Abstract
The degree of intrinsic and interpatient phenotypic heterogeneity and its role in tumour evolution is poorly understood. Phenotypic drifts can be transmitted via inheritable transcriptional programs. Cell-type specific transcription is maintained through the activation of epigenetically-defined regulatory regions including promoters and enhancers. Here we annotated the epigenome of 47 primary and metastatic oestrogen-receptor (ERα)-positive breast cancer clinical specimens and inferred phenotypic heterogeneity from the regulatory landscape, identifying key regulatory elements commonly shared across patients. Shared regions contain a unique set of regulatory information including the motif for the transcription factor YY1. We identify YY1 as a critical determinant of ERα transcriptional activity promoting tumour growth in most luminal patients. YY1 also contributes to the expression of genes mediating resistance to endocrine treatment. Finally, we used H3K27ac levels at active enhancer elements as a surrogate of intra-tumour phenotypic heterogeneity to track the expansion and contraction of phenotypic subpopulations throughout breast cancer progression. By tracking the clonality of SLC9A3R1-positive cells, a bona fide YY1-ERα-regulated gene, we show that endocrine therapies select for phenotypic clones underrepresented at diagnosis. Collectively, our data show that epigenetic mechanisms significantly contribute to phenotypic heterogeneity and evolution in systemically treated breast cancer patients.
Introduction
Breast cancer (BC) is the most common cancer type and the second most frequent cause of cancer related death in women1. 70% of all BC cases contain variable amounts of ERα-positive cells. ERα is central to BC pathogenesis and serves as the target of endocrine therapies (ET) 2. ERα-positive BC is subdivided into ‘intrinsic’ subtypes (luminal A and luminal B3) characterized by distinct prognosis, highlighting functional heterogeneity. Recent analyses demonstrate that inter-patient heterogeneity is more pervasive (reflected by histological 4, genetic architecture 5 and transcriptional differences 6) ultimately influencing long-term response to endocrine treatment7. Indeed, 30-40% of ERα BC patients relapse during or after completion of adjuvant endocrine therapies. At the time of relapse ET resistance is commonplace, partly achieved via treatment-specific genetic evolutionary trajectories8. Yet, recent studies have shown that driver coding-mutations do not significantly change between primary and metastatic luminal breast cancer, with the notable exception of ESR1 mutations9, suggesting that alternative non-genetic mechanisms might contribute to BC progression and drug-resistance. Parallel to genetic evolution, phenotypic/functional changes driven by epigenetic mechanisms can also contribute to breast cancer progression and ET resistance in cell lines10. Epigenetic modifications of histone proteins have been successfully used to map regulatory regions and to annotate the non-coding DNA11,12. Acetylation of lysine 27 on histone 3 (H3K27ac) is strongly associated with promoters and enhancers of transcriptionally active genes 13–15. Increasing evidence suggests that epigenetic information can actively transfer gene transcription states across cell division 16–19. Epigenetic modifications also modulate ERα binding to enhancers by interacting with ERα-associated pioneer factors 20,21. Nevertheless, little is known about the epigenome of BC patients, its influence on intra-tumour phenotypic heterogeneity and its role in breast cancer progression. Here we show the results of the first systematic investigation of the epigenetic landscape of ERα-positive primary and metastatic BC from 47 individuals. Using H3K27ac-ChIP-seq and ad-hoc bioinformatics analyses, we have characterized inter- and intra-patient epigenetic heterogeneity and identified YY1 as a novel key player in ERα-positive BC. Finally, we demonstrate that epigenetic mapping can efficiently estimate phenotypic heterogeneity changes throughout BC progression.
Results
Mapping of regulatory regions in primary and metastatic ERα positive breast cancer
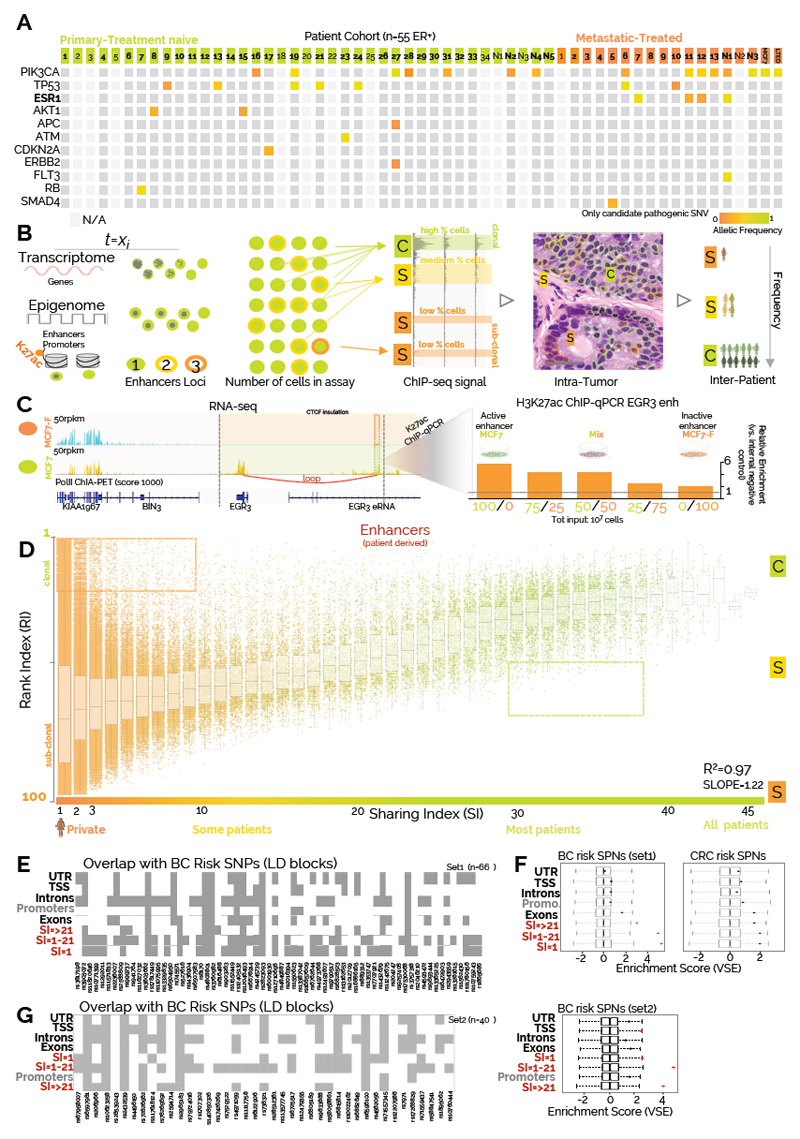
We profiled fifty-five ERα-positive BC samples with H3K27ac ChIP-seq to build a comprehensive compendium of clinically relevant active regulatory regions (Fig. 1A, primary n=39, and metastatic n=16) (Fig. 1A, Supplementary Table 1-2, Supplementary Data 1). H3K27ac-enriched regions were classified into 23,976 proximal-promoters and 326,719 enhancers. 80% of promoters were identified by the profiling of 4 patients, while nearly 40 are needed to reach the same coverage for enhancers, reflecting the 10:1 ratio between captured-enhancers and promoters (Supplementary Figure 1C). These data are in agreement with enhancers being the main determinants of cell-type specific transcriptional differences 13,14,22,23. To gain insights on the penetrance of each regulatory region, we developed a Sharing Index (SI, Supplementary Computational Methods) by annotating all enhancers and promoters in function of the number of patients sharing the H3K27ac signal at each specific location (Supplementary Figure S1D). This analysis shows that a vast proportion of enhancers is patient-specific (SI=1) while active promoters typically display higher SI (Supplementary Figure 1D). Collectively, these data demonstrate that enhancers account for the majority of potential epigenetic heterogeneity in ERα-positive BC.
Figure 1. Assessment of inter- and intra-tumor epigenetic heterogeneity.
A) Mutational analysis for common cancer driver genes in the patient cohort selected for the study B) Main hypothesis of the study. RNA is ultimately an analog signal in which each individual cell, at any given time, can contribute a stochastic amount of RNA while transcriptional data from bulk tissue represent the average over million cells. For chromatin data, at any given time (t=Xi) each cell can only contribute a deterministic value to the bulk signal, generally from two alleles. Therefore, the relative strength of ChIP-seq data is dependent on the number of cells carrying epigenetic signal at discrete loci. C, and S represent strong, and medium/weak signal, respectively (Scale bars, 50 μm). Clonal regulatory regions are commonly shared by BC patients while weak enhancers are more patient specific C) EGR3 mRNA is expressed in MCF7 but not derivative MCF7-F cells. eRNA and Pol-II ChIA-PET show enhancer activity in MC7 but not MCF7-F. CTCF insulated perimeter is shown in yellow. Predicted looping from ChIA-PET is shown in red. The observed ChIP-qPCR signal for H3K27ac at EGR3 enhancers increase with increasing number of MCF7 cells mixed in the sample. Similar results were obtained from three independent experiments D) Linear regression shows that clonal enhancers are commonly shared between breast cancer patients. Of note, a small but discrete proportion of promoters/enhancers escape this general trend having extremely low RI despite being patient-specific or higher RI while being shared (dotted-areas). Y axis=Ranking Index, X axis=Sharing index. Sharing index indicate the number of patients sharing the regulatory region. Each dot represents the median RI (all patients) for each single enhancer. The boxplots represent the median RI value and interquartile ranges for regulatory regions with the same SI E) Overlap between BC risk variants and annotated DNA elements F) Variant Set Enrichment analysis indicates that BC-specific but not CRC-specific GWAS risk variants occur more frequently than expected within the enhancers elements identified in our study G) Overlap with annotated DNA elements and Variant Set Enrichment analysis for the most recent independent set of BC risk variants.
Assessment of phenotypic heterogeneity by enhancer ranking
Genetic heterogeneity is a hallmark of most solid tumours 24 but its impact on phenotypic heterogeneity is characteristically hard to resolve. In agreement, despite extensive inter- and intra-tumoral genetic heterogeneity 25, the majority of ERα-positive patients benefit from systemic ET7. Furthermore, de novo metastatic patients initially respond well to ET, suggesting that genetic heterogeneity on its own cannot explain treatment resistance/response. Of note, phenotypic hierarchies can override genetic hierarchies in brain cancers 26,27, suggesting that inheritable epigenetic programs might contribute to phenotypic heterogeneity and treatment outcome. Phenotypic heterogeneity in breast has been known for decades. For example, immunohistochemistry (IHC) assessment of the proportion of ERα-positive cell in single biopsies varies on a continuum from less than 1% to nearly 100%28. However, IHC has been used to test only few targets whereas deconvolution from bulk transcriptional data is technically unfeasible (Fig 1B). For instance, cells with focal gene amplification have higher bulk gene expression but individual cells contribute stochastic discrete amounts as shown by single-molecule single-cell RNA FISH8. Conversely, recent evidence show that the signal captured by one-way reaction chromatin assays such as ATAC-seq appears to be linearly proportional to the cells contributing to it29. Histone modifications can also be thought of as digital information with each single nucleosome being ON (K27ac) or OFF at any given time (Fig. 1B). Notably, even within genetically clonal cell lines, the H3K27ac signal varies considerably between different regulatory regions. Regulatory regions labelled as super enhancers, for example, have 10-100-times more H3K27ac signal than typical enhancers14. What accounts for the variation in signal is not known, but one possibility is that heterogeneity within the cell population (either clonal or sub-clonal) contribute to signal intensity. While other factors might partially contribute to variation in the signal (local antibody affinity, histone dynamic, cell cycle, sonication efficiency, di-nucleotide content, mappability and copy number aberrations, see Supplementary Computational Methods and Supplementary Figure 2-4), we propose that ChIP-seq signal is robustly positively correlated with the number of contributing cells via a logistic relationship. Super-enhancers might represent regulatory regions active across most cells within a population at any given time (clonal, C-peaks), while “typical” enhancers with lower H3K27ac signal may represent sub-clones (S peaks, Fig. 1B). This interpretation is conceptually similar to using variant allele frequencies (VAF) to infer genetic heterogeneity.
Phenotypical heterogeneity might be the consequence of heterogeneous cell populations (i.e. normal-cancer) or actual cancer-specific epigenetic subclones. As our ChIP-seq data are derived from high tumor burden samples, we hypothesized that H3K27ac signal could allow for a qualitative assessment of phenotypic heterogeneity (Fig. 1B). To test the relationship between clonality and ChIP signal we performed spike-in experiments in which known numbers of cells with well-characterized enhancer activity (MCF7:ON, MCF7-F:OFF) and similar genetic background10 were admixed in incremental proportions prior to H3K27ac ChIP-qPCR. The data shows that H3K27ac enrichment is positively correlated to the number of cells in the absence of genomic differences (Fig. 1C). These findings are corroborated by an independent analysis using a different antibody (ERα) (Supplementary Figure 5). As the signal between different patients is not directly comparable, we quantile-normalized the data assigning to each H3K27ac signal a Rank Index (RI:1-100 strongest to weakest, see Supplementary computational methods and Supplementary Figure 6A). The signal from low RI (C peaks) is then associated with clonal regulatory regions active in almost all cells. Conversely, high RI (S peaks) mark more heterogeneous/sub-clonal enhancer activity. By investigating the relationship between RI and SI (Supplementary computational methods) we found extremely robust correlation between these two parameters (Fig.1D and Supplementary Figure 6B), suggesting that clonal regulatory regions are more common between patients (low RI/high SI) while sub-clonal regulatory elements are more patient-specific (high RI/low SI). For follow up analysis we then split enhancer elements into two main subgroups (SI<21 and SI ≥21) on the hypothesis that SI≥21 might more strongly contribute to the population phenotype.
Enhancers are associated with breast cancer risk-SNP and control gene transcription
Previous analyses from ERα-BC cell lines have shown that genetic predisposition to breast cancer might occur through SNPs that modulate transcription factors binding at enhancers (FOXA1 and ERα30). We then tested the relationship between regulatory regions captured in patients and DNA risk variants specifically associated with BC through GWAS30–32. Almost the totality of known BC risk variants from two independent datasets overlapped with our H3K27ac database. This overlap is highly significant specifically for enhancers but not for other annotations (Fig. 1F-G). Notably, this association is not replicated using colorectal cancer risk variants suggesting that these enhancers might play a specific role in BC development (Fig. 1F). Currently, our patient-derived enhancer dataset represents the most enriched annotation for GWAS variants in breast cancer. Next, we assessed the relationship between estimated enhancers clonality and transcriptional output. As the average expression is function of the number of cells engaged in active transcription and the number of RNA molecule within each cell33, assuming stochastic single-cell contribution, bulk mRNA levels should positively correlate with the number of transcribing cells. We could then test if clonal enhancers active in the majority of cells correlate with higher RNA levels. To do so we linked enhancers to their potential target genes using CTCF insulated boundaries 34. We then analysed three independent BC expression datasets 5,6,35 in function of RI/SI indexes. Our analyses support the hypothesis that genes associated with clonal enhancers have higher bulk RNA levels (Supplementary Figure 7A). We observed more modest associations when analysing the transcriptome from normal breast tissue (Supplementary Figure 7A, small insets) suggesting that our analysis has identified a subset of regulatory regions associated with malignant outgrowth. These data indicate that transcripts identified as dis-regulated in BC might reflect changes in the size of phenotypic subpopulations between the heterogeneous normal tissue and a cancer population dominated by epithelial features. Collectively, our data show that enhancer activity strongly tracks transcriptional changes in breast cancer patients.
Imputed transcription factors landscape of ERα breast cancer patients
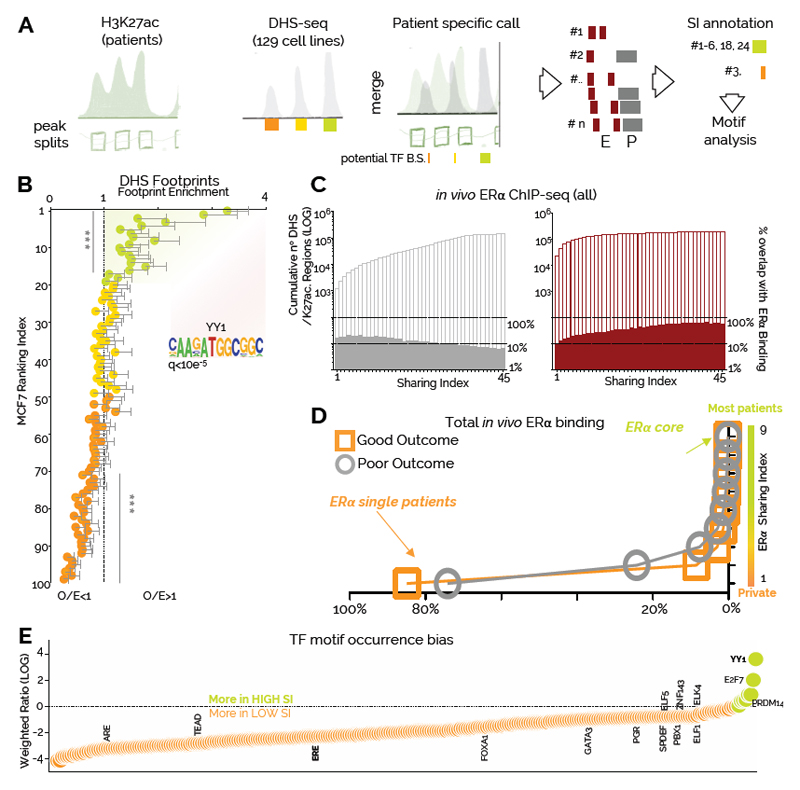
Enhancers store regulatory information in the form of transcription factors (TFs) binding motifs36. The vast majority of TFs require accessible chromatin in order to bind their cognate DNA sequences 37. To extrapolate the TFs landscape from our data we integrated the DNaseI signal (DHS) from 129 cell lines with the inferred nucleosome patterns obtained from the H3K27ac signal (Fig. 2A, Supplementary Computational Methods and Supplementary Figure 7B). As expected, this analysis could identify well-known BC-TFs according to their promoter–enhancer bias (Supplementary Figure 7C). Applying TF motif analysis to regulatory regions defined by the same SI followed by unsupervised clustering identified two major clades (Supplementary Figure 8). Remarkably, high and low SI clustered together suggesting that putative clonal and sub-clonal enhancers contain distinct regulatory information (Supplementary Figure 8). Functional TF binding is often associated with TF leaving a footprint within chromatin accessible regions 36,38. Analyzing footprints in function of RI in ERα-positive MCF7 breast cancer cells reveals that enhancers with RI<20 accumulate more footprints than expected (Fig. 2B). These data show that potentially clonal enhancers might recruit TFs with longer residence time 38. Unexpectedly, we find estrogen-response elements (ERE) motifs significantly enriched only in low SI sub-clonal enhancers (Figure 2E and Supplementary Figure 8). By integrating in vivo ERα binding 39 with our dataset we find that proportion of binding sites increases with the SI for enhancers (Fig. 2C) but not for promoters (Fig. 2C) consistently with ERα preferential binding at enhancer elements40. These data imply that shared enhancers have a strong propensity for ERα binding despite being generally under-represented in EREs. Interestingly, while the bulk of ERα binding events appear to be patient-specific (ERα SI=1), 0.003% of ERα are shared across most patients primary and metastatic patients (484 core-ERα)39 (Fig. 2D). Together, these data support transcription factor imaging data that indicate that only a small fraction of ERα binding events with longer-residency time is functional38. We therefore conclude that the largest portion of ERα binding identified in patients occurs at patient specific, sub-clonal enhancers and might reflect transient ERα-DNA interactions occurring while the receptor scans the genome38. The discrepancy between the small number of highly shared ERα core binding and the observation of ERE-poor clonal enhancers led us to hypothesize that other TFs might collaborate with ERα to increase its transcriptional efficiency at clonal enhancers. Examining TF-motifs bias toward high SI enhancers we identified YY1 as the top candidate (Fig. 2E). YY1 is also the top ranked motif within the footprints of clonal MCF7 enhancers (Fig. 2B). YY1 has been recently implied in de novo formation of enhancer promoter looping during neural development 41,42 and MYC-like ability to potentiate gene expression43 indicating a potential role in modulating the enhancer landscape in ERα-positive BC.
Figure 2. Clonal and sub-clonal regulatory regions contain distinct regulatory information.
A) Bioinformatic framework of the analyses. H3K27ac calls were split to identify approximate nucleosome-level enrichment (sub-peaks). Sub-peaks data were integrated with ENCODE-derived DHS-seq calls to identify potential sites of TF binding. Individual imputed DHS regions were assigned SI values based on the number of patient sharing the region B) Clonal enhancers in MCF7 cells (RI<20) are characterized by a higher number of TF footprints, while sub-clonal enhancers (RI>70) have less footprint than expected. Each bin contains 34 nucleosome free regions. The number of footprint (from Wellington) was normalized for enhancer size. Observed/Expected values were calculated by dividing the number of normalised footprints in each enhancer by the overall average (2.7 footprints per enhancer). Each value and error bar represent the average footprint and 95% CI. Asterisks represent a pValue of <0.001 in a Wilcoxon Signed Rank Test C) Overlap of imputed DHS regions with in vivo derived ERα binding sites. The left Y axis indicates cumulative DHS regions. The right Y axes indicate the percentage of overlap based on total DHS in each SI bin D) Distribution plot of in vivo derived ERα binding sites versus the number of patients in which they were observed E) YY1 motif is enriched in putatively clonal enhancers in luminal breast cancer patients. TF motifs within imputed DHS are plotted based on their bias toward highly shared enhancers (green) or more private enhancers (orange).
YY1 enhancer activity marks a dominant phenotypic clone in breast cancer
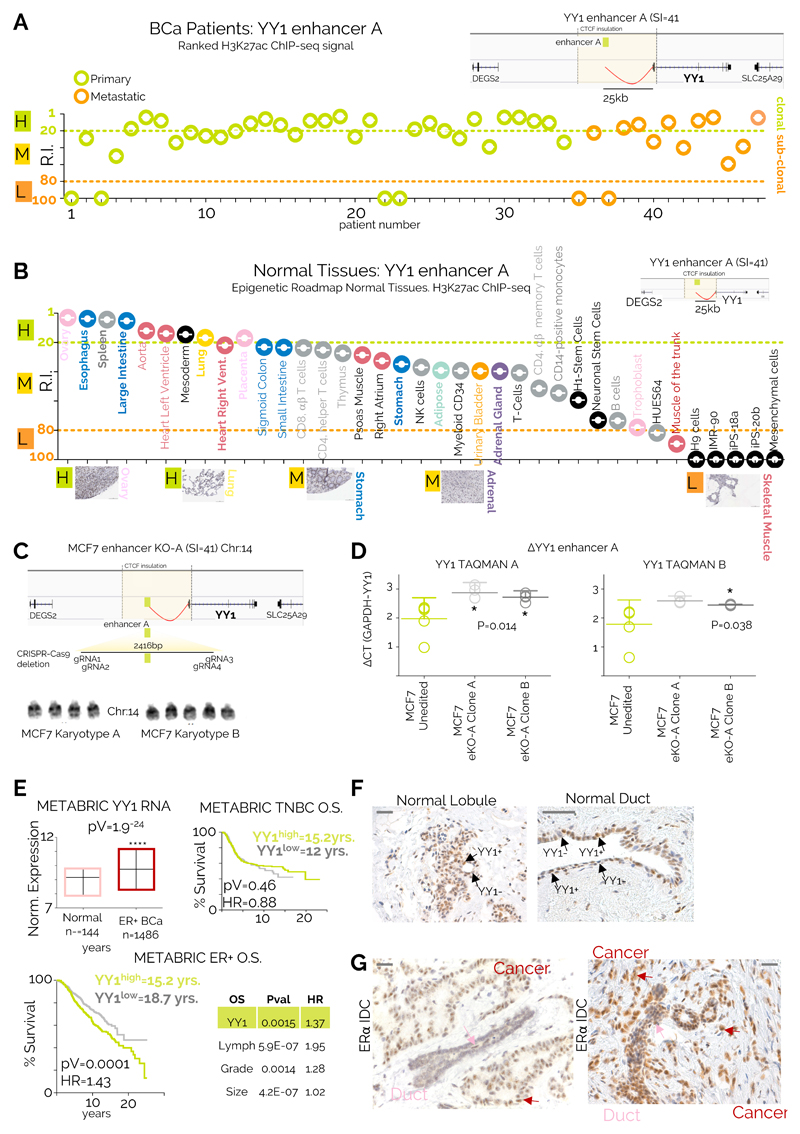
YY1 is a ubiquitously expressed TF (Supplementary Figure 9A-B) that can act as an activator or repressor by binding DNA, RNA and chromatin modifiers44,45. Interestingly, YY1 drosophila homolog PhoRC is involved in epigenetic memory by recruiting of Polycomb repressor complex to sequence specific regions46, but YY1’s role in mammals is only partially understood. Collectively, our analyses predict that most BC cells should be YY1-positive, consequently the enhancer driving YY1 should be clonal. To test this, we identified three bona fide enhancers looping at YY1 promoter using 3D chromatin data 47 (Supplementary Fig. 10A). Enhancer A (SI=41) directly interacts with Enhancer B-C, suggesting a multi-enhancers interaction with YY1 promoter. Enhancer A consistently ranks among the most clonal enhancer in our dataset (Fig. 3A). By comparison, YY1 enhancer A activity is more variable in most normal tissues profiled by H3K27ac within the Epigenome Roadmap consortium11, implying that some tissues might harbour YY1-subclonal subpopulations (Fig 3B). Consistent with these predictions, immunocytochemistry (IHC) meta-analysis (Fig 3B) show sub-clonal YY1-positive populations in tissue with high RI (Fig 3B and Supplementary Figure 10B). To directly test the regulatory potential of enhancer A, we used CRIPR-Cas9 mediated deletion to generate enhancer-KO ERα positive MCF7 cells (eKO cells, Fig. 3C). Deletion of 2/5 alleles directly reduce YY1 mRNA level by 30-35% (Fig. 3D). Collectively, these data show that enhancer ranking can capture qualitative changes in intra-tumoral heterogeneity, and that YY1-enhancer activity marks a dominant phenotypic clone in ERα-positive BC.
Figure 3. YY1 identify a dominant phenotypic clone in ERα BC.
A) RIs for the YY1 enhancer within all the individual patients included in the current study YY1 enhancer location with its 3D interactions are shown in the top right inset B) YY1 enhancer ranking analysis of available Epigenome Roadmap H3K27ac datasets. Tissues are displayed from the strongest to the weakest YY1 enhancer activity (based on RI). Representative IHC analysis of normal tissues stained with a YY1 antibody are shown C) eKO cell lines were generated by deleting a 2.4kb containing YY1-A enhancers in MCF7 cells. Actual karyotyping is shown in the bottom panel was performed on 10 individual cells D) YY1 expression in control and eKO cell lines was measured using RT-qPCR. Lines and error bars represent average and 95% CI of five independent experiments. Significance was calculated with a one-way ANOVA followed by Tukey’s test E) Top left: YY1 expression in ERα-positive breast cancer compared to normal breast tissue. Median, lowest and highest values are reported. Top right: YY1 prognostic value in triple negative breast cancers. Bottom left: YY1 prognostic value in luminal breast cancers. Confidence interval (1.19-1.76). Bottom right: multivariate correction for the luminal breast cancer dataset is shown. Analyses included 1476 ERα-positive and 432 ERα-negative patients. Comparison of survival curves was performed using a Log-rank (Mantel-Cox) test. F) IHC analysis of normal breast tissues highlights YY1 functional subclones in normal breast. Similar results were observed in 10 independent clinical specimens from independent individuals G) IHC analysis of ERα positive invasive ductal carcinomas identify YY1 positive clones as the dominant clonal population (Scale bars, 50 μm).
Tumour tissues generally have significantly higher expression level for YY1 as compared to normal tissues (Supplementary Figure 11A). This observation was replicated in an independent BC dataset (Fig. 3E and Supplementary Figure 11B). These data suggest that BC lesions might contain a larger fraction of YY1-positive as compared to normal breast tissue (Fig. 3B). Meta-analysis of the METABRIC5 datasets shows that ERα-positive patients with higher bulk YY1 mRNA at diagnosis have significantly worse outcome, while this does not hold true for ERα-negative (Fig. 3E). The prognostic value for YY1 in ERα-positive patients is maintained when adjusting for other clinical features (Fig. 3E). To test if increased YY1 mRNA levels could be driven by an expansion of YY1-positive cells from a more heterogeneous population we stained normal breast tissue sections with IHC. Our data show that lobules and ducts contain distinct YY1 positive sub-clonal populations while the nearby tumour tissue is overwhelmingly YY1 positive, (Fig. 3F-G). Interestingly, YY1 staining was absent or limited in several triple negative breast cancer patients (Supplementary Figure 11C).
YY1 modulates functional ERα binding at enhancer regions
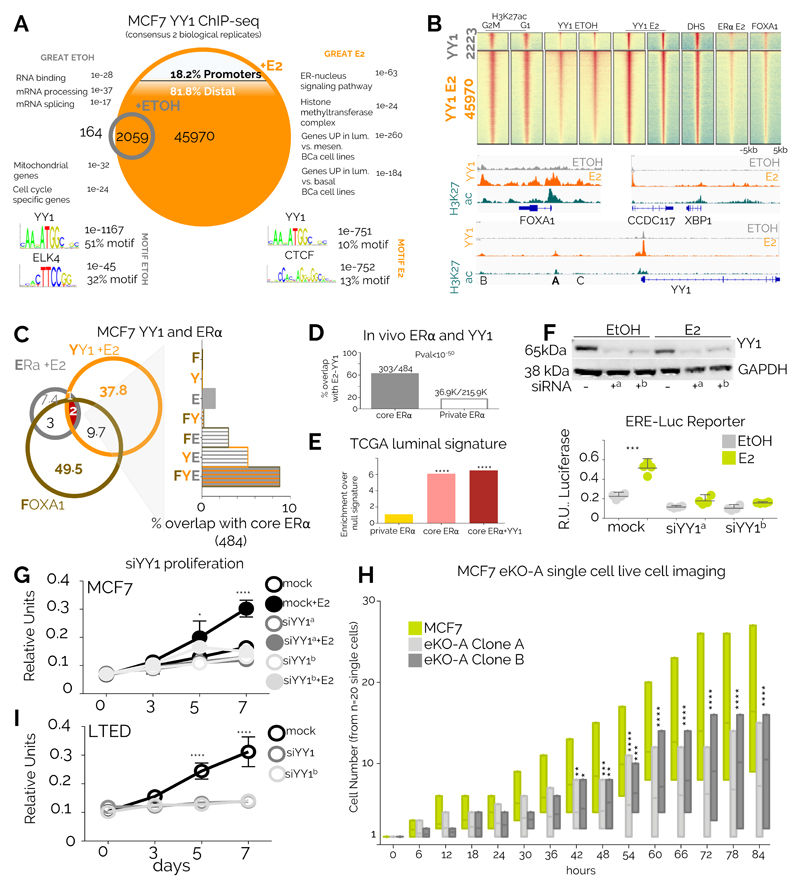
To gain mechanistic insights on the role of YY1 we performed ChIP-Seq in estrogen-deprived and estrogen-stimulated luminal BC MCF7 cells. In absence of estrogen, YY1 occupies a small set of enhancers and promoters near housekeeping genes (Fig. 4A). Strikingly, estrogen stimulation induced a 23-fold expansion of the YY1 binding repertoire, mostly at enhancer regions associated with ERα-BC signatures (Fig. 4A). Orthogonal analyses show that induced YY1 binding involves almost all MCF7 active regulatory regions and is strongly associated with H3K27ac marks (Fig. 4B). Conversely, YY1 binding is absent from silenced genes (Supplementary Figure 12A), demonstrating that YY1 does not associate with PRC2 mediated repression in BC cancer cells. Our in vivo analyses suggest that YY1-motif enriched enhancers are generally deprived of EREs (Fig 2B). In agreement, our in vitro data show only marginal overlap between YY1 and ERα or its pioneer factor FOXA1 (Fig. 4B-C). Nevertheless, YY1, ERα and FOXA1 co-localization becomes significant at core-ERα loci in MCF7 cells (Fig. 4C). Similar observations were made by comparing YY1 overlap with patient-derived ERα binding site analyses (Fig. 4D). In addition, we find that genes defining the luminal subtype in TCGA patients are significantly associated with YY1-ERα core binding but not patient-unique ERα (Fig. 4E). Overall, these data further suggest that YY1 might contribute to ERα binding transcriptional output at a small subset of enhancers captured in most tumour cells and most patients. We further show that YY1 depletion is sufficient to abrogate transcription from an ERα-driven reporter (Fig. 4F). YY1 depletion also abrogates cell proliferation in response to estrogen stimulation in MCF7 (Fig. 4G) suggesting that YY1 is a direct driver of the clonal proliferation observed in BC (Fig. 3D-E). These observations were replicated in independent luminal BC cell models (ZR75 and T47D, Supplementary Figure 12B-C). YY1 depletion leads to significant downregulation of core-ERα target genes in luminal BC cell line models (Supplementary Figure 12D). Finally, monitoring cell proliferation at the single cell level using eKO cell lines, we show that deletion of YY1 enhancer A is sufficient to reduce MCF7 growth in estrogen-supplemented conditions (Fig. 4H). Collectively these data identify YY1 as a novel essential transcription factor significantly contributing to ERα regulatory network transcriptional activity.
Figure 4. YY1 marks critical enhancers in breast cancer cells.
A) ChIP-seq data from ERα-positive MCF7 for YY1 in quiescent or 17ß -estradiol (E2) stimulated cells B) Heatmaps showing global enrichment profiles of several chromatin markers associated with active regulatory regions in MCF7 cells C) Overlap between ERα, YY1 and FOXA1 in MCF7 cells. The right panel shows the potential overlap with in vivo- derived core ERα binding sites D) ERα core binding sites are strongly enriched for YY1 binding in MCF7 cells while patient-specific ERα bindings are generally YY1-free. Proportion were compared using Fisher’s Exact test. E) Genes used to classify luminal breast cancer patients are strongly enriched for ERα-YY1 binding sites. Asterisks represent p<10-5 in a Fisher’s Exact test vs. private ERα F) YY1 depletion leads to transcriptional shut-down of an ERE-driven luciferase reporter. Blot has been cropped, full blot is shown in and Supplementary Figure 12B. Bars and error bars represent the average and 95% CI of 4 independent experiments. Asterisks represent significance at P<0.001 after ANOVA with Dunnet’s correction. G) Silencing YY1 blocks estrogen-induced growth in MCF7 cells. Proliferation assays were conducted in three independent biological replicates. Symbol and error bars indicate average and 95% confidence intervals. Significance was calculated using a two-way ANOVA with Bonferroni’s correction H) YY1-A enhancer deletion directly leads to reduced proliferation in MCF7 cells. Bars represent highest, lowest and median count for cell number from individual colonies (n=20) monitored individually using single-cell live imaging. Significance was calculated using a two-way ANOVA with Bonferroni’s correction Asterisks represent significant differences after ANOVA followed by Dunnet’s test *<0.05, **<0.01, ***<0.001, ****<0.0001. I) Silencing YY1 blocks growth in LTED cells. Proliferation assays were conducted in three independent biological replicates. Symbol and error bars indicate average and 95% confidence intervals. Significance was calculated using a two-way ANOVA with Bonferroni’s correction
YY1 contributes to endocrine resistance in luminal breast cancer
YY1-positive cells appear to dominate both primary and metastatic lesions in luminal patients suggesting that might remain important even after ET (Fig 3A). YY1 depletion is indeed sufficient to abrogate proliferation in LTED cells, an MCF7-derivative mimicking AI-treated breast cancer cells10(Fig. 4I). Interestingly, LTED cells have an expanded repertoire of ERα binding compared to MCF7, fuelled by endogenous ligands8,10. Nonetheless, YY1 and ERα overlap remains restricted to a minority of sites (Supplementary Figure 13A). Intriguingly, the set of enhancers engaged by ERα and YY1 in LTED cells is radically different compared to MCF7, with the majority of ERα-YY1 being specific to each cell type (Supplementary Figure 13A). ERα-YY1 bound enhancers in LTED strongly associates with the transcription of genes involved with acquired endocrine therapy, suggesting that during epigenetic reprogramming, YY1 might stabilize ERα to LTED specific enhancers (Supplementary Figure 13B). Previous studies have shown that the transcription of a small set of estrogen-activated genes is not antagonized by current endocrine therapies 48. Examining the regulatory landscape near these genes we found an ever-increasing association with ERα-YY1 bound enhancers, especially with core ERα-YY1 (Supplementary Figure 13C). Collectively, these data strongly support the role of YY1 in ERα BC growth and progression.
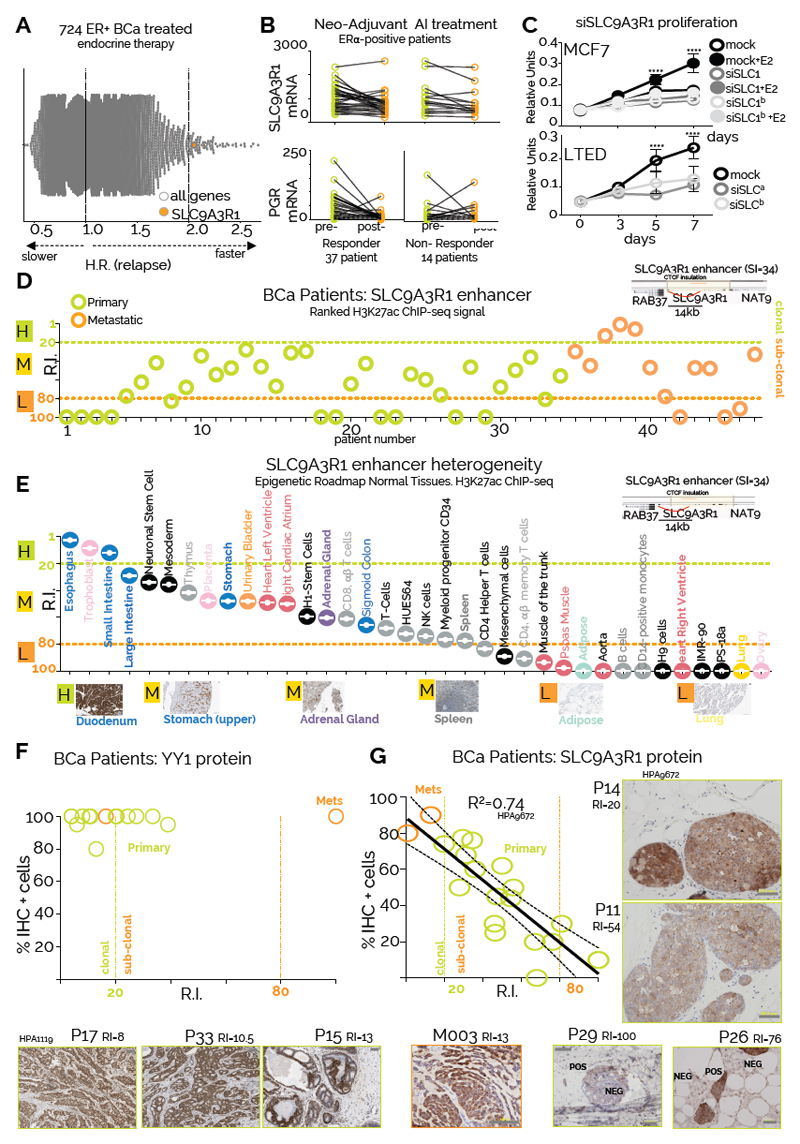
YY1-ERα promote SCL9A3R1 expression despite endocrine treatment
By ranking the set of endocrine unresponsive genes bound by YY1-ERα for gene-specific prognostic power calculated in patient treated with endocrine therapies 35 we identified SLC9A3R1 as a potential driver of endocrine therapy resistance (Fig. 5A). SLC9A3R1 (NHERF1/EBP50) encodes a Na/H exchanger regulatory cofactor with a potential role in metastatic invasion49. High expression of SLC9A3R1 independently correlates with poor survival in additional ERα-BC datasets (Supplementary Figure 14A). Despite being an ERα target, SLC9A3R1 expression is not suppressed by Tamoxifen in MCF7 cells 48. Additionally, SLC9A3R1 remains transcriptionally active in most endocrine therapy resistant BC cell lines that retain ERα expression (Supplementary Figure 14B-E) demonstrating that ERα activity remains critical for SLC9A3R1 expression. In vivo SLC9A3R1 expression is also unaffected by neo-adjuvant AI treatment (Fig. 5B). Notably, bulk RNA-seq data from a panel of cancer cell lines demonstrate that ERα-positive BC cells have the highest levels of SLC9A3R1 mRNA (Supplementary Figure 15A). More importantly, TCGA RNA-seq analysis shows that SLC9A3R1 expression is higher specifically in ERα-positive BC patients compared to normal tissue or other subtypes (Supplementary Figure 15B). Chromatin analyses of MCF7 and LTED cells identify 3 potential enhancers within the insulated SLC9A3R1 locus (E1-E3). Interestingly, E1-E2 enhancers loop to SLC9A3R1 promoter and are characterized by a high SI, YY1/core-ERα binding sites (Supplementary Figure 15C). In vivo transcriptional analysis demonstrates that SLC9A3R1 is the only gene near the E1-E2 enhancers that shows a significant increase in bulk-RNA level when comparing normal breast tissue with ERα–positive BC (Supplementary Figure 15D). Remarkably, enhancer-activity appears to be resistant to endocrine therapies (Supplementary Figure 15C). Furthermore, SLC9A3R1 expression is dependent on YY1 (Supplementary Figure 16A), demonstrating that both ERα and YY1 are essential for full enhancer activity. Collectively, these data demonstrate that SLC9A3R1 expression is driven by a breast cancer specific YY1-ERα bound enhancer. Silencing SLC9A3R1 is sufficient to reduce oestrogen-induced growth in ERα-positive cells (Fig 5C). Intriguingly, SLC9A3R1 is not essential for a second ERα-positive model (T47D) but appears to be a critical gene for both AI-resistant cells models (Fig. 5C and Supplementary Figure S16B). Overall, these data identify SLC9A3R1 as a novel player involved in ET resistance which function remains to be elucidated.
Figure 5. Epigenomic mapping predicts the size of phenotypic clones in patients.
A) Global Kaplan-Meier analysis summarize univariate analysis for 22278 genes included in the Affymetrix microarray platform. Hazard Ratios are plotted in the X axis B) SLC9A3R1 RNA levels pre- and post- short-term aromatase inhibitor treatment in responder and non-responder patients61. Oestrogen-dependent expression of progesterone receptor mRNA is shown as comparison C) Silencing SLC9A3R1 leads to proliferation arrest in response to estrogen stimulation in MCF7 and estrogen independent growth in LTED cells. Proliferation assays were conducted in biological triplicate. Symbol and error bars indicate average and 95% confidence intervals. Asterisks represent significance at P<0.05, 0.01, 0.001 and 0.0001 after two-way ANOVA with Bonferroni’s correction D) RIs for the SLC9A3R1 enhancer within all the individual patients included in the current study. SLC9A3R1 enhancer location and its 3D interactions are shown in the top right inset E) SLC9A3R1 enhancer ranking analysis of available Epigenome Roadmap H3K27ac datasets. Tissues are displayed from the strongest to the weakest SLC9A3R1 enhancer activity (based on RI). Representative IHC analysis of normal tissues stained with a SLC9A3R1 antibody are shown (Scale bars, 50 μm). F-G) YY1 and SLC9A3R1 IHC analysis of BC patients profiled using H3K27ac ChIP-seq. Predicted activity (RI) of YY and SLC9A3R1 enhancers is shown on the X axis. The number of cells positively stained for YY1 and SLC9A3R1 protein is indicated on the Y axis. Representative images are shown in the inbox. We stained one slide for each patient. Linear regression R square, confidence intervals and representative staining are also shown.
Mapping phenotypic heterogeneity using YY1 and SLC9AR1 enhancer activity
SLC9A3R1 enhancer activity (E1-2, SI=34, RI≥20) indicates that SLC9A3R1 marks sub-clonal populations in most primary patients (Fig 5D). Meta-analysis of SLC9A3R1 enhancer activity (RI) within the ENCODE H3K27ac datasets indicates that MCF7 are the only cancer cells contain a clonal SLC9A3R1 population (Supplementary Figure 16C). Of note, the size of the sub-clonal population correlates with total RNA content for the cells contained in both assays, suggesting that the decreasing bulk RNA signal is driven by a progressively smaller subpopulation (Supplementary Figure 16C). Similar analyses of YY1 enhancers indicate that cancer cell lines are prevalently clonal for YY1 expression (Supplementary Figure 16D) while both YY1 and SLC9A3R1 RIs in mammary epithelial cells predict for smaller sub-clonal populations. These observations fit extremely well with experimental data from IHC profiles from normal and malignant breast (Fig. 3D and Supplementary Figure 11C). Meta-analyses of Epigenome Roadmap predict for mainly SLC9A3R1-positive sub-clonal populations with the exception of gastro-intestinal tissues and these data fit well with RNA-seq measurement from independent cohorts (Fig. 5E and Supplementary Figure 17A). Analogously to YY1, meta-analysis of IHC data identifies decreasing SLC9A3R1-positive with increasing RI scores (Fig. 5E and Supplementary Figure 17B). To validate that RI index can estimate phenotypic clones, we retrospectively collected available biopsies for the BC patients profiled with H3K27ac ChIP-seq (n=19). IHC analysis of YY1 (Fig. 5F) shows that with the exception of one metastatic sample (M3), YY1 staining robustly correlate with RI, confirming large clonal YY1 positive populations in all examined tissues (Fig. 5F). In parallel, SLC9A3R1 enhancer activity correctly estimated the size of the sub-clonal subpopulations in individual patients (Fig 5G). Additional meta-analyses on Protein Atlas data support these findings by identifying YY1 clonal populations and SLC9A3R1 sub-clonal populations in most ERα BC samples (Supplementary Figure 18). Overall, these data show that enhancer activity can be used to qualitatively deconvolute heterogeneous populations into phenotypical subclones.
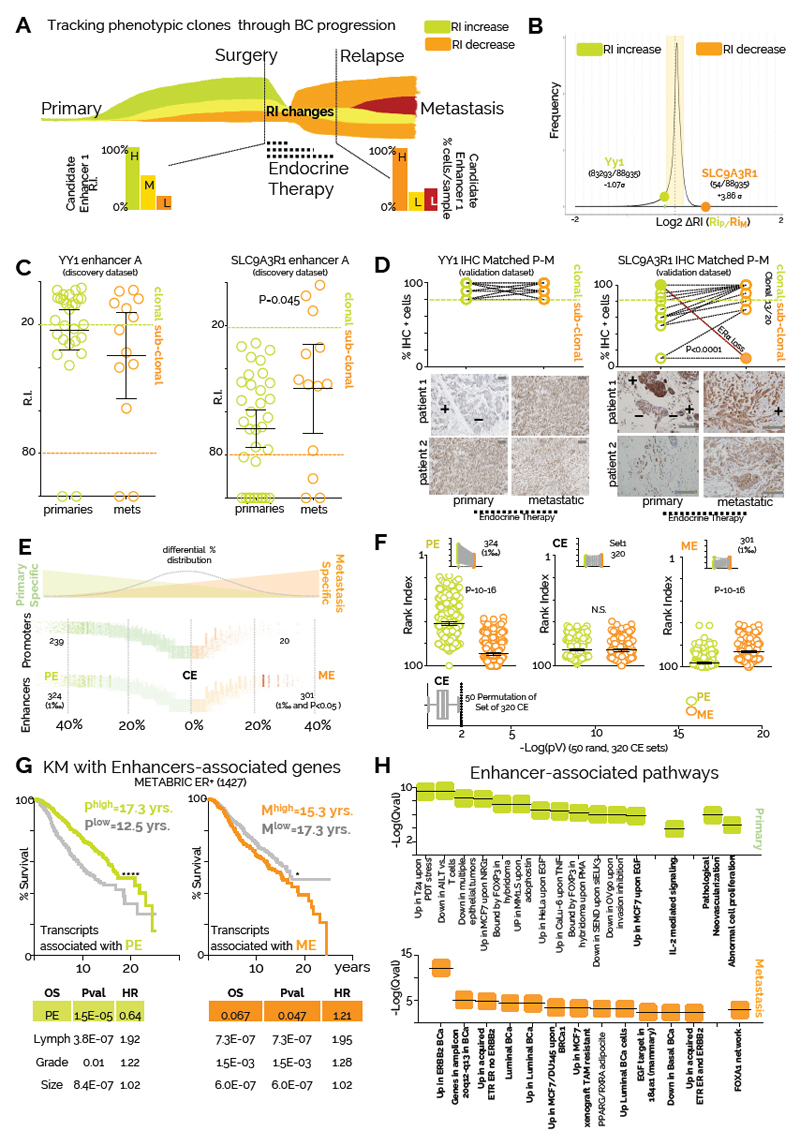
Phenotypic evolution during breast cancer progression is shaped by endocrine treatment
Tumor evolution studies have primarily focused on treatment naïve patients, taking advantage of multi-regional sampling to monitor changes in clonality50,51. Clonal tracking is dependent in part on passenger mutations, and the effect of therapy has been rarely accounted for8,52. More importantly, clonality has been traced using genetic variants, with the intrinsic limitation of correlating genetic changes to phenotypic ones. For example, genetic sub-clones might be phenotypically equivalent, while a recent study using barcoded glioblastoma cells shows that phenotypic clones might evolve independently from genetic clones26. The few studies that looked at driver coding mutations changes in BC show relatively similar mutational landscapes9 (Fig. 1A), suggesting a potential role for epigenetically-driven phenotypic evolution. We thus leveraged our ability to infer phenotypic clones trough enhancer activity to interrogate our patient’s dataset focusing on events occurring between treatment-naïve primaries and treatment-resistant metastatic BC (Fig. 6A). We hypothesized that phenotypic clonal evolution might be driven by a coordinated activation/selection of groups of enhancers during BC progression and this could be influenced by treatment. Our previous results suggest that YY1+ cells remain clonal during progression (Fig 3A). Conversely, we show that SLC9A3R1 expression is not antagonized by endocrine treatment suggesting that SLC9A3R1-positive clones could expand during progression. We then calculated changes in RI (∆RI) for all enhancers captured in at least three patients (SI>3, n=88,935) between primary and metastatic samples (Fig. 6B). SLC9A3R1 ranks amongst the enhancers with the strongest increase in predicted clonality going from primary to metastatic samples (Fig. 6B-C). Conversely, YY1 enhancer activity remains relatively unchanged (Fig. 6B-C). To substantiate these data, we mapped the size of YY1 and SLC9A3R1-positive phenotypic clones in an independent cohort of 20 primary tumour and metastasis-matched longitudinal biopsies. We found YY1-positive cells remain clonal in both settings, while SLC9A3R1-positive subclones significantly expand during metastatic progression(Fig. 6D). Interestingly, the only metastatic case in which we have observed a contraction of the SLC9A3R1+ clone also showed a concomitant loss of ERα and PR positivity, demonstrating that SLC9A3R1 remains an ERα-dependent target despite being ET insensitive in vivo (Fig. 6D). Overall, these data demonstrate that changes in enhancer ranking can estimate functional evolution during breast cancer progression.
Figure 6. Endocrine treatment shapes phenotypic evolution.
A) Theoretical framework of the analysis. The relative size of phenotypic clones can be tracked using enhancer ranking (RIs). Phenotypic clones can be positively or negatively selected during BC progression in response to endocrine therapies. B) Expanding or contracting phenotypic clones were defined based on the RI-ratio in primary and metastatic samples (RIP/RIM). Distribution of RI-ratio shows that YY1 enhancers RI does not change significantly during progression compared to other enhancers, while SLC9A3R1 RI ranks among the enhancers with stronger increase in activity during progression. Vertical bars represent 1σ (Standard Deviation) increments from the population median C) Scatterplot of YY1 and SLC9A3R1 enhancer ranking according to patient stage. Bars indicate mean and 95% confidence intervals. Asterisks represent significance at P<0.05 after students two-tail T-Test D) IHC staining for YY1 and SLC9A3R1 positive cells in an independent matched longitudinal cohort of 22 ERα breast cancer patients (Scale bars, 100 μm). All normal and primaries are treatment naïve. All metastatic have received endocrine therapies (Tamoxifen or Aromatase inhibitors). Statistical significance was calculated using a pair-wise, two-tail T-test. Representative images are also shown E) Enhancer and promoter stratification based on frequency of usage in primary and metastatic patients. Percentages were calculated for each regulatory region for each stage (primary and metastatic) and differential was then derived and plotted on the X-axis. All enhancers and promoters called in figure 1 were used. PE and ME were called by taking the top 1/1000 in the distribution that also satisfied a Fisher-exact test p<0.05. F) Dot-plot represent RI indexes for all PE (324) and ME (301) are plotted. As a control, RI for common enhancers (CE=320) were also plotted. Bottom plot: permutation was used to assess changes in RI in 50 randomly selected sets of 320 CE. Bow and whiskers represent median and 1-99 percentile for P-Value distribution. A Wilcoxon matched-pairs signed rank test was used to test for statistical significance G) Kaplan-Meier analysis using 1427 ERα-positive patients and averaged RNA expression of genes associated with PE or ME regulatory regions. Confidence interval for PE (0.39-0.61). Confidence interval for ME (1.1-1.67). Comparison of survival curves was performed using a Log-rank (Mantel-Cox) test. Genes were assigned considering CTCF insulated perimeters. Multivariate correction for the comparisons is also shown H) Pathway analysis for genes associated with PE or ME regulatory regions. Pathways were identified using GREAT and are listed in order of significance (symbols indicate qValue).
To gain insight on functional evolution, we systematically annotated all regulatory regions based on bias in detection between primary and metastatic patients (Fig 6E). As expected, the bulk of enhancers and promoters do not show bias toward primary and metastatic BC patients (common enhancers, CEs). However, we could identify two distinct sets of regulatory regions which activity is stronger in primary (primary enhancers, PEs) or metastatic (metastatic enhancers, MEs) patients (Fig. 6F). We next explored the potential causes and functional consequences driving these coordinated epigenetic changes by identifying the associated transcriptional targets of MEs and PEs 34. Strikingly, we find that PE-driven gene-transcription is associated with significantly better outcome while ME-associated gene-transcription in primary samples is associated with poor prognosis (Fig 6G). These data imply that primary samples containing larger subpopulations of phenotypic clones with metastatic features relapse earlier. PEs are associated with abnormal proliferation and vascularization, two key events in early tumorigenesis. Remarkably, ME are associated with genes promoting BC progression (FOXA139) or endocrine therapy resistance (Fig. 6H). Altogether, these data suggest that endocrine therapies play a central role in shaping phenotypic clonal evolution. Additional in-depth studies are needed to dissect the temporal events triggered during phenotypic clonal evolution. Phenotypic subclones could evolve by early coordinated activation and decommissioning of epigenetically defined regulatory regions (acquired), selection of the fittest pre-existent epigenomic landscape (de novo) or a combination of both.
Discussion
While genomic profiling of breast cancer patients has revealed extensive clonal heterogeneity and evolution24,53, it remains difficult to link genotype to actual phenotypes. Most RNA-based analyses, which may better reflect the phenotypic state of cancer cells, cannot inform on the existence of distinct subpopulations. Finally, molecular pathology can inform on the relative amount of protein abundance at the single-cell level but is laborious and not suitable for testing multiple targets simultaneously. In this work, we used epigenomic analyses to extrapolate phenotypic heterogeneity in solid tumour samples. Our analysis reveals that histone-based ChIP-seq signals, similarly to ATAC-seq29, generally correlates with the number of cells in a population carrying the specific epigenetic information. Our predictions using YY1 and SLC9A3R1 enhancer fit extremely well with experimental data derived from normal tissues or BC patients. The findings that clonal regulatory regions dominating the landscape of individual tumour samples are shared across many patients, parallel recent genomic evidences showing that truncal (high allele frequency) mutations are also the most common mutations within cancer cohorts.
Our work reveals several critical principles underlying phenotypic-functional heterogeneity and its role in breast cancer progression. First, by comparing samples from drug-resistant metastatic patients with drug-naïve primary samples, we uncovered a set of enhancers marking phenotypic clones that significantly expand during breast cancer progression. A set of enhancers expanding in metastatic samples point at progressive activation of FOXA1 and its network. It was recently reported that FOXA1 levels are increased in metastatic samples39. Our data predict that, similarly to SLC9A3R1, FOXA1 positivity increases as a consequence of the expansion of a phenotypic clone marked by an active FOXA1 enhancer. It is tempting to speculate that this paradigm might be valid for other genes. If correct it might signify that during cancer evolution, the proportion of cells activating transcription is more important than the absolute changes in transcription at the single-cell levels. Interestingly, a set of enhancers deactivated during progression involve IL-2 signalling (Fig. 6H). Reduction in IL-2 signalling was identified as a potential marker of relapse54. Whether the IL-2 signal source is the BC cells themselves or it is due to a small contamination of immune cells, needs to be defined. Equally, it will be important to measure real-time activation/selection of enhancers in appropriate systems to ultimately establish if phenotypic cancer evolution can be driven by Lamarckian events.
Additionally, our analysis has identified two novel drivers of luminal BC. Firstly, we identified YY1 as a key TF associated with clonal enhancers and promoters in BC patients. Our data strongly support the idea that YY1 acts as a global co-activator associating with the entire active epigenetic landscape in BC cells. Several lines of evidence indicate that YY1 might interact directly with modified nucleosomes, possibly through its partner INO8055. YY1 widespread association with clonal enhancer suggests it might play a role in epigenetic memory. Intriguingly, a positive screen for factors that improve induced pluripotent cells formation (iPS), identified YY1 as the top hit, further supporting its potential role as enhancer gatekeeper 56. More specifically to ERα BC, we hypothesize that YY1 plays a critical role to stabilize ERα binding at the transcriptionally productive core- ERα enhancers. Single-molecule imaging shows that estrogen activated ERα increases its residency time on the chromatin38 and recent evidence has shown that eRNA can trap YY1 on the chromatin 45. Altogether, these data raise the intriguing hypothesis that YY1 might contribute to increased ERα residency at clonal enhancers (Supplementary Figure 19). This could explain why some ERα occupancy is captured in most patients, as longer residency time would increase chances of being captured by ChIP-Seq39. Longer residency might also explain the increased transcriptional activity (Fig. 4D) and increased TF footprints (Fig. 2C) of these enhancers. Another possibility is that YY1 defines the set of ERα-bound enhancers with transcriptionally productive looping at target genes 41,42,57. Further studies will investigate these hypotheses. Future studies are also required to investigate the exact mechanisms through which SLC9A3R1 contribute to BC and efficient strategies to antagonize its transcription. We recently demonstrated that individual endocrine therapies can drive parallel genetic evolution in vivo 8 and epigenetic reprogramming in vitro10. Our data now strongly support the notion that therapeutic interventions also play an essential role driving specific epigenetic evolution during BC progression in the clinic.
Online Materials and Methods
Tumour tissue processing
Breast cancer sample for ChIP-seq were collected by Imperial Tissue Bank (project ethic approval R15021) and from Breast Cancer Now Tissue Bank (BCNTB- TR000053-MTA & TR000040). Breast cancer fresh frozen tissue samples each undergo aseptic macroscopic adipose tissue dissection. The dissected tumour tissue is sectioned into 2mm x 2mm fragments in a petri-dish placed over dry ice. Tumour fragments are then fixed using 1% formaldehyde solution for 10 minutes. Cold glycine (1M) is added to the formaldehyde-fixed tissue for 10 minutes. The tumour fragments are then pulverised using pestle and mortar and homogenised using liquid nitrogen. We used samples with high tumour burden to minimize the introduction of noise from non-tumour tissues (>70%, Supplementary Figure 1A). Wherever possible, we profiled patients for known cancer drivers using targeted enrichment sequencing (Fig. 1A and Supplementary Data 1). 85% of samples yielded satisfactory results (47/55, Supplementary Figure 1B and Table S2).
Cancer hotspots mutations
Chromatin immunoprecipitation (ChIP)
The ChIP protocol was conducted as described by Schmidt et al.58 with few modifications. In summary, following fixation, the tumour tissue undergoes chromatin extraction and sonication using the Bioruptor Pico sonication device (Diagenode; B01060001) using 20 cycles (30s on and 30s off) at maximum intensity. Purified chromatin was then separated for 1. Immunoprecipitation using 4ug of H3k27ac antibodies (Abcam; ab4729) per ChIP experiment or using 4ug of YY1 antibodies (Santa Cruz; sc-281 X). ChIP-seq experiment for YY1 were performed in biological duplicates. Cells were stimulated with estrogen for 45 minutes, upon which maximum ERα-binding to chromatin occurs. Biological replicates showed very high correlation (R2=0.98), thus only consensus loci were kept for further analyses. 2. Non-immunoprecipitated chromatin, used as Input control and 3. Assessment of sonication efficiencies using a 1% agarose gel. Before construction of ChIP-seq libraries (NEB Ultra II kit, see supplementary methods), enrichment of the immunoprecipitated sample was ascertained using positive and negative controls for ChIP-qPCR. Library preparation was performed using 10 – 50 ng of immunoprecipitated and Input samples. Before sequencing, libraries were again re-tested to confirm enrichment using positive and negative controls.
ChIP-qPCR
Briefly, reactions were carried out in 10 ul volume containing 5 ul of Sybr-green mix (ABI; 4472918), 0.5 ul of primer (5 uM final concentration), 2.5 ul of genomic DNA and 2 ul of DNASE/RNASE –free water. A three-step cycle programme and a melting analysis were applied. The cycling steps were as follows: 10s at 95 oC, 30s at 60 oC and 30s at 72 oC, repeated 40 times.
Ranking and Sharing Index
VSE
DHS imputations and TF motif analyses
Imputed DHS with vivo ERα binding Overlap
ERα binding from in breast cancer patients were obtained from39. ERα sharing index was calculated as before (see Supplementary Computational Methods). Overlap with imputed DHS was calculated using BedTools calculating the overlap (at least one base pair) via Cistrome Pipeline Analysis Suite (http://cistrome.org/Cistrome/Cistrome_Project.html). The percentage of overlap were calculated using binned DHS as variable first dataset and all the concatenated in vivo ERα as second dataset.
Footprint analysis
Encode and Epigenomic Roadmap Ranking
Immunocytochemistry
Hematoxylin and eosin staining of clinical samples was performed to calculate tumor burden prior to ChIP-seq. Briefly, 4-μm-thick sections were obtained from formalin-fixed and paraffin-embedded specimens. After de-waxing in xylene and graded ethanol, sections were incubated in 3% H2O2 solution for 25 minutes to block endogenous peroxidase activities and then subjected to microwaving in EDTA buffer for antigen retrieval. For YY1 (Protein Atlas HPA001119, Atlas Antibodies Cat#HPA001119, RRID:AB_1858930) the flowing conditions were used: tissue sections were incubated with the primary monoclonal. overnight at 4°C, and chromogen development was performed using the Envision system (DAKO Corporation, Glostrup, Denmark). A minimum of 500 tumor cells were scored with the percentage of tumor cell nuclei in each category recorded. For SLC9A3R1 (HPA9672 and HPA27247, Atlas Antibodies Cat#HPA009672, RRID:AB_1857215 and Atlas Antibodies Cat#HPA027247, RRID:AB_10601162 respectively) the following conditions were used. HPA9672 was diluted 1:400 and HPA27247 was diluted 1:1500. Staining was automatized with a Ventana Benchmark-Ultra using epitope retrieval ER2 for 20 minutes. ER and PgR immunoreactivity was assessed by the FDA-approved ER/PR PharmDX kit (Dako). The prevalence of ER/PgR positive invasive cancer cells, independent of their staining intensity, was quantitatively annotated in the original reports. In accordance with ASCO/CAP guidelines, tumors with ≥1% of immunoreactivity was considered positive
Cell culture
MCF7 was cultured using Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal calf serum (FCS) and 100 U penicillin/0.1 mg ml-1 streptomycin, 2mM L-glutamine plus 10-8 17-β-estradiol (SIGMA E8875). MCF7 long term oestrogen deprived (MCF7-LTED) cells were grown in phenol-free DMEM with 10% charcoal-stripped FCS (DCFCS) and 100 U penicillin/0.1 mg ml-1 streptomycin and 2mM L-glutamine. T47D and T47D-LTED cells were passaged using DMEM containing 10% FCS and 100 U penicillin/0.1 mg ml-1 streptomycin, 2mM L-glutamine and phenol-free DMEM with 10% DCFCS and 100 U penicillin/0.1 mg ml-1 streptomycin and 2mM L-glutamine, respectively. ZR75-1 cells were grown in DMEM containing 10% FCS and 100 U penicillin/0.1 mg ml-1 streptomycin, 2mM L-glutamine.
sIRNA
Small interfering RNA (siRNA) against SLC9A3R1 (Gene ID; 9368: Ambion; s17919, s17920), YY1 (Gene ID; 7528: Ambion; s14958, s14959, s14960) and Silencer negative control (Ambion; AM4611). 1.5 x 105 cells were seeded per well using a 6-well plate. MCF7 cells were seeded in phenol-free DMEM with 10% DCFCS and 100 U penicillin/0.1 mg ml-1 streptomycin and 2mM L-glutamine. Following 24 hours, the cells were then transfected with siRNA using Lipofectamine 3000 (Invitrogen; L3000015). T47D and ZR75-1 cells were seeded in DMEM containing 10% FCS and 100 U penicillin/0.1 mg ml-1 streptomycin, 2mM L-glutamine. Following 24 hours, the cells were then transfected with siRNA using Lipofectamine 3000 (Invitrogen; L3000015). Cells were harvested for analysis following at least 48 hours of transfection.
CRISPR/Cas9 Enhancer Knockout
Live Cell Imaging
MCF7 and YY1-EKO clones cells were plated at a density of 3 x 103 in a 96 well plate in FluoroBrite DMEM media (ThermoFisher) supplemented with 1*10-8M oestradiol. Cells were culture in an Incucyte Zoom (EssenBioscience) programmed to capture images every 6 hours. Twenty single cells for each cell line were followed over the course of 84 hours and their doubling time recorded and plotted.
Cell lysis and Western blot
Cells were washed twice in ice-cold PBS and lysed in RIPA (Sigma-Aldrich; R02780) buffer supplemented with protease (Roche 11697498001) and phosphastase (Sigma-Aldrich 93482) inhibitors for 30 minutes with intermittent vortexing. Samples were centrifuged at 4°C at maximum speed for 30 minutes after which, the supernatant is transferred to a clean Eppendorf. Protein concentrations for each sample was ascertained using the bicinchoninic acid (BCA) assay (ThermoFisher Scientific; 23227). Equal amounts of lysates were loaded into BOLT 4-12% Bis-Tris Plus Gel (Invitrogen; NW04120BOX). Proteins were transferred to a Biotrace nitrocellulose membrane (VWR; PN66485) and incubated with primary antibodies overnight. Proteins were then visualised using goat anti-mouse (ThermoFisher Scientific; 31446) and anti-rabbit (ThermoFisher Scientific; 31462) HRP conjugated secondary antibodies. Amersham ECL start Western Blotting Detection reagent (GE Healthcare Life Sciences; RPN3243) was used for chemiluminescent imaging using the Fusion solo (Vilber; Germany) imager. For SLC9A3R1 we used HPA027247 (protein atlas) at 1:1000 dilution, for YY we used Santa Cruz; sc-281 at 1:500 dilution. For GAPDH we used Abcam #ab9385 at 1:5000 dilution.
Transcriptional profiling
Following 48 hours of transfection, MCF7 cells were either treated with 10-8 17-β-estradiol (SIGMA E8875) or control treatment for 6 hours prior to RNA extraction. T47D and ZR75-1 cells lines were harvested for RNA following 48 hours of transfection. No treatments were added.
RNA extraction and real-time PCR
Total RNA was extracted using RNeasy Mini Kit (Qiagen; 74106), and the cDNA was reverse transcribed from 1ug of RNA using iScript cDNA synthesis kit (Bio-Rad; #1708891). Real time-qPCR (RT-qPCR) reactions were carried out in 10 uL volume containing 5 uL of sybergreen mix (ABI; 4472918), 0.5 ul of primer (2.5 uM final concentration), 2.5 ul of genomic DNA and 2 ul of DNASE/RNASE–free water. A three-step cycle programme and a melting analysis were applied. The cycling steps were as follows: 10s at 95 °C, 30s at 60 °C and 30s at 72 °C, repeated 40 times.
Luciferase reporter assay
MCF7 cells were seeded in a 24-well plate at 5 x 104 cells per well in phenol-free DMEM with 10% DCFCS and 100 U penicillin/0.1 mg ml-1 streptomycin and 2mM L-glutamine. After 24 hours of incubation, transfection of plasmid DNA was performed using Lipofectamine 3000 (Invitrogen; L3000015). Cells were transfected with 100ng of ERE_Luciferase reporter, 10ng of the renilla luciferase control plasmid (pRL-CMV), 10ng of pSG5_ER-α, 15 nm of siRNA and 280ng of Bluescribe DNA (BSM) per well; totalling 400ng of DNA/well. After 12 hours of transfection the media was replaced with fresh phenol-free DMEM with 10% DCFCS and 100 U penicillin/0.1 mg ml-1 streptomycin and 2mM L-glutamine. Treatment with 10-8 17-β-estradiol (SIGMA E8875) or control treatment was administered and the cells incubated for 24 hours. Cell lysates are then obtained using Passive lysis 5X buffer (Promega; E1941). The firefly and renilla luciferase activity was determined using DualGlo luciferase assay kit (Promega; E2920) according to the manufacturer protocol. The renilla luciferase activity measurement was utilised as control for transfection efficiency and therefore the ERE_Luciferase activity was normalised to the reading obtained for the renilla luciferase activity.
SRB assay
Briefly, the sulphorhodamine B (SRB) assay was used to monitor the effects of silencing either SLC9A3R1 or YY1, using siRNAs, on cell proliferation monolayer cultures. Cells were seeded in flat-bottomed 96-well plates (Costar; CLS3585) at a density of 2 x 103. Cells were allowed to attach overnight after which, the first plate (Day 0) is assayed after the cells have become adherent. Prospective plates are assayed sequentially after 3 days, 5 days and 7 days. The cells are fixed by adding 200uL of cold 40% (weight/volume) of trichloroacetic acid (TCA) to each well for at least 60 minutes. The plates were washed five times with distilled water and then 100 uL/well of SRB (0.4% wt/vol SRB in 1% wt/vol acetic acid) reagent is added to each well and the plates are allowed to incubate for 30 minutes. The plates were then washed five times in 1% (wt/vol) acetic acid and allowed to dry overnight. SRB solubilisation was performed by adding 100 uL/well of 10 mM Tris HCl to the plates and allowed to shake for 30 minutes. Optical density was then measured using the Sunrise microplate reader (Tecan; Sunrise) at 492 nm. Cell proliferation is then calculated over the 7-day period using Day 0 as a baseline measurement.
Enrichment scores
RI-IHC correlation
FFPE sections for the patients used in the ChIP-seq section were retrieved from Imperial Tissue bank. Sections were stained with YY1 or SLC9A3R1 antibodies. Stained sections were divided in 20 sectors. 5 sectors with high tumor burden were scored for the number of IHC+ cells and results averaged. The number of IHC+ cells and the matched RI was analyzed using linear regression using Prism 5 (GraphPad software Inc.).
∆RI
YY1 and SLC9A3R1 Pan cancer expression analysis
YY1 and SLC9A3R1 expression profile for matched Normal vs. Cancer samples was obtained using TIMER diff.exp option (https://cistrome.shinyapps.io/timer/). YY1 transcriptional analyses of breast cancer subtypes was performed in the Metabric Dataset (Curtis Breast) using probe ILMN_1770892 or TCGA dataset using Oncomine (https://www.oncomine.org/resource/login.html).
SLC9A3R1 Meta-analyses
SLC9A3R1 expression profile in drug resistant cell lines was performed by analysis of RNA-seq data from10. SLC9A3R1 expression profile in MCF7 cells transfected with siRNA against ERα was performed by analysis of microarray data from GSE27473. SLC9A3R1 expression profile in additional LTED models was performed by analysis of microarray data from E-GEOD-19639. All statistical analyses were performed using Prism 5 (GraphPad software Inc.). Kaplan-Meier analysis using SLC9A3R1 expression were performed by re-analysis of 23 independent microarray datasets (KMPLOT), TCGA RNA-seq data or the combined Metabric Dataset. Multivariate Cox proportional hazards survival analysis was performed using gene expression and clinical variables including nodal status, grade, and size in the Metabric and Affymetrix datasets using Winstat for Excel 2017. A multivariate analysis in the TCGA dataset employing available clinical data including TNM, histology, menopausal status, and race did not deliver significant result for any of the included parameters probably due to the short follow-up combined with limited number of events. SLC9A3R1 transcriptional profile in breast cancer cell lines was obtained from the HPA RNA-seq dataset (http://www.proteinatlas.org/about/download). SLC9A3R1 transcriptional profile from tissues was obtained from the HPA, GTEx and FANTOM5 RNA-seq datasets (http://www.proteinatlas.org/about/download).
Supplementary Material
Acknowledgments
We want to acknowledge and thanks all patients and their families for the support and for donating the research samples. We thank Breast Cancer Now Tissue Bank (project TR0121), Imperial Tissue Bank and the LEGACY study for contributing tissues. The authors gratefully acknowledge infrastructure support from the Cancer Research UK Imperial Centre, the Imperial Experimental Cancer Medicine Centre and the National Institute for Health Research Imperial Biomedical Research Centre. L.M was supported by a CRUK fellowship (C46704/A23110) and an Imperial Junior Fellowship (G53019). D.P was supported by a Wellcome Trust PhD studentship (103034/Z/13/Z). G.C was supported by a Marie Skłodowska Curie Training Grant (642691, EpiPredict). We acknowledge J.A. Buendia, Lorna Watson and Jason Carrol for their constructive comments on the manuscript.
Footnotes
Data Availability
H3K27ac data for all patients’ samples have been deposited at the ENA (http://www.ebi.ac.uk/ena) under project number PRJEB22757.
Author Contribution
L.M. conceived the study. D.K.P., E.E, N.S., and Y.P. performed the experiments. L.M., G.C., B.G., A.S., L.S.P., I.B., and P.S., developed and performed bioinformatic analyses. K.G., organised tissue collection. D.J.H., G.S., P.B., C.P., R.C.C., recruited patients and supplied tissues. S.S., performed pathology assessment of ChIP-seq processed samples. G.P., provided matched material. A.V. and G.P., performed IHC staining and scoring. All authors read and approved the manuscript.
Competing Financial Interests Statement
The authors declare no competing interests.
References
- 1.Ferlay J, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. 136:E359–86. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Ali S, Buluwela L, Coombes C. Antiestrogens and Their Therapeutic Applications in Breast Cancer and Other Diseases. 62:217–232. doi: 10.1146/annurev-med-052209-100305. [DOI] [PubMed] [Google Scholar]
- 3.Perou C, et al. Molecular portraits of human breast tumours. Nature. 406:747–752. doi: 10.1038/35021093. Article. [DOI] [PubMed] [Google Scholar]
- 4.Genestie, et al. Comparison of the prognostic value of Scarff-Bloom-Richardson and Nottingham histological grades in a series of 825 cases of breast cancer: major importance of the mitotic count as a component of both grading systems. 18:571–576. [PubMed] [Google Scholar]
- 5.Curtis C, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. 486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koboldt D, et al. Comprehensive molecular portraits of human breast tumours. 490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.EBCTCG, (EBCTCG) et al. Aromatase inhibitors versus tamoxifen in early breast cancer: patient-level meta-analysis of the randomised trials. The Lancet. 2015;386:1341–1352. doi: 10.1016/S0140-6736(15)61074-1. [DOI] [PubMed] [Google Scholar]
- 8.Magnani, et al. Acquired CYP19A1 amplification is an early specific mechanism of aromatase inhibitor resistance in ERα metastatic breast cancer. Nature Genetics. 2017;49:444–450. doi: 10.1038/ng.3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yates L, et al. Genomic Evolution of Breast Cancer Metastasis and Relapse. 2017;32:169–184.e7. doi: 10.1016/j.ccell.2017.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nguyen V, et al. Differential epigenetic reprogramming in response to specific endocrine therapies promotes cholesterol biosynthesis and cellular invasion. Nature Communications. 2015;6 doi: 10.1038/ncomms10044. ncomms10044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Consortium, R et al. Integrative analysis of 111 reference human epigenomes. 518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Consortium, T et al. An integrated encyclopedia of DNA elements in the human genome. 488:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ernst J, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. 473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Whyte W, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. 153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heintzman N, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. 39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 16.Falahi, et al. Towards Sustained Silencing of HER2/neu in Cancer By Epigenetic Editing. 11:1029–1039. doi: 10.1158/1541-7786.MCR-12-0567. [DOI] [PubMed] [Google Scholar]
- 17.Laprell F, Finkl K, Müller J. Propagation of Polycomb-repressed chromatin requires sequence-specific recruitment to DNA. doi: 10.1126/science.aai8266. eaai8266. [DOI] [PubMed] [Google Scholar]
- 18.Wang, Moazed DNA sequence-dependent epigenetic inheritance of gene silencing and histone H3K9 methylation. Science (New York, N.Y.) 2017 doi: 10.1126/science.aaj2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coleman, Struhl Causal role for inheritance of H3K27me3 in maintaining the OFF state of a Drosophila HOX gene. 2017 doi: 10.1126/science.aai8236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Magnani, Eeckhoute, Lupien Pioneer factors: directing transcriptional regulators within the chromatin environment. Trends in Genetics. 2011;27:465–474. doi: 10.1016/j.tig.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 21.Jozwik K, Carroll J. Pioneer factors in hormone-dependent cancers. :1–5. doi: 10.1038/nrc3263. [DOI] [PubMed] [Google Scholar]
- 22.Hnisz D, et al. Convergence of developmental and oncogenic signaling pathways at transcriptional super-enhancers. 58:362–370. doi: 10.1016/j.molcel.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heintzman N, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. 459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yates L, et al. Subclonal diversification of primary breast cancer revealed by multiregion sequencing. 21:751–759. doi: 10.1038/nm.3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Williams M, Werner B, Barnes C, Graham T, Sottoriva A. Identification of neutral tumor evolution across cancer types. 48:238–244. doi: 10.1038/ng.3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lan X, et al. Fate mapping of human glioblastoma reveals an invariant stem cell hierarchy. Nature. 2017 doi: 10.1038/nature23666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tirosh I, et al. Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma. Nature. 2016;539:309–313. doi: 10.1038/nature20123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harvey, Clark, Osborne, Allred Estrogen receptor status by immunohistochemistry is superior to the ligand-binding assay for predicting response to adjuvant endocrine therapy in breast cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 1999;17:1474–81. doi: 10.1200/JCO.1999.17.5.1474. [DOI] [PubMed] [Google Scholar]
- 29.Buenrostro, et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature. 2015;523:486–490. doi: 10.1038/nature14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.lari R, et al. Breast cancer risk-associated SNPs modulate the affinity of chromatin for FOXA1 and alter gene expression. 44:1191–1198. doi: 10.1038/ng.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cohen A, et al. Hotspots of aberrant enhancer activity punctuate the colorectal cancer epigenome. 8 doi: 10.1038/ncomms14400. 14400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Michailidou K, et al. Association analysis identifies 65 new breast cancer risk loci. Nature. 2017;551:92–94. doi: 10.1038/nature24284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Levsky, Singer Gene expression and the myth of the average cell. Trends in Cell Biology. 2003;13:4–6. doi: 10.1016/s0962-8924(02)00002-8. [DOI] [PubMed] [Google Scholar]
- 34.Wang S, et al. Target analysis by integration of transcriptome and ChIP-seq data with BETA. 8:2502–2515. doi: 10.1038/nprot.2013.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gyorffy B, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. 123:725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 36.Neph S, et al. An expansive human regulatory lexicon encoded in transcription factor footprints. 489:83–90. doi: 10.1038/nature11212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thurman R, et al. The accessible chromatin landscape of the human genome. 489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paakinaho, et al. Single-molecule analysis of steroid receptor and cofactor action in living cells. Nature communications. 2017;8 doi: 10.1038/ncomms15896. 15896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ross-Innes C, et al. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. 481:389–393. doi: 10.1038/nature10730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carroll, et al. Chromosome-Wide Mapping of Estrogen Receptor Binding Reveals Long-Range Regulation Requiring the Forkhead Protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 41.Beagan, et al. YY1 and CTCF orchestrate a 3D chromatin looping switch during early neural lineage commitment. Genome Research. 2017 doi: 10.1101/gr.215160.116. gr.215160.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weintraub A, et al. YY1 Is a Structural Regulator of Enhancer-Promoter Loops. Cell. 2017;171:1573–1588.e28. doi: 10.1016/j.cell.2017.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vella, Barozzi, Cuomo, Bonaldi, Pasini Yin Yang 1 extends the Myc-related transcription factors network in embryonic stem cells. 40:3403–3418. doi: 10.1093/nar/gkr1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jeon Y, Lee J. YY1 tethers Xist RNA to the inactive X nucleation center. 146:119–133. doi: 10.1016/j.cell.2011.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sigova A, et al. Transcription factor trapping by RNA in gene regulatory elements. 350:978–981. doi: 10.1126/science.aad3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Klymenko, et al. A Polycomb group protein complex with sequence-specific DNA-binding and selective methyl-lysine-binding activities. Genes & Development. 2006;20:1110–1122. doi: 10.1101/gad.377406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang, et al. CTCF-Mediated Human 3D Genome Architecture Reveals Chromatin Topology for Transcription. Cell. 2015;163:1611–1627. doi: 10.1016/j.cell.2015.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hurtado A, Holmes K, Ross-Innes C, Schmidt D, Carroll J. FOXA1 is a key determinant of estrogen receptor function and endocrine response. 43:27–33. doi: 10.1038/ng.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cardone, Casavola, Reshkin The role of disturbed pH dynamics and the Na+/H+ exchanger in metastasis. Nature Reviews Cancer. 2005;5 doi: 10.1038/nrc1713. nrc1713. [DOI] [PubMed] [Google Scholar]
- 50.Gerlinger, et al. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nature Genetics. 2014;46:225–233. doi: 10.1038/ng.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McGranahan, Swanton Biological and therapeutic impact of intratumor heterogeneity in cancer evolution. Cancer cell. 2015;27:15–26. doi: 10.1016/j.ccell.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 52.Juric D, et al. Convergent loss of PTEN leads to clinical resistance to a PI(3)Kα inhibitor. 518:240–244. doi: 10.1038/nature13948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shah S, et al. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. 461:809–813. doi: 10.1038/nature08489. [DOI] [PubMed] [Google Scholar]
- 54.Arduino, et al. Reduced IL-2 level concentration in patients with breast cancer as a possible risk factor for relapse. 17:535–537. [PubMed] [Google Scholar]
- 55.Cai, et al. YY1 functions with INO80 to activate transcription. Nature Structural & Molecular Biology. 2007;14:872–874. doi: 10.1038/nsmb1276. [DOI] [PubMed] [Google Scholar]
- 56.Onder T, et al. Chromatin-modifying enzymes as modulators of reprogramming. 483:598–602. doi: 10.1038/nature10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Whalen, Truty, Pollard Enhancer–promoter interactions are encoded by complex genomic signatures on looping chromatin. Nature Genetics. 2016;48 doi: 10.1038/ng.3539. ng.3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schmidt, et al. ChIP-seq: Using high-throughput sequencing to discover protein–DNA interactions. Methods. 2009;48:240–248. doi: 10.1016/j.ymeth.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.