor variable | % relative influence | Predictor variable | % relative influence |
| Oxygen | 33.5 | Salinity | 34.3 | Si | 17.0 |
| NH4+ | 20.3 | Nanoplankton | 21.8 | Salinity | 14.9 |
| Salinity | 14.8 | NH4+ | 8.4 | Oxygen | 11.9 |
| Nanoplankton | 11.9 | PP | 8.2 | Microplankton | 10.8 |
| PP | 6.5 | Diatoms | 6.5 | NH4+ | 7.9 |
| Temperature | 2.9 | Temperature | 6.5 | Nanoplankton | 6.8 |
| Diatoms | 2.9 | Oxygen | 5.9 | Diatoms | 5.6 |
| TChl_a | 2.0 | Si | 3.4 | Temperature | 5.3 |
| Microplankton | 1.4 | TChl_a | 1.5 | Picoplankton | 5.0 |
| Si | 1.0 | Picoplankton | 0.9 | PP | 3.9 |
| Picoplankton | 0.9 | PO43− | 0.9 | MLD | 3.3 |
| MLD | 0.8 | MLD | 0.5 | Phosphate | 2.9 |
| PO43− | 0.7 | Microplankton | 0.5 | TChl_a | 2.4 |
| NO2− | 0.3 | NO2− | 0.5 | NO3− | 1.2 |
| Dinoflagellates | 0.2 | NO3− | 0.1 | NO2− | 1.0 |
| NO3− | 0.1 | Dinoflagellates | 0.1 | Dinoflagellates | 0.1 |
| Training data correlation | 0.88 | Training data correlation | 0.973 | Training data correlation | 0.928 |
| Cv correlation ± SE | 0.779 ± 0.056 | Cv correlation ±SE | 0.921 ± 0.023 | Cv correlation ± SE | 0.773 ± 0.061 |
Cv, coefficient of variation; NOx, NO3− + NO2−; PP, primary productivity; TChl_a, total chlorophyll a. Phytoplankton size classes (microplankton, 20–200 μm; nanoplankton, 2–20 μm; picoplankton, 0.2–2 μm) and taxonomic groups (diatoms and dinoflagellates) were derived from HPLC analysis.
In the MLD, models showed that temperature, primary productivity measured in the overlaying surface waters, and NO2− concentrations explained 50% of the trends in archaeal and bacterial richness along the transect (SI Appendix, Table S2). Primary productivity revealed positive relationships, and NO2− revealed negative relationships for both archaeal and bacterial richness (SI Appendix, Fig. S2B). Patterns in MLD eukaryotic richness were primarily explained by strong negative relationships of Si and NO3− and a positive relationship with temperature (SI Appendix, Fig. S2B and Table S2).
Overall, all nutrients except NO2− showed a significant negative correlation with richness for all domains in the surface waters. On the other hand, we note that all nutrients showed a positive correlation with total chlorophyll a in the surface waters. NO2− just below the MLD was the only nutrient which displayed a positive relationship with latitude and temperature along with a negative relationship with oxygen (SI Appendix, Fig. S2 A and B).
Salinity revealed an equally strong positive correlation for the archaeal and bacterial richness in the surface (SFC) waters, with a stronger correlation for eukaryotic richness. Salinity is the main physical parameter structuring oceanic fronts such as the STF (Fig. 1D), which relates to our third hypothesis that stable fronts and hydrographic features can act as control mechanisms on microbial assemblages.
Richness vs. Productivity in Surface Waters of Distinct Oceanic Provinces.
The strong positive salinity correlation for eukaryotic richness (Table 1), along with the clear primary productivity peaks near the fronts (Fig. 3) and the sharp dissolved inorganic nutrient inflection points along the transect, encouraged us to explore our richness and primary productivity data from surface samples further. To do so, we divided our data according to the oceanic provinces defined by Longhurst (25). These provinces overlap where we measured sharp NO3− concentration inflection points near the STF (∼43°S) and the equatorial upwelling (∼10°S) (Fig. 2A). Spearman correlations between richness and primary productivity in surface waters were higher for all domains (correlations are reported below) when we divided our data along the Longhurst provinces. When we analyzed these significant correlations in more detail, we found strong positive linear and cubic relationships for archaeal, bacterial, and eukaryotic richness: archaea (Fig. 5 A and B) all linear regressions r2 = 0.27, r2 = 0.39; r2 = 0.25, and r2 = 0.75, all P < 0.001 for the Southern Ocean, the STF, the tropical oligotrophic, and the equatorial upwelling provinces, respectively; bacteria (Fig. 5 C and D): linear regressions for the Southern Ocean, the tropical oligotrophic, and the equatorial upwelling provinces with r2 = 0.44, r2 = 0.22, and r2 = 0.67, all P < 0.0001, respectively, and a cubic regression for the STF with r2 = 0.09, P < 0.05; eukaryotes (Fig. 5 E and F): linear regressions for the Southern Ocean, the tropical oligotrophic, and the equatorial upwelling provinces with r2 = 0.03, r2 = 0.10, and r2 = 0.19, all P < 0.05, respectively, and a cubic regression for the STF with r2 = 0.12, P < 0.05.
The STF province showed the absolute highest richness for the archaea and bacteria. The highest richness for the eukaryotes was measured in the tropical oligotrophic province (Fig. 5 E and F). The stations with the highest eukaryotic richness and highest productivity were found just north of the STF (see also Fig. 3). Richness showed the highest values for all three domains in the tropical oligotrophic province for the productivity range <20 nmol⋅L−1⋅h−1 (denoted by asterisks in Fig. 5 B, D, and F).
Spearman correlations for the surface data within the distinct oceanic provinces (SI Appendix, Fig. S3) revealed that latitude and temperature showed positive and negative relationships with pro- and eukaryotic richness. Dissolved inorganic nutrients revealed negative correlations with pro- and eukaryotic richness in the Southern Ocean and the STF. In the tropical oligotrophic region lower levels of dissolved inorganic nutrients resulted in a positive correlation for NO3−. These results complement those from the overall latitudinal correlation plot (SI Appendix, Fig. S2A). In the equatorial upwelling, however, most nutrients showed a positive correlation with pro- and eukaryotic richness. NH4+ showed a negative correlation with pro- and eukaryotic richness in the Southern Ocean, the STF, and the tropical oligotrophic province but a positive correlation in the equatorial province. We note that in the correlation plots (SI Appendix, Fig. S3 B–D) primary productivity was not always significantly correlated with the pro- and eukaryotic richness in the oceanic provinces, as we used the average C-fixation rates (from three replicates). However, primary productivity always showed significant regressions with pro- and eukaryotic richness when we included all replicates (Fig. 5). The nano- and picophytoplankton revealed significantly positive relationships with productivity in the Southern Ocean. The different phytoplankton size classes did not show significant relationships between productivity within the STF. The micro- and nanophytoplankton fractions showed positive relationships in the tropical oligotrophic waters, whereas the picophytoplankton fraction showed a negative correlation. All phytoplankton size classes showed a positive correlation with productivity in the equatorial upwelling region (SI Appendix, Fig. S3).
Beta-Diversity from the Ice Edge to the Equator.
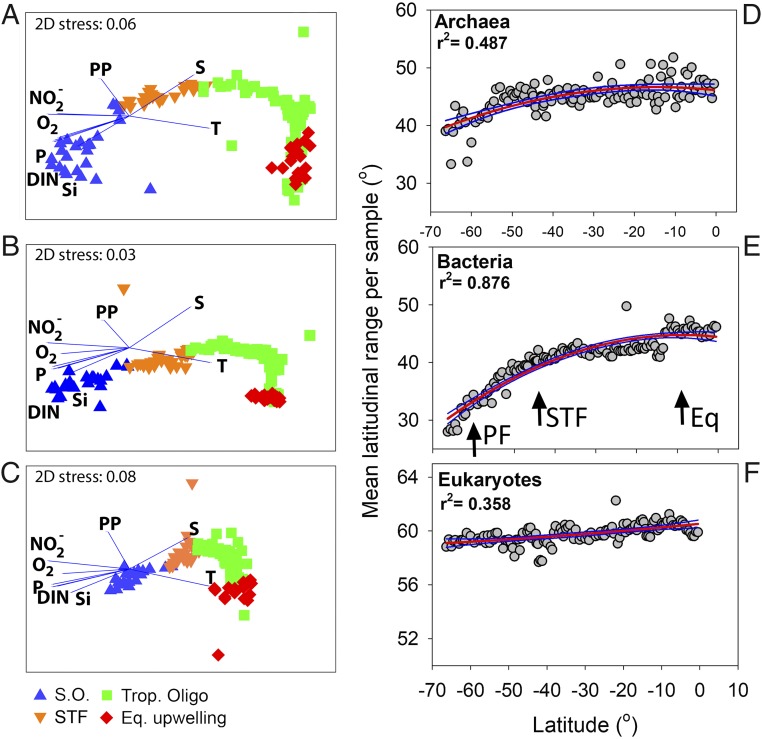
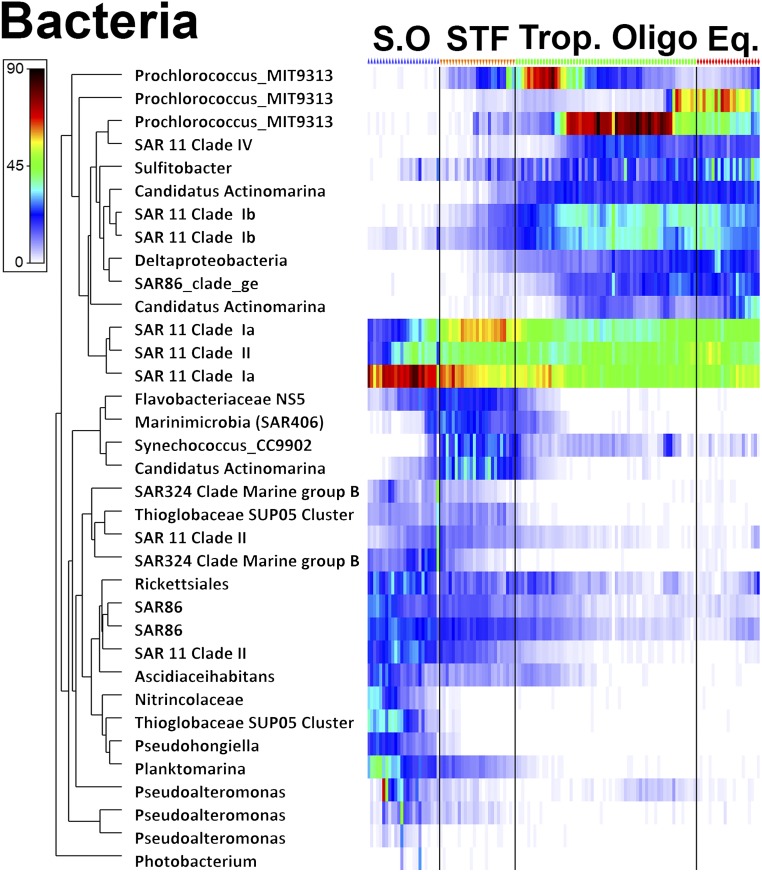
Changes in community composition for all domains (presented as nonmetric multidimensional scaling plots in Fig. 6 A–C) showed strong significant differences between all oceanographic biomes except between the tropical oligotrophic and the equatorial upwelling [see analysis of similarities (ANOSIM) in SI Appendix, Table S3]. The absolute latitudinal range (the difference between the maximum and minimum latitudes where an OTU was present) increased for all domains from south to north (r2 = 0.487, 0.876, and 0.358 for the archaea, bacteria, and eukaryotes, respectively, all P < 0.001). The slope for the eukaryotes was very weak, however, and showed only a 3° increase from south to north. The latitudinal range of the bacteria, on the other hand, increased by 19° from south to north, with the steepest increases in the Southern Ocean (covering both the PF and STF) and near the equatorial upwelling (Fig. 6 D–F). Heatmaps of the most abundant OTUs for all domains confirmed strong differences in community composition in the different biomes (SI Appendix, Fig. S4). In the archaeal domain a key finding was a high abundance of nitrifying organisms such as Candidatus Nitrosopumilus and Nitrosopelagicus in the Southern Ocean. Overall, we saw clear changes in the archaeal beta-diversity, but, because the taxonomy for archaea is not as well resolved as for the bacterial domain, most of the dominant archaeal OTUs belonged to Thermoplasmatales Marine Group II. In the bacterial domain, sequences from SAR11 clade 1a, SAR86, Rickettsiales, Thioglobaceae, Planktomarina, and Pseudoaltermonas revealed an abundant presence in the Southern Ocean. The Flavobacteriaceae, Marinimicrobia, Syncechococcus CC9902, and Candidatus Actinomarina showed a higher abundance in the STF. Procholoroccus, SAR11 claded IV and Ib, Deltaproteobacteria, and Candidatus Actinomarina, on the other hand, showed a high abundance in the tropical biomes (Fig. 7). Key findings for the eukaryotic domain were a high abundance of Dinophyceae and Copepoda in the Southern Ocean and STF, along with a high presence of Phaeocystis (which was also visually confirmed by a bloom). In the Southern Ocean we also noted a very high-read abundance of Collodaria, phylum Radiolaria. Copepods also showed a high-read abundance in the tropical oligotrophic and the equatorial regions.
Fig. 6.
(A–C) nMDS plots for archaea (A), bacteria (B), and eukaryotes (C). Environmental parameters are overlaid as vectors. DIN, nitrate and ammonium; P, phosphorus; PP, primary productivity; S, salinity; T, temperature. (D–F) Rapoport’s analysis. The mean latitudinal range sizes showed significant increases toward the equator for archaea (D), bacteria (E), and eukaryotes (F) (all P < 0.001). All samples were from the surface. Quadratic fits for all plots are shown by red lines. Blue lines present 95% confidence bands. Note the difference in the y axis for the eukaryotes. Black arrows in E highlight the PF, the STF, and the equatorial upwelling (Eq).
Fig. 7.
Heatmap of the bacterial domain. The most abundant sequences of the rarefied OTU matrix are displayed. Sequence reads were square root transformed (note: chloroplasts included in the16S amplicon data were removed from the analysis). The color bar denotes sequence abundance. Blue, orange, green, and red symbols above the heatmap denote CTD stations in the respective oceanographic biomes: the Southern Ocean (S.O), the STF, the tropical oligotrophic (Trop. Oligo.), and the equatorial upwelling (Eq.).
Discussion
Our dataset describing pro- and eukaryotic diversity from the ice edge to the equator along 170°W longitude in the South Pacific Ocean has reconfirmed that (H1) temperature is not always a primary driver of richness gradients, complementing the global model data from Ladau et al. (16); (H2) archaeal and bacterial richness increased significantly with productivity; and (H3) oceanographic features, such as fronts and upwelling, can influence pro- and eukaryotic richness. Our data complement the work by Ladau et al. (16) and Gilbert et al. (19) and, because of our high-resolution spatial sampling, add to our understanding of the marine microbial diversity in ways that were not possible with previous datasets.
Overall, archaeal, bacterial, and eukaryotic richness were all positively correlated to each other and displayed bimodal distributions along our transect, bounded by key oceanographic features including the PF, the STF, and the equatorial upwelling region. These physical features were associated with strong changes in temperature, salinity, dissolved inorganic N concentrations, and PO43− concentrations. Both archaeal and bacterial richness peaked at borders of the STF and near the equatorial upwelling. Archaea and bacteria were significantly correlated with primary productivity in these regions. These results show that permanent oceanographic features, such as the STF and the equatorial upwelling region, can act as ecological boundaries.
Our data suggest a different latitudinal trend in archaeal and bacterial diversity than previously thought. Ladau et al. (16) predicted a decrease in winter from ∼30° toward the equator. To the best of our knowledge, no research has shown or modeled the steady increase for both archaea and bacteria toward the equatorial upwelling region. It is worth noting that the increase in eukaryotic richness toward the equator was more modest than that seen in archaea and bacteria. Our findings also show that temperature is not, as suggested on a global scale by Ladau et al. (16), a primary driver for pro- or eukaryotic richness in the South Pacific Ocean. These findings complement those from Marañón et al. (26), who showed that variability in phytoplankton growth is driven by changes in resource supply rather than by seawater temperature. Boosted regression trees analysis showed that patterns of archaeal and bacterial richness in the surface waters could be largely explained by salinity (a main identifier for fronts along our transect) and by NH4+, the nanophytoplankton fraction, and primary productivity.
The nanophytoplankton fraction was dominated by Dinophyceae and prymnesiophytes, which reaffirms that these groups could be an important player in CO2 fixation, as mentioned by Jardillier et al. (27), in subtropical and tropical ecosystems. NH4+ is a preferred substrate for a wide range of organisms, and the link between NH4+ and primary productivity is explained below in more detail. The nanophytoplankton fraction showed the strongest positive correlation with productivity of the three phytoplanktonic size fractions. Overall, our data suggest that further exploration of whether primary productivity supports greater richness or whether a higher richness stimulates greater primary productivity would be useful.
Eukaryotic richness was influenced up to 32% by salinity and Si in the surface waters and 35% by Si alone just below the MLD. These data suggest that eukaryotic richness is strongly controlled by the sharp abiotic inflection points seen near the oceanographic features in the South Pacific Ocean and, again, that oceanographic features can structure eukaryotic richness, as seen elsewhere (28). The negative correlation with Si also suggests that a more diverse eukaryotic community depletes the Si from the water.
Richness for all domains was higher just below the MLD, which is supported by the global data analysis of Ladau et al. (16) in which proximity to the nutricline, also a hot spot for organic and inorganic particle accumulation (29), was presented as a strong predictor for bacterial richness. In particular, particles and marine aggregates (often at fronts and at density interfaces such as the MLD) are able to provide extra niches, such as anaerobic microhabitats in oxygenated waters, for prokaryotes (30), which in turn can increase their diversity. Near the equatorial upwelling, relatively elevated NO2− and NH4+ concentrations coincided with higher primary productivity rates [note that NO2− and NH4+ concentrations were also high in the Southern Ocean, but there iron limits primary productivity (31, 32)]. Near the equator NO2−, NH4+, and oxygen showed positive relationships with archaeal richness.
Oceanographic fronts are generally characterized by a transition between mixed and stratified waters, and we suggest that they can act as ecological boundaries for pro- and eukaryotes in the ocean as supported by recent research (33). Oceanographic features, such as the STF and the equatorial upwelling region, stimulate primary productivity, which drives enhanced recycling processes [such as ammonification and NH4+ oxidation (34, 35)]. Fronts provide a source of buoyancy through mesoscale and submesoscale instabilities (36), which enhance nutrient supply (via upwelling) and light availability [through mixing (37)]. Salt fingering, which occurs when warm, salty water intrudes into cooler and fresher water, could also clearly be noted in the STF (Fig. 2D). Here, layers of warm, salty subtropical water intrude into the cooler and fresher Southern Ocean waters. Salt fingers have the capacity to transport dissolved organic matter and nutrients horizontally and vertically through molecular and turbulence-driven diffusivities (38, 39). This can play an important role in the supply of new N, which stimulates phytoplankton productivity and eukaryotic richness, as seen in our dataset and by others (40). These results complement reports that productive (high-fertility) regions support a greater richness (22), as seen in regional studies in the eastern Indian Ocean (41).
Fronts are also zones of (seasonal) convergences (25), which lead to the passive accumulation of organic material (e.g., high biomass and particle loads; see SI Appendix, Fig. S1). This in turn increases the recycling of particulate organic material and dissolved organic material through bacterial degradation (42). These processes will increase the NO2− and NH4+ concentrations, which in our data could clearly be noted near the STF and near the equatorial upwelling, where we measured elevated NH4+ concentrations and high chlorophyll a concentrations. We note that the highest primary productivity rates near the STF were measured at low NH4+ concentrations, as expected because NH4+ is rapidly assimilated and as also confirmed by the negative Spearman correlation. It is interesting that dissolved inorganic nutrients showed a negative correlation with productivity (except in the equatorial upwelling), whereas pro- and eukaryotic richness was positively correlated with productivity. These findings suggest that the conversion rates of resources to biomass is more important than the resources themselves. Increasing productivities were noted near two key oceanographic features along the P15 transect: the STF and the equatorial upwelling. Near the equatorial upwelling the positive correlation of NH4+ with prokaryotic richness can be related to increased eukaryotic primary productivity. We note that we did not see distinct changes in the primary productivity rates or in the beta-diversity when we crossed the PF.
Within each oceanic province pro- and eukaryotic richness increased linearly within the 0–20 nmol C⋅L−1⋅h−1 productivity range, and richness saturated at higher primary productivity. Productive ecosystems have been reported to support high species richness (43); in our data we note that eukaryotic richness was highest in the least productive oceanic province, the tropical oligotrophic region. Eukaryotic diversity increased from the polar waters to the STF. Overall, richness did not peak at the highest productivity rate measurements. A possible explanation is that productive regions tend to be associated with lower eukaryotic diversity, possibly due to being dominated by fast-growing organisms which can outcompete other species through resource competition (44). Generally, we need more information to disentangle the question whether primary productivity controls richness or whether higher richness stimulates greater primary productivity.
We observed clear patterns in the richness across the three domains (alpha-diversity) but also in the diversity among communities (beta-diversity). From south to north we noted distinct differences in the community structures and in the abundance of distinct genera for the pro- and eukaryotic domains in the different biomes. Our data showed that oceanographic boundaries can constrain beta-diversity gradients in the South Pacific Ocean. Overall we noted strong differences among all biomes, except between the tropical oligotrophic region and the equatorial upwelling. Interestingly, our high-resolution data revealed an increase in the latitudinal ranges from the most southern to the most northern stations for the archaea and bacteria. The mean latitudinal ranges of archaea and bacteria decreased with increasing latitude, thus not supporting the existence of a Rapoport’s effect in early winter in the South Pacific Ocean. Rapoport’s rule suggests that range sizes are larger near the poles and decrease near the equator. Amend et al. (17) and Sul et al. (45) did confirm a global Rapoport’s effect for marine bacterioplankton. Gaston et al. (46) and Rohde (47), on the other hand, note that the Rapoport’s rule is a strictly local phenomenon. Our data do show a latitudinal trend, but one opposite the reports in the current literature. These data, especially in the Southern Ocean where the latitudinal range was the smallest, highlight the strong need to understand the correlations between the different biotic and abiotic parameters that structure marine pro- and eukaryotic richness, so we can build a mechanistic understanding of patterns of global marine microbial diversity. The Southern Ocean is a region that stands out as a sentinel for responses to climate change (48). Changes in the physical and biogeochemical properties of the Southern Ocean (49) will likely impact pro- and eukaryotic richness and thereby vital ecosystem services. Therefore a good understanding of the alpha- and beta-diversity at the base of the food web is of important.
Taxonomic heatmaps demonstrated strong differences in community structure and revealed that distinct taxa for all domains were present in the different oceanographic biomes. The Alphaproteobacteria and in particular the SAR11 clades showed distinct differences in the Southern Ocean, the STF, and the tropical oligotrophic biomes, as also seen by others (50), across frontal zones in the Southern Ocean. From a functional ecosystem perspective the higher read abundance of nitrifying archaeal organisms in the Southern Ocean raises the interesting question of whether we can link nitrification rates to these higher abundances in the Southern Ocean. In the eukaryotic domain the Calanoida showed different genera across and at opposing ends of the transect. Copepods have been used as an indicator of changes in ocean and climate conditions (51). Continued high-resolution DNA sampling along repeated transect lines in the Ocean, using different eukaryotic marker genes [see the topic of environmental DNA (52)] could potentially help in teasing apart differences in community structures in response to climatic changes.
Conclusion
Our high-resolution data gave insights into the trends in pro- and eukaryotic richness along a vast latitudinal transect, complementing the model data of Ladau et al. (16) showing that temperature is not always a prime control in richness gradients. Here we show that archaeal and bacterial diversity increases from around 10°S to the equator and that the richness of these domains shows a strong and positive correlation with primary productivity. These results update the findings of Pommier et al. (15) and Fuhrman et al. (18), in which bacterial richness was only weakly related to primary productivity (which was calculated from satellite data). Our data showed that permanent oceanographic features can act as ecological boundaries which can influence pro- and eukaryotic richness, as reported by Zinger et al. (53), Wilkins et al. (54), Amend et al. (17), Wolf et al. (55), and Baltar and Arístegui (33). Interestingly our high-resolution data did not confirm the effect of Rapoport’s rule in early winter in the South Pacific Ocean. Our data showed that the mean latitudinal ranges of archaea and bacteria decreased with latitude. Overall, our data support a potential link between productivity and diversity gradients and highlight the need for more information on how local environmental forces can modulate the correlation between diversity and productivity within ocean provinces.
Materials and Methods
Study Region.
Samples were collected along the latitudinal P15S GO-SHIP transect, a 7,000-km decadally repeated transect from the ice edge (∼66°S) to the equator (0°S) (Fig. 1A), from 23 April to 29 June (late austral autumn/early winter) in 2016 (www.go-ship.org/; voyage 096U20160426). The four main oceanographic provinces along the transect are defined by Orsi et al. (56) and Longhurst (25). In this dataset we refer to these oceanic provinces as (i) Southern Ocean waters between 66°S and 52°S, including the PF at 60°S; (ii) the STF between 52°S and 40°S; (iii) tropical oligotrophic waters between 40°S and 10°S; and (iv) equatorial upwelling, between 10°S and 0°S.
Biophysical and Chemical Metadata.
Biophysical and chemical parameters were collected at 140 stations at 36 depths. Samples for dissolved inorganic nutrient concentrations included Si, PO43−, NO3−, NO2−, and NH4+. All were assayed on a Bran+Luebbe AA3 HR segmented flow analyzer at sea by the Commonwealth Scientific and Industrial Research Organisation (CSIRO) Hydrochemistry group, following standard spectrophotometric methods (57). Detection limits were 0.2, 0.01, 0.015, and 0.015 µmol⋅L−1, respectively. Detailed methodology for detection limits and data accessibility are provided in the SI Appendix.
Genomics Analyses.
For DNA analysis 2 L of seawater from the surface and just below the MLD (SI Appendix, Fig. S5) were filtered through a 0.22-μm pore Sterivex GP filter (catalog no. SVGPL10RC; Millipore) with a peristaltic pump. A modified organic (phenol:chloroform:isoamyl-based) DNA extraction protocol (58) was used with the PowerWater Sterivex DNA isolation kit (Mo Bio Laboratories; see SI Appendix for the DNA extraction protocol). Amplicons targeting the bacterial 16S rRNA gene (27F–519R) (59, 60), archaeal 16S rRNA gene (A2F–519R) (60, 61), and eukaryotic 18S rRNA gene [18SV4F and 18SV4R; modified from Stoeck, et al. (62)] were generated and sequenced for each sample at the Ramaciotti Centre for Genomics (University of New South Wales, Sydney). 16S rRNA gene amplicons were sequenced using 300-bp paired-end sequencing, while 18S rRNA gene amplicon reads were generated using 250-bp paired-end sequencing. Sequence OTU tables were prepared after Bissett et al. (63). Briefly, paired-end reads merged using FLASH (64) sequences were clustered into OTUs at 97% sequence similarity using the open reference OTU-picking pipeline in USEARCH 64-bit v8.0.1517 (65) with the UPARSE algorithm (66). Reads were mapped to the resultant OTU representative sequences using USEARCH (97% identity) to calculate read abundances per OTU. OTU tables were subsampled to a constant sampling depth of 13,400, 26,000, and 24,500 sequences per sample for archaea, bacteria, and eukaryotes, respectively (rarefaction curves are shown in SI Appendix, Figs. S7 and S8). Taxonomy was derived from SILVA database version 132 (https://www.arb-silva.de/). Genomic data are available at https://www.ncbi.nlm.nih.gov/bioproject/385736 under the accession no. PRJNA385736.
Pigment Analysis and C Assimilation.
Chlorophyll a extractions were carried out according to Parsons et al. (67) on 0.525 L of sample water via gentle vacuum filtration (pressure drop <10 kPa) using 25-mm GF/F filters at five sampling depths within the mixed layer. Samples were measured on a Turner Trilogy laboratory fluorometer. Four liters of sample water from the surface at each station were filtered in dim light on 25-mm Whatman GF/F filters for analysis of taxonomically significant chlorophylls and carotenoids. Samples were snap-frozen and stored in liquid N. Photosynthetic pigments were analyzed using HPLC according to Hooker et al. (68). HPLC data were analyzed using diagnostic pigments of dominant phytoplankton functional guilds as well as size classes according to Hirata et al. (69); see SI Appendix, Supplementary Information Text and Table S1 for detailed methodology.
Water samples to measure C-assimilation rates were taken from the clean underway flowthrough system (intake at 6 m). Triplicate C-assimilation experiments were initiated by adding 20 µmol⋅L−1 of NaH13CO3 to polycarbonate 1-L incubation bottles. The natural abundance of particulate organic C, used as t-zero values to calculate assimilation rates, was obtained by filtering 4 L of water onto precombusted GF/F filters for each station. Determination of total C and δ13C was carried out using a continuous-flow system consisting of a Sercon 20–22 mass spectrometer connected with an Automated C analyzer at the Stable Isotope Facility of the University of California, Davis. See SI Appendix, Supplementary Information Text for detailed protocols.
Richness and Statistical Analyses.
All analyses utilized subsampled OTU tables as described above. Pro- and eukaryotic richness were calculated as the number of 97% OTUs observed per sample (18). Absolute latitudinal ranges of OTUs were calculated as the difference between the maximum and minimum latitudes where an OTU was present (17). Correlations were calculated using the Spearman coefficient rs as a measure of rank correlation using the corrplot and vegan packages in R studio (70). Nonmetric multidimensional scaling (nMDS) analysis and ANOSIM were performed in Primer v.7 (71). BRT Predictor variable influence on pro- and eukaryotic richness was assessed using BRT (72); see SI Appendix for further information.
Supplementary Material
Acknowledgments
We thank Dr. Susan Wijffels and Dr. Bernadette Sloyan for the opportunity to piggyback on the P15 GO-SHIP transect voyage number IN2016_V03; Bernhard Tschitschko, Nicole Gail Hellessey, and Gabriela Paniagua Cabarrus for their sampling efforts at sea; the officers and crew of the R/V Investigator during cruise IN2016_V03 for their technical assistance while at sea; and the Marine Microbes Project consortium (https://data.bioplatforms.com) for contributions to the generation of data used in this publication. The Marine Microbes Project is supported by funding from Bioplatforms Australia and Integrated Marine Observing System through the Australian Government National Collaborative Research Infrastructure Strategy and the Education Investment Fund Super Science Initiative. This work was supported by CSIRO, the Australian Climate Change Science Program, and the Marine National Facility. A.M.W. was supported by grants from Alfred Wegener Institute and University of Western Australia. Work at CSIRO was supported by CSIRO Office of Community Engagement Science Leader Fellowship R-04202 (to L.B.) and by CSIRO Oceans and Atmosphere Environmental Genomics Grant R-02412.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The genomic data have been deposited to the National Center for Biotechnology Information database, https://www.ncbi.nlm.nih.gov/bioproject/385736 (accession no. PRJNA385736). All physical and chemical data are available at The Clivar & Carbon Hydrographic Data Office (CCHDO; https://cchdo.ucsd.edu/ under GO-SHIP program; voyage P15S; expocode: 096U20160426). Links to the genomics and physical data are also stored at PANGAEA Data Publisher, https://doi.pangaea.de/10.1594/PANGAEA.887803.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1719335115/-/DCSupplemental.
References
- 1.Burrows MT, et al. The pace of shifting climate in marine and terrestrial ecosystems. Science. 2011;334:652–655. doi: 10.1126/science.1210288. [DOI] [PubMed] [Google Scholar]
- 2.Pachauri RK, Meyer LA, editors. 2014. Climate change 2014: Synthesis report. Contribution of working groups I, II and III to the fifth assessment report of the Intergovernmental Panel on Climate Change (IPCC, Geneva)
- 3.Sunagawa S, et al. Tara Oceans coordinators Ocean plankton. Structure and function of the global ocean microbiome. Science. 2015;348:1261359. doi: 10.1126/science.1261359. [DOI] [PubMed] [Google Scholar]
- 4.Hutchins DA, Fu F. Microorganisms and ocean global change. Nat Microbiol. 2017;2:17058. doi: 10.1038/nmicrobiol.2017.58. [DOI] [PubMed] [Google Scholar]
- 5.Barnosky AD, et al. Has the Earth’s sixth mass extinction already arrived? Nature. 2011;471:51–57. doi: 10.1038/nature09678. [DOI] [PubMed] [Google Scholar]
- 6.Richardson AJ, Schoeman DS. Climate impact on plankton ecosystems in the Northeast Atlantic. Science. 2004;305:1609–1612. doi: 10.1126/science.1100958. [DOI] [PubMed] [Google Scholar]
- 7.Frederiksen M, Edwards M, Richardson AJ, Halliday NC, Wanless S. From plankton to top predators: Bottom-up control of a marine food web across four trophic levels. J Anim Ecol. 2006;75:1259–1268. doi: 10.1111/j.1365-2656.2006.01148.x. [DOI] [PubMed] [Google Scholar]
- 8.Johnson CR, et al. Climate change cascades: Shifts in oceanography, species’ ranges and subtidal marine community dynamics in eastern Tasmania. J Exp Mar Biol Ecol. 2011;400:17–32. [Google Scholar]
- 9.Brown MV, et al. Global biogeography of SAR11 marine bacteria. Mol Syst Biol. 2012;8:595. doi: 10.1038/msb.2012.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barton AD, Dutkiewicz S, Flierl G, Bragg J, Follows MJ. Patterns of diversity in marine phytoplankton. Science. 2010;327:1509–1511. doi: 10.1126/science.1184961. [DOI] [PubMed] [Google Scholar]
- 11.Brown MV, et al. Microbial community structure in the North Pacific Ocean. ISME J. 2009;3:1374–1386. doi: 10.1038/ismej.2009.86. [DOI] [PubMed] [Google Scholar]
- 12.Hillebrand H. On the generality of the latitudinal diversity gradient. Am Nat. 2004;163:192–211. doi: 10.1086/381004. [DOI] [PubMed] [Google Scholar]
- 13.Tittensor DP, et al. Global patterns and predictors of marine biodiversity across taxa. Nature. 2010;466:1098–1101. doi: 10.1038/nature09329. [DOI] [PubMed] [Google Scholar]
- 14.Finlay BJ. Global dispersal of free-living microbial eukaryote species. Science. 2002;296:1061–1063. doi: 10.1126/science.1070710. [DOI] [PubMed] [Google Scholar]
- 15.Pommier T, et al. Global patterns of diversity and community structure in marine bacterioplankton. Mol Ecol. 2007;16:867–880. doi: 10.1111/j.1365-294X.2006.03189.x. [DOI] [PubMed] [Google Scholar]
- 16.Ladau J, et al. Global marine bacterial diversity peaks at high latitudes in winter. ISME J. 2013;7:1669–1677. doi: 10.1038/ismej.2013.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amend AS, et al. Macroecological patterns of marine bacteria on a global scale. J Biogeogr. 2013;40:800–811. [Google Scholar]
- 18.Fuhrman JA, et al. A latitudinal diversity gradient in planktonic marine bacteria. Proc Natl Acad Sci USA. 2008;105:7774–7778. doi: 10.1073/pnas.0803070105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gilbert JA, et al. Defining seasonal marine microbial community dynamics. ISME J. 2012;6:298–308. doi: 10.1038/ismej.2011.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mittelbach GG, et al. Evolution and the latitudinal diversity gradient: Speciation, extinction and biogeography. Ecol Lett. 2007;10:315–331. doi: 10.1111/j.1461-0248.2007.01020.x. [DOI] [PubMed] [Google Scholar]
- 21.Rusch DB, et al. The Sorcerer II Global Ocean Sampling expedition: Northwest Atlantic through eastern tropical Pacific. PLoS Biol. 2007;5:e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mittelbach GG, et al. What is the observed relationship between species richness and productivity? Ecology. 2001;82:2381–2396. [Google Scholar]
- 23.Burrows MT, et al. Geographical limits to species-range shifts are suggested by climate velocity. Nature. 2014;507:492–495. doi: 10.1038/nature12976. [DOI] [PubMed] [Google Scholar]
- 24.Karageorgis AP, et al. Particle dynamics in the Eastern Mediterranean Sea: A synthesis based on light transmission, PMC, and POC archives (1991–2001) Deep Sea Res Part I Oceanogr Res Pap. 2008;55:177–202. [Google Scholar]
- 25.Longhurst AR. Ecological Geography of the Sea. 2nd Ed Elsevier; Amsterdam: 2010. [Google Scholar]
- 26.Marañón E, et al. Resource supply overrides temperature as a controlling factor of marine phytoplankton growth. PLoS One. 2014;9:e99312. doi: 10.1371/journal.pone.0099312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jardillier L, Zubkov MV, Pearman J, Scanlan DJ. Significant CO2 fixation by small prymnesiophytes in the subtropical and tropical northeast Atlantic Ocean. ISME J. 2010;4:1180–1192. doi: 10.1038/ismej.2010.36. [DOI] [PubMed] [Google Scholar]
- 28.Rodríguez J, et al. Mesoscale vertical motion and the size structure of phytoplankton in the ocean. Nature. 2001;410:360–363. doi: 10.1038/35066560. [DOI] [PubMed] [Google Scholar]
- 29.MacIntyre S, Alldredge AL, Gotschalk CC. Accumulation of marines now at density discontinuities in the water column. Limnol Oceanogr. 1995;40:449–468. [Google Scholar]
- 30.Woebken D, Fuchs BM, Kuypers MM, Amann R. Potential interactions of particle-associated anammox bacteria with bacterial and archaeal partners in the Namibian upwelling system. Appl Environ Microbiol. 2007;73:4648–4657. doi: 10.1128/AEM.02774-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geider RJ, La Roche J. The role of iron in phytoplankton photosynthesis, and the potential for iron-limitation of primary productivity in the sea. Photosynth Res. 1994;39:275–301. doi: 10.1007/BF00014588. [DOI] [PubMed] [Google Scholar]
- 32.Church MJ, Hutchins DA, Ducklow HW. Limitation of bacterial growth by dissolved organic matter and iron in the Southern Ocean. Appl Environ Microbiol. 2000;66:455–466. doi: 10.1128/aem.66.2.455-466.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baltar F, Arístegui J. Fronts at the surface ocean can shape distinct regions of microbial activity and community assemblages down to the bathypelagic zone: The Azores Front as a case study. Front Mar Sci. 2017;4:252. [Google Scholar]
- 34.Azam F, et al. The ecological role of water-column microbes in the sea. Mar Ecol Prog Ser. 1983;10:257–263. [Google Scholar]
- 35.Smith S, Mackenzie F. The ocean as a net heterotrophic system: Implications from the carbon biogeochemical cycle. Global Biogeochem Cycles. 1987;1:187–198. [Google Scholar]
- 36.Kahru M, Mitchell B, Gille S, Hewes C, Holm‐Hansen O. Eddies enhance biological production in the Weddell‐Scotia confluence of the Southern Ocean. Geophys Res Lett. 2007 doi: 10.1029/2007GL030430. [DOI] [Google Scholar]
- 37.Floodgate G, Fogg G, Jones D, Lochte K, Turley C. Microbiological and zooplankton activity at a front in Liverpool Bay. Nature. 1981;290:133–136. [Google Scholar]
- 38.Gregg MC, Sanford TB. Shear and turbulence in thermohaline staircases. Deep Sea Res A Oceanogr Res Pap. 1987;34:1689–1696. [Google Scholar]
- 39.Fernández-Castro B, et al. Importance of salt fingering for new nitrogen supply in the oligotrophic ocean. Nat Commun. 2015;6:8002. doi: 10.1038/ncomms9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oschlies A, Dietze H, Kähler P. Salt-finger driven enhancement of upper ocean nutrient supply. Geophys Res Lett. 2003 doi: 10.1029/2003GL018552. [DOI] [Google Scholar]
- 41.Raes EJ, Bodrossy L, van de Kamp J, Bissett A, Waite AM. Marine bacterial richness increases towards higher latitudes in the eastern Indian Ocean. Limnol Oceanogr Lett. 2018;3:10–19. [Google Scholar]
- 42.Ward BB. 2000. Nitrification and the marine nitrogen cycle. Microbial Ecology of the Oceans ed Kirchman DL (Wiley, New York), 427–454.
- 43.Rosenzweig ML. Species Diversity in Space and Time. Cambridge Univ Press; 1995. [Google Scholar]
- 44.Irigoien X, Huisman J, Harris RP. Global biodiversity patterns of marine phytoplankton and zooplankton. Nature. 2004;429:863–867. doi: 10.1038/nature02593. [DOI] [PubMed] [Google Scholar]
- 45.Sul WJ, Oliver TA, Ducklow HW, Amaral-Zettler LA, Sogin ML. Marine bacteria exhibit a bipolar distribution. Proc Natl Acad Sci USA. 2013;110:2342–2347. doi: 10.1073/pnas.1212424110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gaston KJ, Blackburn TM, Spicer JI. Rapoport’s rule: Time for an epitaph? Trends Ecol Evol. 1998;13:70–74. doi: 10.1016/s0169-5347(97)01236-6. [DOI] [PubMed] [Google Scholar]
- 47.Rohde K. Rapoport’s rule is a local phenomenon and cannot explain latitudinal gradients in species diversity. Biodiversity Lett. 1996;3:10–13. [Google Scholar]
- 48.Liu W, Lu J, Xie S-P, Fedorov A. Southern Ocean heat uptake, redistribution, and storage in a warming climate: The role of meridional overturning circulation. J Clim. 2018;31:4727–4743. [Google Scholar]
- 49.Matear R, Hirst A, McNeil B. Changes in dissolved oxygen in the Southern Ocean with climate change. Geochem Geophys Geosyst. 2000 doi: 10.1029/2000GC000086. [DOI] [Google Scholar]
- 50.Giebel HA, Brinkhoff T, Zwisler W, Selje N, Simon M. Distribution of Roseobacter RCA and SAR11 lineages and distinct bacterial communities from the subtropics to the Southern Ocean. Environ Microbiol. 2009;11:2164–2178. doi: 10.1111/j.1462-2920.2009.01942.x. [DOI] [PubMed] [Google Scholar]
- 51.Richardson AJ. In hot water: Zooplankton and climate change. ICES J Mar Sci. 2008;65:279–295. [Google Scholar]
- 52.Thomsen PF, Willerslev E. Environmental DNA–An emerging tool in conservation for monitoring past and present biodiversity. Biol Conserv. 2015;183:4–18. [Google Scholar]
- 53.Zinger L, et al. Global patterns of bacterial beta-diversity in seafloor and seawater ecosystems. PLoS One. 2011;6:e24570. doi: 10.1371/journal.pone.0024570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wilkins D, et al. Biogeographic partitioning of Southern Ocean microorganisms revealed by metagenomics. Environ Microbiol. 2013;15:1318–1333. doi: 10.1111/1462-2920.12035. [DOI] [PubMed] [Google Scholar]
- 55.Wolf C, Frickenhaus S, Kilias ES, Peeken I, Metfies K. Protist community composition in the Pacific sector of the Southern Ocean during austral summer 2010. Polar Biol. 2014;37:375–389. [Google Scholar]
- 56.Orsi AH, Whitworth T, III, Nowlin WD., Jr On the meridional extent and fronts of the Antarctic Circumpolar Current. Deep Sea Res Part I Oceanogr Res Pap. 1995;42:641–673. [Google Scholar]
- 57.Hansen HP, Koroleff F. Determination of nutrients. In: Grasshoff K, Kremling K, Ehrhardt M, editors. Methods of Seawater Analysis. 3rd Ed. Wiley-VCH; Weinheim, Germany: 2009. pp. 159–228. [Google Scholar]
- 58.Appleyard S, Abell G, Watson R. Seafood CRC Final Report (2011/729) April 2013. Australian Seafood Cooperative Research Centre; Deakin, Australia: 2013. Tackling microbial related issues in cultured shellfish via integrated molecular and water chemistry approaches; p. 89. [Google Scholar]
- 59.Lane D. Nucleic Acid Techniques in Bacterial Systematics. John Wiley and Sons; New York: 1991. 16S/23S rRNA sequencing. [Google Scholar]
- 60.Lane DJ, et al. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci USA. 1985;82:6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.DeLong EF. Archaea in coastal marine environments. Proc Natl Acad Sci USA. 1992;89:5685–5689. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stoeck T, et al. Multiple marker parallel tag environmental DNA sequencing reveals a highly complex eukaryotic community in marine anoxic water. Mol Ecol. 2010;19:21–31. doi: 10.1111/j.1365-294X.2009.04480.x. [DOI] [PubMed] [Google Scholar]
- 63.Bissett A, et al. Introducing BASE: The Biomes of Australian Soil Environments soil microbial diversity database. Gigascience. 2016;5:21. doi: 10.1186/s13742-016-0126-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Magoč T, Salzberg SL. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 66.Edgar RC. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 67.Parsons TR, Takahashi M, Hargrave B. Biological Oceanographic Processes. Elsevier; Oxford: 2013. [Google Scholar]
- 68.Hooker SB, et al. 2005. The second SeaWiFS HPLC analysis round-robin experiment (SeaHARRE-2) (NASA Goddard Space Flight Center, Greenbelt, MD), Tech. Memo 212785, p 124.
- 69.Hirata T, et al. Synoptic relationships between surface Chlorophyll-a and diagnostic pigments specific to phytoplankton functional types. Biogeosciences. 2011;8:311–327. [Google Scholar]
- 70.Team R 2015 RStudio: Integrated Development for R (RStudio, Inc., Boston), Version 1.0.136. Available at https://www.rstudio.com/
- 71.Clarke KR, Warwick R. Change in Marine Communities: An Approach to Statistical Analysis and Interpretation. PRIMER-E, Ltd., Plymouth Marine Laboratory; Plymouth, UK: 2001. [Google Scholar]
- 72.Elith J, Leathwick JR, Hastie T. A working guide to boosted regression trees. J Anim Ecol. 2008;77:802–813. doi: 10.1111/j.1365-2656.2008.01390.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.