Abstract
Histone modifications are key epigenetic regulatory features that have important roles in many cellular events. Lysine methylations mark various sites on the tail and globular domains of histones and their levels are precisely balanced by the action of methyltransferases (‘writers’) and demethylases (‘erasers’). In addition, distinct effector proteins (‘readers’) recognize specific methyl-lysines in a manner that depends on the neighboring amino-acid sequence and methylation state. Misregulation of histone lysine methylation has been implicated in several cancers and developmental defects. Therefore, histone lysine methylation has been considered a potential therapeutic target, and clinical trials of several inhibitors of this process have shown promising results. A more detailed understanding of histone lysine methylation is necessary for elucidating complex biological processes and, ultimately, for developing and improving disease treatments. This review summarizes enzymes responsible for histone lysine methylation and demethylation and how histone lysine methylation contributes to various biological processes.
Introduction
In eukaryotic cells, genetic information stored in DNA is present in a highly organized chromatin structure. The nucleosome, the basic unit of chromatin, is composed of two copies of each core histone, H2A, H2B, H3 and H4, wrapped by about two turns of DNA.1 Protruding unstructured N-terminal tails as well as structured globular domains of each histone are subject to post-translational modifications that include methylation, acetylation, phosphorylation and ubiquitylation, among others.2, 3 These histone modifications affect chromatin structure and also provide binding platforms for diverse transcription factors, such as chromatin remodelers, histone chaperones, DNA/histone-modifying enzymes and general transcription factors.2, 3 Thus, histone modifications have important roles in many cellular events, including gene expression, DNA replication and repair, chromatin compaction and cell-cycle control.2, 3 Misregulation of histone modifications has been implicated in the pathogenesis of cancer and in developmental defects, further emphasizing the importance of the regulation of histone modifications.4, 5
Although histone methylation and its involvement in transcription were first reported in the 1960s,6 it was only about 15 years ago that the first histone methyltransferase, SUV39H1, containing a catalytic SET (Su(var)3–9, Enhancer of Zeste, and Trithorax) domain, was identified,7 igniting discoveries of numerous histone methyltransferases based on SET-domain homology searches.8 Until the discovery of an H3K4 demethylase LSD1 (lysine-specific histone demethylase 1),9 histone methylations had been thought to turn over more slowly than other histone modifications. The subsequent discovery of the JmjC (jumonji C) domain as a key signature of demethylating enzymes10 has substantially broadened our repertoire of histone demethylases.
There are three lysine methylation states—mono-, di- and trimethylation (me1, me2 and me3, respectively)—none of which changes the electronic charge of the amino-acid side chain; therefore, histone lysine methylation functions are considered to be mainly exerted by effector molecules that specifically recognize the methylated site.11 These ‘reader’ proteins contain methyl-lysine-binding motifs, including PHD, chromo, tudor, PWWP, WD40, BAH, ADD, ankyrin repeat, MBT and zn-CW domains, and also have the ability to distinguish target methyl-lysines based on their methylation state and surrounding amino-acid sequence.12
Unlike other histone modifications, which simply specify active or repressed chromatin states, histone lysine methylations confer active or repressive transcription depending on their positions and methylation states.13 Generally, H3K4, H3K36 and H3K79 methylations are considered to mark active transcription, whereas H3K9, H3K27 and H4K20 methylations are thought to be associated with silenced chromatin states.13 These histone lysine methylations also interact with other histone modifications as well as DNA methylation to regulate precisely gene expression. For example, H3K4 and H3K79 methylations are known to require prior H2B ubiquitylation in yeast.14 Also, bivalent chromatins marked simultaneously by H3K4 and H3K27 methylations have an important role in shifting gene expression from a poised state to active or inactive states in embryonic stem cells (ESCs).15
Numerous studies have shown that mutation or misregulation of histone methylation, methyltransferases, demethylases and methyl-lysine-binding proteins are associated with various diseases.16 Therefore, many histone methylation-related proteins are being studied as potential therapeutic targets.17 Recent advances in next-generation sequencing, mass spectrometry, X-ray crystallography and cryo-EM techniques for analyzing histone modification-related proteins have allowed a more detailed understanding of relationship between histone methylation and diseases.3, 18
In this review, we summarize how histone lysine methylations are regulated by histone methyltransferases (‘writers’) and demethylases (‘erasers’), as depicted in Figure 1. We also discuss the biological roles of histone lysine methylations and associated diseases caused by misregulation of histone lysine methylations, as summarized in Table 1.
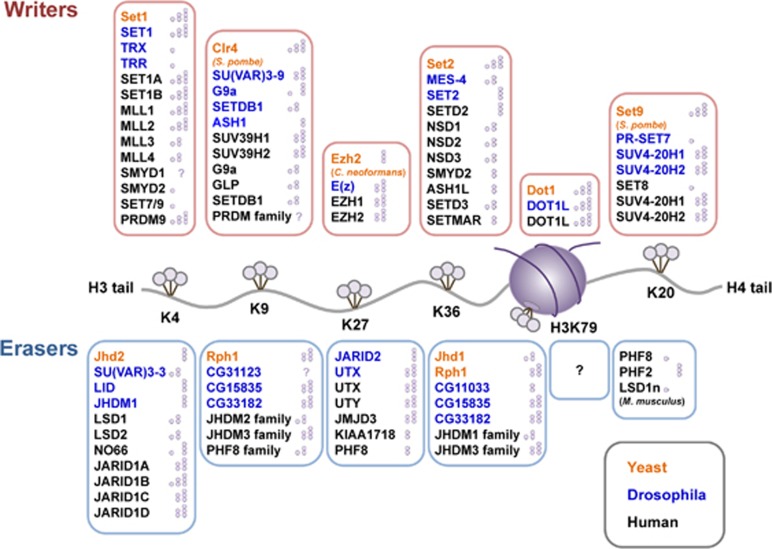
Figure 1.
A schematic depiction of a nucleosome showing principal lysine methylation sites on histones H3 and H4. The reported writers (methyltransferases) and erasers (demethylases) for each lysine methylation are also depicted with their methylation state specificities: single circle ( ), me1; double circle (
), me1; double circle ( ), me2; triple circle (
), me2; triple circle ( ), me3.
), me3.
Table 1. Summary of principal writer (methyltransferases), eraser (demethylases) and reader proteins for lysine methylation on histones H3 and H4 and their associated diseases.
| Writer Name | Diseases | Erasers name | Diseases | Reader name | Domains | Functions | Diseases | |
|---|---|---|---|---|---|---|---|---|
| H3K4 | SET1A/KMT2E | Hepatocarcinogenesis293 | LSD1/KDM1A | Prostate cancer294 | CHD1 | Chromo | Chromatin remodeler67 | Prostate cancer295 |
| SET1B/KMT2F | — | LSD2/KDM1B | Lung cancer296 | BPTF | PHD | NURF complex subunit68 | Alzheimer disease297 | |
| MLL1/KMT2A | Leukemia81 | NO66/MAPJD | Lung carcinoma50 | TAF3 | PHD | TFIID complex subunit69 | — | |
| MLL2/KMT2B | — | JARID1A/KDM5A | Ankylosing spondylitis50 | Sgf29 | Tudor | Crosstalk between H3K4me2/3 and H3 acetylation71 | Hepatocarcinogenesis298 | |
| MLL3/KMT2C | Urinary bladder299 | JARID1B/KDM5B | Breast cancer300 | ING4 | PHD | G/M cell cycle arrest301 | Breast cancer302 | |
| MLL4/KMT2D | Lymphomagenesis299 | JARID1C/KDM5C | Intellectual disability50 | CFP1 | PHD | Binds to unmethylated CpG30 | Involved in hematopoietic cell differentiation303 | |
| SMYD1/KMT3D | Rhabdomyosarcoma304 | JARID1D/KDM5D | Prostate cancer50 | PHF2/KDM7C | PHD | Oxygen sensor in normoxia305 | Breast carcinoma306 | |
| SMYD2/KMT3C | Pancreatic and gastric cancers307, 308 | SPIN1 | PHD | Senescence and apoptosis309 | Liposarcoma310 | |||
| SET7/9/KMT7 | Gastric cancer311 | PHF23 | PHD | Autophagy312 | Myeloid leukaemia5 | |||
| PRDM9/KMT8B | Childhood leukemogenesis313 | PYGO2 | PHD | Self-renewal of mammary progenitor cells5 | Malignant breast tumor5 | |||
| H3K9 | SUV39H/KMT1A–B | Blood cell defects,314 chromosome instability315 | JHDM2A/KDM3A | Malignant colorectal cancer,316 prostate adenocarcinoma metastasis,317 renal cell carcinoma318 | HP1α/β | Chromo | Heterochromatin formation121 | Breast cancer,319 medulloblastoma,320 papillary thyroid carcinoma321 |
| G9a/KMT1C | Breast and ovarian cancers149, 152 | JHDM2B/KDM3B | Myeloid leukemia322 | HP1γ | Chromo | Transcription elongation137 | — | |
| GLP/KMT1D | Kleefstra syndrome323 | JHDM2C/KDM3C | — | UHRF1 | TTD | DNA methylation115 | Gastric cancer324 | |
| SETDB1/KMT1E | Huntington disease148 | JHDM3A/KDM4A | Bladder and breast cancers325, 326 | |||||
| PRDM family | Breast and liver cancers,327 AML,328 myelodysplastic syndrome329 | JHDM3B/KDM4B | Peripheral nerve sheath tumor330 | |||||
| JHDM3C/KDM4C | Esophageal and breast cancers,331, 332 medulloblastoma,333 lymphoma330 | |||||||
| JHDM3D/KDM4D | — | |||||||
| PHF8/KDM7B | X-linked mental retardation334 | |||||||
| PHF2/KDM7C | Breast carcinoma306 | - | ||||||
| H3K27 | EZH1/KMT6B | — | UTX/KDM6A | Kabuki syndrome50 | CBX7 | Chromo | Transcriptional depression of PRC complex target gene.180 | Involved in glioma cell growth and migration335 |
| EZH2/KMT6A | Prostate cancer336 | UTY/KDM6C | Prostate cancer337 | EED | WD40 | Spreads H3K27 methylation162 | Colorectal cancer338 | |
| JMJD3/KDM6B | Hodgkin’s lymphoma50 | BAHD1 | BAH | Facultative heterochromatin formation339 | — | |||
| KIAA1718/KDM7A | — | NSD2 | PWWP | Transcriptional elongation340 | Wolf–Hirschhorn syndrome,186 myeloma207 | |||
| PHF8/KDM7B | X-linked mental retardation334 | |||||||
| H3K36 | SETD2/KMT3A | Renal cell carcinoma341 | JHDM1A/KDM2A | — | DNMT3A | PWWP | DNA methylation195 | AML342 |
| NSD1/KMT3B | Sotos syndrome,205 myelodysplastic syndrome,343 AML,206 neuroblastoma344 | JHDM1B/KDM2B | Leukemia,345 bladder carcinoma346 | LEDGF | PWWP | DNA repair (HR)201 | Medulloblastoma,347 prostate cancer348 | |
| NSD2/KMT3G | Wolf–Hirschhorn syndrome,186 myeloma207 | JHDM3A/KDM4A | Bladder and breast cancers325, 326 | NBS1 | — | DNA repair (NHEJ)200 | Gastric cancer349 | |
| NSD3/KMT3F | Breast cancer,350 AML,351 myelodysplastic syndrome352 | JHDM3B/KDM4B | Peripheral nerve sheath tumor330 | Ku70 | — | DNA repair (NHEJ)200 | Colorectal cancer353 | |
| SMYD2/KMT3C | Pancreatic and gastric cancers307, 308 | JHDM3C/KDM4C | Esophageal cancer,331 breast cancer,332 medulloblastoma,333 lymphoma330 | MRG15 | Chromo | RNA splicing139 | — | |
| ASH1L/KMT2H | Leukemia354 | JHDM3D/KDM4D | — | ZMYND11 | PWWP | RNA splicing203 | Breast cancer355 | |
| SETD3 | B-cell lymphoma356 | |||||||
| SETMAR | AML357 | |||||||
| H3K79 | DOT1L/KMT4 | MLL-rearranged leukemia245, 251, 252, 253, 254 | — | — | TP53BP1 | Tudor | DNA damage response232 | B-cell lymphoma,358 breast cancer359, 360 |
| H4K20 | SET8/KMT5A | Breast cancer361 | PHF8/KDM7B | X-linked mental retardation334 | TP53BP1 | Tudor | DNA damage response276 | B-cell lymphoma358 breast cancer359, 360 |
| SUV4-20H1/KMT5B | — | PHF2/KDM7C | Breast carcinoma306 | L3MBTL1 | MBT | Chromatin compaction279 | — | |
| SUV4-20H2/KMT5C | Liver and lung cancers290, 362 | LSD1n | — | ORC1 | BAH | Replication282, 283 | Microcephalic primordial dwarfism363 | |
| ORCA | WD repeat | Replication283 | — | |||||
| Pdp1 | PWWP | Chromatin localization of H4K20 methyltransferase364 | — |
Abbreviations: AML, acute myeloid leukemia; HR, homologous recombination; MLL, mixed-lineage leukemia 1; NHEJ, non-homologous end joining; NURF, nucleosome remodeling factor; PRC, polycomb repressive complex; TFIID, transcription factor IID.
H3K4 methylation
H3K4 methyltransferases
H3K4 methylation is an evolutionarily conserved histone modification that marks active transcription and is highly enriched at the promoter region and transcription start site.19 In yeast, all H3K4 methylations are carried out by Set1 methyltransferase, which forms a multisubunit Set1complex, also known as COMPASS, with seven other subunits: Swd1, Swd3, Bre2, Sdc1, Swd2, Spp1 and Shg1.20, 21, 22 Set1 contains a catalytic SET domain, in which H3K4 methyltransferase activity is assisted by associated Swd1, Swd3, Bre2 and Sdc1 subunits. The loss of individual Set1 complex subunits differentially affects Set1 stability, complex integrity, global H3K4 methylation level and distribution of H3K4 methylation along active genes.23 H3K4 methyltransferases are highly conserved from yeast to human. Drosophila melanogaster contain three Set1 homologs (SET1, TRX and TRR), whereas mammals have six such homologs (SET1A/KMT2F, SET1B/KMT2G, MLL1 (mixed-lineage leukemia 1)/KMT2A, MLL2/KMT2B, MLL3/KMT2C and MLL4/KMT2D).19 Each Set1 homolog, which functions as a scaffold protein within the complexes, associates with four common subunits (WRAD: WDR5 (WD repeat domain 5), RbBP5 (retinoblastoma-binding protein 5), ASH2L (absent, small or homeotic-2 like) and DPY30), as well as unique subunits that specify distinct functions.19
The association of WRAD with the SET domain of each SET1/MLL family protein (core complex) produces distinct enzymatic properties. The SET domain of MLL1 alone exhibits weak H3K4 monomethylation activity, but complex formation with WRAD allows it to predominantly mono- and dimethylate H3K4 in vitro.24 The interaction of WDR5 with the MLL1 SET domain is crucial for association of ASH2L and RbBP5 with the MLL1 SET domain and H3K4 dimethylation activity of the assembled MLL1 core complex.25, 26 Biochemical analyses with purified proteins have further shown that MLL1/2 core complexes can catalyze H3K4 mono- and dimethylation, whereas the specificity of MLL3/4 core complexes is restricted to H3K4me1.27 These observations indicate that WRAD proteins participate differentially in H3K4 methylation process in each SET1/MLL complex. How SET1/MLL family complexes differentially regulate H3K4 methylation is not precisely known. One possible mechanism is that distinct amino acids within the SET domain of each SET1/MLL proteins, in conjunction with WRAD proteins, create different active sites that differentially modulate H3K4 methyltransferase activity.
In addition to WRAD proteins, unique subunits in each SET1/MLL complex also have roles in regulating H3K4 methylation. For example, it has been shown that WDR82 (WD repeat domain 82) and CFP1 (CXXC finger protein 1) are required for appropriate levels of SET1A/B complex-mediated H3K4 trimethylation.28, 29 More specifically, CFP1 directly binds to unmethylated CpG islands through its CXXC domain and regulates the genome-wide distribution of H3K4me3 in ESCs.30, 31 H3K4me3 levels in specific genes are dependent on the MLL1/2-specific subunit menin during initiation and progression of sporadic pancreatic endocrine tumors.32 PTIP (Pax transactivation domain-interacting protein), a unique subunit of MLL3/MLL4, has been shown to regulate H3K4me3 levels at the Ntrk3 (neurotrophic tyrosine kinase receptor, type 3) locus, whose function is important for podocyte foot process patterning.33 These results indicate that unique subunits in SET1/MLL complexes interact with distinct transcription factors and thus have important roles in the expression of specific target genes for given SET1/MLL complexes.
Structural analyses have recently begun to aid our understanding of the detailed molecular mechanism of H3K4 methylation. A cryo-EM analysis of partial yeast Set1 (SET domain plus Swd1, Swd3, Bre2 and Sdc1) and human MLL1 (SET domain plus WRAD) complexes revealed that the two subunits, Swd1 (RbBP5) and Swd3 (WDR5), are positioned in the top lobe of the Y-shaped structure, whereas Bre2 (ASH2L) and Sdc1 (DPY30) occupy the bottom base.34 A recent X-ray crystallographic analysis of human MLL1-SET/RbBP5/ASH2L and MLL3-SET/RbBP5/ASH2L complexes found that association of heterodimeric RbBP5/ASH2L with the MLL SET scaffold stabilizes the catalytic SET domain and further showed that substrate binding induces a conformational change in the active site that facilitates H3K4 methylation.35 A structural understanding of H3K4 methyltransferase complexes has only begun to be established. Notably, difficulties in biochemical purification of high-molecular-weight and multisubunit complexes have hampered structural analyses of any holo-H3K4 methyltransferase complex. Continuing efforts should ultimately afford a detailed understanding of the mechanism of action of H3K4 methyltransferase complexes.
H2B ubiquitylation-dependent H3K4 methylation in yeast was the first-discovered histone trans-tail relationship, where H3K4 di- and trimethylation were shown to require prior monoubiquitylation at lysine 123 (corresponding to lysine 120 in mammalian cells) of histone H2B.36, 37, 38, 39 Following up on this interesting relationship, two groups reported that Swd2 is a key player in this process, although through different mechanisms.40, 41 However, this assertion was challenged by a biochemical analysis showing that the Set1 complex lacking Swd2 exhibits even higher H2B ubiquitylation-dependent H3K4 methylation activity.42 Instead, the authors of this latter study showed that the n-SET domain within Set1 is essential for H2B ubiquitylation-mediated H3K4 methylation activity of the Set1 complex.42 The H2B ubiquitylation dependence of human H3K4 methyltransferases is unclear, because reducing H2B ubiquitylation by knocking down the human homologs of Bre1 (BRE1A/RNF20 and BRE1B/RNF40) results in only a partial decrease in H3K4 methylation in human cells.43, 44, 45 The incompleteness of this decrease is probably attributable to inefficient knockdown of Bre1 proteins. Interestingly, however, these studies also suggest that six human H3K4 methyltransferase complexes may differentially require H2B ubiquitylation for their H3K4 methylation activity.
H3K4 demethylases
Until the identification of the FAD (flavin adenine dinucleotide)-dependent nuclear amine oxidase LSD1 (also known as KDM1A), the first histone H3K4 demethylase discovered,9 histone methylation was believed to be stable and inheritable. LSD1, and the related LSD2/KDM1B, can demethylate H3K4me1 and H3K4me2.9, 46 It has been shown that LSD1 is recruited to target genes by CoREST-, BHC80- and SFMBT1-containing repressive complexes and members of the zinc-finger transcription factor family, Snail.47, 48, 49 In addition to amine oxidases, JARID1 (jumonji AT-rich interactive domain-1) family proteins (JARID1A/KDM5A, JARID1B/KDM5B, JARID1C/KDM5C and JARID1D/KDM5D) and the JmjC domain-containing protein NO66 (also known as MAPJD)50 were found to demethylate H3K4. NO66 is able to demethylate all three states of H3K4 methylation using α-ketoglutarate and Fe(II) as cofactors,51 whereas JARID1A, JARID1B, JARID1C and JARID1D were shown to be specific for demethylation of H3K4me2 and H3K4me3.52, 53, 54, 55 These proteins function as transcriptional corepressors by demethylating H3K4 or recruiting other corepressors.56, 57
Distribution of H3K4 methylation
Gene expression is associated with the position of histone methyl-lysine residues within genes and their degree of methylation. H3K4me1, H3K4me2 and H3K4me3 have been shown to differentially mark actively transcribing genes. H3K4me1 is highly enriched at enhancers, H3K4me2 is highest toward the 5′ end of transcribing genes, and H3K4me3 is a hallmark of the promoters of actively transcribing and poised genes.58, 59, 60, 61 Although the strong correlation between H3K4 methylation and active transcription is well documented, how SET1/MLL methyltransferase complexes are recruited to specific gene loci is still an open question. However, it has been shown that SET1/MLL complex subunits HCF-1 (host cell factor-1) and menin are required for proper recruitment of these complexes to herpesvirus immediate early promoters and HOX genes, respectively.62, 63 In addition, cell-type-specific transcription factors and cofactors were also shown to mediate recruitment of H3K4 methyltransferases. For example, the transcription factors bZIP28 and bZIP60 bring SET1/MLL complexes to endoplasmic reticulum stress-responsive genes through interactions with Ash2 and WDR5a.64 A direct interaction with p53 was shown to be responsible for recruitment of the SET1 complex to DNA damage-responsive genes.65 In addition, it was reported that the Paf1 transcription elongation complex mediates interactions between Set1 and the C-terminal domain of RNA polymerase II such that the Set1 complex can be recruited to transcribing genes.66
Transcriptional coactivators that recognize H3K4 methylation
Each state of H3K4 methylation recruits distinct downstream effectors containing specific ‘reader’ domains that further regulate gene expression. Several chromatin remodelers are known to read H3K4 methylation and participate in the regulation of gene expression. For example, the ATP-dependent chromatin-remodeling enzyme, CHD1, recognizes H3K4me2 and H3K4me3 through its two N-terminal chromodomains.67 In addition, BPTF (bromodomain PHD finger transcription factor), a subunit of the ATP-dependent chromatin remodeling complex NURF (nucleosome remodeling factor), was shown to interact with H3K4me3 via its PHD domain.68 As an example of a general transcription factor that binds to H3K4 methylation, transcription factor IID was shown to be recruited to H3K4me3 through its PHD domain-containing TAF3 subunit, resulting in more efficient preinitiation complex formation.69, 70
The activity of several histone-modifying enzymes is modulated by recognition of H3K4 methylation. For instance, the yeast SAGA complex binds to H3K4me2 or H3K4me3 through its Sgf29 subunit, which contains a C-terminal H3K4me2- or H3K4me3-binding tudor domain, and thus efficiently acetylates neighboring histones.71 The acetyltransferase activity of the NuA3 histone acetyltransferase complex can be efficiently targeted to H3K14 through recognition of H3K4me3 by the Yng1 subunit, which contains a PHD domain.72 In addition, the HBO1 histone acetyltransferase complex was reported to acetylate histone H3 in a manner that depends on the PHD domain-containing ING4 (inhibitor of growth family member 4) subunit and facilitates apoptosis by enhancing the expression of genotoxic stress-responsive genes.73
In addition, H3K4 methyl-binding domains within H3K4 methyltransferase complexes further contribute to the precise regulation of their enzymatic activities. For example, the PHD domain-dependent binding of CFP1 to H3K4me3 is responsible for recruitment of SET1A/B complexes to H3K4me3-containing chromatin regions.74 CFP1 was also found to be required for deposition of H3K4me3 near the promoters of DNA damage-responsive genes in ESCs.75 Among MLL family H3K4 methyltransferases, only MLL1 is recruited through its PHD domain to regulate HOX gene expression. The roles of PHD domains within other MLL family proteins remain to be characterized.76, 77
H3K4 methylation and cancers
Mutations in H3K4 methyltransferases highly increase the susceptibility to various cancers.78 About 70% of infant leukemia is related to chromosomal translocation of MLL1 genes, which results in fusion of its N-terminal fragment to more than 50 partner proteins. MLL1 translocations are also frequently found in mixed-lineage leukemia, acute lymphoblastic leukemia and acute myeloid leukemia.79 Interestingly, aberrant H3K4 methylation caused by MLL1-AF9 fusion proteins was shown to require an intact MLL1 protein.80 Therefore, considerable effort has been devoted to developing inhibitors that target MLL1 as a cancer therapy strategy. Although inhibitors that directly target the MLL1 SET domain have not yet been discovered, the chemical compounds MM102 and MM-401, which specifically disrupt the interaction between WDR5 and MLL1, but not other SET1/MLL family methyltransferases, has been reported to inhibit proliferation of leukemia cells.81, 82
Mutations in H3K4 demethylases are also related to a number of diseases. JARID1 family proteins often function as transcriptional corepressors that are important for expression of development-related genes. JARID1A was shown to be important in regulating HOX gene expression in Caenorhabditis elegans.52 It has also been reported that cryptic fusion of NUP98 (nucleoporin 98) and JARID1A causes HOXA/B gene overexpression and results in pediatric acute leukemia in humans.83 Increased expression of JARID1B, often found in breast carcinomas and testicular cancer, causes misregulation of 14-3-3σ, BRCA1 (breast cancer 1, early onset), CAV1 (caveolin 1) and HOXA5 (homeobox A5) genes.53 X-linked mental retardation patients have a large number of sense or missense mutations in JARID1C genes, implying an important role for the encoded protein during brain development.84 In addition, a number of inhibitors have been developed for LSD1, which is involved in embryonic development and hematopoiesis, and many types of cancer. Some of these inhibitors were designed to irreversibly deactivate LSD1 by forming a covalent adduct with the cofactor FAD within LSD1.85
H3K4 and H3K27 methylations and bivalent domains in ESCs
Bivalent chromatin has an important role in regulating changes in gene expression from poised to active or inactive states in ESCs.15 In bivalent promoters, coenrichment of active H3K4me3 and repressive H3K27me3 marks was shown be responsible for differentiation into specific cell types.61, 86 It was further found that MLL2 is mainly responsible for H3K4 methylation on bivalent promoters.87, 88 H3K27 methylation is an abundant modification that is crucial for fate determination in ESCs. In embryonic fibroblast cells, H3K27me3 was shown to be highly enriched at the promoters of thousands of genes that are responsible for embryonic development and differentiation.89 Subunits of the H3K4 methyltransferase complex also participate in ESC fate determination. For example, WDR5 interacts with Oct4 (octamer-binding transcription factor 4) and mediates H3K4 methylation at key development loci in ESCs.90 During ESC differentiation, ASH2L downregulation correlates with decreased expression of pluripotent transcription factors and increased expression of differentiation-related genes.91 In addition, a specific role for DPY30 in the differentiation of ESCs, but not the maintenance of their self-renewal capacity, has also been reported.92
H3K9 methylation
H3K9 methyltransferases
H3K9 methylation is a histone modification that is a well-known indicator of silenced transcription and heterochromatin structure.86 Fission yeast has a single H3K9 methyltransferase (Clr4/KMT1) that is responsible for all three states of H3K9 methylation93 and regulates silencing at pericentromere and mating-type loci.94, 95 In mammalian cells, several H3K9 methyltransferases—SUV39H1/KMT1A, SUV39H2/KMT1B, SETDB1/KMT1E, dimeric G9a/KMT1C-GLP (G9a-like protein)/KMT1D and PRDM family—with different catalytic activities and target genes, have roles in diverse cellular events.96 SUV39H1 and SUV39H2 catalyze H3K9 di- and trimethylation in constitutive heterochromatin, including the pericentromeric region.7, 97 Recombinant SUV39H1/2 proteins were shown to possess H3K9 mono-, di- and trimethylation activity;98 however, a cell line lacking SUV39H1/2 proteins was shown to lose H3K9me2 and H3K9me3, but not H3K9me1, marks.99 SETDB1 catalyzes H3K9 monomethylation at the pericentromeric region and provides a substrate for SUV39H1/2 to produce H3K9me3.100 Another H3K9 methyltransferase, a heterodimer of G9a and GLP (G9a-GLP), mono- and dimethylates H3K9 in euchromatin regions to repress gene expression.101 When either G9a or GLP is deleted, H3K9me1 and H3K9me2 levels are reduced in euchromatin. Interestingly, in vitro analyses have shown that G9a and GLP can individually form homodimers that exhibit H3K9 mono-, di- and trimethylation activity.98, 102 In a related observation, the multi-zinc-finger-containing protein Wiz interacts with the G9a-GLP heterodimer and stabilizes its conformation,103 indicating that Wiz-assisted G9a-GLP heterodimer formation is crucial for modulation of G9a-GLP enzymatic activity in vivo.104 Several PRDM (PRDI-BF1 and RIZ homology domain) proteins also contribute to H3K9 methylation. Among 17 members of the PRDM family (PRDM1–17), all of which contain a PR domain similar to the SET domain,105 several are known to possess intrinsic H3K9 methyltransferase activity, whereas the remaining members regulate H3K9 methylation by interacting with other H3K9 methyltransferases, such as G9a.106 However, whether PRDM protein-mediated regulation of H3K9 methylation is direct or indirect is still controversial, and more detailed biochemical studies are necessary to clarify this issue.
H3K9 demethylases
Three classes of mammalian proteins—JHDM2/KDM3, JHDM3(JMJD2)/KDM4 and PHF8/KDM7—have H3K9 demethylation activity. Three JHDM2 (jumonji domain-containing histone demethylase-2) family proteins (JHDM2A–C) have the ability to demethylate H3K9me1 and H3K9me2107 and regulate hormone-dependent transcriptional activation.108 JHDM3 family proteins can demethylate H3K9me2 and H3K9me3 in addition to H3K36me2 and H3K36me3 in vitro.107, 108, 109 PHF8, a member of the PHF (PHD finger) protein family that acts as a demethylase for H3K9me1 and H3K9me2,110 is a mononuclear Fe(II)-dependent hydroxylase that uses 2-oxoglutarate and oxygen as cosubstrates.111 Because PHF8, like other members of the PHF family, contains a PHD domain, it preferentially removes H3K9 methylations from H3K4me3-containing histone peptide substrates.112 This activity would largely account for the mutually exclusive distribution of H3K4 and H3K9 methylations.
Cross-talk between H3K9 methylation and DNA methylation
A number of studies have reported physical and functional interactions between H3K9 methyltransferases and DNA methyltransferases (DNMTs). For example, SUV39H1/2 and DNMT3A/B interact which each other and can be recruited via their interaction with HP1 (heterochromatin protein 1) to methylate H3K9-enriched constitutive heterochromatin regions, thereby reinforcing the condensed chromatin structure.113, 114 In addition, DNMT3A/B interacts with G9a-GLP and is involved in facultative heterochromatin formation in ESCs.115, 116 Moreover, the fact that G9a, DNMT1 and PCNA (proliferating cell nuclear antigen) are colocalized at the replication fork117 implicates H3K9 methylation in the maintenance of DNA methylation during DNA replication. UHRF1 (ubiquitin-like, containing PHD and ring finger domain 1) also has been reported to have a role in maintaining DNA methylation by bringing DNMT1 to the replication fork through its interaction with methylated H3K9, hemimethylated CpG and DNMT1.117, 118 Furthermore, the methyl-CpG-binding protein MBD1 was shown to recruit SETDB1 to the chromatin assembly complex CAF-1, facilitating SETDB1-mediated methylation of H3K9 on newly deposited nucleosomes.119
Establishment of pericentromeric heterochromatin
SETDB1 and SUV39H1/2 are recruited to pericentromeric heterochromatin and catalyze H3K9 methylation.7, 100, 120 HP1α and HP1β bind to H3K9me3 through their chromodomains and form multimers that interact with SUV39H1/2. As SUV39H1/2 also contains a chromodomain, they can be further recruited to methylated H3K9 at the pericentromeric region by HP1α/β as well as by themselves.97, 121 These multiple interactions enable SUV39H1/2 to spread H3K9me3 to neighboring nucleosomes.122 HP1α/β contributes to heterochromatin formation by recruiting many other proteins involved in heterochromatin formation, such as histone deacetylase, transcriptional repressors and chromatin remodelers.123, 124, 125 H3K9 methylation also increases nucleosome occupancy and has an important role in maintaining transposon repression in pericentromeric heterochromatin.126 In fission yeast and plants, heterochromatin formation mediated by interactions of methylated H3K9 with the RNA interference machinery has been well established.127, 128
H3K9 methylation-mediated transcriptional repression
The G9a-GLP heterodimer deposits H3K9me1 and H3K9me2 and represses target gene expression.101 G9a-GLP can be recruited to target gene promoters through direct interactions with diverse DNA-binding proteins.103, 129, 130, 131 Once it binds and methylates H3K9 on target genes, G9a-GLP recruits additional dimers through its ankyrin repeat domain and spreads H3K9me1 and H3K9me2 to neighboring nucleosomes.101 The resulting repression of gene expression is crucial in many biological processes, such as memory formation, immune responses and differentiation.132, 133, 134 During ESC differentiation, G9a/GLP-mediated facultative heterochromatin formation silences Oct3/4 and Nanog.101 Furthermore, G9a-GLP regulates H3K9me2 and represses different sets of genes in a tissue-specific manner. G9a/GLP-mediated silencing also represses non-neuronal genes in neurons135 and skeletal muscle genes in brown adipose tissue.136 In the immune system, H3K9me2 inhibits uncontrolled interferon induction and regulates naïve T-helper cell differentiation.133 Moreover, H3K9me2 is dynamically altered in the hippocampus and entorhinal cortex in response to contextual fear conditioning, and mediates memory formation.132 These observations indicate that the distribution of H3K9me2 in euchromatin is dynamically altered according to cell type and external stimuli. Collectively, the results of these studies imply that G9a-GLP interacts with a large number of DNA-binding proteins in different contexts. Numerous studies have revealed that diverse biological processes are regulated by H3K9 methylation. However, only a few downstream target genes, and no upstream regulators of H3K9 methyltransferases, have been identified. Clarifying the detailed molecular mechanisms by which H3K9 methyltransferases mediate transcriptional repression will require a greater effort to identify upstream regulators that bring H3K9 methyltransferases to target genes.
Interestingly, it has been reported that H3K9me3 and HP1γ are enriched in the coding region of certain active genes in several cell lines.137 In contrast to HP1α/β, HP1γ recruits elongating RNA polymerase II and induces gene expression.137 Consistent with this, the D. melanogaster HP1γ homolog, HP1c, recruits the histone chaperone FACT to RNA polymerase II and enhances transcription elongation of heat-shock genes.138
H3K9 methylation and alternative splicing
Alternative splicing is regulated by nucleosome occupancy and post-translational modifications in transcribing genes.139, 140 A recent study has revealed that local increases in H3K9me2 and H3K9me3 enhance exon inclusion, whereas H3K9 demethylation correlates with exon skipping. Moreover, a genome-wide study showed that H3K9me2 and H3K9me3 are enriched in internal exons.141 Although how H3K9 methylation facilitates exon inclusion is not fully understood, two hypotheses have been proposed. First, H3K9 methylation-bound HP1γ could recruit the splicing regulatory protein SRSF1 for efficient splicing.142 Alternatively, H3K9 methylation would increase nucleosome occupancy and slow down RNA polymerase II elongation, prolonging the time for RNA splicing.143, 144
H3K9 methylation-related diseases
H3K9 methylation is often misregulated in various diseases, such as neurodegenerative diseases, drug addiction and cancer.145, 146 In a mouse Alzheimer disease model, aberrantly increased H3K9 methylation levels in the BDNF (brain-derived neurotrophic factor) gene leads to downregulation of BDNF expression in neurons. BDNF is critical for synaptic plasticity; hence, reduced BDNF levels are thought to be an important contributor to the pathogenesis of Alzheimer disease.147 In addition, overexpression of SETDB1 and elevated levels of H3K9 methylation are often found in Huntington disease patients.148 Although a correlation between abnormal H3K9 methylation and neurodegenerative diseases is well established, more studies are required to identify target genes through which misregulated H3K9 methylation causes these diseases.
Uncontrolled H3K9 methylation is also linked to cancer. In particular, G9a is often overexpressed in various types of cancer.149, 150, 151 Overexpression of G9a hypermethylates H3K9 on tumor suppressor genes and thus represses their expression. The tumor suppressors, DSC3 (desmocollin 3) and MASPIN (mammary serine protease inhibitor) in breast cancer152 and CDH1 (cadherin 1), DUSP5 (dual specificity phosphatase 5) and SPRY4 (sprouty homolog 4) in ovarian cancer,149 are often silenced by G9a. These results suggest that H3K9 methyltransferases could be good therapeutic targets in cancer treatment.
H3K27 methylation
H3K27 methyltransferases
H3K27me3 is a hallmark of transcriptional repression. The EZH2 (enhancer-of-zest homolog 2) subunit (also known as KMT6A) within PRC2 (polycomb repressive complex 2) complex, an evolutionarily conserved class of polycomb group proteins, is an H3K27 methyltransferase153 responsible for all three states of H3K27 methylation.154, 155, 156 The mammalian PRC2 complex is composed of four core subunits: EZH1/2, SUZ12, EED and RbAp46/48.157, 158 PRC2 can also associate with other accessory proteins, such as AEBP2 (AE binding protein 2), JARID2 and PCLs (polycomb-like proteins).159 These accessory proteins are thought to be involved in recruiting PRC2 to target genes and regulating its activity.158 EZH1/2 alone has no enzyme activity, but incorporation into a PRC2 complex with other subunits enables it to methylate H3K27.160, 161 The PRC2 subunits SUZ12 and EED have the ability to bind the histone H3 N-terminal tail and H3K27me3, respectively. Thus, a positive feedback mechanism is used to spread H3K27me3-repressive marks to adjacent gene loci.162, 163, 164
H3K27 demethylases
UTX/KDM6A, UTY/KDM6C and JMJD3/KDM6B demethylate H3K27me2 and H3K27me3 and are primarily involved in gene derepression.165, 166, 167 UTX contains six TPR (tetratricopeptide repeat) domains and one JmjC domain. The evolutionarily conserved TPR domain mediates multisubunit complex assembly.165 UTX, one of the subunits in the MLL4 H3K4 methyltransferase complex, mediates crosstalk between H3K4 and H3K27 methylations.168 Interestingly, UTX target genes responsible for cancer proliferation and invasiveness are also regulated by MLL4, as evidenced by the fact that individual knockdown of UTX or MLL4 phenocopies the traits of breast cancer cells.169 JMJD3, a JmjC-domain-containing protein that catalyzes demethylation of H3K27me2 and H3K27me3,165 activates transcription of development-related genes and directs differentiation of ESCs into definitive endoderm.170 Microbial stimuli cause nuclear factor-κB-mediated enrichment of JMJD3 at the transcription start site of lipopolysaccharide-responsive genes in macrophages.171 PHF subfamily proteins, including KIAA1718/KDM7A and PHF8, also demethylate H3K27 using α-ketoglutarate and iron as cofactors. KIAA1718 contains PHD and JmjC domains, which allow selective demethylation of H3K27me2 on H3K4-trimethylated nucleosomes.112 The JmjC domain-containing PHF8 is specific for H3K27me2.110
Maintenance of gene repression
The PRC2 subunit EED also binds to H3K27me3 through its WD40 domain, and disruption of this interaction leads to reduced H3K27 methylation and developmental defects.162 Binding of PRC2 to H3K27me3 spreads H3K27 methylation to neighboring nucleosomes, and thus has and important role in the maintenance of gene-expression status. Consistent with a repressive role of H3K27 methylation, the inactivated X chromosome in mammalian cells is highly enriched for H3K27 methylation, which stabilizes the inactive chromatin structure.172
H3K27 methylation and cancers
Misregulation of H3K27 methylation is associated with tumorigenesis as well as metastasis. In this context, overexpression of the PRC2 subunit EZH2 has been reported in human breast and prostate cancers as well as lymphoma.173, 174, 175, 176 For instance, an Y641F mutation in the EZH2 SET domain increases H3K27me3 levels and contributes to the pathogenesis of germinal center B-cell lymphomas.177 In addition, another EZH2 mutant lymphoma cell line harboring an A677G mutation shows elevated levels of H3K27me3 and decreased levels of H3K27me2 and H3K27me1. A structural study reported that the A677G mutation enlarges the lysine tunnel, enhancing the ability of PRC2 to catalyze H3K27 dimethylation.178 These results suggest that inhibition of hyperactive PRC2 could be a potential treatment strategy for specific types of lymphoma. In this regard, several EZH2 inhibitors that target B-cell and follicular lymphomas have been developed.17 For example, the EZH2 inhibitor GSK126 was shown to decrease global levels of H3K27 methylation, reactivate PCR2 target genes, and decrease tumor progression in a mouse model.179
H3K27 methylation can be recognized by chromodomain- and WD40 domain-containing proteins. Among several chromodomain-containing proteins that bind to H3K27 methylation, the PRC1 complex subunit CBX7 (chromobox 7) has received attention as a therapeutic target owing to its involvement in tumorigenesis as well as stem cell self-renewal and differentiation. The recently developed chemical compound, MS37452, binds the methyl-binding pocket of the chromodomain of CBX7, resulting in transcriptional derepression of PRC1 complex target genes and inhibition of the proliferation of prostate cancers.180
H3K36 methylation
H3K36 methyltransferases
In yeast, a single H3K36 methyltransferase, Set2/KMT3, catalyzes all three states of H3K36 methylation.181 The SRI (Set2 Rpb1 interacting) domain in Set2 enables it to interact with the S2- and S5-phosphorylated C-terminal domain of RNA polymerase II and methylate H3K36 during transcriptional elongation.181 Mammalian cells contain at least eight H3K36 methyltransferases: NSD1/KMT3B, NSD2/KMT3G, NSD3/KMT3F, SETD2/KMT3A, SETD3, SETMAR, SMYD2/KMT3C and ASH1L/KMT2H.182 Among these, NSD1–3 and SETD2 are considered major H3K36 methyltransferases. Only SETD2 can catalyze H3K36 trimethylation, whereas the methyltransferase activity of the other seven enzymes is restricted to H3K36 mono- and/or dimethylation.183 NSD (nuclear receptor-binding SET domain) enzymes have additional methylation sites on histones as well as non-histone target proteins.184 Biochemical analyses have shown that NSD enzymes lose their H3K36 specificity if histone octamers are used as a substrate instead of physiologically relevant nucleosomes.184 From a structural perspective, NSD1–3 enzymes have an autoinhibitory loop that blocks the substrate-binding site. Interaction of a short segment of nucleosomal DNA appears to interact with the autoinhibitory loop, dislodging it from the substrate-binding site. Therefore, H3K36 located close to the nucleosome core region can enter the substrate-binding site, thus imparting H3K36 specificity on NSD enzymes.185 The H3K36 trimethyltransferase SETD2 has important roles in many biological processes.186, 187, 188, 189 Although recombinant SETD2 was shown to generate all three states of H3K36 methylation in vitro, only H3K36me3 levels were found to decrease in SETD2-knockdown cells.186, 187 In addition, it was reported that not only H3K36me1 and H3K36me2 levels but also H3K36me3 levels are decreased in NSD1–3-deficient cells.190 These results suggest that NSD enzymes provide H3K36me1 and H3K36me2 to SETD2, which then subsequently generates H3K36me3 in vivo.
H3K36 demethylases
There are two H3K36 demethylase families in mammalian cells: JHDM1/KDM2A-B and JHDM3/JMJD2/KDM4A-D. The H3K36me1- and H3K36me2-specific demethylase, JHDM1, contains multiple histone-binding domains, including a PHD domain,10, 191 and it accounts for mutually exclusive distribution of H3K4me3 and H3K36me3 in same genes.60, 192 JHDM3 is specific for H3K36me2 and H3K36me3 demethylation and has also been shown to demethylate H3K9me2 and H3K9me3 in vitro.109, 111
Regulation of transcription initiation and elongation
H3K36 methylation has been implicated in preventing abortive initiation of transcription within the gene body and in regulation of transcription elongation. In yeast, Set2 binds to the phosphorylated C-terminal domain of RNA polymerase II and catalyzes H3K36 methylation in newly deposited nucleosomes.182 The RPD3 deacetylase complex is recruited to nucleosomes through recognition of H3K36me1 and H3K36me2, and then deacetylates histones. Local deacetylation maintains a repressive chromatin state that prevents aberrant transcription initiation.193 In mammalian cells, different players participate in this process. NSD3, LSD2 (lysine demethylase 2) and G9a form a complex that interacts with transcription-elongation factors and the S2-phosphorylated C-terminal domain of RNA polymerase II. This complex, in turn, maintains newly incorporated nucleosomes in a repressed state by methylating H3K9 and H3K36, and demethylating H3K4.194 Moreover, PWWP domain-containing NSD2 and DNMT3A bind to H3K36-methylated nucleosomes and further contribute to repression of aberrant transcription.194, 195 Collectively, these observations indicate that H3K36 methylation, H3K9 methylation and DNA methylation simultaneously accumulate in newly incorporated nucleosomes to control accurate transcription elongation in mammalian cells.
In addition to its preferential localization to the gene body, H3K36 methylation is also enriched in the promoter region of several genes.196 Promoter-enriched H3K36 methylation inhibits activity of the PRC2 complex and prevents PRC2-mediated expansion of H3K27 methylation.197 This observation is further supported by chromatin immunoprecipitation sequencing analyses showing a mutually exclusive distribution H3K27 methylation and H3K36 methylation.198
DNA damage responses
Several studies have demonstrated roles of H3K36 methylation in DNA damage repair. SETD2 induces DNA mismatch repair by catalyzing H3K36me3 at mismatch sites.199 In addition, the H3K36 dimethyltransferase, SETMAR, facilitates non-homologous end joining at DNA double-strand break (DSB) sites.200 When a DSB occurs, SETMAR is recruited to DSB sites and catalyzes H3K36 methylation. NBS1, a subunit of the MRN complex, and Ku70 recognize H3K36 methylation and stabilize DSB until other non-homologous end-joining -related proteins are recruited. Homologous recombination is also known to be regulated by H3K36me3.201 If a DSB occurs, the H3K36me3-interacting protein LEDGF/p75, also known as PSIP1 (PC4- and SFRS1-interacting protein 1), recruits CtIP (C-terminal-binding protein-interacting protein), which carries out DSB resection. RPA (replication protein A) and RAD51 are then recruited to the site and promote homologous recombination repair.201 In the absence of SETD2, homologous recombination repair cannot be performed properly because reduced H3K36me3 levels lead to dissociation of LEDGF from chromatin. This explains why SETD2 functions as a tumor suppressor. Homologous recombination repair is a very accurate DNA-repair process, and it is usually carried out in H3K36me3-enriched coding regions.183 Therefore, H3K36me3 seems to function as surveillance for DNA damage in coding regions to maintain genome stability.
H3K36 methylation and exon exclusion
Similar to H3K9 methylation, H3K36 methylation also has a role in alternative splicing.139 For example, SETD2-mediated H3K36 trimethylation stimulates exon exclusion.202 H3K36me3-bound MORF4L1 (mortality factor 4-like 1; also known as MRG15) recruits PTB (polypyrimidine tract binding protein), which is a well-known exon-inclusion repressor.139 In addition, a recent study showed that H3.3K36me3-recognizing ZMYND11 protein causes large-scale intron retention.203 These studies imply that H3K36me3 exclusively marks exons in the alternative splicing process. In support of this, H3K36me3 levels are very low in intron-less genes.204 Alternative splicing is also regulated by the level of SETD2.189 Specifically, downregulation of SETD2 levels by polyubiquitylation mediated by the E3 ubiquitin ligase complex, SPOP/CUL3, reduces H3K36me3 on SETD2 target genes and induces their splicing alternatively. Taken together with studies on H3K9 methylation, these results indicate that various histone modifications participate actively in RNA splicing processes.
H3K36 methylation-related diseases
Misregulation of H3K36 methylation often leads to various diseases.182 A defect in NSD1 was reported to cause Sotos syndrome, a neurological disorder characterized by macrocephaly and cognitive and motor skill deficiencies.205 In addition, Ndh2-knockout mice die shortly after birth with symptoms of Wolf-Hirschhorn syndrome.186 These studies suggest that a deficiency of H3K36 methyltransferases disrupts neuronal development, although additional investigation will be required to elucidate the detailed molecular mechanism. The NSD genes also often function as oncogenes when overexpressed or translocated to other genes.206, 207 For instance, a fusion protein caused by joining of the NSD1 gene with the NUP98 gene causes acute myeloid leukemia.208 This NSD1–NUP98 fusion protein facilitates H3K36 methylation and induces inappropriate activation of HOX genes.206 In addition, NSD2 overexpression induces multiple myelomas by promoting the expression of several oncogenes, such as TGFA (transforming growth factor alpha), MET and p21.207 Thus, their causal role in such distinctive diseases strongly suggests that NSD1 and NSD2 have different downstream target genes, despite sharing a common methylation site on histones. Moreover, the fact that a loss-of-function mutation of SETD2 causes renal cell carcinoma development implies that SETD2 acts as a tumor suppressor209, 210 reflecting its roles in DNA repair and alternative splicing.
H3K79 methylation
H3K79 methyltransferases
Unlike other histone lysine methylations, which are located on unstructured histone tail domains and are catalyzed by SET domain-containing methyltransferases, H3K79 methylation occurs on the globular domain of histone H3 and is mediated by Dot1, which lacks a SET domain.211 Dot1 was originally identified as a disruptor of telomeric-silencing genes in yeast,212 and subsequent genetic and biochemical studies showed that Dot1, as well as evolutionarily conserved homologs, including human DOT1L (DOT1-like)/KMT4, are responsible for all three states of H3K79 methylation.213, 214, 215, 216, 217
On the basis of structural studies, Dot1 homologs are categorized as a class I S-adenosyl methionine-dependent methyltransferase.218, 219 Despite structural similarities between human DOT1L and the arginine methyltransferase PRMT1,211 Dot1 homologs exhibit methyltransferase specificity toward lysine rather than arginine.213, 214, 220 In this context, it will be interesting to determine whether Dot1 homologs methylate arginine rather than lysine residues, if present, on novel target proteins.
Intriguingly, in vitro histone methyltransferase assays have demonstrated that Dot1 proteins preferentially methylate nucleosomal substrates rather than free histone H3.214, 220, 221 In addition, it has been shown that a positively charged region within human DOT1L (amino-acid residues 390–407) is required for direct interaction with nucleosomes and histone methyltransferase activity.218 These observations suggest that a newly created surface on the nucleosome produced by participation of histones and DNA provides an environment preferential for recognition and methylation by Dot1 proteins.
Another interesting feature of H3K79 methylation, like that of H3K4 methylation, is the trans-tail histone modification relationship with H2B ubiquitylation. Yeast genetic studies have demonstrated that defects in H2B ubiquitylation cause the complete disappearance of H3K79me2 and H3K79me3.39, 222, 223 Importantly, biochemical approaches have further shown that H2B-ubiquitylated recombinant nucleosomes serve as a preferential substrate for human DOT1L,217, 224 indicating that H2B ubiquitylation directly stimulates the H3K79 methylation activity of human DOT1L. The Muir group has made a number of important observations that have helped elucidate the molecular mechanism underlying this interesting trans-tail histone modification. An enzyme kinetic analysis suggested that human DOT1L undergoes a conformational change in the presence of H2B-ubiquitylated nucleosomes.217 In addition, mutation analyses of the surface of ubiquitin linked to nucleosomes have suggested communication of specific amino-acid residues on ubiquitin with human DOT1L.225 Furthermore, a targeted photocrosslinking study provided evidence that H2B ubiquitylation ‘corrals’ human DOT1L into an H3K79-proximal orientation.226 Collectively, these studies support the proposition that installation of ubiquitin on nucleosomes converts dominant, but unproductive, interactions between DOT1L and the nucleosome into less dominant, but productive, interactions that allow H3K79 methylation.
In addition to H2B ubiquitylation, another trans-tail mechanism for regulating H3K79 methylation has been reported. An in vitro analysis showed that a direct physical interaction between Dot1 and the histone H4 N-terminal tail is required for Dot1-mediated H3K79 methylation.227 In a related observation, increased H4K16 acetylation induced by SAS2 (something about silencing 2) overexpression was shown to cause upregulation of H3K79 methylation by inhibiting the interaction between Sir3 (silent information regulator 3) and the histone H4 tail.227 Collectively, these observations suggest that, not only are structural features of the nucleosome important, surrounding histone modifications also affect the overall H3K79-methylation activity of Dot1.
Unidentified H3K79 demethylase
Although many demethylating enzymes responsible for histone lysine methylations have been reported, the reversibility of H3K79 methylations has not been clarified. The fact that H3K79me2 is reduced to a lesser extent on non-replicating extrachromosomal DNA than on chromosomal loci suggests that removal of H3K79 methylation merely depends on replication-dependent histone exchange.228 However, several studies have shown that global H3K79 methylation levels change dynamically during the G1/S transition.214, 229 The observed disappearance of H3K79 methylation in the G2 phase also suggests the existence of a demethylation process. Furthermore, treatment of U-87MG cells with the demethylase inhibitor 2-hydroxyglutarate was shown to result in increased H3K79 methylation, strongly suggesting the presence of an H3K79 demethylase.230 Given that Dot1 homologs preferentially methylate nucleosomal substrates over free histone H3, it is possible that an unidentified H3K79 demethylase may also target only chromatinized H3K79 methylation. These observations argue for the need for a biochemical approach using H3K79-methylated nucleosome substrates to identify the H3K79 demethylase.
Disruption of telomeric silencing
Dot1 was initially identified as a disrupter of telomeric silencing.212 Sir proteins, important players in heterochromatin spreading, are mislocalized to the telomeric region following mutation of H3K79 or overexpression or deletion of Dot1.213, 220 More direct regulatory evidence was provided by Côté and co-workers,227 who showed that an H3K79-methylated peptide exhibits reduced binding to Sir3. Furthermore, a yeast genetic study showed that Dot1 deletion stimulates efficient binding between nucleosome and Sir3.231 These studies imply that proper H3K79 methylation level is crucial for regulation of heterochromatin formation and stability.
DNA damage responses
Dot1/DOT1L and H3K79 methylation also serve important functions in DNA damage responses. Recruitment of human TP53BP1 (tumor protein p53 binding protein 1) to DSBs is dependent on H3K79 methylation.232 Furthermore, DOT1 deletion or H3K79 mutations cause defects in G1/S checkpoint arrest following UV irradiation, ionizing radiation or treatment with a genotoxic agent.233, 234, 235 On the other hand, H3K79 methylation inhibits cell survival following treatment with the alkylating agent, methyl methanesulfonate, by inhibiting translesion synthesis, thereby preventing the bypass of DNA lesions.236, 237 Collectively, these observations support the conclusion that H3K79 methylation has a critical role in DNA damage responses; however, further investigation is necessary to elucidate the molecular details of the function of H3K79 methylation in these processes.238
Transcription elongation
H3K79 methylation is a well-known histone modification that strongly correlates with active transcription. Numerous studies have shown that H3K79me2 and H3K79me3 are enriched in coding regions of actively transcribed genes in many organisms.39, 239, 240, 241, 242 This preferential localization implicates H3K79 methylation in transcription elongation.241, 243, 244 In addition, DOT1L is often associated with the transcription-elongation factor, ENL (eleven-nineteen leukemia), in some human cancer cell lines, and knockdown of ENL in HEK293 cells decreases H3K79 methylation and transcription-elongation efficiency.245 Biochemical approaches in mammalian cells have further shown that DOT1L is present in a number of complexes that also contain transcription-elongation factors.246, 247, 248 The presence of various DOT1L-containing complexes suggests that DOT1L may regulate transcription elongation in certain subset of genes through distinct mechanisms.
DOT1L: a key player in MLL-rearranged leukemia
MLL-fusion proteins generated by chromosomal translocation are common causes of acute leukemia.249 Interestingly, DOT1L-interacting proteins such as AF10, also known as MLLT10 (myeloid/lymphoid or mixed-lineage leukemia translocated to 10), and ENL are often found to be fused to MLL in leukemia, which delivers DOT1L to MLL-regulated genes.245, 250 For example, H3K79me2 is enriched at the MLL target gene HOXA9 in MLL-AF10-transformed cells; as a result of this aberrant localization of DOT1L, expression of the HOXA9 gene and leukemic transformation are highly upregulated.250 Subsequent studies have demonstrated that DOT1L also causes CALM-AF10-, MLL-ENL-, MLL-AF4-, MLL-AF9- and MLL-AF10-mediated leukemogenesis through a similar mechanism.245, 251, 252, 253, 254 Therefore, misregulation of H3K79 methylation is thought to be a crucial cue in MLL-rearranged leukemic cells. In this context, regulation of the enzymatic activity of DOT1L is considered a potential therapeutic target in the treatment of leukemia. Several chemicals that specifically disrupt the interaction between the methyl donor S-adenosyl methionine and the hydrophobic cavity of DOT1L have been shown to inhibit tumor growth and increase animal survival rates.255, 256, 257
Direct role of H3K79 methylation in transcription
Despite the strong correlation between H3K79 methylation and active transcription, whether H3K79 methylation directly stimulates transcription is an unanswered question. A recent study using in vitro transcription assays revealed that transcription was markedly enhanced using a nucleosome template with fully methylated H3K79 compared with an unmethylated nucleosome, suggesting a direct stimulatory effect of H3K79 methylation on transcription.258 However, because the entire transcription unit, including the promoter region, was methylated in this in vitro study, the specific regulatory role of H3K79 methylation at each step of the transcription process needs to be further investigated. In addition, it would be interesting to test whether this stimulatory effect of H3K79 methylation is mediated by binding of as yet unidentified H3K79 methylation-specific reader proteins.
H4K20 methylation
H4K20 methyltransferases
H4K20 methylation is catalyzed by several enzymes whose activities are restricted to specific methylation states. The first identified H4K20 methyltransferase, SET8/KMT5A (also known as PR-SET7), is a monomethylation-specific enzyme.259, 260 H4K20me1 can be further methylated to H4K20me2 and H4K20me3 by SUV4-20H1/KMT5B and SUV4-20H2/KMT5C.261, 262, 263 From a structural perspective, an H4K20me1-containing peptide was shown to fit better into the SET domain of SUV4-20H2 compared with an unmethylated peptide.264 On the other hand, in vitro peptide methyltransferase assays have shown that the enzymatic activity of recombinant SUV4-20H2 protein is restricted to H4K20me2, suggesting the presence of additional factors that enable SUV4-20H2 to trimethylate H4K20 in vivo.264
Interestingly, H4K20 methylation is also regulated by a histone trans-tail mechanism. The C-terminal region of SUV4-20H2 interacts with HP1 protein, which recognizes H3K9 methylation.261 Thus, deletion of H3K9 methyltransferases (Suv39h1 and Suv39h2 in mouse; SU(VAR)3-9 in D. melanogaster) or HP1 causes decreased heterochromatic targeting of SUV4-20H2 and a global reduction in H4K20me3.261, 263 These results suggest that targeting of H4K20 methyltransferases to heterochromatin regions is modulated by H3K9 methylation.265 Moreover, it was found that p300-dependent H4K16 acetylation decreases SET8-mediated H4K20me1.260 Similarly, reduction of H4K20me3 by knockdown of SUV4-20H2 was shown to increase MOF1-dependent H4K16 acetylation and relieve promoter proximal pausing of RNA polymerase II.266
H4K20 demethylases
Several distinct demethylases are involved in the removal of specific H4K20 methylation states. PHD8/KDM7B was first identified as an H4K20me1-specific demethylase in a biochemical analysis using a nucleosomal substrate.267, 268 Intriguingly, the JmjC family protein, PHD8, also contains a PHD domain, enabling it to be recruited to target genes through interactions with H3K4me2 and H3K4me3. Another study found that LSD1n, an alternatively spliced form of LSD1/KDM1A that is exclusively expressed in neurons, exhibits demethylase activity towards H4K20me1 and H4K20me2 in vitro, and at least towards H4K20me1 in vivo.269 In addition, it has been shown that PHF2 is recruited to target genes in an nuclear factor-κB-dependent manner and demethylates H4K20me3.270
Genome stability
H4K20 methylation has important roles in genomic integrity.271 Genomic deletion of SET8 causes lethality in mice and D. melanogaster.259, 272 Knockdown experiments in cell lines have revealed critical roles of SET8 in cell-cycle progression, DNA replication and genome stability.273, 274, 275 Perinatal lethality and cell cycle defects resulting from Suv4-20h1/h2 double-knockout mice also support the function of H4K20 methylation in ensuring genomic integrity.262
DNA damage responses
A structural study has demonstrated a direct interaction between H4K20me2 and tandem tudor domains in TP53BP1.276 A cell-based assay has further shown that a catalytically inactive SET8 mutant fails to restore TP53BP1 recruitment to DNA damage sites.277 Moreover, formation of TP53BP1 foci following irradiation is reduced in Suv4-20h1/h2 double-knockout mice.262 In addition, in fission yeast, recruitment of the TP53BP1 homolog, Crb2, to DSBs is also regulated by the H4K20 methyltransferase, Set9.278 These studies suggest that H4K20 methylation is an evolutionarily conserved regulator of DNA-damage responses.
Chromatin compaction
Another interesting H4K20 methylation-binding protein is L3MBTL1 (L(3)mbt-like 1), a transcriptional repressor that contains three MBT domains.279 Using an electron microscopic analysis followed by sucrose gradient centrifugation, Reinberg and co-workers280 showed that direct recruitment of L3MBTL1 by H4K20 methylation is sufficient for chromatin compaction in vitro. Consistent with this, RNA interference-mediated knockdown of SET8 in HEK293 cells was shown to result in the formation of aberrant nuclei and decondensation of chromatin.274 Furthermore, recombinant nucleosome arrays containing H4K20me3 have provided direct evidence for H4K20 methylation-mediated chromatin condensation.281
DNA replication
The BAH domain in ORC1 (origin recognition complex subunit 1) is known to be a specific reader of H4K20me2.282, 283 Moreover, the WD-repeat domain-containing protein ORCA, also known as LRWD1 (leucine-rich repeats and WD repeat domain-containing 1), has been shown to interact with H4K20me3.283 These observations indicate that H4K20 methylation is involved in recruitment of the ORC complex to the origin of replication. However, how H4K20 di- and trimethylation are regulated at the replication origin requires further investigation. Some studies have revealed that H4K20 monomethylation mediated by SET8 affects S-phase progression and assembly of the pre-RC complex during the DNA replication process.284, 285 However, the fact that deletion of SET8 causes more severe defects than deletion of other subunits in the ORC complex suggests that H4K20me1 may serve more pivotal and diverse functions in DNA replication.
Nucleosome turnover during transcription
A recent study found that H4K20me1 correlates positively with transcription turnover, primarily at transcription start and termination sites.286 Interestingly, another study showed that H4K20 methylation also marks low-turnover nucleosomes in coding DNA sequences.287 This latter observation suggests that transcription-dependent nucleosome recycling allows time for progressive H4K20 methylation in coding DNA sequences. The accumulation of H4K20 methylation in old nucleosomes supports this hypothesis.288 Whether higher levels of H4K20 methylation in old nucleosomes subsequently affect transcription-mediated nucleosome recycling by functioning as a binding platform for other chromatin readers or by affecting intrinsic chromatin compaction functions remains to be elucidated.
H4K20 methylation and cancers
Several lines of evidence have implicated H4K20 methylation in cancer. The loss of H4K20me3 and H4K16 acetylation on repetitive DNA sequences and transposons in human cancer cell lines suggests that aberrant H4K20 methylation and the resulting misregulated transcription contribute to cancer development.289 Subsequent cell line and animal studies have further reported that misregulation of H4K20 methylation is strongly correlated with various types of cancer.290, 291, 292 However, the molecular basis for linkages between H4K20 methylation and tumorigenesis needs to be explored in greater detail.
Concluding remarks
Conserved histone lysine methylation, methyltransferases, demethylases and methyl-lysine-binding proteins are important in development, reprogramming and cancer development. Recent years have seen an explosion of reports on these important epigenetic regulators. Given the importance of histone lysine methylation in the precise regulation of gene expression and as a cause of various diseases, targeting histone lysine methylation regulators has been in the spotlight as a therapeutic strategy. Realizing this goal will require greater insight into structural aspects of histone lysine methylation-related proteins so as to obtain a more detailed understanding of the molecular mechanisms of actions of drugs. In addition to histone H3 and H4 lysine methylations described here are several as yet unstudied lysine methylations at the N-terminal tail of histone H2B and in the globular domains of histones. Our mechanistic understanding of downstream effectors of these lysine methylations is also limited. In particular, lysine methylations in histone globular domains surrounded by DNA have so far been largely overlooked, despite the richness of their multiple modifications. The use of nucleosomes with homogeneously methylated lysine, generated using recently developed genetic and chemical engineering techniques,258 will provide powerful tools for identifying and characterizing novel methyltransferases, demethylases and binding proteins targeting methyl-lysines located in the histone globular domain.
Acknowledgments
This work was supported by grants from the National Research Foundation of Korea (2012M3A9B4027956, 2012M3A9C6049938 and 2015R1A1A1A05001593 to JK). KP is a recipient of the TJ Park Doctoral Fellowship.
Footnotes
The authors declare no conflict of interest.
References
- Luger K, Hansen JC. Nucleosome and chromatin fiber dynamics. Curr Opin Struct Biol 2005; 15: 188–196. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell 2007; 128: 693–705. [DOI] [PubMed] [Google Scholar]
- Allis CD, Jenuwein T. The molecular hallmarks of epigenetic control. Nat Rev Genet 2016; 17: 487–500. [DOI] [PubMed] [Google Scholar]
- Wang GG, Allis CD. Misinterpretation″ of a histone mark is linked to aberrant stem cells and cancer development. Cell Cycle 2009; 8: 1982–1983. [PubMed] [Google Scholar]
- Chi P, Allis CD, Wang GG. Covalent histone modifications—miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer 2010; 10: 457–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allfrey VG, Mirsky AE. Structural modifications of histones and their possible role in the regulation of RNA synthesis. Science 1964; 144: 559. [DOI] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M et al. Regulation of chromatin structure by site specific histone H3 methyltransferases. Nature 2000; 406: 593–599. [DOI] [PubMed] [Google Scholar]
- Dillon SC, Zhang X, Trievel RC, Cheng X. The SET-domain protein superfamily: protein lysine methyltransferases. Genome Biol 2005; 6: 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 2004; 119: 941–953. [DOI] [PubMed] [Google Scholar]
- Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature 2006; 439: 811–816. [DOI] [PubMed] [Google Scholar]
- Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol 2005; 6: 838–849. [DOI] [PubMed] [Google Scholar]
- Musselman CA, Khorasanizadeh S, Kutateladze TG. Towards understanding methyllysine readout. Biochim Biophys Acta 2014; 1839: 686–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black JC, Van Rechem C, Whetstine JR. Histone lysine methylation dynamics: establishment, regulation, and biological impact. Mol Cell 2012; 48: 491–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekharan MB, Huang F, Sun ZW. Histone H2B ubiquitination and beyond: regulation of nucleosome stability, chromatin dynamics and the trans-histone H3 methylation. Epigenetics 2010; 5: 460–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vastenhouw NL, Schier AF. Bivalent histone modifications in early embryogenesis. Curr Opin Cell Biol 2012; 24: 374–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet 2012; 13: 343–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Wu F, Wu J. Targeting histone methylation for cancer therapy: enzymes, inhibitors, biological activity and perspectives. J. Hematol Oncol 2016; 9: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Han Y, Fan E, Zhang K. Analytical strategies used to identify the readers of histone modifications: a review. Anal Chim Acta 2015; 891: 32–42. [DOI] [PubMed] [Google Scholar]
- Shilatifard A. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu Rev Biochem 2012; 81: 65–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs SD, Bryk M, Strahl BD, Cheung WL, Davie JK, Dent SY et al. Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev 2001; 15: 3286–3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller T, Krogan NJ, Dover J, Erdjument-Bromage H, Tempst P, Johnston M et al. COMPASS: a complex of proteins associated with a trithorax-related SET domain protein. Proc Natl Acad Sci USA 2001; 98: 12902–12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roguev A, Schaft D, Shevchenko A, Pijnappel WW, Wilm M, Aasland R et al. The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J 2001; 20: 7137–7148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehé PM, Géli V. The multiple faces of Set1. Biochem Cell Biol 2006; 84: 536–548. [DOI] [PubMed] [Google Scholar]
- Patel A, Dharmarajan V, Vought VE, Cosgrove MS. On the mechanism of multiple lysine methylation by the human mixed lineage leukemia protein-1 (MLL1) core complex. J Biol Chem 2009; 284: 24242–24256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A, Vought VE, Dharmarajan V, Cosgrove MS. A conserved arginine-containing motif crucial for the assembly and enzymatic activity of the mixed lineage leukemia protein-1 core complex. J Biol Chem 2008; 283: 32162–32175. [DOI] [PubMed] [Google Scholar]
- Shinsky SA, Hu M, Vought VE, Ng SB, Bamshad MJ, Shendure J et al. A non-active-site SET domain surface crucial for the interaction of MLL1 and the RbBP5/Ash2L heterodimer within MLL family core complexes. J Mol Biol 2014; 426: 2283–2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinsky SA, Monteith KE, Viggiano S, Cosgrove MS. Biochemical reconstitution and phylogenetic comparison of human SET1 family core complexes involved in histone methylation. J Biol Chem 2015; 290: 6361–6375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Skalnik DG. Wdr82 is a C-terminal domain-binding protein that recruits the Setd1A Histone H3-Lys4 methyltransferase complex to transcription start sites of transcribed human genes. Mol Cell Biol 2008; 28: 609–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Skalnik DG. CpG-binding protein (CXXC finger protein 1) is a component of the mammalian Set1 histone H3-Lys4 methyltransferase complex, the analogue of the yeast Set1/COMPASS complex. J Biol Chem 2005; 280: 41725–41731. [DOI] [PubMed] [Google Scholar]
- Xu C, Bian C, Lam R, Dong A, Min J. The structural basis for selective binding of non-methylated CpG islands by the CFP1 CXXC domain. Nat Commun 2011; 2: 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouaire T, Webb S, Skene P, Illingworth R, Kerr A, Andrews R et al. Cfp1 integrates both CpG content and gene activity for accurate H3K4me3 deposition in embryonic stem cells. Genes Dev 2012; 26: 1714–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal SK, Jothi R. Genome-wide characterization of menin-dependent H3K4me3 reveals a specific role for menin in the regulation of genes implicated in MEN1-like tumors. PLoS ONE 2012; 7: e37952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefevre GM, Patel SR, Kim D, Tessarollo L, Dressler GR. Altering a histone H3K4 methylation pathway in glomerular podocytes promotes a chronic disease phenotype. PLoS Genet 2010; 6: e1001142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi YH, Westfield GH, Oleskie AN, Trievel RC, Shilatifard A, Skiniotis G. Structural analysis of the core COMPASS family of histone H3K4 methylases from yeast to human. Proc Natl Acad Sci USA 2011; 108: 20526–20531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Han J, Zhang Y, Cao F, Liu Z, Li S et al. Structural basis for activity regulation of MLL family methyltransferases. Nature 2016; 530: 447–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun ZW, Allis CD. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 2002; 418: 104–108. [DOI] [PubMed] [Google Scholar]
- Dover J, Schneider J, Tawiah-Boateng MA, Wood A, Dean K, Johnston M et al. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J Biol Chem 2002; 277: 28368–28371. [DOI] [PubMed] [Google Scholar]
- Dehé PM, Pamblanco M, Luciano P, Lebrun R, Moinier D, Sendra R et al. Histone H3 lysine 4 mono-methylation does not require ubiquitination of histone H2B. J Mol Biol 2005; 353: 477–484. [DOI] [PubMed] [Google Scholar]
- Shahbazian MD, Zhang K, Grunstein M. Histone H2B ubiquitylation controls processive methylation but not monomethylation by Dot1 and Set1. Mol Cell 2005; 19: 271–277. [DOI] [PubMed] [Google Scholar]
- Lee JS, Shukla A, Schneider J, Swanson SK, Washburn MP, Florens L et al. Histone crosstalk between H2B monoubiquitination and H3 methylation mediated by COMPASS. Cell 2007; 131: 1084–1096. [DOI] [PubMed] [Google Scholar]
- Vitaliano-Prunier A, Menant A, Hobeika M, Géli V, Gwizdek C, Dargemont C. Ubiquitylation of the COMPASS component Swd2 links H2B ubiquitylation to H3K4 trimethylation. Nat Cell Biol 2008; 10: 1365–1371. [DOI] [PubMed] [Google Scholar]
- Kim J, Kim JA, McGinty RK, Nguyen UT, Muir TW, Allis CD et al. The n-SET domain of Set1 regulates H2B ubiquitylation-dependent H3K4 methylation. Mol Cell 2013; 49: 1121–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Hake SB, Roeder RG. The human homolog of yeast BRE1 functions as a transcriptional coactivator through direct activator interactions. Mol Cell 2005; 20: 759–770. [DOI] [PubMed] [Google Scholar]
- Zhu B, Zheng Y, Pham AD, Mandal SS, Erdjument-Bromage H, Tempst P et al. Monoubiquitination of human histone H2B: the factors involved and their roles in HOX gene regulation. Mol Cell 2005; 20: 601–611. [DOI] [PubMed] [Google Scholar]
- Kim J, Guermah M, McGinty RK, Lee JS, Tang Z, Milne TA et al. RAD6-mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell 2009; 137: 459–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccone DN, Su H, Hevi S, Gay F, Lei H, Bajko J et al. KDM1B is a histone H3K4 demethylase required to establish maternal genomic imprints. Nature 2009; 461: 415–418. [DOI] [PubMed] [Google Scholar]
- Lee MG, Wynder C, Cooch N, Shiekhattar R. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature 2005; 437: 432–435. [DOI] [PubMed] [Google Scholar]
- Lan F, Collins RE, De Cegli R, Alpatov R, Horton JR, Shi X et al. Recognition of unmethylated histone H3 lysine 4 links BHC80 to LSD1-mediated gene repression. Nature 2007; 448: 718–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Wu Y, Li J, Dong C, Ye X, Chi YI et al. The SNAG domain of Snail1 functions as a molecular hook for recruiting lysine-specific demethylase 1. EMBO J 2010; 29: 1803–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Højfeldt JW, Agger K, Helin K. Histone lysine demethylases as targets for anticancer therapy. Nat Rev Drug Discov 2013; 12: 917–930. [DOI] [PubMed] [Google Scholar]
- Sinha KM, Yasuda H, Coombes MM, Dent SY, de Crombrugghe B. de Regulation of the osteoblast-specific transcription factor Osterix by NO66, a Jumonji family histone demethylase. EMBO J 2010; 29: 68–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen J, Agger K, Cloos PA, Pasini D, Rose S, Sennels L et al. RBP2 belongs to a family of demethylases, specific for tri-and dimethylated lysine 4 on histone 3. Cell 2007; 128: 1063–1076. [DOI] [PubMed] [Google Scholar]
- Yamane K, Tateishi K, Klose RJ, Fang J, Fabrizio LA, Erdjument-Bromage H et al. PLU-1 is an H3K4 demethylase involved in transcriptional repression and breast cancer cell proliferation. Mol Cell 2007; 25: 801–812. [DOI] [PubMed] [Google Scholar]
- Tahiliani M, Mei P, Fang R, Leonor T, Rutenberg M, Shimizu F et al. The histone H3K4 demethylase SMCX links REST target genes to X-linked mental retardation. Nature 2007; 447: 601–605. [DOI] [PubMed] [Google Scholar]
- Lee MG, Villa R, Trojer P, Norman J, Yan KP, Reinberg D et al. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science 2007; 318: 447–450. [DOI] [PubMed] [Google Scholar]
- Iwase S, Lan F, Bayliss P, de la Torre-Ubieta L, Huarte M, Qi HH et al. The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell 2007; 128: 1077–1088. [DOI] [PubMed] [Google Scholar]
- Klose RJ, Yan Q, Tothova Z, Yamane K, Erdjument-Bromage H, Tempst P et al. The retinoblastoma binding protein RBP2 is an H3K4 demethylase. Cell 2007; 128: 889–900. [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 2007; 39: 311–318. [DOI] [PubMed] [Google Scholar]
- Kim T, Buratowski S. Dimethylation of H3K4 by Set1 recruits the Set3 histone deacetylase complex to 5′ transcribed regions. Cell 2009; 137: 259–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC. Active genes are tri-methylated at K4 of histone H3. Nature 2002; 419: 407–411. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G et al. Genome-wide maps of chromatin state in pluripotent and lineagecommitted cells. Nature 2007; 448: 553–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan A, Ruyechan WT, Kristie TM. The coactivator host cell factor-1 mediates Set1 and MLL1 H3K4 trimethylation at herpesvirus immediate early promoters for initiation of infection. Proc Natl Acad Sci USA 2007; 104: 10835–10840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama A, Wang Z, Wysocka J, Sanyal M, Aufiero DJ, Kitabayashi I et al. Leukemia proto-oncoprotein MLL forms a SET1-like histone methyltransferase complex with menin to regulate Hox gene expression. Mol Cell Biol 2004; 24: 5639–5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song ZT, Sun L, Lu SJ, Tian Y, Ding Y, Liu JX. Transcription factor interaction with COMPASS-like complex regulates histone H3K4 trimethylation for specific gene expression in plants. Proc Natl Acad Sci USA 2015; 112: 2900–2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Chen WY, Shimada M, Nguyen UT, Kim J, Sun XJ et al. SET1 and p300 act synergistically, through coupled histone modifications, in transcriptional activation by p53. Cell 2013; 154: 297–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell 2003; 11: 709–719. [DOI] [PubMed] [Google Scholar]
- Sims RJ III, Chen CF, Santos-Rosa H, Kouzarides T, Patel SS, Reinberg D. Human but not yeast CHD1 binds directly and selectively to histone H3 methylated at lysine 4 via its tandem chromodomains. J Biol Chem 2005; 280: 41789–41792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature 2006; 442: 86–90. [DOI] [PubMed] [Google Scholar]
- Vermeulen M, Mulder KW, Denissov S, Pijnappel WW, van Schaik FM, Varier RA et al. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell 2007; 131: 58–69. [DOI] [PubMed] [Google Scholar]
- Lauberth SM, Nakayama T, Wu X, Ferris AL, Tang Z, Hughes SH et al. H3K4me3 interactions with TAF3 regulate preinitiation complex assembly and selective gene activation. Cell 2013; 152: 1021–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian C, Xu C, Ruan J, Lee KK, Burke TL, Tempel W et al. Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation. EMBO J 2011; 30: 2829–2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna SD, Ilin S, Rogers RS, Tanny JC, Lavender H, Li H et al. Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs. Mol Cell 2006; 24: 785–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung T, Binda O, Champagne KS, Kuo AJ, Johnson K, Chang HY et al. ING4 mediates crosstalk between histone H3K4 trimethylation and H3 acetylation to attenuate cellular transformation. Mol Cell 2009; 33: 248–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberl HC, Spruijt CG, Kelstrup CD, Vermeulen M, Mann M. A map of general and specialized chromatin readers in mouse tissues generated by label-free interaction proteomics. Mol Cell 2013; 49: 368–378. [DOI] [PubMed] [Google Scholar]
- Clouaire T, Webb S, Bird A. Cfp1 is required for gene expression-dependent H3K4 trimethylation and H3K9 acetylation in embryonic stem cells. Genome Biol 2014; 15: 451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Song J, Milne TA, Wang GG, Li H, Allis CD et al. Pro isomerization in MLL1 PHD3-bromo cassette connects H3K4me readout to CyP33 and HDAC-mediated repression. Cell 2010; 141: 1183–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali M, Hom RA, Blakeslee W, Ikenouye L, Kutateladze TG. Diverse functions of PHD fingers of the MLL/KMT2 subfamily. Biochim Biophys Acta 2014; 1843: 366–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao RC, Dou Y. Hijacked in cancer: the KMT2 (MLL) family of methyltransferases. Nat Rev Cancer 2015; 15: 334–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenney K, Shilatifard A. A COMPASS in the voyage of defining the role of trithorax/MLL-containing complexes: linking leukemogensis to covalent modifications of chromatin. J Cell Biochem 2005; 95: 429–436. [DOI] [PubMed] [Google Scholar]
- Thiel AT, Blessington P, Zou T, Feather D, Wu X, Yan J et al. MLL-AF9-induced leukemogenesis requires coexpression of the wild-type Mll allele. Cancer Cell 2010; 17: 148–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karatas H, Townsend EC, Cao F, Chen Y, Bernard D, Liu L et al. High-affinity, small-molecule peptidomimetic inhibitors of MLL1/WDR5 protein–protein interaction. J Am Chem Soc 2013; 135: 669–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao F, Townsend EC, Karatas H, Xu J, Li L, Lee S et al. Targeting MLL1 H3K4 methyltransferase activity in mixed-lineage leukemia. Mol Cell 2014; 53: 247–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Rooij JD, Hollink IH, Arentsen-Peters ST, van Galen JF, Berna Beverloo H, Baruchel A et al. NUP98/JARID1A is a novel recurrent abnormality in pediatric acute megakaryoblastic leukemia with a distinct HOX gene expression pattern. Leukemia 2013; 27: 2280–2288. [DOI] [PubMed] [Google Scholar]
- Jensen LR, Amende M, Gurok U, Moser B, Gimmel V, Tzschach A et al. Mutations in the JARID1C gene, which is involved in transcriptional regulation and chromatin remodeling, cause X-linked mental retardation. Am J Hum Genet 2005; 76: 227–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Liu Y, Lau JL, Min J. Epigenetic targets and drug discovery. Part 2: histone demethylation and DNA methylation. Pharmacol Ther 2015; 151: 121–140. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z et al. High-resolution profiling of histone methylations in the human genome. Cell 2007; 129: 823–837. [DOI] [PubMed] [Google Scholar]
- Hu D, Garruss AS, Gao X, Morgan MA, Cook M, Smith ER et al. The Mll2 branch of the COMPASS family regulates bivalent promoters in mouse embryonic stem cells. Nat Struct Mol Biol 2013; 20: 1093–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denissov S, Hofemeister H, Marks H, Kranz A, Ciotta G, Singh S et al. Mll2 is required for H3K4 trimethylation on bivalent promoters in embryonic stem cells, whereas Mll1 is redundant. Development 2014; 141: 526–537. [DOI] [PubMed] [Google Scholar]
- Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genomewide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev 2006; 20: 1123–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang YS, Tsai SY, Lee DF, Monk J, Su J, Ratnakumar K et al. Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell 2011; 145: 183–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan M, Liang J, Xiong Y, Shi F, Zhang Y, Lu W et al. The trithorax group protein Ash2l is essential for pluripotency and maintaining open chromatin in embryonic stem cells. J Biol Chem 2013; 288: 5039–5048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Shukla A, Wang X, Chen WY, Bernstein BE, Roeder RG. Role for Dpy-30 in ES cell-fate specification by regulation of H3K4 methylation within bivalent domains. Cell 2011; 144: 513–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 2001; 292: 110–113. [DOI] [PubMed] [Google Scholar]
- Ekwall K, Nimmo ER, Javerzat JP, Borgstrom B, Egel R, Cranston G et al. Mutations in the fission yeast silencing factors clr4+ and rik1+ disrupt the localisation of the chromo domain protein Swi6p and impair centromere function. J Cell Sci 1996; 10: 2637–2648. [DOI] [PubMed] [Google Scholar]
- Jia S, Yamada T, Grewal SI. Heterochromatin regulates cell type-specific long-range chromatin interactions essential for directed recombination. Cell 2004; 119: 469–480. [DOI] [PubMed] [Google Scholar]
- Brower-Toland B, Riddle NC, Jiang H, Huisinga KL, Elgin SC. Multiple SET methyltransferases are required to maintain normal heterochromatin domains in the genome of Drosophila melanogaster. Genetics 2009; 181: 1303–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 2001; 410: 116–120. [DOI] [PubMed] [Google Scholar]
- Collins RE, Tachibana M, Tamaru H, Smith KM, Jia D, Zhang X et al. In vitro and in vivo analyses of a Phe/Tyr switch controlling product specificity of histone lysine methyltransferases. J Biol Chem 2005; 280: 5563–5570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice JC, Briggs SD, Ueberheide B, Barber CM, Shabanowitz J, Hunt DF et al. Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol Cell 2003; 12: 1591–1598. [DOI] [PubMed] [Google Scholar]
- Loyola A, Tagami H, Bonaldi T, Roche D, Quivy JP, Imhof A et al. The HP1alpha-CAF1-SetDB1-containing complex provides H3K9me1 for Suv39-mediated K9me3 in pericentric heterochromatin. EMBO Rep 2009; 10: 769–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M, Sugimoto K, Nozaki M, Ueda J, Ohta T, Ohki M et al. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev 2002; 16: 1779–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubicek S, O'Sullivan RJ, August EM, Hickey ER, Zhang Q, Teodoro ML et al. Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Mol Cell 2007; 25: 473–481. [DOI] [PubMed] [Google Scholar]
- Ueda J, Tachibana M, Ikura T, Shinkai Y. Zinc finger protein Wiz links G9a/GLP histone methyltransferases to the co-repressor molecule CtBP. J Biol Chem 2006; 281: 20120–20128. [DOI] [PubMed] [Google Scholar]
- Tachibana M, Ueda J, Fukuda M, Takeda N, Ohta T, Iwanari H et al. Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes Dev 2005; 19: 815–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumasoni I, Meani N, Rambaldi D, Scafetta G, Alcalay M, Ciccarelli FD. Family expansion and gene rearrangements contributed to the functional specialization of PRDM genes in vertebrates. BMC Evol Biol 2007; 7: 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohenauer T, Moore AW. The Prdm family: expanding roles in stem cells and development. Development 2012; 139: 2267–2282. [DOI] [PubMed] [Google Scholar]
- Yamane K, Toumazou C, Tsukada Y, Erdjument-Bromage H, Tempst P, Wong J et al. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell 2006; 125: 483–495. [DOI] [PubMed] [Google Scholar]
- Okada Y, Scott G, Ray MK, Mishina Y, Zhang Y. Histone demethylase JHDM2A is critical for Tnp1 and Prm1 transcription and spermatogenesis. Nature 2007; 450: 119–123. [DOI] [PubMed] [Google Scholar]
- Cloos PA, Christensen J, Agger K, Maiolica A, Rappsilber J, Antal T et al. The putative oncogene GASC1 demethylates tri- and dimethylated lysine 9 on histone H3. Nature 2006; 442: 307–311. [DOI] [PubMed] [Google Scholar]
- Loenarz C, Ge W, Coleman ML, Rose NR, Cooper CD, Klose RJ et al. PHF8, a gene associated with cleft lip/palate and mental retardation, encodes for an Nepsilon-dimethyl lysine demethylase. Hum Mol Genet 2010; 19: 217–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Yamane K, Bae Y, Zhang D, Erdjument-Bromage H, Tempst P et al. The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature 2006; 442: 312–316. [DOI] [PubMed] [Google Scholar]
- Horton JR, Upadhyay AK, Qi HH, Zhang X, Shi Y, Cheng X. Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases. Nat Struct Mol Biol 2010; 17: 38–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuks F, Hurd PJ, Deplus R, Kouzarides T. The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucleic Acids Res 2003; 31: 2305–2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnertz B, Ueda Y, Derijck AA, Braunschweig U, Perez-Burgos L, Kubicek S et al. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr Biol 2003; 13: 1192–1200. [DOI] [PubMed] [Google Scholar]
- Feldman N, Gerson A, Fang J, Li E, Zhang Y, Shinkai Y et al. G9a-mediated irreversible epigenetic inactivation of Oct-3/4 during early embryogenesis. Nat Cell Biol 2006; 8: 188–194. [DOI] [PubMed] [Google Scholar]
- Epsztejn-Litman S, Feldman N, Abu-Remaileh M, Shufaro Y, Gerson A, Ueda J et al. De novo DNA methylation promoted by G9a prevents reprogramming of embryonically silenced genes. Nat Struct Mol Biol 2008; 15: 1176–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbart SB, Krajewski K, Nady N, Tempel W, Xue S, Badeaux AI et al. Association of UHRF1 with methylated H3K9 directs the maintenance of DNA methylation. Nat Struct Mol Biol 2012; 19: 1155–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babbio F, Pistore C, Curti L, Castiglioni I, Kunderfranco P, Brino L et al. The SRA protein UHRF1 promotes epigenetic crosstalks and is involved in prostate cancer progression. Oncogene 2012; 31: 4878–4887. [DOI] [PubMed] [Google Scholar]
- Sarraf SA, Stancheva I. Methyl-CpG binding protein MBD1 couples histone H3 methylation at lysine 9 by SETDB1 to DNA replication and chromatin assembly. Mol Cell 2004; 15: 595–605. [DOI] [PubMed] [Google Scholar]
- Dodge JE, Kang YK, Beppu H, Lei H, Li E. Histone H3-K9 methyltransferase ESET is essential for early development. Mol Cell Biol 2004; 24: 2478–2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 2001; 410: 120–124. [DOI] [PubMed] [Google Scholar]
- Wang T, Xu C, Liu Y, Fan K, Li Z, Sun X et al. Crystal structure of the human SUV39H1 chromodomain and its recognition of histone H3K9me2/3. PLoS ONE 2012; 7: e52977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall IM, Shankaranarayana GD, Noma K, Ayoub N, Cohen A, Grewal SI. Establishment and maintenance of a heterochromatin domain. Science 2002; 297: 2232–2237. [DOI] [PubMed] [Google Scholar]
- Lechner MS, Schultz DC, Negorev D, Maul GG, Rauscher FJ. The mammalian heterochromatin protein 1 binds diverse nuclear proteins through a common motif that targets the chromoshadow domain. Biochem Biophys Res Commun 2005; 331: 929–937. [DOI] [PubMed] [Google Scholar]
- Yamada T, Fischle W, Sugiyama T, Allis CD, Grewal SI. The nucleation and maintenance of heterochromatin by a histone deacetylase in fission yeast. Mol Cell 2005; 20: 173–185. [DOI] [PubMed] [Google Scholar]
- Penke TJ, McKay DJ, Strahl BD, Matera AG, Duronio RJ. Direct interrogation of the role of H3K9 in metazoan heterochromatin function. Genes Dev 2016; 30: 1866–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SI. RNAi-dependent formation of heterochromatin and its diverse functions. Curr Opin Genet Dev 2010; 20: 134–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaru H. Confining euchromatin/heterochromatin territory: jumonji crosses the line. Genes Dev 2010; 24: 1465–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa H, Ishiguro K, Gaubatz S, Livingston DM, Nakatani Y. A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science 2002; 296: 1132–1136. [DOI] [PubMed] [Google Scholar]
- Duan Z, Zarebski A, Montoya-Durango D, Grimes HL, Horwitz M. Gfi1 coordinates epigenetic repression of p21Cip/WAF1 by recruitment of histone lysine methyltransferase G9a and histone deacetylase 1. Mol Cell Biol 2005; 25: 10338–10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida M, Kato M, Kato Y, Sasai N, Ueda J, Tachibana M et al. Identification of ZNF200 as a novel binding partner of histone H3 methyltransferase G9a. Genes Cells 2007; 12: 877–888. [DOI] [PubMed] [Google Scholar]
- Gupta-Agarwal S, Franklin AV, Deramus T, Wheelock M, Davis RL, McMahon LL et al. G9a/GLP histone lysine dimethyltransferase complex activity in the hippocampus and the entorhinal cortex is required for gene activation and silencing during memory consolidation. J Neurosci 2012; 32: 5440–5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antignano F, Burrows K, Hughes MR, Han JM, Kron KJ, Penrod NM et al. Methyltransferase G9A regulates T cell differentiation during murine intestinal inflammation. J Clin Invest 2014; 124: 1945–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Zhang Z, Wu H, Jiang Y, Meng L, Xiong J et al. Recognition of H3K9 methylation by GLP is required for efficient establishment of H3K9 methylation, rapid target gene repression, and mouse viability. Genes Dev 2015; 29: 379–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer A, Sampath SC, Intrator A, Min A, Gertler TS, Surmeier DJ et al. Control of cognition and adaptive behavior by the GLP/G9a epigenetic suppressor complex. Neuron 2009; 64: 678–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Xu S, Lee JE, Baldridge A, Grullon S, Peng W et al. Histone H3K9 methyltransferase G9a represses PPARgamma expression and adipogenesis. EMBO J 2013; 32: 45–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakoc CR, Mandat SA, Olenchock BA, Blobel GA. Histone H3 lysine 9 methylation and HP1gamma are associated with transcription elongation through mammalian chromatin. Mol Cell 2005; 19: 381–391. [DOI] [PubMed] [Google Scholar]
- Kwon SH, Florens L, Swanson SK, Washburn MP, Abmayr SM, Workman JL. Heterochromatin protein 1 (HP1) connects the FACT histone chaperone complex to the phosphorylated CTD of RNA polymerase II. Genes Dev 2010; 24: 2133–2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luco RF, Pan Q, Tominaga K, Blencowe BJ, Pereira-Smith OM, Misteli T. Regulation of alternative splicing by histone modifications. Science 2010; 327: 996–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilgner H, Knowles DG, Johnson R, Davis CA, Chakrabortty S, Djebali S et al. Deep sequencing of subcellular RNA fractions shows splicing to be predominantly co-transcriptional in the human genome but inefficient for lncRNAs. Genome Res 2012; 22: 1616–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieberstein NI, Kozakova E, Huranova M, Thakur PK, Krchnakova Z, Krausova M et al. TALE-directed local modulation of H3K9 methylation shapes exon recognition. Sci Rep 2016; 6: 29961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salton M, Voss TC, Misteli T. Identification by high-throughput imaging of the histone methyltransferase EHMT2 as an epigenetic regulator of VEGFA alternative splicing. Nucleic Acids Res 2014; 42: 13662–13673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Andre V, Batsche E, Rachez C, Muchardt C. Histone H3 lysine 9 trimethylation and HP1gamma favor inclusion of alternative exons. Nat Struct Mol Biol 2011; 18: 337–344. [DOI] [PubMed] [Google Scholar]
- Kwak H, Fuda NJ, Core LJ, Lis JT. Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science 2013; 339: 950–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casciello F, Windloch K, Gannon F, Lee JS. Functional role of G9a histone methyltransferase in cancer. Front Immunol 2015; 6: 487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer JM. Regulation of cell differentiation and function by the euchromatin histone methyltranserfases G9a and GLP. Biochem Cell Biol 2016; 94: 26–32. [DOI] [PubMed] [Google Scholar]
- Walker MP, LaFerla FM, Oddo SS, Brewer GJ. Reversible epigenetic histone modifications and Bdnf expression in neurons with aging and from a mouse model of Alzheimer's disease. Age (Dordr) 2013; 35: 519–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu H, Lee J, Hagerty SW, Soh BY, McAlpin SE, Cormier KA et al. ESET/SETDB1 gene expression and histone H3 (K9) trimethylation in Huntington's disease. Proc Natl Acad Sci USA 2006; 103: 19176–19181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak RJ, Klimecki WT, Lau SS, Feinstein Y, Futscher BW. 5-Aza-2'-deoxycytidine-mediated reductions in G9A histone methyltransferase and histone H3 K9 di-methylation levels are linked to tumor suppressor gene reactivation. Oncogene 2007; 26: 77–90. [DOI] [PubMed] [Google Scholar]
- Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013; 6: pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua KT, Wang MY, Chen MW, Wei LH, Chen CK, Ko CH et al. The H3K9 methyltransferase G9a is a marker of aggressive ovarian cancer that promotes peritoneal metastasis. Mol Cancer 2014; 13: 189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechanic S, Raynor K, Hill JE, Cowin P. Desmocollins form a distinct subset of the cadherin family of cell adhesion molecules. Proc Natl Acad Sci USA 1991; 88: 4476–4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirrotta V. Polycombing the genome: PcG, trxG, and chromatin silencing. Cell 1998; 93: 333–336. [DOI] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P et al. Role of histone H3 lysine 27 methylation in polycomb-group silencing. Science 2002; 298: 1039–1043. [DOI] [PubMed] [Google Scholar]
- Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal polycomb sites. Cell 2002; 111: 185–196. [DOI] [PubMed] [Google Scholar]
- Müller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B et al. Histone methyltransferase activity of a Drosophila polycomb group repressor complex. Cell 2002; 111: 197–208. [DOI] [PubMed] [Google Scholar]
- Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the enhancer of zeste protein. Genes Dev 2002; 16: 2893–2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Reinberg D. The polycomb complex PRC2 and its mark in life. Nature 2011; 469: 343–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alekseyenko AA, Gorchakov AA, Kharchenko PV, Kuroda MI. Reciprocal interactions of human C10orf12 and C17orf96 with PRC2 revealed by BioTAP-XL cross-linking and affinity purification. Proc Natl Acad Sci USA 2014; 111: 2488–2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Jensen MR, Lazzerini Denchi E, Helin K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J 2004; 23: 4061–4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasov M, Wild B, Müller J. Nucleosome binding and histone methyltransferase activity of Drosophila PRC2. EMBO Rep 2005; 6: 348–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ III et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature 2009; 461: 762–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KH, Bracken AP, Pasini D, Dietrich N, Gehani SS, Monrad A et al. A model for transmission of the H3K27me3 epigenetic mark. Nat Cell Biol 2008; 10: 1291–1300. [DOI] [PubMed] [Google Scholar]
- Yuan W, Wu T, Fu H, Dai C, Wu H, Liu N et al. Dense chromatin activates Polycomb repressive complex 2 to regulate H3 lysine 27 methylation. Science 2012; 337: 971–975. [DOI] [PubMed] [Google Scholar]
- Agger K, Cloos PA, Christensen J, Pasini D, Rose S, Rappsilber J et al. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 2007; 449: 731–734. [DOI] [PubMed] [Google Scholar]
- De Santa F, Totaro MG, Prosperini E, Notarbartolo S, Testa G, Natoli G. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell 2007; 130: 1083–1094. [DOI] [PubMed] [Google Scholar]
- Walport LJ, Hopkinson RJ, Vollmar M, Madden SK, Gileadi C, Oppermann U et al. Human UTY(KDM6C) is a male-specific Nϵ-methyl lysyl demethylase. J Biol Chem 2014; 289: 18302–18313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issaeva I, Zonis Y, Rozovskaia T, Orlovsky K, Croce CM, Nakamura T et al. Knockdown of ALR (MLL2) reveals ALR target genes and leads to alterations in cell adhesion and growth. Mol Cell Biol 2007; 27: 1889–1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Sharma A, Dhar SS, Lee SH, Gu B, Chan CH et al. UTX and MLL4 coordinately regulate transcriptional programs for cell proliferation and invasiveness in breast cancer cells. Cancer Res 2014; 74: 1705–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Wang J, Zhang Y. Histone H3K27me3 demethylases KDM6A and KDM6B modulate definitive endoderm differentiation from human ESCs by regulating WNT signaling pathway. Cell Res 2013; 23: 122–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santa F, Narang V, Yap ZH, Tusi BK, Burgold T, Austenaa L et al. Jmjd3 contributes to the control of gene expression in LPS-activated macrophages. EMBO J 2009; 28: 3341–3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J, Mak W, Zvetkova I, Appanah R, Nesterova TB, Webster Z et al. Establishment of histone h3 methylation on the inactive X chromosome requires transient recruitment of Eed-Enx1 polycomb group complexes. Dev Cell 2003; 4: 481–495. [DOI] [PubMed] [Google Scholar]
- Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci USA 2003; 100: 11606–11611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 2002; 419: 624–629. [DOI] [PubMed] [Google Scholar]
- Bracken AP, Pasini D, Capra M, Prosperini E, Colli E, Helin K. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J 2003; 22: 5323–5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velichutina I, Shaknovich R, Geng H, Johnson NA, Gascoyne RD, Melnick AM et al. EZH2-mediated epigenetic silencing in germinal center B cells contributes to proliferation and lymphomagenesis. Blood 2010; 116: 5247–5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap DB, Chu J, Berg T, Schapira M, Cheng SW, Moradian A et al. Somatic mutations at EZH2 Y641 act dominantly through a mechanism of selectively altered PRC2 catalytic activity, to increase H3K27 trimethylation. Blood 2011; 117: 2451–2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe MT, Graves AP, Ganji G, Diaz E, Halsey WS, Jiang Y et al. Mutation of A677 in histone methyltransferase EZH2 in human B-cell lymphoma promotes hypertrimethylation of histone H3 on lysine 27 (H3K27). Proc Natl Acad Sci USA 2012; 109: 2989–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe MT, Ott HM, Ganji G, Korenchuk S, Thompson C, Van Aller GS et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature 2012; 492: 108–112. [DOI] [PubMed] [Google Scholar]
- Ren C, Morohashi K, Plotnikov AN, Jakoncic J, Smith SG, Li J et al. Small-molecule modulators of methyl-lysine binding for the CBX7 chromodomain. Chem Biol 2015; 22: 161–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kizer KO, Phatnani HP, Shibata Y, Hall H, Greenleaf AL, Strahl BD. A novel domain in Set2 mediates RNA polymerase II interaction and couples histone H3 K36 methylation with transcript elongation. Mol Cell Biol 2005; 25: 3305–3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner EJ, Carpenter PB. Understanding the language of Lys36 methylation at histone H3. Nat Rev Mol Cell Biol 2012; 13: 115–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmunds JW, Mahadevan LC, Clayton AL. Dynamic histone H3 methylation during gene induction: HYPB/Setd2 mediates all H3K36 trimethylation. EMBO J 2008; 27: 406–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Trojer P, Xu CF, Cheung P, Kuo A, Drury WJ et al. The target of the NSD family of histone lysine methyltransferases depends on the nature of the substrate. J Biol Chem 2009; 284: 34283–34295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Q, Li Y, Chen Z, Wang M, Reinberg D, Xu RM. The structure of NSD1 reveals an autoregulatory mechanism underlying histone H3K36 methylation. J Biol Chem 2011; 286: 8361–8368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimura K, Ura K, Shiratori H, Ikawa M, Okabe M, Schwartz RJ et al. A histone H3 lysine 36 trimethyltransferase links Nkx2-5 to Wolf–Hirschhorn syndrome. Nature 2009; 460: 287–291. [DOI] [PubMed] [Google Scholar]
- Yuan W, Xie J, Long C, Erdjument-Bromage H, Ding X, Zheng Y et al. Heterogeneous nuclear ribonucleoprotein L is a subunit of human KMT3a/Set2 complex required for H3 Lys-36 trimethylation activity in vivo. J Biol Chem 2009; 284: 15701–15707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho TH, Park IY, Zhao H, Tong P, Champion MD, Yan H et al. High-resolution profiling of histone h3 lysine 36 trimethylation in metastatic renal cell carcinoma. Oncogene 2016; 35: 1565–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu K, Lei PJ, Ju LG, Wang X, Huang K, Yang B et al. SPOP-containing complex regulates SETD2 stability and H3K36me3-coupled alternative splicing. Nucleic Acids Res 2016; 45: 92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucio-Eterovic AK, Singh MM, Gardner JE, Veerappan CS, Rice JC, Carpenter PB. Role for the nuclear receptor-binding SET domain protein 1 (NSD1) methyltransferase in coordinating lysine 36 methylation at histone 3 with RNA polymerase II function. Proc Natl Acad Sci USA 2010; 107: 16952–16957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ilin S, Wang W, Duncan EM, Wysocka J, Allis CD et al. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature 2006; 442: 91–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Schneider R, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T. Spatial distribution of di- and tri-methyl lysine 36 of histone H3 at active genes. J Biol Chem 2005; 280: 17732–17736. [DOI] [PubMed] [Google Scholar]
- Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 2005; 123: 581–592. [DOI] [PubMed] [Google Scholar]
- Fang R, Barbera AJ, Xu Y, Rutenberg M, Leonor T, Bi Q et al. Human LSD2/KDM1b/AOF1 regulates gene transcription by modulating intragenic H3K4me2 methylation. Mol Cell 2010; 39: 222–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhayalan A, Rajavelu A, Rathert P, Tamas R, Jurkowska RZ, Ragozin S et al. The Dnmt3a PWWP domain reads histone 3 lysine 36 trimethylation and guides DNA methylation. J Biol Chem 2010; 285: 26114–26120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman S, Sowa ME, Ottinger M, Smith JA, Shi Y, Harper JW et al. The Brd4 extraterminal domain confers transcription activation independent of pTEFb by recruiting multiple proteins, including NSD3. Mol Cell Biol 2011; 31: 2641–2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W, Xu M, Huang C, Liu N, Chen S, Zhu B. H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. J Biol Chem 2011; 286: 7983–7989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young MD, Willson TA, Wakefield MJ, Trounson E, Hilton DJ, Blewitt ME et al. ChIP-seq analysis reveals distinct H3K27me3 profiles that correlate with transcriptional activity. Nucleic Acids Res 2011; 39: 7415–7427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Mao G, Tong D, Huang J, Gu L, Yang W et al. The histone mark H3K36me3 regulates human DNA mismatch repair through its interaction with MutSalpha. Cell 2013; 153: 590–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fnu S, Williamson EA, De Haro LP, Brenneman M, Wray J, Shaheen M et al. Methylation of histone H3 lysine 36 enhances DNA repair by nonhomologous end-joining. Proc Natl Acad Sci USA 2011; 108: 540–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister SX, Ahrabi S, Zalmas LP, Sarkar S, Aymard F, Bachrati CZ et al. SETD2-dependent histone H3K36 trimethylation is required for homologous recombination repair and genome stability. Cell Rep 2014; 7: 2006–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Kim H, Fong N, Erickson B, Bentley DL. Pre-mRNA splicing is a determinant of histone H3K36 methylation. Proc Natl Acad Sci USA 2011; 108: 13564–13569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo R, Zheng L, Park JW, Lv R, Chen H, Jiao F et al. BS69/ZMYND11 reads and connects histone H3.3 lysine 36 trimethylation-decorated chromatin to regulated pre-mRNA processing. Mol Cell 2014; 56: 298–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Almeida SF, Grosso AR, Koch F, Fenouil R, Carvalho S, Andrade J et al. Splicing enhances recruitment of methyltransferase HYPB/Setd2 and methylation of histone H3 Lys36. Nat Struct Mol Biol 2011; 18: 977–983. [DOI] [PubMed] [Google Scholar]
- Kurotaki N, Imaizumi K, Harada N, Masuno M, Kondoh T, Nagai T et al. Haploinsufficiency of NSD1 causes Sotos syndrome. Nat Genet 2002; 30: 365–366. [DOI] [PubMed] [Google Scholar]
- Wang GG, Cai L, Pasillas MP, Kamps MP. NUP98-NSD1 links H3K36 methylation to Hox-A gene activation and leukaemogenesis. Nat Cell Biol 2007; 9: 804–812. [DOI] [PubMed] [Google Scholar]
- Kuo AJ, Cheung P, Chen K, Zee BM, Kioi M, Lauring J et al. NSD2 links dimethylation of histone H3 at lysine 36 to oncogenic programming. Mol Cell 2011; 44: 609–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaju RJ, Fidler C, Haas OA, Strickson AJ, Watkins F, Clark K et al. A novel gene, NSD1, is fused to NUP98 in the t(5;11)(q35;p15.5) in de novo childhood acute myeloid leukemia. Blood 2001; 98: 1264–1267. [DOI] [PubMed] [Google Scholar]
- Dalgliesh GL, Furge K, Greenman C, Chen L, Bignell G, Butler A et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature 2010; 463: 360–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, He F, Zeng H, Ling S, Chen A, Wang Y et al. Identification of functional cooperative mutations of SETD2 in human acute leukemia. Nat Genet 2014; 46: 287–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen AT, Zhang Y. The diverse functions of Dot1 and H3K79 methylation. Genes Dev 2011; 25: 1345–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer MS, Kahana A, Wolf AJ, Meisinger LL, Peterson SE, Goggin C et al. Identification of high-copy disruptors of telomeric silencing in Saccharomyces cerevisiae. Genetics 1998; 150: 613–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen F, Gafken PR, Gottschling DE. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell 2002; 109: 745–756. [DOI] [PubMed] [Google Scholar]
- Feng Q, Wang H, Ng HH, Erdjument-Bromage H, Tempst P, Struhl K et al. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr Biol 2002; 12: 1052–1058. [DOI] [PubMed] [Google Scholar]
- Shanower GA, Muller M, Blanton JL, Honti V, Gyurkovics H, Schedl P. Characterization of the grappa gene, the Drosophila histone H3 lysine 79 methyltransferase. Genetics 2005; 169: 173–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones B, Su H, Bhat A, Lei H, Bajko J, Hevi S et al. The histone H3K79 methyltransferase Dot1L is essential for mammalian development and heterochromatin structure. PLoS Genet 2008; 4: e1000190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinty RK, Kim J, Chatterjee C, Roeder RG, Muir TW. Chemically ubiquitylated histone H2B stimulates hDot1L-mediated intranucleosomal methylation. Nature 2008; 453: 812–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min J, Feng Q, Li Z, Zhang Y, Xu RM. Structure of the catalytic domain of human DOT1L, a non-SET domain nucleosomal histone methyltransferase. Cell 2003; 112: 711–723. [DOI] [PubMed] [Google Scholar]
- Sawada K, Yang Z, Horton JR, Collins RE, Zhang X, Cheng X. Structure of the conserved core of the yeast Dot1p, a nucleosomal histone H3 lysine 79 methyltransferase. J Biol Chem 2004; 279: 43296–43306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng HH, Feng Q, Wang H, Erdjument-Bromage H, Tempst P, Zhang Y et al. Lysine methylation within the globular domain of histone H3 by Dot1 is important for telomeric silencing and Sir protein association. Genes Dev 2002; 16: 1518–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacoste N, Utley RT, Hunter JM, Poirier GG, Côté J. Disruptor of telomeric silencing-1 is a chromatin-specific histone H3 methyltransferase. J Biol Chem 2002; 277: 30421–30424. [DOI] [PubMed] [Google Scholar]
- Briggs SD, Xiao T, Sun ZW, Caldwell JA, Shabanowitz J, Hunt DF et al. Gene silencing: trans-histone regulatory pathway in chromatin. Nature 2002; 418: 498. [DOI] [PubMed] [Google Scholar]
- Ng HH, Xu RM, Zhang Y, Struhl K. Ubiquitination of histone H2B by Rad6 is required for efficient Dot1-mediated methylation of histone H3 lysine 79. J Biol Chem 2002; 277: 34655–34657. [DOI] [PubMed] [Google Scholar]
- McGinty RK, Köhn M, Chatterjee C, Chiang KP, Pratt MR, Muir TW Structure–activity analysis of semisynthetic nucleosomes: mechanistic insights into the stimulation of Dot1L by ubiquitylated histone H2B ACS Chem Biol 2009; 4: 958–968. [DOI] [PMC free article] [PubMed]
- Holt MT, David Y, Pollock S, Tang Z, Jeon J, Kim J et al. Identification of a functional hotspot on ubiquitin required for stimulation of methyltransferase activity on chromatin. Proc Natl Acad Sci USA 2015; 112: 10365–10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Holt MT, Ohashi N, Zhao A, Müller MM, Wang B et al. Evidence that ubiquitylated H2B corrals hDot1L on the nucleosomal surface to induce H3K79 methylation. Nat Commun 2016; 7: 10589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altaf M, Utley RT, Lacoste N, Tan S, Briggs SD, Côté J. Interplay of chromatin modifiers on a short basic patch of histone H4 tail defines the boundary of telomeric heterochromatin. Mol Cell 2007; 28: 1002–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katan-Khaykovich Y, Struhl K. Heterochromatin formation involves changes in histone modifications over multiple cell generations. EMBO J 2005; 24: 2138–2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W, Kim R, Park G, Park JW, Kim JE. Deficiency of H3K79 histone methyltransferase Dot1-like protein (DOT1L) inhibits cell proliferation. J Biol Chem 2012; 287: 5588–5599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell 2011; 19: 17–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onishi M, Liou GG, Buchberger JR, Walz T, Moazed D. Role of the conserved Sir3-BAH domain in nucleosome binding and silent chromatin assembly. Mol Cell 2007; 28: 1015–1028. [DOI] [PubMed] [Google Scholar]
- Huyen Y, Zgheib O, DiTullio RA Jr, Gorgoulis VG, Zacharatos P, Petty TJ et al. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature 2004; 432: 406–411. [DOI] [PubMed] [Google Scholar]
- Wysocki R, Javaheri A, Allard S, Sha F, Côté J, Kron SJ. Role of Dot1-dependent histone H3 methylation in G1 and S phase DNA damage checkpoint functions of Rad9. Mol Cell Biol 2005; 25: 8430–8443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannattasio M, Lazzaro F, Plevani P, Muzi-Falconi M. The DNA damage checkpoint response requires histone H2B ubiquitination by Rad6-Bre1 and H3 methylation by Dot1. J Biol Chem 2005; 280: 9879–9886. [DOI] [PubMed] [Google Scholar]
- Game JC, Williamson MS, Spicakova T, Brown JM. The RAD6/BRE1 histone modification pathway in Saccharomyces confers radiation resistance through a RAD51-dependent process that is independent of RAD18. Genetics 2006; 173: 1951–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conde F, Ontoso D, Acosta I, Gallego-Sánchez A, Bueno A, San-Segundo PA. Regulation of tolerance to DNA alkylating damage by Dot1 and Rad53 in Saccharomyces cerevisiae. DNA Repair 2010; 9: 1038–1049. [DOI] [PubMed] [Google Scholar]
- Lévesque N, Leung GP, Fok AK, Schmidt TI, Kobor MS. Loss of H3 K79 trimethylation leads to suppression of Rtt107-dependent DNA damage sensitivity through the translesion synthesis pathway. J Biol Chem 2010; 285: 35113–35122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farooq Z, Banday S, Pandita TK, Altaf M. The many faces of histone H3K79 methylation. Mutat Res Rev Mutat Res 2016; 768: 46–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng HH, Ciccone DN, Morshead KB, Oettinger MA, Struhl K. Lysine-79 of histone H3 is hypomethylated at silenced loci in yeast and mammalian cells: a potential mechanism for position-effect variegation. Proc Natl Acad Sci USA 2003; 100: 1820–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schübeler D, MacAlpine DM, Scalzo D, Wirbelauer C, Kooperberg C, van Leeuwen F et al. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev 2004; 18: 1263–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 2005; 122: 517–527. [DOI] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet 2008; 40: 897–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouskouti A, Talianidis I. Histone modifications defining active genes persist after transcriptional and mitotic inactivation. EMBO J 2005; 24: 347–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steger DJ, Lefterova MI, Ying L, Stonestrom AJ, Schupp M, Zhuo D et al. DOT1L/KMT4 recruitment and H3K79 methylation are ubiquitously coupled with gene transcription in mammalian cells. Mol Cell Biol 2008; 28: 2825–2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller D, Bach C, Zeisig D, Garcia-Cuellar MP, Monroe S, Sreekumar A et al. A role for the MLL fusion partner ENL in transcriptional elongation and chromatin modification. Blood 2007; 110: 4445–4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitoun E, Oliver PL, Davies KE. The mixed-lineage leukemia fusion partner AF4 stimulates RNA polymerase II transcriptional elongation and mediates coordinated chromatin remodeling. Hum Mol Genet 2007; 16: 92–106. [DOI] [PubMed] [Google Scholar]
- Mueller D, García-Cuéllar MP, Bach C, Buhl S, Maethner E, Slany RK. Misguided transcriptional elongation causes mixed lineage leukemia. PLoS Biol 2009; 7: e1000249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan M, Herz HM, Takahashi YH, Lin C, Lai KC, Zhang Y et al. Linking H3K79 trimethylation to Wnt signaling through a novel Dot1-containing complex (DotCom). Genes Dev 2010; 24: 574–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayton PM, Cleary ML. Molecular mechanisms of leukemogenesis mediated by MLL fusion proteins. Oncogene 2001; 20: 5695–5707. [DOI] [PubMed] [Google Scholar]
- Okada Y, Feng Q, Lin Y, Jiang Q, Li Y, Coffield VM et al. hDOT1L links histone methylation to leukemogenesis. Cell 2005; 121: 167–178. [DOI] [PubMed] [Google Scholar]
- Okada Y, Jiang Q, Lemieux M, Jeannotte L, Su L, Zhang Y. Leukaemic transformation by CALM–AF10 involves upregulation of Hoxa5 by hDOT1L. Nat Cell Biol 2006; 8: 1017–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivtsov AV, Feng Z, Lemieux ME, Faber J, Vempati S, Sinha AU et al. H3K79 methylation profiles define murine and human MLL-AF4 leukemias. Cancer Cell 2008; 14: 355–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang MJ, Wu H, Achille NJ, Reisenauer MR, Chou CW, Zeleznik-Le NJ et al. Histone H3 lysine 79 methyltransferase Dot1 is required for immortalization by MLL oncogenes. Cancer Res 2010; 70: 10234–10242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen AT, Taranova O, He J, Zhang Y. DOT1L, the H3K79 methyltransferase, is required for MLL-AF9-mediated leukemogenesis. Blood 2011; 117: 6912–6922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle SR, Olhava EJ, Therkelsen CA, Majer CR, Sneeringer CJ, Song J et al. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell 2011; 20: 53–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle SR, Olhava EJ, Therkelsen CA, Basavapathruni A, Jin L, Boriack-Sjodin PA et al. Potent inhibition of DOT1L as treatment of MLL-fusion leukemia. Blood 2013; 122: 1017–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Deng L, Chen F, Yao Y, Wu B, Wei L et al. Inhibition of histone H3K79 methylation selectively inhibits proliferation, self-renewal and metastatic potential of breast cancer. Oncotarget 2014; 5: 10665–10677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A, Ha S, Ahn J, Kim R, Kim S, Lee Y et al. A chemical biology route to site-specific authentic protein modifications. Science 2016; 354: 623–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J, Feng Q, Ketel CS, Wang H, Cao R, Xia L et al. Purification and functional characterization of SET8, a nucleosomal histone H4-lysine 20-specific methyltransferase. Curr Biol 2002; 12: 1086–1099. [DOI] [PubMed] [Google Scholar]
- Nishioka K, Rice JC, Sarma K, Erdjument-Bromage H, Werner J, Wang Y et al. PR-Set7 is a nucleosome-specific methyltransferase that modifies lysine 20 of histone H4 and is associated with silent chromatin. Mol Cell 2002; 9: 1201–1213. [DOI] [PubMed] [Google Scholar]
- Schotta G, Lachner M, Sarma K, Ebert A, Sengupta R, Reuter G et al. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev 2004; 18: 1251–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotta G, Sengupta R, Kubicek S, Malin S, Kauer M, Callén E et al. A chromatin-wide transition to H4K20 monomethylation impairs genome integrity and programmed DNA rearrangements in the mouse. Genes Dev 2008; 22: 2048–2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Pesavento JJ, Starnes TW, Cryderman DE, Wallrath LL, Kelleher NL et al. Preferential dimethylation of histone H4 lysine 20 by Suv4-20. J Biol Chem 2008; 283: 12085–12092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southall SM, Cronin NB, Wilson JR. A novel route to product specificity in the Suv4-20 family of histone H4K20 methyltransferases. Nucleic Acids Res 2014; 42: 661–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nuland R, Gozani O. Histone H4 lysine 20 (H4K20) methylation, expanding the signaling potential of the proteome one methyl moiety at a time. Mol Cell Proteomics 2016; 15: 755–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor-Vazirani P, Kagey JD, Vertino PM. SUV420H2-mediated H4K20 trimethylation enforces RNA polymerase II promoter-proximal pausing by blocking hMOF-dependent H4K16 acetylation. Mol Cell Biol 2011; 31: 1594–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi HH, Sarkissian M, Hu GQ, Wang Z, Bhattacharjee A, Gordon DB et al. Histone H4K20/H3K9 demethylase PHF8 regulates zebrafish brain and craniofacial development. Nature 2010; 466: 503–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Tanasa B, Tyurina OV, Zhou TY, Gassmann R, Liu WT et al. PHF8 mediates histone H4 lysine 20 demethylation events involved in cell cycle progression. Nature 2010; 466: 508–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Telese F, Tan Y, Li W, Jin C, Basnet H et al. LSD1n is an H4K20 demethylase regulating memory formation via transcriptional elongation control. Nat Neurosci 2015; 18: 1256–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stender JD, Pascual G, Liu W, Kaikkonen MU, Do K, Spann NJ et al. Control of proinflammatory gene programs by regulated trimethylation and demethylation of histone H4K20. Mol Cell 2012; 48: 28–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen S, Schotta G, Sørensen CS. Histone H4 lysine 20 methylation: key player in epigenetic regulation of genomic integrity. Nucleic Acids Res 2013; 41: 2797–2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda H, Okamoto I, Murphy N, Chu J, Price SM, Shen MM et al. Monomethylation of histone H4-lysine 20 is involved in chromosome structure and stability and is essential for mouse development. Mol Cell Biol 2009; 29: 2278–2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huen MS, SM-H Sy, van Deursen JM, Chen J. Direct interaction between SET8 and proliferating cell nuclear antigen couples H4-K20 methylation with DNA replication. J Biol Chem 2008; 283: 11073–11077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houston SI, McManus KJ, Adams MM, Sims JK, Carpenter PB, Hendzel MJ et al. Catalytic function of the PR-Set7 histone H4 lysine 20 monomethyltransferase is essential for mitotic entry and genomic stability. J Biol Chem 2008; 283: 19478–19488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Armstrong RL, Duronio RJ, MacAlpine DM. Methylation of histone H4 lysine 20 by PR-Set7 ensures the integrity of late replicating sequence domains in Drosophila. Nucleic Acids Res 2016; 44: 7204–7218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botuyan MV, Lee J, Ward IM, Kim JE, Thompson JR, Chen J et al. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell 2006; 127: 1361–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda H, Hübner MR, Beck DB, Vermeulen M, Hurwitz J, Spector DL et al. Regulation of the histone H4 monomethylase PR-Set7 by CRL4 Cdt2-mediated PCNA-dependent degradation during DNA damage. Mol Cell 2010; 40: 364–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SL, Portoso M, Mata J, Bähler J, Allshire RC, Kouzarides T. Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell 2004; 119: 603–614. [DOI] [PubMed] [Google Scholar]
- Boccuni P, MacGrogan D, Scandura JM, Nimer SD. The human L(3)MBT polycomb group protein is a transcriptional repressor and interacts physically and functionally with TEL (ETV6). J Biol Chem 2003; 278: 15412–15420. [DOI] [PubMed] [Google Scholar]
- Trojer P, Li G, Sims RJ, Vaquero A, Kalakonda N, Boccuni P et al. L3MBTL1, a histone-methylation-dependent chromatin lock. Cell 2007; 129: 915–928. [DOI] [PubMed] [Google Scholar]
- Lu X, Simon MD, Chodaparambil JV, Hansen JC, Shokat KM, Luger K. The effect of H3K79 dimethylation and H4K20 trimethylation on nucleosome and chromatin structure. Nat Struct Mol Biol 2008; 15: 1122–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo AJ, Song J, Cheung P, Ishibe-Murakami S, Yamazoe S, Chen JK et al. The BAH domain of ORC1 links H4K20me2 to DNA replication licensing and Meier–Gorlin syndrome. Nature 2012; 484: 115–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck DB, Burton A, Oda H, Ziegler-Birling C, Torres-Padilla ME, Reinberg D. The role of PR-Set7 in replication licensing depends on Suv4-20h. Genes Dev 2012; 26: 2580–2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen S, Elvers I, Trelle MB, Menzel T, Eskildsen M, Jensen ON et al. The histone methyltransferase SET8 is required for S-phase progression. J Cell Biol 2007; 179: 1337–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardat M, Brustel J, Kirsh O, Lefevbre C, Callanan M, Sardet C et al. The histone H4 Lys 20 methyltransferase PR-Set7 regulates replication origins in mammalian cells. Nat Cell Biol 2010; 12: 1086–1093. [DOI] [PubMed] [Google Scholar]
- Yang H, Kwon CS, Choi Y, Lee D. Both H4K20 mono-methylation and H3K56 acetylation mark transcription-dependent histone turnover in fission yeast. Biochem Biophys Res Commun 2016; 476: 515–521. [DOI] [PubMed] [Google Scholar]
- Svensson JP, Shukla M, Menendez-Benito V, Norman-Axelsson U, Audergon P, Sinha I et al. A nucleosome turnover map reveals that the stability of histone H4 Lys20 methylation depends on histone recycling in transcribed chromatin. Genome Res 2015; 25: 872–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf AN, Barth TK, Imhof A. Establishment of histone modifications after chromatin assembly. Nucleic Acids Res 2009; 37: 5032–5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraga MF, Ballestar E, Villar-Garea A, Boix-Chornet M, Espada J, Schotta G et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet 2005; 37: 391–400. [DOI] [PubMed] [Google Scholar]
- Van Den Broeck A, Brambilla E, Moro-Sibilot D, Lantuejoul S, Brambilla C, Eymin B et al. Loss of histone h4k20 trimethylation occurs in preneoplasia and influences prognosis of non-small cell lung cancer. Clin Cancer Res 2008; 14: 7237–7245. [DOI] [PubMed] [Google Scholar]
- Elsheikh SE, Green AR, Rakha EA, Powe DG, Ahmed RA, Collins HM et al. Global histone modifications in breast cancer correlate with tumor phenotypes, prognostic factors, and patient outcome. Cancer Res 2009; 69: 3802–3809. [DOI] [PubMed] [Google Scholar]
- Bagnyukova TV, Tryndyak VP, Montgomery B, Churchwell MI, James SR, Muskhelishvili L et al. Genetic and epigenetic changes in rat preneoplastic liver tissue induced by 2-acetylaminofluorene. Carcinogenesis 2008; 29: 638–646. [DOI] [PubMed] [Google Scholar]
- Li T, Zheng Q, An J, Wu M, Li H, Gui X et al. SET1A cooperates with CUDR to promote liver cancer growth and hepatocyte-like stem cell malignant transformation epigenetically. Mol Ther 2016; 24: 261–275. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Lim S, Metzger E, Schüle R, Kirfel J, Buettner R. Epigenetic regulation of cancer growth by histone demethylases. Int J Cancer 2010; 127: 1991–1998. [DOI] [PubMed] [Google Scholar]
- Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature 2012; 487: 239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Yin X, Yang H, Xu Y. Histone demethylase LSD2 acts as an E3 ubiquitin ligase and inhibits cancer cell growth through promoting proteasomal degradation of OGT. Mol Cell 2015; 58: 47–59. [DOI] [PubMed] [Google Scholar]
- Bowser R, Giambrone A, Davies P. FAC1, a novel gene identified with the monoclonal antibody Alz50, is developmentally regulated in human brain. Dev Neurosci 1995; 17: 20–37. [DOI] [PubMed] [Google Scholar]
- Kurabe N, Murakami S, Tashiro F. SGF29 and Sry pathway in hepatocarcinogenesis. World J Biol Chem 2015; 6: 139–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piunti A, Shilatifard A. Epigenetic balance of gene expression by Polycomb and COMPASS families. Science 2016; 352: aad9780. [DOI] [PubMed] [Google Scholar]
- Lu PJ, Sundquist K, Baeckstrom D, Poulsom R, Hanby A, Meier-Ewert S. A novel gene (PLU-1) containing highly conserved putative DNA/chromatin binding motifs is specifically up-regulated in breast cancer. J Biol Chem 1999; 274: 15633–15645. [DOI] [PubMed] [Google Scholar]
- Zhang X, Xu LS, Wang ZQ, Wang KS, Li N, Cheng ZH et al. ING4 induces G2/M cell cycle arrest and enhances the chemosensitivity to DNA-damage agents in HepG2 cells. FEBS Lett 2004; 570: 7–12. [DOI] [PubMed] [Google Scholar]
- Kim S, Welm AL, Bishop JM. A dominant mutant allele of the ING4 tumor suppressor found in human cancer cells exacerbates MYC-initiated mouse mammary tumorigenesis. Cancer Res 2010; 70: 5155–5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young SR, Skalnik DG. CXXC finger protein 1 is required for normal proliferation and differentiation of the PLB-985 myeloid cell line. DNA Cell Biol 2007; 26: 80–90. [DOI] [PubMed] [Google Scholar]
- Coda DM, Lingua MF, Morena D, Foglizzo V, Bersani F, Ala U et al. SMYD1 and G6PD modulation are critical events for miR-206-mediated differentiation of rhabdomyosarcoma. Cell Cycle 2015; 14: 1389–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaelin WG Jr, Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell 2008; 30: 393–402. [DOI] [PubMed] [Google Scholar]
- Sinha S, Singh RK, Alam N, Roy A, Roychoudhury S, Panda CK. Alterations in candidate genes PHF2, FANCC, PTCH1 and XPA at chromosomal 9q22.3 region: pathological significance in early- and late-onset breast carcinoma. Mol Cancer 2008; 7: 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynoird N, Mazur PK, Stellfeld T, Flores NM, Lofgren SM, Carlson SM et al. Coordination of stress signals by the lysine methyltransferase SMYD2 promotes pancreatic cancer. Genes Dev 2016; 30: 772–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu S, Ichikawa D, Hirajima S, Nagata H, Nishimura Y, Kawaguchi T. Overexpression of SMYD2 contributes to malignant outcome in gastric cancer. Br J Cancer 2015; 112: 357–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H, Zhang P, Qin L, Chen L, Shi S, Lu Y et al. Overexpression of SPINDLIN1 induces cellular senescence, multinucleation and apoptosis. Gene 2008; 410: 67–74. [DOI] [PubMed] [Google Scholar]
- Franz H, Greschik H, Willmann D, Ozretić L, Jilg CA, Wardelmann E et al. The histone code reader SPIN1 controls RET signaling in liposarcoma. Oncotarget 2015; 6: 4773–4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama Y, Koda Y, Byeon SJ, Shimada S, Nishikawaji T, Sakamoto A et al. Reduced expression of SET7/9, a histone mono-methyltransferase, is associated with gastric cancer progression. Oncotarget 2016; 7: 3966–3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Hu J, Li G, Qu L, He Q, Lou Y et al. PHF23 (plant homeodomain finger protein 23) negatively regulates cell autophagy by promoting ubiquitination and degradation of E3 ligase LRSAM1. Autophagy 2014; 10: 2158–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussin J, Sinnett D, Casals F, Idaghdour Y, Bruat V, Saillour V et al. Rare allelic forms of PRDM9 associated with childhood leukemogenesis. Genome Res 2013; 23: 419–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braig M, Lee S, Loddenkemper C, Rudolph C, Peters AH, Schlegelberger B et al. Oncogene-induced senescence as an initial barrier in lymphoma development. Nature 2005; 436: 660–665. [DOI] [PubMed] [Google Scholar]
- Song K, Jung Y, Jung D, Lee I. Human Ku70 interacts with heterochromatin protein 1alpha. J Biol Chem 2001; 276: 8321–8327. [DOI] [PubMed] [Google Scholar]
- Uemura M, Yamamoto H, Takemasa I, Mimori K, Hemmi H, Mizushima T et al. Jumonji domain containing 1A is a novel prognostic marker for colorectal cancer: in vivo identification from hypoxic tumor cells. Clin Cancer Res 2010; 16: 4636–4646. [DOI] [PubMed] [Google Scholar]
- Qi J, Nakayama K, Cardiff RD, Borowsky AD, Kaul K, Williams R et al. Siah2-dependent concerted activity of HIF and FoxA2 regulates formation of neuroendocrine phenotype and neuroendocrine prostate tumors. Cancer Cell 2010; 18: 23–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Shi M, Sun L, Wang Y, Gui Y, Cai Z et al. The expression of histone demethylase JMJD1A in renal cell carcinoma. Neoplasma 2011; 58: 153–157. [DOI] [PubMed] [Google Scholar]
- Kirschmann DA, Seftor EA, Nieva DR, Mariano EA, Hendrix MJ. Differentially expressed genes associated with the metastatic phenotype in breast cancer. Breast Cancer Res Treat 1999; 55: 127–136. [DOI] [PubMed] [Google Scholar]
- Pomeroy SL, Tamayo P, Gaasenbeek M, Sturla LM, Angelo M, McLaughlin ME et al. Prediction of central nervous system embryonal tumour outcome based on gene expression. Nature 2002; 415: 436–442. [DOI] [PubMed] [Google Scholar]
- Wasenius VM, Hemmer S, Kettunen E, Knuutila S, Franssila K, Joensuu H. Hepatocyte growth factor receptor, matrix metalloproteinase-11, tissue inhibitor of metalloproteinase-1, and fibronectin are up-regulated in papillary thyroid carcinoma: a cDNA and tissue microarray study. Clin Cancer Res 2003; 9: 68–75. [PubMed] [Google Scholar]
- Kim JY, Kim KB, Eom GH, Choe N, Kee HJ, Son HJ et al. KDM3B is the H3K9 demethylase involved in transcriptional activation of lmo2 in leukemia. Mol Cell Biol 2012; 32: 2917–2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemsen MH, Vulto-van Silfhout AT, Nillesen WM, Wissink-Lindhout WM, van Bokhoven H, Philip N et al. Update on Kleefstra Syndrome. Mol Syndromol 2012; 2: 202–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babacan NA, Egilmez HR, Yucel B, Ilknur P, Seker MM, Kacan T et al. The prognostic value of UHRF-1 and p53 in gastric cancer. Saudi J Gastroenterol 2016; 22: 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman EC, Robinson BD, Downes MJ, Powell LG, Lee MM, Scherr DS et al. Role of androgen receptor and associated lysine-demethylase coregulators, LSD1 and JMJD2A, in localized and advanced human bladder cancer. Mol Carcinogen 2011; 50: 931–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patani N, Jiang WG, Newbold RF, Mokbel K. Histone-modifier gene expression profiles are associated with pathological and clinical outcomes in human breast cancer. Anticancer Res 2011; 31: 4115–4125. [PubMed] [Google Scholar]
- Steele-Perkins G, Fang W, Yang XH, Van Gele M, Carling T, Gu J et al. Tumor formation and inactivation of RIZ1, an Rb-binding member of a nuclear protein-methyltransferase superfamily. Genes Dev 2001; 15: 2250–2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki O, Meguro K, Tohmiya Y, Funato T, Shibahara S, Sasaki T. Altered expression of retinoblastoma protein-interacting zinc finger gene, RIZ, in human leukaemia. Br J Haematol 2002; 119: 940–948. [DOI] [PubMed] [Google Scholar]
- Xie W, Li X, Chen X, Huang S, Huang S. Decreased expression of PRDM2 (RIZ1) and its correlation with risk stratification in patients with myelodysplastic syndrome. Br J Haematol 2010; 150: 242–244. [DOI] [PubMed] [Google Scholar]
- Pryor JG, Brown-Kipphut BA, Iqbal A, Scott GA. Microarray comparative genomic hybridization detection of copy number changes in desmoplastic melanoma and malignant peripheral nerve sheath tumor. Am J Dermatopathol 2011; 33: 780–785. [DOI] [PubMed] [Google Scholar]
- Yang ZQ, Imoto I, Fukuda Y, Pimkhaokham A, Shimada Y, Imamura M et al. Identification of a novel gene, GASC1, within an amplicon at 9p23-24 frequently detected in esophageal cancer cell lines. Cancer Res 2000; 60: 4735–4739. [PubMed] [Google Scholar]
- Liu G, Bollig-Fischer A, Kreike B, van de Vijver MJ, Abrams J, Ethier SP et al. Genomic amplification and oncogenic properties of the GASC1 histone demethylase gene in breast cancer. Oncogene 2009; 28: 4491–4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrbrecht A, Muller U, Wolter M, Hoischen A, Koch A, Radlwimmer B et al. Comprehensive genomic analysis of desmoplastic medulloblastomas: identification of novel amplified genes and separate evaluation of the different histological components. J Pathol 2006; 208: 554–563. [DOI] [PubMed] [Google Scholar]
- Laumonnier F, Holbert S, Ronce N, Faravelli F, Lenzner S, Schwartz CE et al. Mutations in PHF8 are associated with X linked mental retardation and cleft lip/cleft palate. J Med Genet 2005; 42: 780–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawaz Z, Patil V, Arora A, Hegde AS, Arivazhagan A, Santosh V et al. Cbx7 is epigenetically silenced in glioblastoma and inhibits cell migration by targeting YAP/TAZ-dependent transcription. Sci Rep 2016; 6: 27753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Wu ZJ, Groner AC, He HH, Cai C, Lis RT et al. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science 2012; 338: 1465–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta A, Le Magnen C, Mitrofanova A, Ouyang X, Califano A, Abate-Shen C. Identification of an NKX3.1-G9a-UTY transcriptional regulatory network that controls prostate differentiation. Science 2016; 352: 1576–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YL, Gao X, Jiang Y, Zhang G, Sun ZC, Cui BB et al. Expression and clinicopathological significance of EED, SUZ12 and EZH2 mRNA in colorectal cancer. J Cancer Res Clin Oncol 2015; 141: 661–669. [DOI] [PubMed] [Google Scholar]
- Bierne H, Tham TN, Batsche E, Dumay A, Leguillou M, Kernéis-Golsteyn S et al. Human BAHD1 promotes heterochromatic gene silencing. Proc Natl Acad Sci USA 2009; 106: 13826–13831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Carpizo V, Sarmentero J, Han B, Graña O, Ruiz-Llorente S, Pisano DG et al. NSD2 contributes to oncogenic RAS-driven transcription in lung cancer cells through long-range epigenetic activation. Sci Rep 2016; 6: 32952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duns G, van den Berg E, van Duivenbode I, Osinga J, Hollema H, Hofstra RM et al. Histone methyltransferase gene SETD2 is a novel tumor suppressor gene in clear cell renal cell carcinoma. Cancer Res 2010; 70: 4287–4291. [DOI] [PubMed] [Google Scholar]
- Berenstein R, Blau IW, Suckert N, Baldus C, Pezzutto A, Dorken B et al. Quantitative detection of DNMT3A R882H mutation in acute myeloid leukemia. J Exp Clin Cancer Res 2015; 34: 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Starza R, Gorello P, Rosati R, Riezzo A, Veronese A, Ferrazzi E et al. Cryptic insertion producing two NUP98/NSD1 chimeric transcripts in adult refractory anemia with an excess of blasts. Genes Chromosomes Cancer 2004; 41: 395–399. [DOI] [PubMed] [Google Scholar]
- Berdasco M, Ropero S, Setien F, Fraga MF, Lapunzina P, Losson R et al. Epigenetic inactivation of the Sotos overgrowth syndrome gene histone methyltransferase NSD1 in human neuroblastoma and glioma. Proc Natl Acad Sci USA 2009; 106: 21830–21835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Nguyen AT, Zhang Y. KDM2b/JHDM1b, an H3K36me2-specific demethylase, is required for initiation and maintenance of acute myeloid leukemia. Blood 2011; 117: 3869–3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kottakis F, Polytarchou C, Foltopoulou P, Sanidas I, Kampranis SC, Tsichlis PN. FGF-2 regulates cell proliferation, migration, and angiogenesis through an NDY1/KDM2B-miR-101-EZH2 pathway. Mol Cell 2011; 43: 285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan TS, Hawkins C, Krieger JR, McGlade CJ, Huang A. JPO2/CDCA7L and LEDGF/p75 are novel mediators of PI3K/AKT signaling and aggressive phenotypes in medulloblastoma. Cancer Res 2016; 76: 2802–2812. [DOI] [PubMed] [Google Scholar]
- Basu A, Cajigas-Du Ross CK, Rios-Colon L, Mediavilla-Varela M, Daniels-Wells TR, Leoh LS et al. LEDGF/p75 overexpression attenuates oxidative stress-induced necrosis and upregulates the oxidoreductase ERP57/PDIA3/GRP58 in prostate cancer. PLoS ONE 2016; 11: e0146549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altan B, Yokobori T, Ide M, Bai T, Yanoma T, Kimura A et al. High expression of MRE11-RAD50-NBS1 is associated with poor prognosis and chemoresistance in gastric cancer. Anticancer Res 2016; 36: 5237–5247. [DOI] [PubMed] [Google Scholar]
- Angrand PO, Apiou F, Stewart AF, Dutrillaux B, Losson R, Chambon P. NSD3, a new SET domain-containing gene, maps to 8p12 and is amplified in human breast cancer cell lines. Genomics 2001; 74: 79–88. [DOI] [PubMed] [Google Scholar]
- Rosati R, La Starza R, Veronese A, Aventin A, Schwienbacher C, Vallespi T et al. NUP98 is fused to the NSD3 gene in acute myeloid leukemia associated with t(8;11)(p11.2;p15). Blood 2002; 99: 3857–3860. [DOI] [PubMed] [Google Scholar]
- Taketani T, Taki T, Nakamura H, Taniwaki M, Masuda J, Hayashi Y. NUP98-NSD3 fusion gene in radiation-associated myelodysplastic syndrome with t(8;11)(p11;p15) and expression pattern of NSD family genes. Cancer Genet Cytogenet 2009; 190: 108–112. [DOI] [PubMed] [Google Scholar]
- Lu Y, Gao J, Lu Y. Down-expression pattern of Ku70 and p53 coexisted in colorectal cancer. Med Oncol 2015; 32: 98. [DOI] [PubMed] [Google Scholar]
- Zhu L, Li Q, Wong SH, Huang M, Klein BJ, Shen J et al. ASH1L links histone H3 lysine 36 dimethylation to MLL leukemia. Cancer Discov 2016; 6: 770–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen H, Li Y, Xi Y, Jiang S, Stratton S, Peng D et al. ZMYND11 links histone H3.3K36me3 to transcription elongation and tumour suppression. Nature 2014; 508: 263–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Yan CT, Dou Y, Viboolsittiseri SS, Wang JH. The role of a newly identified SET domain-containing protein, SETD3, in oncogenesis. Haematologica 2013; 98: 739–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeyaratnam DC, Baduin BS, Hansen MC, Hansen M, Jorgensen JM, Aggerholm A et al. Delineation of known and new transcript variants of the SETMAR (Metnase) gene and the expression profile in hematologic neoplasms. Exp Hematol 2014; 42: 448–456. [DOI] [PubMed] [Google Scholar]
- Jankovic M, Feldhahn N, Oliveira TY, Silva IT, Kieffer-Kwon KR, Yamane A et al. 53BP1 alters the landscape of DNA rearrangements and suppresses AID-induced B cell lymphoma. Mol Cell 2013; 49: 623–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwman P, Aly A, Escandell JM, Pieterse M, Bartkova J, van der Gulden H et al. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat Struct Mol Biol 2010; 17: 688–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Xu B, Moran MS, Zhao Y, Su P, Haffty BG et al. 53BP1 functions as a tumor suppressor in breast cancer via the inhibition of NF-κB through miR-146a. Carcinogenesis 2012; 33: 2593–2600. [DOI] [PubMed] [Google Scholar]
- Yang F, Sun L, Li Q, Han X, Lei L, Zhang H et al. SET8 promotes epithelial–mesenchymal transition and confers TWIST dual transcriptional activities. EMBO J 2012; 31: 110–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogribny IP, Ross SA, Tryndyak VP, Pogribna M, Poirier LA, Karpinets TV. Histone H3 lysine 9 and H4 lysine 20 trimethylation and the expression of Suv4-20h2 and Suv-39h1 histone methyltransferases in hepatocarcinogenesis induced by methyl deficiency in rats. Carcinogenesis 2006; 27: 1180–1186. [DOI] [PubMed] [Google Scholar]
- Bicknell LS, Walker S, Klingseisen A, Stiff T, Leitch A, Kerzendorfer C et al. Mutations in ORC1, encoding the largest subunit of the origin recognition complex, cause microcephalic primordial dwarfism resembling Meier–Gorlin syndrome. Nat Genet 2011; 43: 350–355. [DOI] [PubMed] [Google Scholar]
- Wang Y, Reddy B, Thompson J, Wang H, Noma K, Yates JR 3rd et al. Regulation of Set9-mediated H4K20 methylation by a PWWP domain protein. Mol Cell 2009; 33: 428–437. [DOI] [PMC free article] [PubMed] [Google Scholar]