Abstract
Introduction: Lupus nephritis (LN) is a major cause of mortality and morbidity in the patients with lupus, a chronic autoimmune disease. The role of genetic and epigenetic factors is emphasized in the pathogenesis of LN. The aim of the present study was to evaluate the levels of immune-regulatory microRNAs (e.g., miR-31, miR-125a, miR-142-3p, miR-146a, and miR-155) in plasma samples of patients with LN.
Methods: In this study, 26 patients with LN and 26 healthy individuals were included. The plasma levels of the microRNAs were evaluated by a quantitative real-time PCR. Moreover, the correlation of circulating plasma microRNAs with disease activity and pathological findings along with their ability to distinguish patients with LN were assessed.
Results: Plasma levels of miR-125a (P = 0.048), miR-146a (P = 0.005), and miR-155 (P< 0.001) were significantly higher in comparison between the cases and controls. The plasma level of miR-146a significantly correlated with the level of anti-double strand-DNA antibody and proteinuria. Moreover, there was a significant correlation between miR-142-3p levels and disease chronicity and activity index (P <0.05). The multivariate ROC curve analysis indicated the plasma circulating miR-125a, miR-142-3p, miR-146, and miR-155 together could discriminate most of the patients with LN from controls with area an under curve (AUC) of 0.89 [95% CI, 0.80-0.98, P<0.001], 88% sensitivity, and 78% specificity.
Conclusion: Based on the findings of the present study, the studied microRNAs may be involved in the pathogenesis and development of LN and have the potential to be used as diagnostic and therapeutic markers in LN.
Keywords: Autoimmunity, Biomarkers, Circulating microRNAs, Systemic Lupus Erythematosus
Introduction
Lupus is an inflammatory multisystem disease with an unknown cause that affects many organs in the body, in which body antibodies and immune complexes cause tissue and cellular damage.1 Different organs especially kidneys are involved in the course of the disease, the most serious symptoms of the lupus.2
Lupus nephritis (LN) is the major cause of mortality and morbidity in patients with lupus, affecting up to the 70% of the patients.3 The accumulation of immune complexes within the glomeruli of the kidneys is the earliest step in the development of LN. This event is associated with increased mesangial cells, excessive production of cellular matrix, and infiltration of inflammatory cells that lead to fibrosis and sclerosis. Depending on the severity of the disease, 15%-30% of LN patients shift towards the end-stage renal disease (ESRD).4 Genetic factors have been identified to be associated with the known and sometimes unknown environmental factors in developing LN. Epigenetic defects have also been shown to play an important role in the pathogenesis of lupus.5-7 Apparently, increasing information on the cellular, molecular, and biological behavior of the disease has an important role in determining diagnostic methods and therapeutic strategies.
Over the last few years, many studies have focused on the role of the microRNAs in various diseases, including cancer and autoimmune diseases.8,9 microRNAs are short, conserved endogenous non-coding RNA molecules that are involved in the regulation of gene expression.8 Recently, the role of microRNAs in regulating immune responses has been approved.9-11 In several studies, microRNAs have been demonstrated to be involved in the pathogenesis of lupus by altering intrinsic immune responses, lymphocyte function, and the toll-like receptors (TLRs) and NF-κB signaling pathways.12 Moreover, microRNAs can stimulate the expression of proinflammatory cytokines and determine the severity of immune responses.13 Given the fact that the microRNAs are highly regulated to maintain the hemostasis and normal function of the immune system, their dysregulated expression can result in the initiation/progression of autoimmune diseases.9
Considering the importance of LN and the significance of determining the prognosis of the disease in patients; this study aimed to evaluate the expression levels of immune-associated microRNAs. These known microRNAs (miR-31, miR-125, 142-3p, miR-146, and miR-155) are important in the differentiation and development of immune responses as well as creating a malfunction in immune system responses.14-18 Moreover, the association of the studied microRNAs with disease chronicity and activity index, and pathological findings were investigated in the patients with LN.
Materials and Methods
Subjects
The current cross-sectional study included 26 consecutive lupus patients with biopsy-proven nephritis enrolled from Kidney Ward of Imam Reza Hospital, Tabriz, Iran. All Azari-Turkish cases were collected from September 2015 to December 2016. Cases met systemic lupus erythematosus (SLE) diagnostic criteria according to the American College of Rheumatology (ACR). According to the National Institutes of Health (NIH) system, an activity index of 0 to 24 is calculated by grading the biopsy on a scale of 0 to 3+ for 6 histologic characteristics comprising endocapillary proliferation, wire loop deposits, glomerular leukocyte infiltration, interstitial inflammation, karyorrhexis, cellular crescents, and fibrinoid necrosis.19 Similarly, the sum of tubular atrophy, glomerulosclerosis, interstitial fibrosis, and fibrous crescents was used for a chronicity index of 0 to 12. Patients with active infection, previous malignancy, and diabetes mellitus were excluded. Moreover, healthy individuals were included as controls (N = 26).
miRNAs extraction and reverse transcription
The blood samples (~2 mL) were collected in EDTA-containing tubes and centrifuged for 10 min (at 1000 × g). Then, was transferred to a new microtube and the circulating RNAs were isolated from 200 μL of plasma samples using miRCURY™ RNA Isolation kit, Biofluid, (Exiqon, Vedbaek, Denmark). The isolated RNAs were reverse-transcribed using the miRCURY LNA™ Universal RT cDNA Synthesis Kit (Exiqon, Vedbaek, Denmark), following the manufacturer recommended protocol.
Quantitative real-time PCR
Quantitative real-time PCR (qPCR) was completed using specific LNA (locked nucleic acid) primers in triplicate (Table 1) based on the protocol of the SYBR® Green master mix kits (Exiqon, Vedbaek, Denmark) by the iCycler iQ system (Bio-Rad). An internal control (miR-191-5p) was used. The efficiency of qPCR was verified using the 10-fold serial dilution method (10-1-10-5 dilutions) to create standard curves. Threshold cycle (Ct) values were used to calculate the relative expression using the ΔΔCt method where relative expression equals to 2-ΔΔCt, and ΔΔCt for each patient = (ΔCt of the related miR for a patient) – (Mean of ΔCt values of healthy subjects).
Table 1. List of lucked nucleic acid primers .
| miRNAs | Mature sequence | Primer's Product No. |
| hsa-miR-31-5p | AGGCAAGAUGCUGGCAUAGCU | 204236 |
| hsa-miR-125a-5p | UCCCUGAGACCCUUUAACCUGUGA | 204339 |
| hsa-miR-142-3p | UGUAGUGUUUCCUACUUUAUGGA | 204291 |
| hsa-miR-155-5p | UUAAUGCUAAUCGUGAUAGGGGU | 204308 |
| hsa-miR-146a-5p | UGAGAACUGAAUUCCAUGGGUU | 204688 |
| has-miR-191-5p | CAACGGAAUCCCAAAAGCAGCUG | 204306 |
Statistical analysis
The data were presented as mean (SD) and median (interquartile range) for normally and not normally distributed values, respectively. For parametric variables, statistical significance between groups was analyzed by the student’s t test while the Kruskal-Wallis H and Mann–Whitney U tests were used for nonparametric values. Spearman rank order correlation was used to analyze the correlation between clinicopathological parameters and levels of the circulating miRNAs. Receiver operating characteristic (ROC) analysis and the area under the curve (AUC) with 95% CI were applied to test the diagnostic values of the miRNAs. Moreover, a binary logistic regression with inter-method was performed to assay the discriminating effects of the studied microRNAs together. miRNAs variables entered into the model (inter-method) as the main effect. Then, the diagnostic cut-off value was determined using Youden J Statistic to evaluate the discriminating ability of the chosen miRNAs. online Centre for Evidence-Based Medicine, Toronto was used for determining the sensitivity and specificity of the miRNAs.20,21 Statistical analyses were performed using the IBM SPSS software version 17.0. A P value < 0.05 was considered statistically significant.
Results
Clinical and demographic characteristics of the patients are presented in Table 2.
Table 2. Demographic and baseline clinical data .
| Characteristics | LN patients | Control group | P value |
| No. of cases | 26 | 26 | - |
| No. of male/female | 6/20 | 9/17 | 0.89 |
| Age, mean ± SD (y) | 32.61 ± 9 | 29.9 ± 8 | 0.40 |
| C3 (mg/dL) | 27 ± 9.2 | 89.9 ± 17.3 | <0.001 |
| C4 (mg/dL) | 13.27 ± 6 | 45.8 ± 16.2 | <0.001 |
| ANA | 7.32 ± 3 | 0.6 ± 0.2 | <0.001 |
| Anti-dsDNA antibody | 61 ± 25 | 14.1 ± 4.2 | <0.001 |
| Creatinine, (mg/dL) | 1.46 ± 0.32 | 0.91 ± 0.10 | <0.001 |
| Proteinuria (mg/24 h) | 2107 ± 1094 | 94.6 ± 15.5 | <0.001 |
| ESR | 33.61 ± 12 | 12.7 ± 3.7 | <0.001 |
| Chronicity index | 6.46 ± 3.1 | 0 | - |
| SLEDAI | 10.0 ± 3.4 | 0 | - |
| Stages (3/4/5) | 9/11/6 | - | - |
ANA: anti-nucleic acid, anti-dsDNA: anti-double strand DNA, ESR: erythrocyte sedimentation rate, SLEDAI: Systemic Lupus Erythematosus Disease Activity Index. The quantity data are expressed as mean ± SD
Different plasma levels of microRNAs between LN and control groups
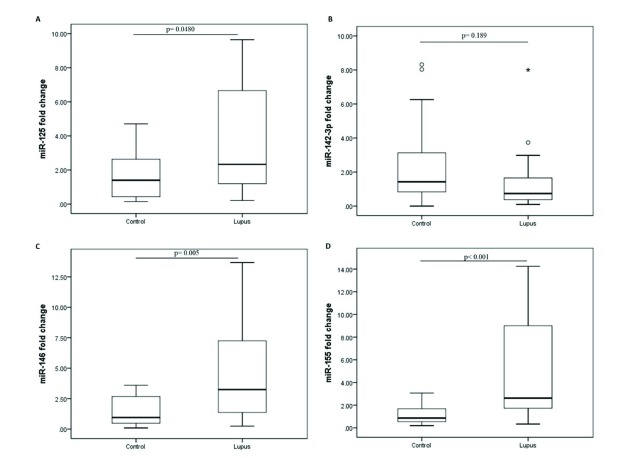
The plasma levels of miR-31, miR-125, miR-142, miR-146, and miR-155 were assessed in LN group and healthy controls. A significant increase was observed in plasma cell-free miR-125, miR-146, and miR-155 of LN patients when compared to controls. However, circulating miR-142-3p (P < 0.001) did not significantly change between the studied groups (Fig. 1). After 40 cycles of amplification, miR-31 was not detected in any of the plasma samples and the related data were not analyzed.
Fig. 1.
Altered expression of plasma miRNAs in lupus nephritis group when compare to controls. (A) miR-125a, (B) miR-142-3p, (C) miR-146, and (D) miR-155 levels in patients with lupus nephritis. The 2-ΔΔCt method was used to calculate the fold change between the study groups. miR-191 was used as normalizing endogenous control.
The relationship between plasma microRNAs and the clinical parameters in LN patients
A linear correlation analysis was accomplished to examine the relationship between the studied miRNAs and clinical parameters in the cases. Significant correlations were detected between miR-142 and activity index (r= -0.573, P = 0.002), chronicity index (r= 0.530, P = 0.005), and serum creatinine (r= 0.497, P = 0.010). Additionally, there were significant correlations between miR-146 and anti-dsDNA (r= 0.475, P = 0.014) and proteinuria (r= -0.389, P = 0.049) (Table 3).
Table 3. Correlations between studied miRNAs and clinical parameters in LN patients .
| Biochemical tests | miR-125 | miR-142 | miR-146 | miR-155 |
| C3 |
r= -0.015 P= 0.940 |
r= 0.175 P= 0.393 |
r= -0.195 P= 0.341 |
r= 0.244 P= 0.230 |
| C4 |
r= 0.097 P= 0.638 |
r= 0.223 P= 0.274 |
r= -0.118 P= 0.566 |
r= -0.124 P= 0.546 |
| Age |
r= 0.202 P= 0.323 |
r= 0.362 P= 0.069 |
r= 0.327 P= 0.103 |
r= 0.006 P= 0.976 |
| ANA |
r= -0.075 P= 0.720 |
r= 0.118 P= 0.575 |
r= 0.088 P= 0.676 |
r= 0.161 P= 0.442 |
| Anti-dsDNA |
r= 0.224 P= 0.272 |
r= 0.263 P= 0.195 |
r= 0.475
P = 0.014 |
r= -0.288 P= 0.154 |
| ESR |
r= 0.118 P= 0.565 |
r= 0.293 P= 0.146 |
r= 0.205 P= 0.316 |
r= -0.293 P= 0.146 |
| SLEDAI |
r= -0.244 P= 0.230 |
r= -0.573
P = 0.002 |
r= -0.175 P= 0.393 |
r= -0.186 P= 0.363 |
| Chronicity index |
r= 0.260 P= 0.200 |
r= 0.530
P = 0.005 |
r= 0.176 P= 0.390 |
r= -0.103 P= 0.617 |
| Stage |
r= -0.172 P= 0.421 |
r= -0.373 P= 0.072 |
r= -0.043 P= 0.843 |
r= 0.186 P= 0.385 |
| Pro24 |
r= -0.035 P= 0.864 |
r= -0.133 P= 0.517 |
r= -0.389
P = 0.049 |
r= 0.182 P= 0.373 |
| Creatinine |
r= 0.157 P= 0.442 |
r= 0.497
P = 0.010 |
r= 0.089 P= 0.665 |
r= -0.115 P= 0.577 |
r: correlation coefficient, ANA: anti-nucleic acid, anti-dsDNA: anti-double strand DNA, ESR: erythrocyte sedimentation rate, SLEDAI: Systemic Lupus Erythematosus Disease Activity Index.
Correlations between the studied microRNAs in LN patients
Correlations between the expression levels of the selected miRNAs were examined in individuals by Spearman correlation. Correlations were found between miR-125 and miR-142 (r= 0.531, P = 0.005) as well as miR-125 and miR-155 (r= 0.390, P = 0.049) fold changes but not others (P ≥ 0.153). Moreover, significant correlations were found between the tested miRNAs delta Ct values (Table 4).
Table 4. Internal correlation between plasma miRNA delta Ct .
| miR-142 | miR-146 | miR-155 | |
| miR-125 |
r= 0.611 P= 0.001 |
r= 0.512 P= 0. 008 |
r= 0.680 P< 0.001 |
| miR-142 |
r= 0.380 P= 0.055 |
r= 0.521 P= 0.006 |
|
| miR-146 |
r= 0.443 P= 0.023 |
r: correlation coefficient.
ROC curve analysis
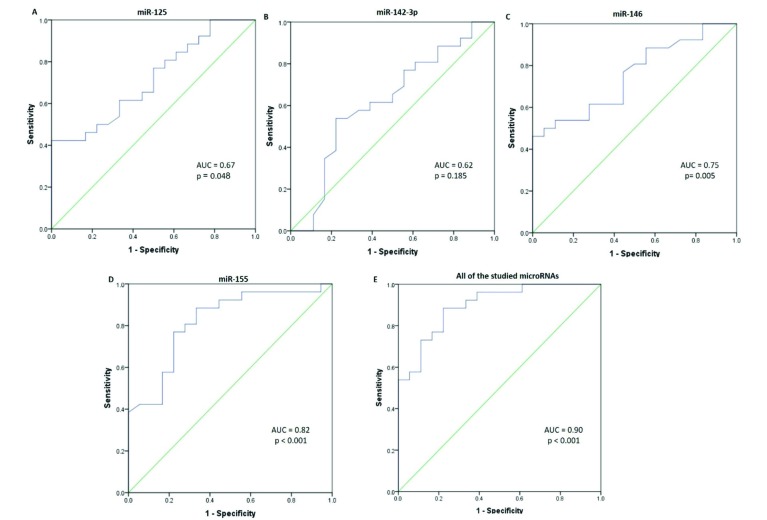
The ROC analysis was performed to evaluate the diagnostic value of miRNAs. Fold change values of microRNAs were used to discriminate cases from controls. miR-125a presented a sensitivity of 92% (0.76 to 0.98), specificity of 34% (0.09 to 0.45), and AUC value of 0.67 (0.52 to 0.83). miR-142-3p produced the AUC value of 0.62 (0.44 to 0.79) with 80% (0.62 to 0.92) sensitivity and 55% (0.13 to 0.74) specificity (Fig. 2B). The AUC of 0.75 (0.61 to 0.89) was yielded for miR-146 with sensitivity and specificity of respectively 56 and 96%. miR-155 presented a sensitivity of 88% and specificity of 67%, and AUC of 0.82 (0.69 to 0.95). As shown in Fig. 2D, all the studied miRNAs exert the high accuracy (AUC= 0.89, CI: 0.80 to 0.98, P < 0.001), 88% (0.71 to 0.96) sensitivity, and 78% (0.548 to 0.91) specificity for discriminating LN patients from the controls.
Fig. 2.
ROC curve analysis of the studied plasma miRNAs. ROC curves analysis based on q-PCR data display the diagnostic power of the following plasma miRNAs; (A) miR-125a (B) miR-142-3p (C) miR-146, (D) miR-155, and (E) the effects of the studied miRNAs (e.g., miR-125, miR-142-3p, miR-146, and miR-155) in distinguishing patients with lupus nephritis from healthy individuals.
Discussion
Advances in molecular science and the role of microRNAs in the development of LN along with their relation to the severity of inflammation, have suggested a significant role for microRNAs.9,22 Their expression can be used to evaluate the severity of the disease and monitor the patients. This assay may make fundamental changes in the management of the disease.14 In accordance with the results of the present study, the miRNAs are likely to contribute to the pathogenesis and progression of LN and may be employed as diagnostic markers for LN.
Different studies described a significant down-regulation of miR-31 in SLE and LN patients.23 Furthermore, the correlation of miR-31 with the SLEDAI score and proteinuria of SLE patients was reported.23 Fan et al demonstrated that miR-31 regulates the IL-2 production in T cells by targeting RhoA. In SLE, decreased levels of the miR-31 result in the unusual production of IL-2 in lupus T cells that leads to altered expression of the nuclear factor of activated T-cells (NFAT) and IL-2 promoter activity.15 These data suggest that the expression of miR-31 may be related to LN activity and the degree of renal injury.15 In the present study, miR-31 was not detectable in all plasma samples (Ct > 40) and its related data was not analyzed. It may be due to the form of miR-31 (cellular vs. secretory), type of miR-31 (-5p vs. -3p), or type of the primer (stem-loop vs. locked nucleic acid).
The miR-125a contributes to upregulated levels of the RANTES, an inflammatory chemokine that is needed for detrimental effects in inflammatory processes, in patients with SLE.24 In T cells of patients with SLE, diminished levels of miR-125a have been reported.24 However, an elevated serum miR-125a was detected in patients with LN that contributed to the upregulation of inflammatory cytokines (e.g., TNF-α, and IL-β, -6).25 Likewise, in the present study, increased levels of circulating miR-125a were observed in LN group when compared to controls that may represent active inflammation.
The miR-142-3p controls the functions of CD4+ T and CD4+ CD25+ Treg cells and its dysregulation leads to the development of autoimmune diseases.26 Down-regulation of miR-142 plays an important role in the pathogenesis of SLE since it increases the levels of autoimmune-related target genes (e.g., signalling lymphocytic activation molecule-associated protein (SAP), CD84, and interleukin-10); resulted in T cell activity and B cell hyperstimulation.26 Moreover, the diminished levels of miR-142 in SLE CD4+ T cells are reported to be associated with histone modifications and DNA methylation changes in upstream of the miR-142 precursor sequence.26 Likewise, in this study, a diminished level of circulating miR-142-3p was observed in patients with LN in comparison to controls; however, it was not significant. An increased level of miR-142-3p was also reported in plasma samples of SLE patients.27
The miR-146a is involved in renal inflammation and fibrosis and its dysregulation is correlated with the occurrence of multiple autoimmune diseases including SLE8 and rheumatoid arthritis.28 During LN, miR-146a prevents the transcriptional activity of NF-κB via inhibiting TNF receptor-associated factor 6 (TRAF6), an intermediate mediator between toll-like receptor 4 (TLR4) and NF-κB.29 Although its detailed mechanism is still unclear, controversial data on dysregulation of miR-146a has been reported. A decrease in the expression of miR-146a has been reported in renal biopsy and peripheral blood leukocytes of patients with LN.29-31 However, a higher expression of miR-146a has been also reported in the glomerulus and urinary exosomes of patients with LN32,33 as well as in the urinary sediment of SLE patients.34 Likewise, in the present study, increased levels of miR-146a were observed in LN group when compared to controls. All these findings indicate a possible role for miR-146a in the immune system or immunity dysregulation and pathogenesis of LN.30,35
The miR-155 is an essential modulator of immune responses. It is required for proper development and maturation of the lymphocytes and antibody production.34,36 Moreover, miR-155 presents a central role in the function of immune cells (e.g., dendritic cells, B cells, and Th17 cells).37 Abnormal expression of miR-155 is detected in many human autoimmune conditions (e.g., SLE and RA).28,34,36 The urinary sediment levels of miR-155 in patients with SLE were significantly higher than that in healthy controls,34 while its serum levels were reduced.36 Moreover, miR-155 was correlated with SLEDAI scores and different SLE clinical parameters.34,36 Consistent with the above-mentioned studies, in the present study, an elevated level of this microRNA was detected in plasma samples of LN group that may point out an active inflammation, inflammation-mediated glomerular endothelial injury, and fibrosis.20
Abnormalities in microRNA have been shown in SLE and LN patients but the data are inconsistent.38,39 The variance between miRNA levels among LN patients in different studies is possibly due to the different ethnicity of patients and hence, various genetic background, small sample size, and different sample sources (e.g., different cell types vs. cell-free). Moreover, different environmental factors may affect the expression of miRNAs such as lifestyle, dietary habits, and exposure to infections.40,41 Changes in miRNA expression levels as a result of LN therapy would be another possible explanation.42 In spite of some studies, at the time of sampling, our patients did not receive any LN-related therapy that may affect miRNAs expression.
Our study had some limitations. Small sample size along with the exclusion of SLE patients to compare the results between groups were the possible limitations of the present study.
Conclusion
Since over- or down-regulation of these miRNAs is associated with the pathogenesis of LN, appropriate levels of the miRNAs may be important for maintaining normal immune responses. Cell-free miR-125a, miR-142-3p, miR-146, and miR-155 are correlated with clinical parameters and together present a diagnostic value to distinguish patients with LN from controls. The identification and validation of novel diagnostic biomarkers would be helpful to develop therapeutic strategies. Further molecular and clinical follow-up and large sample size studies are required to confirm these results.
Ethical approval
The study was institutionally approved by the Clinical Research Ethics Committee of the Tabriz University of Medical Sciences, Tabriz, Iran (Ethical code: TBZMED.REC.1395.494). All patients provided a written informed consent to participate.
Competing interests
There is no conflict of interests to be reported.
Research Highlights
What is current knowledge?
√ Lupus nephritis (LN) is the major cause of mortality and morbidity in patients with lupus, an autoimmune disease.
√ Epigenetic defects, especially microRNAs, have been shown to play an important role in the pathogenesis of lupus.
√ Despite renal biopsy being a gold standard for degree classifying in the patients with LN, it suffers from being invasive.
√ There is a clinical need to discover novel biomarkers for continuous immune monitoring of patients.
What is new here?
√ Levels of circulating miR-125a, miR-146, and miR-155 were evaluated in plasma samples of patients with LN.
√ Circulating miR-142 was significantly correlated with creatinine along with disease activity index and chronicity index.
√ Cell-free miR-125a, miR-142-3p, miR-146, and miR-155 together have diagnostic value to distinguish patients with LN from controls.
References
- 1.Schwartz N, Goilav B, Putterman C. The pathogenesis, diagnosis and treatment of lupus nephritis. Curr Opin Rheumatol. 2014;26:502–9. doi: 10.1097/bor.0000000000000089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Flores-Mendoza G, Sanson SP, Rodriguez-Castro S, Crispin JC, Rosetti F. Mechanisms of Tissue Injury in Lupus Nephritis. Trends Mol Med 2018. [DOI] [PubMed]
- 3.Kwok SK, Tsokos GC. New insights into the role of renal resident cells in the pathogenesis of lupus nephritis. Korean J Intern Med. 2018;33:284–9. doi: 10.3904/kjim.2017.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maroz N, Segal MS. Lupus nephritis and end-stage kidney disease. Am J Med Sci. 2013;346:319–23. doi: 10.1097/MAJ.0b013e31827f4ee3. [DOI] [PubMed] [Google Scholar]
- 5.Wu H, Zhao M, Tan L, Lu Q. The key culprit in the pathogenesis of systemic lupus erythematosus: aberrant DNA methylation. Autoimmun Rev. 2016;15:684–9. doi: 10.1016/j.autrev.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 6.Wang Z, Lu Q, Wang Z. Epigenetic Alterations in Cellular Immunity: New Insights into Autoimmune Diseases. Cell Physiol Biochem. 2017;41:645–60. doi: 10.1159/000457944. [DOI] [PubMed] [Google Scholar]
- 7.Wang Z, Chang C, Peng M, Lu Q. Translating epigenetics into clinic: focus on lupus. Clin Epigenetics. 2017;9:78. doi: 10.1186/s13148-017-0378-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Husakova M. MicroRNAs in the key events of systemic lupus erythematosus pathogenesis. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2016;160:327–42. doi: 10.5507/bp.2016.004. [DOI] [PubMed] [Google Scholar]
- 9.Le X, Yu X, Shen N. Novel insights of microRNAs in the development of systemic lupus erythematosus. Curr Opin Rheumatol. 2017;29:450–7. doi: 10.1097/bor.0000000000000420. [DOI] [PubMed] [Google Scholar]
- 10.Lai NS, Koo M, Yu CL, Lu MC. Immunopathogenesis of systemic lupus erythematosus and rheumatoid arthritis: the role of aberrant expression of non-coding RNAs in T cells. Clin Exp Immunol. 2017;187:327–36. doi: 10.1111/cei.12903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simpson LJ, Ansel KM. MicroRNA regulation of lymphocyte tolerance and autoimmunity. J Clin Invest. 2015;125:2242–9. doi: 10.1172/jci78090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stypinska B, Paradowska-Gorycka A. Cytokines and MicroRNAs as Candidate Biomarkers for Systemic Lupus Erythematosus. Int J Mol Sci. 2015;16:24194–218. doi: 10.3390/ijms161024194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zan H, Tat C, Casali P. MicroRNAs in lupus. Autoimmunity. 2014;47:272–85. doi: 10.3109/08916934.2014.915955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leiss H, Salzberger W, Jacobs B, Gessl I, Kozakowski N, Bluml S. et al. MicroRNA 155-deficiency leads to decreased autoantibody levels and reduced severity of nephritis and pneumonitis in pristane-induced lupus. PLoS One. 2017;12:e0181015. doi: 10.1371/journal.pone.0181015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fan W, Liang D, Tang Y, Qu B, Cui H, Luo X. et al. Identification of microRNA-31 as a novel regulator contributing to impaired interleukin-2 production in T cells from patients with systemic lupus erythematosus. Arthritis Rheum. 2012;64:3715–25. doi: 10.1002/art.34596. [DOI] [PubMed] [Google Scholar]
- 16.Sebastiani G, Ventriglia G, Stabilini A, Socci C, Morsiani C, Laurenzi A. et al. Regulatory T-cells from pancreatic lymphnodes of patients with type-1 diabetes express increased levels of microRNA miR-125a-5p that limits CCR2 expression. Sci Rep. 2017;7:6897. doi: 10.1038/s41598-017-07172-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mann M, Mehta A, Zhao JL, Lee K, Marinov GK, Garcia-Flores Y. et al. An NF-kappaB-microRNA regulatory network tunes macrophage inflammatory responses. Nat Commun. 2017;8:851. doi: 10.1038/s41467-017-00972-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Birlik M, Koçak A, Harmancı D. Role of MicroRNAs in Rheumatoid Arthritis. New Developments in the Pathogenesis of Rheumatoid Arthritis. InTech; 2017.
- 19.Ines L, Silva C, Galindo M, Lopez-Longo FJ, Terroso G, Romao VC. et al. Classification of Systemic Lupus Erythematosus: Systemic Lupus International Collaborating Clinics Versus American College of Rheumatology Criteria A Comparative Study of 2,055 Patients From a Real-Life, International Systemic Lupus Erythematosus Cohort. Arthritis Care Res (Hoboken) 2015;67:1180–5. doi: 10.1002/acr.22539. [DOI] [PubMed] [Google Scholar]
- 20.Zununi Vahed S, Poursadegh Zonouzi A, Ghanbarian H, Ghojazadeh M, Samadi N, Omidi Y. et al. Differential expression of circulating miR-21, miR-142-3p and miR-155 in renal transplant recipients with impaired graft function. Int Urol Nephrol. 2017 doi: 10.1007/s11255-017-1602-2. [DOI] [PubMed] [Google Scholar]
- 21.Zununi Vahed S, Poursadegh Zonouzi A, Mahmoodpoor F, Samadi N, Ardalan M, Omidi Y. Circulating miR-150, miR-192, miR-200b, and miR-423-3p as Non-invasive Biomarkers of Chronic Allograft Dysfunction. Arch Med Res. 2017;48:96–104. doi: 10.1016/j.arcmed.2017.03.004. [DOI] [PubMed] [Google Scholar]
- 22.Cardenas-Gonzalez M, Srivastava A, Pavkovic M, Bijol V, Rennke HG, Stillman IE. et al. Identification, Confirmation, and Replication of Novel Urinary MicroRNA Biomarkers in Lupus Nephritis and Diabetic Nephropathy. Clin Chem. 2017;63:1515–26. doi: 10.1373/clinchem.2017.274175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amr KS, Bayoumi FS, Elgengehy FT, Abdallah SO, Ahmed HH, Eissa E. The role of microRNA-31 and microRNA-21 as regulatory biomarkers in the activation of T lymphocytes of Egyptian lupus patients. Rheumatol Int. 2016;36:1617–25. doi: 10.1007/s00296-016-3550-z. [DOI] [PubMed] [Google Scholar]
- 24.Zhao X, Tang Y, Qu B, Cui H, Wang S, Wang L. et al. MicroRNA-125a contributes to elevated inflammatory chemokine RANTES levels via targeting KLF13 in systemic lupus erythematosus. Arthritis Rheum. 2010;62:3425–35. doi: 10.1002/art.27632. [DOI] [PubMed] [Google Scholar]
- 25.Li H, Ding G. Elevated Serum Inflammatory Cytokines in Lupus Nephritis Patients, in Association with Promoted hsa-miR-125a. Clin Lab. 2016;62:631–8. doi: 10.7754/Clin.Lab.2015.150812. [DOI] [PubMed] [Google Scholar]
- 26.Ding S, Liang Y, Zhao M, Liang G, Long H, Zhao S. et al. Decreased microRNA-142-3p/5p expression causes CD4+ T cell activation and B cell hyperstimulation in systemic lupus erythematosus. Arthritis Rheum. 2012;64:2953–63. doi: 10.1002/art.34505. [DOI] [PubMed] [Google Scholar]
- 27.Carlsen AL, Schetter AJ, Nielsen CT, Lood C, Knudsen S, Voss A. et al. Circulating microRNA expression profiles associated with systemic lupus erythematosus. Arthritis Rheum. 2013;65:1324–34. doi: 10.1002/art.37890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kriegsmann M, Randau TM, Gravius S, Lisenko K, Altmann C, Arens N. et al. Expression of miR-146a, miR-155, and miR-223 in formalin-fixed paraffin-embedded synovial tissues of patients with rheumatoid arthritis and osteoarthritis. Virchows Arch. 2016;469:93–100. doi: 10.1007/s00428-016-1939-4. [DOI] [PubMed] [Google Scholar]
- 29.Zheng CZ, Shu YB, Luo YL, Luo J. The role of miR-146a in modulating TRAF6-induced inflammation during lupus nephritis. Eur Rev Med Pharmacol Sci. 2017;21:1041–8. [PubMed] [Google Scholar]
- 30.Tang Y, Luo X, Cui H, Ni X, Yuan M, Guo Y. et al. MicroRNA-146A contributes to abnormal activation of the type I interferon pathway in human lupus by targeting the key signaling proteins. Arthritis Rheum. 2009;60:1065–75. doi: 10.1002/art.24436. [DOI] [PubMed] [Google Scholar]
- 31.Zhu Y, Xue Z, Di L. Regulation of MiR-146a and TRAF6 in the Diagnose of Lupus Nephritis. Med Sci Monit. 2017;23:2550–7. doi: 10.12659/MSM.900667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu J, Kwan BC, Lai FM, Tam LS, Li EK, Chow KM. et al. Glomerular and tubulointerstitial miR-638, miR-198 and miR-146a expression in lupus nephritis. Nephrology (Carlton) 2012;17:346–51. doi: 10.1111/j.1440-1797.2012.01573.x. [DOI] [PubMed] [Google Scholar]
- 33.Perez-Hernandez J, Forner MJ, Pinto C, Chaves FJ, Cortes R, Redon J. Increased Urinary Exosomal MicroRNAs in Patients with Systemic Lupus Erythematosus. PLoS One. 2015;10:e0138618. doi: 10.1371/journal.pone.0138618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang G, Tam LS, Kwan BC, Li EK, Chow KM, Luk CC. et al. Expression of miR-146a and miR-155 in the urinary sediment of systemic lupus erythematosus. Clin Rheumatol. 2012;31:435–40. doi: 10.1007/s10067-011-1857-4. [DOI] [PubMed] [Google Scholar]
- 35.Luo X, Yang W, Ye DQ, Cui H, Zhang Y, Hirankarn N. et al. A functional variant in microRNA-146a promoter modulates its expression and confers disease risk for systemic lupus erythematosus. PLoS Genet. 2011;7:e1002128. doi: 10.1371/journal.pgen.1002128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang G, Tam LS, Li EK, Kwan BC, Chow KM, Luk CC. et al. Serum and urinary cell-free MiR-146a and MiR-155 in patients with systemic lupus erythematosus. J Rheumatol. 2010;37:2516–22. doi: 10.3899/jrheum.100308. [DOI] [PubMed] [Google Scholar]
- 37.Leng R-X, Pan H-F, Qin W-Z, Chen G-M, Ye D-Q. Role of microRNA-155 in autoimmunity. Cytokine Growth Factor Rev. 2011;22:141–7. doi: 10.1016/j.cytogfr.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 38.Te JL, Dozmorov IM, Guthridge JM, Nguyen KL, Cavett JW, Kelly JA. et al. Identification of unique microRNA signature associated with lupus nephritis. PLoS One. 2010;5:e10344. doi: 10.1371/journal.pone.0010344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dai Y, Huang YS, Tang M, Lv TY, Hu CX, Tan YH. et al. Microarray analysis of microRNA expression in peripheral blood cells of systemic lupus erythematosus patients. Lupus. 2007;16:939–46. doi: 10.1177/0961203307084158. [DOI] [PubMed] [Google Scholar]
- 40.Chung JW, Jeong SH, Lee SM, Pak JH, Lee GH, Jeong JY. et al. Expression of MicroRNA in Host Cells Infected with Helicobacter pylori. Gut Liver. 2017;11:392–400. doi: 10.5009/gnl16265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Palmer JD, Soule BP, Simone BA, Zaorsky NG, Jin L, Simone NL. MicroRNA expression altered by diet: can food be medicinal? Ageing Res Rev. 2014;17:16–24. doi: 10.1016/j.arr.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 42.Tang Q, Yang Y, Zhao M, Liang G, Wu H, Liu Q. et al. Mycophenolic acid upregulates miR-142-3P/5P and miR-146a in lupus CD4+T cells. Lupus. 2015;24:935–42. doi: 10.1177/0961203315570685. [DOI] [PubMed] [Google Scholar]