Significance
The requirement of maintaining balanced expression of a large number of gene products represents a major challenge to the engineering of nitrogen fixation in cereal crops, necessitating reiterative combinatorial assembly cycles to optimize monocistronic gene expression. In this study, we have explored a “fuse-and-cleave” virus-derived polyprotein strategy to reduce gene numbers and achieve balanced expression of protein components required for nitrogenase biosynthesis and activity. After testing and regrouping assemblies on the basis of expression profiles, cleavage patterns, and activity, 14 essential genes were selectively assembled into 5 giant genes that enable growth on dinitrogen. This strategy has potential advantages, not only for transferring nitrogen fixation to plants, but also for the engineering of other complex systems of profound agronomic and ecological importance.
Keywords: nitrogen fixation, polyprotein, synthetic biology
Abstract
Re-engineering of complex biological systems (CBS) is an important goal for applications in synthetic biology. Efforts have been made to simplify CBS by refactoring a large number of genes with rearranged polycistrons and synthetic regulatory circuits. Here, a posttranslational protein-splicing strategy derived from RNA viruses was exploited to minimize gene numbers of the classic nitrogenase system, where the expression stoichiometry is particularly important. Operon-based nif genes from Klebsiella oxytoca were regrouped into giant genes either by fusing genes together or by expressing polyproteins that are subsequently cleaved with Tobacco Etch Virus protease. After several rounds of selection based on protein expression levels and tolerance toward a remnant C-terminal ENLYFQ-tail, a system with only five giant genes showed optimal nitrogenase activity and supported diazotrophic growth of Escherichia coli. This study provides an approach for efficient translation from an operon-based system into a polyprotein-based assembly that has the potential for portable and stoichiometric expression of the complex nitrogenase system in eukaryotic organisms.
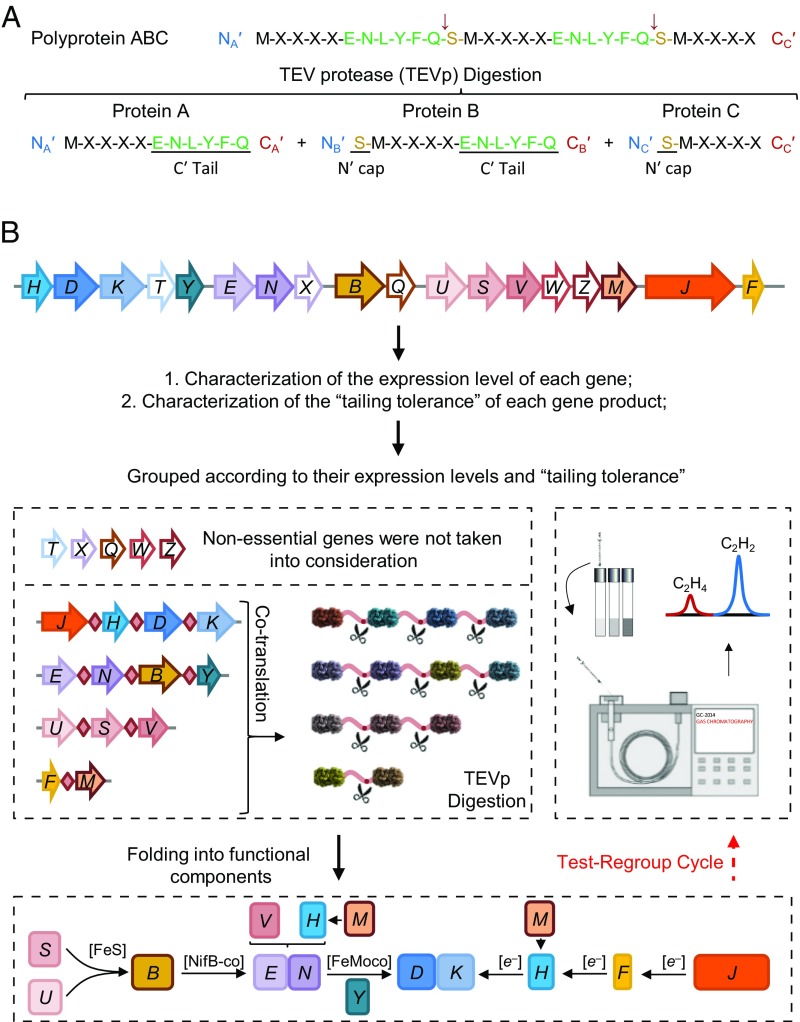
Complex biological systems (CBS) encode useful functions of interest for biotechnology, including nutrient assimilation, metabolic pathways, and natural product biosynthesis (1). However, genetic engineering of such systems with large numbers of genetic components is difficult, particularly when there is a stoichiometric requirement for balanced expression of the encoded protein components. To date, one approach toward engineering CBS has involved the complete refactoring of each individual gene in which all of the original native regulatory components have been removed (2–4). The disadvantage of this approach is the increased fragility of refactored systems, particularly in cases where there is an essential requirement to maintain stoichiometry of the protein-encoded components. An alternative approach is to reassemble the system as polycistronic modules, which maintain protein complex stoichiometry (5). However, large polycistronic operons cannot easily be utilized to express bacterial CBS in eukaryotic cells. One way to solve this problem is to retain the advantage of bacterial operon coexpression by expressing polyproteins that can be subsequently cleaved by a site-specific protease to release individual enzyme components. A natural example is provided by some RNA viruses that express a single giant gene, which is translated as a polyprotein and is subsequently cleaved by viral protease to form functionally viral components with consistent stoichiometry (6). Here, the well-studied Tobacco Etch Virus protease (TEVp) was used for engineering a CBS using a polyprotein-based strategy. TEVp is able to cleave its substrate ENLYFQS between the Q and S residues with high specificity and efficiency, leaving an ENLYFQ-tail on the C terminus of the protein encoded upstream and a serine residue on the N terminus of the downstream-encoded protein (Fig. 1A) (6).
Fig. 1.
Process for reconstructing biological nitrogen fixation using the posttranslational protein splicing strategy. (A) Digestion of polyproteins by TEVp leads to a C-terminal ENLYFQ-tail for the lead protein A, an N-terminal S-cap for the last protein C, and both an N-terminal S-cap and a C-terminal ENLYFQ-tail for the sandwiched protein B. (B) Arrangement of structural and accessory nif genes in the reconstituted operon-based system (pKU7017) from K. oxytoca (5). The regulatory nifLA operon, which controls transcription from σ54-dependent nif promoters, is not included in this diagram (a detailed description of pKU7017 is provided in Materials and Methods). Essential nif genes were regrouped and assembled into giant genes based on their relative expression levels and tailing tolerance. Test-regroup cycles were carried out to select for optimal combinations of the polyprotein-based system that yielded nitrogenase activity as measured by the acetylene reduction assay.
There has been a long-standing interest in reducing dependence on industrial nitrogen fertilizers through engineering nonlegume crops that can ‘‘fix’’ their own nitrogen (7, 8). This could be achieved by transferring nitrogen fixation (nif) genes to plant organelles (e.g., chloroplasts, root plastids, or mitochondria) (9–13). The main challenge to achieving this goal is balanced expression of the many gene products involved in the biological nitrogen fixation (BNF) process (2, 5, 14). Nitrogenase is a complex enzyme consisting of two metalloprotein components: the Fe protein (dinitrogenase reductase) and the MoFe protein (dinitrogenase) (15). Although only three genes—nifH, nifD, and nifK—are required to encode the structural subunits of the enzyme, nitrogenase maturation requires the assembly and insertion of three different metal cofactors in a complex multistep process (16). The functionality of the Fe protein is conferred by a [Fe4S4] cluster (synthesized by NifU and NifS) that bridges the NifH subunits in the Fe protein homodimer and is also dependent on the maturase protein NifM (17, 18). The mature MoFe protein holoenzyme is a heterotetramer formed from the NifD (α) and NifK (β) structural subunits that contains an [Fe8S7] cluster at the α–β interfaces, known as the P cluster, and a complex heterometallic cofactor, known as FeMo-co, that has an interstitial carbon atom at its core and also contains an organic moiety, homocitrate [Fe7-S9-C-Mo-homocitrate] (19). The assembly pathway for FeMo-co biosynthesis, which contains one of the most complex heterometal clusters in biology, is highly complex, requiring at least nine nif genes in vivo (20). The heterotetramer formed by NifEN, which is structurally and functionally related to NifDK, plays a crucial role as a node in the FeMo-co maturation pathway (21). Maintaining the stoichiometry of the NifEN and NifDK tetrameric complexes and the requirement of balancing expression ratios of all of the nif gene products required for nitrogenase synthesis and activity is a vital prerequisite for engineering nitrogenase in nondiazotrophic hosts (2, 3, 5).
In this study, we have chosen a representative BNF system from Klebsiella oxytoca to design a polyprotein-based strategy for stoichiometric expression of components required for the biosynthesis and activity of nitrogenase (Fig. 1). Eighteen nif genes required for nitrogen fixation in recombinant Escherichia coli (5) were grouped according to their expression levels and tolerance of their respective products to the presence of tails remaining after TEVp cleavage. Giant genes were assembled from individual nif-coding sequences linked by TEVp recognition sites. After several rounds of testing expression and functionality of cleaved proteins, we regrouped genes as necessary to optimize each polyprotein and established a nitrogen fixation system containing five giant genes. Following TEVp processing of the polyproteins in vivo, their cleavage products enabled synthesis of active nitrogenase, allowing E. coli to grow on dinitrogen as the sole nitrogen source. Thus, the polyprotein-based strategy provides an effective solution for stoichiometric expression of nitrogenase components from a single precursor protein, thus reducing the number of genes required to engineer BNF in foreign hosts.
Results
Assessment of Nif Components for Polyprotein Assembly.
First, to utilize the polyprotein-based strategy, it is necessary to assess the expression levels of each component to determine which proteins are suitable for grouping together in terms of expression stoichiometry. Second, since both N-terminal and C-terminal tails can remain after TEVp cleavage, depending on positioning within the precursor polyprotein (Fig. 1A), it is necessary to determine the tailing tolerance of each gene product to design the arrangement of the coding sequences within the giant genes. Expression levels of each nif gene in their native operon locations were quantitated using in-frame translational fusions under steady-state diazotrophic conditions (SI Appendix, Fig. S1 and Table S1). Although this assay does not take into account the stability of the native nif-encoded proteins, we observed that the ratio of nifH to nifDK expression was 2:1 as demonstrated for their respective accumulated protein products in Azotobacter vinelandii (22). These relative expression levels suggest polyprotein designs in which the coding sequences of NifHDKJ, NifENBY, NifUSV, and NifFM can be grouped into giant genes that express cleavable polyproteins. Since the presence of tails after TEVp cleavage may introduce additional constraints on giant gene design, we assessed the tolerance of each Nif protein to the presence of a C-terminal ENLYFQ tail by assaying the nitrogenase activities of constructs in which the coding sequence for this tail was added to individual genes in the 18-gene reconstituted Nif system (Fig. 1B and SI Appendix, Table S2). This revealed that NifK cannot tolerate the tail and therefore can be located only at the C terminus of a polyprotein. Most of the other components were tailing-tolerant although this reduced the activity of NifB by about 30%.
To minimize the BNF system and simplify the reassignment of genes encoding polyproteins, we omitted nifT, nifX, nifW, and nifZ, which are not essential for BNF in E. coli, as single mutations in these genes do not influence nitrogenase activity (SI Appendix, Fig. S2B). Similarly, nifQ was also omitted because the function of its gene product can be recovered in the presence of a high concentration of molybdenum (SI Appendix, Fig. S2C), as observed previously (23). Guided by the expression groupings defined above and the tailing tolerance of protein components, giant genes were assembled to encode polyproteins with coding sequences flanked by TEVp recognition sites (Fig. 1B). (These giant genes are annotated from here on as nifJǒHǒDǒK, nifEǒNǒBǒY, nifUǒSǒV, and nifFǒM, where ǒ indicates the presence of a TEVp-processing site.)
Activity-Based Test and Regroup Cycles.
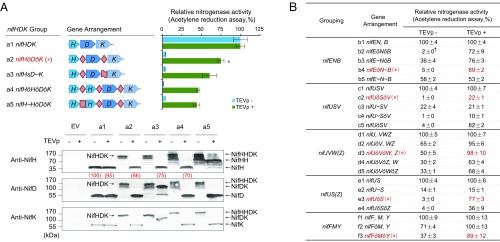
The functionality of our first-generation polyproteins, both before and after TEVp cleavage, was determined by measuring nitrogenase activities obtained from each giant gene when complemented with the remainder of the nif genes, respectively. Cleavage was achieved by introducing a cassette for expressing TEVp under the control of the Ptac promoter after induction with isopropyl β-d-1-thiogalactopyranoside (IPTG). Induction of TEVp expression did not influence the functionality of native Nif proteins. When assayed for acetylene reduction, the giant nifJǒHǒDǒK gene, expressed from the nifH promoter, resulted in only 5% activity after TEVp induction (SI Appendix, Fig. S3A). The nifUǒSǒV and nifEǒNǒBǒY genes also exhibited low activities of 22 and 19%, respectively (Fig. 2B and SI Appendix, Fig. S3C), in contrast to the nifFǒM gene, which showed almost full activity (99%) after TEVp induction (Fig. 2B).
Fig. 2.
Assessment of giant genes for complementation of nitrogenase activity and cleavage of their encoded polyproteins. In all cases, giant genes were expressed from native nif promoters specific to the first gene located in their coding sequences (Materials and Methods). Giant genes taken forward for assembly of the complete polyprotein-based system are indicated in red with an additional asterisk. (A) Example of a dataset for the nifHDK group. The gene arrangement for each giant gene is displayed as colored arrows with letters representing the corresponding nif gene product. TEVp sites are shown as diamonds, and linkers within fusion proteins are indicated by wavy ribbons. The acetylene reduction assay was used to measure complementation by each giant gene of the remaining nif genes in the operon-based system either in the absence of the TEVp-coding sequence (blue bars) or when TEVp expression was induced with 20 μM of IPTG (green bars). Acetylene reduction activities by the reconstituted operon-based system in E. coli were assigned as 100% (specific activities in the absence and presence of TEV were 30.4 ± 2.6 and 29.2 ± 1.7 nmol C2H4/min/mg total protein, respectively). Error bars indicate the SD observed from at least two biological replicates. Samples were immediately collected after the acetylene reduction assay for Western blotting (antibodies used are listed in Materials and Methods. Complete gels of the Western blots are provided in SI Appendix, Fig. S9). “EV” represents empty vector, used as a negative control. Image J software was used for quantification of NifH protein, and relative expression levels are shown in red as a percentage (in parentheses). (B) Summary data for the nifENB, nifUSV, nifJVWZ, nifUSZ, and nifFMY groups. The gene arrangement is shown in short format with rows b1, c1, d1, e1, and f1 representing the original operon-based system in E. coli. Commas indicate separate operons, “ǒ” indicates a TEVp site, and “∼” indicates a fusion protein. “†” indicates SD values lower than 0.5. Complete datasets for these groupings including the associated Western blots are shown in SI Appendix, Fig. S4.
The low activities of proteins following cleavage from polyproteins prompted us to test multiple gene combinations in a series of regrouping and retesting cycles. We observed that coexpression of NifJ with NifHDK led to decreased protein levels of NifH, NifD, and NifK compared with proteins expressed from the native nifHDK operon, although the cleavage products appeared similar (SI Appendix, Fig. S3B). Consequently, when nifJ was excluded from the giant gene, the resultant nifHǒDǒK assembly enabled 75% nitrogenase activity after TEVp cleavage of its polyprotein product, with a similar NifD:NifK protein stoichiometry to that expressed from the native genes (Fig. 2A). As NifB showed weak tolerance (71%) to the C-terminal ENLYFQ-tail (SI Appendix, Table S2), we removed nifY from nifEǒNǒBǒY, and the resultant nifEǒNǒB gene restored 72% of nitrogenase activity, following TEVp induction (Fig. 2B). When the NifY-coding sequence was reassigned to nifFǒM, cleavage of the larger polyprotein encoded by nifFǒMǒY resulted in 89% of the nitrogenase activity exhibited by the native components, and increased levels of NifY were observed (Fig. 2B and SI Appendix, Fig. S4E). For the nifUSV group, posttranslational splicing of the NifUǒSǒV polyprotein led to decreased levels of NifU and nearly undetectable amounts of NifS, but the level of NifV did not apparently change (compare lanes c1 and c2 in SI Appendix, Fig. S4B). In an attempt to improve this, we removed the nifV sequence from the giant gene, but retained NifUS coexpression with nifUǒS. Surprisingly, this restored native levels of NifU and NifS after cleavage and recovered nitrogenase activity to 82% (compare lanes c2 and c5 in SI Appendix, Fig. S4B). To assign nifV to another giant gene, we regrouped it with nifJ to form nifJǒV. Interestingly, in this case the polyprotein product was active even before protease cleavage, resulting in 65% nitrogenase activity, which increased to 95% after splicing with TEVp. Although native levels of NifV were released under these conditions, the amount of NifJ decreased (compare lanes d1 and d2 in SI Appendix, Fig. S4C).
To further optimize activity, we carried out additional regrouping of genes encoding polyproteins and also tested the incorporation of fused genes as a means to simplify the ensembles. Since splicing of NifHǒDǒK by TEVp resulted in a decreased amount of NifH (66%), but in wild-type levels of NifD and NifK (compare lanes a1 and a2 in Fig. 2A), we attempted to restore the optimal 2:1 ratio of NifH:NifDK (22) through assembly of a giant gene (nifHǒHǒDǒK) expressing two copies of nifH. Although this ensemble slightly increased the level of NifH after cleavage (70%), it did not result in increased nitrogenase activity (compare lanes a2 and a4 in Fig. 2A). To further attempt optimization of NifHDK levels, we also incorporated fusion proteins with different linkers guided by previous studies and natural existing examples (24–27). Fused NifD∼K proteins showed broad tolerance to different lengths of GGGGS linkers, with a maximum activity of 91% when 5× GGGGS linkers were added (SI Appendix, Fig. S5A). We also found that two copies of the NifH protein could be functionally fused with an ArsA linker retaining 89% nitrogenase activity (SI Appendix, Fig. S5B). However, integrating NifD∼K or NifH∼H fusions into two further ensembles (nifHǒD∼K and nifH∼HǒDǒK) did not result in higher activities than the original nifHǒDǒK gene (compare lane a2 with lanes a3 and a5 in Fig. 2A).
For the nifENB group, functional fusions of NifE∼N and NifN∼B were obtained using 5× and 3×GGGGS linkers, which exhibited activities of 91 and 115%, respectively (SI Appendix, Fig. S5 C and D). We replaced the corresponding parts in the nifEǒNǒB gene to generate three more assemblies (nifE∼NǒB, nifEǒN∼B, and nifE∼N∼B). The incorporation of either the NifE∼N or NifN∼B fusion resulted in higher nitrogenase activities (76 and 89%, respectively) compared with the nifEǒNǒB gene (compare lanes b3 and b4 with b2 in SI Appendix, Fig. S4A). However, when all three genes were fused to express the NifE∼N∼B protein, only 50% nitrogenase activity was obtained. This decrease may reflect the presence of truncated NifE∼N translation products expressed from nifE∼N∼B (see lane b5 in SI Appendix, Fig. S4A). We also attempted fusion of NifU with NifS and obtained 50% nitrogenase activity with a 5× GGGGS linker when expressed in the native operon arrangement nifU∼SVWZM (SI Appendix, Fig. S5E). However, this fusion protein had lower activity than that obtained after cleavage of the NifUǒS polyprotein (compare lanes c3 and c5 in SI Appendix, Fig. S4B and lanes e2 and e3 in SI Appendix, Fig. S4D).
Assembly and Characterization of Complete Polyprotein-Based Nitrogenase Systems.
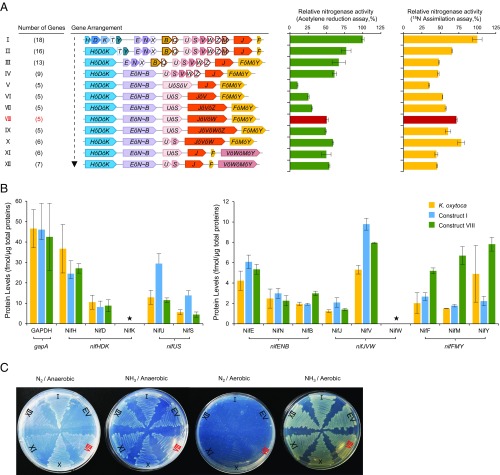
To combine polyproteins into a functional Nif system, we sequentially replaced the native gene parts with giant gene assemblies (Fig. 3A). Sequential combination of nifHǒDǒK with nifFǒMǒY and nifEǒN∼B (thus reducing the number of genes from 16 to 9) resulted in relatively small decreases in nitrogenase activity as measured both by acetylene reduction and 15N assimilation (Fig. 3A, rows I–IV). However, replacement of the native nifUSVWZ genes with nifUǒSǒV (thus reducing the number of genes to 5) resulted in a dramatic decrease in activity (10% of the native system) when acetylene reduction was measured (compare rows IV and V in Fig. 3A). The greater influence on acetylene reduction is probably the result of a kinetic effect as the 15N assay was conducted over a longer time window. Nitrogenase activity improved when nifV was switched from nifUǒSǒV to nifJǒV in the five-gene system (Fig. 3A, compare rows V and VI) as anticipated from the analysis of single polyproteins (Fig. 2B). Nevertheless, the decreased activity observed in the absence of nifW and nifZ prompted us to reconsider their involvement. Although the nifWZ gene products do not apparently have an impact on the activity of our reconstituted system (SI Appendix, Fig. S2B), previous studies suggest that they are required for full activity of the MoFe protein (28, 29). Bearing in mind the native location and expression levels of nifW and nifZ, we assembled additional giant genes designed to express their gene products as polyproteins with NifJ and NifV (nifJǒVǒZ, nifJǒVǒW, and nifJǒVǒWǒZ). When assayed to complement the native genes, the highest activity was obtained with the polyprotein expressing NifJVW (98%), and no benefit was obtained by incorporating NifZ (SI Appendix, Fig. S4C, compare lanes d3, d4, and d5). These activities were mirrored when these giant genes were complemented with the other four genes encoding polyproteins, with nifJǒVǒW again giving the highest level of activity (51% for acetylene reduction; compare arrangements VI, VII, VIII, and IX in Fig. 3A).
Fig. 3.
Assembly and characterization of the polyprotein-based nitrogenase system. (A) Schematic diagram showing the process of assembly by replacing native genes with regrouped giant genes. Numbers in parentheses on the left represent gene numbers (including giant genes and native genes) for each construct. Each ensemble was analyzed by both acetylene reduction and 15N assimilation to measure nitrogen fixation activity. The activities exhibited by the reconstituted operon-based system in E. coli [the pKU7017 plasmid is assembled with seven operon-based biobricks (5)] were assigned as 100% (29.1 ± 0.8 nmol C2H4/min/mg total protein for acetylene reduction assay and 1,172 ± 75 δ15N/14N‰ for the 15N assimilation assay) (row I). Error bars indicate the SD observed from at least two biological replicates. (B) Mass spectrometry analysis of protein levels from samples taken immediately after the acetylene reduction assays. The yellow bars represent protein samples from a nitrogen-fixing culture of K. oxytoca, grown under the same conditions as the E. coli cultures; the blue bars represent protein samples from the reconstituted operon-based nif system in E. coli (construct I in A), and the green bars represent protein samples from the polyprotein-based nif system (construct VIII in A); the GAPDH protein encoded by the E. coli gapA gene (EcgapA) was assigned as the internal reference. Asterisks mark proteins that could not be assigned (for NifK, chemical synthesis of the internal standard peptides failed, and, for NifW, no peptide with a detectable signal was identified). Error bars indicate the SD observed from three biological replicates. (C) Diazotrophic growth promoted by polyprotein-based nitrogenase systems in E. coli in the presence of 20 μM of IPTG. Roman numerals represent the corresponding assemblies in A. EV represents empty vector, used as a negative control. Control plates with strains grown in the absence of IPTG are shown in SI Appendix, Fig. S7.
Quantitative analysis of protein levels by Selected Reaction Monitoring (SRM) mass spectrometry revealed that, overall, the stoichiometry of most components from the polyprotein-based system matched remarkably well with the respective levels from the reconstituted operon-based system in E. coli and the native system in the original K. oxytoca host (Fig. 3B; compare green, blue, and yellow bars, respectively). This was particularly true for the NifHDK and NifENB proteins, where stoichiometry is important for nitrogenase biosynthesis and activity [the level of NifK was determined by quantification of Western blots (SI Appendix, Fig. S6)]. Unexpectedly, NifU and NifS levels in the polyprotein system were more similar to those in the native K. oxytoca host, compared with the original recombinant system in E. coli (Fig. 3B and SI Appendix, Fig. S6). Although expression of NifU and NifS as a single polyprotein limits complementation of a nifUS deletion (Fig. 2B), only a small benefit was achieved when nifU and nifS were included as separate genes, with the other four polyproteins (compare rows VIII and X in Fig. 3A). Although NifJ and NifV were located on the same polyprotein, the level of NifV was fivefold higher than that of NifJ, which is similar to the ratio obtained from the native system. This suggests that NifJ may have a relatively short half-life, which is not reflected when expression is measured at the transcriptional level (SI Appendix, Table S1). However, the rearrangement of nifF, nifM, and nifY into a giant gene (nifFǒMǒY) resulted in increased levels of all three protein products, as anticipated from their inclusion in the same polyprotein (Fig. 3B). This may have consequences for the level of NifH, which, unexpectedly, did not decrease in the polyprotein-based system in contrast to the data obtained in Fig. 2A. The increased level of NifM resulting from posttranslational splicing of the NifFMY polyprotein may be responsible for the increased NifH accumulation since NifM is required for the maturation of NifH (17).
Since the combined five-gene (Fig. 3A, row VIII) and six-gene (Fig. 3A, row X) polyprotein systems exhibited 72 and 75% 15N assimilation activity, respectively, we anticipated that these combinations of giant genes could support diazotrophic growth by E. coli. As in the case of the initial single-gene system (Fig. 3A, row I), the five-gene and six-gene arrangements (assemblies VIII and X, respectively) enabled E. coli to grow on solid media with dinitrogen as the sole nitrogen source, when the expression of TEVp was induced by IPTG. In contrast, assemblies IX and XI, which exhibited lower nitrogenase activates, grew less well under these conditions (Fig. 3C). Control experiments in the absence of IPTG, which results in basal levels of TEVp, resulted in poor growth of assemblies VIII and X as anticipated (SI Appendix, Fig. S7).
Discussion
The use of synthetic biology to re-engineer complex systems has transformed our understanding of fundamental mechanisms and has driven a variety of applications, e.g., materials construction, chemical processing, and energy production (30–33). However, the ability to reliably engineer complex biological systems that behave as expected remains limited due to their intrinsic complexity. Thankfully, biology itself provides natural resources that can be exploited to help satisfy the requirements for re-engineering. In the current study, by employing a posttranslational splicing strategy prevalent in RNA viruses, we have established a method for re-engineering CBS using a polyprotein-based strategy. Although others have used TEVp-based posttranslational cleavage to codeliver a small number of proteins into eukaryotic cells (e.g., refs. 34 and 35), to our knowledge, this strategy has not been used to date to engineer complex biological systems such as BNF. This strategy provides an important approach for efficient adaptation from an operon-based system into a polyprotein-based assembly. Balanced expression of CBS components can be achieved by organizing genes with similar expression levels into giant genes expressed as polyproteins, followed by subsequent cleavage to release single components. The advantages of this strategy are particularly useful for transforming complex systems from prokaryotes into eukaryotes. Eukaryotic promoters and terminators often span hundreds of base pairs, nearly 10 times the amount of their bacterial counterparts (36). Monocistronic expression of large numbers of heterologous genes in eukaryotes requires complex combinatorial design and the screening of many gene expression parts to surmount difficulties in coordinating the expression level of each component (14). Taken together, translation of genes into polyproteins reduces the number of translational units and maintains stoichiometric expression, thus simplifying the approach for adapting prokaryotic CBS for expression in eukaryotic cells. Although other approaches are available for coordinating expression of multiple genes in eukaryotes in an operon-like format—for example, using internal ribosome entry sites to direct translational initiation within the mRNA—this may result in substantially lower expression levels in comparison with normal Cap-dependent translation (37). Another possibility is to introduce the viral polyprotein self-cleavage peptide 2A to generate discrete translation products, although this results in an 18-aa tail, which may influence protein functionality (38).
We have successfully applied the TEV-based posttranslational slicing method for re-engineering the classical molybdenum nitrogenase system. Surprisingly, however, this strategy is less amenable to the minimal iron-only nitrogenase system (39) since the structural AnfH, AnfG, and AnfK proteins are relatively intolerant to the presence of the C-terminal ENLYFQ-tail (SI Appendix, Table S3). For the molybdenum system, the polyprotein strategy has the obvious advantage of maintaining the stoichiometry of the NifDK and NifEN protein complexes, although it cannot alleviate stability issues arising after polyprotein cleavage. Nevertheless, it appears to be remarkably robust to gene rearrangements that result in cotranslation of other nif components, even though this can significantly alter expression levels from those observed in the native system. Tolerance to protein-level changes was particularly notable when we tested the functionality of individual polyproteins in complementing nitrogenase activity in E. coli. For example, decreased levels of NifJ when cotranslated with NifV, or increased levels of NifY when cotranslated with NifM, did not greatly influence nitrogenase activity (SI Appendix, Fig. S4).
However, some protein combinations did not give rise to the expected stoichiometry when assembled as polyproteins. For example, cotranslation of NifU and NifS with NifV, or with NifZ, led to severe decreases in NifUS levels. In both cases, this had a detrimental effect on nitrogenase activity, emphasizing the crucial role played by NifU and NifS in [Fe-S] cluster biosynthesis for the three metalloclusters present in the nitrogenase component proteins (40–42). We also observed some spontaneous processing of NifJ and NifU when cotranslated as polyproteins independent from TEVp cleavage (SI Appendix, Fig. S4). We reason that this may result either from ribosome drop-off (43) or polyprotein instability.
We examined the potential advantages of incorporating fusion proteins into our polyprotein designs as an alternative to splicing and release of each individual component. In accordance with previous studies (25, 44, 45), significant activities were obtained with NifD∼NifK, NifE∼NifN, and NifN∼NifB protein fusions after optimization of linker lengths. However, among the polyprotein combinations that we tested, the only advantage conferred by this strategy was provided by the NifN∼B fusion when located in a polyprotein with NifE (nifEǒN∼B), which exhibited increased activity compared with NifENB spliced into single components (nifEǒNǒB). This enhancement may result from more efficient transfer of the NifB-co cluster from NifB to NifEN as reported previously (25). Interestingly, some polyproteins exhibited activity even in the absence of TEV protease cleavage. These functional polyproteins may provide initiatives for directed evolution of proteins with multiple functions, thus reducing the number of components required to re-engineer a CBS. Previous reports have suggested that NifW and NifZ function in MoFe protein maturation (28, 29), with NifZ acting as a chaperone in the assembly of P clusters (28, 29, 46). However, in our hands, in the heterologous E. coli background, single or double mutants of nifW and nifZ had negligible effects on the activity of the reconstituted operon-based nitrogenase system. In contrast, the polyprotein-based nitrogenase system was more fragile in this respect, with decreased activity being observed in the absence of NifW. It is possible that this represents a kinetic effect since nifW and nifZ mutants delay the induction of nitrogenase activity in K. oxytoca (29). Recently, it has been demonstrated that NifW and NifZ bind to immature forms of apo-NifDK and are likely to function as assembly factors required for P-cluster maturation (47). Since diazotrophic growth is slower in nifW mutants, it has been suggested that either the function of this gene can be partially complemented by another gene or nifW plays a kinetic role in MoFe protein maturation. These observations imply that the operon-based system in E. coli enables optimal maturation of the MoFe protein in the absence of NifW, whereas the requirement for this protein in the polyprotein-based system may reflect a slower rate of P-cluster formation or maturation. Thus, although our polyprotein combination appears to faithfully reproduce the protein stoichiometry observed with native nif operons, further iterations of this approach may be necessary to fully optimize nitrogenase biosynthesis and activity.
In recent studies, we and others have demonstrated that homologs of the [Fe-S] cluster biosynthesis machinery and electron transport modules present in eukaryotes are compatible with the nitrogenase system (10–12). If the Nif-specific components encoding iron-sulfur cluster biosynthesis (nifU, nifS) and electron transfer (nifJ, nifF) can be functionally replaced by their eukaryotic counterparts, the number of components required, for example, to assemble active nitrogenase in plant organelles could be significantly reduced (8, 10–12). One of our polyprotein-based nitrogenase systems, consisting of three giant genes (nifHǒDǒK, nifEǒN∼B, and nifVǒWǒMǒY) with nifU, nifS and nifJ, nifF expressed as separate components, exhibits 50% nitrogenase activity and permits slow growth of E. coli with N2 as the sole nitrogen source (assembly XII in Fig. 3). Our results therefore imply that only three giant genes might be required to engineer diazotrophy in eukaryotic organelles, with components for iron-sulfur cluster assembly and electron donation being supplied by the host.
In addition to complex biological systems such as nitrogen fixation, there is considerable interest in engineering bacterial metabolic pathways in plants, for example, to produce antifungal and antibacterial secondary metabolites that provide resistance against pathogens and for effective degradation of xenobiotics. The polyprotein strategy thus may provide a useful approach to engineering complex bacterial pathways for agronomic, ecological, and economic purposes.
Materials and Methods
Strains and Media.
E. coli Top10 was used for routine cloning and plasmid propagation. E. coli JM109 was the strain background used to measure nitrogenase activity either by acetylene reduction or 15N assimilation. E. coli NCM3722 was used as host for the diazotrophic growth experiments. Luria-Bertani broth for E. coli growth contained 10 g/L of tryptone, 5 g/L of yeast extract, and 10 g/L of NaCl. All nitrogen fixation assays were performed in KPM minimal medium [10.4 g/L of Na2HPO4, 3.4 g/L of KH2PO4, 26 mg/L of CaCl2·2H2O, 30 mg/L of MgSO4, 0.3 mg/L of MnSO4, 36 mg/L of ferric citrate, 10 mg/L of para-aminobenzoic acid, 5 mg/L of biotin, 1 mg/L vitamin B1, 0.05% Casamino acids, and 0.8% (wt/vol) glucose], and 30 nmol/L of Na2MoO4 (according to results from SI Appendix, Fig. S2C), supplied with 10 mM ammonium sulfate (KPM-HN) for pregrowth or 10 mM glutamate (KPM-LN) for nitrogenase activity assays. Diazotrophic growth experiments were carried out with solid KPM-HN medium or solid KPM medium without 0.05% Casamino acids. Antibiotics were used at the following concentration: 25 μg/mL for kanamycin, 25 μg/mL for chloramphenicol, and 10 μg/mL for tetracycline.
Plasmids.
Plasmids used in this study are listed in Dataset S1. All plasmids were verified by sequencing before use for further experiments. The pKU7017 plasmid is assembled with seven operon-based biobricks (nifJ, nifHDKTY, nifENX, nifUSVWZM, nifF, nifBQ, and nifLA) from K. oxytoca, in which regulation is dependent on the native σ54-dependent promoters and is regulated by the nifL and nifA genes (5). In this study, each of the operon-based Biobricks, except for the nifLA operon, were removed from the pKU7017 plasmid, respectively, resulting in six different nif-deficient plasmids: pKU7870 (ΔnifJ), pKU7871 (ΔnifHDKTY), pKU7872 (ΔnifENX), pKU7873 (ΔnifUSVWZM), pKU7874 (ΔnifBQ), and pKU7875 (ΔnifF). The removed operon-based Biobricks were cloned into pBDS402K (BsaI) or pBDS1024K (BpiI) plasmids via Golden Gate assembly (48) to generate six plasmids (pKU7876∼pKU7881) that complement their pKU7017-deficient derivatives (pKU7870∼pKU7875), respectively. The pBDS402K and pBDS1024K plasmids were assembled with a kanamycin resistance gene, pBR322 origin, Rop sequences, and lacZα-coding sequences, flanked with either BsaI for pBDS402K or BpiI for pBDS402K, using the Gibson Assembly kit (#E5520S; NEB) (plasmids maps are provided in SI Appendix, Fig. S6). All giant genes were assembled via Golden Gate assembly, with the coding sequences for TEVp recognition sites or additional specific linkers added by PCR. The TEV protease-coding sequences were chemically synthesized, carrying a S219V substitution to lower auto-inactivation (49). The synthesized TEVp genes were in-frame–fused to the MBP gene to generate a chimeric gene MBP-TEVp. Subsequently, a chimeric gene was assembled with an inducible Ptac promoter, a strong ribosome-binding site from the araB gene of E. coli, and the artificial strong terminator L3S2P21 (50) via Gibson assembly. In each case, 20 μM of IPTG was used to induce TEV protease expression. The complete sequence of the polyprotein-based nitrogenase system with optimal activity (construct VIII in Fig. 3A) is shown in Dataset S2.
Expression-Level Assays.
Primers used for PCR of promoter regions are listed in Dataset S1. To determine the expression level of each nif gene, PCR products carrying the promoter region extending to the eighth codon were fused in-frame with the lacZ-coding sequence on plasmid pBD020 (the plasmid map is provided in SI Appendix, Fig. S1). Plasmid pBD020 is a pRW50 (51) derivative in which the trpAB partial sequences of E. coli have been deleted and an EcoRV restriction site has been introduced just before the ATG codon of the lacZYA operon. The pBD020 derivatives carrying each nif-lacZYA translational fusion were verified by sequencing and cotransformed into a JM109 strain carrying the pKU7017 plasmid. Transformants were grown overnight in KPM-HN medium, and the cells were then diluted into 2 mL KPM-LN medium in 20-mL sealed tubes to a final OD600 of ∼0.3. Air in the tubes was repeatedly evacuated and replaced with argon. After incubation at 30 °C for 16 h, cells were collected and β-galactosidase activities were measured according to Miller (52).
Tailing-Tolerance Assay.
To determine the tolerance of each gene product to the C-terminal tail, each nif gene, carrying an extended ENLYFQ-coding sequence, was used to replace the original gene located in each operon. The resultant new operons encoding proteins with the ENLYFQ-tail were verified by sequencing and cotransformed with their corresponding deficient pKU7017 derivatives into the JM109 strain. Transformants were assayed for acetylene reduction activity. Activities obtained from operons without tagged genes were considered as 100% in each case (SI Appendix, Table S2). For genes that overlap with upstream genes (nifX, nifW, nifZ, nifM, and nifQ), the region of overlap together with their respective ribosome-binding sites was amplified and added just downstream of the stop codon of the upstream genes to avoid frameshifts resulting from the addition of the ENLYFQ-coding sequences to the upstream genes.
Acetylene Reduction and 15N2 Assimilation Assay.
The C2H2 reduction assay and 15N2 assimilation assay were carried out as described previously (53). To measure the activity of E. coli JM109 derivatives expressing reassembled nitrogenase systems, cells were initially grown overnight in KPM-HN medium. The cells were then diluted into 2 mL KPM-LN medium in 20-mL sealed tubes to a final OD600 of ∼0.3. For the acetylene reduction assay, air in the tubes was repeatedly evacuated and replaced with argon. After incubation at 30 °C for 6–8 h, 2 mL C2H2 was injected and the gas phase was analyzed ∼16 h later with a Shimadzu GC-2014 gas chromatograph. For the 15N2 assimilation assay, air in the tubes was repeatedly evacuated and replaced with high-purity nitrogen gas, and then 3 mL gas was removed and 2 mL 15N2 (99%+, Shanghai Engineering Research Center for Stable Isotopes) was injected. After 48 h of incubation at 30 °C, the cultures were collected and then freeze-dried. Isotope ratios are expressed as δ15N where values are a linear transform of the 15N/14N isotope ratios, representing the per thousand difference between the isotope ratios in a sample and in atmospheric N2 (54). Data presented are mean values based on at least two biological replicates for both the C2H2 reduction assays and the 15N2 assimilation assays.
Diazotrophic Growth.
E. coli JM109 was the strain background used to measure nitrogenase activity either by acetylene reduction or 15N assimilation. However, as this strain carries many genetic deficiencies and requires Casamino acids and vitamin B1 for normal growth, it is not suitable for studying diazotrophic growth of E. coli. In this case, the prototrophic E. coli K-12 strain NCM3722 was used for diazotrophic growth, which is more robust than strains of the popular MG1655/W3110 lineage (55). Reconstructed plasmids were transformed into the E. coli NCM3722 strain, and the transformants were spread on LB plates with appropriate concentration of antibiotics. After incubation at 37 °C for 16 h, single colonies were picked and streaked onto KPM-HN and KPM-LN plates. For aerobic growth, the plates were directly incubated at 30 °C. For anaerobic growth, the plates were moved to a 2.5-L anaerobic jar (Oxoid #AG0025A; Thermo Scientific), and subsequently anaerobic gas-generating sachets (Oxoid #AN0025A; Thermo) and an oxygen indicator (Oxoid #BR0055B; Thermo) were added. Anaerobic jars were immediately locked and incubated at 30 °C for 3–4 d.
Western Blot Assay.
In each case, 2-mL cultures combined from four parallel samples (8 mL total volume, OD600 = ∼0.6) were collected immediately after the acetylene reduction assay and suspended in 240 μL PBS buffer supplied with 60 μL 5× SDS buffer (#C508320; Sangon). After boiling for 10 min, samples were cooled to room temperature and centrifuged at 12,000 × g for 2 min. Twenty microliters of each sample were loaded on 15% SDS-polyacrylamide gels with 5 μL of PageRuler Prestained Protein Ladder (#26616; Thermo) as a marker. Proteins on the gels were subsequently transferred to PVDF membranes (#IPVH00010; Merck). The membranes were blocked with 5% skimmed milk (#232100; BD Difco) in PBS. The antibodies for each Nif protein were used at a dilution of 1:3,000. The secondary antibody goat anti-rabbit IgG-HRP (#ZB-2301; ZSGB-BIO) or goat anti-mouse IgG-HRP (#ZB-2305; ZSGB-BIO) was also used at 1:3,000. Development was accomplished by using two different methods: (i) an enhanced chemiluminescent substrate for HRP (#34077; Thermo) captured by Automatic Chemiluminescence Imaging Analysis System (Tanon-4200) or (ii) directly developed on the PVDF membranes by using the DAB Chromogen/HRP Substrate kit (#ZLI-9018; ZSGB-BIO). Antibodies used for immunoblotting and quantification in this study were as follows: (i) antibodies detecting NifH, NifD, NifN, NifU, NifS, NifV, NifM, and NifJ were raised against purified preparations of the corresponding K. oxytoca proteins in rabbits; (ii) antibodies detecting NifK and NifE were raised in rabbits against synthetic peptides corresponding to residues 501–514 (KLDSDTSQLGKTDY) of K. oxytoca NifK protein and residues 1–14 (MKGNEILALLDEPA) of K. oxytoca NifE protein; (iii) antibodies raised in rabbits against synthetic peptides corresponding to residues 179–192 (DGERYSGREAGEIL) of K. oxytoca NifB and residues 31–44 (AQESGETLTPERLA) of K. oxytoca NifY showed no affinity to the corresponding K. oxytoca proteins. Therefore, a 10× His-tag was added to the C terminus of the NifB protein for detection with an anti-His antibody (#TA-02; ZSGB-BIO), and an Myc-tag was added to the C terminus of the NifY protein for detecting the NifY protein with an anti-Myc antibody (#TA-01; ZSGB-BIO); and (iv) anti-MBP antibody (#15089–1; Proteintech) was used to detect the MBP-TEVp. Antibodies raised against purified Nif proteins and synthetic peptides were produced by the GenScript Company.
SRM-Based Quantification of Targeted Proteins by Mass Spectrometry.
Cells were immediately collected after the acetylene reduction assay. Samples were subsequently prepared according to Wiśniewski et al. (56) and then preanalyzed by high-resolution Orbitrap Liquid Chromatograph Mass Spectrometer (LC-MS). Peptides that are unique to the targeted Nif proteins and that show a high mass spectrometry signal response to maximize the sensitivity in the Orbitrap LC-MS assay were selected as references for determining the absolute quantity. The selected peptides were chemically synthesized (BANKPEPTIDE LTD) in both stable isotope-labeled (13C615N2 lysine and 13C615N4 arginine) and unlabeled forms. The synthesized peptides were subjected to LC-SRM, and an optimal method based on each reference peptide was developed. The absolute quantity of targeted proteins in samples was determined by liquid chromatography–isotope dilution mass spectrometry (57). Stable isotope-labeled peptides were mixed with different amounts of standard peptides, and peptide mixtures were then subjected to LC-SRM analysis. A linear response curve was plotted for each targeted peptide. Each sample was added in equal amounts to the stable isotope-labeled peptides. The LC-SRM of each sample was performed on the 6500 QTRAP (AB Sciex) with the established SRM method. Results were analyzed using Skyline (58). The concentration of target protein in the sample was calculated with its linear response curve (detailed methods for mass spectrometry are provided in SI Appendix).
Supplementary Material
Acknowledgments
We thank Professor Jilun Li (China Agriculture University, Beijing) for the antisera against nitrogenase proteins. This research was supported by the National Science Foundation of China (NSFC) (31530081); the 973 National Key Basic Research Program in China (2015CB755700 and 2010CB126503); the China Postdoctoral Science Foundation (2016T90013); the SLS-Qidong Innovation Fund; and the State Key Laboratory of Protein and Plant Gene Research, no. B02. Y.-P.W. is a recipient of a grant from the National Science Fund for Distinguished Young Scholars (NSFC, 39925017). R.D. was funded by the UK Biotechnology and Biological Sciences Research Council (BBS/E/J/000PR9797).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 8849.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1804992115/-/DCSupplemental.
References
- 1.Fischbach M, Voigt CA. Prokaryotic gene clusters: A rich toolbox for synthetic biology. Biotechnol J. 2010;5:1277–1296. doi: 10.1002/biot.201000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Temme K, Zhao D, Voigt CA. Refactoring the nitrogen fixation gene cluster from Klebsiella oxytoca. Proc Natl Acad Sci USA. 2012;109:7085–7090. doi: 10.1073/pnas.1120788109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smanski MJ, et al. Functional optimization of gene clusters by combinatorial design and assembly. Nat Biotechnol. 2014;32:1241–1249. doi: 10.1038/nbt.3063. [DOI] [PubMed] [Google Scholar]
- 4.Song M, et al. Control of type III protein secretion using a minimal genetic system. Nat Commun. 2017;8:14737. doi: 10.1038/ncomms14737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang X, et al. Using synthetic biology to distinguish and overcome regulatory and functional barriers related to nitrogen fixation. PLoS One. 2013;8:e68677. doi: 10.1371/journal.pone.0068677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Palmenberg AC. Proteolytic processing of picornaviral polyprotein. Annu Rev Microbiol. 1990;44:603–623. doi: 10.1146/annurev.mi.44.100190.003131. [DOI] [PubMed] [Google Scholar]
- 7.Beatty PH, Good AG. Plant science. Future prospects for cereals that fix nitrogen. Science. 2011;333:416–417. doi: 10.1126/science.1209467. [DOI] [PubMed] [Google Scholar]
- 8.Vicente EJ, Dean DR. Keeping the nitrogen-fixation dream alive. Proc Natl Acad Sci USA. 2017;114:3009–3011. doi: 10.1073/pnas.1701560114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oldroyd GED, Dixon R. Biotechnological solutions to the nitrogen problem. Curr Opin Biotechnol. 2014;26:19–24. doi: 10.1016/j.copbio.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 10.López-Torrejón G, et al. Expression of a functional oxygen-labile nitrogenase component in the mitochondrial matrix of aerobically grown yeast. Nat Commun. 2016;7:11426. doi: 10.1038/ncomms11426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang J, Xie X, Yang M, Dixon R, Wang Y-P. Modular electron-transport chains from eukaryotic organelles function to support nitrogenase activity. Proc Natl Acad Sci USA. 2017;114:E2460–E2465. doi: 10.1073/pnas.1620058114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ivleva NB, Groat J, Staub JM, Stephens M. Expression of active subunit of nitrogenase via integration into plant organelle genome. PLoS One. 2016;11:e0160951. doi: 10.1371/journal.pone.0160951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burén S, Rubio LM. State of the art in eukaryotic nitrogenase engineering. FEMS Microbiol Lett. 2018;365:fnx274. doi: 10.1093/femsle/fnx274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burén S, et al. Formation of nitrogenase NifDK tetramers in the mitochondria of Saccharomyces cerevisiae. ACS Synth Biol. 2017;6:1043–1055. doi: 10.1021/acssynbio.6b00371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Georgiadis MM, et al. Crystallographic structure of the nitrogenase iron protein from Azotobacter vinelandii. Science. 1992;257:1653–1659. doi: 10.1126/science.1529353. [DOI] [PubMed] [Google Scholar]
- 16.Rubio LM, Ludden PW. Maturation of nitrogenase: A biochemical puzzle. J Bacteriol. 2005;187:405–414. doi: 10.1128/JB.187.2.405-414.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Howard KS, et al. Klebsiella pneumoniae nifM gene product is required for stabilization and activation of nitrogenase iron protein in Escherichia coli. J Biol Chem. 1986;261:772–778. [PubMed] [Google Scholar]
- 18.Dos Santos PC, et al. Iron-sulfur cluster assembly: NifU-directed activation of the nitrogenase Fe protein. J Biol Chem. 2004;279:19705–19711. doi: 10.1074/jbc.M400278200. [DOI] [PubMed] [Google Scholar]
- 19.Rubio LM, Ludden PW. Biosynthesis of the iron-molybdenum cofactor of nitrogenase. Annu Rev Microbiol. 2008;62:93–111. doi: 10.1146/annurev.micro.62.081307.162737. [DOI] [PubMed] [Google Scholar]
- 20.Hu Y, Ribbe MW. Biosynthesis of the iron-molybdenum cofactor of nitrogenase. J Biol Chem. 2013;288:13173–13177. doi: 10.1074/jbc.R113.454041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaiser JT, Hu Y, Wiig JA, Rees DC, Ribbe MW. Structure of precursor-bound NifEN: A nitrogenase FeMo cofactor maturase/insertase. Science. 2011;331:91–94. doi: 10.1126/science.1196954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poza-Carrión C, Jiménez-Vicente E, Navarro-Rodríguez M, Echavarri-Erasun C, Rubio LM. Kinetics of Nif gene expression in a nitrogen-fixing bacterium. J Bacteriol. 2014;196:595–603. doi: 10.1128/JB.00942-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Imperial J, Ugalde RA, Shah VK, Brill WJ. Role of the nifQ gene product in the incorporation of molybdenum into nitrogenase in Klebsiella pneumoniae. J Bacteriol. 1984;158:187–194. doi: 10.1128/jb.158.1.187-194.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lahiri S, Pulakat L, Gavini N. Functional participation of a nifH-arsA2 chimeric fusion gene in arsenic reduction by Escherichia coli. Biochem Biophys Res Commun. 2008;368:311–317. doi: 10.1016/j.bbrc.2008.01.086. [DOI] [PubMed] [Google Scholar]
- 25.Wiig JA, Hu Y, Ribbe MW. NifEN-B complex of Azotobacter vinelandii is fully functional in nitrogenase FeMo cofactor assembly. Proc Natl Acad Sci USA. 2011;108:8623–8627. doi: 10.1073/pnas.1102773108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thiel T. Characterization of genes for an alternative nitrogenase in the cyanobacterium Anabaena variabilis. J Bacteriol. 1993;175:6276–6286. doi: 10.1128/jb.175.19.6276-6286.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suh M-H, Pulakat L, Gavini N. Functional expression of a fusion-dimeric MoFe protein of nitrogenase in Azotobacter vinelandii. J Biol Chem. 2003;278:5353–5360. doi: 10.1074/jbc.M208969200. [DOI] [PubMed] [Google Scholar]
- 28.Jacobson MR, et al. Biochemical and genetic analysis of the nifUSVWZM cluster from Azotobacter vinelandii. Mol Gen Genet. 1989;219:49–57. doi: 10.1007/BF00261156. [DOI] [PubMed] [Google Scholar]
- 29.Paul W, Merrick M. The roles of the nifW, nifZ and nifM genes of Klebsiella pneumoniae in nitrogenase biosynthesis. Eur J Biochem. 1989;178:675–682. doi: 10.1111/j.1432-1033.1989.tb14497.x. [DOI] [PubMed] [Google Scholar]
- 30.Cameron DE, Bashor CJ, Collins JJ. A brief history of synthetic biology. Nat Rev Microbiol. 2014;12:381–390. doi: 10.1038/nrmicro3239. [DOI] [PubMed] [Google Scholar]
- 31.Smanski MJ, et al. Synthetic biology to access and expand nature’s chemical diversity. Nat Rev Microbiol. 2016;14:135–149. doi: 10.1038/nrmicro.2015.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ausländer S, Ausländer D, Fussenegger M. Synthetic biology: The synthesis of biology. Angew Chem Int Ed Engl. 2017;56:6396–6419. doi: 10.1002/anie.201609229. [DOI] [PubMed] [Google Scholar]
- 33.Mathur M, Xiang JS, Smolke CD. Mammalian synthetic biology for studying the cell. J Cell Biol. 2017;216:73–82. doi: 10.1083/jcb.201611002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marcos JF, Beachy RN. Transgenic accumulation of two plant virus coat proteins on a single self-processing polypeptide. J Gen Virol. 1997;78:1771–1778. doi: 10.1099/0022-1317-78-7-1771. [DOI] [PubMed] [Google Scholar]
- 35.Chen X, Pham E, Truong K. TEV protease-facilitated stoichiometric delivery of multiple genes using a single expression vector. Protein Sci. 2010;19:2379–2388. doi: 10.1002/pro.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Redden H, Alper HS. The development and characterization of synthetic minimal yeast promoters. Nat Commun. 2015;6:7810. doi: 10.1038/ncomms8810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Siddiqui MS, Thodey K, Trenchard I, Smolke CD. Advancing secondary metabolite biosynthesis in yeast with synthetic biology tools. FEMS Yeast Res. 2012;12:144–170. doi: 10.1111/j.1567-1364.2011.00774.x. [DOI] [PubMed] [Google Scholar]
- 38.de Felipe P, et al. E unum pluribus: Multiple proteins from a self-processing polyprotein. Trends Biotechnol. 2006;24:68–75. doi: 10.1016/j.tibtech.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 39.Yang J, Xie X, Wang X, Dixon R, Wang Y-P. Reconstruction and minimal gene requirements for the alternative iron-only nitrogenase in Escherichia coli. Proc Natl Acad Sci USA. 2014;111:E3718–E3725. doi: 10.1073/pnas.1411185111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kennedy C, Dean D. The nifU, nifS and nifV gene products are required for activity of all three nitrogenases of Azotobacter vinelandii. Mol Gen Genet. 1992;231:494–498. doi: 10.1007/BF00292722. [DOI] [PubMed] [Google Scholar]
- 41.Johnson DC, Dos Santos PC, Dean DR. NifU and NifS are required for the maturation of nitrogenase and cannot replace the function of isc-gene products in Azotobacter vinelandii. Biochem Soc Trans. 2005;33:90–93. doi: 10.1042/BST0330090. [DOI] [PubMed] [Google Scholar]
- 42.Zhao D, Curatti L, Rubio LM. Evidence for nifU and nifS participation in the biosynthesis of the iron-molybdenum cofactor of nitrogenase. J Biol Chem. 2007;282:37016–37025. doi: 10.1074/jbc.M708097200. [DOI] [PubMed] [Google Scholar]
- 43.Sin C, Chiarugi D, Valleriani A. Quantitative assessment of ribosome drop-off in E. coli. Nucleic Acids Res. 2016;44:2528–2537. doi: 10.1093/nar/gkw137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suh M-H, Pulakat L, Gavini N. Functional expression of the FeMo-cofactor-specific biosynthetic genes nifEN as a NifE-N fusion protein synthesizing unit in Azotobacter vinelandii. Biochem Biophys Res Commun. 2002;299:233–240. doi: 10.1016/s0006-291x(02)02620-7. [DOI] [PubMed] [Google Scholar]
- 45.Lahiri S, Pulakat L, Gavini N. Functional NifD-K fusion protein in Azotobacter vinelandii is a homodimeric complex equivalent to the native heterotetrameric MoFe protein. Biochem Biophys Res Commun. 2005;337:677–684. doi: 10.1016/j.bbrc.2005.09.105. [DOI] [PubMed] [Google Scholar]
- 46.Hu Y, Fay AW, Lee CC, Ribbe MW. P-cluster maturation on nitrogenase MoFe protein. Proc Natl Acad Sci USA. 2007;104:10424–10429. doi: 10.1073/pnas.0704297104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jimenez-Vicente E, et al. Sequential and differential interaction of assembly factors during nitrogenase MoFe protein maturation. J Biol Chem. 2018;293:9812–9823. doi: 10.1074/jbc.RA118.002994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Engler C, Gruetzner R, Kandzia R, Marillonnet S. Golden gate shuffling: A one-pot DNA shuffling method based on type IIs restriction enzymes. PLoS One. 2009;4:e5553. doi: 10.1371/journal.pone.0005553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kapust RB, et al. Tobacco etch virus protease: Mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng. 2001;14:993–1000. doi: 10.1093/protein/14.12.993. [DOI] [PubMed] [Google Scholar]
- 50.Chen Y-J, et al. Characterization of 582 natural and synthetic terminators and quantification of their design constraints. Nat Methods. 2013;10:659–664. doi: 10.1038/nmeth.2515. [DOI] [PubMed] [Google Scholar]
- 51.Lodge J, Fear J, Busby S, Gunasekaran P, Kamini NR. Broad host range plasmids carrying the Escherichia coli lactose and galactose operons. FEMS Microbiol Lett. 1992;74:271–276. doi: 10.1016/0378-1097(92)90441-p. [DOI] [PubMed] [Google Scholar]
- 52.Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press; New York: 1972. Assay of β-galactosidase. [Google Scholar]
- 53.Cannon FC, Dixon RA, Postgate JR. Derivation and properties of F-prime factors in Escherichia coli carrying nitrogen fixation genes from Klebsiella pneumoniae. J Gen Microbiol. 1976;93:111–125. doi: 10.1099/00221287-93-1-111. [DOI] [PubMed] [Google Scholar]
- 54.Montoya JP, Voss M, Kahler P, Capone DG. A simple, high-precision, high-sensitivity tracer assay for N(inf2) fixation. Appl Environ Microbiol. 1996;62:986–993. doi: 10.1128/aem.62.3.986-993.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brown SD, Jun S. Complete genome sequence of Escherichia coli NCM3722. Genome Announc. 2015;3:e00879-15. doi: 10.1128/genomeA.00879-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wiśniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 57.Kirkpatrick DS, Gerber SA, Gygi SP. The absolute quantification strategy: A general procedure for the quantification of proteins and post-translational modifications. Methods. 2005;35:265–273. doi: 10.1016/j.ymeth.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 58.MacLean B, et al. Skyline: An open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.