Abstract
The Mongolian gerbil (Meriones unguiculatus) is a member of the rodent family that displays several features not found in mice or rats, including sensory specializations and social patterns more similar to those in humans. These features have made gerbils a valuable animal for research studies of auditory and visual processing, brain development, learning and memory, and neurological disorders. Here, we report the whole gerbil annotated genome sequence, and identify important similarities and differences to the human and mouse genomes. We further analyze the chromosomal structure of eight genes with high relevance for controlling neural signaling and demonstrate a high degree of homology between these genes in mouse and gerbil. This homology increases the likelihood that individual genes can be rapidly identified in gerbil and used for genetic manipulations. The availability of the gerbil genome provides a foundation for advancing our knowledge towards understanding evolution, behavior and neural function in mammals.
Keywords: genome assembly, gene prediction, fragile X syndrome, oxytocin receptor, hearing, social interaction, plasma membrane calcium ATPase
1. Introduction
The Mongolian gerbil or jird (Meriones unguiculatus, Fig. 1A) belongs to the muridae family of rodentia, along with mice and rats, and originated in the steppes of Mongolia [1, 2]. The Gerbillinae subfamily includes 14 genera [3], and a DNA sequence analysis of two complete mitochondrial genes suggests a split with lineage leading to mice and rats approximately 13 million years ago [4]. This split is associated with certain specializations that make the gerbil of interest to a broad range of scientists.
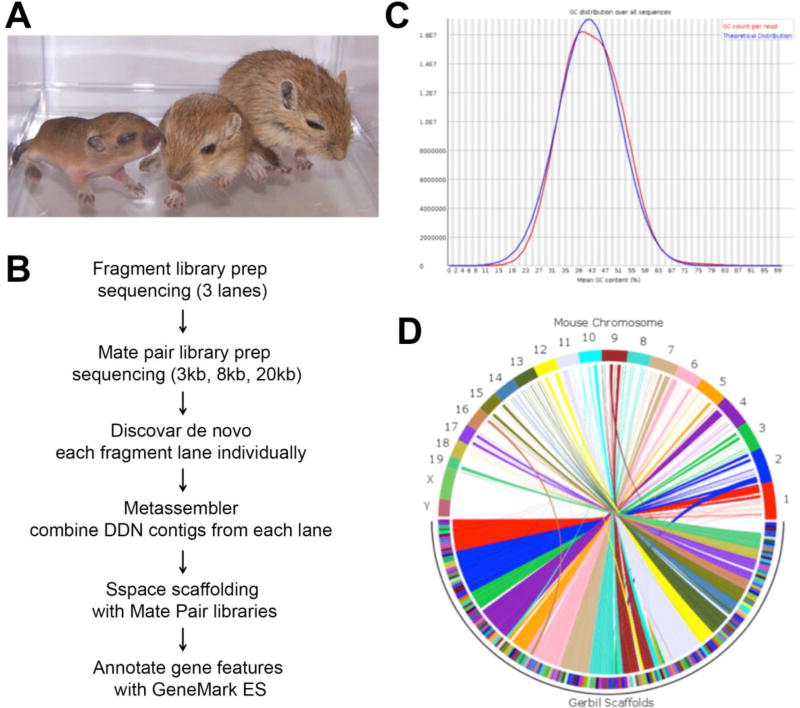
Figure 1. Gerbil genome assembly.
A. Mongolian gerbil (Meriones unguiculatus) at ages of postnatal day (P) 9, 16 and 21 from left to right. Scale bar is 10 cm. B. Workflow of genome assembly. C. GC content of Mongolian Gerbil genome, determined from fragment library reads using FastQC. The distribution shows a slightly skewed shape and is consistent with the overall assembly GC content of 42.09%. D. Synteny map of Mouse GRCm38 chromosomes versus 1 Mbp scaffolds of Gerbil assembly. In the circle diagram, gerbil scaffolds (bottom half) are reordered according to syntenic matches with the mouse chromosomes (top half). No syntenic blocks are observed for the mouse Y chromosome, and very little for the mouse X chromosome. Blocks corresponding to scaffolds with potential translocations are also evident as off-center arcs: blue from mouse chromosome 2; brown from mouse chromosome 9, yellow from mouse chromosome 12 and tan from mouse chromosome 16.
Gerbils have many sensory characteristics that make them a favorable species for studies of vision and audition. For example, they are primarily diurnal [5] and possess superior acuity and photopic vision, as compared to mice or rats [6]. Their retinal structure is more analogous to humans, having a relatively high percentage of cone photoreceptors, as compared to mice [7, 8]. For this reason, gerbils have been used to study retinal physiology [9, 10] and for developing therapeutic drugs and gene delivery approaches following retinal damage [11, 12]. Gerbils also display human-like sensitivity to the low sound frequency range that supports speech perception [13, 14], whereas mice and rats are more sensitive to very high frequency sounds [15]. Because of this specialization, the gerbil auditory pathway has been intensively studied for its structural and functional specializations, and it serves as a popular model for understanding the neural basis of auditory processing in normal and hearing-impaired animals [16– 25]. Examples include middle and inner ear function [26– 28], binaural processing in the auditory brainstem [29], parallel information streams of ascending auditory pathway [30– 33], auditory perception and integration with other sensory modalities in the primary auditory cortex [18, 22], vocal behaviors [34– 36] as well as age-dependent hearing loss [37, 38].
Gerbils are well suited to study a range of pathological conditions, including epilepsy (seizures) and cerebral ischemia (stroke). Gerbils are known to have high susceptibility to seizures that can be induced by simple external stimuli [39, 40]. Studies in gerbils have identified abnormal GABAergic dependent synaptic transmission as an important underlying mechanism of seizures [41– 43]. Investigations using gerbils as a stroke model have shown that “stroke-prone” and “stroke-resistant” gerbils are associated with the conditions of posterior communicating arteries in the circle of Willis [44– 48]. In addition, the gerbil has been commonly used for studying a number of parasitic, viral and bacterial diseases [reviewed in 2]. For example, humans have benefitted with the development of serologic tests and treatment regimens against lymphatic filariasis by studying gerbils infected with filarid nematodes [49– 50]. Gerbils are also used to study gastric ulcers caused by Helicobacter pylori infections as they develop severe gastritis and ulcers [51].
To advance these and other research areas where the gerbil serves as an appropriate animal model, the whole gerbil genome is needed in order to enable further advances at the genetic and molecular levels. We report the whole gerbil genome sequence and initial annotation, providing a fundamental database for gene analyses as well as the development of genetic editing approaches. We compare the gerbil genome to the human and mouse genomes, with the goal of identifying both important similarities and differences across species. As a first application of this gerbil genome database, we also analyzed the chromosomal structure of eight individual genes that are extensively studied in mammalian neural signaling and demonstrate the possibility of further studying these genes at previously unachieved levels.
2. Material and methods
2.1. Animals and tissue preparation
Mongolian gerbils (Meriones unguiculatus), strain 243, were purchased from the Charles River Laboratories (Wilmington, MA). One male animal of 6 weeks of age was used for tissue extraction for this project. All procedures were approved by the Florida State University Institutional Animal Care and Use Committee and conformed to NIH guidelines. Tissues from leg muscle were extracted, immediately flash frozen in liquid nitrogen and stored at −80°C until further processing.
2.2. DNA extraction
For high molecular-weight genomic DNA extraction, leg muscle tissue was thawed and minced finely with a razor blade, then refrozen on dry ice and ground to fine powder with liquid nitrogen. Powdered tissue (1–2 g) was resuspended in 600 µl gDNA extraction buffer (60 mM Trisaminomethane hydrochloride (Tris-HCl) pH 8.0, 100 mM Ethylenediaminetetraacetic acid (EDTA), 0.5% SDS) plus 50 µl Proteinase K (800 U/ml, New England Biolabs) and incubated at 50°C for 3 hours followed by 37°C overnight. Debris were removed by centrifugation and the supernatant was extracted with phenol followed by chloroform:isoamyl alcohol (96:4). The recovered aqueous phase was treated with RNase cocktail for 30 min at 37°C, and the DNA was precipitated by the addition of 1:10 volume sodium acetate (3 M, pH 5.0) and two volumes of isopropanol. DNA pellets were washed with 70% ethanol and resuspended in 100 µl elution buffer (10 mM Tris, 0.5 mM EDTA). Genomic DNA was quantified using Qubit High Sensitivity reagents.
2.3. Library construction, sequencing and assembly
Genomic DNA was ultrasonically sheared to 300 base pairs (bp) in micro-TUBE strips (Covaris, LE220 instrument) for fragment libraries. For mate pair libraries, DNA was sheared to 3-, 8- and 20-kilo bases (kb) using Covaris gTubes. Fragment libraries were constructed with the NxSeq AmpFree kit (Lucigen), and mate pair libraries with the NxSeq Long Mate Pair kit (Lucigen) following manufacturers’ instructions. Fragment libraries were sequenced on three lanes of HiSeq X (Illumina) with 2× 150 paired end (PE) chemistry at the Hudson Alpha Institute for Biotechnology (Huntsville, AL). Mate pair libraries were sequenced on MiSeq (Illumina) 2× 150 PE with V2 chemistry. Mate pair data was processed with Python scripts Illumina-Chimera-Clean5.py and IlluminaJunctionSplit9.py (available from Lucigen) to remove chimeric mate pairs and to trim the right and left mates by detection of the Junction Code sequence.
Initial assembly was performed with Discovar De Novo (ftp://ftp.broadinstitute.org/pub/crd/DiscovarDeNovo/latest_source_code/LATEST_VERSION.tar.gz; Accessed 4 November 2016.) using untrimmed fragment data as suggested in the Discovar manual. Each lane of HiSeq X data was assembled individually. The three sets of final Discovar De Novo contigs were then merged into a single assembly by using Metassembler v1.5. [https://sourceforge.net/projects/metassembler/files/latest/download] [52]. Metassembler contigs were then scaffolded sequentially with the 3-, 8- and 20-kb mate pair libraries using a stand-alone scaffolder of pre-assembled contigs using paired-read data (SSPACE) Basic v2.0 [https://www.baseclear.com/services/bioinformatics/basetools/sspace-standard/] [53]. Repetitive sequences were identified by using Repeat Modeler v1.08 [http://www.repeatmasker.org/RepeatModeler/] [54]; Accessed 7 March 2017] for de novo repeat discovery. The unmasked assembly was filtered to remove all contigs smaller than 1 kb following the National Center for Biotechnology Information (NCBI) Eukaryotic Genome Annotation Pipeline guidelines [55; Accessed 17 May 2017] and the remainder was deposited with GenBank and submitted for annotation by the NCBI Eukaryotic Genome Annotation Pipeline [https://www.ncbi.nlm.nih.gov/genome/annotation_euk/].
Syntenic blocks between mouse GRCm38 chromosomes and gerbil scaffolds were identified using Symap 4.2 [http://www.agcol.arizona.edu/software/symap/v4.2/download] [56], after filtering the gerbil assembly for scaffolds of over 1 mega base pairs (Mbp) (658 scaffolds total). Orthologous gene groups were identified between the Gerbil protein set, the human proteome (UP00005640_9606) and the mouse proteome (UP00000589_10090) from UniProt [http://www.uniprot.org/downloads; Accessed 20 April 2017]. The TriFusion v0.5.0 pipeline [https://pypi.python.org/pypi/trifusion/0.5.0.post3; Accessed 7 April 2017] [57], which incorporates Usearch for protein-protein comparisons and the OrthoMCL pipeline [58], was used for proteome comparisons and generation of orthologous protein families. Gene functional annotation clustering was performed with DAVID 6.8 [https://david.ncifcrf.gov; Accessed 24 April 2017] [59] using 194 human Uniprot accession numbers for unique orthologs shared between human and gerbil. The mouse Uniprot accession numbers were used for 760 unique orthologs shared between mouse and gerbil, and for 538 unique orthologs shared between mouse and human (Supplementary file ‘multi-list.txt’).
3. Results and Discussion
3.1. Gerbil genome assembly
Genomic DNA extracted from leg muscle tissue of an adult male Meriones unguiculates of 6 weeks of age was used to construct fragment and mate pair libraries for sequencing on the Illumina platform, following the work flow shown in Fig. 1B. Fragment libraries were sequenced on 3 lanes of HiSeqX, generating 407.4 Gigabases (Gbases) of raw data (163X genomic coverage based on a genome size of 2.5 Gbases). Unfiltered reads from each lane were assembled with Discovar De Novo to generate 3 initial assemblies that were merged into a single assembly with Metassembler. The merged assembly was then scaffolded sequentially with multiple mate pair libraries (3, 8 and 20 kb insert size) using SSPACE. The final assembly was filtered for contigs and scaffolds >= 1kb resulting in 68,793 scaffolds with N50 of 374,687 bp and a total length of 2.523 Gbp (Table. 1). This corresponds to 98.1% of the size of the current reference mouse genome (GRCm38.p4) of 2.671 Gbp. The average GC content (the percent of guanine and cytosine bases present on the DNA) of the gerbil assembly was 42.09% (Fig. 1C), similar to the mouse at 42.49%.
Table 1. Summary of assembly statistics showing number of contigs/scaffolds, N50, NG50 and % GC content.
The assembly was filtered for compliance with NCBI annotation guidelines. Scaffolds and unplaced contigs of <1kb length were filtered, which removed 316,114 short contig/scaffolds (average length 309 bp) accounting for 3.7% of the initial assembly length (97.7 Mbp).
| Total contigs/scaffolds | 384,902 |
| Total bp | 2,620,810,971 |
| Scaffolds/contigs >= 1kb | 68,788 |
| Total bp >= 1kb | 2,523,112,562 (96.3% of total bp) |
| (excluding Ns) | 2,402,558,981 (4.77% gaps = N) |
| Scaffold N50 | 351,937 bp |
| Scaffold L50 | 13,038 |
| Maximum scaffold | 6,569,692 bp |
| GC content | 42.09% |
Repeats were identified de novo using RepeatModeler, resulting in masking of 33.82% of the genome (853 megabases (Mbases) in 3.8 million elements) (Table S1). These repeats were mainly consisting of LINE1 type elements (13.48% of genome) and unclassified elements (15.78% of genome) with a smaller contribution of LTR elements (1.54% of genome). For comparison, the NCBI annotation masked 35.94% of the genome with RepeatMasker and 30.74% with WindowMasker. The genome masked with WindowMasker was used for subsequent alignment of transcripts and proteins during the NCBI annotation run. The relatively low percentage (34%) of the gerbil genome as repetitive sequences, as compared to the mouse reference genome (44.16%), is likely due to known difficulties with assembly of repetitive sequences leading to the exclusion of small contigs and highly repetitive regions from the draft assembly [66]. Further improvement of the gerbil assembly will allow more detailed analysis by resolving these differences with long read sequencing approach [67].
The masked genome was annotated by the NCBI pipeline mainly by alignment of transcripts, proteins, and existing Gerbil RNA-Seq data from the Sequence Read Archive (SRA) from NCBI. A total of 38,750 mRNAs composed of 227,097 exons were annotated for a total of 23,273 genes (Table 2). Completeness of the assembly was assessed by comparing the 38,750 encoded proteins to the Euarchontoglire single copy orthologous protein database (6192 sequences) with Benchmarking Universal Single-Copy Orthologs (BUSCO) [60] (Table S2). This analysis found 93.6% complete Buscos with 40.1% duplicated. An additional 4.9% of Buscos found were fragmented, and 1.5% were missing.
Table 2. De novo gene annotations.
Annotations were generated by the NCBI eukaryotic annotation pipeline, using existing gerbil transcripts, proteins, and RNA-Seq data from the Sequence Read Archive (SRA) as evidence.
| NCBI annotation | mean length (bp) |
median length (bp) |
minimal length (bp) |
maximum length (bp) |
|
|---|---|---|---|---|---|
| start_codon | 38,750 | ||||
| exon | 227,097 | 262 | 135 | 1 | 17,106 |
| intron | 201,876 | 3,803 | 1,312 | 30 | 452,462 |
| CDS | 38,750 | 1,809 | |||
| all transcripts | 40,519 | 2,708 | 2,212 | 55 | 99,751 |
| mRNA | 59,721 | 2,774 | 2,266 | 117 | 99,751 |
| misc_RNA | 487 | 2,678 | 2,449 | 211 | 10,553 |
| tRNA | 396 | 74 | 72 | 69 | 84 |
| lncRNA | 886 | 995 | 648 | 55 | 8,241 |
| gene | 23,273 | 31,095 | 14,417 | 69 | 753,783 |
Since this is the first report on genome assembly and annotation of the gerbil genome we compared it with other short assembled genomes from other close related species. The assembly metrics (contig N50 of 46.5 kb; annotated protein count 38,763; total length 2523 Mb and GC content 42.1%) are comparable to other short read-based Muroidea de novo genome assemblies such as Cricetulus griseus (Chinese hamster, https://www.ncbi.nlm.nih.gov/assembly/GCF_000223135.1. Accessed 20 Feb 2018) and Peromyscus maniculatus (prairie deer mouse, https://www.ncbi.nlm.nih.gov/assembly/GCF_000500345.1. Accessed 20 Feb 2018). Further analysis revealed not surprisingly, that the majority of identified gerbil genes (81%) are shared between mouse and human. Similarly, it has been found that the complete mitochondrial genome sequence of M. unguiculatus (GenBank accession Nos. KF425526 and NC_023263) displays the typical complement of 37 genes, and a similar base composition and codon usage as compared to several other rodent species [61, 62] Almost identical sequence of gene and protein for 8 individual genes further indicate that these shared genes are highly conserved among these species. For the remaining 19% of genes, 6% are shared between gerbil and mouse, while only 1.5% are shared with human. This observation is not surprising as gerbils and mice belong to the muridae family of rodentia and are far closer evolutionarily than gerbils and humans. Interestingly, the genes shared only between human and gerbil, appear to be mainly involved in gene expression and gene regulation, while the genes shared only by mouse and gerbil are dominated by olfactory and gustatory sensing. Although a more detailed analysis of the particular gene regulatory networks shared by human and gerbil is needed to understand the basis of these differences, one plausible explanation could be shared characteristics of social structure such as communal living.
3.2. Comparison to mouse genome
Meriones unguiculatus belongs to the Muridae family, which includes mouse and rat. The gerbil karyotype, like the rat, contains 21 chromosomes compared to 20 for the mouse [63]. We examined synteny with the mouse genome by mapping gerbil scaffolds of over 1 Mbp length (683 scaffolds totaling 556 Mbp, equal to 22% of the draft gerbil genome; Fig. 1D; Table 3). Overall, 346 syntenic blocks were found among the mouse autosomes, amounting to 27% coverage of the mouse genome with 1% doubled coverage. Syntenic blocks corresponding to scaffolds with translocations were evident for mouse chromosomes 2, 12 and 16. Coverage of the mouse autosomes varied between 14% (chromosome 16) to 43% (chromosome 11). Only 2% of the mouse X chromosome was covered in 12 syntenic blocks, and no syntenic regions were found on the mouse Y chromosome. This low percentage of syntenic blocks between mouse chromosomes and gerbil scaffolds is not unexpected given divergence of Gerbillidae sex chromosomes and apparently autosomal translocations into the X and Y chromosomes [68– 71].
Table 3. Synteny mapping against mouse GRCm38 reference genome.
658 gerbil scaffolds of length > 1 Mpb were mapped against the reference mouse chromosomes, and the relative coverage of the mouse chromosomes is indicated (% Mouse Covered) along with count of syntenic blocks found (#Synteny blocks). A total of 346 syntenic blocks were detected.
| Mouse Chromosome |
Mouse Chr Length | % Mouse Covered |
% Double Coverage |
# Synteny Blocks |
|---|---|---|---|---|
| 1 | 195472000 | 25% | 2% | 56 |
| 2 | 182113000 | 30% | 1% | 58 |
| 3 | 160039000 | 24% | 0% | 44 |
| 4 | 156508000 | 30% | 2% | 40 |
| 5 | 151834000 | 25% | 1% | 43 |
| 6 | 149736000 | 32% | 1% | 63 |
| 7 | 145441000 | 27% | 0% | 54 |
| 8 | 129401000 | 35% | 1% | 48 |
| 9 | 124595000 | 30% | 0% | 43 |
| 10 | 130695000 | 19% | 2% | 44 |
| 11 | 122082000 | 43% | 1% | 56 |
| 12 | 120129000 | 27% | 0% | 38 |
| 13 | 120421000 | 26% | 2% | 44 |
| 14 | 124902000 | 20% | 0% | 33 |
| 15 | 104043000 | 20% | 1% | 26 |
| 16 | 98207000 | 14% | 1% | 31 |
| 17 | 94987000 | 30% | 2% | 33 |
| 18 | 90702000 | 20% | 2% | 25 |
| 19 | 61431000 | 36% | 0% | 22 |
| X | 171031000 | 2% | 0% | 12 |
| Y | 91744000 | 0% | 0% | 0 |
3.3. Comparison to mouse and human proteomes
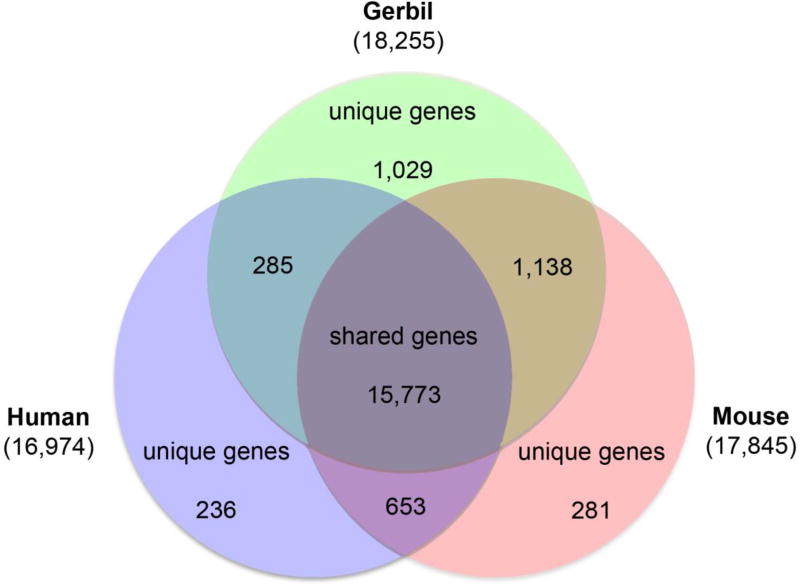
The 38,750 gerbil proteins encoded by the consensus gene set were compared to the human and mouse reference proteomes (GRCh38 and GRCm38 from UniProt) using TriFusion to determine orthologous groups. From a total dataset of 1,728,973 protein sequences for all three species, 19,395 total orthologs were detected as shown in Figure 2 as numbers inside Venn diagram. These numbers include 11,225 single-copy orthologs. 81.3% of the total orthologs were shared between all three taxa (15,773 total), while 84.7% were shared only between human and mouse (16,426 total). Gerbil shared 87.2% with mouse (16,911 total) and 82.8% with human (16,058). 1,029 of orthologs were unique to gerbil, compared to 281 and 236 unique to mice and human respectively. The 1,029 ortholog groups unique to gerbil contained a total of 2,652 annotated protein sequences, and 734 of the ortholog groups (71%) contained only 2 members with the remainder containing 3 to 12 members. The majority of the 1,029 gerbil ortholog groups were attributable to annotated protein isoforms (780, 76%).
Figure 2. Common genes shared among human, mouse and Mongolian gerbil.
The 38,750 protein sequences of gerbil annotated by the NCBI pipeline were compared to the mouse and human reference proteomes (GRCh38 and GRCm38) to identify orthologs. A total of 19,395 ortholog groups were identified, the majority of which are shared by all three species (15,773; 81.3%). Gerbil shared 87.2% with mouse (16,911 total) and 82.8% with human (16,058), while 84.7% were shared only between human and mouse (16,426 total). 1,029 ortholog groups were unique to gerbil (5.3%), compared to 281 unique to mouse and 236 unique human ortholog groups. The majority of the 1,029 gerbil ortholog groups were attributable to annotated protein isoforms (780, 76%).
DAVID analysis of functional annotation enrichment for 760 genes shared by gerbil and mouse but not human resulted in 60 enriched clusters, with the highest cluster enrichment score of 71.71 (DAVID enrichment score equals the geometric mean of the annotation term’s modified Fisher Exact p-values expressed as -log [https://david.ncifcrf.gov/helps/functional_annotation.html#fisher]) for the cluster containing GO terms ‘sensory perception of smell’ (6.1-fold enrichment, p-value 7.61e-115) and ‘olfactory receptor activity’ (5.5-fold enrichment, p-value 2.19e-107). The next highest cluster had an enrichment score of 3.60 and contained GO terms ‘response to stimulus’ (3.9-fold enrichment, p-value 1.18e-8) and ‘bitter taste receptor activity’ (8.2-fold enrichment, p-value 9.12e-6). In contrast, analysis of 194 genes shared by gerbil and human but not mouse resulted in 25 enriched term clusters with enrichment scores of less than 2, with the highest enrichment score of 1.69 for the cluster containing GO term ‘RNA binding’ (2.9-fold enrichment, p-value 9.99e-4) followed by a cluster with enrichment score of 1.52 containing GO term ‘sequence specific DNA binding’ (2.4-fold enrichment, p-value 1.5e-2). Mouse-human shared gene analysis (538 genes) yielded 76 enriched clusters with maximum enrichment score of 2.9 for the cluster containing for GO term ‘synapse’ (2.7-fold enrichment, p-value 1.50e-4), enrichment score 2.9 for the cluster containing GO term ‘GTP-binding’ (3.5-fold enrichment, p-value 7.34e-7), and enrichment score 2.49 for the cluster containing GO term ‘ion transport’ (which includes neurotransmitter-gated ion channels and extracellular ligand-gated ion channels; 12.4-fold enrichment, p-value 9.77e-5). Blastp [61] and ProSplign [https://www.ncbi.nlm.nih.gov/sutils/static/prosplign/prosplign.html] [62]. (Accessed 22 Feb 2018) alignments against human and mouse RefSeq proteomes yielded average protein sequence identity of 73.2 and 76.03% respectively, and average query alignment of 80.25 and 84.55% respectively (Table S3).
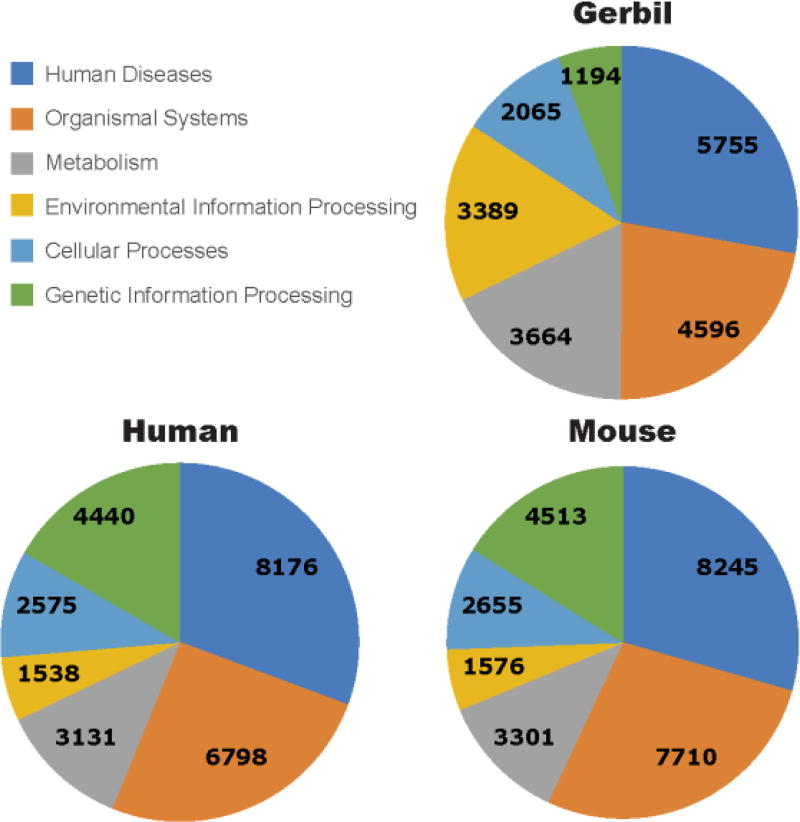
The annotated gerbil protein sequences were classified into the Kyoto Encyclopedia of Genes and Genomes (KEGG) functional categories using Ghost-Koala [GhostKoala [http://www.kegg.jp/ghostkoala/]. Accessed 27 April 2017] and compared to the mouse and human reference protein sets (Fig. 3). A total of 70.1% (27,182 out of 38,750) gerbil proteins were assigned to KEGG categories. The top two categories were Human Diseases (5,755 hits) and Organismal Systems (4,596 hits), similar to human and mouse. Metabolism and Environmental Information Processing were next (3,664 hits and 3,389 hits, respectively), followed by Cellular Processes (2,065 hits) and Genetic Information Processing (1,195 hits). Altogether these top six categories accounted for 20,664 (76%) of the 27,182 proteins assigned.
Figure 3. Venn Diagram comparing protein functional assignments among human, mouse and Mongolian gerbil.
Protein sequences of gerbil were annotated by the NCBI pipeline were assigned to KEGG orthology groups and functional category counts were compared to the mouse and human references. As seen in the pie charts, the two largest categories for all three species are Human Diseases and Organismal Systems. Gerbil however, has a larger number of hits under the Environmental Information Processing and Metabolism categories when compared to humans and mouse. In contrast, gerbil shows relatively fewer hits in Genetic Information Processing. Numbers in the pie chart show the actual number of hits represented in GO terms in each category for that species
3.4. Detailed comparison to eight mouse genes
To refine the comparison to known mouse genes at a finer level, we searched the assembly for scaffolds corresponding to 8 specific genes with great interests in the field of sensory processing and social interaction; two areas where gerbil physiology and behavior resemble human characteristics more closely than mice characteristics. These 8 genes can be divided into two groups. Genes in the first group (ATP2B2, Gabra1, Gabrb2, Kcna1, Kcnc1 and Gphn) have important function in regulating neuronal activity and some are particularly critical for the survival and normal function of fast-spiking auditory cells and neurons. The gene ATP2B2 encodes the type 2 of the plasma membrane calcium ATPase (PMCA2). PMCA is a major calcium efflux system that sets the resting calcium concentration [72– 74]. PMCA2, the most efficient type of PMCA, is necessary for hair cell survival in the cochlea. Spontaneous and induced mutations in ATP2B2 are associated with hearing loss in both humans and mice [75– 82]. In addition, PMCA2 is highly expressed in auditory neurons and is involved in the tonotopic organization of auditory cell groups [83, 84]. Gabra1 and Gabrb2 encode the two essential subunits, alpha 1 and beta 2, of GABA receptors that underlie the chief inhibitory neurotransmission in the brain. In mice and human, Gabra1 and Gabrb2 mutations have been associated with generalized and syndromic epilepsy as well as with intellectual disabilities [85– 87]. Gephyrin, encoded by Gphn, is a neuronal assembly protein that anchors inhibitory neurotransmitter receptors, including GABA receptors, to the postsynaptic cytoskeleton [88]. Kcna1 and Kcnc1 are the genes for voltage-gated potassium channels Kv1.1 and Kv3.1, respectively. These two potassium channels are necessary and kinetically optimized for high-frequency action potential generation and temporal processing with submillisecond temporal resolution [89– 91]. The availability of the gerbil genome enables genetic approaches for further determining the roles of these genes in the gerbil, an advantageous model for studying hearing, especially with regard to temporal processing and for studying epilepsy.
The second group contains Fmr1 and Oxtr, two genes extensively involved in neurological disorders associated with communication and social deficits. Transcriptional silencing of Fmr1 and the resultant loss of its product, the fragile x mental retardation protein (FMRP), are responsible for the fragile X syndrome (FXS) [92, 93]. FXS is characterized with prominent auditory dysfunction and autism-like social difficulties. Examination of human FXS and/or autism brains reveal dramatically disorganized auditory brainstem in particular the medial superior olive (MSO), a center for auditory temporal processing [94– 98]. Gerbils, but not mice, display a well-developed MSO that is structurally and functionally comparable to human [99]. An FMRP knockout gerbil strain can serve as a disease model for FXS and help determine the pathology of auditory dysfunction especially those associated with temporal processing.
The oxytocin receptor (OXTR) is a G-protein coupled receptor for the hormone and neurotransmitter oxytocin [100]. Oxytocin receptors are expressed by the myoepithelial cells of the mammary gland, playing an important role as an inducer of uterine contractions during parturition and of milk ejection. Oxytocin receptors are also present in the central nervous system, modulating a variety of behaviors, including stress and anxiety, social memory and recognition, sexual and aggressive behaviors, bonding (affiliation) and maternal behavior [101– 104]. The prominent bonding and maternal behaviors in gerbils provide an excellent model for studying OXTR mediated social interactions at the genetic and molecular levels.
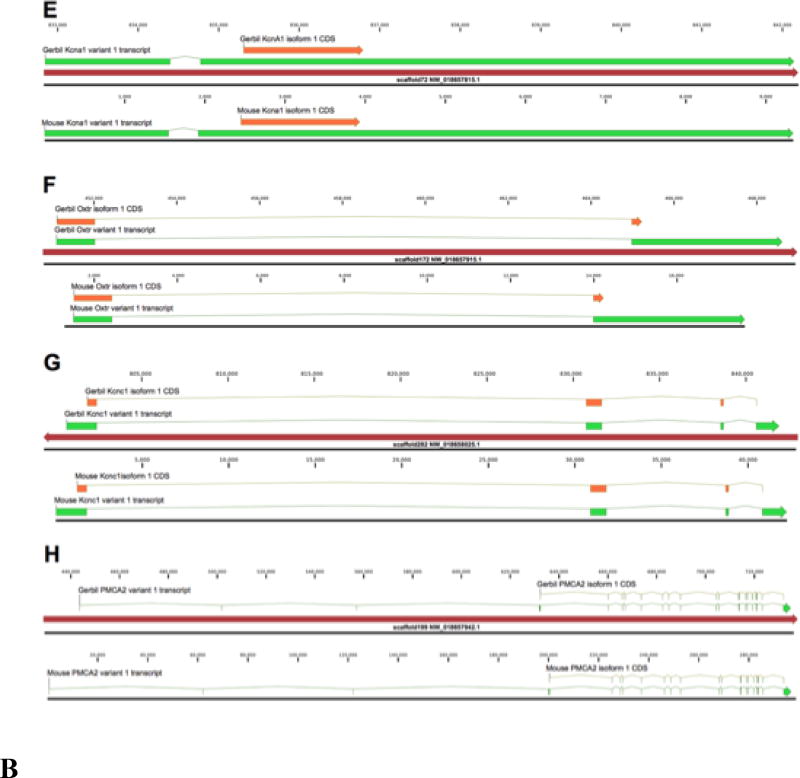
Blastn searches using the full mouse gene loci and their major transcripts allowed identification of 2 to 8 scaffolds covering the entire coding regions of each of the eight genes (Fig. 4). Gerbil exons and CDSs of the major transcript for each gene were annotated by direct genomic sequence comparison of gerbil to mouse. The overall structure of the 8 gerbil genes is nearly identical to that in mouse, with complete conservation of exon counts and specific splice junctions. Minor variation was seen in average exon length, with the most variation occurring in intronic regions. Comparison of the encoded proteins showed 100% identity for Gabrb2, and only 1 to 4 amino acid differences (99.7%-99.8% identity) for Gabra1, Kcna1, Kcnc1 and ATP2B2. Gphn, Fmr1, and Oxtr were also highly conserved with 6 (99.22% identity), 16 (97.40%) and 18 (97.37%) amino acid differences from mouse proteins, respectively (Table 4). Four of the eight genes were assembled as single scaffolds (Kcna1, Oxtr, Kcnc1 and ATP2B2), while Fmr1, Gabra1 and Gabrb2 each spanned two scaffolds. The Gphn gene has exons found on four individual scaffolds. Inter-scaffold breaks always occurred within introns, typically in highly repetitive di- or tri-nucleotide repeats.
Figure 4. Comparative gene structure for 8 orthologs between gerbil and mouse.
The mouse gene and mRNA sequences (green) were used to identify homologous Mongolian gerbil scaffolds (brown arrows) using blast. Comparison at the nucleotide level enabled identification of splice junctions and protein coding sequences (CDSs, illustrated in orange), due to the high level of relatedness between the two species. A, Fragile X-mental retardation gene (FMR1). B, Gamma amino butyric acid receptor alpha 1 gene (Gabra1). Note that mouse and gerbil genes are drawn to different scales due to large size of the penultimate intron in the gerbil Gabra1 gene. However, the significance of the apparent intron enlargement is unclear and could be attributable to the de novo assembly artifacts. C, Gamma amino butyric acid receptor beta 2 gene (Gabrb2). D, Gephyrin gene (Gphn). A flanking Fam71 gene is downstream of the Gphn gene in both gerbil (showing here) and mouse (not illustrated). E, Potassium voltage-gated channel subfamily a member 1 (Kcna1). F, Oxytocyn receptor (Oxtr). G, Potassium voltage-gated channel subfamily C member 1 (Kcnc1). H, ATPase plasma membrane Ca2+ transporting 2 (PMCA2).
Table 4. Gene structure comparison of 8 genes between gerbil and mouse.
Overall, the gerbil genes exhibited extremely similar structure in comparison to mouse. No differences in Exon (E) count were detected, although minor differences in average E length were apparent in all genes except PMCA2. Major differences were seen in average Intron (I) length in Gabra1 and Gpnh, however the significance of the apparent intron expansion is unclear and may be attributable to assembly artifacts. G, Gerbil; M, mouse; Avg., Average; bp, base pairs; ID, Identity.
| Fmr1 | Gabra1 | Gabrb2 | Gpnh | Kcna1 | Oxtr | Kcnc1 | PMCA2 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| species | G | M | G | M | G | M | G | M | G | M | G | M | G | M | G | M |
| exoncount | 17 | 17 | 10 | 10 | 10 | 10 | 23 | 23 | 2 | 2 | 2 | 2 | 4 | 4 | 22 | 22 |
| avg. E length (bp) | 260 | 258 | 749 | 754 | 142 | 143 | 142 | 143 | 4463 | 4484 | 2280 | 2283 | 1060 | 1083 | 195 | 195 |
| intron count | 16 | 16 | 9 | 9 | 9 | 9 | 22 | 22 | 1 | 1 | 1 | 1 | 3 | 3 | 21 | 21 |
| avg. I length (bp) | 2235 | 2190 | 19563 | 5257 | 25259 | 22810 | 15509 | 2068 | 376 | 370 | 12960 | 11558 | 12354 | 12635 | 24419 | 25019 |
| amino acids | 615 | 614 | 455 | 455 | 474 | 474 | 772 | 772 | 495 | 495 | 387 | 388 | 585 | 585 | 1198 | 1198 |
| ID | 599/615 (97.40%) | 454/455 (99.78%) | 474/474 (100%) | 766/772 (99.22%) | 494/495 (99.80%) | 370/388 (97.37%) | 583/585 (99.66%) | 1194/1198 (99.67%) | ||||||||
In summary we have sequenced the Meriones unguiculatus genome and reported here its initial sequence annotation and characterization. We have compared our data set with human and mouse genomes. As expected we have found some similarities and some differences among these data sets. We specifically compared the chromosomal structure of eight genes with high relevance for controlling neural signaling and demonstrate a high degree of homology between these genes in mouse and gerbil. Taken together, the information generated in this study provides an extreme valuable resource that will help researchers advance our knowledge in realms of both behavior and neural function at the molecular level.
Supplementary Material
Acknowledgments
We would like to thank Brendan P. Keough and Brandon J. Converse for technical assistance during the initial set up in the De Novo sequencing of the genome. We thank Dr. Sukant Khurana for providing the photograph in Fig. 1A. This research project is funded in part by NIH grants DC013074 to YW, DC006877 and DC 016169 to NLG, DC011284 to DHS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession number
The Whole Genome Shotgun sequence data from this project has been deposited at DDBJ/ENA/GenBank under the accession NHTI00000000. The version described in this paper is version NHTI01000000. The fragment reads, and mate pair reads have been deposited in the Sequence Read Archive under BioSample accession SAMN06897401.
Conflict of interests
The authors declare no conflict of interests.
Author contributions
Conceived and designed the experiments: YW, DARZ, EWR, DS, NG. Contributed Materials: DARZ, YW. Performed the experiments: SM, DARZ. Analyzed the data: SM, DARZ. Wrote the paper: YW, DARZ, SM, DA, NG, EWR.
References
- 1.Schwentker V. The gerbil. A new laboratory animal. Illinois Veterinarian. 1963;6:5–9. [Google Scholar]
- 2.Batchelder M, Keller LS, Sauer MB, West WL. The Laboratory Rabbit, Guinea Pig, Hamster, and Other Rodents. Academic Press. Chapter. 2012;52:1132–1155. [Google Scholar]
- 3.Nowak RM. Walker’s Mammals of the World. Sixth. II. The John Hopkins University Press; London and Baltimore: 1999. [Google Scholar]
- 4.Chevret P, Dobigny G. Systematics and evolution of the subfamily Gerbillinae (Mammalia, Rodentia, Muridae) Mol. Phylogenet. Evol. 2005;35(3):674–688. doi: 10.1016/j.ympev.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 5.Yang S, Luo X, Xiong G, So KF, Yang H, Xu Y. The electroretinogram of Mongolian gerbil (Meriones unguiculatus): comparison to mouse. Neurosci Lett. 2015;589:7–12. doi: 10.1016/j.neulet.2015.01.018. [DOI] [PubMed] [Google Scholar]
- 6.Baker AG, Emerson VF. Grating acuity of the Mongolian gerbil (Meriones unguiculatus) Behav Brain Res. 1983;8(2):195–209. doi: 10.1016/0166-4328(83)90054-2. [DOI] [PubMed] [Google Scholar]
- 7.Govardovskii VI, Röhlich P, Szél A, Khokhlova TV. Cones in the retina of the Mongolian gerbil, Meriones unguiculatus: an immunocytochemical and electrophysiological study. Vision Res. 1992;32:19–27. doi: 10.1016/0042-6989(92)90108-u. [DOI] [PubMed] [Google Scholar]
- 8.Bytyqi AH, Layer PG. Lamina formation in the Mongolian gerbil retina (Meriones unguiculatus) Anat Embryol (Berl) 2005;209:217–225. doi: 10.1007/s00429-004-0443-9. [DOI] [PubMed] [Google Scholar]
- 9.Zhang T, Huang L, Zhang L, Tan M, Pu M, Pickard GE, So KF, Ren C. ON and OFF retinal ganglion cells differentially regulate serotonergic and GABAergic activity in the dorsal raphe nucleus. Sci Rep. 2016;6:26060. doi: 10.1038/srep26060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garbers C, Henke J, Leibold C, Wachtler T, Thurley K. Contextual processing of brightness and color in Mongolian gerbils. J Vis. 2015;15(1):15.1.13. doi: 10.1167/15.1.13. [DOI] [PubMed] [Google Scholar]
- 11.Delbarre B, Delbarre G, Rochat C, Calinon F. Effect of piribedil, a D-2 dopaminergic agonist, on dopamine, amino acids, and free radicals in gerbil brain after cerebral ischemia. Mol Chem Neuropathol. 1995;26(1):43–52. doi: 10.1007/BF02814940. [DOI] [PubMed] [Google Scholar]
- 12.Mauck MC, Mancuso K, Kuchenbecker JA, Connor TB, Hauswirth WW, Neitz J, Neitz M. Longitudinal evaluation of expression of virally delivered transgenes in gerbil cone photoreceptors. Vis Neurosci. 2008;25(3):273–82. doi: 10.1017/S0952523808080577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lay DM. The anatomy, physiology, functional significance and evolution of specialized hearing organs of gerbilline rodents. J Morphol. 1972;138(1):41–120. doi: 10.1002/jmor.1051380103. [DOI] [PubMed] [Google Scholar]
- 14.Ryan A. Hearing sensitivity of the mongolian gerbil, Meriones unguiculatis. J Acoust Soc Am. 1976;59(5):1222–6. doi: 10.1121/1.380961. [DOI] [PubMed] [Google Scholar]
- 15.Heffner RS, Koay G, Heffner HE. Audiograms of five species of rodents: implications for the evolution of hearing and the perception of pitch. Hear Res. 2001;157(1–2):138–52. doi: 10.1016/s0378-5955(01)00298-2. [DOI] [PubMed] [Google Scholar]
- 16.Gleich O, Strutz J. The Mongolian Gerbil as a Model for the Analysis of Peripheral and Central Age-Dependent Hearing Loss. In: Sadaf Naz., editor. Hearing Loss. 2012. pp. 67–92. Chapter 4. [Google Scholar]
- 17.Belliveau LA, Lyamzin DR, Lesica NA. The neural representation of interaural time differences in gerbils is transformed from midbrain to cortex. J Neurosci. 2014;34(50):16796–808. doi: 10.1523/JNEUROSCI.2432-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Budinger E, Scheich H. Anatomical connections suitable for the direct processing of neuronal information of different modalities via the rodent primary auditory cortex. Hear Res. 2009;258(1–2):16–27. doi: 10.1016/j.heares.2009.04.021. [DOI] [PubMed] [Google Scholar]
- 19.Wang Y, Sakano H, Beebe K, Brown MR, de Laat R, Bothwell M, Kulesza RJ, Jr, Rubel EW. Intense and specialized dendritic localization of the fragile X mental retardation protein in binaural brainstem neurons: a comparative study in the alligator, chicken, gerbil, and human. J Comp Neurol. 2014;522(9):2107–28. doi: 10.1002/cne.23520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson SL. Membrane properties specialize mammalian inner hair cells for frequency or intensity encoding. Elife. 2015;4 doi: 10.7554/eLife.08177. pii: e08177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Franken TP, Roberts MT, Wei L, Golding NL, Joris PX. In vivo coincidence detection in mammalian sound localization generates phase delays. Nat Neurosci. 2015;18(3):444–52. doi: 10.1038/nn.3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mowery TM, Kotak VC, Sanes DH. The onset of visual experience gates auditory cortex critical periods. Nat Commun. 2016;7:10416. doi: 10.1038/ncomms10416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.von Trapp G, Buran BN, Sen K, Semple MN, Sanes DH. A Decline in Response Variability Improves Neural Signal Detection during Auditory Task Performance. J Neurosci. 2016;36(43):11097–11106. doi: 10.1523/JNEUROSCI.1302-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson SL, Olt J, Cho S, von Gersdorff H, Marcotti W. The Coupling between Ca2+ Channels and the Exocytotic Ca2+ Sensor at Hair Cell Ribbon Synapses Varies Tonotopically along the Mature Cochlea. J Neurosci. 2017;37(9):2471–2484. doi: 10.1523/JNEUROSCI.2867-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stange-Marten A, Nabel AL, Sinclair JL, Fischl M, Alexandrova O, Wohlfrom H, Kopp-Scheinpflug C, Pecka M, Grothe B. Input timing for spatial processing is precisely tuned via constant synaptic delays and myelination patterns in the auditory brainstem. Proc Natl Acad Sci U S A. 2017;114(24):E4851–E4858. doi: 10.1073/pnas.1702290114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosowski JJ, Ravicz ME, Teoh SW, Flandermeyer D. Measurements of middle-ear function in the Mongolian gerbil, a specialized mammalian ear. Audiol Neurootol. 1999;4(3–4):129–36. doi: 10.1159/000013831. [DOI] [PubMed] [Google Scholar]
- 27.Chan WX, Lee SH, Kim N, Shin CS, Yoon YJ. Mechanical model of an arched basilar membrane in the gerbil cochlea. Hear Res. 2017;345:1–9. doi: 10.1016/j.heares.2016.12.003. [DOI] [PubMed] [Google Scholar]
- 28.Risoud M, Sircoglou J, Dedieu G, Tardivel M, Vincent C, Bonne NX. Imaging and cell count in cleared intact cochlea in the Mongolian gerbil using laser scanning confocal microscopy. Eur Ann Otorhinolaryngol Head Neck Dis. 2017;134(4):221–224. doi: 10.1016/j.anorl.2017.01.001. [DOI] [PubMed] [Google Scholar]
- 29.Winters BD, Jin SX, Ledford KR, Golding NL. Amplitude Normalization of Dendritic EPSPs at the Soma of Binaural Coincidence Detector Neurons of the Medial Superior Olive. J Neurosci. 2017;37(12):3138–3149. doi: 10.1523/JNEUROSCI.3110-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cant NB. Patterns of convergence in the central nucleus of the inferior colliculus of the Mongolian gerbil: organization of inputs from the superior olivary complex in the low frequency representation. Front Neural Circuits. 2013;6(7):29. doi: 10.3389/fncir.2013.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cant NB, Benson CG. Organization of the inferior colliculus of the gerbil (Meriones unguiculatus): differences in distribution of projections from the cochlear nuclei and the superior olivary complex. J Comp Neurol. 2006;495(5):511–28. doi: 10.1002/cne.20888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cant NB, Benson CG. Multiple topographically organized projections connect the central nucleus of the inferior colliculus to the ventral division of the medial geniculate nucleus in the gerbil, Meriones unguiculatus. J Comp Neurol. 2007;503(3):432–53. doi: 10.1002/cne.21391. [DOI] [PubMed] [Google Scholar]
- 33.Cant NB, Benson CG. Organization of the inferior colliculus of the gerbil (Meriones unguiculatus): projections from the cochlear nucleus. Neuroscience. 2008;154(1):206–17. doi: 10.1016/j.neuroscience.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holman SD, Seale WT. Ontogeny of sexually dimorphic ultrasonic vocalizations in Mongolian gerbils. Dev Psychobiol. 1991;24(2):103–15. [Google Scholar]
- 35.Kobayasi KI, Riquimaroux H. Classification of vocalizations in the Mongolian gerbil, Meriones unguiculatus. J Acoust Soc Am. 2012;131(2):1622–31. doi: 10.1121/1.3672693. [DOI] [PubMed] [Google Scholar]
- 36.Ter-Mikaelian M, Yapa WB, Rubsamen R. Vocal behavior of the Mongolian gerbil in a seminatural enclosure. Behaviour. 2012;149:461–492. [Google Scholar]
- 37.Hellstrom LI, Schmiedt RA. Measures of tuning and suppression in single-fiber and whole-nerve responses in young and quiet-aged gerbils. J Acoust Soc Am. 1996;100(5):3275–85. doi: 10.1121/1.417211. [DOI] [PubMed] [Google Scholar]
- 38.Hamann I, Gleich O, Klump GM, Kittel MC, Boettcher FA, Schmiedt RA, Strutz J. Behavioral and evoked-potential thresholds in young and old Mongolian gerbils (Meriones unguiculatus) Hear Res. 2002;171(1–2):82–95. doi: 10.1016/s0378-5955(02)00454-9. [DOI] [PubMed] [Google Scholar]
- 39.Loskota WJ, Lomax P, Rich ST. The gerbil as a model for the study of the epilepsies. Seizure patterns and ontogenesis. Epilepsia. 1974;15:109–119. doi: 10.1111/j.1528-1157.1974.tb04000.x. [DOI] [PubMed] [Google Scholar]
- 40.Ludvig N, Farias PA, Ribak CE. An analysis of various environmental and specific sensory stimuli on the seizure activity of the Mongolian gerbil. Epilepsy Res. 1991;8(1):30–5. doi: 10.1016/0920-1211(91)90033-c. [DOI] [PubMed] [Google Scholar]
- 41.Kang TC, Park SK, Bahn JH, Jeon SG, Jo SM, Cho SW, Choi SY, Won MH. The alteration of gamma-aminobutyric acid-transaminase expression in the gerbil hippocampus induced by seizure. Neurochem Int. 2001;38(7):609–14. doi: 10.1016/s0197-0186(01)00002-x. [DOI] [PubMed] [Google Scholar]
- 42.Hwang IK, Park SK, An SJ, Yoo KY, Kim DS, Jung JY, Won MH, Choi SY, Kwon OS, Kang TC. GABAA, not GABAB, receptor shows subunit- and spatial-specific alterations in the hippocampus of seizure prone gerbils. Brain Res. 2004;1003(1–2):98–107. doi: 10.1016/j.brainres.2003.12.026. [DOI] [PubMed] [Google Scholar]
- 43.Kwak SE, Kim JE, Kim DS, Jung JY, Won MH, Kwon OS, Choi SY, Kang TC. Effects of GABAergic transmissions on the immunoreactivities of calcium binding proteins in the gerbil hippocampus. J Comp Neurol. 2005;485(2):153–64. doi: 10.1002/cne.20482. [DOI] [PubMed] [Google Scholar]
- 44.Levine S, Payan H. Effects of ischemia and other procedures on the brain and retina of the gerbil (Meriones unguiculatus) Exp.Neurol. 1966;16:255–262. doi: 10.1016/0014-4886(66)90062-8. [DOI] [PubMed] [Google Scholar]
- 45.Delbarre G, Delbarre B, Barrau Y. A suitable method to select gerbils with incomplete circle of Willis. Stroke. 1988;19(1):126. doi: 10.1161/01.str.19.1.126. [DOI] [PubMed] [Google Scholar]
- 46.Pelliccioli GP, Gambelunghe C, Ottaviano PF, Iannaccone S, Ambrosini MV. Variable response of the Mongolian gerbil to unilateral carotid occlusion: magnetic resonance imaging and neuropathological characterization. Ital J Neurol Sci. 1995;16(8):517–26. doi: 10.1007/BF02282909. [DOI] [PubMed] [Google Scholar]
- 47.Small DL, Buchan AM. Animal Models. Br Med Bull. 2000;56(2):307–17. doi: 10.1258/0007142001903238. [DOI] [PubMed] [Google Scholar]
- 48.Lim BH, Noordin R, Nor ZM, Rahman RA, Abdullah KA, Sinnadurai S. Brugia malayi infection in Meriones unguiculatus: antibody response to recombinant BmR1. Exp Parasitol. 2204;108(1–2):1–6. doi: 10.1016/j.exppara.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 49.Shigeno S, Fujimaki Y, Toriyama K, Ichinose A, Mitsui Y, Aoki Y, Kimura E. Temporary shift of microfilariae of Brugia pahangi from the lungs to muscles in Mongolian jirds, Meriones unguiculatus, after a single injection of diethylcarbamazine. J Parasitol. 2006;92(5):1075–80. doi: 10.1645/GE-842R.1. [DOI] [PubMed] [Google Scholar]
- 50.Hübner MP, Torrero MN, McCall JW, Mitre E. Litomosoides sigmodontis: a simple method to infect mice with L3 larvae obtained from the pleural space of recently infected jirds (Meriones unguiculatus) Exp Parasitol. 2006;123(1):95–8. doi: 10.1016/j.exppara.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peek RM. Helicobacter pylori infection and disease: from humans to animal models. Dis Model Mech. 2008;1(1):50–5. doi: 10.1242/dmm.000364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wences AH, Schatz MC. Metassembler: merging and optimizing de novo genome assemblies. Genome Biol. 2008;16:207. doi: 10.1186/s13059-015-0764-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics. 2011;27(4):578–9. doi: 10.1093/bioinformatics/btq683. [DOI] [PubMed] [Google Scholar]
- 54.Smit AFA, Hubley R. RepeatModeler Open-1.0. 2008–2015 http://www.repeatmasker.org.
- 55.NCBI Eukaryotic Genome Annotation Pipeline guidelines. https://www.ncbi.nlm.nih.gov/genome/annotation_euk/process/
- 56.Soderlund C, Nelson W, Shoemaker A, Paterson A. SyMAP: A system for discovering and viewing syntenic regions of FPC maps. Genome Res. 2006;16(9):1159–68. doi: 10.1101/gr.5396706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.TriFusion v0.5.0 pipeline. http://odiogosilva.github.io/TriFusion/
- 58.Fischer S, Brunk BP, Chen F, Gao X, Harb OS, Iodice JB, Shanmugam D, Roos DS, Stoeckert CJ., Jr Using OrthoMCL to assign proteins to OrthoMCL-DB groups or to cluster proteomes into new ortholog groups. Curr Protoc Bioinformatics. 2011 doi: 10.1002/0471250953.bi0612s35. Chapter 6: Unit 6.12.1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 60.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31(19):3210–2. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 61.Kim EB, Lee SG. The complete mitochondrial genome of the Mongolian gerbil, Meriones unguiculatus (Rodentia: Muridae: Gerbillinae) Mitochondrial DNA A DNA Mapp. Seq. Anal. 2016;27:1457–1458. doi: 10.3109/19401736.2014.953091. [DOI] [PubMed] [Google Scholar]
- 62.Li CL, Du XY, Gao J, Wang C, Guo HG, Dai FW, Sa XY, An W, Chen ZW. Phylogenetic analysis of the Mongolian gerbil (Meriones unguiculatus) from China based on mitochondrial genome. Genet. Mol. Res. 2016;15(3) doi: 10.4238/gmr.15037703. [DOI] [PubMed] [Google Scholar]
- 63.Painter TS. A comparison of the chromosomes of the rat and mouse with reference to the question of chromosome homology in mammals. Genetics. 1928;13:181–189. doi: 10.1093/genetics/13.2.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Biscotti MA, Olmo E, Heslop-Harrison JS. Repetitive DNA in eukaryotic genomes. Chromosome Res. 2015;23(3):415–20. doi: 10.1007/s10577-015-9499-z. [DOI] [PubMed] [Google Scholar]
- 65.Mostovoy Y, Levy-Sakin M, Lam J, Lam ET, Hastie AR, Marks P, Lee J, Chu C, Lin C, Džakula Ž, Cao H, Schlebusch SA, Giorda K, Schnall-Levin M, Wall JD, Kwok PY. A hybrid approach for de novo human genome sequence assembly and phasing. Nat Methods. 13(7):587–90. doi: 10.1038/nmeth.3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ratomponirina C, Viegas-Péquignot E, Dutrillaux B, Petter F, Rumpler Y. Synaptonemal complexes in Gerbillidae: probable role of intercalated heterochromatin in gonosome-autosome translocations. Cytogenet Cell Genet. 1986;43(3–4):161–7. doi: 10.1159/000132315. [DOI] [PubMed] [Google Scholar]
- 67.Viegas-Péquignot E, Benazzou T, Dutrillaux B, Petter F. Complex evolution of sex chromosomes in Gerbillidae (Rodentia) Cytogenet Cell Genet. 1982;34(1–2):158–67. doi: 10.1159/000131804. [DOI] [PubMed] [Google Scholar]
- 68.Wahrman J, Richler C, Neufeld E, Friedmann A. The origin of multiple sex chromosomes in the gerbil Gerbillus gerbillus (Rodentia: Gerbillinae) Cytogenet Cell Genet. 1983;35(3):161–80. doi: 10.1159/000131863. [DOI] [PubMed] [Google Scholar]
- 69.de la Fuente R, Parra MT, Viera A, Calvente A, Gómez R, Suja JA, Rufas JS, Page J. Meiotic pairing and segregation of achiasmate sex chromosomes in eutherian mammals: the role of SYCP3 protein. PLoS Genet. 2007;3(11):e198. doi: 10.1371/journal.pgen.0030198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. BLAST+: architecture and applications. BMC Bioinformatics. 2008;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kiryutin B, Souvorov A, Tatusova T. ProSplign - Protein to Genomic Alignment Tool. https://www.ncbi.nlm.nih.gov/sutils/static/prosplign/prosplign.html.
- 72.Carafoli E. Intracellular calcium homeostasis. Annu Rev Biochem. 1987;56:395–433. doi: 10.1146/annurev.bi.56.070187.002143. [DOI] [PubMed] [Google Scholar]
- 73.Thayer SA, Usachev YM, Pottorf WJ. Modulating Ca2+ clearance from neurons. Front Biosci. 2002;1(7):d1255–79. doi: 10.2741/A838. [DOI] [PubMed] [Google Scholar]
- 74.Duman JG, Chen L, Hille B. Calcium transport mechanisms of PC12 cells. J Gen Physiol. 2008;2131(4):307–23. doi: 10.1085/jgp.200709915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dumont RA, Lins U, Filoteo AG, Penniston JT, Kachar B, Gillespie PG. Plasma membrane Ca2+-ATPase isoform 2a is the PMCA of hair bundles. J Neurosci. 2001;21(14):5066–78. doi: 10.1523/JNEUROSCI.21-14-05066.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kozel PJ, Friedman RA, Erway LC, Yamoah EN, Liu LH, Riddle T, Duffy JJ, Doetschman T, Miller ML, Cardell EL, Shull GE. Balance and hearing deficits in mice with a null mutation in the gene encoding plasma membrane Ca2+-ATPase isoform 2. J Biol Chem. 1998;273(30):18693–6. doi: 10.1074/jbc.273.30.18693. [DOI] [PubMed] [Google Scholar]
- 77.Kozel PJ, Davis RR, Krieg EF, Shull GE, Erway LC. Deficiency in plasma membrane calcium ATPase isoform 2 increases susceptibility to noise-induced hearing loss in mice. Hear Res. 2002;164(1–2):231–9. doi: 10.1016/s0378-5955(01)00420-8. [DOI] [PubMed] [Google Scholar]
- 78.Street VA, McKee-Johnson JW, Fonseca RC, Tempel BL, Noben-Trauth K. Mutations in a plasma membrane Ca2+-ATPase gene cause deafness in deafwaddler mice. Nat Genet. 1998;19(4):390–4. doi: 10.1038/1284. [DOI] [PubMed] [Google Scholar]
- 79.Penheiter AR, Filoteo AG, Croy CL, Penniston JT. Characterization of the deafwaddler mutant of the rat plasma membrane calcium-ATPase 2. Hear Res. 2001;62(1–2):19–28. doi: 10.1016/s0378-5955(01)00356-2. [DOI] [PubMed] [Google Scholar]
- 80.Schultz JM, Yang Y, Caride AJ, Filoteo AG, Penheiter AR, Lagziel A, Morell RJ, Mohiddin SA, Fananapazir L, Madeo AC, Penniston JT, Griffith AJ. Modification of human hearing loss by plasma-membrane calcium pump PMCA2. N Engl J Med. 2005;352(15):1557–64. doi: 10.1056/NEJMoa043899. [DOI] [PubMed] [Google Scholar]
- 81.Brini M, Di Leva F, Domi T, Fedrizzi L, Lim D, Carafoli E. Plasmamembrane calcium pumps and hereditary deafness. Biochem Soc Trans. 2007;35:913–8. doi: 10.1042/BST0350913. [DOI] [PubMed] [Google Scholar]
- 82.Tempel BL, Shilling DJ. The plasma membrane calcium ATPase and disease. Subcell Biochem. 2007;45:365–83. doi: 10.1007/978-1-4020-6191-2_13. [DOI] [PubMed] [Google Scholar]
- 83.Wang Y, Cunningham DE, Tempel BL, Rubel EW. Compartment-specific regulation of plasma membrane calcium ATPase type 2 in the chick auditory brainstem. J Comp Neurol. 2009;514(6):624–40. doi: 10.1002/cne.22045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Weatherstone JH, Kopp-Scheinpflug C, Pilati N, Wang Y, Forsythe ID, Rubel EW, Tempel BL. Maintenance of neuronal size gradient in MNTB requires sound-evoked activity. J Neurophysiol. 2017;117(2):756–766. doi: 10.1152/jn.00528.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Srivastava S, Cohen J, Pevsner J, Aradhya S, McKnight D, Butler E, Johnston M, Fatemi A. A novel variant in GABRB2 associated with intellectual disability and epilepsy. Am J Med Genet A. 2014;164A(11):2914–21. doi: 10.1002/ajmg.a.36714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kodera H, Ohba C, Kato M, Maeda T, Araki K, Tajima D, Matsuo M, Hino-Fukuyo N, Kohashi K, Ishiyama A, Takeshita S, Motoi H, Kitamura T, Kikuchi A, Tsurusaki Y, Nakashima M, Miyake N, Sasaki M, Kure S, Haginoya K, Saitsu H, Matsumoto N. De novo GABRA1 mutations in Ohtahara and West syndromes. Epilepsia. 2016;57(4):566–73. doi: 10.1111/epi.13344. [DOI] [PubMed] [Google Scholar]
- 87.Gontika MP, Konialis C, Pangalos C, Papavasiliou A. Novel SCN1A and GABRA1 Gene Mutations With Diverse Phenotypic Features and the Question on the Existence of a Broader Spectrum of Dravet Syndrome. Child Neurol Open. 2017;4 doi: 10.1177/2329048X17706794. 2329048X17706794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Choii G, Ko J. Gephyrin: a central GABAergic synapse organizer. Exp Mol Med. 2015;47:e158. doi: 10.1038/emm.2015.5. [DOI] [PubMed] [Google Scholar]
- 89.Gan L, Kaczmarek LK. When, where, and how much? Expression of the Kv3.1 potassium channel in high-frequency firing neurons. J Neurobiol. 1998;37(1):69–79. doi: 10.1002/(sici)1097-4695(199810)37:1<69::aid-neu6>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 90.Rudy B, McBain CJ. Kv3 channels: voltage-gated K+ channels designed for high-frequency repetitive firing. Trends Neurosci. 2001;24(9):517–26. doi: 10.1016/s0166-2236(00)01892-0. [DOI] [PubMed] [Google Scholar]
- 91.Mathews PJ, Jercog PE, Rinzel J, Scott LL, Golding NL. Control of submillisecond synaptic timing in binaural coincidence detectors by K(v)1 channels. Nat Neurosci. 2010;13(5):601–9. doi: 10.1038/nn.2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hagerman RJ, Berry-Kravis E, Kaufmann WE, Ono MY, Tartaglia N, Lachiewicz A, Kronk R, Delahunty C, Hessl D, Visootsak J, Picker J, Gane L, Tranfaglia M. Advances in the treatment of fragile X syndrome. Pediatrics. 2009;123(1):378–90. doi: 10.1542/peds.2008-0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Raspa M, Wheeler AC, Riley C. Public Health Literature Review of Fragile X Syndrome. Pediatrics. 2017;139(Suppl 3):S153–S171. doi: 10.1542/peds.2016-1159C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Beebe K, Wang Y, Kulesza R. Distribution of fragile X mental retardation protein in the human auditory brainstem. Neuroscience. 2014;273:79–91. doi: 10.1016/j.neuroscience.2014.05.006. [DOI] [PubMed] [Google Scholar]
- 95.Kulesza RJ, Jr, Lukose R, Stevens LV. Malformation of the human superior olive in autistic spectrum disorders. Brain Res. 2011;1367:360–71. doi: 10.1016/j.brainres.2010.10.015. [DOI] [PubMed] [Google Scholar]
- 96.Kulesza RJ, Mangunay K. Morphological features of the medial superior olive in autism. Brain Res. 2008;1200:132–7. doi: 10.1016/j.brainres.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 97.Lukose R, Beebe K, Kulesza RJ., Jr Organization of the human superior olivary complex in 15q duplication syndromes and autism spectrum disorders. Neuroscience. 2015;286:216–30. doi: 10.1016/j.neuroscience.2014.11.033. 2015 Feb 12. [DOI] [PubMed] [Google Scholar]
- 98.Rodier PM, Ingram JL, Tisdale B, Nelson S, Romano J. Embryological origin for autism: developmental anomalies of the cranial nerve motor nuclei. J Comp Neurol. 1996;370(2):247–61. doi: 10.1002/(SICI)1096-9861(19960624)370:2<247::AID-CNE8>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 99.Wang Y, Sakano H, Beebe K, Brown MR, de Laat R, Bothwell M, Kulesza RJ, Jr, Rubel EW. Intense and specialized dendritic localization of the fragile X mental retardation protein in binaural brainstem neurons: a comparative study in the alligator, chicken, gerbil, and human. J Comp Neurol. 522(9):2107–28. doi: 10.1002/cne.23520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gimpl G, Fahrenholz F. The oxytocin receptor system: structure, function, and regulation. Physiol Rev. 2001;81(2):629–83. doi: 10.1152/physrev.2001.81.2.629. [DOI] [PubMed] [Google Scholar]
- 101.Caldwell H, Young W. Oxytocin and Vasopressin: genetics and behavioral implications. In: Lim R, editor. Structure. New York, NY: Springer; 2006. pp. 573–607. [Google Scholar]
- 102.Kiss A, Mikkelsen JD. Oxytocin--anatomy and functional assignments: a minireview. Endocr Regul. 2005;39(3):97–105. [PubMed] [Google Scholar]
- 103.Veenema AH, Neumann ID. Central vasopressin and oxytocin release: regulation of complex social behaviours. Prog Brain Res. 2008;170:261–76. doi: 10.1016/S0079-6123(08)00422-6. [DOI] [PubMed] [Google Scholar]
- 104.Pobbe RL, Pearson BL, Defensor EB, Bolivar VJ, Young WS3rd, Lee HJ, Blanchard DC, Blanchard RJ. Oxytocin receptor knockout mice display deficits in the expression of autism-related behaviors. Horm Behav. 2012;61(3):436–44. doi: 10.1016/j.yhbeh.2011.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mouse Genome Sequencing Consortium. Initial sequencing and comparative analysis of the mouse genom. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.