Fumigation of Arabidopsis with the gaseous signaling molecule NO2 triggers basal pathogen resistance that is dependent on early callose deposition.
Abstract
Nitrogen dioxide (NO2) forms in plants under stress conditions, but little is known about its physiological functions. Here, we explored the physiological functions of NO2 in plant cells using short-term fumigation of Arabidopsis (Arabidopsis thaliana) for 1 h with 10 µL L−1 NO2. Although leaf symptoms were absent, the expression of genes related to pathogen resistance was induced. Fumigated plants developed basal disease resistance, or pattern-triggered immunity, against the necrotrophic fungus Botrytis cinerea and the hemibiotrophic bacterium Pseudomonas syringae. Functional salicylic acid and jasmonic acid (JA) signaling pathways were both required for the full expression of NO2-induced resistance against B. cinerea. An early peak of salicylic acid accumulation immediately after NO2 exposure was followed by a transient accumulation of oxophytodienoic acid. The simultaneous NO2-induced expression of genes involved in jasmonate biosynthesis and jasmonate catabolism resulted in the complete suppression of JA and JA-isoleucine (JA-Ile) accumulation, which was accompanied by a rise in the levels of their catabolic intermediates 12-OH-JA, 12-OH-JA-Ile, and 12-COOH-JA-Ile. NO2-treated plants emitted the volatile monoterpene α-pinene and the sesquiterpene longifolene (syn. junipene), which could function in signaling or direct defense against pathogens. NO2-triggered B. cinerea resistance was dependent on enhanced early callose deposition and CYTOCHROME P450 79B2 (CYP79B2), CYP79B3, and PHYTOALEXIN DEFICIENT3 gene functions but independent of camalexin, CYP81F2, and 4-OH-indol-3-ylmethylglucosinolate derivatives. In sum, exogenous NO2 triggers basal pathogen resistance, pointing to a possible role for endogenous NO2 in defense signaling. Additionally, this study revealed the involvement of jasmonate catabolism and volatiles in pathogen immunity.
Plants face many challenges from phytopathogenic bacteria, fungi, and oomycetes. These pathogenic organisms have evolved various feeding strategies. Biotrophic pathogens such as powdery mildew nourish on nutrients from living cells, while necrotrophic pathogens such as Botrytis cinerea kill the host to feed on dead cell contents (Glazebrook, 2005; Mengiste, 2012). Hemibiotrophs including Pseudomonas syringae, on the other hand, can pursue both feeding strategies (Glazebrook, 2005).
The plant perceives the invading pathogen by recognizing conserved pathogen- and damage-associated molecular patterns (PAMPs and DAMPs), including the bacterial flagellin, fungal chitin, and oligogalacturans derived from damaged plant cell walls (Boller and Felix, 2009; Heil and Land, 2014). The binding of such elicitors to specific pattern-recognition receptors initiates PAMP-triggered immunity (PTI), also referred to as basal pathogen resistance (Boller and Felix, 2009; Couto and Zipfel, 2016). Immediate cellular responses upon PAMP recognition are the rapid influx of calcium ions into the cytosol and the production of reactive oxygen species such as superoxide or hydrogen peroxide (Boller and Felix, 2009; Bigeard et al., 2015). Additionally, reactive nitrogen species (RNS), such as nitric oxide (NO), are crucial for pathogen-induced signal transduction (Gaupels et al., 2011; Mur et al., 2013).
The phytohormones salicylic acid (SA), jasmonic acid (JA), and the bioactive JA-Ile conjugate are considered to be major mediators of plant defense (Browse, 2009; Vlot et al., 2009; Pieterse et al., 2012; Wasternack and Hause, 2013). NONEXPRESSOR OF PR GENES1 (NPR1) and CORONATINE INSENSITIVE1 (COI1) are central transcriptional regulators of SA- and JA-responsive genes, respectively. The SA and JA/ethylene pathways are interconnected via complex regulatory networks and commonly antagonize each other, with SA being a potent antagonist of JA signaling (Robert-Seilaniantz et al., 2011; Caarls et al., 2015). Several NPR1-regulated TGA and WRKY transcription factors have been implicated in SA/JA cross talk (Pieterse et al., 2012; Caarls et al., 2015). The JA pathway also is controlled on the level of jasmonate catabolism. In response to wounding and pathogen attack, excess JA and JA-Ile are inactivated by hydroxylation and carboxylation, forming tuberonic acid (12-OH-JA), hydroxyl-JA-Ile (12-OH-JA-Ile), and dicarboxy-JA-Ile (12-COOH-JA-Ile; Heitz et al., 2016; Caarls et al., 2017; Smirnova et al., 2017). The jasmonate catabolism pathway is inducible by JA in the course of a negative feedback regulation (Caarls et al., 2017).
Pathogens can be prevented from spreading by PAMP-triggered formation of the (1,3)-β-glucan polymer callose, which is deposited between the plasma membrane and cell wall at infection sites (Luna et al., 2011; Ellinger and Voigt, 2014). Callose deposition is induced after B. cinerea infection of Arabidopsis (Arabidopsis thaliana; García-Andrade et al., 2011). POWDERY MILDEW RESISTANT4 (PMR4) is the predominant callose synthase during pathogen infection (Jacobs et al., 2003; Nishimura et al., 2003; Ellinger et al., 2013). Other well-studied components of the plant’s arsenal against pathogens are indole glucosinolates and the phytoalexin camalexin (3-thiazol-2′yl-indole) found in Arabidopsis (Glawischnig, 2007). In planta, camalexin is synthesized upon the detection of various PAMPs and DAMPs (Kliebenstein et al., 2005; Rauhut et al., 2009; Ahuja et al., 2012), and its antimicrobial activity against P. syringae and B. cinerea has been confirmed in vitro (Rogers et al., 1996; Kliebenstein et al., 2005). Indole glucosinolates such as 4-OH-indol-3-ylmethylglucosinolate (4-OH-I3M) have important functions in antifungal defense after activation by the P450 monooxygenase CYP81F2 and the atypical myrosinase PENETRATION RESISTANCE2 (PEN2; Bednarek et al., 2009; Clay et al., 2009).
The RNS nitrogen dioxide (NO2) arises during stress-induced signaling by the oxidation of NO, reduction of nitrite, or decomposition of peroxynitrite (Pryor et al., 2006; Groß et al., 2013). Chloroplastic nitrite reductase activity utilizes electrons diverted from photosynthesis for the multistep reduction of nitrite to ammonia (Beevers and Hageman, 1969). Accordingly, treatment of soybean (Glycine max) with a photosynthesis-inhibiting herbicide or incubation in darkness leads to the accumulation of nitrite and the subsequent emission of NO2 that is derived from nitrite by an unknown mechanism (Klepper, 1979, 1990). In vitro experiments demonstrate that the heme-containing horseradish peroxidase can produce NO2 through one-electron reduction of nitrite in the presence of hydrogen peroxide (Shibata et al., 1995; Sakihama et al., 2003). Additionally, HEMOGLOBIN1 of Arabidopsis and alfalfa (Medicago sativa) can produce NO2 mechanistically similar to horseradish peroxidase (Sakamoto et al., 2004; Maassen and Hennig, 2011).
NO2 is a highly reactive compound that can exert specific physiological functions by the nitration (-NO2 group) of nucleophiles such as fatty acids (FAs), nucleotides, and proteins. The nitration of FAs (nitro-FAs) has been observed in Arabidopsis exposed to abiotic stresses (Mata-Pérez et al., 2016b), and nitro-FAs are proposed to act as signaling molecules (Schopfer et al., 2011; Mata-Pérez et al., 2016b). The nitration of cyclic guanosine monophosphate (cGMP) to give 8-nitro-cGMP triggers stomatal closure, whereas unmodified cGMP mediates stomatal opening (Joudoi et al., 2013). Moreover, increased protein Tyr nitration is a common event during plant defense responses (Arasimowicz-Jelonek and Floryszak-Wieczorek, 2011; Gaupels et al., 2011; Mata-Pérez et al., 2016a; Kolbert et al., 2017). This protein modification is mediated directly by NO2 or via the decomposition of peroxynitrite to NO2, which subsequently binds to accessible protein Tyr residues (Pryor et al., 2006; Groß et al., 2013; Radi, 2013; Kolbert et al., 2017). NO2-modified proteins often are inhibited irreversibly, as described for several antioxidant enzymes and the abscisic acid receptor PYRABACTIN RESISTANCE1 (PYR1)/PYR1-LIKE/REGULATORY COMPONENTS OF ABA RECEPTORS (Gaupels et al., 2011; Groß et al., 2013; Castillo et al., 2015; Mata-Pérez et al., 2016a). Together, these examples illustrate how NO2 can participate in defense signaling. On the other hand, high endogenous levels of RNS also can result in excessive oxidation and nitration of biomolecules, severe metabolic perturbations, and even structural injuries of cells (Corpas and Barroso, 2013; Groß et al., 2013). Dependent on the severity of the inflicted nitrooxidative stress, cells either trigger defense and repair mechanisms or die (Thomas et al., 2008; Groß et al., 2013). In this scenario, NO2 and other RNS would act as inducers of defense signaling rather than signals themselves.
To date, the investigation of NO2 in vivo is hampered by the fact that no specific dyes and donors are commercially available. For this reason, nothing is known about endogenous levels of NO2 under stress conditions. Nevertheless, the functions of NO2 in plants have been explored frequently by fumigations with gaseous NO2 as a donor treatment. After stomatal uptake, the lipophilic NO2 and its more water-soluble dimer N2O4 readily penetrate cell membranes and diffuse into the cytosol (Wellburn, 1990). In the aqueous environment of the leaf, NO2 disproportionates to nitrite and nitrate that are reduced further to ammonia by nitrite and nitrate reductases (Beevers and Hageman, 1969; Zeevaart, 1976; Sparks, 2009). Nitrite levels are correlated positively with NO2-induced leaf damage in a number of plant species (Zeevaart, 1976; Kasten et al., 2016). Plants generally accumulate high nitrite levels and show strong leaf damage after NO2 fumigation in the dark (Zeevaart, 1976; Yoneyama and Sasakawa, 1979; Shimazaki et al., 1992) because, as mentioned above, nitrite reductase activity is dependent on photosynthesis. However, nitrite levels also increase strongly in pea (Pisum sativum) and Arabidopsis after NO2 fumigation in the light, probably because they exceed the enzymatic capacity of nitrite reductase (Zeevaart, 1976; Kasten et al., 2016).
Long-term exposure to nL L−1 levels of NO2 has beneficial effects on plant growth and development (Srivastava et al., 1994; Takahashi et al., 2014), whereas NO2 concentrations in the µL L−1 range cause the induction of antioxidant defense and other stress responses (Xu et al., 2010; Liu et al., 2015; Kasten et al., 2016). In this work, Arabidopsis was exposed to 10 µL L−1 NO2 for 1 h, which did not cause visible leaf symptoms or ion leakage as a measure of membrane damage (Kasten et al., 2016). Responses of Arabidopsis to NO2 were investigated by microarray analysis, pathogen assays, and measurements of phytohormones, volatiles, camalexin, and callose.
RESULTS
NO2 Triggers the Expression of Genes Related to Pathogen Defense
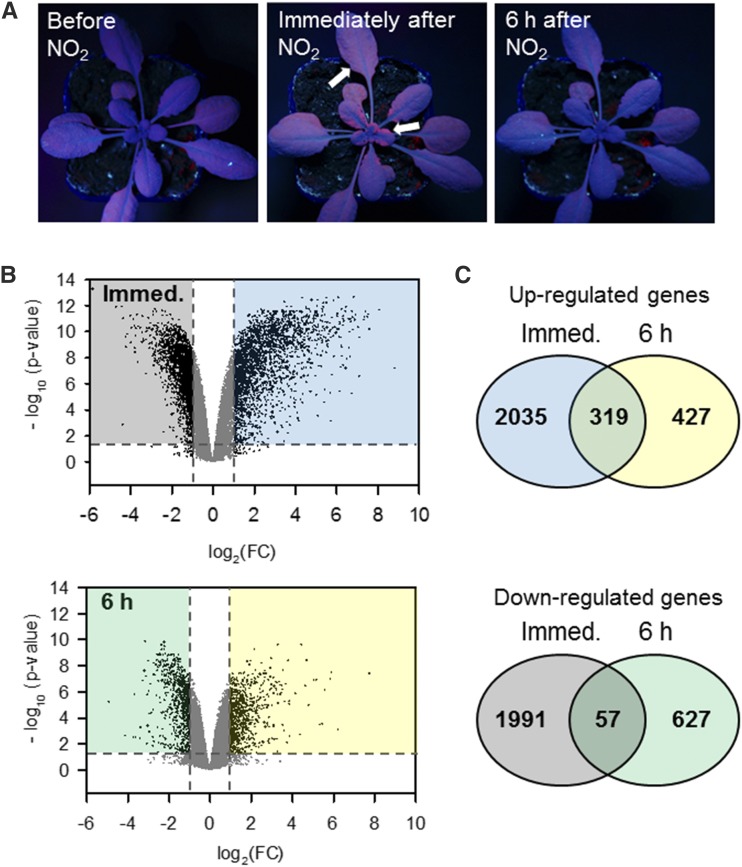
Exposure of Arabidopsis Columbia-0 (Col-0) plants to 10 µL L−1 NO2 for 1 h did not cause visible symptoms (Supplemental Fig. S1), which is in agreement with previous data showing that ion leakage as a measure of membrane damage does not increase after this treatment (Kasten et al., 2016). However, close examination under UV light revealed the emission of red chlorophyll fluorescence immediately after fumigation that faded at the 6-h time point (Fig. 1A), indicative of photoprotective energy dissipation due to a transient stress-induced metabolic perturbation (Lichtenthaler and Miehe, 1997; Chaerle and Van Der Straeten, 2000).
Figure 1.
NO2 triggers a rapid and transient defense response. Arabidopsis Col-0 plants were fumigated with 10 µL L−1 NO2 or air for 1 h. A, NO2 caused no visible leaf damage (Supplemental Fig. S1) but a transient increase in red chlorophyll autofluorescence under UV light (white arrows) indicative of stress-induced photoprotective energy dissipation. B, Leaf material was harvested in quadruplicates for microarray analysis immediately or 6 h after fumigation. Volcano plots visualize the changes in gene expression at 0 and 6 h after fumigation by plotting the adjusted P value over the fold change. Horizontal dashed lines mark P = 0.05; vertical dashed lines indicate log2 fold change [log2(FC)] ± 1. Data points represent the expression of individual genes. The expression of genes appearing in the colored left sections was significantly down-regulated [P < 0.05, log2(FC) < −1], whereas the expression of genes within the colored right sections showed significant up-regulation [P < 0.05, log2(FC) > 1]. C, Venn diagrams illustrate the number of genes that were significantly up-regulated (top) or down-regulated (bottom) after NO2 exposure with P < 0.05 and log2(FC) ± 1. The color code is consistent in B and C, indicating genes down-regulated immediately (gray) and 6 h (green) after fumigation or up-regulated immediately (blue) and 6 h (yellow) after fumigation.
Microarray analysis was performed with leaf material sampled immediately or 6 h after fumigation with air or 10 µL L−1 NO2 for 1 h (Supplemental Data Set S1). Volcano plots illustrated that both the up- and down-regulation of gene expression was more pronounced directly after NO2 fumigation than after 6 h (Fig. 1B). Approximately 4,400 genes were significantly regulated immediately after fumigation, whereas 6 h later, only 1,430 genes were differentially regulated (Fig. 1C). The regulated genes scarcely overlapped among both time points (Fig. 1B). Only 11.5% of all up-regulated genes and 2.1% of all down-regulated genes were affected at both time points, which suggested discrete time-dependent responses of the plant to NO2.
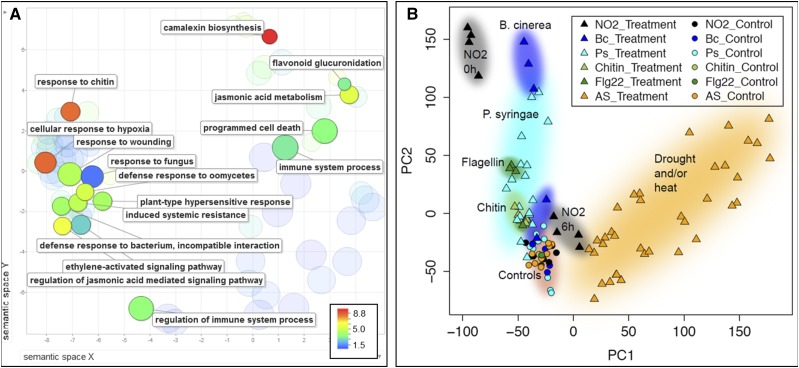
Gene Ontology (GO) term enrichment analysis (Supplemental Fig. S2) was applied to assess biological processes underlying the observed NO2-induced gene regulation. Directly after fumigation, 122 GO terms were enriched significantly in the up-regulated gene set (Supplemental Data Set S1). The majority of GO terms were related to plant defense, including responses to wounding, the fungal elicitor chitin, and fungal as well as bacterial pathogen attacks (Fig. 2A). NO2 also activated genes involved in the JA and ethylene signaling pathways, camalexin biosynthesis, flavonoid glucuronidation, and programmed cell death. Principal component analysis was used for comparison of the NO2-regulated genes with previously published microarray data sets obtained after treatment of plants with B. cinerea (Ferrari et al., 2007), P. syringae (Lewis et al., 2015), chitin (Ramonell et al., 2005), the bacterial elicitor flg22 (Zipfel et al., 2004; Boudsocq et al., 2010), and exposure to the abiotic stresses drought and/or heat (Georgii et al., 2017; Fig. 2B). Although the biotic stress studies were conducted on a different microarray platform (Affymetrix ATH1), their posttreatment expression samples are closer to the NO2-fumigated samples directly after treatment than to the abiotic stress samples sharing the same platform with the NO2 study (Agilent At8x60K; identifier 29132).
Figure 2.
NO2-induced genes are related to pathogen defense. A, GO term enrichment of genes up-regulated directly (0 h) after fumigation. Enriched GO terms (P < 0.05) were identified using the PANTHER 11.0 overrepresentation test and visualized in scatterplots using the REVIGO tool. Each circle represents a GO term, and circle size represents the number of genes encompassed. The color code depicts the fold enrichment of the respective GO term within the data set compared with the PANTHER Arabidopsis reference list. Circles are clustered according to the distance of the respective GO terms within the GO hierarchical tree. Highly enriched or interesting GO terms were labeled. B, Principal component analysis of Arabidopsis gene expression responses to NO2 fumigation, biotic stress, and abiotic stress. Data from microarray analysis after NO2 fumigation were combined with previously published data sets representing responses to different stresses and elicitors (115 samples in total). The overall expression response similarities between samples of the combined data set are visualized using the top two principal components (PC1 and PC2), capturing 22% and 14% of the total variation, respectively. NO2, NO2 fumigation; Bc, B. cinerea infection, ArrayExpress accession number E-GEOD-5684; Ps, P. syringae infection, E-GEOD-6176; Chitin, chitin treatment, E-GEOD-2538; Flg22, flagellin epitope 22 treatment, E-GEOD-17382; AS, abiotic stress treatment study, E-MTAB-4867. For each study, treated samples are marked by triangles and controls by circles.
In summary, the microarray analysis revealed that exposure to 10 µL L−1 NO2 specifically up-regulated the expression of genes associated with defense against fungal and bacterial pathogens.
NO2 Triggers Basal Pathogen Resistance
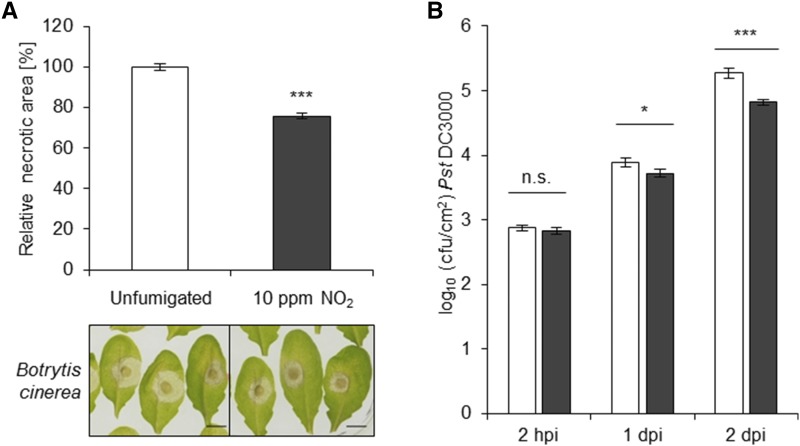
To investigate whether NO2 induces resistance against necrotrophic fungi, as suggested by the gene expression data, NO2-fumigated plants were infected with B. cinerea. Arabidopsis Col-0 plants were fumigated with 10 µL L−1 NO2 for 1 h, followed by droplet infection of detached leaves with B. cinerea 6 h later. The areas of the developing necrotic lesions were then analyzed to assess if NO2 provides resistance against this pathogen. In Figure 3A, representative examples of the necrotic lesions formed on NO2-fumigated and nontreated plants are illustrated. Quantification of the necrotic areas revealed that the average sizes of necrotic lesions formed on NO2-fumigated leaves were reduced significantly by ∼30% when compared with unfumigated leaves (Fig. 3A). Therefore, these results confirmed that NO2 induces resistance against the necrotrophic fungus B. cinerea.
Figure 3.
NO2 induces resistance against B. cinerea and P. syringae. A, Col-0 plants were fumigated or not (control) with 10 µL L−1 NO2 for 1 h, followed by droplet infection of detached leaves with approximately 1,000 spores of B. cinerea 6 h after fumigation. Necrotic lesion area was measured 3 d later using ImageJ. Columns represent means of 18 independent experiments ± se (n = 624–640). Asterisks indicate significant differences from the control according to the Mann-Whitney rank-sum test (***, P < 0.001). Representative photographs of necrotic lesions 3 d after droplet infection with B. cinerea are shown. Bars = 5 mm. B, Col-0 plants were fumigated with 10 µL L−1 NO2 for 1 h and syringe infiltrated with 1 × 105 cfu mL−1 P. syringae pv tomato DC3000 4 h after fumigation. Leaf discs from infected leaves were obtained 2 h or 1 and 2 d after infection to determine the bacterial titer (cfu cm−2 leaf material). Columns represent means ± se from seven independent experiments (n [2 h post infection] = 26–27, n [1 dpi] = 72, and n [2 dpi] = 66). Asterisks indicate significant differences of all pairwise comparisons via two-way ANOVA plus the Holm-Sidak posthoc test (*, P < 0.05 and ***, P < 0.001); n.s., not significant. White columns, unfumigated; black columns, 10 µL L−1 NO2.
The GO term enrichment analysis and principal component analysis suggested that NO2 also elicits defense responses effective against bacterial pathogens. Therefore, plants were fumigated with NO2 followed by syringe infiltration with 1 × 105 colony-forming units (cfu) mL−1 P. syringae pv tomato DC3000 4 h later. The bacterial titers in the infected leaves were determined 2 h, 1 d, and 2 d post infection (dpi) to determine if NO2 fumigation influenced bacterial growth. As shown in Figure 3B, infected leaves pretreated with 10 µL L−1 NO2 harbored fewer bacteria than their unfumigated counterparts. Therefore, it can be concluded that NO2-induced signaling also decreased the susceptibility of Arabidopsis to the hemibiotrophic bacterium P. syringae.
Together, the findings above imply that NO2 initiated the onset of basal pathogen resistance similar to the induction of PTI by PAMPs such as chitin and flagellin.
NO2 Triggers Signaling by SA and Oxophytodienoic Acid, While JA and JA-Ile Are Catabolized
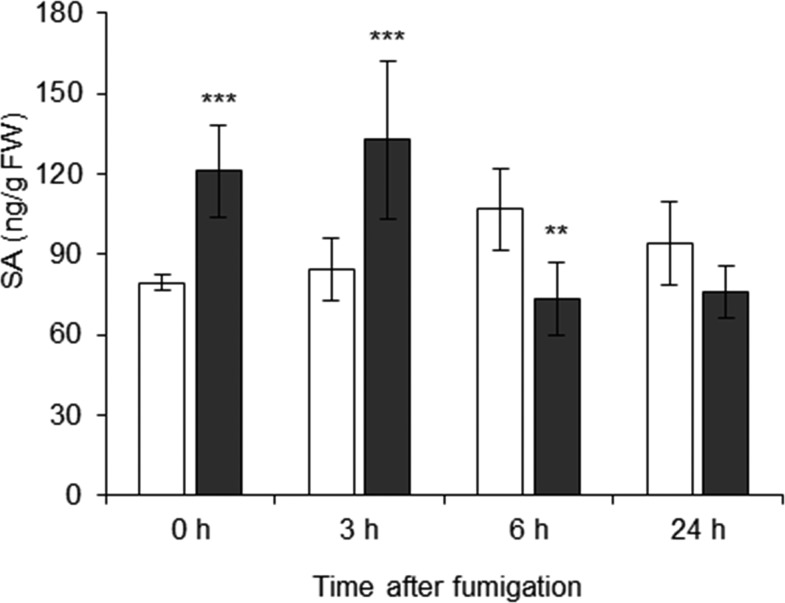
SA biosynthesis and signaling genes were enhanced following NO2 exposure (Supplemental Data Set S1; Supplemental Fig. S3). Therefore, levels of this hormone were determined by liquid chromatography-tandem mass spectrometry (LC-MS/MS) after fumigation with 10 µL L−1 NO2 (Fig. 4). SA levels were approximately 90 ng g−1 fresh weight in air-fumigated leaves when averaged across time points but increased to 121 and 133 ng g−1 fresh weight directly or 3 h after fumigation with NO2, respectively. At the 6-h time point, the SA content declined rapidly again to 73 ng g−1 fresh weight, resulting in a significant decrease of 31% when compared with the concentration in the respective air-fumigated control. This is in line with the observation that transcript levels of the biosynthetic genes declined at this time point as well (Supplemental Data Set S1). In summary, exposure to 10 µL L−1 NO2 provoked a rapid, but transient, accumulation of SA.
Figure 4.
NO2 induces signaling by SA. SA levels at different time points after fumigation with air or 10 µL L−1 NO2 were measured via LC-MS/MS and normalized to the samples’ fresh weight (FW). Columns represent means ± sd (n = 5). Asterisks indicate significant differences within the time points as determined by two-way ANOVA plus the Holm-Sidak posthoc test (**, P < 0.01 and ***, P < 0.001). White columns, air; black columns, 10 µL L−1 NO2.
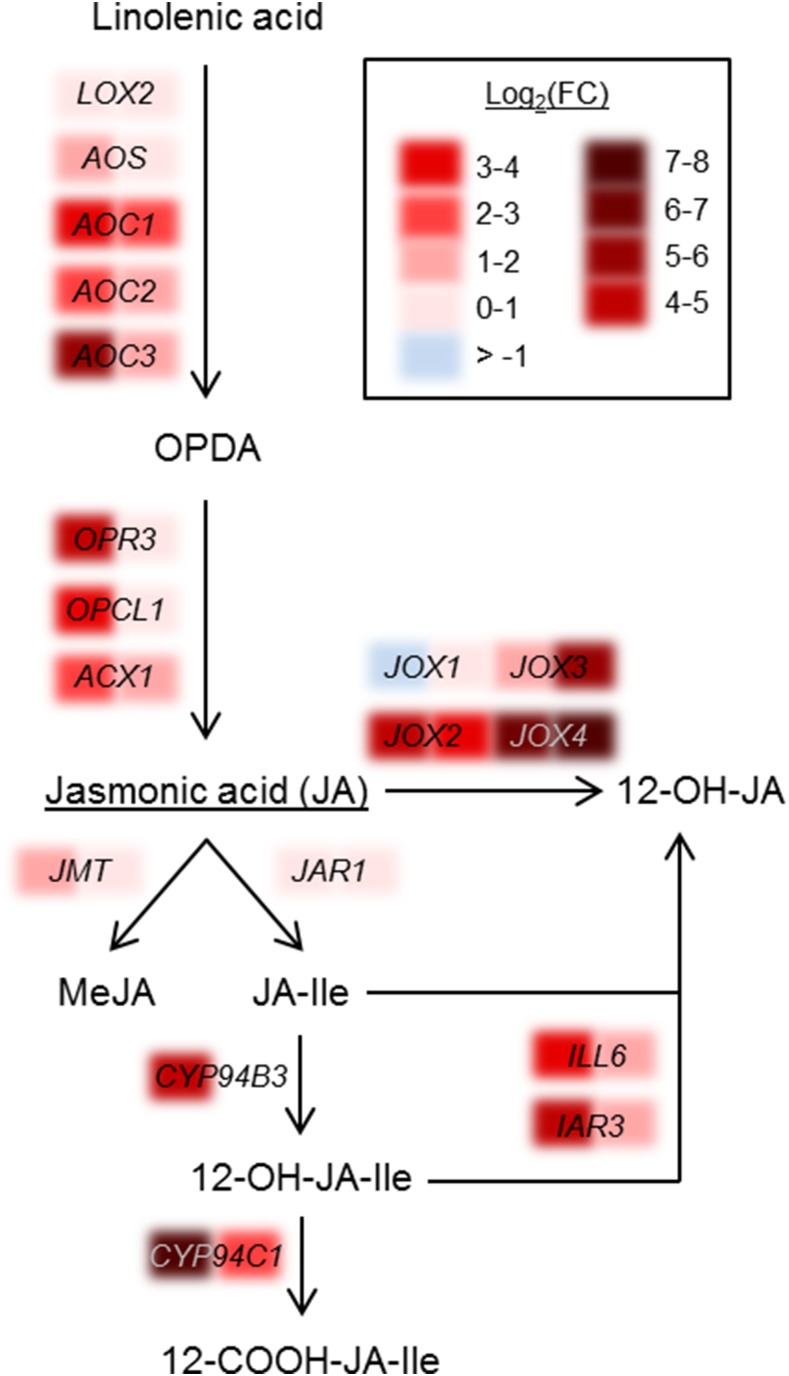
Jasmonates derive from the FA linolenic acid, which undergoes oxidation via lipoxygenases, dehydration via the allene oxide synthase (AOS), followed by subsequent cyclization to oxophytodienoic acid (OPDA) via the allene oxide cyclases (AOC). After cis-OPDA is reduced by OPDA-reductase (OPR3), three rounds of β-oxidation (e.g. via acyl-CoA oxidase and OPC-8:0 CoA ligase) are necessary to form JA. JA, in turn, can be modified to JA-Ile or methyl JA via jasmonate-amido synthetase and JA-carboxyl methyltransferase, respectively (Browse, 2009; Wasternack and Hause, 2013). This biosynthetic pathway is outlined in Figure 5. The majority of depicted genes were significantly up-regulated directly after fumigation, with a log2(FC) of up to 5.9 for AOC3, whereas 6 h after fumigation, the expression levels generally declined.
Figure 5.
JA biosynthesis and degradation pathways are up-regulated simultaneously in response to NO2. The schematic pathway of jasmonate metabolism illustrates the change in expression levels [log2(FC)] of the respective genes obtained from the microarray analysis immediately (0 h, left part of the colored sections) or 6 h (right part of the colored sections) after fumigation with 10 µL L−1 NO2. Expression levels of all depicted genes can be found in Supplemental Table S1. JAR1, Jasmonate-amido synthetase; JMT, JA-carboxyl methyltransferase; MeJA, methyl jasmonate
A major step during the catabolic turnover of active jasmonates is the oxidation of JA-Ile by members of the cytochrome P450 94 (CYP94) family (Fig. 5), resulting in biologically inactive 12-OH-JA-Ile and 12-COOH-JA-Ile (Kitaoka et al., 2011; Koo et al., 2011; Heitz et al., 2012). JA-Ile and its hydroxylated form can be further catabolized to tuberonic acid (12-OH-JA) by the amidohydrolases IAA-ALANINE RESISTANT3 and IAA-LEUCINE RESISTANT-LIKE6 (Widemann et al., 2013). Moreover, jasmonate-induced oxygenases (JOXs) hydroxylate JA to its inactive 12-OH-JA derivative (Caarls et al., 2017; Smirnova et al., 2017), which represents yet another pathway of jasmonate catabolism. Directly after fumigation, the majority of genes involved in these catabolic reactions were highly up-regulated, with fold changes to the respective air controls ranging from log2(FC) 1.3 to 7.4. The genes encoding for the CYP94 enzymes and members of the JOXs were highly induced. The transcript levels of JOXs were still elevated significantly up to a log2(FC) of 7.8 at 6 h after NO2 treatment. Besides the JOXs, the gene transcripts of most of the above-mentioned catabolic enzymes also were still highly abundant at this time point after fumigation. All expression levels of the depicted genes can be found in Supplemental Data Set S1.
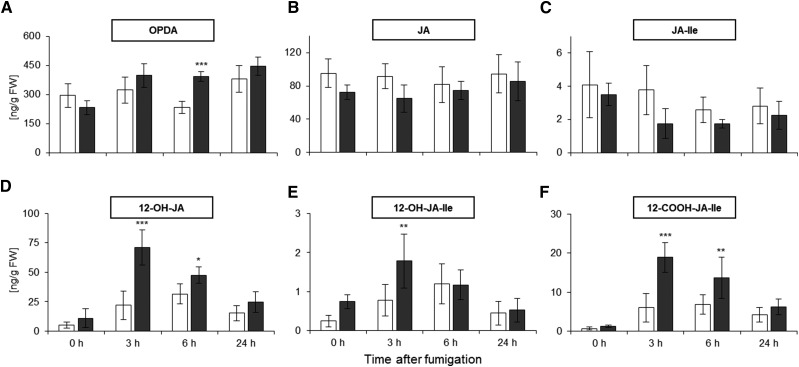
The gene expression data suggested the simultaneous induction of jasmonate biosynthesis and catabolism. LC-MS/MS revealed that OPDA levels were elevated by 69% at 6 h after fumigation compared with leaf extracts from air-fumigated plants (Fig. 6A), which was associated with the enhanced expression of defensin-coding genes, including PLANT DEFENSIN1.2A (Supplemental Data Set S1). By contrast, significant changes were not detected for JA or JA-Ile (Fig. 6, B and C). The rapid and extensive NO2-induced transcription of genes whose products are necessary for jasmonate catabolism encourages the hypothesis that NO2 stimulates rapid jasmonate turnover. Accordingly, all catabolic intermediates of JA and JA-Ile increased and peaked in their concentrations at 3 h after NO2 fumigation. 12-OH-JA increased significantly by a factor of 3.2 from 22 ng g−1 fresh weight in air-fumigated plants to 71 ng g−1 fresh weight after NO2 treatment (Fig. 6D), while 12-OH-JA-Ile levels increased significantly by 2.3-fold at 3 h after fumigation when compared with the air-fumigated control (Fig. 6E). A 3.1-fold increase was observed for 12-COOH-JA-Ile from 6 ng g−1 fresh weight (air) to 19 ng g−1 fresh weight (NO2; Fig. 6F). After the concentrations of all intermediates peaked at 3 h after treatment, their accumulation declined gradually to baseline levels 24 h after NO2 treatment.
Figure 6.
JA degradation products accumulate in response to NO2. Various jasmonates were measured by LC-MS/MS at different time points after fumigation with air or 10 µL L−1 NO2. Concentrations were normalized to the leaf sample fresh weight (FW). A, OPDA. B, JA. C, JA-Ile. D, 12-OH-JA. E, 12-OH-JA-Ile. F, 12-COOH-JA-Ile. A to C show products of the JA biosynthesis pathway, and D to F show JA catabolism products. Columns represent means ± sd (n = 5). Asterisks indicate significant differences within the time points according to two-way ANOVA plus the Holm-Sidak posthoc test (*, P < 0.05; **, P < 0.01; and ***, P < 0.001). White columns, air; black columns, 10 µL L−1 NO2.
Together, these results suggest that exposure to NO2 triggered consecutive peaks of SA and OPDA. The simultaneous induction of jasmonate production and catabolism pathways resulted in the accumulation of 12-OH-JA, 12-OH-JA-Ile, and 12-COOH-JA-Ile. The latter process might be controlled by genes involved in SA/JA antagonism cross talk that were strongly up-regulated upon NO2 exposure (Supplemental Data Set S1).
The SA and JA Signaling Pathways Are Both Crucial for NO2-Induced B. cinerea Resistance
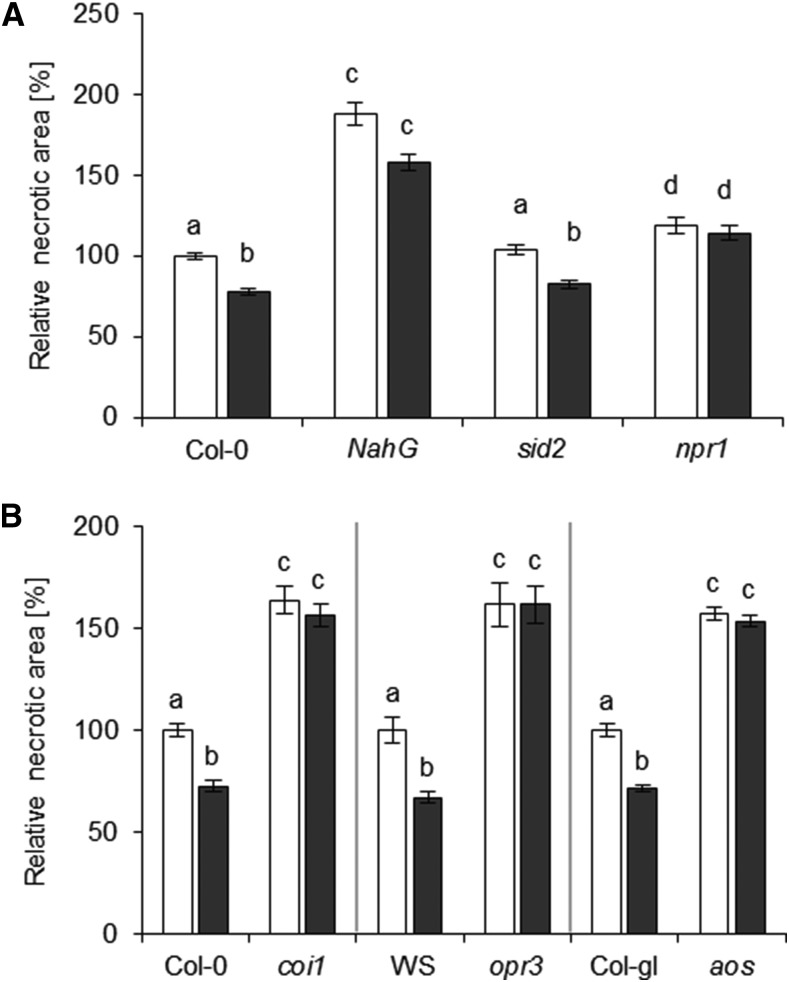
Since SA biosynthesis was up-regulated upon NO2 fumigation, the role of SA in the NO2-induced resistance against B. cinerea was examined by utilizing mutants defective in SALICYLIC ACID INDUCTION DEFICIENT2 (SID2) and plants expressing the Pseudomonas putida NahG gene. The sid2 mutant is impaired in the main SA biosynthesis pathway and, therefore, does not accumulate SA upon pathogen infection (Nawrath and Métraux, 1999; Wildermuth et al., 2001), whereas NahG transgenic plants express a bacterial SA hydroxylase that degrades SA to catechol (Delaney et al., 1994). The B. cinerea infection assay after NO2 fumigation revealed that, in Col-0 plants, the lesion size was reduced by 22% upon NO2 pretreatment (Fig. 7A). The relative necrotic area of NO2-fumigated sid2 also was reduced by 18.4% when compared with the lesion size of its nonfumigated counterpart. However, NO2 pretreatment provoked only a 14% reduction of the relative necrotic area of NahG-expressing plants. This decrease was not significantly different (P > 0.05) from the average lesion size measured in unfumigated NahG plants (Fig. 7A), indicating that the NO2-induced resistance against B. cinerea was compromised. Furthermore, the SA-insensitive npr1 mutant was included in the B. cinerea infection assay after NO2 fumigation. Interestingly, NO2 pretreatment of npr1 plants did not result in a reduction of B. cinerea-induced necrotic lesions. The basal resistance of unfumigated plants was not strongly altered in sid2 and npr1 (+18.7% relative necrotic area compared with Col-0 in untreated trials) but was markedly compromised in NahG transgenic plants (+88% relative necrotic area). Similar results have been reported for these plant lines (Ferrari et al., 2003). Taken together, these results suggest that the NO2-induced resistance against B. cinerea is mediated by NPR1. However, it did not require SA synthesis via SID2, whereas the degradation of SA by bacterial NahG partially abolished the NO2-induced resistance phenotype.
Figure 7.
SA and JA function in NO2-induced resistance against B. cinerea. Mutants were subjected to B. cinerea droplet infection 6 h after fumigation with 10 µL L−1 NO2 for 1 h. Necrotic areas were measured 3 d later and were normalized to the mean necrotic area of the respective unfumigated wild type. A, SA-deficient (NahG and sid2) or SA-signaling (npr1) mutants and the corresponding Col-0 wild type. Columns represent means of at least three independent experiments ± se (n = 95–331). B, JA-deficient (aos and opr3) or JA-signaling (coi-1) mutants and corresponding wild-types (Col-gl for aos, Wassilewskija [WS] for opr3, and Col-0 for coi-1). Columns represent means of three independent experiments ± se (n = 66–126). Letters indicate significant differences of all pairwise comparisons via the Kruskal-Wallis test plus Dunn’s posthoc test (P < 0.05). White columns, unfumigated; black columns, 10 µL L−1 NO2.
NO2 exposure caused a strong rearrangement of jasmonate metabolism. To investigate whether jasmonates or components of the JA signaling pathway were implicated in the NO2-induced pathogen resistance, several Arabidopsis knockout mutants impaired in JA biosynthesis and signaling were subjected to the B. cinerea infection assay. The aos and opr3 knockout mutants were utilized, since they are impeded in JA accumulation upon wounding or B. cinerea infection (Stintzi and Browse, 2000; Park et al., 2002). As shown in Figure 7B, the size of the B. cinerea-induced lesions was not affected by NO2 treatment in both JA-deficient mutants, whereas the necrotic lesions on NO2-treated Col-gl (the aos parental line) were reduced by 28.6% and Wassilewskija (the wild type of opr3) displayed a 33.1% reduction in lesion size upon NO2 fumigation. Knockout mutants that were impaired in JA signaling also were examined for their NO2-induced resistance phenotype. The JA-insensitive coi1 mutant did not show any significant differences in the size of the necrotic lesions that developed on NO2-fumigated or untreated leaves upon B. cinerea infection. These results indicated that the NO2-induced resistance against B. cinerea is dependent on JA accumulation and COI1-mediated signaling. It is noteworthy that the three tested JA mutants all were more susceptible than the respective wild-type lines, confirming the importance of JA in basal resistance against B. cinerea (Thomma et al., 1998).
Collectively, the results above argue for a crucial role of SA and jasmonates during NO2-induced pathogen resistance. However, further phytohormone measurements revealed that the levels of SA, JA, and JA-Ile at 16, 24, and 48 h after B. cinerea infection were not influenced by NO2 pretreatment (Supplemental Fig. S4). This would suggest that SA, OPDA, and possibly the accumulating JA/JA-Ile catabolites function in the NO2-mediated induction of defense responses before but not during B. cinerea infection.
The Volatile Organic Compounds α-Pinene and Longifolene Are Induced after NO2 Exposure
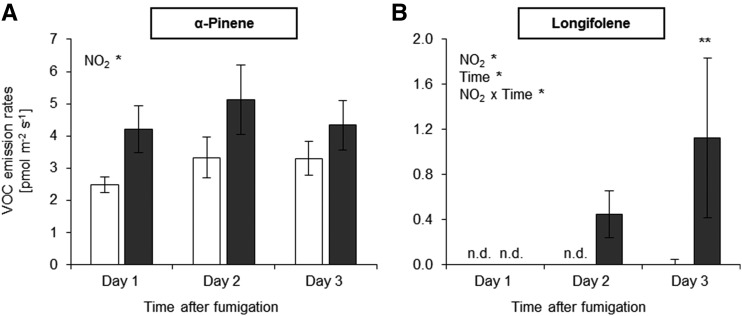
Under stress conditions, plants emit a wide range of volatile organic compounds (VOCs; Niinemets, 2010). Among several detected VOCs (Supplemental Fig. S5), only the emission of the monoterpene α-pinene and the sesquiterpene longifolene were increased significantly after exposing plants to NO2 (Fig. 8; Supplemental Figs. S5 and S6). α-Pinene acts as a signal in the plant-to-plant communication of systemic acquired resistance (SAR; Riedlmeier et al., 2017), while sesquiterpenes often are released after the occurrence of abiotic/biotic stress (Ghirardo et al., 2012, 2016). NO2 induced the emission of α-pinene between 1 and 9 h (day 1) after NO2 fumigation (Fig. 8A), although the expression of the monoterpene biosynthetic gene GERANYLGERANYL REDUCTASE (GGR) was not enhanced immediately or 6 h after NO2 exposure (Supplemental Data Set S1). By comparison, increases of longifolene (syn. junipene) were not detectable until the day after NO2 exposure (day 2) and increased significantly (P < 0.05) the following day (day 3; Fig. 8B). The sesquiterpene-related gene CYP81D11 was found to be up-regulated immediately after the NO2 treatment (Supplemental Data Set S1). Similar to α-pinene, longifolene emission rates were very low.
Figure 8.
NO2 exposure induces volatile emissions. A, Emission of the monoterpene α-pinene. B, Emission of the sesquiterpene longifolene. After 1 h of fumigation with 10 µL L−1 NO2, Arabidopsis Col-0 plants were enclosed in a flow-through cuvette system and volatile emissions were collected and analyzed successively by thermal desorption-gas chromatography-mass spectrometry and multivariate data analysis (Supplemental Figs. S5 and S6). Columns represent means ± se (n = 10–12). Significant main effects (NO2 and Time) and interactions (NO2 × Time) are shown (two-way ANOVA, all pairwise multiple comparison Holm-Sidak posthoc test). *, P < 0.05, **, P < 0.01; and n.d., not detected. White columns, control (air); black columns, 10 µL L−1 NO2.
NO2-Induced B. cinerea Resistance Involves CYP79B2/B3 and PHYTOALEXIN DEFICIENT3 But Not Camalexin
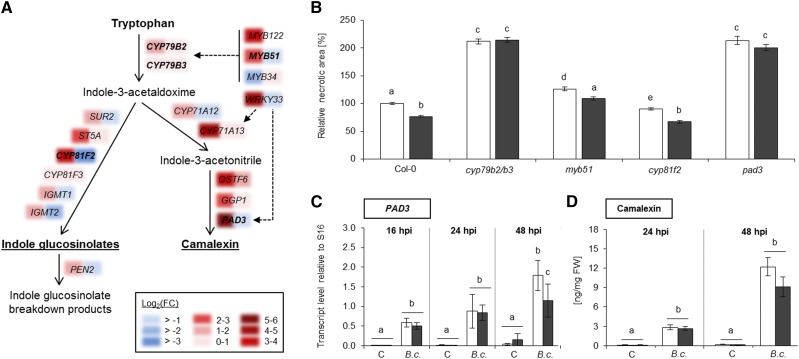
NO2 exposure triggered the expression of genes involved in the biosynthesis of Trp-derived secondary metabolites (Fig. 9A; Supplemental Data Set S1). In the initial step of this pathway, CYP79B2 and CYP79B3 convert Trp into indole-3-acetaldoxime, which serves as a precursor for both indole glucosinolates as well as camalexin (Hull et al., 2000; Glawischnig et al., 2004). The expression of CYP79B2 increased immediately after NO2 fumigation, with a log2(FC) of 1.6, but was not strongly altered at the 6-h time point, while CYP79B3 was generally less responsive to NO2 (Fig. 9A). In the indole glucosinolate pathway, CYP81F2 and CYP81F3 catalyze the hydroxylation of indol-3-ylmethylglucosinolate (I3M; glucobrassicin) to 4-OH-I3M. The expression of CYP81F2 was up-regulated, with a log2(FC) of 3.3, immediately after NO2 fumigation but was down-regulated at the 6-h time point. By comparison, CYP81F3 expression was altered only marginally after the NO2 treatment. Camalexin biosynthesis is dependent on the enzyme PHYTOALEXIN DEFICIENT3 (PAD3), which synthesizes both camalexin and the precursor dihydrocamalexin (Schuhegger et al., 2006; Böttcher et al., 2009). PAD3 expression was enhanced, with a log2(FC) of 5.5, immediately after NO2 fumigation but dropped to wild-type levels at the 6-h time point.
Figure 9.
NO2-induced resistance against B. cinerea is dependent on CYP79B2, CYP79B3, and PAD3 but independent of camalexin. A, The expression of genes related to the biosynthesis of Trp-derived indole glucosinolates and camalexin was strongly up-regulated after fumigation with 10 µL L−1 NO2 for 1 h. Colored areas indicate gene expression [log2(FC)] immediately (left) or 6 h (right) after the NO2 treatment. Genes that were investigated further are highlighted in boldface letters. Gene regulation by transcription factors is indicated by dashed-line arrows. B, The cyp79b2/b3 double mutant and the myb51, cyp81f2, and pad3 mutants were subjected to B. cinerea droplet infection 6 h after fumigation with 10 µL L−1 NO2 for 1 h. Necrotic areas were measured 3 d later and were normalized to the mean necrotic area of the unfumigated Col-0 wild type. Columns represent means of three independent experiments ± se (n = 81–418). Letters indicate significant differences of all pairwise comparisons via the Kruskal-Wallis test plus Dunn’s posthoc test (P < 0.01). C, NO2-exposed or control (unfumigated) Col-0 plants were spray infected with 2 × 105 B. cinerea spores 6 h after fumigation. PAD3 transcript levels were quantified 16, 24, or 48 h after infection relative to housekeeping gene expression via RT-qPCR. Columns represent means of two independent experiments ± sd (n = 5). Letters indicate significant differences of all pairwise comparisons within the time points via two-way ANOVA plus the Holm-Sidak posthoc test (P < 0.01). D, Plants were spray infected with B. cinerea at 6 h after NO2 or air fumigation. Camalexin levels were measured by HPLC-MS 24 and 48 h after infection. Columns represent means ± se (n = 12). Letters indicate significant differences of all pairwise comparisons within the time points via two-way ANOVA plus the Holm-Sidak posthoc test (P < 0.01). FW, Fresh weight.
The above-mentioned genes all function in plant resistance against fungal pathogens; therefore, their possible involvement in NO2-induced resistance against B. cinerea was investigated further using appropriate mutants. Upon B. cinerea infection, the cyp79b2/b3 double mutant displayed a 112% increase in necrotic area formation compared with wild-type plants, which was not influenced by pretreatment with 10 µL L−1 NO2 6 h before inoculation (Fig. 9B). Hence, CYP79B2/B3 play an important role in basal and NO2-induced resistance against B. cinerea. This conclusion was corroborated by the fact that myb51 mutant plants lacking the MYB51 positive regulator of CYP79B2/B3 expression were significantly more susceptible to B. cinerea than Col-0 wild-type plants. Upon NO2 fumigation, the necrotic area was reduced only by 12% as compared with 23% in Col-0 plants, suggesting that the NO2-induced resistance was partially compromised (Fig. 9B).
Additional experiments with the cyp81f2 and pad3 mutants were aimed at determining the specific contributions of indole glucosinolates and camalexin to basal and NO2-induced pathogen immunity. B. cinerea infection of the cyp81f2 mutant caused necrotic lesions with 10% smaller areas than in wild-type plants. Pretreatment with NO2 before infection resulted in a 33% reduction in lesion size, demonstrating that the cyp81f2 mutant was capable of establishing NO2-induced pathogen resistance (Fig. 9B). By contrast, the camalexin-deficient pad3 mutant displayed an enhanced susceptibility toward B. cinerea, as reported earlier (Ferrari et al., 2003). This became apparent by the 113% increase in necrotic lesion size that developed on unfumigated pad3 plants compared with unfumigated wild-type plants (Fig. 9B). At 3 dpi, the necrotic lesions on NO2-pretreated leaves of pad3 did not differ significantly in their size in comparison with their unfumigated control, suggesting that pad3 did not develop NO2-induced B. cinerea immunity (Fig. 9B). Regarding the compromised basal and NO2-induced B. cinerea resistance, pad3 had a similar phenotype to the cyp79b2/b3 mutant.
These findings led us to conclude that the induction of camalexin biosynthesis genes contributed to the NO2-induced resistance against B. cinerea. Surprisingly, however, no accumulation of camalexin was observed upon NO2 exposure (Supplemental Fig. S7). Moreover, the NO2 treatment did not alter PAD3 expression or camalexin levels upon B. cinerea infection (Fig. 9, C and D). Reverse transcription quantitative PCR (RT-qPCR) analysis 16 and 24 h after infection demonstrated that PAD3 transcript levels increased significantly to the same extent upon B. cinerea infection in unfumigated and NO2-treated Col-0 plants (Fig. 9C). Accordingly, no statistical differences in the camalexin content of air- and NO2-treated Col-0 plants were detected after B. cinerea infection (Fig. 9D), although B. cinerea infection led to a significant gradual increase in camalexin from basal 0.1 to 12.2 ng mg−1 fresh weight after 48 h in NO2-treated plants.
Taken together, these results indicated that NO2 fumigation rapidly induced the expression of camalexin biosynthesis genes but did not result in camalexin accumulation. It was shown further that NO2-induced B. cinerea resistance was dependent on CYP79B2/B3 and PAD3 but independent of camalexin, CYP81F2, and indole glucosinolates.
Trp-Derived Secondary Metabolites Accumulate after NO2 Fumigation
Nontargeted Fourier transform ion cyclotron resonance mass spectrometry (FT-ICR-MS) was employed to identify Trp-derived secondary metabolites that could function as signals or defensive compounds in NO2-induced B. cinerea resistance. To this end, Arabidopsis Col-0 plants were exposed to 10 µL L−1 NO2 for 1 h, and leaves were sampled 6 h later because this was the time point at which plants were usually inoculated with B. cinerea spores. Leaf extracts from untreated cyp79b2/b3 plants served as a zero reference because they are devoid of Trp-derived indole glucosinolates and camalexin (Hull et al., 2000; Glawischnig et al., 2004). The following criteria were applied to filter the FT-ICR-MS results for candidate CYP79B2/B3-dependent metabolites involved in NO2-induced pathogen immunity: (1) metabolites were not detected in cyp79b2/b3 extracts but in all 10 leaf extracts from wild-type plants; and (2) metabolites showed significantly up-regulated levels at 6 h after NO2 fumigation. Three of nine identified metabolites had exact masses corresponding to the indole glucosinolate glucobrassicin (I3M), its degradation product ascorbigen, and the methoxylated ascorbigen derivative 1,4-dimethoxyindol-3-ylmethylascorbate (Table 1; Supplemental Table S1), confirming that the experimental approach identified Trp-derived compounds. Two other metabolites contained no sulfur atom but at least one nitrogen atom and, thus, could represent indolic compounds. Further experiments are needed to specify if and how glucobrassicin, ascorbigen, 1,4-dimethoxyindol-3-ylmethylascorbate, and the other identified CYP79B2/B3-dependent metabolites are involved in NO2-induced B. cinerea immunity.
Table 1. CYP79B2/B3-dependent accumulation of metabolites 6 h after fumigation with 10 µL L−1 NO2.
Metabolites were not detected in the cyp79b2/b3 double mutant. NO2-induced up-regulation in wild-type plants is given as fold change of median spectral count. See Supplemental Table S1 for the complete data set, including statistics. Formulae and tentative annotations were deduced from the exact masses as determined by FT-ICR-MS.
| Mass m/z [M-H]− | Up-Regulation by 10 µL L−1 NO2 | Formula [M-H] | Tentative Annotation | |
|---|---|---|---|---|
| Measured | Δ µL L−1 | |||
| 447.0537 | −0.05 | 1.8 | C16H20N2O9S2 | Glucobrassicin, indol-3-ylmethylglucosinolate |
| 367.0783 | ±0.00 | 3.0 | C15H16N2O9 | Unknown CYP79B2/B3-dependent metabolite |
| 364.1038 | ±0.00 | 2.7 | C17H19NO8 | 1,4-Dimethoxyindol-3-ylmethylascorbate |
| 304.0826 | −0.04 | 2.8 | C15H15NO6 | Ascorbigen, indol-3-ylmethylascorbate |
| 232.0463 | ±0.00 | 2.2 | C8H11NO7 | Unknown CYP79B2/B3-dependent metabolite |
Enhanced Callose Formation Is Essential for NO2-Induced B. cinerea Resistance
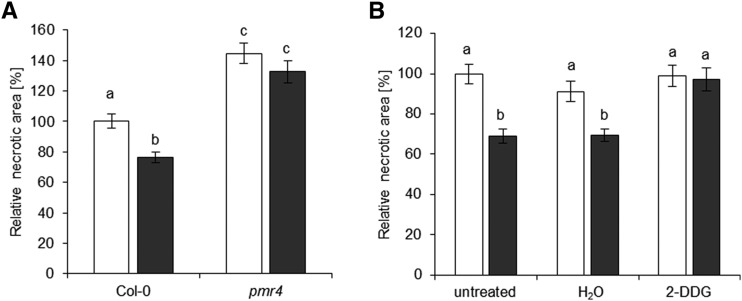
Callose deposition at infection sites is an effective plant defense mechanism against various pathogens (Ellinger and Voigt, 2014). The pmr4 mutant is defective in the GLUCAN SYNTHASE-LIKE5 gene and does not deposit callose upon pathogen infection (Jacobs et al., 2003; Nishimura et al., 2003). This mutant was subjected to the B. cinerea infection assay after NO2 fumigation (Fig. 10A). The size of necrotic lesions did not differ between NO2-treated and unfumigated pmr4 plants, whereas the NO2-treated Col-0 wild type exhibited a 23.7% reduction of the necrotic area. Additionally, Col-0 leaves were infiltrated with the callose deposition inhibitor 2-deoxy-d-glucose (2-DDG; Bayles et al., 1990) or water 24 h before NO2 treatment, followed by B. cinerea droplet infection 6 h after fumigation (Fig. 10B). Water-infiltrated plants developed a 31% lesion size reduction when compared with the lesions formed on unfumigated, noninfiltrated leaves. Importantly, NO2-induced resistance was suppressed in NO2-fumigated, 2-DDG-treated leaves (Fig. 10B). Hence, PMR4-mediated callose deposition is essential for NO2-induced resistance.
Figure 10.
Plants impaired in callose formation display a loss in NO2-induced resistance against B. cinerea. A, Col-0 and callose-deficient pmr4 plants were subjected to B. cinerea droplet infection 6 h after fumigation with 10 µL L−1 NO2 for 1 h. Necrotic areas formed on fumigated leaves after 3 d were normalized to the mean necrotic areas of the respective unfumigated leaves. Columns represent means of four independent experiments ± se (n = 135–145). B, Relative necrotic areas determined on Col-0 plants that were infiltrated with 1.2 mm of the callose synthesis inhibitor 2-DDG (water as a control) 24 h before fumigation followed by B. cinerea infection. Columns represent means ± se (n = 70–130). Letters indicate significant differences of all pairwise comparisons via the Kruskal-Wallis test plus Dunn’s posthoc test (P < 0.05). White columns, unfumigated; black columns, 10 µL L−1 NO2.
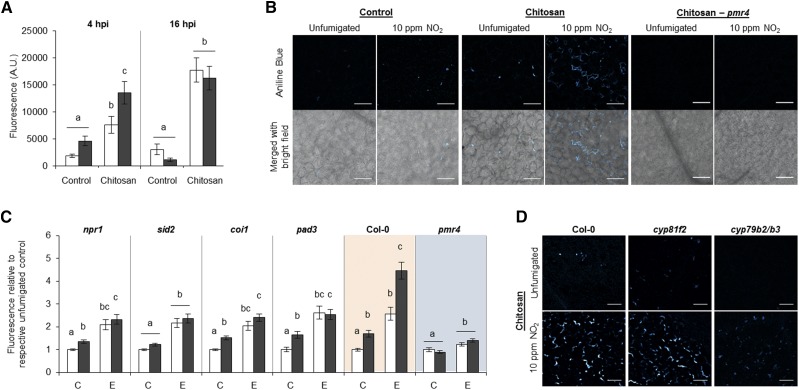
The autofluorescence of B. cinerea interfered with the Aniline Blue staining of callose. Therefore, chitosan was employed as a potent elicitor of callose deposition (Köhle et al., 1985). Arabidopsis Col-0 plants were fumigated with NO2 followed by leaf infiltration of 500 µg mL−1 chitosan 4 h later (Fig. 11). The photometric Aniline Blue assay revealed that chitosan triggered a 4- to 6-fold increase in fluorescence at 4 and 16 h after the elicitor treatment, respectively (Fig. 11A). Exclusively at the earlier time point, the callose-dependent fluorescence was enhanced further in NO2-pretreated plants. Aniline Blue fluorescence was localized in the extracellular space but was absent in pmr4, confirming the specificity of the callose detection (Fig. 11B). The NO2-enhanced callose formation at 4 h after chitosan treatment was suppressed in the SA mutants npr1 and sid2 and in the JA signaling mutant coi1, although the chitosan-induced callose formation was observed in all mutants (Fig. 11C). As expected, almost no chitosan-induced callose formation was detected in the pmr4 mutant. The NO2-enhanced early callose formation upon elicitor treatment was strongly diminished in the camalexin-deficient pad3 mutant and in the cyp79b2/b3 double mutant but was unaffected in the indole glucosinolate mutant cyp81f2 (Fig. 11, C and D). Only in the cyp79b2/b3 double mutant were no chitosan-triggered callose depositions detected by microscopy (Fig. 11D). In many experiments, NO2 alone already stimulated weak callose deposition, which also was seen in the tested mutants except for sid2 and cyp79b2/b3.
Figure 11.
NO2 pretreatment enhances early callose deposition upon treatment with the fungal elicitor chitosan. Plants were fumigated with 10 µL L−1 NO2 for 1 h and infiltrated with 500 µg mL−1 chitosan (0.04% acetic acid as a control) 4 h after fumigation. Leaf discs were obtained for callose quantification with Aniline Blue 4 or 16 h after chitosan treatment. A, Callose quantification in Col-0. Columns represent means ± se (n = 34–44 from 10 plants per time point and treatment). A.U., Arbitrary units; hpi, hours after infection. B, Detection of Aniline Blue-stained callose by confocal laser scanning microscopy. Fluorescence and bright-field channels were merged using ImageJ software. Representative photographs were taken of NO2-fumigated or unfumigated Col-0 or pmr4 (right section) 4 h after treatment with chitosan. Bars = 100 μm. C, Callose quantification in mutants impaired in SA synthesis (sid2), SA signaling (npr1), JA signaling (coi1), camalexin synthesis (pad3), and callose deposition (pmr4). Columns represent means ± se (n = 103–159 for Col-0 and pmr4; n = 57–65 for other mutants). White columns, unfumigated; black columns, 10 µL L−1 NO2. C, Infiltration control; E, elicitor chitosan. Letters indicate significant differences of all pairwise comparisons within time points via the Kruskal-Wallis test plus Dunn’s posthoc test (P < 0.05). White columns, unfumigated; black columns, 10 µL L−1 NO2. D, Detection of Aniline Blue-stained callose in NO2-fumigated or unfumigated cyp81f2 and cyp79b2/b3 mutant plants 4 h after treatment with chitosan. Col-0 stained in the same experiment is shown for comparison. Bars = 100 μm.
Two lines of evidence support an important role of callose in NO2-induced pathogen resistance. (1) The resistance was suppressed in the callose-deficient pmr4 mutant and in plants treated with the callose inhibitor 2-DGG. (2) Mutants that did not exhibit NO2-induced resistance also were impaired in NO2-enhanced callose deposition upon chitosan elicitation, with the exception of sid2, which exhibited NO2-induced pathogen immunity but impaired callose formation.
DISCUSSION
Under physiological conditions, NO2 can arise from the oxidation of NO, reduction of nitrite, or decomposition of peroxynitrite (Pryor et al., 2006; Groß et al., 2013). Although the formation of NO2 under stress conditions is well supported by direct and indirect evidence, less is known about the physiological functions of NO2. To address this issue, we fumigated Arabidopsis plants with µL L−1 levels of NO2 as a donor treatment. Previous experiments showed that 1 h of exposure of Arabidopsis to 30 µL L−1 NO2 triggered rapid cell death, whereas 10 µL L−1 NO2 did not cause visible leaf symptoms or ion leakage as a marker of cell damage (Kasten et al., 2016). However, immediately after NO2 exposure, plants displayed the enhanced chlorophyll autofluorescence (Fig. 1A; Supplemental Fig. S1) indicative of photoprotective energy dissipation in the course of a transient defense response (Lichtenthaler and Miehe, 1997; Chaerle and Van Der Straeten, 2000). In this study, the NO2-induced defense response was investigated in detail.
Short-term exposure to 10 µL L−1 NO2 induced an up-regulation of more than 2,300 genes immediately after the 1-h treatment period. The number of up-regulated genes decreased to approximately 750 at 6 h after fumigation, indicating that many genes were rapidly and transiently induced by NO2 (Fig. 1, B and C). GO term enrichment and cluster analysis revealed that predominantly genes involved in pathogen resistance were expressed strongly after NO2 fumigation (Fig. 2). Many of these genes are induced by flg22 (Zipfel et al., 2004), chitin (Ramonell et al., 2005), B. cinerea (Ferrari et al., 2007), and P. syringae (Lewis et al., 2015; Fig. 2), suggesting that NO2 triggered basal pathogen resistance or PTI.
Accordingly, NO2-pretreated plants showed resistance against the fungal pathogen B. cinerea and the bacterial pathogen P. syringae (Fig. 3). The fact that plants could fend off pathogens of distinct life styles confirmed that NO2 conferred PTI. How the rather simple molecule NO2 can specifically evoke pathogen resistance is not yet known. NO2 might activate signaling cascades by the nitration of electrophiles. Particularly, nitro-FAs such as nitrolinolenic acid have reported functions in defense and antiinflammatory signaling (Schopfer et al., 2011; Mata-Pérez et al., 2016b). Alternatively, endogenous elicitors possibly derived from NO2-induced cell wall or membrane modifications are formed within the leaf. For instance, NO2 can cause both the oxidation as well as nitration of lipids (Pryor et al., 2006; Schopfer et al., 2011), which could lead to membrane damage and the subsequent formation of DAMPs. Such nitro signals and endogenous elicitors also could arise when NO2 is formed under natural stress conditions.
Plant defense responses to pathogen assaults often are orchestrated by SA and JA, although the exact interactions of these phytohormones in PTI are not fully understood (Couto and Zipfel, 2016). SA levels increased 0 to 3 h after NO2 fumigation, which was accompanied by the increased expression of genes involved in the early SA response (Fig. 4; Supplemental Fig. S3; Blanco et al., 2009) and SAR, including METHYL ESTERASE9, FLAVIN-DEPENDENT MONOOXYGENASE1, AZELAIC ACID INDUCED1 (AZI1), AZI2, DEFECTIVE IN INDUCED RESISTANCE1 (DIR1/AZI6), and AGD2-LIKE DEFENSE RESPONSE PROTEIN1 (Supplemental Data Set S1). NO2 also activated the jasmonate biosynthesis pathway. The accumulation of OPDA 6 h after fumigation could be responsible for the enhanced expression of 13 genes coding for antimicrobial defensins at this time point (Figs. 5 and 6; Supplemental Data Set S1), as reported earlier (Stintzi et al., 2001).
Notably, NO2 not only initiated jasmonate biosynthesis but also JA and JA-Ile catabolism (Fig. 5). As a result, the levels of JA and its bioactive derivative JA-Ile were unchanged, whereas their degradation products 12-OH-JA, 12-OH-JA-Ile, and 12-COOH-JA-Ile increased up to 3-fold after fumigation (Figs. 5 and 6). Several genes involved in jasmonate catabolism, including JOX1 to JOX4, are inducible by jasmonates as a means of terminating the defense response (Caarls et al., 2017; Smirnova et al., 2017). However, jasmonate-induced JA catabolism is not a likely scenario after NO2 exposure because neither JA nor JA-Ile levels were altered significantly under these conditions. 12-OH-JA has reported functions in the down-regulation of JA biosynthesis and JA-mediated defense responses (Miersch et al., 2008; Patkar et al., 2015; Caarls et al., 2017; Smirnova et al., 2017), whereas the biological activities of other jasmonate hydroxylation and carboxylation products are yet unexplored. Genes coding for proteins involved in SA/JA cross talk, such as several WRKY transcription factors, GLUTAREDOXIN480, UDP-DEPENDENT GLYCOSYLTRANSFERASE76B1, and jasmonate-zim-domain transcriptional repressors, were strongly up-regulated (Supplemental Data Set S1; von Saint Paul et al., 2011; Caarls et al., 2015). Therefore, it is tempting to speculate that the NO2-induced SA peak and proteins functioning in SA/JA cross talk control both the repression of JA-responsive genes as well as JA/JA-Ile degradation, but this remains to be elucidated.
NO2 fumigation triggered SA and OPDA signaling and defense gene expression. Further experiments were aimed at detailing the role of phytohormones during the NO2-induced basal pathogen immunity. NO2-induced B. cinerea resistance was compromised in plants expressing the SA hydrolase NahG and in the SA signaling mutant npr1 but was not altered in the SA biosynthesis mutant sid2 (Fig. 7). These results are in accordance with previous findings showing that SA produced by Phe ammonia lyase but not the SID2 pathway contributes to the establishment of B. cinerea resistance in Arabidopsis (Ferrari et al., 2003). Further mutant analyses revealed that JA biosynthesis and signaling via the COI1 transcriptional activator was essential for NO2-induced resistance against B. cinerea, as reported earlier (Thomma et al., 1998). Ethylene is well known for its role in PTI (Boller and Felix, 2009; Couto and Zipfel, 2016). The GO term enrichment of genes involved in ethylene-activated signaling pathways indicated that this gaseous defense hormone contributes to NO2-induced immunity. However, this will need to be proven by future experiments.
The emission of the monoterpene α-pinene and the sesquiterpene longifolene was increased significantly after exposing plants to NO2 (Fig. 8). It has been demonstrated that monoterpenes, including α-pinene, play an essential role in the SA-dependent establishment of SAR (Riedlmeier et al., 2017). Likewise, the transient NO2-induced SA peak was followed by the emission of α-pinene, which might trigger SAR within and between plants, as shown recently (Riedlmeier et al., 2017). α-Pinene production was not regulated at the transcriptional level because NO2 exposure had no effect on the expression of GGR1, which codes for an enzyme involved in the biosynthesis of the monoterpene precursor geranyl diphosphate (Tholl and Lee, 2011; Supplemental Data Set S1). NO2-dependent increases of terpenoid emissions might originate from changes of metabolic pool size and fluxes (Ghirardo et al., 2014) and enzyme activities (Ghirardo et al., 2010). These results suggest the induction of local and systemic pathogen resistance by NO2 analogous to the induction of a local PTI and the subsequent establishment of SAR following the treatment with pathogen-derived elicitors (Mishina and Zeier, 2007).
Longifolene occurs commonly in plant species of the genus Pinus, where it is produced by longifolene synthases and stored in (oleo)resin (Martin et al., 2004). Treatment with methyl JA caused an enhanced accumulation of longifolene in Douglas fir (Pseudotsuga menziesii) stem and root samples (Huber et al., 2005). In the resin, longifolene could act as an antimicrobial compound (Himejima et al., 1992). The biosynthesis and functions of longifolene remain undocumented in Arabidopsis. However, it was reported that CYP81D11-overexpressing Arabidopsis lines emitted the sesquiterpene isolongifolene in the context of cis-jasmone-regulated tritrophic interactions between plants, aphids, and parasitoids (Bruce et al., 2008). Noteworthy, CYP81D11 was strongly induced by NO2 (Supplemental Data Set S1).
The indole alkaloid camalexin and indole glucosinolates are both derived from Trp, and their biosynthesis pathways are closely interconnected (Glawischnig et al., 2004). NO2 fumigation triggered the expression of several genes involved in the production of these compounds. CYP79B2, CYP79B3, MYB51, CYP81F2, and PAD3 were investigated further for their possible functions in NO2-induced B. cinerea resistance because these genes have been associated previously with immunity against fungal pathogens. The cyp79b2/b3 mutant is deficient in both camalexin as well as indole glucosinolates (Bednarek et al., 2009). Experiments with this mutant confirmed the reported high susceptibility to B. cinerea (Nafisi et al., 2007) and additionally revealed a complete loss of NO2-induced resistance (Fig. 9B). Moreover, myb51 plants, which have reduced levels of indolic compounds (Frerigmann et al., 2016), were partially compromised in basal and NO2-induced B. cinerea resistance. Together, these results suggest an essential role of indolic secondary metabolites in NO2-induced B. cinerea immunity. cyp81f2 mutant plants produce glucobrassicin but not 4-OH-I3M and its derivatives, which are essential for the basal resistance of Arabidopsis against biotrophic powdery mildews and the necrotrophic fungal pathogen Plectosphaerella cucumerina (Bednarek et al., 2009). However, in this study, cyp81f2 plants did not show a resistance-related phenotype, indicating that CYP81F2-dependent indole glucosinolates are dispensable for basal and NO2-induced resistance against B. cinerea (Fig. 9B).
Camalexin inhibits the growth of B. cinerea (Kliebenstein et al., 2005; Glawischnig, 2007). NO2 exposure triggered the induction of the camalexin biosynthesis gene PAD3, and the pad3 mutant did not develop NO2-induced resistance against B. cinerea (Fig. 9). Regarding the compromised basal and NO2-induced B. cinerea resistance, pad3 had a similar phenotype to the cyp79b2/b3 mutant. Therefore, it was hypothesized that the cyp79b2/b3 phenotype was likely caused by camalexin deficiency rather than a defect in indole glucosinolate biosynthesis. Unexpectedly, during B. cinerea infection, neither PAD3 expression nor camalexin production was influenced by NO2 pretreatment. These findings resemble a previous study showing that PAD3 expression but not camalexin levels were increased strongly upon elicitor treatment with plant cell wall-derived oligogalacturans (Ferrari et al., 2007). Thus, rather than camalexin itself, a downstream metabolite (Böttcher et al., 2009) with so far obscure physiological functions might be involved in NO2-induced B. cinerea resistance.
In a pioneering attempt to identify such Trp-derived metabolites, we analyzed leaf extracts from NO2-treated plants using nontargeted direct infusion FT-ICR-MS. Nine candidate metabolites accumulated significantly at 6 h after NO2 fumigation but were not detectable in leaf samples from cyp79b2/b3 plants that are devoid of indolic compounds (Supplemental Table S1). Five metabolites could represent indole derivatives because they contained at least eight carbon atoms and one nitrogen atom (Table 1). Neither camalexin nor known camalexin-related metabolites (Böttcher et al., 2009) were found among the NO2-regulated CYP79B2/B3-dependent metabolites. Instead, measured exact masses were annotated as the indole glucosinolate glucobrassicin (I3M), its degradation product ascorbigen, and the methoxylated ascorbigen derivative 1,4-dimethoxyindol-3-ylmethylascorbate. If and how these candidate metabolites are linked to NO2-induced B. cinerea resistance will be defined by future FT-ICR-MS runs with leaf extracts from air- and NO2-exposed mutants including pad3 and cyp81f2.
Cell wall fortification by callose deposition is a frequently used readout of PTI induction (Boller and Felix, 2009). In response to pathogens and pathogen-derived elicitors, callose is mostly synthesized by the callose synthase PMR4 (Jacobs et al., 2003; Nishimura et al., 2003; Clay et al., 2009; Ellinger and Voigt, 2014). NO2-induced pathogen resistance was compromised in pmr4 and in wild-type plants treated with a callose synthase inhibitor, implying a major role of callose in NO2-induced B. cinerea immunity (Fig. 10). The fungal elicitor chitosan triggers callose formation (Köhle et al., 1985; Ramonell et al., 2005) and was used here to mimic the infection by a fungal pathogen. NO2 alone already induced a slight increase in callose formation, which was increased further 4 h after chitosan application as compared with unfumigated plants (Fig. 11). Hence, preformed and more rapidly occurring callose deposition contributed to the NO2-induced resistance against B. cinerea. The stimulatory effect of NO2 on chitosan-induced callose formation was not seen in npr1, sid2, and coi1, indicating that SA and JA signaling were essential for the induction of callose formation. However, given that the sid2 mutant was capable of establishing NO2-induced B. cinerea resistance (Fig. 7), this form of immunity is not based solely on callose depositions but can be compensated by other defense mechanisms.
It was reported that, in Arabidopsis, a yet uncharacterized CYP81F2- and PEN2-dependent 4-OH-I3M breakdown product functions as a signal or coactivator for Flg22-induced callose deposition (Clay et al., 2009). However, in this study, chitosan-triggered callose formation was not altered in cyp81f2, which is in line with the previous finding that Flg22- but not chitosan-triggered callose synthesis was affected in the pen2 mutant (Luna et al., 2011). Hence, the regulation of callose buildup is specific to the perceived elicitor. Flg22-induced callose formation was not compromised in the pad3 mutant, suggesting that this defense response was not dependent on camalexin (Clay et al., 2009). Accordingly, Aniline Blue fluorescence was enhanced significantly in pad3 after chitosan treatment (Fig. 11C). By contrast, the enhancing effect of NO2 on the early chitosan-triggered callose deposition was suppressed in pad3 and cyp79b2/b3 (Fig. 10, C and D). The latter mutant showed a reduced ability to produce callose in response to chitosan, although this has to be confirmed by quantitative measurements. Together, these findings argue for a role of PAD3-produced metabolites other than camalexin in the NO2-enhanced early callose deposition. These results also suggest that chitosan-induced callose formation and NO2-enhanced callose formation are controlled by different signaling pathways.
The lack of NO2-enhanced callose formation 4 h after chitosan treatment in pad3 and all tested phytohormone mutants (except sid2) was associated with the inability of these mutants to establish NO2-induced B. cinerea resistance. Callose synthesis in response to NO2 alone was not altered in most of the tested mutants, suggesting that only the NO2-enhanced callose formation upon the perception of pathogen-derived elicitors was decisive for the NO2-induced immunity against B. cinerea.
CONCLUSION
Donor treatments of Arabidopsis with 10 µL L−1 NO2 triggered basal disease resistance against B. cinerea and P. syringae. Transcriptomics suggested that NO2 fumigation led to the onset of phytohormone signaling and the biosynthesis of indolic compounds such as camalexin. Therefore, these biological processes were investigated in more detail. The NO2-induced resistance was dependent on SA and jasmonate signaling. An early peak of SA immediately after the NO2 treatment was followed by the transient accumulation of OPDA and JA catabolites. Particularly interesting was the finding that the activation of JA catabolism represents a mechanism for the complete suppression of JA signaling, presumably in the course of SA/JA antagonism. Mutants impaired in SA or jasmonate signaling were compromised in NO2-induced B. cinerea resistance, confirming that the coordinated action of both signaling pathways is required for this form of pathogen immunity.
The cyp79b2/b3 double mutant that is deficient in indole phytoalexins did not establish NO2-induced B. cinerea resistance. Further investigations of the pad3 mutant combined with biochemical measurements specified that unknown camalexin-derived metabolites but not camalexin itself function in the resistance induction by NO2. The SA and jasmonate signaling mutants as well as the camalexin-deficient mutants all were more susceptible to B. cinerea, suggesting that basal resistance in untreated plants and NO2-induced resistance likely are mediated by similar defense mechanisms. The inability of these mutants to establish NO2-induced immunity was associated with the loss of NO2-enhanced callose formation upon perception of the fungal elicitor chitosan. Therefore, timely callose deposition seems to be an essential defense mechanism during the NO2-induced B. cinerea resistance. Further defense mechanisms could be related to the observed emission of α-pinene and longifolene from NO2-fumigated plants.
The exact mechanism by which NO2 triggers PTI remains to be investigated. NO2 might function as a dedicated signal (e.g. via the nitration of electrophilic target molecules). However, NO2 also could act more indirectly as a defense elicitor by causing nitrooxidative stress.
MATERIALS AND METHODS
Plant Material and NO2 Fumigation
The utilized Arabidopsis (Arabidopsis thaliana) genotypes and their descriptions and origins are summarized in Supplemental Table S2. Plants were grown and fumigated for 1 h with 10 µL L−1 NO2 as described previously (Kasten et al., 2016). A fumigation system was used as detailed in Supplemental Figure S8 (Kasten et al., 2017). The only difference was the installation of a NO2 mixing cylinder (1.5 L) containing Raschig rings, in which 15% (v/v) NO reacted with 100% oxygen to give NO2. Up to 100 plants were fumigated with NO2 in parallel. The light conditions within the fumigation chamber were adjusted to the settings in the growth chamber (65–85 μmol m–2 s–1 light intensity) where the plants were raised to avoid any light artifacts on nitrogen metabolism (Beevers and Hageman, 1969) and plant-pathogen interactions (Roden and Ingle, 2009).
Autofluorescence Detection
UV autofluorescence was detected using a hand-held UV lamp (Blak-Ray B-100AP; UVP) and documented with a Nikon DC300 camera. Camera settings were kept consistently at an exposure time of 2 s at ISO-3200 with an aperture of F/18.
Statistics
SigmaPlot 12.0 (Systat Software) was used for the statistical evaluation of all data sets as described earlier (Kasten et al., 2016). When comparing two independent groups, Student’s t test was used, in cases where the Shapiro-Wilk normality test (P > 0.05) was passed. If the normality test failed, the analysis was done with the nonparametric Mann-Whitney rank-sum test. The comparison of more than two independent groups that passed the Shapiro-Wilk normality test (P > 0.05) was done by one-way ANOVA and subsequent Holm-Sidak posthoc tests for all pairwise comparisons or comparisons against a control group. When the normality assumption of ANOVA failed on original or log-transformed data (Shapiro-Wilk test), the nonparametric Kruskal-Wallis test with subsequent Dunn’s method posthoc test was performed to test for differences between the groups.
Pseudomonas syringae pv tomato DC3000
P. syringae pv tomato (Pst) DC3000 was cultivated at 28°C for 2 d on selective nutrient-yeast extract glycerol agar (0.5% [w/v] bactoprotease pepton, 0.3% [w/v] yeast extract, 2% [v/v] glycerol, and 1.8% [w/v] agar) supplemented with rifampicin and kanamycin (50 µg mL−1). Five-week-old plants were fumigated with 10 µL L−1 NO2 (unfumigated plants as a control) and inoculated 4 h after fumigation with 1 × 105 cfu mL−1 Pst DC3000 in 10 mm MgCl2. Three to four leaves per plant were infiltrated with the bacterial suspension from their abaxial side using a 1-mL needleless syringe. The Pst DC3000 bacterial titer within the leaves was determined 2 h (bacterial load control) or 1 and 2 d after infection. At the indicated time points, 6-mm leaf discs were obtained from each infected leaf and, at the 2-h time point, surface sterilized for 30 s in 80% ethanol. Three leaf discs from different plants were merged into one biological replicate, which was then homogenized for 20 s in 200 µL of 10 mm MgCl2 using a Silamat S6 Tissue Homogenizer (Ivoclar Vivadent) and 1.7- to 2-mm glass beads. The resulting bacterial suspension was diluted in 10 mm MgCl2 in a serial logarithmic dilution (10-fold) ranging from 10° to 105. Subsequently, 20 µL of each dilution was spotted onto selective nutrient-yeast extract glycerol agar before incubating them for 2 d at 28°C. Bacterial colonies were counted in spots containing between 10 and 100 colonies, and the bacterial titer (cfu cm−2) per biological replicate was calculated as follows: cfu cm−2 = colony count × dilution factor × volume total/volume spotted × 1.18 cm−1 (leaf disc factor).
Botrytis cinerea
B. cinerea (strain SAS 56) was cultivated on halves of canned apricots (Prunus armeniaca) that were soaked for several hours in distilled, deionized water to reduce their sugar content. After cultivating B. cinerea on the apricots for approximately 1 week, the spores were used for infection experiments. Leaves of 4-week-old Arabidopsis plants were harvested 6 h after fumigation with 10 µL L−1 NO2 and placed with their abaxial side down onto 0.8% (w/v) agar. Droplets (maximum of 10 µL) containing a maximum of 1,000 spores of B. cinerea in one-half-strength grape juice were spotted onto the leaves, avoiding the middle vein. After a 3-d incubation in a long-day climate chamber, the necrotic lesions were documented with a camera and their areas were determined via ImageJ 1.49m. The areas of the necrotic lesions developed on fumigated leaves were normalized to those formed on unfumigated leaves. NO2-treated and untreated wild-type plants were always included during the evaluation of mutants. For the phytohormone, camalexin, and RT-qPCR analyses, entire plants were infected with B. cinerea 6 h after fumigation with 10 µL L−1 NO2 (controls as indicated) by spraying a one-half-strength grape juice suspension containing 2 × 105 fungal spores mL−1 and 0.01% (v/v) Silwet L-77 (Lehle Seeds) onto the plants until runoff. The negative spray control contained no fungal spores. The infected plants were covered with a clear lid to ensure high humidity for proper infection.
Phytohormone Measurements
To quantify SA, JA, cis-OPDA, OH-JA, OH-JA-Ile, and COOH-JA-Ile, approximately 250 mg of leaf material of 4-week-old Col-0 plants that were fumigated with 10 µL L−1 NO2 or air was harvested 0, 3, 6, and 24 h after fumigation. Similarly, leaf material from plants that were spray infected with B. cinerea 6 h after fumigation was collected 16, 24, and 48 h after infection. The LC-MS/MS analyses were performed as described previously (Vadassery et al., 2012; Kasten et al., 2016).
Camalexin Measurements
Four-week-old Col-0 plants were fumigated with 10 µL L−1 NO2 or air, and approximately 100 mg of leaf material was collected and frozen in liquid nitrogen 6 h after fumigation. At the same time, the remaining plants were spray infected with B. cinerea and harvested 24 and 48 h after infection as described above. Camalexin extraction and quantification were performed as described previously (Frerigmann et al., 2015; Müller et al., 2015).
FT-ICR-MS
A total of 450 mg of leaf material was frozen in liquid nitrogen, homogenized using a Silamat S6 tissue homogenizer (Ivoclar Vivadent) and 1.7- to 2-mm glass pearls, and incubated subsequently in 1.5 mL of extraction buffer (2% acetic acid and 80% ethanol) for 30 min at 4°C. After centrifugation for 20 min at 15,000g and 4°C, the supernatant was collected and the pellet was extracted again with 1.5 mL of extraction buffer. An Oasis WAX 6cc solid-phase extraction column (Waters) was rinsed with 1 mL of methanol and 1 mL of water before addition of the 3-mL pooled leaf extract. The column was then washed with 2 mL of 2% acetic acid. Metabolites were recovered from the solid-phase extraction columns by consecutive elutions with 2 mL of methanol and 2 mL of 5% NH4OH in methanol. Samples were dried under vacuum, dissolved in 1 mL of 70% methanol, centrifugated, and 200-fold diluted in 70% methanol before the MS run.
A Solarix FT-ICR mass spectrometer (Bruker Daltonics) coupled to a 12-Tesla magnet (Magnex) with an Infinity ICR cell was used for the experimental study. A time-domain transient was obtained with 4M Words size (4 million 32-integers) and was Fourier transformed to obtain a frequency spectrum, which was then converted by the Solarix Control program (Bruker Daltonics) into a mass spectrum. All ion excitations were performed in broadband mode (frequency sweep radial ion excitation). Three hundred scans were accumulated for each mass spectrum. Ions were accumulated in the collision cell for 300 ms for thermalization and enrichment prior to ICR ion detection. The base pressure in the ICR vacuum chamber was 7 × 10−10 mbar. The electrospray ionization source (Apollo II; Bruker Daltonics) was used in the negative ionization mode to ionize the studied analytes in 70% methanolic solution (Lichrosolv; Sigma-Aldrich). The sample solutions were injected directly to the ionization source by the use of a microliter pump at a flow rate of 2 µL min−1. A source heater temperature of 200°C was maintained, and no nozzle-skimmer fragmentation was performed in the ionization source. The instrument was calibrated previously by the use of Arg negative cluster ions starting from a methanolic Arg solution of 5 mg L−1.
Results of the FT-ICR-MS runs were subjected to normalization. Wilcoxon rank-sum tests for differential analysis between samples from NO2-fumigated plants and samples from air-fumigated plants were performed in R (version 3.3.3) using wilcox.test (R Core Team, 2014). Accurate masses corresponding to regulated metabolites were searched against public databases with Metlin (Smith et al., 2005) and MassTRIX (Suhre and Schmitt-Kopplin, 2008).
Chitosan Elicitation and Callose Quantification
Four- to 5-week-old plants were treated with the fungal elicitor chitosan (medium Mr; Sigma-Aldrich) 4 h after they were fumigated with 10 µL L−1 NO2 (unfumigated plants as a control). Here, three to four leaves per plant were infiltrated from their abaxial side with 500 µg mL−1 chitosan in 0.04% acetic acid using a 1-mL needleless syringe. As a negative control, plants were treated with 0.04% acetic acid.
Leaf discs (6 mm) were obtained from treated leaves with a cork borer at the indicated time points and incubated overnight in 96% ethanol to remove chlorophyll. The destained leaf discs were gently dried off and then incubated for 1 h in 150 mm K2HPO4 buffer (pH 9.5) at room temperature and with mild agitation. Meanwhile, an Aniline Blue (Sigma-Aldrich) staining solution (0.01% [w/v] Aniline Blue in 150 mm K2HPO4 buffer, pH 9.5) was prepared and stirred until decolorized while protecting it from light. The samples were stained overnight in the dark at room temperature and with gentle agitation. After rinsing the leaf discs in the K2HPO4 buffer, they were transferred into wells of a black flat-bottom 96-well plate containing 50 µL of the same buffer. Callose was quantified by measuring the Aniline Blue fluorescence (mean of nine reads per leaf disc) with the Infinite M1000 Pro plate reader (Tecan) after adjusting the Z-positioning of the fluorescence top optics. Aniline Blue fluorescence was excited with 405 nm (5-nm bandwidth), and the emission wavelength was set to 490 nm (20-nm bandwidth). To minimize the noise of potential autofluorescence, the fluorescence of leaf discs that were incubated overnight in 150 mm K2HPO4 buffer (pH 9.5) without Aniline Blue was subtracted from the values of stained samples for each treatment.
For the microscopic inspection of callose depositions, Aniline Blue-stained leaf discs were mounted in 50% glycerol and analyzed with the TCS SP8 X confocal laser scanning microscope (Leica) using the HC PL APO CS2 20×/0.75 IMM objective. The samples were excited with a Diode 405 Laser (laser line UV 405 nm) at 0.1% laser intensity. The emitted fluorescence was detected with a photomultiplier at 480 to 500 nm (gain 800), whereas bright-field micrographs were taken at gain 400 using the transmission photomultiplier.
In some experiments, leaves of 4-week-old Col-0 plants were syringe infiltrated with 1.2 mm of the callose synthesis inhibitor 2-DDG (Sigma-Aldrich) or distilled, deionized water 24 h before fumigation with 10 µL L−1 NO2 (unfumigated as a control). Six hours after fumigation, the infiltrated leaves were detached, placed on 0.8% agar, and droplet infected with B. cinerea, and the necrotic lesions were analyzed after 3 d. The necrotic areas were compared with those formed on unfumigated and noninfiltrated leaves (= 100%).
VOC Collection and Analysis
Three to four biological replicates were collected in each of the three independent experiments (ntotal = 10–12). For each replicate, 50 Arabidopsis plants were enclosed in glass cuvettes flushed continuously with 200 mL min−1 VOC-free synthetic air containing 400 µL L−1 CO2 and ∼9,000 µL L−1 water. The dynamic cuvette system and the experimental procedure have been described in detail previously (Riedlmeier et al., 2017). Sample collection was 8 h at 60 mL min−1. To ensure the collection of plant volatiles under steady-state conditions of net assimilation (Ghirardo et al., 2014), sampling started 1 h after plants were exposed to NO2 and 2 h after the light was switched on in the morning for the days following the NO2 fumigation. An overflow of ∼140 mL min−1 was maintained during the VOC collection to avoid any contaminations. Background measurements were performed for NO2 and control samples separately, in exactly the same way as the collection of samples, but plants were removed immediately after NO2 or air fumigation for the treated and control plants, respectively. Quantitative and qualitative analyses of VOCs were achieved by thermal desorption-gas chromatography-mass spectrometry analysis following established methods (Ghirardo et al., 2011, 2012, 2016; Weikl et al., 2016). The breaking of VOCs through the polydimethylsiloxane adsorbent was negligible (0.08% ± 0.06% [sd]; n = 8) at ∼10 nL L−1 of an 11-VOC standard mixture containing α-pinene (Apel-Riemer Environmental). Longifolene was quantified using isolongifolene as a pure standard. Fluxes of plant volatiles were calculated after background correction and normalized to biomass dried weight (dw) of leaves. Successively, dw was converted in leaf area (la) by using the factor of 26.6 g m−2 (dw la−1) calculated from previous experiments (Riedlmeier et al., 2017).
Microarray Analysis
Four-week-old Arabidopsis Col-0 plants were fumigated with 10 µL L−1 NO2 or air. Approximately 50 mg of pooled leaf material sampled from at least two different plants was harvested immediately and 6 h after fumigation and frozen in liquid nitrogen. Four biological replicates per treatment were collected. The samples were homogenized twice for 10 s using a Silamat S6 Tissue Homogenizer (Ivoclar Vivadent) and 1.7- to 2-mm glass beads. RNA was extracted, and any potentially remaining DNA was digested using the RNeasy Plant Mini Kit (Qiagen) according to the manufacturer’s protocol. The gene expression measurements were performed using Agilent one-color microarrays as described recently (Riedlmeier et al., 2017). The Agilent Feature Extraction software was used with the template GE1_1100_Jul11. Gene expression levels were determined by the limma software (version 3.18.13; Smyth, 2005) using the TAIR10 genome annotation (Berardini et al., 2015). The differential expression between NO2 and air treatments for each time point was computed using the limma software (version 3.18.13) with a nested interaction model (Ritchie et al., 2015). Genes with adjusted P < 0.05 (based on the false discovery rate method for adjustment) and absolute log2(FC) > 1 were selected for further analysis. The differential expression results were visualized via volcano plots generated by SigmaPlot 12.0 (Systat Software) and Venn diagrams created with jvenn (Bardou et al., 2014). Differentially expressed genes were subjected to GO term overrepresentation analysis with PANTHER 11.0 (release date, July 15, 2016) using the annotation from the GO database (release date, December 28, 2016) and the Arabidopsis reference list from PANTHER (Mi et al., 2017). The obtained enriched GO terms (P < 0.05) were visualized in semantic similarity-based scatterplots generated with the REVIGO tool (Supek et al., 2011).
For the meta-analysis of stress-related expression responses, raw data and sample annotation from five Arabidopsis experiments (accession nos. E-GEOD-5684, E-GEOD-6176, E-GEOD-2538, E-GEOD-17382, and E-MTAB-4867) were downloaded from the ArrayExpress database (http://www.ebi.ac.uk/arrayexpress; Kolesnikov et al., 2015). The abiotic stress data set (E-MTAB-4867) was selected because it was measured with the same microarray platform as the NO2 fumigation data (Agilent At8x60K one-color microarrays; design identifier 29132). The pathogen and pathogen elicitor data sets (E-GEOD-5684, E-GEOD-6176, E-GEOD-2538, and E-GEOD-17382) were found by keyword search. Due to the unavailability of Agilent one-color microarray measurements, Affymetrix ATH1-121501 data sets were chosen for these conditions. The combined Affymetrix data were preprocessed using the R package affy (version 1.40.0; Gautier et al., 2004). The combined Agilent data were preprocessed as stated for the NO2 data set. Based on TAIR10 annotation (Berardini et al., 2015), average log2 gene expression levels were computed and subsequently centered for each experiment relative to the mean of its controls to focus on treatment responses (log fold changes relative to the mean of controls). Principal component analysis across all expression response profiles was performed in R (version 3.0.3) using prcomp (R Core Team, 2014).
RT-qPCR
RNA was isolated from approximately 100 mg of leaf material using the RNeasy Plant Mini Kit (Qiagen) according to the manufacturer’s instructions. If necessary, samples were subjected to the RNA Clean Up Protocol of the RNeasy Mini Kit (Qiagen). Reverse transcription of 1 µg of total RNA to cDNA was performed using the QuantiTect Reverse Transcription Kit (Qiagen) according to the manufacturer’s instructions. cDNA was diluted 1:16 in distilled, deionized water prior to RT-qPCR, which was performed using the SensiMix SYBR Low-ROX Kit (Bioline) and the following primers: RIBOSOMAL PROTEIN S16 housekeeping gene; S16fwd, 5′-TTTACGCCATCCGTCAGAGTAT-3′; S16rev, 5′-TCTGGTAACGAGAACGAGCAC-3′; PAD3fwd, 5′-TACTTGTTGAGATGGCATTGTTGAA-3′; and PAD3rev, 5′-CTTCCTCCTGCTTCGCCAAT-3′. The annealing temperature was 60°C for all primers.
Accession Numbers
The microarray data have been deposited in the ArrayExpress database at EMBL-EBI (https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-6522).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. NO2 fumigation does not cause visible leaf symptoms.
Supplemental Figure S2. GO term enrichment analysis of genes regulated by 10 µL L−1 NO2.
Supplemental Figure S3. Venn diagram showing that the expression of early SA response genes is induced after NO2 fumigation.
Supplemental Figure S4. B. cinerea-induced SA and jasmonate accumulation is not altered by NO2 pretreatment.
Supplemental Figure S5. Overview of volatiles emitted from Arabidopsis following the fumigation experiment.
Supplemental Figure S6. Effects of NO2 fumigation on the volatile emissions.
Supplemental Figure S7. NO2 treatment does not alter camalexin levels.
Supplemental Figure S8. NO2 fumigation system.
Supplemental Table S1. Candidate Trp-derived metabolites involved in the plant response to NO2.
Supplemental Table S2. Arabidopsis mutant lines used in this study.
Supplemental Data Set S1. Microarray analysis of gene expression after fumigation of Arabidopsis for 1 h with 10 µL L−1 NO2.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Michael Reichelt for support with the phytohormone measurements. We acknowledge the donation of the cyp81f2-2 mutant by Henning Frerigmann.
Footnotes
E.Gl. was supported by a Deutsche Forschungsgemeinschaft Heisenberg Fellowship (GL346/5) and the TUM Junior Fellow Fund. E.Ge. was supported by the German Plant Phenotyping Network funded by the German Federal Ministry of Education and Research (DPPN, no. 031A053C).
Articles can be viewed without a subscription.
References
- Ahuja I, Kissen R, Bones AM (2012) Phytoalexins in defense against pathogens. Trends Plant Sci 17: 73–90 [DOI] [PubMed] [Google Scholar]
- Arasimowicz-Jelonek M, Floryszak-Wieczorek J (2011) Understanding the fate of peroxynitrite in plant cells: from physiology to pathophysiology. Phytochemistry 72: 681–688 [DOI] [PubMed] [Google Scholar]
- Bardou P, Mariette J, Escudié F, Djemiel C, Klopp C (2014) jvenn: an interactive Venn diagram viewer. BMC Bioinformatics 15: 293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayles CJ, Ghemawat MS, Aist JR (1990) Inhibition by 2-deoxy-D-glucose of callose formation, papilla deposition, and resistance to powdery mildew in an ml-o barley mutant. Physiol Mol Plant Pathol 36: 63–72 [Google Scholar]
- Bednarek P, Pislewska-Bednarek M, Svatos A, Schneider B, Doubsky J, Mansurova M, Humphry M, Consonni C, Panstruga R, Sanchez-Vallet A, et al. (2009) A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science 323: 101–106 [DOI] [PubMed] [Google Scholar]
- Beevers L, Hageman RH (1969) Nitrate reduction in higher plants. Annu Rev Plant Physiol 20: 495–522 [Google Scholar]
- Berardini TZ, Reiser L, Li D, Mezheritsky Y, Muller R, Strait E, Huala E (2015) The Arabidopsis Information Resource: making and mining the “gold standard” annotated reference plant genome. Genesis 53: 474–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigeard J, Colcombet J, Hirt H (2015) Signaling mechanisms in pattern-triggered immunity (PTI). Mol Plant 8: 521–539 [DOI] [PubMed] [Google Scholar]
- Blanco F, Salinas P, Cecchini NM, Jordana X, Van Hummelen P, Alvarez ME, Holuigue L (2009) Early genomic responses to salicylic acid in Arabidopsis. Plant Mol Biol 70: 79–102 [DOI] [PubMed] [Google Scholar]
- Boller T, Felix G (2009) A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 60: 379–406 [DOI] [PubMed] [Google Scholar]
- Böttcher C, Westphal L, Schmotz C, Prade E, Scheel D, Glawischnig E (2009) The multifunctional enzyme CYP71B15 (PHYTOALEXIN DEFICIENT3) converts cysteine-indole-3-acetonitrile to camalexin in the indole-3-acetonitrile metabolic network of Arabidopsis thaliana. Plant Cell 21: 1830–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudsocq M, Willmann MR, McCormack M, Lee H, Shan L, He P, Bush J, Cheng SH, Sheen J (2010) Differential innate immune signalling via Ca2+ sensor protein kinases. Nature 464: 418–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browse J. (2009) Jasmonate passes muster: a receptor and targets for the defense hormone. Annu Rev Plant Biol 60: 183–205 [DOI] [PubMed] [Google Scholar]
- Bruce TJ, Matthes MC, Chamberlain K, Woodcock CM, Mohib A, Webster B, Smart LE, Birkett M, Pickett J, Napier J (2008) cis-Jasmone induces Arabidopsis genes that affect the chemical ecology of multitrophic interactions with aphids and their parasitoids. Proc Natl Acad Sci USA 105: 4553–4558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caarls L, Pieterse CMJ, Van Wees SCM (2015) How salicylic acid takes transcriptional control over jasmonic acid signaling. Front Plant Sci 6: 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caarls L, Elberse J, Awwanah M, Ludwig NR, de Vries M, Zeilmaker T, Van Wees SCM, Schuurink RC, Van den Ackerveken G (2017) Arabidopsis JASMONATE-INDUCED OXYGENASES down-regulate plant immunity by hydroxylation and inactivation of the hormone jasmonic acid. Proc Natl Acad Sci USA 114: 6388–6393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo M-C, Lozano-Juste J, González-Guzmán M, Rodriguez L, Rodriguez PL, León J (2015) Inactivation of PYR/PYL/RCAR ABA receptors by tyrosine nitration may enable rapid inhibition of ABA signaling by nitric oxide in plants. Sci Signal 8: ra89. [DOI] [PubMed] [Google Scholar]
- Chaerle L, Van Der Straeten D (2000) Imaging techniques and the early detection of plant stress. Trends Plant Sci 5: 495–501 [DOI] [PubMed] [Google Scholar]
- Clay NK, Adio AM, Denoux C, Jander G, Ausubel FM (2009) Glucosinolate metabolites required for an Arabidopsis innate immune response. Science 323: 95–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpas FJ, Barroso JB (2013) Nitro-oxidative stress vs oxidative or nitrosative stress in higher plants. New Phytol 199: 633–635 [DOI] [PubMed] [Google Scholar]
- Couto D, Zipfel C (2016) Regulation of pattern recognition receptor signalling in plants. Nat Rev Immunol 16: 537–552 [DOI] [PubMed] [Google Scholar]
- Delaney TP, Uknes S, Vernooij B, Friedrich L, Weymann K, Negrotto D, Gaffney T, Gut-Rella M, Kessmann H, Ward E, et al. (1994) A central role of salicylic acid in plant disease resistance. Science 266: 1247–1250 [DOI] [PubMed] [Google Scholar]
- Ellinger D, Voigt CA (2014) Callose biosynthesis in Arabidopsis with a focus on pathogen response: what we have learned within the last decade. Ann Bot 114: 1349–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellinger D, Naumann M, Falter C, Zwikowics C, Jamrow T, Manisseri C, Somerville SC, Voigt CA (2013) Elevated early callose deposition results in complete penetration resistance to powdery mildew in Arabidopsis. Plant Physiol 161: 1433–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari S, Plotnikova JM, De Lorenzo G, Ausubel FM (2003) Arabidopsis local resistance to Botrytis cinerea involves salicylic acid and camalexin and requires EDS4 and PAD2, but not SID2, EDS5 or PAD4. Plant J 35: 193–205 [DOI] [PubMed] [Google Scholar]
- Ferrari S, Galletti R, Denoux C, De Lorenzo G, Ausubel FM, Dewdney J (2007) Resistance to Botrytis cinerea induced in Arabidopsis by elicitors is independent of salicylic acid, ethylene, or jasmonate signaling but requires PHYTOALEXIN DEFICIENT3. Plant Physiol 144: 367–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frerigmann H, Glawischnig E, Gigolashvili T (2015) The role of MYB34, MYB51 and MYB122 in the regulation of camalexin biosynthesis in Arabidopsis thaliana. Front Plant Sci 6: 654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frerigmann H, Piślewska-Bednarek M, Sánchez-Vallet A, Molina A, Glawischnig E, Gigolashvili T, Bednarek P (2016) Regulation of pathogen-triggered tryptophan metabolism in Arabidopsis thaliana by MYB transcription factors and indole glucosinolate conversion products. Mol Plant 9: 682–695 [DOI] [PubMed] [Google Scholar]
- García-Andrade J, Ramírez V, Flors V, Vera P (2011) Arabidopsis ocp3 mutant reveals a mechanism linking ABA and JA to pathogen-induced callose deposition. Plant J 67: 783–794 [DOI] [PubMed] [Google Scholar]
- Gaupels F, Kuruthukulangarakoola GT, Durner J (2011) Upstream and downstream signals of nitric oxide in pathogen defence. Curr Opin Plant Biol 14: 707–714 [DOI] [PubMed] [Google Scholar]
- Gautier L, Cope L, Bolstad BM, Irizarry RA (2004) affy: analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20: 307–315 [DOI] [PubMed] [Google Scholar]
- Georgii E, Jin M, Zhao J, Kanawati B, Schmitt-Kopplin P, Albert A, Winkler JB, Schäffner AR (2017) Relationships between drought, heat and air humidity responses revealed by transcriptome-metabolome co-analysis. BMC Plant Biol 17: 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghirardo A, Koch K, Taipale R, Zimmer I, Schnitzler JP, Rinne J (2010) Determination of de novo and pool emissions of terpenes from four common boreal/alpine trees by 13CO2 labelling and PTR-MS analysis. Plant Cell Environ 33: 781–792 [DOI] [PubMed] [Google Scholar]
- Ghirardo A, Gutknecht J, Zimmer I, Brüggemann N, Schnitzler JP (2011) Biogenic volatile organic compound and respiratory CO2 emissions after 13C-labeling: online tracing of C translocation dynamics in poplar plants. PLoS ONE 6: e17393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghirardo A, Heller W, Fladung M, Schnitzler JP, Schroeder H (2012) Function of defensive volatiles in pedunculate oak (Quercus robur) is tricked by the moth Tortrix viridana. Plant Cell Environ 35: 2192–2207 [DOI] [PubMed] [Google Scholar]
- Ghirardo A, Wright LP, Bi Z, Rosenkranz M, Pulido P, Rodríguez-Concepción M, Niinemets Ü, Brüggemann N, Gershenzon J, Schnitzler JP (2014) Metabolic flux analysis of plastidic isoprenoid biosynthesis in poplar leaves emitting and nonemitting isoprene. Plant Physiol 165: 37–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghirardo A, Xie J, Zheng X, Wang Y, Grote R, Block K, Wildt J, Mentel T, Kiendler-Scharr A, Hallquist M, et al. (2016) Urban stress-induced biogenic VOC emissions and SOA-forming potentials in Beijing. Atmos Chem Phys 16: 2901–2920 [Google Scholar]
- Glawischnig E. (2007) Camalexin. Phytochemistry 68: 401–406 [DOI] [PubMed] [Google Scholar]
- Glawischnig E, Hansen BG, Olsen CE, Halkier BA (2004) Camalexin is synthesized from indole-3-acetaldoxime, a key branching point between primary and secondary metabolism in Arabidopsis. Proc Natl Acad Sci USA 101: 8245–8250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J. (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 43: 205–227 [DOI] [PubMed] [Google Scholar]
- Groß F, Durner J, Gaupels F (2013) Nitric oxide, antioxidants and prooxidants in plant defence responses. Front Plant Sci 4: 419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heil M, Land WG (2014) Danger signals: damaged-self recognition across the tree of life. Front Plant Sci 5: 578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitz T, Widemann E, Lugan R, Miesch L, Ullmann P, Désaubry L, Holder E, Grausem B, Kandel S, Miesch M, et al. (2012) Cytochromes P450 CYP94C1 and CYP94B3 catalyze two successive oxidation steps of plant hormone jasmonoyl-isoleucine for catabolic turnover. J Biol Chem 287: 6296–6306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitz T, Smirnova E, Widemann E, Aubert Y, Pinot F, Ménard R (2016) The rise and fall of jasmonate biological activities. In Nakamura Y, Li-Beisson Y, eds, Lipids in Plant and Algae Development. Springer, pp 405–426 [DOI] [PubMed] [Google Scholar]
- Himejima M, Hobson KR, Otsuka T, Wood DL, Kubo I (1992) Antimicrobial terpenes from oleoresin of ponderosa pine tree Pinus ponderosa: a defense mechanism against microbial invasion. J Chem Ecol 18: 1809–1818 [DOI] [PubMed] [Google Scholar]
- Huber DPW, Philippe RN, Madilao LL, Sturrock RN, Bohlmann J (2005) Changes in anatomy and terpene chemistry in roots of Douglas-fir seedlings following treatment with methyl jasmonate. Tree Physiol 25: 1075–1083 [DOI] [PubMed] [Google Scholar]
- Hull AK, Vij R, Celenza JL (2000) Arabidopsis cytochrome P450s that catalyze the first step of tryptophan-dependent indole-3-acetic acid biosynthesis. Proc Natl Acad Sci USA 97: 2379–2384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs AK, Lipka V, Burton RA, Panstruga R, Strizhov N, Schulze-Lefert P, Fincher GB (2003) An Arabidopsis callose synthase, GSL5, is required for wound and papillary callose formation. Plant Cell 15: 2503–2513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joudoi T, Shichiri Y, Kamizono N, Akaike T, Sawa T, Yoshitake J, Yamada N, Iwai S (2013) Nitrated cyclic GMP modulates guard cell signaling in Arabidopsis. Plant Cell 25: 558–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasten D, Mithöfer A, Georgii E, Lang H, Durner J, Gaupels F (2016) Nitrite is the driver, phytohormones are modulators while NO and H2O2 act as promoters of NO2-induced cell death. J Exp Bot 67: 6337–6349 [DOI] [PubMed] [Google Scholar]
- Kasten D, Durner J, Gaupels F (2017) Gas alert: the NO2 pitfall during NO fumigation of plants. Front Plant Sci 8: 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitaoka N, Matsubara T, Sato M, Takahashi K, Wakuta S, Kawaide H, Matsui H, Nabeta K, Matsuura H (2011) Arabidopsis CYP94B3 encodes jasmonyl-L-isoleucine 12-hydroxylase, a key enzyme in the oxidative catabolism of jasmonate. Plant Cell Physiol 52: 1757–1765 [DOI] [PubMed] [Google Scholar]
- Klepper L. (1979) Nitric oxide (NO) and nitrogen dioxide (NO2) emissions from herbicide-treated soybean plants. Atmos Environ 13: 537–542 [Google Scholar]
- Klepper L. (1990) Comparison between NOx evolution mechanisms of wild-type and nr1 mutant soybean leaves. Plant Physiol 93: 26–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein DJ, Rowe HC, Denby KJ (2005) Secondary metabolites influence Arabidopsis/Botrytis interactions: variation in host production and pathogen sensitivity. Plant J 44: 25–36 [DOI] [PubMed] [Google Scholar]
- Köhle H, Jeblick W, Poten F, Blaschek W, Kauss H (1985) Chitosan-elicited callose synthesis in soybean cells as a Ca-dependent process. Plant Physiol 77: 544–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolbert Z, Feigl G, Bordé Á, Molnár Á, Erdei L (2017) Protein tyrosine nitration in plants: present knowledge, computational prediction and future perspectives. Plant Physiol Biochem 113: 56–63 [DOI] [PubMed] [Google Scholar]
- Kolesnikov N, Hastings E, Keays M, Melnichuk O, Tang YA, Williams E, Dylag M, Kurbatova N, Brandizi M, Burdett T, et al. (2015) ArrayExpress update: simplifying data submissions. Nucleic Acids Res 43: D1113–D1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo AJK, Cooke TF, Howe GA (2011) Cytochrome P450 CYP94B3 mediates catabolism and inactivation of the plant hormone jasmonoyl-L-isoleucine. Proc Natl Acad Sci USA 108: 9298–9303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis LA, Polanski K, de Torres-Zabala M, Jayaraman S, Bowden L, Moore J, Penfold CA, Jenkins DJ, Hill C, Baxter L, et al. (2015) Transcriptional dynamics driving MAMP-triggered immunity and pathogen effector-mediated immunosuppression in Arabidopsis leaves following infection with Pseudomonas syringae pv tomato DC3000. Plant Cell 27: 3038–3064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler H, Miehe J (1997) Fluorescence imaging as a diagnostic tool for plant stress. Trends Plant Sci 2: 6–10 [Google Scholar]
- Liu X, Hou F, Li G, Sang N (2015) Effects of nitrogen dioxide and its acid mist on reactive oxygen species production and antioxidant enzyme activity in Arabidopsis plants. J Environ Sci (China) 34: 93–99 [DOI] [PubMed] [Google Scholar]
- Luna E, Pastor V, Robert J, Flors V, Mauch-Mani B, Ton J (2011) Callose deposition: a multifaceted plant defense response. Mol Plant Microbe Interact 24: 183–193 [DOI] [PubMed] [Google Scholar]
- Maassen A, Hennig J (2011) Effect of Medicago sativa Mhb1gene expression on defense response of Arabidopsis thaliana plants. Acta Biochim Pol 58: 427–432 [PubMed] [Google Scholar]
- Martin DM, Fäldt J, Bohlmann J (2004) Functional characterization of nine Norway spruce TPS genes and evolution of gymnosperm terpene synthases of the TPS-d subfamily. Plant Physiol 135: 1908–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata-Pérez C, Begara-Morales JC, Chaki M, Sánchez-Calvo B, Valderrama R, Padilla MN, Corpas FJ, Barroso JB (2016a) Protein tyrosine nitration during development and abiotic stress response in plants. Front Plant Sci 7: 1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata-Pérez C, Sánchez-Calvo B, Padilla MN, Begara-Morales JC, Luque F, Melguizo M, Jiménez-Ruiz J, Fierro-Risco J, Peñas-Sanjuán A, Valderrama R, et al. (2016b) Nitro-fatty acids in plant signaling: nitro-linolenic acid induces the molecular chaperone network in Arabidopsis. Plant Physiol 170: 686–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengiste T. (2012) Plant immunity to necrotrophs. Annu Rev Phytopathol 50: 267–294 [DOI] [PubMed] [Google Scholar]
- Mi H, Huang X, Muruganujan A, Tang H, Mills C, Kang D, Thomas PD (2017) PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res 45: D183–D189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miersch O, Neumerkel J, Dippe M, Stenzel I, Wasternack C (2008) Hydroxylated jasmonates are commonly occurring metabolites of jasmonic acid and contribute to a partial switch-off in jasmonate signaling. New Phytol 177: 114–127 [DOI] [PubMed] [Google Scholar]
- Mishina TE, Zeier J (2007) Pathogen-associated molecular pattern recognition rather than development of tissue necrosis contributes to bacterial induction of systemic acquired resistance in Arabidopsis. Plant J 50: 500–513 [DOI] [PubMed] [Google Scholar]
- Müller TM, Böttcher C, Morbitzer R, Götz CC, Lehmann J, Lahaye T, Glawischnig E (2015) TRANSCRIPTION ACTIVATOR-LIKE EFFECTOR NUCLEASE-mediated generation and metabolic analysis of camalexin-deficient cyp71a12 cyp71a13 double knockout lines. Plant Physiol 168: 849–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mur LAJ, Mandon J, Persijn S, Cristescu SM, Moshkov IE, Novikova GV, Hall MA, Harren FJM, Hebelstrup KH, Gupta KJ (2013) Nitric oxide in plants: an assessment of the current state of knowledge. AoB Plants 5: pls052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nafisi M, Goregaoker S, Botanga CJ, Glawischnig E, Olsen CE, Halkier BA, Glazebrook J (2007) Arabidopsis cytochrome P450 monooxygenase 71A13 catalyzes the conversion of indole-3-acetaldoxime in camalexin synthesis. Plant Cell 19: 2039–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath C, Métraux JP (1999) Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell 11: 1393–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niinemets U. (2010) Mild versus severe stress and BVOCs: thresholds, priming and consequences. Trends Plant Sci 15: 145–153 [DOI] [PubMed] [Google Scholar]
- Nishimura MT, Stein M, Hou BH, Vogel JP, Edwards H, Somerville SC (2003) Loss of a callose synthase results in salicylic acid–dependent disease resistance. Science 301: 969–972 [DOI] [PubMed] [Google Scholar]
- Park JH, Halitschke R, Kim HB, Baldwin IT, Feldmann KA, Feyereisen R (2002) A knock-out mutation in allene oxide synthase results in male sterility and defective wound signal transduction in Arabidopsis due to a block in jasmonic acid biosynthesis. Plant J 31: 1–12 [DOI] [PubMed] [Google Scholar]
- Patkar RN, Benke PI, Qu Z, Chen YY, Yang F, Swarup S, Naqvi NI (2015) A fungal monooxygenase-derived jasmonate attenuates host innate immunity. Nat Chem Biol 11: 733–740 [DOI] [PubMed] [Google Scholar]
- Pieterse CMJ, Van der Does D, Zamioudis C, Leon-Reyes A, Van Wees SCM (2012) Hormonal modulation of plant immunity. Annu Rev Cell Dev Biol 28: 489–521 [DOI] [PubMed] [Google Scholar]
- Pryor WA, Houk KN, Foote CS, Fukuto JM, Ignarro LJ, Squadrito GL, Davies KJ (2006) Free radical biology and medicine: it’s a gas, man! Am J Physiol Regul Integr Comp Physiol 291: R491–R511 [DOI] [PubMed] [Google Scholar]
- Radi R. (2013) Protein tyrosine nitration: biochemical mechanisms and structural basis of functional effects. Acc Chem Res 46: 550–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramonell K, Berrocal-Lobo M, Koh S, Wan J, Edwards H, Stacey G, Somerville S (2005) Loss-of-function mutations in chitin responsive genes show increased susceptibility to the powdery mildew pathogen Erysiphe cichoracearum. Plant Physiol 138: 1027–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauhut T, Luberacki B, Seitz HU, Glawischnig E (2009) Inducible expression of a Nep1-like protein serves as a model trigger system of camalexin biosynthesis. Phytochemistry 70: 185–189 [DOI] [PubMed] [Google Scholar]
- R Core Team (2014) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna [Google Scholar]
- Riedlmeier M, Ghirardo A, Wenig M, Knappe C, Koch K, Georgii E, Dey S, Parker JE, Schnitzler JP, Vlot C (2017) Monoterpenes support systemic acquired resistance within and between plants. Plant Cell 29: 1440–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK (2015) limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43: e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert-Seilaniantz A, Grant M, Jones JDG (2011) Hormone crosstalk in plant disease and defense: more than just jasmonate-salicylate antagonism. Annu Rev Phytopathol 49: 317–343 [DOI] [PubMed] [Google Scholar]
- Roden LC, Ingle RA (2009) Lights, rhythms, infection: the role of light and the circadian clock in determining the outcome of plant-pathogen interactions. Plant Cell 21: 2546–2552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers EE, Glazebrook J, Ausubel FM (1996) Mode of action of the Arabidopsis thaliana phytoalexin camalexin and its role in Arabidopsis-pathogen interactions. Mol Plant Microbe Interact 9: 748–757 [DOI] [PubMed] [Google Scholar]
- Sakamoto A, Sakurao SH, Fukunaga K, Matsubara T, Ueda-Hashimoto M, Tsukamoto S, Takahashi M, Morikawa H (2004) Three distinct Arabidopsis hemoglobins exhibit peroxidase-like activity and differentially mediate nitrite-dependent protein nitration. FEBS Lett 572: 27–32 [DOI] [PubMed] [Google Scholar]
- Sakihama Y, Tamaki R, Shimoji H, Ichiba T, Fukushi Y, Tahara S, Yamasaki H (2003) Enzymatic nitration of phytophenolics: evidence for peroxynitrite-independent nitration of plant secondary metabolites. FEBS Lett 553: 377–380 [DOI] [PubMed] [Google Scholar]
- Schopfer FJ, Cipollina C, Freeman BA (2011) Formation and signaling actions of electrophilic fatty acids. Chem Rev 111: 5997–6021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuhegger R, Nafisi M, Mansourova M, Petersen BL, Olsen CE, Svatos A, Halkier BA, Glawischnig E (2006) CYP71B15 (PAD3) catalyzes the final step in camalexin biosynthesis. Plant Physiol 141: 1248–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata H, Kono Y, Yamshita S, Sawa Y, Ochiai H, Tanaka K (1995) Degradation of chlorophyll by nitrogen dioxide generated from the peroxidase reaction. Biochim Biophys Acta 1230: 45–50 [Google Scholar]
- Shimazaki KI, Yu SW, Sakaki T, Tanaka K (1992) Differences between spinach and kidney bean plants in terms of sensitivity to fumigation with NO2. Plant Cell Physiol 33: 267 [Google Scholar]
- Smirnova E, Marquis V, Poirier L, Aubert Y, Zumsteg J, Ménard R, Miesch L, Heitz T (2017) Jasmonic Acid Oxidase 2 hydroxylates jasmonic acid and represses basal defense and resistance responses against Botrytis cinerea infection. Mol Plant 10: 1159–1173 [DOI] [PubMed] [Google Scholar]
- Smith CA O’Maille G, Want EJ, Qin C, Trauger SA, Brandon TR, Custodio DE, Abagyan R, Siuzdak G (2005) METLIN: a metabolite mass spectral database. Therapeutic Drug Monitoring 27: 747–751 [DOI] [PubMed] [Google Scholar]
- Smyth GK. (2005) Limma: linear models for microarray data. In Gentleman R, Carey VJ, Huber W, Irizarry RA, Dudoit S, eds, Bioinformatics and Computational Biology Solutions Using R and Bioconductor: Statistics for Biology and Health. Springer, New York, pp 397–420 [Google Scholar]
- Sparks JP. (2009) Ecological ramifications of the direct foliar uptake of nitrogen. Oecologia 159: 1–13 [DOI] [PubMed] [Google Scholar]
- Srivastava HS, Ormrod DP, Hale BA (1994) Responses of greening bean seedling leaves to nitrogen dioxide and nutrient nitrate supply. Environ Pollut 86: 37–42 [DOI] [PubMed] [Google Scholar]
- Stintzi A, Browse J (2000) The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc Natl Acad Sci USA 97: 10625–10630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stintzi A, Weber H, Reymond P, Browse J, Farmer EE (2001) Plant defense in the absence of jasmonic acid: the role of cyclopentenones. Proc Natl Acad Sci USA 98: 12837–12842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhre K, Schmitt-Kopplin P (2008) MassTRIX: mass translator into pathways. Nucleic Acids Res 36: W481–W484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek F, Bošnjak M, Škunca N, Šmuc T (2011) REVIGO summarizes and visualizes long lists of Gene Ontology terms. PLoS ONE 6: e21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M, Furuhashi T, Ishikawa N, Horiguchi G, Sakamoto A, Tsukaya H, Morikawa H (2014) Nitrogen dioxide regulates organ growth by controlling cell proliferation and enlargement in Arabidopsis. New Phytol 201: 1304–1315 [DOI] [PubMed] [Google Scholar]
- Tholl D, Lee S (2011) Terpene specialized metabolism in Arabidopsis thaliana. The Arabidopsis Book 9: e0143, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas DD, Ridnour LA, Isenberg JS, Flores-Santana W, Switzer CH, Donzelli S, Hussain P, Vecoli C, Paolocci N, Ambs S, et al. (2008) The chemical biology of nitric oxide: implications in cellular signaling. Free Radic Biol Med 45: 18–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma BPHJ, Eggermont K, Penninckx IAMA, Mauch-Mani B, Vogelsang R, Cammue BPA, Broekaert WF (1998) Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc Natl Acad Sci USA 95: 15107–15111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadassery J, Reichelt M, Hause B, Gershenzon J, Boland W, Mithöfer A (2012) CML42-mediated calcium signaling coordinates responses to Spodoptera herbivory and abiotic stresses in Arabidopsis. Plant Physiol 159: 1159–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlot AC, Dempsey DA, Klessig DF (2009) Salicylic acid, a multifaceted hormone to combat disease. Annu Rev Phytopathol 47: 177–206 [DOI] [PubMed] [Google Scholar]
- von Saint Paul V, Zhang W, Kanawati B, Geist B, Faus-Kessler T, Schmitt-Kopplin P, Schäffner AR (2011) The Arabidopsis glucosyltransferase UGT76B1 conjugates isoleucic acid and modulates plant defense and senescence. Plant Cell 23: 4124–4145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack C, Hause B (2013) Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann Bot 111: 1021–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weikl F, Ghirardo A, Schnitzler JP, Pritsch K (2016) Sesquiterpene emissions from Alternaria alternata and Fusarium oxysporum: effects of age, nutrient availability, and co-cultivation. Sci Rep 6: 22152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellburn AR. (1990) Tansley Review No. 24. Why are atmospheric oxides of nitrogen usually phytotoxic and not alternative fertilizers? New Phytol 115: 395–429 [DOI] [PubMed] [Google Scholar]
- Widemann E, Miesch L, Lugan R, Holder E, Heinrich C, Aubert Y, Miesch M, Pinot F, Heitz T (2013) The amidohydrolases IAR3 and ILL6 contribute to jasmonoyl-isoleucine hormone turnover and generate 12-hydroxyjasmonic acid upon wounding in Arabidopsis leaves. J Biol Chem 288: 31701–31714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildermuth MC, Dewdney J, Wu G, Ausubel FM (2001) Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414: 562–565 [DOI] [PubMed] [Google Scholar]
- Xu Q, Zhou B, Ma C, Xu X, Xu J, Jiang Y, Liu C, Li G, Herbert SJ, Hao L (2010) Salicylic acid-altering Arabidopsis mutants response to NO2 exposure. Bull Environ Contam Toxicol 84: 106–111 [DOI] [PubMed] [Google Scholar]
- Yoneyama T, Sasakawa H (1979) Transformation of atmospheric NO2 absorbed in spinach leaves. Plant Cell Physiol 20: 263–266 [Google Scholar]
- Zeevaart AJ. (1976) Some effects of fumigating plants for short periods with NO2. Environ Pollut 11: 97–108 [Google Scholar]
- Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JDG, Felix G, Boller T (2004) Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428: 764–767 [DOI] [PubMed] [Google Scholar]