Abstract
Current environmental monitoring approaches focus primarily on chemical occurrence. However, based on concentration alone, it can be difficult to identify which compounds may be of toxicological concern and should be prioritized for further monitoring, in-depth testing, or management. This can be problematic because toxicological characterization is lacking for many emerging contaminants. New sources of high-throughput screening (HTS) data like the ToxCast database, which contains information for over 9,000 compounds screened through up to 1,100 bioassays, are now available. Integrated analysis of chemical occurrence data with HTS data offers new opportunities to prioritize chemicals, sites, or biological effects for further investigation based on concentrations detected in the environment linked to relative potencies in pathway-based bioassays. As a case study, chemical occurrence data from a 2012 study in the Great Lakes Basin along with the ToxCast effects database were used to calculate exposure-activity ratios (EARs) as a prioritization tool. Technical considerations of data processing and use of the ToxCast database are presented and discussed. EAR prioritization identified multiple sites, biological pathways, and chemicals that warrant further investigation. Prioritized bioactivities from the EAR analysis were linked to discrete adverse outcome pathways to identify potential adverse outcomes and biomarkers for use in subsequent monitoring efforts.
Introduction
Environmental monitoring has traditionally relied heavily upon analysis of different matrices (water, sediment, soil, etc.) for potential contaminants through targeted analytical methods. Advances in analytical instrumentation have increased the sensitivity of targeted methods, with compounds routinely measured in water at low part per trillion levels.1, 2 Additionally, expansion of high resolution instruments in environmental monitoring has led to nontargeted analytical methods being implemented, with hundreds to thousands of compounds detected and dozens to hundreds of compounds tentatively identified in some instances.3–6 Despite advances in the ability to detect contaminants in the environment, understanding the biological implications from exposure to these compounds has not kept pace. Thus, while chemicals can be identified and quantified in the environment, the potential biological effects of hundreds to thousands of individual contaminants remain poorly defined.7
Advances of high throughput toxicology have led to the development of biological effects data for a large number of chemicals. For example, the Tox21 Program8, 9 and USEPA ToxCast Program10 together provide high-throughput screening (HTS) data for over 9,000 unique substances, including many industrial and environmentally relevant chemicals for which traditional human health or ecological effects data are lacking. Chemicals are prepared and screened in a standardized manner, and each chemical is tested using a consistent dose-response design across assays. This allows derivation of point of departure estimates such as the chemical-specific half-maximal activity concentration (AC50) or activity concentration at cutoff (ACC) for each chemical-assay combination, and chemicals can be subsequently ranked in terms of potency for a given assay target. Together the programs utilize a number of in vitro and in vivo HTS assays to identify a range of pathway-specific and nonspecific biological interactions providing a final database covering hundreds of specific biological pathways and processes.
To effectively leverage the breadth of biological and chemical space covered within these databases, new approaches and tools are being developed. A method for exposure-activity profiling using exposure-activity ratios (EARs) represents one approach for screening and prioritizing chemicals using both chemical occurrence and HTS data,11 and has been previously highlighted with potential for application to environmental monitoring.12, 13 Exposure-activity ratios, similar in concept to toxic units,14 incorporate both the dose (i.e., environmental concentration) and the relative potency (i.e., point of departure estimate) for a given chemical-assay pair, allowing for prioritization of chemicals or sites based on expected biological activity. Assays identified as higher priority based on EARs can be further linked through adverse outcome pathways (AOPs) to identify hazards to organisms and/or ecosystems,15 and measurable endpoints associated with perturbation of specific biological processes identified for confirmation or monitoring of predicted site-specific hazards.
The EAR approach utilizing the ToxCast database offers a currently-viable, standardized method to integrate chemical occurrence and biological effects for prioritization of environmental monitoring datasets. The present work applies this approach to a study focused on identifying and characterizing emerging contaminants across tributaries and Areas of Concern (AOCs) within the Great Lakes,1 specifically demonstrating the use of EAR analysis as a rapid, screening level tool for prioritization of existing chemical occurrence datasets. This study was part of an on-going, multiagency effort directed at effects-based tool development for application to monitoring and surveillance of contaminants of emerging concern and their potential impacts on resident biota and ecosystems in the Great Lakes.16 The present case study demonstrates how EARs can be used to prioritize environmental sites, identify potential biological activity(ies) of concern, and highlight chemicals likely contributing to the biological activity. Further, prioritized biological activities are linked to existing AOPs to identify potential adverse effects and strategies for monitoring predicted hazards in exposed organisms. Issues that may commonly be faced when using HTS databases for EAR calculations are detailed to focus on the application of this approach for utility in environmental monitoring.
Experimental
Study Locations and Sample Collection
Data presented in this study were obtained from a publicly available U.S. Geological Survey (USGS) report.1 In 2012, surface water samples were collected at 66 sites across six watersheds in the Great Lakes Basin, including the St. Louis River/Duluth Harbor, MN; Fox River/Green Bay, WI; Detroit River, MI; Raisin River, MI; Maumee River, OH; and Irondequoit Bay, NY. Sampling locations varied from those with low anticipated anthropogenic impact to sites in close proximity to municipal wastewater treatment plants (WWTPs). Samples were collected from all watersheds in Spring (April-May), and additional samples were collected from the Maumee and St. Louis Rivers in summer (September). A total of 140 water samples were collected for chemical analysis (excluding blanks and duplicates). Samples were collected as either 1 L depth-integrated grab samples or 96 h temporally-integrated samples.17 Site features including depth, flow, and water quality parameters were recorded at the time of sample collection.1 Sample numbers varied across watersheds ranging from 3 to 76. The most intensive efforts were focused at the St. Louis River and Maumee River watersheds with collection of 76 and 37 samples, respectively. Full collection methods are detailed elsewhere.1
Analytical Chemistry
Surface water samples (both grab and composite) were submitted to the USGS National Water Quality Laboratory (Denver, CO, USA) and analyzed for three broad suites of contaminants: wastewater indicators18, pharmaceuticals19, and steroids and sterols20 (see SI Table S1 for complete list of contaminants and method). In total, 134 unique organic compounds were measured using these three GC-MS based analytical methods. In cases where compounds were measured across multiple analytical methods (3β-coprostanol, bisphenol A [BPA], cholesterol), only the result from the method with the lowest laboratory reporting limit (LRL) was retained for our analysis. Further details including complete analyte lists, quality control procedures, and concentrations of analytes were reported previously.1
Chemical Concentration Dataset Processing
Analytical methods used in this study are reported by USGS as information-rich methods,21 meaning additional qualifying information is included alongside analyte quantification. As such, reported concentrations of some compounds are below the given LRL. All reported values, including those below LRLs, were used for EAR calculations without adjustment. Non-detections (reported as <LRL) were assigned a value of zero to prevent highly bioactive compounds (i.e., from the HTS dataset) from skewing EAR results.
In Vitro Bioassays
A limited number of water samples were further screened for total estrogenic activity using the T47D-KBluc cell line, which is stably transfected with an ERα-reporter gene construct.22 Samples were tested in triplicate, independent wells. The full methodology for this bioassay is available in the SI.
ToxCast Database
The ToxCast database23 (including both ToxCast and Tox21 information) contains HTS data for thousands of chemicals screened through multiple assay platforms. The database currently includes 12 platforms covering a variety of cell-based, cell-free, and whole organism HTS assays, encompassing over 300 unique signaling pathways (e.g., estrogen receptor [ER], aryl hydrocarbon receptor, glucocorticoid receptor, etc.) and nonspecific endpoints (oxidative stress, cytotoxicity, organelle conformation, etc.). ToxCast assay sources and operating procedures are described in detail elsewhere.24–41 All ToxCast data used for this study were from the October 2015 data release and are publicly available online through the ToxCast website.23
Exposure-Activity Ratios
To support screening and prioritization, EARs comparing surface water chemical concentration to ACC estimates for all chemicals available in the ToxCast database were calculated (Eq. 1). Assuming non-interactive concentration addition within assays, a summed EAR of environmental mixtures containing n number of chemicals can be calculated (Eq. 2). While the AC50 has previously been used as the point of departure estimate for EAR calculations;11–13 we chose the ACC for the present analysis. ACC estimates the chemical concentration at which a defined threshold of response (the activity cutoff) is achieved within an assay42 (see SI Figure S1). Unlike the AC50 where the concentration estimate may align with different magnitudes of bioassay response for each chemical tested in an assay, the activity cutoff threshold is uniform for all chemicals tested in the same assay. Thus, the ACC is less prone to violating assumptions underlying relative potency estimation.43 The activity cutoff for each assay is based on a multiplier of the baseline median absolute deviation (BMAD; 3x,5x,6x, or 10x), which is calculated as the median absolute deviation for each assay endpoint using data points from the two lowest tested concentrations of all chemicals within a given assay. The multiple of the BMAD was generally selected specific to assay platforms (sometimes individual assays) to account for the baseline variability and signal window with a goal of achieving a high sensitivity for true actives while minimizing false positives due to baseline variability to the extent possible. The two lowest concentrations were used for determining BMAD as they are below bioactive concentrations for the great majority of samples. The use of median over mean provides for protection from extreme values from highly potent chemicals. Also, use of sample wells rather than solvent controls for background activity measurement has an advantage of being derived from wells treated identically to all other sample-treated wells. For assays that are normalized to a positive control, a cutoff threshold of 20% response may also be used to ensure the response is in a reasonable range of the response of a known, reference chemical to provide some test for biological significance in addition to statistical significance. Assay specific cutoff threshold criteria are given in supporting information (SI Table S2).
While the EAR is a simple calculation conceptually, processing of the extensive chemical dataset (154 samples x 134 chemicals) and even larger ToxCast database (1192 assays x 9076 chemicals) was expedited by development of a custom, R-based program to facilitate processing, calculation, and visualization of the EAR dataset (details in SI).
| Eq. 1 |
| Eq. 2 |
Quality Assurance Considerations
During the process of generating EARs, multiple issues with chemical occurrence and ToxCast datasets were identified, which could lead to artifacts and errors in interpretation and use of the EARs. The first issue was the use of chemical salts for toxicity testing in ToxCast assays. Chemical occurrence data are reported as free compound (e.g., methadone, CAS 76–99-3) while some compounds, especially pharmaceuticals, are tested in ToxCast assays as ion pairs (e.g., methadone hydrochloride, CAS 1095–90-5). Compounds from the chemical occurrence dataset that were not identified in the ToxCast database through a search for a matching CAS number were manually screened for tested chemical alternatives. Among 27 analyzed compounds initially identified as not present in the ToxCast database, nine actually were present as chemical analogs. In these cases, CAS numbers were updated in order to better align chemical occurrence data with the ToxCast database for EAR generation (SI Table S3). Additionally, within the ToxCast data, molecular weights for these CAS numbers were updated to match the measured compound and maintain correct µM calculations. For the purposes of EAR generation, free and salt forms were considered to be equipotent across the assays.
In the ToxCast database, assays are grouped by platform and target (as defined by annotation fields from the database).23 Some of these assays were purposefully excluded from EAR consideration. Specifically, since a goal of this case study was to link EARs to potential adverse outcomes, most nonspecific cell-based or cell-free assay endpoints were excluded. For example, we excluded assays from the platforms Apredica (APR) and Bioseek (BSK), which largely target nonspecific endpoints44 such as cell cycle status, cell proliferation, organelle conformation, DNA damage, and stress response, and are difficult to interpret though specific pathway-based toxicity assessment. Assays categorized as ‘background measurement’, ‘cell cycle’, and ‘cell morphology’ (based on ‘intended_target_family’ annotation field) also were excluded from EAR calculations as these monitor baseline assay performance, not specific effects. The ‘intended_target_family’ annotation was used for subsequent grouping of assay results, and several assays designated ‘NA’ were manually assigned to an existing category (SI Table S2). For all zebrafish (Danio rerio) related assays, a new category ‘ZF’ was included. Attagene (ATG) assays reporting in the ‘loss’ direction and Novascreen (NVS) assays reporting in the ‘gain’ direction (based on ‘signal_direction’ annotation field) also were removed from the final EAR results, as these platforms are not optimized or designed to report for the given assay direction.23 Overall, 528 of 1192 assay endpoints were considered for the final EAR calculations (SI Table S2).
Chemical-assay pairs were further censored from the final dataset using ToxCast data quality flags. These flags, explicitly noted in the ToxCast database, were implemented to identify potential false positive and false negative results based on dose-response irregularities.42 Chemical-assay pairs indicating false positives were removed from the final results, specifically, data with flag identifications 7,10,11,15, and 17 (SI Table S4). Employing these flags enabled removal of the majority of false positive chemical-assay pairs from the final EAR results. One specific chemical-assay pair, fluoranthene in the ‘Tox21_p53_BLA_p3_ratio’ assay, was observed to result in very high EAR values (>100). Upon closer examination, the dose response curve was found to be flat and was identified as an unflagged, false positive. This chemical-assay pair also was removed from the final dataset. Other unflagged, false positive chemical-assay pairs may be present in the dataset, but no other individual pairs exhibited the very high EAR values that may greatly skew the final interpretation of results. No flagged, potential false negatives were identified after evaluating actual concentration-response curves, a typical finding given that the “hit-calling” algorithm is designed to be conservative (i.e., favoring false positives) in the interest of identifying all potential hazards.
A final observation was associated with generation of best-fit dose-response curves and corresponding ACC values using the automated ToxCast data analysis pipeline.42 For a number of assays, ACCs of specific chemicals were reported below the minimum concentration tested within the assay. Closer examination of dose-response curves showed that the minimum tested concentration for many strong agonists was at or near saturation for receptor targets (e.g., 17β-estradiol and ER related assays), thus full dose-response curves are estimated from the upper “plateau” of the curve. For example, the lowest concentration of 17β-estradiol tested in the ‘ATG_ERE_CIS-up’ assay (0.09 µM) induces approximately 98% of the maximum 17β-estradiol response, and an ACC value of 3.1e-5 µM is extrapolated from the model fit. For compounds detected in the chemical occurrence dataset, 85 chemical-assay pairs (comprised of 14 chemicals and 34 assays) have ACCs below the minimum tested concentration (SI Table S5). Eleven of the identified chemicals are steroids or xenobiotics known to interact with nuclear receptors, and 27 of 38 assays are related to nuclear receptors. This suggests the dose range generally screened in ToxCast is insufficient to fully characterize strong agonists of some nuclear receptors. Moreover, ACC estimates extrapolated over orders of magnitude in concentration will inherently be less accurate or unreliable compared to those estimated from a full dose-response curve. No adjustments were made for these chemical-assay pairs, and consequently, EARs for these instances may be underestimated.
Results and Discussion
Chemical occurrence summary
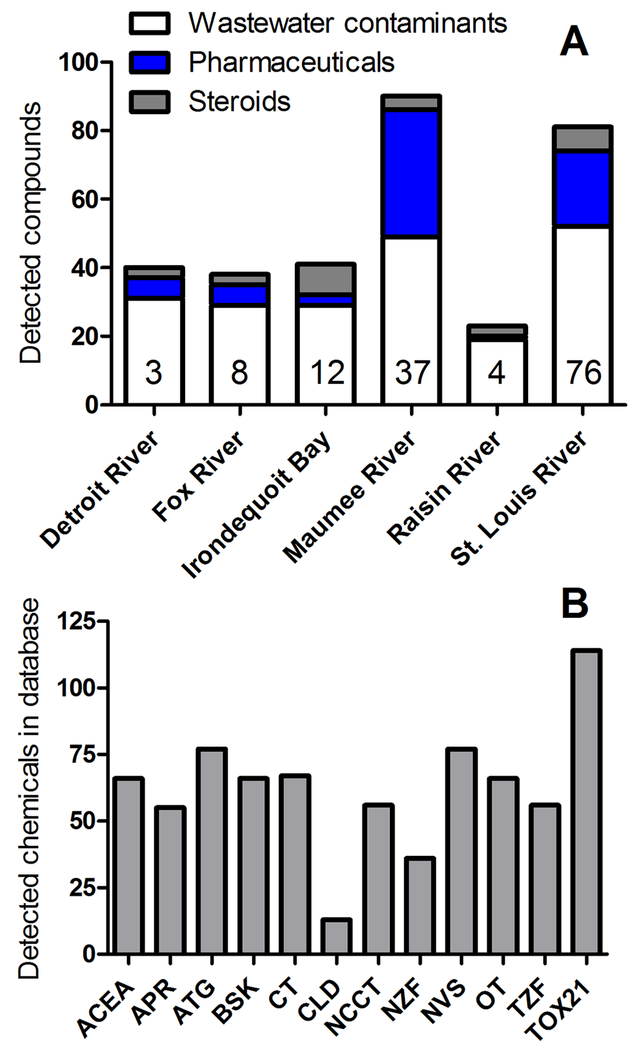
Of 134 contaminants analyzed across the three analytical methods, 109 were reported above LRLs in one or more surface water samples (SI Table S1). By analytical method, 60 of 67 (90%) wastewater contaminants, 39 of 48 (81%) pharmaceuticals, and 10 of 19 (53%) steroids and sterols were detected at one or more sites. By watershed, the greatest number of wastewater contaminants, pharmaceuticals, and steroids and sterols were detected within the St. Louis River, Maumee River, and Irondequoit Bay watersheds, respectively (Figure 1). Of note, the St. Louis River and Maumee River watersheds had more intensive sampling efforts centered on WWTPs, and as such, it could be reasonably expected to observe a greater number of compounds at these sites. Based on frequency and concentration alone, several chemicals can be highlighted from within the dataset. Cholesterol, DEET, fluoranthene, and pyrene, which are all within the wastewater indicators method, were detected most frequently, in 98%, 93%, 71%, and 69% of all samples, respectively. Cholesterol, 3β-coprostanol, β-sitosterol, and BPA, again all within the wastewater indicators method, were detected at the highest single concentrations, with maximum concentrations of 120 µg/L, 95.2 µg/L, 64.4 µg/L, and 60.5 µg/L, respectively. This abbreviated list again demonstrates the need to consider effects concentrations of compounds, as several chemicals present at high concentrations (e.g., the naturally derived sterols) would be expected to have limited biological effects.
Figure 1.
A) Number of detected chemicals within each analytical method at each watershed reported in the chemical occurrence dataset. The value within each column represents the total number of samples from a watershed. B) Number of detected chemicals with available data in each ToxCast assay platform. See SI Figure S2 for more detailed information on chemical coverage across assay platforms.
ACEA = ACEA Biosciences; APR = Apredica; ATG = Attagene; BSK = BioSeek; CT = CeeTox; CLD = CellzDirect; NCCT = National Center for Computational Toxicology; NZF = NHEERL zebrafish; NVS = NovaScreen; OT = Odyssey Thera; TZF = Tanguay Lab zebrafish; Tox21 = Tox21 Initiative
ToxCast coverage of chemicals
Compounds in ToxCast were screened in multiple phases, with different batteries of assays used for some phases of the overall testing program (for overview see 45); thus, the assay coverage for all chemicals in ToxCast is not consistent. For chemicals measured within the current study, 116 of 134 (86%) analyzed chemicals and 96 of 109 (88%) detected chemicals had been evaluated in one or more assays (SI Table S1, SI Table S6). Tox21 had the greatest coverage of detected chemicals (87%), followed by ATG and NVS (61%), and then BSK and CEETOX (51%) (Figure 1). Considering assay coverage of detected chemicals, 48 (44%) have data in 10 or more batteries, 66 (61%) have data in three or more batteries, and 30 chemicals (28%) have data only for the Tox21 assay suite (SI Figure S2).
Exposure-Activity Ratio Based Prioritization
Site Prioritization
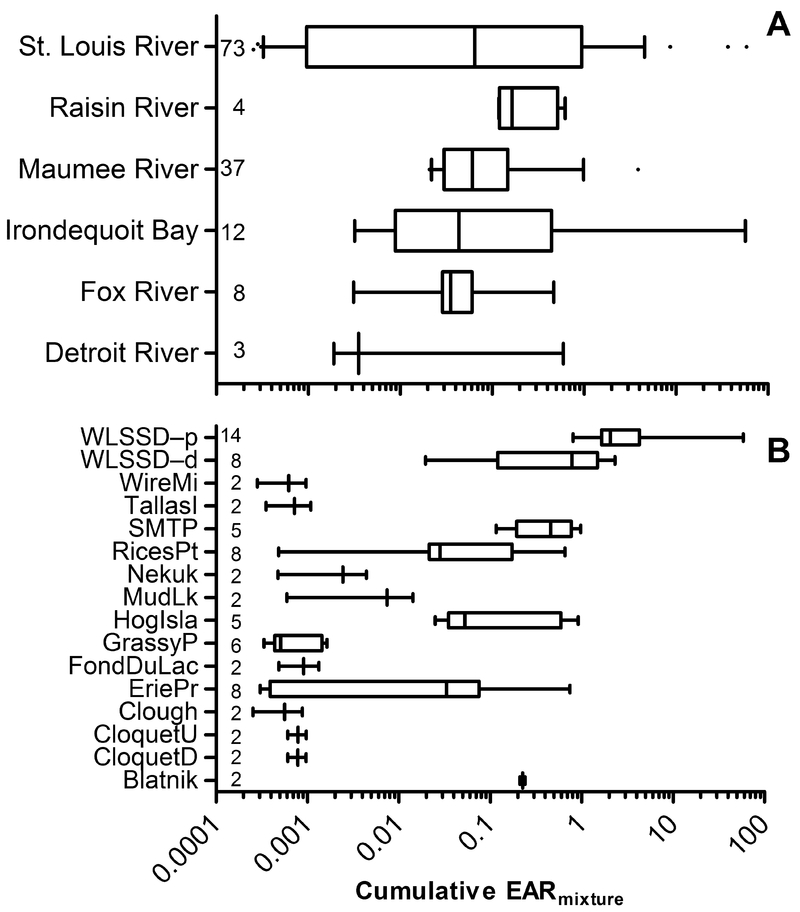
As an initial screen of the chemical occurrence dataset, EARmixture values (Eq. 2) were calculated for all sites (SI Table S7). No active chemicals were detected for 299 of 528 assays leaving 229 total ToxCast assays with calculated EAR values in one or more samples. Similarly, three of 140 water samples had no detected compounds that were active in ToxCast leaving a total of 137 samples with calculated EAR values. To better summarize the EAR dataset by watershed and individual sites, EARmixture values across all 229 assays were summed for each site, as a cumulative EARmixture. Since the data matrix of chemicals and assays is consistent for all samples, this provides a value of estimated activity at each site irrespective of individual assay target. Through this approach, the St. Louis River watershed, which was most intensively sampled, showed the highest values, and the largest range of cumulative EARmixture of any watershed (Figure 2A). Median EAR was highest at the Raisin River, followed by Maumee River, Fox River, and St. Louis River watersheds. Focusing on the St. Louis River (representing the watershed with the maximum EAR values and greatest sample coverage), three sites with the greatest potential impact, WLSSD-Proximal (WLSSD-P), and WLSSD-Distal (WLSSD-D), and SMTP were identified (Figure 2B). These three sites all are notably in close proximity to two WWTPs and reasonably could be hypothesized as having a relatively greater potential for biological effects. WLSSD-P and WLSSD-D are impacted by a larger WWTP receiving both municipal and industrial waste, with WLSSD-P nearest to the WWTP outflow and WLSSD-D at a more distal location with a greater dilution of the effluent.17 SMTP is impacted by a separate, smaller WWTP receiving exclusively municipal waste. Accordingly, the highest cumulative EARs are observed at WLSSD-P followed by WLSSD-D and SMTP. Though details of specific chemicals and relevant biological pathways are not captured at this level of site prioritization, the ability to condense chemical occurrence and biological activity datasets into a single output allowed for a rapid comparison and prioritization of watersheds or individual sites from the dataset.
Figure 2.
A) Cumulative EARmixture (i.e., sum of EARmixture values across all assays) values within each watershed. The value within each row represents the total number of samples from a watershed. For graphical purposes, sites with EARmixture equal to 0 (three samples; all from St. Louis River) were removed. B) Cumulative EARmixture values for each site within the St. Louis River watershed. The value within each row represents the total number of samples from a site. For graphical purposes, sites with EARmixture equal to 0 or with only one sample are not shown. Site information is available from the original data source.1
As an alternative strategy for site prioritization, exceptionally high cumulative EARmixture values were identified across watersheds to highlight unique sites or potential point sources of contamination. Only four sites in total, located in the St. Louis River, Maumee River, and Irondequoit Bay watersheds, had cumulative EARmixture values greater than 2 (Figure 2). As noted above, both sites within the St. Louis River (WLSSD-P and WLSSD-D), as well as the single site in the Maumee River watershed (MX-WWTP) are in close proximity to WWTPs, which likely explains the elevated EARmixture values from these sites. The single site in Irondequoit Bay (IB-06), however, has no obvious point sources of contamination. This site is unique in the composition of detected contaminants, with many steroidal compounds observed only at this site. Several naturally occurring androgenic and estrogenic steroids were detected, as well as two synthetic estrogens, mestranol and diethylstilbestrol, all of which contribute heavily to the high EAR values observed in this sample. The presence of many steroids in this single sample likely indicates an unknown contamination source upstream of the site. Mestranol is a component of some oral contraceptives,46 indicating a possible human waste source; conversely, the presence of 17α-estradiol is generally associated with animal waste or animal production facilities.47, 48 Also, although diethylstilbestrol is no longer used as a human pharmaceutical, its environmental occurrence could be associated with illicit use in livestock operations,49 again suggesting agricultural sources may be contributing to the estrogenic signal. Further investigation would be required to identify potential sources of the observed contamination. Given the elevated EAR values and unique chemical signature at IB-06, further monitoring at this site is warranted.
Biological Activity Prioritization
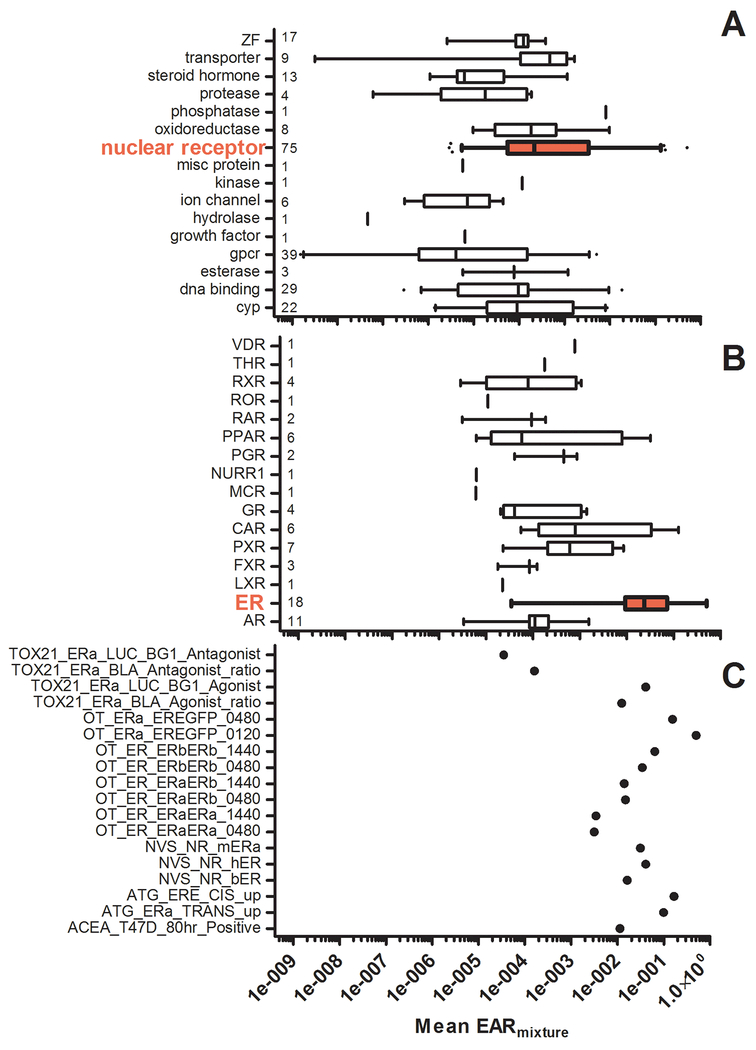
To prioritize molecular targets and biological pathways most likely to be perturbed by chemicals broadly observed in the Great Lakes samples and/or at specific sites, mean EARmixture values were calculated within each assay across all sites (SI Table S8). Assays were then grouped according to the ToxCast database ‘intended_target_family’ annotation23 resulting in 15 assays groups containing one to 75 individual assays (Figure 3A). Of categories containing two or more assays, the greatest median mean EARmixture values were observed for ‘transporter’, ‘nuclear receptor’, and ‘oxidoreductase’ at 4.8e−4, 2.1e−4, and 1.8e−4, respectively. Maximum mean EARs of 0.50, 0.018, and 0.0097 were observed for ‘nuclear receptor’, ‘DNA binding’, and ‘oxidoreductase’ assay groupings. Other endpoints including ‘cyp’, ‘esterase’, and ‘ZF’ assay groupings are also elevated (and likely not statistically different) from assay groupings highlighted above, so these groupings could be further investigated. Because the ‘intended_target_family’ level of assay organization does not inform as to specific molecular targets of the detected chemicals, broad categories such as ‘nuclear receptor’ and ‘DNA binding’ must be further “dissected” to reveal relevant biological pathways. For example, exploring ‘nuclear receptor’ assays in more detail by using the ‘intended_target_gene_symbol’ annotation field of the ToxCast database reveals assay gene targets which would be most relevant for linking biological activity to adverse effects through AOP constructs (Figure 3B). The primary targets driving the high response in ‘nuclear receptor’ assays are ER, constitutive androstane receptor (NR1I3; CAR), and peroxisome proliferator-activated receptor (PPAR) assays (Figure 3B). Elevated EARs observed in ‘oxidoreductase’ and ‘DNA binding’ are driven by assays measuring thyroid peroxidase (TPO) inhibition and increased SOX1 expression, respectively. Since both TPO inhibition and SOX1 expression are measured by only a single assay, it is not possible to evaluate the veracity of this response across multiple test systems.
Figure 3.
A) Mean EARmixture values within each assay category as defined by the ‘intended_target_family’ annotation field (see SI Table S2). The number in each row is the number of assays in a category. B) Mean EARmixture values within the ‘nuclear receptor’ assay category as defined by the ‘intended_target_gene_symbol’ annotation field (see SI Table S2). The number in each row is the number of assays in a category. C) Mean EARmixture values for individual assays under the ‘ESR’ category.
For targets measured by multiple assays, such as ER (which is reflected in some manner in a total of 18 different assay endpoints), the range of EARmixture values from individual assays can be dissected further to compare individual assay endpoints. For the ER-based assays, mean EARmixture values across all sites ranged from 3.5e−5 to 0.50 (Figure 3C). Aside from two antagonist assays, the bulk of the mean EARmixture values for the various ER assays fall between 0.001 and 0.1. However, slightly greater EARs were detected for the Odyssey Thera (OT) ‘OT_ERE_GFP’ assays and the ATG ER assays. The range of EARs observed in the ER assays reinforces that not all assays are equivalent in performance and design. ER assay endpoints are measured at different points along the signaling pathway, including receptor binding (NVS), receptor dimer formation (OT), receptor-DNA interaction (OT), mRNA transcription (ATG), protein production (Tox21), and cell proliferation (ACEA). Additionally, possible errors from extrapolating ACC values below tested concentrations may be contributing to inter-assay variability. The use of aggregate or average values may eliminate much variability for targets with a rich assay coverage (e.g., ER), but for targets with few or only a single assay, caution should be exercised in establishing a predefined EAR screening threshold without first considering the range of responses observed in a dataset.
Chemical Prioritization
Individual chemicals can also be prioritized by EAR calculations. Here, chemicals were prioritized based on EARs calculated using the maximum concentrations reported in the Great Lakes dataset. The full matrix of chemical-assay pairs results in a total of 50688 theoretically possible EARs (96 detected chemicals x 528 considered assays; SI Table S9). However, not all possible EARs can be calculated due to chemicals not being tested in all assay batteries or chemicals being inactive in tested assays. In total, EARs could be calculated for 1245 (2.5%) chemical-assay pairs, with values ranging from 3.5e−8 to 44.9. It should be again noted that chemicals in the ToxCast database have been tested in multiple phases, using variable assay platforms across phases; thus, chemicals present in the database are not necessarily tested in all the same assays (see SI Figure S2, Table S1, Table S6). Of detected chemicals that were present in the ToxCast database, 73 of 96 were active in one or more assay(s). Focusing on EARs ≥0.01, representing approximately the upper 10% of EARs, the chemical list can be narrowed to 20 compounds and 146 chemical-assay pairs (SI Table S9). BPA, at a maximum detected concentration of 60.5 µg/L, had the greatest number of non-zero EARs (89), and the greatest number of EARs >1 (9). BPA is known to be an estrogen receptor agonist,50 and 8 of 9 EARs >1 are from ER related assays. Another assay yielding a high EAR for BPA is a CAR antagonist assay. The CAR is involved in xenobiotic metabolism and energy homeostasis and can increase expression of cytochrome P450 (CYP) enzymes.51 In addition to BPA, five other chemicals (17α-estradiol, 17β-estradiol, diethylstilbestrol, estriol, estrone) have EARs>1; all are natural or synthetic estrogens. Fourteen additional chemicals have one or more EARs>0.01 (see SI Table S9), including androgenic steroids (androstenedione, epitestosterone), pesticides (atrazine, metolachlor), and several other common wastewater and urban contaminants (4-nonylphenol, methadone, triclosan, triphenyl phosphate). Methadone shows the greatest activity in a NVS opiate receptor binding assay, which is consistent with the therapeutic use of methadone as an opiate pain reliever. Triphenyl phosphate shows the greatest activity in a PPARγ receptor binding assay and has been demonstrated to interact with the human PPARγ receptor.52, 53 The known androgen receptor (AR) agonist androstenedione surprisingly has a higher EAR value in the ‘ATG_ERa_trans_up’ assay (EAR = 0.017) than in the AR responsive assay endpoints (maximum EAR = 0.007). In this case, ACC extrapolation error may explain the higher predicted ER activity since the extrapolated ACC within the ‘ATG_ERa_trans_up’ is more than 50 times below the lowest tested concentration. Another possible reason for this elevated ER activity could relate to assay platform differences. Two other ER-responsive assays, ‘TOX21_ERa_LUC_BG1_Agonist’ and ‘ACEA_T47D_80hr_Positive’, predict androstenedione to have much lower ER activity, with EARs of 3.0e−4 and 5.3e−7, respectively. Additionally, androstenedione is not active in two NVS ER assays, which directly measure competitive receptor-binding, suggesting androstenedione has low affinity for direct binding to the ER. Androstenedione is, however, a direct precursor to estrone, a potent ER agonist, and can be converted to such by CYP-aromatase (CYP19A1).54 ATG assays use a modified HepG2 cell line,36 which plausibly could be more metabolically active than other assay cell lines, leading to an elevated ER signal from androstenedione through direct formation of estrone. Taken together, this example highlights the value of considering chemical response across multiple assays (if available) when elevated EAR values are observed for a chemical.
Recently, computational models have been developed to aid in separating true positive and false positive chemicals for ER, AR, and TPO assays within the ToxCast database.55–57 Development of such models requires multiple orthogonal assays, which are not currently available for many assay targets. Nevertheless, if chemicals for these select pathways are identified through EAR analysis, the developed models could be applied to confirm that compounds are predicted to be active in the given biological pathway, further increasing confidence in EAR prioritization of select chemicals and biological pathways.
Exposure-activity Prioritization Validation
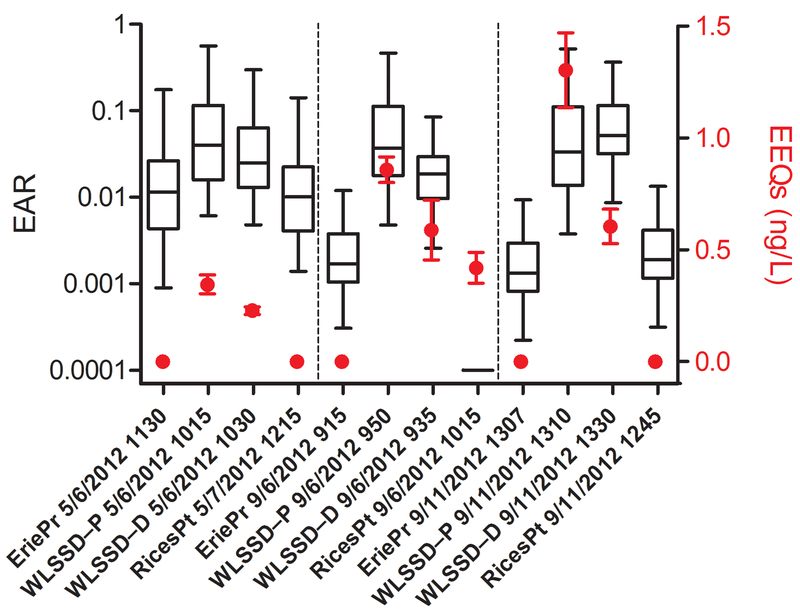
As part of the development of methods for an effects-based monitoring program,16 a set of 12 water samples from the St. Louis River watershed for which we had generated EAR values were screened for estrogenic activity using the T47D-KBluc cell line (SI Table S10; see SI for methods details). This provided an opportunity to directly compare measured ER-mediated activity and EAR prioritization in these samples. The samples were collected both in the Spring and Fall of 2012 along a wastewater gradient with the EriePr site upstream, WLSSD-P at the WWTP outfall, WLSSD-D downstream of the outfall, and RicesPt farthest downstream. Derived 17α-ethinylestradiol equivalents (EEQs) from the T47D-KBluc assay were compared to the range of EARs generated from the 16 ER agonist assays present in the ToxCast database (Fig. 4). Derived EEQs were above detection threshold at the two sites nearest the WWTP (WLSSD-P, WLSSD-D) in all three samples and below the detection threshold at the upstream and downstream site, aside from one sample (RicesPt 9/6/2012 1015). If a median EAR cutoff of 0.01 is applied to the dataset, four samples are excluded, three of which had EEQs below detection. The remaining site (RicesPt 9/6/2012 1015) had no detection of compounds active in the ToxCast ER assays, but had 0.4 ng/L EEQs. For the eight sites with median EAR values above 0.01, two had estimated EEQs below detection. Overall, EAR prioritization identified all sites with EEQs>0.5 ng/L and all but one site with detectable EEQs, demonstrating high sensitivity (0.86) for predicting positive results and thereby supporting EARs as a useful screening level prioritization tool. Incidentally, the concordance of ER results also suggests that the most influential ER agonists are being captured by the targeted analytical methods. We note, however, that the ER is the most well represented assay target in the ToxCast database and endocrine active compounds are also well represented in the suite of chemicals tested through the HTS assays. Biological targets with less assay and chemical coverage will need to be further investigated to verify the accuracy of other EAR estimates; nevertheless, chemical and assay coverage will only increase in the future.
Figure 4.
Comparison of exposure-activity ratios (EARs) generated from 16 estrogen receptor (ER) related assays in the ToxCast database and in 17α-ethinylestradiol equivalents (EEQs; ng/L, in red) derived from in vitro screening with T47D-Kbluc cell line (mean±SEM; n=3). Samples are grouped by collection date and ordered (left to right) from upstream to downstream along the wastewater gradient. For graphical purposes, a site with no EAR calculated was assigned a value of 0.001, and EEQs below detection threshold were assigned a value of 0. Site information is available from the original data source.1
Integrating EARs with Adverse Outcome Pathways
The AOP framework provides a means to link molecular level data (i.e., biological targets or pathways associated with high EARs generated from this study) to apical endpoints of regulatory concern (i.e., reproduction, growth, survival). Assay targets can reflect defined molecular initiating events (MIEs) or key events (KEs) whose perturbation has been credibly linked to adverse outcomes (AOs). Key events downstream of identified assay targets can also serve as potential biomarkers for subsequent monitoring efforts. Biological pathways prioritized through EAR analysis, including ER, TPO, and PPARγ, are associated with established AOPS and were further investigated to explore potential AOs and to identify KEs which could serve as verification of hazard in impacted ecosystems.
Assays associated with ER activation consistently showed the highest EARs across the dataset. This is not unexpected for sites in close proximity to WWTPs; for example, several previous studies have demonstrated the estrogenic nature of WWTP effluent within the St. Louis River.17, 58, 59 An AOP for estrogen receptor activation leading to altered reproduction in adult fishes and subsequent declining population trajectory has been detailed by Ankley et. al.15 At the organismal level, multiple AOs are identified, including reduced fecundity in female fish, altered gamete ratio in spawning males, and impaired spawning behavior in both male and female fishes. Though data gaps exist in the full mechanistic linkage of ER activation to some KEs in the AOP, biomarkers of effect are identified and could be applied to follow-up monitoring efforts. For example, production of the egg yolk precursor protein, vitellogenin (VTG), is linked to ER activation, and measures of VTG mRNA or plasma VTG have historically been used as a biomarker of estrogenic activity in both laboratory and field studies.59–61 In normal, unimpaired male fishes, VTG is not present or present only at very low concentrations, making this an ideal biomarker for identification of exposure to exogenous ER agonists in male fishes. The development of intersex male fish is another potential histological indicator of ER activation. Intersex in fishes has been reported at wastewater impacted sites, and has previously been used as an indicator of estrogenicity of effluents.62, 63 Though the described ER activation AOP is specific to fish, aspects of it likely are applicable to other aquatic egg-laying animals as well (e.g., amphibian, avian species) since ER signaling pathways are highly conserved across vertebrate classes.64 Conversely, a functional ER has not been identified in invertebrates, suggesting they would be of limited utility for biomonitoring for the occurrence of estrogens in the field.
Another assay endpoint that was identified by elevated EARs was TPO inhibition. TPO is an enzyme involved in the synthesis of thyroid hormone from mono- and di-iodotyrosines, so its inhibition leads directly to reduced thyroid hormone production.65 TPO inhibition is a defined MIE in the publicly-available AOPWiki (https://aopwiki.org), linked to two ecologically relevant AOPs: TPO inhibition leading to reduced young of year survival via anterior swim bladder inflation (https://aopwiki.org/aops/159) and TPO inhibition leading to altered amphibian metamorphosis (https://aopwiki.org/aops/175). In both, the final AO from an ecosystem level would be population decline, an endpoint of regulatory importance. Thyroid hormone concentrations in whole body66 or specific tissues65, 67 of fish and amphibians have been used for verification of TPO inhibition in laboratory studies and could serve as a biomarker for follow-up monitoring at specific sites in wild populations68–70 or in situ exposed organisms. Aquatic vertebrates would be considered the most susceptible class to TPO inhibition, as the thyroid axis is well conserved across vertebrates.71 Thyroid hormones have also been demonstrated to play a role in bivalves,71 indicating that some invertebrate taxa could potentially be affected by TPO inhibition. However, it is not currently known how well invertebrate TPO orthologs relate to rat TPO, which is the target for the TPO assay in the ToxCast database.
One final biological response prioritized through the EAR analysis was PPARγ activation. At present, there is one AOP related to PPARγ activation in the AOPWiki: PPARγ activation leading to sarcomas in rodents (https://aopwiki.org/aops/163). Cancer is not generally considered an ecologically relevant endpoint, and other potential effects of PPARγ agonists in aquatic vertebrates are not well-defined. However, PPARγ is involved in energy metabolism, namely adipogenesis (fatty acid storage), and glucose metabolism,72 so putative AOPs relevant to aquatic species such as fish could be proposed. Alterations to normal, homeostatic fatty acid storage or metabolism in aquatic vertebrates may adversely result in altered energy resource storage or allocation, which could plausibly impact individual fitness, leading to decreased resources available for survival or reproduction. For the purposes of possible follow-up environmental monitoring, an obvious, easily measured KE is not currently available for PPARγ activation, so the development of AOPs—and associated biomarkers—for PPAR activation is a research priority for our team.
The three highlighted biological targets (ER, TPO, PPARγ) were prioritized since concentrations of one or more chemicals detected in the environment were near concentrations known to induce biological activity in vitro. This suggests the potential for observed effects in wildlife at impacted sites; however, the study reporting environmental concentrations of compounds across the Great Lakes was designed as a surveillance study, capturing contaminant data from many locations but with little replication at most sites.1 As such, it would be premature to interpret these results through the lens of risk assessment. The results do provide potential targeted endpoints that can be examined in more detailed follow-up monitoring to identify whether specific biological activities are actually a concern in the prioritized sites. The EAR approach will always be limited by both occurrence and effects data, thus the potential to underestimate the biological effects of complex mixtures remains. Incorporating other effects-based monitoring methods, such as targeted bioassays or ‘omics approaches, in a tiered-approach can provide a secondary means to confirm predicted effects or identify novel chemicals or biological pathways of concern that may not be captured through EAR analysis.12
To conclude, the presented case study highlights the potential of using HTS effects databases as a tool for environmental monitoring. Exposure-activity ratios provide a rapid, efficient tool for screening existing chemical monitoring data to prioritize sites, chemicals, and bioactivities of potential concern by leveraging HTS datasets to cast a wide-reaching net in terms of chemical availability and biological targets. While currently presented as a screening level assessment, the approach could be refined by including models to better characterize exposure of ecological receptors to environmental contaminants (e.g., bioaccumulation, metabolism) and to better characterize dosimetry of in vitro HTS test systems. Further refinements, along with an expected increase in HTS data sources, should only continue to increase the future utility of EAR screening for environmental monitoring.
Supplementary Material
Acknowledgments
We thank Jon Doering for providing valuable comments on a previous draft of this paper. This research was performed as part of the Chemical Safety for Sustainability (CSS) research program of the US Environmental Protection Agency (EPA) Office of Research and Development (ORD). Research was supported in part by the Great Lakes Restoration Initiative (GLRI). This manuscript has been reviewed in accordance with the requirements of EPA ORD. The views expressed in this work are those of the authors and do not necessarily reflect the views or policies of the US EPA, nor does the mention of trade names or commercial products constitute endorsement or recommendation for use.
Footnotes
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/ACS.est.xxxxx.
Additional T47D-Kbluc method details, R code used for data processing and calculations, ten tables, and two figures (PDF, XLSX)
References
- 1.Lee KE, Langer SK, Menheer MA, Hansen DS, Foreman WT, Furlong ET, Jorgenson ZG, Choy SJ, Moore JN, Banda, JoAnn, and Gefell DJ, 2015, Chemicals of emerging concern in water and bottom sediment in the Great Lakes Basin, 2012—Collection methods, analytical methods, quality assurance, and study data: U.S. Geological Survey Data Series 910, 14 p., 10.3133/ds910. [DOI] [Google Scholar]
- 2.Kolpin DW; Furlong ET; Meyer MT; Thurman EM; Zaugg SD; Barber LB; Buxton HT, Pharmaceuticals, hormones, and other organic wastewater contaminants in U.S. streams, 1999–2000: a national reconnaissance. Environ. Sci. Technol. 2002, 36, (6), 1202–11. [DOI] [PubMed] [Google Scholar]
- 3.Hug C; Ulrich N; Schulze T; Brack W; Krauss M, Identification of novel micropollutants in wastewater by a combination of suspect and nontarget screening. Environ. Pollut. 2014, 184, 25–32. [DOI] [PubMed] [Google Scholar]
- 4.Peng H; Chen C; Saunders DM; Sun J; Tang S; Codling G; Hecker M; Wiseman S; Jones PD; Li A; Rockne KJ; Giesy JP, Untargeted identification of organo-bromine compounds in lake sediments by ultrahigh-resolution mass spectrometry with the data-independent precursor isolation and characteristic fragment method. Anal. Chem. 2015, 87, (20), 10237–46. [DOI] [PubMed] [Google Scholar]
- 5.Peng H; Chen C; Cantin J; Saunders DM; Sun J; Tang S; Codling G; Hecker M; Wiseman S; Jones PD; Li A; Rockne KJ; Sturchio NC; Giesy JP, Untargeted screening and distribution of organo-bromine compounds in sediments of Lake Michigan. Environ. Sci. Technol. 2016, 50, (1), 321–30. [DOI] [PubMed] [Google Scholar]
- 6.Schymanski EL; Singer HP; Longree P; Loos M; Ruff M; Stravs MA; Ripolles Vidal C; Hollender J, Strategies to characterize polar organic contamination in wastewater: exploring the capability of high resolution mass spectrometry. Environ. Sci. Technol 2014, 48, (3), 1811–8. [DOI] [PubMed] [Google Scholar]
- 7.Judson R; Richard A; Dix DJ; Houck K; Martin M; Kavlock R; Dellarco V; Henry T; Holderman T; Sayre P; Tan S; Carpenter T; Smith E, The toxicity data landscape for environmental chemicals. Environ. Health Perspec.t 2009, 117, (5), 685–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Attene-Ramos MS; Miller N; Huang R; Michael S; Itkin M; Kavlock RJ; Austin CP; Shinn P; Simeonov A; Tice RR; Xia M, The Tox21 robotic platform for the assessment of environmental chemicals--from vision to reality. Drug Discov. Today. 2013, 18, (15–16), 716–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tice RR; Austin CP; Kavlock RJ; Bucher JR, Improving the human hazard characterization of chemicals: a Tox21 update. Environ. Health Perspect. 2013, 121, (7), 756–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dix DJ; Houck KA; Martin MT; Richard AM; Setzer RW; Kavlock RJ, The ToxCast program for prioritizing toxicity testing of environmental chemicals. Toxicol. Sci. 2007, 95, (1), 5–12. [DOI] [PubMed] [Google Scholar]
- 11.Becker RA; Friedman KP; Simon TW; Marty MS; Patlewicz G; Rowlands JC, An exposure:activity profiling method for interpreting high-throughput screening data for estrogenic activity--proof of concept. Regul. Toxicol. Pharmacol. 2015, 71, (3), 398–408. [DOI] [PubMed] [Google Scholar]
- 12.Schroeder AL; Ankley GT; Houck KA; Villeneuve DL, Environmental surveillance and monitoring-The next frontiers for high-throughput toxicology. Environ. Toxicol. Chem. 2016, 35, (3), 513–25. [DOI] [PubMed] [Google Scholar]
- 13.Li S; Villeneuve DL; Berninger JP; Blackwell BR; Cavallin JE; Hughes MN; Jensen KM; Jorgenson Z; Kahl MD; Schroeder AL; Stevens KE; Thomas LM; Weberg MA; Ankley GT, An integrated approach for identifying priority contaminant in the Great Lakes Basin - Investigations in the Lower Green Bay/Fox River and Milwaukee Estuary areas of concern. Sci. Total Environ. 2017, 579, 825–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sprague JB; Ramsay BA, Lethal levels of mixed copper–zinc solutions for juvenile salmon. J. Fish. Res. Board Can 1965, 22, (2), 425–432. [Google Scholar]
- 15.Ankley GT; Bennett RS; Erickson RJ; Hoff DJ; Hornung MW; Johnson RD; Mount DR; Nichols JW; Russom CL; Schmieder PK; Serrrano JA; Tietge JE; Villeneuve DL, Adverse outcome pathways: a conceptual framework to support ecotoxicology research and risk assessment. Environ. Toxicol. Chem 2010, 29, (3), 730–41. [DOI] [PubMed] [Google Scholar]
- 16.Ekman DR; Ankley GT; Blazer VS; Collette TW; Garcia-Reyero N; Iwanowicz LR; Jorgenson ZG; Lee KE; Mazik PM; Miller DH; Perkins EJ; Smith ET; Tietge JE; Villeneuve DL, Environmental reviews and case studies: Biological effects–based tools for monitoring impacted surface waters in the Great Lakes: A multiagency program in support of the Great Lakes Restoration Initiative. Environ. Pract 2013, 15, (04), 409–426. [Google Scholar]
- 17.Kahl MD; Villeneuve DL; Stevens K; Schroeder A; Makynen EA; LaLone CA; Jensen KM; Hughes M; Holmen BA; Eid E; Durhan EJ; Cavallin JE; Berninger J; Ankley GT, An inexpensive, temporally integrated system for monitoring occurrence and biological effects of aquatic contaminants in the field. Environ. Toxicol. Chem 2014, 33, (7), 1584–95. [DOI] [PubMed] [Google Scholar]
- 18.Zaugg SD, Smith SG, and Schroeder MP, Determination of wastewater compounds in whole water by continuous liquid–liquid extraction and capillary-column gas chromatography/mass spectrometry: U.S. Geological Survey Techniques and Methods, book 5, chap. B4, 30 p. 2006. [Google Scholar]
- 19.Zaugg SD, Phillips PJ, and Smith SG, Analysis of pharmaceutical and other organic wastewater compounds in filtered and unfiltered water samples by gas chromatography/mass spectrometry: U.S. Geological Survey Open-File Report 2013–1297, 24 p. 2014. [Google Scholar]
- 20.Foreman WT, Gray JL, ReVello RC, Lindley CE, Losche SA, and Barber LB, Determination of steroid hormones and related compounds in filtered and unfiltered water by solid-phase extraction, derivatization, and gas chromatography with tandem mass spectrometry: U.S. Geological Survey Techniques and Methods, book 5, chap. B9, 118 p. 2012. [Google Scholar]
- 21.Childress C; Foreman W; Connor B; Maloney T New reporting procedures based on long-term method detection levels and some considerations for interpretations of water-quality data provided by the U.S. Geological Survey National Water Quality Laboratory; U.S. Geological Survey Open-File Report 1999–193. 1999. [Google Scholar]
- 22.Wilson VS; Bobseine K; Gray LE Jr., Development and characterization of a cell line that stably expresses an estrogen-responsive luciferase reporter for the detection of estrogen receptor agonist and antagonists. Toxicol. Sci. 2004, 81, (1), 69–77. [DOI] [PubMed] [Google Scholar]
- 23.USEPA, ToxCast & Tox21 Summary Files from invitrodb_v2. October 2015. ed. https://www.epa.gov/chemical-research/toxicity-forecaster-toxcasttm-data
- 24.Attene-Ramos MS; Huang R; Michael S; Witt KL; Richard A; Tice RR; Simeonov A; Austin CP; Xia M, Profiling of the Tox21 chemical collection for mitochondrial function to identify compounds that acutely decrease mitochondrial membrane potential. Environ. Health Perspect. 2015, 123, (1), 49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giuliano KA; Gough AH; Taylor DL; Vernetti LA; Johnston PA, Early safety assessment using cellular systems biology yields insights into mechanisms of action. J. Biomol. Screen. 2010, 15, (7), 783–97. [DOI] [PubMed] [Google Scholar]
- 26.Houck KA; Dix DJ; Judson RS; Kavlock RJ; Yang J; Berg EL, Profiling bioactivity of the ToxCast chemical library using BioMAP primary human cell systems. J. Biomol. Screen. 2009, 14, (9), 1054–66. [DOI] [PubMed] [Google Scholar]
- 27.Huang R; Xia M; Cho MH; Sakamuru S; Shinn P; Houck KA; Dix DJ; Judson RS; Witt KL; Kavlock RJ; Tice RR; Austin CP, Chemical genomics profiling of environmental chemical modulation of human nuclear receptors. Environ. Health Perspect. 2011, 119, (8), 1142–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang R; Sakamuru S; Martin MT; Reif DM; Judson RS; Houck KA; Casey W; Hsieh JH; Shockley KR; Ceger P; Fostel J; Witt KL; Tong W; Rotroff DM; Zhao T; Shinn P; Simeonov A; Dix DJ; Austin CP; Kavlock RJ; Tice RR; Xia M, Profiling of the Tox21 10K compound library for agonists and antagonists of the estrogen receptor alpha signaling pathway. Sci. Rep. 2014, 4, 5664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karmaus AL; Toole CM; Filer DL; Lewis KC; Martin MT, High-throughput screening of chemical effects on steroidogenesis using H295R human adrenocortical carcinoma cells. Toxicol. Sci 2016, 150, (2), 323–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kleinstreuer NC; Yang J; Berg EL; Knudsen TB; Richard AM; Martin MT; Reif DM; Judson RS; Polokoff M; Dix DJ; Kavlock RJ; Houck KA, Phenotypic screening of the ToxCast chemical library to classify toxic and therapeutic mechanisms. Nat. Biotechnol 2014, 32, (6), 583–91. [DOI] [PubMed] [Google Scholar]
- 31.Knudsen TB; Houck KA; Sipes NS; Singh AV; Judson RS; Martin MT; Weissman A; Kleinstreuer NC; Mortensen HM; Reif DM; Rabinowitz JR; Setzer RW; Richard AM; Dix DJ; Kavlock RJ, Activity profiles of 309 ToxCast chemicals evaluated across 292 biochemical targets. Toxicology 2011, 282, (1–2), 1–15. [DOI] [PubMed] [Google Scholar]
- 32.MacDonald ML; Lamerdin J; Owens S; Keon BH; Bilter GK; Shang Z; Huang Z; Yu H; Dias J; Minami T; Michnick SW; Westwick JK, Identifying off-target effects and hidden phenotypes of drugs in human cells. Nat. Chem. Biol. 2006, 2, (6), 329–37. [DOI] [PubMed] [Google Scholar]
- 33.Martin MT; Dix DJ; Judson RS; Kavlock RJ; Reif DM; Richard AM; Rotroff DM; Romanov S; Medvedev A; Poltoratskaya N; Gambarian M; Moeser M; Makarov SS; Houck KA, Impact of environmental chemicals on key transcription regulators and correlation to toxicity end points within EPA’s ToxCast program. Chem. Res. Toxicol. 2010, 23, (3), 578–90. [DOI] [PubMed] [Google Scholar]
- 34.Padilla S; Corum D; Padnos B; Hunter DL; Beam A; Houck KA; Sipes N; Kleinstreuer N; Knudsen T; Dix DJ; Reif DM, Zebrafish developmental screening of the ToxCast Phase I chemical library. Reprod. Toxicol 2012, 33, (2), 174–87. [DOI] [PubMed] [Google Scholar]
- 35.Paul KB; Hedge JM; Rotroff DM; Hornung MW; Crofton KM; Simmons SO, Development of a thyroperoxidase inhibition assay for high-throughput screening. Chem. Res. Toxicol 2014, 27, (3), 387–99. [DOI] [PubMed] [Google Scholar]
- 36.Romanov S; Medvedev A; Gambarian M; Poltoratskaya N; Moeser M; Medvedeva L; Gambarian M; Diatchenko L; Makarov S, Homogeneous reporter system enables quantitative functional assessment of multiple transcription factors. Nat. Methods 2008, 5, (3), 253–60. [DOI] [PubMed] [Google Scholar]
- 37.Rotroff DM; Beam AL; Dix DJ; Farmer A; Freeman KM; Houck KA; Judson RS; LeCluyse EL; Martin MT; Reif DM; Ferguson SS, Xenobiotic-metabolizing enzyme and transporter gene expression in primary cultures of human hepatocytes modulated by ToxCast chemicals. J. Toxicol. Environ. Health B Crit. Rev 2010, 13, (2–4), 329–46. [DOI] [PubMed] [Google Scholar]
- 38.Rotroff DM; Dix DJ; Houck KA; Kavlock RJ; Knudsen TB; Martin MT; Reif DM; Richard AM; Sipes NS; Abassi YA; Jin C; Stampfl M; Judson RS, Real-time growth kinetics measuring hormone mimicry for ToxCast chemicals in T-47D human ductal carcinoma cells. Chem. Res. Toxicol 2013, 26, (7), 1097–107. [DOI] [PubMed] [Google Scholar]
- 39.Sipes NS; Martin MT; Kothiya P; Reif DM; Judson RS; Richard AM; Houck KA; Dix DJ; Kavlock RJ; Knudsen TB, Profiling 976 ToxCast chemicals across 331 enzymatic and receptor signaling assays. Chem. Res. Toxicol 2013, 26, (6), 878–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stossi F; Bolt MJ; Ashcroft FJ; Lamerdin JE; Melnick JS; Powell RT; Dandekar RD; Mancini MG; Walker CL; Westwick JK; Mancini MA, Defining estrogenic mechanisms of bisphenol A analogs through high throughput microscopy-based contextual assays. Chemi. Biol 2014, 21, (6), 743–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Truong L; Reif DM; St Mary L; Geier MC; Truong HD; Tanguay RL, Multidimensional in vivo hazard assessment using zebrafish. Toxicol. Sci 2014, 137, (1), 212–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Filer DL; Kothiya P; Setzer RW; Judson RS; Martin MT, tcpl: The ToxCast pipeline for high-throughput screening data. Bioinformatics 2017, 33, (4), 618–20. [DOI] [PubMed] [Google Scholar]
- 43.Putzrath RM, Estimating relative potency for receptor-mediated toxicity: reevaluating the toxicity equivalence factor (TEF) model. Regul. Toxicol. Pharmacol. 1997, 25, (1), 68–78. [DOI] [PubMed] [Google Scholar]
- 44.Kavlock R; Chandler K; Houck K; Hunter S; Judson R; Kleinstreuer N; Knudsen T; Martin M; Padilla S; Reif D; Richard A; Rotroff D; Sipes N; Dix D, Update on EPA’s ToxCast program: providing high throughput decision support tools for chemical risk management. Chem. Res. Toxicol 2012, 25, (7), 1287–302. [DOI] [PubMed] [Google Scholar]
- 45.Richard AM; Judson RS; Houck KA; Grulke CM; Volarath P; Thillainadarajah I; Yang C; Rathman J; Martin MT; Wambaugh JF; Knudsen TB; Kancherla J; Mansouri K; Patlewicz G; Williams AJ; Little SB; Crofton KM; Thomas RS, ToxCast chemical landscape: Paving the road to 21st Century toxicology. Chem. Res. Toxicol 2016, 29, (8), 1225–51. [DOI] [PubMed] [Google Scholar]
- 46.Dickinson JH; Smith GG, A new and practical oral contraceptive agent: norethindrone with mestranol. Can. Medi. Assoc. J 1963, 89, 242–5. [PMC free article] [PubMed] [Google Scholar]
- 47.Blackwell BR; Brown TR; Broadway PR; Buser MD; Brooks JC; Johnson BJ; Cobb GP; Smith PN, Characterization of trenbolone acetate and estradiol metabolite excretion profiles in implanted steers. Environ. Toxicol. Chem 2014, 33, (12), 2850–8. [DOI] [PubMed] [Google Scholar]
- 48.Hanselman TA; Graetz DA; Wilkie AC, Manure-borne estrogens as potential environmental contaminants: A review. Environ. Sci. Technol 2003, 37, (24), 5471–5478. [DOI] [PubMed] [Google Scholar]
- 49.Raun AP; Preston RL, History of diethylstilbestrol use in cattle. J. Anim. Sci 2002, 80, 1–7. [Google Scholar]
- 50.Krishnan AV; Stathis P; Permuth SF; Tokes L; Feldman D, Bisphenol-A: an estrogenic substance is released from polycarbonate flasks during autoclaving. Endocrinology 1993, 132, (6), 2279–86. [DOI] [PubMed] [Google Scholar]
- 51.Kachaylo EM; Pustylnyak VO; Lyakhovich VV; Gulyaeva LF, Constitutive androstane receptor (CAR) is a xenosensor and target for therapy. Biokhim., Biochem. (Mosc.) 2011, 76, (10), 1087–97. [DOI] [PubMed] [Google Scholar]
- 52.Pillai HK; Fang M; Beglov D; Kozakov D; Vajda S; Stapleton HM; Webster TF; Schlezinger JJ, Ligand binding and activation of PPARgamma by Firemaster(R) 550: effects on adipogenesis and osteogenesis in vitro. Environ. Health Perspect 2014, 122, (11), 1225–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fang M; Webster TF; Ferguson PL; Stapleton HM, Characterizing the peroxisome proliferator-activated receptor (PPARgamma) ligand binding potential of several major flame retardants, their metabolites, and chemical mixtures in house dust. Environ. Health Perspect 2015, 123, (2), 166–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miller WL, Molecular biology of steroid hormone synthesis. Endocr. Rev 1988, 9, (3), 295–318. [DOI] [PubMed] [Google Scholar]
- 55.Judson RS; Magpantay FM; Chickarmane V; Haskell C; Tania N; Taylor J; Xia M; Huang R; Rotroff DM; Filer DL; Houck KA; Martin MT; Sipes N; Richard AM; Mansouri K; Setzer RW; Knudsen TB; Crofton KM; Thomas RS, Integrated model of chemical perturbations of a biological pathway using 18 in vitro high-throughput screening assays for the estrogen receptor. Toxicol. Sci 2015, 148, (1), 137–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kleinstreuer NC; Ceger P; Watt ED; Martin M; Houck K; Browne P; Thomas RS; Casey WM; Dix DJ; Allen D; Sakamuru S; Xia M; Huang R; Judson R, Development and validation of a computational model for androgen receptor activity. Chem. Res. Toxicol 2017, 30, (4), 946–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Paul Friedman K; Watt ED; Hornung MW; Hedge JM; Judson RS; Crofton KM; Houck KA; Simmons SO, Tiered high-throughput screening approach to identify thyroperoxidase inhibitors within the ToxCast Phase I and II chemical libraries. Toxicol. Sci. 2016, 151, (1), 160–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Davis JM; Ekman DR; Teng Q; Ankley GT; Berninger JP; Cavallin JE; Jensen KM; Kahl MD; Schroeder AL; Villeneuve DL; Jorgenson ZG; Lee KE; Collette TW, Linking field-based metabolomics and chemical analyses to prioritize contaminants of emerging concern in the Great Lakes basin. Environ. Toxicol. Chem. 2016, 35, (10), 2493–502. [DOI] [PubMed] [Google Scholar]
- 59.Cavallin JE; Jensen KM; Kahl MD; Villeneuve DL; Lee KE; Schroeder AL; Mayasich J; Eid EP; Nelson KR; Milsk RY; Blackwell BR; Berninger JP; LaLone CA; Blanksma C; Jicha T; Elonen C; Johnson R; Ankley GT, Pathway-based approaches for assessment of real-time exposure to an estrogenic wastewater treatment plant effluent on fathead minnow reproduction. Environ. Toxicol. Chem. 2016, 35, (3), 702–16. [DOI] [PubMed] [Google Scholar]
- 60.Kramer VJ; Miles-Richardson S; Pierens SL; Giesy JP, Reproductive impairment and induction of alkaline-labile phosphate, a biomarker of estrogen exposure, in fathead minnows (Pimephales promelas) exposed to waterborne 17 beta-estradiol. Aquatic Toxicology 1998, 40, (4), 335–360. [Google Scholar]
- 61.Routledge EJ; Sheahan D; Desbrow C; Brighty GC; Waldock M; Sumpter JP, Identification of estrogenic chemicals in STW effluent. 2. In vivo responses in trout and roach. Environ. Sci. Technol. 1998, 32, (11), 1559–1565. [Google Scholar]
- 62.Woodling JD; Lopez EM; Maldonado TA; Norris DO; Vajda AM, Intersex and other reproductive disruption of fish in wastewater effluent dominated Colorado streams. Comp Biochem Physiol C Toxicol Pharmacol : CBP 2006, 144, (1), 10–5. [DOI] [PubMed] [Google Scholar]
- 63.Vajda AM; Barber LB; Gray JL; Lopez EM; Woodling JD; Norris DO, Reproductive disruption in fish downstream from an estrogenic wastewater effluent. Environ. Sci. Technol 2008, 42, (9), 3407–14. [DOI] [PubMed] [Google Scholar]
- 64.Eick GN; Thornton JW, Evolution of steroid receptors from an estrogen-sensitive ancestral receptor. Mol. Cell Endocrinol 2011, 334, (1–2), 31–8. [DOI] [PubMed] [Google Scholar]
- 65.Tietge JE; Degitz SJ; Haselman JT; Butterworth BC; Korte JJ; Kosian PA; Lindberg-Livingston AJ; Burgess EM; Blackshear PE; Hornung MW, Inhibition of the thyroid hormone pathway in Xenopus laevis by 2-mercaptobenzothiazole. Aquat. Toxicol 2013, 126, 128–36. [DOI] [PubMed] [Google Scholar]
- 66.Nelson KR; Schroeder AL; Ankley GT; Blackwell BR; Blanksma C; Degitz SJ; Flynn KM; Jensen KM; Johnson RD; Kahl MD; Knapen D; Kosian PA; Milsk RY; Randolph EC; Saari T; Stinckens E; Vergauwen L; Villeneuve DL, Impaired anterior swim bladder inflation following exposure to the thyroid peroxidase inhibitor 2-mercaptobenzothiazole part I: Fathead minnow. Aquat. Toxicol 2016, 173, 192–203. [DOI] [PubMed] [Google Scholar]
- 67.Hornung MW; Kosian PA; Haselman JT; Korte JJ; Challis K; Macherla C; Nevalainen E; Degitz SJ, In Vitro, Ex Vivo, and In Vivo determination of thyroid hormone modulating activity of benzothiazoles. Toxicol. Sci 2015, 146, (2), 254–64. [DOI] [PubMed] [Google Scholar]
- 68.Brar NK; Waggoner C; Reyes JA; Fairey R; Kelley KM, Evidence for thyroid endocrine disruption in wild fish in San Francisco Bay, California, USA. Relationships to contaminant exposures. Aquat. Toxicol. 2010, 96, (3), 203–15. [DOI] [PubMed] [Google Scholar]
- 69.Simmons DB; McMaster ME; Reiner EJ; Hewitt LM; Parrott JL; Park BJ; Brown SB; Sherry JP, Wild fish from the Bay of Quinte Area of Concern contain elevated tissue concentrations of PCBs and exhibit evidence of endocrine-related health effects. Environ. Int 2014, 66, 124–37. [DOI] [PubMed] [Google Scholar]
- 70.Couderc M; Marchand J; Zalouk-Vergnoux A; Kamari A; Moreau B; Blanchet-Letrouve I; Le Bizec B; Mouneyrac C; Poirier L, Thyroid endocrine status of wild European eels (Anguilla anguilla) in the Loire (France). Relationships with organic contaminant body burdens. Sci. Total Environ 2016, 550, 391–405. [DOI] [PubMed] [Google Scholar]
- 71.Bonett RM, Analyzing endocrine system conservation and evolution. Gen. Comp. Endocrinol. 2016, 234, 3–9. [DOI] [PubMed] [Google Scholar]
- 72.Kersten S; Desvergne B; Wahli W, Roles of PPARs in health and disease. Nature 2000, 405, (6785), 421–4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.