Abstract
Bacterial flagella are rotary nanomachines that contribute to bacterial fitness in many settings, including host colonization. The flagellar motor relies on the multiprotein flagellar motor-switch complex to govern flagellum formation and rotational direction. Different bacteria exhibit great diversity in their flagellar motors. One such variation is exemplified by the motor-switch apparatus of the gastric pathogen Helicobacter pylori, which carries an extra switch protein, FliY, along with the more typical FliG, FliM, and FliN proteins. All switch proteins are needed for normal flagellation and motility in H. pylori, but the molecular mechanism of their assembly is unknown. To fill this gap, we examined the interactions among these proteins. We found that the C-terminal SpoA domain of FliY (FliYC) is critical to flagellation and forms heterodimeric complexes with the FliN and FliM SpoA domains, which are β-sheet domains of type III secretion system proteins. Surprisingly, unlike in other flagellar switch systems, neither FliY nor FliN self-associated. The crystal structure of the FliYC–FliNC complex revealed a saddle-shaped structure homologous to the FliN–FliN dimer of Thermotoga maritima, consistent with a FliY–FliN heterodimer forming the functional unit. Analysis of the FliYC–FliNC interface indicated that oppositely charged residues specific to each protein drive heterodimer formation. Moreover, both FliYC–FliMC and FliYC–FliNC associated with the flagellar regulatory protein FliH, explaining their important roles in flagellation. We conclude that H. pylori uses a FliY–FliN heterodimer instead of a homodimer and creates a switch complex with SpoA domains derived from three distinct proteins.
Keywords: molecular motor, protein structure, protein-protein interaction, protein assembly, protein export, bacterial motility, flagellar motor switch complex, SpoA domain, type III secretion system, virulence factor, flagellum, Fli protein, Helicobacter pylori, SpoA domain, FliY
Introduction
Bacterial flagella are rotary nanomachines that contribute to bacterial fitness in a variety of settings, including mammalian and plant colonization (1, 2). Although the basic function of flagella as a motor organelle is conserved, substantial variation exists among microbes in the components used to build and operate key aspects of the flagella. For example, we now know that there are diverse motor structures from cryoelectron tomography studies (3, 4) and that bacterial motors consist of FliG, FliM, and either FliN, FliY, or the combination of both FliN and FliY (5). Although motor diversity is well appreciated, how motors are built from the various combinations of FliN and FliY and how these two proteins interact with the constant motor proteins FliM and FliG remain unknown.
The flagellar motor-switch complex, also called the C-ring, is found at the base of each flagellum and resides within the cytoplasm (6). It plays an important role in flagellum assembly, torque generation, and rotational switching. Numerous studies have dissected the composition, arrangement, and structure of the switch proteins with a focus on those from Escherichia coli and Salmonella enterica serovar Typhimurium that possess FliG, FliM, and FliN. The motor C-rings of these bacteria contain ∼26 copies of FliG, ∼34 copies of FliM, and >110 copies of FliN (7–9). Most of these structures were determined using proteins from other organisms, and their assembly models have been recently proposed (10–22). Electron microscopic and tomographic studies have shown that the organization of the switch proteins is similar across different species such that FliG is the closest to the cytoplasmic membrane followed by FliM, and FliN is found in a distant membrane location toward the cytoplasm (see Fig. 1A) (4, 23, 24). FliG interacts electrostatically with the flagellar proton channel MotA4MotB2 complexes to drive the motor rotation (25). FliM controls the bidirectional motor switching by thermodynamically binding to the phosphorylated response regulator CheY to confer the flagellar response (16, 26). FliN forms a distinctive donut-shaped structure at the base of the switch complex and is critical for protein export via association with the flagellar type III secretion apparatus (11).
Figure 1.
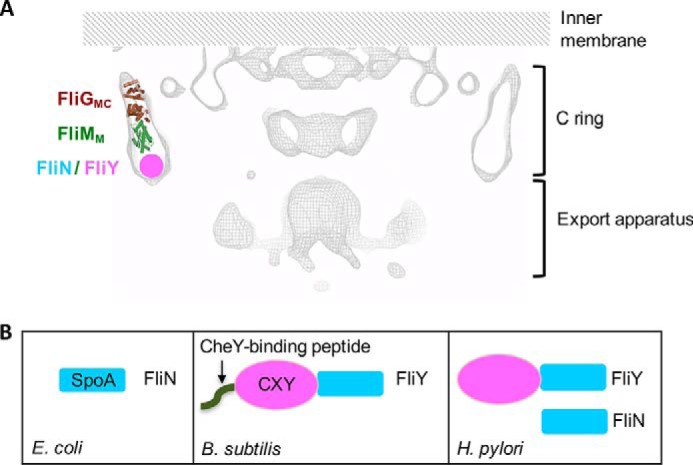
Organization of motor switch proteins. A, cross-sectional view of the H. pylori motor adapted from a recently reported cryoelectron tomographic map (EMD-8459) (24). Only the cytoplasmic portion covering the C-ring and the export apparatus is shown. One FliGMC–FliMM complex is drawn, and the putative position of FliN and/or FliY is indicated. B, domain organization of the FliN/FliY proteins in E. coli, B. subtilis, and H. pylori.
The surface presentation of the antigen (SpoA)5 domain of FliN shares high structural homology to YscQ-C and HrcQB-C, which are the secretion apparatus proteins of the type III secretion systems (T3SSs) from Yersinia pseudotuberculosis and Pseudomonas syringae, respectively (27, 28). The flagellar export apparatus is composed of an export gate complex containing six membrane proteins, FlhA, FlhB, FliO, FliP, FliQ, and FliR, and a cytoplasmic ATPase complex consisting of FliH, FliI, and FliJ. The motor C-ring acts as a platform for the efficient assembly of the ATPase complex to the export gate. There is an intimate connection between the flagellar motor proteins and those of the flagellar secretion apparatus, which drives the assembly of the flagellum. Flagellum assembly begins with the formation of the MS-ring (FliF) followed by the attachment of FliG, FliM, and FliN. The deletion of any of these genes halts flagellum formation (29). The T3SS export gate complex is formed within the FliF MS-ring and is assisted by the ATPase complex, which facilitates the unfolding of the secretion substrates and their delivery through the channel of the export apparatus. The N-terminal segment of FliH recognizes the surface hydrophobic region of dimeric FliN–FliN or FliN–FliM, and the binding concentrates the FliH–FliI complex near the export gate (22, 30–32). Disrupting the FliH–FliN binding causes mislocalization of FliI inside a bacterial cell and impairs flagellar protein export (33).
The main form of variation in the flagellar motors occurs because of FliN. The first recognized variation was identified in Bacillus subtilis and was a substitution of FliY for FliN. FliY consists of FliN-like domains plus a CheY-binding N-terminal peptide and a CheC/CheX/FliY (CXY)-like middle domain (see Fig. 1B). The heterologous expression of FliY in the S. enterica Typhimurium fliN mutant restores its motility and suggests that FliY accomplishes the role of FliN in flagellation (34). In addition, FliY exhibits phosphatase activity toward phosphorylated CheY and therefore helps to regulate the concentration of phospho-CheY (35). The structure of the FliY middle (FliYM) domain from T. maritima was resolved recently, highlighting its structural homology to FliM. One difference between FliM and FliYM is that the latter evolved to contain two EXXN phosphatase motifs, and both motifs display phosphatase activity toward CheY (21).
Interestingly, the gastric pathogen H. pylori and related ϵ-proteobacteria encode both FliN and FliY (5, 36). Previous studies have verified that both genes are necessary for motility in H. pylori. The deletion of FliN or FliY alone allows for partial flagellation, whereas deletion of both aborts flagellation, indicating that these proteins might be partially functionally redundant (37). H. pylori FliY contains the CXY-like domain (FliYN) and the FliN-like domain (FliYC), but it lacks the CheY-binding sequence (Fig. 1B). FliYC shows high sequence conservation to FliN in terms of the functional residues involved in flagellar protein export and homodimerization, but the exact amino acids present are different, which suggests that FliYC and FliN might have distinct roles. Although it has been shown that the H. pylori FliM and FliG counterparts carry similar functions to other bacteria, the functions of FliN and FliY and their interactions with other switch members remain elusive (5, 18, 20). We investigated the function of the H. pylori FliY subdomains and the interaction of the subdomains with other switch components. We demonstrated that FliYC was required for flagellum formation, whereas FliYN regulated the H. pylori chemotactic response. The interaction studies we conducted suggest that FliYC formed functional complexes with the SpoA domain–containing proteins FliN and FliM. The atomic details underlying the formation of the FliYC–FliNC complex were revealed by crystallographic studies. Both the FliY–FliN and FliY–FliM complexes interacted with FliH. Therefore, we determined that H. pylori distinctively incorporated three SpoA domains in the motor C-ring to ensure proper flagellum assembly and functioning.
Results
Both domains of FliY were needed for normal flagellar function
H. pylori FliY contains an N-terminal FliM-like domain and a C-terminal SpoA domain (5). Although it has been shown that FliY is crucial for normal flagellation, the specific role of these two domains remains unclear. We therefore created strains that expressed either the full-length FliY or just the N- or C-terminal domains. This experiment was accomplished by cloning the full-length fliY gene and its truncated fragments into pILL2157 (38) and expressing these in a fliY-null mutant (5). The protein expressions of His6-FliY, His6-FliYN, and FliYC-His6 were confirmed by Western blotting (Fig. S1). We first examined flagellation using EM of negatively stained bacterial cells. The strain that expressed FliYN (FliYN strain) was nonflagellated (Fig. 2A), and the bacteria were immotile (Fig. 2B), consistent with the role of the SpoA domain in flagellum biogenesis. The strain expressing FliYC (FliYC strain), by contrast, displayed WT flagellation with distinctive terminal bulbs (Fig. 2B), implying that FliYN was not required for flagellum assembly. However, the bacterial soft agar migration halo in the FliYC strain was significantly smaller than that of the WT (57.5%), suggesting that the N-terminal domain of FliY was needed for normal flagellar function (Fig. 2B). The reduced soft agar migration could be due to impaired chemotactic behavior (clockwise or counterclockwise bias), reduced growth rate, or defects in flagellum formation (39). Because the FliYN domain belongs to the CXY family that is involved in regulating chemotaxis (21), we examined the swimming behavior of the FliYC strain by fixed-time diffusion analysis (5, 18). The diffusion exponent reflects how close the swimming behavior is to pure diffusion (α = 1). A tumbling bacterium will have an α value close to 1, whereas a bacterium that moves relatively straight will show an α value close to 2. We found that the FliYC strain had an α value of 1.57 ± 0.02, which is closer to 1 when compared with the FliY+ strain (1.67 ± 0.02) (Fig. 2C). The diffusion analysis suggested that the deletion of the FliY N-terminal domain might cause tumbling bias. These data support the idea that FliYN is not needed for flagellation but plays a role in a normal chemotactic response.
Figure 2.
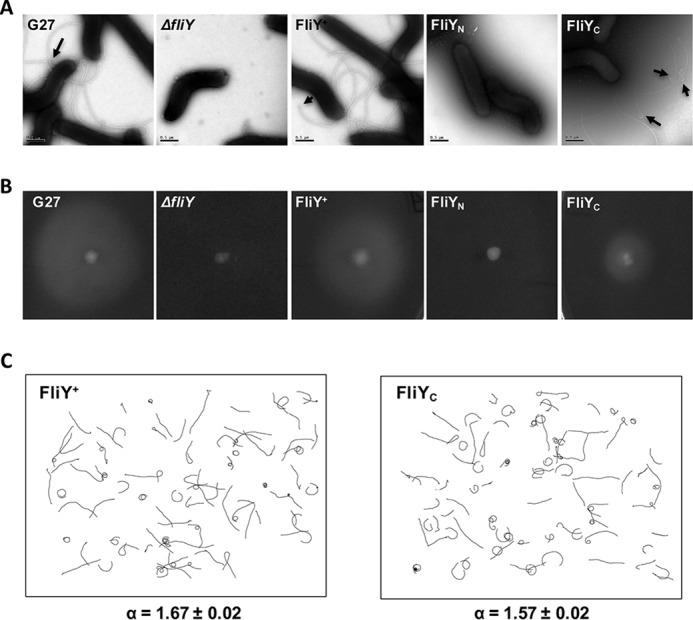
Both FliYN and FliYC were needed for normal flagellar function. A, electron micrograph of bacterial cells, demonstrating the effect of N-terminal or C-terminal truncations of FliY on flagellation. Arrows mark the terminal bulb structure. Scale bars, 0.5 μm. B, soft agar assay. Representative images show colony diameter in WT (n = 18), ΔfliY (n = 26), FliY+ (n = 17), FliYN (n = 27), and FliYC (n = 25) strains measured after 5–7 days. The colony diameter of FliY+ and FliYC strains was 90.6 ± 11 and 57.5 ± 13.4% of WT strain, respectively. C, swimming tracks of complemented FliY+ and FliYC strains used to calculate the diffusion coefficient. Four-second swimming tracks are plotted using the same scale. The diffusion exponent, α, is given at the bottom. The number of tracks analyzed for FliY+ and FliYC strains was 76 and 80, respectively.
FliY associated with both the FliN and FliM switch proteins
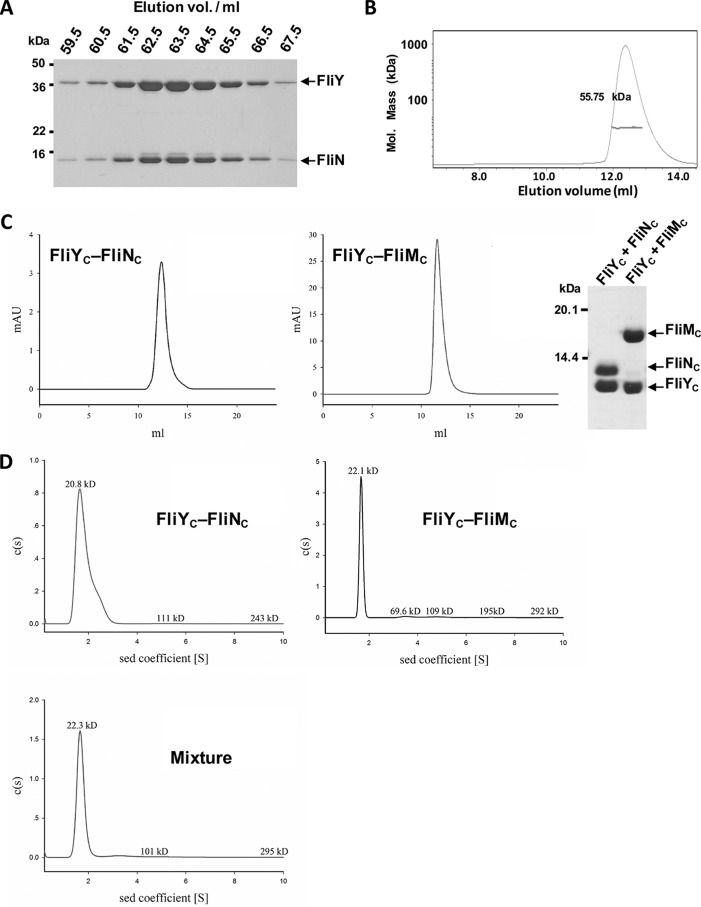
We next examined the protein–protein interactions of FliY. In the flagellar T3SS of E. coli and T. maritima, the SpoA domain within the FliN C-terminal portion participates in two types of protein–protein interactions. It can self-associate to form a homodimer or associate with FliM to form a heterodimer. Both dimeric forms further assemble into macroprotein ringlike complexes (11, 22, 40). The H. pylori FliY, FliM, and FliN each carry a SpoA domain and have the potential to self-assemble or assemble with each other. To assess their interactions, we first attempted to isolate the individual proteins of the full-length or C-terminal domains of FliN, FliY, or FliM. In contrast to E. coli FliN that exists as a dimer or tetramer in solution (11), all three proteins tended to form aggregates when expressed and purified individually, precluding further analysis. Because the SpoA domain could require its cognate interacting partner to become stable (11, 22), we conducted coexpression and copurification assays to examine their interactions. The recombinant GST-tagged FliY and His6-tagged FliN were coexpressed in E. coli. The expressed proteins were purified by Ni-NTA chromatography followed by GSH-Sepharose at which point the GST fusion tag was removed. FliY and FliN were found to be coeluted from the size exclusion chromatography experiment (Fig. 3A). The stoichiometry of the FliY–FliN interaction was also measured by static light scattering. The molecular mass of the complex was determined to be 55.75 kDa, which is close to a FliY:FliN ratio of 1:1 (Fig. 3B).
Figure 3.
FliY formed complexes with FliN and FliM. A, copurification analysis. GST-FliY was coexpressed with His6-FliN in E. coli. The protein mixtures were purified by Ni-NTA and GSH-Sepharose chromatography in which the GST fusion tag was removed by 3C protease. The eluted proteins were separated by Superdex S200 gel filtration chromatography and analyzed by SDS-PAGE. B, static light scattering of FliY–FliN. The curve is the refractive interference signal. The value shown is the native molecular mass of FliY–FliN. The line under the peak indicates the calculated molecular mass of the eluted FliY–FliN complex throughout the peak. C, interactions between FliYC and FliNC or FliMC. GST-FliYC was coexpressed with His6-FliNC or His6-FliMC. The proteins were finally separated by Superdex S75 chromatography. The purified complexes were analyzed by SDS-PAGE. D, the sedimentation velocity analysis of FliYC–FliNC, FliYC–FliMC, and a mixture of the two complexes. All data were collected using an absorbance optical system at a wavelength of 280 nm. Data analysis was performed with SEDFIT, and data were also analyzed using the sedimentation (sed) coefficient distribution model, c(s). mAU, milliabsorbance units.
We next studied the coexpression of the GST-tagged FliYC (residues 208–285) or FliM with either His6-tagged FliN, FliNC (residues 44–122), or FliMC (residues 244–354). FliNC and FliMC were not soluble when coexpressed, indicating these two proteins might not form a stable complex (data not shown). However, we isolated homogeneous stable complexes of FliYC with FliNC and of FliYC with FliMC from the gel filtration column (Fig. 3C). We further carried out a sedimentation-velocity experiment to assess the stoichiometry of the individual components in the FliYC–FliNC and FliYC–FliMC complexes. Both protein complexes behaved dominantly as a single species in solution with sedimentation coefficients of 1.65S and 1.68S, respectively (Fig. 3D). We noted that a fraction of the purified FliYC–FliNC tended to self-associate, especially when the experiment was conducted at a higher protein concentration (data not shown).
We further investigated the interaction between the FliYC–FliNC and FliMC–FliYC complexes by sedimentation velocity (Fig. 3D) and gel filtration analysis (Fig. S2). There was no obvious peak shift in either experiment, suggesting that the two protein complexes did not associate to form a higher-order oligomer. Alternatively, the tripartite association of the full-length FliY, FliN (His6-tagged), and FliM (GST-tagged) was tested by coexpression and copurification. The full-length FliN, FliY, and FliM were coeluted after a two-step affinity chromatography purification, indicating that these three proteins associated to form higher-order protein complexes in the presence of their N-terminal domains. However, in a gel filtration analysis, we found that FliY–FliN–FliM ternary complex was eluted in a broad region, suggesting that multiple oligomeric species were present (Fig. S3). It was estimated that FliY, FliN, and FliM were in an equal stoichiometric ratio in the earlier eluted fractions. However, the ratio shifted to 2:1:1 in the later eluted fractions.
FliY did not interact with FliG
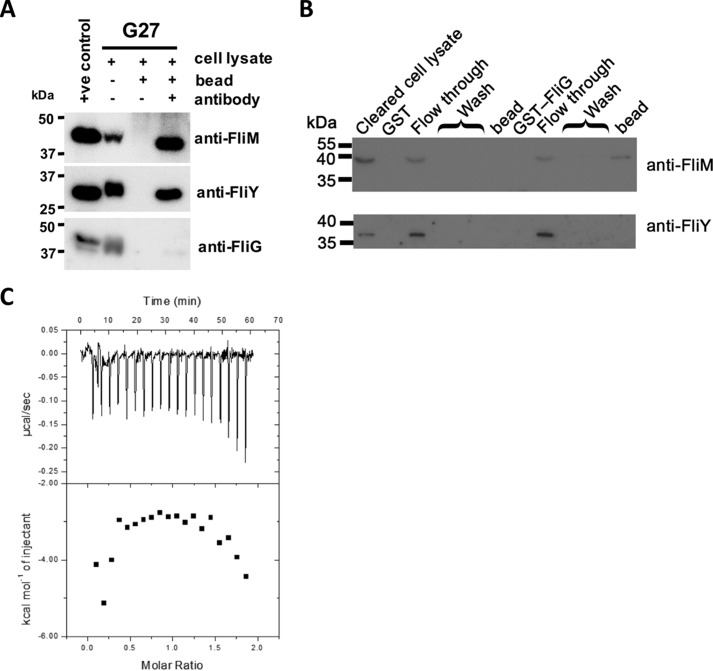
FliYN shares a common protein fold with the middle domain of FliM in that they both contain a GGXG motif that is known to be important in mediating the formation of the FliM–FliG complex (16, 20). The presence of this motif raises the possibility that FliYN might also bind FliG. We conducted a coimmunoprecipitation assay to isolate the potential endogenous FliY–FliG complex. The anti-FliY antibody was immobilized to pull down FliY and its complexes from the cell lysate of H. pylori strain G27. The coprecipitated FliG and FliM were detected by anti-FliG and anti-FliM antibodies, respectively. Our results showed that FliM, but not FliG, was coprecipitated with FliY (Fig. 4A). The biophysical association of FliY and FliG was also tested by using GST pulldown assays. The cell lysate of the G27 fliG-null mutant strain was loaded onto preimmobilized GST-FliG. The results from the immunoblot showed that FliM, but not FliY, was captured by GST-FliG (Fig. 4B). We considered that FliY and FliG might have weak interaction, so we conducted an isothermal titration calorimetry experiment. When FliG was titrated against FliYN, a heat change was not observed (Fig. 4C). Taken together, our data strongly support the idea that FliY does not directly interact with FliG and that these two proteins form a multiprotein complex indirectly through FliM.
Figure 4.
FliY interacted with FliM but not FliG. A, immunoprecipitation assay. The cell lysate of H. pylori G27 strain was mixed with protein G beads with or without preimmobilized anti-FliY antibody. The bound proteins were probed with anti-FliM, anti-FliY, or anti-FliG antibody. Positive controls using recombinant full-length FliM, FliY, and FliG were included. B, pulldown experiment. The bacterial cell lysate of the G27 ΔfliG strain was loaded onto beads preimmobilized with purified GST-FliG or GST. The fractions of flow-through, wash, and protein bound on the beads were analyzed by probing with anti-FliM and anti-FliY antibodies. C, ITC experiment. FliYN was titrated into the cell containing FliG.
Crystal structure of the FliYC–FliNC complex
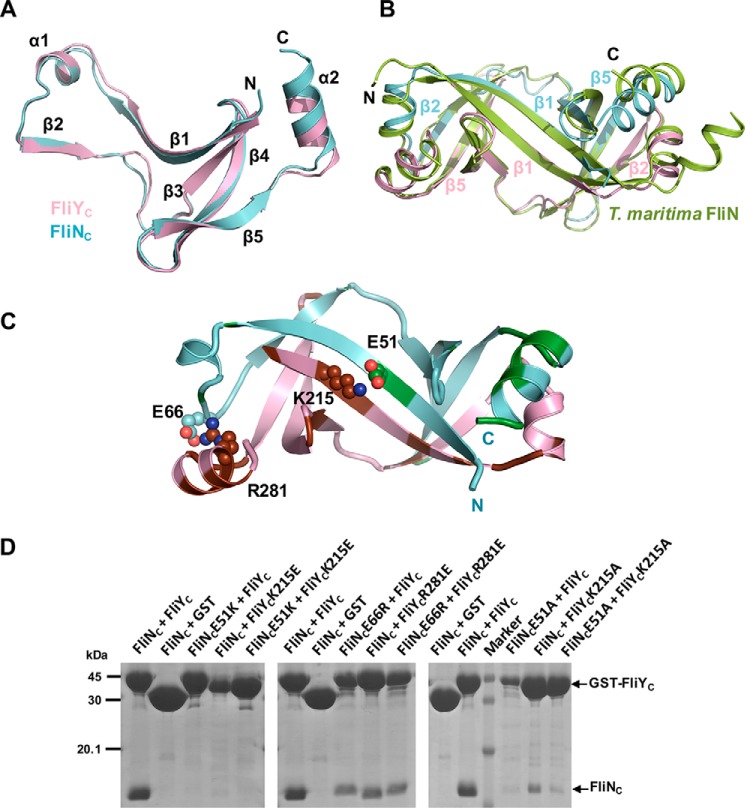
We next probed the molecular basis of the FliY–FliN interactions. We purified the FliYC–FliNC complex, crystallized it, and resolved the structure at 2.5 Å with an R factor and Rfree of 20.26 and 22.86%, respectively (Table S1). There are two FliYC–FliNC complexes per asymmetric unit, and they are virtually identical. Individually, FliYC and FliNC resembled a typical SpoA fold, and they contained a β-sheet comprising four antiparallel β-strands and a protrusion (β1-α1-β2) (Fig. 5A). They formed a heterodimer that buried an extensive interfacial area of 2,288 Å2 per molecule (PDBePISA) (Fig. 5B). The structural comparison by PDBeFold indicated that both FliYC and FliNC shared the highest structural homology to T. maritima FliN with a root mean square deviation of 0.87 and 1.5 Å, respectively. FliYC shared higher sequence identity to T. maritima FliN (52%) than did H. pylori FliN (38%). The complex showed a characteristic saddle-shaped structure formed by the “protrusions” of one monomer packed against the “platform” of the other monomer, forming two symmetric antiparallel β-barrels by extensive hydrophobic interactions. The complex was further stabilized by hydrogen bonding between the main chain of the antiparallel β-strands and salt bridges between the side chains: Lys215FliY::Glu51FliN, Lys222FliY::Glu45FliN, Asp274FliY::Lys67FliN, and Arg281FliY::Glu66FliN.
Figure 5.
Crystal structure of FliNC–FliYC. A, FliNC (pink) and FliYC (cyan) share high structural homology, comprising a platform of four antiparallel β-strands and a β1-α1-β2 protrusion. B, the superimposition of FliNC–FliYC to FliN homodimer of T. maritima (PDB code 1O6A). Heterodimer formation was mediated via the protrusion of FliYC or FliNC packed against the platform of FliNC or FliYC, especially via the β1–β1 and β2–β5 interactions. C, interaction of FliYC and FliNC. FliN- and FliY-specific residues are colored in green and brown, respectively. The residues that formed salt bridges are shown as spheres. D, effect of FliN or FliY mutations on complex formation. The GST-FliYC WT or mutants were coexpressed with His6-FliNC, and the FliY–FliN complexes were captured by GST resin.
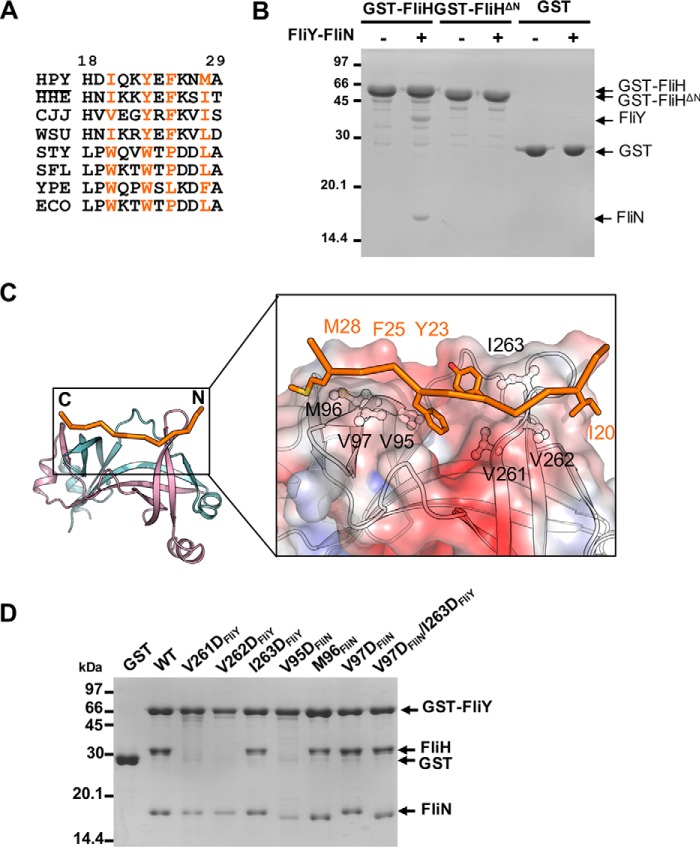
Given the high structural and sequence homology of FliYC and FliNC with their counterparts in other bacterial species, it is intriguing why they specifically form a heterodimer instead of a homodimer. We speculate that the specificity could be attributable to the intermolecular surface complementarity of the proteins. The electrostatic surface calculation of FliYC and FliNC at the interacting interface showed that FliYC was markedly electropositive compared with FliNC (Fig. S4A). Based on the multiple sequence alignment of FliYC and FliNC from the ϵ-proteobacteria containing both FliY and FliN proteins (5), we identified several FliY- and FliN-specific residues at the interface (Fig. S4, A and B). Notably, H. pylori FliY β1 carried several positively charged residues that were replaced either by hydrophobic, polar, or negatively charged residues in H. pylori FliN. For example, Lys215FliY and Arg281FliY were replaced by Ser49FliN and Ile115FliN, and these two FliY residues formed salt bridges with Glu51FliN and Glu66FliN, respectively (Fig. 5C). We therefore suspected these protein-specific interacting residue(s) could be important for the formation of the heterodimer. Accordingly, we mutated these residues into oppositely charged residues to test their importance in maintaining the interaction by a pulldown experiment using GST-FliYC and His6-FliNC (Fig. 5D). We found that K215EFliY and E51KFliN abolished the interaction with their WT FliN and FliY, respectively, and R281EFliY and E66RFliN also showed mild impairment. We concluded that the oppositely charged residues at the FliY–FliN interface contributed to the complementary interaction.
Both FliYC–FliNC and FliYC–FliMC interacted with FliH
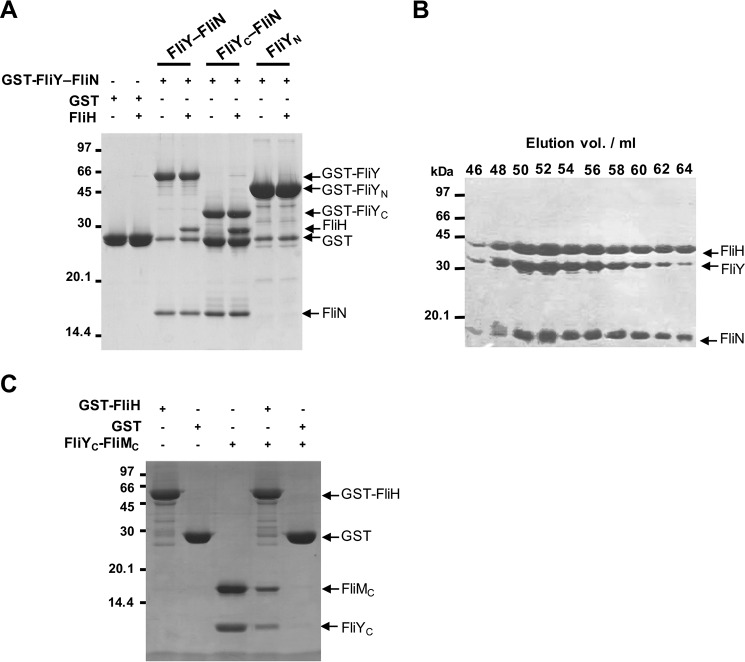
FliN and FliM participate in flagellar protein export by recruiting FliH–FliI to the export gate through protein docking of the FliH N-terminal peptide (FliHN) to their SpoA domains (22, 31–33). FliHN recognizes a hydrophobic surface formed by the homodimeric FliN or heterodimeric FliN–FliM. In H. pylori, FliY, FliN, and FliM are all required for normal flagellation, and they contain conserved sequences for FliH binding (5). It is possible that the analogous FliY–FliN and FliY–FliM in H. pylori identified in this study might be capable of interacting with FliH. To test this possibility, we analyzed the association of FliY–FliN with FliH by pulldown and gel filtration assays. The purified FliH was trapped by the GST-FliY–FliN complex in the GST pulldown experiment (Fig. 6A). FliH also formed a stable complex with FliY–FliN, and the three proteins coeluted with a peak shift of 52 ml compared with the elution peak of the FliY–FliN complex at 63 ml (Figs. 6B and 3A). To determine the domain responsible for the binding, we performed the pulldown experiment using GST-FliYC–FliN or GST-FliYN alone. FliH only bound to GST-FliYC–FliN, showing that the C-terminal SpoA domains mediated the interaction. In addition, we found that purified FliYC–FliMC was also pulled down by GST-FliH (Fig. 6C). Taken together, these results suggest that both FliYC–FliNC and FliYC–FliMC in H. pylori functioned as a protein-docking platform for FliH.
Figure 6.
Both FliY–FliN and FliY–FliM associated with FliH. A, pulldown experiment. The purified GST, GST-FliY–FliN, GST-FliYN, and GST-FliYC–FliN were immobilized on GST beads, and the resins were incubated with FliH in a 1:1.5 molar ratio. B, the purified FliH was preincubated with FliY–FliN at 4 °C for 1 h and injected onto a Superdex S200 column. The elution peak fractions were analyzed by SDS-PAGE. C, the resin preimmobilized with GST or GST-FliH was incubated with FliYC–FliMC. The beads were washed three times with the binding buffer, and the proteins were denatured with SDS loading dye for analysis.
FliY and FliN shared a common docking site for FliH
FliHN is crucial for the association of FliH with the switch proteins, specifically for the recognition of the conserved hydrophobic patch on FliN–FliN or FliN–FliM (22, 31–33). We determined the alignment of FliH from multiple species and noted a conserved pattern of hydrophobic residues consisting of HXXHXHXXH where H denotes hydrophobic residues despite the variability in the length and amino acid composition of FliHN (Fig. 7A). Previous work identified two tryptophan residues that play a crucial role in the interaction of FliH with FliN–FliM by forming an aromatic clamp that inserts into the hydrophobic pocket of S. enterica Typhimurium FliN–FliM (22, 33). Notably, these two residues were not conserved in FliH of the ϵ-proteobacteria as compared with FliH from other bacteria (Fig. 7A). Thus, it is unclear how FliHN in H. pylori interacts with FliN–FliM. To address this question, we created a truncation mutant of FliH by deleting the N-terminal 27 residues, including most of the conserved segment (FliHΔN). The truncated protein was well expressed, but consistent with our prediction, it completely abolished the FliH interaction with FliY–FliN (Fig. 7B). This finding suggests that H. pylori FliH used its N-terminal domain to recognize the SpoA domains, although the exact sequence and recognition mechanism appear to differ.
Figure 7.
Mapping the binding interface between FliH and FliY-FliN. A, excerpted T-COFFEE alignment of H. pylori (HPY) FliH with its homologues from Helicobacter hepaticus (HHE), C. jejuni (CJJ), Wolinella succinogenes (WSU), S. enterica Typhimurium (STY), Shigella flexneri (SFL), Yersinia pestis (YPE), and E. coli (ECO). The conserved hydrophobic residues are colored orange (H. pylori numbering). B, effect of FliH truncation on the binding to FliY-FliN. GST, GST-FliH, or GST-FliHΔ27 was immobilized, and the beads were incubated with purified FliY-FliN. C, model of H. pylori FliHN (residues 18–29) docked onto FliYC–FliNC. The initial model of FliHN was generated by Modeler using FliHN of S. enterica Typhimurium (PDB code 4YXC) as a template (44). The peptide was docked onto the FliYC–FliNC complex using the flexible docking protocol Rosetta FlexPepDock (55). The model with the highest score was chosen. The binding site for FliHN is shown as electrostatic surface in the boxed-in region. The molecular surface was calculated by APBS (Adaptive Poisson-Boltzmann Solver) and contoured at keT = ±3. The conserved hydrophobic residues of FliHN and FliYC–FliNC that participated in the interaction are represented as stick and ball-and-stick models, respectively. D, effect of FliY–FliN mutations on FliH interaction. The GST-FliY–FliN mutants were immobilized on resin followed by incubation with purified FliH.
We next modeled the H. pylori FliHN, composed of residues 18–29, based on the available structure from S. enterica Typhimurium, and docked the peptide onto the FliYC–FliNC structure (Fig. 7C). Notably, the conserved sequence with two HXXH motifs formed two hydrophobic clamps holding the FliH peptide onto the hydrophobic ridges of the C-terminal end of the β4 strand of FliY and FliN. The two aromatic residues docked into the central pocket, while the aliphatic residues bound to the side of the ridges. The conserved residues at the hydrophobic ridges, including Val95, Met96, and Val97 of FliN and Val261, Val262, and Ile263 of FliY, were likely involved in this association. To test this possibility, we conducted a mutagenesis study by substituting these residues with aspartic acid and assessing the impact on the binding to FliH (Fig. 7D). We found that V261DFliY, V262DFliY, and V95DFliN drastically impaired the interaction, whereas the I263DFliY, M96DFliN, V97DFliN, and V97DFliN/I263DFliY double mutant did not have a significant effect. This outcome agrees with a model in which both FliY and FliN contribute to the interaction with FliH. The side chains of Val95FliN and Val261FliY pointed inward to the hydrophobic pocket, which the bulky aromatic side chains of Tyr23FliH and Phe25FliH occupied, and this orientation could explain why both of these mutations abolished the interaction. Although Val262FliY and Met96FliN faced the outside of the pockets, Val262FliY, but not Met96FliN, interfered with the binding. This outcome could be due to the specific orientation of the FliH peptide on FliY–FliN, which still must be resolved by their cocrystal structure.
Discussion
The presence of both FliY and FliN as switch proteins is a common feature within the ϵ-proteobacteria, and their coexistence could be associated with the adaptation of the motility system in these species (5). Somewhat surprisingly, we found that FliY, but not FliN, complexed with FliM, and these proteins selectively interacted to produce FliYC–FliNC and FliYC–FliMC complexes as functional units. This finding agrees with the notion that FliY and FliM are likely structurally and functionally associated. This idea is further supported by the fact that the fliY and fliM genes are co-operonic, whereas fliN is cotranscribed with other genes that are not known for flagellar function (5). Recent interactome studies in H. pylori and Campylobacter jejuni have also found the binary complexes FliY–FliM and FliY–FliN but not FliN–FliM (41, 42). The idea that the FliY–FliN and FliY–FliM heterodimers form the functional units of the flagellar motor is also supported by the experimental evidence that both complexes interacted with FliH. Our data suggest that FliYC–FliNC formed a common FliH-docking surface, and the deletion of either the fliY or fliN gene was likely to affect the synthesis of the protein export platform (5). FliY could play a more significant role in flagellation than FliN because it is involved in the formation of both the FliY–FliN and FliY–FliM complexes for FliH docking, and this hypothesis is supported by a previous in vivo study (5). This scenario is distinct from Bacillus cereus, which also carries all three genes, but the fliY-null mutant remains fully flagellated (43). We speculated that both FliY and FliN were likely able to form a stable complex with FliM in this species to support flagellation.
What is the functional significance of H. pylori carrying two heterotypic SpoA units? These units could facilitate the placement of the correct numbers of the FliYN module in the motor unit, which is important to chemotactic regulation. The impaired expression of FliYN causes a flagellar rotational bias that could be detrimental to the survival of the bacteria in the host (36, 43). Moreover, the excess FliY could cause an overcrowding of more than 30 copies of ∼25-kDa FliYN domains in the C-ring that hinders the docking of other proteins. Interestingly, other bacteria appear to have evolved different approaches to enable the proper assembly of FliY and FliN. Although T. maritima carries only the fliY and fliM genes, there is an alternative translation initiation site in the fliY gene that allows the expression of both FliY- and FliN-like proteins (40). Alternatively, the accumulated protein–protein interaction data in H. pylori and C. jejuni suggest that FliY and FliN might interact further with a different set of proteins (41, 42). FliY might also interact with the flagellar biosynthesis protein FlhF and nonflagellar proteins, suggesting that the presence of both FliY and FliN could contribute to an additional level of control for flagellum formation in the proper spatial and temporal manner. However, this idea requires verification.
FliYN, however, is dispensable for flagellation in H. pylori. Although FliYN shares a similar topology with FliMM and carries a conserved GGXG motif that is important for FliG interaction (21), our data suggest that FliYN and FliMM had distinct functions. Similar to the T. maritima FliY (21), the H. pylori FliYN did not associate with FliG. Hence, FliYN was unlikely a structural component of the C-ring and was not critical for flagellum assembly. In an attempt to explain the differences between FliYN and FliMM, we built a homology model of FliYN based on the T. maritima FliYM structure (PDB code 4HYN) (44). The key hydrophobic residue I149 in FliM was replaced by the charged residue Asp92 in FliY (Fig. S5), and the I149D substitution in FliMM completely abolished FliG–FliM association (20). Thus, the subtle variation at the FliM–FliG interface likely differentiated the role of FliY from FliM. FliYN belongs to CheC/CheX/FliY phosphatase family. The FliY truncation mutant showed a clockwise rotational bias, which seems consistent with its potential role as a CheY phosphatase. However, the H. pylori FliY lacks a CheY-binding sequence that is required for CheY dephosphorylation in B. subtilis (21, 35). A sequence analysis showed that FliYN contained one putative phosphatase consensus motif, -EXXXN-, which is slightly different from the consensus -EXXN- motif in the CheC/CheX/FliY family and the -EXXXQ- motif in CheZ (45). The chemotactic pathway of H. pylori contains multiple CheY-containing CheVs and CheA (5, 46). It is possible that FliYN might target other response regulators and act as a phosphatase toward these proteins.
We also proposed a model of C-ring assembly in H. pylori (Fig. 8) that represents the ϵ-proteobacteria that are distinct from bacteria possessing only FliN or FliY. We previously reported that the FliM–FliG interaction is conserved and that they form the upper part of the C-ring (20). The FliY–FliN interaction is predicted to assemble at the bottom part of the switch complex through the interaction with FliY–FliM, resulting in a donut-shaped dimer of heterodimers (21). The FliYN domain did not interact with other switch proteins and is likely located at the periphery of the C-ring. The comparison of the C-rings between H. pylori and S. enterica Typhimurium, however, did not differ significantly except that the H. pylori C-ring was larger in diameter (57 nm compared with 40 nm) (24). The H. pylori cryo-ET map did not provide information about the position of the FliYN domain. We speculate that the linkage between the N- and C-terminal domains of FliY is flexible, and therefore the density is seemingly invisible in the cryo-ET map. The higher protein content, attributed to FliY, might be partially related to the increased size of the H. pylori motor C-ring. In contrast, a recent cryo-ET map from Leptospira interrogans, which harbors both fliY and fliN genes in the genome, revealed extra density at the bottom outer part of the C-ring that might correspond to the FliYN domain (47). A higher-resolution cryo-EM study of the isolated switch protein complex of H. pylori is definitely needed to further characterize the structure of this bacterial motor.
Figure 8.
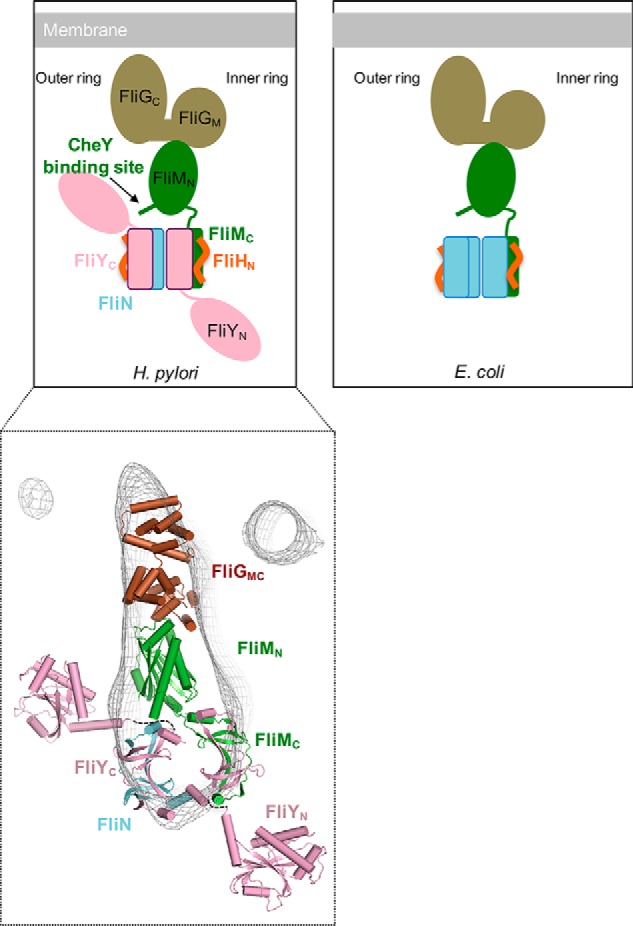
Diagram comparing the switch complex model of H. pylori with E. coli. The molecular interactions of FliG–FliM and FliM–CheY in H. pylori were verified in previous studies (5, 18, 20). We studied the associations among FliYC, FliMC, FliNC, and FliHN. The binding between FliY–FliM and FliY–FliN and the stoichiometry of the switch proteins remain unknown. A proposed model of FliGMC–FliM–FliY–FliN manually docked into the cryoelectron tomography map (EMD-8459) is shown. The dotted lines indicate potential flexible linkers connecting FliYN to FliYC. The models of FliMC and FliYN were generated by MODELLER using FliMC from S. enterica Typhimurium (PDB code 4YXB) and FliYM from T. maritima (PDB code 4HYN) as templates, respectively.
In summary, our data provide a basis to understand the macromolecular assembly of the distinctive C-ring complex of ϵ-proteobacteria. This species-specific macromolecular assembly prompts a different genetic control of flagellum biogenesis in response to environmental cues and awaits further investigation.
Materials and methods
Cloning, expression, and purification
cDNA encoding FliY (hp1030), FliY N-terminal domain (FliYN; residues 1–195), FliY C-terminal domain (FliYC; residues 108–285), FliM (hp1031), FliH (hp0353), and N-terminally truncated FliH (FliHΔN; residues 28–258) were cloned into pGEX-6p-1 vector. FliN (hp0584), FliNC (residues 45–123), and FliMC (residues 244–354) were cloned into pAC28 vector (48).
pGEX-6p-1-FliY, -FliYC, or -FliMC was cotransformed with pAC28-FliN or -FliNC in E. coli BL21 (DE3) strain and coexpressed by induction with 0.3 mm IPTG under growth conditions of 16 °C for 20 h. pGEX-FliYN was expressed under the same growth conditions. The expression of pGEX-FliH was performed at 25 °C instead. For the copurification of His6-FliY–FliN, His6-FliYC–FliN, crude cell extract was first applied to a GST column in buffer (500 mm NaCl, 20 mm HEPES, pH 7.3). After the removal of the GST tag by Prescission protease, the eluted proteins were further purified with Ni-NTA resin in buffer (150 mm NaCl, 20 mm HEPES, pH 7.3, 20 mm imidazole). The eluted protein was subjected to Superdex 200 size exclusion chromatography. FliYN and FliH were purified by a GST column followed by gel filtration. For the purification of proteins carrying the GST tag, proteins were eluted with buffer containing 20 mm reduced glutathione.
For the purification of His6-FliNC–FliYC or His6-FliYC–FliMC, the cell pellet was resuspended in lysis buffer containing 20 mm HEPES, pH 7.5, 250 mm NaCl and lysed by sonication. Clear lysate was loaded onto Ni-NTA. After washing with buffer containing 20 mm HEPES, pH 7.5, 250 mm NaCl, 20 mm imidazole, the target proteins were eluted by buffer containing 20 mm HEPES, pH 7.5, 250 mm NaCl, 250 mm imidazole. Before binding onto GSH-Sepharose, 4 mm DTT and 20 mm l-Arg were added into the elution fraction. After binding and washing, the GST tag was removed by Precission protease (for FliYC-FliMC, 200 mm l-Arg was added before adding Precission protease). Eluted proteins were further purified by a Superdex 75 column in lysis buffer with 4 mm DTT.
For the copurification of FliY–FliN–FliM complexes, fliN and fliY were cloned into pAC28 vector with fliN fused with an upstream His6 tag sequence, and fliM was cloned into pGEX-6p-1 vector. A ribosomal binding sequence was inserted at upstream of fliY for coexpression. Proteins were coexpressed in E. coli Rossetta2 strain by induction with 0.1 mm IPTG and incubated at 20 °C overnight. The complex was purified by Ni-NTA and GST-affinity chromatography followed by Superdex S200 chromatography.
For the H. pylori functional complementation studies, fliY (HPG27_398), fliYN (residues 1–195), and fliYC (residues 108–285) were amplified from the genomic DNA of H. pylori G27 strain using the primers fliYF (5′-ggggggcatatgcatcaccatcaccatcaccaagattttattaagatttttattcaagaggttgtt), fliYR (5′-ggggggggatccttaatgtttcaattgttctaagcgttcttttttagtg), fliYNR (5′-ggggggggatccttaagacgcatcgtgggttttaatct), fliYCF (5′-ggggggcatatgaacatagaaatccgcaatatcagcatgc), and fliYCR (5′-ggggggggatccttagtgatggtgatggtgatgatgtttcaattgttctaagcgttcttttttagtg). His6 tag sequences were included in the forward primers of fliY and fliYN and the reverse primer of fliYC. The genomic DNA was extracted from H. pylori according to the DNeasy Blood and Tissue kit (Qiagen). After amplification, the PCR products were cut with NdeI and BamHI and cloned into similarly cut pILL2157 (kindly provided by Prof. Ivo Boneca) (39) to generate plasmids pILL2157-fliY, -fliYN, and -fliYC. These plasmids replicate exogenously in H. pylori and allow IPTG control of the cloned genes.
H. pylori growth conditions
H. pylori G27 strain was cultured at 37 °C on Columbia blood agar with 5% defibrinated horse blood and H. pylori-selective antibiotics (including trimethoprim, amphotericin, vancomycin, cycloheximide, cefsulodin, polymyxin, and β-cyclodextrin) under microaerobic conditions (5% CO2, 4% O2, and 91% N2) produced by AnaeroGen gas packs (Oxoid). 5 μg/ml chloramphenicol was included for the selection of H. pylori transformants. For liquid culture of H. pylori, Brucella broth containing 10% (v/v) fetal bovine serum (BB10) was used.
H. pylori transformation
The H. pylori G27 deletion strain (ΔfliY::aphA3; called ΔfliY here) was created in a previous study (5). The plasmids pILL2157-fliY, -fliYN, and -fliYC were introduced into ΔfliY strain by natural transformation, generating FliY+, FliYN, and FliYC strains, respectively. ∼20 μg of plasmids were methylated by the addition of H. pylori G27 cell-free extract as described in Donahue et al. (49). For transformation, H. pylori was streaked onto a blood agar plate and allowed to grow for 1 day. The cells were collected and restreaked as a small patch and further grown for 6 h. ∼20 μg of cell-free extract–treated plasmids were stirred with the cells, and the plate was incubated overnight. The cells were recollected and streaked onto a blood agar plate with chloramphenicol. After incubation for 3 days, eight isolated colonies were selected and restreaked on selective plates twice. Positive transformants were verified by PCR analysis using a forward primer complementary to the target gene and a reverse primer complementary to the pILL2157 vector (5′-cagggcggggcgtaattttt-3′).
Immunoblot detection of FliY
To produce anti-FliY antibody, a mouse was immunized with purified recombinant FliYN protein. To detect the expression of FliY, H. pylori G27, ΔfliY strain, and ΔfliY complemented strains transformed with the plasmid (pILL2157-fliY, -fliYN, or -fliYC) were grown in BB10 for 1 day. Cells were pelleted and resuspended in PBS. Total cellular proteins were prepared by boiling with Laemmli sample buffer. The expression level of FliY in complemented strains was probed by anti-FliY or anti-His6 antibody (GE Healthcare).
Characterization of flagellum formation by EM
H. pylori cells were grown in BB10 until A600 reached ∼1.0. Cells were diluted with BB10 to an A600 of ∼0.05 and allowed to grow for 16 h. Cells were pelleted by centrifugation at 8,000 rpm for 3 min. The pellet was resuspended with fresh BB10. A Formvar carbon–coated grid was floated on a droplet of cell suspension for 1 min. The cells were immediately fixed by floating the grid on a drop of 2.5% glutaraldehyde, 2.5% formaldehyde, 100 mm sodium cacodylate, pH 6.5, for 1 min followed by negative staining with 1% phosphotungstate for 1 min. Excess stain was removed with Whatman paper. Flagellum formation was examined by a Tecnai 12 Biotwin transmission electron microscope (FEI/Philips).
Characterization of motility activity by soft agar assay
H. pylori cells were cultured on a Columbia blood agar plate for 2 days. Strains were inoculated into a soft agar plate (Brucella broth with 5% fetal bovine serum, 0.4% agar, chloramphenicol) using a pipette tip. The colony diameter was measured after 5–7-day incubation.
Sedimentation velocity analysis
The experiments were performed at 40,000 rpm at 16 °C in a Beckman-Coulter XL-I analytical ultracentrifuge. Samples containing FliYC–FliNC, FliYC–FliMC, and the mixture of the two protein complexes at a concentration of 2 mg/ml in 20 mm HEPES, pH 7.5, 250 mm NaCl, 4 mm DTT, 20 mm l-arginine were loaded into double-sector centerpieces. All data were collected using an absorbance optical system at a wavelength of 280 nm. Data analysis was performed with SEDFIT, and data were analyzed using a sedimentation coefficient distribution model, c(s).
Interaction studies
0.54 μm GST, GST-FliY–FliN, GST-FliYC–FliN and GST–FliYN were incubated with GST resin pre-equilibrated with buffer (150 mm NaCl, 10 mm HEPES, pH 7.3, 4 mm DTT, 0.15% Tween 20) for 1 h at 25 °C. After washing, FliH (FliH:bait = 1.5:1 molar ratio) was incubated with the immobilized beads for 1 h at 25 °C with gentle shaking. The beads were washed three times and subjected to boiling and SDS-PAGE analysis. For the interaction studies of FliH with FliY–FliN mutants, 50 μg of partially purified His6-FliN–GST-FliY or mutants were immobilized on GST resin followed by incubating the beads with 100 μg of FliH. The GST-FliH–FliY–FliN pulldown experiment was performed using FliH:FliY–FliN at a molar ratio of 1:2.5.
For the FliH–FliN–FliY interaction study by gel filtration, a 1.5:1 molar ratio of FliH was incubated with FliN–FliY in buffer at 4 °C for 1 h. The complex was separated by Superdex 200 gel filtration.
The GST-FliG pulldown experiment was performed as described (20). Proteins were probed with anti-FliY, and anti-FliM (20) was used as a control.
Coimmunoprecipitation
H. pylori G27 and ΔfliY strains were grown in BB10 overnight. Cells were harvested and lysed by sonication in buffer (150 mm NaCl, 20 mm HEPES, pH 7.3, 0.15% Tween 20). Unlysed cells were removed by centrifugation at 8,000 rpm for 3 min. Lysate was loaded onto Dynabeads (Invitrogen) with or without preimmobilized anti-FliY and incubated at room temperature for 1 h. The beads were washed three times with the same buffer and boiled with SDS loading dye. Samples were loaded for SDS-PAGE and probed with anti-FliY, anti-FliM, or anti-FliG antibody. Rabbit anti-FliM and rabbit anti-FliG were generated in previous studies (18, 20). All Western blotting experiments were carried out in duplicates.
Static light scattering
FliY–FliN was subjected to static light scattering using a miniDAWN triangle (45°, 90°, and 135°) light scattering detector (Wyatt Technology Corp., Santa Barbara, CA) connected to an Optilab DSP interferometric refractometer (Wyatt Technology Corp.). This system was connected to a Superdex 200 column (GE Healthcare) controlled by an ÄKTAexplorer chromatography system (GE Healthcare). Before sample injection, the miniDAWN detector system was equilibrated with 150 mm NaCl, 10 mm HEPES, pH 7.5, for at least 2 h to ensure a stable baseline signal. The flow rate was set to 0.5 ml/min, and the sample volume was 100 μl. The laser scattering (687 nm) and the refractive index (690 nm) of the respective protein solutions were recorded. Wyatt Technology ASTRA software was used to evaluate all data obtained.
Isothermal titration calorimetry
The interaction between FliG and FliYN was measured using a MicroCal iTC200 calorimeter (GE Healthcare) as described previously (20). 0.5 mm FliYN was titrated into the cell containing 50 μm FliG proteins in buffer containing 137 mm NaCl, 2.7 mm KCl, 20 mm HEPES, pH 7.5, 2 mm β-mercaptoethanol, 0.1 mm EDTA.
Crystallization, data collection, and structure determination
Crystals of FliYC–FliNC were obtained under conditions with 0.2 m sodium acetate, 0.1 m Tris-HCl, pH 8.5, 30% PEG 4000 using the sitting drop vapor diffusion method. The crystals were soaked briefly in crystallization buffer containing 10% glycerol and cooled by plunging into liquid nitrogen. The X-ray data sets were collected with beamline 13B1 at the National Synchrotron Radiation Research Center, Taiwan. The data sets were processed using the HKL2000 and iMOSFLM packages (1, 50) and scaled and reduced with SCALA from the Collaborative Computational Project, Number 4 (CCP4) suite (51). Crystals of FliYC–FliNC were in the P1 space group. The structure was solved by molecular replacement using FliN from T. maritima (PDB code 1O6A) as a search model. The molecular replacement program Phaser (52), in the CCP4 suite, was used with data in the resolution range. Rounds of refinements and manual rebuilding were performed using the programs REFMAC, Coot, and PHENIX (53, 54). Statistics for data collection and refinement are summarized in Table S1. The coordinates and structure factors of FliYC–FliNC have been deposited in the Protein Data Bank (PDB code 5XRW). All figures were prepared using PyMOL (The PyMOL Molecular Graphics System, Schrödinger, LLC).
Author contributions
K. H. L., C. X., K. M. O., and S. W. N. A. conceptualization; K. H. L., C. X., K. S., H. Z., W. W. L. L., Z. Z., and S. W. N. A. data curation; K. H. L. and S. W. N. A. funding acquisition; K. H. L. and S. W. N. A. validation; K. H. L., C. X., K. S., H. Z., W. W. L. L., Z. Z., J. T. Y. N., W. E. S., P. L., K. F. L., K. M. O., and S. W. N. A. investigation; K. H. L., C. X., K. S., H. Z., W. W. L. L., Z. Z., J. T. Y. N., W. E. S., P. L., K. F. L., K. M. O., and S. W. N. A. methodology; K. H. L. and S. W. N. A. writing-original draft; C. X., K. S., H. Z., W. W. L. L., and S. W. N. A. formal analysis; K. F. L., K. M. O., and S. W. N. A. resources; K. M. O. and S. W. N. A. supervision; K. M. O. and S. W. N. A. writing-review and editing; S. W. N. A. visualization; S. W. N. A. project administration.
Supplementary Material
Acknowledgments
We thank the staff at beamline 13B1 National Synchrotron Radiation Research Center, Taiwan. We thank Prof. Ivo Boneca for the pILL2157 complementation vector.
This work was supported by National Natural Science Foundation of China Grant 31370713, Hong Kong Research Grants Council Grant N_CUHK454/13, and University Grants Committee One-Off Special Equipment Grant SEG CUHK08. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S5 and Table S1.
The atomic coordinates and structure factors (code 5XRW) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- SpoA
- surface presentation of the antigen
- T3SS
- type III secretion system
- PDB
- Protein Data Bank
- CXY
- CheC/CheX/FliY
- Ni-NTA
- nickel-nitrilotriacetic acid
- cryo-ET
- cryo-electron tomography
- IPTG
- isopropyl 1-thio-β-d-galactopyranoside.
References
- 1. Otwinowski Z., and Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 10.1016/S0076-6879(97)76066-X [DOI] [PubMed] [Google Scholar]
- 2. Josenhans C., Jung K., Rao C. V., and Wolfe A. J. (2014) A tale of two machines: a review of the BLAST meeting, Tucson, AZ, 20–24 January 2013. Mol. Microbiol. 91, 6–25 10.1111/mmi.12427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Beeby M., Ribardo D. A., Brennan C. A., Ruby E. G., Jensen G. J., and Hendrixson D. R. (2016) Diverse high-torque bacterial flagellar motors assemble wider stator rings using a conserved protein scaffold. Proc. Natl. Acad. Sci. U.S.A. 113, E1917–E1926 10.1073/pnas.1518952113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen S., Beeby M., Murphy G. E., Leadbetter J. R., Hendrixson D. R., Briegel A., Li Z., Shi J., Tocheva E. I., Müller A., Dobro M. J., and Jensen G. J. (2011) Structural diversity of bacterial flagellar motors. EMBO J. 30, 2972–2981 10.1038/emboj.2011.186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lowenthal A. C., Hill M., Sycuro L. K., Mehmood K., Salama N. R., and Ottemann K. M. (2009) Functional analysis of the Helicobacter pylori flagellar switch proteins. J. Bacteriol. 191, 7147–7156 10.1128/JB.00749-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Minamino T., and Imada K. (2015) The bacterial flagellar motor and its structural diversity. Trends Microbiol. 23, 267–274 10.1016/j.tim.2014.12.011 [DOI] [PubMed] [Google Scholar]
- 7. Zhao R., Pathak N., Jaffe H., Reese T. S., and Khan S. (1996) FliN is a major structural protein of the C-ring in the Salmonella typhimurium flagellar basal body. J. Mol. Biol. 261, 195–208 10.1006/jmbi.1996.0452 [DOI] [PubMed] [Google Scholar]
- 8. Thomas D. R., Morgan D. G., and DeRosier D. J. (1999) Rotational symmetry of the C ring and a mechanism for the flagellar rotary motor. Proc. Natl. Acad. Sci. U.S.A. 96, 10134–10139 10.1073/pnas.96.18.10134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Thomas D., Morgan D. G., and DeRosier D. J. (2001) Structures of bacterial flagellar motors from two FliF-FliG gene fusion mutants. J. Bacteriol. 183, 6404–6412 10.1128/JB.183.21.6404-6412.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brown P. N., Hill C. P., and Blair D. F. (2002) Crystal structure of the middle and C-terminal domains of the flagellar rotor protein FliG. EMBO J. 21, 3225–3234 10.1093/emboj/cdf332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brown P. N., Mathews M. A., Joss L. A., Hill C. P., and Blair D. F. (2005) Crystal structure of the flagellar rotor protein FliN from Thermotoga maritima. J. Bacteriol. 187, 2890–2902 10.1128/JB.187.8.2890-2902.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Park S.-Y., Lowder B., Bilwes A. M., Blair D. F., and Crane B. R. (2006) Structure of FliM provides insight into assembly of the switch complex in the bacterial flagella motor. Proc. Natl. Acad. Sci. U.S.A. 103, 11886–11891 10.1073/pnas.0602811103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Brown P. N., Terrazas M., Paul K., and Blair D. F. (2007) Mutational analysis of the flagellar protein FliG: sites of interaction with FliM and implications for organization of the switch complex. J. Bacteriol. 189, 305–312 10.1128/JB.01281-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Minamino T., Imada K., Kinoshita M., Nakamura S., Morimoto Y. V., and Namba K. (2011) Structural insight into the rotational switching mechanism of the bacterial flagellar motor. PLoS Biol. 9, e1000616 10.1371/journal.pbio.1000616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lee L. K., Ginsburg M. A., Crovace C., Donohoe M., and Stock D. (2010) Structure of the torque ring of the flagellar motor and the molecular basis for rotational switching. Nature 466, 996–1000 10.1038/nature09300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Paul K., Gonzalez-Bonet G., Bilwes A. M., Crane B. R., and Blair D. (2011) Architecture of the flagellar rotor. EMBO J. 30, 2962–2971 10.1038/emboj.2011.188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vartanian A. S., Paz A., Fortgang E. A., Abramson J., and Dahlquist F. W. (2012) Structure of flagellar motor proteins in complex allows for insights into motor structure and switching. J. Biol. Chem. 287, 35779–35783 10.1074/jbc.C112.378380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lam K. H., Ip W. S., Lam Y. W., Chan S. O., Ling T. K., and Au S. W. (2012) Multiple conformations of the FliG C-terminal domain provide insight into flagellar motor switching. Structure 20, 315–325 10.1016/j.str.2011.11.020 [DOI] [PubMed] [Google Scholar]
- 19. Stock D., Namba K., and Lee L. K. (2012) Nanorotors and self-assembling macromolecular machines: the torque ring of the bacterial flagellar motor. Curr. Opin. Biotechnol. 23, 545–554 10.1016/j.copbio.2012.01.008 [DOI] [PubMed] [Google Scholar]
- 20. Lam K. H., Lam W. W., Wong J. Y., Chan L. C., Kotaka M., Ling T. K., Jin D. Y., Ottemann K. M., and Au S. W. (2013) Structural basis of FliG-FliM interaction in Helicobacter pylori. Mol. Microbiol. 88, 798–812 10.1111/mmi.12222 [DOI] [PubMed] [Google Scholar]
- 21. Sircar R., Greenswag A. R., Bilwes A. M., Gonzalez-Bonet G., and Crane B. R. (2013) Structure and activity of the flagellar rotor protein FliY: a member of the CheC phosphatase family. J. Biol. Chem. 288, 13493–13502 10.1074/jbc.M112.445171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Notti R. Q., Bhattacharya S., Lilic M., and Stebbins C. E. (2015) A common assembly module in injectisome and flagellar type III secretion sorting platforms. Nat. Commun. 6, 7125 10.1038/ncomms8125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Thomas D. R., Francis N. R., Xu C., and DeRosier D. J. (2006) The three-dimensional structure of the flagellar rotor from a clockwise-locked mutant of Salmonella enterica serovar typhimurium. J. Bacteriol. 188, 7039–7048 10.1128/JB.00552-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Qin Z., Lin W. ting, Zhu S., Franco A. T., and Liu J. (2017) Imaging the motility and chemotaxis machineries in Helicobacter pylori by cryo-electron tomography. J. Bacteriol. 199, e00695–16 10.1128/JB.00695-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kojima S., and Blair D. F. (2004) Solubilization and purification of the MotA/MotB complex of Escherichia coli. Biochemistry 43, 26–34 10.1021/bi035405l [DOI] [PubMed] [Google Scholar]
- 26. Sarkar M. K., Paul K., and Blair D. (2010) Chemotaxis signaling protein CheY binds to the rotor protein FliN to control the direction of flagellar rotation in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 107, 9370–9375 10.1073/pnas.1000935107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fadouloglou V. E., Tampakaki A. P., Glykos N. M., Bastaki M. N., Hadden J. M., Phillips S. E., Panopoulos N. J., and Kokkinidis M. (2004) Structure of HrcQB-C, a conserved component of the bacterial type III secretion systems. Proc. Natl. Acad. Sci. U.S.A. 101, 70–75 10.1073/pnas.0304579101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bzymek K. P., Hamaoka B. Y., and Ghosh P. (2012) Two translation products of Yersinia yscQ assemble to form a complex essential to type III secretion. Biochemistry 51, 1669–1677 10.1021/bi201792p [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Minamino T., Imada K., and Namba K. (2008) Molecular motors of the bacterial flagella. Curr. Opin. Struct. Biol. 18, 693–701 10.1016/j.sbi.2008.09.006 [DOI] [PubMed] [Google Scholar]
- 30. González-Pedrajo B., Minamino T., Kihara M., and Namba K. (2006) Interactions between C ring proteins and export apparatus components: a possible mechanism for facilitating type III protein export. Mol. Microbiol. 60, 984–998 10.1111/j.1365-2958.2006.05149.x [DOI] [PubMed] [Google Scholar]
- 31. McMurry J. L., Murphy J. W., and González-Pedrajo B. (2006) The FliN-FliH interaction mediates localization of flagellar export ATPase FliI to the C ring complex. Biochemistry 45, 11790–11798 10.1021/bi0605890 [DOI] [PubMed] [Google Scholar]
- 32. Paul K., and Blair D. F. (2006) Organization of FliN Subunits in the flagellar motor of Escherichia coli. J. Bacteriol. 188, 2502–2511 10.1128/JB.188.7.2502-2511.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Minamino T., Yoshimura S. D., Morimoto Y. V., González-Pedrajo B., Kami-Ike N., and Namba K. (2009) Roles of the extreme N-terminal region of FliH for efficient localization of the FliH-FliI complex to the bacterial flagellar type III export apparatus. Mol. Microbiol. 74, 1471–1483 10.1111/j.1365-2958.2009.06946.x [DOI] [PubMed] [Google Scholar]
- 34. Bischoff D. S., and Ordal G. W. (1992) Identification and characterization of FliY, a novel component of the Bacillus subtilis flagellar switch complex. Mol. Microbiol. 6, 2715–2723 10.1111/j.1365-2958.1992.tb01448.x [DOI] [PubMed] [Google Scholar]
- 35. Szurmant H., Muff T. J., and Ordal G. W. (2004) Bacillus subtilis CheC and FliY are members of a novel class of CheY-P-hydrolyzing proteins in the chemotactic signal transduction cascade. J. Biol. Chem. 279, 21787–21792 10.1074/jbc.M311497200 [DOI] [PubMed] [Google Scholar]
- 36. Lertsethtakarn P., Ottemann K. M., and Hendrixson D. R. (2011) Motility and chemotaxis in Campylobacter and Helicobacter. Annu. Rev. Microbiol. 65, 389–410 10.1146/annurev-micro-090110-102908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lowenthal A. C., Simon C., Fair A. S., Mehmood K., Terry K., Anastasia S., and Ottemann K. M. (2009) A fixed-time diffusion analysis method determines that the three cheV genes of Helicobacter pylori differentially affect motility. Microbiology 155, 1181–1191 10.1099/mic.0.021857-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Boneca I. G., Ecobichon C., Chaput C., Mathieu A., Guadagnini S., Prévost M. C., Colland F., Labigne A., and de Reuse H. (2008) Development of inducible systems to engineer conditional mutants of essential genes of Helicobacter pylori. Appl. Environ. Microbiol. 74, 2095–2102 10.1128/AEM.01348-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Berg H. C., and Turner L. (1979) Movement of microorganisms in viscous environments. Nature 278, 349–351 10.1038/278349a0 [DOI] [PubMed] [Google Scholar]
- 40. McDowell M. A., Marcoux J., McVicker G., Johnson S., Fong Y. H., Stevens R., Bowman L. A., Degiacomi M. T., Yan J., Wise A., Friede M. E., Benesch J. L., Deane J. E., Tang C. M., Robinson C. V., et al. (2016) Characterisation of Shigella Spa33 and Thermotoga FliM/N reveals a new model for C-ring assembly in T3SS. Mol. Microbiol. 99, 749–766 10.1111/mmi.13267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Häuser R., Ceol A., Rajagopala S. V., Mosca R., Siszler G., Wermke N., Sikorski P., Schwarz F., Schick M., Wuchty S., Aloy P., and Uetz P. (2014) A second-generation protein–protein interaction network of Helicobacter pylori. Mol. Cell. Proteomics. 13, 1318–1329 10.1074/mcp.O113.033571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Parrish J. R., Yu J., Liu G., Hines J. A., Chan J. E., Mangiola B. A., Zhang H., Pacifico S., Fotouhi F., DiRita V. J., Ideker T., Andrews P., and Finley R. L. (2007) A proteome-wide protein interaction map for Campylobacter jejuni. Genome Biol. 8, R130 10.1186/gb-2007-8-7-r130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Senesi S., Celandroni F., Salvetti S., Beecher D. J., Wong A. C., and Ghelardi E. (2002) Swarming motility in Bacillus cereus and characterization of a fliY mutant impaired in swarm cell differentiation. Microbiology 148, 1785–1794 10.1099/00221287-148-6-1785 [DOI] [PubMed] [Google Scholar]
- 44. Webb B., and Sali A. (2016) Comparative protein structure modeling using MODELLER. Curr. Protoc. Protein Sci. 86, 2.9.1–2.9.37 10.1002/cpps.20 [DOI] [PubMed] [Google Scholar]
- 45. Silversmith R. E. (2010) Auxiliary phosphatases in two-component signal transduction. Curr. Opin. Microbiol. 13, 177–183 10.1016/j.mib.2010.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pittman M. S., Goodwin M., and Kelly D. J. (2001) Chemotaxis in the human gastric pathogen Helicobacter pylori: different roles for CheW and the three CheV paralogues, and evidence for CheV2 phosphorylation. Microbiology 147, 2493–2504 10.1099/00221287-147-9-2493 [DOI] [PubMed] [Google Scholar]
- 47. Raddi G., Morado D. R., Yan J., Haake D. A., Yang X. F., and Liu J. (2012) Three-dimensional structures of pathogenic and saprophytic Leptospira species revealed by cryo-electron tomography. J. Bacteriol. 194, 1299–1306 10.1128/JB.06474-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kholod N., and Mustelin T. (2001) Novel vectors for co-expression of two proteins in E. coli. BioTechniques 31, 322–323, 326–328 [DOI] [PubMed] [Google Scholar]
- 49. Donahue J. P., Israel D. A., Peek R. M., Blaser M. J., and Miller G. G. (2000) Overcoming the restriction barrier to plasmid transformation of Helicobacter pylori. Mol. Microbiol. 37, 1066–1074 10.1046/j.1365-2958.2000.02036.x [DOI] [PubMed] [Google Scholar]
- 50. Battye T. G., Kontogiannis L., Johnson O., Powell H. R., and Leslie A. G. (2011) iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D Biol. Crystallogr. 67, 271–281 10.1107/S0907444910048675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., et al. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 10.1107/S0907444910045749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. London N., Raveh B., Cohen E., Fathi G., and Schueler-Furman O. (2011) Rosetta FlexPepDock web server—high resolution modeling of peptide-protein interactions. Nucleic Acids Res. 39, W249–W253 10.1093/nar/gkr431 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.