Abstract
Transcription factor AP-1 is constitutively activated and IRF4 drives growth and survival in ALK+ and ALK– anaplastic large cell lymphoma (ALCL). Here we demonstrate high-level BATF and BATF3 expression in ALCL. Both BATFs bind classical AP-1 motifs and interact with in ALCL deregulated AP-1 factors. Together with IRF4, they co-occupy AP-1-IRF composite elements, differentiating ALCL from non-ALCL. Gene-specific inactivation of BATFs, or global AP-1 inhibition results in ALCL growth retardation and/or cell death in vitro and in vivo. Furthermore, the AP-1-BATF module establishes TH17/group 3 innate lymphoid cells (ILC3)-associated gene expression in ALCL cells, including marker genes such as AHR, IL17F, IL22, IL26, IL23R and RORγt. Elevated IL-17A and IL-17F levels were detected in a subset of children and adolescents with ALK+ ALCL. Furthermore, a comprehensive analysis of primary lymphoma data confirms TH17–, and in particular ILC3-skewing in ALCL compared with PTCL. Finally, pharmacological inhibition of RORC as single treatment leads to cell death in ALCL cell lines and, in combination with the ALK inhibitor crizotinib, enforces death induction in ALK+ ALCL. Our data highlight the crucial role of AP-1/BATFs in ALCL and lead to the concept that some ALCL might originate from ILC3.
Introduction
Transcription factors (TFs) are crucial determinants of hematopoietic malignancies [1, 2]. One example of lymphoid neoplasms with distinct dysregulated TF activities is anaplastic large cell lymphoma (ALCL) [3, 4]. The WHO classification distinguishes ALK-positive (ALK+) ALCL, which is characterized by the NPM–ALK fusion, and ALK-negative (ALK–) ALCL lacking ALK translocations [5]. Both belong to peripheral T-cell lymphomas (PTCLs). Whereas ALK is considered as causative of ALK+ ALCL [6], the pathogenesis of ALK– ALCL is less clarified [7, 8].
Albeit both ALCL entities show differences in genomic alterations or gene and microRNA expression [9–11], phenotypically they are highly similar and share biological and molecular key aspects [12–14]. In particular, their deregulated TF programs overlap. They share STAT3 and NOTCH1 activation and high-level interferon regulatory factor 4 (IRF4) and MYC (v-myc myelocytomatosis viral oncogene homolog, c-MYC) expression and activity [7, 13, 15–17]. Moreover, we revealed a unique AP-1 activation in ALCL [14, 18, 19]. Several lines of evidence point toward a crucial role of AP-1 in ALCL: NPM-ALK induces JUNB and JUN [20–22], genomic gains of JUNB and FRA2 loci are found in ALCL [23, 14], inhibition of AP-1 in ALK+ ALCL results in growth arrest and cell death [18, 21, 24], and JUNB and JUN deletion in mouse models impairs NPM-ALK-driven lymphomagenesis [25]. Finally, expression of the AP-1 interacting TF BATF3 distinguishes ALCL from other PTCL [26] and is involved in growth control and survival of ALCL [27].
BATFs, comprising BATF, BATF2 and BATF3, are basic leucine zipper TFs, which modulate transcription primarily by interaction with JUN proteins [28]. The lack of a transactivation domain [28], their redundancy [29], and the number of interaction partners make functional characterization of BATFs challenging. Initially thought to inhibit transcription, recent work highlighted positive regulatory functions of BATFs [28–30]. IRF4 and BATF enhance each other's DNA binding [31], and they cooperatively bind to so-called AP-1-IRF composite elements (AICEs) [29, 31, 32]. Moreover, STAT3, IRF4, JUNB and BATF TFs initiate the fate of T helper 17 (TH17) cells, which subsequently enforces expression of the key TH17 TF RORC2 (murine RORγt) [33, 34]. Regarding this TF network and TH17-associated genes, characteristic features are shared with group 3 innate lymphoid cells (ILC3) [35].
Given the role of BATF TFs in this regulatory network and expression of STAT3, IRF4, JUNB and BATF3 in ALCL, we investigated expression and function of BATFs in ALCL.
Materials and methods
Cell lines, culture conditions and transfections
ALCL (Karpas-299 [named K299], SU-DHL-1, DEL, JB6, SUP-M2, all ALK+; Mac-1, Mac-2A, FE-PD, DL40, all ALK–), T-cell leukemia-derived (Jurkat, KE-37, Molt-14, H9) and HEK293 cell lines were cultured as described [14]. Where indicated, 1 μg/ml doxycycline (Dox; Sigma), the ALK inhibitor crizotinib (Selleckchem), the RORC antagonists SR2211, SR1903 (both in-house generated, laboratory PRG) and GSK805 (Calbiochem), or dimethylsulfoxide (DMSO) control was added. For transient transfections and generation of A-Fos-inducible cells, see Supplementary Methods.
DNA constructs
CMV500-based A-Fos for constitutive expression has been described [36]. For BATF, BATF3, RORC1, RORC2, and pRTS-1-based [37] inducible A-Fos expression constructs and lentiviral sgRNA and BATF and BATF3 constructs, refer to Supplementary Methods.
siRNA-mediated knock-down and CRISPR/Cas9-mediated deletion of BATF and BATF3 in ALCL cell lines, chromatin immunoprecipitation (ChIP)
Accell small interfering RNAs (siRNAs) were from Dharmacon (Supplementary Table 1) and passively transfected once into K299 and Mac-1 cells using RPMI-1640 and 1% fetal calf serum. Cells were cultivated at 500–750 nM for each siRNA for 72 h (3 days). Knock-down efficiency was controlled by western blot (WB) at day 3. Functional assays were performed in standard medium. For CRISPR/Cas9-mediated deletion of BATF and BATF3, see Supplementary Methods and Supplementary Table 1; for ChIP, see Supplementary Methods and Supplementary Table 2.
Processing and analysis of microarray data; gene set enrichment analysis (GSEA) and principal component (PC) analysis
For generation of TH17 and ILC3 signatures, microarray data for TH17, ILC3 and TH1 cells were obtained from GEO (GSE78897) [35]. Primary ALCL and PTCL data were obtained from GEO (GSE65823, GSE6338 and GSE19069) [38–40]. For microarray analyses of cell lines, supplementation by ILC3 microarray data, GSEA and PC analyses, refer to Supplementary Methods.
Additional materials and methods
Detailed methodology is described in the Supplementary Materials and Methods.
Results
Characterization of BATF DNA binding and physical interactions of BATFs with JUNB in ALCL
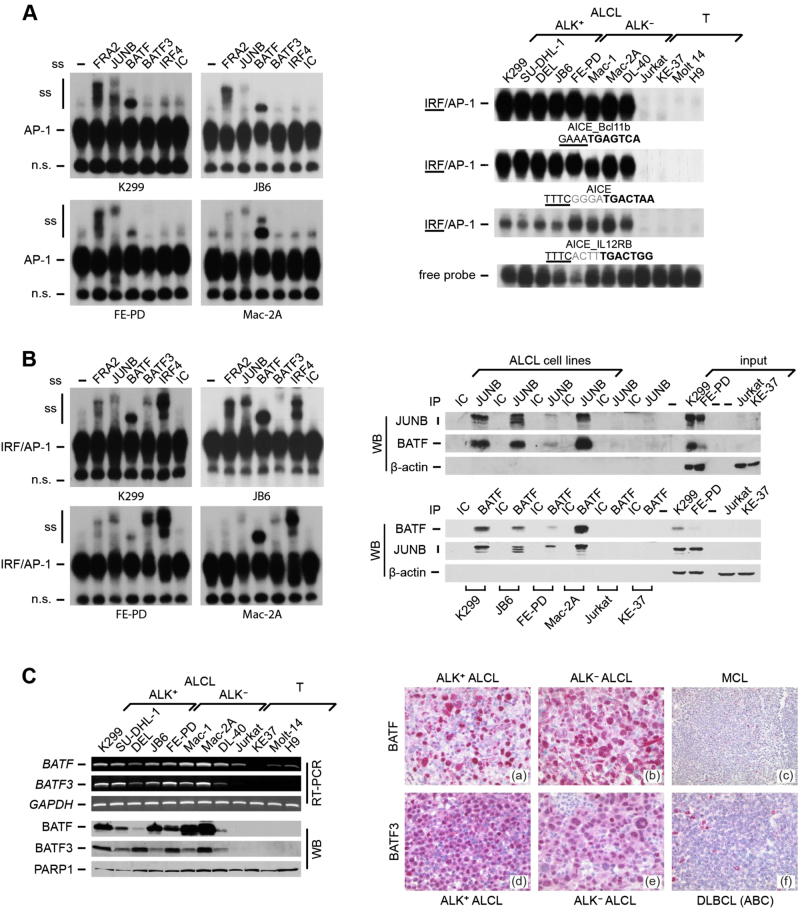
To identify BATF-containing TF complexes in ALCL, we first analyzed AP-1 DNA binding at the classical AP-1 5ʹ-TGA[G/C]TCA-3ʹ motif (Supplementary Figure 1A, top) in a panel of ALK+ and ALK– ALCL and T-cell-derived control (non-ALCL) cell lines already used in our previous studies [13, 18]. We verified ALCL-restricted AP-1 DNA binding activity and high-level JUNB and IRF4 expression (Supplementary Figure 1A) [13, 18]. Supershift analyses revealed DNA binding of the AP-1/FOS members JUNB and FRA2 [14, 18], and in addition strong BATF binding (Fig. 1a, left). In other cellular systems, BATF-JUN drives gene expression together with IRFs from AP-1 IRF composite elements (AICEs) [29, 31, 32]. DNA binding at AICEs was strong in ALCL, in contrast to non-ALCL (Fig. 1a, right). Supershifts demonstrated binding of FRA2 (weaker compared with the classical AP-1 motif), JUNB and, more prominently, BATF, as well as BATF3 and IRF4 (Fig. 1b, left). Immunoprecipitation demonstrated protein–protein interactions between BATF and BATF3 with JUNB specifically in ALCL cell lines (Fig. 1b, right, and Supplementary Figure 1B).
Fig. 1.
BATF and BATF3 DNA binding, co-immunoprecipitation and expression in ALCL. a Left, EMSA of AP-1 TPA responsive element (TRE) complexes without (-) or with addition of specific antibodies, or isotype control (IC). Positions of AP-1 complexes, supershifts (ss) and a nonspecific band (n.s.) are indicated. Right, EMSA of IRF/AP-1 DNA binding at AICEs (AICE_Bcl11b; AICE; AICE_IL12RB). Underlined, IRF motif; bold, AP-1 motif; gray, intervening bases. The free probe of one representative EMSA is shown. b Left, EMSA of AICE_Bcl11b, performed as in a. Right, JUNB and BATF co-immunoprecipitations with anti-JUNB (top), anti-BATF (bottom) or isotype controls (IC). (Co-)immunoprecipitated proteins were detected by immunoblotting (WB). β-Actin and input were controls. c Left, BATF and BATF3 were analyzed at mRNA levels by RT-PCR (top) and at protein level by immunoblotting of nuclear extracts (bottom). GAPDH and PARP1 were controls. Right, BATF and BATF3 IHC of primary lymphomas. Top, BATF IHC of an ALK+ ALCL a, an ALK– ALCL b and a mantle cell lymphoma [MCL; c]. Bottom, BATF3 IHC of an ALK+ ALCL d, an ALK– ALCL e and a DLBCL f
High-level expression of BATF and BATF3 in ALCL
The distinct DNA binding of BATF and BATF3 in ALCL indicated cell-type-specific expression. Indeed, BATF mRNA was largely restricted to, and BATF3 was exclusively expressed in ALCL cell lines (Fig. 1c, upper left). BATF2 was not expressed (data not shown). We confirmed high BATF and BATF3 protein expression in all ALCL cell lines (Fig. 1c, lower left, and Supplementary Figure 1C and 1D). The highest BATF levels in some ALK– cell lines corresponded to their somewhat stronger DNA binding at AICE_IL12RB (see Fig. 1a).
Immunohistochemistry of BATF and BATF3 in human lymphoma specimens demonstrated nuclear localization (Fig. 1c, right). Among 69 non-ALCL B-NHL and T-NHL, none of the mantle cell (MCL; 0/7), follicular (FL; 0/11) and Burkitt lymphomas (BL; 0/11) expressed BATF. expressed BATF, and 15 of 20 DLBCL showed varying numbers of positive lymphoma cells, whereas all CLL cases (9/9; only in proliferative centers), 2/2 NLPHL and 9/9 PTCL (NOS) were BATF positive. We concluded that BATF expression is associated with distinct lymphoma subtypes and cellular subpopulations. Importantly, strong staining was observed in 16/16 ALCL (7 ALK+/9 ALK–) (Fig. 1c right, top) and 8/8 classical Hodgkin lymphoma (cHL) cases.
BATF3 showed a more restricted expression pattern. In total, 16/16 ALCL (7 ALK+/9 ALK–) (Fig. 1c right, bottom) and 8/8 cHL strongly stained positive, whereas among 69 B-NHL and T-NHL (20 DLBCL, 10 MCL, 9 CLL, 11 FL, 8 BL, 9 PTCL, 2 NLPHL) only 1 CLL sample was BATF3 positive. Thus, simultaneous BATF and BATF3 expression was unique to ALCL and cHL.
CRISPR/Cas9-mediated deletion and siRNA knock-down of BATF and/or BATF3 in ALCL
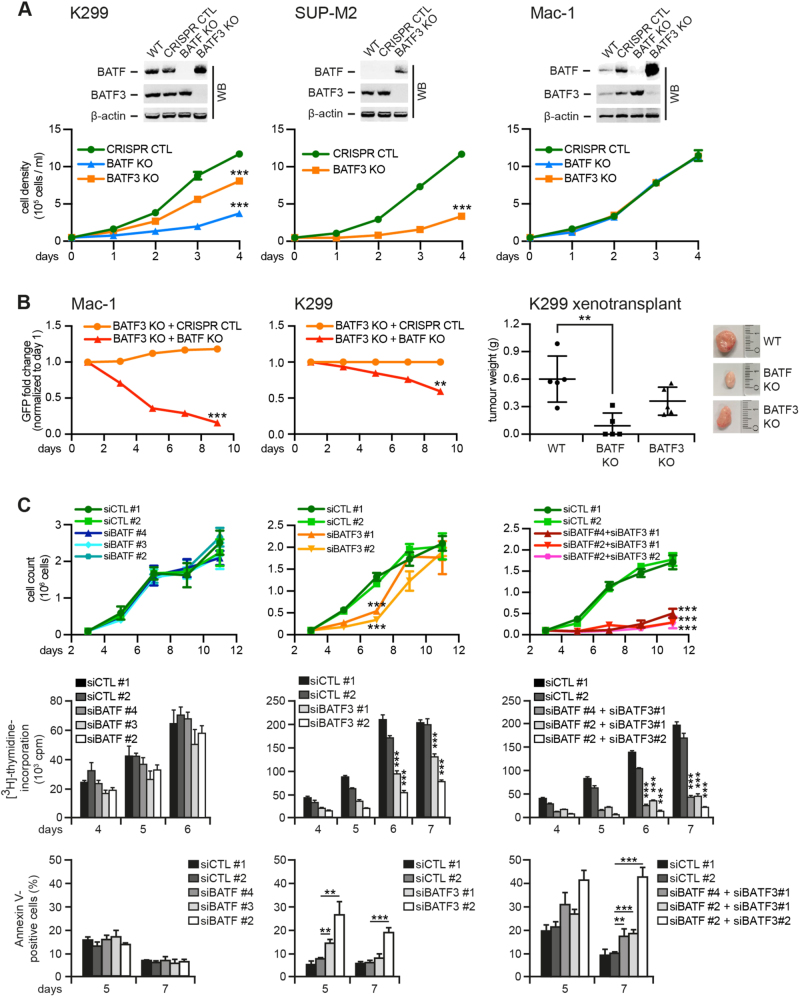
We next defined the role of BATF and BATF3 in ALCL by gene-specific inactivation by CRISPR/Cas9-mediated knock-out (KO) in K299, SUP-M2 and Mac-1 cells (Figs. 2a, b). Interestingly, following BATF3 deletion, BATF was upregulated. This was also observed in SUP-M2 cells, in which BATF was virtually absent at baseline (Supplementary Figure 1D). Deletion of BATF or BATF3 in K299 (Fig. 2a, left) or BATF3 in SUP-M2 cells (Fig. 2a, center) resulted in sustained growth retardation, but did not alter the growth of Mac-1, in which we, however, observed the strongest counterregulation of BATF and BATF3 (Fig. 2a, right).
Fig. 2.
CRISPR/Cas9-mediated deletion and siRNA-mediated knock-down of BATF and BATF3 in ALCL. a CRISPR/Cas9-mediated deletion of BATF and/or BATF3 in K299 (left), SUP-M2 (center) and Mac-1 (right) cells. Top, immunoblotting of wild-type (WT), control-treated (CRISPR CTL), BATF KO and/or BATF3 KO cells for BATF and BATF3. Note the compensatory increase of BATF following BATF3 deletion. Bottom, cell numbers are shown over time. b Left and center, BATF3 single knock-out Mac-1 (left) and K299 (center) cells were transduced with GFP-labeled vector encoding BATF gRNA (BATF KO) or non-targeting control (CRISPR CTL). The percentage of GFP-positive cells over time is indicated. Right, xenotransplantation of K299 WT, BATF KO and BATF3 KO cells on NSG mice. Tumor weight at day 14 is shown in gram (g). Far right, representative tumors at day 14. c siRNA-mediated knock-down of BATF and/or BATF3 in Mac-1. Cells were treated with control siRNAs (siCTL #1 and siCTL #2), siRNAs targeting BATF (siBATF #2, siBATF #3 and siBATF #4; left) or BATF3 (siBATF3 #1 and siBATF3 #2; center), or respective combinations (right). Cell numbers (top), [3H]-thymidine incorporation (center) and percentage of Annexin V-positive cells (bottom) are shown over time. **P < 0.01; ***P < 0.001; n.s., not significant
We were unable to generate BATF and BATF3 double KO ALCL cells, suggesting lethality of double BATF KO to ALCL. To test this, we monitored the fate of CRISPR/Cas9-mediated BATF and BATF3 double KO cells by transducing BATF3 single KO cells with BATF-targeting guide-RNA (gRNA) (Fig. 2b, left and center, Supplementary Figure 2A). In Mac-1 cells, despite no effect of BATF single KO (see Fig. 2a), trans duction of BATF3 single KO cells with BATF-directed gRNA led to a rapid loss of green fluorescent protein (GFP)-positive cells. A similar, somewhat less pronounced effect was observed in K299 cells, which, however, already showed growth retardation in BATF3 single KO cells (see Fig. 2a). Finally, xenotransplantation of K299 BATF KO cells produced significantly smaller tumors compared with WT cells (Fig. 2b, right), with a similar tendency for BATF3 KO tumors.
In a complementary approach, we performed siRNA-mediated knock-down of both BATFs (Fig. 2c and Supplementary Figures 2B and 2C). Single knock-down of BATF did not alter viability of Mac-1 cells (Fig. 2c, left), whereas BATF3 knock-down moderately inhibited growth and induced cell death (Fig. 2c, center). Importantly, simultaneous knock-down of both BATFs resulted in strong growth inhibition and apoptotic death (Fig. 2c, right), which reflected our inability to generate double BATF and BATF3 KO clones using CRISPR techniques. Similar results were obtained with K299 cells (Supplementary Figure 2C). As specificity control, reversion of cellular toxicity was shown by concomitant ectopic BATF and BATF3 expression (Supplementary Figure 3A).
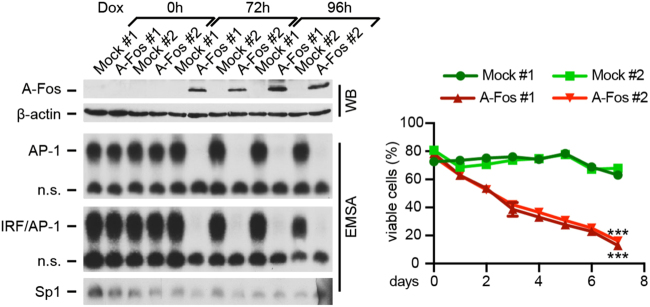
Induction of cell death following global abrogation of AP-1 DNA binding activity in ALCL
In an independent approach, we globally inhibited AP-1 by a dominant repressor of AP-1 and leucine zipper TFs, such as BATFs, named A-Fos [36]. This complemented our approaches targeting BATFs, as BATFs have low DNA affinity and require JUN proteins, mainly JUNB, for heterodimer formation and DNA binding [28]. We generated Dox-inducible A-Fos FE-PD cells (Fig. 3 and Supplementary Figure 3B), in which AP-1 activity was virtually abolished following Dox addition (Fig. 3, left). This inhibited cell growth (Supplementary Figure 3B) and induced cell death (Fig. 3, right). Similar results were obtained following transient A-Fos expression in K299 (Supplementary Figure 3C). Thus, constitutive AP-1/BATF activity is essential for viability of ALK+ and ALK– ALCL cells.
Fig. 3.
Inhibition of global AP-1 activity by its dominant-negative A-Fos in FE-PD cells. Inducible A-Fos expression abrogates constitutive AP-1. Left, analysis of enriched GFP-positive cells by WB for A-Fos expression (top; FLAG antibody) and EMSA for DNA binding to TRE site (center, top) and BCL_11b AICE (center, bottom) after Dox addition for the indicated times. β-Actin and Sp1 DNA binding were controls. Two independent experiments (#1 and #2) are shown. Right, percentage of viable cells following Dox induction measured by PI staining and flow cytometry over time. ***P < 0.001
ALCL shows a gene expression pattern characteristic for TH17 and ILC3 cells
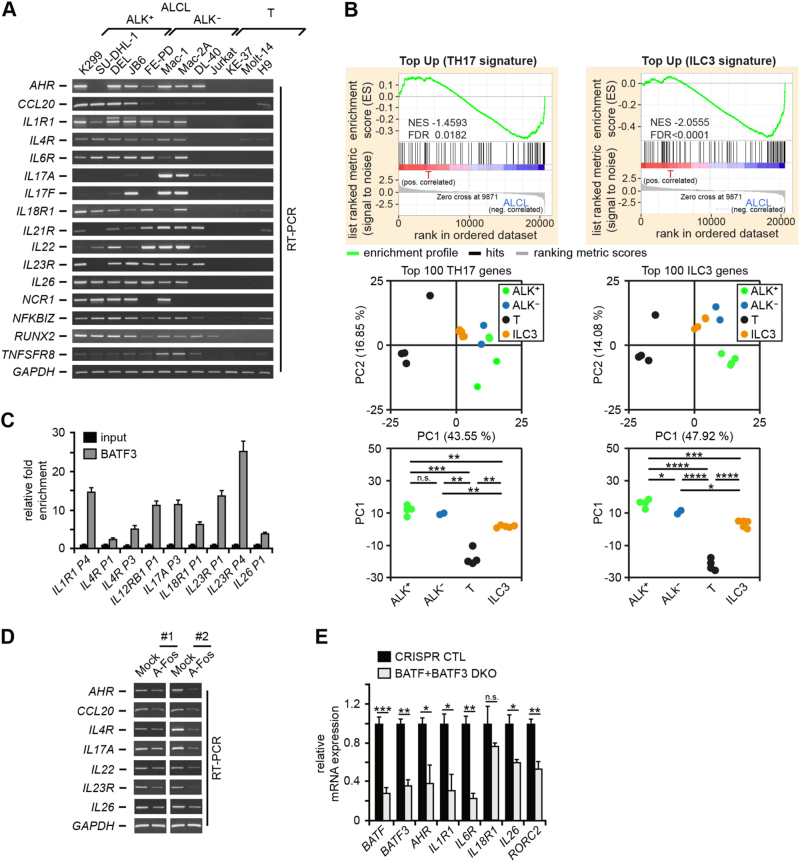
JUN-BATF, IRF4 and STAT3 coordinate TH17 gene expression [33]. As all these TFs are active in ALCL, we hypothesized that their combined activity imposes a TH17 cellular fate in ALCL. Indeed, ALCL cell lines consistently expressed TH17-associated genes (Fig. 4a), including AHR, IL1R1, IL4R, IL18R1, IL22, IL23R and IL26. Given the absence of T-cell receptor (TCR) rearrangement in a (minor) subset of ALCL cases [41, 42], we reasoned that ALCL cells could be derived alternatively from ILC3 cells. These cells are characterized by the absence of BCR or TCR gene rearrangements and have, compared with TH17 cells, an overlapping but distinct gene expression pattern [35].
Fig. 4.
TH17/ILC3 gene expression in ALCL and link to the deregulated BATF/BATF3/AP-1 activity. a mRNA expression of TH17/ILC3-associated genes and, as control, GAPDH were analyzed by RT-PCR. b More global approaches to TH17/ILC3 gene expression in ALCL. Upper panels, GSEA of differentially expressed genes between ALCL (K299, SU-DHL-1, DEL, JB6, FE-PD, Mac-2A) and T control (T) cell lines (Jurkat, KE-37, Molt-14, H9) based on TH17 (left) and ILC3 (right) top 100 upregulated genes. Lower panels, PC analyses of ALK+ and ALK– ALCL and T control samples based on 100 top differentially expressed TH17 (left) or ILC3 (right) genes, separating ALCL and T-cell lines along the PC1 axis. PCAs were supplemented by projection of ILC3 samples [45]. n.s., not significant; *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001. c BATF3 ChIP from K299 cells. Input and precipitated DNA were amplified by qPCR for the indicated promoter or enhancer regions. Combined data of two biological replicates are shown as mean ± SEM. d Inhibition of AP-1 downregulates TH17/ILC3 genes. Enriched A-Fos or Mock transfected K299 cells were analyzed by RT-PCR for TH17/ILC3 genes. Two (#1 and #2) of four independent experiments are shown. e Analysis of TH17/ILC3 genes in K299 cells with double BATF and BATF3 KO at day 13 following lentiviral transduction. qPCR-based expression of the indicated genes in control cells (CRISPR CTL) or cells transduced with a combination of sgRNAs targeting BATF and BATF3 (BATF + BATF3 DKO). BATF and BATF3 were analyzed to verify their KO. The expression level in CRISPR CTL cells was set 1. Error bars denote 95% CIs. One out of three independent experiments is shown. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; n.s., not significant
To test whether a TH17- or ILC3-like signature was an inherent feature of the overall ALCL expression pattern, we performed GSEAs (Fig. 4b, top). Based on published gene expression data [35], we defined TH17 and ILC3 signatures, using the top 100 up- or downregulated genes compared with TH1 cells. Our ALCL cell lines significantly enriched genes upregulated in TH17 cells (Fig. 4b, top, left) and even more prominently ILC3-upregulated genes (Fig. 4b, top, right). ILC3 signature leading edge genes are shown in Supplementary Table 3. Consistently, principal component analysis (PCA) of ALK+ (K299, SU-DHL-1, DEL, JB6), ALK– (FE-PD, Mac-2A) and control (T; Jurkat, KE-37, Molt-14, H9) samples based on the top 100 differentially expressed genes between TH17 and ILC3 and TH1 signatures revealed a clear separation of ALCL and control cells along PC1 (Fig. 4b, bottom). Moreover, ALCL cells were localized closer to projected ILC3, again more significant for ILC3 signatures (P-values ≤ 1.6E–9 for TH17 signature based PCA and 9.5E–17 for ILC3). This suggested an in-between localization of ALCL between the ILC3 and TH17 phenotypes with a stronger skewing towards an ILC3 profile.
Expression of TH17/ILC3 genes in primary ALCL; IL-22, IL-17A and IL-17F are secreted by ALCL cell lines, IL-17A and IL-17F detectable in ALCL patients
We next aimed to confirm expression of TH17- and ILC3-associated genes in primary lymphomas. In the majority of primary ALCL samples, TH17–/ILC3 genes were much stronger or even exclusively expressed compared with NHL controls (Supplementary Figure 4A). Moreover, IL-22 was secreted by three ALK– cell lines and IL-17A and IL-17F by several ALCL cell lines (Supplementary Figure 4B). IL-17A was detectable in three of 21 pretreatment serum/plasma samples of ALK+ ALCL patients and one healthy control, whereas it was undetectable in patients in remission (Supplementary Figure 4C) (P = 0.48). IL-17F levels were high in 4 of 21 ALCL patients, whereas no healthy control or patient in remission exhibited measurable IL-17F (Supplementary Figure 4C). Although there was only a tendency toward a higher mean IL-17F level in ALK+ ALCL patients (P = 0.08), these data suggest specific IL-17F upregulation in distinct ALK+ ALCL patients. We did not observe a significant correlation with other clinical or biological parameters (data not shown).
Recruitment of BATFs and JUNB to regulatory regions of TH17/ILC3 genes and dependency of TH17/ILC3 genes on AP-1/BATFs in ALCL
A direct regulation of TH17-/ILC3-associated genes by AP-1/BATF was investigated by ChIPs of BATF, BATF3 and JUNB. We analyzed promoter or enhancer regions of IL1R1, IL4R, IL12RB, IL17A, IL18R1, IL22, IL23R and IL26 in three ALCL cell lines and Jurkat controls (Fig. 4c and Supplementary Figure 5A and 5B). Most of the regulatory regions showed strong BATF, BATF3 and JUNB recruitment in ALCL cells, which was not observed in Jurkat. Functionally, we confirmed AP-1 involvement in the upregulation of TH17/ILC3 genes, as expression of AHR, CCL20, IL4R, IL17A, IL22, IL23R and IL26 decreased following AP-1 inhibition (Fig. 4d). Furthermore, K299 showed decreased expression of AHR, IL1R1, IL6R, IL26 and RORC2 after enrichment of cells with simultaneous gRNA-mediated double KO of BATF and BATF3 at day 13 after transduction (Fig. 4e).
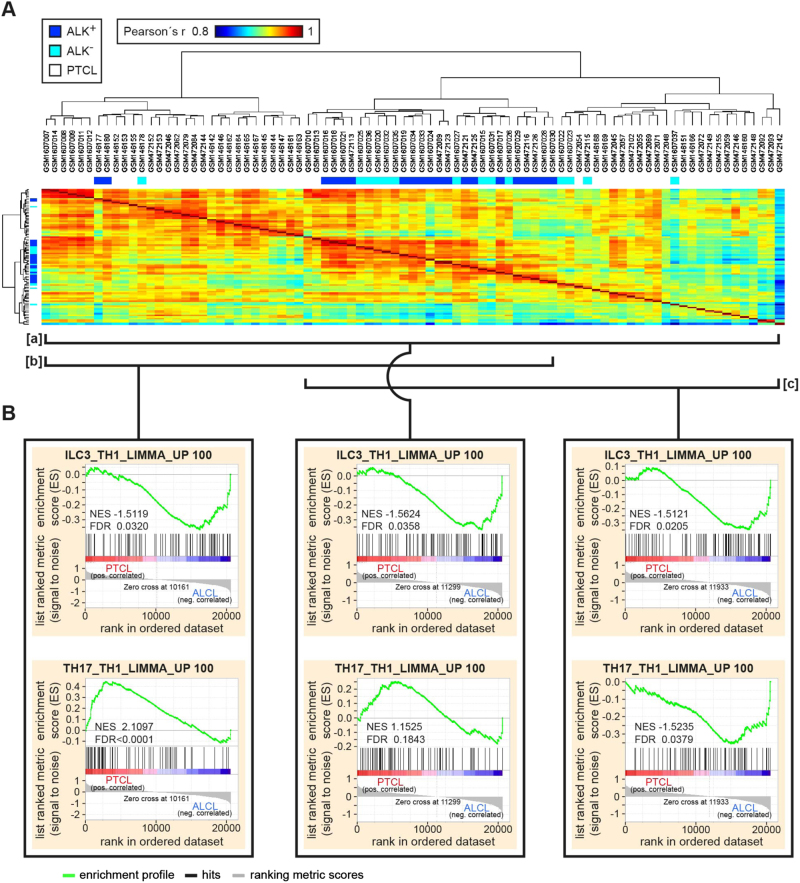
Primary ALCL shows a gene expression pattern characteristic for ILC3, while taking an intermediate position between two groups of PTCL with respect to TH17 signature genes
To globally approach the concept of TH17/ILC3 skewing of primary ALCL, we performed GSEA with microarray data of primary ALCL and PTCL samples used in previous studies (Fig. 5) [38–40]. A correlation analysis of gene expression profiles of all samples (bracket [a]) demonstrated one central ALCL cluster flanked by two distinct PTCL clusters, positioned in bracket [b] and [c] of Fig. 5a. In an overall analysis including all samples (Fig. 5a, bracket [a]), the ILC3 signature enriched in ALCL (Fig. 5b, center top panel). In contrast, the TH17 signature demonstrated a trend toward an enrichment in PTCL (Fig. 5b, center bottom panel). The GSEA between both PTCL clusters revealed a strong TH17 signature enrichment in PTCLs positioned in bracket [b] compared with those positioned in bracket [c] (NES 3.2556; FDR < 0.0001; data not shown). The TH17-like PTCL in bracket [b] also showed an enrichment of the TH17 signature when compared with ALCL (Fig. 5b, left bottom). The ALCL, however, showed an enrichment of the TH17 signature when compared with PTCLs positioned in bracket [c] (Fig. 5b, right bottom). The ILC3 signature was enriched in ALCL when compared with any of the PTCL clusters (Fig. 5b, top). We concluded that gene expression pattern characteristic for ILC3 is a common feature of ALCL, whereas, regarding TH17 signature genes, ALCL takes an intermediate position between PTCL with or without expression of TH17 genes.
Fig. 5.
TH17/ILC3 gene set enrichment analyses of primary ALCL and PTCL. a Pearson correlation heatmap between ALCL and PTCL samples. Samples are clustered by the Euclidean distance and separate into three major clusters. b GSEA of ALCL and PTCL samples from indicated clusters, marked by brackets underneath the heatmap shown in a. Note, although ALCL shows an overall enrichment of the ILC3 signature (GSEA of upper row), the TH17 signature enrichment (GSEA of lower row) is decreasing from left to the right
Expression of RORC2 (RORγt) in ALCL; RORC inhibition results in cell death induction in ALCL and synergizes with ALK inhibitors
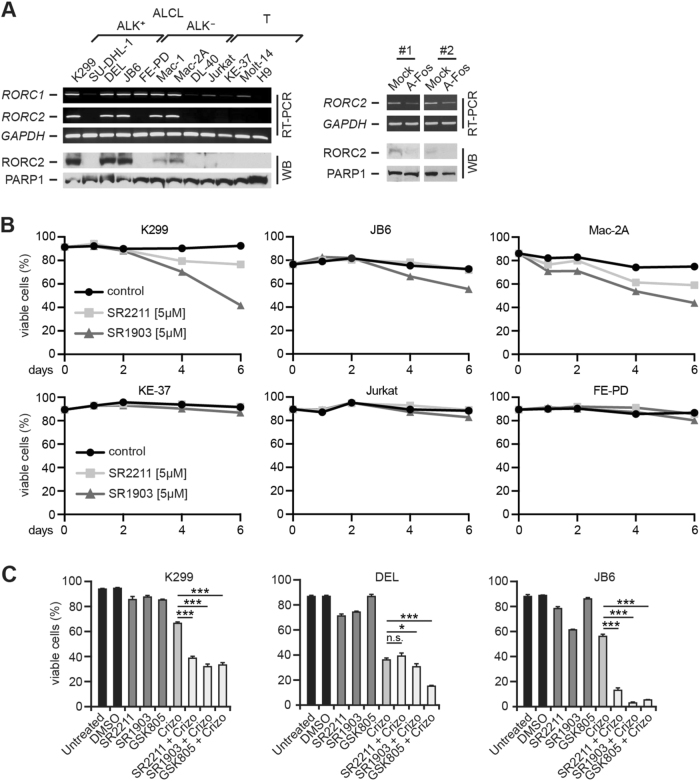
TH17 and ILC3 cells are characterized by a unique expression of RORC2, also known as RORγt [43–45, 46]. RORC2 mRNA was detectable in five of eight ALCL cell lines, but none of the controls (Fig. 6a, upper left). RORC1 mRNA was stronger expressed in ALCL cell lines. At the protein level, we confirmed RORC overexpression and RORC2 restriction to ALCL cell lines (Fig. 6a, lower left, and Supplementary Figure 5C, left). Furthermore, robust RORC2 mRNA expression was detectable in most primary ALCL specimens in contrast to NHL, including five PTCL-NOS (Supplementary Figure 5C, center and right). Functionally, RORC2 expression decreased following AP-1/BATF inhibition by A-Fos (Fig. 6a, right).
Fig. 6.
Expression and inhibition of RORC2 in ALCL. a Left, analysis of RORC1 and RORC2 mRNA by RT-PCR (top) and RORC protein expression by immunoblotting of nuclear extracts (bottom). GAPDH and PARP1 are controls. Right, AP-1 inhibition downregulates RORC2. K299 cell were treated as in Fig. 4d, and RORC2 and GAPDH mRNA were analyzed by RT-PCR (top), and RORC2 expression changes at the protein level by WB (bottom). Two (#1 and #2) of four independent experiments are shown. b RORC inhibition by small compounds induces cell death of ALCL cell lines. Cells were treated with 5 μM of RORC inhibitors SR2211 or SR1903, or the control (DMSO), and the percentage of viable cells was analyzed by PI staining. One of three independent experiments is shown. (c) Enforced cell death induction by crizotinib in combination with RORC inhibitors. The ALK+ cell lines K299, DEL and JB6 cells were left untreated, or treated with DMSO control, the RORC inhibitors SR2211 (7.5 μΜ), SR1903 (7.5 μΜ) and GSK805 (7.5 μM), or 25 nM crizotinib (Crizo) alone, or the different RORC inhibitors together with crizotinib. Cell death was analyzed by Annexin V-FITC/PI staining. The percentage of viable cells is shown. Experiments were performed in triplicates and results are shown as mean ± SD. One of three independent experiments is shown. *P < 0.05; ***P < 0.001; n.s., not significant
Finally, we investigated the effect of pharmacological RORC inhibition in ALCL. Treatment of ALCL cell lines with the inhibitory RORC modulators SR2211 (ref. 47) and SR1903 (a close analog of SR2211) resulted in decreased viable cells over time (Fig. 6b, top), whereas no effect was observed in cells without (KE-37) or low level (Jurkat) RORC expression and in FE-PD cells with RORC1 but no RORC2 expression (Fig. 6b, bottom). Moreover, we investigated the effect of RORC inhibitors SR2211, SR1903 and GSK805 (ref. 48) in combination with ALK inhibition in the ALK+ ALCL cell lines K299, DEL and JB6 (Fig. 6c). The combination of crizotinib with RORC inhibitors enhanced cell death induction.
Discussion
We demonstrate that ALK+ and ALK– ALCL are characterized by an unprecedented activation of the AP-1 family and leucine zipper TFs BATF and BATF3, and that simultaneous high-level expression of both BATFs is a particular feature of ALCL. In accordance with the fact that BATF3 distinguishes ALCL from other PTCL [26, 27], BATF and BATF3 thus add a new layer of complexity to deregulated AP-1 in ALCL [14, 18, 19]. Remarkably, although NPM-ALK induces JUNB [21], ALK+ and ALK– ALCL neither differ in their global AP-1 activity nor in expression of distinct AP-1 factors [refs. 14, 18, 19 and this work], which supports our hypothesis that they share a common pathogenic mechanism [49, 50]. In favor of this, the epigenome between ALK+ and ALK– ALCL is highly similar [49]. Apart from ALCL, we identified a concomitant expression of BATF and, as only recently demonstrated [27], BATF3 in cHL, which is in line with the in cHL known constitutive AP-1 activity and shared key TF activities in cHL and ALCL [16, 18, 19].
AP-1 forms homo- or heterodimers and exerts cell-type and differentiation stage-specific functions [51]. Furthermore, AP-1 effects certainly differ between transient and the long-term activation observed in ALCL, as known, for example, NF-κB [52]. We present evidence that BATF and BATF3 are essential components of the TF network in ALCL. First, deletion of BATF or BATF3 caused growth retardation in vitro and in vivo, in line with a recently published study on BATF3 in ALCL [27]. Our inability to generate BATF and BATF3 double KO cells indicates lethality of combined BATF deletion. Second, we observed a comparable phenotype with siRNA-mediated BATF knock-down, in which the combined BATF and BATF3 knock-down resulted in pronounced growth arrest and cell death induction. The cross-regulation of BATF and BATF3 detected in our cell lines and in genetically manipulated mice and a functional redundancy [28, 29] might attenuate effects of targeting single BATFs. BATFs modify transcriptional activity by interaction with AP-1 TFs like JUNB and JUN [28], both highly activated in ALCL [18, 24]. In line with their concerted activity, global AP-1 inhibition caused death of ALCL cells (this work and ref. 18).
Furthermore, we describe a composite DNA binding activity at AICEs with IRF4, which is known from TH17 cells [30, 32–34]. In ALK+ ALCL, expression of some TH17 genes has been reported [40, 53, 54], and NPM-ALK-induced miR-135b enforces IL-17 production [54]. We demonstrate that TH17 gene expression is a unifying feature of ALK+ and ALK– ALCL, and their expression depends at least in part on AP-1/BATFs. By a comprehensive analysis of the TH17 signature in primary ALCL and PTCL, we identified two PTCL groups, one with strong TH17 signature and one without, whereas ALCL were taking an intermediate position between these PTCL groups. Whereas the biological and clinical relevance of TH17 gene expression in PTCL has to be investigated in future studies, these data confirmed partial TH17 skewing of primary ALCL.
Remarkably, the ILC3 gene set, which overlaps with that of TH17 cells [35], showed a consistent enrichment in ALCL, irrespective of the PTCL comparison. This is of particular relevance regarding the cellular origin of ALCL and opens a new view on ALCL pathogenesis. Approximately 14% of ALCL show germline TCR configuration [41]. The absence of BCR or TCR rearrangements is as much a hallmark of ILC3 as the lack of typical B or T-cell markers [55, 56]. ILC3 are enriched in human tonsils and the intestinal lamina propria, and circulate in the peripheral blood [55]. Whereas nearly all known hematopoietic cell types give rise to malignancies, no ILC3-derived neoplasm is currently known. We propose that a (minor) subfraction of ALCL originates from ILC3. The expression of key ILC3 genes, the lack of B or T lymphoid surface markers, the lack of a genomic lymphoid fingerprint in an ALCL fraction and the frequent expression of IL-22 [53, 57] are in accordance with such a hypothesis. Alternatively, the deregulated TF network might superimpose a TH17/ILC3 cellular fate on ALCL cells.
Our work provides new aspects for targeted treatment strategies for ALCL. Inhibitors of TH17 cells are developed due to TH17 cell involvement in autoimmune and inflammatory diseases [58, 59–61]. Hence, interference with TH17 gene activity might represent a treatment strategy for ALK+ and ALK– ALCL. Among ALK+ ALCL patients, treatment with ALK inhibitors exerts long-term disease control [62], but is obviously unable to eradicate the respective lymphoma clone [63]. The synergistic activity of RORC inhibitors together with ALK inhibitors might eradicate persisting ALCL cells. In addition, we provide the basis for exploring interference with, for example, IL-17 and IL-26 circuits and further evaluation of TH17-related cytokines as diagnostic and prognostic markers for ALCL, as also suggested by an independent study [53].
Overall, we have identified BATF and BATF3 as essential components of the gene regulatory network in ALCL and have demonstrated their pathogenic and therapeutic relevance. Furthermore, we suggest that a subset of ALCL originate from ILC3, a finding that has to be elaborated in future studies.
Electronic supplementary material
Acknowledgements
We thank Simone Lusatis (Berlin) and Brigitte Wollert-Wulf (Berlin) for outstanding technical assistance, Peter Rahn (Berlin) for cell sorting, Dan Littman (New York) for providing RORC antibody and helpful discussion and Georg Bornkamm for helpful discussion. This work was supported in part by grants from the Deutsche Forschungsgemeinschaft to SM and MJ (MA 3313/2-1 and JA 1847/2-1), the Experimental and Clinical Research Center, a joint cooperation between the Charité-Universitätsmedizin Berlin and the MDC, the German Cancer Consortium (DKTK), the Berlin School of Integrative Oncology (BSIO), the Jubiläumsfond der Österreichischen Nationalbank (OM, no. 14856), the Austrian Science Fund (LK, P26011 and 29251), and the Forschungshilfe Peiper (WW and CD-W). MC, LK, H-CL, OM, SM, SDT, WW and CD-W have received funding from the European Union´s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement no. 675712.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Contributor Information
Lukas Kenner, Phone: +43.1.4040051760, Email: lukas.kenner@meduniwien.ac.at.
Stephan Mathas, Phone: +49.30.94062863, Email: stephan.mathas@charite.de.
Electronic supplementary material
The online version of this article (10.1038/s41375-018-0045-9) contains supplementary material, which is available to authorized users.
References
- 1.Rui L, Schmitz R, Ceribelli M, Staudt LM. Malignant pirates of the immune system. Nat Immunol. 2011;12:933–40. doi: 10.1038/ni.2094. [DOI] [PubMed] [Google Scholar]
- 2.Shaffer AL, 3rd, Young RM, Staudt LM. Pathogenesis of human B cell lymphomas. Ann Rev Immunol. 2012;30:565–610. doi: 10.1146/annurev-immunol-020711-075027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stein H, Foss HD, Dürkop H, Marafioti T, Delsol G, Pulford K, et al. CD30(+) anaplastic large cell lymphoma: a review of its histopathologic, genetic, and clinical features. Blood. 2000;96:3681–95. [PubMed] [Google Scholar]
- 4.Piccaluga PP, Gazzola A, Mannu C, Agostinelli C, Bacci F, Sabattini E, et al. Pathobiology of anaplastic large cell lymphoma. Adv Hematol. 2010, Article ID 345053, 10 pages. [DOI] [PMC free article] [PubMed]
- 5.Swerdlow SH, Campo E, Pileri SA, Harris NL, Stein H, Siebert R, et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016;127:2375–90. doi: 10.1182/blood-2016-01-643569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiarle R, Voena C, Ambrogio C, Piva R, Inghirami G. The anaplastic lymphoma kinase in the pathogenesis of cancer. Nat Rev Cancer. 2008;8:11–23. doi: 10.1038/nrc2291. [DOI] [PubMed] [Google Scholar]
- 7.Crescenzo R, Abate F, Lasorsa E, Tabbo F, Gaudiano M, Chiesa N, et al. Convergent mutations and kinase fusions lead to oncogenic STAT3 activation in anaplastic large cell lymphoma. Cancer Cell. 2015;27:516–32. doi: 10.1016/j.ccell.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mereu E, Pellegrino E, Scarfo I, Inghirami G, Piva R. The heterogeneous landscape of ALK negative ALCL. Oncotarget. 2017;8:18525–36. doi: 10.18632/oncotarget.14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lamant L, de Reynies A, Duplantier MM, Rickman DS, Sabourdy F, Giuriato S, et al. Gene-expression profiling of systemic anaplastic large-cell lymphoma reveals differences based on ALK status and two distinct morphologic ALK+ subtypes. Blood. 2007;109:2156–64. doi: 10.1182/blood-2006-06-028969. [DOI] [PubMed] [Google Scholar]
- 10.Merkel O, Hamacher F, Laimer D, Sifft E, Trajanoski Z, Scheideler M, et al. Identification of differential and functionally active miRNAs in both anaplastic lymphoma kinase (ALK)+ and ALK- anaplastic large-cell lymphoma. Proc Natl Acad Sci USA. 2010;107:16228–33. doi: 10.1073/pnas.1009719107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boi M, Rinaldi A, Kwee I, Bonetti P, Todaro M, Tabbo F, et al. PRDM1/BLIMP1 is commonly inactivated in anaplastic large T-cell lymphoma. Blood. 2013;122:2683–93. doi: 10.1182/blood-2013-04-497933. [DOI] [PubMed] [Google Scholar]
- 12.Eckerle S, Brune V, Döring C, Tiacci E, Bohle V, Sundstrom C, et al. Gene expression profiling of isolated tumour cells from anaplastic large cell lymphomas: insights into its cellular origin, pathogenesis and relation to Hodgkin lymphoma. Leukemia. 2009;23:2129–38. doi: 10.1038/leu.2009.161. [DOI] [PubMed] [Google Scholar]
- 13.Weilemann A, Grau M, Erdmann T, Merkel O, Sobhiafshar U, Anagnostopoulos I, et al. Essential role of IRF4 and MYC signaling for survival of anaplastic large cell lymphoma. Blood. 2015;125:124–32. doi: 10.1182/blood-2014-08-594507. [DOI] [PubMed] [Google Scholar]
- 14.Mathas S, Kreher S, Meaburn KJ, Jöhrens K, Lamprecht B, Assaf C, et al. Gene deregulation and spatial genome reorganization near breakpoints prior to formation of translocations in anaplastic large cell lymphoma. Proc Natl Acad Sci USA. 2009;106:5831–6. doi: 10.1073/pnas.0900912106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Falini B, Fizzotti M, Pucciarini A, Bigerna B, Marafioti T, Gambacorta M, et al. A monoclonal antibody (MUM1p) detects expression of the MUM1/IRF4 protein in a subset of germinal center B cells, plasma cells, and activated T cells. Blood. 2000;95:2084–92. [PubMed] [Google Scholar]
- 16.Jundt F, Anagnostopoulos I, Förster R, Mathas S, Stein H, Dörken B. Activated Notch1 signaling promotes tumor cell proliferation and survival in Hodgkin and anaplastic large cell lymphoma. Blood. 2002;99:3398–403. doi: 10.1182/blood.V99.9.3398. [DOI] [PubMed] [Google Scholar]
- 17.Chiarle R, Simmons WJ, Cai H, Dhall G, Zamo A, Raz R, et al. Stat3 is required for ALK-mediated lymphomagenesis and provides a possible therapeutic target. Nat Med. 2005;11:623–9. doi: 10.1038/nm1249. [DOI] [PubMed] [Google Scholar]
- 18.Mathas S, Hinz M, Anagnostopoulos I, Krappmann D, Lietz A, Jundt F, et al. Aberrantly expressed c-Jun and JunB are a hallmark of Hodgkin lymphoma cells, stimulate proliferation and synergize with NF-kappa B. EMBO J. 2002;21:4104–13. doi: 10.1093/emboj/cdf389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Janz M, Hummel M, Truss M, Wollert-Wulf B, Mathas S, Jöhrens K, et al. Classical Hodgkin lymphoma is characterized by high constitutive expression of activating transcription factor 3 (ATF3), which promotes viability of Hodgkin/Reed-Sternberg cells. Blood. 2006;107:2536–9. doi: 10.1182/blood-2005-07-2694. [DOI] [PubMed] [Google Scholar]
- 20.Hsu FY, Johnston PB, Burke KA, Zhao Y. The expression of CD30 in anaplastic large cell lymphoma is regulated by nucleophosmin-anaplastic lymphoma kinase-mediated JunB level in a cell type-specific manner. Cancer Res. 2006;66:9002–8. doi: 10.1158/0008-5472.CAN-05-4101. [DOI] [PubMed] [Google Scholar]
- 21.Leventaki V, Drakos E, Medeiros LJ, Lim MS, Elenitoba-Johnson KS, Claret FX, et al. NPM-ALK oncogenic kinase promotes cell-cycle progression through activation of JNK/cJun signaling in anaplastic large-cell lymphoma. Blood. 2007;110:1621–30. doi: 10.1182/blood-2006-11-059451. [DOI] [PubMed] [Google Scholar]
- 22.Turner SD, Yeung D, Hadfield K, Cook SJ, Alexander DR. The NPM-ALK tyrosine kinase mimics TCR signalling pathways, inducing NFAT and AP-1 by RAS-dependent mechanisms. Cell Signal. 2007;19:740–7. doi: 10.1016/j.cellsig.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 23.Mao X, Orchard G, Lillington DM, Russell-Jones R, Young BD, Whittaker SJ. Amplification and overexpression of JUNB is associated with primary cutaneous T-cell lymphomas. Blood. 2003;101:1513–9. doi: 10.1182/blood-2002-08-2434. [DOI] [PubMed] [Google Scholar]
- 24.Schiefer AI, Vesely P, Hassler MR, Egger G, Kenner L. The role of AP-1 and epigenetics in ALCL. Front Biosci. 2015;7:226–35. doi: 10.2741/s436. [DOI] [PubMed] [Google Scholar]
- 25.Laimer D, Dolznig H, Kollmann K, Vesely PW, Schlederer M, Merkel O, et al. PDGFR blockade is a rational and effective therapy for NPM-ALK-driven lymphomas. Nat Med. 2012;18:1699–704. doi: 10.1038/nm.2966. [DOI] [PubMed] [Google Scholar]
- 26.Agnelli L, Mereu E, Pellegrino E, Limongi T, Kwee I, Bergaggio E, et al. Identification of a 3-gene model as a powerful diagnostic tool for the recognition of ALK-negative anaplastic large-cell lymphoma. Blood. 2012;120:1274–81. doi: 10.1182/blood-2012-01-405555. [DOI] [PubMed] [Google Scholar]
- 27.Lollies A, Hartmann S, Schneider M, Bracht T, Weiss AL, Arnolds J, et al. An oncogenic axis of STAT-mediated BATF3 upregulation causing MYC activity in classical Hodgkin lymphoma and anaplastic large cell lymphoma. Leukemia. 2017; 10.1038/leu.2017.203. [Epub ahead of print]. [DOI] [PubMed]
- 28.Murphy TL, Tussiwand R, Murphy KM. Specificity through cooperation: BATF-IRF interactions control immune-regulatory networks. Nat Rev Immunol. 2013;13:499–509. doi: 10.1038/nri3470. [DOI] [PubMed] [Google Scholar]
- 29.Tussiwand R, Lee WL, Murphy TL, Mashayekhi M, Kc W, Albring JC, et al. Compensatory dendritic cell development mediated by BATF-IRF interactions. Nature. 2012;490:502–7. doi: 10.1038/nature11531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schraml BU, Hildner K, Ise W, Lee WL, Smith WA, Solomon B, et al. The AP-1 transcription factor Batf controls T(H)17 differentiation. Nature. 2009;460:405–9. doi: 10.1038/nature08114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li P, Spolski R, Liao W, Wang L, Murphy TL, Murphy KM, et al. BATF-JUN is critical for IRF4-mediated transcription in T cells. Nature. 2012;490:543–6. doi: 10.1038/nature11530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Glasmacher E, Agrawal S, Chang AB, Murphy TL, Zeng W, Vander Lugt B, et al. A genomic regulatory element that directs assembly and function of immune-specific AP-1-IRF complexes. Science. 2012;338:975–80. doi: 10.1126/science.1228309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ciofani M, Madar A, Galan C, Sellars M, Mace K, Pauli F, et al. A validated regulatory network for Th17 cell specification. Cell. 2012;151:289–303. doi: 10.1016/j.cell.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hasan Z, Koizumi SI, Sasaki D, Yamada H, Arakaki N, Fujihara Y, et al. JunB is essential for IL-23-dependent pathogenicity of Th17 cells. Nat Commun. 2017;8:15628. doi: 10.1038/ncomms15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koues OI, Collins PL, Cella M, Robinette ML, Porter SI, Pyfrom SC, et al. Distinct gene regulatory pathways for human innate versus adaptive lymphoid cells. Cell. 2016;165:1134–46. doi: 10.1016/j.cell.2016.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olive M, Krylov D, Echlin DR, Gardner K, Taparowsky E, Vinson C. A dominant negative to activation protein-1 (AP1) that abolishes DNA binding and inhibits oncogenesis. J Biol Chem. 1997;272:18586–94. doi: 10.1074/jbc.272.30.18586. [DOI] [PubMed] [Google Scholar]
- 37.Bornkamm GW, Berens C, Kuklik-Roos C, Bechet JM, Laux G, Bachl J, et al. Stringent doxycycline-dependent control of gene activities using an episomal one-vector system. Nucleic Acids Res. 2005;33:e137. doi: 10.1093/nar/gni137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scarfo I, Pellegrino E, Mereu E, Kwee I, Agnelli L, Bergaggio E, et al. Identification of a new subclass of ALK-negative ALCL expressing aberrant levels of ERBB4 transcripts. Blood. 2016;127:221–32. doi: 10.1182/blood-2014-12-614503. [DOI] [PubMed] [Google Scholar]
- 39.Piccaluga PP, Agostinelli C, Califano A, Rossi M, Basso K, Zupo S, et al. Gene expression analysis of peripheral T cell lymphoma, unspecified, reveals distinct profiles and new potential therapeutic targets. J Clin Invest. 2007;117:823–34. doi: 10.1172/JCI26833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iqbal J, Weisenburger DD, Greiner TC, Vose JM, McKeithan T, Kucuk C, et al. Molecular signatures to improve diagnosis in peripheral T-cell lymphoma and prognostication in angioimmunoblastic T-cell lymphoma. Blood. 2010;115:1026–36. doi: 10.1182/blood-2009-06-227579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Malcolm TI, Villarese P, Fairbairn CJ, Lamant L, Trinquand A, Hook CE, et al. Anaplastic large cell lymphoma arises in thymocytes and requires transient TCR expression for thymic egress. Nat Commun. 2016;7:10087. doi: 10.1038/ncomms10087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bonzheim I, Geissinger E, Roth S, Zettl A, Marx A, Rosenwald A, et al. Anaplastic large cell lymphomas lack the expression of T-cell receptor molecules or molecules of proximal T-cell receptor signaling. Blood. 2004;104:3358–60. doi: 10.1182/blood-2004-03-1037. [DOI] [PubMed] [Google Scholar]
- 43.Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille J, et al. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–33. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 44.Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 cells. Ann RevImmunol. 2009;27:485–517. doi: 10.1146/annurev.immunol.021908.132710. [DOI] [PubMed] [Google Scholar]
- 45.Glatzer T, Killig M, Meisig J, Ommert I, Luetke-Eversloh M, Babic M, et al. RORgammat(+) innate lymphoid cells acquire a proinflammatory program upon engagement of the activating receptor NKp44. Immunity. 2013;38:1223–35. doi: 10.1016/j.immuni.2013.05.013. [DOI] [PubMed] [Google Scholar]
- 46.He YW, Deftos ML, Ojala EW, Bevan MJ. RORgamma t, a novel isoform of an orphan receptor, negatively regulates Fas ligand expression and IL-2 production in T cells. Immunity. 1998;9:797–806. doi: 10.1016/S1074-7613(00)80645-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kumar N, Lyda B, Chang MR, Lauer JL, Solt LA, Burris TP, et al. Identification of SR2211: a potent synthetic RORgamma-selective modulator. ACS Chem Biol. 2012;7:672–7. doi: 10.1021/cb200496y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xiao S, Yosef N, Yang J, Wang Y, Zhou L, Zhu C, et al. Small-molecule RORgammat antagonists inhibit T helper 17 cell transcriptional network by divergent mechanisms. Immunity. 2014;40:477–89. doi: 10.1016/j.immuni.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hassler MR, Pulverer W, Lakshminarasimhan R, Redl E, Hacker J, Garland GD, et al. Insights into the pathogenesis of anaplastic large-cell lymphoma through genome-wide DNA methylation profiling. Cell Rep. 2016;17:596–608. doi: 10.1016/j.celrep.2016.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roukos V, Mathas S. The origins of ALK translocations. Front Biosci. 2015;7:260–8. doi: 10.2741/s439. [DOI] [PubMed] [Google Scholar]
- 51.Shaulian E. AP-1--the Jun proteins: oncogenes or tumor suppressors in disguise? Cell Signal. 2010;22:894–9. doi: 10.1016/j.cellsig.2009.12.008. [DOI] [PubMed] [Google Scholar]
- 52.Werner SL, Barken D, Hoffmann A. Stimulus specificity of gene expression programs determined by temporal control of IKK activity. Science. 2005;309:1857–61. doi: 10.1126/science.1113319. [DOI] [PubMed] [Google Scholar]
- 53.Savan R, McFarland AP, Reynolds DA, Feigenbaum L, Ramakrishnan K, Karwan M, et al. A novel role for IL-22R1 as a driver of inflammation. Blood. 2011;117:575–84. doi: 10.1182/blood-2010-05-285908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Matsuyama H, Suzuki HI, Nishimori H, Noguchi M, Yao T, Komatsu N, et al. miR-135b mediates NPM-ALK-driven oncogenicity and renders IL-17-producing immunophenotype to anaplastic large cell lymphoma. Blood. 2011;118:6881–92. doi: 10.1182/blood-2011-05-354654. [DOI] [PubMed] [Google Scholar]
- 55.Montaldo E, Juelke K, Romagnani C. Group 3 innate lymphoid cells (ILC3s): origin, differentiation, and plasticity in humans and mice. Eur J Immunol. 2015;45:2171–82. doi: 10.1002/eji.201545598. [DOI] [PubMed] [Google Scholar]
- 56.Simoni Y, Fehlings M, Kloverpris HN, McGovern N, Koo SL, Loh CY, et al. Human innate lymphoid cell subsets possess tissue-type based heterogeneity in phenotype and frequency. Immunity. 2017;46:148–61. doi: 10.1016/j.immuni.2016.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bard JD, Gelebart P, Anand M, Amin HM, Lai R. Aberrant expression of IL-22 receptor 1 and autocrine IL-22 stimulation contribute to tumorigenicity in ALK+ anaplastic large cell lymphoma. Leukemia. 2008;22:1595–603. doi: 10.1038/leu.2008.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huh JR, Leung MW, Huang P, Ryan DA, Krout MR, Malapaka RR, et al. Digoxin and its derivatives suppress TH17 cell differentiation by antagonizing RORgammat activity. Nature. 2011;472:486–90. doi: 10.1038/nature09978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Isono F, Fujita-Sato S, Ito S. Inhibiting RORgammat/Th17 axis for autoimmune disorders. Drug Discov Today. 2014;19:1205–11. doi: 10.1016/j.drudis.2014.04.012. [DOI] [PubMed] [Google Scholar]
- 60.Gordon KB, Blauvelt A, Papp KA, Langley RG, Luger T, Ohtsuki M, et al. Phase 3 trials of ixekizumab in moderate-to-severe plaque psoriasis. N Engl J Med. 2016;375:345–56. doi: 10.1056/NEJMoa1512711. [DOI] [PubMed] [Google Scholar]
- 61.Gege C. RORgammat inhibitors as potential back-ups for the phase II candidate VTP-43742 from Vitae Pharmaceuticals: patent evaluation of WO2016061160 and US20160122345. Expert Opin Ther Pat. 2017;27:1–8. doi: 10.1080/13543776.2017.1262350. [DOI] [PubMed] [Google Scholar]
- 62.Gambacorti Passerini C, Farina F, Stasia A, Redaelli S, Ceccon M, Mologni L, et al. Crizotinib in advanced, chemoresistant anaplastic lymphoma kinase-positive lymphoma patients. J Natl Cancer Inst. 2014;106:djt378. doi: 10.1093/jnci/djt378. [DOI] [PubMed] [Google Scholar]
- 63.Gambacorti Passerini C, Mussolin L, Brugieres L. Abrupt relapse of ALK-positive lymphoma after discontinuation of crizotinib. N Engl J Med. 2016;374:95–96. doi: 10.1056/NEJMc1511045. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.