Significance
Cells have been shown to respond to a host of physical properties of the environments that surround them. However, given that these properties vary considerably across tissues, how these individual properties interact to form unique regulatory environments for cells is largely unknown. This work analyzes the transcriptional responses of cells to unique combinations of microenvironmental material properties to gain broad insights into the coupling among different properties, the magnitude of the transcriptional effects, and the role of cell type. We find significant coupling among these properties, large variation in the magnitude of the transcriptional changes, and qualitative differences in the responses based on cell type, demonstrating the significant context dependence of microenvironmental material sensing.
Keywords: mechanotransduction, biomaterials, RNA-seq, stem cells, systems biology
Abstract
Variations in a multitude of material microenvironmental properties have been observed across tissues in vivo, and these have profound effects on cell phenotype. Phenomenological experiments have suggested that certain of these features of the physical microenvironment, such as stiffness, could sensitize cells to other features; meanwhile, mechanistic studies have detailed a number of biophysical mechanisms for this sensing. However, the broad molecular consequences of these potentially complex and nonlinear interactions bridging from biophysical sensing to phenotype have not been systematically characterized, limiting the overall understanding and rational deployment of these biophysical cues. Here, we explore these interactions by employing a 3D cell culture system that allows for the independent control of culture substrate stiffness, stress relaxation, and adhesion ligand density to systematically explore the transcriptional programs affected by distinct combinations of biophysical parameters using RNA-seq. In mouse mesenchymal stem cells and human cortical neuron progenitors, we find dramatic coupling among these substrate properties, and that the relative contribution of each property to changes in gene expression varies with cell type. Motivated by the bioinformatic analysis, the stiffness of hydrogels encapsulating mouse mesenchymal stem cells was found to regulate the secretion of a wide range of cytokines, and to accordingly influence hematopoietic stem cell differentiation in a Transwell coculture model. These results give insights into how biophysical features are integrated by cells across distinct tissues and offer strategies to synthetic biologists and bioengineers for designing responses to a cell’s biophysical environment.
Biophysical characteristics are key distinguishing features across tissues and at various stages of tissue development and pathology, and significant previous work has provided insights into how such features regulate physiology. For example, microenvironmental stiffness has been implicated in regulation of tumor progression (1) and stem cell fate determination (2), while dynamic adhesions to other cells and to the extracellular matrix (ECM) are critical in controlling force transmission and thus patterning in certain developmental contexts (3). The composition and density of adhesion ligands similarly has been shown to be an important variable in controlling cell adhesion strength (4), cell migration (5), and cell polarization (6) in contexts ranging from cancer to immune cell homing. Recently, significant differences in creep and stress relaxation across different tissues have been reported, and the understanding of the impact and mechanisms of these differences is currently evolving (7, 8).
Customizable hydrogel matrices can be engineered to display bespoke biophysical parameters to cells in 3D cell culture, and thus are useful tools for understanding how cells sense and respond to these cues. Differences in ECM density and composition across tissues can be reflected as differences in hydrogel adhesion ligand densities, viscoelastic behaviors, and elastic moduli. These materials have been instrumental in distilling complex in vivo biophysical environments into their salient features, which can then be perturbed to probe a cell’s response (9). This approach has successfully yielded careful studies of the roles of a multitude of biophysical matrix properties, including stiffness (10), viscoelasticity (8), nanotopography (11), adhesion ligand density (12) and composition (13), and nonlinear elasticity (14) on various cell behaviors, such as morphological changes, proliferation, and stem cell differentiation. However, the particular relevance of these changes to cell behavior across different tissues and between healthy and diseased tissue is poorly understood.
Interestingly, certain recent phenomenological work suggests that various microenvironmental features interact when cells integrate biophysical inputs (10, 12, 15), potentially sensitizing or desensitizing cells to one of these inputs or another. Mechanistic biophysical studies offer a possible explanation, whereby molecular clutches found in the cell’s adhesion complexes are manipulated mechanically by the material’s own mechanical response (16). It is therefore possible that a variety of material environments could induce analogous effects on the cell’s downstream response by perturbing cell–material interactions in mechanically redundant ways. However, we lack a broad view of how cells respond to these covarying biophysical parameters, which limits our understanding of the relative importance of these features in different physiologic contexts and restricts how bioengineers and synthetic biologists deploy biophysical cues. Additionally, the mechanisms by which cells integrate cues from their material microenvironments after the biophysics of sensing is not well understood. While a number of integration mechanisms have been described (17–19), whether these mechanisms integrate into one core substrate-sensing pathway, into individual responses for each biophysical feature, or into a combination of the two approaches is still unclear.
The purpose of this study was twofold: (i) to capture the broad transcriptional changes of cells associated with the sensing of different biophysical features of the microenvironment, and (ii) to test hypotheses surrounding the coupling of these features, the extent of cellular processes affected, and the influence of cell type. To address the uniqueness of the transcriptional effects of different biophysical parameters and how different biophysical features of a cell’s microenvironment couple transcriptionally, we encapsulated cells in ionically cross-linked alginate hydrogels. These gels afford independent control of multiple material properties presented to cells in 3D culture (8), have previously been used to study the effects of mechanosensing (10, 20), and exhibit a zonal cross-linking mechanism that confers consistent transport properties, such as diffusion, even as the mechanical properties of the material are tuned (21). In contrast, many materials systems vary biophysical properties in ways that are coupled to other material properties, or only allow 2D culture. Mouse mesenchymal stem cells (mMSCs) were chosen as a first cell type to study because they have a long history of use in the context of substrate-sensing (22) and have received significant clinical interest (23). Human cortical neuron progenitors (hNPCs) were also studied to explore how these interactions extend to a distinct species and cell lineage. RNA-seq was used to globally map the early transcriptional interactions among stiffness, stress relaxation rate, and adhesion ligand density, demonstrating dramatic transcriptional coupling between these features in both cell types. Finally, motivated by the bioinformatic analysis, we found that the stiffness of hydrogels containing mMSCs modulated hematopoietic stem cell differentiation in a coculture system. Overall, the results demonstrate that different biophysical characteristics of a cell’s microenvironment contextualize each other and are tunable to elicit a wide range of responses. This work additionally demonstrates an approach to marry techniques from bioengineering and next-generation sequencing to gain insights into biophysical principles.
Results
Transcriptomic Comparison of Material Parameter Sensing in mMSCs.
We first developed a workflow to perform RNA-seq on cells cultured in bespoke material microenvironments. Because previous studies have thus far characterized monotonic responses to both adhesion ligand density (12) and stress relaxation (8), we chose low and high values for each of these parameters to capture a physiologically relevant, but wide range. Adhesion ligand density was varied from 200 (±50) μM to 1,500 (±200) μM (SI Appendix, Fig. S1C), spanning an estimated physiologic range (calculations in SI Appendix, Methods). Stress relaxation was found to vary from t1/2 of 35 (±10) s to 790 (±75) s (SI Appendix, Fig. S1D), comparable to stress relaxation values measured in coagulated bone marrow and liver, respectively, and to those previously found to impact long-term MSC differentiation using this same material system (20). While cellular responses to stiffness have been shown to be nonmonotonic in certain cases, we chose to first use a low (3 ± 1 kPa) and a high (30 ± 2 kPa) value of stiffness, again spanning a physiologically relevant and large range to keep the experiment symmetric. To isolate the effects of each of the three parameters and broadly capture any interactions, hydrogels were prepared in eight combinations of the low and high values for each material property (Fig. 1A) and mMSCs were encapsulated. Specifically, we chose to use the D1 cell line (24) to minimize cell–cell heterogeneity. After 40 h, encapsulated cells were isolated and analyzed using RNA-seq. This time point was chosen to allow for the formation of mature cell adhesions and to minimize proliferation, which could confound the results. We confirmed that proliferation as measured by cell number did not significantly change during culture and that the cells were highly viable, and homogeneously distributed throughout the material (SI Appendix, Figs. S2 and S3). Intriguingly, principal component analysis (PCA) of RNA-seq data discriminated among material parameters only if the material was soft (SI Appendix, Fig. S5A), which suggested that the primary sources of variation in transcriptional profiles were not uniquely tied to one material parameter, but to combinations thereof. Next, independent component analysis (ICA) was used to reduce the dimensionality of the transcriptional profiles into statistically independent components, which one would expect to map to one or more material properties if their responses were merely superimposed. However, ICA failed to produce clear separation by any property (SI Appendix, Fig. S5C), suggesting that the primary determinants of the cell’s substrate response are not independent. Pearson correlations of gene expression demonstrated minimal response of the cells to the calcium concentrations in different gels (SI Appendix, Fig. S6), confirming previous reports that the variation in calcium content in this system has minimal impact on encapsulated MSCs (8, 10).
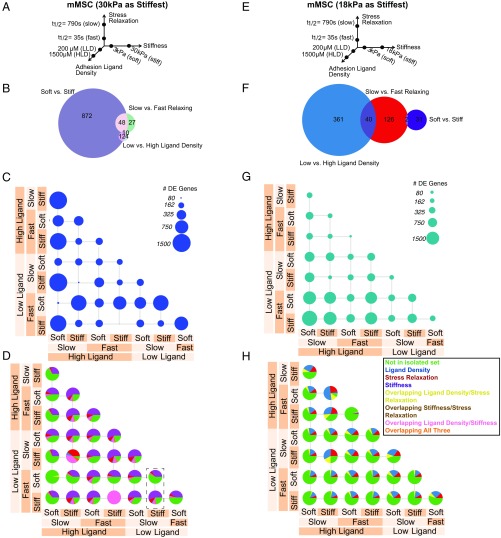
Fig. 1.
Transcriptomic comparison of material parameter sensing in mMSCs. (A–D) Transcriptomic comparison of material parameters sensing with 30 kPa as the high stiffness. (E–H) Transcriptomic comparison of material parameters sensing with 18 kPa as the high stiffness. (A and E) Schematic of experimental conditions for mMSC culture. Hydrogels were fabricated in each of the eight combinations of the low- and high-parameter values and cells were seeded at a density of 10 million cells per milliliter. (B and F) Venn diagrams of DE genes in mMSCs for each material parameter comparison after controlling for other parameters. The numbers of DE genes shared by two parameters are indicated in the overlap in circles. (C and G) Number of DE genes in mMSCs for all pairwise material comparisons. Circle area corresponds to the number of DE genes as indicated in the legend. (D and H) Fraction of DE genes from C and G described by decoupled genes in B and F for all pairwise material comparisons in mMSCs. Green, DE genes not found in the sets from B and F; blue, DE genes from ligand density set from B and F; red, DE genes from stress relaxation set from B and F; purple, DE genes from stiffness set from B and F; yellow, DE genes from overlapping ligand density and stress relaxation set from B and F; brown, DE genes from overlapping stiffness and stress relaxation set from B and F; pink, DE genes from overlapping ligand density and stiffness set from B and F; orange, DE genes from overlapping ligand density, stiffness, and stress relaxation set from B and F. The dashed box in D highlights a comparison in which comparing one material parameter (stress relaxation) results in a different pie chart if the background stiffness is different.
We next used a linear model to extract differentially expressed (DE) genes affected by one of the parameters regardless of the background parameters. For example, this approach reveals DE genes affected by stiffness independently of changes in the stress relaxation or ligand density. A Venn diagram of the resulting decoupled DE genes strikingly finds a large discrepancy in the number of DE genes for the different parameter comparisons (Fig. 1B). Stiffness drove the largest number of DE genes, followed by stress relaxation and ligand density (Fig. 1B). We then performed all pairwise comparisons, and found that the number of DE genes varied between 0 and over 1,500. The latter number far exceeded that of the decoupled gene sets (Fig. 1C), which suggests the superposition of different material sensing mechanisms. We then mapped the decoupled gene sets to those from each material comparison and found that the covariation of multiple material parameters at once gives rise to a large increase in the number of DE genes not noted in the decoupled sets. Moreover, the relative contribution of these gene sets to the DE genes in each comparison varied dramatically based on the background material parameters (Fig. 1D), indicating the presence of coupling and switching mechanics in these material-sensitive gene networks (Fig. 1D). For example, examining the dashed box in Fig. 1D, a different composition of DE genes results when one compares slow to fast stress relaxation in low ligand-density materials at different levels of stiffness.
Because previous data had depicted distinct stiffness values promoting distinct cell fates (25), we repeated this analysis including 18 kPa (±1 kPa) in place of 30 kPa as the high stiffness (Fig. 1E). Strikingly, we found a markedly different relationship among the different parameters, with ligand density inducing the largest number of DE genes, followed by stress relaxation and then stiffness (Fig. 1F). This result is contrasted with stiffness dominating the Venn diagram in the 30 kPa case (Fig. 1B). Thus, the specific stiffness value tested altered the relative ranking of the magnitude of gene-expression changes in response to the three material parameters, signifying that material parameters can be quantitatively tuned to alter the sensitivity of cells to other parameters. The different material parameters were again found to couple in these comparisons, although with different contributions from each parameter (Fig. 1 G and H) than noted in the previous comparison. It should be noted that this tuning does not imply orthogonal parameter sensing mechanisms, but could involve altered signaling in common or convergent pathways.
Transcriptomic Comparison of Material Parameter-Sensing in hNPCs.
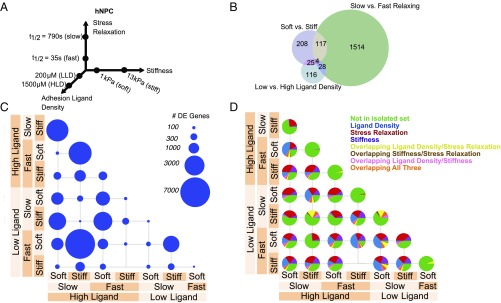
To gauge how relationships among material properties generalize to diverse cell types, we chose another cell type from a different species and developmental lineage and stage than the mMSCs, and next performed the same experiment on human induced pluripotent stem cell (iPSC)-derived NPCs. A stiffness range from 1 to 13 kPa that is physiologically relevant for neural tissues (26, 27) was used in this study (Fig. 2A and SI Appendix, Fig. S1B). Neural lineage cells have been shown to be responsive to substrate stiffness (26, 27), and here hNPCs were generated using an established protocol for producing large and homogeneous hNPC populations (28) (SI Appendix, Figs. S7–S9). As with the mMSCs, Pearson correlations of gene expression demonstrated minimal response of the cells to the calcium concentrations in different gels (SI Appendix, Fig. S10). Similar to the analysis with mMSCs, after PCA the second principal component separated otherwise equivalent conditions by adhesion ligand density only if the materials were soft (SI Appendix, Fig. S5D), consistent with the notion of dependent material property-sensing. We found a large disparity in the number of DE genes, now with stress relaxation inducing the largest change, followed by stiffness and ligand density (Fig. 2B). Individual comparisons between material parameter combinations revealed gene-expression effects that spanned multiple orders-of-magnitude depending on the comparison (Fig. 2C), and these changes again featured genes found in none of the decoupled gene sets, indicating that different parameter combinations elicited distinct gene programs (Fig. 2D). Moreover, enrichment analysis for the sets of DE genes corresponding exclusively to one parameter or another revealed processes important for central nervous system (CNS) regulation (SI Appendix, Fig. S11). For example, ligand density induced DE genes related to regulation of serotonin secretion and synaptic transmission, stiffness induced DE genes related to regulation of Tau pathology in Alzheimer’s disease and dopamine transactivation of PDGFR in the CNS, and stress relaxation induced DE genes tied to neurofilament remodeling and myelination, among others. Moreover, drug target analysis on all DE genes across all parameters noted 48 drug targets that were affected by substrate parameters (SI Appendix, Fig. S12).
Fig. 2.
Transcriptomic comparison of material parameter sensing in hNPCs. (A) Schematic of experimental conditions for hNPC culture. Hydrogels were fabricated in each of the eight combinations of the low- and high-parameter values and seeded at a density of 5 million cells per milliliter. (B) Venn diagram of DE genes in hNPCs for each material parameter comparison after controlling for other parameters. The number of DE genes shared by two parameters are indicated in the overlap in circles. (C) Number of DE genes in hNPCs for all pairwise material comparisons. Circle area corresponds to the number of DE genes as indicated in the legend. (D) Fraction of DE genes from C described by decoupled genes in B (each Venn diagram slice) for all pairwise material comparisons in hNPCs. Green, DE genes not found in the sets from B; blue, DE genes from ligand density set from B; red, DE genes from stress relaxation set from B; purple, DE genes from stiffness set from B; yellow, DE genes from overlapping ligand density and stress relaxation set from B; brown, DE genes from overlapping stiffness and stress relaxation set from B; pink, DE genes from overlapping ligand density and stiffness set from B; orange, DE genes from overlapping ligand density, stiffness, and stress relaxation set from B.
Coexpression Analysis of Material Parameter-Sensing Networks.
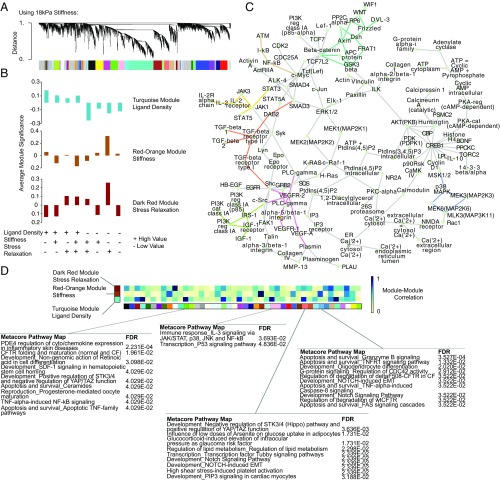
Because certain features of regulatory networks might not be reflected in differential-expression analyses, weighted gene coexpression analysis (29) (WGCNA) was performed to identify modules of highly coexpressed genes that correspond to the sensing of each parameter (Fig. 3A). The three modules with the strongest correlations to our parameters of interest were then chosen for further analysis. We first ran this analysis for the mMSCs using the extreme values for stiffness from Fig. 1 (3 kPa and 30 kPa); however, we only identified modules that consistently corresponded to stiffness (SI Appendix, Fig. S13), likely because the extreme stiffness comparison dominates the variance in the data. However, including the 18-kPa conditions instead of the 30-kPa conditions better matched the sensitivity of cells to all parameters, allowing WGCNA to identify gene modules that correspond to each of the three material parameters (SI Appendix, Fig. S14). We proceeded with this stiffness range to better discriminate modules tied to each material parameter. Plotting the average module significance for each module as a function of the material confirmed the correspondence of each to the parameter of interest (Fig. 3B). Metacore PathwayMap enrichment analysis on the member genes for these modules revealed processes involving cytoskeletal remodeling, cell adhesion, and PDGF signaling (SI Appendix, Fig. S13). Inspection of the top hub genes in these modules revealed modular genes involved in signal transduction and protein transport. Of particular note were Klf6 and Klf4, found in the ligand density and stiffness-associated modules, respectively, both of which have been shown to regulate stem cell differentiation. Also of note in the stiffness module were the immune related-kinase Prkra and the YAP target gene Ankrd26, while Ptges2, a key regulator of MSC immunomodulation, was noted in the stress relaxation module (SI Appendix, Fig. S13). The correlation of each of these modules to the others identified by WGCNA, along with the corresponding ontology annotation, provided a quantitative metric for the inferred regulatory connectedness of the sensing of a material property with other cellular processes (Fig. 3D). In this case, we intriguingly found that certain immune-related processes, such as PDE4 expression, TNF-induced NF-κB signaling, and IL-3 signaling, were enriched in modules with strong correlations to one of the three modules of interest (Fig. 3D). These results were used to inform a putative limited network of material-sensitive genes that again revealed the prominent presence of MAPK, Wnt, and TGF-β signaling pathways. Also notable were genes involved in cell adhesion, such as FAK and integrins (Fig. 3C).
Fig. 3.
WGCNA of material parameter sensing networks in mMSCs. (A) Cluster dendrogram of gene expression showing module identification from WGCNA using an unsigned network and a soft thresholding parameter of 10 for the dataset containing 18-kPa hydrogels as the stiffest condition. (B) Selection of modules that most closely map to ligand density, stiffness, and stress relaxation for the dataset containing 18-kPa hydrogels as the stiffest condition. Average module significance is plotted as a function of each material, showing the correspondence between the module and that parameter of interest. These modules are identified in SI Appendix, Fig. S13. (C) Putative gene network seeded using the top hub genes from each of the modules corresponding to ligand density, stiffness, and stress relaxation (turquoise, red-orange, dark red). Enriched subnetworks were inferred using Metacore software and the three most significantly enriched (highest z-score) subnetworks were chosen and merged to arrive at the network shown. Connections corresponding to the Wnt (teal), TGF-β (orange), VEGF (pink), NF-κB (brown), Jak/STAT (yellow), IGF (green), and MAPK (purple) pathways are highlighted. (D) Heatmap showing absolute value of spearman correlations between the turquoise, orange-red, and dark red modules and all other modules, calculated by correlating the average expression of each module’s member genes. Module member genes for select comparisons showing particularly high or low correlations to the turquoise, orange-red, and dark red modules were selected and the most significant Metacore PathwayMaps were identified using these genes as seeds. Blue corresponds to high correlations and white to low correlations.
WGCNA performed on the hNPC dataset also revealed intriguing relationships (SI Appendix, Fig. S15). Enrichment analysis performed on the module with the highest correlation to stiffness included processes, such as axon development and WNT signaling. The stress-relaxation module was enriched for ECM organization, IL-4 and IL-13 signaling, and Hippo signaling. Finally, the module corresponding to ligand density showed enrichment for morphogenesis processes and neurotransmitter transport.
Functional Testing of Bioinformatic Hypotheses.
To functionally test hypotheses generated by the bioinformatic analysis, we selected a particular comparison between two materials and explored processes predicted by Gene Ontology analysis to be affected. Specifically, we used the DE genes generated from comparing the fast-relaxing, high ligand density 3-kPa hydrogels, to the fast-relaxing, high ligand density 18-kPa hydrogels. Performing Gene Ontology analysis on these DE genes generated several statistically significant processes likely to be affected by the DE genes (SI Appendix, Fig. S17). Among these terms, of note was “hematopoietic progenitor cell differentiation.” MSCs are known to be active in the hematopoietic stem and progenitor cell (HSPC) niche and express HSPC regulatory factors, such as OPN, CXCL12, Tnfrsf1a, Tnfrsf1b, and MCP-1 (30, 31), although the mechanical regulation of this cross-talk and potential mechanical intervention has not been explored. Thus, as a case study of exploring hypotheses concerning material regulation of MSC cytokine secretion that could ultimately have impacts for cell therapies, we examined the effects of the MSC substrate on supporting cultured HSPCs. First, to confirm the relevance of the substrate to secretion of relevant cytokines from MSCs, from day 2–3 of culture, we collected conditioned media from mMSCs cultured in fast-relaxing alginate hydrogels of different ligand densities (200 and 1,500 μM) and stiffnesses (3 and 18 kPa) and analyzed the mMSC secretome using a cytokine antibody array. Numerous cytokines in the array were expressed differentially as stiffness and ligand density were altered. WGCNA modules with high correlations to both stiffness and ligand density were noted to include a number of processes involving secreted cytokines, consistent with this experiment (Fig. 3D). Hierarchical clustering revealed distinct groups of cytokines that associate with the various material conditions (Fig. 4A). Notable were TPO, SDF-1, OPN, IGF1, and IGF-2, which have all been shown to be involved in MSC–HSPC cross-talk (30, 31). The observation that these functionally distinct cytokines separate into different clusters based on substrate properties suggests that the material context of MSCs could potentially regulate different aspects of MSC–HSPC interaction. Interestingly, SDF-1 was found to cluster with IGF1, IGFbp-2, and IGFbp-3, which have been found to enhance SDF-1 expression in MSCs via HIF-1α and PI3K-dependent mechanisms, consistent with these predictions.
Fig. 4.
mMSCs modulate secreted cytokines in response to substrate stiffness. (A) Heatmap representing results of cytokine antibody array performed on conditioned media from mMSCs cultured in hydrogels from days 2–3 of culture. Values were normalized to internal positive controls and the maximum signal for each cytokine across the four materials. Cytokines were hierarchically clustered using a Euclidean distance metric and complete linkage. (B) Schematic of MSC–HSPC coculture set-up. Fast-relaxing hydrogels were used for all experiments. Soft corresponded to 3 kPa and stiff corresponded to 18 kPa. (C) Viable cell number as counted by flow cytometry of cells seeded on Transwell membrane after 1 wk of coculture. Error bars represent SD (one-way ANOVA, Tukey post hoc test, *P < 0.05, **P < 0.01). (D) Number and percentage of CD45+/lin− cells from Transwell membrane after 1 wk of coculture as analyzed by flow cytometry. Error bars represent SD (one-way ANOVA, Tukey post hoc test, *P < 0.05, **P < 0.01).
Using a Transwell coculture system, we then encapsulated mMSCs in alginate hydrogels and cocultured these cells with primary mouse CD45+/Lin−/Ckit+/Sca1+ cells, a putative hematopoietic stem cell population, seeded on the Transwell membrane (Fig. 4B). After 1 wk, HSPCs were collected, counted, and analyzed for maintenance of stemness by flow cytometry. We found that HSPC proliferation was not significantly different across conditions (Fig. 4C), but that the percentage and number of Lin−/CD45+ cells were higher in softer gels independent of ligand density (Fig. 4D), suggesting that altered cytokine secretion profiles mediated by the substrate are associated with functional differences in MSC–HSPC interactions. Moreover, inspection of WGCNA modules with high correlations to changes in stiffness implicated Notch and TNF signaling (Fig. 3D, steel blue module), both of which have been shown to be important factors regulating the HSPC niche (32). Intriguingly, while the coculture system tested here did not allow for direct cell–cell contacts, the Notch prediction also suggests that cell–cell contacts could be altered by the substrate. It should be noted that while the specific material parameter values used for the MSC–HSPC cross-talk experiments were chosen due to their link to the RNA-seq results, additional distinct parameter combinations could potentially yield more dramatic effects.
Discussion
Specific combinations of the three substrate properties dramatically impacted the number of DE genes in both mMSCs and hNPCs. The transcriptional responses from these specific parameter combinations were found to be partially specific to each combination. While for 30-kPa hydrogels stiffness dominated other parameters in mMSCs, and confirmed the implication of previously described substrate-sensitive transcriptional programs, with 18-kPa gels the contribution of stiffness was diminished. This dose-dependence of substrate properties mirrors previous studies of stiffness, stress relaxation, or ligand density individually. For example, osteogenesis has been shown to display a biphasic relationship with stiffness in similar hydrogels, peaking at ∼20 kPa (10). Meanwhile, osteogenic differentiation in mMSCs has been reported to increase with faster stress relaxation times down to t1/2 = 60 s, while adipogenic differentiation displayed the opposite result over the same stress relaxation range (8). Moreover, increasing ligand density from 150 to 1,500 μM nearly tripled markers of osteogenic differentiation in 17-kPa alginate hydrogels with t1/2 between 60 and 300 s (8). In contrast to the mMSC case, stress relaxation dominated the transcriptional response of hNPCs in relatively softer conditions. These findings suggest that because the phenotypic responses to these variables are dose-dependent, the molecular components of the response that are also sensitive to other material parameters will similarly have a dose-dependent response. In this way, it is possible that the specific values of one parameter contextualize the response to other parameters through these common intermediates. The results also follow the biophysical mechanisms of substrate sensing, whereby the ability to cluster adhesion ligands mediates the stability of cell focal adhesions and thus downstream response (33). In this case, each of the substrate parameters of interest is implicated in controlling the ability of cells to cluster these receptors. However, the downstream response of this clustering is cell-type–specific; this work demonstrates the context-dependence of biophysical sensing, not only in terms of the specific combination of parameters present, but also in terms of the cell (species, lineage) that is doing the sensing. The relative differences in the sensitivity of gene expression to substrate properties observed between the mMSCs and hNPCs are also consistent with each cell type’s in vivo biophysical context. MSCs are found throughout the adult body (34) and are thus exposed to a wide range of tissue stiffnesses. NPCs, however, are exposed to a relatively narrow range of stiffnesses in CNS development, as evidenced by the marked similarity in stiffness between the embryonic (35) and adult (36) brain. It is possible that the increased matrix content and cross-linking observed over time in CNS development (35) could alter the viscous properties of the substrate and thus expose these cells to a comparatively large range of stress relaxation values. In any case, because there has been significant interest in using iPSC-derived cells therapeutically, the substrate regulation of iPSC-derived NPCs suggests a strategy of utilizing material carriers to regulate their fate. We should note, however, that the absolute magnitude of gene-expression changes between the mMSCs and hNPCs is likely a function of both species and lineage differences.
While mMSCs and hNPCs were found to be most sensitive to different parameters, the observations that (i) there exists coupling and sensitization of the response to particular material properties, as a function of the values of other properties (Figs. 1 C and G and 2C) and (ii) that these combinations give rise to distinct gene-expression responses (Figs. 1 D and H and 2D), were similar between the two cell types. These results are consistent with previous phenomenological studies showing how certain matrix properties can modulate the response of cells to other matrix properties (12). As a specific illustration of this point in mMSCs, Chaudhuri et al. (8) cultured mMSCs in 9- and 17-kPa alginate hydrogels of similar ligand density and stress relaxation to those used in this study. When they assayed these cells for osteogenic and adipogenic differentiation, the authors found that stiffness acted as a switch that could turn the quantitative stress relaxation dependence on or off. Similar switching effects were seen with the interaction between ligand density and stress relaxation. While Chaudhuri et al. (8) used stiffness values that varied slightly from those used in this study to study stress relaxation and ligand density, our observation of switching dynamics in the magnitude of gene-expression responses (Fig. 1 C and G) is similar to the switching effects that Chaudhuri et al. (8) reported. Generally, it should also be noted that these results demonstrate the contextualization of material sensing, which implies that other important variables, such as cell density, cell history, and ligand composition, could add additional layers of complexity to this sensing behavior. Our hNPC substrate-sensing results complement recent work on NPC multiparametric substrate sensing. For example, Madl et al. (37) showed that NPC substrate remodeling is required for maintenance of NPC stemness, an effect that was independent of substrate stiffness. Varying the stress relaxation in alginate hydrogels has been shown to influence the degree of substrate remodeling (8); thus, our results demonstrating an increased sensitivity in NPC gene expression to stress relaxation versus stiffness is consistent with the results of Madl et al. (37). Tekin et al. (38) performed a transcriptomic analysis of several common NPC culture conditions. While the NPCs were found to be sensitive to Matrigel concentration and stiffness in at least some subset of genes, PCA only discriminately clustered culture conditions with extreme (∼10-fold) differences in stiffness, again consistent with our observed reduced sensitivity of NPCs to stiffness (38).
Inferring a regulatory network in mMSCs using enriched hubs from the WGCNA analysis revealed significant involvement of the canonical signaling pathways MAPK, Wnt, and TGF-β. These results are consistent with studies linking each of these pathways to substrate-sensing (39–41). Because a large number of ligands activate these pathways, including many drug targets, implications for cross-talk between soluble molecular signaling and substrate sensing are potentially far reaching (42–44). Further epistatic analyses would be useful in elucidating such cross-talk.
Our coexpression network analysis revealed modules of genes tightly associated with the sensing of each of the biophysical properties. Annotating each of these modules and correlating them with the parameters of interest provided a score for the potential connection between these parameters and diverse cellular processes. While this approach is correlative, its value lies in both mapping the strength of the responses to these biophysical parameters and generating hypotheses for their effects. Such metrics could prove useful in identifying cellular processes to be targeted using materials or even to identify potential unintended consequences of a biomaterial design.
Using the bioinformatic analysis to generate hypotheses, we found that the substrate stiffness presented to mMSCs modulated the secretion of a host of cytokines, notably ones that affected HSPC differentiation. While MSC–HSPC interactions have been previously described in significant detail (30, 31), the present results have implications for the regulation of stem cell populations in the HSPC niche by mechanical means. For example, enrichment of adipocytes in the bone marrow has been associated with aging and obesity (45), potentially affecting bone marrow mechanics. Thus, it is possible that these sorts of mechanical alterations to the bone marrow could affect the MSC–HSPC interactions described in this work. Moreover, because MSCs have been shown to signal to HSPCs, regulating their homing and retention in the bone marrow (46), it raises the possibility that the variation in HSPC substrate properties in addition could influence HSPC response to that signaling. Our results demonstrating the mechanical regulation of MSC–HSPC interactions suggest avenues for further controlling associated cell therapies, such as through cell delivery via biomaterial carriers or through pharmacologic manipulation of mechanosensitive pathways.
In addition to the predictions concerning MSC–HSPC interactions, our analysis generated additional hypotheses that would be interesting to explore. For example, the prominence of immune processes in modules from the mMSC analysis is particularly intriguing because MSCs have been shown to display an immunomodulatory phenotype (47). In fact, many of the proposed mechanisms for MSC cell therapies are immunological in nature. These results suggest hypotheses for how biophysical aspects of the MSC microenvironment could regulate their immunomodulatory responses. Moreover, while previous studies of mMSCs in similar hydrogels with nearly quantitatively identical properties to those in this work have reported phenotypic outputs, such as proliferation, differentiation, and morphology (8, 10), the diversity of gene-expression changes induced by these materials suggests that these previously assayed phenotypes could represent just a subset of those “reachable” by material interventions. Furthermore, it is possible that some of these distinct molecular responses are overlaid on the phenotypes previously reported. For example, the cells used in previous studies reporting the effects of substrate mechanics on differentiation or proliferation could also potentially display additional molecular phenotypes that could have immunological consequences but were not tested in those studies.
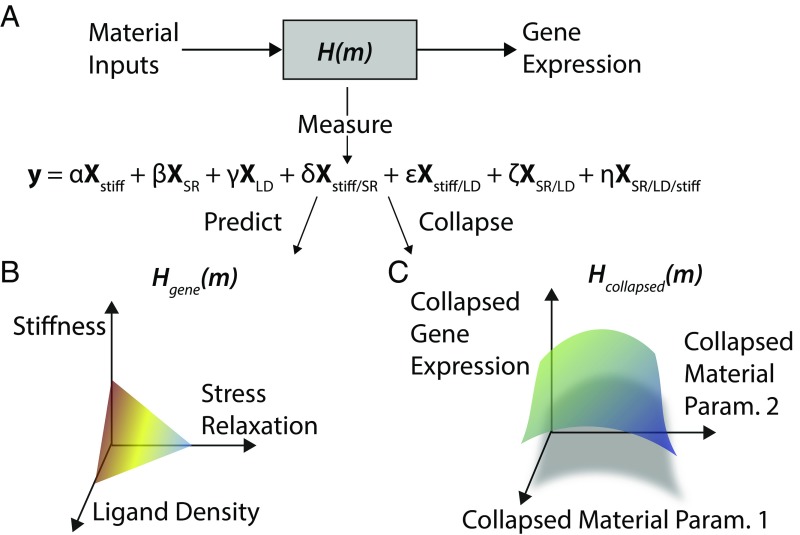
The overall approach described herein of integrating high-dimensional biophysical experiments with high-throughput bioassays to bridge the gap between physical aspects of biology and their biological consequences yields important insights and is likely to become more commonplace. This mapping from substrate input to gene expression can be captured by a linear model that both describes the RNA-seq data as a function of substrate properties and is used to compare the relative significance of changing one or more of these parameters (Fig. 5A). Having such a representation allows for the production of a response surface that describes gene-expression changes as a function of a substrate property coordinate, potentially serving as a predictive tool for biomaterials engineers (Fig. 5B). Additionally, dimensionality reduction techniques, such as PCA used on sequencing data, could be used alongside collapsed representations of substrate properties, such as dimensionless parameters, to generate otherwise difficult visualizations of the global effects on cells in a high-dimensional input and output space (Fig. 5C).
Fig. 5.
Model for gene-expression effects of substrate properties. (A) Model of the substrate response, where cells sense material inputs, process them, and translate them into changes in gene expression. Formulating a statistical model of gene expression as explained by substrate properties and their interaction is loosely analogous to the transfer function between the gene-expression output and the substrate property input. (B) Response surface for gene expression as a function of substrate properties. One of these surfaces exists for each gene. (C) Response surface for collapsed gene expression as a function of collapsed substrate properties. In this approach, the dimensionality of gene-expression space and the substrate property space are collapsed to enable a holistic view of the most prominent substrate effects.
Methods
Casting of Alginate Hydrogels.
Alginate type LF20/40 (FMC Biopolymer) was used as-received for the slow-relaxing hydrogels and was irradiated with an 8-mRad cobalt source to form the fast-relaxing hydrogels. Alginates were modified with GGGGRGDSP peptides (Peptide 2.0) at the reported densities with standard carbodiimide chemistry, as described previously (48). After modification, alginates were dialyzed against a NaCl gradient, treated with activated charcoal, and sterile-filtered. After lyophilization, all alginate was dissolved in serum-free DMEM (Lonza) at 2.5%.
Hydrogels were cast by rapidly mixing the alginate solution with a CaSO4 slurry via two syringes and ejecting the mixture between two glass plates, where it gelled over 1.5 h.
More details on the specific formulations for each condition can be found in SI Appendix.
Hydrogel Mechanical Characterization.
Hydrogels were fabricated as described above at a thickness of 2 mm and subjected to compression testing using a mechanical testing device (Instron). Gels were compressed at a strain rate of 1 mm/min and the Young’s Modulus was calculated as the best-fit slope of the first 5–15% of the resulting stress/strain curve. At 15% strain, the strain was held and the time required for the stress to decay by a factor of two was noted. More details can be found in SI Appendix.
RGD Peptide Quantification.
RGD coupling density was determined using the LavaPep assay following the manufacturer’s instructions. Coupled alginate was dissolved at a concentration of 0.1 mg/mL in PBS before incubation with the LavaPep reagents. A standard curve of GGGGRGDSP peptides was prepared in PBS containing 0.1-mg/mL uncoupled alginate as background. Fluorescence was read using a Biotek plate reader and the resulting concentration was used to find the molar ratio of alginate to peptide. Molar concentration of peptide was calculated assuming a 2% alginate gel from the molar ratio.
mMSC Cell Culture.
D1 mMSCs (ATCC) were encapsulated in the hydrogels during the mixing step at a concentration of 10 million cells per milliliter. After casting and punching, gels were placed in 24-well plates and cultured at 37 °C in DMEM (Lonza) with 10% FBS and 1% penicillin/streptomycin. More details can be found in SI Appendix.
Live/Dead Staining.
Gels were treated with Life Technologies Live/Dead reagent per the manufacturer’s specifications and were then transferred to a microscope slide with a custom-made PDMS well. A coverslip was placed over the hydrated gel and the gels were imaged on a Zeiss LSM 710 upright confocal microscope. Viability was quantified by computing the number of live and dead cells across five representative fields-of-view using ImageJ.
Cell Retrieval from Gels.
After 40 h of culture, gels were removed from the wells and placed into Eppendorf tubes with 50 mM EDTA in Hepes on ice for 10 min. An equal volume of trypsin-EDTA was then added to the tubes for an additional 5 min at 37 °C to ensure the removal of cells from the alginate chains. Cells were centrifuged and rinsed twice before proceeding to additional analysis.
Cell Counting for Proliferation Analysis.
After cell retrieval, as described above, cells were diluted per the manufacturer’s instructions and counted on a Countess FLII automated cell counter (Life Technologies). Cell counts were compared with the original encapsulated cell numbers.
RNA-Seq.
After cell retrieval as described above, cells were lysed and total RNA was extracted per the manufacturer’s instructions with the Qiagen RNeasy Micro kit. Samples were then submitted to the Harvard Medical School Biopolymers Facility, where mRNA enrichment and library preparation was performed. Individual samples were barcoded and run on either an Illumina HiSEq. 2500 Rapid or an Illumina NextSeq.
Statistical Methods.
Statistics for RNA-seq experiments are described in mMSC RNA-Seq DE Analysis and hNPC RNA-Seq Differential Expression Analysis. For flow cytometry for the mMSC immunomodulation and MSC–HSPC cross-talk experiments, Igor Pro software was used to run one-way ANOVA, followed by a Tukey post hoc test.
mMSC RNA-Seq DE Analysis.
Raw reads were aligned to the University of California, Santa Cruz (UCSC) Genome Browser mm10 genome using Subread (49) and counts were aggregated per gene using FeatureCounts (50). After aggregating read counts, we performed TMM normalization. Voom (51) and Limma (52) were then used to perform DE analysis using a multilevel factorial design. Batch was accounted for with an additional factor in the linear model. DE genes were defined as those with a fold-change of at least 2 and a BH-adjusted P value of less than 0.05. For visualization and clustering, Combat was used to remove batch effects. qPCR on selected transcripts and material conditions mirrored the sequencing results (SI Appendix, Fig. S16). More details regarding the creation of Figs. 1 and 2 can be found in SI Appendix.
Neural Progenitor Production.
The human iPSC line 1016a (certified mycoplasma negative and karyotypically normal) was differentiated using a published cortical neuron protocol (28). Cells were plated on a Greiner microclear 96-well plate coated with laminin, polyornithine, and fibronectin for culture. More details regarding hNPC production, marker verification, and staining can be found in SI Appendix.
hNPC Cell Culture.
After production, hNPCs were encapsulated in the hydrogels during the mixing step at a concentration of 5 million cells per milliliter. After casting and punching, gels were placed in 24-well plates and cultured at 37 °C in NIM media (described above). Cells were stained for viability as described for mMSCs and were found to be highly viable and evenly distributed throughout the gel (SI Appendix, Fig. S9).
hNPC RNA-Seq Differential Expression Analysis.
Raw reads were aligned to the UCSC Genome Browser hg38 genome using Subread (49) and counts were aggregated per gene using FeatureCounts (50). Subsequent analysis largely follows that for the mMSCs. More details can be found in SI Appendix.
WGCNA.
WGCNA (29) was run on the Combat-cleaned TPM data. Module significance for stiffness, stress relaxation, and ligand density was computed for each module by correlating the expression of module member genes each parameter encoded as low (0) or high (1) and taking the average gene significance for that module. More details can be found in SI Appendix.
Metacore Network Analysis and Visualization.
For the Fig. 3 analysis, the Metacore web tool was used to identify enriched pathways and to construct the putative substrate sensing network. For the drug target analysis, DE genes for hNPCs corresponding to stress relaxation, stiffness, and ligand density were pooled and fed into the Drug Target pipeline in Metacore. The drug hits in the “therapeutic drug-target interactions” list were taken. More details can be found in SI Appendix.
qPCR.
Cells were retrieved from gels as described above and total RNA was extracted using the Qiagen RNeasy micro kit following the manufacturer’s instructions. Reverse transcription was carried out using Bio-Rad iScript Advanced cDNA synthesis kit and PrimePCR validated primers (SI Appendix, Table S1) along with Bio-Rad sso Advanced Universal SYBR Green Supermix were used for the qPCR assay. More details can be found in SI Appendix.
MSC–HSPC Coculture Experiment.
HSPCs were isolated from the tibia, femur, and pelvis of 6- to 12-wk-old wild-type C57BL/6 mice and seeded on a Transwell membrane. D1s were cocultured with HSPCs in StemSpan SFEM (StemCell Technologies) supplemented with 10% FBS, 1% penicillin/streptomycin, and 10 ng/mL recombinant mouse SCF, FLT3L, and IL-7 (BioLegend). Media change was performed every 2 d, and the coculture was terminated after 1 wk. More details can be found in the SI Appendix.
Conditioned Media Experiment.
Alginate hydrogels containing mMSCs were fabricated as indicated above. We collected the culture media from days 2 to 3. This conditioned media was used per the manufacturer’s specifications in Abcam’s ab193659 mouse 96-target cytokine array and chemiluminescence was read on a FluorChem M imager. More details can be found in SI Appendix.
Supplementary Material
Acknowledgments
We acknowledge NIH/National Institute of Dental and Craniofacial Research Grant R01 DE013033 for funding.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the NCBI Short Read Archive (accession nos. PRJNA419311 and PRJNA419361).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1802568115/-/DCSupplemental.
References
- 1.Levental KR, et al. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell. 2009;139:891–906. doi: 10.1016/j.cell.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murphy WL, McDevitt TC, Engler AJ. Materials as stem cell regulators. Nat Mater. 2014;13:547–557. doi: 10.1038/nmat3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cukierman E, Pankov R, Stevens DR, Yamada KM. Taking cell-matrix adhesions to the third dimension. Science. 2001;294:1708–1712. doi: 10.1126/science.1064829. [DOI] [PubMed] [Google Scholar]
- 4.Balaban NQ, et al. Force and focal adhesion assembly: A close relationship studied using elastic micropatterned substrates. Nat Cell Biol. 2001;3:466–472. doi: 10.1038/35074532. [DOI] [PubMed] [Google Scholar]
- 5.Gardel ML, Schneider IC, Aratyn-Schaus Y, Waterman CM. Mechanical integration of actin and adhesion dynamics in cell migration. Annu Rev Cell Dev Biol. 2010;26:315–333. doi: 10.1146/annurev.cellbio.011209.122036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sackmann E. How actin/myosin crosstalks guide the adhesion, locomotion and polarization of cells. Biochim Biophys Acta. 2015;1853:3132–3142. doi: 10.1016/j.bbamcr.2015.06.012. [DOI] [PubMed] [Google Scholar]
- 7.Sack I, Johrens K, Wurfel J, Braun J. Structure-sensitive elastography: On the viscoelastic powerlaw behavior of in vivo human tissue in health and disease. Soft Matter. 2013;9:5672–5680. [Google Scholar]
- 8.Chaudhuri O, et al. Hydrogels with tunable stress relaxation regulate stem cell fate and activity. Nat Mater. 2016;15:326–334. doi: 10.1038/nmat4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huebsch N, Mooney DJ. Inspiration and application in the evolution of biomaterials. Nature. 2009;462:426–432. doi: 10.1038/nature08601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huebsch N, et al. Harnessing traction-mediated manipulation of the cell/matrix interface to control stem-cell fate. Nat Mater. 2010;9:518–526. doi: 10.1038/nmat2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dalby MJ, Gadegaard N, Oreffo ROC. Harnessing nanotopography and integrin-matrix interactions to influence stem cell fate. Nat Mater. 2014;13:558–569. doi: 10.1038/nmat3980. [DOI] [PubMed] [Google Scholar]
- 12.Engler A, et al. Substrate compliance versus ligand density in cell on gel responses. Biophys J. 2004;86:617–628. doi: 10.1016/S0006-3495(04)74140-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chaudhuri O, et al. Extracellular matrix stiffness and composition jointly regulate the induction of malignant phenotypes in mammary epithelium. Nat Mater. 2014;13:970–978. doi: 10.1038/nmat4009. [DOI] [PubMed] [Google Scholar]
- 14.Hall MS, et al. Fibrous nonlinear elasticity enables positive mechanical feedback between cells and ECMs. Proc Natl Acad Sci USA. 2016;113:14043–14048. doi: 10.1073/pnas.1613058113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wen JH, et al. Interplay of matrix stiffness and protein tethering in stem cell differentiation. Nat Mater. 2014;13:979–987. doi: 10.1038/nmat4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pelham RJ, Jr, Wang Yl. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc Natl Acad Sci USA. 1997;94:13661–13665. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Das A, Fischer RS, Pan D, Waterman CM. YAP nuclear localization in the absence of cell-cell contact is mediated by a filamentous actin-dependent, myosin II- and phospho-YAP-independent pathway during extracellular matrix mechanosensing. J Biol Chem. 2016;291:6096–6110. doi: 10.1074/jbc.M115.708313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dupont S, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 19.Janmey PA, Wells RG, Assoian RK, McCulloch CA. From tissue mechanics to transcription factors. Differentiation. 2013;86:112–120. doi: 10.1016/j.diff.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chaudhuri O, et al. Hydrogels with tunable stress relaxation regulate stem cell fate and activity. Nat Mater. 2015;15:326–334. doi: 10.1038/nmat4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee KY, Mooney DJ. Alginate: Properties and biomedical applications. Prog Polym Sci. 2012;37:106–126. doi: 10.1016/j.progpolymsci.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hao J, et al. Mechanobiology of mesenchymal stem cells: Perspective into mechanical induction of MSC fate. Acta Biomater. 2015;20:1–9. doi: 10.1016/j.actbio.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 23.Aicher WK, et al. Regeneration of cartilage and bone by defined subsets of mesenchymal stromal cells—Potential and pitfalls. Adv Drug Deliv Rev. 2011;63:342–351. doi: 10.1016/j.addr.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 24.Diduch DR, Coe MR, Joyner C, Owen ME, Balian G. Two cell lines from bone marrow that differ in terms of collagen synthesis, osteogenic characteristics, and matrix mineralization. J Bone Joint Surg Am. 1993;75:92–105. doi: 10.2106/00004623-199301000-00012. [DOI] [PubMed] [Google Scholar]
- 25.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 26.Leipzig ND, Shoichet MS. The effect of substrate stiffness on adult neural stem cell behavior. Biomaterials. 2009;30:6867–6878. doi: 10.1016/j.biomaterials.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 27.Saha K, et al. Substrate modulus directs neural stem cell behavior. Biophys J. 2008;95:4426–4438. doi: 10.1529/biophysj.108.132217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rigamonti A, et al. Large-scale production of mature neurons from human pluripotent stem cells in a three-dimensional suspension culture system. Stem Cell Rep. 2016;6:993–1008. doi: 10.1016/j.stemcr.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4:e17. doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
- 30.Saleh M, Shamsasanjan K, Movassaghpourakbari A, Akbarzadehlaleh P, Molaeipour Z. The impact of mesenchymal stem cells on differentiation of hematopoietic stem cells. Adv Pharm Bull. 2015;5:299–304. doi: 10.15171/apb.2015.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Charbord P, et al. A systems biology approach for defining the molecular framework of the hematopoietic stem cell niche. Cell Stem Cell. 2014;15:376–391. doi: 10.1016/j.stem.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 32.Sánchez-Aguilera A, Méndez-Ferrer S. The hematopoietic stem-cell niche in health and leukemia. Cell Mol Life Sci. 2017;74:579–590. doi: 10.1007/s00018-016-2306-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwartz MA, DeSimone DW. Cell adhesion receptors in mechanotransduction. Curr Opin Cell Biol. 2008;20:551–556. doi: 10.1016/j.ceb.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao W, Phinney DG, Bonnet D, Dominici M, Krampera M. Mesenchymal stem cell biodistribution, migration, and homing in vivo. Stem Cells Int. 2014;2014:292109. doi: 10.1155/2014/292109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koser DE, et al. Mechanosensing is critical for axon growth in the developing brain. Nat Neurosci. 2016;19:1592–1598. doi: 10.1038/nn.4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koser DE, Moeendarbary E, Hanne J, Kuerten S, Franze K. CNS cell distribution and axon orientation determine local spinal cord mechanical properties. Biophys J. 2015;108:2137–2147. doi: 10.1016/j.bpj.2015.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Madl CM, et al. Maintenance of neural progenitor cell stemness in 3D hydrogels requires matrix remodelling. Nat Mater. 2017;16:1233–1242. doi: 10.1038/nmat5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tekin H, et al. Effects of 3D culturing conditions on the transcriptomic profile of stem-cell-derived neurons. Nat Biomed Eng. 2018;2:540–554. doi: 10.1038/s41551-018-0219-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoffman BD, Grashoff C, Schwartz MA. Dynamic molecular processes mediate cellular mechanotransduction. Nature. 2011;475:316–323. doi: 10.1038/nature10316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schwartz MA. Integrins and extracellular matrix in mechanotransduction. Cold Spring Harb Perspect Biol. 2010;2:a005066. doi: 10.1101/cshperspect.a005066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mammoto A, Mammoto T, Ingber DE. Mechanosensitive mechanisms in transcriptional regulation. J Cell Sci. 2012;125:3061–3073. doi: 10.1242/jcs.093005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arnsdorf EJ, Tummala P, Jacobs CR. Non-canonical Wnt signaling and N-cadherin related beta-catenin signaling play a role in mechanically induced osteogenic cell fate. PLoS One. 2009;4:e5388. doi: 10.1371/journal.pone.0005388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wozniak MA, et al. Adhesion regulates MAP kinase/ternary complex factor exchange to control a proliferative transcriptional switch. Curr Biol. 2012;22:2017–2026. doi: 10.1016/j.cub.2012.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen JC, Jacobs CR. Mechanically induced osteogenic lineage commitment of stem cells. Stem Cell Res Ther. 2013;4:107. doi: 10.1186/scrt318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ambrosi TH, et al. Adipocyte accumulation in the bone marrow during obesity and aging impairs stem cell-based hematopoietic and bone regeneration. Cell Stem Cell. 2017;20:771–784.e6. doi: 10.1016/j.stem.2017.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu B, Zhang J, Chen J, Li C, Wang X. Molecular biological characteristics of the recruitment of hematopoietic stem cells from bone marrow niche in chronic myeloid leukemia. Int J Clin Exp Pathol. 2015;8:12595–12607. [PMC free article] [PubMed] [Google Scholar]
- 47.Kim N, Cho S-G. Clinical applications of mesenchymal stem cells. Korean J Intern Med. 2013;28:387–402. doi: 10.3904/kjim.2013.28.4.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alsberg E, Anderson KW, Albeiruti A, Franceschi RT, Mooney DJ. Cell-interactive alginate hydrogels for bone tissue engineering. J Dent Res. 2001;80:2025–2029. doi: 10.1177/00220345010800111501. [DOI] [PubMed] [Google Scholar]
- 49.Liao Y, Smyth GK, Shi W. The Subread aligner: Fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013;41:e108. doi: 10.1093/nar/gkt214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liao Y, Smyth GK, Shi W. featureCounts: An efficient general-purpose read summarization program. Bioinformatics. 2013;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 51.Law CW, Chen Y, Shi W, Smyth GK. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.