Long days perceived by photosensory receptors enhance jasmonic acid-dependent resistance in Arabidopsis.
Abstract
Agricultural crops are exposed to a range of daylengths, which act as important environmental cues for the control of developmental processes such as flowering. To explore the additional effects of daylength on plant function, we investigated the transcriptome of Arabidopsis (Arabidopsis thaliana) plants grown under short days (SD) and transferred to long days (LD). Compared with that under SD, the LD transcriptome was enriched in genes involved in jasmonic acid-dependent systemic resistance. Many of these genes exhibited impaired expression induction under LD in the phytochrome A (phyA), cryptochrome 1 (cry1), and cry2 triple photoreceptor mutant. Compared with that under SD, LD enhanced plant resistance to the necrotrophic fungus Botrytis cinerea. This response was reduced in the phyA cry1 cry2 triple mutant, in the constitutive photomorphogenic1 (cop1) mutant, in the myc2 mutant, and in mutants impaired in DELLA function. Plants grown under SD had an increased nuclear abundance of COP1 and decreased DELLA abundance, the latter of which was dependent on COP1. We conclude that growth under LD enhances plant defense by reducing COP1 activity and enhancing DELLA abundance and MYC2 expression.
A given crop species can typically be exposed to a range of different photoperiods, the nature of which depend on sowing date, duration of the cycle, and latitude. Daylength profoundly affects the timing of key developmental transitions, including flowering in many species, tuberization in potato (Solanum tuberosum), and bud set and growth cessation in trees (Jackson, 2009). The ability to respond specifically to current daylength helps to reduce the risk of plants being exposed to severe stressful conditions (Casal et al., 2004). Response to daylength also can enhance the tolerance to seasonal abiotic stress. Short days (SD) anticipate the cold temperatures of winter and increase freezing tolerance (Alonso-Blanco et al., 2005; Lee and Thomashow, 2012). Long days (LD) can induce antioxidative capacities in plants (Becker et al., 2006) and mimic plant acclimation to high light intensities (Lepistö and Rintamäki, 2012) that is typical of summer.
In Arabidopsis (Arabidopsis thaliana), growth under LD maintains the activity of phytochrome A (phyA), cryptochrome 1 (cry1), and cry2 photoreceptors, which promote flowering (Andrés and Coupland, 2012). These photoreceptors stabilize CONSTANS (CO; Valverde et al., 2004) by reducing the activity of the CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1)-SUPRESSOR OF PHYA-105 1 (SPA1)-SPA3-SPA4 complex (Liu et al., 2008). Growth under LD also enhances the expression of CO (Sawa et al., 2007) and the stability of CO protein (Song et al., 2012) via the action of the FLAVIN-BINDING, KELCH REPEAT, F-BOX photoreceptor (Lee et al., 2017). In turn, CO enhances the expression of FLOWERING LOCUS T (FT), which promotes flowering (Andrés and Coupland, 2012). The phyB photoreceptor, PHYTOCHROME INTERACTING FACTOR4 (PIF4), and PIF7 play important roles in repressing the C-repeat-binding factor pathway and freezing tolerance under LD (Lee and Thomashow, 2012). These examples illustrate that different photoreceptors and downstream pathways mediate diverse outputs of photoperiodic signals.
The aim of this work was to explore the occurrence of additional responses to photoperiod mediated by phyA, cry1, and cry2 and to elucidate their key signaling components. To identify and prioritize these responses, we analyzed the transcriptome of plants grown under either SD or LD and tested biological responses guided by overrepresented Gene Ontology (GO) terms. Our results show that growth under LD compared with growth under SD enhances the expression of defense-related genes and plant resistance to the necrotrophic pathogen Botrytis cinerea. Growth under LD does not increase jasmonic acid (JA) levels; however, plants grown in LD had enhanced JA-induced defense by increasing the expression of MYC2 and reducing COP1 nuclear activity, which, in turn, allowed for the increased stability of DELLA proteins (Lorenzo et al., 2004; Wild et al., 2012; Chico et al., 2014).
RESULTS
Transcriptome Responses to LD
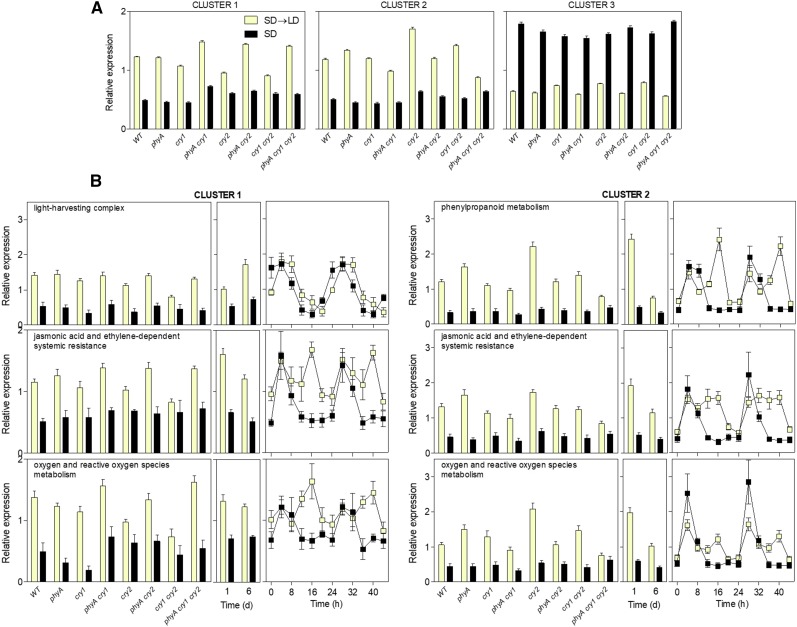
Wild-type plants of Arabidopsis Landsberg erecta and of all the possible combinations among the photoreceptor mutants phyA, cry1, and cry2 (Mazzella et al., 2001) were grown under SD (8 h of white light) for 3 weeks. On day 21, some plants remained under white light beyond the time when the night started in previous days and were harvested when the photoperiod reached 16 h (i.e. at the end of the first LD [SD→LD]). A control group remained under the SD regime and was harvested simultaneously with SD→LD-treated plants but under dim green light (protocol in Supplemental Fig. S1A). Transcriptome analysis revealed that 749 genes showed significant responses to changes in photoperiod, which were grouped into three major clusters (Fig. 1A; Supplemental Table S1).
Figure 1.
Robust responses of the transcriptome to initial LD exposure. A, Three major clusters grouped 749 genes showing statistically significant responses to photoperiod. Plants of Arabidopsis accession Landsberg erecta were grown under SD for 3 weeks, then transferred to LD, and harvested after the end of the first LD photoperiod (experiment 1; Supplemental Fig. S1A). The expression of each gene was normalized to the average for that gene across the genotypes and treatments, and then the cluster average and se were calculated for each genotype and condition. B, For each overrepresented GO term and cluster, average normalized expression and se for each genotype and condition are shown in boxes corresponding to (left to right) experiment 1, experiment 2 (Supplemental Fig. S1A; an independent experiment with accession Columbia following the same protocol as experiment 1 but followed up for 6 d), and publicly available data (Supplemental Fig. S1B; a time course corresponding to plants of accession Landsberg erecta grown continuously under either SD or LD [Michael et al., 2008]). WT, Wild type.
Cluster 1 (163 genes) showed higher expression under SD→LD than under SD. The response to LD was reduced in cry2, cry1, and cry1 cry2, but in these backgrounds, the phyA mutation restored (or overcompensated) the LD response. Exactly the same pattern had been observed for flowering in these mutants under the same light conditions (Mazzella et al., 2001). The enhanced response to LD in the phyA background might reflect the activity of phyB, which can be reduced by phyA (Krzymuski et al., 2014).
Cluster 2 (265 genes) showed higher expression under SD→LD than under SD. The response to LD was reduced significantly in the phyA cry1 cry2 triple mutant but not in the single mutants (for some genes, the phyA and cry2 mutants actually showed an enhanced response), indicating redundancy among phyA, cry1, and cry2.
Cluster 3 (321 genes) showed reduced expression under SD→LD compared with that under SD, but in this cluster, all the mutants showed largely wild-type responses.
To validate the list of genes identified in the wild type as responsive to LD, we conducted a fully independent experiment using the same light protocol in a different growth chamber and plants of the accession Columbia (protocol in Supplemental Fig. S1A). The strong correlation observed between the SD→LD/SD expression ratios of both experiments demonstrates the robustness of the gene expression responses to LD across two different growth conditions and accessions (Supplemental Fig. S2A; Supplemental Table S2).
To test whether these genes also respond in a coordinated manner under different scenarios, we analyzed their expression across samples involving multiple developmental stages and conditions (Obayashi et al., 2011). We observed that the expression of transcription factors present in clusters 1 and 2 tended to positively correlate with the expression of other genes present in these clusters, whereas there was a negative correlation with the expression of genes present in cluster 3 (Supplemental Fig. S3). Conversely, the expression of the transcription factor genes present in cluster 3 positively correlated with the expression of other genes present in cluster 3, and there was a negative correlation of these genes with those present in clusters 1 and 2 (Supplemental Fig. S3). This pattern indicates that changes in photoperiod affect the expression of a set of genes that are part of a robust network.
Daily Gene Expression Responses to LD
To investigate whether the gene expression responses observed initially under LD are largely a transient reaction to the change or represent a daily difference between LD and SD, we compared the SD→LD/SD gene expression ratio of Columbia plants transferred from SD to LD for 1 or 6 d (Supplemental Fig. S1A; Supplemental Table S2). A highly significant correlation indicated that the genes that respond to the first day of exposure to LD tend to respond daily to LD compared with that under SD (Supplemental Fig. S2B).
To challenge the above conclusion, we compared the SD→LD/SD gene expression ratio of our Landsberg erecta plants exposed to a single LD with the LD/SD gene expression ratio calculated for a publicly available time-course data set that was generated using samples from Landsberg erecta plants grown for 7 d under either LD or SD and harvested 16 h after the beginning of the photoperiod (Michael et al., 2008; Supplemental Fig. S1B). The highly significant correlation confirmed and extended the validity of the gene list, further supporting the idea that the genes that respond to the first day of exposure to LD tend to respond daily to LD compared with SD after prolonged exposures to the different photoperiods (Supplemental Fig. S2C).
Specificity of the Gene Expression Responses to LD
Although statistically significant, the correlation observed between the response of the 749-gene set to the first LD and to the first light exposure of fully dark-grown seedlings (Peschke and Kretsch, 2011) was modest (Supplemental Fig. S4A). The list of genes whose expression was at least doubled in both cases was enriched in light-harvesting complexes (P < 10−7) and phenylpropanoid metabolism (P < 10−9). The 749-gene set failed to show correlation between their response to LD and to the transfer of low light-grown plants to high light (Rossel et al., 2002; Kleine et al., 2007; Supplemental Fig. S4B). Therefore, the gene expression response to LD is specific, with restricted similarity to the response to light during deetiolation or during high-light stress.
GO Terms Overrepresented among the Genes Responding to LD
The GO terms enriched (Vandepoele et al., 2009) among the genes that increased their expression in response to LD included light-harvesting complexes (mainly cluster 1), phenylpropanoid metabolism (mainly cluster 2), JA- and ethylene-dependent systemic resistance (clusters 1 and 2), and oxygen and reactive oxygen species metabolism (clusters 1 and 2; Supplemental Table S3). The average expression patterns of these genes in the Landsberg erecta and Columbia SD→LD transition experiments and in the continuous SD or LD time-course experiment demonstrate that their response to LD is robust (Fig. 1B). Furthermore, in all cases, the enhanced expression occurred during the portion of the day when the plants were exposed to light (under LD) versus darkness (i.e. 8–16 h; Fig. 1B).
Analysis of Plant Physiological Outputs and Resistance to B. cinerea under LD
We investigated whether the observed changes in gene expression under LD correlated with rapid changes in physiology (protocol in Supplemental Fig. S1A). No significant differences in leaf chlorophyll or anthocyanin levels were observed 3 d after the SD→LD transfer compared with that in the SD controls (Supplemental Fig. S5). In accordance with other reports (Bermúdez et al., 2010), only a weak increment in oxidative stress was observed after the SD→LD transfer, as indicated by the small differences in the levels of malondialdehyde and the lack of response of catalase activity (Supplemental Fig. S6), which are biological markers of oxidative stress.
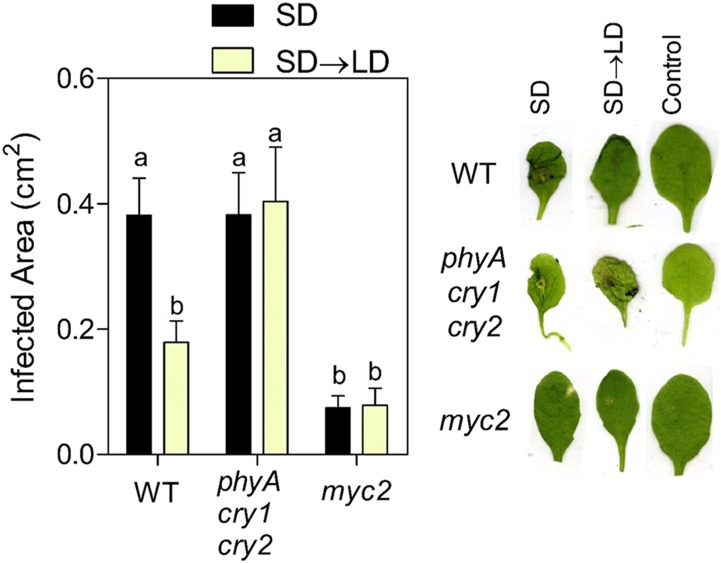
Using reverse transcription quantitative PCR based on independent samples, we confirmed the expression response to SD→LD compared with that under SD of 12 genes present in clusters 1 and 2 and corresponding to the GO term JA- and ethylene-dependent systemic resistance (Supplemental Table S4). Guided by these results, we conducted experiments to test the effects of growth under LD on plant resistance to the necrotrophic fungus B. cinerea. LD significantly reduced the area infected by B. cinerea compared with that in SD-grown plants (Fig. 2). Compared with continuous darkness, a 12-h photoperiod and continuous light also reduce the lesion areas caused by B. cinerea (Canessa et al., 2013). The pathogen-resistance response to LD was not observed in the phyA cry1 cry2 photoreceptor mutant, indicating that extended light acted more as a signal perceived by photoreceptors than as a source of energy via photosynthesis or through alterations of oxidative stress metabolism (Rossi et al., 2017). The phyA cry1 cry2 mutant showed no difference in B. cinerea resistance compared with that in the wild type under SD (Fig. 2), which is consistent with previous reports showing no effects of either lowering blue light or using a cry1 mutant on B. cinerea resistance under SD (Cerrudo et al., 2012). As a negative control, we used the myc2 mutant that is known to have enhanced resistance to B. cinerea (Lorenzo et al., 2004). It must be noted that MYC2 has a dual role as a positive regulator of JA-dependent responses and a negative regulator of ethylene signaling, which, in turn, regulates resistance to necrotrophic fungi synergistically with JA (Song et al., 2014). Due to functional redundancy with MYC3 and MYC4, the phenotype of the myc2 single mutant was dominated by the released repression of the ethylene pathway.
Figure 2.
LD enhances resistance to B. cinerea. Plants of Arabidopsis accession Columbia were grown under SD for 3 weeks and inoculated at 7 h of day 21. One group was transferred to LD while the other remained under SD, and leaves were harvested 48 h after inoculation. Data are means and se of at least 11 plants. Different letters indicate significant differences (P < 0.05) among means determined using Bonferroni posthoc tests. Leaves were photographed individually, and a composite image was produced with representative cases. WT, Wild type.
JA Signaling and Absolute Levels under LD
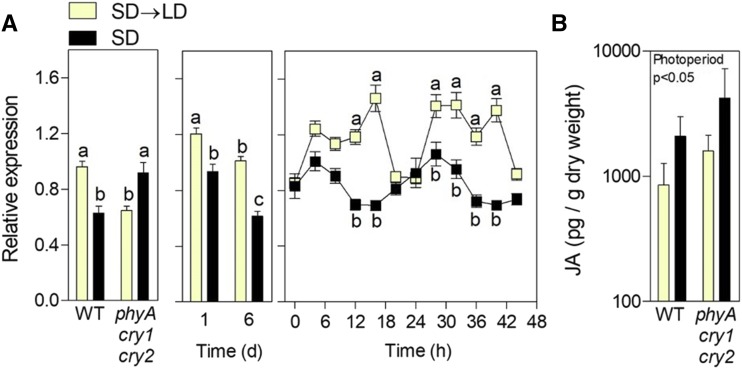
Since 12 out of 13 genes within the GO term JA- and ethylene-dependent systemic resistance also corresponded to response to JA stimulus (Supplemental Table S3), we focused on JA signaling. We analyzed the expression of a set of 100 genes that are known to respond positively to JA (Goda et al., 2008) as a proxy for JA signaling intensity. The index indicated that LD enhanced JA signaling (Fig. 3A). This response to LD could, in principle, be the result of enhanced levels of JA; however, measurements of hormone levels did not support this hypothesis (Fig. 3B). The transcription factor gene MYC2, which is involved in JA signaling (Lorenzo et al., 2004; Chico et al., 2014), showed enhanced expression (cluster 2), and the CACGTG motif, which is the main binding site of MYC2 (Yadav et al., 2005; Dombrecht et al., 2007; Fernández-Calvo et al., 2011), was overrepresented (O’Connor et al., 2005) mainly in cluster 2 (P < 10−10) but also in the three clusters analyzed as a single group (P < 10−10).
Figure 3.
LD enhances JA signaling but not JA levels. A, Expression of a set of 100 genes whose expression is promoted by JA (Goda et al., 2008) was used as a proxy for JA signaling. Left, Experiment 1; middle, experiment 2; right, published data (Michael et al., 2008). B, JA levels in plants exposed to SD→LD. Data are averages ± se. Different letters indicate significant differences (P < 0.05) among means determined using Bonferroni posthoc tests, and the significant effect of photoperiod in factorial ANOVA is shown in B. WT, Wild type.
Correlation between the Effects of COP1 and MYC Transcription Factors on Gene Expression
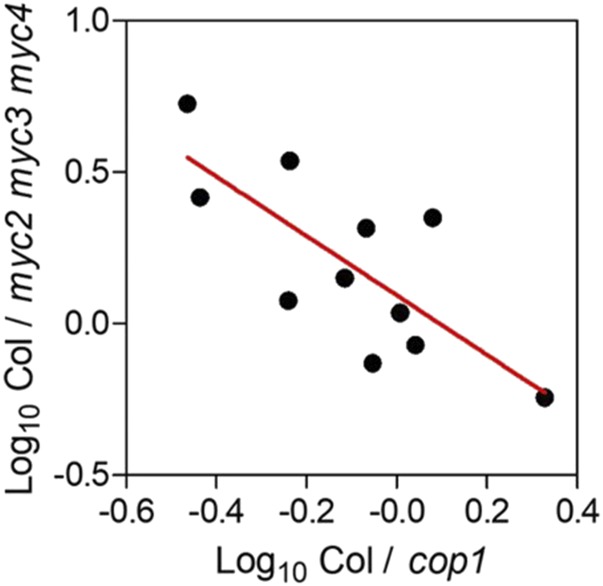
Considering that the LD-specific effect on the genes related to plant defense requires cry1, cry2, and in some cases phyA (Fig. 1A) and that COP1 is a target of these photoreceptors (Lau and Deng, 2012), we investigated the expression of 12 genes present in clusters 1 and 2 that also corresponded to the GO term JA- and ethylene-dependent systemic resistance in cop1 mutant plants (Supplemental Table S4). Compared with that in the wild type, the impact of the cop1 mutations on the expression of 11 of these genes under SD showed a significant inverse correlation with the impact of the myc2 myc3 myc4 mutations (Fernández-Calvo et al., 2011; Fig. 4). The exception was CORI3, which responded more significantly to the cop1 mutations (Supplemental Table S4) than could be predicted by the my2 myc3 myc4 mutant phenotype. These observations suggest that the effect of photoperiod may be mediated by COP1 regulation of MYC2, MYC3, and/or MYC4 activity. Since only the MYC2 gene responded to photoperiod (Supplemental Table S1, cluster 2) and this response was unaffected by the cop1 mutations (Supplemental Table S4), such COP1-mediated regulation of MYC2, MYC3, and/or MYC4 activity likely occurs at the posttranscriptional level.
Figure 4.
Negative correlation between the impact of the myc2 myc3 myc4 (Fernández-Calvo et al., 2011) and cop1 mutations (data in Supplemental Table S4) compared with the wild type. Regression, P < 0.01.
Nuclear Abundance of COP1 under LD and Its Effect on B. cinerea Resistance
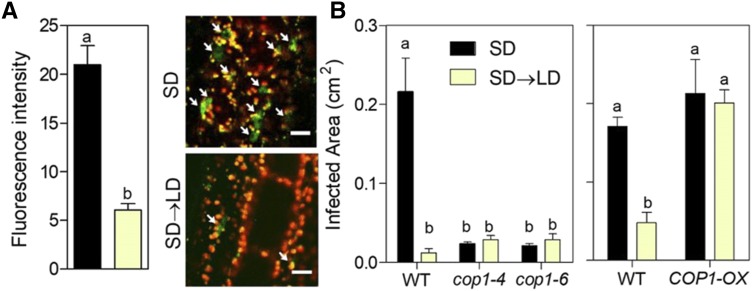
Based on the above observations, we investigated whether LD repressed COP1 activity compared with that under SD. One of the regulatory features of COP1 activity is its nuclear abundance (Lau and Deng, 2012), which is reduced rapidly by dark-to-light transitions (Pacín et al., 2014). Prolonged light exposure under SD→LD reduced the nuclear abundance of COP1 compared with that under SD (Fig. 5A). The expression of COP1 was unaffected by daylength (SD, 792 ± 116; SD→LD, 822 ± 144). Of note, the cop1 mutant showed reduced damage by B. cinerea under SD and failed to respond to LD (Fig. 5B). Furthermore, the COP1 overexpressor showed increased damage by B. cinerea under LD and also failed to respond to LD compared with the response of its Nossen wild type (Fig. 5B).
Figure 5.
COP1 increases the lesions inflicted by B. cinerea under SD, whereas LD reduces COP1 nuclear abundance. A, Nuclear abundance of YFP-COP1 at the end of the first photoperiod under LD and in SD controls. Data are means ± se of eight to nine plant replicates and representative images (arrows point to nuclei with detectable YFP-COP1). Bars = 20 μm. B, Resistance to B. cinerea in cop1 mutants and the COP1 overexpressor (COP1-OX) under SD and LD represented by relative lesion size. Data are means ± se of 13 plant replicates. Different letters indicate significant differences (P < 0.05) determined using Student’s t test (A) or Bonferroni posthoc tests (B). WT, Wild type.
COP1-Dependent DELLA Accumulation under LD
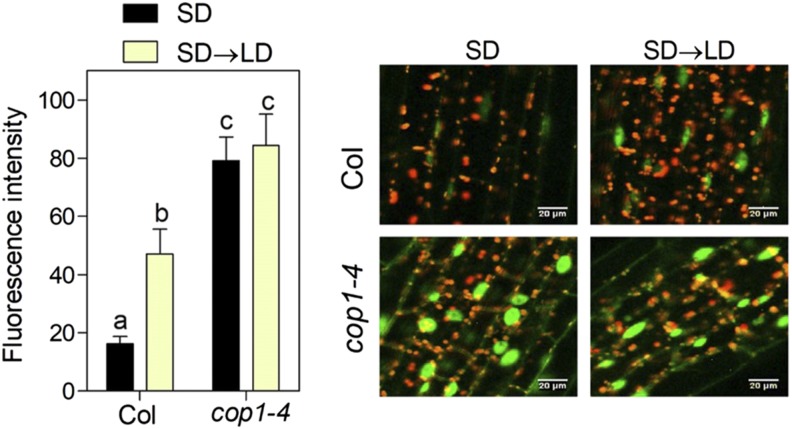
The activity of MYC transcription factors is enhanced by DELLA proteins, which bind JA ZIM-DOMAIN (JAZ) proteins that are negative regulators of MYC2 (Wild et al., 2012). Therefore, we investigated whether COP1 affects the abundance of the DELLA protein REPRESSOR OF ga1-3 (RGA). Confocal microscopy revealed that fluorescence resulting from the pRGA:GFP-RGA transgene increased under SD→LD compared with that under SD in the wild-type background in a COP1-dependent manner (Fig. 6). Moreover, the expression of RGA was unaffected by daylength (SD, 1,231 ± 243; SD→LD, 965 ± 128).
Figure 6.
LD increases RGA abundance in a COP1-dependent manner. Data are means ± se of six plant replicates, and representative images are shown. Different letters indicate significant differences (P < 0.05) among means determined using Bonferroni posthoc tests. Bars = 20 μm.
Function of DELLA Proteins in the Response to Photoperiod
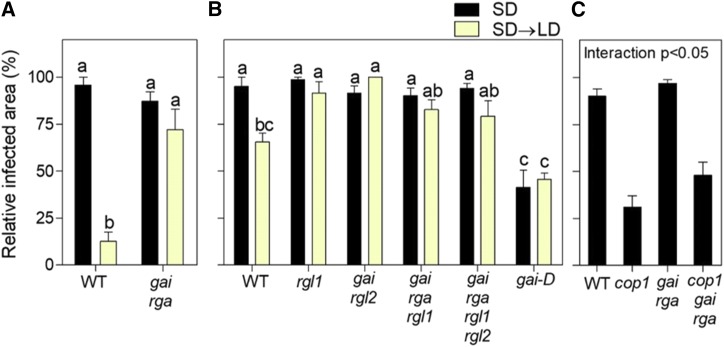
Considering that LD reduces the susceptibility to B. cinerea (Fig. 2) and increases the abundance of RGA (Fig. 6) and that DELLAs are positive regulators of defense against B. cinerea (Wild et al., 2012), we investigated whether the effects of photoperiod on fungal resistance depended on DELLAs. We considered the infected area as a proportion of the total leaf area to compare genotypes of different leaf size. Compared with that in the Columbia wild type, the gai rga double mutant (lacking two DELLAs) of the same background showed increased infection under SD→LD and no response to photoperiod (Fig. 7A). Similarly, compared with that in the Landsberg erecta wild type, the rgl1, gai rgl2, gai rga rgl1, and gai rga rgl1 rgl2 (lacking one to four DELLAs) mutants of the same background showed increased infection under SD→LD, whereas a gai gain-of-function allele showed reduced infection. None of these genotypes responded to photoperiod (Fig. 7B). Of note, even a single loss-of-function mutation resulted in almost the full leaf area affected by the lesion, leaving no room for additional effects in multiple mutants. In other experiments, the cop1 phenotype under SD was rescued partially by the gai rga double mutation (Fig. 7C). This observation provided genetic evidence supporting that the effects of COP1 on the susceptibility to B. cinerea are mediated at least partially by its effects on DELLA proteins. The residual effect of COP1 may be mediated by the remaining DELLA proteins or destabilization of the MYC2 protein (Chico et al., 2014).
Figure 7.
The effect of photoperiod on susceptibility to B. cinerea requires normal DELLA function. A and B, Resistance to B. cinerea infection represented by relative lesion size in mutants affected in DELLA genes in either the Columbia (A) or Landsberg erecta (B) background. C, Resistance to B. cinerea infection in wild-type (WT) and cop1 mutant plants with or without compromised DELLA function conferred by the gai rga double mutation. Data are means ± se of five plant replicates. Different letters indicate significant differences (P < 0.05) among means determined by Bonferroni posthoc tests. The significant interaction between cop1 and gai rga determined by factorial ANOVA is shown in C.
DISCUSSION
To investigate plant processes affected by photoperiod, we analyzed transcriptome responses to SD→LD compared with that under SD, followed by the identification of overrepresented GO terms among responsive genes and a physiological screening. This procedure detected JA-dependent defense as one of the processes enhanced by LD compared with that under SD. We have identified a group of genes that increase their expression immediately in response to LD perceived by cry1 and cry2 (and in some cases also by phyA) and a group of genes that reduce their expression largely independently of these photoreceptors (Fig. 1A). This set of genes is robust (Supplemental Figs. S2 and S3; Supplemental Table S4), does not represent simply a transient response to the SD→LD shift (Supplemental Fig. S2, B and C), and does not normally respond to increased irradiance (Supplemental Fig. S4B). Highly overrepresented GO terms included light-harvesting complexes, phenylpropanoid metabolism, JA- and ethylene-dependent systemic resistance (mainly response to JA stimulus), and oxygen and reactive oxygen species metabolism (Fig. 1B; Supplemental Table S3). The response of light-harvesting complex genes represents a shift of expression toward later hours of the daily cycle induced by SD→LD (Millar and Kay, 1996) without affecting the daily integral (Fig. 1B). No differences in chlorophyll or anthocyanin levels were observed after 3 LD (Supplemental Fig. S5), and the SD→LD transition caused at most modest oxidative stress (Bermúdez et al., 2010; Supplemental Fig. S6). However, compared with SD, LD significantly reduced the lesions caused by the necrotrophic pathogen B. cinerea (Fig. 2), which is consistent with the elevated JA-dependent defense predicted by transcriptome patterns.
Both the transcriptional response of several genes involved in JA-dependent defense (Fig. 1B) and the resistance to B. cinerea infection (Fig. 2) were impaired in the phyA cry1 cry2 triple mutant, indicating that the effects of growth under LD are not simply the result of sustained photosynthesis or oxidative stress (Supplemental Fig. S6; Rossi et al., 2017) driven by the extended daylength. The levels of JA (Goodspeed et al., 2012) and the abundance of MYC2 (Shin et al., 2012) are controlled by the circadian clock. The susceptibility to B. cinerea and the associated transcriptional signature also are clock controlled, causing responses that depend on the time of day at which the plants are inoculated (Ingle et al., 2015). However, the effects of photoperiod reported here do not result from a light-induced shift in the circadian rhythm of sensitivity (gating) because all the plants were inoculated simultaneously before exposure to the different light conditions and gene expression responses occurred during the first day of light extension. LD increased the intensity of JA signaling but not absolute JA levels (Fig. 3), indicating that LD increase the sensitivity to JA by acting downstream of the hormone itself.
cry2 (Zuo et al., 2011) and cry1 (Lian et al., 2011; Liu et al., 2011), activated by blue light, and phyA, activated by far-red light (Sheerin et al., 2015), interact with SPA1 and other SPA proteins reorganizing the COP1/SPA complex. Here, we show that, compared with that under SD, a single photoperiod of LD was enough to significantly reduce the nuclear abundance of COP1 measured at the end of the extended photoperiod (Fig. 5A). Reduced COP1 nuclear abundance is predicted to reduce its activity toward nuclear targets (Pacín et al., 2014). Therefore, we investigated if COP1 was involved in the defense response associated with LD. The cop1 mutant showed elevated defense against B. cinerea under SD and no response to LD (Fig. 5B). The impact of the cop1 mutation on the expression of genes involved in JA-dependent defense showed a negative correlation with the reported impact of the myc2 myc3 myc4 mutation (Fig. 4), indicating that COP1 might act via these transcription factors. Among MYC2, MYC3, and MYC4, only MYC2 was included among the genes that responded to LD (Supplemental Table S1), but this response was largely unaffected by the cop1 mutation (Supplemental Table S4). Therefore, COP1 appears to control the activity of MYC transcription factors downstream of their gene expression levels.
COP1 has been reported to destabilize MYC2 in etiolated seedlings compared with that in young light-grown seedlings; however, MYC2 does not appear to be a direct target of COP1 (Chico et al., 2014). Here, we explored a different possibility involving DELLA proteins that are known to increase JA-dependent defense by binding JAZ proteins, which are negative regulators of MYC2 (Wild et al., 2012). Loss- and gain-of-function mutations in DELLA genes eliminated the response to photoperiod concerning the area of the lesions induced by B. cinerea (Fig. 7), and even low-order mutants displayed clearly increased susceptibility to the pathogen (Wild et al., 2012). Therefore, we investigated whether daylength affected DELLA stability. The levels of RGA increased under SD→LD compared with that under SD in a COP1-dependent manner (Fig. 6). In conclusion, the mechanisms that control JA-dependent defense in response to daylength involve LD perception by cry1, cry2, and phyA, followed by a reduction of COP1 nuclear abundance and a subsequent increase in DELLA abundance. Whether the link between COP1 and DELLA is direct is currently under investigation. In addition, there is a COP1-independent action of daylength on the expression of MYC2.
There is a tight association between the light environment and plant defense (Ballaré, 2014), and light perceived by phyA or phyB increases the responses to JA. The phyA mutant shows reduced JA-induced inhibition of root growth and promotion of gene expression (Robson et al., 2010). Plants exposed to low red/far-red ratios that reduce phyB activity show compromised resistance to B. cinerea and impaired induction of gene expression by either JA or B. cinerea (Cerrudo et al., 2012; de Wit et al., 2013). Conversely, UV-B radiation perceived by UV RESISTANCE LOCUS8 increases the resistance to B. cinerea, but this effect is likely mediated by the increased production of sinapate and not by changes in JA signaling (Demkura and Ballaré, 2012). The reduced responses to JA in plants with low or null phyA or phyB activity are mediated by the enhanced stability of JAZs (Robson et al., 2010; Leone et al., 2014). Therefore, the tradeoff between growth and defense can be uncoupled in a sextuple mutant lacking both phyB and the five JAZs, which shows constitutively high JA responses and no growth reductions (Campos et al., 2016). Low red/far-red ratios also reduce the stability of MYC2 (Chico et al., 2014) and DELLA (Leone et al., 2014), and PIF4 was described recently as a negative regulator of defense (Gangappa et al., 2017). Therefore, although here we have focused on the COP1-DELLA pathway, other aspects of the plant defense network also could be affected by photoperiod.
A priori, there are several reasons why enhanced defense under LD might be advantageous for the plant. These include the potentially higher availability of products of photosynthesis to be invested in defense under LD and the protection of reproductive development initiated under LD. However, it is intriguing that B. cinerea forms conidia in the light (airborne macroconidia are a major source of infection; Canessa et al., 2013) and the concentration of airborne inoculum is significantly higher during day periods than at night (Blanco et al., 2006; Leyronas and Nicot, 2012). Compared with that under SD, LD mainly extends the high expression of genes involved in JA-dependent defense during the period of additional light exposure (i.e. LD does not enhance expression compared with that under SD during the period where both are exposed to light; Fig. 1B). Therefore, the plant response might be an adaptation to the light response of the pathogen under LD.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Plants of Arabidopsis (Arabidopsis thaliana) were grown at 20°C under SD (8 h of light/16 h of darkness) for 3 weeks and then either transferred to LD (16 h of light/8 h of darkness, same lighting) or left as SD controls. White light (300 μmol m−2 s−1 between 400 and 700 nm) was provided by 400-W Philips SON lamps, except in microarray experiment 2 (160 μmol m−2 s−1), where 36-W Philips tubes were used to test the selected gene list under different conditions. The phyA, cry1, and cry2 single and multiple mutants (Mazzella et al., 2001), rgl1-1, gai-t6 rgl2-1, gai-t6 rga-t2 rgl1-1, or gai-t6 rga-t2 rgl1-1 rgl2-1 (Lee et al., 2002; Achard et al., 2006), and the gain-of-function gai-1 mutant (Koornneef et al., 1985) in Landsberg erecta and the myc2-3 (Yadav et al., 2005), phyA cry1 cry2 (Buchovsky et al., 2008), cop1-4 and cop1-6 (McNellis et al., 1994a), and gai-td1 rga-29 (Plackett et al., 2014; Park et al., 2017) mutants in Columbia were compared with their respective wild types. For COP1 overexpression, the p35S:COP1 transgenic line in Nossen (McNellis et al., 1994b) was compared with its Nossen wild type. The lines p35S:YFP-COP1 in Columbia (Oravecz et al., 2006) and pRGA:GFP-RGA in Landsberg erecta (Silverstone et al., 2001) were used for confocal microscopy.
Microarray Experiments
Total RNA was extracted from SD→LD and SD plants in two different experiments (Supplemental Fig. S1) by using the RNeasy Plant Mini Kit (Qiagen). cDNA and cRNA synthesis and hybridization to 22 K (ATH1) Affymetrix Gene Chips were performed according to instructions from Affymetrix. The scaling tab of the Affymetrix microarray suite in the mode all probe sets was used to standardize the trimmed mean signal of each array to the target signal according to the manufacturer’s instructions.
Analysis of Microarray Data
Two different experiments were conducted. In experiment 1 (Supplemental Fig. S1), plants of the wild type and phyA, cry1, cry2, phyA cry1, phyA cry2, cry1 cry2, and phyA cry1 cry2 mutants were harvested at the end of the first LD (SD→LD) or, simultaneously, 8 h after the end of the SD as controls. Expression data for each microarray were first normalized by dividing the expression of each gene by the ratio between the average expression of all the genes in that microarray and the average of all microarray averages. The factor used for normalization ranged between and 0.84 and 1.14, indicating that there were no large differences among microarrays. To investigate the genes that respond to LD and the role played by phyA, cry1, and cry2 in their response, we first used ANOVA and calculated P and q values (Storey and Tibshirani, 2003). Since the experiment was focused on the response to LD and not on the differences among genotypes that could already be present in the SD controls, we pooled the data corresponding to the different genotypes under SD. This procedure offered an objective criterion to eliminate those genes where the differences were mainly present already under SD because these genes showed high error estimates compared with the response to daylength. Therefore, the ANOVA included nine treatments: eight corresponding to each genotype under SD→LD (two biological replicates for each genotype) and one corresponding to the SD control (eight pooled data corresponding to one microarray per genotype). We identified 1,124 genes with P < 0.005 and q < 0.1. We restricted the list to 984 genes by using a wild-type SD→LD/SD gene expression ratio > 1.2 or < 0.8 as a cutoff. By using dChip (Li and Wong, 2003), 805 of the 984 genes were grouped into three major clusters. The clustering step is conservative and reduces the chances that a gene becomes incorporated into the list if it does not share the major patterns of response. For instance, the list does not include FT, which is known to respond to LD, because, although this gene showed significant effects of treatment (P < 0.003, q < 0.08, normalized expression: wild-type, SD→LD = 1.1, SD = 0.3, phyA cry1 cry2, SD→LD = 0.2, SD = 0.3), it was not included in cluster 1 or 2.
Each cluster was restricted further by testing for each gene the statistical significance of the features of each cluster. For cluster 1, we used multiple regression, y = a + b x1 + c x2, where b represents the additive effects of CRY1 and CRY2 wild-type alleles under SD→LD, x1 is 2 for the wild-type SD→LD, 1 for the cry1 or cry2 background under SD→LD, and 0 for the cry1 cry2 background under SD→LD and all genotypes under SD, c represents the effect of the phyA mutant allele in the cry1 and/or cry2 mutant background under SD→LD, and x2 is 1 for the phyA cry1 and phyA cry2 mutants under SD→LD, 2 for the phyA cry1 cry2 mutant under SD→LD, and 0 for all other conditions. For cluster 2, we used simple regression, y = a + b x, where b represents the redundant effect of PHYA, CRY1, and CRY2 wild-type alleles under SD→LD and x assumes 1 for the wild type and the single and double mutants under SD→LD and 0 for the phyA cry1 cry2 triple mutant under SD→LD and all the genotypes under SD. For cluster 3, we used simple regression, y = a + b x, where b represents the effect of SD→LD compared with SD and x is 1 for all genotypes under SD→LD and 0 for all genotypes under SD. Limitation of the clusters by this procedure ensured the homogenous composition of the clusters by statistical criteria. Therefore, 749 genes were grouped among cluster 1 (163 genes), cluster 2 (265 genes), and cluster 3 (321 genes).
Overrepresented functions were investigated for each cluster and for the combination of the two clusters that included genes with expression promoted in SD→LD compared with SD by using ATCOECIS (Vandepoele et al., 2009).
In experiment 2, SD→LD and SD control plants of the wild type were harvested at the end of the first LD and at the end of day 6. Two biological replicates were included in each case. Expression data were normalized as described for experiment 1 and used here to test the robustness of the gene list and the persistence of the effects several days after transition.
Bioassays of Botrytis cinerea Resistance
Plants were grown for 3 weeks under SD. Seven hours after the beginning of day 21, a single droplet of 5 μL of B. cinerea spore suspension (2–3 × 105 spores mL−1) was placed on the adaxial surface of each one of four mature leaves (Muckenschnabel et al., 2002). Pots were enclosed in individual clear polyester chambers to prevent desiccation of the droplets. Forty-eight hours after inoculation, the leaves were harvested and photographed to measure the area of the lesion with the aid of Adobe Photoshop CS3.
Confocal Microscopy
Confocal fluorescence images were taken with an LSM5 Pascal (Zeiss) laser scanning microscope with a water-immersion objective lens (C-Apochromat 40×/1.2; Zeiss). For chloroplast visualization, probes were excited with a HeNe laser (543 nm) and fluorescence was detected using an LP560 filter. For COP1-YFP and RGA-GFP fusion protein visualization, probes were excited with an argon laser (488 nm) and fluorescence was detected using a BP 505 to 530 filter. Fluorescent nuclei were defined as regions of interest, and fluorescence intensity was measured using ImageJ from the National Institutes of Health (Abràmoff et al., 2004). A transmission image also was included to count cells in each image. Representative cells of the leaf parenchyma (first layers beneath the epidermis) were documented by photography during the first 15 min of microscopy analysis.
Reverse Transcription Quantitative PCR
Seedlings were harvested in liquid nitrogen, then total RNA was extracted with the RNeasy Plant Mini Kit (Qiagen) and subjected to a DNase treatment with RQ1 RNase-Free DNase (Promega). cDNA derived from this RNA was synthesized using Invitrogen SuperScript III and an oligo(dT) primer. The synthesized cDNAs were amplified with FastStart Universal SYBR Green Master (Roche) using the 7500 Real Time PCR System (Applied Biosystems) cycler. The UBIQUITIN-CONJUGATING ENZYME2 gene was used as a normalization control (Czechowski et al., 2005). The primers are listed in Supplemental Table S5.
Extraction, Purification, and Estimation of JA Content
JA was extracted from Arabidopsis dry shoots by using a modified version of the protocol of Durgbanshi et al. (2005). Plant material was homogenized and dissolved in 5 mL of ultra-pure water. Fifty nanograms of [2H6]JA (OlChemIm) was added as an internal standard. Extracts were transferred to 50-mL tubes and centrifuged at 1,500g for 15 min. The supernatant was collected, adjusted to pH 2.8 with 15% (v/v) acetic acid, and extracted twice with an equal volume of diethyl ether. The aqueous phase was discarded, and the organic fraction was evaporated under vacuum. Dried extracts were dissolved in 1 mL of methanol. Samples were filtered through a syringe filter tip on a vacuum manifold at a flow rate less than 1 mL min−1, and the eluate was evaporated at 35°C under vacuum in a SpeedVac SC110 (Savant Instruments). Mass spectrometry analysis for JA quantification was performed on a quadruple tandem mass spectrometer (Quattro Ultima; Micromass) outfitted with an electrospray ion source. A mixture containing unlabeled compound and internal standard was separated by reverse-phase HPLC and analyzed by tandem mass spectrometry with multiple reaction monitoring for JA retention time determination. This compound was monitored at mass-to-charge ratio transitions of 209/59 to 15/59 with retention time of 13.5 min. The collision energy used was 20 eV. The cone voltage was 35 V.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AT1G09570 (PHYA), AT4G08920 (CRY1), AT1G04400 (CRY2), AT1G32640 (MYC2), AT5G46760 (MYC3), AT4G17880 (MYC4), AT2G32950 (COP1), AT2G01570 (RGA1), AT1G14920 (GAI), AT1G66350 (RGL1), and AT3G03450 (RGL2).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Experimental protocols.
Supplemental Figure S2. Transcriptome responses to the initial period of LD are robust and persistent.
Supplemental Figure S3. Transcriptional network involving genes present in clusters 1, 2, and 3.
Supplemental Figure S4. Specific signature of gene expression responses to daylength.
Supplemental Figure S5. Chlorophyll and anthocyanin contents do not exhibit rapid responses to daylength.
Supplemental Figure S6. Negligible effects of daylength on oxidative stress markers.
Supplemental Table S1. List of genes corresponding to clusters 1, 2, and 3.
Supplemental Table S2. Expression of genes corresponding to clusters 1, 2, and 3 in experiment 2.
Supplemental Table S3. GO term enrichment.
Supplemental Table S4. Expression of JA- and ethylene-dependent systemic resistance genes in the wild type and cop1 mutants.
Supplemental Table S5. Sequences of primers used for reverse transcription quantitative PCR.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Dr. Sudip Chattopadhyay (National Centre for Plant Genome Research, India) for providing seeds of the myc2 mutant, Dr. Guillermina Abdala for providing facilities for hormone measurements, Dr. Carlos Ballaré (Instituto de Investigaciones Fisiológicas y Ecológicas Vinculadas a la Agricultura) for helpful comments, and Dr. Mercedes Keller (Instituto de Investigaciones Fisiológicas y Ecológicas Vinculadas a la Agricultura) for introducing us to the test with B. cinerea.
Footnotes
This study was supported by a Guggenheim Foundation fellowship (to J.J.C), by Agencia Nacional de Promoción Científica y Tecnológica (PICT-2015-1796), by the University of Buenos Aires (20020100100437, to J.J.C.), by the Howard Hughes Medical Institute (J.I.C.), and by the SIGNAT-Research and Innovation Staff Exchange (H2020-MSCA-RISE-2014, to P.D.C., M.A.B., D.A., and J.J.C.).
References
- Abràmoff MD, Magalhàes PJ, Ram SJ (2004) Image processing with ImageJ. Biophoton Int 11: 36–41 [Google Scholar]
- Achard P, Cheng H, De Grauwe L, Decat J, Schoutteten H, Moritz T, Van Der Straeten D, Peng J, Harberd NP (2006) Integration of plant responses to environmentally activated phytohormonal signals. Science 311: 91–94 [DOI] [PubMed] [Google Scholar]
- Alonso-Blanco C, Gomez-Mena C, Llorente F, Koornneef M, Salinas J, Martínez-Zapater JM (2005) Genetic and molecular analyses of natural variation indicate CBF2 as a candidate gene for underlying a freezing tolerance quantitative trait locus in Arabidopsis. Plant Physiol 139: 1304–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrés F, Coupland G (2012) The genetic basis of flowering responses to seasonal cues. Nat Rev Genet 13: 627–639 [DOI] [PubMed] [Google Scholar]
- Ballaré CL. (2014) Light regulation of plant defense. Annu Rev Plant Biol 65: 335–363 [DOI] [PubMed] [Google Scholar]
- Becker B, Holtgrefe S, Jung S, Wunrau C, Kandlbinder A, Baier M, Dietz KJ, Backhausen JE, Scheibe R (2006) Influence of the photoperiod on redox regulation and stress responses in Arabidopsis thaliana L. (Heynh.) plants under long- and short-day conditions. Planta 224: 380–393 [DOI] [PubMed] [Google Scholar]
- Bermúdez MA, Páez-Ochoa MA, Gotor C, Romero LC (2010) Arabidopsis S-sulfocysteine synthase activity is essential for chloroplast function and long-day light-dependent redox control. Plant Cell 22: 403–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco C, los de Santos B, Romero F (2006) Relationship between concentrations of Botrytis cinerea conidia in air, environmental conditions, and the incidence of grey mould in strawberry flowers and fruits. Eur J Plant Pathol 114: 415–425 [Google Scholar]
- Buchovsky AS, Strasser B, Cerdán PD, Casal JJ (2008) Suppression of pleiotropic effects of functional CRYPTOCHROME genes by TERMINAL FLOWER 1. Genetics 180: 1467–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos ML, Yoshida Y, Major IT, de Oliveira Ferreira D, Weraduwage SM, Froehlich JE, Johnson BF, Kramer DM, Jander G, Sharkey TD, et al. (2016) Rewiring of jasmonate and phytochrome B signalling uncouples plant growth-defense tradeoffs. Nat Commun 7: 12570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canessa P, Schumacher J, Hevia MA, Tudzynski P, Larrondo LF (2013) Assessing the effects of light on differentiation and virulence of the plant pathogen Botrytis cinerea: characterization of the White Collar Complex. PLoS ONE 8: e84223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal JJ, Fankhauser C, Coupland G, Blázquez MA (2004) Signalling for developmental plasticity. Trends Plant Sci 9: 309–314 [DOI] [PubMed] [Google Scholar]
- Cerrudo I, Keller MM, Cargnel MD, Demkura PV, de Wit M, Patitucci MS, Pierik R, Pieterse CMJ, Ballaré CL (2012) Low red/far-red ratios reduce Arabidopsis resistance to Botrytis cinerea and jasmonate responses via a COI1-JAZ10-dependent, salicylic acid-independent mechanism. Plant Physiol 158: 2042–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chico JM, Fernández-Barbero G, Chini A, Fernández-Calvo P, Díez-Díaz M, Solano R (2014) Repression of jasmonate-dependent defenses by shade involves differential regulation of protein stability of MYC transcription factors and their JAZ repressors in Arabidopsis. Plant Cell 26: 1967–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demkura PV, Ballaré CL (2012) UVR8 mediates UV-B-induced Arabidopsis defense responses against Botrytis cinerea by controlling sinapate accumulation. Mol Plant 5: 642–652 [DOI] [PubMed] [Google Scholar]
- de Wit M, Spoel SH, Sanchez-Perez GF, Gommers CMM, Pieterse CMJ, Voesenek LACJ, Pierik R (2013) Perception of low red:far-red ratio compromises both salicylic acid- and jasmonic acid-dependent pathogen defences in Arabidopsis. Plant J 75: 90–103 [DOI] [PubMed] [Google Scholar]
- Dombrecht B, Xue GP, Sprague SJ, Kirkegaard JA, Ross JJ, Reid JB, Fitt GP, Sewelam N, Schenk PM, Manners JM, et al. (2007) MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19: 2225–2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durgbanshi A, Arbona V, Pozo O, Miersch O, Sancho JV, Gómez-Cadenas A (2005) Simultaneous determination of multiple phytohormones in plant extracts by liquid chromatography-electrospray tandem mass spectrometry. J Agric Food Chem 53: 8437–8442 [DOI] [PubMed] [Google Scholar]
- Fernández-Calvo P, Chini A, Fernández-Barbero G, Chico JM, Gimenez-Ibanez S, Geerinck J, Eeckhout D, Schweizer F, Godoy M, Franco-Zorrilla JM, et al. (2011) The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23: 701–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangappa SN, Berriri S, Kumar SV (2017) PIF4 coordinates thermosensory growth and immunity in Arabidopsis. Curr Biol 27: 243–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda H, Sasaki E, Akiyama K, Maruyama-Nakashita A, Nakabayashi K, Li W, Ogawa M, Yamauchi Y, Preston J, Aoki K, et al. (2008) The AtGenExpress hormone and chemical treatment data set: experimental design, data evaluation, model data analysis and data access. Plant J 55: 526–542 [DOI] [PubMed] [Google Scholar]
- Goodspeed D, Chehab EW, Min-Venditti A, Braam J, Covington MF (2012) Arabidopsis synchronizes jasmonate-mediated defense with insect circadian behavior. Proc Natl Acad Sci USA 109: 4674–4677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingle RA, Stoker C, Stone W, Adams N, Smith R, Grant M, Carré I, Roden LC, Denby KJ (2015) Jasmonate signalling drives time-of-day differences in susceptibility of Arabidopsis to the fungal pathogen Botrytis cinerea. Plant J 84: 937–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SD. (2009) Plant responses to photoperiod. New Phytol 181: 517–531 [DOI] [PubMed] [Google Scholar]
- Kleine T, Kindgren P, Benedict C, Hendrickson L, Strand A (2007) Genome-wide gene expression analysis reveals a critical role for CRYPTOCHROME1 in the response of Arabidopsis to high irradiance. Plant Physiol 144: 1391–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Elgersma A, Hanhart CJ, Loenen‐Martinet EP, Rijn L, Zeevaart JAD (1985) A gibberellin insensitive mutant of Arabidopsis thaliana. Physiol Plant 65: 33–39 [Google Scholar]
- Krzymuski M, Cerdán PD, Zhu L, Vinh A, Chory J, Huq E, Casal JJ (2014) Phytochrome A antagonizes PHYTOCHROME INTERACTING FACTOR 1 to prevent over-activation of photomorphogenesis. Mol Plant 7: 1415–1428 [DOI] [PubMed] [Google Scholar]
- Lau OS, Deng XW (2012) The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci 17: 584–593 [DOI] [PubMed] [Google Scholar]
- Lee CM, Thomashow MF (2012) Photoperiodic regulation of the C-repeat binding factor (CBF) cold acclimation pathway and freezing tolerance in Arabidopsis thaliana. Proc Natl Acad Sci USA 109: 15054–15059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BD, Kim MR, Kang MY, Cha JY, Han SH, Nawkar GM, Sakuraba Y, Lee SY, Imaizumi T, McClung CR, et al. (2017) The F-box protein FKF1 inhibits dimerization of COP1 in the control of photoperiodic flowering. Nat Commun 8: 2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Cheng H, King KE, Wang W, He Y, Hussain A, Lo J, Harberd NP, Peng J (2002) Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition. Genes Dev 16: 646–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone M, Keller MM, Cerrudo I, Ballaré CL (2014) To grow or defend? Low red:far-red ratios reduce jasmonate sensitivity in Arabidopsis seedlings by promoting DELLA degradation and increasing JAZ10 stability. New Phytol 204: 355–367 [DOI] [PubMed] [Google Scholar]
- Lepistö A, Rintamäki E (2012) Coordination of plastid and light signaling pathways upon development of Arabidopsis leaves under various photoperiods. Mol Plant 5: 799–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyronas C, Nicot PC (2012) Monitoring viable airborne inoculum of Botrytis cinerea in the South-East of France over 3 years: relation with climatic parameters and the origin of air masses. Aerobiologia 29: 291–299 [Google Scholar]
- Li C, Wong WH (2003) DNA-Chip Analyzer (dChip). In Parmigiani G, Garrett ES, Irizarry R, Zeger SL, eds, The Analysis of Gene Expression Data: Methods and Software. Springer, New York, pp 120–141 [Google Scholar]
- Lian HL, He SB, Zhang YC, Zhu DM, Zhang JY, Jia KP, Sun SX, Li L, Yang HQ (2011) Blue-light-dependent interaction of cryptochrome 1 with SPA1 defines a dynamic signaling mechanism. Genes Dev 25: 1023–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Zuo Z, Liu H, Liu X, Lin C (2011) Arabidopsis cryptochrome 1 interacts with SPA1 to suppress COP1 activity in response to blue light. Genes Dev 25: 1029–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu LJ, Zhang YC, Li QH, Sang Y, Mao J, Lian HL, Wang L, Yang HQ (2008) COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell 20: 292–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O, Chico JM, Sánchez-Serrano JJ, Solano R (2004) JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16: 1938–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzella MA, Cerdán PD, Staneloni RJ, Casal JJ (2001) Hierarchical coupling of phytochromes and cryptochromes reconciles stability and light modulation of Arabidopsis development. Development 128: 2291–2299 [DOI] [PubMed] [Google Scholar]
- McNellis TW, von Arnim AG, Araki T, Komeda Y, Miséra S, Deng XW (1994a) Genetic and molecular analysis of an allelic series of cop1 mutants suggests functional roles for the multiple protein domains. Plant Cell 6: 487–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNellis TW, von Arnim AG, Deng XW (1994b) Overexpression of Arabidopsis COP1 results in partial suppression of light-mediated development: evidence for a light-inactivable repressor of photomorphogenesis. Plant Cell 6: 1391–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael TP, Mockler TC, Breton G, McEntee C, Byer A, Trout JD, Hazen SP, Shen R, Priest HD, Sullivan CM, et al. (2008) Network discovery pipeline elucidates conserved time-of-day-specific cis-regulatory modules. PLoS Genet 4: e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AJ, Kay SA (1996) Integration of circadian and phototransduction pathways in the network controlling CAB gene transcription in Arabidopsis. Proc Natl Acad Sci USA 93: 15491–15496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muckenschnabel I, Goodman BA, Williamson B, Lyon GD, Deighton N (2002) Infection of leaves of Arabidopsis thaliana by Botrytis cinerea: changes in ascorbic acid, free radicals and lipid peroxidation products. J Exp Bot 53: 207–214 [DOI] [PubMed] [Google Scholar]
- O’Connor TR, Dyreson C, Wyrick JJ (2005) Athena: a resource for rapid visualization and systematic analysis of Arabidopsis promoter sequences. Bioinformatics 21: 4411–4413 [DOI] [PubMed] [Google Scholar]
- Obayashi T, Nishida K, Kasahara K, Kinoshita K (2011) ATTED-II updates: condition-specific gene coexpression to extend coexpression analyses and applications to a broad range of flowering plants. Plant Cell Physiol 52: 213–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oravecz A, Baumann A, Máté Z, Brzezinska A, Molinier J, Oakeley EJ, Adám E, Schäfer E, Nagy F, Ulm R (2006) CONSTITUTIVELY PHOTOMORPHOGENIC1 is required for the UV-B response in Arabidopsis. Plant Cell 18: 1975–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacín M, Legris M, Casal JJ (2014) Rapid decline in nuclear constitutive photomorphogenesis1 abundance anticipates the stabilization of its target elongated hypocotyl5 in the light. Plant Physiol 164: 1134–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Oh DH, Dassanayake M, Nguyen KT, Ogas J, Choi G, Sun TP (2017) Gibberellin signaling requires chromatin remodeler PICKLE to promote vegetative growth and phase transitions. Plant Physiol 173: 1463–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschke F, Kretsch T (2011) Genome-wide analysis of light-dependent transcript accumulation patterns during early stages of Arabidopsis seedling deetiolation. Plant Physiol 155: 1353–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plackett ARG, Ferguson AC, Powers SJ, Wanchoo-Kohli A, Phillips AL, Wilson ZA, Hedden P, Thomas SG (2014) DELLA activity is required for successful pollen development in the Columbia ecotype of Arabidopsis. New Phytol 201: 825–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson F, Okamoto H, Patrick E, Harris SR, Wasternack C, Brearley C, Turner JG (2010) Jasmonate and phytochrome A signaling in Arabidopsis wound and shade responses are integrated through JAZ1 stability. Plant Cell 22: 1143–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossel JB, Wilson IW, Pogson BJ (2002) Global changes in gene expression in response to high light in Arabidopsis. Plant Physiol 130: 1109–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi FR, Krapp AR, Bisaro F, Maiale SJ, Pieckenstain FL, Carrillo N (2017) Reactive oxygen species generated in chloroplasts contribute to tobacco leaf infection by the necrotrophic fungus Botrytis cinerea. Plant J 92: 761–773 [DOI] [PubMed] [Google Scholar]
- Sawa M, Nusinow DA, Kay SA, Imaizumi T (2007) FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 318: 261–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheerin DJ, Menon C, zur Oven-Krockhaus S, Enderle B, Zhu L, Johnen P, Schleifenbaum F, Stierhof YD, Huq E, Hiltbrunner A (2015) Light-activated phytochrome A and B interact with members of the SPA family to promote photomorphogenesis in Arabidopsis by reorganizing the COP1/SPA complex. Plant Cell 27: 189–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Heidrich K, Sanchez-Villarreal A, Parker JE, Davis SJ (2012) TIME FOR COFFEE represses accumulation of the MYC2 transcription factor to provide time-of-day regulation of jasmonate signaling in Arabidopsis. Plant Cell 24: 2470–2482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone AL, Jung HS, Dill A, Kawaide H, Kamiya Y, Sun TP (2001) Repressing a repressor: gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. Plant Cell 13: 1555–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Huang H, Gao H, Wang J, Wu D, Liu X, Yang S, Zhai Q, Li C, Qi T, et al. (2014) Interaction between MYC2 and ETHYLENE INSENSITIVE3 modulates antagonism between jasmonate and ethylene signaling in Arabidopsis. Plant Cell 26: 263–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YH, Smith RW, To BJ, Millar AJ, Imaizumi T (2012) FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science 336: 1045–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R (2003) Statistical significance for genomewide studies. Proc Natl Acad Sci USA 100: 9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G (2004) Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303: 1003–1006 [DOI] [PubMed] [Google Scholar]
- Vandepoele K, Quimbaya M, Casneuf T, De Veylder L, Van de Peer Y (2009) Unraveling transcriptional control in Arabidopsis using cis-regulatory elements and coexpression networks. Plant Physiol 150: 535–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild M, Davière JM, Cheminant S, Regnault T, Baumberger N, Heintz D, Baltz R, Genschik P, Achard P, (2012) The Arabidopsis DELLA RGA-LIKE3 is a direct target of MYC2 and modulates jasmonate signaling responses. Plant Cell 24: 3307–3319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav V, Mallappa C, Gangappa SN, Bhatia S, Chattopadhyay S (2005) A basic helix-loop-helix transcription factor in Arabidopsis, MYC2, acts as a repressor of blue light-mediated photomorphogenic growth. Plant Cell 17: 1953–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Z, Liu H, Liu B, Liu X, Lin C (2011) Blue light-dependent interaction of CRY2 with SPA1 regulates COP1 activity and floral initiation in Arabidopsis. Curr Biol 21: 841–847 [DOI] [PMC free article] [PubMed] [Google Scholar]