Abstract
The epigenetics and genomics of glioblastoma (GBM) are complicated. Previous reports indicate that ELFN2 is widely distributed in the cerebral cortex neurons, striatum, and hippocampus cone and in granular cells. However, the function and mechanism of ELFN2, particularly in GBM, have rarely been explored. In this study, we identified ELFN2 as a new hypomethylation gene that acts as an oncogene in GBM. ELFN2 promoted cell autophagy by interacting with AurkA and eIF2α and inhibiting the activation of AurkA. We also demonstrated that aberrantly high ELFN2 expression is obtained due to hypomethylation of its promoter and abnormal miR-101 and LINC00470 expression in GBM. LINC00470 not only enhanced the expression of ELFN2 through adsorption of miR-101 but also affected the methylation level of ELFN2 by decreasing H3K27me3 occupancy. In addition, LINC00470 played a dominant role in the regulation of GBM cell autophagy, even though it upregulated ELFN2 expression. The results indicate that the combination of LINC00470 and ELFN2 has important significance for evaluating the prognosis of astrocytoma patients.
Keywords: long noncoding RNA, miR-101, cell autophagy, AurkA, methylation modification
Wu et al. identified ELFN2 as a hypomethylated gene that interacts with AurkA and eIF2α and promotes cell autophagy. Although LINC00470 mediates epigenetic regulation to contribute to drive ELFN2 expression, it plays a dominant suppressive role in autophagy. LINC00470 and ELFN2 have important significance for astrocytoma patients’ prognosis.
Introduction
Glioblastoma (GBM) is the most malignant brain tumor in adults. Although several studies have increased the understanding of GBM biology, the available of surgical, radiological, chemotherapeutic and adjunctive immunotherapy technologies have greatly improved, the outcomes of GBM patients remain unsatisfactory.1 Therefore, continuing efforts are needed to obtain a comprehensive heterogeneous map of GBM that includes noncoding transcripts and genetic and epigenetic events.
Noncoding RNAs, particularly microRNAs (miRNAs) and long noncoding RNAs (lncRNAs), have been implicated in multiple biological processes in normal and various diseases, including cancer.2, 3 Some of the most commonly overexpressed miRNAs in GBM are miR-21, miR-9, and miR-25,4, 5 and the most commonly decreased miRNAs in GBM include miR-7, miR-34a, and miR-181a.5, 6, 7, 8, 9 A lncRNA microarray analysis indicated that 654 lncRNAs were increased and 654 lncRNAs were decreased in GBM, and CRNDE was identified as the most upregulated lncRNA in GBM.10 Our research has confirmed that the CASC2c lncRNA promotes the proliferation of GBM cells.11 Although the field of circular RNA (circRNA) is in its infancy, a recent deep RNA sequencing study of 10 GBM samples and their paired adjacent normal tissues demonstrated the abnormal expression of various circRNAs in GBM, resulting in the identification of 31,000 circRNA candidates.12
In our previous studies, we first screened and validated nine new hypermethylation genes (LRRC4, SIX3, ANKDD1A, SST, GAD1, PHOX2B, HIST1H3E, PCDHA13, and PCDHA8) and six new hypomethylation genes (F10, POTEH, CPEB1, LMO3, PRDM16, ELFN2) in GBM via high-throughput methylation DNA immunoprecipitation (MeDIP) and CpG island-based promoter chromatin immunoprecipitation (ChIP) assays.13, 14 We have confirmed that the tumor suppressor miR-101 is expressed at a low level in GBM and targets the hypomethylation genes CPEB1, PRDM16, and LMO3 directly or epigenetically to promote GBM cell senescence15 and apoptosis.16, 17 miR-101 also epigenetically restores the expression of the hypermethylation gene LRRC4 to inhibit the interaction between GBM cells and Treg cells.18
ELFN2 (extracellular leucine-rich repeat and fibronectin type III domain-containing 2), is a new hypomethylation gene. This gene, which is also known as protein phosphatase 1 regulatory subunit 29 and as LRRC62, belongs to the leucine-rich repeat family and contains four LRR domains and an FN3 domain.19 Previous studies indicate that ELFN2 is widely distributed in the cerebral cortex neurons, striatum, and hippocampus cone and in granular cells, and expression of this gene is particularly observed in the connections between neurons in the thalamus and cerebral cortex. It has been reported that ELFN2 inhibits the formation of the protein phosphatase complex and inhibits the activity of protein phosphatase 1 by acting as a regulator of protein phosphatase 1.20 However, thus far, the function and mechanism of ELFN2 have rarely been explored, especially in GBM.
In the current study, we observed a high expression of ELFN2 in GBM and found that the LINC00470 lncRNA coordinates the epigenetic regulation of ELFN2 to contribute to its high expression in GBMs. In addition, ELFN2, as a putative oncogene, interacts with AurkA and eukaryotic translation initiation factor 2 subunit alpha (eIF2α) and regulates the kinase activity of AurkA to promote cell autophagy. We also further demonstrated the effects of both LINC00470 and ELFN2 on cell autophagy and identified the important significance of these effects on the prognosis of astrocytoma patients.
Results
Promoter Hypomethylation Contributed to High ELFN2 Expression and Led to Poor Prognosis in Astrocytoma Patients
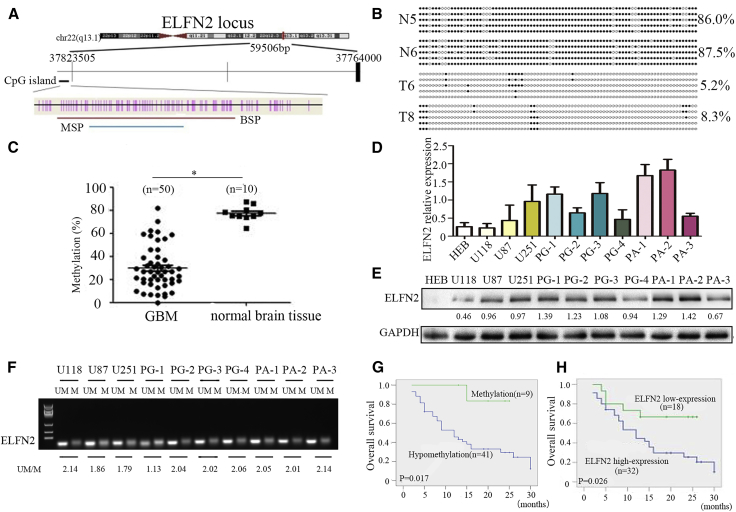
ELFN2, located at 22q13,21 was a new DNA hypomethylation gene that was identified in GBM in our previous work.14 In this study, bisulfite sequencing PCR (BSP) and methylation-specific PCR (MSP) methods were designed to further validate the hypomethylation of ELFN2 in astrocytoma tissues (Figure 1A). BSP showed that the methylation frequency in the CpG dinucleotides of the ELFN2 promoter was 77.3% ± 6.6% in 10 normal brain samples and decreased in astrocytoma samples (29.8% ± 18.7%) (p < 0.05) (Figure 1B). ELFN2 was hypomethylated in 41 (82%) of 50 astrocytoma samples (Table 1; Figure 1C), and this hypomethylation was not significantly correlated with sex, age, or histological grade (Table 1). In addition, a significant correlation was found between ELFN2 promoter hypomethylation and increased ELFN2 expression in astrocytoma (Table 1). ELFN2 was high expression in 32 (64%) of 50 astrocytoma samples compared with that in normal brain tissues and was not correlated with sex, age, or histological grade (Table 2).
Figure 1.
ELFN2 Is Overexpressed due to Promoter Hypomethylation and Is Correlated with a Poor Outcome in Astrocytoma
(A) Schematic diagram of the CpG dinucleotides within the ELFN2 promoter. The nucleotide number is relative to the transcription start site of ELFN2. The red line indicates the region that was tested with BSP, and the blue line indicates the region that was detected with MSP. (B) BSP of the upstream regulatory region of ELFN2 was performed for the representative tissues (N, normal brain tissue; T, GBM sample). (C) The methylation status of ELFN2 in the GBM samples (n = 50) and normal brain tissues (n = 10) was detected using BSP. The data are presented as the means ± SEM of three independent experiments. *p < 0.05. (D) qRT-PCR was used to detect the expression of ELFN2 in GBM cells and primary GBM cells. The data are presented as the means ± SEM of three independent experiments. (E) Western blotting was used to detect the expression of ELFN2 in GBM cells and primary GBM cells. (F) The methylation status of ELFN2 in GBM cell lines was detected by MSP. U, unmethylated primer; M, methylated primer. (G) Kaplan-Meier curve analysis of 50 astrocytoma patients stratified by ELFN2 methylation. Astrocytoma patients with ELFN2 methylation have a better overall survival. (H) Kaplan-Meier curve analysis of 50 astrocytoma patients stratified by ELFN2 expression. Astrocytoma patients with low ELFN2 expression have a better overall survival.
Table 1.
Correlation between ELFN2 Methylation Status, Protein Expression, and Clinical Parameters of Glioma Patients
| Variable | ELFN2 |
p Value | |
|---|---|---|---|
| Hypomethylation | Methylation | ||
| Total (n = 50) | 41 | 9 | – |
| Expression | |||
| <8 (18) | 12 | 6 | 0.034 |
| ≥8 (32) | 29 | 3 | |
| Sex | |||
| Male (34) | 28 | 6 | 0.925 |
| Female (16) | 13 | 3 | |
| Age (years)a | |||
| <42 (27) | 18 | 9 | 0.525 |
| ≥42 (23) | 23 | 0 | |
| Grade | |||
| Low grade (I + II) (20) | 16 | 4 | 0.764 |
| High grade (III + IV) (30) | 25 | 5 | |
ELFN2, extracellular leucine-rich repeat and fibronectin type III domain-containing 2.
Median age is 42 years.
Table 2.
Correlation between ELFN2 Expression and Clinical Parameters of Glioma Patients
| Variable | ELFN2 |
p Value | |
|---|---|---|---|
| <8 | ≥8 | ||
| Total (n = 50) | 18 | 32 | – |
| Sex | |||
| Male (34) | 11 | 23 | 0.925 |
| Female (16) | 7 | 9 | |
| Age (years)a | |||
| <42 (27) | 9 | 18 | 0.670 |
| >42 | 9 | 14 | |
| Grade | |||
| Low grade (I + II) (26) | 10 | 16 | 0.706 |
| High grade (III + IV) (24) | 8 | 16 | |
ELFN2, extracellular leucine-rich repeat and fibronectin type III domain-containing 2.
Median age is 42 years.
To further confirm the relationship between ELFN2 expression and its hypomethylation, we performed qRT-PCR and western blotting assays to test the expression of ELFN2 (Figures 1D and 1E) and an MSP assay to verify the hypomethylation status of the ELFN2 promoter (Figure 1F). The results showed that ELFN2 promoter hypomethylation was closely related to its high protein expression (Figure S1). The correlations among ELFN2 protein expression, methylation status, and overall survival (OS) were statistically significant (Figures 1G and 1H).
LINC00470 Functions as a Sponge for miR-101 to Facilitate ELFN2 Expression
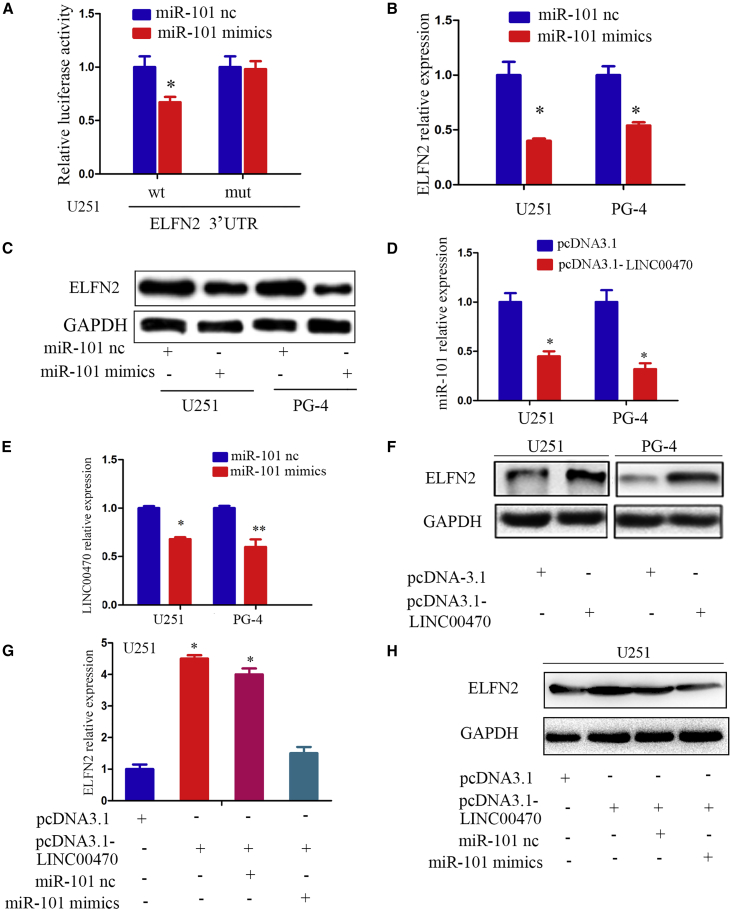
miR-101 has been verified as a suppressive miRNA in multiple cancers, including astrocytoma.15 We performed a luciferase reporter assay and identified and validated ELFN2 as a new direct target of miR-101 (Figure 2A). In addition, the results showed that miR-101 decreased ELFN2 expression in GBM cell lines and primary-cultured GBM cells (Figures 2B and 2C). A bioinformatics analysis using TargetScan and miRanda revealed that miRNA response elements (MREs) of miR-101 were shared by the LINC00470 lncRNA and the 3′ UTR of ELFN2. Therefore, we wondered whether LINC00470 also modulated the expression ELFN2. Our results indicated that LINC00470 and miR-101 inhibited each other (Figures 2D and 2E) and that LINC00470 increased the ELFN2 levels in GBM cells (Figure 2F). The ectopic expression of miR-101 abrogated the increase in ELFN2 expression induced by LINC00470 (Figures 2G and 2H).
Figure 2.
LINC00470 Functions as a Sponge for miR-101 to Facilitate ELFN2 Expression
(A) Luciferase reporter assays were performed to detect the relationship between miR-101 and ELFN2. The data shown are the means ± SEM of six replicates and are representative of three independent experiments. *p < 0.05. (B) qRT-PCR was performed to detect the expression of ELFN2 in GBM cells after transfection with miR-101 mimics. The data are presented as the means ± SEM of three independent experiments. *p < 0.05. (C) Western blotting was performed to detect the expression of ELFN2 in GBM cells after transfection with miR-101 mimics. (D) qRT-PCR was performed to detect the expression of miR-101 in GBM cells after transfection with pcDNA3.1-LINC00470. The data are presented as the means ± SEM of three independent experiments. *p < 0.05. (E) qRT-PCR was performed to detect the expression of LINC00470 in GBM cells after transfection with miR-101. The data are presented as the means ± SEM of three independent experiments. *p < 0.05; **p < 0.01. (F) Western blotting was performed to detect the expression of ELFN2 in GBM cells after transfection with pcDNA3.1-LINC00470. (G) qRT-PCR was performed to detect the expression of ELFN2 in GBM cells after transfection with miR-101 and LINC00470. The data are presented as the means ± SEM of three independent experiments. *p < 0.05. (H) Western blotting was performed to detect the expression of ELFN2 in GBM cells after transfection with miR-101 and LINC00470.
LINC00470 Blocked the Maturation of miR-101 in GBM Cells
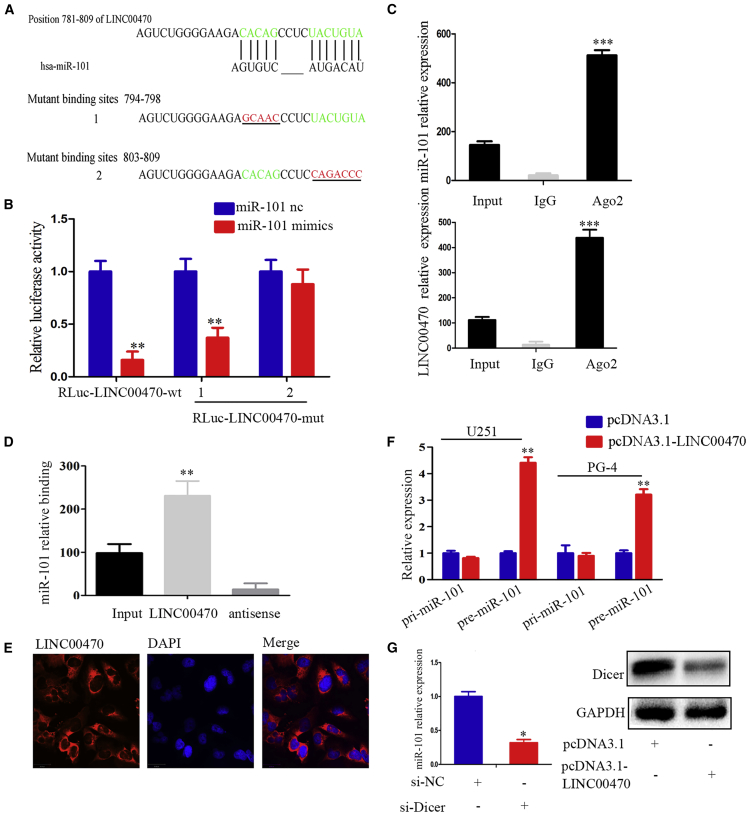
Our results revealed that LINC00470 negatively regulated miR-101 expression in GBM cells (Figures 2D and 2E). We further investigated the relationship between LINC00470 and miR-101 and found that miR-101 had putative binding sites for LINC00470 (Figure 3A). Subsequently, we constructed LINC00470 luciferase reporters that contained miR-101 recognition sequences (RLuc-LINC00470-wt) and mutant derivatives lacking the miR-101 recognition sequences (RLuc-LINC00470-mut1 and -mut2). The miR-101 mimics reduced the luciferase activities of the RLuc-LINC00470-wt reporter but did not affect the luciferase activities of the RLuc-LINC00470-mut2 reporter, indicating that LINC00470 bound to miR-101 through the second binding site (Figure 3B). The miRNAs are known to bind their targets in RNA-induced silencing complex (RISC) components mainly in an AGO2-dependent manner. Therefore, we performed a RIP assay using antibodies against Ago2. Endogenous LINC00470 was specifically enriched in miR-101-transfected U251 cells by AGO2 pull-down (Figure 3C), suggesting that LINC00470 may be a target gene of miR-101.
Figure 3.
LINC00470 Blocks the Maturation of miR-101 in GBM Cells
(A) The alignment of potential LINC00470 base pairing with miR-101 was identified using DIANA LAB (http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php?). Green, binding sites; red, mutation sites. (B) Relative fluorescence activity in U251 cells co-transfected with RLuc-LINC00470-wt/mutant and miR-101 or the negative control. The data are presented as the means ± SEM of three independent experiments. **p < 0.01. (C) Association of LINC00470 and miR-101 with Ago2 in U251 cells. The LINC00470 and miR-101 expression levels were detected by qRT-PCR. The data are presented as the means ± SEM of three independent experiments. ***p < 0.001. (D) The interaction of LINC00470 and miR-101 in U251 cells was detected through RIP assays. The data are presented as the means ± SEM of three independent experiments. **p < 0.01. (E) RNA fluorescence in situ hybridization showing the localization of LINC00470. The nucleus was counterstained with DAPI. Scale bar, 29 μm. (F) The expression of pri-miR-101 and pre-miR-101-1 in U251 and PG-4 cells after transfection with pcDNA3.1-LINC00470 was measured by qRT-PCR. The data are presented as the means ± SEM of three independent experiments. **p < 0.01. (G) Left, the expression of miR-101 in si-Dicer-transfected U251 cells was analyzed by qRT-PCR. The data are presented as the means ± SEM of three independent experiments. *p < 0.05. Right, the expression of Dicer in pcDNA3.1-LINC00470-transfected U251 cells was measured by western blotting.
We subsequently investigated why LINC00470 negatively regulated the expression of miR-101. First, we further validated the interaction between LINC00470 and miR-101 through an RNA pull-down assay. The RNA pulled down with miR-101 was associated with LINC00470 and through a qRT-PCR analysis, we demonstrated that LINC00470 in U251 cells was more significantly associated with miR-101 compared with immunoglobulin G (IgG) (Figure 3D). We synthesized biotin-labeled mature miR-101 and pre-miR-101 probes and found that biotin-labeled mature-miR-101 and pre-miR-101 in U251 cells was more significantly associated with LINC00470 (Figure S2). In situ hybridization assays and qRT-PCR indicated that LINC00470 was localized in the cytoplasm (Figures 3E and S3). miR-101 was enriched in the cytoplasm in GBM cells, consistent with the localization of pre-miR-101, as shown through qRT-PCR (Figure S3). Considering the miRNA biogenesis process, we found that the overexpression of LINC00470 in GBM cells promoted the expression of pre-miR-101 but did not alter the expression of pri-miR-101 (Figure 3F). We also found that the overexpression of LINC00470 inhibited Dicer expression (Figure 3G) and further decreased the Dicer-induced expression of mature miR-101 (Figure 3G). These data demonstrated that LINC00470 blocked the miR-101 maturation process, resulting in reduced levels of mature miR-101.
Knockdown of LINC00470 Reversed the Expression of ELFN2 in GBM Cells through an Epigenetic Regulation Mechanism
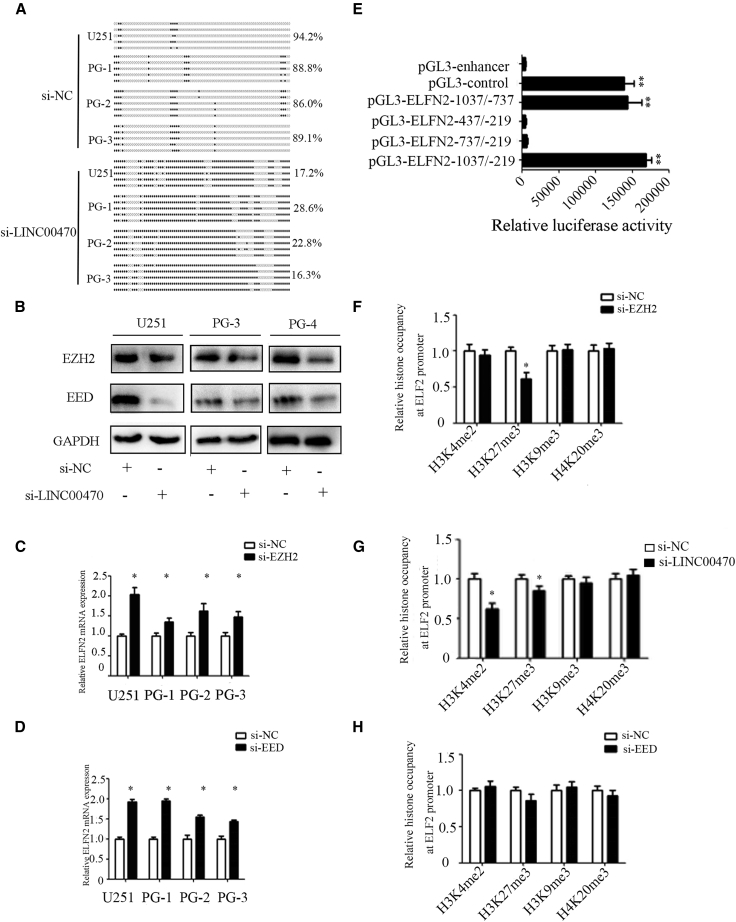
To investigate whether LINC00470 epigenetically regulates ELFN2, BSP assays were performed, and the results indicated that LINC00470 reversed the methylation level of the ELFN2 promoter (Figure 4A). EZH2 and EED, as the core subunits of PRC2, have been shown to correlate with H3K27, H3K4, H3K9, and H4K20. In addition, EZH2 and EED are the direct targets of miR-101.15, 16, 17 We confirmed that the knockdown of LINC00470 decreased the expression of EZH2 and EED (Figure 4B), and the knockdown of both EZH2 and EED promoted the expression of ELFN2 in GBM cells (Figures 4C and 4D). We have performed a “rescue” experiment and found that LINC00470 regulated EZH2 and EED expression via miR-101 (Figure S4). Serial ELFN2 promoter mutants were constructed, and the luciferase reporter assay demonstrated that the core promoter of ELFN2 ranged from 1,037 to 737 (Figure 4E). Subsequently, after the transfection of GBM cells with si-LINC00470, we observed the presence of H3K4me2, H3K27me3, H3K9me3, and H4k20me3 at the core ELFN2 promoter locus. H3K4me2 and H3K27me3 occupancy at the ELFN2 core promoter was decreased in GBM cells transfected with si-LINC00470 compared with the control cells (Figure 4F). Consistently, we observed that H3K27me3 occupancy in the ELFN2 promoter was downregulated in GBM cells in which the expression of EZH2 was knocked down (Figure 4G), whereas the knockdown of EED did not affect the occupancy of H3K4me2, H3K4me2, H3K27me3, H3K9me3, and H4k20me3 in GBM cells (Figure 4H). The above results indicated that the knockdown of LINC00470 reversed the methylation status of ELFN2 by downregulating H3K27me3 occupancy at the core promoter of ELFN2.
Figure 4.
The Knockdown of LINC00470 Reverses the Expression of ELFN2 in GBM Cells through an Epigenetic Regulatory Mechanism
(A) The methylation level of ELFN2 was detected by BSP. (B) Western blotting was performed to detect the expression of EED and EZH2 in GBM cells after LINC00470 knockdown. (C) qRT-PCR was performed to detect the expression of ELFN2 in GBM cells after EZH2 knockdown. The data are presented as the means ± SEM of three independent experiments. *p < 0.05. (D) qRT-PCR was performed to detect the expression of ELFN2 in GBM cells after EED knockdown. The data are presented as the means ± SEM of three independent experiments. *p < 0.05. (E) The promoter activity of ELFN2 core promoter constructs was analyzed by luciferase reporter assays. pGL3-control was used as the positive control, and pGL3-enhancer was used as the negative control. The core promoter region is considered to show higher relative luciferase activity than the pGL3-enhancer. The data are presented as the means ± SEM of three independent experiments. **p < 0.01. (F, G, and H) The histone occupancy of the ELFN2 promoter was affected by si-EZH2 (F), si-LINC00470 (G), and si-EED (H). A ChIP assay was performed to detect the H3K4me2, H3K27me3, H3K9me3, and H4K20me3 occupancy of the ELFN2 core promoter. The data are presented as the means ± SEM of three independent experiments. *p < 0.05.
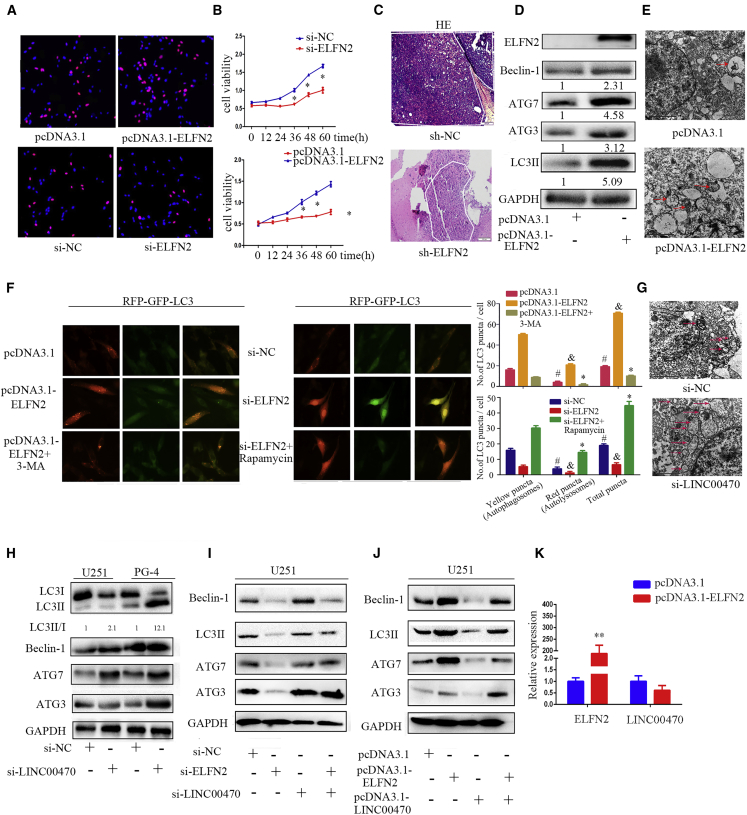
ELFN2 Is a Putative Oncogene, and LINC00470 Inhibits ELFN2-Induced GBM Cell Autophagy
To further understand the biological function of ELFN2 in GBM cells, we performed EdU and CCK8 assays. The knockdown of ELFN2 inhibited the proliferation of GBM cells (Figures 5A, 5B, and S5), and the overexpression of ELFN2 promoted GBM cell proliferation (Figures 5A, 5B, S5). H&E staining revealed that the knockdown of ELFN2 inhibited the growth of intracranial transplanted tumors and that the knockdown of ELFN2 decreased the expression of Ki-67 and ELFN2 in an intracranial orthotopic transplanted model (Figures 5C and S5). We subsequently investigated why ELFN2 promotes GBM cell proliferation and found that ELFN2 promoted the expression of the autophagy biomarkers Beclin-1, ATG7, ATG3, and LC3 II in GBM cells (Figure 5D). Transmission electron microscopy further verified that ELFN2 promoted autophagosome accumulation in the cytoplasm (Figure 5E). We further validated the autophagy flux through an image-based colocalization analysis of RFP-GFP-LC3. GFP fluorescence is quenched by the acidic pH within a lysosome, allowing differentiation between autophagosomes (Figure 5E, yellow) and autolysosomes (Figure 5E, red). The overexpression of ELFN2 resulted in an accumulation of yellow puncta, indicating autophagosome accumulation. Interestingly, we also noted that the ratio of red puncta increased after treatment with ELFN2 (Figure 5F). We used 3-MA, an autophagy inhibitor, as a positive control, and this control showed a low number of red puncta (Figure 5F). To validate the function of ELFN2 in promoting autophagy, we monitored the autophagy flux through the same assay using cells in which ELFN2 was knocked down. In these cells, we observed decreases in the numbers of yellow and red puncta. Rapamycin, an autophagy inductor, reversed the inhibition of cell autophagy inhibited by the knockdown of ELFN2. These results indicated that ELFN2 promoted GBM cell autophagy, and we also detected the expression of autophagy markers after transfection of a miR-101 inhibitor, which indicated that miR-101 inhibits GBM cell autophagy (Figure S6).
Figure 5.
ELFN2 Is a Putative Oncogene and LINC00470 Inhibits ELFN2-Induced GBM Cell Autophagy
(A) An EdU assay was performed to assess the proliferation of GBM cells transfected with pcDNA3.1-ELFN2 or si-ELFN2. The data shown are the means ± SEM of three independent experiments; red scale bars, 200 μm. (B) A CCK8 assay was performed to determine the viability of GBM cells transfected with si-ELFN2 or pcDNA3.1-ELFN2. The data shown are the means ± SEM of three independent experiments. *p < 0.05. (C) H&E staining of shLINC00470-induced tumors in the coronal section of rats. (D) Western blotting was performed to detect the level of autophagy markers in GBM cells transfected with pcDNA3.1-ELFN2. (E) Electron microscopy was performed to detect U251 cell autophagy after transfection with pcDNA3.1-ELFN2. (F) Autophagy flux in ELFN2-induced U251 cells was assessed through an image-based colocalization analysis of RFP-GFP-LC3. #, &, *p < 0.05. (G) Electron microscopy was performed to detect the autophagy of U251 cells transfected with si-LINC00470. (H) Western blotting was performed to detect the level of autophagy markers in GBM cells after knockdown of LINC00470. (I) Western blotting was performed to detect the level of autophagy markers in GBM cells transfected with si-ELFN2 and si-LINC00470. (J) Western blotting was performed to detect the level of autophagy markers in GBM cells transfected with pcDNA3.1-ELFN2 and pcDNA3.1-LINC00470. (K) qRT-PCR was performed to detect the expression of LINC00470 in GBM cells overexpressing ELFN2. The data are presented as the means ± SEM of three independent experiments. **p < 0.01.
We have demonstrated that LINC00470 is a putative onco-RNA that inhibits GBM cell autophagy.22 In the present study, transmission electron microscopy further verified that the knockdown of LINC00470 promoted GBM cell autophagosome accumulation in the cytoplasm (Figure 5G), and the knockdown of LINC00470 increased the LC3 II/LC3 I ratio and the expression of Beclin-1, ATG3, and ATG7 (Figure 5H).
LINC00470 and ELFN2 were identified as oncogenes, and LINC00470 promoted the expression of ELFN2. However, LINC00470 and ELFN2 exerted opposite effects on autophagy. We further co-transfected LINC00470 and ELFN2 and found that the autophagy of GBM cells was restrained by the overexpression of LINC00470 and ELFN2. In addition, the knockdown of both LINC00470 and ELFN2 increased cell autophagy (Figures 5I and 5J), and the overexpression of ELFN2 did not affect LINC00470 expression (Figures 5K).
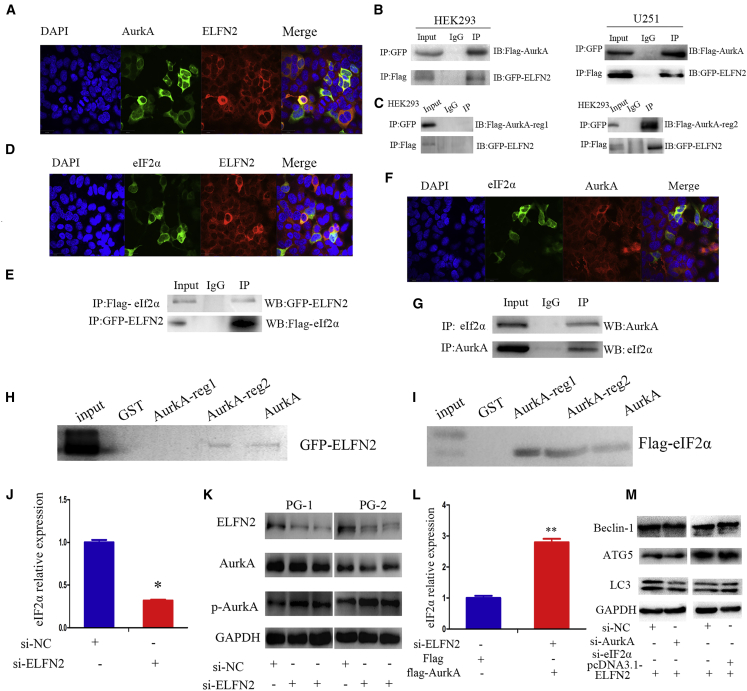
ELFN2 Binds to AurkA and eIF2α and Upregulates eIF2α by Regulating the Kinase Activity of AurkA
To understand the mechanism of ELFN2 on cell autophagy, an interaction between AurkA and ELFN2 was predicted using Scansite 3.0 software. We confirmed that ELFN2 and AurkA were co-located in the cytoplasm of HEK293 cells (Figures 6A and S7). Co-immunoprecipitation (coIP) assays confirmed that ELFN2 interacted with AurkA in HEK293 and U251 cells (Figure 6B; Figure S8). ELFN2 interacted with the reg2 domain of AurkA, which is a kinase domain (Figure 6C, right; Figure S8), but not with the reg1 domain of AurkA, which is a nonkinase domain (Figure 6C, left; and Figure S8).
Figure 6.
ELFN2 Binds to AurkA and eIF2α and Upregulates eIF2α by Regulating the Kinase Activity of AurkA
(A) Representative confocal and immunofluorescence images showing the co-localization of ELFN2 (red) and AurkA (green) in HEK293 cells. (B) coIP analysis showing the interaction between ELFN2 and AurkA in HEK293 and U251 cells. (C) coIP analysis showing the interaction between ELFN2 and AurkA domains in HEK293 cells. (D) Representative confocal and immunofluorescence images showing the co-localization of ELFN2 (red) and eIF2α (green) in HEK293 cells. (E) coIP analysis showing the interaction between ELFN2 and eIF2α in HEK293 cells. (F) Representative confocal and immunofluorescence images showing the co-localization of AurkA (red) and eIF2α (green) in HEK293 cells. (G) coIP analysis showing the interaction between AurkA and eIF2α in HEK293 cells. (H) GST pull-down assays showed that the reg2 domain of AurkA pulled down ELFN2. (I) GST pull-down assays showed that the reg1 and reg2 domains of AurkA pulled down eIF2α. (J) qRT-PCR was performed to detect the expression of eIF2α in GBM cells after the ELFN2 knockdown. The data are presented as the means ± SEM of three independent experiments. *p < 0.05. (K) Western blotting was performed to detect the levels of AurkA and p-AurkA in GBM cells transfected with si-ELFN2. (L) qRT-PCR was performed to detect the expression of eIF2α in GBM cells after ELFN2 knockdown and AurkA overexpression. The data are presented as the means ± SEM of three independent experiments. **p < 0.01. (M) Western blotting was performed to detect the level of autophagy markers in ELFN2-overexpessing GBM cells after AurkA or eIF2α knockdown.
eIF2α is central to the onset of protein synthesis and its modulation in response to a physiological stimulus. Specifically, eIF2α is necessary for the induction of autophagy in response to amino acid deprivation. Scansite software predicted that ELFN2 can interact with eIF2α. Our confocal laser imaging results demonstrated that ELFN2 and eIF2α co-localized in the cytoplasm of HEK293 cells (Figure 6D), and a coIP assay verified their interaction (Figure 6E). We also confirmed that AurkA interacts with eIF2α in HEK293 cells by confocal laser imaging and coIP (Figures 6F and 6G). A GST pull-down assay confirmed that ELFN2 bound to the reg2 domain of AurkA (Figure 6H) and that eIF2α bound to the reg2 and reg1 domains of AurkA (Figure 6I). coIP indicated that we had been unable to confirm the binding between ELFN2 and eIF2α, but when we knocked down AurkA in U251 cells, we found that the combination of ELFN2 and eIF2α was enhanced; these results suggested that AurkA may be used as a blocker to obstruct the binding of ELFN2 and eIF2α (Figure S9).
Further studies indicated that the knockdown of ELFN2 decreased eIF2α (Figure 6J), increased p-AurkA, and decreased AurkA expression in GBM cells (Figure 6K). These results indicated that ELFN2 inhibited the kinase activity of AurkA. We further assessed whether ELFN2 upregulated eIF2α in an AurkA-dependent manner. Because AurkA is a serine/threonine kinase, we reasoned that the activation of AurkA might upregulate eIF2α induced by ELFN2 and found that the ectopic expression of AurkA promoted eIF2α mRNA expression by knockdown of ELFN2 (Figure 6l). These results suggested that ELFN2 upregulated eIF2α through a decrease in the inhibitory effect of ELFN2 on AurkA activity. We subsequently monitored markers of autophagy after the knockdown of AurkA or eIF2α and found that the knockdown of AurkA or eIF2α did not decrease the autophagy level of GBM cells (Figure 6M). These results suggested that ELFN2 affected AurkA- and eIF2α-induced autophagy.
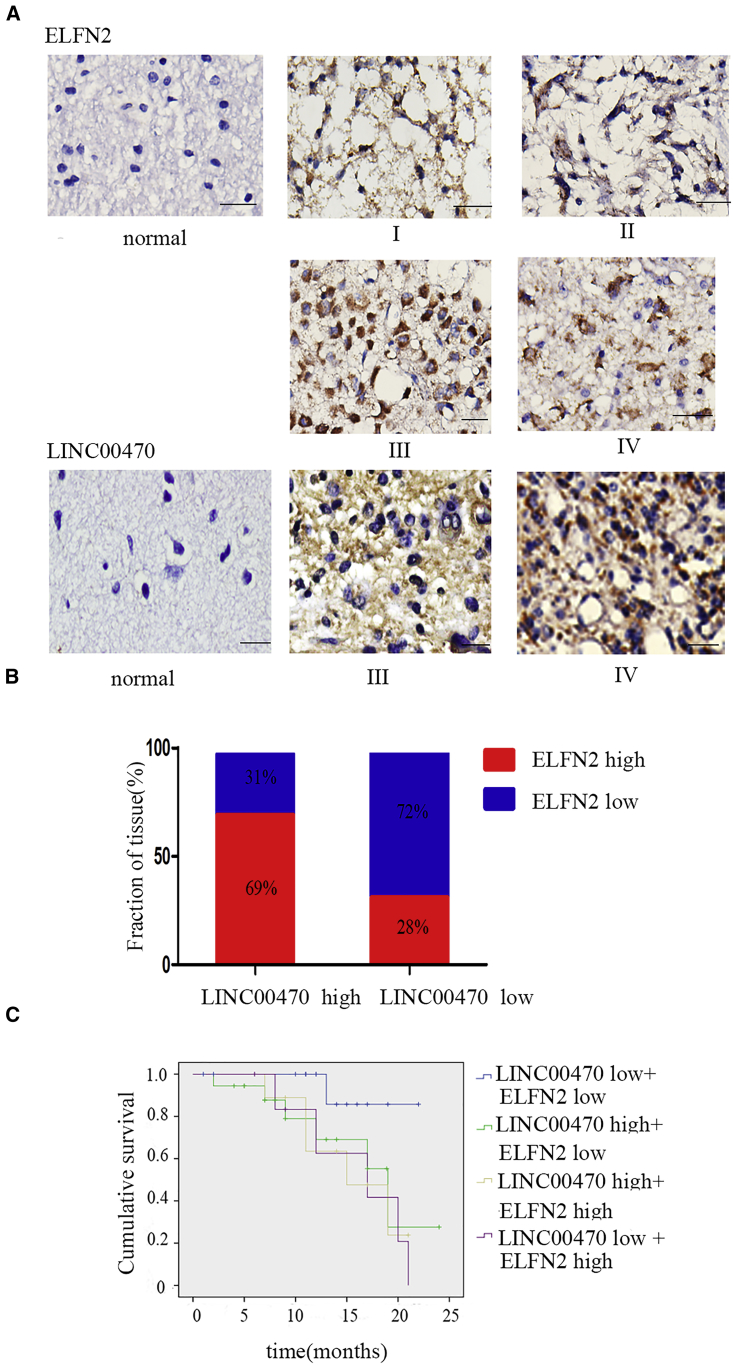
Association between LINC00470 and ELFN2 Expression in Clinical Samples of Astrocytoma
By in situ hybridization (ISH) and immunohistochemistry (IHC), we detected LINC00470 and ELFN2 expression, respectively, in 75 paraffin-embedded astrocytoma tissue samples and 18 normal brain tissues (Figure 7A), indicating that the expression of LINC00470 was positively correlated with ELFN2 expression (Figure 7B). An analysis of the samples in the high-LINC00470-expression group showed that 69% and 31% of the samples showed high and low ELFN2 expression, respectively, whereas 28% and 78% of the samples in the low-LINC00470-expression group showed high and low ELFN2 expression, respectively. Furthermore, through a Kaplan-Meier analysis, we plotted the survival curve of 75 astrocytoma patients. As illustrated in Figure 7C, GBM patients with low ELFN2 expression had a better prognosis. Furthermore, we examined the prognostic value of LINC00470 and ELFN2 on different subgroups of astrocytoma patients. Patients with low expression of LINC00470 and ELFN2 exhibited longer survival than the other subgroups (Figure 7C).
Figure 7.
Association between LINC00470 and ELFN2 Expression in Clinical Samples of Astrocytoma
(A) The expression level of ELFN2 was detected by immunofluorescence, and the expression level of LINC00470 was detected by in situ hybridization. Scale bars, 20 μm. (B) The correlation between LINC00470 expression and ELFN2 levels was analyzed using Spearman’s rank test. (C) Kaplan-Meier analysis of the overall survival of 75 astrocytomas divided into high- and low-risk groups based on the LINC00470 and ELFN2 expression levels.
Discussion
Autophagy is a type of programmed cell death that decides the fate of cells of malignant neoplasms.23 Autophagy is an evolutionarily conserved process in eukaryotes through which the cytoplasmic cargo sequestered inside double-membrane vesicles is delivered to the lysosome for degradation. This process is a crucial mechanism that responds to either extra or intracellular stress.23 Autophagy can perform as a tumor suppressor by activating pro-autophagic genes and blocking antiautophagic genes in oncogenesis.24, 25 However, autophagy can also play a pro-tumor role in carcinogenesis by regulating a number of pathways involving Beclin-1, Bcl-2, phosphatidylinositol 3-kinase (PI3K), and p53.26, 27, 28, 29 Consequently, both autophagy inhibitors, such as chloroquine, and autophagy activators, including proteasome and MTORC1 inhibitors, are currently being investigated in clinical trials.30, 31 Yang and Klionsky32 found that autophagy not only promotes the proliferation of tumor cells but also induces cell death. When analyzing relationships between autophagy and cancer, a common challenge is to determine whether autophagy protects cell survival or contributes to cell death. In this study, we identified ELFN2 as a novel oncogene that promotes cell proliferation by inducing GBM cell autophagy.
Several lines of evidence suggest that the eIFα/ATF4 pathway could play a key role in autophagy regulation.33 ELFN2 is an inhibitor of protein phosphatase 1 (PP1), but the function of ELFN2 has not been studied. We first demonstrated that eIf2α, an autophagy activator, interacted with ELFN2, but ELFN2 did not directly bind to eIf2α. Furthermore, we confirmed that AurkA, an autophagy inhibitor, was a mediator of the interaction between ELFN2 and eIf2α. AurkA bound to ELFN2 by its reg2 kinase domain and bound to eIf2α by its reg2 kinase domain and reg1 nonkinase domain. AurkA is also a serine/threonine kinase that functions as a key regulator of mitosis.34 Abnormal expression or activation of AurkA leads to the development of cancer.35 AURKA inhibition (ALS) promotes cellular apoptosis and autophagy in breast cancer cells through modulation of the p38 mitogen-activated protein kinase (MAPK)/Akt/mammalian target of rapamycin (mTOR) pathway.35 The regulatory relationship between AurkA and eIf2α has not been reported in the literature. Our results first indicated that ELFN2 decreased the expression of p-AurkA, increased the expression of eIf2α, and promoted GBM cell autophagy. The eIF2α expression induced by ELFN2 was dependent on the activation of AurkA.
Although several thousand lncRNAs have been annotated in the human genome, only a limited number of lncRNAs have been functionally characterized thus far.36 Previous studies of these well-characterized lncRNAs have demonstrated that lncRNAs can function as guides for protein-DNA interactions, scaffolds for protein-protein interactions, decoys for proteins or miRNAs, or enhancers of their neighboring genes.37, 38 Consistent with the diverse biochemical functions of lncRNAs, lncRNAs have been shown to regulate various biological processes, such as cell proliferation, differentiation, survival, and migration. Their dysregulation impacts different human diseases, such as cancer and metabolic diseases.39, 40 LncRNAs located in the nucleus always play a role at the level of pre-transcription or transcription, whereas cytoplasmic lncRNAs often function as competing endogenous RNAs and sponge miRNAs, thus regulating the expression of target mRNAs.41 In this study, we first confirmed that ELFN2 is the direct target of miR-101, whereas LINC00470 functions as a sponge of miR-101 to increase ELFN2 expression in GBM cells. LINC00470 was mainly located in the cytoplasm and exerted its regulatory role at the post-transcriptional level. Regulatory interactions were found between LINC00470 and miR-101. LINC00470 was found to be only one of the direct targets of miR-101 but also directly inhibited the expression of miR-101. ELFN2 did not alter the expression of LINC00470. In contrast, the promoter hypomethylation modification of ELFN2 also contributed to the high expression of this gene in GBM. More than half of the astrocytoma samples showed increased ELFN2 expression, but 82% exhibited notable hypomethylation of the promoter, indicating that ELFN2 is subject to pre-transcriptional regulation. LINC00470, as an effective sponge of miR-101, reversed ELFN2 hypomethylation by downregulating H3K27me3 occupancy at the core promoter of ELFN2, and this process enriches the regulation of the expression of a gene by lncRNAs at the epigenetic level. More interestingly, we first demonstrated that in GBM cells in which LINC00470 and ELFN2 co-existed, LINC00470 increased the expression of ELFN2 and distracted the effect of ELFN2 on promoting cell autophagy. The co-existence of LINC00470 and ELFN2 inhibited GBM cell autophagy because ELFN2 was mainly regulated by LINC00470 in GBM cells. LINC00470 was observed to function as a miR-101 sponge to release ELFN2 expression, but ELFN2 did not affect LINC00470 expression. Therefore, LINC00470 was found to play a leading role in the regulation of autophagy in the presence of the oncogenes LINC00470 and ELFN2. The knockdown of AurkA or eIF2α did not decrease the GBM cell autophagy levels. The above-mentioned results indicated that ELFN2 promotes autophagy through AurkA and eIf2α.
Finally, our findings have therapeutic implications. LncRNAs have been reported to be potential biomarkers in human cancers. For example, HOTAIR has been identified as a reliable biomarker of poor prognosis in colorectal cancer and hepatocellular carcinoma.42 Several lncRNAs have been reported to be involved in the progression of OS.43 We found that patients with high LINC00470 levels tended to also exhibit high ELFN2 levels. Low expression of ELFN2 has been associated with a longer survival time in astrocytoma patients. Low expression of LINC00470 and ELFN2 in astrocytoma patients is associated with a better survival.
In conclusion, our research has demonstrated the following. First, we confirmed that ELFN2 is a novel oncogene in GBM that inhibits the activation of AurkA by directly binding to AurkA through its reg2 kinase domain, inhibits the expression of eIF2a, and promotes autophagy in GBM cells. Second, demethylation of the ELFN2 promoter region and high expression of LINC00470 may be one reason for the high expression of ELFN2 observed in GBM cells. LINC00470 not only directly promotes the expression of ELFN2 through adsorption of miR-101 but also affects the methylation level of ELFN2 by regulating the hypomethylation modification of ELFN2. Third, compared with the oncogene ELFN2, LINC00470 plays a dominant role in the regulation of GBM cell autophagy. Therefore, LINC00470 has significant prognostic value in GBM and provides clinical information: low expression of LINC00470 and ELFN2 is associated with better prognosis for GBM patients.
Materials and Methods
Antibodies and Reagents
The reagents, chemicals, and antibodies used in this study are as follows: LC3 (Cell Signaling Technology, 12741), Beclin-1 (Cell Signaling Technology, 3495), Atg5 (Cell Signaling Technology, 12994), Atg12 (Cell Signaling Technology, 4180), Atg7 (Cell Signaling Technology, 8558), Atg3 (Cell Signaling Technology, 3415), GFP (Proteintech, 50430-2-AP), ELFN2 (Sigma, HPA00781), phospho-AurkA (Cell Signaling Technology), AurkA (Cell Signaling Technology, 14475), glyceraldehyde-3-phosphate dehydrogenase (GAPDH; Proteintech, 60004-1-Ig), FLAG (Sigma-Aldrich, F3165), and rapamycin (MedChemExpress, HY-10219).
Astrocytoma Tissues and Cell Lines
We obtained astrocytoma samples and clinical data from Xiangya Hospital, Central South University, and all patients provided informed permission for the collection and use of these samples. We successfully cultivated several primary GBM cells.18 U251 and U87 cells were obtained from the Cell Center of Peking Union Medical College (Beijing, China). The U251 cells were authenticated as originating from the ATCC by short tandem repeat profiling, and U87 cells were found to show 95% similarity with GBM cells from the ATCC by short tandem repeat profiling and 97% similarity with GBM cells from the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ) by short tandem repeat profiling. U251, U118, and primary cells were cultured in DMEM with 10% fetal bovine serum (FBS), and U87 cells were cultured in modified Eagle’s medium (MEM) with 10% FBS at 37°C under 5% CO2 conditions.
Genomic DNA Isolation and Bisulfite DNA Treatment
Genomic DNA isolated from each cell line, primary astrocytoma cells, astrocytoma tissues and normal brain tissues were processed as described previously.16
BSP and MSP
BSP and MSP studies were conducted as described previously.17
miRNA and siRNA Transfection
miR-101 mimics (5′-UACAGUACUGUGAUAACUGAA-3′), ELFN2 siRNA ([1] 5′-GCACAUCAAUAGCACCGUGT-3′, [2] 5′-GCAGUACAACAACAGCUACTT-3′, and [3] 5′-GCCACCAAAGGCAACUAUATT-3′), AURKA siRNA ([1] 5′-GCCAAACTTCGCCGATTCT-3′ and [2] 5′-ATCCTTGAGAACCACAAGT-3′), eIF2α-siRNA ([1] 5′-GGGUCUUUGAUGACAAGUATT-3′ and [2] 5′-CCUCCUCGGUAUGUAAUGATT-3′), and their controls were synthesized by GenePharma (Shanghai, China). Cell transfection was performed using Lipofectamine 3000 (Invitrogen-Life Technologies, Carlsbad, CA, USA) per the manufacturer’s instructions.
Luciferase Reporter Assay
These procedures were conducted as previously described.17 U251 cells were co-transfected with the pGL3 plasmid containing the ELFN2 promoter and pRL-TK (Promega), miR-101 mimics, and RLuc-LINC00470-wt or mutant. Forty-eight hours after transfection, the cells were collected and analyzed using the Dual-Luciferase Reporter Assay System (Promega).
Western Blotting
The cytosol and nuclear proteins in the cells were separated on SDS-polyacrylamide gels, electrophoretically transferred to polyvinylidene difluoride membranes (Merck Millipore, ISEQ00010) and detected with antibodies. ChemicalDoc XRSþ was used to quantify the intensities of the protein fragments (Bio-Rad, Berkeley, CA, USA).
Confocal Microscopy and Immunofluorescence Staining
For analysis of the co-localization of ELFN2, AURKA, and eIF2α, we constructed GFP-ELFN2, GFP-AURKA, FLAG-AURKA, and FLAG-eIF2α plasmids. HEK293 cells were transfected with the GFP-tagged ELFN2 plasmid and the FLAG-tagged AURKA, GFP-AURKA, or FLAG-tagged eIF2α plasmid. Forty-eight hours after transfection, the cells were fixed with 4% paraformaldehyde, permeabilized with 0.25% Triton X-100 in PBS, incubated with anti-FLAG (Sigma-Aldrich, F3165) antibodies for 12 hr at 4°C, and stained with Alexa Fluor 488 antibodies (Thermo Fisher, A11029). The cells were subsequently incubated with DAPI (Beyotime Biotechnology, C1102), and images were captured with a fluorescence microscope (Olympus). The autophagic vacuoles were quantified by counting the numbers of LC3 puncta.
Transmission Electron Microscopy
U251 cells were transfected as previously indicated for 48 hr and then fixed with 1% w/v OsO4 in 0.12 M sodium cacodylate buffer for 24 hr. The samples were dehydrated in a graded series of ethanol, transferred to propylene oxide, embedded in Epon according to standard procedures, cut with a microtome, and collected on copper grids with Formvar-supporting membranes. Sections were stained with uranyl acetate and lead citrate. Imaging was performed using a HITACHI-7500 transmission electron microscope.
Co-immunoprecipitation
For the detection of AURKA, ELFN2, and eIF2α interactions, the cells were prepared and lysed with GLB+ buffer (10 mM Tris-HCl [pH 7.5], 300 mM NaCl, 10 mM EDTA, and 0.5% Triton X-100) containing a protease inhibitor cocktail (Selleck, B14001). The cell lysates were incubated with antibodies for 12 hr at 4°C and then with Protein G beads (Thermo Fisher, 20399) for 4 hr at 4°C. After the beads were washed and 50 μL of GLB+ buffer was boiled, the lysates were subjected to western blotting.
qRT-PCR, Immunohistochemical Staining, and ChIP Assay
These procedures were conducted as previously described.11, 15 The details of the primers are as follows:
BSP:ELFN2, F, 5′-AAGTTTGTTTTTAGTTGTTGAATG-3′; R, 5′-AACAACTCCTCACCCAAATC-3′
ELFN2-M, F, 5′-GAAAGGTTTGCGGAGGGTATC-3′; R, 5′-GCCCGAAAAAAACCAAACGA-3′
ELFN2-UM, F, 5′-GGAAAGGTTTGTGGAGGGTATTG-3′; R, 5′-AAAAACACCCAAAAAAAACCAAACA-3′
ELFN2, F, 5′-CGCAGCACATCAATAGCACC-3′; R, 5′-AGAGGTCGCACTCACAGTTG-3′
EIF2α, F, 5′-CACCTGGACCCCAACCATAC-3′; R, 5′-GCTTCCTGCTTTGCAGCTTT-3′
LINC00470, F, 5′-CGTAAGGTGACGAGGAGCTG-3′; R, 5′-GGGGAATGGCTTTTGGGTCA-3′.
CCK8 Assay and EdU Incorporation Assay
These procedures were described in detail previously.11
RNA Pull-Down Assays
LINC00470 Pull-Down miR-101
Biotin-labeled RNA was transcribed in vitro with the Biotin RNA Labeling Mix (Roche) and T7 RNA polymerase (Ambion). Biotin-labeled full-length LINC00470 RNA or the antisense LINC00470 were heated to 60°C for 10 min, cooled to 4°C, and then incubated with streptavidin beads at 37°C for 3 hr. The RNA was mixed with cellular protein extracts from U251 cells (10-cm dish) overnight at 4°C on a rotator.
miR-101 Pull-Down LINC00470
Control miR (5′-UUGUCAUGUGUGAUAACUGAA-3′-biotin) or 3′ biotin-labeled miR-101 (5′-UACAGUACUGUGAUAACUGAA-3′-biotin) was transfected at 80 nM. Forty-eight hours after transfection, whole-cell lysates were harvested, and for each sample, streptavidin-Dynabeads were coated with 5 μL of yeast tRNA and incubated with rotation at 4°C for 4 hr. The beads were then washed with 500 μL of lysis buffer and resuspended in 50 μL of lysis buffer. The sample lysates were mixed with pre-coated beads (50 μL per sample) and incubated overnight at 4°C on a rotator.
The following day, the lysates were washed three times with 1 mL of lysis buffer. One milliliter of TRIzol (Invitrogen, Carlsbad, CA, USA) was added for RNA extraction using the chloroform-isopropanol method, and detection was then performed by qRT-PCR.
Intracranial Implantation Sprague-Dawley Rat Model
All animal experiments were approved by the Animal Care and Use Committee of Central South University. Sprague-Dawley rat model was performed as previously described.11 Sprague-Dawley rats that were 6 weeks old and 200–250 g were anesthetized by intraperitoneal injection of ketamine (40 mg/kg), and U251 cells were subcutaneously injected into brain parenchyma at a concentration of 1 × 106 cells rat. The cyclophosphamide was injected into Sprague-Dawley rats every 4 days.
Statistical Analyses
The statistical analyses were performed with Student’s t test and ANOVA. A p value less than 0.05 was considered to indicate statistical significance. The data are presented as the means ± SEM from at least three separate experiments. The difference in the ELFN2 promoter methylation status between the normal brain tissues and the glioma tissues was examined using an independent sample t test. The relationships among the ELFN2 methylation status, protein expression, and clinicopathological parameters were examined using the χ2 test. The OS curves were calculated using the Kaplan-Meier method. All the statistical analyses were performed using SPSS16.0 for Windows (SPSS, Chicago, IL, USA).
Author Contributions
M.W. and C.L. designed the experiments and wrote the paper; C.L., H.F., X.L., Q. Lei, Y.Z., and Y.S. conducted the experiments, X.S., Q. Liu, and Q. Lei collected specimens; M.W. and G.L. supervised the study.
Conflicts of Interest
The authors declare no conflict of interest.
Acknowledgments
This study was supported by grants from the National Science Foundation of China (81272297), the National Key Technology Research and Development Program of the Ministry of Science and Technology of China (2014BAI04B02), the 111 Project (111-2), the National Science Foundation of China Youth Fund (81702477), and the Graduate Research and Innovation Projects of Central South University (2017zzts017 and 2017zzts012).
Footnotes
Supplemental Information includes nine figures and can be found with this article online at https://doi.org/10.1016/j.ymthe.2018.06.019.
Supplemental Information
References
- 1.Krauze A.V., Myrehaug S.D., Chang M.G., Holdford D.J., Smith S., Shih J., Tofilon P.J., Fine H.A., Camphausen K. A Phase 2 Study of Concurrent Radiation Therapy, Temozolomide, and the Histone Deacetylase Inhibitor Valproic Acid for Patients With Glioblastoma. Int. J. Radiat. Oncol. Biol. Phys. 2015;92:986–992. doi: 10.1016/j.ijrobp.2015.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Network T.C. Corrigendum: Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2013;494:506. doi: 10.1038/nature11903. [DOI] [PubMed] [Google Scholar]
- 3.Zhang H., Chen Z., Wang X., Huang Z., He Z., Chen Y. Long non-coding RNA: a new player in cancer. J. Hematol. Oncol. 2013;6:37. doi: 10.1186/1756-8722-6-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mathew L.K., Skuli N., Mucaj V., Lee S.S., Zinn P.O., Sathyan P., Imtiyaz H.Z., Zhang Z., Davuluri R.V., Rao S. miR-218 opposes a critical RTK-HIF pathway in mesenchymal glioblastoma. Proc. Natl. Acad. Sci. USA. 2014;111:291–296. doi: 10.1073/pnas.1314341111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Papagiannakopoulos T., Shapiro A., Kosik K.S. MicroRNA-21 targets a network of key tumor-suppressive pathways in glioblastoma cells. Cancer Res. 2008;68:8164–8172. doi: 10.1158/0008-5472.CAN-08-1305. [DOI] [PubMed] [Google Scholar]
- 6.Brennan C.W., Verhaak R.G., McKenna A., Campos B., Noushmehr H., Salama S.R., Zheng S., Chakravarty D., Sanborn J.Z., Berman S.H., TCGA Research Network The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu D.G., Wang Y.Y., Fan L.G., Luo H., Han B., Sun L.H., Wang X.F., Zhang J.X., Cao L., Wang X.R. MicroRNA-7 regulates glioblastoma cell invasion via targeting focal adhesion kinase expression. Chin. Med. J. (Engl.) 2011;124:2616–2621. [PubMed] [Google Scholar]
- 8.Shi L., Cheng Z., Zhang J., Li R., Zhao P., Fu Z., You Y. hsa-mir-181a and hsa-mir-181b function as tumor suppressors in human glioma cells. Brain Res. 2008;1236:185–193. doi: 10.1016/j.brainres.2008.07.085. [DOI] [PubMed] [Google Scholar]
- 9.Li M., Li J., Liu L., Li W., Yang Y., Yuan J. MicroRNA in Human Glioma. Cancers (Basel) 2013;5:1306–1331. doi: 10.3390/cancers5041306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han L., Zhang K., Shi Z., Zhang J., Zhu J., Zhu S., Zhang A., Jia Z., Wang G., Yu S. LncRNA profile of glioblastoma reveals the potential role of lncRNAs in contributing to glioblastoma pathogenesis. Int. J. Oncol. 2012;40:2004–2012. doi: 10.3892/ijo.2012.1413. [DOI] [PubMed] [Google Scholar]
- 11.Liu C., Sun Y., She X., Tu C., Cheng X., Wang L., Yu Z., Li P., Liu Q., Yang H. CASC2c as an unfavorable prognosis factor interacts with miR-101 to mediate astrocytoma tumorigenesis. Cell Death Dis. 2017;8:e2639. doi: 10.1038/cddis.2017.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Song X., Zhang N., Han P., Moon B.S., Lai R.K., Wang K., Lu W. Circular RNA profile in gliomas revealed by identification tool UROBORUS. Nucleic Acids Res. 2016;44:e87. doi: 10.1093/nar/gkw075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Z., Li D., Wu M., Xiang B., Wang L., Zhou M., Chen P., Li X., Shen S., Li G. Promoter hypermethylation-mediated inactivation of LRRC4 in gliomas. BMC Mol. Biol. 2008;9:99. doi: 10.1186/1471-2199-9-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li P., Wu M. Epigenetic Mechanisms of Glioblastoma. In: De Vleeschouwer S., editor. Glioblastoma. Codon Publications; 2017. pp. 43–58. [PubMed] [Google Scholar]
- 15.Xiaoping L., Zhibin Y., Wenjuan L., Zeyou W., Gang X., Zhaohui L., Ying Z., Minghua W., Guiyuan L. CPEB1, a histone-modified hypomethylated gene, is regulated by miR-101 and involved in cell senescence in glioma. Cell Death Dis. 2013;4:e675. doi: 10.1038/cddis.2013.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lei Q., Liu X., Fu H., Sun Y., Wang L., Xu G., Wang W., Yu Z., Liu C., Li P. miR-101 reverses hypomethylation of the PRDM16 promoter to disrupt mitochondrial function in astrocytoma cells. Oncotarget. 2016;7:5007–5022. doi: 10.18632/oncotarget.6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu X., Lei Q., Yu Z., Xu G., Tang H., Wang W., Wang Z., Li G., Wu M. MiR-101 reverses the hypomethylation of the LMO3 promoter in glioma cells. Oncotarget. 2015;6:7930–7943. doi: 10.18632/oncotarget.3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li P., Feng J., Liu Y., Liu Q., Fan L., Liu Q., She X., Liu C., Liu T., Zhao C. Novel Therapy for Glioblastoma Multiforme by Restoring LRRC4 in Tumor Cells: LRRC4 Inhibits Tumor-Infitrating Regulatory T Cells by Cytokine and Programmed Cell Death 1-Containing Exosomes. Front. Immunol. 2017;8:1748. doi: 10.3389/fimmu.2017.01748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hendrickx A., Beullens M., Ceulemans H., Den Abt T., Van Eynde A., Nicolaescu E., Lesage B., Bollen M. Docking motif-guided mapping of the interactome of protein phosphatase-1. Chem. Biol. 2009;16:365–371. doi: 10.1016/j.chembiol.2009.02.012. [DOI] [PubMed] [Google Scholar]
- 20.Dolan J., Walshe K., Alsbury S., Hokamp K., O’Keeffe S., Okafuji T., Miller S.F., Tear G., Mitchell K.J. The extracellular leucine-rich repeat superfamily; a comparative survey and analysis of evolutionary relationships and expression patterns. BMC Genomics. 2007;8:320. doi: 10.1186/1471-2164-8-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y.Q., Zhang J.J., Song H.J., Li D.W. Overexpression of CST4 promotes gastric cancer aggressiveness by activating the ELFN2 signaling pathway. Am. J. Cancer Res. 2017;7:2290–2304. [PMC free article] [PubMed] [Google Scholar]
- 22.Liu C., Zhang Y., She X., Fan L., Li P., Feng J., Fu H., Liu Q., Liu Q., Zhao C. A cytoplasmic long noncoding RNA LINC00470 as a new AKT activator to mediate glioblastoma cell autophagy. J. Hematol. Oncol. 2018;11:77. doi: 10.1186/s13045-018-0619-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levy J.M.M., Towers C.G., Thorburn A. Targeting autophagy in cancer. Nat. Rev. Cancer. 2017;17:528–542. doi: 10.1038/nrc.2017.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Galluzzi L., Bravo-San Pedro J.M., Levine B., Green D.R., Kroemer G. Pharmacological modulation of autophagy: therapeutic potential and persisting obstacles. Nat. Rev. Drug Discov. 2017;16:487–511. doi: 10.1038/nrd.2017.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singh S.S., Vats S., Chia A.Y., Tan T.Z., Deng S., Ong M.S., Arfuso F., Yap C.T., Goh B.C., Sethi G. Dual role of autophagy in hallmarks of cancer. Oncogene. 2018;37:1142–1158. doi: 10.1038/s41388-017-0046-6. [DOI] [PubMed] [Google Scholar]
- 26.Lu Z., Chen C., Wu Z., Miao Y., Muhammad I., Ding L., Tian E., Hu W., Ni H., Li R. A Dual Role of P53 in Regulating Colistin-Induced Autophagy in PC-12 Cells. Front. Pharmacol. 2017;8:768. doi: 10.3389/fphar.2017.00768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Larrue C., Saland E., Boutzen H., Vergez F., David M., Joffre C., Hospital M.A., Tamburini J., Delabesse E., Manenti S. Proteasome inhibitors induce FLT3-ITD degradation through autophagy in AML cells. Blood. 2016;127:882–892. doi: 10.1182/blood-2015-05-646497. [DOI] [PubMed] [Google Scholar]
- 28.Delbridge L.M.D., Mellor K.M., Taylor D.J., Gottlieb R.A. Myocardial stress and autophagy: mechanisms and potential therapies. Nat. Rev. Cardiol. 2017;14:412–425. doi: 10.1038/nrcardio.2017.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kang R., Zeh H.J., Lotze M.T., Tang D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011;18:571–580. doi: 10.1038/cdd.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Amaravadi R.K., Yu D., Lum J.J., Bui T., Christophorou M.A., Evan G.I., Thomas-Tikhonenko A., Thompson C.B. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J. Clin. Invest. 2007;117:326–336. doi: 10.1172/JCI28833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singh P., Godbole M., Rao G., Annarao S., Mitra K., Roy R., Ingle A., Agarwal G., Tiwari S. Inhibition of autophagy stimulate molecular iodine-induced apoptosis in hormone independent breast tumors. Biochem. Biophys. Res. Commun. 2011;415:181–186. doi: 10.1016/j.bbrc.2011.10.054. [DOI] [PubMed] [Google Scholar]
- 32.Yang Z., Klionsky D.J. Eaten alive: a history of macroautophagy. Nat. Cell Biol. 2010;12:814–822. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.B’chir W., Maurin A.C., Carraro V., Averous J., Jousse C., Muranishi Y., Parry L., Stepien G., Fafournoux P., Bruhat A. The eIF2α/ATF4 pathway is essential for stress-induced autophagy gene expression. Nucleic Acids Res. 2013;41:7683–7699. doi: 10.1093/nar/gkt563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu Z., Sun Y., She X., Wang Z., Chen S., Deng Z., Zhang Y., Liu Q., Liu Q., Zhao C. SIX3, a tumor suppressor, inhibits astrocytoma tumorigenesis by transcriptional repression of AURKA/B. J. Hematol. Oncol. 2017;10:115. doi: 10.1186/s13045-017-0483-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li J.P., Yang Y.X., Liu Q.L., Pan S.T., He Z.X., Zhang X., Yang T., Chen X.W., Wang D., Qiu J.X., Zhou S.F. The investigational Aurora kinase A inhibitor alisertib (MLN8237) induces cell cycle G2/M arrest, apoptosis, and autophagy via p38 MAPK and Akt/mTOR signaling pathways in human breast cancer cells. Drug Des. Devel. Ther. 2015;9:1627–1652. doi: 10.2147/DDDT.S75378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goff L.A., Rinn J.L. Linking RNA biology to lncRNAs. Genome Res. 2015;25:1456–1465. doi: 10.1101/gr.191122.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi X., Sun M., Liu H., Yao Y., Song Y. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 2013;339:159–166. doi: 10.1016/j.canlet.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 38.Li C.H., Chen Y. Targeting long non-coding RNAs in cancers: progress and prospects. Int. J. Biochem. Cell Biol. 2013;45:1895–1910. doi: 10.1016/j.biocel.2013.05.030. [DOI] [PubMed] [Google Scholar]
- 39.Yuan J.H., Yang F., Wang F., Ma J.Z., Guo Y.J., Tao Q.F., Liu F., Pan W., Wang T.T., Zhou C.C. A long noncoding RNA activated by TGF-β promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25:666–681. doi: 10.1016/j.ccr.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 40.Hu X., Feng Y., Zhang D., Zhao S.D., Hu Z., Greshock J., Zhang Y., Yang L., Zhong X., Wang L.P. A functional genomic approach identifies FAL1 as an oncogenic long noncoding RNA that associates with BMI1 and represses p21 expression in cancer. Cancer Cell. 2014;26:344–357. doi: 10.1016/j.ccr.2014.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen L.L. Linking Long Noncoding RNA Localization and Function. Trends Biochem. Sci. 2016;41:761–772. doi: 10.1016/j.tibs.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 42.Nie Y., Liu X., Qu S., Song E., Zou H., Gong C. Long non-coding RNA HOTAIR is an independent prognostic marker for nasopharyngeal carcinoma progression and survival. Cancer Sci. 2013;104:458–464. doi: 10.1111/cas.12092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen S.W., Zhu J., Ma J., Zhang J.L., Zuo S., Chen G.W., Wang X., Pan Y.S., Liu Y.C., Wang P.Y. Overexpression of long non-coding RNA H19 is associated with unfavorable prognosis in patients with colorectal cancer and increased proliferation and migration in colon cancer cells. Oncol. Lett. 2017;14:2446–2452. doi: 10.3892/ol.2017.6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.