Abstract
ScARP from the bacterium Streptomyces coelicolor belongs to the pierisin family of DNA-targeting ADP-ribosyltransferases (ARTs). These enzymes ADP-ribosylate the N2 amino groups of guanine residues in DNA to yield N2-(ADP-ribos-1-yl)-2′-deoxyguanosine. Although the structures of pierisin-1 and Scabin were revealed recently, the substrate recognition mechanisms remain poorly understood because of the lack of a substrate-binding structure. Here, we report the apo structure of ScARP and of ScARP bound to NADH and its GDP substrate at 1.50 and 1.57 Å resolutions, respectively. The bound structure revealed that the guanine of GDP is trapped between N-ribose of NADH and Trp-159. Interestingly, N2 and N3 of guanine formed hydrogen bonds with the OE1 and NE2 atoms of Gln-162, respectively. We directly observed that the ADP-ribosylating toxin turn-turn (ARTT)-loop, including Trp-159 and Gln-162, plays a key role in the specificity of DNA-targeting, guanine-specific ARTs as well as protein-targeting ARTs such as the C3 exoenzyme. We propose that the ARTT-loop recognition is a common substrate-recognition mechanism in the pierisin family. Furthermore, this complex structure sheds light on similarities and differences among two subclasses that are distinguished by conserved structural motifs: H-Y-E in the ARTD subfamily and R-S-E in the ARTC subfamily. The spatial arrangements of the electrophile and nucleophile were the same, providing the first evidence for a common reaction mechanism in these ARTs. ARTC (including ScARP) uses the ARTT-loop for substrate recognition, whereas ARTD (represented by Arr) uses the C-terminal helix instead of the ARTT-loop. These observations could help inform efforts to improve ART inhibitors.
Keywords: ADP-ribosylation, substrate specificity, substrate recognition, structure–function, DNA–protein interaction, crystallography, ADP-ribosylating toxin turn-turn motif, ADP-ribosyltransferase, guanine, pierisin
Introduction
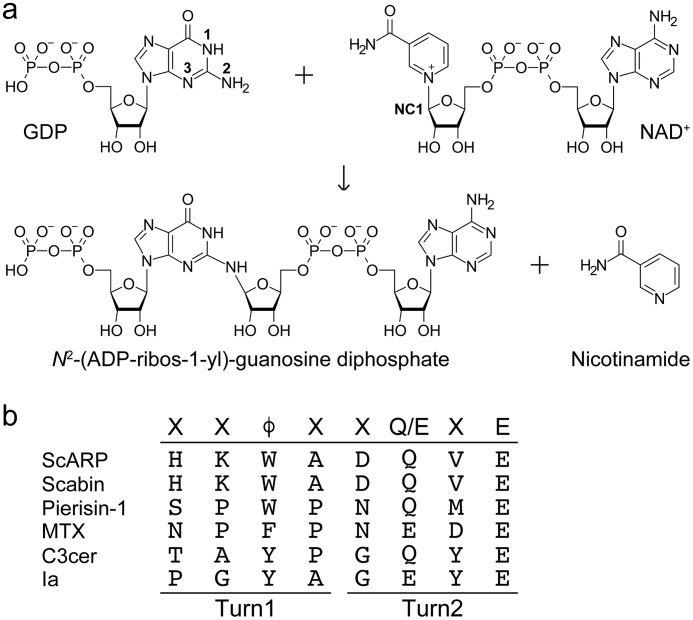
ADP-ribosylation is an important post-translational modification observed in all living organisms. Many bacterial mono-ADP-ribosyltransferases (ARTs)2 are known to attach the ADP-ribosyl moiety to specific target proteins and residues (1, 2). The cholera toxin ADP-ribosylates arginine residues in G proteins (3), whereas the pertussis toxin ADP-ribosylates a cysteine residue (4). Diphtheria toxin and the Pseudomonas aeruginosa exotoxin ADP-ribosylate diphthamide, a modified histidine in elongation factor 2 (5, 6). Furthermore, Clostridium botulinum C3 exoenzyme ADP-ribosylates Asn-41 of RhoA (7, 8) and Clostridium perfringens iota toxin A subunit (Ia) ADP-ribosylates Arg-177 of actin (9, 10). These ARTs fall into two major subclasses that are distinguished by conserved structural motifs: H-Y-E in the ARTD subfamily (related to Diphtheria toxin) and R-S-E in the ARTC subfamily (related to cholera toxin and clostridial toxins (C3 and Ia)). Traditionally, ADP-ribosylation has been considered a protein modification. However, emerging evidence suggests that DNA ADP-ribosylation is also common. The first DNA-targeting ART was found in pierid butterflies and was thus named pierisin (11, 12). Pierisin ADP-ribosylates calf thymus DNA containing dG–dC and N2 amino group of the guanine residue in DNA to yield N2-(ADP-ribos-1-yl)-2′-deoxyguanosine (Fig. 1a) (13, 14). In contrast, the SCO5461 protein (ScARP) from Streptomyces coelicolor was reported to identify an ART that mainly targets mononucleotides and nucleosides and shares 30% homology with pierisin. ScARP ADP-ribosylates deoxyguanosine (dGuo), GMP, dGMP, and cGMP rather than dsDNA (15), whereas pierisin-1 shows weak ADP-ribosylation activity on dGuo. To date, six pierisins (pierisin-1, -1b, and -2–5), ScARP, and Scabin are considered to belong to the pierisin family (15) and are members of the ARTC subfamily. CARP-1, which is present in certain kinds of edible clams, also ADP-ribosylates calf thymus DNA to produce N2-(ADP-ribos-1-yl)-2′-deoxyguanosine (16, 17). However, CARP-1 and pierisins share very little sequence homology, suggesting that they are not derived from a common ancestral gene (16). Other types of DNA-targeting ARTs have also been identified recently. DarT is found as the ADP-ribosyltransferase of thymidines in ssDNA (18). Eukaryotic enzyme poly(ADP-ribose) polymerases (PARPs) can mono-ADP-ribosylate dsDNA ends, which can be reversed by several known ADP-ribosylhydrolases (19–21). This study is the first report of the detailed substrate recognition and ADP-ribosylation mechanisms in the pierisin family DNA-targeting ARTs (22).
Figure 1.
a, ScARP-catalyzed ADP-ribosylation of GDP. b, sequence alignment of ARTT-loop in the pierisin family and other ADP-ribosyltransferases. φ is Tyr, Phe, or Trp.
Only three complex structures of ART toxin with its substrate are available (23–25). Among these, it is well known how C3 exoenzyme recognizes its specific substrate protein (RhoA) and acceptor amino acid via the ARTT-loop. It was proposed that φXXQXE or φXXEXE (where φ is Tyr, Phe, or Trp) known as the ADP-ribosylating toxin turn-turn (ARTT)-loop is important for substrate recognition in C3 and Ia (VIP2), respectively (Fig. 1b) (26). Although it has not been confirmed until recently, we directly observed substrate recognition via the ARTT-loop in the C3–RhoA complex structure (25). Within five loops of the C3 exoenzyme, the ARTT-loop determines the acceptor amino acid. Gln-183 (QXE) on the second turn of the ARTT-loop recognizes Asn-41 of RhoA. Furthermore Tyr-180 on the first turn of the ARTT-loop is also important to recognize the hydrophobic patch of RhoA comprising Val-43, Ala-56, and Trp-58. This interaction facilitates ADP-ribosylation of the specific asparagine of RhoA (25, 27). However, the target residue recognition mechanism of ARTs via the ARTT-loop is still controversial because of the lack of substrate complex structures. Here, we used X-ray crystallography to reveal the ScARP-GDP (substrate)–NADH complex. It shows a common substrate recognition mechanism of pierisin family enzyme and the first direct observation that the ARTT-loop plays a key role in the specificity of DNA-targeting, guanine-specific ART as well as protein-targeting ART. This study also provides the first complete complex structure of small-molecule targeting ART with minimum elements of ADP-ribosylation. Compared with the Arr structure, which is a rifamycin ADP-ribosyltransferase belonging to ARTD, we propose the general significant substrate recognition differences between ARTC and ARTD.
Results
Structures of apo-ScARP and ScARP-GDP (substrate)–NADH
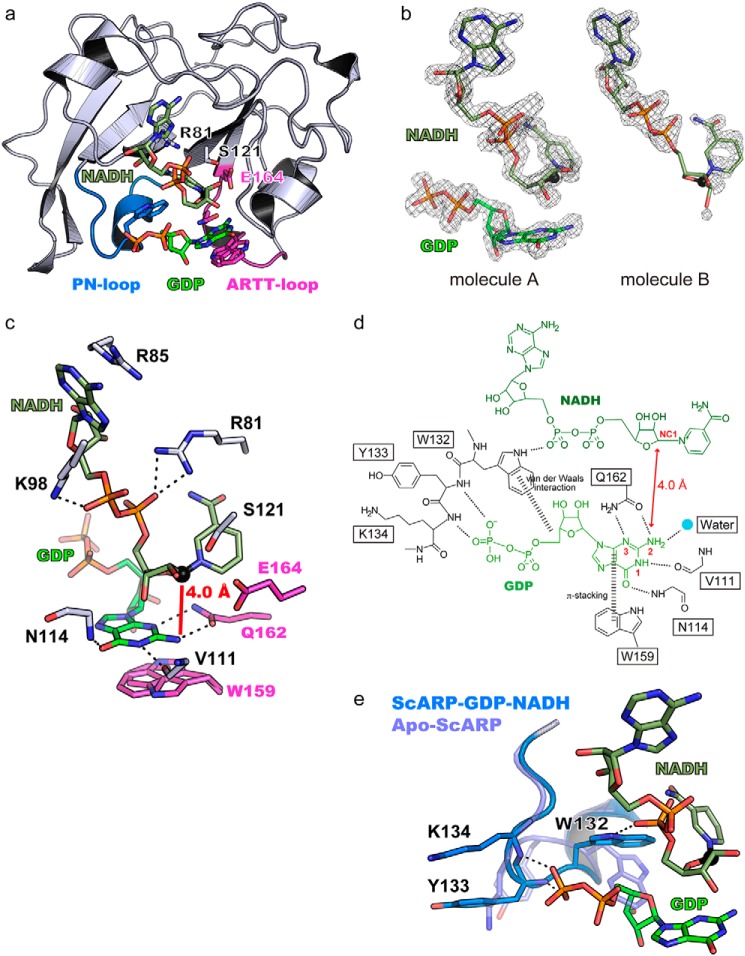
We unraveled the structure of apo-ScARP and ScARP-GDP (substrate)–NADH at 1.50 and 1.57 Å resolutions, respectively (Fig. 2 and Table S1). The apo structure was solved by molecular replacement using another pierisin family protein, Scabin coordinate (PDB code 5daz), which is an ART from Streptomyces scabies (a plant pathogen that causes scab in potatoes). Structural comparison studies revealed similarities between Scabin, pierisin-1, mosquitocidal toxin, and Community-acquired Respiratory Distress Syndrome Toxin (CARDS TX) with RMSD values of 0.4 Å (159 aa), 2.1 Å (146 aa), 2.2 Å (145 aa), and 1.9 Å (127 aa), respectively (28). It also showed less but definite structural similarity to C3 exoenzyme with an RMSD value of 3.7 Å (88 aa). The key features of the ARTC group (R-S-E motif), including cholera toxin, C3, and Ia are conserved in ScARP (Arg-81–Ser-121–Glu-164) and pierisin (Fig. S1) (29). The STS (121–123) motif is just behind the nicotinamide in Fig. 2a. The amide group of nicotinamide is anchored by the Ser-82 backbone carbonyl and amine. The diphosphate of NADH interacts with Arg-81 and Lys-98. Adenine of NADH is retained by cation–π interaction with Arg-85. The unique features of two disulfide bridges (Cys-46-Cys-76, Cys-180–Cys-194) were also conserved as well as those in Scabin (Fig. S2). In the co-crystal structure with two molecules per asymmetric unit, the electron density of NADH and GDP was clearly visible in the A-molecule, but only NADH density was visible in the B-molecule (Fig. 2b). Thus, we described the active site of the A-molecule with NADH and GDP. NAD+ and NADH bind to ART in a similar conformation, but NADH cannot be a substrate to supply the ADP-ribosyl moiety, and thus it works as an inhibitor. The bent conformation of NADH in ScARP was similar to that in other ARTs (30). The GDP (substrate)-bound structure showed that the guanine ring is stacked between the N-ribose of NADH and Trp-159 (two conformations of the indole ring of Trp-159 can be seen, but both of them are parallel to the purine ring of guanine). The O6 and N1 atoms of guanine are fixed by the main-chain amine of Asn-114 (2.8 Å) and the main-chain carbonyl of Val-111 (2.8 Å), respectively. More importantly, the N2 (NH2) and N3 atoms of guanine form hydrogen bonds with OE1 and NE2 of Gln-162, respectively (Fig. 2, c and d). This provides the N2 atom (NH2) of guanine as the acceptor of the ADP-ribosyl moiety with 4.0 Å from the NC1 position of N-ribose. In other words, these binding features guarantee guanine specificity and exclude adenine binding. Notably, Trp-132 on PN-loop is also very important for GDP binding because the PN-loop (Trp-132–Tyr-133–Lys-134–Ser-135–Gly-136) shows the most drastic conformational change upon GDP binding (Fig. 2e). As ribose in GDP (or 2-deoxyribose of DNA) pushes Trp-132, it induces a directional change outward in other PN-loop residues Tyr-133 and Lys-134.
Figure 2.
a, overall structure of ScARP–GDP–NADH. GDP, NADH, the R-S-E motif, and important residues for substrate recognition in the ARTT-loop and PN-loop are shown in a stick model. GDP and NADH are shown in green and faded green, respectively. The ARTT-loop and PN-loop are shown in magenta and marine blue, respectively. b, GDP and NADH Fo − Fc omit electron density maps shown at 3.0σ contour. The GDP electron density map of molecule B is not shown because of the absence of any density. c, residues important for GDP recognition and the R-S-E motif. Dashed lines show hydrogen bonds. The NC1 atom of NADH, which is an electrophile in the ADP-ribosylation reaction, is shown as a black sphere. d, schematic representation of the interactions among ScARP, GDP, and NADH. e, comparison of the PN-loop between apo-ScARP and ScARP–GDP–NADH. The two structures are superimposed.
Structural comparisons with pierisin and Scabin
There is a large structural difference between pierisin and ScARP, in the PN-loop (Fig. S3), suggesting a difference in substrate preference as described under “Discussion.” In pierisin, the long PN-loop and basic cleft were shown to be important for dsDNA by a mutational study (31). Scabin structures were first revealed in apo- and inhibitor-bound forms (32) and very recently in the NADH-bound form (33). A dsDNA (substrate)-bound model was proposed based on the NADH-bound structure. The present GDP–NADH-bound structure contradicts this model as follows: 1) OE1 and NE2 of Gln-162 (QXE of ARTT-loop) form hydrogen bonds with N2 and N3 of guanine (not N2 and N1). 2) The side chain of Trp-132 moves from the apo- to the GDP-bound state (RMSD 4.1 Å) to accept guanosine ribose (Fig. 2, d and e). In other words, the role of Trp-132 seems to be like an adjusting device to accept ribose from guanosine. However, it was suggested that the same tryptophan interacts with other bases in the model of Scabin.
Assay of ADP-ribosylation of GDP using HPLC
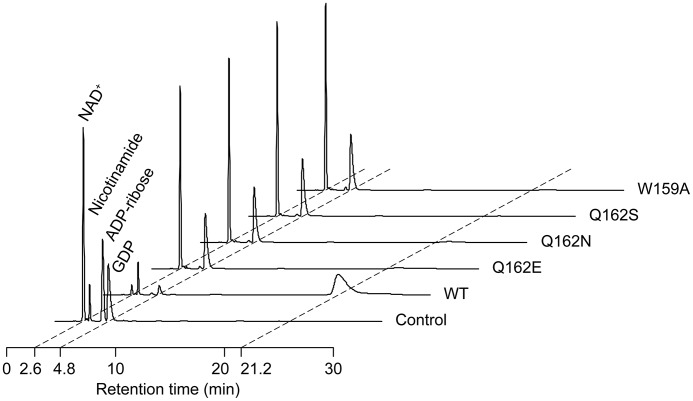
To reveal the role of Gln-162 and Trp-159 on the ARTT-loop, we measured the ADP-ribosylation activity of WT ScARP and the mutants. Single residue mutations of Gln-162 and Trp-159 (Q162E, Q162N, Q162S, and W159A) affected guanine specificity, leading to decreased ADP-ribosylation activity (Fig. 3). Both crystallographic studies and ADP-ribosylation assay revealed the first direct observation that the ARTT-loop is important for specificity in the DNA-targeting guanine-specific ART.
Figure 3.
HPLCs of reaction products with GDP by WT ScARP and the mutants. The retention times of NAD+, nicotinamide, ADP-ribose, and GDP are 2.6, 3.1, 4.3, and 4.8 min, respectively. The reaction product is observed at 21.2 min.
Structural comparison with C3–RhoA complex and ADP-ribosylation reaction mechanism of ScARP
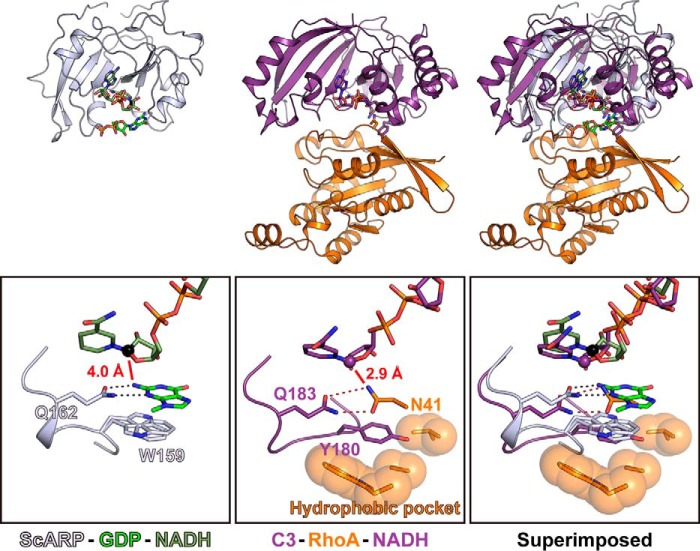
In the case of C3 exoenzyme, the first turn aromatic residue on the ARTT-loop anchors RhoA, and the second turn Gln on the ARTT-loop forms hydrogen bonds with Asn-41 of RhoA, which is the acceptor of ADP-ribose. Surprisingly, the relative positions of NAD(H), the acceptor of ADP-ribose, and Gln of the ARTT-loop are exactly the same position in two different complex structures of C3 and ScARP (Fig. 4). These relative position similarities using the ARTT-loop were conserved between protein-targeting ART and DNA-targeting ART, suggesting their importance for ADP-ribosylation. In guanine-specific ScARP, the reactions are summarized as follows. 1) Trp-159 on the first turn of ARTT-loop and Gln-162 on the second turn of ARTT-loop place the guanine close to the NC1 position of N-ribose, cooperatively. 2) The second turn Glu-164 of the ARTT-loop is important for cleaving NAD+ to produce nicotinamide and the oxocarbenium cation. 3) The oxocarbenium cation is transferred to N2 of guanine and produces N2-(ADP-ribos-1-yl)-2′-deoxyguanosine. It should be noted that the role of Gln-162 is not only to grip N2 of guanine but also to direct the orbital of the lone pair of N2 against NC1. In general, it is reported that an Sn1 reaction occurs via the oxocarbenium cation in ART (34). The short distance (4.0 Å) between NC1 of N-ribose (electrophile) and the N2 atom of guanine (nucleophile) may permit an Sn2 reaction with a direct back attack using the N2 atom as a nucleophile. In the case of Ia-actin, the Sn1 strain-alleviation model was suggested as a possible reaction mechanism based on its complex crystal structure (24, 35). It was reported that ScARP produces only a single isomer of N2-(ADP-ribos-1-yl)-2′-deoxyguanosine and that it anomerized within 4 h. However, which isomer acted as the initial product was an open question (15). This study suggests that the initial product is in the α form, which is then converted to the β form by nonenzymatic anomerization.
Figure 4.
Comparison between ScARP–GDP–NADH and C3–RhoA–NADH (PDB code 4XSH). Close-up views of ARTT-loop are shown in black boxes.
Discussion
Although the overall structures among pierisin family members are similar, their substrate preferences are different. However, the present complex structure provides an important insight into the common guanine specificity in the pierisin family. Recently, the structure of pierisin-1 was revealed (31). In pierisin-1, the key residues (Trp-160, Gln-163, and Glu-165 on the ARTT-loop) are also conserved. Pierisin seems to have a preference for dsDNA, but its final product, N2-(ADP-ribos-1-yl)-2′-deoxyguanosine, is the same as that of ScARP. Scabin was reported as a DNA-targeting mono-ART but shows preference for dsDNA with a single-base overhang rather than a blunt end, with the same guanine specificity (as there is no activity with dI, it was proposed that the product is the same as that of ScARP and pierisin) (33). There might be some varieties with a greater preference for dsDNA, ssDNA, and mononucleotide in each of the pierisin family enzymes. These differences in substrate preference are mainly due to the differences in the PN-loop and its nearby sites in ScARP, Scabin, and pierisin. The electrostatic potentials show large differences between pierisin-1 and Scabin/ScARP (Fig. S2). In pierisin-1, the basic surface is created by the basic residues in the PN-loop (Lys-122, Lys-123, Lys-124, Arg-130, and Arg-134), Arg-67, Arg-181, Arg-187, and Arg-211. In particular, Lys-122, Lys-123, Lys-124, Arg-181, and Arg-187 are indispensable for DNA binding of pierisin-1 (31). These residues are not conserved in ScARP and Scabin, and there are no obvious basic regions in ScARP. Scabin has a small basic region created by Lys-130 in the PN-loop, Lys-180, Arg-183, and Lys-186. It was proposed that the basic region was associated with dsDNA binding (36). Although the substrate preferences are different among the pierisin family, the ARTT-loop that is essential for binding with GDP is conserved among the three enzymes. This means that the described key guanine recognition via the ARTT-loop and ADP-ribosylation mechanism is common in DNA-targeting guanine-specific ARTs. On the basis of this idea, we considered the model structure of pierisin-1 with dsDNA. We built the model of pierisin-1 with dsDNA containing flipped O6-methylguanosine (PDB code 1T38, O6-alkylguanine-DNA alkyltransferase (AGT)) (Fig. S4). We used dsDNA containing a flipped guanosine because guanosine cannot bind with the ARTT-loop unless it is flipped from the base stack, and we superimposed the flipped O6-methylguanosine moiety on the GDP overlaid on pierisin. Although dsDNA partially overlaps with pierisin-1, this rough model gave three important insights. 1) The basic cleft of pierisin-1 is used for binding with dsDNA. 2) Guanosine of dsDNA has to be flipped from the base stack to bind with the ARTT-loop. 3) Arg-130 in PN-loop protrudes into the DNA duplex to promote flipping guanosine out of the base stack in the same way as Arg-128 of AGT (PDB code 1T38) (37).
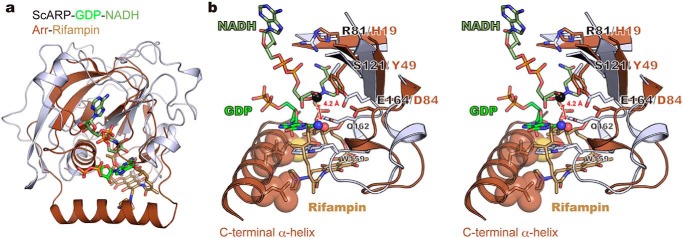
There is another report of small molecule-targeting ART: ADP-ribosylation of antibiotic rifamycin results in antibiotic resistance. The structure of the enzyme Arr from Mycobacterium smegmatis was reported in the rifampin (a semisynthetic derivative of the natural product of rifamycin B)-bound state (38), but ADP-ribosylation mechanism was unclear because of no NAD(H)-bound structure. Arr belongs to the ARTD subclass, which has the H-Y-E motif instead of the R-S-E motif found in ARTC and lacks the ARTT-loop. In another ARTD–substrate complex structure ExoA–eEF2–NAD+, the ADP-ribosylation mechanism was also unclear because the structure exhibited a long distance between electrophile and nucleophile (23, 39). In this case, the conformational change of nucleophile (diphthamide) during the reaction was speculated. The structural comparison with ScARP–GDP–NADH clearly showed the same spatial arrangements of NC1 of N-ribose (electrophile) close to the acceptor oxygen atom of rifampin (nucleophile) in the model of Arr–rifampin–NAD(H) (Fig. 5), providing the first clear insight of ADP-ribosylation mechanism in ARTD. In ARTD, rifampin (substrate) was recognized by the C-terminal α-helix instead of the ARTT-loop in ARTC. By structure-based phylogenetic analysis, it was confirmed that two small molecule-targeting ARTs, ScARP and Arr, localize in ARTC and ARTD, respectively (Fig. S5). Comparing complete coordinates of a small molecule targeting ARTs with minimum elements of ADP-ribosylation, we have identified the substrate recognition difference between ARTD and ARTC for the first time. Our study is not limited to the bacterial ARTCs. This leads to the insights of ARTD, including mammalian PARP. Furthermore, these deeper understandings of substrate recognition of both ARTC and ARTD would lead to the development of better inhibitors of ARTC and ARTD.
Figure 5.
Comparison between ScARP–GDP–NADH and Arr-rifampin (PDB code 2hw2). a, superimposed structures. Arr and rifampin are shown in brown and light brown, respectively. b, stereo view of the active sites. The nucleophilic nitrogen atom in GDP and oxygen atom in rifampin are shown as blue and red spheres, respectively. The electrophilic carbon atom in NADH is shown as a black sphere. Hydrophobic residues interacting with rifampin in the C-terminal α-helix are shown as stick and sphere models.
Experimental procedures
Expression and purification of ScARP
The ScARP gene (UniProt ID: Q9L1E4), sco5461, without the signal peptide (residues 43–204) was cloned into a pRham plasmid with N-terminal hexahistidine and SUMO protein and was overexpressed in the Escherichia coli BL21 Star (DE3) strain. The transformants were cultivated in LB medium containing 50 μg/ml carbenicillin, 0.05% glucose, and 0.2% rhamnose for 24 h at 30 °C. The harvested cells were resuspended in lysis buffer containing 20 mm Tris, pH 8.0, 300 mm NaCl, and 20 mm imidazole and were disrupted by a French press. After centrifugation at 180,000 × g for 40 min, the supernatant was loaded onto a Ni-NTA–agarose column. After washing the column with lysis buffer, the column was again washed with 20 mm Tris, pH 8.0, and 20 mm imidazole for desalting. The His–SUMO tag was then cleaved using SUMO protease with an N-terminal hexahistidine tag on Ni-NTA–agarose for 3 h at room temperature. The flow-through and wash fractions were pooled and loaded onto a HiTrap Q HP 5-ml column (GE Healthcare). The Q column was equilibrated with 20 mm Tris, pH 8.0, and eluted with a linear gradient from 0 to 500 mm NaCl. The ScARP fraction was finally loaded onto a Superdex 75 column (GE Healthcare) with 10 mm Tris, pH 8.0, and 100 mm NaCl, concentrated to 20 mg/ml, rapidly frozen in liquid nitrogen, and stored at −80 °C.
Crystallization
Crystallization was carried out using the hanging drop vapor diffusion method. Single crystals of apo-ScARP were obtained by two-step crystallization. In the first crystallization step, the drop was composed of equal volumes of 20 mg/ml ScARP and a reservoir solution containing 0.1 m HEPES, pH 7.0, and 8% PEG8000. Bad quality crystals appeared within a few days at 20 °C. These crystals were crushed, diluted with the reservoir solution, and used for the second crystallization step as a seed. In the second crystallization step, the drop was composed of 1 μl of 10 mg/ml ScARP, 1 μl of the reservoir solution containing 0.1 m sodium acetate, pH 5.0, and 15% 2-methyl-2,4-pentanediol, and 0.1 μl of the diluted seed solution. This condition was determined by microseed matrix screening (40). The second crystallization generated single crystals at 20 °C. Prior to data collection, the crystals were briefly soaked in the second crystallization reservoir solution, including 20% xylitol. To obtain the ScARP–GDP–NADH complex crystals, a solution containing 17 mg/ml (0.9 mm) ScARP, 9 mm GDP, and 9 mm NADH was incubated for 1 h at 4 °C before crystallization. The drop comprised equal volumes of the protein solution and the reservoir solution containing 0.1 m ammonium acetate, pH 5.0, and 7% PEG10000. Rod-shaped crystals appeared within a few days at 4 °C. Prior to data collection, the crystals were transferred to a mixture of paraffin and paratone oils (Hampton).
Data collection and structure determination
Diffraction data sets were collected at 100 K on a beamline PF BL-5A. The structure of ScARP was solved through molecular replacement using the program PHASER (41) using the Scabin structure (Protein Data Bank code 5DAZ). The model was then iteratively built using COOT (42) and refined using phenix.refine (43). The atomic model and structure factors were deposited at Protein Data Bank under accession code 5ZJ4 (apo-ScARP) and 5ZJ5 (ScARP–GDP–NADH).
Activity measurement
To measure the ADP-ribosylation activity, WT ScARP and the mutants were prepared as follows. The ScARP gene (UniProt ID: Q9L1E4) without the signal peptide (residues 43–204) was cloned into a pET15b plasmid with N-terminal hexahistidine and was overexpressed in the E. coli BL21(DE3) strain. The transformants were cultivated in LB medium containing 50 μg/ml ampicillin at 37 °C until the absorbance at 620 nm reached 0.6. After inducing expression with 0.5 mm isopropyl β-d-1-thiogalactopyranoside, the culture was incubated for 16 h at 16 °C. The harvested cells were resuspended in lysis buffer containing 50 mm Tris, pH 8.0, 300 mm NaCl, and 20 mm imidazole, disrupted by sonication, and centrifuged. The supernatant was loaded onto a Ni-NTA–agarose column. After washing the column with lysis buffer, ScARP was eluted with 50 mm Tris, pH 8.0, 300 mm NaCl, and 400 mm imidazole. The ScARP fractions were finally loaded onto a Superdex 75 column (GE Healthcare) with 10 mm Tris, pH 8.0, and 100 mm NaCl, concentrated to 1.5 mg/ml, rapidly frozen in liquid nitrogen, and stored at −80 °C. Reaction products with ScARP were analyzed by HPLC. The ADP-ribosylation activity was assayed in a reaction mixture of 0.5 mm GDP, 0.5 mm NAD+, 5 nm ScARP, and 10 mm HEPES, pH 7.4, in a final volume of 50 μl for 10 min at room temperature. The control compounds were prepared as a mixture of 0.5 mm GDP, 0.5 mm NAD+, 0.5 mm nicotinamide, 0.5 mm ADP-ribose, and 10 mm HEPES, pH 7.4, in a final volume of 50 μl. The reaction mixture or control compounds of 15 μl were then applied to a ReDual AX-C18 column (Shimadzu) on an LC-2000Plus HPLC system (Jasco) using 100 mm potassium phosphate, pH 6.5, and 20% methanol as the mobile phase at room temperature at a flow rate of 1 ml/min with monitoring at 260 nm.
Author contributions
T. Y. and H. T. conceptualization; T. Y. data curation; T. Y. and H. T. funding acquisition; T. Y. validation; T. Y. and H. T. writing-original draft; H. T. supervision; H. T. project administration.
Supplementary Material
Acknowledgment
We thank the staff at KEK-Photon Factory for data collection.
This work was supported in part by Grants-in-aid for Scientific Research, MEXT of Japan, 17K15095 (to T. Y.) and 15K08289 (to H. T.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S5, Table S1 and supporting Refs. 1–2.
The atomic coordinates and structure factors (codes 5ZJ4 and 5ZJ5) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- ART
- ADP-ribosyltransferase
- ARTT
- ADP-ribosylating toxin turn–turn
- PARP
- enzyme poly(ADP-ribose) polymerase
- aa
- amino acid
- RMSD
- root mean square deviation
- PN
- phosphate nicotinamide
- SUMO
- small ubiquitin-like modifier
- Ni-NTA
- nickel-nitrilotriacetic acid
- ssDNA
- single-stranded DNA
- PDB
- Protein Data Bank
- AGT
- O6-alkylguanine-DNA alkyltransferase.
References
- 1. Simon N. C., Aktories K., and Barbieri J. T. (2014) Novel bacterial ADP-ribosylating toxins: structure and function. Nat. Rev. Microbiol. 12, 599–611 10.1038/nrmicro3310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lüscher B., Bütepage M., Eckei L., Krieg S., Verheugd P., and Shilton B. H. (2018) ADP-ribosylation, a multifaceted posttranslational modification involved in the control of cell physiology in health and disease. Chem. Rev. 118, 1092–1136 10.1021/acs.chemrev.7b00122 [DOI] [PubMed] [Google Scholar]
- 3. Cassel D., and Pfeuffer T. (1978) Mechanism of cholera toxin action: covalent modification of the guanyl nucleotide-binding protein of the adenylate cyclase system. Proc. Natl. Acad. Sci. U.S.A. 75, 2669–2673 10.1073/pnas.75.6.2669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Katada T., and Ui M. (1982) ADP ribosylation of the specific membrane protein of C6 cells by islet-activating protein associated with modification of adenylate cyclase activity. J. Biol. Chem. 257, 7210–7216 [PubMed] [Google Scholar]
- 5. Collier R. J., and Pappenheimer A. M. Jr. (1964) Studies on the mode of action of diphtheria toxin. II. Effect of toxin on amino acid incorporation in cell-free systems. J. Exp. Med. 120, 1019–1039 10.1084/jem.120.6.1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Honjo T., Nishizuka Y., and Hayaishi O. (1968) Diphtheria toxin-dependent adenosine diphosphate ribosylation of aminoacyl transferase II and inhibition of protein synthesis. J. Biol. Chem. 243, 3553–3555 [PubMed] [Google Scholar]
- 7. Aktories K., and Frevert J. (1987) ADP-ribosylation of a 21–24-kDa eukaryotic protein(s) by C3, a novel botulinum ADP-ribosyltransferase, is regulated by guanine nucleotide. Biochem. J. 247, 363–368 10.1042/bj2470363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ohashi Y., and Narumiya S. (1987) ADP-ribosylation of a Mr 21,000 membrane protein by type D botulinum toxin. J. Biol. Chem. 262, 1430–1433 [PubMed] [Google Scholar]
- 9. Aktories K., Bärmann M., Ohishi I., Tsuyama S., Jakobs K. H., and Habermann E. (1986) Botulinum C2 toxin ADP-ribosylates actin. Nature 322, 390–392 10.1038/322390a0 [DOI] [PubMed] [Google Scholar]
- 10. Tsuge H., Nagahama M., Nishimura H., Hisatsune J., Sakaguchi Y., Itogawa Y., Katunuma N., and Sakurai J. (2003) Crystal structure and site-directed mutagenesis of enzymatic components from Clostridium perfringens ι-toxin. J. Mol. Biol. 325, 471–483 10.1016/S0022-2836(02)01247-0 [DOI] [PubMed] [Google Scholar]
- 11. Koyama K., Wakabayashi K., Masutani M., Koiwai K., Watanabe M., Yamazaki S., Kono T., Miki K., and Sugimura T. (1996) Presence in Pieris rapae of cytotoxic activity against human carcinoma cells. Jpn. J. Cancer Res. 87, 1259–1262 10.1111/j.1349-7006.1996.tb03141.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Watanabe M., Kono T., Koyama K., Sugimura T., and Wakabayashi K. (1998) Purification of pierisin, an inducer of apoptosis in human gastric carcinoma cells, from cabbage butterfly, Pieris rapae. Jpn. J. Cancer Res. 89, 556–561 10.1111/j.1349-7006.1998.tb03297.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Takamura-Enya T., Watanabe M., Totsuka Y., Kanazawa T., Matsushima-Hibiya Y., Koyama K., Sugimura T., and Wakabayashi K. (2001) Mono(ADP-ribosyl)ation of 2′-deoxyguanosine residue in DNA by an apoptosis-inducing protein, pierisin-1, from cabbage butterfly. Proc. Natl. Acad. Sci. U.S.A. 98, 12414–12419 10.1073/pnas.221444598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Takamura-Enya T., Watanabe M., Koyama K., Sugimura T., and Wakabayashi K. (2004) Mono(ADP-ribosyl)ation of the N2 amino groups of guanine residues in DNA by pierisin-2, from the cabbage butterfly, Pieris brassicae. Biochem. Biophys. Res. Commun. 323, 579–582 10.1016/j.bbrc.2004.08.132 [DOI] [PubMed] [Google Scholar]
- 15. Nakano T., Matsushima-Hibiya Y., Yamamoto M., Takahashi-Nakaguchi A., Fukuda H., Ono M., Takamura-Enya T., Kinashi H., and Totsuka Y. (2013) ADP-ribosylation of guanosine by SCO5461 protein secreted from Streptomyces coelicolor. Toxicon 63, 55–63 10.1016/j.toxicon.2012.11.019 [DOI] [PubMed] [Google Scholar]
- 16. Nakano T., Matsushima-Hibiya Y., Yamamoto M., Enomoto S., Matsumoto Y., Totsuka Y., Watanabe M., Sugimura T., and Wakabayashi K. (2006) Purification and molecular cloning of a DNA ADP-ribosylating protein, CARP-1, from the edible clam Meretrix lamarckii. Proc. Natl. Acad. Sci. U.S.A. 103, 13652–13657 10.1073/pnas.0606140103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nakano T., Takahashi-Nakaguchi A., Yamamoto M., and Watanabe M. (2015) Pierisins and CARP-1: ADP-ribosylation of DNA by ARTCs in butterflies and shellfish. Curr. Top. Microbiol. Immunol. 384, 127–149 10.1007/82_2014_416 [DOI] [PubMed] [Google Scholar]
- 18. Jankevicius G., Ariza A., Ahel M., and Ahel I. (2016) The toxin-antitoxin system DarTG catalyzes reversible ADP-ribosylation of DNA. Mol. Cell 64, 1109–1116 10.1016/j.molcel.2016.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Munnur D., and Ahel I. (2017) Reversible mono-ADP-ribosylation of DNA breaks. FEBS J. 284, 4002–4016 10.1111/febs.14297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Talhaoui I., Lebedeva N. A., Zarkovic G., Saint-Pierre C., Kutuzov M. M., Sukhanova M. V., Matkarimov B. T., Gasparutto D., Saparbaev M. K., Lavrik O. I., and Ishchenko A. A. (2016) Poly(ADP-ribose) polymerases covalently modify strand break termini in DNA fragments in vitro. Nucleic Acids Res. 44, 9279–9295 10.1093/nar/gkw675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zarkovic G., Belousova E. A., Talhaoui I., Saint-Pierre C., Kutuzov M. M., Matkarimov B. T., Biard D., Gasparutto D., Lavrik O. I., and Ishchenko A. A. (2018) Characterization of DNA ADP-ribosyltransferase activities of PARP2 and PARP3: new insights into DNA ADP-ribosylation. Nucleic Acids Res. 46, 2417–2431 10.1093/nar/gkx1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dölle C., and Ziegler M. (2017) ADP-ribosylation of DNA moving into focus. FEBS J. 284, 3999–4001 10.1111/febs.14326 [DOI] [PubMed] [Google Scholar]
- 23. Jørgensen R., Merrill A. R., Yates S. P., Marquez V. E., Schwan A. L., Boesen T., and Andersen G. R. (2005) Exotoxin A-eEF2 complex structure indicates ADP ribosylation by ribosome mimicry. Nature 436, 979–984 10.1038/nature03871 [DOI] [PubMed] [Google Scholar]
- 24. Tsuge H., Nagahama M., Oda M., Iwamoto S., Utsunomiya H., Marquez V. E., Katunuma N., Nishizawa M., and Sakurai J. (2008) Structural basis of actin recognition and arginine ADP-ribosylation by Clostridium perfringens ι-toxin. Proc. Natl. Acad. Sci. U.S.A. 105, 7399–7404 10.1073/pnas.0801215105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Toda A., Tsurumura T., Yoshida T., Tsumori Y., and Tsuge H. (2015) Rho GTPase recognition by C3 exoenzyme based on C3–RhoA complex structure. J. Biol. Chem. 290, 19423–19432 10.1074/jbc.M115.653220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Han S., Arvai A. S., Clancy S. B., and Tainer J. A. (2001) Crystal structure and novel recognition motif of rho ADP-ribosylating C3 exoenzyme from Clostridium botulinum: structural insights for recognition specificity and catalysis. J. Mol. Biol. 305, 95–107 10.1006/jmbi.2000.4292 [DOI] [PubMed] [Google Scholar]
- 27. Tsuge H., Yoshida T., and Tsurumura T. (2015) Conformational plasticity is crucial for C3–RhoA complex formation by ARTT-loop. Pathog. Dis. 73, ftv094 10.1093/femspd/ftv094 [DOI] [PubMed] [Google Scholar]
- 28. Holmes K. C., Popp D., Gebhard W., and Kabsch W. (1990) Atomic model of the actin filament. Nature 347, 44–49 10.1038/347044a0 [DOI] [PubMed] [Google Scholar]
- 29. Aravind L., Zhang D., de Souza R. F., Anand S., and Iyer L. M. (2015) The natural history of ADP-ribosyltransferases and the ADP-ribosylation system. Curr. Top. Microbiol. Immunol. 384, 3–32 10.1007/82_2014_414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tsuge H., and Tsurumura T. (2015) Reaction mechanism of Mono-ADP-ribosyltransferase based on structures of the complex of enzyme and substrate protein. Curr. Top. Microbiol. Immunol. 384, 69–87 10.1007/82_2014_415 [DOI] [PubMed] [Google Scholar]
- 31. Oda T., Hirabayashi H., Shikauchi G., Takamura R., Hiraga K., Minami H., Hashimoto H., Yamamoto M., Wakabayashi K., Shimizu T., and Sato M. (2017) Structural basis of autoinhibition and activation of the DNA-targeting ADP-ribosyltransferase pierisin-1. J. Biol. Chem. 292, 15445–15455 10.1074/jbc.M117.776641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lyons B., Ravulapalli R., Lanoue J., Lugo M. R., Dutta D., Carlin S., and Merrill A. R. (2016) Scabin, a novel DNA-acting ADP-ribosyltransferase from Streptomyces scabies. J. Biol. Chem. 291, 11198–11215 10.1074/jbc.M115.707653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lyons B., Lugo M. R., Carlin S., Lidster T., and Merrill A. R. (2018) Characterization of the catalytic signature of Scabin toxin, a DNA-targeting ADP-ribosyltransferase. Biochem. J. 475, 225–245 10.1042/BCJ20170818 [DOI] [PubMed] [Google Scholar]
- 34. Berti P. J., Blanke S. R., and Schramm V. L. (1997) Transition state structure for the hydrolysis of NAD catalyzed by diphtheria toxin. J. Am. Chem. Soc. 119, 12079–12088 10.1021/ja971317a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tsurumura T., Tsumori Y., Qiu H., Oda M., Sakurai J., Nagahama M., and Tsuge H. (2013) Arginine ADP-ribosylation mechanism based on structural snapshots of ι-toxin and actin complex. Proc. Natl. Acad. Sci. U.S.A. 110, 4267–4272 10.1073/pnas.1217227110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lugo M. R., Lyons B., Lento C., Wilson D. J., and Merrill A. R. (2018) Dynamics of Scabin toxin. A proposal for the binding mode of the DNA substrate. PLoS ONE 13, e0194425 10.1371/journal.pone.0194425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Daniels D. S., Woo T. T., Luu K. X., Noll D. M., Clarke N. D., Pegg A. E., and Tainer J. A. (2004) DNA binding and nucleotide flipping by the human DNA repair protein AGT. Nat. Struct. Mol. Biol. 11, 714–720 10.1038/nsmb791 [DOI] [PubMed] [Google Scholar]
- 38. Baysarowich J., Koteva K., Hughes D. W., Ejim L., Griffiths E., Zhang K., Junop M., and Wright G. D. (2008) Rifamycin antibiotic resistance by ADP-ribosylation: structure and diversity of Arr. Proc. Natl. Acad. Sci. U.S.A. 105, 4886–4891 10.1073/pnas.0711939105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jørgensen R., Wang Y., Visschedyk D., and Merrill A. R. (2008) The nature and character of the transition state for the ADP-ribosyltransferase reaction. EMBO Rep. 9, 802–809 10.1038/embor.2008.90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. D'Arcy A., Bergfors T., Cowan-Jacob S. W., and Marsh M. (2014) Microseed matrix screening for optimization in protein crystallization: what have we learned? Acta Crystallogr. F Struct. Biol. Commun. 70, 1117–1126 10.1107/S2053230X14015507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 43. Afonine P. V., Grosse-Kunstleve R. W., Echols N., Headd J. J., Moriarty N. W., Mustyakimov M., Terwilliger T. C., Urzhumtsev A., Zwart P. H., and Adams P. D. (2012) Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 10.1107/S0907444912001308 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.