Abstract
Biobanks play an increasing role in contemporary research projects. These units meet all requirements to regard them as a one of the most innovative and up-to-date in the field of biomedical research. They enable conducting wide-scale research by the professional collection of biological specimens and correlated clinical data. Pathology units may be perceived roots of biobanking. The review aims at describing the concept of biobanks, their model of function and scientific potential. It comprises the division of biobanks, sample preservation methods and IT solutions as well as guidelines and recommendations for management of a vast number of biological samples and clinical data. Therefore, appropriate standard operating procedures and protocols are outlined. Constant individualization of diagnostic process and treatment procedures creates the niche for translational units. Thus, the role of biobanks in personalized medicine was also specified. The exceptionality of biobanks poses some new ethical-legal issues which have various solutions, in each legal system, amongst the world. Finally, distribution and activity of European biobanks are mentioned.
Keywords: Biobank, Personalized medicine, Biorepository, Biospecimen, Tissue Banking
Introduction - Concept of Biobanks
Immense development in the field of biomedical research multiplies challenge in the eternal problem of obtaining, preserving and analyzing human samples. Ever since the first experiment had been conducted, a problem of preserving samples for future purposes appeared. Furthermore, the possibility of harvesting biological material during routine activities of medical and biological professionals was the trigger to manage it in an organized manner. Hospital pathology units are limited in storing a large number of preserved samples. Also, they are primarily designed to diagnose the obtained specimens, what implies restricted samples’ legal status (analyses limited to diagnostic purposes, not for research) and certain collection method. Thus, came up a niche for a novel solution – biobanks (BBs).
Definition and Genesis of Biobanks
Wide heterogeneity and still developing central control over such initiatives causes difficulty to give one and sufficient definition. Principally, biobanks are professional repositories of biologic samples. Activities in the scope of a biobank include: storing specimens harvested over time, combined with clinical, epidemiological and general source data. Moreover, collected samples are processed, primarily analyzed and adequately preserved for sharing with the wider scientific community. They also deliver details about the acquisition, handling and storage of each sample – e.g. ischemia time, preservation method, shipping conditions. Summary of definitions of human biobanks and relevant terms are well described by Fransson et al. [1]
Biobank workflow is maintained in a strictly organized manner. Standard operating procedures (SOPs) ensure correct implementation of essential biobanking components (samples’ donors’ anonymization, samples: acquisition, transport, preparation and analysis process faultlessness, proper storage conditions and terms of samples’ sharing (e.g. local/international law)). For these reasons, biobanks combine a wide variety of data mainly for research purposes.
One of the very first attempt to create a detached unit responsible for human specimen storage has begun with Framingham Heart Study (FHS) established to collect blood samples and patient data since 1948 [2]. Construction of the study created a unique opportunity to combine biological data obtained via blood analysis with clinical examinations and lifestyle interviews to elicit risk factors for cardiovascular disorders. However, the beginning of biospecimen repositories may be counted since hospital pathology units started to store harvested samples. So that, the history of BBs is longer than it appears [3].
Types of Biobanks
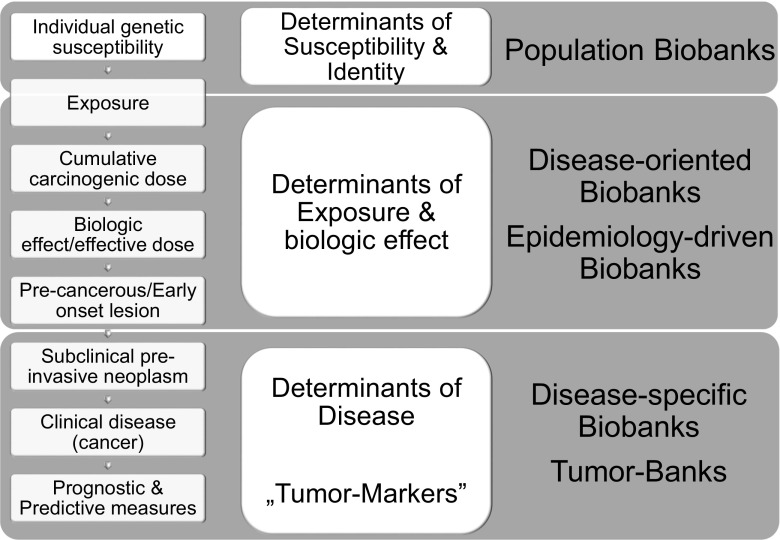
The need for correlating a wide variety of data gave rise to particular types of biorepositories, namely: population-based, disease-oriented, and tissue biobanks (See Fig. 1) [1, 4–6].
Fig. 1.
Types of biobanks based on type of biomarker research. The different stage of the natural history of a disease (left column) defines types of biomarkers that can be utilized (second column). Finally, biobank’s type originates from the biomarker classification (right column). [4, 5]
Population-based units comprise samples and epidemiological/clinical data collected from volunteers without specific inclusion or exclusion criteria – material aims to mirror the status of the general population [7]. One of the best example of this type is population DNA biobank, which was established in order to examine human genome within PHG (Public Health Genomics) projects like HuGENet™, Estonian Biobank in Estonian Genome Center, CARTaGENE [8–10]. For instance, FarGen Project aims at sequencing whole genomes of Faroese residents to obtain more thorough public health data (http://www.fargen.fo, date of access: 27.07.2017). Mentioned BBs not only collect DNA or molecular data, but also combine various clinical data (physical examination, demographic, laboratory test results, miscellaneous questionnaire data etc.). The results of the first human genome sequencing enabled the development of next generation sequencing techniques (NGS). It has enormous influence on biobanks since it decreases the cost and time of analysis per sample and produces complete information about each patient’s genome, epigenome, transcriptome and even more. Samples of specific tissue types are the source of data about the public health status. Cervical cytology biobanks are an example of such units [11].
Many biobanks are established in relation with particular scientific projects. “Children of the 90s” study led to collecting a great amount of placenta tissue from 14,541 pregnant women between 1990 and 1992 in South West England [12]. Population-based biobanks are the source of samples for many research groups. The strength and importance of such studies are enormous thanks to the large-scale character [13–16].
Setting inclusion or exclusion criteria during collection of samples leads to disease-oriented biobanks (or disease-oriented biobanks for epidemiology). The aim is to determine specific exposure factors for a disease by the careful collection of specimen from a patient, often comprising various samples from the subject. For instance, HiWATE (Health impacts of long-term exposure to disinfection byproducts in drinking water) project focuses on determining risks for human health that are brought about byproducts in drinking water. Studies require a wide range of specimens (e.g. semen, biomarkers in the blood) [4–6, 11, 17, 18]. In the EPIC (European Prospective Investigation into Cancer and Nutrition) associated studies BBs play the role as a source of data for e.g. IGF-I role in ovarian cancer development, fish consumption and mortality and lifestyle influence on breast cancer risk [5, 19–21]. BBs also solve a problem of gathering a significant number of samples from the patients with rare diseases. MRC Centre Biobank for Neuromuscular Diseases in Newcastle and London contributed to diagnostics, basic science research, industry, drug development, and therapy of neuromuscular diseases [22]. Disease-oriented biobanks gather also materials collected from patients suffering from infectious diseases. The UCSF ASB (AIDS Specimen Biobank) collects peripheral blood mononuclear cells (PBMC) that contributed to the discovery of factors that cause AIDS and Kaposi’s sarcoma [23]. Recent Ebola outbreak in Africa led WHO to establish new initiative-Ebola Biobank. 100,000 samples of blood, semen, urine and breast milk from confirmed and suspected Ebola patients will be stored and analyzed to increase understanding and predicting Ebola outbreaks [24].
Finally, the tissue biobanks, known also as a tumor banks, aim at exploring the biology of a particular sample by collecting and comparing unaffected tissues and the neoplastic one. Generally, the main purpose is to deepen the knowledge about the molecular basis of the disease and/or determine new applicable biomarkers of an examined disorder. These projects require isolation of intracellular particles such as DNA, RNA and proteins. Ultimately, obtained biological data from high-throughput analyses are combined and correlated with clinical data [4, 5]. To exemplify, following projects are worth mentioning: European Human Tumor Frozen Tissue Bank, the National Cancer Institute Office of Biorepositories and Biospecimen Research, the Canadian Tumor Repository Network (CTRNet) [4, 5].
Basic Components of Biobank
European Commission Joint Research Centre (EC-JRC) provides the community with general feature of biobanks such as [6, 11]:
collection and storage of biological materials combined with medical, epidemiological data,
dynamic development of the biobanks - continuous collection in a long-term prospect,
association with an ongoing research project,
application of specimens’ anonymization for the donors’ privacy sake,
implementation of governance standards and procedures.
Given the exact purpose of a biobank establishment, its characteristics vary for the best suitability to a project (e.g. tissue type, a target of the study).
A combination of professional biological storage solutions, novel bioinformatics data processing systems and firm governance compose a model of biobank for multiple purposes (see Fig. 2). Samples from the model BB are prepared for basic research, drug development and other yet unplanned research on the date of samples collection.
Fig. 2.
Samples Analysis and Preservation Methods
Sample acquisition process starts with obtaining informed consent from the patient (see more 3.3 Ethical and legal issues). Sample’s cycle in biobank starts with harvesting the sample during medical procedures. In the case of tumor biobanks, the research purposes are not allowed to interfere with diagnostic pathway (see Tab 2. MMI). Thus, involving a pathologist into the acquisition of specimen for biobanking may perfectly combine both diagnostic purposes with a proper biobanking harvest (avoiding tumor margin, necrosis, blood clots, irrelevant surrounding tissue). Care must be taken to place the sample, in a sterile manner, in the vial with appropriate media/buffers depending on planned analysis (see Tab. 1). Detached and/or thoroughly-trained staff is recommended. Samples should be shipped under strictly determined and monitored conditions (e.g. temperature, warm/cold ischemia time, maximum transport time). After the arrival at the Biobank unit, a sample is anonymized, labeled and handled to storage/analysis unit. Here, a crucial step – aliquoting is implemented. It comprises the division of sample for a smaller portion which decreases freeze/thaw cycles of a sample of a patient, creates a backup, eases the sharing of samples without thawing. In the UK Biobank, the standard aliquoting procedure for blood and urine from each patient produces 19 aliquots from 5 primary samples [55]. After completing survey data acquisition, the record is ready to be used in a project or shared. This multi-step process requires standardization to keep reproducibility. For this reason, Standard Operating Procedures (SOPs) were established to maintain standardized and replicable protocols regarding every step of sample handling. Molecular Medicine Ireland has published “Guidelines for Standardized Biobanking” [26]. The document outlines SOPs in detail. The authors divided SOPs into 3 sections:
pre-clinical SOPs: Assessment of the Research Participant, Safety Guidelines, Specimen Identification and Labeling,
clinical SOPs: Blood Collection – Venipuncture, Saliva Collection, Urine Collection, Faeces Collection, Buccal Collection, Bronchoalveolar Lavage Collection using Bronchoscopy,
laboratory SOPs: Personal Protective Equipment, Specimen Receipt, Preparation of Serum and Plasma from Blood, DNA Extraction from Blood, RNA Extraction from Blood, Protein Extraction from Blood, Peripheral Blood Mononuclear Cell isolation from Blood, Processing of Urine, Processing of Buccal Swabs for DNA Extraction, Processing of Faeces for DNA extraction, Processing of Tissue Samples, Processing of Cultured Cells, Processing of Bronchoalveolar Lavage [26].
Table 2.
List of crucial guidelines concerning biobanking issues
| Institution | Document | Content | Year | Source (date of access: 27.07.2017) |
|---|---|---|---|---|
| National Cancer Institute | NCI Best Practices for Biospecimen Resources | A. Scope, applicability, and implementation B. Technical and operational best practices C. Ethical, legal, and policy best practices |
2016 | https://biospecimens.cancer.gov/bestpractices/2016-NCIBestPractices.pdf |
| International Society for Biological and Environmental Repositories | Best Practices for Repositories: Collection, Storage, Retrieval and Distribution of Biological Materials for Research | Repository planning considerations, facilities, storage equipment and environments, quality management, safety, training, records management, cost management, biological material tracking, packaging and shipping, specimen collection, processing and retrieval, legal and ethical issues for biospecimens, specimen access, utilization and destruction | 2012 | http://c.ymcdn.com/sites/www.isber.org/resource/resmgr/Files/ISBER_Best_Practices_3rd_Edi.pdf |
| Molecular Medicine Ireland (MMI) | MMI Guidelines for Standardised Biobanking |
Part I: Pre-clinical standard operating procedures Part II: Clinical standard operating procedures Part III: Laboratory standard operating procedures |
2010 | http://www.molecularmedicineireland.ie/wp-content/uploads/2015/09/MMIGuidelinesforStandardisedBiobanking_FINAL160610.pdf |
| Organization for Economic Cooperation & Development | OECD Guidelines on Human Biobanks and Genetic Research Databases |
Part I. Guidelines on Human Biobanks and Genetic Research Databases Part II. Annotations |
2009 | https://www.oecd.org/sti/biotech/44054609.pdf |
Table 1.
Overview of tissue preservation and storage methods for various scientific purpose
| Tissue | Target | Preservation methods (e.g. medium/buffer/vial type/kit) | Short term storage/transport conditions | Long term storage conditions | Special notes | Literature |
|---|---|---|---|---|---|---|
| Blood | DNA | EDTA, whole blood or serum | 4 °C or on wet ice, within 24 h | −80 °C for years | – | [26–28] |
| RNA | Paxgene®/Tempus™ tubes | 4 °C or on wet ice, within 24 h | −80 °C for years | Blood in Tempus™ tubes may be stored up to 6 years in −80 °C. | [26, 29] | |
| Proteomics | Plasma separating tube with heparin and serum separation tube with heparin and Plain tube | 4 °C or on wet ice, within 24 h | Plasma and RBCs in −80 °C | Storage of RBCs is recommended at −80 °C for cell membrane proteomics research. | [26–28, 30, 31] | |
| Biochemistry | Plasma separating tube with heparin and serum separation tub and Plain tube with heparin | 4 °C or on wet ice, within 24 h | Plasma, immediate analysis or − 80 °C for years | – | [26] | |
| PBMC (Peripheral Blood Mononuclear Cells) | acid citrate dextrose, BD CPT™, LeukoSep™ | RT, within 24 h | −80 °C or preferred LN2 with a cryopreservative | – | [31, 32] | |
| Circulating Tumor Cells | Cell-Free DNA™ BCT® tube | for at least 4 days at RT | −80 °C or preferred LN2 with a cryopreservative | – | [33, 34] | |
| Cell-free DNA | Streck® Cell-Free DNA™ Blood Collection Tubes | up to 7 days at ambient temperature, avoid shipping/storage in 4 °C | immediate extraction or − 80 °C for years | Plasma is preferable to serum, cell lysis should be avoided to prevent increase of unspecific cfDNA level. Spike-in should be considered. | [35–39] | |
| Circulating non-coding RNA | Plasma or plasma separating tube | 4 °C or on wet ice, within 24 h | Plasma samples should be frozen immediately at −80 °C (stability) up to 1 year | Archival samples may be used | [40–42] | |
| Exosomes | Plasma or plasma separating tube | 4 °C or on wet ice, within 24 h | −80 °C for years | Special equipment needed for exosomes isolation: ultracentrifuge/chromatographs/filters. | [43, 44] | |
| Platelets (e.g. tumor-educated platelets) | EDTA | 4 °C or on wet ice, within 48 h | Obtain the platelet-rich plasma with series of centrifugation, process within 48 h | The platelet pellet collected on RNAlater® and frozen at −80 °C; plasma stored directly at −80 °C. | [45] | |
| Metabolomics | Plasma separating tube with heparin and serum separation tube and Plain tube | 4 °C or on wet ice, within 24 h | −80 °C for years | – | [26] CEN/TS 16945:2016 | |
| Tumor | DNA | Tumor parts: snap freeze in cooled isopentane | Preserve within an hour from excision/biopsy; transport in closed, sterile container on ice at 4 °C before preservation | −80 °C or liquid nitrogen | – | [26, 46] |
| formalin-fixed, paraffin-embedded (FFPE) | Store at RT, embed within 72 h | RT in dry conditions for years | In case of DNA extracted from FFPE samples - special NGS library preparation needed due to degraded and shredded template). | [47, 48] | ||
| RNA | RNAlater™ - RNA preservation | Preserve within an hour from excision/biopsy; if RNA preservative added – transport within 72 h when stored at 4 °C | Discard preservation medium and store at −80 °C | May be also retrieved from FFPE samples and analyzed with NGS. | [26, 49, 50] | |
| Protein | Tumor part snap freeze in cooled isopentane | Preserve within an hour from excision/biopsy; transport in closed, sterile container on ice at 4 °C before preservation | store at −80 °C | – | [26] | |
| Microscopic morphology | FFPE – 10% buffered formalin | Store at RT, embed within 72 h | RT in dry conditions for years | Samples preserved in RNALater may also serve for microscopic morphology analysis purposes | [26, 50] | |
| OCT (Optimal Cutting Temperature medium)-embedded -snap frozen | Preserve within an hour from excision/biopsy; transport in closed, sterile container on ice at 4 °C before preservation | −80 °C for years | ||||
| Cancer cells for culture | Before cells isolation – culture medium with or without fetal bovine serum (FBS) | 4 °C or on wet ice, within 24 h | LN2 with a cryopreservative or implant in immunodeficient mouse. | Immediate dissociation of tissue is required – enzymatic dissociation (or chemical or mechanical); Specific isolation methods for different tissue [51]. Population of cancer stem cells may be separated. Different culture media additives are required for proper culturing (Table 4 in [52]). | [51–53] | |
| Cervical cytology | DNA, RNA, protein, cells | Liquid biopsies in Thinprep (TP) containing 20 ml PreservCyt | 1–4 weeks RT | −25 °C | – | [54] |
| Urine | Metabolic products, DNA, RNA, protein | e.g. 9 ml in the vacutainer system | 4 °C or on wet ice, within 24 h | Direct storage in −80 °C or immediate analysis | – | [55, 56] CEN/TS 16945:2016 |
| Semen | Semen analysis, DNA, RNA, protein | Sterile container | Immediate analysis: heating chamber 37 °C; for storage - 4 °C or on wet ice, within 24 h | −80 °C or preferred LN2 with a cryopreservative | Semen analysis - within few hours after acquisition | [57, 58] |
| Stool | DNA, RNA, Proteome, Microbiome | Sterile container or Genotek tubes for DNA analyses | RT, within 24 h | Direct storage in −80 °C or immediate extraction | – | [26, 59] |
| Saliva | DNA, biomarkers | Sterile container or collection kit | RT, within 24 h | −80 °C for years | Exceptionally increased value of specimen if provided full record of oral diseases. | [26, 60] |
| Breast milk | Trace of organic pollutants (POP), biochemical composition analysis | Sterile, clean bottles, e.g. with Teflon coating | 4 °C, for no longer than 72 h | at −20 °C, for longer periods. Freezing expressed human milk is safe for at least 3 months. | If the sample cannot be refrigerated, you can add a small tablet of potassium dichromate (POP analysis, document available at: http://www.ccbasilea-crestocolmo.org.uy/wp-content/uploads/2010/11/ing1.final_.SOP-POP-Regional-Sampling-Breast-Milk-1.pdf date of access: 27.07.2017). | [61, 62] |
| Nail and hair | Metal trace, DNA, brominated and organophosphate flame retardants exposure, effects of cosmetic products | Nails should be clipped after a few weeks after recent clipping Clean, labeled envelope | room temperature in the driest condition possible | Like short term, may be stored frozen at −20 °C for long term | Limited usefulness, questionable value, noninvasive collection, status of medium/long term exposure. | [55, 63, 64] |
RT – room temperature; LN2 – liguid nitrogen;
The wide variety of collected human tissue samples requires adequate preparation and preservation procedures for the least quality loss during transportation and storage (see Tab. 1). Even temporary storage in 0 or 4 °C exposes samples to evaporation [65]. Another often neglected issue is storage container selection, when choosing the wrong polypropylene tube type may decrease the yield of protein extraction [66]. The other essential step of pre-analytical samples’ management is to implement a replicable, precise and errorless aliquoting workflow. Except manual techniques combined with trained staff and SOPs, there are novel automated robots e.g. The Sample Array Tube Handler (Thermo Scientific, San José, CA, USA) [67]. Such solution decreases the amount of sample needed and its loss, limits human-dependent pre-analytical errors and simplifies identification and storage system. The other component of a proper identification system is labeling. There are numerous solutions like 1D, 2D barcodes, tubes with laser-etched barcodes or radio-frequency identification based labels (RFID) [68]. Regardless of labeling system type – it must be unique for each sample and its aliquot. It also should be compatible with IT system o f a Biobank to ensure fast access to the sample (decreasing thawing of other samples while searching). Unifying labeling with SPREC results in a straightforward and sharable database of a Biobank collection [69].
IT Systems
An efficient bioinformatics in biobanks is a crucial matter. IT systems have to serve databases in a real-time, easy access and user-friendly manner. Additionally, privacy protection and anonymization components cannot be neglected (see 3.5. Ethical and legal issues subchapter). The enormous number of digital, sensitive data requires optimized both hardware and software. There has been developed international standards for data obtaining and processing to enable compatible data sharing. International organizations such as NCI, ISBER, caHUB, OECD, EC-JRC are providers of recommendation useful in harmonizing every branch of BBs’ functionality [70]. But for the neatness, a consistent database should meet the standards for the international compatibility. The issue is essential in any case of cooperation, which increases the extent and significance of a project and brings profits for the shareholders. Therefore, implementing common standards and participation in international BBs’ networks is a necessity.
Standards implementation regards every step of any information processing either biological or epidemiological. For maintaining a predictable and restorable sample collection Standard Preanalytical Coding for Biospecimens (SPREC) is recommended. This coding system enables biorepositories to describe procedures and status of just obtained sample. This simple coding has an international clear interpretation and its widespread encourages biobankers to utilize the system [71].
Compilation of SPREC, a list of data to be collected (such as BRISQ - Biospecimen Reporting for Improved Study Quality [72], NCRI recommendations) and SOPs (https://www.ctrnet.ca/operating-procedures, date of access: 27.07.2017) creates a complement and worldwide compatible system. Moreover, diligent planning and constant evolution of IT systems ensure the right maintenance of both data and samples collection along with keeping wide international collaborative capabilities (Tables 1 and 2).
Record Constitution
Numerous partners provide ready-to-implement sets of data, distinguishing every piece of information is helpful for further research. NCRI has published such document, the sets of tables comprise the list, which divides information about the patient and collected samples. Each table is extended with a list of particular records. Document available at: (http://ccb.ncri.org.uk/wp-content/uploads/2014/03/CCB-Data-Standard-v1.pdf; date of access: 27.07.2017). Also, BRISQ has a resourceful and worldwide compatible protocol for preparation a novel and up-to-date database in biobank [72].
BBMRI (Biobanking and Biomolecular Resources Research Infrastructure) created in 2016 MIABIS 2.0 (Minimum Information About BIobank data Sharing) – comprehensive and detailed document describing guidelines for sharing data between biobanks within networks. It standardizes minimum information required to initiate collaboration which includes: information on biobank, sample and data collection details, study description [73]. For other relevant guidelines, see Table 2.
Biobanks may also undergo certification of their activity. NF S96–900 is a French norm legislating the requirements for the management system of a Biological Resource Centre (BRC) and the quality of biological resources. The certificate is based on OECD Guidelines and is compatible with the international ISO 9001. NF S96–900 document is available at: (http://www.p3gobservatory.org/download/NFS96-900F.pdf; date of access: 27.07.2017). The other laboratory accreditation procedure (ISO 15189) was implemented by the Swedish Cervical Cytology Biobank (SCCB) as an extension of liquid based cytology procedures at clinical laboratories. It ensures a quality management system (QMS) including quality assurance (QA) and quality control (QC) programs, covering the full spectrum of biobanking operations [54].
Workflow and Online Tools for Biobanking
The wide range of data, such as: demographic, ethnical, medical, environmental, genetic and other [74] collected from the patient trigger a need for the development of complex IT systems. But for the clinical data, there are sets of biological information, which also must be input to the IT system, stored and processed - thus appeared complicated tasks. Holistic IT solutions, compatible with hospital and pathology units are the optimal solution. Many researchers and companies took on the evolution of bioinformatics applications such as MIBBI, MIAME, SysMOSEEK, openBIS, Gaggle-BRM, MIMAS, XperimentR, ISA tools, BASE, LabKey [75]. One of the most interesting approaches was implemented in XTENS, which consists of a web portal, an internal database, and a data grid storage element [75]. There are a few available online tools for researchers seeking advice on legal regulations of data sharing among European countries:
BioMedBridges Legal Assessment Tool (LAT)
Biobanking and Biomolecular Resources Research Infrastructure (BBMRI) legal WIKI
Human Sample Exchange Regulation Navigator (hSERN)
International Policy interoperability and data Access Clearinghouse’ (IPAC) provided by the Public Population Project in Genomics and Society (P3G) [76].
Biobanks IT systems face further described ethical issues by administration of novel solution like short message communication with donors in case of need [77].
Ethical and Legal Issues
Biobanks are unique units placed between a research and clinical unit. Therefore, many legal and ethical issues arose during the evolution of these novel initiatives. Despite a strong resemblance to hospital pathology units, which also store patients’ specimens, BBs have to overcome more sophisticated obstacles. These difficulties divide into groups [1]:
Ownership
Ever since human tissue samples have been used for the R&D (Research and development) purpose, an issue of ownership claims of donors arose. Specimens may be used not only for strictly research goals (biomarkers determination, cells biology analysis and the like) but also for the discovery of new drug targets or a novel treatment. In this case, patients may become eager to participate in the results and benefits of commercialization. Until now contemporary verdicts are in favor of scientist [1]. Nevertheless, exact ownership rights or their deprivation should be established just from the startup of a biobank. Moreover, matters of ownership play a great role in transferring samples between foreign researchers (is a Material Transfer Agreement (MTA) sufficient?) along with specimen utilization or procedures after donor’s death [78].
-
b.
Consent limitation issues
Optimization of consent content is crucial in BBs’ functioning. Appropriate consent must combine both affairs of scientist and patient. Classical informed consent turns out to be insufficient in biobanking due to the limitation of sample use for one, specific project [1, 7, 79, 80]. Additionally, there is no international consensus on the consent issue along with the differences between each legal system of each country. It hinders international sharing of samples. A novel form of consent was propagated - general/broad consent [1, 79, 80]. It comprises a patient’s agreement for the utilization of his sample for current studies and a future one (within a specified framework), without the need for the contact with the patient. But if the framework changes all the consents should be re-applied [79]. Thus, it results in wide-range, general and unspecified consents.
Along with the development of IT tools – a novel solution has been achieved, namely a dynamic consent. This type of consent requires tools for an easy accessible constant contact with the patient in order to manage re-consent for each new research [79]. But for the firm and conscious agreement, it is possible to inform patients about important finding - IF (incidental findings) and IRR (individual research results). IF is defined as “a finding concerning an individual research participant [or here, an individual contributor] that has potential health or reproductive importance and is discovered during conducting research, but is beyond the aims of the study.” [81]. Whilst, an IRR “is a finding concerning an individual contributor that has potential health or reproductive importance and is discovered in the course of research, when the finding is on the focal variables under study in meeting the stated aims of the research project” [82]. A recent study on the Australian population showed that the majority (94,4%) of queried people responded that they would like to receive “specific information obtained from your sample that may be important to your health or treatment” [83].
Biobanks storing materials obtained from children usually require parents’ (or legal guardians) consent. Ethical and legal guidelines indicate that children should be involved in the consent procedure as well. Also, child’s will: assent and dissent must be respected [84]. Another issue is whether the re-consent of pediatric patients, whose tissues in childhood were obtained and granted by parental consent, at the age of majority is needed [85]. The discussion is still ongoing [80].
-
c.
Storage and protection of privacy-anonymization
Long-term storage of biological samples requires adequate anonymization as well as identification procedures. A vast amount of data about the specific sample and their donor needs to be protected according to best-known standards. EU Data Protection Directive obliges investigator and administrators to provide professional and safe data management with full respect to the donor sake (document available at: http://ec.europa.eu/justice/data-protection/; date of access: 27.07.2017). Data anonymization done by simply deleting an identification information is insufficient in the context of their security. Often pseudonymization is implemented (data are secured by assigning a key or cipher instead of personal data) [76]. Therefore, sample coding is done in an ordered manner, universal for all samples. Nonetheless, identification data on donor combined with a coding symbol (number/barcode) or database, enabling decoding should be managed with the highest standards. It enables researchers to contact with the donor in case of obtaining accidental and significant findings for the donor’s health [86]. Another issue, often neglected, is a case of closure of a biobank or necessity to eliminate stored samples and data. The BBs’ founders are rarely prepared for such case [87].
-
d.
Whole genome protection and accessibility
Application of wide and accurate genomic sequencing leads to some new issues concerning security of obtained data. Storage of ones’ whole genome sequence poses a temptation for some third-parties to seize the data for their purposes. Comprehensive pieces of information encapsulated in genome sequence are utilized not only as an identification tool but also as a source of health status and burdens of the patient. Thus, thorough security procedures are implemented in every step of sample processing or analysis. Some donors are concerned about allowing BBs to process their genome information what decreases the number of volunteers [88]. However, researcher’s community opts for increasing the utility of the data by open access to genomic data along with respect to autonomy and anonymization of data. New legislation solutions, like GINA (Genetic Information Nondiscrimination Act), are needed to protect such data from misuse whilst open access [89].
Generally, any ambiguity of laws is discussed and solved by the ethics committees, which are obliged to give an opinion and requirements for a project. Currently, many countries are developing their own legal solutions according to BBMRI and/or OECD recommendations for biobank establishment.
The Role of Biobanks in Personalized Medicine
Personalized medicine (P4) defines a new approach to a patient and the disease. The concept of this personalization comprises 4 features [25, 90]:
Predictive - ability to conduct fast, precise and wide analysis of risk for particular diseases requiring easy access and affordable methods. Recent intensive progression in the field brings us closer to this solution [91]. However, biobanks play a crucial role in discovering new predictive factors like genetic aberrations [14, 92, 93]. In turn, correlating the discoveries with clinical data may facilitate predicting and support next step of P4 - prevention.
Preventive - comprises the idea of avoiding disease progression by an early application of accurate and personalized treatment. It may not seem to be a novel concept because there are yet implemented effective preventive solutions like vaccination, but unlike vaccines, which are recommended for the majority of the population, personalized medicine focuses on individuals. Biobanks-aided advancement can bring us to the higher level. In future, it will be possible to elevate the prognostic value of early symptoms and combine them with genome data what finally will lead physicians to quick and accurate diagnosis and enable to administer the right treatment on time. Yet conducted experiments confirmed the unique role of biobanks - numerous studies presented a new risk for diseases, on the basis of data stored there [13, 94–98].
Personalized - genotypic and phenotypic differences in human population have a significant influence on treatment efficacy. The more individualized it is the more efficient results are obtained. Recently whole genome and whole exome sequencing are widely available and more affordable. Deep knowledge about genetic and environmental circumstances of the patient increases the accuracy of diagnosis and treatment. Biobanks are centers of both types of data [99–101].
Participatory - increasing awareness of both patients and medical professionals and their mutual communication are the basis of P4 medicine. Conversely, in this point importance of IT companies increases, since they mediate the patient-doctor relationship by the development of intuitive, accessible and privacy-safe-oriented systems. Moreover, bioinformatics and new IT solutions are crucial for processing and organizing huge amounts of data collected from a patient.
Each component creates a possibility for more efficient and suitable treatment choice. These units pose a chance to create a core of each part of a personalized medicine approach consistent with evidence-based medicine (EBM).
Oncologic diseases are an especial benefiter of personalized medicine solutions. In the context of P4 medicine, biobanks may significantly develop the process of prevention, diagnosis and finally the treatment dedicated to the individuals. Numerous studies prove the significant role of biobanks in mentioned steps [102–104]:
Screening and prevention – role of PSA level in prostate cancer, circulating miR-196a and miR-196b in oral cancer [105] or chromogranin A in different malignancies e.g. ovarian cancer [106]
Diagnosis – searching for biomarkers in pancreatic cancer [94], thyroid neoplasm [107] or in colorectal cancer [108]
Prediction - patient response to the treatment on the basis of the genetic profile – KIT mutation (N505I) and sensitivity to imatinib [100]
Pharmacological – patients’ reaction to the drug and proper dosage: [109–111].
Biobanks Review among Europe
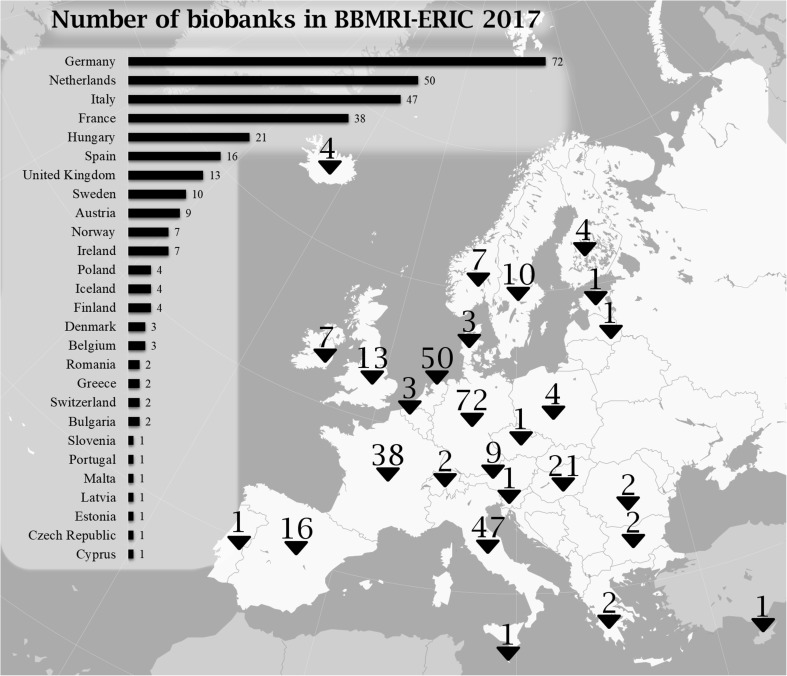
Contemporarily biobanks organize themselves into international networks. Such cooperation increases the impact of research findings. BBMRI (Biobanking and Biomolecular Resources Research Infrastructure) was one of the first European Research Infrastructure projects funded by the European Commission (EC) in January 2011. For the moment, it has 325 members (Biobanks), placed in Europe (https://www.bbmriportal.eu date of access: 27.07.2017). Distribution of BBMRI members is depicted in Fig. 3 (another available https://web.bbmri-eric.eu/Directory-files/directory-map-3-1-big-labels.png, date of access 27.07.2017). In comparison, there were 636 biobanks listed in the United States of America in 2013 [112].
Fig. 3.
Distribution and localization of BBMRI members (July 2017, data from https://www.bbmriportal.eu; map source – Creative Commons)
Another significant organization is EuroBioBank (http://www.eurobiobank.org date of access: 27.07.2017), it has 26 members: 22 biobanks (20 from Europe countries: France, Germany, Hungary, Italy, Malta, Slovenia, Spain, United-Kingdom, Turkey and 2 from Israel and Canada), EuroBioBank Coordinating Team, EURORDIS (European Organization for Rare Diseases), 3C-R: Expertise and consulting organization for biobanks, Telethon organization for genetic diseases research. EuroBioBank’s mission is to gather biological material from the patients affected by rare diseases and provide it to researchers who often have problems with obtaining enough samples. There are 130,000 samples combined with data available in the online catalogue.
Conclusions
The role of biobanks in the constantly developing world keeps increasing over time. Even more research projects incorporate biobank establishment or utilization of samples from the BBs. Possibilities brought by these institutions enable scientists to conduct wide-scale analyses with the unparalleled extent of thoroughness and significance. However, they do not only give benefits to the scientific community, but also for a patients’ health. Lower cost of mass processing and high operation standards implementations produce benefits for the donors in the form of information about the genome, prediction and prevention advice. Constant development is supported by international networks and organizations, which share their longtime experiences and act as a guide during new biobank creation. Thus, international collaboration and support are the keys to future biobanking development. In terms of mentioned tendency, local government and legal system ought to be prepared for the forthcoming evolution of biorepositories. The interdisciplinary character of biobanks opens new pathways (and evolve the old one) for biomedical researchers, clinicians and industrial partners, which cannot be neglected.
Abbreviations
- BBs
biobanks
- SOPs
Standard operating procedures
Funding
This study was supported by The National Centre for Research and Development [grant no. STRATEGMED1/233624/4/NCBR/2014]. All authors declare no other conflict of interest.
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
References
- 1.Fransson MN, Rial-Sebbag E, Brochhausen M, Litton JE (2015) Toward a common language for biobanking. Eur J Hum Genet 23. 10.1038/ejhg.2014.45 [DOI] [PMC free article] [PubMed]
- 2.Kang B, Park J, Cho S, Lee M, Kim N, Min H, et al. Current status, challenges, policies, and bioethics of biobanks. Genomics inform. 2013;11(4):211–217. doi: 10.5808/GI.2013.11.4.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sak J, Pawlikowski J, Goniewicz M, Witt M. Population biobanking in selected European countries and proposed model for a Polish national DNA bank. J Appl Genet. 2012;53(2):159–165. doi: 10.1007/s13353-012-0082-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Riegman PH, Morente MM, Betsou F, de Blasio P, Geary P, Marble Arch International Working Group on Biobanking for Biomedical R Biobanking for better healthcare. Mol Oncol. 2008;2(3):213–222. doi: 10.1016/j.molonc.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goebell PJ, Morente MM. New concepts of biobanks--strategic chance for uro-oncology. Urol Oncol. 2010;28(4):449–457. doi: 10.1016/j.urolonc.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 6.Asslaber M, Zatloukal K. Biobanks: transnational, European and global networks. Brief Funct Genom Proteomics. 2007;6(3):193–201. doi: 10.1093/bfgp/elm023. [DOI] [PubMed] [Google Scholar]
- 7.Swede H, Stone CL, Norwood AR. National population-based biobanks for genetic research. Genet Med. 2007;9(3):141–149. doi: 10.1097/GIM.0b013e3180330039. [DOI] [PubMed] [Google Scholar]
- 8.Leitsalu L, Haller T, Esko T, Tammesoo ML, Alavere H, Snieder H et al (2014) Cohort Profile: Estonian Biobank of the Estonian Genome Center, University of Tartu. Int J Epidemiol. 10.1093/ije/dyt268 [DOI] [PubMed]
- 9.Zawati M, Borry P, Howard H. Closure of population biobanks and direct-to-consumer genetic testing companies. Hum Genet. 2011;130(3):425–432. doi: 10.1007/s00439-011-1019-4. [DOI] [PubMed] [Google Scholar]
- 10.Awadalla P, Boileau C, Payette Y, Idaghdour Y, Goulet JP, Knoppers B et al. (2013) Cohort profile of the CARTaGENE study: Quebec's population-based biobank for public health and personalized genomics. Int J Epidemiol 42 (5):1285–1299. 10.1093/ije/dys160 [DOI] [PubMed]
- 11.Andersson K, Bray F, Arbyn M, Storm H, Zanetti R, Hallmans G, et al. The interface of population-based cancer registries and biobanks in etiological and clinical research--current and future perspectives. Acta Oncol. 2010;49(8):1227–1234. doi: 10.3109/0284186X.2010.496792. [DOI] [PubMed] [Google Scholar]
- 12.Fannin M, Kent J. Origin stories from a regional placenta tissue collection. New Genet Soc. 2015;34(1):25–51. doi: 10.1080/14636778.2014.999153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bonnelykke K, Sleiman P, Nielsen K, Kreiner-Moller E, Mercader JM, Belgrave D, et al. A genome-wide association study identifies CDHR3 as a susceptibility locus for early childhood asthma with severe exacerbations. Nat Genet. 2014;46(1):51–55. doi: 10.1038/ng.2830. [DOI] [PubMed] [Google Scholar]
- 14.Haukkala A, Kujala E, Alha P, Salomaa V, Koskinen S, Swan H, et al. The return of unexpected research results in a biobank study and referral to health care for heritable long QT syndrome. Public Health Genomics. 2013;16(5):241–250. doi: 10.1159/000354105. [DOI] [PubMed] [Google Scholar]
- 15.Wood AR, Esko T, Yang J, Vedantam S, Pers TH, Gustafsson S, et al. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat Genet. 2014;46(11):1173–1186. doi: 10.1038/ng.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shui IM, Lindström S, Kibel AS, Berndt SI, Campa D, Gerke T, et al. Prostate Cancer (PCa) Risk Variants and Risk of Fatal PCa in the National Cancer Institute Breast and Prostate Cancer Cohort Consortium. Eur Urol. 2014;65(6):1069–1075. doi: 10.1016/j.eururo.2013.12.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nieuwenhuijsen MJ, Smith R, Golfinopoulos S, Best N, Bennett J, Aggazzotti G et al. (2009) Health impacts of long-term exposure to disinfection by-products in drinking water in Europe: HIWATE. J Water Health 7 (2):185–18207. 10.2166/wh.2009.073 [DOI] [PubMed]
- 18.Jeong CH, Wagner ED, Siebert VR, Anduri S, Richardson SD, Daiber EJ, et al. Occurrence and toxicity of disinfection byproducts in European drinking waters in relation with the HIWATE epidemiology study. Environ Sci Technol. 2012;46(21):12120–12128. doi: 10.1021/es3024226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McKenzie F, Ferrari P, Freisling H, Chajes V, Rinaldi S, de Batlle J et al (2014) Healthy lifestyle and risk of breast cancer among postmenopausal women in the European Prospective Investigation into Cancer and Nutrition cohort study. Int J Cancer. 10.1002/ijc.29315 [DOI] [PubMed]
- 20.Engeset D, Braaten T, Teucher B, Kuhn T, Bueno-de-Mesquita HB, Leenders M et al (2014) Fish consumption and mortality in the European Prospective Investigation into Cancer and Nutrition cohort. Eur J Epidemiol. 10.1007/s10654-014-9966-4 [DOI] [PMC free article] [PubMed]
- 21.Ose J, Fortner RT, Schock H, Peeters PH, Onland-Moret NC, Bueno-de-Mesquita HB et al (2014) Insulin-like growth factor I and risk of epithelial invasive ovarian cancer by tumour characteristics: results from the EPIC cohort. Br J Cancer. 10.1038/bjc.2014.566 [DOI] [PMC free article] [PubMed]
- 22.Reza M, Cox D, Phillips L, Johnson D, Manoharan V, Grieves M et al (2017) MRC centre neuromuscular biobank (newcastle and london): supporting and facilitating rare and neuromuscular disease research worldwide. Neuromuscul Disord. 10.1016/j.nmd.2017.07.001 [DOI] [PMC free article] [PubMed]
- 23.De Souza YG, Greenspan JS. Biobanking past, present and future: responsibilities and benefits. AIDS (London, England) 2013;27(3):303–312. doi: 10.1097/QAD.0b013e32835c1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Check Hayden E. Proposed Ebola biobank would strengthen African science. Nature. 2015;524(7564):146–147. doi: 10.1038/524146a. [DOI] [PubMed] [Google Scholar]
- 25.Hutchinson E. Towards individualized cancer therapy: Challenges and prospects. Mol Oncol. 2014;8(1):1–8. doi: 10.1016/j.molonc.2013.12.008. [DOI] [Google Scholar]
- 26.Guerin JS, Murray DW, McGrath MM, Yuille MA, McPartlin JM, Doran PP. Molecular medicine ireland guidelines for standardized biobanking. Biopreserv Biobank. 2010;8(1):3–63. doi: 10.1089/bio.2010.8101. [DOI] [PubMed] [Google Scholar]
- 27.Mohamadkhani A, Poustchi H. Repository of Human Blood Derivative Biospecimens in Biobank: Technical Implications. Middle East J Dig Dis. 2015;7(2):61–68. [PMC free article] [PubMed] [Google Scholar]
- 28.Shabihkhani M, Lucey GM, Wei B, Mareninov S, Lou JJ, Vinters HV, et al. The procurement, storage, and quality assurance of frozen blood and tissue biospecimens in pathology, biorepository, and biobank settings. Clin Biochem. 2014;47(4–5):258–266. doi: 10.1016/j.clinbiochem.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duale N, Lipkin WI, Briese T, Aarem J, Ronningen KS, Aas KK, et al. Long-term storage of blood RNA collected in RNA stabilizing Tempus tubes in a large biobank--evaluation of RNA quality and stability. BMC Res Notes. 2014;7:633. doi: 10.1186/1756-0500-7-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.King MJ, Garcon L, Hoyer JD, Iolascon A, Picard V, Stewart G, et al. ICSH guidelines for the laboratory diagnosis of nonimmune hereditary red cell membrane disorders. Int J Lab Hematol. 2015;37(3):304–325. doi: 10.1111/ijlh.12335. [DOI] [PubMed] [Google Scholar]
- 31.Maimuna Mendy EC, Lawlor RT, Wright J, Wild CP (2017) Common minimum technical standards and protocols for biobanks dedicated to cancer research. Int Agency Res Cancer [PubMed]
- 32.Higdon LE, Lee K, Tang Q, Maltzman JS. Virtual Global Transplant Laboratory Standard Operating Procedures for Blood Collection, PBMC Isolation, and Storage. Transplantat Direct. 2016;2(9):e101. doi: 10.1097/TXD.0000000000000613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qin J, Alt JR, Hunsley BA, Williams TL, Fernando MR. Stabilization of circulating tumor cells in blood using a collection device with a preservative reagent. Cancer Cell Int. 2014;14(1):23. doi: 10.1186/1475-2867-14-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Riethdorf S, Fritsche H, Muller V, Rau T, Schindlbeck C, Rack B, et al. Detection of circulating tumor cells in peripheral blood of patients with metastatic breast cancer: a validation study of the CellSearch system. Clin Cancer Res. 2007;13(3):920–928. doi: 10.1158/1078-0432.CCR-06-1695. [DOI] [PubMed] [Google Scholar]
- 35.Oxnard GR, Paweletz CP, Sholl LM. Genomic Analysis of Plasma Cell-Free DNA in Patients With Cancer. JAMA Oncol. 2017;3(6):740–741. doi: 10.1001/jamaoncol.2016.2835. [DOI] [PubMed] [Google Scholar]
- 36.Ignatiadis M, Dawson SJ. Circulating tumor cells and circulating tumor DNA for precision medicine: dream or reality? Ann Oncol. 2014;25(12):2304–2313. doi: 10.1093/annonc/mdu480. [DOI] [PubMed] [Google Scholar]
- 37.Devonshire AS, Whale AS, Gutteridge A, Jones G, Cowen S, Foy CA, et al. Towards standardisation of cell-free DNA measurement in plasma: controls for extraction efficiency, fragment size bias and quantification. Anal Bioanal Chem. 2014;406(26):6499–6512. doi: 10.1007/s00216-014-7835-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wong D, Moturi S, Angkachatchai V, Mueller R, DeSantis G, van den Boom D, et al. Optimizing blood collection, transport and storage conditions for cell free DNA increases access to prenatal testing. Clin Biochem. 2013;46(12):1099–1104. doi: 10.1016/j.clinbiochem.2013.04.023. [DOI] [PubMed] [Google Scholar]
- 39.Hidestrand M, Stokowski R, Song K, Oliphant A, Deavers J, Goetsch M, et al. Influence of temperature during transportation on cell-free DNA analysis. Fetal Diagn Ther. 2012;31(2):122–128. doi: 10.1159/000335020. [DOI] [PubMed] [Google Scholar]
- 40.Callari M, Tiberio P, De Cecco L, Cavadini E, Dugo M, Ghimenti C, et al. Feasibility of circulating miRNA microarray analysis from archival plasma samples. Anal Biochem. 2013;437(2):123–125. doi: 10.1016/j.ab.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 41.Spornraft M, Kirchner B, Haase B, Benes V, Pfaffl MW, Riedmaier I. Optimization of extraction of circulating RNAs from plasma--enabling small RNA sequencing. PLoS One. 2014;9(9):e107259. doi: 10.1371/journal.pone.0107259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sourvinou IS, Markou A, Lianidou ES. Quantification of circulating miRNAs in plasma: effect of preanalytical and analytical parameters on their isolation and stability. J Mol Diagn: JMD. 2013;15(6):827–834. doi: 10.1016/j.jmoldx.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 43.Baranyai T, Herczeg K, Onodi Z, Voszka I, Modos K, Marton N, et al. Isolation of Exosomes from Blood Plasma: Qualitative and Quantitative Comparison of Ultracentrifugation and Size Exclusion Chromatography Methods. PLoS One. 2015;10(12):e0145686. doi: 10.1371/journal.pone.0145686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim JW, Wieckowski E, Taylor DD, Reichert TE, Watkins S, Whiteside TL. Fas ligand-positive membranous vesicles isolated from sera of patients with oral cancer induce apoptosis of activated T lymphocytes. Clin Cancer Res. 2005;11(3):1010–1020. [PubMed] [Google Scholar]
- 45.Best MG, Sol N, Kooi I, Tannous J, Westerman BA, Rustenburg F, et al. RNA-Seq of Tumor-Educated Platelets Enables Blood-Based Pan-Cancer, Multiclass, and Molecular Pathway Cancer Diagnostics. Cancer Cell. 2015;28(5):666–676. doi: 10.1016/j.ccell.2015.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aibaidula A, Lu JF, Wu JS, Zou HJ, Chen H, Wang YQ, et al. Establishment and maintenance of a standardized glioma tissue bank: Huashan experience. Cell Tissue Bank. 2015;16(2):271–281. doi: 10.1007/s10561-014-9459-4. [DOI] [PubMed] [Google Scholar]
- 47.Wang Z, Lebron JA, Wolf JJ. Reliable quantification of mRNA in archived formalin-fixed tissue with or without paraffin embedding. J Pharmacol Toxicol Methods. 2015;71:103–109. doi: 10.1016/j.vascn.2014.09.007. [DOI] [PubMed] [Google Scholar]
- 48.Schweiger MR, Kerick M, Timmermann B, Albrecht MW, Borodina T, Parkhomchuk D, et al. Genome-wide massively parallel sequencing of formaldehyde fixed-paraffin embedded (FFPE) tumor tissues for copy-number- and mutation-analysis. PLoS One. 2009;4(5):e5548. doi: 10.1371/journal.pone.0005548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hedegaard J, Thorsen K, Lund MK, Hein AM, Hamilton-Dutoit SJ, Vang S, et al. Next-generation sequencing of RNA and DNA isolated from paired fresh-frozen and formalin-fixed paraffin-embedded samples of human cancer and normal tissue. PLoS One. 2014;9(5):e98187. doi: 10.1371/journal.pone.0098187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Florell SR, Coffin CM, Holden JA, Zimmermann JW, Gerwels JW, Summers BK, et al. Preservation of RNA for functional genomic studies: a multidisciplinary tumor bank protocol. Modern pathology : an official journal of the United States and Canadian Academy of Pathology. Inc. 2001;14(2):116–128. doi: 10.1038/modpathol.3880267. [DOI] [PubMed] [Google Scholar]
- 51.Langdon SP (2004) Cancer Cell Culture. Methods in Molecular Medicine, vol 88. Humana Press. 10.1385/1592594069
- 52.Mitra A, Mishra L, Li S. Technologies for deriving primary tumor cells for use in personalized cancer therapy. Trends Biotechnol. 2013;31(6):347–354. doi: 10.1016/j.tibtech.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Giannini C, Sarkaria JN, Saito A, Uhm JH, Galanis E, Carlson BL, et al. Patient tumor EGFR and PDGFRA gene amplifications retained in an invasive intracranial xenograft model of glioblastoma multiforme. Neuro-Oncology. 2005;7(2):164–176. doi: 10.1215/S1152851704000821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Perskvist N, Norman I, Eklund C, Litton JE, Dillner J. The Swedish cervical cytology biobank: sample handling and storage process. Biopreserv Biobank. 2013;11(1):19–24. doi: 10.1089/bio.2012.0036. [DOI] [PubMed] [Google Scholar]
- 55.Elliott P, Peakman TC. The UK Biobank sample handling and storage protocol for the collection, processing and archiving of human blood and urine. Int J Epidemiol. 2008;37(2):234–244. doi: 10.1093/ije/dym276. [DOI] [PubMed] [Google Scholar]
- 56.Witon M, Strapagiel D, Glenska-Olender J, Chroscicka A, Ferdyn K, Skokowski J, et al. Organization of BBMRI.pl: The Polish Biobanking Network. Biopreserv Biobank. 2017;15(3):264–269. doi: 10.1089/bio.2016.0091. [DOI] [PubMed] [Google Scholar]
- 57.Vested A, Ramlau-Hansen CH, Olsen SF, Bonde JP, Kristensen SL, Halldorsson TI, et al. Associations of in utero exposure to perfluorinated alkyl acids with human semen quality and reproductive hormones in adult men. Environ Health Perspect. 2013;121(4):453–458. doi: 10.1289/ehp.1205118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee JE, Kim JH, Hong EJ, Yoo HS, Nam HY, Park O. National Biobank of Korea: Quality control Programs of Collected-human Biospecimens. Osong Public Health Res Perspect. 2012;3(3):185–189. doi: 10.1016/j.phrp.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mathay C, Hamot G, Henry E, Georges L, Bellora C, Lebrun L, et al. Method optimization for fecal sample collection and fecal DNA extraction. Biopreserv Biobank. 2015;13(2):79–93. doi: 10.1089/bio.2014.0031. [DOI] [PubMed] [Google Scholar]
- 60.Podzimek S, Vondrackova L, Duskova J, Janatova T, Broukal Z. Salivary Markers for Periodontal and General Diseases. Dis Markers. 2016;2016:9179632. doi: 10.1155/2016/9179632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Eglash A. ABM clinical protocol #8: human milk storage information for home use for full-term infants (original protocol March 2004; revision #1 March 2010) Breastfeed Med. 2010;5(3):127–130. doi: 10.1089/bfm.2010.9988. [DOI] [PubMed] [Google Scholar]
- 62.2012 best practices for repositories collection, storage, retrieval, and distribution of biological materials for research international society for biological and environmental repositories Biopreserv Biobank. 2012;10(2):79–161. doi: 10.1089/bio.2012.1022. [DOI] [PubMed] [Google Scholar]
- 63.He K. Trace elements in nails as biomarkers in clinical research. Eur J Clin Investig. 2011;41(1):98–102. doi: 10.1111/j.1365-2362.2010.02373.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu LY, He K, Hites RA, Salamova A. Hair and Nails as Noninvasive Biomarkers of Human Exposure to Brominated and Organophosphate Flame Retardants. Environ Sci Technol. 2016;50(6):3065–3073. doi: 10.1021/acs.est.5b05073. [DOI] [PubMed] [Google Scholar]
- 65.Willemse EA, Koel-Simmelink MJ, Durieux-Lu S, van der Flier WM, Teunissen CE. Standard biobanking conditions prevent evaporation of body fluid samples. Clin Chim Acta. 2015;442:141–145. doi: 10.1016/j.cca.2015.01.036. [DOI] [PubMed] [Google Scholar]
- 66.Kofanova OA, Mommaerts K, Betsou F. Tube Polypropylene: A Neglected Critical Parameter for Protein Adsorption During Biospecimen Storage. Biopreserv Biobank. 2015;13(4):296–298. doi: 10.1089/bio.2014.0082. [DOI] [PubMed] [Google Scholar]
- 67.Malm J, Lindberg H, Erlinge D, Appelqvist R, Yakovleva M, Welinder C, et al. Semi-automated biobank sample processing with a 384 high density sample tube robot used in cancer and cardiovascular studies. Clin Transl Med. 2015;4(1):67. doi: 10.1186/s40169-015-0067-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nussbeck SY, Skrowny D, O'Donoghue S, Schulze TG, Helbing K. How to Design Biospecimen Identifiers and Integrate Relevant Functionalities into Your Biospecimen Management System. Biopreserv Biobank. 2014;12(3):199–205. doi: 10.1089/bio.2013.0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Riondino S. Sample PReanalytical Code for labeling of biospecimens: an analysis of specimen labeling protocols. J Biorepository Sci Appl Med. 2015;3:15–21. [Google Scholar]
- 70.Artene SA, Ciurea ME, Purcaru SO, Tache DE, Tataranu LG, Lupu M, et al. Biobanking in a constantly developing medical world. TheScientificWorldJOURNAL. 2013;2013:343275. doi: 10.1155/2013/343275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Betsou F, Lehmann S, Ashton G, Barnes M, Benson EE, Coppola D, et al. Standard preanalytical coding for biospecimens: defining the sample PREanalytical code. Cancer Epidemiol Biomark Prev. 2010;19(4):1004–1011. doi: 10.1158/1055-9965.EPI-09-1268. [DOI] [PubMed] [Google Scholar]
- 72.Moore HM, Kelly AB, Jewell SD, McShane LM, Clark DP, Greenspan R, et al. Biospecimen reporting for improved study quality (BRISQ) Cancer Cytopathol. 2011;119(2):92–101. doi: 10.1002/cncy.20147. [DOI] [PubMed] [Google Scholar]
- 73.Merino-Martinez R, Norlin L, van Enckevort D, Anton G, Schuffenhauer S, Silander K, et al. Toward Global Biobank Integration by Implementation of the Minimum Information About BIobank Data Sharing (MIABIS 2.0 Core) Biopreserv Biobank. 2016;14(4):298–306. doi: 10.1089/bio.2015.0070. [DOI] [PubMed] [Google Scholar]
- 74.Zika E, Paci D, Braun A, Rijkers-Defrasne S, Deschenes M, Fortier I, et al. A European survey on biobanks: trends and issues. Public Health Genomics. 2011;14(2):96–103. doi: 10.1159/000296278. [DOI] [PubMed] [Google Scholar]
- 75.Izzo M, Mortola F, Arnulfo G, Fato MM, Varesio L. A digital repository with an extensible data model for biobanking and genomic analysis management. BMC Genomics. 2014;15(Suppl 3):S3. doi: 10.1186/1471-2164-15-S3-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sariyar M, Schluender I, Smee C, Suhr S. Sharing and Reuse of Sensitive Data and Samples: Supporting Researchers in Identifying Ethical and Legal Requirements. Biopreserv Biobank. 2015;13(4):263–270. doi: 10.1089/bio.2015.0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tebbakha R. Biobank–Short Message Service for Linking Patients and Samples. Telemed e-Health. 2013;19(9):717–721. doi: 10.1089/tmj.2012.0231. [DOI] [PubMed] [Google Scholar]
- 78.Ravid R. Standard Operating Procedures, ethical and legal regulations in BTB (Brain/Tissue/Bio) banking: what is still missing? Cell Tissue Bank. 2008;9(2):121–137. doi: 10.1007/s10561-007-9055-y. [DOI] [PubMed] [Google Scholar]
- 79.Steinsbekk KS, Kare Myskja B, Solberg B. Broad consent versus dynamic consent in biobank research: is passive participation an ethical problem? Eur J Hum Gen: EJHG. 2013;21(9):897–902. doi: 10.1038/ejhg.2012.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Colledge F, Persson K, Elger B, Shaw D. Sample and data sharing barriers in biobanking: consent, committees, and compromises. Ann Diagn Pathol. 2014;18(2):78–81. doi: 10.1016/j.anndiagpath.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 81.Wolf SM, Lawrenz FP, Nelson CA, Kahn JP, Cho MK, Clayton EW, et al. Managing incidental findings in human subjects research: analysis and recommendations. J Law Med Ethics. 2008;36(2):219–248. doi: 10.1111/j.1748-720X.2008.00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wolf SM, Crock BN, Van Ness B, Lawrenz F, Kahn JP, Beskow LM, et al. Managing Incidental Findings and Research Results in Genomic Research Involving Biobanks & Archived Datasets. Genet Med. 2012;14(4):361–384. doi: 10.1038/gim.2012.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fleming J, Critchley C, Otlowski M, Stewart C, Kerridge I. Attitudes of the general public towards the disclosure of individual research results and incidental findings from biobank genomic research in Australia. Intern Med J. 2015;45(12):1274–1279. doi: 10.1111/imj.12911. [DOI] [PubMed] [Google Scholar]
- 84.Giesbertz NA, Bredenoord AL, van Delden JJ (2015) Consent procedures in pediatric biobanks. Eur J Hum Gen: EJHG 23 (9):1129–1134. 10.1038/ejhg.2014.267 [DOI] [PMC free article] [PubMed]
- 85.Rush A, Battisti R, Barton B, Catchpoole D. Opinions of Young Adults on Re-Consenting for Biobanking. J Pediatr. 2015;167(4):925–930. doi: 10.1016/j.jpeds.2015.07.005. [DOI] [PubMed] [Google Scholar]
- 86.Virani AH, Longstaff H (2014) Ethical Considerations in Biobanks: How a Public Health Ethics Perspective Sheds New Light on Old Controversies. J Genet Couns. 10.1007/s10897-014-9781-9 [DOI] [PubMed]
- 87.Cadigan RJ, Lassiter D, Haldeman K, Conlon I, Reavely E, Henderson GE. Neglected ethical issues in biobank management: Results from a U.S. study. Life Sci Soc Policy. 2013;9(1):1. doi: 10.1186/2195-7819-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Melas PA, Sjoholm LK, Forsner T, Edhborg M, Juth N, Forsell Y, et al. Examining the public refusal to consent to DNA biobanking: empirical data from a Swedish population-based study. J Med Ethics. 2010;36(2):93–98. doi: 10.1136/jme.2009.032367. [DOI] [PubMed] [Google Scholar]
- 89.Pereira S, Gibbs RA, McGuire AL. Open Access Data Sharing in Genomic Research. Gene. 2014;5(3):739–747. doi: 10.3390/genes5030739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hood L. Systems biology and p4 medicine: past, present, and future. Rambam Maimonides Med J. 2013;4(2):e0012. doi: 10.5041/RMMJ.10112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kreiner T, Irion S. Whole-genome analysis, stem cell research, and the future of biobanks. Cell Stem Cell. 2013;12(5):513–516. doi: 10.1016/j.stem.2013.04.024. [DOI] [PubMed] [Google Scholar]
- 92.Gelpi E, van der Zee J, Turon Estrada A, Van Broeckhoven C, Sanchez-Valle R. TARDBP mutation p.Ile383Val associated with semantic dementia and complex proteinopathy. Neuropathol Appl Neurobiol. 2014;40(2):225–230. doi: 10.1111/nan.12063. [DOI] [PubMed] [Google Scholar]
- 93.Koszarska M, Kucsma N, Kiss K, Varady G, Gera M, Antalffy G, et al. Screening the Expression of ABCB6 in Erythrocytes Reveals an Unexpectedly High Frequency of Lan Mutations in Healthy Individuals. PLoS One. 2014;9(10):e111590. doi: 10.1371/journal.pone.0111590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jenkinson C, Elliott V, Menon U, Apostolidou S, Fourkala OE, Gentry-Maharaj A, et al. Evaluation in pre-diagnosis samples discounts ICAM-1 and TIMP-1 as biomarkers for earlier diagnosis of pancreatic cancer. J Proteome. 2014;113c:400–402. doi: 10.1016/j.jprot.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 95.Guo X, Long J, Zeng C, Michailidou K, Ghoussaini M, Bolla MK, et al. Fine-Scale Mapping of the 4q24 Locus Identifies Two Independent Loci Associated with Breast Cancer Risk. Cancer Epidemiol Biomark Prev. 2015;24(11):1680–1691. doi: 10.1158/1055-9965.EPI-15-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kvistad SS, Myhr KM, Holmoy T, Saltyte Benth J, Wergeland S, Beiske AG, et al. Body mass index influence interferon-beta treatment response in multiple sclerosis. J Neuroimmunol. 2015;288:92–97. doi: 10.1016/j.jneuroim.2015.09.008. [DOI] [PubMed] [Google Scholar]
- 97.Teumer A, Tin A, Sorice R, Gorski M, Yeo NC, Chu AY et al (2015) Genome-wide Association Studies Identify Genetic Loci Associated with Albuminuria in Diabetes. Diabetes. 10.2337/db15-1313 [DOI] [PMC free article] [PubMed]
- 98.Westerlind H, Mellander MR, Bresso F, Munch A, Bonfiglio F, Assadi G et al (2015) Dense genotyping of immune-related loci identifies HLA variants associated with increased risk of collagenous colitis. Gut. 10.1136/gutjnl-2015-309934 [DOI] [PubMed]
- 99.Wakuda K, Kenmotsu H, Serizawa M, Koh Y, Isaka M, Takahashi S, et al. Molecular profiling of small cell lung cancer in a Japanese cohort. Lung Cancer (Amst, Netherlands) 2014;84(2):139–144. doi: 10.1016/j.lungcan.2014.02.013. [DOI] [PubMed] [Google Scholar]
- 100.Allegra M, Giacchero D, Segalen C, Dumaz N, Butori C, Hofman V, et al. A new KIT mutation (N505I) in acral melanoma confers constitutive signaling, favors tumorigenic properties, and is sensitive to imatinib. J Investig Dermatol. 2014;134(5):1473–1476. doi: 10.1038/jid.2013.525. [DOI] [PubMed] [Google Scholar]
- 101.Esplin ED, Oei L, Snyder MP. Personalized sequencing and the future of medicine: discovery, diagnosis and defeat of disease. Pharmacogenomics. 2014;15(14):1771–1790. doi: 10.2217/pgs.14.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lisanti MP, Tanowitz HB. Translational discoveries, personalized medicine, and living biobanks of the future. Am J Pathol. 2012;180(4):1334–1336. doi: 10.1016/j.ajpath.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 103.van Ommen GJ, Tornwall O, Brechot C, Dagher G, Galli J, Hveem K et al (2014) BBMRI-ERIC as a resource for pharmaceutical and life science industries: the development of biobank-based Expert Centres. Eur J Hum Gen: EJHG. 10.1038/ejhg.2014.235 [DOI] [PMC free article] [PubMed]
- 104.Womack C, Mager SR. Human biological sample biobanking to support tissue biomarkers in pharmaceutical research and development. Methods (San Diego, Calif) 2014;70(1):3–11. doi: 10.1016/j.ymeth.2014.01.014. [DOI] [PubMed] [Google Scholar]
- 105.Lu YC, Chang JT, Huang YC, Huang CC, Chen WH, Lee LY et al (2014) Combined determination of circulating miR-196a and miR-196b levels produces high sensitivity and specificity for early detection of oral cancer. Clin Biochem. 10.1016/j.clinbiochem.2014.11.020 [DOI] [PubMed]
- 106.Malaguarnera M, Uccello M, Bellanca S, La Rosa B, Vacante M, Cristaldi E, et al. Elevated chromogranin A serum levels in ovarian carcinoma patients. Indian J Cancer. 2014;51(3):315–318. doi: 10.4103/0019-509X.146776. [DOI] [PubMed] [Google Scholar]
- 107.Nagar S, Ahmed S, Peeples C, Urban N, Boura J, Thibodeau B, et al. Evaluation of genetic biomarkers for distinguishing benign from malignant thyroid neoplasms. Am J Surg. 2014;207(4):596–601. doi: 10.1016/j.amjsurg.2013.06.012. [DOI] [PubMed] [Google Scholar]
- 108.Letellier E, Schmitz M, Baig K, Beaume N, Schwartz C, Frasquilho S, et al. Identification of SOCS2 and SOCS6 as biomarkers in human colorectal cancer. Br J Cancer. 2014;111(4):726–735. doi: 10.1038/bjc.2014.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bielinski SJ, Olson JE, Pathak J, Weinshilboum RM, Wang L, Lyke KJ, et al. Preemptive genotyping for personalized medicine: design of the right drug, right dose, right time-using genomic data to individualize treatment protocol. Mayo Clin Proc. 2014;89(1):25–33. doi: 10.1016/j.mayocp.2013.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Almoguera B, Vazquez L, Connolly JJ, Bradfield J, Sleiman P, Keating B, et al. Imputation of TPMT defective alleles for the identification of patients with high-risk phenotypes. Front Genet. 2014;5:96. doi: 10.3389/fgene.2014.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kawai VK, Cunningham A, Vear SI, Van Driest SL, Oginni A, Xu H, et al. Genotype and risk of major bleeding during warfarin treatment. Pharmacogenomics. 2014;15(16):1973–1983. doi: 10.2217/pgs.14.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Henderson GE, Cadigan RJ, Edwards TP, Conlon I, Nelson AG, Evans JP, et al. Characterizing biobank organizations in the U.S.: results from a national survey. Genome Med. 2013;5(1):3. doi: 10.1186/gm407. [DOI] [PMC free article] [PubMed] [Google Scholar]