Abstract
Mitochondrial genomes of animals have long been considered to evolve under the action of purifying selection. Nevertheless, there is increasing evidence that they can also undergo episodes of positive selection in response to shifts in physiological or environmental demands. Vampire bats experienced such a shift, as they are the only mammals feeding exclusively on blood and possessing anatomical adaptations to deal with the associated physiological requirements (e.g., ingestion of high amounts of liquid water and iron). We sequenced eight new chiropteran mitogenomes including two species of vampire bats, five representatives of other lineages of phyllostomids and one close outgroup. Conducting detailed comparative mitogenomic analyses, we found evidence for accelerated evolutionary rates at the nucleotide and amino acid levels in vampires. Moreover, the mitogenomes of vampire bats are characterized by an increased cytosine (C) content mirrored by a decrease in thymine (T) compared with other chiropterans. Proteins encoded by the vampire bat mitogenomes also exhibit a significant increase in threonine (Thr) and slight reductions in frequency of the hydrophobic residues isoleucine (Ile), valine (Val), methionine (Met), and phenylalanine (Phe). We show that these peculiar substitution patterns can be explained by the co-occurrence of both neutral (mutational bias) and adaptive (positive selection) processes. We propose that vampire bat mitogenomes may have been impacted by selection on mitochondrial proteins to accommodate the metabolism and nutritional qualities of blood meals.
Keywords: Phyllostomidae, hematophagy, synonymous substitutions, radical replacements
Introduction
Animal mitochondrial genomes are considered to evolve according to the nearly neutral theory of molecular evolution with strong purifying selection purging most deleterious mutations, and with fixation of some slightly deleterious mutations modulated by effective population size (Nabholz, Ellegren, et al. 2013; Popadin et al. 2013). There is, however, increasing evidence for episodes of positive selection acting on mitochondrial DNA (Meiklejohn et al. 2007; Garvin et al. 2015; James et al. 2016). Several examples document shifts in life-history traits associated to positive selection on mitogenomes: increased longevity in rockfishes (Hua et al. 2015), modified diets and physiologies in snakes (Castoe et al. 2008), change to the energetically demanding subterranean lifestyle in rodents (Tomasco and Lessa 2011), origin of flight in bats (Shen et al. 2010), colonization of high-altitude ecosystems in caprines (Hassanin et al. 2009), emergence of ecotypes in killer-whales (Foote et al. 2011), and resistance to starvation and ability to recover from cold coma in Drosophila simulans (Ballard et al. 2007).
The occurrence of both neutral and adaptive processes can be inferred from their evolutionary footprints on nucleotide and amino acid sequences. At the DNA level, neutral processes like mutational biases modify the nucleotide composition. In protein-coding sequences, this can result in a bias in codon usage, as synonymous codons enriched in the favored nucleotide(s) will be preferred independently of any direct functional implications (but see Chamary et al. 2006; Hershberg and Petrov 2008). Instead, adaptive episodes often involve an increase of the ratio between non-synonymous (dN) versus synonymous (dS) substitutions. Consequently, second codon positions, and most first codon positions, at which most non-synonymous substitutions occur, are likely targets to detect positive selection.
In proteins, the signature of adaptive processes is rather evaluated in terms of properties of the residues involved by comparing radical versus conservative amino acid replacements. Indeed, while a non-synonymous substitution is defined in terms of changes in the encoded amino acids, some groups of residues share similar physico-chemical properties and can be conservatively exchanged without significant functional impact (Betts and Russel 2007; Weber et al. 2014). Conversely, radical changes are often seen as evidence for the action of positive selection, as slight changes in the local properties of residues can result in an increased stability or efficiency of the folded protein. Besides, the amino acid composition of proteins can also be modified by mutational biases (Foster et al. 1997), and the frequencies of the replaced amino acids have been then shown to covary with those of the favored nucleotide(s) within the corresponding coding genes (Jobson et al. 2010). In some cases, these random substitutions result in a selective advantage, as illustrated by the case of high-altitude adaptation in house wrens (Troglodytes aedon), in which amino acid replacements from valine (Val) to isoleucine (Ile) in hemoglobin—as the byproduct of biased hypermutability from G-to-A that eliminates CpG dinucleotides in the βA-globin gene—resulted in increased hemoglobin affinity for oxygen (Galen et al. 2015). Finally, changes in protein sequences, regardless if they result from mutational biases or from positive selection, can also lead to compensatory replacements either elsewhere in the protein or in its associated partners in order to maintain the selective advantage or to compensate for negative, potentially deleterious effects (Singer and Hickey 2000).
The shift to a blood-based diet (hematophagy or sanguivory) has evolved independently in several clades of metazoans. Some of the more conspicuous blood-feeding organisms include leeches, fleas, sucking lice, kissing bugs, mosquitoes, ticks, lampreys, and vampires. Moreover, opportunistic consumption of blood has been reported in noctuid moths (Zaspel et al. 2012), and in some avian taxa such as members of the family Buphagidae (Weeks 2000) and one of Darwin’s finches (Geospiza difficilis) (Schluter and Grant 1984). A diet based exclusively on blood imposes heavy physiological constraints on the organism as a whole. For example, the evolution of sanguivory in vampire bats was recently suggested to involve hologenomic adaptations (Zepeda Mendoza et al. 2018) in agreement with previous suggestions for several groups of organisms in which blood digestion might be facilitated by the bacterial hemolytic enzymes of their gut microbiota (Müller et al. 1980; Greenhall et al. 1983; Chaverri 2013). Interestingly, several of the physiological adaptations needed to successfully feed on blood demand an increased supply of energy, a function in which mitochondria are deeply involved. This makes mitogenomes readily accessible targets to investigate the potential impact of changes in life-history traits on their molecular evolution.
A comparison among blood-feeding organisms to look for trends in their mitogenomic evolution is, however, not straightforward because of the evolutionary singularities of these phylogenetically distant groups. These peculiarities include the following: 1) non-strict hematophagy, as in mosquitoes (Ribeiro 2000; Takken and Verhulst 2013), and 2) parasitic habits restricted to a particular stage of the life cycle, like in lampreys (Tetlock et al. 2012). Moreover, deep divergence of groups like leeches (636–476 Ma; dos Reis et al. 2015) and ticks (ca. 319 Ma; Mans et al. 2012), in which hematophagy occurs along the entire clade, reduces the possibility of finding non-hematophagous close relatives for appropriate comparative analyses. Additionally, the evolutionary dynamics of mitogenomes in these groups differ greatly from those found in vertebrates with multiple genome rearrangements and different distribution of genes along the two DNA strands (Shao et al. 2001; Dowton et al. 2002; Covacin et al. 2006; Cameron 2014; Weigert et al. 2016), important shifts and reversals in nucleotide composition (Hassanin et al. 2005), as well as strong compositional biases and reorganization into several chromosomes (Shao et al. 2009). The co-occurrence of two or more of these phenomena results in evolutionary patterns that can differentially impact evolutionary rates (Xu et al. 2006), making tricky to distinguish their effects from those due to the shift to hematophagy.
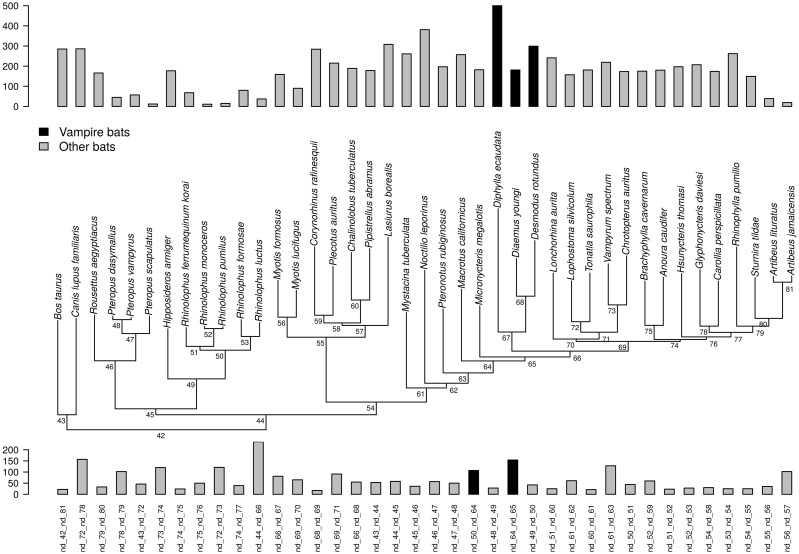
The strictly hematophagous vampire bats offer the possibility to study the impact, if any, of the dietary shift to hematophagy on organismal biology at a narrower phylogenetic scale and in the framework of a conserved organization of the mitochondrial genome. Indeed, the family Phyllostomidae, to which vampire bats belong, also comprise species that do not feed on blood and are instead insectivorous, frugivorous, nectarivorous, and carnivorous (Wetterer et al. 2000; Datzmann et al. 2010; Baker et al. 2012). Phyllostomids diverged from their sister group ∼40 Mya (Baker et al. 2012; Rojas et al. 2016) and the three species of vampires diverged from their closest relatives 20–25 Mya (Baker et al. 2012; Rojas et al. 2016). The three extant species are the common vampire bat (Desmodus rotundus) (Greenhall et al. 1983), the white-winged vampire bat (Diaemus youngi) (Greenhall and Schutt 1996) and the hairy-legged vampire bat (Diphylla ecaudata) (Greenhall et al. 1984). Each of these three species constitutes a monotypic genus classified in the subfamily Desmodontinae, with Desmodus being closer to Diaemus than to Diphylla (Baker et al. 2003).
In order to assimilate each blood meal, which may represent ∼60% of the bat body weight and contain up to 800 times the amount of iron ingested in a human meal, important changes have occurred in the vampire digestive and urinary systems, among others. Adapting to this peculiar diet for example requires dealing with the following: 1) the excess of liquid resulting from blood ingestion that is conveyed to the bladder through active transport of water (Breidenstein 1982; Harlow and Braun 1997), 2) the potential toxicity of iron through its efficient sequestration in the intestinal epithelium both limiting its absorption and minimizing unnecessary losses (Morton and Wimsat 1980; Morton and Janning 1982), and 3) the high protein content of the blood meal that results in higher urea production and increased renal activity (Singer 2002). Given the ATP-dependent nature of active transport and the oxidative conditions created, for example, by the high amount of iron in blood (Whiten et al. 2018), these adaptations can be expected to have direct and indirect consequences on both mitochondrial physiology and the evolution of mitochondrial DNA. First, the increase in energy production is fulfilled through different mechanisms including regulation of the expression of mitochondrial proteins (Mehrabian et al. 2005) and/or the number of mitochondria (Safiulina and Kaasik 2013), metabolic signaling, modulation of mitochondrial enzyme content and kinetic regulation of some respiratory chain complex subunits (Devin and Rigoulet 2007), and, less frequently, positive selection on mitochondrial proteins to favor changes resulting in better performances (Hua et al. 2015). Second, an increase in energy production, and thus on respiration, coupled to an enhanced oxidative metabolism, can be expected to result in an augmented production of radical oxygenated species (ROS), which, as by-products of respiration, are potential sources of damage to cellular components, leading ultimately to senescence (Brunet-Rossinni and Austad 2004; Munshi-South and Wilkinson 2010).
With such a highly oxidative metabolism, which leads to increased amounts of ROS, vampire bats should thus undergo accelerated aging. Paradoxically, instead of aging quickly and exhibiting shorter life spans, vampire bats live as long as or considerably longer than other non-hematophagous bat species. Records include observations of Diphylla living, on average, 8 years in the wild, Desmodus females that are reproductively active at an age of ∼15 years (Tschapka and Wilkinson 1999), and reports of captive individuals of Diaemus and Desmodus that reached respectively 20 (Wilson 2003; Myhrvold et al. 2015) and 29.2 years (Weigl 2005), the maximal longevity values documented for phyllostomid bats. Given the involvement of mitochondrial DNA in aging, this paradox might suggest that mitochondrial proteins of vampire bats have undergone modifications allowing them to counter the negative effects of oxidation while fulfiling the increased energy production requirements.
In this study, we analyze patterns of molecular evolution of the mitochondrial genomes of phyllostomids in a phylogenetic context encompassing representatives of all major clades of Neotropical leaf-nosed bats (Phyllostomidae) including the three species of vampires. The molecular phylogenies inferred from mitogenomic sequences suggest that vampire bats exhibit accelerated evolutionary rates compared with other lineages of phyllostomids, at the nucleotide and the amino acid levels. Both neutral and selective processes are investigated to provide a detailed depiction of the molecular evolution of vampire bats mitogenomes. We then evaluate the possibility that the observed changes are a consequence of the shift to hematophagy and that they can somehow be related to the life-history trait paradox observed in vampire bats.
Materials and Methods
Taxon Sampling and DNA Sequencing
We assembled a data set of available complete mitochondrial genomes for chiropterans, including the common vampire bat Desmodus rotundus, and those of cow (Bos taurus) and dog (Canis lupus familiaris) as non-chiropteran Laurasiatheria outgroups (table 1). With the aim of including at least a member of each subfamily of phyllostomid bats as defined by Baker et al. (2003), we obtained tissue samples for two vampire bats—Diaemus youngi (MSB: Mamm: 56205) and Diphylla ecaudata (MSB: Mamm: 211697)—, and representative members of the subfamilies Macrotinae (Macrotus californicus, MSB: Mamm: 140888), Lonchophyllinae (Hsunycteris thomasi, MNHN: PCD-1243), Phyllostominae (Chrotopterus auritus, MUSM 13196), Lonchorhininae (Lonchorhina aurita, MVZ 185587), and Glyphonycterinae (Glyphonycteris daviesi, ROM: 114050). Additionally, a species of the family Noctilionidae (Noctilio leporinus, MHNG-1939.065) was sampled as a closer outgroup to phyllostomids.
Table 1.
Taxon Sampling, Voucher Information, and Accession Numbers
| Family | Subfamily | Species | Tissue Number | Museumb | Accession Number |
|---|---|---|---|---|---|
| Noctilionidae | – | Noctilio leporinus | T4573 | ISEM | KU743910* |
| Phyllostomidae | Carollinae | Carollia perspicillata | T5412 | ISEM | NC_022422.1 |
| Phyllostomidae | Desmodontinae | Desmodus rotundus | T3418 | ISEM | NC_022423.1 |
| Phyllostomidae | Desmodontinae | Diaemus youngi | NK13129 | MSB | KU743906* |
| Phyllostomidae | Desmodontinae | Diphylla ecaudata | NK13627 | MSB | KU743911* |
| Phyllostomidae | Glossophaginae | Anoura caudifer | T3681 | ISEM | NC_022420.1 |
| Phyllostomidae | Glossophaginae | Brachyphylla cavernarum | T5240 | ISEM | NC_022421.1 |
| Phyllostomidae | Glyphonycterinae | Glyphonycteris daviesi | T3711 | ISEM | KU743912* |
| Phyllostomidae | Lonchophyllinae | Hsunycteris thomasi | T3845 | ISEM | KU743907* |
| Phyllostomidae | Lonchorhininae | Lonchorhina aurita | MVZ185587 | MVZ | KU743908* |
| Phyllostomidae | Macrotinae | Macrotus californicus | NK80553 | MSB | KU743909* |
| Phyllostomidae | Micronycterinae | Micronycteris megalotis | T5539 | ISEM | NC_022419.1 |
| Phyllostomidae | Phyllostominae | Chrotopterus auritus | AMNH109773 | AMNH | KU743905* |
| Phyllostomidae | Phyllostominae | Lophostoma silvicolum | T4497 | ISEM | NC_022424.1 |
| Phyllostomidae | Phyllostominae | Tonatia saurophila | T4488 | ISEM | NC_022428.1 |
| Phyllostomidae | Phyllostominae | Vampyrum spectrum | T3393 | ISEM | NC_022429.1 |
| Phyllostomidae | Rhynophyllinae | Rhinophylla pumilio | T4670 | ISEM | NC_022426.1 |
| Phyllostomidae | Stenodermatinae | Artibeus jamaicensis | – | – | NC_002009.1 |
| Phyllostomidae | Stenodermatinae | Artibeus lituratus | – | – | NC_016871.1 |
| Phyllostomidae | Stenodermatinae | Sturnira tildae | T5299 | ISEM | NC_022427.1 |
| Mormoopidae | – | Pteronotus rubiginosus | T5215 | ISEM | NC_022425.1 |
| Mystacinidae | – | Mystacina tuberculata | – | – | NC_006925.1 |
| Rhinolophidae | – | Rhinolophus ferrumequinum korai | – | – | NC_016191.1 |
| Rhinolophidae | – | Rhinolophus formosae | – | – | NC_011304.1 |
| Rhinolophidae | – | Rhinolophus luctus | – | – | NC_018539.1 |
| Rhinolophidae | – | Rhinolophus monoceros | – | – | NC_005433.1 |
| Rhinolophidae | – | Rhinolophus pumilus | – | – | NC_005434.1 |
| Pteropodidae | – | Pteropus dasymallus | – | – | NC_002612.1 |
| Pteropodidae | – | Pteropus scapulatus | – | – | NC_002612.1 |
| Pteropodidae | – | Pteropus vampyrus | – | – | Ensembl_v57_Scaffold_17814 |
| Pteropodidae | – | Rossetus aegyptiacus | – | – | NC_007393.1 |
| Vespertilionidae | – | Chalinolobus tuberculatus | – | – | NC_002626.1 |
| Vespertilionidae | – | Lasiurus borealis | – | – | NC_016873.1 |
| Vespertilionidae | – | Myotis formosus | – | – | NC_015828.1 |
| Vespertilionidae | – | Myotis lucifugus | – | – | Ensembl_v57_Scaffold_144518 |
| Vespertilionidae | – | Pipistrellus abramus | – | – | NC_005436.1 |
| Vespertilionidae | – | Plecotus auritus | – | – | NC_015484.1 |
| Vespertilionidae | – | Plecotus rafinesquii | – | – | NC_016872.1 |
| Hipposideridae | – | Hipposideros armiger | – | – | NC_018540.1 |
| Bovidae | – | Bos taurus | – | – | NC_006853.1 |
| Canidae | – | Canis lupus familiaris | – | – | NC_002008.4 |
This study.
University of Montpellier (ISEM: Montpellier, FR), Museum of Southwestern Biology (MSB: Albuquerque, NM), Museum of Vertebrate Zoology at the University of California (MVZ: Berkeley, CA), and American Museum of Natural History (AMNH: New York, NY).
For these eight species, total genomic DNA was extracted using the QIAGEN Blood and Tissue Kit and fragmented by sonication using an ultrasonic cleaning unit (Elmasonic). The 3′-ends of the obtained fragments were then repaired and filled before being ligated with adaptors and tagged according to a custom cost-effective version of the Meyer and Kircher (2010) protocol for Illumina libraries preparation (Tilak et al. 2015). Tagged DNA libraries were pooled and sequenced on a single Illumina HiSeq 2000 lane to produce a total of ∼200 millions single-end 101-bp reads. Accession numbers for all sequences used and voucher information for the new mitogenomes are provided in table 1.
Mitochondrial Genome Assembly
Raw reads were first trimmed using Cutadapt 1.3 (Martin 2011) to remove adaptor fragments. Then, a mapping of the reads on the phylogenetically closest available mitochondrial genome was performed for each species using the Geneious R6 read mapper (Kearse et al. 2012) with the following custom parameters: a minimum of 24 consecutive nucleotides matching the reference, a minimum of 95% of nucleotide similarity in overlap region, and a maximum of 3% of gaps with a maximum gap size of three nucleotides. Iterative mapping cycles were performed in order to elongate the sequence until the assembly of the complete mitogenome was accomplished. A high-quality consensus keeping the highest quality base at each column was generated, and the circularity of the mitogenome was verified by the exact superimposition of the 100 nucleotides at the assembly extremities. The number of satellite repetitions in the control region (CR) is estimative as there were Illumina reads containing exclusively repeats, making it difficult to determine the exact copy number. Annotation of the new mitogenomes was made in Geneious R6 by transferring annotations on the basis of sequence similarity from the closest available bat mitogenome, followed by manual adjustment of incomplete and/or overlapping gene boundaries.
Phylogenetic Analyses
Ribosomal RNA (rRNA) and transfer RNA (tRNA) sequences were aligned using MUSCLE (Edgar 2004) with default parameters whereas sequences of protein-coding genes were aligned with MACSE (Ranwez et al. 2011) in order to conserve open reading frames. Stop codons at the 3′-end of protein-coding genes were removed in subsequent concatenations. All individual alignments were cleaned from ambiguously aligned positions using trimAl (Capella-Gutierrez et al. 2009) with the –automated1 option optimizing alignments for maximum likelihood analysis while keeping the codon structure in protein-coding genes. The concatenation of all genes excluding the light-strand encoded nd6 gene and the control region yielded a final alignment of 14,768 nucleotide sites and 3,612 residues for the amino acid concatenation.
We first inferred the phylogeny of phyllostomids from all alignments under the maximum likelihood (ML) criterion using RAxML v7.4.6 (Stamatakis 2006). Sequence evolution was modeled using a GTR matrix of substitution rates along with a gamma distribution and a fraction of invariable sites (GTR + GAMMA + I) in order to incorporate the heterogeneity of evolutionary rate among and within mitochondrial genes. We then chose the best partitioning scheme using PartitionFinder v1.1 (Lanfear et al. 2012). For nucleotides, we considered two independent partitions for rRNAs and for tRNAs, combined with either 12 partitions (one for each protein-coding gene), or three partitions (one for each codon position), or 36 partitions (one per gene and per codon position). For amino acids, the best partitioning scheme was chosen between a single model for the whole concatenation and a distinct model for each of the 12 mitochondrial proteins (table 2). Models were compared using the Akaike Informative Criterion (AIC). The statistical reliability of inferred relationships was evaluated by bootstrap support (BS) obtained by performing 100 replicates under the thorough bootstrap procedure called with the “-f b” option in RAxML.
Table 2.
Statistics on the Assemblies and Base Composition of the Eight Sequenced Mitochondrial Genomes
| Species | Total Reads | mtDNA Reads (%) | Mitogenome Length | Mean Coverage | G + C-Content (%) | A + T-Skew | G + C-Skew |
|---|---|---|---|---|---|---|---|
| Chrotopterus auritus | 5,398,643 | 0.29 | 16,707 | 93.8 | 46.2 | 0.07 | −0.32 |
| Diaemus youngi | 6,954,180 | 4.19 | 16,628 | 1,759.8 | 44.7 | 0.12 | −0.39 |
| Diphylla ecaudata | 11,513,077 | 2.46 | 16,618 | 1,713.9 | 42.4 | 0.14 | −0.38 |
| Glyphonycteris daviesi | 11,358,201 | 0.22 | 16,655 | 151.0 | 39.6 | 0.044 | −0.32 |
| Hsunycteris thomasi | 11,046,978 | 0.17 | 16,654 | 112.6 | 37.7 | 0.043 | −0.29 |
| Lonchorhina aurita | 11,100,715 | 1.78 | 16,661 | 1,192.5 | 40.4 | 0.067 | −0.33 |
| Macrotus californicus | 12,054,161 | 1.06 | 16,691 | 618.7 | 39.5 | 0.069 | −0.32 |
| Noctilio leporinus | 17,560,768 | 0.14 | 16,659 | 143.8 | 38.7 | 0.094 | −0.30 |
Additionally, Bayesian phylogenetic inference (BI) was performed on both data sets under the CAT + GTR + GAMMA mixture model as implemented in PhyloBayes 3.3f (Lartillot et al. 2009). Two independent MCMC chains were run until convergence for each data set with sampling every ten cycles, each cycle being equivalent to a variable number of generations as counted in other programs of Bayesian inference (i.e., in our case 5,500 cycles corresponded to ca. 4,000,000 generations). Chain convergence was checked periodically and considered to be adequate when the bpcomp command estimated a maximum difference < 0.1 for posterior probabilities among the same bipartitions independently identified by the two chains, and the tracecomp program output resulted in effective sample sizes (effsize) > 200 and maxdiff < 0.1 for most of the parameters with the exception of alpha (0.11) and allocent (0.21).
Nucleotide and Amino Acid Compositions
Nucleotide and amino acid compositions were estimated for rRNAs, tRNAs, and protein-coding genes separately, in order to evaluate potential compositional biases in translated versus non-translated portions of the mitogenome. Hereafter, we denote by C1, C2, and C3 the cytosine composition on first, second, and third codon positions, respectively. All calculations were done using MEGA v5.2.2 (Tamura et al. 2011).
Detection of Positive Selection
To avoid the statistical limitations imposed by too short sequences, the analyses that aimed at evaluating the occurrence of positive selection were conducted on the concatenation of the 12 protein-coding genes of the H strand and using the ML topology obtained under the best-fitting model.
We first evaluated the ratio of non-synonymous to synonymous substitutions (ω = dN/dS) along the different lineages of bats using three different approaches: 1) the dsom procedure of the integrated Bayesian approach implemented in the CoEvol program (Lartillot and Poujol 2011); 2) the substitution mapping procedure as implemented in MapNH (Romiguier et al. 2012), in which optimal values for the different parameters of the model are first estimated with the bppML program (Guéguen et al. 2013) and then used to count the dN and the dS per branch, from which ω is approximated by dN/(3×dS). The same procedure was performed in order to obtain estimates for the number of substitutions among pairs of nucleotides (A to T and T to A, C to T and T to C, and so on) to evaluate the existence of mutational biases; and 3) the free ratio branch model of the CODEML program in PAML v4.6 (Yang 2007) allowing to estimate a different ω for each branch of the reference topology.
To further assess the potential occurrence of positive selection episodes in the vampire bat lineage, we used the branch and branch-site models implemented in CODEML. The branch model (Yang 1998; Yang and Nielsen 1998) allows two ω values: one for the branch(es) of interest and one for the rest of the tree. The branch-site model (Yang and Nielsen 2002; Zhang et al. 2005) allows variation in ω for sites along the branches of interest (“foreground” branches). Selective hypotheses are compared with their respective null hypotheses of neutral evolution using likelihood ratio tests (LRT). Given that positive selection can occur at any of the branches of the lineage of interest, each of the models was evaluated with focus either on the entire Desmodontinae clade or along its ancestral branch (fig. 1, branch a), and either on the ancestral branch of [Desmodus + Diaemus] (fig. 1, branch b), or on each of the branches subtending the vampire bat species. As a comparison, we also performed the same set of analyses on the ancestral branch of another C-rich clade including Vampyrum spectrum and Chrotopterus auritus (see later). This allowed us to evaluate the potential impact of nucleotide composition on the detection of positive selection.
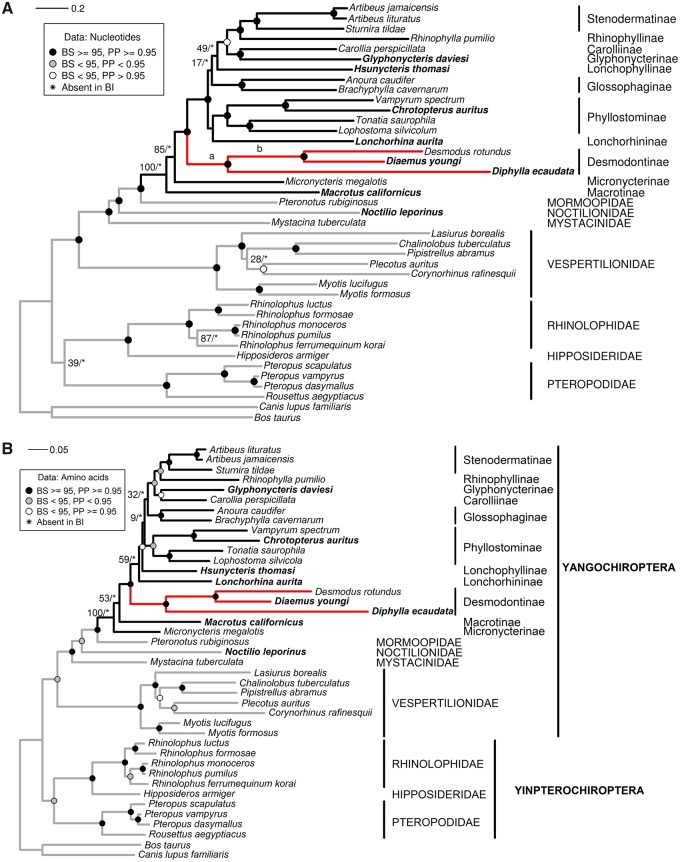
Fig. 1.
—Molecular phylogenies inferred from the mitogenomes of phyllostomid bats and representatives of other chiropteran lineages. Taxa names in bold correspond to the species for which new mitogenomes were produced in this study. In (A), the topology is inferred using an ML approach under the GTR+GAMMA+I model on nucleotide sequences; (a) and (b) indicate the ancestral branches of vampires and of [Diaemus + Desmodus], respectively. In (B), the tree results from amino acid sequences. Symbols on nodes describe the statistical support provided by ML and Bayesian inferences.
Radical Amino Acid Substitutions
Radical amino acid substitutions were evaluated using TREEsaap v3.2 (Woolley et al. 2003) on the concatenation of the 12 proteins encoded by the H strand. A first run including the 32 available protein properties, with a sliding-window of size 1 and a step of 1, was performed with the aim of identifying the properties modified by significant radical substitutions. A second analysis was run using only the properties identified in the first step, with a sliding-window of 20 and a step of 1 to identify the regions and sites likely to be under positive selection for the different branches of the tree. Radical substitutions inferred for the vampire bat lineage were analyzed in terms of the impacted properties, the amino acid residues involved, and in combination with the information provided by the analyses of nucleotide and amino acid compositional biases.
Analysis of Nuclear Genes
To better understand the pattern of molecular evolution of phyllostomid mitochondrial sequences, and, more precisely, the possible impact of the nutritional quality of blood on the amino acid content of proteins, we drew a comparison with nuclear genes of chiropterans. For this, we focused on the orthologous genes available for Myotis lucifugus and Pteropus vampyrus in the OrthoMam v9 database (Douzery et al. 2014). Using a tblastn approach, we isolated the corresponding orthologues from transcriptomic sequences available for Artibeus jamaicensis (SRX176203) and Desmodus rotundus (SRX201167). We then compared the amino acid compositions of nuclear sequences with those of mitochondrial genes using a t-test among average contents of each residue in both species.
Results
Sequencing and Assembly of Mitogenomes
The total number of shotgun Illumina reads sequenced per species ranged from 5,398,643 (Chrotopterus auritus) to 17,560,768 (Noctilio leporinus). From these, 0.14 to 4.19% corresponded to mitochondrial reads, and allowed the successful assembly of eight complete mitogenomes (fig. 1 and table 2). Their total length varied between 16,618 bp for Diphylla ecaudata and 16,707 bp for Chrotopterus auritus, with differences mostly due to variable number of satellite repeats in CR. Mean coverage, defined as the mean number of reads covering a given site, ranged from 94× for Chrotopterus auritus to 1,760× for Diaemus youngi.
For each of these eight mitogenomes, we successfully annotated the 37 genes documented in other mammalian mitogenomes. The gene distribution on both strands and the gene order were conserved as compared with other chiropterans or closely related placentals. These new sequences were aligned with published mitochondrial genomes and provided high-quality mitogenomic data for representatives of all phyllostomid subfamilies including the three species of vampire bats, and a close outgroup belonging to the family Noctilionidae.
Mitogenomic Phylogenetics of Bats
For nucleotides, the partitioning scheme by gene and codon positions yielded the smallest AIC value and was thus the one retained in subsequent ML analyses. For amino acids, the model partitioned for the 12 proteins performed better than the one with a single partition (table 3). In the topologies reconstructed by maximum likelihood and Bayesian inference from the nucleotide and amino acid data sets, most of the nodes were supported by bootstrap support (BS) values of >70% and Bayesian posterior probabilities (PP) >0.95. A notable exception was the sister-group relationship between Pteropodidae and Rhinolophidae (suborder Yinpterochiroptera), which was weakly supported in ML topologies (BS = 39 and BS = 70 for nucleotides and amino acids, respectively) (fig. 1a and b). In the Bayesian approach, this node was absent from the topology inferred from nucleotides, but present in the one inferred from amino acids (PP = 0.96).
Table 3.
Comparison of Partitioned Models in PartitionFinder
| Data | Partitioning | Number of Partitions | Number of Parameters | −lnL | AIC |
|---|---|---|---|---|---|
| Nucleotides | A single partition | 1 | 89 | −247,968.3 | 496,114.5 |
| Genes (rRNA, tRNA, 12 protein-coding genes) | 14 | 232 | −246,756.8 | 493,977.6 | |
| rRNA+tRNA+codon positions | 5 | 133 | −239,705.4 | 479,676.9 | |
| rRNA+tRNA+genes+codon positions | 38 | 496 | −238,352.2 | 477,696.4 | |
| Amino acids | A single partition | 1 | 81 | −58,151. 9 | 116,463.9 |
| Partitioned by gene | 12 | 1,028 | −57,906.3 | 116,016.6 |
Note.—For the protein alignments, the best models per partition were mtMAM + GAMMA + I (at6, at8, co2, co3, cyb, nd1, nd3, nd4, and ndl), mtMAM + GAMMA + I+F (nd2, nd5), and mtREV + GAMMA + I+F (co1), with F meaning empirical amino acid frequencies. The lowest AIC values are in bold for nucleotides and amino acids, which is indicative of a better fit of the alternative, more parameterized models.
Within Noctilionoidea, there was strong support (BS = 100; PP = 1 for nucleotides, and BS = 100; PP = 1 for amino acids) for the node placing the New Zealand short-tailed bat Mystacina tuberculata as the sister-group of all the other sampled noctilionoid bats comprising the families Noctilionidae, Mormoopidae, and Phyllostomidae. Within phyllostomids, the 11 subfamilies as defined by Baker et al. (2003) were retrieved: Macrotinae (big-eared bats), Micronycterinae (little big-eared bats), Desmodontinae (vampire bats), Phyllostominae, Glossophaginae (long-tongued, long-nosed, long-tailed, single-leaf, and banana bats), Lonchorhininae (sword-nosed bats), Lonchophyllinae (nectar bats), Glyphonycterinae (tricolored bats), Carollinae (short-tailed leaf-nosed bats), Rhinophyllinae (little fruit bats), and Stenodermatinae (neotropical fruit-eating bats) (fig. 1a and b).
Focusing on vampire bats, the relationships among the three species were well resolved in all inferred topologies with maximal statistical support (BS = 100 and PP = 1). Furthermore, a remarkable feature common to all topologies was the long branches exhibited by the vampire clade, demonstrating an elevated evolutionary rate relative to other phyllostomids (fig. 1).
Nucleotide and Amino Acid Compositions of Mitochondrial Genes
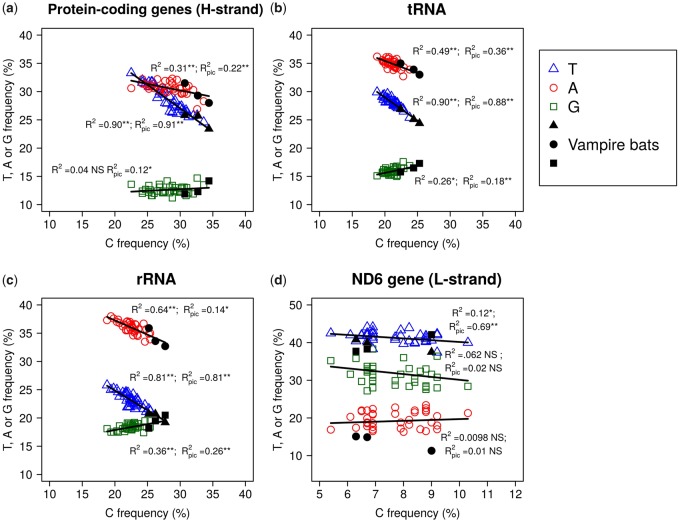
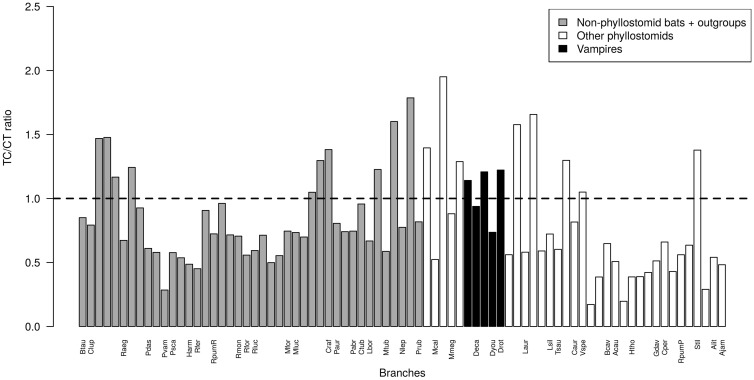
The comparison of the nucleotide compositions among chiropteran mitochondrial genes revealed an increase in C content mirrored by a decrease in T content for vampire bats, resulting in a significant negative correlation between both nucleotide frequencies (fig. 2). This change in base composition was also reflected in the G + C content and A + T-skew and G + C-skew estimates for the mitogenomes of vampires (table 2). This trend was observed for the 12 H-strand protein-coding, the ribosomal RNA, and the tRNA partitions of the mitogenome (fig. 2a–c). These correlations remained significant even after correcting for phylogenetic inertia. For the L-strand encoded nd6 gene, vampire bats were the G-richest taxa, mirroring the increase in C observed on the complementary H-strand (fig. 2d). The trend toward the C-enrichment was partially explained when mapping C-to-T and T-to-C substitutions along the branches of the phylogeny (fig. 3). A group of taxa including two vampires, Diphylla and Desmodus, which exhibited the highest values, and the non-vampires Vampyrum, Chrotopterus, Sturnira, and Micronycteris presented a number of T to C substitutions above the mean of phyllostomids. This trend was accentuated when looking at internal branches, with the two ancestral branches of vampire bats being longer than those leading to other clades of phyllostomids.
Fig. 2.
—Nucleotide compositions of the mitochondrial partitions rRNA, tRNA, the 12 protein coding genes contained in the H strand, and the nd6 gene, encoded by the L strand of the mitogenome. Black symbols correspond to vampire bats and empty symbols to the other chiropteran taxa. Correlation coefficients and pvalues are indicated next to the corresponding regression straight line. R2 = Correlation coefficient for raw data, R2pic = Correlation coefficient after correction by phylogenetically independent contrasts.
Fig. 3.
—Ratio between T-to-C and C-to-T inferred substitutions. Terminal branches (labelled by taxa) and internal branches (not labelled) are ordered following their divergence on the tree. Among phyllostomids, vampire bats (Drot, Dyou, Deca) along with Vampyrum, Chrotopterus, Micronycteris, and Sturnira exhibit a higher number of T-to-C substitutions, supporting the hypothesis of a mutational bias toward these kind of changes in the corresponding lineages.
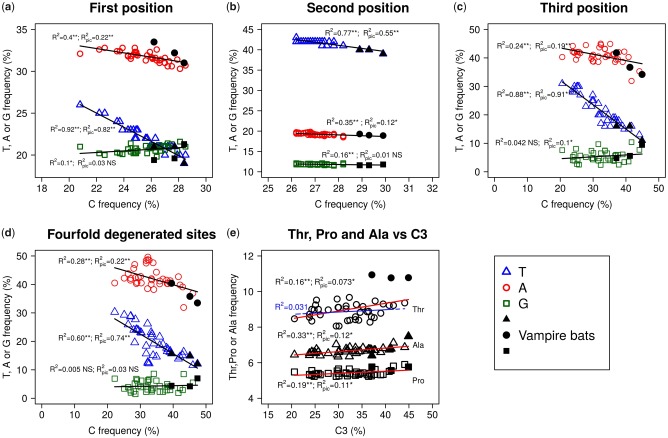
When the nucleotide composition was computed on the first, second, and third codon positions of the 12 H-strand protein-coding genes, the same trends were observed (fig. 4a–d). After correcting for phylogenetic inertia, there was still a negative correlation between C and T contents, and vampire bats presented high-C and low-T values at all three codon positions, as well as at 4-fold degenerated sites. This is in agreement with expectations for the occurrence of a mutational bias. Interestingly, for the selectively constrained second codon positions, vampires fell apart from other bats, with an increase in C2 larger than twice the SD of the observed C2 values for all other chiropterans, and concomitantly present the lowest T values.
Fig. 4.
—Nucleotide compositions for the three positions of the codon – first (a), second (b), third (c) –, four-fold degenerated sites (d), and the linear regression between C3 and Thr, Pro and Ala content (e) for the 12 H-strand protein-coding genes. R2: correlation coefficient for raw data; R2pic: correlation coefficient after correction for phylogeny. Vampire bats (black symbols) appear always toward high C and low T values, suggesting the occurrence of a bias from T to C. The last plot shows that C3 content is not able to predict the Thr content while, as it would be expected in presence of a mutational bias only, it positively correlates with Pro and Ala content. Moreover, vampire bats are clearly much Thr-richer than other taxa with similar C3 contents. This would suggest the existence of a selective component driving the evolution of mitochondrial genomes.
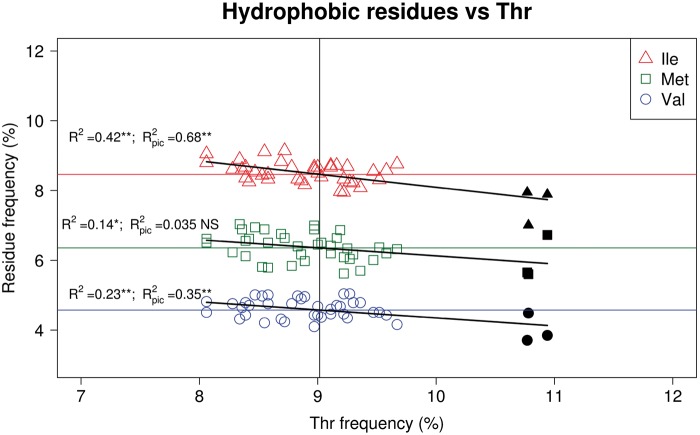
At the amino acid level, vampire bats also set apart from mean values with a significant (greater than twice the SD) increase in threonine (Thr—encoded by ACN codons) while presenting lower frequencies of the hydrophobic residues isoleucine (Ile—ATY codons), methionine (Met—ATR codons), phenylalanine (Phe—TTY codons), and valine (Val—GTN codons) (fig. 5 and supplementary fig. S4, Supplementary Material online). Moreover, after correction using phylogenetically independent contrasts (pic), negative correlations between the frequency of Thr and those of Ile (R2 = 0.42, p = 5.3e-06, R2pic = 0.13, Ppic = 0.02), Val (R2 = 0.23, p = 0.002; R2pic = 0.13, Ppic = 0.02), and Met (R2 = 0.14, p = 0.02, R2pic= 0.1, Ppic = 0.04) were observed. Moreover, although a positive correlation existed between C3 content and Thr frequencies (R2 = 0.16, P = 0.01), Thr values observed for vampire bats were higher than what might be expected following the regression and with respect to other C3-rich taxa (fig. 4e).
Fig. 5.
—Correlations between threonine (Thr), and isoleucine (Ile) (red triangles), methionine (dark green squares), and valine (blue circles) contents. Black symbols correspond to vampire bats, empty symbols to other lineages of chiropterans. R2 = Correlation coefficient for raw data; R2pic = Correlation coefficient after correction by phylogenetically independent contrasts.
Comparison with Nuclear Proteins
To explore the variation of amino acid compositions in nuclear proteins, we obtained 6,595 sequences for Artibeus jamaicensis and Desmodus rotundus. After comparison between the two species, and consistent with observations in mitochondrial proteins, Thr content in Desmodus proteins was, on average, significantly higher (t-test, p < 0.01) than that in Artibeus whereas Ile, Val, and Met contents were significantly (P value < 0.01) lower in Desmodus than in Artibeus proteins (supplementary fig. S5, Supplementary Material online). The same trend was observed when looking only at 75 nuclear OXPHOS genes: proteins of Desmodus were significantly enriched in Thr (P value < 0.01) and had significantly lower contents of the hydrophobic residues Ile, Val, and Met (p < 0.01).
Detection of Positive Selection
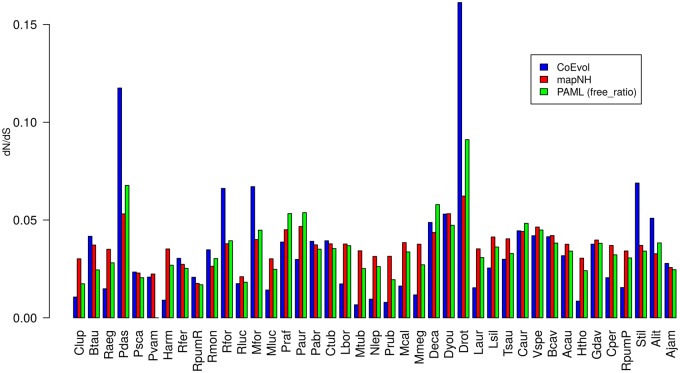
The different methods and models used to estimate the dN/dS ratio showed that vampire bats display higher values compared with other chiropteran lineages (Wilcoxon test P value < 0.05 for all three methods) (fig. 6). Moreover, the dN/dS values estimated for vampire bats were globally higher than those of other phyllostomids (mean value when excluding vampires was 0.03 and P value < 0.05 for all three methods) reaching values close to 0.06 and 0.10 as estimated by ML and Bayesian approaches, respectively. Additionally, a separate analysis of dN and dS values showed that the dN/dS increase in vampire bats is due to a proportionally higher dN compared with dS, and the difference is more marked in the internal branches of the vampire clade (supplementary fig. S2, Supplementary Material online).
Fig. 6.
—dN/dS estimations with three different methods: CoEvol (blue), mapNH(red), and PAML free ratio (green). Taxa are ordered following divergences in the tree. Vampire bats (Drot, Dyou, and Deca) present higher values than most of phyllostomid bats (mean dN/dS = 0.03).
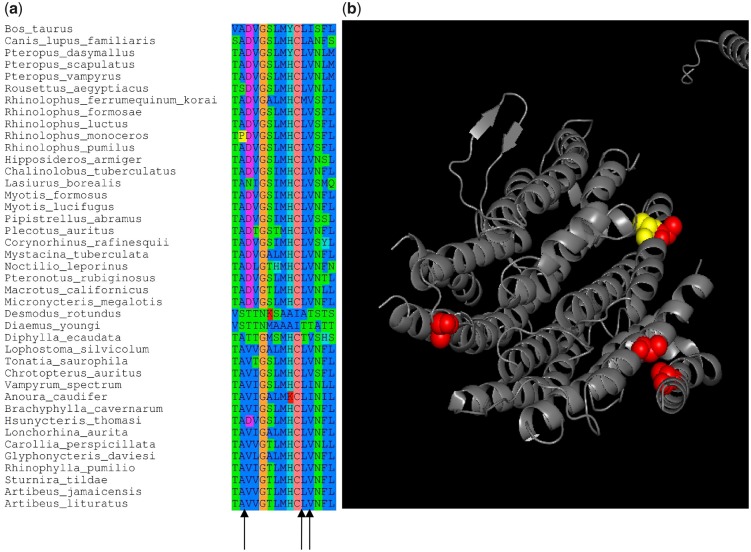
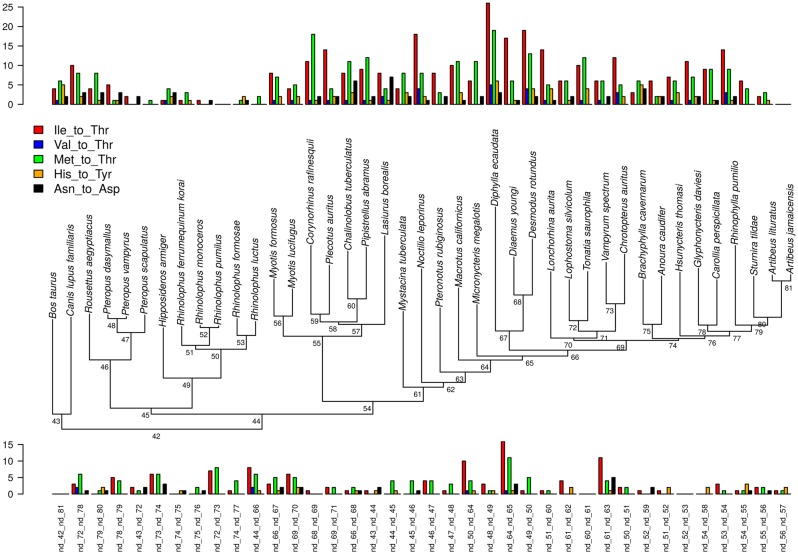
Likelihood ratio tests (LRT) for the branch model showed that a better fit is obtained when assuming different dN/dS values for each of the five branches of the vampire clade, indicating the existence of significant differences with respect to other bat lineages (supplementary table S1, Supplementary Material online). However, when the 12 H-strand protein-coding genes were concatenated, the branch-site model did not detect significant dN/dS variations in vampires, neither along branches nor among sites. Interestingly, when genes were analyzed separately, the LRT became significant for the nd5 gene in the two ancestral branches leading to vampires and to [Diaemus + Desmodus], respectively (table 4). For this gene, the sites identified as undergoing positive selection (Bayes empirical Bayes > 0.95) for both ancestral branches of vampires were highly conserved along the tree, varying only in vampires and corresponding to changes from hydrophobic to Thr residues (fig. 7a). Moreover, when mapped on the 3 D structure of the nd5 molecule, these sites are part of alpha-helices and located in spatially close positions in the folded protein (fig 7b).
Table 4.
Results of the Tests for Detection of Positive Selection in Individual Genes
| Branch | Model parameters | Genes |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| at8 | at6 | nd1 | nd2 | nd3 | nd4 | nd5 | ndl | nd6 | co1 | co2 | co3 | cyb | ||
| All (Free ratio model) | Null_hyp | −4088.54 | −12083.40 | −16448.58 | −20904.62 | −6571.89 | −26216.88 | −36009.42 | −5632.12 | −10755.64 | −22641.04 | −10549.32 | −12359.44 | −18998.22 |
| Free_ratio | −4035.53 | −12007.90 | −16373.49 | −20805.34 | −6518.46 | −26101.33 | −35911.43 | −5569.62 | −10696.58 | −22544.81 | −10459.91 | −12275.49 | −18937.97 | |
| LRT | 106.03 | 151.00 | 150.16 | 198.55 | 106.86 | 231.10 | 195.99 | 125.01 | 118.13 | 192.44 | 178.81 | 167.89 | 120.49 | |
| Ancestor of vampire bats | −lnL 2ω | −4085.55 | −12081.72 | −16448.22 | −20903.13 | −6571.73 | −26214.36 | −36004.92 | −5626.94 | −10748.01 | −22639.58 | −10545.09 | −12356.19 | −18992.30 |
| ω_others | 0.20109 | 0.03197 | 0.02453 | 0.05030 | 0.03651 | 0.03500 | 0.04871 | 0.03040 | 0.05753 | 0.00798 | 0.01887 | 0.02028 | 0.02609 | |
| ω_AncVamp | 999.00 | 978.48635 | 0.01456 | 0.19529 | 0.06373 | 0.21912 | 801.49397 | 999.00 | 999.00 | 0.03032 | 999.00 | 741.58554 | 999.00 | |
| LRT_2ω | 5.98 | 3.38 | 0.73 | 2.99 | 0.31 | 5.05 | 9.02 | 10.36 | 15.26 | 2.90 | 8.45 | 6.50 | 11.84 | |
| ModelA_Ha | −3930.92 | −11980.57 | −16330.21 | −20726.44 | −6472.06 | −25943.00 | −35430.16 | −5629.61 | −10593.33 | −22530.10 | −10507.07 | −12230.03 | −18871.76 | |
| ModelA_Ho | −3931.03 | −11980.57 | −16330.24 | −20726.44 | −6472.04 | −25943.19 | −35431.43 | −5629.81 | −10594.34 | −22530.10 | −10507.16 | −12230.04 | −18874.02 | |
| ω_bg | 0.08 | 0.03 | 0.02 | 0.05 | 0.03 | 0.03 | 0.04 | 0.03 | 0.05 | 0.01 | 0.02 | 0.02 | 0.02 | |
| ω_fg | 3.72 | 1.00 | 1.49 | 1.00 | 1.00 | 3.05 | 7.01 | 999.00 | 999.00 | 1.06 | 999.00 | 999.00 | 1.00 | |
| LRT_ModelA | 0.21 | 0.00 | 0.06 | 0.00 | −0.04 | 0.37 | 2.53 | 0.41 | 2.04 | 0.00 | 0.19 | 0.03 | 4.51 | |
| Ancestor of Desmodus and Diaemus | −lnL 2ω | −4087.00 | −12078.74 | −16448.23 | −20902.61 | −6570.00 | −26215.33 | −36001.10 | −5629.65 | −10755.45 | −22641.01 | −10542.70 | −12358.78 | −18998.99 |
| ω_others | 0.20053 | 0.03165 | 0.02451 | 0.05015 | 0.03596 | 0.03498 | 0.04849 | 0.03052 | 0.05837 | 0.00807 | 0.01855 | 0.02036 | 0.02516 | |
| ω_DesDia | 1.49075 | 1.12454 | 0.01634 | 0.22693 | 999.00 | 0.08028 | 0.66769 | 999.00 | 0.03645 | 0.00953 | 999.00 | 0.03775 | 0.06075 | |
| LRT_2ω | 3.10 | 9.34 | 0.71 | 4.03 | 3.76 | 3.10 | 16.66 | 4.95 | 0.37 | 0.05 | 13.23 | 1.31 | −1.54 | |
| ModelA_Ha | −3930.87 | −11972.50 | −16330.81 | −20725.65 | −6470.43 | −25946.57 | −35417.96 | −5629.88 | −10601.85 | −22530.80 | −10509.31 | −12231.51 | −18880.46 | |
| ModelA_Ho | −3931.15 | −11974.74 | −16330.81 | −20725.68 | −6470.43 | −25946.57 | −35423.66 | −5629.98 | −10602.14 | −22530.80 | −10509.48 | −12231.51 | −18880.46 | |
| ω_bg | 0.08 | 0.03 | 0.02 | 0.05 | 0.03 | 0.03 | 0.04 | 0.03 | 0.05 | 0.01 | 0.02 | 0.02 | 0.02 | |
| ω_fg | 6.63 | 999.00 | 3.22 | 1.40 | 1.00 | 1.78 | 14.00 | 999.00 | 4.36 | 1.00 | 2.96 | 1.00 | 1.00 | |
| LRT_ModelA | 0.54 | 4.49 | 0.00 | 0.05 | 0.00 | 0.00 | 11.40 | 0.21 | 0.58 | 0.00 | 0.34 | 0.00 | 0.00 | |
Note.—This table shows the two branches for which significant positive selection was inferred, the ancestor of vampires (AncVamp) and the ancestor of Desmodus and Diaemus (DesDia). Free ratio = one omega (ω) per branch; 2ω = branch model; ModelA = branch-site model A; ω_bg= omega for background; ω_fg= omega for foreground. Likelihood ratio tests (LRT) were conducted between the branch model and a null hypothesis with a single ω value for all branches and between model A and a null hypothesis considering ω for the background set to 1. LRT values in bold correspond to P values < 0.05.
Fig. 7.
—(a) Positively selected sites (p > 0.90) in mitochondrial genes along the ancestral branch of all vampires and that of [Diaemus + Desmodus]. These sites are highly conserved either in all chiropterans or among phyllostomids varying only in vampires. Most replacements are from hydrophobic residues (Ile, Val, Met) toward Thr. Species are ordered following their divergence in the mitogenomic tree. (b) 3 D representation of the ND5 folded protein of Bos taurus (PBD: 5-COD, chain A). Spheres show the sites positively selected (P > 95%) in the ancestral branch of vampires (yellow) and that of [Diaemus + Desmodus] (red). The 3D protein structure plot was generated using pymol v1.7.5 (DeLano 2002).
Radical Changes in Mitochondrial Proteins
The detailed analysis of the radical substitutions occurring along each branch of the tree and the quantification of the magnitude of change in protein properties showed that vampire bats, as compared with the rest of chiropterans, exhibit a higher number of radical amino acid changes (fig. 8) and a higher number of replacements among hydrophobic residues (Ile, Met, Val), and between them and Thr (fig. 9). In addition, six protein properties were identified as being radically modified by these changes (table 5), with exchanges among hydrophobic and Thr residues simultaneously affecting three of the six earlier-mentioned properties.
Fig. 8.
—Number of radical amino acid substitutions for each branch of the phylogeny. Bars on the top correspond to terminal branches, those at the bottom to internal branches. Black bars belong to vampire bats.
Fig. 9.
—Number of substitutions toward threonine from the hydrophobic residues isoleucine (red), valine (blue) and methionine (green). Additionally, substitutions from histidine to tyrosine (orange) and from asparagine to aspartate (black) are given.
Table 5.
Properties of Mitochondrial Proteins of Vampire Bats Modified by Radical Substitutions between Isoleucine and Threonine, Asparagine and Aspartate, Histidine and Tyrosine, and Methionine and Threonine
| Properties | Magnitude of Change | AA Changes Involved | Direction of Change |
|---|---|---|---|
| Solvent accesibility ratio | 8 | Ile → Thr | Decrease |
| Long-range non-bonded energy | 6 | Ile → Thr | Decrease |
| Equilibrium constant (ionization of COOH) | 6 | Ile → Thr | Increase |
| Power to be at the C-terminal | 6 | Asn → Asp | Increase |
| Power to be in the middle of the alpha-helix | 7 | His → Tyr | Decrease |
| Alpha-helical tendencies | 6 | Met → Thr | Decrease |
Note.—The direction of the induced changes is given.
Discussion
Mitogenomics and the Phylogeny of Leaf-Nosed Bats
Given that all samples were processed using the same protocol, the variations observed in the resulting fractions of mitochondrial reads and the coverage of mitogenomes, along with the absence of correlation with the total number of sequencing reads, suggest the occurrence of tissue sample-specific factors like organ type or preservation treatments. Indeed, it has been shown that the number of mitochondria per cell and thus, of mitochondrial DNA copies, can vary greatly both among and within organs (Robin and Wong 1988; Veltri et al. 1990; Fuke et al. 2011), therefore modifying the ratio of mtDNA and nuDNA from one sample to another and potentially contributing to the observed differences (Botero-Castro et al. 2013).
An important point when assembling mitogenomes is the risk of incorporating nuclear copies of mitochondrial genes (NUMTs), which exhibit evolutionary trends distinct from authentic mitochondrial sequences (Bensasson et al. 2001; Hazkani-Covo et al. 2010). If no controls are implemented to minimize this risk, assembly can result in chimerical mitogenomic sequences containing misleading information on phylogeny and evolutionary rates (Hassanin et al. 2010). The absence of frameshifts and premature stop codons in open reading frames for each of the newly assembled mitogenomes (Calvignac et al. 2011), the high coverage provided by hundreds to thousands of overlapping reads supporting each site, and the circularity of the final sequence assemblies, validate our mitogenome sequences as authentic mitochondrial genomes.
The relationships among the subfamilies of phyllostomid bats inferred from our mitogenomic data set were mostly well supported, and in agreement with previous phylogenetic hypotheses. The weak statistical support obtained for some of the nodes can be attributed to the occurrence of short branches in relation with the rapid evolutionary radiation of phyllostomid bats (Baker et al. 2003; Datzmann et al. 2010; Rojas et al. 2016). Some subfamilies are also represented by a single taxon producing isolated branches and unstable nodes in the current mitogenomic tree. Although the position of Lonchophyllinae is still uncertain, they never branched with Glossophaginae, confirming the independent evolution of nectarivory in phyllostomid bats as suggested by Datzmann et al. (2010).
In the phylograms inferred from the mitochondrial DNA and protein alignments, the long branches observed for vampire bats suggest the occurrence of neutral (shifts in nucleotide and/or amino acid compositions, accelerated synonymous substitution rates) and/or adaptive processes (episodes of positive selection) in this clade (Bromham 2009). These two processes are difficult to disentangle (cf. Kitazoe et al. 2008 and Jobson et al. 2010) and hereafter, we investigate their respective contribution to mitochondrial DNA evolution in vampires, and evaluate whether they are related to the shift to a blood-based diet.
Evidence for Mutational Bias in Vampire Bats
Because mutations occur randomly throughout the mitogenome, the impact of a mutational bias is expected to be observed all along the H or L strands. The trends observed in both the nucleotide and amino acid compositions of vampire bats mitogenomes suggest the occurrence of a mutational bias toward enrichment in C content at the expense of T content. At the nucleotide level, this bias contributes to the increased number of T to C substitutions observed in the vampire clade, contributing to longer branches in the tree (fig. 1a). At the amino acid level, the mutational bias contributes to the observed excess in threonine, as the ACN codons encoding for this residue involve a C at the second position. It is also an explanation for the decrease of hydrophobic residues, isoleucine (ATH codons), and valine (GTN), which instead need a T at the second codon position, in agreement with the observed negative correlations at the nucleotide and amino acid levels (fig. 4). Moreover, the reverse pattern observed for the L-strand encoded nd6 gene indicates that this phenomenon might be strand-specific and possibly associated to the asymmetrical directional mutation pressure affecting mammalian mitogenomes (Reyes et al. 1998). Additionally, the occurrence of a C-enrichment in some other lineages of phyllostomids—for example, Vampyrum, Chrotopterus, Sturnira, and Micronycteris—suggests that such a bias might represent a recurrent trend in this family (fig. 3).
A potential source of bias in base composition of mitochondrial genomes is the asymmetry of mitochondrial replication related to the time spent in a single-stranded state during replication (Faith and Pollock 2003). Under this hypothesis, one would expect to find a direct correlation between the position of the genes along the mitogenome, as measured through the predicted duration times spent in a single-strand state for each of the protein-coding genes of the H-strand (DssH), and the variation in substitution rates along the mitogenome. Contrary to these expectations, we did not find any relationship between the time spent by each gene in the single-strand state and the ratio of T-to-C over C-to-T substitutions, neither over complete sequences of genes (supplementary fig. S1a, Supplementary Material online), nor using only third positions of 4-fold-degenerated codons (supplementary fig. S1b, Supplementary Material online). Instead, vampire bats exhibit higher ratios of T-to-C over C-to-T substitutions compared with other bats all along the mitogenome, which would agree with the expectations derived from the action of a mutational bias that is independent of the replication process of mitochondrial DNA.
Evidence of Positive Selection
Besides the mutational bias, an adaptive component might also be modulating the evolution of the mitogenome in vampire bats. If base composition of the mitogenomes of vampire bats were driven by mutational bias only—that is, in the absence of selective forces—, the evolutionary trends for amino acids encoded by C-rich codons would be expected to follow similar trends within vampires and other taxa exhibiting similar compositional biases. However, this is not the case when comparing, for example, Pro, Ala, and Thr, encoded by CCN, GCN, and ACN codons, respectively, in vampires and in non-hematophagous taxa with C-rich mitogenomic codons (e.g., Chrotopterus and Vampyrum). Whereas Pro and Ala content in mitochondrial proteins is high in both groups, the amount of Thr residues is, as shown in fig. 4e, higher in vampire bats than in non-vampire taxa with similar or higher values of C3 content. Actually, the correlation observed between C3 and Thr seems to be entirely driven by the extreme values of vampire bats, as the same correlation evaluated excluding vampires was not significant (fig 4e, blue line) and showed vampire bats away from the expected values. This might thus indicate that an additional factor, other than mutational bias, is driving the evolution of Thr content, suggesting a role for positive selection.
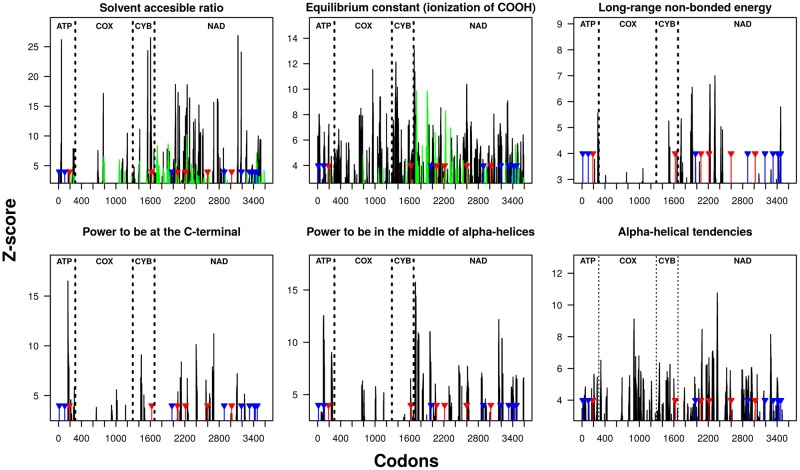
A second set of arguments is provided by the joint analysis of dN/dS and protein properties in a comparative phylogenetic framework. This analysis showed that vampire bats have higher dN/dS than their phyllostomid relatives. At the protein level this is reflected by a higher number of radical replacements in general and, more particularly, by a higher number of replacements between Thr and hydrophobic residues (fig. 9). The observed increase in dN/dS could be the outcome of dS underestimation due to saturation at synonymous positions because of non-detected multiple substitutions in the fast-evolving mitochondrial DNA of vampire bats. However, this possibility can be ruled out as patristic distances involving vampires, and estimated for the first and third codon positions, spanned the whole range of values observed for all species pairs, whereas they were among the highest values for the most constrained, second positions (supplementary fig. S3, Supplementary Material online). A high dN/dS may also reflect the occurrence of either relaxed purifying selection or adaptive episodes. The estimations of ω on individual genes showed values >1 for the vampire branches in several genes (supplementary table S2, Supplementary Material online), but only nd5 was identified as significantly undergoing positive selection (table 4) on the two ancestral branches of the clade. This would point toward the occurrence of positive selection, which is expected to impact only particular genes rather than all of them, as it would be expected under a generalized reduction of selective pressures (Tomasco and Lessa 2011). Moreover, the regions neighboring the sites likely to be positively selected, as identified by the branch-site model, are also identified as likely undergoing radical modifications in the local properties of the protein (fig. 10).
Fig. 10.
—Protein properties and residues inferred as likely to be under positive selection. The regions colored in black and green correspond to those with Z scores > 3.09 (P value < 0.01), susceptible of having undergone radical changes in each property as inferred from the TRESAAP analysis; sticks correspond to the sites likely to be under selection (blue: 0.90 < p < 0.95; red: p > 0.90) following Bayes empirical Bayes in PAML (see also fig. 7).
Altogether, these observations suggest that these sites might play an important role in determining the structure and/or function of the protein, perhaps in relation with the role of the chiral chain on the local properties of the regions where these substitutions took place. For instance, it is known that changes in the size of lateral chains can be detrimental for protein function (Betts and Russel 2007). Concerning the functional impact of these modifications, it has been documented that changes from Ile to Thr simultaneously produce: 1) a decrease in the solvent accessible reduction ratio, making the region more hydrophilic; 2) a decrease of the long-range non-bonded energy, which indicates that the protein is less compact, or more open, in the corresponding surrounding regions; and 3) an increase in equilibrium constant for COOH oxidation, meaning a more product-driven reaction, a lower production of ROS in caloric restriction, and potentially a deceleration of aging (Beckstead et al. 2009).
When estimating positive selection on the basis of dN/dS, some additional considerations are needed before validating the inference. Actually, estimation of ω is known to be affected by increased fixation of slightly deleterious mutations through processes like shifts in population size (e.g., Nabholz, Uwimana, et al. 2013; Subramanian 2013) and variations in nucleotide compositions, both of which can lead to false positives when detecting positive selection. However, several arguments suggest that our results have not been affected in a significant way by these problems. First, a study evaluating the behavior of the branch-site test with respect to extreme values of G + C (< 30% and >80%) showed that the dN/dS test is conservative and robust to these biases (Gharib and Robinson-Rechavi 2013): the number of false positives is not significantly different even in presence of extreme G + C values due to shifts in base composition among branches or genes. Thus, although the unusual nucleotide composition described here for the mitogenomes of vampire bats could at some point violate the assumptions of the model used to perform the tests of detection of positive selection, the values of G + C content for these taxa remain within the limits of robustness for the test (see above). Second, our data set contains taxa that present biases in nucleotide composition similar to those observed in vampire bats. However, neither positively selected sites nor increased amounts of radical amino acid replacements are identified for these taxa when the corresponding tests are applied to the respective branches (e.g., the internal branch subtending the clade formed by Chrotopterus and Vampyrum).
Mitogenomic Evolution and Hematophagy in Vampire Bats
Most of the organisms adapted to highly specialized diets have been documented to exploit alternative resources in order to complement the nutrients missing from their main food supply. For example, hummingbirds and nectarivorous bats complement their carbohydrate-rich diets with protein sources like insects (Stiles 1995; Clare et al. 2014). Instead, and unlike other hematophagous organisms, vampire bats rely entirely on blood feeding, and are thus constrained by the nutrients it contains, including amino acids. Interestingly, all of the residues in the mitochondrial proteins of vampire bats displaying variations in their frequencies with respect to other chiropterans correspond to essential amino acids that cannot be synthesized, and must therefore be acquired from food items (supplementary fig. S4, Supplementary Material online). A clear example of the impact of reduced availability of essential amino acids in blood is provided by mosquitoes, in which isoleucine is essential for egg production and quality. In order to cope with Ile scarcity in blood, mosquitoes are forced to pick repeatedly in order to get the necessary income of this amino acid (Harrington et al. 2001).
In agreement with the patterns described earlier for isoleucine and methionine, studies on their nutritional quality have shown that both avian and mammalian blood—the main preys of vampire bats—have a low content of these two residues (Duarte et al. 1999; Márquez et al. 2005). The small fraction of these amino acids available in blood might thus be imposing vampire bats a constraint on the residues available for biosynthesizing proteins, a hypothesis that seems to be also supported by the amino acid composition of proteins encoded in the nucleus (see below). Furthermore, the limiting nature of isoleucine in mitochondrial proteins might also come from the need to biosynthesize other important metabolites like acetyl-coenzyme A (acetyl-CoA), an essential molecule of the oxidative pathway that is key for energy production. Acetyl-CoA is mainly synthesized in mitochondria from cytosolic pyruvate after oxidation of glucose. However, the fact that vampire bats do not exhibit maltase, sucrose, or trehalase activity (Schondube et al. 2001) suggests that they would not be able neither to produce acetyl-CoA from the carbohydrates present in blood nor to directly use them as a source of energy. A similar situation was described in tse-tse flies (Glossina morsitans), in which no sugar nor glycogen was detected, and a congruent reduction of genes associated to carbohydrate metabolism was documented (International Glossina Genome Initiative 2014). To counter these restrictions, vampire bats were recently suggested to rely on different components of their gut microbiome (Zepeda Mendoza et al. 2018). There is, however, experimental evidence of glucose intolerance in vampires (Freitas et al. 2013), which should not be observed if glucose degradation were performed by gut microbiome.
Being unable to process carbohydrates, vampire bats might thus be forced to synthesize acetyl-CoA from the amino acid pathway, which is accomplished in mitochondria. This alternative metabolic route was already suggested following observations of responses to fasting in Diphylla ecaudata (Gomes 2008) and exists in the common tick (Ixodes scapularis), in which Val and Leu are also used as substrates for the biosynthesis of acetyl-CoA (Cabezas-Cruz et al. 2017). Moreover, from the seven residues yielding acetyl-CoA, isoleucine involves a single reaction, while the other six need more complex pathways (Nelson and Cox 2013).
One way to deal with a deficit of amino acid in relation with diet constraints would be a replacement by a residue that is more abundant in blood, yet with similar physico-chemical properties. As all the exchangeable residues (i.e., hydrophobic ones) are poorly represented in blood, the change toward Thr might correspond to the less radical change among all the remaining possible replacements. Indeed, Thr and Ile (as well as Val) share the property of having a chiral side chain that makes the region more bulky near the protein backbone, and the residues more constrained in the conformations that can be adopted by the main chain (Betts and Russel 2007). Thus, the observed increase in exchange rate between Thr and the hydrophobic Ile and Val residues might correspond and take place at positions of the peptidic chain where chiral side chain is relevant, which is in agreement with inferences from radical changes in protein properties (see above). This might also partly explain the observed increase in C content, mainly at the second codon position, which would fulfill the need for Thr codons (ACN). Moreover, the exchange between these residues is facilitated at the nucleotide level by the similarity among the corresponding codons: Thr codons (ACN) are close to those of the hydrophobic residues, all of which have a T at the second position (ATY for Ile, ATR for Met, and GTN for Val), which is consistent with the negative C versus T correlations observed.
The comparison of the amino acid frequencies in transcriptomic nuclear sequences between Artibeus jamaicensis and Desmodus rotundus provide an answer to the corollary question of whether the trends observed in mitochondrial genes of vampires also apply to genes of the mitochondrial machinery encoded in the nucleus. Even if not all of the nuclear genes that we analyzed are concerned, 3,692 (56%) of them, including some of the OXPHOS subunits, exhibit at least one of the amino acid replacements detected in mitochondrial genes involving Thr and hydrophobic residues. Also, on average, Desmodus proteins have a higher Thr and lower hydrophobic residues contents than those of the non-hematophagous Artibeus, suggesting a possible impact of the reduced availability of some amino acids in blood on the protein synthesis of vampire bats.
Increased Lifespan, Hematophagy, and the Mitochondrial DNA of Vampire Bats
Some of the evolutionary trends of the mitochondrial DNA of vampires suggest hypotheses for their paradoxical increase in lifespan relative to other bats. First, the increase in the frequency of Thr residues in transmembrane domains has been suggested to be linked to the ability of this amino acid to form hydrogen bonds, improving the interaction among helices and conferring enhanced stability to the resulting protein in primates (Kitazoe et al. 2008; Min and Hickey 2008). Although this was initially interpreted as the result of selection for an increase in maximum life span, it was later shown to be the result of a mutational bias from T toward C. This was evidenced by a direct correlation of Thr content with C content on the third position—in which most substitutions are expected to be synonymous—of the 4-fold-degenerated ACN triplets coding for Thr (Jobson et al. 2010; Aledo et al. 2012).
In vampire bats, however, the increase in Thr content is not correlated to C3, suggesting that a mutation-based process is not explanatory of all the trends observed. This is consistent with the position of the residues identified as positively selected within the nd5 protein (fig. 7b), and the potential involvement of the lateral chains of Thr residues that might be associated to an increase in the stability of these proteins. Additionally, a study on a human haplotype proposed that the Ile7Thr replacement in the Cytochrome b protein, and similar to the replacements documented in vampires, modifies the properties associated to the hydrophilic affinity of the protein and improves the efficiency of ubiquinone leading to reduced production of ROS (Beckstead et al. 2009). Both enhanced stability of proteins and reduced ROS production are inversely related to aging, suggesting an association between the enhanced longevity of vampire bats and the increase in Thr content observed in their mitochondrial proteins.
Further support for a relationship between hematophagy and increased longevity in vampires comes from the trends observed for methionine frequencies in the mitochondrial proteins of vampires. Frequencies of methionyl residues in proteins have been documented to be inversely related to an increase in longevity (Aledo et al. 2011), thanks to the property of methionine of reversing oxidation thus acting as an antioxidant and ROS scavenger agent (Levine et al. 1996). There is also experimental evidence showing that a reduction in the amount of ingested methionine results in lower production and leak of ROS in highly aerobic organs like kidney, heart, and brain (Caro et al. 2008; Sánchez-Román et al. 2011), as well as in slower aging of immune system and lens, reduced levels of glucose, insulin, IGF-I, and enhanced resistance to oxidative damage (Miller et al. 2005), with methionine potentially regulating the concentration of complex I in mitochondria (Sanz et al. 2006; Page et al. 2010). Although these mechanisms do not involve directly the mitogenome, they reduce oxidative damage on mitochondrial DNA as they both result in a decrease of the amount of potentially harming ROS. Interestingly, both low Met content in proteins, as compared with closely related species, and reduced methionine ingestion are observed in vampire bats, due to blood being naturally impoverished in this essential residue (Duarte et al. 1999; Márquez et al. 2005). This suggests a relationship between the nutritional restrictions imposed by exclusively feeding on blood and the increased longevity in vampire bats through regulation of the mitochondrial metabolism and oxidative damage.
Conclusions
Our analyses provide evidence that the observed accelerated evolutionary rate of mitochondrial genomes of vampire bats might be due to the co-occurrence of both a neutral mutational bias toward C, and selective changes in both nucleotide and amino acid sequences, possibly to fill and compensate some of the metabolic constraints imposed by hematophagy as an extreme feeding habit. These substitutional changes seem to have occurred all along the divergence of vampire species and their shift to hematophagy, and subsequently during the divergence of the three genera and the specialization on mammalian and avian preys.
Supplementary Material
Acknowledgments
We gratefully thank Neil Duncan (American Museum of Natural History, AMNH, NewYork, NY), Joseph Cook (Museum of Southwestern Biology, MSB, University of New Mexico, NM), Chris Conroy (Museum of Vertebrate Zoology, MVZ, University of California Berkeley, CA), and Robert J. Baker, Heath Garner, and Kathy MacDonald (Texas Tech University, TTU, Lubbock, TX) for generously providing bat tissues. We would like to thank Nicolas Galtier and Benoit Nabholz for helpful discussions and suggestions during the analyses and writing of this manuscript. We also thank three anonymous referees and the associate editor Dennis Lavrov for helpful comments. We thank the Montpellier Bioinformatics Biodiversity platform (MBB) for access to computational resources. This publication is contribution No. 2018-119 of the Institut des Sciences de l’Evolution de Montpellier. This work has benefited from an “Investissements d'Avenir” grant managed by Agence Nationale de la Recherche (CEBA, ref. ANR-10-LABX-25-01).
Literature Cited
- Aledo JC, Li Y, de Magalhães JP, Ruíz-Camacho M, Pérez-Claros JA.. 2011. Mitochondrially encoded methionine is inversely related to longevity in mammals. Aging Cell 10(2):198–207. [DOI] [PubMed] [Google Scholar]
- Aledo JC, Valverde H, de Magalhães JP.. 2012. Mutational bias plays an important role in shaping longevity-related amino acid content in mammalian mtDNA-encoded proteins. J Mol Evol. 74(5–6):332–341. [DOI] [PubMed] [Google Scholar]
- Baker RJ, Bininda-Emonds ORP, Mantilla-Meluk H, Porter CA, Van Den Bussche RA.. 2012. Molecular timescale of diversification of feeding strategy and morphology in New World leaf-nosed bats (Phyllostomidae): a phylogenetic perspective In: Gunnell GF, Simmons NB, editors. Evolutionary history of bats: fossils, molecules and morphology. Cambridge: Cambridge University Press; p. 385–409. [Google Scholar]
- Baker RJ, Hoofer SR, Porter CA, Van Den Bussche RA.. 2003. Diversification among New World leaf-nosed bats: an evolutionary hypothesis and classification inferred form digenomic congruence of DNA sequence. Occas Papers Mus Texas Tech Univ. 230:1–32. [Google Scholar]
- Ballard JWO, Melvin RG, Katewa SD, Maas K.. 2007. Mitochondrial DNA variation is associated with measurable differences in life-history traits and mitochondrial metabolism in Drosophila simulans. Evolution 61(7):1735–1747. [DOI] [PubMed] [Google Scholar]
- Beckstead WA, Ebbert MT, Rowe MJ, McClellan DA.. 2009. Evolutionary pressure on mitochondrial cytochrome b is consistent with a role of CytbI7T affecting longevity during caloric restriction. PLoS One 4(6):e5836.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensasson D, Zhang D-X, Hartl DL, Hewitt GM.. 2001. Mitochondrial pseudogenes: evolution’s misplaced witnesses. Trends Ecol Evol. 16(6):314–321. [DOI] [PubMed] [Google Scholar]
- Betts MJ, Russel RB.. 2007. Amino-Acid properties and consequences of substitutions In: Barnes MR, editor. Bioinformatics for geneticists. Chichester: John Wiley & Sons, Ltd; p. 311–342. [Google Scholar]
- Botero-Castro F, et al. 2013. Next-generation sequencing and phylogenetic signal of complete mitochondrial genomes for resolving the evolutionary history of leaf-nosed bats (Phyllostomidae). Mol Phylogenet Evol. 69(3):728–739. [DOI] [PubMed] [Google Scholar]
- Breidenstein CP. 1982. Digestion and assimilation of bovine blood by a vampire bat (Desmodus rotundus). J Mammal. 63(3):482–484. [Google Scholar]
- Bromham L. 2009. Why do species vary in their rate of molecular evolution? Biol Lett. 5(3):401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet-Rossinni AK, Austad SN.. 2004. Ageing studies on bats: a review. Biogerontology 5(4):211–222. [DOI] [PubMed] [Google Scholar]
- Cabezas-Cruz A, Espinosa PJ, Obregón DA, Alberdi P, de la Fuente J.. 2017. Ixodes scapularis tick cells control Anaplasma phagocytophilum infection by increasing the synthesis of phosphoenolpyruvate from tyrosine. Front Cell Infect Microbiol. 7:375.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvignac S, Konecny L, Malard F, Douady CJ.. 2011. Preventing the pollution of mitochondrial datasets with nuclear mitochondrial paralogs (numts). Mitochondrion 11(2):246–254. [DOI] [PubMed] [Google Scholar]
- Cameron SL. 2014. Insect mitochondrial genomics: implications for evolution and phylogeny. Annu Rev Entomol. 59:95–117. [DOI] [PubMed] [Google Scholar]
- Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T.. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25(15):1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caro P, et al. 2008. Forty percent and eighty percent methionine restriction decrease mitochondrial ROS generation and oxidative stress in rat liver. Biogerontology 9(3):183–196. [DOI] [PubMed] [Google Scholar]
- Castoe TA, Jiang ZJ, Gu W, Wang ZO, Pollock D.. 2008. Adaptive evolution and functional redesign of core metabolic proteins in snakes. PLoS One 3(5):e2201.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamary JV, Parmley JL, Hurst LD.. 2006. Hearing silence: non-neutral evolution at synonymous sites in mammals. Nat Rev Genet. 7(2):98.. [DOI] [PubMed] [Google Scholar]
- Chaverri G. 2013. Flora bacteriana aeróbica del tracto digestivo del vampiro común, Desmodus rotundus (Chiroptera: phyllostomidae). Rev Biol Trop. 54(3):717–724. [PubMed] [Google Scholar]
- Clare EL, et al. 2014. Trophic niche flexibility in Glossophaga soricina: how a nectar seeker sneaks an insect snack. Funct Ecol. 28(3):632–641. [Google Scholar]
- Covacin C, Shao R, Cameron S, Barker SC.. 2006. Extraordinary number of gene rearrangements in the mitochondrial genomes of lice (Phthiraptera: insecta). Insect Mol Biol. 15(1):63–68. [DOI] [PubMed] [Google Scholar]
- Datzmann T, von Helversen O, Mayer F.. 2010. Evolution of nectarivory in phyllostomid bats (Phyllostomidae Gray, 1825, Chiroptera: mammalia). BMC Evol Biol. 10:165.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL. “The PyMOL Molecular Graphics System.” DeLano Scientific, San Carlos, CA, USA. http://www.pymol.org, last accessed 3 July 2018.
- Devin A, Rigoulet M.. 2007. Mechanisms of mitochondrial response to variations in energy demand in eukaryotic cells. Am J Physiol Cell Physiol. 292(1):C52–C58. [DOI] [PubMed] [Google Scholar]
- dos Reis M, et al. 2015. Uncertainty in the timing of origin of animals and the limits of precision in molecular timescales. Curr Biol. 25(22):2939–2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douzery EJP, et al. 2014. OrthoMaM v8: a database of orthologous exons and coding sequences for comparative genomics in mammals. Mol Biol Evol. 31(7):1923–1928. [DOI] [PubMed] [Google Scholar]
- Dowton M, Castro LR, Austin AD.. 2002. Mitochondrial gene rearrangements as phylogenetic characters in the invertebrates: the examination of genome ‘morphology’. Invert Syst. 16:345–356. [Google Scholar]
- Duarte RT, Carvalho Simões MC, Sgarbieri VC.. 1999. Bovine blood components: fractionation, composition, and nutritive value. J Agric Food Chem. 47(1):231–236. [DOI] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith JJ, Pollock DD.. 2003. Likelihood analysis of asymmetrical mutation bias gradients in vertebrate mitochondrial genomes. Genetics 165(2):735–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foote AD, et al. 2011. Positive selection on the killer whale mitogenome. Biol Lett. 7(1):116–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster PG, Jermiin LS, Hickey DA.. 1997. Nucleotide composition bias affects amino acid content in proteins coded by animal mitochondria. J Mol Evol. 44(3):282–288. [DOI] [PubMed] [Google Scholar]
- Freitas MB, et al. 2013. Reduced insulin secretion and glucose intolerance are involved in the fasting susceptibility of common vampire bats. General Comp Endocrinol. 183:1–6. [DOI] [PubMed] [Google Scholar]
- Fuke S, Kubota-Sakashita M, Kasahara T, Shigeyoshi Y, Kato T.. 2011. Regional variation in mitochondrial DNA copy number in mouse brain. Biochim Biophys Acta 1807(3):270–274. [DOI] [PubMed] [Google Scholar]
- Galen SC, et al. 2015. Contribution of a mutational hot spot to hemoglobin adaptation in high-altitude Andean house wrens. Proc Natl Acad Sci USA. 112(45):13958–13963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvin MR, Bielawski JP, Sazanov LA, Gharre AJ.. 2015. Review and meta-analysis of natural selection in mitochondrial complex I in metazoans. J Zool Syst Evol Res. 53(1):1–17. [Google Scholar]
- Gharib WH, Robinson-Rechavi M.. 2013. The branch-site test of positive selection is surprisingly robust but lacks power under synonymous substitution saturation and variation in GC. Mol Biol Evol. 30(7):1675–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes CID. 2008. Metabolismo energético e resposta ao jejum do morcego hematófago Diphylla ecaudata. [Dissertação]. Brasília: Universidade de Brasília. [Google Scholar]
- Greenhall AM, Joermann G, Schmidt U, Seidel MR.. 1983. Desmodus rotundus. Mamm Spec. 202(202):1–6. [Google Scholar]
- Greenhall AM, Schmidt U, Joermann G.. 1984. Diphylla ecaudata. Mamm Spec. 227(227):1–3. [Google Scholar]
- Greenhall AM, Schutt WAJ.. 1996. Diaemus youngi. Mamm Spec. 533(533):1–7. [Google Scholar]
- Guéguen L, et al. 2013. Bio++: efficient, extensible libraries and tools for computational molecular evolution. Mol Biol Evol. 30(8):1745–1750. [DOI] [PubMed] [Google Scholar]
- Harlow HJ, Braun EJ.. 1997. Gastric Na+K+ATPase activity and intestinal urea hydrolysis of the common vampire bat, Desmodus rotundus. Comp Biochem Physiol A Physiol. 118(3):665–669. [DOI] [PubMed] [Google Scholar]
- Harrington LC, Edman JD, Scott TW.. 2001. Why do female Aedes aegypti (Diptera: culicidae) feed preferentially and frequently on human blood? J Med Entomol. 38(3):411–422. [DOI] [PubMed] [Google Scholar]
- Hassanin A, Bonillo C, Nguyen BX, Cruaud C.. 2010. Comparisons between mitochondrial genomes of domestic goat (Capra hircus) reveal the presence of numts and multiple sequencing errors. Mitoch DNA 21(3–4):68–76. [DOI] [PubMed] [Google Scholar]
- Hassanin A, Léger N, Deutsch J.. 2005. Evidence for multiple reversals of asymmetric mutational constraints during the evolution of the mitochondrial genome of metazoa, and consequences for phylogenetic inferences. Syst Biol. 54(2):277–298. [DOI] [PubMed] [Google Scholar]
- Hassanin A, Ropiquet A, Couloux A, Cruaud C.. 2009. Evolution of the mitochondrial genome in mammals living at high altitude: new insights from a study of the tribe Caprini (Bovidae, Antilopinae). J Mol Evol. 68(4):293–310. [DOI] [PubMed] [Google Scholar]
- Hazkani-Covo E, Zeller RM, Martin W.. 2010. Molecular poltergeists: mitochondrial DNA Copies (numts) in sequenced nuclear genomes. PLoS Genet. 6(2):e1000834.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershberg R, Petrov DA.. 2008. Selection on codon bias. Annu Rev Genet. 42:287–299. [DOI] [PubMed] [Google Scholar]
- Hua X, Cowman P, Warren D, Bromham L.. 2015. Longevity is linked to mitochondrial mutation rates in rockfish: a test using Poisson regression. Mol Biol Evol. 32(10):2633–2645. [DOI] [PubMed] [Google Scholar]
- International Glossina Genome Initiative. 2014. Genome sequence of the Tsetse fly (Glossina morsitans): vector of African trypanosomiasis. Science 344:380–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James JE, Piganeau G, Eyre-Walker A.. 2016. The rate of adaptive evolution in animal mitochondria. Mol Ecol. 25(1):67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobson RW, Dehne-Garcia A, Galtier N.. 2010. Apparent longevity-related adaptation of mitochondrial amino acid content is due to nucleotide compositional shifts. Mitochondrion 10(5):540–547. [DOI] [PubMed] [Google Scholar]
- Kearse M, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28(12):1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitazoe Y, et al. 2008. Adaptive threonine increase in transmembrane regions of mitochondrial proteins in higher primates. PLoS One 3(10):e3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfear R, Calcott B, Ho SY, Guindon S.. 2012. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol. 29(6):1695–1701. [DOI] [PubMed] [Google Scholar]
- Lartillot N, Lepage T, Blanquart S.. 2009. PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics 25(17):2286–2288. [DOI] [PubMed] [Google Scholar]
- Lartillot N, Poujol R.. 2011. A phylogenetic model for investigating correlated evolution of substitution rates and continuous phenotypic characters. Mol Biol Evol. 28(1):729–744. [DOI] [PubMed] [Google Scholar]
- Levine RL, Mosoni L, Berlett BS, Stadtman ER. 1996. Methionine residues as endogenous antioxidants in proteins. Proc Natl Acad Sci U S A. 93(26):15036–15040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mans BJ, de Klerk D, Pienaar R, de Castro MH, Latif AA.. 2012. The mitochondrial genomes of Nuttalliella namaqua (Ixodoidea: nuttalliellidae) and Argas africolumbae (Ixodoidae: argasidae): estimation of divergence dates for the major tick lineages and reconstruction of ancestral blood-feeding characters. PLoS One 7(11):e49461.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Márquez E, Bracho M, Archile A, Rangel L, Benítez B.. 2005. Proteins, isoleucine, lysine and methionine content of bovine, porcine and poultry blood and their fractions. Food Chem. 93(3):503–505. [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17(1):10–12. [Google Scholar]
- Mehrabian Z, Liu L-I, Fiskum G, Rapoport SI, Chandrasekaran K.. 2005. Regulation of mitochondrial gene expression by energy demand in neural cells. J Neurochem. 93(4):850–860. [DOI] [PubMed] [Google Scholar]
- Meiklejohn CD, Montooth KL, Rand DM.. 2007. Positive and negative selection on the mitochondrial genome. Trends Genet. 23(6):259–263. [DOI] [PubMed] [Google Scholar]
- Meyer M, Kircher M. 2010. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb Protoc. 2010(6):pdb.prot5448. [DOI] [PubMed] [Google Scholar]
- Miller RA, et al. 2005. Methionine-deficient diet extends mouse lifespan, slows immune and lens aging, alters glucose, T4, IGF-I and insulin levels, and increases hepatocyte MIF levels and stress resistance. Aging Cell 4(3):119–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min XJ, Hickey DA.. 2008. An evolutionary footprint of age-related natural selection in mitochondrial DNA. J Mol Evol. 67(4):412–417. [DOI] [PubMed] [Google Scholar]
- Morton D, Janning JT.. 1982. Iron balance in the common vampire bat Desmodus rotundus. Comp Biochem Physiol A Comp Physiol. 73(3):421–425. [DOI] [PubMed] [Google Scholar]
- Morton D, Wimsat WA.. 1980. Distribution of iron in the gastrointestinal tract of the common vampire bat: evidence for macrophage-linked iron clearance. Anat Rec. 198(2):183–192. [DOI] [PubMed] [Google Scholar]
- Müller HE, Pinus M, Schmidt U.. 1980. Aeromonas hydrophila as a normal intestinal bacterium of the vampire bat Desmodus rotundus. Zentralbl Veterinarmed B 27:419–424. [PubMed] [Google Scholar]
- Munshi-South J, Wilkinson GS.. 2010. Bats and birds: exceptional longevity despite high metabolic rates. Ageing Res Rev. 9(1):12–19. [DOI] [PubMed] [Google Scholar]
- Myhrvold NP, et al. 2015. An amniote life-history database to perform comparative analyses with birds, mammals, and reptiles. Ecological Archives. E096–269. [Google Scholar]
- Nabholz B, Ellegren H, Wolf JBW.. 2013. High levels of gene expression explain the strong evolutionary constraint of mitochondrial protein-coding genes. Mol Biol Evol. 30(2):272–284. [DOI] [PubMed] [Google Scholar]
- Nabholz B, Uwimana N, Lartillot N.. 2013. Reconstructing the phylogenetic history of long-term effective population size and life-history traits using patterns of amino acid replacement in mitochondrial genomes of mammals and birds. Genome Biol Evol. 5(7):1273–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DL, Cox MM.. 2013. Leningher principles of biochemistry. New York: W.H. Freeman and Company. [Google Scholar]
- Page MM, Robb EL, Salway KD, Stuart JA.. 2010. Mitochondrial redox metabolism: aging, longevity and dietary effects. Mech Ageing Dev. 131(4):242–252. [DOI] [PubMed] [Google Scholar]
- Popadin KY, Nikolaev S, Junier T, Baranova M, Antonarakis SE.. 2013. Purifying selection in mammalian mitochondrial protein-coding genes is highly effective and congruent with evolution of nuclear genes. Mol Biol Evol. 30(2):347–355. [DOI] [PubMed] [Google Scholar]
- Ranwez V, Harispe S, Delsuc F, Douzery EJP.. 2011. MACSE: multiple alignment of coding sequences accounting for frameshifts and stop codons. PLoS One 6(9):e22594.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes A, Gissi C, Pesole G, Saccone C.. 1998. Asymmetrical directional mutation pressure in the mitochondrial genome of mammals. Mol Biol Evol. 15(8):957–966. [DOI] [PubMed] [Google Scholar]
- Ribeiro JMC. 2000. Blood-feeding in mosquitoes: probing time and salivary gland anti-haemostatic activities in representatives of three genera (Aedes, Anopheles, Culex). Med Vet Entomol. 14(2):142–148. [DOI] [PubMed] [Google Scholar]
- Robin ED, Wong R.. 1988. Mitochondrial DNA molecules and virtual number of mitochondria per cell in mammalian cells. J Cell Phys. 136(3):507–513. [DOI] [PubMed] [Google Scholar]
- Rojas D, Warsi OM, Dávalos LM.. 2016. Bats (Chiroptera: noctilionoidea) challenge a recent origin of extant neotropical diversity. Syst Biol. 65(3):432–448. [DOI] [PubMed] [Google Scholar]
- Romiguier J, et al. 2012. Fast and robust characterization of time-heterogeneous sequence evolutionary processes using substitution mapping. PLoS One 7(3):e33852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safiulina D, Kaasik A.. 2013. Energetic and dynamic: how mitochondria meet neuronal energy demands. PLoS Biol. 11(12):e1001755.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Román I, et al. 2011. Forty percent methionine restriction lowers DNA methylation, complex I ROS generation, and oxidative damage to mtDNA and mitochondrial proteins in rat heart. J Bioenerg Biomembr. 43(6):699–708. [DOI] [PubMed] [Google Scholar]
- Sanz A, et al. 2006. Methionine restriction decreases mitochondrial oxygen radical generation and leak as well as oxidative damage to mitochondrial DNA and proteins. FASEB J. 20(8):1064–1073. [DOI] [PubMed] [Google Scholar]
- Schluter D, Grant PR.. 1984. Ecological correlates of morphological evolution in a Darwin’s finch, Geospiza difficilis. Evolution 38(4):856–869. [DOI] [PubMed] [Google Scholar]
- Schondube JE, Herrera LG, Martínez del Rio C.. 2001. Diet and the evolution of digestion and renal function in phyllostomid bats. Zoology 104(1):59–73. [DOI] [PubMed] [Google Scholar]
- Shao R, Campbell NJH, Schmidt ER, Barker SC.. 2001. Increased rate of gene rearrangement in the mitochondrial genomes of three orders of hemipteroid insects. Mol Biol Evol. 18(9):1828–1832. [DOI] [PubMed] [Google Scholar]
- Shao R, Kirkness EF, Barker SC.. 2009. The single mitochondrial chromosome typical of animals has evolved into 18 minichromosomes in the human body louse, Pediculus humanus. Genome Res. 19(5):904–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y-Y, et al. 2010. Adaptive evolution of energy metabolism genes and the origin of flight in bats. Proc Natl Acad Sci USA. 107(19):8666–8671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer MA. 2002. Vampire bat, shrew, and bear: comparative physiology and chronic renal failure. Am J Physiol Regul Integr Comp Physiol. 282(6):R1583–R1592. [DOI] [PubMed] [Google Scholar]
- Singer MA, Hickey DA.. 2000. Nucleotide bias causes a genomewide bias in the amino acid composition of proteins. Mol Biol Evol. 17(11):1581–1588. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22(21):2688–2690. [DOI] [PubMed] [Google Scholar]
- Stiles FG. 1995. Behavioral, ecological and morphological correlates of foraging for arthropods by the hummingbirds of a tropical wet forest. Condor 97(4):853–878. [Google Scholar]
- Subramanian S. 2013. Significance of population size on the fixation of nonsynonymous mutations in genes under varying levels of selection pressure. Genetics 193(3):995–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takken W, Verhulst NO.. 2013. Host preferences of blood-feeding mosquitoes. Annu Rev Entomol. 58:433–453. [DOI] [PubMed] [Google Scholar]
- Tamura K, et al. 2011. MEGA5: molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28(10):2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetlock A, Yost CK, Stravrinides J, Manzon RG.. 2012. Changes in the gut microbiome of the sea lamprey during metamorphosis. Appl Environ Microbiol. 78(21):7638–7644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilak M, et al. 2015. A cost-effective straightforward protocol for shotgun Illumina libraries designed to assemble complete mitogenomes from non-model species. Conserv Genet Res. 7(1):37–40. [Google Scholar]
- Tomasco IH, Lessa EP.. 2011. The evolution of mitochondrial genomes in subterranean caviomorph rodents: adaptation against a background of purifying selection. Mol Phylogenet Evol. 61(1):64–70. [DOI] [PubMed] [Google Scholar]
- Tschapka M, Wilkinson GS.. 1999. Free-ranging vampire bats (Desmodus rotundus, Phyllostomidae) survive 15 years in the wild. Mammal Biol. 64:239–240. [Google Scholar]
- Veltri KL, Espiritu M, Singh G.. 1990. Distinct genomic copy number in mitochondria of different mammalian organs. J Cell Phys. 143(1):160–164. [DOI] [PubMed] [Google Scholar]
- Weber CC, Nabholz B, Romiguier J, Ellegren H.. 2014. Kr/Kc but not dN/dS correlates positively with body mass in birds, raising implications for inferring lineage-specific selection. Genome Biol. 15(12):542.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks P. 2000. Red-billed oxpeckers: vampires or tickbirds? Behav Ecol. 11(2):154–160. [Google Scholar]
- Weigert A, et al. 2016. Evolution of mitochondrial gene order in Annelida. Mol Phylogenet Evol. 94(Pt A):196–206. [DOI] [PubMed] [Google Scholar]
- Weigl R. 2005. Longevity of mammals in captivity; from the living collections of the world. Stuttgart: Schweizerbart’sche Verlagsbuchhandlung. [Google Scholar]
- Wetterer AL, Rockman MV, Simmons NB.. 2000. Phylogeny of phyllostomid bats (Mammalia: chiroptera): data from diverse morphological systems, sex chromosomes, and restriction sites. Bull Am Mus Nat Hist. 248:1–200. [Google Scholar]
- Wilson EO. 2003. The encyclopedia of life. Trends Ecol Evolut. 18(2):77–80. [Google Scholar]
- Whiten SR, Eggleston H, Adelman ZN. 2018. Ironing out the details: exploring the role of iron and heme in blood-sucking arthropods. Front. Physiol. 8:1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolley S, Johnson J, Smith MJ, Crandall KA, McClellan DA.. 2003. TreeSAAP: selection on amino acid properties using phylogenetic trees. Bioinformatics 19(5):671–672. [DOI] [PubMed] [Google Scholar]
- Xu W, Jameson D, Tang B, Higgs PG.. 2006. The relationship between the rate of molecular evolution and the rate of genome rearrangement in animal mitochondrial genomes. J Mol Evol. 63(3):375–392. [DOI] [PubMed] [Google Scholar]
- Yang Z. 1998. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol Biol Evol. 15(5):568–873. [DOI] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24(8):1586–1591. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R.. 1998. Synonymous and nonsynonymous rate variation in nuclear genes of mammals. J Mol Evol. 46(4):409–418. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R.. 2002. Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol Biol Evol. 19(6):908–917. [DOI] [PubMed] [Google Scholar]
- Zaspel JM, et al. 2012. A molecular phylogenetic analysis of the vampire moths and their fruit-piercing relatives (Lepidoptera: erebidae: calpinae). Mol Phylogenet Evol. 65(2):786–791. [DOI] [PubMed] [Google Scholar]
- Zepeda Mendoza ML, et al. 2018. Hologenomic adaptations underlying the evolution of sanguivory in the common vampire bat. Nature Ecol Evol. 2(4):659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Nielsen R, Yang Z.. 2005. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol. 22(12):2472–2479. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.