Abstract
Objectives:
The immune environment and the potential for neuroendocrine tumors (NETs) to respond to immune checkpoint inhibitors remain largely unexplored. We assessed immune checkpoint marker expression, lymphocytic infiltrate, and associated mutational profiles in a cohort of small intestine and pancreatic neuroendocrine tumors.
Methods:
We assessed expression of PDCD1 (PD-1), CD274 (PD-L1) and PDCD1LG2 (PD-L2 ) in archival tissue from 64 small intestine (SINET) and 31 pancreatic neuroendocrine tumors (pNET). We additionally assessed T cell infiltrates, categorizing T cell subsets based on expression of the T cell markers CD3, CD8, CD45RO (PTPRC) or FOXP3. Finally, we explored associations between immune checkpoint marker expression, lymphocytic infiltrate and tumor mutational profiles.
Results:
Expression of PD-1 or PD-L1 in small intestine or pancreatic NET was rare, while expression of PD-L2 was common in both NET subtypes. T-cell infiltrates were more abundant in pNET than in SINET. We found no clear associations between immune checkpoint marker expression, immune infiltrates, and specific mutational profile within each tumor type.
Conclusions:
Our findings provide an initial assessment of the immune environment of well-differentiated neuroendocrine tumors. Further studies to define the immunologic differences between pNET and SINET, as well as the role of PD-L2 in these tumors, are warranted.
Keywords: PD-1, PD-L1, PD-L2, immune checkpoint, T-cell markers, neuroendocrine tumor
INTRODUCTION
Treatment options for patients with advanced, well differentiated neuroendocrine tumors remain limited. Currently approved systemic treatments in this setting include somatostatin analogs, everolimus, and sunitinib; however, tumor regression with these agents is rare.1–4 Treatment with immune checkpoint inhibitors targeting PD-L1 or PD-1 has led to significant tumor responses in a broad range of human malignancies.5 The potential for neuroendocrine tumors (NET) to respond to immune checkpoint inhibition remains uncertain.
Response to treatment with checkpoint inhibitors has been associated with immunohistochemical expression of PD-1 and its ligand PD-L1, although clinical benefit and responses have been observed even in the absence of expression.6 PD-L2, like PD-L1, has been shown to inhibit T-cell proliferation and T-cell mediated killing.7 Though less well studied than PD-L1, PD-L2 expression as also been associated with response to checkpoint inhibition in some settings, including head and neck cancer and melanoma.8–10
The degree of T-cell infiltration of tumors has also been considered as both a general prognostic factor and as a specific predictor of response to checkpoint inhibition. Tumor infiltration with effector (i.e CD3/CD8 positive) T-cells and CD45RO positive T-cells, indicative of T-cell exposure to tumor antigen, have been associated with improved host immune response and improved overall survival in colorectal cancer,11 while expression of FOXP3 (Treg) cells, has been associated with the presence of an immunosuppressive immune environment.12–14 Similar findings, showing improved survival associated with high CD8 infiltrates and poor survival with increased FOXP3 (Treg) infiltrates were observed in a study of clear cell renal cell carcinoma.15 The presence of CD8 positive T-cell infiltrates have been associated with improved response to checkpoint inhibitors in patients with melanoma. 16
Both host immune response and response to checkpoint inhibition have, in turn, been associated with mutational burden. The presence of microsatellite instability has been associated with both the presence of a robust immune infiltrate as well and response to checkpoint inhibition across a range of human malignancies, and the PD-1 inhibitor pembrolizumab was recently approved for this indication.17,18 Mutational burden in the absence of microsatellite instability has also been shown to predict a favorable response to PD-1 or PD-L1 blockade.19
Checkpoint marker expression and host immune response in neuroendocrine tumors remains relatively unexplored. Small cell carcinoma, considered to be the most aggressive neuroendocrine tumor subtype, has been shown to express both PD-1 and PD-L1, and the PD-1 inhibitor, Nivolumab, either alone or in combination with Ipilimumab has shown activity in this setting.20 In contrast, few studies have explored expression of immune checkpoint markers in more common, well-differentiated neuroendocrine tumors. Expression of both PD-1 and PD-L1 was reported in one study of well differentiated pulmonary neuroendocrine tumors, while expression of these markers was not identified neuroendocrine tumors of the foregut or hindgut.21–23 No studies to date have correlated checkpoint marker expression, immune infiltrates, and mutational burden in neuroendocrine tumors.
To better characterize both checkpoint marker expression and the more general immune environment of neuroendocrine tumors we examined the expression of the key checkpoint protein PD-1, PD-L1, and PD-L2 on both tumor and surrounding stromal cells in cohorts of small intestine neuroendocrine tumors (SINET) and pancreatic neuroendocrine tumors (pNET). We further evaluated T cell lymphocytic infiltrates and mutational profiles in the tumor specimens, and assessed associations between immune checkpoint marker expression, T cell infiltrates, and tumoral mutational profiles.
MATERIALS AND METHODS
Study Population
Archival NET tumor specimens with associated demographic and clinical information were obtained from the Dana-Farber Cancer Institute Neuroendocrine Tumor Biospecimen database at Dana-Farber Cancer Institute, under an IRB-approved study.
Assessment of Immune Checkpoint Markers and Lymphocytic Infiltrate by Immunohistochemistry (IHC)
Immunostaining for PD-1 (EH33), PD-L1 (405.9A11) and PD-L2 (366C.9E5) was performed using an automated staining system (BOND-III Fully Automated IHC and ISH, Leica Biosystems, Buffalo Grove, Ill) following the manufacturer’s protocol. 4 μm paraffin-embedded sections were pre-baked at 60°C for one hour. Slides were then loaded onto Bond III with “Bond Universal Covertiles” (Leica Biosystems). PD-1 (EH33) immunostaining was performed with 1:1000 dilution using Da Vinci Green Diluent (Biocare Medical, Pacheco, Calif). PD-L1 (405.9A11) Immunostaining was performed with 1:100 dilution (final concentration: 13 μg/ml) using Ab Discovery Diluent (Ventana Medical Systems, Oro Valley, Ariz). PD-L2 (366C.9E5) staining utilized the same reagents. Slides were first dewaxed and rehydrated. Heat induced antigen retrieval was performed using ER2 solution (pH8) (Leica Biosystems) for 20 minutes (PD-1) and 30 minutes (PD-L1, PD-L2). For PD-1, the primary antibody was incubated for 30 minutes, followed by 10 minutes of post primary blocking reagent, 10 minutes of horseradish peroxidase-labeled polymer, 5 minutes of peroxidase block, and 10 minutes of DAB developing. For PD-L1 and PD-L2 the primary antibody was incubated for total of 2 hours with two separate applications, follow by 8 minutes of post-primary blocking reagent, 12 minutes of horseradish peroxidase-labeled polymer, 5 minutes of peroxidase block, and 15 minutes of DAB developing. Slides were counterstained by hematoxylin for 5 minutes. Tonsil sections were used as positive controls for PD-1, PD-L1 and PD-L2.
Two pathologists ADS and YM analyzed and scored each tumor independently, using an Olympus (Tokyo, Japan) microscope model BX43. Tumoral membranous expression of PD-L1 and PD-L2 was recorded as absent if <5% of cells had any intensity of membrane staining and positive if ≥5% of cells had any intensity of membrane staining.24 PD-L1 and PD-L2 cytoplasmic expression in neoplastic cells was reported using a score ranging from 0 to 3 (0-absent, <10% with any intensity; 1-weak, ≥10%; 2-moderate, ≥10% with moderate intensity; 3-strong, ≥10% with strong intensity).24 For the analysis of stromal cells, positive PD-1, PD-L1, or PD-L2 cells were counted in 10 hpf (microscopic high power field) per section and the absolute number was recorded. The average of the number of positive cells per high power field was used to create scores as follows: 0 (absence of positive cells), 1 (1–20 cells/hpf), 2 (20–50 cells/hpf) and score 3 (>50 cells/hpf).24
Immunohistochemical Analysis of T-cell infiltrates
Immunostaining was performed for CD3 (clone 7.2.38, 1:250, Dako Cytomation, Carpinteria, Calif), CD8 (clone C8/144B, 1:100, Dako Cytomation), CD45RO (clone UCHL1, 1:100, Dako Cytomation) and FOXP3 (clone 206D, 1:50, BioLegend, San Diego, Calif). Antigen retrieval was performed in deparaffinized 4 μm thick tissue sections in EDTA Solution (pH 8.0) (Abcam, Cambridge, Mass) and slides were microwaved for 15 minutes for the markers CD3, CD8, and FOXP3. For CD45RO, citrate buffer (pH 6.0) (BioScience, San Jose, Calif) was used for antigen retrieval and slides were microwaved for 17 minutes. Sections were treated by endogenous peroxidase blocker (10 minutes) and protein blocker (20 minutes), and then incubated for 1 hour at room temperature with primary mouse monoclonal antibodies. Envision System HRP-labeled polymer anti-mouse (Dako Cytomation, Glostrup, Demark) was then applied for 30 minutes, followed by visualization using the chromogen 3,3-diaminobenzidine (Dako) and hematoxylin counterstain.
One pathologist (ADS) initially analyzed and scored the T-cell infiltrates using an Olympus microscope model BX43. The location of the lymphocytes was defined as intratumoral (lymphocytes present on top of neoplastic cells, between neoplastic cells and within the stroma immediately surrounding the neoplastic cells) or extratumoral (lymphocytes present at the edge of the tumor or at the interface between normal tissue and the tumor edge or capsule). Sections of each tumor were divided in 4 equal sized areas and the absolute number of lymphocytes was manually counted in at least 2 hpf in each quarter. Areas with highest density of lymphoid cells were prioritized; though we also considered at least two random areas in the center of the tumor, counting 10 hpf per section, for each T-cell category. Intra-vascular lymphocytes were excluded, as were necrotic, ulcerated or heavily hemorrhagic areas. The average of the number of T-cells per high power field was used to create scores as follows: 0 (absence of positive cells), 1 (1–20 cells/hpf), 2 (20–50 cells/hpf) and score 3 (>50 cells/hpf). Cases were categorized as having either low (scores 0 and 1) or high density (scores 2 and 3) lymphocytic infiltrates.11,24,25 A random sample of 45 cases was analyzed by a second pathologist YM to ensure agreement in scores. We confirmed interobserver agreement, with a weighted κ of 0.68 for CD45RO, 0.72 for CD3, 0.78 for CD8, and 0.66 for FOXP3.
RNA- sequencing
RNA-sequencing was used to further characterize the relative expression of PD-L1 and PD-L2 in snap frozen tissue. A small portion from each primary tumor (6 pNET, 23 SINET) was pulverized using the Covaris (Worburn, Mass) T-prep method on dry ice (cat # 520097). The Agencourt RNAdvance (Agencourt Bioscience Corporation, A Beckman Coulter Company, Beverly, Mass) for isolation of nucleic acids from frozen tissue (cat # A32645) was implemented on a Biomek® FXP Laboratory Automation Workstation (Beckman Coulter Life Sciences, Indianapolis, Ind) (Dual arm system with multichannel pipette and span-8 pipetters, cat# A31844). Briefly the tissue is lysed, after which the RNA is immobilized onto the magnetic particles. The RNA is then treated with DNase and the contaminants rinsed away using a simple wash procedure. RNA isolates were eluted in a 55ul volume of RNase/DNase-free H2O. All RNA was stored at −80°C. RNA quality was assessed by Agilent (Santa Clara, Calif) Bioanalyzer using the RNA 6000 pico kit (cat# 5067–1513). 150ng RNA was utilized as input for RNA-Seq library preparation. Removal of ribosomal RNA (rRNA) and human mitochondrial RNA was done using biotinylated, target-specific oligos combined with Ribo-Zero Gold rRNA removal beads from the TruSeq Stranded Total RNA kit (Illumina, San Diego, Calif, cat # RS-122–2301). The method was automated on the Biomek® FXP Laboratory Automation Workstation. The Stranded TruSeq method comprised ribosomal RNA removal, cDNA synthesis and purification and PCR amplification. cDNA libraries were quantified utilizing the Quanti-iT PicoGreen assay (Life Technologies, Carlsbad, Calif, cat# P7589). 1 μl of cDNA was required for quantification. Concentration is measured as ng/ul. Libraries were excited at 480 nm and the fluorescence emission intensity was measured at 520 nm using a Victor X3 spectrophotometer (Perkin Elmer, Waltham, Mass, cat# 2030–0030). Fluorescence intensity was plotted versus concentration over the low calibration range, 0–50 ng/μl. Libraries were also quality checked by Agilent Bioanalyzer using the High Sensitivity DNA kit (cat# 5067–4626). cDNA libraries were then sequenced on the Illumina NextSeq500 platform as 75bp paired end reads. Data was streamed in real-time to the Illumina BaseSpace cloud tool. TopHat Alignment (Version: 1.0.0 using TopHat2 alignment) was utilized to align the data to the genome, Homo sapiens UCSC hg19 (RefSeq gene annotations). FPKM abundance estimates of reference genes and transcripts were produced. Mean FPKM abundance of PD-L1 and PDL-2 were calculated.
DNA Sequencing and Analysis
Assessment of mutational status in SINET has been previously described; data from this study was applied to the cohort of SINET used in this study.26 For assessment of pancreatic NET mutational status, 200 ng of gDNA from 27 tumor specimens was submitted for mutational profiling, together with matched normal tissue from adjacent pancreas or from blood specimens derived from the same patient. Mutational status was assessed using OncoPanel v3.0, which is a custom hybrid capture bait set (Agilent Technologies, Inc.), created by the Center for Cancer Genome Discovery at the Dana-Farber Cancer Institute, covering the exons of 560 genes of known cancer relevance, including MEN1, DAXX, ATRX, and selected mTOR pathway genes. Sequencing libraries were prepared as previously described27, with inclusion of a unique barcode for each sample to enable pooling. Libraries were quantified using a MiSeq Nano flow cell (Illumina Inc.), pooled in equal mass to 500 ng total, and captured using the OncoPanel_v3.0 using the Agilent SureSelect hybrid capture kit (Agilent Technologies, Inc.). Captured libraries were quantitated using qPCR (Kapa Biosystems, Inc., Wilmington, Mass), further pooled, and sequenced on two lanes of an Illumina HiSeq3000. Pooled sample reads were de-convoluted and sorted using the Picard tools (http://broadinstitute.github.io/picard/picard-metric-definitions.html for details). The reads were aligned to the reference sequence b37 edition from the Human Genome Reference Consortium using bwa (http://bio-bwa.sourceforge.net/bwa.shtml) using the following parameters “-q 5 -l 32 -k 2 -o 1”, and duplicate reads were identified and removed using the Picard tools.28 The alignments were further refined using the GATK tool for localized realignment around indel sites: (http://gatkforums.broadinstitute.org/discussion/38/local-realignment-around-indels). Recalibration of the quality scores was also performed using GATK tools:
(http://gatkforums.broadinstitute.org/discussion/44/base-quality-score-recalibration-bqsr)
The average mean target coverage was 165x for samples reaching 80% of target bases covered ≥ 30x. The samples having <80% of the targets covered 30x were excluded from the analysis. Mutation analysis for single nucleotide variants (SNV) was performed using MuTect v1.1.429 in paired mode by pairing the tumor to a matched normal or to CEPH control DNA if no matched normal was available (n = 4), and annotated using Variant Effect Predictor (VEP),30 and the SomaticIndelDetector tool that is part of the GATK was used for indel calling. For tumors without a matching normal, variants were filtered against the 6500 exome release of the Exome Sequencing Project (ESP) database. Variants represented at >1% in either the African American or European-American populations and not in COSMIC > 2x were considered to be likely germline.
Statistical Analysis
SAS software (version 9.2; SAS Institute Inc, Cary, NC) was used. All P values were two-sided at alpha = 0.05. We used the Chi-Square test or Fisher’s or Exact test when applicable, to detect differences in the frequencies of the categorized expression levels among the tumor subgroups. Cox proportional hazards regression models were used to compute event hazard ratio (HR) and 95% confidence interval (CI). Multivariate Cox proportional hazards regression models included sex, age at diagnosis, TNM stage (M0 vs. M1), and MKI67 labeling index. For RNA-seq data, the mean and standard deviation were calculated in Excel.
RESULTS
Clinicopathological Characteristics of Small Intestine (SINET) and Pancreatic Neuroendocrine Tumors (pNET)
We evaluated tumors from 95 patients comprising 64 SINET and 31 pNET (Table 1). All cases had well differentiated histology, and the majority (91/95) were classified as grade 1 or 2 based on the WHO classification.31 Four pancreatic NET were classified as having well differentiated grade 3 tumors based on a proliferative index (MKI-67) of greater than >20%.32 Nearly all tumors evaluated were primary tumors; in 4 cases (2 pNET and 2 SINET) tumor samples were derived from metastatic lesions.
TABLE 1.
Clinicopathological Features of the Patient Cohort
| pNET | SINET | ||
|---|---|---|---|
| N (independent cases) | 31 | 64 | |
| Age, median (range), y | 55.1 (27.3–76.6) | 58.5 (26.5–84.2) | |
| Sex, n (%) | |||
| Female | 14 (45) | 33 (52) | |
| Male | 17 (55) | 31 (48) | |
| Tumor site, n (%) | |||
| Primary | 29 (94) | 62 (97) | |
| Metastatic Lesion | 2 (6) | 2 (3) | |
| Ki-67 labeling index, n (%)* | |||
| ≤2% (Grade 1) | 10 (32) | 39 (61) | |
| >2% and <20% (Grade 2) | 17 (55) | 25 (39) | |
| >20% (Grade 3) | 4 (13) | 0 (0) | |
All tumors were well-differentiated
Expression of PD-1, PD-L1, and PD-L2 in NET
Tumoral expression of PD-L1 was observed in no cases of small intestine NET and in only 7.4% of pancreatic NET (Fig. 1; Table 2). PD-1 expression, limited to the stromal compartment, was also uncommon and was observed at a high level in only a single case of small intestine NET. In contrast, strong expression of PD-L2 was observed in a high proportion of neuroendocrine tumor cells in both the small intestine (82%) and pancreas (97%). Notably, tumoral expression of PD-L2 was in most cases limited to the cytoplasm rather than the membrane. Strong expression of PD-L2 in the stromal compartment was not observed.
FIGURE 1. PD-L1, PD-L2 and PD-1 in tumor and stromal cells.
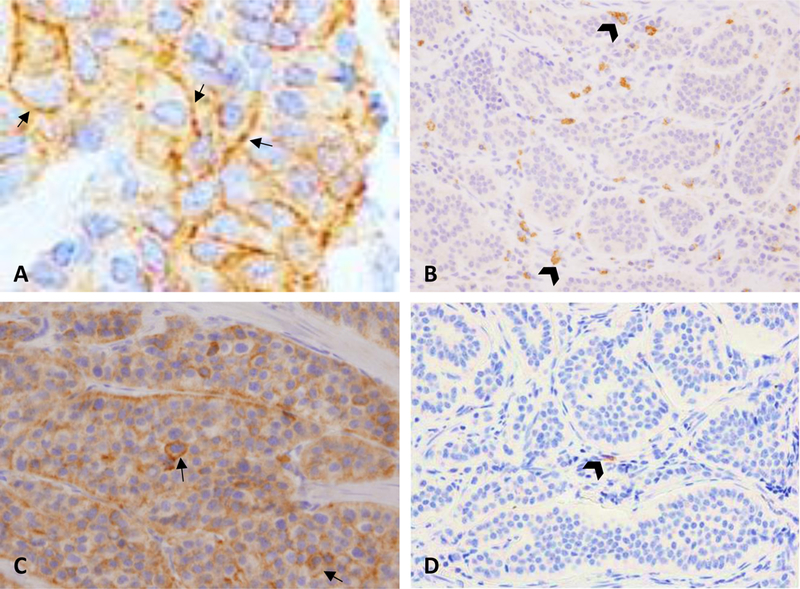
A, Membranous expression of PD-L1 in pNET. High expression was rare and found in only 7% of cases. B, High expression of PD-L1 in immune stromal cells (head arrows) was found in only a single case of SINET. C, High expression of PD-L2 in pNET tumor cells. Expression was predominantly cytoplasmic with approximately 5% of cells also showing membranous expression (arrows). D, Representative low density expression of PD-1 in immune stromal cells in SINET (head arrow).
TABLE 2.
Expression of PD-1, PD-L1, and PD-L2 in Pancreatic and Small Intestine NET
| Pancreatic NET |
Small Intestine NET |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Tumor, n (%) | Stroma, n (%) | Tumor, n (%) | Stroma, n (%) | ||||||
| Negative | Positive | Low Expression | High Expression | Negative | Positive | Low Expression | High Expression | ||
| PD-1 | NA | NA | 28 (100) | 0 (0) | NA | NA | 59 (98) | 1 (2) | |
| PD-L1 | 1 (3) | 28 (97) | 27 (100) | 0 (0) | 59 (100) | 0 (0) | 58 (98) | 1 (2) | |
| PD-L2 | 1 (3) | 28 (97)* | 30 (100) | 0 (0) | 9 (18) | 52 (82)* | 61 (100) | 0 (0) | |
Cytoplasmic expression of PD-L2 classified as strong NA indicates not applicable.
To validate these observations, we assessed relative PD-L1 and PD-L2 expression in an independent cohort of fresh frozen tissue samples comprising 6 pNET and 23 SINET. Consistent with our IHC results, mRNA expression of PD-L2 was higher than mRNA expression of PD-L1 in both tumor types (Fig. 2).
FIGURE 2. mRNA expression of PD-L1 and PD-L2 in pNET and SINET.
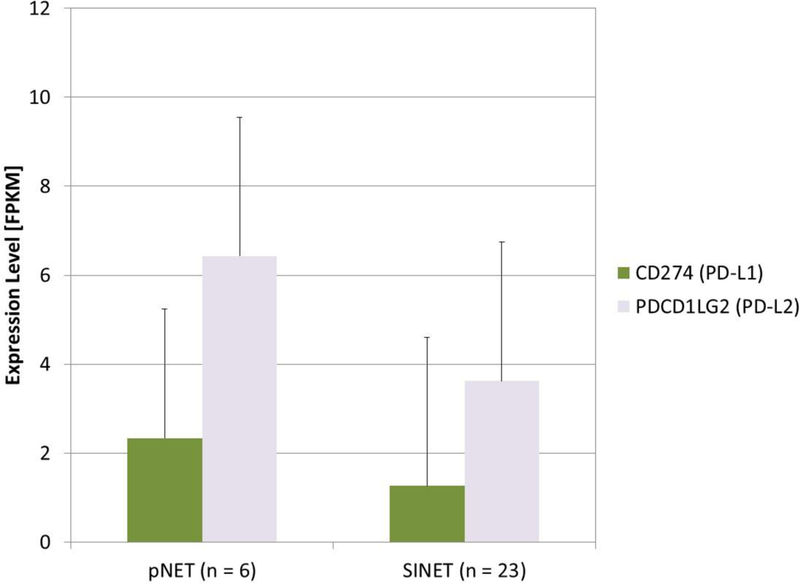
RNA-seq was performed in an independent cohort of 6 Pnet AND 23 SINET tissues, using fresh frozen tissue specimens. Mean expression levels of PD-L1 (CD274) and PD-L2 (PDCD1LG2) were extracted from transcriptome data as FKPM values. Data are presented as FPKM mean with standard deviation.
T-cell Immune Infiltrates in SINET and pNET
We next evaluated T-cell infiltrates in both the intra and extratumoral compartments with a panel of T-cell markers comprising CD3 (general T-cell marker), CD45RO (T memory), CD8 (cytotoxic T-cell), as well as FOXP3 (T regulatory cell) (Table 3; Fig. 3). CD45RO, CD3, and CD8 T cell infiltrates in the intratumoral compartment were relatively similar between small intestine NET and pancreatic NET: across the three populations, 32–65% of pNET and 14–48% of small intestine NET demonstrated high (score 2–3) T-cell infiltrates. In the extratumoral compartment, on the other hand, pancreatic NET demonstrated a much higher degree of infiltration than small intestine with these three populations: 50–70% of pancreatic NET demonstrated high infiltrates, as compared to only 4–19% of small intestine NET. Levels of FOXP3 T regulatory cell infiltration were low in both tumor subtypes.
TABLE 3.
T-Cell Infiltrates in Pancreatic and Small Intestine NET
| Pancreatic NET (N = 31) |
Small Intestine NET (N = 64) |
||
|---|---|---|---|
| CD45RO | Intra-tumoral, n (%) | ||
| Low | 11 (36) | 31 (53) | |
| High | 20 (65) | 28 (48) | |
| Extra-tumoral, n (%) | |||
| Low | 6 (26) | 25 (81) | |
| High | 17 (74) | 6 (19) | |
| CD3 | Intra-tumoral, n (%) | ||
| Low | 16 (52) | 32 (54) | |
| High | 15 (48) | 27 (45) | |
| Extra-tumoral, n (%) | |||
| Low | 10 (50) | 26 (94) | |
| High | 10 (50) | 1 (4) | |
| CD8 | Intra-tumoral, n (%) | ||
| Low | 21 (68) | 51 (86) | |
| High | 10 (32) | 8 (14) | |
| Extra-tumoral, n (%) | |||
| Low | 11 (50) | 28 (96) | |
| High | 11 (50) | 1 (4) | |
| FOXP3 | Intra-tumoral, n (%) | ||
| Low | 30 (97) | 59 (100) | |
| High | 1 (3) | 0 (0) | |
| Extra-tumoral, n (%) | |||
| Low | 22 (100) | 30 (100) | |
| High | 0 (0) | 0 (0) | |
Infiltrates were assessed based on T-cell marker expression and in the intra and extra-tumoral compartments separately. Low and high level infiltrates were defined as infiltrates with scores of 0–1 or 2–3, respectively, as defined in the Methods section.
FIGURE 3. Representative intra and extratumoral lymphocytic infiltrates in pNET and SINET.
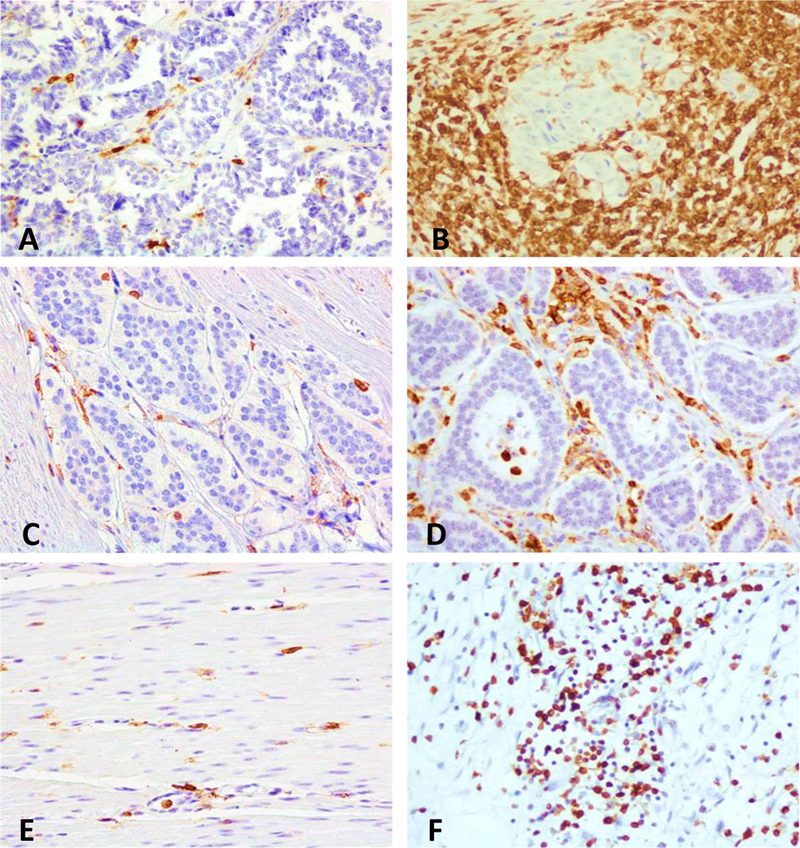
CD45RO+ T-cells are shown in the intratumoral (A-D) and extratumoral compartments (E, F). A, C, and E show a low density of CD45RO+ cells, in the intratumoral compartment of a pNET and SINET, and in the extratumoral compartment of a SINET respectively. B, D, and F show a high density of CD45RO+ cells, in intratumoral compartment of a pNET and SINET, and in the extratumoral area of a pNET, respectively.
Grade 3 Well-differentiated NET
Our cohort included 4 pNET cases with a Ki-67 of >20%, though still well-differentiated by morphology. The characteristics of these tumors did not seem to differ significantly from their lower grade counterparts: none of the tumor cells were positive for PD-L1, while cytoplasmic expression of PD-L2 was observed in 3 of the 4 cases. Low levels of stromal PD-1, PD-L1, and PD-L2 were also observed. Infiltrates of CD3, CD45RO, CD8, and FOXP3 T cells followed a pattern similar to that seen in the lower grade tumors.
Associations Between Mutational Profile, Immune Checkpoint Marker Expression, and T-cell Infiltrate
We finally assessed whether the mutational profiles of the small intestine NET and pNETs in our cohort were associated with immune checkpoint marker expression and/or levels of T cell infiltrates. Twenty-two small intestine NET cases in our cohort had been previously sequenced for CDKN1B, the only gene reported to be recurrently mutated in small intestine NET.26 Of these 22 cases, 5 contained mutations in CDKN1B. We found no clear associations between the presence of CDKN1B mutations, expression of PD-1, PD-L1, PD-L2, or T-cell infiltrates. We evaluated 27 pNET cases from our cohort for mutations in MEN1, DAXX, ATRX, and MTOR pathway genes, which comprise most genes that have been previously reported as being mutated in pNET.33,34 Of these 27 cases, 17 were evaluable for mutations in all of the above genes: mutations were observed in MEN1 9/17 (53%), ATRX 3/17 (18%), DAXX 8/17 (47%), TSC1 3/17 (18%), TSC2 2/17 (12%), PTEN 1/17 (6%), and PIK3CA 1/17 (6%). We found no clear association between the presence of these mutations, expression of checkpoint markers, or degree of T-cell infiltrates. We similarly did not identify clear associations between these variables and patient survival.
DISCUSSION
Our study provides an initial overview of immune checkpoint marker expression and T-cell immune response in well differentiated small intestine and pancreatic neuroendocrine tumors. We found that expression of PD-1 and PD-L1 in both tumor subtypes is uncommon, but that both subtypes express high levels of PD-L2. T-cell immune infiltrates were more prominent in pNET than in small intestine NET, as were known mutations in general, though we did not observe clear correlations between immune infiltrates and mutations in specific genes.
Our observation that well differentiated small intestine and pancreatic neuroendocrine tumors express only low levels of PD-1 and PD-L1 builds on findings in a prior publication focused on well differentiated foregut and hindgut NET, which also found low levels of expression of these markers.22 We observed high levels of expression of PD-L2 in both pNET and SINET, using both immunohistochemical techniques and direct measurement of RNA expression. PD-L2, like PD-L1, binds to the PD-1 receptor although its function has been less well defined. Most studies to date have reported that PD-L2 has an inhibitory function on T-cell proliferation, analogous to the function of PD-L1.7 PD-L1 and PD-L2 are both located at the same locus on ch9, and copy number gains and amplifications, as well as increased protein expression of both ligands, has been reported in Hodgkin’s lymphoma.35 High levels of PD-L2 expression have also been reported in mediastinal large B-cell lymphoma and in esophageal adenocarcinoma.36,37 The expression of PD-L2 in neuroendocrine tumors raises the possibility that use of PD-1 inhibitors (but not PD-L1) inhibitors may have potential therapeutic benefit. However, our concurrent observation that PD-L2 expression was predominantly cytoplasmic rather than membranous raises questions regarding its potential therapeutic significance. Further studies are warranted to confirm the potential role of PD-L2 in NET.
T-cell immune infiltrates have been considered a general marker of effective immune response. A prior study evaluating T-cell infiltrates in patients with resected neuroendocrine tumors found that the presence of a high CD3+ T-cell infiltrate, suggestive of a robust immune response, was associated with improved recurrence-free survival in patient with intermediate grade tumors, whereas the presence of high FOXP3+ T-cell infiltrates, suggestive of an immune suppressive environment, was associated with shorter survival in NET with liver metastases.38 We did not observe clear associations between T-cell infiltrates and survival; but did observe that the prevalence of FOXP3+ (T regulatory) cells was relatively low in both pNET and SINET, suggesting that T regulatory cells may not play a major role in suppressing an immune response in this disease. We further found that high levels of both intra-and extratumoral immune infiltrates, defined by staining with CD45RO (T memory), CD3 (general T-cell), and CD8 (cytotoxic T-cell) markers were present in subsets of both pancreatic and small bowel NET, and were generally more common in pNET than in SINET.
T-cell infiltrates have been broadly associated with both mutational burden and response to immune checkpoint inhibitors.17,39 Our observation of higher T cell infiltrates in pNET compared to small intestine NET is consistent with fact that recurrent mutations are relatively uncommon in SINET; in a previous study, recurrent mutations in CDKN1B were observed in only 8% of tumors.26 In contrast, pancreatic NET have been shown to have a relatively high prevalence of mutations in MEN1, DAXX, and ATRX, with less common recurrent mutations in mTOR pathway genes.33,34 We did not, however, identify associations between T cell infiltrates and mutations in specific genes. We similarly did not identify associations between immune checkpoint marker expression and tumoral mutational profiles.
In conclusion, both pNET and SINET are characterized by low PD-1 and PD-L1 expression. The significance of high cytoplasmic expression PD-L2 in neuroendocrine tumor cells warrants further investigation. Tumor infiltrating T-cells were observed in both small intestine and pancreatic NET, with more prominent infiltrates observed in pancreatic NET. Our observations suggest the presence of a tumor-specific immune response in neuroendocrine tumors that potentially could be harnessed with emerging immunotherapeutic strategies.
ACKNOWLEDGMENTS
The authors thank Gordon Freeman for providing the antibodies for PD-1 (EH33), PD-L1 (405.9A11) and PD-L2 (366C.9E5). The authors are grateful for support from the Gitta and Saul Kurlat Fund for Neuroendocrine Tumor Research, The Meador Fund for Neuroendocrine Tumor Research, the Jane Dybowski Fund of Neuroendocrine Cancer, McIntyre Family Fund for Neuroendocrine Tumor Research, Lipson Family Fund, Goldhirsh-Yellin Foundation Fund for Neuroendocrine Tumor Research, and The Murphy Family Fund for Carcinoid Tumor Research.
Funding: This work was supported in part by a grant from the NET Research Foundation to MHK. This work was also supported by U.S. National Institutes of Health (NIH) grants R01 CA151532 to MHK; R01 CA151993 to SO and R35 CA197735 to SO.
Footnotes
Declaration of interest: The authors declare that they have no conflicts of interest.
REFERENCES
- 1.Caplin ME, Pavel M, Cwikla JB, et al. Lanreotide in metastatic enteropancreatic neuroendocrine tumors. N Engl J Med. 2014;371:224–233. [DOI] [PubMed] [Google Scholar]
- 2.Yao JC, Shah MH, Ito T, et al. Everolimus for advanced pancreatic neuroendocrine tumors. N Engl J Med.2011;364:514–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yao JC, Fazio N, Singh S, et al. Everolimus for the treatment of advanced, non-functional neuroendocrine tumours of the lung or gastrointestinal tract (RADIANT-4): a randomised, placebo-controlled, phase 3 study. Lancet. 2016;387:968–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Raymond E, Dahan L, Raoul JL, et al. Sunitinib malate for the treatment of pancreatic neuroendocrine tumors. N Engl J Med. 2011;364:501–513. [DOI] [PubMed] [Google Scholar]
- 5.Postow MA, Callahan MK, Wolchok JD. Immune Checkpoint Blockade in Cancer Therapy. J Clin Oncol. 2015;33:1974–1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robert C, Schachter J, Long GV, et al. Pembrolizumab versus Ipilimumab in Advanced Melanoma. N Engl J Med. 2015;372:2521–2532. [DOI] [PubMed] [Google Scholar]
- 7.Latchman Y, Wood CR, Chernova T, et al. PD-L2 is a second ligand for PD-1 and inhibits T cell activation. Nat Immunol. 2001;2:261–268. [DOI] [PubMed] [Google Scholar]
- 8.Yearley JH, Gibson C, Yu N, et al. PD-L2 Expression in Human Tumors: Relevance to Anti-PD-1 Therapy in Cancer. Clin Cancer Res. 2017;23:3158–3167. [DOI] [PubMed] [Google Scholar]
- 9.Jessurun CAC, Vos JAM, Limpens J, et al. Biomarkers for Response of Melanoma Patients to Immune Checkpoint Inhibitors: A Systematic Review. Front Oncol. 2017;7:233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Allen EM, Miao D, Schilling B, et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science. 2015;350:207–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nosho K, Baba Y, Tanaka N, et al. Tumour-infiltrating T-cell subsets, molecular changes in colorectal cancer, and prognosis: cohort study and literature review. J Pathol. 2010;222:350–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giannakis M, Mu XJ, Shukla SA, et al. Genomic Correlates of Immune-Cell Infiltrates in Colorectal Carcinoma. Cell Rep. 2016;26;15(4):857–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Masugi Y, Nishihara R, Yang J, et al. Tumour CD274 (PD-L1) expression and T cells in colorectal cancer. Gut. 2017;66:1463–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adams TA, Vail PJ, Ruiz A, et al. Composite analysis of immunological and metabolic markers defines novel subtypes of triple negative breast cancer. Mod Pathol. 2018;31:288–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Senbabaoglu Y, Gejman RS, Winer AG, et al. Tumor immune microenvironment characterization in clear cell renal cell carcinoma identifies prognostic and immunotherapeutically relevant messenger RNA signatures. Genome Biol. 2016;17:231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tumeh PC, Harview CL, Yearley JH, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515:568–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le DT, Uram JN, Wang H, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med. 2015;372:2509–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Le DT, Durham JN, Smith KN, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357:409–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goodman AM, Kato S, Bazhenova L, et al. Tumor Mutational Burden as an Independent Predictor of Response to Immunotherapy in Diverse Cancers. Mol Cancer Ther. 2017;16:2598–2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Antonia SJ, Lopez-Martin JA, Bendell J, et al. Nivolumab alone and nivolumab plus ipilimumab in recurrent small-cell lung cancer (CheckMate 032): a multicentre, open-label, phase 1/2 trial. Lancet Oncol. 2016;17:883–895. [DOI] [PubMed] [Google Scholar]
- 21.Fan Y, Ma K, Wang C, et al. Prognostic value of PD-L1 and PD-1 expression in pulmonary neuroendocrine tumors. Onco Targets Ther. 2016;9:6075–6082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim ST, Ha SY, Lee S, et al. The Impact of PD-L1 Expression in Patients with Metastatic GEP-NETs. J Cancer. 2016;7:484–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schultheis AM, Scheel AH, Ozretic L, et al. PD-L1 expression in small cell neuroendocrine carcinomas. Eur J Cancer. 2015;51:421–426. [DOI] [PubMed] [Google Scholar]
- 24.Calles A, Liao X, Sholl LM, et al. Expression of PD-1 and Its Ligands, PD-L1 and PD-L2, in Smokers and Never Smokers with KRAS-Mutant Lung Cancer. J Thorac Oncol. 2015;10:1726–1735. [DOI] [PubMed] [Google Scholar]
- 25.Galon J, Costes A, Sanchez-Cabo F, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960–1964. [DOI] [PubMed] [Google Scholar]
- 26.Francis JM, Kiezun A, Ramos AH, et al. Somatic mutation of CDKN1B in small intestine neuroendocrine tumors. Nat Genet. 2013;45:1483–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abo RP, Ducar M, Garcia EP, et al. BreaKmer: detection of structural variation in targeted massively parallel sequencing data using kmers. Nucleic Acids Res. 2015;43:e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McLaren W, Pritchard B, Rios D, et al. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26:2069–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bosman FT, Carneiro F, Hruban RH, Theise ND, et al. WHO Classification of Tumours of the Digestive System. Lyon, France: IARC Press;2010. [Google Scholar]
- 32.Basturk O, Yang Z, Tang LH, et al. The high-grade (WHO G3) pancreatic neuroendocrine tumor category is morphologically and biologically heterogenous and includes both well differentiated and poorly differentiated neoplasms. Am J Surg Pathol. 2015;39:683–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiao Y, Shi C, Edil BH, et al. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science. 2011;331:1199–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scarpa A, Chang DK, Nones K, et al. Whole-genome landscape of pancreatic neuroendocrine tumours. Nature. 2017;543:65–71. [DOI] [PubMed] [Google Scholar]
- 35.Roemer MG, Advani RH, Ligon AH, et al. PD-L1 and PD-L2 Genetic Alterations Define Classical Hodgkin Lymphoma and Predict Outcome. J Clin Oncol. 2016;34:2690–2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shi M, Roemer MG, Chapuy B, et al. Expression of programmed cell death 1 ligand 2 (PD-L2) is a distinguishing feature of primary mediastinal (thymic) large B-cell lymphoma and associated with PDCD1LG2 copy gain. Am J Surg Pathol. 2014;38:1715–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Derks S, Nason KS, Liao X, et al. Epithelial PD-L2 Expression Marks Barrett’s Esophagus and Esophageal Adenocarcinoma. Cancer Immunol Res. 2015;3:1123–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Katz SC, Donkor C, Glasgow K, et al. T cell infiltrate and outcome following resection of intermediate-grade primary neuroendocrine tumours and liver metastases. HPB (Oxford). 2010;12:674–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stagg J, Allard B. Immunotherapeutic approaches in triple-negative breast cancer: latest research and clinical prospects. Ther Adv Med Oncol. 2013;5:169–181. [DOI] [PMC free article] [PubMed] [Google Scholar]


