Significance
The human heart is unable to meaningfully recover lost cardiac muscle after injury. As a result, injuries such as myocardial infarction cause irreversible damage that increases the risk for heart failure. Replacing lost or dysfunctional cardiac tissue can potentially reverse or prevent heart failure. Here we demonstrate that experimentally increasing expression of the angiogenic factor vegfaa alone is able to induce a cardiac growth program in zebrafish. We also show evidence that the site of vegfaa expression is important for patterning the cardiac growth response. This work identifies Vegfa as a growth factor capable of activating genetic programs for heart regeneration and has generalizable implications for therapeutic administration of cardiac growth factors.
Keywords: heart, vegfa, angiogenesis, regeneration
Abstract
During heart development and regeneration, coronary vascularization is tightly coupled with cardiac growth. Although inhibiting vascularization causes defects in the innate regenerative response of zebrafish to heart injury, angiogenic signals are not known to be sufficient for triggering regeneration events. Here, by using a transgenic reporter strain, we found that regulatory sequences of the angiogenic factor vegfaa are active in epicardial cells of uninjured animals, as well as in epicardial and endocardial tissue adjacent to regenerating muscle upon injury. Additionally, we find that induced cardiac overexpression of vegfaa in zebrafish results in overt hyperplastic thickening of the myocardial wall, accompanied by indicators of angiogenesis, epithelial-to-mesenchymal transition, and cardiomyocyte regeneration programs. Unexpectedly, vegfaa overexpression in the context of cardiac injury enabled ectopic cardiomyogenesis but inhibited regeneration at the site of the injury. Our findings identify Vegfa as one of a select few known factors sufficient to activate adult cardiomyogenesis, while also illustrating how instructive factors for heart regeneration require spatiotemporal control for efficacy.
For patients with heart failure, new approaches that can replace lost or dysfunctional cardiac tissue through regeneration offer the possibility to improve clinical outcomes. Unlike adult mammals, zebrafish and neonatal mice mount strong regenerative responses following injury and have emerged as model systems to understand mechanisms of heart regeneration (1–3). For zebrafish and neonatal mice, cardiac injury results in the deployment of a developmental growth program and proliferation of spared cardiomyocytes (CMs) (2, 4, 5). Thus, a major strategy for enhancing innate regenerative responses has been to identify CM mitogens. However, heart regeneration is more complex than CM proliferation alone and requires an orchestration of interactions between the multiple cell types that make up the heart. For instance, disruption of signaling between epicardium, endocardium, and CMs by interference with FGF, retinoic acid, NF-κB, or Notch signaling pathways can vitiate zebrafish heart regeneration (6–10). Similarly, alterations to nerve or macrophage responses in neonatal mice or adult zebrafish impair heart regeneration (11–14). A detailed understanding of the mechanisms that coordinate tissue–tissue interactions during regeneration may inspire novel ways to stimulate regeneration in the adult mammalian heart.
During cardiac development and regeneration, angiogenesis shadows tissue growth. In developing mouse and zebrafish hearts, thickening of the ventricular wall by CM hyperplasia is tightly coupled to coronary angiogenesis (15, 16). Accordingly, during zebrafish heart regeneration, impairment of angiogenesis compromises the regenerative program and results in scarring (6, 15, 17). Recent work established angiogenesis as an early event during heart regeneration, while also implicating the Vegfa paralog Vegfaa as a required endothelial mitogen (17). The extent to which an angiogenic factor like Vegfa can influence cardiac repair in mammals on its own is a question of therapeutic significance that has been asked many times, with varying results (18). In a context of innate heart regeneration, this topic is unexplored.
Here, we examined whether and how induction of angiogenesis by increased expression of vegfaa can influence cardiac growth. We describe endogenous expression of zebrafish vegfaa and demonstrate the effects of ectopic vegfaa during multiple stages of cardiac growth, including injury-induced regeneration. Although, to our knowledge, Vegfaa has not been previously described as a cardiac mitogen in the uninjured heart, we found that overexpression of vegfaa drives CM hyperplasia and activates regenerative programs in the epicardium and endocardium. Together, these data place vegfaa in an exceptional group of secreted factors capable of inducing a cardiac regenerative program.
Results
vegfaa Is Expressed in Epicardium During Zebrafish Cardiac Growth and Homeostasis.
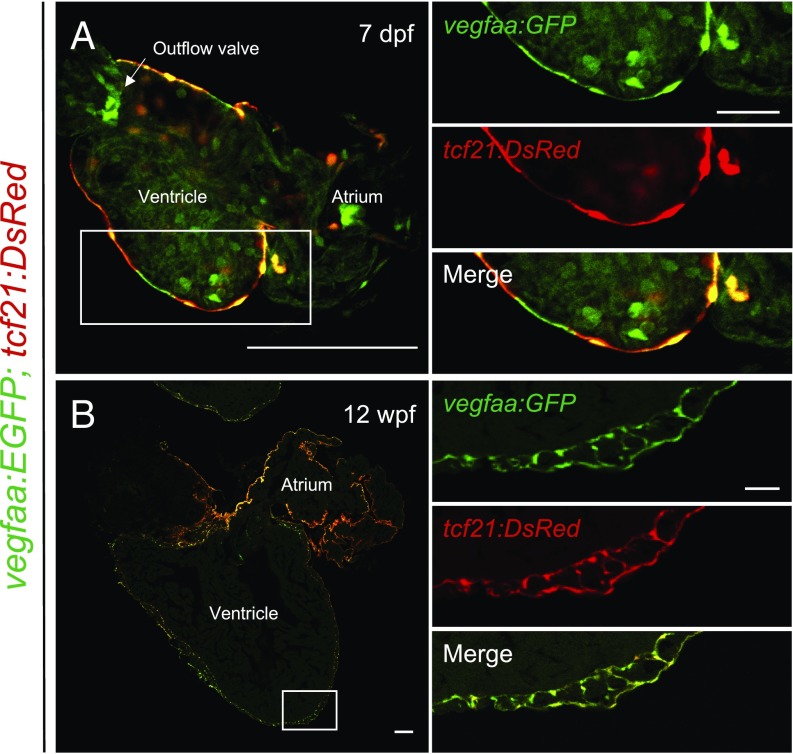
Transcriptional profiling of the zebrafish heart and candidate gene analysis have demonstrated that vegfaa is rapidly up-regulated in the zebrafish heart following injury (17, 19). To define spatiotemporal patterns of vegfaa expression during heart regeneration, we generated a BAC transgenic line with vegfaa regulatory sequences directing the expression of a fluorescent reporter, EGFP [TgBAC(vegfaa:EGFP)pd260, hereafter referred to as vegfaa:EGFP]. Mirroring previous reports of vegfaa expression, vegfaa:EGFP fish displayed EGFP fluorescence in developing somites, indicating that vegfaa regulatory sequences faithfully report domains of endogenous vegfaa (SI Appendix, Fig. S1 A and B) (20–24). During cardiac development, the expression domain for Vegfa varies among species. Whereas assessment in mouse indicates that Vegfa is expressed by CMs and the endocardium, Vegfa is expressed by CMs and the epicardium during development of the quail heart (25, 26). In vegfaa:EGFP zebrafish, we did not detect EGFP in CMs at any stage of zebrafish development. Instead, vegfaa:EGFP hearts displayed colocalization of EGFP with the epicardial reporter tcf21:DsRed during development and homeostasis (Fig. 1). EGFP signal was also present in epicardial-derived cells and tcf21-negative cardiac valves, and, in rare instances, endocardial cells. Our data indicate that the epicardium is the major source of Vegfaa under homeostatic conditions.
Fig. 1.
vegfaa expression during zebrafish heart development and homeostasis. Tiled images of vegfaa:EGFP;tcf21:DsRed cardiac sections during (A) larval and (B) adult stages. Sections are immunostained for EGFP. Boxes correspond to the region magnified in adjacent panels. (Scale bars: A and B, Left, 100 μm; A and B, Right, 25 μm.)
vegfaa Expression Is Dynamic During Zebrafish Heart Regeneration.
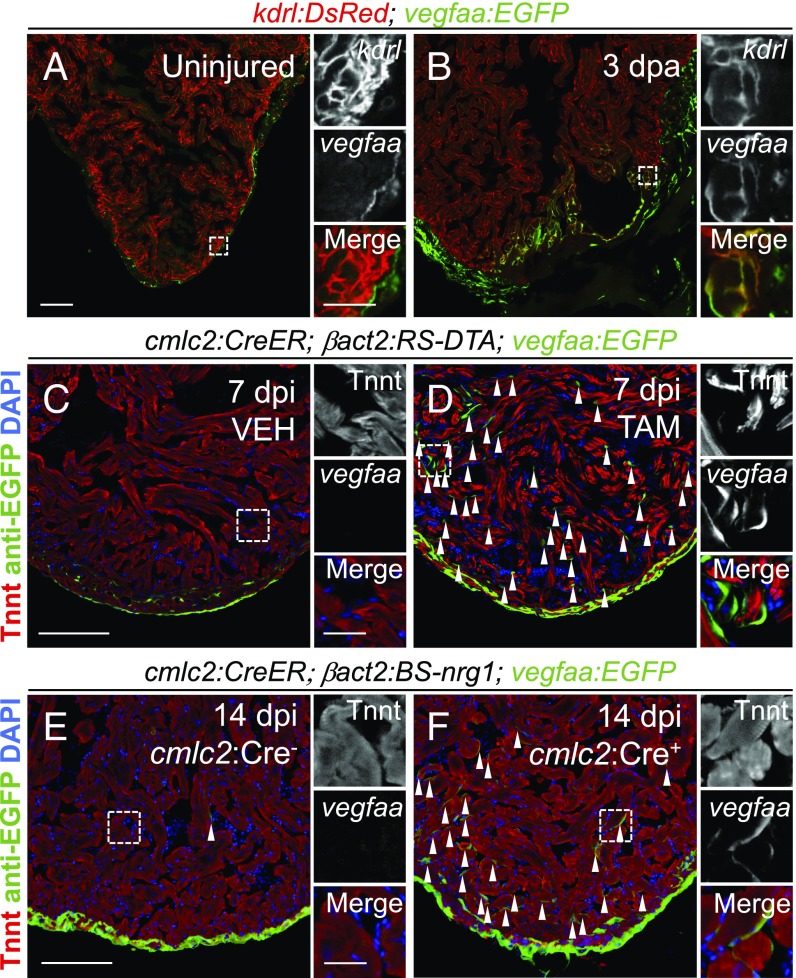
To determine which cardiac cells increase vegfaa expression during heart regeneration, we analyzed vegfaa:EGFP fish following amputation of the ventricular apex. Compared with the uninjured heart, we noted more EGFP+ cells on the surface of the heart. Additionally, we visualized more EGFP+ cells within the wound (SI Appendix, Fig. S1 C and E). These results are consistent with a previous report demonstrating a greater than twofold increase in vegfaa transcripts following cryoinjury in zebrafish (17). To better define the cellular sources of vegfaa during regeneration, we crossed vegfaa:EGFP fish to kdrl:dsRed fish that mark endothelial cells, including the ventricular endocardium (8). EGFP fluorescence colocalized with DsRed fluorescence following apical resection, whereas these markers rarely colocalized in the uninjured heart (Fig. 2 A and B). We also observed vegfaa:EGFP expression following genetic ablation of CMs, indicating that endocardial expression of vegfaa is not unique to the mode of injury (Fig. 2 C and D) (27, 28). vegfaa was also induced by global cardiac expression of the CM mitogen nrg1 in the absence of injury, further relating vegfaa induction to muscle hyperplasia (Fig. 2 E and F) (28). Unlike with apical resection, vegfaa-positive endocardial cells were observed throughout the heart after CM ablation or nrg1 induction, consistent with the global proliferative responses in these models. Together, our results demonstrate the emergence of an endocardial source of vegfaa during zebrafish heart regeneration.
Fig. 2.
vegfaa expression during adult zebrafish heart regeneration. (A and B) Colocalization of vegfaa:EGFP and kdrl:DsRed expression in hearts without injury and at 3 dpa. (C and D) vegfaa:EGFP expression in an uninjured heart and a heart 7 d post CM ablation (dpi). (E and F) vegfaa:EGFP expression in an uninjured heart and a heart after 14 d (dpi) of nrg1 overexpression. Arrows indicate EGFP+ endocardial cells. Boxes correspond to the region magnified in adjacent panels. [Scale bars: A, C, and E, 100 μm (Left) and 25 μm (Right); scale bars correspond to B, D, and F, respectively.]
Induced vegfaa Overexpression Hypervascularizes the Zebrafish Heart and Rapidly Leads to Cardiomegaly.
To determine the influence of vegfaa on cardiac growth, we generated a new transgenic strain of zebrafish to conditionally overexpress vegfaa, Tg(βactin2:loxP-mTagBFP-STOP-loxP-vegfaa)pd262, hereafter referred to as βact2:BS-vegfaa. We crossed βact2:BS-vegfaa fish with cmlc2:CreER fish to enable conditional overexpression of vegfaa from CMs by administration of tamoxifen (SI Appendix, Fig. S2 A–C). Further, we crossed cmlc2:CreER;βact2:BS-vegfaa fish with fli1a:EGFP fish to visualize EGFP+ endothelial cells following vegfaa overexpression. When we induced vegfaa overexpression at 30 d post fertilization (dpf), a time point before the development of coronary vessels, we observed precocious and enhanced coronary vasculogenesis by 6 wk post fertilization (wpf; SI Appendix, Fig. S2 D, F, and G). Thus, vegfaa overexpression is sufficient to induce ectopic coronary vasculature in the zebrafish heart, similar to what has been observed in mice (29).
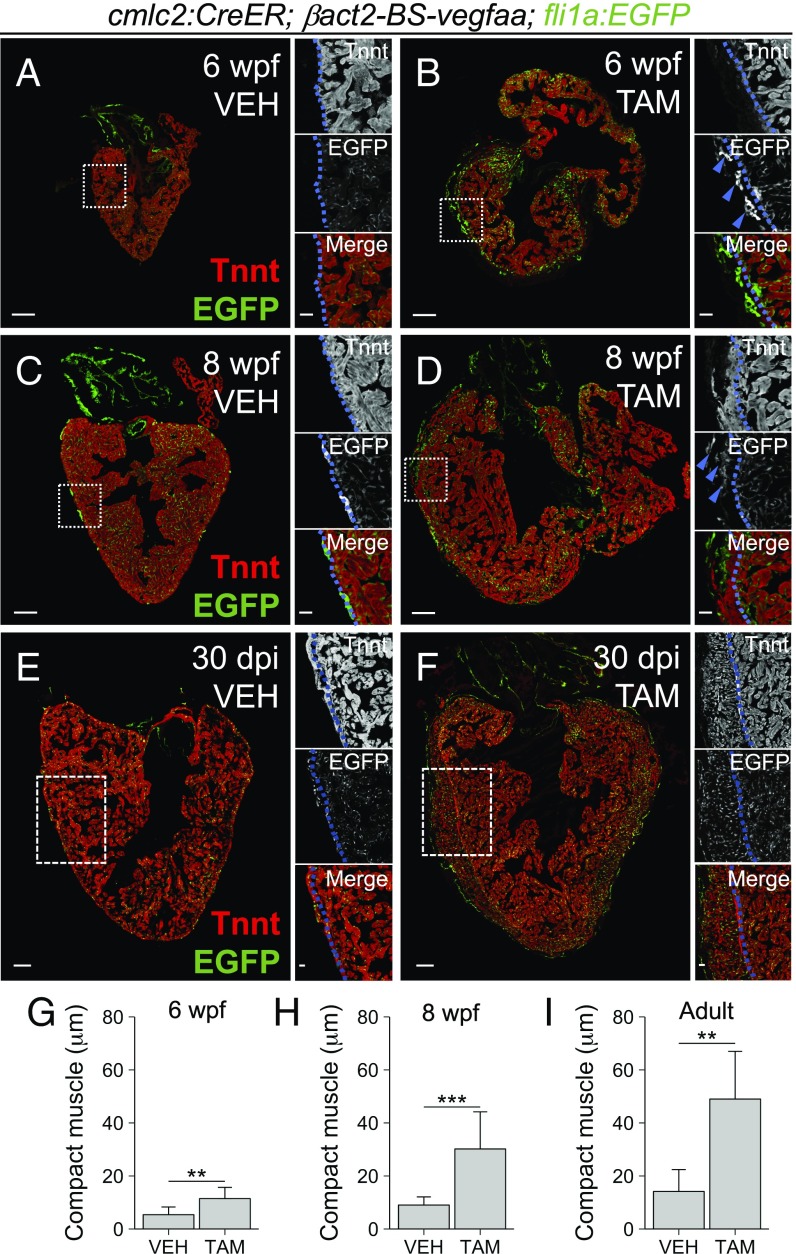
In addition to increased vasculature, we noticed that hearts overexpressing vegfaa as juveniles were dysmorphic, with larger chambers. When examined in cross-section at 6 wpf, we found hearts overexpressing vegfaa to have CMs dispersed sparsely in areas of ectopic vasculature (Fig. 3 A and B). Overall, the maximal thickness of the Tnnt+ compact muscle in hearts overexpressing vegfaa at 6 wpf was more than twice that of control hearts (11.47 ± 4.23 μm vs. 5.40 ± 2.90 μm, P = 0.003, Student’s t test; Fig. 3G). By 8 wpf, the precocious compact muscle layer became even more pronounced (Fig. 3 C and D). Maximal compact muscle thickness was increased by more than threefold with vegfaa overexpression compared with control hearts (30.19 ± 14.00 μm vs. 9.04 ± 3.06 μm, P < 0.001, Student’s t test; Fig. 3H). Together, these results reveal that vegfaa overexpression can stimulate vascular growth and expansion of the muscular ventricular wall during zebrafish heart development.
Fig. 3.
Effects of cardiac vegfaa overexpression in the absence of injury. (A–D) Tiled images of cmlc2:CreER;βact2:BS-vegfaa;fli1a:EGFP hearts at 6 wpf and 8 wpf after treatment with vehicle or tamoxifen at 30 dpf. (E and F) Tiled images of adult cmlc2:CreER;βact2:BS-vegfaa;fli1a:EGFP hearts 30 dpi with vehicle or tamoxifen. Sections are immunostained for Tnnt. Boxes correspond to the region magnified in adjacent panels. Blue lines approximate the CM primordial layer and blue arrows denote ectopic vasculature. (G–I) Maximum thickness of the Tnnt+ compact layer at 6 wpf after treating 30-dpf fish with vehicle (n = 11) or tamoxifen (n = 6); at 8 wpf after treating 30-dpf fish with vehicle (n = 11) or tamoxifen (n = 9); and at 30 dpi after treating adult fish with vehicle (n = 5) or tamoxifen (n = 6). Error bars indicate SD (**P < 0.01 and ***P < 0.001, Student’s t test). (Scale bars: A–F, Left, 100 μm; A–F, Right, 25 μm.)
To determine the extent to which vegfaa overexpression could affect the adult heart, we induced vegfaa overexpression in mature, 4-mo-old zebrafish and assessed animals 30 d later (SI Appendix, Fig. S2E). Strikingly, induced cardiac vegfaa overexpression caused expansion of the coronary vasculature, atrial growth, and thickening of the compact muscle layer in the ventricle (Fig. 3 E and F). Importantly, ectopic blood vessels were filled with nucleated blood cells, suggestive of functional perfusion (SI Appendix, Fig. S2 H and I). Similar to 4 wk of overexpression during development, the maximal thickness of the compact muscle layer after 30 d of vegfaa overexpression in the adult heart was more than three times that of control hearts (49.00 ± 18.00 μm vs. 14.16 ± 8.26 μm, P = 0.003, Student’s t test; Fig. 3I). Thus, ectopic vegfaa is sufficient to stimulate cardiac growth in zebrafish at juvenile and adult stages.
vegfaa Induces an Ectopic Heart Regeneration Program.
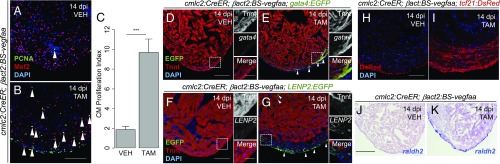
Zebrafish heart regeneration proceeds by induced cell cycle entry and successful division of a predominantly mononucleate, diploid population of CMs (4, 5, 30). Accompanying this proliferative response is a pronounced activation and expansion of the epicardial cells, which cover and integrate into the cardiac wound (6, 31, 32). To determine whether vegfaa could induce cardiomegaly through a regenerative program, we assayed these hallmarks. First, we evaluated CM cell cycle reentry by determining the percentage of Mef2+/PCNA+ cells after vegfaa overexpression, a proxy for CM proliferation used in many studies (1). Although we did not detect a significant increase in the total number of Mef2+ CMs by 14 d post incubation (dpi) with tamoxifen (1,397 ± 243 Mef2+ cells per section vs. 1,113 ± 215 Mef2+ cells per section in control hearts, P = 0.40, Mann–Whitney U test), we identified a significant increase in the number of cycling CMs. In vehicle-treated hearts, 1.86 ± 0.35% of Mef2+ CMs were Mef2+/PCNA+ (mean ± SEM, n = 10). By contrast, CM proliferation was sharply increased in hearts overexpressing vegfaa, with 9.67 ± 1.37% of CMs positive for PCNA, an approximately fivefold increase (mean ± SEM, n = 8; P < 0.001, Mann–Whitney U test; Fig. 4 A–C), indicating that cardiomegaly directed by Vegfaa is hyperplastic. Interestingly, we observed trabecular and compact CM cycling during vegfaa overexpression. However, CM cycling and ectopic muscle was most prominent in the compact layer, suggesting a strong effect of vegfaa overexpression to expand the compact layer. We also noted a large increase in Mef2−/PCNA+ nonmyocardial cells on the outer surface of the heart (Fig. 4 A and B). By using a tcf21:DsRed transgene to visualize epicardial responses, we observed a massive thickening of the tcf21:DsRed+ tissue layer, indicative of epicardial expansion (Fig. 4 H and I). The maximal thickness of the tcf21+ cell layer after 2 wk of vegfaa overexpression (72.46 ± 38.25 μm, mean ± SD, n = 5) was increased by more than threefold compared with control hearts (21.25 ± 5.27 μm, mean ± SD, n = 4; P = 0.03, Student’s t test).
Fig. 4.
Cardiac vegfaa overexpression elicits an ectopic heart regeneration program. (A and B) Tiled images of cmlc2:CreER;βact2:BS-vegfaa hearts 14 dpi with vehicle or tamoxifen. Arrowheads indicate Mef2+/PCNA+ nuclei. Mef2−/PCNA+ nuclei are likely to be cycling epicardial cells, endothelial cells, fibroblasts, or inflammatory cells. (C) CM proliferation 14 dpi with vehicle (n = 10) or tamoxifen (n = 8). Error bars indicate SEM (***P < 0.001, Mann–Whitney U test). (D and E) cmlc2:CreER;βact2:BS-vegfaa;gata4:EGFP hearts 14 dpi with vehicle or tamoxifen. Arrowheads indicate gata4+ CMs. (F and G) Images of cmlc2:CreER;βact2:BS-vegfaa;LENP2:EGFP hearts 14 dpi with vehicle or tamoxifen. Arrowheads indicate LENP2+ cells. Tnnt marks CMs. Boxes correspond to the region magnified in adjacent panels. (H and I) Images of cmlc2:CreER;βact2:BS-vegfaa;tcf21:DsRed hearts 14 dpi with vehicle or tamoxifen. (J and K) In situ hybridization for raldh2 on ventricular sections. Violet staining indicates expression. [Scale bars: B, H, and J, 100 μm (also apply to A, I, and K, respectively); D and F, 100 μm (Left) and 25 μm (Right) (also apply to E and G, respectively).]
In addition to structural features of regeneration, we assessed vegfaa-overexpressing hearts for markers of regenerative signaling. Regenerating CMs, for example, can be marked by activation of regulatory sequences of the cardiogenic transcription factor gata4 and sarcomere disassembly (4, 33). Animals overexpressing vegfaa had gata4 expression in the compact muscle that was not observed in control animals (Fig. 4 D and E). Additionally, we noted that the expanded compact muscle layer in vegfaa-overexpressing hearts stains less prominently for Tnnt and is generally less organized compared with the trabecular muscle (Fig. 3 D and F). As a part of the epicardial response during heart regeneration, epicardial cells undergo epithelial-to-mesenchymal transition (EMT); correspondingly, we observed induction of the EMT markers snai2, twist1, and pdgfrb by in situ hybridization in hearts following vegfaa overexpression (SI Appendix, Fig. S3). Notably, these markers were induced in the region corresponding to expansion of tcf21+ epicardial cells and their derivatives (Fig. 4 H and I). Hence, in zebrafish, vegfaa overexpression results in epicardial expansion and EMT, similar to what has been reported following overexpression of Vegfa in the mouse heart (34). Additionally, during zebrafish heart regeneration, the epicardium and the endocardium are activated to secrete paracrine factors, like retinoic acid, that support cell-cycle reentry of spared CMs (8). We noted increased expression in these tissues of the enzyme responsible for retinoic acid synthesis, raldh2, with vegfaa overexpression (Fig. 4 J and K). Finally, we recently identified an enhancer element upstream of the lepb gene that is activated in endocardial and epicardial tissues during regeneration, but not during embryogenesis (19). We found that vegfaa overexpression was sufficient to trigger expression via this enhancer (Fig. 4 F and G). Together, our data strongly suggest that vegfaa overexpression directs cardiac growth by induction of a regenerative program.
Features of the Vegfaa-Instructed Regeneration Program.
To understand how vegfaa overexpression induces a regenerative program, we compared transcriptional profiles of hearts overexpressing vegfaa vs. control hearts (SI Appendix, Fig. S4A). As might be expected, gene-set enrichment analysis revealed a significant up-regulation of genes related to “endothelial migration.” However, we also observed enrichment of genes related to “immune response” and “epithelial cell migration” (SI Appendix, Fig. S4 B–D). Gene sets down-regulated with vegfaa overexpression were related to “mitochondrion,” “energy derivation by oxidation of organic compounds,” and “generation of precursor metabolites and energy,” suggesting that vegfaa overexpression results in changes to cardiac energy utilization (SI Appendix, Fig. S4 E–G).
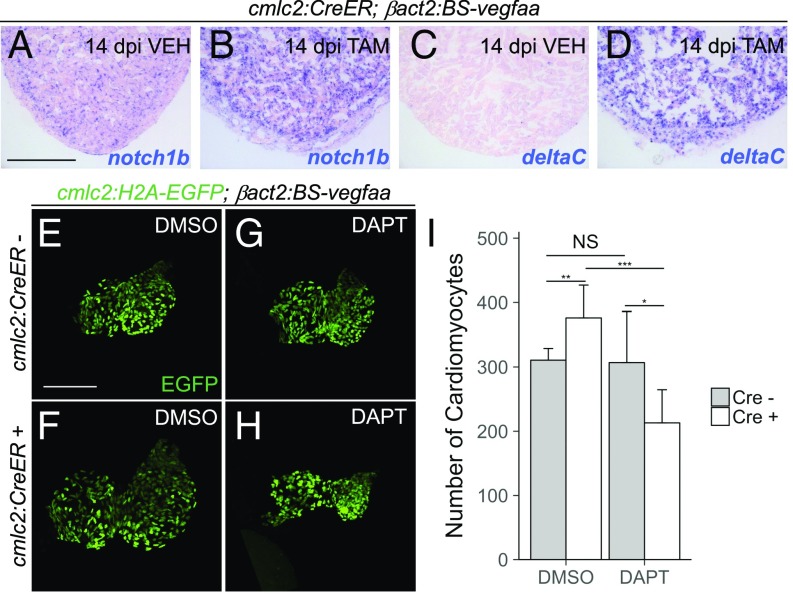
As a possible mediator of endothelial responses, we assessed Notch signaling. Endothelial Notch signaling is induced during zebrafish heart regeneration and is known to modulate CM proliferation (9, 35). Similarly, during mouse heart development, Notch inhibition leads to defects in CM proliferation (36, 37). Consistent with increased Notch signaling, our transcriptional profiling experiments showed hearts overexpressing vegfaa to have increased expression of Notch ligands and Notch receptors (Dataset S1). In situ hybridizations for notch1b and deltaC confirmed strong induction with vegfaa overexpression (Fig. 5 A–D).
Fig. 5.
Dependence of vegfaa-induced CM hyperplasia on Notch signaling. (A–D) In situ hybridization for notch1b and deltaC on cardiac sections from cmlc2:CreER;βact2:BS-vegfaa animals 14 dpi with vehicle or tamoxifen. Violet staining indicates expression. (E–H) Maximum-intensity projections of 7 dpf hearts from cmlc2:H2A-EGFP;βact2:BS-vegfaa fish with or without an additional cmlc2:CreER transgene. Animals were first treated with 4-HT, followed by DMSO (n = 9 for Cre− animals and n = 10 for Cre+ animals) or DAPT (n = 8 for Cre− animals and n = 9 for Cre+ animals). (I) Number of CMs at 7 dpf for treatment groups shown in E–H. Error bars indicate SD (*P < 0.05, **P < 0.01, and ***P < 0.001, Student’s t test). NS, not significant. [Scale bars: A and E, 100 μm (also apply to B–D and F–H, respectively).]
To determine the mechanism by which vegfaa induces CM hyperplasia, we first examined whether Vegfaa effects were the result of increased coronary vasculature vs. other tissues. To distentangle contributions from the coronary circulation, we studied the effects of vegfaa overexpression on CM numbers in zebrafish embryos, not yet having formed coronary vasculature (38). We treated βact2:BS-vegfaa;cmlc2:H2B-EGFP and cmlc2:CreER;βact2:BS-vegfaa;cmlc2:H2B-EGFP fish with 4-HT at 2 dpf and then DMSO until 7 dpf. We found that vegfaa overexpression caused a ∼21% expansion of CM numbers (376.4 ± 50.6 CMs vs. 310.7 ± 18.2 CMs, P = 0.002, Student’s t test; Fig. 5 E, F, and I), suggesting that Vegfaa is able to induce CM hyperplasia independently of the coronary vasculature.
To test whether the effects on CM numbers caused by vegfaa overexpression depend on Notch signaling, we employed the γ-secretase inhibitor DAPT (39). As demonstrated previously, inhibition of Notch signaling with the small molecule DAPT does not directly affect CM numbers and results in hearts with small ventricles (306.9 ± 79.3 CMs vs. 310.7 ± 18.2 CMs, P = 0.89, Student’s t test; Fig. 5 G–I) (40). However, vefgaa overexpression did not increase CM numbers in the presence of DAPT, even lowering counts by ∼30% (213 ± 51.6 CMs vs. 306.9 ± 79.3 CMs, P = 0.01, Student’s t test; Fig. 5 F, G, and I). These data suggest that the positive effects of vegfaa on CM proliferation may act in part through Notch signaling.
Ectopic vegfaa Misdirects Cardiogenesis After Injury.
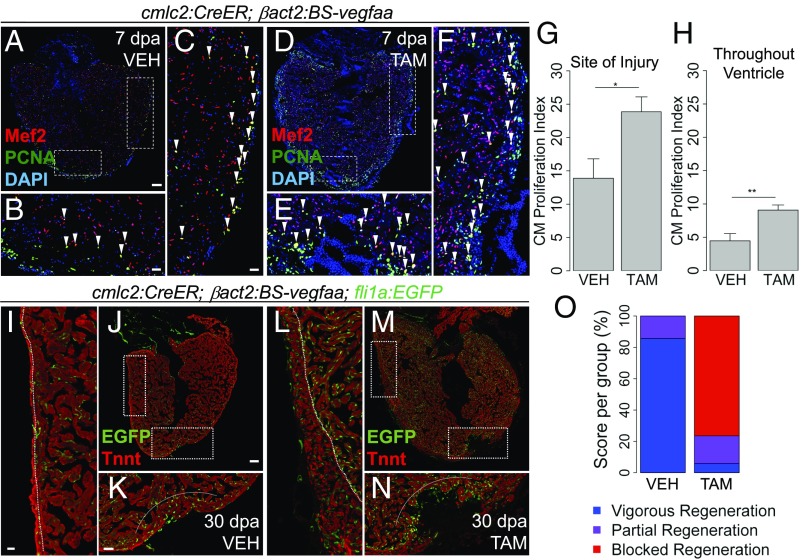
As vegfaa overexpression on its own increased indicators of regeneration, we predicted that supplementing Vegfaa after cardiac injury would accelerate muscle regeneration. To test this idea, we partially resected ventricles 1 wk after inducing myocardial vegfaa expression. Fish overexpressing vegfaa displayed a ∼72% increased percentage of proliferating CMs at the wound (23.87 ± 2.24%, mean ± SEM, n = 12) compared with control animals (13.89 ± 2.91%, mean ± SEM, n = 8; P = 0.02, Mann–Whitney U test; Fig. 6 B, E, and G). Importantly, we also noted an increased percentage of cycling CMs more globally throughout the heart, including away from the wound. In animals overexpressing vegfaa, the number of CMs throughout the ventricle that were positive for PCNA was nearly double that in control hearts (9.08 ± 0.86% vs. 4.47 ± 1.08%, mean ± SEM, P = 0.004, Mann–Whitney U test; Fig. 6 C, F, and H). Thus, vegfaa overexpression increases CM proliferation during heart regeneration.
Fig. 6.
Effect of vegfaa overexpression on heart regeneration. (A–E) Tilescan images of cmlc2:CreER;βact2:BS-vegfaa hearts at 7 dpa after treatment with vehicle or tamoxifen. Boxes mark the regions at the injury site (B and E) and away from the injury site (C and F). Arrowheads indicate Mef2+/PCNA+ nuclei. PCNA+ nuclei that do not colocalize with Mef2 are likely to be cycling epicardial cells, endothelial cells, fibroblasts, or inflammatory cells. (G and H) CM proliferation adjacent to the wound (G) and throughout the ventricle (H) at 7 dpa and after treatment with vehicle (n = 8) or tamoxifen (n = 12). Error bars indicate SEM (*P < 0.05 and **P < 0.01, Mann–Whitney U test). (I–N) cmlc2:CreER;βact2:BS-vegfaa;fli1a:EGFP ventricles at 30 dpa. Tnnt marks CMs. Boxes mark the regions at the injury site (K and N) and away from the injury (I and L). Dashed lines in K and N approximate the resection plane. Dashed lines in I and L indicate the boundary of trabecular muscle and compact muscle. (O) Regeneration scores at 30 dpa after treatment with vehicle (n = 14) or tamoxifen (n = 17; P < 0.001, Kruskal–Wallis test). [Scale bars: A and J, 100 μm (also apply to D and M, respectively); B, C, I, and K, 25 μm (also apply to E, F, L, and N, respectively).]
To determine effects on muscle regeneration, we induced vegfaa overexpression, performed apical resection, and assessed hearts histologically at 30 d post amputation (dpa), when regeneration of the myocardial wall is typically complete (3). Surprisingly, animals globally overexpressing vegfaa had persistent gaps and scarring at the resection site (Fig. 6 J, K, M, and N and SI Appendix, Fig. S5). Blinded, semiquantitative scoring of regenerative responses, as performed previously (7, 11), confirmed this impaired regeneration (mean regeneration score of 2.71 ± 0.59 with vegfaa overexpression vs. 1.14 ± 0.36 in control animals; P < 0.001, Kruskal–Wallis test; Fig. 6O). However, we still noted consistent tissue growth away from the wound, with thicker ventricular walls in vegfaa-overexpressing hearts (Fig. 6 I, J, L, and M). Thus, global vegfaa overexpression prevents regeneration at the resection site despite organ-wide neocardiogenesis, indicating a classic, instructive role for vegfaa signaling in tissue morphogenesis during regeneration.
Discussion
During organogenesis, a dialogue between parenchymal cells and organ-specific vascular endothelium coordinates angiogenesis with organ growth. For instance, Vegfa expression in mice by alveolar epithelial cells and by CMs guides the development of the pulmonary and coronary vasculatures, respectively (41, 42). Reciprocally, specialized vascular endothelium secretes angiocrine factors to support organogenesis. Signals from the endothelium are required for growth of the liver bud and differentiation of pancreatic islets during development (43, 44). More recently, an appreciation of the cross-talk between vascular endothelium and parenchymal cells has been extended to tissue regeneration. Angiocrine factors support hepatic regeneration and expansion of basal cells and alveolar cells during lung regeneration (45). Here, we extend the symbiotic link between angiogenesis and tissue growth to the context of innate heart regeneration in zebrafish.
We identify Vegfa as an instructive factor for hyperplastic cardiac growth. Endocardial expression of vegfaa is spatiotemporally related to regions of regenerative growth after injury. However, global misexpression of vegfaa in cardiac muscle after apical resection abrogates local tissue regeneration despite concomitant ventricular wall neocardiogenesis. In contrast to other manipulations that block heart regeneration in zebrafish, ectopic vegfaa expression formally misdirects growth rather than inhibiting it (7–9, 17). Ultimately, our investigation of vegfaa roles during animal development and regeneration indicate that cardiomyogenic growth and morphogenesis are linked but distinct: a crucial concept in regeneration of intricately patterned structures like limbs, but not intuitive for heart regeneration.
Although Vegfa has been suggested to stimulate low levels of CM cycling, but past literature does not predict such marked effects on cardiomyogenesis (46–48). While our data in larvae suggest that the coronary vasculature is not required for Vegfa to induce CM proliferation, the cardiac endothelium is likely a key mediator of Vegfaa effects. Not only are the primary receptors for Vegfa, kdrl and flt1, expressed by the endocardium and vascular endothelium (Fig. 2B), but vegfaa-induced CM hyperplasia depends on Notch signaling (Fig. 5 E–I) (8, 17). Prior work has shown Notch pathway elements to be induced in the endocardium during development and regeneration in zebrafish (9, 10, 40). Notch inhibition reduces indicators of CM proliferation and impairs heart regeneration after resection or cryoinjury. Interestingly, hyperactivation of Notch signaling was reported to increase CM proliferation after cryoinjury but to reduce CM proliferation after resection injury. This discrepancy may be the result of different genetic tools or injury contexts, yet neither study reported activation of overt ectopic myocardial growth (9, 10). Thus, in addition to inducing Notch signaling, Vegfaa is likely to trigger additional factors that promote cardiomyogenesis. Future work can be directed to identify how and which subsets of the cardiac endothelium might mediate vegfaa effects on cardiac growth.
Our work suggests that Vegfa signaling can be manipulated for therapeutic heart regeneration. In fact, Vegfa signaling might even account for some of the stimulatory effects on CM cell cycling recently reported with chronic hypoxia, as Vegfa is classically induced in hypoxic tissue (49–51). Although we show that misexpression of vegfaa can disrupt regenerative morphogenesis, a more targeted application of Vegfa should better direct and augment regenerative responses. In support of this concept, localized injection of virus overexpressing Vegfa and plasmids encoding Vegfa are reported to increase ventricular function when they are injected into the border zone following experimental myocardial infarction in mammals (18). Unfortunately, despite promising preclinical findings in mammalian models, human clinical trials with VEGFA have not been successful, likely related to the low doses of VEGFA used (18). Indeed, our work suggests a threshold effect for Vegfa. In zebrafish, vegfaa is expressed at low levels by the epicardium, but only with increases in expression above these levels did we observe ectopic cardiogenesis and regenerative responses. Thus, our findings help highlight the need for attention to factor dosage, duration, and site of delivery to translate regenerative biology to therapeutics. Indeed, newer strategies that allow for controlled delivery of Vegfa and other factors via modified RNA, engineered nanofibers, or specialized regulatory elements may be needed to achieve therapeutic heart regeneration (19, 34, 52).
Materials and Methods
Procedures for zebrafish transgenesis and ventricular resection have been previously described (7, 32). Immunostaining and in situ hybridization procedures are described in refs. 7 and 8. Compact muscle thickness, CM proliferation, and regeneration scores were measured as previously described (7, 30). Detailed materials and methods are provided in the SI Appendix.
Supplementary Material
Acknowledgments
We thank the Duke Zebrafish core facility for zebrafish care. This work was supported by National Institutes of Health (NIH) Mentored Clinical Scientist Award K08-HL116485 (to R.K.), NIH Predoctoral Fellowship Award F30 HL126487 (to M.J.F.), NIH Grants R01-HL081674 and R01-HL131319 (to K.D.P.), March of Dimes Grant 1-FY14-205 (to K.D.P.), the Walker P. Inman Endowment (R.K.), the Edna and Fred L. Mandel, Jr., Foundation (R.K.), and an American Heart Association Merit Award 16MERIT27940012 (to K.D.P.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The fastq files for the RNASeq data of Vegf overexpression vs. controls are available on National Center for Biotechnology Information (NCBI) BioProject, https://www.ncbi.nlm.nih.gov/bioproject (accession no. PRJNA475163) and on NCBI Sequence Read Archive, https://www.ncbi.nlm.nih.gov/sra (accession no. SRP150028).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1722594115/-/DCSupplemental.
References
- 1.Karra R, Poss KD. Redirecting cardiac growth mechanisms for therapeutic regeneration. J Clin Invest. 2017;127:427–436. doi: 10.1172/JCI89786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Porrello ER, et al. Transient regenerative potential of the neonatal mouse heart. Science. 2011;331:1078–1080. doi: 10.1126/science.1200708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poss KD, Wilson LG, Keating MT. Heart regeneration in zebrafish. Science. 2002;298:2188–2190. doi: 10.1126/science.1077857. [DOI] [PubMed] [Google Scholar]
- 4.Kikuchi K, et al. Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature. 2010;464:601–605. doi: 10.1038/nature08804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jopling C, et al. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature. 2010;464:606–609. doi: 10.1038/nature08899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lepilina A, et al. A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell. 2006;127:607–619. doi: 10.1016/j.cell.2006.08.052. [DOI] [PubMed] [Google Scholar]
- 7.Karra R, Knecht AK, Kikuchi K, Poss KD. Myocardial NF-κB activation is essential for zebrafish heart regeneration. Proc Natl Acad Sci USA. 2015;112:13255–13260. doi: 10.1073/pnas.1511209112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kikuchi K, et al. Retinoic acid production by endocardium and epicardium is an injury response essential for zebrafish heart regeneration. Dev Cell. 2011;20:397–404. doi: 10.1016/j.devcel.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao L, et al. Notch signaling regulates cardiomyocyte proliferation during zebrafish heart regeneration. Proc Natl Acad Sci USA. 2014;111:1403–1408. doi: 10.1073/pnas.1311705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Münch J, Grivas D, González-Rajal Á, Torregrosa-Carrión R, de la Pompa JL. Notch signalling restricts inflammation and serpine1 expression in the dynamic endocardium of the regenerating zebrafish heart. Development. 2017;144:1425–1440. doi: 10.1242/dev.143362. [DOI] [PubMed] [Google Scholar]
- 11.Mahmoud AI, et al. Nerves regulate cardiomyocyte proliferation and heart regeneration. Dev Cell. 2015;34:387–399. doi: 10.1016/j.devcel.2015.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aurora AB, et al. Macrophages are required for neonatal heart regeneration. J Clin Invest. 2014;124:1382–1392. doi: 10.1172/JCI72181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lavine KJ, et al. Distinct macrophage lineages contribute to disparate patterns of cardiac recovery and remodeling in the neonatal and adult heart. Proc Natl Acad Sci USA. 2014;111:16029–16034, and erratum (2016) 113:E1414. doi: 10.1073/pnas.1406508111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lai SL, et al. Reciprocal analyses in zebrafish and medaka reveal that harnessing the immune response promotes cardiac regeneration. eLife. 2017;6:e25605. doi: 10.7554/eLife.25605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harrison MR, et al. Chemokine-guided angiogenesis directs coronary vasculature formation in zebrafish. Dev Cell. 2015;33:442–454. doi: 10.1016/j.devcel.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sedmera D, McQuinn T. Embryogenesis of the heart muscle. Heart Fail Clin. 2008;4:235–245. doi: 10.1016/j.hfc.2008.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marín-Juez R, et al. Fast revascularization of the injured area is essential to support zebrafish heart regeneration. Proc Natl Acad Sci USA. 2016;113:11237–11242. doi: 10.1073/pnas.1605431113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taimeh Z, Loughran J, Birks EJ, Bolli R. Vascular endothelial growth factor in heart failure. Nat Rev Cardiol. 2013;10:519–530. doi: 10.1038/nrcardio.2013.94. [DOI] [PubMed] [Google Scholar]
- 19.Kang J, et al. Modulation of tissue repair by regeneration enhancer elements. Nature. 2016;532:201–206. doi: 10.1038/nature17644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jensen LD, et al. VEGF-B-neuropilin-1 signaling is spatiotemporally indispensable for vascular and neuronal development in zebrafish. Proc Natl Acad Sci USA. 2015;112:E5944–E5953. doi: 10.1073/pnas.1510245112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.So JH, et al. FIH-1, a novel interactor of mindbomb, functions as an essential anti-angiogenic factor during zebrafish vascular development. PLoS One. 2014;9:e109517. doi: 10.1371/journal.pone.0109517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amali AA, Sie L, Winkler C, Featherstone M. Zebrafish hoxd4a acts upstream of meis1.1 to direct vasculogenesis, angiogenesis and hematopoiesis. PLoS One. 2013;8:e58857. doi: 10.1371/journal.pone.0058857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stahlhut C, Suárez Y, Lu J, Mishima Y, Giraldez AJ. miR-1 and miR-206 regulate angiogenesis by modulating VegfA expression in zebrafish. Development. 2012;139:4356–4364. doi: 10.1242/dev.083774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mouillesseaux K, Chen JN. Mutation in utp15 disrupts vascular patterning in a p53-dependent manner in zebrafish embryos. PLoS One. 2011;6:e25013. doi: 10.1371/journal.pone.0025013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tomanek RJ, Hansen HK, Dedkov EI. Vascular patterning of the quail coronary system during development. Anat Rec A Discov Mol Cell Evol Biol. 2006;288:989–999. doi: 10.1002/ar.a.20365. [DOI] [PubMed] [Google Scholar]
- 26.Miquerol L, Gertsenstein M, Harpal K, Rossant J, Nagy A. Multiple developmental roles of VEGF suggested by a LacZ-tagged allele. Dev Biol. 1999;212:307–322. doi: 10.1006/dbio.1999.9355. [DOI] [PubMed] [Google Scholar]
- 27.Wang J, et al. The regenerative capacity of zebrafish reverses cardiac failure caused by genetic cardiomyocyte depletion. Development. 2011;138:3421–3430. doi: 10.1242/dev.068601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gemberling M, Karra R, Dickson AL, Poss KD. Nrg1 is an injury-induced cardiomyocyte mitogen for the endogenous heart regeneration program in zebrafish. eLife. 2015;4:e05871. doi: 10.7554/eLife.05871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miquerol L, Langille BL, Nagy A. Embryonic development is disrupted by modest increases in vascular endothelial growth factor gene expression. Development. 2000;127:3941–3946. doi: 10.1242/dev.127.18.3941. [DOI] [PubMed] [Google Scholar]
- 30.Wills AA, Holdway JE, Major RJ, Poss KD. Regulated addition of new myocardial and epicardial cells fosters homeostatic cardiac growth and maintenance in adult zebrafish. Development. 2008;135:183–192. doi: 10.1242/dev.010363. [DOI] [PubMed] [Google Scholar]
- 31.González-Rosa JM, Peralta M, Mercader N. Pan-epicardial lineage tracing reveals that epicardium derived cells give rise to myofibroblasts and perivascular cells during zebrafish heart regeneration. Dev Biol. 2012;370:173–186. doi: 10.1016/j.ydbio.2012.07.007. [DOI] [PubMed] [Google Scholar]
- 32.Kikuchi K, et al. tcf21+ epicardial cells adopt non-myocardial fates during zebrafish heart development and regeneration. Development. 2011;138:2895–2902. doi: 10.1242/dev.067041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gupta V, et al. An injury-responsive gata4 program shapes the zebrafish cardiac ventricle. Curr Biol. 2013;23:1221–1227. doi: 10.1016/j.cub.2013.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zangi L, et al. Modified mRNA directs the fate of heart progenitor cells and induces vascular regeneration after myocardial infarction. Nat Biotechnol. 2013;31:898–907. doi: 10.1038/nbt.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raya A, et al. Activation of Notch signaling pathway precedes heart regeneration in zebrafish. Proc Natl Acad Sci USA. 2003;100:11889–11895. doi: 10.1073/pnas.1834204100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grego-Bessa J, et al. Notch signaling is essential for ventricular chamber development. Dev Cell. 2007;12:415–429. doi: 10.1016/j.devcel.2006.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.D’Amato G, et al. Sequential Notch activation regulates ventricular chamber development. Nat Cell Biol. 2016;18:7–20. doi: 10.1038/ncb3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Foglia MJ, Cao J, Tornini VA, Poss KD. Multicolor mapping of the cardiomyocyte proliferation dynamics that construct the atrium. Development. 2016;143:1688–1696. doi: 10.1242/dev.136606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Geling A, Steiner H, Willem M, Bally-Cuif L, Haass C. A gamma-secretase inhibitor blocks Notch signaling in vivo and causes a severe neurogenic phenotype in zebrafish. EMBO Rep. 2002;3:688–694. doi: 10.1093/embo-reports/kvf124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Han P, et al. Coordinating cardiomyocyte interactions to direct ventricular chamber morphogenesis. Nature. 2016;534:700–704. doi: 10.1038/nature18310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ng YS, Rohan R, Sunday ME, Demello DE, D’Amore PA. Differential expression of VEGF isoforms in mouse during development and in the adult. Dev Dyn. 2001;220:112–121. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1093>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 42.Wu B, et al. Endocardial cells form the coronary arteries by angiogenesis through myocardial-endocardial VEGF signaling. Cell. 2012;151:1083–1096. doi: 10.1016/j.cell.2012.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matsumoto K, Yoshitomi H, Rossant J, Zaret KS. Liver organogenesis promoted by endothelial cells prior to vascular function. Science. 2001;294:559–563. doi: 10.1126/science.1063889. [DOI] [PubMed] [Google Scholar]
- 44.Lammert E, Cleaver O, Melton D. Induction of pancreatic differentiation by signals from blood vessels. Science. 2001;294:564–567. doi: 10.1126/science.1064344. [DOI] [PubMed] [Google Scholar]
- 45.Rafii S, Butler JM, Ding BS. Angiocrine functions of organ-specific endothelial cells. Nature. 2016;529:316–325. doi: 10.1038/nature17040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Laguens R, et al. Entrance in mitosis of adult cardiomyocytes in ischemic pig hearts after plasmid-mediated rhVEGF165 gene transfer. Gene Ther. 2002;9:1676–1681. doi: 10.1038/sj.gt.3301844. [DOI] [PubMed] [Google Scholar]
- 47.Vera Janavel G, et al. Plasmid-mediated VEGF gene transfer induces cardiomyogenesis and reduces myocardial infarct size in sheep. Gene Ther. 2006;13:1133–1142. doi: 10.1038/sj.gt.3302708. [DOI] [PubMed] [Google Scholar]
- 48.Laguens R, et al. Cardiomyocyte hyperplasia after plasmid-mediated vascular endothelial growth factor gene transfer in pigs with chronic myocardial ischemia. J Gene Med. 2004;6:222–227. doi: 10.1002/jgm.478. [DOI] [PubMed] [Google Scholar]
- 49.Nakada Y, et al. Hypoxia induces heart regeneration in adult mice. Nature. 2017;541:222–227. doi: 10.1038/nature20173. [DOI] [PubMed] [Google Scholar]
- 50.Ladoux A, Frelin C. Hypoxia is a strong inducer of vascular endothelial growth factor mRNA expression in the heart. Biochem Biophys Res Commun. 1993;195:1005–1010. doi: 10.1006/bbrc.1993.2144. [DOI] [PubMed] [Google Scholar]
- 51.Forsythe JA, et al. Activation of vascular endothelial growth factor gene transcription by hypoxia-inducible factor 1. Mol Cell Biol. 1996;16:4604–4613. doi: 10.1128/mcb.16.9.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin YD, et al. Instructive nanofiber scaffolds with VEGF create a microenvironment for arteriogenesis and cardiac repair. Sci Transl Med. 2012;4:146ra109. doi: 10.1126/scitranslmed.3003841. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.