Abstract
Many intracellular proteins are reversibly modified by O-linked GlcNAc (O-GlcNAc), a post-translational modification that dynamically regulates fundamental cellular processes in response to diverse environmental cues. Accumulating evidence indicates that both excess and deficiency of protein O-GlcNAcylation can have deleterious effects on the cell, suggesting that maintenance of O-GlcNAc homeostasis is essential for proper cellular function. However, the mechanisms through which O-GlcNAc homeostasis is maintained in the physiologic state and altered in the disease state have not yet been investigated. Here, we demonstrate the existence of a homeostatic mechanism involving mutual regulation of the O-GlcNAc–cycling enzymes O-GlcNAc transferase (OGT) and O-GlcNAcase (OGA) at the transcriptional level. Specifically, we found that OGA promotes Ogt transcription through cooperation with the histone acetyltransferase p300 and transcription factor CCAAT/enhancer-binding protein β (C/EBPβ). To examine the role of mutual regulation of OGT and OGA in the disease state, we analyzed gene expression data from human cancer data sets, which revealed that OGT and OGA expression levels are highly correlated in numerous human cancers, particularly in pancreatic adenocarcinoma. Using a KrasG12D-driven primary mouse pancreatic ductal adenocarcinoma (PDAC) cell line, we found that inhibition of extracellular signal–regulated kinase (ERK) signaling decreases OGA glycosidase activity and reduces OGT mRNA and protein levels, suggesting that ERK signaling may alter O-GlcNAc homeostasis in PDAC by modulating OGA-mediated Ogt transcription. Our study elucidates a transcriptional mechanism that regulates cellular O-GlcNAc homeostasis, which may lay a foundation for exploring O-GlcNAc signaling as a therapeutic target for human disease.
Keywords: O-linked N-acetylglucosamine (O-GlcNAc), O-GlcNAcylation, O-linked N-acetylglucosamine (O-GlcNAc) transferase (OGT), E1A binding protein p300 (P300), CCAAT-enhancer-binding protein (C/EBP), extracellular-signal-regulated kinase (ERK), pancreatic cancer, O-GlcNAcase (OGA)
Introduction
O-GlcNAcylation, or the attachment of single O-linked GlcNAc (O-GlcNAc)3 moieties to serine and threonine residues of cytoplasmic, nuclear, and mitochondrial proteins, is an essential post-translational modification in metazoans (1, 2). As a product of nutrient flux through the hexosamine biosynthetic pathway, protein O-GlcNAcylation couples changes in nutrient availability to dynamic regulation of fundamental cellular processes such as gene expression, metabolism, and signal transduction (1–9). Perturbations in cellular O-GlcNAc homeostasis have been linked to human disorders ranging from cancer and diabetes to cardiovascular and neurodegenerative disease, suggesting that maintenance of O-GlcNAc homeostasis is vital for normal cellular and physiological function (2, 10–14). However, what mechanisms exist to preserve cellular O-GlcNAc homeostasis under physiologic conditions is an outstanding question in the field that has remained largely unexplored.
We previously hypothesized that cellular O-GlcNAc homeostasis is maintained by mutual regulation of O-GlcNAc transferase (OGT) and O-GlcNAcase (OGA), the sole enzymes that add and remove O-GlcNAc, respectively (2). Several studies have shown that OGT and OGA mRNA levels are responsive to perturbations in O-GlcNAc signaling and that OGT and OGA are themselves O-GlcNAcylated, suggesting that mutual regulation of OGT and OGA could occur at both the transcriptional and post-translational levels (15–18). These putative homeostatic mechanisms could function to preserve the balance between OGT and OGA activity and expression and thereby maintain cellular O-GlcNAcylation levels within an “optimal zone” (2). Here, we provide evidence to support the existence of mutual regulation of OGT and OGA at the transcriptional level, demonstrating that OGA does indeed modulate Ogt transcription and vice versa. Furthermore, we describe a specific mechanism in which OGA, in conjunction with the histone acetyltransferase p300, promotes Ogt transcription via the transcription factor CCAAT/enhancer-binding protein β (C/EBPβ).
Although the role of aberrant protein O-GlcNAcylation in the pathogenesis of various human disorders has become increasingly well characterized, the molecular mechanisms responsible for altered cellular O-GlcNAc homeostasis in these conditions remain poorly studied. For instance, OGT and OGA expression levels are known to be elevated in numerous human cancers and both OGT and OGA have been shown to promote tumorigenesis (10, 19–25); however, how OGT and OGA expression levels are differentially regulated in cancer versus normal cells is a fundamental question that requires further investigation. Here, we show that OGT and OGA mRNA levels are highly correlated across a wide range of human cancers, particularly in pancreatic adenocarcinoma. Mechanistically, we demonstrate that the OGA–C/EBPβ axis regulates Ogt expression in primary mouse pancreatic ductal adenocarcinoma (PDAC) cells. Furthermore, we show that loss of extracellular signal–regulated kinase (ERK) signaling in these cells decreases OGA glycosidase activity and reduces OGT mRNA and protein levels, raising the possibility that oncogenic ERK signaling may impinge upon OGA-mediated Ogt transcription to increase OGT expression in pancreatic cancer.
Results
OGA regulates Ogt transcription and vice versa
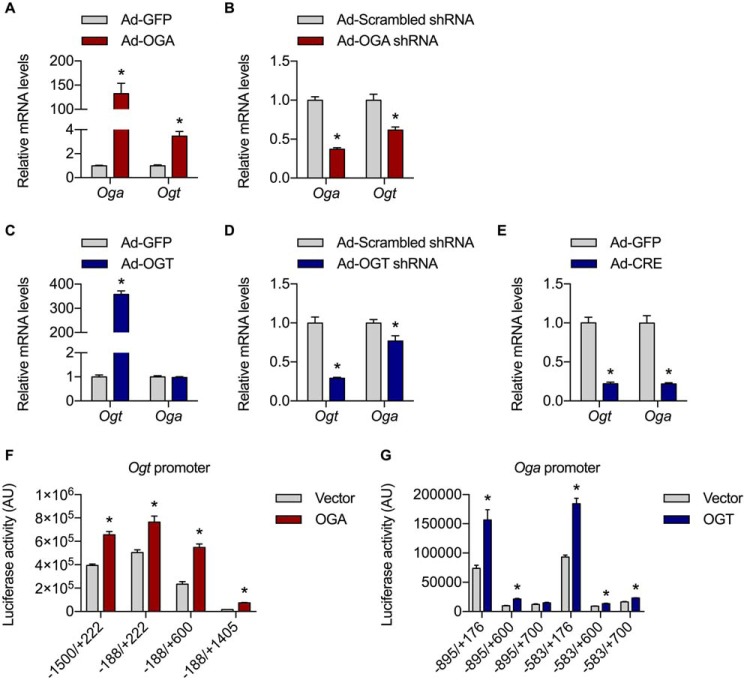
To test the hypothesis that OGT and OGA regulate each other at the transcriptional level, primary hepatocytes were first isolated from WT mice and transduced with adenoviral vectors expressing OGA, OGA shRNA, OGT, or OGT shRNA. Overexpression of OGA produced a concomitant increase in Ogt mRNA levels, whereas knockdown of OGA reduced Ogt expression (Fig. 1, A and B). Although overexpression of OGT failed to induce Oga expression, knockdown of OGT did result in a slight but significant decrease in Oga mRNA levels (Fig. 1, C and D). To determine whether knocking out OGT would produce a more dramatic change in Oga expression, primary hepatocytes were isolated from Ogt-floxed mice and transduced with an adenoviral vector expressing Cre. Deletion of the Ogt gene led to a dramatic reduction in both Ogt and Oga mRNA levels (Fig. 1E). Taken together, these data demonstrate that OGA modulates Ogt expression and vice versa.
Figure 1.
OGA regulates Ogt transcription and vice versa. A–D, primary hepatocytes isolated from WT mice were transduced with adenoviral vectors expressing GFP or OGA (n = 12, 6) (A), scrambled or OGA shRNA (n = 6) (B), GFP or OGT (n = 12, 6) (C), and scrambled or OGT shRNA (n = 6) (D), and Oga and Ogt mRNA levels were measured by RT–qPCR. E, primary hepatocytes isolated from Ogt-floxed mice were transduced with adenoviral vectors expressing GFP or CRE (n = 6), and Ogt and Oga mRNA levels were measured by RT–qPCR. F and G, Ogt (n = 4) (F) and Oga (n = 4) (G) promoter luciferase assays performed in CV-1 cells transfected with the indicated plasmids. All values represent means ± S.E. *, p < 0.05 by multiple t tests with Holm–Sidak correction for multiple comparisons.
To determine whether the observed changes in Ogt and Oga gene expression could be due to altered regulation of Ogt and Oga transcription, luciferase reporter constructs containing various truncations of the mouse Ogt and Oga promoters were generated. Overexpression of OGA in CV-1 cells transiently expressing any of the four Ogt promoter luciferase reporters resulted in a significant increase in Ogt promoter activity; similarly, overexpression of OGT induced Oga promoter activity in five of the six Oga promoter reporter constructs (Fig. 1, F and G). Collectively, these data demonstrate that OGA promotes Ogt transcription and vice versa and suggest that mutual regulation of OGT and OGA does indeed exist at the transcriptional level.
To corroborate these findings, the genes most highly coexpressed with OGT and OGA were obtained from the gene coexpression database COXPRESdb (26). This analysis revealed that OGA was among the top genes coexpressed with OGT in human, mouse, and rat tissues and vice versa (Table S1 and S2). The correlation was particularly notable in human tissues, where OGA was ranked ninth among OGT coexpressed genes, and OGT was ranked fifth among OGA coexpressed genes. These data further support the notion that OGT and OGA expression levels are tightly coupled and suggest that this homeostatic phenomenon is conserved across tissues and species.
OGA and p300 promote Ogt transcription through C/EBPβ
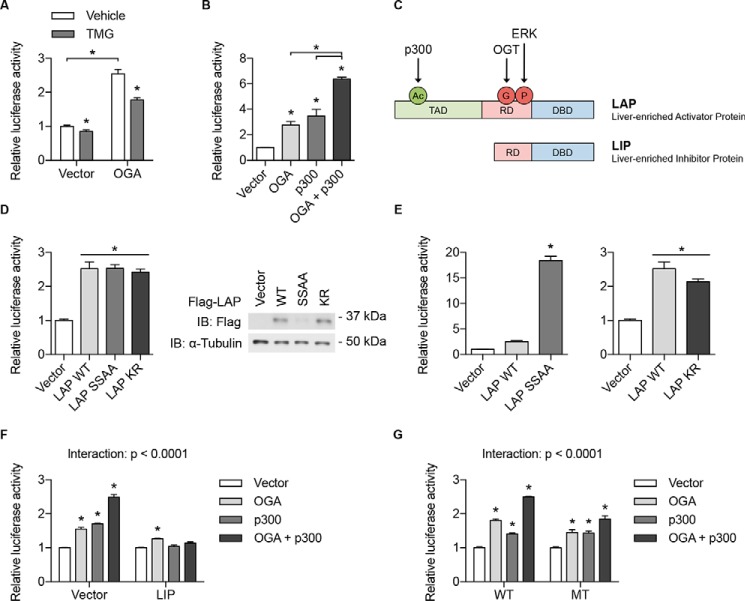
Our next aim was to examine the molecular mechanisms underlying mutual regulation of OGT and OGA at the transcriptional level, with a focus on OGA-mediated Ogt transcription because Ogt mRNA levels were particularly responsive to changes in Oga expression (Fig. 1, A and B). To probe this pathway, the luciferase reporter construct containing the −1500/+222 region of the mouse Ogt promoter was first transiently expressed in human embryonic kidney (HEK) 293T cells. Overexpression of OGA in these cells produced an increase in Ogt promoter activity that was blunted by concomitant administration of the OGA inhibitor Thiamet G (TMG), suggesting that OGA promotes Ogt transcription at least in part through its glycosidase activity (Fig. 2A) (27).
Figure 2.
OGA and p300 promote Ogt transcription through C/EBPβ. A, B, and D–G, Ogt promoter luciferase assays performed in HEK 293T cells transfected with the indicated plasmids. A, HEK 293T cells were also treated with vehicle or TMG (40 μm, 24 h). D, right panel, Western blotting showing the expression level of each protein after transfection of equal amounts of DNA. See Fig. S1B for quantification. E, data from D after normalizing luciferase activity to the relative levels of LAP SSAA and LAP KR. G, luciferase assay was performed using both WT and C/EBPβ-binding site mutation (MT) Ogt promoter luciferase reporter constructs. See Fig. S1, D and E for binding site sequence and mutation. All values represent means ± S.E. (n = 4 for A and D–G and n = 3 for B). *, p < 0.05 by Student's t test (A, vector versus OGA), multiple t tests with Holm–Sidak correction for multiple comparisons (A, vehicle versus TMG), one-way analysis of variance with Tukey's multiple comparisons test (B, D, and E), or two-way analysis of variance with Dunnett's multiple comparisons test (F and G). C, schematic illustration of the domain structures and post-translational modifications of the two main isoforms of C/EBPβ. TAD, transactivation domain; RD, regulatory domain; DBD, DNA-binding domain; Ac, acetylation; G, O-GlcNAcylation; P, phosphorylation; IB, immunoblot.
Because OGA is not known to possess a DNA-binding domain, it likely regulates Ogt transcription by functioning as a coactivator for specific transcription factors (2, 4, 5). To identify candidate coactivators and transcription factors that may regulate Ogt transcription in conjunction with OGA, ChIP sequencing (ChIP-seq) data from the Encyclopedia of DNA Elements (ENCODE) were visualized in the University of California, Santa Cruz Genome Browser. These data revealed that the histone acetyltransferase and transcriptional coactivator p300 is localized to the human OGT promoter (Fig. S1A). Additionally, the OGT promoter was found to be enriched in histone H3 lysine 27 acetylation (H3K27ac) (Fig. S1A), an active promoter and enhancer mark deposited by the p300/CBP coactivator family (28–30). Consistent with these observations, coexpression of OGA and p300 induced Ogt promoter activity to a greater extent than did overexpression of either OGA or p300 alone (Fig. 2B).
Analysis of ENCODE ChIP-seq data also identified C/EBPβ as one of the most abundant transcription factors at the human OGT promoter (Fig. S1A). Previous studies have shown that O-GlcNAcylation of C/EBPβ at serines 180 and 181 blocks its DNA binding and that acetylation of C/EBPβ at lysine 39 by p300 enhances its transcriptional activity, suggesting that OGA and p300 could function as coactivators for C/EBPβ (Fig. 2C) (31, 32). To determine whether C/EBPβ promotes Ogt transcription, liver-enriched activator protein (LAP), one of the two main isoforms of C/EBPβ (Fig. 2C), was overexpressed in HEK 293T cells transiently expressing the Ogt promoter luciferase reporter (31). Overexpression of wildtype LAP (LAP WT) increased Ogt promoter activity, indicating that C/EBPβ does indeed promote Ogt transcription (Fig. 2D). Mutation of the LAP O-GlcNAcylation sites to alanine (LAP SSAA) or the lysine 39 acetylation site to arginine (LAP KR) initially appeared to have no effect on the ability of LAP to regulate Ogt promoter activity; however, Western blotting analysis revealed that LAP SSAA levels were markedly lower and LAP KR levels were slightly higher than those of LAP WT despite transfection of equal amounts of DNA (Fig. 2D and Fig. S1B). This observation could be explained by compensatory degradation and stabilization of highly active (LAP SSAA) and less active (LAP KR) transcription factors, respectively (33). Normalization of luciferase activity to the relative levels of LAP SSAA and LAP KR demonstrated that LAP SSAA was dramatically more active and LAP KR was slightly but not significantly less active than LAP WT at the Ogt promoter (Fig. 2E). Because coexpression of LAP and p300 increased Ogt promoter activity in a cooperative fashion (Fig. S1C), the marginal effect of the KR mutation even after normalization could be attributed to the existence of other LAP acetylation sites that contribute to its transcriptional activity. Alternatively, p300 may stimulate LAP-dependent Ogt transcription through acetylation-independent mechanisms (34). Taken together, these data suggest that OGA and p300 regulate Ogt transcription in conjunction with C/EBPβ, perhaps by reducing its O-GlcNAcylation and increasing its acetylation.
To determine whether C/EBPβ is required for OGA- and p300-mediated regulation of Ogt transcription, liver-enriched inhibitor protein (LIP), the dominant negative isoform of C/EBPβ (Fig. 2C), was coexpressed with OGA and p300 in HEK 293T cells transiently expressing the Ogt promoter luciferase reporter (31, 35). Overexpression of LIP almost completely abolished the effect of OGA and/or p300 on Ogt promoter activity, indicating that LAP activity is essential for OGA- and p300-dependent Ogt transcription (Fig. 2F). To corroborate these results, we next sought to determine whether mutating the C/EBPβ-binding site on the Ogt promoter affects the ability of OGA and p300 to promote Ogt transcription. To address this question, the DNA sequence of the mapped C/EBPβ-binding region was first obtained from the University of California, Santa Cruz Genome Browser (Fig. S1A) and searched for the C/EBPβ consensus sequence 5′-T[TG]NNGNAA[TG]-3′ (36). One DNA element matching the consensus sequence was identified and found to be conserved among the human, mouse, and rat OGT promoters (Fig. S1D). Two consecutive bases of the identified binding motif were then mutated in the Ogt promoter luciferase reporter construct (Fig. S1E), which was subsequently expressed in HEK 293T cells with additional coexpression of OGA and p300. Mutation of the conserved C/EBPβ-binding site significantly blunted the effect of OGA and/or p300 on Ogt promoter activity, suggesting that OGA- and p300-mediated Ogt transcription is at least partially dependent on C/EBPβ binding to the Ogt promoter (Fig. 2G). The incomplete abrogation of Ogt promoter activity by the C/EBPβ-binding site mutation could be attributed to the existence of other C/EBPβ-binding sites on the Ogt promoter, to residual binding of C/EBPβ to the mutated binding site, or to the unperturbed actions of other transcription factors. Collectively, these data demonstrate that OGA and p300 promote Ogt transcription through C/EBPβ.
OGT and OGA expression levels are highly correlated in human cancers
Thus far, we have demonstrated the existence of mutual regulation of OGT and OGA at the transcriptional level and have outlined a potential mechanism that may contribute to this phenomenon in a physiologic system. Because perturbations in cellular O-GlcNAc homeostasis have been linked to numerous human disorders, our next aim was to examine the role of mutual regulation of OGT and OGA in the pathogenesis of a relevant human disease. Altered expression of OGT and OGA has been observed in a wide range of human cancers; thus, we postulated that cancer would be an appropriate system in which to study the relevance of mutual regulation of OGT and OGA to human disease.
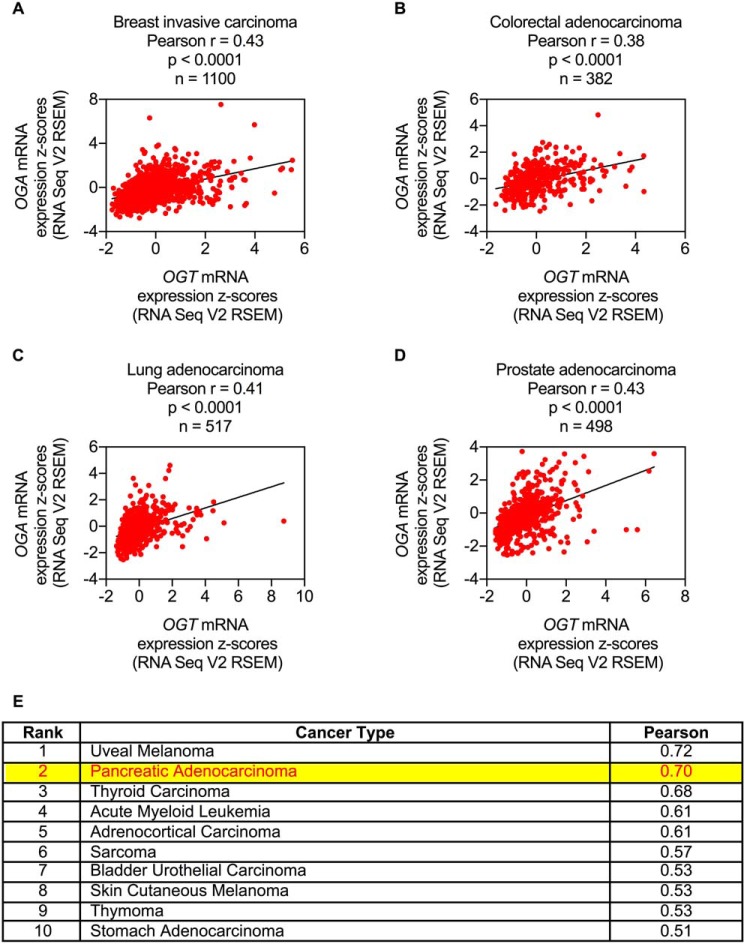
To broadly assess the potential role of mutual regulation of OGT and OGA in cancer pathophysiology, The Cancer Genome Atlas (TCGA) gene expression data for 32 types of human cancer were analyzed for correlations between OGT and OGA expression (37, 38). Remarkably, a significant positive correlation between OGT and OGA mRNA levels was observed in nearly every cancer type analyzed, including the four most prevalent cancers in the United States: breast, colorectal, lung, and prostate (Fig. 3, A–D, and Table S3) (39). Further analysis of TCGA data revealed that genomic alterations (i.e. amplifications, deletions, and/or mutations) in the OGT and OGA genes are relatively rare in human cancers, suggesting that dysregulation of OGT and OGA expression in cancer cells may be driven primarily by transcriptional rather than genomic changes (Fig. S2, A and B). These data demonstrate that OGT and OGA expression levels are highly correlated in human cancers and suggest that altered transcriptional regulation of OGT and OGA may play a particularly important role in cancer biology.
Figure 3.
OGT and OGA expression levels are highly correlated in human cancers. A–D, OGT versus OGA mRNA expression z scores from TCGA RNA Seq V2 RSEM data for breast invasive carcinoma (A), colorectal adenocarcinoma (B), lung adenocarcinoma (C), and prostate adenocarcinoma (D). E, ranked list of cancer types with the highest Pearson correlation coefficients for OGT versus OGA mRNA expression z scores. See Table S3 for complete list.
OGT and OGA expression levels are highly correlated and elevated in human pancreatic cancer
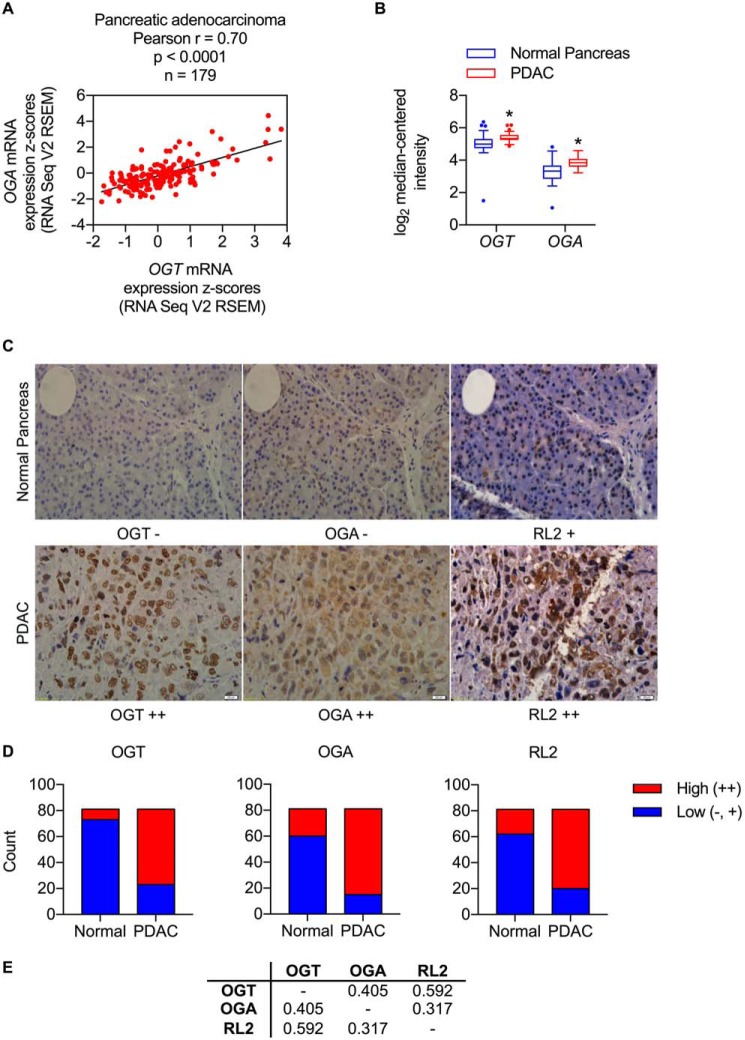
Among the cancer types with the highest correlation coefficients (Fig. 3E), pancreatic adenocarcinoma was selected for further analysis because perturbations in O-GlcNAc homeostasis had previously been described in this form of cancer (40–42). In addition to a strong positive correlation between OGT and OGA expression, pancreatic adenocarcinoma was found to have elevated OGT and OGA mRNA levels relative to normal pancreas (Fig. 4, A and B). To determine whether these transcript-level phenomena were also observable at the protein level, tissue sections of normal human pancreas and PDAC were stained for OGT, OGA, and O-GlcNAc. These immunohistochemical analyses revealed that OGT, OGA, and protein O-GlcNAcylation levels are dramatically elevated in PDAC relative to normal pancreas (Fig. 4, C and D, and Table S4). Notably, significant positive correlations among OGT, OGA, and O-GlcNAc were observed in PDAC, including an unexpected positive correlation between OGA and O-GlcNAc (Fig. 4E and Tables S5–S7). Taken together, these data demonstrate that both mRNA and protein levels of OGT and OGA are highly correlated and elevated in human pancreatic cancer and indicate that this expression pattern is ultimately associated with increased protein O-GlcNAcylation.
Figure 4.
OGT and OGA expression levels are highly correlated and elevated in human pancreatic cancer. A, OGT versus OGA mRNA expression z scores from TCGA RNA Seq V2 RSEM data for pancreatic adenocarcinoma. B, box-and-whisker plot of OGT and OGA mRNA expression levels in normal pancreas versus PDAC (n = 39). *, p < 0.05 by multiple t tests with Holm–Sidak correction for multiple comparisons. C, representative images of normal pancreas and PDAC tissue sections stained for OGT, OGA, and RL2 (O-GlcNAc). D, counts of normal pancreas and PDAC tissue sections with low or high levels of OGT, OGA, and RL2 staining (n = 81). See Table S4 for raw data and statistical analysis. E, table of Spearman correlation coefficients for OGT versus OGA, OGT versus RL2, and OGA versus RL2 staining levels in PDAC. See Tables S5–S7 for raw data and statistical analyses.
OGA, C/EBPβ, and ERK signaling regulate Ogt expression in pancreatic cancer
Our next aim was to examine the potential mechanisms leading to correlation and elevation of OGT and OGA expression levels in pancreatic cancer. We hypothesized that 1) mutual regulation of OGT and OGA at the transcriptional level remains largely intact in pancreatic cancer cells, leading to a positive correlation between OGT and OGA expression, and 2) an aberrantly activated signaling pathway in these cells impinges upon mutual regulation of OGT and OGA to increase the expression of these genes.
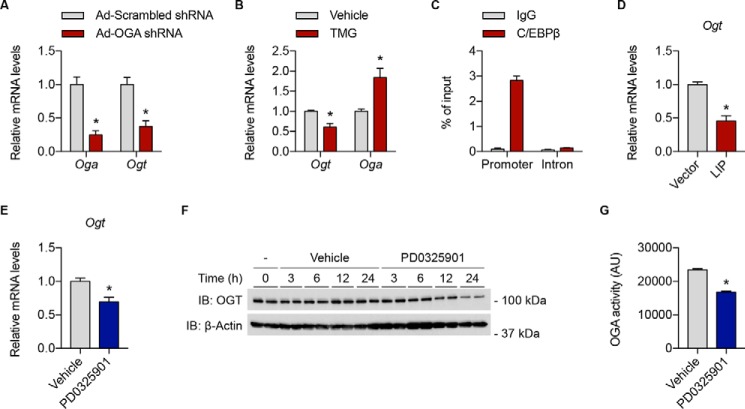
To test these hypotheses in an established in vitro system, primary mouse PDAC cells with doxycycline-inducible expression of KrasG12D and hemizygous deletion of p53 were first transduced with OGA shRNA adenovirus or treated with TMG (40). Genetic knockdown of OGA and pharmacological inhibition of OGA glycosidase activity both reduced Ogt mRNA levels in these cells, suggesting that OGA also regulates Ogt transcription in pancreatic cancer (Fig. 5, A and B). Interestingly, TMG treatment elicited a compensatory increase in Oga mRNA levels, indicating that Oga expression is also responsive to changes in O-GlcNAc homeostasis as reported previously (Fig. 5B) (15). Consistent with a role for C/EBPβ in modulating Ogt transcription in pancreatic cancer, C/EBPβ occupancy was found to be highly and specifically enriched at the Ogt promoter in PDAC cells and overexpression of LIP led to a significant decrease in Ogt mRNA levels (Fig. 5, C and D). Taken together, these data demonstrate that the OGA–C/EBPβ axis also regulates Ogt expression in PDAC cells, supporting our first hypothesis that mutual regulation of OGT and OGA at the transcriptional level remains largely intact in pancreatic cancer.
Figure 5.
OGA, C/EBPβ, and ERK signaling regulate Ogt expression in pancreatic cancer. A, primary mouse PDAC cells were transduced with adenoviral vectors expressing scrambled or OGA shRNA, and Oga and Ogt mRNA levels were measured by RT–qPCR. B, primary mouse PDAC cells were treated with vehicle or TMG (10 μm, 12 h), and Ogt and Oga mRNA levels were measured by RT–qPCR. C, sheared chromatin from primary mouse PDAC cells was immunoprecipitated with IgG or an anti-C/EBPβ antibody and analyzed by RT–qPCR. Promoter primers amplify a 124-bp region containing the C/EBPβ binding site in Fig. S1D, and intron primers target a 73-bp region in intron 11-12. D, primary mouse PDAC cells were transfected with vector or LIP and Ogt mRNA levels were measured by RT–qPCR. E–G, primary mouse PDAC cells were treated with vehicle or PD0325901 (10 μm) for 12 h (E); 0, 3, 6, 12, or 24 h (F); or 6 h (G), and Ogt mRNA levels were measured by RT–qPCR (E), OGT protein levels were measured by Western blotting (F), or total cellular OGA activity was measured by OGA activity assay (G). See Fig. S3B for quantification of F. All values represent means ± S.E. (n = 3 for A–D and G; n = 6 for E, pooled from two independent experiments). *, p < 0.05 by multiple t tests with Holm–Sidak correction for multiple comparisons (A and B) or Student's t test (D, E, and G). IB, immunoblot.
Over 90% of human PDACs possess gain-of-function mutations in the KRAS oncogene, which lead to constitutive activation of downstream signaling pathways such as the mitogen-activated protein kinase (MAPK)/ERK pathway. Because transgenic KrasG12D expression had previously been shown to raise protein O-GlcNAcylation levels in PDAC cells, we hypothesized that aberrant activation of ERK signaling in these cells impinges upon OGA-mediated Ogt transcription to increase OGT expression (40). To test this hypothesis, ERK signaling was inhibited in PDAC cells using PD0325901 (Fig. S3A), a potent and selective inhibitor of the upstream ERK activator MAPK/ERK kinase (MEK) (43, 44). Inhibition of MEK led to a significant decrease in both mRNA and protein levels of OGT, indicating that ERK signaling regulates OGT expression in PDAC cells (Fig. 5, E and F, and Fig. S3B). Because OGA-mediated Ogt transcription is at least partially dependent on OGA glycosidase activity (Fig. 2A), we next sought to determine whether ERK signaling also modulates OGA activity in PDAC cells. Indeed, inhibition of MEK using PD0325901 reduced total cellular OGA activity without altering OGA protein levels (Fig. 5G and Fig. S3C). Collectively, these data suggest that oncogenic ERK signaling may up-regulate OGT expression in pancreatic cancer by increasing OGA glycosidase activity.
Discussion
Aberrant protein O-GlcNAcylation is being identified as a key pathological feature of an increasing number of human disorders; thus, elucidating the mechanisms through which cellular O-GlcNAc homeostasis is maintained in the physiologic state and altered in the disease state is of significant biomedical interest. We previously hypothesized that cellular O-GlcNAcylation levels are maintained within an optimal zone by mutual regulation of the O-GlcNAc–cycling enzymes OGT and OGA (2). Here, using established in vitro systems and large-scale analysis of human cancer data, we demonstrate that 1) OGT and OGA indeed regulate each other at the transcriptional level; 2) OGA promotes Ogt transcription through cooperation with the histone acetyltransferase p300 and transcription factor C/EBPβ; 3) OGT and OGA expression levels are highly correlated across a wide range of human cancers, particularly in pancreatic adenocarcinoma; and 4) OGA, C/EBPβ, and ERK signaling regulate Ogt expression in PDAC.
To demonstrate the existence of mutual regulation of OGT and OGA at the transcriptional level, we used both genetic and pharmacological methods to manipulate OGT and OGA in a variety of cell types, including primary mouse hepatocytes, CV-1 monkey kidney cells, HEK 293T cells, and primary mouse PDAC cells. One important question for future investigation is whether mutual regulation of OGT and OGA at the transcriptional level is a universal phenomenon that exists in all cell types of an organism. Numerous studies in diverse cellular contexts have reported compensatory changes in OGT and OGA mRNA levels in response to manipulation of the O-GlcNAc and hexosamine biosynthetic pathways, suggesting that mutual regulation of OGT and OGA at the transcriptional level may indeed be a fundamental process that is generalizable to many cell types (15, 16, 45–47). However, in contrast to our findings in primary hepatocytes and PDAC cells, Zhang et al. (15) demonstrated in three different cell lines (HeLa cervical adenocarcinoma, K562 chronic myelogenous leukemia, and SH-SY5Y neuroblastoma cells) that OGA but not OGT expression is responsive to manipulation of O-GlcNAc signaling, indicating that the specific mechanisms underlying this process (e.g. the OGA-p300-C/EBPβ axis described here) likely differ among cell types. We speculate that cell type–specific genetic programs ultimately determine the precise mechanisms controlling mutual regulation of OGT and OGA in different cellular contexts.
Mutual regulation of OGT and OGA may also occur at levels beyond transcription. For instance, Park et al. (16) identified a post-transcriptional mechanism that differentially regulates splicing of OGT pre-mRNA in response to changes in O-GlcNAc homeostasis. In this pathway, high cellular O-GlcNA cylation levels promote OGT intron retention and subsequent mRNA degradation, whereas low cellular O-GlcNAcylation levels enhance the formation of fully spliced OGT mRNA. Intriguingly, deletion of the OGT intronic splicing silencer in colon cancer cells increases the expression of both OGT and OGA and renders these cells highly sensitive to TMG treatment, suggesting that compensatory suppression of OGT expression in response to OGA inhibition is essential for cell survival (16). In future studies, it would be interesting to determine whether OGA plays a direct role in regulating OGT pre-mRNA splicing and intron retention as it does in modulating Ogt transcription. In addition to transcriptional and post-transcriptional mechanisms, mutual regulation of OGT and OGA may also occur at the post-translational level because OGT and OGA have themselves been identified as O-GlcNAcylated proteins (17, 18). Post-translational regulation of OGT and OGA activity and/or stability would allow cells to respond more rapidly to perturbations in O-GlcNAc homeostasis than would transcriptional or post-transcriptional regulation of OGT and OGA expression; thus, elucidating these post-translational mechanisms will be an important area for future investigation in the field.
In addition to mutual regulation of OGT and OGA, transcription factors that coregulate OGT and OGA expression may also contribute to maintenance of O-GlcNAc homeostasis. For instance, Muthusamy et al. (48) demonstrated that the transcription factor E2F1 binds to the promoters of both OGT and OGA and suppresses their expression, raising the possibility that E2F1 can concurrently repress OGT and OGA transcription to maintain a balance between OGT and OGA mRNA levels. Notably, ENCODE ChIP-seq analysis also identified E2F1 as one of the most abundant transcription factors at the human OGT promoter (Fig. S1A).
By analyzing TCGA gene expression data, we found that OGT and OGA mRNA levels are highly correlated across a wide range of human cancers. We then used pancreatic cancer as a model to demonstrate that oncogenic signaling pathways such as the MAPK/ERK pathway have the potential to impinge upon OGA-mediated Ogt transcription to increase OGT expression. One important question for future investigation is whether ERK-dependent regulation of OGT expression is a feature of cancer types other than pancreatic adenocarcinoma, which may be particularly reliant on ERK signaling because >90% possess gain-of-function mutations in KRAS (40). Intriguingly, many of the other cancer types with the highest correlation coefficients for OGT versus OGA expression are also characterized by frequent activating mutations in the MAPK/ERK signaling pathway (e.g. thyroid carcinoma, skin cutaneous melanoma) (Fig. 3E) (49, 50). Furthermore, a previous study by Zhang et al. (51) demonstrated that ERK signaling also stimulates OGT expression in lung and prostate cancer cells. These findings suggest that ERK-dependent regulation of OGT expression is not limited to pancreatic adenocarcinoma and may in fact be a feature of a wide range of human cancers.
Because inhibition of MEK reduces total cellular OGA activity in PDAC cells, we propose that ERK signaling regulates OGT expression at least in part by modulating OGA glycosidase activity. In future studies, it will be important to determine whether ERK regulates OGA activity through direct or indirect mechanisms. Interestingly, a chemical genetic screen for ERK2 substrates based on engineered analog-sensitive kinases identified OGA as a novel target of ERK2, raising the possibility that ERK can modulate OGA activity through direct phosphorylation (52). Alternative mechanisms through which ERK signaling may regulate OGT expression include modulation of C/EBPβ DNA-binding activity. Indeed, removal of O-GlcNAc from serines 180 and 181 of C/EBPβ leads to phosphorylation of neighboring threonine 188 by ERK, which in turn promotes C/EBPβ DNA binding (31, 53). Thus, OGA and ERK may cooperatively stimulate OGT transcription by modulating the interplay between O-GlcNAcylation and phosphorylation on the regulatory domain of C/EBPβ. Attenuation of oncogenic ERK signaling in PDAC cells via loss of KrasG12D transgene expression has also been shown to reduce cellular O-GlcNAcylation levels by decreasing Gfpt1 expression, indicating that ERK signaling can alter O-GlcNAc homeostasis in PDAC through a variety of different mechanisms (40).
Increases in OGT expression and decreases in OGA expression have been reported in a number of human cancers, putting forth the idea that uncoupling of OGT and OGA expression is responsible for elevated O-GlcNAcylation levels in cancer cells (21, 24, 41). However, in the present study and our previous work, we utilized large-scale analysis of human cancer data to show that OGT and OGA levels are in fact positively correlated and concomitantly up-regulated in most human cancer types.4 We also demonstrate here that the OGA–C/EBPβ axis that regulates Ogt transcription in normal cells appears to be retained in PDAC cells. These results raise an important question: how are O-GlcNAcylation levels increased in cancer cells if both OGT and OGA levels are elevated? Our previous work suggests that, under high-glucose conditions, OGA may in fact promote the O-GlcNAcylation of specific substrates via its cryptic acetyltransferase activity, raising the possibility that OGT and OGA can act cooperatively on key proteins to promote cell proliferation.4 Consistent with this hypothesis, OGA overexpression enhances tumor growth in a xenograft model,4 and partial loss of OGA suppresses intestinal tumorigenesis in Apcmin/+ mice (25). Thus, we propose that mutual regulation of OGT and OGA at the transcriptional level serves to maintain O-GlcNAc homeostasis in normal cells but can be utilized by cancer cells to drive increased expression of both OGT and OGA and thereby promote tumor growth.
Experimental procedures
Cell culture
Primary hepatocytes were isolated from WT and Ogt-floxed C57BL/6 mice by the Yale Liver Center Core Facility and cultured in Dulbecco's modified Eagle's medium with 10% fetal bovine serum (FBS), 2 mm sodium pyruvate, 1 μm dexamethasone, and 0.1 μm insulin on collagen I-coated plates. CV-1 and HEK 293T cells were cultured in Dulbecco's modified Eagle's medium with 10% FBS. PDAC cells were generously provided by Dr. Alec Kimmelman (New York University) and cultured in RPMI 1640 medium with 10% FBS and 1 μg/ml doxycycline to maintain KrasG12D transgene expression (40). The cells were transfected using FuGENE HD transfection reagent (Promega). The cells were treated with TMG (Carbosynth) and PD0325901 (Sigma) as indicated.
Adenoviruses and plasmids
Recombinant adenoviruses were generated using the AdEasy system (54). Ogt and Oga promoter luciferase reporter constructs were generated by cloning the indicated regions of the mouse Ogt and Oga promoters into the pGL3-Basic luciferase reporter vector (Promega). OGT and OGA expression vectors were generated by cloning FLAG/HA-rOGT and 3*FLAG/2*Myc-mOGA into the pAdTrack-CMV plasmid. pCMVβ-p300-myc (catalog no. 30489) and pCMV-FLAG-LIP (catalog no. 15737) plasmids were purchased from Addgene (55). pcDNA3.1-FLAG-LAP WT-HA and pcDNA3.1-FLAG-LAP SSAA-HA plasmids were generously provided by Dr. Qi-Qun Tang (Fudan University) (31). Plasmids with point mutations (pcDNA3.1-FLAG-LAP KR-HA and MT Ogt promoter luciferase reporter construct) were generated using the QuikChange XL II site-directed mutagenesis kit (Agilent).
Quantitative reverse transcription–PCR
Total RNA was isolated using TRIzol Reagent (Invitrogen). cDNA was synthesized using the iScript cDNA synthesis kit (Bio-Rad). cDNA was amplified and quantified using iQ SYBR Green Supermix (Bio-Rad) and the LightCycler 480 system (Roche). All data were normalized to 36b4 expression. The primer sequences were as follows: 36b4-F, 5′-AGA TGC AGC AGA TCC GCA T-3′; 36b4-R, 5′-GTT CTT GCC CAT CAG CAC C-3′; Ogt-F, 5′-AAG AGG CAC GCA TTT TTG AC-3′; Ogt-R, 5′-ATG GGG TTG CAG TTC GAT AG-3′; Oga-F, 5′-CTC AGA GGC TGA GAA AAT AAT GTT GAG-3′; and Oga-R, 5′-AAG GGA AGT TGG CAA GGA AAG T-3′.
Luciferase assay
The cells were seeded in 48-well plates for 24 h and then transfected with 100 ng of each plasmid (luciferase reporter construct and expression vectors). After 48 h, the cells were lysed using passive lysis buffer (Promega), and luciferase assay was performed according to the manufacturer's protocol.
Western blotting
Anti-FLAG (F3165), anti-α-tubulin (T5168), and anti-β-actin (A5441) antibodies were purchased from Sigma. Anti-OGT (ab96718) and anti-β-tubulin (ab15568) antibodies were purchased from Abcam. Anti-p-ERK (9106) and anti-ERK (9102) antibodies were purchased from Cell Signaling. Anti-OGA (NBP1–96679) antibody was purchased from Novus. The cells were lysed in passive lysis buffer (Promega) or 1% Nonidet P-40, 50 mm Tris-HCl (pH 8.0), 150 mm NaCl, 0.1 mm EDTA, and protease and phosphatase inhibitors. Equal amounts of whole cell lysate were electrophoresed on SDS–PAGE gels and transferred to polyvinylidene difluoride membranes. Membranes were incubated with primary antibodies overnight at 4 °C. Western blotting visualization was performed using horseradish peroxidase–conjugated secondary antibodies and ECL substrate. Western blotting quantification was performed using Adobe Photoshop.
Chromatin immunoprecipitation
ChIP was performed as described previously (56, 57). Briefly, the cells grown to confluence in 10-cm dishes were cross-linked with 1% formaldehyde for 8 min and quenched with glycine (final concentration, 125 mm). The cells were then lysed and sonicated using a Misonix Sonicator 3000 (20 cycles, 7 s on, 45 s off, output level 5). Sheared chromatin was immunoprecipitated by overnight incubation with an anti-C/EBPβ antibody (sc-150, Santa Cruz), and antibody-chromatin complexes were captured by a 1-h incubation with protein A/G beads (sc-2003, Santa Cruz). Immunoprecipitated and input chromatin was de-cross-linked overnight and purified by phenol-chloroform extraction and ethanol precipitation. Chromatin samples were then analyzed by qPCR. All data were normalized to input DNA. Promoter primers amplify a 124-bp region containing the C/EBPβ-binding site in Fig. S1D, and intron primers target a 73-bp region in intron 11-12. The primer sequences were as follows: promoter-F, 5′-CCT AGA CAG GGT TGT CGC AT-3′; promoter-R, 5′-GCG CGT AAC AAG ACT ACC GA-3′; intron-F, 5′-TCG TTG TCA GGT ACG CAA GG-3′; and intron-R, 5′-AGG CAA TCT CCT ACA AGC ATG G-3′.
OGA activity assay
OGA activity assay was performed as described previously (58). Briefly, duplicate 50-μl reactions containing whole cell lysate, 50 mm sodium cacodylate, pH 6.4, 0.3% BSA, 100 mm N-acetyl-d-galactosamine (GalNAc), and 1 mm 4-methylumbelliferyl (4MU)–GlcNAc or 4MU–GalNAc (Sigma) were set up in black flat-bottomed 96-well plates. The reactions were incubated at 37 °C for 1–2 h and quenched with glycine, pH 10.75 (final concentration, 150 mm). Fluorescence intensity was measured using a Molecular Devices Spectramax M5 microplate reader (excitation, 368 nm; emission, 450 nm; sensitivity, 50). OGA activity was calculated by subtracting 4MU–GalNAc fluorescence from 4MU–GlcNAc fluorescence to exclude lysosomal hexoaminidase activity.
Gene expression data set analysis
Analysis of top genes coexpressed with OGT or OGA in human, mouse, and rat tissues was performed using COXPRESdb (26). TCGA gene expression and genomic alteration data were analyzed using cBioPortal for Cancer Genomics (37, 38). Normal pancreas versus PDAC gene expression data were analyzed using Oncomine (Thermo Fisher).
Immunohistochemistry
After baking on a panel at 70 °C, formalin-fixed paraffin-embedded tissue samples were deparaffinized with xylenes and rehydrated through gradient ethanol immersion. Endogenous peroxidase activity was quenched by incubation with 3% hydrogen peroxide in methanol for 12 min, followed by three 3-min washes with PBS. The sections were then blocked with 10% normal goat serum in PBS for 25 min, followed by overnight incubation with primary antibody in a moist chamber at 4 °C. Primary antibodies were diluted in PBS containing 1% BSA and preimmune serum was substituted for primary antibody as a negative control. After three 5-min washes with PBS, the sections were incubated with secondary antibody for 25 min at room temperature, followed by three additional 5-min washes with PBS. Reaction products were visualized with diaminobenzidine (ZLI-9032, ZSGB) at room temperature. After being counterstained with Harris hematoxylin (ZLI-9039, ZSGB) for 4 min and rinsed with tap water, the sections were immediately dehydrated by sequential immersion in gradient ethanol and xylenes and mounted with resin and cover slips. Images were obtained using an Olympus BX51 light microscope equipped with a DP70 digital camera. For evaluation of cell staining, sections were examined by two independent observers without prior knowledge of the clinical or clinicopathological status of the specimens. Antigen expression levels were stratified into negative (−), low (+), or high (++) by optical evaluation. Staining of less than 10% of the cancer cells in a core was classified as negative.
Statistical analysis
Statistical analysis was performed as indicated in the figure legends using GraphPad Prism 7.
Author contributions
K. Q., M.-D. L., H.-B. R., and X. Y. conceptualization; K. Q., S. W., M. F., J. Z., J. P. S., Y. N., and X. Y. data curation; K. Q. software; K. Q., S. W., J. Z., and X. Y. formal analysis; K. Q., S. W., and M. F. validation; K. Q., S. W., M. F., J. Z., J. P. S., M.-D. L., Y. Y., K. Z., J. W., Y. N., H.-B. R., and X. Y. investigation; K. Q., S. W., M. F., J. Z., J. P. S., M.-D. L., Y. Y., K. Z., J. W., Y. N., H.-B. R., and X. Y. methodology; K. Q. and X. Y. writing-original draft; K. Q. and X. Y. writing-review and editing; Y. N. and X. Y. resources; Y. N. and X. Y. funding acquisition; H.-B. R. and X. Y. supervision; X. Y. project administration.
Supplementary Material
Acknowledgments
We thank Dr. Qi-Qun Tang (Fudan University) for generously providing the LAP WT and LAP SSAA plasmids and Dr. Alec Kimmelman (New York University) for generously providing the PDAC cell line. We thank all members of the Yang laboratory for stimulating discussions.
This work was supported by National Institutes of Health Grants R01DK089098, R01DK102648, and P01DK057751; American Cancer Society Grant RSG-14-244-01-TBE; and State of Connecticut Grant DPH2014-0139 (to X. Y.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Tables S1–S7 and Figs. S1–S3.
K. Qian, S. Wang, M. Fu, J. Zhou, J. P. Singh, M.-D. Li, Y. Yang, K. Zhang, J. Wu, Y. Nie, H.-B. Ruan, and X. Yang, unpublished results.
- O-GlcNAc
- O-linked GlcNAc
- OGT
- O-GlcNAc transferase
- OGA
- O-GlcNAcase
- C/EBP
- CCAAT/enhancer-binding protein
- PDAC
- pancreatic ductal adenocarcinoma
- ERK
- extracellular signal–regulated kinase
- HEK
- human embryonic kidney
- TMG
- Thiamet G
- ChIP-seq
- ChIP sequencing
- LAP
- liver-enriched activator protein
- LIP
- liver-enriched inhibitor protein
- TCGA
- The Cancer Genome Atlas
- MAPK
- mitogen-activated protein kinase
- MEK
- MAPK/ERK kinase
- FBS
- fetal bovine serum
- 4MU
- 4-methylumbelliferyl
- GalNAc
- N-acetyl-d-galactosamine
- qPCR
- quantitative PCR.
References
- 1. Hart G. W., Housley M. P., and Slawson C. (2007) Cycling of O-linked β-N-acetylglucosamine on nucleocytoplasmic proteins. Nature 446, 1017–1022 10.1038/nature05815 [DOI] [PubMed] [Google Scholar]
- 2. Yang X., and Qian K. (2017) Protein O-GlcNAcylation: emerging mechanisms and functions. Nat. Rev. Mol. Cell Biol. 18, 452–465 10.1038/nrm.2017.22,10.1038/nrn.2017.89,10.1038/nrn.2017.87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hart G. W., Slawson C., Ramirez-Correa G., and Lagerlof O. (2011) Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annu. Rev. Biochem. 80, 825–858 10.1146/annurev-biochem-060608-102511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hanover J. A., Krause M. W., and Love D. C. (2012) Bittersweet memories: linking metabolism to epigenetics through O-GlcNAcylation. Nat. Rev. Mol. Cell Biol. 13, 312–321 10.1038/nrm3334 [DOI] [PubMed] [Google Scholar]
- 5. Ruan H. B., Singh J. P., Li M. D., Wu J., and Yang X. (2013) Cracking the O-GlcNAc code in metabolism. Trends Endocrinol. Metab. 24, 301–309 10.1016/j.tem.2013.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bond M. R., and Hanover J. A. (2015) A little sugar goes a long way: the cell biology of O-GlcNAc. J. Cell Biol. 208, 869–880 10.1083/jcb.201501101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ruan H. B., Han X., Li M. D., Singh J. P., Qian K., Azarhoush S., Zhao L., Bennett A. M., Samuel V. T., Wu J., Yates J. R. 3rd, and Yang X. (2012) O-GlcNAc transferase/host cell factor C1 complex regulates gluconeogenesis by modulating PGC-1α stability. Cell Metab. 16, 226–237 10.1016/j.cmet.2012.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li M. D., Ruan H. B., Hughes M. E., Lee J. S., Singh J. P., Jones S. P., Nitabach M. N., and Yang X. (2013) O-GlcNAc signaling entrains the circadian clock by inhibiting BMAL1/CLOCK ubiquitination. Cell Metab. 17, 303–310 10.1016/j.cmet.2012.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ruan H. B., Ma Y., Torres S., Zhang B., Feriod C., Heck R. M., Qian K., Fu M., Li X., Nathanson M. H., Bennett A. M., Nie Y., Ehrlich B. E., and Yang X. (2017) Calcium-dependent O-GlcNAc signaling drives liver autophagy in adaptation to starvation. Genes Dev. 31, 1655–1665 10.1101/gad.305441.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yi W., Clark P. M., Mason D. E., Keenan M. C., Hill C., Goddard W. A. 3rd, Peters E. C., Driggers E. M., and Hsieh-Wilson L. C. (2012) Phosphofructokinase 1 glycosylation regulates cell growth and metabolism. Science 337, 975–980 10.1126/science.1222278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yang X., Ongusaha P. P., Miles P. D., Havstad J. C., Zhang F., So W. V., Kudlow J. E., Michell R. H., Olefsky J. M., Field S. J., and Evans R. M. (2008) Phosphoinositide signalling links O-GlcNAc transferase to insulin resistance. Nature 451, 964–969 10.1038/nature06668 [DOI] [PubMed] [Google Scholar]
- 12. Ruan H. B., Dietrich M. O., Liu Z. W., Zimmer M. R., Li M. D., Singh J. P., Zhang K., Yin R., Wu J., Horvath T. L., and Yang X. (2014) O-GlcNAc transferase enables AgRP neurons to suppress browning of white fat. Cell 159, 306–317 10.1016/j.cell.2014.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Erickson J. R., Pereira L., Wang L., Han G., Ferguson A., Dao K., Copeland R. J., Despa F., Hart G. W., Ripplinger C. M., and Bers D. M. (2013) Diabetic hyperglycaemia activates CaMKII and arrhythmias by O-linked glycosylation. Nature 502, 372–376 10.1038/nature12537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yuzwa S. A., Shan X., Macauley M. S., Clark T., Skorobogatko Y., Vosseller K., and Vocadlo D. J. (2012) Increasing O-GlcNAc slows neurodegeneration and stabilizes Tau against aggregation. Nat. Chem. Biol. 8, 393–399 10.1038/nchembio.797 [DOI] [PubMed] [Google Scholar]
- 15. Zhang Z., Tan E. P., VandenHull N. J., Peterson K. R., and Slawson C. (2014) O-GlcNAcase expression is sensitive to changes in O-GlcNAc homeostasis. Front. Endocrinol. (Lausanne) 5, 206 10.3389/fendo.2014.00206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Park S. K., Zhou X., Pendleton K. E., Hunter O. V., Kohler J. J., O'Donnell K. A., and Conrad N. K. (2017) A conserved splicing silencer dynamically regulates O-GlcNAc transferase intron retention and O-GlcNAc homeostasis. Cell Rep. 20, 1088–1099 10.1016/j.celrep.2017.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kreppel L. K., Blomberg M. A., and Hart G. W. (1997) Dynamic glycosylation of nuclear and cytosolic proteins: cloning and characterization of a unique O-GlcNAc transferase with multiple tetratricopeptide repeats. J. Biol. Chem. 272, 9308–9315 10.1074/jbc.272.14.9308 [DOI] [PubMed] [Google Scholar]
- 18. Khidekel N., Ficarro S. B., Clark P. M., Bryan M. C., Swaney D. L., Rexach J. E., Sun Y. E., Coon J. J., Peters E. C., and Hsieh-Wilson L. C. (2007) Probing the dynamics of O-GlcNAc glycosylation in the brain using quantitative proteomics. Nat. Chem. Biol. 3, 339–348 10.1038/nchembio881 [DOI] [PubMed] [Google Scholar]
- 19. Mi W., Gu Y., Han C., Liu H., Fan Q., Zhang X., Cong Q., and Yu W. (2011) O-GlcNAcylation is a novel regulator of lung and colon cancer malignancy. Biochim. Biophys. Acta 1812, 514–519 10.1016/j.bbadis.2011.01.009 [DOI] [PubMed] [Google Scholar]
- 20. Lynch T. P., Ferrer C. M., Jackson S. R., Shahriari K. S., Vosseller K., and Reginato M. J. (2012) Critical role of O-linked β-N-acetylglucosamine transferase in prostate cancer invasion, angiogenesis, and metastasis. J. Biol. Chem. 287, 11070–11081 10.1074/jbc.M111.302547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Krzeslak A., Forma E., Bernaciak M., Romanowicz H., and Brys M. (2012) Gene expression of O-GlcNAc cycling enzymes in human breast cancers. Clin. Exp. Med. 12, 61–65 10.1007/s10238-011-0138-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Champattanachai V., Netsirisawan P., Chaiyawat P., Phueaouan T., Charoenwattanasatien R., Chokchaichamnankit D., Punyarit P., Srisomsap C., and Svasti J. (2013) Proteomic analysis and abrogated expression of O-GlcNAcylated proteins associated with primary breast cancer. Proteomics 13, 2088–2099 10.1002/pmic.201200126 [DOI] [PubMed] [Google Scholar]
- 23. Ferrer C. M., Sodi V. L., and Reginato M. J. (2016) O-GlcNAcylation in cancer biology: linking metabolism and signaling. J. Mol. Biol. 428, 3282–3294 10.1016/j.jmb.2016.05.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ferrer C. M., Lynch T. P., Sodi V. L., Falcone J. N., Schwab L. P., Peacock D. L., Vocadlo D. J., Seagroves T. N., and Reginato M. J. (2014) O-GlcNAcylation regulates cancer metabolism and survival stress signaling via regulation of the HIF-1 pathway. Mol. Cell 54, 820–831 10.1016/j.molcel.2014.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yang Y. R., Jang H. J., Yoon S., Lee Y. H., Nam D., Kim I. S., Lee H., Kim H., Choi J. H., Kang B. H., Ryu S. H., and Suh P. G. (2014) OGA heterozygosity suppresses intestinal tumorigenesis in Apc(min/+) mice. Oncogenesis 3, e109 10.1038/oncsis.2014.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Okamura Y., Aoki Y., Obayashi T., Tadaka S., Ito S., Narise T., and Kinoshita K. (2015) COXPRESdb in 2015: coexpression database for animal species by DNA-microarray and RNAseq-based expression data with multiple quality assessment systems. Nucleic Acids Res. 43, D82–D86 10.1093/nar/gku1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yuzwa S. A., Macauley M. S., Heinonen J. E., Shan X., Dennis R. J., He Y., Whitworth G. E., Stubbs K. A., McEachern E. J., Davies G. J., and Vocadlo D. J. (2008) A potent mechanism-inspired O-GlcNAcase inhibitor that blocks phosphorylation of Tau in vivo. Nat. Chem. Biol. 4, 483–490 10.1038/nchembio.96 [DOI] [PubMed] [Google Scholar]
- 28. Creyghton M. P., Cheng A. W., Welstead G. G., Kooistra T., Carey B. W., Steine E. J., Hanna J., Lodato M. A., Frampton G. M., Sharp P. A., Boyer L. A., Young R. A., and Jaenisch R. (2010) Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. U.S.A. 107, 21931–21936 10.1073/pnas.1016071107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jin Q., Yu L. R., Wang L., Zhang Z., Kasper L. H., Lee J. E., Wang C., Brindle P. K., Dent S. Y., and Ge K. (2011) Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 30, 249–262 10.1038/emboj.2010.318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Josefowicz S. Z., Shimada M., Armache A., Li C. H., Miller R. M., Lin S., Yang A., Dill B. D., Molina H., Park H. S., Garcia B. A., Taunton J., Roeder R. G., and Allis C. D. (2016) Chromatin kinases act on transcription factors and histone tails in regulation of inducible transcription. Mol. Cell 64, 347–361 10.1016/j.molcel.2016.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li X., Molina H., Huang H., Zhang Y. Y., Liu M., Qian S. W., Slawson C., Dias W. B., Pandey A., Hart G. W., Lane M. D., and Tang Q. Q. (2009) O-Linked N-acetylglucosamine modification on CCAAT enhancer-binding protein β: role during adipocyte differentiation. J. Biol. Chem. 284, 19248–19254 10.1074/jbc.M109.005678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ceseña T. I., Cardinaux J. R., Kwok R., and Schwartz J. (2007) CCAAT/enhancer-binding protein (C/EBP) β is acetylated at multiple lysines: acetylation of C/EBPβ at lysine 39 modulates its ability to activate transcription. J. Biol. Chem. 282, 956–967 10.1074/jbc.M511451200 [DOI] [PubMed] [Google Scholar]
- 33. Shen T., Horwitz K. B., and Lange C. A. (2001) Transcriptional hyperactivity of human progesterone receptors is coupled to their ligand-dependent down-regulation by mitogen-activated protein kinase-dependent phosphorylation of serine 294. Mol. Cell. Biol. 21, 6122–6131 10.1128/MCB.21.18.6122-6131.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Giordano A., and Avantaggiati M. L. (1999) p300 and CBP: partners for life and death. J. Cell. Physiol. 181, 218–230 10.1002/(SICI)1097-4652(199911)181:2%3C218::AID-JCP4%3E3.0.CO%3B2-5 [DOI] [PubMed] [Google Scholar]
- 35. Kajimura S., Seale P., Kubota K., Lunsford E., Frangioni J. V., Gygi S. P., and Spiegelman B. M. (2009) Initiation of myoblast to brown fat switch by a PRDM16-C/EBP-β transcriptional complex. Nature 460, 1154–1158 10.1038/nature08262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Akira S., Isshiki H., Sugita T., Tanabe O., Kinoshita S., Nishio Y., Nakajima T., Hirano T., and Kishimoto T. (1990) A nuclear factor for IL-6 expression (NF-IL6) is a member of a C/EBP family. EMBO J. 9, 1897–1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cerami E., Gao J., Dogrusoz U., Gross B. E., Sumer S. O., Aksoy B. A., Jacobsen A., Byrne C. J., Heuer M. L., Larsson E., Antipin Y., Reva B., Goldberg A. P., Sander C., and Schultz N. (2012) The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gao J., Aksoy B. A., Dogrusoz U., Dresdner G., Gross B., Sumer S. O., Sun Y., Jacobsen A., Sinha R., Larsson E., Cerami E., Sander C., and Schultz N. (2013) Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, pl1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jemal A., Siegel R., Xu J., and Ward E. (2010) Cancer statistics, 2010. CA Cancer J. Clin. 60, 277–300 10.3322/caac.20073 [DOI] [PubMed] [Google Scholar]
- 40. Ying H., Kimmelman A. C., Lyssiotis C. A., Hua S., Chu G. C., Fletcher-Sananikone E., Locasale J. W., Son J., Zhang H., Coloff J. L., Yan H., Wang W., Chen S., Viale A., et al. (2012) Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell 149, 656–670 10.1016/j.cell.2012.01.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ma Z., Vocadlo D. J., and Vosseller K. (2013) Hyper-O-GlcNAcylation is anti-apoptotic and maintains constitutive NF-κB activity in pancreatic cancer cells. J. Biol. Chem. 288, 15121–15130 10.1074/jbc.M113.470047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang T., Yu Q., Li J., Hu B., Zhao Q., Ma C., Huang W., Zhuo L., Fang H., Liao L., Eugene Chin Y., and Jiang Y. (2017) O-GlcNAcylation of fumarase maintains tumour growth under glucose deficiency. Nat. Cell Biol. 19, 833–843 10.1038/ncb3562 [DOI] [PubMed] [Google Scholar]
- 43. Bain J., Plater L., Elliott M., Shpiro N., Hastie C. J., McLauchlan H., Klevernic I., Arthur J. S., Alessi D. R., and Cohen P. (2007) The selectivity of protein kinase inhibitors: a further update. Biochem. J. 408, 297–315 10.1042/BJ20070797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhang E. R., Liu S., Wu L. F., Altschuler S. J., and Cobb M. H. (2016) Chemoattractant concentration-dependent tuning of ERK signaling dynamics in migrating neutrophils. Sci. Signal. 9, ra122 10.1126/scisignal.aag0486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Taylor R. P., Parker G. J., Hazel M. W., Soesanto Y., Fuller W., Yazzie M. J., and McClain D. A. (2008) Glucose deprivation stimulates O-GlcNAc modification of proteins through up-regulation of O-linked N-acetylglucosaminyltransferase. J. Biol. Chem. 283, 6050–6057 10.1074/jbc.M707328200 [DOI] [PubMed] [Google Scholar]
- 46. Cheung W. D., and Hart G. W. (2008) AMP-activated protein kinase and p38 MAPK activate O-GlcNAcylation of neuronal proteins during glucose deprivation. J. Biol. Chem. 283, 13009–13020 10.1074/jbc.M801222200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Taylor R. P., Geisler T. S., Chambers J. H., and McClain D. A. (2009) Up-regulation of O-GlcNAc transferase with glucose deprivation in HepG2 cells is mediated by decreased hexosamine pathway flux. J. Biol. Chem. 284, 3425–3432 10.1074/jbc.M803198200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Muthusamy S., Hong K. U., Dassanayaka S., Hamid T., and Jones S. P. (2015) E2F1 transcription factor regulates O-linked N-acetylglucosamine (O-GlcNAc) transferase and O-GlcNAcase expression. J. Biol. Chem. 290, 31013–31024 10.1074/jbc.M115.677534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cohen Y., Xing M., Mambo E., Guo Z., Wu G., Trink B., Beller U., Westra W. H., Ladenson P. W., and Sidransky D. (2003) BRAF mutation in papillary thyroid carcinoma. J. Natl. Cancer Inst. 95, 625–627 10.1093/jnci/95.8.625 [DOI] [PubMed] [Google Scholar]
- 50. Burotto M., Chiou V. L., Lee J. M., and Kohn E. C. (2014) The MAPK pathway across different malignancies: a new perspective. Cancer 120, 3446–3456 10.1002/cncr.28864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhang X., Ma L., Qi J., Shan H., Yu W., and Gu Y. (2015) MAPK/ERK signaling pathway-induced hyper-O-GlcNAcylation enhances cancer malignancy. Mol. Cell. Biochem. 410, 101–110 10.1007/s11010-015-2542-8 [DOI] [PubMed] [Google Scholar]
- 52. Carlson S. M., Chouinard C. R., Labadorf A., Lam C. J., Schmelzle K., Fraenkel E., and White F. M. (2011) Large-scale discovery of ERK2 substrates identifies ERK-mediated transcriptional regulation by ETV3. Sci. Signal. 4, rs11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tang Q. Q., Grønborg M., Huang H., Kim J. W., Otto T. C., Pandey A., and Lane M. D. (2005) Sequential phosphorylation of CCAAT enhancer-binding protein β by MAPK and glycogen synthase kinase 3β is required for adipogenesis. Proc. Natl. Acad. Sci. U.S.A. 102, 9766–9771 10.1073/pnas.0503891102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Luo J., Deng Z. L., Luo X., Tang N., Song W. X., Chen J., Sharff K. A., Luu H. H., Haydon R. C., Kinzler K. W., Vogelstein B., and He T. C. (2007) A protocol for rapid generation of recombinant adenoviruses using the AdEasy system. Nat. Protoc. 2, 1236–1247 10.1038/nprot.2007.135 [DOI] [PubMed] [Google Scholar]
- 55. Gomis R. R., Alarcón C., Nadal C., Van Poznak C., and Massagué J. (2006) C/EBPβ at the core of the TGFβ cytostatic response and its evasion in metastatic breast cancer cells. Cancer Cell 10, 203–214 10.1016/j.ccr.2006.07.019 [DOI] [PubMed] [Google Scholar]
- 56. Ferrari A., Longo R., Fiorino E., Silva R., Mitro N., Cermenati G., Gilardi F., Desvergne B., Andolfo A., Magagnotti C., Caruso D., Fabiani E., Hiebert S. W., and Crestani M. (2017) HDAC3 is a molecular brake of the metabolic switch supporting white adipose tissue browning. Nat. Commun. 8, 93 10.1038/s41467-017-00182-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rajbhandari P., Thomas B. J., Feng A. C., Hong C., Wang J., Vergnes L., Sallam T., Wang B., Sandhu J., Seldin M. M., Lusis A. J., Fong L. G., Katz M., Lee R., Young S. G., et al. (2018) IL-10 signaling remodels adipose chromatin architecture to limit thermogenesis and energy expenditure. Cell 172, 218–233.e17 10.1016/j.cell.2017.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Groves J. A., Maduka A. O., O'Meally R. N., Cole R. N., and Zachara N. E. (2017) Fatty acid synthase inhibits the O-GlcNAcase during oxidative stress. J. Biol. Chem. 292, 6493–6511 10.1074/jbc.M116.760785 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.