Abstract
Motivation
Lipids are divided into fatty acyls, glycerolipids, glycerophospholipids, sphingolipids, saccharolipids, sterols, prenol lipids and polyketides. Fatty acyls and glycerolipids are commonly used as energy storage, whereas glycerophospholipids, sphingolipids, sterols and saccharolipids are common used as components of cell membranes. Lipids in fatty acyls, glycerophospholipids, sphingolipids and sterols classes play important roles in signaling. Although more than 36 million lipids can be identified or computationally generated, no single lipid database provides comprehensive information on lipids. Furthermore, the complex systematic or common names of lipids make the discovery of related information challenging.
Results
Here, we present LipidPedia, a comprehensive lipid knowledgebase. The content of this database is derived from integrating annotation data with full-text mining of 3923 lipids and more than 400 000 annotations of associated diseases, pathways, functions and locations that are essential for interpreting lipid functions and mechanisms from over 1 400 000 scientific publications. Each lipid in LipidPedia also has its own entry containing a text summary curated from the most frequently cited diseases, pathways, genes, locations, functions, lipids and experimental models in the biomedical literature. LipidPedia aims to provide an overall synopsis of lipids to summarize lipid annotations and provide a detailed listing of references for understanding complex lipid functions and mechanisms.
Availability and implementation
LipidPedia is available at http://lipidpedia.cmdm.tw.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Lipids are a group of hydrophobic or amphipathic small molecules, divided into eight well-defined classes: fatty acyls, glycerolipids, glycerophospholipids, sphingolipids, saccharolipids, sterols, prenol lipids and polyketides (Fahy et al., 2009). Studies of cellular lipids in biological systems have been gaining importance in several biomedical fields, such as the study of chronic lymphocytic leukemia (Benakanakere et al., 2014), lung cancer (Bradley et al., 2014), Alzheimer's disease (Cheng et al., 2013; Wood, 2012), myocardial infarction (Kain et al., 2014), breast cancer (Perez et al., 2014), schizophrenia (Wood et al., 2014), prostate cancer (Zhou et al., 2012). Large-scale high-throughput lipid analysis using mass spectrometry (MS)-based techniques, such as LC-MS, GC-MS, or tandem MS is able to detect and identify hundreds of lipids in a single experiment. Lipids identified can help researchers to understand physiological and pathological mechanisms in biomedical studies. However, it is challenging for researchers to understand and interpret each identified lipid, despite the availability of lipid databases and metabolomics databases that include lipids, such as HMDB (Wishart et al., 2007), LipidDAT (Caffrey and Hogan, 1992), LipidBank (Watanabe et al., 2000), LIPID MAPS Structure Database (LMSD) (Sud et al., 2007), LipidHome (Foster et al., 2013) and LipidBlast (Kind et al., 2013). Moreover, the systematic names of lipids are not easy to recognize, as they are generally very long and similar to one another due to the similar chemical structures of lipids in the same lipid class (Taguchi and Ishikawa, 2010). However, the functions or other properties of lipids can be very different, even with small changes in the length of fatty acyl chains or the number or position of double bonds on the fatty acid chain (Taguchi and Ishikawa, 2010). Currently, researchers can only understand the functions or relevant information of a lipid by manually search the literature using identified lipids as keywords along with certain biomedical concepts, such as inflammation and signal transductions. For example, a researcher may need to search or try different combinations of keywords to obtain a rough summary of identified sphingosine 1-phosphate (S1P) and its associated diseases, pathways, or genes. Thus, we have developed LipidPedia to provide an organized, lipid-centered, encyclopedia-like database using a text-mining strategy based on UMLS (Unified Medical Language System) (Bodenreider, 2004) to accelerate the labor-intensive process of reading, summarizing and searching references. Currently, LipidPedia is the first lipid knowledgebase with associated biomedical annotations of lipids.
2 Materials and methods
2.1 Construction of lipid species lists and lipid reference database
The list of lipid species was retrieved from LMSD, January 2018 (Sud et al., 2007). LMSD from LIPID MAPS provides lipid classifications and various kinds of chemical information. The list contained 40 823 lipids with LIPID MAPS database IDs, systematic classifications, systematic chemical names, synonymous names and cross-reference database IDs, such as PubChem Compound IDs (CID) (Wang et al., 2009). Here, only lipids with cross-reference PubChem Substance IDs (SID) were converted to PubChem CID. Relevant references for each lipid on the list were collected through PubChem, which provided PubMed citations and NLM (National Library of Medicine) curated PubMed citations as the reliable reference source for Lipidpedia (Kim et al., 2016).
The lipid list with PubChem CIDs was used to query the PubChem database to extract the reference list. With the application programming interface of PubChem Power User Gateway, reference lists were retrieved from NCBI Entrez search engine through PubChem to PubMed. The R packages XML and RCurl were used to download the list of references with web links. GNU Wget utility was used to retrieve journal full-text files from previously downloaded reference links. Perl Module MOJO::Dom was used to parse full-text html files. The text and data mining API services of Elsevier was used to access references published from Elsevier (https://dev.elsevier.com/). We retrieved 2.6 million citations associated with 3923 lipids.
2.2 Classification of the biomedical terms
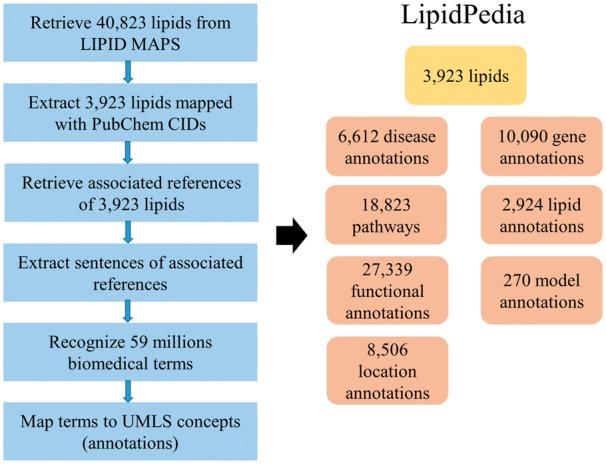
The biomedical terms used in the retrieved references needed to be recognized. For example, asthmatics, airway hyperreactivity or asthma (biomedical terms) refer to the same disease, and consolidations are needed for mapping these terms to the asthma annotation (a unified biomedical concept). Pre-processing of full-text articles was performed with uninformative paragraph removal and tokenization of text. The paragraphs in the cited references and acknowledgments section were removed from the articles. Tokenization of all journal text was performed with the Maxent sentence detector of Apache openNLP library (http://opennlp.apache.org/index.html) (Baldridge, 2005). A total of 1 410 731 articles published from June 1951 to January 2018 were split into 7 million sentences. An in-house Perl script was used to clean up all sentences to filter journal information, author affiliations and page headers and footers. Relevant biomedical terms and concepts associated with the lipids were collected by biomedical entity recognition performed with SemRep version 1.7 (Rindflesch and Fiszman, 2003) with UMLS (Bodenreider, 2004) Specialist Lexicon (version 2015) to extract biological terms and map these terms to UMLS concepts with semantic types (broad categories) in UMLS semantic network (McCray, 1989). For example, the asthma concept is classified to semantic type ‘dsyn’, which means a disease or syndrome. A total of 3 million UMLS concepts and 59 million biological terms from 1410 731 references associated with 3923 lipids (Fig. 1).
Fig. 1.
The overall process of constructing LipidPedia and the biomedical annotations of lipids in LipidPedia
2.3 The service and web application framework
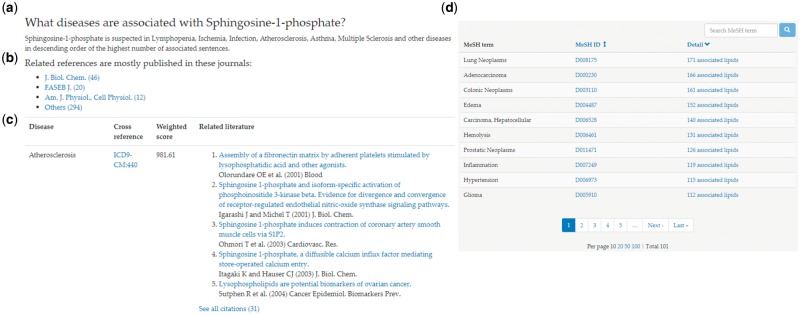
LipidPedia provides biomedical aspects of a specific lipid, including diseases, locations, pathways, functions, genes/proteins, lipids and experimental models. The layout of each biomedical aspect has a general section and a specialized summary for specific biomedical aspects. The general section of each biomedical aspect displays the frequently mapped UMLS concepts. The biomedical terms are recognized and counted in the references from processing named entity recognition in the sentences of all relevant PubMed references provided by PubChem depositors or curated by the National Library of Medicine. The number of associated references is displayed in the table directly as the recognition counts of the concepts. The general section includes the following: (i) a disease-specific paragraph describing the associated annotations with the highest number of related sentences from the information extraction result (Fig. 2a), (ii) statistics of scientific journals with related references (Fig. 2b), (iii) an organized table displaying the annotations, references and links to journal articles or other biomedical databases (Fig. 2c) and (iv) associated disease MeSH terms by collecting disease MeSH terms mapped to the associated references (Fig. 2d).
Fig. 2.
The general composition of each section in LipidPedia. (a) A paragraph describing the concepts with the highest number of related sentences in the references. (b) A list of journals that the associated references are published in. (c) An organized table displaying the concepts with cross-references, a weighted score and associated citations. The weighted score is used to evaluate. (d) Associated disease MeSH terms by collecting disease MeSH terms mapped to the associated references
The organized table of biomedical annotations includes (i) all concept names (annotation names), (ii) cross-references, (iii) a weighted score for evaluation of UMLS concept with all matched terms in LipidPedia (see the Supplementary File) and (iv) a selection of the associated citations with hyperlinks to a detailed view of all associated references (Fig. 2). The mapped annotations are sorted in descending order of the number of associated citations. The detailed layout of each lipid page is as follows:
Disease: The disease section of consists of the general composition with the associated abnormality annotations. The cross-reference of the diseases is ICD-9CM. The cross-reference IDs are listed in the organized table. In addition, Disease MeSH terms list are collected from associated references.
Location: The location section is composed of a visualization describing the associated location annotations with the most related sentences on the cellular structure and in the organized table. The cross-reference of location annotations is the cellular component of GO terms.
Pathway: Current metabolic pathway databases provide few lipid pathways. Pathways cannot be recognized because there are no defined pathway-related concepts in UMLS. As an alternative, ‘pathway’ is searched in all sentences. Each sentence is listed with the PubMed ID for users to search the original articles. NCBI Biosystems and Pathways information through PubChem are provided for additional pathway resources.
Function: In addition to the general composition of each section, the function section has a word cloud display and a hierarchical view. The word cloud visualization displays the frequency of the function annotations. Larger annotations represent more associated sentences. The mapped annotations may have cross-references to GO IDs, and the top 30 GO IDs with the highest number of the related sentences were collected and sent to a web tool (REVIGO) to remove redundant GO terms and return summarized figures and a list of the slimmed GO terms. A hierarchical visualization displays the related functions of slimmed GO terms in a tree view.
Gene/protein: Genes and gene products were included in an organized table. The cross-reference of the gene/protein section is linked to the Entrez Gene or UniProt database.
Lipid: Users may want to know which lipids are commonly present with the lipid of interest in all references. With the help cross-references, lipids can be mapped to ChEBI.
Experimental model: Users may want to know what experimental models are used in related studies for the lipid of interest, so we organized associated experimental models. The models are mapped to MeSH terms.
3 Results and discussion
3.1 Database content
Currently, LipidPedia contains 40 823 lipids from LIPIDMAPS and 3923 of 40 823 lipids are extracted with associated references through PubChem. A total of 1 410 731 references are associated with these 3923 lipids. Among the references, 59 million of biomedical terms are recognized and mapped to UMLS concepts. UMLS concepts can be seen as defined annotations. LipidPedia covers 6612 disease annotations, 18 823 pathways, 27 339 functional annotations, 10 090 gene annotations, 2924 lipid annotations, 8506 location annotations and 270 model annotations (Fig. 1).
3.2 Features of the database
LipidPedia aims to help researchers collect a wide range of relevant annotations on a lipid of interest with a few clicks on a single web page with related citations. Researchers can search lipids by name or by LIPID MAPS class or can browse lipids by LIPID MAPS classes. LipidPedia includes biomedical aspects or so called biomedical annotations of the specific lipid, including diseases, pathways, locations, functions, genes/proteins, lipids and experimental models. The layout of each biomedical aspect has a specialized summary and a general section with cross-references and cross-links to other biomedical databases for specific biomedical aspects (Fig. 2). LipidPedia is built to serve as an encyclopedia of lipids. We will demonstrate the use of LipidPedia by reviewing the roles and functions of sphingosine 1-phosphate (S1P) in vascular inflammation (Galvani et al., 2015).
3.3 Summarization of relevant annotations of sphingosine 1-phosphate
We have summarized a large volume of annotations to generate comprehensive information on different biomedical aspects to help researchers access biomedical annotations (biomedical concepts defined by UMLS) with the most citations.
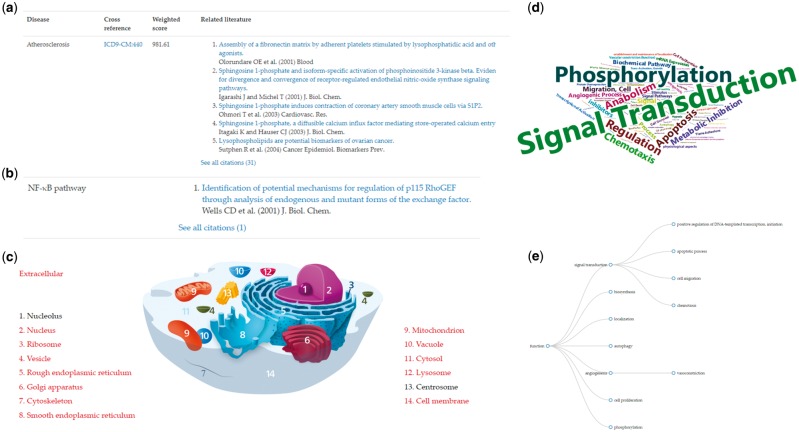
Galvani et al. reviewed the functional antagonist of S1P1 (S1P receptor 1) Fingolimod/Gilenya, which is a structural analog of S1P that has been approved to treat multiple sclerosis (Brinkmann et al., 2010; Galvani et al., 2015). S1P is also known to act as a lipid mediator in vascular inflammation and atherosclerosis (Galvani et al., 2015). With the help of disease summaries in LipidPedia, researchers can see that S1P is associated with atherosclerosis, vascular inflammation and multiple sclerosis (Fig. 3a). The NF-κB pathway, which is known to be associated with inflammation, is also listed in pathway section (Fig. 3b).
Fig. 3.
Screenshots of sphingosine 1-phosphate (S1P) in LipidPedia. The screenshots cover seven biomedical sections, including (a) atherosclerosis and related references in the disease section, (b) NF-κB pathway and related references in the pathway section, (c) cellular visualization in the location section, where subcellular locations in red are associated with S1P, (d) word cloud visualization of functional annotations, where larger words represent more frequent appearance and (e) hierarchical visualization of functional annotations according to Gene Ontology. The gene, lipid and experimental model sections, which only have organized tables, are not shown here
In the location summary, the annotations are mapped to cellular components of Gene Ontology (GO) and lipid locations are visualized on cellular structures. The associated locations of S1P are shown in red, and non-associated locations are shown in black. Hyperlinks to the GO database are also provided by clicking the subcellular locations on the left or right side of the cellular structure image (Fig. 3c). From the cellular visualization, researchers can see the locations of S1P in a cell. In the functional summary, LipidPedia provides a visualization of function annotations in a word cloud format. The word cloud visualization displays the frequency of the functional annotations. Larger annotations represent more associated phrases. S1P signals through G-protein coupled receptors (S1P1-5) to regulate cell response, so phosphorylation is found in the word cloud visualization (Galvani et al., 2015). Researchers can see that S1P is associated with signal transduction and regulation (Blaho and Hla, 2011; Blaho and Hla, 2014) from the word cloud of functions in a few seconds (Fig. 3d). In addition to the word cloud visualization, LipidPedia also provides a hierarchical visualization to display the functional hierarchy of Gene Ontology terms (Fig. 3e). Each circle is an associated function. From the hierarchical visualization, researchers can see that S1P signaling may involve apoptosis, cell migration or transcriptional regulation (Blaho and Hla, 2011).
In addition to biomedical summary of a lipid, LipidPedia also provides a detailed, downloadable, sortable and filterable list of associated biomedical annotations with cross-references and cross-links to other biomedical databases.
3.4 Detailed biomedical information with cross-references and citations
The biomedical summary provides a view of annotations with the most associated references. The organized table of biomedical annotations includes (i) all annotation names (concept names), (ii) cross-references and cross-links to other biomedical databases, (iii) a weighted score for evaluation quality of concepts with all matched terms in LipidPedia and (iv) a selection of associated citations with hyperlinks to detailed views of all associated references (Fig. 2a). The mapped annotations are sorted in descending order of the number of associated citations. Galvani et al. noted that S1P is carried by the ApoM+ subfraction of HDL (Galvani et al., 2015). In LipidPedia, the APOM gene (Apolipoprotein M) is extracted from related references (other than Galvani) and displayed in the gene aspect table on the S1P page. The associated references show that Apolipoprotein M is associated with HDL and binds S1P (Liu et al., 2014). S1P with lipopolysaccharides induced inflammatory molecules and leukocyte adhesion on endothelial cells (Liu et al., 2014). Lipopolysaccharides are also listed in the lipid aspect table of the S1P page in LipidPedia. S1P-induced leukocyte cell-cell adhesion is displayed in the functional aspect of the S1P page. In a review of S1P1, the authors noted that S1P inhibits vascular permeability and is required for blood vessel development (Liu et al., 2000). Both vascular permeability and blood vessel development are listed in the function table of the S1P page with cross-references to the GO database. In a review, an LDL-R-/- mouse model was developed to recapitulate atherosclerotic plaque development and to evaluate a synthetic S1P analog, FTY720 (Nofer et al., 2007). LDL-R-/- is thus associated with the annotation ‘Mouse model’ in the model table. With LipidPedia, biomedical annotations associated with the lipid of interest can be discovered by reading the lipid page in LipidPedia as either a summary or a detailed table of biomedical annotations. Researchers can see the associations of the lipid and biomedical annotations with all related references. LipidPedia provides a convenient, time-saving way to help researchers identify useful information and references without reading through hundreds of abstracts from PubMed to filter out the references that they need.
3.5 Database design and organization
LipidPedia is designed as an updatable database. The contents of LipidPedia are extracted biomedical annotations recognized by SemRep (Rindflesch and Fiszman, 2003) from associated references. The extracted annotations with the lipid of interest presented in the same sentence are stored in the database. Internally, the information on references, lipids, extracted biomedical terms and term-mapped concepts as annotations are organized into tables. The information on associated references of lipids contains author names, journal names, publication dates, abstracts and other. All the sentences extracted from available full text of associated references are stored in the sentence table. The biomedical terms and term-mapped concepts from sentences are extracted and stored with semantic type information in the table. The term table contains a semantic type of mapped concept that is derived from the sentence that contains the extracted terms. The concepts table contains the defined concepts name, definition and associated lipids. For more information on implementation of the database, refer to ‘Implementation of LipidPedia’ in the Supplementary File.
3.6 Database update frequency
Related literature and the text mining data of lipids are updated monthly, and the concepts of UMLS are updated annually following the annual release of UMLS.
3.7 Data availability
Related citations are listed in each section of biomedical information, and all collected references are downloadable in comma-separated values (CSV) format at the bottom of the web page of a lipid. References contain author names, title, journal name, publication date and PubMed ID. Researchers can download the reference list and determine which paper contains useful information through LipidPedia.
4 Conclusion
The LipidPedia database was developed to accelerate the process of understanding quantified lipids from an enormous literature search. Each lipid in LipidPedia has its own record consisting of a text summary organized by annotations. These annotations frequently appear in the biomedical literature, including diseases, pathways, genes, locations, functions, lipids and experimental models. LipidPedia aims to provide an overall synopsis of lipids to summarize lipid annotations and provide a detailed listing of references to understand complex lipid functions and mechanisms. LipidPedia (http://lipidpedia.cmdm.tw) has a user-friendly search interface. All citations associated with a lipid can be downloaded through a link in the reference section. S1P was used as the example to demonstrate what LipidPedia can provide from article mining. S1P is a lipid messenger for important physiological processes in vascular inflammation in several signal transduction pathways. It was shown that information associated with S1P was successfully extracted and organized. It was easy to discover associated biological knowledge for S1P from LipidPedia.
Supplementary Material
Acknowledgements
We thank Dr Lie-Fen Shyur, Dr Po-Hsiu Kuo, Dr Ching-Hua Kuo, Dr Kuo-Ching Wang and Dr San-Yuan Wang for their valuable comments.
Funding
This work was supported by the Taiwan Ministry of Science and Technology [grant numbers MOST 106-2622-B-002-008-, MOST 106-2911-I-002-533, MOST 106-2321-B-002-041-, MOST 105-3011-F-002-010-] and National Taiwan University grants [NTU-CDP-106R7820, NTU-ERP-106R880803 and NTU-107L7820]. Resources of the Laboratory of Computational Molecular Design and Metabolomics and the Department of Computer Science and Information Engineering of National Taiwan University were used in performing these studies.
Conflict of Interest: none declared.
References
- Baldridge J. (2005) The opennlp project. http://opennlp.apache.org/index.html (2 February 2012, date last accessed).
- Benakanakere I. et al. (2014) Targeting cholesterol synthesis increases chemoimmuno-sensitivity in chronic lymphocytic leukemia cells. Exp. Hematol. Oncol., 3, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaho V.A., Hla T. (2014) An update on the biology of sphingosine 1-phosphate receptors. J. Lipid Res., 55, 1596–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaho V.A., Hla T. (2011) Regulation of mammalian physiology, development, and disease by the sphingosine 1-phosphate and lysophosphatidic acid receptors. Chem. Rev., 111, 6299–6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenreider O. (2004) The unified medical language system (UMLS): integrating biomedical terminology. Nucleic Acids Res., 32, 267D–D270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley E. et al. (2014) Critical role of Spns2, a sphingosine-1-phosphate transporter, in lung cancer cell survival and migration. PLoS ONE, 9, e110119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkmann V. et al. (2010) Fingolimod (FTY720): discovery and development of an oral drug to treat multiple sclerosis. Nat. Rev. Drug Disc., 9, 883–897. [DOI] [PubMed] [Google Scholar]
- Caffrey M., Hogan J. (1992) LIPIDAT: a database of lipid phase transition temperatures and enthalpy changes. DMPC data subset analysis. Chem. Phys. Lipids, 61, 1–109. [DOI] [PubMed] [Google Scholar]
- Cheng H. et al. (2013) Specific changes of sulfatide levels in individuals with pre-clinical Alzheimer's disease: an early event in disease pathogenesis. J. Neurochem., 127, 733–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahy E. et al. (2009) Update of the LIPID MAPS comprehensive classification system for lipids. J. Lipid Res., 50, S9–S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster J.M. et al. (2013) LipidHome: a database of theoretical lipids optimized for high throughput mass spectrometry lipidomics. PloS One, 8, e61951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvani S. et al. (2015) HDL-bound sphingosine 1-phosphate acts as a biased agonist for the endothelial cell receptor S1P(1) to limit vascular inflammation. Sci. Signal., 8, ra79–ra79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kain V. et al. (2014) Inflammation revisited: inflammation versus resolution of inflammation following myocardial infarction. Basic Res. Cardiol., 109, 1–17. [DOI] [PubMed] [Google Scholar]
- Kim S. et al. (2016) Literature information in PubChem: associations between PubChem records and scientific articles. J. Cheminf., 8, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kind T. et al. (2013) LipidBlast in silico tandem mass spectrometry database for lipid identification. Nat. Methods, 10, 755–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M. et al. (2014) Hepatic apolipoprotein M (apoM) overexpression stimulates formation of larger apoM/sphingosine 1-phosphate-enriched plasma high density lipoprotein. J. Biol. Chem., 289, 2801–2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. et al. (2000) Edg-1, the G protein–coupled receptor for sphingosine-1-phosphate, is essential for vascular maturation. Journal of Clinical Investigation, 106, 951–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCray A.T. (1989) The UMLS semantic network In: Proc 13th Annu Symp Comput Appl Med Care. pp. 503–507. [Google Scholar]
- Nofer J.-R. et al. (2007) FTY720, a synthetic sphingosine 1 phosphate analogue, inhibits development of atherosclerosis in low-density lipoprotein receptor-deficient mice. Circulation, 115, 501–508. [DOI] [PubMed] [Google Scholar]
- Perez O. et al. (2014) Abstract 3496: breast cancer and obesity impact the lipid composition of breast adipose tissue: a preliminary study using shotgun lipidomics. Cancer Res., 74, 3496. [Google Scholar]
- Rindflesch T.C., Fiszman M. (2003) The interaction of domain knowledge and linguistic structure in natural language processing: interpreting hypernymic propositions in biomedical text. J. Biomed. Inf., 36, 462–477. [DOI] [PubMed] [Google Scholar]
- Sud M. et al. (2007) LMSD: lIPID MAPS structure database. Nucleic Acids Res., 35, D527–D532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi R., Ishikawa M. (2010) Precise and global identification of phospholipid molecular species by an Orbitrap mass spectrometer and automated search engine Lipid Search. J. Chromatogr. A, 1217, 4229–4239. [DOI] [PubMed] [Google Scholar]
- Wang Y. et al. (2009) PubChem: a public information system for analyzing bioactivities of small molecules. Nucleic Acids Res., 37, W623–W633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K. et al. (2000) How to search the glycolipid data in ‘LIPIDBANK for Web’, the newly developed lipid database in Japan. Trends Glycosci. Glycotechnol., 12, 175–184. [Google Scholar]
- Wishart D.S. et al. (2007) HMDB: the Human Metabolome Database. Nucleic Acids Res., 35, D521–D526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood P. (2012) Lipidomics of Alzheimer's disease: current status. Alzheimer's Res. Therapy, 4, 5.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood P.L. et al. (2014) Lipidomics reveals dysfunctional glycosynapses in schizophrenia and the G72/G30 transgenic mouse. Schizophr Res., 159, 365–369. [DOI] [PubMed] [Google Scholar]
- Zhou X. et al. (2012) Identification of plasma lipid biomarkers for prostate cancer by lipidomics and bioinformatics. PLoS ONE, 7, e48889. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.