Significance
Oligodendrocyte precursor cells (OPCs) constitute the main proliferative cells in the adult brain and deregulation of OPC proliferation-differentiation balance results in either glioma formation or defective (re)myelination. Mutations in chromatin remodelers CHD7 and CHD8 are the cause of CHARGE syndrome and some autism spectrum disorders (ASD). Here we show that Chd7 protects OPCs from apoptosis by chromatin closing and gene repression of p53, while Chd7 induces chromatin opening and gene activation of OPC-differentiation regulators. Chd7 is, however, dispensable for oligodendrocyte stage progression, consistent with Chd8 compensatory function, as suggested by their common chromatin-binding profiles, including ASD-risk–associated genes. Our results thus involve oligodendroglia in ASD and CHARGE and offer new avenues to understand and modulate CHD7/CHD8 functions in normal and pathological brain development.
Keywords: oligodendrocyte, chromatin remodeling, CHARGE, autism spectrum disorder, transcription regulation
Abstract
Oligodendrocyte precursor cells (OPCs) constitute the main proliferative cells in the adult brain, and deregulation of OPC proliferation-differentiation balance results in either glioma formation or defective adaptive (re)myelination. OPC differentiation requires significant genetic reprogramming, implicating chromatin remodeling. Mounting evidence indicates that chromatin remodelers play important roles during normal development and their mutations are associated with neurodevelopmental defects, with CHD7 haploinsuficiency being the cause of CHARGE syndrome and CHD8 being one of the strongest autism spectrum disorder (ASD) high-risk–associated genes. Herein, we report on uncharacterized functions of the chromatin remodelers Chd7 and Chd8 in OPCs. Their OPC-chromatin binding profile, combined with transcriptome and chromatin accessibility analyses of Chd7-deleted OPCs, demonstrates that Chd7 protects nonproliferative OPCs from apoptosis by chromatin closing and transcriptional repression of p53. Furthermore, Chd7 controls OPC differentiation through chromatin opening and transcriptional activation of key regulators, including Sox10, Nkx2.2, and Gpr17. However, Chd7 is dispensable for oligodendrocyte stage progression, consistent with Chd8 compensatory function, as suggested by their common chromatin-binding profiles and genetic interaction. Finally, CHD7 and CHD8 bind in OPCs to a majority of ASD risk-associated genes, suggesting an implication of oligodendrocyte lineage cells in ASD neurological defects. Our results thus offer new avenues to understand and modulate the CHD7 and CHD8 functions in normal development and disease.
Beyond the well-known function in saltatory conduction of action potentials, oligodendrocytes (OLs) have been shown to metabolically support axons with lactate shuttling through the myelin sheaths (1, 2). Accumulating evidence over the last years has demonstrated a role for neuronal activity-dependent adaptive myelination in adult brain plasticity, which confers a key neurological function to OL lineage cells (3). Oligodendrocyte precursor cells (OPCs) originate from focal regions of ventricular zones in the brain and spinal cord (4, 5) and, through proliferation and migration, occupy the whole CNS before starting to differentiate into myelinating OLs (6). Unlike most precursor cells, OPCs remain abundant in the adult CNS, being one of its neural cell subtypes (7, 8). OPCs need to keep a tight balance between proliferation, survival, and differentiation. This balance is crucial to maintain the OPC pool while contributing to myelin plasticity in adult life, and to remyelination in diseases, such as multiple sclerosis (MS). A large diversity of extrinsic signals (9), as well as many transcription factors [TFs, reviewed in Emery and Lu (10)], have been involved in OPC proliferation, survival, and differentiation. However, the mechanism for how these signals are integrated in the nucleus to balance OPC behavior is largely unknown.
OPC differentiation requires profound changes in chromatin and gene expression (10, 11). TFs, such as Sox10, Olig2, Nkx2.2, or Ascl1, are key regulators of OL differentiation by directly controlling transcription of genes implicated in this process (12–15) but being already expressed in OPCs, it is still unclear how these TFs control the induction of differentiation. A growing body of evidence suggests that some of these TFs work together with chromatin ` factors during transcriptional initiation/elongation to drive robust transcription (16). Accordingly, Olig2 and Brg1, a SWI/SNF chromatin remodeler expressed in oligodendroglia, have been found to cooperate and promote the expression of OPC differentiation genes (15).
Chd7 and Chd8, ATP-dependent nucleosome remodeling factors belonging to the subgroup III of the chromodomain helicase DNA-binding (CHD) family modulate the chromatin configuration to regulate the temporal and spatial expression of genes during development (17, 18) and, more specifically, neurogenesis (19–23). Importantly, while CHD7 mutations are the main cause of CHARGE syndrome, an autosomal-dominant syndrome that often impairs normal brain development, leading to cognitive disabilities (24), CHD8 is one of the nine high-confidence autism spectrum disorder (ASD)-risk genes (25, 26) and autism features are found in some CHARGE syndrome patients (27). We recently showed that Chd7 expression is highly enriched in OL lineage cells and by Chd7 loss-of-function (LOF) experiments demonstrated that Chd7 is required for myelination and remyelination through cooperation with Sox10 and activation of myelin-associated gene expression (28). In the present study, we have investigated the role of Chd7 in OPC cell fates and explored the possible redundancy with its paralog gene, Chd8. Indeed, Chd8 is another CHD subgroup III protein previously shown to interact with Chd7 (21, 29), and although Chd8 plays an important role in neurodevelopment (22, 23), its function in glial cells has not been investigated so far. Here, using a time-controlled Chd7 deletion in OPCs, we show that Chd7 inactivation leads to decreased OPC survival specifically in noncycling OPCs and that, in parallel, OPC differentiation is reduced without affecting OL stage progression due to Chd8 functional compensation. Combining Chd7/Chd8 chromatin-binding profiles from in vivo OPCs with transcriptome and chromatin accessibility analyses of Chd7-deleted OPCs, we demonstrate that mechanistically, Chd7 promotes OPC survival via chromatin closing and transcriptional repression of p53/Trp53, while conversely, Chd7 binding is required for chromatin opening and transcriptional activation of key regulators of OPC differentiation. Finally, integration of chromatin-binding profiles and associated epigenetic signals from OL lineage cells suggest that Chd7/Chd8 remodelers and Olig2/Sox10 TFs cooperate to activate oligodendroglia stage-specific genes in a time-controlled manner.
Results
Chd7 Binds to Promoters and Enhancers of Genes Involved in OPC Differentiation, Proliferation, and Survival.
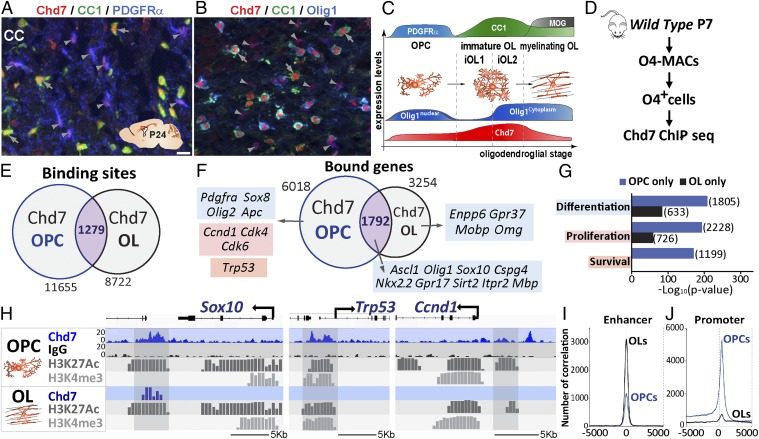
Chd7 is known to be required for the normal onset of myelination (28) but its involvement in early stages of oligodendroglial cell lineage has not been addressed. In support for Chd7’s role in OPC cell-fate decisions, we found that Chd7 protein was expressed at the OPC stage (Fig. 1 A–C, arrowheads, and SI Appendix, Fig. S1) and accumulated in immature OLs (iOLs) (Fig. 1 A–C, arrows, and SI Appendix, Fig. S1) (28). OPCs show a tight balance between proliferation and differentiation by mechanisms that are not completely understood. We therefore investigated the possible regulation of Chd7 in OPC proliferation, survival, and differentiation by generating a chromatin-binding profile of Chd7 in purified OPCs. Using magnetic-assisted cell sorting (MACS) from mouse postnatal day (P) 7 cortices (Fig. 1D), we prepared highly purified O4+ oligodendroglia (98%, being ∼80% of PDGFRα+ OPCs and ∼20% of Nkx2.2+/CNP+ iOLs) (SI Appendix, Fig. S2 A–C), directly used to perform Chd7 chromatin-binding analysis by ChIP combined with high-throughput sequencing (ChIP-seq) (Fig. 1D), and identified 11,655 Chd7-binding sites associated with 6,018 protein-coding genes (Fig. 1 E and F). Remarkably, almost all (94%) of Chd7-bound genes had a dynamic expression pattern in OL lineage cells (SI Appendix, Fig. S2D). To investigate whether Chd7 binding in OPCs was stage-specific, we compared Chd7-binding sites in OPCs to our previous data from OLs (28). We found that Chd7 binds only 10% of common sites at both stages (1,279) (Fig. 1E), corresponding to ∼30% of commonly bound genes (2,168) (Fig. 1F). These genes included key regulators of oligodendrogenesis (Ascl1, Olig1, Sox10, Nkx2.2) (Fig. 1 F and H). Notably, gene ontology (GO) enrichment analysis indicated that, in OPCs, Chd7 binds genes involved in proliferation (such as Ccnd1, Cdk4, and Cdk6) and apoptosis (such as p53/Trp53), while myelin-genes (such as Mobp and Omg) were Chd7-bound only in OLs (Fig. 1 F–H). Therefore, Chd7 binds in a stage-specific manner to genes implicated in oligodendroglial cell survival, proliferation, and differentiation.
Fig. 1.
Chd7 bind to genes involved in OPCs proliferation, differentiation, and survival. (A and B) Immunostaining of Chd7 with PDGFRα and CC1 (A) or Olig1 and CC1 (B) in the CC of P24 mice. Gray arrowheads show OPCs and gray arrows show OLs. (Scale bar, 10 μm.) (C) Scheme representing expression levels of different markers depending on OL stage. (D) Diagram representing MACSorting of O4+ cells of P7 wild-type mice followed by Chd7 ChIP-seq. (E and F) Venn diagrams depicting the overlap of Chd7-binding sites (E) and bound genes (F) in OPCs and OLs, with examples of genes involved in OPC differentiation (blue), cell death (orange), and cell cycle (red). (G) Barplot representing the GO analysis of the OPC- or OL-only Chd7-bound genes. (H) Representative ChIP-seq tracks for Chd7 and control IgG together with active epigenetic marks (H3K27ac and H3K4me3) of Trp53, Ccnd1, and Sox10 genes in OPCs and OLs. (I and J) Graph showing the number of correlations of Chd7 peaks in OPCs (blue) and OLs (black) compared with the position in enhancer regions (I) and promoter regions (J).
We assessed the cell-type specificity of Chd7 chromatin binding by comparing its binding profile in OPCs, embryonic stem cells (ESCs) (30), OLs (28), and neonatal cerebellar granule precursors (GNPs) (21). Interestingly, Chd7-binding profiles in ESCs, OPCs, and OLs overlapped only in 151 peaks (representing 1% of OPC peaks), with 14% of OPC peaks shared with ESCs (SI Appendix, Fig. S2E). Similarly, only 63 peaks overlapped between OPCs, OLs, and GNPs, with 4% of OPC peaks shared with GNPs (SI Appendix, Fig. S2F). The Chd7 chromatin binding is thus mostly cell-type– and lineage-stage–specific.
We next examined whether Chd7 preferentially binds enhancer elements in OPCs, as has been reported in ESCs (17, 30). To identify enhancer elements in OPCs, we took advantage of oligodendroglia-specific ChIP-seq datasets for Sox10 and Olig2, key TFs activating oligodendrogenesis (15, 28). In oligodendroglial cells, Sox10/Olig2-commonly bound regions (12,094) (SI Appendix, Fig. S2G) display strong H3K27ac marks (SI Appendix, Fig. S2I), characteristic of active regulatory elements (31), and are mainly located outside promoter regions (31) (SI Appendix, Fig. S2H) and thus correspond to active enhancers. We found that Chd7 binds many H3K27ac+ Sox10/Olig2-active enhancers both in OPCs and OLs (Fig. 1I). Unexpectedly, in OPCs Chd7 binding was especially enriched in promoter regions defined by either H3K4me3/H3K27ac active promoter marks or the Genomatix portal (Fig. 1J and SI Appendix, Fig. S2 J and K), contrary to rare Chd7 binding to promoters in other cell types, such as OLs, GNPs (21), and ESCs (30). Taken together, our data suggest that Chd7 binding to promoters is temporally restricted to the OPC stage while Chd7 binds to enhancers both in OPCs and OLs.
Chd7 Regulates Genes Involved in OPC Proliferation, Differentiation, and Survival.
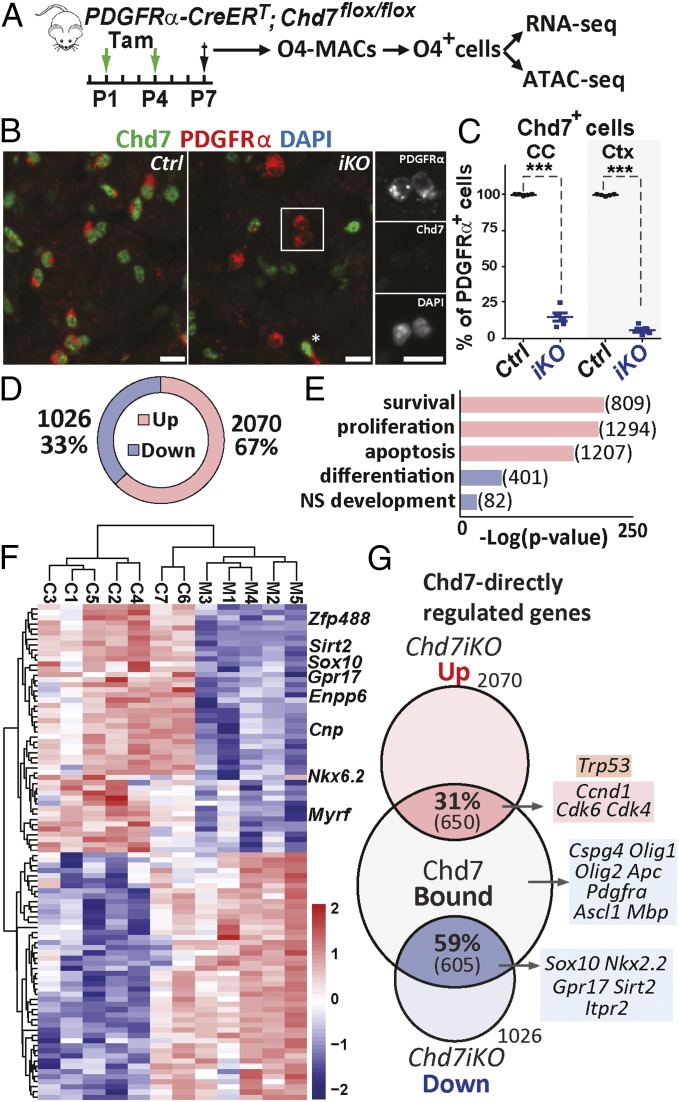
To investigate whether the expression of Chd7-bound genes in OPCs was modified upon Chd7 LOF, we induced Chd7-conditional deletion (Chd7iKO) in neonatal OPCs, using PDGFRa-CreERT and Chd7Flox alleles (32, 33) (Fig. 2A). Neonatal tamoxifen administration allowed us to delete Chd7 in ∼90% of OPCs, as shown by Chd7 immunofluorescence and qRT-PCR at P7 (Fig. 2 B and C and SI Appendix, Fig. S3A). We then purified O4+ cells from Chd7iKO (PDGFRα-CreERT; Chd7Flox/Flox) or control (Chd7Flox/Flox) (Fig. 2A) P7 cortices. Because the proportion of OL stages (80% of OPCs and 20% iOLs) was not altered by the loss of Chd7 (SI Appendix, Fig. S2C), we could collect comparable oligodendroglial cell populations from mutant and control brains.
Fig. 2.
Chd7-regulated genes involved in OPCs proliferation, differentiation, and survival. (A) Diagram representing tamoxifen (Tam) administration to control (Ctrl) and Chd7iKO (iKO) mice at P1 and P4 followed by MACS-sorting of O4+ cells at P7. Cells are then used for either RNA-seq or ATAC-seq. (B) Immunostaining of P7 brain sections showing that all PDGFRα+ OPCs express Chd7 in the CC of Ctrl mice, while only a few OPCs maintain Chd7 expression in iKO mice. Star shows a Chd7-expressing OPC in iKO. (Scale bars, 10 μm.) (C) Quantification of Chd7+ OPCs as a percentage of total PDGFRα+ cells in the CC and cortex (Ctx) of P7 Ctrl and iKO mice. Data are presented as mean ± SEM (n = 5). Exact P values can be found in Dataset S2. ***P < 0.001. (D) Pie chart showing the relative percentage and number of genes that were significantly up-regulated or down-regulated in P7 iKO O4+ cells compared with Ctrl (fold-change > 1.2; P < 0.05). (E) GO analysis of the up-regulated (red) and down-regulated (blue) genes in Chd7iKO cells compared with Ctrl. Numbers indicate the number of genes of each category. (F) Heatmap representing the expression of 100 most different genes in Ctrl and iKO O4+ cells (n = 7 Ctrl and 5 iKO). (G) Venn diagram depicting the overlap of Chd7-binding genes in OPCs with down-regulated (blue) or up-regulated (red) genes in iKO cells with examples of Chd7-bound genes involved in OL differentiation (blue), cell death (orange), and cell cycle (red).
The impact of Chd7 LOF on gene transcription was assessed by genome-wide transcriptome analysis (RNA-seq) (Fig. 2A). Surprisingly, despite Chd7 being described mostly as an activator of transcription (17, 18, 21), many deregulated genes in Chd7iKO OPCs (3,096 genes, fold-change > 1.2 and P < 0.05) were up-regulated (67%, 2,070 genes) (Fig. 2D), while only 33% were down-regulated (1,026 genes) (Fig. 2D). GO analysis indicated that many up-regulated genes were associated with cell survival, apoptosis, and cell proliferation (Fig. 2E and SI Appendix, Fig. S3 B–D). In contrast, genes associated with OL differentiation were exclusively found among down-regulated genes (Fig. 2F and SI Appendix, Fig. S3 C and D). Almost all genes Chd7-bound and deregulated in Chd7iKO OPCs had a dynamic expression pattern during OPC differentiation (SI Appendix, Fig. S3E). On the one hand, one-third (31%) of up-regulated genes were bound by Chd7, including cell-cycle regulators (e.g., Ccnd1, Cdk4 and Cdk6) and regulators of cell survival/apoptosis (e.g., p53/Trp53, Bax, Apaf1) (Fig. 2G). On the other hand, a majority of down-regulated genes (59%) were bound by Chd7, including genes involved in OPC differentiation (such as Sox10, Nkx2.2, Gpr17, and Sirt2) (Fig. 2G). Therefore, Chd7 regulates OPC gene transcription by both repressing apoptosis/proliferation genes and activating differentiation genes.
Chd7 and Chd8 Bind to the Same Regulatory Regions of OPC Differentiation, Proliferation, and Survival Genes.
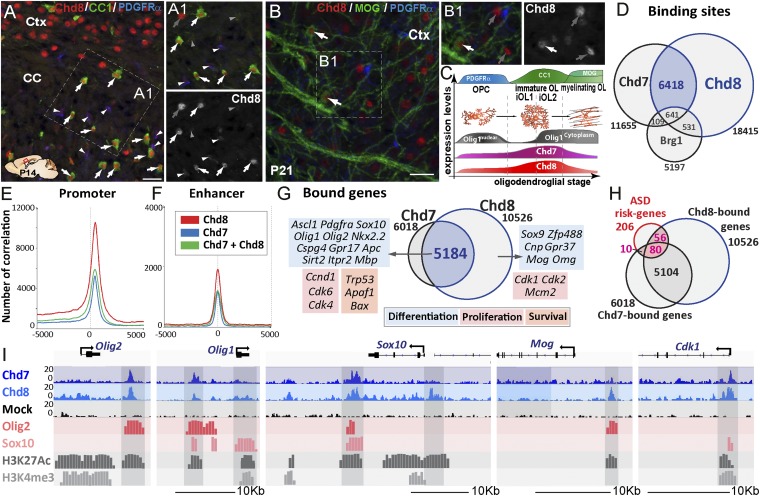
Combined Chd7 chromatin-binding and transcriptomics analysis of Chd7 mutant OPCs indicate that, despite Chd7 binding to many genes involved in different OPC functions, only a fraction of these genes are deregulated upon Chd7 deletion (Fig. 2G). We therefore asked if Chd8 cooperates with Chd7 and compensates for Chd7 LOF in the regulation of Chd7-bound genes. We thus investigated Chd8 protein expression in the postnatal brain. Indeed, Chd7 and Chd8 proteins showed similar expression patterns in postnatal brain oligodendroglia; Chd8 protein was detected in PDGFRα+ OPCs (Fig. 3 A and C, arrowheads) and found at higher levels in differentiating OLs (CC1high cells) (Fig. 3 A and C, arrows), and its levels decreased in mature MOG+ OLs (Fig. 3 B and C, arrows), in agreement to Chd8 transcript pattern in postnatal brain cells shown by RNA-seq databases (34, 35). Furthermore, upon LPC-induced focal demyelination of the adult corpus callosum (CC), Chd8 was detected at high levels in PDGFRα+ OPCs and iOLs (CC1+/Olig1+ cells) in/around lesions during remyelination (SI Appendix, Fig. S4 A–E), similar to Chd7 expression (SI Appendix, Fig. S1 J and K). Chd8 protein may thus compensate for Chd7 LOF in differentiating OPCs/iOLs.
Fig. 3.
Chd8 binds together with Chd7 to OPC differentiation, proliferation, and survival genes. (A) Chd8 immunolabeling of P14 brain sections showing Chd8 expression in all maturing OLs (CC1high-expressing cells, arrows) of the CC, as well as in neurons of the cortex (Ctx). (A1) Detail of the Inset in A showing that, beside the high Chd8 expression in maturing OLs (white arrows) and neurons (gray arrows), a low level of Chd8 expression is detected in few OPCs (PDGFRα+ cells, white arrowheads) but is hardly detectable in astrocytes (CC1low-expressing cells, gray arrowheads). (Scale bar, 20 μm.) (B) Immunofluorescence at P21 showing young mature MOG+ OLs still expressing Chd8 (white arrows). (B1) Detail of the Inset shown in B; gray arrows correspond to Chd8+ neurons. (Scale bar, 20 μm.) (C) Summary representation of Chd8 and Chd7 expression at different stages of the OL cell lineage, as identified by PDGFRα, CC1, and MOG expression. (D) Venn diagrams depicting the overlap of Chd7-, Chd8-, and Brg1-binding sites in OPCs. (E and F) Graph showing the number of correlations of Chd7 (blue), Chd8 (red), and Chd7-Chd8 (green) peaks in OPCs compared with the position in promoter regions (E) or enhancer regions (F). (G) Overlap of Chd7- and Chd8-bound genes in OPCs with examples of genes involved in OL cell differentiation (blue), cell death (orange), and cell cycle (red). (H) Overlap between ASD-risk genes and Chd7- and Chd8-bound genes in OPCs. (I) Representative ChIP-seq tracks for Chd8 and control IgG together with Olig2, Sox10, Chd7, and active epigenetic marks (H3K27ac and H3K4me3) in Olig2, Olig1, Sox10, Mog, and Cdk1 gene regions in OPCs.
To investigate whether Chd7 and Chd8 regulate common OPC genetic programs, a genome-wide Chd8 chromatin-binding profile was established from in vivo purified OPCs and then compared with Chd7-binding sites. Chd8 and Chd7 share many sites (6,418 regions) (Fig. 3D), representing 55% of Chd7-bound regions. A majority (57.6%) of Chd8-binding sites were found in promoter regions (Fig. 3E), consistent with reports of other cell types (22, 36). In addition, Chd8, like Chd7, also bound a large number of OPC active enhancers (Fig. 3F). Interestingly, the majority of Chd7-bound genes were also bound by Chd8 (86%, 5,184 genes), including genes associated with cell differentiation, proliferation, and survival (Fig. 3G). Furthermore, the genes encoding well-known regulators of oligodendroglial development (Ascl1, Nkx2.2, Olig1, Olig2, Sox10), oligodendroglial markers (Pdgfra, Cspg4, Gpr17, Apc, Itpr2, Mbp), R checkpoint regulators (Ccnd1, Cdk4, Cdk6), and apoptotic/survival genes (p53, Bax, Apaf1) were all bound by both Chd7 and Chd8 (Fig. 3 G and I). Of note, Chd7 and Chd8 share very few binding sites with Brg1 (Fig. 3D), another chromatin remodeler that cooperates with Olig2 to control OL differentiation (15), suggesting that Chd7/Chd8 and Brg1 may regulate distinct OPC differentiation processes. Given that we integrated ChIP-seq datasets obtained from rat and mouse, we asked whether binding is conserved between species. To address this question, we compared the Chd8-binding profile between our in vivo mouse-purified O4+ cells with rat OPC primary cultures (37) and observed a large overlap (87%) (SI Appendix, Fig. S5B) of bound genes between species, comparable to the reported overlap of Chd8 ChIP-seq replicates from the same sample (36, 38). Furthermore, global alignment of Chd7-, Chd8-, Olig2-, and Sox10-binding peaks to active regulatory regions found in oligodendroglial cells, but not in other neural cells (Fig. 3I and SI Appendix, Figs. S5–S8), strongly support cell-type–specific binding conservation to regulatory elements in orthologous chromosomic regions, consistent with conservation in TF-to-TF connections and regulatory networks found in mammals by the ENCODE project (39). Taken together, our findings show that Chd7 and Chd8 chromatin remodelers, acting together or functionally compensating for each other, regulate the genetic networks controlling OPC proliferation, survival, and differentiation.
Finally, we assessed whether Chd7 and Chd8 binding to OPC genes was enriched among 201 ASD risk genes described previously (40, 41). A large number of them were Chd7/Chd8-bound genes in OPCs (90 of 201 and 136 of 201, respectively) (Fig. 3H and Dataset S1), but only Chd7 was found to be significantly enriched over its genome-wide binding to protein-coding genes (P < 0.0034, hypergeometric test), suggesting that OPCs may contribute to brain developmental defects in ASD patients displaying mutations in either CHD8, CHD7, or their targets among ASD-risk genes.
Chd7 LOF Reduces Brain OPC Pool Without Impacting OPC Proliferation.
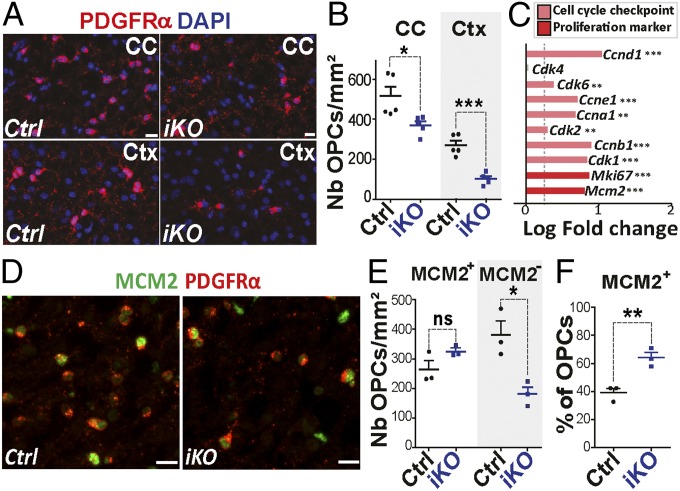
To determine the consequences of Chd7 LOF in OPC population, we quantified PDGFRα+ OPCs in P7 brains and found that Chd7iKO brains display less numerous OPCs in the CC and cortex, compared with controls (∼30% and ∼60%, respectively) (Fig. 4 A and B). Chd7 deletion thus affects either OPC proliferation or survival. To assess the possible impact of Chd7 deletion on OPC proliferation, we examined proliferation-associated genes in our OPC RNA-seq datasets and found an up-regulation of key regulators of both cell-cycle checkpoints (Ccnd1, Cdk6, Ccne1, Ccna1, Cdk2, Ccnb1, Cdk1) and proliferation markers (Mki67, Mcm2) (Fig. 4C). Therefore, we assessed the proliferative status of Chd7iKO OPCs in P7 brains. Surprisingly, the number of Mcm2+ or Ki67+ proliferating OPCs in the CC and cortex was similar between Chd7iKO and controls (Fig. 4 D and E and SI Appendix, Fig. S9 A–E). We then addressed possible changes in cell-cycle length measuring the proportion of OPCs in S-phase (labeled by short pulse of BrdU) among cycling OPCs (MCM+/PDGFRα+ cells) and found no changes in cell-cycle length between Chd7iKO and controls at P7 (SI Appendix, Fig. S9 G and H).
Fig. 4.
Chd7 LOF in OPCs does not affect cell proliferation. (A) Immunolabeling of PDGFRα+ cells in the CC and cortex (Ctx) of P7 Control (Ctrl) and Chd7iKO (iKO) mice. (Scale bars, 10 μm.) (B) Quantification of PDGFRα+ cells (nb/mm2) in the CC and Ctx of P7 Ctrl and iKO mice. Data are presented as mean ± SEM (n = 5). (C) Barplot representing the log fold-change (LogFC) of genes involved in cell cycle and proliferation between iKO and Ctrl mice. Dashed gray line represent FC = 1.2 (n = 7). (D) Immunostaining of MCM2 and PDGFRα in the CC from P7 Ctrl and iKO mice. (Scale bars, 10 μm.) (E) Quantification of the density of MCM2+ and MCM2− PDGFRα+-OPCs (nb/mm2) in the CC of P7 Ctrl and iKO mice. Data are presented as mean ± SEM (n = 3). (F) Quantification of MCM2+ cells as a percentage of total PDGFRα+ cells in the CC of P7 Ctrl and iKO mice. Data are presented as mean ± SEM (n = 3). Exact P values can be found in Dataset S2. *P < 0.05, **P < 0.01, and ***P < 0.001.
These results led us to hypothesize that the up-regulation of cell-cycle genes may result from a selective reduction of nonproliferating OPCs. The number of cycling and noncycling OPCs was counted in Chd7iKO and control brains, showing a reduction of nonproliferative OPCs (Mcm2−/PDGFRα+ cells) (Fig. 4 D and E and SI Appendix, Fig. S9B), leading to an increased MCM2+ fraction of OPCs (Fig. 4F and SI Appendix, Fig. S9C) in Chd7iKO brains. Similar results were obtained upon Chd7 deletion in OPCs at a later time point (SI Appendix, Fig. S9 I and J). Therefore, Chd7-deficient brain OPCs are not impacted in their proliferation or cell-cycle length. In contrast, nonproliferative Chd7-deficient OPCs are reduced in numbers, suggesting that Chd7 may regulate OPC survival.
Chd7 Promotes OPC Survival by Chromatin-Closing and Transcription Repression of p53.
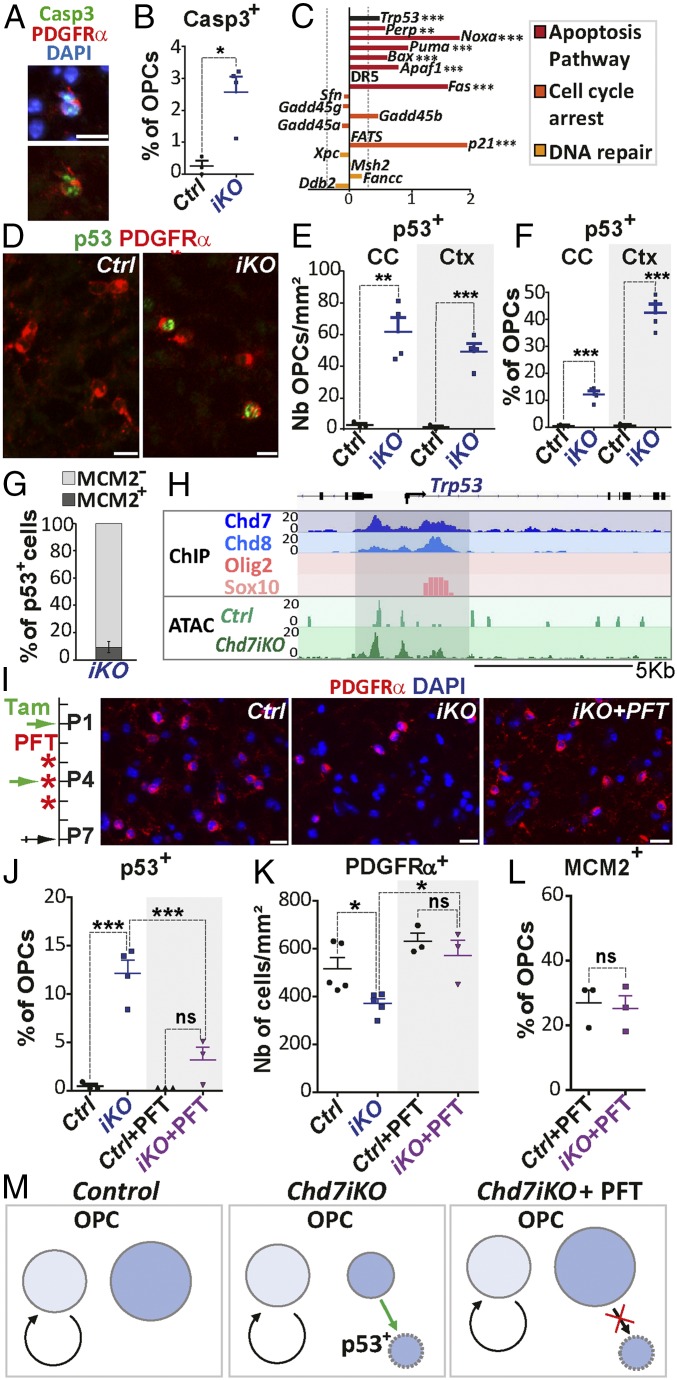
We next investigated whether OPCs underwent abnormal apoptosis in P7 Chd7iKO brains. Despite being actively removed by microglia (SI Appendix, Fig. S9K), apoptotic OPCs labeled with antiactivated Caspase3-recognizing antibodies (Casp3+/PDGFRα+ cells) were observed in Chd7iKO brains, while they were rare in controls (Fig. 5 A and B). Furthermore, the apoptosis GO category was highly enriched among the up-regulated transcripts of purified OPCs from Chd7iKO brains compared with controls (RNA-seq) (Figs. 2E and 5C). Notably, transformation-related protein 53 (Trp53 also named p53), a main regulator of apoptosis, and recently linked to Chd7-mediated defects in CHARGE syndrome (42), was among the up-regulated transcripts (Fig. 5C) and confirmed by qRT-PCR (SI Appendix, Fig. S10A). Several p53 target genes involved in apoptosis (e.g., Bax, Apaf1) were also up-regulated, but not those involved in cell-cycle arrest (expecting p21) or DNA repair (Fig. 5C). We therefore assessed for p53 protein up-regulation in P7 cerebral OPCs and found p53+ OPCs (15% of CC OPCs and 40% of cortical OPCs) in Chd7iKO brains but not in controls (Fig. 5 D–F). Interestingly, 90% of Chd7iKO OPCs expressing p53 were postmitotic (Mcm2−/PDGFRα+ cells) (Fig. 5G), confirming that loss of Chd7 results in specific apoptosis of noncycling OPCs likely mediated by p53 up-regulation (Fig. 5M).
Fig. 5.
Chd7 promotes OPC survival through p53 down-regulation. (A) Immunostaining of Casp3 and PDGFRα in the CC of P7 iKO mice. (Scale bar, 10 μm.) (B) Quantification of Casp3+ OPCs as a percentage of OPCs in P7 Ctrl and iKO mice. Data are presented as mean ± SEM (n = 3 Ctrl and 4 iKO). (C) Barplot representing the LogFC of Trp53 and p53 target-genes involved in apoptosis pathway (red), cell-cycle arrest (orange), or DNA repair (yellow) between iKO and Ctrl mice. Dashed gray line represents FC = 1.2 (n = 7 Ctrl and 5 iKO). (D) Immunostaining of p53 and PDGFRα in the CC from P7 Ctrl and iKO mice. (Scale bars, 10 μm.) (E and F) Quantification of p53+ OPCs (nb/mm2, E) or as a percentage of OPCs (F) in the CC and Ctx of P7 Ctrl and iKO mice. Data are presented as mean ± SEM (n = 3 Ctrl and 4 iKO). (G) Quantification of MCM2+ and MCM2− cells as a percentage of total p53+ cells in the CC of iKO P7 mice. Data are presented as mean ± SEM (n = 3 Ctrl and 4 iKO). (H) Representative tracks for Trp53 locus integrating: (i) ChIP-seq data for Olig2 and Sox10, Chd7, and Chd8 in OPCs and; (ii) ATAC data from Ctrl and iKO P7 OPCs. (I, Left) Diagram representing tamoxifen (Tam) administration at P1 and P4 and PFT injection at P3, P4, and P5 followed by tissue collection at P7. (Right) Immunostaining of PDGFRα in the CC from P7 Ctrl, iKO and iKO+PFT mice. (Scale bars, 10 μm.) (J) Quantification of p53+ cells as a percentage of OPCs in the CC of P7 Ctrl, iKO, Ctrl+PFT, and iKO+PFT mice. Data are presented as mean ± SEM (n = 3 Ctrl, 4 iKO, 3 Ctrl+PFT and 3 iKO+PFT). (K) Quantification of PDGFRα+ cells (nb/mm2) in the CC of P7 Ctrl, iKO, Ctrl+PFT and iKO+PFT mice. Data are presented as mean ± SEM (n= 5 Ctrl, 5 iKO, 3 Ctrl+PFT and 3 iKO+PFT). (L) Quantification of MCM2+ cells as a percentage of OPCs in the CC of P7 Ctrl+PFT and iKO+PFT mice. Data are presented as mean ± SEM (n = 3). (M) Scheme representing Chd7 function in OPC survival. Proliferative OPCs (light blue), noncycling OPCs (blue) in Ctrl, iKO, and iKO+PFT mice. Exact P values can be found in Dataset S2. *P < 0.05, **P < 0.01, and ***P < 0.001.
To obtain more insight into Chd7 regulation of Chd7-bound genes, we compared the chromatin accessibility of these genes between Chd7iKO and control OPCs. Therefore, we generated a genome-wide chromatin accessibility profiling by Tn5 transposase-accessible chromatin with high-throughput sequencing (ATAC-seq) combined with sequencing (43), from purified O4+ cells from either Chd7iKO or control cortices (Fig. 2A). We obtained a high genome-wide correlation between transcript expression and chromatin accessibility in the transcription start sites, indicating efficient open chromatin mapping (SI Appendix, Fig. S10B). Interestingly, the promoter elements bound by Chd7 and Chd8 in the p53/Trp53 locus showed larger chromatin accessibility in Chd7iKO OPCs compared with controls (Fig. 5H), suggesting that Chd7 normally represses p53/Trp53 transcription by binding and inducing chromatin closing at these regulatory elements.
To demonstrate that p53 up-regulation was responsible for Chd7iKO OPCs apoptosis, we used Pifithin-α (PFT) (Fig. 5I), a p53 inhibitor that impedes p53 activation (44). Interestingly, treatment with PFT reduced the number of p53+ OPCs (Fig. 5J) and rescued the normal density of OPCs in Chd7iKO CC (Fig. 5 I and K), indicating that Chd7 promotes OPC survival by inhibiting p53-mediated apoptosis. Notably, rescuing noncycling OPCs permitted to recover a similar proportion of MCM2+ OPCs between control and Chd7iKO brains (Fig. 5L), confirming that Chd7-deletion does not have an impact on brain OPC proliferation. Together, these results indicate that Chd7 contributes to the survival of postmitotic OPCs by inhibiting p53/Trp53 through chromatin closing at its promoter elements (Fig. 5M).
Chd7 Is Required to Induce OPC Differentiation but Not OL Stage Progression, by Chromatin-Opening, and Gene Activation of Sox10, Nkx2.2, and Gpr17.
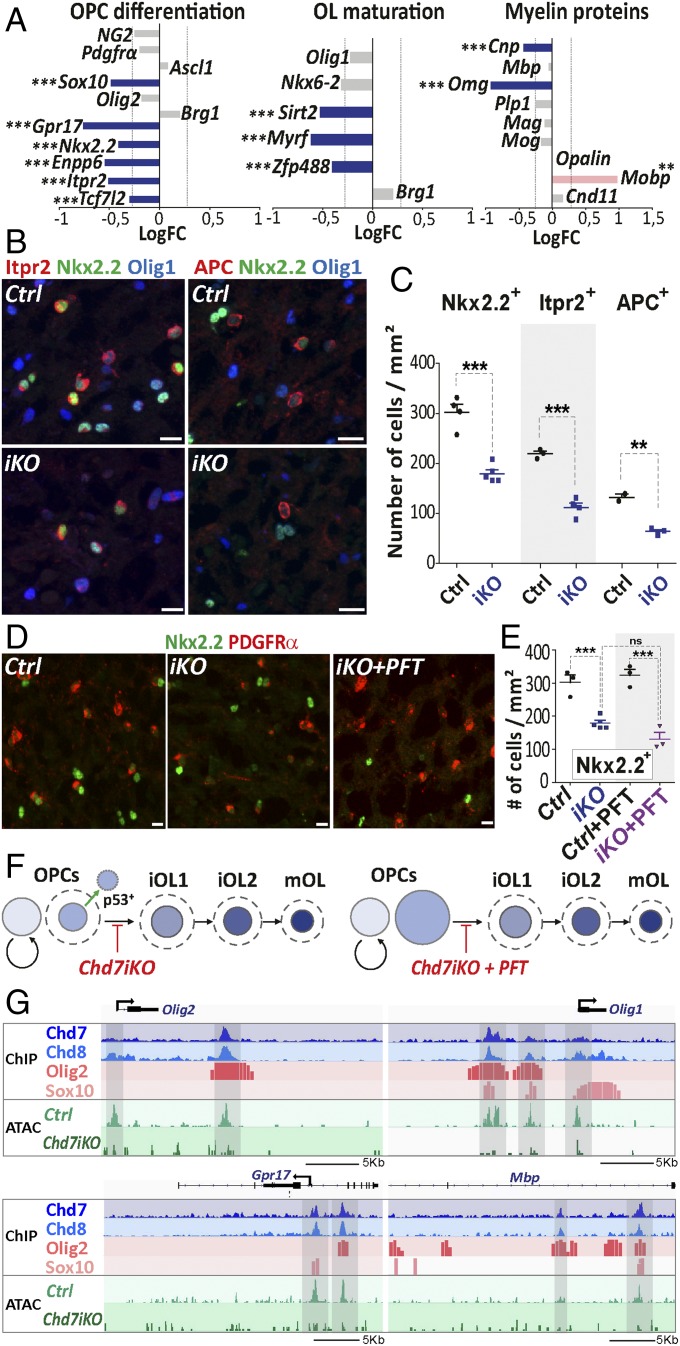
We then assessed for the Chd7 role in OPC differentiation that was not been specifically addressed in our previously study (28). In depth analysis of down-regulated transcripts in Chd7iKO oligodendroglia showed reduced expression of key regulators of OPC differentiation, such as Sox10, Gpr17, Nkx2.2, and Tcf7l2 (Fig. 6A). Chd7 LOF also impacted the expression of some regulators of OL maturation (Sirt2, Myrf, Zfp488) and in myelin protein genes (Cnp, Omg) (Fig. 6A). These results were confirmed by qPCR analysis (SI Appendix, Fig. S10A). Therefore, we quantified the different OL cell types in the CC of Chd7iKO mice at postnatal stage P7, a peak time for cortical OPC differentiation. P7 Chd7iKO mice showed a strong decrease (∼40%) of Nkx2.2+ iOLs compared with control brains (Fig. 6 B and C), indicating that Chd7 deletion in OPCs impairs normal OPC differentiation. The OPC/iOL ratio was, however, not modified in Chd7iKO brains, due to a similar reduction in OPC and iOL populations (SI Appendix, Fig. S10 C and D). The number of Nkx2.2+, Itpr2+ (35), and APC+ (45) cells, which we show by immunofluorescence, mark successive stages of OL cell lineage according to their transcript expression pattern (SI Appendix, Fig. S10 E–H), were similarly decreased (Fig. 6 B and C), indicating that Chd7 loss do not alter OL stage progression once OPC differentiation is started. In agreement with this interpretation, we found a similar CC1/Olig1 immunofluorescence, which distinguishes three different stages of OL differentiation (13), between Chd7iKO mice and controls (SI Appendix, Fig. S11). Taken together, these data indicate that Chd7 deletion impairs OPC capacity to start differentiation but not the normal progression once differentiation has started (Fig. 6F).
Fig. 6.
Chd7 promotes the expression of genes involved in OPC differentiation and maturation. (A) Barplot of the LogFC of genes involved in OPC differentiation, OL maturation, or coding for myelin proteins between P7Chd7iKO (iKO) and Ctrl. The dashed gray line represents fold-change = 1.2 (n = 7 Ctrl and 5 iKO). (B) Immunostaining of Itpr2 and APC together with Nkx2.2 and Olig1 in the CC from P7 control (Ctrl) and iKO (iKO) mice. (Scale bars, 10 μm.) (C) Quantification of Nkx2.2+, Itpr2+, and APC+ cells (nb/mm2) in the CC of P7 Ctrl and iKO mice. Data are presented as mean ± SEM (n = 4 Ctrl and 5 iKO). (D) Immunostaining of PDGFRα and Nkx2.2 in the CC from P7 Ctrl, iKO, and iKO+PFT mice. (Scale bars, 10 μm.) (E) Quantification of Nkx2.2+ cells (nb/mm2) in the CC of P7 Ctrl, iKO, Ctrl+PFT and iKO+PFT mice. Data are presented as mean ± SEM (n = 3 Ctrl, 5 iKO, 3 Ctrl+PFT and 3 iKO+PFT). (F) Summary representation of OL cell lineage progression in P7 iKO and iKO+PFT, mice. Circle size is proportional to the size of OL lineage cell-type population, as quantified in the CC. Dotted lines: OL population size in Ctrl. (G) Representative tracks for Olig1, Olig2, Gpr17, and Mbp locus integrating: (i) ChIP data for main oligodendroglial TFs (Olig2 and Sox10) and chromatin remodeling factors (Chd7, Chd8) in OPCs and; (ii) ATAC data from Ctrl and iKO P7 OPCs. Exact P values can be found in Dataset S2. *P < 0.05, **P < 0.01, and ***P < 0.001.
To assess whether OPC apoptosis was sufficient to account for the reduction of differentiating OLs in Chd7iKO brains, or if Chd7 functions in survival and differentiation could be separated, we rescued OPC survival and numbers by PFT treatment (Fig. 5 I and K). Remarkably, the number of iOLs (Nkx2.2+ cells) in the CC was the same in PFT-treated and untreated Chd7iKO mice (Fig. 6 D and E). Therefore, the rescue of OPC survival is not sufficient to recover normal differentiation of Chd7iKO OPCs, indicating that Chd7 functions in OPC survival and differentiation can be dissociated (Fig. 6F).
To address Chd7 remodeling function in the loci involved in OPC differentiation, we compared their chromatin accessibility in Chd7iKO OPCs and controls. Interestingly, OL differentiation genes bound by Chd7, such as Olig2, Olig1, and Gpr17 (coding for a G protein-coupled receptor involved in OL differentiation), showed a decreased chromatin accessibility at regulatory regions in Chd7 mutant OPCs compared with controls (Fig. 6G). Therefore, our findings indicate that Chd7 promotes chromatin opening of key factors involved in OPC differentiation.
OPC Differentiation Requires both Chd7 and Chd8 Remodelers.
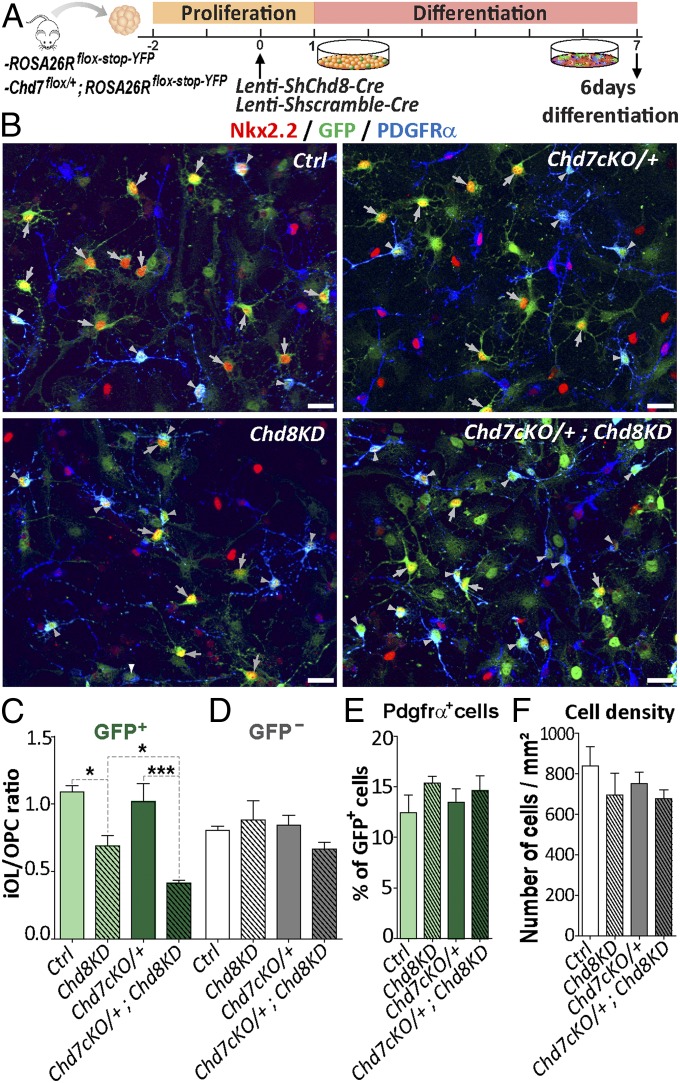
To study whether Chd8 and Chd7 are required during OPC differentiation, we transfected neural progenitors derived from the neonatal SVZ of Rosa26Rflox-stop-YFP or Chd7flox/+;Rosa26Rflox-stop-YFP mice with lentivirus-expressing Cre-recombinase together with either Chd8 shRNA or scramble shRNA (Fig. 7A). After 3 d in differentiation conditions favoring oligodendroglial differentiation (40% of cells) (SI Appendix, Fig. S12 A–C), Chd7 and Chd8 levels were found to be successfully reduced in Chd8KD (shChd8) and Chd7cKO/+ (Chd7flox/+) conditions compared with control (ShScramble), but the number of OPCs was not altered (SI Appendix, Fig. S12 D–H). At 6 d of differentiation, we found many iOLs expressing strong levels of both CNP and Nkx2.2 (SI Appendix, Fig. S12I). Interestingly, while all genotypes showed comparable numbers of PDGFRα+ OPCs and cell densities (Fig. 7 B and F), the ratio iOL/OPCs (Nkx2.2+/PDGFRα+ cells) was reduced by 36% in Chd8KD cells (Fig. 7 B and C), suggesting a requirement of Chd8 in OPC differentiation. Furthermore, while the loss of one Chd7 allele (Chd7cKO/+ cells) did not modify their OPC differentiation, its combination with Chd8KD further reduced iOL/OPC ratio (Fig. 7 B and C). These results thus indicate that Chd7 and Chd8 genetically interact in the process of OPC differentiation and suggest that Chd8 may partially compensate the loss of Chd7 during OPC differentiation.
Fig. 7.
OPC differentiation requires both Chd7 and Chd8 remodelers. (A) Diagram representing transduction protocol used on neural stem cells (NSCs) with a lentivirus-expressing Cre and a scramble shRNA or a shRNA against Chd8 and followed by fixation after 6 d of differentiation. (B) Immunostaining of Nkx2.2 and PDGFRα together with GFP in Ctrl (ROSA26Rstop-floxed-YFP +scramble), Chd8KD (ROSA26Rstop-floxed-YFP +Chd8 shRNA), Chd7cKO/+ (Chd7Flox/+;ROSA26Rstop-floxed-YFP +scramble), or Chd7cKO/+;Chd8KD (Chd7Flox/+;ROSA26Rstop-floxed-YFP +Chd8 shRNA) conditions. White arrowheads represent PDGFRα+-GFP+ OPCs and gray arrowheads represent Nkx2.2+-GFP+ iOLs. (Scale bars, 20 μm.) (C and D) Ratio between Nkx2.2+-GFP+ iOLs and PDGFRα+-GFP+ OPCs (C) and between Nkx2.2+-GFP− iOLs and PDGFRα+-GFP− OPCs (D) in Ctrl, Chd8KD, Chd7cKO/+, and Chd7cKO/+;Chd8KD conditions. Data are presented as mean ± SEM (n = 4). (E) Quantification of PDGFRα+-GFP+ cells as a percentage of total GFP+ cells in Ctrl, Chd8KD, Chd7cKO/+, and Chd7cKO/+;Chd8KD conditions. Data are presented as mean ± SEM (n = 3). (F) Quantification of cells (nb/mm2) in Ctrl, Chd8KD, Chd7cKO/+, and Chd7cKO/+;Chd8KD conditions. Data are presented as mean ± SEM (n = 3). Exact P values can be found in Dataset S2. *P < 0.05, **P < 0.01, and ***P < 0.001.
Chd7/Chd8 and Olig2/Sox10 Bind to Regulatory Elements of Active Stage-Specific Genes in Oligodendroglia.
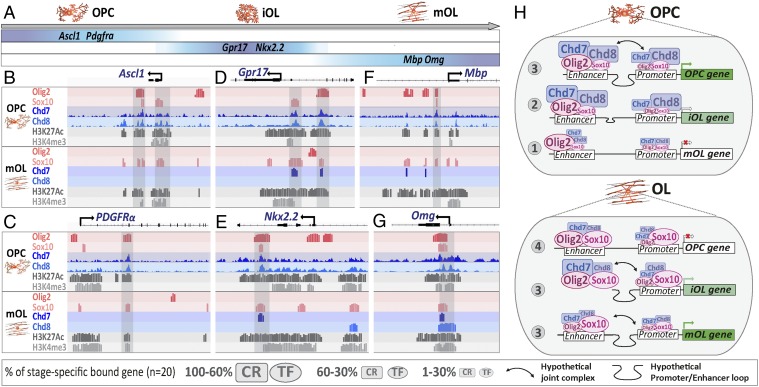
Finally, we further investigated the mechanisms by which Chd7/Chd8 control gene expression in a timely manner during the course of OL cell lineage differentiation, integrating different datasets. An oligodendroglia single-cell transcriptome database (35) was used to classify genes according to their expression timing (Fig. 8A). Focusing on oligodendroglial stage-specific gene expression, we classified oligodendroglial-genes into three groups: (i) “OPC-genes” expressed in OPCs and down-regulated upon differentiation (e.g., Ascl1, Cspg4, Pdgfra); (ii) “iOL-genes” up-regulated and maintained upon early OPC differentiation (COPs of ref. 35; e.g., Nkx2.2, Gpr17, Itpr2); and (iii) “mOL-genes” up-regulated only in maturing OLs (NFOLs and mOLs; e.g., Mbp, Mog, Omg).
Fig. 8.
Chd7/Chd8 and Olig2/Sox10 bind to regulatory elements of active stage-specific genes in oligodendroglia. (A) Diagram representing a set of genes with timely controlled expression (data from ref. 35) divided in three groups: OPC genes (Ascl1 and Pdgfrα), iOL genes (Nkx2.2 and Gpr17), and mOL genes (Mbp and Omg). (B–G) Schematic representation from Genomatix genome browser of Ascl1 (B), Pdgfrα (C), Gpr17 (D), Nkx2.2 (E), Mbp (F), and Omg (G) locus integrating ChIP-seq data for main oligodendroglial TFs (Olig2 and Sox10), chromatin remodeling factors (Chd7, Chd8), and active epigenetic marks (H3K27ac and H3K4me3) in OPCs and OLs. (H) Synthesis representation of the dynamic cooperation between TFs (Sox10, Olig2) and chromatin remodelers (Chd7, Chd8) for the timely regulation of OPC- and OL-gene expression with genes not yet expressed (1), genes starting to express (2), expressed genes (3), and genes not expressed anymore (4).
We then integrated the chromatin-binding profiles for key TFs (Sox10 and Olig2), chromatin remodelers (Chd7 and Chd8), and active histone marks (H3K4me3 and H3K27ac) (Fig. 8 and SI Appendix, Fig. S13) (present study and refs. 15 and 28) to analyze genes of each group (OPC-, iOL-, and mOL-genes’; n = 20) (SI Appendix, Table S1). This integrative approach allowed us to identify four regulatory states, which are recapitulated in Fig. 8H and outlined as follows. State 1: Among not-yet-expressed genes, such as the majority of mOL-genes in OPCs, only a few are bound by Chd7, Chd8, and Sox10, while all are bound by Olig2 in agreement with the pioneer factor function of Olig2 (15). State 2: Among genes at their expression-onset (that start to be expressed), such as iOL-genes in OPCs, all are bound by Chd8, Chd7, and Olig2 (to less extent by Sox10) at enhancers but only a few at their cognate promoters. State 3: Among robustly expressed genes, such as most OPC-genes in OPCs and iOL- and mOL-genes in OLs, all are bound by all factors (Chd7, Chd8, Olig2, and Sox10) both at enhancers and their cognate promoters, suggesting that the presence of all these factors drive active gene transcription, with Sox10 preferentially binding active genes in OLs compared with OPCs. State 4: Among silenced genes (not expressed anymore), such as OPC-genes in OLs, all are still bound by Olig2 and Sox10, while a majority of them have lost Chd7 and Chd8 binding, suggesting that gene silencing is marked by, or results from, the lack of binding of chromatin remodelers, and not of TFs.
We therefore propose a dynamic model of cooperation between Chd7/Chd8 remodelers and Olig2/Sox10 key oligodendrogenic TFs to select and timely tune the gene-expression program at different OL lineage stages, in which the present of chromatin remodelers is key for robust gene transcription (SI Appendix, Fig. S14).
Discussion
Mounting evidence indicates that chromatin remodeling factors play important roles in normal development and neurodevelopmental diseases (46), with CHD7 haploinsuficiency being the cause of CHARGE syndrome (47) and CHD8 being one of the strongest ASD high-risk–associated genes (25). It is thus of relevance for human health to unravel the mechanisms controlled by Chd7/Chd8 chromatin remodelers and to understand how their haploinsuficiency leads to developmental brain pathology involved in these diseases. The present study addresses the mechanisms of gene-expression control by Chd7/Chd8 in OPCs, by generating and integrating genome-wide transcriptome, chromatin binding, and chromatin accessibility profiles of purified OPCs, isolated from control and OPC-specific Chd7-deficient mice. These experiences led to the following conclusions (SI Appendix, Fig. S15). First, Chd7 and Chd8 bind together to regulatory regions of genes involved in OPC differentiation, proliferation, and survival, suggesting that they regulate these different biological processes. Second, Chd7 protects OPCs from cell death through chromatin closing and direct transcriptional repression of the p53/Trp53 promoter. Third, Chd7 is partially required to induce OPC differentiation through chromatin opening and transcriptional activation of key regulators of this process, although being dispensable for OL stage progression, consistent with Chd8 compensatory function, as suggested by Chd7/Chd8 common chromatin-binding profile and genetic synergy during OPC differentiation. Finally, integration of genome-wide occupancy repertoires and associated epigenetic signals from OL lineage cells suggests that Chd7/Chd8 remodelers and Olig2/Sox10 TFs cooperate to activate oligodendroglia stage-specific genes.
Chd7 Promotes OPC Survival Through Direct p53/Trp53 Down-Regulation.
Our timely controlled Chd7 deletion in OPCs provided insights into Chd7 function in proliferation and survival that were not directly addressed in our previous work using this time-controlled Chd7 deletion (29). Interestingly, we found that Chd7 OPC-specific deletion leads a selective apoptosis of noncycling OPCs and that the p53/Trp53 transcripts were up-regulated in Chd7-deleted OPCs. Notably, Trp53 heterozygosity has been recently shown to partially rescue the phenotypes of Chd7-null mouse embryos, and p53 inappropriate activation leads to CHARGE syndrome traits (42). Here, we provide evidence that in vivo, Chd7 directly represses p53/Trp53 expression in OPCs by binding and inducing chromatin closing at its promoter. These results are consistent with recent in vitro evidence for Chd7 binding to the p53/Trp53 promoter in neural crest cells (42). Using a p53 inhibitor (PFT), we were able to prevent p53/Trp53 induction and cell death, thus restoring the pool of OPCs. It is worth noting that rescuing normal OPC numbers was not sufficient to restore normal OPC differentiation, indicating that Chd7 function in OPC survival can be dissociated from its role in promoting OPC differentiation.
The above results point out PFT as a potential treatment to reduce p53 inappropriate expression in CHARGE patients. However, our PFT treatment rescued normal OPC numbers but not normal OPC differentiation, suggesting that targeting gene-networks promoting cell-differentiation may also be required to restore CHD7 function in CHARGE patients. The Chd7 direct repression of p53/Trp53 in OPCs also has a potential interest in MS because MS brain tissues up-regulate p53 expression (48), suggesting that a transient, and if possible brain-restricted, administration of PFT to avoid possible proinflammatory effects (49), may help in the treatment of MS patients.
Diverse Chd7 Activities Regulate Different OPC Functions.
Our ChIP-seq analyses on brain-purified OPCs allowed us to produce an in vivo genome-wide chromatin-binding profiling available for Chd7 and Chd8. In addition, the comparative transcriptome (RNA-seq) and chromatin accessibility (ATAC-seq) analyses performed between control and Chd7-deficient OPCs led us to demonstrate that Chd7 directly binds and opens chromatin at the enhancer and promoter regions of key regulators of OPC differentiation, such as Sox10, Nkx2.2, and Gpr17. The role of Chd7 in OPCs, as a chromatin-opening factor and activator of differentiation, is consistent with Chd7 function in cerebellar granule progenitors (21). Remarkably, Chd7-binding properties differ between cell-types, as Chd7 binds to a completely different set of genes in OPCs and granule neuron progenitors and Chd7 also acts as a transcriptional repressor (closing chromatin) of many target genes in OPCs (e.g., p53/Tpr53). These results raise important questions that remain to be elucidated: first, whether the cell-type–specific recruitment to regulatory elements of CHD remodelers depends on particular histone modifications and on the presence of cell-type–specific TFs; and second, what cofactors determine their chromatin-opening versus -closing activity.
Mechanisms of Time-Controlled Gene Expression During OPC Differentiation.
We showed that in OL lineage cells, the binding of Chd7/8 to OL-lineage TF genes varies upon their lineage stage. This partial requirement of Chd7 during differentiation/maturation of Chd7-deficient OPCs result at least in part from a functional compensation provided by Chd8 which, like Chd7, binds to the majority of OL differentiation genes in OPCs. Together with our previous report on Chd7 expression during OL differentiation and myelination (28), and our complementary study on Chd8 LOF in OL lineage cells (37), the present results underscore CHD chromatin remodelers as key regulators of the progression along the differentiation/maturation pathway in OL lineage cells.
To further investigate the mechanisms regulating transcription during OL differentiation, herein we integrated oligodendroglia-specific chromatin profiling of chromatin remodelers (Chd7 and Chd8), key TFs (Sox10 and Olig2), and epigenetic histone marks (15, 28, 50). This integrative method highlighted a timely controlled binding of Chd7 and Chd8 to stage-specific genes in OPCs and OLs (Fig. 7). This finding leads us to propose a dynamic transcription regulatory model (SI Appendix, Fig. S14), including the following steps: (i) Olig2 pioneer TF (15, 51) binding to chromatin-enhancer elements; (ii) Olig2 binding-mediated recruitment of Chd7 and Chd8 on chromatin-enhancer elements; (iii) Chd7- and Chd8-induced chromatin opening, likely by sliding nucleosomes, results in Sox10 binding; (iv) additional clustering of other regulators together with DNA bending activity of the Sox10 HMG domain, allowing the mediator complex to form a stable promoter–enhancer loop; and (v) eventually, induction of robust transcription of OL differentiation genes. This timing of events could explain the crucial importance of Olig2 and Sox10 in activating OL lineage genes (12, 15), but also the partial compensation of Chd7 by Chd8 in OPC differentiation, cell survival, and likely proliferation. Remarkably, we found that in OLs, most of OPC-specific genes are still bound by Olig2 and Sox10 but not by Chd7/Chd8 remodelers (Fig. 7). This could imply that the loss of Chd7 and Chd8 binding, together with the recruitment of some repressors (e.g., Nkx2.2) (14), is required for down-regulation of OPC genes and subsequent OL differentiation.
Materials and Methods
Mice.
Mice homozygous for Chd7 floxed alleles (Chd7Flox/Flox) (33) were crossed with PDGFRα-CreERT (32) mice to generate the OPC-specific Chd7iKO (iKO) mice. Animals of either sex were used in the study and PDGFRα-CreERT negative littermates were used as controls (Ctrl). All animal studies were conducted following protocols approved by Ethic Committee #5, Charles Darwin and French regulatory authorities (#03860.07).
Tamoxifen Administration.
For conditional Chd7 removal experiments, tamoxifen (T5648; Sigma) was dissolved in corn oil (C-8267; Sigma) and injected subcutaneously at 20-mg/mL concentration at postnatal stages P1 (40 µL) and P4 (30 µL) in Ctrl and Chd7iKO animals. Brains were then collected at P7.
PFT Administration.
PFT (P4359; Sigma) was dissolved in DMSO (less than 10% final) and NaCl and injected subcutaneously at 0.8-mg/mL concentration at postnatal stages P3, P4, and P5 (5 µL per day). Brains were collected at P7.
OPC MACSorting.
Dissociation of cortex and CC from Ctrl, Chd7iKO, or wild-type P7 mice was done using neural tissue dissociation kit (P) (130–093-231; Miltenyi Biotec) and dissociator (130-096-427). Magnetic sorting was done using anti-O4–coupled-beads (130-094-543) and the MultiMACS Cell24 Separator Plus (130-098-637). O4+-cells were collected in PBS before being process to RNA-seq, ATAC-seq or ChIP-seq.
ChIP-Seq, RNA-Seq, ATAC-Seq, and Data Analysis.
A detailed description of the methods used can be found in SI Appendix.
Supplementary Material
Acknowledgments
We thank Grace Houser for her help revising the English writing of the manuscript; Michel Mallat for his advice all along the project; Dwight Bergles for the PDGFRα::CreERT mice; Matthieu Gerard for validated Chd8 shRNA sequence; Damien Ulveling for help generating Bigwig files; and Emmanuelle Huillard for the shScramble lentivirus. All animal work was conducted at the Institut du Cerveau et de la Moelle Épinière (ICM) PHENOPARC Core Facility. Data generated relied on ICM Core Facilities: bioinformatics (ICONICS), sequencing, CELIS, histology, and ICM Quant; we thank all personnel involved for their contribution and help. The Core Facilities were supported by the “Investissements d’avenir” (ANR-10- IAIHU-06 and ANR-11-INBS-0011-NeurATRIS) and the “Fondation pour la Recherche Médicale.” This work was supported by funding by grants from the National Multiple Sclerosis Society (NMSS RG-1501-02851) and the Fondation pour l’Aide à la Recherche sur la Sclérose en Plaques (ARSEP, 2014, 2015, 2017). C.M. was supported by funding from Sorbonne Université (Université Pierre et Marie Curie-Paris6) and Fondation pour la Recherche Médicale (FRM, FDT20160435662). D.M.M. is supported by NIH R01 DC009410 and R01 DC014456. Q.R.L. is supported by NIH Grants R01NS072427 and R01NS075243.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE116601).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1802620115/-/DCSupplemental.
References
- 1.Fünfschilling U, et al. Glycolytic oligodendrocytes maintain myelin and long-term axonal integrity. Nature. 2012;485:517–521. doi: 10.1038/nature11007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee Y, et al. Oligodendroglia metabolically support axons and contribute to neurodegeneration. Nature. 2012;487:443–448. doi: 10.1038/nature11314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mount CW, Monje M. Wrapped to adapt: Experience-dependent myelination. Neuron. 2017;95:743–756. doi: 10.1016/j.neuron.2017.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spassky N, et al. Multiple restricted origin of oligodendrocytes. J Neurosci. 1998;18:8331–8343. doi: 10.1523/JNEUROSCI.18-20-08331.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tekki-Kessaris N, et al. Hedgehog-dependent oligodendrocyte lineage specification in the telencephalon. Development. 2001;128:2545–2554. doi: 10.1242/dev.128.13.2545. [DOI] [PubMed] [Google Scholar]
- 6.Rowitch DH. Glial specification in the vertebrate neural tube. Nat Rev Neurosci. 2004;5:409–419. doi: 10.1038/nrn1389. [DOI] [PubMed] [Google Scholar]
- 7.Ffrench-Constant C, Raff MC. The oligodendrocyte-type-2 astrocyte cell lineage is specialized for myelination. Nature. 1986;323:335–338. doi: 10.1038/323335a0. [DOI] [PubMed] [Google Scholar]
- 8.Suzuki SO, Goldman JE. Multiple cell populations in the early postnatal subventricular zone take distinct migratory pathways: A dynamic study of glial and neuronal progenitor migration. J Neurosci. 2003;23:4240–4250. doi: 10.1523/JNEUROSCI.23-10-04240.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bergles DE, Richardson WD. Oligodendrocyte development and plasticity. Cold Spring Harb Perspect Biol. 2015;8:a020453. doi: 10.1101/cshperspect.a020453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Emery B, Lu QR. Transcriptional and epigenetic regulation of oligodendrocyte development and myelination in the central nervous system. Cold Spring Harb Perspect Biol. 2015;7:a020461. doi: 10.1101/cshperspect.a020461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Küspert M, Wegner M. SomethiNG 2 talk about—Transcriptional regulation in embryonic and adult oligodendrocyte precursors. Brain Res. 2016;1638:167–182. doi: 10.1016/j.brainres.2015.07.024. [DOI] [PubMed] [Google Scholar]
- 12.Stolt CC, et al. Terminal differentiation of myelin-forming oligodendrocytes depends on the transcription factor Sox10. Genes Dev. 2002;16:165–170. doi: 10.1101/gad.215802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakatani H, et al. Ascl1/Mash1 promotes brain oligodendrogenesis during myelination and remyelination. J Neurosci. 2013;33:9752–9768. doi: 10.1523/JNEUROSCI.0805-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qi Y, et al. Control of oligodendrocyte differentiation by the Nkx2.2 homeodomain transcription factor. Development. 2001;128:2723–2733. doi: 10.1242/dev.128.14.2723. [DOI] [PubMed] [Google Scholar]
- 15.Yu Y, et al. Olig2 targets chromatin remodelers to enhancers to initiate oligodendrocyte differentiation. Cell. 2013;152:248–261. doi: 10.1016/j.cell.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zaret KS, Mango SE. Pioneer transcription factors, chromatin dynamics, and cell fate control. Curr Opin Genet Dev. 2016;37:76–81. doi: 10.1016/j.gde.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schnetz MP, et al. CHD7 targets active gene enhancer elements to modulate ES cell-specific gene expression. PLoS Genet. 2010;6:e1001023. doi: 10.1371/journal.pgen.1001023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schulz Y, et al. CHD7, the gene mutated in CHARGE syndrome, regulates genes involved in neural crest cell guidance. Hum Genet. 2014;133:997–1009. doi: 10.1007/s00439-014-1444-2. [DOI] [PubMed] [Google Scholar]
- 19.Feng W, et al. The chromatin remodeler CHD7 regulates adult neurogenesis via activation of SoxC transcription factors. Cell Stem Cell. 2013;13:62–72. doi: 10.1016/j.stem.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 20.Micucci JA, et al. CHD7 and retinoic acid signaling cooperate to regulate neural stem cell and inner ear development in mouse models of CHARGE syndrome. Hum Mol Genet. 2014;23:434–448. doi: 10.1093/hmg/ddt435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feng W, et al. Chd7 is indispensable for mammalian brain development through activation of a neuronal differentiation programme. Nat Commun. 2017;8:14758. doi: 10.1038/ncomms14758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cotney J, et al. The autism-associated chromatin modifier CHD8 regulates other autism risk genes during human neurodevelopment. Nat Commun. 2015;6:6404. doi: 10.1038/ncomms7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Katayama Y, et al. CHD8 haploinsufficiency results in autistic-like phenotypes in mice. Nature. 2016;537:675–679. doi: 10.1038/nature19357. [DOI] [PubMed] [Google Scholar]
- 24.Jongmans MC, et al. CHD7 mutations in patients initially diagnosed with Kallmann syndrome—The clinical overlap with CHARGE syndrome. Clin Genet. 2009;75:65–71. doi: 10.1111/j.1399-0004.2008.01107.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bernier R, et al. Disruptive CHD8 mutations define a subtype of autism early in development. Cell. 2014;158:263–276. doi: 10.1016/j.cell.2014.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O’Roak BJ, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hartshorne TS, Grialou TL, Parker KR. Autistic-like behavior in CHARGE syndrome. Am J Med Genet A. 2005;133A:257–261. doi: 10.1002/ajmg.a.30545. [DOI] [PubMed] [Google Scholar]
- 28.He D, et al. Chd7 cooperates with Sox10 and regulates the onset of CNS myelination and remyelination. Nat Neurosci. 2016;19:678–689. doi: 10.1038/nn.4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Batsukh T, et al. Identification and characterization of FAM124B as a novel component of a CHD7 and CHD8 containing complex. PLoS One. 2012;7:e52640. doi: 10.1371/journal.pone.0052640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schnetz MP, et al. Genomic distribution of CHD7 on chromatin tracks H3K4 methylation patterns. Genome Res. 2009;19:590–601. doi: 10.1101/gr.086983.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Creyghton MP, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kang SH, Fukaya M, Yang JK, Rothstein JD, Bergles DE. NG2+ CNS glial progenitors remain committed to the oligodendrocyte lineage in postnatal life and following neurodegeneration. Neuron. 2010;68:668–681. doi: 10.1016/j.neuron.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hurd EA, Poucher HK, Cheng K, Raphael Y, Martin DM. The ATP-dependent chromatin remodeling enzyme CHD7 regulates pro-neural gene expression and neurogenesis in the inner ear. Development. 2010;137:3139–3150. doi: 10.1242/dev.047894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci. 2014;34:11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marques S, et al. Oligodendrocyte heterogeneity in the mouse juvenile and adult central nervous system. Science. 2016;352:1326–1329. doi: 10.1126/science.aaf6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sugathan A, et al. CHD8 regulates neurodevelopmental pathways associated with autism spectrum disorder in neural progenitors. Proc Natl Acad Sci USA. 2014;111:E4468–E4477. doi: 10.1073/pnas.1405266111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao C, et al. Dual requirement of CHD8 for chromatin landscape establishment and histone methyltransferase recruitment to promote CNS myelination and repair. Dev Cell. 2018;45:753–768.e758. doi: 10.1016/j.devcel.2018.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Subtil-Rodriguez A, et al. The chromatin remodeller CHD8 is required for E2F-dependent transcription activation of S-phase genes. Nucleic Acids Res. 2013;42:2185–2196. doi: 10.1093/nar/gkt1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stergachis AB, et al. Conservation of trans-acting circuitry during mammalian regulatory evolution. Nature. 2014;515:365–370. doi: 10.1038/nature13972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Willsey AJ, et al. Coexpression networks implicate human midfetal deep cortical projection neurons in the pathogenesis of autism. Cell. 2013;155:997–1007. doi: 10.1016/j.cell.2013.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu L, et al. DAWN: A framework to identify autism genes and subnetworks using gene expression and genetics. Mol Autism. 2014;5:22. doi: 10.1186/2040-2392-5-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Van Nostrand JL, et al. Inappropriate p53 activation during development induces features of CHARGE syndrome. Nature. 2014;514:228–232. doi: 10.1038/nature13585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. ATAC-seq: A method for assaying chromatin accessibility genome-wide. Curr Protoc Mol Biol. 2001;109:21.29.1–21.29.9. doi: 10.1002/0471142727.mb2129s109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murphy PJM, et al. Pifithrin-α inhibits p53 signaling after interaction of the tumor suppressor protein with hsp90 and its nuclear translocation. J Biol Chem. 2004;279:30195–30201. doi: 10.1074/jbc.M403539200. [DOI] [PubMed] [Google Scholar]
- 45.Lang J, et al. Adenomatous polyposis coli regulates oligodendroglial development. J Neurosci. 2013;33:3113–3130. doi: 10.1523/JNEUROSCI.3467-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoo AS, Crabtree GR. ATP-dependent chromatin remodeling in neural development. Curr Opin Neurobiol. 2009;19:120–126. doi: 10.1016/j.conb.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vissers LE, et al. Mutations in a new member of the chromodomain gene family cause CHARGE syndrome. Nat Genet. 2004;36:955–957. doi: 10.1038/ng1407. [DOI] [PubMed] [Google Scholar]
- 48.Li J, et al. Inhibition of p53 transcriptional activity: A potential target for future development of therapeutic strategies for primary demyelination. J Neurosci. 2008;28:6118–6127. doi: 10.1523/JNEUROSCI.0184-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gudkov AV, Gurova KV, Komarova EA. Inflammation and p53: A tale of two stresses. Genes Cancer. 2011;2:503–516. doi: 10.1177/1947601911409747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.He D, et al. lncRNA functional networks in oligodendrocytes reveal stage-specific myelination control by an lncOL1/Suz12 complex in the CNS. Neuron. 2017;93:362–378. doi: 10.1016/j.neuron.2016.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Raposo AA, et al. Ascl1 coordinately regulates gene expression and the chromatin landscape during neurogenesis. Cell Rep. 2015;10:1544–1556. doi: 10.1016/j.celrep.2015.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.