Abstract
Huntington’s disease (HD) is a fatal neurodegenerative disorder caused by a CAG trinucleotide repeat expansion in the huntingtin gene. Previously, we showed strong huntingtin reduction and prevention of neuronal dysfunction in HD rodents using an engineered microRNA targeting human huntingtin, delivered via adeno-associated virus (AAV) serotype 5 vector with a transgene encoding an engineered miRNA against HTT mRNA (AAV5-miHTT). One of the challenges of rodents as a model of neurodegenerative diseases is their relatively small brain, making successful translation to the HD patient difficult. This is particularly relevant for gene therapy approaches, where distribution achieved upon local administration into the parenchyma is likely dependent on brain size and structure. Here, we aimed to demonstrate the translation of huntingtin-lowering gene therapy to a large-animal brain. We investigated the feasibility, efficacy, and tolerability of one-time intracranial administration of AAV5-miHTT in the transgenic HD (tgHD) minipig model. We detected widespread dose-dependent distribution of AAV5-miHTT throughout the tgHD minipig brain that correlated with the engineered microRNA expression. Both human mutant huntingtin mRNA and protein were significantly reduced in all brain regions transduced by AAV5-miHTT. The combination of widespread vector distribution and extensive huntingtin lowering observed with AAV5-miHTT supports the translation of a huntingtin-lowering gene therapy for HD from preclinical studies into the clinic.
Keywords: Huntington disease, transgenic minipig, microRNA, gene silencing, AAV
AAV5-miHTT gene therapy demonstrates broad distribution and strong human mutant huntingtin lowering in the brain of a Huntington’s disease minipig model. Evers and colleagues show that one-time intracranial administration of a microRNA-based gene therapy was feasible, well tolerated, and resulted in dose-dependent reduction of toxic human huntingtin protein in the transgenic minipig model of Huntington’s disease. This is the first demonstration of widespread human mutant huntingtin lowering in the large-animal model.
Introduction
Huntington’s disease (HD) is an autosomal dominant neurodegenerative disorder caused by a CAG trinucleotide repeat expansion in the first exon of the huntingtin (HTT) gene. Neurodegeneration starts in the forebrain and specifically affects GABAergic medium spiny neurons of the striatal complex, the caudate nucleus and putamen, thereby disrupting cortico-striatal pathways in the striatum.1 Disruption of these cortico-striatal pathways in HD leads to impairment of cognition, motor function, and behavior.2, 3 HD has a monogenic cause and the mutated HTT gene has full penetrance, such that carriers with alleles containing 40 or more CAG repeats will inevitably develop HD symptoms. The CAG expansion in the HTT gene results in a prolonged polyglutamine repeat in the huntingtin protein with toxic gain-of-function affecting numerous cellular processes.4
Because the culprit of the disease is considered to be the expanded polyglutamine-containing huntingtin protein, strategies to lower the mutant huntingtin protein can potentially modify the progression of the disease. Currently, several strategies are under development to lower huntingtin protein synthesis, and preclinical studies in HD rodent models have demonstrated that lowering mutant huntingtin protein reduces downstream deleterious effects.5, 6, 7 Most approaches of huntingtin lowering aim to decrease translation of huntingtin exploiting the endogenous RNAi mechanism by using synthetic small interfering RNA (siRNA) or by using modified single-stranded antisense oligonucleotides.7, 8, 9, 10, 11, 12, 13, 14 Both siRNAs and antisense oligonucleotides need to be delivered by continuous infusion or repetitive intrathecal injections into the cerebrospinal fluid (CSF), and their therapeutic effect is thought to be most potent in the brain areas adjacent to the ventricular system.7, 15 Based on this rationale of huntingtin lowering as a disease-modifying therapy for HD, a phase I clinical trial with repeated injections of antisense oligonucleotides in early manifest HD patients has been initiated recently.16
Next to repetitive infusion of synthetic oligonucleotides, RNAi-based gene therapy approaches using short hairpin RNA (shRNA) and microRNA (miRNA) have been explored as well. RNAi-based gene therapy comprises a single administration of an adeno-associated virus (AAV) or lentiviral (LV) vector, resulting in continuous expression of shRNA or artificial miRNA precursors and subsequent long-lasting huntingtin lowering.5, 6, 12, 13, 17, 18, 19, 20 Intracranial parenchymal injections of artificial miRNA delivered by AAV vectors resulted in huntingtin reduction in HD animal models without signs of toxicity.21, 22, 23, 24
We have previously reported strong huntingtin protein reduction in humanized HD mice upon intrastriatal injection of an AAV serotype 5 vector with a transgene encoding an engineered miRNA against HTT mRNA (AAV5-miHTT).25, 26 We demonstrated that AAV5-miHTT treatment almost completely prevented mutant huntingtin aggregate formation and ultimately resulted in suppression of neuronal dysfunction in an acute LV rat model of HD.21
Translating preclinical studies in HD animal models to the clinic involves assessing the therapeutic window by evaluating distribution, efficacy, and tolerability. Although in many cases proof-of-concept in HD rodent models has been shown, thus far none of the proposed disease-modifying treatments could be translated fully to the clinic.27, 28 One of the challenges of rodents as a model of neurodegenerative diseases is their relatively small brain, making successful translation to the HD patient difficult, particularly for gene therapy approaches, where the achieved distribution upon local injection in the parenchyma is likely dependent on brain size and structure. To overcome this issue, great effort has been put in the establishment of larger animal models of HD, such as sheep, monkey, and minipig models.29, 30, 31
In the current study, we aimed to demonstrate the translation of a huntingtin-lowering gene therapy approach to a large-animal brain. We investigated the feasibility, efficacy, and tolerability of AAV5-miHTT administration in a transgenic HD (tgHD) minipig model that in addition to porcine huntingtin, ubiquitously expresses a 548-amino acid N-terminal human huntingtin fragment containing 124 glutamines.30 Here, we describe widespread vector distribution in the minipig brain, extensive human mutant huntingtin lowering, long-term expression, and tolerability of AAV5-miHTT, supporting the translation of a huntingtin-lowering gene therapy for HD from preclinical studies into the clinic.
Results
Combined Striatal and Thalamic Administration of AAV5 Results in Widespread Transduction of Minipig Brain
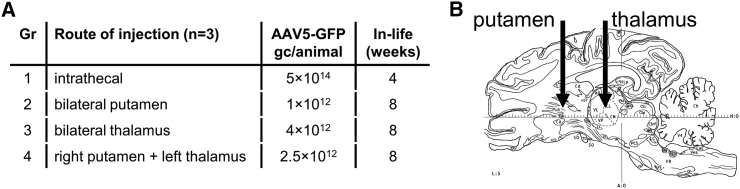
The primary pathology in HD is degeneration of the striatal complex: the caudate nucleus and putamen.1 Therefore, the core target brain structure for an AAV-based huntingtin-lowering gene therapy for HD, to prevent neuronal dysfunction caused by mutant huntingtin, is the striatum. To determine the delivery route needed for transduction of the affected putamen and caudate nucleus in a large-animal brain, a study with AAV5 vectors encoding GFP was conducted in healthy minipigs. Four routes of injection of AAV5-GFP were explored (Figure 1A): (1) intrathecal and parenchymal convection-enhanced delivery (CED), (2) bilateral into the putamen, (3) bilaterally into the thalamus, or (4) in the right putamen and the left thalamus combined (Figure 1B).
Figure 1.
AAV5-GFP Distribution Study in Healthy Minipig Brain
(A) Experimental outline. n = 3 per group. (B) Schematic representation (adapted from Félix et al.72) of intracranial injection routes into the putamen and thalamus.
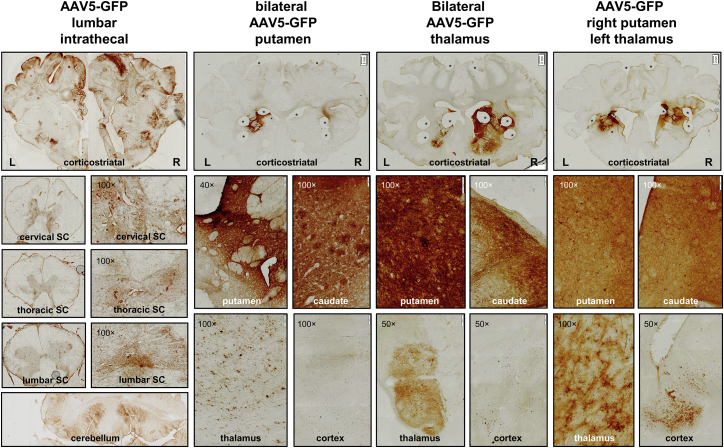
To visualize AAV5 distribution in the brain, GFP immunohistochemical analysis was performed on brain slices (Figure 2). Based on our previous experience in non-human primates, we have observed that the GFP transgene itself can provoke an immune response when injected into the CSF. Previously, it has been shown that steady-state expression levels of an AAV5 encapsulated transgene, as well as a CAG promoter transgene, are maintained from 4 weeks onward.32, 33 Therefore, in light of the animal well-being, it was decided to sacrifice the intrathecal injected group at 4 weeks. After intrathecal administration, GFP-positive cells were found in the cerebral cortex, cerebellum, and spinal cord. Within the spinal cord, the highest transduction was found in the lumbar region, the site of infusion. Very limited numbers of GFP-positive cells were found in the striatum and other deep brain structures. Following injection into the putamen, local GFP expression was observed in the putamen and caudate nucleus, and GFP-positive cells in the cerebral cortex. Thalamic injections resulted in transduction of cells in the thalamus, as well as the striatum and cerebral cortex. GFP expression in the putamen, caudate nucleus, thalamus, and cerebral cortex was also seen after the combined AAV5-GFP delivery into the right putamen and left thalamus.
Figure 2.
AAV5-GFP Distribution after Intrathecal Injection of 5 × 1014 gc or Intracranial Injection into the Bilateral Putamen (1 × 1012 gc), Bilateral Thalamus (4 × 1012 gc), or Right Putamen and Left Thalamus Combined (2.5 × 1012 gc)
Top panels: GFP immunohistochemical staining of cortico-striatal brain slices. Upon intrathecal infusion, positive transduction was found in the cerebral cortex; cervical, thoracic, or lumbar spinal cord (SC); and cerebellum. Bilateral injection into the putamen resulted in strong putamen and caudate, and thalamic transduction, as well as GFP-positive neurons in the cortex. Bilateral injection in the thalamus caused thalamic transduction as well as GFP-positive neurons in striatal and cortical regions. Infusion into the right putamen and left thalamus showed a similar transduction pattern. Magnification factor is between ×1 and ×1.25. Asterisks indicate biopsies taken for biomolecular analyses. Scale bars, 2 mm.
To check for biodistribution of vector DNA outside of the CNS, genomic DNA was isolated from the liver, kidney, and adrenal glands (Figure S1). Intrathecal injection of AAV5-GFP resulted in transduction in all tested peripheral organs, with up to 6.6 × 104 genome copies (gc)/μg DNA in the liver. In contrast, intracranial parenchymal delivery of AAV5-GFP did not result in the presence of vector DNA above background in any of the tested organs.
Based on the GFP immunohistochemical analysis, we concluded that intrathecal injection of AAV5 in minipigs leads to cerebral cortical transduction, with limited transduction of cells in the striatum and other deep brain structures. Combined striatal and thalamic administration resulted in a robust local transduction of the striatum, as well as more distal brain structures such as the cerebral cortex, suggesting that for an AAV-based huntingtin-lowering gene therapy for HD the delivery route of AAV5 is a combined striatal and thalamic injection.
Widespread AAV5-GFP Transduction in tgHD Minipig Brain after Combined Striatal and Thalamic Administration
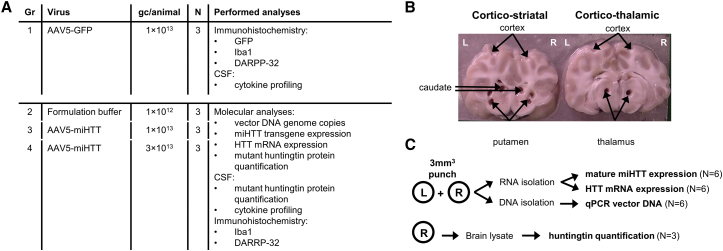
After assessing the route of administration in healthy minipigs, we used tgHD minipigs to study the efficacy of AAV5-miHTT treatment in a large-animal model of the disease.30 In addition to AAV5-miHTT, we also tested AAV5-GFP to compare distribution in tgHD with previously assessed distribution in the healthy minipig brain. To achieve a stronger and more widespread distribution pattern, we increased the dose to 1 × 1013 gc AAV5-GFP per tgHD minipig. In total, 12 tgHD minipigs, three animals per group, were injected bilateral into the putamen and thalamus with a fixed volume to obtain a dose of 1 × 1013 gc AAV5-GFP or 1 × 1013 or 3 × 1013 total gc AAV5-miHTT per animal. As a sham control, another group of animals was injected in the same target areas with the same volume of formulation buffer. For overview of the groups, see Figure 3A and Table S1.
Figure 3.
TgHD Minipig AAV5-miHTT Efficacy Study
(A) Experimental outline and performed bioanalytical procedures per treatment group. Brain slices of group 1 were used for immunohistochemical analysis. (B) From groups 2–4, corticostriatal and corticothalamic slices were obtained and tissue punches were taken from putamen, caudate nucleus, thalamus, and cortex. One animal died 2 days post-surgery because of complications related to the surgical procedure; thus, only two animals could be subsequently analyzed of the 1 × 1013 gc AAV5-miHTT group. (C) From left (L) and right (R) hemisphere punches DNA and total RNA were isolated for vector DNA, mature miHTT expression, and HTT mRNA quantification. From right hemisphere punches, lysate was used for mutant huntingtin protein quantification.
The surgery was generally well tolerated, with no clinical symptoms and normal body weight gain over time (Figure S2). Routine hematology was measured, and no significant differences were measured at the pre- and post-dosing time points (Table S2). One animal treated with 1 × 1013 gc AAV5-miHTT died 2 days post-surgery due to complications related to the surgical procedure. For this reason, in the 1 × 1013 gc/brain of AAV5-miHTT group, only two animals could be subsequently analyzed.
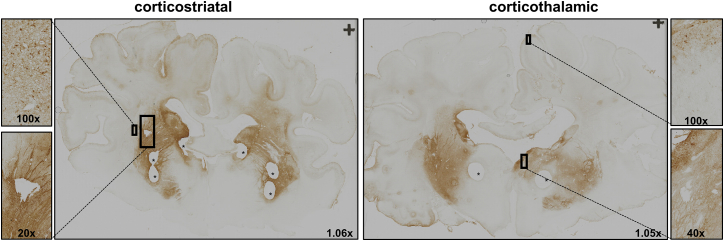
Three months after injections, immunohistochemical analysis on cortico-striatal and cortico-thalamic brain slices showed GFP expression in the striatum, thalamus, and cerebral cortex (Figure 4). The observed transduction pattern of the putamen, caudate, and thalamus was comparable with the previous experiment in healthy minipigs. Because the injected dose was four times higher as compared with the first experiment, the local transduction of the putamen and caudate nucleus, and in particular the thalamus, was much stronger, and more GFP-positive cells were observed in the cerebral cortex. The primary motor cortex is known to degenerate relatively early in the disease process of HD.1 Higher magnification showed positive transduction of neurons in the motor cortex and part of the parietal cortex after bilateral injections of AAV5-GFP into the putamen and thalamus of tgHD minipigs.
Figure 4.
GFP Immunohistochemistry of Cortico-Striatal and Cortico-Thalamic Section after Combined Striatal and Thalamic Injection of 1 × 1013 gc AAV5-GFP in tgHD Minipigs
Bilateral injection into the putamen and thalamus resulted in strong putamen and caudate transduction, as well as thalamic and cortical transduction. Magnification factors are depicted in the panels. Asterisks represent punches taken for biomolecular analyses.
Dose-Dependent AAV5-miHTT Transduction and miRNA Expression in the tgHD Minipig Brain
After confirmation of a widespread transduction with GFP-positive signal in the striatum and more distant in the cerebral cortex of tgHD minipigs, we studied the transduction efficacy and target engagement of AAV5-miHTT. Punches taken from brain tissue around the injection sites, as well as more distal locations, were used for biomolecular analyses (Figure 3B). Samples from both hemispheres were used for: (1) genomic DNA isolation to determine vector DNA and (2) total RNA isolation to assess transgene expression (mature miHTT) and HTT mRNA lowering. Punches from the right hemisphere were used for protein analysis to assess huntingtin protein lowering (Figure 3C).
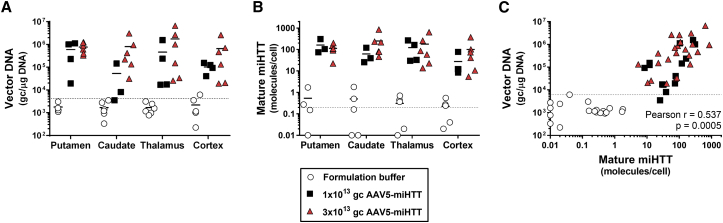
In all four brain areas, a positive transduction of the vector was observed in a dose-dependent manner (Figure 5A): in the striatum, with 6.0 × 105 or 7.8 × 105 gc/μg DNA in the putamen and 5.2 × 104 or 8.1 × 105 gc/μg in the caudate nucleus after a dose of 1 × 1013 and 3 × 1013 gc/brain of AAV5-miHTT, respectively; in the thalamus, we measured 4.6 × 105 or 1.7 × 106 gc/μg DNA and in the cortex 1.0 × 105 or 6.6 × 105 gc/μg after dosing of 1 × 1013 and 3 × 1013 gc/brain of AAV5-miHTT, respectively. Although the acquired genome copies vary among both hemispheres and animals, on average we observed a dose-dependent transduction of AAV5-miHTT, with highest transduction after the highest injected dose.
Figure 5.
AAV5-miHTT Distribution and miHTT Expression 3 Months after Putaminal and Thalamic Injection in tgHD Minipig Brain: Putamen, Caudate Nucleus, Thalamus, and Cortex
One punch was taken per hemisphere per brain structure. Punches from left and right are displayed separately. (A) Vector DNA copies (gc/μg DNA) in the tgHD minipig brain. (B) Mature miHTT expression was determined by custom TaqMan RT-qPCR; values are represented as molecules per cell. (C) Correlation analysis graph plotting vector DNA and miHTT expression from matching brain region. Data from dissected regions from AAV5-miHTT-treated animals were evaluated with non-linear regression log-log line with ordinary fit and Pearson correlation. Formulation buffer samples were excluded from the correlation and fit. Dotted lines represent lower limit of quantification.
As a measure for transgene expression, total RNA was isolated and mature guide miRNA miHTT quantified. In the striatal punches of one hemisphere of a 1 × 1013 gc/brain of AAV5-miHTT-treated animal, the isolated RNA was of low quality and thus could not be further analyzed. Mature miHTT molecules were detected in all tested brain areas from animals treated with AAV5-miHTT (Figure 5B). Levels of miHTT in all dissected regions from tgHD minipigs that were injected with formulation buffer were below background. In the striatum, the miHTT molecules per cell were 161.4 (SD ±126.9) or 113.2 SD ±60.4) in the putamen and 61.2 (SD ±51.0) or 249.8 (SD ±30.4) in the caudate after 1 × 1013 or 3 × 1013 gc/brain of AAV5-miHTT. High levels of miHTT were also measured in the thalamus at 122.5 (SD ±113.8) or 179.1 (SD ±242.6) molecules per cell after a dose of, respectively 1 × 1013 or 3 × 1013 gc/brain of AAV5-miHTT. In line with the above-described positive transduction of neurons in the motor cortex upon AAV5-GFP injection, we also detected miHTT in the cortex from 27.3 (SD ±33.1) miHTT molecules per cell in the 1 × 1013 gc/brain of AAV5-miHTT injected group up to 100.3 (SD ±138.0) miHTT molecules per cell in the 3 × 1013 gc/brain of AAV5-miHTT injected group.
To compare the results obtained from both DNA and RNA isolation, we performed correlation analysis of vector DNA and mature miHTT levels from the tissue punch. Mature miHTT expression significantly correlated with AAV5-miHTT vector DNA measured in the same brain punch with Pearson r = 0.537, p = 0.0005 (Figure 5C). Accordingly, animals that showed the strongest vector DNA transduction displayed the highest miHTT expression in the same brain region, verifying the mechanism-of-action in these large tgHD minipig brains from vector transduction, transgene expression, to processing of the mature miHTT. Because the group sizes were relatively small, we also investigated transduction efficacy in healthy minipig brain (Figure S3). In accordance with the tgHD minipigs, a clear transduction pattern was observed in all four brain areas with highest levels of miHTT molecules measured in the punches that showed strongest vector DNA transduction (Pearson r = 0.9373, p < 0.0001; Figure S3C).
Specific Human Mutant HTT mRNA Reduction in tgHD Minipig Brain
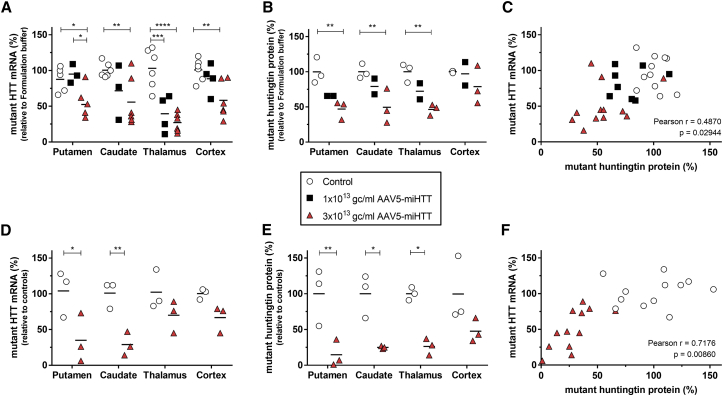
After establishing the mechanism-of-action, we focused on the efficacy of AAV5-miHTT to lower human mutant huntingtin mRNA and protein levels 3 months after one-time treatment. A trend of human mutant HTT mRNA lowering was observed after 1 × 1013 gc/brain of AAV5-miHTT in the putamen, caudate nucleus, and cortex, although it did not reach significance because of low number and variation between punches from different animals (Figure 6A). In the thalamus, a clear significant reduction of human mutant HTT mRNA of on average 41.5% (SD ±26.6%; p = 0.0002) was measured.
Figure 6.
AAV5-miHTT Treatment Demonstrates a Dose-Dependent Human Mutant Huntingtin Target Engagement in tgHD Minipig Brains
(A) Human HTT-specific RT-qPCR of mutant HTT mRNA expression relative to an average of formulation buffer-treated animals in the tgHD minipig brain: putamen, caudate nucleus, thalamus, and cortex. Primer-probe against GAPDH used as reference gene. (B) SMC quantification of soluble human mutant huntingtin protein levels in brain punches from the right hemisphere. (C) Pearson correlation of mutant HTT mRNA and mutant huntingtin protein levels showing a positive correlation between mutant HTT mRNA with mutant huntingtin protein levels in punches taken from the right hemisphere. Formulation buffer-injected punches were not taken along for correlation. (D) Human mutant HTT mRNA expression in the left hemisphere injected with AAV5-miHTT in the striatum. Values relative to an average of same brain punch from control tgHD animals (n = 3). (E) SMC quantification of soluble human mutant huntingtin protein levels in brain punches from the left hemisphere injected with AAV5-miHTT in the striatum. (F) Pearson correlation of mutant HTT mRNA and mutant huntingtin protein levels showing a positive correlation between mutant HTT mRNA with mutant huntingtin protein levels in punches injected with 3 × 1013 gc/brain of AAV5-miHTT in the striatum. Data were evaluated using two-way ANOVA with Tukey’s multiple comparison test: *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
The high dose of 3 × 1013 gc/brain of AAV5-miHTT resulted in a significant reduction of human mutant HTT mRNA expression in all investigated brain regions. The human mutant HTT mRNA was reduced in the putamen by on average 47.5% (SD ±21.8%; p = 0.0387) and 44.2% (SD ±34.8%; p = 0.0041) in the caudate nucleus (Figure 6A). The strongest average reduction in human mutant HTT mRNA was measured in the thalamus, with an average of 72.8% (SD ±13.4%; p < 0.0001). Consistent with the vector DNA and miHTT expression in the cortex, we also detected a significant reduction of 42.7% (SD ±25.4%; p = 0.0066) more distal in the cortex.
To demonstrate the specificity of AAV5-miHTT for human HTT, we analyzed the porcine HTT mRNA expression levels in the transduced areas. The endogenous porcine HTT mRNA, which has a two-nucleotide mismatch with the miHTT target region, was not affected by AAV5-miHTT treatment (Figure S4), demonstrating the specificity of AAV5-miHTT for the human HTT gene.
In sum, we detected a dose-dependent human mutant HTT mRNA lowering with reduction in all brain regions studied, after the highest dose of AAV5-miHTT.
Dose-Dependent Human Mutant Huntingtin Protein Reduction in tgHD Minipig Brain
Next, we investigated soluble human mutant huntingtin protein levels in the brain regions of the right hemisphere using an ultrasensitive single-molecule counting (SMC) immunoassay.34 The SMC immunoassay was specific for human mutant huntingtin because the porcine huntingtin was not detected in healthy control minipigs (data not shown). Upon treatment with AAV5-miHTT, a dose-dependent reduction in soluble human mutant huntingtin protein levels was observed (Figure 6B). The high dose of 3 × 1013 gc/brain of AAV5-miHTT showed strong target engagement with a significant human mutant huntingtin reduction in the putamen of 53.0% (SD ±13.2%; p = 0.0024), in the caudate nucleus of 50.5% (SD ±24.6%; p = 0.0036), and in the thalamus of 53.5% (SD ±7.8%, p = 0.0022). Two out of three tgHD minipigs also showed a mutant huntingtin reduction in the cortex, with an average 21.1% (SD ±27.12%; p = 0.2860) human mutant huntingtin protein lowering. In the tissue punches of the 1 × 1013 gc/brain of AAV5-miHTT animals, we also detected a trend toward human mutant huntingtin protein lowering, although this did not reach significance because of the small group size.
In the biopsies from the right hemisphere the corresponding mutant HTT mRNA and mutant huntingtin protein levels per punch were plotted (Figure 6C). A trend between mutant HTT mRNA with mutant huntingtin protein level was observed (Pearson r = 0.4870; p = 0.02944), with highest lowering of both mutant HTT mRNA and huntingtin protein observed in the punches obtained from the 3 × 1013 gc/brain of AAV5-miHTT-treated tgHD minipigs.
Because the group sizes were relatively small, we injected an additional group of three animals with 3 × 1013 gc/mL AAV5-miHTT in the striatum only. Six months after injection, the animals were sacrificed and punches taken as described previously (Figure 3B). Samples from left hemispheres were used to assess HTT mRNA and protein lowering. Human mutant HTT mRNA lowering was measured in the putamen (69.0%; p = 0.0109), caudate nucleus (72.0%; p = 0.0079), thalamus (33.3%; p = 0.3917), and cortex (33.7%; p = 0.3544) (Figure 6D). In accordance with HTT mRNA levels, a strong mutant huntingtin protein lowering was also observed after 3 × 1013 gc/mL AAV5-miHTT in the putamen (85.3%; p = 0.0042), caudate nucleus (75.0%; p = 0.0116), thalamus (73.7%; p = 0.0132), and cortex (52.0%; p = 0.1042) (Figure 6E). Like the administration into the striatum and thalamus combined, striatal injection resulted only in a robust trend between mutant HTT mRNA with mutant huntingtin protein level (Pearson r = 0.7176; p = 0.00860) in the 3 × 1013 gc/mL AAV5-miHTT-treated tgHD minipigs (Figure 6F).
We detected a strong human mutant HTT mRNA reduction in the transduced brain regions that correlated with a strong human mutant huntingtin protein reduction in the tgHD minipig model. Next to the huntingtin lowering in the transduced regions, we also observed a trend toward cortical reduction in the high-dose-treated tgHD minipigs.
Transient Increase in Mutant Huntingtin after Intracranial Injection in tgHD Minipig CSF
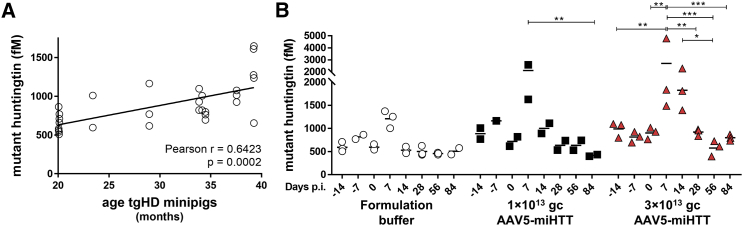
Quantification of disease-associated proteins in CSF of HD patients may be useful for monitoring of disease progression and assessing the effect of therapeutic interventions. One of the best studied biomarkers to track disease progression, and possibly assess huntingtin lowering efficacy, is soluble mutant huntingtin in the CSF. Concomitant with disease progression, mutant huntingtin concentration increases in the CSF of HD patients, suggesting the utility of this measurement as a disease-specific biomarker.34 To assess baseline human mutant huntingtin protein levels in the tgHD minipigs, we measured human mutant huntingtin before treatment with AAV5-miHTT. In pre-dosing CSF samples, we observed high variation in human mutant huntingtin protein levels between animals. Because the tgHD minipigs varied in age (range: 20–39 months), we plotted the age of the animal with its human mutant huntingtin level (Figure 7A). A positive correlation between age of the tgHD minipigs and human mutant huntingtin protein levels was found (Pearson r = 0.6423; p = 0.0002), with highest human mutant huntingtin protein levels in the CSF of the older tgHD minipigs.
Figure 7.
Single Molecule Counting Quantification of Soluble Human Mutant Huntingtin in CSF of tgHD Minipigs before and after AAV5-miHTT Treatment
(A) Correlation plot of age versus human mutant huntingtin levels in measurements at −14, 7, and 0 days pre-treatment. (B) Longitudinal mutant huntingtin levels of individual CSF samples. Data were evaluated using two-way ANOVA with repeated measurements: *p < 0.05; **p < 0.01; ***p < 0.001.
In an effort to assess huntingtin lowering efficacy translationally, we measured human mutant huntingtin protein in longitudinal CSF samples of the tgHD minipigs before and after AAV5-miHTT treatment (Figure 7B). Unexpectedly, we observed a brief increase in mutant huntingtin protein levels 7 days post-injection in CSF in all treatment groups. The high levels of soluble mutant huntingtin at this time point were not caused by blood contamination of the CSF, because hemoglobin levels were not found to be associated with the concentration of mutant huntingtin (Figure S5). The increase of mutant huntingtin most likely was related to the intracranial parenchymal administration procedure, because it was measured in both doses of AAV5-miHTT, as well as the formulation buffer group.
Transient Mild Cytokine Increase after AAV5-miHTT Treatment in tgHD Minipigs
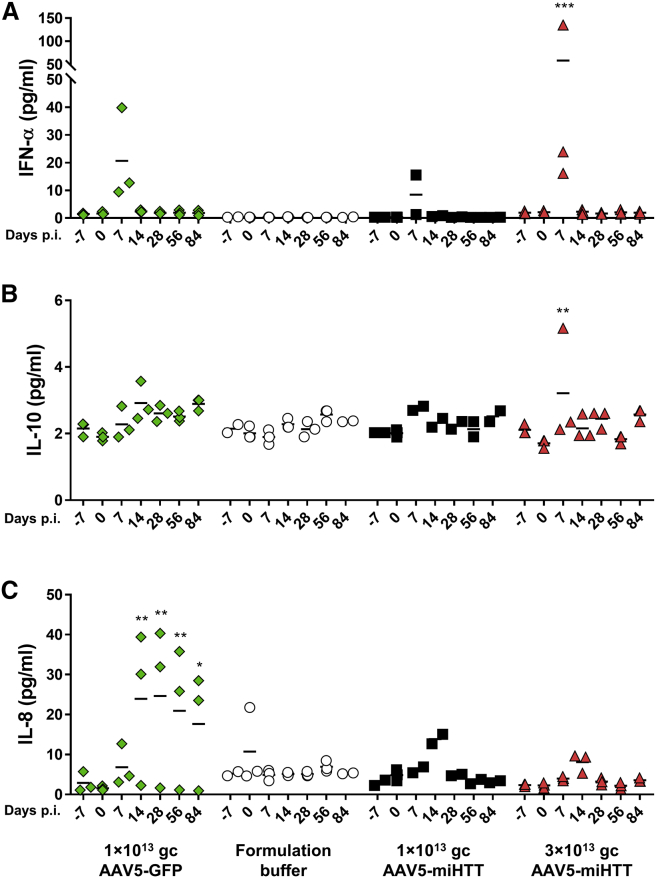
Although the current study was specifically designed to assess the distribution and efficacy, CSF was taken to explore, besides efficacy measures, a putative inflammatory response to the treatment. Pigs have a highly developed immune system, and immune reactions following intraparenchymal injection of AAV5 can be used as potential indicators for human application. Also, tgHD pigs have been shown to have altered cytokine profiles in CSF as compared with healthy pigs, with reduced interferon-α (IFN-α) and interleukin-10 (IL-10) levels, and increased secretion of IL-8 and IL-1β levels.35 To assess a potential immune response to the AAV5-miHTT treatment, cytokine profiling of the CSF from tgHD minipig was performed using a sensitive multiplex assay measuring seven cytokines in parallel.35 Of the seven porcine cytokines, three cytokines were analyzed in detail, being IFN-α, IL-10, and IL-8, whereas the other cytokines showed only background levels, and thus were excluded from the analysis.
Seven days post-injection with AAV5-miHTT, a statistically significant increase of IFN-α from 2.1 (SD ±0.5) to 58.3 pg/mL (SD ±66.4; p = 0.0002) was measured in the CSF of 3 × 1013 gc AAV5-miHTT-treated tgHD minipigs (Figure 8A). Because IFN-α levels were back to baseline on day 14 and the transient reactivity was absent in the formulation buffer-treated tgHD minipigs, it likely represents a response to the AAV administration. No changes were observed for IL-10, except a transient increase in one of the tgHD minipigs treated with 3 × 1013 gc/brain of AAV5-miHTT (Figure 8B). At 14 days post-injection with AAV5-miHTT, a transient IL-8 upregulation was measured, which returned to basal levels at the next time point, 28 days post-treatment (Figure 8C). This brief increase in IL-8 levels in the CSF indicates a mild transient innate response to the expressed transgene or viral capsid. The formulation buffer-treated tgHD minipigs did not show any cytokine elevations, suggesting that the transient response was not related to the surgery.
Figure 8.
Cytokine Multiplexing Luminex in CSF tgHD Animals before and after AAV5-GFP, Formulation Buffer, and AAV5-miHTT Treatment
(A) IFN-α. (B) IL-10. (C) IL-8. Data were evaluated using two-way ANOVA with repeated measurements (*p < 0.05; **p < 0.01; ***p < 0.001) to day 0.
Because the GFP transgene is known to provoke immune responses,36 we also studied tgHD minipigs treated with 1 × 1013 gc/brain of AAV5-GFP. As expected, and in line with a continued immune response to the GFP transgene, IL-8 upregulation was more prominent after AAV5-GFP injection than in any of the other groups and persisted until the termination of the study.
In both AAV5-miHTT- and AAV5-GFP-injected animals, CSF mutant huntingtin protein levels correlated with the IFN-α expression. However, this correlation was mainly driven by increase in both markers at day 7 post-injection, and no significant correlation was observed in the formulation buffer-injected group and in the group with the lower AAV5-miHTT dose. This confirms that increased mutant huntingtin release to CSF is most likely related to tissue injury at the time of intraparenchymal administration, whereas IFN-α levels would be more indicative of a transient immune response to the injected AAV5-miHTT.
Both cytokines and soluble mutant huntingtin levels returned to basal levels at day 14 post-injection, providing further support for the notion of a transient response to the invasive administration procedure.
Intrastriatal Administration of AAV5-miHTT Is Tolerated in tgHD Minipig Brain
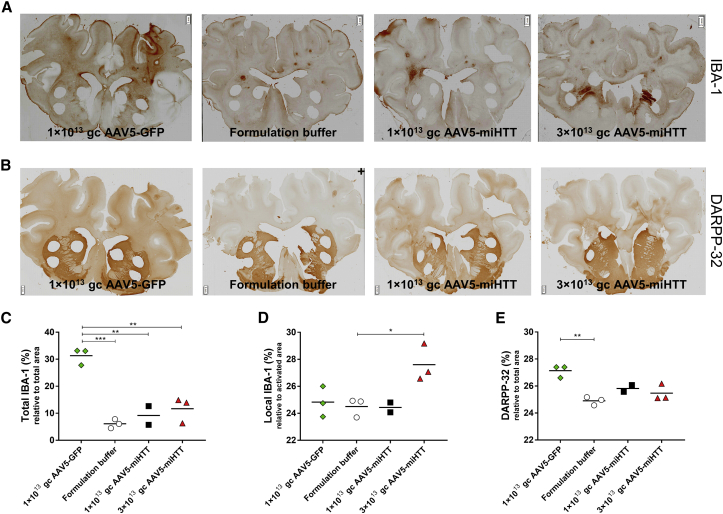
At study termination, brain slices were used for histological examination to investigate a putative response to the surgery and/or AAV5-miHTT treatment. Brain slices were stained with the microglial marker ionized calcium-binding adaptor molecule-1 (IBA-1). In brain slices covering the cortico-striatal area obtained from the formulation buffer-infused animals, very mild microgliosis was observed around the needle track, suggesting no general response to the surgery (Figure 9A). Quantification of IBA-1 staining of cortico-striatal brain slices showed that 1 × 1013 and 3 × 1013 gc/brain of AAV5-miHTT-treated animals did not significantly increase in total IBA-1 expression relative to the whole section area, with levels of, respectively, 9.2% (SD ±4.9%) and 11.7% (SD ±4.7%), as compared with the formulation buffer (6.1%, SD ±1.7%) (Figure 9C). In contrast, and in accordance to the elevated cytokines, 1 × 1013 gc/brain of AAV5-GFP induced a significant increase in IBA-1 expression to 31.3% (SD ±3.1%; p = 0.0002). This broad microglial expression in the AAV-GFP animals probably reflects an innate immune response against the GFP transgene that is linked to the widespread AAV5-GFP distribution pattern. AAV5-miHTT was also distributed and expressed throughout the tgHD minipig brain. Thus, the lack of comprehensive IBA-1 expression in the AAV5-miHTT animals suggests the absence of an immune response to the expressed transgene. Next to total microglial expression, we also quantified local microglial expression around the infusion site. Groups injected with 1 × 1013 gc/brain of AAV5-GFP, the formulation buffer, and 1 × 1013 gc/brain of AAV5-miHTT displayed similar local IBA-1 density, whereas in 3 × 1013 gc/brain of AAV5-miHTT animals a significant 12.6% (p = 0.02) increase in local IBA-1 expression was measured (Figure 9D). Because we did not observe widespread microgliosis, this local response is probably showing a local reaction to the high number of particles around the tip of infusion.
Figure 9.
Histological Examination of tgHD Minipig Cortico-Striatal Brain Slices after Injections of AAV5-GFP, Formulation Buffer, and AAV5-miHTT
(A) Microglial marker IBA1 immunohistochemical staining in representative sections of all groups. (B) Striatal medium-sized spiny neuronal marker DARPP-32 immunohistochemical staining in representative sections of all groups. Magnification factor between ×1 and ×1.25. *Biopsies taken for biomolecular analyses. Scale bars, 2 mm. (C) Quantification of total IBA-1 relative to whole section area. (D) Quantification of local IBA-1 optical density, relative to the activated area. (E) Quantification of DARPP-32 expression relative to manual selected striatal area. Data were evaluated using a one-way ANOVA with Bonferroni’s multiple comparison test: **p < 0.01; ***p < 0.001.
As a marker for neuronal health, dopamine- and cAMP-regulated phosphoprotein, 32-kDa (DARPP-32) staining of GABAergic medium spiny neurons was performed on the cortico-striatal brain slices (Figure 9B). All animals treated with AAV5-miHTT showed consistent DARPP-32 staining, indicating normal striatal functioning (Figure 9E).
Normal striatal functioning in combination with the lack of global microglial expression provide evidence that intracranial injection of AAV5-miHTT is well tolerated in the tgHD minipigs.
Discussion
Here, we aimed to demonstrate the translation of a huntingtin-lowering gene therapy to a large HD animal model brain. Previously, we have shown that intracranial administration of AAV5-miHTT in the striatum of HD rodent models resulted in strong huntingtin lowering and reduction in huntingtin aggregation, with a subsequent prevention of neuronal dysfunction.21, 26 To translate these results acquired in a small rodent brain to a large HD patient brain, great efforts were put in the generation of larger animal models of HD, such as sheep, monkey, and minipig.29, 30, 31 The tgHD minipig and sheep brain weigh approximately 90 and 130 g, respectively, and both have a gross brain structure, blood supply, and immune response similar to humans.37 Because these animals express an N-terminal (tgHD minipig) or full (HD sheep) human HTT transgene, they provide a good model to test target engagement of human sequence-specific therapeutic modalities. The major limitation of both large-animal models is that they have not developed severe neuropathology thus far, and related to the latter point, that a clear phenotype has not yet been established.38 This absence of HD-like symptom manifestation is probably caused by a combination of relatively low mutant HTT transgene expression levels, lifespan up to 20 years, small group sizes, and the fact that behavioral assessments are under development. Nevertheless, these large tgHD animal models are important to close the gap between preclinical research in HD rodents and clinical research in humans.39
We showed successful transduction of the structures mostly affected by HD following administration of an AAV5-based gene therapy. AAV5 has previously been found to distribute via axons and preferentially make use of anterograde transport, whereas retrograde transport could be achieved as well with higher concentration of virus, suggesting the transduction pattern to be dose dependent.40 A layer of complexity is added by the natural disease pathology, with evidence for white matter degeneration as disease progresses, suggested to result in reduced connectivity between the putamen and motor cortex.41 In the current study, we did observe both anterograde (e.g., to caudate) and retrograde (to cortex) viral transport of AAV5-miHTT, suggesting that the connectivity is not altered in the tgHD minipig brain at this age (up to 40 months of age). Although both doses of AAV5-miHTT resulted in a local transduction and subsequently HTT mRNA and huntingtin protein lowering, the highest dose of AAV5-miHTT resulted in vector DNA and miHTT expression in the cortex, suggesting a dose-dependent viral transport.
Next to widespread vector distribution and transgene expression, we demonstrate a successful human mutant HTT mRNA and huntingtin protein lowering in the brain of a large-animal model of HD. Although the sample size in this study was limited, we observed a clear trend toward a dose-dependent reduction of human mutant huntingtin after AAV5-miHTT administration. We achieved HTT mRNA lowering of up to 72.8% and human mutant huntingtin protein up to 85.3% in a large HD animal model brain, similar to knockdown levels achieved in smaller HD rodents.7, 12, 13, 14, 15 For the translatability to the clinic, various studies have been conducted in healthy large animals with gene-lowering therapies for HD.7, 10, 17, 42 Although antisense oligonucleotides have shown to be very promising in pre-clinical HD rodent models, injection into the CSF of species with a larger brain resulted in HTT mRNA target engagement of primarily cortex and spinal cord, and to lesser extent in the deeper brain structures, such as the striatum.7 Another limitation of those studies was that they used non-human primates targeting the monkey huntingtin. Especially for the development of oligonucleotide therapeutics for HD, the use of a full complementary sequence is an advantage because it provides the opportunity to completely examine the mechanism of action and subsequent efficacy. The observed strong human mutant huntingtin protein lowering in the striatum of tgHD minipigs provides additional confidence for future applicability of the AAV5-miHTT huntingtin-lowering gene therapy for HD patients.
Next to the parenchymal mutant huntingtin reduction, we also investigated soluble mutant huntingtin protein levels in the CSF upon AAV5-miHTT treatment. Recently, in vitro and ex vivo evidence suggest that mutant huntingtin is secreted extracellularly.43 Surprisingly, we observed a transient elevation of mutant huntingtin in the CSF shortly after surgery. Because elevated mutant huntingtin protein levels in the CSF have been suggested to be the result of increased cell death,44 and mutant huntingtin levels in the CSF correlate with the neuronal degeneration marker proteins such as tau and neurofilament light chain in HD patients,34 the observed increase in soluble huntingtin in the CSF could have been triggered by local tissue damage due to the administration procedure. In accordance with the huntingtin CSF levels in tgHD minipigs, longitudinal cytokine profiles demonstrated a consistent pattern with a transient mild increase in cytokine levels at 7 and 14 days post AAV5-miHTT injection. Unfortunately, we did not observe a significant reduction in human mutant huntingtin protein in the CSF of tgHD minipigs after treatment with AAV5-miHTT. Although we observed a trend toward huntingtin lowering, the lack of a significant effect is most likely due to the combination of the variability between the age of the animals, the low number of animals, and the transient increase in human mutant huntingtin protein levels after surgery in combination with the restricted in-life of 3 months. To better assess huntingtin lowering in the CSF in the future, more animals should be included in the study, with age matching and longer observation period.
AAV5-miHTT does not discriminate between wild-type and mutant huntingtin and will result in a lower expression of the disease-inducing huntingtin and the wild-type allele. Because of the essential function of huntingtin during development,45, 46, 47, 48 the safety aspect of wild-type huntingtin lowering needs to be carefully evaluated. The tolerated levels of long-term total huntingtin lowering cannot be addressed in the current study, because the porcine HTT mRNA is not lowered by AAV5-miHTT. However, it was demonstrated in non-human primates that 45% reduction of wild-type HTT mRNA in putamen is well tolerated up to 6 months.10, 42, 49 In rodents, non-selective huntingtin reduction up to 50%–75% by antisense oligonucleotides is well tolerated both in HD rats and mice.7 Recently, it was shown that total depletion of huntingtin in the brain of adult mice was well tolerated at all ages, with no motor problems or body weight issues.50 Others showed that total knockdown in the striatum and cerebral cortex in the adult brain was well tolerated up to 14 months after conditional knockout.51 The same study showed that the thalamus and cerebellum are more vulnerable to total huntingtin elimination.51 Likewise, carriers of heterozygous HTT mutations causing partial loss-of-function do not display any symptoms, from which it can be concluded that at least 50% reduction of huntingtin is well tolerated.46, 48 Based on the existing knowledge to date on the huntingtin-lowering tolerability in non-human primates and rodents, as well as identified loss-of-function mutations in humans, an anticipated maximum clinical efficacy rate of 50%–70% total huntingtin lowering in the striatum is anticipated to be well tolerated in adult HD patients.
Immunohistochemical analysis showed no histological pathology in the brain of tgHD minipigs treated with the 1 × 1013 gc/brain dose of AAV5-miHTT. In the high-dose group, we did see microglial reactivity locally around the injection site. tgHD minipigs have previously been suggested to have increased microglial activation30 and altered cytokine profiles as compared with healthy pigs,35 suggesting a primed immune system in tgHD minipigs. This reflects the abnormal immune activation in human HD patients, with mutant huntingtin-related microglial activation in the striatum and subsequently cytokine upregulation in the CSF and peripheral body fluids.52, 53 Although there are only limited reports of histopathological examination after intracranial delivery in neurodegenerative disorders, there is evidence from clinical studies showing mild immune activation after intracranial parenchymal treatments.54 A localized glial response around the needle track was seen after recurrent parenchymal recombinant glial cell line-derived neurotrophic factor (GDNF) injection in Parkinson’s disease.55 Local immune activation was also observed after fetal neural cell transplantation in HD patients56 and transplantation of fibroblasts expressing nerve growth factor (NGF) in Alzheimer’s disease patients.57 In HD patients, it has been shown that implantation of electrodes for chronic stimulation of the globus pallidus is a safe procedure and lacks procedure-related side effects,58 although neuropathological examinations are not available. Likewise, in Parkinson’s disease patients, several clinical trials have been conducted in which putaminal magnetic resonance-guided CED of recombinant proteins and gene therapy was found to be safe and well tolerated.59, 60
Although we describe here a favorable widespread brain distribution after thalamic injection via anterograde transport of AAV5 through brain structures that receive projections from the thalamus, neuropathological examination showed surgery-related lesion in the thalamus in one hemisphere of one tgHD minipig. This single finding in the current study, together with the complexity of the thalamus, a recent study describing that the thalamus is very susceptible to total huntingtin lowering,51 and the fact that the thalamus is only limited affected at later stages of HD,1 makes that the thalamus is not a key structure to treat with AAV5-miHTT in HD patients. Although the thalamus is an attractive structure to infuse from a viral distribution perspective, for clinical application more experiments need to be conducted to assess the tolerability, volume, viral load, trajectory, and the specific thalamic nucleus to inject safely.
The divergence in vector distribution pattern seen between both lateral hemispheres reveal that for further development of a surgical procedure to reliably infuse AAV5-miHTT in these brain structure, magnetic resonance-guidance should be incorporated. Our recent study showed that parenchymal magnetic resonance-guided CED resulted in consistent global transduction of AAV5-GFP in non-human primate brain.40 For development and clinical application, a less complicated and minimally invasive administration procedure, such as intravenous or intrathecal, might at first sight seem to be preferred over an intracranial parenchymal delivery. Recently, data acquired with an engineered AAV capsid showing robust crossing of the blood-brain barrier upon intravenous administration in rodents are very encouraging,61, 62 but its efficacy in larger species and level of transduction in deep brain structures still need to be determined. Here, we show that although intrathecal infusion results in transduction and transgene expression in the CNS, particularly in the cerebral cortex, transgene expression, and thus efficacy, is most likely to be too low in the deeper brain structures to be disease modifying. Furthermore, 40–100 times higher viral titers will be required for intrathecal injection to achieve a very moderate transduction of the basal ganglia. This low transduction in the CNS could have been accompanied by the observed leakage of the AAV5 virus into the periphery. In the case of HD, transduction of peripheral organs is not per se unfavorable, because mutated HTT is widely expressed outside the CNS and several peripheral signs of disease have been reported.63
The present study is the first demonstration of a successful human mutant huntingtin lowering in the brain of a large-animal model of HD. The combination of widespread vector distribution, long-term expression, extensive human huntingtin lowering, and high specificity and tolerability observed with AAV5-miHTT treatment support the continuation of the development of huntingtin-lowering gene therapy for HD patients.
Materials and Methods
AAV5 Vectors
AAV5 vector encoding cDNA of the enhanced GFP transgene and miHTT cassette was packaged into AAV5 by a baculovirus-based AAV production system (uniQure, Amsterdam, the Netherlands) as previously described.64, 65 The complete transcription unit was flanked by two non-coding AAV-derived inverted terminal repeats, and expression was driven by a combination of the cytomegalovirus early enhancer element and chicken β-actin promoter. The expression cassettes were inserted in a recombinant baculovirus vector by homologous recombination, and clones were selected by plague purification. The recombinant baculovirus containing the cassettes was further amplified and clones screened for best production and stability by PCR and qPCR. To generate AAV5, we performed baculovirus infections on insect cells with recombinant baculoviruses encoding rep for replication and packaging, and cap-5 for the AAV5 capsid and the expression cassette. After viral particle assembled, prep purification was performed with AVB Sepharose high-performance affinity medium (GE Healthcare, Piscataway, NJ, USA) using AKTA Explorer purification system (GE Healthcare), and titer was determined by qPCR with primer-probe combinations directed against the CAG promoter transgene. Both AAV5-GFP and AAV5-miHTT vectors have been described previously.26, 40
Anti-AAV Neutralizing Antibody Assay
Prior to the start of the study, minipig plasma samples were tested for anti-AAV neutralizing antibodies as described previously.66 All animals selected were considered seronegative with neutralizing antibody titers lower than 1:50 at baseline.
Minipigs, Anesthesia, Surgical Procedure, and AAV5 Vectors Application
All experiments were carried out according to the guidelines for the care and use of experimental animals and approved by the State Veterinary Administration of the Czech Republic. tgHD minipigs and healthy control subjects30 of both sexes, 20–39 months old, were used. The pilot experiment with AAV5-GFP constructs to determine the optimal delivery route and to study the biodistribution was performed in 12 wild-type minipigs. The second study with AAV5-miHTT was performed in 12 tgHD minipigs. The two hemispheres received 270 μL of virus per hemisphere (54 μL in the putamen and 216 μL in the thalamus).
General anesthesia of minipigs was induced by intramuscular application of tiletamine, zolazepam, ketamine, and xylazine combination (TKX) mixture containing tiletamine 4 mg/kg, zolazepam 4 mg/kg (Zoletil 100; Virbac), ketamine 5 mg/kg (Narketan 10; Chassot), and xylazine 1 mg/kg (Rometar 2%; Spofa) followed by intravenous ear cannulation and intubation. Artificial ventilation and isoflurane/nitrous oxide anesthesia were used during the rest of the procedure. The head of animals was mounted into a stereotaxic localizer box.67 Putamen and thalamus coordinates were histologically based estimated from bone bregma of age- and body-weight-matched animals (post mortem). Craniotomy and dural incision were performed by oscillation saw, neurosurgery mallet, and chisel. AAV5 vectors as well as formulation buffer were delivered by a 500-μL Hamilton syringe with 34G beveled NanoFil needle (World Precision Instruments; NF34BV) with 185 μm optical density (OD), placed into the Injection Robot (Neurostar, Germany) and mounted onto stereotaxic localizer box. Single injection of AAV5 vectors or formulation buffer was administered to each animal, with injection rate increasing every minute until reaching the final volume of 54 μL per putamen and 216 μL per thalamus (first min: 1 μL/min, second min: 2 μl/min, etc.). The skull was closed by a bone graft and cement, periosteum stitching, and skin by suturing. Perioperative as well as postoperative antibiotic (cefazolin) and analgesic (flunixin) were administrated. For intrathecal administration of AAV5 construct, a spinal needle (Yale; 121884; 1.2 × 90 mm) attached to a 5-mL syringe was used. The application started immediately after CSF collection; 5 mL of AAV5 vector was delivered slowly (over approximately 5 min) by hand injection into the lumbar region. The same type of spinal needle was also used for CSF collection in all experiments.
CSF Collection
CSF was collected by a lumbar puncture in both healthy (n = 12) and tgHD animals (n = 12) in the following time intervals: −14, −7, 0 (surgery day), 7, 14, 28, 56, and 84 days. The animals were food deprived for 12 hr and then anaesthetized with a TKX mixture as mentioned above in the Minipigs, Anesthesia, Surgical Procedure, and AAV5 Vectors Application section. After cleaning and disinfection of the lumbar area, approximately 5 mL of CSF was obtained using a spinal needle. The puncture area was covered by liquid dressing and the animal was let to recover. CSF samples were immediately centrifuged at 1,500 × g for 10 min at 4°C, followed by further centrifugation at 2,400 × g for 10 min at 4°C, aliquoted to prevent repeated freeze-thaw cycles and frozen to −80°C.
Necropsy, Sample Collection, and Brain Sectioning
At the end of the experiments the animals were anesthetized by TKX mixture and euthanized by an overdose of thiopental. Immediately after overdosing, the pigs were transcardially perfused with 20 L of ice-cold PBS. The samples of inner organs (5 × 5 × 5 mm necropsies of liver, kidney, spleen, and adrenal glands) were removed first, followed by brain samples (4-mm punch from 4-mm-thick coronal brain slabs) and spinal cord samples (5-mm transversal section). The brain samples (two punches from the cortex, two punches from the striatum, one from caudate nucleus, and one from the thalamus) were collected as shown in Figure 4. The samples (inner organs, brain punches, and spinal cord transversal sections) for qPCR and protein detection were immediately snap-frozen in liquid nitrogen and stored at −80°C. PBS-perfused brains were cut into 4-mm slabs and after collection of punch necropsies post-fixed in 4% paraformaldehyde (pH 7.4) in PBS for 24 hr. The brain slabs were then transferred into 30% sucrose containing 0.01% sodium azide and left until saturation. Well-saturated slabs were subsequently frozen in cryostat (CM1950; Leica Biosystems), sectioned into 20-μm-thick coronal sections, and mounted onto large microscope slides (76 × 51 × 1 mm).
DNA Isolation and Vector Genome Copies Determination
Tissue punches were crushed using CryoPrep System (Covaris, Woburn, MA, USA), and powder was divided for RNA and DNA analyses. For DNA isolation, 20 mg of powdered tissue was used with the DNeasy 96 Blood and Tissue kit (QIAGEN, Germany). Primers specific for the CAG promoter were used to amplify a sequence specific for the transgenes by SYBR Green Fast qPCR (Thermo Fisher Scientific). The amount of vector DNA was calculated from a plasmid standard curve, which was taken along on the same plate. Results were reported as gc per microgram of genomic DNA.
RNA Isolation and qPCR
For RNA isolation, crushed tissue was homogenized in TRIzol using gentleMACS Dissociator (Miltenyi Biotec), and total RNA was isolated according the manufacturer’s protocol (Invitrogen). To remove genomic DNA, we treated RNA with double-stranded DNase (dsDNase; Thermo Scientific, Waltham, MA, USA). RNA samples were reverse transcribed using a DyNAmo cDNA synthesis kit (F-470L; Thermo Fisher Scientific) following manufacturer’s instructions. To detect human HTT mRNA knockdown, we used TaqMan gene expression assays (Applied Biosystems) with primer-probe against human HTT (Hs00918178_m1) and porcine GAPDH (Ss03382302_s1) as reference gene. TaqMan Universal Mastermix II with UNG (Applied Biosystems) was used to perform the qPCR. The expression level of each gene was normalized to endogenous GAPDH levels. HTT mRNA knockdown percentages relative to formulation buffer treatment were calculated based on the 2−ΔΔCt method.68
To examine miHTT miRNA expression, we synthesized cDNA from isolated total RNA with gene-specific RT primers targeting mature miHTT using TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems). A single-stranded mature miHTT RNA standard line was taken along. Next, gene-specific TaqMan qPCR was performed with mature miHTT-specific probes using TaqMan Fast Universal PCR Master Mix (Applied Biosystems). Using the mature miHTT standard line, we determined miHTT molecules per reaction. Subsequently, the amount of miHTT molecules per cell of tissue was calculated based on the assumption that one cell contains 15 pg of total RNA.69
Mutant Huntingtin Protein Quantification
Quantification of mutant huntingtin protein in longitudinal CSF samples and brain regions from tgHD minipigs was generally performed as described previously,34 at Evotec (Germany). Pulverized tgHD minipig brain samples were homogenized in 170 μL ice-cold lysis buffer using a FastPrep24 tissue homogenizer. The homogenates were aliquoted, frozen on dry ice, and stored at −80°C. With 1 aliquot the total protein concentration was determined by bicinchoninic acid assay. Another aliquot of homogenate (one freeze and thaw cycle) was thawed on the same day of the Singulex assay, diluted to 1 μg/mL in aCSF buffer (artificial CSF + 1% Tween 20 + 1 × protease inhibitor) and tested in technical triplicates. The human HTT-Q46_GST, 1-548 recombinant standard protein was used for quantification of HTT spiked into aCSF buffer. Standard protein was applied at a final concentration range between 0.1 and 400 pg/mL. tgHD minipig CSF samples (3 aliquots of 125–130 μL) were stored at −80°C before thawing on ice. Aliquots of each sample were pooled and tested in undiluted technical triplicates, and CSF samples of each animal were run in parallel on the same plate. A final concentration of 8 fM to 6,114 fM human HTT-Q46_GST, 1-548 was used for quantification. The Erenna (Singulex-Millipore) reader was calibrated on the day of the experiment following manufacturer’s protocol. The SMC immunoassay was performed using a combination of 2B7 (Novartis) antibody directed against the first 17 amino acids of the huntingtin protein, and MW1 (Caltech) antibody that binds to the expanded polyglutamine repeat.34 The 2B7-MW1 signal is dependent on both the mutant huntingtin size (i.e., the smaller the size, the higher signal) and the polyglutamine expansion (i.e., the higher the expansion, the higher the signal). Because the standard protein has a shorter poly-glutamine expansion than the protein of the tgHD minipig model (Q46 with respect to Q124), it should be noted that, although relative levels between samples are reliable, the absolute concentration of mutant huntingtin, as depicted for the CSF measurements, is probably an underestimation. As negative control, cortical lysates from wild-type minipigs were taken along, which showed as expected low levels in mutant huntingtin, validating the specificity of the assay.
To evaluate possible blood contamination of the CSF samples, we used a competitive pig hemoglobin ELISA (LS-F8543; LSBio), following the manufacturer’s instructions with minor modifications: the final horseradish peroxidase-3,3',5,5'-tetramethylbenzidine reactions were terminated with stop solution after instead of the 10–20 min suggested in the original protocol in order to increase the sensitivity of the assay. All CSF samples were assayed in duplicate at a 1:5 dilution, along with minipig plasma samples as a positive control.
Cytokine Analysis
To analyze cytokine levels in porcine CSF, we used Swine cytokine Magnetic 7-Plex Panel (catalog no. LSC0001M; Thermo Fisher Scientific) enabling the simultaneous measurement of IL-1β, IL-4, IL-8, IL-10, IFN-α, IFN-γ, and tumor necrosis factor alpha (TNF-α). Kits of the same batch (lot no. 1804247A, expiry 7/2018) were used for all measurements. Each sample was analyzed in duplicate (50 μL of CSF each). CSF samples were thawed on ice and clarified by centrifugation at 16,000 × g for 10 min at 4°C. The xMAP bead-based assay was prepared according to manufacturer’s instructions with slight modifications for CSF as published in Valekova et al.35 Standard curves were extended to comprise nine data points for improved readability of low end curve values. The fluorescence intensity together with the bead identification were recorded using Luminex 200 instrument with xPonent software, version 3.1.871.0 (Luminex, Austin, TX, USA). Protocol parameters were set to quantitative measurement, sample volume 75 μL, doublet discriminator 7,800–20,000, photomultiplier tube default (high), timeout 100 s. The Luminex system was properly calibrated, and its performance was verified according to the manufacturer’s instructions. For each analyte, the fluorescence intensity of a minimum of 100 beads was measured, and the median fluorescence intensity (MFI) was used for quantitation of cytokine concentrations. The csv data files containing raw data were exported from xPonent and utilized for follow-up statistical evaluation. Further data processing and analysis were performed in the R statistical environment.70 Package drLumi71 was used for standard curve fitting and estimation of cytokine concentrations in CSF samples. Options used for standard curve fitting were five-parameter logistic regression (SSL5) with four-parameter logistic regression (SSL4) as a fallback for occasions where the SSL5 model would not converge. Concentrations of two technical replicates of each CSF sample were averaged.
Immunohistochemistry, Scanning, and Image Analysis
In some brain slices (DARPP32) heat-induced epitope retrieval was performed by overnight incubation in retrieval solution in a hybridization oven set to 60°C. The endogenous peroxidase activity was blocked with a solution of 0.3% of hydrogen peroxide in methanol for 20 min, and the brain sections were immunostained using the following rabbit primary antibodies: anti-GFP (1:1,000, ab6556; Abcam), anti-IBA1 (1:400; 234 003; Synaptic Systems), and anti-DARPP32 (1:15,000, ab40801; Abcam). Sections were then treated with a biotinylated donkey anti-rabbit secondary antibody (1:400, RPN 1004V; GE Healthcare Life Sciences) followed by an avidin-peroxidase complex (1:400, A3151; Sigma-Aldrich). The avidin-peroxidase complex was visualized by incubation with solution containing a dissolved 3, 3 -diaminobenzidine tablet (4170; Kementec Diagnostics). The sections were dehydrated and mounted with DePeX (Sigma). Images were acquired using a histological scanner (Virtual Slide Microscope VS120-5 fluorescence; Olympus), and quantitative analysis of IHC-stained brain sections was performed using Fiji ImageJ distribution (https://fiji.sc/). The sections were scaled, calibrated, converted to 8-bit, and thresholded using default (for IBA1, GFP, and DARPP32 positivity) as well as triangle (for whole section) threshold. Manually assigned ROIs were used for striatum selection in DARPP32-stained sections.
Statistical Analysis
For two groups, data were analyzed using Student’s t test or ordinary one-way ANOVA to determine statistically significances between samples. For longitudinal data, two-way ANOVA with repeated measurements and Tukey’s post hoc test (α = 0.05) were applied. The p values were either listed or represented by the following number of asterisks: *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. Statistical analysis of cytokine levels was performed in R statistical environment, version 3.4.0, using software packages tidyverse, stringr, forcats, cowplot, lme4, and broom.
Author Contributions
Conceptualization, M.M.E., Z.E., J. Motlik, P.K.; Methodology, J. Miniarikova, Z.E., J. Motlik; Formal Analysis, A.V., P.V.; Investigation, M.M.E., S.J., B. Bohuslavova, J.J., H.K.S., P.V., C.B.; Writing, M.M.E.; Funding Acquisition, Z.E., J. Motlik; Resources, B. Blits, J.L.; Supervision, M.M.E., H.K., Z.E., S.J., H.P., J. Motlik, P.K.
Conflicts of Interest
M.M.E., A.V., C.B., B.B., J.L., S.V.D., H.P., and P.K. are employees and shareholders at uniQure. J. Motlik and PIGMOD have a collaborative agreement with uniQure. uniQure has sponsored a portion of the research performed in J. Motlik’s lab.
Acknowledgments
The authors would like to thank Raygene Martier and Sonay Keskin for technical assistance. The authors are grateful to Eileen Sawyer and Olivier ter Braake for critically reading the manuscript. Part of this work was supported by PIGMOD Center’s sustainability program: National Sustainability Programme, project number LO1609 (Czech Ministry of Education, Youth and Sports). We are also grateful for the support this project has received from the CHDI Foundation, in particular Douglas MacDonald and David Howland.
Footnotes
Supplemental Information includes five figures and two tables and can be found with this article online at https://doi.org/10.1016/j.ymthe.2018.06.021.
Supplemental Information
References
- 1.Waldvogel H.J., Kim E.H., Tippett L.J., Vonsattel J.P., Faull R.L. The neuropathology of Huntington’s disease. Curr. Top. Behav. Neurosci. 2015;22:33–80. doi: 10.1007/7854_2014_354. [DOI] [PubMed] [Google Scholar]
- 2.Rüb U., Seidel K., Heinsen H., Vonsattel J.P., den Dunnen W.F., Korf H.W. Huntington’s disease (HD): the neuropathology of a multisystem neurodegenerative disorder of the human brain. Brain Pathol. 2016;26:726–740. doi: 10.1111/bpa.12426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ross C.A., Pantelyat A., Kogan J., Brandt J. Determinants of functional disability in Huntington’s disease: role of cognitive and motor dysfunction. Mov. Disord. 2014;29:1351–1358. doi: 10.1002/mds.26012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ross C.A., Tabrizi S.J. Huntington’s disease: from molecular pathogenesis to clinical treatment. Lancet Neurol. 2011;10:83–98. doi: 10.1016/S1474-4422(10)70245-3. [DOI] [PubMed] [Google Scholar]
- 5.Boudreau R.L., McBride J.L., Martins I., Shen S., Xing Y., Carter B.J., Davidson B.L. Nonallele-specific silencing of mutant and wild-type huntingtin demonstrates therapeutic efficacy in Huntington’s disease mice. Mol. Ther. 2009;17:1053–1063. doi: 10.1038/mt.2009.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drouet V., Perrin V., Hassig R., Dufour N., Auregan G., Alves S., Bonvento G., Brouillet E., Luthi-Carter R., Hantraye P., Déglon N. Sustained effects of nonallele-specific Huntingtin silencing. Ann. Neurol. 2009;65:276–285. doi: 10.1002/ana.21569. [DOI] [PubMed] [Google Scholar]
- 7.Kordasiewicz H.B., Stanek L.M., Wancewicz E.V., Mazur C., McAlonis M.M., Pytel K.A., Artates J.W., Weiss A., Cheng S.H., Shihabuddin L.S. Sustained therapeutic reversal of Huntington’s disease by transient repression of huntingtin synthesis. Neuron. 2012;74:1031–1044. doi: 10.1016/j.neuron.2012.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Evers M.M., Pepers B.A., van Deutekom J.C., Mulders S.A., den Dunnen J.T., Aartsma-Rus A., van Ommen G.J., van Roon-Mom W.M. Targeting several CAG expansion diseases by a single antisense oligonucleotide. PLoS ONE. 2011;6:e24308. doi: 10.1371/journal.pone.0024308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu J., Matsui M., Gagnon K.T., Schwartz J.C., Gabillet S., Arar K., Wu J., Bezprozvanny I., Corey D.R. Allele-specific silencing of mutant huntingtin and ataxin-3 genes by targeting expanded CAG repeats in mRNAs. Nat. Biotechnol. 2009;27:478–484. doi: 10.1038/nbt.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McBride J.L., Pitzer M.R., Boudreau R.L., Dufour B., Hobbs T., Ojeda S.R., Davidson B.L. Preclinical safety of RNAi-mediated HTT suppression in the rhesus macaque as a potential therapy for Huntington’s disease. Mol. Ther. 2011;19:2152–2162. doi: 10.1038/mt.2011.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carroll J.B., Warby S.C., Southwell A.L., Doty C.N., Greenlee S., Skotte N., Hung G., Bennett C.F., Freier S.M., Hayden M.R. Potent and selective antisense oligonucleotides targeting single-nucleotide polymorphisms in the Huntington disease gene / allele-specific silencing of mutant huntingtin. Mol. Ther. 2011;19:2178–2185. doi: 10.1038/mt.2011.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stanek L.M., Sardi S.P., Mastis B., Richards A.R., Treleaven C.M., Taksir T., Misra K., Cheng S.H., Shihabuddin L.S. Silencing mutant huntingtin by adeno-associated virus-mediated RNA interference ameliorates disease manifestations in the YAC128 mouse model of Huntington’s disease. Hum. Gene Ther. 2014;25:461–474. doi: 10.1089/hum.2013.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cambon K., Zimmer V., Martineau S., Gaillard M.C., Jarrige M., Bugi A., Miniarikova J., Rey M., Hassig R., Dufour N. Preclinical evaluation of a lentiviral vector for Huntingtin silencing. Mol. Ther. Methods Clin. Dev. 2017;5:259–276. doi: 10.1016/j.omtm.2017.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harper S.Q., Staber P.D., He X., Eliason S.L., Martins I.H., Mao Q., Yang L., Kotin R.M., Paulson H.L., Davidson B.L. RNA interference improves motor and neuropathological abnormalities in a Huntington’s disease mouse model. Proc. Natl. Acad. Sci. USA. 2005;102:5820–5825. doi: 10.1073/pnas.0501507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Datson N.A., González-Barriga A., Kourkouta E., Weij R., van de Giessen J., Mulders S., Kontkanen O., Heikkinen T., Lehtimäki K., van Deutekom J.C. The expanded CAG repeat in the huntingtin gene as target for therapeutic RNA modulation throughout the HD mouse brain. PLoS ONE. 2017;12:e0171127. doi: 10.1371/journal.pone.0171127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ionis Pharmaceuticals, Inc. (2015). Safety, tolerability, pharmacokinetics, and pharmacodynamics of IONIS-HTTRx in patients with early manifest Huntington’s disease. https://clinicaltrials.gov/ct2/show/NCT02519036.
- 17.Grondin R., Ge P., Chen Q., Sutherland J.E., Zhang Z., Gash D.M., Stiles D.K., Stewart G.R., Sah D.W., Kaemmerer W.F. Onset time and durability of Huntingtin suppression in rhesus putamen after direct infusion of antihuntingtin siRNA. Mol. Ther. Nucleic Acids. 2015;4:e245. doi: 10.1038/mtna.2015.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Machida Y., Okada T., Kurosawa M., Oyama F., Ozawa K., Nukina N. rAAV-mediated shRNA ameliorated neuropathology in Huntington disease model mouse. Biochem. Biophys. Res. Commun. 2006;343:190–197. doi: 10.1016/j.bbrc.2006.02.141. [DOI] [PubMed] [Google Scholar]
- 19.Monteys A.M., Wilson M.J., Boudreau R.L., Spengler R.M., Davidson B.L. Artificial miRNAs targeting mutant Huntingtin show preferential silencing in vitro and in vivo. Mol. Ther. Nucleic Acids. 2015;4:e234. doi: 10.1038/mtna.2015.7. [DOI] [PubMed] [Google Scholar]
- 20.Rodriguez-Lebron E., Denovan-Wright E.M., Nash K., Lewin A.S., Mandel R.J. Intrastriatal rAAV-mediated delivery of anti-huntingtin shRNAs induces partial reversal of disease progression in R6/1 Huntington’s disease transgenic mice. Mol. Ther. 2005;12:618–633. doi: 10.1016/j.ymthe.2005.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miniarikova J., Zimmer V., Martier R., Brouwers C.C., Pythoud C., Richetin K., Rey M., Lubelski J., Evers M.M., van Deventer S.J. AAV5-miHTT gene therapy demonstrates suppression of mutant huntingtin aggregation and neuronal dysfunction in a rat model of Huntington’s disease. Gene Ther. 2017;24:630–639. doi: 10.1038/gt.2017.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boudreau R.L., Spengler R.M., Davidson B.L. Rational design of therapeutic siRNAs: minimizing off-targeting potential to improve the safety of RNAi therapy for Huntington’s disease. Mol. Ther. 2011;19:2169–2177. doi: 10.1038/mt.2011.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McBride J.L., Boudreau R.L., Harper S.Q., Staber P.D., Monteys A.M., Martins I., Gilmore B.L., Burstein H., Peluso R.W., Polisky B. Artificial miRNAs mitigate shRNA-mediated toxicity in the brain: implications for the therapeutic development of RNAi. Proc. Natl. Acad. Sci. USA. 2008;105:5868–5873. doi: 10.1073/pnas.0801775105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Monteys A.M., Spengler R.M., Dufour B.D., Wilson M.S., Oakley C.K., Sowada M.J., McBride J.L., Davidson B.L. Single nucleotide seed modification restores in vivo tolerability of a toxic artificial miRNA sequence in the mouse brain. Nucleic Acids Res. 2014;42:13315–13327. doi: 10.1093/nar/gku979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Southwell A.L., Skotte N.H., Villanueva E.B., Østergaard M.E., Gu X., Kordasiewicz H.B., Kay C., Cheung D., Xie Y., Waltl S. A novel humanized mouse model of Huntington disease for preclinical development of therapeutics targeting mutant huntingtin alleles. Hum. Mol. Genet. 2017;26:1115–1132. doi: 10.1093/hmg/ddx021. [DOI] [PubMed] [Google Scholar]
- 26.Miniarikova J., Zanella I., Huseinovic A., van der Zon T., Hanemaaijer E., Martier R., Koornneef A., Southwell A.L., Hayden M.R., van Deventer S.J. Design, characterization, and lead selection of therapeutic miRNAs targeting Huntingtin for development of gene therapy for Huntington’s disease. Mol. Ther. Nucleic Acids. 2016;5:e297. doi: 10.1038/mtna.2016.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Venuto C.S., McGarry A., Ma Q., Kieburtz K. Pharmacologic approaches to the treatment of Huntington’s disease. Mov. Disord. 2012;27:31–41. doi: 10.1002/mds.23953. [DOI] [PubMed] [Google Scholar]
- 28.Travessa A.M., Rodrigues F.B., Mestre T.A., Ferreira J.J. Fifteen years of clinical trials in Huntington’s disease: a very low clinical drug development success rate. J. Huntingtons Dis. 2017;6:157–163. doi: 10.3233/JHD-170245. [DOI] [PubMed] [Google Scholar]
- 29.Morton A.J., Howland D.S. Large genetic animal models of Huntington’s disease. J. Huntingtons Dis. 2013;2:3–19. doi: 10.3233/JHD-130050. [DOI] [PubMed] [Google Scholar]
- 30.Baxa M., Hruska-Plochan M., Juhas S., Vodicka P., Pavlok A., Juhasova J., Miyanohara A., Nejime T., Klima J., Macakova M. A transgenic minipig model of Huntington’s disease. J. Huntingtons Dis. 2013;2:47–68. doi: 10.3233/JHD-130001. [DOI] [PubMed] [Google Scholar]
- 31.Jacobsen J.C., Bawden C.S., Rudiger S.R., McLaughlan C.J., Reid S.J., Waldvogel H.J., MacDonald M.E., Gusella J.F., Walker S.K., Kelly J.M. An ovine transgenic Huntington’s disease model. Hum. Mol. Genet. 2010;19:1873–1882. doi: 10.1093/hmg/ddq063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gasmi M., Herzog C.D., Brandon E.P., Cunningham J.J., Ramirez G.A., Ketchum E.T., Bartus R.T. Striatal delivery of neurturin by CERE-120, an AAV2 vector for the treatment of dopaminergic neuron degeneration in Parkinson’s disease. Mol. Ther. 2007;15:62–68. doi: 10.1038/sj.mt.6300010. [DOI] [PubMed] [Google Scholar]
- 33.Davidson B.L., Stein C.S., Heth J.A., Martins I., Kotin R.M., Derksen T.A., Zabner J., Ghodsi A., Chiorini J.A. Recombinant adeno-associated virus type 2, 4, and 5 vectors: transduction of variant cell types and regions in the mammalian central nervous system. Proc. Natl. Acad. Sci. USA. 2000;97:3428–3432. doi: 10.1073/pnas.050581197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wild E.J., Boggio R., Langbehn D., Robertson N., Haider S., Miller J.R., Zetterberg H., Leavitt B.R., Kuhn R., Tabrizi S.J. Quantification of mutant huntingtin protein in cerebrospinal fluid from Huntington’s disease patients. J. Clin. Invest. 2015;125:1979–1986. doi: 10.1172/JCI80743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Valekova I., Jarkovska K., Kotrcova E., Bucci J., Ellederova Z., Juhas S., Motlik J., Gadher S.J., Kovarova H. Revelation of the IFNα, IL-10, IL-8 and IL-1β as promising biomarkers reflecting immuno-pathological mechanisms in porcine Huntington’s disease model. J. Neuroimmunol. 2016;293:71–81. doi: 10.1016/j.jneuroim.2016.02.012. [DOI] [PubMed] [Google Scholar]
- 36.Samaranch L., Sebastian W.S., Kells A.P., Salegio E.A., Heller G., Bringas J.R., Pivirotto P., DeArmond S., Forsayeth J., Bankiewicz K.S. AAV9-mediated expression of a non-self protein in nonhuman primate central nervous system triggers widespread neuroinflammation driven by antigen-presenting cell transduction. Mol. Ther. 2014;22:329–337. doi: 10.1038/mt.2013.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schramke S., Schubert R., Frank F., Wirsig M., Fels M., Kemper N., Schuldenzucker V., Reilmann R. The Libechov minipig as a large animal model for preclinical research in Huntington’s disease – thoughts and perspectives. Cesk. Neurol. Neurochir. 2015;78/111(Suppl 2):55–60. [Google Scholar]
- 38.Schuldenzucker V., Schubert R., Muratori L.M., Freisfeld F., Rieke L., Matheis T., Schramke S., Motlik J., Kemper N., Radespiel U., Reilmann R. Behavioral testing of minipigs transgenic for the Huntington gene—a three-year observational study. PLoS ONE. 2017;12:e0185970. doi: 10.1371/journal.pone.0185970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Howland D.S., Munoz-Sanjuan I. Mind the gap: models in multiple species needed for therapeutic development in Huntington’s disease. Mov. Disord. 2014;29:1397–1403. doi: 10.1002/mds.26008. [DOI] [PubMed] [Google Scholar]
- 40.Samaranch L., Blits B., San Sebastian W., Hadaczek P., Bringas J., Sudhakar V., Macayan M., Pivirotto P.J., Petry H., Bankiewicz K.S. MR-guided parenchymal delivery of adeno-associated viral vector serotype 5 in non-human primate brain. Gene Ther. 2017;24:253–261. doi: 10.1038/gt.2017.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shaffer J.J., Ghayoor A., Long J.D., Kim R.E., Lourens S., O’Donnell L.J., Westin C.F., Rathi Y., Magnotta V., Paulsen J.S., Johnson H.J. Longitudinal diffusion changes in prodromal and early HD: evidence of white-matter tract deterioration. Hum. Brain Mapp. 2017;38:1460–1477. doi: 10.1002/hbm.23465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grondin R., Kaytor M.D., Ai Y., Nelson P.T., Thakker D.R., Heisel J., Weatherspoon M.R., Blum J.L., Burright E.N., Zhang Z., Kaemmerer W.F. Six-month partial suppression of Huntingtin is well tolerated in the adult rhesus striatum. Brain. 2012;135:1197–1209. doi: 10.1093/brain/awr333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trajkovic K., Jeong H., Krainc D. Mutant Huntingtin is secreted via a late endosomal/lysosomal unconventional secretory pathway. J. Neurosci. 2017;37:9000–9012. doi: 10.1523/JNEUROSCI.0118-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Southwell A.L., Smith S.E., Davis T.R., Caron N.S., Villanueva E.B., Xie Y., Collins J.A., Ye M.L., Sturrock A., Leavitt B.R. Ultrasensitive measurement of huntingtin protein in cerebrospinal fluid demonstrates increase with Huntington disease stage and decrease following brain huntingtin suppression. Sci. Rep. 2015;5:12166. doi: 10.1038/srep12166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Duyao M.P., Auerbach A.B., Ryan A., Persichetti F., Barnes G.T., McNeil S.M., Ge P., Vonsattel J.P., Gusella J.F., Joyner A.L. Inactivation of the mouse Huntington’s disease gene homolog Hdh. Science. 1995;269:407–410. doi: 10.1126/science.7618107. [DOI] [PubMed] [Google Scholar]
- 46.Lopes F., Barbosa M., Ameur A., Soares G., de Sá J., Dias A.I., Oliveira G., Cabral P., Temudo T., Calado E. Identification of novel genetic causes of Rett syndrome-like phenotypes. J. Med. Genet. 2016;53:190–199. doi: 10.1136/jmedgenet-2015-103568. [DOI] [PubMed] [Google Scholar]
- 47.Nasir J., Floresco S.B., O’Kusky J.R., Diewert V.M., Richman J.M., Zeisler J., Borowski A., Marth J.D., Phillips A.G., Hayden M.R. Targeted disruption of the Huntington’s disease gene results in embryonic lethality and behavioral and morphological changes in heterozygotes. Cell. 1995;81:811–823. doi: 10.1016/0092-8674(95)90542-1. [DOI] [PubMed] [Google Scholar]
- 48.Rodan L.H., Cohen J., Fatemi A., Gillis T., Lucente D., Gusella J., Picker J.D. A novel neurodevelopmental disorder associated with compound heterozygous variants in the huntingtin gene. Eur. J. Hum. Genet. 2016;24:1826–1827. doi: 10.1038/ejhg.2016.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stiles D.K., Zhang Z., Ge P., Nelson B., Grondin R., Ai Y., Hardy P., Nelson P.T., Guzaev A.P., Butt M.T. Widespread suppression of huntingtin with convection-enhanced delivery of siRNA. Exp. Neurol. 2012;233:463–471. doi: 10.1016/j.expneurol.2011.11.020. [DOI] [PubMed] [Google Scholar]
- 50.Wang G., Liu X., Gaertig M.A., Li S., Li X.J. Ablation of huntingtin in adult neurons is nondeleterious but its depletion in young mice causes acute pancreatitis. Proc. Natl. Acad. Sci. USA. 2016;113:3359–3364. doi: 10.1073/pnas.1524575113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dietrich P., Johnson I.M., Alli S., Dragatsis I. Elimination of huntingtin in the adult mouse leads to progressive behavioral deficits, bilateral thalamic calcification, and altered brain iron homeostasis. PLoS Genet. 2017;13:e1006846. doi: 10.1371/journal.pgen.1006846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Björkqvist M., Wild E.J., Thiele J., Silvestroni A., Andre R., Lahiri N., Raibon E., Lee R.V., Benn C.L., Soulet D. A novel pathogenic pathway of immune activation detectable before clinical onset in Huntington’s disease. J. Exp. Med. 2008;205:1869–1877. doi: 10.1084/jem.20080178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Crotti A., Benner C., Kerman B.E., Gosselin D., Lagier-Tourenne C., Zuccato C., Cattaneo E., Gage F.H., Cleveland D.W., Glass C.K. Mutant Huntingtin promotes autonomous microglia activation via myeloid lineage-determining factors. Nat. Neurosci. 2014;17:513–521. doi: 10.1038/nn.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cicchetti F., Barker R.A. The glial response to intracerebrally delivered therapies for neurodegenerative disorders: is this a critical issue? Front. Pharmacol. 2014;5:139. doi: 10.3389/fphar.2014.00139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Love S., Plaha P., Patel N.K., Hotton G.R., Brooks D.J., Gill S.S. Glial cell line-derived neurotrophic factor induces neuronal sprouting in human brain. Nat. Med. 2005;11:703–704. doi: 10.1038/nm0705-703. [DOI] [PubMed] [Google Scholar]
- 56.Keene C.D., Chang R.C., Leverenz J.B., Kopyov O., Perlman S., Hevner R.F., Born D.E., Bird T.D., Montine T.J. A patient with Huntington’s disease and long-surviving fetal neural transplants that developed mass lesions. Acta Neuropathol. 2009;117:329–338. doi: 10.1007/s00401-008-0465-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tuszynski M.H., Thal L., Pay M., Salmon D.P., U H.S., Bakay R., Patel P., Blesch A., Vahlsing H.L., Ho G. A phase 1 clinical trial of nerve growth factor gene therapy for Alzheimer disease. Nat. Med. 2005;11:551–555. doi: 10.1038/nm1239. [DOI] [PubMed] [Google Scholar]
- 58.Wojtecki L., Groiss S.J., Hartmann C.J., Elben S., Omlor S., Schnitzler A., Vesper J. Deep brain stimulation in Huntington’s disease—preliminary evidence on pathophysiology, efficacy and safety. Brain Sci. 2016;6:E38. doi: 10.3390/brainsci6030038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lang A.E., Gill S., Patel N.K., Lozano A., Nutt J.G., Penn R., Brooks D.J., Hotton G., Moro E., Heywood P. Randomized controlled trial of intraputamenal glial cell line-derived neurotrophic factor infusion in Parkinson disease. Ann. Neurol. 2006;59:459–466. doi: 10.1002/ana.20737. [DOI] [PubMed] [Google Scholar]
- 60.Mittermeyer G., Christine C.W., Rosenbluth K.H., Baker S.L., Starr P., Larson P., Kaplan P.L., Forsayeth J., Aminoff M.J., Bankiewicz K.S. Long-term evaluation of a phase 1 study of AADC gene therapy for Parkinson’s disease. Hum. Gene Ther. 2012;23:377–381. doi: 10.1089/hum.2011.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Deverman B.E., Pravdo P.L., Simpson B.P., Kumar S.R., Chan K.Y., Banerjee A., Wu W.L., Yang B., Huber N., Pasca S.P., Gradinaru V. Cre-dependent selection yields AAV variants for widespread gene transfer to the adult brain. Nat. Biotechnol. 2016;34:204–209. doi: 10.1038/nbt.3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chan K.Y., Jang M.J., Yoo B.B., Greenbaum A., Ravi N., Wu W.L., Sánchez-Guardado L., Lois C., Mazmanian S.K., Deverman B.E., Gradinaru V. Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems. Nat. Neurosci. 2017;20:1172–1179. doi: 10.1038/nn.4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Carroll J.B., Bates G.P., Steffan J., Saft C., Tabrizi S.J. Treating the whole body in Huntington’s disease. Lancet Neurol. 2015;14:1135–1142. doi: 10.1016/S1474-4422(15)00177-5. [DOI] [PubMed] [Google Scholar]
- 64.Unzu C., Hervás-Stubbs S., Sampedro A., Mauleón I., Mancheño U., Alfaro C., de Salamanca R.E., Benito A., Beattie S.G., Petry H. Transient and intensive pharmacological immunosuppression fails to improve AAV-based liver gene transfer in non-human primates. J. Transl. Med. 2012;10:122. doi: 10.1186/1479-5876-10-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Urabe M., Ding C., Kotin R.M. Insect cells as a factory to produce adeno-associated virus type 2 vectors. Hum. Gene Ther. 2002;13:1935–1943. doi: 10.1089/10430340260355347. [DOI] [PubMed] [Google Scholar]
- 66.Majowicz A., Salas D., Zabaleta N., Rodríguez-Garcia E., González-Aseguinolaza G., Petry H., Ferreira V. Successful repeated hepatic gene delivery in mice and non-human primates achieved by sequential administration of AAV5ch and AAV1. Mol. Ther. 2017;25:1831–1842. doi: 10.1016/j.ymthe.2017.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Glud A.N., Hedegaard C., Nielsen M.S., Søorensen J.C., Bendixen C., Jensen P.H., Mogensen P.H., Larsen K., Bjarkam C.R. Direct MRI-guided stereotaxic viral mediated gene transfer of alpha-synuclein in the Göttingen minipig CNS. Acta Neurobiol. Exp. (Warsz.) 2011;71:508–518. doi: 10.55782/ane-2011-1867. [DOI] [PubMed] [Google Scholar]
- 68.Pfaffl M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen C., Ridzon D.A., Broomer A.J., Zhou Z., Lee D.H., Nguyen J.T., Barbisin M., Xu N.L., Mahuvakar V.R., Andersen M.R. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.R_Core_Team. (2017). R: a language and environment for statistical computing. https://www.R-project.org.
- 71.Sanz, H., Aponte, J., Harezlak, J., Dong, Y., Murawska, M., Valim, C., Ayestaran, A., Aguilar, R., and Moncunill, G. (2015). drLumi: multiplex immunoassays data analysis. R package version 0.1.2 (The R Foundation). https://cran.r-project.org/web/packages/drLumi/index.html.
- 72.Félix B., Léger M.E., Albe-Fessard D., Marcilloux J.C., Rampin O., Laplace J.P. Stereotaxic atlas of the pig brain. Brain Res. Bull. 1999;49:1–137. doi: 10.1016/s0361-9230(99)00012-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.