Abstract
Context
Self-limited delayed puberty (DP) segregates in an autosomal-dominant pattern, but the genetic basis is largely unknown. Although DP is sometimes seen in relatives of patients with hypogonadotropic hypogonadism (HH), mutations in genes known to cause HH that segregate with the trait of familial self-limited DP have not yet been identified.
Objective
To assess the contribution of mutations in genes known to cause HH to the phenotype of self-limited DP.
Design, Patients, and Setting
We performed whole-exome sequencing in 67 probands and 93 relatives from a large cohort of familial self-limited DP, validated the pathogenicity of the identified gene variant in vitro, and examined the tissue expression and functional requirement of the mouse homolog in vivo.
Results
A potentially pathogenic gene variant segregating with DP was identified in 1 of 28 known HH genes examined. This pathogenic variant occurred in HS6ST1 in one pedigree and segregated with the trait in the six affected members with heterozygous transmission (P = 3.01 × 10−5). Biochemical analysis showed that this mutation reduced sulfotransferase activity in vitro. Hs6st1 mRNA was expressed in peripubertal wild-type mouse hypothalamus. GnRH neuron counts were similar in Hs6st1+/− and Hs6st1+/+ mice, but vaginal opening was delayed in Hs6st1+/− mice despite normal postnatal growth.
Conclusions
We have linked a deleterious mutation in HS6ST1 to familial self-limited DP and show that heterozygous Hs6st1 loss causes DP in mice. In this study, the observed overlap in potentially pathogenic mutations contributing to the phenotypes of self-limited DP and HH was limited to this one gene.
A damaging mutation in HS6ST1 was found to cause familial self-limited delayed puberty (DP), and heterozygous Hs6st1 loss produced DP in mice. Mutations in other GnRH deficiency genes were not seen.
Abnormal pubertal timing affects >4% of adolescents and is associated with adverse health and psychosocial outcomes (1–3). However, in most patients with early or late onset of puberty, the underlying pathophysiology is unknown. Self-limited delayed puberty (DP) represents an extreme variant of normal pubertal timing and has been shown to cluster in families (4). Several groups have noted that DP segregates in an autosomal-dominant pattern (4, 5), suggesting monogenic or oligogenic inheritance conferred by haploinsufficiency or a gain-of-function mutation. As such, pedigrees with familial DP represent an invaluable resource to investigate the genetic regulation of puberty onset.
Significant insights into the genetic control of the hypothalamic–pituitary–gonadal (HPG) axis have come from the discovery of rare variants underlying conditions of GnRH deficiency, such as hypogonadotropic hypogonadism (HH) (6). Perturbed embryonic migration of GnRH neurons from the nose to hypothalamus has been shown to cause HH in humans and in animal models (7, 8). Alternatively, HH may arise through mutations that affect GnRH secretion or function in the presence of a normal GnRH neuron number in the hypothalamus (9).
In view of the possible overlap between the pathophysiology of DP and conditions of GnRH deficiency, a few studies have examined the contribution of mutations in genes that cause HH to the pathogenesis of DP (10–12). Potentially pathogenic variants in a small number of genes causing HH (GNRHR, TAC3, TACR3, IL17RD, and SEMA3A) were identified by whole-exome sequencing (WES) in a few cases of self-limited DP, including those with constitutional delay of growth and puberty (13). Moreover, evidence from WES in our own cohort supports the hypothesis that mutations in genes that influence GnRH neuronal migration or development can cause self-limited DP (14). In this study, we assessed the contribution of mutations in genes known to cause HH to the phenotype of familial self-limited DP and have identified a heterozygous HS6ST1 mutation as a novel cause of self-limited DP.
Methods
Patients
The patients selected for this study belong to a previously described and accurately phenotyped Finnish DP cohort, for which diagnosis is based on objective evidence of a delayed pubertal growth spurt rather than self-recall (15). Patients were referred with DP to specialist pediatric care in central and southern Finland from 1982 to 2004. All patients (n = 492) met the diagnostic criteria for self-limited DP, defined as the onset of Tanner genital stage II (testicular volume >3 mL) >13.5 years in boys or Tanner breast stage II >13.0 years in girls (i.e., 2 SD later than average pubertal development) (16). Medical history, clinical examination, and routine laboratory tests were reviewed to exclude those with chronic illness. HH, if suspected, was excluded by spontaneous pubertal development at follow-up. Normal fertility has been demonstrated in affected family members (5). Families of the DP patients were invited to participate via structured interviews and using archived height measurement records. The criteria for DP in probands’ family members were: (1) age at take-off; or (2) peak height velocity occurring 1.5 SD beyond the mean, that is, age at take-off >12.9 and 11.3 years, or age at peak height velocity >14.8 and 12.8 years in males and females, respectively; or (3) age at attaining adult height >18 or 16 years in males and females, respectively (15). Written informed consent was obtained from all participants. The study protocol was approved by the Ethics Committee for Pediatrics, Adolescent Medicine and Psychiatry, Hospital District of Helsinki and Uusimaa (570/E7/2003). UK ethical approval was granted by the London–Chelsea National Research Ethics Service committee (13/LO/0257). The study was conducted in accordance with the guidelines of the Declaration of Helsinki.
Genetic analysis
Genetic analysis was performed in 67 probands with DP from those 67 families with the greatest number of affected individuals in our cohort (male, n = 57; female, n = 10, see Supplemental Fig. 1), 58 affected family members (male, n = 36; female, n = 22), and 35 of their unaffected family members (male, n = 13; female, n = 22). WES was performed on DNA extracted from peripheral blood leukocytes of these 160 individuals, using a Nimblegen v.2 or Agilent v.5 platform and Illumina HiSeq 2000 sequencing. The exome sequences were aligned to the University of California Santa Cruz hg19 reference genome. Picard tools and the Genome Analysis Toolkit were used to mark PCR duplicates, realign around indels, recalibrate quality scores, and call variants.
Variants were analyzed and filtered for potential causal variants using filters for quality control, predicted function, minor allele frequency (MAF), and biological relevance (Fig. 1). Filtering by MAF included only variants with MAF <1% in the 1000 Genomes database, the National Heart, Lung, and Blood Institute exome variant server, and the ExAC and gnomAD databases. Biological relevance filtering allowed prioritization of variants according to our “HH genes” list (Table 1), comprising genes known to be relevant to the phenotype of HH based on previously published studies and using pathway analysis with MetaCore (GeneGo/Thomson Reuters). The segregation with trait filter retained only variants present in ≥n − 1 affected individuals (where n indicates the number of affected individuals in a given pedigree) and not present in more than one unaffected individual (Table 1).
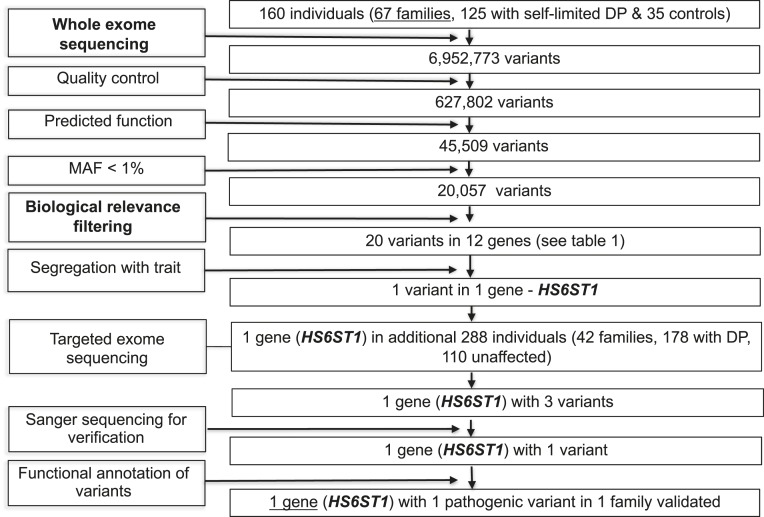
Figure 1.
Flowchart of WES filtering strategy to identify HS6ST1. WES was performed on DNA extracted from peripheral blood leukocytes of 160 individuals from our cohort (67 DP probands, 58 DP relatives, and 35 controls). The exome sequences were aligned to the University of California Santa Cruz hg19 reference genome. Picard tools and the Genome Analysis Toolkit were used to mark PCR duplicates, realign around indels, recalibrate quality scores, and call variants. Potential causal variants were identified using filters for quality control, predicted functional annotation, MAF, biological relevance (i.e., “HH gene” list), and segregation with trait (see Methods and Table 1 for further information on filtering criteria). Targeted exome sequencing using a Fluidigm array of one candidate gene was then performed in a further 42 families from the same cohort (288 individuals, 178 with DP and 110 controls). Variants identified via targeted resequencing were filtered using the same criteria as the WES data, with additional rare variant burden testing. Functional annotation of the variants is as described in “Methods.”
Table 1.
HH Gene Filtering
| Gene |
No. of Rare and Predicated Damaging Variants Filtered From WES Data
|
|
|---|---|---|
| Not Segregating With the DP Trait | Segregating With the DP Trait | |
| ANOS1 | 0 | 0 |
| AXL | 3 | 0 |
| CHD7 | 3 | 0 |
| DAX1 | 0 | 0 |
| FEZF1 | 3 | 0 |
| FGF8 | 0 | 0 |
| FGFR1 | 2 | 0 |
| GNRH1 | 0 | 0 |
| GNRHR | 0 | 0 |
| HESX1 | 0 | 0 |
| HS6ST1 | 1 | 1 |
| KISS1 | 0 | 0 |
| KISS1R | 0 | 0 |
| LEP | 0 | 0 |
| LEPR | 0 | 0 |
| LHX3 | 1 | 0 |
| LHX4 | 1 | 0 |
| NELF | 0 | 0 |
| PCSK1 | 0 | 0 |
| PROK2 | 0 | 0 |
| PROKR2 | 2 | 0 |
| PROP1 | 1 | 0 |
| SEMA3A | 0 | 0 |
| SF1 | 0 | 0 |
| SPRY4 | 1 | 0 |
| TAC3 | 1 | 0 |
| TAC3R | 0 | 0 |
| WDR11 | 1 | 0 |
Twenty-eight HH genes, mutations in which have been identified as causal or potentially causal in patients with GnRH deficiency, used for filtering after WES are shown. Details are given of number of variants in these genes identified in 67 probands with DP after WES. Variants are divided into those that were found to segregate with the DP trait within cohort pedigrees and those that did not segregate.
Targeted exome sequencing (Fluidigm) of the remaining candidate gene postfiltering was performed in a further 42 families from the same cohort (288 individuals, 178 with DP: male, n = 106; female, n = 72; and 110 controls: male, n = 55; female, n = 55; Fig. 1), with filtering as in Howard et al. (14). Whole-gene rare variant burden testing was performed after sequencing. A Fisher’s exact test was used to compare the prevalence of deleterious variants in our cohort with the Finnish population, using the ExAC browser (Exome Aggregation Consortium, Cambridge, MA). For each gene, all variants from the ExAC database with minor allele frequency <2.5%, predicted to be deleterious by both Polyphen-2 (17) and SIFT (18), were included in the analysis, with each family in our cohort represented by the proband only. A multiple comparison adjustment was applied post hoc using the Benjamini–Hochberg method (19), as detailed in Howard et al. (14). All variants identified were confirmed via Sanger sequencing.
In vitro assay of sulfotransferase activity
The human HS6ST1 cDNA (IMAGE consortium accession ID BC099638) was obtained from GeneCopoeia in a pReceiverM14a backbone (pReceiverM14a-WThHS6ST1). The p.Arg375His and p.Met404Val point mutations were introduced by PCR-mediated mutagenesis (QuickChange II, Agilent Technologies), using the following primers: Hs6st1_R375H_FOR, 5′-GCCTGAGGAGCCACGAGGAGCGTCT-3′, Hs6st1_R375H_REV 5′-AGACGCTCCTCGTGGCTCCTCAGGC-3′ for p.Arg375His; and Hs6st1_M404V_FOR, 5′-CCCACCGAGGACTACGTGAGCCACATCATTG-3′, Hs6st1_M404V_REV, 5′-CAATGATGTGGCTCACGTAGTCCTCGGTGGG-3′ for p.Met404Val. Mutations were confirmed by Sanger sequencing. COS7 cells, at 90% confluency in six-well plates, were transfected with 2 μg of wild-type (WT) or mutant plasmids using FuGENE HD (Promega). After 48 hours, cells were lysed with 50 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, and 1% Triton X-100 (pH 7.4). The homogenates were stirred for 1 hour and then centrifuged at 10,000 × g for 30 seconds at 4°C to clear debris. HS6ST1-FLAG fusion proteins were isolated from the supernatant using an anti-FLAG M2 affinity chromatography kit (A2220, Sigma-Aldrich). The purity of the eluted protein was checked by Coomassie brilliant blue (Thermo Fisher) staining of SDS-PAGE electrophoretic gels. Protein relative amounts were calculated by densitometric analysis of Western blots with mouse monoclonal M1 anti-FLAG antibody (1:1000 dilution, Sigma-Aldrich).
Sulfotransferase activity was measured using R&D Systems’ colorimetric assay (EA003). Briefly, different amounts of purified HS6ST1 proteins were added to a mix of 0.2 mM 3′-phosphoadenosine-5′-phosphosulfate (donor), 1 mM N-acetylglucosamine (acceptor), and 10 ng/mL inositol monophosphatase 3 (coupling enzyme) in a total volume of 50 μL of reaction buffer. The elution product of mock-transfected cells was used as negative control. The reaction was allowed to proceed for 20 minutes at 37°C with inorganic phosphate measured immediately after by the malachite green method. Enzymatic activity was represented as percentage of WT activity.
Animals
Animal procedures were performed in accordance with institutional and UK Home Office guidelines. The Hs6st1+/− mice have been described previously (20, 21) and were maintained on a C57BL/6J background by pairing with WT partners. Hs6st1+/+ and Hs6st1+/− female mice were checked daily for vaginal opening (VO) and weight after weaning, that is, from 21 days of age (22).
Histological analyses
Peripubertal and adult mice were perfused with 4% formaldehyde in PBS and then brains and testis were dissected and postfixed overnight at 4°C. Testes were dehydrated, embedded in paraffin, microtome sectioned at 8 μm. and stained with hematoxylin and eosin, as previously described (23). For in situ hybridization (ISH) or immunostaining, brains were cryoprotected in 30% sucrose and frozen in OCT compound (VWR). Twelve-micrometer-thick serial sagittal and coronal cryosections were collected on Superfrost Plus slides (VWR) and incubated with antisense riboprobes for Gnrh (24) or Hs6st1 (21). In some experiments, we combined ISH with GnRH immunofluorescence and used a different mouse Hs6st1 probe, which was generated by PCR amplification from brain cDNA using 5′-ACTGGACCGAACTCACCAAC-3′ and 5′-AACTCAGTGAGGCCGAAGAA-3′ as primers and cloning into the dual promoter vector pGEM-T easy (Promega). Probes were generated as described (25). For immunoperoxidase staining, mouse 25-μm mouse brain cryosections were incubated with Bloxall (Vector Laboratories) to quench endogenous peroxidase activity before incubation with GnRH primary antibody (1:1000 in PBS, ImmunoStar) followed by incubation with biotinylated goat anti-rabbit antibody (1:400 in PBS; Vector Laboratories). Immunostaining was developed with the ABC kit (Vector Laboratories) and diaminobenzidine (Sigma-Aldrich), as described (24).
Image processing and quantification
Images were acquired either using a Leica DM5500B microscope equipped with a DCF295 camera and DCViewer software (Leica) and then processed with Photoshop CS6 and Illustrator CS6 (Adobe) or acquired on an Axioskop 2 Plus microscope (Zeiss) equipped with a TCH-5.0 ICE digital camera (TiEsseLab). In some experiments, bright-field images of ISH patterns were converted to RGB color mode for superimposition onto fluorescent images. GnRH-positive cells were counted on fifty 25-μm sections through the entire medial preoptic area (MPOA) of each animal. To compare the abundance of GnRH-positive neurites at the median eminence (ME), we measured the pixel intensity of GnRH staining in 25-μm-thick coronal sections through the entire ME.
Statistical significance
Data are expressed as mean ± SEM. Differences between groups were evaluated by a two-tailed student t test and considered significant at P < 0.05. Statistical analysis was performed using Prism (GraphPad Software).
Results
Exome sequencing of families with self-limited DP identified an HS6ST1 variant
WES of 67 informative families from our large cohort with self-limited DP identified 20 rare (MAF <1%) and predicted deleterious variants in 12 genes from a list of known HH genes (Table 1). However, after filtering for segregation with trait, only HS6ST1 (ENSG00000136720, gene ID number 9394) was retained as a candidate gene (Fig. 1). One proband and his affected relatives from one pedigree sequenced carried a rare and likely damaging HS6ST1 variant [NM_004807.2: c.1124G>A (rs182882999) p.Arg375His] that causes a nonconservative amino acid substitution in the coding sequence. This proband carried an additional potentially pathogenic variant in a known HH gene, FEZF1 (NM_ NM_001024613.3: c.1010T>A, p.Iso337Lys). However, this variant was not found in any other affected individual from this family and was, therefore, discounted due to lack of familial segregation.
The rare heterozygous missense variant HS6ST1 (p.Arg375His) is predicted to be deleterious to protein function with five of six prediction tools, as the affected amino acid residue resides in a coiled-coil domain that is highly conserved among species, as revealed by the PhyloP and GERP score and a multiple sequence alignment (Supplemental Fig. 2 and Supplemental Table 1). This specific variant was present at low MAF in some public databases, but whole-gene rare variant burden testing showed significant enrichment of HS6ST1 rare predicted pathogenic variants in our cohort as compared with ethnically matched controls from the ExAC database (adjusted P = 3.01 × 10−5).
After targeted exome sequencing, two additional variants in HS6ST1 were identified in three further affected probands [NM_004807.2: c199A>T (rs202247387) p.Lys67* and c585G>A p.Trp195*]. However, Sanger sequencing did not validate the existence of these variants. No further rare, potentially damaging, and trait-segregating variants in HS6ST1 were identified by targeted sequencing.
Pedigree with a potentially pathogenic HS6ST1 variant displayed an autosomal-dominant inheritance pattern and classical self-limited DP
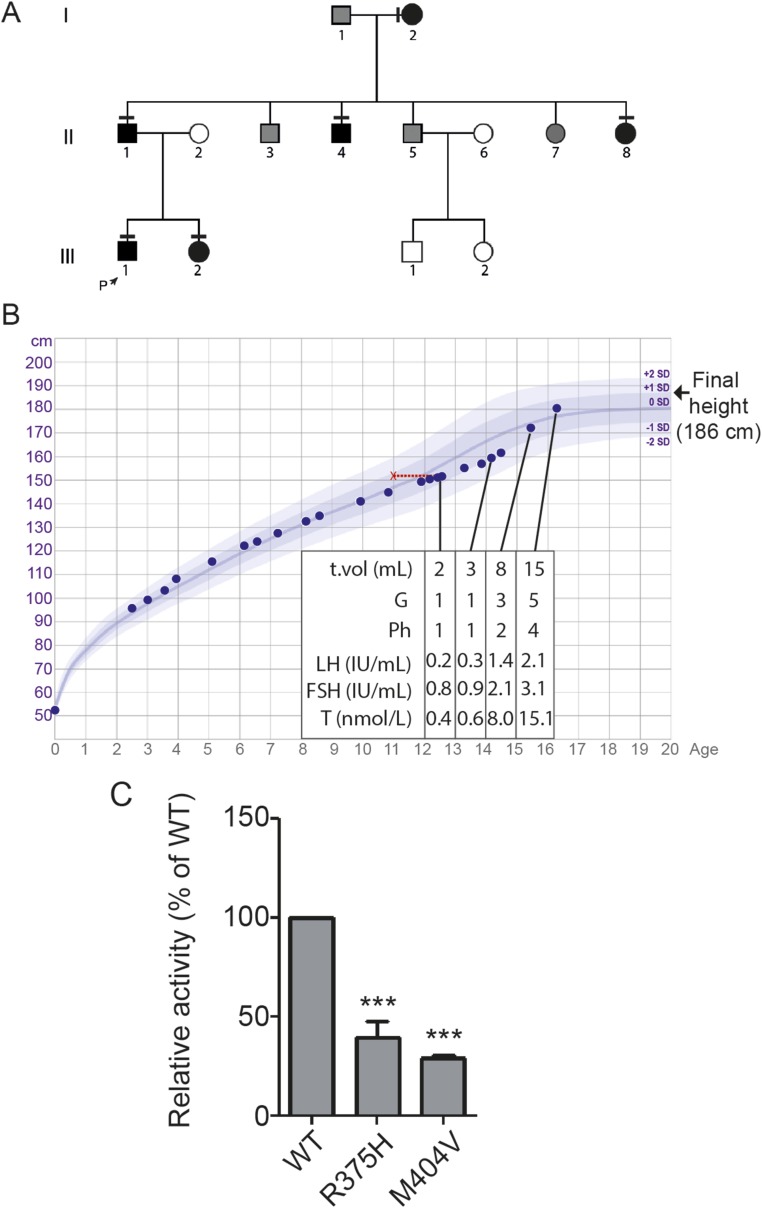
The pedigree with the p.Arg375His variant has several family members with typical features of self-limited DP (Fig. 2A). The proband was first investigated for growth delay at 12.8 years, at which time his bone age was 11 years (Fig. 2B). Examination at this stage found him to be prepubertal, with bilateral testis volumes of 2 mL and no pubic hair development. His blood biochemistry at that time showed a typical picture of functional HH. Spontaneous onset of puberty was observed at ∼14.3 years of age. During the next 2.4 years, he achieved testis volume and testosterone levels within the normal adult range. His sister’s age at menarche was 15 years. Both siblings had normal birth weight and birth length. Their father and paternal uncle and aunt also had DP with delayed pubertal growth spurt. All family members with DP had self-reported normal olfaction.
Figure 2.
Pedigree and clinical details of a patient with a p.Arg375His mutation in HS6ST1 that impairs sulfotransferase activity. (A) Pedigree of the proband with the p.Arg375His mutation. Squares indicate male family members; circles indicate female family members. Black symbols represent clinically affected family members, gray symbols represent family members with unknown phenotype, and clear symbols represent unaffected individuals. The arrow labeled “P” indicates the proband in the family. A horizontal black line above or adjacent to an individual’s symbol indicates that they are heterozygous for the p.Arg375His mutation as identified by either WES or Fluidigm array and verified by Sanger sequencing. (B) Clinical details of the proband with the p.Arg375His mutation. Height chart for the proband shows reduction in growth velocity from 12 y of age, with associated delayed bone age shown in red. The subject was prepubertal until 14.1 y of age. This was followed by spontaneous onset of puberty and subsequent development during 2 y to normal adult testosterone levels and testicular volume. (C) Sulfotransferase activity assay. The p.Arg375His mutation reduces HS6ST1 sulfotransferase activity. Relative specific activity (mean ± SD) of recombinant WT or mutant (p.Arg375His or p.Met404Val) HS6ST1 proteins are shown. All assays were done as three independent biological repeats (n = 3 for each experiment) using equal amounts of protein. ***P < 0.001. G, Tanner genital stage, Ph, Tanner pubic hair stage, T, testosterone; T vol, testicular volume.
The HS6ST1 p.Arg375His mutant protein has reduced sulfotransferase activity in vitro
In vitro analysis of sulfotransferase function of the p.Arg375His mutant HS6ST1 protein (Fig. 2C) showed reduced activity at 39% of the WT HS6ST1 protein (mean ± SD: 39.81 ± 8.21, P < 0.001). This reduction in enzymatic function is within the range seen with other pathogenic mutations in HS6ST1 found in patients with HH (26) and similar to that of the p.Met404Val mutant HS6ST1 protein (mean ± SD: 28.8 ± 1.46). This observation is consistent with the HS6ST1 p.Arg375His mutation as causative in self-limited DP.
Hs6st1 is expressed within the adult hypothalamus and olfactory bulbs
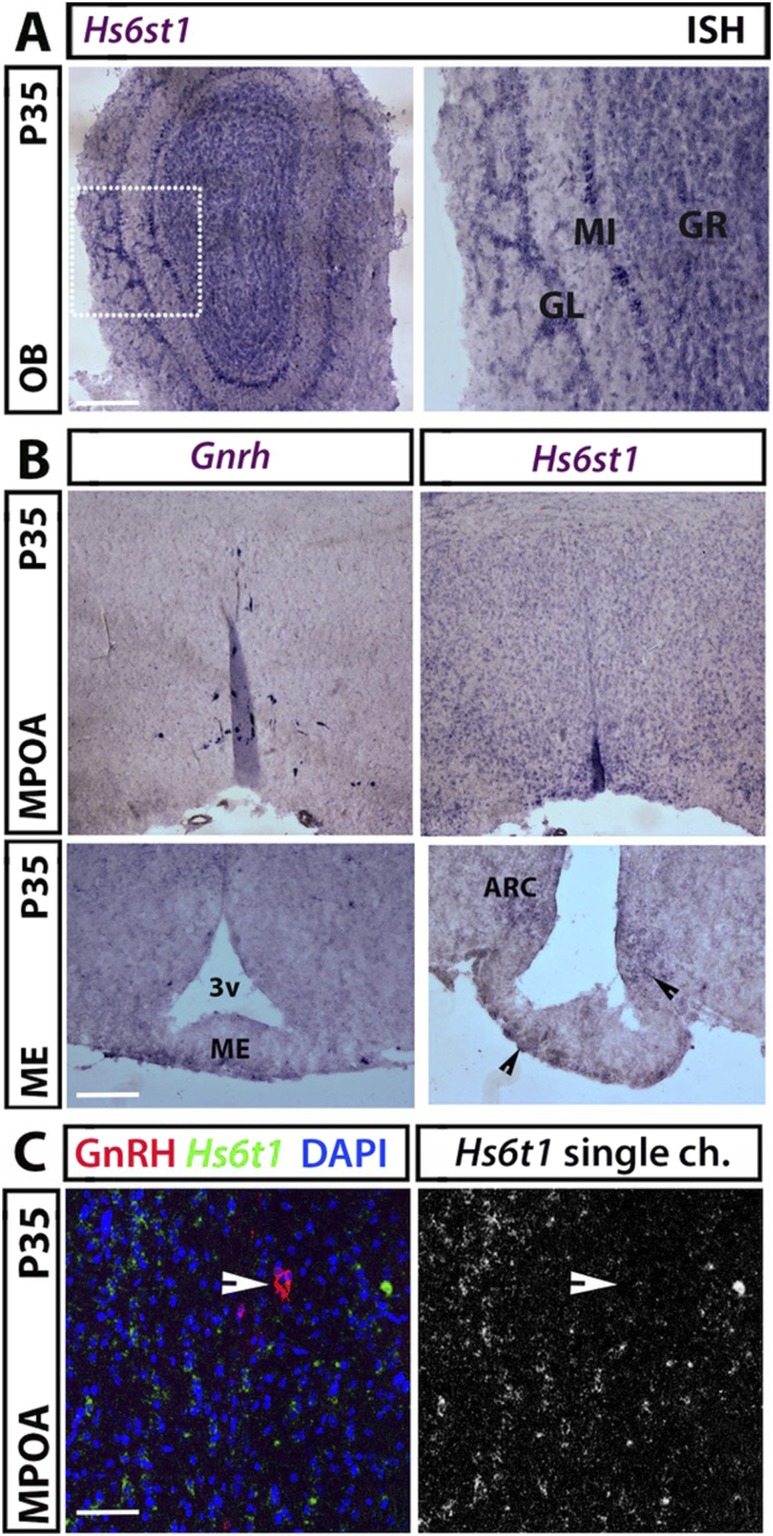
In the adult brain, GnRH neurons are scattered in a bilateral continuum between the olfactory bulb (OB) and the MPOA of the hypothalamus, where most cell bodies reside (27). We found that Hs6st1 is highly expressed in the granular, mitral, and glomerular layers of the adult OB (Fig. 3A) and diffusely in the MPOA (Fig. 3B, upper panel). Hs6st1 expression was also observed in the arcuate nucleus (ARC), which harbors neurons that regulate GnRH secretion (Fig. 3B, lower panel). However, Hs6st1 mRNA did not localize to GnRH+ neurons (Fig. 3C). These observations suggest that Hs6st1 is expressed by hypothalamic cells that do not correspond to GnRH neurons.
Figure 3.
Hs6st1 mRNA expression in GnRH neuronal territories on P35. (A) Coronal sections of adult mouse OB were labeled for Hs6st1 by ISH. The squared box in the left image is shown at higher magnification to the right. (B) Contiguous coronal sections of adult mouse hypothalamus representing the MPOA (upper panels) and the ME (lower panels) were labeled for Gnrh and Hs6st1 by ISH; arrowheads indicate the Hs6st1 expression in the ARC and ME. (C) Hs6st1 ISH (green) of coronal sections from P35 MPOA followed by immunolabeling for GnRH (red) revealed no expression of Hs6st1 in GnRH-positive neurons (example of GnRH neuron is indicated with a solid arrowhead). Scale bars: 125 µm (A and B, upper panels), 50 µm (B, lower panels), 25 µm (C). 3v, third ventricle; GL, glomerular layer; GR, granular layer; MI, mitral layer.
Normal positioning and number of GnRH neurons in the MPOA of Hs6st1 heterozygous mice
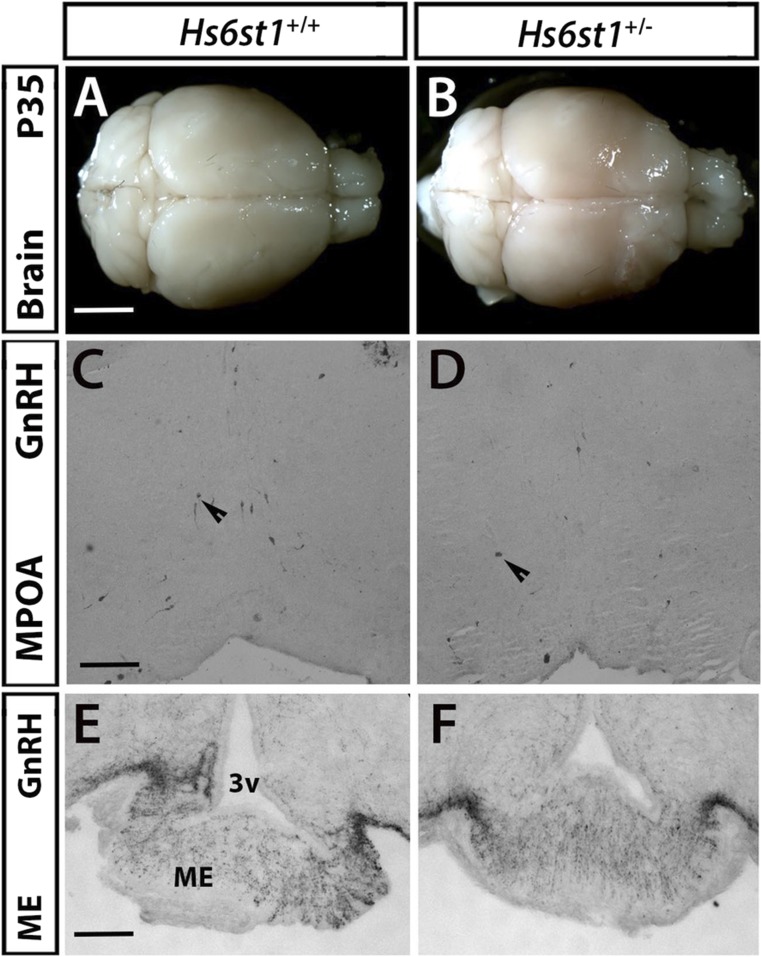
To determine whether HS6ST1 is required for the normal development of the GnRH neuron system, we compared mice heterozygous for a Hs6st1 null allele (Hs6st1+/−) to their WT littermates (Hs6st1+/+). However, there were no gross differences in brain or OB anatomy between the genotypes (Fig. 4A and 4B). Moreover, there were no obvious differences in the positioning and number of GnRH neurons in the MPOA (Fig. 4C and 4D; number of GnRH+ cells: Hs6st1+/− 341.2 ± 24.2 vs Hs6st1+/+ 378.3 ± 30.2, n = 5 each; P = 0.37). GnRH neurons in Hs6st1+/− mice also projected similarly to the ME, the site where they normally release GnRH into the portal blood vessels of the pituitary gland; moreover, the pixel intensity of GnRH-stained neurites was similar in both genotypes (Fig. 4E and 4F; percentage stained neurite area: Hs6st1+/+ 2.9 ± 0.4 vs Hs6st1+/− 3.1 ± 0.3, n = 5 each; P = 0.09).
Figure 4.
Hs6st1+/− mice showed no defects in olfactory bulbs or in the number and projections of GnRH neurons. (A and B) Perfused adult Hs6st1+/+ and Hs6st1+/− brains were dissected, photographed, and showed similar olfactory bulb structure in both genotypes. (C and D) Coronal sections of adult mouse MPOA were immunostained for GnRH; arrowheads indicate examples of GnRH neurons that are normally present both in Hs6st1+/− and WT mice. (E and F) Immunostaining for GnRH in coronal sections of adult mouse ME shows that GnRH neurons project similarly to the ME in Hs6st1+/+ and Hs6st1+/− mice. Scale bars: 3 mm (A and B), 125 µm (C and D), 50 µm (E and F). Arrows demonstrate representative GnRH cell bodies. 3v, third ventricle.
Hs6st1+/− mice show significant DP onset
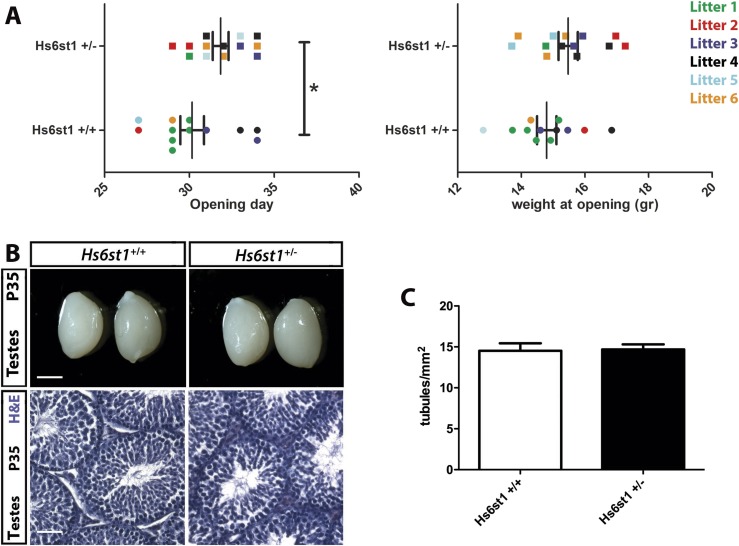
To determine whether heterozygous HS6ST1 deficiency is sufficient to cause DP, we compared the timing of puberty in Hs6st1+/− and Hs6st1+/+ female mice by identifying the day of VO, a proxy measurement for pubertal activation of the HPG axis in mice (22). We found that VO was delayed by on average 1.88 ± 0.82 days in Hs6st1+/− compared with Hs6st1+/+ females (Fig. 5A, left; postnatal day (P) of VO: Hs6st1+/+ 30.2 ± 0.7, n = 12, vs Hs6st1+/− 31.9 ± 0.5, n = 13; P = 0.04; mice were pooled form six different litters). DP in Hs6st1+/− females was not due to overall delayed development, as they were not smaller than Hs6st1+/+ littermate females around the time of VO (weight at P30: Hs6st1+/+ 14.12 ± 0.58 g, n = 12, vs Hs6st1+/−13.67 ± 0.58 g, n = 13; P = 0.29). Instead, and consistent with their DP and thus older age at VO in the Hs6st1+/− females, there was an insignificant trend toward greater weight at the time of VO in Hs6st1+/− compared with Hs6st1+/+ females within the cohort examined (Fig. 5A, right; body weight on the day of VO: Hs6st1+/+ 14.9 ± 0.3 g, n = 12, vs Hs6st1+/− 15.5 ± 0.3 g, n = 13; P = 0.06).
Figure 5.
Peripubertal female Hs6st1+/− mice show delayed VO whereas young adult male Hs6st1+/− mice showed normal testes morphology. (A) Age (left graph) and weight (right graph) at the time of the VO in female Hs6st1+/+ (n = 12) and Hs6st1+/− (n = 13) mice. Values for littermates are shown in the same color. *P < 0.05. (B and C) Hs6st1+/+ and Hs6st1+/− mice have similar testes size (B, upper panels), normal spermatogenesis after H&E staining of paraffin sections (B, lower panels), and similar number of seminiferous tubules (C). Scale bars: 3 mm (B, upper panels), 25 µm (B, lower panels).
Despite DP, the fertility of young adult Hs6st1+/− males and females of both sexes appeared normal, because they produced litters without obvious delay and of normal litter size when paired to WT mice at 4 to 6 months of age (litter size: Hs6st1+/− × Hs6st1+/+: 7.2 ± 0.95 pups per litter in six litters, vs Hs6st1+/+ × Hs6st1+/+ 7.3 ± 0.71 pups per litter in seven litters; P = 0.89; Hs6st1+/− mice were born at the expected Mendelian ratio). In agreement with the ability to father litters of normal size, testes size was similar in young adult Hs6st1+/− and Hs6st1+/+ males (Fig. 5B; P35 testes length (in mm): Hs6st1+/− 8.30 ± 0.16, n = 6, vs Hs6st1+/+ 8.31 ± 0.17, n = 8; P = 0.97). Moreover, testes showed normal organization of germ cells and interstitial Leydig cells and contained a similar number of seminiferous tubules per area (Fig. 5B and 5C; number of seminiferous tubules per mm2: Hs6st1+/+ 14.53 ± 0.91 vs Hs6st1+/− 14.69 ± 0.62; n = 5 each; P = 0.88). Taken together, these findings show that HS6ST1 haploinsufficiency delays puberty in mice without compromising fertility in the adult.
Discussion
The inheritance of self-limited DP is under strong genetic influence, with clear autosomal-dominant segregation in many families with or without complete penetrance (4), and thus represents a useful basis for the investigation of puberty genetics (5). However, only a few genes responsible for self-limited DP have been identified. In view of the possible overlap between the pathophysiology of DP and HH, screening self-limited DP patients for mutations in known HH genes appears a prudent strategy; however, minimal genetic overlap between the two conditions has been identified to date (13). Additionally, there have been some observed differences in inheritance patterns and penetrance between the two conditions (28). In HH, >40 different genes have been identified, and incomplete penetrance is often seen within pedigrees, leading to a wide spectrum of phenotypes (29–31). As the understanding of the genetic basis of both self-limited DP and HH improves, it is likely that genetic testing will be able to help establish a definitive diagnosis in adolescent patients presenting with delayed onset of puberty.
In this study, we have identified a deleterious mutation, p.Arg375His, in the HH gene HS6ST1 as the likely causal factor for self-limited DP in one pedigree from our cohort of patients with familial DP.
HS6ST1 mutations have been previously identified in up to 2% of patients with idiopathic HH (26, 32), but they have not previously been reported in pedigrees segregating with the trait of self-limited DP. Our in vitro analysis showed that the HS6ST1 p.Arg375His mutation had reduced sulfotransferase activity, comparable to previously published mutations in this gene in patients with HH (26). Taken together, these results suggest a working model in which HS6ST1 heterozygosity causes self-limited DP, whereas potentially HS6ST1 homozygosity or a “second hit” in a separate gene would produce the phenotype of HH.
Our studies in a murine model corroborate heterozygous Hs6st1 deficiency as a cause of delayed pubertal timing without compromised fertility. Thus, Hs6st1+/− mice were born at normal Mendelian ratios without obvious defects in GnRH neuron or testes development, but females showed delayed VO. The question of sex bias in self-limited DP is an interesting one, as it is diagnosed in up to 83% of boys and 30% of girls presenting with pubertal delay (33–35). The underlying reasons for this sex difference are not clear. However, this difference may be due, in part, to ascertainment bias. Indeed, a previous study has shown a nearly equal sex ratio consistent with autosomal-dominant inheritance when all of the affected individuals from our large cohort were examined, as opposed to the probands only (5).
In our murine model of Hs6st1+/− mice, we did not find any evidence of a sex difference in anatomical studies. In this study only females were examined for timing of pubertal onset (VO), as VO is a very robust marker of pubertal onset in female mice. Male mice were not assessed for the timing of puberty, but the adult male mice had normal testicular morphology, including the presence of spermatogenesis. Normal spermatogenesis is an excellent marker in males for reproductive competence and excludes GnRH deficiency. In female mice the reproductive competence was assessed as fertility of the females. With these methods GnRH deficiency was excluded both in male and female Hs6st1+/− mice. Importantly, mice deficient in Hs6st1 were not significantly smaller or lighter than their WT littermates, excluding poor growth as the underlying cause for pubertal delay. The viability of Hs6st1+/− mice is in contrast to that of Hs6st1 knockout (Hs6st1−/−) mice, which could not be examined for puberty defects due to embryonic lethality (36).
Our expression studies in mice indicate that Hs6st1 is expressed in several tissues relevant to normal GnRH neuron migration or function, including the OB and the MPOA, ARC, and PVN of the hypothalamus. However, we did not identify any obvious abnormalities in OB morphology, GnRH neuron number in the MPOA, or in GnRH neuron innervation of the ME in Hs6st1+/− mice. Instead, Hs6st1 expression in the ARC and PVN, where kisspeptin neurons and tanycytes modulate GnRH secretion and function (37, 38), raises the possibility that HS6ST1 haploinsufficiency affects the regulation of GnRH neuron activity or other relevant downstream pathways.
Hs6st1 is required for the function of Anos1 (also known as Kal1) and Fgfr1, two genes that are required for normal HPG axis function (26). In a Caenorhabditis elegans model, Hs6st1 regulates neural branching in concert with Anos1 and Fgfr1 (26). Moreover, Anos1 is also able to enhance Fgfr1 signaling in an HS-dependent manner in an immortalized human cell model of GnRH neurons, and it has been proposed that this interaction promotes olfactory and GnRH neuron development (39). However, Hs6st1 mutations may also impact the HPG axis via Anos1-independent pathways, as HS modifications of many different proteoglycans in the extracellular matrix have the potential to affect multiple signaling pathways (40) previously implicated in the neuroendocrine control of fertility (41).
In summary, we have identified a new pathogenic mutation in HS6ST1 as the likely cause of DP in a pedigree from our extensive patient cohort, with no other pathogenic mutations in genes known to cause HH identified in our cohort. These findings suggest that, with the exception of a few genes like HS6ST1, the genetic background of HH and DP is either largely different or is due to mutations in as yet undiscovered genes. Our findings differ from those in previous publications, in which up to 14% of DP probands were reported to carry potentially pathogenic variants in HH genes (13). However, previous results have not been adjusted for segregation with the DP trait within families, nor have the variants identified in these previous studies been tested for pathogenicity, and thus prior estimates of HH mutation rates in DP patients may have been an overestimation. In agreement, prior to adjustment for segregation, our results suggested that 17.9% of DP probands had potentially pathogenic variants in HH genes, highlighting the value of familial data in identification of causal variants in a condition such as DP. Although further pedigrees with familial DP need to be studied to identify additional causative genes and provide accurate estimates of the genetic HH and DP overlap, our findings provide evidence that perturbations in a single allele of a gene regulating the HPG axis is sufficient to cause self-limited DP. In contrast, available evidence suggests that more deleterious alterations in the same gene, or in combination with additional genes, are required to cause more severe HH phenotypes.
Supplementary Material
Acknowledgments
We thank the patients and families who contributed their time, medical information, and DNA samples to this study. We also thank Laura Denti and the staff of the Biological Resources Unit at the University College London Institute of Ophthalmology for help with mouse husbandry.
Financial Support: S.R.H. was funded by the National Institutes for Health Research, Wellcome Trust Award 102745, and by Rosetrees Trust Award M222. A.P. was funded by Barts and The London Charity Strategic Research Grant MEAG1l6R. L.G. was funded by Biotechnology and Biological Sciences Research Council Grant BB/L002671/1. L.D. was partially supported by Academy of Finland Grant 14135. M.R.B. and C.P.C. were funded by the National Institutes for Health Research, and this work forms part of the portfolio of translational research of the National Institutes for Health Research Biomedical Research Unit at Barts. A.C. was funded by Telethon Foundation Grant GGP13142. C.R. was funded by the Biotechnology and Biological Sciences Research Council Grant BB/L002639/1.
Disclosure Summary: The authors have nothing to disclose.
Glossary
Abbreviations:
- ARC
arcuate nucleus
- DP
delayed puberty
- HH
hypogonadotropic hypogonadism
- HPG
hypothalamic–pituitary–gonadal
- ISH
in situ hybridization
- MAF
minor allele frequency
- ME
median eminence
- MPOA
medial preoptic area
- OB
olfactory bulb
- P
postnatal day
- VO
vaginal opening
- WES
whole-exome sequencing
- WT
wild-type
References
- 1. Ritte R, Lukanova A, Tjonneland A, Olsen A, Overvad K, Mesrine S, Fagherazzi G, Dossus L, Teucher B, Steindorf K, Boeing H, Aleksandrova K, Trichopoulou A, Lagiou P, Trichopoulos D, Palli D, Grioni S, Mattiello A, Tumino R, Sacerdote C, Quiros JR, Buckland G, Molina-Montes E, Chirlaque MD, Ardanaz E, Amiano P, Bueno-de-Mesquita B, van Duijnhoven F, van Gils CH, Peeters PH, Wareham N, Khaw KT, Key TJ, Travis RC, Krum-Hansen S, Gram IT, Lund E, Sund M, Andersson A, Romieu I, Rinaldi S, McCormack V, Riboli E, Kaaks R. Height, age at menarche and risk of hormone receptor-positive and -negative breast cancer: a cohort study. Int J Cancer. 2013;132(11):2619–2629. [DOI] [PubMed] [Google Scholar]
- 2. He C, Zhang C, Hunter DJ, Hankinson SE, Buck Louis GM, Hediger ML, Hu FB. Age at menarche and risk of type 2 diabetes: results from 2 large prospective cohort studies. Am J Epidemiol. 2010;171(3):334–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Day FR, Elks CE, Murray A, Ong KK, Perry JR. Puberty timing associated with diabetes, cardiovascular disease and also diverse health outcomes in men and women: the UK Biobank study. Sci Rep. 2015;5(1):11208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sedlmeyer IL, Hirschhorn JN, Palmert MR. Pedigree analysis of constitutional delay of growth and maturation: determination of familial aggregation and inheritance patterns. J Clin Endocrinol Metab. 2002;87(12):5581–5586. [DOI] [PubMed] [Google Scholar]
- 5. Wehkalampi K, Widén E, Laine T, Palotie A, Dunkel L. Patterns of inheritance of constitutional delay of growth and puberty in families of adolescent girls and boys referred to specialist pediatric care. J Clin Endocrinol Metab. 2008;93(3):723–728. [DOI] [PubMed] [Google Scholar]
- 6. Beate K, Joseph N, de Roux N, Wolfram K. Genetics of isolated hypogonadotropic hypogonadism: role of GnRH receptor and other genes. Int J Endocrinol. 2012;2012:147893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wray S. From nose to brain: development of gonadotrophin-releasing hormone-1 neurones. J Neuroendocrinol. 2010;22(7):743–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cadman SM, Kim SH, Hu Y, González-Martínez D, Bouloux PM. Molecular pathogenesis of Kallmann’s syndrome. Horm Res. 2007;67(5):231–242. [DOI] [PubMed] [Google Scholar]
- 9. Seminara SB, Messager S, Chatzidaki EE, Thresher RR, Acierno JS Jr, Shagoury JK, Bo-Abbas Y, Kuohung W, Schwinof KM, Hendrick AG, Zahn D, Dixon J, Kaiser UB, Slaugenhaupt SA, Gusella JF, O’Rahilly S, Carlton MB, Crowley WF Jr, Aparicio SA, Colledge WH. The GPR54 gene as a regulator of puberty. N Engl J Med. 2003;349(17):1614–1627. [DOI] [PubMed] [Google Scholar]
- 10. Lin L, Conway GS, Hill NR, Dattani MT, Hindmarsh PC, Achermann JC. A homozygous R262Q mutation in the gonadotropin-releasing hormone receptor presenting as constitutional delay of growth and puberty with subsequent borderline oligospermia. J Clin Endocrinol Metab. 2006;91(12):5117–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vaaralahti K, Wehkalampi K, Tommiska J, Laitinen EM, Dunkel L, Raivio T. The role of gene defects underlying isolated hypogonadotropic hypogonadism in patients with constitutional delay of growth and puberty. Fertil Steril. 2011;95(8):2756–2758. [DOI] [PubMed] [Google Scholar]
- 12. Pugliese-Pires PN, Fortin JP, Arthur T, Latronico AC, Mendonca BB, Villares SM, Arnhold IJ, Kopin AS, Jorge AA. Novel inactivating mutations in the GH secretagogue receptor gene in patients with constitutional delay of growth and puberty. Eur J Endocrinol. 2011;165(2):233–241. [DOI] [PubMed] [Google Scholar]
- 13. Zhu J, Choa RE, Guo MH, Plummer L, Buck C, Palmert MR, Hirschhorn JN, Seminara SB, Chan YM. A shared genetic basis for self-limited delayed puberty and idiopathic hypogonadotropic hypogonadism. J Clin Endocrinol Metab. 2015;100(4):E646–E654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Howard SR, Guasti L, Ruiz-Babot G, Mancini A, David A, Storr HL, Metherell LA, Sternberg MJ, Cabrera CP, Warren HR, Barnes MR, Quinton R, de Roux N, Young J, Guiochon-Mantel A, Wehkalampi K, André V, Gothilf Y, Cariboni A, Dunkel L. IGSF10 mutations dysregulate gonadotropin-releasing hormone neuronal migration resulting in delayed puberty. EMBO Mol Med. 2016;8(6):626–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wehkalampi K, Widén E, Laine T, Palotie A, Dunkel L. Association of the timing of puberty with a chromosome 2 locus. J Clin Endocrinol Metab. 2008;93(12):4833–4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Palmert MR, Dunkel L. Delayed puberty. N Engl J Med. 2012;366(5):443–453. [DOI] [PubMed] [Google Scholar]
- 17. Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4(7):1073–1081. [DOI] [PubMed] [Google Scholar]
- 19. Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125(1-2):279–284. [DOI] [PubMed] [Google Scholar]
- 20. Leighton PA, Mitchell KJ, Goodrich LV, Lu X, Pinson K, Scherz P, Skarnes WC, Tessier-Lavigne M. Defining brain wiring patterns and mechanisms through gene trapping in mice. Nature. 2001;410(6825):174–179. [DOI] [PubMed] [Google Scholar]
- 21. Tillo M, Charoy C, Schwarz Q, Maden CH, Davidson K, Fantin A, Ruhrberg C. 2- and 6-O-sulfated proteoglycans have distinct and complementary roles in cranial axon guidance and motor neuron migration. Development. 2016;143(11):1907–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Messina A, Langlet F, Chachlaki K, Roa J, Rasika S, Jouy N, Gallet S, Gaytan F, Parkash J, Tena-Sempere M, Giacobini P, Prevot V. A microRNA switch regulates the rise in hypothalamic GnRH production before puberty. Nat Neurosci. 2016;19(6):835–844. [DOI] [PubMed] [Google Scholar]
- 23. Macchi C, Steffani L, Oleari R, Lettieri A, Valenti L, Dongiovanni P, Romero-Ruiz A, Tena-Sempere M, Cariboni A, Magni P, Ruscica M. Iron overload induces hypogonadism in male mice via extrahypothalamic mechanisms. Mol Cell Endocrinol. 2017;454:135–145. [DOI] [PubMed] [Google Scholar]
- 24. Cariboni A, André V, Chauvet S, Cassatella D, Davidson K, Caramello A, Fantin A, Bouloux P, Mann F, Ruhrberg C. Dysfunctional SEMA3E signaling underlies gonadotropin-releasing hormone neuron deficiency in Kallmann syndrome. J Clin Invest. 2015;125(6):2413–2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Guasti L, Paul A, Laufer E, King P. Localization of Sonic hedgehog secreting and receiving cells in the developing and adult rat adrenal cortex. Mol Cell Endocrinol. 2011;336(1–2):117–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tornberg J, Sykiotis GP, Keefe K, Plummer L, Hoang X, Hall JE, Quinton R, Seminara SB, Hughes V, Van Vliet G, Van Uum S, Crowley WF, Habuchi H, Kimata K, Pitteloud N, Bülow HE. Heparan sulfate 6-O-sulfotransferase 1, a gene involved in extracellular sugar modifications, is mutated in patients with idiopathic hypogonadotrophic hypogonadism. Proc Natl Acad Sci USA. 2011;108(28):11524–11529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Merchenthaler I, Görcs T, Sétáló G, Petrusz P, Flerkó B. Gonadotropin-releasing hormone (GnRH) neurons and pathways in the rat brain. Cell Tissue Res. 1984;237(1):15–29. [DOI] [PubMed] [Google Scholar]
- 28. Howard SR. Genes underlying delayed puberty. Mol Cell Endocrinol. 2018;S0303-7207(18)30149-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Raivio T, Falardeau J, Dwyer A, Quinton R, Hayes FJ, Hughes VA, Cole LW, Pearce SH, Lee H, Boepple P, Crowley WF Jr, Pitteloud N. Reversal of idiopathic hypogonadotropic hypogonadism. N Engl J Med. 2007;357(9):863–873. [DOI] [PubMed] [Google Scholar]
- 30. Pitteloud N, Quinton R, Pearce S, Raivio T, Acierno J, Dwyer A, Plummer L, Hughes V, Seminara S, Cheng YZ, Li WP, Maccoll G, Eliseenkova AV, Olsen SK, Ibrahimi OA, Hayes FJ, Boepple P, Hall JE, Bouloux P, Mohammadi M, Crowley W. Digenic mutations account for variable phenotypes in idiopathic hypogonadotropic hypogonadism. J Clin Invest. 2007;117(2):457–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sidhoum VF, Chan YM, Lippincott MF, Balasubramanian R, Quinton R, Plummer L, Dwyer A, Pitteloud N, Hayes FJ, Hall JE, Martin KA, Boepple PA, Seminara SB. Reversal and relapse of hypogonadotropic hypogonadism: resilience and fragility of the reproductive neuroendocrine system. J Clin Endocrinol Metab. 2014;99(3):861–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pitteloud N, Meysing A, Quinton R, Acierno JS Jr, Dwyer AA, Plummer L, Fliers E, Boepple P, Hayes F, Seminara S, Hughes VA, Ma J, Bouloux P, Mohammadi M, Crowley WF Jr. Mutations in fibroblast growth factor receptor 1 cause Kallmann syndrome with a wide spectrum of reproductive phenotypes. Mol Cell Endocrinol. 2006;254-255:60–69. [DOI] [PubMed] [Google Scholar]
- 33. Varimo T, Miettinen PJ, Känsäkoski J, Raivio T, Hero M. Congenital hypogonadotropic hypogonadism, functional hypogonadotropism or constitutional delay of growth and puberty? An analysis of a large patient series from a single tertiary center. Hum Reprod. 2017;32(1):147–153. [DOI] [PubMed] [Google Scholar]
- 34. Lawaetz JG, Hagen CP, Mieritz MG, Blomberg Jensen M, Petersen JH, Juul A. Evaluation of 451 Danish boys with delayed puberty: diagnostic use of a new puberty nomogram and effects of oral testosterone therapy. J Clin Endocrinol Metab. 2015;100(4):1376–1385. [DOI] [PubMed] [Google Scholar]
- 35. Sedlmeyer IL, Palmert MR. Delayed puberty: analysis of a large case series from an academic center. J Clin Endocrinol Metab. 2002;87(4):1613–1620. [DOI] [PubMed] [Google Scholar]
- 36. Habuchi H, Nagai N, Sugaya N, Atsumi F, Stevens RL, Kimata K. Mice deficient in heparan sulfate 6-O-sulfotransferase-1 exhibit defective heparan sulfate biosynthesis, abnormal placentation, and late embryonic lethality. J Biol Chem. 2007;282(21):15578–15588. [DOI] [PubMed] [Google Scholar]
- 37. Parkash J, Messina A, Langlet F, Cimino I, Loyens A, Mazur D, Gallet S, Balland E, Malone SA, Pralong F, Cagnoni G, Schellino R, De Marchis S, Mazzone M, Pasterkamp RJ, Tamagnone L, Prevot V, Giacobini P. Semaphorin7A regulates neuroglial plasticity in the adult hypothalamic median eminence. Nat Commun. 2015;6(1):6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pielecka-Fortuna J, Chu Z, Moenter SM. Kisspeptin acts directly and indirectly to increase gonadotropin-releasing hormone neuron activity and its effects are modulated by estradiol. Endocrinology. 2008;149(4):1979–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hu Y, Guimond SE, Travers P, Cadman S, Hohenester E, Turnbull JE, Kim SH, Bouloux PM. Novel mechanisms of fibroblast growth factor receptor 1 regulation by extracellular matrix protein anosmin-1. J Biol Chem. 2009;284(43):29905–29920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bishop JR, Schuksz M, Esko JD. Heparan sulphate proteoglycans fine-tune mammalian physiology. Nature. 2007;446(7139):1030–1037. [DOI] [PubMed] [Google Scholar]
- 41. Condomitti G, de Wit J. Heparan sulfate proteoglycans as emerging players in synaptic specificity. Front Mol Neurosci. 2018;11:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.