Abstract
Transgenerational inheritance requires mechanisms by which epigenetic information is transferred via gametes. Canonical thought holds that mammalian sperm chromatin would be incapable of carrying epigenetic information as post-translational modifications of histones because of their replacement with protamine proteins. Furthermore, compaction of the sperm genome would hinder DNA accessibility of proteins involved in transcriptional regulation and genome architecture. In this Minireview, we delineate the paternal chromatin remodeling events during spermatogenesis and fertilization. Sperm chromatin is epigenetically modified at various time points throughout its development. This allows for the addition of environment-specific modifications that can be passed from parents to offspring.
Keywords: chromatin, transcription, histone, sperm, epigenetics, CTCF, epigenetic inheritance, protamine, sperm chromatin, transgenerational inheritance
Background
Sperm chromatin in mammals is thought to be structurally distinct from that of somatic cells. In somatic cells, chromatin contains epigenetic information in the form of DNA methylation and post-translational modifications on histones, information that is thought to influence chromosome architecture and gene expression. Methylation at the 5-position of a cytosine base pair is an epigenetic mark predominantly located at cytosine–phosphate–guanine (CpG)2 dinucleotides and is associated with gene silencing when at the promoter and active transcription when in the gene body (1). These site-specific marks are established in sperm by DNA methyltransferases (DNMT) and ten-eleven translocation methylcytosine dioxygenase (TET) proteins performing active DNA methylation and demethylation, respectively (2). Post-translational modifications in histones come primarily in the form of methylation or acetylation of amino acids in the N-terminal tails of these proteins, which in turn affect the structure of chromatin (3). These modifications correlate with either transcriptionally active or silenced regions of the genome.
One considerable difference between sperm and all other cells is its size. Although mammalian sperm possess half the DNA than is contained in a typical somatic cell, the nucleus has 40-fold less volume (4). A comparison of mouse sperm nuclei with that of liver cells, taking into account the nuclear volume, density of DNA packaging, and difference in ploidy of these cell types, uncovered a 6-fold compaction of sperm nuclear DNA (4, 5). It has been hypothesized that this compaction is due to the presence of smaller protamine proteins on sperm DNA replacing larger, more complex histone octamers (Fig. 1A) (6). Protamines are basic 50–60–amino acid sequence proteins with DNA-binding domains of 10–11 amino acids. The protamine binds to the major groove of the DNA and neutralizes its phosphate backbone (7). Early studies assumed that the replacement of nucleosomes with protamines is responsible for the tight packaging of DNA in the sperm nucleus; however, certain organisms such as zebrafish (Danio rerio) have similarly small, dense sperm nuclei and no protamines (8, 9). Based on these reports, it remains unclear what role protamines play in sperm.
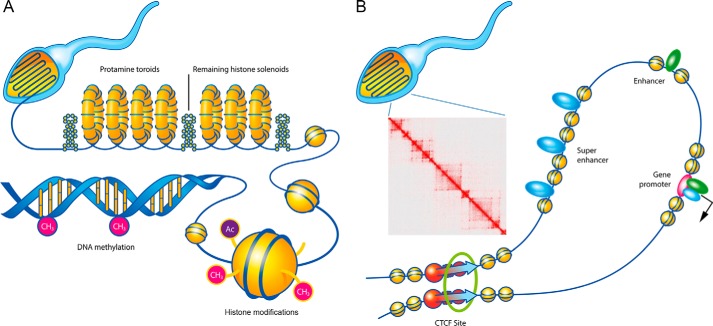
Figure 1.
Old and new views of chromatin structure in mature sperm. A, most sperm DNA was thought to be packaged in protamine toroids (yellow rings) and remaining histone octameres (yellow spheres). DNA is methylated at 5mC (pink), whereas histones are methylated or acetylated in their N-tails. B, recent results suggest the retention of more nucleosomes than previously thought in mouse sperm. These nucleosomes contain most or all histone modifications found in somatic cells (data not shown). Well-positioned nucleosomes flank TSSs and transcription factor-binding sites, including CTCF. Many enhancers and super-enhancers active in embryonic or adult tissues are already specified in the sperm epigenome. CTCF and cohesin mediate long-range chromatin interactions and establish the three-dimensional architecture of the chromatin. This is represented by a Hi-C heatmap displaying sperm chromatin organization into compartments and domains similar to those found in other cell types.
Until recently, it has been thought that the role of sperm in mammals is to deliver the transcriptionally inert paternal DNA, mostly devoid of histones and complexed with protamines, to the egg during fertilization (10, 11). However, recent findings suggest a more conventional view of sperm chromatin, with histones containing typical covalent modifications retained at important genomic sites and a three-dimensional architecture similar to that of somatic cells (Fig. 1B) (12–16). Mammalian sperm may thus be capable of not only carrying epigenetic information, but also passing this information to cells of the early embryo, producing changes that may affect differentiated adult tissues.
Foundations of the epigenetic landscape established in primordial germ cells: Sperm development
Mouse spermatogenesis takes place in three phases, which include self-renewal of spermatogonia through mitosis, followed by meiosis of spermatocytes to form haploid spermatids and transformation of spermatids into spermatozoa by means of spermiogenesis (17). Spermatozoa development begins from primordial germ cells (PGCs) at the base of the emerging allantois in the endoderm of the yolk sac of the mouse embryo (18). PGCs migrating into the genital ridges between E 7.5 and E13.5 (19) undergo extensive epigenetic reprogramming in the form of global demethylation. PGC methylation level reaches its minimum between E10.5 and E12.5 (20). The process of sexual dimorphic development of the post-migration PGCs into male gametes begins at E13.5 and involves extensive remethylation of sperm DNA. Around day 5 after birth, some prospermatogonia within the basement membrane of the pup's seminiferous tubule resume mitotic division to form primary spermatocytes. Secondary spermatocytes are formed by meiotic division, and they later divide once more to become haploid spermatids. The remaining prospermatogonia not forming spermatids will continue to divide mitotically and produce spermatogonial stem cells, to ensure the continuation of spermatogonia supply. Finally, spermiogenesis involves microtubule growth, tail formation, and the tight packaging of DNA into condensed, protamine-bound chromatin (21).
Chromatin state in mature sperm
Methylation
Mature sperm are highly methylated on a global level as compared with somatic cells (Fig. 1A) (22). This methylation, established by DNMTs during spermatogenesis, is necessary for normal embryo development (23, 24). Several groups have found that patterns of paternal methylation are maintained through the early embryonic stages, suggesting that this methylation continues to serve a regulatory role during embryo development (23–25). Furthermore, several oocyte-derived genes in the developing embryo adopt a methylation pattern similar to that of sperm, namely gaining methylation in genes involved in germline specification (25). It appears that the dense global methylation on sperm is highly gene-specific, which may play a role in directing transcription in the early embryo.
Equally important as site-specific methylation in the sperm genome is the absence of this modification from specific sequences. Sites in sperm that are demethylated despite the dense global methylation levels lie mostly in CpG islands, regions of high CpG density, found mostly near promoters (26). One comparison study between human sperm and embryonic stem cells (ESCs) found very similar distributions of methylation in each cell type. They found an enrichment of hypomethylated regions at promoters and highly methylated repeat elements (27). This group also found that sperm-specific hypomethylated regions are located within genes related to germ cell development. Another group confirmed these findings in a comparison study with oocytes and early embryos. They found hypomethylated sequences enriched at high-density CpG promoters, enhancers, and exons. Sperm exhibits a high density of methylation at intergenic regions (28). There is evidence of parent-of-origin–specific methylation patterns in the early embryo, with the male sperm contributing primarily differentially methylated sites in intergenic regions (29). DNA methylation is globally erased and re-established both during spermatogenesis and after fertilization, suggesting that 5mC may not be a good candidate, on its own, to carry epigenetic information between generations in mammals. However, evidence of site-specific methylation in mature sperm suggests there must be complementary or alternative pathways by which epigenetic information can be altered and transmitted through the paternal germline. Other candidate mechanisms are discussed below.
Histone modifications
In 1977, Balhorn et al. (30) examined histones retained in human and mouse sperm by electrophoretic analyses of HCl-extracted proteins from sperm chromatin. They determined that only 1% of the mouse sperm genome is associated with histones (Fig. 1A), and these histones were said to be related to developmental and/or housekeeping genes important for embryo development (31). However, more recent studies using micrococcal nuclease DNA digestion and ATAC-seq (assay for transposase-accessible chromatin using sequencing) techniques revealed that 7.5% of histones present in diploid somatic cells are retained in sperm nuclei when taking ploidy into account. The distribution and location of these histones in the paternal genome are actively debated, with some reports suggesting histones are distributed primarily within distal intergenic and intronic regions, and others have identified enrichment in imprinted genes (14, 15). This discrepancy may be explained by differences in methodology (14).
Results from ATAC-seq and ChIP-seq experiments suggest that sperm histones located in promoter regions possess post-translational modifications that are both a consequence of the transcriptional state of the preceding round spermatid stage as well as a prelude to expression patterns observed in ESCs and adult tissues (Fig. 1B). Modifications on histones at promoters can either be active, repressive, or bivalent. Approximately 60% of sperm promoters are in an active epigenetic state, and the TSSs of these promoters are flanked by three to four nucleosomes upstream and five to six nucleosomes downstream (16). It has been shown that 28% of promoters containing H3K27me3 also have H3K4me2, bivalent marks indicative of a poised promoter state (13). These are surprising findings considering the transcriptionally inert state of mature sperm. However, current studies have found that genes encoding for proteins involved in embryo development, HOX, SOX, FOX, TBX, PAX, CDX, and GATA family transcription factors, are the ones whose promoters possess these bivalent marks (12, 13, 33–35). Perhaps these histone modifications contain epigenetic information that affects transcription in the early embryo.
Sperm not only have promoters in a primed state that correlates with expression in mESCs, but they also appear to have enhancers in a similar state (Fig. 1B). Sperm enhancers were defined by the presence of ATAC-seq signal and the presence of H3K4me1 and H3K27ac (16). Results from ATAC-seq experiments suggest the presence of around 58,000 transposase-hypersensitive sites that may be bound by specific transcription factors, 10,240 of which correspond to enhancers previously identified in embryonic or adult tissues. In addition to typical enhancers, the sperm genome also contains around 645 super-enhancers, most of them in common with mESCs or specific cell lineages found in the adult organism. Interestingly, super-enhancers are present in loops formed by CTCF and cohesin to create insulated neighborhoods as found previously in mESCs (Fig. 1B) (36). These results suggest that enhancer elements are already specified in the sperm and may be primed for subsequent function during embryogenesis and in the establishment of specific cell fates during the formation of differentiated tissues (16).
3D organization of sperm chromatin
The dense compaction of sperm chromatin was thought to result in the establishment of a 3D architecture very different from that of somatic cells. However, recent studies suggest that the three-dimensional organization of sperm chromatin is very similar to that of other cells, in particular the ESCs (16, 37). CTCF is an architectural protein responsible, at least in part, for this organization (38), and binding motifs for CTCF can be detected at micrococcal nuclease and ATAC-seq sites on sperm DNA, suggesting that this protein may be retained in the sperm nucleus (Fig. 1B) (14, 16). The presence of CTCF at sequence motifs in accessible sperm DNA was confirmed via ChIP-seq experiments (16). CTCF has been found to be essential for normal spermiogenesis, sperm fertility, and histone retention in mature sperm (39). Genome-wide Hi-C mapping experiments have uncovered the presence of compartments, CTCF-mediated loops, and topologically associating domains in sperm chromatin similar to those found in somatic cell lines and embryonic stem cells (16, 37). Recent single-nucleus Hi-C experiments have shown that a similar higher-order chromatin organization also exists in the female gamete at the germinal vesicle stage but not in the MII stage, at which time the oocyte is arrested in metaphase (40).
Epigenetic remodeling after fertilization
Histone replacement with protamines
It is now understood that mature sperm DNA is both highly methylated and bound by protamines, modified histones, and transcription factors when it first encounters an oocyte. Fertilization begins with the binding of the sperm head to the oocyte zona pellucida. Sperm fusion with the membrane activates the oocyte and initiates completion of meiosis II. The sperm head and its contents are then engulfed by the oocyte. While the oocyte completes meiosis II, the sperm nucleus undergoes several changes. Its chromatin is primed for later nuclear syngamy and transcription in two ways: the first is through the replacement of protamines with histones, and the second is through active demethylation. Decondensation of the sperm nucleus occurs ∼45–60 min after fertilization. Disulfide bonds, which allow protamines to attach to sperm DNA, are broken upon exposure to oocyte-produced chemicals such as GSH (41); however, it is unclear whether chromatin decondensation is initiated by protamine detachment. Experiments showing a depletion of radiolabeled protamines occurring after chromatin decondensation (41, 42) may be explained by unbound protamine degradation. The paternal chromatin is then loaded with histones produced by the oocyte prior to fertilization, as suggested by immunofluorescence data showing the appearance of histones in the male pronucleus following protamine depletion (43). These histones carry modifications associated with newly-synthesized histones (44), including histone H4 Lys-5 and Lys-12 acetylation (45). One study detected the presence of H3K4me1 in the male pronucleus beginning at stage PN1, and H3K4me3 beginning at stage PN4. Researchers hypothesized that these observed modifications can be explained by histone methyltransferase activity in the zygote (46). Alternatively, the presence of H3K4me1 and H3K4me3 in mature sperm (12, 16) suggests that histone modifications detected in the early embryo may be paternally derived and maintained during fertilization and early embryogenesis.
Demethylation of the paternal pronucleus
It was previously thought that both female and male pronuclear DNA is passively demethylated once replication commences. This is shown to be true of the maternal nucleus (47, 48); however, evidence suggests there is active demethylation of the male pronucleus before the start of DNA replication (49, 50). Immunofluorescence and bisulfite sequencing data show a sharp decrease in methylation in the male pronucleus within 4 h of fertilization (48, 50) Replication begins 7–9 h into the first cell cycle, when pronuclei enter the PN3 stage (51). Therefore, any demethylation occurring prior to the start of replication must be active. One proposed mechanism for this demethylation is the oxidation of 5-methylcytosine by TET proteins present in the oocyte cytoplasm (52–54). One study confirmed the presence of 5-hydroxymethylcytosine (5hmC) in the mouse male pronucleus using antibody staining and the specific expression of TET protein in the early embryo (52). The 5hmC signal increases significantly between stage PN0 and PN5, and a deficiency of Tet3 prevents the oxidation of 5mC into 5hmC and results in developmental abnormalities (55). Another model suggests the presence of sperm chromatin-specific demethylases in the oocyte mediating pronuclear demethylation (42). Enrichment of 5hmC in sperm highlights the differences in the methylation state of male and female pronuclear chromatin before the start of nuclear syngamy.
Recent studies explore the apparent asymmetry in the epigenetic states of male and female pronuclei and its implications in the later stages of embryonic development. Inoue et al. (56) used DNase-sequencing techniques to compare DNase I–hypersensitive sites (DHSs) in the maternal and paternal mature gametes, pronuclei, morula, and blastula stage embryos. In searching for allele specificity and proof of parent-specific inheritance of epigenetic marks, they discovered that most DHSs in the early embryo are maternally derived and prime differential gene expression at the time of zygotic genome activation (56). Certain aspects of the epigenetic state of sperm and oocyte chromatin are maintained throughout early stages of embryonic development, suggesting a role for the transfer of information from the gamete to the embryo.
Conclusions
Current research reveals that sperm chromatin contains far more complex epigenetic information than was previously recognized. The sperm nucleus contains sex-specific methylation patterns, nucleosomes at promoters carrying both active and silencing histone modifications, putative enhancer and super-enhancer elements flanked by nucleosomes and possibly bound by transcription factors, and CTCF/cohesin bound at specific sites to establish highly-organized chromatin interactions within the three-dimensional nuclear space. It is difficult to imagine that the presence of this wealth of information in the sperm is not used during early embryonic development to guide the initial steps controlling gene expression after fertilization. DNA methylation, an obvious candidate to explain the inheritance of epigenetic information through generations, is erased and re-established during spermatogenesis and after fertilization and thus has been discounted as a plausible mechanism underlying transgenerational effects (57). However, the erasure of 5mC in the paternal genome is only partial, suggesting the existence of mechanisms that maintain some of the epigenetic information established by DNA methylation. It is possible that the presence of DNA-bound transcription factors and histone modifications in specific regions of the genome may serve to guide re-methylation of the DNA after the demethylation of the paternal chromosomes that takes place immediately after fertilization. Under this model, although part of the epigenetic information specified by DNA methylation of the paternal chromosomes may be erased after fertilization, some information may be maintained by the interaction of specific DNA-bound transcription factors that preserve a memory of critical regulatory functions encoded by enhancers, super-enhancers, and 3D organization (16).
The information carried in the sperm epigenome has the potential to be transferred transgenerationally, explaining the occurrence of experience-based transmission of epiphenotypes. Environmental factors causing novel transcription during development of the paternal germline may result in the persistence of DNA-binding proteins at new sites in the genome, which in turn would modulate DNA re-methylation after fertilization and transcription in the early embryo. Other possible products of altered transcription as a consequence of environmental signals are small RNA fragments, including tRNAs and miRNAs, whose presence in sperm has also been shown to mediate the transfer of epigenetic information between generations (58–60). Skinner and co-workers (61) made the important discovery that exposure to endocrine disruptors in utero can lead to several phenotypes that persist as far as the F3 generation. Phenotypes include testicular and kidney diseases, tumors, and obesity, among others. Transgenerational transmission of these phenotypes may be mediated by epigenetic changes occurring in gametes upon exposure to these toxins. Other environment-altering elements such as diet, stress, smoking, and even certain medications have been found to have inter- or transgenerational phenotypic implications (32, 62–64). Epigenetic information carried in sperm in the form of DNA and histone modifications, transcription factors, noncoding RNA, and chromatin 3D architecture is one potential explanation for this phenomenon.
This work was supported by National Institutes of Health Award R01 ES027859 from United States Public Health Service. This is the fifth article in the Thematic Minireview series “Chromatin and Transcription.” The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- CpG
- cytosine–phosphate–guanine
- ESC
- embryonic stem cell
- mESC
- mouse ESC
- DNMT
- DNA methyltransferase
- TET
- ten-eleven translocation methylcytosine dioxygenase
- PGC
- primordial germ cell
- E
- embryonic day
- 5mC
- 5-methylcytosine
- miRNA
- microRNA
- seq
- sequencing
- ATAC
- assay for transposase-accessible chromatin
- DHS
- DNase I–hypersensitive site
- TSS
- transcription start site
- CTCF
- CCCTC-binding factor.
References
- 1. Schultz M. D., He Y., Whitaker J. W., Hariharan M., Mukamel E. A., Leung D., Rajagopal N., Nery J. R., Urich M. A., Chen H., Lin S., Lin Y., Jung I., Schmitt A. D., Selvaraj S., et al. (2015) Human body epigenome maps reveal noncanonical DNA methylation variation. Nature 523, 212–216 10.1038/nature14465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hajkova P., Jeffries S. J., Lee C., Miller N., Jackson S. P., and Surani M. A. (2010) Genome-wide reprogramming in the mouse germ line entails the base excision repair pathway. Science 329, 78–82 10.1126/science.1187945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bannister A. J., and Kouzarides T. (2011) Regulation of chromatin by histone modifications. Cell Res. 21, 381–395 10.1038/cr.2011.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wyrobek A. J., Meistrich M. L., Furrer R., and Bruce W. R. (1976) Physical characteristics of mouse sperm nuclei. Biophys. J. 16, 811–825 10.1016/S0006-3495(76)85730-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ward W. S., and Coffey D. S. (1991) DNA packaging and organization in mammalian spermatozoa: comparison with somatic cell. Biol. Reprod. 44, 569–574 10.1095/biolreprod44.4.569 [DOI] [PubMed] [Google Scholar]
- 6. Teperek M., and Miyamoto K. (2013) Nuclear reprogramming of sperm and somatic nuclei in eggs and oocytes. Reprod. Med. Biol. 12, 133–149 10.1007/s12522-013-0155-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Balhorn R. (2007) The protamine family of sperm nuclear proteins. Genome Biol. 8, 227 10.1186/gb-2007-8-9-227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ausió J., González-Romero R., and Woodcock C. L. (2014) Comparative structure of vertebrate sperm chromatin. J. Struct. Biol. 188, 142–155 10.1016/j.jsb.2014.09.004 [DOI] [PubMed] [Google Scholar]
- 9. Carrell D. T. (2011) Epigenetic marks in zebrafish sperm: insights into chromatin compaction, maintenance of pluripotency, and the role of the paternal genome after fertilization. Asian J. Androl. 13, 620–621 10.1038/aja.2011.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pogany G. C., Corzett M., Weston S., and Balhorn R. (1981) DNA and protein content of mouse sperm. Implications regarding sperm chromatin structure. Exp. Cell Res. 136, 127–136 10.1016/0014-4827(81)90044-6 [DOI] [PubMed] [Google Scholar]
- 11. Balhorn R. (1982) A model for the structure of chromatin in mammalian sperm. J. Cell Biol. 93, 298–305 10.1083/jcb.93.2.298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hammoud S. S., Nix D. A., Zhang H., Purwar J., Carrell D. T., and Cairns B. R. (2009) Distinctive chromatin in human sperm packages genes for embryo development. Nature 460, 473–478 10.1038/nature08162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Brykczynska U., Hisano M., Erkek S., Ramos L., Oakeley E. J., Roloff T. C., Beisel C., Schübeler D., Stadler M. B., Peters A. H. (2010) Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nat. Struct. Mol. Biol. 17, 679–687 10.1038/nsmb.1821 [DOI] [PubMed] [Google Scholar]
- 14. Carone B. R., Hung J.-H., Hainer S. J., Chou M.-T., Carone D. M., Weng Z., Fazzio T. G., and Rando O. J. (2014) High-resolution mapping of chromatin packaging in mouse embryonic stem cells and sperm. Dev. Cell 30, 11–22 10.1016/j.devcel.2014.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Samans B., Yang Y., Krebs S., Sarode G. V., Blum H., Reichenbach M., Wolf E., Steger K., Dansranjavin T., and Schagdarsurengin U. (2014) Uniformity of nucleosome preservation pattern in mammalian sperm and its connection to repetitive DNA elements. Dev. Cell 30, 23–35 10.1016/j.devcel.2014.05.023 [DOI] [PubMed] [Google Scholar]
- 16. Jung Y. H., Sauria M. E. G., Lyu X., Cheema M. S., Ausio J., Taylor J., and Corces V. G. (2017) Chromatin states in mouse sperm correlate with embryonic and adult regulatory landscapes. Cell Rep. 18, 1366–1382 10.1016/j.celrep.2017.01.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yao C., Liu Y., Sun M., Niu M., Yuan Q., Hai Y., Guo Y., Chen Z., Hou J., Liu Y., and He Z. (2015) MicroRNAs and DNA methylation as epigenetic regulators of mitosis, meiosis and spermiogenesis. Reproduction 150, R25–R34 10.1530/REP-14-0643 [DOI] [PubMed] [Google Scholar]
- 18. Chiquoine A. D. (1954) The identification, origin, and migration of the primordial germ cells in the mouse embryo. Anat. Rec. 118, 135–146 10.1002/ar.1091180202 [DOI] [PubMed] [Google Scholar]
- 19. Barton L. J., LeBlanc M. G., and Lehmann R. (2016) Finding their way: themes in germ cell migration. Curr. Opin. Cell Biol. 42, 128–137 10.1016/j.ceb.2016.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Teng F., and Zhou Q. (2013) Epigenetic Re-programming during mammalian preimplantation embryogenesis and PGC development. J. Fertil. In Vitro IVF Worldw. Reprod. Med. Genet. Stem Cell Biol. 10.4172/2375-4508.1000114 [DOI] [Google Scholar]
- 21. de Kretser D. M., Loveland K., and O'Bryan M. (2016) in Endocrinology: Adult and Pediatric (Jameson J. L., and De Groot L. J., eds) 7th Ed., pp. 2325–2353.e9, W. B. Saunders, Philadelphia [Google Scholar]
- 22. Kafri T., Ariel M., Brandeis M., Shemer R., Urven L., McCarrey J., Cedar H., and Razin A. (1992) Developmental pattern of gene-specific DNA methylation in the mouse embryo and germ line. Genes Dev. 6, 705–714 10.1101/gad.6.5.705 [DOI] [PubMed] [Google Scholar]
- 23. Hackett J. A., and Surani M. A. (2013) Beyond DNA: programming and inheritance of parental methylomes. Cell 153, 737–739 10.1016/j.cell.2013.04.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jiang L., Zhang J., Wang J.-J., Wang L., Zhang L., Li G., Yang X., Ma X., Sun X., Cai J., Zhang J., Huang X., Yu M., Wang X., Liu F., et al. (2013) Sperm, but not oocyte, DNA methylome is inherited by zebrafish early embryos. Cell 153, 773–784 10.1016/j.cell.2013.04.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Potok M. E., Nix D. A., Parnell T. J., and Cairns B. R. (2013) Reprogramming the maternal zebrafish genome after fertilization to match the paternal methylation pattern. Cell 153, 759–772 10.1016/j.cell.2013.04.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gardiner-Garden M., and Frommer M. (1987) CpG islands in vertebrate genomes. J. Mol. Biol. 196, 261–282 10.1016/0022-2836(87)90689-9 [DOI] [PubMed] [Google Scholar]
- 27. Molaro A., Hodges E., Fang F., Song Q., McCombie W. R., Hannon G. J., and Smith A. D. (2011) Sperm methylation profiles reveal features of epigenetic inheritance and evolution in primates. Cell 146, 1029–1041 10.1016/j.cell.2011.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Guo H., Zhu P., Yan L., Li R., Hu B., Lian Y., Yan J., Ren X., Lin S., Li J., Jin X., Shi X., Liu P., Wang X., Wang W., et al. (2014) The DNA methylation landscape of human early embryos. Nature 511, 606–610 10.1038/nature13544 [DOI] [PubMed] [Google Scholar]
- 29. Smith Z. D., Chan M. M., Mikkelsen T. S., Gu H., Gnirke A., Regev A., and Meissner A. (2012) A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature 484, 339–344 10.1038/nature10960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Balhorn R., Gledhill B. L., and Wyrobek A. J. (1977) Mouse sperm chromatin proteins: quantitative isolation and partial characterization. Biochemistry 16, 4074–4080 10.1021/bi00637a021 [DOI] [PubMed] [Google Scholar]
- 31. Gatewood J. M., Cook G. R., Balhorn R., Bradbury E. M., and Schmid C. W. (1987) Sequence-specific packaging of DNA in human sperm chromatin. Science 236, 962–964 10.1126/science.3576213 [DOI] [PubMed] [Google Scholar]
- 32. Marczylo E. L., Amoako A. A., Konje J. C., Gant T. W., and Marczylo T. H. (2012) Smoking induces differential miRNA expression in human spermatozoa: a potential transgenerational epigenetic concern? Epigenetics 7, 432–439 10.4161/epi.19794 [DOI] [PubMed] [Google Scholar]
- 33. Arpanahi A., Brinkworth M., Iles D., Krawetz S. A., Paradowska A., Platts A. E., Saida M., Steger K., Tedder P., and Miller D. (2009) Endonuclease-sensitive regions of human spermatozoal chromatin are highly enriched in promoter and CTCF binding sequences. Genome Res. 19, 1338–1349 10.1101/gr.094953.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Weber M., Hellmann I., Stadler M. B., Ramos L., Pääbo S., Rebhan M., and Schübeler D. (2007) Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat. Genet. 39, 457–466 10.1038/ng1990 [DOI] [PubMed] [Google Scholar]
- 35. Wu S.-F., Zhang H., and Cairns B. R. (2011) Genes for embryo development are packaged in blocks of multivalent chromatin in zebrafish sperm. Genome Res. 21, 578–589 10.1101/gr.113167.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dowen J. M., Fan Z. P., Hnisz D., Ren G., Abraham B. J., Zhang L. N., Weintraub A. S., Schujiers J., Lee T. I., Zhao K., and Young R. A. (2014) Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell 159, 374–387 10.1016/j.cell.2014.09.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Battulin N., Fishman V. S., Mazur A. M., Pomaznoy M., Khabarova A. A., Afonnikov D. A., Prokhortchouk E. B., and Serov O. L. (2015) Comparison of the three-dimensional organization of sperm and fibroblast genomes using the Hi-C approach. Genome Biol. 16, 77 10.1186/s13059-015-0642-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ong C.-T., and Corces V. G. (2014) CTCF: an architectural protein bridging genome topology and function. Nat. Rev. Genet. 15, 234–246 10.1038/nrg3663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hernández-Hernández A., Lilienthal I., Fukuda N., Galjart N., and Höög C. (2016) CTCF contributes in a critical way to spermatogenesis and male fertility. Sci. Rep. 6, 28355 10.1038/srep28355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Flyamer I. M., Gassler J., Imakaev M., Brandão H. B., Ulianov S. V., Abdennur N., Razin S. V., Mirny L. A., and Tachibana-Konwalski K. (2017) Single-nucleus Hi-C reveals unique chromatin reorganization at oocyte-to-zygote transition. Nature 544, 110–114 10.1038/nature21711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Perreault S. D. (1992) Chromatin remodeling in mammalian zygotes. Mutat. Res. 296, 43–55 10.1016/0165-1110(92)90031-4 [DOI] [PubMed] [Google Scholar]
- 42. Morgan H. D., Santos F., Green K., Dean W., and Reik W. (2005) Epigenetic reprogramming in mammals. Hum. Mol. Genet. 14, Spec. No. 1:R47–58 10.1093/hmg/ddi114 [DOI] [PubMed] [Google Scholar]
- 43. Nonchev S., and Tsanev R. (1990) Protamine–histone replacement and DNA replication in the male mouse pronucleus. Mol. Reprod. Dev. 25, 72–76 10.1002/mrd.1080250113 [DOI] [PubMed] [Google Scholar]
- 44. Parthun M. R. (2007) Hat1: the emerging cellular roles of a type B histone acetyltransferase. Oncogene 26, 5319–5328 10.1038/sj.onc.1210602 [DOI] [PubMed] [Google Scholar]
- 45. Sobel R. E., Cook R. G., Perry C. A., Annunziato A. T., and Allis C. D. (1995) Conservation of deposition-related acetylation sites in newly synthesized histones H3 and H4. Proc. Natl. Acad. Sci. U.S.A. 92, 1237–1241 10.1073/pnas.92.4.1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lepikhov K., and Walter J. (2004) Differential dynamics of histone H3 methylation at positions K4 and K9 in the mouse zygote. BMC Dev. Biol. 4, 12 10.1186/1471-213X-4-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Inoue A., and Zhang Y. (2011) Replication-dependent loss of 5-hydroxymethylcytosine in mouse preimplantation embryos. Science 334, 194 10.1126/science.1212483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Santos F., Hendrich B., Reik W., and Dean W. (2002) Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev. Biol. 241, 172–182 10.1006/dbio.2001.0501 [DOI] [PubMed] [Google Scholar]
- 49. Fulka H., Mrazek M., Tepla O., and Fulka J. Jr. (2004) DNA methylation pattern in human zygotes and developing embryos. Reproduction 128, 703–708 10.1530/rep.1.00217 [DOI] [PubMed] [Google Scholar]
- 50. Oswald J., Engemann S., Lane N., Mayer W., Olek A., Fundele R., Dean W., Reik W., and Walter J. (2000) Active demethylation of the paternal genome in the mouse zygote. Curr. Biol. 10, 475–478 10.1016/S0960-9822(00)00448-6 [DOI] [PubMed] [Google Scholar]
- 51. Adenot P. G., Mercier Y., Renard J. P., and Thompson E. M. (1997) Differential H4 acetylation of paternal and maternal chromatin precedes DNA replication and differential transcriptional activity in pronuclei of 1-cell mouse embryos. Development 124, 4615–4625 [DOI] [PubMed] [Google Scholar]
- 52. Iqbal K., Jin S.-G., Pfeifer G. P., and Szabó P. E. (2011) Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc. Natl. Acad. Sci. U.S.A. 108, 3642–3647 10.1073/pnas.1014033108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wossidlo M., Arand J., Sebastiano V., Lepikhov K., Boiani M., Reinhardt R., Schöler H., and Walter J. (2010) Dynamic link of DNA demethylation, DNA strand breaks and repair in mouse zygotes. EMBO J. 29, 1877–1888 10.1038/emboj.2010.80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Guo F., Li X., Liang D., Li T., Zhu P., Guo H., Wu X., Wen L., Gu T.-P., Hu B., Walsh C. P., Li J., Tang F., and Xu G.-L. (2014) Active and passive demethylation of male and female pronuclear DNA in the mammalian zygote. Cell Stem Cell 15, 447–459 10.1016/j.stem.2014.08.003 [DOI] [PubMed] [Google Scholar]
- 55. Gu T.-P., Guo F., Yang H., Wu H.-P., Xu G.-F., Liu W., Xie Z.-G., Shi L., He X., Jin S.-G., Iqbal K., Shi Y. G., Deng Z., Szabó P. E., Pfeifer G. P., Li J., and Xu G.-L. (2011) The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature 477, 606–610 10.1038/nature10443 [DOI] [PubMed] [Google Scholar]
- 56. Inoue A., Jiang L., Lu F., Suzuki T., and Zhang Y. (2017) Maternal H3K27me3 controls DNA methylation-independent imprinting. Nature 547, 419–424 10.1038/nature23262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Heard E., and Martienssen R. A. (2014) Transgenerational epigenetic inheritance: myths and mechanisms. Cell 157, 95–109 10.1016/j.cell.2014.02.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sharma U., Conine C. C., Shea J. M., Boskovic A., Derr A. G., Bing X. Y., Belleannee C., Kucukural A., Serra R. W., Sun F., Song L., Carone B. R., Ricci E. P., Li X. Z., Fauquier L., et al. (2016) Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science 351, 391–396 10.1126/science.aad6780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chen Q., Yan M., Cao Z., Li X., Zhang Y., Shi J., Feng G.-H., Peng H., Zhang X., Zhang Y., Qian J., Duan E., Zhai Q., and Zhou Q. (2016) Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science 351, 397–400 10.1126/science.aad7977 [DOI] [PubMed] [Google Scholar]
- 60. Shiels P. G., McGuinness D., Eriksson M., Kooman J. P., and Stenvinkel P. (2017) The role of epigenetics in renal ageing. Nat. Rev. Nephrol. 13, 471–482 10.1038/nrneph.2017.78 [DOI] [PubMed] [Google Scholar]
- 61. Manikkam M., Tracey R., Guerrero-Bosagna C., and Skinner M. K. (2013) Plastics derived endocrine disruptors (BPA, DEHP and DBP) induce epigenetic transgenerational inheritance of obesity, reproductive disease and sperm epimutations. PLoS ONE 8, e55387 10.1371/journal.pone.0055387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ng S.-F., Lin R. C., Laybutt D. R., Barres R., Owens J. A., and Morris M. J. (2010) Chronic high-fat diet in fathers programs β-cell dysfunction in female rat offspring. Nature 467, 963–966 10.1038/nature09491 [DOI] [PubMed] [Google Scholar]
- 63. Lumey L. H., Stein A. D., Kahn H. S., and Romijn J. A. (2009) Lipid profiles in middle-aged men and women after famine exposure during gestation: the Dutch Hunger Winter Families Study. Am. J. Clin. Nutr. 89, 1737–1743 10.3945/ajcn.2008.27038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Dias B. G., and Ressler K. J. (2014) Parental olfactory experience influences behavior and neural structure in subsequent generations. Nat. Neurosci. 17, 89–96 10.1038/nn.3594 [DOI] [PMC free article] [PubMed] [Google Scholar]