Significance
Plant vacuoles are essential organelles and occupy up to 90% of the cell volume. Their roles include regulation of stomata movements, protein storage in seeds, gravity sensing, and ion homeostasis. Vacuole or lysosome fusion in eukaryotes is mediated by two multisubunit complexes, SNARE and homotypic fusion and vacuolar protein sorting (HOPS), but only the SNARE complex is well characterized in plants. Here, we show that, similar to other eukaryotes, HOPS mediates vacuole fusion in plants by interaction with SNAREs and that the HOPS subunit VPS33 and the SNARE protein SYP22 display the sites for interaction between these complexes. In contrast to other eukaryotes, however, plant HOPS recruitment to liposomes is inhibited by phosphoinositides, which appear to define strict rules for regulating fusion and fragmentation of dynamic vacuoles.
Keywords: Arabidopsis, vacuole fusion, HOPS, SNARE, phosphoinositides
Abstract
The vacuole is an essential organelle in plant cells, and its dynamic nature is important for plant growth and development. Homotypic membrane fusion is required for vacuole biogenesis, pollen germination, stomata opening, and gravity perception. Known components of the vacuole fusion machinery in eukaryotes include SNARE proteins, Rab GTPases, phosphoinositides, and the homotypic fusion and vacuolar protein sorting (HOPS) tethering complex. HOPS function is not well characterized in plants, but roles in embryogenesis and pollen tube elongation have been reported. Here, we show that Arabidopsis HOPS subunits VPS33 and VPS41 accumulate in late endosomes and that VPS41, but not VPS33, accumulates in the tonoplast via a wortmannin-sensitive process. VPS41 and VPS33 proteins bind to liposomes, but this binding is inhibited by phosphatidylinosiltol-3-phosphate [PtdIns(3)P] and PtdIns(3,5)P2, which implicates a nonconserved mechanism for HOPS recruitment in plants. Inducible knockdown of VPS41 resulted in dramatic vacuole fragmentation phenotypes and demonstrated a critical role for HOPS in vacuole fusion. Furthermore, we provide evidence for genetic interactions between VPS41 and VTI11 SNARE that regulate vacuole fusion, and the requirement of a functional SNARE complex for normal VPS41 and VPS33 localization. Finally, we provide evidence to support VPS33 and SYP22 at the initial stage for HOPS–SNARE interactions, which is similar to other eukaryotes. These results highlight both conserved and specific mechanisms for HOPS recruitment and function during vacuole fusion in plants.
The plant vacuole is an essential organelle with multiple roles including storage, pH homeostasis, and recycling (1). Vacuole fusion is required for stomata opening, pollen fertility, and embryo viability (2–4). Two types of vacuoles exist in plant cells. Embryos in developing seeds contain multiple protein storage vacuoles (PSVs), which have a neutral pH and accumulate storage proteins such as the 7S and 11S globulins. Cells in most mature tissues, instead, contain one large lytic vacuole, which is acidic, abundant in lytic proteases, and critical for ion homeostasis and cell turgor (5). Transition from PSVs to lytic vacuoles occurs during germination (6) and involves PSV fusion and maturation (7). Vacuoles in root meristematic cells are formed by interconnected tubular structures (8), and membrane fusion is necessary for the establishment of the central lytic vacuole (2). Transient and mobile structures such as transvacuolar strands, sheets, and bulbs contribute to the dynamic nature of plant vacuoles (9–12). In vegetative cells, however, very little vacuole fragmentation occurs and most cells contain a single large vacuole. Two exceptions are the changes in guard cell vacuoles during stomata movements (2, 13, 14) and in the leaf motor cells of the mimosa tree (Albizia julibrissin) (15). In both cases, vacuole remodeling occurs during dramatic changes in cellular turgor that drive organ movements. The changes in guard cell vacuoles that occur during stomata opening require homotypic membrane fusion mediated by the vacuolar soluble NSF attachment protein receptor (SNARE) complex containing VTI11 (2).
Vacuole fusion is not well characterized in plants, but in eukaryotic cells it requires SNAREs, RAB GTPases, membrane lipids, and a tethering complex. SNARE proteins mediate membrane fusion by assembly of a four-helix bundle, or trans-SNARE complex, from one SNARE located in the incoming vesicle and three SNAREs located in the target membrane. The SNARE complex brings the two membranes together via a zippering mechanism that is thought to result in membrane fusion (16). A SNARE complex for plant prevacuolar compartments (PVCs) and the vacuole is composed of SYP22/VAM3 (Qa-SNARE), VTI11 (Qb-SNARE, called Vti1p in yeast), SYP51 (Qc-SNARE), and VAMP727 (R-SNARE) (17, 18). The Qabc complex can also associate with the R-SNAREs VAMP711 or VAMP713 (19, 20). A role of these SNAREs in vacuole fusion is evidenced by altered vacuole phenotypes of vti11, syp22, and syp22 vamp727 double mutants (2, 18, 21).
The homotypic fusion and vacuole protein sorting (HOPS) tethering complex was recently shown to mediate homotypic vacuole fusion in plants (3). HOPS is important for the fusion of vacuoles in yeast (16, 22), and fusion of lysosomes in Caenorhabditis elegans (23) and humans (24). The yeast HOPS complex is the best characterized and is formed by six subunits. Vps39p and Vps41p are HOPS specific, while Vps11p, Vps16p, Vps18p, and Vps33p form the “CORE” and are shared with CORVET, a tethering complex that functions at the early endosome (25, 26). HOPS recruitment to yeast vacuole membranes requires phosphatidylinositol 3-phosphate [PtdIns(3)P] and PtdIns(4,5)P2, two important regulatory lipids in vacuole fusion (27). Vps33 belongs to the family of Sec1/Munc18 (SM) proteins, which are implicated in assembly and regulation of SNARE complexes. SM proteins have a three-domain architecture, in which domains 1 and 3 form a cleft that accommodates the SNAREs (28). The X-ray structure of a fungal Vps33 bound to a single SNARE, together with in vitro fusion assays, suggests that VPS33 catalyzes SNARE complex assembly through a specific SNARE motif recognition (29). All other HOPS subunits are predicted to form a β-propeller at the N terminus and an α-solenoid at the C terminus (30). The complete HOPS complex has been detected in Arabidopsis by mass spectrometry (3), and all subunits are encoded in the Arabidopsis genome as single genes (31). VACUOLELESS1 (VCL1), the homolog of yeast Vps16, VPS11, VPS39, and VPS41 are required for vacuole biogenesis and pollen development (3, 4, 32–35). VCL1 interacts with VPS33 and VPS11 and localizes to PVCs and the tonoplast (35). VPS18 and VPS39 were also localized to a subdomain of the tonoplast and to cytosolic organelles (3). The molecular mechanisms for HOPS function and regulation during vacuole fusion in plants are not well characterized.
The RAB5 and RAB7 GTPase families have been implicated in regulation of endosomes maturation and membrane fusion at the vacuole (36). RAB5 and RAB7 localize to partially overlapping endosome populations, with only RAB7 detected at the tonoplast (19, 37–41). The current model is that the SAND-CCZ complex is a RAB5 effector, exerts GEF function on RAB7, and is necessary for the conversion of RAB5 to RAB7 in PVCs (42–44). Active RAB7 is required for vacuolar localization of HOPS subunits VPS18 and VPS39 (3).
Plant homotypic vacuole fusion is also regulated by phosphoinositides. Pollen grains defective in phosphatidylinositol 4-phophate 5-kinase 1 (PI5K1) and PI5K2 have larger vacuoles than the WT, which implicates PtdIns(4,5)P2 in vacuole biogenesis (45). Depletion of PtdIns(3)P after wortmannin (Wm) treatment induces rapid vacuole membrane fusion in root and hypocotyl cells of vti11/zig1 mutants and in guard cells of WT (2). Wm treatment also induces the swelling of plant PVCs, which results in doughnut-shaped structures known as Wm bodies (46). These effects of Wm are due to its inhibitory action against phosphatidylinositol 3-kinase (47) and implicate PtdIns(3)P and other phosphoinositides in vacuole fusion in plants. Interestingly, these effects of Wm have not been observed in yeast or animal cells (48, 49), suggesting the possibility of specific mechanisms for plant vacuole fusion.
Here, we characterized VPS41 and VPS33 using reverse genetics, fluorescently tagged proteins, and biochemical analyses. We show that VPS41 and VPS33 are important for pollen fertility and PSV maturation in embryos, and that VPS41 is required for homotypic vacuole fusion in developing seedlings. We demonstrate that both proteins localize to late endosomes, that only VPS41 is detected in the tonoplast, and that they interact with each other and with the vacuolar SNARE SYP22. The VPS41-GFP localization at the tonoplast, but not that at late endosomes, is Wm sensitive. In vitro lipid–protein binding assays show that while VPS33 and VPS41 bind to phosphoinositides on lipid strips, neither protein binds to PtdIns(3)P or PtdIns(3,5)P2-containing liposomes, even though they do bind to control liposomes. We speculate that specific affinities for membrane lipids together with membrane curvature are key regulatory mechanisms behind vacuole fusion events mediated by HOPS and SNAREs.
Results
VPS41 and VPS33 Partially Colocalize with Late Endosomes, but only VPS41 Is Detected at the Tonoplast.
VPS41 (AT1G08190) and VPS33 (AT3G54860) share 23.29% and 19.17% sequence identity with yeast Vps41p and Vps33p, respectively. Therefore, VPS33 and VPS41 are likely homologs of these yeast HOPS proteins (31, 50). We first investigated whether VPS41 and VPS33 colocalized in vacuolar or prevacuolar membranes, as expected for HOPS. Transient expression of mRFP-VPS41 and VPS33-GFP under control of UBQ10 promoters in Nicotiana benthamiana leaves indicated that these proteins accumulate in the cytosol and nucleus, and associate with membranes in punctate structures (SI Appendix, Fig. S1 A and B). RFP-VPS41 and VPS33-GFP colocalized in the majority of the punctate structures and the nucleus, and partially at the cell periphery (SI Appendix, Fig. S1C), which supports their participation in a complex. These experiments, however, do not provide enough resolution to identify the compartments where these proteins accumulate and they may be influenced by overexpression. To better characterize protein localizations, VPS41-GFP (4) and VPS33-GFP were analyzed in stable Arabidopsis transgenic plants using full-length genomic fusions to GFP under native promoters. Both proteins were again detected in the cytosol, but membrane association was more discernible. Similar to its localization in pollen (4), VPS41-GFP was detected at the tonoplast and punctate organelles in roots (Fig. 1A), and this localization was not affected by the location of the fluorescence tag as the localization of YFP-VPS41 was very similar (SI Appendix, Fig. S1D). Colocalization with known organelle markers was used to identify the structures labeled with VPS41-GFP and VPS33-GFP. VPS41-GFP colocalized with the tonoplast marker RFP-SYP22 (18) and confirmed its tonoplast association (Fig. 1 A–C and SI Appendix, Fig. S1E). Furthermore, VPS41-GFP partially colocalized with mCherry-RAB7, a marker for the late stages of PVC maturation (51) (Fig. 1 D–F), consistent with results from pollen (4). Colocalization with the early PVC marker mCherry-RAB5 (51) was detected in only a few organelles (Fig. 1 G–I). These results indicate that VPS41-GFP localizes to the vacuole and late endosomes. Similar experiments with VPS33-GFP indicate that VPS41 and VPS33 have overlapping but not identical localizations. VPS33-GFP rarely colocalized with RFP-SYP22 at the tonoplast, except for a few puncta (Fig. 1 J–L and SI Appendix, Fig. S1F), indicating that this protein is absent from the vacuole. VPS33-GFP is present in the cytosol and in cytosolic punctate structures, where it partially colocalized with mCherry-RAB7 (Fig. 1 M–O) and shows strong colocalization with mCherry-RAB5 (Fig. 1 P–R). Quantification of these colocalization experiments further validates the overlapping but distinct localization of VPS33 and VPS41 (Fig. 1 S and T). Both proteins show significant colocalization with RAB7 endosomes. While VPS33 has the strongest colocalization with RAB5 endosomes, it is weakly colocalized with the tonoplast marker SYP22. In contrast, VPS41 shows significant accumulation at the tonoplast but is not associated with RAB5 endosomes (Fig. 1 S and T). Interestingly, both VPS41-GFP and VPS33-GFP seem to accumulate in more endosomal compartments when coexpressed with mCherry-RAB5 (compare Fig. 1A with Fig. 1G and Fig. 1J with Fig. 1P). The effect of the mCherry-RAB5 fusion on the recruitment or stability of these proteins at the endosome or the endosomes themselves will require further analysis beyond the scope of this work. Overall, these results suggest that VPS33 and VPS41 are cytosolic proteins that preferentially associate with late endosomal compartments, and this is consistent with a role for HOPS in the tethering of vacuoles or prevacuoles. Interestingly, only VPS41 is detected at the tonoplast, and only VPS33 has significant accumulation in RAB5 endosomes.
Fig. 1.
Subcellular localization of VPS41-GFP and VPS33-GFP. Genomic fusion constructs of VPS41 (VPS41-GFP) and VPS33 (VPS33-GFP) driven by native promoters were cotransformed with subcellular markers into Arabidopsis, and root cells were imaged by confocal microscopy. (A–I) Colocalization of VPS41-GFP (A, D, and G) with mRFP-SYP22 (B), mCherry-RAB7 (E), or mCherry-RAB5 (H) in root cells of stably transformed Arabidopsis seedlings. Corresponding merged images are shown (C, F, and I). Arrowheads indicate sites of marker colocalization at the tonoplast, and arrows point to colocalization in endosomes. (Scale bars: 20 µm.) (J–R) Colocalization of VPS33-GFP (J, M, and P) with mRFP-SYP22 (K), mCherry-RAB7 (N), or mCherry-RAB5 (Q). The merged images are shown (L, O, and R). No apparent accumulation of VPS33-GFP was detected at the tonoplast. Partial colocalization was detected in RAB5 and RAB7 endosomes (arrows). (Scale bars: 20 µm.) (S and T) Pearson and Spearman correlation coefficients from the colocalization analyses between VPS41-GFP (S) or VPS33-GFP (T) with mRFP-SYP22, mCherry-RAB5, or mChery-RAB7. Thirty-seven to 56 images from nine seedlings combined were used for each analysis. Bars represent SE.
VPS41 Tonoplast Association Is Dependent on Phosphoinositides.
The distinct localizations of VPS33-GFP and VPS41-GFP prompted us to test whether the two proteins were characterized by different phospholipid binding affinities. This was tested using an in vitro lipid-binding assay with recombinant proteins. GST-VPS41 and GST-VPS33 fusion proteins were expressed in Escherichia coli and purified by affinity chromatography. Purified proteins were incubated on lipid strips, and protein binding was detected with anti-GST. GST alone was used as negative control (SI Appendix, Fig. S2A). Both GST-VPS41 and GST-VPS33 show affinity toward all phosphoinositides in this assay, including the two that are most abundant at the late endosomes and vacuole, PtdIns(3)P and PtdIns(3,5)P2 (Fig. 2A). These results are consistent with the hypothesis that VPS41 and VPS33 association with membranes may be regulated in part by phosphoinositide binding.
Fig. 2.
Phosphoinositides regulate the membrane association of VPS41. (A) GST-VPS41 and GST-VPS33 bind to phosphoinositide lipids. GST-VPS41 (Center) and GST-VPS33 (Right) were expressed in E. coli and purified by affinity chromatography. Lipid affinity of purified proteins was analyzed using a lipid strip assay. Lipids include the following: LPA, lysophosphatidic acid; LPC, lysophosphocholine; PA, phosphatidic acid; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PS, phosphatidylserine; S1P, sphingosine-1-phosphate. (B) GST-VPS41 and GST-VPS33 bind to liposomes that lack phosphoinositides. Affinity-purified GST-VPS41 and GST-VPS33 proteins were incubated without liposomes (none) or with PC/PE liposomes bearing no phosphoinositides (control), PtdIns(3)P, or PtdIns(3,5)P2, and protein binding was detected by cosedimentation and immunoblot with α-GST. Incubation with PI(3)P-GRIP was used as positive control, and GST alone was used as negative control. (C–H) Wm treatment induces dissociation of VPS41-GFP from the tonoplast, but not from endosomes. Arabidopsis seedlings expressing VPS41-GFP (C and F) and mCherry-RAB7 (D and G) were treated with DMSO (C–E) or 33 µM Wm (F–H) for 3 h and observed by confocal microscopy. Merged images are shown (E and H). Tonoplast localization of VPS41-GFP is indicated with arrowheads. Wm induced the enlargement of RAB7-labeled endosomes (arrows) that retained VPS41-GFP signal. (Scale bar: 20 µm.) (I–N) Wm treatment induces dissociation of VPS41-GFP from the tonoplast. Arabidopsis seedlings expressing VPS41-GFP (I and L) and mRFP-SYP22 (J and M) were treated with DMSO (I–K) or 33 µM Wm (L–N) for 3 h and observed by confocal microscopy. VPS41-GFP signal is depicted using a rainbow LUT (I and L), where high intensity values are represented in red and low intensity values are shown in blue. Red pixels were associated with the tonoplast in the DMSO control only. Merged images are shown using green color for the GFP signal (K and N). (Scale bar: 20 µm.) (O) Quantification of the tonoplast localization of VPS41-GFP from I–N. The ratio between the membrane and cytosolic fluorescence intensity of VPS41-GFP was calculated using ImageJ from DMSO or Wm-treated seedlings (N = 27 images from seven to eight seedlings per treatment; *significant in a t test at P < 0.05).
To characterize lipid binding in a physiological context, we used a liposome-binding assay that better resembles lipid binding in a bilayer. Phosphatidylcholine (PC)/phosphatidylethanolamine (PE) liposomes bearing either no phosphoinositide, PtdIns(3)P, or PtdIns(3,5)P2 were incubated in the presence of recombinant proteins, and binding was assayed by cosedimentation. As shown in Fig. 2B, both GST-VPS41 and GST-VPS33 bound to the control liposomes, indicating that these proteins have strong affinity for neutral lipids. Surprisingly, addition of PtdIns(3)P or PtdIns(3,5)P2 abolished this binding, which suggests that VPS33 and VPS41 membrane binding is tightly controlled. The PI(3)P-GRIP protein which contains a PtdIns(3)P-binding domain was used as a positive control, and GST was used as a negative control. These results underscore VPS41 and VPS33 sensitivities to phosphoinositides and to the different presentation of these lipids between the flat surface of the lipid strip and the curved membrane of the liposomes.
PtdIns(3)P is a negative regulator for homotypic vacuole fusion in plants (2) and recruits HOPS to the vortex ring in yeast (52). Hence, PtdIns(3)P and other phosphoinositides are good candidates to regulate HOPS subunit association with the vacuole and PVC. We therefore asked whether the localizations of GFP-VPS41 was affected by phosphoinositide availability using Wm treatment, which results in PtdIns(3)P depletion from cellular membranes (53). GFP-VPS41 was found at the tonoplast and some mCherry-RAB7-labeled endosomes, in the DMSO control as expected (Fig. 2 C–E). Wm treatment resulted in PVC swelling with the characteristic doughnut-like structures that were decorated with both VPS41-GFP and mCherry-RAB7 (Fig. 2 F–H), which suggests that Wm treatment did not alter the VPS41 association with late endosome membranes. Remarkably, VPS41-GFP was no longer detected at the tonoplast after Wm treatment (Fig. 2 F–H). To quantify the loss of VPS41-GFP from the tonoplast, we used the RFP-SYP22 marker as a proxy for vacuole membrane localization. As shown in Fig. 2 I–K, the sites of highest VPS41-GFP fluorescence intensity in the control colocalize with RFP-SYP22 at the tonoplast. This tonoplast bright signal is lost after the Wm treatment (Fig. 2 L–N). Quantification of these results further indicates that Wm treatment results in a 35% decrease in the membrane/cytosolic ratio of VPS41-GFP (Fig. 2O). These results indicate that tonoplast association of VPS41 is mediated by phosphoinositide binding, but its association with late endosome membranes is independent of phosphoinositides. Major changes in VPS33-GFP localization were not detected after Wm treatment as a few RAB7 endosomes were still labeled with VPS33-GFP (SI Appendix, Fig. S2 B–G).
HOPS Is Essential for Male Fertility and Important for Plant Development.
To define the role of VPS41 and VPS33 in vacuole fusion and dynamics, we identified loss-of-function alleles for VPS41 and VPS33 from the SALK T-DNA collection (54). T-DNA insertions were confirmed for vps41-1 (SALK_076372) and vps41-3 (SALK_036406), which carry the T-DNA insertions in the 13th exon and the promoter region of VPS41, respectively. Three T-DNA insertion alleles were confirmed for VPS33, vps33-3 (SALK_016684C), vps33-4 (SALK_151719), and vps33-5 (SALK_143407). These mutants carry the insertions in the 13th intron, the promoter, and the seventh exon of VPS33 (SI Appendix, Fig. S3A). RT-PCR from homozygous lines showed no altered VPS41 transcript accumulation for vps41-3, and this line was not used further. No homozygous plants were recovered from over 50 progeny of vps41-1/+, vps33-3/+, vps33-4/+, or vps33-5/+, and none had evidence for embryo abortion in siliques from heterozygous plants, which suggested the possibility of gametophyte lethality or sterility. To determine which gametophyte was unfertile, reciprocal crosses were performed between vps41-1/+ or vps33-3/+ and Col-0 WT plants to determine the male and female transmission efficiency (TE) (55) (SI Appendix, Table S1). The results show that transmission through the female gametes is not compromised in either mutant. However, male vps41-1 TE is abolished, indicating that VPS41 is essential for pollen fertility. The vps33-3 pollen, instead, is not completely sterile, but the male TE is strongly reduced to 16.1%. The pollen defects of vps33 and vps41 loss-of-function alleles are consistent with previous reports (4, 32) and, together with the phenotype of vps18 and vps39 mutants recently reported (3), support an important role of the HOPS complex in male gametophyte development. Similar to VPS41-GFP (4), the full-length genomic fusion construct for VPS33-GFP complements the pollen fertility defects of loss-of-function mutants as homozygous vps33-3 can be recovered (SI Appendix, Fig. S3 B–D).
HOPS Is Important for Vacuole Biogenesis and Fusion.
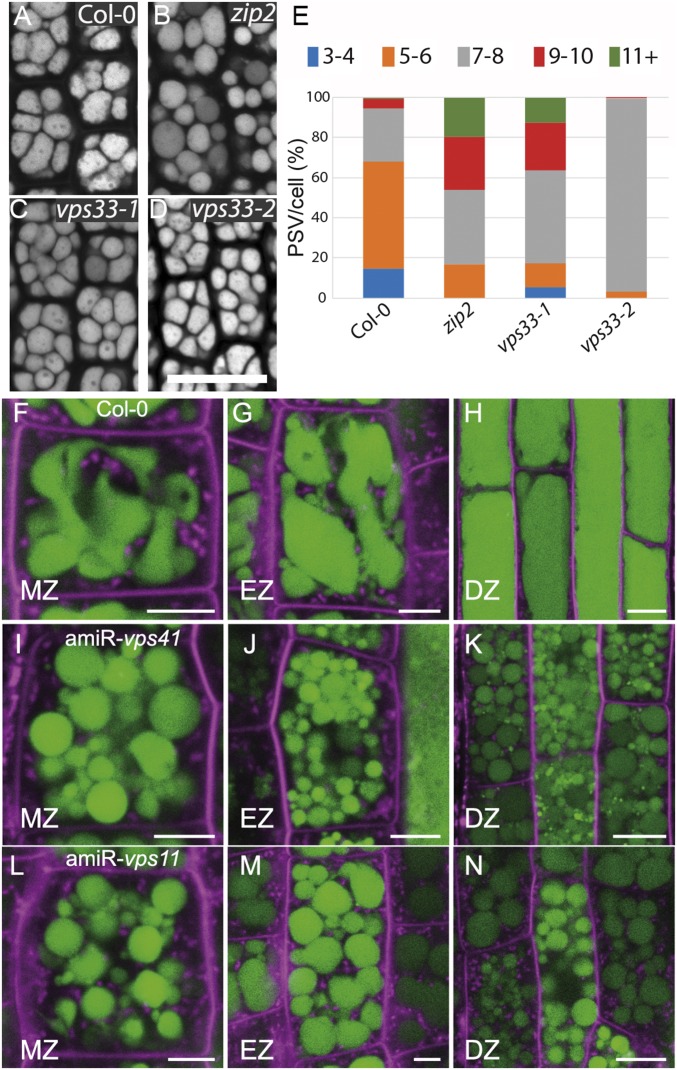
We identified viable weaker alleles to characterize VPS41 and VPS33 (SI Appendix, Fig. S3A). One EMS allele for VPS41, zip2/vps41 (56), and two viable TILLING (57) alleles of VPS33, vps33-1 and vps33-2, are available. The zip2/vps41 allele was identified from a screen of suppressors of zig-1/vti11 in gravitropism (56). The mutation results in a G721R substitution within an annotated clathrin heavy chain repeat (CHCR) of the VPS41 protein that is predicted to mediate protein–protein interactions (58). The mutations in vps33-1 and vps33-2 result in S95F and V136M substitutions in the VPS33 protein, respectively. Both are predicted to occur in domain 1 of the VPS33 protein, which, together with domain 3, has been shown in fungi and yeast to form a cleft that is thought to accommodate SNARE bundles (28). None of the single mutants shows developmental phenotypes in rosettes or inflorescences under normal growth conditions (SI Appendix, Fig. S3E). No vacuole phenotypes were observed in zip2/vps41, vps33-1, or vps33-2 when seedling roots were analyzed using LysoTracker Red staining (SI Appendix, Fig. S3F). However, we observed an increased number of PSVs in mature zip2/vps41, vps33-1, and vps33-2 embryos compared with the WT (Fig. 3 A–D). The number of vacuoles per cell was counted in each genotype to quantify this defect (Fig. 3E). Only cells with a cross-section area between 11 and 15.9 µm2 were included in the analysis. Fifty-three percent of the cells from the WT control have five to six vacuoles per cell, and a smaller fraction (26%) have seven to eight vacuoles. In contrast, only 16.8% of the cells from the zip2/vps41 embryos have five to six vacuoles while the rest (88%) have seven or more. Similar results were detected for vps33-1 where 17% of the cells contain six or fewer vacuoles, while the rest (83%) had seven or more. Finally, almost all of the vps33-2 mutant embryos showed seven to eight vacuoles per cell. These results support the hypothesis that VPS41 and VPS33 are involved in vacuole homotypic fusion and are important for PSV maturation.
Fig. 3.
HOPS is required for protein storage and lytic vacuole morphology. (A–D) Morphology of protein storage vacuoles (PSVs) in mature embryos from the WT control (Col-0) (A), zip2/vps41 (B), vps33-1 (C), and vps33-2 (D). Autofluorescence of the PSVs was acquired by CLSM. (Scale bar: 20 µm.) (E) Vacuole number distribution in mature embryos. Embryos from WT control (Col-0), vps33-1, vps33-2, and zip2/vps41 were imaged as in A–D, and the number of vacuoles per cell was counted. Only cells with a cross-section area between 11 and 15.9 µm2 were included in the analysis. Bars represent the percentage of cells with less than 4 (blue), 5–6 (orange), 7–8 (gray), 9–10 (red), and 11 or more (green) vacuoles per cell in each genotype. Col-0, n = 165 cells; zip2, n = 146 cells; vps33-1, n = 154 cells; vps33-2, n = 118 cells. (F–N) Vacuole morphology of amiRNA lines after DEX induction. Seedlings from Col-0 WT (control, F–H), amiR-vps41 (I–K), and amiR-vps11 (L–N) were incubated for 72 h with DEX and stained with BCECF and FM4-64. Roots were imaged at the meristematic zone (MZ), elongation zone (EZ), and differentiation zone (DZ). (Scale bar: 20 µm.)
A dexamethasone (DEX)-inducible amiRNA system was used to determine the function of HOPS in developing seedlings. The HOPS-CORE subunit VPS11 and the HOPS-specific subunit VPS41 were targeted by the inducible amiRNA system. WT, amiR-vps41, and amiR-vps11 seedlings were treated with DMSO (control) or DEX for 72 h, and vacuole morphology was observed by 2′,7′-bis-(2-carboxyethyl)-5-(and-6)-carboxyfluorescein (BCECF) staining. Normal vacuole morphology was observed in both amiR-vps11 and amiR-vps41 in the absence of DEX compared with the WT (SI Appendix, Fig. S3G) and in the WT after DEX treatment (Fig. 3 F–H). In contrast, dramatic changes in vacuole morphology were observed after DEX induction of the two lines. Both amiR-vps41 (Fig. 3 I–K) and amiR-vps11 (Fig. 3 L–N) seedlings showed highly fragmented vacuoles in meristematic zone (MZ), elongation zone (EZ), and differentiation zone (DZ) of the root after DEX induction. We note that the phenotype was observed only in cells that were not elongated at the time of induction. This phenotype together with the PSV phenotype of allelic mutants demonstrates a critical role of HOPS for vacuole fusion and biogenesis.
VPS41 and VPS33 Interact in Vivo with Each Other and with SYP22 in Specific Foci.
If HOPS is involved in membrane fusion at the vacuole, it is likely to interact with the vacuolar SNARE complex (17, 18), including SYP22 and VTI11. To determine whether VPS41 and VPS33 interact with each other and with the vacuolar SNAREs, split YFP fusion proteins were constructed for bimolecular fluorescence complementation (BiFC) experiments in Nicotiana leaf epidermis (59, 60). Only N-terminal n/cYFP fusions of the SNARE proteins were used because the C termini of vacuolar SNAREs reside in the vacuole lumen. For VPS33 and VPS41, both N- and C-terminal constructs were generated, but none of the C-terminal n/cYFP fusions restored YFP signal in any combination. Coexpression of cYFP-VPS41 with nYFP-VPS33 resulted in a punctate pattern at the vacuole periphery (Fig. 4A), indicating that VPS41 and VPS33 are likely to participate in the HOPS complex. Coexpression of the same cYFP-VPS41 construct with the SNARE fusion nYFP-SYP22 resulted in a punctate pattern as well, indicating that these two proteins interact in vivo (Fig. 4B). In contrast, no YFP fluorescence was detected when cYFP-VPS41 was coexpressed with nYFP-VTI11 (Fig. 4C). Note that broad fluorescent structures (asterisks) in Fig. 4 B and C are likely background signal because they were observed in negative controls (SI Appendix, Fig. S4 A and B). Next, we tested interactions with VPS33. Again, coexpression of cYFP-VPS33 with nYFP-VPS41 resulted in a punctate pattern indicative of protein–protein interaction (Fig. 4D). When cYFP-VPS33 was coexpressed with nYFP-SYP22, a strong YFP signal was detected in punctate foci (Fig. 4E), supporting the hypothesis that HOPS and SNAREs interact to mediate membrane fusion. Similar to the VPS41 interactions, no YFP fluorescence was detected when cYFP-VPS33 and nYFP-VTI11 were coexpressed, suggesting the absence of close interactions between these two proteins in vivo (Fig. 4F). To further validate these results, the c/nYFP-VPS33 was coexpressed with a n/cYFP- fusion to the aquaporin TIP2;1 as a negative control. No YFP signal was detected in either combination (SI Appendix, Fig. S4 D and E), which confirms the lack of interaction for the negative controls. The two vacuolar SNAREs VTI11 and SYP22 were used as positive controls as they have been shown to interact (21) and localize at the PVC (18, 61). Indeed, coexpression of cYFP-VTI11 with nYFP-SYP22 as well as cYFP-SYP22 with nYFP-VTI11 resulted in similar punctate structures at the periphery of the vacuole (SI Appendix, Fig. S4 C and F). Overall, these data show that VPS33 and VPS41 can assemble in subcellular foci and that both proteins can interact in vivo with SYP22, but not with VTI11. Taken together, these data support the hypothesis for HOPS–SNARE interactions mediating membrane fusion at the vacuole.
Fig. 4.
VPS41 and VPS33 interact and can bind the SNARE protein SYP22. (A–F) VPS41 and VPS33 interact in vivo and bind the SNARE protein SYP22. BiFC analysis of protein interactions in infiltrated N. benthamiana leaf cells by confocal microscopy. The N-terminal half (nY) or the C-terminal half (cY) of YFP were fused with VPS41, VPS33, SYP22, or VTI11. (A–C) cYFP-VPS41 interacts with nYFP-VPS33 (A), nYFP-SYP22 (B), but not with nYFP-VTI11 (C). (D–F) cYFP-VPS33 interacts with nYFP-VPS41 (D), nYFP-SYP22 (E), but not with nYFP-VTI11 (F). Asterisks indicate background signal that was observed in the negative control. (G) VPS33 binds the SYP22 SNARE in vitro. Recombinant GST-VPS41, GST-VPS33, HIS-SYP22, and HIS-VTI11 protein fusions were expressed in E. coli and purified by affinity chromatography. The purified proteins were mixed as indicated, and immunoprecipitation was carried out with an α-HIS antibody. Proteins from 5% of the input, immunoprecipitate (IP), or 5% of the flowthrough were separated by SDS/PAGE, and GST-VPS41 or GST-VPS33 were detected by Western blot with an α-GST antibody. Arrows indicate the expected sizes of GST-VPS41 and GST-VPS33. The positions of 150-kDa (Top) and 100-kDa (Bottom) molecular markers are indicated with arrowheads. (H–K) Genetic interaction between hypomorphic HOPS mutant alleles and zig-1. Seedlings were stained for 2 h in LysoTracker Red (Invitrogen) and acquired in LSCM. Abnormal vacuole morphology in the maturation zone was observed in zig-1 (H), vps33-1 zig-1 (J), and vps33-2 zig-1 mutants (K). The vacuole phenotype was complemented in the root differentiation zone (DZ) of the double-mutant zig-1 zip2 (I). (L–N) Loss of the VTI11 SNARE induces abnormal VPS41-GFP localization. VPS41-GFP was imaged in the Col-0 WT (L) or the zig-1 mutant background (M and N). The protein was found in bright aggregates at the junction of zig-1 vacuoles or in clusters in the cytosol as shown with the Airyscan mode of the confocal (N). (O–Q). Loss of the VTI11 SNARE induces abnormal VPS33-GFP localization. VPS33-GFP was imaged in the Col-0 WT (O) or zig-1 (P and Q). Details of the aggregates at the junction between unfused vacuoles can be observed in the Airyscan mode (Q). (Scale bars: 10 µm.)
VSP33 Interacts Directly with the Qa SNARE SYP22.
VPS33 mediates SNARE complex assembly in yeast (29), and we could detect interactions between VPS33, VPS41, and SYP22 by BiFC. Therefore, we tested whether either of the two HOPS subunits could support a direct interaction with SYP22 by in vitro coimmunopreciptation. Proteins were expressed as tagged fusions to either GST (VPS33 and VPS41) or HIS (SYP22 and VTI11), and immunoprecipitation was carried out using an α-HIS antibody. GST-VPS41 was not detected in the α-HIS immunoprecipitate when incubated with either HIS-SYP22 or HIS-VTI11 (Fig. 4G), indicating a lack of direct interaction. In contrast, a direct interaction between GST-VPS33 and HIS-SYP22 was detected using this assay, as GST-VPS33 was detected in the immunoprecipitate (Fig. 4G). This interaction was confirmed in a complementary experiment where α-GST pulled down both HIS-SYP22 and GST-VPS33 in the coimmunoprecipitate (SI Appendix, Fig. S4G). Whether or not there were direct interactions with HIS-VTI11 was less definitive. Incubation of GST-VPS33 and HIS-VTI11 with α-HIS produced detectable signals of GST-VPS33 on immunoblots (Fig. 4G), but no signal was detected for HIS-VTI11 when α-GST was used (SI Appendix, Fig. S4G). Overall, these data provide strong evidence that HOPS–SNARE interactions in plants are mediated by direct interaction of VPS33 and SYP22, most likely due to conserved mechanisms.
The HOPS Subunit VPS41 Genetically Interacts with the VTI11 SNARE.
The evidence for protein–protein interactions between HOPS subunits and the vacuolar SNARE SYP22 prompted us to test for genetic interactions between the two complexes. zip2, a mutant allele of VPS41 (SI Appendix, Fig. S3A), was identified as a suppressor of the agravitropic phenotype of zig-1, a loss-of-function allele of VTI11 (56). To determine whether zip2 suppresses the vacuole phenotype of zig-1, vacuoles from the zig-1 zip2 double mutants were visualized with LysoTracker Red (Fig. 4 H–K). The zig-1 mutant displayed the previously described vacuole phenotype in both the root tip and root maturation zone (2) (Fig. 4H and SI Appendix, Fig. S4H). The zig-1 zip2 still exhibited multiple vacuoles near the root tip (SI Appendix, Fig. S4I), but this phenotype was not observed in the root maturation zone (Fig. 4I). These results indicate that zip2 partially suppresses the zig-1 phenotype at the subcellular level and confirm a genetic interaction between ZIP2/VPS41 and VTI11. Furthermore, this result suggests that the zip2 mutation results in changes in the VPS41 protein structure that may alter the interactions between HOPS and SNAREs. A similar approach was used to test for genetic interactions between VPS33 and VTI11, but unlike zig-1 zip2 doubles, vps33-1 zig-1 and vps33-2 zig-1 double mutants showed the abnormal zig-1 vacuole phenotype in all root cells (Fig. 4 J and K and SI Appendix, Fig. S4 J and K) and the zig-zagged phenotype of the inflorescence. These results indicate that the vps33-1 and vps33-2 mutations failed to complement the vacuole phenotype, perhaps due to their hypomorphic nature, which may have innocuous effects on the VPS33 protein.
The proposed interaction between HOPS and SNAREs led us to predict that loss of the VTI11 SNARE from the vacuole could alter the localization of VPS41 or VPS33. We tested this possibility in zig-1 mutants. VPS41-GFP localization in the WT background showed the cytosolic, punctate, and tonoplast localization previously described (Fig. 4L). In contrast, VPS41-GFP was not detected at the tonoplast and only accumulated in bright aggregated patterns in the zig-1 background (Fig. 4M). When visualized using the Airyscan mode of the microscope, these bright patterns were associated with adjacent vacuoles or clusters of small organelles in the cytosol (Fig. 4N). A similar result was obtained with VPS33-GFP. In contrast to the dispersed puncta of VPS33-GFP in the WT (Fig. 4O), zig-1 mutants showed clustered structures labeled with VPS33-GFP (Fig. 4 P and Q). At least some of these clusters represent small vacuoles as similar clusters were detected in zig-1 mutants labeled with RFP-SYP22 (SI Appendix, Fig. S4 L–N). These results indicate that a functional SNARE complex is required for the normal localization of VPS41 and VPS33.
Discussion
The plant HOPS tethering complex is evolutionarily conserved with other eukaryotes (3, 31, 35), is involved in vacuole traffic via a RAB7-mediated pathway, and is critical for homotypic vacuole fusion (3). HOPS shares four subunits with CORVET, a similar conserved complex that mediates tethering of RAB5 endosomes and vacuoles in a plant-specific manner (3). Our results show that stable VPS33–VPS41 interactions predominantly occur in punctate organelles likely to correspond to endosomes as expected for a tether between endosomes and vacuoles. The possibility remains, however, that the functional HOPS complex also assembles transiently at sites of vacuole homotypic fusions, which are not abundant in the cells used in these experiments.
HOPS is essential for vacuole membrane fusion in plants. VPS33 and VPS41 are essential in pollen fertility, and in fact, loss of VPS41 function interfered with pollen penetration into the stigma (4). This and previous results (3, 4, 33, 34) indicate that HOPS is essential for plant viability. The increased number of PSVs of hypomorphic mutants zip2, vps33-1, and vps33-2 is indicative of HOPS function in PSV fusion or trafficking, as it has been observed in other vacuolar trafficking mutants (2, 18, 43, 62, 63). Moreover, the inducible amiRNA silencing of VPS41 and VPS11 underscores HOPS function in lytic vacuole fusion.
Phosphoinositides and Membrane Curvature Delimit VPS41 and VPS33 Recruitment.
VPS41-GFP and VPS33-GFP localize to late endosomes, which is consistent with a role in membrane fusion at the PVC and vacuole as part of HOPS (31) and with previous results in pollen tubes (4). The exclusive presence of VPS41 at the vacuole, however, was unexpected. One interpretation is that VPS33 recruitment to the vacuole is transient and limited to sites of membrane fusion, while VPS41 binding to the tonoplast is stable and results from specific lipid-binding affinities. Even though both proteins showed affinity for most phosphoinositides in a lipid strip assay, these results do not rule out the possibility of different affinities to specific phosphoinositides or that their recruitment to specific membranes is enhanced by interaction with other proteins. Two other HOPS subunits, VPS18 and VPS39, the latter being HOPS specific, are also absent from the vacuole (3). Given the competing needs of the core subunits VPS33, VPS18, VPS11, and VPS16 for either HOPS or CORVET assembly, the stable recruitment of VPS41 to the tonoplast could serve as a reservoir for this subunit in a way that does not compete for complex assembly.
Phosphoinositides recruit HOPS to the vertex ring between two juxtaposed vacuoles undergoing fusion in yeast (27, 52). We hypothesized that these lipids could fulfill a similar role in plants, and this was first supported by VPS41 and VPS33 binding to immobilized phosphoinositides on a membrane. Surprisingly, both VPS33 and VPS41 also bound to membrane liposomes but only in the absence of PtdIns(3)P or PtdIns(3,5)P2. This result implicates membrane curvature as an additional factor in the recruitment of HOPS to cellular membranes in plants and highlights the sensitivity of these proteins to both membrane topology and phosphoinositide lipid content. The plant vacuole membrane is a flat surface compared with the late endosomes, and membrane curvature could indeed represent a proxy for organelle identity in plant vacuole traffic. We propose that VPS41 binding to the vacuole membrane entails its recruitment by PtdIns(3)P or PtdIns(3,5)P2, since PtdIns(3)P has been detected at the tonoplast, and both are important for vacuolar traffic (64, 65). In contrast, VPS41 binding to curved membranes such as those of late endosomes is independent of and inhibited by phosphoinositides. This model is further supported by the loss of VPS41-GFP from the vacuole membrane but not from late endosomes in Wm-treated cells.
The inhibitory effect of phosphoinositides on VPS33 and VPS41 binding to liposomes suggests that plants have evolved different mechanisms of regulating vacuole fusion compared with yeast. In contrast to plants, the complete yeast HOPS complex and VPS41 alone bind to liposomes containing vacuolar phosphoinositides, but they do not bind strongly to bare liposomes (27, 66, 67). Therefore, while phosphoinositides promote recruitment of HOPS to curved membranes in yeast, they have the opposite effect in plants. Current models for vacuole fusion propose that the site of HOPS function, the vertex of two juxtaposed vacuoles, represents a highly curved membrane (68, 69). Moreover, small liposomes may be used as a proxy for HOPS interactions with the membrane at the vertex ring (68). The liposomes used here are not small enough to represent membrane curvature at sites of vacuole fusion, but if phosphoinositides also abolish HOPS binding at these sites, it would be consistent with yet another phenomenon observed only in plants. Our group has shown that Wm induces vacuole fusion in plants, both in developing roots and in guard cells, and we proposed that phosphoinositides function as negative regulators of vacuole fusion (2). This effect of Wm, however, is not observed in yeast vacuoles (48) or animal lysosomes (49), and therefore, plants seemed to have coerced the regulatory role of phosphoinositides into inhibitors rather than promoters of vacuole fusion. This difference may be due to evolutionary changes in the mechanisms by which VPS41 binds to membranes. VPS41 binding to highly curved membranes in yeast is mediated by an amphipathic lipid packing sensor (ALPS) motif, which also contains two important regulatory phosphorylation sites (67). The ALPS motif is not conserved in plants including Arabidopsis (SI Appendix, Fig. S5), which further supports the existence of plant-specific mechanisms for recruitment of HOPS to highly curved membranes.
PtdIns(3)P and PtdIns(3,5)P2 are important phosphoinositides for vacuole traffic and dynamics (64). PtdIns(3,5)P2 is produced by phosphorylation of the PtdIns(3)P precursor by the PI5 kinases FAB1A and FAB1B (70), and FAB1A has been localized to late endosomes (71). A new sensor of PtdIns(3,5)P2 indicates that this lipid accumulates in late endosomes (72), but it is not clear how much can persist after late endosome fusion with the vacuole. The possibility that PtdIns(3,5)P2 accumulates in the tonoplast is supported by the localization of the PtdIns(3,5)P2 phosphatases SUPPRESSOR OF ACTIN 2–5 (SAC2–5) exclusively to the vacuole membrane (73). Interestingly, Wm treatment resulted in significant dissociation of VPS41-GFP from the tonoplast, which could result from rapid depletion of PtdIns(3,5)P2 specifically in the tonoplast if both synthesis is stalled due to the unavailability of its precursor and active dephosphorylation by SAC phosphatases (73). PtdIns(3,5)P2 has recently emerged as an important regulator of plant growth, development, and stress responses. PtdIns(3,5)P2 is critical for polar growth (74) and is induced under osmotic stress in plants (75) and yeast (76). Loss of FAB1A and FAB1B results in pollen lethality due to the formation of aberrantly large vacuoles during pollen development (77), which implicates PtdIns(3,5)P2 in vacuole biogenesis. Moreover, PtdIns(3,5)P2 was found to be important for vacuole acidification and fragmentation during stomata closing (78), and loss of the PtdIns(3,5)P2 phosphatases SAC2–5, which is expected to increase its accumulation in membranes, induces vacuole fragmentation in Arabidopsis roots (73). It is tempting to speculate that these roles of PtdIns(3,5)P2 in promoting vacuole fragmentation in guard cells during stomata closure result from its inhibitory effect on VPS33 and VPS41 recruitment to sites of apposing vacuole membranes, which would be otherwise available for fusion. Our results suggest that vacuolar phosphoinositides are likely to play an important role in vacuole fusion–fragmentation dynamics by controlling the recruitment of HOPS to apposing vacuole membranes. Further research is necessary to test this hypothesis.
HOPS–SNARE Interactions Are Mediated by the SYP22 Qa SNARE.
The HOPS complex is expected to interact with SNARE proteins to regulate membrane fusion at the vacuole. That zip2/vps41 partially suppressed the fragmented vacuole phenotype of zig-1/vti11 is consistent with functional interactions between HOPS and the vacuole SNARE complexes containing VTI11 and SYP22. The mutation in zip2/vps41 may result in altered HOPS–SNARE interactions or the ability of HOPS to promote or proofread the SNARE complex assembly. Furthermore, the altered localization of VPS41-GFP and VPS33-GFP in the zig-1 mutant is consistent with a functional interaction between HOPS and SNARE that mediates vacuole fusion. We speculate that the bright punctate localizations of the tagged VPS41 and VPS33 in zig-1 may represent nonfunctional fusogenic complexes where SNAREs and HOPS are retained, with delays in their tonoplast accumulation and recycling. This was also observed when the SYP22 SNARE was localized to the same clusters of organelles. Furthermore, we previously hypothesized that the fragmented vacuole phenotype of zig-1 mutants could result from a putative proofreading activity of HOPS upon the mismatched vacuolar SNARE complex missing VTI11 (2), as it has been shown for yeast (79). The aggregation of VPS41 molecules in zig-1 is consistent with such activity, as HOPS could be loaded onto the abnormal SNARE complex while preventing fusion to occur. Further experiments are necessary to confirm this hypothesis. BiFC experiments showed that both HOPS subunits, VPS33 and VPS41, interacted with SYP22 in vivo, but only the VPS33–SYP22 interaction was detected in vitro. This is consistent with a role of HOPS as a chaperone for SNARE complex assembly (see below) and with coimmunoprecipitation between VCL1 and SYP22 in Arabidopsis seedlings (35). These interactions were detected in punctate structures similar to those of VPS33-VPS41 and VTI11-SYP22, suggesting that HOPS–SNARE interactions probably occur in late endosomes. Neither VPS33 or VPS41 interacted with VTI11 in the BiFC assay, which suggests that SNARE–HOPS interactions are not mediated by VTI11 protein surfaces. Overall, our results are consistent with the current model for HOPS–SNARE interactions that mediate membrane fusion in yeast. In this model, VPS33 binding to the Qa-SNARE Vam3 is the first step for SNARE complex assembly. The VPS33–Qa-SNARE interaction is thought to promote a conformational change, which exposes binding surfaces for other SNAREs on the VPS33 subunit (80). The interactions between VPS33 and SYP22, both in BiFC and in vitro, suggest that this mechanism is conserved in plants. Therefore, initial binding of VPS33 and SYP22 may be the first step in the HOPS–SNARE interactions that mediate vacuole fusion in plants.
Methods
Plant Materials, Growth Conditions, and Transformation.
Arabidopsis thaliana plants were grown as described (2). The ecotype Columbia (Col-0) was used as WT control. The genomic VPS41-GFP fusion (VPS41p::VPS41-GFP) was described elsewhere (4). Organelle markers mRFP-SYP22 (18), mCherry-RAB5 (mCherry-RabF2a), and mCherry-RAB7 (mCherry-RabG3f) (51) have been previously described. SALK T-DNA insertion lines, including vps41-1 (SALK_076372), vps41-2 (SALK_035406), vps33-3 (SALK_016684C), vps33-4 (SALK_151719), and vps33-5 (SALK_143407), were obtained from the Arabidopsis Biological Resource Center (ABRC) and genotyped by PCR (SI Appendix, Table S4). The zip2 zig-1 mutant was previously characterized (56). The TILLING lines (57) vps33-1 (CS92162) and vps33-2 (CS93595) were obtained from ABRC and backcrossed three times with Col-0 WT. Homozygous vps33-1 and vps33-2 were genotyped by PCR amplification (SI Appendix, Table S4) and restriction digestion with BsmBI (vps33-1) or TspRI (vps33-2). Nicotiana benthamiana plants were grown at room temperature for 3–4 wk in Metro-Mix 300 soil with a 16-h light/8-h dark photoperiod.
Arabidopsis transformation was carried out by floral dip (81) using Agrobacterium tumefaciens C58C1 and GV3101. Transient plant Agro-infiltration of 3- to 4-wk-old N. benthamiana leaves was carried out as described (82). GFP or RFP fluorescence was observed by confocal laser-scanning microscopy (CLSM) 36–48 h after infiltration.
Molecular Cloning.
The full-length genomic fusion of VPS33 with GFP driven by either 3- or 1-kb native promoters were generated using the GoldenBraid system (83). Primers for VPS33 genomic sequences (SI Appendix, Table S4) were generated by the GB domesticator software (https://gbcloning.upv.es/tools/domestication/), and each part was amplified by PCR (83, 84). Each transcriptional unit (TU) was assembled by restriction–ligation reaction into pUPD2 and sequenced. The full fusion construct containing the VPS33 promoter and genomic sequence, GFP, and the VPS33 terminator sequence was then assembled into pDGB3_alpha1. This cassette was finally inserted into pDGB3_omega1 together with the kanamycin resistance cassette from pGB0184 to generate pVPS33::gVPS33-GFP (VPS33-GFP). All experiments were carried out with the 3-kb promoter, except for Fig. 1 P–R, where the 1-kb promoter was used.
UBIQUITIN10 (UBQ10) promoter-driven and BiFC constructs were generated by Gateway cloning. Coding sequences of VPS41 (At1g08190), VPS33 (At3G54860), VTI11 (At5G39510), SYP22 (AT5G146860), and AtTIP2;1 (AT3G16240) were obtained by RT-PCR. Briefly, total mRNA was extracted from Col-0 WT seedlings using TRI reagent (Ambion) and treated with DNase I (New England Biolabs) according to manufacturer’s instruction. cDNAs were synthesized using iScript cDNA Synthesis Kit (Bio-Rad) and amplified by PCR using gene-specific primers (SI Appendix, Table S4) compatible for Gateway cloning. Entry clones were generated in pENTR4 or pENTR/D-TOPO (Invitrogen) (SI Appendix, Table S3). Expression clones (SI Appendix, Table S2) were generated by recombination between the entry clones and multiple destination vectors using LR clonase II (Invitrogen). Vectors driven by the pUBQ10 promoter were generated after substitution of the short UBQ promoter of the vector set described in ref. 85 with the full pUBQ10 promoter (1,596 bp) from pNIGEL07 (51). pDEST15 and pDEST17 destination vectors (Invitrogen) were used for protein expression in bacteria, and N- and C-terminal nEYFP and cEYFP pSITE vectors (86) were used to generate constructs for BiFC. All expression constructs are described in SI Appendix, Table S2.
The amiR-vps constructs were cloned using the GreenGate system (87). Specific amiRNA sequences against VPS41 and VPS11 (AT2G05170) were identified with the Web MicroRNA Designer (WMD3) tool (88) at wmd3.weigelworld.org/cgi-bin/webapp.cgi. The amiR-vps sequences were amplified in four independent PCRs (SI Appendix, Table S5) with the pRS300 plasmid containing the miR319a precursor sequence. To be suited for the GreenGate cloning system, primers “A” and “B” were modified with specific overhangs. The final PCR products (fourth reaction) were first digested with Eco31I and then ligated into the preopened intermediate vectors (SI Appendix, Table S5). The resulting vectors (supermodule A and B for each amiR-vps) were sequenced and combined in consecutive GreenGate reactions as described (87). The DEX induction system is based on the p6xOP/LhG4-GR transcription system (89, 90). Primers and vectors used in these experiments are reported in SI Appendix, Tables S3–S5.
Microscopy.
Zeiss LSM 710 or Zeiss LSM880 with Airyscan confocal microscopes were used for all imaging experiments unless specified. The excitation/emission wavelengths for imaging were 488 nm/492–570 nm for GFP and YFP, 561 nm/588–696 nm for mCherry and mRFP, and 561 nm/566–690 nm for LysoTracker Red. Three-day-old seedlings were analyzed with a 40×/1.2 N.A. water-immersion objective. To image PSVs, seeds were imbibed in water for 1 h, and seed coats were peeled off with a sharp needle. Vacuoles in the mature embryos were imaged by capturing PSV autofluorescence (excitation/emission wavelengths were 405/450–510 nm) from a single Z plane per sample. PSV quantification was carried out using Fiji (Fiji Is Just ImageJ, based on ImageJ 1.51d; fiji.sc/) (91). Linear (Pearson) and the nonlinear (Spearman’s) correlation coefficients for colocalization analysis were obtained with the PSC colocalization plug-in (92). The threshold level for the noise was set at 4. Membrane/cytosolic ratios of VPS41-GFP were calculated in Fiji by extracting intensity values of VPS41-GFP only from the vacuole membrane. This was accomplished by generating a membrane signal mask by thresholding of the RFP-SYP22 marker in the red channel to 40 or above. The mask was then applied to the GFP channel to extract the histograms of intensity values for VPS41-GFP at the membrane. A “cytosolic” intensity average was calculated by measuring intensity values from four cytosolic regions of interest per image.
Live-cell CLSM of vacuoles from amiRNA lines was done using a Leica TCS SP5II microscope (Leica Microsystems) equipped with a Leica HCX PL APO lambda blue 63×/1.20 N.A. UV water-immersion objective. Images were acquired with a lateral pixel resolution between 50 and 96 nm at a speed of 400 Hz with bidirectional scanning and five times line averaging. To reduce background fluorescence, the pinhole was adjusted between 0.75 and 1.0 airy unit (AU). The excitation/emission wavelengths for imaging were 488 nm/495–550 nm for BCECF and 561 nm/600–680 nm for FM4-64. Image postprocessing operations (Gaussian blur between 0.5 and 0.75; adjustment of brightness and contrast of the whole image) were done with Fiji.
Chemical Treatments and Fluorescent Dyes.
Three- or 4-d-old seedlings were used in all chemical treatments and fluorescent dye stainings. Wm (W3144; Sigma) was diluted in liquid AGM to a final concentration of 33 μM, and the same volume of DMSO was added to the control (1% final). Seedlings were treated for 3 h in the dark at 22 °C. A minimum of three seedlings was analyzed for each treatment, and the experiment was repeated three times. LysoTracker Red staining was carried out by incubation of seedlings in liquid AGM supplemented with 2 μM LysoTracker Red (Invitrogen) for 2 h. For amiRNA induction with DEX, 5 d-old seedlings were incubated for 72 h in liquid AGM supplemented with 30 µM DEX, or DMSO (control). Seedlings were stained with 10 µM BCECF, acetoxymethyl ester, and 1 µM FM 4-64 as described (8).
Protein Purification.
BL21-AI E. coli cells (Invitrogen) were transformed with T7::GST-VPS41, T7::GST-VPS33, T7::GST, T7::HIS-SYP22, or T7::HIS-VTI11. Recombinant bacteria were cultured in 300 mL of LB broth supplemented with 50 μg/mL carbenicillin and 1% glucose at 37 °C to an OD of 0.4–0.6. Expression of the tagged proteins was induced with 0.2% arabinose. Cultures were incubated at 28 °C for 5 h, except for T7::HIS-SYP22, which was incubated at 37 °C for 4 h, and HIS-VTI11, which was incubated at 16 °C for 24 h. Cells were harvested by centrifugation at 16,000 × g for 15 min and the pellet was resuspended in lysis buffer (20 mM Tris⋅HCl, pH 7.5, 150 mM NaCl, 0.1% β-mercaptoethanol, 1% Triton X-100, 1× protease inhibitors; Roche; #04693132001) and 12 μg/mL lysozyme (Sigma). The extract was cleared by centrifugation at 11,200 × g for 20 min, and the supernatant was then added to either a Gluthatione Sepharose 4B resin (GE Healthcare) or a Ni Sepharose 6 Fast Flow resin (GE Healthcare), accordingly. The recombinant proteins were eluted after an overnight incubation at 4 °C in slow rotation following the manufacturer’s recommendations.
Lipid–Protein Binding Assays.
Phospholipid-coated PIP strips (Echelon) were blocked with TBST buffer [10 mM Tris/HCl, pH 8.0, 150 mM NaCl, and 0.1% (vol/vol) Tween 20] containing 3% (wt/vol) BSA for 1 h at room temperature. Strips were then incubated with 3 mL of 0.33 nM purified GST-VPS41, GST-VPS33, or GST in TBST and 3% (wt/vol) BSA at 4 °C overnight. Strips were washed three times and incubated with mouse anti-GST primary antibody [1:1,000 dilution in 3% (wt/vol) BSA; Thermo Fisher]. A secondary HRP-conjugated anti-mouse antibody [diluted 1:2,000 in 3% (wt/vol) BSA; Thermo Fisher Scientific] was used next, and signal was detected by western ECL substrate (Bio-Rad) using a G:BOX Chemi XRQ gel documentation system (Syngene). This experiment was repeated three times with similar results.
The liposome binding assay was modified from refs. 93 and 94 using PolyPIPosomes (Echelon Biosciences) containing no phosphoinositides (control; catalog #Y-0000), PI(3)P (catalog #Y-003), or PI(3,5)P2 (catalog #Y-035). Proteins were first clarified by ultracentrifugation at 16,000 × g for 20 min. Then, 2.5 µg of GST-VPS41, 1.7 µg of GST-VPS33, 500 ng of GST alone, and 600 ng of PI(3)P Grip (p40 PX) (Echelon Biosciences) were diluted in a final volume of 95 µL of 1× binding buffer (20 mM Hepes, pH 7.4; 120 mM NaCl, 1 mM EDTA, 1 mM MgCl2, 1 mg/mL BSA; 0.2 mM CaCl2, 5 mM KCl, and 1× protease inhibitors from Roche; #04693132001). Five microliters of the PolyPIPosome was added, and the final 100-µL volume was incubated at room temperature for 30 min with slow rotation. Liposomes were then pelleted by ultracentrifugation at 16,000 × g for 20 min at room temperature and washed one time in 1× binding buffer. The pellet was resuspended in 30 µl of Laemmli buffer and resolved together with 30% of the supernatant by SDS/PAGE using a 3–8% Tris-acetate protein gel (Invitrogen). Similar results were obtained in three different experiments.
In Vitro Protein–Protein Interaction.
Mouse anti-GST (catalog #MA4-004; Thermo Fisher Scientific) or mouse Penta-HIS (Qiagen 34660) antibodies were bound to Dynabeads Protein G (Thermo Scientific) according to the manufacturer’s guidelines. To avoid coelution of the antibodies, we treated the beads with BS3 (Thermo Scientific) cross-linking agent. Beads and antibodies were incubated for 30 min at room temperature, treated with 1 M Tris⋅HCl, pH 7.5, quenching buffer, and incubated for 15 min at room temperature. After three washes, the different protein combinations were added. Samples were incubated overnight at 4 °C with rotation. Beads were washed three times with PBS buffer containing protease inhibitors, and bound proteins were eluted by 5-min denaturation at 90 °C in Laemmli buffer. Proteins were resolved in 10% acrylamide gels or 3–8% Tris-acetate protein gels (Invitrogen).
Supplementary Material
Acknowledgments
We thank J. Fernandez, J. Gillikin and D. Selote for technical assistance, and J. Lohmann and J. Forner for providing entry modules for the DEX induction cassette. We thank M. T. Morita (Nagoya University) and L.-J. Qu (Peking University) for the sharing of seed materials. We also thank reviewers for excellent feedback. This work was supported by National Aeronautics and Space Administration Space Life and Physical Sciences Division [NNX13AM49G (to M.R.-P.)], the National Science Foundation [DBI-1624613 and MCB-1244354 (to M.R.-P.)], and the Deutsche Forschungsgemeinschaft [DFG SFB1101 TPA02 (to K.S.)].
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1807763115/-/DCSupplemental.
References
- 1.Eisenach C, Francisco R, Martinoia E. Plant vacuoles. Curr Biol. 2015;25:R136–R137. doi: 10.1016/j.cub.2014.11.056. [DOI] [PubMed] [Google Scholar]
- 2.Zheng J, Han SW, Rodriguez-Welsh MF, Rojas-Pierce M. Homotypic vacuole fusion requires VTI11 and is regulated by phosphoinositides. Mol Plant. 2014;7:1026–1040. doi: 10.1093/mp/ssu019. [DOI] [PubMed] [Google Scholar]
- 3.Takemoto K, et al. Distinct sets of tethering complexes, SNARE complexes, and Rab GTPases mediate membrane fusion at the vacuole in Arabidopsis. Proc Natl Acad Sci USA. 2018;115:E2457–E2466. doi: 10.1073/pnas.1717839115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hao L, Liu J, Zhong S, Gu H, Qu LJ. AtVPS41-mediated endocytic pathway is essential for pollen tube-stigma interaction in Arabidopsis. Proc Natl Acad Sci USA. 2016;113:6307–6312. doi: 10.1073/pnas.1602757113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zouhar J, Rojo E. Plant vacuoles: Where did they come from and where are they heading? Curr Opin Plant Biol. 2009;12:677–684. doi: 10.1016/j.pbi.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 6.Hunter PR, Craddock CP, Di Benedetto S, Roberts LM, Frigerio L. Fluorescent reporter proteins for the tonoplast and the vacuolar lumen identify a single vacuolar compartment in Arabidopsis cells. Plant Physiol. 2007;145:1371–1382. doi: 10.1104/pp.107.103945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng H, Staehelin LA. Protein storage vacuoles are transformed into lytic vacuoles in root meristematic cells of germinating seedlings by multiple, cell type-specific mechanisms. Plant Physiol. 2011;155:2023–2035. doi: 10.1104/pp.110.170159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Viotti C, et al. The endoplasmic reticulum is the main membrane source for biogenesis of the lytic vacuole in Arabidopsis. Plant Cell. 2013;25:3434–3449. doi: 10.1105/tpc.113.114827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ruthardt N, Gulde N, Spiegel H, Fischer R, Emans N. Four-dimensional imaging of transvacuolar strand dynamics in tobacco BY-2 cells. Protoplasma. 2005;225:205–215. doi: 10.1007/s00709-005-0093-7. [DOI] [PubMed] [Google Scholar]
- 10.Uemura T, Yoshimura SH, Takeyasu K, Sato MH. Vacuolar membrane dynamics revealed by GFP-AtVam3 fusion protein. Genes Cells. 2002;7:743–753. doi: 10.1046/j.1365-2443.2002.00550.x. [DOI] [PubMed] [Google Scholar]
- 11.Han SW, Alonso JM, Rojas-Pierce M. REGULATOR OF BULB BIOGENESIS1 (RBB1) is involved in vacuole bulb formation in Arabidopsis. PLoS One. 2015;10:e0125621. doi: 10.1371/journal.pone.0125621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ibl V, Stoger E. Live cell imaging during germination reveals dynamic tubular structures derived from protein storage vacuoles of barley aleurone cells. Plants (Basel) 2014;3:442–457. doi: 10.3390/plants3030442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao XQ, et al. The dynamic changes of tonoplasts in guard cells are important for stomatal movement in Vicia faba. Plant Physiol. 2005;139:1207–1216. doi: 10.1104/pp.105.067520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanaka Y, et al. Intra-vacuolar reserves of membranes during stomatal closure: The possible role of guard cell vacuoles estimated by 3-D reconstruction. Plant Cell Physiol. 2007;48:1159–1169. doi: 10.1093/pcp/pcm085. [DOI] [PubMed] [Google Scholar]
- 15.Campbell NA, Garber RC. Vacuolar reorganization in the motor cells of Albizzia during leaf movement. Planta. 1980;148:251–255. doi: 10.1007/BF00380035. [DOI] [PubMed] [Google Scholar]
- 16.Wickner W. Membrane fusion: Five lipids, four SNAREs, three chaperones, two nucleotides, and a Rab, all dancing in a ring on yeast vacuoles. Annu Rev Cell Dev Biol. 2010;26:115–136. doi: 10.1146/annurev-cellbio-100109-104131. [DOI] [PubMed] [Google Scholar]
- 17.Sanderfoot AA, Kovaleva V, Bassham DC, Raikhel NV. Interactions between syntaxins identify at least five SNARE complexes within the Golgi/prevacuolar system of the Arabidopsis cell. Mol Biol Cell. 2001;12:3733–3743. doi: 10.1091/mbc.12.12.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ebine K, et al. A SNARE complex unique to seed plants is required for protein storage vacuole biogenesis and seed development of Arabidopsis thaliana. Plant Cell. 2008;20:3006–3021. doi: 10.1105/tpc.107.057711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ebine K, et al. A membrane trafficking pathway regulated by the plant-specific RAB GTPase ARA6. Nat Cell Biol. 2011;13:853–859. doi: 10.1038/ncb2270. [DOI] [PubMed] [Google Scholar]
- 20.Fujiwara M, et al. Interactomics of Qa-SNARE in Arabidopsis thaliana. Plant Cell Physiol. 2014;55:781–789. doi: 10.1093/pcp/pcu038. [DOI] [PubMed] [Google Scholar]
- 21.Yano D, et al. A SNARE complex containing SGR3/AtVAM3 and ZIG/VTI11 in gravity-sensing cells is important for Arabidopsis shoot gravitropism. Proc Natl Acad Sci USA. 2003;100:8589–8594. doi: 10.1073/pnas.1430749100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hickey CM, Wickner W. HOPS initiates vacuole docking by tethering membranes before trans-SNARE complex assembly. Mol Biol Cell. 2010;21:2297–2305. doi: 10.1091/mbc.E10-01-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Delahaye JL, et al. Caenorhabditis elegans HOPS and CCZ-1 mediate trafficking to lysosome-related organelles independently of RAB-7 and SAND-1. Mol Biol Cell. 2014;25:1073–1096. doi: 10.1091/mbc.E13-09-0521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zlatic SA, Tornieri K, L’hernault SW, Faundez V. Metazoan cell biology of the HOPS tethering complex. Cell Logist. 2011;1:111–117. doi: 10.4161/cl.1.3.17279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Balderhaar HJ, Ungermann C. CORVET and HOPS tethering complexes—coordinators of endosome and lysosome fusion. J Cell Sci. 2013;126:1307–1316. doi: 10.1242/jcs.107805. [DOI] [PubMed] [Google Scholar]
- 26.Solinger JA, Spang A. Tethering complexes in the endocytic pathway: CORVET and HOPS. FEBS J. 2013;280:2743–2757. doi: 10.1111/febs.12151. [DOI] [PubMed] [Google Scholar]
- 27.Stroupe C, Collins KM, Fratti RA, Wickner W. Purification of active HOPS complex reveals its affinities for phosphoinositides and the SNARE Vam7p. EMBO J. 2006;25:1579–1589. doi: 10.1038/sj.emboj.7601051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baker RW, Jeffrey PD, Hughson FM. Crystal structures of the Sec1/Munc18 (SM) protein Vps33, alone and bound to the homotypic fusion and vacuolar protein sorting (HOPS) subunit Vps16*. PLoS One. 2013;8:e67409. doi: 10.1371/journal.pone.0067409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baker RW, et al. A direct role for the Sec1/Munc18-family protein Vps33 as a template for SNARE assembly. Science. 2015;349:1111–1114. doi: 10.1126/science.aac7906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nickerson DP, Brett CL, Merz AJ. Vps-C complexes: Gatekeepers of endolysosomal traffic. Curr Opin Cell Biol. 2009;21:543–551. doi: 10.1016/j.ceb.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vukašinović N, Žárský V. Tethering complexes in the Arabidopsis endomembrane system. Front Cell Dev Biol. 2016;4:46. doi: 10.3389/fcell.2016.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tan X, Wei J, Li B, Wang M, Bao Y. AtVps11 is essential for vacuole biogenesis in embryo and participates in pollen tube growth in Arabidopsis. Biochem Biophys Res Commun. 2017;491:794–799. doi: 10.1016/j.bbrc.2017.07.059. [DOI] [PubMed] [Google Scholar]
- 33.Rojo E, Gillmor CS, Kovaleva V, Somerville CR, Raikhel NV. VACUOLELESS1 is an essential gene required for vacuole formation and morphogenesis in Arabidopsis. Dev Cell. 2001;1:303–310. doi: 10.1016/s1534-5807(01)00024-7. [DOI] [PubMed] [Google Scholar]
- 34.Hicks GR, Rojo E, Hong S, Carter DG, Raikhel NV. Geminating pollen has tubular vacuoles, displays highly dynamic vacuole biogenesis, and requires VACUOLESS1 for proper function. Plant Physiol. 2004;134:1227–1239. doi: 10.1104/pp.103.037382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rojo E, Zouhar J, Kovaleva V, Hong S, Raikhel NV. The AtC-VPS protein complex is localized to the tonoplast and the prevacuolar compartment in Arabidopsis. Mol Biol Cell. 2003;14:361–369. doi: 10.1091/mbc.E02-08-0509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uemura T, Ueda T. Plant vacuolar trafficking driven by RAB and SNARE proteins. Curr Opin Plant Biol. 2014;22:116–121. doi: 10.1016/j.pbi.2014.10.002. [DOI] [PubMed] [Google Scholar]
- 37.Limpens E, et al. Medicago N2-fixing symbiosomes acquire the endocytic identity marker Rab7 but delay the acquisition of vacuolar identity. Plant Cell. 2009;21:2811–2828. doi: 10.1105/tpc.108.064410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ueda T, Yamaguchi M, Uchimiya H, Nakano A. Ara6, a plant-unique novel type Rab GTPase, functions in the endocytic pathway of Arabidopsis thaliana. EMBO J. 2001;20:4730–4741. doi: 10.1093/emboj/20.17.4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cui Y, et al. Biogenesis of plant prevacuolar multivesicular bodies. Mol Plant. 2016;9:774–786. doi: 10.1016/j.molp.2016.01.011. [DOI] [PubMed] [Google Scholar]
- 40.Sohn EJ, et al. Rha1, an Arabidopsis Rab5 homolog, plays a critical role in the vacuolar trafficking of soluble cargo proteins. Plant Cell. 2003;15:1057–1070. doi: 10.1105/tpc.009779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kotzer AM, et al. AtRabF2b (Ara7) acts on the vacuolar trafficking pathway in tobacco leaf epidermal cells. J Cell Sci. 2004;117:6377–6389. doi: 10.1242/jcs.01564. [DOI] [PubMed] [Google Scholar]
- 42.Cui Y, et al. Activation of the Rab7 GTPase by the MON1-CCZ1 complex is essential for PVC-to-vacuole trafficking and plant growth in Arabidopsis. Plant Cell. 2014;26:2080–2097. doi: 10.1105/tpc.114.123141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ebine K, et al. Plant vacuolar trafficking occurs through distinctly regulated pathways. Curr Biol. 2014;24:1375–1382. doi: 10.1016/j.cub.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 44.Singh MK, et al. Protein delivery to vacuole requires SAND protein-dependent Rab GTPase conversion for MVB-vacuole fusion. Curr Biol. 2014;24:1383–1389. doi: 10.1016/j.cub.2014.05.005. [DOI] [PubMed] [Google Scholar]
- 45.Ugalde JM, et al. Phosphatidylinositol 4-phosphate 5-kinases 1 and 2 are involved in the regulation of vacuole morphology during Arabidopsis thaliana pollen development. Plant Sci. 2016;250:10–19. doi: 10.1016/j.plantsci.2016.05.014. [DOI] [PubMed] [Google Scholar]
- 46.Wang J, Cai Y, Miao Y, Lam SK, Jiang L. Wortmannin induces homotypic fusion of plant prevacuolar compartments. J Exp Bot. 2009;60:3075–3083. doi: 10.1093/jxb/erp136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stack JH, Emr SD. Vps34p required for yeast vacuolar protein sorting is a multiple specificity kinase that exhibits both protein kinase and phosphatidylinositol-specific PI 3-kinase activities. J Biol Chem. 1994;269:31552–31562. [PubMed] [Google Scholar]
- 48.Boeddinghaus C, Merz AJ, Laage R, Ungermann C. A cycle of Vam7p release from and PtdIns 3-P-dependent rebinding to the yeast vacuole is required for homotypic vacuole fusion. J Cell Biol. 2002;157:79–89. doi: 10.1083/jcb.200112098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fernandez-Borja M, et al. Multivesicular body morphogenesis requires phosphatidyl-inositol 3-kinase activity. Curr Biol. 1999;9:55–58. doi: 10.1016/s0960-9822(99)80048-7. [DOI] [PubMed] [Google Scholar]
- 50.Koumandou VL, Dacks JB, Coulson RM, Field MC. Control systems for membrane fusion in the ancestral eukaryote; evolution of tethering complexes and SM proteins. BMC Evol Biol. 2007;7:29. doi: 10.1186/1471-2148-7-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Geldner N, et al. Rapid, combinatorial analysis of membrane compartments in intact plants with a multicolor marker set. Plant J. 2009;59:169–178. doi: 10.1111/j.1365-313X.2009.03851.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fratti RA, Jun Y, Merz AJ, Margolis N, Wickner W. Interdependent assembly of specific regulatory lipids and membrane fusion proteins into the vertex ring domain of docked vacuoles. J Cell Biol. 2004;167:1087–1098. doi: 10.1083/jcb.200409068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jung JY, et al. Phosphatidylinositol 3- and 4-phosphate are required for normal stomatal movements. Plant Cell. 2002;14:2399–2412. doi: 10.1105/tpc.004143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alonso JM, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 55.Howden R, et al. Selection of T-DNA-tagged male and female gametophytic mutants by segregation distortion in Arabidopsis. Genetics. 1998;149:621–631. doi: 10.1093/genetics/149.2.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Niihama M, Takemoto N, Hashiguchi Y, Tasaka M, Morita MT. ZIP genes encode proteins involved in membrane trafficking of the TGN-PVC/vacuoles. Plant Cell Physiol. 2009;50:2057–2068. doi: 10.1093/pcp/pcp137. [DOI] [PubMed] [Google Scholar]
- 57.Till BJ, et al. Large-scale discovery of induced point mutations with high-throughput TILLING. Genome Res. 2003;13:524–530. doi: 10.1101/gr.977903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Plemel RL, et al. Subunit organization and Rab interactions of Vps-C protein complexes that control endolysosomal membrane traffic. Mol Biol Cell. 2011;22:1353–1363. doi: 10.1091/mbc.E10-03-0260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Walter M, et al. Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 2004;40:428–438. doi: 10.1111/j.1365-313X.2004.02219.x. [DOI] [PubMed] [Google Scholar]
- 60.Bracha-Drori K, et al. Detection of protein-protein interactions in plants using bimolecular fluorescence complementation. Plant J. 2004;40:419–427. doi: 10.1111/j.1365-313X.2004.02206.x. [DOI] [PubMed] [Google Scholar]
- 61.Niihama M, et al. Conversion of functional specificity in Qb-SNARE VTI1 homologues of Arabidopsis. Curr Biol. 2005;15:555–560. doi: 10.1016/j.cub.2005.02.021. [DOI] [PubMed] [Google Scholar]
- 62.Reguera M, et al. pH regulation by NHX-type antiporters is required for receptor-mediated protein trafficking to the vacuole in Arabidopsis. Plant Cell. 2015;27:1200–1217. doi: 10.1105/tpc.114.135699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu X, Ebine K, Ueda T, Qiu Q-S. AtNHX5 and AtNHX6 are required for the subcellular localization of the SNARE complex that mediates the trafficking of seed storage proteins in Arabidopsis. PLoS One. 2016;11:e0151658. doi: 10.1371/journal.pone.0151658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Noack LC, Jaillais Y. Precision targeting by phosphoinositides: How PIs direct endomembrane trafficking in plants. Curr Opin Plant Biol. 2017;40:22–33. doi: 10.1016/j.pbi.2017.06.017. [DOI] [PubMed] [Google Scholar]
- 65.Simon ML, et al. A multi-colour/multi-affinity marker set to visualize phosphoinositide dynamics in Arabidopsis. Plant J. 2014;77:322–337. doi: 10.1111/tpj.12358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hickey CM, Stroupe C, Wickner W. The major role of the Rab Ypt7p in vacuole fusion is supporting HOPS membrane association. J Biol Chem. 2009;284:16118–16125. doi: 10.1074/jbc.M109.000737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cabrera M, et al. Phosphorylation of a membrane curvature-sensing motif switches function of the HOPS subunit Vps41 in membrane tethering. J Cell Biol. 2010;191:845–859. doi: 10.1083/jcb.201004092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ho R, Stroupe C. The HOPS/class C Vps complex tethers high-curvature membranes via a direct protein-membrane interaction. Traffic. 2016;17:1078–1090. doi: 10.1111/tra.12421. [DOI] [PubMed] [Google Scholar]
- 69.D’Agostino M, Risselada HJ, Lürick A, Ungermann C, Mayer A. A tethering complex drives the terminal stage of SNARE-dependent membrane fusion. Nature. 2017;551:634–638. doi: 10.1038/nature24469. [DOI] [PubMed] [Google Scholar]
- 70.Meijer HJ, Munnik T. Phospholipid-based signaling in plants. Annu Rev Plant Biol. 2003;54:265–306. doi: 10.1146/annurev.arplant.54.031902.134748. [DOI] [PubMed] [Google Scholar]
- 71.Hirano T, Munnik T, Sato MH. Phosphatidylinositol 3-phosphate 5-kinase, FAB1/PIKfyve kinase mediates endosome maturation to establish endosome-cortical microtubule interaction in Arabidopsis. Plant Physiol. 2015;169:1961–1974. doi: 10.1104/pp.15.01368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hirano T, Stecker K, Munnik T, Xu H, Sato MH. Visualization of phosphatidylinositol 3,5-bisphosphate dynamics by a tandem ML1N-based fluorescent protein probe in Arabidopsis. Plant Cell Physiol. 2017;58:1185–1195. doi: 10.1093/pcp/pcx011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nováková P, et al. SAC phosphoinositide phosphatases at the tonoplast mediate vacuolar function in Arabidopsis. Proc Natl Acad Sci USA. 2014;111:2818–2823. doi: 10.1073/pnas.1324264111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.van Gisbergen PA, Li M, Wu SZ, Bezanilla M. Class II formin targeting to the cell cortex by binding PI(3,5)P(2) is essential for polarized growth. J Cell Biol. 2012;198:235–250. doi: 10.1083/jcb.201112085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Meijer HJG, Divecha N, van den Ende H, Musgrave A, Munnik T. Hyperosmotic stress induces rapid synthesis of phosphatidyl-d-inositol 3,5-bisphosphate in plant cells. Planta. 1999;208:294–298. [Google Scholar]
- 76.Dove SK, et al. Osmotic stress activates phosphatidylinositol-3,5-bisphosphate synthesis. Nature. 1997;390:187–192. doi: 10.1038/36613. [DOI] [PubMed] [Google Scholar]
- 77.Whitley P, Hinz S, Doughty J. Arabidopsis FAB1/PIKfyve proteins are essential for development of viable pollen. Plant Physiol. 2009;151:1812–1822. doi: 10.1104/pp.109.146159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bak G, et al. Rapid structural changes and acidification of guard cell vacuoles during stomatal closure require phosphatidylinositol 3,5-bisphosphate. Plant Cell. 2013;25:2202–2216. doi: 10.1105/tpc.113.110411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Starai VJ, Hickey CM, Wickner W. HOPS proofreads the trans-SNARE complex for yeast vacuole fusion. Mol Biol Cell. 2008;19:2500–2508. doi: 10.1091/mbc.E08-01-0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Baker RW, Hughson FM. Chaperoning SNARE assembly and disassembly. Nat Rev Mol Cell Biol. 2016;17:465–479. doi: 10.1038/nrm.2016.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 82.Goodin MM, Dietzgen RG, Schichnes D, Ruzin S, Jackson AO. pGD vectors: Versatile tools for the expression of green and red fluorescent protein fusions in agroinfiltrated plant leaves. Plant J. 2002;31:375–383. doi: 10.1046/j.1365-313x.2002.01360.x. [DOI] [PubMed] [Google Scholar]
- 83.Sarrion-Perdigones A, et al. GoldenBraid 2.0: A comprehensive DNA assembly framework for plant synthetic biology. Plant Physiol. 2013;162:1618–1631. doi: 10.1104/pp.113.217661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sarrion-Perdigones A, Palaci J, Granell A, Orzaez D. Design and construction of multigenic constructs for plant biotechnology using the GoldenBraid cloning strategy. Methods Mol Biol. 2014;1116:133–151. doi: 10.1007/978-1-62703-764-8_10. [DOI] [PubMed] [Google Scholar]
- 85.Grefen C, et al. A ubiquitin-10 promoter-based vector set for fluorescent protein tagging facilitates temporal stability and native protein distribution in transient and stable expression studies. Plant J. 2010;64:355–365. doi: 10.1111/j.1365-313X.2010.04322.x. [DOI] [PubMed] [Google Scholar]
- 86.Martin K, et al. Transient expression in Nicotiana benthamiana fluorescent marker lines provides enhanced definition of protein localization, movement and interactions in planta. Plant J. 2009;59:150–162. doi: 10.1111/j.1365-313X.2009.03850.x. [DOI] [PubMed] [Google Scholar]
- 87.Lampropoulos A, et al. GreenGate—a novel, versatile, and efficient cloning system for plant transgenesis. PLoS One. 2013;8:e83043. doi: 10.1371/journal.pone.0083043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D. Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell. 2006;18:1121–1133. doi: 10.1105/tpc.105.039834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Craft J, et al. New pOp/LhG4 vectors for stringent glucocorticoid-dependent transgene expression in Arabidopsis. Plant J. 2005;41:899–918. doi: 10.1111/j.1365-313X.2005.02342.x. [DOI] [PubMed] [Google Scholar]
- 90.Moore I, Samalova M, Kurup S. Transactivated and chemically inducible gene expression in plants. Plant J. 2006;45:651–683. doi: 10.1111/j.1365-313X.2006.02660.x. [DOI] [PubMed] [Google Scholar]
- 91.Schindelin J, et al. Fiji: An open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.French AP, Mills S, Swarup R, Bennett MJ, Pridmore TP. Colocalization of fluorescent markers in confocal microscope images of plant cells. Nat Protoc. 2008;3:619–628. doi: 10.1038/nprot.2008.31. [DOI] [PubMed] [Google Scholar]
- 93.Islam KT, Velivelli SLS, Berg RH, Oakley B, Shah DM. A novel bi-domain plant defensin MtDef5 with potent broad-spectrum antifungal activity binds to multiple phospholipids and forms oligomers. Sci Rep. 2017;7:16157. doi: 10.1038/s41598-017-16508-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Julkowska MM, Rankenberg JM, Testerink C. Plant Lipid Signaling Protocols. Humana Press; Totowa, NJ: 2013. Liposome-binding assays to assess specificity and affinity of phospholipid–protein interactions; pp. 261–271. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.