Significance
The outer membrane of Gram-negative bacteria prevents the entry of many antibiotics and limits treatment options for Gram-negative infections. This unique membrane is effective due to its asymmetric lipid composition, with the glycolipid lipid A [LPS or lipooligosaccharide (LOS)] in the outer leaflet at the cell surface and glycerophospholipids in the inner leaflet. Furthering our understanding of how outer membrane asymmetry is maintained is critical for the development of novel therapeutics to target multidrug-resistant bacteria. Here, we used a Gram-negative bacterium without LOS to probe for factors that impact cell-envelope maintenance in the absence of LOS. Our approach enabled us to explore fundamental mechanisms of cell-envelope biology and expand our holistic view of the asymmetrical, Gram-negative outer membrane.
Keywords: lipopolysaccharide, Mla, PldA, glycerophospholipid, outer membrane
Abstract
The outer membrane of Gram-negative bacteria is a critical barrier that prevents entry of noxious compounds. Integral to this functionality is the presence of lipopolysaccharide (LPS) or lipooligosaccharide (LOS), a molecule that is located exclusively in the outer leaflet of the outer membrane. Its lipid anchor, lipid A, is a glycolipid whose hydrophobicity and net negative charge are primarily responsible for the robustness of the membrane. Because of this, lipid A is a hallmark of Gram-negative physiology and is generally essential for survival. Rare exceptions have been described, including Acinetobacter baumannii, which can survive in the absence of lipid A, albeit with significant growth and membrane permeability defects. Here, we show by an evolution experiment that LOS-deficient A. baumannii can rapidly improve fitness over the course of only 120 generations. We identified two factors which negatively contribute to fitness in the absence of LOS, Mla and PldA. These proteins are involved in glycerophospholipid transport (Mla) and lipid degradation (PldA); both are active only on mislocalized, surface-exposed glycerophospholipids. Elimination of these two mechanisms was sufficient to cause a drastic fitness improvement in LOS-deficient A. baumannii. The LOS-deficient double mutant grows as robustly as LOS-positive wild-type bacteria while remaining resistant to the last-resort polymyxin antibiotics. These data provide strong biological evidence for the directionality of Mla-mediated glycerophospholipid transport in Gram-negative bacteria and furthers our knowledge of asymmetry-maintenance mechanisms in the context of the outer membrane barrier.
The asymmetric outer membrane found in the Gram-negative cell envelope is a fundamental organelle that protects the cell from the entry of many noxious compounds (1). This asymmetry is defined by the enrichment of lipopolysaccharide (LPS) or lipooligosaccharide (LOS) in the outer leaflet of the outer membrane and glycerophospholipids in the inner leaflet (2, 3). The canonical LPS molecule consists of three moieties, the lipid A anchor, a short series of core sugars, and O-antigen (1). LOS, an analog of LPS, lacks the lengthy O-antigen and instead has a few additional sugars attached to the core (SI Appendix, Fig. S1) (4). The lipid anchor of LPS, lipid A, is a disaccharide of glucosamine that is fatty acylated with a net negative charge due to phosphorylation of the sugar backbone (SI Appendix, Fig. S1) (5). Additional sites of phosphorylation within the core oligosaccharide further increase the anionic nature of the LPS molecule (6). LPS is deposited directly to the outer leaflet of the outer membrane by the lipopolysaccharide transport (Lpt) system, establishing the basis for the asymmetric outer membrane (7–11). The negatively charged lipid A interacts via cross-bridging with divalent cations in the environment (12, 13). While these interactions stabilize the outer membrane, the anionic nature of lipid A also makes the cell uniquely susceptible to a class of antibiotics—cationic antimicrobial peptides (CAMPs).
CAMPs are ubiquitous in nature (14) and are defined by their amphipathic structure, a characteristic essential for their mechanism of action. Gram-negative bacteria evolved multiple mechanisms of resistance to these peptides, the primary mechanism being the modification or removal of the phosphate groups on lipid A, thereby reducing the net negative charge of the bacterial surface (15). Curiously, Acinetobacter baumannii is one of three species of bacteria (along with Neisseria meningitidis and Moraxella catarrhalis) that can survive in the total absence of lipid A (16–19). In A. baumannii, this phenotype is selected for in the presence of high concentrations of the CAMPs colistin or polymyxin B (17), both of which are currently used to treat multidrug-resistant Gram-negative pathogens (20). Colonies that form on these plates have genetic mutations in one of the three first steps of lipid A biosynthesis (lpxA, lpxC, or lpxD) that entirely disrupt lipid A biosynthesis.
When LOS-deficient A. baumannii are initially isolated, a number of phenotypes are observed in regards to transcriptomic changes that may contribute to its ability to survive in the absence of LOS (21–23). Additional studies have compared virulence phenotypes using in vivo models and found that the virulence of LOS-deficient A. baumannii is attenuated (24). While these studies have been broadly informative, it has been difficult to link gene expression to any molecular model showing the way in which the LOS-deficient cell envelope has been altered and the subsequent impacts on physiology.
One of the common themes from the transcriptomic datasets was the up-regulation of both lipoproteins and the lipoprotein transport pathway, Lol (23, 25–27). These studies suggested a role for outer membrane lipoproteins in LOS-deficient cell envelopes. Several of these lipoproteins were found to localize to the outer leaflet of the outer membrane (26). This lends support to the hypothesis that perhaps lipoproteins were in part responsible for stabilizing a LOS-deficient cell envelope. Another up-regulated operon related to the cell envelope is the capsular polysaccharide, poly N-acetylglucosamine (PNAG) (23, 28). This is particularly interesting, considering work done on LPS-deficient N. meningitidis suggesting that the capsule plays an important role in the viability of an LPS-deficient cell (19, 29).
Despite the ability of A. baumannii to survive with inactivated lipid A biosynthesis and the concurrent changes in gene expression that are thought to permit this phenotype, it remains severely defective for growth. When grown on agar plates, LOS-deficient A. baumannii are incapable of forming single colonies. In liquid, both growth rate and overall growth yield are significantly reduced. These observations were perhaps expected, due to the typical essentiality of lipid A and LOS for Gram-negative organisms. However, it raised several critically important questions regarding the unidentified factors that contribute to this growth phenotype and their link to LOS deficiency.
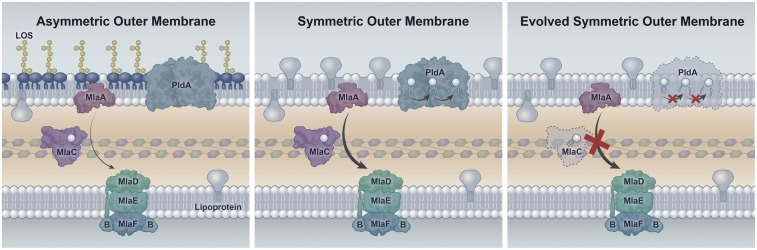
In this study, we used an evolution experiment to identify factors implicated in the fitness of LOS-deficient A. baumannii. After only 120 generations, the LOS-deficient cultures grew significantly better. We utilized whole-genome sequencing and RNA-sequencing (RNA-seq) to identify genes that were involved in the fitness of these LOS-deficient strains. To our surprise, the two asymmetry-maintenance systems in A. baumannii, Mla (maintain lipid asymmetry) retrograde glycerophospholipid transport and the outer membrane phospholipase PldA, were frequently disrupted. By deleting genes for both systems, we show they are the only mutations necessary to restore growth fitness to that of wild-type levels. These data present compelling biological evidence that Mla functions in a retrograde manner to extract mislocalized glycerophospholipids from the outer leaflet of the outer membrane and return them to the inner membrane (see Fig. 6, Left). Overall, our data show that in the absence of asymmetry these pathways are not merely unnecessary but are deleterious to the cell.
Fig. 6.
Model of the deleterious role of Mla and PldA in LOS-deficient cells. (Left) An asymmetric membrane with the two systems involved in glycerophospholipid removal of the outer leaflet of the outer membrane. These proteins are responsible either for extracting and removing intact glycerophospholipids from the outer leaflet (Mla) or for degrading mislocalized glycerophospholipids (PldA). In a cell with intact asymmetry, these systems are inactive or have a low level of activity due to a low frequency of mislocalized glycerophospholipids. (Center) Upon the loss of LOS, glycerophospholipids occupy both leaflets of the outer membrane. The abundance of glycerophospholipids in the outer leaflet stimulates both Mla and PldA activity, causing constitutive removal of glycerophospholipids from a glycerophospholipid bilayer. Surface-exposed lipoproteins are up-regulated, hinting at their potential role as space-fillers. (Right) In the evolved LOS-deficient cells, the loss of a functional Mla system (loss of MlaC function is shown as an example) and lack of PldA prevents the constant removal of glycerophospholipids from the bilayer. This allows homeostasis of membrane components to be achieved.
Results
LOS-Deficient A. baumannii Improve in Vivo Fitness Rapidly During a Short-Term Evolution Experiment.
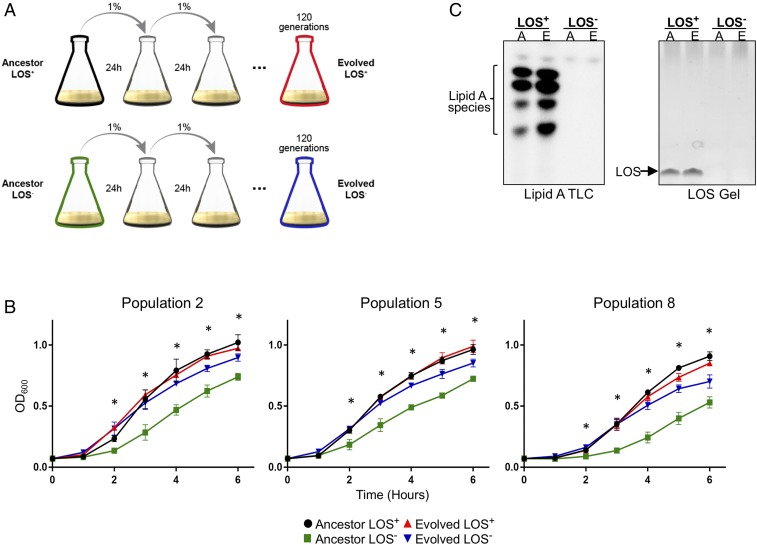
LOS deficiency in multiple strains of A. baumannii was generated by selecting for growth on 10 µg/mL polymyxin B. Each of the isolated LOS-deficient cells accumulated a mutation in either lpxA or lpxC that disrupted function. While we are readily able to isolate LOS-deficient A. baumannii, they all exhibited severe growth defects (Fig. 1B, green curves). We hypothesized that compensatory mutations may rescue the fitness defect and further our understanding of cell-envelope physiology in Gram-negative organisms. To this end, we used an evolution experiment. Ten independent populations of three different strains of A. baumannii (four AB5075 populations, three ATCC19606 populations, and three AYE populations) were grown in lysogeny broth (LB) with polymyxin B (LOS−) or without polymyxin B (LOS+). We utilized a complex medium with no nutritional limitations to remove any nutritional selection pressure while polymyxin B was present to prevent potential reversion of LOS− strains to LOS+ strains. Both the LOS+ and LOS− bacteria were passaged daily by transferring 1% of stationary-phase culture into fresh medium at an approximate starting OD600 of 0.05 (Fig. 1A). The evolution experiment was carried out until we saw significantly improved growth in our populations (Fig. 1B). To our surprise, this required only ∼120 generations of passaging. Generations were defined by the serial dilution factor (1:100) and the carrying capacity of the growth medium, as the cells are capable of undergoing ∼6.6 generations per cycle. During the 24-h growth period cells go through a lag, exponential, and stationary phase before the next dilution. Although the presence of polymyxin B should maintain LOS deficiency, we confirmed that the populations remained LOS deficient throughout the evolution period by measuring detectable LOS and lipid A as explained in Materials and Methods. In both cases, samples lacked LOS (Fig. 1C and SI Appendix, Fig. S2) and lipid A (Fig. 1C and SI Appendix, Fig. S3). This was expected due to the lpxA or lpxC mutations that initially occurred upon isolation (SI Appendix, Tables S2 and S3).
Fig. 1.
A short-term evolution experiment results in improved growth fitness. (A) A schematic of the short-term evolution experiment in which populations of A. baumannii, either LOS+ or LOS−, were passaged daily for 120 generations. For LOS− cultures, polymyxin B was supplemented to maintain selection pressure for LOS deficiency. (B) Growth curves of three representative populations show that evolved LOS− cultures (blue) grow significantly better than the ancestor LOS-deficient counterpart (green). All growth curves were performed in triplicate. Asterisks indicate significantly different values between evolved LOS-deficient and ancestor LOS-deficient as determined by a multiple-comparisons Student’s t test, α < 0.05. n = 3. (C) The presence or absence of lipid A/LOS was confirmed by 32P radiolabeling and isolation of lipid A or by staining for intact LOS molecules. A, ancestor; E, evolved. Representative images showing LOS+ versus LOS− strains for both of these methods are depicted here; the complete datasets are presented in SI Appendix, Figs. S2 and S3.
Evolved LOS-Deficient A. baumannii Exhibit Improved Outer Membrane Integrity.
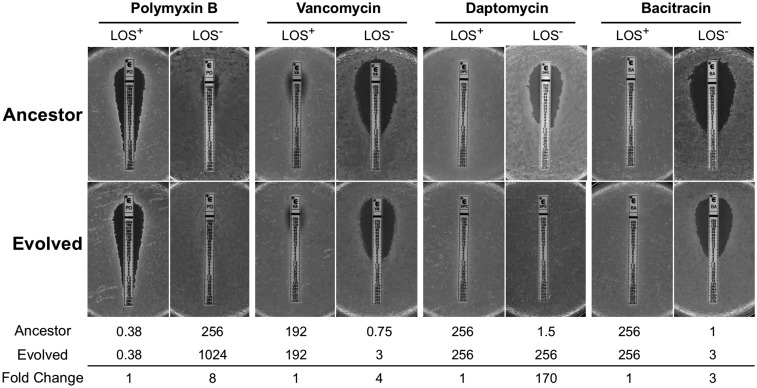
While LOS-deficient A. baumannii are highly resistant to polymyxin B (Fig. 2), they become more permeable and therefore more sensitive to antibiotics that are normally ineffective against Gram-negative bacteria. We hypothesized that changes during the evolution period would compensate for the lack of LPS by altering the cell envelope in some manner. To characterize the outer membrane integrity of the evolved populations, we assessed the resistance profiles to vancomycin, daptomycin, and bacitracin using E-test strips of population 2. These antibiotics are typically unable to permeate the Gram-negative outer membrane and as such allow us to probe quickly for cell-envelope integrity. Several populations tested exhibited increased minimum inhibitory concentrations (MICs) to these antibiotics (Fig. 2 and SI Appendix, Table S1). Resistance to vancomycin and bacitracin increased approximately three or four times for population 2 compared with its isogenic LOS-deficient ancestor. Resistance to daptomycin appeared to change the most. In the evolved population 2, it had increased 170-fold, from 1.5 to 256 μg/mL, restoring resistance to wild-type levels. Daptomycin has a unique mechanism of action by interacting with the membrane in an anionic glycerophospholipid-dependent manner (Fig. 2 and SI Appendix, Table S1) (30). When we began the evolution experiment, we could not predict whether the cell would increase fitness by altering the cell envelope or through more global changes in metabolism and physiology. These data were a clear indication that the cell envelope and/or outer membrane specifically changed over the course of the evolution experiment. While the outer membrane provides the barrier function, it is also attached to the peptidoglycan sacculus, which impacts cell shape. To determine whether these cells also exhibited any morphological changes, we used phase-contrast microscopy. The ancestor LOS-deficient cells exhibited a wide array of irregular morphologies compared with the LOS+ wild-type cells (SI Appendix, Fig. S4A). When we examined the evolved LOS-deficient cells, they appeared morphologically consistent and more comparable to a wild-type LOS+ cell (SI Appendix, Fig. S4A).
Fig. 2.
Resistance profiles of evolved populations suggest alterations to the cell envelope. The MICs of four antibiotics were tested against ancestor and evolved cultures for population 2. LOS+ resistance profiles do not change after evolution, whereas LOS− resistance increased for all antibiotics, including polymyxin B, to which the ancestor is already highly resistant. Upon loss of LOS, A. baumannii becomes susceptible to hydrophobic antibiotics (shown here: vancomycin, daptomycin, and bacitracin). After the evolution period, resistance to these antibiotics increased. For antibiotics targeting peptidoglycan components, the fold change was between three and four times. The resistance to daptomycin, which targets glycerophospholipids, was more drastic, although the reason for these differences is currently unknown. MIC values as determined by the E-test (representative of three biological replicates) and fold changes are shown below each set of images. Additional populations tested in this manner are summarized in SI Appendix, Table S1. All MIC values are in micrograms per milliliter.
Whole-Genome Sequencing Reveals That Cell-Envelope Maintenance Mechanisms Are Deleterious to Fitness in LOS Deficiency.
Knowing that the evolved populations exhibited increased growth fitness, altered resistance to hydrophobic antibiotics, and altered cellular morphology, we sought to understand whether any genomic changes could be attributed to these phenotypes. We sequenced the genomes of all 10 populations (both ancestor LOS+/− and evolved LOS+/−) for a total of 40 genome sequences and analyzed the genomes for SNPs, small insertions, and small deletions using an established pipeline (26, 31). We sequenced and compared our evolved strains with both the isogenic LOS+ ancestor and the evolved LOS+ strain. This allowed us to identify SNPs that accumulated during our experiment and account for changes that may be due simply to the daily passaging and not specific to LOS deficiency. Strikingly, seven of the 10 evolved LOS-deficient populations acquired mutations that inactivate mla genes (Table 1).
Table 1.
Nucleotide polymorphisms detected by whole-genome sequencing reveal cell envelope-related genes are frequent targets of disruption
| Gene* | Strain | Population | Mutation | Sequence | Impact† | Frequency‡, % |
| mlaABCDEF | 5075 | 1 | Insertion (mlaA) | CCAGCTAATT | Gln136fs | 98.91 |
| 2 | Deletion (mlaE) | 519C | Leu175fs | 99.93 | ||
| 3 | Insertion (mlaD) | T | Ile161fs | 100 | ||
| AYE | 5 | Deletion (mlaC) | 363T | Tyr122fs | 94.3 | |
| 6 | Deletion (mlaC) | 363T | Tyr122fs | 100 | ||
| 7 | Deletion (mlaC) | 272C | Thr91fs | 100 | ||
| 19606 | 8 | Transcriptional§ | – | ↓296X mlaA | – | |
| 10 | Deletion (mlaC) | 363–364TT | Tyr122fs | 100 | ||
| pldA | 5075 | 2 | Replacement | TT > AAAG | Leu22fs | 90.34 |
| 3 | Deletion | 84T | Tyr28fs | 95.65 | ||
| 4 | Deletion | 731T | Val246fs | 100 | ||
| ponA (PBP1A) | 5075 | 1 | Insertion | CTTGGTGA | Val349fs | 97.89 |
| 4 | Deletion | 2007T | Asn669fs | 100 | ||
| ompR/envZ | 19606 | 9 | SNP (ompR) | C > T | Arg31Cys | 100 |
| 9 | SNP (ompR) | C > A | Pro10Tyr | 100 | ||
| 10 | SNP (envZ) | G > T | Met264Ile | 100 | ||
| 10 | SNP (envZ) | C > A | Synonymous | 98.53 |
For simplicity, all mla genes are grouped together.
“fs” indicates frameshift.
Frequency refers to the number of times the SNP was identified relative to the total number of reads available for that sequence.
In population 8, while no genomic mutation was detected in an mla gene, we detected a 296× change in transcript level of mlaA relative to the ancestor LOS− strain.
The Mla pathway is a glycerophospholipid transport system that has been predicted to maintain lipid asymmetry by removing mislocalized glycerophospholipids from the outer leaflet of the outer membrane and returning them to the inner membrane. Previous work by our laboratory and others has shown that this pathway is initially up-regulated upon loss of LOS (23, 26); however, our results here clearly show that Mla-mediated lipid transport is ultimately deleterious to fitness in the absence of LOS. Supporting this conclusion is the fact that the second most frequently disrupted gene is pldA, which codes for an outer membrane phospholipase (3). PldA exclusively degrades mislocalized glycerophospholipids. Additional SNPs were found in other cell-envelope genes, namely the ompR/envZ two-component system and penicillin-binding protein 1A (Table 1). A full summary of genomic changes in the evolved LOS-deficient populations is shown in SI Appendix, Table S2, and all genomic changes detected in the ancestor LOS+, evolved LOS+, and ancestor LOS-deficient bacteria are shown in SI Appendix, Table S3.
Strains Defective for Retrograde Glycerophospholipid Transport and Degradation Exhibit Increased Fitness Independent of Evolution.
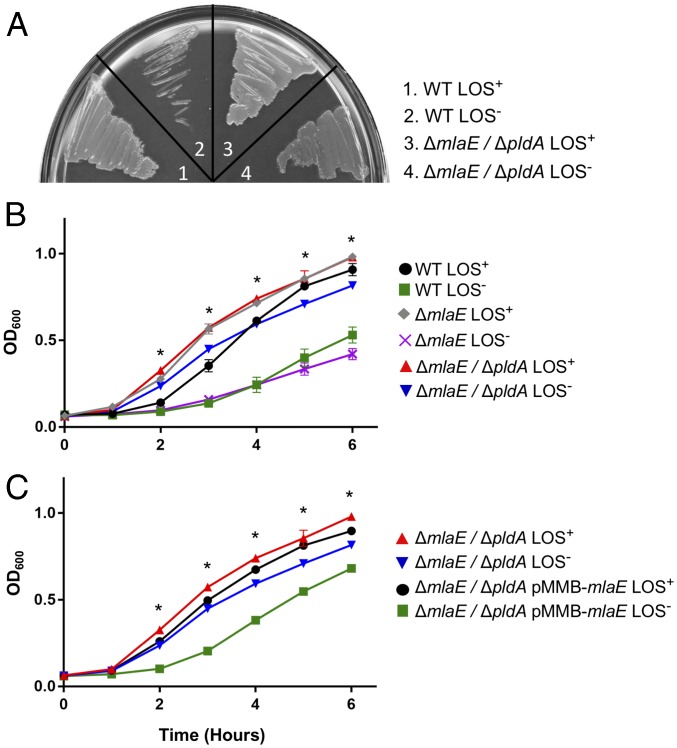
We were curious whether the improvements in fitness could be attributed exclusively to these genomic mutations. We hypothesized that introducing these mutations into A. baumannii independent of the evolution experiment would result in an inherently more fit LOS-deficient strain. To determine this, we introduced clean deletions of mlaE and pldA in a naive strain of 19606. These mutations were chosen as they mimic the mutations accumulated in population 2 (Table 1) and represent the two most common mutated genes in our experiment. MlaE functions as an inner membrane permease, although it is important to note that inactivation of any of the mla genes has been shown to inactivate the function of the system (32). If our hypothesis is correct, a lack of both genes, and therefore a lack of any asymmetry-maintenance mechanisms, should confer a fitness benefit independent of any laboratory-derived passaging. The double mutant was plated at 10 µg/mL on polymyxin B, and LOS-deficient individual colonies were selected for overnight. Indeed, we found that the LOS-deficient mlaE, pldA double mutant grows significantly better both on solid medium (Fig. 3A) and in broth culture (Fig. 3B). This strain was confirmed to be LOS-deficient via both LOS staining and lipid A isolation (SI Appendix, Fig. S5). An LOS-deficient single mutant for mlaE isolated in an identical manner was insufficient to improve growth (Fig. 3B). Despite multiple attempts to isolate a LOS-deficient pldA mutant, we were unsuccessful. The reason for the inability to select for pldA LOS− colonies is currently under investigation. To confirm that the increased fitness was due to the deletion in mlaE and pldA, we complemented mlaE on a plasmid. The complemented strain once again had a growth defect (Fig. 3C). This complementation confirms that the decrease in fitness was directly due to a functional Mla system in a ΔpldA background. It also shows that having only a singular pldA deletion is insufficient to restore growth to that of the double mutant.
Fig. 3.
Two mutations in glycerophospholipid maintenance systems restore wild-type growth to LOS-deficient A. baumannii. (A and B) Growth of the 19606 ΔmlaE/ΔpldA double mutant is highly robust relative to an LOS-deficient strain lacking any additional mutations both on plates (A) and in liquid broth (B). Asterisks indicate significantly different values between the ΔmlaE/ΔpldA LOS− and the 19606 LOS− strains as determined by a multiple-comparisons Student’s t test, α < 0.05, n = 3. (C) Complementation of mlaE on a plasmid decreases fitness in the ΔmlaE/ΔpldA double mutant. Asterisks indicate significantly different values between the ΔmlaE/ΔpldA LOS− and the ΔmlaE/ΔpldA pMMB-mlaE LOS− strains as determined by a multiple-comparisons Student’s t test, α < 0.05, n = 3.
We questioned whether the changes we found in the antibiotic-resistance profile of the evolved populations were also attributable to mutations in mla and pldA. In short, it appears that is the case. The LOS-deficient double mutant was significantly more resistant to both vancomycin and daptomycin than a wild-type LOS-deficient strain (Table 2). This furthers the argument that the presence of these two asymmetry-maintenance mechanisms negatively impacts LOS-deficient membrane integrity.
Table 2.
Antibiotic resistance profiles of the LOS− ΔmlaE/ΔpldA mimic that of the evolved LOS− populations
| Genotype | Polymyxin B | Vancomycin | Daptomycin | Bacitracin |
| 19606 wild type | 0.25 | 195 | 256 | 256 |
| 19606 LOS− | 256 | 0.38 | 3 | 0.5 |
| 19606 LOS− evolved (population 8) | 1,024 | 4 | 96 | 6 |
| ΔmlaE ΔpldA LOS+ | 0.38 | 256 | 256 | 256 |
| ΔmlaE ΔpldA LOS− | 64 | 4 | 48 | 3 |
All values are in micrograms per milliliter.
While this genotype confers improved growth and resistance to hydrophobic compounds, it is not sufficient to restore a normal cellular morphology, as detected by phase-contrast microscopy (SI Appendix, Fig. S4B). This differs from population 2 that accumulated a set of mutations in the same two genes (SI Appendix, Fig. S4A). It would have been surprising if Mla and PldA had such a strong impact on cellular morphology, as these have no known direct relationship to the peptidoglycan sacculus. Instead, it suggests that underlying impacts on peptidoglycan or peptidoglycan-modifying factors occurred during the evolution experiment that we have yet to identify. In the context of LOS deficiency, these asymmetry-maintenance mechanisms are deleterious to membrane integrity, but they are independent of the cell-shape defects observed in LOS-deficient A. baumannii.
Down-Regulated mlaA Transcript in Population 8 Mimics an mla Mutation.
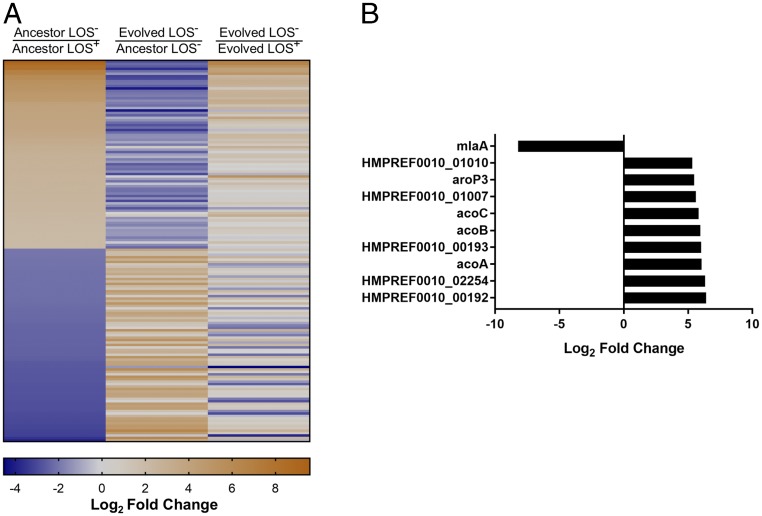
Of the 10 evolved populations, population 8 had no detectable SNPs but showed increased fitness. One explanation for this was that there were transcriptional differences likely caused by an unidentified mutation. We performed RNA-seq on the LOS+ ancestor and evolved strains and on the LOS− ancestor and evolved strains of population 8. The first comparison we made was between the ancestor LOS− strain and the ancestor LOS+ strain. Between these two strains, there were 156 genes with values that differed by a log2 fold change of ±2 with a false discovery rate (FDR)-corrected P value of < 0.05 (Fig. 4A, Left). While this is not a conservative cutoff, it allows us to detect the variety of changes that occur upon the loss of LOS for this strain. We next wanted to compare the evolved LOS− strain with the ancestor LOS− strain to identify transcriptional changes that occurred during evolution. From this analysis we noticed that the trend for many of the genes that were initially up- or down-regulated in the ancestor LOS− strain was reversed after the evolution period (Fig. 4A, Center). While the transcription levels of these genes may more closely approximate wild-type levels after the evolution period, the extent of change remained to be determined. When we directly compared the evolved LOS− strain with the evolved LOS+ strain, which allowed us to account for LOS-independent changes that may have accumulated during the evolution period, only 49 genes of the initial 156 genes differed by a log2 of ±2, FDR-corrected P value < 0.05 (Fig. 4A, Right). The fact that these genes remain different from the LOS+ genes even after the evolution period suggests that they may play a fundamental role in the physiology of LOS-deficient A. baumannii. Of these 49 genes, 13 are conserved, up-regulated genes in LOS-deficient cells regardless of the evolution status, suggesting a critical role in LOS deficiency. When separated by clusters of orthologous groups (COG) category, a majority of the genes, both up- or down-regulated, had no predicted function (SI Appendix, Fig. S6). Up-regulated genes with known function were enriched for lipid metabolism and cell-envelope biogenesis; however, a clear connection among these genes remains to be determined.
Fig. 4.
The evolved LOS− population reverses the transcriptomic changes initially caused by LOS deficiency. (A, Left) A heatmap of the 156 differentially regulated genes from the ancestor LOS− versus ancestor LOS+ condition from population 8. (Center) For this same set of genes, the log2 fold-change values were also calculated for the evolved LOS− versus ancestor LOS− strains to determine changes that occurred after the evolution period. (Right) A comparison between evolved LOS− versus evolved LOS+. Only 49 genes were significantly different. This serves as a direct comparison with the ancestor LOS− versus ancestor LOS+ column. (B) The top 10 up- or down-regulated genes in the evolved LOS−/ancestor LOS−. The most down-regulated gene (a log2 of −8.21 or a fold change of ∼296) was mlaA. For both A and B, the scale is based on log2 fold-change values.
The increased fitness of the evolved population presented a unique opportunity to examine cell-envelope components that were initially predicted to be important for LOS deficiency. We looked at genes for previously identified lipoproteins (SI Appendix, Fig. S7A), PNAG biosynthesis (SI Appendix, Fig. S7B), lipoprotein transport (SI Appendix, Fig. S7C), and the baeRS two-component system (SI Appendix, Fig. S7D) and compared the log2 fold change for both the ancestor LOS−/LOS+ strains (green bars) and the evolved LOS−/LOS+ strains (blue bars). For each of the lipoproteins, we saw a decrease in the expression. However, each of these lipoproteins in the evolved LOS− is still significantly up-regulated compared to the LOS+ strain. We saw a similar trend with the periplasmic lipoprotein transport chaperone LolA. Transcription for lolA remained up-regulated in the evolved LOS−, albeit to a lesser extent than in the ancestral LOS− strain (SI Appendix, Fig. S7C). In contrast, PNAG biosynthesis is no longer up-regulated in the evolved LOS− strain (SI Appendix, Fig. S7B). Curiously, only one known stress response is up-regulated upon LOS deficiency. This system, baeRS, is no longer up-regulated in the evolved LOS− compared with the LOS+ strain (SI Appendix, Fig. S7D). These data suggest that, while PNAG is most likely an artifact of an initial stress response to LOS deficiency, outer membrane lipoproteins play a specific and important role in the cell envelope in the absence of LOS.
Our initial transcriptomic analysis in Fig. 4A was restricted to genes that were differentially regulated in the ancestor LOS−/ancestor LOS+ dataset. This analysis was informative as to the plasticity of a LOS-deficient transcriptome; however, it proved insufficient to address the question of factors that could explain improved fitness in the evolved LOS− strain of population 8. To better identify changes that occurred during evolution, we examined all genes that were differentially regulated in the evolved LOS−/ancestor LOS− comparison with a log2 of ±2 with a FDR-corrected P value of < 0.05. When we looked at the top 10 differentially regulated genes from this set, the most down-regulated gene with a log2 fold change of −8.21 (a fold change of ∼296), was mlaA (Fig. 4B). Sanger sequencing of the mlaA coding and upstream promoter regions confirmed that there were no SNPs or indels from the −440 to +841 nucleotides of the mlaA ORF. This mlaA down-regulation phenotype, in light of our whole-genome sequencing (Table 1), was the likely explanation for the improved fitness we saw in the evolved LOS-deficient population 8 and reinforces its role as a factor deleterious to fitness.
Mla-Mediated Glycerophospholipid Transport Has Wide-Spread Impacts on LOS-Deficient Physiology Independent of the Evolution Experiment.
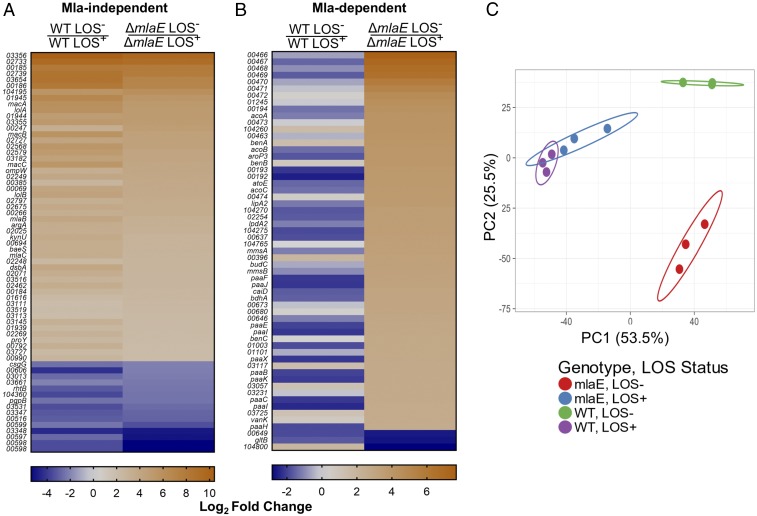
To characterize the impact of Mla-mediated glycerophospholipid transport, we generated a clean-deletion mutant in mlaE, the inner membrane permease protein of the Mla complex. Comparative RNA-seq was done on wild-type LOS+, wild-type LOS−, mlaE LOS+, and mlaE LOS− strains. This experiment would reveal if Mla plays a specific, localized biochemical role in the outer membrane or if it has a broader impact on LOS-deficient cellular physiology. To elucidate this, we compared the ΔmlaE LOS− and ΔmlaE LOS+ transcriptome with the wild-type LOS− and wild-type LOS+ transcriptome using our previously established cutoff of a log2 fold change of ±2 with an FDR-corrected P value of < 0.05. This revealed two distinct clusters of genes. The first cluster were categorized as Mla-independent, as the transcription of these genes was up- or down-regulated in the LOS− strain versus the LOS+ strain regardless of functional Mla (Fig. 5A). The second cluster, and the one of most interest, comprised the Mla-dependent genes. These genes were up- or down-regulated in the ΔmlaE LOS− background but not in wild-type LOS− cells as determined by our cutoff of log2 fold change of ±2 with an FDR-corrected P value of < 0.05. In all, 120 genes were differentially regulated in LOS-deficient cells in an Mla-dependent manner (SI Appendix, Fig. S8). For simplicity of viewing, the top 60 genes are shown in Fig. 5B; the complete datasets for the Mla-independent and Mla-dependent are shown in SI Appendix, Tables S4 and S5, respectively. A comparable number of genes were differentially regulated only in the wild-type LOS− background compared with its isogenic LOS+ parent, with the transcription of 65 genes conserved across both genotypes (SI Appendix, Fig. S8). These differences are easily visualized via principal components analysis (PCA) (Fig. 5C). LOS+ transcriptomes are not altered regardless of functional Mla, as they are shown clustered together by PCA. In contrast, the transcriptomes vary drastically between the LOS− wild type versus LOS− ∆mlaE. Together, these data demonstrate the huge degree of transcriptomic change caused by a nonfunctional Mla-transport system in the context of LOS deficiency.
Fig. 5.
Mla-mediated phospholipid transport drastically alters LOS-deficient transcriptomes. (A) Genes categorized as Mla-independent based on the RNA-seq of 19606 ΔmlaE LOS−/LOS+ strains versus wild-type 19606 LOS−/LOS+ strains. In this case, genes exhibited up- or down-regulation regardless of the mlaE genotype. (B) Genes shown to be Mla-dependent. These genes were up- or down-regulated in the ΔmlaE LOS−/LOS+ and not the wild-type LOS−/LOS+ strains based on a log2 fold change of ±2. Only the top 60 genes out of 120 are shown in B. When possible, loci with gene annotations were updated. For nonannotated genes the prefix HMPREF0010_ is excluded from the figure. For both A and B, the scale is based on log2 fold-change values. Complete datasets for A and B are in SI Appendix, Tables S4 and S5. (C) The variation in the transcriptomes between the wild-type and ΔmlaE strains as visualized by PCA analysis. Each circle represents a biological replicate for the indicated strain. This PCA was generated using ClustVis (42).
Discussion
Here we report that LOS-deficient A. baumannii are capable of rapidly improving fitness in an evolution experiment. After only 120 generations, we obtained populations of LOS-deficient A. baumannii that grew at a very similar rate to the isogenic wild type. These evolved populations exhibited a rapid rate of evolution in the absence of any exogenous selective pressure, reinforcing the idea that these cells are initially very sick in the absence of LOS. The two major genetic loci that were disrupted during this evolution experiment were mlaABCDEF and pldA, both of which are implicated in cell-envelope maintenance. By disrupting these systems, the cell alleviates both known mechanisms in A. baumannii that are involved in removing mislocalized glycerophospholipids from the outer leaflet of the outer membrane.
The Mla pathway consists of six proteins found within different cellular compartments: MlaA in the outer membrane, MlaC in the periplasm, and the MlaBDEF complex localized to the cytoplasmic membrane. Removal of any one Mla protein results in the loss of resistance to a number of stressors that disrupt outer membrane integrity (32, 33). In Escherichia coli, the Mla proteins are dispensable under standard laboratory growth (34). When E. coli cells are stressed with compounds that perturb the outer membrane, such as SDS and EDTA, Mla activity is essential for survival (32, 35). SDS/EDTA destabilizes the outer membrane by both stripping away cross-bridging between LPS molecules (EDTA) and causing pore formation via detergent activity (SDS). Under this condition, glycerophospholipids will flip into the outer leaflet, resulting in symmetric lipid rafts and increased membrane permeability (3, 36). Additionally, a spontaneously generated mutant, mlaA*, which has an internal two-amino-acid deletion, appears to reverse the system: Strains harboring this variant “leak” glycerophospholipids into the outer leaflet (33). Suppressor analysis identified another glycerophospholipid maintenance protein, outer membrane phospholipase A (PldA), as a suppressor for both the Δmla and mla* genotypes (33). In the former, a multicopy suppressor of pldA increases PldA levels sufficiently to prevent lethal accumulation of glycerophospholipids in the outer leaflet. Conversely, in a ΔpldA background, the mlaA* dominant allele was no longer lethal in a stationary-phase phenotype (33). These findings, combined with recent work establishing a link between PldA byproducts as a feedback system to modify LPS levels (37), establishes PldA as a sensor of outer membrane asymmetry and playing an important role in cell-envelope maintenance.
While these activities are advantageous with asymmetry, in its absence the systems would constitutively disrupt the outer leaflet of the outer membrane, causing obvious defects to the cell envelope (Fig. 6, Center). These findings were somewhat surprising at first, as multiple studies had shown that mla genes were up-regulated upon the initial loss of LOS (23, 26). However, when we take into consideration the function of the Mla pathway and the strong conservation of LPS and asymmetry, these observations begin to make more sense. When asymmetry is intact, the cell requires robust systems to extract mislocalized glycerophospholipids to preserve asymmetry. When we select for LOS-deficient A. baumannii in the laboratory, we assume that the cell is “blind” to its inability to produce LPS. In this context, Mla and PldA would function as they have evolved to do, i.e., efficiently extract or degrade glycerophospholipids from the outer leaflet of the outer membrane. In the case of a LOS-deficient cell envelope, this activity would severely hinder membrane homeostasis and cell division.
The data presented here are strong biological evidence in support of Mla as a retrograde transport system. This is the only directionality in which its activity would be deleterious in the absence of asymmetry. If it functioned in an anterograde manner, we would expect up-regulation to be beneficial, as it would increase the flow of glycerophospholipids to the outer membrane. Supporting this conclusion is the fact that pldA was inactivated in several evolved populations. This would remove the second molecular mechanism of glycerophospholipid removal in the absence of asymmetry. The Silhavy group (32) considers Mla the predominant system because an Mla-null SDS/EDTA sensitivity phenotype is far more drastic than a PldA-null sensitivity phenotype, and the data we have presented here support this position. The fact that we could reconstitute only two mutations (mlaE and pldA) into A. baumannii and generate a LOS-deficient strain with robust growth confirms the critical role these two pathways play in outer membrane maintenance.
The transcriptomic data presented here allow us to draw several conclusions in both an evolution-dependent and independent context. For the evolution-dependent context, the transcriptomic changes that are initially detected upon the loss of LOS are largely reversed after the evolution period. We attribute a large majority of these changes to the disruption of Mla activity, which would be constant in a symmetric outer membrane. Genes that remain differentially regulated after evolution likely play a much more important role in the physiology of a LOS-deficient cell and warrant further exploration. Additional factors with known roles in LOS deficiency were also altered after evolution. In particular, the expression of a number of surface-exposed lipoproteins was down-regulated after the evolution experiment. The hypothesized role of these lipoproteins is that they are space fillers for a membrane that is constantly having glycerophospholipids removed from the outer leaflet in the absence of LOS (Fig. 6, Center). That these lipoproteins remain up-regulated, albeit to a lesser extent, suggests that they remain important in an LOS-deficient membrane. However, in the absence of Mla-mediated glycerophospholipid removal they are needed at a lower abundance. On the contrary, PNAG biosynthesis and capsule production is not necessary for LOS deficiency. At the end of the evolution experiment, the evolved LOS− strain no longer up-regulated pgaABCD. By continuing this evolution experiment beyond the current study, we may identify additional factors that contribute positively or negatively to the cell envelope of Gram-negative organisms.
By using a clean deletion mutant independent of the evolution experiment, we show that the presence of Mla has a large impact on the global transcriptome of LOS-deficient A. baumannii, which we would not expect, as it has no clear connection with transcriptional regulators. This suggests that a sensor for Mla activity, which has yet to be identified, connects Mla activity with transcription. Additionally, the down-regulation of mlaA in the absence of any genomic SNPs is an exciting avenue to identify transacting regulatory factors for mla gene regulation. Taken together, these observations support the notion that the activity of Mla in a LOS-deficient cell has wide-spread consequences on bacterial physiology and fitness.
These data lend themselves to a model which provides unique insight into the roles of asymmetry-maintenance mechanisms in A. baumannii, particularly that of the Mla pathway. In a LOS+ cell, these mechanisms robustly remove mislocalized glycerophospholipids to preserve asymmetry (Fig. 6, Left). In the absence of LOS, these pathways are hyperactive in a “misguided” attempt to preserve asymmetry. The result is that glycerophospholipids are consistently removed from the outer leaflet of the outer membrane with no lipid A/LOS available to refill these gaps (Fig. 6, Center). In the absence of Mla, and to some extent PldA, this glycerophospholipid removal ceases, allowing homeostasis to be achieved in a LOS/lipid A-deficient outer membrane (Fig. 6, Right).
While these genes were the ones most frequently inactivated, other cell-envelope systems were also targets of disruption in our populations. One of these, PBP1A, is a penicillin-binding protein (PBP) involved in de novo peptidoglycan biosynthesis (38). It is one of two major high molecular weight PBPs in A. baumannii, and its function has previously been implicated as being deleterious to LOS deficiency (26). Current evidence suggests that certain strains of A. baumannii can survive with inactivated LOS while having PBP1A, while others cannot survive unless PBP1A is inactivated. The fact that PBP1A was inactivated in cells that can already lose LOS despite the presence of PBP1A suggests that PBP1A is ubiquitously deleterious to fitness in the absence of LOS; the reason for this remains enigmatic and is currently under investigation. PBP1A remains the only known peptidoglycan-related protein we have identified as playing a role in fitness in LOS-deficient A. baumannii. While the mla-null, pldA-null LOS-deficient strain exhibits increased growth, it remained morphologically distinct from the wild type. This supports the idea that PBP1A, and potentially other unknown peptidoglycan-related factors, play a role in LOS-deficient A. baumannii morphology.
Another system that was altered in two populations was the EnvZ/OmpR two-component system. This two-component system is involved in regulating cell-envelope stress responses. The mutations that were detected are SNPs that resulted in coding sequence changes; however, the effect of these remains to be determined. The SNPs accrued in the ompR coding region cause substitutions in the response regulatory domain. The envZ mutation occurred several amino acids before the histidine kinase domain. Given these observations, it is possible these mutations alter the sensing ability of EnvZ or the DNA-binding activity of OmpR. These mutations could alter porin regulation, either negatively or positively, which could be important for membrane integrity in the absence of LOS (39).
We were able to restore wild-type growth to a highly polymyxin B-resistant A. baumannii strain by the introduction of two mutations which we had identified through the evolution experiment. This speaks to the power of this technique for identifying additional novel factors that may play a role in cell-envelope maintenance in the absence of asymmetry. While these cells sustain high levels of polymyxin B resistance, we rarely see full restoration of resistance to vancomycin or other hydrophobic antibiotics. Thus, these could become useful therapeutics if LOS-deficient A. baumannii become more prominent in clinical settings. This is a newfound concern, considering that these mutations that confer fitness benefits are null mutations, which eliminate the need for complicated evolutionary processes such as gene duplication and subsequent neofunctionalization to occur.
Taken together, this evolution experiment highlighted fundamental aspects of cell-envelope maintenance within Gram-negative bacteria and provided insight into preexisting outer membrane-maintenance systems. Further, it has given us a real look at cell-envelope maintenance during LOS deficiency. We identified two membrane-maintenance mechanisms (Mla and PldA) that function deleteriously in the absence of asymmetry, confirming the retrograde directionality of Mla transport. This work establishes LOS-deficient A. baumannii as a robust in vivo system for the identification and confirmation of both known and unknown factors that may contribute to cell-envelope biogenesis in Gram-negative bacteria.
Materials and Methods
Bacterial Strains and Growth.
All strains and plasmids used in this study are listed in SI Appendix, Table S6. Strains were grown in either LB or LB + 1.5% agar at 37 °C. Selective antibiotics were used when indicated at the following concentrations: kanamycin, 15 μg/mL or 30 μg/mL; polymyxin B, 10 μg/mL; tetracycline, 10 μg/mL; and vancomycin, 10 μg/mL. For growth curves, cultures were grown in 5 mL of LB at a starting OD600 of ∼0.05. For a given time point, 200 μL of culture was transferred to a polystyrene 96-well plate. OD600 values were measured by a plate reader (BioTek). Growth curves were generated in GraphPad Prism. A multiple-comparisons Students t test was performed using GraphPad Prism with α < 0.05.
Construction of Deletion Mutants in A. baumannii.
Primers used in this study are listed in SI Appendix, Table S7. A. baumannii mutants were generated by recombineering methodology (40, 41). Strains carrying recombineering machinery on a plasmid were grown overnight in LB and subcultured into 50 mL of fresh LB broth at an OD600 of ∼0.05. Cultures were grown at 37 °C for 2 h before induction with 2 mM isopropyl β-d-1-thiogalactopyranoside (IPTG). Cells were grown for an additional hour to a final OD600 of ∼0.5–0.7. Cells were pelleted by centrifugation at 5,000 × g and were washed three times with ice-cold 10% glycerol. The resulting pellet was resuspended in ∼500 μL of 10% glycerol. Approximately 2 μg of linear recombineering PCR product was incubated with 50 μL of electrocompetent cells before electroporation at 1.8 mV. Cells were recovered in 4 mL of LB with 2 mM IPTG for 4 h before plating on LB + 15 μg/mL of kanamycin.
To generate a markerless deletion mutant, the recombineering machinery plasmid was removed via serial passaging of a positive transformant on LB agar plates. Loss of the plasmid was determined via sensitivity to tetracycline while maintaining resistance to kanamycin. Subsequently, pMMB67EH carrying FLP recombinase machinery was introduced via electroporation of competent cells. After recovery, cells were plated on tetracycline. The following day, positive transformants were passaged on LB + 1 mM IPTG. After overnight incubation, a majority of colonies became kanamycin sensitive. To generate double or triple mutants, this process was repeated iteratively.
Isolation of LOS-Deficient A. baumannii.
To isolate LOS-deficient A. baumannii, overnight cultures were normalized to an OD600 of ∼1.0. Cells were pelleted and washed twice with 1× PBS before being resuspended in 100 μL of 1× PBS. This volume was plated on LB plates with polymyxin B. After 24–30 h of growth, colonies were tested for polymyxin resistance and vancomycin sensitivity (26). These colonies were confirmed to be LOS-deficient via LOS staining using the ProQ Emerald 300 lipopolysaccharide staining kit (Molecular Probes, Inc.) with proteinase K-treated lysates.
Lipid A Radiolabeling.
Isolation of 32P-radiolabeled lipid A and glycerophospholipids was carried out as described previously (26, 40). Briefly, a culture (starting OD600 of ∼0.05) was grown with 2.5 mCi of 32P ortho-Phosphoric acid (Perkin-Elmer) to an OD600 of 0.8–1.0. Cells were pelleted and washed with PBS. Lipid A was extracted via mild-acid hydrolysis and Bligh–Dyer solvent extraction. TLC analysis of lipid A samples was done in a pyridine, chloroform, 88% formic acid, aqueous (50:50:16:4) tank, and plates were exposed to a phosphor screen overnight before imaging.
Next-Generation Sequencing and Analysis.
Cultures for either whole-genome sequencing or RNA-seq were grown to an OD600 of ∼0.5. Cells were pelleted and either were resuspended in 1 mL of RNAlater (Invitrogen) for RNA-seq or DNA was extracted using an Easy-DNA gDNA purification kit (Thermo Fisher). RNA extraction, library building, and sequencing were contracted out with Genewiz, Inc. Genomic sequences were mapped to the respective published genomes using CLC Genomic Workbench software (Qiagen). These reads were locally realigned, and high-frequency variants were detected. Impacts on coding regions were predicted. For RNA-seq, reads were mapped to the respective published genomes using CLC Genomic Workbench software (Qiagen). After local alignment, the reads per kilobase of transcript per million mapped reads (RPKM) was calculated for each gene/coding sequence. For statistical analysis, comparisons were made between RNA-seq datasets with a log2 fold-change cutoff greater than |3|. A PCA plot was generated using ClustVis (42).
Determination of MICs.
MICs were determined by E-strip (BioMerieux). One hundred fifty microliters of cultures was spread on LB plates and allowed to dry for 30 min. A sterile E-strip was added to the plate and was incubated overnight at 37 °C. The MIC was assigned as the value where the zone of inhibition intersected with the strip.
Short-Term Evolution Experiment.
Cultures of populations to be evolved were grown in LB broth without polymyxin B (LOS+ populations) or with polymyxin (LOS− populations). Every 24 h, 1% of the culture was subcultured into fresh medium, resulting in an OD600 of ∼0.05. These populations cycle through a lag phase, exponential growth, and stationary phase daily. Periodically, cultures were frozen in LB + 20% glycerol.
Phase Microscopy.
Cultures were grown in 5 mL of LB at a starting OD600 of ∼0.05 to an OD600 of 0.5–0.7. One milliliter of culture was washed in PBS and concentrated twofold. Five microliters was spotted on an agar pad and imaged using a wide-field upright microscope (Nikon Eclipse 80i).
Sanger Sequencing.
PCR products for mlaA plus 500 bp upstream were purified with a Qiagen PCR Purification Kit. Sanger sequencing was performed by Genewiz Inc.
Supplementary Material
Acknowledgments
We thank Alex Crofts for insightful comments. This work was funded by NIH Grant AI138576 (to M.S.T.) and National Science Foundation Graduate Research Fellowship 049347-06 (to M.J.P.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data discussed in this publication have been deposited in the National Center for Biotechnology’s Gene Expression Omnibus (Bioproject ID PRJNA450804).
See Commentary on page 8852.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1806714115/-/DCSupplemental.
References
- 1.Whitfield C, Trent MS. Biosynthesis and export of bacterial lipopolysaccharides. Annu Rev Biochem. 2014;83:99–128. doi: 10.1146/annurev-biochem-060713-035600. [DOI] [PubMed] [Google Scholar]
- 2.Funahara Y, Nikaido H. Asymmetric localization of lipopolysaccharides on the outer membrane of Salmonella typhimurium. J Bacteriol. 1980;141:1463–1465. doi: 10.1128/jb.141.3.1463-1465.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kamio Y, Nikaido H. Outer membrane of Salmonella typhimurium: Accessibility of phospholipid head groups to phospholipase c and cyanogen bromide activated dextran in the external medium. Biochemistry. 1976;15:2561–2570. doi: 10.1021/bi00657a012. [DOI] [PubMed] [Google Scholar]
- 4.Preston A, Mandrell RE, Gibson BW, Apicella MA. The lipooligosaccharides of pathogenic Gram-negative bacteria. Crit Rev Microbiol. 1996;22:139–180. doi: 10.3109/10408419609106458. [DOI] [PubMed] [Google Scholar]
- 5.Henderson JC, et al. The power of asymmetry: Architecture and assembly of the Gram-negative outer membrane lipid bilayer. Annu Rev Microbiol. 2016;70:255–278. doi: 10.1146/annurev-micro-102215-095308. [DOI] [PubMed] [Google Scholar]
- 6.Tamayo R, et al. Identification of cptA, a PmrA-regulated locus required for phosphoethanolamine modification of the Salmonella enterica serovar typhimurium lipopolysaccharide core. J Bacteriol. 2005;187:3391–3399. doi: 10.1128/JB.187.10.3391-3399.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Okuda S, Sherman DJ, Silhavy TJ, Ruiz N, Kahne D. Lipopolysaccharide transport and assembly at the outer membrane: The PEZ model. Nat Rev Microbiol. 2016;14:337–345. doi: 10.1038/nrmicro.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sherman DJ, et al. Lipopolysaccharide is transported to the cell surface by a membrane-to-membrane protein bridge. Science. 2018;359:798–801. doi: 10.1126/science.aar1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qiao S, Luo Q, Zhao Y, Zhang XC, Huang Y. Structural basis for lipopolysaccharide insertion in the bacterial outer membrane. Nature. 2014;511:108–111. doi: 10.1038/nature13484. [DOI] [PubMed] [Google Scholar]
- 10.Botos I, et al. Structural and functional characterization of the LPS transporter LptDE from Gram-negative pathogens. Structure. 2016;24:965–976. doi: 10.1016/j.str.2016.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li X, Gu Y, Dong H, Wang W, Dong C. Trapped lipopolysaccharide and LptD intermediates reveal lipopolysaccharide translocation steps across the Escherichia coli outer membrane. Sci Rep. 2015;5:11883. doi: 10.1038/srep11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murata T, Tseng W, Guina T, Miller SI, Nikaido H. PhoPQ-mediated regulation produces a more robust permeability barrier in the outer membrane of Salmonella enterica serovar typhimurium. J Bacteriol. 2007;189:7213–7222. doi: 10.1128/JB.00973-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schindler M, Osborn MJ. Interaction of divalent cations and polymyxin B with lipopolysaccharide. Biochemistry. 1979;18:4425–4430. doi: 10.1021/bi00587a024. [DOI] [PubMed] [Google Scholar]
- 14.Bahar AA, Ren D. Antimicrobial peptides. Pharmaceuticals (Basel) 2013;6:1543–1575. doi: 10.3390/ph6121543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Needham BD, Trent MS. Fortifying the barrier: The impact of lipid A remodelling on bacterial pathogenesis. Nat Rev Microbiol. 2013;11:467–481. doi: 10.1038/nrmicro3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Powers MJ, Trent MS. Expanding the paradigm for the outer membrane: Acinetobacter baumannii in the absence of endotoxin. Mol Microbiol. 2018;107:47–56. doi: 10.1111/mmi.13872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moffatt JH, et al. Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide production. Antimicrob Agents Chemother. 2010;54:4971–4977. doi: 10.1128/AAC.00834-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peng D, Hong W, Choudhury BP, Carlson RW, Gu X-X. Moraxella catarrhalis bacterium without endotoxin, a potential vaccine candidate. Infect Immun. 2005;73:7569–7577. doi: 10.1128/IAI.73.11.7569-7577.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steeghs L, et al. Meningitis bacterium is viable without endotoxin. Nature. 1998;392:449–450. doi: 10.1038/33046. [DOI] [PubMed] [Google Scholar]
- 20.Nation RL, Velkov T, Li J. Colistin and polymyxin B: Peas in a pod, or chalk and cheese? Clin Infect Dis. 2014;59:88–94. doi: 10.1093/cid/ciu213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moffatt JH, et al. Lipopolysaccharide-deficient Acinetobacter baumannii shows altered signaling through host Toll-like receptors and increased susceptibility to the host antimicrobial peptide LL-37. Infect Immun. 2013;81:684–689. doi: 10.1128/IAI.01362-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bojkovic J, et al. Characterization of an Acinetobacter baumannii lptD deletion strain: Permeability defects and response to inhibition of lipopolysaccharide and fatty acid biosynthesis. J Bacteriol. 2015;198:731–741. doi: 10.1128/JB.00639-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henry R, et al. Colistin-resistant, lipopolysaccharide-deficient Acinetobacter baumannii responds to lipopolysaccharide loss through increased expression of genes involved in the synthesis and transport of lipoproteins, phospholipids, and poly-β-1,6-N-acetylglucosamine. Antimicrob Agents Chemother. 2012;56:59–69. doi: 10.1128/AAC.05191-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beceiro A, et al. Biological cost of different mechanisms of colistin resistance and their impact on virulence in Acinetobacter baumannii. Antimicrob Agents Chemother. 2014;58:518–526. doi: 10.1128/AAC.01597-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grabowicz M, Silhavy TJ. Redefining the essential trafficking pathway for outer membrane lipoproteins. Proc Natl Acad Sci USA. 2017;114:4769–4774. doi: 10.1073/pnas.1702248114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boll JM, et al. A penicillin-binding protein inhibits selection of colistin-resistant, lipooligosaccharide-deficient Acinetobacter baumannii. Proc Natl Acad Sci USA. 2016;113:E6228–E6237. doi: 10.1073/pnas.1611594113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsuyama Si, Yokota N, Tokuda H. A novel outer membrane lipoprotein, LolB (HemM), involved in the LolA (p20)-dependent localization of lipoproteins to the outer membrane of Escherichia coli. EMBO J. 1997;16:6947–6955. doi: 10.1093/emboj/16.23.6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Choi AHK, Slamti L, Avci FY, Pier GB, Maira-Litrán T. The pgaABCD locus of Acinetobacter baumannii encodes the production of poly-beta-1-6-N-acetylglucosamine, which is critical for biofilm formation. J Bacteriol. 2009;191:5953–5963. doi: 10.1128/JB.00647-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bos MP, Tommassen J. Viability of a capsule- and lipopolysaccharide-deficient mutant of Neisseria meningitidis. Infect Immun. 2005;73:6194–6197. doi: 10.1128/IAI.73.9.6194-6197.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller WR, Bayer AS, Arias CA. Mechanism of action and resistance to daptomycin in Staphylococcus aureus and Enterococci. Cold Spring Harb Perspect Med. 2016;6:a026997. doi: 10.1101/cshperspect.a026997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crofts AA, et al. Campylobacter jejuni transcriptional and genetic adaptation during human infection. Nat Microbiol. 2018;3:494–502. doi: 10.1038/s41564-018-0133-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malinverni JC, Silhavy TJ. An ABC transport system that maintains lipid asymmetry in the Gram-negative outer membrane. Proc Natl Acad Sci USA. 2009;106:8009–8014. doi: 10.1073/pnas.0903229106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sutterlin HA, et al. Disruption of lipid homeostasis in the Gram-negative cell envelope activates a novel cell death pathway. Proc Natl Acad Sci USA. 2016;113:E1565–E1574. doi: 10.1073/pnas.1601375113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jia W, et al. Lipid trafficking controls endotoxin acylation in outer membranes of Escherichia coli. J Biol Chem. 2004;279:44966–44975. doi: 10.1074/jbc.M404963200. [DOI] [PubMed] [Google Scholar]
- 35.Isom GL, et al. MCE domain proteins: Conserved inner membrane lipid-binding proteins required for outer membrane homeostasis. Sci Rep. 2017;7:8608. doi: 10.1038/s41598-017-09111-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.May KL, Silhavy TJ. The Escherichia coli Phospholipase PldA regulates outer membrane homeostasis via lipid signaling. MBio. 2018;9:e00379-18. doi: 10.1128/mBio.00379-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Di Guilmi AM, Dessen A, Dideberg O, Vernet T. Bifunctional penicillin-binding proteins: Focus on the glycosyltransferase domain and its specific inhibitor moenomycin. Curr Pharm Biotechnol. 2002;3:63–75. doi: 10.2174/1389201023378436. [DOI] [PubMed] [Google Scholar]
- 39.Slauch JM, Silhavy TJ. Genetic analysis of the switch that controls porin gene expression in Escherichia coli K-12. J Mol Biol. 1989;210:281–292. doi: 10.1016/0022-2836(89)90330-6. [DOI] [PubMed] [Google Scholar]
- 40.Boll JM, et al. Reinforcing lipid A acylation on the cell surface of Acinetobacter baumannii promotes cationic antimicrobial peptide resistance and desiccation survival. MBio. 2015;6:e00478-15. doi: 10.1128/mBio.00478-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tucker AT, et al. Defining gene-phenotype relationships in Acinetobacter baumannii through one-step chromosomal gene inactivation. MBio. 2014;5:e01313-14. doi: 10.1128/mBio.01313-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Metsalu T, Vilo J. ClustVis: A web tool for visualizing clustering of multivariate data using principal component analysis and heatmap. Nucleic Acids Res. 2015;43:W566–W570. doi: 10.1093/nar/gkv468. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.